- 1Department of Human Genetics, McGill University, Montreal, QC, Canada
- 2Centre of Genomics and Policy, Victor Phillip Dahdaleh Institute of Genomic Medicine, McGill University, Montreal, QC, Canada
The rapid evolution of genomic knowledge has made reanalysis and reinterpretation of clinical genetic testing results an ethical imperative to ensure optimal patient care. However, significant discrepancies persist between policies, laboratory practices, and stakeholder perspectives regarding the responsibility for initiating and communicating reclassified variants. This perspective examines the current landscape of ethical, legal, and practical challenges for laboratories, clinicians, and patients. We highlight the tension between the duty of care and resource constraints, finding that while the ethical importance of reinterpretation is acknowledged, the lack of standardized guidelines and legal clarity fuels uncertainty and discordant stakeholder views. To address these challenges, we propose an actionable, shared-responsibility framework that aligns duties with expertise. In this model, diagnostic laboratories are positioned to monitor new evidence and initiate updates for reinterpretation, while clinicians manage patient recontact and initiate case-level reanalysis, and health systems provide the necessary infrastructure. Realizing this framework through multidisciplinary collaboration and investment is crucial for establishing equitable best practices and integrating reinterpretation into the evolving standard of care.
1 Introduction
Clinical genetic testing has revolutionized diagnostic capabilities for genetic disorders, offering diagnostic yields of 25%–40% in rare conditions (Wojcik et al., 2023). However, the dynamic nature of genomic knowledge necessitates periodic reinterpretation of genetic data to ensure clinical relevance. Systematic reanalysis can provide diagnoses for an additional 13%–22% of previously unsolved cases, underscoring its potential to improve patient care (Hartley et al., 2023; Tan et al., 2020; Machini et al., 2019). Variant classification, governed by guidelines from the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP), as well as ClinGen specifications, remains inherently fluid, with classifications evolving as new evidence emerges (Rehm et al., 2015; Harrison et al., 2019; Richards et al., 2015). Because diagnostic laboratories maintain the infrastructure for genomic data management, gene and variant curation, and evidence synthesis, they are best equipped to conduct ongoing variant surveillance, whereas clinicians remain responsible for ensuring that updated results are communicated to patients through recontact when clinically indicated.
The implications of variant reinterpretation are profound. Reclassification can alter clinical management strategies, including treatment plans, surveillance protocols, and preventive interventions such as cancer risk-reducing surgeries (Murray et al., 2011; El Mecky et al., 2019; Walsh et al., 2024). A recent scoping review highlighted the fluid nature of variant classification, reporting that proactive reinterpretation changed the classifications for an average of 31% of variants (ranging from 4.7% to 100%), while routine clinical practice led to reclassification in approximately 20% of cases, predominantly affecting variants of uncertain significance (VUS) (Thummala et al., 2024). For patients receiving genetic testing for hereditary cancer syndromes, 7% had reclassified variants and of those reclassifications about 12% had the potential to significantly change clinical care (Turner et al., 2019). Despite these benefits, the field lacks consensus on the optimal frequency and mechanisms for initiating reinterpretation. Reported reclassification rates vary (3.6%–58.8%), with most occurring within 2 years of initial reporting (Walsh et al., 2024; Wright et al., 2018). Longitudinal analysis of hereditary cancer genetic testing (n > 1.9 million) revealed a 4.7% variant reclassification rate over two decades (Esterling et al., 2020). Notably, while most were downgrades, approximately 20% of these reclassifications represented upgrades to pathogenic or likely pathogenic status. The clinical significance of variant reinterpretation is demonstrated by a study in arrhythmogenic right ventricular cardiomyopathy, where over 40% of reclassified variants had implications for patient management and 10% of patients had a variant downgrade that led to a loss of their disease status (Costa et al., 2021). This emphasizes the importance of an accurate genetic diagnosis and periodic reassessment of variant pathogenicity classifications.
Responsibility for initiating reinterpretation remains a contentious issue. Laboratories are generally not obligated to perform reinterpretation unless prompted by clinicians or patients (Clayton et al., 2021). This reactive approach contrasts with calls for systematic frameworks that integrate reinterpretation into routine practice. However, significant barriers persist, including resource constraints, inconsistent policies, and unclear legal obligations. These challenges are exacerbated by discordant stakeholder perspectives on who bears responsibility for initiating and communicating reclassified results.
Clinical genetic testing presents a persistent ethical challenge: as variant interpretation evolves, who is responsible for ensuring patients receive updated information that could impact their care? Despite the clear benefits of reanalysis and reinterpretation, significant gaps remain, including inconsistent guidelines, unclear responsibilities, discordant stakeholder perspectives, and systemic barriers (Chisholm et al., 2018; Appelbaum et al., 2023). This perspective critically examines these ethical, legal, and policy dimensions, highlighting the need for standardized frameworks to align ethical imperatives with practical constraints and proposing actionable recommendations to ensure equitable and effective practices in genomic medicine.
