Precision medicine helps healthcare professionals design medical treatment for individuals for decades. And biomarker matching therapies has shown effective for survival rate improvement (1). In a study of metastatic tumors, 15.6% of CGP-tested patients received targeted therapy and CGP-guided treatment was correlated with prolonged progression-free survival [pooled hazard ratio (HR) = 0.63] (2). Today, we are facing an important turning point where the convergence of genomics and multi-modal artificial intelligence is transforming precision medicine into new ways to diagnose, treat, and prevent disease.
Genomics as the cornerstone of precision medicine
Genomics are the foundations of precision medicine. The completion of the Human Genome Project in 2003 marked the beginning of a new era in healthcare, revealing how genetic variations influence disease diagnose, drug metabolism, and treatment response.
The impact of genomics on clinics is profound. Pharmacogenomic testing can guide dosing decisions for critical medications, preventing adverse reactions and treatment failures (3, 4). In oncology, tumor genomic profiling has revolutionized cancer care, enabling targeted therapies that specifically attack cancer cells based on their genetics (5). Hereditary cancer syndromes, and other genetic diseases can now be diagnosed by genetic techniques, allowing for early intervention and family screening (6). In neurology, researchers developed technique to detect biomarker of demyelinating disease (7). And for an example in infectious disease, researchers developed VirCapSeq-VERT, a positive selection system based on genetics for detecting RNA and DNA viruses (8).
However, genomics also showed the limitations of genetic determinism. The complex interactions between genes, environment, and human body means that genetic information alone cannot fully predict health conditions or optimal treatments. This realization has driven the start toward multi-modal approaches that consider genomics as the cornerstone while recognizing the need for other layers of biological and clinical information.
Not only the genome: the multi-modal imperative
While genomics is foundation for precision medicine, we believe that genes alone can tell only part of the story. The human body is an intricate system where genetic intersects with environmental factors, lifestyle choices, and organs. Multi-modal AI can be one of our most powerful techniques to explore this complexity.
Unlike traditional AI systems that focus on single data types, multi-modal AI can simultaneously process and integrate genomics sequences, medical imaging, electronic health records, and even social variants of health (see Figure 1). To be more specific, multi-modal AI models incorporate different types of data source and transform them into embedding by joining independent embedding of data sources (9). More recent models using data fusion layer to fuse multiple embedding into one embedding for downstream tasks (10), such as predicting diagnostics. This approach mirrors how physicians naturally think about patients—not as isolated genetic data, but as complex individuals whose health comes from multiple interacting factors.
Figure 1
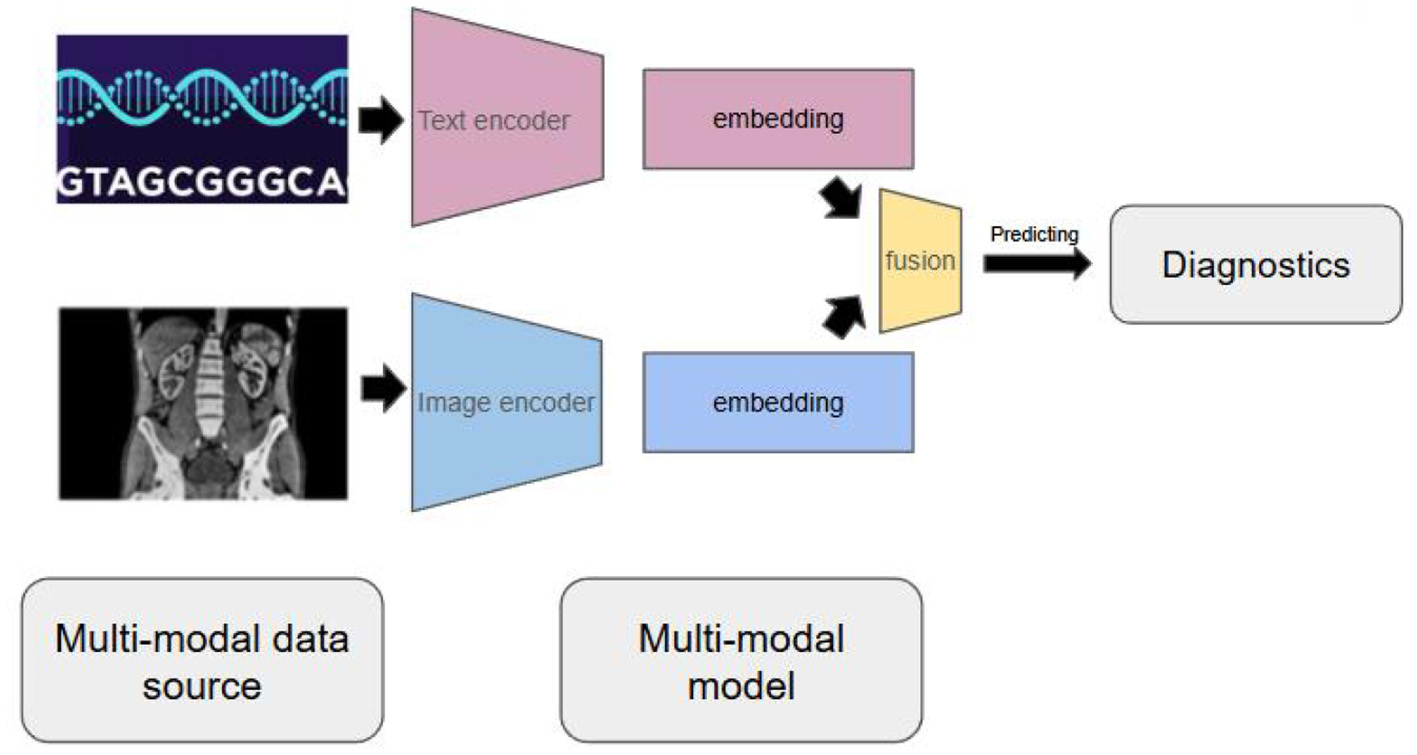
Multi-modal AI models can incorporate different data source and transform them into embedding and perform downstream tasks, such as predicting diagnostics.
