- 1Hangzhou Plastic Surgery Hospital (The Affiliated Hospital of the College of Mathematical Medicine, Zhejiang Normal University), Hangzhou, Zhejiang, China
- 2College of Mathematical Medicine, Zhejiang Normal University, Jinhua, Zhejiang, China
- 3School of Computer Science and Technology (School of Artificial Intelligence), Zhejiang Normal University, Jinhua, Zhejiang, China
- 4School of Mathematical Sciences, Zhejiang Normal University, Jinhua, Zhejiang, China
- 5Puyang Institute of Big Data and Artificial Intelligence, Puyang, Henan, China
Skin scars, resulting from the natural healing cascade following cutaneous injury, impose enduring physiological and psychological burdens on patients. This review first summarizes the biological classification of scars, their formation mechanisms, and conventional clinical assessment techniques. We then introduce core concepts of artificial intelligence, contrasting traditional machine learning algorithms with modern deep learning architectures, and review publicly available dermatology datasets. Standardized quantitative evaluation metrics and benchmarking protocols are presented to enable fair comparisons across studies. In the Methods Review section, we employ a systematic literature search strategy. Traditional machine learning methods are classified into unsupervised and supervised approaches. We examine convolutional neural networks (CNNs) as an independent category. We also explore advanced algorithms, including multimodal fusion, attention mechanisms, and self-supervised and generative models. For each category, we outline the technical approach, emphasize performance benefits, and discuss inherent limitations. Throughout, we also highlight key challenges related to data scarcity, domain shifts, and privacy legislation, and propose recommendations to enhance robustness, generalizability, and clinical interpretability. By aligning current capabilities with unmet clinical needs, this review offers a coherent roadmap for future research and the translational deployment of intelligent scar diagnosis systems.
1 Introduction
Scarring is a natural part of the skin healing process after injury, where permanent fibrous tissue replaces damaged skin. This process occurs when the body produces either an excessive or insufficient amount of collagen during wound healing, resulting in visible marks or traces on the skin's surface (1). Scars represent the skin's attempt to restore structure and function by replacing damaged tissue. However, these scar tissues differ from normal skin in terms of structure and function, often manifesting as changes in color, texture, or elasticity. Scars can vary widely in type, depending on the underlying cause, and typically include normal scars, hypertrophic scars, keloids, and atrophic scars, among others (2–4). Several examples of clinical images typical skin scars are illustrated in Figure 1.
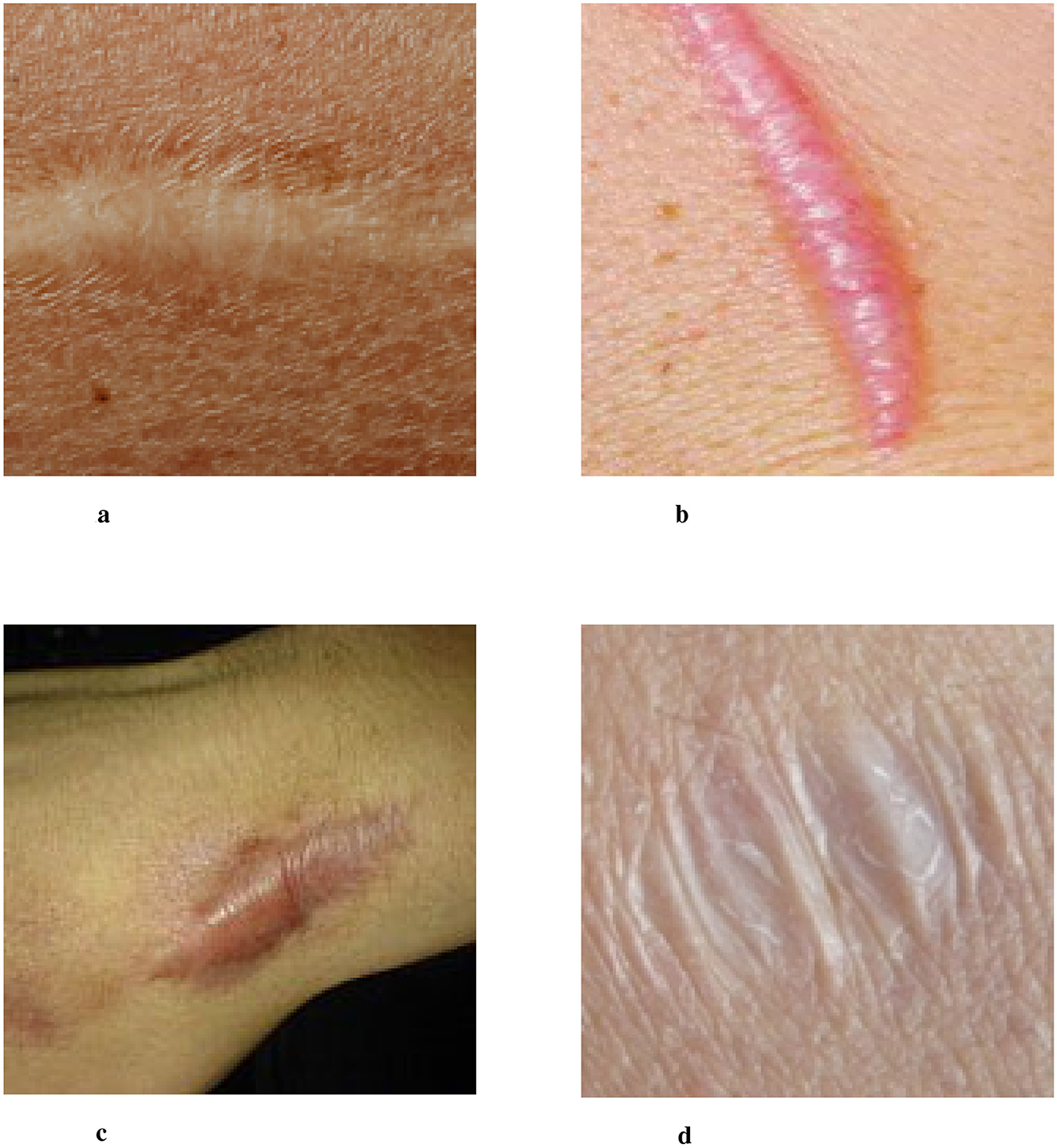
Figure 1. Four images of different scar types labeled a, b, c, and d. Image a shows a flat, pale scar. Image b depicts a raised, pinkish scar. Image c features a wide, darkened scar. Image d shows a wrinkled, white scar. Examples of clinical images of typical skin scars. The image data is sourced from publicly available datasets (Fitzpatrick 17k, etc.) and the Wikipedia entry “Scar.” (a) Normal Scar: Flat in appearance, with coloration closely resembling the surrounding skin, and a smooth surface texture. (b) Hypertrophic scar: characterized by a red or pink raised appearance that remains confined within the boundaries of the original wound. (c) Keloid: prominently elevated scar tissue that extends beyond the original wound margins, often darker in color. (d) Atrophic scar: marked by skin depression or indentation, commonly observed following the healing of acne or varicella (chickenpox) lesions.
The impact of scarring extends beyond the skin's surface, profoundly affecting the psychological and emotional wellbeing of patients, particularly when scars are located on visible areas such as the face. Scarring can lead to self-esteem issues, social anxiety, and even depression (5–8). Furthermore, certain types of scars, such as keloids, may also cause physical discomfort, including pain or itching, which can significantly impair the quality of daily life (9).
