- 1Division Research and Landsteiner Laboratory, Department of Immunopathology, Sanquin Blood Supply, Amsterdam University Medical Center (UMC), Amsterdam, Netherlands
- 2Department of Pediatric Immunology, Rheumatology and Infectious Diseases, Emma Children’s Hospital, Amsterdam UMC, Amsterdam, Netherlands
- 3Division Research and Landsteiner Laboratory, Department of Blood Cell Research, Sanquin Blood Supply, Amsterdam UMC, Amsterdam, Netherlands
Febrile patients, suffering from an infection, inflammatory disease or autoimmunity may present with similar or overlapping clinical symptoms, which makes early diagnosis difficult. Therefore, biomarkers are needed to help physicians form a correct diagnosis and initiate the right treatment to improve patient outcomes following first presentation or admittance to hospital. Here, we review the landscape of novel biomarkers and approaches of biomarker discovery. We first discuss the use of current plasma parameters and whole blood biomarkers, including results obtained by RNA profiling and mass spectrometry, to discriminate between bacterial and viral infections. Next we expand upon the use of biomarkers to distinguish between infectious and non-infectious disease. Finally, we discuss the strengths as well as the potential pitfalls of current developments. We conclude that the use of combination tests, using either protein markers or transcriptomic analysis, have advanced considerably and should be further explored to improve current diagnostics regarding febrile infections and inflammation. If proven effective when combined, these biomarker signatures will greatly accelerate early and tailored treatment decisions.
Introduction
Fever is a common symptom in children and adults experiencing a wide variety of conditions. Fever in children signifies systemic inflammation, typically in response to a bacterial, viral, and sometimes parasitic infection, or less commonly, a non-infectious etiology (1).
Additional causes of fever must be considered for adults, malignancies in particular (2). To discriminate between a broad range of infectious and non-infectious febrile diseases, physicians must rely on longstanding experience, interviewing skills and thorough physical examination, as well as the right diagnostic tools such as microbiological tests and imaging (3). In many cases the exact trigger for the febrile condition is not found immediately at the first consultation or admittance to the hospital (4) and antibiotics, sometimes combined with antiviral medication, are started (5). For optimal treatment of disease, rapid diagnosis is of the essence and early diagnostic biomarkers that can reliably indicate the cause of fever within hours by rapid testing, are still lacking.
As mentioned, antibiotics are often prescribed whilst a physician is uncertain whether a bacterial or viral infection is present. This is particular true for children with little capacity to express themselves and in whom disease progression may take place more rapidly than in adults. This extensive use of antimicrobials comes with a price: inducing antimicrobial resistance which is increasing worldwide (5–8). The first important goal for using biomarkers in diagnostics is to discriminate between bacterial and viral infections in febrile patients, so limiting antibiotic use. Additional to bacterial and viral infection, parasitic infections cause a tremendous burden of disease in both the tropics and subtropics. Malaria, is not included here because of the many tests available to rapidly diagnose this infection (9), despite being the only parasite infestation that does cause fever in the acute stage of infection (10)
Beyond the distinction between bacterial versus viral disease, fever can be part of the early presentation of many autoinflammatory or autoimmune diseases. These are rare, more difficult to diagnose and is often mistaken for an infectious disorder. In the most severe presentations of an infection, we observe that almost all septic patients also suffer from systemic inflammation. However, not all patients with a systemic inflammatory syndrome are septic or suffer from a systemic viral disease. Autoinflammatory diseases are characterized by prolonged or recurrent febrile episodes, with a predominant dysregulation of the innate immune system and absence of autoantibody production (11), whereas autoimmune diseases are mostly driven by the adaptive immunity with involvement of dysregulated T- and B-cell responses (12). In such conditions, fever is a hallmark of disease and can be easily misjudged for, or even triggered by, an infection (13, 14). Correct treatment varies per disease, but involves immunosuppressive drugs or biologic therapies targeting specific cellular mechanisms, immune cells or cytokines of the immune system (15). Therefore, the second important goal of biomarkers is to distinguish between infectious and non-infectious disease.
As described, it is difficult to distinguish between infection, inflammation and autoimmunity. Biomarkers can, in combination with the presenting symptoms of the patient, help physicians to consider a proper diagnosis and treatment. In addition, specific biomarkers might also make it possible to bypass laborious microbiological tests and avoid invasive diagnostic procedures (such as bronchoalveolar lavage, biopsies or surgical procedures), while starting effective therapies for non-infectious inflammatory or autoimmune disease more promptly upon its onset (16).
CRP and PCT as Current Biomarkers in the Clinic
Upon infection, in particular bacterial infection, pro-inflammatory cytokines such as IL-1 and IL-6 are released into the circulation in response to pathogen-associated molecular patterns (PAMPs) (17). Additionally, during inflammation and tissue injury, damage-associated molecular patterns (DAMPs) can be released from injured tissue cells, activating immune cells by Toll Like Receptor (TLR) signaling to generate and release IL-1 and IL-6. Both these cytokines have been shown to induce fever by binding respectively to the IL-1 receptor (18) and IL-6 receptor (19) on endothelial cells in the brain, especially in the hypothalamus. IL-1 induces ubiquitous IL-6 synthesis in many cells including hepatocytes and macrophages in the liver (i.e. Kupffer cells), which in turn stimulates CRP production in the liver as part of the acute phase response (Figure 1). CRP regulates important inflammatory processes, such as activating the complement system, stimulating apoptosis, inducing release of proinflammatory cytokines (20). In the case of uncommon primary genetic defects in these signaling cascades, or by blocking either IL-1 or IL-6 signaling, fever is largely prevented and no CRP is released (21, 22).
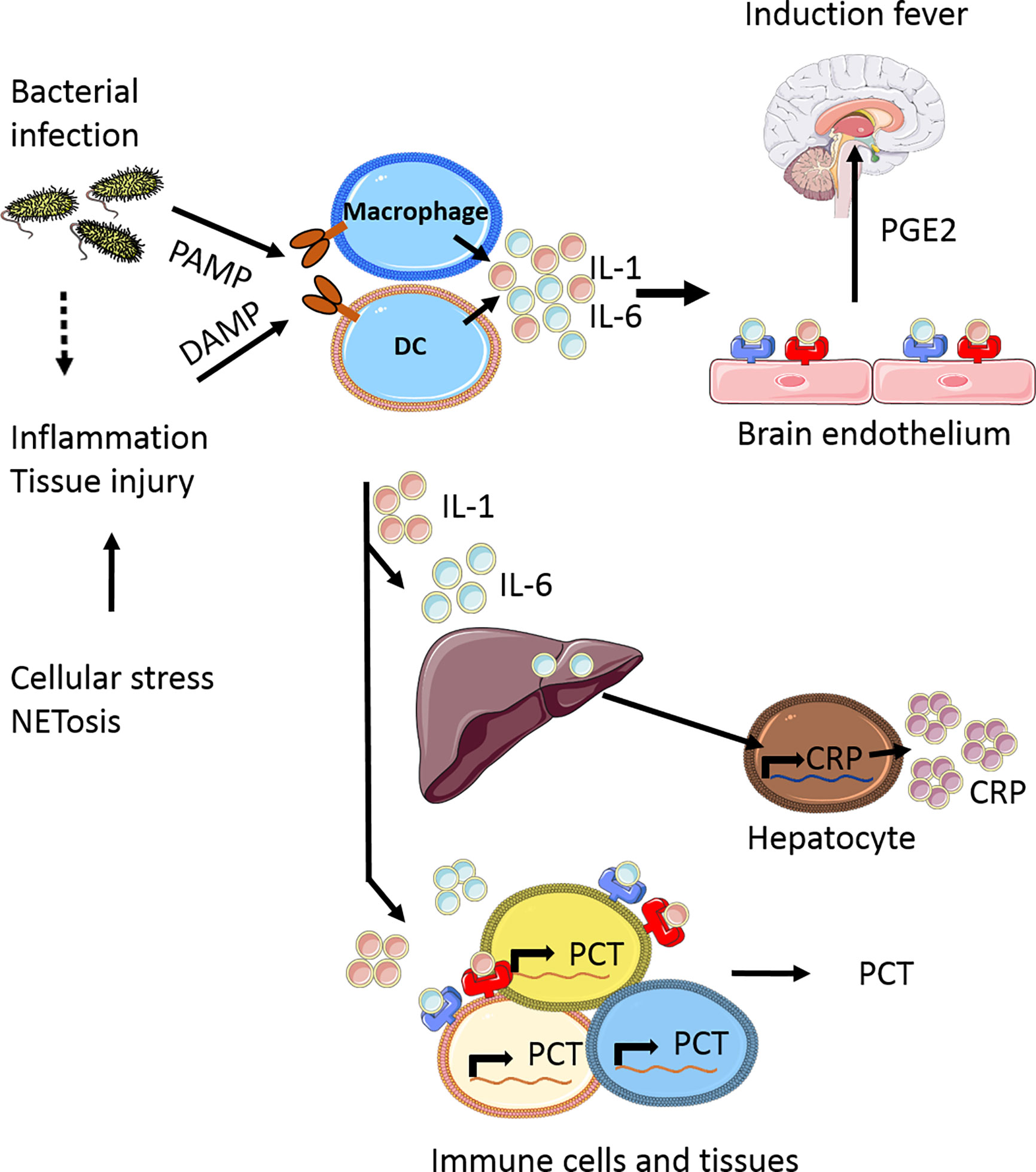
Figure 1 Simplified scheme of the induction of fever and release acute phase response proteins. Upon bacterial infection pathogen-associated molecular patterns (PAMPs) are released into the circulations which can be recognized by various cells, including macrophages and dendritic cells (DC). At the same time inflammation can occur, either as result of the infection or by tissue injury due to NETosis, leading to a release of damage-associated molecular patterns (DAMPs). In response to DAMPs and PAMPs recognition by several pattern recognition receptors (PPRs) receptor families, being ultimately involved in the production of pro-inflammatory cytokines, including IL-1 and IL-6. These cytokines are released and can bind to specialized regions of brain endothelium, which in response produces prostaglandin E2 (PGE2) as one of the major mediators to induce fever (17). In addition to fever, IL-1 and IL-6 also induce C-reactive protein (CRP) production in the liver and procalcitonin (PCT) production by various immune cells (macrophages, monocytes) and tissues (liver, spleen, lung), which are the most common biomarkers currently measured in the clinical setting of febrile disease, as part of the acute phase response. This figure is made using Sevier Medical Art.
IL-1 and IL-6 have been studied as biomarkers for bacterial infection but fail to show sufficient specificity and sensitivity in both children (16, 23) and adults (24), most likely because of the extremely short half-life of both cytokines (25). In contrast, CRP is more stable with a half-life of 18-20 hours and has become the most commonly used biomarker for bacterial infection and inflammation and is routinely measured in the clinic nowadays for both children and adults. Next to CRP, procalcitonin (PCT) is often used as biomarker for bacterial infections and inflammation. Under normal conditions, PCT is produced by neuroendocrine C cells of the thyroid as the 116-amino acid precursor of calcitonin. During bacterial infection, PCT is upregulated and consequently expressed in all cells of the body (26), but mainly by hepatocytes (27), leading to the release of elevated amounts of PCT in the circulation (26). PCT levels rise quickly after infection, reaching a peak between 12-24 hours. This is earlier than CRP, which peaks after 2-3 days (20). With a half-life of 24-30 hours PCT can stimulate the release of pro-inflammatory cytokines, amplifying inflammatory responses (28). PCT is a better discriminator between viral or bacterial infections compared to CRP (29). This is due to the fact that many inflammatory cytokines, including IL-1 and IL-6, contribute to the up-regulation of PCT. An exception is interferon-gamma (IFN-γ) which reduces PCT expression, therefore resulting in the lower concentrations of PCT found in viral infections (30). In addition, it has been suggested that PCT is particularly useful in detecting Gram-negative infections, with better discrimination compared to CRP (31, 32). Gram-negative and Gram-positive bacteria activate mostly TLR-4 or TLR-2, respectively (33), leading to the activation of different signaling pathways and distinct cytokine profiles (34).
