- 1Department of Dermatology, Fuzhou First General Hospital, Fuzhou, China
- 2Department of Respiratory and Critical Care, Xinxiang Central Hospital, Xinxiang, Henan, China
- 3Department of Dermatology, Venereology and Allergology, Charité-Universitätsmedizin Berlin, Berlin, Germany
The balance between proteases and their inhibitors is essential for maintaining the structural and functional homeostasis of the skin. Numerous studies have shown that serine protease inhibitors are highly expressed in the skin and play diverse roles in preserving its physiological integrity. Among them, SERPINs have been closely linked to various skin disorders—for instance, mutations in SERPINB7 are associated with palmoplantar keratoderma, while SERPINA1 has been implicated in the pathogenesis of adult-onset immunodeficiency syndrome and generalized pustular psoriasis, both of which currently have limited treatment options. This review focuses on the biological functions of SERPINs in the skin, aiming to provide insights into the mechanisms underlying SERPIN-related skin diseases and to facilitate the development of targeted therapeutic strategies.
1 Introduction
Skin, being the largest organ of the human body, relies on the function of proteases to maintain the stability of its barrier structure and physiological condition (Kishibe, 2019). Positioned on the surface of the body, the skin acts as a natural defense against harmful substances from the external environment (De Veer et al., 2014; Steele et al., 2020). The process of desquamation is intricate, involving the gradual migration of basal keratinocytes towards the stratum corneum under the influence of various regulatory factors. During this process, several proteases hydrolyze structural proteins such as filaggrin, corneodesmosin and desmoglein in the upper epidermis, contributing to desquamation and barrier function. Any abnormalities in the activity of proteases and their inhibitors can disrupt the synthesis and breakdown of these structural proteins, leading to premature or delayed shedding and resulting in parakeratosis or hyperkeratosis (Kishibe, 2019).
SERPINs are the largest protease inhibitor family, with many members and complex functions. They are divided into 16 distinct subfamilies (A-P), with the first 9 classes (A-I) in humans (Gettins, 2002). SERPINs have a highly conserved secondary structure, containing 7-9 α-helices, 3 β-helices, and an exposed reactive center loop (RCL). The tertiary structure of SERPINs is metastable: when the protease cleaves RCL, SERPINs undergo a conformational change from a stressed state to a relaxed state, which is critical to the function of SERPINs (Law et al., 2006). The change in RCL configuration causes a structural change in the active site of SERPINs. Thus, the function of serine protease is inhibited, leading to the loss of the activity of protease hydrolysis. Because the binding of SERPINs and proteases is irreversible, this process is also known as the suicide substrate mechanism (Huntington, 2011). In this case, inhibition relies on the inability of the ends of the scissile bond (P1-P1', according to Schechter and Berger’s nomenclature) to separate after proteolytic cleavage. Consequently, the inhibitor binds tightly, yet reversibly, to a protease without either protein undergoing conformational change. In contrast, serpin RCLs are typically long (20–24 residues) and flexible, resembling a substrate loop (Janciauskiene et al., 2024).
SERPINs are widely expressed in animals, plants, viruses, and other living organisms. So far, more than 1,500 species have been found, 37 of which are found in the human body. A host of SERPIN genes are clustered on the same chromosome, especially chromosomes 14 and 18 (Aslam and Yuan, 2020). Within each cluster, all SERPINs genes belong to the same clade. For instance, the clusters of genes on chromosomes 18 entirely belong to clade B, which indicates these genes evolved from a common ancestor by chromosomal duplications. Interestingly, despite their chromosomal proximity, the functions of these genes vary greatly. SERPINs can be secreted into the cytoplasm, nucleus, and even the extracellular environment and exert a protease inhibition effect in the main suicide substrate mechanism in the human body. However, some SERPINs do not act as protease inhibitors but participate in inflammation, anticoagulation, antitumor, and other processes. Of note, numerous skin diseases have been attributed to SERPINs monogenetic mutations, most of which are deleterious.
Emerging evidence has highlighted the involvement of SERPINs in the regulation of key intracellular signaling pathways that orchestrate inflammatory and stress responses in the skin. For instance, SERPINB3 and SERPINB4 have been shown to suppress the activation of the NF-κB and MAPK pathways in keratinocytes, thereby limiting the expression of pro-inflammatory cytokines such as IL-6 and IL-8 (Sun et al., 2017). Additionally, SERPINA1 (α1-antitrypsin) has been reported to inhibit STAT3 phosphorylation in models of cutaneous inflammation, suggesting broader immune-modulatory effects (Kantaputra P. et al., 2021). In vivo studies further substantiate these findings. Transgenic mice overexpressing SERPINB3 exhibit reduced epidermal inflammation and improved barrier recovery following irritant exposure, whereas SERPINB3 knockout models demonstrate exacerbated skin inflammation and increased protease activity (Sivaprasad et al., 2015). Likewise, SERPINE1-deficient mice show delayed wound healing and enhanced matrix metalloproteinases (MMPs) activation, reinforcing its role in protease balance and tissue remodeling (Chen et al., 2021). These in vivo models provide mechanistic insight into how SERPINs maintain skin integrity and modulate inflammatory responses under physiological and pathological conditions.
A recent integrative study combining tissue proteomics and the public cutaneous squamous cell carcinoma(cSCC) transcriptomic dataset GSE32628 identified 20 overlapping genes and validated eight proteins (TNC, FSCN1, SERPINB1, ACTN1, RAB31, COL3A1, COL1A1, CD36) as differentially expressed between Bowen’s disease, cSCC and normal skin (Biao et al., 2022); importantly, SERPINB1 was functionally interrogated by siRNA in A431 cells, where its knockdown reduced migration and invasion, providing direct proteomic-to-functional evidence linking a SERPIN to invasive behaviour. Independent transcriptomic and proteomic literature further supports involvement of the SERPIN family across cutaneous disorders: elevated SERPINB3/B4 expression has been reproducibly observed in inflammatory skin diseases and implicated in barrier dysfunction and early inflammation, consistent with transcript-level dysregulation observed in RNA-seq studies (Sivaprasad et al., 2015). Genetic and transcriptomic analyses in neutrophil-rich pustular dermatoses have also identified SERPINA1 and SERPINA3 as disease-relevant (including loss-of-function variants that perturb protease inhibition and IL-36 regulation), illustrating that both sequence variation and expression changes of SERPINs can contribute to cutaneous inflammatory pathology (Yang et al., 2023). More broadly, proteomic profiling of keratinocytic lesions and recent multi-omics reviews demonstrate that proteome and transcriptome datasets can discriminate lesion types (actinic keratosis, Bowen’s disease, cSCC) and uncover pathways (ECM–receptor interaction, focal adhesion, PI3K-Akt signaling) in which SERPINs participate, thereby providing systems-level context for the specific SERPIN alterations highlighted above (Rusiñol and Puig, 2024). Taken together, these transcriptomic, proteomic and genetic studies—centered on the Tang et al. proteomics–GEO integration and supported by independent omics and genetic reports—supply the high-throughput evidence the reviewers requested and justify the inclusion of SERPINB1, SERPINB3/4, SERPINA1/3 and related proteins in our review as candidate mediators and biomarkers of progression from Bowen’s disease to invasive cSCC (Biao et al., 2022).
An intricate regulatory relationship exists between serine protease inhibitors (SERPINs) and MMPs in maintaining skin homeostasis and responding to pathological stimuli. MMPs, including MMP-1, MMP-2, and MMP-9, are essential for extracellular matrix (ECM) remodeling, keratinocyte migration, and inflammation resolution. Dysregulated MMP activity has been implicated in various skin disorders, such as chronic wounds, psoriasis, and atopic dermatitis, often resulting in excessive ECM degradation and barrier dysfunction (Ohtomo et al., 2008). SERPINs, traditionally recognized for their roles in inhibiting serine proteases, have emerged as indirect regulators of MMPs. For example, SERPINE1 can modulate MMP activation by limiting plasmin generation, which is required for the proteolytic activation of latent MMPs (Chen et al., 2021). Moreover, studies suggest that SERPINB3 and SERPINB4 may influence MMP expression through modulation of oxidative stress and inflammatory pathways (Sun et al., 2017). The loss of balance between SERPIN-mediated inhibition and MMP-driven proteolysis may contribute to exaggerated inflammation, impaired barrier repair, and tissue remodeling in inflammatory skin diseases. Further elucidation of the SERPIN–MMP axis may offer novel therapeutic opportunities for modulating protease activity in skin pathology.
