- 1Department of Public Health, College of Applied Medical Sciences, Qassim University, Buraydah, Saudi Arabia
- 2Department of Physiology, Faculty of Medicine, University of Tabuk, Tabuk, Saudi Arabia
- 3Defence Against Weapons of Destruction Department, Armed Forces Medical Services, Riyadh, Saudi Arabia
- 4Department of Basic Oral Sciences and Dental Education, College of Dentistry, Qassim University, Buraydah, Saudi Arabia
- 5Department of Food Service, King Fahad Armed Forces Hospital, Jeddah, Saudi Arabia
- 6Family Medicine Department, King Fahad Armed Hospital, Jeddah, Saudi Arabia
- 7Health Informatics Department, Prince Sultan Military Medical City, Riyadh, Saudi Arabia
- 8Education and Training Department, Prince Sultan Military College of Health Sciences, Dammam, Saudi Arabia
- 9Pharmaceutical Department, Prince Sultan Military Medical City, Riyadh, Saudi Arabia
- 10Department of Public Health, College of Applied Medical Science, King Khalid University, Abha, Saudi Arabia
- 11Department of Veterinary Preventive Medicine, College of Veterinary Medicine, Qassim University, Buraydah, Saudi Arabia
- 12Orthopedic Department, King Abdulaziz Hospital, Jeddah, Saudi Arabia
- 13Department of Pathology and Laboratory Diagnosis, College of Veterinary Medicine, Qassim University, Buraydah, Saudi Arabia
Tuberculosis (TB) remains one of the leading causes of infectious disease mortality worldwide, increasingly complicated by the emergence of drug-resistant strains and limitations in existing diagnostic and therapeutic strategies. Despite decades of global efforts, the disease continues to impose a significant burden, particularly in low- and middle-income countries (LMICs) where health system weaknesses hinder progress. This comprehensive review explores recent advancements in TB diagnostics, antimicrobial resistance (AMR surveillance), treatment strategies, and vaccine development. It critically evaluates cutting-edge technologies including CRISPR-based diagnostics, whole-genome sequencing, and digital adherence tools, alongside therapeutic innovations such as shorter multidrug-resistant TB regimens and host-directed therapies. Special emphasis is placed on the translational gap—highlighting barriers to real-world implementation such as cost, infrastructure, and policy fragmentation. While innovations like the Xpert MTB/RIF Ultra, BPaLM regimen, and next-generation vaccines such as M72/AS01E represent pivotal progress, their deployment remains uneven. Implementation science, cost-effectiveness analyses, and health equity considerations are vital to scaling up these tools. Moreover, the expansion of the TB vaccine pipeline and integration of AI in diagnostics signal a transformative period in TB control. Eliminating TB demands more than biomedical breakthroughs—it requires a unified strategy that aligns innovation with access, equity, and sustainability. By bridging science with implementation, and integrating diagnostics, treatment, and prevention within robust health systems, the global community can accelerate the path toward ending TB.
1 Introduction
Tuberculosis (TB) is a bacterial disease primarily caused by Mycobacterium tuberculosis (M. tuberculosis) and related species (1). It remains a major global health concern due to its high morbidity and mortality rates (2). In 2022, TB was responsible for an estimated 1.3 million deaths, and projections suggest it could cause up to 31 million deaths in the coming years (3). Although TB primarily affects the lungs, it can also involve other organs, resulting in extrapulmonary manifestations (4, 5). Approximately 15–20% of TB cases present as extrapulmonary TB, especially among individuals co-infected with HIV (6). Furthermore, M. tuberculosis has been associated with tumor-like formations, with some studies linking pulmonary TB to lung neoplasms, emphasizing the importance of effective management to reduce this risk (7–12).
Transmission occurs through airborne particles expelled when an infected individual coughs or sneezes, making air a crucial vector for the spread of TB (13). Delayed treatment of active TB significantly increases the risk of transmission (14). Diagnosing TB can be particularly challenging due to misdiagnosis, non-specific symptoms, and limited laboratory capacity in many settings (15). The COVID-19 pandemic has exacerbated these challenges by overwhelming healthcare systems, interrupting TB services, and diverting essential resources. This disruption has led to delays in diagnosis and treatment, contributing to increased transmission and worsened clinical outcomes. Although the Global TB Report highlights partial recovery of TB services, the pandemic's residual impact continues to impede progress (16).
Additionally, the growing incidence of non-tuberculous mycobacterial infections, especially in elderly and immunocompromised individuals, complicates differential diagnosis and clinical management (17). Globally, ~23% of the population (95% CI: 20.4–26.4%), corresponding to 1.7–1.9 billion people, are estimated to harbor latent M. tuberculosis infection (LTBI), rather than active disease (18). In 2022, an estimated 7.5 million new TB cases and 1.5 million deaths were reported (16). Early detection remains vital for initiating timely treatment and curbing disease transmission (7, 19). According to the WHO Global Tuberculosis Report 2024, an estimated 10.8 million people (95% uncertainty interval [UI]: 10.1–11.7 million) developed active TB in 2023, corresponding to 134 new cases per 100,000 population (95% UI: 125–145) (20).
Since 2000, global efforts to combat TB have led to measurable progress. However, these gains were significantly undermined by the COVID-19 pandemic, which disrupted TB services worldwide. The reallocation of healthcare resources to COVID-19, combined with lockdowns and mobility restrictions, caused widespread delays in diagnosis and treatment. Furthermore, ongoing armed conflicts and deteriorating socioeconomic conditions—particularly in high-burden regions—have compounded these difficulties (21, 22). Economic instability, rising living costs, and declining public health funding have disproportionately affected impoverished communities already at heightened risk for TB (23).
According to WHO data, global TB case notifications dropped by 18% in 2020 due to limited access to health services. Although a partial rebound occurred in 2021, it failed to fully address the backlog of missed cases. Countries such as India, Indonesia, and the Philippines recorded the largest declines in notifications, resulting in a significant number of untreated cases and sustained community-level transmission. Between 2019 and 2021, TB-related deaths rose, with 1.6 million deaths reported in 2021 alone-−1.4 million among HIV-negative individuals and 187,000 among those living with HIV. These figures highlight the heightened vulnerability of HIV-positive individuals and the critical importance of uninterrupted TB services.
The pandemic also intensified pre-existing disparities in access to diagnostic and treatment tools. LMICs, which account for nearly 80% of TB cases globally, continue to face substantial challenges in adopting WHO-endorsed technologies (23). In 2022, TB incidence climbed to an estimated 10.6 million new cases—a 4.5% increase from 2020—sustaining the trend sparked by the pandemic. Simultaneously, drug-resistant TB remained a significant challenge, with ~410,000 new cases of rifampicin-resistant or multidrug-resistant TB reported (24). Studies indicate that MDR-TB disproportionately affects regions with low socio-demographic indices, adding complexity to control strategies (25).
Although molecular diagnostics such as GeneXpert MTB/RIF and line probe assays have transformed TB detection, their widespread implementation remains limited in many resource-constrained settings. Key obstacles include underdeveloped laboratory infrastructure, inefficient specimen transport systems, and shortages of trained personnel (26, 27). In contrast, high-income countries benefit from advanced diagnostic networks, enabling timely detection and robust drug resistance profiling that support effective patient care. Despite diagnostic advancements, only about 7.5 million of the estimated 10.6 million TB cases in 2022 were reported, reflecting a diagnostic gap of ~3.1 million cases. This persistent shortfall, worsened by pandemic-related disruptions, continues to delay treatment, perpetuate transmission, and increase mortality.
Effective TB control requires a multipronged approach that integrates conventional methods—such as microscopy and culture—with cutting-edge molecular diagnostics (28, 29). Clinical microbiology laboratories play a central role in diagnosis, and recent genetic technologies now allow for rapid identification of both the pathogen and its resistance profile. In countries where TB testing is predominantly conducted by general health or private sectors, it is essential to assess whether service expansion improves health outcomes or merely increases capacity. Evidence suggests that the quality of TB laboratory services, rather than their independence, is what determines their public health value (29).
Furthermore, distinguishing latent from active TB and ensuring equitable access to diagnostic services remain urgent priorities (30). Reliable drug susceptibility testing and a better understanding of the molecular mechanisms of resistance in M. tuberculosis are also critical. As multidrug-resistant and extensively drug-resistant TB pose growing threats, the need for improved detection and treatment strategies is more pressing than ever. Recent genetic innovations have accelerated the diagnosis of TB and its resistance traits. However, low-resource countries still urgently need affordable, rapid, and accurate diagnostic solutions (31). This review aims to provide a comprehensive overview of both standard and advanced diagnostic methods, while also exploring emerging therapeutic strategies to combat TB, with special emphasis on multidrug-resistant forms of the disease.
2 History and pathogenesis of TB
TB, caused by M. tuberculosis, remains one of the most lethal infectious diseases globally. First identified by Robert Koch in 1882 (32), the bacillus is transmitted via aerosols and primarily infects the lungs (pulmonary TB), although extrapulmonary forms also occur (33, 34). TB's ability to establish latent infection in an estimated two billion people worldwide contributes to its persistence (33). Latent TB infection can reactivate, particularly in immunocompromised individuals, including those with HIV, who have an 18-fold increased risk of disease progression (35, 36).
The infection begins when inhaled bacilli reach the alveoli and are phagocytosed by resident macrophages (37). Instead of being destroyed, M. tuberculosis manipulates the host immune system using the ESX-1 secretion system to release ESAT-6, facilitating escape from the phagosome into the cytoplasm (38, 39). The bacilli avoid degradation, inhibit phagosome maturation, and modulate cytokine responses, thereby creating an environment conducive to survival (40). This early evasion strategy is central to the pathogen's persistence and underlies the challenges in early TB diagnosis.
The host's immune response typically culminates in granuloma formation—a hallmark of TB pathogenesis—where bacilli are sequestered but not eradicated (41). Within these structures, M. tuberculosis can enter a dormant state, resisting immune clearance and pharmacological treatment (42). In individuals with compromised immunity, granulomas can break down, leading to bacterial reactivation, tissue necrosis, and contagious active TB (43, 44). Recent studies highlight that lipid metabolism, particularly the formation of foamy macrophages and caseous necrosis, plays a crucial role in long-term bacterial survival and disease progression (43, 45).
Understanding TB pathogenesis is no longer solely of academic interest—it informs the development of new diagnostic tools and therapeutic strategies. For instance, insights into the ESX-1 system and granuloma biology have prompted the exploration of immunomodulatory therapies and biomarkers for early detection (46). Moreover, caseum-resident bacilli represent a significant challenge to drug penetration, guiding the design of new drug regimens with improved tissue distribution. In summary, TB pathogenesis illustrates a complex interplay between bacterial virulence and host immunity. A deeper mechanistic understanding is essential not only for accurate diagnosis—particularly distinguishing active from latent infection—but also for innovating treatment strategies that can overcome the barriers posed by immune evasion and granuloma-mediated bacterial persistence. The progression and immune interactions in TB pathogenesis are illustrated in Figure 1.
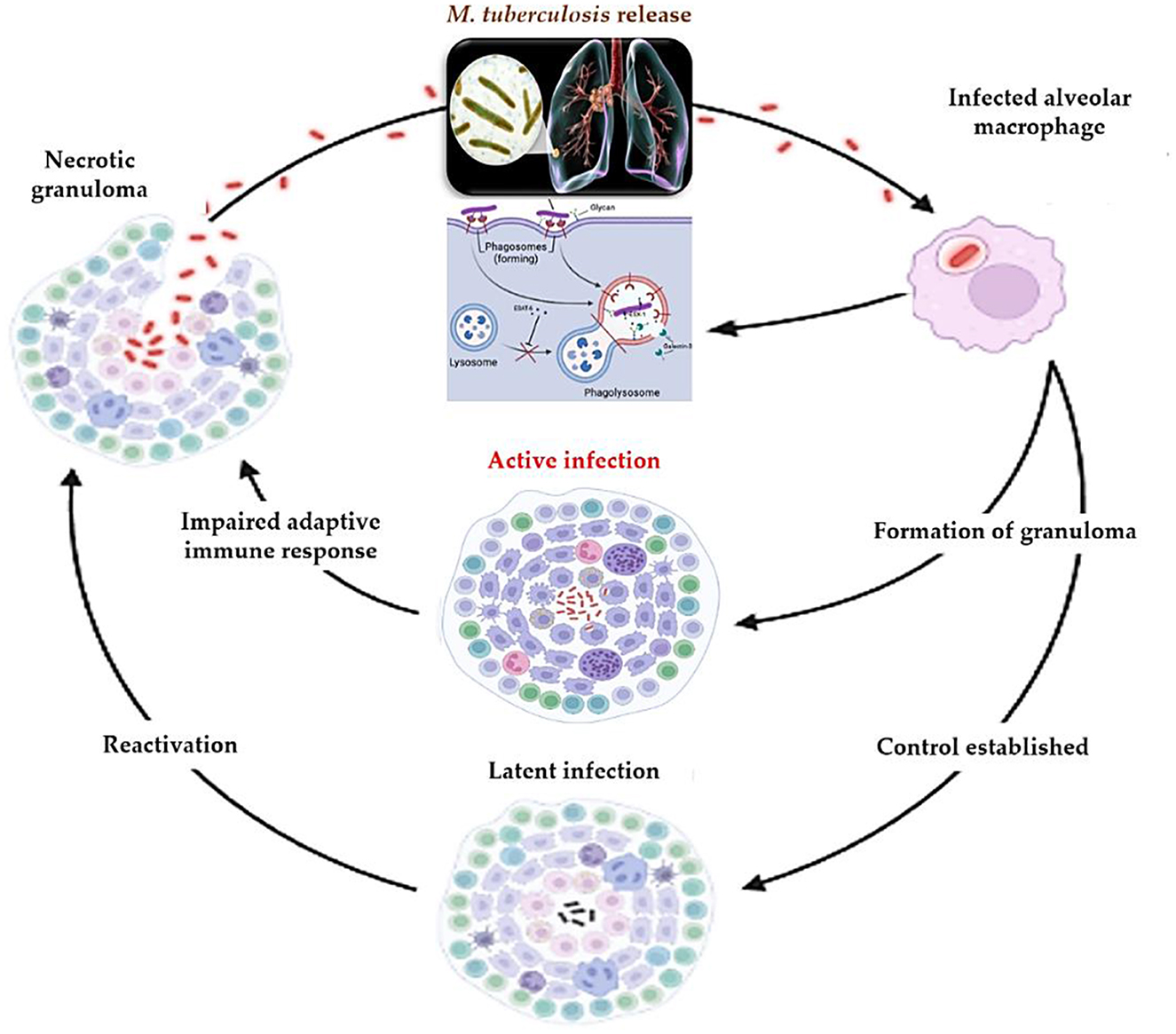
Figure 1. Pathogenesis of M. tuberculosis: from initial infection to reactivation. Following inhalation, M. tuberculosis is phagocytosed by alveolar macrophages, where it employs the ESX-1 secretion system to escape phagosomal destruction. This triggers granuloma formation as the host attempts to contain the infection. Control may be established, leading to latent infection, or the host may progress to active disease. Impaired immune responses or external stressors can disrupt granuloma integrity, leading to reactivation. Necrotic granulomas facilitate bacillary dissemination and renewed transmission.
3 Innovations in tuberculosis diagnostics
Effective TB control hinges on accurate, rapid diagnosis, yet the disease continues to be underdiagnosed, especially in resource-limited settings. While conventional methods such as smear microscopy and culture remain important, they lack sensitivity or are too slow for timely intervention. In recent years, molecular and next-generation diagnostic tools have dramatically enhanced the landscape of TB diagnostics, offering improved sensitivity, specificity, and speed. The main diagnostic platforms for TB, including conventional and modern approaches, are summarized in Figure 2.
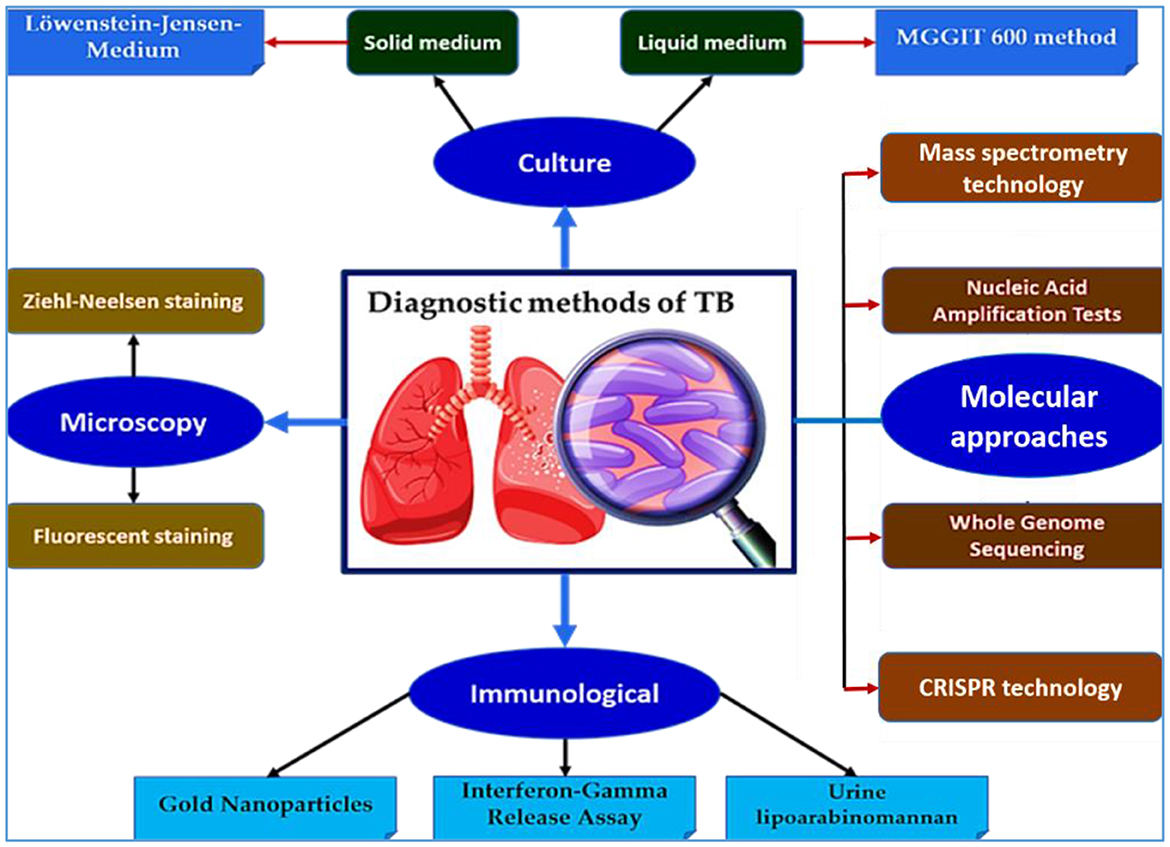
Figure 2. Overview of diagnostic methods for TB. This schematic illustrates the major diagnostic approaches for TB, including culture-based methods (solid and liquid media), microscopy (Ziehl-Neelsen and fluorescence staining), molecular diagnostics—such as nucleic acid amplification tests (NAATs), whole genome sequencing (WGS), CRISPR-based assays, and mass spectrometry—as well as immunological tools (e.g., interferon-gamma release assays, lipoarabinomannan (LAM) detection, and gold nanoparticles (GNPs)-based tests). These platforms enable early detection, resistance profiling, and improved disease management, especially in high-burden settings.
3.1 Culturing-based mycobacterial detection
Culture remains the definitive standard for diagnosing M. tuberculosis infection and assessing drug susceptibility (47). Although M. tuberculosis exhibits a slow growth rate, dividing every 18–20 h, culture remains vital for confirming active disease and resistance patterns (48). Despite limitations in rapid detection (49), culture-based techniques are WHO-endorsed due to their high specificity and utility in confirming TB and its resistance profile (50). Culture-based methods outperform traditional microscopy, which detects only 10–100 bacteria per milliliter (51). Solid media such as Lowenstein-Jensen (LJ), Ogawa, and Middlebrook 7H10/7H11 are routinely employed for isolating M. tuberculosis (52). Middlebrook agars promote faster colony development, whereas LJ slopes allow reliable assessment of growth dynamics (53). However, the protracted growth period on solid media—typically up to 6 weeks—limits their practicality in urgent clinical scenarios.
To address this, automated liquid culture systems like the BACTEC MGIT 960 have been increasingly adopted. These systems improve time to detection—often reducing it to 10–15 days—and demonstrate enhanced sensitivity and specificity compared to solid culture (54–56). Liquid media such as 7H9 broth facilitate optimal bacterial proliferation and long-term strain preservation (57). Nonetheless, differentiating M. tuberculosis from non-tuberculous species, especially in HIV co-infected patients, requires supplemental biochemical or molecular confirmation (58). Although both media types differ in contamination rates and detection speed, the WHO recommends employing both in tandem where feasible (59). In resource-limited settings, this dual approach is often hindered by financial and logistical constraints (60). Despite these challenges, culture retains its importance in detecting persistent infections and evaluating recurrence risk (61). Recent innovations such as Thin-Layer Agar (TLA) culture offer promising alternatives. A 1-year study by Battaglioli et al. (62) in Indonesia demonstrated that TLA outperformed LJ media in both sensitivity and detection time, making it a potential tool for faster TB confirmation in high-burden areas.
The BACTEC MGIT 960 system supports rapid detection by culturing mycobacteria in a closed liquid environment with oxygen-sensitive fluorescent sensors (63, 64). A positive MGIT result, however, requires subsequent identification of the M. tuberculosis complex. Immunochromatographic tests detecting MPT-64 antigens are WHO-recommended tools that deliver rapid species-level identification with high sensitivity (98.1–98.6%) and specificity (99.2–100%) (65–67). While liquid culture offers reduced turnaround times (~2 weeks), it carries a higher risk of contamination (68). Drug susceptibility testing using MGIT 960 has shown high accuracy-−90% for isoniazid and 99.4% for rifampicin—with excellent specificity, outperforming nitrate reductase assays (69). Nonetheless, false negatives remain a concern in low-bacterial-load samples, especially during treatment monitoring. Cultures must therefore be conducted in well-equipped reference laboratories with trained personnel and biosafety precautions.
3.2 Microscopy
Microscopy remains a foundational tool in TB diagnosis, particularly in low-resource settings. For over a century, sputum smear microscopy has been the principal method for detecting M. tuberculosis, providing rapid, low-cost diagnostic support (29, 70). Despite being superseded in some regions by molecular assays, it remains essential for frontline diagnosis and infectiousness assessment in high-burden, low-income countries (71, 72). Two main staining approaches are widely used: the Ziehl-Neelsen (ZN) stain, which highlights acid-fast bacilli (AFB) in red against a blue background (73), and fluorescence microscopy (FM) using auramine dyes, which offers improved sensitivity but requires more specialized equipment (74). ZN microscopy is especially valuable in resource-limited environments where culture facilities are lacking (68, 75). It allows for the detection of high bacillary loads (>104 bacilli/mL), correlating with greater transmission risk (76). However, the technique suffers from low sensitivity, a high false-negative rate, and an inability to differentiate between viable/non-viable or tuberculous/non-tuberculous mycobacteria (29, 30).
A recent implementation of the ZEISS Axio Scan platform demonstrated a sensitivity of 97.06% and specificity of 86.44% for detecting and enumerating acid-fast bacilli, offering a valuable enhancement in diagnostic accuracy and operational efficiency for laboratory personnel (77). In pulmonary tissue specimens, the combination of fluorescent antibody labeling and laser confocal microscopy has proven especially effective in identifying M. tuberculosis, particularly when traditional ZN staining produces suboptimal results (78). Additionally, the integration of digital pathology systems, such as the Pat-Scan platform, with paraffin-embedded ZN-stained tissues, enables rapid and reliable identification and quantification of microorganisms, thereby reducing turnaround time for TB diagnosis (79).
