- 1Department of Ophthalmology, The Affiliated Hospital of Guizhou Medical University, Guiyang, China
- 2Xiamen Eye Center and Eye Institute of Xiamen University, School of Medicine, Xiamen, China
- 3Fujian Provincial Key Laboratory of Ophthalmology and Visual Science, Fujian Engineering and Research Center of Eye Regenerative Medicine, School of Medicine, Xiamen University, Xiamen, Fujian, China
- 4Department of Ophthalmology, The Qinglong County People’s Hospital, Qinglong, Guizhou, China
- 5National Engineering Research Center of Ophthalmology and Optometry, Eye Hospital, Wenzhou Medical University, Wenzhou, China
- 6State Key Laboratory of Ophthalmology, Optometry and Visual Science, Eye Hospital, Wenzhou Medical University, Wenzhou, China
- 7National Clinical Research Center for Ocular Diseases, Eye Hospital, Wenzhou Medical University, Wenzhou, China
- 8Department of Biomedical Sciences, School of Infection, Inflammation and Immunology, College of Medicine and Health, University of Birmingham, Birmingham, United Kingdom
Ocular surface tissues, primarily consisting of the cornea, meibomian glands, conjunctiva and lacrimal glands, are crucial components of the eyes and are in direct contact with external environment. Various ocular surface abnormalities can lead to ocular surface diseases, and in severe cases, blindness. The intricate diversity of cell types and states, along with the absence of definitive biomarkers for ocular surface tissues, has posed significant challenges to fully understanding corneal stability, disease mechanisms, and therapeutic development. Single-cell RNA sequencing (scRNA-seq) is an advanced analytical technique used to examine the transcriptomes of individual cells. It enables detailed analysis of complex cellular dynamics, the distinction of various cell types, and the discovery of new biomarkers, thus deepening our insight into diverse cellular behaviors. Currently, scRNA-seq is mainly applied to study the developmental processes of ocular surface cells and to explore the pathogenic mechanisms of related diseases, such as dry eye disease, pterygium, keratoconus, Fuchs corneal endothelial dystrophy, ocular graft-versus-host disease, and primary acquired nasolacrimal duct obstruction, which involve the cornea, conjunctiva, and lacrimal gland. This review summarizes the principles and applications of the scRNA-seq technique, including its mechanism, effects, limitations, and applications in ocular surface research, aiming to bridge the gap between incomplete understanding and rapid technological progress of scRNA-seq.
1 Introduction
The ocular surface is formed by continuous epithelial tissues with distinct regional specialization, such as the cornea, meibomian glands, conjunctiva, and lacrimal glands (Cher, 2014; Ou et al., 2022). These components serve as crucial protective barriers for the eye. The cornea, which is transparent and lacks vasculature, plays a vital role in providing both transparency and refraction, thereby enabling precise focusing of light onto retinal photoreceptors (Sridhar, 2018). The conjunctiva, in contrast to the cornea, is a semi-translucent mucous layer lining the inner eyelid and extending across the anterior sclera to the corneal boundary (Weingeist, 1973). Characterized by heightened vascularity, it hosts conjunctival goblet cells, which can synthesize and secrete soluble mucin within the tear film (Swamynathan and Wells, 2020; Dong et al., 2009). This mucin, in turn, supports lubrication and protective functions. The tear film, a thin protective liquid layer atop the ocular surface’s epithelial cells, is composed of several constituents (Pflugfelder and Stern, 2020; Li et al., 2022). It comprises a lipid layer from the meibomian glands, an aqueous layer primarily secreted by the lacrimal glands and partly by the conjunctival epithelium, and a mucin layer originating from conjunctival goblet cells (Swamynathan and Wells, 2020). Beyond their structural and biochemical roles, these ocular surface components function as an integrated unit. Their homeostasis relies on coordinated regulation by neural, vascular, endocrine, and immune pathways, which are essential for preserving corneal clarity and stable vision. When these regulatory mechanisms are disrupted, clinical manifestations often follow, most commonly in the form of dry eye disease or keratitis. Moreover, because ocular surface tissues are directly exposed to the external environment, they remain particularly vulnerable to injury and environmental insults. As a result, the prevalence of ocular surface disorders, including dry eye disease, pterygium, and keratoconus, has been steadily increasing. These conditions not only produce symptoms such as redness, irritation, and dryness, but in severe cases may compromise corneal integrity, impair vision, and even lead to blindness (Craig et al., 2017). Therefore, a comprehensive exploration of the heterogeneity and complexity of different ocular surface cells is imperative.
Single-cell RNA sequencing (scRNA-seq) technology has become a leading powerful tool for uncovering the heterogeneity and complexity of RNA transcripts at the single-cell level (den Braanker et al., 2021; Papalexi and Satija, 2018). It also enables the identification of various cell types and their functions within a single tissue or organ (Borner et al., 2021; Jovic et al., 2022). Since its inception, scRNA-seq research has provided substantial insights across various fields, yielding significant advancements in our understanding of cell composition and function in humans, animals, and plants (Tang et al., 2009). Recently, Ahsanuddin and Wu (2023) conducted an extensive review of single-cell transcriptomics studies focusing on the ocular anterior segment, summarizing datasets, experimental considerations, and clinical relevance. Similarly, Arts et al. (2023) reviewed scRNA-seq applications specifically in corneal biology, highlighting the current understanding of corneal cell states and key biomarker genes. Our review builds upon these prior works by not only summarizing scRNA-seq studies of normal ocular surface tissues, such as the cornea, lacrimal gland and conjunctiva, but also extending the discussion to include a disease-oriented perspective. We critically analyze how scRNA-seq has contributed to uncovering cellular and molecular changes in ocular surface diseases, including dry eye disease, keratoconus, and lacrimal gland dysfunction. Furthermore, we provide an in-depth evaluation of technical limitations and biological challenges specific to ocular surface tissues and propose future directions including integrating spatial transcriptomics and multi-omics approaches for improving the understanding of ocular surface health and disease.
2 scRNA-seq technology
2.1 The working principle of scRNA-seq technology
Cells are the fundamental units of structure and functionality in organisms. Remarkably, each cell possesses its own distinct identity, even within a homogeneous tissue, potentially expressing a unique set of genes. While traditional methods such as microarray analysis (Yao et al., 2022) and bulk RNA sequencing (Roodnat et al., 2022) have been employed to ascertain the average gene expression levels across all cells, these techniques fall short in discerning the intricate disparities in gene expression at the individual cell level. Consequently, they fail to unveil the underlying complexities of cellular changes. The advent of scRNA-seq technology marks a significant stride in research methodology, offering the capacity to illuminate the diversity of each cell, even though their presence within the same tissue and close spatial proximity (Voigt et al., 2019). By enabling biological investigations at a microscopic level, this technique resolves previously insurmountable queries, such as detecting subtle transcriptional changes within heterogeneous cell populations and uncovering the unique molecular identity of individual cells (Hedlund and Deng, 2018).
The scRNA-seq workflow involves steps such as single-cell isolation, cell lysis, reverse transcription, cDNA amplification, and preparation of sequencing libraries (Kolodziejczyk et al., 2015). Among these steps, the initial one, the isolation and capture of single cells, assumes paramount importance. This process commences with the isolation of individual cells, achieved through enzyme treatment, laser capture microdissection, or patch clamping (Hedlund and Deng, 2018). Single-cell capture is a pivotal endeavor aimed at obtaining cells of exceptional quality. A range of capture techniques are commonly employed, such as fluorescence-activated cell sorting (Polisetti et al., 2022), limiting dilution (Kawakita et al., 2008), microfluidic system (Junker and van Oudenaarden, 2015), magnetic-activated cell sorting (Du H. et al., 2023), and laser microdissection (Sutherland et al., 2018). Following isolation and capture, reverse transcription converts cellular RNA into first-strand cDNA. It is then subjected to amplification via PCR or in vitro transcription, resulting in second-strand synthesis (Kolodziejczyk et al., 2015). After barcoded cDNAs are produced from single cells or single nuclei, they can be sequenced using various sequencing platforms (Kleino et al., 2022; Shi et al., 2023). Subsequently, the sequencing data undergo quality control, normalization, and dimensionality reduction, followed by clustering and differential expression analyses, through which cell subpopulations are identified and disease-related molecular signatures in ocular tissues are revealed.
2.2 The importance of scRNA-seq technology
First discovered in 2009 (Tang et al., 2009), scRNA-seq has become an important technology in biomedical and clinical research fields. It now serves as a robust technology for detecting and characterizing novel or rare cell types and subpopulations in tissues and organs from different species. To date, it has provided invaluable insights encompassing developmental processes (Tang et al., 2024), tumorigenesis (Amin et al., 2024), immunology (Xia et al., 2025), microbiology (Jia et al., 2024), and a multitude of other domains within the spectrum of health and disease. Despite its achievements, the clinical research via scRNA-seq remains nascent, centering predominantly on augmenting our comprehension of disease processes and the identification of biomarkers for diagnosis and therapeutic intervention.
3 Application of scRNA-seq on the ocular surface
3.1 Cornea
Structurally, the cornea is divided into the epithelium, Bowman’s layer, stroma, Descemet’s membrane, and the endothelial cell layer. Due to the development of scRNA-seq, the fate of each layer of corneal cells can now be explored, enabling researchers to further understand their similarities and differences at the molecular level (Arts et al., 2023; Sun et al., 2023). One example is the study by Wu et al. (2023), who performed unbiased clustering of scRNA-seq data from 14,732 cells across 40 corneas, identifying 17 clusters corresponding to six major cell types: nine epithelial, three keratocyte, two corneal endothelial, and one cluster each of immune, vascular endothelial, and fibroblast cells. Swarup et al. (2023) used scRNA-seq to study corneal organoids during development and determined the specific time points at which these different cell types appear in fetal corneal development. They found that certain pluripotent cell clusters are committed to the epithelial cell lineage at 1 month, with corneal epithelial, endothelial, and stromal cell biomarkers being expressed. By 3 months, keratocytes become more specified, and by 4 months, epithelial cells are further differentiated. Building upon these findings, the following sections delve into single-cell RNA sequencing analyses of individual corneal layers.
3.1.1 Corneal epithelium
3.1.1.1 XYZ theory of corneal epithelium
Serving as the eye’s first line of defense, the corneal epithelium protects against external pathogens and environmental exposure. It is regularly replenished throughout life, primarily mediated by corneal epithelial stem cells (Schermer et al., 1986). These cells, also known as limbal stem cells (LSCs), reside at the base of the limbus at the junction of the cornea and conjunctiva and are characterized by slow cell cycle, nucleoplasmic ratio, small size and high proliferative potential (Sun et al., 2023; Long et al., 2024). Through proliferation and differentiation, LSCs generate corneal transient amplifying cells (TACs) and limbal progenitor cells (LPCs). TACs move toward the center and surface of the corneal epithelium, eventually developing into adult corneal epithelial cells (Davanger and Evensen, 1971), as defined by the XYZ theory (Thoft and Friend, 1983). ScRNA-seq technology has been utilized to study various aspects of the corneal epithelium, particularly in elucidating the developmental trajectories within this intricate tissue.
