- 1Department of Vector Genomics and Vector Biology, ICMR- National Institute of Malaria Research, New Delhi, India
- 2Academy of Scientific and Innovative Research (AcSIR), Ghaziabad, India
- 3ICMR-National Institute of Nutrition, Hyderabad, Telangana, India
- 4ICMR-National Institute of Malaria Research, Field Unit, Chennai, India
Aim: The insect cuticle, vital for structural maintenance, forms their exoskeleton. It is mainly composed of an intermesh of – structural cuticle proteins (CPs) with polysaccharide chitin. The insect CPs encoded by CP genes are indispensable for morphology, development and adaptation to various ecological niches across all life stages. The number of CPs may vary across genera and species, with almost 150 proteins in Bombyx mori and more than 298 CPs found in Anopheles gambiae. While they have been extensively studied in insects such as agricultural pests, limited studies have been conducted on mosquitoes, particularly those relevant to public health, such as the Anopheles a key malaria vector.
Objective: This review recapitulates current knowledge on CPs in insects, while also underscoring vital knowledge gaps regarding regulation and metabolic crosstalk of CPs with other signaling and/or metabolic pathways.
Methods: We performed a comprehensive review of published studies and extracted data from databases including Vectorbase and NCBI with the aim of retrieving information on cuticular proteins, their gene families, abundance and associated functions. Additionally, we identified and analyzed the gaps in the available information. A literature search was conducted between (2000 and 2025) in an electronic database using PubMed, Scopus and Google Scholar. The search keywords were: cuticular proteins, cuticular genes, Anopheles, mosquito cuticle proteins, insecticide resistance, and CP gene families.Inclusion criteria: peer-reviewed research articles and review papers particularly focused on CPs in insects and Anopheles mosquito species.
Results: In the present review, we provide comprehensive analysis of cuticle protein families across insects including mosquitoes based on available data. We further highlight their basic constituents and protein domain structure, offering insight into their role in insect physiology. We have effectively integrated insect studies with mosquito-specific research on CPs (bridging the gap between insect and mosquito-specific research). This holistic approach would facilitate a broader comprehension of CPs in both insect and mosquito vectors.
Main conclusions: The goal of this study is to enhance our understanding of insects and Anopheles biology and how studies on CPs could be leveraged to develop novel strategy for management of pest and combat vector-borne diseases (VBDs).
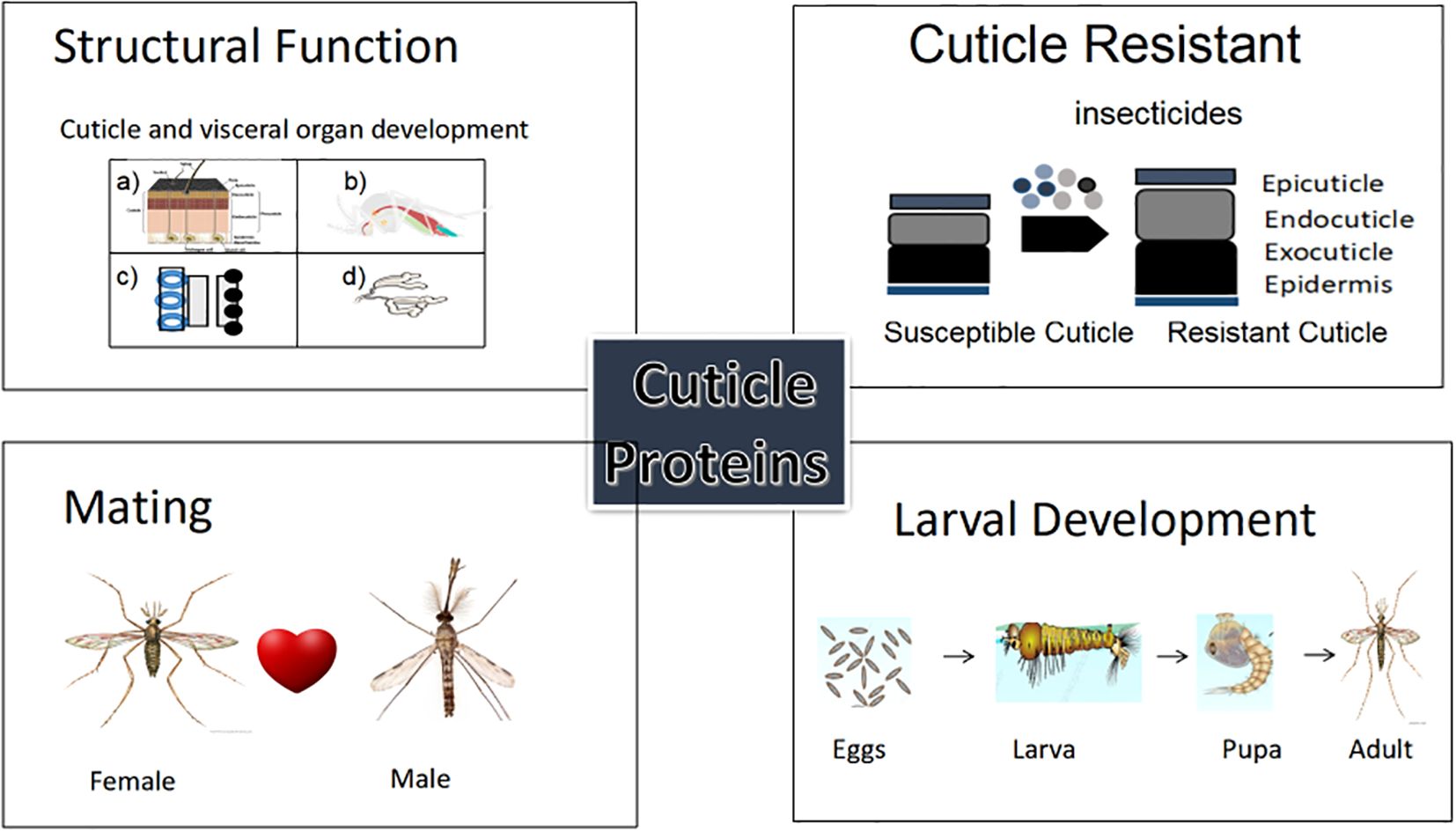
Graphical Abstract. The figure illustrates the wide-ranging functional roles of cuticular proteins (CPs) in insects and mosquito vectors. Each panel showcases a distinct biological function (1) structural development (2) insecticide resistance, (3) mating behavior, and (4) metamorphic development.
1 Introduction
The Phylum Arthropoda, the largest phylum under metazoa, encompasses the class Insecta, characterized by distinct features such as hard exoskeletons, chitin, and jointed appendages (1). Insects are renowned for playing crucial roles in ecosystems such as decomposing organic substances, participating in predator-prey and host-parasite interaction as well as maintaining ecological balances, however they also represent significant threat to public health and the agricultural industry (2). The cuticle of insect is central to their success, playing a multitude of roles essential for their survival. The insect cuticle offers a broad range of function including protection against water desiccation and retention, providing immunity, aid to locomotory movement and adhesion, facilitate morphogenetic shedding of the cuticle during their life cycle development, and temporary storage of food during starving or any unfavorable condition (3, 4). These research studies underscore the importance of CPs in insect biology. Targeting cuticular proteins offers a promising approach for developing novel insecticides.
The cuticle of insects consists of two primary layers: the epicuticle (without chitin) and the procuticle. The epicuticle is further subdivided into the outer epicuticle and the inner epicuticle (5). The outer layer of the epicuticle, cuticulin, consist of various protective layers, including a tanned thin layer made up of polyphenols and lipids whereas the inner epicuticle comprises lipids and proteins that form a barrier, rendering insects impermeable to water, preventing from desiccation and enzymatic digestion. Additionally, this layer facilitates the gradual release of hydrocarbon compounds (pheromones) during mate and colony selection (6). The inner layer of the cuticle, the procuticle, is further divided into the exocuticle and the endocuticle. The exocuticle, the outermost layer, also referred to as the envelope of the membrane, forms a physically harder layer. The endocuticle, formed by softer poly-N-acetylglucosamine (chitin), is composed of characteristically distinct cuticular proteins (CPs) known as arthropodin (7, 8). The CPs and chitin filaments act as functional partners to facilitate the shedding of old cuticles and the formation of new cuticle during the molting process.
The insect cuticle, the outermost complex and essential protective exoskeleton layer, varies in composition among genera and species (9). Furthermore, its composition alters at every metamorphic stage of insects (10). For instance, the cuticular layer of the Drosophila melanogaster undergoes alteration during its various development stages accordingly (11). The process of pigmentation and sclerotization eventuates as the cuticle ages wherein CPs are cross-linked to chitin filaments through quinones or quinone methides produced by laccase -2 enzyme, which oxidizes N-acetyl catechols (12). During sclerotization, CPs participate in the formation of histidine-β-dopamine, the primary adduct of hard cuticles, which is formed by the histidine amino acid residues in CPs (13). CPs, encoded by CP genes, make up approximately 1–2% of an insect’s genome (14). Consequently, these proteins offer diverse functionalities tailored to each developmental stage of the insect (5, 11, 15–17). The world incurs heavy economic losses due to insect pests which also jeopardize food security (18). Additionally, mosquitoes through transmission of parasites and pathogens cause several VBDs, significantly contributing to morbidity and mortality in public across the globe (According to 19). Given that the CPs play vital roles at each developmental stage of the insects including mosquito, from the larval to the adult stage- and some are even activated in the egg stage. The study of the CPs is crucial for comprehending the life cycle, morphology, and physiology of insects. Evaluating the mechanistic insight into the role of CPs can empower us to comprehend the underlying molecular mechanisms governing the insect/mosquito physiology and may open a new avenue in discovering novel targeted pest and vector control tools.
2 Cuticle protein
The CPs have been characterized with unique protein motifs, signal peptides, and a chitin-binding domain. Nearly every CP has glycine, proline, and tyrosine amino acid residues with distinct features and functions. Overall, the CPs have been categorized into different families based on their conserved motifs (Table 1). Certain protein families are unique to specific insect groups (Figure 1), such as the CPLC family found in Dipterans while others are exclusive to particular insect species, like Apidermins in honey bees and Dumpy in fruit flies (26, 33).
