Abstract
Objective:
This study aims to assess the mesenchymal-epithelial transition factor’s (c-MET) prognostic value in oesophageal carcinoma (ESCA) through a meta-analysis and bioinformatics.
Methods:
We analysed c-MET expression in ESCA tissues using data from The Cancer Genome Atlas (TCGA) and conducted a meta-analysis to evaluate its association with clinicopathological factors and survival outcomes. The meta-analysis included studies reporting hazard ratios (HRs) and odds ratios (ORs) for survival and metastatic outcomes.
Results:
The Cancer Genome Atlas analysis revealed elevated c-MET expression in ESCA, which was significantly correlated with lymph node metastasis, tumour grade and stage, though not with overall survival (OS). In the meta-analysis, 278 publications were identified, and 89 duplicates were removed. After screening, 176 articles were excluded, leaving 13 for full-text review. Of these, 5 studies lacked sufficient survival data, resulting in 8 eligible studies with a total of 1,488 patients. Meta-analysis findings indicated that high c-MET expression was associated with worse OS (HR = 1.54, 95% confidence interval [CI]: 1.17–2.01; p = 0.002), distant metastasis (OR = 1.97, 95% CI: 1.14–3.40; p = 0.02) and advanced stage (OR = 2.23, 95% CI: 1.41–3.53; p = 0.0006).
Conclusion:
High c-MET expression is associated with poor prognosis and advanced disease in ESCA, highlighting its potential as a biomarker for risk stratification. Further studies are needed to confirm its prognostic value and explore therapeutic implications.
1 Introduction
Oesophageal carcinoma (ESCA) is one of the deadliest malignancies globally, with a significant increase in incidence. According to the IARC, 604,100 new ESCA cases and 544,076 deaths were reported in 2020 (1). Oesophageal carcinoma ranks seventh in incidence and sixth in mortality worldwide. Oesophageal squamous cell carcinoma (ESCC), the most prevalent histological type, accounts for approximately 90% of ESCA cases (2). Treatment options including surgery, radiotherapy, chemotherapy, endoscopy and traditional Chinese medicine have been implemented widely; however, the 5-year survival rate for patients with ESCA remains poor (3). The increasing incidence and low survival rate underscore the urgent need for improved diagnostic and predictive biomarkers.
The mesenchymal-epithelial transition factor (c-MET), a receptor tyrosine kinase found on the surface of various epithelial cells, typically plays a crucial role in wound recovery and tissue remodelling in humans. However, abnormal activation of the c-MET signaling pathway often occurs during cancer development, promoting the growth, invasion and metastasis of tumour cells (4). Numerous studies have confirmed that c-MET is overexpressed or mutated in various solid tumours, including lung, gastric, liver, breast, skin and colorectal cancers, with significant effects on tumour formation and progression (5, 6). The activation of c-MET is frequently associated with high-grade and advanced-stage tumours. The overexpression of c-MET has been shown to correlate with pathological stage, tumour grade, muscle invasion and lymph node involvement in bladder cancer (7). High c-MET expression is linked to an increased risk of lymph node metastasis in various tumours (8–10). Activation of c-MET has also been associated with tumour angiogenesis. By activating downstream signaling pathways, c-MET promotes the proliferation and migration of vascular endothelial cells, thereby supporting tumour growth and metastasis (11, 12). Mesenchymal-epithelial transition with different mutations can have varying effects; METex14 tumours exhibit differences in immuno-oncology biomarkers and the somatic landscape compared with non-METex14 NSCLC tumours, with variations in immune profiles potentially influencing immunotherapy selection in MET-altered NSCLC (13). Germline or somatic mutations, chromosomal rearrangements, gene amplification, transcriptional upregulation in MET or alterations in autocrine or paracrine c-MET signalling have been associated with cancer cell proliferation and survival, including in renal cell carcinoma, and linked to disease progression (14). Recently, small molecule inhibitors targeting c-MET, such as capmatinib and tepotinib, have entered clinical trials and shown positive results in patients with non-small cell lung cancer (15). Nonetheless, the specific role of c-MET in ESCC has not been well documented. Although earlier studies have identified c-MET expression in ESCC, its prognostic value and therapeutic potential as a therapeutic target remain unclear, mainly due to differences in methodology and research priorities across studies, which have led to divergent understandings of c-MET’s action.
Addressing the unmet need in ESCC management, it is crucial to explore the clinical significance of c-MET and its association with patient prognosis. Further investigation into c-MET expression in ESCC and its correlation with clinical outcomes could provide valuable insights for future clinical applications. We aimed to assess the prognostic impact of c-MET expression in ESCC, contributing to the foundation of potential therapeutic strategies and advancing clinical practise in this challenging area.
2 Materials and methods
2.1 UALCAN and GEPIA online cancer data analysis
The expression level of c-MET mRNA and its prognostic value in ESCA can be assessed by analysing data from The Cancer Genome Atlas (TCGA) database. This analysis can be performed using two online tools: UALCAN1 and GEPIA.2 The comprehensive cancer data research platform UALCAN offers convenient access to multiple public cancer omics datasets, including TCGA, MET500, CPTAC and CBTTC (16). In contrast, the GEPIA database integrates research data from TCGA and GTEx projects, covering RNA sequencing information from more than 9,736 tumour samples and 857 normal samples. Using these tools, researchers have been able to investigate the expression pattern of c-MET in oesophageal cancer and its impact on prognosis (17).