Apart from CRP, PCT is routinely measured in some but not all clinics. Whereas CRP is broadly used as diagnostic biomarkers for bacterial infections, PCT is predominantly applied as a diagnostic biomarker in neonatal sepsis (35). Under circumstances where viral infection is less likely both CRP and PCT are good indicators for the need to treat with antibiotics, such as in febrile neonates and very young infants when disease is often caused by Streptococcus agalactiae or Escherichia coli infection postpartum (36). However, the absence of standardized algorithms for the use of PCT – also in the field of neonatology – makes interpretation difficult (37, 38). In addition, also in some viral infections CRP and PCT can be increased, as observed in enteroviral or respiratory syncytial virus infections, as well as in severe SARS-CoV-2 infections with concomitant inflammation (39–42). Irrespective of age, both CRP and PCT concentrations rise in response to both bacterial infection and non-infectious inflammation (43–45). Although potentially useful for disease-monitoring purposes (46–48), these two biomarkers may be sensitive but not specific to correctly discriminate early on between the type of infection or inflammation in a febrile patient.
Potential New Biomarkers for Infection
Several studies have tried to identify other biomarkers that are more suitable to determine bacterial or viral infection, as extensively reviewed for studies until 2015 (49). Here, we discuss the most promising candidates including the latest data. A summary of this data, including study details, is displayed in Supplementary Table 1.
One of these markers is soluble CD14, also known as presepsin. CD14 is a receptor on monocytes and macrophages that recognizes different surface structures on Gram-positive and Gram-negative bacteria, including lipoteichoic acid and proteoglycans or lipopolysaccharides, respectively. After binding bacterial antigens, CD14 is cleaved and released as presepsin (50). As a biomarker, presepsin has a higher sensitivity but a lower specificity than PCT or CRP in diagnosing sepsis in children (51). In adults PCT and presepsin have similar diagnostic accuracy (52). In another study it was shown that presepsin alone had a high sensitivity and specificity to diagnose neonatal sepsis, which did not change in combination with CRP and/or PCT (53). Irrespective of age, pancreatic stone protein (PSP) which is secreted by the pancreas in response to stress during systemic infection, has been studied in both infants and adults to predict sepsis (54) and was developed into a point-of-care test by Abionic. However, proper validation is needed because PSP levels are elevated in various other diseases as well (55–57).
Upon viral infection, cells start producing interferons (IFNs) to inhibit viral replication as their first anti-viral response (58). Although protein assays have been developed for measuring interferon levels at extremely low (i.e. attomolar) concentrations (59), these tests are experimental and routine detection remains challenging. Hence, surrogate proteins induced by interferon are also investigated. Expression of the two Myxovirus resistance genes in humans are strictly controlled by IFNs, acting as a cytoplasmic GTPase that has activity against a wide range of viruses. In humans, MX1 and MX2 (also known as MxA and MxB respectively) are, in contrast to many other interferon-induced proteins, strictly induced by IFN and found to be increased in the plasma of patients suffering from viral infections (60, 61). MX1 is induced 1-2 hours after infection with a half-life of more than 2 days. Although seen as an interesting biomarker to indicate viral infection, its sensitivity questionable. A commercially available combination immunoassay for CRP and MX1, called FebriDx, was unable to discriminate between viral and bacterial infections in a prospective cohort of upper respiratory infections (62). To date, the test has not been extensively studied in other infection cohorts.
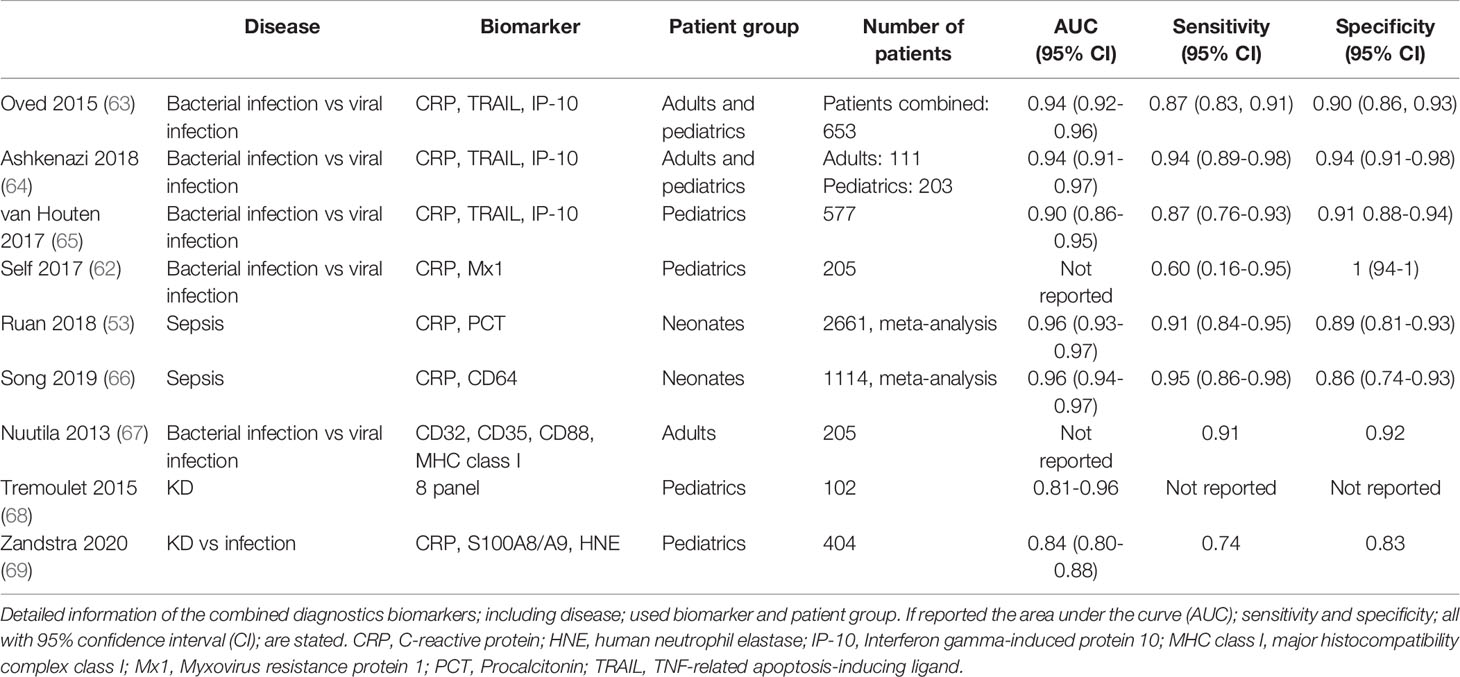
To date, further studies are being conducted to combine plasma biomarkers to discriminate between bacterial from viral infection, of which a selection of promising studies is shown in Table 1. For example, soluble TNF-related apoptosis-inducing ligand (TRAIL), IFN-γ-induced protein-10 (IP-10, CXCL10) and CRP were used in combination to distinguish between bacterial and viral infection in children and adults (63, 65). In contrast to CRP, the biomarkers TRAIL and IP-10 showed increased protein levels in viral infections (63–65, 70). Known as the ImmunoXpert test, the assay has the potential to be developed into a ready-to-use assay to prevent inappropriate antibiotic use in patients with infections, but remains to be confirmed in other cohorts to validate its performance and accuracy (63).
In addition to soluble plasma markers of cell-derived surface proteins, cellular expression of surface proteins themselves have been investigated as biomarkers linked to cell activation during infectious disease (49). Most studies include surface proteins on neutrophils activated during bacterial infections, or monocytes which can increase in number during viral infections. An accurate diagnosis of bacterial or viral infection within 1 hour using flow cytometry was possible using a set of surface proteins on neutrophils and monocytes, including Fcγ receptor I (FcγRI/CD64), FcγRII/CD32, complement receptor 1 (CR1/CD35), HLA-class-I, and the receptor for complement-derived anaphylatoxin C5a (C5aR/CD88) (67). CD32, CD35 and CD88 all have a higher expression on both neutrophils and monocytes in bacterial infections compared to viral infections, whereas HLA-class-I and CD169 on monocytes were increased in viral infections rather than bacterial infections (71, 72). These studies only included adults, no data on the pediatric population is available.
In neonatal sepsis, other neutrophil membrane bound proteins such as CD11b and CD64 were studied (73, 74). The meta-analysis for CD11b showed a very wide range in sensitivity and specificity between 0.56-1.00. A comparable range between sensitivity and specificity was observed for CD64. This variation might be due to the different cut-off values used in the studies, making it difficult to compare the overall sensitivity and specificity. Further studies are needed before implementation of such cellular assays as routine diagnostic test in the clinic can be considered. We also need to take into account that cell-based assays will require robust logistics and expertise making high-throughput application and interpretation much more demanding than a straight-forward ELISA system that can be easily repeated on the same sample when failed.
To conclude, a single protein marker that can distinguish bacterial from viral infections has not yet been identified. Promising results are shown by combining different markers. At the moment, large-scale studies using a heterogeneous research population are needed to bridge the gap to transform research findings into clinical diagnostics.
Biomarkers in Inflammatory Diseases
Separating infectious from non-infectious disease is key, as the diagnosis and treatment of non-infectious disease is totally different from that of infections (11, 75). As discussed, detection of most cytokines is challenging because of their very short half-life. One notable exception is the prolonged presence of strongly increased levels of circulating IL-18 in systemic non-infectious inflammatory disorders. Initially described as an IFN-γ inducing factor, IL-18 is involved in Th1 response, NK cell activation, and results in macrophage activation. IL-18, a member of the IL-1 family, is processed by caspase-1 to an active mature form that binds to its specific receptor (76). IL-18 can be determined in plasma as a biomarker because it is protected from clearance by associating with a plasma protein, IL-18 binding protein (IL-18BP), that can also be therapeutically applied to neutralize the biological activity of IL-18 (77).
IL-18 and IL-1 are involved in classic autoinflammatory diseases including Familial Mediterranean Fever, cryopyrinopathies, and hyperimmunoglobulin D Syndrome (11), in which the enzyme protein complex known as the inflammasome that processes the pro-forms of IL-18 and IL-1 is dysregulated by genetic mutations. These diseases are often considered because of the recurrence of non-infectious febrile episodes, which often start during early childhood. Biomarker screening that includes IL-18 may help to confirm severe autoinflammation. These autoinflammatory disorders encompass non-inherited systemic onset juvenile idiopathic arthritis (soJIA) (78, 79), adult-onset systemic arthritis (Still’s disease) (80), or hemophagocytic syndrome (81, 82), the latter being also known as Macrophage Activation Syndrome (MAS) secondary to rheumatic diseases like soJIA and Still’s disease. Although the exact mechanisms involved in the development of MAS are not fully understood, increasing evidence demonstrate the role of IL-18 in upregulating the production of IFN-γ (83). In such severe autoinflammatory disorders, circulating IL-18 is readily measured at high levels while IL-1 cannot easily be detected even though it is actively involved in the disease process. These severe febrile disorders are preferably treated as early as possible with the recombinant IL-1 Receptor antagonists (84) or, still experimentally, IL-18BP (85), so avoiding the administration of antimicrobials. Classic inflammasome defects are not associated with MAS (11), which seems to correspond with much lower IL-18 levels, suggesting that high levels of IL-18 may be the necessary link between inflammasome hyperactivity and secondary HLH/MAS (86). Further to this, IFN-γ-induced biomarkers like CXCL10 (79) or CXCL9 (87) may become important biomarkers to include in multiplexed diagnostic tests.