Due to the complexity of scars and their profound impact on individuals, developing precise and objective scar assessment methods is of paramount importance. Traditional scar assessment relies on clinicians' experience and subjective judgment. Training a physician capable of accurately diagnosing dermatological conditions requires many years of education and clinical practice, involving exposure to thousands of patients (10). With advancements in artificial intelligence (AI) and deep learning technologies, intelligent recognition and diagnostic systems have emerged as a powerful tool in research and clinical practice, offering an efficient and standardized approach to scar assessment.
Intelligent diagnostic systems analyze skin images to automatically identify scar types and severity, providing clinicians with accurate and objective diagnostic information. This technology not only accelerates the diagnostic process but also improves accuracy and consistency, allowing more personalized and targeted treatment plans for patients. More importantly, intelligent recognition techniques offer a non-invasive and convenient evaluation method, significantly enhancing patient experience and satisfaction (11).
Although AI and machine learning have achieved remarkable improvements in medical image diagnosis, such as skin cancer detection and dermatological lesion analysis (12, 13), research on the intelligent recognition and diagnosis of skin scars remains relatively scarce. Our comprehensive review of the existing literature confirmed this gap. This research gap may be attributed to several factors:
1. Limited availability of high-quality datasets: Compared to other medical imaging domains, systematically collecting and annotating high-quality scar images poses significant challenges. Standardization, privacy concerns, and ethical considerations further complicate the process. Unlike imaging modalities such as computed tomography (CT) or magnetic resonance imaging (MRI), which follow strict acquisition protocols, scar images can be highly variable due to differences in lighting conditions, camera devices, angles, and distances. Additionally, since scars may appear in private or sensitive areas of the body, patient privacy concerns and ethical constraints pose barriers to dataset acquisition.
2. Disparity in clinical research priorities: Medical research resources are often allocated to conditions deemed more urgent or life-threatening. While scars can significantly affect a patient's quality of life, they may not always be prioritized as a critical medical issue, leading to relatively limited research efforts in this domain.
In addition to the scar-focused intelligent recognition methods reviewed herein, several representative studies in related domains have emerged. Li et al. (14) proposed a skin lesion classification model that combines multi-scale feature enhancement with an interaction Transformer module; Wang et al. (15) developed a segmentation network that fuses edge and region cues to improve lesion boundary delineation; Wang et al. (16) demonstrated a wide-field quantitative phase imaging approach using phase-manipulating Fresnel lenses to enhance tissue contrast; and Wu et al. (17) introduced a Dynamic Security Computing Framework based on zero-trust privacy-domain prevention and control theory to secure privacy data. Although these works do not directly target scars, their innovations in network architecture design, imaging modality enhancement, and system-level security offer valuable, transferable insights for the future development of intelligent scar analysis systems. We hope that this review will inspire further research and technological advancements, driving the application of intelligent medical technologies in scar diagnosis and management. By improving diagnostic accuracy and efficiency, these innovations have the potential to provide more effective, personalized, and patient-centered treatment solutions.
2 Classification, mechanism and traditional diagnosis of scars
2.1 Classification and formation mechanism
The formation of scars is a complex biomedical process that involves multiple stages of the skin's self-repair mechanism. Each stage is influenced by various factors, leading to different types of scars. This section introduces the formation mechanisms of various scar types, including normal scars, hypertrophic scars, keloids, and atrophic scars. Understanding these mechanisms is essential for leveraging AI and machine learning (ML) technologies to improve scar recognition and assessment, ultimately enabling more precise and personalized treatment strategies for patients.
Scars are generally classified into the following types:
1. Normal scars: these are the most common type of scars, typically resulting from minor cuts or incisions. Over time, they tend to fade and become less noticeable.
2. Hypertrophic scars (2): these scars form due to excessive collagen production during the healing process, resulting in thickened and raised tissue. However, unlike keloids, hypertrophic scars remain confined to the original wound boundaries.
3. Keloids (4): keloids are an overgrown form of scar tissue that extends beyond the original wound margins. They are typically firmer than normal skin and may be accompanied by pain or itching. Certain individuals are genetically predisposed to keloid formation, making them more susceptible to this condition.
4. Atrophic scars: characterized by a sunken appearance, atrophic scars form when the healing process leads to tissue loss. They are commonly seen as residual scars from chickenpox or acne (18, 19).
The process of scar formation follows the skin's natural wound healing mechanism, which occurs in several key phases (20, 21):
1. Inflammation phase: this phase begins immediately after an injury and lasts for several days. The affected area exhibits redness, swelling, heat, and pain as part of the inflammatory response. Immune cells, such as white blood cells and macrophages, infiltrate the wound site to remove dead cells, pathogens, and foreign debris. Additionally, inflammatory mediators release cytokines and growth factors that play a crucial role in stimulating subsequent cell proliferation and tissue formation.
2. Proliferation phase: during this phase, fibroblasts rapidly proliferate and synthesize extracellular matrix proteins, such as collagen, to establish a new tissue framework. Concurrently, new blood vessels form (a process known as angiogenesis) to supply nutrients and oxygen to the developing tissue. However, excessive fibroblast activity and collagen deposition can lead to the overgrowth of scar tissue, resulting in hypertrophic scars or keloids.
3. Remodeling phase: this final phase of wound healing can last from several months to years. Newly synthesized collagen undergoes structural rearrangement and maturation, making the scar tissue more closely resemble normal skin. Over time, scars may become flatter and softer, although in some cases, suboptimal healing can result in persistent depressions or protrusions.
Scar formation is a dynamic and ongoing process, and extensive research has been conducted on the different stages of scar development, shown in Figure 2. A deeper understanding of these processes can significantly enhance the application of AI-driven technologies for intelligent scar recognition and diagnosis, ultimately leading to improved patient outcomes.