2 Legal considerations
Currently, no laws explicitly mandate the routine reinterpretation of clinical genetic test results, and courts have yet to impose liability for failing to reanalyze or recontact patients with updated genetic findings (Clayton et al., 2021). While ethical imperatives to preserve and reassess genetic data are widely acknowledged, legal frameworks remain ambiguous, leaving laboratories and clinicians uncertain about their obligations. For example, the ACMG recommends that laboratories establish protocols for variant re-evaluation and case-level reanalysis (Deignan et al., 2019), but practices vary across institutions (El Mecky et al., 2019). In Canada, the Canadian College of Medical Geneticists (CCMG) similarly proposes reinterpretation as a shared responsibility among healthcare professionals rather than a legal duty (Goh et al., 2024).
The courts have not imposed a legal obligation, as highlighted by Williams v. Quest/Athena, where a laboratory’s initial classification of a variant later reclassified as pathogenic was central to litigation following a child’s death (GenomeWeb, 2016). The plaintiff argued that if the variant had been correctly classified initially, alternative treatment could have been pursued, potentially preventing the child’s death. The court ruled in favor of the laboratory, citing insufficient evidence to establish causation between the misclassification and patient outcomes (GenomeWeb, 2020). This case highlights the unresolved legal questions surrounding liability for variant misclassification or delayed reinterpretation (McGrath et al., 2021).
Laboratory obligations for reinterpretation requests often depend on regional data protection laws. Laboratories are generally required to provide existing reports or raw data upon patient request but are not obligated to perform reinterpretation unless prompted by clinicians (Clayton et al., 2021). Reinterpretation is typically reactive, initiated by clinicians in response to new clinical information or evolving patient phenotypes (Clayton et al., 2021). While laboratories and physicians do not have a legal obligation to routinely reassess genetic test results, this is an emerging area of focus within the field.
Marchant et al. underscore the importance of revisiting legal liabilities in the context of clinical genetics to ensure that guidelines evolve with technological advances (Marchant et al., 2020). The concept of treating genomic data as a lifetime resource raises concerns about future liability for unreported findings or secondary variants (Marchant et al., 2020). To mitigate these risks, many laboratories currently favor targeted testing approaches limited to well-characterized genes immediately relevant to diagnosis. Conversely, Roberts and Foulkes argue that clinically meaningful variant reclassifications should carry a legal duty for the laboratory and the ordering clinician, presenting a potential framework for liability (Roberts and Foulkes, 2020). Unsurprisingly, there is a high degree of concern for liability of reinterpretation and recontact among both clinical genetic and laboratory genetic providers (Berger et al., 2022).
The existing consensus holds that laboratories are not legally compelled to undertake reanalysis or reinterpretation of clinical genetic tests (Vears et al., 2018; Matthijs et al., 2016; Geest et al., 2024). Although routine reanalysis is not the standard of care, practices may evolve to establish reinterpretation as a legal obligation (Clayton et al., 2021; Marchant et al., 2020; Foulkes et al., 2020). This lack of clarity creates uncertainty for laboratories and clinicians.
3 Ethical imperatives
The ethical debate surrounding the reinterpretation of genetic test results centers on the responsibilities of laboratories, clinicians, and healthcare systems in ensuring patients benefit from updated knowledge and new technology. Beneficence, the moral obligation to act in the patient’s best interest, is frequently invoked to argue for reanalysis and reinterpretation to improve clinical care. For example, reclassification of variants can lead to actionable changes in treatment plans or surveillance strategies, underscoring the potential for significant patient benefit (Supplementary Table S1) (Appelbaum et al., 2020). Non-maleficence, the duty to avoid harm, is critical, as failing to reinterpret variants or communicate updated results may prevent patients from having opportunities for improved care or expose them to unnecessary interventions based on inaccurate information.
While there is no recognized legal duty to take action on reclassified genetic test results, there is an ongoing debate regarding the extent of the responsibilities borne by the laboratories, clinicians, healthcare system, and patients (Roberts and Foulkes, 2020).
Appelbaum et al. argue that ordering genetic tests creates an ethical imperative to continually reinterpret variants as knowledge evolves, obligating providers to update and communicate new findings (Appelbaum et al., 2020). Although logistical and resource constraints exist, they maintain that ethical duties outweigh these barriers and predict that advancing technologies will soon make reinterpretation a standard of care (Appelbaum et al., 2020). Furthermore, they assert that laboratories carry an ongoing duty (beyond legal requirements) to proactively reanalyze results, warning that failure to do so risks abandoning patients (Appelbaum et al., 2020).
Watts and Newson contend that universal, systematic reinterpretation is impractical given current logistical and financial constraints and instead endorse reactive, clinician-triggered reinterpretation (Watts and Newson, 2023). They argue that although clinicians have a continuing duty of care to patients that are undergoing clinical genetic testing, this does not extend to routine reassessment of all VUSs (Watts and Newson, 2023). They argue that despite offering genetic tests that have the potential to yield clinically meaningful results in the future if reliably reinterpreted, these genetic tests are not currently offered with this condition (Watts and Newson, 2023). Instead, laboratories should focus on reevaluating pathogenic variants, especially those with a high risk of false positives or those common in underrepresented populations, while making two exceptions: VUSs identified as potentially clinically relevant and cases where laboratories receive explicit payment for reinterpretation services (Watts and Newson, 2023).