In 2019, Kaplan et al. (2019) applied scRNA-seq to categorize mouse corneal limbal/corneal epithelial cells into distinct classes: stem/early transient amplifying (TA) cells, mature TA cells, and relatively differentiated cells. This categorization was based on variations in gene expression related to stemness, proliferation, and differentiation. Following this, Li DQ. et al. (2021) extracted 16,360 single cells from the basal limbus of the human cornea for sequencing. Subsequent heterogeneity analysis facilitated the classification of these cells into five primary types: LSCs, LPCs, TACs, postmitotic cells (PMCs, biomarker: HTRA1), and terminally differentiated cells (TDCs, biomarkers: KRT3 and KRT12). Subsequently, Altshuler et al. (2021) combined scRNA-seq with quantitative lineage tracing to delineate mouse LSC populations located in the limbal subregion, identifying them as “outer” and “inner” LSCs. The “outer” LSC population contains slow-dividing quiescent LSCs (qLSCs), involved in wound healing and boundary formation, as well as in eco-t-cell regulation. The “inner” LSC population contains actively renewing LSCs (aLSCs), playing an important role in maintaining corneal epithelial homeostasis. Around the same time, Song et al. (2022) mapped the heterogeneous cell populations of corneal limbal cells under normal homeostatic conditions and during the wound healing process. They found that two different types of LSCs might participate in the damage repair process: “putative active LSCs” that actively divide during damage and “putative quiescent LSCs” that remain quiescent. As noted earlier, the recent report by Wu et al. (2023) identified nine epithelial cell subtypes, including qLSCs, TACs, differentiated cells from the cornea, and two minor conjunctival epithelial clusters. These findings are corroborated by other publications mentioned above. These works contribute to refining the XYZ theory of corneal epithelium and establish a foundational understanding of the differentiation trajectory of corneal LSCs (Figure 1).
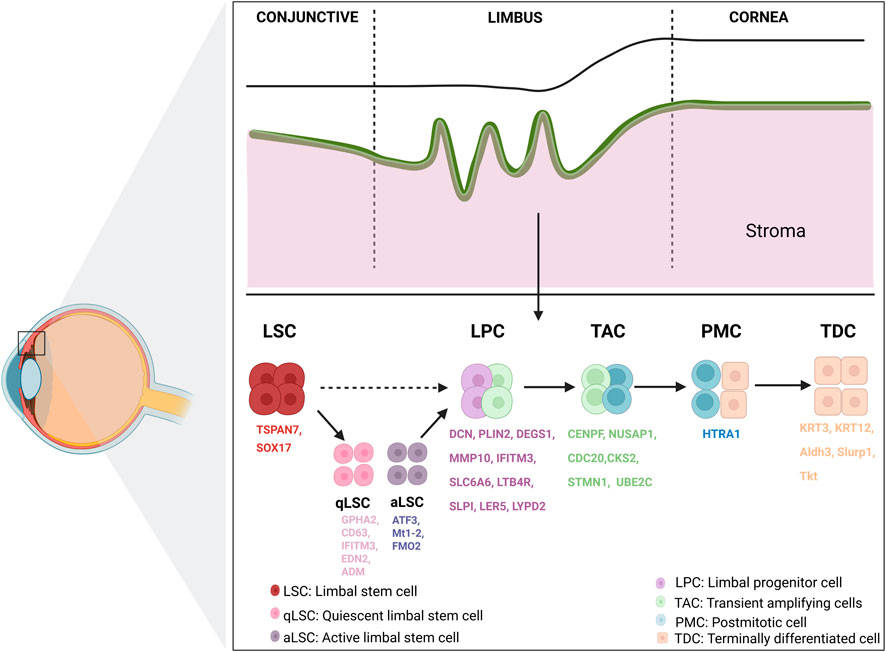
Figure 1. Overview of limbal cornea structure and biomarkers. The limbal region harbors LSCs, which play a central role in maintaining corneal epithelial homeostasis according to the XYZ theory. LSCs, residing in the basal layer of the limbus, give rise to a hierarchical lineage of progeny including qLSCs, aLSCs, LPCs, TACs, PMCs, and TDCs. LSC, limbal stem cell; qLSC, quiescent limbal stem cell; aLSC, active limbal stem cell; LPC, limbal progenitor cell; TAC, transient amplifying cell; PMC, post-mitotic cell; TDC, terminally differentiated cell. Created in BioRender. Wu, Y. (2025).
However, the designation of “LSCs” in many studies warrants careful interpretation. Current evidence primarily relies on gene expression profiles rather than functional validation of self-renewal or multipotency. It remains possible that the so-called LSCs might represent a spectrum of progenitor cells at different stages of activation rather than a distinct stem cell population. This distinction is critical, particularly in the context of limbal stem cell deficiency, where the true absence of a pluripotent stem cell population has not been definitively established.
3.1.1.2 New biomarkers of different developmental stages of corneal epithelial stem cells
Although some biomarkers have been identified for corneal limbal basal cells, distinguishing LSCs from other epithelial cell types continues to be challenging. The advent of scRNA-seq technology has opened new avenues for the accurate identification of LSCs and their diverse developmental stages. Li DQ. et al. (2021) validated the outcomes of human corneal limbal scRNA-seq through RNA scope HiPlex and antibody staining, discovering that SOX17+ and TSPAN7+ cells were primarily confined to corneal limbal basal epithelium. TSPAN7 and SOX17 were identified as novel LSC markers, functionally verified through siRNA-mediated knockdown assays that impaired proliferation and epithelial regeneration. Additionally, LPC-enriched genes were identified, including SLPI, LTB4R, SLC6A6, IFITM3, MMP10, DEGS1, PLIN2 and DCN. In addition, Català et al. (2021) identified possible biomarkers of the human corneal limbal stem cell ecological niche: CAV1, CPVL, HOMER3, CXCL14, and genes unique to TACs: CKS2, STMN1, and UBE2C. Two differentiated (GPHA2+ and KRT6B+) and two highly stem-like (TP63+ and CCL20+) cell states of corneal limbal stem/progenitor cells were further defined by Dou et al. (2021). In the same year, CDC20, UBE2C, NUSAP1 and CENPF were discovered for the first time as biomarkers for TACs by Li JM. et al. (2021) using scRNA-seq on 16,360 limbal epithelial basal cells from humans. Further analysis of scRNA-seq also identified potential initiators of these makers changes. Zhu et al. (2023) found the FEZ1-DKK1 axis is crucial in regulating proliferation and senescence of corneal epithelial cells. Sun et al. (2024) found Creb5, a transcription factor, is present in LSCs and markedly increases in expression after injury. Recently, Vattulainen et al. (2024) used scRNA-seq to investigate the heterogeneity in the process of differentiating human pluripotent stem cells toward LSCs. Their study identified distinct cell clusters, including a robust induced LSC population expressing important LSC biomarkers such as TP63 and KRT14. Importantly, they proposed AREG and ITGA6 as novel surface biomarkers to enrich for LSC-like cells and improve differentiation consistency across different hPSC lines.
The scRNA-seq results of qLSCs and aLSCs further reveal the changes in makers during the transitional phase. Applying in situ hybridization probes to label mouse conjunctival and corneal basal and suprabasal cells, respectively, Altshuler et al. (2021) found that Gpha2 staining could divide the corneal limbus into qLSCs and aLSCs, which were labeled by Gpha2, Cd63, Ifitm3, and Atf3, Mt1-2, respectively. Song et al. (2022) applied scRNA-seq to identify heterogeneous cell populations at the corneal limbus under normal homeostasis and following injury. Their investigation unveiled “putative quiescent LSCs” (biomarkers: Edn2 and Adm) and “putative active LSCs” (biomarkers: Fmo2). They also identified diverse LPC subtypes through distinct biomarkers such as Ler5 and Lypd2. Additionally, they noted a progressive increase in the expression of biomarkers associated with mature corneal epithelium (Aldh3, Slurp1, and Tkt) during corneal epithelial maturation.
Nevertheless, the specificity and functional significance of these biomarkers require further investigation. Many of the biomarkers proposed for LSCs and their progeny, such as TSPAN family members and UBE2C, are also associated with general epithelial proliferation in other tissues (Liu et al., 2020; Zhang H. et al., 2024). Therefore, it is essential to distinguish whether these biomarkers define distinct cell populations or merely reflect dynamic cellular states during differentiation or injury response.
3.1.1.3 scRNA-seq for corneal epithelial wound healing
In scRNA-seq studies of the cornea, analyses focusing on corneal epithelial wound healing are common, as this process directly captures the key cellular players involved as well as the differentiation trajectory from LSCs to corneal epithelial cells (Sun et al., 2024; Mirmoeini et al., 2023; Zhou et al., 2024; Yin et al., 2025; Lu et al., 2025). In an earlier study, Mirmoeini et al. (2023) employed scRNA-seq on dissociated healthy, de-epithelialized, and denervated corneal limbi to investigate the cellular composition of the limbal niche and gene expression changes linked to innervation-dependent epithelial renewal. Their results revealed that Schwann cells, glial cells associated with tissue-innervating axon terminals, play a central role as regulators of corneal epithelial regeneration. Subsequent studies have identified additional biomarkers and biological processes involved in corneal epithelial wound healing. Beyond the previously reported biomarker Creb5 discovered in a mouse model (Sun et al., 2024), Zhou et al. (2024) conducted scRNA-seq for the first time on corneal epithelial wound-healing samples derived from non-human primates (Cynomolgus monkeys). Their analysis highlighted the critical roles of limbal epithelial cells and basal epithelial cells in extracellular matrix formation and wound repair, as well as the importance of suprabasal epithelial cells in epithelial differentiation. Moreover, they identified Thbs1 as a key biomarker of a transit-amplifying cell subcluster that promotes early stages of healing. Lu et al. (2025), from the same research group as Zhou, employed single-cell multiomics analysis, including scRNA-seq and single-cell assay for transposase-accessible chromatin using sequencing to characterize the early transcriptomic and chromatin accessibility changes of early wound response in mouse corneal epithelium, revealing a reduction in cell type-specific genes accompanied by an increase in common transcriptional responses, widespread alterations in chromatin accessibility across epithelial cell types, and a marked enrichment of Fosl1/AP-1 binding sites within wound-induced open regions.
3.1.2 Corneal stroma
Comprising about 90% of the cornea’s thickness, the stroma is rich in collagen layers and populated mainly by keratocytes, which are key to preserving corneal biomechanics and transparency (Català et al., 2021; Ligocki et al., 2021; Hertsenberg and Funderburgh, 2015). Corneal stromal cells have been extensively examined in scRNA-seq studies. LUM, KERA, and DCN (Ligocki et al., 2021) are commonly used biomarkers for identifying human stromal cells. However, these studies have also identified biomarkers in corneal stromal cells that include stromal fibroblasts, keratocytes, and corneal stromal stem cells, with biomarkers varying across different studies. Collin et al. (2021) found MMP3, and ENG (CD105) as biomarkers for human corneal stromal stem cells. ALDH3A1, LUM, and KERA has also been discerned as a human keratocyte biomarker (Català et al., 2021). Additionally, van Zyl et al. (2022) used MME and KERA to pinpoint corneal stromal fibroblasts, and ANGPTL7 to identify corneal stromal keratocytes (Figure 2). Human keratocyte function is also supported by high levels of matrix metalloproteinases MMP2 and MMP3, metalloproteinase inhibitors TIMP1 and TIMP2, adhesion molecules THSB4, THSB1, ITGB4 and ITGB1, along with collagen-related biomarkers COL12A1, COL6A3, COL1A2, and PCOLCE2 (Ligocki et al., 2021).

Figure 2. Application of scRNA-seq in healthy ocular surface structure and biomarkers. The cornea panel summarizes epithelium, stroma, CSSCs, keratocytes, stromal fibroblasts, and endothelium with representative biomarkers. The conjunctiva panel highlights epithelial, wing cells, and basal/superficial layers with representative biomarkers. The meibomian-gland panel depicts meibocytes enriched for lipid-synthesis genes. The lacrimal-gland panel shows glandular epithelium, fibroblasts, myeloid-derived immune cells, lymphoid-derived immune cells, vascular-associated cells, and tear-component with representative biomarkers. scRNA-seq, single-cell RNA sequencing; CSSC, corneal stromal stem cell. Created in BioRender. Wu, Y. (2025).
