Abstract
Objective:
To develop a mammography (MG)-based post-fusion model combined with Clinical, Radiomics, and Deep Learning Models to evaluate the status of sentinel lymph node (SLN) in patients with breast cancer.
Methods:
A total of 290 breast cancer patients who underwent MG were randomly divided into a training set (n = 203) and an internal validation set (n = 87), with an additional 82 patients included in the test set for independent validation. From the MG images of mediolateral oblique (MLO) and craniocaudal (CC) views, 1726 radiomic (Rad) features and 1,024 deep learning (DL) features were extracted for each patient. After the feature fusion and selection, the single-modal models and pre-fusion models were established by stochastic gradient descent (SGD). Using the probabilities of single-modal models, the post-fusion models were developed by support vector machine (SVM). The area under the receiver operating characteristic curve (AUC) was used for accessing the performance of models. The clinical net benefit and predictive accuracy were evaluated through decision curve analysis (DCA) and calibration curves.
Results:
The post-fusion model Clinical+Rad+DL combined probabilities of single modal models, showed the best discrimination ability in the internal validation set (AUC [95%CI]: 0.845 [0.769–0.921]) and test set (AUC [95%CI]: 0.825 [0.812–0.932]).
Conclusion:
The proposed post-fusion model Clinical+Rad+DL demonstrated the method of probabilities fusion was effective and showed promise for predicting SLN metastasis in breast cancer.
1 Introduction
Breast cancer is the most commonly diagnosed cancer and the leading cause of cancer-related death in women (1). Axillary lymph node (ALN) status is critical in staging breast cancer and guiding treatment decisions (2, 3). Sentinel lymph node biopsy (SLNB) has become the preferred method for assessing ALN metastasis in early-stage breast cancer patients because SLN is recognized as the primary site for tumor spread to the axillary region (4). However, it’s important to note that SLNB is an invasive procedure that can lead to complications such as axillary wound infection, seroma formation, and paresthesias (5). That being said, ultrasound (6, 7), mammography (MG) (8, 9), and magnetic resonance imaging (MRI) (10) detect lymph node metastasis by identifying morphological and functional characteristics, but their sensitivity and specificity do not meet clinical needs.
Radiomics (Rad) is a non-invasive method that involves the high-throughput extraction of large amounts of image features from radiographic images to predict tumor diagnosis and prognosis (11). Several studies have applied Rad features to predict SLN metastasis in breast cancer (12, 13). Moreover, it is worth noting that deep learning (DL) has been widely employed in breast MRI (14–17) and breast ultrasound (18–20) for various tasks, including segmentation, diagnosis, grading, and metastasis prediction. DL features have the potential to provided more comprehensive information than Rad features, as they can capture complex and subtle features within images. The combination of Rad and DL features may potentially enhance the model’s performance. Various methods for fusion have been proposed, including feature fusion (pre-fusion) and probability fusion (post-fusion). In a study by Xie et al. (21), an approach was proposed that integrates decision-level texture, shape, and DL features for classifying lung nodules. Furthermore, Li et al. (22) utilized a probabilistic fusion technique to create a model based on MRI for forecasting ALN metastasis, which yielded an AUC of 0.91. This level of performance exceeded that of both the Rad and DL models. These studies indicate that the use of post-fusion techniques, such as probability fusion, to construct predictive models for breast cancer SLN metastasis exhibits potential.
Thus, our study aimed to develop and compare pre-fusion and post-fusion models encompassing clinical, Rad, and DL features of MG to predict SLN metastasis in breast cancer.
2 Materials and methods
2.1 Patient population
The research was conducted in accordance with the ethical guidelines established in the 2013 revision of the Declaration of Helsinki for studies involving human participants. Approval for the study was obtained from the institutional review committee of our hospital. Since the study had a retrospective design, the requirement for informed consent was exempt.
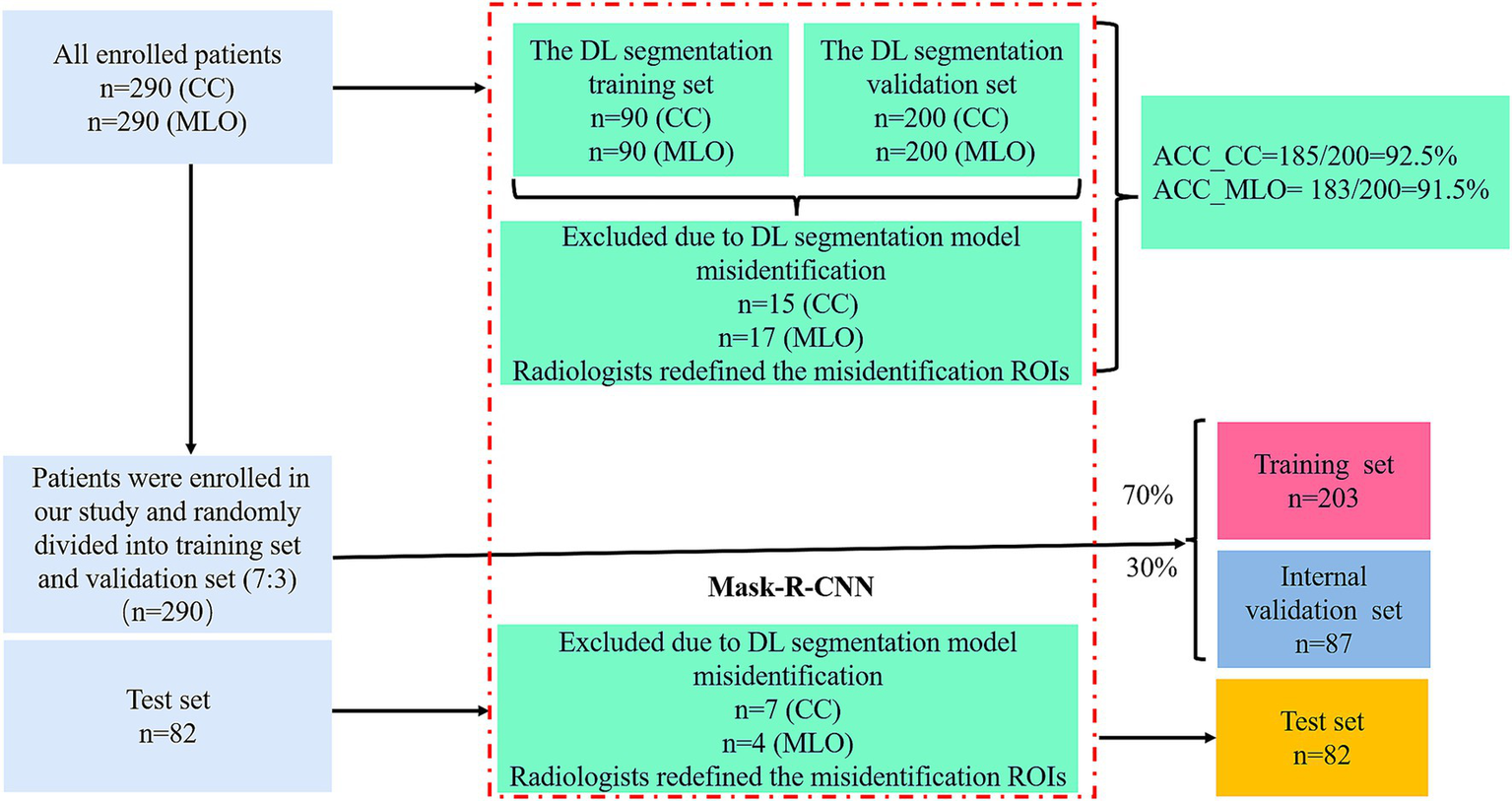