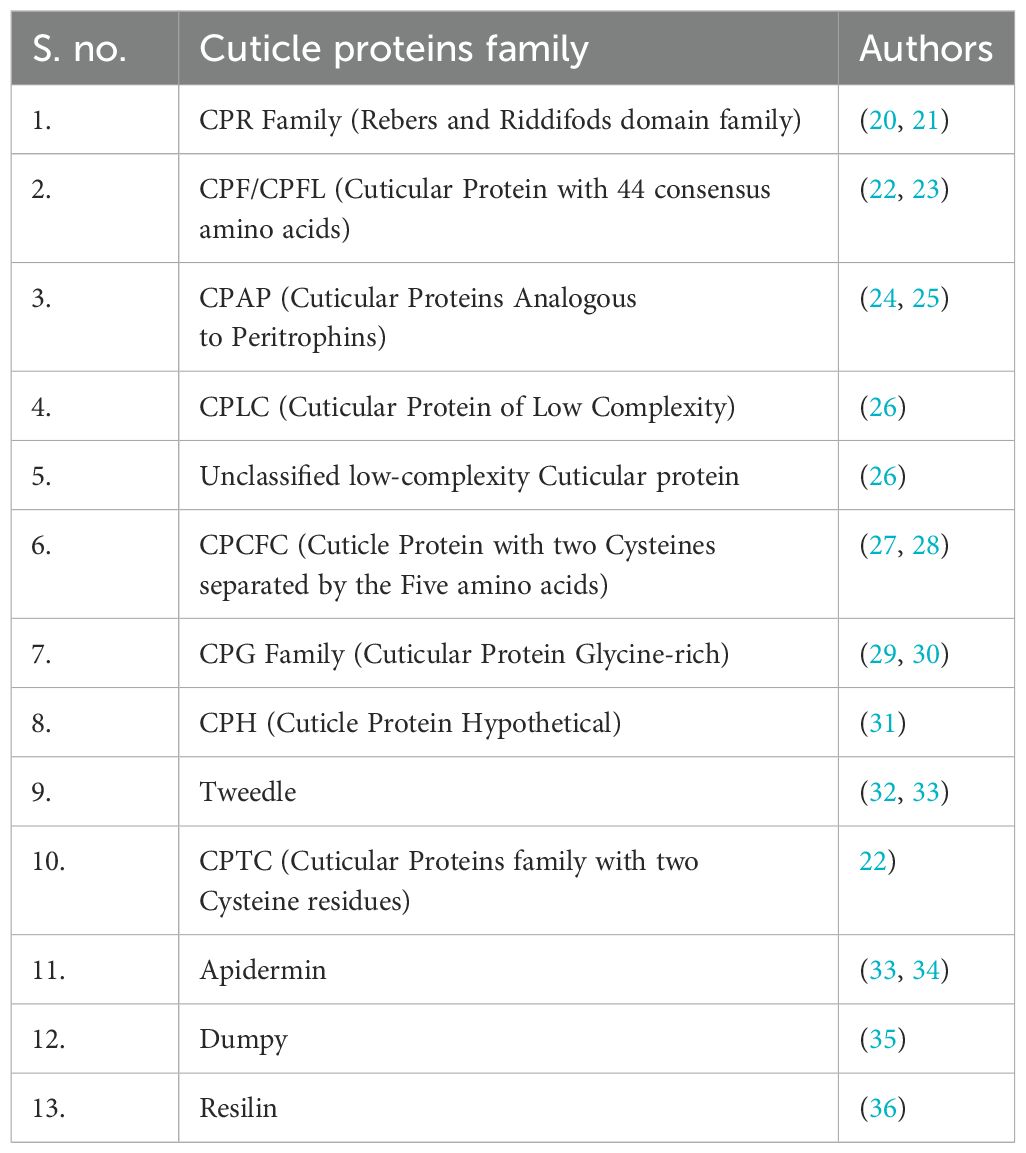
Table 1. The cuticular proteins family is divided into 13 major groups (as mentioned in Figure 1).
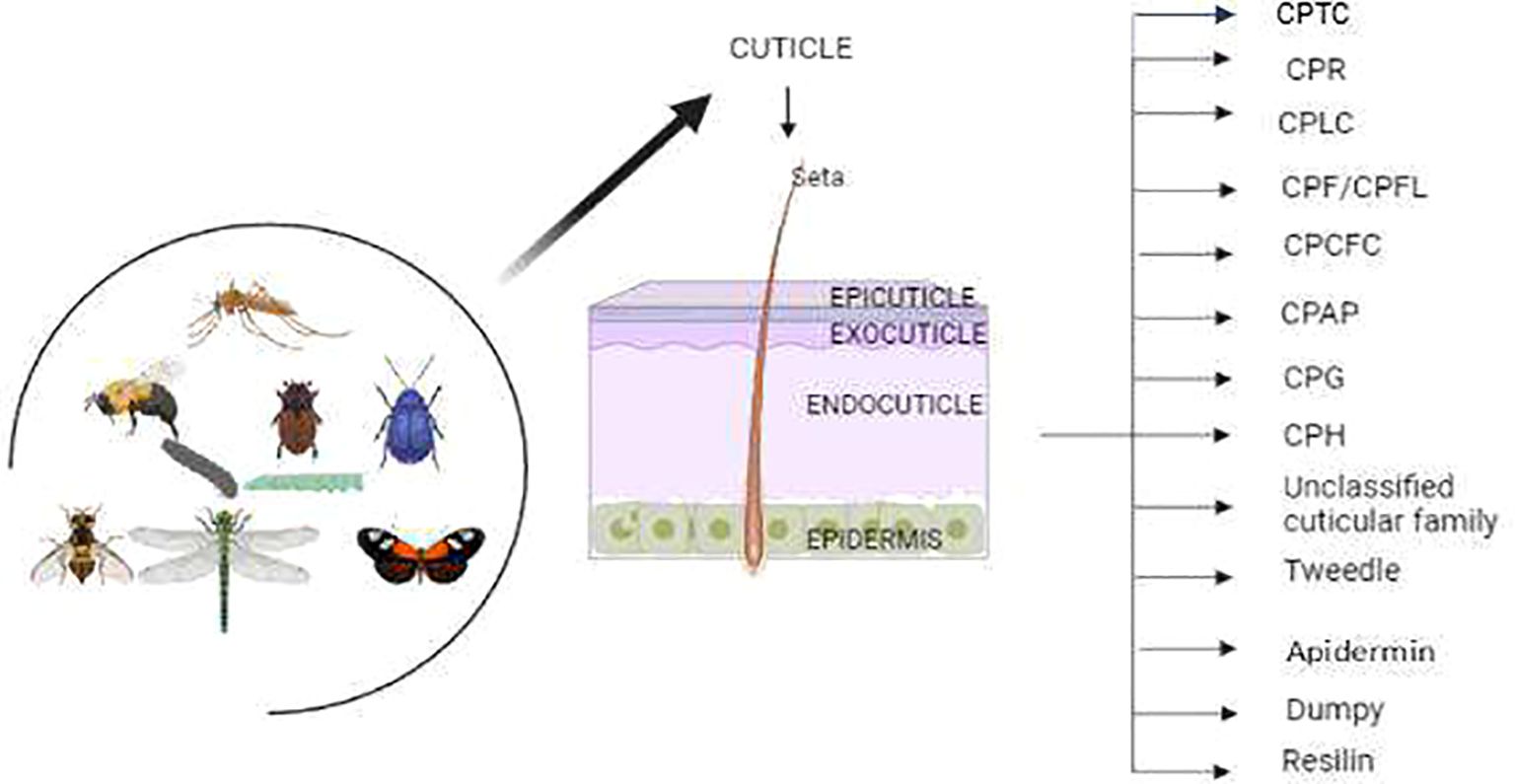
Figure 1. The insects have two major cuticle layers 1) Exocuticle and 2) Procuticle, where procuticle layers are subdivided into epicuticle and exocuticle. The Procuticle layers have 13 major cuticle proteins in insects including moths, beetles, flies and mosquitoes.
3 Common domain structure of the Cuticle Protein
The cuticular protein is a large family of proteins that also have various common amino acid residues and motifs in their peptides. The occurrence of the motifs depends on the function of the peptides (33, 37, 38). In many CP families, various cuticular genes are found to have a single signal peptide (158/160) at the N-terminus (mentioned in Figure 2), repeats of GGX or AAP (AV), and at least one chitin-binding domain as described in Figure 2, except the CPF/CPFL gene family (13, 38, 39). The cuticle proteins are normally secreted by epidermal cells and categorized into protein families based on their conserved motifs (40).

Figure 2. General diagram of cuticle proteins having signal peptides at the N – terminal and at least a chitin-binding domain in peptide.
4 Cuticular protein families
4.1 CPR family
CPR family represent the Chitin-binding type R&R consensus with the Interpro ID: IPR031311 (https://www.ebi.ac.uk/interpro/search/text/IPR031311, Pfam ID PF00379). It is the largest cuticular family. This family derives its name from the discoverers of the RR motif- Rebers and Riddiford (20, 21). The RR domain possesses 35–36 amino acid consensus sequences and is present in the cuticular layers of the insects (Table 2). The extended consensus sequence having 53 amino acids is critical for chitin binding. All the RR family proteins have an RR-2 domain with the distinct sequence GFNAVV in the C-terminal of the peptide (37, 42) (refer to Figure 3). The family divides into two major sub families i.e., RR-1 and RR-2. RR-1 is present exclusively in exocuticle while RR-2 is present in both the endocuticle and exocuticle and is more histidine rich compared to RR-1 (39, 41). The conserved sequence (34–36 amino acids) extended to the 65 amino acids long stretch and was further divided into the three families, RR-1, RR-2 and RR-3. RR-3 is a small sub family, and the distinct features of this family of CPs have not been defined (44). The RR family has a high amount of histidine residues which are functionally supposed to facilitate cuticle-hardening (13).
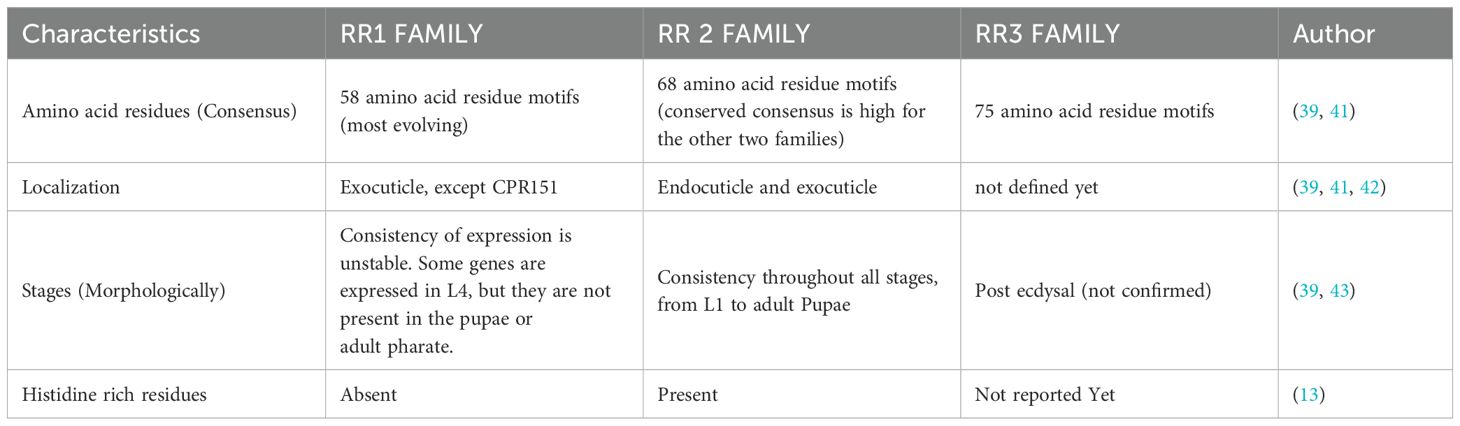
Table 2. The CPR family has three different types of domains, differentiated by each group’s characteristics.
