Abstract
Purpose:
Burkholderia multivorans, a Gram-negative bacterium, often infect patients with severe immunocompromised and cystic fibrosis. B. multivorans infection is challenging to treat due to its ability to disrupt the action of multiple antimicrobial agents through intrinsic and acquired resistance mechanisms. A better understanding of the pulmonary microbial spectrum of B. multivorans infection is crucial for the prevention and treatment of B. multivorans.
Case presentation:
This case series reviewed the respiratory microbiome structure and alternations during the treatment of B. multivorans infection through metagenomic next-generation sequencing (mNGS). Analysis of mNGS data of 19 pharyngeal secretion samples collected from the 3 COVID-19 patients at different time points showed that the relative abundance of B. multivorans was fluctuated and eventually increased, indicating the possible development of drug resistance. A total of 40 antibiotic-resistant genes (ARGs) were detected. Significantly, the levels of CEOA, CEOB, and OPCM were consistent with the trends in the relative abundance of B. multivorans. Besides, we described nine previously uncharacterized non-synonymous mutations in PenA of B. multivorans. These mutations lead to amino acid changes Thr32Ala, Ala43Ser, Gln105Arg, Asn202Ser, Gln219Arg, Gly241Ala, Val259Ala, Thr279Ala, and Ser298Ile that may associate with resistance to β-lactam antibiotics.
Conclusion:
This report shed light on the importance of rapidly diagnosis and treatment of B. multivorans infection. mNGS serve as a powerful microbial detection tool that provides a comprehensive, sensitive, and rapid method for pathogen detection and drug resistance analysis.
Introduction
Burkholderia multivorans, a member of the Burkholderia cepacia complex (BCC), is an opportunistic pathogen, responsible for severe infections in cystic fibrosis and immunocompromised individuals (1). B. multivorans is associated with “cepacia syndrome”, a rapidly progressive necrotizing pneumonia (2). Infections caused by B. multivorans are often difficult to treat as this pathogen has a large genome (three chromosomes of ∼7.0 Mbp) that carries multiple antibiotic resistance genes, and is inherently resistant to a multitude of antibiotics. A major antibiotic resistance determinant of B. multivorans is an inducible class A β-lactamase of the Pen family, PenA. It possesses a very wide range of substrates, including carbapenems and β-lactamase inhibitors (3). Trimethoprim-sulfamethoxazole, minocycline, doxycycline, doripenem, meropenem, and ceftazidime have recommended as preferred therapies for BCC infections (4). However, resistance to ceftazidime can develop rapidly during treatment of infections caused by B. multivorans. Besides, PenA has a free-moving loop at the entrance to the active site, called the Ω-loop (5). It is a motif composed of sixteen amino acids (residues 164 to 179). The Ω-loop of PenA emerges as a “hot spot” for obtaining single amino acid substitution that extends the substrate heterogeneity of PenA in clinically isolated B. multivorans (3). Substitutions at residues 164, 167, 169, and 179 of PenA were often observed in clinical isolates, conferring resistance to ceftazidime (5).
COVID-19 is a global pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). It has been reported that bacteria and fungi may cause co-infection in critically ill COVID-19 patients, elevating its morbidity and mortality (6). Despite previous studies reporting on the epidemiology and antibiotic resistance profiles of B. multivorans infection (7), the rapid diagnosis and treatment of B. multivorans infection in severe COVID-19 patients are not well-defined. The present study interpreted the results of metagenomic sequencing to determine the respiratory microbiome structure and alternations in 3 COVID-19 patients infected by B. multivorans. Importantly, we described nine previously uncharacterized non-synonymous mutations in PenA of B. multivorans that may associated with resistance to β-lactam antibiotics.
