- 1Senior Department of Otolaryngology Head and Neck Surgery, The 6th Medical Center of Chinese PLA General Hospital, Chinese PLA Medical School, Beijing, China
- 2State Key Laboratory of Hearing and Balance Science, Beijing, China
- 3National Clinical Research Center for Otolaryngologic Diseases, Beijing, China
- 4Key Laboratory of Hearing Science, Ministry of Education, Beijing, China
- 5Beijing Key Laboratory of Hearing Impairment Prevention and Treatment, Beijing, China
- 6School of Life Sciences, Tsinghua University, Beijing, China
- 7Chinese Institute for Brain Research, Beijing, China
- 8Department of Otolaryngology, PLA Rocket Force Characteristic Medical Center, Beijing, China
The solute carriers (SLCs) are important membrane-bound transporters that regulate cellular nutrition, metabolism, homeostasis and survival. Emerging evidence highlights the critical involvement of SLCs in auditory physiology. To date, over ten SLC family members have been linked to hearing function. MFSD3 (also known as SLC33A2), is a putative plasma membrane-localized acetyl-CoA transporter regulating lipid metabolism and energy homeostasis. It has been found to be associated with the pathogenesis of neurodegenerative dementia and tumor progression. Nevertheless, its potential role in hearing remains unexplored. In this study, through qRT-PCR, we demonstrated that mfsd3 was predominantly expressed during early embryonic development in zebrafish. Morpholino-mediated mfsd3 knockdown in zebrafish induced inner ear malformations (hypoplastic otic vesicles, reduced otolith size) and hair cells loss in lateral line neuromasts. Additionally, Mfsd3 deficiency led to developmental defects (pericardial edema, body axis curvature) and impaired locomotor activity in zebrafish. The qRT-PCR analysis further revealed significant upregulation of key Wnt/β-catenin pathway components (dkk1b, wnt8a, lrp6, frzb and COX2) in mfsd3 knockdown zebrafish. Our findings suggest MFSD3 as a potential participant in auditory function and embryogenesis, with implications for understanding hearing loss pathogenesis.
1 Introduction
Hearing loss (HL) affects over 1.5 billion people worldwide, with profound impacts on speech development, communication, cognition, education and mental health (Chadha et al., 2021). Genetic factors play a major role, as evidenced by a recent mouse study identifying over 1,000 genes potentially involved in hearing maintenance (Ingham et al., 2019). Given this complexity, the discovery of novel hearing-related genes and their functions remains critical.
Membrane transporters, particularly solute carriers (SLCs) proteins, are physiologically important for cellular homeostasis and survival (Hediger et al., 2013). Over ten SLCs genes have been found to be associated with hearing impairment, including SLC26A4 (MIM *605646), SLC44A4 (MIM *606107), SLC7A14 (MIM *615720), SLC12A2 (MIM *600840), SLC17A8 (MIM *607557) and SLC22A4 (MIM *604190) (Qian et al., 2022). The major facilitator superfamily domain containing protein 3 (MFSD3) encoded by MFSD3 (MIM *620308, NM_138431) gene, also known as SLC33A2, belongs to the SLC superfamily and is predicted to function as an acetyl-CoA transporter localized in the plasma membrane (Perland et al., 2017a; Perland et al., 2017b). Heterozygous variants in MFSD3 have been associated with dementia with Lewy bodies (MIM #127750), while its overexpression has been linked to metastatic progression in uveal melanoma (MIM #155720) (Li et al., 2019; Shigemizu et al., 2022). However, the role of MFSD3 in hearing has not been explored. We previously identified a missense variant in MFSD3 that co-segregated with HL in a Chinese Han family using whole-genome sequencing (Supplementary Figure S1). According to the HL-specific ACMG/AMP guidelines (Oza et al., 2018), this variant was classified as a variant of uncertain significance (VUS). And no variant in the MFSD3 gene has been detected in other known deafness families. Therefore, it is essential to investigate the potential association between MFSD3 and HL through functional experiments.
The zebrafish is an ideal model for hearing research due to its transparent body, rapid development, genetic tractability and conserved orthologs of human deafness genes (Vona et al., 2020). They possess two mechanosensory systems, the inner ear and the lateral line. The inner ear shares structural and functional similarities with that of mammals and is essential for auditory and vestibular functions. In contrast, the lateral line system detects local water motion and facilitates behaviors including prey detection, predator avoidance and social interactions. Anatomically, the lateral line is organized into two components: the cranial lateral line (anterior lateral line) consisting of the head neuromasts, and the trunk lateral line (posterior lateral line), which is composed of a continuous canal housing multiple neuromasts. The neuromast contains a superficial cluster of hair cells surrounded by supporting cells. These lateral line hair cells share structural and functional similarities with the inner ear hair cells, making it an excellent model to studying hair cell development and function (Sheets et al., 2021). Additionally, morpholino oligonucleotides (MOs), a rapid and efficient gene-editing tool, have been widely used to validate candidate deafness genes (Gao et al., 2018; Li et al., 2023). However, the role of mfsd3 in zebrafish auditory function and embryonic development remains unexplored.
In this study, we examined the expression levels of mfsd3 throughout various embryonic developmental stages in zebrafish. We further demonstrated that MO-mediated knockdown of mfsd3 in zebrafish led to auditory organ dysfunction, developmental defects and potential disruption of the Wnt/β-catenin signaling pathway.
2 Materials and methods
2.1 RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from 4–10 embryos per group using Trizol (Roche) according to the manufacturer’s protocols. cDNA was synthesized using the PrimeScript RT reagent Kit with gDNA Eraser (Takara). qPCR was performed in triplicates on a Realplex system (Eppendorf) using iQ SYBR Green Supermix (Bio-rad), with ef1α as the endogenous control. Relative expression levels were calculated using the 2−ΔΔCT method. Primer sequences are listed in Supplementary Table S2.