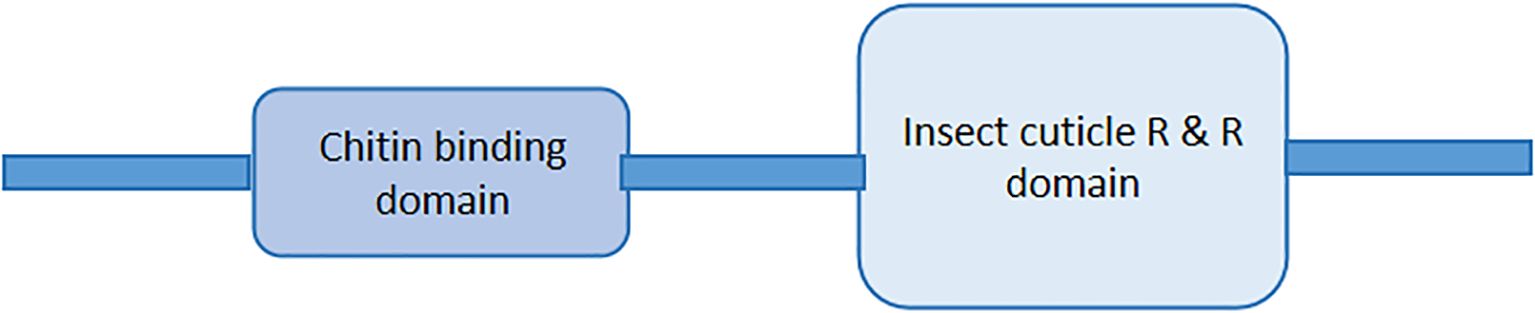
Figure 3. The RR domain structure of all three CPR proteins - The CPR family have a chitin-binding domain at the C-terminal and one Conserved RR - domain at the N-terminal.
4.1.1 CPR functions
In insects, CPs undergo repeated cycles of formation and degradation, essential for their transition from larval stages to adult stage, in the molting process. The CPR genes, accountable for cuticle formation, are significantly expressed in Tetranychus urticae possibly under the regulatory network of juvenile hormone and ecdysterone hormone dynamic during the development process (15, 45). RR1, a subtype of CPR, is remarkably associated with flexible and less sclerotized cuticle regions (for example, hind wings and intersegmental membrane) while RR2 subtype is related to rigid cuticle regions (for example, head capsules (14, 46). The functional studies based on the RR-3 subfamily have not yet been reported (44),
Many studies have evidenced that the CPR genes (e.g. CPR63 and CPR47), expressed in the exocuticle as well as endocuticle segment of the cuticle layer in insects, attribute to resistance against insecticide and confer protection from the hostile environmental during unfavorable conditions (43, 47, 48). Research on Tribolium castaneum suggests that the TcCPR4 gene is responsible for the rigidity of the cuticle and that silencing of this gene results in malfunctioning in pharate adults (12). One of the study also highlighted that transcripts corresponding to RR-2 proteins were widely distributed and highly expressed in the ovary of the Bombyx mori (29).
4.2 CPF/CPFL
The family have 44 conserved amino acid motifs and a C-terminal motif at the end. The reason CPFL is named CPFL-like is that the protein group shares the same C-terminal motif after the sequence but lacks exact homology with a 44 amino acid motifs. The conserved regions contain three aromatic amino acids, and the two families do not bind with chitin (22, 23, 39). The Pfam and InterPro IDs are PF11018 (https://www.ebi.ac.uk/interpro/entry/pfam/PF11018) /IPR022727 (https://www.ebi.ac.uk/interpro/entry/InterPro/IPR022727). Unlike other CPs, the CPF/CPFL Family proteins lack a chitin-binding domain, demonstrating that these proteins may not be associated directly with chitin. Alternatively, they may confer a non- structural role or any auxiliary role in insect cuticle.
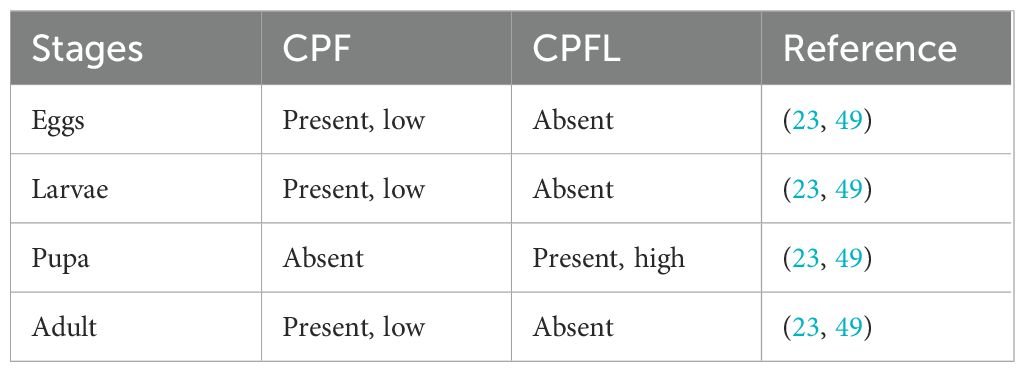
According to (23), the CPFL expression was higher during molting of the larvae. The mRNA of CPFL genes 2–7 are significantly upregulated just after ecdysis in Anopheles gambiae mosquito (Table 3).
4.2.1 Function of CPF/CPFL
Despite the presence across all insects, the function of CPF/CPFL proteins is still unclear. These proteins lack the chitin-binding domain and are not associated with chitin within the cuticle. However, their gene expression pattern was shown very active during the larval-to-adult transition, possibly underscoring their functional role in the molting process (39, 50).
4.3 CPAP family
The CPAP family comprises the three chitin-binding domains (CHBD2) with conserved cysteine residues, initially identified as obstructor protein (obsA) in Drosophila melanogaster (refer to Figure 4) (24, 51). Later, the protein family was renamed as the CPAP family characterized by having six cysteine residue rich CHBD2 domains (24, 25, 51). Overall, the CPAP genes, based on the number of CHBD2 they contain, are categorized into two families CPAP1 (one domain) and CPAP3 (three domains).

Figure 4. The common Domain structure of the CPAP family has three chitin-binding domains (https://www.ebi.ac.uk/interpro/result/InterProScan/iprscan5-R20231117-095218-0179-21412921-p1m/).
4.3.1 CPAP function
The CPAP genes are primarily localized to and expressed in tissue derived from ectoderm including the epidermis, trachea, hindgut, and foregut (24, 39). These genes are primarily known for scaffolding the cuticle structure (39), The Obs-A protein, belonging to the CPAP3 family, interacts with proteins such as, Knickkopf, Serpentine (chitin deacetylase) and chitin within the cuticle extracellular matrix (ECM) to form a complex, termed as obs-A complex. This complex reinforces the cuticle ECM and shields the cuticle from digestion and premature degradation during development (52). Silencing of CPAP1 and CPAP3 genes in T. casteneum results in abnormalities in the development stages, and specifically silencing of the CPAP3 genes lead to abnormal development of wings, walking defects, and increased post-eclosion adult mortality (53). One of the study also reported their possible role in cuticle resistance mechanism. The CPAP3 genes (CPAP3-A1, CPAP3-C1, CPAP3-D1, and CPAP3-E2) expression increased significantly in Bactrocera dorsali when exposed to malathion (54).
4.4 CPLC
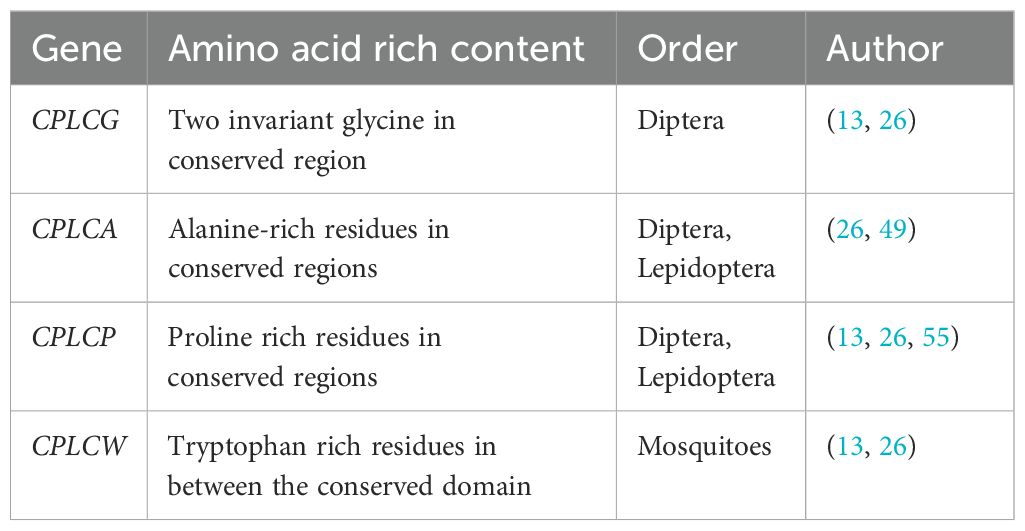
The family includes the CPLCG, CPLCW, CPLCP, and CPLCA families (Table 4). The CPLC stands for the high proportion of low complexity sequence, and the last suffix stands for the single letter of each family’s most repeated amino acid (26).
4.4.1 CPLCG
The first proteins of the gene family were identified as Dacp-1 and Dacp-2 in Drosophila melanogaster (11). The suffix ‘G’ at the end of the CPLCG denotes the two invariant glycine residues in the conserved region separated by a few amino acids and are considered to be exclusive to the order Diptera (13, 26).
4.4.2 CPLCA
The subfamily named as the CPs consists of two or more conserved alanine residues in their conserved region. The first two members of the gene were found by He et al. (22) in An. gambiae. The An. gambiae consists a conserved homologous sequences of the retinin domain for CPLCA proteins (13, 26). (13, 26). The gene family members are also identified in Lepidoptera (for example, Bombyx mori and Dendrolimus punctatus) (49).