While scRNA-seq studies suggest heterogeneity within corneal stromal cells, it remains uncertain whether the identified populations represent distinct cell types or reflect varying activation states of keratocytes. For instance, biomarkers such as ENG are often used to define stromal stem cells, yet their expression is also associated with vascular cells (Mid et al., 2005). Similarly, elevated MMP and TIMP expression may indicate a dynamic response to tissue stress (Rudra et al., 2022).
3.1.3 Corneal endothelium
Situated in the innermost layer of the cornea, the corneal endothelium is composed of a monolayer of hexagonal cells essential for maintaining corneal transparency (Chen et al., 2017). scRNA-seq of various human corneal tissues has revealed biomarkers for corneal endothelial cells, including ATP1A1, CD166 (ALCAM), sPrdx1 (PRDX1), ZO-1 (TJP1), SLC4A11, and CA3 (Català et al., 2021; Ligocki et al., 2021). Beyond these biomarkers, a cell expressing the myofibroblast biomarker ACTA2 (α-sma) was identified as a fibroblastic corneal endothelial cell from human in Collin’s study (Collin et al., 2021). Additionally, genes involved in human corneal endothelial cell signaling pathways, such as FZD2, FGF7, and FGFR1, were identified (Ligocki et al., 2021) (Figure 2).
However, whether the observed transcriptional differences within corneal endothelial cells represent true biological subtypes or merely reflect variations in cell state, age, or experimental conditions remains unclear. For instance, the identification of ACTA2-expressing endothelial-like cells may suggest a population undergoing endothelial-to-mesenchymal transition (Wu et al., 2017), a process known to occur in disease states or during stress.
3.2 Conjunctiva
Several sequencing analyses of ocular tissues have now identified the presence of conjunctival cells. The utilization of a combination of biomarkers, notably KRT14 (Collin et al., 2021; van Zyl et al., 2022), has proven instrumental in identifying conjunctival epithelial cells. Moreover, distinct biomarkers including MUC5AC, AQP5, KRT13, and KRT19 have been applied for recognizing conjunctival epithelial cells (Li DQ. et al., 2021; Ligocki et al., 2021; Collin et al., 2021; van Zyl et al., 2022; Maiti et al., 2022; Gautam et al., 2021). In addition, biomarkers for basal conjunctival epithelium and superficial conjunctival epithelium, such as S100A8, BCAS1, and S100A9, were also identified. van Zyl et al. (2022) further used different biomarkers for basal (KRT14, PDGFC), superficial (KRT7, LCN2, WFDC2, ENTPD2), and wing (KRT13, KRT15, BCAS1, RARRES1) conjunctival epithelium (Figure 2).
However, it remains hard to distinguish true conjunctival epithelial subtypes from transitional or activated cell states based solely on scRNA-seq data. Several biomarkers, such as KRT19, are broadly expressed in stratified epithelial tissues (Natesan et al., 2019). Furthermore, biomarkers like S100A8 and S100A9 are known to be upregulated under other inflammatory conditions (Jukic et al., 2021), which may confound interpretations regarding basal or superficial epithelial identity. The identification of wing conjunctival epithelial cells based on WFDC2 expression also requires further validation, as these genes could be induced by environmental stress or microbial exposure (Ebihara et al., 2023).
3.3 Meibomian glands
Given that one of the principal functions of the meibomian glands is the secretion of lipids that contribute to the ocular surface tear film, Butovich and Wilkerson (2022) combined scRNA-seq with lipidomic and transcriptomic analyses for development and maturation of meibomian glands of C57Bl/6J founder mice to explore how gene expression and lipid composition change. In single-cell transcriptomic profiling of mouse tarsal plates, 14 cell clusters were identified in adult (about 80 days) samples, with 4 clusters (cluster IV, VI, IX, and XIII) showing high expression of meibogenesis-related genes, such as Elovl1/3/4 (fatty acid elongation), Far1/2 (fatty acyl reduction to fatty alcohols), Scd1/3/4 (fatty acid desaturation), Awat1/2 (acyl-CoA:wax alcohol acyltransferases producing wax esters), Soat1 (cholesteryl ester synthesis), and cholesterol biosynthesis genes including Hsd17b2, Msmo, Hmgcs, and Dhcr24. These clusters were interpreted as meibocytes at different stages of differentiation or their progenitors. In the tarsal plates of pups (about 7 days), meibogenesis-rich cells were confined to a single cluster (cluster 16), with markedly fewer cells than in adults, reflecting the lower proportion of mature meibum lipids at this stage. Differential expression patterns were observed even within the same cluster, for example, Far1 vs. Far2 or Scd3 vs. Scd4, suggesting subpopulation-specific specialization in lipid production.
However, at present, scRNA-seq studies on the meibomian glands remain limited, with no existing studies based on human tissue. This is likely because human meibomian glands have poor regenerative capacity, making living individuals unlikely to donate such low-regeneration tissues, and the number of cells obtainable from deceased individuals is extremely limited. Further research on healthy human samples may provide additional guidance for the management of meibomian gland-related diseases.
3.4 Lacrimal glands
scRNA-seq has been expanded to studies of the lacrimal gland, a critical organ responsible for tear production, ocular surface protection, lubrication, and maintaining surface homeostasis (Lin Y. et al., 2023). Tear secretion in the mature lacrimal gland is driven by the coordinated activity of acinar, ductal, and myoepithelial cells, all terminally differentiated and responsive to neural signals (Figure 2).
Several reports have studied the signaling mechanisms that control early lacrimal branching morphogenesis, revealing various mechanisms for the initial development of lacrimal glands (Dean et al., 2004; Ma et al., 2023; Hayashi et al., 2022). However, the exact developmental timeline of various cell types, their transcriptional characteristics, and progenitor cells in the lacrimal gland has yet to be determined. In 2017, Farmer et al. (2017) performed the first scRNA-seq of the lacrimal gland at two time points spanning key morphological changes during lacrimal gland development. They used scRNA-seq for the first time to reveal cellular composition, cellular dynamics, and lineage relationships in the developing lacrimal gland. Tracing different epithelial populations by lineage revealed novel features of epithelial homeostasis, providing the first direct evidence of a pool of progenitor cells in the lacrimal gland. Furthermore, Huang et al. (2024) and Fan et al. (2024) explored the heterogeneity and complexity (such as immune cell diversity) of murine extraorbital lacrimal gland through scRNA-seq and found over 37 subclasses of cells, including seven types of glandular epithelial cells (Acinar cells: co-expressing Agr2, Bhlha15 and Sval2; Myoepithelial cells: co-expressing Acta2 (α-sma); Ductal epithelial cells: co-expressing Na+/K+-ATPase), three types of fibroblasts (co-expressing Col1a2, Col1a1 and Pdgfra), ten types of myeloid-derived immune cells (Macrophages: expressing Itgax or Folr2; Monocyte cells: two clusters co-express Mafb, Cebpb and Csf1r; Dendritic cells: the makers depend on their subtype; Mast cells: co-expressing Kit, Ccl2, Gata2, and Cpa3), at least eleven types of lymphoid-derived immune cells (the makers depend on their cell types), and five types of vascular-associated cell subsets (co-expressing Cyyr1, Cdh5, Emcn, and Pecam1) (Figure 2).
Recently, Bannier-Hélaouët et al. (2021) applied scRNA-seq technology to study human lacrimal gland tissues and organoids. They identified new tear component biomarkers WFDC2, PRR27, SMR3A, and SMR3B in lacrimal gland tissues. Through heterogeneity analysis of lacrimal gland acinar cells, various tear components were identified in addition to the expression of the acinar cell biomarkers LYZ, LTF, LCN1, PRR27, SMR3A, and SMR3B. Ductal cells predominantly expressed LCN2, C3, and WFDC2. Furthermore, they found that lacrimal gland organoid cells originated predominantly from ductal progenitor cells expressing KRT.
Despite the detailed cellular mapping provided by scRNA-seq studies of the lacrimal gland, several challenges remain in interpreting these findings. Many of the identified biomarkers, such as LCN2 and WFDC2, are not unique to the lacrimal gland and are expressed in other secretory tissues (Han et al., 2018). Additionally, the observed immune cell heterogeneity may be influenced by tissue processing or environmental stimuli rather than representing a stable resident population. It remains unclear whether the diverse immune cell subsets reflect steady-state physiology or potential inflammatory activation during sample preparation.
4 Application of single-cell RNA sequencing in the study of ocular surface diseases
Beyond its contributions to understanding physiological processes, scRNA-seq also plays critical role in investigating ocular surface disorders. Conditions like dry eye disease, keratoconus, Fuchs corneal endothelial dystrophy, corneal transplant rejection, and various other ocular surface diseases have been researched through this technology.
4.1 Dry eye disease
Dry eye disease, a multifactorial ocular surface disease, poses a significant threat to quality of life and may even lead to vision impairment or blindness (Craig et al., 2017; Kai et al., 2024; Qu et al., 2019). Numerous investigations have explored the pathogenesis of dry eye disease. Currently, tear film hyperosmolarity and ocular surface inflammation are considered the primary etiologies of dry eye disease (Hessen and Akpek, 2014; Yang et al., 2018; Lin S. et al., 2023), although comprehensive research remains essential. Recently, scRNA-seq tool has been used to the in-depth work within dry eye disease (Figure 3).

Figure 3. Application of scRNA-seq in ocular surface diseases and their biomarkers. These include dry eye disease, where immune and epithelial interactions are highlighted; pterygium, with altered epithelial, stromal, and immune cell populations; keratoconus, involving corneal stromal dysregulation; FECD, showing endothelial cell heterogeneity and regulatory pathways; and corneal transplantation rejection, marked by immune-mediated changes; Lacrimal gland-associated diseases, such as oGVHD and PANDO. FECD, Fuchs endothelial corneal dystrophy; oGVHD, ocular graft-versus-host disease; PANDO, primary acquired nasolacrimal duct. Created in BioRender. Wu, Y. (2025).
Alam et al. (2022) were the first to analyse the expression profile of conjunctival cells using scRNA-seq in C57BL/6J and RXRα mutant mice. Their investigation revealed distinct clusters of conjunctival immune cells in C57BL/6J mice. They compared gene expression in conjunctival immune cells, finding that γδ T cells were the main population expressing Il-17. Furthermore, they identified RXRα as an inhibitor of Il-17 production by conjunctival lymphocytes, suggesting RXRα as a potential therapeutic target for dry eye disease. Following this, Alam et al. (2024) reported the scRNA-seq results of short-term dry eye disease-induced changes in corneal immune cell populations. The transition from monocytes to terminal resident macrophages was characterized, revealing significant differential expression across 1,365 genes. Resident macrophages exhibited increased expression of the Tam receptor (Mertk), inflammatory biomarkers (Junb, Adam17, and Vcam), and cytokines and chemokines (Il1rn, Tnf, Cxcl1, and Ccl12), along with decreased expression of complement and MHCII genes (Figure 3). Additionally, elevated levels of Cxcl1 in the cornea intensified irritation and pain responses to topical hypertonic saline application. These findings suggest that these molecular changes contribute to the sensorineural alterations observed in dry eye disease. The similar scRNA-seq work by Liu et al. (2024) revealed a dry eye disease-associated pro-inflammatory microenvironment in the conjunctiva associated with dry eye disease. This microenvironment is centered around epithelial cells and involves interactions with macrophages and CD4 T cells, including Th1, Th17, and regulatory T cells (Treg).
