- 1Guizhou University of Traditional Chinese Medicine, Guiyang, Guizhou, China
- 2The Second Affiliated Hospital of Guizhou University of Traditional Chinese Medicine, Guiyang, Guizhou, China
- 3Department of Internal Medicine, The No. 944 Hospital of Logistic Support Force of PLA, Jiuquan, Gansu, China
- 4School of Computing and Information Systems, The University of Melbourne, Melbourne, VC, Australia
Introduction: Predicting interactions between microRNAs (miRNAs) and competing endogenous RNAs (ceRNAs), including long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs), is essential for understanding gene regulation. With the development of Graph Neural Networks (GNNs), existing works have demonstrated the ability to capture information from miRNA-ceRNA interactions to predict unseen associations. However, current deep GNNs only leverage node-node pairwise features, neglecting the information inherent in the RNA chains themselves, as different RNAs possess chains of varying lengths.
Methods: To address this issue, we propose a novel model termed the BERT-based ceRNA Graph Predictor (BCGP), which leverages both RNA sequence information and the heterogeneous relationships among lncRNAs, circRNAs, and miRNAs. Our BCGP method employs a transformer-based model to generate contextualized representations that consider the global context of the entire RNA sequence. Subsequently, we enrich the RNA interaction graph using these contextualized representations. Furthermore, to improve the performance of association prediction, BCGP utilizes the Neural Common Neighbour (NCN) technique to capture more refined node features, leading to more informative and flexible representations.
Results: Through comprehensive experiments on two real-world datasets of lncRNA-miRNA and circRNA-miRNA associations, we demonstrate that BCGP outperforms competitive baselines across various evaluation metrics and achieves higher accuracy in association predictions. In our case studies on two types of miRNAs, we show BCGP’s remarkable performance in predicting both miRNA-lncRNA and miRNA-circRNA associations.
Discussion: Our findings demonstrate that by integrating RNA sequence information with interaction relationships within the graph, the BCGP model significantly enhances the accuracy of association prediction. This provides a new computational tool for understanding complex gene regulatory networks.
1 Introduction
MicroRNAs (miRNAs) are a class of small, non-coding RNA molecules that play a crucial role in the regulation of gene expression (Ye et al., 2019). MiRNAs are present in plants, animals and some viruses and they can significantly affect a broad range of biological processes (Bartel, 2018). Specifically, they primarily regulate gene expression through binding to the 3′ untranslated regions (UTRs) of target mRNAs, leading to their degradation or inhibition of translation. The degree of complementarity between the miRNA and the target mRNA is crucial, as it determines the mechanism of repression. MiRNA sponges, also known as competing endogenous RNAs (ceRNAs), embody a sophisticated biological mechanism that serves to regulate the activity of miRNAs within cellular environments (Alkan and Akgül, 2022). Such a mechanism leverages RNA molecules containing multiple miRNA binding sites to effectively “absorb” or “sponge” certain miRNAs. As a result, it diminishes the suppressive impacts of these miRNAs on their intended target mRNAs. There are numerous previous studies that have employed machine learning to forecast the miRNA-disease associations, achieving satisfactory results (Chen et al., 2021; Ha et al., 2020).
Long non-coding RNAs (lncRNAs), a unique type of RNA with over 200 nucleotides, do not have protein-coding capacity. Circular RNAs (circRNAs) constitute another category of non-coding RNA, distinguished by their unique covalently closed-loop configuration. Unlike linear RNAs, circRNAs lack both a 5′ cap and a 3′ poly-A tail in their sequences. They are synthesized through a mechanism known as back-splicing, wherein a splice donor site downstream is connected to a splice acceptor site upstream, resulting in the circularization of the RNA molecule (Kristensen et al., 2022). Both lncRNA and circRNA are considered as the major types of ceRNAs, where they can sponge specific miRNAs when they have miRNA binding sites, which can potentially alleviate the inhibitory effects these miRNAs exert on their target mRNAs.
Inspired by methods for predicting miRNA-disease associations (Ha et al., 2019), several approaches for ceRNA association prediction have emerged in recent years. For example, Zhu et al. (2017) discovered that lnc-mg promotes myogenesis by sponging microRNA-125b to regulate the abundance of the IGF2 protein. Furthermore, Zhang et al. (2018) identified that lncRNA MAR1 acts as a miR-487b sponge to promote skeletal muscle differentiation and regeneration. Similarly, Yang et al. (2018) found that circ-ITCH can sponge miR-17 and miR-224, thereby upregulating the expression of p21 and PTEN, which in turn suppresses the aggressive biological behaviors associated with bladder cancer.
Traditionally, determining the associations between miRNA and ceRNAs requires the use of technologies such as High-Throughput Sequencing (Li et al., 2018), RNA Immunoprecipitation (Gawronski et al., 2018) and Dual-Luciferase Reporter (Cao et al., 2016). However, these methods can cost a large investment of time and resources. With the advancement of deep learning technologies especially the deep Graph Neural Networks (GNNs) (Kipf and Welling, 2016) and the accumulation of historical experimental data, there has been a surge in efforts to predict lncRNA-miRNA and circRNA-miRNA associations using computational techniques. For example, Wang W. et al. (2022) proposed a model, named GCNCRF, to predict lncRNA-miRNA associations based on the graph convolutional network (GCN) and conditional random field. Furthermore, a recent work (Wang Z. et al., 2023) has employed a sequence pre-training-based Graph Neural Network to predict associations between lncRNAs and miRNAs.