Equity is a particularly critical issue in the context of variant reinterpretation. Individuals from minority populations are disproportionately affected by VUSs due to limited representation in genomic databases (Walsh et al., 2024; Plon and Rehm, 2018; Slavin et al., 2018; Makhnoon et al., 2023; Caswell-Jin et al., 2018). These disparities result in fewer definitive diagnoses, a higher burden of VUSs, and higher rates of genetic misclassification, exacerbating health inequities (Manrai et al., 2016). Slavin et al. demonstrated that rates of variant re-classification differ significantly across ancestries; for example, 18% of BRCA1/2 variants were reclassified over 20 years, with minority populations benefiting most from these updates (Slavin et al., 2018). This highlights the essential role of VUS reinterpretation in upholding effective clinical care (Popejoy and Fullerton, 2016). Addressing these inequities requires expanding genomic datasets to include diverse populations and integrating routine reinterpretation into clinical practice (Veenstra et al., 2021). Doing so would improve diagnostic accuracy and reduce disparities in access to actionable genetic information (Lee et al., 2022).
Despite the significant practical limitations, there is mounting pressure for laboratories to assume the responsibility of actively or proactively reinterpreting variants, and that this ethical responsibility should inform policymaking and the development of best practices (Watts and Newson, 2023; Appelbaum et al., 2020). The ethical debate surrounding the reinterpretation of genetic test results highlights the tension between idealistic commitments to beneficence, non-maleficence, autonomy, and equity, and the practical realities faced by laboratories and clinicians.
4 Policy gaps
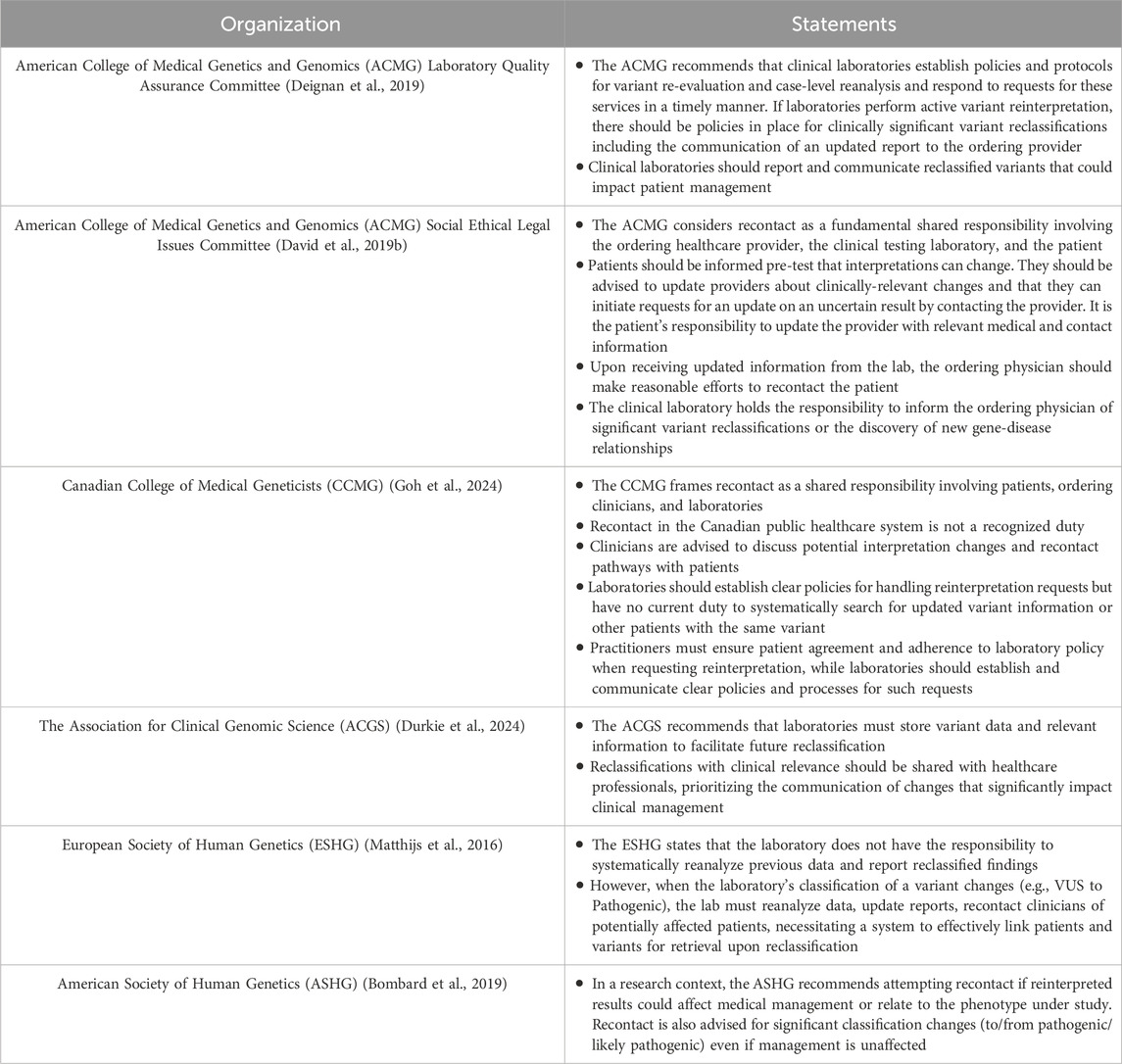
Reanalysis and reinterpretation of clinical genetic test results are guided by recommendations from professional societies, including the American College of Medical Genetics and Genomics (ACMG), the Canadian College of Medical Geneticists (CCMG), and the Association for Clinical Genomic Science (ACGS). While the organizations emphasize an ethical importance, there remains a lack of clarity and discordance on the responsibilities for reinterpretation. The key guidelines and position statements are summarized in Table 1.
