- 1Hoplite Research Lab, Department of Pathology and Microbiology, Atlantic Veterinary College, University of Prince Edward Island, Charlottetown, PE, Canada
- 2Huntsman Marine Science Centre, St. Andrews, NB, Canada
- 3Department of Infectious Diseases and Public Health, Jockey Club College of Veterinary Medicine and Life Sciences, City University of Hong Kong, Hong Kong, Hong Kong SAR, China
- 4Onda, Souris, PE, Canada
Sea lice (Lepeophtheirus salmonis) infestation continues to pose a persistent and escalating challenge to the global salmon aquaculture industry. Given the complexity of host-parasite interactions, family-based transcriptomic studies provide crucial insights into genetic variation in host responses to sea lice, potentially guiding the development of selective breeding programs to manage parasite resistance in Atlantic salmon. This study investigated global gene expression (transcriptomic) responses of the skin and head kidney of Atlantic salmon (Salmo salar) from different families following infestation at two distinct stages of sea lice, chalimus II and adult, under varying temperature conditions (10°C and 20°C). RNA sequencing results revealed consistent expression of lice-responsive genes across different families under varying thermal conditions, which allowed the identification of potential biomarkers associated with adult-stage compared to chalimus-stage infestations. Our findings highlight critical physiological disruptions in salmon infested with advanced (adult) stages of lice, including uncontrolled and persistent inflammation, dampened/dysregulated immune responses, and impaired tissue repair at attachment sites. This study provides a comprehensive analysis of the transcriptomic responses of Atlantic salmon to different developmental stages of sea lice under specific temperature conditions (10°C and 20°C), and identifies several novel molecular markers from RNA-seq analysis that may be instrumental in developing targeted control strategies for this economically important parasite.
1 Introduction
Atlantic salmon (Salmo salar) aquaculture has witnessed remarkable growth over the past few decades, becoming a critical contributor to global seafood production (FAO, 2024). This growth, however, has been accompanied by persistent challenges that threaten the sustainability and economic viability of the industry. The ectoparasitic Lepeophtheirus salmonis, commonly known as the salmon louse, is currently the most significant threat to the health and productivity of Atlantic salmon farming (Whyte et al., 2014; Ugelvik and Dalvin, 2022). The economic impact of sea lice infestations on the global salmon farming industry is estimated to exceed $1 billion annually, encompassing reduced growth performance, expenses associated with treatment and prevention strategies, and direct losses through morbidity (Barker, et al., 2019). Sea lice infestation has also raised significant concerns regarding welfare of infested fish on the farm due to practices associated with delousing and potential ecological impacts on wild salmon populations adjacent to farming operations (Larsen and Vormedal, 2021).
The life cycle of L. salmonis comprises free-swimming planktonic stages (nauplius I, nauplius II) followed by the transitionary planktonic and first parasitic stage, the copepodid. Additional parasitic stages exist (chalimus I, chalimus II, pre-adult I, pre-adult II and adult) in which the parasite attaches to the host fish and feeds on the mucus, skin cells, and blood (Hamre et al., 2013). The chalimus stages are distinguished by a frontal filament that enables their secure attachment to the host, whereas the mobile pre-adult and adult stages are capable of moving across the fish’s surface, inflicting more substantial tissue damage through their aggregatory feeding activities (Fast and Braden, 2022). These various developmental stages exhibit stage-specific patterns of host-parasite interaction, with varying impacts on the host’s physiological and immunological responses (Dalvin et al., 2020; Øvergård et al., 2023). The progressive damage caused during sea lice development ultimately results in skin lesions, secondary infections, osmoregulatory disruption, and although rarer, mortality in heavily infested fish (Rodger et al., 2022).
Previous studies have revealed complex gene expression patterns that evolve throughout the course of infestation with sea lice and vary according to the parasite developmental stage (Sutherland et al., 2014; Tadiso et al., 2011). As the primary interface between the host and parasite, the salmon skin exhibits pronounced transcriptomic alterations following sea lice attachment (Krasnov et al., 2012; Sutherland et al., 2014; Holm et al., 2015; Robledo et al., 2018; Umasuthan et al., 2020; Caballero-Solares et al., 2022; Cai et al., 2022; Cai et al., 2024; Ugelvik and Dalvin, 2022; Øvergård et al., 2023). Local responses at mucosal surfaces, particularly the skin, involve upregulation of innate immune genes such as interleukins (e.g., il1b, il8), acute phase proteins, and antimicrobial peptides, often accompanied by inflammation and epithelial disruption (Skugor et al., 2008; Braden et al., 2015). These responses are typically more pronounced during the early chalimus stage, when the lice are attached to the epithelium, but they may be suppressed or dysregulated as the lice mature. In addition to localized responses at attachment sites, sea lice infestation induces systemic alterations, including modulation of immune markers, stress-related genes (e.g., heat shock proteins), and metabolic pathways, in internal tissues such as the head kidney, which functions as the primary hematopoietic organ in teleost fish (Skugor et al., 2008; Tadiso et al., 2011; Robledo et al., 2019; Øvergård et al., 2023; Valenzuela-Muñozet al., 2023; Zhong et al., 2023). Comparative transcriptomic studies have identified remarkable variation in response patterns between resistant and susceptible salmon species, providing insights into potential genetic determinants of resistance (Robledo et al., 2018; Robledo et al., 2019; Braden et al., 2023; Valenzuela-Muñoz et al., 2023; Cáceres et al., 2024; Salisbury et al., 2024). These findings hold significant implications for selective breeding programs focused on improving natural resistance to sea lice in farmed salmon populations.
In addition to the direct effects of sea lice on salmon, environmental stressors, particularly temperature fluctuations, can compound the impacts of sea lice on salmon health and immune function (Alfonso et al., 2021; Islam et al., 2022; Scharsack and Franke, 2022). Global warming and increased water temperatures are anticipated to reshape the dynamics of salmon aquaculture by simultaneously affecting fish physiology and sea lice biology (Hamre et al., 2019; Beemelmanns et al., 2021). Earlier research has indicated that elevated temperatures may exacerbate the severity of infectious diseases in Atlantic salmon, potentially altering salmon fitness, immune function and metabolic responses (Shi et al., 2019; Medcalf et al., 2021; Franke et al., 2024). In the context of L. salmonis, higher temperature has been shown to influence the parasite’s life cycle, development, and settlement success while modifying host-parasite interactions and efficacy of louse reduction, leading to enhanced infestation rates and more detrimental impacts on salmon health (Overton et al., 2019; Godwin et al., 2020; Ugelvik et al., 2022; Nilsson et al., 2023; Oldham et al., 2023). Furthermore, high temperatures can alter the stress signaling pathways and the expression of immune-related genes, potentially compromising the salmon’s immunological defenses against sea lice (Jensen et al., 2015; Nuez-Ortín et al., 2018).
The effects of infestation with different developmental stages of sea lice have been previously investigated on salmon individuals (infested versus healthy) using gene expression analyses (Skugor et al., 2008; Tadiso et al., 2011; Sutherland et al., 2014; Ugelvik and Dalvin, 2022; Øvergård et al., 2023). Prior studies suggest that systemic responses are often delayed or subdued, potentially reflecting immune evasion strategies employed by the parasite (Skugor et al., 2008; Tadiso et al., 2011; Ugelvik and Dalvin, 2022; Øvergård et al., 2023). These stage-specific and compartmentalized host responses underscore the complexity of the salmon-lice interaction and highlight the need for integrated analyses to identify meaningful biomarkers associated with the parasite’s developmental stages. However, transcriptomic differences between fish parasitized with chalimus and adult stages of L. salmonis have not been directly compared, particularly across multiple salmon families to identify consistently expressed genes associated with sea lice infestation. The present study therefore aims to remove inherent variation in this system by profiling the transcriptomic responses of the skin and head kidney of lice-infested Atlantic salmon at the family level under normal and elevated temperature conditions. The findings of this study are expected to significantly contribute to our understanding of the molecular mechanisms and biological pathways underlying the salmon’s response to sea lice infestation.
2 Materials and methods
2.1 Fish husbandry
The experimental population consisted of North American Atlantic salmon from 20 families (100 fish per family), produced from 16 sires and 16 dams from three different year classes in a larger breeding program with anticipated variable performance for sea lice resistance. All the smolts were bred and reared at the Huntsman Marine Science Centre (Huntsman Marine) in St. Andrews, New Brunswick, Canada. At approximately 2900–3000 degree days (dd, 16.5 g mean weight), all fish were anesthetized with tricaine methane sulphonate (MS-222, 100 mg/L) before insertion of a passive integrated transponder (PIT) tag to distinguish between the families. Fish were allowed to recover in oxygenated water, transferred to a single tank (7.5 m3) with additional PIT tagged salmon and reared in a semi-closed recirculating freshwater system with ambient light and water temperature maintained 1°C–3°C above ambient. Once fish had reached smolt-size (>50 g mean weight, ∼4476 dd), they were anesthetized (MS-222; 150 mg/L) and intraperitoneally vaccinated (100 µL/fish) with Micro Forte V II™ (multivalent vaccine containing inactivated ISA) and Renogen™ (Merck Animal Health, Canada). These fish were subsequently smolted remaining in a single 7.5 m3 tank.
Fish were fed a commercial diet (Skretting, Canada) using automatic feeders with ambient photoperiod. Using the post-smolt body mass and families, they were randomly distributed to 8 tanks (1.3 m3) according to their body mass and family groups at a stocking density of 120 fish per tank (6 fish per family, 20 families per tank). Fish were maintained at 6°C then 2°C for ∼6 months (19 March to 15 August 2020) due to COVID-19 shut down (constraints in sourcing sea lice for the challenge). At this point, the salmon were acclimated to ambient temperature from 16 August to 31 October 2020 (ranging from approximately 14°C in mid-August to 11°C in later October) and then gradually adjusted to 10°C ± 1°C for 10 days (01 November to 10 November 2020). After initial acclimation for 10 days, there was a daily increase of 1°C in the temperature of 4 out of the 8 tanks until it reached 20°C ± 1°C (11 November to 21 November 2020). Two temperature conditions were selected to investigate the transcriptomic responses of Atlantic salmon infested with adult versus chalimus stages of sea lice: 10°C, which falls within the species’ optimal physiological range, and 20°C, a temperature known to induce thermal stress (Beemelmanns et al., 2021). Lepeophtheirus salmonis has the potential to infest Atlantic salmon across a temperature range of approximately 6°C–22°C (Hamre et al., 2019; Godwin et al., 2020).
2.2 Lice challenge and sample collection
Sea lice (i.e., gravid females) were collected from Atlantic salmon reared in a marine cage site in the Bay of Fundy, New Brunswick, Canada. Egg strings were removed from the females and incubated in two incubators at 12.3°C for 8 days to reach the infective copepodid stage. On the 9th day (113.1 dd), the incubation temperature was raised to 15°C in both incubators and then adjusted by 1°C per hour to 10°C or 20°C in either incubator, corresponding to the temperatures in fish tanks. The copepodids were then used to infest the Atlantic salmon at 100 copepodids per fish for 1 h in each tank. The status of lice development was monitored throughout the experimental challenge. When the copepodids developed to the chalimus II larvae (110 dd), fish in half of the tanks (2 tanks per temperature) were subjected to biometric assessment and lice counting. The lice were allowed to develop to the adult stage (360 dd) in the remaining four tanks, after which the same procedure was conducted (2 tanks per temperature). Lice were counted on all body surfaces of fish including gills (removed for counting with a dissecting scope in the chalimus stage). All fish in each tank were included in lice counting (at individual sampling events) except mortalities, which occurred outside of those dates. Immediately following lice counting, two tissue samples (skin and head kidney) were collected from fish parasitized with intermediate/adult forms and preserved in RNAlater solution for further molecular analyses. Tissue samples were collected from the anterior head kidney on the right side and skin located immediately behind the dorsal fin (10 mm × 10 mm) of each Atlantic salmon. Lice abundance (lice per fish) was separately calculated for fish parasitized with chalimus or adult stages.
2.3 RNA extraction and library preparation
Portions of the tissues (∼30 mg) collected were each subjected to RNA extraction. Total RNA extraction was carried out using TRIzol-chloroform phase separation followed by DNase treatment and purification of the aqueous phase with the RNA Clean & Concentrator Kit (Zymo Research, CA, United States) according to the manufacturer’s instructions. The concentration and purity of isolated RNA samples were measured using NanoDrop spectrophotometry (ND 2000; Thermo Scientific), after which the RNA integrity was assessed initially by 1.5% agarose gel electrophoresis and subsequently using a Bioanalyzer (BioRad) (Cai et al., 2024). RNA samples with A260/A280 ≥ 1.9 and A260/A230 ≥ 1.7 were used for subsequent analyses. RNA samples from individuals of five families of different lice burden (F175, F265, F292, F361, F419) were selected for RNA sequencing in this study (3–5 samples per family). The RNA samples were standardized to 100 ng/μL and then used for RNA-Seq library construction using the TruSeq Stranded mRNA Library Prep Kit (Illumina, CA, United States) as recommended by manufacturer’s instructions. All libraries were sequenced (2 × 100 bp) by the Illumina NovaSeq 6000 S4 platform according to the standard Illumina protocol at Genome Québec (Montreal, Canada). The raw sequencing reads generated were deposited in Sequence Read Archive (SRA) database of NCBI (BioProject accession number: PRJNA1256531).
2.4 RNA sequencing data analysis
Quality control and preprocessing of the raw sequence (fastq) data were performed using FastQC v0.15.3. Residual adaptor sequences and low-quality bases were trimmed using Trimmomatic v0.36 (phred33, illuminaclip:TruSeq3-PE.fa:2:30:10, leading:3, trailing:3, slidingwindow:4:15 minlen:36). STAR v.2.7.9a was applied to map the trimmed reads to the latest Atlantic salmon genome assembly (Ssal_v3.1, GenBank accession no: GCF_905237065.1) and sort the obtained binary alignment map (BAM) files (Salisbury et al., 2024). In order to generate a count matrix from the estimated reads mapped to individual genes, featureCounts was used as part of the Subread package v1.6.5 (Liao et al., 2014). The edgeR package (classic approach) was used to perform pairwise comparisons between representatives of families parasitized with adult versus chalimus stages of sea lice (3–5 samples per family). The following workflow was implemented in edgeR: calcNormFactors (method = “TMM”) for normalization, estimateDisp () for dispersion estimation, and exactTest () for differential expression analysis. Genes were considered significantly differentially expressed based on the following thresholds: |log2-fold change| > 1.25, adjusted p-value <0.05 and |log-transformed counts per million reads| > 1.00. Moreover, interactions between differentially expressed genes (DEGs) obtained from pairwise comparisons were also investigated. Principal component analysis (PCA) was used using the prcomp function in R to identify variations among RNA-seq samples corresponding to each pairwise comparison. The RNA-seq datasets that were identified as outliers were excluded from downstream analyses. The hierarchical clustering of shared DEGs among different pairwise comparisons was performed using “pheatmap” function in R.
2.5 Gene ontology and KEGG enrichment analyses
Gene annotation was performed using the R package AnnotationHub (orgDb: S. salar, database no: AH114250), and gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses of fully annotated up- and downregulated DEGs were conducted using ClueGO plug-in (v2.5.10) in Cytoscape v3.10.2 to visualize the non-redundant biological terms in a functionally grouped network (Shannon et al., 2003; Bindea et al., 2009). The ClueGO enrichment analyses were implemented using the following options/adjustments: pathway p-value cut-off <0.05, kappa score: 0.4 and right-sided hypergeometric test with Benjamini–Hochberg p value correction. To reduce complexity, the GO term fusion option was applied in most comparisons. However, a non-fusion approach was used to generate a more comprehensive overview of leading and sub-leading GO terms associated with shared DEGs across pairwise comparisons. The leading GO terms coupled with all genes associated with each GO group were also extracted for further analyses. The enriched (over-represented) leading GO terms and KEGG pathways obtained for comparisons related to temperature (for both paired-related and shared DEGs) and infestation stage of lice (for only shared DEGs) were categorized into different functional themes based on their parent GO terms as well as DEGs and sub-leading GO terms and KEGG pathways involved in each GO group: 1) metabolism, protein interaction and gene regulation, 2) cellular and developmental dynamics, 3) signal transduction and stimulus response, 4) enzymatic functions, and 5) molecular localization and transport. Terms/pathways associated with metabolic (either anabolic or catabolic) processes/pathways, transcriptional and replication regulation, protein-containing complex and protein-protein interactions were grouped under the functional theme “metabolism, protein interaction and gene regulation.” The GO terms “organelle envelope” and “intracellular organelle,” which were interconnected with metabolism-related terms and involved genes associated with metabolic processes, were also classified under this functional theme. Terms/pathways related to cellular process, cellular anatomical structure, developmental process and tissue repair were categorized as ‘cellular and developmental dynamics’. The theme ‘signal transduction and stimulus response’ included terms/pathways related to cell signaling pathways and responses to various stimuli and stressors. The functional theme “enzymatic functions” encompassed terms/pathways correlated with catalytic enzyme activities, while terms/pathways involved in intracellular localization and molecular transport mechanisms were classified under “molecular localization and transport.” The gene ontology and KEGG databases used for grouping the leading GO terms included Gene Ontology Browser (https://www.informatics.jax.org), Gene Ontology and GO Annotations (https://www.ebi.ac.uk) and KEGG Pathway Database (https://www.genome.jp/kegg/pathway.html).
2.6 Quantitative real-time PCR (qPCR) assay
Samples (head kidney and skin) collected from representatives of 12 families (infested with adult or chalimus stages of lice at 10°C or 20°C) were included in qPCR analyses. Of these, representatives from four families were analyzed under both 10°C and 20°C conditions (i.e., F175, F229, F361, F419) and those of eight families were analyzed at either 10°C (F153, F154, F185, F265) or 20°C (F215, F292, F397, F415). For qPCR analyses, cluster of differentiation 83 (cd83), c-type lectin domain family 4 (clec4), collagen alpha-1(I) chain (col1a1), matrix metalloproteinase 9 (mmp9), neutrophil cytosolic factor 2 (ncf2), serum amyloid A5 (saa5), tissue inhibitor of metalloproteinase 2 (timp2) and thioredoxin b (txnb) were selected as genes of interest (GOIs) alongside three reference genes including eukaryotic translation initiation factor 3 subunit 6 (eif3), elongation factor 1 alpha (ef1a) and RNA polymerase I (rpl1) (Sutherland et al., 2014; Skugor et al., 2008; Todorčević et al., 2008; Leong et al., 2010; Braden et al., 2015; Braden et al., 2018; Chalmers et al., 2018; Carvalho et al., 2020; Xue et al., 2021). The selection of these genes was based on previous studies that demonstrated their relevance to host responses during sea lice infestation (Braden et al., 2015; Braden et al., 2018; Carvalho et al., 2020). The selected genes along with the primer sequences used in this study are listed in Supplementary Table S1.
A total of 1 µg of extracted RNA samples was used for synthesis of complementary DNA (cDNA, 20 µL reactions) using a commercially available kit (High Capacity cDNA Reverse Transcription Kit, Applied Biosystems, CA, United States) as per manufacturer’s instructions. The qPCR reactions were conducted on CFX96 Touch Real-Time PCR System (BioRad) in 12 µL reaction mixture comprised of 5 µL of Sso Advanced™ Universal SYBR® Green Supermix (Bio-Rad), 0.5 µL of 10 mM forward and reverse primers, 4 µL of nuclease-free water and 2 µL of cDNA template using the cycling conditions: 95°C for 30 s, 95°C for 15 s, 60°C for 15 s, for 39 cycles, followed by a dissociation curve analysis ramping from 65°C to 95°C with continuous fluorescence detection every 0.5 s with a ramp rate of 0.5°C. qPCR reactions were performed in technical triplicates for each sample, alongside a calibrator pool (an equimolar mix of all cDNA samples for each tissue type), and two control reactions: a no-template control (NTC) and a no-reverse transcriptase control (no-RT). Gene expression levels for all genes of interest (GOIs) were normalized to the geometric mean of three housekeeping genes (eif3, ef1a, rpl1), incorporating gene-specific amplification efficiencies. Relative quantification (RQ) values were calculated using the comparative Ct (ΔΔCt) method, with the calibrator pool serving as the reference sample (Bustin et al., 2009). RQ values were then log2-transformed and statistically compared between groups infested with adult and chalimus stages of sea lice.
