- 1Department of Biological Pharmacy, Faculty of Pharmacy, Universitas Padjadjaran, Sumedang, Indonesia
- 2Unit of Pharmacology, Clinical Pharmacy and Community, Faculty of Science, Universitas Garut, Garut, Indonesia
- 3Division of Rheumatology, Department of Internal Medicine, Faculty of Medicine, Universitas Padjadjaran, Hasan Sadikin Hospital, Bandung, Indonesia
- 4Department of Pharmacology and Clinical Pharmacy, Faculty of Pharmacy, Universitas Padjadjaran, Bandung, Indonesia
- 5Center of Excellence for Pharmaceutical Care Innovation, Universitas Padjadjaran, Bandung, Indonesia
Systemic Lupus Erythematosus (SLE) is an autoimmune disease that often requires treatment with immunosuppressant drugs to manage symptoms and prevent organ damage. However, the use of immunosuppressant can be associated with various adverse effects. The spectrum of immunosuppressant toxicity is influenced by various factors such as organ function and medication interval, but genetic variations—particularly single nucleotide polymorphisms—have emerged as critical determinants due to their direct impact on the drug’s pharmacokinetics and pharmacodynamics alteration, also on patient susceptibility to adverse reactions. This review summarizes the current knowledge on gene polymorphisms associated with immunosuppressant adverse effects in SLE patients, focusing on commonly used drugs such as Methotrexate (MTX), Azathioprine (AZA), Cyclophosphamide (CYC), and Mycophenolate Mofetil (MMF). A total of 23 relevant studies published in the last decade were identified through a comprehensive literature search, specifically investigating the relationship between gene polymorphisms and adverse drug reactions in SLE patients. The findings reveal that gene polymorphisms are frequently associated with adverse effects for each immunosuppressant, including MTX (MTHFR and ATIC), AZA (TPMT, NUDT15, ITPA, ABCC4), CYC (CYP2C19, GSTM1, GSTT1, GSTP1, ALDH), and MMF (SLCO1B1, IMPDH1, UGT2B7). Understanding the functional implications of these gene polymorphisms contributes to the application of precision medicine, as they can serve as potential markers for drug selection and dosage adjustment during initiation treatment of immunosuppressant to enhance treatment efficacy, minimize toxicity, and improve outcomes for SLE patients.
1 Introduction
Systemic Lupus Erythematosus (SLE) is a chronic autoimmune inflammatory disease featuring complex pathogenesis that affects various organ systems, leading to significant mortality and morbidity (Basta et al., 2020; Cattaneo et al., 2008). The incidence and prevalence of SLE vary widely across different ethnicities and regions, including the United States, Europe, the Middle East, and Asia (Ju et al., 2016). Treatment management strategies depend on disease type and severity, with mild to moderate cases typically treated using nonsteroidal anti-inflammatory drugs (NSAIDs), antimalarial agents (e.g., hydroxychloroquine), and corticosteroids. As the disease severity increases, high-dose corticosteroids and immunosuppressive agents, including Methotrexate (MTX), azathioprine (AZA), cyclophosphamide (CYC), and mycophenolate mofetil (MMF), are often used to control symptoms. The selection of immunosuppressant in SLE patients depends on disease manifestations, organ involvement, patient age, childbearing potential, safety considerations, and cost. However, their clinical utility is frequently constrained by serious adverse effects due to their narrow therapeutic index. These adverse effects not only increase the risk of long-term organ damage and treatment failure but also contribute to higher mortality rates and reduced quality of life, even during remission phases (Basta et al., 2020).
Among the many factors influencing immunosuppressant-related serious adverse effects, genetic variability—particularly in the form of single nucleotide polymorphisms (SNPs)—has gained considerable attention. These genetic differences can affect drug metabolism, efficacy, and the likelihood of adverse effects. Pharmacogenomics, the study of the role of genetics in drug response, was introduced to optimize the treatment while minimizing drug-related toxicity using SNPs as molecular markers, forming the basis for precision medicine (Cattaneo et al., 2008). A growing number of pharmacogenomic studies in SLE have explored the role of SNPs in determining the safety and effectiveness of immunosuppressants (Petri et al., 2012). This narrative review aims to summarize and discuss the current knowledge on gene polymorphisms associated with adverse effects from the most commonly used immunosuppressants in SLE: MTX, AZA, CYC, and MMF. Although these agents are administered within relatively standardized therapeutic dose ranges, the incidence and severity of adverse effects vary significantly among individuals. This variability can be attributed, in part, to genetic polymorphisms that affect drug metabolism, transport, and cellular targets. Therefore, elucidating the role of genetic polymorphisms is essential for understanding the underlying mechanisms of immunosuppressant-induced adverse effects. By highlighting the associations between specific genetic variants and drug toxicities, this review provides a foundation for integrating genetic screening into clinical decision-making. Such an approach may enhance treatment efficacy and safety, ultimately leading to improved SLE patient outcomes. The subsequent sections of this review will explore each immunosuppressant in detail. For each drug, we will discuss its pharmacological mechanism, common adverse effects, and the genetic polymorphisms known to influence its toxicity. This structure is intended to offer a practical, drug-centered understanding of how pharmacogenetics can inform and refine SLE treatment.
2 Method
This manuscript is a narrative review article. The search strategy and inclusion criteria were conducted using Google Scholar and PubMed databases, which include the use of Boolean operators for keyword combinations. Specifically, these keywords were combined using Boolean operators (AND, OR) as follows: “genetic polymorphism” OR “SNPs” AND “immunosuppressant drugs” OR “azathioprine” OR “thiopurine” OR “methotrexate” OR “cyclophosphamide” OR “mycophenolate mofetil” OR “SLE therapy” AND “Adverse effects”. We included studies published in English, excluding narrative reviews, communication studies, and unpublished manuscripts. A total of 23 articles from the past 10 years were included in the review, categorized by drug: 7 on MTX, 8 on AZA, 6 on CYC, and 2 on MMF.
3 Gene polymorphisms and adverse effects of immunosuppressants in SLE
SLE is a highly diverse autoimmune condition, showing a wide array of symptoms and affecting various organs (Basta et al., 2020; Cattaneo et al., 2008; Ju et al., 2016; Petri et al., 2012; Hochberg, 1997). This disease arises when the immune system erroneously attacks healthy tissues, leading to inflammation and harm, such as visceral damage, flare-ups, neuropsychiatric lupus, and many more. The severity and specific expressions of SLE can differ significantly from one individual to another. Patients with mild SLE are primarily given a low dose of glucocorticoids (GCs) as therapy because of effectivity in controlling SLE activity rapidly and reducing exacerbation (Katarzyna et al., 2023). A higher dose of GCs is used during more severe SLE activity or in some life-threatening conditions, such as lupus nephritis (LN). However, the adverse effects of GCs are dose-dependent, suggesting that an increase in administered GCs dose leads to higher risk of adverse effects, such as infection, cardiovascular disease, cancer, osteoporosis, and many more (McKeon and Jiang, 2020). To mitigate these risks, combination therapy involving GCs and immunosuppressive agents is often employed. As reviewed in the literature, most patients receiving immunosuppressive treatment also remain on concurrent GC therapy to achieve synergistic therapeutic effects.
Immunosuppressant is used when the disease progresses from moderate to severe condition and the administration of GCs as SLE first-line treatment cannot sustain clinical remission (Gatto et al., 2019). In general, the immunosuppressant mechanism of action requires suppressing and decreasing the autoimmune responses, which can target various organs and systems in the body. This serves to minimize damage in various organs, thereby preventing life-threatening conditions. The use of the immunosuppressant is based on the organs engaged in SLE activity and the conditions of patients. Moderately active lupus and joints involved are treated with MTX, while LN and other severe cases are treated primarily with MMF and CYC (Mohamed et al., 2019; Fraenkel et al., 2021).
Immunosuppressant administration can decrease GCs exposure, stabilize SLE, and increase the probability of better survival than using GCs alone but toxicity incidence is high, ranging from 42.8% to 97.3% (Oglesby et al., 2013). Several immunosuppressive agents of the drugs can cause complications, such as liver dysfunction, bone marrow suppression, pulmonary toxicity, and many more. As the study of pharmacogenetics is advancing, variations in genomic diversity, including SNPs, are found to be a potential crucial factor affecting toxicity incidence due to the alteration of pharmacokinetics and pharmacodynamics of the administered drugs (Meng et al., 2018). The summary of all studies discussed, including the sample size, drug toxicity manifestation, and the most common gene polymorphisms associated with immunosuppressant adverse effects are shown in Table 1. Given the comparable spectrum of therapeutic doses and the potential for adverse reaction across diseases such as SLE, studies concerning genes implicated in the adverse effects of immunosuppressant drugs for treatment are also included.
3.1 Methotrexate
MTX is an antifolate antimetabolite commonly used in the treatment of RA, cancer, as well as SLE, and associated with a significant decrease in GCs dose used in adult patients. A systematic study showed that MTX appeared to offer significant benefits for individuals experiencing active arthritis or cutaneous symptoms in SLE (Sakthiswary and Suresh, 2014). The entry of MTX into the cell is facilitated by human reduced folate carriers (hFRC), a major importer of folates, known as SLC19A1. Following cellular uptake, MTX passes through polyglutamation by folylpolyglutamate synthase, leading to the retention in the cell. The mechanism of action includes folate metabolism, specifically inhibition of dihydrofolate reductase (DHFR), an enzyme essential in converting dihydrofolate to tetrahydrofolate (THF) active form. THF is indispensable for various cellular processes, including the synthesis of DNA and RNA nucleotides. MTX disrupts these processes by impeding DHFR, causing intracellular depletion of THF, particularly in rapidly dividing cells. Beyond DHFR inhibition, MTX and the polyglutamate forms impede de novo purine synthesis and thymidylate synthase, intensifying the cytotoxic effects that affect cell proliferation and growth (Mohamed et al., 2019).
Despite the efficacy of MTX, related toxicity has been reported in several studies, including liver toxicity (increase of liver function, risk of liver failure), kidney toxicity (renal impairment, renal failure), hematological toxicity (pancytopenia, myelosuppression, leukopenia, neutropenia, megaloblastic anemia), pulmonary toxicity (wheezing, asthma), dermatological toxicity (skin lesion), and gastrointestinal (GI) effects (diarrhea, nausea, vomiting, etc.) (Hamed et al., 2022). The most common major toxicity of low-dose MTX was pancytopenia, followed by oral mucositis, hypoalbuminemia, acute renal failure, and pneumonitis, while minor toxicity included diarrhea, abdominal pain, and fever (Kivity et al., 2014). The duration of MTX use may affect the severity of the toxicity manifestation. Skin lesions and mucosal ulcers were reported in patients with less than 7 days of consumption, while more severe toxicity such as leukopenia, thrombocytopenia, and anemia were more common in those exposed to more than 7 days of consumption (Ahmadzadeh et al., 2019). Gene polymorphisms are suggested to affect the incidence of MTX-related toxicity, as several studies show that MTHFR c.667C>T, MTHFR c.1298A>C, and ATIC c.347C>G are the most common genes responsible for MTX toxicity, in both the treatment of SLE and other diseases including RA. Figure 1 presents effects of these gene polymorphisms on MTX-related toxicity.
