- 1Institute of Plant Protection, Inner Mongolia Academy of Agricultural and Animal Husbandry Sciences, Hohhot, China
- 2Institute of Grassland Research, Chinese Academy of Agricultural Science, Hohhot, China
- 3Research Department, Inner Mongolia University of Finance and Economics, Hohhot, China
- 4College of Grassland Science, Inner Mongolia Agricultural University, Hohhot, China
Introduction: Caucasian clover (Trifolium ambiguum Bieb.), a perennial legume forage grass, exhibits strong adaptability and resistance to adverse conditions. Owing to its rhizome specificity, it cannot nodulate or fix nitrogen outside of its original location, which limits its promotion and cultivation. The phenomenon of spontaneous nodulation of Caucasian clover was observed for the first time in Inner Mongolia, and investigation of its nodulation and nitrogen fixation mechanisms assists in the subsequent promotion of Caucasian clover cultivation from a theoretical perspective.
Methods: In this study, rhizobia extracted from Caucasian clover were inoculated into the field, and the nodulation part of the root system was investigated during the regreening stage of the second year using transcriptomics and proteomics techniques.
Results and Discussion: The study identified 70,280 differentially expressed genes (DEGs) and 770 differentially expressed proteins (DEPs) in total, among which carbonic anhydrase, cyanate lyase, phenylalanine, caffeate/5-hydroxyferulate 3-O-methyltransferase, caffeoyl CoA 3-O methyltransferase, chalcone synthase, and chalcone isomerase may have been the main factors affecting the nodulation and nitrogen fixation of Caucasian clover. This study theoretically contributes to the future genetic validation and selection of Caucasian clover varieties with a strong ability to nodulate and fix nitrogen.
1 Introduction
Caucasian clover (Trifolium ambiguum Bieb.) is a long-rooted tiller legume forage native to Eastern Europe and the Caucasus region and exhibits excellent adaptability, cold resistance, and other natural endowments owing to the harsh local environment. Since the 1970s, Australia, New Zealand, the United States, Canada, and other countries have introduced and domesticated Caucasian clover and conducted numerous theoretical and applied studies, including studies on seed development characteristics (Hay et al., 2010), vegetative propagation characteristics (Baker, 2012), rhizobia and nitrogen fixation (Beauregard et al., 2003), and response to fertilizer (Sheaffer and Seguin, 2011).
Caucasian clover has the same nitrogen-fixing capacity and similar efficiency to that of white clover (Black et al., 2006). However, because of the selective specificity of Caucasian clover for rhizobacteria (Andrews et al., 2007; Miller et al., 2007), it can only be nodulated for nitrogen fixation with rhizobacteria from its places of origin, i.e., the Caucasus and parts of Eastern Europe, which affects its widespread cultivation. The Caucasian clover cultivar “Trifolium ambiguum Bieb. cv. Mengnong No. 1” (Inner Mongolia Agricultural University) can be tumor-fixed with rhizobacteria in Inner Mongolia, suggesting that Caucasian clover has great potential for nitrogen fixation via tumor fixation. Therefore, in this study, “T. ambiguum Bieb. cv. Mengnong No.1” was inoculated in the field, and its rhizosphere was analyzed using transcriptomics and a 4D label-free quantitative (LFQ) proteomic method with the aim of examining the gene and protein variation in the rhizosphere after rhizobial inoculation to guide the subsequent improvement of Caucasian clover. These findings are theoretically instrumental in the improvements in the nitrogen-fixation capacity of Caucasian clover.
Establishing a symbiotic nitrogen-fixing system involves a complex series of processes, including signal recognition and infection between the legume host crop and rhizobia, infestation line formation, rhizobial primordium formation, and root nodule maturation. Establishment of a symbiotic nitrogen-fixation relationship begins with the release of signaling substances and mutual recognition between the legumes and rhizobia. Legumes release metabolite flavonoid substances in the inter-root environment through the root system (Redmond et al., 1986), and when rhizobia successfully and specifically sense the flavonoid signal, those in the vicinity of the root system activate the expression of their own nodulation genes (nod genes) and release nodulation factors (Nod factors) around the crop roots (Caetano-Anollés et al., 1991; Spaink, 2000). Nod factors can induce nodule development in legume roots (Truchet et al., 1991). Thus, flavonoid compounds crucially impact the interactions between legumes and rhizobia. In Lotus japonicus, Nod factors may interact with the DFR-like protein, LjDFL1, to regulate flavonoid synthesis. Overexpression of DFL1 increases root invasion and promotes rhizobial infestation (Duan et al., 2019). In Medicago truncatula, downregulation of MtFNSII expression causes the depletion of flavonoids and interferes with rhizome formation. Flavonoids are important compounds in the early nodulation process, and phenylpropanoids also play an inducing role (Stafford, 1997).
Transcriptomic technology has emerged as a powerful tool for identifying key genes involved in symbiotic nitrogen fixation in legumes. By exploring the gene abundance in root nodules during the symbiotic interaction between legumes and rhizobia, this approach enables a comprehensive understanding of the molecular mechanisms underlying their mutualistic relationship. For instance, Hayashi et al. (2012) performed transcriptomic analysis on the nodulation zone of soybean roots inoculated with either wild-type rhizobia or a nodC mutant strain, identifying 2,915 genes that are specifically regulated by Nod factors produced by rhizobia. Similarly, Yuan et al. (2016) used RNA-seq to investigate differentially expressed genes (DEGs) in soybean roots interacting with different rhizobial strains. Their findings revealed that these genes primarily encode resistance proteins, nodulation factors, nodulins, immune defense proteins, and enzymes involved in flavonoid/flavone/flavonol biosynthesis and plant–pathogen interactions.
Proteomics has been extensively employed to study various aspects of legume crops, including growth and development, stress responses, and symbiotic interactions with rhizobia, thereby providing a solid foundation for elucidating the underlying molecular mechanisms. LV (2023) used proteomic techniques to demonstrate that the deletion of bjaR1 suppresses the expression of proteins associated with the nitrogen regulatory system. Chen et al. (2011) analyzed the impact of phosphorus (P) on the soybean nodule proteome, identifying 44 proteins that are responsive to P starvation—27 from the plant and 17 from the rhizobia. Their findings highlight the collaborative effort between soybean and rhizobia in adapting to P deficiency. Additionally, Salavati et al. (2012) compared the root proteomes of three soybean cultivars with varying nitrogen-fixing abilities (efficient: En-b0-1; inefficient: En1282; normal: Enrei) after rhizobial inoculation. They identified 56 differentially expressed proteins (DEPs) that are primarily involved in energy metabolism and signal transduction pathways.
Therefore, in this study, we inoculated T. ambiguum Bieb. ‘Mengnong No. 1’ with rhizobia under field conditions and employed transcriptomic and 4D label-free quantitative (LFQ) proteomic analyses to investigate changes in gene expression and protein abundance in the rhizosphere. Our aim was to characterize the molecular mechanisms underlying the symbiotic interaction between T. ambiguum and rhizobia, providing insights to guide the genetic improvement in this legume species. The findings of this study are expected to contribute theoretically to enhancing the nitrogen fixation efficiency in T. ambiguum.