Materials and methods
Three patients with COVID-19 infected by B.multivorans who were hospitalized between February and April 2020 were included. Demographic, laboratory, radiological data and therapeutic management were obtained from the medical records. Pharyngeal secretion samples were collected at different time points during hospitalization according to standard procedures (8) and delivered to the laboratory for metagenomic next-generation sequencing (mNGS) detection within 24 h. DNA was extracted using the MAPMI sample preparation Kit (CapitalBio Corporation, Beijing, China). Sequencing libraries were constructed through enzymatic DNA fragmentation (200–300bp), end repair, adapters ligation, and polymerase chain reaction amplification. Sequencing was performed using the BioelectronSeq 4000 sequencer (CapitalBio, Beijing, China). Quality control was carried out on the original sequencing data, and reads with a length of less than 50 bp, low quality and low complexity were removed. The remaining sequencing data were mapped to the human reference genome GRCh38 to remove the interference of the host sequence by Bowtie2 software (9). The microorganism composition of the samples was annotated, the microbial genomic sequences were assembled using SPAdes, and the abundance of antibiotic-resistant genes (ARGs) was analyzed in combination with the comparison results and the ARGs annotation database. Changes in the pulmonary microbiota and related drug-resistant genes during treatment were analyzed in combination with clinical medication information of the patients. The schedule of pharyngeal secretion samples collection from the same patients at different time points was indicated in Supplementary Table 1. A total of nineteen pharyngeal secretion samples were collected from the three patients. Besides, the antibiotic medications of the patient during hospitalization were summarized in Supplementary Table 2.
Study protocols were reviewed and approved by the Ethics Committee of Peking University First Hospital (approval No. 2020-258), Beijing, China. All procedures followed were in accordance with the ethical standards of the responsible committees and with Declaration of Helsinki. Written informed consent for the publication of case-related details was obtained from each participant or legal guardians.
Results
Their age ranged from 59 to 77 years and two of them were male. Analysis of the mNGS data of nineteen pharyngeal secretion samples collected from three severe COVID-19 patients at different time points showed a higher relative abundance of B. multivorans. Besides, the relative abundance of B. pseudomallei, B. thailandensis, and B. cenocepacia of the same genus was also ranked high across different samples (Figure 1).
FIGURE 1

The abundance of B. multivorans in infected patients. (a) The relative abundance of B. multivorans during the treatment by metagenomic sequencing of patient 1; (b) the relative abundance of B. multivorans during the treatment by metagenomic sequencing of patient 2; (c) the relative abundance of B. multivorans during the treatment by metagenomic sequencing of patient 3.
Patient 1, a 59-year-old man, was admitted for COVID-19 on Feb 4 2020. On the sixth day after hospitalization, he was transferred to the intensive care unit due to respiratory failure and received mechanical ventilation. PCR testing for SARS-CoV-2 became negative on day 17 of admission. Nevertheless, his condition deteriorated tenaciously. Data on metagenomic next-generation sequencing (mNGS) of pharyngeal secretion collected at post-sulfamethoxazole Co. and ceftazidime-avibactam treatment time point showed an increased relative abundance of B. multivorans. Subsequently, the therapeutic regimen was adjusted to amikacin and levofloxacin maintained for 1 week, and ceftazidime maintained for 2 weeks. Sputum culture examination became positive for BCC on march 30, at which time his therapy was switched to levofloxacin and piperacillin-tazobactam according to the results of antimicrobial susceptibility testing of the isolated BCC. There was a fluctuated relative abundance of B. multivorans and eventually increased (Figure 1a). Besides, the changes of inflammatory markers (white blood cell count, neutrophil count, and C-reactive protein) and procalcitonin of the patient during hospitalization were shown in Supplementary Figure 1. On May 18, the patient was discharged.
Patient 2, a 74-year-old woman with a history of hypertension, was admitted to the hospital due to fever, shortness of breath, and respiratory failure. Nasopharyngeal swab PCR tested positive for SARS-CoV-2 and the chest CT showed diffuse lesions in both lungs. The patient was subsequently diagnosed with severe COVID-19. On the third day after admission, she underwent endotracheal intubation and mechanical ventilatory support due to acute respiratory distress syndrome. PCR testing for SARS-CoV-2 became negative after 15 days of hospitalization. On Feb 27, pharyngeal secretion mNGS revealed B. multivorans. One day later, sputum culture was positive for BCC. Subsequently, the patient was successively treated with cefoperazone-sulbactam, ceftazidime-averbactam, amikacin, sulfamethoxazole Co., and ceftazidime (Supplementary Table 2). Supplementary Figure 2 presented the changes of inflammatory markers and procalcitonin. Nevertheless, data on pharyngeal secretion mNGS at different time points showed fluctuated relative abundance of B. multivorans (Figure 1b). Despite broadening antimicrobials, her condition exacerbation and succumbed by severe sepsis on day 78 of admission.
