- 1Medical Laboratory CSD, Kyiv, Ukraine
- 2Institute of Molecular Biology and Genetics NASU, Kyiv, Ukraine
- 3Department of Endocrinology, Bogomolets National Medical University, Kyiv, Ukraine
- 4Department of Pathology, Kyiv Medical University, Kyiv, Ukraine
- 5Communal Nonprofit Enterprise of Lviv Regional District “Lviv Regional Oncology Medical Diagnostic Centre”, Lviv, Ukraine
- 6Denys Clinics, Kyiv, Ukraine
- 7Department of Oncology, European International University, Kyiv, Ukraine
- 8Department of Oncology, Sumy State University, Sumy, Ukraine
- 9Specialized Mammalogical Center, Kyiv, Ukraine
- 10Audubon Bioscience, Kyiv, Ukraine
- 11Ukrainian Association of Research Biobanks, Kyiv, Ukraine
- 12Ukrainian Association for Precision Medicine, Kyiv, Ukraine
Introduction: Although the role of various genetic alterations was highlighted among factors affecting the response to immunotherapy in non-small cell lung cancer (NSCLC), the relations between oncogenic driver variants and changes in the cancer immunity cycle are still unclear.
Aim: The study aimed to discover the links between the molecular and immune context of NSCLC.
Materials and methods: This cohort study included 254 cases of NSCLC (193 Lung Adenocarcinomas) (LUAD; 76%), and 61 squamous cell carcinomas (SCC; 24%), with pathology reports and next-generation sequencing (NGS) data available. First, the rate and spectrum of genetic alterations were assessed in the Ukrainian cohort. Second, we uncovered the relationship between the oncogenic driver mutations and PD-L1 expression in NSCLC. Finally, T-cytotoxic lymphocytes (CD8+) and tumor-associated macrophages (CD163+) were evaluated in samples with EGFR and KRAS mutations, ALK rearrangements and LUAD with no genetic findings. Immune desert, immune excluded and inflamed types of tumor immune microenvironment (TME) were defined according to the cancer immunity cycle.
Results: More than half (52%) of the observed NSCLC cases harbored single (48.03%) or concomitant (3.94%) genetic alterations in oncogenes. The Ukrainian cohort demonstrated a high rate of EGFR (18.5%) and ALK rearrangements (9.4%) with a relatively moderate frequency of KRAS mutations (16.9%). NSCLC tumors with alterations in EGFR and ALK demonstrated a high incidence of PD-L1 expression and specific immune contexture. The number of CD8+ cells varied significantly between oncogene-driven and wild-type LUAD (p = 0.019). Non-oncogene-addicted NSCLC demonstrated the prevalence of Inflamed TME rich in CD163+ macrophages. In contrast, over half of EGFR mutant LUAD cases possessed immune desert TME type, while ALK-rearranged and KRAS mutant NSCLC showed mostly immune excluded TME.
Conclusion: The high rate of PD-L1 expression in NSCLC driven by EGFR and ALK alterations was accompanied by a prevalence of low immunogenicity with a shift toward ID TME in EGFR mutant tumors and IE TME in ALK-rearranged and KRAS mutant NSCLC. Further discovery of mechanisms affecting tumor immune contexture is needed for tailoring patient management in line with particular mechanisms of immune evasion.
Introduction
Non-small cell lung cancer (NSCLC) remains the leading cause of death among various malignancies globally (1). The exploration of the molecular landscape of NSCLC has revealed numerous clinically actionable genetic alterations defining a wide spectrum of biomarker-directed targeted therapies, tailoring personalized patient management and improving outcomes (2). An alternative approach relying on reactivating patient immune response against cancer cells facilitated the implementation of immunotherapy that has revolutionized NSCLC treatment (3). The discovery of CTLA-4 and PD-1/PD-L1 at the beginning of the 21st century resulted in practice-changing implementation of immune checkpoint inhibitors (ICI) (4). Several studies demonstrated the efficiency of immunotherapy in patients with NSCLC (5, 6), emphasizing the clear benefits of ICI for NSCLC patient survival and quality of life. Immunotherapy has become a part of standard treatment for patients with NSCLC (7). The benefits of ICI have now been recognized in the early stages of the disease, resulting in the durable reinvigoration of the patient’s immune system and the emergence of long-term responders (4, 8). Nevertheless, not all patients respond to immunotherapy, and only a few patients achieve long-term survival under ICI application.
The role of various genetic alterations was highlighted among factors that affect response to immunotherapy (9). Existing evidence suggests a severe limitation to the efficacy of ICI in patients with EGFR-mutant NSCLC, especially after developing resistance to tyrosine kinase inhibitor (TKI) (10, 11). Similarly, real-world data demonstrated weak response to ICI in ALK-positive NSCLC patients, particularly compared to those treated with approved ALK TKIs (12). Although various mutations are associated with upregulation of PD-L1 expression in tumor cells, the mechanisms of decreased efficacy of ICI on EGFR-and ALK-driven NSCLC remains unclear (13, 14). Alternatively, KRAS mutations in NSCLC were shown to coincide with a better response to ICI, which might be related to T-cell infiltration or expression of PD-L1 (15). Besides, most KRAS-mutant tumors are found in tobacco smokers with corresponding signatures and a high tumor mutational burden (TMB) (16). Hereby, the interplay between tumor molecular profile and response to ICI is still under debate, being actively researched and varying significantly between different populations (17).
Genetic alterations in oncogenic drivers such as EGFR, ALK, KRAS, BRAF, MET and others can modify immunogenicity and impact the tumor microenvironment (TME) (18). Recent studies highlighted the importance of CD8+ tumor-infiltrating lymphocytes and other immune cells in defining the response to ICI (19, 20). Indeed, CD8+ cells are a crucial component of antitumor immune response and a backbone of currently approved cancer immunotherapies (21). An entire framework of sequential events generating anti-tumor immune response is embedded in the concept of the cancer-immunity cycle, which defines specific mechanisms of tumor-immune coevolution and its immunophenotypical repercussions. Furthermore, a high infiltration by CD8+ cells is prognostically beneficial in patients with NSCLC and many other cancers (22, 23). However, little is known about the impact of targetable oncogenic driver mutations on the cancer-immunity cycle in NSCLC. This defines an interest in associating oncogenic driver variants with tumor immune contexture, which can help to identify mechanisms of tumor immune evasion and predict sensitivity to various classes of immunotherapy.
The study aimed to assess the rate and spectrum of genetic alterations in NSCLC in the Ukrainian cohort and to discover the links between molecular and immune contexture of NSCLC.
Materials and methods
Setting and participants
A total of 254 patients were enrolled in this cohort study. Descriptive information on the patients is presented in Table 1. The observed cohort consisted of 157 males (61.8%) aged 61 (54.8–66.3), and 97 females (38.2%) aged 56 (46–67). 193 patients had Lung Adenocarcinoma (LUAD; 76%), and 61 patients were diagnosed with Squamous cell carcinoma (SCC; 24%).
The study included three phases addressing three particular research questions. In the first stage, we assessed the rate and spectrum of genetic alterations in known oncogenes in NSCLC in the Ukrainian population. This stage included 254 NSCLC patient tumors with pathology reports and next-generation sequencing (NGS) data available. All the enrolled cases underwent NGS testing with a 10-gene panel for detecting single nucleotide variants (SNVs) and indel in EGFR, ALK, ROS1, RET, KRAS, NRAS, PIK3CA, BRAF, ERBB2 and MET; fusions involving ALK, ROS1 and RET, as well as exon 14 CNV analysis in MET gene (Amoy Dx Essential panel, China) using NextSeq 550Dx (Illumina, US).
In the second stage, we assessed the relationship between the oncogenic driver mutations and PD-L1 expression in NSCLC. This stage included 180 formalin-fixed paraffin-embedded (FFPE) tumor samples of patients who had NGS testing performed and PD-L1 immunohistochemical (IHC) results available.
In the third stage, 40 samples from the primary cohort were analyzed via IHC to assess the immune contexture of LUAD harboring targetable genetic alterations in oncogenes (10 samples with EGFR mutations, 10 samples with KRAS mutations, 10 samples with ALK rearrangements) compared to LUAD tumors with no genetic findings (WT).
Molecular analysis
Extraction of DNA was performed using Omega Bio-tek E.Z.N.A.® FFPE DNA Kit (US). The quantity of DNA was assessed via DeNovix dsDNA Broad Range Assay (US). DNA fragmentation levels were measured with ArcherDX PreSeq DNA QC Assay (US). AmoyDx Essential NGS Panel from AmoyDx (China) was used to test FFPE samples via NGS. Library preparation was performed according to the manufacturer’s protocol. No less than 30 ng of DNA input material was used for library preparation. Normalization to 4 nM concentration and sample pooling was performed after quantifying each library using Roche KAPA Library Quantification Kit (Switzerland) and fragment size analysis via Agilent TapeStation (US).
Sequencing was performed using the Illumina NextSeq 550 Mid-Output Kit on an Illumina NextSeq 550Dx (US) platform. NGS data analysis and annotation of genetic variants (SNV, indel, gene fusions) was performed using AmoyDx ANDAS server analysis module ADXLC10, version 3.3.0 (AmoyDx).
Methodology of immune contexture assessment in LUAD
In order to uncover the possible mechanisms of immune evasion in LUAD, T-cytotoxic lymphocytes (CD8+) and tumor-associated macrophages (CD163+) were visualized via IHC in samples with EGFR and KRAS mutations, ALK rearrangements and samples with no genetic findings (10 samples per group).
Several types of TME were defined: Immune desert (ID), immune excluded (IE) and inflamed (Inf). This was done to assess tumor-host interplay within the cancer immune cycle (24–26). The ID type reflects a lack of pre-existing immunity and demonstrates a low number of T-cells inside and around the tumor. The IE type shows prominent peritumoral infiltration but few intratumoral T-lymphocytes and is indicative of a failure in T-cell trafficking. The Inf TME type displays severe lymphocytic infiltration, reflecting the activation of antitumor T-cells with impaired antitumor activity (27). The number and spatial distribution of CD8+ cells were analyzed with respect to heterogeneity within tumor clusters (TC) and tumor stroma (TS) in 10 different fields of view (28). Next, the number of CD163+ cells corresponding to M2 macrophages was assessed at TC and TS in samples with respect to every TME type. The count of immune cells was recalculated per 1 mm2.