While solid-state microscopy cannot reliably distinguish between viable and non-viable bacilli or differentiate M. tuberculosis from non-tuberculous mycobacteria, ZN staining continues to provide valuable morphological insight, especially in previously treated patients with AFB (80). The use of FM not only improves diagnostic sensitivity but also increases throughput and reduces labor demands (81). However, it is associated with limitations, including transient staining effects and the risk of false positives due to non-specific binding of fluorochrome dyes (82, 83).
To overcome these drawbacks, Light-Emitting Diode (LED) microscopy has emerged as a sustainable and practical alternative, particularly in resource-limited settings. Its extended battery life and low maintenance requirements make it suitable for peripheral laboratories where conventional light sources may be unreliable. In parallel, the fluorescein diacetate (FDA) staining method has gained attention for its ability to detect viable M. tuberculosis bacilli. This metabolic staining technique converts non-fluorescent FDA into green fluorescence within live cells, allowing for rapid assessment of bacterial viability and potentially predicting culture positivity within 1 h (84, 85). Fully automated imaging systems can now insert, focus, and scan smears, classifying them as positive or negative using algorithm-driven analysis (86).
Supporting these technical advances, Dzodanu et al. (87) compared ZN staining and FM in 100 suspected pulmonary TB patients at Kade Government Hospital. Of 200 sputum samples, 35.5% tested positive via FM, 23.2% via ZN staining, and 42% via the Xpert MTB/RIF assay. FM outperformed ZN in sensitivity (84.5% vs. 54.8%) with similar specificity (100%). Masali et al. (88) also reported superior performance for FM, detecting 42.3% positive cases vs. 18.2% by ZN, with FM achieving 98% sensitivity and a negative predictive value of 99%. These findings support FM as a more sensitive alternative to conventional staining techniques in TB diagnostics.
3.3 Molecular diagnostics for tuberculosis
Molecular diagnostics have significantly advanced TB detection by enabling rapid, sensitive, and specific identification of M. tuberculosis and associated drug resistance mutations. Unlike microscopy or culture—which are limited by either low sensitivity or long processing times—molecular tools offer results within hours, playing a vital role in early diagnosis and treatment decisions. This section outlines key molecular platforms used in TB diagnostics, including NAATs, real-time PCR (RT-PCR), WGS, mass spectrometry, and CRISPR-based diagnostics.
3.3.1 Nucleic acid amplification tests (NAATs)
Over recent decades, various NAATs have been developed to improve the detection of M. tuberculosis complex (89–92). These tests show high specificity (74–99.3%) and variable sensitivity (64–100%), which can decrease to 40–84% in smear-negative samples or those with low bacillary loads (93). While many NAATs perform well in acid-fast smear-positive cases (~95% sensitivity), their sensitivity drops significantly in paucibacillary samples (94). Additionally, the presence of other microbes (e.g., non-tuberculous mycobacteria or fungal pathogens) may interfere with amplification, potentially leading to false-positive or false-negative outcomes (95, 96). Thus, enhancing diagnostic performance in smear-negative and extrapulmonary TB remains a key priority.
Insertion sequences (ISs) are widely used in NAATs to improve sensitivity (93). Targets such as IS986, IS987, IS1081, and particularly IS6110, are highly repetitive in the M. tuberculosis genome, allowing for effective amplification in multiplex PCR assays (97–99). However, certain strains (e.g., M. bovis BCG) may harbor few or no IS6110 copies, which may reduce test sensitivity. The Xpert MTB/RIF Ultra assay, endorsed by WHO, incorporates IS6110 and IS1081 as targets and is now widely used in clinical practice. This cartridge-based test detects TB DNA and rifampicin resistance with a sensitivity of 87.5% and a detection limit of 15.6 CFU/mL (100, 101). In lung specimens, it achieves 88% sensitivity and 96% specificity; in extrapulmonary TB, it yields 98.5% sensitivity and 97% specificity (100, 102). However, its sensitivity decreases to 78.9% in smear-negative samples. Despite its clinical utility, its cost (~$9.98/cartridge) poses challenges in low-resource settings (103), prompting a shift toward in-house real-time PCR protocols that are more affordable and adaptable.
Nested PCR, a subtype of NAAT, improves analytical sensitivity by using two rounds of amplification. Although effective for extrapulmonary TB (sensitivity: 72.2% for blood/urine vs. 33.3% for pleural fluid) (104–106), it is labor-intensive, prone to contamination, and costly. To overcome these drawbacks, single-tube nested PCR was developed, incorporating outer primers with higher annealing temperatures than inner primers. This method offers a simplified workflow and enhanced accuracy—achieving up to 89% sensitivity in pulmonary TB and 42% in extrapulmonary forms. Notably, single-tube nested RT-PCR has demonstrated even higher performance, with 97.2% sensitivity and 99.7% specificity (107). In a comparative analysis, Choi et al. reported 94.6% sensitivity for IS6110 RT-PCR and 100% for single-tube nested RT-PCR in sputum samples (108). Despite these advances, diagnostic performance for smear-negative and extrapulmonary TB remains suboptimal, reinforcing the urgent need for more robust, accessible, and affordable molecular platforms. Figure 3 illustrates the standard NAATs currently employed in TB diagnostics and their role in expediting accurate clinical decision-making.
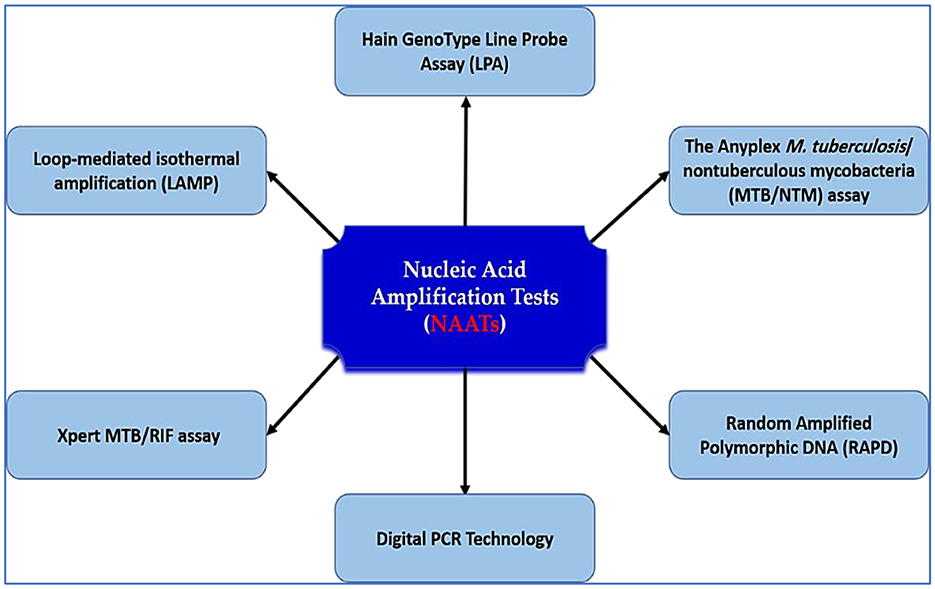
Figure 3. NAATs for tuberculosis diagnosis. This schematic illustrates the major nucleic acid amplification platforms used for TB detection, including Loop-mediated isothermal amplification (LAMP), Xpert MTB/RIF, digital PCR, Hain GenoType Line Probe Assay, Anyplex MTB/NTM assay, and RAPD. These assays improve diagnostic accuracy, speed, and resistance profiling, and are central to early TB case detection and treatment guidance.
3.3.1.1 Loop-mediated isothermal amplification (LAMP)
LAMP, developed by Eiken Chemical Co., Ltd. (Tokyo, Japan), is an innovative nucleic acid amplification technique for TB detection that has been endorsed by the WHO (109). LAMP offers a cost-effective alternative to the Xpert MTB/RIF assay, with studies in China reporting diagnostic cost savings of 50–70% (110, 111). One of its major advantages is the visual readout of results, eliminating the need for sophisticated equipment and enabling deployment in decentralized, resource-limited settings. Multiple studies in China have evaluated LAMP's diagnostic performance for pulmonary TB, though reported accuracy has varied (112–114). Lin et al. (109) conducted a multicenter evaluation using respiratory samples from suspected TB patients across six sites in China between June 2018 and December 2019. LAMP showed good concordance with Xpert MTB/RIF, fluorescent smear microscopy, and BACTEC MGIT 960, with a sensitivity of 78.6% and specificity of 88.7%. The study concluded that LAMP is a rapid, user-friendly, and affordable alternative for TB detection in clinical practice.
Further validation came from Zaber et al. (115), who assessed a multiplex LAMP assay in 130 sputum samples in Bangladesh. Compared to qPCR, which detected M. tuberculosis in 56.92% of cases, LAMP identified 53.85%. LAMP demonstrated a sensitivity of 95% and specificity of 81.4% vs. culture, outperforming ZN staining by 16.93% and FM by 13.08%. Importantly, LAMP failed to detect non-tuberculous mycobacteria that were identified by qPCR in 7.69% of cases, suggesting high specificity for M. tuberculosis. These findings support LAMP as a WHO-recommended, low-cost, and effective diagnostic tool for TB control, especially in LMICs aiming to meet End TB Strategy targets by 2035 (115).
3.3.1.2 Xpert MTB/RIF assay
The emergence of multidrug-resistant (MDR) M. tuberculosis strains has underscored the urgency for rapid diagnostic tools. In response, the Centers for Disease Control and Prevention (CDC) recommends integrating molecular diagnostics with traditional testing to overcome the limitations of conventional culture-based methods. The Xpert MTB/RIF assay (Cepheid, Sunnyvale, CA, USA) represents a transformative advancement, enabling direct detection of M. tuberculosis and rifampicin resistance from clinical specimens in under 2 h (116). In Brazil, the National Tuberculosis Control Program introduced the Xpert platform in 92 high-burden municipalities between 2014 and 2015. Subsequently, the Clinics Hospital of the University of São Paulo adopted the Xpert MTB/RIF assay for diagnosing both pulmonary and extrapulmonary TB.
In a study by Feliciano et al. (117), 2,148 respiratory samples collected between 2015 and 2018 were analyzed. The assay demonstrated a sensitivity of 94%, specificity of 98%, positive predictive value (PPV) of 89%, and negative predictive value (NPV) of 99%. Agreement with phenotypic drug susceptibility testing was 94.1%, while concordance with WGS was 78.9%. More recently, Terzi et al. (118) evaluated the performance of Xpert MTB/RIF in 2,082 specimens (1,526 respiratory and 556 non-respiratory). M. tuberculosis was cultured in 153 samples (7.3%), while 203 (9.7%) were Xpert-positive. The assay achieved 89.5% sensitivity, 96.6% specificity, 67.5% PPV, and 99.1% NPV. Its speed, simplicity, and ability to detect resistance in a single step make it a vital tool in TB diagnostics, especially in high-burden and resource-constrained settings.
3.3.1.3 Line probe assay (LPA)
Line Probe Assays (LPAs), endorsed by the WHO in 2008, enable rapid molecular detection of M. tuberculosis and assessment of resistance to rifampicin and isoniazid—the two cornerstone drugs in TB treatment (119). A widely used commercial version was originally developed by Hain Lifescience (now acquired by Bruker). Based on DNA-STRIP technology, LPA improves diagnostic speed and precision compared to conventional phenotypic methods, though it requires greater technical expertise than the Xpert MTB/RIF assay (120, 121). The second-generation LPA (version 2) accommodates a wider variety of sample types and provides detailed resistance profiles, including detection of both low- and high-level resistance mutations (122).
In a 2024 study, Nandwani et al. (123) tested 196 sputum samples from suspected pulmonary TB patients using LPA. TB was confirmed in 104 positive cases. Sensitivity for smear-negative samples varied by target: 47.36% for TB detection, 72.72% for rifampicin resistance, and 88.88% for isoniazid resistance, with specificities ranging from 86.96% to 95.65%. In smear-positive cases, LPA achieved high sensitivity-−89.09% for TB, 95.83% for rifampicin resistance, and 98.07% for isoniazid resistance—with specificities exceeding 98% for all targets. These results support the use of LPA as a reliable and efficient tool for first- and second-line drug resistance detection, particularly in smear-positive patients, and as a complement to other rapid molecular tests in TB diagnostic algorithms.
3.3.1.4 Anyplex MTB/NTM real-time detection assay
The Anyplex™ MTB/NTM assay (Seegene Inc., South Korea) utilizes real-time PCR with dual priming oligonucleotide (DPO™) technology to simultaneously detect M. tuberculosis complex and differentiate it from nontuberculous mycobacteria, offering enhanced specificity and multiplexing capabilities. It targets highly specific genetic markers, including the insertion sequence IS6110 (99) and the MPB64 gene (124), allowing for precise identification of clinically relevant mycobacterial species. In a large-scale evaluation by Sawatpanich et al. (125), the assay was applied to 9,575 clinical specimens. The test demonstrated a sensitivity of 79.7% and specificity of 94.5% for detecting MTBC, while for NTM, sensitivity was lower at 44.9% but specificity remained high at 97.7%. Among AFB smear-positive samples, the assay achieved significantly improved sensitivity-−97.7% for MTBC and 80% for NTM. These results support the utility of Anyplex MTB/NTM as a rapid and accurate tool, particularly valuable in high-throughput laboratory settings where distinguishing MTBC from NTM is crucial for patient management.
In parallel, Random Amplified Polymorphic DNA (RAPD)—also known as Arbitrary Primed PCR (AP-PCR)—is a molecular fingerprinting method that requires no prior sequence information. By employing randomly selected primers ranging from 5 to 50 base pairs, RAPD generates DNA profiles that can reveal inter-strain polymorphisms (126). Although some variability in results is attributed to technical reproducibility concerns (127), RAPD remains a valuable tool for analyzing strain diversity among NTM species. For instance, RAPD has been successfully used to genotype M. abscessus and M. chelonae, two species prone to DNA fragmentation during electrophoresis and difficult to analyze via pulsed-field gel electrophoresis (PFGE) (128). Furthermore, RAPD has proven effective in characterizing genetic diversity within other clinically significant NTMs such as M. phocaicum, M. gordonae, M. szulgai, and M. malmoense (129–132). The method's affordability and simplicity make it particularly appealing for resource-constrained laboratories aiming to monitor mycobacterial diversity and trace epidemiological patterns.
3.3.1.5 Droplet digital polymerase chain reaction (ddPCR)
Droplet digital polymerase chain reaction (ddPCR) is an advanced molecular technique that enables absolute quantification of nucleic acids without requiring a standard curve (133). This method partitions the sample into thousands of nanoliter-sized droplets, where amplification occurs independently, improving detection sensitivity and reducing variability. ddPCR has emerged as a powerful tool for detecting low-abundance genetic targets, making it particularly suitable for infectious diseases such as tuberculosis (134). Recent studies have demonstrated the utility of ddPCR in TB diagnostics. Devonshire et al. (135) assessed ddPCR performance using M. tuberculosis DNA templates and confirmed its robustness and accuracy in quantifying bacterial DNA. In follow-up research, ddPCR successfully identified M. tuberculosis in artificially prepared sputum specimens, supporting its applicability in clinical settings (136). The method showed strong reproducibility and precision, even in samples with low bacterial loads.
Due to its superior sensitivity, ddPCR holds promise for various TB-related applications, including early diagnosis, quantification of drug resistance mutations, and monitoring of bacterial burden during treatment (137). Future innovations are expected to yield minimally invasive, rapid, and highly accurate ddPCR-based platforms for the detection of M. tuberculosis-specific genetic sequences, which could substantially improve disease surveillance and management strategies.
3.3.2 Whole genome sequencing (WGS)
WGS has emerged as a transformative tool for tuberculosis diagnostics, offering unparalleled resolution in identifying M. tuberculosis complex strains and their resistance profiles. Thanks to reduced costs and rapid technological advances, WGS is now transitioning from research laboratories into clinical workflows for TB detection, drug resistance prediction, and epidemiological surveillance (138). WGS surpasses traditional genotyping techniques such as PCR and microarrays by enabling comprehensive analysis of nearly all genomic mutations associated with drug resistance, as well as differentiating closely related Mycobacterium subspecies (139). It offers significant value in tracking transmission chains and understanding pathogen evolution (140). Advances in sequencing platforms, including Illumina MiSeq™ and HiSeq 4000, have greatly increased throughput—generating between 15 and 1,500 gigabytes of sequence data—making WGS increasingly cost-effective in diagnostic settings (141).
The utility of WGS has been underscored by several key studies (142–144). The WHO issued a practical guide in 2018 promoting WGS to characterize treatment-resistant TB strains, highlighting platforms such as Ion Personal Genome Machine®, Nanopore MinION®, and GeneReader. In a large-scale study, Campbell et al. (145) sequenced nine drug-resistance loci in 314 clinical isolates, reporting a sensitivity of 90.8% and specificity of 94.7% for multidrug resistance, but only 40% sensitivity for extensively drug-resistant (XDR) strains. These findings highlight the promise and current limitations of WGS, especially for detecting rare resistance mutations (63). Emerging research illustrates that WGS can uncover low-level resistance mutations often missed by conventional phenotypic drug susceptibility tests (DST). For example, genome analysis has identified ethambutol resistance-associated mutations that remain undetected in phenotypic assays (146). Similarly, certain isoniazid-resistant isolates may yield negative WGS results, reflecting challenges in correlating genotype and phenotype (147).
In a study by Sun et al. (148), WGS was evaluated in newly diagnosed multidrug-resistant TB cases in China. The method showed high concordance with phenotypic DST for amikacin/kanamycin and rifampicin (97.7%) but lower agreement for rifabutin and ethambutol (67.2% and 79.1%, respectively). Pyrazinamide resistance-associated mutations were detected in 27.9% of isolates. No resistance mutations were found for newer drugs such as linezolid, bedaquiline, or clofazimine, affirming WGS's role in guiding individualized therapy in high-burden settings.
Recognizing the diagnostic potential of next-generation sequencing, the WHO issued guidelines in October 2023 to support its integration into national TB programs, particularly for tracking drug-resistant strains. However, real-world implementation faces numerous barriers. Vogel et al. (149) reported logistical and financial challenges in Kyrgyzstan, while Ness et al. (150) and others have emphasized the need for robust infrastructure, expert training, and standardized quality control measures (151, 152). Recent innovations, such as direct sputum WGS (bypassing culture), show promise for reducing turnaround times (153, 154). Yet, obstacles such as low DNA yield, high costs, limited database access, and technical complexity continue to hinder widespread clinical adoption (155). Despite these constraints, WGS remains a powerful tool in the global fight against TB, with future applications likely to enhance precision diagnostics and public health interventions.
3.4 Immunological approaches
3.4.1 Gold nanoparticles (GNPs)
GNPs have emerged as promising tools in TB diagnostics due to their nanoscale size, ease of synthesis, high stability, and biocompatibility (156, 157). Their unique optical behavior—particularly localized surface plasmon resonance—is influenced by particle size, shape, and interparticle spacing, allowing colorimetric detection without the need for complex instrumentation (158). In point-of-care (POC) platforms, GNPs function as optical tags for antigen–antibody detection, offering a rapid, visual signal for TB diagnosis (159, 160). GNPs possess the unique ability to change color via a mechanism known as localized surface plasmon resonance, which is influenced by the particle's shape, size, and local refractive index (161, 162). To date, only one GNP-based diagnostic platform—the TB-LAMP assay developed by Eiken Chemical Co.—has received endorsement by the WHO. This approval was issued in 2016, recommending its use as a molecular alternative to sputum smear microscopy in resource-limited settings for the detection of M. tuberculosis (163).
Several studies have demonstrated the diagnostic efficacy of GNPs in TB. Becerra et al. (164) developed a plasmonic system using GNPs coated with mycobacterial lipid glycans to detect anti-lipid antibodies in patient sera. The system showed measurable shifts in LSPR (up to 2 nm), enabling sensitive antigen–antibody detection confirmed across multiple clinical samples (165). Other biosensor platforms targeting the M. tuberculosis IS6110 gene reported sensitivities ranging from 84.7% to 100% and specificities approaching 100%, with detection limits from 5 pg to 81 ng per 25 μL reaction (166–170).
Dahiya et al. (171) developed a magnetic bead-coupled GNP immuno-PCR (MB-GNP-I-PCR) assay for detecting TB antigens in clinical fluids, demonstrating 89.3% sensitivity for pulmonary TB and 78.1% for extrapulmonary TB, with specificity exceeding 97.9%. This approach outperformed conventional assays such as Magneto-ELISA and GeneXpert. Similarly, Kooti et al. (158) validated a GNP biosensor assay that reliably detected M. tuberculosis in sputum samples, underscoring its utility as a simple and scalable method adaptable to various diagnostic settings. In summary, GNP-based diagnostics represent a powerful and cost-effective tool for rapid TB detection. While further standardization is needed, their sensitivity, adaptability, and compatibility with low-resource environments support their future integration into global TB control programs.
3.4.2 Interferon gamma release assay (IGRA)
Interferon-gamma release assays (IGRAs) have emerged as valuable tools in the diagnosis of LTBI, offering improved specificity over the traditional tuberculin skin test (TST). The TST is prone to false-positive results, particularly in individuals vaccinated with Bacille Calmette-Guérin (BCG) or exposed to non-tuberculous mycobacteria (172). IGRAs circumvent this issue by utilizing M. tuberculosis-specific antigens, such as ESAT-6 and CFP-10, which are absent from BCG strains and most environmental mycobacteria. Two widely used IGRAs are the QuantiFERON-TB Gold Plus (QFT-Plus) and T-SPOT.TB, developed by Qiagen and Oxford Immunotec, respectively (173, 174). QFT-Plus, a fourth-generation IGRA, detects IFN-γ released by T cells in response to M. tuberculosis complex-specific antigens, including M. tuberculosis, M. bovis, and M. africanum (175). It serves as a more specific alternative to the traditional TST for identifying latent TB infection. Compared to earlier versions like QuantiFERON-TB Gold In-Tube (QFT-GIT), QFT-Plus incorporates both CD4+ and CD8+ T-cell responses, potentially enhancing sensitivity.
Although IGRA results can indicate M. tuberculosis infection, they do not distinguish between latent and active TB (176). Furthermore, variables such as overnight incubation and host immune status may affect test reliability, especially in immunocompromised individuals (177, 178). In a 2019 study by Hong et al. comparing 33 active TB patients and 57 controls with LTBI, QFT-Plus demonstrated a sensitivity of 93.9% and a specificity of 92.6%, while QFT-GIT showed identical sensitivity but slightly higher specificity at 100%. Notably, IFN-γ levels were lower in latent TB cases using QFT-Plus, although the distinction between active and latent TB remained inconclusive (179). Similarly, Venkatappa et al. (180) evaluated the concordance between QFT-Plus, QFT-GIT, T-SPOT.TB, and TST across 506 high-risk individuals in a multicenter study. QFT-Plus and QFT-GIT exhibited 94% overall agreement, with 19% positivity and 75% negativity, reinforcing their equivalence in clinical utility. Despite their diagnostic strengths, IGRAs face limitations in differentiating disease stages and in implementation across low-resource settings. Nonetheless, they remain a vital component of TB screening protocols, especially in BCG-vaccinated populations and high-risk groups.
3.4.3 Urine lipoarabinomannan assay (LAM)
Urine-based antigen detection offers a noninvasive diagnostic option for TB, eliminating the need for aerosol-generating procedures during specimen collection (181). One promising biomarker in this approach is LAM, a glycolipid component of the M. tuberculosis cell wall that can be excreted in urine. Although the exact mechanism of LAM excretion remains unclear, it is hypothesized to result from bacterial degradation during infection (182–184). The Determine™ TB LAM Ag assay (formerly AlereLAM), developed by Alere Inc. and now marketed by Abbott Laboratories, is a lateral flow immunochromatographic test designed for point-of-care use, particularly in resource-limited settings. It provides rapid, bedside detection of urinary LAM, with utility in diagnosing extrapulmonary TB cases that may yield negative sputum results by conventional methods such as GeneXpert (181). The performance of the Determine™ TB LAM test varies significantly based on patient characteristics, including the presence of TB symptoms, hospitalization status, and CD4+ T-cell count (185).