A total of 290 patients diagnosed with invasive breast carcinoma between March 2016 and June 2023 were enrolled, while 82 patients diagnosed between January 2014 and February 2016 served as an independent test set. The inclusion criteria were as follows: (1) Underwent MG examination within the 2 weeks before surgery, and the images met the diagnostic requirements; (2) Underwent SLNB during surgery to assess the status of the SLN. The exclusion criteria were as follows: (1) Had chemotherapy, radiotherapy, or endocrine therapy before surgery; (2) Received treatment or biopsy before MG examination; (3) Diagnosed with bilateral, multicentric, multifocal breast cancer, or evidence of distant metastasis. The flowchart for enrolled patients is illustrated in Supplementary Figure 1.
The patient data included the following datasets: (a) A regions of interest (ROIs) training set used to train a DL segmentation model for identifying MG lesions; (b) An ROIs validation set used to assess the DL segmentation model’s performance in ROIs segmentation; (c) A radiomics dataset where patients were randomly divided into a training set and an internal validation set at a 7:3 ratio; (d) A test set from a separate cohort was included to further evaluate the model’s performance. Ultimately, 290 patients and 82 test patients were enrolled in the study. The cohort selection flowchart is shown in Figure 1.
Figure 1

Flow chart of cohort selection.
2.2 MG examination and image acquisition
The Hologic Selenia full digital MG camera (Hologic Medical Systems, Boston, MA) was utilized to conduct bilateral digital MG examinations, acquiring digital MG images in mediolateral oblique (MLO) and craniocaudal (CC) views. The images were analyzed using a Hologic breast computer-aided diagnosis workstation (SecureViewDx; Hologic) equipped with two 5-megapixel monitors, each with a resolution of 1792 × 2048.
2.3 Assessment of conventional semantic features in MG and clinicopathologic characteristics
The evaluation of the conventional semantic features of MG was carried out by two experienced breast imaging radiologists, Radiologists 1 and Radiologists 2, who have 30 and 10 years of expertise in MG diagnosis, respectively. The assessment was conducted using the workstation without prior knowledge of the pathological outcomes. The study examined the conventional semantic features of MG based on the American College of Radiology Breast Imaging Reporting and Data System (ACR BI-RADS) 5th edition standard. This included analyzing diameter, shape (round or oval/irregular), glandular type (non-dense breast/dense breast), margin (spiculated/non-spiculated), mass density (high density/equal density), suspicious morphology of calcifications (absent/present). In addition, suspicious lymph node signs in MG included rounded or irregular shape, absence of fatty hilum, small diameter ≤1 cm, and increased density. If any of these signs were present, the MG-reported abnormal lymph node (MG_reported_LN) was recorded as positive8, 9.
The agreement of the conventional semantic features of MG was analyzed using the Kappa test.
Clinicopathologic features included the patient’s age, weight, height, body mass index (BMI), neutrophil-to-lymphocyte ratio (NLR), estrogen receptor (ER) status (positive/negative), progesterone receptor (PR) status (positive/negative), human epidermal growth factor receptor-2 (HER-2) status (positive/negative), Ki-67 (≥30%/<30%) status, and histological grading (I/II/III).
The conventional semantic features and clinicopathologic features were defined as the Clinical features.
2.4 CNN-based MG images segmentation
The workflow is shown in Figures 1, 2. Radiologists first randomly selected 90 patients, including their MLO and CC view images, and performed manual segmentation using 3D Slicer (version 5.2.2), a tool widely used for accurate medical image annotation, to generate labeled data for the segmentation task. This labeled dataset formed the basis for training a DL segmentation model using the Mask-R-Convolutional Neural Network (Mask-R-CNN) architecture, chosen for its proven effectiveness in medical image segmentation. The convolutional layers of the Mask-R-CNN model were initially pre-trained on the Microsoft Common Objects in Context (COCO) dataset to acquire general feature representations, with a learning rate of 0.001, a batch size of 10, and a total of 100 epochs. Subsequently, the model was fine-tuned using the DL segmentation training set and validated on the DL segmentation validation set to evaluate and optimize its segmentation performance. Finally, images excluded due to DL segmentation model misidentification (e.g., failure to identify tumor boundaries or incorrect ROIs placement) were re-annotated with ROIs by Radiologists. The corrected data were then integrated back into datasets.
Figure 2
The design of workflow for the study.
2.5 Radiomic feature extraction and DL feature extraction
Feature extraction was carried out using the open-source software PyRadiomics (version 3.1.0). A total of 1726 Rad features were extracted from two regions of interest (CC and MLO views) for each patient. These features included shape, intensity, textural, and wavelet features.