Immunohistochemistry
For immunohistochemistry, 4 μm thick sections were cut, mounted on positively charged slides, and stained according to the manufacturer’s protocol. Visualization of anti-tumor T-cytotoxic lymphocytes was done using antibodies to CD8 (Clone C8/144B, Agilent, United States). Assessment of M2-macrophages was done with CD163 antibodies (Cell Marque, Clone MRQ-26). Additionally, PD-L1 (Dako, 22C3) expression was assessed in all of the selected cases to investigate tumor immune escape mechanisms. PD-L1 expression was assessed via Tumor Proportion Score (TPS). PD-L1 positivity was considered in the at TPS ≥ 1%. Tumors with PD-L1 expression levels of 1–49% were annotated PD-L1-low positive, and PD-L1 expression of 50% and higher were considered PD-L1 high. Two independent pathologists conducted histological and IHC evaluations for each case.
Statistical analysis
Statistical analysis was conducted using MedCalc® Statistical Software version 22.0161 (MedCalc Software Ltd., Ostend, Belgium; 2023) and GraphPad Prism [GraphPad Prism Version 10.4.02 (621 GraphPad Software, San Diego, California, United States)]. Descriptive statistics were provided as Median and interquartile range (IQR; QI – QIII). The Kruskal-Wallis test was applied to compare continuous variables between subgroups. Categorical data were assessed as frequency (%). χ2 or Fisher’s exact tests were used to compare frequencies. The p value of < 0.05 was considered statistically significant.
Results
Aligned with the world NSCLC statistics, there were significant gender differences between histological subtypes of NSCLC (p = 0.002). LUAD cohort included 86 females (44.6%) and 107 males (55.4%). Female patients were diagnosed with cancer at a younger age [55 (46–67) years old] compared to males [60 (55.0–62.8) years old], although these differences were not significant (p = 0.09). In contrast, SCC was more prevalent in males (50; 82.0%) than females (11; 18.0%).
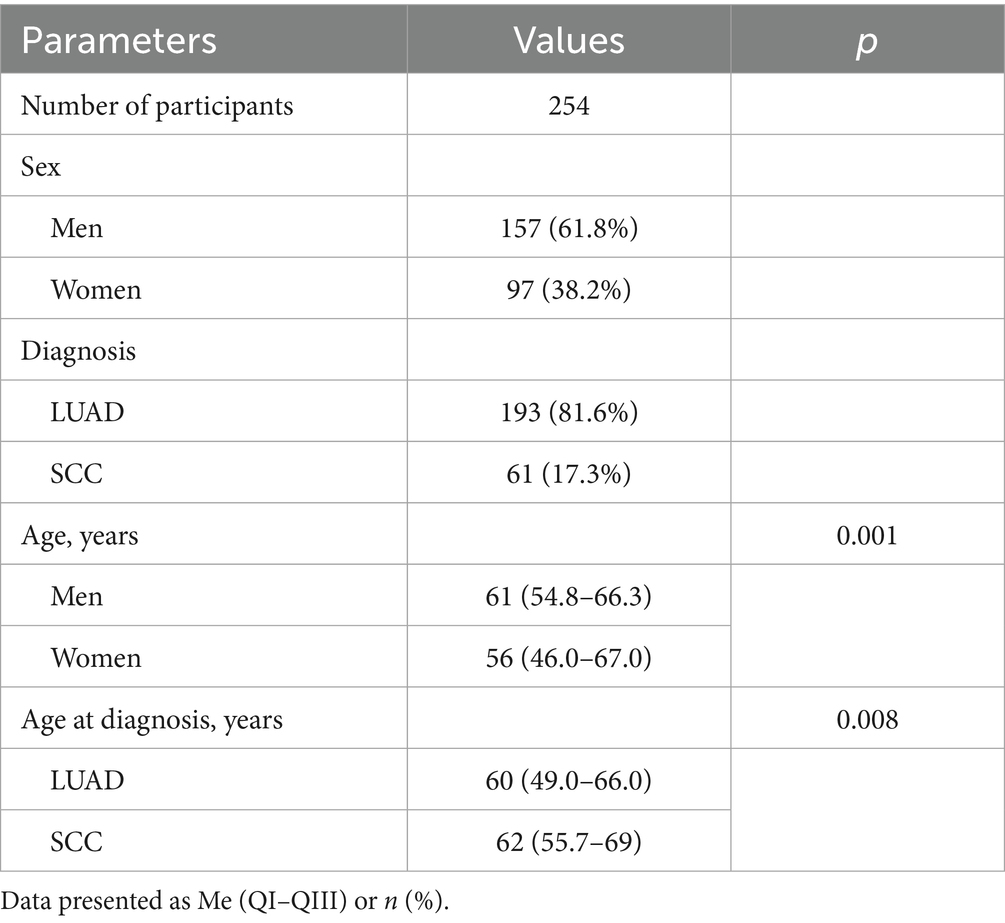
Rate of genetic alterations in Ukrainian patients with NSCLC
More than half of the observed NSCLC cases harbored genetic alterations (132 out of 254 patients; 52%) in oncogenes. Among the observed cohort, 122 patients harbored a single alteration in oncogene (48.03% of the cohort), and 10 patient samples had co-existing mutations in one or more genes comprising 7.58% of oncogene-driven NSCLC cases and 3.94% of the whole sample size (Figure 1). All the co-occurring mutations were found in LUAd cases.
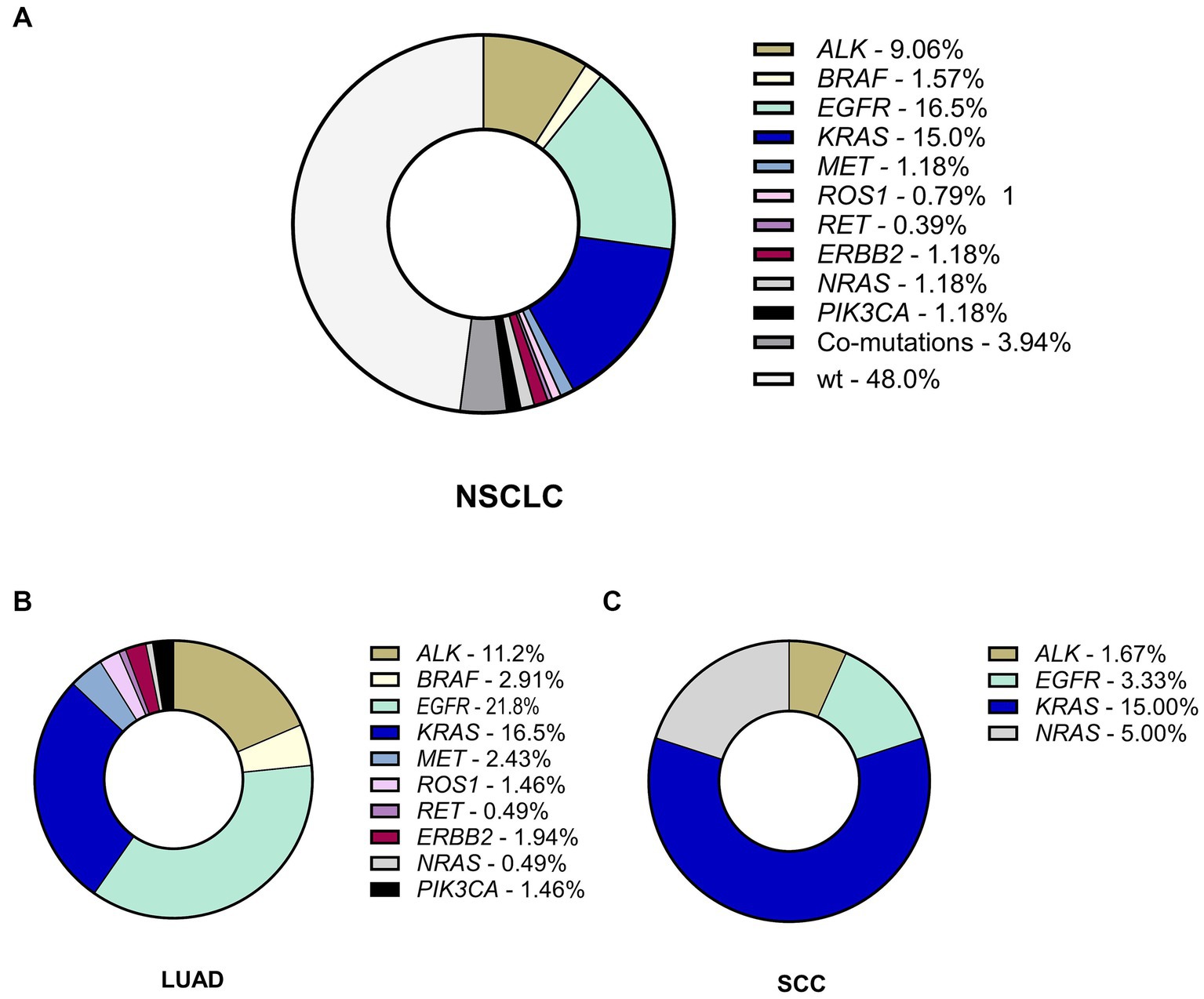
Figure 1. The incidence and spectrum of genetic alterations in NSCLC. (A) The frequency of genetic alterations in the whole Ukrainian cohort of patients with NSCLC. (B) The spectrum and rate of genetic findings in LUAD. (C) The spectrum and rate of genetic findings in SCC. LUAD, lung adenocarcinoma; SCC, squamous cell carcinoma.
Aligning with previous reports, the rate of EGFR genetic variants was much higher in females (31 of 97 patients; 32.0%, Supplementary Table S1) compared to males (16 of 157 patients; 10.2%; p < 0.001). Similar sex differences were typical for ALK rearrangements (p = 0.014). ALK rearrangements were associated with a younger age of onset (48; 44–53) compared to WT-NSCLC (61; 53.0–67.5). Although the rate of KRAS mutations was almost as twice as high in males with NSCLC compared to females (32 of 157 cases, 20.4% in males vs 11 of 97 cases; 11.3% in females), this difference was not found significant (p = 0.084). Other genetic alterations also did not demonstrate statistically significant differences (Supplementary Table S2).