Patient 3, a 77-year-old man, presented with intermittent fever and oxygen saturation of 93%. Throat swab PCR tested positive for SARS-CoV-2. The patient was transferred to intensive care unit due to lung function deterioration and respiratory failure. Sputum samples collected post-meropenem, vancomycin, and caspofungin treatment exhibited culture-positive for BCC. Pharyngeal secretion mNGS test showed B. multivorans, at which time his therapy was switched to piperacillin-tazobactam and maintained for 1 week. Drug susceptibility testing on Mar 7 revealed that clinically isolated BCC was sensitive to ceftazidime (MIC: 2 ug/mL) and ciprofloxacin (MIC: ≤0.25 ug/mL), and was resistant to ampicillin, ampicillin-sulbactam, cefazolin, and imipenem. Ceftazidime-averbactam was administered between the second and third mNGS tests. Then, his therapeutic regimen was switched to ceftazidime and levofloxacin according to the results of pharyngeal secretion mNGS on March 11 (Figure 1c). However, there was no significant improvement after active anti-infective treatment and the patient died 45 days after admission (Supplementary Figure 3 and Supplementary Table 2).
According to the results of the antimicrobial susceptibility testing of clinically isolated B. cepacia and literature reports, the three patients successively treated with sulfamethoxazole Co., ceftazidime, ceftazidime-avibactam, levofloxacin, and other first-line treatment for B. multivorans infection (Figure 1). The relative abundance of B. multivorans with a similar trend were observed, indicating the possible development of drug resistance. Two of the three patients died during hospitalization despite modification of antimicrobial therapy accordingly to the results of mNGS and drug sensitivity tests. They responded poorly to antimicrobial treatment. Therefore, a retrospective analysis of drug resistance in these patients was performed through the mNGS workflow. A total of 40 ARGs were detected, including CEOA, CEOB, OPCM, TETC, TEM, NORM, EFMA, and AMRB (Figure 2). Significantly, the levels of CEOA, CEOB, and OPCM were consistent with the trends in the relative abundance of B. multivorans. CEOA