2 Materials and methods
2.1 Experimental materials
The Caucasian clover variety “Trifolium ambiguum Bieb. cv. Mengnong No.1” bred by Inner Mongolia Agricultural University was used as the experimental material in this study.
The experimental strain was a rhizobacterium isolated from the root nodules of the variety and was identified as Rhizobium leguminosarum based on 16S rDNA analysis (Jiawei et al., 2023).
2.2 Experimental method
2.2.1 Sampling method
Isolated rhizobia, after being inoculated into yeast mannitol agar (YMA) liquid medium, were cultured at at 180 r·min−1 and 28 C until the optical density value (OD600) exceeded 0.7, followed by centrifugation at 10,000 r·min−1 for 10 min. The resultant supernatant was resuspended in sterile water to achieve an optical density (OD600) of 0.7, which was then prepared for subsequent use (Duan et al., 2018).
The experiment consisted of two treatments: inoculation with rhizobia (NN) and a control group (CK). For the NN treatment, the prepared rhizobial suspension was inoculated into the root system of Caucasian clover (planted in July 2020) in August 2020. No rhizobial inoculation was applied to the CK treatment. Caucasian clover was sampled during its second year of re-greening.
2.2.2 Determination of plant nutritional indices
The aboveground parts of plants were excised at the soil surface, first treated at 105 °C for 30 min, and then dried at 65° C to a constant weight. Nutritional indices, including neutral detergent fiber (NDF), acid detergent fiber (ADF), and crude protein (CP), were determined, with specific methods referenced from Duan et al. (2018). Additionally, the data were analyzed using a t-test.
2.2.3 Root sampling method of plants
After being rinsed with distilled water and blotted dry using clean filter paper, the underground roots were treated with phosphate-buffered saline to remove surface soil. Roots (0.5 g) were sampled from each treatment, with three biological replicates. Following 3–4 h of incubation in liquid nitrogen, the roots were stored at −80 °C for subsequent transcriptomic and proteomic analyses.
2.2.4 Total RNA extraction and data acquisition
Purification of total plant RNA relied on ethanol precipitation and the CTAB-pBIOZOL reagent as per the producer’s protocol. A NanoDrop Spectrophotometer and an Agilent 2100 Bioanalyzer (Thermo Fisher Scientific, MA, United States), respectively, were used for the identification and quantification of total RNA. Jingjie PTM BioLab Co. Inc. (Hangzhou, China) was responsible for purifying, reverse transcribing, and sequencing the obtained RNA, along with constructing the relevant library as per the manufacturer’s protocol (Illumina, San Diego, CA).
The RNA-seq root transcript library was prepared using the Illumina® Stranded mRNA Kit with 1 µg of total RNA. In brief, messenger RNA isolated using oligo (dT) beads by the poly (A) selection method was fragmented. A SuperScript Double-stranded cDNA Synthesis Kit (Invitrogen, Carlsbad, CA, United States) was responsible for the synthesis of double-stranded cDNA using random hexamer primers (Illumina). The cDNA was then end-repaired, phosphorylated, and adenylated with the ‘A’ base as per the library construction protocol (Illumina). A library of 300 Pb cDNA target fragments was selected, fusion DNA polymerase (NEB) was used to select a low-range overlap of 2% (lower range 2%), and then an amplification cycle of 15 polymerase chains (PCR) was performed. Qubit 4.0 quantification was followed by the sequencing of the paired-end RNA-seq library using a NovaSeq 6000 Sequencer (2 × 150 bp read length).
fastp with default parameters was used to control the quality of the trimmed raw paired-end reads (Chen et al., 2018). Clean data were subjected to de novo assembly with the assistance of the Trinity (Grabherr et al., 2011). CD-HIT and TransRate were used to filter all assembled sequences to improve assembly quality, and DIAMOND was then used to search the assembled transcripts against the NCBI non-redundant (NR), COG, and Kyoto Encyclopedia of Genes and Genomes (KEGG) protein databases, thereby retrieving the functional annotations for the most similar sequences, with a cut-off E-value <1.0 × 10−5. The Blast2GO program helped acquire the GO annotations of the specially assembled transcripts for the description of the biological processes (BPs), molecular functions (MFs), and cellular components (CCs) (Conesa et al., 2005). Metabolic pathway analyses relied on KEGG (Kanehisa and Goto, 2000). The raw sequencing data were submitted to the NCBI BioProject database (project No. PRJNA907928 and PRJNA907870).
The expression level of each transcript was calculated using the transcripts per million method, and the expansion between the two samples was determined. RSEM served for gene abundance quantification (Li and Dewey, 2011). Differential expression analyses relied on DESeq2 or DEGseq, confirming DEGs with |log2FC| ≥ 1 and FDR ≤0.05 (DESeq2) or FDR ≤0.001 (DEGseq) as DEGs with significance. In addition, Gene Ontology (GO) and KEGG analyses, respectively, by virtue of GOATools and KOBAS, determined DEGs presenting a significant enrichment in metabolic pathways and GO terms (Bonferroni-corrected p-value ≤0.05) versus the whole-transcriptome sequencing (Xie et al., 2011).
2.2.5 Quantitative real-time PCR analysis
Nine genes significantly influencing nodule nitrogen fixation were selected from the transcriptomic data and validated via quantitative real-time polymerase chain reaction (qRT-PCR). First-strand cDNA was synthesized using the FastKing RT Kit with gDNase (TIANGEN Biotech Co., LTD., Beijing, China) following the manufacturer’s protocol. Quantitative real-time PCR was performed using the SYBR Green SuperReal PreMix Plus (TIANGEN Biotech Co., LTD., Beijing, China) on an Applied Biosystems 7500 Fast Real-Time PCR System (Monad Biotech Co., Ltd., Guangzhou, China). The thermal cycling conditions were as follows: initial denaturation at 95 °C for 15 min, followed by 40 cycles of 95 °C for 10 s and 66 °C for 32 s. Melting curve analysis was conducted from 66 °C to 95 °C after the final cycle to verify primer specificity. All reactions were run in triplicate. Relative gene expression levels were calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001). Primers were designed using Primer Premier 5 software (PREMIER Biosoft, United States), and their sequences are provided in Supplementary Table S1. The RCD-1 gene was selected as an internal reference Yin et al. (2021).