In the United States, the ACMG Laboratory Quality Assurance Committee recommends that laboratories establish protocols for variant re-evaluation and case-level reanalysis but does not mandate routine reinterpretation or specify whether laboratories or clinicians should initiate the process (Deignan et al., 2019). However, a statement from the ACMG Social, Ethical, and Legal Issues (SELI) Committee suggests the laboratory bears responsibility for informing the ordering physician of significant variant reclassifications or newly established gene-disease relationships (David et al., 2019b), a position potentially incongruent with other guidance from the ACMG. Overall, the ACMG framework leans towards a shared responsibility model involving the laboratory, provider, and patient, noting that recontact often depends on patient initiative (David et al., 2019a). Similarly, Canadian guidance from the CCMG affirms the ethical responsibility and highlights a shared responsibility model among laboratories, clinicians, and patients but acknowledges that resource constraints and the absence of established protocols (Goh et al., 2024). The CCMG advisory document outlines a push and pull model for genetic result reinterpretation. Practitioners pull updates by requesting them based on time or clinical changes, while labs push significant reclassifications, prompting reasonable practitioner efforts to recontact patients (Goh et al., 2024). Patients should be made aware of reinterpretation opportunities and are advised to initiate contact if re-evaluation becomes relevant. Ordering clinicians are advised to proactively request variant reinterpretation from laboratories when clinical indications arise or when sufficient time has elapsed to warrant reanalysis. Diagnostic laboratories are recommended to develop specific policies for handling reinterpretation requests, including criteria for unsolicited reclassification updates. In cases where laboratories report clinically significant changes, clinicians are expected to make reasonable efforts to recontact the patient.
In the United Kingdom, the ACGS emphasizes the importance of storing variant data to facilitate future reclassification and advises laboratories to prioritize clinically significant updates (Durkie et al., 2024). Notably, reclassifications that affect clinical relevance should be shared with relevant healthcare professionals, and for variants impacting clinical management decisions, rapid dissemination of this information is advised (e.g., CanVar-UK database for cancer-susceptibility gene variants). However, like the ACMG and CCMG, it does not explicitly mandate proactive reinterpretation.
In contrast, research-focused organizations such as the European Society of Human Genetics (ESHG) and the American Society of Human Genetics (ASHG) adopt a more general stance on the ethical responsibility. While ESHG guidelines state laboratories are not obligated to proactively reinterpret results based on new evidence, they do mandate that if a lab reclassifies a variant, it must reanalyze, report, and recontact clinicians. This creates a significant ethical duty for reactive reinterpretation prompted by external triggers like clinician requests (Table 1) (Matthijs et al., 2016). The ASHG focuses on research contexts, recommending recontact when reinterpretation yields findings with clinical relevance (Bombard et al., 2019).
A notable gap in existing policies is the absence of clear guidance on defining the scope of reanalysis. While the shared ethical responsibilities for patients, ordering clinicians, and laboratories are broadly acknowledged, effective implementation requires standardized guidelines and improved infrastructure and resources (Goh et al., 2024).
5 Stakeholder discordance
There is discordance among laboratory providers, clinicians, and patients regarding roles, responsibilities, and expectations of reinterpretation and reanalysis in clinical genetic testing. While diagnostic laboratory directors largely view the responsibility of initiating reinterpretation as belonging to the ordering clinician and the patient (El Mecky et al., 2019), this view is not always shared; one study found that while 59% of clinicians believe labs should trigger reinterpretation, only 39% of lab directors agreed (Berger et al., 2022).
Most patients and clinical or laboratory providers were in favor of reporting all new findings, while the non-genetic providers prioritized new results that could change the patient’s management. Within the laboratory providers, the laboratory directors were less inclined to report a new result compared to laboratory genetic counselors (Berger et al., 2022). Most respondents across groups, including 74% of laboratory providers, believe there should not be a fixed end-point for the laboratory’s responsibility to reinterpret previously reported genetic variants (Berger et al., 2022). However, this raises questions about the systems and practices that will need to be implemented to allow for the long-term storage of clinical genomic data should this become the expected standard.