2.2 Literature searching
A comprehensive literature search was conducted according to the PRISMA guidelines (18) to identify studies published up to September 2022. The following databases were searched: PubMed, Web of Science, Cochrane Library, Wanfang and CNKI. The search strategy included the following key phrases: ‘esophageal squamous cell carcinoma’ OR ‘oesophageal squamous cell carcinoma’ OR ‘esophageal carcinoma’ OR ‘ESCC’ OR ‘ESCA’ AND ‘c-MET’ OR ‘MET’ OR ‘hepatocyte growth factor receptor’ OR ‘HGFR’.
For each database, the search strategy was tailored to fit the specific syntax and indexing terms used by the database. For example, in PubMed, the search query was constructed as follows: (‘esophageal squamous cell carcinoma’ [MeSH Terms] OR ‘oesophageal squamous cell carcinoma’ [MeSH Terms] OR ‘esophageal carcinoma’ [MeSH Terms] OR ‘ESCC’ [All Fields] OR ‘ESCA’ [All Fields]) AND (‘c-MET’ [MeSH Terms] OR ‘MET’ [MeSH Terms] OR ‘hepatocyte growth factor receptor’ [MeSH Terms] OR ‘HGFR’ [All Fields]). In Web of Science and Scopus, the search was conducted using the ‘Topic’ or ‘Title/Abstract/Keywords’ fields to ensure broad coverage. The search terms were combined using Boolean operators (AND/OR) to refine the search results. The search results were then exported to reference management software (e.g., EndNote) for deduplication and initial screening.
To enhance the reproducibility of this study, the search was restricted to titles and abstracts and supplemented by manual searches of reference lists from relevant articles. Searches were limited to studies in English and Chinese, and all identified articles were evaluated for eligibility through a multi-step screening process. The search timeframe extended up to September 2022, including studies from all years prior, to ensure comprehensive coverage of existing literature.
2.3 Selection criteria
The inclusion and exclusion process was conducted independently by two reviewers to ensure accuracy and minimise bias. Studies included in the meta-analysis had to meet the following conditions: (1) histopathological confirmation of ESCC was required for the study participants; (2) immunohistochemistry or fluorescence in situ hybridisation (FISH) techniques were used to assess c-MET protein expression in the study; (3) the relationship between c-MET expression and DSS, DFS, PFS or overall survival (OS) was investigated; and (4) the full text of the study had to be available for review. If the study did not utilise Kaplan–Meier survival curves for analysis or if data were incomplete and hazard ratios (HRs) could not be calculated, the study was excluded. Two reviewers independently screened articles based on titles and abstracts and then reviewed the full text to determine if inclusion criteria were met. During the review, disagreements between reviewers were resolved by discussion; if no agreement could be reached, a third reviewer was invited to participate in the discussion to ensure rigour and transparency throughout the selection process.
2.4 Data extraction and quality assessment
Literature was initially screened by reading the title and abstract, and the uncertain literature was determined by further reading the full text. Literature selection was completed by two investigators independently, and when opinions were inconsistent, they were resolved through discussion or consultation with the third investigator. To assess the scientific quality of the included studies, we used the Newcastle-Ottawa Scale (NOS). This scale is designed to evaluate the quality of randomised, case–control and cohort studies and scored by examining the selection, comparability and assessment of exposure or outcomes of the study participants. The NOS uses a semiquantitative scoring system with a maximum total score of nine, of which six and above are considered high-quality studies. For each article, the following details were extracted: first author’s name, publication year, article title, sample size, study site, tumour type, clinicopathological characteristics (including number of patients by sex, median or mean age, TNM stage, distant metastasis, tumour differentiation and clinical stage), c-MET protein expression detection technique, the criteria for high expression determination and survival data (including HRs for OS and its 95% confidence interval [CI]).
2.5 Statistical analysis
Review Manager 5.3 software (RevMan, Cochrane Collaborative, Oxford, UK) was used, which graphically presented results and facilitated meta-analysis. To assess heterogeneity across studies, the Q test and I2 statistic were used based on chi-squared (χ2) statistics. Significant heterogeneity amongst studies was considered if p < 0.05 in the Q test and I2 > 50%. If homogeneity across studies was confirmed, a fixed-effects model was used to calculate the combined treatment effect and HR with its 95% CI. If heterogeneity amongst studies was significant, a random-effects model was used instead. Additionally, funnel plots and Egger’s test were used to assess publication bias, with p < 0.1 considered statistically significant, indicating publication bias.
3 Results
3.1 C-MET mRNA expression was up-regulated in ESCA
By analysing the public cancer database TCGA, we investigated c-MET mRNA expression levels in adjacent and tumour tissues from patients with ESCA. The results of the analysis showed that c-MET mRNA expression was higher in tumour tissues than in adjacent non-cancerous tissues in most cases, particularly in oesophageal cancer, and this difference was statistically significant (p < 0.001; Figure 1A). In addition, this finding was validated by the GEPIA database, which integrated datasets from TCGA and GTEx and included more oesophageal cancer samples, further confirming differences in c-MET mRNA expression between tumours and adjacent non-cancerous tissues (p < 0.05; Figure 1B). Furthermore, we investigated the association of c-MET mRNA expression with patient characteristics in ESCA samples using data from TCGA database. As shown in Figures 1C–F, the expression level of c-MET mRNA was associated with lymph node metastasis, tumour grade and stage but not significantly with the gender of the patients (Figures 1E, F). Finally, we analysed the relationship between c-MET mRNA expression in TCGA database and the prognosis of patients with ESCA and found no significant difference in survival time between patients with high c-MET expression and those with low or moderate c-MET expression (p = 0.71; Figure 1G).
