- Centre for Soil, Agrifood and Biosciences, Cranfield University, Cranfield, United Kingdom
Protoplast-based systems have been utilised in a wide variety of plant species to enable genome editing without chromosomal introgression of foreign DNA into plant genomes. DNA-free genome editing followed by protoplast regeneration allows elite cultivars to be edited without further genetic segregation, preserving their unique genetic composition and their regulatory status as non-transgenic. However, protoplast isolation presents a barrier to the development of advanced breeding technologies in raspberry and no protocol has been published for DNA-free genome editing in the species. Pre-assembled ribonucleoprotein complexes (RNPs) do not require cellular processing and the commercial availability of Cas9 proteins and synthetic guide RNAs has streamlined genome editing protocols. This study presents a novel high-yielding protoplast isolation protocol from raspberry stem cultures and RNP-mediated transfection of protoplast with CRISPR-Cas9. Targeted mutagenesis of the phytoene desaturase gene at two intragenic loci resulted in an editing efficiency of 19%, though estimated efficiency varied depending on the indel analysis technique. Only amplicon sequencing was sensitive enough to confirm genome editing in a low efficiency sample. To our knowledge, this study constitutes the first use of DNA-free genome editing in raspberry protoplast. This protocol provides a valuable platform for understanding gene function and facilitates the future development of precision breeding in this important soft fruit crop.
Introduction
Raspberry (Rubus idaeus; RRID:NCBITaxon_32247) is a high-value horticultural crop that, alongside other Rubus berries, has undergone a substantial increase in consumption worldwide in recent decades (Foster et al., 2019). Globally, production increased 48% between 2011 and 2021 (FAO, 2024). Often sold as a “superfood,” raspberries contain high levels of anthocyanins, flavanols and phenolic acids that are essential for normal metabolism. Lack of dietary sources of these bioactive nutrients has been associated with cancer, stroke, Alzheimer’s and autoimmune diseases (Derrick et al., 2021; Hancock et al., 2007). Raspberry is a highly heterozygous species that is not true to seed, meaning both sexual reproduction and selfing can introduce allelic variation that alters the berry phenotype of elite cultivars. Commercial raspberry production thus relies on vegetative propagation. This challenge, in addition to a 2-year fruiting cycle in floricanes, has contributed to a lack of progress on next-generation methods for raspberry breeding.
Improving agricultural sustainability is a key priority for reaching net zero (FAO. Emissions due to agriculture, 2020). Food production systems must undergo sustainable intensification, and gains can be made in raspberry production through improving efficiency while reducing food waste. Advanced breeding to improve traits such as fungal resistance or fruit firmness would substantially improve shelf life (Simpson et al., 2017). Enhanced plant and fruiting architecture could reduce labour demands for crop management and harvesting while eliminating genetic disorders, such as crumbly fruit syndrome, would prevent widespread yield losses (Scolari et al., 2023). Such improvements can also minimise chemical inputs and decrease overall land use, leading to more productive and sustainable raspberry cultivation. Greater access to this nutritious species would also support the UN’s 2030 Sustainable Development Goals of zero hunger and good health and wellbeing (United Nations, 2024).
The advent of CRISPR (clustered regularly interspaced short palindromic repeats) genome editing has greatly increased the scope and speed of plant breeding (Jinek et al., 1979). Through the formation of ribonucleoprotein complexes (RNPs), composed of Cas nucleases and programmable guide RNA (gRNA), targeted mutagenesis of any genomic region of interest can be achieved (Gasiunas et al., 2012; Sternberg and Doudna, 2015; Wright et al., 2016). The most commonly used nuclease, Cas9, induces a double strand break (DSB) in genomic DNA at a specific locus identified by complementary binding of the gRNA (Zhan et al., 2021). Often, error-prone DNA repair subsequently creates insertions and/or deletions (indels) at the specified locus, leading to gene knockout (KO).
Applying this system to plant species is challenging as the plant cell wall is impermeable to nucleic acids encoding Cas9/gRNA or pre-assembled RNPs. Inbreeding, annual, seed-propagated crops can utilize Agrobacterium-mediated transformation or biolistics to transfer DNA encoding CRISPR/Cas9 into intact cells and to integrate this into chromosomes. After genome editing has occurred, transgenes can be easily removed by backcrossing and genetic segregation as transgenic crops have historically been contentious from regulatory and public perception standpoints (Chan et al., 2020; Turnbull et al., 2021). However, to realise the benefits of genome editing in the advanced breeding of allogamous, clonally-propagated crops, such as raspberry, editing must take place without chromosomal introgression of foreign DNA. This allows elite clones to be edited without the need for further genetic segregation, preserving their unique genetic composition and their regulatory status as non-transgenic. Where Agrobacterium-mediated transformation cannot be used, the resurgence of protoplast-based techniques offers a transgene-free alternative (Yue et al., 2021; Yoo et al., 2007; Barceló et al., 2019; Reed and Bargmann, 2021; Hsu et al., 2021; Woo et al., 2015).
Enzymatic digestion of the plant cell wall (enzymolysis) liberates single-celled protoplasts (Yoo et al., 2007; Cocking, 1960; Davey et al., 2005) with membranes permeable to plasmids and pre-assembled RNPs in the presence of polyethylene glycol (PEG) (Woo et al., 2015). After editing, protoplast can be regenerated into whole plants (Reed and Bargmann, 2021; Jeong et al., 2021; Scintilla et al., 2022), although this is technically challenging and species dependent. Cas9 and gRNA genes encoded within plasmids are expressed within the protoplast nucleus, triggering genome editing, but do not integrate into the genome. Pre-assembled RNPs, exogenously synthesised and formed extracellularly, are also able to directly translocate the protoplast membrane to initiate editing (Woo et al., 2015). RNPs are then degraded by normal cellular processes. This form of DNA-free genome editing is rapid, highly specific and indistinguishable from natural mutagenesis–factors that have recently found favour with regulators (Genetic Technology (Precision Breeding) Act, 2023; European Commission, 2021). Pre-assembled RNPs are of particular interest in crop species like raspberry as they eliminate the chance of random integration of DNA into the genome, which is a risk with plasmid transfection. The commercial availability of synthetic gRNAs and Cas nucleases from many vendors, and evidence for higher editing efficiencies than plasmid-based techniques (Zhang Y. et al., 2022), also favour the use of DNA-free RNPs.
Genome editing and genetic engineering in raspberry have not been extensively developed (Foster et al., 2019). Agrobacterium-mediated transformation has been achieved by several groups (Mathews et al., 1995; Mezzetti et al., 2004; Kim and Dai, 2022; Zhang J. et al., 2022; Khadgi, 2020), however transgenic raspberries have never been commercialised. To our knowledge, no method for DNA-free CRISPR genome editing in raspberry has been published, however there have been attempts to use CRISPR transgenically (Khadgi, 2020; Miller, 2019), with some potential success with biolistic delivery. Nonetheless, there is a substantial research gap in this field. Pre-assembled RNPs present an excellent opportunity to rapidly test gRNAs if reliable and modern protoplast-based methods can be elucidated. Existing protoplast research on raspberry is limited in scope and predates the advent of genome editing (Phan et al., 1997; Nita-Lazar et al., 2000; Mezzetti et al., 1999), but provides evidence that raspberry protoplasts can be isolated and cultured. A high-yielding tissue source of protoplast is critical, but high heterozygosity in raspberry complicates the issue as tissue-cultured seedlings, used in many other protocols [e.g. (Yoo et al., 2007)], would result in allele segregation and genetically variable regenerated plants and compromise the elite berry phenotype of commercial raspberry cultivars. Nonetheless, many raspberry cultivars are suitable for tissue culture; vegetative propagation is common industry practice (Funt and Hall, 2013) and raspberry produces shoots vigorously, including directly from roots, suggesting a propensity for regeneration. As there is no selection in protoplast regeneration, reporter genes such as phytoene desaturase (PDS) are useful to assess the success of genome editing in regenerated plants, particularly during initial development of protoplast regeneration. KO of PDS, which plays a critical role in carotenoid biosynthesis, results in a distinctive albino phenotype that is visible during the early stages of regeneration (Lin et al., 2018). Functional targets are also of interest, particularly those that could be beneficial to raspberry shelf life and where gene KO results in an improved phenotype. For example, evidence from other fruit species indicates that polygalacturonase (PG) can increase fruit firmness (López-Casado et al., 2023; Atkinson et al., 2012; Posé et al., 2013), WRKY52 can improve resistance to B. cinerea (Jia et al., 2020) and powdery mildew (Wang et al., 2017), and nonexpressor of pathogenesis-related genes 1 (NPR1) can also improve resistance to B. cinerea (Li et al., 2020; Li et al., 2021).