Numerous other biomarkers are studied for diagnosis and monitoring of autoinflammatory diseases (88, 89). S100A8/A9 and S100A12 are good biomarkers for diagnostic purposes, monitoring disease activity, predicting treatment response, or risk for relapse in such non-infectious febrile diseases (90), including soJIA, adult Still’s disease and the above mentioned classic fever syndromes. S100A8/A9 protein dimers are also known as the MRP8/14 heterodimer or more commonly calprotectin. This biomarker has already been used for more than a decade as a diagnostic test in fecal samples from patients with inflammatory bowel disease (91). S100A8/A9 as well as the homodimer S100A12 proteins are released from activated innate cells, mostly neutrophils, and are of direct pathophysiologic interest (92). As extensively reviewed (92), S100 proteins have direct inflammation-propagating activity once released from neutrophils (or monocytes) as local or systemic activating signals of the innate cells by binding to the TLR4 and the ‘Receptor for Advanced Glycation End products’ (RAGE), respectively. TLR4 is a multiligand receptor for PAMPs of various bacterial pathogens as well as DAMPs, while RAGE is a multiligand member of the immunoglobulin superfamily of cell surface molecules, interacts with distinct molecules implicated in homeostasis and inflammation. Both are abundantly expressed on immune cells, endothelial cells and other extravascular tissue cells, which may contribute to initiate and propagate systemic disease and multiorgan failure during invasive infections or inflammation.
Autoimmune diseases are mostly driven by the adaptive immune system, with the predominant involvement of autoreactive T- and B-cell responses (12). There are at least 80 types of autoimmune diseases (12). However, a strict separation between autoimmunity and autoinflammation may sometimes be very difficult. Autoimmune diseases are to some extent overlapping disorders because of concomitant inflammation in which a role for innate immune cells is more prominent. For example, while systemic lupus erythematosus (SLE) is an autoantibody and immune complex disease by nature, most of its organ manifestations are in fact inflammatory. This may also apply to IBD and autoantibody-associated vasculitis. Hence, proinflammatory cytokines are not only involved in autoinflammatory disease but also autoimmune diseases, as shown by strong upregulation of IL-1 and IL-18 in active SLE (93).
One example of vasculitis that usually presents as a febrile disease in childhood is Kawasaki disease (KD). Apart from the high and persistent fever, the clinical symptoms in KD mimic both bacterial and viral disease presenting with lymphadenopathy, skin rash, red eyes, red tongue and/or red hands and feet, in the presence of a raised CRP. KD is often misdiagnosed for streptococcal infection or a measles-like viral disease. No blood test for KD is available to date and the diagnosis is based on clinical criteria (94). Treatment with a single high-dose intravenous immunoglobulin (IVIG) helps to quickly recover from and protects against the development of coronary artery aneurysms. This complication is why the disease is feared and should be recognized swiftly to administer the IVIG as soon as possible. To discriminate KD from infectious disease, many different biomarkers have been studied over the years but lack good specificity and sensitivity, with many studies lacking proper control groups (95). Recently, we showed that a combination of three biomarkers; CRP, neutrophil elastase and S100A8/A9, reached high sensitivity and specificity in KD to efficiently differentiate KD from infectious disease in febrile controls (69). To validate these findings, a new study comparing children with KD and other types of vasculitis or inflammatory syndromes is necessary before developing a point-of-care test to implement this combination of biomarkers in the clinic.
In summary, when febrile episodes mimic infections tests should not only separate bacterial from viral disease, but also discriminate infectious from inflammatory disorders.
Transcriptomics
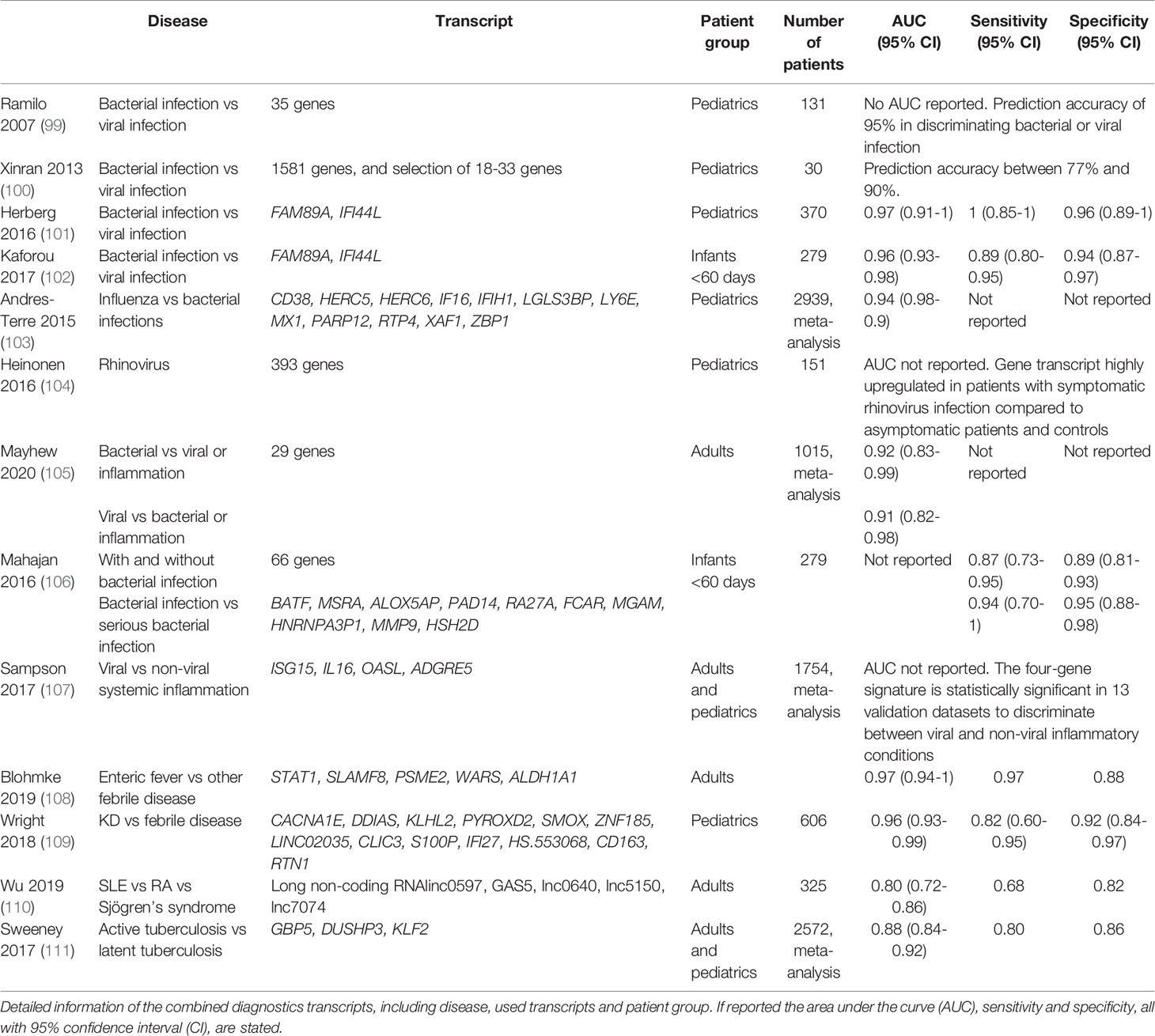
Recent studies show that highly sensitive and specific biomarkers can be found using RNA transcript analysis. RNA transcriptional activity changes during the course of disease leading to different transcript levels, new transcripts or splice variants when compared to the healthy state or among different diseases (96). As discussed, there is a serious advantage for the use of multiple protein markers at the same time to distinguish the cause of fever. In this respect, unbiased transcriptome analysis may account for thousands of transcripts, allowing an even more in-depth refinement of the possible cause of a given febrile disease. In agreement with previous results, many gene transcripts that are upregulated are related to interferon signaling in viral disease (97) while integrin-related gene transcripts are more prominent in bacterial disease (98) (Table 2). The overwhelming number of transcripts obtained by transcriptomics need to be reduced to make a potentially rapid test result available in the near future. This has led to a 2-transcript host RNA signature of combining FAM89A and IFI44L to discriminate between bacterial or viral infection in children (101), which has been modified to the combination of EMR1-ADGRE1 and IFI44L when the signature was further developed into a point-of-care test (112). Using these RNA signatures it was shown that while >90% of patients present in the group of unknown infections received antibiotic, only 46% of this group had a bacterial infection (101). This study again shows the excessive use of antibiotics in infections and the need to improve diagnostics for febrile children.
The next challenge lies in the fact that biomarkers are needed to distinguish between viral infections and inflammatory disease. Whereas the abovementioned IFI44L acts as viral biomarker, it is part of a more extensive set of interferon-stimulated genes (ISGs), or so-called ISG signature (58, 113). Other ISGs can be used to identify systemic autoinflammatory diseases, including SLE (114–116). The differences in viral and autoinflammatory ISGs are illustrated in Figure 2.
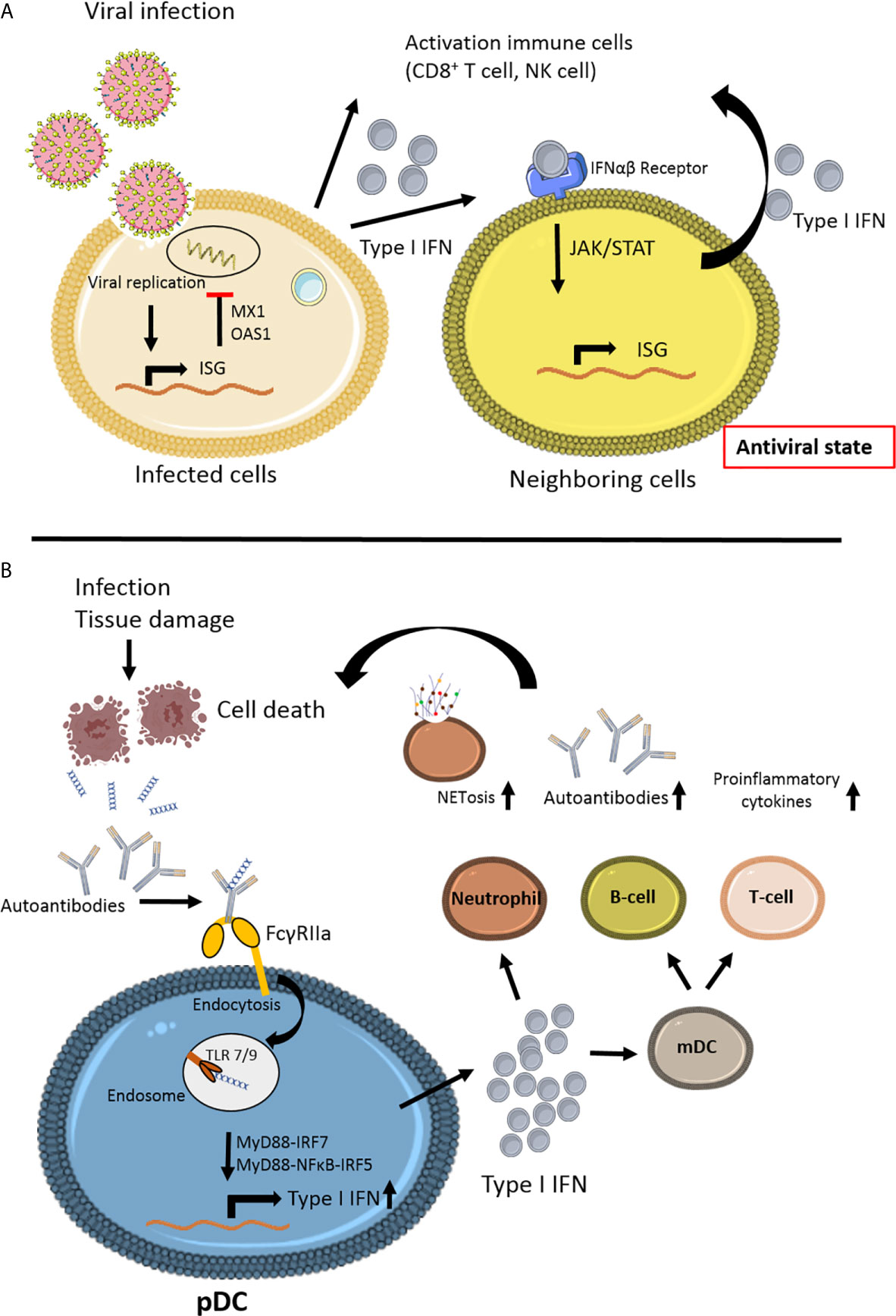
Figure 2 Schematic view of the different steps of interferon signaling involved in viral infection and autoimmune disease. (A) Viral infection induces interferon stimulated gene (ISG) stimulation (58). Antiviral proteins (MX1, OAS1) will inhibit viral replication in the cell. Secretion of Type I IFN by the infected cell will lead to paracrine signaling to the neighboring cell via JAK/STAT signaling, thereby stimulating ISG in non-infecting cells, leading to an antiviral state, preventing further viral infection. The anti-viral immune response is induced by activating various immune cells, especially CD8+ T cell survival and natural killer (NK) cell activation. (B) In autoimmune disease, such as Systemic Lupus Erythematosus (SLE), a variety of environmental factors can trigger the disease in genetically predisposed individuals (12). Infection or tissue damage can be an example of a trigger for autoimmune disease by the formation of autoantibody complexes with DNA released from apoptotic cells. The antibodies bind to the IgG receptor FcγRIIa and via endocytosis can activate endosomal Toll like receptor (TLR)7 and TLR9 (117). Due to mutations in JAK/STAT and/or interferon regulating factors IRFs the release of Type I IFN is upregulated in plasmacytoid dendritic cells (pDC). High levels of Type I IFNs can activate T- and B-cells via myeloid DCs, leading to more autoantibodies and proinflammatory cytokines. Type I IFN can promote cytotoxicity NK cells, phagocytosis in macrophages and NETosis in neutrophils (118, 119). This figure is made using Sevier Medical Art.