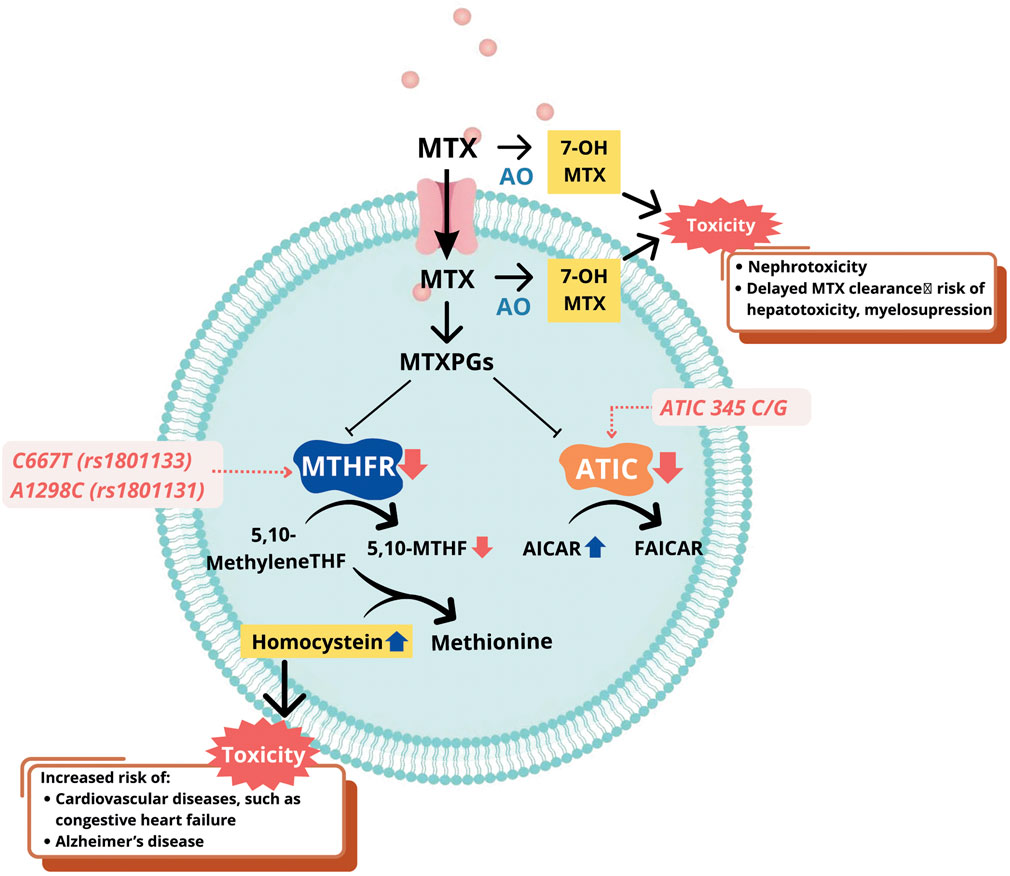
Figure 1. Mechanism of gene polymorphisms affecting MTX-related toxicity. Abbreviations: MTX, methotrexate; AO, aldehyde oxidase; 7-OH-MTX, 7-hydroxy methotrexate; ATIC, 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase; AICAR, aminoimidazole carboxamide ribonucleotide transformylase/IMP cyclohydrolase; MTHFR, methylenetetrahydrofolate reductase; 5,10-methylene THF, 5,10-methylenetetrahydrofolate; 5-THF, 5-methyltetrahydrofolate; MTXPGs, methotrexate polyglutamates; FAICAR, formyl-AICAR.
3.1.1 MTHFR polymorphisms
The MTHFR (Methylenetetrahydrofolate reductase) gene encodes a 77-kDa MTHFR enzyme which participates in folate metabolism, specifically in the conversion of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate (5-MTHF), and this process is crucial for the synthesis of nucleotides and DNA. Several studies, including meta-analyses, have reported an association between the two most common SNPs of the MTHFR gene, namely c.667C>T (rs1801133) and c.1298A>C (rs1801131), with MTX toxicity (Song et al., 2014; Dwivedi et al., 2020; Yang et al., 2023; Martusevich et al., 2020; Von Feldt et al., 2006; Juster-switlyk et al., 2017; Zhao et al., 2016; Campbell et al., 2016). In c.667C>T polymorphism, alanine is substituted with valine due to a change from C to T at nucleotide 677 (Rosenberg et al., 2002). Meanwhile, in c.1298A>C polymorphism, A is replaced with C at position 1,298, leading to the substitution of alanine to glutamine (Sharaki et al., 2018). Both SNPs lead to the lower enzymatic activity of MTHFR, affecting MTX pharmacodynamics (Van Der Put et al., 1998). Consequently, there is a decrease in the production of 5-MTHF, which serves as a methyl donor in the re-methylation of homocysteine to methionine (Figure 1). This leads to homocysteine accumulation in the blood, a condition known as hyperhomocysteinemia. Elevated homocysteine levels have been associated with increased toxicity of MTX and elevated cardiovascular risks, such as coronary artery calcification, high blood pressure, and many more (Von Feldt et al., 2006; Gande et al., 2023). The exact mechanism underlying MTX toxicity caused by lower MTHFR enzymatic activity remains unclear and requires further investigation.
3.1.2 ATIC polymorphism
ATIC gene is located at chromosome 2q35 and encodes aminoimidazole carboxamide adenosine ribonucleotide transformylase (ATIC) that participates in the de novo purine synthesis and transforms aminoimidazole carboxamide adenosine ribonucleotide (AICAR) into formyl- AICAR. MTX mechanisms of action includes inhibiting ATIC after entering the cells, causing AICAR intracellular accumulation (Figure 1). This leads to the release of adenine into extracellular which inhibits the functions of several immune cells, such as monocytes, T-lymphocytes, and NK cells, initiating anti-inflammatory activities (Lee and Bae, 2016). The most commonly explored ATIC polymorphism is ATIC c.347C>G, with the C to G variation prompting the change of threonine to serine at position 116 of gene. Several studies showed that patients with the ATIC c.347G allele had a higher risk of MTX-related toxicity, specifically GI toxicity (Muralidharan et al., 2016; Grabar et al., 2010; Huang et al., 2020; Londono et al., 2020). Other studies on the same polymorphism did not report a relationship between gene and toxicity, but associated ATIC c.347C>G with the efficacy or non-responsiveness of MTX (Lee and Bae, 2016; Sha et al., 2022). A meta-analysis consisting of nine comparative studies showed that ATIC c.347C>G polymorphism might be associated with MTX toxicity in Caucasians, compared to Asian patients. This scientific finding remains uncertain and tends to be associated with a higher frequency of the allele in Caucasians (Lee and Bae, 2016).
3.2 Azathioprine
AZA is an immunosuppressant used in managing SLE as second-line treatment. In the liver, AZA is initially converted to 6-mercaptopurine (6-MP) and then passes through three metabolism pathways. In the first pathway, 6-MP can be metabolized into 6-methylmercaptopurine (6-MMP) by thiopurine methyltransferase (TPMT). In the second pathway, 6-MP is oxidized by Xanthine Oxidase (XO) into 6-Thiouric Acid (6-TUA) which is an inactive metabolite. In the third pathway, metabolic processes of mercaptopurine nucleotide lead to the production of Thio inosine 5′-monophosphate (TIMP) by hypoxanthine phosphoribosyl transferase (HPRT). TIMP is converted by nudix hydrolase 15 (NUDT15) into thioguanine nucleotides (TGNs), including thioguanosine monophosphate (TGMP), thioguanosine diphosphate (TGDP), and thioguanosine triphosphate (TGTP). TGNs are the active metabolites of 6-MP which produce cytotoxic activity, specifically by inducing apoptosis in active T-cells. TGDP is converted to Thio deoxyguanosine triphosphate (TdGTP), which is subsequently incorporated into DNA. This process involves Thio deoxyguanosine diphosphate (TdGDP) as a metabolic intermediate. TGTP integrates into RNA disrupting the normal functions of these nucleic acids and leading to cell death (Mohamed et al., 2019; Fraenkel et al., 2021; Oglesby et al., 2013).
TIMP passes through alternative metabolic pathways and is transformed into 6-thio inosine 5′-triphosphate (6-TITP), a toxic metabolite. An enzyme called inosine triphosphate pyrophosphatase (ITPA) converts TITP into TIMP to restrict the accumulation. Another pathway is TIMP conversion into methyl thio inosine 5′-monophosphate (Me-TIMP) by TMPT. Me-TIMP has a role to impede the de novo synthesis of purine nucleotides, further compromising cellular processes crucial for T cell survival. Additionally, it inhibits Ras-related C3 botulinum toxin substrate, a protein essential in cell signaling pathways (Wright et al., 2004).
The use of AZA for SLE treatment is limited due to the drug-related toxicity reported, such as myelosuppression, leukopenia, pancreatic toxicity, and many more (Rashid et al., 2020; Liu et al., 2015; Fei et al., 2018b). The association between AZA toxicity and several SNPs has been investigated. The most common SNPs studied are TPMT gene polymorphisms [TPMT*2 G238C (rs1800462), TPMT*3B (rs1800460), and TPMT*3C (rs1142345)], NUDT15 R139C, ITPA c.94C>A, and ABCC4 c.2269G>A. Figure 2 shows effects of these gene polymorphisms on AZA-related toxicity.