This study demonstrates successful, DNA-free genome editing of raspberry protoplasts using CRISPR-Cas9 for the first time. We have applied state-of-the-art protoplast methods to a new species and achieved a high genome editing efficiency. These are critical steps in the development of next-generation breeding technologies (NGBTs) in raspberry and will hopefully stimulate further work in an underserved yet economically and nutritionally valuable species.
Materials and methods
Plant cultivation
Cold-treated (4°C, ≥50 days, ∼15 × 20 cm) roots of R. idaeus cultivar BWP102 (supplied by a commercial propagator) were planted in 7.5 L pots containing peat-rich soil (Sinclair All Purpose Growing Medium) with perlite and fertilised with Hoagland’s solution (Hoagland and Arnon, 1950) twice a week. Highly vigorous, 1.5 cm diameter, 1 m tall canes produced from cold-treated roots were cut down after approximately 1 month in a partially environmentally controlled glasshouse (set points of 20°C, 16:8 hrs day: night, supplemented with high-pressure sodium lamps). Petioles and leaves were removed, and cane stems cut into 50–70 mm segments containing one axillary bud. Cuttings were sterilised in 70% v/v ethanol for 30 s and 10% v/v sodium hypochlorite (with 0.002% Tween 20) for 4 minutes (Funt and Hall, 2013). Exposed stem ends were cut off and stem explants were planted in Murashige and Skoog (MS) basal media with Gamborg B5 vitamins (Melford; pH 5.8, 30 g L-1 sucrose, 7.5 g L-1 agar, 1 mg L-1 6-benzyl-aminopurine, 0.1 mg L-1 indole-3-butyric acid) within autoclaved Magenta™ boxes (Merck).
Tissue culture material was grown in a Sanyo MLR-350 growth cabinet with 45 μmol m-2 s -1 light from fluorescent tubes (Toshiba FL40SSW/37) at 25°C with a 16: 8h day: night cycle.
Protoplast isolation
Protoplasts were isolated from plantlets (stem cultures) grown from the axial bud cane cuttings. This material was chosen as a protoplast source as it is vegetatively grown, sterile, soft and young; therefore, it is amenable to enzymatic digestion. Plantlets (0.3 g, 20–30 mm tall) from axillary buds were harvested after 10 days of culture and cut with a sterile scalpel blade into 0.5–1 mm strips, then immediately immersed in 13 mL enzymolysis solution (20 mM MES pH 5.8, 0.4 M mannitol, 20 mM KCl, 10 mM CaCl2, 1.5% Cellulase R-10 (Duchefa), 0.5% Macerozyme R-10 (Duchefa)) in a 90 mm Petri dish (Yoo et al., 2007). The tissue was vacuum infiltrated for 30 min and then incubated on an orbital shaker at 50 rpm for 16 h in the dark at 28°C. Protoplast were harvested by adding 10 mL of W5 media (2 mM MES pH 5.8, 154 mM NaCl, 125 mM CaCl2, 5 mM KCl) to the Petri dish and agitating gently, then filtering the suspension through a 70 µM cellulose filter (Fisher Scientific) into a 50 mL centrifuge tube. Protoplast were pelleted by centrifugation (Sigma 3-16 KL) at 200 rcf for 4 minutes with slow centrifugal acceleration and breaking (both set at level 6). The supernatant was removed, and protoplasts were resuspended in W5 for a second wash at 200 rcf for 4 minutes. Finally, the supernatant was removed, and 2 mL 0.4 M mannitol (pH 5.8) was added to resuspend the protoplast.
Protoplast were then purified through a sucrose cushion (Jeong et al., 2021): 6 mL of 0.6 M sucrose solution was gently overlaid with the 2 mL protoplast-mannitol suspension in a 15 mL centrifuge tube and centrifuged at 80 rcf for 5 minutes. Protoplast suspended at the interface were carefully aspirated into a new 15 mL tube and then centrifuged at 200 rcf for 4 minutes and resuspended in 5 mL MMG (4 mM MES pH 5.8, 0.4 M mannitol, 15 mM MgCl2) solution to form a highly concentrated pellet of viable protoplast. Sucrose purification was only successful when initial protoplast yields exceeded 1 × 106 protoplast mL-1.
In vitro cleavage test and gRNA design
PCR product of BWP102 phytoene desaturase (PDS) was used to test the efficacy of two synthetic gRNAs that targeted separate regions of the gene in vitro. PDS in raspberry was identified by BLAST mapping strawberry (Fragaria vesca) PDS sequence to a novel genome assembly of raspberry cultivar BWP102 (Kevei et al., unpub.) in Geneious Prime v.2024.0.7 (Dotmatics, RRID:SCR_010519). gRNAs and primers were designed using the same genome assembly to amplify two separate coding regions of PDS (PDS1, PDS2) with gRNA cut sites producing uneven cleavage products distinguishable by gel electrophoresis (Table 1). gRNA1 and gRNA2 targeted separate loci in exon 13 and eight of PDS respectively. Custom Invitrogen TrueGuide gRNAs and Invitrogen TrueCut Cas9 were commercially synthesised by Thermo Fisher Scientific. Additional, functionally relevant gRNAs and primers were also designed targeting PG, WRKY52 and NPR1 and tested through in vitro cleavage tests (Supplementary Table S1).
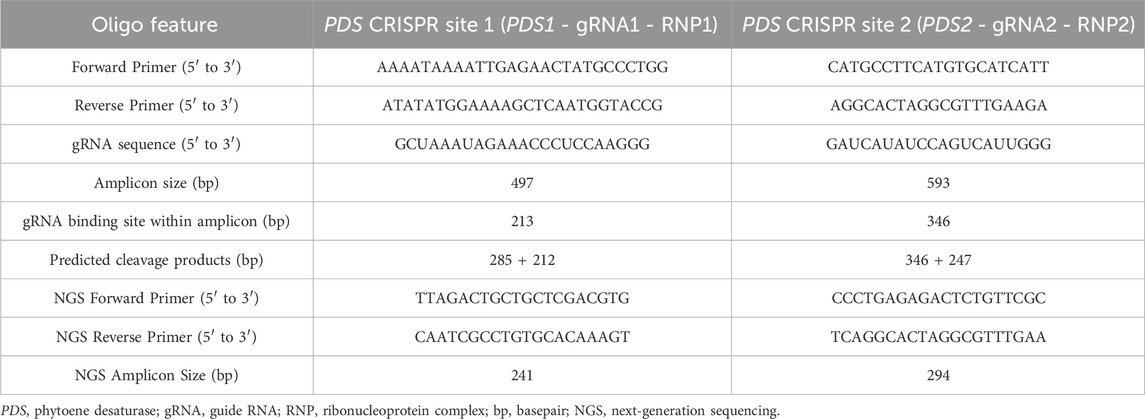
Table 1. Oligonucleotides and gRNAs used in this study. Predicted cleavage products were deduced assuming DSB 3bp upstream from PAM site. NGS primer pairs flank the same respective gRNA binding site.