Recently, a panel of only 5 long non-coding RNAs was used to distinguish SLE patients from patients with rheumatoid arthritis and primary Sjögren’s syndrome (110). In the same way, it was shown that a 13-gene transcript signature was able to distinguish KD patients from both infectious and other inflammatory patients (109), demonstrating the power of multiplexed analysis as a leading principle. The strength of this study was the inclusion of a wide range of disorders with overlapping symptoms of KD, infectious and inflammatory patients. In addition, the proposed model was validated with an independent group of patients to strengthen their findings. Results from various studies on infectious as well as inflammatory diseases, indicate the value of using RNA transcriptome analyses for diagnostic purposes, as summarized in Table 2.
Although transcriptomic approaches look promising, they come with limitations. Apart from the lack of routine availability (120), another important disadvantage of transcriptomics is its cell dependence (112). Often the cellular composition may not be stable making the signatures less robust. For example, a leukopenic blood sample might have a very limited yield, most likely limiting the discriminatory results, preventing its application in oncology patients admitted with febrile neutropenia. Despite these difficulties, the implementation of transcriptomic biomarker tests into the clinic is one step closer. For the 2-transcript signature (101) a fast and highly accurate RT-qPCR was developed which efficiently distinguished between bacterial and viral infections (121), as well as a point-of-care test with the improved transcript signature (112), showing potential for rapid testing in the near future. In adults though, distinguishing bacterial disease and sepsis from viral disease or inflammation using transcriptomics was shown to be challenging if not ineffective (122–126), showing the limitations.
Biomarkers via Mass Spectrometry or Multiplexed Protein Microarrays
As described for RNA signatures, recent techniques allow for further and more dynamic profiling which might enhance diagnostic power. Blood is estimated to contain over 10,000 distinct proteins (irrespective post-translationally modifications), with concentrations spanning a dynamic range up to 12 orders of magnitude (127). To date, the number of novel biomarkers that have been identified from biomarker discovery projects involving plasma profiling are still limited. Using this technique, the acute phase serum amyloid A (SAA) proteins as well as the S100 proteins have been identified as biomarker for rheumatic disease (128), confirming previous protein tests (129, 130). Moreover, in a targeted cohort of the deadly Ebola virus infection a multi/omics approach including plasma profiling showed upregulated HLA-class-I and molecules found on endothelial cells (e.g. VCAM1), macrophages (e.g. CD14 [presepsin], CD163, FCGR3A [FcgRIIIa]), and neutrophils (e.g. FCGR3B, FcgRIIIb) (131). Activation was more pronounced in fatalities, and were consistent with metabolomics and lipidomics signatures that predicted immune cell activation and sepsis-like immune responses in Ebola virus fatalities. Of interest is the increase in soluble VSIG4, a co-stimulatory molecule expressed on macrophages, which was only observed in fatalities and may thus act as a completely novel biomarker of a severe and fatal outcome. These studies show that the use of plasma profiling might result in identification of novel biomarkers that can help in the future to advance clinical management.
Although unbiased Mass Spectrometry has been used for plasma profiling and biomarker identification, there is a balance in the yield and the expectations. While fractionation at the peptide level greatly increases the depth of analysis, a major drawback has been the significant increase in the amount of time it takes to analyze a single sample. This trade-off between depth and throughput means that plasma proteomics for diagnosis and monitoring with relatively large numbers of samples results in shallow depth of coverage, whereas studies emphasizing depth of coverage typically have small sample numbers (132). For discovery of biomarkers plasma proteomics is preferred but the analysis takes days to weeks as shown for murine and human plasma samples (133, 134). Taken together, plasma profiling studies are still rare but they might potentially result in discovery of novel biomarkers to distinguish between more complex diseases or disease outcomes.
Another method for plasma protein discovery consists of the expanded aptamer-based multiplex proteins assay (SOMAscan) which can quantify more than 3000 proteins (135–139). Multiplexed detection approaches using antibodies as part of targeted protein panels such as current Luminex (140) or Olink platforms (141), are able to measure smaller number of analytes and are cheaper to use. Overall these multiplex microarray assays have been designed as discovery platforms and measure relative protein concentrations which cannot give absolute values without having internal controls and standard curves for accurate measurements within the known linear dynamic range of the aptamers used.
The Complexity of Overlapping Disease
Often bacterial and viral infections are presented as separable disease categories, but in reality, these categories often overlay and cannot be regarded as sheer single entities. For instance, co-infections, such as Respiratory Syncytial Virus (RSV)-related bacterial bronchopneumonia in infants (142), influenza-associated Staphylococcus aureus pneumonia (143), or chickenpox-related streptococcal impetiginization of the skin (144), will influence diagnostic test results and interpretation (145). In fact, we may learn from the application of combination tests of biomarkers or transcripts, that co-infections of different pathogens may be more common than we previously believed as based on routine microbiology cultures and serology. The cut-off values in the interpretation of novel diagnostic biomarker approaches may be used to differentiate true co-infection from colonization, or innocent prolonged viral shedding from true co-infection, as mere examples in which interpretation of current PCR-based microbiology results are not straight-forward.
Another level of complexity consists of the infectious triggers that can initiate autoinflammatory derailment or autoimmunity, as is illustrated in the recent COVID-19 pandemic (146). Severe COVID-19 infection has a large overlap with MAS, with a systemic inflammatory reaction, concomitant high serum macrophage-derived ferritin and a life-threatening hyper-inflammation sustained by a cytokine storm which eventually leads to multi-organ failure. IL-6 is used a predictive marker of mortality (147). In the current COVID-19 era, post-viral Kawasaki-like multi-inflammatory syndrome in children (MIS-C) (148–152) or adults (MIS-A) (153) has been linked to cytokine release syndrome some weeks following an often asymptomatic SARS-CoV-2 infection. High levels of CRP as well as increased levels of cytokines including IL-6 can be detected which, as discussed earlier, are very unusual to find elevated during a viral infection (154). Some of the disease parameters [including ferritin (147)] are reminiscent of MAS, or of secondary HLH (155) in which bone marrow suppression is clearly present. Secondary forms of HLH occur more common in the context of infections (typically Epstein-Barr virus), malignancies, systemic autoimmunity or severe sepsis in otherwise genetically normal individuals (82, 156, 157). All these forms of HLH are clinically characterized by persistent fever, severe malaise, variable degrees of cytopenia and hepatosplenomegaly. Taken together, biomarkers alone can be misleading. Care should be taken in their interpretation when trying to diagnose complex clinical disease because of overlap between clinical syndromes with totally different etiologies.
Conclusion
In this review we discuss the current landscape of biomarkers with a focus on, but not restricted to, febrile pediatric patients. Currently, apart from the common use of CRP in both adults and children, biomarkers like PCT, and IL-18 are frequently used in clinic practice. PCT is specifically used to differentiate between bacterial and viral infections in neonatal sepsis (29). IL-18 is a good biomarker for various auto-inflammatory diseases (78, 80, 81).
A rapid increase in the development of combination tests is observed. The latest developments regarding transcriptome analysis shows promising results, although proper validation is often needed to confirm diagnostic results. It was shown that a 2-transcript host RNA signature of combining EMR1-ADGR and IFI44L could discriminate between bacterial or viral infection in children (112), confirmed by the development of a point-of-care test, see Table 2. Further studies will need to show whether transcriptome analysis is able to distinguish between bacterial subgroups (i.e. Gram-positive and Gram-negative, atypical, or mycobacterial infections). In addition, both transcriptome analysis, and whole plasma profiling might result in the identification of novel plasma markers, expanding the repertoire of currently tested plasma markers.
To conclude, combining biomarkers, either plasma markers or transcriptomics, will improve diagnostics in febrile children similar as in the adult population. Further studies are needed to determine the exact combination of markers and techniques that together will lead to a solid diagnostic tool and its general application in infectious and non-infectious disease. We should emphasize that in this review, we have focused on the biomarker landscape for the western world. It should not be forgotten that infectious disease constitutes the major cause of childhood death worldwide. Most deaths in 2015 occurred in South Asia and sub-Saharan Africa (75% of the total burden) and underscores the need of a better understanding of the cause of infection in a febrile child (158). Therefore, there is still an urgent need to identify biomarkers that can be implemented throughout the world, adapted to local epidemiology, logistics and costs.
Author Contributions
JZ wrote the manuscript. IJ and TK reviewed and revised the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
Supported by the European Union’s Horizon 2020 research and innovation program under Grant Agreement No. 668303.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Sophie Bliss for carefully editing our manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.631308/full#supplementary-material
Supplementary Table 1 | Single biomarkers mentioned in the manuscript. Detailed information of the single diagnostics biomarkers, including disease, used biomarker and patient group. If reported the area under the curve (AUC), sensitivity and specificity, all with 95% confidence interval (CI), are stated. IL- 6, Interleukin- 6; NT-proBNP, N-Terminal Pro-B Type Natriuretic Peptide; Mx1, Myxovirus resistance protein 1; SuPAR, soluble urokinase-type plasminogen activator receptor; PSP, pancreatic stone protein.
References
1. Wing R, Dor MR, McQuilkin PA. Fever in the Pediatric Patient. Emergency Med Clinics North America (2013) 31:1073–96. doi: 10.1016/j.emc.2013.07.006
2. Foggo V, Cavenagh J. Malignant Causes of Fever of Unknown Origin. Clin Med J R Coll Physicians London (2015) 15(3):292–4. doi: 10.7861/clinmedicine.15-3-292
3. Miller JM, Binnicker MJ, Campbell S, Carroll KC, Chapin KC, Gilligan PH, et al. A Guide to Utilization of the Microbiology Laboratory for Diagnosis of Infectious Diseases: 2018 Update by the Infectious Diseases Society of America and the American Society for Microbiology. Clin Infect Dis (2018) 67(6):e1–94. doi: 10.1093/cid/ciy381
4. Martinón-Torres F, Salas A, Rivero-Calle I, Cebey-López M, Pardo-Seco J, Herberg JA, et al. Life-Threatening Infections in Children in Europe (the EUCLIDS Project): A Prospective Cohort Study. Lancet Child Adolesc Heal (2018) 2(6):404–14. doi: 10.1016/S2352-4642(18)30113-5
5. Hagedoorn NN, Borensztajn DM, Nijman R, Balode A, von Both U, Carrol ED, et al. Variation in Antibiotic Prescription Rates in Febrile Children Presenting to Emergency Departments Across Europe (Mofiche): A Multicentre Observational Study. PloS Med (2020) 17(8). doi: 10.1371/journal.pmed.1003208
6. Cassini A, Högberg LD, Plachouras D, Quattrocchi A, Hoxha A, Simonsen GS, et al. Attributable Deaths and Disability-Adjusted Life-Years Caused by Infections With Antibiotic-Resistant Bacteria in the EU and the European Economic Area in 2015: A Population-Level Modelling Analysis. Lancet Infect Dis (2019) 19(1):56–66. doi: 10.1016/S1473-3099(18)30708-4
7. CDC. Antibiotic Resistance Threats in the United States Vol. 2019. Atlanta, GA: U.S. Department of Health and Human Services, CDC (2019).