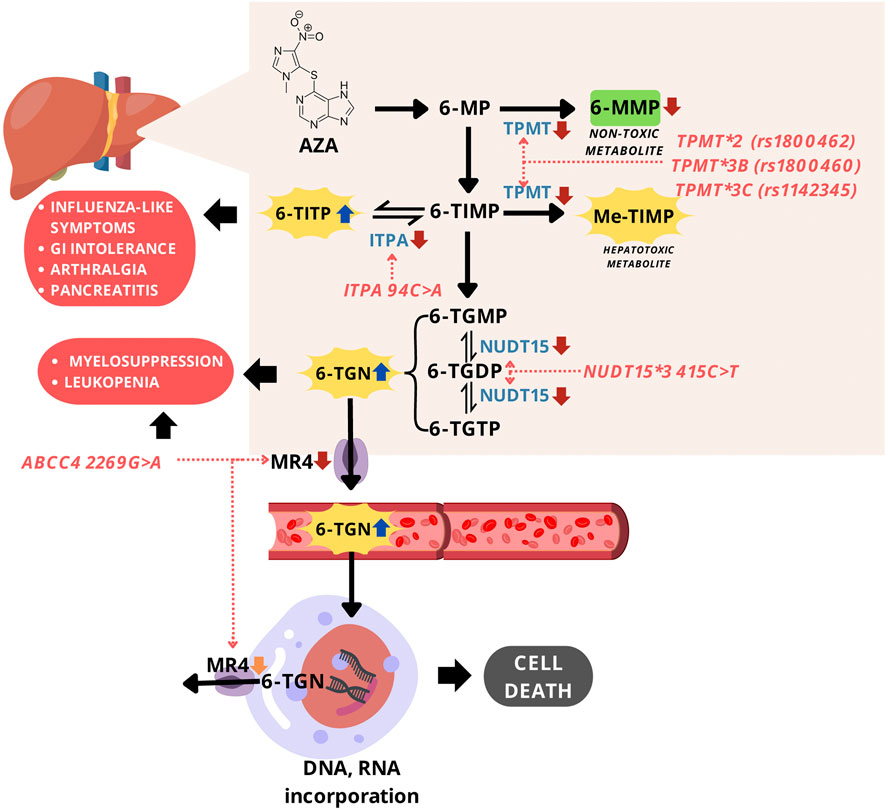
Figure 2. Mechanism of gene polymorphisms affecting AZA-related toxicity. Abbreviations: AZA, azathioprine; 6-MP, 6-mercaptopurine; 6-MMP, 6-methylmercaptopurine; 6-TIMP, 6-thioinosine 5'-monophosphate; 6-TITP, 6-thioinosine 5'-triphosphate; Me-TIMP, methyl-thioinosine 5'-monophosphate; 6-TGMP, 6-thioguanosine monophosphate; 6-TGDP, 6-thioguanosine diphosphate; 6-TGTP, 6-thioguanosine triphosphate; TGN, thioguanine nucleotides; TPMT, thiopurine S-methyltransferase; ITPA, inosine triphosphate pyrophosphatase; NUDT15, nudix hydrolase 15; MRP4, multidrug resistance-associated protein 4.
3.2.1 TPMT polymorphisms
TPMT gene is responsible for encoding thiopurine S-methyltransferase (TPMT), an enzyme that contributes to thiopurine drugs metabolism in cells. TPMT catalyzes the S-methylation of thiopurine including AZA into inactive and non-toxic forms. There are several alleles of the TPMT gene (TPMT*2 to TPMT*20) which affect TMPT activity, but the most frequently studied polymorphisms associated with AZA toxicity are TPMT*2 c.238G>C (rs1800462), TPMT*3B c.460G>A (rs1800460), and TPMT*3C c.719A>G (rs1142345). TPMT*2, TPMT*3B, and TPMT*3C are more common in Caucasians and Africans, while TPMT*3C is the most frequent in the Asian population (Gu et al., 2023). TPMT*2 c.238G>C were reported to prompt the substitution of alanine to proline. TPMT*3B c.460G>A and TPMT*3C c.719A>G are responsible for the deficiency or the loss of TPMT activity leading to AZA-related toxicity because of a decrease in the drug metabolism (Figure 2) (Steponaitiene et al., 2016). The reduction of AZA metabolism causes excessive accumulation of cytotoxic compounds, TGNs, inducing severe toxicity such as bone marrow toxicity and GI manifestations (Murugesan et al., 2009).
The data reported for the association of TMPT alleles and AZA-induced hepatotoxicity were conflicting. A meta-analysis showed that TPMT variations were not associated with hepatotoxic (Liu et al., 2015). Other studies found a higher risk of hepatotoxicity due to these polymorphisms, hence individualized AZA dosing could minimize the risk (Sheu et al., 2022). There was no correlation between the TPMT genotype and leukopenia incidence (p = 0.95) in Chinese autoimmune patients (Fei et al., 2018a). In the case of myelotoxicity, the data reported were conflicting because the rate of myelosuppression was significantly higher in Chinese patients with TPMT*2 than TPMT*3B and TPMT*3C polymorphisms, which could lead to clinical failure of AZA treatment (Fei et al., 2018a). Significant relationship was not observed between myelotoxicity and the TPMT polymorphisms (p = 0.973) in 70 Chinese patients receiving AZA (Su et al., 2020). Therefore, the presence of TPMT polymorphisms alone may not serve as a universally reliable predictor for AZA-induced myelotoxicity, particularly in certain populations such as East Asians, where additional genetic variants (e.g., NUDT15) have shown greater clinical relevance.
3.2.2 NUDT15 polymorphism
NUDT15 gene encodes NUDT15 enzymes, which function to dephosphorylate thiopurine triphosphate to monophosphate. This is included among the metabolism pathways of thiopurine drugs, such as AZA. The most common alleles studied and found to have an association with AZA-related toxicity consisted of NUDT15*3. This variant c.415C>T (rs116855232) causes the change of arginine to cysteine at position 139 and leads to function loss of NUDT15 enzyme due to a decrease in the thermal stability (Yang et al., 2014). The loss of NUDT15 function induces the excessive amount of thiopurine triphosphate and increases the number of TGNs incorporated into DNA and RNA, leading to severe AZA-related cytotoxicity. Among all polymorphisms related to AZA toxicity, NUDT15*3 was reported to have the strongest association with myelosuppression and leukopenia in patients, specifically Asians (Song et al., 2014; Martusevich et al., 2020; Wang et al., 2022). This is due to the frequency of the allele being higher in Asians population than in Caucasians (Tanaka and Saito, 2021). The genetic screening of NUDT15*3 gene has been implemented for personalizing AZA doses to decrease the risk of AZA-induced leukopenia in China (Wang et al., 2022).
3.2.3 ITPA polymorphism
ITPA gene encodes ITPA which contributes to thiopurine metabolism, such as NUDT15 and TPMT enzymes. This has a role in preventing toxicity by restricting the accumulation of toxic thiopurine metabolites, 6-thioinosine-5-triphosphate, through conversion to TIMP (Nagamine et al., 2012). Gene variant of interest is ITPA c.94C>A (rs1127354), a missense mutation reported to be responsible for the decrease or even the loss of ITPA enzymatic activity (Cao and Hegele, 2002; Sumi et al., 2002). This impairs AZA metabolism process, leading to toxicity in patients with SLE and other diseases requiring AZA treatment (Nagamine et al., 2012). Compared to TPMT variants and NUDT15, ITPA c.94C>A was not associated with myelosuppression and hepatotoxicity (Steponaitiene et al., 2012; Chen et al., 2019), but influenza-like symptoms, digestive intolerance, pancreatitis, and arthralgia (Nagamine et al., 2012; Chen et al., 2021). The exact reason and mechanism of the phenomena remain inconclusive.
3.2.4 ABCC4 polymorphism
ABCC4 gene is responsible for encoding MRP4, an ATP-binding cassette transporter that functions as a transmembrane efflux pump to transfer 6-TGN out of the cell. ABCC4 c.2269G>A (rs3765534) variant is suspected to be related to the side effects of leukopenia in patients given thiopurine because of 6-TGN accumulation (Song et al., 2014; Martusevich et al., 2020). MRP4/ABCC4 c.2269G>A (rs3765534) decreases MRP4 function, might be responsible for myelosuppression (Milosevic et al., 2018), and is rarely found in the Caucasian race. However, the allele has been found at a higher frequency among the Asian population, including 14.7%–23% in the Japanese and 8.3% in the Han Chinese (Juster-switlyk et al., 2017; Zhao et al., 2016; Campbell et al., 2016).
3.3 Cyclophosphamide
CYC is a well-established alkylating agent widely used in the treatment of severe manifestations of SLE, particularly lupus nephritis (LN) (Zhang et al., 2014). Its efficacy lies in its potent immunosuppressive activity, which involves the inhibition of DNA replication and induction of cell death in rapidly proliferating immune cells. CYC is a prodrug that undergoes hepatic conversion to its active metabolite, 4-hydroxycyclophosphamide (4-OH-CPA), primarily facilitated by cytochrome P450 (CYP) enzymes, including CYP2B6, CYP2C9, and CYP3A4, with additional contributions from CYP2A6, CYP2C8, and CYP2C19 (Lamba et al., 2014). 4-OH-CPA is the major circulating metabolite and is in equilibrium with aldophosphamide, which subsequently breaks down into phosphoramide mustard, the active cytotoxic compound, and acrolein, a toxic byproduct responsible for bladder toxicity. While CYP enzymes are responsible for activation, enzymes such as glutathione S-transferase (GST) contribute to the detoxification of reactive metabolites, particularly acrolein (Alnasser, 2025). The active alkylating component, phosphoramide mustard, forms alkyl adducts with DNA through a phosphoramide aziridinium intermediate, while DNA alkylation induces a damage leading to cell death (Juster-switlyk et al., 2017). However, the use of CYC is frequently limited by its substantial adverse drug reactions. Well-documented CYC-related adverse effects include myelosuppression, urotoxicity (e.g., hemorrhagic cystitis), gonadotoxicity (e.g., ovarian failure and infertility), hepatotoxicity, and secondary malignancies (Mok, 2016; Mok et al., 1998). The occurrence and severity of CYC-related adverse effects can vary markedly among individuals, even with similar dosing regimens. It is increasingly recognized to be influenced by genetic differences, particularly single nucleotide polymorphisms (SNPs) in genes encoding CYC-metabolizing enzymes, transporters, and detoxification proteins. Genetic polymorphisms can affect the formation and clearance of both therapeutic and toxic CYC metabolites, ultimately altering CYC’s safety and efficacy profile. Polymorphisms in GST (e.g., GSTM1 and GSTP1), CYPs, and aldehyde dehydrogenase (ALDH) genes have been associated with altered metabolism and increased risk of CYC-induced adverse effects in SLE patients (Illustrated in Figure 3) (Audemard-Verger et al., 2016; Indrawijaya et al., 2023; Hajdinák et al., 2020; Indrawijaya et al., 2024).
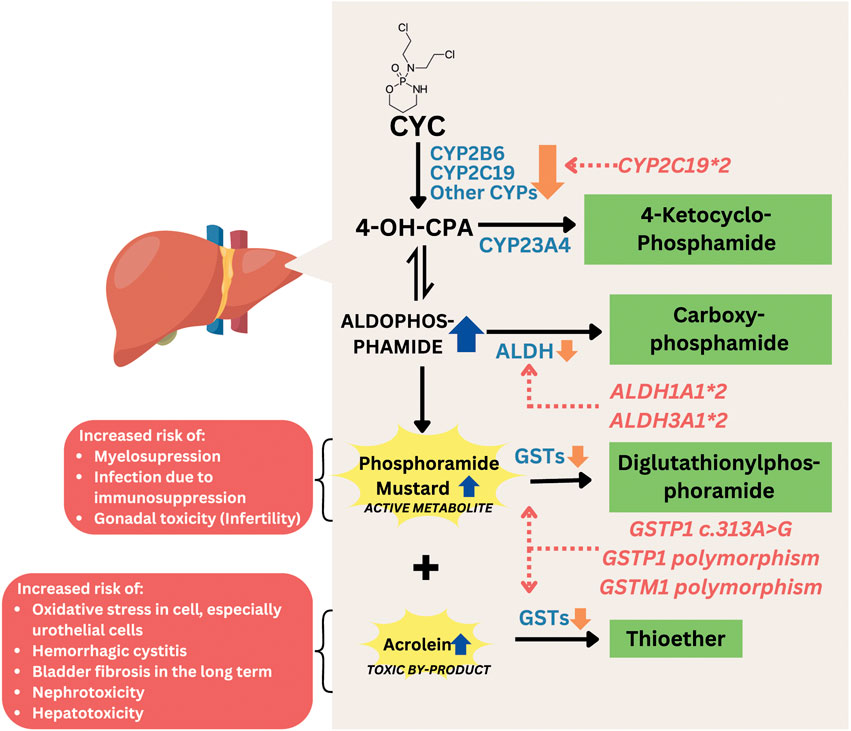
Figure 3. Mechanism of gene polymorphisms affecting CYC-related toxicity. Abbreviations: CYC, cyclophosphamide; CYP, cytochrome P450; 4-OH-CPA, 4-hydroxycyclophosphamide; ALDH, aldehyde dehydrogenase; GST, glutathione S-transferase.