In this study, we reviewed (Table 1) the clinical manifestation and pathogenesis of skin diseases related to SERPINs in order to improve our understanding of the potential role of SERPINs (Maas and de Maat, 2021). Due to numerous biological functions and pathological states associated with SERPINs, further characterization of the structure and mechanism information of SERPINs will shed light on the therapeutic targets of SERPIN-related skin diseases.
2 SERPINs related to keratosis
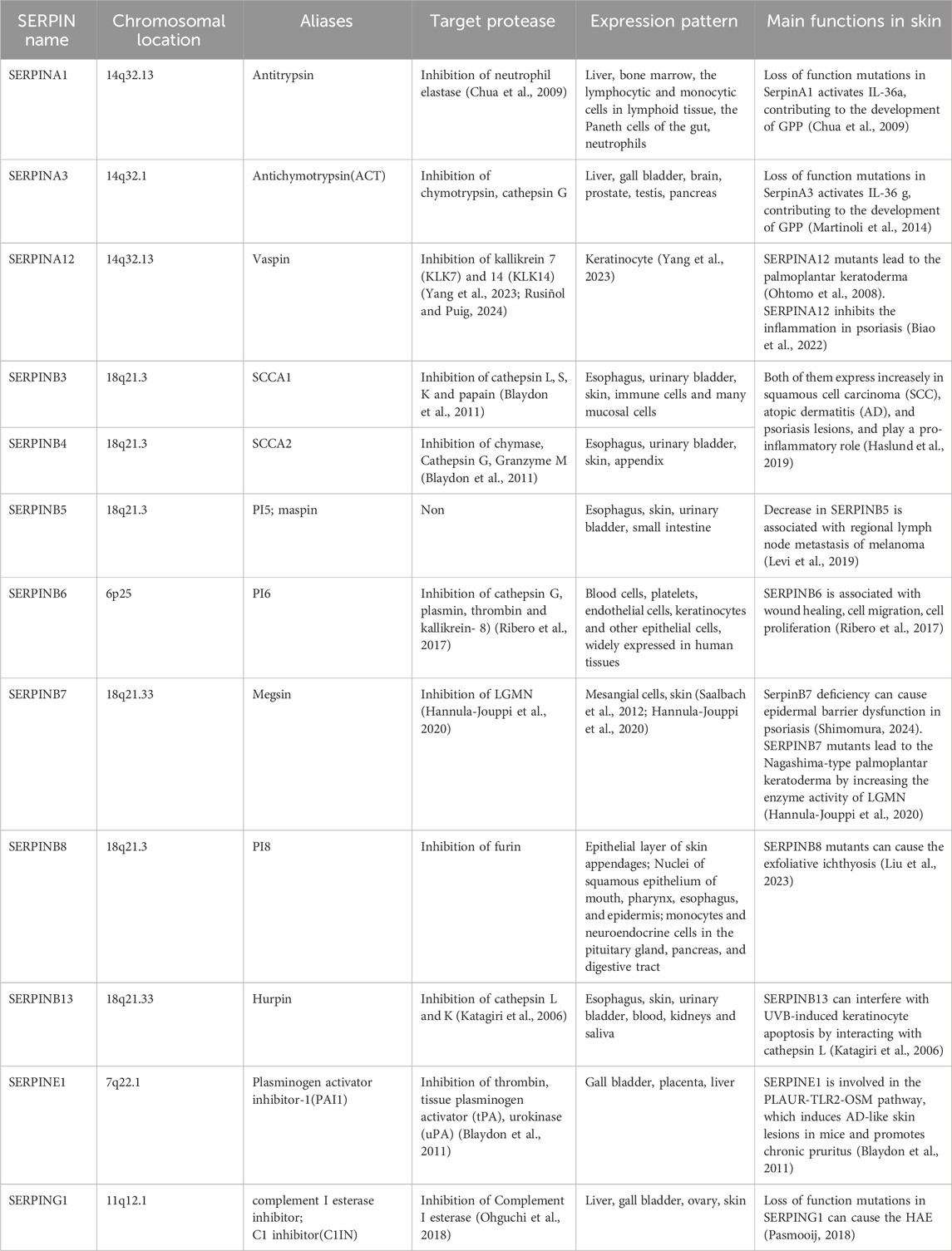
SERPINA12 encodesthe VASPIN (Visceral Adipose Tissue-Derived Serine Protease Inhibitor) protein, which consists of 414 amino acids. VASPIN comprises 3 beta folds, 9 alpha helices, and a RCL structure located between amino acids G364 and P381 at the C-terminal (Figure 1). The RCL region is identified and cleaved between the M378 (P) and E379 (P1′) sites to form a stable complex (Weiner et al., 2019; Kurowska et al., 2021). Although VASPIN was initially discovered in human fat tissue and its association with diabetes, its presence in skin was first reported in 2012 (Hida et al., 2005). VASPIN is predominantly expressed in the suprabasal layerof the epidermis. Studies by Saalbach et al. (2012) revealed lower VASPIN expression in psoriatic skin lesions compared to non-psoriatic skin lesions. However, VASPIN expression was found to be higher in differentiated cells within psoriatic skin lesions. When HaCaT cells were co-cultured with immune cells, VASPIN treatment demonstrated the ability to modulate inflammatory phenotypes in psoriasis models. These findings suggest that VASPIN may play a role in regulating inflammation during the development of psoriasis, making it a potential target for psoriasis treatment. VASPINfunctions as an inhibitor of serine proteases, but it does not inhibit common serine proteases such as pancreatic enzymes. It is known to inhibit kallikrein 7 (KLK7) and KLK14 (Heiker et al., 2013; Ulbricht et al., 2018). In 2020, Mohamad et al. (2020) originally reported two cases of autosomal recessive palmoplantar keratoderma (PPK) patients with SERPINA12 mutations, including c.631C>T(p.Arg211Ter) and c.1051G>T(p.Glu351Ter). Both of the variants are homozygous nonsense variants, leading to the lower expression and the loss of its function. Mohamad et al found that VASPINregulates the process of skin desquamation by inhibiting the protease activity of KLK7. KLK7 activity is markedly increased after the loss of SERPINA12 mutation and increases the proteolytic degradation of DSG1 and CDSN, which is necessary for normal cornification and epidermal differentiation (Steele et al., 2020; Mohamad et al., 2020). Subsequently, additional studies have described cases of SERPINA12-related PPK. In 2022, Liu et al. (2022) reported six autosomal recessive PPK patients caused by SERPINA12 mutations and identified four new mutations: c.970_971del, c.662delA, c.656A>G, and c.635-7A>G. In population genetics, a founder effect refers to the reduced genetic diversity that results when a population is descended from a small number of ancestors. This can lead to the increased frequency of rare genetic variants, including pathogenic mutations, within specific populations. Haplotype analysis results of the c.970_971del mutation, which had the highest frequency among these mutations, suggest that SERPINA12 c.970_971del may be a founder mutation. Patients carrying this mutation share a common haplotype with a distance of at least 17kb. Recently, Hannula-Jouppi (Brandt et al., 2024) lab identified a new SERPINA12 c.1100G>A (p.Gly367Glu) missense variant. And a previously reported SERPINA12 c.631C>T (p.Arg211*) variant is carried by all three patients, and they share a 53 kilobases haplotype, which suggesting c.631C>T may be a founder mutation. Collectively, seven distinct SERPINA12 variants have been reported as the cause of PPK, including c.970_971del, c.662delA, c.656A>G, c.635-7A>G, c.631C>T, c.1051G>T, c.1100G>A. Interestingly, the SERPINA12-related PPK patients reported by Hannula-Jouppi et al. (2020) all carry a heterozygous SERPINB7 c.1136G>A (p.Cys379Tyr) variant, which suggests the potential connection between SERPINA12 and SERPINB7.