Figure 1

Expression level of c-MET mRNA in ESCA. (A) c-MET mRNA expression was remarkably overexpressed in ESCA tissues compared with normal peritumoral tissues in TCGA database. (B) The expression of c-MET mRNA in ESCA tissues was significantly overexpressed compared with that in normal peritumoral tissues by GEPIA (*p < 0.05). (C) The mRNA expression level of c-MET in different lymph node metastasis status based on TCGA database. (D) The mRNA expression level of c-MET in different tumor grades based on TCGA database. (E) The mRNA expression level of c-MET in different genders based on TCGA database. (F) The mRNA expression level of c-MET in different cancer stages based on TCGA database. (G) OS of ESCA patients in c-MET-low/medium expression group and c-MET-high expression group based on TCGA database. c-MET, cellular-mesenchymal epithelial transition factor; ESCA, esophageal carcinoma; TCGA, the cancer genome atlas; GEPIA, gene expression profiling interactive analysis; OS, overall survival.
3.2 Identification and characterisation of relevant studies
To further investigate the association between c-MET expression and ESCA prognosis, we performed a meta-analysis. A total of 278 relevant articles were initially identified through databases and hand searches. After removing 89 duplicate publications, we excluded 176 articles by reviewing the titles and abstracts. Subsequently, we read and assessed the full text of the remaining 13 studies. Unfortunately, five studies (19–23) were excluded because they lacked the necessary survival data for the analysis, and a total of eight publications (24–31) eventually met the inclusion criteria. Figure 2 shows the literature screening process in detail. The total number of patients included in the meta-analysis was 1,488, with a mean sample size of 186, ranging from 90 to 495. Literature quality was assessed using the NOS, and the mean score of the included studies was 7.25 points (score range: 6–8 points), indicating high study quality. We extracted HRs and 95% CIs from the 8 articles that met the criteria. Table 1 presents the main characteristics of these studies.
Figure 2

Flowchart of the literature searching in this meta-analysis.
Table 1
| Study year | Country | Cancer type | Technology | Sample size | Median or mean age (y) | Gender (F/M) | c-MET (H/L) | Median or mean follow-up time (months) | Outcome | HR (95%CI) | p value | Cut-off value | NOS score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hara 2019 | Japan | ESCC | IHC | 147 | 66 | 25/122 | 73/74 | NA | OS | 1.79 (1.03–3.04) | p = 0.038 | H-score ≥ 90 | 7 |
| Kim 2016 | South Korea | ESCC | IHC | 200 | 65 (41–83) | 12/188 | 42/158 | 33.2 (0.6–178.7) | OS | 1.12 (0.73–1.72) | p = 0.601 | H-score ≥ 50 | 7 |
| Ozawa 2015 | Japan | ESCC | IHC | 104 | 64 | 18/86 | 72/32 | NA | OS | 2.237 (1.066–5.190) | p = 0.017 | H-score ≥ 40 | 7 |
| Shi 2022 | China | ESCC | IHC | 172 | 61.8 (32–83) | 54/118 | 98/74 | 34 (2–108) | OS | 0.371(0.237–0.582) | P < 0.001 | IHC Score > 4 | 8 |
| Wang 2019 | China | ESCC | FISH | 495 | 61 (34–83) | 87/408 | 28/467 | 35 (3–102) | OS | 1.926 (1.243–2.983) | p = 0.003 | Copy number ≥ 5 and a MET/CEP7 ratio ≥ 2 | 8 |
| Xu 2015 | China | ESCC | IHC | 90 | 59 | 17/73 | 39/51 | 26.3 | OS | 1.805(1.045–3.117) | p = 0.034 | H-score ≥ 20 | 8 |
| Xu 2016 | China | ESCC | IHC | 180 | 59 (37–80) | 16/164 | 84/96 | 46.4 | OS | 0.459(0.287–0.733) | p = 0.001 | H-score ≥ 160 | 7 |
| Zhou 2016 | China | ESCC | IHC | 100 | 59 | 33/67 | 49/51 | NA | OS | 2.34 (1.63–4.45) | p < 0.05 | NA | 6 |
Main characteristics of the 8 included studies in the meta-analysis.
F, female; M, male; H, high; L, low; HR, hazard ratio; CI, confidence interval; NOS, Newcastle-Ottawa Scale; ESCC, esophageal squamous cell carcinoma; IHC, immunohistochemistry; NA, not availiable; OS, overall survival; FISH, Fluorescence in situ hybridization.
3.3 Relationship between c-MET expression and ESCC survival
In the collated dataset, a total of six studies provided data on OS, whereas two studies (27, 28) reported both OS and DFS. Therefore, we used OS as the primary outcome to evaluate the effect of c-MET expression levels on survival in patients with ESCC. We first analysed the relationship between c-MET expression and OS in eight studies. Preliminary analysis showed that patients with high c-MET expression had a worse prognosis than those with low c-MET expression (HR = 1.23, 95% CI: 1.04–1.45; p = 0.02; Figure 3A). However, because the I2 value exceeded 50% in the heterogeneity test, we reperformed the analysis using a random-effects model. The results of this analysis showed no significant correlation between c-MET expression levels and OS in patients ESCC (HR = 1.24, 95% CI: 0.74–2.09; p = 0.41; Figure 3B).