8. Friedman ND, Temkin E, Carmeli Y. The Negative Impact of Antibiotic Resistance. Clin Microbiol Infect (2016) 22(5):416–22. doi: 10.1016/j.cmi.2015.12.002
9. Mathison BA, Pritt BS. Update on Malaria Diagnostics and Test Utilization. J Clin Microbiol (2017) 55:2009–17. doi: 10.1128/JCM.02562-16
10. Talapko J, Škrlec I, Alebić T, Jukić M, Malaria VčevA. The Past and the Present. Microorganisms (2019) 7(6):179. doi: 10.3390/microorganisms7060179
11. de Jesus AA, Goldbach-Mansky R. Monogenic Autoinflammatory Diseases: Concept and Clinical Manifestations. Clin Immunol (2013) 147(3):155–74. doi: 10.1016/j.clim.2013.03.016
12. Wang L, Wang FS, Gershwin ME. Human Autoimmune Diseases: A Comprehensive Update. J Intern Med (2015) 278(4):369–95. doi: 10.1111/joim.12395
13. Getts DR, Chastain EML, Terry RL, Miller SD. Virus Infection, Antiviral Immunity, and Autoimmunity. Immunological Rev (2013) 255(1):197–209. doi: 10.1111/imr.12091
14. Qiu CC, Caricchio R, Gallucci S. Triggers of Autoimmunity: The Role of Bacterial Infections in the Extracellular Exposure of Lupus Nuclear Autoantigens. Front Immunol (2019) 10:2608. doi: 10.3389/fimmu.2019.02608
15. Havnaer A, Han G. Autoinflammatory Disorders: A Review and Update on Pathogenesis and Treatment. Am J Clin Dermatol (2019) 20(4):539–64. doi: 10.1007/s40257-019-00440-y
16. Panato APR, Tomasi LT, Simon CS, Madeira K, Simoes LR, Medeiros LR, et al. Meta-Analysis Identifies Tumor Necrosis Factor-Alpha and Interleukin-1 Beta as Diagnostic Biomarkers for Bacterial and Aseptic Meningitis. Curr Neurovascular Res (2014) 11(4):340–8. doi: 10.2174/1567202611666140912120940
17. Evans SS, Repasky EA, Fisher DT. System Feels the Heat. Nat Rev Immunol (2016) 15(6):335–49. doi: 10.1038/nri3843
18. Ching S, Zhang H, Belevych N, He L, Lai W, Pu XA, et al. Endothelial-Specific Knockdown of Interleukin-1 (IL-1) Type 1 Receptor Differentially Alters CNS Responses to IL-1 Depending on Its Route of Administration. J Neurosci (2007) 27(39):10476–86. doi: 10.1523/JNEUROSCI.3357-07.2007
19. Eskilsson A, Mirrasekhian E, Dufour S, Schwaninger M, Engblom D, Blomqvist A. Immune-Induced Fever is Mediated by IL-6 Receptors on Brain Endothelial Cells Coupled to Stat3-Dependent Induction of Brain Endothelial Prostaglandin Synthesis. J Neurosci (2014) 34(48):15957–61. doi: 10.1523/JNEUROSCI.3520-14.2014
20. Sproston NR, Ashworth JJ. Role of C-Reactive Protein At Sites of Inflammation and Infection. Front Immunol (2018) 9(APR):1–11. doi: 10.3389/fimmu.2018.00754
21. Kramer F, Torzewski J, Kamenz J, Veit K, Hombach V, Dedio J, et al. Interleukin-1β Stimulates Acute Phase Response and C-Reactive Protein Synthesis by Inducing an Nfκb- and C/Ebpβ-Dependent Autocrine Interleukin-6 Loop. Mol Immunol (2008) 45(9):2678–89. doi: 10.1016/j.molimm.2007.12.017
22. Spencer S, Bal SK, Egner W, Allen HL, Raza SI, Ma CA, et al. Loss of the Interleukin-6 Receptor Causes Immunodeficiency, Atopy, and Abnormal Inflammatory Responses. J Exp Med (2019) 216(9):1986–98. doi: 10.1084/jem.20190344
23. Iwase S, Nakada Ta, Hattori N, Takahashi W, Takahashi N, Aizimu T, et al. Interleukin-6 as a Diagnostic Marker for Infection in Critically Ill Patients: A Systematic Review and Meta-Analysis. Am J Emerg Med (2019) 37(2):260–5. doi: 10.1016/j.ajem.2018.05.040
24. Carlyn CJ, Andersen NJ, Baltch AL, Smith R, Reilly AA, Lawrence DA. Analysis of Septic Biomarker Patterns: Prognostic Value in Predicting Septic State. Diagn Microbiol Infect Dis (2015) 83(3):312–8. doi: 10.1016/j.diagmicrobio.2015.07.003
25. Woensel JBM, Biezeveld MH, Hack CE, Bos AP, Kuijpers TW. Elastase and Granzymes During Meningococcal Disease in Children: Correlation to Disease Severity. Intensive Care Med (2005) 31(9):1239–47. doi: 10.1007/s00134-005-2720-3
26. Dong R, Wan B, Lin S, Wang M, Huang J, Wu Y, et al. Procalcitonin and Liver Disease: A Literature Review. J Clin Trans Hepatology (2019) 7:51–5. doi: 10.14218/JCTH.2018.00012
27. Bai Y, Lu J, Cheng Y, Zhang F, Fan X, Weng Y, et al. NF-Kb Increases LPS-Mediated Procalcitonin Production in Human Hepatocytes. Sci Rep (2018) 8(1):8913. doi: 10.1038/s41598-018-27302-7
28. Dahaba AA, Metzler H. Procalcitonin’s Role in the Sepsis Cascade. Is procalcitonin sepsis marker mediator? Minerva Anestesiol. (2009) 75(7-8):447–52.
29. Simon L, Gauvin F, Amre DK, Saint-Louis P, Lacroix L. Serum Procalcitonin and C-Reactive Protein Levels as Markers of Bacterial Infection: A Systematic Review and Meta-Analysis. Clin Infect Dis (2004) 39(2):206–17. doi: 10.1086/421997
30. Matwiyoff GN, Prahl JD, Miller RJ, Carmichael JJ, Amundson DE, Seda G, et al. Immune Regulation of Procalcitonin: Abiomarker and Mediator of Infection. Inflammation Res (2012) 61(5):401–9. doi: 10.1007/s00011-012-0439-5
31. Koizumi Y, Sakanashi D, Ohno T, Nakamura A, Yamada A, Shibata Y, et al. Plasma Procalcitonin Levels Remain Low At the Onset of Gram-Positive Bacteremia Regardless of Severity or the Presence of Shock: A Retrospective Analysis of Patients With Detailed Clinical Characteristics. J Microbiol Immunol Infect (2020) S1684–1182(20):30213–9. doi: 10.1016/j.jmii.2020.08.015
32. Bassetti M, Russo A, Righi E, Dolso E, Merelli M, D’Aurizio F, et al. Role of Procalcitonin in Predicting Etiology in Bacteremic Patients: Report From a Large Single-Center Experience. J Infect Public Health (2020) 13(1):40–5. doi: 10.1016/j.jiph.2019.06.003
33. Elson G, Dunn-Siegrist I, Daubeuf B, Pugin J. Contribution of Toll-Like Receptors to the Innate Immune Response to Gram-Negative and Gram-Positive Bacteria. Blood. (2007) 109(4):1574–83. doi: 10.1182/blood-2006-06-032961
34. Surbatovic M, Popovic N, Vojvodic D, Milosevic I, Acimovic G, Stojicic M, et al. Cytokine Profile in Severe Gram-Positive and Gram-Negative Abdominal Sepsis. Sci Rep (2015) 5:11355. doi: 10.1038/srep11355
35. Vouloumanou EK, Plessa E, Karageorgopoulos DE, Mantadakis E, Falagas ME. Serum Procalcitonin as a Diagnostic Marker for Neonatal Sepsis: A Systematic Review and Meta-Analysis. Intensive Care Med (2011) 37(5):747–62. doi: 10.1007/s00134-011-2174-8
36. Stoll BJ, Hansen NI, Sánchez PJ, Faix RG, Poindexter BB, Van Meurs KP, et al. Early Onset Neonatal Sepsis: The Burden of Group B Streptococcal and E. Coli Disease Continues. Pediatrics (2011) 127(5):817–26. doi: 10.1542/peds.2010-2217
37. Chu DC, Mehta AB, Walkey AJ. Practice Patterns and Outcomes Associated With Procalcitonin Use in Critically Ill Patients With Sepsis. Clin Infect Dis (2017) 64(11):1509–15. doi: 10.1093/cid/cix179
38. Ross RK, Keele L, Kubis S, Lautz AJ, Dziorny AC, Denson AR, et al. Effect of the Procalcitonin Assay on Antibiotic Use in Critically Ill Children. J Pediatr Infect Dis Soc (2018) 7(2):E43–6. doi: 10.1093/jpids/piy004
39. Heiskanen-Kosma T, Korppi M. Serum C-Reactive Protein Cannot Differentiate Bacterial and Viral Aetiology of Community-Acquired Pneumonia in Children in Primary Healthcare Settings. Scand J Infect Dis (2000) 32(4):399–402. doi: 10.1080/003655400750044971
40. Gautam S, Cohen AJ, Stahl Y, Valda Toro P, Young GM, Datta R, et al. Severe Respiratory Viral Infection Induces Procalcitonin in the Absence of Bacterial Pneumonia. Thorax (2020) 75(11):974–981. doi: 10.1164/ajrccm-conference.2020.201.1_MeetingAbstracts.A6232
41. Russo A, Venditti M, Ceccarelli G, Mastroianni CM, d’Ettorre G. Procalcitonin in Daily Clinical Practice: An Evergreen Tool Also During a Pandemic. Intern Emerg Med (2021) 16(3):541–43. doi: 10.1007/s11739-021-02659-2
42. Garcia-Vidal C, Sanjuan G, Moreno-García E, Puerta-Alcalde P, Garcia-Pouton N, Chumbita M, et al. Incidence of Co-Infections and Superinfections in Hospitalized Patients With COVID-19: A Retrospective Cohort Study. Clin Microbiol Infect (2021) 27(1):83–8. doi: 10.1016/j.cmi.2020.07.041
43. Pope JE. Choy EH. C-Reactive Protein and Implications in Rheumatoid Arthritis and Associated Comorbidities. Semin Arthritis Rheumatism (2021) 51(1):219–29. doi: 10.1016/j.semarthrit.2020.11.005
44. Dharmajaya R, Sari DK. Role and Value of Inflammatory Markers in Brain Tumors: A Case Controlled Study. Ann Med Surg (2021) 63:102107. doi: 10.1016/j.amsu.2021.01.055
45. Tian F, Li H, Wang L, Li B, Aibibula M, Zhao H, et al. The Diagnostic Value of Serum C-Reactive Protein, Procalcitonin, Interleukin-6 and Lactate Dehydrogenase in Patients With Severe Acute Pancreatitis. Clin Chim Acta (2020) 510:665–70. doi: 10.1016/j.cca.2020.08.029
46. Prkno A, Wacker C, Brunkhorst FM, Schlattmann P. Procalcitonin-Guided Therapy in Intensive Care Unit Patients With Severe Sepsis and Septic Shock - a Systematic Review and Meta-Analysis. Crit Care (2013) 17(6):R291. doi: 10.1186/cc13157
47. de Jong E, van Oers JA, Beishuizen A, Vos P, Vermeijden WJ, Haas LE, et al. Efficacy and Safety of Procalcitonin Guidance in Reducing the Duration of Antibiotic Treatment in Critically Ill Patients: A Randomised, Controlled, Open-Label Trial. Lancet Infect Dis (2016) 16(7):819–27. doi: 10.1016/S1473-3099(16)00053-0
48. Jensen JU, Hein L, Lundgren B, Bestle MH, Mohr TT, Andersen MH, et al. Procalcitonin-Guided Interventions Against Infections to Increase Early Appropriate Antibiotics and Improve Survival in the Intensive Care Unit: A Randomized Trial. Crit Care Med (2011) 39(9):2048–58. doi: 10.1097/CCM.0b013e31821e8791