2.7 Statistical analysis
The normality of lice abundance and gene expression data was checked using the Shapiro-Wilk test. One-way analysis of variance (ANOVA), followed by Tukey’s post hoc test, was performed to assess significant differences in lice abundance among salmon families infested with either chalimus or adult stages of sea lice under each temperature condition. An independent samples t-test (or Mann-Whitney U test, where appropriate) was used to assess significant differences in gene expression levels between families infested with chalimus versus adult stages of lice. The same tests were also applied to compare lice abundance between temperature conditions for families infested with either chalimus or adult stages. A significance level of p < 0.05 was applied for all statistical comparisons. The qPCR and lice abundance data analyses were performed using GraphPad Prism version 10 (GraphPad Software Inc., United States).
3 Results
3.1 Differences in lice burden
The mean lice abundance calculated for families infested by chalimus or adult stages of sea lice at either temperature (10°C and 20°C) has been summarized in Figures 1A–D. The mean lice abundance (calculated here for both chalimus and adult stages) for families included in RNA-seq analyses (F361, F175, F419, F265, F292) was respectively as follows: F361 (22.83 ± 5.52 at 10°C; 30.94 ± 11.57 at 20°C), F175 (30.70 ± 8.34 at 10°C; 45.38 ± 24.95 at 20°C), F419 (25.68 ± 8.71 at 10°C; 28.30 ± 10.27 at 20°C), F265 (20.74 ± 5.41 at 10°C; 31.53 ± 12.70 at 20°C), F292 (27.64 ± 9.80 at 10°C; 32.00 ± 10.94 at 20°C). The mean lice abundance was higher in most families infested at 20°C compared to 10°C, with significant differences observed in F175, F185 and F265 for chalimus-infested fish (Figure 1E), and in F153, F154, F265, F361, F413 and F414 for adult-infested fish (Figure 1F). Greater variability in lice abundance among families infested with chalimus stages was observed at the higher temperature condition (Figure 1E).
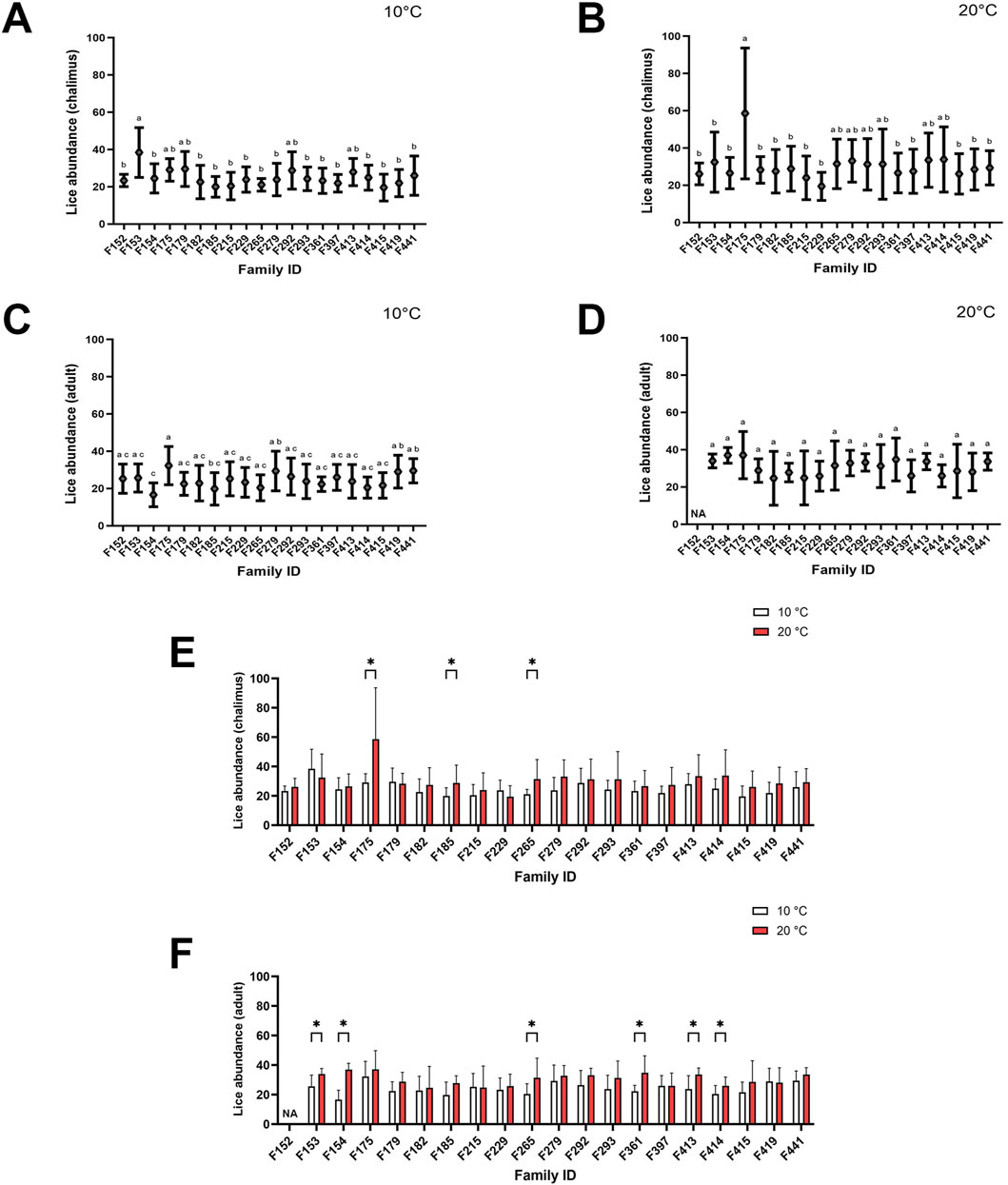
Figure 1. Average lice abundance (mean ± SD) in twenty salmon families infested with chalimus and adult stages of sea lice. Lice abundance was calculated for families infested with chalimus stages at 10°C (A) and 20°C (B), and for families infested with adult stages at 10°C (C) and 20°C (D). Average lice abundance was compared between temperature conditions for families infested with chalimus (E) and adult (F) lice stages. Different lowercase letters denote significant differences (p < 0.05) in lice abundance among families infested with chalimus or adult stages at each temperature (A–D). Asterisks indicate significant differences (p < 0.05) in lice abundance based on within-family comparisons between the two temperature conditions following infestation with either chalimus or adult lice stages (E,F). Representatives of five families (F175, F265, F292, F361 and F419) were included in RNA-seq analyses.
3.2 Transcriptomic responses
Pairwise comparisons within specific families (F361, F175, F419, F265, F292) infested with two different developmental stages of lice (adult vs. chalimus), which were separately carried out at two different temperatures (T10A vs. T10C and T20A vs. T20C, see Table 1), resulted in a remarkable number of DEGs in the skin (Supplementary Files S1–3) compared to the head kidney (Supplementary Files S4, 5). Venn diagrams generated using DEGs identified based on pairwise comparisons within families showed 562 and 3288 shared DEGs among all families at 10°C (T10A vs. T10C) and 20°C (T20A vs. T20C) in the skin, respectively (Figures 2A,B). There were 400 shared DEGs (299 consistently upregulated and 101 uniformly downregulated) among all families at both 10°C and 20°C in the skin (T10A vs. T10C | T20A vs. T20C). The heatmaps created using shared DEGs identified in the skin of fish infested at 10°C (n = 562) and 20°C (n = 3288), and those common at both temperatures (n = 400) resulted in complete clustering of their associated samples based on parasite infestation stage (Figure 2C). PCA conducted on the expression data associated with comparisons between infested families did not result in distinct segregation of head kidney samples in most comparisons (p > 0.05 for PC1 or PC2), suggesting inconsiderable differences between transcriptomic responses of fish infested with adult compared to chalimus stages of lice in this tissue (Figures 3A–H). On the other hand, PCA (Figures 3I–P) revealed distinct grouping (p < 0.05 for PC1 or PC2) of skin samples based on infestation stages of lice (T10A vs. T10C and T20A vs. T20C). The more frequent/significant genes identified based on family-level comparisons between infestation stages (expressed in the skin of most or all families) are listed in Supplementary File S6.
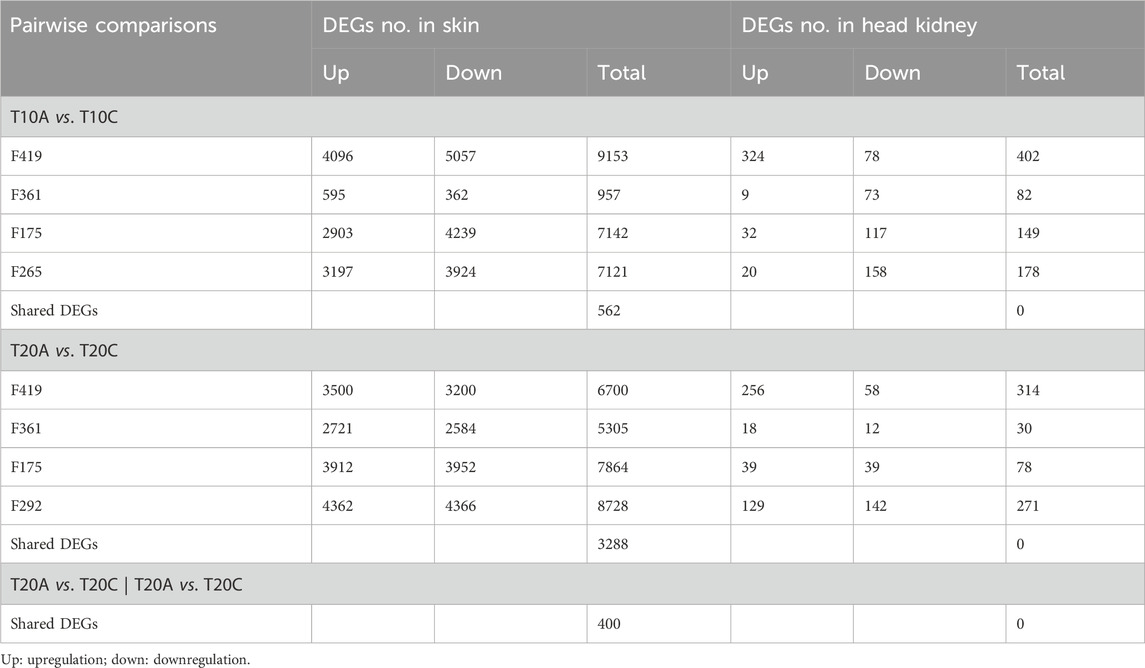
Table 1. Differentially expressed genes (DEGs) resulted from pairwise comparisons within specific salmon families parasitized with adult compared to chalimus stages of sea lice at a particular temperature for each tissue.
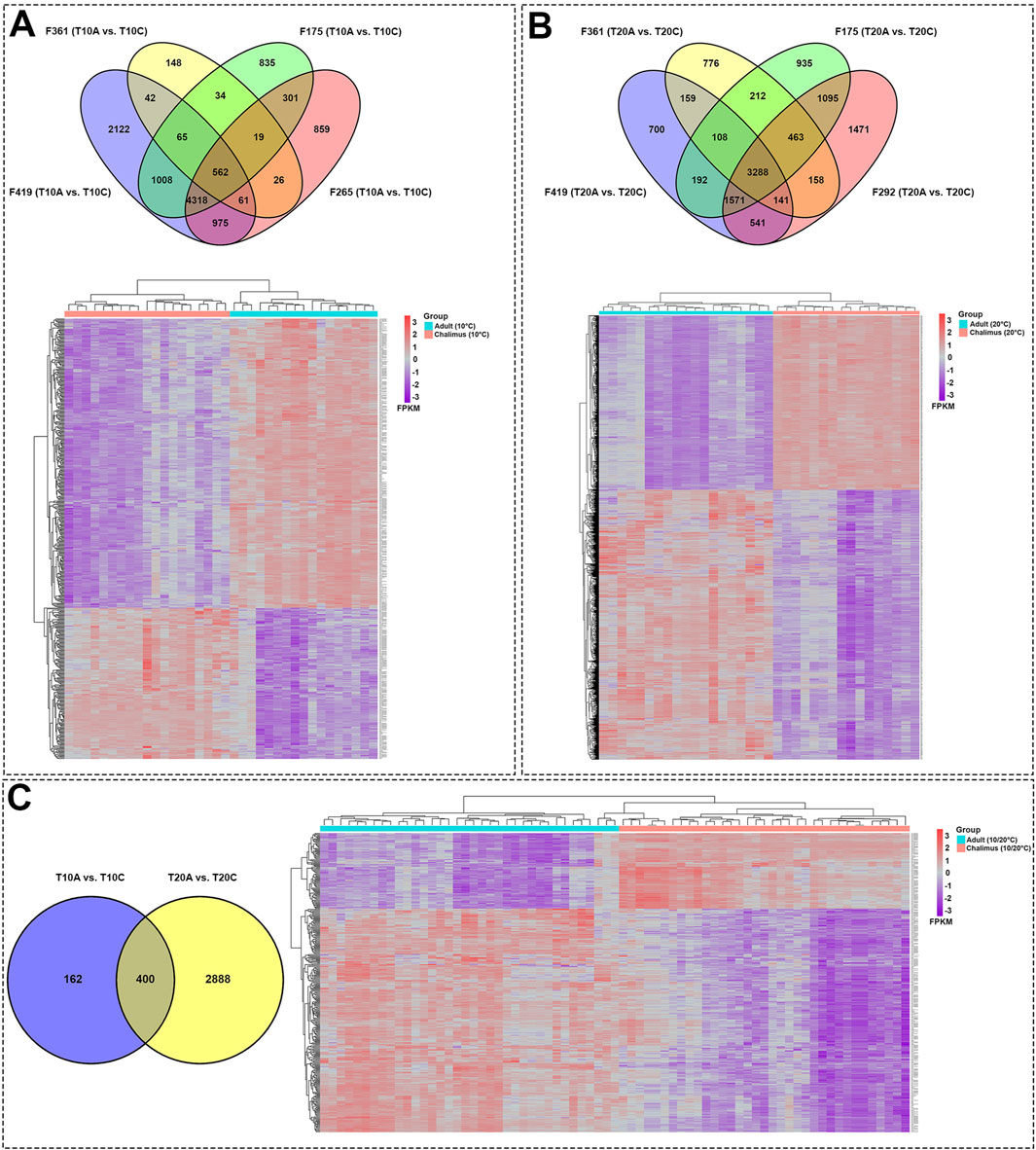
Figure 2. Skin transcriptome responses of Atlantic salmon infested with adult compared to chalimus stages of sea lice at normal and elevated temperature conditions. (A) The top Venn diagram shows the distribution of DEGs identified based on specific-family pairwise comparisons (F361, F175, F419, F265) in response to different infestation stages of lice at 10°C (T10A vs. T10C). The bottom heatmap illustrates the hierarchical clustering of shared DEGs (n = 562) among all families following infestation with different developmental stages of lice at 10°C. (B) The top Venn diagram represents the distribution of DEGs resulting from within-family pairwise comparisons (F361, F175, F419, F292) in response to different lice infestation stages at 20°C (T20A vs. T20C). The bottom heatmap displays the hierarchical clustering of shared DEGs (n = 3288) among all families following infestation with different developmental stages of lice at 20°C. (C) The left Venn diagram depicts the interaction of shared DEGs among all families infested with adult versus chalimus stages of lice at 10°C, in comparison to those infested at 20°C (T10A vs. T10C | T20A vs. T20C). The bottom heatmap presents the hierarchical clustering of shared DEGs (n = 400) across all families infested with adult versus chalimus lice stages at both 10°C and 20°C. Heatmaps were generated using FPKM-normalized gene expression values.
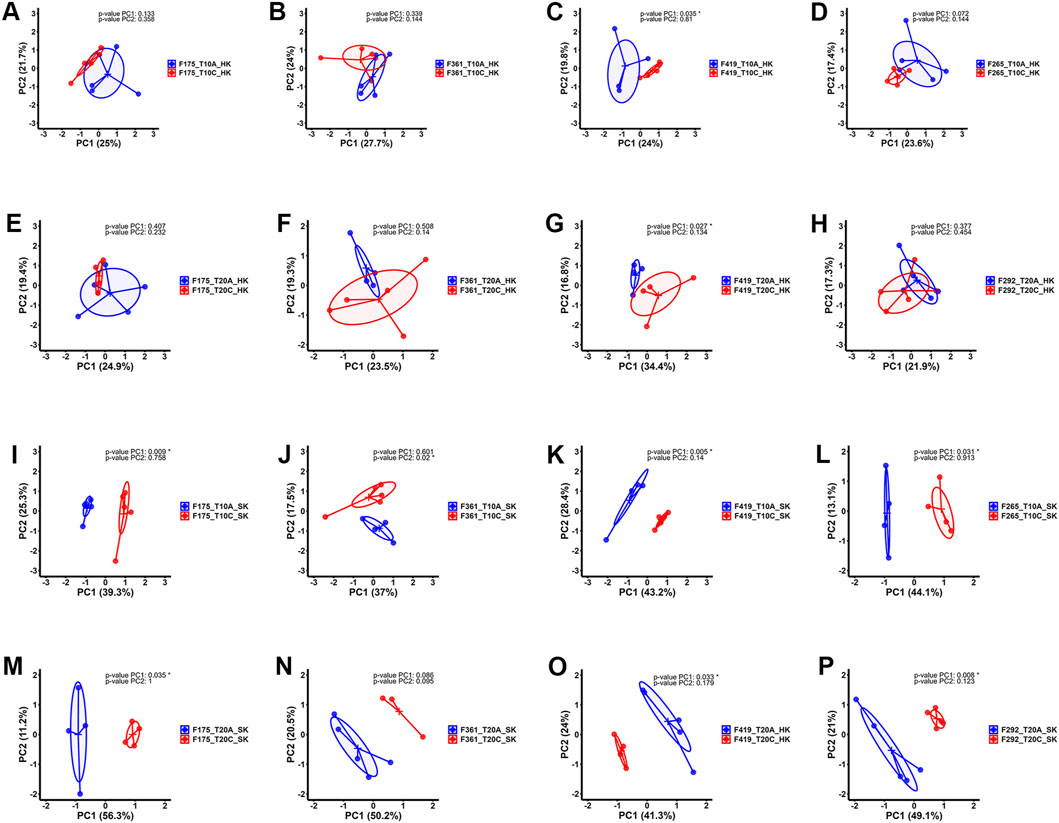
Figure 3. Principal component analysis (PCA) plots illustrating the expression data distribution of RNA-seq samples. (A–D) PCA plots based on the expression data comparing head kidney samples from families infested with adult (blue) versus chalimus (red) stages of sea lice at 10°C. (E–H) PCA plots based on the expression data related to comparisons between head kidney samples of families infested with adult (blue) compared to chalimus (red) stages of sea lice at 20°C. (I–L) PCA plots presenting the expression data comparing skin samples from families infested with adult (blue) versus chalimus (red) stages of sea lice at 10°C. (M–P) PCA plots illustrating the expression data from skin samples of families parasitized with adult (blue) versus chalimus (red) stages of sea lice at 20°C.
3.3 GO/KEGG enrichment analyses of DEGs
The GO terms and KEGG pathways associated with shared skin DEGs across all families comparing adult and chalimus stages at each temperature and their interaction (T10A vs. T10C, T20A vs. T20C and T10A vs. T10C | T20A vs. T20C) are provided in Supplementary Files S7–9. The leading GO terms and KEGG pathways were classified into five functional themes (Figures 4, 5A,B). Overall, the theme ‘metabolism, protein interaction and gene regulation’ represented higher number of GO terms and KEGG pathways compared to other functional themes. The GO and KEGG enrichment analyses revealed several terms/pathways involved in metabolism of sugars (fructose, mannose), lipids (arachidonic acid, cholesterol, steroid/brassinosteroid, sphingolipid, glycerophospholipids), proteins (proteinogenic amino acids, phenylalanine) and/or other organic compounds (secondary alcohols, small molecules, isoprenoids, pyrimidine, one-carbon molecules, carboxylic acid, dicarboxylic acid, tetrahydrofolic acid, retinol) in the skin of fish infested with adult versus chalimus stages. Notably, metabolism-related GO terms and KEGG pathways exhibited greater interconnectedness based on infestation stage comparisons, particularly under elevated temperature (Figure 4).