In recent years, ResNet has been shown to have excellent performance and good applications in medical imaging tasks (23, 24). We adopted a pre-trained ResNet18 model by maintaining the original kernel size, stride, and padding settings, allowing for direct application of deep learning feature extraction from medical images. We used SimpleITK (version 2.2.0) to read images and ROIs and convert them into Numpy arrays. We then normalized and standardized these arrays. To make the model output features rather than classification results, we removed the last full connection layer of the model to obtain the intermediate features from the penultimate layer. In total, 1,024 features were extracted from 2 ROIs (CC and MLO views) for each patient. All features were normalized using the z-score method, converting them to a standardized range of values.
2.6 Feature fusion and selection
To construct the pre-fusion model, we combined three types of features (Clinical, Rad, DL features) separately to get 3 types of pre-fusion features (Clinical+Rad, Clinical+DL, Clinical+Rad+DL) (Figure 2).
To obtain the features most closely associated with SLN metastasis in the training set, a three-step selection process was performed. First, we used differential analysis (Mann–Whitney U-test or independent t-test was performed for quantitative features, while chi-squared test or Fisher’s exact test was applied for categorical features) with a p-value threshold of 0.05 to obtain the features associated with SLN status. Then, taking into account the correlations between features, we calculated the correlation coefficients between features using Pearson or Spearman correlation. If the correlation coefficient between two features exceeded 0.75, one of the features was eliminated. Finally, the Least Absolute Shrinkage and Selection Operator (LASSO) with fivefold cross-validation to tune the parameters with the minimum lambda was used to select the optimal features.
2.7 Models’ development
In our study, we developed three types of models using the selected features: 1. Single-modal models (Clinical model, Rad model, and DL model); 2. Pre-fusion models: These models used the fusion features to construct an integrated model (pre-fusion model Clinical+Rad, pre-fusion model Clinical+DL, and pre-fusion model Clinical+Rad+DL); 3. Post-fusion models: these models integrated probabilities from meta-classifiers constructed separately on single-modal models to build an integrated model (post-fusion model Clinical+Rad, post-fusion model Clinical+DL, and post-fusion model Clinical+Rad+DL). The stochastic gradient descent (SGD) was utilized to construct single-modal models and pre-fusion models, and the support vector machine (SVM) was employed to develop the post-fusion models. These machine-learning algorithms have been proven to have good applications in the medical field (25, 26).
The AUC of the receiver operating characteristic (ROC) curves, accuracy, sensitivity, and specificity were used to evaluate the performance of the models. The AUCs were compared using the Delong test. Calibration curves were performed to evaluate the goodness of fit of the models. In addition, the clinical benefits of the models were assessed using the decision curve analysis (DCA) (27).
2.8 Statistical analysis
All statistical work, feature extraction of Rad and DL as well as model construction were conducted in Python (version 3.12.3),1 along with open source packages such as PyTorch, Scipy, and scikit-learn. Quantitative features were presented as means with standard deviations or as medians with the 25th and 75th percentiles. The independent sample t-test or the Mann–Whitney U-test was used for analyzing the quantitative features, while the chi-square test or the Fisher’s exact test was used for analyzing the categorical features. All tests were two-sided, with p-values <0.05 considered statistically significant.
3 Results
3.1 Clinical features
The clinical features of the patients are shown in Table 1. In both the training, internal validation and test sets, margin, MG_reported_LN showed the most significant differences in distribution between the SLN+ and SLN− groups (p < 0.05), indicating that these three features have certain differences in their ability to predict SLNM. Although no differences were observed in the internal validation set, shape and diameter showed statistically significant differences in the training set. In addition, no statistical differences were found for NLR, ER status, PR status, HER-2 status, Ki-67 and other clinical features.
Table 1
| Clinical features | Training set (n = 203) | Internal validation set (n = 87) | Test set (n = 82) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| SLN− (N = 147) | SLN+ (N = 56) | p-value | SLN− (N = 63) | SLN+ (N = 24) | p-value | SLN− (N = 59) | SLN+ (N = 23) | p-value | |
| Age | 56 (46, 64) | 54 (46, 60) | 0.47 | 53 (42, 60) | 57 (46, 64) | 0.24 | 57 (46, 65) | 54 (45, 61) | 0.34 |
| Diameter | 2.00 (1.70, 2.50) | 2.35 (1.90, 3.00) | 0.009 | 2.10 (1.85, 2.50) | 2.10 (1.80, 2.40) | 0.94 | 2.10 (1.73, 2.50) | 2.10 (1.78, 2.63) | 0.49 |