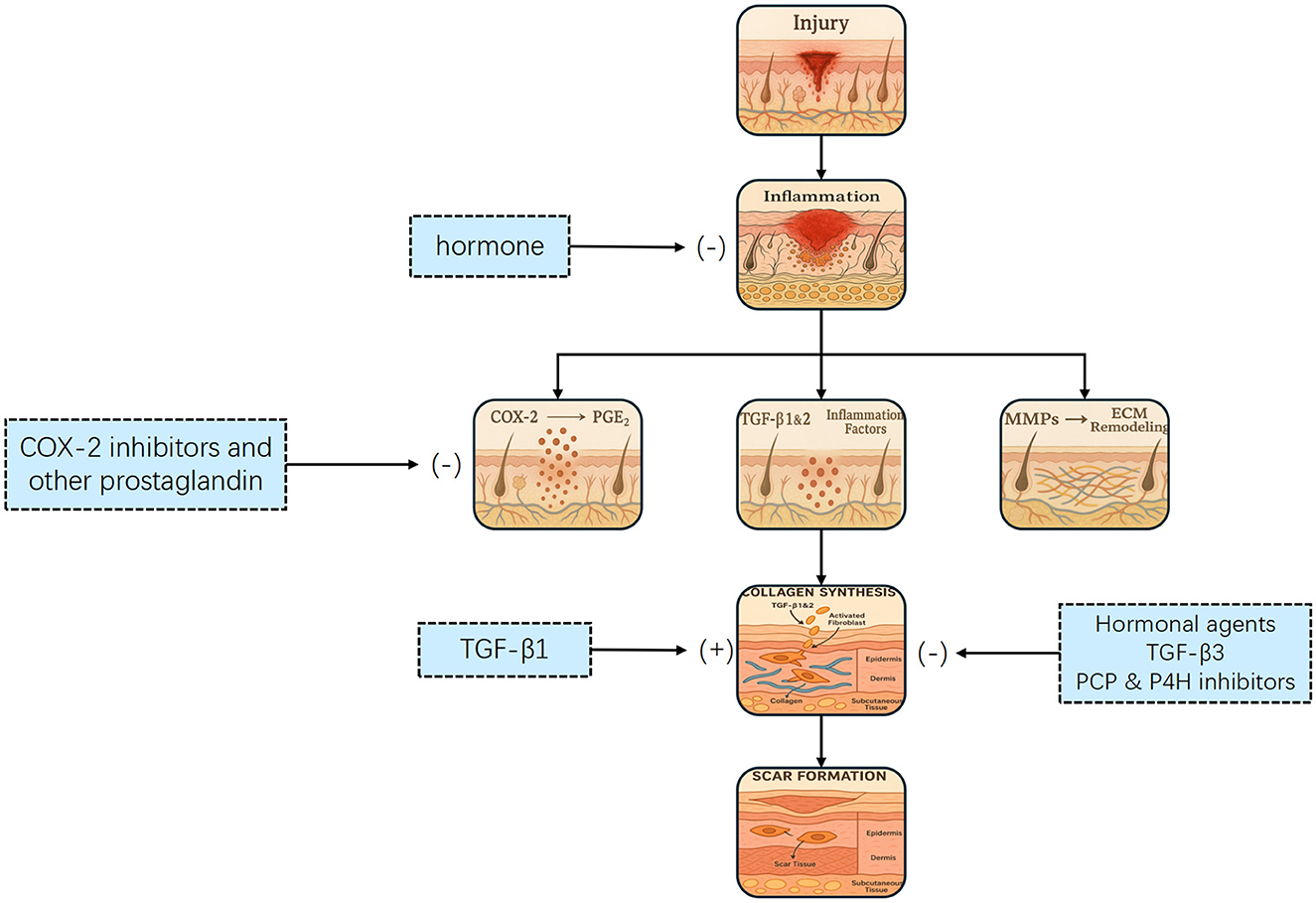
Figure 2. Flowchart illustrating the wound healing process, showing pathways from injury to scar formation. It includes inflammation, collagen synthesis, and ECM remodeling stages. Hormones, COX-2 inhibitors, and hormonal agents influence the process. The process of scar formation. Tissue injury initiates an inflammatory response that activates transforming growth factor-β (TGF–β) and other mediators, leading to fibroblast proliferation, migration, and differentiation. This promotes collagen synthesis and deposition, ultimately resulting in scar formation. Multiple therapeutic interventions targeting key steps—such as cyclooxygenase-2 (COX-2), TGF-β signaling, and fibroblast activity—may attenuate or prevent excessive scarring.
2.2 Traditional diagnostic methods
Before delving into intelligent scar recognition and diagnosis, it is essential to understand the foundation laid by traditional methods. Conventional scar assessment depends primarily on clinicians' experience and intuitive judgment, with visual and tactile examinations forming the core of the evaluation. Physicians first observe scar color, size, shape and contrast with surrounding skin to judge potential functional or aesthetic impact. Palpation then assesses hardness, texture, elasticity and temperature differences, helping to detect underlying inflammation or circulatory issues. Beyond these basic examinations, clinicians perform pain and sensory function tests and evaluate any functional limitations—for example, reduced joint range of motion due to perijoint scars. Standardized scales such as the Vancouver Scar Scale (VSS) and the Patient and Observer Scar Assessment Scale (POSAS) lend additional structure: the VSS scores vascularity, pigmentation, pliability and height, while the POSAS combines patient-reported symptoms with observer-rated scar characteristics (22, 23).
However, several key studies have quantified substantial inter-rater variability in these traditional scales. Draaijers et al. (23) evaluated 49 burn scar areas and reported single-observer reliability coefficients of r = 0.73 for the POSAS observer scale vs. r = 0.69 for the VSS (Cronbach's α = 0.69 and 0.49, respectively), indicating only moderate agreement among raters. Nedelec et al. (24) demonstrated that individual mVSS subscales yielded ICCs ≤ 0.30 and total mVSS scores ≤ 0.50, highlighting poor reproducibility of subjective metrics. More recently, Lee et al. (25) confirmed that both mVSS and POSAS fell below the acceptable Intraclass Correlation Coefficient (ICC) threshold of 0.70, whereas objective devices (e.g., ultrasound, colorimetry) achieved ICCs > 0.90.
These quantitative findings vividly illustrate the limitations of traditional visual and tactile assessment—namely, their reliance on subjective judgment and limited reproducibility. Consequently, there is a clear and growing need for AI-driven diagnostic approaches that can provide objective, consistent and fine-grained analysis of scar characteristics.
3 What is AI?
AI is a multidisciplinary field of computer science that aims to develop systems capable of performing tasks that typically require human intelligence. These tasks include reasoning, learning, problem-solving, perception, and language understanding. AI has evolved significantly over the past decades, driven by advances in computational power, data availability, and algorithmic innovations.
The field of AI encompasses several subdomains, including machine learning, natural language processing (NLP), computer vision (CV), expert systems, and robotics. Among these, machine learning—which enables systems to learn from data and improve their performance without being explicitly programmed—is one of the most transformative approaches, particularly in medical applications. With the development of deep learning, a subset of machine learning that utilizes neural networks to model complex patterns, AI has achieved remarkable breakthroughs in medical imaging, diagnosis, and personalized treatment (26).
As AI continues to advance, its integration into healthcare, including dermatology and scar assessment, holds great promise. The ability to automate medical image analysis and enhance diagnostic accuracy has positioned AI as a powerful tool in modern medicine, paving the way for more precise, efficient, and accessible healthcare solutions. The theoretical framework and methodologies of AI, as well as commonly used algorithms and network frameworks in ML and deep learning (DL), are illustrated in Figure 3.