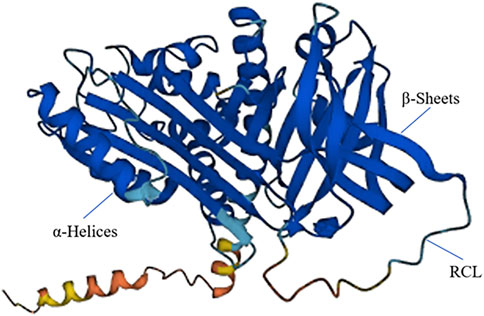
Figure 1. The 3D structure of SERPINA12. SERPINA12 with labeled structural elements: α-helices, β-sheet and reactive center loop (RCL).
SERPINB7 encodes the MEGSIN protein, which consists of 380 amino acids (Figure 2). In 1998, Kurokawa (Miyata et al., 1998) first cloned the SERPINB7 gene from human mesangial cells. By comparing its amino acid sequence with other proteins, it was determined that SERPINB7 belongs to the serine protease inhibitor family and exhibits the characteristic structural features of the SERPIN family, including the RCL structure. MEGSIN is primarily expressed in human mesangial cells, and its expression is notably elevated in the mesangial cells of patients with IgA nephropathy. In 2002, Miyata et al. (2002) created a mouse model that overexpressed Serpinb7. This led to distinct changes in the mesangial matrix within the kidneys of the mice. There was an expansion in the matrix and an increase in the number of mesangial cells, and in vitro experiments confirmed that Megsin could inhibit plasminase.
Figure 2. The 3D structure of SERPINB7. SERPINB7 with labeled structural elements: α-helices, β-sheet and reactive center loop (RCL).
The inhibitory function of MEGSINon proteases suggests its potential role in regulating fibrinolysis, a process involved in the pathogenesis of IgA nephropathy. In 2008, Toshio Miyata et al. (Ohtomo et al., 2008) discovered that MEGSIN can reduce the protease activity of MMP-1 and MMP-9. MMP-1 and MMP-9 are metalloproteinases that can degrade essential components of the mesangial extracellular matrix, including type IV collagen, laminin, and fibronectin, which play a role in regulating the synthesis and degradation of the extracellular matrix. MMP-1 and MMP-9 exist in an inactive proenzyme state and require activation by plasminase. MEGSINinhibits plasminase, and the inability of MMP-9 to activate may contribute to the mechanisms by which MEGSINis involved in diabetic nephropathy. In 2013, Kubo et al. (2013) identified SERPINB7 as the causing gene of autosomal recessive Nagashima-type Palmoplantar Keratoderma (NPPK) through whole exon sequencing of samples from three unrelated Japanese patients, and confirming SERPINB7 c.796C>T as a founder mutation through haplotype analysis. NPPK is characterized by the presence of clear erythema in the palmar and plantar areas, often accompanied by mild diffuse hyperkeratosis that may extend to the backs of the hands, feet, and the inside of the wrists. NPPK is more prevalent in east Asia and is the most common type of palmoplantar keratoderma in the Chinese Han population (Shimomura, 2024). Hannula-Jouppi et al. (2020) indicates that NPPK, an autosomal recessive diffuse PPK caused by SERPINB7 variants resulting in excessive protease activity, exhibits resemblances to SERPINA12-related PPK. SERPINB7 may lead to PPK through the similar mechanism of PPK caused by the SERPINA12 mutation. A group of total 234 Chinese NPPK patients were established, which would enhance the current understanding of NPPK, expand the mutation spectrum of SERPINB7, and offer evidence-based recommendations for future clinical management, diagnosis, and genetic counseling of NPPK patients (Liu et al., 2023). At present, seventeen different SERPINB7 variants has been identified, including c.122_127del, c.218_219delins, c.271delC, c.336+2T>G, c.382C>T, c.434G>C, c.455-1G>A, c.455G>A, c.455G>T, c.522dup, c.530T>C, c.635del, c.643A>G, c.650_653del, c.796C>T, c.830C>T, c.1136G>T (Brandt et al., 2024). Among these mutations, c.796C>T is the most frequent and proposed to be enriched in east Asian populations for founder effect. Besides, c.218_219delins and c.830C>T are relatively more common than other variants. Recent study (Liu et al., 2024) describes the potential digenic inheritance of SERPINB7 and SERPINA12 variants, shed light on the relationship between these two important serine protease inhibitors.
One recent study (Li et al., 2024) showed that the mutation of SERPINB7 is the cause of NPPK through inhibiting the protease legumain. MEGSINbound directly with legumain and inhibited legumain activity in a ‘protease-substrate' manner at the cleavage sites of MEGSINas Asn71 and Asn343. This research demonstrated that MEGSINdeficiency results in the overactivation of legumain, which may further compromise the epidermal barrier, leading to NPPK phenotypes. These findings suggest that legumain could be a promising therapeutic target for NPPK. Recent studies have contributed to a better understanding of the regulation of protease and inhibitor networks in the skin, shedding light on their potential roles in the pathogenesis of keratoderma.
Current treatments for this condition typically involve the topical use of moisturizer for symptom relief. Ohguchi et al. (2018) conducted clinical trials and confirmed that the administration of gentamicin can induce ribosome read-through of the SERPINB7 c.796C>T nonsense mutation. Gentamicin causes the ribosome to read through the stop codon and the production of full-length MEGSINprotein, leading to a significant improvement in patients’ phenotypes. The mechanism involves nonsense mutation-mediated mRNA decay (NMD) and 26S proteasome-mediated protein degradation, preventing the production of truncated proteins (Pasmooij, 2018; Wang et al., 2023).
SERPINB7 has also been reported to function as a key signaling molecule during the pathogenesis of psoriasis (Zheng et al., 2022), which is abundantly expressed in the lesions of psoriasis patients and imiquimod-induced psoriatic mouse model. SERPINB7 deficiency inhibited keratinocyte differentiation and increased inflammation by diminishing the calcium concentration in the cytosol and mitochondria, contributing to the development of psoriasis. The deficiency of SerpinB7 impairs skin barrier function and modulates the expression of inflammatory mediators in mice, thereby exacerbating psoriasis-like lesions. Transcriptomic and proteomic analyses reveal that the absence of SerpinB7 disrupts keratinocyte differentiation and alters the expression of genes associated with the calcium signaling pathway. Experimental data confirm that the loss of SerpinB7 reduces calcium ion concentration in keratinocytes, leading to inhibited differentiation of these cells. This inhibition subsequently promotes the expression of keratinocytes and inflammatory mediators, thereby influencing the onset and progression of psoriasis.
SERPINB8 encodes a protein consisting of 374 amino acids. In 2016, Pigors et al. (2016) reported a series of cases of exfoliative ichthyosis in three unrelated families caused by loss-of-function mutations in SERPINB8. These mutations included SERPINB8 c.947delA (p.Lys316Serfs90), c.850C>T (p.Arg284), and c.2T>C (p.Met1?). Cell experiments confirmed an upregulation in the expression of desmoplakin isoforms I and II as well as desmoglein-1 proteins in HaCaT cells after SERPINB8 knockdown. The specific molecular mechanisms through which SERPINB8 mutations lead to exfoliative ichthyosis remain to be explored. It's worth noting that CSTA mutation of c.67-2A>T and c.256C>T can also lead to exfoliative ichthyosis (Blaydon et al., 2011), indicating a correlation between SERPINB8 and CSTA, which could involve an upstream or downstream relationship, or they may share the same enzyme substrates, potentially contributing to a similar disease.