2.2.6 Protein extraction
Jingjie PTM BioLab (Hangzhou, China) Co. Inc. carried out the proteomic sequencing analysis on NN and CK samples. After thorough grinding under nitrogen, the root samples were placed in a phenol extraction buffer (ratio: 1:4) and subjected to ultrasonic fragmentation. The mixture of the samples and Tris underwent 10 min of centrifugation at 5,500 g and 4 °C. The supernatants, combined with 0.1 M ammonium acetate/methanol (volume ratio: 1:5), underwent precipitation. After methanol and acetone washes, protein precipitates were re-dissolved in 8 M urea. A BCA kit was used to detect protein concentration. The amount used for enzymatic hydrolysis was kept consistent for each protein. After gradual addition of trichloroacetic acid to a final concentration of 20%, the mixture was vortexed and precipitated for 2 h at 4 °C. This was followed by 5 min of centrifugation at 4,500 g and disposal of the supernatant. The pellet underwent 2–3 rounds of precipitation in precooled acetone. Following the drying of the precipitate, TEAB was added to a final concentration of 200 mM. After ultrasound dispersion, the mixture was incubated with trypsin (1:50) for overnight enzymatic hydrolysis. After the addition of DTT at 5 mM, the mixture was subjected to half an hour of incubation at 56°C, and after the addition of IAA to 11 mM, the mixture was cultured for 15 min at room temperature in the dark.
With regard to liquid chromatography, we employed a NanoElute ultra-performance liquid system to isolate the peptides after dissolution in mobile phase A (Sun et al., 2021). Mobile phase A consisted of an aqueous solution containing 0.1% formic acid and 2% acetonitrile; mobile phase B consisted of an aqueous solution containing 0.1% formic acid and 100% acetonitrile. The liquid phase gradient was as follows: 0–70 min, 5–22% B; 70–84 min, 22–32% B; 84–87 min, 32–80% B; and 87–90 min, 80% B; the flow rate was set at 300 nL/min. The isolated peptides were injected into the capillary ion source for ionization (voltage: 2.0 kV), followed by analysis using timsTOF Pro 3 mass spectrometry. High-resolution time-of-flight mass spectrometry was used to detect and analyze the parent ions of the peptides and their corresponding secondary fragments. For the secondary mass spectrum, the scanning range was 100–1,700. The parallel accumulation–serial fragmentation (PASEF) mode was used for data collection. The collection of primary spectrograms was followed by 10 rounds of the collection of secondary spectrograms, capturing 0–5 parent ions in the PASEF mode. For tandem mass spectrometry, we set the dynamic exclusion time to 30 s to prevent the parent ions from being scanned repeatedly (Cox et al., 2009).
Protein annotation included KEGG annotation and subcellular localization. Protein pathways were annotated using the KEGG databases. The identified proteins underwent BLAST comparison (blastp, e-value ≤ 1e−4), with those possessing the highest scores being annotated. In addition, the KEGG online service tool KAAS was used to assist in protein annotation within the KEGG database, and the results were integrated into the KEGG pathway database using KEGG mapper. WoLF PSORT, an updated version of PSORT/PSORT II, was used to predict subcellular localization.
2.2.7 PRM analysis
The extracted proteins were subjected to proteomic analysis, following the previous description. After dissolution in mobile phase A, they were isolated using the EASY-nLC 1000 UPLC system. Mobile phase A consisted of an aqueous solution containing 0.1% formic acid and 2% acetonitrile; mobile phase B consisted of an aqueous solution containing 0.1% formic acid and 90% acetonitrile. The liquid phase gradient was as follows: 0–16 min, 8–30% B; 16–22 min, 30–40% B; 22–26 min, 40–80% B; and 26–30 min, 80% B. The flow rate was 500 nL/min. The peptides isolated under an ultra-performance liquid phase system were injected into an NSI ion source for ionization (ion source voltage: 2.1 kV), and a Q Exactive Plus Mass Spectrometer was used for relevant analysis. Orbitrap, with a scanning resolution of 17,500, was used to detect and analyze the peptide parent ions and the corresponding secondary fragments. For the primary mass spectrometer, the scanning range was 382–953 m/z, and the scanning resolution was 70,000. A data-dependent scan program was adopted for data collection, and the HCD collision pool presented a fragmentation energy of 27. For the primary mass spectrometer, the automatic gain control (AGC) was set to 3E6, and the maximum ion implantation time (IT) was 50 ms, while for the secondary mass spectrometer, the two parameters were set to 1E5 and 220 ms, respectively, with an isolation window of 1.6 m/z. Regarding peptide parameters, experimenters set the protease to trypsin [Kr/P], with a maximum of 0 missed cleavage sites; the allowed peptide length was 7–25 amino acid residues, and cysteine alkylation was treated as a fixed modification. For transition parameters, the charge states of parent and fragment ions were set to 2–3 and 1, respectively, with B and Y ion types selected. Fragment ion selection ranged from the third ion to the last, and ion matching was performed with a mass error tolerance of 0.02 Da (Glen et al., 2010). In the experiment, quantitative analysis of each protein was based on more than two unique peptides. For PRM verification of certain proteins, a single unique peptide was used due to sensitivity limitations. A heavy isotope-labeled peptide was employed to normalize of the quantitative information, followed by the relative quantitative analysis (using three biological replicates) on the target proteins. Microsoft Excel 2007 and SPSS 16.00 were used to analyze the relative protein expression. Quantitative real time-PCR and PRM data analysis were analyzed using ANOVA, and the least significant difference test was used to compare treatment means at a probability level of p = 0.05.
3 Results
3.1 Effect of inoculation with rhizobia on nutritional quality
Pairwise t-tests were conducted to analyze the crude protein (CP), NDF, and ADF contents of Caucasian clover. The results showed that, compared with the control group (without rhizobium inoculation; CK), the CP content of Caucasian clover in the NN treatment (inoculated with rhizobia) was significantly increased, while the NDF and ADF contents were significantly reduced. (Figure 1).
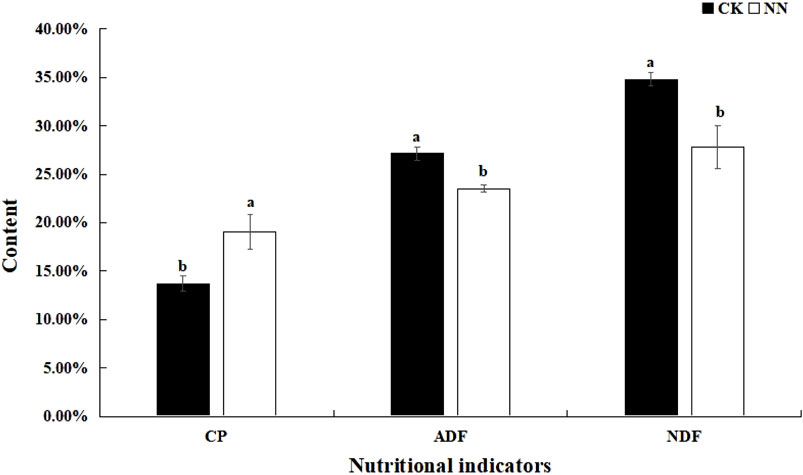
Figure 1. Effect of rhizobia inoculation on Caucasian clover nutritional quality. CK, control; NN, inoculation with rhizobia; CP, crude protein; NDF, neutral detergent fiber; ADF, acid detergent fiber.