There are differing views on the consenting process in reinterpretation and recontact. While most genetics professionals did not feel specific re-consent was necessary prior to reinterpretation, patients and non-genetic providers favored explicit consent before reinterpretation occurs (Berger et al., 2022). Patient understanding of the reclassification process itself is often limited, however, most patients value being recontacted despite the associated challenges and generally view healthcare providers as responsible for initiating recontact (Otten et al., 2014). Genetic counselors express conflicting views on their role, with some accepting primary responsibility (Scherr et al., 2015) and others viewing it as shared or patient-led (Mueller et al., 2019). The validity and durability of initial consent obtained years prior, often without detailed discussion of future reanalysis possibilities, presents a significant challenge, particularly given that current practices sometimes involve reinterpretation without updated patient agreement (El Mecky et al., 2019; Geest et al., 2024). This highlights a critical need for a dynamic consent process and improved patient education regarding the evolving nature of genomic knowledge (El Mecky et al., 2019; David et al., 2019b; Appelbaum et al., 2023; Deignan et al., 2019).
Recontact is viewed as a shared duty, where clinicians handle results for their active patients, but healthcare system-level solutions are needed to ensure variant updates reach all affected individuals (David et al., 2019b; Goh et al., 2024). Clayton et al. suggest that laboratories should be cautious about returning reinterpreted results directly to patients, as this establishes a clinical relationship that carries potentially unforeseen responsibilities (Clayton et al., 2021). Marchant et al. further emphasize that laboratories do not necessarily have the means to recontact patients with reports of variant reclassifications. Rather, they suggest that if the practice of active variant reinterpretation emerges, then the responsibility may lie with the laboratory to contact the ordering physician with the reinterpreted results, where the responsibility would then remain with them to communicate the results to the patient (Marchant et al., 2020).
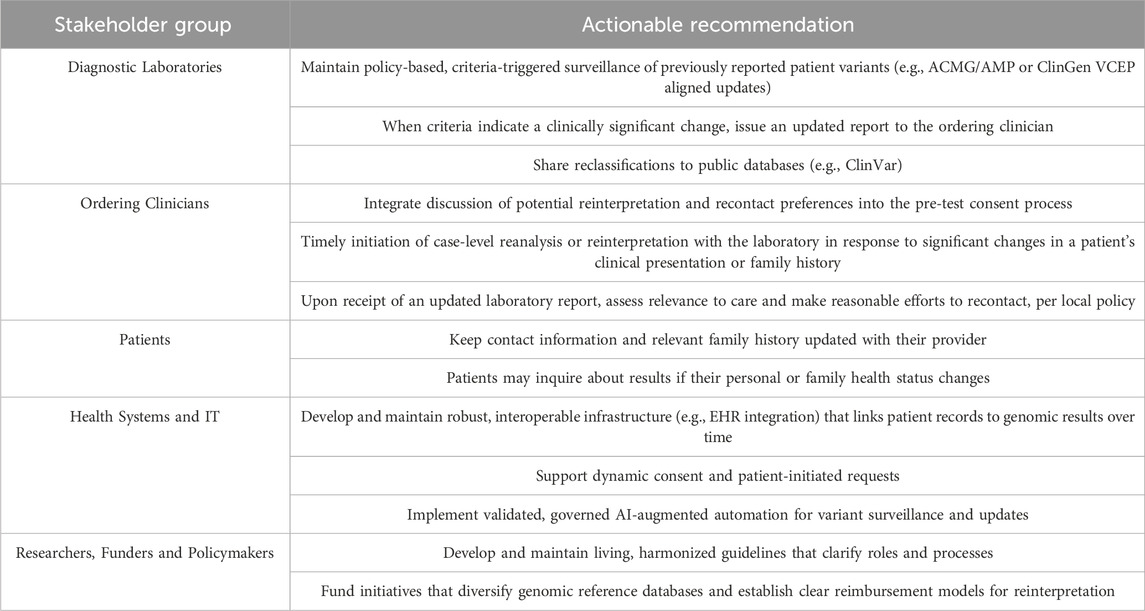
6 Actionable recommendations for a shared responsibility framework
The stakeholder discordance, conflicting expectations, and operational gaps described above demonstrate that while a shared responsibility framework is broadly endorsed in principle, it remains to be realized. To operationalize a shared responsibility model, roles must align with expertise. Our proposed framework clarifies this by distinguishing between proactive, variant-level reinterpretation initiated by diagnostic laboratories and reactive, case-level reanalysis initiated by clinicians in response to new patient information. Table 2 outlines these responsibilities, designed to support the integration of reinterpretation as a standard of care without asserting a new legal duty.
7 Conclusion
Discordance persists over who is responsible for initiating reinterpretation and recontact. Combined with legal ambiguity, lack of harmonized policies, and equity gaps, these challenges limit the potential of genomic medicine. While a shared responsibility framework is broadly endorsed, operational and role-designation gaps remain. The actionable framework proposed here aims to resolve these issues by aligning duties with expertise. Bridging these gaps through collaboration and infrastructure innovation is essential to making reanalysis and reinterpretation an equitable standard of care.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
ZS: Writing – original draft, Conceptualization, Writing – review and editing. MHZ: Conceptualization, Supervision, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. ZS would like to acknowledge the support of the Canadian Institutes of Health Research (CIHR) Doctoral Research Award. MHZ would like to acknowledge the support of the FRQS through the Junior 2 Career Award.