49. Kapasi AJ, Dittrich S, Gonzalez IJ, Rodwell TC. Host Biomarkers for Distinguishing Bacterial From Non-Bacterial Causes of Acute Febrile Illness: A Comprehensive Review. PloS One (2016) 11(8):1–29. doi: 10.1371/journal.pone.0160278
50. Chenevier-Gobeaux C, Borderie D, Weiss N, Mallet-Coste T, Claessens YE. Presepsin (Scd14-ST), an Innate Immune Response Marker in Sepsis. Clin Chim Acta (2015) 450:97–103. doi: 10.1016/j.cca.2015.06.026
51. Yoon SH, Kim EH, Kim HY, Ahn JG. Presepsin as a Diagnostic Marker of Sepsis in Children and Adolescents: A Systemic Review and Meta-Analysis. BMC Infect Dis (2019) 19(1):1–11. doi: 10.1186/s12879-019-4397-1
52. Kondo Y, Umemura Y, Hayashida K, Hara Y, Aihara M, Yamakawa K. Diagnostic Value of Procalcitonin and Presepsin for Sepsis in Critically Ill Adult Patients: A Systematic Review and Meta-Analysis. J Intensive Care (2019) 7:22. doi: 10.1186/s40560-019-0374-4
53. Ruan L, Chen GY, Liu Z, Zhao Y, Xu GY, Li SF, et al. The Combination of Procalcitonin and C-Reactive Protein or Presepsin Alone Improves the Accuracy of Diagnosis of Neonatal Sepsis: A Meta-Analysis and Systematic Review. Crit Care (2018) 22(1):1–9. doi: 10.1186/s13054-018-2236-1
54. Eggimann P, Que YA, Rebeaud F. Measurement of Pancreatic Stone Protein in the Identification and Management of Sepsis. Biomarkers Med (2019) 13(2):135–45. doi: 10.2217/bmm-2018-0194
55. Yang J, Li L, Raptis D, Li X, Li F, Chen B, et al. Pancreatic Stone Protein/Regenerating Protein (PSP/Reg): A Novel Secreted Protein Up-Regulated in Type 2 Diabetes Mellitus. Endocrine (2015) 48(3):856–62. doi: 10.1007/s12020-014-0427-3
56. Cui C, Fu M, Gao B. Procalcitonin and Pancreatic Stone Protein Function as Biomarkers in Early Diagnosis of Pediatric Acute Osteomyelitis. Med Sci Monit (2017) 23:5211–7. doi: 10.12659/MSM.904276
57. Scherr A, Graf R, Bain M, Christ-Crain M, Müller B, Tamm M, et al. Pancreatic Stone Protein Predicts Positive Sputum Bacteriology in Exacerbations of COPD. Chest (2013) 143(2):379–87. doi: 10.1378/chest.12-0730
58. Schoggins JW, Rice CM. Interferon-Stimulated Genes and Their Antiviral Effector Functions. 1 Curr Opin Virol (2011) 1(6):519–25. doi: 10.1016/j.coviro.2011.10.008
59. Rodero MP, Decalf J, Bondet V, Hunt D, Rice GI, Werneke S, et al. Detection of Interferon Alpha Protein Reveals Differential Levels and Cellular Sources in Disease. J Exp Med (2017) 214(5):1547–55. doi: 10.1084/jem.20161451
60. Engelmann I, Dubos F, Lobert PE, Houssin C, Degas V, Sardet A, et al. Diagnosis of Viral Infections Using Myxovirus Resistance Protein a (Mxa). Pediatrics (2015) 135(4):e985–93. doi: 10.1542/peds.2014-1946
61. Piri R, Ivaska L, Yahya M, Toivonen L, Lempainen J, Kataja J, et al. Prevalence of Respiratory Viruses and Antiviral Mxa Responses in Children With Febrile Urinary Tract Infection. Eur J Clin Microbiol Infect Dis (2020) 39(7):1239–44. doi: 10.1007/s10096-020-03836-5
62. Self W, Rosen J, Sharp S, Filbin M, Hou P, Parekh A, et al. Diagnostic Accuracy of Febridx: A Rapid Test to Detect Immune Responses to Viral and Bacterial Upper Respiratory Infections. J Clin Med (2017) 6(10):94. doi: 10.3390/jcm6100094
63. Oved K, Cohen A, Boico O, Navon R, Friedman T, Etshtein L, et al. A Novel Host-Proteome Signature for Distinguishing Between Acute Bacterial and Viral Infections. PloS One (2015) 10(3):e0120012. doi: 10.1371/journal.pone.0120012
64. Ashkenazi-Hoffnung L, Oved K, Navon R, Friedman T, Boico O, Paz M, et al. A Host-Protein Signature is Superior to Other Biomarkers for Differentiating Between Bacterial and Viral Disease in Patients With Respiratory Infection and Fever Without Source: A Prospective Observational Study. Eur J Clin Microbiol Infect Dis (2018) 37(7):1361–71. doi: 10.1007/s10096-018-3261-3
65. van Houten CB, de Groot JAH, Klein A, Srugo I, Chistyakov I, de Waal W, et al. A Host-Protein Based Assay to Differentiate Between Bacterial and Viral Infections in Preschool Children (OPPORTUNITY): A Double-Blind, Multicentre, Validation Study. Lancet Infect Dis (2016) 3099(16):1–10. doi: 10.1016/S1473-3099(16)30519-9
66. Song Y, Chen Y, Dong X, Jiang X. Diagnostic Value of Neutrophil CD64 Combined With CRP for Neonatal Sepsis: A Meta-Analysis. Am J Emerg Med (2019) 37(8):1571–6. doi: 10.1016/j.ajem.2019.05.001
67. Nuutila J, Jalava-Karvinen P, Hohenthal U, Kotilainen P, Pelliniemi TT, Nikoskelainen J, et al. A Rapid Flow Cytometric Method for Distinguishing Between Febrile Bacterial and Viral Infections. J Microbiol Methods (2013) 92(1):64–72. doi: 10.1016/j.mimet.2012.11.005
68. Tremoulet AH, Dutkowski J, Sato Y, Kanegaye JT, Ling XB, Burns JC. Novel Data-Mining Approach Identifies Biomarkers for Diagnosis of Kawasaki Disease. Pediatr Res (2015) 78(5):547–53. doi: 10.1038/pr.2015.137
69. Zandstra J, van de Geer A, Tanck MWT, van Stijn-Bringas Dimitriades D, Aarts CEM, Dietz SM, et al. Biomarkers for the Discrimination of Acute Kawasaki Disease From Infections in Childhood. Front Pediatrics (2020) 8:355. doi: 10.3389/fped.2020.00355
70. Srugo I, Klein A, Stein M, Golan-Shany O, Kerem N, Chistyakov I, et al. Validation of a Novel Assay to Distinguish Bacterial and Viral Infections. Pediatrics. (2017) 140(4):e20163453. doi: 10.1542/peds.2016-3453
71. Bourgoin P, Lediagon G, Arnoux I, Bernot D, Morange PE, Michelet P, et al. Flow Cytometry Evaluation of Infection-Related Biomarkers in Febrile Subjects in the Emergency Department. Future Microbiol (2020) 15(3):189–201. doi: 10.2217/fmb-2019-0256
72. Bourgoin P, Soliveres T, Barbaresi A, Loundou A, Belkacem IA, Arnoux I, et al. CD169 and CD64 Could Help Differentiate Bacterial From Covid-19 or Other Viral Infections in the Emergency Department. Cytom Part A (2021) 25:10.1002/cyto.a.24314. doi: 10.1101/2020.10.28.20221259
73. Qiu X, Li J, Yang X, Tang J, Shi J, Tong Y, et al. Is Neutrophil CD11b a Special Marker for the Early Diagnosis of Sepsis in Neonates a Systematic Review and Meta-Analysis. BMJ Open (2019) 9(4):1–7. doi: 10.1136/bmjopen-2018-025222
74. Dai J, Jiang W, Min Z, Yang J, Tan Y, Ma T, et al. Neutrophil CD64 as a Diagnostic Marker for Neonatal Sepsis: Meta-Analysis. Adv Clin Exp Med (2017) 26(2):327–32. doi: 10.17219/acem/58782
75. Herlihy JM, D’Acremont V, Burgess DCH, Hamer DH. Diagnosis and Treatment of the Febrile Child. in: Disease Control Priorities, Third Edition. Reproductive Maternal Newborn Child Health (2016) 2:137–61. doi: 10.1596/978-1-4648-0348-2_ch8
76. Boraschi D, Italiani P, Weil S, Martin MU. The Family of the Interleukin-1 Receptors. Immunological Rev (2018) 281:197–232. doi: 10.1111/imr.12606
77. Dinarello CA, Novick D, Kim S, Kaplanski G. Interleukin-18 and IL-18 Binding Protein. Front Immunol (2013) 4:289. doi: 10.3389/fimmu.2013.00289
78. Shimizu M, Nakagishi Y, Inoue N, Mizuta M, Ko G, Saikawa Y, et al. Interleukin-18 for Predicting the Development of Macrophage Activation Syndrome in Systemic Juvenile Idiopathic Arthritis. Clin Immunol (2015) 160(2):277–81. doi: 10.1016/j.clim.2015.06.005
79. Bracaglia C, De Graaf K, Marafon DP, Guilhot F, Ferlin W, Prencipe G, et al. Elevated Circulating Levels of Interferon-Γ and Interferon-Γ-Induced Chemokines Characterize Patients With Macrophage Activation Syndrome Complicating Systemic Juvenile Idiopathic Arthritis. Ann Rheum Dis (2017) 76(1):166–72. doi: 10.1136/annrheumdis-2015-209020
80. Girard C, Rech J, Brown M, Allali D, Roux-Lombard P, Spertini F, et al. Elevated Serum Levels of Free Interleukin-18 in Adult-Onset Still’s Disease. Rheumatol (United Kingdom) (2016) 55(12):2237–47. doi: 10.1093/rheumatology/kew300
81. Mazodier K, Marin V, Novick D, Farnarier C, Robitail S, Schleinitz N, et al. Severe Imbalance of IL-18/IL-18BP in Patients With Secondary Hemophagocytic Syndrome. Blood (2005) 106(10):3483–9. doi: 10.1182/blood-2005-05-1980
82. Put K, Vandenhaute J, Avau A, van Nieuwenhuijze A, Brisse E, Dierckx T, et al. Inflammatory Gene Expression Profile and Defective Interferon-Γ and Granzyme K in Natural Killer Cells From Systemic Juvenile Idiopathic Arthritis Patients. Arthritis Rheumatol (2017) 69(1):213–24. doi: 10.1002/art.39933
83. Prencipe G, Bracaglia C, De Benedetti F. Interleukin-18 in Pediatric Rheumatic Diseases. Curr Opin Rheumatol (2019) 31(5):421–7. doi: 10.1097/BOR.0000000000000634
84. Vastert SJ, De Jager W, Noordman BJ, Holzinger D, Kuis W, Prakken BJ, et al. Effectiveness of First-Line Treatment With Recombinant Interleukin-1 Receptor Antagonist in Steroid-Naive Patients With New-Onset Systemic Juvenile Idiopathic Arthritis: Results of a Prospective Cohort Study. Arthritis Rheumatol (2014) 66(4):1034–43. doi: 10.1002/art.38296
85. Belkaya S, Michailidis E, Korol CB, Kabbani M, Cobat A, Bastard P, et al. Inherited IL-18BP Deficiency in Human Fulminant Viral Hepatitis. J Exp Med 2019/06/18 Ed (2019) 216(8):1777–90. doi: 10.1084/jem.20190669
86. Bracaglia C, Prencipe G, De Benedetti F. Macrophage Activation Syndrome: Different Mechanisms Leading to a One Clinical Syndrome. Pediatr Rheumatol (2017) 15(1):5. doi: 10.1186/s12969-016-0130-4