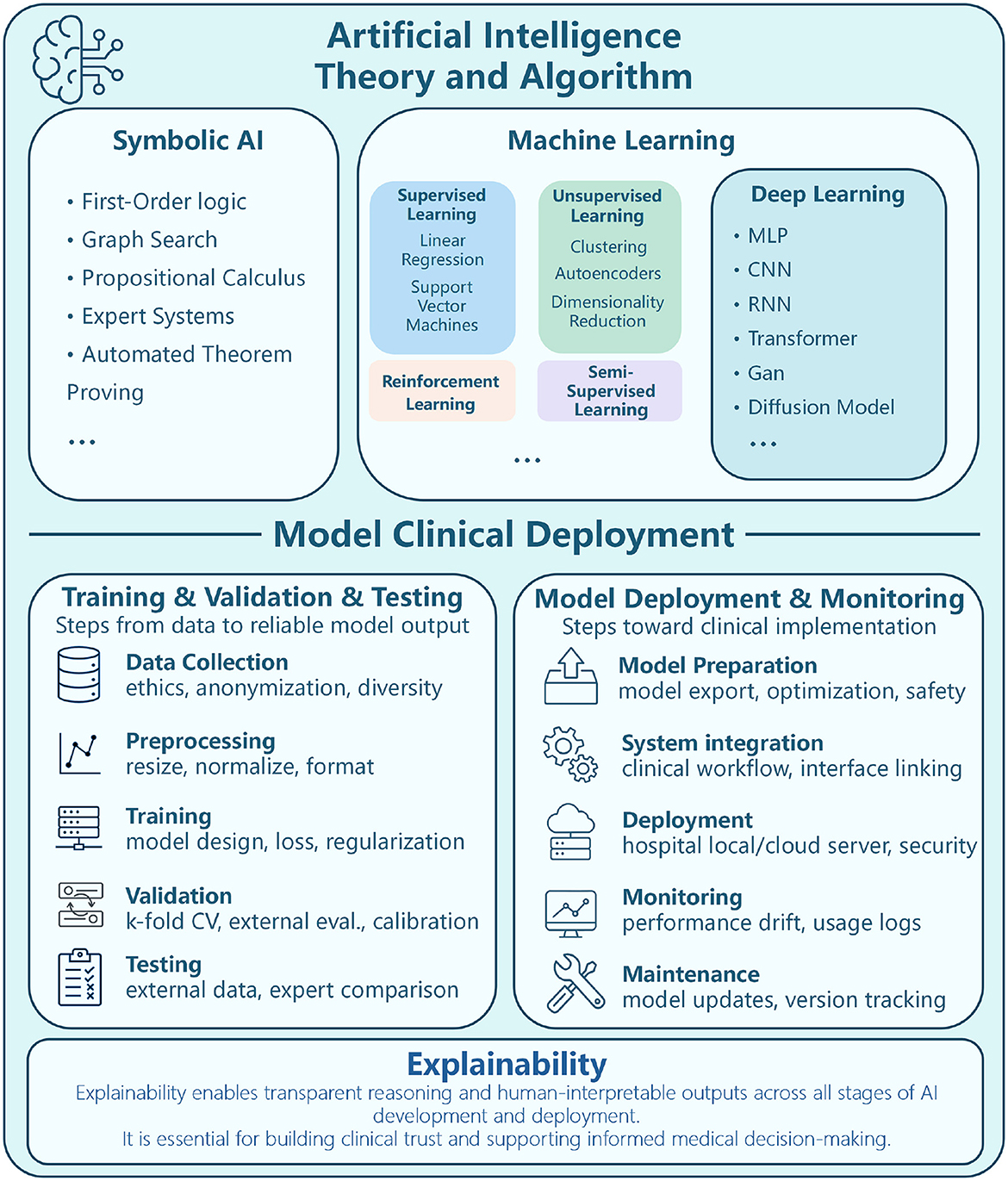
Figure 3. Diagram depicting AI theory, algorithm, and clinical deployment. Upper section outlines AI categories Symbolic AI (first-order logic, graph search), Machine Learning (supervised, unsupervised, reinforcement), and Deep Learning (MLP, CNN, RNN). Lower section details model clinical deployment, divided into Training/Validation/Testing (data collection, preprocessing, training, validation, testing) and Model Deployment/Monitoring (preparation, system integration, deployment, monitoring, maintenance). Emphasis on explainability for transparent reasoning and informed medical decision-making. Theoretical framework and methods of AI, ML algorithms, DL network framework.
3.1 Machine learning
Machine learning (ML) is a branch of artificial intelligence that enables systems to learn from data and improve their performance over time, without being explicitly programmed (27). ML models can be broadly categorized into three types based on how the data is used to train the model: supervised learning (28), unsupervised learning (29), and semi-supervised learning (30).
Supervised learning is the most common type of machine learning, where models are trained on labeled data, meaning each input data point has a corresponding output label. The goal is for the model to learn a mapping between inputs and outputs, so that it can predict the labels of new, unseen data. Common algorithms in supervised learning include linear regression (31), support vector machines (SVM) (32), k-nearest neighbors (KNN) (33), and decision trees (34). These algorithms are widely applied in tasks such as classification and regression, including applications like medical image classification (e.g., distinguishing benign from malignant tumors) and predicting patient outcomes (35). Supervised learning is essential when there is a large, labeled dataset available for training.
In contrast, unsupervised learning involves training models on data that does not have labeled outputs. The model's objective is to uncover the hidden structure or patterns within the data. Clustering and dimensionality reduction are typical examples of unsupervised learning tasks (36, 37). Algorithms such as k-means clustering (38), hierarchical clustering (39), and principal component analysis (PCA) are often used (40). In medical applications, unsupervised learning is helpful for segmenting medical images or identifying unknown patterns in complex datasets, such as detecting new disease subtypes based on genetic data (41).
Semi-supervised learning lies between supervised and unsupervised learning, where the model is trained on a combination of labeled and unlabeled data. This approach proves to be especially valuable when acquiring large labeled datasets is either difficult or costly, a situation that frequently arises in medical fields due to the scarcity of expert annotations. Semi-supervised learning can significantly improve the performance of the model by leveraging the abundance of unlabeled data. Techniques such as self-training and graph-based models are often employed in this approach. In healthcare, semi-supervised learning is increasingly used in medical image analysis, where only a small portion of the images may be annotated by experts, yet vast amounts of unannotated data are available (42).
3.2 Deep learning
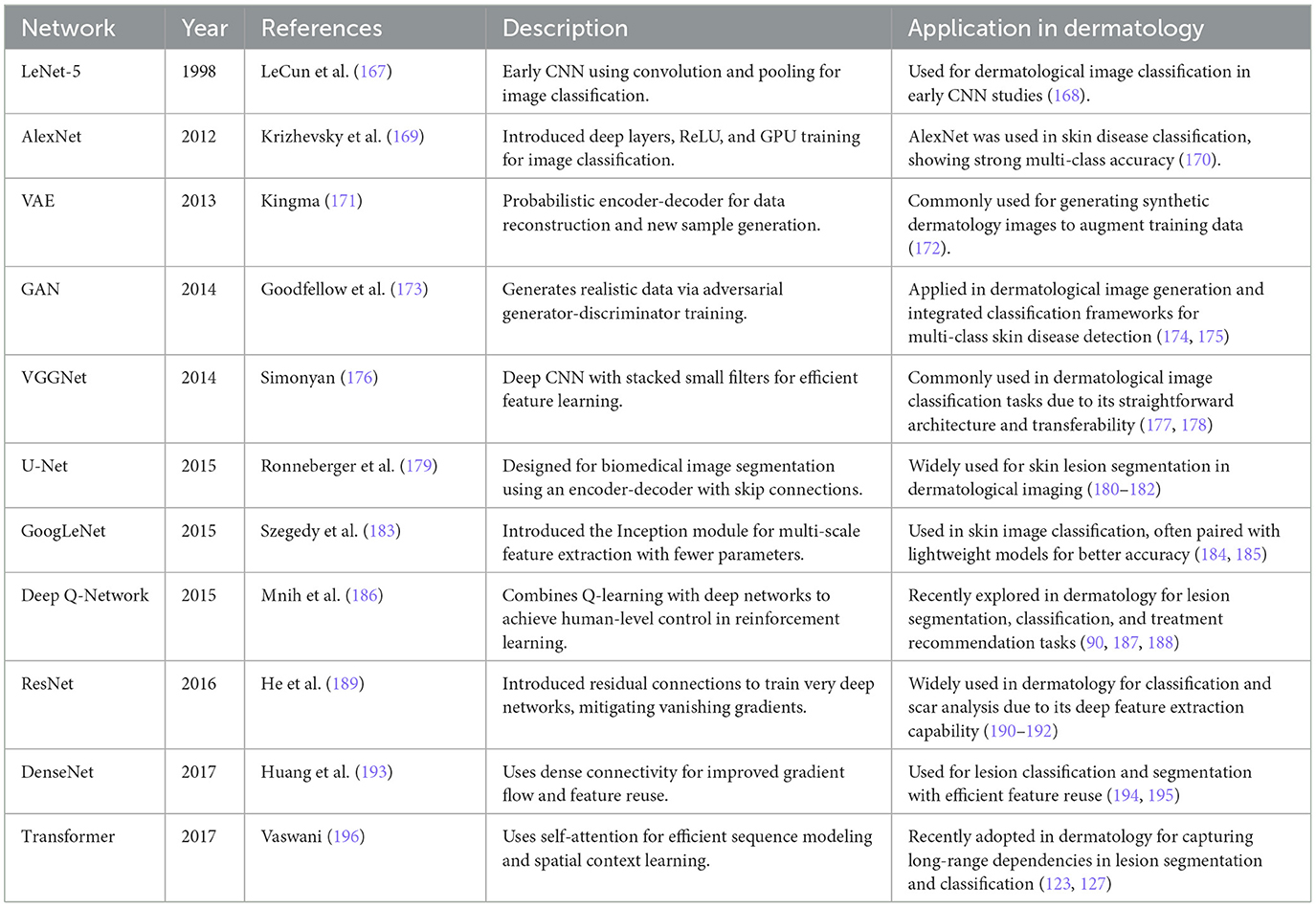
Deep learning has emerged as a transformative advancement in artificial intelligence, enabling machines to perform complex tasks that traditionally required human expertise. As a subfield of machine learning, deep learning utilizes multi-layered neural networks to automatically extract hierarchical features from raw data, thereby obviating the need for manual feature selection. This ability to learn directly from data allows deep learning models to generalize across diverse applications. At its core, deep learning processes information through interconnected layers, with early layers capturing low-level features (e.g., edges, textures) and deeper layers identifying more complex patterns, such as object structures or diagnostic markers in medical data (43). This hierarchical representation learning allows deep learning to achieve superior performance across domains.
