- 1Department of Animal Science, Federal University of Bahia, Salvador, Brazil
- 2Department of Animal Sciences, Purdue University, West Lafayette, IN, United States
- 3Department of Animal Science, University of Nebraska-Lincoln, Lincoln, NE, United States
- 4United States Department of Agriculture - Agricultural Research Service, Roman L. Hruska U.S. Meat Animal Research Center, Clay Center, NE, United States
- 5United States Department of Agriculture - Agricultural Research Service, Range Sheep Production Efficiency Research Unit, Dubois, ID, United States
- 6United States Department of Agriculture - Agricultural Research Service, Dale Bumpers Small Farms Research Center, Booneville, AR, United States
Ewe longevity indicators are complex traits that are lowly heritable, expressed late in life, and sex-limited, making them challenging to include in breeding programs. In this context, genome-wide association studies (GWASs) can provide more information on the complex genetic control of these traits. Therefore, the primary objective of this study was to carry out association analyses for 8 longevity-related traits in 12,734 Katahdin ewes. A total of 126 associations at the chromosome-wide level and 3 at genome-wide level were found. These associations involved 86 single-nucleotide polymorphisms (SNPs) located across 22 chromosomes, with 24 of these SNPs associated with two or more traits. The variants overlapped with genes previously associated with prolificacy (APOH, NLRP9, H3PXD2A, CKB, and HERC4), ovarian follicle pool (GALNT13, TMEM150B, and BRSK1), synthesis and release of reproductive hormones (SULT1B1, LEF1, and EIF5), and early pregnancy events (ITGAV, HADH, ZNFX1, ZSCAN4, EPN1, FBXW8, NOS1, ST3GAL4, and GFRA1). Moreover, genes related to response to stress or pathological conditions (ADCY5, HADH, ATRNL1, LEP, IL11, NLRP9, PRKCG, PRKCA, NEDD4L, FECH, CTNNA3, HECTD1, LRRTM3, and zinc-finger proteins), growth performance (GRID2, MED13L, DCPS, and LEP), and carcass traits (CMYA5 and SETD3) were also implicated. Metabolic pathways such as oxytocin signaling and cardiac-related pathways were enriched. These findings suggest that longevity indicators in Katahdin ewes are highly polygenic traits influenced by a combination of voluntary and involuntary culling reasons. Candidate genes and metabolic pathways influencing reproductive performance and health may play a key role in the functional longevity of Katahdin ewes.
1 Introduction
Ewe culling or death may be due to multiple causes, which can be classified as voluntary and involuntary culling (McLaren et al., 2020). Voluntary culling occurs when a breeder decides to cull a healthy and fertile ewe due to, for instance, low prolificity or old age. Voluntary culling is often necessary and contributes to genetic improvement of the flock as older ewes may be replaced with higher genetic merit ewe lambs. In contrast, involuntary culling occurs when a breeder culls (or the ewe dies) due to involuntary reasons such as disease or infertility, which results in ewe wastage. Involuntary culling is a major issue in many sheep flocks (Flay et al., 2021). Ewe wastage results in economic losses for the breeder due to the costs of raising more ewe lambs to maintain a consistent number of ewes in the flock, a reduction in the number of female lambs to sell, and loss of high production expected from mature ewes. A 10% reduction in ewe wastage could increase farm profitability by 17% (Farrell et al., 2019).
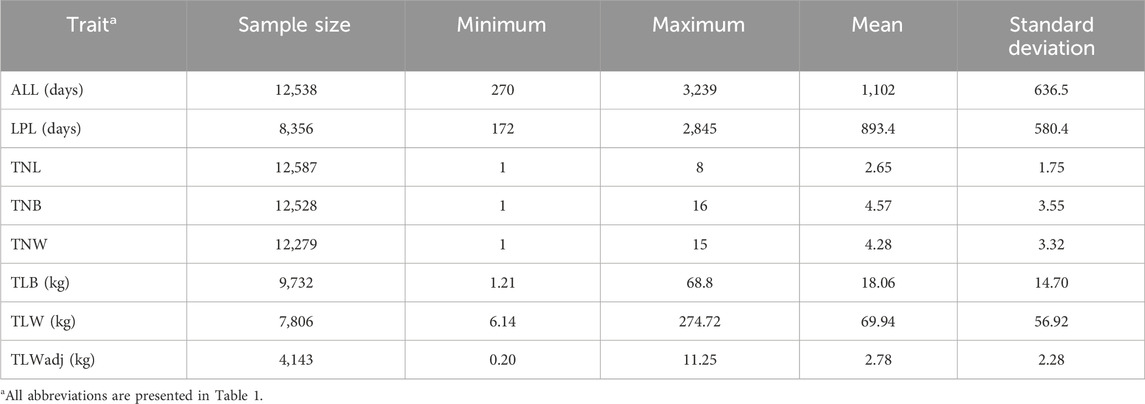
There are some direct indicators of ewe longevity, such as age at last lambing (ALL) and length of productive life (LPL), which measure total and functional lifetime, respectively. These traits have small additive genetic variances that can be used in selection schemes. For instance, heritability estimates of 0.05 for ALL were reported in both the U.S. Targhee sheep (Borg et al., 2009) and African Dorper sheep (Zishiri et al., 2013). In Spanish Churra dairy ewes, heritability estimates from 0.02 to 0.05 were reported for direct indicators of longevity (El-Saied et al., 2005). In New Zealand, longevity was defined as ALL minus 2 years, and heritability estimates of 0.10 (in ram breeder flocks) and 0.13 (in commercial flocks) were reported (Lee et al., 2015).
Accumulated ewe performance traits, such as the total number of litters (TNL), total number of lambs born (TNB), and weaned (TNW), as well as total litter body weight of the lamb measured at birth (TLB) and weaning (TLW), can be used as indirect longevity indicators. Furthermore, these traits seem to have slightly greater heritability estimates than ALL and LPL. For instance, heritability estimates of 0.10 (TNL), 0.11 (TNB), and 0.10 (TLW) were reported for Suffolk ewes (Milerski et al., 2018), while estimates of 0.13 (TLB) and 0.14 (TLW) were reported for Malpura sheep in India (Gowane et al., 2014). For South African Dorper sheep, low heritability estimates were reported for TNB (0.10) and TNW (0.09) (Zishiri et al., 2013), while moderate heritability estimates were reported for TNB (0.23), TNW (0.17), and TLW (0.20) in Merino sheep (Duguma et al., 2002). Therefore, genetic selection for improved longevity could significantly reduce ewe wastage.
Regardless of the differences in heritability estimates across studies, longevity-related traits in sheep have a heritable component that may be exploited in breeding schemes. However, many ewe longevity indicators tend to be recorded late in life and are sex-limited (female trait). Therefore, predicting accurate breeding values for selection candidates is difficult, especially for younger animals. Genomic selection has revolutionized genetic progress for such traits. However, in addition to performing genomic prediction of breeding values, there is a need to investigate the genomic architecture of complex traits such as ewe longevity indicators. Still, only one recent genome-wide association study (GWAS) investigated ewe longevity, which included Rambouillet, Polypay, Suffolk, and Columbia breeds (Smitchger et al., 2024). This study reported several single-nucleotide polymorphisms (SNPs) associated with traits directly and indirectly related to ewe longevity. Moreover, several genes previously associated with reproduction, dentition, and the immune system were reported. Nevertheless, this study was performed based on a small sample size (less than 500 animals per breed) from a single production system. Therefore, additional studies could validate the genomic regions previously reported or contribute to the identification of new genomic regions associated with ewe longevity.
The Katahdin is a composite sheep breed that is growing in popularity in the United States and has the largest reference population for genomic studies in the country. This breed has been directly selected for improving body weight at different ages, body composition, reproduction performance, and gastrointestinal parasite resistance (Notter and Lewis, 2018). However, U.S. Katahdin breeders have not directly selected for longevity indicators in breeding schemes. A previous study showed that a high percentage of primiparous ewes were culled before the second lambing (Pinto et al., 2025a). Then, variance components were estimated, and genomic predictions were carried out for eight longevity indicators in U.S. Katahdin sheep (Pinto et al., 2025b), which can be used as selection criteria to improve the U.S. Katahdin ewe longevity, but the genetic architecture of these traits remained unknown. Previous studies were performed to identify variants associated with resistance or susceptibility to gastrointestinal nematodes in Katahdin sheep (Becker et al., 2020; Becker et al., 2022; Notter et al., 2022). However, no previous studies were carried out to identify genomic variants associated with longevity-related traits in Katahdin sheep. Thus, the primary objective of this study was to carry out GWAS analyses to identify SNPs associated with eight ewe longevity indicators in this breed.
2 Materials and methods
2.1 Ethics approval
Approval from the Animal Use and Ethics Committee was not needed for this study as pre-existing datasets were provided by the National Sheep Improvement Program (NSIP) and recorded by U.S. Katahdin producers during routine husbandry activities.
2.2 Population and traits
The complete Katahdin pedigree from the NSIP contained information from 127,535 lambs born between 1985 and 2023. A total of 26,392 unique ewes with lambing records were identified, producing 118,510 lambs in 67,397 litters. Ewes (n = 1,233) with lambing intervals shorter than 150 days or longer than 720 days and ewes (n = 2,666) with age at first lambing younger than 270 days or older than 1,095 days were removed from further analyses. After this data filtering, 22,493 ewes from 231 flocks, which produced 100,981 lambs in 57,436 litters, remained.
We then removed flocks with few years of recorded data, inconsistent data reporting, or few records submitted to NSIP per year. Only data from flocks that reported lambing events for more than six consecutive years were retained. This threshold ensured that each flock was enrolled in NSIP long enough to capture the upper end of the expected productive life of ewes (McLaren et al., 2020; Hanna et al., 2023). Some flocks reported lambing records for more than 6 years but with a subsequent large and abrupt reduction in submitted lamb records in the last few years before leaving the NSIP; these were considered inconsistent flocks. We calculated the average number of lambing records across years for each flock. We found 20 lambing records per year to be the minimal value for excluding all the inconsistent flocks. This filter also enabled the exclusion of very small flocks, which reduced overall environmental noise. After the flock filters, data from 17,712 ewes from 58 flocks remained to be analyzed. These ewes produced 85,591 lambs in 48,533 litters.
Currently, the NSIP members do not record culling reasons and culling dates for all or the majority of their ewes. This prevents us from including the culling types (voluntary or involuntary) as a classification factor in our analyses, as done in studies for other species such as cattle (Oliveira et al., 2020). We recommend that sheep breeders start recording such information to facilitate future studies on ewe longevity based on culling reasons. As a consequence of the lack of this information in the studied population, we assumed that ewes were dead or culled if they had no lambing records during the last 2 years for which the flock reported data. Thus, ultimately, 12,734 ewes born between 1989 and 2020 in 58 flocks were identified as culled and included in subsequent GWAS analyses. The ewes contributing records were daughters of 1,245 sires and 6,325 dams and produced 61,178 lambs in 34,796 litters.
Eight longevity indicator traits were derived, and their definitions and abbreviations are presented in Table 1. Individual body weights at birth, weaning, and post-weaning required adjustment before calculating TLB, TLW, and an adjusted TLW (TLWadj). Lamb birth weight was adjusted to a female equivalent (
where BW represents the lamb’s actual birth weight and
where
The LPL can be defined as the number of days between the first lambing and culling (or death) (Ducrocq et al., 1988). As there were no records of culling or death dates in the Katahdin datasets provided by NSIP, we defined LPL as the difference (in days) between the first and last lambing records for each ewe (Milerski et al., 2018). For this reason, all ewes with LPL data had two or more lambing records. Descriptive statistics for all longevity indicator traits are presented in Table 2.
2.3 Genomic data
The NSIP Katahdin population had 10,032 animals genotyped. This population was genotyped with three SNP arrays: two versions of the Ovine 50K BeadChip and the Illumina Ovine Infinium HD BeadChip (Illumina Inc., San Diego, CA, United States). All SNP positions were remapped to the ARS-UI_Ramb_v2.0 reference using the National Center for Biotechnology Information Genome Remapping Service. Only shared positions were kept, and duplicate positions were reduced to those with the highest call rate. The distribution of genotyped animals across years is shown in Supplementary Figure S1. Quality control (QC) criteria were used to exclude SNPs with a minor allele frequency (MAF) lower than 0.05, SNPs and animals with a call rate lower than 0.90, SNPs with a difference greater than 0.15 between the observed and expected heterozygosity (extreme departure from Hardy–Weinberg equilibrium as an indication of genotyping errors), and non-autosomal SNPs. After QC, a panel of 30,408 SNPs genotyped in 10,032 animals remained for posterior analyses.
We also performed a principal component analysis of genomic information using the plotPCA option of postGSf90 software from the BLUPF90 suite (Misztal et al., 2022) to identify potential population stratification (Supplementary Figure S2). Supplementary Figure S2 is a Cartesian plot of the first two principal components of the genomic data of animals included in the study, which explains the largest percentage of total variance in the genotype data. A single cluster was observed and, therefore, we assumed no significant stratification in this population.
2.4 Association analyses
The GWAS analyses were performed based on the single-step GWAS (ssGWAS) approach (Misztal et al., 2009). The model can be described as follows:
where
In this MME, new terms include
where
The animal effect was fitted as a random effect and assumed as
PostGSf90 software (Misztal et al., 2022) was used to estimate the SNP effect. The SNP effects were derived as
The Bonferroni correction for multiple tests assumes that all hypothesis tests are independent. In the GWAS context, the association tests are not independent as there is linkage disequilibrium among SNPs. Therefore, significant threshold levels were calculated for each chromosome as
2.5 Functional annotation analyses
The function {find_genes_qtls_around_markers} from the GALLO R-package version 1.5 (Fonseca et al., 2020) was used to identify candidate genes and quantitative trait loci (QTL) within a 100 Kbp upstream and 100 Kbp downstream region surrounding each significant SNP position. The gene map (ARS-UI_Ramb_v2.0.112. gtf) retrieved from the Ensembl database (https://ftp.ensembl.org/pub/release-112/gtf/ovis_aries_rambouillet/) and the QTL map (QTLdb_sheepOAR_rambo2. gff, release #53) retrieved from the Sheep QTLdb (www.animalgenome.org/cgi-bin/QTLdb/OA/index) were used as inputs in the GALLO R package version 1.5 (Fonseca et al., 2020). Functional genomic annotation and enrichment analyses were carried out on the DAVID (Database for Annotation, Visualization, and Integrated Discovery) web server (Sherman et al., 2022), while the STRING database (Szklarczyk et al., 2023) was used to analyze protein–protein association networks.
3 Results
3.1 Association analyses
Genome-wide association analyses were performed for eight longevity-related traits in Katahdin ewes. The distributions of the -log10 of p-values are presented in Figure 1 (ALL and LPL), Figure 2 (TNL, TNB, and TNW), and Figure 3 (TLB, TLW, and TLWadj). Green points in these figures indicate significant chromosome-wide associations, which were distributed as follows: 22 (ALL; Table 3), 22 (LPL; Table 4), 16 (TNL; Table 5), 18 (TNB; Table 6), 17 (TNW; Table 7), 11 (TLB; Table 8), 11 (TLW; Table 9), and 12 (TLWadj; Table 9). The number of significant associations per Ovis aries chromosomes (OAR) was as follows: 5 (OAR1), 14 (OAR2), 7 (OAR3), 7 (OAR4), 5 (OAR6), 3 (OAR7), 2 (OAR8), 4 (OAR9), 3 (OAR10), 8 (OAR11), 7 (OAR13), 11 (OAR14), 3 (OAR15), 25 (OAR17), 8 (OAR18), 5 (OAR19), 1 (OAR20), 1 (OAR21), 2 (OAR22), 2 (OAR23), 3 (OAR25), and 3 (OAR26). Moreover, the genome-wide threshold was indicated by a red horizontal line in Figures 1–3. Three genome-wide associations (P < 2.12 × 10−05) of the SNP rs399310772 with ALL, LPL, and TNL were found. This SNP is on 12.2 Mb of OAR4, has a MAF of 0.39, and is an intron variant in the novel genes (ENSOARG00020028395 and ENSOARG00020033537). The Q–Q plots of each association analysis are also shown in Figures 1–3. The lambda values were approximately 1.00 for all traits as the p-values were adjusted for the genomic inflation factor.
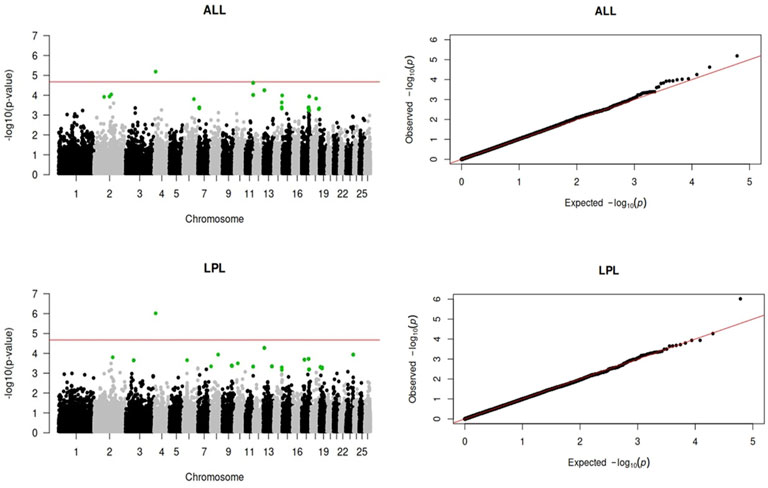
Figure 1. Manhattan and Q–Q plots of the genome-wide association analysis for age at last lambing (ALL) and length of productive life (LPL) in U.S. Katahdin sheep. The significant SNP at the chromosome-wide threshold are highlighted in green. The horizontal red line indicates the significance of the genome-wide threshold.