4.4.3 CPLCP
Cornman and Willis (26) described the CPLCP gene family as enriched with proline. The four members of this family have been found in Drosophila, which encodes proline, lysine, valine, and tyrosine. However, in recent years, CPLCP has also been discovered in B. mori (55, 56).
4.4.4 CPLCW
The protein is rich in tryptophan, and the gene family is exclusively expressed only in mosquitoes; CPLCW refers to an invariant tryptophan within the conserved domain (13, 26).
4.4.5 Function of the CPLC gene
CPLC gene has been reported to contribute to insecticide resistance (IR) in different genera of mosquitoes. It has been found that the CPLCG subgroup has been particularly conferring IR in Culex and Anopheles mosquitoes, by facilitating cuticle thickening and thereby preventing cuticle penetration. However, the roles of other subgroups of the CPLC Family in the IR mechanism remain unknown. The reduced expression of CPLC genes along with CPFs and CPR genes has been shown to delay ecdysis and cause deformities in adult mosquitoes (57).
4.5 Unclassified low-complexity Cuticular protein
The Unclassified low-complexity Cuticular protein, initially discovered in An. gambiae by Cornman and Willis (26) and Willis (33), is also a low-complexity sequence but is not assigned to any CPLC family. The Cornnan named this family “CPLCX’’ as it corresponds to the chromosome number at which they have expressed. They are rich in alanine, proline, and tyrosine, and they almost have the same length and amino acid composition (26). The function is not known, but the family is exclusive to mosquitoes, particularly An. gambiae (26).
4.6 CPCFC family
Jensen et al. (27) discovered the first CPCFC gene in the cuticle of the cockroach (Blaberus craniifer) in the nymph stage as Bc-NCP1. The CPCFC name was designated by Willis et al. (28). The name CPCFC was described as a protein that has a motif with two cysteines separated by the five amino acids, i.e., C-X(5)-C, and these motifs are repeated by three times within16 amino acids (27, 28, 58).
4.6.1 Function of CPCFC
Experimental data from An. gambiae shows that the expression of agamCPCFC1 transcripts increases immediately following molting. The study revealed that CPCFC1 protein is transferred to the endocuticle where it promotes cuticle hardening (58). In Manduca sexta, the CPCFC gene has been found in the head of post-molt larvae but is absent in adults (59).
4.7 CPG family
Glycine-rich protein family with conserved sequence repeats of GXGX or GGXG named as Ld-GRPs in Lepidoptera found in epidermal cells or epidermis (16, 29, 30, 33). The first CPG gene was discovered by Suzuki et al. (60) as CPG1, and Futahashi et al. (29) found 29 whole new CPG genes in B. mori and named them CPG (cuticular protein glycine-rich).
4.7.1 Function of CPG genes
The CPG genes are richly expressed during pupal stages. In B. mori, transcript of CPG genes have been reported to be present in the hard cuticle structure including the pheromone gland, compound eye, and maxillary galea (29).
During the insect molting process, the cuticle layer undergoes a cuticle renewal process (repeated cycles of synthesis and degradation) managed by the coordinated action of ecdysteroid and juvenile hormone (JH). Suzuki et al. (60) reported that transient surge of ecdysteroid hormone during molt induces heightened expression of CPG in epidermis in B. mori. The protein, encoded by CPG1, serves as important constituent of epicuticle and is possibly implicated in cross-linking by its GGY repeats.
4.8 Apidermin
This CPs gene family is exclusive to the Apis genus and was first identified in the Apis mellifera. The family consists of three hydrophobic proteins distinguished by the high alanine content (33, 34).
4.8.1 Function of apidermins
The spatial-temporal analysis of apidermin gene family proteins revealed different expression patterns in different tissues. Apd-1 gene was predominantly expressed in the epidermis underlying the cuticle layer possibly bound for sclerotization whereas apd-2 as well as apd-3 genes were expressed in the trachea and digestive tract. Apd-3 gene was also found to be expressed in the outer epidermal structure such as eyes (33).
4.9 CPH
The protein family was also discovered by Futahashi et al. (29) in B. mori and designated a group of 34 proteins as cuticular protein hypothetical (CPH) characterized by a signal peptide and possessing sequence similarity to known CPs gene. Some of the family members are characterized by the presence of AAP (A/V) motifs specific to cuticle protein genes (31). The members of these families are highly active during the larval stage (61).
4.9.1 The function of CPH
The CPH plays a major role in the larval molting process, contributing to the cuticle thickening and conferring resistance to deltamethrin in Lepidopterans larvae (61). Gan et al. (62) discovered a midgut-specific CPH like gene in B.mori which plays a vital role in histogenesis.
4.10 Tweedle family
The Tweedle gene was named after its discovery in the mutant Drosophila TweedleD, which showed drastic changes in body shape and size in the larval stage and resembled the corpulent Tweedledee (short and stout) (32, 33). Guan et al. (32) proposed that the observed body alteration is due to the Tweedle gene and is associated with changes in the cuticle. The tweedle gene is thought to have evolved 300 million years ago through insects (26, 33). The tweedle protein does not have RR-conserved sequences. They have aromatic acids that facilitate the formation of bonds with chitin in the ECM. Tweedle has four conserved blocks, and the tweedle motif is made up of a beta-barrel structure of multiple beta-strands, which are necessary for the binding of the cuticle protein with chitin (32, 63). The conserved blocks consist of (a) the consensus of KX2–5YX3–4P in Blocks 1 and 2; (b) the consensus of Block 3 is TX2YVLXK; and (c) the consensus of Block 4 is KPEVXFX2Y, which is most conserved among insects, and the amino acids separated by blocks 1, 2, and 3, 4 are variable and proline-rich among insects and then there is abundance of proline in block 1 and 2 also (X represents the non-conserved amino acid residues within the consensus region site) (mentioned in Figure 5) (64).
4.10.1 The function of the tweedle family
It has been discovered that members of this CP family significantly impact insect cuticle development and their molting process. It is evident from the study demonstrated in Locusta migratoria where silencing of the LmTwdl1 gene, belonging to the tweedle family, led to high mortality before being molted to the next phase and thinner cuticle development (65). Soares et al. (66) studied the cross-talk of ecdysteroid hormones with tweedle genes in honey bees and their expression was found to be higher in the outer integument after ecdysteroid peak during adult exoskeleton development. Separately, heightened expression of the tweedle gene in the epidermis of Drosophila and B.mori also pinpoints their role in cuticle development. Apart from cuticle formation, higher expression of tweedle genes in the gut tissue of honey bees underscore their possible role in the formation of PM (peritrophic matrix). In a recent finding, Wang et al. (67) investigated the molecular factor conferring tolerance to exposed cold and hypoxia in Drosophila suzukii larvae. The study revealed that expression of structural cuticle genes, especially from the tweedle family, along with ATP-dependent proton pump provided biological adaptation to withstand cold and hypoxic conditions.
4.11 Dumpy
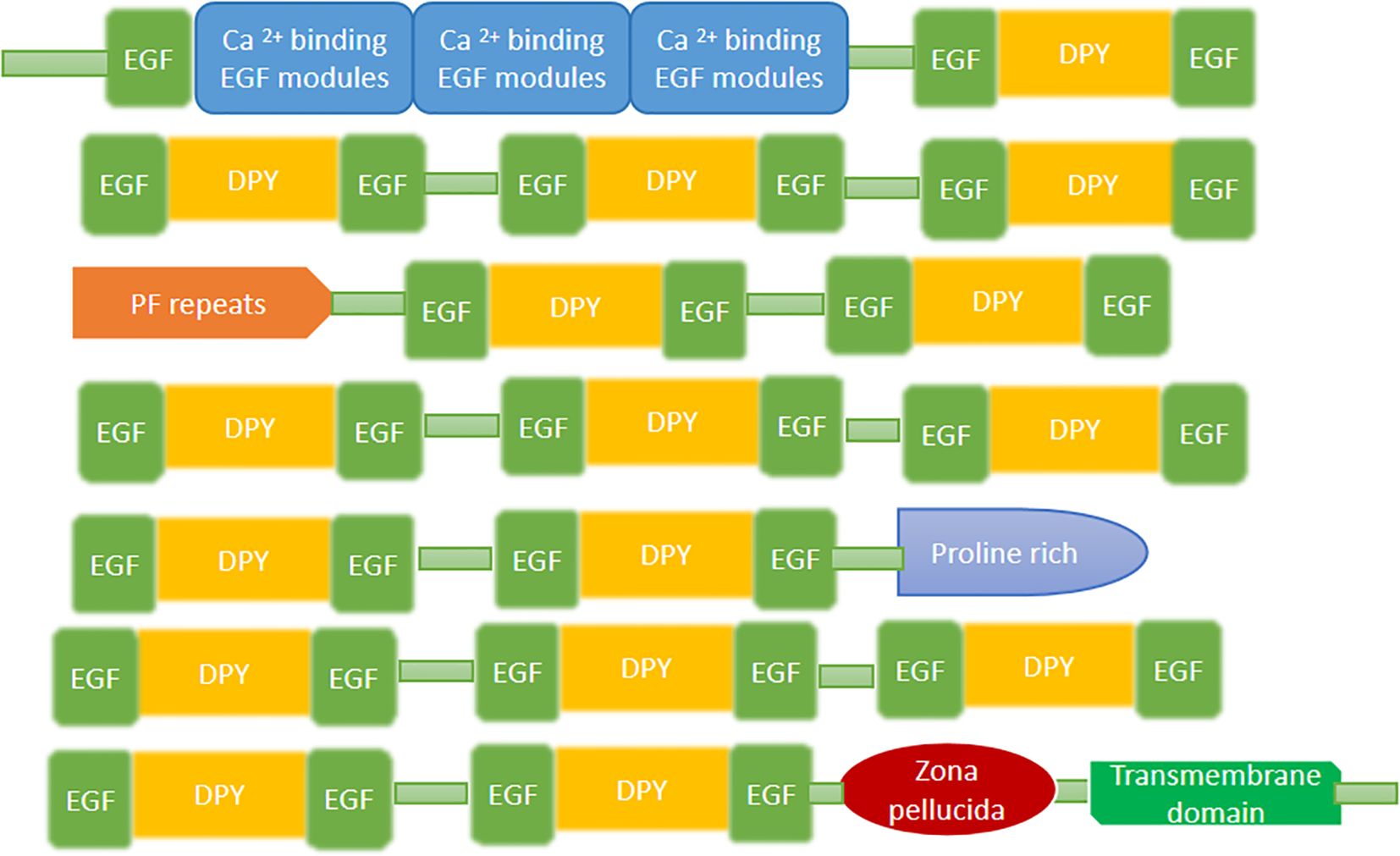
The Dumpy protein, characterized by having cysteine residue in its conserved region, was identified in Drosophila melanogaster and it is one of the largest protein found in D. melanogaster. The dumpy (DPY) protein is found in the ECM with EGF factors in its structure (35). The structure contains the multiple EGF-DPY-EGF triad domains; DPY has the cysteine-spacing pattern, with two repeated regions containing conserved tandem repeats of serine/threonine-rich P-F repeat (PF) and proline-rich repeats altogether with the PGVINIPSVPQP consensus motif. The N-terminus is enriched in calcium-binding EGF repeats, and the C-terminus contains a transmembrane domain and a Zona Pellucida (ZP) domain (mentioned in Figure 6) (35, 68).