In a recent publication (Lin JB. et al., 2023), scRNA-seq was employed to comprehensively profile the cellular landscape of the cornea under different physiological and pathological conditions, including healthy, dry eye disease, aging, and diabetic mouse models. This high-resolution analysis revealed five distinct cell populations that were specifically enriched or altered in the corneas of mice with dry eye disease, highlighting the cellular heterogeneity associated with the disorder. Notably, the study identified the matricellular protein SPARC as a key regulator during the adaptive regeneration of the corneal epithelium. SPARC was shown to modulate extracellular matrix remodeling and influence cell–matrix interactions, thereby facilitating epithelial cell migration and wound closure. Loss- and gain-of-function experiments further demonstrated that SPARC deficiency impaired epithelial repair, whereas its presence was indispensable for restoring barrier integrity.
4.2 Pterygium
Pterygium is a common, benign, wedge-shaped, fleshy tissue growth of the conjunctiva extending onto the cornea (Zhong et al., 2016; Zhang Y. et al., 2022). Zhang X. et al. (2024) utilized scRNA-seq to investigate the cellular transcriptional landscape of pterygium, revealing insights into its underlying pathogenesis and potential therapeutic targets. By comparing the conjunctival tissues of patients with primary pterygium to those of healthy individuals, significant differences in cell lineages were revealed. The study identified 9 major cell lineages in pterygium and normal conjunctival tissue: melanocytes (MLANA, PMEL, TYRP1), proliferating cells (MKI67, BIRC5, PCNA), B cells (CD19, MS4A1, BANK1), myeloid cells (TYROBP, LYZ, CD68), plasma cells (MZB1, JCHAIN, IGHG1), T cells and natural killer (NK) cells (CD3D, CD3E, TRAC, NCR1), epithelial cells (KRT19, KRT5, CLDN4), endothelial cells (DCN, VWF, RAMP3), and fibroblasts (DCN, COL1A2, COL1A1) (Figure 3). In addition, the study identified distinct sub-clusters of endothelial cells, epithelial cells, and fibroblasts. Various immune cell types were also analyzed, highlighting their potential roles in driving vascularization, inflammation, and immunosuppression in pterygium through dense cell-cell interactions. Notably, macrophages and ACKR1+ endothelial cells were found to potentially contribute to pterygium progression via intercellular communication. The findings further suggest that macrophages, recruited by ACKR1-activated vascular endothelial cells, may promote angiogenesis, immune suppression, and inflammatory responses, playing a pivotal role in pterygium development.
4.3 Keratoconus
Keratoconus is a progressive corneal disorder and the most prevalent form of primary ectasia, characterized by corneal thinning, irregular astigmatism, and reduced visual acuity (Dong et al., 2022). However, the factors contributing to the pathogenesis and progression of keratoconus are still not fully understood. The use of scRNA-seq technology offers a powerful tool to explore cellular heterogeneity in keratoconus.
In 2021, Collin et al. (2021) conducted the first scRNA-seq analysis of central corneal samples from individuals with keratoconus, comparing them to healthy central corneas. Their investigation revealed an increase in corneal stromal keratocytes in conical corneal samples, primarily driven by EIF2 and mTOR signaling pathways, oxidative phosphorylation, and mitochondrial dysfunction. Recently, Dou et al. (2022) compared scRNA-seq analyses of central corneal cells from healthy individuals and those with keratoconus. They elucidated the cellular composition of the central cornea in both groups and confirmed the central role of stromal cells in the disease, highlighting the imbalance of collagen and extracellular matrix. They also identified potential new biomarkers of keratoconus, YAP1, TEAD1, CTSD, and CTSK (Figure 3).
4.4 Fuchs corneal endothelial dystrophy
The human corneal endothelium, a non-proliferative layer, typically remains quiescent. However, pathological conditions or specific ophthalmic procedures can damage the corneal endothelium, leading to endothelial loss and consequent visual impairment (Zhang et al., 2023). While single-cell transcription maps of endothelial cells across various tissues exist (Dumas et al., 2020; Kalucka et al., 2020), it remains uncertain whether corneal endothelial cells exhibit alterations in functional status at the single-cell level. Using scRNA-seq technology to reveal the heterogeneity of corneal endothelial cells at the single-cell level may provide insights into their functionality, pathological conditions, and the origins and mechanisms underlying corneal endothelial-related disorders (Yang et al., 2024).
Wang et al. (2022) performed the first high-resolution single-cell study of the human corneal endothelium. The sequencing results showed that the endothelium consists of heterogeneous cells with different transcriptional signals and functions. These endothelial cells were categorized into four clusters: C0-C3. These clusters were associated with oxygen level response and redox processes, translation elongation and peptide metabolism, cell cycle and DNA replication, and immune responses, respectively. Importantly, the study found that long noncoding RNA (lncRNA) NEAT1 was highly enriched in the C0 subpopulation but significantly downregulated in Fuchs endothelial corneal dystrophy (FECD). Further investigations confirmed the deletion of NEAT1 and its antioxidant role in the FECD model. Their findings suggest that NEAT1 may be a potential therapeutic target for FECD (Figure 3).
4.5 Corneal transplantation rejection
Corneal transplantation is a critical treatment modality for various ocular pathologies but faces limitations due to insufficient corneal donors and postoperative immune rejection (Li et al., 2024). Efforts to address these challenges have focused on developing alternative therapies that reduce reliance on donor corneas.
In 2015, Peh et al. (2015) pioneered a novel dual-media approach for propagating primary human corneal endothelial cells, making it possible to replace corneal transplantation with cell injection therapy. Subsequently, Kinoshita et al. (2018) successfully treated herpetiform keratitis by injecting human corneal endothelial primary cells supplemented with ROCK inhibitors, marking an important milestone in corneal endothelial cell injection therapy. However, the culture of corneal endothelial cells presents challenges. The culture process may lead to alterations in cell characteristics, and there is no standardized cellular benchmark for endothelial injections. Thus, understanding the heterogeneity of corneal endothelial cells across different generations is crucial. Català et al. (2023) recently performed single-cell sequencing of human primary corneal endothelial cells of different generations and culture times. This study revealed various cellular subtypes of human corneal endothelium generated from primary cultures and identified CGNL1, NCAM1, and CD166 as biomarkers of therapy-grade primary corneal endothelial cells. These findings provide important references for cellular therapies for endothelial diseases of the cornea.
Additionally, rejection induced by allogeneic corneal transplantation remains a significant issue. Further studies on the mechanisms of this immune rejection are necessary. Lai et al. (2023) applied scRNA-seq towards corneas from mice transplanted with allogeneic corneas and found that the T-cell biomarker genes Ctla4, Ccl5, and Tcf7 may play key roles in corneal transplant rejections, which may be achieved by affecting the activation of CD4+ T cells (Figure 3).
4.6 Lacrimal gland related diseases
4.6.1 Ocular graft-versus-host disease
Ocular graft-versus-host disease (oGVHD) represents the most frequent complication following allogeneic hematopoietic stem cell transplantation (Bonelli et al., 2022). He et al. (2024) were the first to apply scRNA-seq to characterize lacrimal gland cell populations implicated in mouse oGVHD model, identifying 23 cell populations belonging to 11 cell types. They found that myoepithelial cells showed reduced secretion function, with lower expression of genes related to lipid metabolism and calcium channel activity. These cells also exhibited decreased extracellular matrix synthesis, indicated by downregulation of Abi3bp, Sparc, and Lgals1, and increased expression of chemokine-related genes Ccl7, Ackr3, Ccl2, and Cxcl10. Similarly, fibroblasts showed a reduction in extracellular matrix production, reflected by lower levels of Lum, Col14a1, Col3a1, and Col1a1. In contrast, they displayed an inflammatory gene expression pattern, characterized by elevated expression of cytokine and chemokine genes (Ccl7, Ccl8, Cxcl13, Il11) and increased levels of MHC class I-related genes (H2-K1, H2-D1, B2m) as well as the MHC class II-associated gene Cd74. Additionally, a newly identified epithelial population, referred to as Lrg1-high epithelial cells, characterized by increased levels of Acot1, Lcn2, Cxcl17, Lrg1, and Epcam, was found exclusively in the oGVHD lacrimal gland (Figure 3).
4.6.2 Primary acquired nasolacrimal duct obstruction
Primary acquired nasolacrimal duct obstruction (PANDO) is a prevalent lacrimal drainage disorder in adults that severely impairs the lacrimal pump function responsible for transporting tears from the ocular surface to the nasal cavity (Ali, 2023). In scRNA-seq results of 5 PANDO patients lacrimal sac samples, Zhang W. et al. (2024) observed several key findings from 11 cell types were identified among 25,791 cells. They noted that T cells, which varied greatly in number, played an active role in the inflammatory response, showing higher expression of HLA-DRB1, IGLC3, IGHG4, IGHG3, and IGHG1. B cells were found to regulate the migration and proliferation of epithelial cells during the late inflammatory stage, with genes related to epithelial cell proliferation, such as ZFP36, TNF, IGFBP5, HMOX1, EGR3, and CCL2, being downregulated in memory B cells. Epithelial cells expressed SASP and upregulated inflammatory genes, including MMP2, MMP10, CXCL3, CXCL1, TNF, and CXCL8 (IL-8) during the late inflammatory stage. Additionally, neutrophils were recruited by epithelial cells through interactions between chemokines (CXCL8, CXCL6, CXCL5, CXCL3, and CXCL1) and their receptors (CXCR2 and CXCR1), which were increased during the late inflammatory stage (Figure 3).
5 Summary and future perspectives
The integration of scRNA-seq technology has been used recently in a wide range of ocular tissues, including iris (Gautam et al., 2021), ciliary body (Youkilis and Bassnett, 2021), lens (Giannone et al., 2023), uvea (Zhang X. et al., 2022), choroid (Tong et al., 2024), retina (Mao et al., 2023), as well as cornea, conjunctiva, lacrimal gland, and lacrimal sac mentioned earlier. With the advancement of this technology, our understanding of the physiological mechanisms of ocular cells and the molecular pathology of ocular diseases will improve. This is due to the ability of scRNA-seq to identify not only key and typical biomarker genes of ocular cell types but also new and promising biomarker genes. Additionally, these studies have reported both common and rare cell states across different cell types and identified differences in gene expression among them.
Because the maintenance of ocular surface homeostasis is indispensable for clear vision, it is particularly important to characterize the normal functions of ocular surface cells and their alterations in disease at single-cell resolution. These cells are constantly exposed to the external environment and therefore highly susceptible to environmental insults, especially under extreme living or working conditions. Given this background, we comprehensively report the current application of scRNA-seq to the ocular surface, including the molecular identification of normal ocular surface cells and the molecular changes of these cells in disease states.
However, we have noted that similar scRNA-seq studies may present different insights about the same cells. This inconsistency may originate from various parts of the scRNA-seq procedures, including single-cell isolation and capture, cell lysis, reverse transcription, cDNA amplification, and sequencing library preparation. Another important source of variability among ocular surface scRNA-seq studies is the choice of sequencing platform. Although droplet-based methods such as 10x Genomics Chromium, Drop-seq, and inDrop share a similar overall workflow, their performances differ considerably in sensitivity, capture efficiency, and technical noise. Systematic benchmarking demonstrated that 10x Genomics offers the highest mRNA detection sensitivity and lower dropout rates, which is particularly advantageous when profiling rare or fragile cell types such as limbal stem cells or goblet cells, albeit at a higher per-cell cost (Zhang et al., 2019; Yamawaki et al., 2021). Drop-seq and inDrop provide more cost-effective solutions with scalable throughput, but suffer from reduced capture efficiency and increased barcode errors, potentially limiting their utility for small ocular tissues with low RNA content. Other platforms, such as ICELL8 or ddSEQ, can achieve high library efficiency or reduced multiplet rates, but generally detect fewer transcripts per cell compared to the latest 10x chemistries (Zhang et al., 2019; Yamawaki et al., 2021). For ocular surface applications, where sample quantity is often limiting and the preservation of delicate subpopulations is essential, the balance between sensitivity, throughput, and cost should be carefully considered when selecting a platform.