Rate of genetic alterations in LUAD and SCC
The rate of genetic alterations in LUAD (116 cases with alterations of 193 LUADs; 60.1%) was almost three times higher than that of SCC (16 of 61; 26.2% in SCC; p < 0.001), demonstrating higher frequency of EGFR and ALK genetic findings (Supplementary Table S2). EGFR mutations were found in 47 patients (18.5%), 45 of which had LUAD (23.3%) and 2 SCC (3.3%; p < 0,001). Similarly, ALK mutations found in 24 patients (9.4%) were also more common in LUAD (23 of 194 cases in LUAD; 11.9%) than SCC (1 of 61 cases; 1.6%; p = 0.012). The most common fusion partner for ALK was EML4, although we found 2 NSCLC cases with DCTN1 (exon 26)-ALK (exon 20) fusions.
Alterations in KRAS-oncogene were detected in 43 patients (16.9%), who represented single gene mutations (n = 38) and co-occurring mutations in KRAS and other genes (n = 5). The rate of KRAS mutations was comparable among NSCLC of different histology reaching 17.6% in LUAD (34 of 193) and 14.8% in SCC (9 of 61; p = 0.689). Finally, 6 patients with LUAD (3.1%) harbored BRAF mutations, 5 (1.97%)—MET alterations, 3 (1.18%)—ROS1 rearrangements, 4 (1.57%)—ERBB2 mutations, 1 (0.39%)—a RET gene fusion. The frequency of alterations in BRAF, MET, ROS1, ERBB2, PIK3CA and RET genes was not linked to gender or histological type of NSCLC (Supplementary Tables S1, S2). Besides 4 patients (1.57%) harbored NRAS mutation as a single gene alteration (n = 1) or as a co-existing mutation (n = 3), and these alterations were more common in SCC (p = 0.044). Thus, the Ukrainian cohort demonstrated a relatively high rate of EGFR and ALK alterations in LUAD with a moderate frequency of KRAS mutations compared to other European cohorts.
Spectrum of oncogene-driven genetic alterations in NSCLC
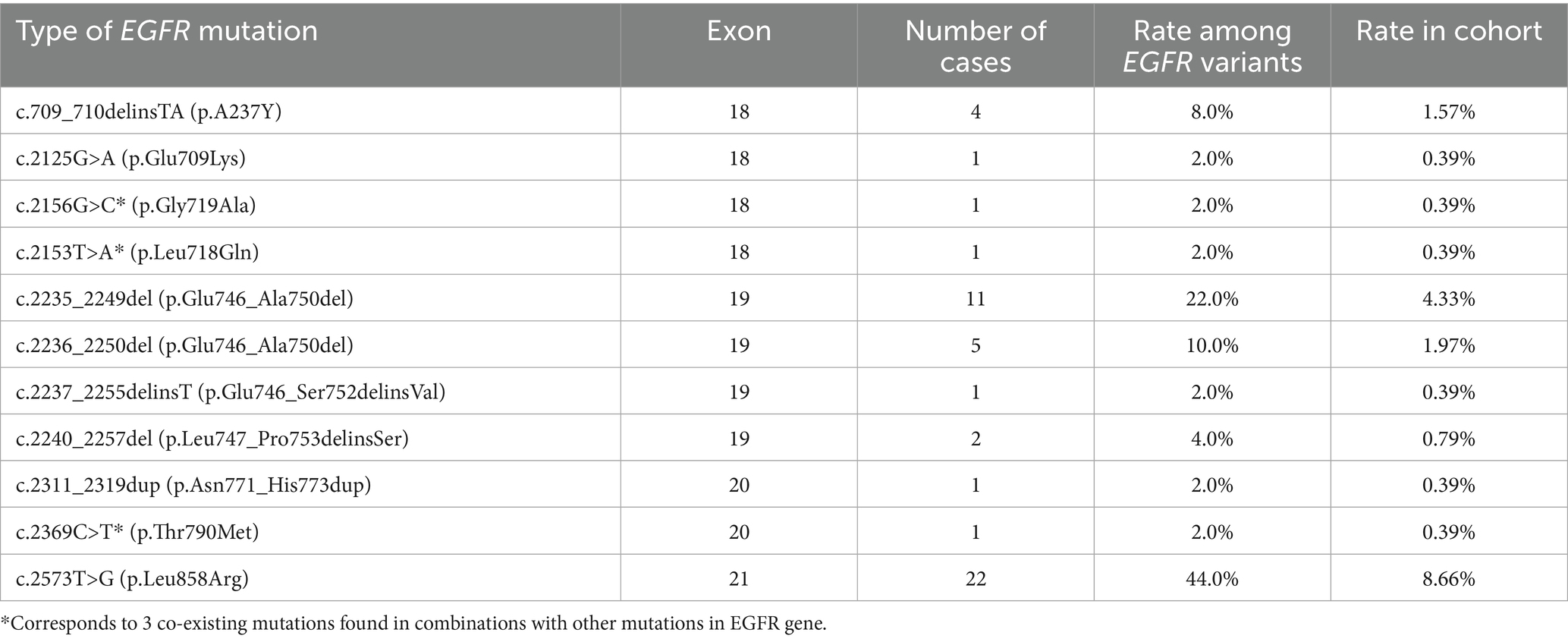
Analyzing the spectrum of genetic variants in EGFR gene, we found 50 primary and secondary EGFR mutations in 47 NSCLC patients (including 3 mutations co-occurring with other EGFR alterations) (Table 2). The most prominent hotspots for EGFR mutation were exons 21 and 19 including p.Leu858Arg (44.0%) and exon 19 deletions (38.0%), representing the main anti-EGFR TKI susceptibility mutations. Rare mutations in exon 18 accounted for 14% of EGFR genetic findings. Among them, a rare mutation p.Leu718Gln associated with resistance to osimertinib (29) was found in one patient (2%). An exon 20 insertion was detected in one patient and represents 2% of EGFR mutation spectrum in this cohort. A secondary p.Thr790Met variant associated with acquired resistance to first-and second-generation anti-EGFR TKI was also detected in one patient (2% of EGFR mutation spectrum). Among the observed EGFR alterations 2 mutations found in SCC included L858R and deletion in exon 19, while LUAD possessed a much wider spectrum of EGFR mutations.
There were 5 cases with co-occurring primary and secondary mutations in EGFR or other genes. One of them combined EGFR L858R with two mutations of resistance (T790M and L718Q), and another patient demonstrated a complex of p.Glu709Lys combined with p.Leu747_Pro753delinsSer. There was a case of co-existing p.Glu709Lys and p.Gly719Ala alterations in EGFR. Besides we found one case with co-mutations in EGFR and BRAF genes (EGFR L858R + BRAFc1799T>A) and one NSCLC harboring EGFR c.2235_2249del (p.Glu746_Ala750del) combined with MET amplification.
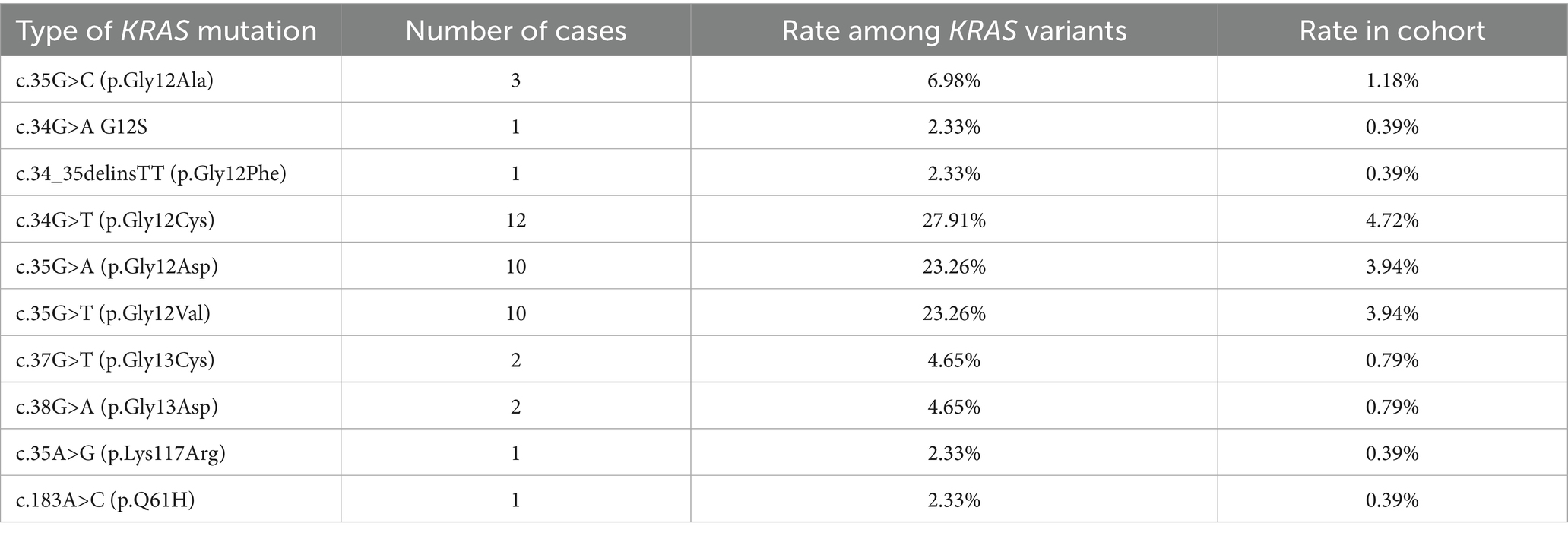
The spectrum of KRAS point mutations comprised single nucleotide variants and indels in clinically relevant exons 2 and 4. p.G12C substitution, associated with sensitivity to KRAS small molecule inhibitors, was the most common and comprised more than one-fourth (12 of 43; 27.91%) of mutations in KRAS (Table 3). Among exon 4 mutations, codon 146 substitution was detected in one patient (2.56%) and a point p.Lys117Arg mutation was detected in 2 patients with NSCLC (5.12%).