4.11.1 The function of dumpy
Dumpy, a membrane anchored extracellular matrix protein, the majority portion of it extending into ECM, plays an important role in the epithelial tissue attachment with adjacent ECM fibers. The protein is well anchored, through its cytoplasmic domain, in epithelial tissue enabling it to bear the mechanical strength. 68 reported that dumpy protein, through its motif (repeated modular unit) anchors the epithelial tissue to overlying cuticle structure. It plays a critical role in maintaining the infrastructure and integrity of cuticle structure and drives the process of morphogenesis during development. In addition, dumpy protein also involved in wing morphogenesis (68).
4.12 Resilin
Resilins are elastomeric proteins that were first identified in the D. melanogaster (36). Weis-Fogh (69) discovered a rubber-like protein within the ECM of the thorax, wings, and hinge ligaments of dragonflies and locusts, designated them as resilin proteins. These proteins are enriched with glycine and proline and are cross-linked with di-and tri-tyrosine residues which provide them mechanical elasticity. The resilin sequence contains two isoforms called pro-resilins isoform A and isoform B. Ardell and Andersen (36)Lyons et al. (70) concluded that Drosophila melanogaster, Flea and Dragon fly have EPPVXXXYLPP consensus residues after a signal peptide at the N-terminal and GYSGGRPGGQDLG consensus sequence present before the C-terminal. This family proteins also consists of RR-2 conserved regions in the complete protein sequence. A tyrosine residue present at C - terminus and YGAP residues present at N- terminus in the resilin block arrangements Figure 7). Tyrosine residues function to cross-link with ECM (36). The RR sequence is responsible for the binding of resilin to the cuticle.
4.12.1 Function of resilin
Resilins are present in high amounts in the joints and hinges of insects, where they provide elasticity and resiliency to these structures and ward off muscle fatigueness (71). Resilins are capable of storing kinetic energy, providing high resilience, low stiffness and large strain in the tendons of insects, attributing to efficient movement and locomotion (72). All the arginine and lysine residues in the peptide chain are followed by a proline residue, which makes the peptide bond resistant to trypsin hydrolysis (36).
4.13 CPTC family
An. sinensis is also frequently discovered in dipterans, and the name implies that the cuticle protein has the two conserved cysteines that share the cysteine-containing CBDs and determined four CPTCs (CPTC 1-4) in An. gambiae (22, 73). The same sequence is found in the gene GASP (Gene Analogous to Small Peritrophins), which is related to the Drosophila cys-rich gene and is located in the tracheal epidermis (22).
4.13.1 Function of CPTC
According to Adams et al. (74), CPTC proteins are involved in the chitin-binding cuticle formation process. The CPTC functions as a serine protease inhibitor and provide protection to the old cuticle from the proteases activities. The CPTC family plays a major role in tracheae during the metamorphosis and development (73).
5 Role of cuticle in mosquitoes
Beyond maintaining structural integrity, CPs in mosquitoes also affect physiological and life history behavioral traits that are essential for survival, adaptability and reproduction. This section discusses about emerging role of cuticle proteins, particularly focus on their implication in IR, mating biology and developmental stage-specific functions (Figure 8).
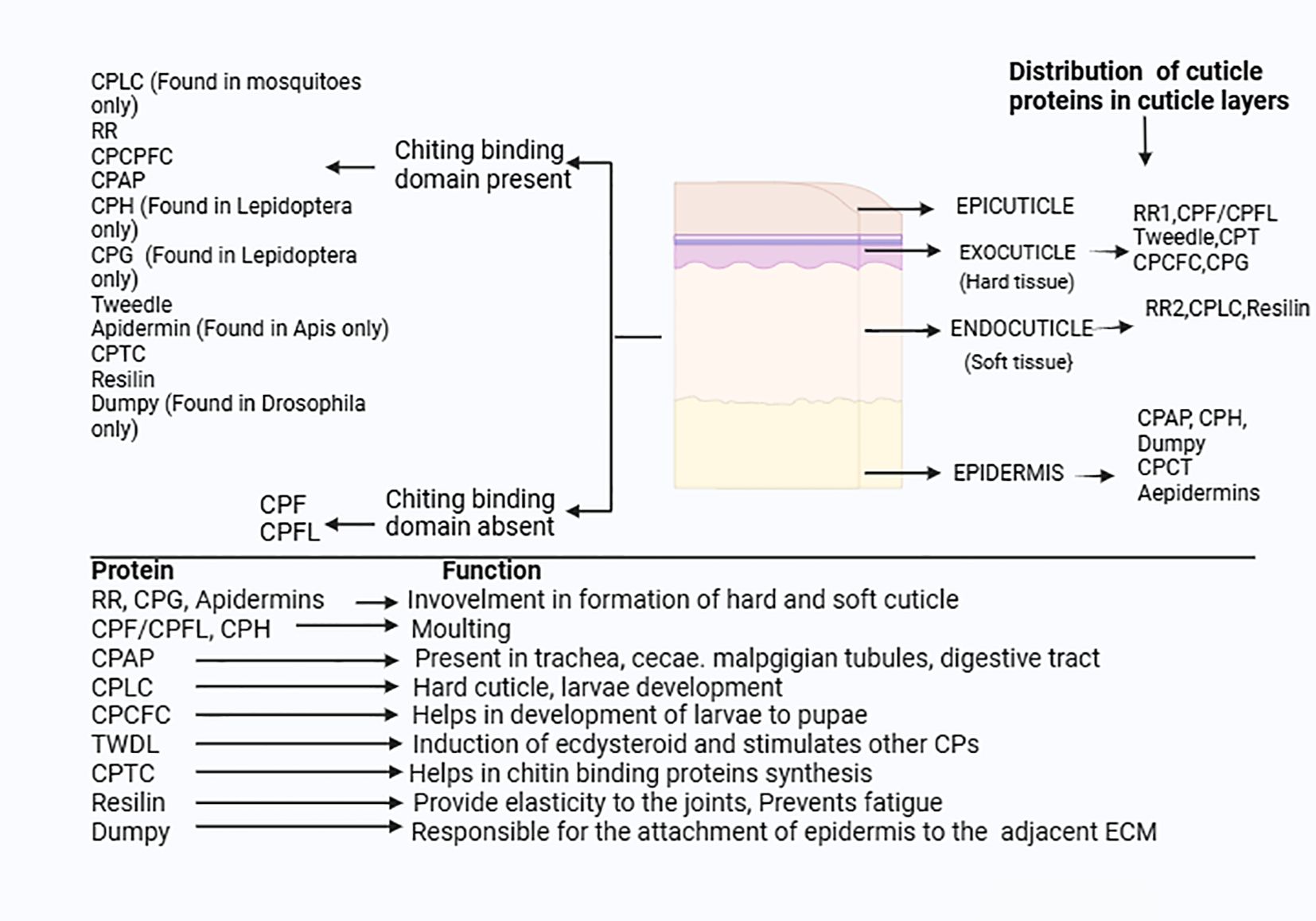
Figure 8. Distribution and function of CPs in insects: The CPs families are divided into the two major group based on presence and absence of the chitin binding domain.
5.1 Role in insecticide resistance
The use of insecticide is one of the first and most efficient methods for vector control in order to combat VBD. However, prolonged and over use of these chemical toxins designed against mosquito vectors (MVs) led to development of resistance. There are several mechanisms such as genetic, metabolic, behavioral, microbial, physiological and penetration resistance through which MVs have acquired resistance against conventionally used insecticide.
Contact dependent insecticide such as pyrethroid must traverse the cuticle before reaching their target, nervous system. Mosquitoes like other insects have developed penetration resistance mechanisms, to reduce the insecticide penetration. The cuticle modification in An. gambiae and Ae. aegypti is considered one of the key mechanisms conferring insecticide resistance (IR). Cuticular resistance is far less well known than other IR mechanisms such as CYP enzyme mediated detoxification and Knockdown resistance (kdr). The emerging evidence demonstrates that CPs attributes to insecticide resistance primarily by altering the structural and biochemical properties of the mosquito cuticle, thereby preventing permeability to insecticides. One key strategy is thickening of cuticle layers, facilitated by enhanced expression of certain CPs, like CPLCG3, identified in resistant An. gambiae. (75). In Culex pipiens pallens, the CPLCG5 gene is differentially expressed in mosquito legs which reduces the penetration of insecticides by hardening the extracellular matrix (ECM) of the cuticle during insecticide exposure in mosquitoes. It is CPLCGs that contribute to insecticide resistance by facilitating the formation of a rigid matrix and increasing the thickness of the cuticle (76). Another study reported heightened expression of two CPLC genes in resistant strains of Anopheles stephensi compared to susceptible strain mosquitoes (77). The relatively high expression of both CPLCG3 and CPLCG4 genes in the appendages of An. gambiae indicates that they are possibly accountable for conferring resistance to pyrethroids (78). These proteins are presumed to modify the cuticle structure by increasing deposition of cuticle material, thereby reducing the permeability to contact insecticides.
In addition to cuticle thickening, alteration in the structural component of cuticle layer, such as increased content of chitin polysaccharide(structural constituent of cuticle), led to slow or reduced absorption of pyrethroids in mosquito (79). Though, cuticle alteration mechanisms attribute low level of protection but become significant when synergies with other resistance mechanisms (79–81). Yahouédo et al. (75) reported synergy of cuticle and metabolic process conferring resistance in MRS strain of An. gambiae (lack kdr mutation). The overexpression of cuticle genes leads to thickening of the cuticle layer which slows the permeation of insecticide and culminates in extended time for metabolic enzymes to detoxify and excrete out toxins.
A study on Anopheles albimanus also highlight that insecticide exposure changes the bacterial diversity on both cuticle surface and internal tissues, promoting selection and abundance of Pantoea agglomerans and Pseudomonas fragi, well known insecticide metabolizing enzyme, thus attributing to IR (82, 83). This study demonstrates the possible link of host symbiotic bacteria with insecticide resistance.