2.2 Zebrafish husbandry and mfsd3 MO knockdown
Wild-type AB strain and Tg (Brn3c:mGFP) S356T transgenic zebrafish (provided by Prof. Hua-Wei Li, Fudan University) were maintained at 28.5 °C under a 14 h/10 h light/dark cycle. For the latter, membrane-targeted GFP is specifically expressed in hair cells under control of the brn3c promoter. Antisense MO (Gene Tools, LLC, United States of America) was microinjected into fertilized embryos at one-cell-stage following standard protocols (Nasevicius and Ekker, 2000). Two specific morpholino antisense strategies were employed to target zebrafish mfsd3 RNA, preventing either translation of the gene (ATG-MO) or proper splicing of exon2 (E2I2-MO). The sequences for mfsd3-ATG-MO and mfsd3-E2I2-MO were 5′-GAACACCAGCTTGTCGTTCATCATG-3′ and 5′-ACAACAAAACACACTCCCTACCTGT-3′, respectively. The sequence for the standard control morpholino was 5′-CCTCTTACCTCAGTTACAATTTATA-3’ (Gene Tools). The optimal dose (4ng/embryo) of MOs was determined through dose-response experiments. Knockdown efficacy was verified by reverse transcription PCR (RT-PCR) using primers spanning mfsd3 exons 1–4 (forward: 5′-GTGTGGAGGAAGCTGTTATC-3’; reverser: 5′-AGCACCGCCATAACTTTC-3′).
2.3 Inner ear morphology and hair cells quantification
At 3 dpf, Tg (Brn3c:mGFP) zebrafish embryos were anesthetized using 0.016% MS-222 (tricaine methanesulfonate; Sigma-Aldrich, St. Louis, MO, United States of America). The anesthetized larvae were then laterally positioned (anterior to the left, posterior to the right, and dorsal side up) and immobilized in 3% methylcellulose within a depression slide for fluorescence microscopic observation. The larvae were divided into three groups: Control-MO, mfsd3-E2I2-MO and mfsd3-ATG-MO. The inner ear morphology and hair cells in lateral line neuromasts were quantified analyzed: The number of hair cells in lateral line neuromasts, otic vesicle area (×103 μm2), otolith area (μm2) and diameter (μm). The maximum diameter or cross-sectional area (the maximum projection) of both otoliths from each zebrafish was measured, using NIS-Elements D4.6 measurement software integrated into the Nikon SMZ18. The average value was then calculated to represent the otolith diameter or area for each individual. Ten zebrafish per group were randomly selected for measurement, followed by quantitative statistical analysis.
2.4 Behavioral analysis
5-dpf larvae (n = 10/group) were acclimated for 15 min in 96-well plates (0.2 mL/well) and tracked for 30 min using Ethovision XT software (Noldus) (Zhao et al., 2012). Movement parameters analyzed included: Total distance traveled, average velocity, mobility percentage, maximum acceleration. A 0.2 mm movement threshold was applied to filter system noise. The larvae were divided into three groups: Control-MO, mfsd3-E2I2-MO and mfsd3-ATG-MO. Ten zebrafish per group were randomly selected for monitoring, followed by quantitative statistical analysis.
2.5 Image acquisition and statistical analysis
Images were acquired using a Nikon SMZ18 fluorescence microscope and processed with Adobe Photoshop 7.0 software (Adobe, San Jose, California) for optimize visualization. Quantitative analyses were performed using NIS-Elements D4.6 (Japan) and ImageJ (U.S. National Institutes of Health, Bethesda, MD, United States of America). Inverted fluorescent images were utilized for processing. Data were presented as mean ± SEM. Ten animals from each treatment group underwent quantification with averaging performed on total signal per animal. And GraphPad Prism 5.0 (GraphPad Software, San Diego, CA) was used for statistical analysis and graphical representation of the data. Statistical analyses of the comparison between two groups (control MO and E2I2-MO) were performed by Student’s t-test. One-way analysis of variance (ANOVA) was used for comparison among three or more groups. For further pairwise comparisons among groups, the Bonferroni’s multiple comparison test (among control MO, ATG-MO and E2I2-MO) and Tukey’s multiple comparisons test (mfsd3 expression level among different developmental stages) were utilized. Statistical significance is indicated as *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001.
3 Results
3.1 Knockdown of mfsd3 causes inner ear defects in zebrafish
We first confirmed mfsd3 in zebrafish via qRT-PCR at multiple time points prior to 6 dpf. Additionally, we obtained the relative expression level (TPM) of mfsd3 across 18 developmental stages from the Expression Atlas database (http://www.ebi.ac.uk/gxa/experiments/E-ERAD-475), which were derived from poly(A)-selected RNA-seq data (White et al., 2017). Both datasets revealed a significant decline in mfsd3 mRNA levels after 6 hpf, suggesting its potential involvement in early zebrafish development (Supplementary Figure S2). The efficiency of mfsd3 knockdown was verified by RT-PCR (Figures 1A,B).