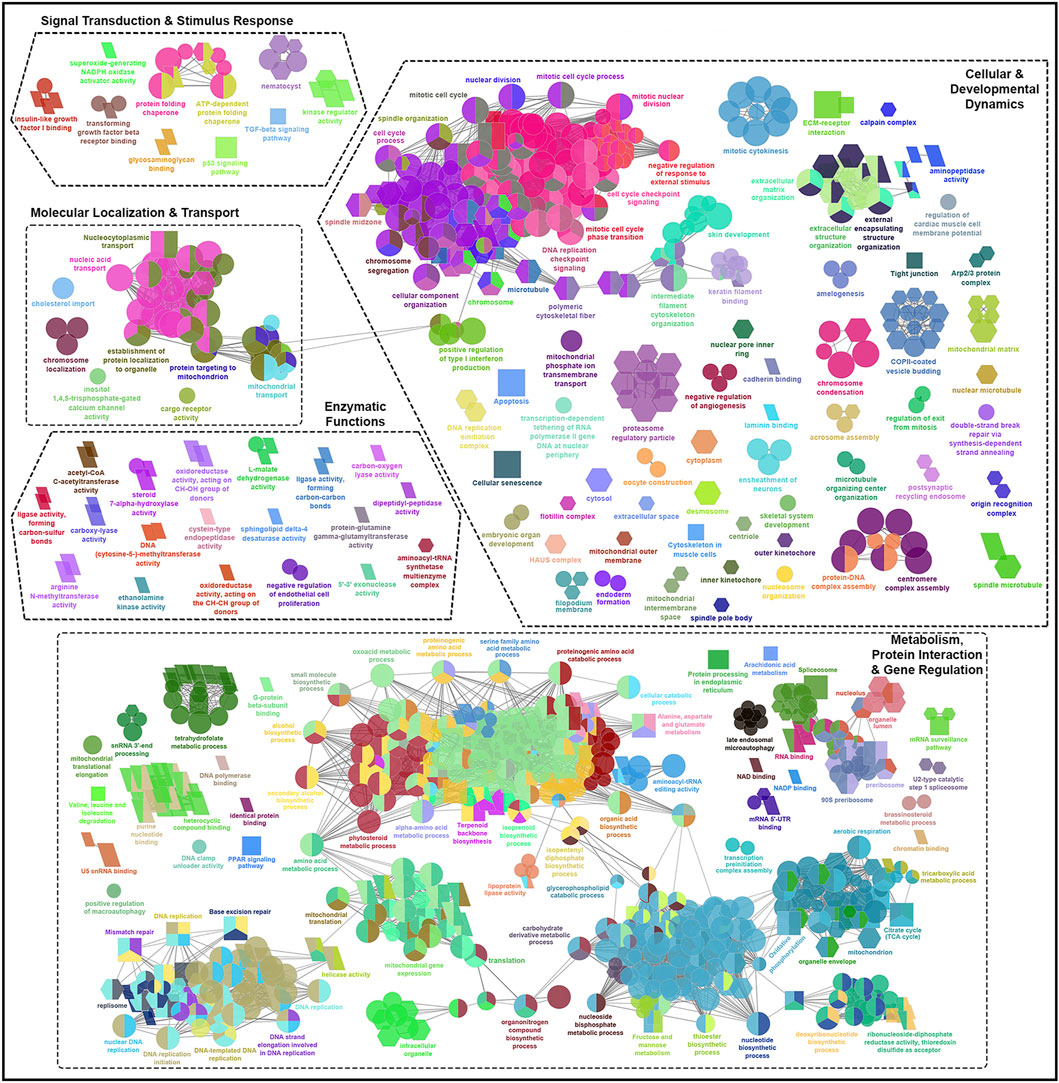
Figure 4. Functionally grouped network visualization of enriched GO terms and KEGG pathways based on infestation stage comparisons under elevated temperature condition. GO terms and KEGG pathways enriched based on shared DEGs identified in the skin of all families in response to infestation with adult versus chalimus stages of sea lice at 20°C (T20A vs. T20C). The leading GO terms and KEGG pathways were classified into 5 functional themes including metabolism, protein interaction and gene regulation (1), cellular and developmental dynamics (2), signal transduction and stimulus response (3), enzymatic functions (4), and molecular localization and transport (5). The GO domains and KEGG pathways are represented as follows: GO biological process (ellipse), GO cellular component (hexagon), GO molecular function (parallelogram), KEGG pathways (rectangle). Connections between GO terms and KEGG pathways represent shared genes among categories, grouped based on functional similarity using ClueGO’s kappa statistics (kappa score: 0.4). Only terms with Benjamini–Hochberg corrected p-values <0.05 were included.
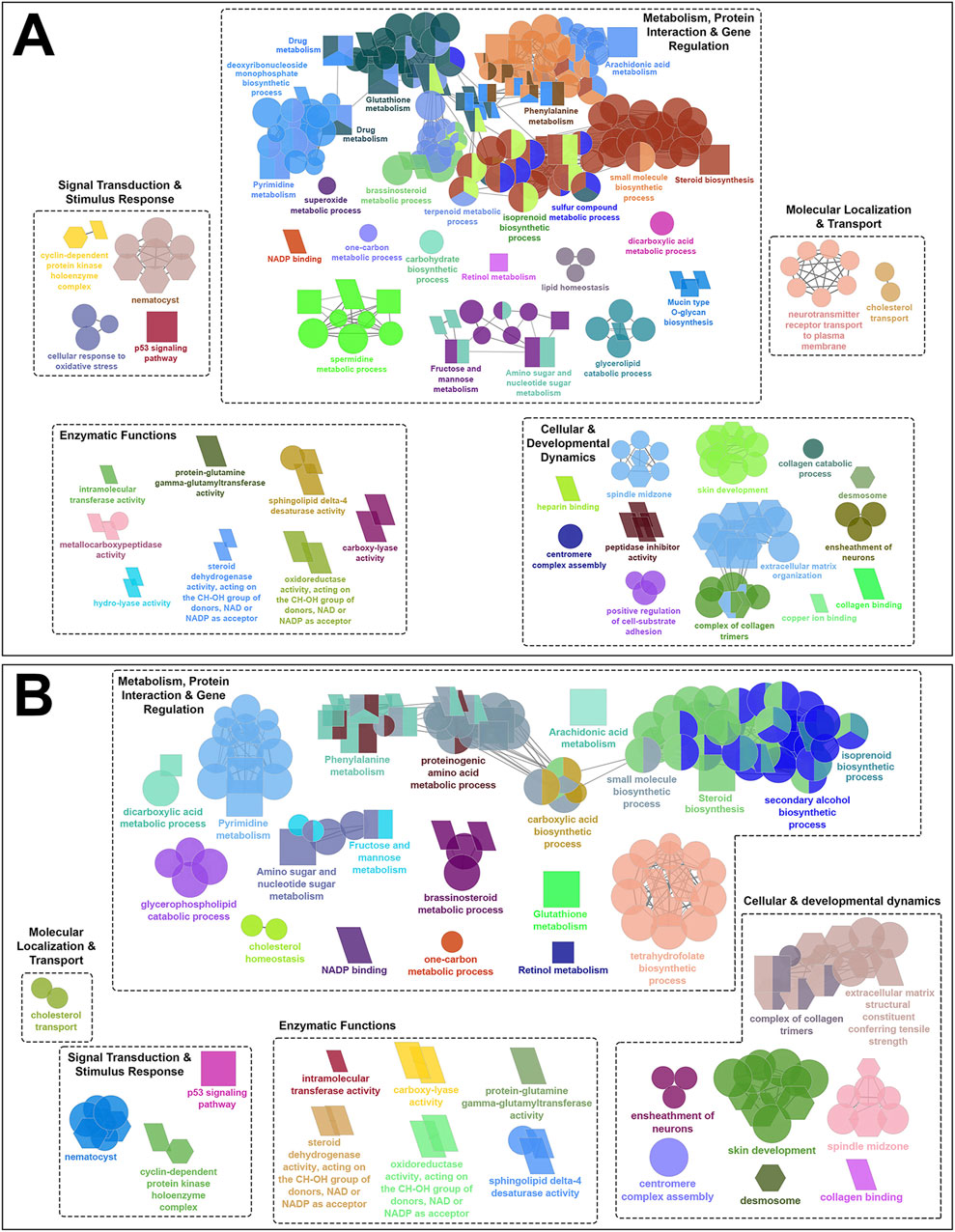
Figure 5. Functionally grouped network visualization of enriched GO terms and KEGG pathways based on infestation stage comparisons under different temperature conditions. (A) GO terms and KEGG pathways enriched based on shared DEGs identified in the skin of all families following infestation with adult compared to chalimus stages of sea lice at 10°C (T10A vs. T10C). (B) GO terms and KEGG pathways enriched based on shared DEGs across all families infested with adult versus chalimus lice stages at both 10°C and 20°C (T10A vs. T10C | T20A vs. T20C). The leading GO terms and KEGG pathways were classified into 5 functional themes including metabolism, protein interaction and gene regulation (1), cellular and developmental dynamics (2), signal transduction and stimulus response (3), enzymatic functions (4), and molecular localization and transport (5). The GO domains and KEGG pathways are represented as follows: GO biological process (ellipse), GO cellular component (hexagon), GO molecular function (parallelogram), KEGG pathways (rectangle). Connections between GO terms and KEGG pathways represent shared genes among categories, grouped based on functional similarity using ClueGO’s kappa statistics (kappa score: 0.4). Only terms with Benjamini–Hochberg corrected p-values <0.05 were included.
Our findings suggest enhanced metabolic demands and stress-induced metabolic reprogramming in infested fish maintained at 20°C compared to those held at 10°C (Figure 4). This is supported by the enrichment of several over-represented GO terms and KEGG pathways related to DNA repair and processing (e.g., “chromatin binding” [GO:0003682], “DNA replication” [KEGG:03030]), RNA processing (e.g., “RNA binding” [GO:0016071], “spliceosome” [GO:0000398]), and protein-protein interactions (e.g., “translation” [GO:0090079], “protein processing in endoplasmic reticulum” [KEGG:04141]). Furthermore, broader metabolic processes/pathways involving a greater number of genes were enriched, including “carbohydrate derivative metabolic process” (GO:1901135), “small molecule biosynthetic process” (GO:0032787), “organonitrogen compound biosynthetic process” (GO:0006412) and “PPAR signaling pathway” (KEGG:03320). Examples of GO terms and KEGG pathways enriched based on shared DEGs related to T10A vs. T10C (Figure 5A) included “carbohydrate biosynthetic process” (GO:0016051), “terpenoid metabolic process” (GO:0042445) and “mucin type O-glycan biosynthesis” (KEGG:00512), while terms/pathways associated with T10A vs. T10C | T20A vs. T20C (Figure 5B) were represented by “carboxylic acid biosynthetic process” (GO:0016053), “glycerophospholipid catabolic process” (GO:0046434), “glutathione metabolism” (KEGG:00480) and “arachidonic acid metabolism” (KEGG:00590).
The infestation stage-related leading GO terms and/or KEGG pathways classified under “cellular and developmental dynamics” were more frequent and diverse at elevated temperature condition (Figure 4). For instance, there was a notable enrichment in terms associated with cell cycle control (e.g., “cell cycle process” [GO:0022402]), cellular and structural organization (e.g., “cytoplasm” [GO:0005737], “cellular component organization” [GO:0016043], “extracellular structure organization” [GO:0030020]), and organelle function and assembly (e.g., “mitochondrial outer membrane” [GO:0005741]). Moreover, the enrichment of KEGG pathways such as “apoptosis” (KEGG:04210), “cellular senescence” (KEGG:04218) and “tight junction” (KEGG:04530), with predominantly upregulated associated genes, likely reflects an intensified host effort to counteract parasite-induced damage under higher temperature stress. Comparatively, the narrower enrichment of GO terms and KEGG pathways related to localized tissue remodeling at 10°C (Figure 5A), such as “ensheathment of neurons” (GO:0007272), “skin development” (GO:0008544), “extracellular matrix organization” (KEGG:04512), suggests a more controlled developmental and structural response at this thermal condition.
Compared to the previously described functional themes, GO terms and/or KEGG pathways associated with ‘signal transduction and stimulus response’, ‘enzymatic functions’ and ‘molecular localization and transport’ were less abundant and exhibited lower levels of functional connectivity (Figure 4; Figures 5A,B). However, within these themes, the associated GO terms and/or KEGG pathways also contained a broader and more frequently occurring set of genes under elevated temperature conditions. Among the GO terms classified under ‘signal transduction and stimulus response’, “protein-glutamine gamma-glutamyltransferase activity” (GO:0003810), “carboxy-lyase activity” (GO:0016830), oxidoreductase activity (GO:0016614) and “sphingolipid delta-4 desaturase activity” (GO:0016717) were consistently enriched at both 10°C and 20°C (Figure 5B). This pattern suggests a core enzymatic response of Atlantic salmon to lice infestation (adult vs. chalimus) regardless of temperature, likely contributing to tissue remodeling and repair (via transglutaminase activity), lipid metabolism and membrane remodeling (via sphingolipid desaturase), and redox homeostasis (via oxidoreductases).
The leading GO terms and KEGG pathways linked to the functional theme ‘signal transduction and stimulus response’ that were enriched under both temperature conditions (Figure 5B) included those associated with pore formation (“nematocyst” [GO:0044218]) and with stress signaling and cell cycle regulation response (“p53 signaling pathway” [KEGG:04115], “cyclin-dependent protein kinase holoenzyme complex” [GO:0016538]). A notable expansion of GO terms and KEGG pathways related to growth factor signaling (e.g., “insulin-like growth factor I binding” [GO:0005520], “TGF-beta signaling pathway” [KEGG:04350]), kinase regulation (“kinase regulator activity” [GO:0019207]), and chaperone-mediated folding (e.g., “protein folding chaperone” [GO:0140662]) indicates the enhanced signal transduction complexity at elevated temperature (Figure 4). Likewise, a broader range of GO terms and KEGG pathways under the theme ‘molecular localization and transport’ were enriched at 20°C (Figure 4), including those associated with organelle-specific transport (e.g., “mitochondrial transport” [GO:0006839]), macromolecule localization (e.g., “chromosome localization” [GO:0050000], “nucleic acid transport” [KEGG:03013], “nucleocytoplasmic transport” [KEGG:03013]) and trafficking mechanisms (e.g., cargo receptor activity [GO:0038024], establishment of protein localization to organelle [GO:0006839]). These findings suggest that compartmental communication, intracellular trafficking, and molecular localization were impacted under enhanced temperature conditions.
The infestation stage-associated leading GO terms and KEGG pathways enriched based on shared DEGs identified in the skin of all families at both thermal conditions along with their corresponding up- and downregulated DEGs have been summarized in Table 2 (see Supplementary Table S2 for the complete list). tgm2, tgm3, emx1, camp, tubb2a, ddit4l, il1b, iff6, mslna, mal, casp3b, lbh, krt8, krt13 and cell surface glycoprotein 1 (unavailable symbol) were among shared DEGs with high transcript levels that were expressed in all families infested by adult compared to chalimus stages of lice at both temperatures. By comparison, entpd5a, fndc1, chad, ccdc3b, prss23, ncana, col10a1a, col1a2, col16a1 and col1a1 were among shared DEGs with lowest expressions in all families parasitized with adults compared to chalimus stages at both thermal conditions.
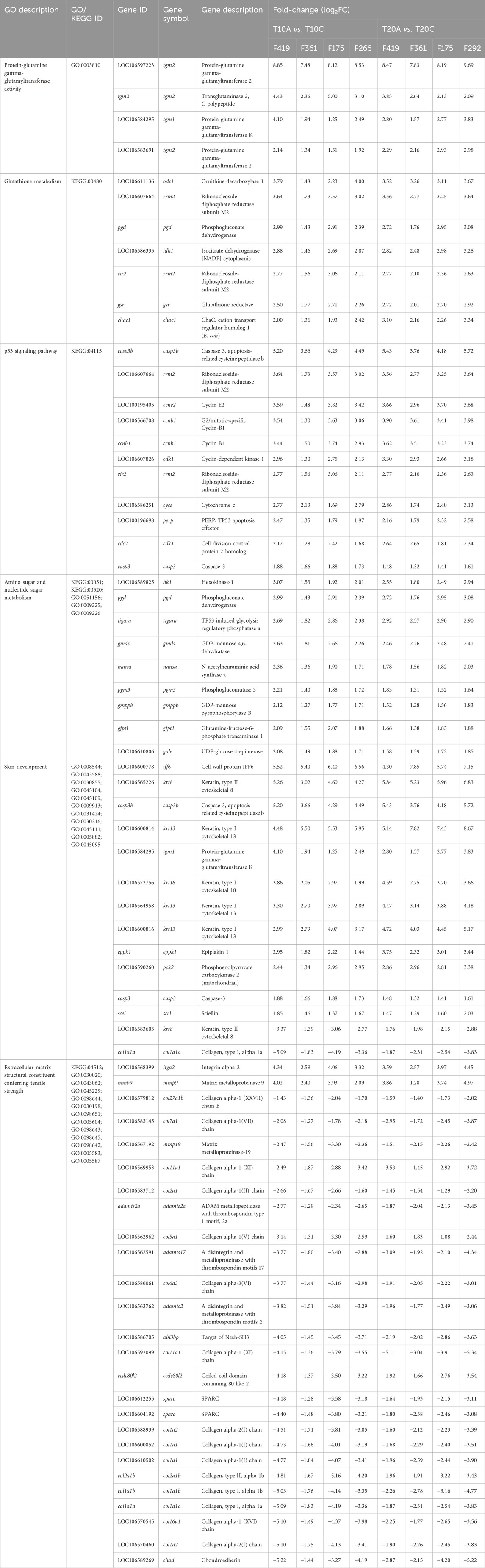
Table 2. Selected leading GO terms and KEGG pathways enriched based on shared DEGs associated with pairwise comparisons within specific salmon families (F419, F361, F175, F265, F292) infested with adult compared to chalimus stages of lice at 10°C and 20°C in the skin (see Supplementary Table S2 for the complete list of enriched leading GO terms).
In the head kidney, contrary to skin, there were no shared DEGs among families infested with different developmental stages of lice. However, certain within-family comparisons were found to be associated with GO terms involved in detoxification (e.g., “peroxidase activity” and “hydrogen peroxide catabolic process”), heat shock response (“ATP-dependent protein folding chaperone” and “response to heat”), transporter activity (“bicarbonate transmembrane transporter activity” and “ammonium transmembrane transporter activity”) and immune response (“defense response to virus” and “lymphocyte differentiation”) (Supplementary Files S10, 11).
3.4 qPCR analysis of selected genes
Multiple samples from different families (n = 8 at each temperature [10 or 20°C] for head kidney [HK] and skin [SK]; HK-T10, HK-T20; SK-T10, SK-T20) were included in qPCR analyses. Of the eight GOIs (cd83, clec4, col1a1, mmp9, ncf2, saa5, timp2 and txnb) selected for qPCR analyses, col1a1 and mmp9 revealed the most significant changes compared to other genes (Figure 6), particularly in fish infested at 10°C (see below). qPCR results for all GOIs analyzed in this study are presented in Supplementary Figures S1–S4. The expression of col1a1 decreased in the skin and head kidney of all families at 10°C, with significant changes (p < 0.05) identified in F153, F154, F175, F185, F229 and F265 in SK-10, and F154, F185, F265, F361 and F419 in HK-10. Based on RNA-seq analyses, the transcript level of this gene (LOC106610502) was reduced in the skin of all families at both temperatures (F419, F175, F361, F265 and/or F292 in SK-10/20), but its lowered expression was detected in the head kidney of few families (F265 in HK-10 and F292 in HK-20). On the other hand, the mRNA level of mmp9 increased in the skin of almost all families (except F361) at both temperatures, although these changes were significant (p < 0.05) in F153, F154, F185, F229 and F265 in SK-10, and F397 in SK-20 (Figure 6). RNA-seq analyses showed enhanced expression of mmp9 in the skin of all families at both temperatures (F419, F175, F361, F265 and/or F292 in SK-10/20), with decreased upregulation in F361 compared to other families at both temperatures. qPCR results revealed increased expression of mmp9 in the head kidney of certain families only at 10°C (F153, F229 and F361), but this gene was not differentially expressed at either temperature based on RNA-seq analyses. Supplementary Figures S1–S4.