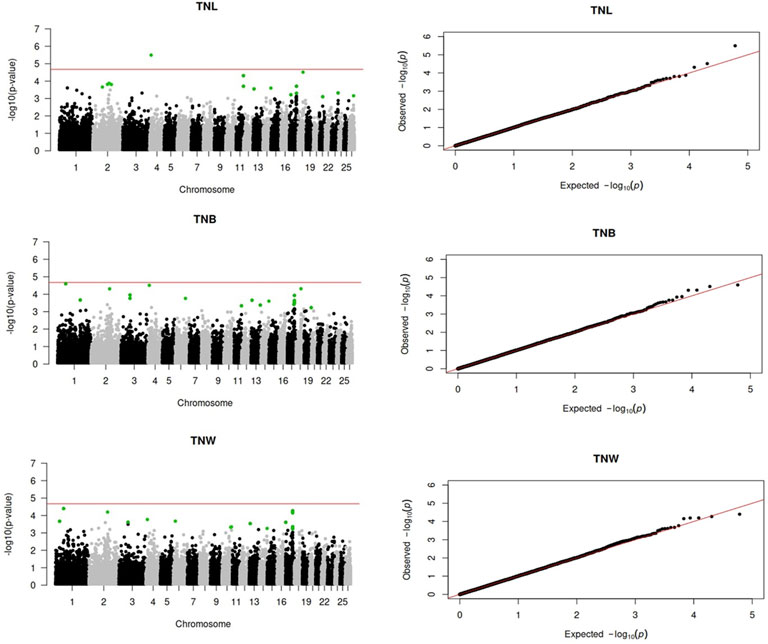
Figure 2. Manhattan and Q–Q plots of the genome-wide association analysis for total number of litters (TNL), lambs born (TNB), and lambs weaned (TNW) over ewe lifetime in U.S. Katahdin sheep. The significant SNP at the chromosome-wide threshold are highlighted in green. The horizontal red line indicates the significance of the genome-wide threshold.
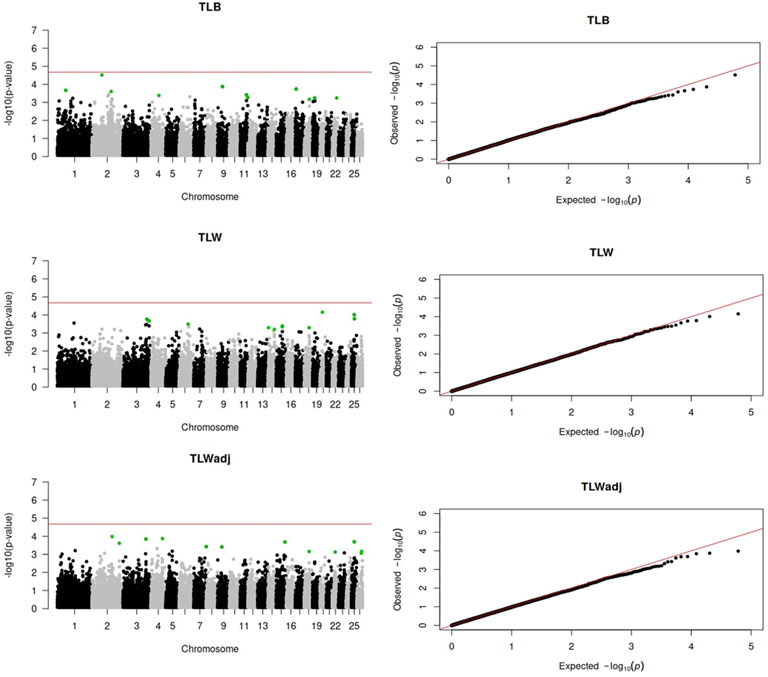
Figure 3. Manhattan and Q–Q plots of the genome-wide association analysis for total lamb weight at birth (TLB), total lamb weight at weaning (TLW), and total lamb weight at weaning divided by the ewe’s post-weaning weight (TLWadj) in U.S. Katahdin sheep. The significant SNP at the chromosome-wide threshold are highlighted in green. The horizontal red line indicates the significance of the genome-wide threshold.
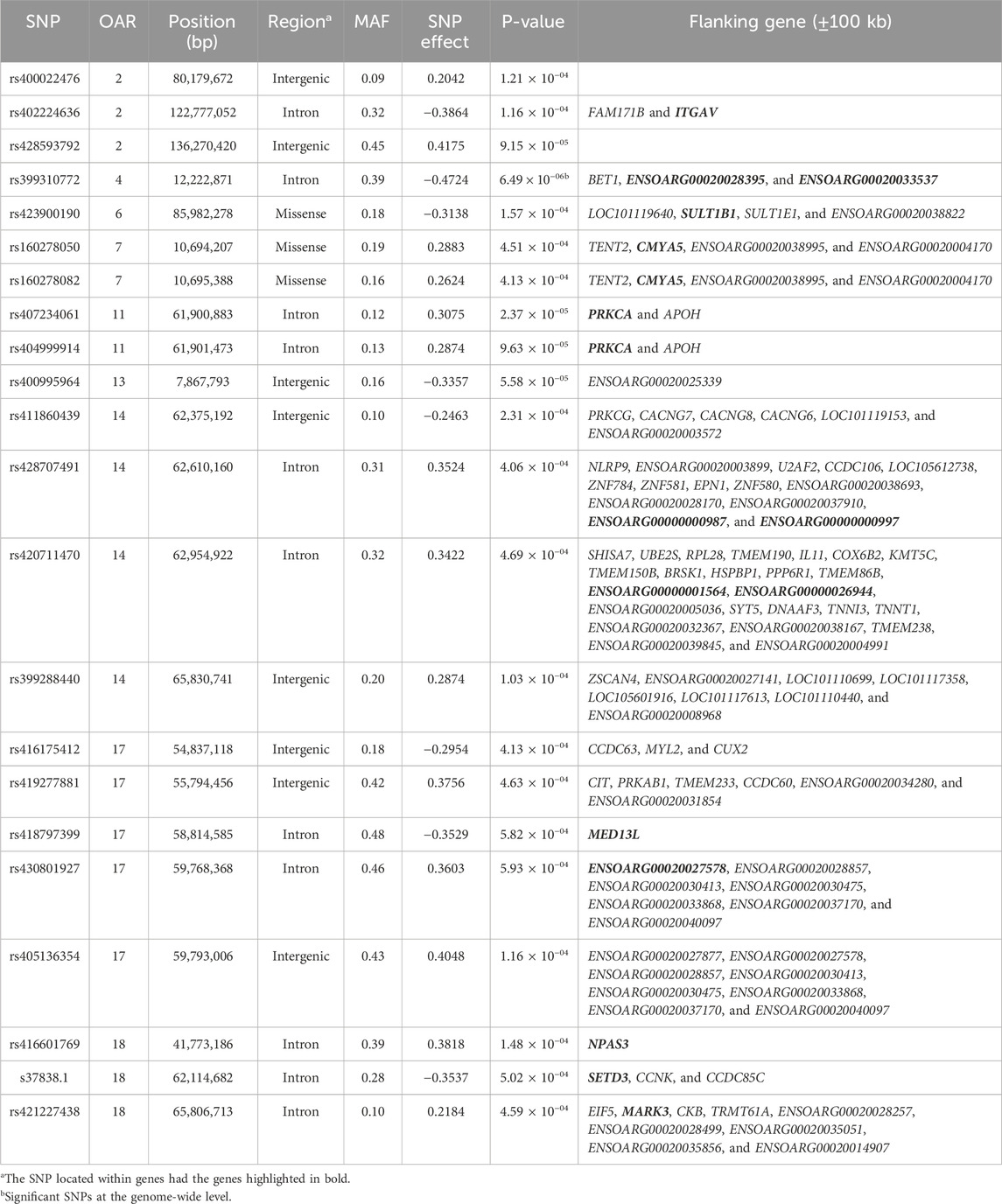
Table 3. SNPs associated with the age at last lambing (ALL) in U.S. Katahdin sheep, including chromosome (OAR) position, genomic region, MAF, and p-value of the association test.
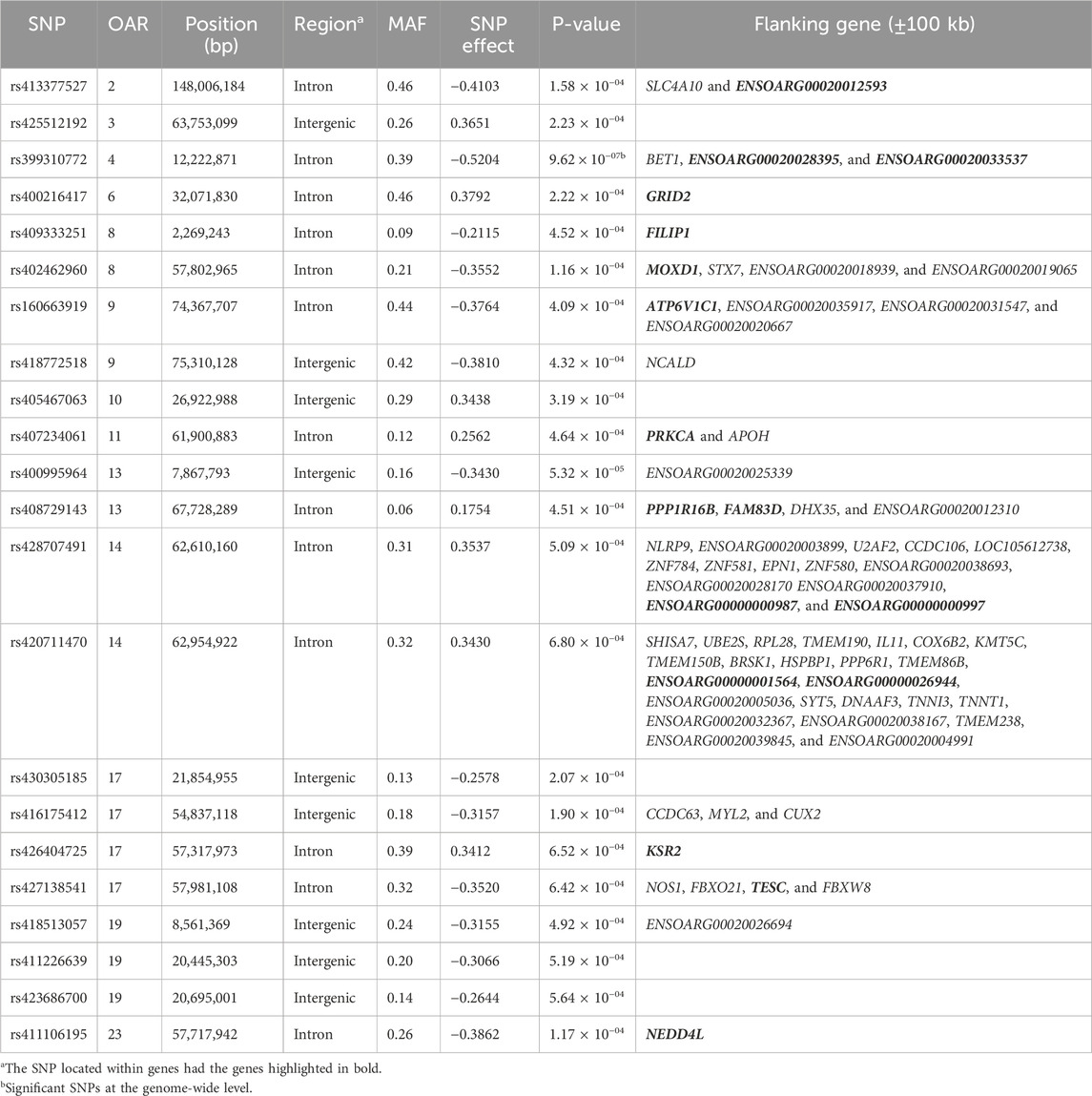
Table 4. SNPs associated with the lenght of productive life (LPL) in U.S. Katahdin sheep, including chromosome (OAR) position, genomic region, MAF, and p-value of the association test.
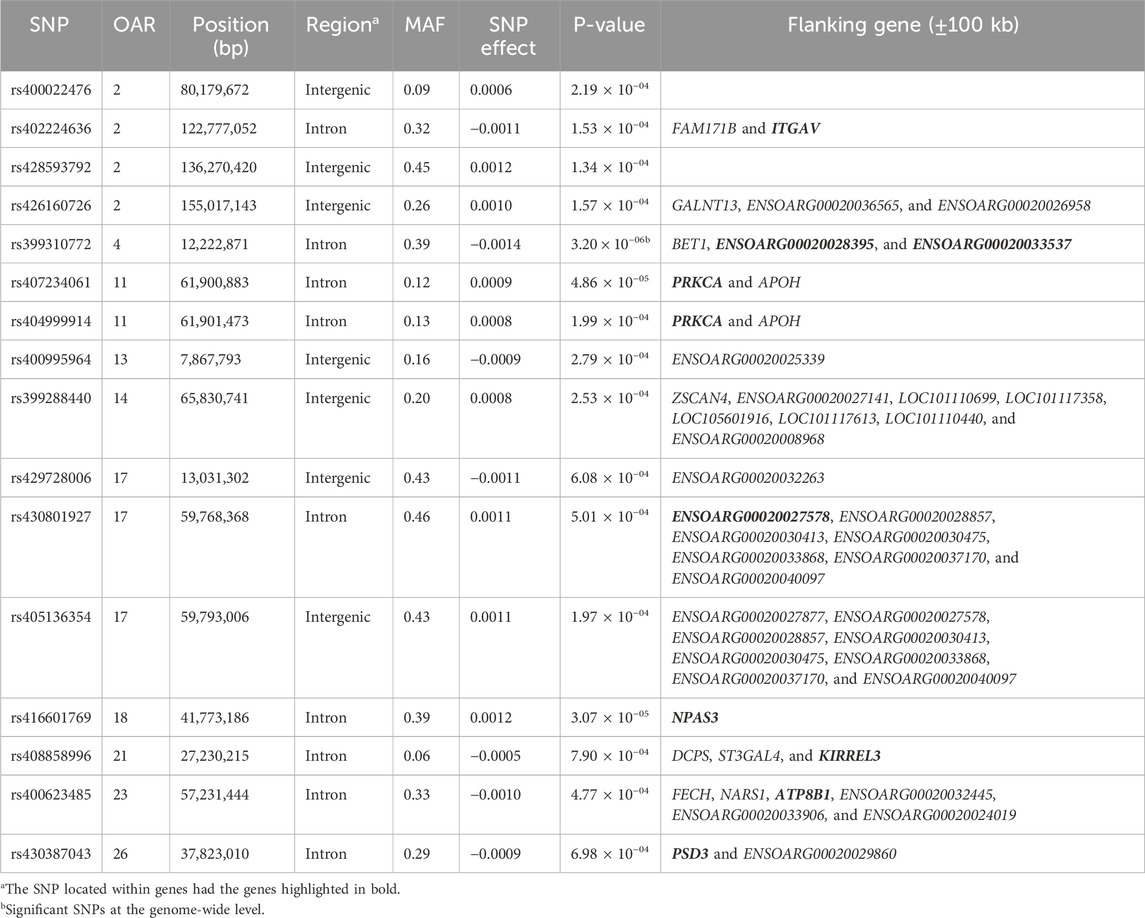
Table 5. SNPs associated with the total number of litters (TNL) over ewe lifetime in U.S. Katahdin sheep, including chromosome (OAR) position, genomic region, MAF, and p-value of the association test.