Although effective, existing methods still have several limitations. First, most of the graph-based models have not effectively utilized the information contained within RNA sequences. As the foundational elements, an RNA sequence contains nearly all the information of the RNA (Charles Richard and Eichhorn, 2018). Using an appropriate sequential model to analyze these RNA sequences could uncover enormous characteristics of each RNA. Furthermore, most previous studies used only homogeneous or bipartite graphs to predict the associations between lncRNA-miRNA or circRNA-miRNA. However, according to the mechanism of ceRNAs (Zhong et al., 2018), circRNAs and lncRNAs, although two different types of RNAs, both function as miRNA sponges within this network. Therefore, a heterogeneous graph can be used to construct the ceRNA network among circRNA, lncRNA, and miRNA to further boost the performance.
To address the aforementioned challenges, we propose a novel framework, the BERT-based ceRNA Graph Predictor (BCGP), which uniquely integrates sequence-level and structural information for comprehensive ceRNA interaction prediction. Unlike existing methods, BCGP innovatively combines Bidirectional Encoder Representations from Transformers (BERT) for sequence pre-training and a heterogeneous graph model for fine-tuning. Specifically, our framework incorporates heterogeneous relations between lncRNAs, miRNAs, and circRNAs to capture both contextual and relational dependencies. In the pre-training stage, BCGP uses BERT with Masked Language Modeling (MLM) as the training objective, a choice justified by extensive comparative analysis, to derive high-quality embeddings for different types of RNA sequences. These embeddings are then seamlessly integrated into a heterogeneous graph, where nodes represent RNAs and edges capture their intricate relationships. In the fine-tuning stage, BCGP leverages the Neural Common Neighbour (NCN) method (Wang X. et al., 2023), further enhancing the expressive power of the graph embeddings by incorporating relational patterns. Extensive experiments have demonstrated the effectiveness of our novel BCGP framework, consistently outperforming other state-of-the-art models. Our ablation studies confirm the critical contributions of each component within BCGP, while case studies on two specific miRNAs, hsa-miR-143 and hsa-miR-6808-5p, validate the superior practicality and real-world relevance of our approach. By bridging the gap between sequence-level and structural modeling, our work establishes a new paradigm for ceRNA interaction prediction.
2 Materials and methods
2.1 Overview
In this section, we will describe our proposed method BCGP that we use to generate pre-trained embeddings for lncRNAs, circRNAs, or miRNAs and fine-tune with general Graph Neural Networks to recover the unseen associations between them. Additionally, we integrate a novel training method, Neural Common Neighbour (NCN) (Wang X. et al., 2023) to further enhance the performance of the fine-tuning GNNs. In Section 2.2, we will present the notations used in this article and briefly describe our research problem formulation. Second, in Section 2.3, we demonstrate the pre-training stage of BCGP, where we use the
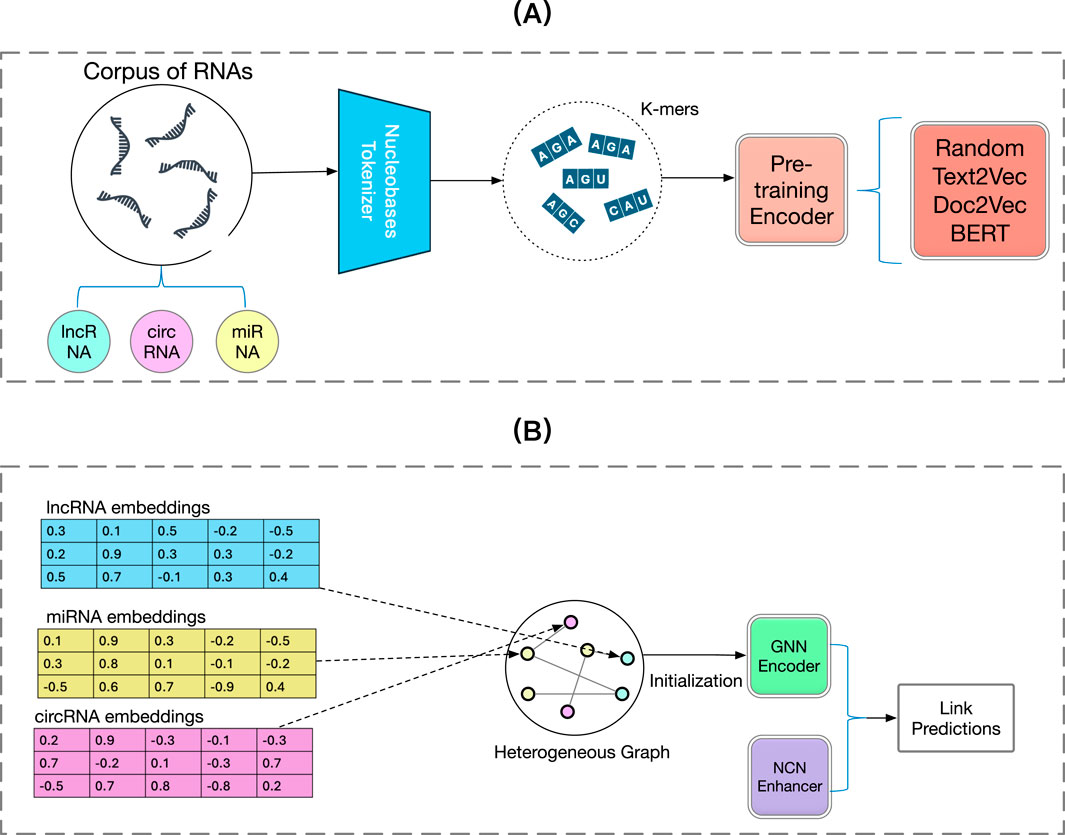
Figure 1. Overview framework of the proposed BCGP model. In the pre-training stage, BCGP first tokenizes an RNA sequence with
2.2 Preliminaries
To initialize our task, we first define all entities and their associated information. In particular, we denote sets of lncRNAs, circRNAs and miRNAs as
where
In this next section, we will provide a detailed description of our proposed method, including the techniques in the pre-training and fine-tuning stages.