87. Mizuta M, Shimizu M, Inoue N, Nakagishi Y, Yachie A. Clinical Significance of Serum CXCL9 Levels as a Biomarker for Systemic Juvenile Idiopathic Arthritis Associated Macrophage Activation Syndrome. Cytokine (2019) 119:182–7. doi: 10.1016/j.cyto.2019.03.018
88. Gohar F, Kessel C, Lavric M, Holzinger D, Foell D. Review of Biomarkers in Systemic Juvenile Idiopathic Arthritis: Helpful Tools or Just Playing Tricks? Arthritis Res Ther (2016) 18(1):163. doi: 10.1186/s13075-016-1069-z
89. Feist E, Mitrovic S, Fautrel B. Mechanisms, Biomarkers and Targets for Adult-Onset Still’s Disease. Nat Rev Rheumatol (2018) 14(10):603–18. doi: 10.1038/s41584-018-0081-x
90. Shoop-Worrall SJW, Wu Q, Davies R, Hyrich KL, Wedderburn LR. Predicting Disease Outcomes in Juvenile Idiopathic Arthritis: Challenges, Evidence, and New Directions. Lancet Child Adolesc Heal (2019) 3(10):725–33. doi: 10.1016/S2352-4642(19)30188-9
91. Degraeuwe PLJ, Beld MPA, Ashorn M, Canani RB, Day AS, Diamanti A, et al. Faecal Calprotectin in Suspected Paediatric Inflammatory Bowel Disease. J Pediatr Gastroenterol Nutr (2015) 60(3):339–46. doi: 10.1097/MPG.0000000000000615
92. Austermann J, Spiekermann C, Roth J. S100 Proteins in Rheumatic Diseases. Nat Rev Rheumatol (2018) 14(9):528–41. doi: 10.1038/s41584-018-0058-9
93. Mende R, Vincent FB, Kandane-Rathnayake R, Koelmeyer R, Lin E, Chang J, et al. Analysis of Serum Interleukin (Il)-1β and IL-18 in Systemic Lupus Erythematosus. Front Immunol (2018) 9:1250. doi: 10.3389/fimmu.2018.01250
94. McCrindle BW, Rowley AH, Newburger JW, Burns JC, Bolger AF, Gewitz M, et al. Diagnosis, Treatment, and Long-Term Management of Kawasaki Disease: A Scientific Statement for Health Professionals From the American Heart Association. Circulation (2017) 135(17):e927–99. doi: 10.1161/CIR.0000000000000484
95. Chaudhary H, Nameirakpam J, Kumrah R, Pandiarajan V, Suri D, Rawat A, et al. Biomarkers for Kawasaki Disease: Clinical Utility and the Challenges Ahead. Front Pediatr (2019) 18(7):1–10. doi: 10.3389/fped.2019.00242
96. Mutz KO, Heilkenbrinker A, Lönne M, Walter JG, Stahl F. Transcriptome Analysis Using Next-Generation Sequencing. Curr Opin Biotechnol (2013) 24(1):22–30. doi: 10.1016/j.copbio.2012.09.004
97. Lazear HM, Schoggins JW, Diamond MS. Shared and Distinct Functions of Type I and Type Iii Interferons. Immunity (2019) 50(4):907–23. doi: 10.1016/j.immuni.2019.03.025
98. Pizarro-Cerdá J, Cossart P. Bacterial Adhesion and Entry Into Host Cells. Cell (2006) 124:715–27. doi: 10.1016/j.cell.2006.02.012
99. Ramilo O, Allman W, Chung W, Mejias A, Ardura M, Glaser C, et al. Gene Expression Patterns in Blood Leukocytes Discriminate Patients With Acute Infections. Blood (2007) 109(5):2066–77. doi: 10.1182/blood-2006-02-002477
100. Hu X, Yu J, Crosby SD, Storch GA. Gene Expression Profiles in Febrile Children With Defined Viral and Bacterial Infection. Proc Natl Acad Sci USA (2013) 110(31):12792–7. doi: 10.1073/pnas.1302968110
101. Herberg JA, Kaforou M, Wright VJ, Eleftherohorinou H, Hoggart CJ, Cebey- M, et al. Diagnostic Test Accuracy of a 2-Transcript Host RNA Signature for Discriminating Bacterial Vs Viral Infection in Febrile Children. JAMA (2016) 316: (8):835–45. doi: 10.1001/jama.2016.11236
102. Kaforou M, Herberg JA, Wright VJ, Coin LJM, Levin M. Diagnosis of Bacterial Infection Using a 2-Transcript Host RNA Signature in Febrile Infants 60 Days or Younger. JAMA - J Am Med Assoc (2017) 317(15):1577–8. doi: 10.1001/jama.2017.1365
103. Andres-Terre M, McGuire HM, Pouliot A, Bongen E, Sweeney TE, Tato CM, et al. Integrated, Multi-Cohort Analysis Identifies Conserved Transcriptional Signatures Across Multiple Respiratory Viruses. Immunity (2015) 43(6):1199–211. doi: 10.1016/j.immuni.2015.11.003
104. Heinonen S, Jartti T, Garcia C, Oliva S, Smitherman C, Anguiano E, et al. Rhinovirus Detection in Symptomatic and Asymptomatic Children Value of Host Transcriptome Analysis. Am J Respir Crit Care Med (2016) 193(7):772–82. doi: 10.1164/rccm.201504-0749OC
105. Mayhew MB, Buturovic L, Luethy R, Midic U, Moore AR, Roque JA, et al. A Generalizable 29-Mrna Neural-Network Classifier for Acute Bacterial and Viral Infections. Nat Commun (2020) 11(1):1177. doi: 10.1038/s41467-020-14975-w
106. Mahajan P, Kuppermann N, Mejias A, Suarez N, Chaussabel D, Casper TC, et al. Association of RNA Biosignatures With Bacterial Infections in Febrile Infants Aged 60 Days or Younger. JAMA - J Am Med Assoc (2016) 316(8):846–57. doi: 10.1001/jama.2016.9207
107. Sampson DL, Fox BA, Yager TD, Bhide S, Cermelli S, McHugh LC, et al. A Four-Biomarker Blood Signature Discriminates Systemic Inflammation Due to Viral Infection Versus Other Etiologies. Sci Rep (2017) 7(1):1–17. doi: 10.1038/s41598-017-02325-8
108. Blohmke CJ, Muller J, Gibani MM, Dobinson H, Shrestha S, Perinparajah S, et al. Diagnostic Host Gene Signature for Distinguishing Enteric Fever From Other Febrile Diseases. EMBO Mol Med (2019) 11(10):1–16. doi: 10.15252/emmm.201910431
109. Wright VJ, Herberg JA, Kaforou M, Shimizu C, Eleftherohorinou H, Shailes H, et al. Diagnosis of Kawasaki Disease Using a Minimal Whole-Blood Gene Expression Signature. JAMA Pediatr (2018) 172(10):1–10. doi: 10.1001/jamapediatrics.2018.2293
110. Wu GC, Hu Y, Guan SY, Ye DQ, Pan HF. Differential Plasma Expression Profiles of Long Non-Coding Rnas Reveal Potential Biomarkers for Systemic Lupus Erythematosus. Biomolecules (2019) 9(6):206. doi: 10.3390/biom9060206
111. Sweeney TE, Braviak L, Tato CM, Khatri P. Genome-Wide Expression for Diagnosis of Pulmonary Tuberculosis: A Multicohort Analysis. Lancet Respir Med (2016) 4(3):213–24. doi: 10.1016/S2213-2600(16)00048-5
112. Pennisi I, Rodriguez-Manzano J, Moniri A, Kaforou M, Herberg JA, Levin M, et al. Translation of a Host Blood RNA Signature Distinguishing Bacterial From Viral Infection Into a Platform Suitable for Development as a Point-of-Care Test. JAMA Pediatrics (2021) 175(4):417–19. doi: 10.1001/jamapediatrics.2020.5227
113. Kyogoku C, Smiljanovic B, Grün JR, Biesen R, Schulte-Wrede U, Häupl T, et al. Cell-Specific Type I IFN Signatures in Autoimmunity and Viral Infection: What Makes the Difference? PloS One (2013) 8(12):e83776. doi: 10.1371/journal.pone.0083776
114. de Jesus A, Hou Y, Malle L, Canna S, Brooks SR, Kim H, et al. High Interferon (IFN) Signatures and Overlapping Clinical Features Characterize Subgroups of Patients With Presumed IFN-Mediated Autoinflammatory Diseases [Abstract]. Arthritis Rheumatol (2017) 69(suppl 10)
115. de Jesus AA, Hou Y, Brooks S, Malle L, Biancotto A, Huang Y, et al. Distinct Interferon Signatures and Cytokine Patterns Define Additional Systemic Autoinflammatory Diseases. J Clin Invest (2020) 130(4):1669–82. doi: 10.1172/JCI129301
116. Crow MK, Ronnblom L. Type I Interferons in Host Defence and Inflammatory Diseases. Lupus Sci Med (2019) 6(1):1–10. doi: 10.1136/lupus-2019-000336
117. Marongiu L, Gornati L, Artuso I, Zanoni I, Granucci F. Below the Surface: The Inner Lives of TLR4 and TLR9. J Leukocyte Biol (2019) 106:147–60. doi: 10.1002/JLB.3MIR1218-483RR
118. Barrat FJ, Crow MK, Ivashkiv LB. Interferon Target-Gene Expression and Epigenomic Signatures in Health and Disease. Nat Immunol (2019) 20:1574–83. doi: 10.1038/s41590-019-0466-2
119. Chyuan IT, Tzeng HT, Chen JY. Signaling Pathways of Type I and Type III Interferons and Targeted Therapies in Systemic Lupus Erythematosus. Cells (2019) 8:963. doi: 10.3390/cells8090963
120. Byron SA, Van Keuren-Jensen KR, Engelthaler DM, Carpten JD, Craig DW. Translating RNA Sequencing Into Clinical Diagnostics: Opportunities and Challenges. Nat Rev Genet (2016) 17:257–71. doi: 10.1038/nrg.2016.10
121. Gómez-Carballa A, Cebey-López M, Pardo-Seco J, Barral-Arca R, Rivero-Calle I, Pischedda S, et al. A Qpcr Expression Assay of IFI44L Gene Differentiates Viral From Bacterial Infections in Febrile Children. Sci Rep (2019) 9(1):1–12. doi: 10.1038/s41598-019-48162-9
122. McHugh L, Seldon TA, Brandon RA, Kirk JT, Rapisarda A, Sutherland AJ, et al. A Molecular Host Response Assay to Discriminate Between Sepsis and Infection-Negative Systemic Inflammation in Critically Ill Patients: Discovery and Validation in Independent Cohorts. PloS Med (2015) 12(12):e1001916. doi: 10.1371/journal.pmed.1001916
123. Sweeney TE, Shidham A, Wong HR, Khatri P. A Comprehensive Time-Course-Based Multicohort Analysis of Sepsis and Sterile Inflammation Reveals a Robust Diagnostic Gene Set. Sci Transl Med (2015) 7(287):287ra71. doi: 10.1126/scitranslmed.aaa5993
124. Sweeney TE, Wong HR, Khatri P. Robust Classification of Bacterial and Viral Infections Via Integrated Host Gene Expression Diagnostics. Sci Transl Med (2016) 8(346):346ra91. doi: 10.1126/scitranslmed.aaf7165
125. Davenport EE, Burnham KL, Radhakrishnan J, Humburg P, Hutton P, Mills TC, et al. Genomic Landscape of the Individual Host Response and Outcomes in Sepsis: A Prospective Cohort Study. Lancet Respir Med (2016) 4(4):259–71. doi: 10.1016/S2213-2600(16)00046-1
126. Scicluna BP, Wiewel MA, Van Vught LA, Hoogendijk AJ, Klarenbeek AM, Franitza M, et al. Molecular Biomarker to Assist in Diagnosing Abdominal Sepsis Upon ICU Admission. Am J Respir Crit Care Med (2018) 197:1070–3. doi: 10.1164/rccm.201707-1339LE
127. Hortin GL, Jortani SA, Ritchie JC, Valdes R, Chan DW. Proteomics: A New Diagnostic Frontier. Clin Chem (2006) 52(7):1218–22. doi: 10.1373/clinchem.2006.067280
