Abstract
Background:
Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal malignancies, with limited treatment options and poor prognosis. Recent advances in cancer genomic analysis enable the identification of actionable gene alterations, opening new opportunities for personalized therapy. Among these, homologous recombination DNA repair (HRR) gene alterations are associated with distinct biological behavior, favorable prognosis, and increased sensitivity to platinum-based chemotherapy. However, the prognostic impact of coexisting mutations in key driver genes—KRAS, TP53, CDKN2A, and SMAD4—within HRR-altered PDAC remains poorly understood.
Methods:
We retrospectively analyzed PDAC patients who underwent genomic profiling testing with FoundationOne® CDx between June 2019 and December 2021 through the Center for Cancer Genomics and Advanced Therapeutics (C-CAT) database. We compared the prevalence and prognostic significance of key gene alterations between HRR-altered and HRR–wild-type (WT) tumors.
Results:
Of 2,381 PDAC patients, 274 (11.5%) harbored HRR alterations. These patients showed significantly longer overall survival (OS) than those with HRR-WT tumors (HR = 0.66, p = 0.002). The frequencies of KRAS, TP53, and CDKN2A mutations were less frequent in HRR-altered tumors. TP53 mutation was independently associated with poorer OS across both HRR subgroups, while CDKN2A alteration was a poor prognostic factor in HRR-WT tumors. Interestingly, SMAD4 alteration was linked to improved survival in the HRR-altered group.
Conclusion:
HRR-altered PDAC has a distinct genomic profile and is associated with a favorable prognosis. Our findings demonstrate that coexisting alterations are significant prognostic factors in both HRR-altered and HRR–wild-type tumors. These results highlight the clinical relevance of incorporating comprehensive genomic profiling into routine care to stratify patient prognosis better and inform individualized treatment strategies in PDAC.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the sixth leading cause of cancer-related deaths worldwide (1). The prognosis of metastatic and advanced PDAC remains poor, with a 5-year survival rate of less than 10% (2). The integration of molecular profiling, including next-generation sequencing (NGS), into routine clinical practice has significantly advanced our understanding of genetic alterations associated with PDAC. KRAS, TP53, CDKN2A, and SMAD4 are the most frequently altered genes in PDAC. Although these gene alterations contribute to aggressive tumor characteristics and lead to treatment resistance and poor prognosis (3–6), they have remained undruggable for decades. However, recent scientific advances have enabled the targeting of these gene alterations. For example, structural and biochemical analyses can now be used to design drugs targeting KRAS mutations and p53 (7, 8). Moreover, rapid progress in proteolysis-targeting chimera (PROTAC) technology has provided a new strategy for targeting activated KRAS proteins (9), providing hope for improving the prognosis of PDAC.
Homologous recombination DNA repair (HRR) gene alterations have been identified in 14–16.5% of PDAC patients (10, 11), and these patients have a more favorable prognosis than those with wild-type (WT) (12, 13). Further, PDAC patients with HRR gene mutations demonstrate greater sensitivity to platinum-based therapies than do those with HRR-WT PDAC (14). PARP inhibitors have been evaluated in PDAC patients with HRR gene mutations based on the concept of synthetic lethality (15); however, their efficacy is limited to germline BRCA1/2 mutations in maintenance therapy after platinum-based treatment (16). With the recent progress in molecularly targeted therapies for PDAC, a comprehensive understanding of the genetic landscape and its influence on survival outcomes has become an important area of investigation. Previous studies have noted that the mutation frequencies of KRAS, TP53, and CDKN2A in PDAC differ depending on the presence or absence of HRR gene alterations (17, 18); however, the prognostic significance of these genetic alterations when stratified by HRR gene alteration status has not yet been explored.
Thus, this study aimed to evaluate the prevalence and prognostic impact of coexisting gene alterations according to the HRR gene alteration status in PDAC. Toward this goal, we analyzed the data of patients with PDAC registered in the Center for Cancer Genomics and Advanced Therapeutics (C-CAT) (19), the largest real-world comprehensive genomic profiling registry in Japan.
Methods
Ethical approval
This study was approved by the Medical Ethics Review Committee of Kindai University of Medicine (Approval Number: R05-029) and the C-CAT review board (C-CAT Control Number: CDU2023-021 N) and was conducted according to the tenets of the Declaration of Helsinki.
Study design and patients
This retrospective observational study was conducted using data from the C-CAT database (19), a repository of clinical and genomic information on Japanese patients with cancer who underwent genomic profiling tests as part of the insurance system. This study focused on patients with pancreatic cancer who underwent Foundation One® CDx (F1CDx) testing (Foundation Medicine Inc., Cambridge, USA) (20). All reported pathogenic and likely pathogenic mutations met the quality control criteria defined by Foundation Medicine. The limit of detection (LOD) of F1CDx varies depending on the type of variant. The LOD ranges from 1.8 to 7.9% for base substitutions and from 7.1 to 11.7% for insertions and deletions (indels) (20, 21). All patients registered between June 2019 and December 2021 were included whose clinical and genomic data available at the time of the data update in April 2022. PDAC patients were identified as those documented with pancreatic adenocarcinoma according to the OncoTree cancer classification platform (November 2, 2021), within the C-CAT database (22). Briefly, the C-CAT provides the following information: age, sex, Eastern Cooperative Oncology Group performance status (ECOG PS), smoking history, drinking habit, cancer type, pathological diagnosis, site of metastasis, site of the specimen used for genomic testing, type of specimen used for genomic testing, date of death, last confirmed date of survival, and information related to chemotherapy (e.g., regimens, first and last date of administration, best response, date of progression, and serious adverse events). Genomic information provided by C-CAT includes genetic alterations detected, allele frequencies, copy number (CN), and pathogenicity.