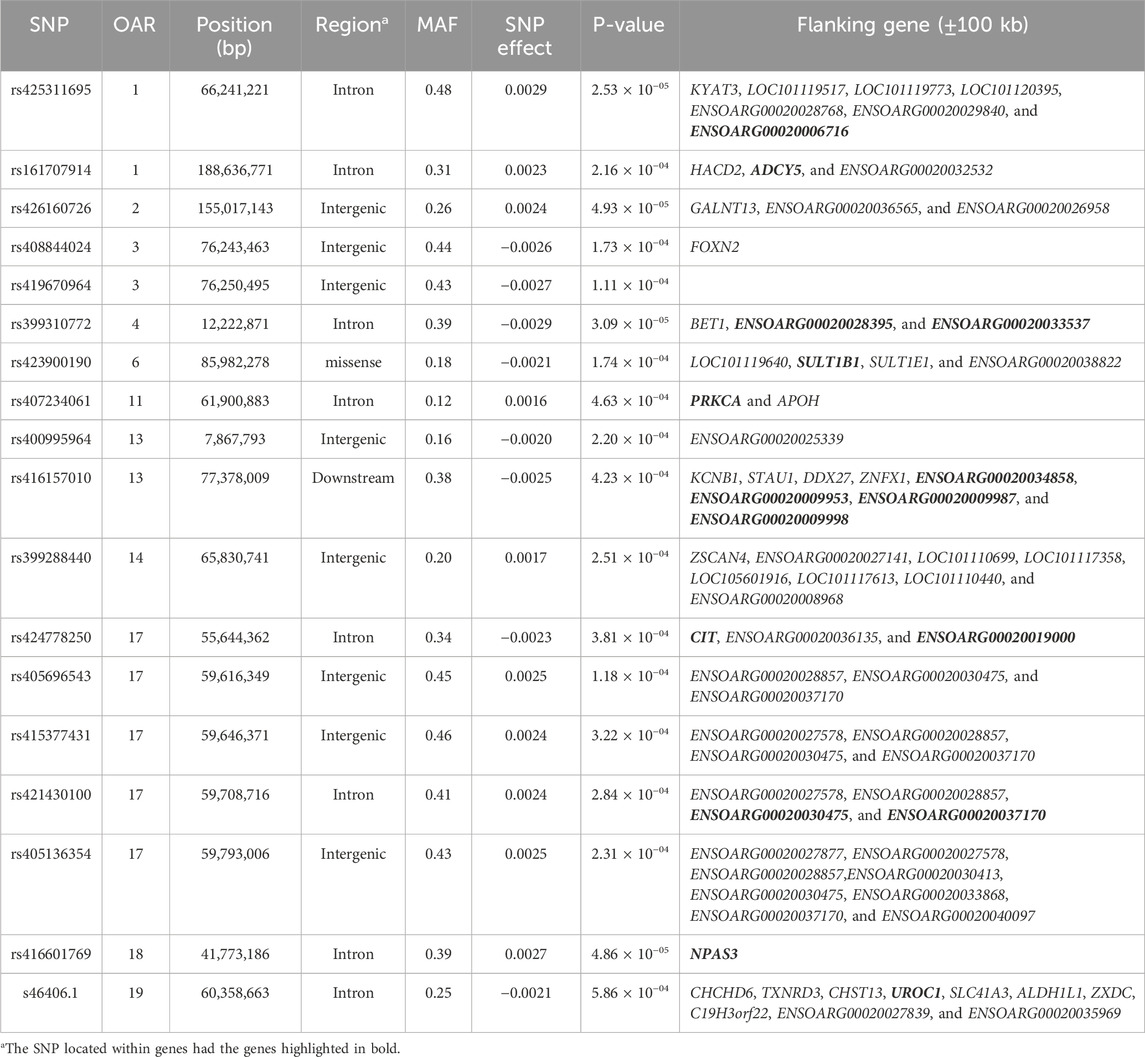
Table 6. SNPs associated with the total number of lambs born (TNB) over ewe lifetime in U.S. Katahdin sheep, including chromosome (OAR) position, genomic region, MAF, and p-value of the association test.
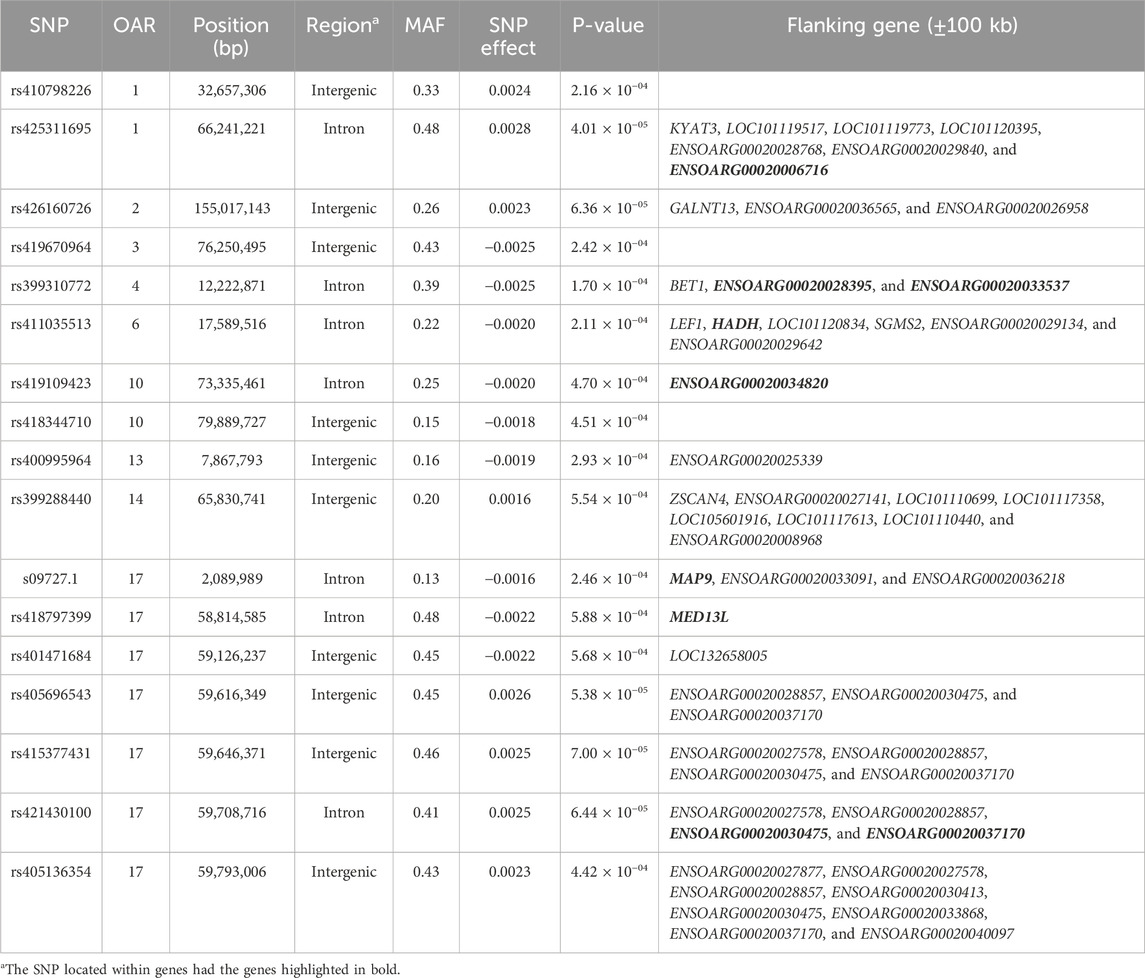
Table 7. SNPs associated with the total number of lambs weaned (TNW) over ewe lifetime in U.S. Katahdin sheep, including chromosome (OAR) position, genomic region, MAF, and p-value of the association test.
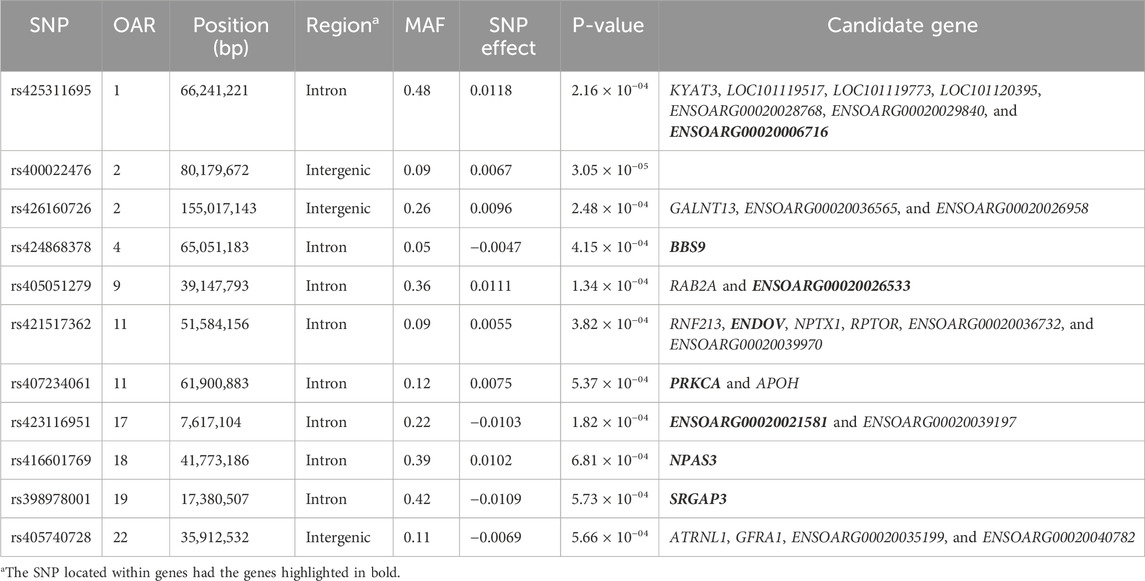
Table 8. SNPs associated with total lamb birth weight (TLB) over ewe lifetime in U.S. Katahdin sheep, including chromosome (OAR) position, genomic region, MAF, and p-value of the association test.
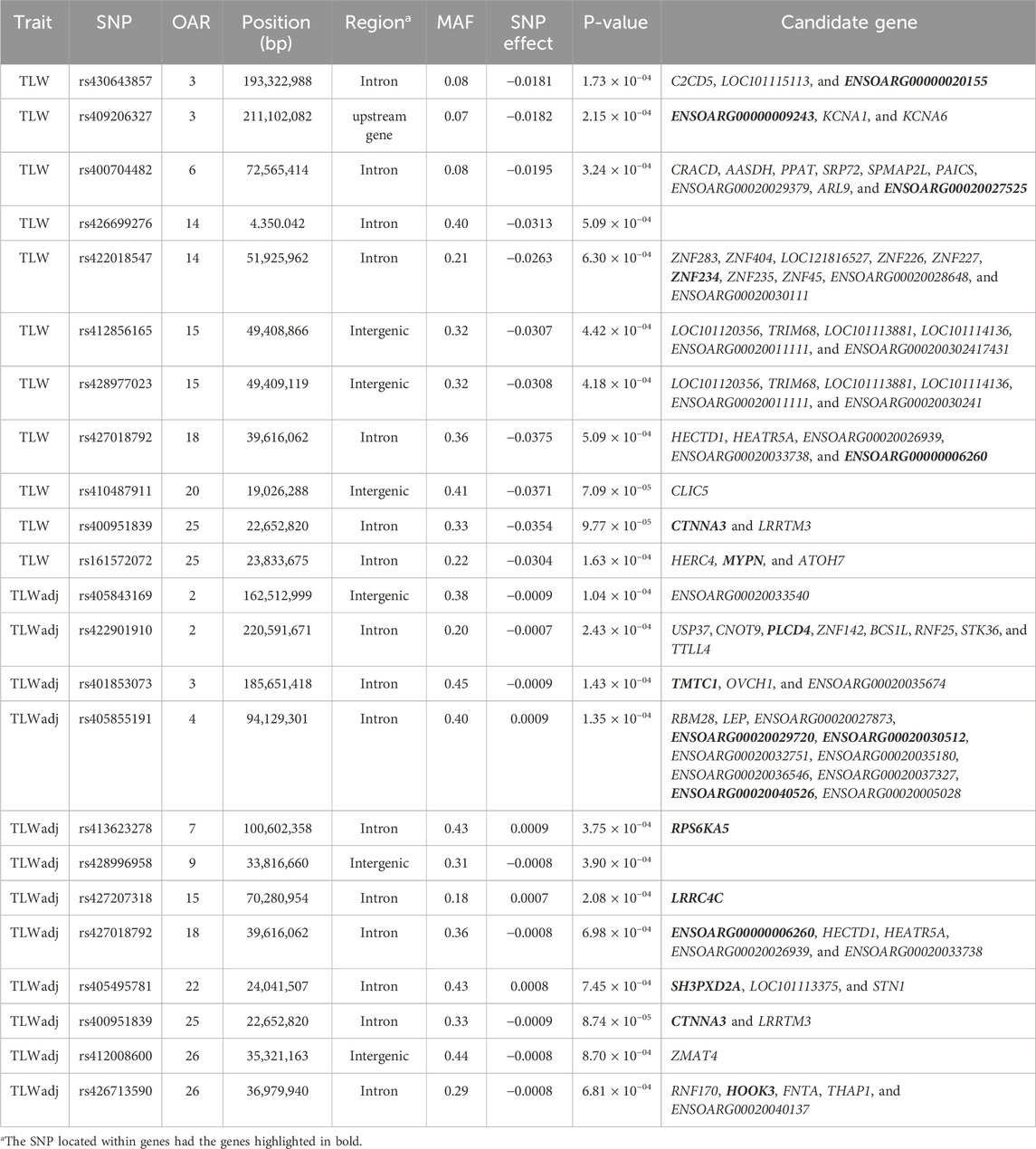
Table 9. SNPs associated with total lamb weight at weaning over ewe lifetime (TLW) and TLW divided by the ewe weight adjusted for 120 days (TLWadj) in U.S. Katahdin sheep, including chromosome (OAR) position, genomic region, MAF, and p-value of the association test.
The significant associations involved 86 unique SNPs as 24 of them were associated with two or more longevity-related traits. For instance, the variants rs399310772 on OAR4:12,222,871, rs407234061 on OAR11:61,900,883, and rs400995964 on OAR13:7,867,793 were associated with five traits. The intron variant rs399310772 is located in the genes ENSOARG00020028395 and ENSOARG00020033537, which encode long non-coding RNA (lncRNA) and are close to the protein-coding gene BET1 (Bet1 Golgi vesicular membrane trafficking protein), while the SNP rs400995964 is close to the novel protein-coding gene ENSOARG00020025339. Both SNPs were associated with ALL, LPL, TNL, TNB, and TNW. The SNP rs407234061, an A>G change in intron 1 of the PRKCA (protein kinase C alpha), was found to be associated with ALL, LPL, TNL, TNB, and TLB.
The 86 variants associated with the longevity-related traits are distributed in different genomic regions, including intergenic (31 SNP), intron (50 SNP), exon (3 SNP), upstream gene (1 SNP), and downstream gene (1 SNP) variants. The MAF of these variants ranged from 0.05 to 0.48 in the current Katahdin population. The missense SNP rs423900190 on OAR6:85,982,278 was associated with both ALL (Table 3) and TNB (Table 6). This is a T>C variant in exon 6 of the SULT1B1 (sulfotransferase family 1B member 1) gene, which results in an amino acid change from isoleucine to valine at position 136. The variants (Table 3) rs160278050 (OAR7:10,694,207) and rs160278082 (OAR7:10,695,388), also associated with ALL, are T>G and A>G substitutions, respectively, located in exon 2 of the CMYA5 (cardiomyopathy-associated 5) gene. These variants result in amino acid changes from valine to glycine (at position 2,654) and lysine to glutamic acid (at position acid 3,048).
3.2 Functional annotation analyses
Genome windows of 100 Kbp downstream and 100 Kbp upstream of each significant SNP were selected for functional analyses, and 272 genes were found. Of these, 203 are protein- and 69 RNA-coding genes (Supplementary File S1). Of the protein-coding genes, 174 have a biological function known, and 29 are novel genes. Of the RNA-coding genes, 52 encode long non-coding RNAs (lncRNAs), eight encode small nucleolar RNAs (snoRNAs), six encode small nuclear RNAs (snRNAs), and three encode microRNAs (miRNAs).
A list of 272 Ensembl gene identification codes was submitted to the DAVID tool, and 183 genes were retrieved. Three genes (ADCY5, PRKAB1, and RPTOR) are key components in the longevity-regulating pathway. Twenty-three genes (HACD2, ATP6V1C1, ST3GAL4, LOC101119640, ADCY5, ALDH1L1, CKB, LOC101120834, LOC101121036, LOC101113375, LOC105608607, FECH, HADH, KYAT3, KMT5C, NOS1, PPAT, PAICS, GALNT13, SGMS2, LOC101120331, TMEM86B, and UROC1) are associated with metabolic pathways. Seven genes (ADCY5, CACNG6, CACNG7, CACNG8, PRKAB1, PRKCA, and PRKCG) are associated with the oxytocin signaling pathway. Some genes play important roles in health, such as ADCY5, CACNG6, CACNG7, CACNG8, CTNNA3, ITGAV, LEF1, COX6B2, MYL2, NOS1, PRKAB1, PRKCA, RPS6KA5, and TNNI3, which are components of cardiac pathways, such as hypertrophic cardiomyopathy, dilated cardiomyopathy, arrhythmogenic right ventricular cardiomyopathy, adrenergic signaling in cardiomyocytes, and cardiac muscle contraction. Other genes are related to the immune system (LOC101110699, LOC101110440, LOC105601916, ZNF227, ZNF235, ZNF283, ZNF404, LOC101117358, LOC101117613, CTNNA3, MYL2, PRKCA, and PRKCG) and are components of pathways such as the herpes simplex virus 1 infection pathway and leukocyte transendothelial migration. There were also genes related to signaling pathways such as ITGAV, MED13L, PRKCA, and PRKCG (thyroid hormone signaling pathway); RAB2A, LEP, PRKAB1, and RPTOR (AMPK signaling pathway); and CACNG6, CACNG7, CACNG8, PRKCA, PRKCG, and RPS6KA5 (MAPK signaling pathway). The genes LOC101119773, LOC101119517, and LOC101120395 are members of the guanylate-binding protein family and are related to immunity and innate immunity biological processes.