3.3.1 GST polymorphisms
GST genes encode glutathione S-transferase (GST), an important enzyme involved in the detoxification of cyclophosphamide metabolites. GST catalyzes the conjugation of reactive metabolites, such as phosphoramide mustard, into less toxic compounds like 4-glutathionyl-cyclophosphamide (Dirven et al., 1994). A reduction or loss of GST activity can lead to the accumulation and prolonged exposure to these toxic metabolites, thereby increasing the risk of adverse effects and CYC-related toxicity (Hajdinák et al., 2020; Conklin et al., 2015). The well-known GST genes variants associated with occurrence of adverse effects-related to CYC are GSTM1, GSTP1 and GSTT1 (Audemard-Verger et al., 2016; Hahn et al., 2010). GSTA1 is not included in this discussion, as its polymorphisms have been reported to influence the efficacy of CYC treatment rather than its toxicity or adverse effects (Indrawijaya et al., 2024; Wang et al., 2015).
GSTM1 gene encodes Glutathione S-transferase Mu 1 (GSTM1), a key phase II detoxification enzyme that catalyzes the conjugation of glutathione to electrophilic compounds, including toxic metabolites of cyclophosphamide such as acrolein and phosphoramide mustard (NCBI, 2025a) GSTM1 null genotype is the common polymorphism in GSTM1. Individuals with this genotype do not produce functional GSTM1, leading to impaired detoxification capacity and accumulation of acrolein and phosphoramide mustards. There is a strong evidence that the GSTM1 null genotype increases the risk of adverse effects from cyclophosphamide, regardless of other patient characteristics (age, gender, kidney function, and total CYC dose), with the odds ratio was 3.345, compared to those with functioning GSTM1 gene (Audemard-Verger et al., 2016). While some studies have reported a significant association between the GSTM1 null genotype and increased CYC-related adverse effects, particularly when adjusted for clinical variables, others have found no relationship in the context of short-term high-dose regimens (Zhong et al., 2006). These discrepancies may reflect differences in study design, treatment protocol, population genetics, or definitions of toxicity.
GSTP1 gene encodes glutathione S-transferase Pi 1, an enzyme involved in the phase II detoxification of reactive drug metabolites, including those generated during cyclophosphamide (CYC) metabolism. GSTP1 catalyzes the conjugation of electrophilic CYC byproducts with glutathione, facilitating their elimination (NCBI, 2025b; Hayes et al., 2005). A commonly studied variant of this gene is Ile105Val polymorphism (rs1695), which involves a single nucleotide change from A to G, resulting in the substitution of isoleucine (Ile) with valine (Val) at codon 105 of the enzyme (Hasni et al., 2016). This amino acid change alters the structure and function of GSTP1, potentially reducing its catalytic efficiency (Gorukmez et al., 2016). As GSTP1 is only active in its dimer form, both heterozygous and homozygous variant causes reduction in GTSP1 activity, causing an increase of acrolein and phosphoramide mustard concentration. Although increased levels of phosphoramide mustard are associated with improved cyclophosphamide efficacy in cancer patients, in SLE patients this increase has been linked to a higher incidence of myelotoxicity, particularly at higher cyclophosphamide doses. In contrast, gastrointestinal (GI) toxicity tends to be more frequent at lower doses of cyclophosphamide in SLE, likely due to its non-linear pharmacokinetic profile, which results in greater biliary excretion of toxic metabolites following low-dose administration compared to high-dose administration (Zhong et al., 2006).
GSTT1 gene encodes for the enzyme glutathione S-transferase theta 1, which also plays a role in the detoxification of toxic metabolites of CYC. A common polymorphism involves a homozygous deletion of the GSTT1 gene, referred to as the GSTT1 null genotype, resulting in the absence of functional enzyme activity. While the GSTT1 null genotype has been associated with increased chemotherapy-related toxicities in oncology settings (Cho et al., 2010; Wang et al., 2016; Aguiar et al., 2012), this variant does not appear to significantly affect the risk of cyclophosphamide-related adverse effects—such as myelosuppression and gastrointestinal toxicity—in SLE patients (Zhong et al., 2006). This may be due to higher, more frequent doses of CYC, and different patient characteristics in cancer patients compared to SLE patients.
3.3.2 CYP polymorphisms
CYP genes are responsible for encoding a family of Cytochrome P450 enzymes which activate CYC to the active metabolite, 4-OH-CPA. It is generated mainly by CYP2B6, CYP3A4, and CYP2C9, with additional contributions from CYP2C19, CYP2A6, and CYP2C8. Genetic polymorphisms in these enzymes can significantly influence CYC’s pharmacokinetics, leading to interindividual differences in efficacy and toxicity. Excessive amounts or prolonged duration of active metabolites exposure to body cells may be associated with CYC-related adverse effects, such as ovarian toxicity. Among these enzymes, the most studied polymorphism variants are CYP2B6, which has been studied in relation to CYC metabolism and toxicity. Three notable variants of CYP2B6 include c.516G>T (rs3745274), c.785A>G (rs2279343), and −750T>C (rs4802101). The c.516G>T variant, which defines the CYP2B6*6, results in the substitution of glutamine with histidine at codon 172 (Q172H), leading to reduced enzyme expression and enzyme (Abuelsoud et al., 2021). The c.785A>G (rs2279343) variant causes a lysine-to-arginine substitution at position 262 (K262R) and often co-occurs with c.516G>T as part of the CYP2B6*6, further contributing to impaired metabolic activity (Tran et al., 2008). This may reduce the formation of active metabolites, potentially lowering toxicity but also decreasing efficacy. Meanwhile, the −750T>C (rs4802101) variant is located in the promoter region, which may influence gene expression levels, though its clinical impact remains inconclusive (Zanger and Klein, 2013). For instance, a study involving 116 Chinese patients, individuals carrying the C allele had a significantly lower incidence of gastrointestinal toxicity and leukocytopenia compared to those with the wild-type TT genotype (Shu et al., 2016). However, other studies have reported that there was no association between CYP2B6 polymorphisms and reduced CYC-related adverse effects (Kumaraswami et al., 2017; Singh et al., 2007).
CYP3A4 is another major to CYC metabolism. One notable polymorphism of CYP3A4 is CYP3A4*1B, characterized by an A>G transition in the 5′-flanking region of the gene. This variant has been associated with altered gene expression and enzymatic activity (Su et al., 2010). While there is still no study investigating the functional impact of this variant on CYC-related adverse effects in SLE patients, a study on cancer patients found no significant association between CYP3A4*1B and the pharmacokinetics of CYC or its active metabolite, 4-OH-CPA (Ekhart et al., 2008). CYP2C9 also plays a role in cyclophosphamide (CYC) metabolism, but contributes only minimally to its activation into 4-hydroxycyclophosphamide (4-OH-CPA) (Griskevicius et al., 2003).
CYP2C19, CYP2A6, and CYP2C8 play ancillary roles in CYC metabolism (Muñiz et al., 2022). Among these, the CYP2C19*2 is one of the most extensively studied variants due to its high frequency in many populations, especially in Asian population, and its association with reduced enzymatic activity (Audemard-Verger et al., 2016). The CYP2C19*2 c.681G>A (rs4244285) in exon 5 is responsible for a decrease in CYP enzyme activity. There are conflicting data regarding whether CYP2C19 is associated with a higher risk of CYC toxicity. Several studies stated that the presence of this allele is related to ovarian toxicity and the risk of CYC treatment failure (Kumaraswami et al., 2017; Lee et al., 2016; Ngamjanyaporn et al., 2011). However, a study associated CYP2C19*2 presence with a lower risk of toxicity, specifically ovarian toxicity in Indian patients (Singh et al., 2007). Another study reported that CYP2C19*2 had no association with CYC-related toxicity (Audemard-Verger et al., 2016). Further investigation is required to understand more about the reason for these results. Additionally, studies on genetic polymorphisms of CYP2A6 and CYP2C8 in the context of CYC metabolism are limited, particularly in patients with systemic lupus erythematosus (SLE). Thus, their clinical relevance in modulating CYC efficacy or toxicity remains unclear and warrants further investigation.
3.3.3 ALDH polymorphisms
ALDH encodes Aldehyde Dehydrogenase (ALDH), an enzyme that contributes to CYC metabolism and detoxification, such as GST. ALDH converts aldophosphamide to a non-toxic metabolite, carbophosphamide. The presence of two variants (ALDH3A1*2 and ALDH1A1*2) is known to reduce the activity of the ALDH enzyme, which affects the detoxification capacity of cyclophosphamide (CYC), theoretically increasing the risk of toxicity from CYC (Ekhart et al., 2008). A study involving 113 Caucasian patients receiving high-dose chemotherapy with a combination of CYC, thiotepa, and carboplatin showed that the ALDH1A1*2 variant, located in the promoter region and potentially having significant gene regulatory effects, was associated with an increased risk of liver toxicity and hemorrhagic cystitis (Ekhart et al., 2008). Another study at Michigan Hospital involving 846 patients receiving CYC-based chemotherapy regimens indicated that the presence of the ALDH1A1 c.1234A>G (rs8187996) variant was actually associated with a reduced risk of ≥3 toxicity or the need for treatment modification due to toxicity. The rs8187996 variant is located in the intron (non-coding) region, which has a minor effect on gene regulation (Hwang et al., 2022). The differences in the location and functional effects of the genetic variants studied, as well as the differences in chemotherapy regimens used, may explain the differing results between the two studies.