| Weight | 61 (57, 70) | 60 (55, 70) | 0.69 | 60 (55, 65) | 62 (55, 70) | 0.25 | 60 (57, 69) | 62 (57, 69) | 0.76 |
| Height | 1.60 (1.58, 1.65) | 1.60 (1.58, 1.62) | 0.31 | 1.60 (1.58, 1.62) | 1.60 (1.59, 1.64) | 0.51 | 1.60 (1.60, 1.66) | 1.60 (1.59, 1.63) | 0.16 |
| BMI | 23.8 (22.0, 26.6) | 23.6 (21.7, 26.6) | 0.79 | 23.4 (22.03, 24.99) | 24.1 (22.48, 26.03) | 0.23 | 23.44 (21.49, 25.24) | 23.53 (22.53, 26.73) | 0.28 |
| NLR | 1.82 (1.42, 2.27) | 1.83 (1.46, 2.27) | 0.74 | 1.74 (1.31, 2.36) | 1.82 (1.52, 2.33) | 0.73 | 1.87 (1.43 2.37) | 1.82 (1.47, 2.13) | 0.76 |
| Breast Composition | 0.9 | 0.91 | 0.88 | ||||||
| Non-dense breast | 75 (51%) | 28 (50%) | 35 (56%) | 13 (54%) | 28 (48.3%) | 12(50.0%) | |||
| Dense breast | 72 (49%) | 28 (50%) | 28 (44%) | 11 (46%) | 30 (51.7%) | 12(50.0%) | |||
| Density | 0.3 | 0.91 | 0.18 | ||||||
| Equal density | 88 (60%) | 29 (52%) | 35 (56%) | 13 (54%) | 36 (62.1%) | 11(45.8%) | |||
| High density | 59 (40%) | 27 (48%) | 28 (44%) | 11 (46%) | 22(37.9%) | 13 (54.2%) | |||
| Shape | 0.02 | 0.54 | 0.14 | ||||||
| Round or Oval | 42 (29%) | 7 (13%) | 13 (21%) | 3 (13%) | 19 (32.8%) | 4 (16.7%) | |||
| Irregular | 105 (71%) | 49 (88%) | 50 (79%) | 21 (88%) | 39 (67.2%) | 20 (83.3%) | |||
| Margin | <0.001 | 0.03 | 0.03 | ||||||
| Non-spiculated | 37 (25%) | 36 (64%) | 21 (33%) | 14 (58%) | 37 (63.8%) | 9 (37.5%) | |||
| Spiculated | 110 (75%) | 20 (36%) | 42 (67%) | 10 (42%) | 21 (36.2%) | 15 (62.5%) | |||
| Calcifications | 0.04 | 0.71 | 0.05 | ||||||
| Absent | 94 (64%) | 27 (48%) | 42 (67%) | 15 (63%) | 42 (72.4%) | 12 (50.0%) | |||
| Present | 53 (36%) | 29 (52%) | 21 (33%) | 9 (38%) | 16 (27.6%) | 12 (50.0%) | |||
| MG_reported_LN | <0.001 | <0.001 | 0.02 | ||||||
| Negative | 118 (80%) | 24 (43%) | 55 (87%) | 12 (50%) | 52 (89.7%) | 16 (66.7%) | |||
| Positive | 29 (20%) | 32 (57%) | 8 (13%) | 12 (50%) | 6 (10.3%) | 20 (83.3%) | |||
| ER | 0.5 | 0.02 | 0.77 | ||||||
| Negative | 23 (16%) | 11 (20%) | 17 (27%) | 1 (4%) | 12 (20.7%) | 4 (16.7%) | |||
| Positive | 124 (84%) | 45 (80%) | 46 (73%) | 23 (96%) | 46 (79.3%) | 170 (81%) | |||
| PR | 0.55 | 0.009 | 0.89 | ||||||
| Negative | 36 (24%) | 16 (29%) | 23 (37%) | 2 (8%) | 16 (27.6%) | 7 (29.2%) | |||
| Positive | 111 (76%) | 40 (71%) | 40 (63%) | 22 (92%) | 42 (72.4%) | 17 (70.8%) | |||
| HER-2 | 0.66 | 0.16 | 0.12 | ||||||
| Negative | 122 (83%) | 45 (80%) | 51 (81%) | 16 (67%) | 50 (86.2%) | 17 (70.8%) | |||
| Positive | 25 (17%) | 11 (20%) | 12 (19%) | 8 (33%) | 8 (13.8%) | 7 (29.2%) | |||
| KI-67 | 0.98 | 0.68 | 0.95 | ||||||
| <0.3 | 79 (54%) | 30 (54%) | 31 (49%) | 13 (54%) | 31 (53.4%) | 13 (54.2%) | |||
| ≥0.3 | 68 (46%) | 26 (46%) | 32 (51%) | 11 (46%) | 27 (46.6%) | 11 (45.8%) | |||
| Histological Grading | 0.43 | 0.69 | 0.98 | ||||||
| I | 19 (13%) | 4 (7%) | 4 (6%) | 2 (8%) | 6 (10.3%) | 2 (8.3%) | |||
| II | 73 (50%) | 32 (57%) | 37 (59%) | 12 (50%) | 33 (56.9%) | 14 (58.3%) | |||
| III | 55 (37%) | 20 (36%) | 22 (35%) | 10 (42%) | 19 (32.8%) | 8 (33.3%) | |||
Clinical features of the patients.
ER, estrogen receptor; HER-2, human epidermal growth factor receptor-2; NLR, neutrophil-to-lymphocyte ratio; PR, progesterone receptor; SLN+, sentinel lymph node with metastasis; SLN−, sentinel lymph node without metastasis.
3.2 MG images segmentation and feature selection
185 CC views and 183 MLO views from 400 images of 200 patients were accurately segmented, with an accuracy of 92.5% for the CC set and 91.5% for the MLO set, respectively.
The kappa values for conventional semantic features of MG by two radiologists were all >0.80.
We implemented an independent feature selection approach for each feature set within the training set. After feature selection, the features of the single-modal models and the pre-fusion models are shown in Supplementary Table 1. For the Clinical model in the single-modal models, the features diameter, shape, margin, and MG_reported_LN and were selected.
3.3 Model construction and performance
Three single-modal models were built based on the selected features. The AUC of these models (Clinical model, Rad model, and DL model) were 0.797, 0.834, and 0.744 in the training set and 0.732, 0.793 and 0.726 in the internal validation set (Table 2; Supplementary Table 2). Second, the selected pre-fusion features were used to build the pre-fusion models: Clinical+Rad, Clinical+DL, and Clinical+Rad+DL. Among these pre-fusion models, the Clinical+Rad+DL model achieved the best performance with an AUC, accuracy, sensitivity and specificity of 0.873, 0.847, 0.768 and 0.878, respectively, in the training set and 0.776, 0.701, 0.791 and 0.667, respectively, in the internal validation set (Table 2; Figure 3). Finally, the prediction probabilities of the three single-modal models were further fused using SVM to build post-fusion models. The prediction probabilities of the Clinical model and the Rad model were fused to construct the post-fusion model Clinical+Rad; the prediction probabilities of the Clinical model and the DL model were combined to construct the post-fusion model Clinical+DL; and the prediction probabilities of the Clinical model, the Rad model, and the DL model were integrated to develop the post-fusion model Clinical+Rad+DL.