Outcomes
Clinical and genomic data were extracted on January 2024. HRR-altered PDAC patients was identified as those with pathogenic or likely pathogenic alterations in the following genes: ATM, BAP1, BARD1, BLM, BRCA1, BRCA2, BRIP1, CHEK2, FAM175A, FANCA, FANCC, NBN, PALB2, RAD50, RAD51, RAD51C, and RTELI. The prevalence of major pathogenic alterations in the frequently mutated genes KRAS, TP53, SMAD4, and CDKN2A, referred to as the “Big 4” driver mutations, was then compared between HRR-altered and HRR-WT PDAC patients. The prevalences of mutations and CN loss were analyzed separately. Patients with mutations and CN loss in the same gene (one patient with CDKN2A mutations and two patients with SMAD4 mutations) were excluded. Treatment outcomes were evaluated in patients who received the folinic acid, fluorouracil, irinotecan hydrochloride, and oxaliplatin (FFX) regimen including modified FFX or the gemcitabine and nab-paclitaxel (GA) regimen in the first-line setting. Patients who received perioperative chemotherapy were excluded. Overall survival (OS) and time to treatment failure (TTF) were analyzed. OS was defined as the duration (days) from the date of first-line chemotherapy initiation for locally advanced or metastatic PDAC to the date of death or the last confirmed date of survival. TTF was defined as the duration from the start of treatment to the date of treatment discontinuation or death from any cause. Progression free survival, overall response rate, and adverse events were not evaluated in this study because relevant data were not available in the C-CAT registry.
Statistical analysis
The prognostic significance of HRR gene alteration status was evaluated with respect to OS and TTF. The HRR-altered group was compared to the HRR wild-type (HRR-WT) group using Kaplan–Meier method with the log-rank test. Additionally, interactions between TP53, CDKN2A, KRAS and SMAD4 alteration status and OS were analyzed within each HRR group. Univariate and multivariate analyses were performed by using the Cox proportional hazard model. The prevalence of these genetic alterations within the two HRR groups was summarized, and differences between the groups were calculated. We reported p values in addition to confidence intervals and statistical significance was set at p < 0.05; however, there were not to be interpreted as hypothesis tests. The results should be interpreted with caution.
All statistical analyses were performed using EZR version 4.3.1 (Saitama Medical Center, Jichi Medical University, Saitama, Japan) (23), and using GraphPad Prism version 10.2.2 for Windows (GraphPad Software, Boston, Massachusetts USA, www.graphpad.com).
Results
Clinical and molecular characteristics based on HRR gene alteration status
Among the 2,381 patients with PDAC, 274 patients (11.5%) had HRR alteration (Figure 1). The most frequently altered HRR genes were ATM (n = 87, 32%) and BRCA2 (n = 87, 32%), followed by PALB2 (n = 40, 15%) (Supplementary Figure 1). The clinical characteristics of the patients in the HRR-altered and HRR-WT groups are shown in Table 1. The median patient age was 65 years (range, 30–83 years) in the HRR-altered group and 67 years (range, 26–88 years) in the HRR-WT group. The median (range) ECOG PS was 0 (0–3) in the HRR-altered group and 0 (0–3) in the HRR-WT group. Sex distribution and the number of metastatic sites were comparable between the two groups. With respect to the prevalence of gene alterations, KRAS mutations (77% vs. 97%, p < 0.001), TP53 mutations (48% vs. 79%, p < 0.001), and CDKN2A mutations (10% vs. 19%, p < 0.001) were significantly less prevalent in the HRR-altered group than in the HRR-WT group (Table 2).
Figure 1

CONSORT diagram of the study.
Table 1
| Characteristic | HRR-altered group | HRR-WT group |
|---|---|---|
| n = 274 | n = 2,107 | |
| Age at registration, years | ||
| median (range) | 65 (30–83) | 67 (26–88) |
| Sex, n (%) | ||
| Female | 124 (45) | 933 (44) |
| Male | 150 (55) | 1,174 (56) |
| Unknown | 0 (0) | 0 (0) |
| Smoking, n (%) | ||
| Yes | 127 (46) | 1,001 (48) |
| No | 132 (48) | 1,014 (48) |
| Unknown | 15 (5) | 92 (4) |
| Heavy alcohol consumption, n (%) | ||
| Yes | 35 (13) | 253 (12) |
| No | 206 (75) | 1,638 (78) |
| Unknown | 33 (12) | 216 (10) |
| ECOG PS, n (%) | ||
| 0–1 | 259 (95) | 2,006 (95) |
| ≥ 2 | 6 (2) | 38 (2) |
| Unknown | 9 (3) | 63 (3) |
| Metastatic sites, n (%) | ||
| 1 | 143 (52) | 1,165 (55) |
| ≥ 2 | 100 (36) | 689 (33) |
| Unknown | 31 (11) | 253 (12) |
| Sampling methods, n (%) | ||
| Surgery | 137 (50) | 1,146 (54) |
| Biopsy | 135 (49) | 936 (44) |
| Unknown | 2 (1) | 25 (1) |
Patient characteristics by HRR alteration status.