DNA was extracted from BWP102 leaf tissue with E.Z.N.A Plant DNA kit (Omega Bio-tek) and amplified with Phusion high-fidelity polymerase (Thermo Fisher Scientific), followed by purification with QIAquick PCR purification kit (QIAGEN). The in vitro cleavage test was followed as in Brandt et al. (2020) (Brandt et al., 2020). In brief, 1 µg gRNA and 1 µg Cas9 were pre-mixed and incubated with 200 ng PCR product for 2 hours. The sample was run on a 2% agarose gel (0.5% Tris Borate EDTA buffer, 0.005% Safeview (NBS Biologicals)). RNP activity was identified by the cleavage of the PCR product into two distinct smaller bands of expected sizes.
Protoplast transfection
The protoplast suspension was centrifuged at 200 rcf for 4 minutes with low acceleration and deceleration after sucrose cushion filtration. The supernatant was removed as much as possible; 0.05% fluorescein diacetate (FDA) was added to a 10 µL subsample and protoplast yield was determined via haemocytometry on a fluorescent microscope (Leica Microsystems, DM2500, RRID:SCR_020224). Protoplast concentration was standardised to 1 × 106 cells mL-1 in MMG and 200 µL (2 × 105 protoplast total) was transferred to a sterile 1.5 mL microcentrifuge tube using wide-bore pipette tips.
RNPs were formed by mixing 10 µg of gRNA, 10 µg Cas9, 2 µL lipofectamine CRISPRMAX (Thermo Fisher Scientific), 2 µL Plus reagent, 2 µL NEB 3.1 buffer (New England Biolabs) up to 20 µL with ultrapure H2O in a PCR tube and incubated for 10 min at 25°C. Negative controls were identical but excluded gRNA. RNPs were added to the protoplast suspension followed immediately by 220 µL 40% PEG4000 (Merck) solution (0.2 M mannitol, 0.1 M CaCl2, 2 g PEG4000, up to 5 mL ddH2O) (Scintilla et al., 2022; Wang et al., 2022; González et al., 2020; Gou et al., 2020; Lee et al., 2024). The RNP-protoplast suspension was pipetted 5–10 times with a wide-bore tip and incubated for 10 min (Scintilla et al., 2022; Lee et al., 2024; Wu et al., 2017) at 25°C. Transfection was terminated by the addition of 800 µL of W5 and the suspension transferred to a clean 15 mL tube and centrifuged for 2 minutes at 200 rcf. The supernatant was discarded and 800 µL W5 added for a second wash through centrifugation for 2 minutes at 200 rcf. The supernatant was removed and 500 µL of basic culture media (MS pH 5.8, 0.4 M mannitol, 30 g L-1 sucrose) was added to resuspend the protoplast and the suspension was left for 24 h (Park et al., 2019). Protoplast were lysed by centrifugation at 10,000 rcf for 1 minute, the culture media was aspirated, and DNA was then extracted with the E.Z.N.A Plant DNA kit (Omega Bio-tek).
Determination of editing efficiency with T7EI and deconvolution
Editing efficiency was determined both in vitro and in silico for PDS samples. Protoplast DNA was amplified by PCR with Phusion polymerase with the same respective primer pairs used for the in vitro cleavage test. Amplification was visualised on a 2% agarose gel. PCR products were purified with QIAquick PCR purification kit and the T7 endonuclease I (T7EI) digestion protocol was followed (New England Biolabs, 2023) with minor modifications. In brief, PCR amplicons were denatured at 98°C for 30 s and then annealed by decreasing the temperature gradually by −2°C s-1–85°C, then −0.1°C s-1–25°C. T7EI (New England Biolabs) was added and incubated for 1 hour at 37°C (Brandt et al., 2020). Annealed amplicons were purified again and then run on a 2% agarose gel for 45 min. PCR product from the same genome edited protoplast DNA samples was purified and sent to GENEWIZ (Azenta Life Sciences) for Sanger sequencing. Files from negative control and experimental samples were uploaded onto Tracking of Indels by Decomposition (TIDE) and deconvoluted (Brinkman et al., 2014) to estimate editing efficiency.
Determination of editing efficiency with NGS
PCR product from the same PDS genome edited protoplast DNA samples described for T7EI/deconvolution and for PG, WRKY52 and NPR1 was purified and also sent to GENEWIZ (Azenta Life Sciences) for short-read Illumina next-generation sequencing (NGS; Amplicon-EZ). Different primer pairs flanking the same gRNA binding sites were used for PDS NGS as Amplicon-EZ requires primer pairs <450 bp (Table 1). FASTQ. files from the amplicon sequencing (≤70k sequences) were paired, trimmed and merged. Total wild-type and variant sequence percentages were compared to identify CRISPR indels in Geneious Prime (Prime, 2024).
Results
Tissue culture and protoplast isolation
The quality of the initial raspberry canes was a critical factor in protoplast isolation (Figures 1A,B). Cuttings from highly vigorous, 1-month old canes with no signs of senescence (no wilting/discolouring of any leaves, deep red thorns, thick bright green stem) generated fast growing plantlets (Figure 1C) that yielded between 1 × 106 to 1.2 × 107 protoplasts ml-1, (3.3 × 106 g-1 to 3.6 × 107 g-1) with an average of ∼5 × 106 protoplast ml-1 (1.5 × 107 g-1) (Figures 1F,G).
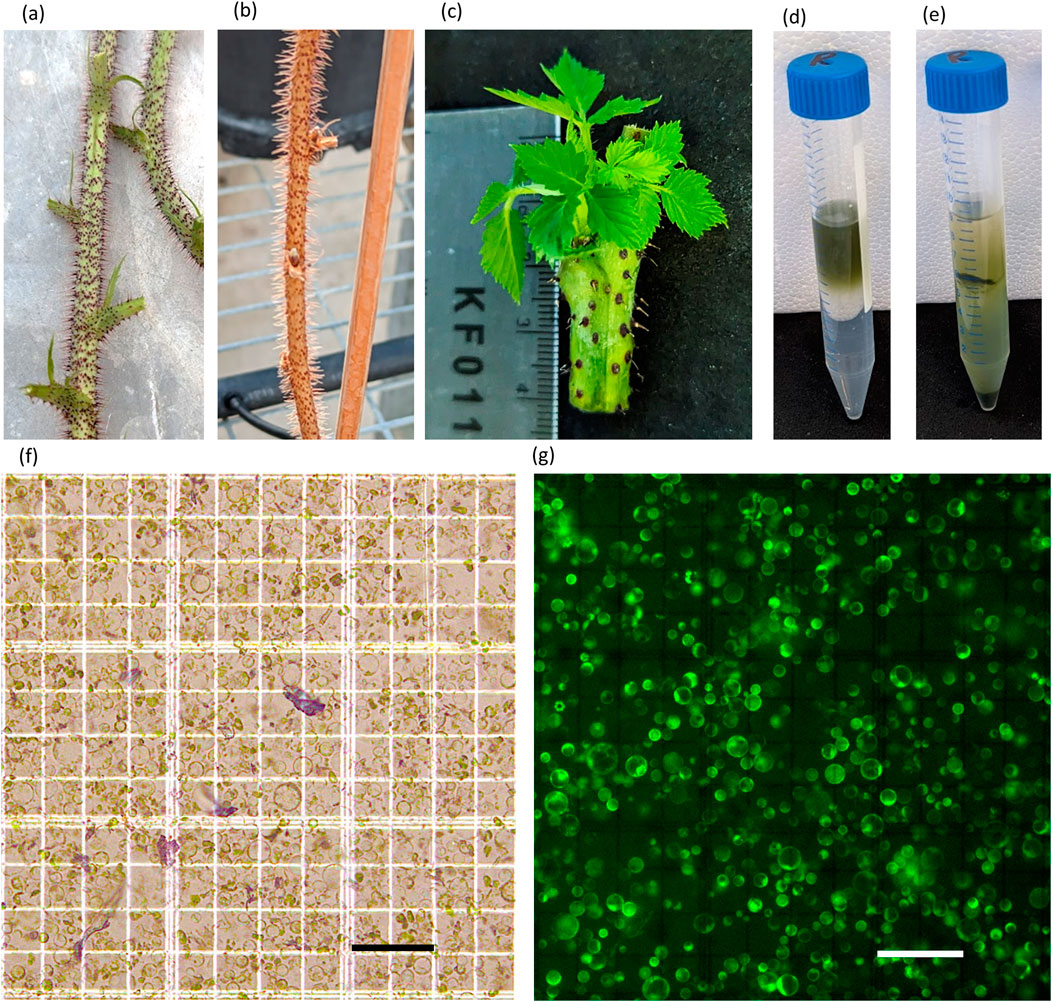
Figure 1. Stages of protoplast isolation in raspberry. (A) High-quality canes used for shoot culture, (B) low-quality cane, (C) young plantlets formed from shoot cultures, (D,E) purification of protoplast before (D) and after (E) sucrose cushion, (F,G) protoplast isolated from plantlets under bright-field (F) and fluorescent (G) illumination (bars = 100 µM).