128. Nys G, Cobraiville G, Servais AC, Malaise MG, de Seny D, Fillet M. Targeted Proteomics Reveals Serum Amyloid a Variants and Alarmins S100A8-S100A9 as Key Plasma Biomarkers of Rheumatoid Arthritis. Talanta (2019) 204:507–17. doi: 10.1016/j.talanta.2019.06.044
129. De Seny D, Fillet M, Ribbens C, Marée R, Meuwis MA, Lutteri L, et al. Monomeric Calgranulins Measured by SELDI-TOF Mass Spectrometry and Calprotectin Measured by ELISA as Biomarkers in Arthritis. Clin Chem (2008) 54(6):1066–75. doi: 10.1373/clinchem.2007.099549
130. De Seny D, Fillet M, Meuwis MA, Geurts P, Lutteri L, Ribbens C, et al. Discovery of New Rheumatoid Arthritis Biomarkers Using the Surface-Enhanced Laser Desorption/Ionization Time-of-Flight Mass Spectrometry Proteinchip Approach. Arthritis Rheumatol (2005) 52(12):3801–12. doi: 10.1002/art.21607
131. Eisfeld AJ, Halfmann PJ, Wendler JP, Kyle JE, Burnum-Johnson KE, Peralta Z, et al. Multi-Platform ‘Omics Analysis of Human Ebola Virus Disease Pathogenesis. Cell Host Microbe (2017) 22(6):817–29.e8. doi: 10.1016/j.chom.2017.10.011
132. Geyer PE, Holdt LM, Teupser D, Mann M. Revisiting Biomarker Discovery by Plasma Proteomics. Mol Syst Biol (2017) 13(9):942. doi: 10.15252/msb.20156297
133. Whiteaker JR, Lin C, Kennedy J, Hou L, Trute M, Sokal I, et al. A Targeted Proteomics-Based Pipeline for Verification of Biomarkers in Plasma. Nat Biotechnol (2011) 29(7):625–34. doi: 10.1038/nbt.1900
134. Addona TA, Shi X, Keshishian H, Mani DR, Burgess M, Gillette MA, et al. A Pipeline That Integrates the Discovery and Verification of Plasma Protein Biomarkers Reveals Candidate Markers for Cardiovascular Disease. Nat Biotechnol (2011) 29(7):635–43. doi: 10.1038/nbt.1899
135. Di Narzo AF, Telesco SE, Brodmerkel C, Argmann C, Peters LA, Li K, et al. High-Throughput Characterization of Blood Serum Proteomics of IBD Patients With Respect to Aging and Genetic Factors. PloS Genet (2017) 13(1):e1006565. doi: 10.1371/journal.pgen.1006565
136. Sun W, Kechris K, Jacobson S, Drummond MB, Hawkins GA, Yang J, et al. Common Genetic Polymorphisms Influence Blood Biomarker Measurements in COPD. PloS Genet (2016) 12(8):e1006011. doi: 10.1371/journal.pgen.1006011
137. Candia J, Cheung F, Kotliarov Y, Fantoni G, Sellers B, Griesman T, et al. Assessment of Variability in the Somascan Assay. Sci Rep (2017) 7(1):14248. doi: 10.1038/s41598-017-14755-5
138. Sun BB, Maranville JC, Peters JE, Stacey D, Staley JR, Blackshaw J, et al. Genomic Atlas of the Human Plasma Proteome. Nature. (2018) 558(7708):73–9. doi: 10.1038/s41586-018-0175-2
139. Penn-Nicholson A, Hraha T, Thompson EG, Sterling D, Mbandi SK, Wall KM, et al. Discovery and Validation of a Prognostic Proteomic Signature for Tuberculosis Progression: A Prospective Cohort Study. PloS Med (2019) 16(4):e1002781. doi: 10.1371/journal.pmed.1002880
140. Lin A, Salvador A, Carter JM. Multiplexed Microsphere Suspension Array-Based Immunoassays. Methods Mol Biol (2015) 1318:107–18. doi: 10.1007/978-1-4939-2742-5_11
141. Lundberg M, Eriksson A, Tran B, Assarsson E, Fredriksson S. Homogeneous Antibody-Based Proximity Extension Assays Provide Sensitive and Specific Detection of Low-Abundant Proteins in Human Blood. Nucleic Acids Res (2011) 39(15):e102. doi: 10.1093/nar/gkr424
142. Miriam CL, Jethro H, Jacobo PS, Alberto GC, Nazareth MT, Antonio S, et al. Does Viral Co-Infection Influence the Severity of Acute Respiratory Infection in Children? PloS One (2016) 11(4):e0152481. doi: 10.1371/journal.pone.0152481
143. Campigotto A, Mubareka S. Influenza-Associated Bacterial Pneumonia; Managing and Controlling Infection on Two Fronts. Expert Rev Anti-Infective Ther (2015) 13:55–68. doi: 10.1586/14787210.2015.981156
144. Patel RA, Binns HJ, Shulman ST. Reduction in Pediatric Hospitalizations for Varicella-Related Invasive Group a Streptococcal Infections in the Varicella Vaccine Era. J Pediatr (2004) 144(1):68–74. doi: 10.1016/j.jpeds.2003.10.025
145. McArdle AJ, Turkova A, Cunnington AJ. When Do Co-Infections Matter? Curr Opin Infect Diseases. (2018) 31:209–15. doi: 10.1097/QCO.0000000000000447
146. Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LFP. The Trinity of COVID-19: Immunity, Inflammation and Intervention. Nat Rev Immunol (2020) 20:363–74. doi: 10.1038/s41577-020-0311-8
147. Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical Course and Risk Factors for Mortality of Adult Inpatients With COVID-19 in Wuhan, China: A Retrospective Cohort Study. Lancet (2020) 395(10229):1054–62. doi: 10.1016/S0140-6736(20)30566-3
148. Whittaker E, Bamford A, Kenny J, Kaforou M, Jones CE, Shah P, et al. Clinical Characteristics of 58 Children With a Pediatric Inflammatory Multisystem Syndrome Temporally Associated With SARS-Cov-2. JAMA - J Am Med Assoc (2020) 324(3):259–69. doi: 10.1001/jama.2020.10369
149. Verdoni L, Mazza A, Gervasoni A, Martelli L, Ruggeri M, Ciuffreda M, et al. An Outbreak of Severe Kawasaki-Like Disease At the Italian Epicentre of the SARS-Cov-2 Epidemic: An Observational Cohort Study. Lancet (2020) 395(10239):1771–78. doi: 10.1016/S0140-6736(20)31103-X
150. Toubiana J, Poirault C, Corsia A, Bajolle F, Fourgeaud J, Angoulvant F, et al. Kawasaki-Like Multisystem Inflammatory Syndrome in Children During the Covid-19 Pandemic in Paris, France: Prospective Observational Study. BMJ (2020) 369:m2094. doi: 10.1136/bmj.m2094
151. Dufort EM, Koumans EH, Chow EJ, Rosenthal EM, Muse A, Rowlands J, et al. Multisystem Inflammatory Syndrome in Children in New York State. N Engl J Med (2020) 383(4):347–358. doi: 10.1056/NEJMoa2021756
152. Feldstein LR, Rose EB, Horwitz SM, Collins JP, Newhams MM, Son MBF, et al. Multisystem Inflammatory Syndrome in U.S. Children and Adolescents. N Engl J Med (2020) 383(4):34–346. doi: 10.1056/NEJMoa2021680
153. Morris SB, Schwartz NG, Patel P, Abbo L, Beauchamps L, Balan S, et al. Case Series of Multisystem Inflammatory Syndrome in Adults Associated With SARS-Cov-2 Infection — United Kingdom and United States, March–August 2020. MMWR Morb Mortal Wkly Rep (2020) 69(40):1450–6. doi: 10.15585/mmwr.mm6940e1
154. Merad M, Martin JC. Pathological Inflammation in Patients With COVID-19: A Key Role for Monocytes and Macrophages. Nat Rev Immunol (2020) 20:355–62. doi: 10.1038/s41577-020-0331-4
155. Alunno A, Carubbi F, Rodríguez-Carrio J. Storm, Typhoon, Cyclone or Hurricane in Patients With COVID-19? Beware same storm that has different origin. RMD Open (2020) 6(1):e001295. doi: 10.1136/rmdopen-2020-001295
156. Kaufman KM, Linghu B, Szustakowski JD, Husami A, Yang F, Zhang K, et al. Whole-Exome Sequencing Reveals Overlap Between Macrophage Activation Syndrome in Systemic Juvenile Idiopathic Arthritis and Familial Hemophagocytic Lymphohistiocytosis. Arthritis Rheumatol (2014) 66(12):3486–95. doi: 10.1002/art.38793
157. Grandemange S, Sanchez E, Louis-Plence P, Tran Mau-Them F, Bessis D, Coubes C, et al. A New Autoinflammatory and Autoimmune Syndrome Associated With NLRP1 Mutations: NAIAD (NLRP1- Associated Autoinflammation With Arthritis and Dyskeratosis). Ann Rheum Dis (2017) 76(7):1191–8. doi: 10.1136/annrheumdis-2016-210021
158. Kassebaum N, Kyu HH, Zoeckler L, Olsen HE, Thomas K, Pinho C, et al. Child and Adolescent Health From 1990 to 2015: Findings From the Global Burden of Diseases, Injuries, and Risk Factors 2015 Study. JAMA Pediatr (2017) 171:573–92. doi: 10.1001/jamapediatrics.2017.0250
159. Siahanidou T, Margeli A, Tsirogianni C, Charonu S, Giannaki M, Vavourakis E, et al. Clinical value of soluble urokinase-type plasminogen activator receptor levels in term neonates with infection or sepsis: a prospective study. Mediators Inflamm (2014) 2014:375702. doi: 10.1155/2014/375702
160. Ni W, Han Y, Zhao J, et al. Serum soluble urokinase-Type plasminogen activator receptor as a biological marker of bacterial infection in adults: A systematic review and meta-Analysis. Sci Rep (2016) 6. doi: 10.1038/srep39481
161. Schlapbach L, Graf R, Woerner A, et al. Pancreatic stone protein as a novel marker for neonatal sepsis. Crit Care (2012) 16(S3). doi: 10.1186/cc11765
162. Toivonen L, Schuez-Havupalo L, Rulli M, Ilonen J, Pelkonen J, Melen K, et al. Blood Mxa Protein as a Marker for Respiratory Virus Infections in Young Children. J Clin Virol (2015) 62:8–13. doi: 10.1016/j.jcv.2014.11.018
Keywords: biomarker, febrile children, bacterial infection, viral infection, inflammation
Citation: Zandstra J, Jongerius I and Kuijpers TW (2021) Future Biomarkers for Infection and Inflammation in Febrile Children. Front. Immunol. 12:631308. doi: 10.3389/fimmu.2021.631308
Received: 19 November 2020; Accepted: 12 April 2021;
Published: 17 May 2021.
Edited by:
Luregn J. Schlapbach, University Children’s Hospital Zurich, SwitzerlandReviewed by:
Elizabeth Alpern, Ann & Robert H. Lurie Children’s Hospital of Chicago, United StatesThierry Roger, Centre Hospitalier Universitaire Vaudois (CHUV), Switzerland
Copyright © 2021 Zandstra, Jongerius and Kuijpers. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Taco W. Kuijpers, t.w.kuijpers@amsterdamumc.nl
 Judith Zandstra
Judith Zandstra Ilse Jongerius
Ilse Jongerius Taco W. Kuijpers
Taco W. Kuijpers