Acknowledgments
We would like to thank Chiara S. Spino, Administrative Coordinator at the Centre of Genomics and Policy, for her editorial assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2025.1685854/full#supplementary-material
References
Appelbaum, P. S., Parens, E., Berger, S. M., Chung, W. K., and Burke, W. (2020). Is there a duty to reinterpret genetic data? The ethical dimensions. Genet. Med. Official J. Am. Coll. Med. Genet. 22 (3), 633–639. doi:10.1038/s41436-019-0679-7
Appelbaum, P. S., Berger, S. M., Brokamp, E., Brown, H. S., Burke, W., Clayton, E. W., et al. (2023). Practical considerations for reinterpretation of individual genetic variants. Genet. Med. Official J. Am. Coll. Med. Genet. 25 (5), 100801. doi:10.1016/j.gim.2023.100801
Berger, S. M., Appelbaum, P. S., Siegel, K., Wynn, J., Saami, A. M., Brokamp, E., et al. (2022). Challenges of variant reinterpretation: opinions of stakeholders and need for guidelines. Genet. Med. Official J. Am. Coll. Med. Genet. 24 (9), 1878–1887. doi:10.1016/j.gim.2022.06.002
Bombard, Y., Brothers, K. B., Fitzgerald-Butt, S., Garrison, N. A., Jamal, L., James, C. A., et al. (2019). The responsibility to recontact research participants after reinterpretation of genetic and genomic research results. Am. J. Hum. Genet. 104 (4), 578–595. doi:10.1016/j.ajhg.2019.02.025
Caswell-Jin, J. L., Gupta, T., Hall, E., Petrovchich, I. M., Mills, M. A., Kingham, K. E., et al. (2018). Racial/ethnic differences in multiple-gene sequencing results for hereditary cancer risk. Genet. Med. 20 (2), 234–239. doi:10.1038/gim.2017.96
Chisholm, C., Daoud, H., Ghani, M., Mettler, G., McGowan-Jordan, J., Sinclair-Bourque, L., et al. (2018). Reinterpretation of sequence variants: one diagnostic laboratory’s experience, and the need for standard guidelines. Genet. Med. 20 (3), 365–368. doi:10.1038/gim.2017.191
Clayton, E. W., Appelbaum, P. S., Chung, W. K., Marchant, G. E., Roberts, J. L., and Evans, B. J. (2021). Does the law require reinterpretation and return of revised genomic results? Genet. Med. Official J. Am. Coll. Med. Genet. 23 (5), 833–836. doi:10.1038/s41436-020-01065-x
Costa, S., Medeiros-Domingo, A., Gasperetti, A., Akdis, D., Berger, W., James, C. A., et al. (2021). Impact of genetic variant reassessment on the diagnosis of arrhythmogenic right ventricular cardiomyopathy based on the 2010 task force criteria. Circulation Genomic Precis. Med. 14 (1), e003047. doi:10.1161/CIRCGEN.120.003047
David, K. L., Best, R. G., Manace Brenman, L., Bush, L., Deignan, J. L., Flannery, D., et al. (2019a). Response to Knoppers et Al. Genet. Med. 21 (10), 2403. doi:10.1038/s41436-019-0496-z
David, K. L., Best, R. G., Manace Brenman, L., Bush, L., Deignan, J. L., Flannery, D., et al. (2019b). Patient Re-Contact after revision of genomic test results: points to Consider—A statement of the American college of medical genetics and genomics (ACMG). Genet. Med. 21 (4), 769–771. doi:10.1038/s41436-018-0391-z
Deignan, J. L., Chung, W. K., Kearney, H. M., Monaghan, K. G., Rehder, C. W., Chao, E. C., et al. (2019). Points to consider in the reevaluation and reanalysis of genomic test results: a statement of the American college of medical genetics and genomics (ACMG). Genet. Med. Official J. Am. Coll. Med. Genet. 21 (6), 1267–1270. doi:10.1038/s41436-019-0478-1
Durkie, M., Cassidy, E.-J., Berry, I., Owens, M., Turnbull, C., Taylor, R. W., et al. (2024). ACGS best practice guidelines for variant classification in rare disease 2024.
El Mecky, J., Johansson, L., Plantinga, M., Fenwick, A., Lucassen, A., Dijkhuizen, T., et al. (2019). Reinterpretation, reclassification, and its downstream effects: challenges for clinical laboratory geneticists. BMC Med. Genomics 12, 170. doi:10.1186/s12920-019-0612-6
Esterling, L., Wijayatunge, R., Brown, K., Morris, B., Hughes, E., Pruss, D., et al. (2020). Impact of a cancer gene variant reclassification program over a 20-Year period. JCO Precis. Oncol. 4, 944–954. doi:10.1200/PO.20.00020
Foulkes, A. L., Roberts, J. L., Appelbaum, P. S., Chung, W. K., Clayton, E. W., Evans, B., et al. (2020). Can clinical genetics laboratories be sued for medical malpractice? Ann. Health Law Life Sci. 29 (1), 153–172. Available online at: https://lawecommons.luc.edu/annals/vol29/iss1/5/.