Deep learning has revolutionized fields like computer vision (44), natural language processing (45), and biomedical research (46). In healthcare, it has enhanced medical imaging, enabling automated disease detection, segmentation, and classification (47–49). It has also driven advancements in drug discovery, genomics, and personalized treatment strategies.
The rapid adoption of deep learning can be attributed to three main factors:
1. Powerful feature extraction: deep learning's capability to learn representations directly from raw data eliminates manual engineering, allowing models to capture complex patterns;
2. Growth in data and computational power: the surge in digital data and advancements in computational resources have fueled deep learning's success;
3. Continuous evolution of architectures: innovations in model architectures and training techniques, coupled with open-source frameworks, have accelerated the deployment of deep learning solutions.
As deep learning continues to evolve, ongoing research aims to improve model interpretability, reduce data dependency, and enhance architecture efficiency. Its continued integration into healthcare and other industries is paving the way for intelligent automation, improved decision-making, and new scientific breakthroughs.
To illustrate the architectural diversity and historical evolution of deep learning, several representative models are summarized in Table 1, and a visual overview of a typical deep learning workflow shown in Figure 4.
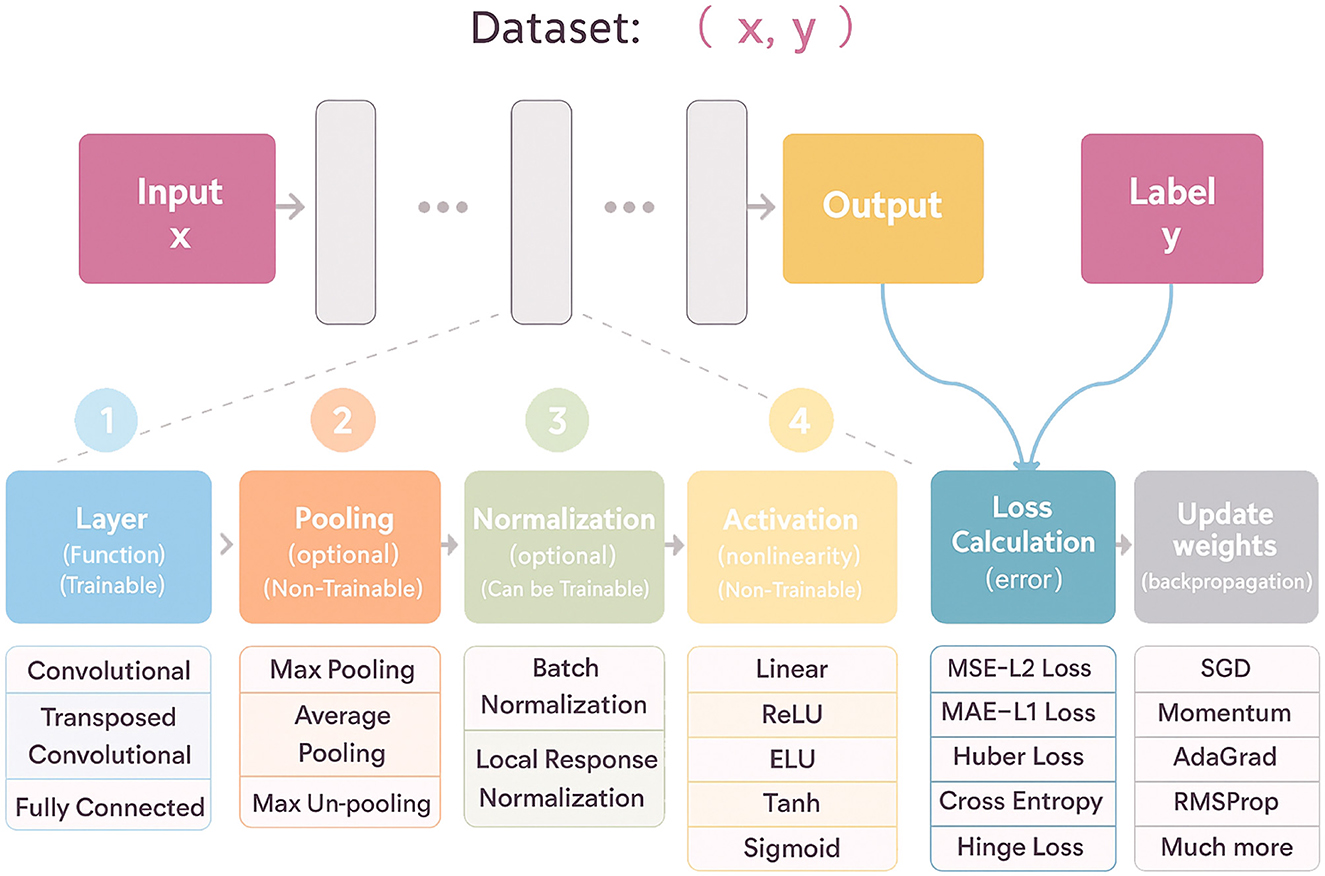
Figure 4. Flowchart illustrating a machine learning dataset process. Input x leads to an output and label y through stages: trainable layers with options like convolutional; optional non-trainable pooling; optional normalization; non-trainable activation functions; loss calculation; and weight updates via gradient descent methods. A visual overview of a typical deep learning workflow, illustrating the flow from input data through trainable and non-trainable components—such as convolutional layers, pooling, normalization, activation functions—to loss calculation and weight updates via backpropagation.
4 Dataset
Datasets are foundational to artificial intelligence, acting as carriers of information and knowledge that determine both the ceiling and the failure modes of downstream models (Figure 5). In current scar recognition research, however, most datasets are private and originate from hospital-affiliated projects with strict privacy and use restrictions. While such datasets may contain rich clinical detail, limited accessibility constrains reproducibility and the external validity of published findings.