Protoplast isolated from plantlets were 10–40 µM in diameter and bright green in colour. The sucrose cushion allowed the concentration of viable protoplast and removal of debris and dead cells (Figures 1D,E). FDA staining demonstrated that the vast majority of protoplast were viable for transfection.
In vitro cleavage of DNA amplicons by RNPs
Both PDS RNPs showed a high level of activity against PCR amplicons with two bands clearly visible below the original band (Figures 2A,B). Compared to negative controls, the original PCR product was much reduced in intensity–indicating almost complete cleavage of the amplicons. PDS RNP1 and RNP2 cleavage resulted in predicted bands at approximately 212 bp + 285 bp and 247 bp + 346 bp, respectively. Cleavage by the RNPs was highly specific as no other bands were visible (Figures 2A,B). Successful in vitro cleavage was also seen in amplicons of PG, WRKY52 and NPR1 by their respective RNPs (Supplementary Figure S1).
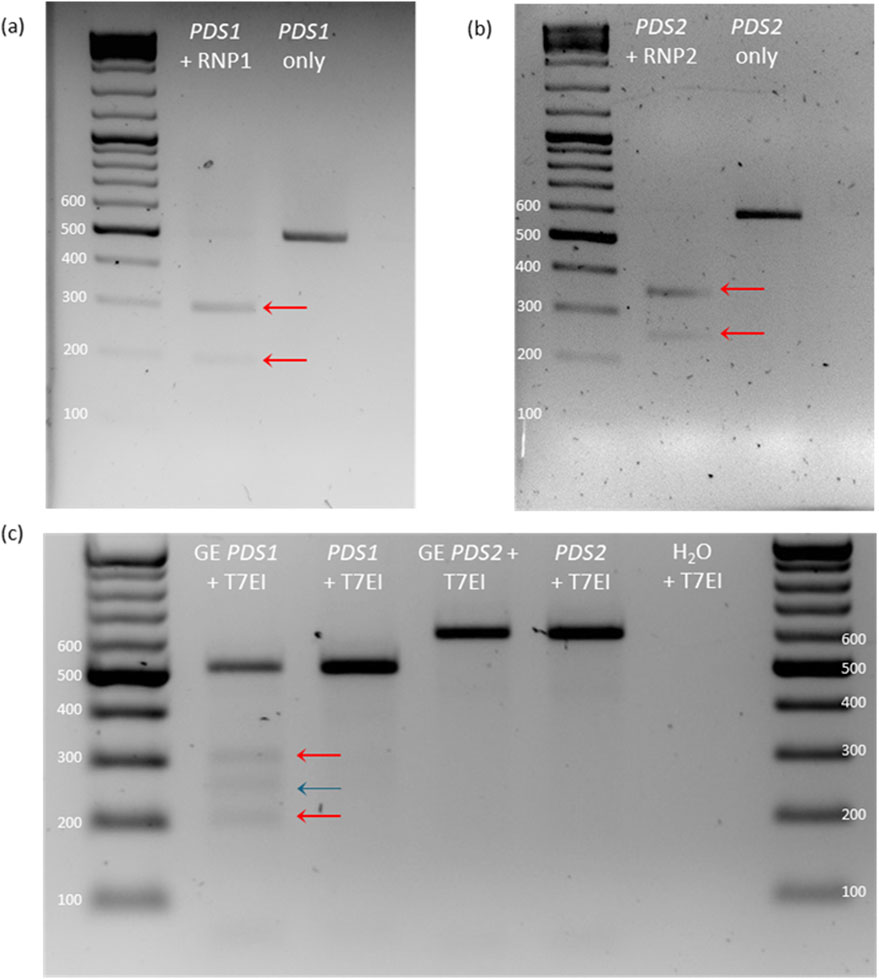
Figure 2. Detection of in vitro RNP activity and genome editing in protoplasts. (A,B) In vitro cleavage test of RNP1 (A) and RNP2 (B) on leaf-derived PDS PCR product. (C) T7EI assay of PDS PCR product of gene edited (GE) protoplast transfected with RNP1 and RNP2. Red arrows indicate predicted cleavage products, and the blue line indicates the additional cleavage product.
Detection of genome editing in protoplasts
The T7EI assay confirmed the presence of mutations in the PDS1 sample through the presence of cleavage products beneath the WT amplicon band. The T7 endonuclease cleaves mismatched DNA, therefore degrading and gradually reannealing a mixed pool of protoplast-derived WT and edited amplicons results in mismatched double-stranded DNA. One strand contains indels produced through genome editing, and the other strand contains the WT sequence, creating a cleavable mismatch. The two bands were approximately the predicted sizes at 212 bp and 285 bp, however there was an additional band at 250 bp. Furthermore, the WT amplicon band was also higher than expected at approximately 510 bp. However, PDS2 showed no cleavage by T7EI (Figure 2C).
Sanger sequencing followed by deconvolution algorithmically estimates genome editing efficiency by aligning the sequence trace files of a gene-edited sequence to a WT sequence. Using the gRNA sequence to determine the probable cut site, deconvolution programmes compare the individual basepair confidence of the two sequences to estimate what indels are present in the gene-edited sample and their frequencies (Brinkman et al., 2014; Bloh et al., 2021). TIDE deconvolution estimated editing efficiencies of 14% for PDS1 with predicted indels ranging from +1 to −5 (Figure 3A). Despite the T7EI assay not producing any visible cleavage bands, editing efficiency was estimated at 6.2% for PDS2 with the most common predicted indel of +1 (Figure 3C). Aberrant signal increased directly downstream of the gRNA binding site, particularly for PDS1, which was also visible on the chromatogram (Supplementary Figures S3A, C).

Figure 3. Estimated indels predicted by Sanger sequencing/TIDE deconvolution and actual indels detected by NGS. (A,B) PDS1 indels (C,D) PDS2 indels. N refers to estimated insertions of unknown nucleotides from TIDE analysis. Vertical dashed lines indicate gRNA cut sites, red arrows indicate gRNA sequences and green lines indicate PAM sequences. All sequences are shown in the 5′ to 3′ direction. Note that gRNA1 binds in the 3′ to 5′ direction.
Amplicon sequencing followed by CRISPR analysis revealed 19.0% editing efficiency at PDS1 with 12 variants (minimum presence in sequence population of 0.1%) ranging from −28 bp to +1 bp (Figure 3B). The most common indel was a 3 bp deletion present at 6% and there was some variation in where the DSB occurred. Some variation may be genuine, but for the most common mutations, DSB variance could be a sequencing artifact due to the start and end of the deletion both being a guanine nucleotide. PDS2 had a 2.3% editing efficiency, with four variants (present at >0.1% of sequences) ranging from −4 bp to +1 bp (Figure 3D). The highest frequency indel was a 1 bp deletion present at 1.2%. CRISPR analysis of PG, WRKY52, NPR1 revealed an editing efficiency of 1.2%, 0.3% and 6.0% respectively (Supplementary Figure S2).