3.2 Transcriptomic data analysis of Caucasian clover under different treatments
The study obtained 166,528 unigenes in total, and the average length was 1,223 bp. The N50, N70, and N90 values of the unigenes were 1896, 1,300 and 586 bp, respectively (Supplementary Table S2). Supplementary Table S2 illustrates the sequence assembly after Illumina sequencing.
In total, 89,567 genes were divided into three ontologies (Figure 2). In terms of the BP category, genes related to cellular processes (21,945), “biological regulation” (9,192), “cellular component organization or biogenesis” (5,731), and localization (4,749) were highly represented. With regard to the CC category, genes related to “membrane part” (27,103), cell (21,369), “organelle part” (10,272), and “extracellular region” (1,174) were highly represented. The highly represented GO terms (level 2) were ‘catalytic activity” (47,002), “binding” (46,570), “transporter activity” (5,075), and “structural molecule activity” (2,208) in the MF category.
In total, there were 70,280 DEGs (33,692 upregulated and 36,588 downregulated) in the roots of Caucasian clover in the NN and control groups, respectively (Supplementary Figure S1).
3.3 DEG functional annotation in KEGG enrichment
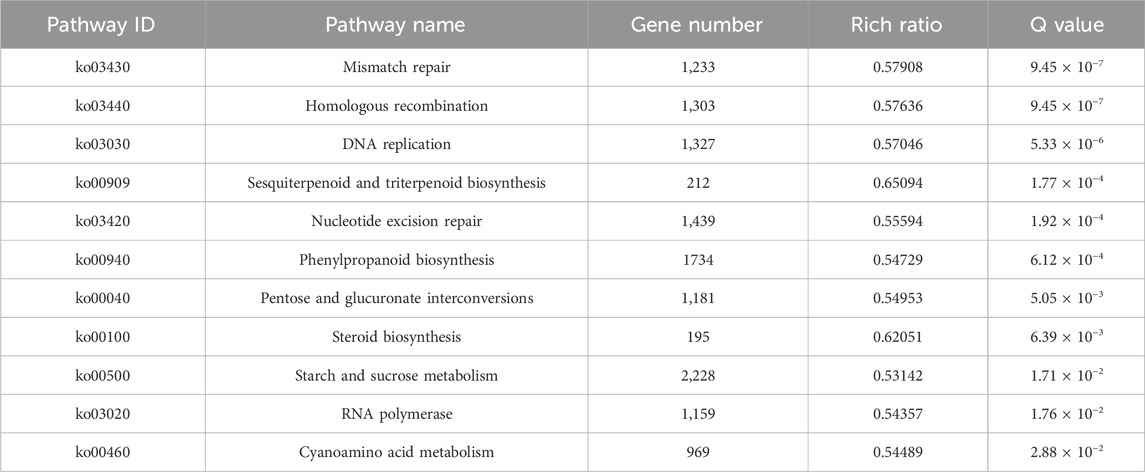
According to KEGG enrichment analysis on DEGs, the top five pathways were “sesquiterpenoid and triterpenoid biosynthesis” (ko00909), “steroid biosynthesis” (ko00100), “mismatch repair” (ko03430), “homologous recombination” (ko03440), and “DNA replication” (ko03030) (Table 1). They were also significantly enriched in “phenylpropanoid biosynthesis” (ko00940), “pentose and glucuronate interconversion” (ko00040), “starch and sucrose metabolism” (ko00500), and “cyanoamino acid metabolism” (ko00460) pathways.
3.4 Protein profiles of Caucasian clover at different developmental stages
Nodular and non-nodular roots of Caucasian clover were subjected to proteomic analysis. Liquid chromatography‒mass spectrometry identified approximately 39,570 peptides, corresponding to 6,468 proteins. A 2.0-fold-change cut-off was used to determine differential expression, resulting in the identification of 770 DEPs, including 406 upregulated and 364 downregulated proteins during the comparison between nodular and non-nodular roots (Figure 3).
3.5 KEGG analysis of DEPs
In total, 528 proteins were assigned to 105 pathways. The pathways associated with “phenylpropanoid biosynthesis” (ko00940), “tyrosine metabolism” (ko00350), “isoquinoline alkaloid biosynthesis” (ko00950), “phenylalanine metabolism” (ko00360), “nitrogen metabolism” (ko00910), and “flavonoid biosynthesis” (ko00941) were predominantly enriched in the NN vs. CK comparison (Figure 4).
3.6 Transcriptomics and proteomics crosstalk analysis
In the NN vs. CK comparison, 6,468 proteins were identified in the proteomics dataset and 166,528 transcripts were identified in the transcriptomics dataset. The transcript and protein expression levels of 157 genes were upregulated, and the mRNA and protein levels of 160 genes were downregulated (Figure 5A). The expression levels of mRNA and protein show a positive correlation (Figure 5B).

Figure 5. Proteomic and transcriptomic differential analysis. (A) Comparative analysis of differentially expressed proteins and transcripts in the comparison group Venn diagram. (B) Scatter plot of transcripts and their corresponding protein expressions.
GO enrichment analysis of upregulated DEPs revealed that in MF processes, DEP enrichment occurred in “intramolecular lyase activity” (GO:0016872), “chalcone isomerase activity” (GO:0045430), and “oxidoreductase activity” (GO:0016491); in the “CC” category, DEP enrichment could be observed in “thylakoid lumen” (GO:0031977), “thylakoid part” (GO:0044436), and “thylakoid” (GO:0009579); and in the “BP” category, the “P450 epoxygenase pathway” (GO:0019373), “monocarboxylic acid biosynthetic process” (GO:0072330), “monocarboxylic acid metabolic process” (GO:0032787), etc., presented DEG enrichment (Figure 6).