HRR, homologous recombination DNA repair; WT, wild type; ECOG PS, Eastern Cooperative Oncology Group performance status.
Table 2
| Gene alteration | Status | HRR-altered group (n = 274) | HRR-WT group (n = 2,107) | Percentage difference (%) | p Value |
|---|---|---|---|---|---|
| KRAS mutation, n (%) | + | 212 (77) | 2,039 (97) | 20 | < 0.001 |
| − | 62 (23) | 68 (3) | − | ||
| TP53 mutation, n (%) | + | 132 (48) | 1,656 (79) | 31 | < 0.001 |
| − | 142 (52) | 451 (21) | − | ||
| CDKN2A mutation, n (%) | + | 28 (10) | 410 (19) | 9 | < 0.001 |
| − | 246 (90) | 1,697 (81) | − | ||
| CDKN2A Loss, n (%) | + | 84 (31) | 684 (32) | 1 | 0.583 |
| − | 190 (69) | 1,423 (68) | − | ||
| SMAD4 mutation, n (%) | + | 54 (20) | 421 (20) | 0 | 1.000 |
| − | 220 (80) | 1,686 (80) | − | ||
| SMAD4 Loss, n (%) | + | 13 (5) | 166 (8) | 3 | 0.085 |
| − | 261 (95) | 1,941 (92) | − |
Prevalence of KRAS, TP53, SMAD4, and CDKN2A gene alterations based on HRR gene alteration status.
HRR, homologous recombination DNA repair; WT, wild type.
Fisher’s exact test was used to compare the proportions between the groups.
Clinical outcomes according to HRR gene alteration status
A total of 976 patients who were treated with chemotherapy only for palliative intent and had available survival data were included in the survival analysis (Figure 1). The clinical characteristics of the patients are shown in Supplementary Table 1. In total, 113 and 863 patients had HRR-altered and HRR-WT PDAC. Patients with HRR-altered PDAC tend to have a better prognosis and obtain greater survival benefits from platinum-based agents than HRR-WT patients. Therefore, we analyzed survival outcomes based on the HRR gene alteration status. The results showed that OS was significantly better in the HRR-altered group than in the HRR-WT group (median OS: 24.7 months [95% CI: 20.5–33.6 months] vs. 19.2 months [95% CI: 17.7–20.8 months], p = 0.002; Figure 2).
Figure 2

Kaplan–Meier analysis of overall survival (OS) based on homologous recombination DNA repair (HRR) gene alteration status.
Meanwhile, 308 patients who received FFX and 472 patients who received GA as the first-line therapy were included in the comparison of TTF and OS between the HRR-altered and HRR-WT groups. The clinical characteristics of the patients are shown in Supplementary Table 2. In patients treated with FFX as the first-line therapy, TTF was significantly prolonged in the HRR-altered group (median TTF: 7.8 months [95% CI: 5.5–9.6 months] vs. 5.4 months [95% CI: 4.3–5.8 months], p = 0.012; Supplementary Figure 2A). Although OS was not significantly different, there was a favorable trend in the HRR-altered group (median OS: 24.3 months [95% CI: 13.5–36.1 months] vs. 16.8 months [95% CI: 15.5–19.5 months], p = 0.100; Supplementary Figure 2B). In patients treated with GA as the first-line therapy, both TTF (median: 5.4 months [95% CI: 4.4–6.9 months] vs. 5.2 months [95% CI: 4.7–6.0 months], p = 0.859) and OS (median: 24.5 months [95% CI: 17.1–38.8 months] vs. 20.8 months [95% CI: 18.8–22.9 months], p = 0.094) were comparable according to the HRR gene alteration status (Supplementary Figures 3A,B).
Distinct impact of KRAS, TP53, SMAD4, and CDKN2A gene alterations by HRR alteration status
We investigated the impact of KRAS, TP53, SMAD4, and CDKN2A alterations on the survival of HRR-altered and HRR-WT PDAC, respectively.