However, the regulatory molecular mechanism governing the CP gene expression during IR remains poorly explored demonstrating the need for future research at molecular and functional level.
5.2 Role in mating behavior
In addition to maintaining structural functions, the cuticle hydrocarbons associated with CPs also play a vital role in sexual communication in mosquitoes. Understanding how these compounds influence mating dynamics offers insights into vector population maintenance and opportunities for novel control strategies. A study in Anopheles coluzzii reported increased cuticle hydrocarbons in mated male compared to unmated male reflect the possible role of CHC in the mating behavior (84). Furthermore, Wang et al. (85), study demonstrated that when Anopheles stephensi male was treated with cuticle hydrocarbon heptacosane, it led to increased rate of insemination compared to untreated male highlighting its role in sexual behavior. These findings suggest the importance of cuticle associated hydrocarbons in mosquito reproductive success, which may be leveraged to develop mating behavior intervention vector control strategies.
5.3 Stage-specific functions
CPs are differentially expressed in stage-specific manner, facilitating mosquitoes to adapt in different environmental conditions at each phase of its life cycle. In Aedes aegypti, CPR100, a key constituent of the cuticle layer in the procuticle is essential for survival, egg development and hatching during aquatic developmental stages. Knockdown of the CPR100 and/or its associated protein result in mortality, reduced egg hatching rate, impaired egg shell formation and enhanced sensitivity to low temperature. (86). These findings highlight that CPs also contribute in developmental transition of mosquito not only structural components but also play active roles in regulating developmental transitions.
Given that CPs cater to a wide range of functions, further exploring the role of CPs and its associated auxiliary protein catering to different physiological processes could reveal potential targets to control mosquito vectors and VBDs.
6 Research gap
1. The CPFL/CPF Protein Family, which was first discovered in lepidopterans, is still poorly understood in other insect orders. There is currently very few research available. Similarly, the CPCFC protein family has also been least explored across different arthropods species. The existing research on CPFL/CPF and CPCFC is confined to specific insect species, leaving a knowledge gap about their role characterization and distribution across other insect species. Understanding the physiological roles of these underexplored cuticle protein genes families may unravel new molecular targets for disrupting pest or mosquito cuticle integrity.
2. The cuticle proteins such as dumpy, tweedle, resilin, and CPH have been characterized only in model organisms such as Drosophila melanogaster and Bombyx mori. Their role needs to be explored in other insects. Their functional roles need to be explored in mosquito vector species, especially Anopheles and Aedes. Further deciphering their functional role in vector mosquitoes may unravel vector-specific functions that may be exploited in targeted vector control strategies.
3. The RR families cuticle protein and resilins proteins share consensus RR cuticle protein domains with few amino acids differences. Not enough studies have been conducted to determine the evolutionary link between the RR family and resilin, including their origin and diversion or possible categorization of resilins as a subgroup within the RR family.
4. Microbiota directly or indirectly through its metabolites via gut- brain-axis affect the wide array of insect biology including survival, development, immunity and cuticle formation (87). Insecticide exposure changes the bacterial community inclusive of cuticle surface bacteria. However, how this alteration in bacterial communities influence the metabolite profile and how this altered metabolite affects the cuticle surface- its structure, composition and resistance is not explored. This information gap emphasizes the importance of further inquiry into how altered bacterial populations affect metabolite profile and sequentially structure of cuticle and IR mechanism and thereby knowledge could be leveraged to reduced cuticle mediated resistance in vector mosquitoes.
5. Different MVs inhabiting different geography exhibit specific cuticle compositions that influence IR. Identifying common CPs providing resistance may reveal a shared cuticle resistance mechanism. Identifying common CPs associated with cuticle resistance may aid in developing molecular markers to detect cuticle mediated resistance.
6. Insect employs a multifaceted approach including genetic, behavioral, physiological and cuticle resistance to neutralize insecticide toxicity. While each strategy has been examined independently, there is a need to explore whether there is a regulatory mechanism that synchronizes all these processes to work in tandem. This lack of knowledge limits our understanding on interplay of cuticle resistance with different resistance mechanisms. Exploring this mechanistic interaction of cuticle resistance with different resistance mechanisms may reveal new molecular targets that can be leveraged to reduce the overall resistance mechanism.
7. The CPs offer a multitude of functions across diverse insect species ranging from structural support, contributing to immunity, preventing severe environmental stress and insecticides, to facilitating development essential for their survival and development. Although, role of different CPs have been decoded in insects and mosquitoes, leaving a significant knowledge gap on molecular signaling pathway, regulatory process involved in their gene expression and the molecular cross talk of CPs with other metabolic networks to drive any insect behavior remains unexplored.
The role of CPs goes beyond just being a structural protein. We emphasized the necessity of further research to unravel the underlying molecular intricacies of CPs in order to narrow down the current knowledge gap in this vital field of insect biology. Addressing these research gaps might open new avenues for effective pest and vector control.
7 Conclusions
This review focuses on providing comprehensive information on cuticle proteins and emphasizes their importance in insect and mosquito vectors, especially Anopheles. The diversity of cuticular proteins across different insect species reflects adaptations to various ecological niches and environmental conditions. Investigating the diversity, expression patterns, and functions of cuticular proteins can provide insights into the evolutionary processes that shaped insect morphology and physiology. With the rapid emergence of insecticide resistance presenting a significant threat to pest and vector control strategies, it is imperative to develop novel innovative strategic initiatives for sustainable pest and vector control management.
The majority of research studies have just focused on comprehending the role of CP, leaving a research gap on molecular mechanisms. Future research deciphering regulatory pathways, molecular signaling and molecular cross-with metabolic pathways may reveal new molecular target for gene based interventions like CRISPR-mediated knockdowns or RNA silencing, also known as RNA interference (RNAi). Furthermore, integration of these molecular mechanistic insights with advanced genomic and bioinformatic tools might unravel the key molecular targets for developing species specific vector or insect pest control strategies.
Author contributions
YT: Formal analysis, Data curation, Validation, Methodology, Writing – original draft, Investigation. ST: Writing – review & editing, Data curation. GK: Writing – review & editing. MJ: Writing – review & editing. VV: Writing – review & editing. RD: Writing – review & editing. SP: Project administration, Conceptualization, Writing – review & editing, Supervision. AE: Writing – review & editing. JK: Supervision, Project administration, Writing – review & editing, Conceptualization, Formal Analysis, Resources.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. The authors acknowledge the institutional support from ICMR-NIMR, to carry the work. Also, the work is part of Ph.D work for which YT received fellowship from University Grants Commission (UGC).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
CPs, Cuticle Proteins; Dpy, Dumpy; TF, Transcriptional factors; ECM, Extracellular matrix; CBD, Chitin binding domain; EGF, Ectodermal growth factor; PCP, Pupal cuticle protein; CHC, Cuticle hydrocarbons; FTZ, F1, Fushi tarazu factor 1 (Ftz-f1); Ae-aaNAT1, Aedes, Arylalkalamine N-acetyltransferase-1; JH, Juvenile Hormone; Ecd, Ecdysterone; VBDs, Vector Control Diseases; MV, Mosquitoes Vectors.
References
1. Ruggiero MA, Gordon DP, Orrell TM, Bailly N, Bourgoin T, Brusca RC, et al. A higher level classification of all living organisms. PloS One. (2015) 10:e0119248. doi: 10.1371/journal.pone.0119248
2. Eggleton P. The state of the world’s insects. Annu Rev Environ Resources. (2020) 45:61–82. doi: 10.1146/annurev-environ-012420-050035
3. Andersen SO. Biochemistry of insect cuticle. Annu Rev Entomology. (1979) 24:29–59. doi: 10.1146/annurev.en.24.010179.000333
4. Vincent JF and Wegst UG. Design and mechanical properties of insect cuticle. Arthropod structure Dev. (2004) 33:187–99. doi: 10.1016/j.asd.2004.05.006
5. Andersen SO, Hojrup P, and Roepstorff P. Insect cuticular proteins. Insect Biochem Mol Biol. (1995) 25:153–76. doi: 10.1016/0965-1748(94)00052-J
6. Sugumaran M. Complexities of cuticular pigmentation in insects. Pigment Cell melanoma Res. (2009) 22:523–25. doi: 10.1111/j.1755-148X.2009.00608.x
7. Wigglesworth VB. Croonian lecture: the insect as a medium for the study of physiology. Proc R Soc London. Ser B-Biological Sci. (1948) 135:430–46. doi: 10.1098/rspb.1948.0021
8. Locke M. The Wigglesworth Lecture: Insects for studying fundamental problems in biology. J Insect Physiol. (2001) 47:pp.495–507. doi: 10.1016/S0022-1910(00)00123-2
9. Rajabi H, Jafarpour M, Darvizeh A, Dirks JH, and Gorb SN. Stiffness distribution in insect cuticle: a continuous or a discontinuous profile? J R Soc Interface. (2017) 14:20170310. doi: 10.1098/rsif.2017.0310
10. Jafarpour M, Eshghi S, Darvizeh A, Gorb S, and Rajabi H. Functional significance of graded properties of insect cuticle supported by an evolutionary analysis. J R Soc Interface. (2020) 17:20200378.
11. Qiu J and Hardin PE. Temporal and spatial expression of an adult cuticle protein gene from Drosophila suggests that its protein product may impart some specialized cuticle function. Dev Biol. (1995) 167:pp.416–425. doi: 10.1006/dbio.1995.1038
12. Noh MY, Muthukrishnan S, Kramer KJ, and Arakane Y. Tribolium castaneum RR-1 cuticular protein TcCPR4 is required for formation of pore canals in rigid cuticle. PloS Genet. (2015) 11:e1004963. doi: 10.1371/journal.pgen.1004963
13. Liu W, Chang T, Zhao K, Sun X, Qiao H, Yan C, et al. Genome-wide annotation of cuticular protein genes in non-biting midge Propsilocerus akamusi and transcriptome analysis of their response to heavy metal pollution. Int J Biol Macromolecules. (2022) 223:pp.555–566. doi: 10.1016/j.ijbiomac.2022.10.279
14. Zhou Y, Badgett MJ, Billard L, Bowen JH, Orlando R, and Willis JH. Properties of the cuticular proteins of Anopheles Gambiae as revealed by serial extraction of adults. PloS One. (2017) 12:e0175423. doi: 10.1371/journal.pone.0175423
15. Charles JP. The regulation of expression of insect cuticle protein genes. Insect Biochem Mol Biol. (2010) 40:205–13. doi: 10.1016/j.ibmb.2009.12.005
16. Pan PL, Ye YX, Lou YH, Lu JB, Cheng C, Shen Y, et al. A comprehensive omics analysis and functional survey of cuticular proteins in the brown planthopper. Proc Natl Acad Sci. (2018) 115:pp.5175–5180. doi: 10.1073/pnas.1716951115
17. Zhou S, Zhou Y, Wang Y, Chen J, Pang L, Pan Z, et al. The developmental transcriptome of Trichopria drosophilae (Hymenoptera: Diapriidae) and insights into cuticular protein genes. Comp Biochem Physiol Part D: Genomics Proteomics. (2019) 29:245–54. doi: 10.1016/j.cbd.2018.12.005
18. Venette RC and Hutchison WD. Invasive insect species: Global challenges, strategies & opportunities. Front Insect Science. (2021) 1:650520. doi: 10.3389/finsc.2021.650520
19. World Health Organization. WHO guidelines for malaria, 14 March 2023. WHO Guidelines for Malaria (2023). Available online at: https://www.who.int/publications/i/item/guidelines-for-malaria.