Figure 3

Forest plots of HRs for the association between c-MET expression and OS in patients with ESCC. Forest plots of the overall association between c-MET expression and OS in ESCC were performed using (A) fixed effect model and (B) random effect model. (C) After excluding some of the included literatures, forest plot of the overall association between c-MET expression and OS in ESCC was performed using fixed effect model. HRs, hazard ratios; c-MET, cellular-mesenchymal epithelial transition factor; OS, overall survival; ESCC, esophageal squamous cell carcinoma; CI, confidence interval.
Given that the cut-off value, antibody and technical method may affect the results of the meta-analysis, we further analysed the judgement of the cut-off value, the source of the antibody and the technical method used in the eight studies (Supplementary Table 1). The study by Shi et al. (27) was excluded due to differences in the definition of the cut-off value. The study by Zhou et al. (31) was excluded because it did not clarify the specific cut-off value. The study by Xu et al. (30) was excluded as the cut-off value was defined as H-score ≥ 160, which was significantly higher than in other studies. The study by Wang et al. (28) was also excluded because c-MET expression was detected using the FISH method. We then performed a new meta-analysis after excluding these four studies, and the results suggested that patients with ESCC with high c-MET expression had a worse prognosis (HR = 1.54, 95% CI: 1.17–2.01; p = 0.002; Figure 3C).
3.4 Association between c-MET expression and clinicopathological parameters
Pooled ORs showed that c-MET expression was associated with distant metastasis (odds ratio [OR] = 1.97, 95% CI: 1.14–3.40; p = 0.02; Figure 4) and clinical stage (OR = 2.23, 95% CI: 1.41–3.53; p = 0.0006; Figure 4), although some missing information resulted in the inclusion of few studies. However, c-MET expression was not associated with lymph node metastasis (OR = 1.21, 95% CI: 0.89–1.64; p = 0.23; Figure 4), tumour differentiation (OR = 1.30, 95% CI: 0.94–1.81; p = 0.12; Figure 4), sex (OR = 1.04, 95% CI: 0.73–1.48; p = 0.21; Figure 4) or T classification (OR = 1.88, 95% CI: 0.87–4.08; p = 0.11; Figure 4). These results indicate that patients with ESCC with high c-MET expression are more likely to develop distant metastasis and experience accelerated tumour progression.
Figure 4

Forest plots of ORs for the association between c-MET expression and clinicopathological parameters in patients with ESCC. Forest plots for all data sets analysis based on (A) lymph node metastasis, (B) tumor differentiation, (C) gender, (D) distant metastasis, (E) clinical stage, and (F) T classification. ORs, odds ratios; c-MET, cellular-mesenchymal epithelial transition factor; ESCC, esophageal squamous cell carcinoma; CI, confidence interval.
3.5 Heterogeneity analysis and publication bias
To verify the robustness of the analytical results, we used the Q test and I2 statistic based on chi-squared statistics to assess heterogeneity across studies. Significant heterogeneity was detected when investigating the association between c-MET expression and tumour T stage, so we chose a random-effects model for analysis (p < 0.05; I2 = 70%). We also constructed Begg’s funnel plots to assess possible publication bias. Based on the shape of the funnel plot, we did not find any indication of publication bias in the meta-analysis (Figure 5). The results of Egger’s test were consistent with the observations from the funnel plots (Table 2).
Figure 5
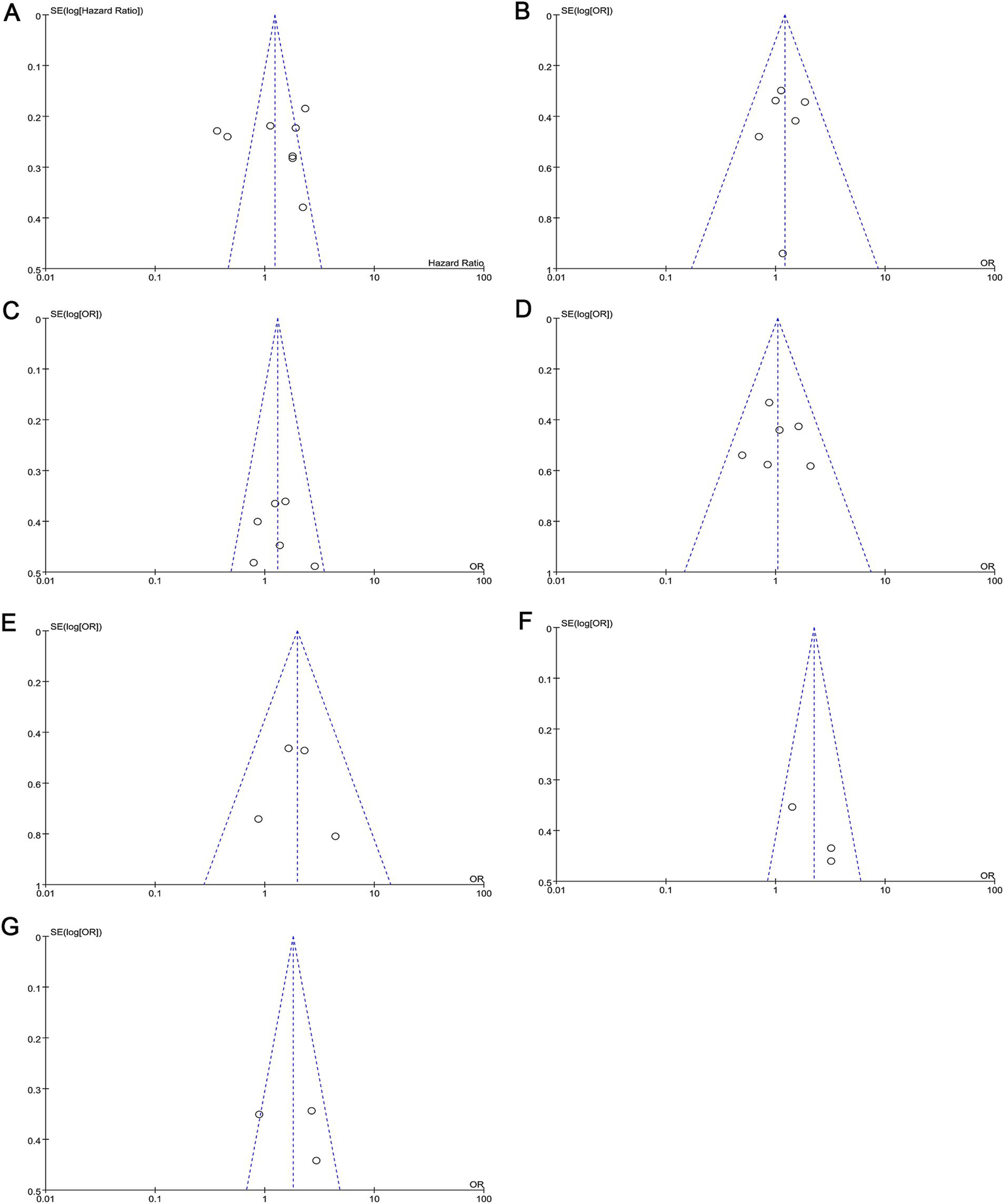
Funnel plots for the assessment of publication bias. Funnel plots for publication bias based on (A) OS, (B) lymph node metastasis, (C) tumor differentiation, (D) gender, (E) distant metastasis, (F) clinical stage, and (G) T classification. OS, overall survival.
Table 2
| Comparison | t | p-value | 95%CI |
|---|---|---|---|
| OS | 0.14 | 0.895 | −14.713–16.4586 |
| Lymph node metastasis | −0.30 | 0.777 | −3.975–3.194 |
| Tumor differentiation | 0.30 | 0.778 | −9.148–1.381 |
| Sex | 0.14 | 0.898 | −5.839–6.441 |