Patients with TP53 mutation had significantly worse prognosis in both the HRR-altered (median OS: 17.1 months [95% CI: 13.6–24.5 months] vs. 34.0 months [95% CI: 24.7–39.0 months], p < 0.001) and HRR-WT (median OS: 17.6 months [95% CI: 16.3–19.2 months] vs. 24.3 months [95% CI: 21.2–30.1 months], p < 0.001) groups (Figures 3A,B). In the HRR-altered group, patients with CDKN2A alteration tended to have a poor OS compared to CDKN2A WT patients (median OS: 21.4 months [95% CI: 13.6–35.7 months] vs. 28.9 months [95% CI: 20.7–33.7 months], p = 0.095). In the HRR-WT group, patients with CDKN2A alteration had significantly shorter OS compared to patients with CDKN2A WT (median OS: 17.6 months [95% CI: 15.9–19.2 months] vs. 22.4 months [95% CI: 19.7–25.8 months], p < 0.001; Figures 4A,B).
Figure 3

Kaplan–Meier analysis of overall survival (OS) by TP53 mutation status in PDAC patients. Data for the homologous recombination DNA repair (HRR)-altered group (A) and the HRR wild-type (WT) group (B) are shown.
Figure 4

Kaplan–Meier analysis of overall survival (OS) by CDKN2A alteration status in PDAC patients. Data for the homologous recombination DNA repair (HRR)-altered group (A) and the HRR wild-type (WT) group (B) are shown.
Regarding the presence or absence of KRAS mutations, OS was not significantly different between the HRR-altered (median OS: 24.5 months [95% CI: 16.6–34.0 months] vs. 24.7 months [95% CI: 17.8–49.8 months], p = 0.878) and HRR-WT groups (median OS: 19.2 months [95% CI: 17.6–20.8 months] vs. 19.7 months [95% CI: 14.4–26.4 months], p = 0.715) (Figures 5A,B).
Figure 5
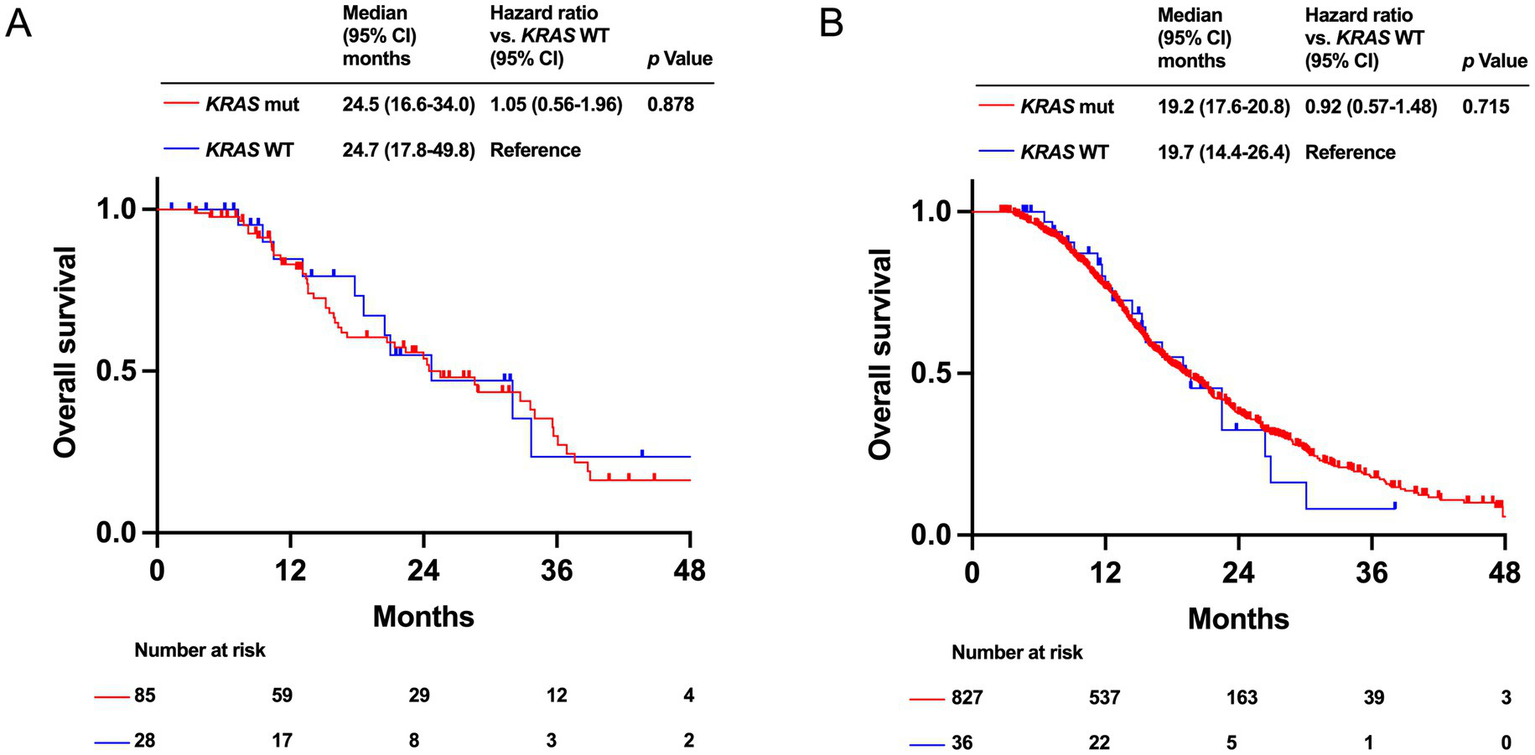
Kaplan–Meier analysis of overall survival (OS) by KRAS mutation status in PDAC patients with homologous recombination DNA repair (HRR) alteration group (A) and with HRR wild-type (WT) group (B).