Since 2015, the WHO has recommended AlereLAM for TB screening in HIV-positive patients, particularly those with advanced immunosuppression (186). Meta-analyses report a pooled sensitivity of 42%, increasing to 54% among individuals with CD4+ counts below 100 cells/mm3, but falling to just 17% in those with higher counts (185). Despite its relatively modest sensitivity, AlereLAM remains a cost-effective diagnostic option in low-resource settings and contributes valuable clinical information, especially in advanced HIV infection (187, 188). The Fujifilm SILVAMP TB LAM assay (FujiLAM) represents a second-generation test that addresses several limitations of AlereLAM. FujiLAM incorporates high-affinity monoclonal antibodies and a silver amplification step to improve signal detection, thereby enhancing sensitivity and lowering the detection threshold for LAM in urine (189–192).
Clinical studies have reported that FujiLAM can achieve diagnostic sensitivities of up to 85% in HIV-positive adults, with significantly higher specificity than AlereLAM (193–195). In a 2022 study conducted in Indonesia involving 62 patients, FujiLAM demonstrated a sensitivity of 61% and specificity of 92.31%, outperforming AlereLAM and other diagnostic methods in identifying extrapulmonary TB cases (196). In the same cohort, the general urine LAM test showed a sensitivity of 75% and specificity of 73.91%. Despite being three to four times more affordable than nucleic acid amplification tests (NAATs), AlereLAM's low sensitivity, particularly in patients with CD4+ T-cell counts above 200 cells/mm3, limits its broader clinical use (197). Additionally, ambiguities in the interpretation of AlereLAM results, as noted in the WHO 2019 guidelines, further complicate its clinical utility due to variability in host immune status and bacillary burden (185). A generalized schematic of the urine LAM assay principle is presented in Figure 4, illustrating the noninvasive detection of LAM antigens through a lateral flow immunoassay format. This process involves urine sample application, antigen-antibody binding, and visual interpretation of results via colored test and control lines.
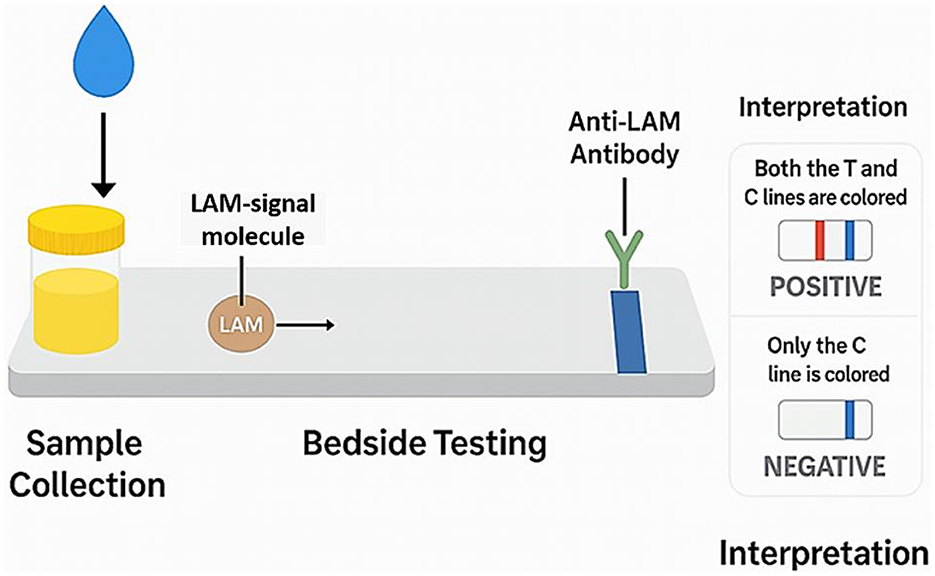
Figure 4. Schematic illustration of the LAM Assay principle. This diagram outlines the core steps of the LAM-based lateral flow immunoassay used for TB diagnosis. Following urine collection, LAM antigens bind to anti-LAM antibodies conjugated with a detection label. As the sample migrates along the test strip, the presence of LAM is indicated by a colored test line, while a control line ensures assay validity. The interpretation panel distinguishes between positive (both lines visible) and negative (only control line visible) results. This generic schematic avoids proprietary device branding and reflects the underlying principle applicable across LAM assay platforms.
3.5 Mass spectrometry technology
Matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) has become an essential tool for microbial identification through protein profiling (198–200). While highly effective for many bacteria, its application to mycobacterial species has been more limited due to challenges in protein extraction from their lipid-rich, rigid cell walls (201, 202). These barriers complicate cell lysis and cytoplasmic protein release, both of which are critical for reliable MALDI-TOF MS analysis (203). Moreover, the slow growth rate and lower ribosomal content of MTBC, along with the need for biosafety level 3 (BSL-3) inactivation protocols, further hinder protein detection (204–206).
Recent advances in sample preparation have enhanced MALDI-TOF MS performance in mycobacterial diagnostics. Mechanical and chemical lysis techniques—particularly those employing silica/zirconia beads—have improved protein extraction efficiency (207), Bruker Daltonics' MycoEx protocol incorporates bead beating and acetonitrile extraction, showing improved performance in identifying mycobacteria, including M. bovis and M. tuberculosis strains (201, 205, 208, 209). Bacanelli et al. (210) demonstrated that high-powered homogenization (MycoLyser method), compared to vortexing (MycoEx), produced significantly higher Biotyper log scores (1.800 vs. lower scores), indicating enhanced identification accuracy.
Beyond species identification, MALDI-TOF MS is now applied for antimicrobial resistance profiling. The MassARRAY platform enables detection of hundreds of single nucleotide polymorphisms (SNPs) from 15 drug resistance-associated genes, generating comprehensive drug susceptibility profiles (211). In a clinical study of 201 pulmonary TB patients, Shi et al. (212) reported that MALDI-TOF MS had a detection rate of 94.3%, outperforming smear microscopy (43.2%), LAMP, 68.2% and Xpert MTB/RIF (85.2%). Notably, in culture-negative samples, MALDI-TOF MS still achieved a sensitivity of 78.8%, far exceeding other methods. Among rifampicin-resistant cases, MALDI-TOF MS identified 96.72%, compared to 81.97% with Xpert. These findings support MALDI-TOF MS as a robust and scalable platform for both rapid TB detection and drug resistance analysis. Continued refinement of sample processing protocols and broader clinical validation could solidify its role in frontline TB diagnostics, especially in settings requiring high-throughput, accurate, and rapid testing.
3.6 CRISPR-based diagnostics
The application of CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) and CRISPR-associated protein (Cas) systems in TB diagnostics represents a novel and promising molecular approach. CRISPR/Cas systems utilize sequence-specific cleavage activity, guided by short RNA molecules, to recognize and cleave complementary DNA sequences (213). In addition to their cis-cleavage capabilities at the target site, many Cas enzymes exhibit trans-cleavage activity—wherein abundant, non-target reporter oligonucleotides are repeatedly cleaved in proportion to target presence—allowing intrinsic signal amplification (214, 215).
This dual-cleavage mechanism enables ultra-sensitive detection of nucleic acids, including targets at low copy numbers or those differentiated by single nucleotide polymorphisms (SNPs) (216–219). Furthermore, CRISPR systems have demonstrated utility in detecting SNPs associated with drug resistance in M. tuberculosis, making them highly applicable for both diagnosis and antimicrobial resistance profiling (220). Their compatibility with portable, point-of-care platforms further enhances their diagnostic appeal (221–223).
Most CRISPR-based TB assays incorporate nucleic acid amplification (NAA), such as recombinase polymerase amplification (RPA) or LAMP, to enhance sensitivity (215, 224). These platforms can detect trace quantities of M. tuberculosis DNA with high accuracy in under an hour (225–227). However, total assay time often exceeds 60 min due to multi-step workflows (226, 228–232), which may also increase the risk of cross-contamination. Additionally, fluorescent signal detection in many CRISPR systems requires external devices, limiting field applicability in resource-limited settings until simpler readout systems become available. Lateral flow strips, however, offer promising visual alternatives for qualitative interpretation (227, 232, 233).
Most CRISPR-based TB diagnostics target multicopy genomic sequences such as IS6110 and IS1081, which are highly conserved and present in multiple copies across M. tuberculosis complex strains, thereby improving diagnostic sensitivity (225–227, 229, 232–234). Some systems have also been designed to detect mutations in the rifampicin resistance-determining region of the rpoB gene—mutated in over 95% of rifampicin-resistant and more than 78% of multidrug-resistant TB strains (228, 235, 236). Additional targets, such as 16S rRNA, rpsL, and gyrB, may also indicate TB presence or drug resistance. While genomic conservation across M. tuberculosis complex species exceeds 99% (237), IS6110 remains a universal and highly specific target for CRISPR-based detection platforms (238).
In 2023, Zhang et al. introduced a CRISPR/Cas12a-based system combined with recombinase-aided amplification for detecting M. tuberculosis, achieving rapid, equipment-free molecular identification (239). The entire workflow—from DNA extraction to signal detection—was completed within 2 h, including 20 min for extraction, 30 min for amplification, and 30 min for CRISPR-based readout. Compared to Xpert MTB/RIF (120 min) and conventional culture (up to 30 days) (240), the CRISPR approach offers significant time and cost advantages, with expenses estimated to be nearly 50% lower than standard PCR methods (241).
Despite its potential, several challenges remain. The variability in repeatability and the amplification duration may affect detection sensitivity and complicate quantification of bacterial load. Furthermore, the limitations of traditional culture-based reference standards (e.g., low positivity in older samples) suggest that aligning CRISPR results with clinical diagnoses may improve diagnostic accuracy (239). Nonetheless, CRISPR-based diagnostics represent a powerful emerging tool in the early and precise detection of TB and drug resistance mutations, particularly in resource-limited and high-burden settings.
3.7 The emerging role of artificial intelligence in TB diagnosis and resistance prediction
The integration of artificial intelligence (AI) and machine learning (ML) in TB diagnostics represents a transformative frontier in clinical microbiology and global health (242). These technologies are increasingly being used to optimize image interpretation, predict drug resistance, and integrate diverse diagnostic data streams for improved clinical decision-making. In microscopy, AI algorithms have demonstrated strong potential in automating the detection of AFB in stained sputum smears. For instance, convolutional neural network (CNN)-based models have been trained to identify AFB with high accuracy, thereby minimizing manual effort and inter-observer variability. Hwang et al. (243) developed a deep learning model that achieved over 96% sensitivity and 98% specificity for detecting TB-positive smears, enabling rapid, high-throughput analysis suitable for decentralized laboratories where trained personnel are limited.
In the domain of chest radiography, AI-driven computer-aided detection (CAD) systems such as CAD4TB, Lunit INSIGHT CXR, and qXR have become integral to TB screening programs in community and clinical settings. These systems analyze digital chest X-rays in real time and assign TB likelihood scores to assist in triaging suspected cases. Qin et al. (244) conducted a multicenter study demonstrating that CAD4TB version 6 performed comparably to expert radiologists in identifying TB, especially in asymptomatic and HIV-positive individuals. Similarly, the WHO evaluated multiple AI-based CAD tools and reported that Lunit and qXR achieved sensitivities exceeding 90% and met or surpassed the WHO's target product profile for TB triage tests (245). The latest versions of these systems have shown area under the curve (AUC) values nearing 0.90, supporting their deployment in mobile clinics and high-volume screening programs.
Beyond imaging, AI is playing a transformative role in the prediction of drug resistance using WGS data. ML models, including ensemble techniques like XGBoost, have been applied to genomic datasets to forecast resistance to first-line anti-TB drugs. Walker et al. (246) demonstrated that such models could predict resistance to rifampicin and isoniazid with over 90% accuracy, offering a rapid alternative to conventional phenotypic susceptibility testing. This innovation is particularly critical for timely management of multidrug-resistant TB. Furthermore, AI is being incorporated into clinical decision support systems that integrate diverse data inputs—including patient history, lab results, radiographic findings, and genetic profiles—to guide diagnosis and therapy in real time. These systems represent a move toward precision medicine in TB care.
Nonetheless, several barriers remain. These include the requirement for large annotated datasets, variability in model performance across populations, concerns over algorithm transparency, and data privacy regulations that complicate implementation. Additionally, infrastructure limitations and regulatory uncertainty pose challenges to the large-scale adoption of AI tools in TB programs. Despite these limitations, the expanding evidence base supports the transformative potential of AI in TB diagnostics. By improving detection speed, enhancing diagnostic precision, and enabling personalized treatment approaches, AI is poised to become a cornerstone of global TB control strategies. Figure 5 provides a visual summary of the integration of AI across key TB diagnostic platforms, including microscopy, chest radiography, and WGS.
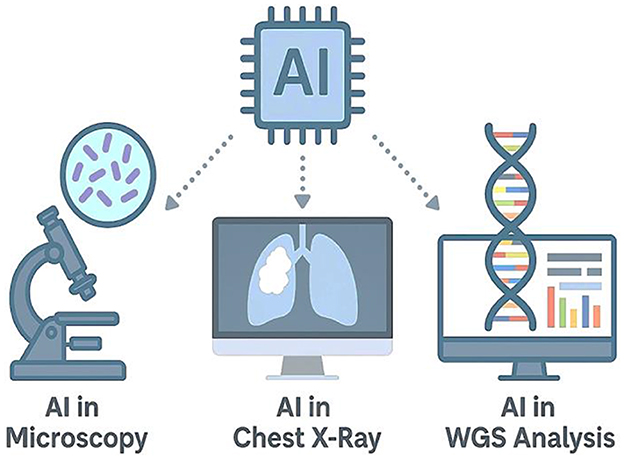
Figure 5. Artificial intelligence applications in tuberculosis diagnostics. This infographic illustrates the integration of AI across three major TB diagnostic platforms. Left: AI-assisted microscopy uses deep learning to detect AFB in smear samples. Center: AI-driven radiographic systems, such as CAD4TB, analyze chest X-rays for automated TB screening. Right: AI algorithms applied to WGS data enable rapid prediction of drug resistance mutations in M. tuberculosis. At the center, an AI microchip symbolizes the convergence of machine intelligence in advancing TB diagnostics across modalities.
3.8 Influence of host genetics and microbiome composition on TB susceptibility and treatment outcomes
Emerging research has highlighted the critical role of host genetic variation and microbiome composition in modulating susceptibility to TB, response to treatment, and risk of disease progression. These host-specific factors represent promising frontiers for the development of precision medicine approaches in TB management. Host genetics play a pivotal role in determining individual risk of TB infection, latency, and treatment response. Genome-wide association studies have identified multiple susceptibility loci, such as variants in the TLR1, VDR, IFNGR1, and IL12B genes, which affect immune recognition and cytokine signaling during M. tuberculosis infection. For instance, Curtis et al. (247) found that polymorphisms in HLA class II genes were significantly associated with progression from latent to active TB in diverse populations. Similarly, Quistrebert et al. (248) identified rare monogenic variants in TYK2 and STAT1 pathways linked to early-onset extrapulmonary TB, particularly in children.
Genetic differences also influence treatment outcomes. Polymorphisms in NAT2, which encodes N-acetyltransferase 2, affect isoniazid metabolism, leading to variations in drug efficacy and hepatotoxicity risk. In a meta-analysis by Huang et al. (249), slow acetylators had significantly higher risks of isoniazid-induced liver injury, suggesting the potential utility of pharmacogenomic-guided dosing. Beyond host DNA, the gut and lung microbiomes have emerged as important modulators of host immunity during TB infection and treatment. Dysbiosis—characterized by reduced microbial diversity and loss of commensal species—is commonly observed in TB patients, potentially impairing immune homeostasis. Naidoo et al. (250) demonstrated that anti-TB therapy induces long-term gut microbiome alterations, including depletion of Clostridiales and Bifidobacterium spp., which correlated with systemic inflammation and altered immune profiles.
Alterations in the lung microbiota have been increasingly linked to TB severity. Studies show that patients with active TB exhibit reduced microbial diversity and a shift from commensal genera like Streptococcus and Prevotella to potentially pathogenic taxa such as Rothia and Veillonella, reflecting a dysbiotic state that may worsen inflammation and lung damage (251). These findings suggest that the lung microbiome not only influences host immunity but may also serve as a biomarker or therapeutic target in TB management. Figure 6 illustrates how host genetic factors and microbiome composition interact to influence TB susceptibility, progression, and treatment outcomes.
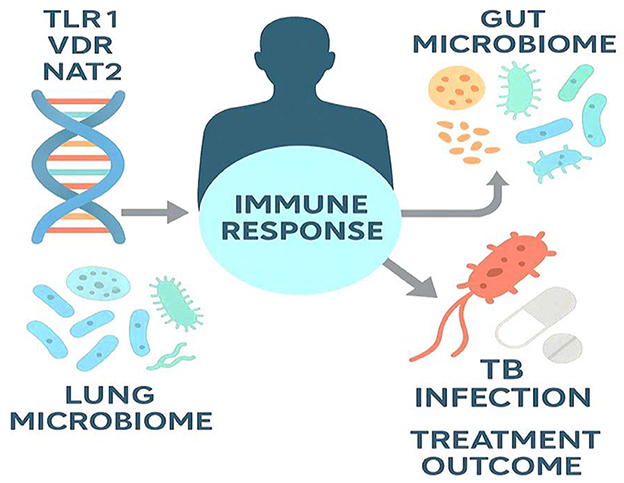
Figure 6. Host genetics and microbiome interactions shaping TB outcomes. The schematic highlights how host genetic variants (e.g., TLR1, VDR, NAT2) and the composition of the gut and lung microbiomes modulate the immune response to M. tuberculosis. These host-specific factors influence infection risk, disease severity, and the success of pharmacologic therapy, underscoring the potential of precision-guided TB management that integrates genomics and microbiome profiling.
4 Treatment options of TB
Accurate diagnosis of TB is crucial for guiding treatment strategies, determining drug selection, and defining therapy duration. Techniques such as sputum smear microscopy, molecular assays, and immunological tests are essential for distinguishing between drug-sensitive and drug-resistant strains of M. tuberculosis (252). Effective diagnostics improve therapeutic outcomes, reduce transmission, and enhance public health interventions. While 85% of patients with drug-sensitive TB benefit from current treatment regimens, only 57% of those with drug-resistant TB achieve successful outcomes (253), highlighting the need for optimized, cost-effective oral therapies (254).
4.1 Therapy for drug-susceptible TB
The standard 6-month regimen for drug-susceptible tuberculosis, established through decades of clinical trials by the British Medical Research Council, remains the global benchmark (255). It comprises a 2-month intensive phase using rifampicin, isoniazid, and pyrazinamide, followed by a 4-month continuation phase with rifampicin and isoniazid. The inclusion of pyrazinamide significantly reduced the treatment duration from 9 to 6 months, enhancing global treatment adherence (255). While this short-course chemotherapy has saved millions of lives, its 6-month duration presents logistical and adherence challenges for both patients and TB programs (256). To address this, efforts have focused on shortening therapy without compromising efficacy. Fluoroquinolones such as moxifloxacin and gatifloxacin have demonstrated accelerated sputum sterilization (257, 258), but three pivotal phase III trials in 2014 failed to prove non-inferiority compared to the standard regimen (259, 260). Nonetheless, subgroup analyses suggest that certain patient populations may benefit from abbreviated regimens (261, 262).
Rifamycin optimization is another approach to treatment shortening. High-dose rifampicin (up to 40 mg/kg/day) is well tolerated and may improve sterilizing activity (263), particularly for TB meningitis. A recent phase III trial demonstrated that a 4-month regimen containing rifapentine, isoniazid, pyrazinamide, and moxifloxacin was non-inferior to the standard 6-month protocol, prompting provisional WHO endorsement in 2022 for drug-susceptible pulmonary TB (264, 265). Newer agents such as pretomanid, a nitroimidazole compound, have also shown promise. A phase II trial reported faster sputum culture conversion compared to standard therapy, though its use was associated with a higher incidence of hepatotoxicity, especially when combined with pyrazinamide (266).
In pediatric cases, shorter therapy may also be effective (267). The SHINE trial, a phase III open-label study, revealed that 16-week regimens were non-inferior to 6-month protocols in children with non-severe, smear-negative TB (268). Similarly, the 2023 TRUNCATE-TB trial evaluated 2-month regimens in adults. Among 674 participants, the 8-week bedaquiline-linezolid group achieved non-inferiority with no significant safety concerns, while the rifampin-linezolid group did not meet the non-inferiority margin (269). These findings highlight a growing opportunity for individualized, shorter-course therapy in selected cases of drug-susceptible TB.
4.2 Isoniazid-resistant, rifampicin-susceptible TB treatment
Isoniazid has served as a cornerstone for both active and latent TB treatment for over five decades (270). However, increasing rates of isoniazid resistance have become a major clinical concern. According to the WHO, ~7.4% of newly diagnosed TB patients and 11.4% of previously treated individuals are resistant to isoniazid while remaining susceptible to rifampicin—a condition classified as isoniazid-resistant, rifampicin-susceptible TB (Hr-TB) (271). This form of resistance is more common than multidrug-resistant TB (MDR-TB), highlighting the urgency of appropriate therapeutic strategies. Failure to manage isoniazid resistance according to guidelines significantly elevates the risk of acquiring additional resistance, particularly to rifampicin and other first-line agents (272, 273). Recognizing this, the WHO currently recommends a 6-month regimen comprising rifampicin, ethambutol, pyrazinamide, and levofloxacin for the treatment of Hr-TB. However, this recommendation remains conditional due to the limited strength of supporting evidence and the absence of randomized controlled trials validating its efficacy (272, 274, 275).
Alternative international guidelines, such as those from European and American expert panels, suggest using pyrazinamide only during the initial 2 months of therapy in selected cases to minimize hepatotoxicity (276). In scenarios where fluoroquinolone resistance is confirmed or suspected, a 6-month combination of rifampicin, ethambutol, and pyrazinamide is typically administered. However, such recommendations are largely based on expert consensus rather than high-quality clinical data. Additionally, the WHO has not yet addressed the potential role of high-dose isoniazid in managing Hr-TB, despite emerging evidence suggesting that its effectiveness may depend on specific resistance-conferring mutations and individual acetylator status (277). Most existing data on Hr-TB treatment stem from observational studies rather than randomized trials, underscoring the critical need for rigorously designed clinical research to inform optimal therapeutic regimens (278).
In a comprehensive analysis involving WHO surveillance data from 156 countries between 2003 and 2017, Dean et al. (271) estimated that Hr-TB prevalence was 7.4% in newly diagnosed cases and 11.4% in previously treated patients. Resistance to pyrazinamide and levofloxacin was comparatively rare, being reported in only 1.8% and 5.3% of the assessed countries, respectively. Whole-genome sequencing (WGS) data from 4,563 clinical samples showed that 78.6% of isoniazid-resistant strains harbored mutations in the katG gene, particularly the Ser315Thr substitution, which is known to confer high-level resistance. A recent 2023 study by Liu et al. (279) explored the genetic mechanisms of isoniazid resistance in M. tuberculosis isolates from China. Out of 4,922 clinical isolates analyzed using WGS, 384 (7.8%) exhibited resistance to isoniazid while remaining rifampicin-sensitive. The katG Ser315Thr mutation was observed in 63.0% of cases, and fabG1 mutations were found in 29.9%. Importantly, resistance rates for pyrazinamide (0.8%), ethambutol, fluoroquinolones (2.3%), and amikacin (0.5%) were low, whereas resistance to streptomycin was significantly higher at 39.6%. These findings support the use of rifampicin, ethambutol, pyrazinamide, and levofloxacin as an effective combination regimen in managing Hr-TB, provided resistance to companion drugs is excluded.