| Distant metastasis | 0.12 | 0.914 | −14.713–16.458 |
| Clinical stage | 4.74 | 0.132 | −14.560–31.914 |
| T classification | 0.49 | 0.711 | −150.95–163.00 |
Results of Egger’s test.
OS, overall survival; CI, confidence interval.
4 Discussion
The c-MET protein, encoded by the MET proto-oncogene, possesses tyrosine kinase activity and functions as a receptor for HGF (15). In cellular function, c-MET plays a key role in regulating cell signalling and cytoskeletal reorganisation, processes essential for cell proliferation, differentiation and motility (32). Recently, with the development of targeted therapeutic strategies against c-MET, it has gained renewed attention due to its central role in tumour development. It has been reported that abnormal activation of c-MET can prompt normal cells to transform into tumour cells and further enhance the invasiveness, metastatic ability and spread of cancer cells (33, 34).
Given the important role of c-MET in tumour development, it has been extensively investigated as a prognostic indicator in a variety of cancers. Previous meta-analyses have shown that c-MET is a poor prognostic marker in head and neck squamous cell carcinoma (35, 36) and non-small cell lung cancer (37, 38). However, for ESCA, especially ESCC, there are few relevant studies, leaving a significant gap in understanding the prognostic value of c-MET in this type of cancer. Analysis of online data showed no significant association between c-MET mRNA levels in ESCA and overall prognosis, but there was an association with lymph node metastasis, tumour grade and stage. Considering that ESCC is the predominant subtype of oesophageal cancer in China (accounting for approximately 90%), we performed a meta-analysis to assess the prognostic and clinicopathological significance of c-MET protein expression in ESCC. Our findings showed that high c-MET expression was associated with a worse prognosis, an increased risk of distant metastasis and an advanced clinical stage in patients with ESCC. In this study, there was no statistically significant difference in OS between patients with high and low c-MET expression. We speculate that this may be due to the small sample size included in the database, which highlights the need for a larger sample size and more prognostic information for analysis.
Our study’s findings are consistent with existing evidence regarding c-MET overexpression in other cancer types. Specifically, c-MET overexpression is known to be prevalent in solid tumours, even when MET gene mutations or amplifications are rare (18, 28). For example, Liberati et al. (18) found that although MET mutations and amplifications are uncommon in ESCC (affecting only 5–6% of cases), 84% of cases exhibit at least a twofold increase in c-MET protein expression. Similarly, Wang et al. (28) reported that true MET amplification was present in only 1% of ESCC cases, whereas Hu et al. demonstrated frequent c-MET overexpression, particularly in well-to-moderately differentiated ESCC tumours. These findings align with our results and indicate that MET gene amplification is not the primary driver of c-MET overexpression in ESCC. Instead, c-MET protein upregulation may occur due to transcriptional mechanisms or post-transcriptional modifications, often following the activation of other driver genes that contribute to tumour progression. Although this study does not fully reveal the specific mechanisms underlying c-MET’s role in ESCC, it highlights c-MET’s significance in disease prognosis and the need for further research on its molecular pathways. Xu YP et al. (30) showed that OS was significantly different between patients with high MET expression and those with low or negative MET expression, and high MET expression was the only prognostic factor for OS. Shi Y et al. (27) reported that MACC1 may affect the prognosis of ESCC by regulating the expression of the MET/cyclin D1 axis. Yuan H et al. (39) reported that ISG15 promotes ESCC tumourigenesis via the MET/Fyn/β-catenin signalling pathway. However, the molecular mechanism of high MET expression in ESCC remains to be further investigated.