Interestingly, 5 cases with KRAS mutations demonstrated co-existing genetic alterations in other genes. c.350A>G (p.Lys117Arg) in KRAS co-occurred with c.1781A>G (p.Asp594Gly) in BRAF and EML4 (exon13) → ALK (exon20) translocation in metastatic LUAD. Besides we found 3 cases with co-occurring mutations in KRAS (G12S) and NRAS (Q61R), one case harboring co-existing mutations in KRAS (c.35G>A, p.Gly12Asp) and ERBB2 (c.2440C>T; p.Arg814Cys), and one case demonstrating co-occurrence of KRAS mutation (G12C) with MET substitution (c.2975C>T; p.Thr992Ile). In addition, one sample combined KRAS mutation (c.35G>C; p.Gly12Ala) with ROS1-rearrangement [ROS1 (exon33)–SYNPO2 (exon2)].
PD-L1 expression in oncogene-addicted NSCLC
Oncogene-addicted NSCLC demonstrated a significantly higher rate of PD-L1 expression (p < 0.001, Figure 2). Among 180 cases with known genetic alterations and PD-L1 expression, 96 were PD-L1 positive (53.3%) and 84 cases (46.7%) did not demonstrate PD-L1 expression. PD-L1 expression with TPS 1–49% was found in 34.4% (62 of 180) and high PD-L1 expression was identified in 18.9% (34 of 180) NSCLC. We found no significant differences in PD-L1 expression between LUAD and SCC (p = 0.076; Supplementary Table S3).
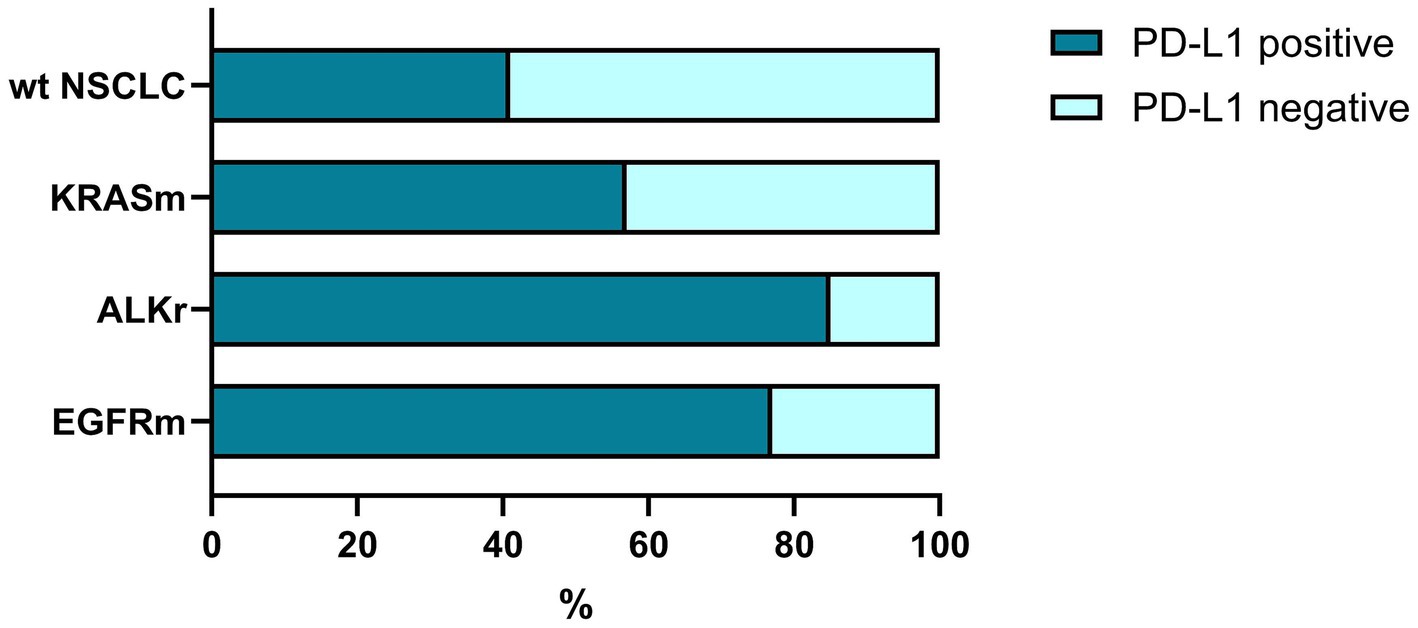
Figure 2. The rate of PD-L1 expression in oncogene-addicted and non-oncogene-addicted NSCLC. Tumors with genetic alterations in EGFR and ALK demonstrated were prone to PD-L1 expression that reached 85–87% of all carcinomas. In contrast, non-oncogene addicted NSCLC demonstrated a significantly lower rate of PD-L1 expression.
Among this set 91 cases were oncogene addicted, and about 2/3 of them (60; 65.9%) demonstrated PD-L1 positivity (Figure 2). In contrast, among non-oncogene addicted NSCLC, only 40.4% (36 of 89) showed positive expression of PD-L1 while the rest of the cases were PD-L1 negative (53 of 89 wild-type cases, 59.6%).
At the same time, tumors harboring EGFR mutations demonstrated a higher rate of PD-L1 positivity compared to NSCLC with no genetic findings (p = 0.029), specifically in patients with LUAD (p = 0.015), but not SCC (p = 0.441; Supplementary Table S4).
Similarly, ALK-positive NSCLC was associated with a higher expression of PD-L1 (p = 0,048) in both PD-L1 low and PD-L1 high groups (Supplementary Table S3). At the same time, there was no link between KRAS mutation status and PD-L1 expression within the cohort (Figure 2; Supplementary Table S4).
Thus, NSCLC tumors with genetic alterations in EGFR and ALK demonstrated a high prevalence of PD-L1 expression reflecting a predisposition to oncogene-driven immune escape. The concept of the cancer-immunity cycle was applied for further analysis of the tumor immune landscape with regard to genetic changes.
Links between genetic alterations and cancer immunity cycle in lung adenocarcinoma
In this study, oncogene-driven LUAD demonstrated distinguishable peculiarities of tumor-host immunity interplay. The density and distribution of tumor-infiltrating lymphocytes (TILs) differed depending on tumor genetic findings. The number of intra-tumorous CD8+ cells varied significantly between oncogene-driven and wild-type LUAD (p = 0.019). Wild-type LUAD demonstrated a higher number of tumor-infiltrating CD8+ T-cells, while in tumors with EGFR and ALK alterations, the number of CD8+ T-cells was much lower (Figure 3), and KRASm LUAD was heterogeneous in terms of tumor-infiltrating CD8+ cells count. This finding correlated with the prevalence of ID tumor microenvironment in EGFR mutant LUAD and IE immunophenotype among ALK-rearranged and KRAS mutant adenocarcinomas.
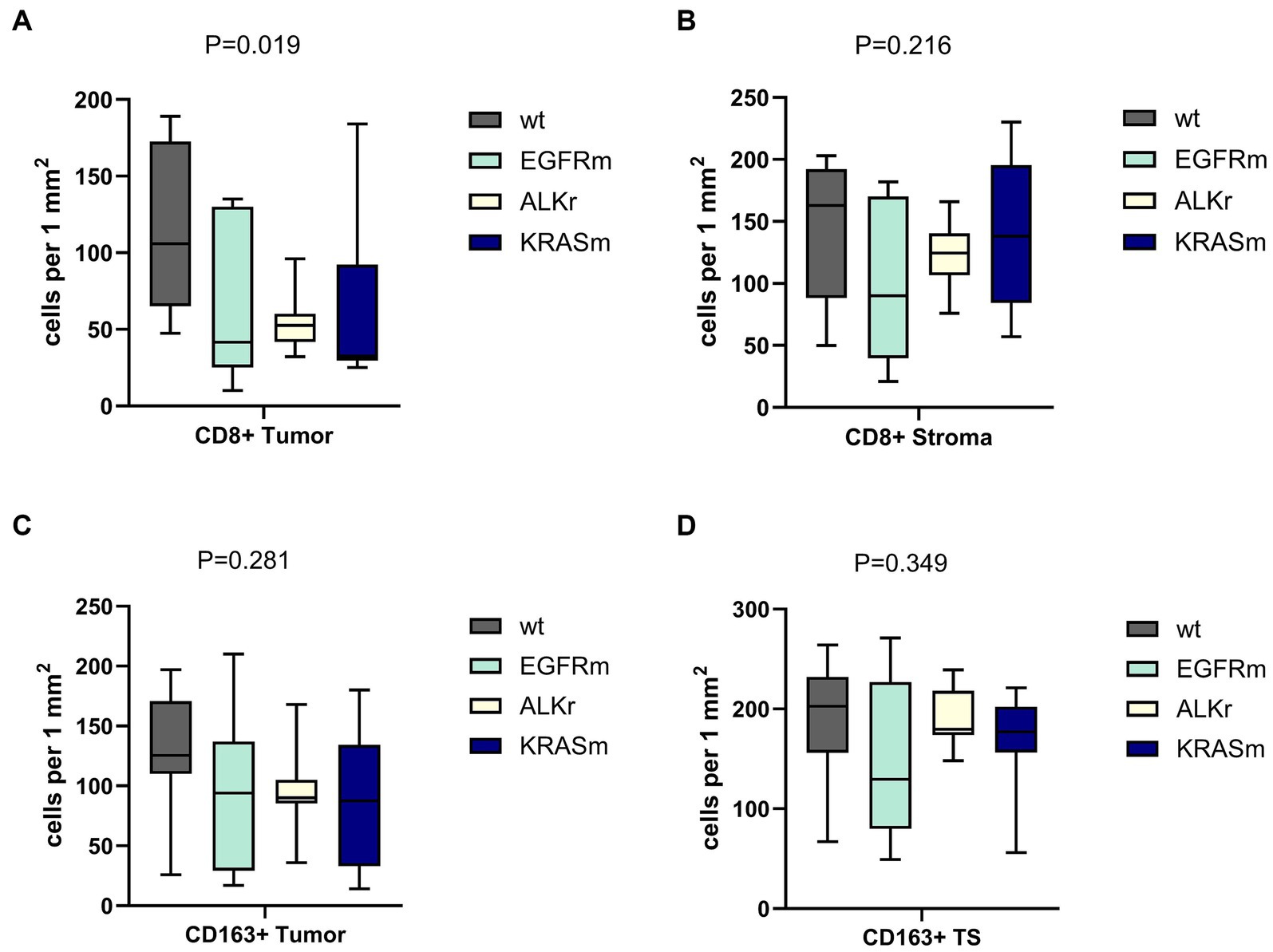
Figure 3. Density of CD8+ and CD163+ immune cell presence within tumor and tumor stroma of wild-type (WT), EGFR mutant, ALK-rearranged, and KRAS mutant LUAD. Panels (A,B) demonstrate the number of CD8+ cells within the tumor and in the peritumor stroma, respectively. Panels (C,D) showed the amount of CD163+ macrophages within the tumor and in the peritumor stroma, respectively. Oncogene-addicted LUAD demonstrated a significantly lower number of CD8+ cells infiltrating the tumor.