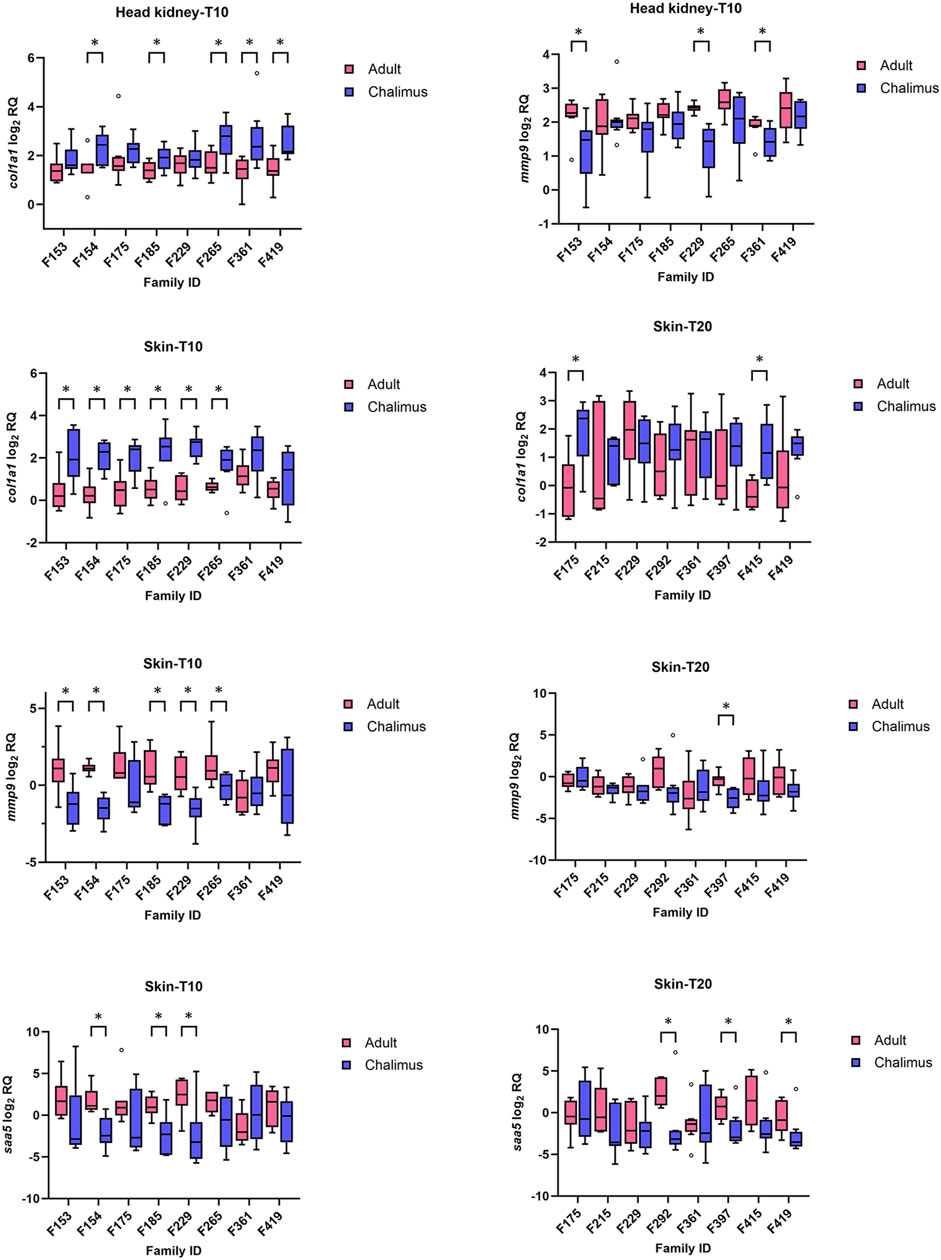
Figure 6. qPCR results for selected genes (col1a1, mmp9 and saa5) with significant alterations in the skin and/or head kidney of different families infested by adult compared to chalimus stages of sea lice at 10°C (T10) or 20 (T20) °C. Data are presented as box plots with median and Tukey whiskers (fences). Significant differences (p < 0.05) in gene expression responses between infestation stages within each family are indicated by asterisks. qPCR results for all genes of interest (GOIs) tested in this study are presented in Supplementary Figures S1–S4.
The expression of cd83 was mostly reduced in both the skin and head kidney of families infested by adult compared to chalimus at 10°C (Supplementary Figures S1, S3), although these changes were significant (p < 0.05) in F175 and F265 in HK-T10, and F185, F361 and F419 in SK-T10 (p < 0.05). The transcript level of cd83 was unaffected in the skin and head kidney of most families at 20°C, and RNA-seq data showed no modulation of this gene in either tissue. The expression level of clec4 was not impacted in the skin and head kidney of most families at both temperatures (Supplementary Figures S1–S4). According to RNA-seq data, different members of clec4 family had mostly reduced expression in the skin of adult-compared to chalimus-infested families at both temperatures. RNA-seq analysis showed no modulation of ncf2 in either tissue, although qPCR data indicated inconsistent modulation of this gene in a few families. Both RNA-seq and qPCR data did not show significant expression of txnb, timp2 and saa5 in the head kidney at both thermal conditions (Supplementary Figures S1, S2). Based on qPCR results, the transcript level of txnb increased in the skin of all families at 10°C, with significant changes (p < 0.05) in F154, F229 and F265. RNA-seq analysis revealed enhanced expression of this gene in some/all families at both temperatures. The expression of timp2 was significantly (p < 0.05) higher in the skin of all families parasitized with adult compared to chalimus stages of lice in RNA-seq analysis. Based on qPCR data, increased expression of this gene was observed in the majority of families, although with significant alteration only in F154 and F229 at 10°C (Supplementary Figure S3). RNA-seq analysis revealed increased transcript levels of saa5 in most families at both thermal profiles (Figure 6). Consistently with these findings, qPCR results indicated the enhanced expression of saa5 in the skin of all families (except F361), where significant changes (p < 0.05) were observed in F154, F185 and F229 in SK-T10, and F292, F397 and F419 in SK-T20.
4 Discussion
This is the first comprehensive family-based transcriptome study in Atlantic salmon in response to infestation with different developmental stages of L. salmonis. Beyond its relevance to salmon aquaculture, this study provides broader insights into the molecular mechanisms underlying host-parasite interactions in lower vertebrates. The identification of host responses to distinct developmental stages of sea lice enhances our understanding of how parasites modulate host immunity at different phases of infestation. Moreover, these findings may inform the development of targeted strategies for breeding parasite-resistant fish, improving fish welfare, and enhancing the sustainability of aquaculture systems globally. Based on within-family pairwise comparisons (T10A vs. T10C, T20A vs. T20C), a large number of genes (e.g., muc2, muc5ac, litaf, krt13, ccl20, lcn, cd22, map4k4, pbx1, trim16) were represented by paralogs showing either similar or contrasting transcriptional patterns. This observation aligns with previous findings of transcriptional divergence among duplicated genes in Atlantic salmon and may reflect functional specialization following whole-genome duplication events (Macqueen and Johnston, 2014; Lien et al., 2016; Umasuthan et al., 2020; Ignatz et al., 2024). The GO and KEGG enrichment analyses revealed terms/pathways represented by DEGs (nearly all upregulated) encoding a diverse range of enzymes/biomolecules involved in metabolism of various organic substances in the skin of fish infested with adult versus chalimus stages. These metabolic shifts facilitate the mobilization of energy substrates/stores to cope with the stress caused during infestation. In addition, the enhanced expression of several genes directly or indirectly associated with organismal stress response (ercc6, xrcc5, xrcc6, brca1, lepb2, ddb1, gadd45a, hspd1, hspa14, hsp90b1, cct8, cct6a, hif1ab, cirbp, hspa8 [hsc70], ube2v1, ube2n, sumo2) suggested a response to the more severe skin damage induced by adult forms compared to chalimus stages. These changes are consistent with elevated plasma cortisol levels and increased epithelial cell apoptosis and necrosis observed following infestation with advanced stages of sea lice (Johnson and Fast, 2004; Nolan et al., 1999).
The transcriptomic changes associated with infestation with adult versus chalimus stages of lice were remarkable in the skin. This variation was not visible in the head kidney samples from the same salmon between the two different stages. In the head kidney, no shared DEGs were identified between all families at either 10 or 20°C, and GO analyses revealed a limited number of GO terms. Based on within-family pairwise comparisons, the most frequent DEGs in the head kidney of fish infested with adult versus chalimus at both temperatures included fcrl5, cat, hbb, hbb1 and hba4 (downregulated). On the other hand, a total of 384 annotated genes (286 upregulated and 97 downregulated) were identified to be differentially expressed in the skin of Atlantic salmon infested with adult versus chalimus stages of sea lice at both 10°C and 20°C. Based on within-family comparisons, more abundant genes (present at least in 5 out of 8 within-family comparisons at either temperature, unless otherwise specified) with significant functions that were expressed in the skin of most or all families at one or both temperatures are discussed in this study, considering their expression trends/levels (see Supplementary File S6). It should be noted that one family (F265 at 10°C and F292 at 20°C) differed between temperature conditions. However, as each comparison was conducted within the same family (adult vs. chalimus stages), and the primary aim was to identify genes consistently expressed across families within each temperature, the impact of this substitution on the overall findings is likely minimal. Nonetheless, the potential influence of family-specific responses cannot be fully excluded and should be considered in the interpretation of shared gene expression patterns.
The salmon louse is known to dampen various immune-related responses in susceptible salmonids such as Atlantic salmon (Fast et al., 2006; Wagner et al., 2008; Umasuthan et al., 2020; Øvergård et al., 2023; Øvergård et al., 2025). In this study, immune responses were substantially suppressed/modulated in adult- versus chalimus-infested families. For example, the altered transcription of certain suppressor of cytokine signaling (SOCS) proteins (up: socs1; down: socs2; up-down paralogs: socs3) implied that the salmon louse could manipulate cytokine receptor signaling by recruiting SOCS proteins to circumvent the host’s immune defenses (Salisbury et al., 2024). In addition, the transcription levels of multiple genes belonging to the semaphorin family showed significant changes in the skin of families infested by adult versus chalimus stages, most of which showed lowered expressions at both temperatures (e.g., sema3ab, sema3c, sema3fb, sema4d, sema6d). On the other hand, sema3f and sema7a tended to have different paralogs with reverse expression patterns. The expression levels of plexins (plxna2, plxnb2 [LOC106584474], plxdc1 and plxdc2) and neuropilins (nrp1a and nrp2), which are known as transmembrane proteins serving as cell surface receptors for semaphorins, were also reduced at both temperatures. The enrichment of GO terms predominantly consisted of semaphorin family members (chemorepellent activity [GO:0045499] and negative chemotaxis [GO:0050919]) that are involved in stimulation of antigen-presenting cells and/or neuroinflammation (Lee et al., 2019; Rajabinejad et al., 2020; Thomas and Yang, 2023) highlighted the immunosuppression and immunoinflammatory disturbance induced by adult forms. Further immunosuppression/modulation resulting from infestation with adult compared to chalimus stages of L. salmonis at the attachment sites was supported by differential expression of several immune-related components including representatives of genes related to Toll/NOD-like receptors (up: sigirr, nlrc3l; down: tlr21, tlr5, nlrp1, nlrp12, card8, nlrc3, bcl2), nuclear factors (down: nfatc2, ikbip, nfatc1 [in 3 families]), CD molecules (up: cd276, cd44b, cd9; down: cd2, cd6, cd3e, cd8a, cd28, cd40, cd44, cd59, cd63, cd81, cd82, cd93; up-down paralogs: cd22, cd55), scavenger receptors (down: scara3, scara5, cd163) and protein kinases/phosphatases (up: tab3, mapk13, dusp4, dusp5, ptpn2b; down: map3k11, map3k20, map3k3; up-down paralogs: map3k5).
The proteinaceous (i.e., trypsin and cathepsin L) and non-proteinaceous (i.e., prostaglandin E2, PGE2) products secreted by the salmon louse are known to interfere with the host’s immune responses and facilitate the flow of blood to the attachment sites (Johnson et al., 2002; Fast et al., 2004; Fast et al., 2007; Firth et al., 2000; McCarthy et al., 2012). Based on within-family comparisons, three subtypes of prostaglandin E2 (PGE2) receptors (EP1, EP2 and EP4 encoded by the genes ptger1 [down], ptger2 [up-down paralogs] and ptger4 [down]) along with prostaglandin D2 receptor 2 (ptgdr2, up) and prostaglandin F2 receptor negative regulator (ptgfrn, up) were impacted in families infested with adult compared to chalimus stages of lice. Upregulation of the subtype EP2 of PGE2 receptor has been previously reported in the skin of Atlantic salmon infested with copepodids of lice compared to non-infested fish (Øvergård et al., 2023). The reduced expression of EP receptors in fish infested by adult lice may be a consequence of chronic exposure to secretary products (i.e., PGE2) of the salmon louse, resulting in their desensitization or internalization (Malty et al., 2016). The enhanced expression of pla2g4c, ptgis, ptgs2b, lta4h, and ltb4r, which are involved in lipid mediator synthesis, vasodilation, and/or leukotriene signaling, was observed in nearly all families, indicating a heightened inflammatory response at sites infested by adult lice compared to chalimus stages (Murakami et al., 2003; Ricciotti and FitzGerald, 2011). Dampened expression of apc, encoding angiotensin-converting enzyme that promotes vasoconstriction, in all families seemed to be a feedback mechanism to control excessive inflammation. This, however, could allow the parasite to cause chronic infestation by impairing the host’s immune ability. In addition, several other genes correlated with inflammation and inflammasome signaling were also differentially expressed in the skin of most families (up: gpr84; down: nlrp1, nlrp12, card8, nlrc3, up-down: nlrp3, usp7 and usp47). The increased expression of gpr84 that promotes the release of inflammatory cytokines and reduced expression of nlrp12, card8 and nlrc3 that negatively regulates inflammation was indicative of excessive inflammatory responses (Zheng et al., 2020; Zhang et al., 2022). On the other hand, a decreased level of nlrp1, which is involved in normal release of inflammatory cytokines, accompanied with fluctuated expression of nlrp3 and its regulators (usp7 and usp47) could lead to a dysregulated status contributing to chronic inflammation or suppressed immune responses (Palazón-Riquelme et al., 2018).
The expression of several mediators associated with acute inflammation were found to be reduced in the skin of fish parasitized with adult compared to chalimus stages of lice (crp, hp, a2m, ltc4s). Conversely, saa5 showed higher expression in the skin of most families at both temperatures. Earlier findings highlighted the upregulation of saa5 in Atlantic salmon infested by various stages of lice (Sutherland et al., 2014; Braden et al., 2015; Ugelvik and Dalvin, 2022), suggesting that persistent sea lice infestation could result in chronic inflammation, characterized by prolonged expression of pro-inflammatory cytokines such as saa5. In addition, higher transcript levels of il1b (LOC100136449 and LOC106570815), il4i1, il1r2 and sigirr (il-1r8) were noticed in most/all families at both temperatures, while il4/13a and il17d showed reduced transcript expression. Increased mRNA levels of il1b and il8 have been previously shown using qPCR analysis in the skin of both Atlantic salmon and rainbow trout following infestation with juvenile to advanced stages of sea lice including those with no blood-feeding activity (copepodid and/or chalimus) (Dalvin et al., 2020; Ugelvik and Dalvin, 2022). The upregulation of il1b (pre-adult-infested vs. non-infested) and reduced expression of il4/13a (adult vs. chalimus) was also reported in the skin of Atlantic salmon in a previous study (Øvergård et al., 2023). This unregulated expression of il1b could lead to excessive or prolonged inflammation, as augmented transcriptional levels of il1r2 (a decoy receptor that antagonizes il1b) and sigirr (a negative regulator of TLR/IL-1R signalling) were detected in almost all families, possibly to dampen il-1β-mediated signaling and protect skin tissue from potential damage (Qin et al., 2005; Schlüter et al., 2018). Interleukin-4 induced 1 (il4i1) is an L-amino acid oxidase that produces immunosuppressive metabolites by catabolizing essential amino acids required for T-cell activation/proliferation, and thus, its higher transcript level could be regarded as immunosuppression caused by advanced stages of sea lice (Hirose et al., 2024). The lowered expression of il4/13a and il17d, which are involved in protection against parasites, was indicative of compromised immunity, tissue healing and barrier function (Sequeida et al., 2020; Huangfu et al., 2023). The expression profile of il8 was found to be different among families (down in 3 families at 10 and up in 1 family at 20). This could be due to the varied expression level of il8 in fish infested by adult or chalimus stages, since this gene was shown to be expressed (upregulated) in both fish infested by chalimus and adult stages of lice (compared to un-infested fish) in a previous study (Ugelvik and Dalvin, 2022).
The transcription of several chemokines and their ligands/receptors (up: ackr4b, ccr3, cxcr1 and cxcr2; down: ackr3, ackr3b, ackr4, ccr7, ccl8, ccl13, ccl17, ccl21, ccl25 [only at 20], ccl25a, ccl27a and cxcl12a; up-down paralogs: ccl4 and ccl20) significantly differed between families infested by adult compared to chalimus stages. Similar chemokine-related transcriptomic changes have been previously documented for fish infested by advanced (pre-adult female and adult male) forms of sea lice (Cai et al., 2024). Contrary to inconsistent expression of il8, the transcription of il8 receptors (cxcr1 and cxcr2) that are primary stimulants of neutrophils was enhanced (3-6-fold changes) in most families, potentially contributing to uncontrolled neutrophil responsiveness. Two gene copies related to atypical chemokine receptor 4 represented opposite transcription patterns (up: ackr4b; down: ackr4 [LOC106568919]). Atypical chemokine receptor 4 (ackr4), which is expressed primarily in non-immune cells such as keratinocytes, is known to bind and internalize the chemokines ccl19, ccl20, ccl21 and ccl25, thus regulating the migration and positioning of immune cells (dendritic and T cells) expressing the chemokine receptors ccr6 (ligand: ccl20), ccr7 (ligands: ccl19 and ccl21) and ccr9 (ligand: ccl25) (Meyrath et al., 2021). Likewise, ackr3 internalizes and serves as a scavenger receptor for cxcl11 (receptor: cxcr3) and cxcl12 (receptors: cxcr4 and cxcr7), modulating their availability (Cuesta-Margolles et al., 2024). In addition, the ccr3 gene encodes a protein primarily found on eosinophils and a subset of Th2 lymphocytes, serving as a receptor for chemokines such as eotaxins (ccl11 and ccl26), monocyte chemoattractant protein-3/4 (ccl7, ccl13) and RANTES (ccl5) (Manousou et al., 2010). The dysregulated/contrasting expression of several members of the chemokine family, known for their involvement in directing immune cell chemotaxis, in the skin of fish infested by adult versus chalimus stages of lice could favor the recruitment of specific immune cells that aid sea lice persistence. The immunomodulatory effects of salivary products of blood-feeding ticks on chemokine activity of immune cells have been previously reported in terrestrial-associated models in vivo and in vitro (Vančová et al., 2007; Wang et al., 2007; Oliveira et al., 2008; Langhansova et al., 2015). The association of ackr4 and ccr9 in resistance against the ectoparasite Ichthyophthirius multifiliis has been also suggested in Colossoma macropomum using a genome-wide association study (Lira et al., 2023). These results highlight the importance of identifying louse-derived secretary products that influence the chemokine signalling system in Atlantic salmon to develop possible parasite-protective vaccines.