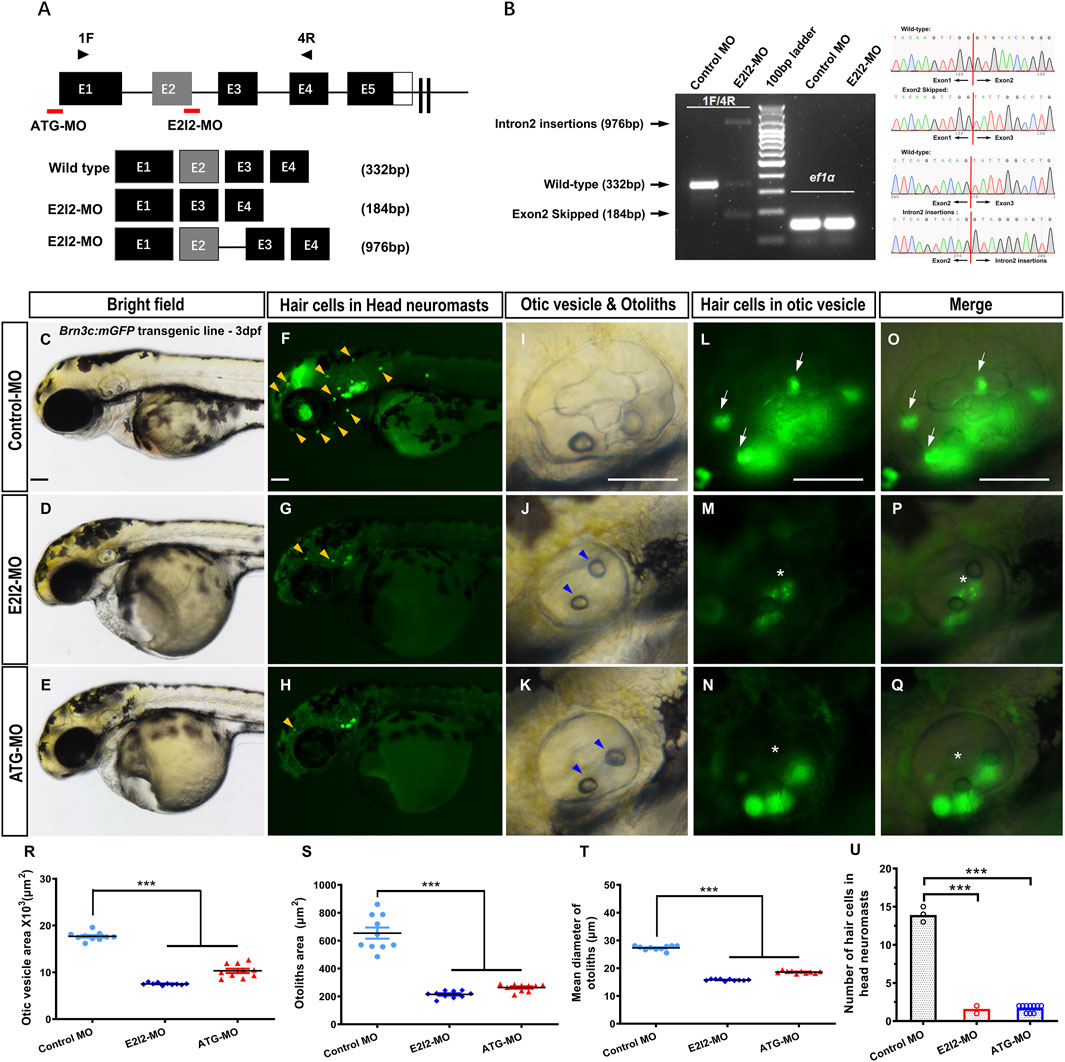
Figure 1. Phenotypes of auditory system in mfsd3 zebrafish morphants. (A) Schematic of two MO targeting strategies: ATG-MO blocks translation initiation while E2I2-MO disrupts exon 2-intron 2 splicing. Primers pairs (1F/4R) detect wild-type transcripts or aberrant splicing products containing retained intron 2 or skipped exon 2. (B) Left: RT-PCR analysis of mfsd3 transcript from 1 dpf embryos injected with control-MO or E2I2-MO (4 ng). Aberrant splicing product (intron 2 retention and exon2 skipping) are indicated by shifted band patterns. Right: Sanger sequencing confirmation of wild-type sequence, intron 2-inserted transcript and exon2-skipped transcript. (C–Q) Gross morphology of head neuromasts and inner ear in Tg (Brn3c: mGFP) embryos at 3-dpf. Compared to control MO, Mfsd3 deficiency led to reduced hair cell numbers in head neuromasts ((G,H), yellow arrowheads), hypoplastic otic vesicle (J,K), malformed otoliths ((J,K), blue arrowheads) and damaged otic vesicle hair cells ((M–N), (P–Q), asterisk). (R–T) Quantitative measurements demonstrated significant reduction in: otic vesicle area (R), otoliths area (S) and mean otolith diameter (T). ***P < 0.001 (n = 10; ANOVA). (U) Quantitative of hair cell numbers in head neuromasts showing significant decrease in morphants. Scale bar, 100 μm; Scatter plot with bars; ***P < 0.001 (n = 10; ANOVA). dpf, days post fertilization.
Compared to wild-type (WT) controls, mfsd3 morphants exhibited a reduced number of hair cells in both head neuromasts and posterior lateral line neuromasts (Figures 1F–H,L–Q,U; Figures 2D–L,R). At 3 dpf, the average hair cell counts in head neuromasts were: 1.6 ± 0.52 (E2I2-MO), 1.7 ± 0.48 (ATG-MO) and 13.9 ± 0.57 (WT); the average hair cell counts in posterior lateral line neuromasts were: 0.1 ± 0.32 (E2I2-MO and ATG-MO) and 10.2 ± 0.92 (WT). Smaller otic vesicles and otoliths were observed in the morphant than in the WT (Figures 1C–E, I–K, R–T). The average areas (×103, μm2) of otic vesicles were 7.5 ± 0.25 (E2I2-MO), 10.3 ± 1.45 (ATG-MO) and 17.7 ± 0.89 (WT). The average areas (μm2) of otoliths were 214.4 ± 24.79 (E2I2-MO), 263.3 ± 25.90 (ATG-MO) and 654.0 ± 126.50 (WT). The average diameters (μm) of otoliths were 15.8 ± 0.32 (E2I2-MO), 18.6 ± 0.45 (ATG-MO) and 27.3 ± 0.82 (WT). These data indicate that Mfsd3 deficiency leads to otic vesicle and otolith hypoplasia, as well as hair cells loss in lateral line system.