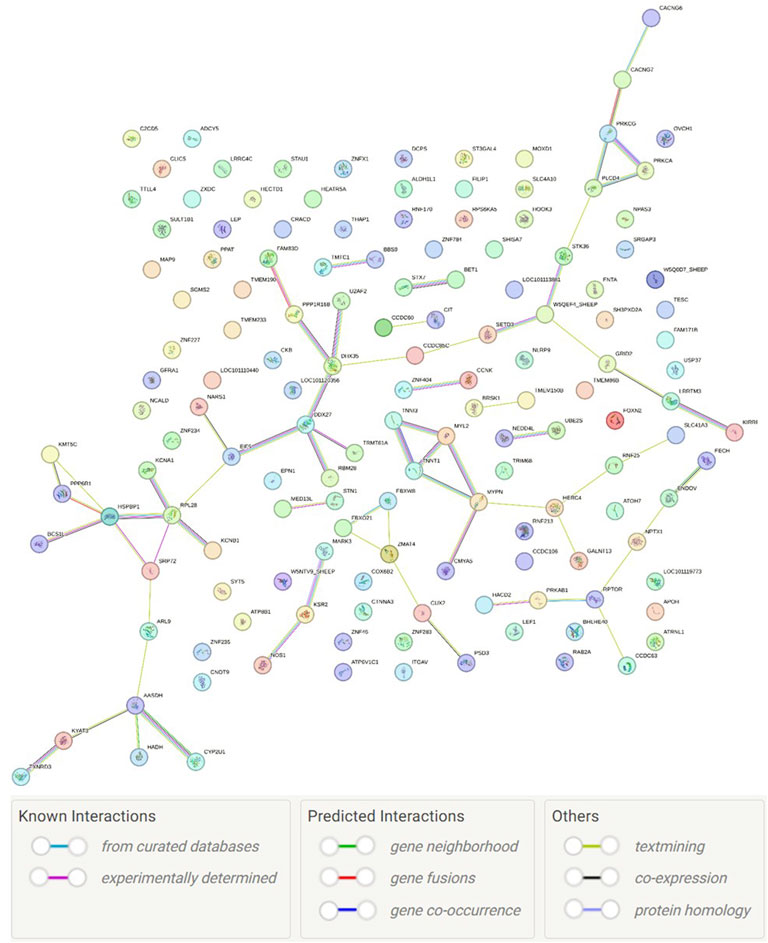
Although the genes are involved in many pathways, only six enriched pathways were found, namely, hypertrophic cardiomyopathy pathway (P = 0.00002), dilated cardiomyopathy pathway (P = 0.000028), arrhythmogenic right ventricular cardiomyopathy (P = 0.000084), adrenergic signaling in cardiomyocytes (P = 0.00039), oxytocin signaling pathway (P = 0.0020), and cardiac muscle contraction (P = 0.0023). A UniProt term called transferase (KW-0808; P = 0.0032; with 17 genes: BRSK1, HECTD1, NEDD4L, ST3GAL4, LOC101119640, CIT, LOC105608607, FNTA, KSR2, KYAT3, KMT5C, PPAT, GALNT13, PRKCA, PRKCG, RPS6KA5, RNF170, STK36, SGMS2, LOC101120084, LOC101120331, TRMT61A, TENT2, and UBE2S) was found to be enriched. Finally, the protein–protein interaction network (Figure 4) had significantly more interactions than expected (P = 0.0371).
The search for QTLs in the genome windows ±100 Kbp also found interesting results (Table 10). For instance, there were QTLs from OAR10:26,866,363 to OAR10:26,995,420 previously associated with horn type and horn circumference. QTLs for health-related traits such as pleurisy, mean corpuscular hemoglobin concentration, and Haemonchus contortus resistance have been previously reported on OAR2:136,270,247–136,270,251 bp, OAR6:17,589,514–17,589,518 bp, OAR8:2,217,907–2,217,911 bp, OAR10:73,366,801–73,366,805 bp, and OAR17:55,671,815–59,533,215 bp. For lactation-related traits such as milk, fat, and protein yields, there were QTLs located on OAR3:63664566–63664570 bp, OAR4:94162840–94162844 bp, OAR13:67636466–67769899 bp, OAR17:58854886–58854890 bp, and on OAR21:27132598–27132602 bp, while QTLs for growth-related traits such as average daily gain and body weight were identified on OAR4:94162840–94162844 bp and OAR14:62350077–62350081 bp. The enrichment analyses found an enriched QTL for H. contortus resistance on OAR17 (P = 0.0007) and another on OAR6 for mean corpuscular hemoglobin concentration (P = 0.0076).
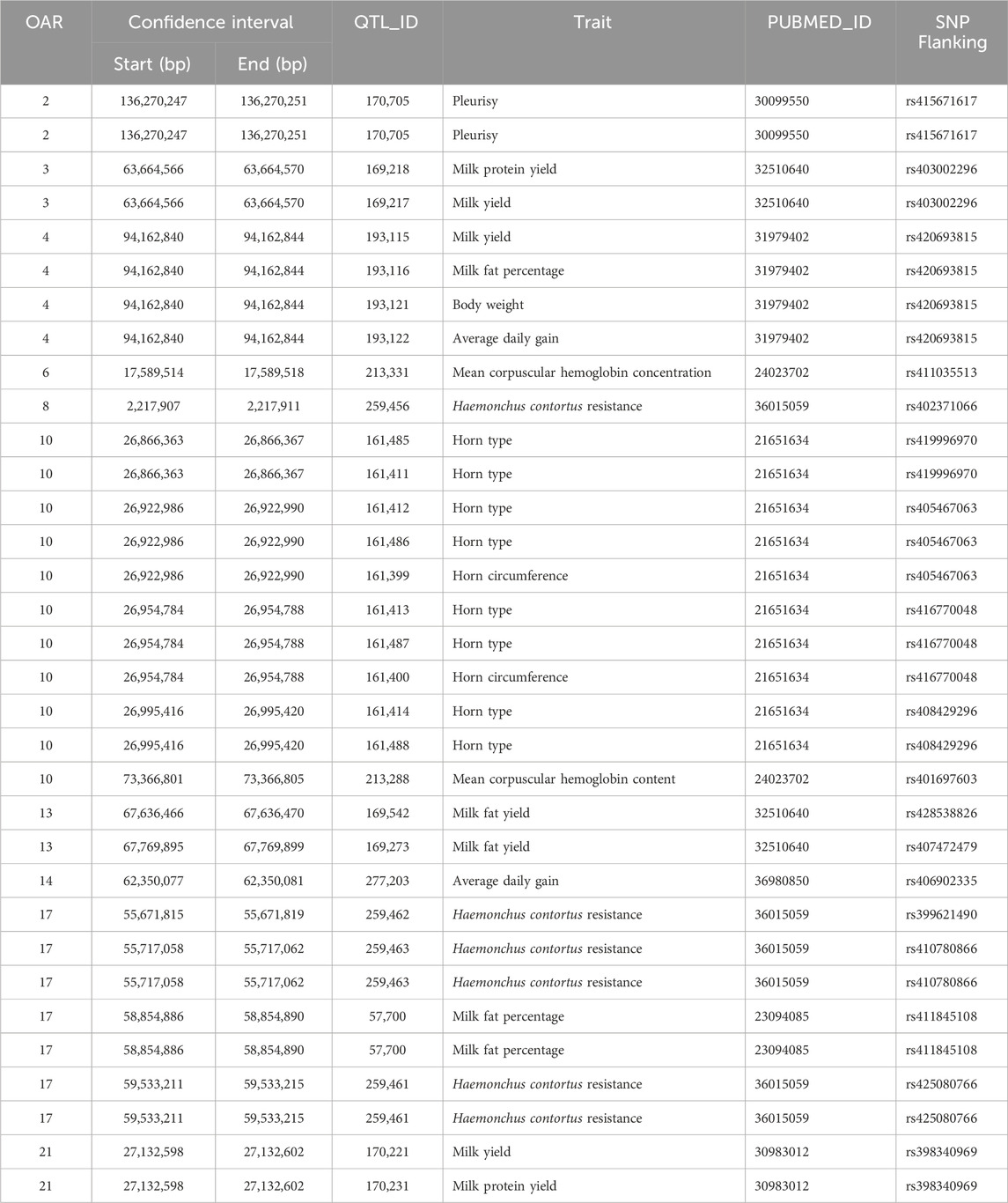
Table 10. Quantitative trait loci (QTL) previously mapped close to the SNPs associated with longevity-related traits in the U.S. Katahdin population.
4 Discussion
4.1 Association analyses
Ewe longevity has a major economic impact (Krupová et al., 2009; Wolfová et al., 2009b; 2009a), but longevity-related traits are not often directly included in sheep breeding schemes. This occurs because traits such as ALL and LPL often have low heritability estimates (Riggio et al., 2009; Milerski et al., 2018; Pelmuş et al., 2020; Medrado et al., 2021) and are expressed late in an ewe’s life. These issues allow the additional use of genomics for longevity traits to have a greater impact in breeding programs. This was the first study to perform GWASs for longevity-related traits in the U.S. Katahdin sheep. Moreover, as far as we know, only one recent previous study reported GWASs for longevity traits in sheep (Smitchger et al., 2024), which evaluated ALL, TNL, TNB, TNW, and TLW recorded in 1,130 ewes from four U.S. sheep breeds (Rambouillet, Polypay, Suffolk, and Columbia). The present study performed GWAS analyses using a breed with a larger genotypic dataset and additional traits such as LPL, TLB, and TLWadj. Moreover, the previous study analyzed data from a single production system, while in the current study, a wide array of production systems and flock locations were analyzed. A greater number of genome-wide associations (n = 25) was reported than that in the present study (Smitchger et al., 2024). Moreover, the genomic regions reported in the previous study did not overlap the positions reported in the current study. Differences between GWAS results from different studies and populations may be a consequence of the variability (genotypic and phenotypic) observed for longevity-related traits across breeds (Annett et al., 2011; Hanna et al., 2023), trait definition, statistical models used, data editing and quality control procedures, and stringency of multiple testing correction. Moreover, longevity indicators show a complex polygenic control, where each locus has a small effect on the total variability of the trait, which makes it more difficult to identify the same locus explaining variance in longevity traits across breeds and even in different datasets from the same breed.
We identified 24 markers associated with two or more traits, which is strong evidence that a high genetic correlation between these traits exists. Genetic correlations greater than 0.8 between these eight longevity traits have been observed (Pinto et al., 2025b). In a multi-breed U.S. population, Pearson correlations ranging from 0.82 to 0.97 between ALL, TNL, TNB, TNW, and TLW were reported (Smitchger et al., 2024). In Spanish Churra ewes, genetic correlations of 0.96 (ALL x LPL), 0.97 (ALL x TNL), and 0.96 (LPL x TNL) were also reported (El-Saied et al., 2005). In Dorper sheep, the genetic correlation between TNB and TNW was 0.85 (Zishiri et al., 2013). Therefore, these longevity indicators are likely to have a similar genetic architecture.
Ewe longevity depends on several factors (McLaren et al., 2020). In general, highly productive ewes have longer lifespans than low-producing ewes because this second group is voluntarily culled earlier. However, productive ewes may die, or breeders may need to cull highly productive ewes due to disease or reproductive issues, which are involuntary culling reasons. Unfortunately, the culling reasons for Katahdin ewes were not recorded within the NSIP. However, previous sheep studies have reported disease and reproductive issues as some of the main causes of involuntary culling (McLaren et al., 2020; Ridler et al., 2024). Furthermore, Katahdin breeders tend to use estimated breeding values for the number of lambs born and the number of lambs weaned to select replacement animals (Notter and Lewis, 2018), which is expected to impact ewe lifetime, as low genetic merit ewes for these traits have increased chances of being culled earlier compared to high genetic merit ewes. Therefore, it is expected that genes related to factors such as reproduction, diseases, immunity, and growth performance will be identified when GWASs are performed for ewe longevity indicators.
4.2 Reproduction-related genes
In U.S. Katahdin sheep, the number of lambs born and the number of weaned are two key selection criteria used to increase ewe reproductive performance (Notter and Lewis, 2018). Thus, high prolificacy ewes have increased chances of being retained in the flock for a longer time than less prolific ewes. Many genes have been reported as candidates for litter size in various sheep breeds (Abdoli et al., 2016) and some of these genes were found close to the SNPs associated with longevity. For instance, the SNP rs407234061 on OAR11:61,900,883 was associated with five traits (ALL, LPL, TNL, TNB, and TLB), and close to this variant is the APOH (apolipoprotein H) gene, which is involved in lipoprotein metabolism. Previous studies reported a role of this gene in both women (Miettinen et al., 2001) and cow (Minozzi et al., 2013) fertility. Furthermore, APOH has been reported as a candidate gene for prolificacy in sheep (Wang et al., 2024). The SNP rs428707491 on OAR14:62,610,160 was associated with ALL and LPL, and it is close to the NLRP9 (NLR family pyrin domain containing 9) gene, which is a member of a family that encodes intracellular proteins with critical roles in inflammatory response, early mammalian embryogenesis, and reproduction (Van Gorp et al., 2014). NLRP9 was suggested as a candidate gene for prolificacy in both sheep (Zhang et al., 2020) and pigs (Zhang Y. et al., 2023). The variants rs421227438 on OAR18:65,806,713, which are also associated with ALL, are close to the CKB (creatine kinase B) gene. A previous study reported that CKB may be associated with a better ability to defend against oxidative stress, resulting in good-quality embryos (Lee et al., 2010). Moreover, a homozygous region close to the CKB gene was associated with litter size in Hu sheep (Tao et al., 2021). The variant rs405495781 on OAR22:24,041,507 was associated with TLWadj, and close to this position was found the SH3PXD2A (SH3 And PX Domains 2A) gene, which acts as an organizer protein that facilitates NOX1-or NOX3-dependent reactive oxygen species (ROS) generation and ROS localization. SH3PXD2A was previously reported as a candidate gene for litter size in pigs (Mo et al., 2022). On OAR25:23,833,675 was found the SNP rs161572072, which was found to be associated with TLW, and close to this position is the HERC4 (HECT and RLD domain containing E3 ubiquitin protein ligase 4) gene. HERC4 was overexpressed in ovary of the hyper prolific Small Tail Han sheep compared to Dorset sheep (Miao et al., 2016), suggesting a potential role in sheep prolificacy.
Our study also found other genes that, although do not have evidence of a direct relationship with prolificacy, were reported as candidates for enhancing mammalian fertility. In several mammalian species, aging promotes a continuous decline in the ovarian follicle pool until reaching the end of the female reproductive stage (Winkler and Goncalves, 2023), a physiological process called menopause in humans. The SNP rs426160726 on OAR2:155,017,143 was found to be associated with TNL, TNB, TNW, and TLB, and close to this variant is the GALNT13 (polypeptide N-acetylgalactosaminyltransferase 13) gene, which is involved in the metabolism of proteins and was reported as a candidate gene for age at menopause in women (Lunetta et al., 2007). Another SNP, rs420711470 on OAR14:62,954,922, was associated with ALL and LPL, and close to this variant was found the TMEM150B (transmembrane protein 150B) gene. TMEM150B increases cell survival by triggering autophagy when cells are in a stressful environment (Mrschtik and Ryan, 2016). Autophagy may be a cell survival mechanism to maintain the provision of female germ cells before establishing primordial follicle pools in the ovary (Gawriluk et al., 2011). BRSK1 (BR serine/threonine kinase 1) is another gene close to the SNP rs420711470, which affects the release of the gonadotropin-releasing hormone from the hypothalamic–pituitary–ovarian axis (Qin et al., 2012). Variants in both TMEM150B and BRSK1 genes were associated with age at menopause and length of reproductive lifespan in women (He et al., 2009; Stolk et al., 2009; Chen et al., 2012). Moreover, BRSK1 was reported as a candidate gene for conceptus elongation in sheep (Brooks et al., 2015).
Female reproductive activity depends on a complex physiological process that involves multiple hormones, and some genes identified in the current study are involved with the synthesis and release of reproductive hormones. The SNP rs423900190 on OAR6:85,982,278 was associated with ALL and TNB, and close to this variant were found the genes SULT1B1 (sulfotransferase 1B1) and SULT1E1 (sulfotransferase 1E1), two sulfotransferases involved in the metabolism of endogenous compounds such as thyroid and steroid hormones, including estradiol inactivation (Yi et al., 2021), which play a key role in mammalian pregnancy (Bairagi et al., 2018). In another region of OAR6 was found the variant rs411035513 (OAR6:17,589,516), which was found to be associated with TNW, which is close to the LEF1 (lymphoid enhancer binding factor 1) gene. LEF1 was required for the formation of endometrial glands in the mouse uterus, and its expression varied cyclically with the mouse estrus cycle (Shelton et al., 2012). Moreover, LEF1 was reported as a candidate gene for fertility-related traits in Hu sheep by regulating the function of the reproductive axis (Yao et al., 2022). On OAR18:65,806,713 was found the SNP rs421227438, which was found to be associated with ALL, which is close to the EIF5 (eukaryotic translation initiation factor 5) gene. EIF5 may increase circRNAs to promote the expression of some other genes related to hormone activities such as GnRH, with a potential impact on sheep reproduction (Zhang et al., 2019) and goat puberty precocity (Liu Y. et al., 2023).