4.3 Multidrug-resistant and rifampicin-resistant TB treatment
Multidrug-resistant tuberculosis (MDR-TB) and rifampicin-resistant TB (RR-TB) continue to pose significant public health challenges globally, with an estimated 450,000 new cases of RR-TB expected in 2021 (280). The global treatment success rate for patients treated for MDR/RR-TB improved from ~50 % in 2012 to 60 % in 2019, rising further to 63 % in 2020 (281). However, it is concerning that 15% of patients diagnosed with MDR/RR-TB do not survive. In December 2022, the WHO released the WHO Consolidated Guidelines on TB, Module 4: Treatment—Drug-Resistant TB Treatment, which replaces the 2020 edition and expands on previous recommendations (265). The 2022 update outlines seven critical areas relevant to the treatment of MDR-TB (282). These areas include strategies for managing MDR/RR-TB, handling isoniazid-resistant TB, monitoring patient responses to therapy, determining the optimal timing for initiating antiretroviral therapy in patients co-infected with HIV, and considering surgical interventions for patients with MDR/RR-TB (265).
The 2022 guidelines recommend two new treatments for MDR/RR-TB (283). First, a 6-month regimen of bedaquiline, pretomanid, linezolid (600 mg), and moxifloxacin is proposed as an alternative to longer regimens (284). Second, an all-oral regimen for 9 months is advised if fluoroquinolone resistance is eliminated, though extended regimens may still be an option (285). In 2018, more than 12,000 patients with MDR/RR-TB (286, 287) experienced a reduction in treatment duration from 18–20 months to 9–12 months. The STREAM Stage 2 trial revealed that 71% of patients on injectable regimens and 83% in the all-oral group had positive outcomes (288), with lower rates of grade 3/4 hearing loss in the all-oral group (2% vs. 9%).
The WHO recommended a 9–12 month bedaquiline regimen for TB cases without fluoroquinolone resistance (265). The TB-PRACTECAL study (289) demonstrated that the BPaLM regimen (bedaquiline, linezolid, pretomanid, moxifloxacin) achieved 89% positive outcomes with fewer side effects than standard treatment, leading to the study's early termination. The NExT trial further reduced treatment duration to 6 months with bedaquiline, linezolid, and fluoroquinolones (290). An interim analysis of the BEAT Tuberculosis trial reported 87% effectiveness with a 6-month regimen (291). The MDR-END trial accomplished 75% success with a non-bedaquiline regimen, showing non-inferiority to the previous 20–24-month treatment duration recommended by the WHO in 2014 (292). Progress in managing MDR/RR-TB is hindered by inadequate drug resistance testing (293). The lack of standardized testing limits diagnostics and undermines clinician trust, while high drug costs restrict availability in many countries (294).
4.4 Treatment of multidrug-resistant/rifampicin-resistant and fluoroquinolone-resistant TB
The treatment of pre-extensively drug-resistant tuberculosis (pre-XDR-TB)—which includes multidrug-resistant (MDR) and rifampicin-resistant (RR) TB with additional fluoroquinolone (FQ) resistance—remains a major clinical challenge. This is largely due to limited therapeutic options, high drug costs, frequent adverse effects, and historically poor outcomes (256). The BEAT-India trial demonstrated promising results, achieving a 91% treatment success rate among 153 patients treated with a 6–9-month regimen containing bedaquiline, linezolid (600 mg daily), clofazimine, and delamanid. Despite the high efficacy, linezolid-related toxicities were significant, though some patients tolerated a reduced dose of 300 mg (295). The NiX-TB trial evaluated a three-drug BPaL regimen (bedaquiline, pretomanid, and linezolid 1,200 mg daily) for 6 months, reporting a 90% success rate but with high rates of adverse effects-−81% developed peripheral neuropathy, and 48% experienced myelosuppression. The subsequent ZeNix trial tested lower linezolid doses (600 or 1200 mg for 2 or 6 months) and showed success rates between 84% and 93%, with improved safety at the 600 mg dose (296, 297).
Based on these findings, the WHO recommends the BPaL regimen for the treatment of fluoroquinolone-resistant TB (298). However, evidence supporting the 600 mg dose recommendation is still evolving, and the optimal duration and dosage of linezolid remain subjects of ongoing investigation (299, 300). A retrospective cohort study conducted by Lee et al. in South Korea between 2005 and 2017 included 129 MDR-TB patients, of whom 30.2% were FQ-resistant and 69.8% were FQ-sensitive. Linezolid was the most frequently prescribed drug in the FQ-resistant group (51.3%), followed by bedaquiline (20.5%) and delamanid (10.3%). Although no significant difference in treatment success was observed between FQ-sensitive and FQ-resistant patients, the study emphasized that individualized regimens incorporating new drugs can improve treatment outcomes for difficult-to-treat TB cases (301).
In support of this approach, Nehru et al. conducted a genomic epidemiological study using the WHO-endorsed GenoType MTBDRsl Ver 2.0 assay to evaluate fluoroquinolone resistance in various TB subtypes. The study found FQ resistance in 33% of MDR-TB cases, 16.5% of RR-TB cases, and 5.4% of non-MDR-TB isolates. The most prevalent mutation was D94G in the gyrA gene, accounting for 49.5% of resistance-related mutations. Alarmingly, 5.12% of isoniazid mono-resistant isolates also exhibited FQ resistance, and isolates harboring the S450L mutation in the rpoB gene were associated with increased risk. These findings underscore the critical need for routine resistance testing before initiating treatment, especially in high-burden regions (302).
4.5 Host-directed therapies
In the ongoing pursuit of more effective TB treatments, two parallel strategies have gained prominence: the development of antimycobacterial drugs and host-directed therapies (HDTs), which aim to enhance the host's immune response (303, 304). HDTs reduce disease burden and potentially counteract antibiotic resistance by minimizing reliance on antimicrobials and enhancing the efficacy of existing drugs (305). These therapies often involve the repurposing of immune-modulating agents that improve pathogen clearance, suppress harmful inflammation, and limit tissue destruction (304, 305). HDT targets key aspects of TB immunopathogenesis, including excessive inflammation (306), host metabolic processes (307), and the immune evasion mechanisms employed by M. tuberculosis (308). By modulating these host factors, HDTs offer an adjunctive approach to conventional anti-TB treatment. As illustrated in Figure 7, HDTs exert their effects via four primary pathways: reducing lung inflammation and tissue damage; enhancing antimicrobial immune responses; promoting direct bactericidal activity; and disrupting granulomas to expose intracellular bacteria to host defenses and antibiotics.
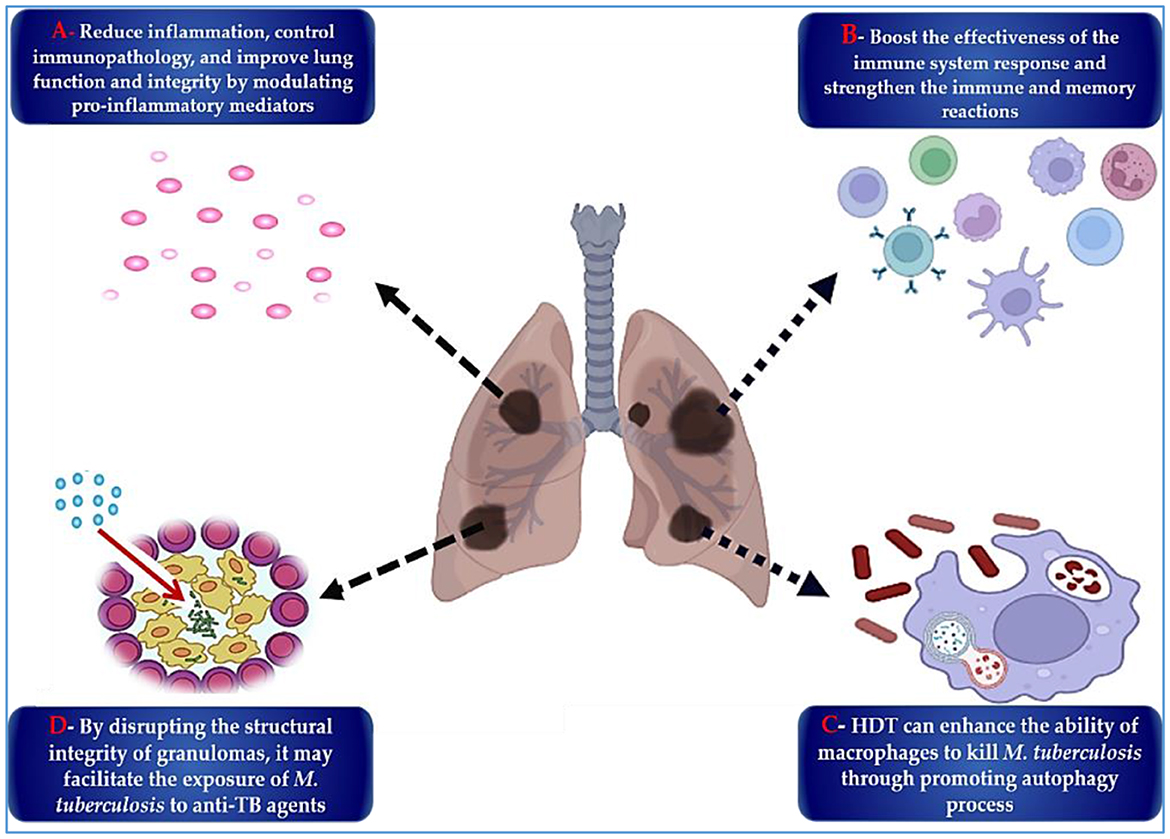
Figure 7. The key HDTs that may enhance the efficacy of M. tuberculosis treatment are as follows: (A) Regulating inflammatory mechanisms and mediators is crucial for reducing inflammation and preventing lung tissue damage, ultimately enhancing lung integrity. (B) Strengthening the host immune and memory responses is essential for overall health. (C) Enhancing host bactericidal mechanisms, such as macrophage-mediated killing of M. tuberculosis and inhibiting bacillary growth, is vital for an effective immune response. (D) The disintegration of granulomas and the release of M. tuberculosis bacilli increase exposure to anti-TB medications.
The overarching objective of HDT is to amplify protective host responses while limiting detrimental ones that contribute to bacterial persistence and lung pathology (309). If successfully implemented, HDT may improve treatment adherence, reduce treatment duration, lower the risk of resistance development, and ultimately improve cure rates with fewer long-term complications (310). Many HDTs currently under investigation are repurposed drugs being tested in preclinical or clinical trials as adjuncts to standard therapy. Notably, agents such as acetylsalicylic acid and statins offer potential advantages due to their established safety profiles and affordability, which may facilitate rapid clinical integration if proven effective (311). Moreover, HDTs may reduce the dependency on prolonged use of repurposed antibiotics such as oxazolidinones and fluoroquinolones, thereby lowering the risk of resistance development in M. tuberculosis and other pathogens (312). Additionally, their anti-inflammatory properties may mitigate pulmonary damage and improve patient outcomes (309). Nonetheless, HDT development faces key challenges. In vitro studies remain essential for initial pharmacological screening (313), while animal models provide valuable insights into immune modulation, treatment efficacy, and disease progression. These models help assess therapeutic effects on bacterial load, tissue pathology, and survival (314). Ultimately, findings from experimental models must be validated in clinical trials to determine real-world applicability and address safety, efficacy, and regulatory considerations (315).
4.6 Digital health tools and telemedicine in TB management
The emergence of digital health technologies and telemedicine platforms has opened new avenues for improving TB care, particularly in settings with limited healthcare access or disrupted services. These tools are increasingly utilized for treatment adherence monitoring, contact tracing, remote diagnostics, and patient education—forming a critical component of modern TB control strategies. One of the most impactful digital innovations is the digital adherence technology (DAT) platform, which includes tools such as 99DOTS, evriMED smart pillboxes, and video directly observed therapy (vDOT) (316). These systems track medication intake in real-time and alert healthcare providers to missed doses. A multicountry evaluation by Thomas et al. (317) found that 99DOTS, a low-cost cellphone-based system, improved medication adherence among TB patients in India, with 93% of doses recorded on the platform and significantly fewer missed doses compared to standard DOT.
Similarly, vDOT platforms like SureAdhere have been adopted by TB programs in the U.S. and Europe, enabling patients to record and submit videos of themselves taking their medications. A randomized controlled trial by Story et al. (318) demonstrated that vDOT was non-inferior to in-person DOT, with higher patient satisfaction and reduced logistical burden on health workers. In rural or crisis-affected regions, telemedicine has played a vital role in bridging access gaps. Real-time video consultations, chat-based triage systems, and remote expert panels facilitate timely diagnosis and clinical decision-making. For example, during the COVID-19 pandemic, the TB REACH initiative implemented a telehealth-supported TB triage model in Pakistan that reduced diagnostic delays by over 40% (319).
Mobile applications (e.g., Nikshay in India, TB eHealth in Uganda) now offer centralized digital platforms to track patient records, laboratory results, and treatment progress, allowing health authorities to monitor outbreaks in real time and improve linkage to care (320). These platforms also integrate AI-based analytics to identify high-risk patients and predict treatment interruptions. Despite their potential, digital TB tools face challenges such as poor internet connectivity, digital illiteracy, privacy concerns, and lack of integration into national health information systems. Nonetheless, as part of the WHO's End TB Strategy, digital health is recognized as a cornerstone for improving TB program efficiency, enhancing patient-centered care, and achieving universal health coverage.
4.7 Advances in TB vaccine development: challenges and new frontiers
The development of an effective TB vaccine remains a pressing global health priority. While the BCG vaccine, first introduced in 1921, is still in use, its protective efficacy is highly variable—especially against pulmonary TB in adolescents and adults (321). With increasing rates of drug-resistant TB and the global burden of latent infections, the need for improved preventive strategies has gained renewed momentum. Recent advances in immunology, systems biology, and vaccine technology have led to the emergence of over 17 vaccine candidates currently in various phases of clinical development (322). Among the most promising candidates is M72/AS01E, a subunit vaccine developed by GSK and Aeras. In a pivotal Phase IIb trial involving 3,573 latently infected adults in Kenya, South Africa, and Zambia, the vaccine demonstrated ~50% efficacy in preventing active TB over a 3-year follow-up period (323). This result marked the first successful demonstration of vaccine-mediated protection against TB in latently infected adults and has led to plans for a large-scale Phase III efficacy trial.
VPM1002, a recombinant BCG vaccine engineered to express listeriolysin O and delete the ureC gene, has shown enhanced immunogenicity and safety in comparison to traditional BCG. Phase II trials conducted in India and South Africa have reported favorable results, and Phase III trials are currently underway in both infants and adults (324). Another advanced candidate is MTBVAC, the first live-attenuated vaccine derived directly from M. tuberculosis rather than M. bovis. Developed by Biofabri and the University of Zaragoza, MTBVAC has completed Phase I and II studies with promising safety and immunogenicity profiles. It is currently in Phase III trials in multiple countries, including high-burden settings in Africa (325). In addition to these, ID93+GLA-SE, a protein–adjuvant vaccine, has shown robust immune responses in early trials and completed Phase IIa evaluation (326).
Ad5Ag85A, a recombinant adenoviral-vectored vaccine, is being evaluated for its ability to enhance mucosal immunity—a key defense mechanism in pulmonary TB. Notably, mRNA-based TB vaccines, such as those developed by BioNTech, have recently entered Phase I clinical testing following strong preclinical evidence of immunogenicity and protection (327). These candidates represent a promising shift toward faster, more adaptable vaccine development strategies. A summary of the leading TB vaccine candidates currently in clinical development is provided in Table 1. These candidates span diverse technological platforms and clinical stages, underscoring the momentum and innovation in the TB vaccine pipeline.
Despite the growing diversity of vaccine strategies, several challenges continue to hinder progress. These include the absence of validated immune correlates of protection, the high costs and lengthy timelines associated with efficacy trials, and the biological complexity of latent TB infection. Furthermore, regulatory uncertainties, limited commercial incentives, and diminished prioritization in high-income countries where TB incidence is low contribute to underinvestment in vaccine development (328). Nonetheless, this surge of innovation marks a turning point in TB vaccine research. Continued support for translational science, robust public–private partnerships, and equitable implementation planning will be essential. With strategic investments, the global community may finally be on the cusp of introducing the first new TB vaccine in over a century—one that can significantly alter the trajectory of the epidemic by preventing disease, reducing relapse, and mitigating the spread of drug-resistant strains.
5 Future directions for diagnosis and treatment of TB
Significant strides have been made in the diagnosis and treatment of TB, driven by advances in point-of-care technologies, innovative therapeutics, and integrated public health frameworks. The incorporation of molecular assays, immunodiagnostics, mass spectrometry, and CRISPR-based platforms has markedly improved diagnostic precision and speed, enabling real-time detection of M. tuberculosis and its resistance profiles. These advancements support earlier and more targeted treatment initiation, particularly in high-burden settings. Therapeutically, the emergence of host-directed therapies aimed at modulating the immune response holds promise for enhancing treatment efficacy, reducing tissue damage, and shortening therapy duration. In parallel, vaccine development has progressed with the design of novel candidates intended to bolster both preventive and therapeutic immunity against TB.
Public health strategies are also evolving to emphasize community-centered approaches and integrated care models, particularly for individuals with co-morbid conditions such as HIV and diabetes. These models aim to improve patient retention, treatment adherence, and overall outcomes. Moreover, increased global investment in TB research and the formation of public-private partnerships are accelerating innovation, facilitating the translation of scientific breakthroughs into scalable interventions. Together, these developments signify a shift toward a more holistic and forward-thinking TB control paradigm—one that prioritizes early detection, personalized treatment, and equitable access to care in the global effort to reduce TB incidence and mortality.
6 Implementation challenges and cost-effectiveness of advanced TB technologies
The effective integration of diagnostic and therapeutic innovations into TB control programs hinges not only on technological readiness but also on their cost-effectiveness, health system compatibility, and equitable access—key concerns addressed by implementation science (Figure 8) (329). This discipline evaluates how well evidence-based tools function in real-world clinical environments, particularly in LMICs. Despite the promise of technologies such as WGS, CRISPR-based diagnostics, and new regimens like BPaLM, their widespread adoption remains limited. Logistical, infrastructural, and financial constraints often impede implementation. For example, although WGS provides comprehensive and rapid drug-resistance profiling, its use in resource-limited settings is curtailed by high setup costs, the requirement for skilled personnel, and limited bioinformatics infrastructure. A study in the Kyrgyz Republic reported an initial capital investment of over $220,000, with early per-sample costs averaging $277—figures that present major barriers to scale-up (149).
Figure 8. Implementation challenges and strategic solutions for advanced TB technologies. This infographic outlines four major barriers to implementing innovative TB diagnostics and treatments—high costs, limited infrastructure and workforce, fragmented healthcare systems, and regulatory or funding hurdles—and pairs each with corresponding solutions. These include cost-effectiveness analysis, training programs, service integration, and policy advocacy, all of which are critical to ensuring the real-world success of TB innovations in diverse health system contexts.
Cost-effectiveness analyses are vital for policy-making in settings with limited resources. Vassall et al. (330) found that molecular diagnostics like Xpert MTB/RIF outperform smear microscopy in both effectiveness and cost, particularly in high-burden settings by reducing diagnostic delays. Similarly, Isaacs et al. (331) showed that digital adherence tools—such as video-observed therapy and SMS reminders—can be cost-saving, especially when accounting for patient-incurred costs, although effectiveness varies by context. Healthcare system fragmentation, especially in countries with parallel public and private sectors, further complicates implementation. Pai and Pakdil (332) noted that expanding diagnostic access without concurrent investments in healthcare quality, supply chains, workforce capacity, and accountability mechanisms yields limited benefits.
Sustainable adoption is also hindered by regulatory barriers, fragmented funding streams, and donor-driven priorities that may neglect long-term system strengthening. As a result, validated technologies like Line Probe Assays or video-DOT platforms often remain underutilized. To overcome these challenges, national TB strategies must incorporate implementation research, health technology assessments, and economic modeling. Embedding these components in TB innovation pipelines will help ensure that emerging tools are not only scientifically effective but also affordable, scalable, and contextually appropriate for diverse health systems.
7 Conclusions
Despite over a century of medical progress, tuberculosis remains a formidable global health challenge, exacerbated by the rise of antimicrobial resistance and the limitations of existing diagnostic and therapeutic tools. This review has highlighted how recent innovations—ranging from advanced molecular diagnostics and next-generation sequencing to novel therapeutics and promising vaccine candidates—are reshaping the landscape of TB control. However, technological advancement alone is insufficient. The true impact of these innovations depends on their integration into health systems, especially in LMICs where the burden is highest. Effective implementation must be guided by cost-effectiveness analyses, health equity considerations, and robust infrastructure development. Furthermore, the urgency to overcome regulatory, logistical, and funding barriers cannot be overstated if we are to bridge the gap between scientific discovery and public health impact. Encouragingly, the TB vaccine pipeline has expanded significantly, offering hope for a long-overdue replacement or complement to the BCG vaccine. To turn the tide against TB, a multifaceted approach is essential—one that harmonizes scientific innovation, political will, cross-sectoral partnerships, and community engagement. Only through such coordinated global efforts can we hope to achieve the goals of the End TB Strategy and finally relegate this ancient disease to history.
Author contributions
AE: Supervision, Formal analysis, Writing – original draft, Visualization, Data curation, Methodology, Investigation, Validation, Writing – review & editing. EM: Writing – review & editing, Validation, Writing – original draft, Formal analysis, Methodology, Visualization, Data curation, Investigation. HE: Visualization, Validation, Writing – review & editing, Writing – original draft. RA: Writing – review & editing, Visualization, Validation, Writing – original draft. ATE: Visualization, Writing – original draft, Writing – review & editing, Validation. FA: Funding acquisition, Writing – review & editing, Writing – original draft, Visualization, Validation. SA: Visualization, Writing – original draft, Validation, Writing – review & editing. AAlg: Writing – review & editing, Writing – original draft, Validation, Visualization. KAlm: Writing – original draft, Visualization, Writing – review & editing, Validation. IA: Validation, Writing – review & editing, Writing – original draft, Visualization. AAlb: Visualization, Validation, Writing – review & editing, Writing – original draft. KAla: Writing – original draft, Visualization, Validation, Writing – review & editing. MI: Writing – original draft, Visualization, Validation, Writing – review & editing. AAlm: Investigation, Writing – review & editing, Writing – original draft, Validation, Visualization. FD: Writing – review & editing, Data curation, Funding acquisition, Visualization. AA-O: Writing – review & editing, Data curation, Funding acquisition, Visualization.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The researchers would like to thank the Deanship of Graduate Studies and Scientific Research at Qassim University for financial support (QU-APC-2025).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Gagneux S. Ecology and evolution of mycobacterium tuberculosis. Nat Rev Microbiol. (2018) 16:202–13. doi: 10.1038/nrmicro.2018.8
2. Huang Y, Ai L, Wang X, Sun Z, Wang F. Review and updates on the diagnosis of tuberculosis. J Clin Med. (2022) 11:5826. doi: 10.3390/jcm11195826
3. Silva S, Arinaminpathy N, Atun R, Goosby E, Reid M. Economic impact of tuberculosis mortality in 120 countries and the cost of not achieving the sustainable development goals tuberculosis targets: a full-income analysis. Lancet Global Health. (2021) 9:e1372–e9. doi: 10.1016/S2214-109X(21)00299-0
4. Bhalla AS, Goyal A, Guleria R, Gupta AK. Chest tuberculosis: radiological review and imaging recommendations. Indian J Radiol Imag. (2015) 25:213–25. doi: 10.4103/0971-3026.161431
5. Natarajan A, Beena PM, Devnikar AV, Mali S. A systemic review on tuberculosis. Indian J Tuberc. (2020) 67:295–311. doi: 10.1016/j.ijtb.2020.02.005
6. Sharma SK, Mohan A, Kohli M. Extrapulmonary tuberculosis. Expert Rev Respir Med. (2021) 15:931–48. doi: 10.1080/17476348.2021.1927718
7. Pai M, Behr M, Dowdy D, Dheda K, Divangahi M, Boehme C, et al. Nature reviews disease primers. Tuberculosis. (2016) 2:16076. doi: 10.1038/nrdp.2016.76
8. Cronan MR. In the thick of it: formation of the tuberculous granuloma and its effects on host and therapeutic responses. Front Immunol. (2022) 13:820134. doi: 10.3389/fimmu.2022.820134
9. Alsayed SS, Gunosewoyo H. Tuberculosis: pathogenesis, current treatment regimens and new drug targets. Int J Mol Sci. (2023) 24:5202. doi: 10.3390/ijms24065202
10. Qin Y, Chen Y, Chen J, Xu K, Xu F, Shi J. The relationship between previous pulmonary tuberculosis and risk of lung cancer in the future. Infect Agent Cancer. (2022) 17:20. doi: 10.1186/s13027-022-00434-2
11. Hwang SY, Kim JY, Lee HS, Lee S, Kim D, Kim S, et al. Pulmonary tuberculosis and risk of lung cancer: a systematic review and meta-analysis. J Clin Med. (2022) 11:765. doi: 10.3390/jcm11030765
12. Ho L-J, Yang H-Y, Chung C-H, Chang W-C, Yang S-S, Sun C-A, et al. Increased risk of secondary lung cancer in patients with tuberculosis: a nationwide, population-based cohort study. PLoS ONE. (2021) 16:e0250531. doi: 10.1371/journal.pone.0250531
13. WHO. Global Tuberculosis Report 2021 (2021). Available online at: https://www.who.int/publications/digital/global-tuberculosis-report-2021/tb-diagnosis-treatment/treatment (Accessed March 11, 2025).