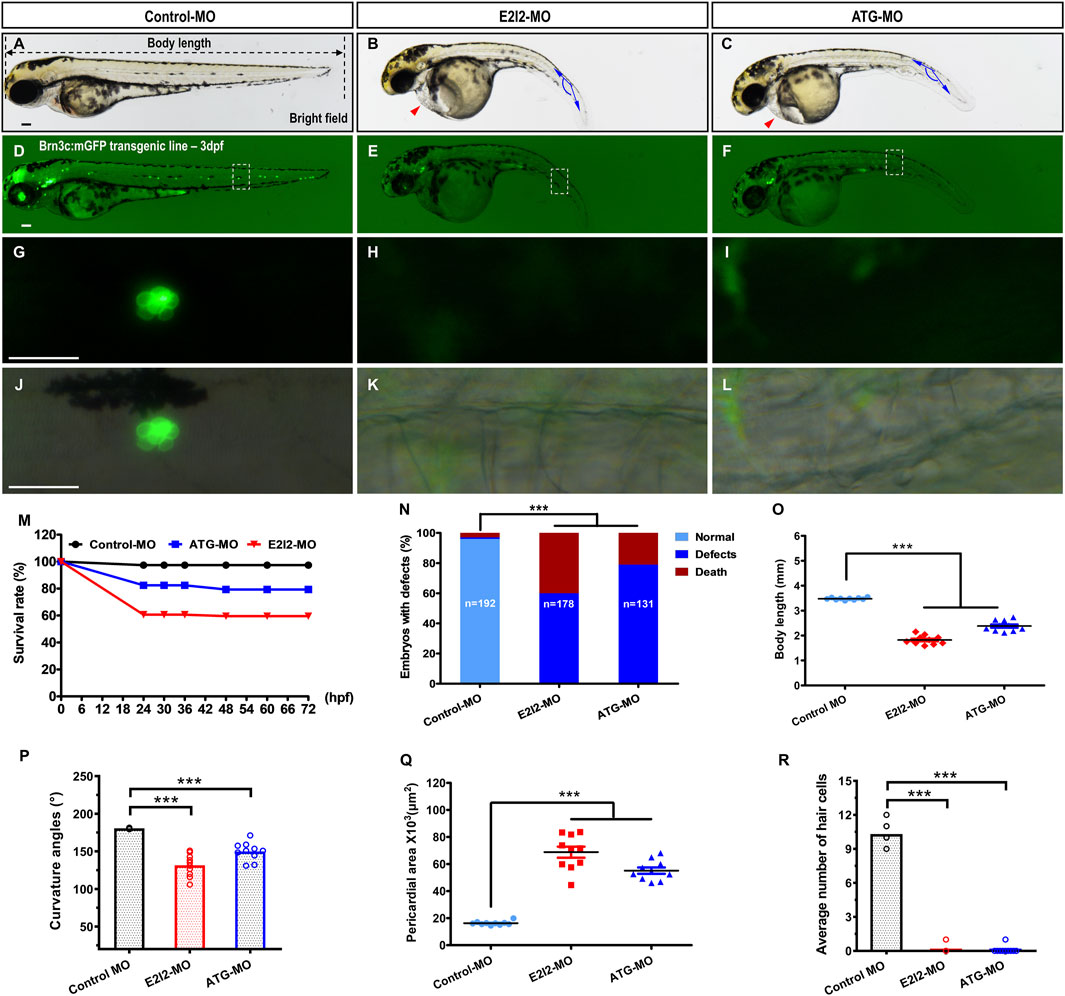
Figure 2. Morphological defects and hair cells loss in Mfsd3-deficient zebrafish. (A–C) Compared with control MO, the mfsd3 knockdown MO showing significant body shortening (B,C), axis curvature (B,C) and pericardial edema ((B,C), red arrowheads). (D–L) Live imaging of Brn3c: mGFP transgenic embryo at 3-dpf showed GFP expression from RGCs (retinal ganglion cells) and neuromasts (green dots) of the posterior lateral line and head. Zebrafish under control exhibited normal hair cells number (D). Conversely, significantly decreased hair cells of the posterior lateral line were observed in mfsd3 morphants (E,F). The white boxed regions are presented in amplified form in the lower panels (G–L) GFP fluorescence signal alone (G–I) and GFP fluorescence signal merged with bright field image (J–L). Morphometric analyses were utilized to quantify the fluorescence particle signal of hair cells. (M) A time-course plot of survival rates in control vs. mfsd3 morphants for 3 days. (N) The percentage of embryos with development defects in control vs. mfsd3 morphants. (O,P) Quantitative analysis of body length (O) and curvature angle (P) of embryos. (n = 10; ANOVA; ***P < 0.001). (Q) Quantitative analysis of the pericardial area of embryos. (R) Quantitative analysis of the average number of hair cells at posterior lateral line. Scale bar, 100 μm; Scatter plot with bars (n = 10; ANOVA; **P < 0.0001). dpf, days post fertilization.
3.2 mfsd3 knockdown results in decreased survival, morphological defects and pericardial edema
At 3 dpf, both mfsd3-E2I2 and mfsd3-ATG morphants exhibited nearly identical phenotypes, including morphological abnormalities and pericardial edema (Figure 2; Supplementary Figure S3), along with significantly lower survival rates: 59.55% (E2I2-MO, n = 178 embryos), 79.39% (ATG-MO, n = 131 embryos) and 97.40% (WT controls, n = 192 embryos) (Figure 2M). Loss of Mfsd3 also caused pronounced developmental defects (Figure 2N), including shortened body length, curved body axis and pericardial edema (Figures 2A–C, O–Q). The mean body lengths (mm) were 1.826 (E2I2-MO), 2.382 (ATG-MO) and 3.477 (WT, n = 10). The mean body axis curvatures (°) were 131.5 (E2I2-MO), 149.9 (ATG-MO) and 180.6 (WT, n = 10). The mean pericardial areas (×103, μm2) were 68.81 (E2I2-MO), 55.16 (ATG-MO) and 16.29 (WT, n = 10). These findings indicate that the mfsd3 gene plays a critical role in early zebrafish development.
3.3 mfsd3 knockdown results in locomotor defects
Given the morphological abnormalities observed in mfsd3 morphants at 3 dpf, we further assessed their swimming behavior at 5 dpf. Both E2I2-MO and ATG-MO morphants exhibited significant reduction in total swimming distance, velocity, mobility and maximum acceleration compared to WT (Figure 3, n = 10). Mean distances (mm) were 1708 (E2I2-MO), 1,639 (ATG-MO) and 5,185 (WT). Mean velocities (μm/s) were 949.5 (E2I2-MO), 911.2 (ATG-MO) and 2,925 (WT). Mean mobilities (%) were 7.25 (E2I2-MO), 6.73 (ATG-MO) and 14.38 (WT). The average maximum accelerations (mm/s2) were 139.9 (E2I2-MO), 201.7 (ATG-MO) and 1,396 (WT). These deficits likely stem from tail dyskinesia, which may be secondary to the abnormal body axis morphology.

Figure 3. Locomotor capacity was reduced in mfsd3 zebrafish morphants. (A,B) The digital tracks and heatmap image in larvae from control-MO and mfsd3-MO injected groups at 5-dpf. Each behavioral test consisted of ten larvae each placed into individual wells. (C–F) The statistical analyses showed significantly reduced total movement distance (C), velocity (D), mobility (E) and maximum acceleration (F) in mfsd3 morphants compared to control-MO injected groups (n = 10; ANOVA; ***P < 0.001). dpf, days post fertilization.