We also found some genes that may affect the early stages of pregnancy. On OAR2:122,777,052 was found the SNP rs402224636, which was found to be associated with ALL and TNL, and close to this SNP is the ITGAV (integrin subunit alpha V) gene, which encodes essential cell adhesion molecules that improve endometrial receptivity in pregnancy success (Ai et al., 2021). Moreover, ITGAV promotes endoderm differentiation (Brafman et al., 2013), which is central to embryonic development. In a different region of OAR2 is the SNP rs413377527 (OAR2:148,006,184) associated with LPL. Close to this SNP is the SLC4A10 (solute carrier family 4 member 10) gene, a member of a family of sodium/bicarbonate cotransporters, which assists in the regulation of intracellular pH with impacts on mammalian reproduction, including early embryo development (Liu et al., 2012). SLC4A10 was also reported as a candidate gene for TLW in South African Merino (Snyman et al., 2020). Therefore, SLC4A10 may also play a potential role in ewe maternal ability, increasing offspring weight, with a positive effect on ewe longevity. Close to the SNP rs411035513 (on OAR6:17,589,516), the HADH (hydroxyacyl-CoA dehydrogenase) gene was found, which may play a role in inhibiting the proliferation of mouse embryonic fibroblasts (Xia et al., 2022). On OAR13:77,378,009 was found the SNP rs416157010, which was found to be associated with TNB, and in this region is the ZNFX1 (zinc finger NFX1-type containing 1) gene, a component of the regulation of the telomerase pathway, and is required against some bacterial infections. Although its role in reproduction is unclear, ZNFX1 had increased expression in pregnant compared with non-pregnant cows (Forde et al., 2012), suggesting that genes involved in the immune response may affect uterine receptivity to implantation. On OAR14:65,830,741 was found the SNP rs399288440, which was found to be associated with ALL, TNL, TNB, and TNW, and close to this SNP, another zinc finger gene, ZSCAN4 (zinc finger and SCAN domain containing 4), was found. ZSCAN4 is a member of the pre-implantation embryo pathway, which is essential for the early embryonic development of bovine (Liu B. et al., 2023) and swine (Zhang et al., 2022). On OAR14:62,610,160 was found the SNP rs428707491, which was found to be associated with ALL and LPL, and close to this region was found the EPN1 (Epsin 1) gene, which is a component of the membrane trafficking pathway and was found to be differentially expressed in uterine epithelial tissue during the peri-implantation period of pregnancy in sheep (Brooks et al., 2016). On OAR17:57,981,108 was found the SNP rs427138541, which was associated with LPL. Close to this SNP is the FBXW8 (F-box and WD repeat domain containing 8) gene, which plays an important role in the mid-to-late stage of placenta development in mice (Kinterová et al., 2022), and it has been suggested as a candidate gene for goat fertility (Sun et al., 2022). Also close to the SNP rs427138541 is the NOS1 (nitric oxide synthase 1) gene, a member of the enzyme family that synthesizes nitric oxide; this gene plays several roles in animal reproduction, including spermatogenesis, follicle development, steroidogenesis, and ovulation (Zhang W. et al., 2023). Moreover, NOS1 was identified as a candidate gene for heat tolerance in sheep (Li et al., 2019), and heat stress can reduce ewe reproductive performance (Van Wettere et al., 2021). On OAR21:27,230,215 was found the SNP rs408858996, which was found to be associated with TNL, and in this region is the ST3GAL4 (ST3 beta-galactoside alpha-2,3-sialyltransferase 4) gene, which had lower expression in the cervical mucus of Suffolk ewes compared with high-fertility sheep breeds (Abril-Parreño et al., 2022), suggesting a potential effect on ewe fertility. On OAR22:35,912,532 was found the SNP rs405740728, which was found to be associated with TLB, and close to this region is the GFRA1 (GDNF family receptor alpha 1) gene, a receptor of glial cell-line-derived neurotrophic factor, which promotes primordial follicle development and mediates autocrine and paracrine cell–cell interactions required during folliculogenesis (Dole et al., 2008). GFRA1 has been reported as an important gene in bovine oocyte maturation and early embryo development (Kamalludin et al., 2018; Wang et al., 2018). The current study also identified 52 lncRNA-encoding genes on different chromosomes. A differential expression study in the ovaries of low- and high-fecundity Hanper sheep identified the lncRNA Xist (loc101112291) and Gtl2 (loc101123329) genes to be highly expressed, which suggests an effect on the regulation of follicular development by mediating methylation processes (Liu et al., 2021). Moreover, lncRNAs may be a key regulator of the functions in sheep oviductal tissue (Chen et al., 2024).
Some genes that can indirectly affect ewe reproductive performance were also found. For instance, on OAR1:188,636,771 was found SNP rs161707914, which was found to be associated with TNB, and close to this SNP is the ADCY5 (adenylate cyclase 5) gene, which encodes an enzyme that catalyzes the conversion of ATP to cAMP, and plays crucial roles in normal biological functions (e.g., lipolysis, gluconeogenesis, and respiration) and pathophysiological states (e.g., diabetes and obesity) (Vatner et al., 2013). ADCY5 has been suggested as a candidate gene for longevity in mammalian species (Yan et al., 2007; Cash et al., 2014) and for fertility-related traits in multiple livestock species, including sheep (Martinez-Royo et al., 2017; Du et al., 2022), cattle (Li et al., 2021), pigs (Sun et al., 2023), and ducks (Bello et al., 2021). On OAR4:65,051,183 was found the SNP rs424868378, which was found to be associated with TLB, and close to this SNP is the BBS9 (Bardet–Biedl syndrome 9) gene, a member of a family of genes that cause Bardet–Biedl syndrome, which is characterized by various hypogonadism or genitourinary abnormalities such as a malformed uterus, hydrometrocolpos, and vaginal atresia, resulting in irregular menstrual cycle and polycystic ovaries (Melluso et al., 2023). In pigs, a recessive lethal deletion in BBS9 was assumed to cause fetal lethality in mutant homozygotes, which has antagonistic pleiotropic effects on fertility and growth (Derks and Steensma, 2021).
4.3 Health-related genes
Diseases are one of the main causes of involuntary culling in many ruminant species, including dairy and beef cattle (Aleri et al., 2021; Souza et al., 2023), goats (Malher et al., 2001; Davachi, 2019), and sheep (McLaren et al., 2020). Haemonchosis is a critical infectious disease in sheep, which is caused by the gastrointestinal nematode H. contortus (Flay et al., 2022). We found significant SNPs on OAR8:2,269,243 (rs409333251) and OAR17 (rs405696543—OAR17:59,616,349, rs419277881—OAR17:55,794,456, and rs424778250—OAR17:55,644,362) close to regions where QTLs were reported for H. contortus resistance (Niciura et al., 2022). Moreover, we also found SNPs on OAR6:17,589,516 (rs411035513) and OAR10:73,335,461 (rs419109423), where QTLs for mean corpuscular hemoglobin concentration/content (MCHC) were reported (Gonzalez et al., 2013). MCHC is an indicator of anemia and can be used to identify sheep that are more resistant to gastrointestinal nematodes (Jiménez-Penago et al., 2021). Our study found an association between the intron variant rs411035513 in HADH (hydroxyacyl-CoA dehydrogenase) and TNW, while an association of this same SNP with MCHC was also reported (Gonzalez et al., 2013), suggesting a possible pleiotropic QTL in this region of OAR6. We found a variant (rs405740728) on OAR22:35,912,532 associated with TLB, and close to this position is the ATRNL1 (attractin like 1) gene. Variation in ATRNL1 was associated with fecal egg count in Akkaraman sheep (Arzik et al., 2022), which is an indicator of parasite resistance, but there is no QTL reported for fecal egg count in this region of OAR22 in the Sheep QTLdb (www.animalgenome.org/cgi-bin/QTLdb/OA/index). Moreover, long non-coding RNA genes may also be associated with response against parasite infections in sheep (Chitneedi et al., 2021), and the present study found 52 lncRNA genes.
Other chromosomal regions identified in the current study are not close to the previous health QTLs reported in Sheep QTLdb (www.animalgenome.org/cgi-bin/QTLdb/OA/index), but they may also be related to physiological responses to health issues. On OAR4:94,129,301 is the SNP rs405855191, which was associated with TLWadj, and close to this position was found the LEP (leptin) gene. This gene regulates the stem cell numbers and functions in hematopoiesis under homeostasis or stress-associated situations and pathological conditions (Trinh and Broxmeyer, 2015), and continuous blood cell production is a key response against anemia caused by H. contortus infection. On OAR14:62,954,922 was found the SNP rs420711470, which was found to be associated with ALL and LPL, and close to this SNP is the IL11 (interleukin 11) gene, another gene with hematopoietic effects and potential therapeutic value in anemia conditions (Du and Williams, 1997). Moreover, the interleukin-11 signaling pathway plays a key role in maintaining adequate blood volume levels, which is especially important after sheep are infected by H. contortus (Al Kalaldeh et al., 2019). Another SNP in OAR14 is rs428707491 (OAR14:62,610,160), which was also associated with ALL and LPL, and close to this SNP is the NLRP9 (on OAR14) gene. This gene initiates cytosolic multiprotein complexes of the innate immune system to activate an inflammatory response to intestinal infection and could also impact responses to gastrointestinal nematode infections (Mullins and Chen, 2021). The intron variant rs407234061 in the PRKCA gene (OAR11:61,900,883) was associated with five longevity traits, while on OAR14:62,375,192 was found the SNP rs411860439, which was found to be associated with ALL, and close to this SNP is the PRKCG (protein kinase C gamma) gene. These are two gene members of the large family of protein kinase C, which play key roles in hematopoietic and immune responses (Altman and Kong, 2016). In sheep, PRKCA was found as a differentially expressed gene during subclinical paratuberculosis infection (Purdie et al., 2019).
On OAR18:39,616,062 was found the SNP rs427018792, which was found to be associated with TLW and TLWadj, and close to this SNP is the HECTD1 (HECT domain E3 ubiquitin protein ligase 1) gene. HECTD1 is involved in multiple pathways such as adaptive immune system, antigen processing, ubiquitination and proteasome degradation, class I MHC-mediated antigen processing and presentation, and immune system pathways. Moreover, HECTD1 was reported as a candidate gene for somatic cell count in Frizarta dairy sheep (Kominakis et al., 2019). On OAR23:57,717,942 is the SNP rs411106195, an intron variant in NEDD4L (NEDD4-like E3 ubiquitin protein ligase) gene, which was associated with LPL. NEDD4L is a component of several pathways related to immunity such as infectious disease and innate immune system pathways. Moreover, NEDD4L was suggested as a candidate gene for gastrointestinal nematode infection-related traits in cattle (Wolf et al., 2021). On OAR23:57,231,444 is the SNP rs400623485, which was associated with TNL. Close to this SNP is the FECH (ferrochelatase) gene, which encodes an enzyme that catalyzes the terminal step of the heme biosynthesis pathway, which is essential for many hemoproteins, including hemoglobin (Medlock and Dailey, 2022). In humans, FECH variants were associated with mean corpuscular hemoglobin volume and concentration, red blood cell distribution width, and mean corpuscular volume (Sakaue et al., 2021), i.e., blood traits that may play a role against H. contortus infection.
On OAR25:22,652,820 was found the SNP rs400951839, which was associated with TLW and TLWadj, and close to this SNP are the CTNNA3 (Catenin Alpha 3) and LRRTM3 (leucine-rich repeat transmembrane neuronal 3) genes, which have been associated with several brain disorders in humans according to the GeneCards database. A recent study reported the region of OAR25 close to the genes CTNNA3 and LRRTM3 as being prone to escape reprogramming and a potential candidate for transgenerational epigenetic inheritance, suggesting a potential genetic overlap between brain and infertility disorders (Braz et al., 2024). Our study also found SNPs close to various genes that encode zinc-finger proteins (ZNF genes), which play a known role in the development and differentiation of several tissues and are involved in the onset of several diseases (Cassandri et al., 2017).
4.4 Growth- and milk-related genes
Increasing the growth rate is a major selection objective in meat sheep breeds, and lamb growth depends on direct and maternal genetic components (Pech et al., 2012). Growth-related selection criteria such as birth and weaning weights are currently used in U.S. Katahdin sheep (Notter and Lewis, 2018). Thus, it is expected that ewes with low-EBV for these traits will likely be culled earlier than ewes with higher genetic merit. Our study found genomic regions close to QTL reported for growth- and milk-related traits, which may impact Katahdin lamb growth and have a consequent impact on ewe longevity. Functional annotation analyses enabled the identification of candidate genes in these regions.
The intron variant rs400216417 on OAR6:32,071,830 was associated with LPL. This variant encodes the GRID2 (glutamate ionotropic receptor delta type subunit 2) gene, a component of intracellular calcium signaling, which plays a role in calcium homeostasis. Calcium is required for animal growth and milk production (Ni et al., 2024). The intron variant rs418797399 on OAR17:58,814,585 was associated with ALL and TNW. This variant encodes the MED13L (mediator complex subunit 13L) gene, a component of metabolic pathways, and was reported as a candidate gene for growth-related traits in cattle (Ma et al., 2023). The variant rs405855191 on OAR4:94,129,301 was associated with TLWadj, and close to this position was found the LEP gene. LEP is associated with energy homeostasis, neuroendocrine and immune functions, and the metabolism of glucose, lipids, and bone (Park and Ahima, 2015). In sheep, LEP was reported as a candidate gene associated with many traits, including carcass-related traits (Meira et al., 2018), body fat reserves (Macé et al., 2022), morphology, and body weight (Shojaei et al., 2010; Machado et al., 2020; Darwish et al., 2023). Leptin is also involved in skeletal development as it increases the abundance of the insulin-like growth factor 1 receptor (IGF1R) within the chondrocyte and progenitor cell populations (Reid et al., 2018). This receptor has been reported as a candidate gene for both sheep growth (Proskura et al., 2014) and longevity (Byun et al., 2012). Therefore, LEP gene may have an indirect effect on longevity by increasing the IGF1R level. On OAR21:27,230,215 was found the variant rs408858996, which was associated with TNL, and close to this variant is the DCPS (decapping enzyme, scavenger) gene, which has been suggested as a candidate gene for milk and protein yields in both sheep (Sutera et al., 2019) and cattle (Suchocki et al., 2016); therefore, DCPS may have a potential effect on animal growth by affecting milk-related traits.
4.5 Carcass-related genes
The improvement of carcass traits is also the target of selective schemes in meat sheep breeding programs, and at least two genes identified in the current study were also reported to be associated with carcass traits in sheep. On OAR7:10,694,207 and OAR7:10,695,388 were found two missense variants, rs160278050 and rs160278082, respectively, in exon 2 of the CMYA5 gene, which encodes myospryn, a protein expressed predominantly in skeletal and cardiac muscles (Benson et al., 2017). Myospryn is an inhibitor of the calcineurin signaling pathway in skeletal muscle, playing an important regulatory function in muscle differentiation, fiber-type determination, hypertrophy, and muscle regeneration (Kielbasa et al., 2011). Some previous studies reported CMYA5 as a candidate gene for carcass- and meat quality-related traits in cattle (Bruscadin et al., 2021) and pigs (Xu et al., 2011). On OAR18:62,114,682 is the SNP s37838.1, an intron variant in the SETD3 (SET domain containing 3 actin N3 (tau)-histidine methyltransferase) gene. SETD3 is associated with multiple other genes, regulating various biological processes such as cell cycle and apoptosis (Witecka et al., 2021). In sheep, SETD3 was suggested as a candidate gene for carcass-related traits (Souza et al., 2022).
4.6 Metabolic pathways
Our study also identified more protein–protein interactions than expected (Figure 4). Protein–protein interactions mediate essentially all biological processes, especially in the disease context (Lage, 2014). These interactions indicate that many of the candidate genes found in the current study may be acting together on phenotype expression. For instance, the genes PRKCG and PRKCA are components of multiple pathways, such as GnRH secretion and gastric acid secretion, while PRKCA and PLCD4 are components of the thyroid hormone and calcium signaling pathways. Physiological mechanisms associated with longevity have been more extensively studied in species such as humans and rodents since livestock animals are usually culled before reaching old age. Several genes (HACD2, ATP6V1C1, ST3GAL4, LOC101119640, ADCY5, ALDH1L1, CKB, LOC101120834, LOC101121036, LOC101113375, LOC105608607, FECH, HADH, KYAT3, KMT5C, NOS1, PPAT, PAICS, GALNT13, SGMS2, LOC101120331, TMEM86B, and UROC1) found in the current study are components of metabolic pathways, which are known to play a key function in feed intake (Jorge-Smeding et al., 2021). These genes can be involved in maintaining a good nutritional status of mammalians, especially based on low-calorie diets, which has been suggested as an efficient mechanism for attenuating aging (Flanagan et al., 2020), because excessive energy intake and adiposity can cause systemic inflammation (Kökten et al., 2021). Moreover, some signaling pathways associated with longevity were also found. For instance, the thyroid hormone signaling pathway may modulate lifespan in mammals as thyroid hormones are involved with activating, inhibiting, or modulating the gene expression of key regulators of metabolism, growth, and inflammation (Bowers et al., 2013). The AMPK signaling pathway may also be related to aging as AMPK activation extends the lifespan in both Caenorhabditis elegans and rodents (Yu et al., 2021), which may be a consequence of AMPK roles in the regulation of cellular homeostasis, resistance to stress, cell survival and growth, cell death, and autophagy. MAPK signaling is another essential pathway for many biological processes (e.g., proliferation, differentiation, apoptosis, and stress response), and MAPK enzymes may be upregulated with aging (Cano et al., 2019). Finally, three genes (ADCY5, PRKAB1, and RPTOR) found in the current study are components of the longevity-regulating pathway, indicating that the genomic regions close to the positions OAR1:188,636,771, OAR17:55,794,456, and OAR11:51,584,156 may be associated with ewe longevity.