Discussion
These results represent the first instance of DNA-free RNP-mediated genome editing in raspberry protoplast. The transfection efficiency of 19% with pre-assembled CRISPR RNPs is substantial and in line with existing research; recent meta-analysis revealed that the average transfection efficiency of protoplast transfection with PEG was 17% (Rustgi et al., 2022). If gene edited protoplasts can be regenerated into raspberry plants, a 19% transfection efficiency would likely result in higher recovery rate of transformed plants than Agrobacterium-based methods in raspberry, which is currently between 0.5%–2% (Mathews et al., 1995; Georgieva et al., 2008). However, the editing efficiencies identified here are highly variable between different gRNAs (0.2%–19%), and while similar variation has been found elsewhere (Rustgi et al., 2022), more consistency in editing efficiency would be beneficial. Differences in gRNA efficiency is hypothesised to be due to variance in the accessibility of chromatin at the binding locus (Lee et al., 2024; Van Campenhout et al., 2019; Uusi-Mäkelä et al., 2018), therefore large-scale screening of gRNAs may be unavoidable. Nonetheless, successful DNA-free editing in PG, WRKY52 and NPR1 demonstrates the versatility of this protocol and its potential with agriculturally relevant functional targets. The development of novel protoplast-based transfection protocols in crop species has increased rapidly in recent years, with new methods in grapevine (Scintilla et al., 2022; Najafi et al., 2023; Tricoli and Debernardi, 2024), tomato (Lin et al., 2022; Liu et al., 2022), strawberry (Barceló et al., 2019; Gou et al., 2020), lettuce (Park et al., 2019), and potato (Moon et al., 2021), reflecting widespread interest. As genome-editing research accelerates in Arabidopsis, tobacco, wheat and rice (Shan et al., 2020), it will be vital to invest in method development in a wider range of crops such as raspberry to ensure that NGBTs can be fully deployed. With established DNA-free methods, raspberry breeders could target gene orthologs involved in firmness (e.g., PG (Atkinson et al., 2012; Posé et al., 2013)), B. cinerea resistance (e.g., WRKY52 (Jia et al., 2020); NPR1 [Li et al., 2020; Li et al., 2021)], and powdery mildew resistance [e.g., WRY52 (Wang et al., 2017)] that may improve shelf life, a major breeding target (Funt and Hall, 2013). Fruit flavour/taste, cane habit, plant architecture, aphid resistance and local climate adaptation (drought/flood tolerance) could also be broader targets for raspberry breeding through genome editing if associated genomic and phenomic links can be elucidated. This study demonstrates the amenability of raspberry to protoplast isolation, with considerably higher protoplast yields (average 1.5 × 107 protoplast g-1) than previously reported (3.93 × 106 g-1) (Phan et al., 1997). Yields from BWP102 are also higher than recent reports in woody species such as grapevine (6.6 × 106 g-1) (Scintilla et al., 2022) and comparable to yields of 1 × 107 protoplast g-1 seen in Arabidopsis (Yoo et al., 2007). The most important variable for successful protoplast isolation in raspberry, as found elsewhere (Yoo et al., 2007; Barceló et al., 2019), is the tissue quality used as a protoplast source. The outbreeding nature and high heterozygosity of raspberry adds complexity, compared to many other inbreeding species, as seeds cannot be used to grow sterile, soft, young plants for enzymolysis. As such, each raspberry seed is genetically different, and the genetic backgrounds of elite cultivars must be preserved to maintain berry consistency and quality. Therefore, vegetative stem cultures are a good alternative protoplast source. However, stem cultures are time and energy intensive, susceptible to contamination, and the health/quality of the canes is hugely influential on the protoplast yield. Although not systematically explored here, anecdotal evidence suggests indicators that improve protoplast yield include bright green stems that had deep red thorns and healthy lower leaves, alongside rapid growth of plantlets (within 10 days) from axillary buds in tissue culture. In future, isolation from callus could be an alternative protoplast source (Kang et al., 2020) to solve issues such as contamination and yield inconsistency.
Raspberry tissue remained mostly undigested despite 16 h of digestion, in contrast to Arabidopsis where leaves almost fully disintegrate into the enzymolysis solution (Jeong et al., 2021). As a woody species, raspberry likely has higher concentrations of structural polymers, such as lignin and cellulose, compared to model species. We hypothesise that levels of structural polymers in raspberry have a positive correlation to age and abiotic stress (Wang et al., 2016). Therefore, high levels of structural polymers may interfere with protoplast isolation (Brandt et al., 2020), resulting in lower yields from plantlets derived from older canes in poor health.
Pre-assembled RNPs were used in this study for several reasons. Primarily, non-transgenic crops are preferable as new legislation in England (Genetic Technology, 2023) permits only precision bred crops with mutations that could theoretically be produced by natural mutagenesis. Similar legislation is planned in the European Union (European Commission, 2021). The genomes of genetically modified crops inevitably contain foreign DNA (Agrobacterium T-DNA, Cas9/gRNA DNA or plasmid sequences) which need to be removed through backcrossing. As previously stated, backcrossing is not practical in raspberry, and in many other soft fruits, as the species is highly heterozygous and outbreeding. Public perception of transgenic food also remains a major issue (Woźniak-Gientka et al., 2022) and would likely influence marketability and eventual sales. Transgene-free cultivars generated by pre-assembled RNPs with enhanced phenotypes present a solution that research shows is more acceptable with consumers when used to improve sustainability or for societal benefit (Sprink et al., 2022). Furthermore, the ability to rapidly introduce targeted non-transgenic mutations directly in clones with elite genetic backgrounds would be a significant advantage compared to the years of selection and trials required for conventional raspberry breeding. Floricane cultivars of raspberry take 2 years to fruit, thus RNP-based methods represent a great increase in breeding speed and accuracy. However, the above advantages still rely upon the elucidation of a protoplast regeneration protocol in raspberry (Phan et al., 1997).
Plasmid-derived RNPs would likely be effective in this protocol, and we have successfully tested plasmid YFP expression in BWP102 protoplasts (Supplementary Figure S4). However, the risk of genomic plasmid integration (Kim et al., 2014) resulted in a preference for pre-assembled RNPs. Commercially synthesised Cas9 and gRNA enable high purity and sequence accuracy, which promotes better editing efficiencies. Furthermore, it reduces the technical knowledge required to construct new plasmids for transfection. However, synthetic gRNAs and Cas9 are expensive, therefore once gRNAs are validated, it may be cost-efficient for further work to synthesise RNPs in-house.
The cleavage test validated the efficacy of the gRNAs in vitro. Screening of gRNAs through cleavage tests is recommended prior to protoplast transfection (Brandt et al., 2020) to validate the activity of gRNAs using minimal components. Availability of cultivar-specific genome assemblies were crucial for the design of specific gRNAs and oligonucleotides with no sequence mismatches or off-target complementarity. The assembly used in this study is intended for publication (Kevei et al., unpub.), however other raspberry cultivar genome sequences are available (Davik et al., 2022; Price et al., 2023).
Quantifying genome editing efficiency in an amplicon pool is complex. T7EI and Sanger sequencing/deconvolution were used initially, which revealed positive results for PDS1, therefore amplicon sequencing was additionally conducted for comprehensive analysis. Sanger sequencing followed by computational deconvolution offers a low-cost alternative when NGS is financially unfeasible. Many free-to-use programs exist, namely, TIDE (Brinkman et al., 2014), ICE (Conant et al., 2022) and DECODR (Bloh et al., 2021). While useful, discrepancies between different programs mean they cannot be singularly relied upon (Brockman et al., 2023). Thus, if only using Sanger sequencing, editing estimates should be complemented with in vitro proof with T7EI digestion. Taken together, these two analysis techniques estimate what genotypes are present in the population and their relative frequencies at low cost. In this study, the T7EI assay demonstrated successful editing with RNP1, however did not validate the evidence of editing with RNP2 from TIDE, likely because editing was at a much lower frequency. The additional third band present in the T7EI PDS1 sample could be explained by the endonuclease binding to a proportion of the DNA, reducing mobility through the agarose gel (New England Biolabs, 2024). Only amplicon sequencing was sensitive enough to detect the low editing efficiency of RNP2. Comparing the results of deconvolution and NGS reveals the former to be a rough estimation of true indels and their frequencies. Many correct indels were detected, however frequencies within the sequence pool were not accurate. Until recently, amplicon sequencing has been prohibitively expensive, and so methods such as T7EI and deconvolution were appealing. However, the decreasing cost of sequencing technologies, use of short-read (<500 bp) sequencing and the lack of high levels of accuracy with alternative methods, as identified here, indicates that amplicon NGS is the best choice for the detection of genome editing.