Some members of the tumor necrosis factor (TNF) cytokine family represented modulated expressions in almost all families (up: tnfrsf6b and tnfrsf9 [LOC106572390]; down: cd40, fas, tnfsf11 [LOC106582173], tnfsf12, tnfrsf14 and traf3ip3). Paralogs with opposite transcription patterns were identified in all families for litaf, a DNA-binding protein that contributes to the progression of inflammation by altering cytokine levels and plays a role in the p53-induced apoptotic pathway (Zou et al., 2015). The transcription of several DEGs associated with apoptosis (up: aifm1, bax, casp3b, casp3, perp, malt1, tp53, api5, tigara; down: bcl2, bcl11ba, bcl6b, prf1, gzma, phlda1, camk4), coupled with enrichment of the GO term “p53 signaling pathway”, indicated the induced/dysregulated apoptotic response in damaged/infested cells in families infested (adult vs. chalimus) (Wang H. et al., 2023). The transcript levels of genes associated with interferons showed varied expressions in adult- vs. chalimus-infested families (up: ifnar1a, ifngr1a, ifngr1b, irf6, irf7, irf8, ifi44, ifit1, ifrd2, sting1; down: ifi30; up-down: ifit5). Genes related to DEAD-box helicases (up: ddx3x, ddx5, ddx19a, ddx55, ddx56), which act as sensors for dsRNA, cytosolic DNA and viral RNAs (Taschuk and Cherry, 2020), exhibited higher expression in families infested by adult forms. In addition, some TRIM proteins involved in host antiviral defense and/or regulation of interferon signaling (up: ftr82, ftr83, trim14, trim39, isg15, uba7; down: trim9, trim21, trim25, trim35) showed altered expressions (van Gent et al., 2018; Wang and Ning, 2021). Modulation of viral-responsive genes has been previously reported in fish infested by various developmental stages of lice (Umasuthan et al., 2020; Cai et al., 2022; Caballero-Solares et al., 2022; Øvergård et al., 2023). Motile-stage sea lice are known to carry divergent RNA viruses, which might account for modulation of interferon-related markers in the skin of Atlantic salmon (Økland et al., 2014; Chang et al., 2023). In addition, two paralogs related to cyclic GMP-AMP synthase gene (cgas and cgasa) showed higher expression in almost all families. cgas is a cytosolic DNA sensor that detects aberrant/foreign DNA from invading pathogens, resulting in type-I interferon production via the cGAS-STING pathway (Ou et al., 2021).
Two paralogs (LOC100136453 and LOC100136439) of the cathelicidin antimicrobial peptide (camp or ll37) gene were identified to have higher expression in the skin of fish infested with adult compared to chalimus stages, one of which was shared among all families at both temperatures (LOC100136453). In contrast, defensin, beta-like 1 (defb1) gene showed lower expression in most families. Similar changes in expression have been also observed in Atlantic salmon following sea lice infestation (Krasnov et al., 2015). Increased expression of camp (asCATH2) in lice-infested fish was suggested to be a host signal that facilitates the recognition of host by the parasite (Núñez-Acuña et al., 2018). The association of elevated level of camp with modulated expression of B-cell lymphoma 3 (bcl3), nuclear factor kappa B (NF-κB) and vitamin D receptor (VDR) has been already demonstrated in human keratinocytes (Büchau et al., 2009; Park et al., 2011). A variety of secreted gel-forming and transmembrane mucins (up: muc17; down: muc19; up-down: muc2, muc3a and muc5ac) were differentially expressed in most/all families. In addition, inconsistent transcript levels of fcgbp encoding a mucin-like protein that interacts with muc2 to preserve the integrity of the mucosal layer were detected among families (Liu et al., 2022). The modulation of muc2, muc4 and muc5 has been previously reported in lice-infested Atlantic salmon (Caballero-Solares et al., 2022; Gao et al., 2024; Salisbury et al., 2024). The mucus layer contains protective and signal-delivering mucin glycoproteins that are typically secreted upon exposure to various stressors and pathogens (Kufe, 2009). The glycan-rich molecules present on the surface of lice or in their secretions may interact with host mucins by modifying and/or mimicking their structures to persist on the host. Representatives of lectin-associated genes differentially expressed in the skin of families infested with adult versus chalimus stages of lice included: asal (up); mrc2, pla2r1, colec12, clec10a [mgl], clec3ba, clec4e [mincle], clec4m [cd209l], cd209a, cd209c, cd209e, zg16, lgals4, lgalsl and lgalsla (down); cd22, mrc1 and siglec1 (up-down). Despite enhanced expression of the asal gene encoding a poorly-characterized mannose-specific lectin, the majority of C-type (CLECs), S-type (galectins) and I-type (siglecs) lectins that are involved in recognition/presentation of pathogen-specific biomolecules such as carbohydrate and chitin represented reduced expression in most families, suggesting persistent immune evasion by the parasite during infestation (Varki and Angata, 2006; Cote et al., 2017; Sousa et al., 2024). In lice-resistant salmonid species (i.e., Coho salmon), multiple classes of C-type lectin receptors (clec6a, clec4e, and cd209) are strongly overexpressed in early stages of infestation, which facilitates the rejection of the attached parasite (Braden et al., 2023). The expression of nccrp1, serving as a receptor-like protein expressed in non-specific cytotoxic cells (NCCs), was enhanced in almost all families. nccrp1 is believed to play a role in the recognition of cells infected by pathogens and activation of cytotoxic responses by activating intracellular signaling pathways, notably the JAK-STAT pathway (Sukeda et al., 2023). This gene has been reported to bind to the receptors of some protozoan parasites and bacterial pathogens (Sukeda et al., 2023; Teng et al., 2023). Several genes related to the complement system were modulated in adult-compared to chalimus-infested families (up: c1qbp, cfh and c5ar1; down: c1r, cd93, c4, c4b and cfd; up-down: c3 and cd55). c1qbp encodes a multifunctional protein involved in mitochondrial fitness, inflammation, apoptosis regulation and ribosome biogenesis (Wang et al., 2022). The elevated expression of c1qbp may represent a regulatory mechanism for cGAS activation, as its higher expression was observed in all families (Song et al., 2021). Complement factor H (cfh) is an essential inhibitory regulator of the alternative pathway activation, whereas c5ar1 stimulates chemotaxis in response to C5a (Coulthard et al., 2018; Saxena et al., 2024). All complement-relevant genes with lowered expression were found to be involved in activation of the complement system and innate immune responses. Taken together, these changes implicated the possible suppression/dysfunction of components correlated with complement pathways.
Genes encoding major histocompatibility (MHC) antigens/molecules (down: bl3-7, h2-q10, mr1, rt1-b; up-down: h2aa) showed differential expression in adult- versus chalimus-infested families. MHC class I and II proteins play a pivotal role in the cell-mediated adaptive immune responses, and their reduced/modulated expression suggest diminished immune function (Wieczorek et al., 2017). The expression of immunoglobulin-associated genes was modulated in families infested with adult versus chalimus stages (up: vsig10l and vsig8b; down: kbas, mopc-321, lrig1 and ighd; up-down: pigr). Many of the members of the V-Set and immunoglobulin (Ig) domain-containing (VSIG) protein superfamily represent immunosuppressive properties. For example, both vsig8b and vsig10 can suppress T-cell proliferation and cytokine production (Zhou et al., 2022). Depressed transcript levels of lrig1 and ighd may induce chronic inflammation and reduce antigen binding activity, respectively (Nakamura et al., 2013; Qiu et al., 2021). The gene pigr encodes a receptor protein that transports polymeric immunoglobulins (IgM) to the mucosal surface of epithelial cells, and its regulation is controlled by interferons and cytokines (Kong et al., 2018). Some genes belonging to the butyrophilin family showed lower expression in most/all families (btn1a1, btn2a2, btn3a1, btn3a2, btnl2). Butyrophilins belong to the immunoglobulin superfamily and are associated with the MHC complex, primarily serving as co-signaling molecules involved in T lymphocyte regulation (Malinowska et al., 2017). Most aforementioned butyrophilins (except btn3a1) represent inhibitory functions on T-cell proliferation and/or cytokine/interferon production, and their reduced expression may impair tissue repair and contribute to chronic inflammation (Lebrero-Fernández et al., 2016). The expression of T-cell (up: vtcn1, mal and pbk; down: gzma, cd2, cd6, cd28, nfatc2, rftn1, rftn2, trac, trbc2, trb and cabin1) and B-cell (up: bcl3 and bcl10; down: bcl6b, pbx1 and pbxip1; up-down: bcl11b and cd22) relevant transcripts was modulated in the skin of salmon infested by adult versus chalimus. As an immune checkpoint molecule, vtcn1 negatively regulates the immune reactions by inhibiting T-cell activation/proliferation and cytokine production (Vaishnav et al., 2022). Reduced expression of receptors/antigens essential for T-cell activation, proliferation and/or differentiation (gzma, cd2, cd6, cd28, trac, trbc2, trb) and nfatc2 that is a transcription factor implicated in induced expression of cytokine genes in T-cells indicated the repressed adaptive immune responses (Peng et al., 2001; Antonacci et al., 2020; Tian et al., 2022). bcl10 is a central component of the card11-bcl10-malt1 (CBM) signaling complex, which links antigen receptor signaling to NF-κB activation. Likewise, bcl3 is a context-dependent regulator of T cell responses that controls NF-κB activity (Herrington and Nibbs, 2016). bcl3 was reported to alter cytokine secretion profile by dendritic cells in response to carbohydrate-expressing pathogens like Schistosoma mansoni (through interaction of the parasite sugar ligands with cd209 [dc-sign]), leading to inhibited expression of pro-inflammatory cytokines but enhanced expression of il10 and chemokines capable of attracting Th2 cells (Gringhuis et al., 2014). Similar anti-parasitic responses were also reported in Coho salmon, where Th1-type response was associated with lice rejection (Braden et al., 2023). The lack of such responses in Atlantic salmon makes this species prone to infestation with sea lice (Sveen et al., 2025).
Representatives of markers associated with other immune cells (neutrophils, monocytes/macrophages, mast cells and dendritic cells) that were differentially expressed in families infested by different developmental stages of lice included: lect2, ncf2, arg1, arg2 and lifr (up); lsp1a, aif1, kit, kita, csf1a and mrc2 (down); serpinb1, mrc1 and mst1r (up-down). Of the two lect2 genes existing in teleost fish, lect2-b is highly expressed in mucosal organs, such as the skin and intestine (Hu et al., 2022). Enhanced expression of lect2, a multifunctional protein that acts as a chemotactic factor (Lao et al., 2024), was also reported in Atlantic salmon infested with early to advanced stages of sea lice (Skugor et al., 2008; Gallardi et al., 2019; Øvergård et al., 2023). Co-stimulation of arg1 and arg2 may contribute to T-cell hyporesponsiveness as a result of depletion of L-arginine required for T-cell proliferation and function (Martí i Líndez and Reith, 2021). mst1r (ron), mainly expressed in the tissue-resident macrophages, inhibits inflammation and promotes wound healing (Dai et al., 2016). The reduced expression of mst1r (loss of ron signaling) in most families could result in prolonged inflammation, impaired tissue repair and increased susceptibility to infestation (Hunt et al., 2023). Unlike mcr1 which is more specialized for pathogen recognition, mrc2 is involved in collagen turnover process (Melander et al., 2015). The reduced expression of mrc2 concurrent with modulation of its receptor, i.e., plaur (up-down at 10°C and up at 20°C in all families) suggested the potential changes in extracellular matrix (ECM) remodeling and cellular migration (Zhao et al., 2022). serpinb1 is a protease inhibitor that regulates neutrophil serine proteases, protecting tissues from excessive damage during inflammation (Choi et al., 2019). The transcript levels of several genes corresponding to melanogenesis, melanocyte regulation, melanocyte survival, melanosome transportation and/or skin pigmentation (e.g., mc5r, mlph, mreg, mitf, sox4a, sox10, pax3, pax7, pax7a, adamts20, rab27b, rab38, kitlg, kitlga, kit and kita) were found to be reduced in adult-compared to chalimus-infested fish (Kawakami and Fisher, 2017; Xu et al., 2020; Myung et al., 2021; Sevilla and Grichnik, 2024). The kit receptor tyrosine kinase and kit ligand have been duplicated in teleost, where kita and kitlga are the paralogs associated with melanogenesis (Braasch et al., 2007; Hultman et al., 2007). Melanin-based pigmentation was suggested to be correlated with immunocompetence and resistance to ectoparasites in Atlantic salmon (Kittilsen et al., 2012). kit/kita receptors are mainly expressed on melanocytes and mast cells, and their decreased expression might be associated with enhanced levels of il4 and adrenocorticotropic hormone (ACTH) that suppress melanin production (Pillaiyar et al., 2017).
The wound healing process in fish involves four overlapping phases: re-epithelialization, inflammation, granulation (repair) tissue formation and tissue remodelling (Sveen et al., 2020). Prolonged parasitic infestations can lead to a sustained inflammatory response, which hinders the process of wound healing and promotes the development of chronic wounds (Skugor et al., 2008; Gumede et al., 2024). Chronic wounds are characterized by persistent inflammation (elevated levels of neutrophils and pro-inflammatory macrophages), a reduced proliferative phase and increased proteolytic activity (Wilkinson and Hardman, 2020). The transcription level of a large number of genes involved in ECM architecture and wound healing process/regulation was modulated in adult-compared to chalimus-infested families (up: tmprss13b, anxa1, panx1, cd44b, plat, ctnnb1, errfi1, pim1, met, myl6 and tpm3; down: eln, ogn, krt18a, krt20, krt8.2.l, hpse2, prss16, cd44, ctsw, ctsl, serpine1, dact1, raf1, adra2b, ret, mras, mdkb, ccdc80l2, ppara and pparb2b, myh10; up-down: col1a1, col6a2, krt13, krt18, krt8, mylk and myo1c). The majority of structural and non-structural components of ECM revealed reduced expression. For instance, there were 13 DEGs belonging to the collagen family (col1a1a, col1a1b, col1a2, col2a1, col2a1b, col5a1, col6a3, col7a1, col10a1, col10a1a, col11a1, col16a1 and col27a1b) that consistently showed reduced expression in all families at both temperatures. In addition, matricellular proteins of different families (CCN, SPARC, ENPP, fasciclin, fibulin, olfactomedin, R-spondin, tenascin, thrombospondin) mainly had decreased transcription levels (down: ccn1, ccn2, ccn4, thbs2a, thbs2, tnn, tnxb, postna, postn, sparc, smoc2, sparcl1, rspo3, spon1, spon2, fbln7, abi3bp, enpp2 and adgrl2a; up-down: thbs1 and smoc1). Unlike structural ECM proteins, matricellular proteins primarily function as modulators of angiogenesis, inflammation and ECM assembly, with their expression significantly increasing at the site of injury to facilitate the repair process (Cárdenas-León et al., 2022). The reduced expression of both structural and non-structural ECM components in the skin can lead to compromised skin integrity, promoted chronic inflammation and delayed wound healing (Cárdenas-León et al., 2022). Transglutaminases (TGs), also known as protein-glutamine γ-glutamyltransferases, catalyze the formation of amide bonds between proteins to form insoluble crosslinked protein structures resistant to proteases degradation (Martins and Choupina, 2018). The expression of some genes related to the transglutaminase family was modulated in most/all families at both temperatures (up: tgm2, tgm3, tgm1l1 and tgm5l; up-down: tgm1). tgm1, tgm3 and tgm5 are expressed in epithelial tissue and involved in cell-envelope formation during keratinocyte differentiation, whereas tgm2 is produced in different tissues and plays a role in multiple processes such as wound healing and apoptosis. The induced expression of TGs is associated with extremely stressful conditions and mucosal inflammation/epithelial lesions (Lorand and Graham, 2003; Mehta, 2005). The enrichment of the GO term “keratinization” consisted of representatives of TGs and keratins (krt8.2.l and krt8) revealed the possible protective role of epidermal keratinization against infestation caused by adult lice. In addition, some genes encoding proteins with important roles in skin barrier function (up: degs2, cldn7b, aloxe3; down: cldn3c, cldn4, cldn5, cldn5a, pnpla1 [only at 10°C]) were modulated in most/all families. degs2 encodes a bifunctional enzyme that produces ceramides, and aloxe3 encodes the eLOX3 enzyme that is involved in the production of oxylipins (Krieg et al., 2013; Song et al., 2023). The enhanced expression of degs2 and aloxe3 could maintain lipid balance and contribute to the permeable barrier function of the skin against infestation with adult stages. The altered expression of claudins that play a crucial role in maintaining the integrity of tight junctions could result in dysregulated epithelial permeability (Angelow et al., 2008).
A group of matrix metalloproteinase enzymes (up: mmp9 and mmp13; down: mmp15, mmp16 and mmp23bb; up-down: mmp14 and mmp19) demonstrated significant transcriptional changes in the skin infested by adult versus chalimus stages. mmp9 and mmp13 respectively degrade type IV and type II collagens as their preferred substrates (Yabluchanskiy et al., 2013; Hu and Ecker, 2021). mmp9 (gelatinase B) is secreted by a variety of cell types, including neutrophils, macrophages, and skin keratinocytes/fibroblasts (He et al., 2023), and it can increase mRNA levels of pro-inflammatory cytokines and modulates chemokine signaling, resulting in impaired vascular remodelling and delayed resolution of skin inflammation (Yabluchanskiy et al., 2013). mmp13 (collagenase 3) which is produced by keratinocytes, fibroblasts and macrophages is abundantly expressed in chronic wounds but not in acute wounds and the normal skin epidermis, suggesting its role in delayed wound healing process (Vaalamo et al., 1997; Saarialho-Kere, 1998). The systematic increase of mmp9 and mmp13 has been suggested as a hallmark of transit from acute to chronic inflammation following sea lice infestation (Skugor et al., 2008; Tadiso et al., 2011). Wound-mediated induction of proteinases can not only break down several components of the ECM but also modulate the activity of growth factors (Stamenkovic, 2003). In this regard, multiple genes encoding growth factor-related peptides (TGF-β, IGFs or other growth factors) that have important roles in various phases of wound healing exhibited variable expression levels in adult- versus chalimus-infested families (e.g., tgfb1 [in 4 families], fgfbp1, hbegf, vegfaa [up]; tgfb2, tgfb3, tgfbr1, ltbp4, igf1, igfbp5, pdgfc, pdgfa [down]). In addition, some genes associated with BMPs, SMAD and WFIKKN proteins (e.g., bmp2, bmp4, bmp7, smad6, smad7, wfikkn2) represented lower expression. bmp2 and bmp4 promote angiogenesis and tissue remodeling, whereas smad6 and smad7 respectively function as inhibitory regulators of BMP and TGF-β signaling pathways to maintain tissue homeostasis (Goto et al., 2007; Dyer et al., 2014). wfikkn2 encodes a companion protein that regulates the balance and signaling activity of growth factors such as tgfb1, bmp2 and bmp4 (Szláma et al., 2010; Monestier and Blanquet, 2016). A variety of genes encoding proteins belonging to ADAMs (up: adam8, adam9 and adam28; up-down: adam10), ADAMTSs (down: adamts2, adamts2a, adamts5, adamts10, adamts17, adamts20), TIMPs (up: timp2; down: timp3) and TRIM (e.g., trim9 [down]; trim16 [up-down]) were differentially expressed in the skin of adult- versus chalimus-infested families. The increased expression of adam9 may impair keratinocytes migration/proliferation and wound re-epithelialization (Chou et al., 2020), delaying wound closure. adam10 sheds cell surface proteins such as Notch receptors and mediates E-cadherin shedding in fibroblasts and keratinocytes (Maretzky et al., 2005; Sezin et al., 2022). The elevated levels of ADAMs may contribute to delayed healing by excessive tissue breakdown, promoting sustained inflammation and dysregulated signaling (Wang Z. et al., 2023). Reduced levels of the abovementioned ADAMTSs proteins that are involved in collagen maturation (adamts2), regulation of ECM structure (adamts5) and formation of microfibrils (adamts10 and adamts17) can lead to chronic wounds, ultimately reducing tissue integrity and delaying the healing process (Kelwick et al., 2015; Karoulias et al., 2020). trim9 regulates cytoskeletal dynamics and cell migration (Menon et al., 2021), which are crucial for the reorganization and closure of wounds. trim16 has been shown to be an important regulator of keratinocyte differentiation and proinflammatory cytokine secretion (Ren et al., 2020). The misregulation of TRIM proteins can contribute to improper wound healing and chronic wound pathology.