The current study also found seven genes that are components of the oxytocin signaling pathway, which were enriched in our analyses. The oxytocin hormone plays a key role during the peripartum period by stimulating physiological events (e.g., myometrial contractions and prostaglandin production), which makes the exogenous application of oxytocin a veterinary practice often used to induce parturition (Marcet-Rius et al., 2023). Oxytocin also has functions in sheep lactation, such as stimulating milk ejection, sustaining mammary cells, and increasing lactation persistency (Nezamidoust et al., 2015). Oxytocin is considered an essential component to stimulate maternal behaviors, which include nest-building, reluctance to leave the nest, genital and overall licking of the newborn, nursing, and direct contact with the litter (Mota-Rojas et al., 2023). Assuming that lambing issues and reduced maternal behavior may be influencing culling of ewes, our results suggest that the oxytocin signaling pathway may be involved with Katahdin ewe longevity.
Our study found enriched pathways related to the cardiovascular system. Cardiovascular diseases are the leading cause of death in humans aged 65 years or older (North and Sinclair, 2012), indicating a relationship between the deterioration of cardiac function and aging. The health of the cardiovascular system contributes to delivering oxygenated blood to other body tissues, improving their functions and resulting in a positive impact on mammalian longevity. Therefore, cardiovascular aging and longevity share common pathophysiological mechanisms, and delaying cardiovascular aging increases the likelihood of greater longevity (Pietri and Stefanadis, 2021). Many ewes are culled at a young age as the selection process requires that the generation interval be as short as possible. Therefore, heart diseases are not expected for young and fertile ewes, as in the current Katahdin population. However, Katahdin ewes are selected for increased fertility, and this selection may have some impact on the cardiovascular system as estrogen modulates multiple cardiovascular functions (Murphy, 2011). We did not find evidence of this possible positive or negative pleiotropic effect between reproductive performance and cardiovascular health in livestock. However, other traits such as residual feed intake seem to have some relationship with cardiac function (Munro et al., 2019). Moreover, gene expression in cardiac muscle from yaks and cattle suggests an involvement of cardiac function with high-altitude adaptation (Wang et al., 2021). Therefore, future studies should investigate whether ewe reproductive performance is associated with cardiovascular function, which would explain a possible effect on Katahdin ewe longevity.
4.7 Final considerations
We found genomic regions harboring several genes that may play key roles in growth, reproductive performance, health, milk production, and carcass quality in U.S. Katahdin sheep; these factors will directly impact breeders’ decision to retain ewes in the flock, thereby affecting ewe longevity. Ewe longevity differs from their life expectancy because ewes in commercial flocks are often culled prematurely due to many voluntary and involuntary reasons before their natural life expectancy. For instance, the current Katahdin ewe population had an average ALL of 1,102 days, i.e., on average, they were culled approximately 3 years old. Even healthy and fertile ewes rarely reach more than 7 years of productive life (McLaren et al., 2020; Hanna et al., 2023). This occurs because breeders annually replace some ewes with ewe-lambs to improve the genetic merit of their flocks. This practice contributes to reducing the generation interval on the female side and improves annual genetic gain. Therefore, the candidate genes identified for ewe productive longevity related to other important traits such as fertility and reproduction are unsurprising as these traits directly impact ewes’ ability to remain in a flock.
Longevity is still a poorly studied topic in sheep, with few studies reporting candidate genes directly associated with longevity traits. FOXO3 (forkhead box O3) (Byun et al., 2011) and IGF1R (IGF1 receptor) (Byun et al., 2012) were two genes reported for sheep longevity, which were identified in candidate gene studies, while only one GWAS was reported for ewe longevity (Smitchger et al., 2024). Therefore, the current study provides more information by revealing novel genomic regions and candidate genes for longevity-related traits in sheep. Our genomic regions do not overlap with those previously reported for ewe longevity in other U.S. sheep breeds (Smitchger et al., 2024), which may be a consequence of the low-density SNP map used in both studies. Thus, we recommend that future studies use higher-density maps. Another hypothesis is the small polygenic effect and large environmental effects as longevity-related traits often show low heritability estimates (El-Saied et al., 2005; Borg et al., 2009; Zishiri et al., 2013; Lee et al., 2015). In a previous study with the current Katahdin population, we estimated heritability between 0.07 and 0.15 for these longevity indicators (Pinto et al., 2025b). It must be noted that the heritability estimates for longevity indicators may be moderate as culling reasons are considered in the analyses (Mekkawy et al., 2009). This could also improve GWAS analyses by identifying candidate genes most closely related to specific culling reasons. Therefore, NSIP and Katahdin breeders are encouraged to record the culling reasons to allow future genetic studies to separate voluntary or involuntary culling to improve the precision of association analyses.
5 Conclusion
We performed the first GWAS analyses for ewe longevity-related traits in U.S. Katahdin sheep and identified novel candidate genes and pathways associated with ewe longevity. Our results suggested that ewe longevity is under a complex polygenic control where candidate genes may be involved with prolificacy (RORA, APOH, NLRP9, CKB, and HERC4), a decline of the ovarian follicle pool (GALNT13, TMEM150B, and BRSK1), synthesis and release of some reproductive hormones (SULT1B1, LEF1, and EIF5), early pregnancy events (ITGAV, HADH, ZNFX1, ZSCAN4, EPN1, FBXW8, NOS1, ST3GAL4, GFRA1, and multiple lncRNAs), disease or syndromes affecting reproduction (ADCY5 and BBS9), response to stress or pathological conditions (ADCY5, HADH, ATRNL1, LEP, IL11, NLRP9, PRKCG, PRKCA, NEDD4L, FECH, CTNNA3, LRRTM3, and zinc-finger proteins), growth performance (GRID2, MED13L, DCPS, and LEP), and carcass traits (CMYA5 and SETD3). Moreover, our study found enriched pathways that may be influencing ewe longevity, such as oxytocin signaling and cardiac pathways. Taken together, this information suggests that Katahdin ewe longevity depends on a complex combination of voluntary and involuntary culling reasons, along with their underlying biological mechanisms. Identifying genes that are associated with early culling is an important step toward improving ewe longevity and enhancing the economic prosperity and sustainability of the U.S. sheep industry.
Data availability statement
The datasets presented in this article are not readily available because they are the property of the U.S. sheep producers, and the information in these datasets is commercially sensitive. Requests to access the datasets should be directed to the corresponding author.
Ethics statement
Ethical approval was not required for the studies involving animals in accordance with the local legislation and institutional requirements because approval from the Animal Use and Ethics Committee was not needed for this study as pre-existing datasets were provided by the National Sheep Improvement Program (NSIP) and recorded by U.S. Katahdin producers during routine husbandry activities. Written informed consent was not obtained from the owners for the participation of their animals in this study because Approval from the Animal Use and Ethics Committee was not needed for this study as pre-existing datasets were provided by the National Sheep Improvement Program (NSIP) and recorded by U.S. Katahdin producers during routine husbandry activities.
Author contributions
LP: Conceptualization, Formal analysis, Methodology, Writing – original draft, Writing – review and editing. RL: Data curation, Funding acquisition, Resources, Writing – review and editing. AR: Writing – review and editing. BF: Resources, Writing – review and editing. TM: Resources, Writing – review and editing. CW: Resources, Writing – review and editing. SN: Resources, Writing – review and editing. JB: Resources, Writing – review and editing. LB: Conceptualization, Supervision, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Organic Agriculture Research and Extension Initiative (grant no. 2016-51300-25723/project accession no. 1010329) and the Agriculture and Food Research Initiative Competitive Grant (grant no. 2022-67015-36073/project accession no. 1027785) from the USDA National Institute of Food and Agriculture. Luis F. B. Pinto was supported by the CAPES-Print program (grant no: 88887.834214/2023-00).
Acknowledgments
The authors thank the National Sheep Improvement Program, and its member Katahdin sheep producers, for their contributions to this research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2025.1600587/full#supplementary-material
References
Abdoli, R., Zamani, P., Mirhoseini, S. Z., Ghavi Hossein-Zadeh, N., and Nadri, S. (2016). A review on prolificacy genes in sheep. Reprod. Domest. Anim. 51, 631–637. doi:10.1111/rda.12733
Abril-Parreño, L., Morgan, J., Krogenæs, A., Druart, X., Cormican, P., Gallagher, M. E., et al. (2022). Biochemical and molecular characterization of sialylated cervical mucins in sheep. Biol. Reprod. 107, 419–431. doi:10.1093/biolre/ioac077
Aguilar, I., Legarra, A., Cardoso, F., Masuda, Y., Lourenco, D., and Misztal, I. (2019). Frequentist p-values for large-scale-single step genome-wide association, with an application to birth weight in American Angus cattle. Genet. Sel. Evol. 51, 28. doi:10.1186/s12711-019-0469-3
Ai, J., Ebrahimi-Barough, S., Nouri, M., Ziadi, M., Pashaiefar, H., Ahmadvand, M., et al. (2021). Influence of follicular fluid and seminal plasma on the expression of endometrial receptivity genes in endometrial cells. Cell J. 22, 457–466. doi:10.22074/cellj.2021.6851
Aleri, J. W., Lyons, A., Laurence, M., Coiacetto, F., Fisher, A. D., Stevenson, M. A., et al. (2021). A descriptive retrospective study on mortality and involuntary culling in beef and dairy cattle production systems of Western Australia (1981–2018). Aust. Vet. J. 99, 395–401. doi:10.1111/avj.13096
Al Kalaldeh, M., Gibson, J., Lee, S. H., Gondro, C., and Van Der Werf, J. H. J. (2019). Detection of genomic regions underlying resistance to gastrointestinal parasites in Australian sheep. Genet. Sel. Evol. 51, 37. doi:10.1186/s12711-019-0479-1
Altman, A., and Kong, K. F. (2016). Protein kinase C enzymes in the hematopoietic and immune systems. Annu. Rev. Immunol. 34, 511–538. doi:10.1146/annurev-immunol-041015-055347
Annett, R. W., Carson, A. F., Dawson, L. E. R., Irwin, D., Gordon, A. W., and Kilpatrick, D. J. (2011). Comparison of the longevity and lifetime performance of Scottish Blackface ewes and their crosses within hill sheep flocks. Animal 5, 347–355. doi:10.1017/S1751731110002107
Arzik, Y., Kizilaslan, M., White, S. N., Piel, L. M. W., and Çınar, M. U. (2022). Genomic analysis of gastrointestinal parasite resistance in Akkaraman sheep. Genes 13, 2177. doi:10.3390/genes13122177
Bairagi, S., Grazul-Bilska, A. T., Borowicz, P. P., Reyaz, A., Valkov, V., and Reynolds, L. P. (2018). Placental development during early pregnancy in sheep: progesterone and estrogen receptor protein expression. Theriogenology 114, 273–284. doi:10.1016/j.theriogenology.2018.04.002
Becker, G. M., Burke, J. M., Lewis, R. M., Miller, J. E., Morgan, J. L. M., Rosen, B. D., et al. (2022). Variants within genes EDIL3 and ADGRB3 are associated with divergent fecal egg counts in Katahdin sheep at weaning. Front. Genet. 13, 817319. doi:10.3389/fgene.2022.817319
Becker, G. M., Davenport, K. M., Burke, J. M., Lewis, R. M., Miller, J. E., Morgan, J. L. M., et al. (2020). Genome-wide association study to identify genetic loci associated with gastrointestinal nematode resistance in Katahdin sheep. Anim. Genet. 51, 330–335. doi:10.1111/age.12895
Bello, S. F., Xu, H., Guo, L., Li, K., Zheng, M., Xu, Y., et al. (2021). Hypothalamic and ovarian transcriptome profiling reveals potential candidate genes in low and high egg production of white Muscovy ducks (Cairina moschata). Poult. Sci. 100, 101310. doi:10.1016/j.psj.2021.101310
Benson, M. A., Tinsley, C. L., Waite, A. J., Carlisle, F. A., Sweet, S. M. M., Ehler, E., et al. (2017). Ryanodine receptors are part of the myospryn complex in cardiac muscle. Sci. Rep. 7, 6312. doi:10.1038/s41598-017-06395-6
Borg, R. C., Notter, D. R., and Kott, R. W. (2009). Genetic analysis of Ewe stayability and its association with lamb growth and adult production. J. Anim. Sci. 87, 3515–3524. doi:10.2527/jas.2008-1623
Bowers, J., Terrien, J., Clerget-Froidevaux, M. S., Gothie, J. D., Rozing, M. P., Westendorp, R. G. J., et al. (2013). Thyroid hormone signaling and homeostasis during aging. Endocr. Rev. 34, 556–589. doi:10.1210/er.2012-1056
Brafman, D. A., Phung, C., Kumar, N., and Willert, K. (2013). Regulation of endodermal differentiation of human embryonic stem cells through integrin-ECM interactions. Cell Death Differ. 20, 369–381. doi:10.1038/cdd.2012.138
Braz, C. U., Passamonti, M. M., and Khatib, H. (2024). Characterization of genomic regions escaping epigenetic reprogramming in sheep. Environ. Epigenet 10, dvad010–12. doi:10.1093/eep/dvad010
Brooks, K., Burns, G. W., Moraes, J. G. N., and Spencer, T. E. (2016). Analysis of the uterine epithelial and conceptus transcriptome and luminal fluid proteome during the peri-implantation period of pregnancy in sheep. Biol. Reprod. 95, 88. doi:10.1095/biolreprod.116.141945
Brooks, K. E., Burns, G. W., and Spencer, T. E. (2015). Peroxisome proliferator activator receptor gamma (PPARG) regulates conceptus elongation in sheep. Biol. Reprod. 92, 42. doi:10.1095/biolreprod.114.123877
Bruscadin, J. J., de Souza, M. M., de Oliveira, K. S., Rocha, M. I. P., Afonso, J., Cardoso, T. F., et al. (2021). Muscle allele-specific expression QTLs may affect meat quality traits in Bos indicus. Sci. Rep. 11, 7321. doi:10.1038/s41598-021-86782-2
Byun, S. O., Forrest, R. H., Frampton, C. M., Zhou, H., and Hickford, J. G. H. (2012). An association between lifespan and variation in insulin-like growth factor I receptor in sheep. J. Anim. Sci. 90, 2484–2487. doi:10.2527/jas.2011-4148
Byun, S. O. K., Zhou, H., and Hickford, J. G. H. (2011). Characterization of genetic variation in the forkhead box class O3 gene (FOXO3) in sheep. DNA Cell Biol. 30, 449–452. doi:10.1089/dna.2010.1193
Cano, M., Guerrero-Castilla, A., Nabavi, S. M., Ayala, A., and Argüelles, S. (2019). Targeting pro-senescence mitogen activated protein kinase (Mapk) enzymes with bioactive natural compounds. Food Chem. Toxicol. 131, 110544. doi:10.1016/j.fct.2019.05.052
Cash, T. P., Pita, G., Domínguez, O., Alonso, M. R., Moreno, L. T., Borrás, C., et al. (2014). Exome sequencing of three cases of familial exceptional longevity. Aging Cell 13, 1087–1090. doi:10.1111/acel.12261
Cassandri, M., Smirnov, A., Novelli, F., Pitolli, C., Agostini, M., Malewicz, M., et al. (2017). Zinc-finger proteins in health and disease. Cell Death Discov. 3, 17071. doi:10.1038/cddiscovery.2017.71
Chen, C. T. L., Fernández-Rhodes, L., Brzyski, R. G., Carlson, C. S., Chen, Z., Heiss, G., et al. (2012). Replication of loci influencing ages at menarche and menopause in Hispanic women: the women’s health initiative SHARe study. Hum. Mol. Genet. 21, 1419–1432. doi:10.1093/hmg/ddr570