3.4 mfsd3 knockdown potentially disrupts the wnt/β-catenin signaling pathway
In this study, targeted knockdown of mfsd3 in zebrafish embryos resulted in multiple developmental defects, including pericardial edema, body axis curvature, and hair cells loss. These phenotypes suggest potential dysregulation of the Wnt/β-catenin pathway (Bellipanni et al., 2006; Ueno et al., 2007; Ma and Raible, 2009). Since both morphant groups exhibited similar defects, we focused on E2I2-MO for pathway analysis.
qRT-PCR revealed upregulation of key Wnt/β-catenin components in mfsd3 morphants, including dkk1b, wnt8a, wnt9a, lrp5, lrp6, frzb, fzd7a, fzd7b, gsk-3β, axin1, axin2, mycn, lef1, myca, β-catenin and COX2. Notably, dkk1b, wnt8a, lrp6, frzb and COX2 showed the most pronounced changes (Figure 4). These results suggest that Mfsd3 deficiency may disrupt Wnt/β-catenin signaling through direct or indirect mechanisms.
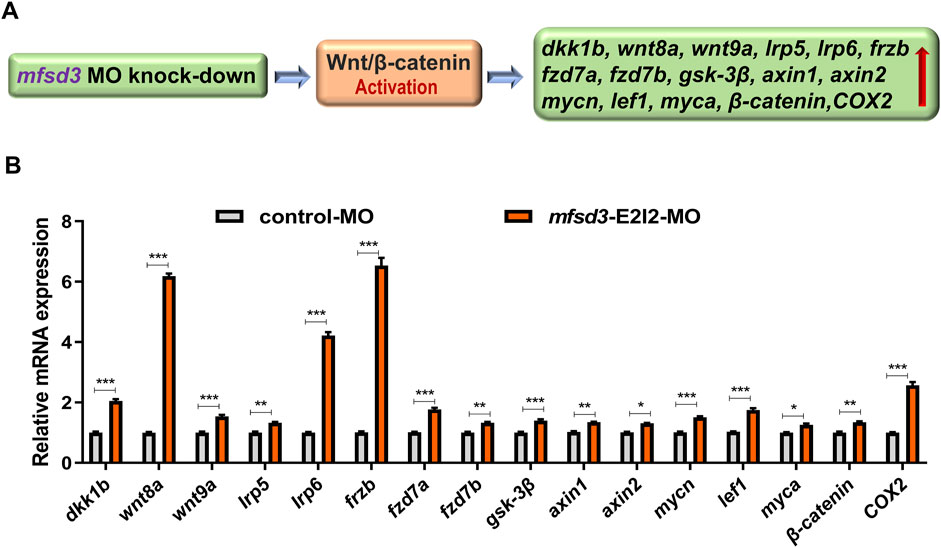
Figure 4. Zebrafish with Mfsd3 deficiency exhibited upregulated Wnt/β-catenin signaling activity. (A) Schematic diagram depicting the potential regulatory action of mfsd3 on the Wnt/β-catenin signaling pathway. (B) Wnt/β-catenin signaling pathway was upregulated in mfsd3 morphants. Endogenous dkk1b, wnt8a, wnt9a, lrp5, lrp6, frzb, fzd7a, fzd7b, gsk-3β, axin1, axin2, mycn, lef1, myca, β-catenin and COX2 in control and mfsd3 morphants measured by qRT-PCR assay (n = 6–10 individual embryos). The dkk1b, wnt8a, lrp6, frzb and COX2 were all upregulated significantly in the mfsd3 morphants group. ***P < 0.001; **P < 0.01; *P < 0.05.
4 Discussion
MFSD3 belongs to the acetyl-CoA transporter family SLC33 and shares 19.5% sequence homology with SLC33A1 (Perland et al., 2017b). The National Center for Biotechnology Information (NCBI) database annotations predict MFSD3 to function as a solute: proton symporter involved in proton transmembrane transport while serving as an integral membrane component. The Kyoto Encyclopaedia of Genes and Genomes (KEGG) database further supports its potential role as an acetyl-CoA transporter. Expression data from Mouse Genome Informatics (MGI) and Shared Harvard Inner Ear Laboratory (SHIELD) databases revealed Mfsd3 expression in mouse cochlea structures, including the organ of Corti, otic progenitor cells, spiral ganglion and vestibular ganglion. Additionally, we analyzed single-cell RNA-sequencing data from three independent published studies via Seurat in R 4.3.3 (Petitpré et al., 2018; Shrestha et al., 2018; Petitpré et al., 2022). This analysis further demonstrated an age-dependent increase in Mfsd3 expression in the spiral ganglion neurons of mice cochlea (Supplementary Figure S4).
Current research on MFSD3 remains limited, with only five international studies published to date (Perland et al., 2017b; Li et al., 2019; Nicoletti et al., 2020; Xu et al., 2021; Shigemizu et al., 2022). Among these, three suggest potential disease associations through clinical data screening, though the protein’s exact function and physiological mechanism remain undefined. Key findings demonstrated that Mfsd3 shows abundant expression in the nervous system, liver, kidneys and ovaries of WT mice (Perland et al., 2017b), with potential links to neurodegenerative disorders and tumors (Li et al., 2019; Shigemizu et al., 2022). Notably, MFSD3 localized specifically to neuronal plasma membranes in mice brains (Perland et al., 2017b), with Mfsd3 mRNA levels decreasing throughout the brain under high-fat diets yet showing brainstem-specific upregulation during starvation (Perland et al., 2017b). Transkingdom Network analysis connects Mfsd3 to liver fatty acid metabolism in mouse (Rodrigues et al., 2021), while clinical evidence associates MFSD3 hypermethylation with post-bariatric weight regain (Nicoletti et al., 2020). These collective findings position MFSD3 as a potential regulator of nutrition intake, lipid metabolism and energetic homeostasis. Given the exceptional sensitivity of the cochlea to local lipid metabolism, disturbances in this metabolic process can induce hearing impairment via the degeneration or apoptosis of hair cells (Saito et al., 1986; Wu et al., 2012; Yao et al., 2019). This underpins the potential involvement of MFSD3 in auditory function.