The importance of single-cell isolation and capture as the initial steps in scRNA-seq is self-evident. Therefore, we previously analysed the current main methods and proposed an optimized single-cell suspension for preparing mouse corneas based on previous attempts to ensure the high activity of cells (Liu et al., 2023). Arts et al. (2023) aimed to develop a comprehensive approach beginning with the final sequencing library, focusing on constructing a corneal meta-atlas by integrating all publicly available human corneal scRNA-seq datasets. This would provide a comprehensive biomarker gene and cell state reference, albeit at the cost of significant data storage and human maintenance. Furthermore, it is important to recognize that cell type identification in scRNA-seq studies inherently relies on clustering algorithms and the use of known biomarker genes, which may introduce classification biases (Chen et al., 2024). In many cases, the observed clusters might represent transitional cell states, stress-induced artifacts, or technical noise rather than distinct biological cell types (Livne and Efroni, 2024; Denisenko et al., 2020). The absence of universally specific biomarkers for certain populations further complicates accurate classification. Additionally, most studies infer cell identities solely based on transcriptomic similarity without further validation at the protein or functional level, increasing the risk of misinterpretation. This issue becomes particularly critical when analyzing rare cell types or disease-related subpopulations, where technical variation or batch effects may distort biological conclusions (Su et al., 2022). Another challenge limiting the use of scRNA-seq on the ocular surface is the relatively high cost. An entire scRNA-seq process could cost between $2,000 and $3,000 (Ali, 2023), depending on the supplier’s equipment and technical expertise, as well as the level of economic development in the location where it is performed. This financial barrier has significantly impacted the development of RNA-seq in the field of ocular surface research, reducing the number of related publications. Additionally, the complexity of data analysis requires researchers to invest substantial effort, highlighting the need for user-friendly analysis software.
Beyond the general challenges of scRNA-seq, several unique limitations arise when applying this technology specifically to ocular surface tissues. The ocular surface is anatomically thin, highly specialized, and composed of heterogeneous cell populations distributed across distinct regions such as the cornea, limbus, conjunctiva, and lacrimal glands. Due to the limited amount of biological material, particularly in small structures like the human limbus or conjunctiva, achieving sufficient cell numbers for robust single-cell analysis can be technically challenging. Additionally, the isolation of ocular surface cells generally requires enzymatic dissociation, for instance, the optimized corneal protocol (Liu et al., 2023), or alternative approaches such as magnetic bead-activated cell sorting and post-sort culture optimization (Vattulainen et al., 2024). Nevertheless, these methods may still perturb cellular states or activate stress-responsive genes, thereby complicating downstream transcriptomic analyses. Some fragile cell types, including goblet cells or rare limbal progenitors, are especially prone to loss during dissociation or under-representation in sequencing outputs. Another concern is that most of the newly proposed biomarkers, for example, SOX17+/TSPAN7+ for LSCs, are supported solely by transcriptomic evidence and lack validation through in vitro assays. In addition, recently identified disease-associated biomarkers (CTSD and CTSK) may have limited relevance for clinical diagnosis, as only one publication has reported them as biomarkers for keratoconus, and no follow-up studies have been published in the past 3 years; therefore, further comparative analyses against established clinical indicators are warranted. Furthermore, the ocular surface exhibits pronounced spatial heterogeneity-cells of the central cornea, peripheral limbus, and conjunctiva perform distinct functions but may express overlapping biomarkers, complicating cell identity assignment when spatial context is lost. This spatial limitation is further compounded by the absence of positional information in most scRNA-seq datasets, which hinders the ability to distinguish true cell type differences from regional expression variability.
Future integration of spatial transcriptomics, multi-omics approaches, and advanced computational deconvolution methods may help address these issues (Du J. et al., 2023), enabling more precise characterization of ocular surface cell types and states. Spatial transcriptomics complements scRNA-seq by preserving the positional context of cells within intact tissue, thereby enabling the distinction between transcriptional variation driven by intrinsic cell-type heterogeneity and that shaped by the surrounding microenvironment (Longo et al., 2021). In ocular surface research, this is particularly critical for distinguishing limbal stem cells from morphologically similar epithelial progenitors and for mapping immune cell infiltration patterns in disease states. Furthermore, multi-omics approaches, which integrate genomic, transcriptomic, epigenomic, proteomic, and metabolomic profiles at the single-cell level, offer a holistic view of cellular identity and function (Fan et al., 2025). Such integration can validate transcriptome-derived cell-type classifications by linking them to protein expression or metabolic activity, thereby reducing misclassification caused by reliance on RNA biomarkers alone.
In addition to these integrative approaches, a major technical confounder in ocular surface scRNA-seq might be dissociation-induced stress, which rapidly triggers immediate-early response genes (such as Fos, Jun, Arc) (Denisenko et al., 2020; Machado et al., 2021; Lacar et al., 2016), heat-shock proteins (Denisenko et al., 2020; Neuschulz et al., 2023), and inflammatory cytokines (O'Flanagan et al., 2019), potentially masking native transcriptional states. Mitigation strategies include minimizing enzymatic digestion time, performing low-temperature dissociation with cold-active proteases, applying in situ fixation or transcriptional inhibitors, and optimizing mechanical handling to reduce cellular activation (Machado et al., 2021). One effective approach to circumvent dissociation-induced artifacts is to bypass whole-cell isolation altogether. For delicate and low-yield ocular tissues, single-nucleus RNA-seq can avoid enzymatic and mechanical dissociation and better preserve native signatures (Machado et al., 2021); however, it has trade-offs such as lower transcript capture, loss of cytoplasmic RNAs, and underrepresentation of certain cell types (Bakken et al., 2018). The choice between scRNA-seq and snRNA-seq should be guided by tissue fragility, target cell abundance, and the resolution required for the research question.
When these challenges are alleviated by future technologies, it will be possible to report in detail on more common ocular surface diseases affecting visual acuity that are currently underreported. Examples include various ocular surface diseases, such as corneal neovascularization after surgery or associated simultaneously with other ocular surface diseases, and meibomian gland dysfunction affecting ocular surface comfort and tear film stability (Wang et al., 2016). In addition, the growing repertoire of validated cell-type-specific and disease-associated biomarkers identified through scRNA-seq might hold substantial clinical translational potential, for example, the ACKR1+ endothelial cells in pterygium (Zhang X. et al., 2024) could point toward novel therapeutic targets. Such biomarkers could facilitate early diagnosis of ocular surface disorders, especially as short-term symptoms may lead to transcriptome changes in dry eye disease (Alam et al., 2024), enable more precise patient stratification, and inform targeted therapeutic strategies, including cell-based regenerative approaches and molecular interventions aimed at restoring ocular surface function.
6 Method of literature search
A comprehensive search was performed in the PubMed and Web of Science databases (up to September 2024), without limitations on publication date or type. Articles not published in English or lacking peer review were excluded. This narrative review incorporated a range of relevant keywords and phrases, including but not limited to: “single cell sequencing,” “single-cell sequencing,” “ocular surface,” “cornea,” “conjunctiva,” “meibomian glands,” “lacrimal gland,” “keratoconus,” and “pterygium.”
Author contributions
SO: Funding acquisition, Writing – review and editing, Supervision, Writing – original draft, Project administration, Methodology. MC: Conceptualization, Writing – review and editing, Writing – original draft, Data curation. YF: Writing – review and editing, Software, Writing – original draft, Visualization. SL: Investigation, Writing – review and editing. XZ: Writing – review and editing. SZ: Writing – review and editing. HG: Project administration, Writing – review and editing, Funding acquisition, Supervision. YW: Writing – original draft, Funding acquisition, Writing – review and editing, Supervision.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by the Guizhou Provincial Basic Research Program (QKHJC-ZK[2024]ZD043 and QKHJC-ZK[2025]MS473), Medical Research Union Fund for High-quality health development of Guizhou Province (2024GZYKYJJKM0043), Fujian Provincial Science Fund for Distinguished Young Scholars (2023J06053), the Natural Science Foundation of China (82101084), National Natural Science Foundation of China Cultivation Project of Guizhou Medical University (gyfynsfc[2024]-01) and China Scholarship Council (202306310049).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
aLSCs, actively renewing limbal stem cells; DCs, Dendritic cells; FECD, Fuchs endothelial corneal dystrophy; lncRNA, long noncoding RNA; LPCs, limbal progenitor cells; LSCs, limbal stem cells; oGVHD, ocular graft-versus-host disease; PANDO, primary acquired nasolacrimal duct obstruction; PCR, polymerase chain reaction; PMCs, postmitotic cells; qLSCs, quiescent limbal stem cells; scRNA-seq, single-cell RNA sequencing; TA, transient amplifying; TACs, transient amplifying cells; TDCs, terminally differentiated cells; Treg, regulatory T cells.