and CEOB were related to drug and biocide resistance, which contains a variety of antibiotic efflux pumps. Moreover, OPCM was associated with multi-drug resistance that resistant to two or more different antibiotics. Resistance nodulation cell division drug efflux pump CeoAB-OpcM has been reported to cause resistance to fluoroquinolones and aminoglycosides (10).
FIGURE 2
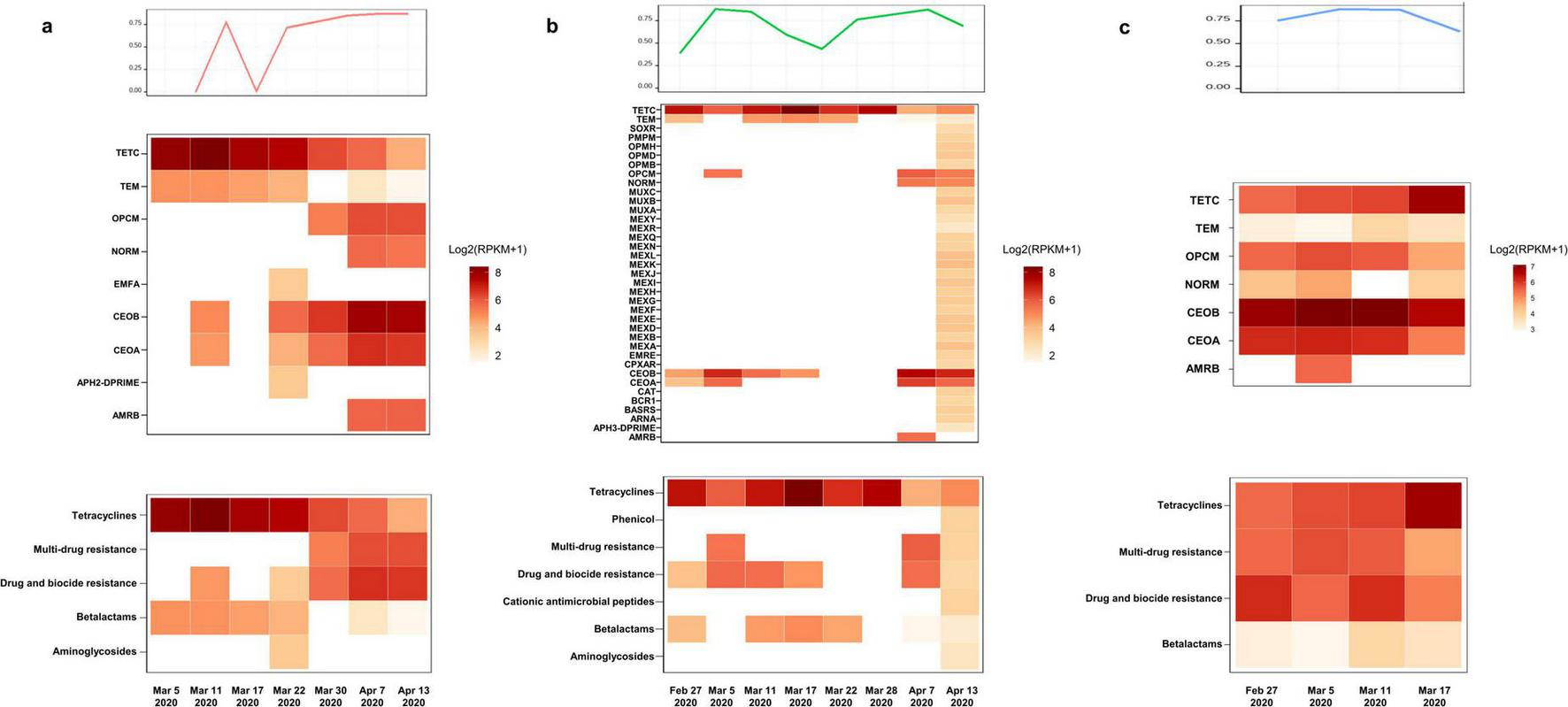
Antibiotic-resistant genes (ARGs) detection through metagenomic sequencing workflow. The levels of CEOA, CEOB, and OPCM were consistent with the trends in the relative abundance of B. multivorans. The related proteins expressed by these genes constitute the CeoAB-OpcM efflux pump, which may cause resistance to fluoroquinolones such as levofloxacin and aminoglycosides such as amikacin. (a) A total of 9 antibiotic-resistant genes (ARGs) were detected in 7 pharyngeal secretion samples collected from patient 1 at different time points through metagenomic sequencing workflow; (b) a total of 37 ARGs were detected in 8 pharyngeal secretion samples collected from patient 2; (c) a total of 7 ARGs were detected in 4 pharyngeal secretion samples collected from patient 3.
Single nucleotide polymorphism (SNP) analysis of PenA through SAMtools showed that the SNP sites of PenA in pharyngeal secretion samples collected from divergent patient at different time points were consistent, prompting that B. multivorans probably originated from the same strain and no new mutations emerged during the treatment period (Figure 3a). Most notably, we identified nine previously uncharacterized non-synonymous mutations in PenA, which codes for a class A β-lactamase and was involved in resistance to β-lactam antibiotics. These mutations lead to amino acid changes Thr32Ala, Ala43Ser, Gln105Arg, Asn202Ser, Gln219Arg, Gly241Ala, Val259Ala, Thr279Ala, and Ser298Ile compared to B. multivorans ATCC 17616, that may associate with the decreased sensitivity of B. multivorans to ceftazidime and ceftazidime-avibactam (Figure 3b).
FIGURE 3

(a) Single nucleotide polymorphism analysis of PenA through SAMtools; (b) nine novel non-synonymous mutations in PenA of B. multivorans.