14. Tawfick MM, Badawy MSE, Taleb MH, Menofy NGE. Tuberculosis diagnosis and detection of drug resistance: a comprehensive updated review. J Pure Appl Microbiol. (2023) 17:2849–66. doi: 10.22207/JPAM.17.4.56
15. MacGregor-Fairlie M, Wilkinson S, Besra GS, Goldberg Oppenheimer P. Tuberculosis diagnostics: overcoming ancient challenges with modern solutions. Emerg Top Life Sci. (2020) 4:435–48. doi: 10.1042/ETLS20200335
16. WHO. Global Tuberculosis Report 2023 (2023). Available online at: https://www.who.int/publications/i/item/9789240083851 [Accessed July 09, 2025].
17. Ahmed I, Tiberi S, Farooqi J, Jabeen K, Yeboah-Manu D, Migliori GB, et al. Non-tuberculous mycobacterial infections—a neglected and emerging problem. Int J Infect Dis. (2020) 92:S46–50. doi: 10.1016/j.ijid.2020.02.022
18. Houben RM, Dodd PJ. The global burden of latent tuberculosis infection: a re-estimation using mathematical modelling. PLoS Med. (2016) 13:e1002152. doi: 10.1371/journal.pmed.1002152
19. Lewinsohn DM, Leonard MK, LoBue PA, Cohn DL, Daley CL, Desmond E, et al. Official American Thoracic Society/Infectious Diseases Society of America/Centers for Disease Control and prevention clinical practice guidelines: diagnosis of tuberculosis in adults and children. Clin Infect Dis. (2017) 64:e1–e33. doi: 10.1093/cid/ciw694
20. WHO. Global Tuberculosis Report 2024 (2024). Available online at: https://www.who.int/teams/global-programme-on-tuberculosis-and-lung-health/tb-reports/global-tuberculosis-report-2024/tb-disease-burden/1-1-tb-incidence (Accessed July 25, 2025).
21. Network GT, Casco N, Jorge AL, Palmero DJ, Alffenaar J-W, Fox GJ, et al. Long-term outcomes of the global tuberculosis and COVID-19 co-infection cohort. Eur Respir J. (2023) 62:2300563. doi: 10.1183/13993003.00563-2023
22. García-García J-M, Blanc F-X, Buonsenso D, Centis R, Codecasa LR, D'Ambrosio L, et al. COVID-19 hampered diagnosis of TB infection in France, Italy, Spain and the United Kingdom. Arch Bronconeumol. (2022) 58:783. doi: 10.1016/j.arbres.2022.07.013
23. Goletti D, Al-Abri S, Migliori GB, Arlehamn CL, Haldar P, Sundling C, et al. World tuberculosis day 2024 theme “Yes! We Can End Tb” can be made a reality through concerted global efforts that advance detection, diagnosis, and treatment of tuberculosis infection and disease. Int J Infect Dis. (2024) 141:106993. doi: 10.1016/j.ijid.2024.106993
24. Mancuso G, Midiri A, De Gaetano S, Ponzo E, Biondo C. Tackling drug-resistant tuberculosis: new challenges from the old pathogen mycobacterium tuberculosis. Microorganisms. (2023) 11:2277. doi: 10.3390/microorganisms11092277
25. Soto J, Linsley C, Song Y, Chen B, Fang J, Neyyan J, et al. Engineering materials and devices for the prevention, diagnosis, and treatment of COVID-19 and infectious diseases. Nanomaterials. (2023) 13:2455. doi: 10.3390/nano13172455
26. Teibo TKA. Andrade RLdP, Rosa RJ, de Abreu PD, Olayemi OA, Alves YM, et al. Barriers that interfere with access to tuberculosis diagnosis and treatment across countries globally: a systematic review. ACS Infect Dis. (2024) 10:2600–14. doi: 10.1021/acsinfecdis.4c00466
27. Karanja PW, Mulaku MN, Ochodo EA, A. Synthesis of qualitative evidence of barriers and facilitators in implementing guidelines for TB testing in healthcare settings. Implement Sci Commun. (2024) 5:30. doi: 10.1186/s43058-024-00565-0
28. Procop GW. Laboratory diagnosis and susceptibility testing for mycobacterium tuberculosis. In: Tuberculosis and Nontuberculous Mycobacterial Infections. Washington, DC: ASM Press (2017). p. 45–58.
29. Azadi D, Motallebirad T, Ghaffari K, Shojaei H. Mycobacteriosis and tuberculosis: laboratory diagnosis. Open Microbiol J. (2018) 12:41. doi: 10.2174/1874285801812010041
30. Asadi L, Croxen M, Heffernan C, Dhillon M, Paulsen C, Egedahl ML, et al. How much do smear-negative patients really contribute to tuberculosis transmissions? Re-examining an old question with new tools. EClinicalMedicine. (2022) 43:101250. doi: 10.1016/j.eclinm.2021.101250
31. WHO. Noncommercial Culture and Drug-Susceptibility Testing Methods for Screening Patients at Risk for Multidrug-Resistant Tuberculosis: Policy Statement (2011). Available online at: https://iris.who.int/bitstream/handle/10665/44601/9789241501620_eng.pdf (Accessed March 17, 2025).
32. Kaufmann SH, Schaible UE. 100th anniversary of robert koch's nobel prize for the discovery of the Tubercle Bacillus. Trends Microbiol. (2005) 13:469–75. doi: 10.1016/j.tim.2005.08.003
33. WHO. Global Tuberculosis Report 2013 (2013). Available online at: https://www.who.int/publications/i/item/9789241564656 (Accessed March 12, 2025).
34. Gopalaswamy R, Dusthackeer VA, Kannayan S, Subbian S. Extrapulmonary tuberculosis—an update on the diagnosis, treatment and drug resistance. J Respir. (2021) 1:141–64. doi: 10.3390/jor1020015
35. Flynn JL, Chan J. Tuberculosis: latency and reactivation. Infect Immun. (2001) 69:4195–201. doi: 10.1128/IAI.69.7.4195-4201.2001
36. Philips JA, Ernst JD. Tuberculosis pathogenesis and immunity. Ann Rev Pathol Mech Dis. (2012) 7:353–84. doi: 10.1146/annurev-pathol-011811-132458
37. Leung AN. Pulmonary tuberculosis: the essentials. Radiology. (1999) 210:307–22. doi: 10.1148/radiology.210.2.r99ja34307
38. Conrad WH, Osman MM, Shanahan JK, Chu F, Takaki KK, Cameron J, et al. Mycobacterial Esx-1 secretion system mediates host cell lysis through bacterium contact-dependent gross membrane disruptions. Proc Nat Acad Sci. (2017) 114:1371–6. doi: 10.1073/pnas.1620133114
39. Gao LY, Guo S, McLaughlin B, Morisaki H, Engel JN, Brown EJ, et al. Mycobacterial virulence gene cluster extending rd1 is required for cytolysis, bacterial spreading and Esat-6 secretion. Mol Microbiol. (2004) 53:1677–93. doi: 10.1111/j.1365-2958.2004.04261.x
40. Tomlin H, Piccinini AM, A. Complex interplay between the extracellular matrix and the innate immune response to microbial pathogens. Immunology. (2018) 155:186–201. doi: 10.1111/imm.12972
41. Luies L, Du Preez I. The echo of pulmonary tuberculosis: mechanisms of clinical symptoms and other disease-induced systemic complications. Clin Microbiol Rev. (2020) 33:e00036–20. doi: 10.1128/CMR.00036-20
42. Schluger NW. The pathogenesis of tuberculosis: the first one hundred (and twenty-three) years. Am J Respir Cell Mol Biol. (2005) 32:251–6. doi: 10.1165/rcmb.F293
43. Russell DG, Cardona P-J, Kim M-J, Allain S, Altare F. Foamy macrophages and the progression of the human tuberculosis granuloma. Nat Immunol. (2009) 10:943–8. doi: 10.1038/ni.1781
44. Huszar S, Chibale K, Singh V. The quest for the holy grail: new antitubercular chemical entities, targets and strategies. Drug Discov Today. (2020) 25:772–80. doi: 10.1016/j.drudis.2020.02.003
45. Marrakchi H, Lanéelle M-A, Daffé M. Mycolic acids: structures, biosynthesis, and beyond. Chem Biol. (2014) 21:67–85. doi: 10.1016/j.chembiol.2013.11.011
46. Grosset J. Mycobacterium tuberculosis in the extracellular compartment: an underestimated adversary. Antimicrobial Agents Chemother. (2003) 47:833–6. doi: 10.1128/AAC.47.3.833-836.2003
47. Gholoobi A, Masoudi-Kazemabad A, Meshkat M, Meshkat Z. Comparison of culture and Pcr methods for diagnosis of mycobacterium tuberculosis in different clinical specimens. Jundishapur J Microbiol. (2014) 7:e8939. doi: 10.5812/jjm.8939
48. Rageade F, Picot N, Blanc-Michaud A, Chatellier S, Mirande C, Fortin E, et al. Performance of solid and liquid culture media for the detection of mycobacterium tuberculosis in clinical materials: meta-analysis of recent studies. Eur J Clin Microbiol Infect Dis. (2014) 33:873–83. doi: 10.1007/s10096-014-2105-z
49. Campelo TA, Cardoso de. Sousa PR, Nogueira LdL, Frota CC, Zuquim Antas PR. Revisiting the methods for detecting mycobacterium tuberculosis: what has the new millennium brought thus far? Access microbiology. (2021) 3:000245. doi: 10.1099/acmi.0.000245
50. Acharya B, Acharya A, Gautam S, Ghimire SP, Mishra G, Parajuli N, et al. Advances in diagnosis of tuberculosis: an update into molecular diagnosis of mycobacterium tuberculosis. Mol Biol Rep. (2020) 47:4065–75. doi: 10.1007/s11033-020-05413-7
51. Hobby GL, Holman AP, Iseman MD, Jones JM. Enumeration of tubercle bacilli in sputum of patients with pulmonary tuberculosis. Antimicrobial Agents Chemother. (1973) 4:94–104. doi: 10.1128/AAC.4.2.94
52. Joloba ML, Johnson JL, Feng P-JI, Bozeman L, Goldberg SV, Morgan K, et al. What is the most reliable solid culture medium for tuberculosis treatment trials? Tuberculosis. (2014) 94:311–6. doi: 10.1016/j.tube.2014.03.002
53. Naveen G. PeeraPur Bv. Comparison of the lowenstein-jensen medium, the middlebrook 7h10 medium and mb/bact for the isolation of mycobacterium tuberculosis (Mtb) from clinical specimens. J Clin Diagn Res. (2012) 6:1704. doi: 10.7860/JCDR/2012/4603.2635
54. Bartolomeu-Gonçalves G. Souza JMd, Fernandes BT, Spoladori LFA, Correia GF, Castro IMd, et al. Tuberculosis diagnosis: current, ongoing, and future approaches. Diseases. (2024) 12:202. doi: 10.3390/diseases12090202
55. Ma Y, Fan J, Li S, Dong L, Li Y, Wang F, et al. Comparison of lowenstein-jensen medium and mgit culture system for recovery of mycobacterium tuberculosis from abscess samples. Diagn Microbiol Infect Dis. (2020) 96:114969. doi: 10.1016/j.diagmicrobio.2019.114969
56. Kumar H, Singh VA, Mehta S, Nagpal S, Bala R, Biswas D. Comparative evaluation of conventional media with bactec mgit 960 for detection of mycobacterium tuberculosis in clinically suspected cases of pulmonary and extra-pulmonary tuberculosis. Indian J Public Health. (2020) 11:818–22. doi: 10.37506/v11/i2/2020/ijphrd/194913
57. Abdelhay A, Magnin J-P, Gondrexon N, Baup S, Willison J. Adaptation of a mycobacterium strain to phenanthrene degradation in a biphasic culture system: influence on interfacial area and droplet size. Biotechnol Lett. (2009) 31:57–63. doi: 10.1007/s10529-008-9832-0
58. Bammann RH, Zamarioli LA, Pinto VS, Vázquez CM, Litvoc MN, Klautau GB, et al. High prevalence of drug-resistant tuberculosis and other mycobacteria among HIV-infected patients in Brazil: a systematic review. Memórias do Instituto Oswaldo Cruz. (2010) 105:838–41. doi: 10.1590/S0074-02762010000600019
59. Asmar S, Drancourt M. Rapid culture-based diagnosis of pulmonary tuberculosis in developed and developing countries. Front Microbiol. (2015) 6:1184. doi: 10.3389/fmicb.2015.01184
60. WHO. Policy Framework for Implementing New Tuberculosis Diagnostics (2010). Available online at: https://iris.who.int/bitstream/handle/10665/162712/9789241508612_eng.pdf (Accessed Juli 25, 2025).
61. Horne DJ, Royce SE, Gooze L, Narita M, Hopewell PC, Nahid P, et al. Sputum monitoring during tuberculosis treatment for predicting outcome: systematic review and meta-analysis. Lancet Infect Dis. (2010) 10:387–94. doi: 10.1016/S1473-3099(10)70071-2
62. Battaglioli T, Rintiswati N, Martin A, Palupi K, Bernaerts G, Dwihardiani B, et al. Comparative performance of thin layer agar and Löwenstein–Jensen culture for diagnosis of tuberculosis. Clin Microbiol Infect. (2013) 19:E502–E8. doi: 10.1111/1469-0691.12265
63. Zaporojan N, Negrean RA, Hodişan R, Zaporojan C, Csep A, Zaha DC. Evolution of laboratory diagnosis of tuberculosis. Clin Pract. (2024) 14:388–416. doi: 10.3390/clinpract14020030
64. Srivastava S, Chapagain M, Gumbo T. Effect of specimen processing, growth supplement, and different metabolic population on mycobacterium tuberculosis laboratory diagnosis. PLoS ONE. (2020) 15:e0230927. doi: 10.1164/ajrccm-conference.2020.201.1_MeetingAbstracts.A6086
65. Hasan M, Munshi SK, Momi MSB, Rahman F, Noor R. Evaluation of the effectiveness of bactec Mgit 960 for the detection of mycobacteria in Bangladesh. Int J Mycobacteriol. (2013) 2:214–9. doi: 10.1016/j.ijmyco.2013.09.001
66. Brent AJ, Mugo D, Musyimi R, Mutiso A, Morpeth S, Levin M, et al. Performance of the Mgit Tbc identification test and meta-analysis of mpt64 assays for identification of the mycobacterium tuberculosis complex in liquid culture. J Clin Microbiol. (2011) 49:4343–6. doi: 10.1128/JCM.05995-11
67. Oommen S, Banaji N. Laboratory diagnosis of tuberculosis: advances in technology and drug susceptibility testing. Indian J Med Microbiol. (2017) 35:323–31. doi: 10.4103/ijmm.IJMM_16_204
68. Ryu YJ. Diagnosis of pulmonary tuberculosis: recent advances and diagnostic algorithms. Tuberc Respir Dis. (2015) 78:64. doi: 10.4046/trd.2015.78.2.64
69. Zabaleta-Vanegas A, Llerena-Polo C, Orjuela-Gamboa D, Valbuena-Arias Y, García-González L, Mejía-Restrepo G, et al. Evaluation of Bactec™ Mgit™ 960 and the nitrate reductase assay in the national laboratory network of Colombia. Int J Tuberc Lung Dis. (2013) 17:125–8. doi: 10.5588/ijtld.11.0661
70. Chopra K, Singh S. Tuberculosis: newer diagnostic tests: applications and limitations. Indian J Tuberc. (2020) 67:S86–90. doi: 10.1016/j.ijtb.2020.09.025
71. Evelina L, Olga C, Alina M, Adriana N, Stela K, Alexandru C. Clinical and paraclinical similarities and differences between pulmonary tuberculosis and community-acquired pneumonia. Public Health Econ Manag Med. (2022) 93-S:258–64. doi: 10.1007/s10389-022-01732-x
72. Shah MI, Mishra S, Yadav VK, Chauhan A, Sarkar M, Sharma SK, et al. Ziehl–Neelsen sputum smear microscopy image database: a resource to facilitate automated bacilli detection for tuberculosis diagnosis. J Med Imag. (2017) 4:027503. doi: 10.1117/1.JMI.4.2.027503
73. Zurac S, Mogodici C, Poncu T, Trăscău M, Popp C, Nichita L, et al. A new artificial intelligence-based method for identifying mycobacterium tuberculosis in Ziehl–Neelsen stain on tissue. Diagnostics. (2022) 12:1484. doi: 10.3390/diagnostics12061484
74. Ulukanligil M, Aslan G, Tasçi S, A. Comparative study on the different staining methods and number of specimens for the detection of acid fast bacilli. Memórias do Instituto Oswaldo Cruz. (2000) 95:855–8. doi: 10.1590/S0074-02762000000600019
75. Parsons LM, Somoskövi Á, Gutierrez C, Lee E, Paramasivan C. Abimiku Al, et al. Laboratory diagnosis of tuberculosis in resource-poor countries: challenges and opportunities. Clin Microbiol Rev. (2011) 24:314–50. doi: 10.1128/CMR.00059-10
76. Caulfield AJ, Wengenack NL. Diagnosis of active tuberculosis disease: from microscopy to molecular techniques. J Clin Tuberc Other Mycobact Dis. (2016) 4:33–43. doi: 10.1016/j.jctube.2016.05.005
77. Zingue D, Weber P, Soltani F, Raoult D, Drancourt M. Automatic microscopic detection of mycobacteria in sputum: a proof-of-concept. Sci Rep. (2018) 8:11308. doi: 10.1038/s41598-018-29660-8
78. Erokhina MV, Lepekha LN, Voronezhskaya EE, Nezlin LP, Avdienko VG, Ergeshov AE. Application of laser scanning confocal microscopy for the visualization of M. Tuberculosis in Lung Tissue samples with weak Ziehl–Neelsen staining. J Clin Med. (2019) 8:1185. doi: 10.3390/jcm8081185
79. Sua LF, Bolaños JE, Maya J, Sánchez A, Medina G, Zúñiga-Restrepo V, et al. Detection of mycobacteria in paraffin-embedded Ziehl–Neelsen-stained tissues using digital pathology. Tuberculosis. (2021) 126:102025. doi: 10.1016/j.tube.2020.102025
80. Dong B, He Z, Li Y, Xu X, Wang C, Zeng J. Improved conventional and new approaches in the diagnosis of tuberculosis. Front Microbiol. (2022) 13:924410. doi: 10.3389/fmicb.2022.924410
81. Steingart KR, Henry M, Ng V, Hopewell PC, Ramsay A, Cunningham J, et al. Fluorescence vs. conventional sputum smear microscopy for tuberculosis: a systematic review. Lancet Infect Dis. (2006) 6:570–81. doi: 10.1016/S1473-3099(06)70578-3
82. Boyd JC, Marr JJ. Decreasing reliability of acid-fast smear techniques for detection of tuberculosis. Ann Intern Med. (1975) 82:489–92. doi: 10.7326/0003-4819-82-4-489
83. Minion J, Shenai S, Vadwai V, Tipnis T, Greenaway C, Menzies D, et al. Fading of auramine-stained mycobacterial smears and implications for external quality assurance. J Clin Microbiol. (2011) 49:2024–6. doi: 10.1128/JCM.00507-11
84. Kanade S, Nataraj G, Ubale M, Mehta P. Fluorescein diacetate vital staining for detecting viability of acid-fast bacilli in patients on antituberculosis treatment. Int J Mycobacteriol. (2016) 5:294–8. doi: 10.1016/j.ijmyco.2016.06.003
85. Datta S, Sherman JM, Bravard MA, Valencia T, Gilman RH, Evans CA. Clinical evaluation of tuberculosis viability microscopy for assessing treatment response. Clin Infect Dis. (2015) 60:1186–95. doi: 10.1093/cid/ciu1153
86. Lewis JJ, Chihota VN, van der Meulen M, Fourie PB, Fielding KL, Grant AD, et al. “Proof-of-Concept” evaluation of an automated sputum smear microscopy system for tuberculosis diagnosis. PLoS ONE. (2012) 7:e50173. doi: 10.1371/journal.pone.0050173
87. Dzodanu EG, Afrifa J, Acheampong DO, Dadzie I. Diagnostic yield of fluorescence and Ziehl-Neelsen staining techniques in the diagnosis of pulmonary tuberculosis: a comparative study in a district health facility. Tuberc Res Treat. (2019) 2019:4091937. doi: 10.1155/2019/4091937
88. Masali HT, Takpere A, Shahapur P. A comparative study of Ziehl-Neelsen Stain and fluorescent stain microscopy in the diagnosis of pulmonary tuberculosis. J Pure Appl Microbiol. (2021) 15:2027–33. doi: 10.22207/JPAM.15.4.24
89. Mor P, Dahiya B, Parshad S, Gulati P, Mehta PK. Recent updates in diagnosis of abdominal tuberculosis with emphasis on nucleic acid amplification tests. Expert Rev Gastroenterol Hepatol. (2022) 16:33–49. doi: 10.1080/17474124.2022.2021068
90. Khandekar P, Amin A, Reddi P, Banwalikar J, Shivannaure T, Katoch V. Evaluation of Pcr based test for the detection of mycobacterium tuberculosis in coded sputum specimens. Indian J Med Res. (1994) 100:167–71.