References
Ahsanuddin, S., and Wu, A. Y. (2023). Single-cell transcriptomics of the ocular anterior segment: a comprehensive review. Eye (Lond) 37 (16), 3334–3350. doi:10.1038/s41433-023-02539-3
Alam, J., Yazdanpanah, G., Ratnapriya, R., Borcherding, N., de Paiva, C. S., Li, D., et al. (2022). Il-17 producing lymphocytes cause dry eye and corneal disease with aging in Rxrα mutant mouse. Front. Med. (Lausanne) 9, 849990. doi:10.3389/fmed.2022.849990
Alam, J., Yaman, E., Silva, G. C. V., Chen, R., de Paiva, C. S., Stepp, M. A., et al. (2024). Single cell analysis of short-term dry eye induced changes in cornea immune cell populations. Front. Med. (Lausanne) 11, 1362336. doi:10.3389/fmed.2024.1362336
Ali, M. J. (2023). Etiopathogenesis of primary acquired nasolacrimal duct obstruction (Pando). Prog. Retin Eye Res. 96, 101193. doi:10.1016/j.preteyeres.2023.101193
Altshuler, A., Amitai-Lange, A., Tarazi, N., Dey, S., Strinkovsky, L., Hadad-Porat, S., et al. (2021). Discrete limbal epithelial stem cell populations mediate corneal homeostasis and wound healing. Cell Stem Cell 28 (7), 1248–61.e8. doi:10.1016/j.stem.2021.04.003
Amin, M. T., Coussement, L., and De Meyer, T. (2024). Characterization of loss-of-imprinting in breast cancer at the cellular level by integrating single-cell full-length transcriptome with bulk rna-seq data. Biomolecules 14 (12), 1598. doi:10.3390/biom14121598
Arts, J. A., Laberthonniere, C., Lima Cunha, D., and Zhou, H. (2023). Single-cell rna sequencing: opportunities and challenges for studies on corneal biology in health and disease. Cells 12 (13), 1808. doi:10.3390/cells12131808
Bakken, T. E., Hodge, R. D., Miller, J. A., Yao, Z., Nguyen, T. N., Aevermann, B., et al. (2018). Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PLoS One 13 (12), e0209648. doi:10.1371/journal.pone.0209648
Bannier-Hélaouët, M., Post, Y., Korving, J., Trani Bustos, M., Gehart, H., Begthel, H., et al. (2021). Exploring the human lacrimal gland using organoids and single-cell sequencing. Cell Stem Cell 28 (7), 1221–32.e7. doi:10.1016/j.stem.2021.02.024
Bonelli, F., Lasagni Vitar, R. M., Merlo Pich, F. G., Fonteyne, P., Rama, P., Mondino, A., et al. (2022). Corneal endothelial cell reduction and increased Neurokinin-1 receptor expression in a graft-versus-host disease preclinical model. Exp. Eye Res. 220, 109128. doi:10.1016/j.exer.2022.109128
Borner, K., Teichmann, S. A., Quardokus, E. M., Gee, J. C., Browne, K., Osumi-Sutherland, D., et al. (2021). Anatomical structures, cell types and biomarkers of the human reference atlas. Nat. Cell Biol. 23 (11), 1117–1128. doi:10.1038/s41556-021-00788-6
Butovich, I. A., and Wilkerson, A. (2022). Dynamic changes in the gene expression patterns and lipid profiles in the developing and maturing Meibomian glands. Int. J. Mol. Sci. 23 (14), 7884. doi:10.3390/ijms23147884
Català, P., Groen, N., Dehnen, J. A., Soares, E., van Velthoven, A. J. H., Nuijts, R., et al. (2021). Single cell transcriptomics reveals the heterogeneity of the human cornea to identify novel markers of the limbus and stroma. Sci. Rep. 11 (1), 21727. doi:10.1038/s41598-021-01015-w
Català, P., Groen, N., LaPointe, V. L. S., and Dickman, M. M. (2023). A single-cell rna-seq analysis unravels the heterogeneity of primary cultured human corneal endothelial cells. Sci. Rep. 13 (1), 9361. doi:10.1038/s41598-023-36567-6
Chen, J., Li, Z., Zhang, L., Ou, S., Wang, Y., He, X., et al. (2017). Descemet's membrane supports corneal endothelial cell regeneration in rabbits. Sci. Rep. 7 (1), 6983. doi:10.1038/s41598-017-07557-2
Chen, Z., Wang, C., Huang, S., Shi, Y., and Xi, R. (2024). Directly selecting cell-type marker genes for single-cell clustering analyses. Cell Rep. Methods 4 (7), 100810. doi:10.1016/j.crmeth.2024.100810
Cher, I. (2014). Ocular surface concepts: development and citation. Ocul. Surf. 12 (1), 10–13. doi:10.1016/j.jtos.2013.10.004
Collin, J., Queen, R., Zerti, D., Bojic, S., Dorgau, B., Moyse, N., et al. (2021). A single cell atlas of human cornea that defines its development, limbal progenitor cells and their interactions with the immune cells. Ocul. Surf. 21, 279–298. doi:10.1016/j.jtos.2021.03.010
Craig, J. P., Nichols, K. K., Akpek, E. K., Caffery, B., Dua, H. S., Joo, C. K., et al. (2017). Tfos dews Ii definition and classification report. Ocul. Surf. 15 (3), 276–283. doi:10.1016/j.jtos.2017.05.008
Davanger, M., and Evensen, A. (1971). Role of the pericorneal papillary structure in renewal of corneal epithelium. Nature 229 (5286), 560–561. doi:10.1038/229560a0
Dean, C., Ito, M., Makarenkova, H. P., Faber, S. C., and Lang, R. A. (2004). Bmp7 regulates branching morphogenesis of the lacrimal gland by promoting mesenchymal proliferation and condensation. Development 131 (17), 4155–4165. doi:10.1242/dev.01285
den Braanker, H., van Stigt, A. C., Kok, M. R., Lubberts, E., and Bisoendial, R. J. (2021). Single-cell rna sequencing reveals heterogeneity and functional diversity of lymphatic endothelial cells. Int. J. Mol. Sci. 22 (21), 11976. doi:10.3390/ijms222111976
Denisenko, E., Guo, B. B., Jones, M., Hou, R., de Kock, L., Lassmann, T., et al. (2020). Systematic assessment of tissue dissociation and storage biases in single-cell and single-nucleus rna-seq workflows. Genome Biol. 21 (1), 130. doi:10.1186/s13059-020-02048-6
Dong, N., Li, W., Lin, H., Wu, H., Li, C., Chen, W., et al. (2009). Abnormal epithelial differentiation and tear film alteration in pinguecula. Invest. Ophthalmol. Vis. Sci. 50 (6), 2710–2715. doi:10.1167/iovs.08-2905
Dong, X. X., Liu, K. F., Zhou, M., Liang, G., and Pan, C. W. (2022). Diabetes Mellitus and keratoconus: a systematic review and meta-analysis. Cornea 41 (11), 1398–1404. doi:10.1097/ICO.0000000000002876
Dou, S., Wang, Q., Qi, X., Zhang, B., Jiang, H., Chen, S., et al. (2021). Molecular identity of human limbal heterogeneity involved in corneal homeostasis and privilege. Ocul. Surf. 21, 206–220. doi:10.1016/j.jtos.2021.04.010
Dou, S., Wang, Q., Zhang, B., Wei, C., Wang, H., Liu, T., et al. (2022). Single-Cell atlas of keratoconus corneas revealed aberrant transcriptional signatures and implicated mechanical stretch as a trigger for keratoconus pathogenesis. Cell Discov. 8 (1), 66. doi:10.1038/s41421-022-00397-z
Du, H., Li, S., Lu, J., Tang, L., Jiang, X., He, X., et al. (2023a). Single-cell Rna-Seq and bulk-seq identify Rab17 as a potential regulator of angiogenesis by human dermal microvascular endothelial cells in diabetic foot ulcers. Burns Trauma 11, tkad020. doi:10.1093/burnst/tkad020
Du, J., Yang, Y. C., An, Z. J., Zhang, M. H., Fu, X. H., Huang, Z. F., et al. (2023b). Advances in spatial transcriptomics and related data analysis strategies. J. Transl. Med. 21 (1), 330. doi:10.1186/s12967-023-04150-2
Dumas, S. J., Meta, E., Borri, M., Goveia, J., Rohlenova, K., Conchinha, N. V., et al. (2020). Single-cell rna sequencing reveals renal endothelium heterogeneity and metabolic adaptation to water deprivation. J. Am. Soc. Nephrol. 31 (1), 118–138. doi:10.1681/asn.2019080832
Ebihara, T., Matsubara, T., Togami, Y., Matsumoto, H., Tachino, J., Matsuura, H., et al. (2023). Combination of Wfdc2, Chi3l1, and Krt19 in plasma defines a clinically useful molecular phenotype associated with prognosis in critically ill Covid-19 patients. J. Clin. Immunol. 43 (2), 286–298. doi:10.1007/s10875-022-01386-3
Fan, Q., Yan, R., Li, Y., Lu, L., Liu, J., Li, S., et al. (2024). Exploring immune cell diversity in the lacrimal glands of healthy mice: a single-cell Rna-Sequencing atlas. Int. J. Mol. Sci. 25 (2), 1208. doi:10.3390/ijms25021208
Fan, B. L., Chen, L. H., Chen, L. L., and Guo, H. (2025). Integrative multi-omics approaches for identifying and characterizing biological elements in crop traits: current progress and future prospects. Int. J. Mol. Sci. 26 (4), 1466. doi:10.3390/ijms26041466
Farmer, D. T., Nathan, S., Finley, J. K., Shengyang Yu, K., Emmerson, E., Byrnes, L. E., et al. (2017). Defining epithelial cell dynamics and lineage relationships in the developing lacrimal gland. Development 144 (13), 2517–2528. doi:10.1242/dev.150789
Gautam, P., Hamashima, K., Chen, Y., Zeng, Y., Makovoz, B., Parikh, B. H., et al. (2021). Multi-species single-cell transcriptomic analysis of ocular compartment regulons. Nat. Commun. 12 (1), 5675. doi:10.1038/s41467-021-25968-8
Giannone, A. A., Sellitto, C., Rosati, B., McKinnon, D., and White, T. W. (2023). Single-cell rna sequencing analysis of the early postnatal mouse lens epithelium. Invest. Ophthalmol. Vis. Sci. 64 (13), 37. doi:10.1167/iovs.64.13.37
Han, Y., You, X., Xing, W., Zhang, Z., and Zou, W. (2018). Paracrine and endocrine actions of bone-the functions of secretory proteins from osteoblasts, osteocytes, and osteoclasts. Bone Res. 6, 16. doi:10.1038/s41413-018-0019-6
Hayashi, R., Okubo, T., Kudo, Y., Ishikawa, Y., Imaizumi, T., Suzuki, K., et al. (2022). Generation of 3d lacrimal gland organoids from human pluripotent stem cells. Nature 605 (7908), 126–131. doi:10.1038/s41586-022-04613-4
He, J., Zheng, F., Zhang, L., Cai, J., Ogawa, Y., Tsubota, K., et al. (2024). Single-cell Rna-Sequencing reveals the transcriptional landscape of lacrimal gland in gvhd mouse model. Ocul. Surf. 33, 50–63. doi:10.1016/j.jtos.2024.04.006
Hedlund, E., and Deng, Q. (2018). Single-cell Rna sequencing: technical advancements and biological applications. Mol. Asp. Med. 59, 36–46. doi:10.1016/j.mam.2017.07.003
Hertsenberg, A. J., and Funderburgh, J. L. (2015). Stem cells in the cornea. Prog. Mol. Biol. Transl. Sci. 134, 25–41. doi:10.1016/bs.pmbts.2015.04.002
Hessen, M., and Akpek, E. K. (2014). Dry eye: an inflammatory ocular disease. J. Ophthalmic Vis. Res. 9 (2), 240–250.