2.3 Pre-training stage
During the pre-training stage, our goal is to effectively project the information of RNAs from their nucleotide sequences to latent embeddings. Specifically, we aim to simplify computational demands while enriching the semantic content of the embeddings, which captures the intrinsic patterns and relations within RNA sequences. Such embeddings not only preserve the biological significance and genetic information of RNA sequences but also serve as the initial features of nodes for the subsequent fine-tuning stage so that those sequence information can be encoded into the graph representation.
Instead of treating each base (A, C, G, U) as an individual token, given an RNA sequence
where
After tokenizing each RNA sequence into the overlapping segment using
Given a sequence of
where
Building upon the aforementioned self-attention mechanism, we adapt the Masked Language Modeling (MLM) to train the BERT model on RNA sequences. For each RNA sequence, we randomly select regions constituting
where
where
2.4 Fine-tuning stage
After obtaining the initial RNA embeddings for lncRNAs, circRNAs and miRNAs from BERT during the pre-training stage, we construct a heterogeneous graph. This graph integrates known associations between lncRNAs and miRNAs, as well as the high-quality associations between circRNAs and miRNAs, with these relationships represented as edges. The pre-trained RNA sequence embeddings are utilized as node features in this graph. To obtain effective node embeddings, we leverage GNNs to capture the intricate and complex relationships between entities.
The current de facto design of GNNs follows the message passing framework (Kipf and Welling, 2016), which is based on the core idea of recursive neighborhood aggregation. Specifically, for an
where
To further boost the performance of association prediction, we leverage the Neural Common Neighbour (Wang X. et al., 2023) to capture more refined node features such as multi-hop structure and attribute information. Specifically, after obtaining the representation
where
where
To train the model on the link prediction (LP) task, we use the binary cross-entropy loss, which is denoted as:
where
3 Results
In this section, we present the evaluation performance of our BCGP. We begin by describing the datasets used in our experiments. Following this, we introduce the experimental results, including the evaluation of pre-training, fine-tuning, performance comparisons with baselines, hyperparameter analysis, and case studies. For more details on the evaluation metrics and experimental settings, please refer to Sections 1, 2 of the Supplementary Material.
3.1 Datasets
In this study, we focus on leveraging a comprehensive dataset to model lncRNA-miRNA and circRNA-miRNA associations. We have constructed a total of four datasets: lncRNA-miRNA association 1 (LMA1), circRNA-miRNA associations 1 (CMA1), lncRNA-miRNA association 2 (LMA2), and circRNA-miRNA associations 2 (CMA2). For the lncRNA-miRNA associations in LMA1, based on previous research (Wang Z. et al., 2023), we utilized the LncACTdb 3.0 (Wang P. et al., 2022) database. From this database, we extracted 1,057 experimentally verified lncRNA-miRNA associations, containing 284 lncRNAs and 520 miRNAs. Regarding the circRNA-miRNA associations in CMA1, building upon prior studies (Guo et al., 2024), we used CircBank (Liu et al., 2019) as the primary data source, obtaining 20,771 high-quality associations involving 3,802 circRNAs and 1,273 miRNAs. The lncRNA-miRNA associations in LMA2 were sourced from lncRNASNP V3.0 (Yang et al., 2023), comprising a total of 8,502 lncRNA-miRNA associations, including 467 lncRNAs and 254 miRNAs. As for the circRNA-miRNA associations in CMA2, they were derived from the dataset 1 of the KGANCDA (Lan et al., 2022), with a total of 702 circRNA-miRNA associations, encompassing 471 circRNAs and 439 miRNAs. The sequences of lncRNAs were sourced from LNCipedia (Volders et al., 2019) and NONCODE (Zhao et al., 2021), the sequences of circRNAs were obtained from CircBase (Glažar et al., 2014), and the sequences of miRNAs were acquired from miRBase (Griffiths-Jones et al., 2007).