91. O'Sullivan CE, Miller DR, Schneider PS, Roberts GD. Evaluation of gen-probe amplified mycobacterium tuberculosis direct test by using respiratory and nonrespiratory specimens in a tertiary care center laboratory. J Clin Microbiol. (2002) 40:1723–7. doi: 10.1128/JCM.40.5.1723-1727.2002
92. MacLean E, Kohli M, Weber SF, Suresh A, Schumacher SG, Denkinger CM, et al. Advances in molecular diagnosis of tuberculosis. J Clin Microbiol. (2020) 58:e01582–19. doi: 10.1128/JCM.01582-19
93. Lin C-R, Wang H-Y, Lin T-W, Lu J-J, Hsieh JC-H, Wu M-H. Development of a two-step nucleic acid amplification test for accurate diagnosis of the mycobacterium tuberculosis complex. Sci Rep. (2021) 11:5750. doi: 10.1038/s41598-021-85160-2
94. Nikam C, Jagannath M, Narayanan MM, Ramanabhiraman V, Kazi M, Shetty A, et al. Rapid diagnosis of mycobacterium tuberculosis with truenat Mtb: a near-care approach. PLoS ONE. (2013) 8:e51121. doi: 10.1371/journal.pone.0051121
95. Pai M, Flores LL, Pai N, Hubbard A, Riley LW, Colford JM. Diagnostic accuracy of nucleic acid amplification tests for tuberculous meningitis: a systematic review and meta-analysis. Lancet Infect Dis. (2003) 3:633–43. doi: 10.1016/S1473-3099(03)00772-2
96. Flores LL, Pai M, Colford JM, Riley LW. In-house nucleic acid amplification tests for the detection of mycobacterium tuberculosis in sputum specimens: meta-analysis and meta-regression. BMC Microbiol. (2005) 5:1–10. doi: 10.1186/1471-2180-5-55
97. Bisognin F, Lombardi G, Lombardo D, Re MC, Dal Monte P. improvement of mycobacterium tuberculosis detection by xpert mtb/rif ultra: a head-to-head comparison on xpert-negative samples. PLoS ONE. (2018) 13:e0201934. doi: 10.1371/journal.pone.0201934
98. Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. (1998) 62:725–74. doi: 10.1128/MMBR.62.3.725-774.1998
99. Alonso H, Samper S, Martín C, Otal I. Mapping is 6110 in high-copy number mycobacterium tuberculosis strains shows specific insertion points in the Beijing genotype. BMC Genom. (2013) 14:1–11. doi: 10.1186/1471-2164-14-422
100. Dorman SE, Schumacher SG, Alland D, Nabeta P, Armstrong DT, King B, et al. Xpert Mtb/Rif ultra for detection of mycobacterium tuberculosis and rifampicin resistance: a prospective multicentre diagnostic accuracy study. Lancet Infect Dis. (2018) 18:76–84. doi: 10.1016/S1473-3099(17)30691-6
101. Zhang M, Xue M, He J-q. diagnostic accuracy of the new xpert Mtb/Rif ultra for tuberculosis disease: a preliminary systematic review and meta-analysis. Int J Infect Dis. (2020) 90:35–45. doi: 10.1016/j.ijid.2019.09.016
102. Allahyartorkaman M, Mirsaeidi M, Hamzehloo G, Amini S, Zakiloo M, Nasiri MJ. Low diagnostic accuracy of xpert Mtb/Rif assay for extrapulmonary tuberculosis: a multicenter surveillance. Sci Rep. (2019) 9:18515. doi: 10.1038/s41598-019-55112-y
103. Atherton RR, Cresswell FV, Ellis J, Kitaka SB, Boulware DR. Xpert Mtb/Rif ultra for tuberculosis testing in children: a mini-review and commentary. Front Pediatr. (2019) 7:34. doi: 10.3389/fped.2019.00034
104. Kaur J, Singh J, Mishra P. Comparative evaluation of Cfx96tm real time Pcr with conventional Pcr for rapid diagnosis of mycobacterium tuberculosis complex in clinical isolates. Arch Clin Microbiol. (2016) 7:1–9. doi: 10.4172/1989-8436.100053
105. Makiani MJ, Davoodian P, Baghershiroodi M, Nejatizadeh AA, Fakkhar F, Zangeneh M, et al. Urine-based nested Pcr for the diagnosis of mycobacterium tuberculosis: a comparative study between HIV-positive and HIV-negative patients. Jundishapur J Microbiol. (2016) 9:e35634. doi: 10.5812/jjm.35634
106. Montenegro LML, Silva BCd, Lima JFdC, Cruz HLAd, Montenegro RdA, Lundgren FLC, et al. The Performance of an in-House Nested-Pcr Technique for Pleural Tuberculosis Diagnoses Revista da Sociedade Brasileira de Medicina Tropical. (2013) 46:594–9. doi: 10.1590/0037-8682-0127-2013
107. Leung KS-S, Siu GK-H, Tam KK-G, Ho P-L, Wong SS-Y, Leung EK-C, et al. Diagnostic evaluation of an in-house developed single-tube, duplex, nested Is6110 real-time Pcr assay for rapid pulmonary tuberculosis diagnosis. Tuberculosis. (2018) 112:120–5. doi: 10.1016/j.tube.2018.08.008
108. Choi Y, Jeon B-Y, Shim TS, Jin H, Cho S-N, Lee H. Development of a highly sensitive one-tube nested real-time pcr for detecting mycobacterium tuberculosis. Diagn Microbiol Infect Dis. (2014) 80:299–303. doi: 10.1016/j.diagmicrobio.2014.08.009
109. Lin J-J, Xi X-H, Xia L, Tan Y-J, Chen Y, Di H-Q, et al. Diagnostic accuracy of loop-mediated isothermal amplification for pulmonary tuberculosis in China. Front Trop Dis. (2022) 3:1046948. doi: 10.3389/fitd.2022.1046948
110. Mitarai S, Okumura M, Toyota E, Yoshiyama T, Aono A, Sejimo A, et al. Evaluation of a simple loop-mediated isothermal amplification test kit for the diagnosis of tuberculosis. Int J Tuber Lung Dis. (2011) 15:1211–7. doi: 10.5588/ijtld.10.0629
111. World Health Organization. The Use of Loop-Mediated Isothermal Amplification (Tb-Lamp) for the Diagnosis of Pulmonary Tuberculosis: Policy Guidance. Geneva: WHO (2016).
112. Ou X, Wang S, Dong H, Pang Y, Li Q, Xia H, et al. Multicenter evaluation of a real-time loop-mediated isothermal amplification (Realamp) test for rapid diagnosis of mycobacterium tuberculosis. J Microbiol Methods. (2016) 129:39–43. doi: 10.1016/j.mimet.2016.07.008
113. Deng S, Sun Y, Xia H, Liu Z, Gao L, Yang J, et al. Accuracy of commercial molecular diagnostics for the detection of pulmonary tuberculosis in China: a systematic review. Sci Rep. (2019) 9:4553. doi: 10.1038/s41598-019-41074-8
114. Ou X, Li Q, Xia H, Pang Y, Wang S, Zhao B, et al. Diagnostic accuracy of the pure-lamp test for pulmonary tuberculosis at the county-level laboratory in China. PLoS ONE. (2014) 9:e94544. doi: 10.1371/journal.pone.0094544
115. Zaber M, Hoque F, Paean IM, Tarafder S. Evaluation of multiplex loop-mediated isothermal amplification assay for the detection of mycobacterium tuberculosis complex from clinically suspected cases of pulmonary tuberculosis. Heliyon. (2024) 10:e39847. doi: 10.1016/j.heliyon.2024.e39847
116. Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, Krapp F, et al. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med. (2010) 363:1005–15. doi: 10.1056/NEJMoa0907847
117. Feliciano CS, Menon LJB, Anselmo LMP, Dippenaar A, Warren RM, Silva Jr WA, et al. Xpert Mtb/Rif performance to diagnose tuberculosis and rifampicin resistance in a reference centre in Southern Brazil. ERJ Open Res. (2019) 5:00043–2019. doi: 10.1183/23120541.00043-2019
118. Terzi Ha, Aydemir Ö, Karakeçe E, Demiray T, Köroglu M. Evaluation of the Xpert Mtb/Rif test performance in the diagnosis of suspected M. tuberculosis in pulmonary and extrapulmonary clinical specimens. Evaluation. (2024) 13:13. doi: 10.4274/mjima.galenos.2024.23055.13
119. Nathavitharana RR, Cudahy PG, Schumacher SG, Steingart KR, Pai M, Denkinger CM. Accuracy of line probe assays for the diagnosis of pulmonary and multidrug-resistant tuberculosis: a systematic review and meta-analysis. Eur Respir J. (2017) 49:1601075. doi: 10.1183/13993003.01075-2016
120. Tayal D, Sethi P, Jain P. Diagnostic modalities of tuberculosis-then and now. Asian J Med Health. (2022) 20:33–45. doi: 10.9734/ajmah/2022/v20i230436
121. Ling DI, Zwerling AA, Pai M. Rapid diagnosis of drug-resistant Tb using line probe assays: from evidence to policy. Expert Rev Respir Med. (2008) 2:583–8. doi: 10.1586/17476348.2.5.583
122. Yadav RN, Singh BK, Sharma R, Chaubey J, Sinha S, Jorwal P. Comparative performance of line probe assay (Version 2) and xpert Mtb/Rif assay for early diagnosis of rifampicin-resistant pulmonary tuberculosis. Tuberc Respir Dis. (2021) 84:237. doi: 10.4046/trd.2020.0171
123. Nandwani S, Singhal S, Gothi D, Singhal R, Perween N, Gupta K, et al. Evaluation of diagnostic performance of line probe assay on smear-negative samples of pulmonary tuberculosis. Cureus. (2024) 16:e58298. doi: 10.7759/cureus.58298
124. Harboe M, Nagai S, Patarroyo ME, Torres M, Ramirez C, Cruz N. Properties of proteins Mpb64, Mpb70, and Mpb80 of mycobacterium bovis Bcg. Infect Immun. (1986) 52:293–302. doi: 10.1128/iai.52.1.293-302.1986
125. Sawatpanich A, Petsong S, Tumwasorn S, Rotcheewaphan S. Diagnostic performance of the anyplex Mtb/Ntm real-time Pcr in detection of mycobacterium tuberculosis complex and nontuberculous mycobacteria from pulmonary and extrapulmonary specimens. Heliyon. (2022) 8:e11935. doi: 10.1016/j.heliyon.2022.e11935
126. Zhang Y, Rajagopalan M, Brown BA, Wallace Jr RJ. Randomly amplified polymorphic DNA Pcr for comparison of mycobacterium abscessus strains from nosocomial outbreaks. J Clin Microbiol. (1997) 35:3132–9. doi: 10.1128/jcm.35.12.3132-3139.1997
127. Risjani Y, Abidin G. Genetic diversity and similarity between green and brown morphotypes of kappaphycus alvarezii using rapd. J Appl Phycol. (2020) 32:2253–60. doi: 10.1007/s10811-020-02223-z
128. Mougari F, Guglielmetti L, Raskine L, Sermet-Gaudelus I, Veziris N, Cambau E. Infections caused by mycobacterium abscessus: epidemiology, diagnostic tools and treatment. Expert Rev Anti Infect Ther. (2016) 14:1139–54. doi: 10.1080/14787210.2016.1238304
129. Cooksey RC, Jhung MA, Yakrus MA, Butler WR, Adékambi T, Morlock GP, et al. Multiphasic approach reveals genetic diversity of environmental and patient isolates of mycobacterium mucogenicum and mycobacterium phocaicum associated with an outbreak of bacteremias at a texas hospital. Appl Environ Microbiol. (2008) 74:2480–7. doi: 10.1128/AEM.02476-07
130. Vogiatzakis E, Stefanou S, Skroubelou A, Anagnostou S, Marinis E, Matsiota-Bernard P. Molecular markers for the investigation of mycobacterium gordonae epidemics. J Hosp Infect. (1998) 38:217–22. doi: 10.1016/S0195-6701(98)90277-8
131. van Ingen J, Boeree MJ, de Lange WC, de Haas PE, Dekhuijzen PR, van Soolingen D. Clinical relevance of mycobacterium Szulgai in the Netherlands. Clin Infect Dis. (2008) 46:1200–5. doi: 10.1086/529443
132. Kauppinen J, Mäntyjärvi R, Katila M-L. Random amplified polymorphic DNA genotyping of mycobacterium malmoense. J Clin Microbiol. (1994) 32:1827–9. doi: 10.1128/jcm.32.7.1827-1829.1994
133. Colafigli G, Scalzulli E, Di Prima A, Pepe S, Loglisci MG, Diverio D, et al. Digital droplet pcr as a predictive tool for successful discontinuation outcome in chronic myeloid leukemia: is it time to introduce it in the clinical practice? Crit Rev Oncol Hematol. (2021) 157:103163. doi: 10.1016/j.critrevonc.2020.103163
134. Cheng X, Sun L, Zhao Q, Mi Z, Yu G, Wang Z, et al. Development and evaluation of a droplet digital pcr assay for the diagnosis of paucibacillary leprosy in skin biopsy specimens. PLoS Negl Trop Dis. (2019) 13:e0007284. doi: 10.1371/journal.pntd.0007284
135. Devonshire AS, Honeyborne I, Gutteridge A, Whale AS, Nixon G, Wilson P, et al. Highly reproducible absolute quantification of mycobacterium tuberculosis complex by digital Pcr. Anal Chem. (2015) 87:3706–13. doi: 10.1021/ac5041617
136. Devonshire AS, O'Sullivan DM, Honeyborne I, Jones G, Karczmarczyk M, Pavšič J, et al. The use of digital pcr to improve the application of quantitative molecular diagnostic methods for tuberculosis. BMC Infect Dis. (2016) 16:1–10. doi: 10.1186/s12879-016-1696-7
137. Fan Y, Chen J, Liu M, Xu X, Zhang Y, Yue P, et al. Application of droplet digital Pcr to detection of mycobacterium tuberculosis and mycobacterium leprae infections: a narrative review. Infect Drug Resist. (2022):1067-76. doi: 10.2147/IDR.S349607
138. Meehan CJ, Goig GA, Kohl TA, Verboven L, Dippenaar A, Ezewudo M, et al. Whole genome sequencing of mycobacterium tuberculosis: current standards and open issues. Nat Rev Microbiol. (2019) 17:533–45. doi: 10.1038/s41579-019-0214-5
139. Zhang H, Tang M, Li D, Xu M, Ao Y, Lin L. Applications and advances in molecular diagnostics: revolutionizing non-tuberculous mycobacteria species and subspecies identification. Front Public Health. (2024) 12:1410672. doi: 10.3389/fpubh.2024.1410672
140. Loman NJ, Pallen MJ. Xdr-Tb Genome Sequencing: A Glimpse of the Microbiology of the Future. Routledge: Taylor & Francis (2008). p. 111–3.
141. Witney AA, Cosgrove CA, Arnold A, Hinds J, Stoker NG, Butcher PD. Clinical use of whole genome sequencing for mycobacterium tuberculosis. BMC Med. (2016) 14:1–7. doi: 10.1186/s12916-016-0598-2
142. Miller S, Chiu C. The role of metagenomics and next-generation sequencing in infectious disease diagnosis. Clin Chem. (2022) 68:115–24. doi: 10.1093/clinchem/hvab173
143. Di Resta C, Galbiati S, Carrera P, Ferrari M. Next-generation sequencing approach for the diagnosis of human diseases: open challenges and new opportunities. Ejifcc. (2018) 29:4.
144. Lecuit M, Eloit M. The diagnosis of infectious diseases by whole genome next generation sequencing: a new era is opening. Front Cell Infect Microbiol. (2014) 4:25. doi: 10.3389/fcimb.2014.00025
145. Campbell PJ, Morlock GP, Sikes RD, Dalton TL, Metchock B, Starks AM, et al. Molecular detection of mutations associated with first-and second-line drug resistance compared with conventional drug susceptibility testing of mycobacterium tuberculosis. Antimicrobial Agents Chemother. (2011) 55:2032–41. doi: 10.1128/AAC.01550-10
146. Safi H, Lingaraju S, Amin A, Kim S, Jones M, Holmes M, et al. Evolution of high-level ethambutol-resistant tuberculosis through interacting mutations in decaprenylphosphoryl-B-D-arabinose biosynthetic and utilization pathway genes. Nat Genet. (2013) 45:1190–7. doi: 10.1038/ng.2743
147. Clark TG, Mallard K, Coll F, Preston M, Assefa S, Harris D, et al. Elucidating emergence and transmission of multidrug-resistant tuberculosis in treatment experienced patients by whole genome sequencing. PLoS ONE. (2013) 8:e83012. doi: 10.1371/journal.pone.0083012
148. Sun W, Gui X, Wu Z, Zhang Y, Yan L. Prediction of drug resistance profile of multidrug-resistant mycobacterium tuberculosis (Mdr-Mtb) isolates from newly diagnosed case by whole genome sequencing (Wgs): a study from a high tuberculosis burden country. BMC Infect Dis. (2022) 22:499. doi: 10.1186/s12879-022-07482-4
149. Vogel M, Utpatel C, Corbett C, Kohl TA, Iskakova A, Ahmedov S, et al. Implementation of whole genome sequencing for tuberculosis diagnostics in a low-middle income, high Mdr-Tb burden country. Sci Rep. (2021) 11:15333. doi: 10.1038/s41598-021-94297-z
150. Ness TE, DiNardo A, Farhat MR. High throughput sequencing for clinical tuberculosis: an overview. Pathogens. (2022) 11:1343. doi: 10.3390/pathogens11111343
151. Tafess K, Ng TTL, Lao HY, Leung KSS, Tam KKG, Rajwani R, et al. Targeted-sequencing workflows for comprehensive drug resistance profiling of mycobacterium tuberculosis cultures using two commercial sequencing platforms: comparison of analytical and diagnostic performance, turnaround time, and cost. Clin Chem. (2020) 66:809–20. doi: 10.1093/clinchem/hvaa092
152. Papaventsis D, Casali N, Kontsevaya I, Drobniewski F, Cirillo D, Nikolayevskyy V. Whole genome sequencing of mycobacterium tuberculosis for detection of drug resistance: a systematic review. Clin Microbiol Infect. (2017) 23:61–8. doi: 10.1016/j.cmi.2016.09.008
153. Doyle RM, Burgess C, Williams R, Gorton R, Booth H, Brown J, et al. Direct whole-genome sequencing of sputum accurately identifies drug-resistant mycobacterium tuberculosis faster than mgit culture sequencing. J Clin Microbiol. (2018) 56(8):10.1128/jcm. 00666-18. doi: 10.1128/JCM.00666-18
154. Iketleng T, Lessells R, Dlamini MT, Mogashoa T, Mupfumi L, Moyo S, et al. Mycobacterium tuberculosis next-generation whole genome sequencing: opportunities and challenges. Tuberc Res Treat. (2018) 2018:1298542. doi: 10.1155/2018/1298542
155. Smith C, Halse TA, Shea J, Modestil H, Fowler RC, Musser KA, et al. Assessing nanopore sequencing for clinical diagnostics: a comparison of next-generation sequencing (Ngs) methods for mycobacterium tuberculosis. J Clin Microbiol. (2020) 59:e00583–20. doi: 10.1128/JCM.00583-20
156. Chang C-C, Chen C-P, Wu T-H, Yang C-H, Lin C-W, Chen C-Y. Gold nanoparticle-based colorimetric strategies for chemical and biological sensing applications. Nanomaterials. (2019) 9:861. doi: 10.3390/nano9060861
157. Bansal SA, Kumar V, Karimi J, Singh AP, Kumar S. Role of gold nanoparticles in advanced biomedical applications. Nanoscale Adv. (2020) 2:3764–87. doi: 10.1039/D0NA00472C
158. Kooti S, Kadivarian S, Abiri R, Mohajeri P, Atashi S, Ahmadpor H, et al. Modified gold nanoparticle colorimetric probe-based biosensor for direct and rapid detection of mycobacterium tuberculosis in sputum specimens. World J Microbiol Biotechnol. (2023) 39:118. doi: 10.1007/s11274-023-03564-w
159. McNerney R, Daley P. Towards a point-of-care test for active tuberculosis: obstacles and opportunities. Nat Rev Microbiol. (2011) 9:204–13. doi: 10.1038/nrmicro2521
160. Valdemar-Aguilar CM, Manisekaran R, Acosta-Torres LS, López-Marín LM. Spotlight on mycobacterial lipid exploitation using nanotechnology for diagnosis, vaccines, and treatments. Nanomed Nanotechnol Biol Med. (2023) 48:102653. doi: 10.1016/j.nano.2023.102653
161. Amendola V, Pilot R, Frasconi M, Maragò OM, Iatì MA. Surface plasmon resonance in gold nanoparticles: a review. J Phys Condensed Matter. (2017) 29:203002. doi: 10.1088/1361-648X/aa60f3
162. Zhang Z, Wang H, Chen Z, Wang X, Choo J, Chen L. Plasmonic colorimetric sensors based on etching and growth of noble metal nanoparticles: strategies and applications. Biosens Bioelectron. (2018) 114:52–65. doi: 10.1016/j.bios.2018.05.015
163. WHO. The Use of Loop-Mediated Isothermal Amplification (Tb-Lamp) for the Diagnosis of Pulmonary Tuberculosis: Policy Guidance. Geneva: WHO (2016). Available online at: https://www.who.int/publications/i/item/9789241511186
164. Becerra AH, Flores MG, Palma-Nicolas JP, Ramírez-García G, López-Marín LM. Mycosome-coated gold nanoparticles for plasmonic detection of tuberculosis-associated antibodies. ACS Appl Nano Mat. (2024) 7:11203–13. doi: 10.1021/acsanm.4c00706
165. Eivazzadeh-Keihan R, Saadatidizaji Z, Mahdavi M, Maleki A, Irani M, Zare I. Recent advances in gold nanoparticles-based biosensors for tuberculosis determination. Talanta. (2024) 275:126099. doi: 10.1016/j.talanta.2024.126099
166. Soo P-C, Horng Y-T, Chang K-C, Wang J-Y, Hsueh P-R, Chuang C-Y, et al. A simple gold nanoparticle probes assay for identification of mycobacterium tuberculosis and mycobacterium tuberculosis complex from clinical specimens. Mol Cell Probes. (2009) 23:240–6. doi: 10.1016/j.mcp.2009.04.006
167. Kaewphinit T, Santiwatanakul S, Chansiri K. Colorimetric DNA based biosensor combined with loop-mediated isothermal amplification for detection of mycobacterium tuberculosis by using gold nanoprobe aggregation. Sens Transduc. (2013) 149:123.
168. Liandris E, Gazouli M, Andreadou M, Comor M, Abazovic N, Sechi LA, et al. Direct detection of unamplified DNA from pathogenic mycobacteria using DNA-derivatized gold nanoparticles. J Microbiol Methods. (2009) 78:260–4. doi: 10.1016/j.mimet.2009.06.009
169. Hussain MM, Samir TM, Azzazy HM. Unmodified gold nanoparticles for direct and rapid detection of mycobacterium tuberculosis complex. Clin Biochem. (2013) 46:633–7. doi: 10.1016/j.clinbiochem.2012.12.020
170. Veigas B, Pedrosa P, Couto I, Viveiros M, Baptista PV. Isothermal DNA amplification coupled to au-nanoprobes for detection of mutations associated to rifampicin resistance in mycobacterium tuberculosis. J Nanobiotechnology. (2013) 11:1–6. doi: 10.1186/1477-3155-11-38
171. Dahiya B, Prasad T, Singh V, Khan A, Kamra E, Mor P, et al. Diagnosis of tuberculosis by nanoparticle-based immuno-Pcr assay based on mycobacterial Mpt64 and Cfp-10 detection. Nanomedicine. (2020) 15:2609–24. doi: 10.2217/nnm-2020-0258
172. Salmanzadeh S, Abbasissifar H, Alavi SM. Comparison study of quantiferon test with tuberculin skin testing to diagnose latent tuberculosis infection among nurses working in teaching hospitals of Ahvaz, Iran. Caspian J Internal Med. (2016) 7:82.
173. Pai M, Riley LW, Colford JM. Interferon-Γ assays in the immunodiagnosis of tuberculosis: a systematic review. Lancet Infect Dis. (2004) 4:761–76. doi: 10.1016/S1473-3099(04)01206-X
174. Ferrara G, Losi M, D'Amico R, Roversi P, Piro R, Meacci M, et al. Use in routine clinical practice of two commercial blood tests for diagnosis of infection with mycobacterium tuberculosis: a prospective study. Lancet. (2006) 367:1328–34. doi: 10.1016/S0140-6736(06)68579-6
175. Yancey JR, Melchert VE. Quantiferon-Tb Gold+ for the diagnosis of mycobacterium tuberculosis infection. Am Fam Physician. (2021) 103:177–8.
176. Qiagen. Quantiferon-Tb Gold Plus (Qft-Plus) for Use as an Aid in the Detection of Mycobacterium Tuberculosis (Tb) Infection. Available online at: https://www.qiagen.com/us/products/diagnostics-and-clinical-research/tb-management/quantiferon-tb-gold-plus-us 2018 (Accessed March 9, 2025).