Patients with SMAD4 alteration had a significantly better OS in the HRR-altered group (median OS: 36.4 months [95% CI: 14.1-NA months] vs. 24.0 months [95% CI: 17.1–32.0 months], p = 0.025). In the HRR-WT group, there is no significantly difference in OS between SMAD4 alteration and SMAD4 WT (median OS: 19.2 months [95% CI: 16.2–21.3 months] vs. 22.4 months [95% CI: 19.7–25.8 months], p = 0.912; Figures 6A,B).
Figure 6

Kaplan–Meier analysis of overall survival (OS) by SMAD4 alteration status in PDAC patients. Data for the homologous recombination DNA repair (HRR)-altered group (A) and the HRR-wild type (WT) group (B) are shown.
Finally, we examined the prognostic factors associated with OS using univariate and multivariate analyses for the HRR-altered and HRR-WT groups, respectively (Supplementary Tables 3, 4). Therefore, these analyses were conducted including age, sex, and coexisting gene alterations. In HRR-altered group, TP53 mutation was identified as a statistically significant independent predictor of OS (HR = 2.91; 95% CI 1.64–5.16, p < 0.001), while SMAD4 alteration was associated with better OS (HR = 0.42; 95% CI 0.20–0.89, p = 0.023). In HRR WT group, TP53 mutation (HR = 1.51; 95% CI 1.19–1.91, p < 0.001) and CDKN2A alteration (HR = 1.41; 95% CI 1.18–1.70, p < 0.001) were identified as a statistically significant independent predictor of OS.
Discussion
The current study showed the prognostic significance of HRR status in a large Japanese PDAC cohort, showing that PDAC patients with HRR alteration had significantly longer TTF with the first-line FFX than PDAC patients with HRR-WT. In contrast, no survival benefit was observed with GA. Also, our study demonstrated the prognostic impact of coexisting KRAS, TP53, CDKN2A, and SMAD4 alterations according to HRR status in PDAC.
Our results underscore previous findings of a correlation between HRR gene alterations and favorable prognosis, as well as the clinical benefit of platinum agents in PDAC (13, 17). Preclinical research has indicated that HRR gene-altered tumor cells have defective DNA repair, making them more vulnerable to platinum-induced DNA damage (24). The GENERATE trial demonstrated the superior survival benefit of GA over mFFX as a frontline treatment for locally advanced or metastatic PDAC patients in Japan (25). Therefore, GA is the preferred first-line treatment in Japan. The combination of irinotecan liposomes and fluoropyrimidine is preferred over mFFX following GA (26), limiting the subsequent use of mFFX and reduces platinum exposure throughout the clinical course. Although NGS is not currently recommended in the management guidelines for patients with PDAC (27), our findings highlight the importance of NGS in identifying patients with HRR-altered PDAC who are likely to benefit from platinum agents.
The current study demonstrated that KRAS, TP53, and CDKN2A mutations were significantly less prevalent in HRR-altered patients than in HRR-WT patients, consistent with previous findings (17, 18). The exact reasons for the differences in these gene mutations between HRR-altered and HRR-WT PDAC remain unclear. However, considering the carcinogenesis in BRCA1/2-mutated tumors (28), the development of HRR-altered PDAC may be different from the stepwise mutations of KRAS, CDKN2A, TP53, and SMAD4 typically observed in general PDAC (29).
Although alterations in TP53, CDKN2A, KRAS, and SMAD4 affect the prognosis of PDAC (3–6, 30), the influence of these gene alterations on the prognosis of HRR-altered PDAC has not been previously reported. To our knowledge, this study is the first to report on the prognostic impact of these gene alterations according to HRR alteration status in PDAC. The results demonstrated that TP53 mutation was a poor prognostic factor, regardless of the HRR gene alteration status. Tumors harboring TP53 mutations have higher genomic instability, resulting in the evasion of apoptosis, plasticity, and reduced lethality (31). Although TP53 mutation is a well-known negative prognostic marker in PDAC (4, 6), our findings further confirm that is also associated with poor outcomes even in HRR-altered PDAC.
In the HRR-WT group, patients with CDKN2A alteration had significantly shorter OS than those with CDKN2A WT. In the HRR-altered group, patients with CDKN2A altered group had a trend toward worse prognosis than those with CDKN2A WT. These results were consistent with the previous studies (30, 32), while we indicated that CDKN2A alteration was a poor prognostic factor regardless of HRR alteration status in PDAC for the first time. The difference in prognosis between HRR-altered and WT group is probably due to the limited number of HRR-altered PDAC patients with CDKN2A alterations, despite our cohort being the largest in Japan, remains a challenge for this analysis. Understanding the status of CDKN2A alteration is important as CDKN2A loss frequently accompanies methylthioadenosine phosphorylase (MTAP) deletion (33) that is the target of AMG193, a protein arginine methyltransferase 5 (PRMT5) inhibitor (34).