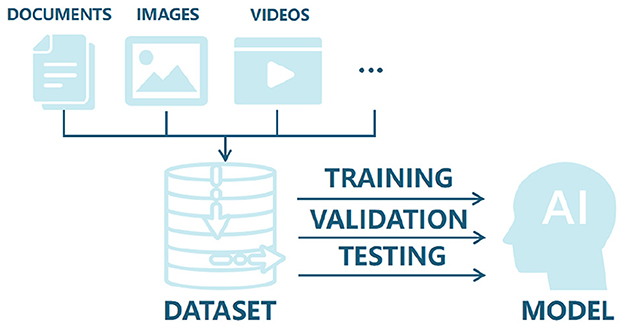
Figure 5. Diagram showing documents, images, and videos feeding into a dataset, which is used for training, validation, and testing an AI model. The process leads to a final AI model output. The foundational role of datasets in developing AI: from data collection to training, validation, and testing of intelligent systems.
Private datasets are typically collected and annotated by medical professionals. Their size and quality depend on patient volume, acquisition workflows, and the expertise of annotators. Although private collections may exhibit heterogeneous imaging conditions and granular labels, restricted access prevents independent validation and hampers community-wide progress.
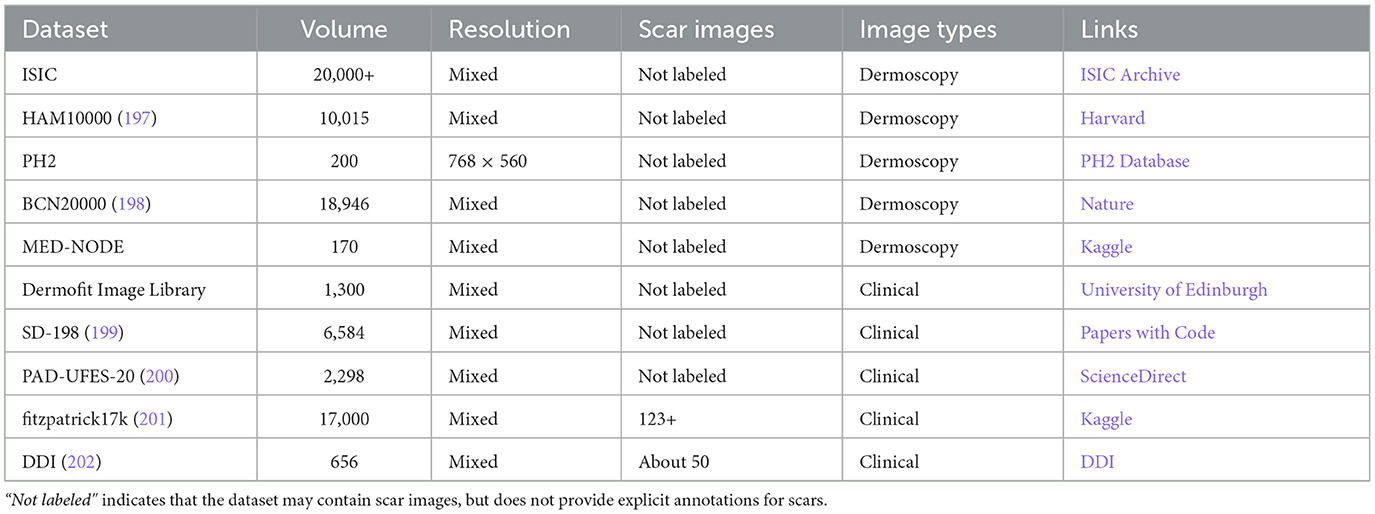
Beyond scar-specific corpora, the broader dermatology field maintains several well-established public datasets (Table 2), some of which incidentally include scar images (see examples from the ISIC repository in Figure 6). These resources, however, were seldom curated with scars as a primary target, and often lack the metadata necessary to study fairness and generalization in scar analysis. Such metadata gaps extend beyond technical parameters and include clinically and technically salient variables-such as patient phenotype, scar architecture, and imaging conditions-whose omission can hinder comprehensive bias and generalization assessments.
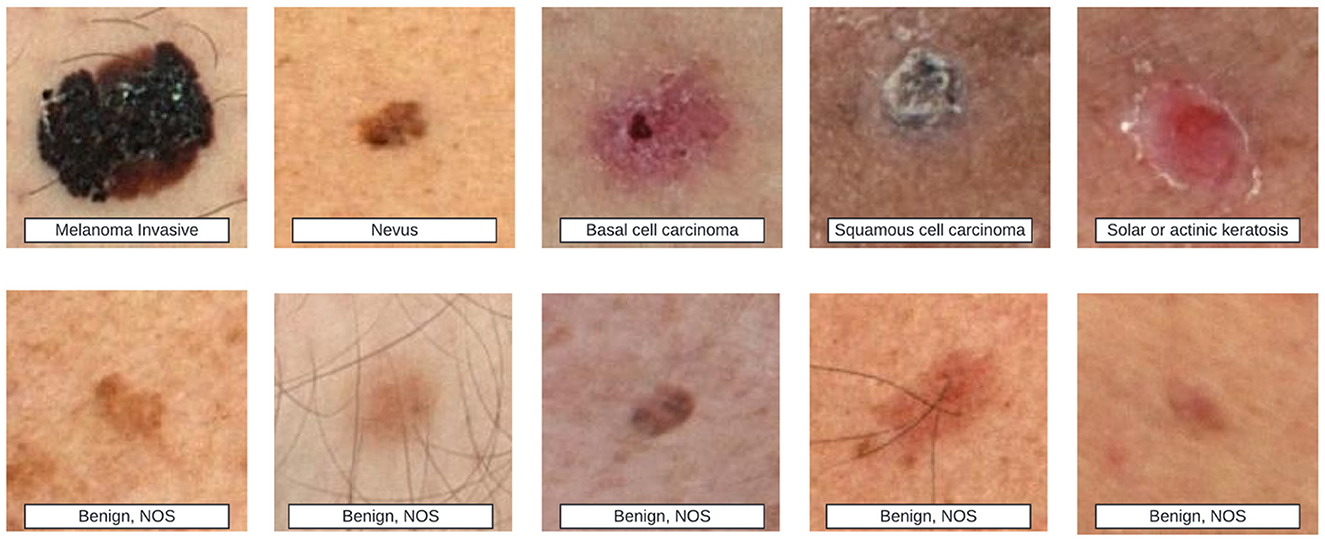
Figure 6. Top row shows various skin lesions labeled as melanoma invasive, nevus, basal cell carcinoma, squamous cell carcinoma, and solar or actinic keratosis. Bottom row contains different benign skin lesions labeled as benign, NOS. Example images from the ISIC dataset. The top row shows strong-labeled images, where detailed disease types are annotated. The bottom row shows weak-labeled images, where only benign or malignant status is provided. Some of these images may contain scar-like features, indicating their potential relevance to skin scar analysis.
4.1 Representational diversity (skin tones)
A growing body of evidence shows that widely used dermatology datasets are skewed toward lighter skin tones (Fitzpatrick I–III), resulting in performance disparities on darker phenotypes (50). For instance, generative or discriminative models trained on imbalanced data can systematically underperform on Fitzpatrick IV–VI even when sample size is controlled (51, 52). To support fair evaluation in scar analysis, future datasets should (i) record skin phenotype explicitly (e.g., Fitzpatrick I–VI or validated proxies), (ii) target balanced sampling across tone strata, and (iii) require subgroup reporting (per-tone sensitivity/specificity, balanced accuracy, worst-group accuracy, and calibration).