Chen, W., Li, Z., Zhong, R., Sun, W., and Chu, M. (2024). Expression profiles of oviductal mRNAs and lncRNAs in the follicular phase and luteal phase of sheep (Ovis aries) with 2 fecundity gene (FecB) genotypes. Genes Genomes Genet. 14, jkad270. doi:10.1093/g3journal/jkad270
Chitneedi, P. K., Weikard, R., Arranz, J. J., Martínez-Valladares, M., Kuehn, C., and Gutiérrez-Gil, B. (2021). Identification of regulatory functions of LncRNAs associated with T. circumcincta infection in adult sheep. Front. Genet. 12, 685341. doi:10.3389/fgene.2021.685341
Darwish, A. M., Abdelhafez, M. A., Abdel-Hamid, Z. G., Othman, S. I., Mohamed, I. E., and Allam, A. A. (2023). Correlation analysis between polymorphism of leptin and IGFI genes and body measurements in Barki and Farafra sheep. Beni Suef Univ. J. Basic Appl. Sci. 12, 119. doi:10.1186/s43088-023-00450-0
Davachi, D., Vatandoost, M., Dirandeh, E., and Dadashpour Davachi, N. (2019). Characterization and pattern of culling in goats. Arch. Razi Inst. 74, 441–446. doi:10.22092/ari.2019.125298.1301
Derks, M. F. L., and Steensma, M. (2021). Review: balancing selection for deleterious alleles in livestock. Front. Genet. 12, 761728. doi:10.3389/fgene.2021.761728
Dole, G., Nilsson, E. E., and Skinner, M. K. (2008). Glial-derived neurotrophic factor promotes ovarian primordial follicle development and cell-cell interactions during folliculogenesis. Reprod 135, 671–682. doi:10.1530/REP-07-0405
Du, X., He, X., Liu, Q., Di, R., Liu, Q., and Chu, M. (2022). Comparative transcriptomics reveals the key lncRNA and mRNA of Sunite sheep adrenal gland affecting seasonal reproduction. Front. Vet. Sci. 9, 816241. doi:10.3389/fvets.2022.816241
Du, X., and Williams, D. A. (1997). Interleukin-11: review of molecular, cell biology, and clinical use. Blood 45, 3897–3908. doi:10.1182/blood.v89.11.3897
Ducrocq, V., Quaas, R. L., Pollak, E. J., and Casella, G. (1988). Length of productive life of dairy cows. 1. Justification of a Weibull model. J. Dairy Sci. 71, 3061–3070. doi:10.3168/jds.S0022-0302(88)79906-3
Duguma, G., Schoeman, S. J., Cloete, S. W. P., and Jordaan, G. F. (2002). Genetic and environmental parameters for Ewe productivity in Merinos. S Afr. J. Anim. Sci. 32, 154–159. doi:10.4314/sajas.v32i3.3740
El-Saied, U. M., De La Fuente, L. F., Carriedo, J. A., and San Primitivo, F. (2005). Genetic and phenotypic parameter estimates of total and partial lifetime traits for dairy ewes. J. Dairy Sci. 88, 3265–3272. doi:10.3168/jds.S0022-0302(05)73009-5
Farrell, L. J., Tozer, P. R., Kenyon, P. R., Ramilan, T., and Cranston, L. M. (2019). The effect of Ewe wastage in New Zealand sheep and beef farms on flock productivity and farm profitability. Agric. Syst. 174, 125–132. doi:10.1016/j.agsy.2019.04.013
Flanagan, E. W., Most, J., Mey, J. T., and Redman, L. M. (2020). Calorie restriction and aging in humans. Annu. Rev. Nutr. 40, 105–133. doi:10.1146/annurev-nutr-122319-034601
Flay, K. J., Hill, F. I., and Muguiro, D. H. (2022). A review: Haemonchus contortus infection in pasture-based sheep production systems, with a focus on the pathogenesis of anaemia and changes in haematological parameters. Animals 12, 1238. doi:10.3390/ani12101238
Flay, K. J., Ridler, A. L., Compton, C. W. R., and Kenyon, P. R. (2021). Ewe wastage in New Zealand commercial flocks: extent, timing, association with hogget reproductive outcomes and BCS. Animals 11, 779. doi:10.3390/ani11030779
Fonseca, P. A. S., Suárez-Vega, A., Marras, G., and Cánovas, Á. (2020). GALLO: an R package for genomic annotation and integration of multiple data sources in livestock for positional candidate loci. Gigascience 9, giaa149. doi:10.1093/gigascience/giaa149
Forde, N., Duffy, G. B., McGettigan, P. A., Browne, J. A., Mehta, J. P., Kelly, A. K., et al. (2012). Evidence for an early endometrial response to pregnancy in cattle: both dependent upon and independent of interferon tau. Physiol. Genomics 44, 799–810. doi:10.1152/physiolgenomics.00067.2012
Gawriluk, T. R., Hale, A. N., Flaws, J. A., Dillon, C. P., Green, D. R., and Rucker, E. B. (2011). Autophagy is a cell survival program for female germ cells in the murine ovary. Reprod 141, 759–765. doi:10.1530/REP-10-0489
Georgiopoulos, G., and Evangelou, E. (2016). Power considerations for λ inflation factor in meta-analyses of genome-wide association studies. Genet. Res. 98, e9. doi:10.1017/S0016672316000069
Gonzalez, M. V., Mousel, M. R., Herndon, D. R., Jiang, Y., Dalrymple, B. P., Reynolds, J. O., et al. (2013). A divergent artiodactyl MYADM-like repeat is associated with erythrocyte traits and weight of lamb weaned in domestic sheep. PLoS One 8, e74700. doi:10.1371/journal.pone.0074700
Gowane, G. R., Prince, L. L. L., Paswan, C., Misra, S. S., Sharma, R. C., and Naqvi, S. M. K. (2014). Genetic analysis of reproductive and fitness traits of Malpura sheep in semi-arid tropics of India. Agric. Res. 3, 75–82. doi:10.1007/s40003-014-0091-0
Hanna, L. L. H., Taylor, J. B., Holland, P. W., Vonnahme, K. A., Reynolds, L. P., and Riley, D. G. (2023). Effect of Ewe birth litter size and estimation of genetic parameters on Ewe reproductive life traits. Animal 17, 100900. doi:10.1016/j.animal.2023.100900
He, C., Kraft, P., Chen, C., Buring, J. E., Paré, G., Hankinson, S. E., et al. (2009). Genome-wide association studies identify loci associated with age at menarche and age at natural menopause. Nat. Genet. 41, 724–728. doi:10.1038/ng.385
Jiménez-Penago, G., Hernández-Mendo, O., González-Garduño, R., Torres-Hernández, G., Torres-Chablé, O. M., and Maldonado-Simán, E. (2021). Mean corpuscular haemoglobin concentration as haematological marker to detect changes in red blood cells in sheep infected with Haemonchus contortus. Vet. Res. Commun. 45, 189–197. doi:10.1007/s11259-021-09800-8
Jorge-Smeding, E., Bonnet, M., Renand, G., Taussat, S., Graulet, B., Ortigues-Marty, I., et al. (2021). Common and diet-specific metabolic pathways underlying residual feed intake in fattening Charolais yearling bulls. Sci. Rep. 11, 24346. doi:10.1038/s41598-021-03678-x
Kamalludin, M. H., Garcia-Guerra, A., Wiltbank, M. C., and Kirkpatrick, B. W. (2018). Trio, a novel high fecundity allele: I. Transcriptome analysis of granulosa cells from carriers and noncarriers of a major gene for bovine ovulation rate. Biol. Reprod. 98, 323–334. doi:10.1093/biolre/iox133
Kielbasa, O. M., Reynolds, J. G., Wu, C., Snyder, C. M., Cho, M. Y., Weiler, H., et al. (2011). Myospryn is a calcineurin-interacting protein that negatively modulates slow-fiber-type transformation and skeletal muscle regeneration. FASEB J. 25, 2276–2286. doi:10.1096/fj.10-169219
Kinterová, V., Kaňka, J., Bartková, A., and Toralová, T. (2022). SCF ligases and their functions in oogenesis and embryogenesis—summary of the most important findings throughout the animal kingdom. Cells 11, 234. doi:10.3390/cells11020234
Kökten, T., Hansmannel, F., Ndiaye, N. C., Heba, A. C., Quilliot, D., Dreumont, N., et al. (2021). Calorie restriction as a new treatment of inflammatory diseases. Adv. Nutr. 12, 1558–1570. doi:10.1093/advances/nmaa179
Kominakis, A., Saridaki, A., and Antonakos, G. (2019). Novel candidate genes for somatic cell count in Frizarta dairy sheep. Int. J. Genet. Genom 7, 103. doi:10.11648/j.ijgg.20190704.13
Krupová, Z., Wolfová, M., Wolf, J., Oravcová, M., Margetín, M., Peskovicová, D., et al. (2009). Economic values for dairy sheep breeds in Slovakia. Asian-Australas J. Anim. Sci. 22, 1693–1702. doi:10.5713/ajas.2009.90054
Lage, K. (2014). Protein-protein interactions and genetic diseases: the interactome. Biochim. Biophys. Acta Mol. Basis Dis. 1842 (10), 1971–1980. doi:10.1016/j.bbadis.2014.05.028
Lee, M. A., Cullen, N. G., Newman, S. A. N., Dodds, K. G., McEwan, J. C., and Shackell, G. H. (2015). Genetic analysis and genomic selection of stayability and productive life in New Zealand ewes. J. Anim. Sci. 93, 3268–3277. doi:10.2527/jas.2014-8259
Lee, M. S., Liu, C. H., Lee, T. H., Wu, H. M., Huang, C. C., Huang, L. S., et al. (2010). Association of creatin kinase B and peroxiredoxin 2 expression with age and embryo quality in cumulus cells. J. Assist. Reprod. Genet. 27, 629–639. doi:10.1007/s10815-010-9459-7
Li, J., Shen, C., Zhang, K., Niu, Z., Liu, Z., Zhang, S., et al. (2021). Polymorphic variants of bovine ADCY5 gene identified in GWAS analysis were significantly associated with ovarian morphological related traits. Gene 766, 145158. doi:10.1016/j.gene.2020.145158
Li, Y., Feng, X., Wang, H., Meng, C., Zhang, J., Qian, Y., et al. (2019). Transcriptome analysis reveals corresponding genes and key pathways involved in heat stress in Hu sheep. Cell Stress Chaperones 24, 1045–1054. doi:10.1007/s12192-019-01019-6
Liu, A., Liu, M., Li, Y., Chen, X., Zhang, L., and Tian, S. (2021). Differential expression and prediction of function of lncRNAs in the ovaries of low and high fecundity Hanper sheep. Reprod. Domest. Anim. 56, 604–620. doi:10.1111/rda.13898
Liu, B., Yan, J., Li, J., and Xia, W. (2023a). The role of BDNF, YBX1, CENPF, ZSCAN4, TEAD4, GLIS1 and USF1 in the activation of the embryonic genome in bovine embryos. Int. J. Mol. Sci. 24, 16019. doi:10.3390/ijms242216019
Liu, Y., Cao, G., Xie, Y., and Chu, M. (2023b). Identification of differentially expressed genes associated with precocious puberty by suppression subtractive hybridization in goat pituitary tissues. Anim. Biotechnol. 34, 619–632. doi:10.1080/10495398.2021.1990940
Liu, Y., Wang, D. K., and Chen, L. M. (2012). The physiology of bicarbonate transporters in mammalian reproduction. Biol. Reprod. 86, 99. doi:10.1095/biolreprod.111.096826
López-Cortegano, E. (2022). PurgeR: inbreeding and purging in pedigreed populations. Bioinformatics 38, 564–565. doi:10.1093/bioinformatics/btab599
Lourenco, D., Legarra, A., Tsuruta, S., Masuda, Y., Aguilar, I., and Misztal, I. (2020). Single-step genomic evaluations from theory to practice: using snp chips and sequence data in blupf90. Genes 11, 790. doi:10.3390/genes11070790
Lunetta, K. L., D’Agostino, R. B., Karasik, D., Benjamin, E. J., Guo, C. Y., Govindaraju, R., et al. (2007). Genetic correlates of longevity and selected age-related phenotypes: a genome-wide association study in the framingham study. BMC Med. Genet. 8, S13. doi:10.1186/1471-2350-8-S1-S13
Ma, Z., Chang, Y., Brito, L. F., Li, Y., Yang, T., Wang, Y., et al. (2023). Multitrait meta-analyses identify potential candidate genes for growth-related traits in Holstein heifers. J. Dairy Sci. 106, 9055–9070. doi:10.3168/jds.2023-23462
Macé, T., González-García, E., Foulquié, D., Carrière, F., Pradel, J., Durand, C., et al. (2022). Genome-wide analyses reveal a strong association between LEPR gene variants and body fat reserves in ewes. BMC Genomics 23, 412. doi:10.1186/s12864-022-08636-z
Machado, A. L., Meira, A. N., Jucá, A. de F., Azevedo, H. C., Muniz, E. N., Coutinho, L. L., et al. (2020). Variants in gh, igf1, and lep genes associated with body traits in Santa Inês sheep. Sci. Agric. 78. doi:10.1590/1678-992x-2019-0216
Malher, X., Seegers, H., and Beaudeau, F. (2001). Culling and mortality in large dairy goat herds managed under intensive conditions in western France. Livest. Prod. Sci. 71, 75–86. doi:10.1016/s0301-6226(01)00242-1
Marcet-Rius, M., Bienboire-Frosini, C., Lezama-García, K., Domínguez-Oliva, A., Olmos-Hernández, A., Mora-Medina, P., et al. (2023). Clinical experiences and mechanism of action with the use of oxytocin injection at parturition in domestic animals: effect on the myometrium and fetuses. Animals 13, 768. doi:10.3390/ani13040768
Martinez-Royo, A., Alabart, J. L., Sarto, P., Serrano, M., Lahoz, B., Folch, J., et al. (2017). Genome-wide association studies for reproductive seasonality traits in Rasa Aragonesa sheep breed. Theriogenology 99, 21–29. doi:10.1016/j.theriogenology.2017.05.011
McLaren, A., McHugh, N., Lambe, N. R., Pabiou, T., Wall, E., and Boman, I. A. (2020). Factors affecting Ewe longevity on sheep farms in three European countries. Small Rumin. Res. 189, 106145. doi:10.1016/j.smallrumres.2020.106145
Medlock, A. E., and Dailey, H. A. (2022). New avenues of heme synthesis regulation. Int. J. Mol. Sci. 23, 7467. doi:10.3390/ijms23137467
Medrado, B. D., Pedrosa, V. B., and Pinto, L. F. B. (2021). Meta-analysis of genetic parameters for economic traits in sheep. Livest. Sci. 247, 104477. doi:10.1016/j.livsci.2021.104477
Meira, A. N., Moreira, G. C. M., Coutinho, L. L., Mourão, G. B., Azevedo, H. C., Muniz, E. N., et al. (2018). Carcass and commercial cut yield of Santa Ines sheep affected by polymorphisms of the LEP gene. Small Rumin. Res. 166, 121–128. doi:10.1016/j.smallrumres.2018.06.012
Mekkawy, W., Roehe, R., Lewis, R. M., Davies, M. H., Bünger, L., Simm, G., et al. (2009). Genetic relationship between longevity and objectively or subjectively assessed performance traits in sheep using linear censored models. J. Anim. Sci. 87, 3482–3489. doi:10.2527/jas.2008-1398
Melluso, A., Secondulfo, F., Capolongo, G., Capasso, G., and Zacchia, M. (2023). Bardet-Biedl syndrome: current perspectives and clinical outlook. Ther. Clin. Risk Manag. 19, 115–132. doi:10.2147/TCRM.S338653
Miao, X., Luo, Q., Zhao, H., and Qin, X. (2016). Ovarian proteomic study reveals the possible molecular mechanism for hyperprolificacy of Small Tail Han sheep. Sci. Rep. 6, 27606. doi:10.1038/srep27606
Miettinen, H. E., Rayburn, H., and Krieger, M. (2001). Abnormal lipoprotein metabolism and reversible female infertility in HDL receptor (SR-BI)–deficient mice. J. Clin. Invest 108, 1717–1722. doi:10.1172/JCI13288
Milerski, M., Zavadilová, L., Schmidová, J., Junkuszew, A., and Bojar, W. (2018). Analysis of longevity in Suffolk sheep in the Czech republic. Med. Weter. 74, 493–496. doi:10.21521/mw.6109
Minozzi, G., Nicolazzi, E. L., Stella, A., Biffani, S., Negrini, R., Lazzari, B., et al. (2013). Genome wide analysis of fertility and production traits in Italian Holstein cattle. PLoS One 8, e80219. doi:10.1371/journal.pone.0080219
Misztal, I., Legarra, A., and Aguilar, I. (2009). Computing procedures for genetic evaluation including phenotypic, full pedigree, and genomic information. J. Dairy Sci. 92, 4648–4655. doi:10.3168/jds.2009-2064
Misztal, I., Tsuruta, S., Lourenco, D., Masuda, Y., Aguilar, I., Legarra, A., et al. (2022). Manual for BLUPF90 family of programs. 1–149. Available online at: http://nce.ads.uga.edu/wiki/lib/exe/fetch.php?media=blupf90_all8.pdf
Mo, J., Lu, Y., Zhu, S., Feng, L., Qi, W., Chen, X., et al. (2022). Genome-wide association studies, runs of homozygosity analysis, and copy number variation detection to identify reproduction-related genes in bama Xiang pigs. Front. Vet. Sci. 9, 892815. doi:10.3389/fvets.2022.892815
Mota-Rojas, D., Marcet-Rius, M., Domínguez-Oliva, A., Martínez-Burnes, J., Lezama-García, K., Hernández-Ávalos, I., et al. (2023). The role of oxytocin in domestic animal’s maternal care: parturition, bonding, and lactation. Animals 13, 1207. doi:10.3390/ani13071207
Mrschtik, M., and Ryan, K. M. (2016). Another DRAM involved in autophagy and cell death. Autophagy 12, 603–605. doi:10.1080/15548627.2015.1137412
Mullins, B., and Chen, J. (2021). NLRP9 in innate immunity and inflammation. Immunology 162, 262–267. doi:10.1111/imm.13290
Munro, J. C., Physick-Sheard, P. W., Pyle, W. G., Schenkel, F. S., Miller, S. P., and Montanholi, Y. R. (2019). Cardiac function and feed efficiency: increased right-heart workload in feed inefficient beef cattle. Livest. Sci. 229, 159–169. doi:10.1016/j.livsci.2019.09.029
Murphy, E. (2011). Estrogen signaling and cardiovascular disease. Circ. Res. 109, 687–696. doi:10.1161/CIRCRESAHA.110.236687
Nezamidoust, M., Razzaghzadeh, S., Ezati, E., and Ghorbani, R. (2015). Impact of oxytocin-milking method on lactation performance and lactation length of sheep. Iran. J. Appl. Anim. Sci. 5, 105–113. Available online at: https://www.researchgate.net/publication/286711312.