Geest, M.A. van der, Maeckelberghe, E. L. M., Gijn, M.E. van, Lucassen, A. M., Swertz, M. A., Langen, I.M. van, et al. (2024). Systematic reanalysis of genomic data by diagnostic laboratories: a scoping review of ethical, economic, legal and (psycho)Social implications. Eur. J. Hum. Genet., 1–9. doi:10.1038/s41431-023-01529-z
GenomeWeb (2016). Mother’s negligence suit against quest’s athena could broadly impact genetic testing labs. Available online at: https://www.genomeweb.com/molecular-diagnostics/mothers-negligence-suit-against-quests-athena-could-broadly-impact-genetic.
GenomeWeb (2020). Quest diagnostics win in wrongful death case reveals ongoing challenges for variant classification. Available online at: https://www.genomeweb.com/molecular-diagnostics/quest-diagnostics-win-wrongful-death-case-reveals-ongoing-challenges-variant.
Goh, E. S.-Y., Chad, L., Richer, J., Bombard, Y., Mighton, C., Agatep, R., et al. (2024). Canadian college of medical geneticists: clinical practice advisory document – responsibility to recontact for reinterpretation of clinical genetic testing. J. Med. Genet. 61 (12), 1123–1131. doi:10.1136/jmg-2024-110330
Harrison, S. M., Biesecker, L. G., and Rehm, H. L. (2019). Overview of specifications to the ACMG/AMP variant interpretation guidelines. Curr. Protoc. Hum. Genet. 103 (1), e93. doi:10.1002/cphg.93
Hartley, T., Soubry, É., Acker, M., Osmond, M., Couse, M., Gillespie, M. K., et al. (2023). Bridging clinical care and research in Ontario, Canada: maximizing diagnoses from reanalysis of clinical exome sequencing data. Clin. Genet. 103 (3), 288–300. doi:10.1111/cge.14262
Lee, S.S.-J., Appelbaum, P. S., and Chung, W. K. (2022). Challenges and potential solutions to health disparities in genomic medicine. Cell 185 (12), 2007–2010. doi:10.1016/j.cell.2022.05.010
Machini, K., Ceyhan-Birsoy, O., Azzariti, D. R., Sharma, H., Rossetti, P., Mahanta, L., et al. (2019). Analyzing and reanalyzing the genome: findings from the MedSeq project. Am. J. Hum. Genet. 105 (1), 177–188. doi:10.1016/j.ajhg.2019.05.017
Makhnoon, S., Levin, B., Ensinger, M., Mattie, K., Volk, R. J., Zhao, Z., et al. (2023). A multicenter study of clinical impact of variant of uncertain significance reclassification in breast, ovarian and colorectal cancer susceptibility genes. Cancer Med. 12 (3), 2875–2884. doi:10.1002/cam4.5202
Manrai, A. K., Funke, B. H., Rehm, H. L., Olesen, M. S., Maron, B. A., Szolovits, P., et al. (2016). Genetic misdiagnoses and the potential for health disparities. N. Engl. J. Med. 375 (7), 655–665. doi:10.1056/NEJMsa1507092
Marchant, G., Barnes, M., Evans, J. P., LeRoy, B., and Wolf, S. M.LawSeq Liability Task Force (2020). From genetics to genomics: facing the liability implications in clinical care. J. Law, Med. and Ethics 48 (1), 11–43. doi:10.1177/1073110520916994
Matthijs, G., Souche, E., Alders, M., Corveleyn, A., Eck, S., Feenstra, I., et al. (2016). Guidelines for diagnostic next-generation sequencing. Eur. J. Hum. Genet. 24 (1), 2–5. doi:10.1038/ejhg.2015.226
McGrath, S. P., Peabody, A. E., Walton, D., and Walton, N. (2021). Legal challenges in precision medicine: what duties arising from genetic and genomic testing does a physician owe to patients? Front. Med. 8, 663014. doi:10.3389/fmed.2021.663014
Mueller, A., Dalton, E., Enserro, D., Wang, C., and Flynn, M. (2019). Recontact practices of cancer genetic counselors and an exploration of professional, legal, and ethical duty. J. Genet. Couns. 28 (4), 836–846. doi:10.1002/jgc4.1126
Murray, M. L., Cerrato, F., Bennett, R. L., and Jarvik, G. P. (2011). Follow-up of carriers of BRCA1 and BRCA2 variants of unknown significance: variant reclassification and surgical decisions. Genet. Med. 13 (12), 998–1005. doi:10.1097/GIM.0b013e318226fc15
Otten, E., Plantinga, M., Birnie, E., Verkerk, M. A., Lucassen, A. M., Ranchor, A. V., et al. (2014). Is there a duty to recontact in light of new genetic technologies? A systematic review of the literature. Genet. Med. 17 (8), 668–678. doi:10.1038/gim.2014.173
Plon, S. E., and Rehm, H. L. (2018). The ancestral pace of variant reclassification. JNCI J. Natl. Cancer Inst. 110 (10), 1133–1134. doi:10.1093/jnci/djy075
Popejoy, A. B., and Fullerton, S. M. (2016). Genomics is failing on diversity. Nature 538 (7624), 161–164. doi:10.1038/538161a
Rehm, H. L., Berg, J. S., Brooks, L. D., Bustamante, C. D., Evans, J. P., Landrum, M. J., et al. (2015). ClinGen--the clinical genome resource. N. Engl. J. Med. 372 (23), 2235–2242. doi:10.1056/NEJMsr1406261
Richards, S., Aziz, N., Bale, S., Bick, D., Das, S., Gastier-Foster, J., et al. (2015). Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet. Med. Official J. Am. Coll. Med. Genet. 17 (5), 405–424. doi:10.1038/gim.2015.30
Roberts, J. L., and Foulkes, A. L. (2020). Genetic duties. William Mary Law Rev. 62 (1), 143–211. Available online at: https://scholarship.law.wm.edu/wmlr/vol62/iss1/4/.