Our investigation of zebrafish mfsd3 revealed a distinct temporal expression pattern, with peak expression occurring between 0.75 and 6 hpf. Given that zygotic genome activation (ZGA) initiates at 2.75-3hpf (Shen et al., 2022), we interpret transcripts before 3hpf as maternal in origin, with zygotic transcription commencing thereafter. The subsequent sharp decline after 6 hpf followed by a sustained low-level expression implies coordinated contributions from both maternal and zygotic mfsd3 during early development. However, the mechanism driving the pronounced expression surge during the early zygotic stage (0–0.75 hpf) requires further elucidation.
Functional characterization using two distinct MOs (ATG blocker and E2I2 splice blocker) yielded consistent phenotypes in zebrafish, including otic vesicle and otoliths hypoplasia, hair cells loss in lateral line, and motor deficit. The observed hair cells reduction might be attributed to the complete absence of entire neuromasts, a hypothesis that necessitates validation through future immunostaining with cell-type specific markers like SOX2 (supporting cells) and BrdU (proliferated cells). These auditory developmental defects suggest MFSD3’s potential role in auditory function, while acknowledging possible interspecies differences between zebrafish and mammalian systems. Concurrent phenotypes of reduced embryo viability, morphological abnormalities and pericardial edema underscore the mfsd3’s importance in early development of zebrafish. Notably, our use of low-dose MOs (4ng/injection) minimizes potential off-target effects, as background phenotypes typically require higher doses (≥40 ng) (Söllner et al., 2003). This represents the first reported zebrafish model of Mfsd3 deficiency. Building on its predicted acetyl-CoA transporter activity (Perland et al., 2017b), we propose that Mfsd3 loss may impair acetyl-CoA membrane transport, potentially disrupting energy production and lipid metabolism while promoting pathological lipid accumulation in auditory structures, which was a hypothesis requiring further experimental validation.
The evolutionarily conserved Wnt/β-catenin pathway has been confirmed to regulate both development and oncogenesis (Moon et al., 2002). Studies have shown that the alterations to Wnt/β-catenin signaling induced body axis and cardiac defects in zebrafish (Bellipanni et al., 2006; Wan et al., 2021), which was also observed in our study. Growing evidence highlights this pathway’s critical role in auditory system development (Chai et al., 2012; Munnamalai and Fekete, 2013), particularly in cochlea sensory epithelium differentiation and neuromast progenitor proliferation (Shi et al., 2014; Tang et al., 2019). While our work implicates potential mfsd3 in Wnt/β-catenin signaling regulation, definitive confirmation awaits further rescue-based functional validation.
Several limitations and future directions merit consideration. First, potential MO-induced mosaicism necessitates CRISPR/Cas9-generated knockout models for phenotype confirmation. Second, phenotypic analysis requires refinement through: 1) left-right patterning assessment via of Kupffer’s vesicle and cardiac looping analysis (Essner et al., 2005; Bowley et al., 2022); 2) lateral line primordium development assessment and migration studies using immunohistochemistry; and 3) comprehensive evaluation of motor dysfunction through neuromuscular assessment. Third, given the extensive crosstalk among Wnt, FGF, Notch, BMP and SHH pathways during inner ear development (Munnamalai and Fekete, 2013; Roccio and Edge, 2019), future RNA-seq analysis should explore mfsd3’s broader signaling impacts beyond Wnt/β-catenin. In addition, to characterize the expression pattern of Mfsd3 in cochlea tissue, we initially tested a commercially available MFSD3 antibody (AV51707, Sigma-Aldrich, species reactivity: human and mouse) in HEK293T cells transfected with the pcDNA3.1-CMV-MFSD3 (human-NM_138431.3)-EGFP plasmid. However, the antibody exhibited poor specificity (Supplementary Figure S5). Future studies are essential to examine the Mfsd3 spatiotemporal expression pattern in the mice inner ears through RNA in situ hybridization, as well as to identify whether the inner ear structures and hearing function are impaired in murine models with aberrant Mfsd3.
5 Conclusion
Our study implicates MFSD3 in auditory function and zebrafish embryogenesis. We identified predominant early embryonic expression of mfsd3 gene in zebrafish. Functional knockdown of mfsd3 in zebrafish produced diverse abnormalities, affecting auditory structures, embryonic morphology, cardiac development and motor behavior, along with potential Wnt/β-catenin pathway upregulation. These findings establish foundational insights while highlighting the need for deeper mechanistic exploration of MFSD3’s role in auditory and developmental biology.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics statement
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used. The animal study was approved by Chinese People’s Liberation Army General Hospital Research Ethics Committee (reference number S2016–120–02). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
YM: Methodology, Writing – original draft. S-wQ: Writing – review and editing, Methodology. W-QW: Methodology, Writing – review and editing. H-FF Methodology, Writing – review and editing. LZ: Writing – review and editing, Methodology. G-GW: Writing – review and editing, Software. H-YN: Writing – review and editing, Formal Analysis. J-YY: Formal Analysis, Writing – review and editing. Y-JC: Writing – review and editing, Data curation. PD: Writing – review and editing, Conceptualization. XG: Writing – review and editing, Conceptualization. Y-YY: Conceptualization, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was funded by grants from National key Research and development project of China (2022YFC2703602), National Natural Science Foundation of China (82271177, 82171158, 82271185, 81873704, 82171155 and 81900953) and Natural Science Foundation of Beijing (7242137). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgments
We would like to thank the Busch-Nentwich lab for providing RNA-seq data.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2025.1634493/full#supplementary-material
Abbreviations
MFSD3, Major facilitator superfamily domain containing protein 3; SLC, solute carrier; SLC33A2, solute carrier family 33 member 2; MFS, major facilitator superfamily; HL, hearing loss; SNHL, sensorineural hearing loss; ACMG, American College of Medical Genetics and Genomics; AMP, Association for Molecular Pathology; VUS, variant of uncertain significance; SEM, standard error of the mean; NCBI, National Center for Biotechnology Information; KEGG, Kyoto Encyclopaedia of Genes and Genomes; MGI, Mouse Genome Informatics; SHIELD, Shared Harvard Inner Ear Laboratory Database; ZGA, zygotic genome activation; KV, Kupffer’s vesicle; FGF, fibroblast growth factor; BMP, bone morphogenetic protein; SHH, sonic hedgehog.