Indeed, genetic alterations in oncogenes were linked to the prevalence of various TME types in LUAD (p = 0.002). Non-oncogene-addicted NSCLC demonstrated the prevalence of Inflamed TME (Figure 4). In contrast, over half of EGFR mutant LUAD cases demonstrated immune desert TME type, 30% possessed immune inflamed TME and 10% had an immune excluded type. In contrast, adenocarcinoma with ALK rearrangement demonstrated mostly IE-TME, with few cases having Inflamed TME. KRASm NSCLC were the most heterogenous demonstrating 50% cases with IE phenotype and the rest were of ID and Inflamed TME.
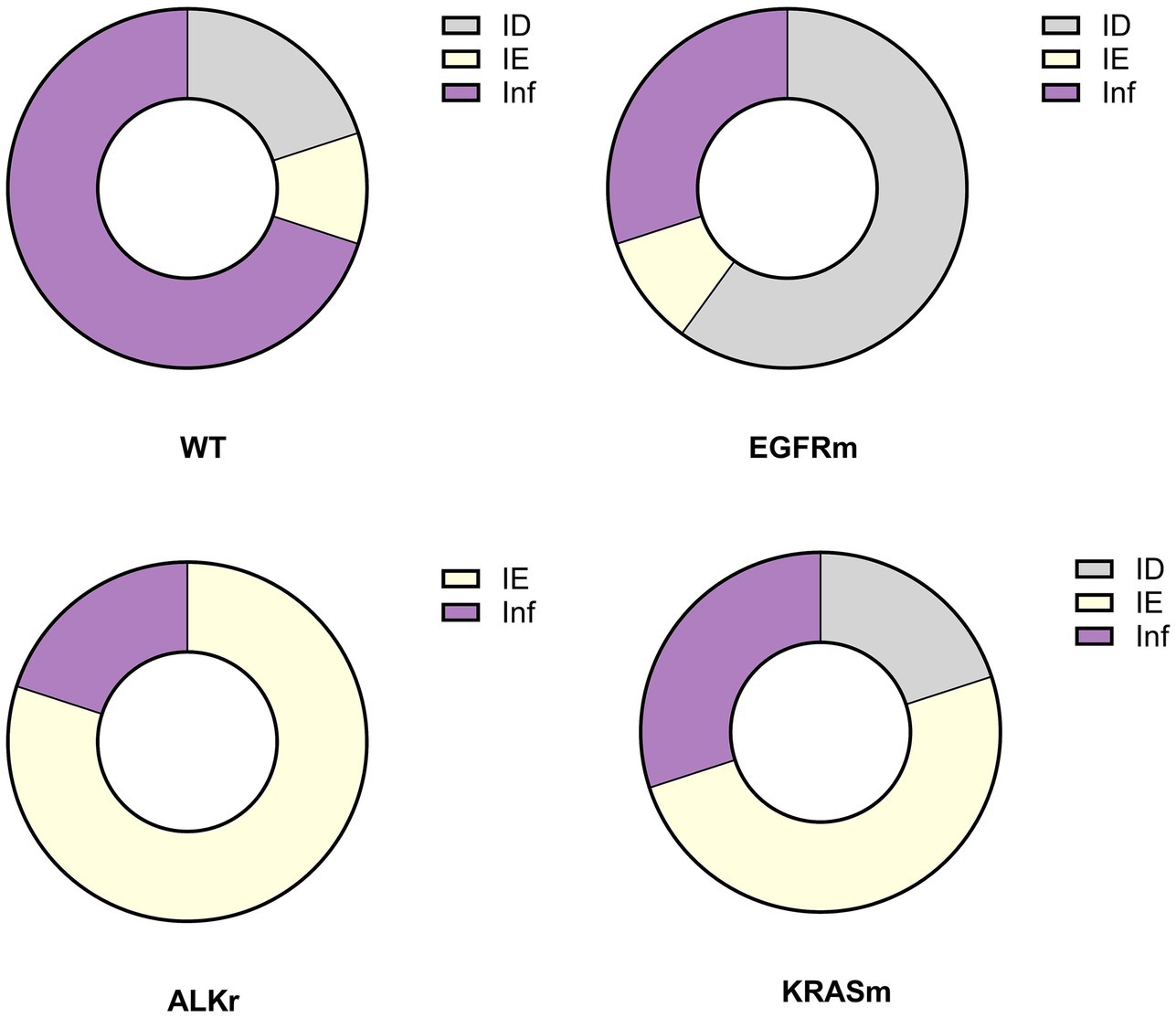
Figure 4. Prevalence of immune phenotypes in tumors with no detected genetic variants (wild-type, WT), EGFR mutant (EGFRm), ALK-rearranged (ALKr), and KRAS mutant LUAD (KRASm).
Notably, the tumor-associated macrophage (TAM) number was significantly higher than CD8+ cells in LUAD. The count of TAMs was comparable in ID and IE cases but was significantly increased in tumors with immune-inflamed TME (p < 0.001; Figure 5). The high number of CD163+ cells within the tumor was associated with positive PD-L1 status (p = 0.01), although there was no statistically significant link between the number of CD8+ cells and PD-L1 expression.
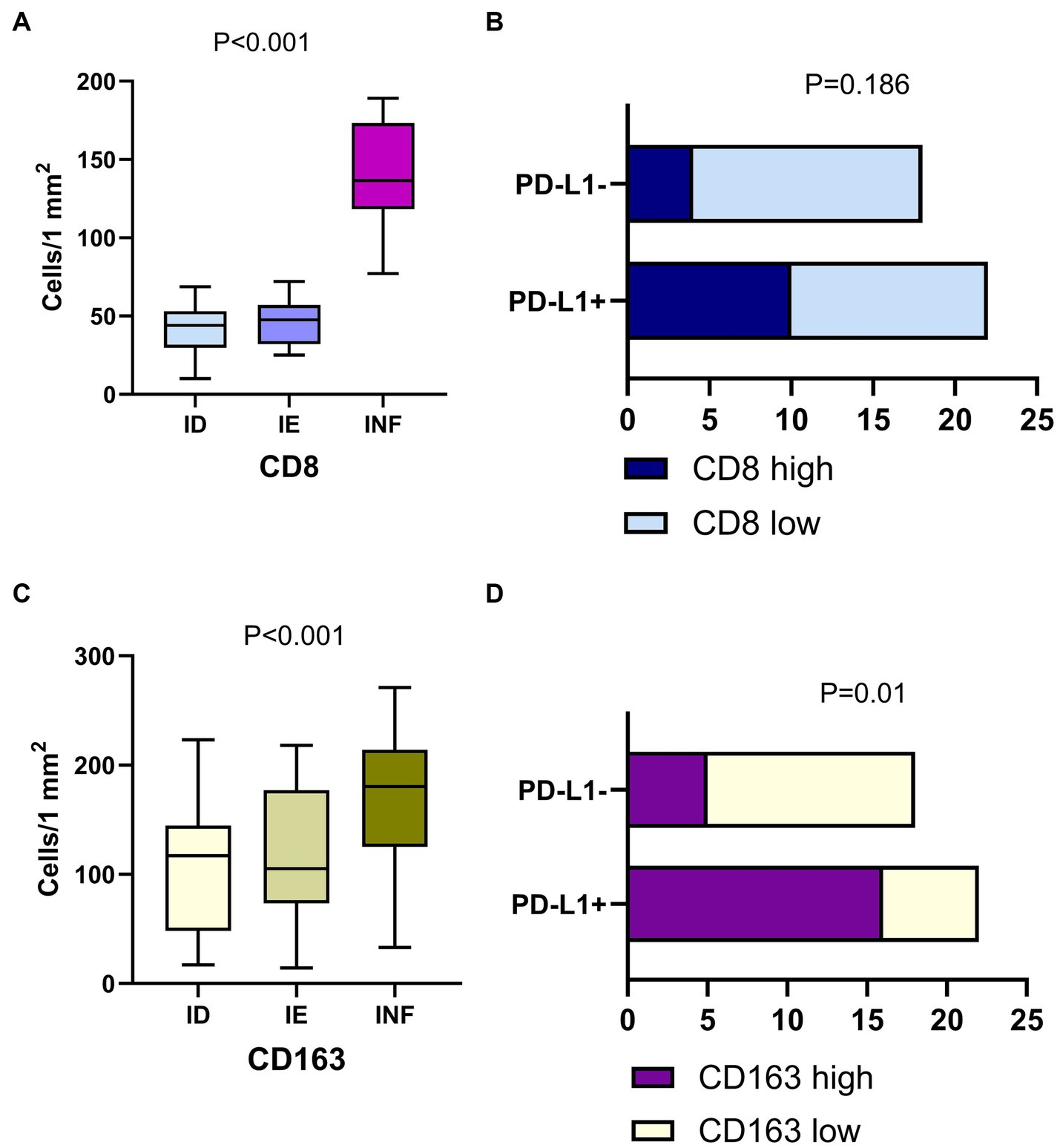
Figure 5. Immune cells in LUAD of with different immune phenotype and their interplay with PD-L1 expression. Panels (A,C) demonstrate the count of CD8+ cells and CD163 macrophages in LUAD of the immune desert (ID), immune excluded (IE) and Inflamed (INF) type of tumor microenvironment. Panels (B,D) show the link between PD-L1 expression and density of CD8+ cells and CD163+ macrophages. Positive tumor PD-L1 status is related to a high density of CD163+ cells, but not CD8+ cells.