Scherr, C. L., Lindor, N. M., Malo, T. L., Couch, F. J., and Vadaparampil, S. T. (2015). Genetic counselors’ practices and confidence regarding variant of uncertain significance results and reclassification from BRCA testing. Clin. Genet. 88 (6), 523–529. doi:10.1111/cge.12563
Slavin, T. P., Tongeren, L.R. van, Behrendt, C. E., Solomon, I., Rybak, C., Nehoray, B., et al. (2018). Prospective study of cancer genetic variants: variation in rate of reclassification by ancestry. JNCI J. Natl. Cancer Inst. 110 (10), 1059–1066. doi:10.1093/jnci/djy027
Tan, N. B., Stapleton, R., Stark, Z., Delatycki, M. B., Yeung, A., Hunter, M. F., et al. (2020). Evaluating systematic reanalysis of clinical genomic data in rare disease from single center experience and literature review. Mol. Genet. and Genomic Med. 8 (11), e1508. doi:10.1002/mgg3.1508
Thummala, A., Sudhakaran, R., Gurram, A., Mersch, J., Badalamenti, A., Gottaway, G., et al. (2024). Variant reclassification and recontact research: a scoping review. Genet. Med. Open 2, 101867. doi:10.1016/j.gimo.2024.101867
Turner, S. A., Rao, S. K., Morgan, R. H., Vnencak-Jones, C. L., and Wiesner, G. L. (2019). The impact of variant classification on the clinical management of hereditary cancer syndromes. Genet. Med. 21 (2), 426–430. doi:10.1038/s41436-018-0063-z
Vears, D. F., Sénécal, K., Clarke, A. J., Jackson, L., Laberge, A. M., Lovrecic, L., et al. (2018). Points to consider for laboratories reporting results from diagnostic genomic sequencing. Eur. J. Hum. Genet. 26 (1), 36–43. doi:10.1038/s41431-017-0043-9
Veenstra, D. L., Rowe, J., Pagán, J. A., Brown, H. S., Schneider, J., Gupta, A., et al. (2021). Reimbursement for genetic variant reinterpretation: five questions payers should ask. Am. J. Manag. Care 27 (10), e336–e338. doi:10.37765/ajmc.2021.88763
Walsh, N., Cooper, A., Dockery, A., and O’Byrne, J. J. (2024). Variant reclassification and clinical implications. J. Med. Genet. 61 (3), 207–211. doi:10.1136/jmg-2023-109488
Watts, G., and Newson, A. J. (2023). Is there a duty to routinely reinterpret genomic variant classifications? J. Med. Ethics 49 (12), 808–814. doi:10.1136/jme-2022-108864
Wojcik, M. H., Reuter, C. M., Marwaha, S., Mahmoud, M., Duyzend, M. H., Barseghyan, H., et al. (2023). Beyond the exome: what’s next in diagnostic testing for Mendelian conditions. Am. J. Hum. Genet. 110 (8), 1229–1248. doi:10.1016/j.ajhg.2023.06.009
Keywords: clinical genetic testing, medical genetics, variant classification, reanalysis, variant reinterpretation, ethical considerations, legal, policy
Citation: Sentell ZT and Zawati MH (2025) Ethical, legal, and policy dimensions and contentions for reanalysis and reinterpretation of clinical genetic testing results. Front. Genet. 16:1685854. doi: 10.3389/fgene.2025.1685854
Received: 14 August 2025; Accepted: 29 September 2025;
Published: 08 October 2025.
Edited by:
Go Yoshizawa, Kwansei Gakuin University, JapanReviewed by:
Amanda Courtright-Lim, Mayo Clinic, United StatesCopyright © 2025 Sentell and Zawati. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ma’n H. Zawati, bWFuLnphd2F0aUBtY2dpbGwuY2E=
 Zachary T. Sentell
Zachary T. Sentell Ma’n H. Zawati
Ma’n H. Zawati