Discussion
Burkholderia multivorans, a Gram-negative bacterium, is a common etiologic agent causing nosocomial infection and diseases in immunocompromised individuals and cystic fibrosis sufferers (11, 12). A prospective analysis indicated that B. multivorans infection was strongly associated with adverse clinical outcomes. Hanulik et al. indicated that of the 46 patients isolated with B. multivorans, 23.9% cases died, of which 8 deaths were attribute to hospital-acquired pneumonia (13). The present study highlights the extreme complexity of B. multivorans infection in patients with COVID-19 in the intensive care unit. However, the interaction between SARS-CoV-2 and B. multivorans infection is poorly understood. Within the first few days after SARS-CoV-2 infection, critically ill patient with COVID-19 generally develop distorted respiratory tract or an imbalance in the lung flora, which can further progress into secondary bacterial or fungal infection few weeks later (14). Moreover, patients infected by B. multivorans usually exhibit a severe systemic inflammatory response, resulting in deterioration of the condition and triggering critical acute respiratory distress syndrome (2).
Antimicrobial resistance poses a significant threat to global health and is projected to cause 10 million deaths each year by 2050 (15). The resistance of B. multivorans to β -lactam is mainly driven by the generation of β-lactamases, which can hydrolyze β-lactam and prevent them from reaching the targets of penicillin-binding proteins (3). B. multivorans possess two types of chromosomal β-lactamases, PenA and a class C β-lactamases (AmpC). The PenA1 β -lactamase from B. multivorans ATCC 17616 is responsible for extending the spectrum of β -lactam resistance, while AmpC1 from B. multivorans ATCC 17616 has a very narrow spectrum and contributes very little to β -lactam resistance (16). Besides, PenA enzyme is similar to KPC-2, the most clinically important serine carbapenemase (17). Infections caused by B. multivorans is challenging to treat due to its ability to evade the action of multiple antimicrobial agents through intrinsic and acquired resistance mechanisms. On one hand, BCC microorganisms are intrinsically resistant to a variety of antibiotics, including β-lactams, fluoroquinolones, aminoglycosides, and polymyxins (18). Additionally, efflux pumps are crucial players in B. multivorans drug resistance. The synergistic effect between efflux and reduced outer membrane permeability was recognized as a common theme in the increased resistance exhibited by non-fermentative Gram-negative bacteria such as BCC (10). An early report indicated an outer membrane lipoprotein, OpcM, was implicated in resistance to several antibiotics (ciprofloxacin, trimethoprim, and chloramphenicol) (19). Notably, OpcM is the outer membrane channel of the efflux pump of the resistance nodulation cell division family. In the present study, we observed a higher abundance of resistance genes, among which CEOA, CEOB, and OPCM were associated with BCC, which was consistent with the trends in the relative abundance of B. multivorans. The related proteins expressed by these genes constitute the CeoAB-OpcM efflux pump, lead to resistance to fluoroquinolones such as levofloxacin and aminoglycosides such as amikacin (10). Moreover, B. multivorans is resistant to polymyxins. The hpnN gene of B. multivorans encodes the integrated membrane protein of the hpnN transporter family, which is responsible for shuttling hopanoids to the outer membrane (18). Hopanoid biosynthesis is involved in regulating outer membrane stability and permeability, contributing to polymyxin resistance (20). On the other, drug target alternations are key players of drug resistance. The Ω-loop of PenA emerges as a “hot spot” for obtaining single amino acid substitution that extends the substrate heterogeneity of PenA in clinically isolated B. multivorans (3). Substitutions at residues 164, 167, 169, and 179 of PenA were often observed in clinical isolates, conferring resistance to ceftazidime (5).