The expression of genes related to signaling pathways involved in wound healing such as TGF-β, Notch, Wnt/β-catenin, ErbB, Ephrin/Eph, PI3K/Akt, HIF-1, JAK/STAT, MAPK and focal adhesion has been reported to be dysregulated in chronic wounds (Shi et al., 2015; Leyane et al., 2021; Basu and Martins-Green, 2022; Bonnici et al., 2023). In this study, representatives of genes directly or indirectly associated with JNK/Wnt (up: hif1ab; down: wnt3, wnt5b, wnt6, wnt7a, mapk8 [jnk1], mycb, up-down: lbh), HIF-1 (up: hif1ab, vegfaa, tgfb1 and nos2b [inosb, in 4 families]; down: usp20), Ephrin/Eph (up: ephb4a, epha2 and efna5; down: epha3, epha4, epha7, ephb2, efnb3, efna2 and efna5b; up-down: efnb1), ErbB (down: erbb3 [in 4 families]), Notch (down: dtx1), JAK/STAT (down: stat1, stat5a, stat5b), MAPK (up-down: mapkapk2 and map4k4) and focal adhesion (up: lama3, lamb3, lamc2, itga2, itgb3; down: lama4, itga5, itga1, itgal, itgb1 [cd29]) signaling pathways were identified in adult-compared to chalimus-infested families. In addition, several members of semaphorins, ephrins, netrins (ntn1, ntn1a, ntn2 and ntn3) and slit proteins (slit1, slitrk6, srgap), represented mainly by reduced transcript levels, were enriched in GO/KEGG pathways associated with axon guidance. Axon guidance molecules have been shown to regulate cell migration, angiogenesis and tissue remodeling, and their dysregulation contributes to delayed/impaired wound healing processes (Chilton, 2006; Nakanishi et al., 2022). The increased expression of hif1ab that is stabilized during hypoxia concurrent with deceased expression of hemoglobin subunits (hbb, hba4, hba1, hbb1) that are involved in oxygen transport and binding suggested the possible hypoxic environment in infested sites, which may contribute to wound persistence and delayed healing. Several genes encoding lysyl oxidase (lox) and lox-like (loxl) proteins showed decreased expression in families infested by adult compared to chalimus stages (loxa, lox, loxl2a, loxl2b, loxl3b, loxl4). Lysyl oxidase (lox) proteins facilitate the cross-linking of elastin and collagen fibers, and their altered/dysregulated expression is linked to skin pathologies (Szauter et al., 2005; Laczko and Csiszar, 2020).
As indicated by enrichment analyses, GO terms and KEGG pathways obtained based on comparisons under elevated temperature conditions encompassed a more diverse and frequent set of genes. While this study provides important insights, further research is required to determine whether these gene modulations are driven by elevated temperature, increased lice pathogenicity, or both, as this could not be fully addressed within the scope of the present work. Based on RNA-seq analysis, the transcriptomic responses of salmon families with different lice burden to infestation with adult compared to chalimus stages of lice were not highly dependent to temperature (10/20°C), as the majority of the identified genes were significantly expressed at both temperatures. In addition, the expression patterns resulted from comparisons between families infested by adult versus chalimus stages of sea lice were mostly identical at both thermal conditions. On the other hand, the expression level of genes identified based on these comparisons showed important differences. For example, among the genes expressed across all families, several (e.g., gsr, cox2, ptgis, il1b, pla2g4c, c1qbp, mmp9, mmp13, casp3b, perp, tigara, ftr83, camp, itga2, tubb2a, tubd1 and gdpd3) exhibited reduced upregulation, while others (e.g., tgfb3, adamts17, adamts20, camk1a, trim9, col16a1, col2a1b, col10a1a, fndc1, ncana and ltbp4) represented less pronounced downregulation in F361 at both temperatures. serpinb1 and hspa8 were among DEGs with paralogs exhibiting opposite expression patterns, which showed reduced up- and downregulation in F361 compared to other families. Moreover, several genes related to tissue repair (e.g., mrc2, lama3, lamc2, efna5b, serpine1, wnt5b, itga1, ret, ptk2ba, ptk6, ptprj), growth factor signalling (stat5a, stat5b, smad6, smad7, s100a13), inflammatory response (e.g., nlrp1, bmp7, usp7), melanogenesis (kita), stress/DNA damage response (e.g., mao, maoa, api5, dnaja, dnajb6b, hsp10, trim39, scara3, mgst3) and immune system (e.g., cd209e, sema3a, sema4d, sema6a, sema7a, plxdc1, btn2a2, btn3a1, nfatc2, c1r, cd40, tnfsf10, tnfsf11, tnfsf12, tnfrsf14) were not expressed in F361, while they had modulated (up or down) expression in other families. The qPCR analysis also revealed important differences in the expression level and pattern of col1a1, mmp9 and saa5 at the attachment site (skin) between F361 and other families at both temperatures. These results collectively suggest that genes associated with immune-inflammatory response, tissue repair, apoptosis, cell adhesion and heat shock response are associated with potential differences among salmon families to sea lice infestation.
5 Conclusion
This study presents a novel transcriptomic investigation of Atlantic salmon responses to sea lice infestation, utilizing RNA-seq analysis across multiple salmon families to unravel the complex molecular mechanisms underlying host-parasite interactions. By analyzing the global gene expression profiles of salmon families in response to different sea lice developmental stages (adult vs. chalimus) under both normal and elevated temperatures, we have unveiled a substantially more intricate molecular response than previously recognized. The complexity of transcriptomic responses to sea lice infestation in Atlantic salmon is further reflected by the contrasting transcriptional patterns observed among multiple gene paralogs, suggesting possible functional diversification among closely related genes. This genomic complexity further complicates the molecular response mechanisms employed by Atlantic salmon when confronting pathogens such as sea lice. Our findings reveal significant dysregulation of multiple genes and biological pathways, most notably those associated with inflammatory responses, immune system functioning, and wound healing processes. These disruptions fundamentally compromise the host’s ability to mount an effective defense, ultimately contributing to the persistence and survival of sea lice on susceptible hosts. The induced transcriptomic alterations suggest a multifaceted strategy adopted by sea lice that undermines the host’s defensive capabilities.
Interestingly, our analysis indicates that the transcriptomic response to sea lice infestation was not highly temperature-dependent, although elevated temperature potentiated metabolic and stress responses and may potentially exacerbate the impact of infestation over prolonged exposure. This study demonstrates that family-based transcriptomic analysis is a powerful tool for identifying specific molecular markers associated with sea lice infestation and host resistance, which could be used for breeding more resilient salmon populations with improved defense against sea lice infestation. Overall, this research has enhanced our understanding of the molecular intricacies underlying host-parasite interactions in Atlantic salmon, providing a foundation for future research and potential practical applications in sea lice control. The RNA-seq data generated in this study can support genomic selection for sea lice resistance in salmon breeding programs through integration with genome-wide association studies (GWAS) and the application of weighted correlation network analysis (WGCNA) to identify gene modules associated with infestation.
Data availability statement
The RNA-seq data have been deposited in the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA1256531 (https://www.ncbi.nlm.nih.gov/bioproject/1256531).
Ethics statement
The animal study was reviewed and approved by UPEI Animal Care Committee (Protocol # 16-051). All animal procedures were conducted in accordance with approved protocols under animal use protocol (AUP, no. 20-31), reviewed and approved by the Fisheries and Oceans Canada (Maritimes & Gulf) and Huntsman Marine Science Centre Regional Animal Care Committee.
Author contributions
RG-M: Formal analysis, Data curation, Visualization, Writing – original draft, Writing – review and editing. WC: Formal analysis, Methodology, Writing – review and editing. JDP: Formal analysis, Methodology, Writing – review and editing. SKW: Project administration, Methodology, Writing – review and editing. AFG: Conceptualization, Supervision, Investigation, Resources, Funding acquisition, Writing – review and editing. MDF: Conceptualization, Supervision, Investigation, Validation, Funding acquisition, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was possible through the Mitigating the Impact of Climate-Related Challenges on Salmon Aquaculture (MICCSA) project with financial support from the Atlantic Canada Opportunities Agency (ACOA) – Atlantic Innovation Fund (AIF) (ACOA-AIF program Project # 781-9658-205222), the Huntsman Marine Science Centre (Huntsman Marine) and the Center for Aquaculture Technologies Canada (CATC) and Innovation PEI; a graduate research stipend from AVC-UPEI (post-doctoral support to RG from Mitacs Accelerate program to AVC-UPEI with Huntsman Marine partner organization, MITACS IT30843 - Fast).
Acknowledgments
The authors would like to thank the Huntsman Marine Science Centre staff who were involved in family production, general husbandry and setting up and executing the sea lice challenge. We would also like to thank Chris Bridger (Huntsman Marine) for his comments related to the MICCSA project. We gratefully acknowledge the support of Genome Canada and Genome Atlantic. We would also like to thank Atlantic Computational Excellence Network (ACENET) and Compute Canada for providing computational resources for RNA-Seq data analyses.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2025.1633603/full#supplementary-material
References
Alfonso, S., Gesto, M., and Sadoul, B. (2021). Temperature increase and its effects on fish stress physiology in the context of global warming. J. Fish. Biol. 98 (6), 1496–1508. doi:10.1111/jfb.14599
Angelow, S., Ahlstrom, R., and Yu, A. S. L. (2008). Biology of claudins. Am. J. Physiol. Ren. Physiol. 295 (4), F867–F876. doi:10.1152/ajprenal.90264.2008
Antonacci, R., Massari, S., Linguiti, G., Caputi Jambrenghi, A., Giannico, F., Lefranc, M. P., et al. (2020). Evolution of the T-cell receptor (TR) loci in the adaptive immune response: the tale of the TRG locus in mammals. Genes. 11 (6), 624. doi:10.3390/genes11060624
Barker, S. E., Bricknell, I. R., Covello, J., Purcell, S., Fast, M. D., Wolters, W., et al. (2019). Sea lice, Lepeophtheirus salmonis (krøyer 1837), infected Atlantic salmon (Salmo Salar L.) are more susceptible to infectious salmon anemia virus. PLoS One 14 (1), e0209178. doi:10.1371/journal.pone.0209178
Basu, P., and Martins-Green, M. (2022). Signaling pathways associated with chronic wound progression: a systems biology approach. Antioxidants 11 (8), 1506. doi:10.3390/antiox11081506
Beemelmanns, A., Zanuzzo, F. S., Xue, X., Sandrelli, R. M., Rise, M. L., and Gamperl, A. K. (2021). The transcriptomic responses of Atlantic salmon (Salmo Salar) to high temperature stress alone, and in combination with moderate hypoxia. BMC Genomics 22, 261. doi:10.1186/s12864-021-07464-x
Bindea, G., Mlecnik, B., Hackl, H., Charoentong, P., Tosolini, M., Kirilovsky, A., et al. (2009). ClueGO: a cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25 (8), 1091–1093. doi:10.1093/bioinformatics/btp101
Bonnici, L., Suleiman, S., Schembri-Wismayer, P., and Cassar, A. (2023). Targeting signalling pathways in chronic wound healing. Int. J. Mol. Sci. 25 (1), 50. doi:10.3390/ijms25010050
Braasch, I., Schartl, M., and Volff, J. N. (2007). Evolution of pigment synthesis pathways by gene and genome duplication in fish. BMC Evol. Biol. 7, 74. doi:10.1186/1471-2148-7-74
Braden, L. M., Koop, B. F., and Jones, S. R. M. (2015). Signatures of resistance to Lepeophtheirus salmonis include a TH2-type response at the louse-salmon interface. Dev. Comp. Immunol. 48 (1), 178–191. doi:10.1016/j.dci.2014.09.015
Braden, L. M., Michaud, D., Groman, D., Byrne, P., Hori, T. S., and Fast, M. D. (2023). Rejection of Lepeophtheirus salmonis driven in part by chitin sensing is not impacted by seawater acclimitization in coho salmon (Oncorhynchus kisutch). Sci. Rep. 13 (1), 9685. doi:10.1038/s41598-023-36632-0
Braden, L. M., Rasmussen, K. J., Purcell, S. L., Ellis, L., Mahony, A., Cho, S., et al. (2018). Acquired protective immunity in Atlantic salmon Salmo Salar against the myxozoan kudoa Thyrsites involves induction of MHIIβ+ CD83+ antigen-presenting cells. Infect. Immun. 86 (1), e00556-17–17. doi:10.1128/iai.00556-17
Büchau, A. S., MacLeod, D. T., Morizane, S., Kotol, P. F., Hata, T., and Gallo, R. L. (2009). Bcl-3 acts as an innate immune modulator by controlling antimicrobial responses in keratinocytes. J. Investig. Dermatol. 129 (9), 2148–2155. doi:10.1038/jid.2009.49
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55 (4), 611–622. doi:10.1373/clinchem.2008.112797
Caballero-Solares, A., Umasuthan, N., Xue, X., Katan, T., Kumar, S., Westcott, J. D., et al. (2022). Interacting effects of sea louse (Lepeophtheirus salmonis) infection and formalin-killed Aeromonas salmonicida on Atlantic salmon skin transcriptome. Front. Immunol. 13, 804987. doi:10.3389/fimmu.2022.804987
Cáceres, P., Lopéz, P., Garcia, B., Cichero, D., Ødegård, J., Moen, T., et al. (2024). Meta-analysis of GWAS for sea lice load in Atlantic salmon. Aquaculture 584, 740543. doi:10.1016/j.aquaculture.2024.740543
Cai, W., Kumar, S., Umasuthan, N., Caballero-Solares, A., Carvalho, L. A., Whyte, S. K., et al. (2022). Transcriptome analysis of Atlantic salmon (Salmo Salar) skin in response to sea lice and infectious salmon anemia virus co-infection under different experimental functional diets. Front. Immunol. 12, 787033. doi:10.3389/fimmu.2021.787033
Cai, W., Zhong, L., Parrish, K., Kumar, S., Eslamloo, K., Fajei, E., et al. (2024). Synergies of co-infecting pathogens, sea lice (Lepeophtheirus salmonis) and Moritella viscosa, are impacted by exposure order, and host response to initial infection. Aquaculture 591, 741115. doi:10.1016/j.aquaculture.2024.741115
Cárdenas-León, C. G., Mäemets-Allas, K., Klaas, M., Lagus, H., Kankuri, E., and Jaks, V. (2022). Matricellular proteins in cutaneous wound healing. Front. Cell. Dev. Biol. 10, 1073320. doi:10.3389/fcell.2022.1073320
Carvalho, L. A., Whyte, S. K., Braden, L. M., Purcell, S. L., Manning, A. J., Muckle, A., et al. (2020). Impact of co-infection with Lepeophtheirus salmonis and Moritella viscosa on inflammatory and immune responses of Atlantic salmon (Salmo Salar). J. Fish. Dis. 43 (4), 459–473. doi:10.1111/jfd.13144
Chalmers, L., Vera, L. M., Taylor, J. F., Adams, A., and Migaud, H. (2018). Comparative ploidy response to experimental hydrogen peroxide exposure in Atlantic salmon (Salmo Salar). Fish. Shellfish Immunol. 81, 354–367. doi:10.1016/j.fsi.2018.07.017
Chang, T., Hunt, B. P. V., Hirai, J., and Suttle, C. A. (2023). Divergent RNA viruses infecting sea lice, major ectoparasites of fish. PLoS Pathog. 19 (6), e1011386. doi:10.1371/journal.ppat.1011386
Chilton, J. K. (2006). Molecular mechanisms of axon guidance. Dev. Biol. 292 (1), 13–24. doi:10.1016/j.ydbio.2005.12.048
Choi, Y. J., Kim, S., Choi, Y., Nielsen, T. B., Yan, J., Lu, A., et al. (2019). SERPINB1-mediated checkpoint of inflammatory caspase activation. Nat. Immunol. 20 (3), 276–287. doi:10.1038/s41590-018-0303-z
Chou, C.-W., Huang, Y.-K., Kuo, T.-T., Liu, J.-P., and Sher, Y.-P. (2020). An overview of ADAM9: structure, activation, and regulation in human diseases. Int. J. Mol. Sci. 21 (20), 7790. doi:10.3390/ijms21207790
Cote, R., Eggink, L. L., and Hoober, J. K. (2017). CLEC receptors, endocytosis and calcium signaling. AIMS Allergy Immunol. 1 (4), 207–231. doi:10.3934/Allergy.2017.4.207
Coulthard, L. G., Hawksworth, O. A., and Woodruff, T. M. (2018). “C3aR1,” in The complement FactsBook (London: Academic Press), 385–396. doi:10.1016/B978-0-12-810420-0.00036-5
Cuesta-Margolles, G., Schlecht-Louf, G., and Bachelerie, F. (2024). ACKR3 in skin homeostasis, an overlooked player in the CXCR4/CXCL12 axis. J. Investig. Dermatol. 145, 1039–1049. doi:10.1016/j.jid.2024.08.022
Dai, W., Zheng, H., Cheung, A. K. L., Tang, C. S. M., Ko, J. M. Y., Wong, B. W. Y., et al. (2016). Whole-exome sequencing identifies MST1R as a genetic susceptibility gene in nasopharyngeal carcinoma. Proc. Natl. Acad. Sci. U.S.A. 113 (12), 3317–3322. doi:10.1073/pnas.1523436113
Dalvin, S., Jørgensen, L. v. G., Kania, P. W., Grotmol, S., Buchmann, K., and Øvergård, A. C. (2020). Rainbow trout Oncorhynchus mykiss skin responses to salmon louse lepeophtheirus Salmonis: from copepodid to adult stage. Fish. Shellfish Immunol. 103, 200–210. doi:10.1016/j.fsi.2020.05.014
Dyer, L. A., Pi, X., and Patterson, C. (2014). The role of BMPs in endothelial cell function and dysfunction. Trends Endocrinol. Metab. 25 (9), 472–480. doi:10.1016/j.tem.2014.05.003
FAO (2024). The state of world fisheries and aquaculture 2024 - blue transformation in action. Rome. doi:10.4060/cd0683en
Fast, M., and Braden, L. (2022). “Salmon-louse interaction and immunological consequences for the host,” in Sea lice biology and control (Wallingford, UK: CABI), 272–293. doi:10.1079/9781800629301.0013
Fast, M. D., Johnson, S. C., Eddy, T. D., Pinto, D., and Ross, N. W. (2007). Lepeophtheirus salmonis secretory/excretory products and their effects on Atlantic salmon immune gene regulation. Parasite Immunol. 29 (4), 179–189. doi:10.1111/j.1365-3024.2007.00932.x
Fast, M. D., Muise, D. M., Easy, R. E., Ross, N. W., and Johnson, S. C. (2006). The effects of Lepeophtheirus salmonis infections on the stress response and immunological status of Atlantic salmon (Salmo Salar). Fish. Shellfish Immunol. 21 (3), 228–241. doi:10.1016/j.fsi.2005.11.010
Fast, M. D., Ross, N. W., Craft, C. A., Locke, S. J., MacKinnon, S. L., and Johnson, S. C. (2004). Lepeophtheirus Salmonis: characterization of prostaglandin E2 in secretory products of the salmon louse by RP-HPLC and mass spectrometry. Exp. Parasitol. 107 (1-2), 5–13. doi:10.1016/j.exppara.2004.04.001
Firth, K. J., Johnson, S. C., and Ross, N. W. (2000). Characterization of proteases in the skin mucus of Atlantic salmon (Salmo Salar) infected with the salmon louse (Lepeophtheirus salmonis) and in whole-body louse homogenate. J. Parasitol. 86 (6), 1199–1205. doi:10.1645/0022-3395(2000)086[1199:COPITS]2.0.CO;2
Franke, A., Beemelmanns, A., and Miest, J. J. (2024). Are fish immunocompetent enough to face climate change? Biol. Lett. 20 (2), 20230346. doi:10.1098/rsbl.2023.0346
Gallardi, D., Xue, X., Hamoutene, D., Lush, L., and Rise, M. L. (2019). Impact of origin (Wild vs. farmed) and sea lice (Lepeophtheirus salmonis) infestation on expression of immune-relevant genes in Atlantic salmon (Salmo Salar L.) skin. Aquaculture 499, 306–315. doi:10.1016/j.aquaculture.2018.09.036
Gao, S., Tan, S., Purcell, S. L., Whyte, S. K., Parrish, K., Zhong, L., et al. (2024). A comparative analysis of alternative splicing patterns in Atlantic salmon (Salmo Salar) in response to Moritella viscosa and sea lice (Lepeophtheirus salmonis) infection. Fish. Shellfish Immunol. 149, 109606. doi:10.1016/j.fsi.2024.109606
Godwin, S. C., Fast, M. D., Kuparinen, A., Medcalf, K. E., and Hutchings, J. A. (2020). Increasing temperatures accentuate negative fitness consequences of a marine parasite. Sci. Rep. 10 (1), 18467. doi:10.1038/s41598-020-74948-3
Goto, K., Kamiya, Y., Imamura, T., Miyazono, K., and Miyazawa, K. (2007). Selective inhibitory effects of Smad6 on bone morphogenetic protein type I receptors. J. Biol. Chem. 282 (28), 20603–20611. doi:10.1074/jbc.M702100200
Gringhuis, S. I., Kaptein, T. M., Wevers, B. A., Mesman, A. W., and Geijtenbeek, T. B. H. (2014). Fucose-specific DC-SIGN signalling directs T helper cell type-2 responses via IKKε- and CYLD-Dependent Bcl3 activation. Nat. Commun. 5 (1), 3898. doi:10.1038/ncomms4898
Gumede, D. B., Abrahamse, H., and Houreld, N. N. (2024). Targeting Wnt/β-catenin signaling and its interplay with TGF-β and notch signaling pathways for the treatment of chronic wounds. Cell. Commun. Signal. 22 (1), 244. doi:10.1186/s12964-024-01623-9
Hamre, L. A., Bui, S., Oppedal, F., Skern-Mauritzen, R., and Dalvin, S. (2019). Development of the salmon louse Lepeophtheirus salmonis parasitic stages in temperatures ranging from 3 to 24°C. Aquac. Environ. Interact. 11, 429–443. doi:10.3354/aei00320
Hamre, L. A., Eichner, C., Caipang, C. M. A., Dalvin, S. T., Bron, J. E., Nilsen, F., et al. (2013). The salmon louse Lepeophtheirus salmonis (copepoda: caligidae) life cycle has only two chalimus stages. PLoS One 8 (9), e73539. doi:10.1371/journal.pone.0073539
He, L., Kang, Q., Chan, K. I., Zhang, Y., Zhong, Z., and Tan, W. (2023). The immunomodulatory role of matrix metalloproteinases in colitis-associated cancer. Front. Immunol. 13, 1093990. doi:10.3389/fimmu.2022.1093990
Herrington, F. D., and Nibbs, R. J. B. (2016). Regulation of the adaptive immune response by the IκB family protein Bcl-3. Cells 5 (2), 14. doi:10.3390/cells5020014
Hirose, S., Mashima, T., Yuan, X., Yamashita, M., Kitano, S., Torii, S., et al. (2024). Interleukin-4 induced 1-mediated resistance to an immune checkpoint inhibitor through suppression of CD8+ T cell infiltration in melanoma. Cancer Sci. 115 (3), 791–803. doi:10.1111/cas.16073
Holm, H., Santi, N., Kjøglum, S., Perisic, N., Skugor, S., and Evensen, Ø. (2015). Difference in skin immune responses to infection with salmon louse (Lepeophtheirus salmonis) in Atlantic salmon (Salmo Salar L.) of families selected for resistance and susceptibility. Fish. Shellfish Immunol. 42 (2), 384–394. doi:10.1016/j.fsi.2014.10.038
Hu, Q., and Ecker, M. (2021). Overview of MMP-13 as a promising target for the treatment of osteoarthritis. Int. J. Mol. Sci. 22 (4), 1742. doi:10.3390/ijms22041742
Hu, Y. Z., Ma, Z. Y., Wu, C. S., Wang, J., Zhang, Y. A., and Zhang, X. J. (2022). LECT2 is a novel antibacterial protein in vertebrates. J. Immunol. 208 (8), 2037–2053. doi:10.4049/jimmunol.2100812
Huangfu, L., Li, R., Huang, Y., and Wang, S. (2023). The IL-17 family in diseases: from bench to bedside. Signal Transduct. Target. Ther. 8 (1), 402. doi:10.1038/s41392-023-01620-3
Hultman, K. A., Bahary, N., Zon, L. I., and Johnson, S. L. (2007). Gene duplication of the zebrafish kit ligand and partitioning of melanocyte development functions to kit ligand a. PLoS Genet. 3 (1), e17. doi:10.1371/journal.pgen.0030017
Hunt, B. G., Fox, L. H., Davis, J. C., Jones, A., Lu, Z., and Waltz, S. E. (2023). An introduction and overview of RON receptor tyrosine kinase signaling. Genes. 14 (2), 517. doi:10.3390/genes14020517
Ignatz, E. H., Hall, J. R., Eslamloo, K., Gamperl, A. K., and Rise, M. L. (2024). Characterization and transcript expression analyses of four Atlantic salmon (Salmo Salar) serpinh1 paralogues provide evidence of evolutionary divergence. Gene 894, 147984. doi:10.1016/j.gene.2023.147984
Islam, M. J., Kunzmann, A., and Slater, M. J. (2022). Responses of aquaculture fish to climate change-induced extreme temperatures: a review. J. World Aquac. Soc. 53 (2), 314–366. doi:10.1111/jwas.12853
Jensen, L. B., Wahli, T., McGurk, C., Eriksen, T. B., Obach, A., Waagbø, R., et al. (2015). Effect of temperature and diet on wound healing in Atlantic salmon (Salmo Salar L.). Fish. Physiol. Biochem. 41, 1527–1543. doi:10.1007/s10695-015-0105-2
Johnson, S., Ewart, K., Osborne, J., Delage, D., Ross, N., and Murray, H. (2002). Molecular cloning of trypsin cDNAs and trypsin gene expression in the salmon louse Lepeophtheirus salmonis (copepoda: caligidae). Parasitol. Res. 88, 789–796. doi:10.1007/s00436-002-0656-x
Johnson, S. C., and Fast, M. D. (2004). “Interactions between sea lice and their hosts,” in Host-parasite interactions, 131–159.