177. U.S. Preventive Services Task Force. Final Recommendation Statement. Latent Tuberculosis Infection: Screening (2023). Available online at: https://www.uspreventiveservicestaskforce.org/uspstf/recommendation/latent-tuberculosis-infection-screening (Accessed March 9, 2025).
178. Kobashi Y. Current status and future landscape of diagnosing tuberculosis infection. Respir Investig. (2023) 61:563–78. doi: 10.1016/j.resinv.2023.04.010
179. Hong JY, Park SY, Kim A, Cho S-N, Hur Y-G. Comparison of Qft-plus and Qft-git tests for diagnosis of M. Tuberculosis infection in immunocompetent Korean subjects. J Thorac Dis. (2019) 11:5210. doi: 10.21037/jtd.2019.12.11
180. Venkatappa TK, Punnoose R, Katz DJ, Higgins MP, Banaei N, Graviss EA, et al. Comparing quantiferon-Tb gold plus with other tests to diagnose mycobacterium tuberculosis infection. J Clin Microbiol. (2019) 57:e00985–19. doi: 10.1128/JCM.00985-19
181. Lawn SD. Point-of-care detection of lipoarabinomannan (Lam) in urine for diagnosis of Hiv-associated tuberculosis: a state of the art review. BMC Infect Dis. (2012) 12:1–12. doi: 10.1186/1471-2334-12-103
182. Choudhry V, Saxena R. Detection of mycobacterium tuberculosis antigens in urinary proteins of tuberculosis patients. Eur J Clin Microbiol Infect Dis. (2002) 21:1–5. doi: 10.1007/s10096-001-0651-7
183. Achkar JM, Lawn SD, Moosa M-YS, Wright CA, Kasprowicz VO. Adjunctive tests for diagnosis of tuberculosis: serology, elispot for site-specific lymphocytes, urinary lipoarabinomannan, string test, and fine needle aspiration. J Infect Dis. (2011) 204:S1130–S41. doi: 10.1093/infdis/jir450
184. Shivakoti R, Sharma D, Mamoon G, Pham K. Association of Hiv infection with extrapulmonary tuberculosis: a systematic review. Infection. (2017) 45:11–21. doi: 10.1007/s15010-016-0960-5
185. Bjerrum S, Schiller I, Dendukuri N, Kohli M, Nathavitharana RR, Zwerling AA, et al. Lateral flow urine lipoarabinomannan assay for detecting active tuberculosis in people living with HIV. Cochr Database Syst Rev. (2019) 10:CD011420. doi: 10.1002/14651858.CD011420.pub3
186. WHO. The Use of Lateral Flow Urine Lipoarabinomannan Assay (Lf-Lam) for the Diagnosis and Screening of Active Tuberculosis in People Living with HIV: Policy Guidance. Geneva: WHO (2015).
187. Gupta-Wright A, Corbett EL, van Oosterhout JJ, Wilson D, Grint D, Alufandika-Moyo M, et al. Rapid urine-based screening for tuberculosis in hiv-positive patients admitted to hospital in Africa (Stamp): a pragmatic, multicentre, parallel-group, double-blind, randomised controlled trial. Lancet. (2018) 392:292–301. doi: 10.1016/S0140-6736(18)31267-4
188. Grant AD, Charalambous S, Tlali M, Karat AS, Dorman SE, Hoffmann CJ, et al. Algorithm-guided empirical tuberculosis treatment for people with advanced HIV (Tb Fast Track): an open-label, cluster-randomised trial. The lancet HIV. (2020) 7:e27–37. doi: 10.1016/S2352-3018(19)30266-8
189. Bulterys MA, Wagner B, Redard-Jacot M, Suresh A, Pollock NR, Moreau E, et al. Point-of-care urine lam tests for tuberculosis diagnosis: a status update. J Clin Med. (2019) 9:111. doi: 10.3390/jcm9010111
190. Nicol MP, Schumacher SG, Workman L, Broger T, Baard C, Prins M, et al. Accuracy of a novel urine test, fujifilm silvamp tuberculosis lipoarabinomannan, for the diagnosis of pulmonary tuberculosis in children. Clin Infect Dis. (2021) 72:e280–e8. doi: 10.1093/cid/ciaa1052
191. Reddy KP, Denkinger CM, Broger T, McCann NC, Gupta-Wright A, Kerkhoff AD, et al. Cost-effectiveness of a novel lipoarabinomannan test for tuberculosis in patients with human immunodeficiency virus. Clin Infect Dis. (2021) 73:e2077–e85. doi: 10.1093/cid/ciaa1698
192. Sigal GB, Pinter A, Lowary TL, Kawasaki M, Li A, Mathew A, et al. A Novel sensitive immunoassay targeting the 5-methylthio-D-xylofuranose–lipoarabinomannan epitope meets the who's performance target for tuberculosis diagnosis. J Clin Microbiol. (2018) 56:e01338–18. doi: 10.1128/JCM.01338-18
193. Broger T, Nicol MP, Székely R, Bjerrum S, Sossen B, Schutz C, et al. Diagnostic accuracy of a novel tuberculosis point-of-care urine lipoarabinomannan assay for people living with HIV: a meta-analysis of individual in-and outpatient data. PLoS Med. (2020) 17:e1003113. doi: 10.1371/journal.pmed.1003113
194. Broger T, Sossen B, du Toit E, Kerkhoff AD, Schutz C, Reipold EI, et al. Novel lipoarabinomannan point-of-care tuberculosis test for people with HIV: a diagnostic accuracy study. Lancet Infect Dis. (2019) 19:852–61. doi: 10.1016/S1473-3099(19)30001-5
195. Broger T, Muyoyeta M, Kerkhoff AD, Denkinger CM, Moreau E. Tuberculosis test results using fresh vs. biobanked urine samples with Fujilam. Lancet Infect Dis. (2020) 20:22–3. doi: 10.1016/S1473-3099(19)30684-X
196. Indirawati NN, Yunihastuti E, Yulianti M, Nasir UZ, Wulandari D, Rinaldi I. Lateral flow urine lipoarabinomannan assay for extrapulmonary tuberculosis diagnosis in adults who are HIV-positive. Int J Infect Dis. (2022) 122:415–9. doi: 10.1016/j.ijid.2022.06.007
197. Kerkhoff AD, Lawn SD. A breakthrough urine-based diagnostic test for Hiv-associated tuberculosis. Lancet. (2016) 387:1139–41. doi: 10.1016/S0140-6736(16)00146-X
198. Kostrzewa M, Nagy E, Schröttner P, Pranada AB. How Maldi-Tof mass spectrometry can aid the diagnosis of hard-to-identify pathogenic bacteria–the rare and the unknown. Expert Rev Mol Diagn. (2019) 19:667–82. doi: 10.1080/14737159.2019.1643238
199. Shi J, He G, Ning H, Wu L, Wu Z, Ye X, et al. Application of matrix-assisted laser desorption ionization time-of-flight mass spectrometry (Maldi-Tof Ms) in the detection of drug resistance of mycobacterium tuberculosis in re-treated patients. Tuberculosis. (2022) 135:102209. doi: 10.1016/j.tube.2022.102209
200. Elbehiry A, Aldubaib M, Abalkhail A, Marzouk E, ALbeloushi A, Moussa I, et al. How Maldi-Tof mass spectrometry technology contributes to microbial infection control in healthcare settings. Vaccines. (2022) 10:1881. doi: 10.3390/vaccines10111881
201. Rodriguez-Temporal D, Perez-Risco D, Struzka EA, Mas M, Alcaide F. Evaluation of two protein extraction protocols based on freezing and mechanical disruption for identifying nontuberculous mycobacteria by matrix-assisted laser desorption ionization–time of flight mass spectrometry from liquid and solid cultures. J Clin Microbiol. (2018) 56:e01548–17. doi: 10.1128/JCM.01548-17
202. Pastrone L, Curtoni A, Criscione G, Scaiola F, Bottino P, Guarrasi L, et al. Evaluation of two different preparation protocols for Maldi-Tof Ms nontuberculous mycobacteria identification from liquid and solid media. Microorganisms. (2023) 11:120. doi: 10.3390/microorganisms11010120
203. Gordon SV, Parish T. Microbe profile: mycobacterium tuberculosis: humanity's deadly microbial foe. Microbiology. (2018) 164:437–9. doi: 10.1099/mic.0.000601
204. Pranada AB, Schwarz G, Kostrzewa M. Maldi biotyping for microorganism identification in clinical microbiology. In: Advances in MALDI and Laser-Induced Soft Ionization Mass Spectrometry (2016). p. 197–225.
205. Rotcheewaphan S, Lemon JK, Desai UU, Henderson CM, Zelazny AM. Rapid one-step protein extraction method for the identification of mycobacteria using Maldi-Tof Ms. Diagn Microbiol Infect Dis. (2019) 94:355–60. doi: 10.1016/j.diagmicrobio.2019.03.004
206. Morales MPF, Lim CK, Shephard L, Weldhagen GF. Mycobacterial inactivation protein extraction protocol for matrix-assisted laser desorption ionization time-of-flight characterization of clinical isolates. Int J Mycobacteriol. (2018) 7:217–21. doi: 10.4103/ijmy.ijmy_104_18
207. Saleeb PG, Drake SK, Murray PR, Zelazny AM. Identification of mycobacteria in solid-culture media by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol. (2011) 49:1790–4. doi: 10.1128/JCM.02135-10
208. Robinne S, Saad J, Morsli M, Hamidou ZH, Tazerart F, Drancourt M, et al. Rapid identification of mycobacterium tuberculosis complex using mass spectrometry: a proof of concept. Front Microbiol. (2022) 13:753969. doi: 10.3389/fmicb.2022.753969
209. Bacanelli G, Olarte LC, Silva MR, Rodrigues RA, Carneiro PA, Kaneene JB, et al. Matrix assisted laser desorption ionization-time-of-flight mass spectrometry identification of mycobacterium bovis in Bovinae. J Vet Med Sci. (2019) 81:1400–8. doi: 10.1292/jvms.19-0214
210. Bacanelli G, Araujo FR, Verbisck NV. Improved Maldi-Tof Ms Identification of mycobacterium tuberculosis by use of an enhanced cell disruption protocol. Microorganisms. (2023) 11:1692. doi: 10.3390/microorganisms11071692
211. Williams GR, Cook L, Lewis LD, Tsongalis GJ, Nerenz RD. Clinical validation of a 106-Snv Maldi-Tof Ms pharmacogenomic panel. J Appl Lab Med. (2020) 5:454–66. doi: 10.1093/jalm/jfaa018
212. Shi H-m, Wang Z-k, Wu H, Li J-k, Mo Y-y, Liu X, et al. Clinical application of time-of-flight mass spectrometry nucleic acid detection technology in diagnosis of drug-resistant pulmonary tuberculosis. Diag Microbiol Infect Dis. (2025) 111:116686. doi: 10.1016/j.diagmicrobio.2025.116686
213. Huang Z, Zhang G, Lyon CJ, Hu TY, Lu S. Outlook for crispr-based tuberculosis assays now in their infancy. Front Immunol. (2023) 14:1172035. doi: 10.3389/fimmu.2023.1172035
214. Fonfara I, Richter H, Bratovič M, Le Rhun A, Charpentier E. The Crispr-associated DNA-cleaving enzyme Cpf1 also processes precursor crispr Rna. Nature. (2016) 532:517–21. doi: 10.1038/nature17945
215. Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, et al. Nucleic acid detection with Crispr-Cas13a/C2c2. Science. (2017) 356:438–42. doi: 10.1126/science.aam9321
216. Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. (2018) 360:439–44. doi: 10.1126/science.aaq0179
217. Yue H, Shu B, Tian T, Xiong E, Huang M, Zhu D, et al. Droplet Cas12a assay enables DNA quantification from unamplified samples at the single-molecule level. Nano Lett. (2021) 21:4643–53. doi: 10.1021/acs.nanolett.1c00715
218. Xiong Y, Zhang J, Yang Z, Mou Q, Ma Y, Xiong Y, et al. Functional DNA regulated Crispr-Cas12a sensors for point-of-care diagnostics of non-nucleic-acid targets. J Am Chem Soc. (2019) 142:207–13. doi: 10.1021/jacs.9b09211
219. Shen J, Zhou X, Shan Y, Yue H, Huang R, Hu J, et al. Sensitive detection of a bacterial pathogen using allosteric probe-initiated catalysis and Crispr-Cas13a amplification reaction. Nat Commun. (2020) 11:267. doi: 10.1038/s41467-019-14135-9
220. Wang S, Li H, Kou Z, Ren F, Jin Y, Yang L, et al. Highly sensitive and specific detection of hepatitis B virus DNA and drug resistance mutations utilizing the Pcr-based Crispr-Cas13a system. Clin Microbiol Infect. (2021) 27:443–50. doi: 10.1016/j.cmi.2020.04.018
221. Nguyen PQ, Soenksen LR, Donghia NM, Angenent-Mari NM, de Puig H, Huang A, et al. Wearable materials with embedded synthetic biology sensors for biomolecule detection. Nat Biotechnol. (2021) 39:1366–74. doi: 10.1038/s41587-021-00950-3
222. Arizti-Sanz J, Bradley AD, Zhang YB, Boehm CK, Freije CA, Grunberg ME, et al. Simplified Cas13-based assays for the fast identification of SARS-CoV-2 and its variants. Nat Biomed Eng. (2022) 6:932–43. doi: 10.1038/s41551-022-00889-z
223. Chandrasekaran SS, Agrawal S, Fanton A, Jangid AR, Charrez B, Escajeda AM, et al. Rapid detection of SARS-CoV-2 Rna in saliva via Cas13. Nat Biomed Eng. (2022) 6:944–56. doi: 10.1038/s41551-022-00917-y
224. Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, et al. Crispr-Cas12a target binding unleashes indiscriminate single-stranded dnase activity. Science. (2018) 360:436–9. doi: 10.1126/science.aar6245
225. Li H, Cui X, Sun L, Deng X, Liu S, Zou X, et al. High concentration of Cas12a effector tolerates more mismatches on Ssdna. FASEB J. (2021) 35:e21153. doi: 10.1096/fj.202001475R
226. Sam IK. Chen Y-y, Ma J, Li S-y, Ying R-y, Li L-x, et al. Tb-Quick: Crispr-Cas12b-assisted rapid and sensitive detection of mycobacterium tuberculosis. J Infect. (2021) 83:54–60. doi: 10.1016/j.jinf.2021.04.032
227. Huang Z, LaCourse SM, Kay AW, Stern J, Escudero JN, Youngquist BM, et al. Crispr detection of circulating cell-free mycobacterium tuberculosis DNA in adults and children, including children with HIV: a molecular diagnostics study. Lancet Microbe. (2022) 3:e482–e92. doi: 10.1016/S2666-5247(22)00087-8
228. Xiao G, He X, Zhang S, Liu Y, Liang Z, Liu H, et al. Cas12a/Guide Rna-based platform for rapid and accurate identification of major mycobacterium species. J Clin Microbiol. (2020) 58:e01368–19. doi: 10.1128/JCM.01368-19
229. Xu H, Zhang X, Cai Z, Dong X, Chen G, Li Z, et al. An isothermal method for sensitive detection of mycobacterium tuberculosis complex using clustered regularly interspaced short palindromic repeats/Cas12a cis and trans cleavage. J Mol Diag. (2020) 22:1020–9. doi: 10.1016/j.jmoldx.2020.04.212
230. Bai X, Gao P, Qian K, Yang J, Deng H, Fu T, et al. A highly sensitive and specific detection method for mycobacterium tuberculosis fluoroquinolone resistance mutations utilizing the Crispr-Cas13a System. Front Microbiol. (2022) 13:847373. doi: 10.3389/fmicb.2022.847373
231. Huang J, Liang Z, Liu Y, Zhou J, He F. Development of an Mspqc nucleic acid sensor based on Crispr/Cas9 for the detection of mycobacterium tuberculosis. Anal Chem. (2022) 94:11409–15. doi: 10.1021/acs.analchem.2c02538
232. Thakku SG, Lirette J, Murugesan K, Chen J, Theron G, Banaei N, et al. Genome-wide tiled detection of circulating mycobacterium tuberculosis cell-free DNA using Cas13. Nat Commun. (2023) 14:1803. doi: 10.1038/s41467-023-37183-8
233. Wang Y, Li J, Li S, Zhu X, Wang X, Huang J, et al. Lamp-Crispr-Cas12-based diagnostic platform for detection of mycobacterium tuberculosis complex using real-time fluorescence or lateral flow test. Microchimica Acta. (2021) 188:1–9. doi: 10.1007/s00604-021-04985-w
234. Ai J-W, Zhou X, Xu T, Yang M, Chen Y, He G-Q, et al. Crispr-based rapid and ultra-sensitive diagnostic test for mycobacterium tuberculosis. Emerg Microbes Infect. (2019) 8:1361–9. doi: 10.1080/22221751.2019.1664939
235. Augustin L, Agarwal N. Designing a Cas9/Grna-assisted quantitative real-time Pcr (Carp) assay for identification of point mutations leading to rifampicin resistance in the human pathogen mycobacterium tuberculosis. Gene. (2023) 857:147173. doi: 10.1016/j.gene.2023.147173
236. Zaw MT, Emran NA, Lin Z. Mutations inside Rifampicin-Resistance Determining Region of Rpob Gene Associated with Rifampicin-Resistance in Mycobacterium Tuberculosis. J Infect Public Health. (2018) 11:605–10. doi: 10.1016/j.jiph.2018.04.005
237. Brosch R, Gordon SV, Pym A, Eiglmeier K, Garnier T, Cole ST. Comparative genomics of the mycobacteria. Int J Med Microbiol. (2000) 290:143–52. doi: 10.1016/S1438-4221(00)80083-1
238. Wang M, Zhang R, Li J. Crispr/Cas systems redefine nucleic acid detection: principles and methods. Biosens Bioelectron. (2020) 165:112430. doi: 10.1016/j.bios.2020.112430
239. Zhang X, He X, Zhang Y, Chen L, Pan Z, Huang Y, et al. New Method for the detection of mycobacterium tuberculosis based on the Crispr/Cas system. BMC Infect Dis. (2023) 23:680. doi: 10.1186/s12879-023-08656-4
240. Yan L, Xiao H, Zhang Q. Systematic review: comparison of Xpert Mtb/Rif, Lamp and Sat methods for the diagnosis of pulmonary tuberculosis. Tuberculosis. (2016) 96:75–86. doi: 10.1016/j.tube.2015.11.005
241. Maxmen A. Faster, better, cheaper: the rise of crispr in disease detection. Nature. (2019) 566:437–8. doi: 10.1038/d41586-019-00601-3
242. Li L-S, Yang L, Zhuang L, Ye Z-Y, Zhao W-G, Gong W-P. From immunology to artificial intelligence: revolutionizing latent tuberculosis infection diagnosis with machine learning. Milit Med Res. (2023) 10:58. doi: 10.1186/s40779-023-00490-8
243. Hwang S, Kim H-E, Jeong J, Kim H-J, editors. A novel approach for tuberculosis screening based on deep convolutional neural networks. In: Medical Imaging 2016: Computer-Aided Diagnosis. San Diego, CA: SPIE (2016).
244. Qin ZZ, Ahmed S, Sarker MS, Paul K, Adel ASS, Naheyan T, et al. Tuberculosis detection from chest X-rays for triaging in a high tuberculosis-burden setting: an evaluation of five artificial intelligence algorithms. Lancet Digital Health. (2021) 3:e543–e54. doi: 10.1016/S2589-7500(21)00116-3
245. Hua D, Nguyen K, Petrina N, Young N, Cho J-G, Yap A, et al. Benchmarking the diagnostic test accuracy of certified Ai products for screening pulmonary tuberculosis in digital chest radiographs: preliminary evidence from a rapid review and meta-analysis. Int J Med Inform. (2023) 177:105159. doi: 10.1016/j.ijmedinf.2023.105159
246. Walker TM, Kohl TA, Omar SV, Hedge J, Elias CDO, Bradley P, et al. Whole-genome sequencing for prediction of mycobacterium tuberculosis drug susceptibility and resistance: a retrospective cohort study. Lancet Infect Dis. (2015) 15:1193–202. doi: 10.1016/S1473-3099(15)00062-6
247. Curtis J, Luo Y, Zenner HL, Cuchet-Lourenço D, Wu C, Lo K, et al. Susceptibility to tuberculosis is associated with variants in the Asap1 gene encoding a regulator of dendritic cell migration. Nat Genet. (2015) 47:523–7. doi: 10.1038/ng.3248
248. Quistrebert J, Orlova M, Kerner G, Ton LT, Luong NT, Danh NT, et al. Genome-wide association study of resistance to mycobacterium tuberculosis infection identifies a locus at 10q26.2 in three distinct populations. PLoS Genet. (2021) 17:e1009392. doi: 10.1371/journal.pgen.1009392
249. Huang Y-S, Tseng S-Y, Chang T-E, Perng C-L, Huang Y-H. Sulfamethoxazole-trimethoprim-induced liver injury and genetic polymorphisms of Nat2 and Cyp2c9 in Taiwan. Pharmacogenet Genom. (2021) 31:200–6. doi: 10.1097/FPC.0000000000000441
250. Naidoo K, Moodley MC, Hassan-Moosa R, Dookie N, Yende-Zuma N, Perumal R, et al. Recurrent subclinical tuberculosis among antiretroviral therapy–accessing participants: incidence, clinical course, and outcomes. Clin Infect Dis. (2022) 75:1628–36. doi: 10.1093/cid/ciac185
251. Ma R, Cheng L, Song Y, Sun Y, Gui W, Deng Y, et al. Altered lung microbiome and metabolome profile in children with pulmonary arterial hypertension associated with congenital heart disease. Front Med. (2022) 9:940784. doi: 10.3389/fmed.2022.940784
252. Gill CM, Dolan L, Piggott LM, McLaughlin AM. New developments in tuberculosis diagnosis and treatment. Breathe. (2022) 18:220149. doi: 10.1183/20734735.0149-2021
253. WHO. Global Tuberculosis Report 2020 (2020). Available online at: https://www.who.int/publications/i/item/9789240013131 (Accessed March 11, 2025).
254. Migliori GB, Tiberi S, Zumla A, Petersen E, Chakaya JM, Wejse C, et al. Mdr/Xdr-Tb management of patients and contacts: challenges facing the new decade. the 2020 clinical update by the global tuberculosis network. Int J Infect Dis. (2020) 92:S15–25. doi: 10.1016/j.ijid.2020.01.042
255. Fox W, Ellard GA, Mitchison DA. Studies on the treatment of tuberculosis undertaken by the british medical research council tuberculosis units, 1946–1986, with relevant subsequent publications. Int J Tuberc Lung Dis. (1999) 3:S231–S79.