The impact of KRAS mutation on survival outcomes has been inconsistent in previous studies (35, 36). However, our analysis found no significant differences in OS between patients with and without KRAS mutations in both the HRR-altered and WT groups. Although the prevalence of KRAS mutations was significantly lower in the HRR-altered group than in the HRR-WT group, it remains relatively high. As the development of pan-KRAS inhibitors (37), represents promising strategies to improve the prognosis of patients with PDAC, it is important to understand the presence and absence of KRAS mutations.
In the HRR-altered group, patients with SMAD4 alteration had a significantly longer OS than those of SMAD4 WT. Meanwhile, in the HRR-WT group, there was no significantly difference on survival between SMAD4 altered and SMAD4 WT groups. The impact of SMAD4 alteration on survival outcomes has been obscure in previous study (5, 6). Our finding suggests a possibility that SMAD4 alteration had a different prognostic impact according to HRR alteration status, while it is difficult to explain the reason at this point.
This study has some limitations. First, we could not differentiate between germline and somatic HRR gene alterations, and homologous recombination deficiency (HRD) status was not incorporated into the analysis as it was not reported by in the F1CDx testing. The HRD score, which serves as a genomic scar score and predicts treatment response to platinum agents in advanced PDAC, is generally higher in tumors with germline HRR alterations than in those with somatic alterations (38). Second, clinical data in the C-CAT database are limited with respect to both quantity and quality, with a notable number of missing entries. Thus, we were unable to perform propensity score matching to adjust for detailed patient characteristics when comparing the survival outcomes.
In conclusion, HRR gene alteration is a favorable prognostic factor and provides clinical benefit from platinum-based chemotherapy in PDAC. HRR-altered PDAC has a distinct molecular profile. Additionally, coexisting gene alterations affect the prognosis in both HRR-altered and HRR-WT PDAC patients. Collectively, these results emphasize the practical value of routine comprehensive genomic profiling to predict prognosis and guide treatment decisions in patients with PDAC.
Statements
Data availability statement
The data analyzed in this study is subject to the following licenses/restrictions: data are available upon reasonable request. Requests to access these datasets should be directed to Yusuke Kawanaka, 1954c0@med.kindai.ac.jp.
Ethics statement
The studies involving humans were approved by Medical Ethics Review Committee of Kindai University of Medicine and C-CAT review board. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
YK: Data curation, Formal analysis, Writing – original draft, Writing – review & editing. CI: Conceptualization, Writing – original draft, Writing – review & editing, Data curation, Formal analysis. MO: Writing – review & editing. SM: Writing – review & editing. TT: Conceptualization, Data curation, Writing – review & editing, Formal analysis. KY: Writing – review & editing. YC: Writing – review & editing, Methodology. KN: Writing – review & editing. HK: Writing – review & editing. HH: Supervision, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
We express our gratitude to the members of the Medical Oncology Department at Kindai University for critically reviewing our study. We would also like to thank C-CAT for their continuous efforts in data management and for providing the clinical and genomic data. We would like to thank Editage (www.editage.jp) for English language editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2025.1570731/full#supplementary-material
References
1.
Bray F Laversanne M Sung H Ferlay J Siegel RL Soerjomataram I et al . Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2024) 74:229–63. doi: 10.3322/caac.21834
2.
Rawla P Sunkara T Gaduputi V . Epidemiology of pancreatic Cancer: global trends, etiology and risk factors. World J Oncol. (2019) 10:10–27. doi: 10.14740/wjon1166
3.
Yousef A Yousef M Chowdhury S Abdilleh K Knafl M Edelkamp P et al . Impact of KRAS mutations and co-mutations on clinical outcomes in pancreatic ductal adenocarcinoma. NPJ Precis Oncol. (2024) 8:27. doi: 10.1038/s41698-024-00505-0
4.
Pan M Jiang C Zhang Z Achacoso N Alexeeff S Solorzano AV et al . TP53 gain-of-function and non-gain-of-function mutations are associated with differential prognosis in advanced pancreatic ductal adenocarcinoma. JCO Precis Oncol. (2023) 7:e2200570. doi: 10.1200/PO.22.00570
5.
Blackford A Serrano OK Wolfgang CL Parmigiani G Jones S Zhang X et al . Smad4 gene mutations are associated with poor prognosis in pancreatic cancer. Clin Cancer Res. (2009) 15:4674–9. doi: 10.1158/1078-0432.CCR-09-0227
6.
McIntyre CA Lawrence SA Richards AL Chou JF Wong W Capanu M et al . Alterations in driver genes are predictive of survival in patients with resected pancreatic ductal adenocarcinoma. Cancer. (2020) 126:3939–49. doi: 10.1002/cncr.33038
7.
Wei D Wang L Zuo X Maitra A Bresalier RS . A small molecule with big impact: MRTX1133 targets the KRASG12D mutation in pancreatic Cancer. Clin Cancer Res. (2024) 30:655–62. doi: 10.1158/1078-0432.CCR-23-2098
8.