20. Rebers JE and Riddiford LM. Structure and expression of a Manduca sexta larval cuticle gene homologous to Drosophila cuticle genes. J Mol Biol. (1988) 203:pp.411–423. doi: 10.1016/0022-2836(88)90009-5
21. Vaclaw MC, Sprouse PA, Dittmer NT, Ghazvini S, Middaugh CR, Kanost MR, et al. Self-assembled coacervates of chitosan and an insect cuticle protein containing a Rebers–Riddiford motif. Biomacromolecules. (2018) 19:pp.2391–2400. doi: 10.1021/acs.biomac.7b01637
22. He N, Botelho JM, McNall RJ, Belozerov V, Dunn WA, Mize T, et al. Proteomic analysis of cast cuticles from Anopheles Gambiae by tandem mass spectrometry. Insect Biochem Mol Biol. (2007) 37:135–46. doi: 10.1016/j.ibmb.2006.10.011
23. Togawa T, Dunn WA, Emmons AC, and Willis JH. CPF and CPFL, two related gene families encoding cuticular proteins of Anopheles Gambiae and other insects. Insect Biochem Mol Biol. (2007) 37:675–88. doi: 10.1016/j.ibmb.2007.03.011
24. Behr M and Hoch M. Identification of the novel evolutionary conserved obstructor multigene family in invertebrates. FEBS Lett 579(30). (2005) pp:6827–33. doi: 10.1016/j.febslet.2005.11.021
25. Jasrapuria S, Specht CA, Kramer KJ, Beeman RW, and Muthukrishnan S. Gene families of cuticular proteins analogous to peritrophins (CPAPs) in Tribolium castaneum have diverse functions. PloS One. (2012) 7:p.e49844. doi: 10.1371/journal.pone.0049844
26. Cornman RS and Willis J. Annotation and analysis of low-complexity protein families of Anopheles Gambiae that are associated with cuticle. Insect Mol Biol. (2009) 18:607–22. doi: 10.1111/j.1365-2583.2009.00902.x
27. Jensen UG, Rothmann A, Skou L, Andersen SO, Roepstorff P, and Højrup P. Cuticular proteins from the giant cockroach, Blaberus craniifer. Insect Biochem Mol Biol. (1997) 27:109–20. doi: 10.1016/S0965-1748(96)00074-4
28. Willis JH, Papandreou NC, Iconomidou VA, and Hamodrakas SJ. Cuticular proteins. Insect Mol Biol Biochem. (2012) 4:134–66. doi: 10.1016/B978-0-12-384747-8.10005-4122
29. Futahashi R, Okamoto S, Kawasaki H, Zhong YS, Iwanaga M, Mita K, et al. Genome-wide identification of cuticular protein genes in the silkworm, Bombyx mori. Insect Biochem Mol Biol. (2008) 38:1138–46. doi: 10.1016/j.ibmb.2008.05.007
30. Zhang J, Goyer C, and Pelletier Y. Environmental stresses induce the expression of putative glycine-rich insect cuticular protein genes in adult Leptinotarsa decemlineata (Say). Insect Mol Biol. (2008) 17:209–16. doi: 10.1111/j.1365-2583.2008.00796.x
31. Liu J, Li S, Li W, Peng L, Chen Z, Xiao Y, et al. Genome-wide annotation and comparative analysis of cuticular protein genes in the noctuid pest Spodoptera litura. Insect Biochem Mol Biol. (2019) 110:pp.90–97. doi: 10.1016/j.ibmb.2019.04.012
32. Guan X, Middlebrooks BW, Alexander S, and Wasserman SA. Mutation of TweedleD, a member of an unconventional cuticle protein family, alters body shape in Drosophila. Proc Natl Acad Sci. (2006) 103:16794–9. doi: 10.1073/pnas.0607616103
33. Willis JH. Structural cuticular proteins from arthropods: annotation, nomenclature, and sequence characteristics in the genomics era. Insect Biochem Mol Biol. (2010) 40:189–204. doi: 10.1016/j.ibmb.2010.02.001
34. Kucharski R, Maleszka J, and Maleszka R. Novel cuticular proteins revealed by the honey bee genome. Insect Biochem Mol Biol. (2007) 37:pp.128–134. doi: 10.1016/j.ibmb.2006.10.009
35. Carmon A, Guertin MJ, Grushko O, Marshall B, and MacIntyre R. A molecular analysis of mutations at the complex dumpy locus in Drosophila melanogaster. PloS One. (2010) 5:e12319. doi: 10.1371/journal.pone.0012319
36. Ardell DH and Andersen SO. Tentative identification of a resilin gene in Drosophila melanogaster. Insect Biochem Mol Biol. (2001) 1:965–70. doi: 10.1016/S0965-1748(01)00044-3
37. Rebers JE and Willis JH. A conserved domain in arthropod cuticular proteins binds chitin. Insect Biochem Mol Biol. (2001) 31:pp.1083–1093. doi: 10.1016/S0965-1748(01)00056-X
38. Cornman RS, Lopez D, and Evans JD. Transcriptional response of honey bee larvae infected with the bacterial pathogen Paenibacillus larvae. PloS One. (2013) 8:e65424. doi: 10.1371/journal.pone.0065424
39. Zhao X, Gou X, Qin Z, Li D, Wang Y, Ma E, et al. Identification and expression of cuticular protein genes based on Locusta migratoria transcriptome. Sci Rep. (2017) 7:45462. doi: 10.1038/srep45462
40. Liu W, An S, Cheng P, Zhang K, Gong M, Zhang Z, et al. Whole-transcriptome profiling across different developmental stages of Aedes albopictus (Diptera: Culicidae) provides insights into chitin-related non-coding RNA and competing endogenous RNA networks. Parasites Vectors. (2023) 16:33. doi: 10.1186/s13071-022-05648-2
41. Vannini L and Willis JH. Immunolocalization of cuticular proteins in Johnston’s organ and the corneal lens of Anopheles Gambiae. Arthropod structure Dev. (2016) 45:519–35. doi: 10.1016/j.asd.2016.10.006
42. Andersen SO. Characteristic properties of proteins from pre-ecdysial cuticle of larvae and pupae of the mealworm Tenebrio molitor. Insect Biochem Mol Biol. (2002) 32:1077–87. doi: 10.1016/S0965-1748(02)00045-0
43. Cornman RS, Togawa T, Dunn WA, He N, Emmons AC, and Willis JH. Annotation and analysis of a large cuticular protein family with the R&R Consensus in Anopheles Gambiae. BMC Genomics. (2008) 9:1–16. doi: 10.1186/1471-2164-9-22
44. Andersen SO. Studies on proteins in post-ecdysial nymphal cuticle of locust, Locusta migratoria, and cockroach, Blaberus craniifer. Insect Biochem Mol Biol. (2000) 30:569–77. doi: 10.1016/S0965-1748(00)00029-1
45. Li G, Gu X, Gui S, Guo J, Yi T, and Jin D. Transcriptome analysis of hormone-and cuticle-related genes in the development process of deutonymph in Tetranychus urticae. Insects. (2021) 12:p.736. doi: 10.3390/insects12080736
46. Dittmer NT, Hiromasa Y, Tomich JM, Lu N, Beeman RW, Kramer KJ, et al. Proteomic and transcriptomic analyses of rigid and membranous cuticles and epidermis from the elytra and hindwings of the red flour beetle, Tribolium castaneum. J Proteome Res. (2012) 11:269–78. doi: 10.1021/pr2009803
47. Sun X, Guo J, Ye W, Guo Q, Huang Y, Ma L, et al. Cuticle genes CpCPR63 and CpCPR47 may confer resistance to deltamethrin in Culex pipiens pallens. Parasitol Res. (2017) 116:2175–9. doi: 10.1007/s00436-017-5521-z
48. Xu Y, Xu J, Zhou Y, Li X, Meng Y, Ma L, et al. CPR63 promotes pyrethroid resistance by increasing cuticle thickness in Culex pipiens pallens. Parasites Vectors. (2022) 15:54. doi: 10.1186/s13071-022-05175-0
49. Yang CH, Yang PC, Zhang SF, Shi ZY, Kang L, and Zhang AB. Identification, expression pattern, and feature analysis of cuticular protein genes in the pine moth Dendrolimus punctatus (Lepidoptera: Lasiocampidae). Insect Biochem Mol Biol. (2017) 83:94–106. doi: 10.1016/j.ibmb.2017.03.003
50. Masson V, Arafah K, Voisin S, and Bulet P. Comparative proteomics studies of insect cuticle by tandem mass spectrometry: application of a novel proteomics approach to the pea aphid cuticular proteins. Proteomics. (2018) 18:p.1700368. doi: 10.1002/pmic.201700368
51. Abehsera S, Zaccai S, Mittelman B, Glazer L, Weil S, Khalaila I, et al. CPAP3 proteins in the mineralized cuticle of a decapod crustacean. Sci Rep. (2018) 8:2430. doi: 10.1038/s41598-018-20835-x
52. Petkau G, Wingen C, Jussen LC, Radtke T, and Behr M. Obstructor-A is required for epithelial extracellular matrix dynamics, exoskeleton function, and tubulogenesis. J Biol Chem. (2012) 287:21396–405. doi: 10.1074/jbc.M112.359984
53. Jasrapuria S, Arakane Y, Osman G, Kramer KJ, Beeman RW, and Muthukrishnan S. Genes encoding proteins with peritrophin A-type chitin-binding domains in Tribolium castaneum are grouped into three distinct families based on phylogeny, expression and function. Insect Biochem Mol Biol. (2010) 40:pp.214–227. doi: 10.1016/j.ibmb.2010.01.011
54. Zhu JY, Li L, Xiao KR, He SQ, and Gui FR. Genomic and transcriptomic analysis reveals cuticular protein genes responding to different insecticides in fall armyworm Spodoptera frugiperda. Insects. (2021) 12:p.997. doi: 10.3390/insects12110997
55. Xiong G, Tong X, Gai T, Li C, Qiao L, Monteiro A, et al. Body shape and coloration of silkworm larvae are influenced by a novel cuticular protein. Genetics. (2017) 207:pp.1053–1066. doi: 10.1534/genetics.117.300300