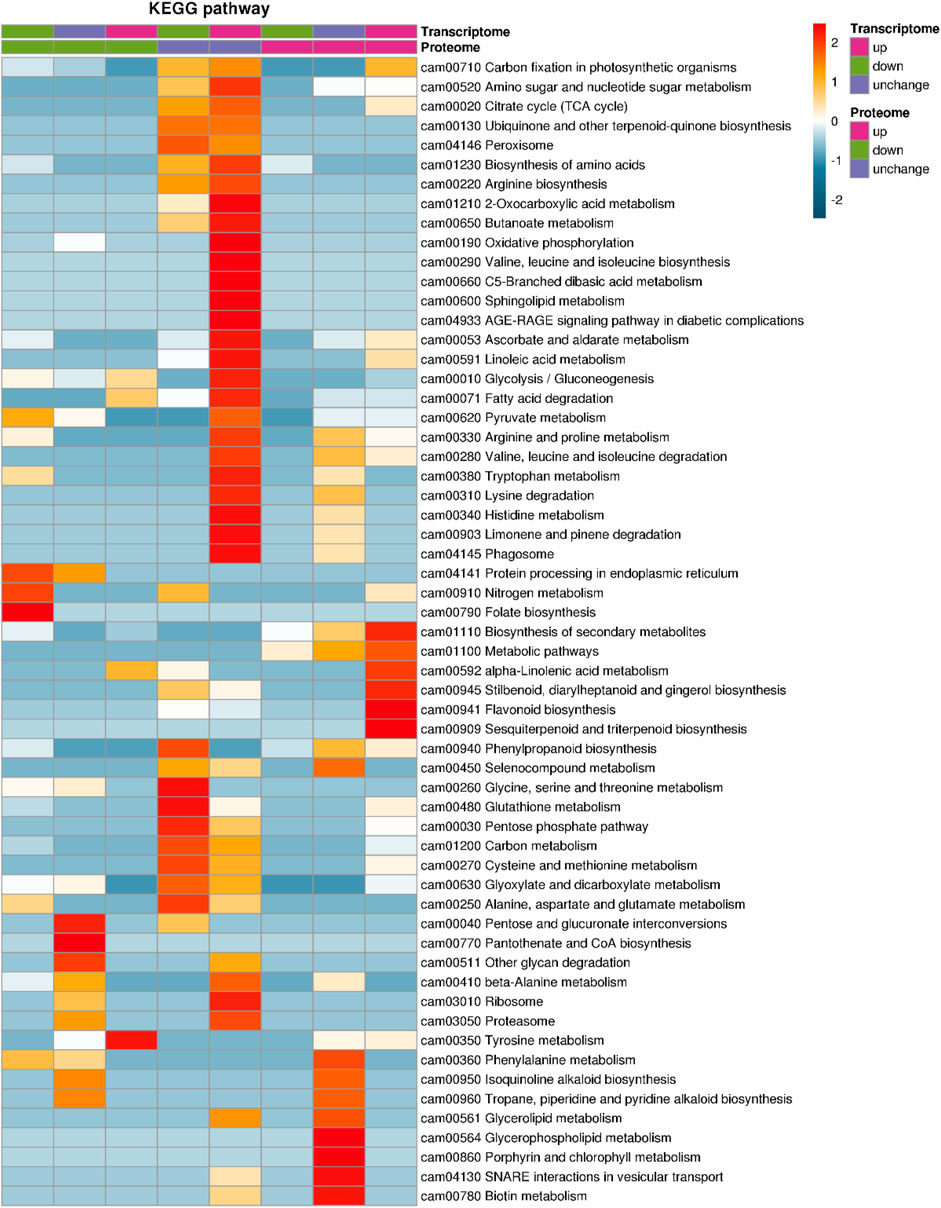
The enriched KEGG pathway for co-upregulated genes in transcriptomes and proteomics in NN vs. CK comparison were “flavonoid biosynthesis” (ko00941), “biosynthesis of secondary metabolites” (ko01110), “metabolic pathways” (ko01100), “α-linolenic acid metabolism” (ko00592), “stilbenoid, diarylheptanoid, and gingerol biosynthesis” (ko00945), and “sesquiterpenoid and triterpenoid biosynthesis” (ko00909), and the co-downregulated gene/protein-enriched pathways were “nitrogen metabolism” (ko00910), “folate biosynthesis” (ko00790), and “protein processing in endoplasmic reticulum” (ko04141) (Figure 7).
3.7 Expression of key pathway-related genes
To investigate the nitrogen-fixation mechanism in Caucasian clover, this study focused on genes associated with the nitrogen metabolism, phenylpropanoid metabolism, and flavonoid biosynthesis pathways. During nitrogen metabolism, the expression of DEGs encoding Nrt, GS, NADH-GOGAT, and FdGOGAT was downregulated, and the DEP content was similarly reduced; meanwhile, the expression of DEGs encoding CA and CYN presented an obvious increase and a similar upregulation in the proteome. The expression of DEGs encoding PAL, COMT, HCT, CAD, CHS, CCoAOMT, CHI, and F3H was upregulated during phenylpropanoid metabolism and flavonoid biosynthesis, with a similar upregulation observed at the proteomic level (Figure 8).

Figure 8. Pathways associated with nitrogen fixation in nodules of Caucasian clover. (A) Simplified model of nitrogen metabolic processes in Caucasian clover. (B) Simplified model diagram of phenylpropanoid biosynthesis and flavonoid biosynthesis in Caucasian clover. The red module expresses upregulation, the green module expresses downregulation, and the blue module indicates both up- and downregulation. Nrt, nitrate/nitrite transporter; GS, glutamine synthetase; FdGOGAT, glutamate synthase (ferredoxin); NADH-GOGAT, glutamate synthase (NADPH/NADH); CA, carbonic anhydrase; CYN, cyanate lyase; PAL, phenylalanine ammonia lyase; F3H, flavanone 3-hydroxylase; COMT, caffeate/5-hydroxyferulate 3-O-methyltransferase; CAD, cinnamyl alcohol dehydrogenase; REF, coniferyl-aldehyde dehydrogenase; HCT, hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase; CCoAOMT, caffeoyl CoA 3-O-methyltransferase; CHS, chalcone synthase; CHI, chalcone isomerase.
3.8 Protein validation by PRM
To remove possible errors, we randomly selected seven differential proteins (GS2, FdGOGAT, CYN, CFEP1, At5g28840, BGLU40, and PER42) for PRM analysis to validate the LFQ results. The results of the targeted proteins detected by PRM were in agreement with those analyzed in the test, suggesting that the proteomics sequencing results were highly reliable and reproducible (Figure 9).
3.9 Accuracy of qRT-PCR verification results
In this study, we identified genes associated with nitrogen metabolism and flavonoid biosynthesis during symbiotic nitrogen fixation in Trifolium ambiguum Bieb. The reliability of RNA-seq results and the DEGs selected was validated using qRT-PCR (Figure 10).

Figure 10. Comparison of qRT-PCR verification and FPKM value. The bar graph indicates the relative expression levels of 10 differentially expressed genes in qRT-PCR, and the folded line indicates the FPKM values of 10 differentially expressed genes in the transcriptome.
4 Discussion
Previous studies showed that Caucasian clover could only form a nitrogen-fixing symbiosis with certain rhizobia, a characteristic that limits its establishment (Hely, 1957; Kannenberg and Elliott, 1962). Caucasian clover is more vigorous and resistant to adversity than white clover (Watson et al., 1996), and because of its selective specificity for rhizobia, it must be inoculated with specific rhizobia in areas outside its native range to produce nodule fixation. More studies have been conducted on nodule nitrogen fixation in legumes than on Caucasian clover. Building upon this foundation, our study is the first one examining nitrogen fixation by nodulation in Caucasian clover using transcriptomic and proteomic techniques.
The nutritional quality of forage crops directly influences livestock growth, development, and the quality of animal products. Our findings revealed that rhizobial inoculation significantly increased the crude protein content and significantly decreased the NDF and ADF content in Caucasian clover. These results align with previous studies on other legumes (Li et al., 2016), indicating a high degree of compatibility between the inoculated rhizobia and T. ambiguum, thereby enhancing its nutritional quality.