Karoulias, S. Z., Taye, N., Stanley, S., and Hubmacher, D. (2020). The ADAMTS/Fibrillin connection: insights into the biological functions of ADAMTS10 and ADAMTS17 and their respective sister proteases. Biomolecules 10 (4), 596. doi:10.3390/biom10040596
Kawakami, A., and Fisher, D. E. (2017). The master role of microphthalmia-associated transcription factor in melanocyte and melanoma biology. Lab. Investig. 97 (6), 649–656. doi:10.1038/labinvest.2017.9
Kelwick, R., Desanlis, I., Wheeler, G. N., and Edwards, D. R. (2015). The ADAMTS (A disintegrin and metalloproteinase with thrombospondin motifs) family. Genome Biol. 16, 113. doi:10.1186/s13059-015-0676-3
Kittilsen, S., Johansen, I. B., Braastad, B. O., and Øverli, Ø. (2012). Pigments, parasites and personality: towards a unifying role for steroid hormones? PLoS One 7 (4), e34281. doi:10.1371/journal.pone.0034281
Kong, X., Wang, L., Pei, C., Zhang, J., Zhao, X., and Li, L. (2018). Comparison of polymeric immunoglobulin receptor between fish and mammals. Vet. Immunol. Immunopathol. 202, 63–69. doi:10.1016/j.vetimm.2018.06.002
Krasnov, A., Skugor, S., Todorcevic, M., Glover, K. A., and Nilsen, F. (2012). Gene expression in Atlantic salmon skin in response to infection with the parasitic copepod Lepeophtheirus salmonis, cortisol implant, and their combination. BMC Genomics 13, 130. doi:10.1186/1471-2164-13-130
Krasnov, A., Wesmajervi Breiland, M. S., Hatlen, B., Afanasyev, S., and Skugor, S. (2015). Sexual maturation and administration of 17β-estradiol and testosterone induce complex gene expression changes in skin and increase resistance of Atlantic salmon to ectoparasite salmon louse. Gen. Comp. Endocrinol. 212, 34–43. doi:10.1016/j.ygcen.2015.01.002
Krieg, P., Rosenberger, S., De Juanes, S., Latzko, S., Hou, J., Dick, A., et al. (2013). Aloxe3 knockout mice reveal a function of epidermal lipoxygenase-3 as hepoxilin synthase and its pivotal role in barrier formation. J. Investig. Dermatol. 133 (1), 172–180. doi:10.1038/jid.2012.250
Kufe, D. W. (2009). Mucins in cancer: function, prognosis and therapy. Nat. Rev. Cancer 9 (12), 874–885. doi:10.1038/nrc2761
Laczko, R., and Csiszar, K. (2020). Lysyl oxidase (LOX): functional contributions to signaling pathways. Biomolecules 10 (8), 1093. doi:10.3390/biom10081093
Langhansova, H., Bopp, T., Schmitt, E., and Kopecký, J. (2015). Tick saliva increases production of three chemokines including monocyte chemoattractant protein-1, a histamine-releasing cytokine. Parasite Immunol. 37 (2), 92–96. doi:10.1111/pim.12168
Lao, J., Zhu, H., You, Q., Nie, M., and Pathak, J. L. (2024). Updates on the role of leukocyte cell-derived chemotaxin-2 in inflammation regulation and immunomodulation. Cytokine 181, 156697. doi:10.1016/j.cyto.2024.156697
Larsen, M. L., and Vormedal, I. (2021). The environmental effectiveness of sea lice regulation: compliance and consequences for farmed and wild salmon. Aquaculture 532, 736000. doi:10.1016/j.aquaculture.2020.736000
Lebrero-Fernández, C., Wenzel, U. A., Akeus, P., Wang, Y., Strid, H., Simrén, M., et al. (2016). Altered expression of butyrophilin (BTN) and BTN-like (BTNL) genes in intestinal inflammation and Colon cancer. Immun. Inflamm. Dis. 4 (2), 191–200. doi:10.1002/iid3.105
Lee, W. S., Lee, W. H., Bae, Y. C., and Suk, K. (2019). Axon guidance molecules guiding neuroinflammation. Exp. Neurobiol. 28, 311–319. doi:10.5607/en.2019.28.3.311
Leong, J. S., Jantzen, S. G., von Schalburg, K. R., Cooper, G. A., Messmer, A. M., Liao, N. Y., et al. (2010). Salmo Salar and Esox lucius full-length cDNA sequences reveal changes in evolutionary pressures on a post-tetraploidization genome. BMC Genomics 11, 279. doi:10.1186/1471-2164-11-279
Leyane, T. S., Jere, S. W., and Houreld, N. N. (2021). Cellular signalling and photobiomodulation in chronic wound repair. Int. J. Mol. Sci. 22 (20), 11223. doi:10.3390/ijms222011223
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30 (7), 923–930. doi:10.1093/bioinformatics/btt656
Lien, S., Koop, B. F., Sandve, S. R., Miller, J. R., Kent, M. P., Nome, T., et al. (2016). The Atlantic salmon genome provides insights into rediploidization. Nature 533 (7602), 200–205. doi:10.1038/nature17164
Lira, L. V. G., Mastrochirico-Filho, V. A., Mendes, N. J., Ariede, R. B., Yáñez, J. M., and Hashimoto, D. T. (2023). Genome-wide association study of host resistance to the ectoparasite Ichthyophthirius multifiliis in the amazon fish Colossoma macropomum. Mol. Biol. Rep. 50 (1), 599–607. doi:10.1007/s11033-022-08062-0
Liu, Q., Niu, X., Li, Y., Zhang, J., Zhu, S., Yang, Q., et al. (2022). Role of the mucin-like glycoprotein FCGBP in mucosal immunity and cancer. Front. Immunol. 13, 863317. doi:10.3389/fimmu.2022.863317
Lorand, L., and Graham, R. M. (2003). Transglutaminases: crosslinking enzymes with pleiotropic functions. Nat. Rev. Mol. Cell. Biol. 4 (2), 140–156. doi:10.1038/nrm1014
Macqueen, D. J., and Johnston, I. A. (2014). A well-constrained estimate for the timing of the salmonid whole genome duplication reveals major decoupling from species diversification. Proc. R. Soc. B Biol. Sci. 281 (1778), 20132881. doi:10.1098/rspb.2013.2881
Malinowska, M., Tokarz-Deptuła, B., and Deptuła, W. (2017). Butyrophilins: an important new element of resistance. Cent. Eur. J. Immunol. 42 (4), 399–403. doi:10.5114/ceji.2017.72806
Malty, R. H., Hudmon, A., Fehrenbacher, J. C., and Vasko, M. R. (2016). Long-term exposure to PGE2 causes homologous desensitization of receptor-mediated activation of protein kinase A. J. Neuroinflammation 13, 181. doi:10.1186/s12974-016-0645-0
Manousou, P., Kolios, G., Valatas, V., Drygiannakis, I., Bourikas, L., Pyrovolaki, K., et al. (2010). Increased expression of chemokine receptor CCR3 and its ligands in ulcerative colitis: the role of colonic epithelial cells in in vitro studies. Clin. Exp. Immunol. 162 (2), 337–347. doi:10.1111/j.1365-2249.2010.04248.x
Maretzky, T., Reiss, K., Ludwig, A., Buchholz, J., Scholz, F., Proksch, E., et al. (2005). ADAM10 mediates E-cadherin shedding and regulates epithelial cell-cell adhesion, migration, and β-catenin translocation. Proc. Natl. Acad. Sci. U.S.A. 102 (26), 9182–9187. doi:10.1073/pnas.0500918102
Martí i Líndez, A. A., and Reith, W. (2021). Arginine-dependent immune responses. Cell. Mol. Life Sci. 78 (13), 5303–5324. doi:10.1007/s00018-021-03828-4
Martins, I. M., and Choupina, A. (2018). “Protein-glutamine gamma-glutamyltransferase,” in Encyclopedia of signaling molecules. Editor S. Choi (Cham: Springer). doi:10.1007/978-3-319-67199-4_101758
McCarthy, E., Cunningham, E., Copley, L., Jackson, D., Johnston, D., Dalton, J. P., et al. (2012). Cathepsin L proteases of the parasitic copepod, Lepeophtheirus salmonis. Aquaculture 356, 264–271. doi:10.1016/j.aquaculture.2012.05.007
Medcalf, K. E., Hutchings, J. A., Fast, M. D., Kuparinen, A., and Godwin, S. C. (2021). Warming temperatures and ectoparasitic sea lice impair internal organs in juvenile Atlantic salmon. Mar. Ecol. Prog. Ser. 660, 161–169. doi:10.3354/meps13610
Mehta, K. (2005). “Mammalian transglutaminases: a family portrait. In: transglutaminases: family of enzymes with diverse functions,”Prog. Exp. Tumor Res., 38. Basel: S. Karger AG, 1–18. doi:10.1159/000084229
Melander, M. C., Jürgensen, H. J., Madsen, D. H., Engelholm, L. H., and Behrendt, N. (2015). The collagen receptor uPARAP/Endo180 in tissue degradation and cancer (review). Int. J. Oncol. 47 (4), 1177–1188. doi:10.3892/ijo.2015.3120
Menon, S., Goldfarb, D., Ho, C. T., Cloer, E. W., Boyer, N. P., Hardie, C., et al. (2021). The TRIM9/TRIM67 neuronal interactome reveals novel activators of morphogenesis. Mol. Biol. Cell. 32 (4), 314–330. doi:10.1091/mbc.E20-10-0622
Meyrath, M., Reynders, N., Uchański, T., Chevigné, A., and Szpakowska, M. (2021). Systematic reassessment of chemokine-receptor pairings confirms CCL20 but not CXCL13 and extends the spectrum of ACKR4 agonists to CCL22. J. Leukoc. Biol. 109 (2), 373–376. doi:10.1002/JLB.2AB0520-275R
Monestier, O., and Blanquet, V. (2016). WFIKKN1 and WFIKKN2: “companion” proteins regulating TGFB activity. Cytokine Growth Factor Rev. 32, 75–84. doi:10.1016/j.cytogfr.2016.06.003
Murakami, M., Masuda, S., and Kudo, I. (2003). Arachidonate release and prostaglandin production by group IVC phospholipase A2 (cytosolic phospholipase A2gamma). Biochem. J. 372 (3), 695–702. doi:10.1042/bj20030061
Myung, C. H., Lee, J. E., Jo, C. S., Park, J. I., and Hwang, J. S. (2021). Regulation of melanophilin (mlph) gene expression by the glucocorticoid receptor (GR). Sci. Rep. 11 (1), 16813. doi:10.1038/s41598-021-96276-w
Nakamura, T., Hamuro, J., Takaishi, M., Simmons, S., Maruyama, K., Zaffalon, A., et al. (2013). LRIG1 inhibits STAT3-dependent inflammation to maintain corneal homeostasis. J. Clin. Investig. 124 (1), 385–397. doi:10.1172/JCI71488
Nakanishi, Y., Kang, S., and Kumanogoh, A. (2022). Axon guidance molecules in immunometabolic diseases. Inflamm. Regen. 42 (1), 5. doi:10.1186/s41232-021-00189-0
Nilsson, J., Barrett, L. T., Mangor-Jensen, A., Nola, V., Harboe, T., and Folkedal, O. (2023). Effect of water temperature and exposure duration on detachment rate of salmon lice (lepeophtheirus Salmonis); testing the relevant thermal spectrum used for delousing. Aquaculture 562, 738879. doi:10.1016/j.aquaculture.2022.738879
Nolan, D. T., Reilly, P., and Wendelaar Bonga, S. E. (1999). Infection with low numbers of the sea louse Lepeophtheirus salmonis induces stress-related effects in postsmolt Atlantic salmon (Salmo Salar). Can. J. Fish. Aquat. Sci. 56 (6), 947–959. doi:10.1139/f99-021
Nuez-Ortín, W. G., Carter, C. G., Nichols, P. D., Cooke, I. R., and Wilson, R. (2018). Liver proteome response of pre-harvest Atlantic salmon following exposure to elevated temperature. BMC Genomics 19, 1–13. doi:10.1186/s12864-018-4517-0
Núñez-Acuña, G., Gallardo-Escárate, C., Fields, D. M., Shema, S., Skiftesvik, A. B., Ormazábal, I., et al. (2018). The Atlantic salmon (Salmo Salar) antimicrobial peptide cathelicidin-2 is a molecular host-associated cue for the salmon louse (Lepeophtheirus salmonis). Sci. Rep. 8 (1), 13738. doi:10.1038/s41598-018-31885-6
Økland, A. L., Nylund, A., Øvergård, A. C., Blindheim, S., Watanabe, K., Grotmol, S., et al. (2014). Genomic characterization and phylogenetic position of two new species in rhabdoviridae infecting the parasitic copepod, salmon louse (Lepeophtheirus salmonis). PLoS One 9 (11), e112517. doi:10.1371/journal.pone.0112517
Oldham, T., Simensen, B., Trengereid, H., and Oppedal, F. (2023). Environmentally responsive parasite prevention halves salmon louse burden in commercial marine cages. Aquaculture 563, 738902. doi:10.1016/j.aquaculture.2022.738902
Oliveira, C. J. F., Cavassani, K. A., Moré, D. D., Garlet, G. P., Aliberti, J. C., Silva, J. S., et al. (2008). Tick saliva inhibits the chemotactic function of MIP-1alpha and selectively impairs chemotaxis of immature dendritic cells by down-regulating cell-surface CCR5. Int. J. Parasitol. 38 (6), 705–716. doi:10.1016/j.ijpara.2007.10.006
Ou, L., Zhang, A., Cheng, Y., and Chen, Y. (2021). The cGAS-STING pathway: a promising immunotherapy target. Front. Immunol. 12, 795048. doi:10.3389/fimmu.2021.795048
Øvergård, A. C., Borchel, A., Eichner, C., Hjertaker, S., Nagata, J., Doherty Midtbø, H. M., et al. (2025). The generalist Caligus elongatus is better at dampening the Atlantic salmon immune response than the salmonid specialist Lepeophtheirus salmonis. Fish. Shellfish Immunol. 160, 110225. doi:10.1016/j.fsi.2025.110225
Øvergård, A. C., Eichner, C., Nuñez-Ortiz, N., Kongshaug, H., Borchel, A., and Dalvin, S. (2023). Transcriptomic and targeted immune transcript analyses confirm localized skin immune responses in Atlantic salmon towards the salmon louse. Fish. Shellfish Immunol. 138, 108835. doi:10.1016/j.fsi.2023.108835
Overton, K., Dempster, T., Oppedal, F., Kristiansen, T. S., Gismervik, K., and Stien, L. H. (2019). Salmon lice treatments and salmon mortality in Norwegian aquaculture: a review. Rev. Aquac. 11 (4), 1398–1417. doi:10.1111/raq.12299
Palazón-Riquelme, P., Worboys, J. D., Green, J., Valera, A., Martín-Sánchez, F., Pellegrini, C., et al. (2018). USP7 and USP47 deubiquitinases regulate NLRP3 inflammasome activation. EMBO Rep. 19 (10), e44766. doi:10.15252/embr.201744766
Park, K., Elias, P. M., Oda, Y., Mackenzie, D., Mauro, T., Holleran, W. M., et al. (2011). Regulation of cathelicidin antimicrobial peptide expression by an endoplasmic reticulum (ER) stress signaling, vitamin D receptor-independent pathway. J. Biol. Chem. 286 (39), 34121–34130. doi:10.1074/jbc.M111.250431
Peng, S. L., Gerth, A. J., Ranger, A. M., and Glimcher, L. H. (2001). NFATc1 and NFATc2 together control both T and B cell activation and differentiation. Immunity 14 (1), 13–20. doi:10.1016/S1074-7613(01)00085-1
Pillaiyar, T., Manickam, M., and Jung, S. H. (2017). Recent development of signaling pathways inhibitors of melanogenesis. Cell. Signal 40, 99–115. doi:10.1016/j.cellsig.2017.09.004
Qin, J., Qian, Y., Yao, J., Grace, C., and Li, X. (2005). SIGIRR inhibits interleukin-1 receptor- and toll-like receptor 4-mediated signaling through different mechanisms. J. Biol. Chem. 280 (26), 25233–25241. doi:10.1074/jbc.M501363200
Qiu, Q., Zhang, P., Zhang, N., Shen, Y., Lou, S., and Deng, J. (2021). Development of a prognostic nomogram for acute myeloid leukemia on IGHD gene family. Int. J. Gen. Med. 14, 4303–4316. doi:10.2147/IJGM.S317528
Rajabinejad, M., Asadi, G., Ranjbar, S., Afshar Hezarkhani, L., Salari, F., Gorgin Karaji, A., et al. (2020). Semaphorin 4A, 4C, and 4D: function comparison in the autoimmunity, allergy, and cancer. Gene 746, 144637. doi:10.1016/j.gene.2020.144637
Ren, X., Yu, J., Guo, L., and Ma, H. (2020). TRIM16 protects from OGD/R-induced oxidative stress in cultured hippocampal neurons by enhancing Nrf2/ARE antioxidant signaling via downregulation of Keap1. Exp. Cell. Res. 391 (1), 111988. doi:10.1016/j.yexcr.2020.111988
Ricciotti, E., and FitzGerald, G. A. (2011). Prostaglandins and inflammation. Arterioscler. Thromb. Vasc. Biol. 31 (5), 986–1000. doi:10.1161/ATVBAHA.110.207449
Robledo, D., Gutiérrez, A. P., Barría, A., Lhorente, J. P., Houston, R. D., and Yáñez, J. M. (2019). Discovery and functional annotation of quantitative trait loci affecting resistance to sea lice in Atlantic salmon. Front. Genet. 10, 56. doi:10.3389/fgene.2019.00056