References
Bellipanni, G., Varga, M., Maegawa, S., Imai, Y., Kelly, C., Myers, A. P., et al. (2006). Essential and opposing roles of zebrafish beta-catenins in the formation of dorsal axial structures and neurectoderm. Development 133 (7), 1299–1309. doi:10.1242/dev.02295
Bowley, G., Kugler, E., Wilkinson, R., Lawrie, A., van Eeden, F., Chico, T. J. A., et al. (2022). Zebrafish as a tractable model of human cardiovascular disease. Br. J. Pharmacol. 179 (5), 900–917. doi:10.1111/bph.15473
Chadha, S., Kamenov, K., and Cieza, A. (2021). The world report on hearing, 2021. Bull. World Health Organ 99 (4), 242–242a. doi:10.2471/blt.21.285643
Chai, R., Kuo, B., Wang, T., Liaw, E. J., Xia, A., Jan, T. A., et al. (2012). Wnt signaling induces proliferation of sensory precursors in the postnatal mouse cochlea. Proc. Natl. Acad. Sci. U. S. A. 109 (21), 8167–8172. doi:10.1073/pnas.1202774109
Essner, J. J., Amack, J. D., Nyholm, M. K., Harris, E. B., and Yost, H. J. (2005). Kupffer's vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development 132 (6), 1247–1260. doi:10.1242/dev.01663
Gao, X., Yuan, Y. Y., Lin, Q. F., Xu, J. C., Wang, W. Q., Qiao, Y. H., et al. (2018). Mutation of IFNLR1, an interferon lambda receptor 1, is associated with autosomal-dominant non-syndromic hearing loss. J. Med. Genet. 55 (5), 298–306. doi:10.1136/jmedgenet-2017-104954
Hediger, M. A., Clémençon, B., Burrier, R. E., and Bruford, E. A. (2013). The ABCs of membrane transporters in health and disease (SLC series): introduction. Mol. Asp. Med. 34 (2-3), 95–107. doi:10.1016/j.mam.2012.12.009
Ingham, N. J., Pearson, S. A., Vancollie, V. E., Rook, V., Lewis, M. A., Chen, J., et al. (2019). Mouse screen reveals multiple new genes underlying mouse and human hearing loss. PLoS Biol. 17 (4), e3000194. doi:10.1371/journal.pbio.3000194
Li, Y., Yang, X., Yang, J., Wang, H., and Wei, W. (2019). An 11-gene-based prognostic signature for uveal melanoma metastasis based on gene expression and DNA methylation profile. J. Cell Biochem. 120 (5), 8630–8639. doi:10.1002/jcb.28151
Li, Y., Ning, G., Kang, B., Zhu, J., Wang, X. Y., Wang, Q., et al. (2023). A novel recessive mutation in OXR1 is identified in patient with hearing loss recapitulated by the knockdown zebrafish. Hum. Mol. Genet. 32 (5), 764–772. doi:10.1093/hmg/ddac229
Ma, E. Y., and Raible, D. W. (2009). Signaling pathways regulating zebrafish lateral line development. Curr. Biol. 19 (9), R381–R386. doi:10.1016/j.cub.2009.03.057
Moon, R. T., Bowerman, B., Boutros, M., and Perrimon, N. (2002). The promise and perils of Wnt signaling through beta-catenin. Science 296 (5573), 1644–1646. doi:10.1126/science.1071549
Munnamalai, V., and Fekete, D. M. (2013). Wnt signaling during cochlear development. Semin. Cell Dev. Biol. 24 (5), 480–489. doi:10.1016/j.semcdb.2013.03.008
Nasevicius, A., and Ekker, S. C. (2000). Effective targeted gene 'knockdown' in zebrafish. Nat. Genet. 26 (2), 216–220. doi:10.1038/79951
Nicoletti, C. F., Pinhel, M. S., Noronha, N. Y., Jácome, A., Crujeiras, A. B., and Nonino, C. B. (2020). Association of MFSD3 promoter methylation level and weight regain after gastric bypass: assessment for 3 y after surgery. Nutrition 70, 110499. doi:10.1016/j.nut.2019.04.010
Oza, A. M., DiStefano, M. T., Hemphill, S. E., Cushman, B. J., Grant, A. R., Siegert, R. K., et al. (2018). Expert specification of the ACMG/AMP variant interpretation guidelines for genetic hearing loss. Hum. Mutat. 39 (11), 1593–1613. doi:10.1002/humu.23630
Perland, E., Bagchi, S., Klaesson, A., and Fredriksson, R. (2017a). Characteristics of 29 novel atypical solute carriers of major facilitator superfamily type: evolutionary conservation, predicted structure and neuronal co-expression. Open Biol. 7 (9), 170142. doi:10.1098/rsob.170142
Perland, E., Hellsten, S. V., Lekholm, E., Eriksson, M. M., Arapi, V., and Fredriksson, R. (2017b). The novel membrane-bound proteins MFSD1 and MFSD3 are putative SLC transporters affected by altered nutrient intake. J. Mol. Neurosci. 61 (2), 199–214. doi:10.1007/s12031-016-0867-8
Petitpré, C., Wu, H., Sharma, A., Tokarska, A., Fontanet, P., Wang, Y., et al. (2018). Neuronal heterogeneity and stereotyped connectivity in the auditory afferent system. Nat. Commun. 9 (1), 3691. doi:10.1038/s41467-018-06033-3
Petitpré, C., Faure, L., Uhl, P., Fontanet, P., Filova, I., Pavlinkova, G., et al. (2022). Single-cell RNA-sequencing analysis of the developing mouse inner ear identifies molecular logic of auditory neuron diversification. Nat. Commun. 13 (1), 3878. doi:10.1038/s41467-022-31580-1
Qian, F., Jiang, X., Chai, R., and Liu, D. (2022). The roles of solute carriers in auditory function. Front. Genet. 13, 823049. doi:10.3389/fgene.2022.823049
Roccio, M., and Edge, A. S. B. (2019). Inner ear organoids: new tools to understand neurosensory cell development, degeneration and regeneration. Development 146 (17), dev177188. doi:10.1242/dev.177188