4.2 Scar architecture coverage
Clinical scars are heterogeneous in type, etiology, maturity, and anatomical site. Representative types include hypertrophic, keloid, atrophic, and contracture scars. Common etiologies include surgical wounds, burns, and trauma. Using labels aligned with established clinical instruments such as POSAS and VSS (23, 53), and recording item-level attributes-thickness, vascularity, and pliability-improves both learning and interpretability. Dataset splits should be stratified by patient identity as well as by scar type and anatomical site to prevent shortcut learning, where background skin texture or body region inadvertently serves as a proxy.
4.3 Imaging settings and acquisition variability
Generalization in clinical use hinges on robustness to illumination and equipment variability. We recommend recording: device class (smartphone/DSLR/dermoscope), sensor and lens, optical setting (polarized vs. non-polarized, flash/ring light), resolution and compression, white-balance/exposure mode, use of color charts, scene context (rulers, dressings, tattoos, hair), and capture protocol. Such metadata enables (i) cross-device/lighting analyses, (ii) leave-one-device/site-out validation, and (iii) targeted data augmentation (color constancy, exposure jitter) evaluated against held-out domains rather than the training distribution (54, 55).
4.4 Multimodal and metadata-rich datasets (clinical photos, dermoscopy, and 3D)
Beyond routine photographic images, scar categorization benefits substantially from complementary modalities and structured metadata. Clinical photographs capture global color and texture together with contextual cues; dermoscopic images (polarized/non-polarized) reveal vascular and pigment structures that aid in distinguishing hypertrophic from keloid scars (56, 57). Three-dimensional surface imaging (e.g., stereophotogrammetry or laser profilometry) provides height and volume maps for objective quantification and treatment monitoring (58, 59). Cross-sectional modalities such as optical coherence tomography (OCT) and high-frequency ultrasound (HFUS, with elastography where available) capture subsurface morphology, thickness, and stiffness associated with activity and maturity (60–62). In parallel, aggregated meta-datasets in dermatology increasingly pair clinical and dermoscopic photographs or integrate multi-institution, multi-modality collections with standardized metadata, which improves skin-tone-stratified analyses and cross-site/device generalization (60, 63, 64). When paired with well-defined fields (anatomical site, etiology and maturity, Fitzpatrick or Monk Skin Tone, device/illumination/polarization, calibration targets), such resources provide stronger supervision for differentiating scar architectures, quantifying activity, and disentangling confounders due to lighting or device variability.
Despite these advantages, most existing scar datasets either lack the above modalities or do not release consistent metadata schemas, limiting fairness assessments across skin tones and external validity across clinics and equipment. We therefore advocate curating aggregated, multi-institution datasets that (i) include harmonized clinical photos, dermoscopy, and-where feasible-3D or cross-sectional imaging; (ii) adopt standardized acquisition protocols and per-image metadata fields (see Table 3); and (iii) support evaluation protocols that explicitly test cross-modality generalization, leave-one-site/device-out splits, worst-group performance (e.g., Fitzpatrick types IV-VI), and probability calibration. These recommendations align dataset design with downstream clinical reliability.
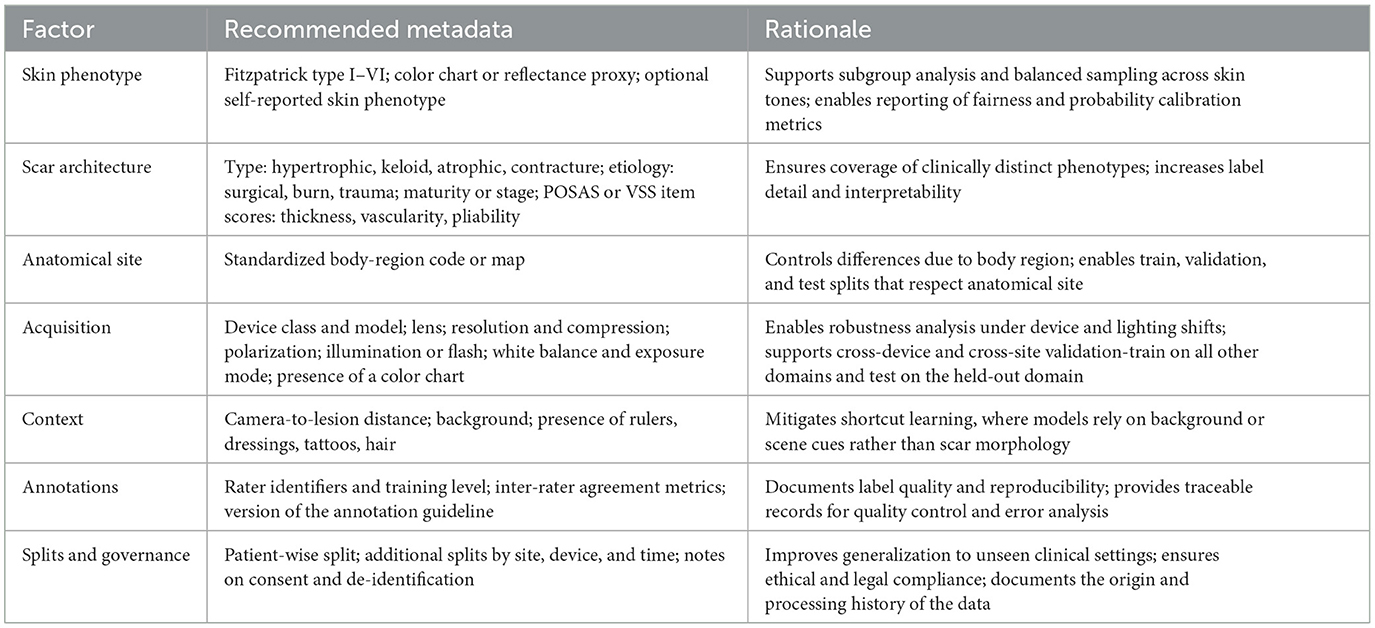
Table 3. Recommended metadata fields for scar-image datasets to support fairness assessment and generalization.
Building on these considerations, future research should aim to develop standardized, multimodal, and metadata-rich meta-datasets, together with bias-aware evaluation frameworks.
5 Privacy constraints and ethical AI training
As noted above regarding dataset privacy and ethical approvals, contemporary medical artificial intelligence development must navigate stringent privacy regulations, e.g., general data protection regulation (GDPR), health insurance portability and accountability act (HIPAA), and ethical review processes, which restrict data sharing and centralization. To address these challenges, researchers have developed a range of privacy-preserving techniques, including federated learning, synthetic data augmentation, differential privacy, and encryption-based methods, each of which balances data utility, privacy guarantees, and computational overhead in its own way.
5.1 Federated learning for decentralized model training
Federated Learning (FL) enables multiple institutions to collaboratively train a global model by exchanging local model updates rather than raw patient data, thus minimizing privacy risks associated with central data aggregation (65). In medical imaging, FL frameworks have been successfully applied to histopathology and radiology datasets, maintaining performance comparable to centralized training while respecting data sovereignty (66, 67). Recent advances integrate transfer learning and adaptive aggregation to further improve accuracy across heterogeneous sites without compromising privacy (68).
5.2 Synthetic data augmentation
When real-world medical datasets are scarce or cannot be shared due to privacy constraints, synthetic data generated by Generative Adversarial Networks (GANs) can augment training sets. GAN-based augmentation has been shown to improve CNN performance in tasks such as liver lesion classification and chest X-ray analysis, boosting sensitivity and specificity on underrepresented classes (69, 70). Comprehensive reviews demonstrate that synthetic data not only increases data diversity but can also serve as an anonymization tool, enabling model training without exposing patient-identifiable images (71, 72).