3 Hereditary angioedema caused by SERPING1 mutation
SERPING1 encodes the complement 1 esterase inhibitor (C1 esterase inhibitor, C1-INH) protein, which plays a crucial role in the regulation of complement activity. Classic hereditary angioedema (HAE) is primarily caused by mutations in SERPING1, making it the most common type of HAE (Haslund et al., 2019). As previously reviewed, more than 700 distinct SERPING1 variants have been reported to result from HAE. Among these mutations (Sinnathamby et al., 2023; Santacroce et al., 2021), a host of genotypes are observed, including missense variants (32.1%) and nonsense variants (9.0%), short deletions, duplications, and delins variants (36.2%), splice defects (13.6%), large deletions and duplications (8.2%), and a few variants affecting 5′-untranscribed and 3′-untranslated region (3′-UTR) sequences (0.9%). HAE is characterized by acute, recurrent subcutaneous and submucosal edema. Clinical manifestations can affect various parts of the body, including the face, limbs, trunk, digestive tract, and respiratory tract, and can be life-threatening in severe cases. SERPING1 mutations can lead to two main types of HAE: type I HAE, accounting for approximately 85% of classic HAE cases, results from reduced expression of C1-INH, leading to insufficient function, and type II HAE, accounting for about 15% of classic HAE cases, is characterized by normal expression but lacking function of C1-INH (Betschel et al., 2023). Complement C1 is a vital serum protein involved in regulating phagocyte function and mediating inflammatory responses, including increasing vascular permeability. SERPING1 mutations decrease the inhibitory function of C1-INH, leading to overactivation of C1. Activated C1 can cleave downstream substrates, including complement C2 and C4, resulting in reduced blood vessel concentrations of C2 and C4 (Levi et al., 2019).
Loss of C1-INH function also affects coagulation factor XII, which activates clotting factor XIIa, promoting the conversion of prokallikrenase into kallikrenase. Through positive feedback, this leads to increased levels of clotting factor XIIa. Kallikrein binds to the bradykinin β2 receptor, leading to vasodilation and increased blood vessel permeability, which can result in skin and mucosal edema. In addition to SERPING1 mutations, HAE can also be caused by mutations in the F12, PLG, and ANGPT1 genes (Levi et al., 2019). However, the precise pathogenesis of HAE is still not fully understood. Understanding the genetic basis of HAE, including the role of genes such as SERPING1, is crucial for accurate diagnosis and subsequent treatment of this condition. HAE is a life-threatening disease required immediate and effective therapies especially when it involves the upper airway.
HAE is primarily driven by the pro-inflammatory mediator bradykinin and results from a deficiency of the liver-produced acute-phase reactant C1-inhibitor protein. The C1-inhibitor protein limits the activation of C1r, C1s, MASP-1, and MASP-2, thereby inhibiting the complement system. When C1-inhibitor protein is nonfunctional or present in low levels, unrestrained complement activity occurs, leading to increased vascular permeability and edema (Sinnathamby et al., 2023). Furthermore, the C1-inhibitor protein also inhibits plasma kallikrein, the enzyme responsible for converting high-molecular-weight kininogen (HMWK) to bradykinin. Low levels of C1-inhibitor allow plasma kallikrein levels to rise, subsequently increasing bradykinin levels. Notably, plasma kallikrein enhances the activation of factor XII to factor XIIa, which further promotes the conversion of prekallikrein to kallikrein, thus boosting bradykinin production. Bradykinin is responsible for many HAE symptoms, as it induces endothelial contraction, nociceptor activation, and bronchoconstriction, leading to edema, pain, and dry cough typical in HAE patients. It is worth noting that angiotensin-converting enzyme (ACE) reduces bradykinin levels. Therefore, ACE inhibitors, which are effective anti-hypertensive agents, can induce HAE, a condition known as ACE-induced HAE (Sinnathamby et al., 2023; Busse and Christiansen, 2020).
4 SERPINs associated with skin tumors
Squamous cell carcinoma antigen 1 (SCCA1) and squamous cell carcinoma antigen 2 (SCCA2) were encoded by SERPINB3 and SERPINB4, respectively. The base sequences of SERPINB3and SERPINB4are highly homologous. However, the RCL sequences of the two genes are distinct, and the inhibitory substrates are also different. SERPINB3 mainly inhibits cathepsin K, L, S, and V, while SERPINB4 primarily inhibits CTSG, Granzyme M and chymase (Sun et al., 2017). Studies have shown that the expression of SERPINB3 and SERPINB4 increases in SCC, atopic dermatitis, and psoriasis lesions, and they can play a pro-inflammatory role (Ren et al., 2020). Research conducted by Katagiri et al. revealed that the expression of SERPINB3/4 increased in UV-irradiated epidermis. This increase inhibited the phosphatase activity of JNK1 and ultimately suppressed UV-induced autophagy (Katagiri et al., 2006). Sivaprasad et al. (2015) observed an elevation in SERPINB3/4 expression in the skin lesions of patients with AD. Furthermore, the knockout of SERPINB3/4 inhibited the acute inflammatory response in mouse models of AD, indicating their proinflammatory effect in chronic inflammatory skin diseases.
SERPINB5 encodes the MASPIN protein which consists of 375 amino acids. It functions as a regulator to suppress the tumor metastasis, such as melanoma, without serine protease activity inhibitory effect (Chua et al., 2009). Melanoma is a malignant skin tumor characterized by frequent local recurrence and early metastasis. A study of mass spectrometry-based proteomics on a variety of melanoma samples shows that the decrease in SERPINB5 is associated with regional lymph node metastasis of melanoma (Martinoli et al., 2014). In primary melanomas, nuclear MASPIN expression was correlated with disease stage and melanoma prognostic factors (tumor thickness, mitotic rate, nodular histotype, and ulceration). In contrast, cytoplasmic MASPIN was more frequently seen in thin superficial spreading melanomas that did not undergo mitosis (Ribero et al., 2017).
5 SERPINs associated with common inflammatory skin diseases
SERPINs play a role in mediating the release of inflammatory factors during the development of common inflammatory skin diseases, such as AD and psoriasis. Besides the SERPINA12, SERPINB7, SERPINB3, SERPINB4, et al., other SERPINs are also correlated with the pathogenesis of AD and psoriasis (Morizane et al., 2022). SERINE11 and SERPINB13 are involved in the pathogenesis of inflammatory skin diseases while no diseases caused by the monogenic mutation of these two genes have been determined.
SERPINE1 encodes a plasminogen activator inhibitor type-1(PAI-1) comprising 402 amino acids. SERPINE1 is dermally overexpressed in the lesions of AD patients, and TRPV3 activation induces the release of SERPINE1 from the primary keratinocyte (Sun et al., 2017). As previously reported, intradermal injection of SerpinE1 induced an itch-like behavior in AD mouse model, and its antagonist ameliorated itch, implicating its role in AD patients with chronic itch. In addition, SERPINE1 is involved in the PLAUR-TLR2-OSM pathway, which induces AD-like skin lesions in mice and promotes chronic pruritus. PAI-1 promotes fibrosis through the lytic activation of tissue plasminogen activator (t-PA) and urokinase plasminogen activator (u-PA). Elevated expression of SERPINE1 temporally and spatially plays a role in proteolytic activity, facilitating the invasive potential of squamous cell carcinoma, and is involved in wound healing, cell migration, cell proliferation, and other processes. PAI-1 is increasingly expressed in the skin, especially the epidermis and microvessel endothelium, in patients with systemic sclerosis. PAI-1 neutralization can induce plasmin-mediated metalloproteinase 1 activation and then reduce the initial vascular injury and subsequent tissue inflammation, leading to the improvement of skin fibrosis.
In addition, PAI-1 upregulates the intercellular adhesion molecule type 1 in skin fibroblasts and binds to mast cells. The fibroblast–mast cell interactions result in the activation of the adherent mast cells, which secret cytokines compromising IL-4 and IL-13, contributing to fibrogenesis. These studies highlight the potential role of PAI-1 in fibrosis-related diseases such as systemic sclerosis, wound healing, et al.