256. Motta I, Boeree M, Chesov D, Dheda K, Günther G, Horsburgh Jr CR, et al. Recent advances in the treatment of tuberculosis. Clin Microbiol Infect. (2024) 30:1107–14. doi: 10.1016/j.cmi.2023.07.013
257. Rustomjee R, Lienhardt C, Kanyok T, Davies G, Levin J, Mthiyane T, et al. A Phase II study of the sterilising activities of ofloxacin, gatifloxacin and moxifloxacin in pulmonary tuberculosis. Int J Tuber Lung Dis. (2008) 12:128–38. doi: 10.5588/ijtld.08.0003
258. Conde MB, Efron A, Loredo C, De Souza GRM, Graça NP, Cezar MC, et al. Moxifloxacin vs. ethambutol in the initial treatment of tuberculosis: a double-blind, randomised, controlled phase II trial. Lancet. (2009) 373:1183–9. doi: 10.1016/S0140-6736(09)60333-0
259. Merle CS, Fielding K, Sow OB, Gninafon M, Lo MB, Mthiyane T, et al. A four-month gatifloxacin-containing regimen for treating tuberculosis. N Engl J Med. (2014) 371:1588–98. doi: 10.1056/NEJMoa1315817
260. Jindani A, Harrison TS, Nunn AJ, Phillips PP, Churchyard GJ, Charalambous S, et al. High-dose rifapentine with moxifloxacin for pulmonary tuberculosis. N Engl J Med. (2014) 371:1599–608. doi: 10.1056/NEJMoa1314210
261. Imperial MZ, Nahid P, Phillips PP, Davies GR, Fielding K, Hanna D, et al. A patient-level pooled analysis of treatment-shortening regimens for drug-susceptible pulmonary tuberculosis. Nat Med. (2018) 24:1708–15. doi: 10.1038/s41591-018-0224-2
262. Imperial MZ, Phillips PP, Nahid P, Savic RM. Precision-enhancing risk stratification tools for selecting optimal treatment durations in tuberculosis clinical trials. Am J Respir Crit Care Med. (2021) 204:1086–96. doi: 10.1164/rccm.202101-0117OC
263. Te Brake LH, de Jager V, Narunsky K, Vanker N, Svensson EM, Phillips PP, et al. Increased bactericidal activity but dose-limiting intolerability at 50 Mg Kg−1 rifampicin. Eur Respir J. (2021) 58:2000955. doi: 10.1183/13993003.00955-2020
264. Dorman SE, Nahid P, Kurbatova EV, Phillips PP, Bryant K, Dooley KE, et al. Four-month rifapentine regimens with or without moxifloxacin for tuberculosis. N Engl J Med. (2021) 384:1705–18. doi: 10.1056/NEJMoa2033400
265. WHO. WHO Consolidated Guidelines on Tuberculosis. Module 4: Treatment-Drug-Resistant Tuberculosis Treatment, 2022 Update. Geneva: WHO (2022). Available online at: https://www.who.int/publications/i/item/9789240007048
266. Dooley KE, Hendricks B, Gupte N, Barnes G, Narunsky K, Whitelaw C, et al. Assessing pretomanid for tuberculosis (Apt), a randomized phase 2 trial of pretomanid-containing regimens for drug-sensitive tuberculosis: 12-week results. Am J Respir Crit Care Med. (2023) 207:929–35. doi: 10.1164/rccm.202208-1475OC
267. Tiemersma EW, van der Werf MJ, Borgdorff MW, Williams BG, Nagelkerke NJ. Natural history of tuberculosis: duration and fatality of untreated pulmonary tuberculosis in HIV negative patients: a systematic review. PLoS ONE. (2011) 6:e17601. doi: 10.1371/journal.pone.0017601
268. Turkova A, Wills GH, Wobudeya E, Chabala C, Palmer M, Kinikar A, et al. Shorter treatment for nonsevere tuberculosis in African and Indian children. N Engl J Med. (2022) 386:911–22. doi: 10.1056/NEJMoa2104535
269. Paton NI, Cousins C, Suresh C, Burhan E, Chew KL, Dalay VB, et al. Treatment strategy for rifampin-susceptible tuberculosis. N Engl J Med. (2023) 388:873–87. doi: 10.1056/NEJMoa2212537
270. Chakraborty S, Rhee KY. Tuberculosis drug development: history and evolution of the mechanism-based paradigm. Cold Spring Harb Perspect Med. (2015) 5:a021147. doi: 10.1101/cshperspect.a021147
271. Dean AS, Zignol M, Cabibbe AM, Falzon D, Glaziou P, Cirillo DM, et al. Prevalence and genetic profiles of isoniazid resistance in tuberculosis patients: a multicountry analysis of cross-sectional data. PLoS Med. (2020) 17:e1003008. doi: 10.1371/journal.pmed.1003008
272. Fregonese F, Ahuja SD, Akkerman OW, Arakaki-Sanchez D, Ayakaka I, Baghaei P, et al. Comparison of different treatments for isoniazid-resistant tuberculosis: an individual patient data meta-analysis. Lancet Respir Med. (2018) 6:265–75. doi: 10.1016/S2213-2600(18)30078-X
273. Gegia M, Winters N, Benedetti A, van Soolingen D, Menzies D. Treatment of isoniazid-resistant tuberculosis with first-line drugs: a systematic review and meta-analysis. Lancet Infect Dis. (2017) 17:223–34. doi: 10.1016/S1473-3099(16)30407-8
274. Jhun BW, Koh W-J. Treatment of isoniazid-resistant pulmonary tuberculosis. Tuberc Respir Dis. (2020) 83:20–30. doi: 10.4046/trd.2019.0065
275. Rahman SMM, Samina P, Rahman T, Adel ASS, Nasrin R, Uddin MKM, et al. Isoniazid resistance pattern among pulmonary tuberculosis patients in Bangladesh: an exploratory study. Clin Microbiol Infect. (2025) 31:220–5. doi: 10.1016/j.cmi.2024.09.027
276. Nahid P, Mase SR, Migliori GB, Sotgiu G, Bothamley GH, Brozek JL, et al. Treatment of drug-resistant tuberculosis. An Official Ats/Cdc/Ers/Idsa clinical practice guideline. Am J Respir Crit Care Med. (2019) 200:e93–e142. doi: 10.1164/rccm.201909-1874ST
277. Dooley KE, Miyahara S, von Groote-Bidlingmaier F, Sun X, Hafner R, Rosenkranz SL, et al. Early bactericidal activity of different isoniazid doses for drug-resistant tuberculosis (inhindsight): a randomized, open-label clinical trial. Am J Respir Crit Care Med. (2020) 201:1416–24. doi: 10.1164/rccm.201910-1960OC
278. Stagg H, Lipman M, McHugh T, Jenkins H. Isoniazid-resistant tuberculosis: a cause for concern? Int J Tuberc Lung Dis. (2017) 21:129–39. doi: 10.5588/ijtld.16.0716
279. Liu D, Zhao B, Zheng Y, Ou X, Wang S, Zhou Y, et al. Characterization of isoniazid resistance and genetic mutations in isoniazid-resistant and rifampicin-susceptible mycobacterium tuberculosis in China. Infect Med. (2024) 3:100129. doi: 10.1016/j.imj.2024.100129
280. Migliori G, Tiberi S. Who drug-resistant Tb guidelines 2022: what is new. Int J Tuberc Lung Dis. (2022) 26:590–1. doi: 10.5588/ijtld.22.0263
281. WHO. Tuberculosis: Multidrug-Resistant (Mdr-Tb) or Rifampicin-Resistant Tb (Rr-Tb) (2024). Available online at: https://www.who.int/news-room/questions-and-answers/item/tuberculosis-multidrug-resistant-tuberculosis-(mdr-tb) (Accessed July 25, 2025).
282. WHO. Rapid Communication: Key Changes to the Treatment of Drug-Resistant Tuberculosis. Geneva: WHO (2022). Available online at: https://www.who.int/southeastasia/publications/i/item/WHO-UCN-TB-2022-2
283. Vanino E, Granozzi B, Akkerman OW, Munoz-Torrico M, Palmieri F, Seaworth B, et al. Update of drug-resistant tuberculosis treatment guidelines: a turning point. Int J Infect Dis. (2023) 130:S12–S5. doi: 10.1016/j.ijid.2023.03.013
284. Chang KC, Nuermberger E, Sotgiu G, Leung CC. New drugs and regimens for tuberculosis. Respirology. (2018) 23:978–90. doi: 10.1111/resp.13345
285. Pontali E, Raviglione MC, Migliori GB. Regimens to treat multidrug-resistant tuberculosis: past, present and future perspectives. Eur Respir Rev. (2019) 28:190035. doi: 10.1183/16000617.0035-2019
286. Ahmad N, Ahuja SD, Akkerman OW, Alffenaar J-WC, Anderson LF, Baghaei P, et al. Treatment correlates of successful outcomes in pulmonary multidrug-resistant tuberculosis: an individual patient data meta-analysis. Lancet. (2018) 392:821–34. doi: 10.1016/S0140-6736(18)31644-1
287. Schnippel K, Ndjeka N, Maartens G, Meintjes G, Master I, Ismail N, et al. Effect of bedaquiline on mortality in south african patients with drug-resistant tuberculosis: a retrospective cohort study. Lancet Respir Med. (2018) 6:699–706. doi: 10.1016/S2213-2600(18)30235-2
288. Goodall RL, Meredith SK, Nunn AJ, Bayissa A, Bhatnagar AK, Bronson G, et al. Evaluation of two short standardised regimens for the treatment of rifampicin-resistant tuberculosis (stream stage 2): an open-label, multicentre, randomised, non-inferiority trial. Lancet. (2022) 400:1858–68. doi: 10.1016/S0140-6736(22)02078-5
289. Nyang'wa B-T, Berry C, Kazounis E, Motta I, Parpieva N, Tigay Z, et al. A 24-week, all-oral regimen for rifampin-resistant tuberculosis. N Engl J Med. (2022) 387:2331–43. doi: 10.1056/NEJMoa2117166
290. Esmail A, Oelofse S, Lombard C, Perumal R, Mbuthini L, Goolam Mahomed A, et al. An all-oral 6-month regimen for multidrug-resistant tuberculosis: a multicenter, randomized controlled clinical trial (the next study). Am J Respir Crit Care Med. (2022) 205:1214–27. doi: 10.1164/rccm.202107-1779OC
291. Conradie F, Phillips P, Badat T, Poswa A, Rajaram S, Nosipho N, et al. High rate of successful outcomes treating Rr-Tb with a delamanid–bedaquiline regimen in beat tuberculosis: an interim analysis. In: Union World Conference on Lung Health (2022).
292. Mok J, Lee M, Kim DK, Kim JS, Jhun BW, Jo K-W, et al. 9 Months of delamanid, linezolid, levofloxacin, and pyrazinamide vs. conventional therapy for treatment of fluoroquinolone-sensitive multidrug-resistant tuberculosis (Mdr-End): a multicentre, randomised, open-label phase 2/3 non-inferiority trial in South Korea. Lancet. (2022) 400:1522–30. doi: 10.1016/S0140-6736(22)01883-9
293. Günther G, Guglielmetti L, Leu C, Lange C, van Leth F, Hafizi H, et al. Availability and costs of medicines for the treatment of tuberculosis in Europe. Clin Microbiol Infect. (2023) 29:77–84. doi: 10.1016/j.cmi.2022.07.026
294. Saluzzo F, Cirillo DM. Mind the Gap. Rolling out new drug resistant tuberculosis regimens with limited diagnostic tools. J Clin Tuberc Other Mycobact Dis. (2023) 32:100350. doi: 10.1016/j.jctube.2023.100350
295. Padmapriyadarsini C, Vohra V, Bhatnagar A, Solanki R, Sridhar R, Anande L, et al. Bedaquiline, delamanid, linezolid, and clofazimine for treatment of pre-extensively drug-resistant tuberculosis. Clin Infect Dis. (2023) 76:e938–e46. doi: 10.1093/cid/ciac528
296. Conradie F, Bagdasaryan TR, Borisov S, Howell P, Mikiashvili L, Ngubane N, et al. Bedaquiline–pretomanid–linezolid regimens for drug-resistant tuberculosis. N Engl J Med. (2022) 387:810–23. doi: 10.1056/NEJMoa2119430
297. Conradie F, Diacon AH, Ngubane N, Howell P, Everitt D, Crook AM, et al. Treatment of highly drug-resistant pulmonary tuberculosis. N Engl J Med. (2020) 382:893–902. doi: 10.1056/NEJMoa1901814
298. WHO. Who Antenatal Care Recommendations for a Positive Pregnancy Experience. Maternal and Fetal Assessment Update: Imaging Ultrasound before 24 Weeks of Pregnancy. Geneva: World Health Organization (2022). Available online at: https://www.who.int/publications/i/item/9789241549912
299. Guglielmetti L, Ardizzoni E, Atger M, Baudin E, Berikova E, Bonnet M, et al. Evaluating newly approved drugs for multidrug-resistant tuberculosis (ENDTB): study protocol for an adaptive, multi-country randomized controlled trial. Trials. (2021) 22:1–15. doi: 10.1186/s13063-021-05491-3
300. Patil S, Tamirat M, Khazhidinov K, Ardizzoni E, Atger M, Austin A, et al. Evaluating newly approved drugs in combination regimens for multidrug-resistant tuberculosis with fluoroquinolone resistance (endtb-q): study protocol for a multi-country randomized controlled trial. Trials. (2023) 24:773. doi: 10.1186/s13063-023-07701-6
301. Lee EH, Yong SH, Leem AY, Lee SH, Kim SY, Chung KS, ., editors. Improved fluoroquinolone-resistant and extensively drug-resistant tuberculosis treatment outcomes. Open Forum Infect Dis. (2019) 6:S525. doi: 10.1093/ofid/ofz118
302. Nehru VJ, Jose Vandakunnel M, Brammacharry U, Ramachandra V, Pradhabane G, Mani BR, et al. Risk assessment and transmission of fluoroquinolone resistance in drug-resistant pulmonary tuberculosis: a retrospective genomic epidemiology study. Sci Rep. (2024) 14:19719. doi: 10.1038/s41598-024-70535-y
303. van den Boogaard J, Kibiki GS, Kisanga ER, Boeree MJ, Aarnoutse RE. New Drugs against Tuberculosis: Problems, Progress, and Evaluation of Agents in Clinical Development. Antimicrob Agents Chemother. (2009) 53:849–62. doi: 10.1128/AAC.00749-08
304. Cubillos-Angulo JM, Nogueira BM, Arriaga MB, Barreto-Duarte B, Araújo-Pereira M, Fernandes CD, et al. Host-directed therapies in pulmonary tuberculosis: updates on anti-inflammatory drugs. Front Med. (2022) 9:970408. doi: 10.3389/fmed.2022.970408
305. Ayodele S, Kumar P, van Eyk A, Choonara YE. Advances in immunomodulatory strategies for host-directed therapies in combating tuberculosis. Biomed Pharmacother. (2023) 162:114588. doi: 10.1016/j.biopha.2023.114588
306. Zumla A, Rao M, Parida SK, Keshavjee S, Cassell G, Wallis R, et al. Inflammation and tuberculosis: host-directed therapies. J Intern Med. (2015) 277:373–87. doi: 10.1111/joim.12256
307. Kim J-S, Kim Y-R, Yang C-S. Host-directed therapy in tuberculosis: targeting host metabolism. Front Immunol. (2020) 11:1790. doi: 10.3389/fimmu.2020.01790
308. Ahmed S, Raqib R, Guð*mundsson GH, Bergman P, Agerberth B, Rekha RS. Host-directed therapy as a novel treatment strategy to overcome tuberculosis: targeting immune modulation. Antibiotics. (2020) 9:21. doi: 10.3390/antibiotics9010021
309. Kaufmann SH, Dorhoi A, Hotchkiss RS, Bartenschlager R. Host-directed therapies for bacterial and viral infections. Nat Rev Drug Discov. (2018) 17:35–56. doi: 10.1038/nrd.2017.162
310. Wallis RS, Hafner R. Advancing host-directed therapy for tuberculosis. Nat Rev Immunol. (2015) 15:255–63. doi: 10.1038/nri3813
311. Kolloli A, Subbian S. Host-directed therapeutic strategies for tuberculosis. Front Med. (2017) 4:171. doi: 10.3389/fmed.2017.00171
312. Young C, Walzl G, Du Plessis N. Therapeutic host-directed strategies to improve outcome in tuberculosis. Mucosal Immunol. (2020) 13:190–204. doi: 10.1038/s41385-019-0226-5
313. Yang H-J, Wang D, Wen X, Weiner DM, Via LE. One Size Fits All? Not in in vivo modeling of tuberculosis chemotherapeutics. Front Cell Infect Microbiol. (2021) 11:613149. doi: 10.3389/fcimb.2021.613149
314. Rojas-Caraballo J, López-Abán J. Pérez del Villar L, Vizcaíno C, Vicente B, Fernández-Soto P, et al. In vitro and in vivo studies for assessing the immune response and protection-inducing ability conferred by fasciola hepatica-derived synthetic peptides containing B-and T-cell epitopes. PLoS ONE. (2014) 9:e105323. doi: 10.1371/journal.pone.0105323
315. Tezera LB, Reichmann MT, Al Shammari B, Elkington PT. In vitro models of human granuloma formation to analyze host-directed therapies. Adv Host-Direct Ther Against Tuberc. (2021):259-65. doi: 10.1007/978-3-030-56905-1_17
316. Fielding K, Subbaraman R, Khan A, Celan C, Charalambous S, Franke MF, et al. The Use of Digital Technologies in Adherence to Anti-Tuberculosis Treatment. Digit Respir Healthcare (ERS Monograph) Sheffield: Eur Respir Soc. (2023):170-84. doi: 10.1183/2312508X.10002223
317. Thomas BE, Kumar JV, Onongaya C, Bhatt SN, Galivanche A, Periyasamy M, et al. Explaining differences in the acceptability of 99dots, a cell phone–based strategy for monitoring adherence to tuberculosis medications: qualitative study of patients and health care providers. JMIR mHealth uHealth. (2020) 8:e16634. doi: 10.2196/16634
318. Story A, Aldridge RW, Smith CM, Garber E, Hall J, Ferenando G, et al. Smartphone-enabled video-observed vs. directly observed treatment for tuberculosis: a multicentre, analyst-blinded, randomised, controlled superiority trial. Lancet. (2019) 393:1216–24. doi: 10.1016/S0140-6736(18)32993-3
319. Fatima R, Akhtar N, Yaqoob A, Harries AD, Khan MS. Building better tuberculosis control systems in a post-covid world: learning from Pakistan during the COVID-19 pandemic. Int J Infect Dis. (2021) 113:S88–90. doi: 10.1016/j.ijid.2021.03.026
320. Needamangalam Balaji J, Prakash S, Park Y, Baek JS, Shin J, Rajaguru V, et al. Scoping review on accentuating the pragmatism in the implication of mobile health (Mhealth) technology for tuberculosis management in India. J Pers Med. (2022) 12:1599. doi: 10.3390/jpm12101599
321. Nguyen HB, Quang Vo LN, Forse RJ, Wiemers AMC, Huynh HB, Dong TTT, et al. Is convenience really king? Comparative evaluation of catastrophic costs due to tuberculosis in the public and private healthcare sectors of viet nam: a longitudinal patient cost study. Infect Dis Pov. (2024) 13:17–28. doi: 10.1186/s40249-024-01196-2
322. Zhuang L, Ye Z, Li L, Yang L, Gong W. Next-generation Tb vaccines: progress, challenges, and prospects. Vaccines. (2023) 11:1304. doi: 10.3390/vaccines11081304
323. Tait DR, Hatherill M, Van Der Meeren O, Ginsberg AM, Van Brakel E, Salaun B, et al. Final analysis of a trial of M72/As01e vaccine to prevent tuberculosis. N Engl J Med. (2019) 381:2429–39. doi: 10.1056/NEJMoa1909953
324. Nieuwenhuizen NE, Kulkarni PS, Shaligram U, Cotton MF, Rentsch CA, Eisele B, et al. The recombinant bacille Calmette–Guérin vaccine Vpm1002: ready for clinical efficacy testing. Front Immunol. (2017) 8:1147. doi: 10.3389/fimmu.2017.01147
325. Spertini F, Audran R, Chakour R, Karoui O, Steiner-Monard V, Thierry A-C, et al. Safety of human immunisation with a live-attenuated mycobacterium tuberculosis vaccine: a randomised, double-blind, controlled phase I trial. Lancet Respir Med. (2015) 3:953–62. doi: 10.1016/S2213-2600(15)00435-X
326. Day TA, Penn-Nicholson A, Luabeya AKK, Fiore-Gartland A, Du Plessis N, Loxton AG, et al. Safety and immunogenicity of the adjunct therapeutic vaccine id93+ gla-se in adults who have completed treatment for tuberculosis: a randomised, double-blind, placebo-controlled, phase 2a trial. Lancet Respir Med. (2021) 9:373–86. doi: 10.1016/S2213-2600(20)30319-2
327. Ogongo P, Broderick A. Broader evaluation of vaccine-induced T cell immunity against tuberculosis. Front Tuberc. (2024) 2:1435344. doi: 10.3389/ftubr.2024.1435344
328. Kaufmann SH, Dockrell HM, Drager N, Ho MM, McShane H, Neyrolles O, et al. Tbvac2020: advancing tuberculosis vaccines from discovery to clinical development. Front Immunol. (2017) 8:1203. doi: 10.3389/fimmu.2017.01203
329. Olawade DB, Eberhardt J, David-Olawade AC, Balogun MA, Bolarinwa OA, Esan DT. Transforming multidrug-resistant tuberculosis care: the potentials of telemedicine in resource-limited settings. Health Sci Rev. (2024) 12:100185. doi: 10.1016/j.hsr.2024.100185
330. Vassall A, Siapka M, Foster N, Cunnama L, Ramma L, Fielding K, et al. Cost-effectiveness of xpert Mtb/Rif for tuberculosis diagnosis in south africa: a real-world cost analysis and economic evaluation. Lancet Global Health. (2017) 5:e710–e9. doi: 10.1016/S2214-109X(17)30205-X
331. Isaacs K, Shifflett A, Patel K, Karpisek L, Cui Y, Lawental M, et al. Women empowered to connect with addiction resources and engage in evidence-based treatment (We-Care): a usability and feasibility study of a mobile health application for the universal screening of alcohol, substance use, depression and anxiety. JMIR Form Res. (2025) 9:e62915. doi: 10.2196/62915
332. Pai DR, Pakdil F. Emergency departments: the gatekeepers of admissions in Pennsylvania's rural hospitals. Am J Emerg Med. (2022) 57:138–48. doi: 10.1016/j.ajem.2022.05.002
333. Penn-Nicholson A, Geldenhuys H, Burny W, van der Most R, Day CL, Jongert E, et al. Safety and immunogenicity of candidate vaccine M72/As01e in adolescents in a Tb endemic setting. Vaccine. (2015) 33:4025–34. doi: 10.1016/j.vaccine.2015.05.088
Keywords: tuberculosis, diagnostic innovation, therapeutic strategies, vaccine development, public health
Citation: Elbehiry A, Marzouk E, Edrees HM, AlShaqi R, Ellethy AT, Alzaben F, Anagreyyah S, Algarni A, Almuhaydili K, Alotaibi I, Albaqami A, Alamri K, Ibrahem M, Almuzaini AM, Dhahri F and Abu-Okail A (2025) Advancing the fight against tuberculosis: integrating innovation and public health in diagnosis, treatment, vaccine development, and implementation science. Front. Med. 12:1596579. doi: 10.3389/fmed.2025.1596579
Received: 19 March 2025; Accepted: 01 August 2025;
Published: 25 August 2025.
Edited by:
Guofang Deng, Shenzhen Third People's Hospital, ChinaReviewed by:
Newton Valerio Verbisck, Brazilian Agricultural Research Corporation (EMBRAPA), BrazilZheng Yuan, China Academy of Chinese Medical Sciences, China
Copyright © 2025 Elbehiry, Marzouk, Edrees, AlShaqi, Ellethy, Alzaben, Anagreyyah, Algarni, Almuhaydili, Alotaibi, Albaqami, Alamri, Ibrahem, Almuzaini, Dhahri and Abu-Okail. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ayman Elbehiry, YXIuZWxiZWhpcnlAcXUuZWR1LnNh
 Ayman Elbehiry
Ayman Elbehiry Eman Marzouk
Eman Marzouk Husam M. Edrees
Husam M. Edrees Riyad AlShaqi3
Riyad AlShaqi3 Abousree T. Ellethy
Abousree T. Ellethy Khalid Almuhaydili
Khalid Almuhaydili Ibrahim Alotaibi
Ibrahim Alotaibi Abdulrahman Albaqami
Abdulrahman Albaqami Mai Ibrahem
Mai Ibrahem Abdulaziz M. Almuzaini
Abdulaziz M. Almuzaini Akram Abu-Okail
Akram Abu-Okail