Although it has been recognized that B. multivorans infection is associated with an accelerated decline in respiratory function and increased morbidity and mortality, there is a lack of evidence-based antibiotic therapy for this infection. Ceftazidime and trimethoprim-sulfamethoxazole are recommended as the preferred treatment for infections caused by B. multivorans. Unfortunately, resistance to ceftazidime is increasing and can develop rapidly during the treatment, that poses a major threat to populations vulnerable to B. multivorans infection (5). Here, we identified nine previously uncharacterized non-synonymous mutations in PenA. These mutations lead to amino acid changes Thr32Ala, Ala43Ser, Gln105Arg, Asn202Ser, Gln219Arg, Gly241Ala, Val259Ala, Thr279Ala, and Ser298Ile, that may associate with the decreased sensitivity of B. multivorans to ceftazidime and ceftazidime-avibactam. As monotherapy may occur drug resistance, some experts would consider initial treatment with multiple antibiotics followed by a combination of two or three drugs depending on the drug susceptibility results. B. multivorans infection might require prolonged antibiotic courses and need to be closely monitored. In addition, bacteriophage (phage) therapy is a promising and potential alternative treatment for Burkholderia infections (21, 22). However, three documented cases of phage therapy for Burkholderia infection were unsuccessful and all three patients died (23–25). The prophages existing in the genomes of the Burkholderia spp. are potential useful starting points for the isolation and development of novel phages for phage therapy (21).
In the present study, we applied mNGS to pharyngeal secretion and the results showed B. multivorans. Subsequent conventional microbiological examination confirmed these results. Generally, mNGS allows for the direct detection of a wide range of pathogens. The wide detection spectrum, fast, and high sensitivity of mNGS are critical in the management of severe infections, where timely diagnosis can significantly impact patient outcomes (26). Generally, mNGS has the potential to sequence antibiotic resistance genes, providing valuable information on resistance mechanisms. This capability is essential to guide treatment regimen and improve antibiotic stewardship by helping to select the most effective antibiotics and preventing the spread of resistance. Moreover, the technology is transforming the landscape of clinical microbiology laboratories by providing a more comprehensive view of the microbial landscape in infections, which can lead to more accurate diagnoses and better patient management. Overall, mNGS is a powerful microbial detection tool that provides a comprehensive, sensitive, and rapid method for pathogen detection and drug resistance analysis, which is critical for improving diagnostic accuracy and guiding clinical decision-making in infectious disease management.
Conclusion
In conclusion, this case series describes the respiratory microbiome alternations and antibiotic-resistant genes during the treatment of B. multivorans infection through mNGS. Additionally, we reported nine previously uncharacterized non-synonymous mutations in PenA of B. multivorans. These mutations lead to amino acid changes Thr32Ala, Ala43Ser, Gln105Arg, Asn202Ser, Gln219Arg, Gly241Ala, Val259Ala, Thr279Ala, and Ser298Ile that may associate with resistance to β-lactam antibiotics. As monotherapy may occur drug resistance, B. multivorans infection should be initially treated with multiple antibiotics, followed by a combination of two or three drugs depending on the drug susceptibility results. Moreover, B. multivorans infection might require prolonged antibiotic courses and need to be closely monitored. Moreover, for critically ill patients or immunocompromised individuals facing difficult or complex infectious diseases, it is recommended to use mNGS to optimize antibiotic treatment strategies, thereby improving therapeutic outcomes and reducing the unnecessary use of antibiotics.
Statements
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by the Ethics Committee of Peking University First Hospital (approval number: 2020-258), Beijing, China. The studies were conducted in accordance with the local legislation and institutional requirements. The participants or their guardian provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) or their guardian for the publication of any potentially identifiable images or data included in this article.
Author contributions
HX: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Resources, Writing – original draft, Writing – review and editing. RZ: Data curation, Investigation, Methodology, Writing – review and editing. XZ: Data curation, Investigation, Methodology, Writing – review and editing. ZZ: Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Supervision, Writing – review and editing. YF: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Supervision, Writing – review and editing. LL: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Supervision, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Key Research and Development Program of China (2020YFC2005401 and 2023YFC0872400) and National High Level Hospital Clinical Research Funding (Scientific Research Fund of Peking University First Hospital) (2024CX14).
Conflict of interest
ZZ was employed by CapitalBio Corporation. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2025.1577363/full#supplementary-material
References
1.
Wang G Zhao J Zhang X Li S Sun C Gu G . Immunological studies of Burkholderia multivorans O-antigen oligosaccharide-rsScpA193 conjugates as potential candidates for vaccine development.Int J Biol Macromol. (2025) 287:138570. 10.1016/j.ijbiomac.2024.138570
2.