Rhizobial inoculation induced numerous DEGs and DEPs in Caucasian clover, suggesting the involvement of specific genes and proteins in the nodulation and nitrogen fixation processes. DEG enrichment primarily occurred in sesquiterpenoid, triterpenoid, and steroid biosynthesis. Signal-regulating substances, such as flavonoids and terpenoids, are produced during the interaction between legumes and rhizobia, and terpenoids are not only involved in plant defense and growth (Mumm et al., 2008; Umehara et al., 2008; Zhou et al., 2015) but also induce nod genes to be expressed, promoting the interaction between legumes and rhizobacteria (Ali et al., 2021), which conforms to the findings of our study. Nitrogen fixation must occur under microaerobic conditions because oxygen generated by photosynthesis inactivates the oxygen-labile nitrogenase enzyme, hindering the process. Some rhizobial strains form specialized structures, such as bacteroid-containing vesicles, concomitant with nitrogen fixation within nodules (Fay, 1992).
Following symbiosis establishment, the host plant supplies sucrose to the rhizobia, which convert it into energy and organic acids for their metabolism. In return, the rhizobia fix atmospheric nitrogen and provide reduced nitrogen compounds to the host (Day et al., 2001). We observed a significant increase in CA activity in inoculated Caucasian clover, which is consistent with findings during symbiotic nitrogen fixation in soybean (Kavroula et al., 2000). This increase is likely due to the high demand for bicarbonate during symbiotic nitrogen fixation; elevated CA expression and activity facilitate the conversion of CO2 to bicarbonate, sustaining the process. Future research should focus on the mechanisms of bicarbonate supply in symbiotic nitrogen fixation.
Inorganic nitrogen in plants mainly originates from nitrate, and nitrate uptake in soil occurs through the combined action of specific nitrate transporters. Nrt is an important component of the NO3− transport system for root uptake (Li et al., 2007). Interestingly, rhizobial inoculation downregulated Nrt expression in Caucasian clover. Previous research indicates that in Lotus japonicus, Nrt proteins positively regulate symbiotic nitrogen fixation at low nitrate concentrations (Valkov et al., 2020), suggesting that low levels of nitrate (NO3−) can promote nodule development and nitrogen fixation, although the underlying mechanisms remain largely unknown. Whether this downregulation results from the initiation of symbiotic nitrogen fixation requires further investigation.
Root nodule formation in legumes is a process of interaction between the host plant and rhizobia and involves the expression and regulation of several genes in a complex series of regulatory networks. The interaction between legumes and rhizobia usually begins in nitrogen-deficient soils (Redmond et al., 1986; Peters et al., 1986), where legumes secrete flavonoid metabolites and rhizobia use flavonoids as recognition signals for synthesis and release through nod genes (D’Haeze and Holsters, 2002). Flavonoids in legume root secretions are chemo-attractive to rhizobia under low-nitrogen conditions (Cooper, 2004; Graham, 1991). Flavonoids are a class of plant phenolic compounds present in all land plants, playing vital roles in different aspects of plant life, including UV protection, pollinator attraction, seed dispersal through pigmentation, defense, and signaling between plants and microorganisms. Flavonoid is a secondary plant metabolite synthesized by the phenylpropanoid pathway (Ferrer et al., 2008). Phenylpropanoid is a physical barrier and mechanical support for the cell wall and a major component of lignin, the accumulation of which is necessary for vascular bundle development in root nodules. Therefore, the expression of the phenylpropanoid pathway genes is responsible for vascular bundle synthesis in root nodules (Takanashi et al., 2012). In the present study, white clover inoculated with rhizobacteria showed significant increases in PAL, HCT, COMT, and CCoAOMT levels. Inoculation with rhizobacteria leads to elevated plant lignin content, which is primarily caused by the expression of PAL and HCT (Zhang et al., 2016), conforming to the findings of our study. COMT is a downstream product of the lignin biosynthesis pathway that acts on F5H, and COMT expression may increase the plant lignin content (Guo et al., 2001).
CHS and CHI catalyze the first and second steps, respectively, in flavonoid biosynthesis and are key enzymes in this pathway. During the Caucasian clover nodulation nitrogen fixation, CHI and CHS contents were significantly increased, and the rhizobial factor was a key factor in promoting CHS expression and flavonoid accumulation in legumes. Nod-factor-mediated signal transduction causes CHS production in plant roots, thus promoting flavonoid production (Khandual, 2007). Silencing CHS genes in Medicago truncatula reduced root flavonoid synthesis and abolished nodulation, which could be restored by exogenous flavonoid application, demonstrating the critical role of CHS in symbiotic nitrogen fixation (Wasson et al., 2006). Similarly, overexpression of the soybean CHI gene significantly increased nodule number and isoflavonoid content. In our study, the significant increases in CHI and CHS levels during nodulation and nitrogen fixation in Caucasian clover indicate their pivotal roles in the symbiotic process.
5 Conclusion
After inoculation with rhizobia in Caucasian clover, we found that DEGs and DEPs were mainly enriched in metabolic pathways such as flavonoid biosynthesis, secondary metabolite biosynthesis, and nitrogen metabolism. Among them, CA, CYN, PAL, COMT, CCoAOMT, CHS, and CHI in nitrogen metabolism, flavonoid biosynthesis, and phenylpropanoid biosynthesis pathways may serve as key factors affecting nodulation and nitrogen fixation in Trifolium ambiguum. Follow-up studies can continue to conduct detailed analyses of the genes and proteins identified in this research.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA907928.
Author contributions
JL: Methodology, Writing – original draft, Writing – review and editing. XH: Conceptualization, Data curation, Investigation, Writing – original draft, and Writing – review and editing. YG: Supervision, Writing – original draft, and Writing – review and editing. MW: Data curation, Funding acquisition, Methodology, Writing – original draft, and Writing – review and editing. FH: Conceptualization, Data curation, Methodology, Project administration, Writing – original draft, and Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the Natural Science Foundation of Inner Mongolia, grant number 2022MS03010; the Science and Technology planning of Inner Mongolia, grant number 2023YFHH0029; and the Science and Technology planning of Inner Mongolia, grant number 2023YFDZ0078.