3.2 Examination of pre-training
To rigorously evaluate the performance of pre-training within our method, we compare several pre-training methods commonly used for initializing node embedding in GNN. We include two baselines, namely, the Random Embedding and the Adjacency Matrix Embedding methods which do not consider any RNA sequence information. The Random Embedding method initializes node embeddings with random values generated from a Gaussian distribution, while the Adjacency Matrix Embedding method leverages the adjacency matrix, representing the relationships between nodes in a lncRNA-miRNA-circRNA association graph, to generate node embeddings. Furthermore, we also compare three sequence-specific pre-training methods, including Text2vec (Mikolov et al., 2013), Doc2vec (Le and Mikolov, 2014) and HyenaDNA (Nguyen et al., 2024). Firstly, Text2vec employs a “bag of words” model for the
The experimental results on the LMA1 and CMA1 datasets presented in Table 1, demonstrate the effectiveness of BCGP-BERT. For lncRNA-miRNA prediction, BCGP-BERT achieves the highest scores across all evaluation metrics, with an F1 score of 0.439, AUC of 0.904, AP of 0.435, and NDCG of 0.816, outperforming all other pre-training methods. Similarly, for circRNA-miRNA prediction, BCGP-BERT excels in all metrics, with an F1 score of 0.572, AUC of 0.948, AP of 0.712, and NDCG of 0.957. These results suggest that BCGP-BERT enhances association prediction by learning contextualized representations that capture the global context of entire sequences. Furthermore, the findings also highlight the effectiveness of using the BERT model trained on task-related data through Masked Language Modeling, enabling BCGP-BERT to discover the intricate pattern of RNA sequences and demonstrate its robustness in predicting RNA interactions. Additional pre-training results on the LMA2 and CMA2 datasets are provided in Supplementary Appendix S3.
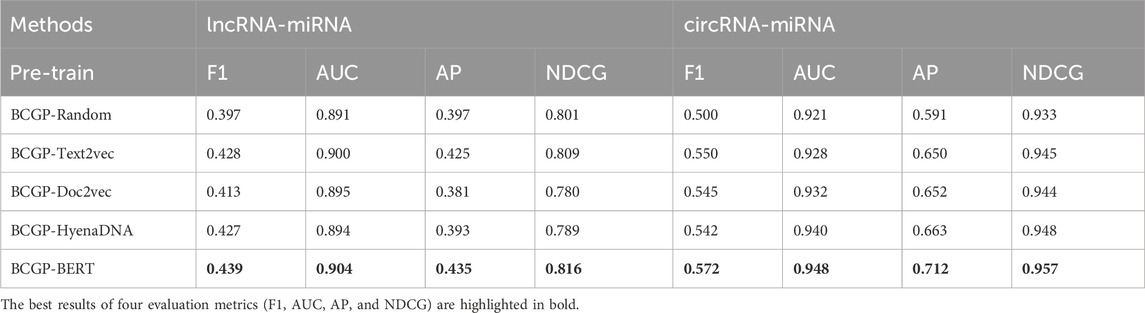
Table 1. Overall performance comparison of different pre-training methods fine-tuned with GCN on lncRNA-miRNA and circRNA-miRNA association prediction tasks using the LMA1 and CMA1 datasets.
3.3 Examination of fine-tuning
To evaluate the performance of BCGP integrated with different fine-tuning methods, we conduct association prediction experiments using six different GNNs. Specifically, we compare GAT (Veličković et al., 2017), GATv2 (Brody et al., 2021), FiLM (Brockschmidt, 2020), GraphSAGE (Hamilton et al., 2017), SGC (Wu et al., 2019), and GCN (Kipf and Welling, 2016) using the BCGP-BERT pre-training framework. All methods share the same embedding dimension. As detailed in Table 2, for the lncRNA-miRNA association prediction, GCN outperforms all other fine-tuning methods, achieving an F1 score of 0.439, AUC of 0.904, AP of 0.435, and NDCG of 0.816. For the circRNA-miRNA association prediction, GCN also demonstrates a robust performance with an F1 score of 0.572, AUC of 0.948, AP of 0.712, and NDCG of 0.957, while SGC closely follows with competitive performance. These results demonstrate that GCN is the most effective fine-tuning method on BCGP-BERT for predicting lncRNA-miRNA and circRNA-miRNA interactions. Further fine-tuning results on the LMA2 and CMA2 datasets are provided in Supplementary Appendix S3.
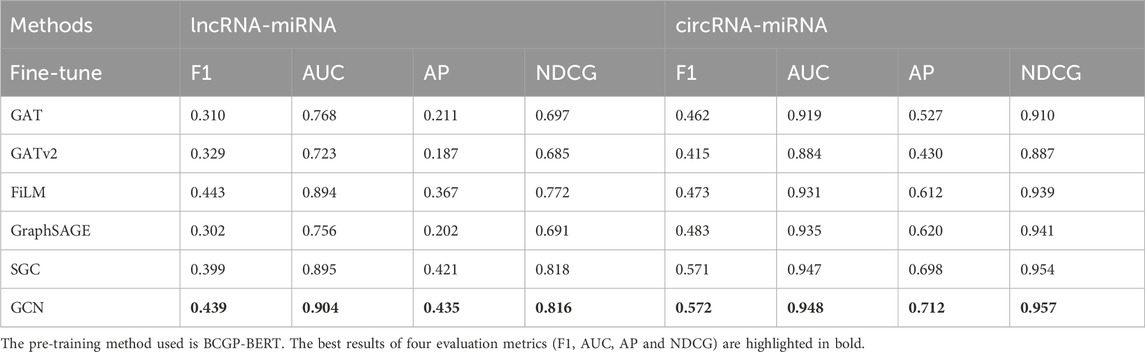
Table 2. Overall performance comparison of different fine-tuning GNN methods on lncRNA-miRNA and circRNA-miRNA association prediction tasks using the LMA1 and CMA1 datasets.