Ho SSC Nashid N Waters VJ LiPuma JJ Zlosnik JEA Otley A et al Burkholderia multivorans septicemia in a pediatric liver transplant patient. Am J Transplant. (2019) 19:933–8. 10.1111/ajt.15065
3.
Becka SA Zeiser ET Marshall SH Gatta JA Nguyen K Singh I et al Sequence heterogeneity of the PenA carbapenemase in clinical isolates of Burkholderia multivorans. Diagn Microbiol Infect Dis. (2018) 92:253–8. 10.1016/j.diagmicrobio.2018.06.005
4.
Sfeir MM . Burkholderia cepacia complex infections: More complex than the bacterium name suggest.J Infect. (2018) 77:166–70. 10.1016/j.jinf.2018.07.006
5.
Papp-Wallace KM Becka SA Taracila MA Zeiser ET Gatta JA LiPuma JJ et al Exploring the role of the Ω-loop in the evolution of ceftazidime resistance in the PenA β-lactamase from Burkholderia multivorans, an important cystic fibrosis pathogen. Antimicrob Agents Chemother. (2017) 61:e1941–1916. 10.1128/AAC.01941-16
6.
Yang S Hua M Liu X Du C Pu L Xiang P et al Bacterial and fungal co-infections among COVID-19 patients in intensive care unit. Microbes Infect. (2021) 23:104806. 10.1016/j.micinf.2021.104806
7.
Diaz Caballero J Clark ST Wang PW Donaldson SL Coburn B Tullis DE et al A genome-wide association analysis reveals a potential role for recombination in the evolution of antimicrobial resistance in Burkholderia multivorans. PLoS Pathog. (2018) 14:e1007453. 10.1371/journal.ppat.1007453
8.
Xu Y Jiang Y Wang Y Meng F Qin W Lin Y . Metagenomic next-generation sequencing of bronchoalveolar lavage fluid assists in the diagnosis of pathogens associated with lower respiratory tract infections in children.Front Cell Infect Microbiol. (2023) 13:1220943. 10.3389/fcimb.2023.1220943
9.
Chen H Bai X Gao Y Liu W Yao X Wang J . Profile of bacteria with ARGs among real-world samples from ICU admission patients with pulmonary infection revealed by metagenomic NGS.Infect Drug Resist. (2021) 14:4993–5004. 10.2147/IDR.S335864
10.
Podnecky NL Rhodes KA Schweizer HP . Efflux pump-mediated drug resistance in Burkholderia.Front Microbiol. (2015) 6:305. 10.3389/fmicb.2015.00305
11.
Lood C Peeters C Lamy-Besnier Q Wagemans J De Vos D Proesmans M et al Genomics of an endemic cystic fibrosis Burkholderia multivorans strain reveals low within-patient evolution but high between-patient diversity. PLoS Pathog. (2021) 17:e1009418. 10.1371/journal.ppat.1009418
12.
Doubravská L Htoutou Sedláková M Fišerová K Pudová V Urbánek K Petrželová J et al Bacterial resistance to antibiotics and clonal spread in COVID-19-positive patients on a tertiary hospital intensive care unit, Czech Republic. Antibiotics (Basel). (2022) 11:783. 10.3390/antibiotics11060783
13.
Hanulik V Webber MA Chroma M Uvizl R Holy O Whitehead RN et al An outbreak of Burkholderia multivorans beyond cystic fibrosis patients. J Hosp Infect. (2013) 84:248–51. 10.1016/j.jhin.2013.04.001
14.
Rawson TM Moore LSP Zhu N Ranganathan N Skolimowska K Gilchrist M et al Bacterial and fungal coinfection in individuals with coronavirus: A rapid review to support COVID-19 antimicrobial prescribing. Clin Infect Dis. (2020) 71:2459–68. 10.1093/cid/ciaa530
15.