SERPINB13 encodes the HURPIN protein, which comprises 391 amino acids. In normal human skin, SERPINB13 is expressed in the basal layer. However, in disease lesions like AD, psoriasis, and squamous cell carcinoma, its increased expression is observed in the spinous and granular layers. HURPINspecifically inhibited cathepsins K and L with a serpin-like mechanism. Through differential gene expression analysis, researchers have observed the downregulation of SERPINB13 after UVB irradiation of HaCaT cells and SERPINB13 can interfere with UVB-induced keratinocyte apoptosis by interacting with cathepsinL. Deletion of SERPINB13 serves to induce the overactivation of CTSL (Welss et al., 2003), which generates caspase 3 and thus contributes to apoptosis. In addition, SERPINB3 is also designated as the differentiation marker in psoriasis, squamous cell carcinoma, and other skin diseases.
6 The role of SERPINs in other skin diseases
Generalized pustular psoriasis(GPP) is a pustular autoinflammatory skin disease characterized by acute generalized erythema and scaling with numerous aseptic pustules. It shares a similar skin manifestation with the autoimmune disease adult-onset immunodeficiency (AOID), which is induced by the overproduction and overresponse of IFN-γ. AOID is characterized by patients always having high titers of anti-IFN-γ autoantibodies, strongly correlated with higher severity.
Numerous gene mutations have been identified to cause the AOID and GPP, including IL36RN, CARD14, IL1RN, AP1S3, MPO, TNIP1, and SERPINs. Variants in SERPIN genes resulting in AOID and GPP indicate that the function of SERPINs is involved in the pathogenesis of these diseases, and genetic investigation is necessary for further diagnosis and treatment of these two diseases.
SERPINA1 encodes a protein comprising 418 amino acids. Two unrelated patients were reported, one with adult-onset immunodeficiency syndrome (AOID) and a pustular skin reaction and the other with GPP, both of whom carried a heterozygous missense SERPINA1 mutation, c.718G>A (p.Val240Met) (Kantaputra P. et al., 2021). SERPINA1, one significant member of the serine protease family, plays a vital role in proteolytic activity. The SERPINA1 mutation leads to reduced inhibition of elastase and cathepsin G, resulting in subsequent amplification of inflammatory reactions with neutrophil recruitment. Besides inhibiting protease activity, SERPINA1 can also regulate neutrophil-driven autoimmunity. Deficiency of SERPINA1 function has been determined to increase the level of neutrophilic TNF-α, resulting in neutrophil degranulation. Collectively, these mechanisms could explain the pathogenesis of the SERPINA1 mutation leading to AOIDand GPP, respectively. SERPINA1 shares almost 40%–45% of its amino acids and inhibitory functions with SERPINA3, which is also associated with AOIDand GPP.
SERPINA3 encodes alpha-1-antichymotrypsin (ACT), comprising 423 amino acids. The RCL region of SERPINA3 encompasses amino acid sites 378–395. SERPINA3 inhibits cathepsin G and chymotrypsin. SERPINA3 c.966delT(p.Tyr322Ter) was determined to be the cause of GPP patients through nonsense-mediated mRNA decay and therefore haploinsufficiency of SERPINA3 (Kantaputra P. N. et al., 2021). The loss of SERPINA3 inhibitory function causes the overactivation of neutrophilic serine proteases, especially cathepsin G. In addition, four rare variants of SERPINA3 resulting in skin pustules and adult-onset immunodeficiency have been reported, including c.684G>C, c.923T>C, c.1240A>G, and c.1246_1247del. The first three are missense mutations, and the last one is a deletion mutation. The inhibitory effect of SERPINA3 with loss-of-function variants on cathepsin G decreased, contributing to the GPP.
In 2023, Kantaputra P. N. et al. (2021) reported that the heterozygous missense variant SERPINA1 c.438G>T (p.Lys146Asn) was determined in two patients with AOID and GPP, respectively. In addition, SERPINA3 c.917A>G (p.Asp306Gly) is also reported with AOID (Kantaputra P. et al., 2021). Immunohistochemistry shows increased expression of SERPINA1 and SERPINA3 in the skin of patients, compared to a lower level in normal people. Due to the same causing gene of SERPINA3, GPP and AOID appear to share pathogenetic mechanisms. The main substrate of SERPINA3 is cathepsin L, which can inactivate SERPINA1 and lead to the subsequent inflammatory response and autoimmune reaction.
SERPINB6 encodes a protein containing 376 amino acids, which is namedplasminogen activator inhibitor 2 (PAI-2). Plasminogen activator inhibitor type-2 (PAI-2) co-activates tissue plasminogen activator (t-PA) and urokinase plasminogen activator (u-PA). It also plays a role in the pathogenesis of wound healing, cell migration, cell proliferation, and other processes. In 2007, Scott et al. (2007) discovered that SERPINB6 is co-expressed with KLK8 in the basal layer of human skin. Co-immunoprecipitation experiments confirmed that SERPINB6 can bind to KLK8, forming a complex that inhibits KLK8 protease activity. In 2010, Sirmaci et al. (2010) reported a patient with sensorineural deafness due to a SERPINB6 p.E245X (c.733G>T) mutation. It's worth noting that skin diseases caused by SERPINB6 mutations have not been reported.
7 Systematic evaluation of potential adverse effects associated with SERPIN modulation
Although targeting SERPINs represents a promising therapeutic strategy in skin disorders characterized by dysregulated protease activity, chronic inflammation, or impaired barrier repair, growing evidence suggests that modulation of SERPIN activity may also carry unintended consequences. These adverse effects can arise due to the pleiotropic and context-dependent functions of SERPINs across tissues and cell types (Janciauskiene et al., 2024).
For instance, overexpression of SERPINE1, while beneficial in limiting matrix degradation and protease-driven inflammation, has been implicated in pathological fibrosis, thrombotic complications, and delayed tissue remodeling (Ghosh and Vaughan, 2012; Flevaris and Vaughan, 2017). In animal models, PAI-1 overactivity led to excessive extracellular matrix accumulation and impaired wound healing, raising concerns about potential pro-fibrotic side effects in clinical settings. Similarly, systemic administration of SERPINA1 is under clinical investigation for inflammatory diseases, but its broad immunosuppressive effects may increase the risk of infections or impair host defense mechanisms, especially in the skin where microbial surveillance is critical (Stockley and Turner, 2014; Greene et al., 2016).
Conversely, suppression or genetic deficiency of certain SERPINs may exacerbate tissue injury. For example, loss of SERPINB3/4 in mice has been associated with heightened sensitivity to oxidative stress and amplified inflammatory cytokine responses (Wang et al., 2022). Given that some SERPINs play a protective role against apoptosis and oxidative damage, their inhibition may lead to collateral tissue injury, particularly under conditions of chronic inflammation or environmental stress such as UV exposure.
Importantly, SERPINs often participate in complex protease networks involving serine, cysteine, and metalloproteinases. Perturbing this delicate balance through exogenous modulation—such as gene therapy, recombinant protein administration, or small-molecule inhibitors—may disrupt homeostatic protease-antiprotease interactions, potentially leading to paradoxical effects like unrestrained proteolysis, impaired keratinocyte differentiation, or abnormal angiogenesis (Gettins and Olson, 2009). Moreover, the effects of SERPINs modulation may vary across skin compartments (e.g., epidermis vs dermis) and between physiological states (e.g., healthy vs inflamed skin), emphasizing the need for tissue-specific delivery systems and careful dose titration.
Therefore, while SERPINs constitute attractive therapeutic targets, particularly in diseases such as atopic dermatitis, psoriasis, and chronic wounds, a comprehensive risk-benefit analysis is crucial. Future research should prioritize in vivo safety studies using tissue-specific knockout or overexpression models, alongside rigorous pharmacokinetic and toxicological assessments of SERPINs-based therapeutics. Only through such systematic evaluation can the clinical potential of SERPINs modulation be realized while minimizing unintended harm.