3.4 Method comparison
Next, we compare our BCGP to the state-of-the-art SPGNN (Wang Z. et al., 2023), which utilizes the
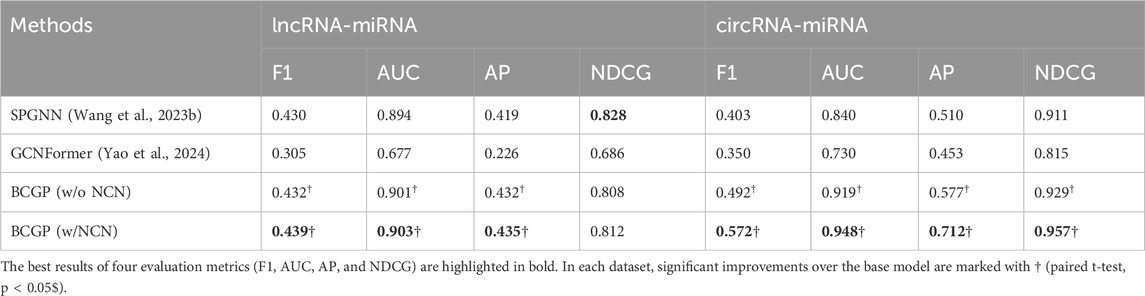
Table 3. Overall performance comparison of SPGNN and our BCGP method on lncRNA-miRNA-circRNA association prediction task.
3.5 Analysis of hyperparameters
In this section, we investigate the impacts of two important parameters of the BCGP, including the
3.5.1 Examination of k-value
In our BCGP method, we employ the
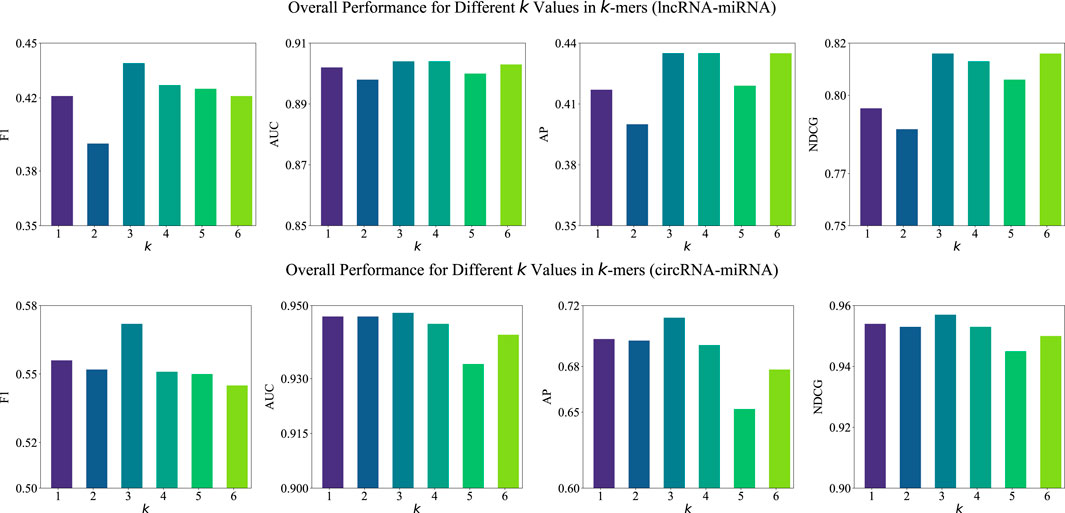
Figure 2. Overall performance (F1, AUC, AP, and NDCG) of BCGP-BERT-GCN across varying k values in k-mers for lncRNA-miRNA (first row) and circRNA-miRNA (second row) association predictions.
3.5.2 Examination of embedding size in the pre-training stage
After analyzing the impact of
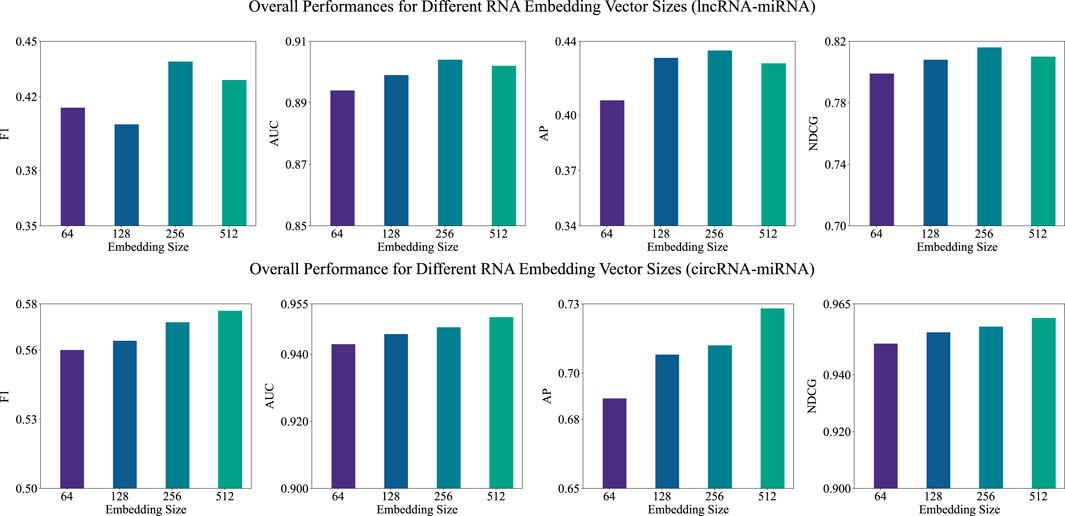
Figure 3. Overall performance (F1, AUC, AP, and NDCG) of BCGP-BERT-GCN across varying embedding sizes for lncRNA-miRNA (first row) and circRNA-miRNA (second row) association predictions.