Table 2
| Cohort | Model | AUC (95%CI) | Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|---|
| Training set | Clinical model | 0.797 (0.741–0.852) | 0.793 | 0.571 | 0.878 |
| Pre-fusion model | |||||
| Clinical+Rad | 0.853 (0.805–0.902) | 0.852 | 0.75 | 0.891 | |
| Clinical+DL | 0.849 (0.800–0.898) | 0.833 | 0.679 | 0.891 | |
| Clinical+Rad+DL | 0.873 (0.827–0.919) | 0.847 | 0.768 | 0.878 | |
| Post-fusion model | |||||
| Clinical+Rad | 0.854 (0.806–0.903) | 0.828 | 0.696 | 0.878 | |
| Clinical+DL | 0.827 (0.774–0.879) | 0.852 | 0.554 | 0.966 | |
| Clinical+Rad+DL | 0.881 (0.836–0.925) | 0.833 | 0.804 | 0.844 | |
| Internal validation set | Clinical model | 0.732 (0.639–0.825) | 0.816 | 0.5 | 0.937 |
| Pre-fusion model | |||||
| Clinical+Rad | 0.762 (0.672–0.851) | 0.805 | 0.625 | 0.873 | |
| Clinical+DL | 0.74 (0.648–0.832) | 0.667 | 0.792 | 0.619 | |
| Clinical+Rad+DL | 0.776 (0.688–0.863) | 0.701 | 0.791 | 0.667 | |
| Post-fusion model | |||||
| Clinical+Rad | 0.78 (0.693–0.867) | 0.759 | 0.833 | 0.73 | |
| Clinical+DL | 0.776 (0.688–0.863) | 0.851 | 0.542 | 0.968 | |
| Clinical+Rad+DL | 0.845 (0.769–0.921) | 0.782 | 0.875 | 0.746 | |
| Test set | Post-fusion model | ||||
| Clinical+Rad+DL | 0.825 (0.812–0.932) | 0.862 | 0.779 | 0.883 | |
Performance of the different models in training set and validation set.
AUC, Area under the curve; DL, deep learning; Rad, radiomics.
Figure 3

Receiver operating characteristic (ROC) curves of the Clinical model and pre-fusion models in the training (A) and validation set (B). ROC curves of the post-fusion models in the training (C) and validation set (D).
The post-fusion model Clinical+Rad+DL showed the best performance among all models (Table 2; Figure 3). Table 3 shows the DeLong test results comparing the Clinical model, the pre-fusion model Clinical+Rad+DL, and the post-fusion model Clinical+Rad+DL. In the training set, this model achieved the highest AUC of 0.881, which was statistically significantly higher than both the Clinical model (p < 0.001) and the pre-fusion model Clinical+Rad+DL (p = 0.03). Similarly, in the internal validation set, the AUC (0.845) of the post-fusion Clinical+Rad+DL model was the highest and statistically significant when compared to the Clinical model (p = 0.04) and the pre-fusion Clinical+Rad+DL model (p = 0.04).
Table 3
| Model vs model | p-value |
|---|---|
| Training set | |
| Clinical vs. Pre-fusion Clinical+Rad+DL | 0.032 |
| Clinical vs. Post-fusion Clinical+Rad+DL | 0.044 |
| Pre-fusion Clinical+Rad+DL vs. Post-fusion Clinical+Rad+DL | 0.03 |
| Internal validation set | |
| Clinical vs. Pre-fusion Clinical+Rad+DL | 0.78 |
| Clinical vs. Post-fusion Clinical+Rad+DL | 0.038 |
| Pre-fusion Clinical+Rad+DL vs. Post-fusion Clinical+Rad+DL | 0.027 |
Comparison of diagnostic performance between different models.
DL, deep learning; Rad, radiomics.
The calibration curves (Figure 4) indicated that the true statement of SLN was consistent with the result of the post-fusion model Clinical+Rad+DL in the training and internal validation sets. The DCA for the post-fusion models is shown in Figure 4. When an individual’s threshold probability is <0.77, the post-fusion model Clinical+Rad+DL would add net benefit compared to the treat-all or treat-none tactics. The calibration curves, and DCA curves of the other models are shown in Supplementary Figure 4.
Figure 4

The calibration curves of post-fusion models in the training (A) and validation set (B). Calibration curves demonstrate the goodness-of-fit of models. Decision curves analysis (DCA) for post-fusion models are showed in the training (C) and validation set(D); the y-axis indicates the net benefit, the x-axis indicates threshold probability.
Finally, we applied the most optimal model, the post-fusion model Clinical+Rad+DL, to the test set, which also demonstrated good discrimination (AUC = 0.825), calibration, and clinical applicability (Figure 5).
Figure 5
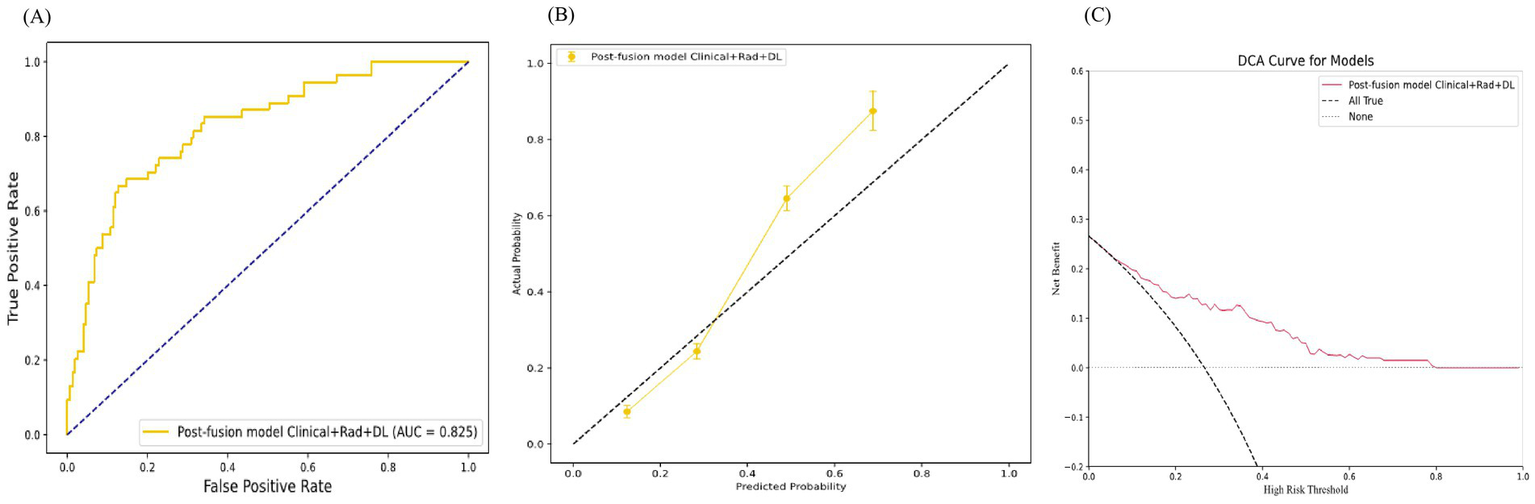
The test set performance of the Post-Fusion Model Clinical+Rad+DL. (A) Receiver operating characteristic (ROC) curves. (B) Calibration curves. (C) Decision curves analysis (DCA).