Robledo, D., Gutiérrez, A. P., Barría, A., Yáñez, J. M., and Houston, R. D. (2018). Gene expression response to sea lice in Atlantic salmon skin: RNA sequencing comparison between resistant and susceptible animals. Front. Genet. 9, 287. doi:10.3389/fgene.2018.00287
Rodger, H., Herrero, A., and Matthews, C. (2022). “Pathologies associated with sea lice,” in Sea lice biology and control (Wallingford, UK: CABI), 134–149. doi:10.1079/9781800629301.0007
Saarialho-Kere, U. K. (1998). Patterns of matrix metalloproteinase and TIMP expression in chronic ulcers. Arch. Dermatol. Res. 290, S47–S54. doi:10.1007/PL00007453
Salisbury, S. J., Ruiz Daniels, R., Monaghan, S. J., Bron, J. E., Villamayor, P. R., Gervais, O., et al. (2024). Keratinocytes drive the epithelial hyperplasia key to sea lice resistance in coho salmon. BMC Biol. 22 (1), 160. doi:10.1186/s12915-024-01952-8
Saxena, R., Gottlin, E. B., Campa, M. J., Bushey, R. T., Guo, J., Patz Jr, E. F., et al. (2024). Complement factor H: a novel innate immune checkpoint in cancer immunotherapy. Front. Cell. Dev. Biol. 12, 1302490. doi:10.3389/fcell.2024.1302490
Scharsack, J. P., and Franke, F. (2022). Temperature effects on teleost immunity in the light of climate change. J. Fish. Biol. 101 (4), 780–796. doi:10.1111/jfb.15163
Schlüter, T., Schelmbauer, C., Karram, K., and Mufazalov, I. A. (2018). Regulation of IL-1 signaling by the decoy receptor IL-1R2. J. Mol. Med. 96 (10), 983–992. doi:10.1007/s00109-018-1684-z
Sequeida, A., Castillo, A., Cordero, N., Wong, V., Montero, R., Vergara, C., et al. (2020). The Atlantic salmon interleukin 4/13 receptor family: structure, tissue distribution and modulation of gene expression. Fish. Shellfish Immunol. 98, 773–787. doi:10.1016/j.fsi.2019.11.030
Sevilla, A., and Grichnik, J. (2024). Therapeutic modulation of KIT ligand in melanocytic disorders with implications for mast cell diseases. Exp. Dermatol. 33 (5), e15091. doi:10.1111/exd.15091
Sezin, T., Selvakumar, B., and Scheffold, A. (2022). The role of a disintegrin and metalloproteinase (ADAM)-10 in T helper cell biology. Biochim. Biophys. Acta Mol. Cell. Res. 1869 (4), 119192. doi:10.1016/j.bbamcr.2021.119192
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13 (11), 2498–2504. doi:10.1101/gr.1239303
Shi, K. P., Dong, S. L., Zhou, Y. G., Li, Y., Gao, Q. F., and Sun, D. J. (2019). RNA-Seq reveals temporal differences in the transcriptome response to acute heat stress in the Atlantic salmon (Salmo Salar). Comp. Biochem. Physiol. D. Genomics Proteomics 30, 169–178. doi:10.1016/j.cbd.2018.12.011
Shi, Y., Shu, B., Yang, R., Xu, Y., Xing, B., Liu, J., et al. (2015). Wnt and notch signaling pathway involved in wound healing by targeting c-Myc and Hes1 separately. Stem Cell. Res. Ther. 6, 120–13. doi:10.1186/s13287-015-0103-4
Skugor, S., Glover, K. A., Nilsen, F., and Krasnov, A. (2008). Local and systemic gene expression responses of Atlantic salmon (Salmo Salar L.) to infection with the salmon louse (Lepeophtheirus salmonis). BMC Genomics 9, 498. doi:10.1186/1471-2164-9-498
Song, K., Wu, Y., Fu, B., Wang, L., Hao, W., Hua, F., et al. (2021). Leaked mitochondrial C1QBP inhibits activation of the DNA sensor cGAS. J. Immunol. 207 (8), 2155–2166. doi:10.4049/jimmunol.2100392
Song, R., Fond, A., Li, X., Tang, M., Zhan, X., Gordillo, R., et al. (2023). The dual lipid desaturase/hydroxylase DEGS2 controls phytoceramide levels necessary to counter intestinal inflammation. Dis. Model. Mech. 16 (9), dmm050043. doi:10.1242/dmm.050043
Sousa, C. R., Yamasaki, S., and Brown, G. D. (2024). Myeloid C-type lectin receptors in innate immune recognition. Immunity 57 (4), 700–717. doi:10.1016/j.immuni.2024.03.005
Stamenkovic, I. (2003). Extracellular matrix remodelling: the role of matrix metalloproteinases. J. Pathol. 200 (4), 448–464. doi:10.1002/path.1400
Sukeda, M., Prakash, H., Nagasawa, T., Nakao, M., and Somamoto, T. (2023). Non-specific cytotoxic cell receptor protein-1 (NCCRP-1) is involved in anti-parasite innate CD8+ T cell-mediated cytotoxicity in ginbuna crucian carp. Fish. Shellfish Immunol. 139, 108904. doi:10.1016/j.fsi.2023.108904
Sutherland, B. J. G., Koczka, K. W., Yasuike, M., Jantzen, S. G., Yazawa, R., Koop, B. F., et al. (2014). Comparative transcriptomics of Atlantic Salmo Salar, chum Oncorhynchus keta and pink salmon O. gorbuscha during infections with salmon lice Lepeophtheirus salmonis. BMC Genomics 15, 200. doi:10.1186/1471-2164-15-200
Sveen, L., Fast, M. D., Tengs, T., Kline, R. A., Marti, J. A., Kurian, D., et al. (2025). Local inflammation at the salmon louse (Lepeophtheirus salmonis) attachment site contributes to copepodid rejection in coho salmon (Oncorhynchus kisutch). Cell. Tissue Res., 1–31. doi:10.1007/s00441-025-03976-0
Sveen, L., Karlsen, C., and Ytteborg, E. (2020). Mechanical induced wounds in fish - a review on models and healing mechanisms. Rev. Aquac. 12 (4), 2446–2465. doi:10.1111/raq.12443
Szauter, K. M., Cao, T., Boyd, C. D., and Csiszar, K. (2005). Lysyl oxidase in development, aging and pathologies of the skin. Pathol. Biol. 53 (7), 448–456. doi:10.1016/j.patbio.2004.12.033
Szláma, G., Kondás, K., Trexler, M., and Patthy, L. (2010). WFIKKN1 and WFIKKN2 bind growth factors TGFβ1, BMP2 and BMP4 but do not inhibit their signalling activity. FEBS J. 277 (24), 5040–5050. doi:10.1111/j.1742-4658.2010.07909.x
Tadiso, T. M., Krasnov, A., Skugor, S., Afanasyev, S., Hordvik, I., and Nilsen, F. (2011). Gene expression analyses of immune responses in Atlantic salmon during early stages of infection by salmon louse (Lepeophtheirus salmonis) revealed bi-phasic responses coinciding with the copepod-chalimus transition. BMC Genomics 12, 141. doi:10.1186/1471-2164-12-141
Taschuk, F., and Cherry, S. (2020). DEAD-Box helicases: sensors, regulators, and effectors for antiviral defense. Viruses 12 (2), 181. doi:10.3390/v12020181
Teng, J., Cui, M. Y., Zhao, Y., Chen, H. J., Du, W. J., Xue, L. Y., et al. (2023). Expression changes of non-specific cytotoxic cell receptor (NCCRP1) and proliferation and migration of NCCs Post-nocardia Seriolae infection in northern snakehead. Dev. Comp. Immunol. 139, 104576. doi:10.1016/j.dci.2022.104576
Thomas, R., and Yang, X. (2023). Semaphorins in immune cell function, inflammatory and infectious diseases. Curr. Res. Immunol. 4, 100060. doi:10.1016/j.crimmu.2023.100060
Tian, H. F., Xing, J., Tang, X. Q., Chi, H., Sheng, X. Z., and Zhan, W. B. (2022). Cluster of differentiation antigens: essential roles in the identification of teleost fish T lymphocytes. Mar. Life Sci. Technol. 4 (3), 303–316. doi:10.1007/s42995-022-00136-z
Todorčević, M., Vegusdal, A., Gjøen, T., Sundvold, H., Torstensen, B. E., Kjær, M. A., et al. (2008). Changes in fatty acids metabolism during differentiation of Atlantic salmon preadipocytes; effects of n-3 and n-9 fatty acids. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 1781 (6-7), 326–335. doi:10.1016/j.bbalip.2008.04.014
Ugelvik, M. S., and Dalvin, S. (2022). The effect of different intensities of the ectoparasitic salmon lice (Lepeophtheirus salmonis) on Atlantic salmon (Salmo Salar). J. Fish. Dis. 45 (8), 1133–1147. doi:10.1111/jfd.13649
Ugelvik, M. S., Mæhle, S., and Dalvin, S. (2022). Temperature affects settlement success of ectoparasitic salmon lice (Lepeophtheirus salmonis) and impacts the immune and stress response of Atlantic salmon (Salmo Salar). J. Fish. Dis. 45 (7), 975–990. doi:10.1111/jfd.13619
Umasuthan, N., Xue, X., Caballero-Solares, A., Kumar, S., Westcott, J. D., Chen, Z., et al. (2020). Transcriptomic profiling in fins of Atlantic salmon parasitized with sea lice: evidence for an early imbalance between chalimus-induced immunomodulation and the host’s defense response. Int. J. Mol. Sci. 21 (7), 2417. doi:10.3390/ijms21072417
Vaalamo, M., Mattila, L., Johansson, N., Kariniemi, A. L., Karjalainen-Lindsberg, M. L., Kähäri, V. M., et al. (1997). Distinct populations of stromal cells express collagenase-3 (MMP-13) and collagenase-1 (MMP-1) in chronic ulcers but not in normally healing wounds. J. Investig. Dermatol. 109 (1), 96–101. doi:10.1111/1523-1747.ep12276722
Vaishnav, J., Khan, F., Yadav, M., Parmar, N., Buch, H., Jadeja, S. D., et al. (2022). V-set domain containing T-cell activation inhibitor-1 (VTCN1): a potential target for the treatment of autoimmune diseases. Immunobiology 227 (6), 152274. doi:10.1016/j.imbio.2022.152274
Valenzuela-Muñoz, V., Gallardo-Escárate, C., Valenzuela-Miranda, D., Nuñez-Acuña, G., Benavente, B. P., Alert, A., et al. (2023). Transcriptome signatures of Atlantic Salmon—Resistant phenotypes against sea lice infestation are associated with tissue repair. Genes. 14 (5), 986. doi:10.3390/genes14050986
Vančová, I., Slovák, M., Hajnicka, V., Labuda, M., Šimo, L., Peterková, K., et al. (2007). Differential anti-chemokine activity of Amblyomma variegatum adult ticks during blood-feeding. Parasite Immunol. 29 (4), 169–177. doi:10.1111/j.1365-3024.2006.00931.x
van Gent, M., Sparrer, K. M. J., and Gack, M. U. (2018). TRIM proteins and their roles in antiviral host defenses. Annu. Rev. Virol. 5 (1), 385–405. doi:10.1146/annurev-virology-092917-043323
Varki, A., and Angata, T. (2006). Siglecs—The major subfamily of I-type lectins. Glycobiology 16 (1), 1R-27R–27R. doi:10.1093/glycob/cwj008
Wagner, G. N., Fast, M. D., and Johnson, S. C. (2008). Physiology and immunology of Lepeophtheirus salmonis infections of salmonids. Trends Parasitol. 24 (4), 176–183. doi:10.1016/j.pt.2007.12.010
Wang, H., Guo, M., Wei, H., and Chen, Y. (2023a). Targeting p53 pathways: mechanisms, structures and advances in therapy. Signal Transduct. Target Ther. 8 (1), 92. doi:10.1038/s41392-023-01347-1
Wang, L., and Ning, S. (2021). TRIMming type I interferon-mediated innate immune response in antiviral and antitumor defense. Viruses 13 (2), 279. doi:10.3390/v13020279
Wang, Q., Chai, D., Sobhani, N., Sun, N., Neeli, P., Zheng, J., et al. (2022). C1QBP regulates mitochondrial plasticity to impact tumor progression and antitumor immune response. Front. Physiol. 13, 1012112. doi:10.3389/fphys.2022.1012112
Wang, Y. H., Reverter, A., Kemp, D., McWilliam, S. M., Ingham, A., Davis, C. A., et al. (2007). Gene expression profiling of Hereford shorthorn cattle following challenge with boophilus Microplus tick larvae. Aust. J. Exp. Agric. 47 (12), 1397–1407. doi:10.1071/EA07012
Wang, Z., Li, W., Chen, S., and Tang, X. X. (2023b). Role of ADAM and ADAMTS proteases in pathological tissue remodeling. Cell. Death Discov. 9 (1), 447. doi:10.1038/s41420-023-01744-z
Whyte, S. K., Westcott, J. D., Jimenez, D., Revie, C. W., and Hammell, K. L. (2014). Assessment of sea lice (Lepeophtheirus salmonis) management in New Brunswick, Canada using deltamethrin (AlphaMax®) through clinical field treatment and laboratory bioassay responses. Aquaculture 422, 54–62. doi:10.1016/j.aquaculture.2013.11.027
Wieczorek, M., Abualrous, E. T., Sticht, J., Álvaro-Benito, M., Stolzenberg, S., Noé, F., et al. (2017). Major histocompatibility complex (MHC) class I and MHC class II proteins: conformational plasticity in antigen presentation. Front. Immunol. 8, 292. doi:10.3389/fimmu.2017.00292
Wilkinson, H. N., and Hardman, M. J. (2020). Wound healing: cellular mechanisms and pathological outcomes. Open Biol. 10 (9), 200223. doi:10.1098/rsob.200223
Xu, Y., Guan, X., Zhou, R., and Gong, R. (2020). Melanocortin 5 receptor signaling pathway in health and disease. Cell. Mol. Life Sci. 77, 3831–3840. doi:10.1007/s00018-020-03511-0
Xue, X., Caballero-Solares, A., Hall, J. R., Umasuthan, N., Kumar, S., Jakob, E., et al. (2021). Transcriptome profiling of Atlantic salmon (Salmo Salar) parr with higher and lower pathogen loads following Piscirickettsia salmonis infection. Front. Immunol. 12, 789465. doi:10.3389/fimmu.2021.789465
Yabluchanskiy, A., Ma, Y., Iyer, R. P., Hall, M. E., and Lindsey, M. L. (2013). Matrix metalloproteinase-9: many shades of function in cardiovascular disease. Physiology 28 (6), 391–403. doi:10.1152/physiol.00029.2013
Zhang, Q., Chen, L., Yang, H., Fang, Y., Wang, S., Wang, M., et al. (2022). GPR84 signaling promotes intestinal mucosal inflammation via enhancing NLRP3 inflammasome activation in macrophages. Acta Pharmacol. Sin. 43 (8), 2042–2054. doi:10.1038/s41401-021-00825-y
Zhao, Z., Yang, Y., Liu, Z., Chen, H., Guan, X., Jiang, Z., et al. (2022). Prognostic and immunotherapeutic significance of mannose receptor C type II in 33 cancers: an integrated analysis. Front. Mol. Biosci. 9, 951636. doi:10.3389/fmolb.2022.951636
Zheng, D., Liwinski, T., and Elinav, E. (2020). Inflammasome activation and regulation: toward a better understanding of complex mechanisms. Cell. Discov. 6, 36. doi:10.1038/s41421-020-0167-x
Zhong, L., Carvalho, L. A., Gao, S., Whyte, S. K., Purcell, S. L., Fast, M. D., et al. (2023). Transcriptome analysis revealed immune responses in the kidney of Atlantic salmon (Salmo Salar) co-infected with sea lice (Lepeophtheirus salmonis) and infectious salmon anemia virus. Fish. Shellfish Immunol. 143, 109210. doi:10.1016/j.fsi.2023.109210
Zhou, X., Khan, S., Huang, D., and Li, L. (2022). V-Set and immunoglobulin domain containing (VSIG) proteins as emerging immune checkpoint targets for cancer immunotherapy. Front. Immunol. 13, 938470. doi:10.3389/fimmu.2022.938470
Keywords: parasitic infestation, family-specific transcriptomics, genetic background, infestation stage, selective breeding
Citation: Ghanei-Motlagh R, Cai W, Poley JD, Whyte SK, Garber AF and Fast MD (2025) Sea lice (Lepeophtheirus salmonis) life stage impacts atlantic salmon transcriptomic responses under different thermal profiles. Front. Genet. 16:1633603. doi: 10.3389/fgene.2025.1633603
Received: 23 May 2025; Accepted: 02 July 2025;
Published: 29 July 2025.
Edited by:
Suxu Tan, Qingdao University, ChinaReviewed by:
Konrad Taube, Estonian University of Life Sciences, EstoniaAntonio Casuso, Universidad de Concepción, Chile
Copyright © 2025 Ghanei-Motlagh, Cai, Poley, Whyte, Garber and Fast. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Reza Ghanei-Motlagh, cmdoYW5laS1tb3RsYWdoQHVwZWkuY2E=; Mark D. Fast, bWZhc3RAdXBlaS5jYQ==
 Reza Ghanei-Motlagh
Reza Ghanei-Motlagh Wenlong Cai
Wenlong Cai Jordan D. Poley
Jordan D. Poley Shona K. Whyte
Shona K. Whyte Amber F. Garber
Amber F. Garber Mark D. Fast
Mark D. Fast