8 Current progress and challenges in SERPIN-Targeted drug development
In recent years, SERPINs have emerged as promising therapeutic targets due to their central roles in regulating protease activity, inflammation, and tissue remodeling. Drug development efforts targeting SERPINs fall into several categories: recombinant SERPINs protein replacement (e.g., α1-antitrypsin for AAT deficiency), gene therapy to restore or silence SERPINs expression, small molecule modulators, and more recently, RNA-based therapeutics such as siRNA or antisense oligonucleotides (Cohen et al., 2019).
Recombinant SERPINA1 is among the most clinically advanced examples, approved for intravenous infusion in patients with genetic AAT deficiency-associated emphysema. However, its broader application in inflammatory skin diseases remains investigational (Yan et al., 2023). Other candidates, such as SERPINE1 inhibitors (e.g., tiplaxtinin/PAI-039), have shown promise in preclinical models of fibrosis and thrombosis, although translation to dermatological indications has yet to be explored.
Despite encouraging progress, multiple challenges hinder the advancement of SERPIN-targeted therapies. A key concern is specificity: many SERPINs exhibit context-dependent and cell-type-specific functions, making broad inhibition or overexpression potentially harmful (Chen et al., 2025). Additionally, recombinant SERPINs are often large and unstable proteins, complicating delivery and limiting tissue penetration. For gene therapies, viral vectors (e.g., AAV) pose immunogenicity risks and require long-term safety validation (Kryvalap and Czyzyk, 2022). Small molecule modulators of SERPINs activity remain scarce, largely due to the structural complexity of SERPINs and the allosteric nature of their regulation. Furthermore, off-target effects and protease network compensation can undermine therapeutic efficacy.
The development of skin-targeted delivery systems (e.g., nanoparticles, microneedles, or topical formulations) and engineered SERPINs variants with improved stability or selectivity are actively under investigation (Wu et al., 2024; Kelly-Robinson et al., 2021). Another promising direction lies in modulating SERPINs expression through epigenetic or transcriptional control, which may allow finer, disease-state-specific intervention. Ultimately, translating SERPIN biology into effective dermatological therapeutics will require a multidisciplinary approach integrating molecular design, bioengineering, pharmacokinetics, and clinical trial innovation.
9 Conclusion
Serine protease inhibitors are critical regulators of proteolytic activity in the skin, playing essential roles in maintaining epidermal integrity, modulating inflammation, and contributing to barrier homeostasis. This review has synthesized current evidence linking mutations in specific SERPIN genes with distinct dermatological phenotypes, including ichthyosis, palmoplantar keratoderma, and psoriasis-like conditions. Beyond genetic associations, we highlighted how SERPINs influence key signaling pathways (e.g., NF-κB, MAPK, JAK/STAT) and interact with protease networks such as MMPs to regulate keratinocyte behavior and immune responses.
Although several SERPINs have emerged as potential therapeutic targets, drug development remains challenged by issues of specificity, delivery, and off-target effects. Furthermore, in vivo studies and transcriptomic analyses reveal cell- and context-specific expression patterns that underscore the complexity of their biological roles in skin physiology and pathology. A deeper understanding of SERPIN-regulated pathways and genotype–phenotype correlations will be essential for translating this knowledge into clinical interventions. Future work should also address the functional consequences of rare variants, clarify the pathogenic mechanisms of SERPIN dysregulation, and explore their potential as diagnostic or therapeutic biomarkers in skin diseases.
Author contributions
ZX: Writing – original draft. YK: Writing – original draft. YZ: Writing – original draft. RL: Writing – original draft. YT: Writing – original draft, Writing – review and editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aslam, M. S., and Yuan, L. (2020). Serpina3n: potential drug and challenges, mini review. J. Drug Target 28 (4), 368–378. doi:10.1080/1061186X.2019.1693576
Betschel, S. D., Banerji, A., Busse, P. J., Cohn, D. M., and Magerl, M. (2023). Hereditary angioedema: a review of the current and evolving treatment landscape. J. Allergy Clin. Immunol. Pract. 11 (8), 2315–2325. doi:10.1016/j.jaip.2023.04.017
Biao, T., Cai-Feng, H., Xiao-Hong, L., Xiao-Li, C., Wen-Bei, L., Jun, W., et al. (2022). From bowen disease to cutaneous squamous cell carcinoma: eight markers were verified from transcriptomic and proteomic analyses. J. Transl. Med. 20 (1), 416. doi:10.1186/s12967-022-03622-1
Blaydon, D. C., Nitoiu, D., Eckl, K.-M., Cabral, R. M., Bland, P., Hausser, I., et al. (2011). Mutations in CSTA, encoding cystatin A, underlie exfoliative ichthyosis and reveal a role for this protease inhibitor in cell-cell adhesion. Am. J. Hum. Genet. 89 (4), 564–571. doi:10.1016/j.ajhg.2011.09.001
Brandt, E., Harjama, L., Elomaa, O., Saarela, J., Donner, K., Lappalainen, K., et al. (2024). A novel SERPINA12 variant and first European patients with diffuse palmoplantar keratoderma. J. Eur. Acad. Dermatol Venereol. 38 (2), 413–418. doi:10.1111/jdv.19498
Busse, P. J., and Christiansen, S. C. (2020). Hereditary angioedema. N. Engl. J. Med. 382 (12), 1136–1148. doi:10.1056/NEJMra1808012
Chen, T.-Y., Zhou, M., Lin, M.-Q., Liang, S. T., Yan, Y., Wang, S. M., et al. (2021). Research progress on the SERPINE1 protein and chronic inflammatory diseases of the upper respiratory tract: a literature review. Int. Arch. Allergy Immunol. 182 (11), 1097–1102. doi:10.1159/000516195
Chen, J., Wang, Z., Wang, S., Lyu, J., Fang, Z., Qi, W., et al. (2025). Probing the familial ties between serpin members kallistatin and PEDF: a comparative analysis review. Life Sci. 362, 123333. doi:10.1016/j.lfs.2024.123333
Chua, R., Setzer, S., Govindarajan, B., Sexton, D., Cohen, C., and Arbiser, J. L. (2009). Maspin expression, angiogenesis, prognostic parameters, and outcome in malignant melanoma. J. Am. Acad. Dermatol 60 (5), 758–766. doi:10.1016/j.jaad.2009.01.018
Cohen, M., Davydov, O., and Fluhr, R. (2019). Plant serpin protease inhibitors: specificity and duality of function. J. Exp. Bot. 70 (7), 2077–2085. doi:10.1093/jxb/ery460
De Veer, S. J., Furio, L., Harris, J. M., and Hovnanian, A. (2014). Proteases: common culprits in human skin disorders. Trends Mol. Med. 20 (3), 166–178. doi:10.1016/j.molmed.2013.11.005