4 Discussion
In this study, the post-fusion model Clinical+Rad+DL, which integrated the probabilities of the Clinical, Rad, and DL models, achieved the best performance in distinguishing SLN metastasis status. Our results indicated that the post-fusion model Clinical+Rad+DL demonstrated promising predictive performance, with important implications for surgical planning in breast cancer patients.
After feature selection, clinical features such as diameter, shape, margin, and MG_reported_LN were incorporated into the Clinical model. Many previous studies have confirmed their association with lymph node metastasis. Lyu et al. (28) found that tumor size is an independent risk factor for SLN metastasis in breast cancer. In the study by Yuan et al. (29), patients with spiculated margins on MG images were more likely to have SLN metastasis. Breast cancer shape on MG showed no statistical difference in the validation set, likely due to the small sample size and the division between training and validation sets. Although there is no literature suggesting that irregularly shaped breast cancers are more prone to lymph node metastasis on MG, breast cancers with irregular shapes on ultrasound (30) are more likely to undergo lymph node metastasis.
This Clinical model incorporating the MG_reported_LN feature showed lower sensitivity (0.5), with some studies also confirming the drawback of MG for accessing lymph node status29. One possible reason is that some patients’ axillae may not be fully exposed in the standard positions (CC and MLO views). In our study, the use of a model with the post-fusion mode Clinical+Rad+DL can compensate for this drawback (sensitivity:0.875) and also avoid errors arising from radiologists’ subjectivity and reliance on experience. Previous research on the prediction of lymph node metastasis by radiomics has mainly focused on the characteristics of the primary tumor (30, 31). Lymph node metastasis in breast cancer is a complex process, typically associated with changes in the immune microenvironment of the primary tumor region (32). Rad features have been shown to reflect the heterogeneity of the primary tumor site and the degree of immune cell infiltration (33, 34). Consequently, models based on features extracted from the primary tumor may improve model performance and serve as one of the strategies to overcome the limitations of MG.
Previous studies have shown that traditional Rad research based on MG shows promising results, with AUCs ranging from 0.767 to 0.87635-37. Compared to these previous studies, we further integrated features from ResNet18, either through pre- or post-fusion models, both of which yielded satisfactory results and demonstrated certain advantages (Table 2) in predicting SLN metastasis. In contrast to the quantified features of Rad features, DL models can extract more abstract and higher-dimensional information from images. Combining DL features with Rad features allows the complementary integration of information from both sources, enabling a more comprehensive analysis of images and thus improving the predictive ability.
In the current study, the performance of single-modal models was unsatisfactory. However, the post-fusion models using probabilistic fusion outperformed the pre-fusion models using feature fusion. Specifically, the post-fusion model combining Clinical+Rad+DL had a higher AUCs with values of 0.881 on the training set and 0.845 on the validation set. Such models using the post-fusion strategy of probabilistic fusion will perform better than the pre-fusion model, and the same conclusion has been reached in other studies (22, 35). The post-fusion model offers several advantages. First, since different models may excel in different aspects, model fusion can leverage the strengths of different models to achieve more accurate prediction results. Second, combining multiple models can mitigate the risk of overfitting associated with individual models, thereby improving the robustness and stability of the model. In addition, multi-model fusion can improve the generalization ability of the model by reducing its variance, leading to better performance on test data.
Our study has several limitations. First, it was a retrospective analysis with data collected from a single center and a relatively small sample size, and it lacked an independent external dataset for validation, which may introduce selection bias and limit the generalizability of the findings. To address this issue, future work should involve larger patient cohorts and multicenter prospective studies, which would help validate our results and enhance the robustness and clinical utility of the proposed model. Moreover, our patient cohort was heterogeneous, including different pathological subtypes and clinical stages. A more precise selection of patient subgroups may yield better predictive performance and should be further explored in future studies. Finally, our study was based solely on Rad features derived from MG images. Beyond Rad, genomics can provide rich complementary information for the diagnosis, classification, and prognosis of breast cancer (36–39). Future research should focus on integrating genomics with Rad. Genomics can provide complementary biological information to improve the interpretability of Rad features, while combining the two to construct multi-omics models may further enhance diagnostic performance and facilitate more precise breast cancer management.
5 Conclusion
In this study, the proposed post-fusion model Clinical+Rad+DL gets the best performance, which may be potential and perspective for patients with breast cancer to avoid ALN dissection.
Statements
Data availability statement
The datasets presented in this article are not readily available because protection of patient privacy. Requests to access the datasets should be directed to Bo Gao, gaobo72519@hrbmu.edu.cn.
Ethics statement
The studies involving humans were approved by the Ethics Committee of the Second Affiliated Hospital of Harbin Medical University. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
XL: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. YR: Data curation, Formal analysis, Methodology, Writing – review & editing. SC: Data curation, Investigation, Writing – review & editing. MZ: Data curation, Software, Writing – review & editing. ZS: Investigation, Software, Writing – review & editing. YJ: Data curation, Investigation, Writing – review & editing. YW: Data curation, Investigation, Writing – review & editing. BG: Conceptualization, Funding acquisition, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (62172129).