Fan D Cao Y Cao M Wang Y Cao Y Gong T . Nanomedicine in cancer therapy. Signal Transduct Target Ther. (2023) 8:293. doi: 10.1038/s41392-023-01536-y
9.
Yang J Wang QL Wang GN Ye JC Li ZQ Wang JY et al . A pan-KRAS degrader for the treatment of KRAS-mutant cancers. Cell Discov. (2024) 10:70. doi: 10.1038/s41421-024-00699-4
10.
Singhi AD George B Greenbowe JR Chung J Suh J Maitra A et al . Real-time targeted genome profile analysis of pancreatic ductal adenocarcinomas identifies genetic alterations that might be targeted with existing drugs or used as biomarkers. Gastroenterology. (2019) 156:2242–53.e4. doi: 10.1053/j.gastro.2019.02.037
11.
Pishvaian MJ Blais EM Brody JR Rahib L Lyons E Arbeloa PD et al . Outcomes in patients with pancreatic adenocarcinoma with genetic mutations in DNA damage response pathways: results from the know your tumor program. JCO Precis Oncol. (2019) 3:1–10. doi: 10.1200/PO.19.00115
12.
Yamai T Ikezawa K Sugimoto N Urabe M Kai Y Takada R et al . Utility of comprehensive genomic profiling tests for patients with incurable pancreatic Cancer in clinical practice. Cancers. (2023) 15:970. doi: 10.3390/cancers15030970
13.
Yadav S Kasi PM Bamlet WR Ho TP Polley EC Hu C et al . Effect of germline mutations in homologous recombination repair genes on overall survival of patients with pancreatic adenocarcinoma. Clin Cancer Res. (2020) 26:6505–12. doi: 10.1158/1078-0432.CCR-20-1788
14.
Kondo T Kanai M Kou T Sakuma T Mochizuki H Kamada M et al . Association between homologous recombination repair gene mutations and response to oxaliplatin in pancreatic cancer. Oncotarget. (2018) 9:19817–25. doi: 10.18632/oncotarget.24865
15.
Tan E Hicks JK Blue K Kim RD Carballido EM Kim DW . Efficacy of olaparib therapy in metastatic pancreatic ductal adenocarcinoma (PDAC) with homologous recombination deficiency (HRD). J Clin Oncol. (2021) 39:e16266. doi: 10.1200/JCO.2021.39.15_suppl.e16266
16.
Kindler HL Hammel P Reni M Van Cutsem E Macarulla T Hall MJ et al . Overall survival results from the POLO trial: a phase III study of active maintenance Olaparib versus placebo for germline BRCA-mutated metastatic pancreatic Cancer. J Clin Oncol. (2022) 40:3929–39. doi: 10.1200/JCO.21.01604
17.
Chen KT Madison R Moore J Jin D Fleischmann Z Newberg J et al . A novel HRD signature is predictive of FOLFIRINOX benefit in metastatic pancreatic Cancer. Oncologist. (2023) 28:691–8. doi: 10.1093/oncolo/oyad178
18.
Golan T O'Kane GM Denroche RE Raitses-Gurevich M Grant RC Holter S et al . Genomic features and classification of homologous recombination deficient pancreatic ductal adenocarcinoma. Gastroenterology. (2021) 160:2119–2132.e9. doi: 10.1053/j.gastro.2021.01.220
19.
Kohno T Kato M Kohsaka S Sudo T Tamai I Shiraishi Y et al . C-CAT: the National Datacenter for Cancer genomic medicine in Japan. Cancer Discov. (2022) 12:2509–15. doi: 10.1158/2159-8290.CD-22-0417
20.
Milbury CA Creeden J Yip WK Smith DL Pattani V Maxwell K et al . Clinical and analytical validation of FoundationOne®CDx, a comprehensive genomic profiling assay for solid tumors. PLoS One. (2022) 17:e0264138. doi: 10.1371/journal.pone.0264138
21.
Woodhouse R Li M Hughes J Delfosse D Skoletsky J et al . Clinical and analytical validation of FoundationOne liquid CDx, a novel 324-gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS One. (2020) 15:e0237802. doi: 10.1371/journal.pone.0237802
22.
Kundra R Zhang H Sheridan R Sirintrapun SJ Wang A Ochoa A et al . OncoTree: a Cancer classification system for precision oncology. JCO Clin Cancer Inform. (2021) 5:221–30. doi: 10.1200/CCI.20.00108
23.
Kanda Y . Investigation of the freely available easy-to-use software 'EZR' for medical statistics. Bone Marrow Transplant. (2013) 48:452–8. doi: 10.1038/bmt.2012.244
24.
Turner N Tutt A Ashworth A . Hallmarks of 'BRCAness' in sporadic cancers. Nat Rev Cancer. (2004) 4:814–9. doi: 10.1038/nrc1457
25.
Ohba A Ozaka M Ogawa G Okusaka T Kobayashi S Yamashita T et al . 1616O nab-paclitaxel plus gemcitabine versus modified FOLFIRINOX or S-IROX in metastatic or recurrent pancreatic cancer (JCOG1611, GENERATE): a multicentred, randomized, open-label, three-arm, phase II/III trial. Ann Oncol. (2023) 34:S894. doi: 10.1016/j.annonc.2023.09.2565
26.