Consider cancer treatment, where this convergence is already yielding remarkable results. Multi-modal AI systems can analyze tumor genomics alongside histopathological images, treatment history, and biomarkers to predict which patients will respond to specific immunotherapies (11, 12). Companies such as Tempus and Foundation Medicine are pioneering platforms that combine genomic profiling with clinical data analysis, enabling oncologists to make treatment decisions based on comprehensive molecular information. In cardiology, researchers have developed models that integrate genetics with risk variants of patients (13, 14). In neurology, researchers developed a multi-modal screening system for elderly neurological diseases (15).
Multi-modal approaches are particularly powerful for addressing health disparities. By incorporating social determinants of health alongside genomic data, these systems can better account for the environmental and socioeconomic factors that significantly influence health outcomes in different populations (16, 17). This is crucial for ensuring that precision medicine benefits all patients, not just those in affluent healthcare systems.
Discussion
Challenges
Despite the great potential, significant challenges remain. Data privacy and security concerns exist when integrating such comprehensive personal information. Patients' genomic and health data meed to be protected while still enabling the data sharing necessary for AI model development and validation. Governments aware the importance of user privacy in AI applications and proposed standards such as GDPR and HIPAA to protect users, and federated learning enable AI model training without disclosing original training data but still under the risk of privacy attack in parameter exchanges (18).
The interpretability of multi-modal AI models presents another critical challenge. As these systems become more sophisticated, understanding how they arrive at specific recommendations becomes important, but complex. Healthcare providers need AI tools that not only provide accurate predictions but can also offer clear explanations to patients and incorporated into clinical decision-making.
Future directions
Looking ahead, the convergence of genomics and multi-modal AI will likely transform healthcare in ways we cannot imagine today. Chronic diseases could be predicted and prevented even before symptoms appear. Mental health conditions might be detected through subtle patterns in genomic data and behavioral indicators captured by wearable devices.
The integration of real-world evidence with genomic analysis will enable continuous refinement of treatment protocols, creating a learning healthcare system thatimproves outcomes for patients. Pharmacogenomics will become routine, reducing trial-and-error prescribing and reducing adverse drug reactions.
To advance precision medicine, we need sustained collaboration between healthcare providers, researchers, policymakers, and patients. Academic institutions need to train clinicians who can work effectively at the intersection of genomics, AI, and clinical care. Healthcare systems must develop the infrastructure necessary to capture, store, and analyze multi-modal health data securely.
Most importantly, we must keep patients at the center of this transformation. The ultimate measure of success will not be the accuracy of our algorithms or the value of our datasets, but whether we can deliver better health outcomes for individuals and communities.
The convergence of genomics and multi-modal AI represents more than a technological advancement—it embodies our evolving understanding of human health as a complex, dynamic system. By embracing this complexity rather than oversimplifying it, we can finally deliver on the promise of personalized medicine and even personalized healthcare. The revolution is already underway, and we need to ensure that the power of precision medicine serves humanity's highest aspirations for health and healing.
Statements
Author contributions
HZ: Writing – review & editing, Writing – original draft.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that Gen AI was used in the creation of this manuscript. Generative AI was used to summarize literature and make suggestions.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1.
Malone ER Oliva M Sabatini PJB Stockley TL Siu LL . Molecular profiling for precision cancer therapies. Genome Med. (2020) 12:8. 10.1186/s13073-019-0703-1
2.