Flevaris, P., and Vaughan, D. (2017). The role of plasminogen activator inhibitor Type-1 in fibrosis. Semin. Thromb. Hemost. 43 (2), 169–177. doi:10.1055/s-0036-1586228
Gettins, P. G. W. (2002). Serpin structure, mechanism, and function. Chem. Rev. 102 (12), 4751–4804. doi:10.1021/cr010170+
Gettins, P. G. W., and Olson, S. T. (2009). Exosite determinants of serpin specificity. J. Biol. Chem. 284 (31), 20441–20445. doi:10.1074/jbc.R800064200
Ghosh, A. K., and Vaughan, D. E. (2012). PAI-1 in tissue fibrosis. J. Cell Physiol. 227 (2), 493–507. doi:10.1002/jcp.22783
Greene, C. M., Marciniak, S. J., Teckman, J., Ferrarotti, I., Brantly, M. L., Lomas, D. A., et al. (2016). α1-Antitrypsin deficiency. Nat. Rev. Dis. Prim. 2, 16051. doi:10.1038/nrdp.2016.51
Hannula-Jouppi, K., Harjama, L., Einarsdottir, E., Elomaa, O., Kettunen, K., Saarela, J., et al. (2020). Nagashima-type palmoplantar keratosis in Finland caused by a SERPINB7 founder mutation. J. Am. Acad. Dermatol 83 (2), 643–645. doi:10.1016/j.jaad.2019.11.004
Haslund, D., Ryø, L. B., Seidelin, M. S., Rose, I., Skipper, K. A., Fryland, T., et al. (2019). Dominant-negative SERPING1 variants cause intracellular retention of C1 inhibitor in hereditary angioedema. J. Clin. Invest 129 (1), 388–405. doi:10.1172/JCI98869
Heiker, J. T., Klöting, N., Kovacs, P., Kuettner, E. B., Sträter, N., Schultz, S., et al. (2013). Vaspin inhibits kallikrein 7 by serpin mechanism. Cell Mol. Life Sci. 70 (14), 2569–2583. doi:10.1007/s00018-013-1258-8
Hida, K., Wada, J., Eguchi, J., Zhang, H., Baba, M., Seida, A., et al. (2005). Visceral adipose tissue-derived serine protease inhibitor: a unique insulin-sensitizing adipocytokine in obesity. Proc. Natl. Acad. Sci. U. S. A. 102 (30), 10610–10615. doi:10.1073/pnas.0504703102
Huntington, J. A. (2011). Serpin structure, function and dysfunction. J. Thromb. Haemost. 9 (Suppl. 1), 26–34. doi:10.1111/j.1538-7836.2011.04360.x
Janciauskiene, S., Lechowicz, U., Pelc, M., Olejnicka, B., and Chorostowska-Wynimko, J. (2024). Diagnostic and therapeutic value of human serpin family proteins. Biomed. Pharmacother. 175, 116618. doi:10.1016/j.biopha.2024.116618
Kantaputra, P., Chaowattanapanit, S., Kiratikanon, S., Chaiwarith, R., Choonhakarn, C., Intachai, W., et al. (2021). SERPINA1, generalized pustular psoriasis, and adult-onset immunodeficiency. J. Dermatol 48 (10), 1597–1601. doi:10.1111/1346-8138.16081
Kantaputra, P. N., Chuamanochan, M., Kiratikanon, S., Chiewchanvit, S., Chaiwarith, R., Intachai, W., et al. (2021). A truncating variant in SERPINA3, skin pustules and adult-onset immunodeficiency. J. Dermatol 48 (8), e370–e371. doi:10.1111/1346-8138.15942
Katagiri, C., Nakanishi, J., Kadoya, K., and Hibino, T. (2006). Serpin squamous cell carcinoma antigen inhibits UV-induced apoptosis via suppression of c-JUN NH2-terminal kinase. J. Cell Biol. 172 (7), 983–990. doi:10.1083/jcb.200508064
Kelly-Robinson, G. A., Reihill, J. A., Lundy, F. T., McGarvey, L. P., Lockhart, J. C., Litherland, G. J., et al. (2021). The serpin superfamily and their role in the regulation and dysfunction of serine protease activity in COPD and other chronic lung diseases. Int. J. Mol. Sci. 22 (12), 6351. doi:10.3390/ijms22126351
Kishibe, M. (2019). Physiological and pathological roles of kallikrein-related peptidases in the Epidermis. J. Dermatol Sci. 95 (2), 50–55. doi:10.1016/j.jdermsci.2019.06.007
Kryvalap, Y., and Czyzyk, J. (2022). The role of proteases and serpin protease inhibitors in β-Cell biology and diabetes. Biomolecules 12 (1), 67. doi:10.3390/biom12010067
Kubo, A., Shiohama, A., Sasaki, T., Nakabayashi, K., Kawasaki, H., Atsugi, T., et al. (2013). Mutations in SERPINB7, encoding a member of the serine protease inhibitor superfamily, cause Nagashima-type palmoplantar keratosis. Am. J. Hum. Genet. 93 (5), 945–956. doi:10.1016/j.ajhg.2013.09.015
Kurowska, P., Mlyczyńska, E., Dawid, M., Jurek, M., Klimczyk, D., Dupont, J., et al. (2021). Review: Vaspin (SERPINA12) expression and function in endocrine cells. Cells 10 (7), 1710. doi:10.3390/cells10071710
Law, R. H. P., Zhang, Q., McGowan, S., Buckle, A. M., Silverman, G. A., Wong, W., et al. (2006). An overview of the serpin superfamily. Genome Biol. 7 (5), 216. doi:10.1186/gb-2006-7-5-216
Levi, M., Cohn, D. M., and Zeerleder, S. (2019). Hereditary angioedema: linking complement regulation to the coagulation system. Res. Pract. Thromb. Haemost. 3 (1), 38–43. doi:10.1002/rth2.12175
Li, S., Wu, Y., Bu, D., Hu, L., Liu, Y., Liu, J., et al. (2024). SERPINB7 deficiency increases legumain activity and impairs the epidermal barrier in nagashima-type palmoplantar keratoderma. J. Invest Dermatol 145, 359–369.e8. doi:10.1016/j.jid.2024.05.025
Liu, Y., Tan, Y., Liu, J., Song, Z., Hu, L., Mo, R., et al. (2022). Novel and founder variants of SERPINA12 in Chinese patients with autosomal recessive palmoplantar keratoderma. Br. J. Dermatol 187 (2), 267–270. doi:10.1111/bjd.21064
Liu, J., Chen, Z., Hu, L., Song, Z., Mo, R., Tsang, L. S. L., et al. (2023). Investigation of Nagashima-type palmoplantar keratoderma in China: a cross-sectional study of 234 patients. J. Dermatol 50 (3), 375–382. doi:10.1111/1346-8138.16621
Liu, Y., Liu, J., Chen, Y., Mo, R., Xiang, R., Song, Z., et al. (2024). Potential digenic inheritance of SERPINB7 and SERPINA12 variants in Chinese patients with Nagashima-type palmoplantar keratosis. Br. J. Dermatol 191 (1), 136–138. doi:10.1093/bjd/ljae134
Maas, C., and de Maat, S. (2021). Therapeutic SERPINs: improving on nature. Front. Cardiovasc Med. 8, 648349. doi:10.3389/fcvm.2021.648349
Martinoli, C., Gandini, S., Luise, C., Mazzarol, G., Confalonieri, S., Giuseppe Pelicci, P., et al. (2014). Maspin expression and melanoma progression: a matter of sub-cellular localization. Mod. Pathol. 27 (3), 412–419. doi:10.1038/modpathol.2013.157
Miyata, T., Nangaku, M., Suzuki, D., Inagi, R., Uragami, K., Sakai, H., et al. (1998). A mesangium-predominant gene, megsin, is a new serpin upregulated in IgA nephropathy. J. Clin. Invest 102 (4), 828–836. doi:10.1172/JCI2450
Miyata, T., Inagi, R., Nangaku, M., Imasawa, T., Sato, M., Izuhara, Y., et al. (2002). Overexpression of the serpin megsin induces progressive mesangial cell proliferation and expansion. J. Clin. Invest 109 (5), 585–593. doi:10.1172/JCI14336
Mohamad, J., Sarig, O., Malki, L., Rabinowitz, T., Assaf, S., Malovitski, K., et al. (2020). Loss-of-Function variants in SERPINA12 underlie autosomal recessive palmoplantar keratoderma. J. Invest Dermatol 140 (11), 2178–2187. doi:10.1016/j.jid.2020.02.030
Morizane, S., Sunagawa, K., Nomura, H., and Ouchida, M. (2022). Aberrant serine protease activities in atopic dermatitis. J. Dermatol Sci. 107 (1), 2–7. doi:10.1016/j.jdermsci.2022.06.004