Acknowledgments
The authors also thank Jingjie PTM BioLab (Hangzhou) Co. Inc. for technical assistance. They thank the Key Laboratory of Grassland Resources, Ministry of Education of P.R. of China, for providing facilities.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2025.1600377/full#supplementary-material
References
Ali, M., Miao, L., Hou, Q., Darwish, D. B., Alrdahe, S. S., Ali, A., et al. (2021). Overexpression of terpenoid biosynthesis genes from garden sage (Salvia officinalis) modulates rhizobia interaction and nodulation in soybean. Front. Plant Sci. 12, 783269. doi:10.3389/fpls.2021.783269
Andrews, M., Scholefield, D., Abberton, M. T., McKenzie, B. A., Hodge, S., and Raven, J. A. (2007). Use of white clover as an alternative to nitrogen fertiliser for dairy pastures in nitrate vulnerable zones in the UK: productivity,environmental impact and economic considerations. Ann. Appl. Biol. 151 (1), 11–23. doi:10.1111/j.1744-7348.2007.00137.x
Baker, J. M. (2012). Vegetative propagation of kura clover: a field-scale test. Can. J. Plant Sci. 92, 1245–1251. doi:10.4141/cjps2012-014
Beauregard, M. S., Seguin, P., Sheaffer, C. C., and Graham, P. H. (2003). Characterization and evaluation of North American trifolium ambiguum-nodulating rhizobia. Biol. Fertil. Soils 38, 311–318. doi:10.1007/s00374-003-0661-y
Black, A. D., Moot, D. J., and Lucas, R. J. (2006). Development and growth characteristics of Caucasian and white clover seedlings, compared with perennial ryegrass. Grass Forage Sci. 61, 442–453. doi:10.1111/j.1365-2494.2006.00553.x
Caetano-Anollés, G., Paparozziz, E. T., and Gresshoff, P. M. (1991). Mature nodules and root tips control nodulation in soybean. J. plant physiology 137 (4), 389–396. doi:10.1016/s0176-1617(11)80306-8
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34 (17), i884–i890. doi:10.1093/bioinformatics/bty560
Chen, Z., Cui, Q., Liang, C., Tian, J., and Liao, H. (2011). Identification of differentially expressed proteins in soybean nodules under phosphorus deficiency through proteomic analysis. Proteomics 11 (24), 4648–4659. doi:10.1002/pmic.201100231
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21 (18), 3674–3676. doi:10.1093/bioinformatics/bti610
Cooper, J. E. (2004). Multiple responses of rhizobia to flavonoids during legume root infection. Adv. botanical Res. 41, 1–62. doi:10.1016/s0065-2296(04)41001-5
Cox, J., Matic, I., Hilger, M., Nagaraj, N., Selbach, M., Olsen, J. V., et al. (2009). A practical guide to the MaxQuant computational platform for SILAC-Based quantitative proteomics. Nat. Protoc. 4 (5), 698–705. doi:10.1038/nprot.2009.36
Day, D. A., Kaiser, B. N., Thomson, R., Udvardi, M. K., Moreau, S., and Puppo, A. (2001). Nutrient transport across symbiotic membranes from legume nodules. Funct. Plant Biol. 28 (7), 669–676. doi:10.1071/pp01028
D’Haeze, W., and Holsters, M. (2002). Nod factor structures, responses, and perception during initiation of nodule development. Glycobiology 12 (6), 79R–105R. doi:10.1093/glycob/12.6.79r
Duan, L., Pei, J., Ren, Y., Li, H., Zhou, X., Zhu, H., et al. (2019). A dihydroflavonol-4-reductase-like protein interacts with nfr5 and regulates rhizobial infection in lotus Japonicus. Mol. Plant-Microbe Interact. 32 (4), 401–412. doi:10.1094/MPMI-04-18-0104-R
Duan, R., Wei, X., and An, C. (2018). Growth of Ormosia henryi seedlings and photosynthetic and physiological characteristics with rhizobia inoculation. J. Zhejiang A and F Univ. 35 (06), 1098–1106. doi:10.11833/j.issn.2095-0756.2018.06.013
Fay, P. (1992). Oxygen relations of nitrogen fixation in Cyanobacteria. Microbiol. Rev. 56 (2), 340–373. doi:10.1128/mr.56.2.340-373.1992
Ferrer, J. L., Austin, M. B., Stewart, Jr C., and Noel, J. P. (2008). Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiology Biochem. 46 (3), 356–370. doi:10.1016/j.plaphy.2007.12.009
Glen, A., Evans, C. A., Gan, C. S., Cross, S. S., Hamdy, F. C., Gibbins, J., et al. (2010). Eight-plex iTRAQ analysis of variant metastatic human prostate cancer cells identifies candidate biomarkers of progression: an exploratory study. Prostate 70 (12), 1313–1332. doi:10.1002/pros.21167
Grabherr, M. G., Haas, B. J., Moran, Y., Levin, J. Z., Thompson, D. A., Ido, A., et al. (2011). Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 29 (7), 644–652. doi:10.1038/nbt.1883
Graham, T. L. (1991). Flavonoid and isoflavonoid distribution in developing soybean seedling tissues and in seed and root exudates. Plant physiol. 95 (2), 594–603. doi:10.1104/pp.95.2.594
Guo, D., Chen, F., Inoue, K., Blount, J. W., and Dixon, R. A. (2001). Downregulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa: impacts on lignin structure and implications for the biosynthesis of G and S lignin. plant cell 13 (1), 73–88. doi:10.1105/tpc.13.1.73
Hay, F. R., Smith, R. D., Ellis, R. H., and Butler, L. H. (2010). Developmental changes in the germinability, desiccation tolerance, hardseededness, and longevity of individual seeds of trifolium Ambiguum. Ann. Bot. 105, 1035–1052. doi:10.1093/aob/mcq037
Hayashi, S., Reid, D. E., Lorenc, M. T., Stiller, J., Edwards, D., Gresshoff, P. M., et al. (2012). Transient nod factor-dependent gene expression in the nodulation-competent zone of soybean (glycine Max [L.] merr.) roots. Plant Biotechnol. J. 10 (8), 995–1010. doi:10.1111/j.1467-7652.2012.00729.x
Hely, F. W. (1957). Symbiotic variation in trifolium ambiguum M. Bleb. With special reference to the nature of resistance. Aust. J. Biol. Sci. 10 (1), 1–16. doi:10.1071/bi957001
Jiawei, L., Mingjiu, W., Yan, Z., Kefan, A., Lijun, C, Xinyan, H., et al. (2023). Transcriptomics and metabolomics profiling reveals involvement of flavonoids in early nodulation of Caucasian clover (trifolium Ambiguum). Plant physiology Biochem. PPB, 203108050–108050. doi:10.1016/j.plaphy.2023.108050
Kanehisa, M., and Goto, S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic acids Res. 28 (1), 27–30. doi:10.1093/nar/28.1.27
Kannenberg, L. W., and Elliott, F. C. (1962). Ploidy in trifolium ambiguum M. Bieb. In relation to some morphological and physiological characters 1. Crop Sci. 2 (5), 378–381. doi:10.2135/cropsci1962.0011183x000200050004x
Kavroulakis, N., Flemetakis, E., Aivalakis, G., and Katinakis, P. (2000). Carbon metabolism in developing soybean root nodules: the role of carbonic anhydrase. Mol. plant-microbe Interact. 13 (1), 14–22. doi:10.1094/MPMI.2000.13.1.14
Khandual, S. (2007). Flavonoids as signaling molecules and regulators of root nodule development. Dyn. soil, Dyn. plant 1 (2), 83–94.