Ni, X., Zhao, X., Danzeng, B., Li, Y., Larbi, A., Yang, H., et al. (2024). Calcium requirement of Yunnan Semi-fine wool rams (Ovis aries) based on growth performance, calcium utilization, and selected serum biochemical indexes. Animals 14, 1681. doi:10.3390/ani14111681
Niciura, S. C. M., Benavides, M. V., Okino, C. H., Ibelli, A. M. G., Minho, A. P., Esteves, S. N., et al. (2022). Genome-wide association study for Haemonchus contortus resistance in morada nova sheep. Pathogens 11, 939. doi:10.3390/pathogens11080939
North, B. J., and Sinclair, D. A. (2012). The intersection between aging and cardiovascular disease. Circ. Res. 110, 1097–1108. doi:10.1161/CIRCRESAHA.111.246876
Notter, D. R., Heidaritabar, M., Burke, J. M., Shirali, M., Murdoch, B. M., Morgan, J. L. M., et al. (2022). Single nucleotide polymorphism effects on lamb fecal egg count estimated breeding values in progeny-tested Katahdin sires. Front. Genet. 13, 866176. doi:10.3389/fgene.2022.866176
Oliveira, H. R., Brito, L. F., Miller, S. P., and Schenkel, F. S. (2020). Using random regression models to genetically evaluate functional longevity traits in North American Angus cattle. Animals 10, 2410. doi:10.3390/ani10122410
Park, H. K., and Ahima, R. S. (2015). Physiology of leptin: energy homeostasis, neuroendocrine function and metabolism. Metabolism 64, 24–34. doi:10.1016/j.metabol.2014.08.004
Pech, C. I. V. M., Vázquez, J. A. T., Zepeda, A. B., Utrera, Á. R., Rodríguez, J. J. B., Velázquez, G. M., et al. (2012). Estimación de parámetros genéticos para características de crecimiento en borregos Katahdin usando diferentes modelos. Rev. Mex. Cienc. Pecu. 3, 487–500. doi:10.22319/rmcp.v3i4.4878
Pelmuş, R. Ş., Grosu, H., Rotar, M. C., Gras, M. A., Lazăr, C., and Popa, F. (2020). Analysis of Ewe longevity and lamb survival in Teleorman Black Head sheep. Asian J. Dairy Food Res. 39, 207–211. doi:10.18805/ajdfr.DR-164
Pietri, P., and Stefanadis, C. (2021). Cardiovascular aging and longevity: JACC state-of-the-art review. J. Am. Coll. Cardiol. 77, 189–204. doi:10.1016/j.jacc.2020.11.023
Pinto, L. F. B., Lewis, R. M., Rocha, A. O., Nilson, S. M., Murphy, T. W., Wilson, C. S., et al. (2025a). Factors affecting the length of productive life in U.S. Katahdin ewes. J. Anim. Sci. 103, skae361. doi:10.1093/jas/skae361
Pinto, L. F. B., Lewis, R. M., Rocha, A. O., Nilson, S. M., Murphy, T. W., Wilson, C. S., et al. (2025b). Estimation of genetic parameters and genetic trends for Ewe longevity indicators in U.S. Katahdin sheep. J. Anim. Sci., skaf125. Online ahead of print. doi:10.1093/jas/skaf125
Proskura, W., Proskura, W. S., and Szewczuk, M. (2014). The polymorphism in the IGF1R gene is associated with body weight and average daily weight gain in Pomeranian Coarsewool ewes. Pak Vet. J. 34, 514–517. Available online at: www.pvj.com.pk.
Purdie, A. C., Plain, K. M., Begg, D. J., de Silva, K., and Whittington, R. J. (2019). Gene expression profiles during subclinical Mycobacterium avium subspecies paratuberculosis infection in sheep can predict disease outcome. Sci. Rep. 9, 8245. doi:10.1038/s41598-019-44670-w
Qin, Y., Sun, M., You, L., Wei, D., Sun, J., Liang, X., et al. (2012). ESR1, HK3 and BRSK1 gene variants are associated with both age at natural menopause and premature ovarian failure. Orphanet J. Rare Dis. 7, 5. doi:10.1186/1750-1172-7-5
R Core Team (2022). R: a language and environment for statistical computing. Available online at: https://www.R-project.org/(Accessed October 11, 2023).
Reid, I. R., Baldock, P. A., and Cornish, J. (2018). Effects of leptin on the skeleton. Endocr. Rev. 39, 938–959. doi:10.1210/er.2017-00226
Ridler, A. L., Kenyon, P. R., Greer, A. W., Logan, C. M., Morgan, S., and Corner-Thomas, R. A. (2024). Ewe culling in New Zealand: an interview study of 38 farmers. N. Z. J. Agric. Res. 67, 361–371. doi:10.1080/00288233.2023.2280624
Riggio, V., Maizon, D. O., Portolano, B., Bovenhuis, H., and van Arendonk, J. A. M. (2009). Effect of somatic cell count level on functional longevity in Valle del Belice dairy sheep assessed using survival analysis. J. Dairy Sci. 92, 6160–6166. doi:10.3168/jds.2008-1316
Sakaue, S., Kanai, M., Tanigawa, Y., Karjalainen, J., Kurki, M., Koshiba, S., et al. (2021). A cross-population atlas of genetic associations for 220 human phenotypes. Nat. Genet. 53, 1415–1424. doi:10.1038/s41588-021-00931-x
Schaeffer, L. R. (2018). Necessary changes to improve animal models. J. Anim. Breed. Genet. 135, 124–131. doi:10.1111/jbg.12321
Shelton, D. N., Fornalik, H., Neff, T., Park, S. Y., Bender, D., DeGeest, K., et al. (2012). The role of LEF1 in endometrial gland formation and carcinogenesis. PLoS One 7, e40312. doi:10.1371/journal.pone.0040312
Sherman, B. T., Hao, M., Qiu, J., Jiao, X., Baseler, M. W., Lane, H. C., et al. (2022). DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 50, W216–W221. doi:10.1093/nar/gkac194
Shojaei, M., Mohammadabadi, M. R., Fozi, M. A., and Dayani, O. (2010). Association of growth trait and Leptin gene polymorphism in Kermani sheep. J. Cell Mol. Res. 2, 67–73. doi:10.22067/jcmr.v2i2.3117
Smitchger, J. A., Taylor, J. B., Mousel, M. R., Schaub, D., Thorne, J. W., Becker, G. M., et al. (2024). Genome-wide associations with longevity and reproductive traits in U.S. rangeland ewes. Front. Genet. 15, 1398123. doi:10.3389/fgene.2024.1398123
Snyman, M., Olivier, W., and Visser, C. (2020). Identification of genes targeted in South African Merino and Afrino Sheep populations under long-term selection for reproduction and body weight. Res. Sq., 1–31. doi:10.21203/rs.3.rs-128951/v1
Souza, T. C., Pinto, L. F. B., Cruz, V. A. R., Oliveira, H. R., Pedrosa, V. B., Oliveira, G. A., et al. (2023). A comprehensive characterization of longevity and culling reasons in Canadian Holstein cattle based on various systematic factors. Transl. Anim. Sci. 7, txad102. doi:10.1093/tas/txad102
Souza, T. C., Souza, T. C., Cruz, V. A. R., Mourão, G. B., Pedrosa, V. B., Rovadoscki, G. A., et al. (2022). Estimates of heritability and candidate genes for primal cuts and dressing percentage in Santa Ines sheep. Livest. Sci. 264, 105048. doi:10.1016/j.livsci.2022.105048
Stolk, L., Zhai, G., Van Meurs, J. B. J., Verbiest, M. M. P. J., Visser, J. A., Estrada, K., et al. (2009). Loci at chromosomes 13, 19 and 20 influence age at natural menopause. Nat. Genet. 41, 645–647. doi:10.1038/ng.387
Suchocki, T., Wojdak-Maksymiec, K., and Szyda, J. (2016). Using gene networks to identify genes and pathways involved in milk production traits in Polish Holstein dairy cattle. Czech J. Anim. Sci. 61, 526–538. doi:10.17221/43/2015-cjas
Sun, J., Xiao, J., Jiang, Y., Wang, Y., Cao, M., Wei, J., et al. (2023). Genome-wide association study on reproductive traits using imputation-based whole-genome sequence data in Yorkshire pigs. Genes 14, 861. doi:10.3390/genes14040861
Sun, Z., Liu, Y., He, X., Di, R., Wang, X., Ren, C., et al. (2022). Integrative proteomics and transcriptomics profiles of the oviduct reveal the prolificacy-related candidate biomarkers of goats (Capra hircus) in estrous periods. Int. J. Mol. Sci. 23, 14888. doi:10.3390/ijms232314888
Sutera, A. M., Riggio, V., Mastrangelo, S., Di Gerlando, R., Sardina, M. T., Pong-Wong, R., et al. (2019). Genome-wide association studies for milk production traits in Valle del Belice sheep using repeated measures. Anim. Genet. 50, 311–314. doi:10.1111/age.12789
Szklarczyk, D., Kirsch, R., Koutrouli, M., Nastou, K., Mehryary, F., Hachilif, R., et al. (2023). The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51, D638–D646. doi:10.1093/nar/gkac1000
Tao, L., He, X., Wang, X., Di, R., and Chu, M. (2021). Litter size of sheep (Ovis aries): inbreeding depression and homozygous regions. Genes 12, 109–111. doi:10.3390/genes12010109
Trinh, T., and Broxmeyer, H. E. (2015). Role for leptin and leptin receptors in stem cells during health and diseases. Stem Cell Rev. Rep. 17, 511–522. doi:10.1007/s12015-021-10132-y
Turner, S. D. (2018). qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. J. Open Source Softw. 3, 731. doi:10.21105/joss.00731
Van Gorp, H., Kuchmiy, A., Van Hauwermeiren, F., and Lamkanfi, M. (2014). NOD-like receptors interfacing the immune and reproductive systems. FEBS J. 281, 4568–4582. doi:10.1111/febs.13014
VanRaden, P. M. (2008). Efficient methods to compute genomic predictions. J. Dairy Sci. 91, 4414–4423. doi:10.3168/jds.2007-0980
Van Wettere, W. H. E. J., Kind, K. L., Gatford, K. L., Swinbourne, A. M., Leu, S. T., Hayman, P. T., et al. (2021). Review of the impact of heat stress on reproductive performance of sheep. J. Anim. Sci. Biotechnol. 12, 26. doi:10.1186/s40104-020-00537-z
Vatner, S. F., Park, M., Yan, L., Lee, G. J., Lai, L., Iwatsubo, K., et al. (2013). Adenylyl cyclase type 5 in cardiac disease, metabolism, and aging. Am. J. Physiol. Heart Circ. Physiol. 305, H1–H8. doi:10.1152/ajpheart.00080.2013
Wang, D. H., Zhou, H. X., Liu, S. J., Zhou, C. J., Kong, X. W., Han, Z., et al. (2018). Glial cell line-derived neurotrophic factor supplementation promotes bovine in vitro oocyte maturation and early embryo development. Theriogenology 113, 92–101. doi:10.1016/j.theriogenology.2018.02.015
Wang, H., Zhong, J., Wang, J., Chai, Z., Zhang, C., Xin, J., et al. (2021). Whole-transcriptome analysis of yak and cattle heart tissues reveals regulatory pathways associated with high-altitude adaptation. Front. Genet. 12, 579800. doi:10.3389/fgene.2021.579800
Wang, X., Guo, X., He, X., Di, R., Zhang, X., Zhang, J., et al. (2024). Proteomic analysis identifies distinct protein patterns for high ovulation in FecB mutant Small Tail Han sheep granulosa cells. Animals 14, 11. doi:10.3390/ani14010011
Winkler, I., and Goncalves, A. (2023). Do mammals have menopause? Cell 186, 4729–4733. doi:10.1016/j.cell.2023.09.026
Witecka, A., Kwiatkowski, S., Ishikawa, T., and Drozak, J. (2021). The structure, activity, and function of the setd3 protein histidine methyltransferase. Life 11, 1040. doi:10.3390/life11101040
Wolf, M. J., Yin, T., Neumann, G. B., Korkuć, P., Brockmann, G. A., König, S., et al. (2021). Genome-wide association study using whole-genome sequence data for fertility, health indicator, and endoparasite infection traits in German black pied cattle. Genes 12, 1163. doi:10.3390/genes12081163
Wolfová, M., Wolf, J., Krupová, Z., and Margetín, M. (2009a). Estimation of economic values for traits of dairy sheep: II. Model application to a production system with one lambing per year. J. Dairy Sci. 92, 2195–2203. doi:10.3168/jds.2008-1412
Wolfová, M., Wolf, J., and Milerski, M. (2009b). Calculating economic values for growth and functional traits in non-dairy sheep. J. Anim. Breed. Genet. 126, 480–491. doi:10.1111/j.1439-0388.2009.00815.x
Xia, G., Gao, Y., Wu, C., Pan, H., Hou, J., Su, J., et al. (2022). A novel isoform of hydroxyacyl-CoA dehydrogenase inhibits cell proliferation. Biochem. Biophys. Res. Commun. 606, 75–79. doi:10.1016/j.bbrc.2022.03.102
Xu, X., Xu, X., Yin, Q., Sun, L., Liu, B., and Wang, Y. (2011). The molecular characterization and associations of porcine cardiomyopathy asssociated 5 (CMYA5) gene with carcass trait and meat quality. Mol. Biol. Rep. 38, 2085–2090. doi:10.1007/s11033-010-0334-5
Yan, L., Vatner, D. E., O’Connor, J. P., Ivessa, A., Ge, H., Chen, W., et al. (2007). Type 5 adenylyl cyclase disruption increases longevity and protects against stress. Cell 130, 247–258. doi:10.1016/j.cell.2007.05.038
Yao, X., Yang, F., El-Samahy, M. A., Liu, B., Zhao, B., Gao, X., et al. (2022). Identification and characterization of unique and common lncRNAs and mRNAs in the pituitary, ovary, and uterus of Hu sheep with different prolificacy. Genomics 114, 110511. doi:10.1016/j.ygeno.2022.110511
Yi, M., Negishi, M., and Lee, S. J. (2021). Estrogen sulfotransferase (SULT1E1): its molecular regulation, polymorphisms, and clinical perspectives. J. Pers. Med. 11, 194. doi:10.3390/jpm11030194
Yu, M., Zhang, H., Wang, B., Zhang, Y., Zheng, X., Shao, B., et al. (2021). Key signaling pathways in aging and potential interventions for healthy aging. Cells 10, 660. doi:10.3390/cells10030660
Zhang, T., Zheng, Y., Han, R., Kuang, T., Min, C., Wang, H., et al. (2022). Effects of pyruvate on early embryonic development and zygotic genome activation in pigs. Theriogenology 189, 77–85. doi:10.1016/j.theriogenology.2022.06.013
Zhang, W., Chen, S. J., Guo, L. Y., Zhang, Z., Zhang, J. B., Wang, X. M., et al. (2023a). Nitric oxide synthase and its function in animal reproduction: an update. Front Physiol. 14, 1288669. doi:10.3389/fphys.2023.1288669
Zhang, Y., Song, Y., Zhang, W., Xiao, T., and Peng, H. (2023b). Effect of NLR family pyrin domain containing 9 gene polymorphism on litter size in large white pigs. Anim. Biotechnol. 34, 4547–4552. doi:10.1080/10495398.2023.2166840
Zhang, Z., Tang, J., He, X., Di, R., and Chu, M. (2020). Mutations in NLRP5 and NLRP9 are associated with litter size in Small Tail Han sheep. Animals 10, 689. doi:10.3390/ani10040689
Zhang, Z., Tang, J., He, X., Zhu, M., Gan, S., Guo, X., et al. (2019). Comparative transcriptomics identify key hypothalamic circular rnas that participate in sheep (Ovis aries) reproduction. Animals 9, 557. doi:10.3390/ani9080557
Keywords: genes, genome-wide associate study, lifespan, longevity, ovine, productive life
Citation: Pinto LFB, Lewis RM, Rocha AO, Freking BA, Murphy TW, Wilson CS, Nilson SM, Burke JM and Brito LF (2025) Genome-wide association and functional annotation analyses reveal candidate genes and pathways associated with various ewe longevity indicators in U.S. Katahdin sheep. Front. Genet. 16:1600587. doi: 10.3389/fgene.2025.1600587
Received: 26 March 2025; Accepted: 16 May 2025;
Published: 03 July 2025.
Edited by:
Jesús Fernández, Instituto Nacional de Investigación y Tecnología Agroalimentaria (INIA), SpainReviewed by:
Malena Serrano, Instituto Nacional de Investigación y Tecnología Agroalimentaria (INIA), SpainHosein Salehian Dehkordi, Chinese Academy of Sciences (CAS), China
Copyright © 2025 Pinto, Lewis, Rocha, Freking, Murphy, Wilson, Nilson, Burke and Brito. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luiz F. Brito, YnJpdG9sQHB1cmR1ZS5lZHU=
†ORCID: Tom W. Murphy, https://orcid.org/0000-0003-3527-8814; Sara M. Nilson, https://orcid.org/0000-0003-3213-5658
 Luis F. B. Pinto
Luis F. B. Pinto Ronald M. Lewis
Ronald M. Lewis Artur O. Rocha
Artur O. Rocha Brad A. Freking
Brad A. Freking Tom W. Murphy4†
Tom W. Murphy4† Carrie S. Wilson
Carrie S. Wilson Joan M. Burke
Joan M. Burke Luiz F. Brito
Luiz F. Brito