Acknowledgments
We would like to express our sincere gratitude to Dr. Gao for her invaluable contributions in conceptualizing the study, designing the research, drafting the manuscript, and making the final decision to submit the article for publication.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2025.1659422/full#supplementary-material
Footnotes
References
1.
Sung H Ferlay J Siegel RL Laversanne M Soerjomataram I Jemal A et al . Global Cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
2.
Andersson Y Bergkvist L Frisell J de Boniface J . Long-term breast cancer survival in relation to the metastatic tumor burden in axillary lymph nodes. Breast Cancer Res Treat. (2018) 171:359–69. doi: 10.1007/s10549-018-4820-0
3.
van la Parra RF Francissen CM Peer PG Ernst MF de Roos WK Van Zee KJ et al . Assessment of the Memorial Sloan-Kettering Cancer Center nomogram to predict sentinel lymph node metastases in a Dutch breast cancer population. Eur J Cancer. (2013) 49:564–71. doi: 10.1016/j.ejca.2012.04.025
4.
Lyman GH Somerfield MR Bosserman LD Perkins CL Weaver DL Giuliano AE . Sentinel lymph node biopsy for patients with early-stage breast Cancer: American Society of Clinical Oncology clinical practice guideline update. J Clin Oncol. (2017) 35:561–4. doi: 10.1200/JCO.2016.71.0947
5.
Wilke LG McCall LM Posther KE Whitworth PW Reintgen DS Leitch AM et al . Surgical complications associated with sentinel lymph node biopsy: results from a prospective international cooperative group trial. Ann Surg Oncol. (2006) 13:491–500. doi: 10.1245/ASO.2006.05.013
6.
Park SH Kim MJ Park BW Moon HJ Kwak JY Kim EK . Impact of preoperative ultrasonography and fine-needle aspiration of axillary lymph nodes on surgical management of primary breast cancer. Ann Surg Oncol. (2011) 18:738–44. doi: 10.1245/s10434-010-1347-y
7.
Nori J Vanzi E Bazzocchi M Bufalini FN Distante V Branconi F et al . Role of axillary ultrasound examination in the selection of breast cancer patients for sentinel node biopsy. Am J Surg. (2007) 193:16–20. doi: 10.1016/j.amjsurg.2006.02.021
8.
Liu Q Xing P Dong H Zhao T Jin F . Preoperative assessment of axillary lymph node status in breast cancer patients by ultrasonography combined with mammography: a STROBE compliant article. Medicine (Baltimore). (2018) 97:e11441. doi: 10.1097/MD.0000000000011441
9.
Valente SA Levine GM Silverstein MJ Rayhanabad JA Weng-Grumley JG Ji L et al . Accuracy of predicting axillary lymph node positivity by physical examination, mammography, ultrasonography, and magnetic resonance imaging. Ann Surg Oncol. (2012) 19:1825–30. doi: 10.1245/s10434-011-2200-7
10.
Kvistad KA Rydland J Smethurst HB Lundgren S Fjosne HE Haraldseth O . Axillary lymph node metastases in breast cancer: preoperative detection with dynamic contrast-enhanced MRI. Eur Radiol. (2000) 10:1464–71. doi: 10.1007/s003300000370
11.
Lambin P Rios-Velazquez E Leijenaar R Carvalho S van Stiphout RG Granton P et al . Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. (2012) 48:441–6. doi: 10.1016/j.ejca.2011.11.036
12.
Liu C Ding J Spuhler K Gao Y Serrano Sosa M Moriarty M et al . Preoperative prediction of sentinel lymph node metastasis in breast cancer by radiomic signatures from dynamic contrast-enhanced MRI. J Magn Reson Imaging. (2019) 49:131–40. doi: 10.1002/jmri.26224
13.
Zhao M Zheng Y Chu J Liu Z Dong F . Ultrasound-based radiomics combined with immune status to predict sentinel lymph node metastasis in primary breast cancer. Sci Rep. (2023) 13:16918. doi: 10.1038/s41598-023-44156-w
14.
Cong C Li X Zhang C Zhang J Sun K Liu L et al . MRI-based breast Cancer classification and localization by multiparametric feature extraction and combination using deep learning. J Magn Reson Imaging. (2024) 59:148–61. doi: 10.1002/jmri.28713
15.
Sun R Wei L Hou X Chen Y Han B Xie Y et al . Molecular-subtype guided automatic invasive breast cancer grading using dynamic contrast-enhanced MRI. Comput Methods Prog Biomed. (2023) 242:107804. doi: 10.1016/j.cmpb.2023.107804
16.
Yang X Fan X Lin S Zhou Y Liu H Wang X et al . Assessment of Lymphovascular invasion in breast Cancer using a combined MRI morphological features, Radiomics, and deep learning approach based on dynamic contrast-enhanced MRI. J Magn Reson Imaging. (2024) 59:2238–49. doi: 10.1002/jmri.29060
17.
Zhang B Yu Y Mao Y Wang H Lv M Su X et al . Development of MRI-based deep learning signature for prediction of axillary response after NAC in breast cancer. Acad Radiol. (2024) 31:800–11. doi: 10.1016/j.acra.2023.10.004
18.
Harrison P Hasan R Park K . State-of-the-art of breast Cancer diagnosis in medical images via convolutional neural networks (CNNs). J Healthc Inform Res. (2023) 7:387–432. doi: 10.1007/s41666-023-00144-3
19.
Huang Y Yao Z Li L Mao R Huang W Hu Z et al . Deep learning radiopathomics based on preoperative US images and biopsy whole slide images can distinguish between luminal and non-luminal tumors in early-stage breast cancers. EBioMedicine. (2023) 94:104706. doi: 10.1016/j.ebiom.2023.104706
20.
Wang C Zhao Y Wan M Huang L Liao L Guo L et al . Prediction of sentinel lymph node metastasis in breast cancer by using deep learning radiomics based on ultrasound images. Medicine (Baltimore). (2023) 102:e35868. doi: 10.1097/md.0000000000035868
21.
Xie Y Zhang J Xia Y Fulham M Zhang Y . Fusing texture, shape and deep model-learned information at decision level for automated classification of lung nodules on chest CT. Inform Fusion. (2018) 42:102–10. doi: 10.1016/j.inffus.2017.10.005
22.