3.6 Case study
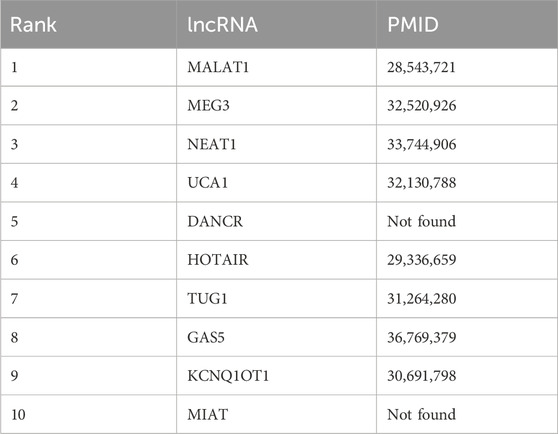
Prediction of new circRNA-miRNA and lncRNA-miRNA interactions can reveal new biomarkers, identify therapeutic targets, and enhance understanding of the regulatory mechanisms of biological networks. To validate the practicality of our method, we select two miRNAs, namely hsa-miR-143 and hsa-miR-6808-5p, to verify the prediction results of miRNA-lncRNA and miRNA-circRNA associations generated by BCGP. External literature is employed to validate the predictions for miRNA-lncRNA associations, while associations recorded in CircBank (Liu et al., 2019) are used for verifying miRNA-circRNA associations. For the target miRNAs, 8 out of the top-10 predicted miRNA-lncRNA associations are confirmed in Pubmed, and 9 out of the top 10 miRNA-circRNA associations are validated in Circbank.
Using the BCGP model, we predict lncRNAs linked to hsa-miR-143. As illustrated in Table 4, the top-10 predicted lncRNAs are MALAT1, MEG3, NEAT1, UCA1, DANCR, HOTAIR, TUG1, GAS5, KCNQ1OT1, and MIAT, with 8 of these associations being validated in external literature. For instance, MALAT1 the top-ranked lncRNA, was shown in a study by Chen et al. (2017) to regulate ZEB1 expression by sponging miR-143-3p and promoting the progression of Hepatocellular Carcinoma. Additionally, Dong et al. (2020) has demonstrated that MEG3 overexpression inhibited LPS-induced injury in PDLCs by deactivating the AKT/IKK pathway by sponging miR-143-3p.
Similarly, we predict circRNAs that are potentially associated with hsa-miR-6808-5p validated by CircBank. The results presented in Table 5 reveal that the top-10 ranked circRNAs as hsa_circ_0082878, hsa_circ_0020316, hsa_circ_0049111, hsa_circ_0057955, hsa_circ_0037997, hsa_circ_0000726, hsa_circ_0049109, hsa_circ_0049112, hsa_circ_0016773, and hsa_circ_0085900. Upon searching CircBank for hsa-miR-6808-5p, we find 9 out of the top 10 predicted results in the CircBank dataset. The results of the case study indicate that BCGP possesses commendable practicality.
4 Conclusion
In this article, we propose a novel method named BCGP, to leverage RNA sequence information and heterogeneous relationships to enhance the prediction of lncRNA-miRNA and circRNA-miRNA associations. To comprehensively capture contextual and structural information, BCGP integrates BERT in the pre-training stage to consider the global context of the entire sequence. To further enhance the performance of association prediction, BCGP leverages the Neural Common Neighbour technique in the fine-tuning stage to learn more informative and flexible representations. Extensive experiments on two real-world benchmark datasets demonstrate the effectiveness of our BCGP, showing that it significantly improves prediction accuracy by capturing complex interactions in both lncRNA-miRNA and circRNA-miRNA association prediction tasks compared with competitive baselines.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
ZX: Data curation, Writing – review and editing, Methodology, Formal Analysis, Writing – original draft, Resources, Visualization, Conceptualization. TY: Software, Investigation, Writing – review and editing, Resources, Validation, Writing – original draft. GJ: Formal Analysis, Conceptualization, Writing – review and editing, Resources, Writing – original draft. SL: Writing – original draft, Writing – review and editing, Methodology, Investigation, Conceptualization. JL: Formal Analysis, Writing – original draft, Data curation, Methodology, Software, Writing – review and editing. LT: Project administration, Methodology, Supervision, Writing – original draft, Writing – review and editing, Resources, Funding acquisition.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The project is supported by the Guizhou Provincial Science and Technology Program (Basic Research - ZK (2024) General 414).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2025.1606016/full#supplementary-material
References
Alkan, A. H., and Akgül, B. (2022). “Endogenous mirna sponges,” in miRNomics: MicroRNA Biology and computational analysis, 91–104.
Brockschmidt, M. (2020). “Gnn-film: graph neural networks with feature-wise linear modulation,” in International conference on machine learning (PMLR), 1144–1152.
Brody, S., Alon, U., and Yahav, E. (2021). “How attentive are graph attention networks?,” in International conference on learning representations.