3.4 Mycophenolate mofetil
MMF is a pro-drug for mycophenolic acid (MPA), which hinders inosine monophosphate (IMP), thereby suppressing the production of guanosine monophosphate (GMP) to initiate reduced proliferation of B and T cells, as well as diminished production of antibodies. This pro-drug is used to treat SLE, specifically LN, due to the immunosuppressive activity. MMF experiences rapid absorption in the GI system and is transformed into MPA by esterase enzymes, particularly carboxylesterase 2 (CES2). MPA then engages in an enterohepatic cycle facilitated by an organic anion-carrying polypeptide. In this cycle, glucuronidation transforms MPA into the inactive forms, namely, 7-O-glucoside and acyl-glucuronide. The liver, kidney, and GI tract use UDP-glucuronosyltransferase 1A9 (UGT1A9) and other UGT1A superfamily enzymes for this glucuronidation process, leading to the formation of MPA 7-O-glucuronide (MPAG). The pharmacologically active metabolite, acyl-glucuronide of MPA (AcMPAG), is thought to contribute to the typical adverse effects of MMF. Following glucuronidation, MPAG is eliminated from the body through organic anion transporters (OATs) (Liu et al., 2015). Several MMF-related toxicities have been reported, such as GI manifestation, infections, anemia, low platelet count, leukopenia, and many more (Mok, 2015; Riskalla et al., 2003). The most common gene polymorphisms associated with MMF toxicity are IMPDH, UGT2B7, and SLCO1B1 polymorphisms. For example, IMPDH2 c.3757T>C (rs11706052) has been associated with gastrointestinal toxicity (OR = 3.05, 95% CI: 1.22–7.60, p = 0.02), UGT2B7 -900A>G (rs7438135) with GI toxicity (OR = 2.34, 95% CI: 1.14–4.79, p = 0.02), and SLCO1B1 c.521T>C (rs4149056) with hematologic toxicity (OR = 3.10, 95% CI: 1.23–7.82, p = 0.02) (Na Takuathung et al., 2021). Figure 4 shows effects of these gene polymorphisms on MMF-related toxicity.
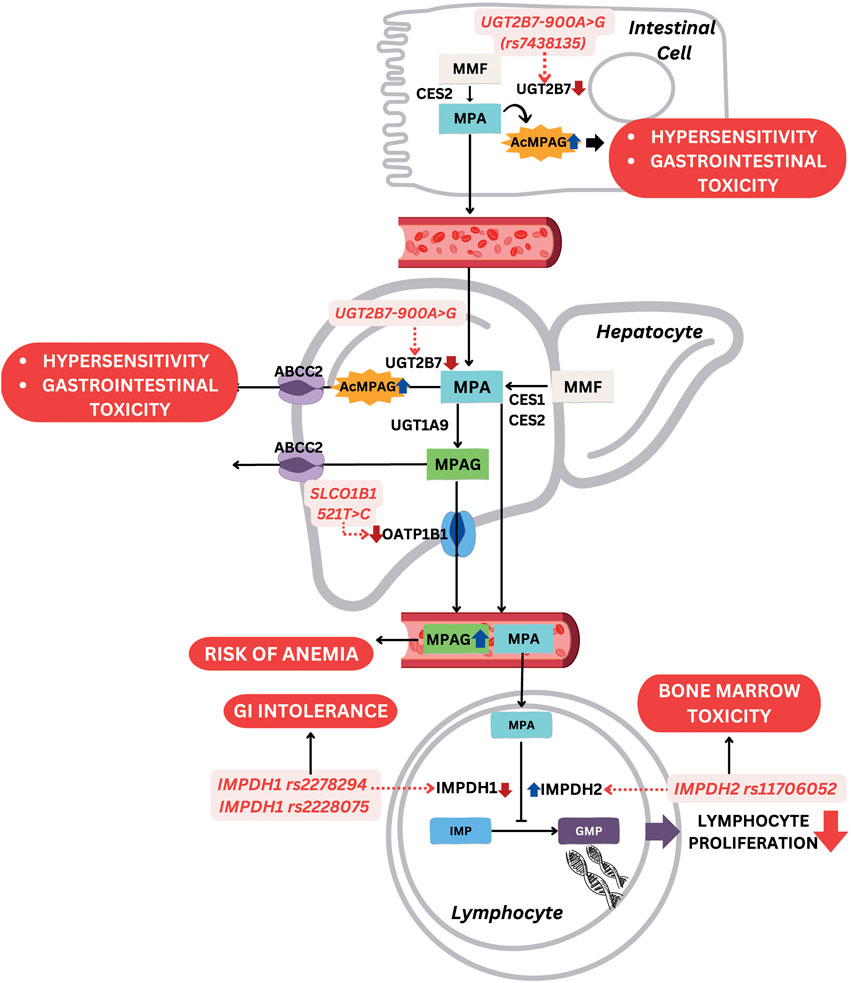
Figure 4. Mechanism of gene polymorphisms affecting MMF-related toxicity. Abbreviations: MMF, mycophenolate mofetil; MPA, mycophenolic acid; AcMPAG, acyl-glucuronide of mycophenolic acid; MPAG, mycophenolic acid 7-O-glucuronide; IMP, inosine monophosphate; GMP, guanosine monophosphate; CES2, carboxylesterase 2; UGT, UDP-glucuronosyltransferases; ABCC2, ATP-binding cassette sub-family C member 2; OAT, organic anion transporters; SLCO1B1, solute carrier organic anion transporter family member 1B1; IMPDH, inosine 5'-monophosphate dehydrogenase; GI, gastrointestinal.
3.4.1 IMPDH polymorphisms
IMPDH encodes an enzyme called inosine 5′-monophosphate dehydrogenase 1 (IMPDH), which functions to produce the guanosine required in the lymphocytes proliferation process. MPA works by inhibiting IMPDH activity, while IMPDH1 and IMPDH2 variants influence the incidence of MMF toxicity. The variant of IMPDH1 -106C>A (rs2278294) and c.1575G>A (rs2228075) were strongly associated with infection susceptibility and GI intolerance, such as nausea, vomiting, and diarrhea (Ohmann et al., 2010; Shu et al., 2021). IMPDH1 encodes inosine monophosphate dehydrogenase type I, a key enzyme involved in the de novo synthesis of guanine nucleotides, which is essential for the proliferation of T and B lymphocytes. Mycophenolic acid (MPA), the active metabolite of mycophenolate mofetil (MMF), exerts its immunosuppressive effects by inhibiting this enzyme. Certain IMPDH1 polymorphisms, such as nonsynonymous variants including rs2278294 and rs2228075, have been reported to reduce enzymatic activity through altered protein stability and impaired tetramer formation, without significantly affecting mRNA levels, indicating a post-translational regulatory mechanism. This reduction in IMPDH1 activity may enhance the pharmacodynamic effects of MPA, thereby intensifying immunosuppression and predisposing patients to infection. Additionally, enhanced local MPA effects in the gastrointestinal tract may contribute to increased risk of nausea, vomiting, and diarrhea. Ethnic differences in the distribution of IMPDH1 haplotypes may also partially explain interindividual variability in toxicity profiles (Wu et al., 2010). Meanwhile, IMPDH2 c.3757T>C (rs11706052) was associated with bone marrow toxicity due to an increase in IMPDH activity (Neerman and Boothe, 2003).
3.4.2 UGT2B7 polymorphisms
UGT encodes UDP-glucuronosyltransferases (UGTs), such as UGT1A9 and UGT1A8, which convert MPA to the inactive MPAG. However, one variant of UGT known as UGT2B7 produces a minor highly reactive metabolite, AcMPAG, which is associated with drug toxicity (Bernard et al., 2006). A UGT2B7 variant called UGT2B7 -900A>G (rs7438135) was associated with a higher risk of leukopenia and anemia. In this variant, adenine (A) at coding DNA position 900 is replaced by guanine (G), causing a decrease in UGT2B7 activity and leading to an accumulation of toxic metabolite, AcMPAG. Another genetic variant, UGT2B7 c.802C>T (rs7439366), was identified with contributions to increased susceptibility to infections, particularly Pneumocystis carinii pneumonia (Shu et al., 2021).
3.4.3 SLCO1B1 polymorphism
SLCO1B1 or solute carrier OATs family member 1B1, is a gene that encodes a membrane transporter protein primarily found in the liver. This transporter plays a crucial role in facilitating various endogenous and exogenous substances, including MMF, across cell membranes. SLCO1B1 participates in the clearance of numerous drugs, and the most common alleles studied include SLCO1B1 c.521T>C (rs4149056) that causes the substitution of valine to alanine at position 174. The substitution leads to a decrease in the ability of the transporter to facilitate MPA intake, increasing MPA plasma concentration. SLCO1B1 c.521T>C was associated with MMF-induced anemia (Shu et al., 2021), but a previous study did not find a relationship between this polymorphism and toxicity (Neerman and Boothe, 2003).
4 Discussion
4.1 Immunosuppressant-related toxicity
Immunosuppressant is used when the disease progresses from moderate to severe condition and the administration of GCs as SLE first-line treatment cannot sustain clinical remission (Gatto et al., 2019). In general, immunosuppressant mechanism of action requires suppressing and decreasing the autoimmune responses, which can target various organs and systems in the body. This serves to minimize damage in various organs, thereby preventing life-threatening conditions. The use of immunosuppressant is based on the organs engaged in SLE activity and the conditions of patients. Moderately active lupus and joints involved are treated with MTX, while LN and other severe cases are treated primarily with MMF and CYC (Mohamed et al., 2019; Fraenkel et al., 2021). Immunosuppressants contribute to reduced GC exposure, improved disease stabilization, and enhanced long-term survival. Nonetheless, their use is associated with a high incidence of adverse effects, with reported toxicity rates ranging from 42.8% to 97.3%. Common toxicities include infections, gastrointestinal disturbances, amenorrhea, ovarian dysfunction, hematologic cytopenia, hepatic dysfunction, bone marrow suppression, pulmonary toxicity, and others. The specific toxicity profile often depends on the pharmacological agent used and the patient’s genetic predisposition (Oglesby et al., 2013).
4.2 Pharmacogenomics and personalized treatment
The integration of pharmacogenomic strategies into SLE management—particularly regarding immunosuppressive therapy—has gained increasing attention. Advances in pharmacogenetic research have identified single nucleotide polymorphisms (SNPs) as critical determinants of interindividual variability in drug response and toxicity (Meng et al., 2018). Genetic polymorphisms in genes involved in drug metabolism and enzymatic activity—such as TPMT*2, *3B, *3C variants in azathioprine (AZA) recipients and MTHFR c.667C>T and c.1298A>C in MTX-treated patients—have been associated with increased susceptibility to drug-induced toxicity (Van Der Put et al., 1998; Gu et al., 2023; Murugesan et al., 2009) (see Table 1).
Furthermore, ethnic variability contributes significantly to the genetic landscape of SLE, affecting the distribution and impact of pharmacogenomic markers. For instance, the ATIC c.347C>G polymorphism has shown differential toxicity outcomes across racial groups in MTX users (Lee and Bae, 2016). Similarly, the ABCC4 c.2269G>A variant has been linked to AZA toxicity, with the highest incidence observed in Asian populations and minimal occurrence in Caucasians (Juster-switlyk et al., 2017; Zhao et al., 2016; Campbell et al., 2016; Milosevic et al., 2018). These findings underscore the need for population-specific pharmacogenomic research to develop equitable and evidence-based treatment strategies across diverse demographic groups.