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinforma. 12, 323–16. doi:10.1186/1471-2105-12-323
Li, S., Zhang, Z., and Wang, Y. (2016). Effect of symbiotic rhizobium in alfalfa on PhysiologicalChange under cold stress. Acta Agrestia Sin. 24 (02), 377–383. doi:10.11733/j.issn.1007-0435.2016.02.020
Li, W., Wang, Y., Okamoto, M., Crawford, N. M., Siddiqi, M. Y., and Glass, A. D. M. (2007). Dissection of the AtNRT2. 1: atNRT2. 2 inducible high-affinity nitrate transporter gene cluster. Plant physiol. 143 (1), 425–433. doi:10.1104/pp.106.091223
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25 (4), 402–408. doi:10.1006/meth.2001.1262
Lv, E. M. (2023). Proteomic analysis mediated by the quorum sensing system bjaR1-bjaI in Bradyrhizobium diazoefficiens, a soybean rhizobium. China: Lanzhou University.
Miller, S. H., Elliot, R. M., Sullivan, J. T., and Ronson, C. W. (2007). Host-specific regulation of symbiotic nitrogen fixation in Rhizobium leguminosarum biovar trifolii. Microbiology 153, 3184–3195. doi:10.1099/mic.0.2007/006924-0
Mumm, R., Posthumus, M. A., and Dicke, M. (2008). Significance of terpenoids in induced indirect plant defence against herbivorous arthropods. Plant, Cell and Environ. 31 (4), 575–585. doi:10.1111/j.1365-3040.2008.01783.x
Peters, N. K., Frost, J. W., and Long, S. R. (1986). A plant flavone, luteolin, induces expression of Rhizobium meliloti nodulation genes. Science 233 (4767), 977–980. doi:10.1126/science.3738520
Redmond, J., Batley, M., Djordjevic, M., Innes, R. W., Kuempel, P. L., and Rolfe, B. G. (1986). Flavones induce expression of nodulation genesin rhizobium. Nature 323, 632–635. doi:10.1038/323632a0
Salavati, A., Khatoon, A., Nanjo, Y., and Komatsu, S. (2012). Analysis of proteomic changes in roots of soybean seedlings during recovery after flooding. J. proteomics 75 (3), 878–893. doi:10.1016/j.jprot.2011.10.002
Sheaffer, C. C., and Seguin, P. (2011). Kura clover response to potassium fertilization. Commun. Soil Sci. Plant Analysis 42, 450–456. doi:10.1080/00103624.2011.542221
Spaink, H. (2000). Root nodulation and infection factors produced by rhizobial bacteria. Annu. Rev. Microbiol. 54, 257–288. doi:10.1146/annurev.micro.54.1.257
Stafford, H. A. (1997). Roles of flavonoids in symbiotic and defense functions in legume roots. Botanical Rev. 63, 27–39. doi:10.1007/bf02857916
Sun, Y., Wang, X., Chen, Z., Qin, L., Li, B., Ouyang, L., et al. (2021). Quantitative proteomics and transcriptomics reveals differences in proteins during anthers development in Oryza longistaminata. Front. Plant Sci. 12, 744792. doi:10.3389/fpls.2021.744792
Takanashi, K., Takahashi, H., Sakurai, N., Sugiyama, A., Suzuki, H., Shibata, D., et al. (2012). Tissue-specific transcriptome analysis in nodules of lotus Japonicus. Mol. plant-microbe Interact. 25 (7), 869–876. doi:10.1094/MPMI-01-12-0011-R
Truchet, G., Roche, P., Lerouge, P., Vasse, J., Camut, S., Billy, F., et al. (1991). Sulphated lipo-oligosaccharide signals of Rhizobium meliloti elicit root nodule organogenesis in alfalfa. Nature 351, 670–673. doi:10.1038/351670a0
Umehara, M., Hanada, A., Yoshida, S., Akiyama, K., Arite, T., Takeda-Kamiya, N., et al. (2008). Inhibition of shoot branching by new terpenoid plant hormones. Nature 455 (7210), 195–200. doi:10.1038/nature07272
Valkov, V. T., Sol, S., Rogato, A., and Chiurazzi, M. (2020). The functional characterization of LjNRT2. 4 indicates a novel, positive role of nitrate for an efficient nodule N2-fixation activity. New Phytol. 228 (2), 682–696. doi:10.1111/nph.16728
Wasson, A. P., Pellerone, F. I., and Mathesius, U. (2006). Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by rhizobia. Plant Cell 18 (7), 1617–1629. doi:10.1105/tpc.105.038232
Watson, R. N., Neville, F. J., Bell, N. L., and Harris, S. (1996). Caucasian clover as a pasture legume for dryland dairying in the coastal Bay of Plenty. Proc. N. Z. Grassl. Assoc., 183–188. doi:10.33584/jnzg.1996.58.2212
Xie, C., Mao, X., Huang, J., Ding, Y., Wu, J., Dong, S., et al. (2011). KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic acids Res. 39 (Suppl. l_2), W316–W322. doi:10.1093/nar/gkr483
Yin, X., He, T., Yi, K., Cui, G., Hu, Y., Liu, J., et al. (2021). Comprehensive evaluation of candidate reference genes for quantitative real-time PCR-Based analysis in Caucasian clover. Sci. Rep. 11 (1), 3269. doi:10.1038/s41598-021-82633-2
Yuan, S., Li, R., Chen, S., Chen, H., Zhang, C., Hao, Q., et al. (2016). RNA-seq analysis of differential gene expression responding to different rhizobium strains in soybean (glycine Max) roots. Front. plant Sci. 7, 721. doi:10.3389/fpls.2016.00721
Zhang, Z., Shao, L., Chang, L., Cao, Y., Zhang, T., Wang, Y., et al. (2016). Effect of rhizobia symbiosis on lignin levels and forage quality in alfalfa (Medicago sativa L.). Agric. Ecosyst. and Environ. 233, 55–59. doi:10.1016/j.agee.2016.08.035
Zhou, Y., Stuart-Williams, H., Grice, K., Kayler, Z. E., Zavadlav, S., Vogts, A., et al. (2015). Allocate carbon for a reason: priorities are reflected in the ¹³C/¹²C ratios of plant lipids synthesized via three independent biosynthetic pathways. Phytochemistry 111, 14–20. doi:10.1016/j.phytochem.2014.12.005
Keywords: Caucasian clover, nodulation, nitrogen fixation, transcriptomics, proteomics
Citation: Liu J, Hao X, Guo Y, Wang M and Huang F (2025) Mechanism of nodulation and nitrogen fixation in Caucasian clover (Trifolium ambiguum Bieb.) based on transcriptomics and proteomics analyses. Front. Genet. 16:1600377. doi: 10.3389/fgene.2025.1600377
Received: 26 March 2025; Accepted: 27 June 2025;
Published: 24 July 2025.
Edited by:
Weibo Ren, Inner Mongolia University, ChinaCopyright © 2025 Liu, Hao, Guo, Wang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingjiu Wang, d2FuZ21qXzA1NDBAMTYzLmNvbQ==; Fan Huang, aHVhbmdmYW5AY2Fhcy5jbg==
 Jiawei Liu
Jiawei Liu Xinyan Hao1
Xinyan Hao1 Yue Guo
Yue Guo Mingjiu Wang
Mingjiu Wang