Cao, Y., Xu, R., Xu, X., Zhou, Y., Cui, L., and He, X. (2016). Downregulation of lncrna casc2 by microrna-21 increases the proliferation and migration of renal cell carcinoma cells. Mol. Med. Rep. 14, 1019–1025. doi:10.3892/mmr.2016.5337
Charles Richard, J. L., and Eichhorn, P. J. A. (2018). Platforms for investigating lncrna functions. SLAS Technol. Transl. Life Sci. Innov. 23, 493–506. doi:10.1177/2472630318780639
Chen, L., Yao, H., Wang, K., and Liu, X. (2017). Long non-coding rna malat1 regulates zeb1 expression by sponging mir-143-3p and promotes hepatocellular carcinoma progression. J. Cell. Biochem. 118, 4836–4843. doi:10.1002/jcb.26158
Chen, X., Li, T.-H., Zhao, Y., Wang, C.-C., and Zhu, C.-C. (2021). Deep-belief network for predicting potential mirna-disease associations. Briefings Bioinforma. 22, bbaa186. doi:10.1093/bib/bbaa186
Devlin, J., Chang, M.-W., Lee, K., and Toutanova, K. (2018). Bert: pre-training of deep bidirectional transformers for language understanding. arXiv Prepr. doi:10.18653/v1/N19-1423
Dong, Y., Feng, S., and Dong, F. (2020). Maternally-expressed gene 3 (meg3)/mir-143-3p regulates injury to periodontal ligament cells by mediating the akt/inhibitory κb kinase (ikk) pathway. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 26 (e922486–1), e922486. doi:10.12659/MSM.922486
Dutta, A., Dubey, T., Singh, K. K., and Anand, A. (2018). Splicevec: distributed feature representations for splice junction prediction. Comput. Biol. Chem. 74, 434–441. doi:10.1016/j.compbiolchem.2018.03.009
Fey, M., and Lenssen, J. E. (2019). “Fast graph representation learning with pytorch geometric,” in ICLR 2019 (RLGM workshop).
Gawronski, A. R., Uhl, M., Zhang, Y., Lin, Y.-Y., Niknafs, Y. S., Ramnarine, V. R., et al. (2018). Mechrna: prediction of lncrna mechanisms from rna–rna and rna–protein interactions. Bioinformatics 34, 3101–3110. doi:10.1093/bioinformatics/bty208
Glažar, P., Papavasileiou, P., and Rajewsky, N. (2014). circbase: a database for circular rnas. Rna 20, 1666–1670. doi:10.1261/rna.043687.113
Griffiths-Jones, S., Saini, H. K., Van Dongen, S., and Enright, A. J. (2007). mirbase: tools for microrna genomics. Nucleic acids Res. 36, D154–D158. doi:10.1093/nar/gkm952
Guo, L.-X., Wang, L., You, Z.-H., Yu, C.-Q., Hu, M.-L., Zhao, B.-W., et al. (2024). Likelihood-based feature representation learning combined with neighborhood information for predicting circrna–mirna associations. Briefings Bioinforma. 25, bbae020. doi:10.1093/bib/bbae020
Ha, J., Park, C., and Park, S. (2019). Pmamca: prediction of microrna-disease association utilizing a matrix completion approach. BMC Syst. Biol. 13, 33–13. doi:10.1186/s12918-019-0700-4
Ha, J., Park, C., Park, C., and Park, S. (2020). Imipmf: inferring mirna-disease interactions using probabilistic matrix factorization. J. Biomed. Inf. 102, 103358. doi:10.1016/j.jbi.2019.103358
Hamilton, W., Ying, Z., and Leskovec, J. (2017). Inductive representation learning on large graphs. Adv. neural Inf. Process. Syst. 30. doi:10.5555/3294771.3294869
Järvelin, K., and Kekäläinen, J. (2002). Cumulated gain-based evaluation of ir techniques. ACM Trans. Inf. Syst. (TOIS) 20, 422–446. doi:10.1145/582415.582418
Kingma, D. P., and Ba, J. (2015). “Adam: a method for stochastic optimization,” in International conference on learning representations.
Kipf, T. N., and Welling, M. (2016). “Semi-supervised classification with graph convolutional networks,” in International conference on learning representations.
Kristensen, L. S., Jakobsen, T., Hager, H., and Kjems, J. (2022). The emerging roles of circrnas in cancer and oncology. Nat. Rev. Clin. Oncol. 19, 188–206. doi:10.1038/s41571-021-00585-y
Lan, W., Dong, Y., Chen, Q., Zheng, R., Liu, J., Pan, Y., et al. (2022). Kgancda: predicting circrna-disease associations based on knowledge graph attention network. Briefings Bioinforma. 23, bbab494. doi:10.1093/bib/bbab494
Le, Q., and Mikolov, T. (2014). “Distributed representations of sentences and documents,” in International conference on machine learning (PMLR), 1188–1196.