Wang-Gillam A Li C-P Bodoky G Dean A Shan Y-S Jameson G et al . Nanoliposomal irinotecan with fluorouracil and folinic acid in metastatic pancreatic cancer after previous gemcitabine-based therapy (NAPOLI-1): a global, randomised, open-label, phase 3 trial. Lancet. (2016) 387:545–57. doi: 10.1016/S0140-6736(15)00986-1
27.
Mosele MF Westphalen CB Stenzinger A Barlesi F Bayle A Bièche I et al . Recommendations for the use of next-generation sequencing (NGS) for patients with advanced cancer in 2024: a report from the ESMO precision medicine working group. Ann Oncol. (2024) 35:588–606. doi: 10.1016/j.annonc.2024.04.005
28.
Roy R Chun J Powell SN . BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer. (2011) 12:68–78. doi: 10.1038/nrc3181
29.
Pelosi E Castelli G Testa U . Pancreatic Cancer: molecular characterization, clonal evolution and Cancer stem cells. Biomedicine. (2017) 5:65. doi: 10.3390/biomedicines5040065
30.
Doyle A Kubler M Harris A López A Govindaraj P Prins P et al . The impact of CDKN2A mutations on overall survival in pancreatic adenocarcinoma. J Clin Oncol. (2019) 37:278–8. doi: 10.1200/JCO.2019.37.4_suppl.278
31.
Amelio I Melino G . Context is everything: extrinsic signalling and gain-of-function p53 mutants. Cell Death Discov. (2020) 6:16. doi: 10.1038/s41420-020-0251-x
32.
Lin JC Liu TP Yang PM . CDKN2A-inactivated pancreatic ductal adenocarcinoma exhibits therapeutic sensitivity to paclitaxel: a bioinformatics study. J Clin Med. (2020) 9:4019. doi: 10.3390/jcm9124019
33.
Han G Yang G Hao D Lu Y Thein K Simpson BS et al . 9p21 loss confers a cold tumor immune microenvironment and primary resistance to immune checkpoint therapy. Nat Commun. (2021) 12:5606. doi: 10.1038/s41467-021-25894-9
34.
Rodon J Prenen H Sacher A Villalona-Calero M Penel N El Helali A et al . First-in-human study of AMG 193, an MTA-cooperative PRMT5 inhibitor, in patients with MTAP-deleted solid tumors: results from phase I dose exploration. Ann Oncol. (2024) 35:1138–47. doi: 10.1016/j.annonc.2024.08.2339
35.
Bournet B Muscari F Buscail C Assenat E Barthet M Hammel P et al . KRAS G12D mutation subtype is a prognostic factor for advanced pancreatic adenocarcinoma. Clin Transl Gastroenterol. (2016) 7:e157. doi: 10.1038/ctg.2016.18
36.
Kim ST Lim DH Jang KT Lim TK Lee J Choi YL et al . Impact of KRAS mutations on clinical outcomes in pancreatic cancer patients treated with first-line gemcitabine-based chemotherapy. Mol Cancer Ther. (2011) 10:1993–9. doi: 10.1158/1535-7163.MCT-11-0269
37.
Jiang J Jiang L Maldonato BJ Wang Y Holderfield M Aronchik I et al . Translational and therapeutic evaluation of RAS-GTP inhibition by RMC-6236 in RAS-driven cancers. Cancer Discov. (2024) 14:994–1017. doi: 10.1158/2159-8290.CD-24-0027
38.
Qing T Wang X Jun T Ding L Pusztai L Huang K-L . Genomic determinants of homologous recombination deficiency across human cancers. Cancer. (2021) 13:4572. doi: 10.3390/cancers13184572
Summary
Keywords
homologous recombination DNA repair, pancreatic ductal adenocarcinoma, comprehensive genomic profiling, real-world data, chemotherapy
Citation
Kawanaka Y, Inagaki C, Okura M, Mitani S, Takahama T, Yonesaka K, Chiba Y, Nakagawa K, Kawakami H and Hayashi H (2025) Prognostic impact of gene alterations via homologous recombination DNA repair gene alteration status in pancreatic ductal adenocarcinoma. Front. Med. 12:1570731. doi: 10.3389/fmed.2025.1570731
Received
08 February 2025
Accepted
30 July 2025
Published
25 August 2025
Volume
12 - 2025
Edited by
Alessio G. Morganti, University of Bologna, Italy
Reviewed by
Jiayue Qin, Acornmed Biotechnology Co., Ltd., China
Paula Dobosz, Poznan University of Medical Sciences, Poland
Updates
Copyright
© 2025 Kawanaka, Inagaki, Okura, Mitani, Takahama, Yonesaka, Chiba, Nakagawa, Kawakami and Hayashi.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chiaki Inagaki, Chiaki_inagaki@med.kindai.ac.jp
†These authors have contributed equally to this work and share first authorship
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.