56. Xin L, Yang X, Xu Y, Ye X, Zhang Y, and Zhao G. Pyrethroid pesticide-induced differential gene expression in midguts of two silkworm (Bombyx mori) strains of different susceptibilities. ScienceAsia. (2023) 49:737–43. doi: 10.2306/scienceasia1513-1874.2023.073
57. Mang’era CM, Khamis FM, Awuoche EO, Hassanali A, Ombura FLO, and Mireji PO. Transcriptomic response of Anopheles Gambiae sensu stricto mosquito larvae to Curry tree (Murraya koenigii) phytochemicals. Parasites Vectors. (2021) 14:pp.1–14. doi: 10.1186/s13071-020-04505-4
58. Vannini L, Bowen JH, Reed TW, and Willis JH. The CPCFC cuticular protein family: anatomical and cuticular locations in Anopheles Gambiae and distribution throughout Pancrustacea. Insect Biochem Mol Biol. (2015) 65:57–67. doi: 10.1016/j.ibmb.2015.07.002
59. Dittmer NT, Tetreau G, Cao X, Jiang H, Wang P, and Kanost MR. Annotation and expression analysis of cuticular proteins from the tobacco hornworm, Manduca sexta. Insect Biochem Mol Biol. (2015) 62:100–13. doi: 10.1016/j.ibmb.2014.12.010
60. Suzuki Y, Matsuoka T, Iimura Y, and Fujiwara H. Ecdysteroid-dependent expression of a novel cuticle protein gene BMCPG1 in the silkworm, Bombyx mori. Insect Biochem Mol Biol. (2002) 32:599–607. doi: 10.1016/S0965-1748(01)00136-9
61. Zhao C, Huang Q, Qian Y, Zhao X, Guo S, Kan Y, et al. The hypothetical cuticular protein, CPH19, is involved in cuticle formation during molt of silkworm Bombyx mori. J Asia-Pacific Entomology. (2023) 26:102111. doi: 10.1016/j.aspen.2023.102111
62. Gan L, Zhuo W, Li J, Wang Y, Sima Y, and Xu S. A novel Cph-like gene involved in histogenesis and maintenance of midgut in Bombyx mori. Pest Manage science. (2013) 69:1298–306. doi: 10.1002/ps.3501
63. Hamodrakas SJ, Willis JH, and Iconomidou VA. A structural model of the chitin-binding domain of cuticle proteins. Insect Biochem Mol Biol. (2002) 32:pp.1577–1583. doi: 10.1016/S0965-1748(02)00079-6
64. Liang J, Wang T, Xiang Z, and He N. Tweedle cuticular protein BmCPT1 is involved in innate immunity by participating in recognition of Escherichia coli. Insect Biochem Mol Biol. (2015) 58:pp.76–88. doi: 10.1016/j.ibmb.2014.11.004
65. Song TQ, Yang ML, Wang YL, Liu Q, Wang HM, Zhang J, et al. Cuticular protein LmTwdl1 is involved in molt development of the migratory locust. Insect Science. (2016) 23:520–30.
66. Soares MP, Silva-Torres FA, Elias-Neto M, Nunes FM, Simoes ZL, and Bitondi MM. Ecdysteroid-dependent expression of the tweedle and peroxidase genes during adult cuticle formation in the honey bee, Apis mellifera. PloS One. (2011) 6:e20513. doi: 10.1371/journal.pone.0020513
67. Wang X, Liu L, Guo S, Liu B, Zhai Y, Yan S, et al. Tweedle gene family of Drosophila suzukii (Matsumura) larva enhances the basal tolerance to cold and hypoxia. Pest Manage Science. (2023) 79:3012–21. doi: 10.1002/ps.7476
68. Wilkin MB, Becker MN, Mulvey D, Phan I, Chao A, Cooper K, et al. Drosophila Dumpy is a gigantic extracellular protein required to maintain tension at epidermal–cuticle attachment sites. Curr Biol. (2000) 10:pp.559–567. doi: 10.1016/S0960-9822(00)00482-6
69. Weis-Fogh T. A rubber-like protein in insect cuticle. J Exp Biol. (1960) 37:889–907. doi: 10.1242/jeb.37.4.889
70. Lyons RE, Wong DC, Kim M, Lekieffre N, Huson MG, Vuocolo T, et al. Molecular and functional characterisation of resilin across three insect orders. Insect Biochem Mol Biol. (2011) 41:pp.881–890. doi: 10.1016/j.ibmb.2011.08.002
71. Lerch S, Zuber R, Gehring N, Wang Y, Eckel B, Klass KD, et al. Resilin matrix distribution, variability and function in Drosophila. BMC Biol. (2020) 18:pp.1–15. doi: 10.1186/s12915-020-00902-4
72. Elvin CM, Carr AG, Huson MG, Maxwell JM, Pearson RD, Vuocolo T, et al. Synthesis and properties of crosslinked recombinant pro-resilin. Nature. (2005) 437:999–1002. doi: 10.1038/nature04085
73. Liu BQ, Qiao L, He QY, Zhou Y, Ren S, and Chen B. Genome-wide identification, characterization and evolution of cuticular protein genes in the malaria vector Anopheles sinensis (Diptera: Culicidae). Insect science. (2018) 25:pp.739–750. doi: 10.1111/1744-7917.12483
74. Adams M, Bosco G, Denlinger D, Dhadialla T, Field L, Hildebrand J, et al. Abstracts of the fifth international symposium on molecular insect science: may 20–24, 2006, tucson, arizona USA, organizing committee. J Insect Sci. (2006) 6:46. doi: 10.1673/2006_06_45.1
75. Yahouédo GA, Chandre F, Rossignol M, Ginibre C, Balabanidou V, Mendez NGA, et al. Contributions of cuticle permeability and enzyme detoxification to pyrethroid resistance in the major malaria vector Anopheles Gambiae. Sci Rep. (2017) 7:11091. doi: 10.1038/s41598-017-11357-z
76. Huang Y, Guo Q, Sun X, Zhang C, Xu N, Xu Y, et al. Culex pipiens pallens cuticular protein CPLCG5 participates in pyrethroid resistance by forming a rigid matrix. Parasites vectors. (2018) 11:1–10. doi: 10.1186/s13071-017-2567-9
77. Vontas J, David JP, Nikou D, Hemingway J, Christophides GK, Louis C, et al. Transcriptional analysis of insecticide resistance in Anopheles stephensi using cross-species microarray hybridization. Insect Mol Biol. (2007) 16:315–24. doi: 10.1111/j.1365-2583.2007.00728.x
78. Vannini L, Reed TW, and Willis JH. Temporal and spatial expression of cuticular proteins of Anopheles Gambiae implicated in insecticide resistance or differentiation of M/S incipient species. Parasites vectors. (2014) 7:1–11. doi: 10.1186/1756-3305-7-24
79. Jacobs E, Chrissian C, Rankin-Turner S, Wear M, Camacho E, Broderick NA, et al. Cuticular profiling of insecticide resistant Aedes Aegypti. Sci Rep. (2023) 13:10154. doi: 10.1038/s41598-023-36926-3
80. Jones CM, Haji KA, Khatib BO, Bagi J, Mcha J, Devine GJ, et al. The dynamics of pyrethroid resistance in Anopheles arabiensis from Zanzibar and an assessment of the underlying genetic basis. Parasites vectors. (2013) 6:pp.1–13. doi: 10.1186/1756-3305-6-343
81. Balabanidou V, Kefi M, Aivaliotis M, Koidou V, Girotti JR, and Mijailovsky SJ. Mosquitoes cloak their legs to resist insecticides. Proc Biol Sci. (2019) 286(1907):20191091. doi: 10.1098/rspb.2019.1091
82. Dada N, Lol JC, Benedict AC, López F, Sheth M, Dzuris N, et al. Pyrethroid exposure alters internal and cuticle surface bacterial communities in Anopheles albimanus. ISME J. (2019) 13:2447–64. doi: 10.1038/s41396-019-0445-5
83. Dada N, Benedict AC, López F, Lol JC, Sheth M, Dzuris N, et al. Comprehensive characterization of internal and cuticle surface microbiota of laboratory-reared F 1 Anopheles albimanus originating from different sites. Malaria J. (2021) 20:1–18. doi: 10.1186/s12936-021-03934-5
84. Adams KL, Sawadogo SP, Nignan C, Niang A, Paton DG, Robert SW, et al. Cuticular hydrocarbons are associated with mating success and insecticide resistance in malaria vectors. Commun Biol. (2021) 4:911. doi: 10.1038/s42003-021-02434-1
85. Wang G, Vega-Rodríguez J, Diabate A, Liu J, Cui C, Nignan C, et al. Clock genes and environmental cues coordinate Anopheles pheromone synthesis, swarming, and mating. Science. (2021) 371:411–5. doi: 10.1126/science.abd4359
86. Chen J, Wu Y, Chen J, Lu H, Cheng G, Tu ZJ, et al. Roles of a newly lethal cuticular structural protein, AaCPR100A, and its upstream interaction protein, G12-like, in Aedes aegypti. Int J Biol Macromol. (2024) 268(Pt 1):131704. doi: 10.1016/j.ijbiomac.2024.131704
Keywords: insect, mosquitoes, cuticle proteins, cuticle genes, protein domain, anopheles
Citation: Thakur Y, Tevatiya S, Kumar G, Jeena M, Verma V, Dixit R, Pasi S, Eapen A and Kaur J (2025) Panoramic view of diversity and function of cuticular proteins in insects and mosquitoes biology. Front. Insect Sci. 5:1602055. doi: 10.3389/finsc.2025.1602055
Received: 28 March 2025; Accepted: 28 July 2025;
Published: 29 August 2025.
Edited by:
Sengodan Karthi, Manonmaniam Sundaranar University, IndiaReviewed by:
Ruchir Mishra, University of Florida, United StatesNishtha Nayyar, University of Kentucky, United States
Copyright © 2025 Thakur, Tevatiya, Kumar, Jeena, Verma, Dixit, Pasi, Eapen and Kaur. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jaspreet Kaur, a2F1ci5qYXNwcmVldDA3MDhAZ21haWwuY29tShweta Pasi, c2h3ZXRhLnBhc2lAZ21haWwuY29t
 Yamini Thakur1,2
Yamini Thakur1,2 Sanjay Tevatiya
Sanjay Tevatiya Gaurav Kumar
Gaurav Kumar Alex Eapen
Alex Eapen Jaspreet Kaur
Jaspreet Kaur