Li X Yang L Jiao X . Comparison of traditional radiomics, deep learning radiomics and fusion methods for axillary lymph node metastasis prediction in breast cancer. Acad Radiol. (2023) 30:1281–7. doi: 10.1016/j.acra.2022.10.015
23.
Zhang B Chen Z Yan R Lai B Wu G You J et al . Development and validation of a feature-based broad-learning system for opportunistic osteoporosis screening using lumbar spine radiographs. Acad Radiol. (2024) 31:84–92. doi: 10.1016/j.acra.2023.07.002
24.
Zhang W Yan Z Peng J Zhao S Ran L Yin H et al . Magnetic resonance imaging and deoxyribonucleic acid methylation-based radiogenomic models for survival risk stratification of glioblastoma. Med Biol Eng Comput. (2024) 62:853–64. doi: 10.1007/s11517-023-02971-3
25.
Ye JY Fang P Peng ZP Huang XT Xie JZ Yin XY . A radiomics-based interpretable model to predict the pathological grade of pancreatic neuroendocrine tumors. Eur Radiol. (2024) 34:1994–2005. doi: 10.1007/s00330-023-10186-1
26.
Mohammadi SM Moniri S Mohammadhoseini P Hanafi MG Farasat M Cheki M . A computed tomography-based Radiomics analysis of low-energy proximal femur fractures in the elderly patients. Curr Radiopharm. (2023) 16:222–32. doi: 10.2174/1874471016666230321120941
27.
Wu S Zheng J Li Y Yu H Shi S Xie W et al . A radiomics nomogram for the preoperative prediction of lymph node metastasis in bladder Cancer. Clin Cancer Res. (2017) 23:6904–11. doi: 10.1158/1078-0432.CCR-17-1510
28.
Lyu W Guo Y Peng H Xie N Gao H . Analysis of the influencing factors of sentinel lymph node metastasis in breast cancer. Evid Based Complement Alternat Med. (2022) 2022:5775971. doi: 10.1155/2022/5775971
29.
Yuan C Xu G Zhan X Xie M Luo M She L et al . Molybdenum target mammography-based prediction model for metastasis of axillary sentinel lymph node in early-stage breast cancer. Medicine (Baltimore). (2023) 102:e35672. doi: 10.1097/MD.0000000000035672
30.
Guo Q Dong Z Zhang L Ning C Li Z Wang D et al . Ultrasound features of breast cancer for predicting axillary lymph node metastasis. J Ultrasound Med. (2018) 37:1354–3. doi: 10.1002/jum.14469
31.
Chen Y Li J Zhang J Yu Z Jiang H . Radiomic nomogram for predicting axillary lymph node metastasis in patients with breast cancer. Acad Radiol. (2024) 31:788–99. doi: 10.1016/j.acra.2023.10.026
32.
Nathanson SD Krag D Kuerer HM Newman LA Brown M Kerjaschki D et al . Breast cancer metastasis through the lympho-vascular system. Clin Exp Metastasis. (2018) 35:443–54. doi: 10.1007/s10585-018-9902-1
33.
Han X Guo Y Ye H Chen Z Hu Q Wei X et al . Development of a machine learning-based radiomics signature for estimating breast cancer TME phenotypes and predicting anti-PD-1/PD-L1 immunotherapy response. Breast Cancer Res. (2024) 26:18. doi: 10.1186/s13058-024-01776-y
34.
Qian H Ren X Xu M Fang Z Zhang R Bu Y et al . Magnetic resonance imaging-based radiomics was used to evaluate the level of prognosis-related immune cell infiltration in breast cancer tumor microenvironment. BMC Med Imaging. (2024) 24:31. doi: 10.1186/s12880-024-01212-9
35.
Liang X Tang K Ke X Jiang J Li S Xue C et al . Development of an MRI-based comprehensive model fusing clinical, Radiomics and deep learning models for preoperative histological stratification in intracranial solitary fibrous tumor. J Magn Reson Imaging. (2024) 60:523–33. doi: 10.1002/jmri.29098
36.
Foruzandeh Z Alivand MR Ghiami-Rad M Zaefizadeh M Ghorbian S . Identification and validation of miR-583 and mir-877-5p as biomarkers in patients with breast cancer: an integrated experimental and bioinformatics research. BMC Res Notes. (2023) 16:72. doi: 10.1186/s13104-023-06343-w
37.
Nourolahzadeh Z Houshmand M Mohammad FM Ghorbian S . Correlation between Lsp1 (Rs3817198) and Casc (Rs4784227) polymorphisms and the susceptibility to breast cancer. Rep Biochem Mol Biol. (2020) 9:291–6. doi: 10.29252/rbmb.9.3.291
38.
Ghorbian S Nargesian M Talaneh S Asnaashari O Sharifi R . Association of genetic variations in XRCC1 and ERCC1 genes with sporadic breast cancer. Gene Cell Tissue. (2018) 5:166. doi: 10.5812/gct.80166
39.
Sabour L Sabour M Ghorbian S . Clinical applications of next-generation sequencing in cancer diagnosis. Pathol Oncol Res. (2017) 23:225–34. doi: 10.1007/s12253-016-0124-z
Summary
Keywords
breast cancer, radiomics, sentinel lymph node, machine learning, full-field digital mammography, information fusion
Citation
Liu X, Ruan Y, Cao S, Zhao M, Shi Z, Jin Y, Wang Y and Gao B (2025) Development and internal validation of a mammography-based model fusing clinical, radiomics, and deep learning models for sentinel lymph node metastasis prediction in breast cancer. Front. Med. 12:1659422. doi: 10.3389/fmed.2025.1659422
Received
04 July 2025
Accepted
22 August 2025
Published
09 September 2025
Volume
12 - 2025
Edited by
Yuhua Yao, Hainan Normal University, China
Reviewed by
Saeid Ghorbian, Islamic Azad University of Ahar, Iran
Ujjwal Agarwal, Tata Memorial Hospital, India
Updates
Copyright
© 2025 Liu, Ruan, Cao, Zhao, Shi, Jin, Wang and Gao.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Gao, gaobo72519@hrbmu.edu.cn
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.