Thus, here we report the prevalence of ID TME in EGFR mutant tumors and IE TME in ALK-rearranged and KRAS mutant LUAD, while wild-type LUAD mostly displayed immune-inflamed TME type. Inflamed TME was associated with higher M2 macrophages affecting the rate of PD-L1 expression. This data suggests an interaction between genetic alterations in LUAD and molecular mechanisms regulating the cancer-immunity cycle. With these findings, further investigation into the specific mechanisms regulating TME is necessary to apply the observed markers of immune evasion to patient management and diagnostics.
Discussion
This study first described the molecular landscape of NSCLC in the Ukrainian population. We found a high rate of genetic alterations in LUAD and a significantly lower rate of actionable mutations in SCC. In general, this data aligns with the global reports on the molecular landscape of NSCLC (30, 31).
At the same time, the Ukrainian cohort observed in this study demonstrated peculiar features. First, the rate of alterations in EGFR and ALK was relatively high, while the incidence of KRAS mutations was lower than the average reported frequency worldwide. Various populations demonstrate differences in the spectrum of oncogenic driver mutations. For instance, in an Irish study on 2052 NSCLC patients, actionable genetic alterations were found in 53% of cases, with the highest rate of KRAS mutation reported to be 32%, while the rate of EGFR mutation was only 8.8%, and ALK rearrangements were found in only 2.1% cases (32). Alternatively, in the Indian cohort of 325 patients with NSCLC, the rate of EGFR mutation reached 45.8%, and ALK rearrangements were found in 11.4% of cases, however, KRAS mutations were present in only 10.2% (33). The availability of such population data in cancer research is essential to understanding the ethnic peculiarities of NSCLC, planning clinical trials and introducing novel precision medicines.
The rate of genetic mutation in NSCLC also depends on various extrinsic (tobacco smoking, air pollution, exposure to radiation or occupational) factors as well as intrinsic factors, including population distribution, mean / median age, gender, histological tumor type and stage, etc (34). There are well-known facts on the association of NSCLC genotypes with such factors, e.g., EGFR mutations and ALK fusions prevail in female patients with adenocarcinoma of younger age and no history of smoking, while KRAS mutations are more common in smokers (35). Besides, the role of alterations in genes affecting chromatin remodeling and intercellular crosstalk can facilitate immune evasion mechanisms affecting susceptibility to immunotherapy (9). Importantly, NGS testing affordability is also a factor impacting the rate of detected genetic alterations, especially in developing countries where these expensive services are paid mostly out of pocket (36).
According to this study’s results, alterations in EGFR and ALK were associated with high PD-L1 expression in NSCLC. The high rate of PD-L1 positive NSCLC aligns with previous data, demonstrating positive PD-L1 status in up to 65% of NSCLC tumors (37). Indeed, the increased rate of PD-L1 positive tumors was previously shown for EGFR mutant LUAD (38, 39). Tumors harboring EGFR variants can evade immune response by exposing PD-L1 on tumor cells and inhibiting T-cell antitumor activity (40). EGFR activation in cancer cells can upregulate PD-L1 through the MAPK signaling pathway that leads to increased activity of ERK1/2 and AKT which facilitate cell proliferation and invasion. However, high expression of PD-L1 in EGFR mutant NSCLC, in theory, should make the tumor susceptible to ICI, shaping the possibility to reactivate the immune system against tumor cells. However, as it was previously shown, anti-PD-1/PD-L1 therapy demonstrates limited success in NSCLC with activating EGFR and ALK genetic alterations (41). Importantly, we found that increased PD-L1 expression was linked to high numbers of M2 macrophages, which is consistent with the data from other cancer types (25, 26).
At the same time, in our study, KRAS mutant carcinoma was also found to be heterogeneous with the prevalence of IE TME and less frequent inflamed immune contexture. This data at least partly contradicts the widely accepted concept of high immunogenicity of KRAS mutant NSCLC. A recent study illuminated that KRAS mutation status in NSCLC positively correlated with TMB, higher neoantigens generation, PD-L1 expression, and T-cell infiltration (16). On the other hand, the impact of KRAS mutations on the immunity responses remains disputable in NSCLC patients (42). Moreover, it was shown that KRAS status did not affect patients outcomes under nivolumab application (43). Interpreting the prevalence of various TME types in oncogene-driven NSCLC, it seems essential to understand the bidirectional nature of the interplay between the mutational landscape and immune contexture of tumors. On one hand, genetic alterations can be associated with the expression of neoantigens improving the recognition of tumor cells and activation of the cancer immunity cycle. On the other hand, tumor-secreted exosomes, hypoxia, and acidic milieu can promote immunosuppressive TME (42). The complexity of tumor-host interplay also relies on chronological dimension, cancer cells evolution and immune cells involvement in clone selection.
Although EGFR and KRAS are closely linked within their signaling pathways, harboring oncogenic mutations in these genes differently affects prognosis, tumor biology, histological differentiation and immunogenicity (44, 45). Contrary to the immune contexture of EGFR and ALK altered tumors, the KRAS driven NSCLC displays predominantly IE and inflamed TME types, which suggests a different route to cancer immune evasion. The cancer-immunity cycle describes the following steps to efficient anti-tumor response: release of cancer cell antigens, antigen presentation by dendritic cells, priming and activation of T cells in lymph nodes, trafficking of effector T cells through the bloodstream, infiltration of T cells into the tumor, recognition of cancer cells by the T cells, and elimination of cancer cells (24, 27). From this perspective, KRAS-and EGFR-mutant tumors obviously diverge at multiple points in anti-tumor response, leading to distinct immune microenvironment phenotypes.
First, KRAS-mutant tumors typically exhibit a higher tumor mutational burden (TMB) generating more neoantigens which can be captured by antigen presenting cells (APC) (46). According to the study by Li T et al., EGFR-mutant NSCLC exhibit lower TMB (mean ~3.9 vs. ~6.1 mutations/Mb in KRAS, p < 0.001), resulting in fewer neoantigens to be available to APC to prime T cells. Next, via IL-6/JAK/STAT3 axis and NF-kB activation, KRAS mutant tumors promote inflammation and immune invasion. At the same time however, KRAS activation in lung cancer is also linked to MEK/ERK/AP-1 mediated secretion of IL-10 and TGF-β1, which regulate recruitment of CD4+, myeloid-derived suppressor cells (MDSCs) and regulatory T cells (Treg) resulting in an immunosuppressive effect via downregulation of MHC-I expression on tumor cells, preventing antigen presentation and T-cells priming (47, 48). Moreover, KRAS-mutant cancer cells are prone to activating cancer-associated fibroblasts (CAFs) via MAPK and PI3K signaling, promoting desmoplasia and deposition of collagen around tumor clusters (47). Such stromal rearrangements might form a barrier physically excluding T cells from penetrating into the tumor core, despite activated inflammatory IL-6 signaling and CXCR2 chemokine gradient (49). Such changes could explain the prevalence of immune excluded phenotype in KRASm NSCLC. Interestingly, EGFR signaling can also induce extracellular matrix remodeling via PKCδ-activation which leads to enhanced collagen deposition around tumor cells, reducing T-cell entry into the tumor (50). Nevertheless, it seems that in complex with low TMB and compromised tumor antigen recognition, these changes can manifest into “immune desert” or “cold” immunophenotype, defining a poor response to immune checkpoint blockade.
Finally, co-occurring genetic alterations which typically accompany KRAS mutation in NSCLC could also impact TME. KRAS co-mutations with STK11/LKB1 were shown to impair innate immune sensing by downregulating STING pathway to interferon production and diminish the expression of vascular adhesion molecule-1, thereby blocking T-cell extravasation. Additionally, the combination of KRASm with KEAP1 loss suppresses dendritic cell maturation via NRF2-mediated antioxidant programs.
Applying the concept of the cancer-immunity cycle in this study allowed us to reveal the potential mechanisms of non-responsiveness to ICI relying on low immunogenicity of tumors with altered ALK and EGFR. Most of them demonstrated ID or IE TME reflecting the inefficiency of primary tumor cell recognition.
Thus, early events in cancer cell recognition and T-cell priming are compromised. Recognition of tumor cells by the immune system relies on the functioning of APC, including dendritic cells and macrophages, able to distinguish self from nonself via neoantigens and the major histocompatibility complex (MHC). Further priming of T cells leads to their activation and is regulated by several antigen-processing pathways involving proteins of MHC class II and B7 (51). Expression of cytotoxic T-lymphocytes-associated antigen 4 (CTLA-4) instead of B7 contributes to immunosuppression at an early stage, defining peripheral immune tolerance and preventing T-cell activation. Indeed, activating EGFR mutations can upregulate signal transducer and activator of the transcription 3 (STAT3), which is involved in STAT1 regulation (52). The latter is essential for HLA1 expression. This can affect antigen presentation machinery and cancer cell recognition by APC (53). In addition, STAT3 regulates the production of various cytokines and growth factors, modulating the expression of vascular endothelial growth factor (VEGF), IL-6, and IL-10, promoting anti-inflammatory effects and inhibiting APC differentiation and maturation (54).
Limitations of the study
This study included a limited number of cases with known genetic alterations in a confined number of genes analyzed using NGS technology. The panel utilized for NGS testing did not include such genes as KEAP1, STK11, etc. which did not allow to consider the potential impact of other genetic alterations in shaping the immunotype of tumors. No information about therapy, its efficacy and patients` outcomes was included in this study. When assessing the immune contexture of NSCLC only LUAD with alterations in EGFR, ALK and KRAS were included in the analysis. Tumors with alterations in other genes were not analyzed due to a low incidence and a small number of cases with verified oncogenic variants. Further prospective studies are required to discover molecular mechanisms defining the interplay between oncogene-driven cancer cells and immune response with respect to the tumor immunity cycle.