4.3 Current research gaps and future directions
Current pharmacogenomic studies in SLE predominantly focus on single-drug and single-gene associations, with limited exploration into polygenic interactions or the effects of gene–drug combination therapies on toxicity profiles. Broader research encompassing gene-gene and gene-environment interactions is essential to fully understand the complexity of treatment responses in SLE. The incorporation of advanced genomic technologies such as next-generation sequencing (NGS) holds promise for generating more comprehensive datasets. Such data could inform clinical decision-making and facilitate the development of personalized medicine approaches tailored to the genetic and clinical characteristics of individual patients.
5 Conclusion
Understanding the association between immunosuppressant-related adverse effects and gene polymorphisms is crucial for assessing patient risk, enabling individualized drug therapy, and enriching global pharmacogenetic knowledge. Insights from pharmacogenetics can support the prediction and prevention of adverse reactions to drugs such as methotrexate (MTX), azathioprine (AZA), cyclophosphamide (CYC), and mycophenolate mofetil (MMF). Nevertheless, current evidence is limited by small sample sizes, underrepresentation of specific populations (e.g., pediatric and ethnically diverse groups), and methodological challenges in genotyping and data interpretation.
Author contributions
SH: Writing – original draft, Investigation, Data curation, Visualization. LH: Methodology, Supervision, Conceptualization, Writing – review and editing. RA: Writing – review and editing, Methodology, Supervision. MB: Supervision, Writing – review and editing, Methodology, Conceptualization.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by Ministry of Education, Culture, Research, and Technology, Republic of Indonesia under Doctoral Dissertation Research scheme grant.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abuelsoud, N., Fayed, H., and Elkateeb, E. (2021). The association between azathioprine genetic polymorphisms, clinical efficacy and adverse drug reactions among egyptian patients with autoimmune diseases. Pharmacogenomics Pers. Med 14, 179–187. doi:10.2147/PGPM.S285033
Aguiar, E. S. de, Giacomazzi, J., Schmidt, A. V., Bock, H., Saraiva-Pereira, M. L., Schuler-Faccini, L., et al. (2012). GSTM1, GSTT1, and GSTP1 polymorphisms, breast cancer risk factors and mammographic density in women submitted to breast cancer screening. Rev. Bras. Epidemiol. 15 (2), 246–255. doi:10.1590/s1415-790x2012000200002
Ahmadzadeh, A., Zamani, N., Hassanian-Moghaddam, H., Hadeiy, S. K., and Parhizgar, P. (2019). Acute versus chronic methotrexate poisoning; A cross-sectional study. BMC Pharmacol. Toxicol. 20 (1), 39–7. doi:10.1186/s40360-019-0316-8
Alnasser, S. M. (2025). The role of glutathione S-transferases in human disease pathogenesis and their current inhibitors. Genes Dis. 12 (4), 101482. doi:10.1016/j.gendis.2024.101482
Audemard-Verger, A., Silva, N. M., Verstuyft, C., Costedoat-Chalumeau, N., Hummel, A., Le Guern, V., et al. (2016). Glutathione S transferases polymorphisms are independent prognostic factors in lupus nephritis treated with cyclophosphamide. PLoS One 11 (3), 1–9. doi:10.1371/journal.pone.0151696
Basta, F., Fasola, F., Triantafyllias, K., and Schwarting, A. (2020). Systemic lupus erythematosus (SLE) therapy: the old and the New. Rheumatol. Ther. 7 (3), 433–446. doi:10.1007/s40744-020-00212-9
Bernard, O., Tojcic, J., Journault, K., Perusse, L., and Guillemette, C. (2006). Influence of nonsynonymous polymorphisms of UGT1A8 and UGT2B7 metabolizing enzymes on the formation of phenolic and acyl glucuronides of mycophenolic acid. Drug Metab. Dispos. 34 (9), 1539–1545. doi:10.1124/dmd.106.010553
Campbell, J. M., Bateman, E., Stephenson, M. D., Bowen, J. M., Keefe, D. M., and Peters, M. D. J. (2016). Methotrexate-induced toxicity pharmacogenetics: an umbrella review of systematic reviews and meta-analyses. Cancer Chemother. Pharmacol. 78 (1), 27–39. doi:10.1007/s00280-016-3043-5
Cao, H., and Hegele, R. A. (2002). DNA polymorphisms in ITPA including basis of inosine triphosphatase deficiency. J. Hum. Genet. 47 (11), 620–622. doi:10.1007/s100380200095
Cattaneo, D., Baldelli, S., and Perico, N. (2008). Pharmacogenetics of immunosuppressants: progress, pitfalls and promises. Am. J. Transpl. 8 (7), 1374–1383. doi:10.1111/j.1600-6143.2008.02263.x
Chen, Z. Y, Zhu, Y. H., Zhou, L. Y., Shi, W. Q., Qin, Z., Wu, B., et al. (2021). Association between genetic polymorphisms of metabolic enzymes and azathioprine-induced myelosuppression in 1,419 Chinese patients: a retrospective study. Front Pharmacol 12, 672769. doi:10.3389/fphar.2021.672769
Cho, H. J., Eom, H. S., Kim, H. J., Kim, I. S., Lee, G. W., and Kong, S. Y. (2010). Glutathione-S-transferase genotypes influence the risk of chemotherapy-related toxicities and prognosis in Korean patients with diffuse large B-cell lymphoma. Cancer Genet. Cytogenet 198 (1), 40–46. doi:10.1016/j.cancergencyto.2009.12.004
Conklin, D. J., Haberzettl, P., Jagatheesan, G., Baba, S., Merchant, M. L., Prough, R. A., et al. (2015). Glutathione S-transferase P protects against cyclophosphamide-induced cardiotoxicity in mice. Toxicol. Appl. Pharmacol. 285 (2), 136–148. doi:10.1016/j.taap.2015.03.029
Dirven, C. M. F., Van Ommen, B., Van Bladeren, P. J., and Muller, M. (1994). Inhibition of glutathione conjugation of electrophilic compounds by ethacrynic acid. Biochem. Pharmacol., 47 (3), 495–502. Available online at: https://pubmed.ncbi.nlm.nih.gov/7954469/.
Dwivedi, A., Jha, A. K., and Gupta, V. (2020). Impact of MTHFR C677T and A1298C gene polymorphisms on MTX drug toxicity and efficacy profile of RA patients in North India. Meta Gene 24, 100705. doi:10.1016/j.mgene.2020.100705
Ekhart, C., Rodenhuis, S., Smits, P. H. M., Beijnen, J. H., and Huitema, A. D. R. (2008). Relations between polymorphisms in drug-metabolising enzymes and toxicity of chemotherapy with cyclophosphamide, thiotepa and carboplatin. Pharmacogenet Genomics 18 (11), 1009–1015. doi:10.1097/FPC.0b013e328313aaa4
Fei, X., Shu, Q., Hua, B. Z., Wang, S. Y., Chen, Z. Y., Ge, W. H., et al. (2018b). NUDT15 R139C variation increases the risk of azathioprine-induced toxicity in Chinese subjects. Medicine (Baltimore) 97 (17), e0301. doi:10.1097/MD.0000000000010301
Fei, X., Shu, Q., Zhu, H., Hua, B., Wang, S., Guo, L., et al. (2018a). NUDT15 R139C variants increase the risk of Azathioprine-induced leukopenia in Chinese autoimmune patients. Front. Pharmacol. 9, 460–468. doi:10.3389/fphar.2018.00460
Fraenkel, L., Bathon, J. M., England, B. R., St Clair, E. W., Arayssi, T., Carandang, K., et al. (2021). 2021 American college of rheumatology guideline for the treatment of rheumatoid arthritis. Arthritis Care Res. 73 (7), 924–939. doi:10.1002/acr.24596
Gande, N., Hochmayr, C., Staudt, A., Bernar, B., Stock, K., Kiechl, S. J., et al. (2023). Plasma homocysteine levels and associated factors in community-dwelling adolescents: the EVA-TYROL study. Front. Cardiovasc Med. 10, 1140990. doi:10.3389/fcvm.2023.1140990
Gatto, M., Zen, M., Iaccarino, L., and Doria, A. (2019). New therapeutic strategies in systemic lupus erythematosus management. Nat. Rev. Rheumatol. 15 (1), 30–48. doi:10.1038/s41584-018-0133-2
Gorukmez, O., Yakut, T., Gorukmez, O., Sag, S. O., Topak, A., Sahinturk, S., et al. (2016). Glutathione S-transferase T1, M1 and P1 genetic polymorphisms and susceptibility to colorectal cancer in Turkey. Asian Pac J. Cancer Prev. 17, 3855–3859. doi:10.1016/j.genrep.2019.100365
Grabar, P. B., Rojko, S., Logar, D., and Dolžan, V. (2010). Genetic determinants of methotrexate treatment in rheumatoid arthritis patients: a study of polymorphisms in the adenosine pathway. Ann. Rheum. Dis. 69 (5), 931–932. doi:10.1136/ard.2009.111567
Griskevicius, L., Yasar, Ü., Sandberg, M., Hidestrand, M., Eliasson, E., Tybring, G., et al. (2003). Bioactivation of cyclophosphamide: the role of polymorphic CYP2C enzymes. Eur. J. Clin. Pharmacol. 59 (2), 103–109. doi:10.1007/s00228-003-0590-6
Gu, J., Lin, Y., and Wang, Y. (2023). Case report: NUDT15 polymorphism and severe azathioprine-induced myelosuppression in a young Chinese female with systematic lupus erythematosus: a case analysis and literature review. Front. Pharmacol. 14, 1001559. doi:10.3389/fphar.2023.1001559
Hahn, T., Zhelnova, E., Sucheston, L., Demidova, I., Savchenko, V., Battiwalla, M., et al. (2010). A deletion polymorphism in glutathione-S-transferase mu (GSTM1) and/or theta (GSTT1) is associated with an increased risk of toxicity after autologous blood and marrow transplantation. Biol. Blood Marrow Transpl. 16 (6), 801–808. doi:10.1016/j.bbmt.2010.01.001
Hajdinák, P., Szabó, M., Kiss, E., Veress, L., Wunderlich, L., and Szarka, A. (2020). Genetic polymorphism of GSTP-1 affects cyclophosphamide treatment of autoimmune diseases. Molecules 25 (7), 1542. doi:10.3390/molecules25071542
Hamed, K. M., Dighriri, I. M., Baomar, A. F., Alharthy, B. T., Alenazi, F. E., Alali, G. H., et al. (2022). Overview of methotrexate toxicity: a comprehensive literature review. Cureus 14 (9), e29518. doi:10.7759/cureus.29518
Hasni, D., Siregar, K. B., and Lim, H. (2016). The influence of glutathion S-transferase P-1 polymorphism A313G rs1695 on the susceptibility to cyclophosphamide hematologic toxicity in Indonesian patients. Med. J. Indones. 25 (2), 118–126. doi:10.13181/mji.v25i2.1308
Hayes, J. D., Flanagan, J. U., and Jowsey, I. R. (2005). Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 45, 51–88. doi:10.1146/annurev.pharmtox.45.120403.095857
Hochberg, M. C. (1997). Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Reum. 40 (9), 1725. doi:10.1002/art.1780400928
Huang, J., Fan, H., Qiu, Q., Liu, K., Lv, S., Li, J., et al. (2020). Are gene polymorphisms related to adverse events of methotrexate in patients with rheumatoid arthritis? A retrospective cohort study based on an updated meta-analysis. Ther. Adv. Chronic Dis. 11, 2040622320916026. doi:10.1177/2040622320916026
Hwang, M., Kidwell, K. M., and Hertz, D. L. (2022). Association of ALDH1A1 polymorphism with toxicity from cyclophosphamide treatment. Durham, NC: Research Square.