PDS was chosen as a proof-of-concept reporter gene as null mutants would present a distinctive albino phenotype early in protoplast regeneration. We also demonstrate editing in three agriculturally-relevant functional genes, null mutants of which have been shown to have increased firmness (PG) (Atkinson et al., 2012; Posé et al., 2013) and resistance to B. cinerea (WRKY52 and NPR1) (Jia et al., 2020; Li et al., 2021) and powdery mildew (WRKY52) (Wang et al., 2017; Li et al., 2021) in other fruit species. In tandem with genome assemblies, this method enables rapid editing of any loci within the raspberry genome. However, as widely documented (Reed and Bargmann, 2021; Liu et al., 2022), protoplast regeneration of gene edited elite lines remains the greatest challenge to commercial implementation.
Reed and Bargmann (2021); Liu et al. (2022) This study aims to instigate further research into precision breeding in Rubus species by providing a straightforward, effective and reproducible protoplast isolation and quantifiable DNA-free genome editing protocol. Avenues have been opened for future work on gene expression and function. If protoplast regeneration methods can be elucidated in raspberry, genetic improvement of many raspberry traits will become feasible, with substantial benefits for the sustainability and efficiency of raspberry production.
Data availability statement
Datasets are available in a publicly accessible repository: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA1245325.
Author contributions
RC: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Writing – original draft, Writing – review and editing. AT: Funding acquisition, Project administration, Resources, Supervision, Writing – review and editing. ZK: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by BerryWorld Plus™.
Acknowledgments
We thank Dr Yaomin Cai (Earlham Institute) and Dr Mehmet Fatih Kara (University of York) for their advice on protoplast isolation and transfection and Dr Tomasz Kurowski (Cranfield University) for his work on the genome assembly. This manuscript is also available on BioRxiv (Creeth et al., 2025).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgeed.2025.1589431/full#supplementary-material
References
Atkinson, R. G., Sutherland, P. W., Johnston, S. L., Gunaseelan, K., Hallett, I. C., Mitra, D., et al. (2012). Down-regulation of POLYGALACTURONASE1 alters firmness, tensile strength and water loss in apple (Malus x domestica) fruit. BMC Plant Biol. 12, 129. doi:10.1186/1471-2229-12-129
Barceló, M., Wallin, A., Medina, J. J., Gil-Ariza, D. J., López-Casado, G., Juarez, J., et al. (2019). Isolation and culture of strawberry protoplasts and field evaluation of regenerated plants. Sci. Hortic. 256, 108552. doi:10.1016/j.scienta.2019.108552
Bloh, K., Kanchana, R., Bialk, P., Banas, K., Zhang, Z., Yoo, B.-C., et al. (2021). Deconvolution of complex DNA repair (DECODR): establishing a novel deconvolution algorithm for comprehensive analysis of CRISPR-edited sanger sequencing data. CRISPR J. 4, 120–131. doi:10.1089/crispr.2020.0022
Brandt, K. M., Gunn, H., Moretti, N., and Zemetra, R. S. (2020). A streamlined protocol for wheat (Triticum aestivum) protoplast isolation and transformation with CRISPR-cas ribonucleoprotein complexes. Front. Plant Sci. 11, 769. doi:10.3389/fpls.2020.00769
Brinkman, E. K., Chen, T., Amendola, M., and van Steensel, B. (2014). Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168. doi:10.1093/nar/gku936
Brockman, Q. R., Scherer, A., McGivney, G. R., Gutierrez, W. R., Rytlewski, J., Sheehan, A., et al. (2023). Discrepancies in indel software resolution with somatic CRISPR/Cas9 tumorigenesis models. Sci. Rep. 13, 14798. doi:10.1038/s41598-023-41109-1
Chan, R. L., Trucco, F., and Otegui, M. E. (2020). Why are second-generation transgenic crops not yet available in the market? J. Exp. Bot. 71, 6876–6880. doi:10.1093/jxb/eraa412
Cocking, E. C. (1960). A method for the isolation of plant protoplasts and vacuoles. Nature 187, 962–963. doi:10.1038/187962a0
Conant, D., Hsiau, T., Rossi, N., Oki, J., Maures, T., Waite, K., et al. (2022). Inference of CRISPR edits from sanger trace data. CRISPR J. 5, 123–130. doi:10.1089/crispr.2021.0113
Creeth, R., Thompson, A., and Kevei, Z. (2025). DNA-Free CRISPR genome editing in raspberry (Rubus idaeus) through RNP-mediated protoplast transfection and comparison of indel analysis techniques. BioRxiv. doi:10.1101/2025.01.14.632935
Davey, M. R., Anthony, P., Power, J. B., and Lowe, K. C. (2005). Plant protoplasts: status and biotechnological perspectives. Biotechnol. Adv. 23, 131–171. doi:10.1016/j.biotechadv.2004.09.008
Davik, J., Røen, D., Lysøe, E., Buti, M., Rossman, S., Alsheikh, M., et al. (2022). A chromosome-level genome sequence assembly of the red raspberry (Rubus idaeus L.). PLoS One 17, e0265096. doi:10.1371/journal.pone.0265096
Derrick, S. A., Kristo, A. S., Reaves, S. K., and Sikalidis, A. K. (2021). Effects of dietary red raspberry consumption on pre-diabetes and type 2 diabetes mellitus parameters. Int. J. Environ. Res. Public Health 18, 9364. doi:10.3390/ijerph18179364
European Commission (2021). Proposal for a regulation of the European parliament and of the council on plants obtained by certain new genomic techniques and their food and feed, and amending Regulation (EU).
FAO (2024). Crops and livestock products. (Accessed December 4, 2023). Available online at: https://www.fao.org/faostat/en/#data/QCL.
FAO. Emissions due to agriculture (2020). Global, regional and country trends 2000–2018, 18. Rome: FAOSTAT Analytical Brief Series No.
Foster, T. M., Bassil, N. V., Dossett, M., Leigh Worthington, M., and Graham, J. (2019). Genetic and genomic resources for Rubus breeding: a roadmap for the future. Hortic. Res. 6, 116. doi:10.1038/s41438-019-0199-2
Gasiunas, G., Barrangou, R., Horvath, P., and Siksnys, V. (2012). Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. 109, E2579–E2586. doi:10.1073/pnas.1208507109
Georgieva, M., Kondakova, K., Djilyanov, D., Badjakov, I., and Yancheva, S. (2008). Genetic transformation of raspberries by means of Agrobacterium tumefaciens. Analele universitatii din craiova, seria biologie, horticultura, tehnologia prelucra rii produselor agricole. Ing. Mediu. 13, 5–14.
González, M. N., Massa, G. A., Andersson, M., Turesson, H., Olsson, N., Fält, A. S., et al. (2020). Reduced enzymatic browning in potato tubers by specific editing of a polyphenol oxidase gene via ribonucleoprotein complexes delivery of the CRISPR/Cas9 system. Front. Plant Sci. 10, 497481. doi:10.3389/fpls.2019.01649
Gou, Y. J., Li, Y. L., Bi, P. P., Wang, D. J., Ma, Y. Y., Hu, Y., et al. (2020). Optimization of the protoplast transient expression system for gene functional studies in strawberry (Fragaria vesca). Plant Cell. Tissue Organ Cult. 141, 41–53. doi:10.1007/s11240-020-01765-x
Hancock, R. D., McDougall, G. J., and Stewart, D. (2007). Berry fruit as ‘superfood’: hope or hype. Biologist 54, 73–79.