Conclusion
This study revealed a high rate of EGFR and ALK rearrangements in the Ukrainian cohort of NSCLC patients with a relatively moderate frequency of KRAS mutations. The high rate of PD-L1 expression in NSCLC driven by EGFR and ALK alterations was accompanied by a prevalence of low immunogenicity with the prevalence of ID TME in EGFR mutant tumors and IE TME in ALK-rearranged and KRAS mutant tumors. Further discovery of mechanisms affecting tumor immune contexture is needed for tailoring patient management in line with particular mechanisms of immune evasion.
Data availability statement
The data presented in the study are deposited in the NCBI Sequence Read Archive (SRA), submission: SUB15436110, accession number: PRJNA1287077.
Ethics statement
The studies involving humans were approved by Research Ethics Committee of Medical Laboratory CSD LAB (protocol # 12/22 on December 13, 2022). The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from a by-product of routine care or industry. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
DK: Conceptualization, Supervision, Writing – original draft. NK: Conceptualization, Data curation, Project administration, Supervision, Writing – original draft. SL: Formal analysis, Methodology, Visualization, Writing – review & editing. OSe: Formal analysis, Methodology, Writing – review & editing. OKos: Formal analysis, Methodology, Writing – review & editing. AM: Formal analysis, Methodology, Writing – review & editing. YS: Formal analysis, Methodology, Writing – review & editing. OKol: Formal analysis, Methodology, Writing – review & editing. YM: Formal analysis, Methodology, Writing – review & editing. OV: Formal analysis, Methodology, Writing – review & editing. RM: Formal analysis, Methodology, Writing – review & editing. SK: Formal analysis, Methodology, Writing – review & editing. AK: Formal analysis, Methodology, Writing – review & editing. BS: Formal analysis, Methodology, Writing – review & editing. OSu: Conceptualization, Data curation, Funding acquisition, Project administration, Supervision, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was partly covered by grants of the external aid instrument of the European Union for the fulfillment of Ukraine’s obligations in the Framework Program of the European Union for Scientific Research and Innovation “Horizon 2020” No. RN/11-2023 “The role of the DNA repair system in the pathogenesis and immunogenicity of lung cancer.”
Conflict of interest
BS was employed by company Audubon Bioscience.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2025.1558016/full#supplementary-material
Footnotes
References
1. Fitzmaurice, C, Abate, D, Abbasi, N, Abbastabar, H, Abd-Allah, F, Abdel-Rahman, O, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2017: a systematic analysis for the global burden of disease study. JAMA Oncol. (2019) 5:1749–68. doi: 10.1001/JAMAONCOL.2019.2996
2. Tan, AC, and Tan, DSW. Targeted therapies for lung Cancer patients with oncogenic driver molecular alterations. J Clin Oncol. (2022) 40:611–25. doi: 10.1200/JCO.21.01626
3. Reck, M, Remon, J, and Hellmann, MD. First-line immunotherapy for non-small-cell lung Cancer. J Clin Oncol. (2022) 40:586–97. doi: 10.1200/JCO.21.01497
4. Stanley, R, Flanagan, S, Reilly, DO, Kearney, E, Naidoo, J, and Dowling, CM. Immunotherapy through the lens of non-small cell lung cancer. Cancers. (2023) 15:2996. doi: 10.3390/CANCERS15112996
5. Chaft, JE, Oezkan, F, Kris, MG, Bunn, PA, Wistuba, II, Kwiatkowski, DJ, et al. Neoadjuvant atezolizumab for resectable non-small cell lung cancer: an open-label, single-arm phase II trial. Nat Med. (2022) 28:2155–61. doi: 10.1038/S41591-022-01962-5
6. Borghaei, H, Gettinger, S, Vokes, EE, Chow, LQM, Burgio, MA, de, J, et al. Five-year outcomes from the randomized, phase III trials CheckMate 017 and 057: nivolumab versus docetaxel in previously treated non-small-cell lung cancer. J Clin Oncol. (2021) 39:723–33. doi: 10.1200/JCO.20.01605
7. Uprety, D, Mandrekar, SJ, Wigle, D, Roden, AC, and Adjei, AA. Neoadjuvant immunotherapy for NSCLC: current concepts and future approaches. J Thorac Oncol. (2020) 15:1281–97. doi: 10.1016/J.JTHO.2020.05.020
8. Lazzari, C, Spagnolo, CC, Ciappina, G, Di Pietro, M, Squeri, A, Passalacqua, MI, et al. Immunotherapy in early-stage non-small cell lung Cancer (NSCLC): current evidence and perspectives. Curr Oncol. (2023) 30:3684–96. doi: 10.3390/CURRONCOL30040280
9. Cucchiara, F, Crucitta, S, Petrini, I, de Miguel Perez, D, Ruglioni, M, Pardini, E, et al. Gene-network analysis predicts clinical response to immunotherapy in patients affected by NSCLC. Lung Cancer. (2023) 183:107308. doi: 10.1016/J.LUNGCAN.2023.107308
10. Soo, RA, Lim, SM, Syn, NL, Teng, R, Soong, R, Mok, TSK, et al. Immune checkpoint inhibitors in epidermal growth factor receptor mutant non-small cell lung cancer: current controversies and future directions. Lung Cancer. (2018) 115:12–20. doi: 10.1016/J.LUNGCAN.2017.11.009
11. Jia, Y, Li, X, Jiang, T, Zhao, S, Zhao, C, Zhang, L, et al. EGFR-targeted therapy alters the tumor microenvironment in EGFR-driven lung tumors: implications for combination therapies. Int J Cancer. (2019) 145:1432–44. doi: 10.1002/IJC.32191
12. Jahanzeb, M, Lin, HM, Pan, X, Yin, Y, Baumann, P, and Langer, CJ. Immunotherapy treatment patterns and outcomes among ALK-positive patients with non-small-cell lung cancer. Clin Lung Cancer. (2021) 22:49–57. doi: 10.1016/J.CLLC.2020.08.003
13. Shi, C, Wang, Y, Xue, J, and Zhou, X. Immunotherapy for EGFR-mutant advanced non-small-cell lung cancer: current status, possible mechanisms and application prospects. Front Immunol. (2022) 13:288. doi: 10.3389/FIMMU.2022.940288
14. Gainor, JF, Shaw, AT, Sequist, LV, Fu, X, Azzoli, CG, Piotrowska, Z, et al. EGFR mutations and ALK rearrangements are associated with low response rates to PD-1 pathway blockade in non-small cell lung Cancer: A retrospective analysis. Clin Cancer Res. (2016) 22:4585–93. doi: 10.1158/1078-0432.CCR-15-3101
15. Dong, ZY, Zhong, WZ, Zhang, XC, Su, J, Xie, Z, Liu, SY, et al. Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin Cancer Res. (2017) 23:3012–24. doi: 10.1158/1078-0432.CCR-16-2554
16. Liu, C, Zheng, S, Jin, R, Wang, X, Wang, F, Zang, R, et al. The superior efficacy of anti-PD-1/PD-L1 immunotherapy in KRAS-mutant non-small cell lung cancer that correlates with an inflammatory phenotype and increased immunogenicity. Cancer Lett. (2020) 470:95–105. doi: 10.1016/J.CANLET.2019.10.027
17. Coelho, MA, de Carné Trécesson, S, Rana, S, Zecchin, D, Moore, C, Molina-Arcas, M, et al. Oncogenic RAS signaling promotes tumor immunoresistance by stabilizing PD-L1 mRNA. Immunity. (2017) 47:1083–1099.e6. doi: 10.1016/J.IMMUNI.2017.11.016
18. Li, X, Lian, Z, Wang, S, Xing, L, and Yu, J. Interactions between EGFR and PD-1/PD-L1 pathway: implications for treatment of NSCLC. Cancer Lett. (2018) 418:1–9. doi: 10.1016/J.CANLET.2018.01.005
19. Yu, Y, Zeng, D, Ou, Q, Liu, S, Li, A, Chen, Y, et al. Association of Survival and Immune-Related Biomarkers with Immunotherapy in patients with non-small cell lung Cancer: A Meta-analysis and individual patient-level analysis. JAMA Netw Open. (2019) 2:e196879. doi: 10.1001/JAMANETWORKOPEN.2019.6879
20. Lee, CK, Man, J, Lord, S, Cooper, W, Links, M, Gebski, V, et al. Clinical and molecular characteristics associated with survival among patients treated with checkpoint inhibitors for advanced non–small cell lung carcinoma: a systematic review and meta-analysis. JAMA Oncol. (2017) 4:210–6. doi: 10.1001/JAMAONCOL.2017.4427
21. Raskov, H, Orhan, A, Christensen, JP, and Gögenur, I. Cytotoxic CD8+ T cells in cancer and cancer immunotherapy. Br J Cancer. (2020) 124:359–67. doi: 10.1038/s41416-020-01048-4
22. George, JT, and Levine, H. Implications of tumor-immune coevolution on Cancer evasion and optimized immunotherapy. Trends Cancer. (2021) 7:373–83. doi: 10.1016/J.TRECAN.2020.12.005
23. Hiraoka, K, Miyamoto, M, Cho, Y, Suzuoki, M, Oshikiri, T, Nakakubo, Y, et al. Concurrent infiltration by CD8+ T cells and CD4+ T cells is a favourable prognostic factor in non-small-cell lung carcinoma. Br J Cancer. (2006) 94:275–80. doi: 10.1038/SJ.BJC.6602934