In the context of clinical applications, the findings reinforce c-MET’s potential as a valuable prognostic biomarker for ESCC. However, implementing c-MET as a routine prognostic marker or therapeutic target faces several challenges, including technical feasibility and cost-effectiveness, as c-MET testing often requires advanced, costly equipment and expertise. Furthermore, although c-MET-targeted therapies have shown promising results in other cancers, particularly non-small cell lung cancer (40), there remains a paucity of clinical trials and data on c-MET inhibitors in ESCC. Kashyap et al. (41) noted the absence of published clinical data on c-MET tyrosine kinase inhibitors for ESCC, emphasising a significant gap in translational research for this cancer subtype (41). Our study may provide foundational data to guide future clinical trials, potentially paving the way for ESCC-specific therapeutic strategies targeting c-MET.
It is important to acknowledge some limitations in our meta-analysis. This study included only four studies in the final analysis due to heterogeneity in cut-off values and detection methods, which may impact the robustness of our conclusions. Furthermore, some included studies had small sample sizes, which can influence the reliability of the results due to reduced statistical power. Larger, multicentre studies are needed to validate our findings and establish c-MET’s role more definitively in the prognosis of ESCC.
5 Conclusion
In conclusion, this study suggests that c-MET expression is a significant risk factor in ESCC; elevated c-MET levels are associated with poor survival outcomes, later clinical stages and increased distant metastasis. This knowledge may aid in identifying high-risk patients through c-MET expression assessment, thereby contributing to risk stratification efforts in ESCC. Based on these findings, c-MET expression holds promise as a prognostic marker in clinical practise, but further studies on c-MET in ESCC are warranted to solidify its role and advance therapeutic development. Future research should focus on larger sample sizes, exploration of molecular mechanisms and the assessment of clinical feasibility, including cost and accessibility, to ensure the efficient integration of c-MET as a prognostic tool in ESCC management.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
QZ: Conceptualization, Data curation, Investigation, Writing – original draft, Writing – review & editing. XL: Investigation, Methodology, Project administration, Visualization, Writing – original draft, Writing – review & editing. JL: Formal analysis, Investigation, Methodology, Resources, Writing – original draft, Writing – review & editing. ZZ: Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the Natural Science Foundation of Xinjiang Uygur Autonomous Region, China (grant number 2023D01C97), the State Key Laboratory of Pathogenesis, Prevention and Treatment of High Incidence Diseases in Central Asia Fund, China (grant number SKL-HIDCA-2024-43, SKL-HIDCA-2024-SG4), the Xinjiang Uygur Autonomous Region “Tianchi Talents” Introduction Program-Young Doctoral Program for Qiqi Zhang and the Technology Innovation Team (Tianshan Innovation Team) Project, China (grant number 2022TSYCTD0018).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2025.1548160/full#supplementary-material
Abbreviations
CI, confidence interval; c-MET, Cellular-mesenchymal epithelial transition factor; DFS, disease-free survival; ESCC, esophageal squamous cell carcinoma; ESCA, esophageal carcinoma; HR, hazard ratio; IHC, immunohistochemistry; OR, odds ratio; OS, overall survival; TCGA, The Cancer Genome Atlas.
References
1.
Sung H Ferlay J Siegel RL Laversanne M Soerjomataram I Jemal A et al . Global Cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
2.
Yang T Hui R Nouws J Sauler M Zeng T Wu Q . Untargeted metabolomics analysis of esophageal squamous cell cancer progression. J Transl Med. (2022) 20:127. doi: 10.1186/s12967-022-03311-z
3.
Li X Chen L Luan S Zhou J Xiao X Yang Y et al . The development and progress of nanomedicine for esophageal cancer diagnosis and treatment. Semin Cancer Biol. (2022) 86:873–85. doi: 10.1016/j.semcancer.2022.01.007
4.
Dong Y Xu J Sun B Wang J Wang Z . MET-targeted therapies and clinical outcomes: a systematic literature review. Mol Diagn Ther. (2022) 26:203–27. doi: 10.1007/s40291-021-00568-w
5.
Fujita R Blot V Wong E Stewart C Lieuw V Richardson R et al . A novel non-agonist c-met antibody drug conjugate with superior potency over a c-met tyrosine kinase inhibitor in c-met amplified and non-amplified cancers. Cancer Biol Ther. (2020) 21:549–59. doi: 10.1080/15384047.2020.1737490
6.
Yang X Liao HY Zhang HH . Roles of MET in human cancer. Clin Chim Acta. (2022) 525:69–83. doi: 10.1016/j.cca.2021.12.017
7.