Indrawijaya, Y. Y., Artarini, A., Hamijoyo, L., and Iwo, M. I. (2024). GSTA1 gene polymorphisms are associated with cyclophosphamide effectiveness in lupus nephritis patients: A case-control study in Indonesia. Narra J., 4 (3), e1144. doi:10.52225/narra.v4i3.1144
Indrawijaya, Y. Y. A., Hamijoyo, L., Artarini, A. A., and Iwo, M. I. (2023). Genetic polymorphisms associated with cyclophosphamide outcome and risk of toxicity in patients with lupus nephritis. Acta Med. Indones. 55 (3), 343–349.
Jung, J. H., Soh, M. S., Ahn, Y. H., Um, Y. J., Jung, J. Y., Suh, C. H., et al. (2016). Thrombocytopenia in systemic lupus erythematosus. Med. (United States) 95 (6), 1–7. doi:10.1097/MD.0000000000002818
Juster-Switlyk, K., Smith, A. G., Kovacsovics, T., Stephens, D., Glenn, M., Palmer, C. A., et al. (2017). MTHFR C677T polymorphism is associated with methotrexate-induced myelopathy risk. Neur. 88 (6), 1–2. doi:10.1212/WNL.0000000000003590
Katarzyna, P. B., Wiktor, S., Ewa, D., and Piotr, L. (2023). Current treatment of systemic lupus erythematosus: a clinician’s perspective. Rheumatol. Int. 43 (8), 1395–1407. doi:10.1007/s00296-023-05306-5
Kivity, S., Zafrir, Y., Loebstein, R., Pauzner, R., Mouallem, M., and Mayan, H. (2014). Clinical characteristics and risk factors for low dose methotrexate toxicity: a cohort of 28 patients. Autoimmun. Rev. 13 (11), 1109–1113. doi:10.1016/j.autrev.2014.08.027
Kumaraswami, K., Katkam, S. K., Aggarwal, A., Sharma, A., Manthri, R., Kutala, V. K., et al. (2017). Epistatic interactions among CYP2C19*2, CYP3A4 and GSTP1 on the cyclophosphamide therapy in lupus nephritis patients. Pharmacogenomics 18 (15), 1401–1411. doi:10.2217/pgs-2017-0069
Lamba, V., Sangkuhl, K., Sanghavia, K., Fish, A., Altman, R. B., and Klein, T. E. (2014). PharmGKB summary: mycophenolic acid pathway. NIH Public Access 24 (1), 73–79. doi:10.1097/FPC.0000000000000010
Lee, J. Y., Vinayagamoorthy, N., Han, K., Kwok, S. K., Ju, J. H., Park, K. S., et al. (2016). Association of polymorphisms of cytochrome P450 2D6 with blood hydroxychloroquine levels in patients with systemic lupus erythematosus. Arthritis Rheumatol. 68 (1), 184–190. doi:10.1002/art.39402
Lee, Y. H., and Bae, S. C. (2016). Association of the ATIC 347 C/G polymorphism with responsiveness to and toxicity of methotrexate in rheumatoid arthritis: a meta-analysis. Rheumatol. Int. 36 (11), 1591–1599. doi:10.1007/s00296-016-3523-2
Liu, Y. P., Xu, H. Q., Li, M., Yang, X., Yu, S., Fu, W. L., et al. (2015). Association between thiopurine S-methyltransferase polymorphisms and azathioprine-induced adverse drug reactions in patients with autoimmune diseases: a meta-analysis. PLoS One 10 (12), e0144234. doi:10.1371/journal.pone.0144234
Londono, J., Saldarriaga, E. L., Rueda, J. C., Giraldo-Bustos, R., Angarita, J. I., Restrepo, L., et al. (2020). Pharmacogenetic aspects of methotrexate in a cohort of colombian patients with rheumatoid arthritis. Biomed. Rep. 13 (4), 34–37. doi:10.3892/br.2020.1341
Martusevich, N., Aksenova, E., and Gudkevich, K. (2020). Rheumatoid arthritis treatment: the earlier the better to prevent joint damage. Ann. Rheum. Dis. 79 (Suppl 1), 1311. doi:10.1136/rmdopen-2015-000057
McKeon, K. P., and Jiang, S. H. (2020). Treatment of systemic lupus erythematosus. Aust. Prescr. 43 (3), 85–90. doi:10.18773/austprescr.2020.022
Meng, H. Y., Luo, Z. H., Hu, B., Jin, W. L., Yan, C. K., Li, Z. B., et al. (2018). SNPs affecting the clinical outcomes of regularly used immunosuppressants. Pharmacogenomics 19 (5), 495–511. doi:10.2217/pgs-2017-0182
Milosevic, G., Kotur, N., Krstovski, N., Lazic, J., Zukic, B., Stankovic, B., et al. (2018). Variants in TPMT, ITPA, ABCC4 and ABCB1 genes as predictors of 6-mercaptopurine induced toxicity in children with acute lymphoblastic leukemia. J. Med. Biochem. 37 (3), 320–327. doi:10.1515/jomb-2017-0060
Mohamed, A., Chen, Y., Wu, H., Liao, J., Cheng, B., and Lu, Q. (2019). Therapeutic advances in the treatment of SLE. Int. Immunopharmacol. 72, 218–223. doi:10.1016/j.intimp.2019.03.010
Mok, C. C. (2015). Mycophenolate mofetil for lupus nephritis: an update. Expert Rev. Clin. Immunol. 11 (12), 1353–1364. doi:10.1586/1744666X.2015.1087314
Mok, C. C. (2016). Con: cyclophosphamide for the treatment of lupus nephritis. Nephrol. Dial. Transpl. 31 (7), 1053–1057. doi:10.1093/ndt/gfw068
Mok, C. C., Lau, C., and Wong, R. (1998). Risk factors for ovarian failure in patients with systemic lupus erythematosus receiving cyclophosphamide therapy. Arthritis Reum. 41 (5), 831–837. doi:10.1002/1529-0131(199805)41:5<831::AID-ART9>3.0.CO;2-1
Muñiz, P., Andrés-Zayas, C., Carbonell, D., Chicano, M., Bailén, R., Oarbeascoa, G., et al. (2022). Association between gene polymorphisms in the cyclophosphamide metabolism pathway with complications after haploidentical hematopoietic stem cell transplantation. Front. Immunol. 13, 1–12. doi:10.3389/fimmu.2022.1002959
Muralidharan, N., Mariaselvam, C. M., Jain, V. K., Gulati, R., and Negi, V. S. (2016). ATIC 347C>G gene polymorphism may be associated with methotrexate-induced adverse events in south Indian Tamil rheumatoid arthritis. Pharmacogenomics 17 (3), 241–248. doi:10.2217/pgs.15.170
Murugesan, R., Vahab, S. A., Patra, S., Rao, R., Rao, J., Rai, P., et al. (2009). Thiopurine S-methyltransferase alleles, TPMT* 2, * 3B and * 3C, and genotype frequencies in an Indian population. Exp. Ther. Med. 1 (1), 121–127. doi:10.3892/etm_00000021
Nagamine, A., Takenaka, M., Aomori, T., Okada, Y., Hiromura, K., Nojima, Y., et al. (2012). Effect of genetic polymorphisms on effectiveness of low-dose azathioprine in Japanese patients with systemic lupus erythematosus. Am. J. Heal Pharm. 69 (23), 2072–2078. doi:10.2146/ajhp120179
Na Takuathung, M., Sakuludomkan, W., and Koonrungsesomboon, N. (2021). The impact of genetic polymorphisms on the pharmacokinetics and pharmacodynamics of mycophenolic acid: systematic review and meta-analysis. Clin. Pharmacokinet. 60 (10), 1291–1302. doi:10.1007/s40262-021-01037-7
NCBI (2025a). GSTM1 glutathione S-transferase mu 1 [Homo sapiens (human)] – Gene ID: 2944. Available online at: https://www.ncbi.nlm.nih.gov/gene/2944.
NCBI (2025b). GSTP1 glutathione S-transferase pi 1 [Homo sapiens (human)] – Gene ID: 2950. Available online at https://www.ncbi.nlm.nih.gov/gene/2950.