Hoagland, D., and Arnon, D. (1950). The water-culture method for growing plants without soil. Calif. Agric. Exp. Stn. 347, 35–37.
Hsu, C.-T., Lee, W.-C., Cheng, Y.-J., Yuan, Y.-H., Wu, F.-H., and Lin, C.-S. (2021). Genome editing and protoplast regeneration to study plant–pathogen interactions in the model plant nicotiana benthamiana. Front. Genome 2, 627803. doi:10.3389/fgeed.2020.627803
Jeong, Y. Y., Lee, H.-Y., Kim, S. W., Noh, Y.-S., and Seo, P. J. (2021). Optimization of protoplast regeneration in the model plant Arabidopsis thaliana. Plant Methods 17, 21. doi:10.1186/s13007-021-00720-x
Jia, S., Wang, Y., Zhang, G., Yan, Z., and Cai, Q. (2020). Strawberry FaWRKY25 transcription factor negatively regulated the resistance of strawberry fruits to Botrytis cinerea. Genes. (Basel) 12, 56–17. doi:10.3390/GENES12010056
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (1979)2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi:10.1126/science.1225829
Kang, H. H., Naing, A. H., and Kim, C. K. (2020). Protoplast isolation and shoot regeneration from protoplast-derived callus of petunia hybrida cv. Mirage rose. Biol. (Basel) 9, 228. doi:10.3390/biology9080228
Khadgi, A. (2020). Uncovering genetic and transcriptomic regulation of prickle development in red raspberry. Cornell University.
Kim, C., and Dai, W. (2022). Agrobacterium-mediated transformation of purple raspberry (Rubus occidentalis × R. idaeus) with the PtFIT (FER-like iron deficiency–induced transcription factor 1) gene. Vitro Cell. and Dev. Biol. - Plant 58, 343–350. doi:10.1007/s11627-021-10228-7
Kim, S., Kim, D., Cho, S. W., Kim, J., and Kim, J.-S. (2014). Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 24, 1012–1019. doi:10.1101/gr.171322.113
Lee, S., Park, S. H., Jeong Jeong, Y., Kim, S., Kim, B. R., Ha, B. K., et al. (2024). Optimization of CRISPR/Cas9 ribonucleoprotein delivery into cabbage protoplasts for efficient DNA-free gene editing. Plant Biotechnol. Rep. 18, 415–424. doi:10.1007/s11816-024-00901-9
Li, R., Li, Y., Zhang, Y., Sheng, J., Zhu, H., and Shen, L. (2021). Transcriptome analysis reveals that SlNPR1 mediates tomato fruit resistance against Botrytis cinerea by modulating phenylpropanoid metabolism and balancing ROS homeostasis. Postharvest Biol. Technol. 172, 111382. doi:10.1016/J.POSTHARVBIO.2020.111382
Li, R., Wang, L., Li, Y., Zhao, R., Zhang, Y., Sheng, J., et al. (2020). Knockout of SlNPR1 enhances tomato plants resistance against Botrytis cinerea by modulating ROS homeostasis and JA/ET signaling pathways. Physiol. Plant 170, 569–579. doi:10.1111/PPL.13194
Lin, C.-S., Hsu, C.-T., Yang, L.-H., Lee, L.-Y., Fu, J.-Y., Cheng, Q.-W., et al. (2018). Application of protoplast technology to CRISPR/Cas9 mutagenesis: from single-cell mutation detection to mutant plant regeneration. Plant Biotechnol. J. 16, 1295–1310. doi:10.1111/pbi.12870
Lin, C. S., Hsu, C. T., Yuan, Y. H., Zheng, P. X., Wu, F. H., Cheng, Q. W., et al. (2022). DNA-free CRISPR-Cas9 gene editing of wild tetraploid tomato Solanum peruvianum using protoplast regeneration. Plant Physiol. 188, 1917–1930. doi:10.1093/plphys/kiac022
Liu, Y., Andersson, M., Granell, A., Cardi, T., Hofvander, P., and Nicolia, A. (2022). Establishment of a DNA-free genome editing and protoplast regeneration method in cultivated tomato (Solanum lycopersicum). Plant Cell. Rep. 41, 1843–1852. doi:10.1007/s00299-022-02893-8
López-Casado, G., Sánchez-Raya, C., Ric-Varas, P. D., Paniagua, C., Blanco-Portales, R., Muñoz-Blanco, J., et al. (2023). CRISPR/Cas9 editing of the polygalacturonase FaPG1 gene improves strawberry fruit firmness. Hortic. Res. 10, uhad011. doi:10.1093/hr/uhad011
Mathews, H., Wagoner, W., Cohen, C., Kellogg, J., and Bestwick, R. (1995). Efficient genetic transformation of red raspberry, Rubus ideaus L. Plant Cell. Rep. 14, 471–476. doi:10.1007/BF00232777
Mezzetti, B., Costantini, E., Chionchetti, F., Landi, L., Pandolfini, T., and Spena, A. (2004). Genetic transformation in strawberry and raspberry for improving plant productivity and quality. Acta Hortic., 107–110. doi:10.17660/ActaHortic.2004.649.19
Mezzetti, B., Landi, L., Phan, H. B., Mantovani, I., Ruggieri, S., Rosati, P., et al. (1999). Protoplast technology and regeneration studies for Rubus breeding. Acta Hortic., 215–222. doi:10.17660/ActaHortic.1999.505.27
Miller, S. (2019). Gene editing of red raspberry (Rubus ideaus L.) with CRISPR/Cas9 knocking out F3’H. Ås, Norway: Norwegian University of Life Sciences.
Moon, K.-B., Park, J.-S., Park, S.-J., Lee, H.-J., Cho, H.-S., Min, S.-R., et al. (2021). A more accessible, time-saving, and efficient method for in vitro plant regeneration from potato protoplasts. Plants 10, 781. doi:10.3390/plants10040781
Najafi, S., Bertini, E., D’Incà, E., Fasoli, M., and Zenoni, S. (2023). DNA-free genome editing in grapevine using CRISPR/Cas9 ribonucleoprotein complexes followed by protoplast regeneration. Hortic. Res. 10, uhac240. doi:10.1093/hr/uhac240
New England Biolabs (2024). Quick Tips - why don’t my bands on a gel look correct after T7 digestion?
Nita-Lazar, M., Iwahara, S., Takegawa, K., and Lienart, Y. (2000). The active oxygen response of raspberry protoplasts to O-glycans of Fusarium sp. M7-1. J. Plant Physiol. 156, 306–311. doi:10.1016/S0176-1617(00)80066-8
Park, J., Choi, S., Park, S., Yoon, J., Park, A. Y., and Choe, S. (2019). “DNA-free genome editing via ribonucleoprotein (RNP) delivery of CRISPR/Cas in lettuce,” in Plant genome editing with CRISPR systems. Editor Y. Qi, 1917. 1st ed., 337–354. doi:10.1007/978-1-4939-8991-1_25
Phan, B. H., Lucia, L., Taruschio, L., Mott, D., Mezzetti, B., and Rosati, P. (1997). Protoplast isolation, culture, and cell differentiation in raspberry and blackberry cultivars (Rubus spp.). Angew. Bot. 71, 131–137.
Posé, S., Paniagua, C., Cifuentes, M., Blanco-Portales, R., Quesada, M. A., and Mercado, J. A. (2013). Insights into the effects of polygalacturonase FaPG1 gene silencing on pectin matrix disassembly, enhanced tissue integrity, and firmness in ripe strawberry fruits. J. Exp. Bot. 64, 3803–3815. doi:10.1093/JXB/ERT210
Price, R. J., Davik, J., Fernandéz Fernandéz, F., Bates, H. J., Lynn, S., Nellist, C. F., et al. (2023). Chromosome-scale genome sequence assemblies of the ‘Autumn Bliss’ and ‘Malling Jewel’ cultivars of the highly heterozygous red raspberry (Rubus idaeus L.) derived from long-read Oxford Nanopore sequence data. PLoS One 18, e0285756. doi:10.1371/journal.pone.0285756
Prime, G. (2024). Analyze CRISPR editing. Available online at: https://www.geneiouscom/tutorials/analyze-crispr-editing.