24. Chen, DS, and Mellman, I. Oncology meets immunology: the cancer-immunity cycle. Immunity. (2013) 39:1–10. doi: 10.1016/J.IMMUNI.2013.07.012
25. Mashukov, A, Shapochka, D, Seleznov, O, Kobyliak, N, Sulaieva, O, Falalyeyeva, T, et al. Histological differentiation impacts the tumor immune microenvironment in gastric carcinoma: relation to the immune cycle. World J Gastroenterol. (2021) 27:5259–71. doi: 10.3748/WJG.V27.I31.5259
26. Stakhovskyi, O, Kobyliak, N, Voylenko, O, Stakhovskyi, E, Ponomarchuk, R, and Sulaieva, O. Immune microenvironment of muscular-invasive urothelial carcinoma: the link to tumor immune cycle and prognosis. Cells. (2022) 11:1802. doi: 10.3390/CELLS11111802
27. Mellman, I, Chen, DS, Powles, T, and Turley, SJ. The cancer-immunity cycle: indication, genotype, and immunotype. Immunity. (2023) 56:2188–205. doi: 10.1016/J.IMMUNI.2023.09.011
28. Sulaieva, O, Chernenko, O, Selesnov, O, Nechay, O, Maievskyi, O, Falalyeyeva, T, et al. Mechanisms of the impact of Hashimoto thyroiditis on papillary thyroid carcinoma progression: relationship with the tumor immune microenvironment. Endocrinol Metab. (2020) 35:443–55. doi: 10.3803/ENM.2020.35.2.443
29. Li, M, Qin, J, Xie, F, Gong, L, Han, N, and Lu, H. L718Q/V mutation in exon 18 of EGFR mediates resistance to osimertinib: clinical features and treatment. Discov Oncol. (2022) 13:72. doi: 10.1007/S12672-022-00537-7
30. Pao, W, Miller, V, Zakowski, M, Doherty, J, Politi, K, Sarkaria, I, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. (2004) 101:13306–11. doi: 10.1073/PNAS.0405220101
31. Rotow, J, and Bivona, TG. Understanding and targeting resistance mechanisms in NSCLC. Nat Rev Cancer. (2017) 17:637–58. doi: 10.1038/NRC.2017.84
32. Keogh, RJ, Barr, MP, Keogh, A, McMahon, D, O’Brien, C, Finn, SP, et al. Genomic landscape of NSCLC in the Republic of Ireland. JTO Clin Res Rep. (2023) 5:627. doi: 10.1016/J.JTOCRR.2023.100627
33. Jha, P, Joshi, A, Mishra, R, Biswal, RP, Kulkarni, PM, Limaye, S, et al. Landscape of clinically relevant genomic alterations in the Indian non-small cell lung Cancer patients. Clin Lung Cancer. (2024) 25:e420–e430.e20. doi: 10.1016/J.CLLC.2024.07.011
34. Kimbrough, E, Dada, HI, Drusbosky, L, Zhao, Y, Manochakian, R, and Lou, Y. Genomic landscape differences in patients with advanced non-small cell lung cancer by sex and age. J Clin Oncol. (2021) 39:9108–8. doi: 10.1200/JCO.2021.39.15_SUPPL.9108
35. Lee, JJK, Park, S, Park, H, Kim, S, Lee, J, Lee, J, et al. Tracing oncogene rearrangements in the mutational history of lung adenocarcinoma. Cell. (2019) 177:1842–1857.e21. doi: 10.1016/J.CELL.2019.05.013
36. Sulaieva, O, Falalyeyeva, T, Kobyliak, N, Pellicano, R, and Dudin, O. Precision oncology: ethical challenges and justification. Minerva Med. (2022) 113:603–5. doi: 10.23736/S0026-4806.22.08063-6
37. Cronin-Fenton, D, Dalvi, T, Movva, N, Pedersen, L, Hansen, H, Fryzek, J, et al. PD-L1 expression, EGFR and KRAS mutations and survival among stage III unresected non-small cell lung cancer patients: a Danish cohort study. Sci Rep. (2021) 11:16892. doi: 10.1038/S41598-021-96486-2
38. Tang, Y, Fang, W, Zhang, Y, Hong, S, Kang, S, Yan, Y, et al. The association between PD-L1 and EGFR status and the prognostic value of PD-L1 in advanced non-small cell lung cancer patients treated with EGFR-TKIs. Oncotarget. (2015) 6:14209–19. doi: 10.18632/ONCOTARGET.3694
39. Azuma, K, Ota, K, Kawahara, A, Hattori, S, Iwama, E, Harada, T, et al. Association of PD-L1 overexpression with activating EGFR mutations in surgically resected nonsmall-cell lung cancer. Annals Oncol. (2014) 25:1935–40. doi: 10.1093/ANNONC/MDU242
40. Sznol, M, and Chen, L. Antagonist antibodies to PD-1 and B7-H1 (PD-L1) in the treatment of advanced human cancer. Clin Cancer Res. (2013) 19:1021–34. doi: 10.1158/1078-0432.CCR-12-2063
41. Chen, X, Gao, A, Zhang, F, Yang, Z, Wang, S, Fang, Y, et al. ILT4 inhibition prevents TAM-and dysfunctional T cell-mediated immunosuppression and enhances the efficacy of anti-PD-L1 therapy in NSCLC with EGFR activation. Theranostics. (2021) 11:3392–416. doi: 10.7150/THNO.52435
42. Guo, X, Song, J, Liu, M, Ou, X, and Guo, Y. The interplay between the tumor microenvironment and tumor-derived small extracellular vesicles in cancer development and therapeutic response. Cancer Biol Ther. (2024) 25:2356831. doi: 10.1080/15384047.2024.2356831
43. Passiglia, F, Cappuzzo, F, Alabiso, O, Bettini, AC, Bidoli, P, Chiari, R, et al. Efficacy of nivolumab in pre-treated non-small-cell lung cancer patients harbouring KRAS mutations. Br J Cancer. (2019) 120:57–62. doi: 10.1038/S41416-018-0234-3
44. Amanam, I, Mambetsariev, I, Gupta, R, Achuthan, S, Wang, Y, Pharaon, R, et al. Role of immunotherapy and co-mutations on KRAS-mutant nonsmall cell lung cancer survival. J Thorac Dis. (2020) 12:5086–95. doi: 10.21037/JTD.2020.04.18
45. Marks, JL, Broderick, S, Zhou, Q, Chitale, D, Li, AR, Zakowski, MF, et al. Prognostic and therapeutic implications of EGFR and KRAS mutations in resected lung adenocarcinoma. J Thorac Oncol. (2008) 3:111–6. doi: 10.1097/JTO.0b013e318160c607
46. Li, T, Pang, X, Wang, J, Wang, S, Guo, Y, He, N, et al. Exploration of the tumor-suppressive immune microenvironment by integrated analysis in EGFR-mutant lung adenocarcinoma. Front Oncol. (2021) 11:922. doi: 10.3389/FONC.2021.591922
47. Hamarsheh, S, Groß, O, Brummer, T, and Zeiser, R. Immune modulatory effects of oncogenic KRAS in cancer. Nat Commun. (2020) 11:5439. doi: 10.1038/S41467-020-19288-6
48. de Visser, KE, and Joyce, JA. The evolving tumor microenvironment: from cancer initiation to metastatic outgrowth. Cancer Cell. (2023) 41:374–403. doi: 10.1016/J.CCELL.2023.02.016
49. Zhou, Y, Kuang, Y, Wang, C, Yu, Y, Pan, L, and Hu, X. Impact of KRAS mutation on the tumor microenvironment in colorectal cancer. Int J Biol Sci. (2024) 20:1947–64. doi: 10.7150/IJBS.88779
50. Zuo, YH, Gao, WN, Xie, YJ, Yang, SY, Zhou, JT, Liang, HH, et al. Tumor PKCδ instigates immune exclusion in EGFR-mutated non–small cell lung cancer. BMC Med. (2022) 20:470. doi: 10.1186/S12916-022-02670-0
51. Buchbinder, EI, and Desai, A. CTLA-4 and PD-1 pathways: similarities, differences, and implications of their inhibition. Am J Clin Oncol. (2016) 39:98–106. doi: 10.1097/COC.0000000000000239
52. Rubin Grandis, J, Drenning, SD, Chakraborty, A, Zhou, MY, Zeng, Q, Pitt, AS, et al. Requirement of Stat3 but not Stat1 activation for epidermal growth factor receptor-mediated cell growth in vitro. J Clin Invest. (1998) 102:1385–92. doi: 10.1172/JCI3785
53. Concha-Benavente, F, Srivastava, RM, Trivedi, S, Lei, Y, Chandran, U, Seethala, RR, et al. Identification of the cell-intrinsic and-extrinsic pathways downstream of EGFR and IFNγ that induce PD-L1 expression in head and neck cancer. Cancer Res. (2016) 76:1031–43. doi: 10.1158/0008-5472.CAN-15-2001
Keywords: non-small cell lung cancer, adenocarcinoma, genetic alterations, oncogenes, immune contexture, cancer immunity cycle
Citation: Kozakov D, Kobyliak N, Livshun S, Seleznov O, Koshyk O, Matvieieva A, Shparyk Y, Kolesnik O, Moskalenko Y, Vynnychenko O, Moskalenko R, Kropyvko S, Khmel A, Shkarupii B and Sulaieva O (2025) Genetic alterations affect immune contexture of non-small cell lung cancer: Ukrainian study. Front. Med. 12:1558016. doi: 10.3389/fmed.2025.1558016
Edited by:
Luigi M. Terracciano, University of Basel, SwitzerlandReviewed by:
Zhaohua Hou, Memorial Sloan Kettering Cancer Center, United StatesGian Marco Leone, University of Catania, Italy
Copyright © 2025 Kozakov, Kobyliak, Livshun, Seleznov, Koshyk, Matvieieva, Shparyk, Kolesnik, Moskalenko, Vynnychenko, Moskalenko, Kropyvko, Khmel, Shkarupii and Sulaieva. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Denys Kozakov, ZC5rb3pha292QGNzZC5jb20udWE=
 Denys Kozakov
Denys Kozakov Nazarii Kobyliak
Nazarii Kobyliak Sofiia Livshun
Sofiia Livshun Oleksii Seleznov
Oleksii Seleznov Olena Koshyk
Olena Koshyk Alina Matvieieva
Alina Matvieieva Yaroslav Shparyk
Yaroslav Shparyk Oleksii Kolesnik6,7
Oleksii Kolesnik6,7 Yuliia Moskalenko
Yuliia Moskalenko Oleksandr Vynnychenko
Oleksandr Vynnychenko Roman Moskalenko
Roman Moskalenko Serhii Kropyvko
Serhii Kropyvko Bogdana Shkarupii
Bogdana Shkarupii Oksana Sulaieva
Oksana Sulaieva