Odoom A Osman AH Dzuvor CKO . Recent advances in immunotherapeutic and vaccine-based approaches for the treatment of drug-resistant bacterial infections.ACS Infect Dis. (2025). 10.1021/acsinfecdis.5c00001[Epub ahead of print].
16.
Mojica MF Nukaga M Becka SA Zeiser ET Hoshino T LiPuma JJ et al Frameshift mutations in genes encoding PBP3 and PBP4 trigger an unusual, extreme β-lactam resistance phenotype in Burkholderia multivorans. ACS Infect Dis. (2024) 10:3810–20. 10.1021/acsinfecdis.4c00330
17.
Papp-Wallace KM Taracila MA Gatta JA Ohuchi N Bonomo RA Nukaga M . Insights into β-lactamases from Burkholderia species, two phylogenetically related yet distinct resistance determinants.J Biol Chem. (2013) 288:19090–102. 10.1074/jbc.M113.458315
18.
Kumar N Su CC Chou TH Radhakrishnan A Delmar JA Rajashankar KR et al Crystal structures of the Burkholderia multivorans hopanoid transporter HpnN. Proc Natl Acad Sci USA. (2017) 114:6557–62. 10.1073/pnas.1619660114
19.
Burns JL Wadsworth CD Barry JJ Goodall CP . Nucleotide sequence analysis of a gene from Burkholderia (Pseudomonas) cepacia encoding an outer membrane lipoprotein involved in multiple antibiotic resistance.Antimicrob Agents Chemother. (1996) 40:307–13. 10.1128/AAC.40.2.307
20.
Malott RJ Steen-Kinnaird BR Lee TD Speert DP . Identification of hopanoid biosynthesis genes involved in polymyxin resistance in Burkholderia multivorans.Antimicrob Agents Chemother. (2012) 56:464–71. 10.1128/AAC.00602-11
21.
Nordstrom HR Griffith MP Rangachar Srinivasa V Wallace NR Li A Cooper VS et al Harnessing the diversity of Burkholderia spp. prophages for therapeutic potential. Cells. (2024) 13:428. 10.3390/cells13050428
22.
Strathdee SA Hatfull GF Mutalik VK Schooley RT . Phage therapy: From biological mechanisms to future directions.Cell. (2023) 186:17–31. 10.1016/j.cell.2022.11.017
23.
Haidar G Chan BK Cho ST Hughes Kramer K Nordstrom HR Wallace NR et al Phage therapy in a lung transplant recipient with cystic fibrosis infected with multidrug-resistant Burkholderia multivorans. Transplant Infect Dis. (2023) 25:e14041. 10.1111/tid.14041
24.
Aslam S Courtwright AM Koval C Lehman SM Morales S Furr CL et al Early clinical experience of bacteriophage therapy in 3 lung transplant recipients. Am J Transplant. (2019) 19:2631–9. 10.1111/ajt.15503
25.
Smith M. Salt in my soul: An unfinished life, Random House Trade Paperbacks. New York, NY: Random House Publishing Group (2020).
26.
Diao Z Han D Zhang R Li J . Metagenomics next-generation sequencing tests take the stage in the diagnosis of lower respiratory tract infections.J Adv Res. (2022) 38:201–12. 10.1016/j.jare.2021.09.012
Summary
Keywords
B. multivorans , infection, metagenomic sequencing, antibiotic resistance, Burkholderia cepacia complex
Citation
Xu H, Zhang R, Zhang X, Zhang Z, Feng Y and Lin L (2025) Pulmonary microbial spectrum of Burkholderia multivorans infection identified by metagenomic sequencing. Front. Med. 12:1577363. doi: 10.3389/fmed.2025.1577363
Received
15 February 2025
Accepted
21 May 2025
Published
17 June 2025
Volume
12 - 2025
Edited by
Binod Rayamajhee, University of New South Wales, Australia
Reviewed by
Pawan Kumar Kanaujia, Mahayogi Gorakhnath University, India
Wang Zhang, Sir Run Run Shaw Hospital, China
Updates
Copyright
© 2025 Xu, Zhang, Zhang, Zhang, Feng and Lin.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yingmei Feng, yingmeif13@ccmu.edu.cnLianjun Lin, 06474@pkufh.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.