Ohguchi, Y., Nomura, T., Suzuki, S., Takeda, M., Miyauchi, T., Mizuno, O., et al. (2018). Gentamicin-induced readthrough and nonsense-mediated mRNA decay of SERPINB7 nonsense mutant transcripts. J. Invest Dermatol 138 (4), 836–843. doi:10.1016/j.jid.2017.10.014
Ohtomo, S., Nangaku, M., Izuhara, Y., Yamada, N., Dan, T., Mori, T., et al. (2008). The role of megsin, a serine protease inhibitor, in diabetic mesangial matrix accumulation. Kidney Int. 74 (6), 768–774. doi:10.1038/ki.2008.302
Pasmooij, A. M. G. (2018). Topical gentamicin for the treatment of genetic skin diseases. J. Invest Dermatol 138 (4), 731–734. doi:10.1016/j.jid.2017.12.008
Pigors, M., Sarig, O., Heinz, L., Plagnol, V., Fischer, J., Mohamad, J., et al. (2016). Loss-of-Function mutations in SERPINB8 linked to exfoliative ichthyosis with impaired mechanical stability of intercellular adhesions. Am. J. Hum. Genet. 99 (2), 430–436. doi:10.1016/j.ajhg.2016.06.004
Ren, C., Liu, Q., Ma, Y., Wang, A., Yang, Y., and Wang, D. (2020). TEAD4 transcriptional regulates SERPINB3/4 and affect crosstalk between keratinocytes and T cells in psoriasis. Immunobiology 225 (5), 152006. doi:10.1016/j.imbio.2020.152006
Ribero, S., Senetta, R., Osella-Abate, S., Scalzo, M. S., Castellano, I., Lentini, F., et al. (2017). Prognostic role of maspin expression in melanoma: probably far from clinical use. Histopathology 71 (1), 158–162. doi:10.1111/his.13188
Rusiñol, L., and Puig, L. (2024). Multi-omics approach to improved diagnosis and treatment of atopic dermatitis and psoriasis. Int. J. Mol. Sci. 25 (2), 1042. doi:10.3390/ijms25021042
Saalbach, A., Vester, K., Rall, K., Tremel, J., Anderegg, U., Beck-Sickinger, A. G., et al. (2012). Vaspin--a link of obesity and psoriasis? Exp. Dermatol 21 (4), 309–312. doi:10.1111/j.1600-0625.2012.01460.x
Santacroce, R., D'Andrea, G., Maffione, A. B., Margaglione, M., and d'Apolito, M. (2021). The genetics of hereditary angioedema: a review. J. Clin. Med. 10 (9), 2023. doi:10.3390/jcm10092023
Scott, F. L., Sun, J., Whisstock, J. C., Kato, K., and Bird, P. I. (2007). SerpinB6 is an inhibitor of kallikrein-8 in keratinocytes. J. Biochem. 142 (4), 435–442. doi:10.1093/jb/mvm156
Shimomura, Y. (2024). SERPINA12-associated palmoplantar keratoderma May be prevalent across different populations. J. Eur. Acad. Dermatol Venereol. 38 (2), 247–248. doi:10.1111/jdv.19628
Sinnathamby, E. S., Issa, P. P., Roberts, L., Norwood, H., Malone, K., Vemulapalli, H., et al. (2023). Hereditary angioedema: diagnosis, clinical implications, and pathophysiology. Adv. Ther. 40 (3), 814–827. doi:10.1007/s12325-022-02401-0
Sirmaci, A., Erbek, S., Price, J., Huang, M., Duman, D., Cengiz, F. B., et al. (2010). A truncating mutation in SERPINB6 is associated with autosomal-recessive nonsyndromic sensorineural hearing loss. Am. J. Hum. Genet. 86 (5), 797–804. doi:10.1016/j.ajhg.2010.04.004
Sivaprasad, U., Kinker, K. G., Ericksen, M. B., Lindsey, M., Gibson, A. M., Bass, S. A., et al. (2015). SERPINB3/B4 contributes to early inflammation and barrier dysfunction in an experimental murine model of atopic dermatitis. J. Invest Dermatol 135 (1), 160–169. doi:10.1038/jid.2014.353
Steele, L., Tawfik, S. S., and O'Toole, E. A. (2020). The proteolytic network in palmoplantar keratoderma: SERPINA12 joins the family. J. Invest Dermatol 140 (11), 2111–2113. doi:10.1016/j.jid.2020.06.031
Stockley, R. A., and Turner, A. M. (2014). α-1-Antitrypsin deficiency: clinical variability, assessment, and treatment. Trends Mol. Med. 20 (2), 105–115. doi:10.1016/j.molmed.2013.11.006
Sun, Y., Sheshadri, N., and Zong, W.-X. (2017). SERPINB3 and B4: from biochemistry to biology. Semin. Cell Dev. Biol. 62, 170–177. doi:10.1016/j.semcdb.2016.09.005
Ulbricht, D., Tindall, C. A., Oertwig, K., Hanke, S., Sträter, N., and Heiker, J. T. (2018). Kallikrein-related peptidase 14 is the second KLK protease targeted by the serpin vaspin. Biol. Chem. 399 (9), 1079–1084. doi:10.1515/hsz-2018-0108
Wang, S., Luke, C. J., Pak, S. C., Shi, V., Chen, L., Moore, J., et al. (2022). SERPINB3 (SCCA1) inhibits cathepsin L and lysoptosis, protecting cervical cancer cells from chemoradiation. Commun. Biol. 5 (1), 46. doi:10.1038/s42003-021-02893-6
Wang, S., Yang, Z., Liu, Y., Zhang, H., Liu, Z., Wang, X., et al. (2023). Application of topical gentamicin ointment in the treatment of Nagashima-type palmoplantar keratosis in children with a nonsense mutation. Pediatr. Investig. 7 (3), 163–167. doi:10.1002/ped4.12389
Weiner, J., Zieger, K., Pippel, J., and Heiker, J. T. (2019). Molecular mechanisms of vaspin action - from adipose tissue to skin and bone, from blood vessels to the brain. Adv. Exp. Med. Biol. 1111, 159–188. doi:10.1007/5584_2018_241
Welss, T., Sun, J., Irving, J. A., Blum, R., Smith, A. I., Whisstock, J. C., et al. (2003). Hurpin is a selective inhibitor of lysosomal cathepsin L and protects keratinocytes from ultraviolet-induced apoptosis. Biochemistry 42 (24), 7381–7389. doi:10.1021/bi027307q
Wu, S., Yang, Y., Zhang, M., Khan, A. U., Dai, J., and Ouyang, J. (2024). Serpin peptidase inhibitor, clade E, member 2 in physiology and pathology: recent advancements. Front. Mol. Biosci. 11, 1334931. doi:10.3389/fmolb.2024.1334931
Yan, B., Luo, L., Liu, L., Wang, Z., Chen, R., Wu, Y., et al. (2023). Serpin family proteins as potential biomarkers and therapeutic drugs in stroke: a systematic review and meta-analysis on clinical/preclinical studies. CNS Neurosci. Ther. 29 (7), 1738–1749. doi:10.1111/cns.14205
Yang, S.-F., Lin, M.-H., Chou, P.-C., Hu, S. K., Shih, S. Y., Yu, H. S., et al. (2023). Genetics of generalized pustular psoriasis: current understanding and implications for future therapeutics. Genes (Basel) 14 (6), 1297. doi:10.3390/genes14061297
Keywords: serpins, reactive center loop, dermatoses, palmoplantar keratoderma, therapeutic strategies
Citation: Xiao Z, Kang Y, Zhuo Y, Li R and Tan Y (2025) The roles of serine protease inhibitors in dermatoses. Front. Genet. 16:1624512. doi: 10.3389/fgene.2025.1624512
Received: 07 May 2025; Accepted: 09 September 2025;
Published: 03 October 2025.
Edited by:
Rogério Saad Vaz, Federal University of Paraná, BrazilReviewed by:
Yuhang Liu, Children’s Hospital of Chongqing Medical University, ChinaHans Christian Hennies, Staffordshire University, United Kingdom
Copyright © 2025 Xiao, Kang, Zhuo, Li and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rui Li, MjkyNDMwMzg0QHFxLmNvbQ==; Yingjian Tan, RHIueWluZ2ppYW4udGFuQGdtYWlsLmNvbQ==
†These authors have contributed equally to this work
 Zhenzhen Xiao
Zhenzhen Xiao Yue Kang
Yue Kang Yunqian Zhuo
Yunqian Zhuo Rui Li1*
Rui Li1* Yingjian Tan
Yingjian Tan