Naguib EM Ismail EF Badran DI Sherief MH El-Abaseri TB . Clinicopathological significance of c-MET and HER2 altered expression in bladder cancer. J Egypt Natl Canc Inst. (2024) 36:42. doi: 10.1186/s43046-024-00250-2
8.
Tsujio G Yashiro M Sakuma T Aoyama R Maruo K Yamamoto Y et al . Impact of SMAD2 and MET expression on lymph node metastasis of HER2-positive gastric Cancer cells. Anticancer Res. (2023) 43:4359–64. doi: 10.21873/anticanres.16631
9.
Van Boxtel W MJM U Verhaegh GW Willems SM Jonker MA PALGA Group et al . Prognostic value of PSMA, c-MET and E-cadherin in salivary duct carcinoma. Oral Oncol. (2020) 110:105018. doi: 10.1016/j.oraloncology.2020.105018
10.
Ray S Saha D Alam N Mitra Mustafi S Mandal S Sarkar A et al . Exposure to chewing tobacco promotes primary oral squamous cell carcinoma and regional lymph node metastasis by alterations of SDF1α/CXCR4 axis. Int J Exp Pathol. (2021) 102:80–92. doi: 10.1111/iep.12386
11.
Demkova L Matuskova M Gercakova K Kozovska Z Smolkova B Kucerova L . The impact of c-met inhibition on molecular features and metastatic potential of melanoma cells. Neoplasma. (2024) 71:417–27. doi: 10.4149/neo_2024_240523N232
12.
Mohan CD Shanmugam MK Gowda SGS Chinnathambi A Rangappa KS Sethi G . C-MET pathway in human malignancies and its targeting by natural compounds for cancer therapy. Phytomedicine. (2024) 128:155379. doi: 10.1016/j.phymed.2024.155379
13.
Minne RL Luo NY Traynor AM Huang M DeTullio L Godden J et al . Genomic and immune landscape comparison of MET exon 14 skipping and MET-amplified non-small cell lung Cancer. Clin Lung Cancer. (2024) 25:567–576.e1. doi: 10.1016/j.cllc.2024.05.001
14.
Silva Paiva R Gomes I Casimiro S Fernandes I Costa L . C-met expression in renal cell carcinoma with bone metastases. J Bone Oncol. (2020) 25:100315. doi: 10.1016/j.jbo.2020.100315
15.
Mathieu LN Larkins E Akinboro O Roy P Amatya AK Fiero MH et al . FDA approval summary: Capmatinib and Tepotinib for the treatment of metastatic NSCLC harboring MET exon 14 skipping mutations or alterations. Clin Cancer Res. (2022) 28:249–54. doi: 10.1158/1078-0432.CCR-21-1566
16.
Chandrashekar DS Karthikeyan SK Korla PK Patel H Shovon AR Athar M et al . UALCAN: an update to the integrated cancer data analysis platform. Neoplasia. (2022) 25:18–27. doi: 10.1016/j.neo.2022.01.001
17.
Li C Tang Z Zhang W Ye Z Liu F . GEPIA2021: integrating multiple deconvolution-based analysis into GEPIA. Nucleic Acids Res. (2021) 49:W242–6. doi: 10.1093/nar/gkab418
18.
Liberati A Altman DG Tetzlaff J Mulrow C Gøtzsche PC Ioannidis JP et al . The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ. (2009) 339:b2700. doi: 10.1136/bmj.b2700
19.
Abboud HS Camuzi D Rapozo DC Fernandes PV Nicolau-Neto P Guaraldi S et al . MET overexpression and intratumor heterogeneity in esophageal squamous cell carcinoma. Braz J Med Biol Res. (2021) 54:e10877. doi: 10.1590/1414-431X2020e10877
20.
Hu YC Lam KY Law S Wong J Srivastava G . Profiling of differentially expressed cancer-related genes in esophageal squamous cell carcinoma (ESCC) using human cancer cDNA arrays: overexpression of oncogene MET correlates with tumor differentiation in ESCC. Clin Cancer Res. (2001) 7:3519–25. PMID:
21.
Liang M Yang M Wang F Wang X He B Mei C et al . Near-infrared fluorescence-guided resection of micrometastases derived from esophageal squamous cell carcinoma using a c-met-targeted probe in a preclinical xenograft model. J Control Release. (2021) 332:171–83. doi: 10.1016/j.jconrel.2021.02.019
22.
Mesteri I Schoppmann SF Preusser M Birner P . Overexpression of CMET is associated with signal transducer and activator of transcription 3 activation and diminished prognosis in oesophageal adenocarcinoma but not in squamous cell carcinoma. Eur J Cancer. (2014) 50:1354–60. doi: 10.1016/j.ejca.2014.01.022
23.
Wang H Jiang D Song Q Xu C Shi Y Li X et al . Prognostic impact and potential interaction of EGFR and c-met in the progression of esophageal squamous cell carcinoma. Tumour Biol. (2016) 37:9771–9. doi: 10.1007/s13277-015-4692-4
24.
Hara T Makino T Yamasaki M Tanaka K Miyazaki Y Takahashi T et al . Effect of c-met and CD44v6 expression in resistance to chemotherapy in esophageal squamous cell carcinoma. Ann Surg Oncol. (2019) 26:899–906. doi: 10.1245/s10434-018-07126-5
25.
Kim R Keam B Kwon D Ock CY Kim M Kim TM et al . Programmed death ligand-1 expression and its prognostic role in esophageal squamous cell carcinoma. World J Gastroenterol. (2016) 22:8389–97. doi: 10.3748/wjg.v22.i37.8389
26.