Reed, K. M., and Bargmann, B. O. R. (2021). Protoplast regeneration and its use in new plant breeding technologies. Front. Genome 3, 734951. doi:10.3389/fgeed.2021.734951
Rustgi, S., Naveed, S., Windham, J., Zhang, H., and Demirer, G. S. (2022). Plant biomacromolecule delivery methods in the 21st century. Front. Genome 4, 1011934. doi:10.3389/fgeed.2022.1011934
Scintilla, S., Salvagnin, U., Giacomelli, L., Zeilmaker, T., Malnoy, M. A., Rouppe van der Voort, J., et al. (2022). Regeneration of non-chimeric plants from DNA-free edited grapevine protoplasts. Front. Plant Sci. 13, 1078931. doi:10.3389/fpls.2022.1078931
Scolari, L. M., Hancock, R. D., Graham, J., Freitag, S., Verrall, S., Moreno-Mellado, A. R., et al. (2023). Hormone profiling in artificially induced ‘crumbly’ fruit in raspberry (Rubus idaeus L.) at two different development stages. J. Hortic. Sci. Biotechnol. 98, 384–393. doi:10.1080/14620316.2022.2141140
Shan, S., Soltis, P. S., Soltis, D. E., and Yang, B. (2020). Considerations in adapting CRISPR/Cas9 in nongenetic model plant systems. Appl. Plant Sci. 8, e11314. doi:10.1002/aps3.11314
Simpson, C. G., Cullen, D. W., Hackett, C. A., Smith, K., Hallett, P. D., McNicol, J., et al. (2017). Mapping and expression of genes associated with raspberry fruit ripening and softening. Theor. Appl. Genet. 130, 557–572. doi:10.1007/s00122-016-2835-7
Sprink, T., Wilhelm, R., and Hartung, F. (2022). Genome editing around the globe: an update on policies and perceptions. Plant Physiol. 190, 1579–1587. doi:10.1093/plphys/kiac359
Sternberg, S. H., and Doudna, J. A. (2015). Expanding the biologist’s toolkit with CRISPR-cas9. Mol. Cell. 58, 568–574. doi:10.1016/j.molcel.2015.02.032
Tricoli, D. M., and Debernardi, J. M. (2024). An efficient protoplast-based genome editing protocol for Vitis species. Hortic. Res. 11, uhad266. doi:10.1093/hr/uhad266
Turnbull, C., Lillemo, M., and Hvoslef-Eide, T. A. K. (2021). Global regulation of genetically modified crops amid the gene edited crop boom – a review. Front. Plant Sci. 12, 630396. doi:10.3389/fpls.2021.630396
United Nations (2024). The 17 Goals. Sustain. Dev. Available online at: https://sdgs.un.org/goals (Accessed February 27, 2025).
Uusi-Mäkelä, M. I. E., Barker, H. R., Bäuerlein, C. A., Häkkinen, T., Nykter, M., and Rämet, M. (2018). Chromatin accessibility is associated with CRISPR-Cas9 efficiency in the zebrafish (Danio rerio). PLoS One 13, e0196238. doi:10.1371/JOURNAL.PONE.0196238
Van Campenhout, C., Cabochette, P., Veillard, A. C., Laczik, M., Zelisko-Schmidt, A., Sabatel, C., et al. (2019). Guidelines for optimized gene knockout using CRISPR/Cas9. Biotechniques 66, 295–302. doi:10.2144/BTN-2018-0187
Wang, T., McFarlane, H. E., and Persson, S. (2016). The impact of abiotic factors on cellulose synthesis. J. Exp. Bot. 67, 543–552. doi:10.1093/jxb/erv488
Wang, X., Guo, R., Tu, M., Wang, D., Guo, C., Wan, R., et al. (2017). Ectopic expression of the wild grape WRKY transcription factor VqWRKY52 in Arabidopsis thaliana enhances resistance to the biotrophic pathogen powdery mildew but not to the necrotrophic pathogen Botrytis cinerea. Front. Plant Sci. 8, 97. doi:10.3389/FPLS.2017.00097
Wang, Y., Zhang, Y., Dong, Y., Li, D., Shi, S., Li, S., et al. (2022). A highly efficient mesophyll protoplast isolation and PEG-mediated transient expression system in eggplant. Sci. Hortic. 304, 111303. doi:10.1016/j.scienta.2022.111303
Woo, J. W., Kim, J., Kwon, S., Corvalán, C., Cho, S. W., Kim, H., et al. (2015). DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 33, 1162–1164. doi:10.1038/nbt.3389
Woźniak-Gientka, E., Agata, T., Milica, P., Anna, B., Dennis, E., Nick, V., et al. (2022). Public perception of plant gene technologies worldwide in the light of food security. Gm. Crops Food 13, 218–241. doi:10.1080/21645698.2022.2111946
Wright, A. V., Nuñez, J. K., and Doudna, J. A. (2016). Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell. 164, 29–44. doi:10.1016/J.CELL.2015.12.035
Wu, J. Z., Liu, Q., Geng, X. S., Li, K. M., Luo, L. J., and Liu, J. P. (2017). Highly efficient mesophyll protoplast isolation and PEG-mediated transient gene expression for rapid and large-scale gene characterization in cassava (Manihot esculenta Crantz). BMC Biotechnol. 17, 29–8. doi:10.1186/s12896-017-0349-2
Yoo, S.-D., Cho, Y.-H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi:10.1038/nprot.2007.199
Yue, J.-J., Yuan, J.-L., Wu, F.-H., Yuan, Y.-H., Cheng, Q.-W., Hsu, C.-T., et al. (2021). Protoplasts: from isolation to CRISPR/Cas genome editing application. Front. Genome 3, 717017. doi:10.3389/FGEED.2021.717017
Zhan, X., Lu, Y., Zhu, J., and Botella, J. R. (2021). Genome editing for plant research and crop improvement. J. Integr. Plant Biol. 63, 3–33. doi:10.1111/jipb.13063
Zhang, J., Yan, X., Huang, T., Liu, H., Liu, F., Yang, M., et al. (2022b). Overexpressing 4-coumaroyl-CoA ligase and stilbene synthase fusion genes in red raspberry plants leads to resveratrol accumulation and improved resistance against Botrytis cinerea. J. Plant Biochem. Biotechnol. 32, 85–91. doi:10.1007/s13562-022-00784-3
Keywords: CRISPR-Cas9, genome editing, Rubus idaeus, DNA-free, RNPs, protoplasts
Citation: Creeth R, Thompson A and Kevei Z (2025) DNA-free CRISPR genome editing in raspberry (Rubus idaeus) protoplast through RNP-mediated transfection. Front. Genome Ed. 7:1589431. doi: 10.3389/fgeed.2025.1589431
Received: 07 March 2025; Accepted: 13 June 2025;
Published: 30 June 2025.
Edited by:
Yunpeng Gai, Beijing Forestry University, ChinaReviewed by:
Lida Fuentes-Viveros, Pontifical Catholic University of Valparaíso, ChileJunjie Tan, Nanjing Agricultural University, China
Copyright © 2025 Creeth, Thompson and Kevei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ryan Creeth, cnlhbi5jcmVldGhAY3JhbmZpZWxkLmFjLnVr; Zoltan Kevei, ei5sLmtldmVpQGNyYW5maWVsZC5hYy51aw==
 Ryan Creeth
Ryan Creeth Andrew Thompson
Andrew Thompson Zoltan Kevei
Zoltan Kevei