Rodrigues, R. R., Gurung, M., Li, Z., García-Jaramillo, M., Greer, R., Gaulke, C., et al. (2021). Transkingdom interactions between lactobacilli and hepatic mitochondria attenuate western diet-induced diabetes. Nat. Commun. 12 (1), 101. doi:10.1038/s41467-020-20313-x
Saito, T., Sato, K., and Saito, H. (1986). An experimental study of auditory dysfunction associated with hyperlipoproteinemia. Arch. Otorhinolaryngol. 243 (4), 242–245. doi:10.1007/bf00464438
Sheets, L., Holmgren, M., and Kindt, K. S. (2021). How zebrafish can drive the future of genetic-based hearing and balance research. J. Assoc. Res. Otolaryngol. 22 (3), 215–235. doi:10.1007/s10162-021-00798-z
Shen, W., Gong, B., Xing, C., Zhang, L., Sun, J., Chen, Y., et al. (2022). Comprehensive maturity of nuclear pore complexes regulates zygotic genome activation. Cell 185 (26), 4954–4970.e20. doi:10.1016/j.cell.2022.11.011
Shi, F., Hu, L., Jacques, B. E., Mulvaney, J. F., Dabdoub, A., and Edge, A. S. (2014). β-Catenin is required for hair-cell differentiation in the cochlea. J. Neurosci. 34 (19), 6470–6479. doi:10.1523/jneurosci.4305-13.2014
Shigemizu, D., Asanomi, Y., Akiyama, S., Higaki, S., Sakurai, T., Ito, K., et al. (2022). Network-based meta-analysis and the candidate gene association studies reveal novel ethnicity-specific variants in MFSD3 and MRPL43 associated with dementia with lewy bodies. Am. J. Med. Genet. B Neuropsychiatr. Genet. 189 (5), 139–150. doi:10.1002/ajmg.b.32908
Shrestha, B. R., Chia, C., Wu, L., Kujawa, S. G., Liberman, M. C., and Goodrich, L. V. (2018). Sensory neuron diversity in the inner ear is shaped by activity. Cell 174 (5), 1229–1246. doi:10.1016/j.cell.2018.07.007
Söllner, C., Burghammer, M., Busch-Nentwich, E., Berger, J., Schwarz, H., Riekel, C., et al. (2003). Control of crystal size and lattice formation by starmaker in otolith biomineralization. Science 302 (5643), 282–286. doi:10.1126/science.1088443
Tang, D., He, Y., Li, W., and Li, H. (2019). Wnt/β-catenin interacts with the FGF pathway to promote proliferation and regenerative cell proliferation in the zebrafish lateral line neuromast. Exp. Mol. Med. 51 (5), 1–16. doi:10.1038/s12276-019-0247-x
Ueno, S., Weidinger, G., Osugi, T., Kohn, A. D., Golob, J. L., Pabon, L., et al. (2007). Biphasic role for Wnt/beta-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc. Natl. Acad. Sci. U. S. A. 104 (23), 9685–9690. doi:10.1073/pnas.0702859104
Vona, B., Doll, J., Hofrichter, M. A. H., Haaf, T., and Varshney, G. K. (2020). Small fish, big prospects: using zebrafish to unravel the mechanisms of hereditary hearing loss. Hear Res. 397, 107906. doi:10.1016/j.heares.2020.107906
Wan, M., Huang, L., Liu, J., Liu, F., Chen, G., Ni, H., et al. (2021). Cyclosporine A induces cardiac developmental toxicity in zebrafish by Up-Regulation of wnt signaling and oxidative stress. Front. Pharmacol. 12, 747991. doi:10.3389/fphar.2021.747991
White, R. J., Collins, J. E., Sealy, I. M., Wali, N., Dooley, C. M., Digby, Z., et al. (2017). A high-resolution mRNA expression time course of embryonic development in zebrafish. Elife 6, e30860. doi:10.7554/eLife.30860
Wu, S., Cao, X., He, R., and Xiong, K. (2012). Detrimental impact of hyperlipidemia on the peripheral nervous system: a novel target of medical epidemiological and fundamental research study. Neural Regen. Res. 7 (5), 392–399. doi:10.3969/j.issn.1673-5374.2012.05.011
Xu, Y., Chen, X., Yu, L., Wang, Y., Wang, H., Wu, Z., et al. (2021). SLC4A11 and MFSD3 gene expression changes in deoxynivalenol treated IPEC-J2 cells. Front. Genet. 12, 697883. doi:10.3389/fgene.2021.697883
Yao, J., Zeng, H., Zhang, M., Wei, Q., Wang, Y., Yang, H., et al. (2019). OSBPL2-disrupted pigs recapitulate dual features of human hearing loss and hypercholesterolaemia. J. Genet. Genomics 46 (8), 379–387. doi:10.1016/j.jgg.2019.06.006
Keywords: MFSD3, morpholino knockdown, zebrafish, auditory system, wnt/β-catenin
Citation: Ma Y, Qiu S-W, Wang W-Q, Feng H-F, Zheng L, Wei G-G, Nie H-Y, Yang J-Y, Chen Y-J, Dai P, Gao X and Yuan Y-Y (2025) Functional characterization of MFSD3 in auditory system and zebrafish embryogenesis. Front. Genet. 16:1634493. doi: 10.3389/fgene.2025.1634493
Received: 24 May 2025; Accepted: 27 August 2025;
Published: 15 September 2025.
Edited by:
Mara Marongiu, National Research Council (CNR), ItalyReviewed by:
Yazhi Xing, Shanghai Jiao Tong University, ChinaOlga Strelkova, Harvard University, United States
Copyright © 2025 Ma, Qiu, Wang, Feng, Zheng, Wei, Nie, Yang, Chen, Dai, Gao and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xue Gao, bWl4dWVlcjAxMTBAMTI2LmNvbQ==; Yong-Yi Yuan, eXl5bXpoQDE2My5jb20=
 Ying Ma
Ying Ma Shi-Wei Qiu
Shi-Wei Qiu Wei-Qian Wang
Wei-Qian Wang Hai-Feng Feng
Hai-Feng Feng Lu Zheng
Lu Zheng Ge-Ge Wei1,2,3,4,5
Ge-Ge Wei1,2,3,4,5 Pu Dai
Pu Dai Xue Gao
Xue Gao Yong-Yi Yuan
Yong-Yi Yuan