Ozawa Y Nakamura Y Fujishima F Felizola SJ Takeda K Okamoto H et al . C-met in esophageal squamous cell carcinoma: an independent prognostic factor and potential therapeutic target. BMC Cancer. (2015) 15:451. doi: 10.1186/s12885-015-1450-3
27.
Shi Y Li MY Wang H Li C Liu WY Gao YM et al . The relationship between MACC1/c-met/cyclin D1 Axis expression and prognosis in ESCC. Anal Cell Pathol (Amst). (2022) 2022:9651503–19. doi: 10.1155/2022/9651503
28.
Wang Y Jiang Z Xu C Wang H Tan L Su J et al . Increased MET gene copy number negatively affects the survival of esophageal squamous cell carcinoma patients. BMC Cancer. (2019) 19:240. doi: 10.1186/s12885-019-5450-6
29.
Xu Y Peng Z Li Z Lu M Gao J Li Y et al . Expression and clinical significance of c-met in advanced esophageal squamous cell carcinoma. BMC Cancer. (2015) 15:6. doi: 10.1186/s12885-014-1001-3
30.
Xu YP Lin G Sun XJ Yan MH Zhang G Hu JL et al . C-met as a molecular marker for esophageal squamous cell carcinoma and its association with clinical outcome. J Cancer. (2016) 7:587–94. doi: 10.7150/jca.13687
31.
Zhou Z Chen G Xie Z Tang J Ben X Xie L et al . Predictive value of c-met for long-term mortality in patients with esophageal squamous cell carcinoma. J South Med Univ. (2016) 36:1153–6. doi: 10.3969/j.issn.1673-4254.2016.08.23
32.
Stanislovas J Kermorgant S . C-met-integrin cooperation: mechanisms, tumorigenic effects, and therapeutic relevance. Front Cell Dev Biol. (2022) 10:994528. doi: 10.3389/fcell.2022.994528
33.
Mori S Akita H Kobayashi S Iwagami Y Yamada D Tomimaru Y et al . Inhibition of c-MET reverses radiation-induced malignant potential in pancreatic cancer. Cancer Lett. (2021) 512:51–9. doi: 10.1016/j.canlet.2021.04.029
34.
Xing F Zhao D Wu SY Tyagi A Wu K Sharma S et al . Epigenetic and posttranscriptional modulation of SOS1 can promote breast Cancer metastasis through obesity-activated c-met signaling in African-American women. Cancer Res. (2021) 81:3008–21. doi: 10.1158/0008-5472.CAN-19-4031
35.
Li L Sun Z Huang X Li X Sun L Zhang L et al . Role of c-met expression on prognosis of head and neck cancer: a literature review and meta-analysis. Head Neck. (2019) 41:1999–2006. doi: 10.1002/hed.25655
36.
Vsiansky V Gumulec J Raudenska M Masarik M . Prognostic role of c-met in head and neck squamous cell cancer tissues: a meta-analysis. Sci Rep. (2018) 8:10370. doi: 10.1038/s41598-018-28672-8
37.
Vuong HG Ho ATN Altibi AMA Nakazawa T Katoh R Kondo T . Clinicopathological implications of MET exon 14 mutations in non-small cell lung cancer - a systematic review and meta-analysis. Lung Cancer. (2018) 123:76–82. doi: 10.1016/j.lungcan.2018.07.006
38.
Ye S Li J Hao K Yan J Zhou H . The efficacy and risk profile of c-met inhibitors in non-small cell lung Cancer: a Meta-analysis. Sci Rep. (2016) 6:35770. doi: 10.1038/srep35770
39.
Yuan H Zhou W Yang Y Xue L Liu L Song Y . ISG15 promotes esophageal squamous cell carcinoma tumorigenesis via c-MET/Fyn/β-catenin signaling pathway. Exp Cell Res. (2018) 367:47–55. doi: 10.1016/j.yexcr.2018.03.017
40.
Guo R Luo J Chang J Rekhtman N Arcila M Drilon A . MET-dependent solid tumours molecular diagnosis and targeted therapy. Nat Rev Clin Oncol. (2020) 17:569–87. doi: 10.1038/s41571-020-0377-z
41.
Kashyap MK Abdel-Rahman O . Expression, regulation and targeting of receptor tyrosine kinases in esophageal squamous cell carcinoma. Mol Cancer. (2018) 17:54. doi: 10.1186/s12943-018-0790-4
Summary
Keywords
proto-oncogene proteins mesenchymal-epithelial transition factor, oesophageal squamous cell carcinoma, prognosis, meta-analysis, biomarkers
Citation
Zhang Q, Li X, Li J and Zhang Z (2025) Prognostic value of c-MET in oesophageal squamous cell carcinoma: a study based on the mRNA expression in TCGA database and a meta-analysis. Front. Med. 12:1548160. doi: 10.3389/fmed.2025.1548160
Received
19 December 2024
Accepted
07 February 2025
Published
26 February 2025
Volume
12 - 2025
Edited by
Iain Brownlee, Northumbria University, United Kingdom
Reviewed by
Giulia Maria Stella, University of Pavia, Italy
Yunhuan Liu, Tongji University, China
Updates
Copyright
© 2025 Zhang, Li, Li and Zhang.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiqiang Zhang, drzhiqiang@163.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.