Li, M., Liu, Y., Zhang, X., Liu, J., and Wang, P. (2018). Transcriptomic analysis of high-throughput sequencing about circrna, lncrna and mrna in bladder cancer. Gene 677, 189–197. doi:10.1016/j.gene.2018.07.041
Liu, M., Wang, Q., Shen, J., Yang, B. B., and Ding, X. (2019). Circbank: a comprehensive database for circrna with standard nomenclature. RNA Biol. 16, 899–905. doi:10.1080/15476286.2019.1600395
Mikolov, T., Chen, K., Corrado, G., and Dean, J. (2013). Efficient estimation of word representations in vector space. arXiv Prepr. doi:10.48550/arXiv.1301.3781
Nguyen, E., Poli, M., Faizi, M., Thomas, A., Wornow, M., Birch-Sykes, C., et al. (2024). Hyenadna: long-range genomic sequence modeling at single nucleotide resolution. Adv. neural Inf. Process. Syst. 36. doi:10.48550/arXiv.2306.15794
Paszke, A., Gross, S., Chintala, S., Chanan, G., Yang, E., DeVito, Z., et al. (2017). Automatic differentiation in pytorch
Veličković, P., Cucurull, G., Casanova, A., Romero, A., Lio, P., and Bengio, Y. (2017). Graph attention networks. arXiv Prepr. doi:10.17863/CAM.48429
Volders, P.-J., Anckaert, J., Verheggen, K., Nuytens, J., Martens, L., Mestdagh, P., et al. (2019). Lncipedia 5: towards a reference set of human long non-coding rnas. Nucleic acids Res. 47, D135–D139. doi:10.1093/nar/gky1031
Wang, P., Guo, Q., Qi, Y., Hao, Y., Gao, Y., Zhi, H., et al. (2022a). Lncactdb 3.0: an updated database of experimentally supported cerna interactions and personalized networks contributing to precision medicine. Nucleic acids Res. 50, D183–D189. doi:10.1093/nar/gkab1092
Wang, W., Zhang, L., Sun, J., Zhao, Q., and Shuai, J. (2022b). Predicting the potential human lncrna–mirna interactions based on graph convolution network with conditional random field. Briefings Bioinforma. 23, bbac463. doi:10.1093/bib/bbac463
Wang, X., Yang, H., and Zhang, M. (2023a). Neural common neighbor with completion for link prediction. arXiv Prepr. doi:10.48550/arXiv.2302.00890
Wang, Z., Liang, S., Liu, S., Meng, Z., Wang, J., and Liang, S. (2023b). Sequence pre-training-based graph neural network for predicting lncrna-mirna associations. Briefings Bioinforma. 24, bbad317. doi:10.1093/bib/bbad317
Wu, F., Souza, A., Zhang, T., Fifty, C., Yu, T., and Weinberger, K. (2019). “Simplifying graph convolutional networks,” in International conference on machine learning (PMLR), 6861–6871.
Yang, C., Yuan, W., Yang, X., Li, P., Wang, J., Han, J., et al. (2018). Circular rna circ-itch inhibits bladder cancer progression by sponging mir-17/mir-224 and regulating p21, pten expression. Mol. cancer 17, 19–12. doi:10.1186/s12943-018-0771-7
Yang, Y., Wang, D., Miao, Y.-R., Wu, X., Luo, H., Cao, W., et al. (2023). Lncrnasnp v3: an updated database for functional variants in long non-coding rnas. Nucleic Acids Res. 51, D192–D198. doi:10.1093/nar/gkac981
Yao, D., Li, B., Zhan, X., Zhan, X., and Yu, L. (2024). Gcnformer: graph convolutional network and transformer for predicting lncrna-disease associations. BMC Bioinforma. 25 (5), 5. doi:10.1186/s12859-023-05625-1
Ye, J., Xu, M., Tian, X., Cai, S., and Zeng, S. (2019). Research advances in the detection of mirna. J. Pharm. analysis 9, 217–226. doi:10.1016/j.jpha.2019.05.004
Zhang, Z.-K., Li, J., Guan, D., Liang, C., Zhuo, Z., Liu, J., et al. (2018). A newly identified lncrna mar1 acts as a mir-487b sponge to promote skeletal muscle differentiation and regeneration. J. cachexia, sarcopenia muscle 9, 613–626. doi:10.1002/jcsm.12281
Zhang, Y., Liu, Y., Xu, J., Wang, X., Peng, X., Song, J., et al. (2021). Leveraging the attention mechanism to improve the identification of dna n6-methyladenine sites. Briefings Bioinforma. 22, bbab351. doi:10.1093/bib/bbab351
Zhao, L., Wang, J., Li, Y., Song, T., Wu, Y., Fang, S., et al. (2021). Noncodev6: an updated database dedicated to long non-coding rna annotation in both animals and plants. Nucleic acids Res. 49, D165–D171. doi:10.1093/nar/gkaa1046
Zhong, Y., Du, Y., Yang, X., Mo, Y., Fan, C., Xiong, F., et al. (2018). Circular rnas function as cernas to regulate and control human cancer progression. Mol. cancer 17, 79–11. doi:10.1186/s12943-018-0827-8
Keywords: lncRNA, circRNA, miRNA, ceRNA, pre-train, graph neural network
Citation: Xie Z, Ying T, Jing G, Liang S, Liu J and Tang L (2025) Integrating BERT pre-training with graph common neighbours for predicting ceRNA interactions. Front. Genet. 16:1606016. doi: 10.3389/fgene.2025.1606016
Received: 04 April 2025; Accepted: 08 July 2025;
Published: 03 September 2025.
Edited by:
Martina Calore, University of Padua, ItalyReviewed by:
Zhihao Ma, East China Normal University, ChinaMorteza Kouhsar, University of Exeter, United Kingdom
Jie Ni, Changzhou University, China
Copyright © 2025 Xie, Ying, Jing, Liang, Liu and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lianghua Tang, NjkzOTAwNkBxcS5jb20=
†These authors have contributed equally to this work and share first authorship
 Zhengxing Xie
Zhengxing Xie Tianping Ying2†
Tianping Ying2† Shiyang Liang
Shiyang Liang