Huang, D., Jiao, X., Huang, S., Liu, J., Si, H., Qi, D., et al. (2024). Analysis of the heterogeneity and complexity of murine extraorbital lacrimal gland via single-cell rna sequencing. Ocul. Surf. 34, 60–95. doi:10.1016/j.jtos.2024.06.005
Jia, M., Zhu, S., Xue, M. Y., Chen, H., Xu, J., Song, M., et al. (2024). Single-cell transcriptomics across 2,534 microbial species reveals functional heterogeneity in the rumen microbiome. Nat. Microbiol. 9 (7), 1884–1898. doi:10.1038/s41564-024-01723-9
Jovic, D., Liang, X., Zeng, H., Lin, L., Xu, F., and Luo, Y. (2022). Single-cell rna sequencing technologies and applications: a brief overview. Clin. Transl. Med. 12 (3), e694. doi:10.1002/ctm2.694
Jukic, A., Bakiri, L., Wagner, E. F., Tilg, H., and Adolph, T. E. (2021). Calprotectin: from biomarker to biological function. Gut 70 (10), 1978–1988. doi:10.1136/gutjnl-2021-324855
Junker, J. P., and van Oudenaarden, A. (2015). Single-cell transcriptomics enters the age of mass production. Mol. Cell 58 (4), 563–564. doi:10.1016/j.molcel.2015.05.019
Kai, J. Y., Wu, Y. B., Shi, B., Li, D. L., Dong, X. X., Wang, P., et al. (2024). Dry eye symptoms and health-related quality of life among Chinese individuals: a national-based study. Br. J. Ophthalmol. 108, 1500–1507. doi:10.1136/bjo-2023-324677
Kalucka, J., de Rooij, L., Goveia, J., Rohlenova, K., Dumas, S. J., Meta, E., et al. (2020). Single-cell transcriptome atlas of murine endothelial cells. Cell 180 (4), 764–779. doi:10.1016/j.cell.2020.01.015
Kaplan, N., Wang, J., Wray, B., Patel, P., Yang, W., Peng, H., et al. (2019). Single-cell rna transcriptome helps define the limbal/corneal epithelial stem/early transit amplifying cells and how autophagy affects this population. Invest. Ophthalmol. Vis. Sci. 60 (10), 3570–3583. doi:10.1167/iovs.19-27656
Kawakita, T., Shimmura, S., Hornia, A., Higa, K., and Tseng, S. C. (2008). Stratified epithelial sheets engineered from a single adult murine corneal/limbal progenitor cell. J. Cell Mol. Med. 12 (4), 1303–1316. doi:10.1111/j.1582-4934.2008.00297.x
Kinoshita, S., Koizumi, N., Ueno, M., Okumura, N., Imai, K., Tanaka, H., et al. (2018). Injection of cultured cells with a rock inhibitor for bullous keratopathy. N. Engl. J. Med. 378 (11), 995–1003. doi:10.1056/NEJMoa1712770
Kleino, I., Frolovaitė, P., Suomi, T., and Elo, L. L. (2022). Computational solutions for spatial transcriptomics. Comput. Struct. Biotechnol. J. 20, 4870–4884. doi:10.1016/j.csbj.2022.08.043
Kolodziejczyk, A. A., Kim, J. K., Svensson, V., Marioni, J. C., and Teichmann, S. A. (2015). The technology and biology of single-cell rna sequencing. Mol. Cell 58 (4), 610–620. doi:10.1016/j.molcel.2015.04.005
Lacar, B., Linker, S. B., Jaeger, B. N., Krishnaswami, S. R., Barron, J. J., Kelder, M. J. E., et al. (2016). Nuclear Rna-Seq of single neurons reveals molecular signatures of activation. Nat. Commun. 7, 11022. doi:10.1038/ncomms11022
Lai, Q., Hu, L., Zhang, W., Jiang, Z., Zeng, C., and Hu, J. (2023). Single-Cell rna sequencing highlights the regulatory role of T cell marker genes Ctla4, Ccl5 and Tcf7 in corneal allograft rejection of mouse model. Int. Immunopharmacol. 117, 109911. doi:10.1016/j.intimp.2023.109911
Li, D. Q., Kim, S., Li, J. M., Gao, Q., Choi, J., Bian, F., et al. (2021a). Single-Cell transcriptomics identifies limbal stem cell population and cell types mapping its differentiation trajectory in limbal basal epithelium of human cornea. Ocul. Surf. 20, 20–32. doi:10.1016/j.jtos.2020.12.004
Li, J. M., Kim, S., Zhang, Y., Bian, F., Hu, J., Lu, R., et al. (2021b). Single-cell transcriptomics identifies a unique entity and signature markers of transit-amplifying cells in human corneal limbus. Invest. Ophthalmol. Vis. Sci. 62 (9), 36. doi:10.1167/iovs.62.9.36
Li, Y., Ou, S., Lin, S., Qian, H., Zhao, Z., Zhang, M., et al. (2022). Meibomian gland alteration in patients with systemic lupus erythematosus. Lupus 31 (4), 407–414. doi:10.1177/09612033221079760
Li, S., Zhang, P., Li, A., Bao, J., Pan, Z., and Jie, Y. (2024). Rho-kinase inhibitor alleviates Cd4(+)T cell mediated corneal graft rejection by modulating its Stat3 and Stat5 activation. Exp. Eye Res. 242, 109857. doi:10.1016/j.exer.2024.109857
Ligocki, A. J., Fury, W., Gutierrez, C., Adler, C., Yang, T., Ni, M., et al. (2021). Molecular characteristics and spatial distribution of adult human corneal cell subtypes. Sci. Rep. 11 (1), 16323. doi:10.1038/s41598-021-94933-8
Lin, Y., Zhang, Y., Shi, K., Wu, H., and Ou, S. (2023a). Advances in clinical examination of lacrimal gland. Front. Med. (Lausanne) 10, 1257209. doi:10.3389/fmed.2023.1257209
Lin, S., Cai, M., Zhang, L., Mao, Y., Wu, H., Liu, X., et al. (2023b). Limbal stem cell dysfunction induced by severe dry eye via activation of the P38 mapk signaling pathway. Am. J. Pathol. 193 (11), 1863–1878. doi:10.1016/j.ajpath.2023.08.003
Lin, J. B., Shen, X., Pfeifer, C. W., Shiau, F., Santeford, A., Ruzycki, P. A., et al. (2023c). Dry eye disease in mice activates adaptive corneal epithelial regeneration distinct from constitutive renewal in homeostasis. Proc. Natl. Acad. Sci. U. S. A. 120 (2), e2204134120. doi:10.1073/pnas.2204134120
Liu, Y., Zhao, R., Chi, S., Zhang, W., Xiao, C., Zhou, X., et al. (2020). Ube2c is upregulated by estrogen and promotes epithelial-mesenchymal transition via P53 in endometrial cancer. Mol. Cancer Res. 18 (2), 204–215. doi:10.1158/1541-7786.MCR-19-0561
Liu, X., Zhang, S., Mao, Y., Lin, S., Wu, H., and Ou, S. (2023). Optimization of method for achieving a single-cell suspension from mouse corneas. Exp. Eye Res. 233, 109544. doi:10.1016/j.exer.2023.109544
Liu, Z., Xie, H., Li, L., Jiang, D., Qian, Y., Zhu, X., et al. (2024). Single-cell landscape reveals the epithelial cell-centric pro-inflammatory immune microenvironment in dry eye development. Mucosal Immunol. 17 (3), 491–507. doi:10.1016/j.mucimm.2023.11.008
Livne, D., and Efroni, S. (2024). Pathway metrics accurately stratify T cells to their cells states. BioData Min. 17 (1), 60. doi:10.1186/s13040-024-00416-7
Long, Q., Huang, C., Zhang, L., Jiang, H., Zhao, S., Zhang, L., et al. (2024). A novel tissue-engineered corneal epithelium based on ultra-thin amniotic membrane and mesenchymal stem cells. Sci. Rep. 14 (1), 17407. doi:10.1038/s41598-024-68219-8
Longo, S. K., Guo, M. G., Ji, A. L., and Khavari, P. A. (2021). Integrating single-cell and spatial transcriptomics to elucidate intercellular tissue dynamics. Nat. Rev. Genet. 22 (10), 627–644. doi:10.1038/s41576-021-00370-8
Lu, Z. J., Ye, J. G., Li, J. N., Liang, J. B., Zhou, M., Hu, Q. L., et al. (2025). Single-cell multiomics analysis of early wound response programs in the mouse corneal epithelium. Invest. Ophthalmol. Vis. Sci. 66 (3), 9. doi:10.1167/iovs.66.3.9
Ma, B., Zhou, Y., Hu, Y., Duan, H., Sun, Z., Wang, P., et al. (2023). Mapping resident immune cells in the murine ocular surface and lacrimal gland by flow cytometry. Ocular Immunol. Inflamm. 31 (4), 748–759. doi:10.1080/09273948.2023.2182327
Machado, L., Relaix, F., and Mourikis, P. (2021). Stress relief: emerging methods to mitigate dissociation-induced artefacts. Trends Cell Biol. 31 (11), 888–897. doi:10.1016/j.tcb.2021.05.004
Maiti, G., Monteiro de Barros, M. R., Hu, N., Dolgalev, I., Roshan, M., Foster, J. W., et al. (2022). Single cell rna-seq of human cornea organoids identifies cell fates of a developing immature cornea. PNAS Nexus 1 (5), pgac246. doi:10.1093/pnasnexus/pgac246
Mao, P., Shen, Y., Mao, X., Liu, K., and Zhong, J. (2023). The single-cell landscape of alternative transcription start sites of diabetic retina. Exp. Eye Res. 233, 109520. doi:10.1016/j.exer.2023.109520
Middleton, J., Americh, L., Gayon, R., Julien, D., Mansat, M., Mansat, P., et al. (2005). A Comparative Study of Endothelial Cell Markers Expressed in Chronically Inflamed Human Tissues: meca-79, Duffy Antigen Receptor for Chemokines, Von Willebrand Factor, Cd31, Cd34, Cd105 and Cd146. J. Pathol. 206 (3), 260–268. doi:10.1002/path.1788
Mirmoeini, K., Tajdaran, K., Zhang, J., Gordon, T., Ali, A., Kaplan, D. R., et al. (2023). Schwann cells are key regulators of corneal epithelial renewal. Invest. Ophthalmol. Vis. Sci. 64 (4), 7. doi:10.1167/iovs.64.4.7
Natesan, S., Wrice, N. L., and Christy, R. J. (2019). Peroxisome proliferator-activated receptor-alpha agonist and all-trans retinoic acid induce epithelial differentiation of subcutaneous adipose-derived stem cells from debrided burn skin. J. Cell Biochem. 120 (6), 9213–9229. doi:10.1002/jcb.28197
Neuschulz, A., Bakina, O., Badillo-Lisakowski, V., Olivares-Chauvet, P., Conrad, T., Gotthardt, M., et al. (2023). A single-cell rna labeling strategy for measuring stress response upon tissue dissociation. Mol. Syst. Biol. 19 (2), e11147. doi:10.15252/msb.202211147
O'Flanagan, C. H., Campbell, K. R., Zhang, A. W., Kabeer, F., Lim, J. L. P., Biele, J., et al. (2019). Dissociation of solid tumor tissues with cold active protease for single-cell rna-seq minimizes conserved collagenase-associated stress responses. Genome Biol. 20 (1), 210. doi:10.1186/s13059-019-1830-0
Ou, S. K., Jeyalatha, M. V., Mao, Y., Wang, J. Q., Chen, C., Zhang, M. J., et al. (2022). The role of ectodysplasin a on the ocular surface homeostasis. Int. J. Mol. Sci. 23 (24), 15700. doi:10.3390/ijms232415700
Papalexi, E., and Satija, R. (2018). Single-cell rna sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol. 18 (1), 35–45. doi:10.1038/nri.2017.76
Peh, G. S., Chng, Z., Ang, H. P., Cheng, T. Y., Adnan, K., Seah, X. Y., et al. (2015). Propagation of human corneal endothelial cells: a novel dual media approach. Cell Transpl. 24 (2), 287–304. doi:10.3727/096368913x675719
Pflugfelder, S. C., and Stern, M. E. (2020). Biological functions of tear film. Exp. Eye Res. 197, 108115. doi:10.1016/j.exer.2020.108115
Polisetti, N., Sharaf, L., Schlotzer-Schrehardt, U., Schlunck, G., and Reinhard, T. (2022). Efficient isolation and functional characterization of niche cells from human corneal limbus. Int. J. Mol. Sci. 23 (5), 2750. doi:10.3390/ijms23052750
Qu, M., Qi, X., Wang, Q., Wan, L., Li, J., Li, W., et al. (2019). Therapeutic effects of Stat3 inhibition on experimental murine dry eye. Invest. Ophthalmol. Vis. Sci. 60 (12), 3776–3785. doi:10.1167/iovs.19-26928
Roodnat, A. W., Callaghan, B., Doyle, C., Henry, M., Goljanek-Whysall, K., Simpson, D. A., et al. (2022). Genome-wide RNA sequencing of human trabecular meshwork cells treated with TGF-β1: relevance to pseudoexfoliation glaucoma. Biomolecules 12 (11), 1693. doi:10.3390/biom12111693
Rudra, D. S., Pal, U., Chowdhury, N., Maiti, N. C., Bagchi, A., and Swarnakar, S. (2022). Omeprazole prevents stress induced gastric ulcer by direct inhibition of Mmp-2/Timp-3 interactions. Free Radic. Biol. Med. 181, 221–234. doi:10.1016/j.freeradbiomed.2022.02.007
Schermer, A., Galvin, S., and Sun, T. T. (1986). Differentiation-related expression of a major 64k corneal keratin in vivo and in culture suggests limbal location of corneal epithelial stem cells. J. Cell Biol. 103 (1), 49–62. doi:10.1083/jcb.103.1.49
Shi, Z. X., Chen, Z. C., Zhong, J. Y., Hu, K. H., Zheng, Y. F., Chen, Y., et al. (2023). High-throughput and high-accuracy single-cell rna isoform analysis using pacbio circular consensus sequencing. Nat. Commun. 14 (1), 2631. doi:10.1038/s41467-023-38324-9
Song, Z., Chen, B., Tsai, C. H., Wu, D., Liu, E., Hawkins, I. S., et al. (2022). Differentiation trajectory of limbal stem and progenitor cells under normal homeostasis and upon corneal wounding. Cells 11 (13), 1983. doi:10.3390/cells11131983
Sridhar, M. S. (2018). Anatomy of cornea and ocular surface. Indian J. Ophthalmol. 66 (2), 190–194. doi:10.4103/ijo.IJO_646_17
Su, M., Pan, T., Chen, Q. Z., Zhou, W. W., Gong, Y., Xu, G., et al. (2022). Data analysis guidelines for single-cell rna-seq in biomedical studies and clinical applications. Mil. Med. Res. 9 (1), 68. doi:10.1186/s40779-022-00434-8
Sun, D., Shi, W. Y., and Dou, S. Q. (2023). Single-cell rna sequencing in cornea research: insights into limbal stem cells and their niche regulation. World J. Stem Cells 15 (5), 466–475. doi:10.4252/wjsc.v15.i5.466
Sun, D., Zhang, X., Chen, R., Sang, T., Li, Y., Wang, Q., et al. (2024). Decoding cellular plasticity and niche regulation of limbal stem cells during corneal wound healing. Stem Cell Res. Ther. 15 (1), 201. doi:10.1186/s13287-024-03816-y
Sutherland, C., Wang, Y., Brown, R. V., Foley, J., Mahler, B., Janardhan, K. S., et al. (2018). Laser capture microdissection of highly pure trabecular meshwork from mouse eyes for gene expression analysis. J. Vis. Exp. JoVE (136), 57576. doi:10.3791/57576
Swamynathan, S. K., and Wells, A. (2020). Conjunctival goblet cells: ocular surface functions, disorders that affect them, and the potential for their regeneration. Ocul. Surf. 18 (1), 19–26. doi:10.1016/j.jtos.2019.11.005
Swarup, A., Phansalkar, R., Morri, M., Agarwal, A., Subramaniam, V., Li, B., et al. (2023). Single-cell transcriptomic analysis of corneal organoids during development. Stem Cell Rep. 18 (12), 2482–2497. doi:10.1016/j.stemcr.2023.10.022
Tang, F., Barbacioru, C., Wang, Y., Nordman, E., Lee, C., Xu, N., et al. (2009). Mrna-seq whole-transcriptome analysis of a single cell. Nat. Methods 6 (5), 377–382. doi:10.1038/nmeth.1315
Tang, X., Chen, C., Yan, S., Yang, A., Deng, Y., Chen, B., et al. (2024). Single-nucleus rna-seq reveals spermatogonial stem cell developmental pattern in shaziling pigs. Biomolecules 14 (6), 607. doi:10.3390/biom14060607
Thoft, R. A., and Friend, J. (1983). The X, Y, Z hypothesis of corneal epithelial maintenance. Invest. Ophthalmol. Vis. Sci. 24 (10), 1442–1443.