Zerdes I Filis P Fountoukidis G El-Naggar AI Kalofonou F D'Alessio A et al . Comprehensive genome profiling for treatment decisions in patients with metastatic tumors: real-world evidence meta-analysis and registry data implementation. J Natl Cancer Inst. (2025) 117:1117–24. 10.1093/jnci/djaf015
3.
Johnson JA Caudle KE Gong L Whirl-Carrillo M Stein CM Scott SA et al . Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for pharmacogenetics-guided warfarin dosing. Clin Pharmacol Ther. (2017) 102:397–404. 10.1002/cpt.668
4.
Scott SA Sangkuhl K Stein CM Hulot JS Mega JL Roden DM et al . Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy. Clin Pharmacol Ther. (2013) 94:317–23. 10.1038/clpt.2013.105
5.
Tsimberidou AM Fountzilas E Nikanjam M Kurzrock R . Review of precision cancer medicine: evolution of the treatment paradigm. Cancer Treat Rev. (2020) 86:102019. 10.1016/j.ctrv.2020.102019
6.
Manickam K Buchanan AH Schwartz ML Hallquist ML Williams JL Rahm K et al . Exome sequencing-based screening for BRCA1/2 expected pathogenic variants among adult biobank participants. JAMA Netw Open. (2018) 4:e2035312. 10.1001/jamanetworkopen.2018.2140
7.
Zelek WM Fathalla D Morgan A Touchard S Loveless S Tallantyre E et al . Cerebrospinal fluid complement system biomarkers in demyelinating disease. Mult Scler. (2020) 26:1929–37. 10.1177/1352458519887905
8.
Kapoor V Briese T Ranjan A Donovan WM Mansukhani MM Chowdhary R et al . Validation of the VirCapSeq-VERT system for differential diagnosis, detection, and surveillance of viral infections. J Clin Microbiol. (2024) 62:e0061223. 10.1128/jcm.00612-23
9.
Radford A Kim JW Hallacy C Ramesh A Goh G Agarwal S et al . Learning transferable visual models from natural language supervision. arXiv (2021). 10.48550/arXiv.2103.00020
10.
Li LH Zhang P Zhang H Yang J Li C Zhong Y et al . Grounded language-image pre-training. arXiv (2022). 10.1109/CVPR52688.2022.01069
11.
Kather JN Pearson AT Halama N Jäger D Krause J Loosen SH et al . Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med. (2019) 26:1054–6. 10.1038/s41591-019-0462-y
12.
Schmauch B Romagnoni A Pronier E Saillard C Maillé P Calderaro J et al . A deep learning model to predict RNA-Seq expression of tumours from whole slide images. Nat Commun. (2020) 11:3877. 10.1038/s41467-020-17678-4
13.
Khera AV Chaffin M Aragam KG Haas ME Roselli C Choi SH et al . Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. (2018) 50:1219–24. 10.1038/s41588-018-0183-z
14.
Aragam KG Jiang T Goel A Kanoni S Wolford BN Atri DS et al . Discovery and systematic characterization of risk variants and genes for coronary artery disease in over a million participants. Nat Genet. (2022) 54:1803–15. 10.1038/s41588-022-01233-6
15.
Park S No C Kim S Han K Jung J-M Kwon K-Y et al . A multimodal screening system for elderly neurological diseases based on deep learning. Sci Rep. (2023) 13:21013. 10.1038/s41598-023-48071-y
16.
Popejoy AB Fullerton SM . Genomics is failing on diversity. Nature. (2016) 538:161–4. 10.1038/538161a
17.
Sirugo G Williams SM Tishkoff SA . The missing diversity in human genetic studies. Cell. (2019) 177:26–31. 10.1016/j.cell.2019.02.048
18.
Truong N Sun K Wang S Guitton F Guo Y . Privacy preservation in federated learning: an insightful survey from the GDPR perspective. Comput Sec. (2021) 110:102402. 10.1016/j.cose.2021.102402
Summary
Keywords
precision medicine, multi-modal AI, disease, genomics, diagnostics
Citation
Zhuang H (2025) How genomics and multi-modal AI are reshaping precision medicine. Front. Med. 12:1660889. doi: 10.3389/fmed.2025.1660889
Received
07 July 2025
Accepted
12 August 2025
Published
26 August 2025
Volume
12 - 2025
Edited by
Wen-Jing Wang, BGI Life Sciences Institute, China
Reviewed by
Ghizal Fatima, ERA's Lucknow Medical College, India
Yong Bai, Beijing Genomics Institute (BGI), China
Updates
Copyright
© 2025 Zhuang.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Han Zhuang han.zhuang.leon@gmail.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.