Neerman, M. F., and Boothe, D. M. (2003). A possible mechanism of gastrointestinal toxicity posed by mycophenolic acid. Pharmacol. Res. 47 (6), 523–526. doi:10.1016/S1043-6618(03)00055-0
Ngamjanyaporn, P., Thakkinstian, A., Verasertniyom, O., Chatchaipun, P., Vanichapuntu, M., Nantiruj, K., et al. (2011). Pharmacogenetics of cyclophosphamide and CYP2C19 polymorphism in Thai systemic lupus erythematosus. Rheumatol. Int., 31 (9), 1215–1218. doi:10.1007/s00296-010-1420-7
Oglesby, A., Shaul, A. J., Pokora, T., Paramore, C., Cragin, L., Dennis, G., et al. (2013). Adverse event burden, resource use, and costs associated with immunosuppressant medications for the treatment of systemic lupus erythematosus: a systematic literature review. Int. J. Rheumatol. 2013, 347520. doi:10.1155/2013/347520
Ohmann, E. L., Burckart, G. J., Chen, Y., Pravica, V., Brooks, M. M., Zeevi, A., et al. (2010). Inosine 5-monophosphate dehydrogenase 1 haplotypes and association with mycophenolate mofetil gastrointestinal intolerance in pediatric heart transplant patients. Pediatr. Transpl. 14 (7), 891–895. doi:10.1111/j.1399-3046.2010.01367.x
Petri, M., Orbai, A. M., Alarcõn, G. S., Gordon, C., Merrill, J. T., Fortin, P. R., et al. (2012). Derivation and validation of the systemic lupus international collaborating clinics classification criteria for systemic lupus erythematosus. Arthritis Rheum. 64 (8), 2677–2686. doi:10.1002/art.34473
Rashid, M. M. U., Ahmed, I., Islam, M. A., Tasnim, T., Nahid, N. A., Apu, M. N. H., et al. (2020). Influence of TPMT polymorphisms on azathioprine-induced myelosuppression in Bangladeshi patients with systemic lupus erythematosus. Drugs Ther. Perspect. 36 (5), 202–207. doi:10.1007/s40267-020-00716-y
Riskalla, M. M., Somers, E. C., Fatica, R. A., and McCune, W. J. (2003). Tolerability of mycophenolate mofetil in patients with systemic lupus erythematosus. J. Rheumatol, 30 (7), 1508–1512. Available online at: https://pubmed.ncbi.nlm.nih.gov/12858449/
Rosenberg, N., Murata, M., Ikeda, Y., Opare-Sem, O., Zivelin, A., Geffen, E., et al. (2002). The frequent 5,10-methylenetetrahydrofolate reductase C677T polymorphism is associated with a common haplotype in whites, Japanese, and Africans. Am. J. Hum. Genet. 70 (3), 758–762. doi:10.1086/338932
Sakthiswary, R., and Suresh, E. (2014). Methotrexate in systemic lupus erythematosus: a systematic review of its efficacy. Lupus. 23 (3), 225–235. doi:10.1177/0961203313519159
Sha, H. X., Veerapen, K., Chow, S. K., Gun, S. C., Lau, I. S., Lim, R. L. H., et al. (2022). Genetic variations in methotrexate metabolic pathway genes influence methotrexate responses in rheumatoid arthritis patients in Malaysia. Sci. Rep. 12 (1), 11844–13. doi:10.1038/s41598-022-15991-0
Sharaki, O. A., Elgerby, A. H., Nassar, E. S., and Khalil, S. S. E. (2018). Impact of methylenetetrahydrofolate reductase (MTHFR) A1298C gene polymorphism on the outcome of methotrexate treatment in a sample of Egyptian rheumatoid arthritis patients. Alex. J. Med. 54 (4), 633–638. doi:10.1016/j.ajme.2017.11.008
Sheu, H. S., Chen, Y. M., Liao, Y. J., Wei, C. Y., Chen, J. P., Lin, H. J., et al. (2022). Thiopurine S-methyltransferase polymorphisms predict hepatotoxicity in azathioprine-treated patients with autoimmune diseases. J. Pers. Med. 12 (9), 1399. doi:10.3390/jpm12091399
Shu, Q., Fan, Q., Hua, B., Liu, H., Wang, S., Liu, Y., et al. (2021). Influence of slco1b1 521t>c, ugt2b7 802c>t and impdh1 −106g>a genetic polymorphisms on mycophenolic acid levels and adverse reactions in Chinese autoimmune disease patients. Pharmgenomics Pers. Med. 14, 713–722. doi:10.2147/PGPM.S295964
Shu, W., Guan, S., Yang, X., Liang, L., Chen, Z., Liang, L., et al. (2016). Genetic markers in CYP2C19 and CYP2B6 for prediction of cyclophosphamide’s 4-hydroxylation, efficacy and side effects in Chinese patients with systemic lupus erythematosus. Br. J. Clin. Pharmacol. 81 (2), 327–340. doi:10.1111/bcp.12800
Singh, G., Saxena, N., Aggarwal, A., and Misra, R. (2007). Cytochrome P450 polymorphism as a predictor of ovarian toxicity to pulse cyclophosphamide in systemic lupus erythematosus. J. Rheumatol. 34 (4), 731–733. doi:10.1016/s0973-3698(10)60212-9
Song, G. G., Bae, S. C., and Lee, Y. H. (2014). Association of the MTHFR C677T and A1298C polymorphisms with methotrexate toxicity in rheumatoid arthritis: a meta-analysis. Clin. Rheumatol. 33 (12), 1715–1724. doi:10.1007/s10067-014-2645-8
Steponaitiene, R., Kupcinskas, J., Survilaite, S., Varkalaite, G., Jonaitis, L., Kiudelis, G., et al. (2016). TPMT and ITPA genetic variants in Lithuanian inflammatory bowel disease patients: prevalence and azathioprine-related side effects, Adv. Med. Sci., 61, 135–140. doi:10.1016/j.advms.2015.09.008
Su, H. I., Sammel, M. D., Velders, L., Horn, M., Stankiewicz, C., Matro, J., et al. (2010). Association of cyclophosphamide drug-metabolizing enzyme polymorphisms and chemotherapy-related ovarian failure in breast cancer survivors. Fertil. Steril. 94 (2), 645–654. doi:10.1016/j.fertnstert.2009.03.034
Su, S. S., Lin, Y. F., and Zhou, H. (2020). Association of thiopurine S -methyltransferase and NUDT15 polymorphisms with azathioprine-induced myelotoxicity in Chinese patients with rheumatological disease. Chin. Med. J. Engl. 133 (8), 1002–1004. doi:10.1097/CM9.0000000000000756
Sumi, S., Marinaki, A. M., Arenas, M., Fairbanks, L., Shobowale-Bakre, M., Rees, D. C., et al. (2002). Genetic basis of inosine triphosphate pyrophosphohydrolase deficiency. Hum. Genet. 111 (4-5), 360–367. doi:10.1007/s00439-002-0798-z
Tanaka, Y., and Saito, Y. (2021). Importance of nudt15 polymorphisms in thiopurine treatments. J. Pers. Med. 11 (8), 778–10. doi:10.3390/jpm11080778
Tran, A., Bournerias, F., Le Beller, C., Mir, O., Rey, E., Pons, G., et al. (2008). Serious haematological toxicity of cyclophosphamide in relation to CYP2B6, GSTA1 and GSTP1 polymorphisms. Br. J. Clin. Pharmacol. 65 (2), 279–280. doi:10.1111/j.1365-2125.2007.03020.x
Van Der Put, N. M. J., Gabreëls, F., Stevens, E. M. B., Smeitink, J. A., Trijbels, F. J., Eskes, T. K., et al. (1998). A second common mutation in the methylenetetrahydrofolate reductase gene: an additional risk factor for neural-tube defects? Am. J. Hum. Genet. 62 (5), 1044–1051. doi:10.1086/301825
Von Feldt, J. M., Scalzi, L. V., Cucchiara, A. J., Morthala, S., Kealey, C., Flagg, S. D., et al. (2006). Homocysteine levels and disease duration independently correlate with coronary artery calcification in patients with systemic lupus erythematosus. Arthritis Rheum. 54 (7), 2220–2227. doi:10.1002/art.21967
Wang, C. W., Chi, M. H., Tsai, T. F., Yu, K. H., Kao, H. W., Chen, H. C., et al. (2022). Implementation of NUDT15 genotyping to prevent azathioprine-induced leukopenia for patients with autoimmune disorders in Chinese population. Clin. Pharmacol. Ther. 112 (5), 1079–1087. doi:10.1002/cpt.2716
Wang, H. N., Zhu, X. Y., Zhu, Y., Xie, Q. H., Lai, L. Y., Zhao, M., et al. (2015). The GSTA1 polymorphism and cyclophosphamide therapy outcomes in lupus nephritis patients. Clin. Immunol. 160 (2), 342–348. doi:10.1016/j.clim.2015.07.010
Wang, Y., He, J., Ma, T. J., Lei, W., Li, F., Shen, H., et al. (2016). GSTT1 null genotype significantly increases the susceptibility to urinary system cancer: evidences from 63,876 Subjects. J. Cancer 7 (12), 1680–1693. doi:10.7150/jca.15494
Wright, S., Sanders, D. S., Lobo, A. J., and Lennard, L. (2004). Clinical significance of azathioprine active metabolite concentrations in inflammatory bowel disease. Gut 53 (8), 1123–1128. doi:10.1136/gut.2003.032896
Wu, T. Y., Peng, Y., Pelleymounter, L. L., Moon, I., Eckloff, B. W., Wieben, E. D., et al. (2010). Pharmacogenetics of the mycophenolic acid targets inosine monophosphate dehydrogenases IMPDH1 and IMPDH2: gene sequence variation and functional genomics. Br. J. Pharmacol. 161 (7), 1584–1598. doi:10.1111/j.1476-5381.2010.00987.x
Yang, F. Y., Xu, L. H., Wang, J., Zhang, Y. T., Lin, S. F., Wang, K. M., et al. (2023). Relationship between MTHFR gene polymorphism(C677T) and adverse reactions of high-dose methotrexate in pediatric patients with acute lymphoblastic leukemia. Zhongguo. Shi. Yan. Xue. Ye. Xue. Za. Zhi. 31 (4), 967–972. doi:10.19746/j.cnki.issn.1009-2137.2023.04.006
Yang, S. K., Hong, M., Baek, J., Choi, H., Zhao, W., Jung, Y., et al. (2014). A common missense variant in NUDT15 confers susceptibility to thiopurine-induced leukopenia. Nat. Genet. 46 (9), 1017–1020. doi:10.1038/ng.3060
Zanger, U. M., and Klein, K. (2013). Pharmacogenetics of cytochrome P450 2B6 (CYP2B6): advances on polymorphisms, mechanisms, and clinical relevance. Front. Genet. 4, 24–12. doi:10.3389/fgene.2013.00024
Zhang, X. W., Li, C., Ma, X. X., Zhao, J. X., An, Y., Liu, S., et al. (2014). Short-interval lower-dose intravenous cyclophosphamide as induction and maintenance therapy for lupus nephritis: a prospective observational study. Clin. Rheumatol. 33 (7), 939–945. doi:10.1007/s10067-014-2590-6
Zhao, M., Liang, L., Ji, L., Chen, D., Zhang, Y., Zhu, Y., et al. (2016). MTHFR gene polymorphisms and methotrexate toxicity in adult patients with hematological malignancies: a meta-analysis. Pharmacogenomics 17 (9), 1005–1017. doi:10.2217/pgs-2016-0004
Zhong, S., Huang, M., Yang, X., Liang, L., Wang, Y., Romkes, M., et al. (2006). Relationship of glutathione S-transferase genotypes with side-effects of pulsed cyclophosphamide therapy in patients with systemic lupus erythematosus. Br. J. Clin. Pharmacol. 62 (4), 457–472. doi:10.1111/j.1365-2125.2006.02690.x
Keywords: single nucleotide polymorphism, adverse effect, methotrexate, azathioprine, cyclophosphamide, mycophenolate mofetil
Citation: Hamdani S, Hamijoyo L, Amalia R and Barliana MI (2025) Gene polymorphisms associated with immunosuppressant adverse effects in systemic lupus erythematosus: a narrative review. Front. Genet. 16:1594648. doi: 10.3389/fgene.2025.1594648
Received: 20 March 2025; Accepted: 29 May 2025;
Published: 24 June 2025.
Edited by:
Simone L. Cree, University of Otago, New ZealandReviewed by:
Eng Wee Chua, National University of Malaysia, MalaysiaYen Yen Ari Indrawijaya, Universitas Islam Negeri Maulana Malik Ibrahim, Indonesia
Copyright © 2025 Hamdani, Hamijoyo, Amalia and Barliana. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Siva Hamdani, c2l2YTIxMDAyQG1haWwudW5wYWQuYWMuaWQ=; Melisa I. Barliana, bWVsaXNhLmJhcmxpYW5hQHVucGFkLmFjLmlk
 Siva Hamdani1,2*
Siva Hamdani1,2* Laniyati Hamijoyo
Laniyati Hamijoyo Riezki Amalia
Riezki Amalia Melisa I. Barliana
Melisa I. Barliana