Tong, M., Bai, Y., Han, X., Kong, L., Ren, L., Zhang, L., et al. (2024). Single-cell profiling transcriptomic reveals cellular heterogeneity and cellular crosstalk in choroidal neovascularization model. Exp. Eye Res. 242, 109877. doi:10.1016/j.exer.2024.109877
van Zyl, T., Yan, W., McAdams, A. M., Monavarfeshani, A., Hageman, G. S., and Sanes, J. R. (2022). Cell atlas of the human ocular anterior segment: tissue-specific and shared cell types. Proc. Natl. Acad. Sci. U. S. A. 119 (29), e2200914119. doi:10.1073/pnas.2200914119
Vattulainen, M., Smits, J. G. A., Arts, J. A., Lima Cunha, D., Ilmarinen, T., Skottman, H., et al. (2024). Deciphering the heterogeneity of differentiating Hpsc-Derived corneal limbal stem cells through single-cell rna sequencing. Stem Cell Rep. 19 (7), 1010–1023. doi:10.1016/j.stemcr.2024.06.001
Voigt, A. P., Whitmore, S. S., Flamme-Wiese, M. J., Riker, M. J., Wiley, L. A., Tucker, B. A., et al. (2019). Molecular characterization of foveal versus peripheral human retina by single-cell rna sequencing. Exp. Eye Res. 184, 234–242. doi:10.1016/j.exer.2019.05.001
Wang, S., Zhao, H., Huang, C., Li, Z., Li, W., Zhang, X., et al. (2016). Impact of chronic smoking on Meibomian gland dysfunction. PLoS One 11 (12), e0168763. doi:10.1371/journal.pone.0168763
Wang, Q., Dou, S., Zhang, B., Jiang, H., Qi, X., Duan, H., et al. (2022). Heterogeneity of human corneal endothelium implicates lncrna Neat1 in fuchs endothelial corneal dystrophy. Mol. Ther. Nucleic acids 27, 880–893. doi:10.1016/j.omtn.2022.01.005
Weingeist, T. A. (1973). The conjunctiva. Int. Ophthalmol. Clin. 13 (3), 85–91. doi:10.1097/00004397-197301330-00008
Wu, Q., Ouyang, C., Xie, L., Ling, Y., and Huang, T. (2017). The rock inhibitor, thiazovivin, inhibits human corneal endothelial-to-mesenchymal transition/epithelial-To-mesenchymal transition and increases ionic transporter expression. Int. J. Mol. Med. 40 (4), 1009–1018. doi:10.3892/ijmm.2017.3103
Wu, Y. F., Chang, N. W., Chu, L. A., Liu, H. Y., Zhou, Y. X., Pai, Y. L., et al. (2023). Single-cell transcriptomics reveals cellular heterogeneity and complex cell-cell communication networks in the mouse cornea. Invest. Ophthalmol. Vis. Sci. 64 (13), 5. doi:10.1167/iovs.64.13.5
Xia, M., Li, Y., Liu, Y., Dong, Z., and Liu, H. (2025). Single-cell Rna-Sequencing analysis provides insights into iga nephropathy. Biomolecules 15 (2), 191. doi:10.3390/biom15020191
Yamawaki, T. M., Lu, D. R., Ellwanger, D. C., Bhatt, D., Manzanillo, P., Arias, V., et al. (2021). Systematic comparison of high-throughput single-cell rna-Seq methods for immune cell profiling. BMC Genomics 22 (1), 66. doi:10.1186/s12864-020-07358-4
Yang, Y., Huang, C., Lin, X., Wu, Y., Ouyang, W., Tang, L., et al. (2018). 0.005% preservative-free latanoprost induces dry eye-like ocular surface damage via promotion of inflammation in mice. Invest. Ophthalmol. Vis. Sci. 59 (8), 3375–3384. doi:10.1167/iovs.18-24013
Yang, G. N., Sun, Y. B. Y., Roberts, P. K., Moka, H., Sung, M. K., Gardner-Russell, J., et al. (2024). Exploring single-cell rna sequencing as a decision-making tool in the clinical management of fuchs' endothelial corneal dystrophy. Prog. Retin Eye Res. 102, 101286. doi:10.1016/j.preteyeres.2024.101286
Yao, W., Luo, D., Lv, Z., Yang, Y., Wang, L., Ma, B., et al. (2022). The Rabep1-Mediated endocytosis and activation of trypsinogen to promote pancreatic stellate cell activation. Biomolecules 12 (8), 1063. doi:10.3390/biom12081063
Yin, Y., Jiao, L., Wang, J., Song, J., Yang, Z., and Bi, H. (2025). Single-Cell rna sequencing of mouse cornea during wound healing after infrared laser irradiation. Exp. Eye Res. 260, 110615. doi:10.1016/j.exer.2025.110615
Youkilis, J. C., and Bassnett, S. (2021). Single-cell Rna-Sequencing analysis of the ciliary epithelium and contiguous tissues in the mouse eye. Exp. Eye Res. 213, 108811. doi:10.1016/j.exer.2021.108811
Zhang, X., Li, T., Liu, F., Chen, Y., Yao, J., Li, Z., et al. (2019). Comparative analysis of droplet-based ultra-high-throughput single-cell Rna-Seq systems. Mol. Cell 73 (1), 130–142. doi:10.1016/j.molcel.2018.10.020
Zhang, Y., Fang, X., Lin, Z., Xie, Z., Wu, H., and Ou, S. (2022a). Histopathology-based diagnosis of mooren's ulcer concealed beneath the pterygium on eye. J. Histotechnol. 45 (4), 195–201. doi:10.1080/01478885.2022.2137666
Zhang, X., Qiu, J., Huang, F., Han, P., Shan, K., and Zhang, C. (2022b). Construction and verification of a hypoxia-related nine-gene prognostic model in uveal melanoma based on integrated single-cell and bulk rna sequencing analyses. Exp. Eye Res. 223, 109214. doi:10.1016/j.exer.2022.109214
Zhang, J., Dai, Y., Li, Y., and Xu, J. (2023). Integrative analysis of gene expression datasets in corneal endothelium samples of fuchs endothelial corneal dystrophy. Exp. Eye Res. 237, 109712. doi:10.1016/j.exer.2023.109712
Zhang, H., Song, Q., Shang, K., Li, Y., Jiang, L., and Yang, L. (2024a). Tspan protein family: focusing on the occurrence, progression, and treatment of cancer. Cell Death Discov. 10 (1), 187. doi:10.1038/s41420-024-01961-0
Zhang, X., Han, P., Qiu, J., Huang, F., Luo, Q., Cheng, J., et al. (2024b). Single-cell rna sequencing reveals the complex cellular niche of pterygium. Ocul. Surf. 32, 91–103. doi:10.1016/j.jtos.2024.01.013
Zhang, W., Huang, H., Liu, X., Zhang, L., Li, L., Ding, Y., et al. (2024c). Scrna-seq: First atlas and cellular landscape of lacrimal sac: implications in primary acquired nasolacrimal duct obstruction pathogenesis. Invest. Ophthalmol. Vis. Sci. 65 (3), 38. doi:10.1167/iovs.65.3.38
Zhong, H., Chen, Q., Li, J., Shen, W., Sheng, X., Niu, Z., et al. (2016). Ethnic variations in pterygium in a rural population in Southwestern China: the Yunnan minority eye studies. Ophthalmic Epidemiol. 23 (2), 116–121. doi:10.3109/09286586.2015.1099685
Zhou, M., Shi, Z. X., Liu, Z., Ke, S. R., Wang, C. Y., Liang, X. L., et al. (2024). Single-cell transcriptomic analysis reveals dynamic cellular processes in corneal epithelium during wound healing in cynomolgus monkeys. Invest. Ophthalmol. Vis. Sci. 65 (11), 43. doi:10.1167/iovs.65.11.43
Keywords: biomarker, cornea, conjunctiva, Fuchs corneal endothelial dystrophy, lacrimal gland, meibomian gland, ocular surface, single-cell RNA sequencing
Citation: Ou S, Cai M, Feng Y, Lin S, Zheng X, Zhao S, Gu H and Wu Y (2025) Ocular surface health and disease: insight from single-cell RNA sequencing. Front. Genet. 16:1683167. doi: 10.3389/fgene.2025.1683167
Received: 29 August 2025; Accepted: 08 October 2025;
Published: 11 November 2025.
Edited by:
Pasquale Aragona, University of Messina, ItalyReviewed by:
George Maiti, New York University Health, United StatesLudovico Alisi, Sapienza University of Rome, Italy
Copyright © 2025 Ou, Cai, Feng, Lin, Zheng, Zhao, Gu and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shangkun Ou, b3VzaGFuZ2t1bkBnbWMuZWR1LmNu; Hao Gu, Z3VoYW9AZ21jLmVkdS5jbg==; Yiming Wu, eS53dS4xM0BwZ3IuYmhhbS5hYy51aw==
†These authors have contributed equally to this work
 Shangkun Ou
Shangkun Ou Minqing Cai2,3†
Minqing Cai2,3† Yiming Wu
Yiming Wu