5.3 Differential privacy and encryption techniques
Differential Privacy (DP) introduces carefully calibrated noise into model updates or outputs, providing quantifiable privacy guarantees against inference attacks. DP-enabled FL frameworks have demonstrated practical viability in complex medical image analysis, achieving performance on par with non-private methods while bounding privacy loss (66, 73). Encryption approaches, particularly Homomorphic Encryption (HE), allow computations to be performed directly on encrypted data, ensuring that raw data remain confidential throughout training and inference (74). Fully Homomorphic Encryption (FHE) schemes, though computationally intensive, have been successfully prototyped for optical coherence tomography(OCT) image classification and chest CT nodule detection, marking a step toward “zero-trust” AI in healthcare (75, 76).
5.4 Other emerging strategies
Beyond these core methods, secure multi-party computation (SMPC) and zero-knowledge proofs (ZKP) are gaining attention for enabling privacy-preserving analytics without revealing sensitive inputs (77). Concurrently, the development of synthetic cohort generation via diffusion models and advance in privacy-balanced data sharing agreements hold promise for ethically ground AI research while safeguarding patient rights.
By embedding these privacy-centric techniques into the AI lifecycle—from data augmentation to model deployment—researchers can better balance clinical innovation with ethical and regulatory imperatives, fostering trust and enabling broader adoption of AI in medicine.
6 Intelligent scar recognition and diagnosis
6.1 Search strategy
To ensure this review encompasses all relevant research on “Intelligent Recognition and Diagnosis of Skin Scars,” a multi-step search strategy was employed. Initially, a comprehensive search was conducted in databases such as Google Scholar, PubMed, Web of Science, and Science Direct. Keywords were systematically combined, including terms such as “skin scars,” “scarring,” “burn,” “wound,” “hypertrophic,” “keloids,” “atrophic,” “dermatology,” “intelligent,” “automatic,” “recognition,” “diagnosis,” “segmentation,” “detection,” and “image analysis.” Additionally, to broaden the search scope, auxiliary keywords like “computer vision,” “machine learning,” “deep learning,” and “artificial intelligence” were also included.
The search was limited to English-language publications from the past 5–10 years to ensure the inclusion of the most recent advancements in the field. The inclusion criteria for the selected papers were: (i) research on skin scars related to the detection, recognition, segmentation, and classification of prior damage, (ii) traditional image processing methods, (iii) conventional machine learning methods, (iv) deep learning methods, (v) digital image modalities, and (vi) articles published in well-defined, reputable journals.
The initial search yielded 67,500 papers. The results were then refined through several rounds of screening: (1) removal of duplicate articles and inclusion based on the above criteria, (2) a thorough review of full-text papers to exclude studies with inadequate methodologies or irrelevant data, (3) manual examination of reference lists to ensure no relevant studies were overlooked. After these steps, a total of 33 articles were selected for inclusion. These articles comprehensively address all aspects of the topic, ranging from image acquisition and preprocessing to the application of traditional image processing and AI-based methods, as well as the evaluation of experimental results. The overall process is shown in the Figure 7.
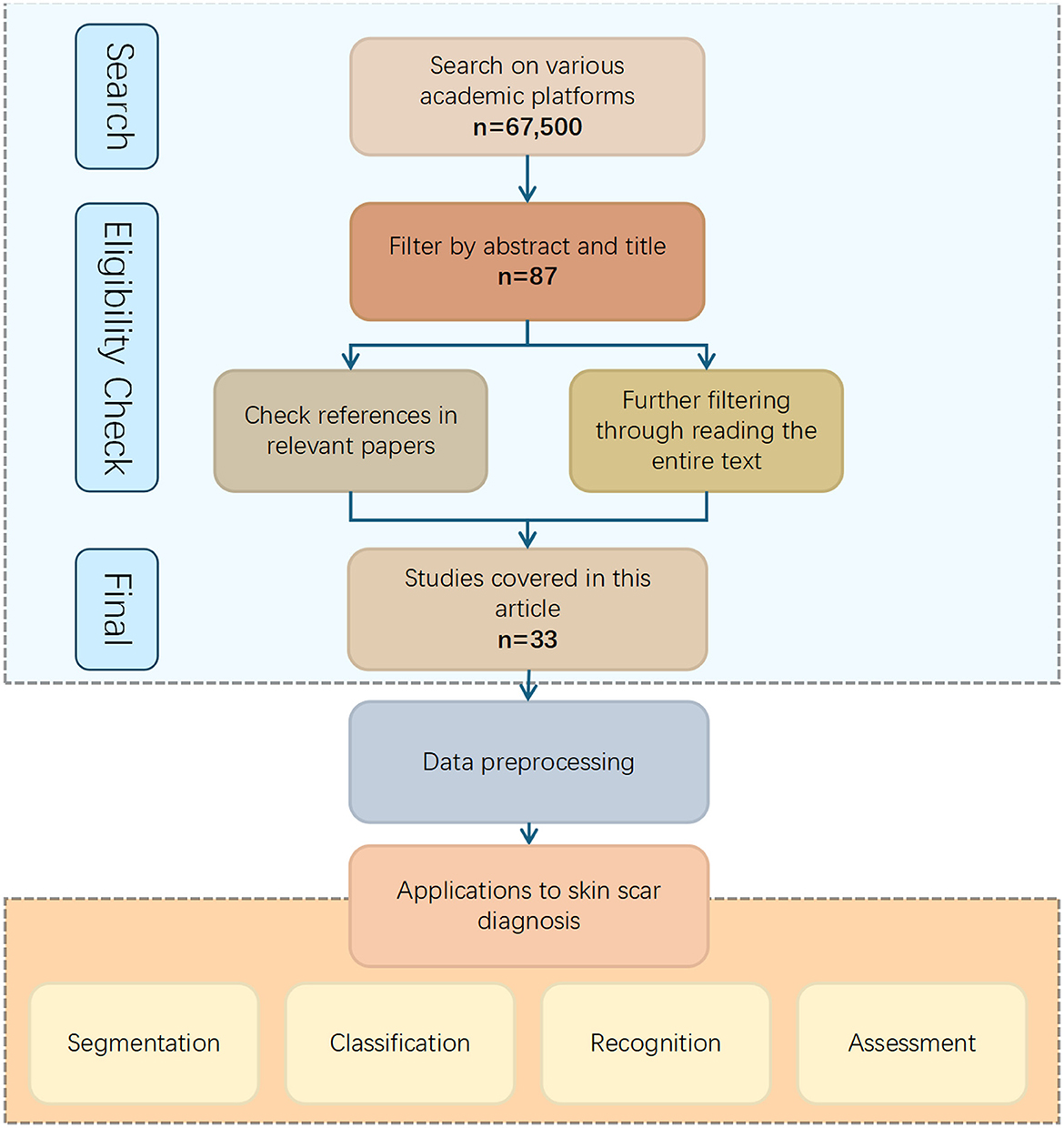
Figure 7. Flowchart depicting the process of selecting studies for skin scar diagnosis applications. Starting with a search on academic platforms yielding sixty-seven thousand five hundred results, filtering by abstract and title reduces it to eighty-seven, and further reference and text checks lead to thirty-three studies. These are used for data preprocessing, resulting in applications for segmentation, classification, recognition, and assessment of skin scars. Search strategy.
This broad coverage of literature ensures the comprehensiveness and depth of this review, providing a solid foundation for future research directions.
6.2 Quantitative evaluation metrics for intelligent diagnosis
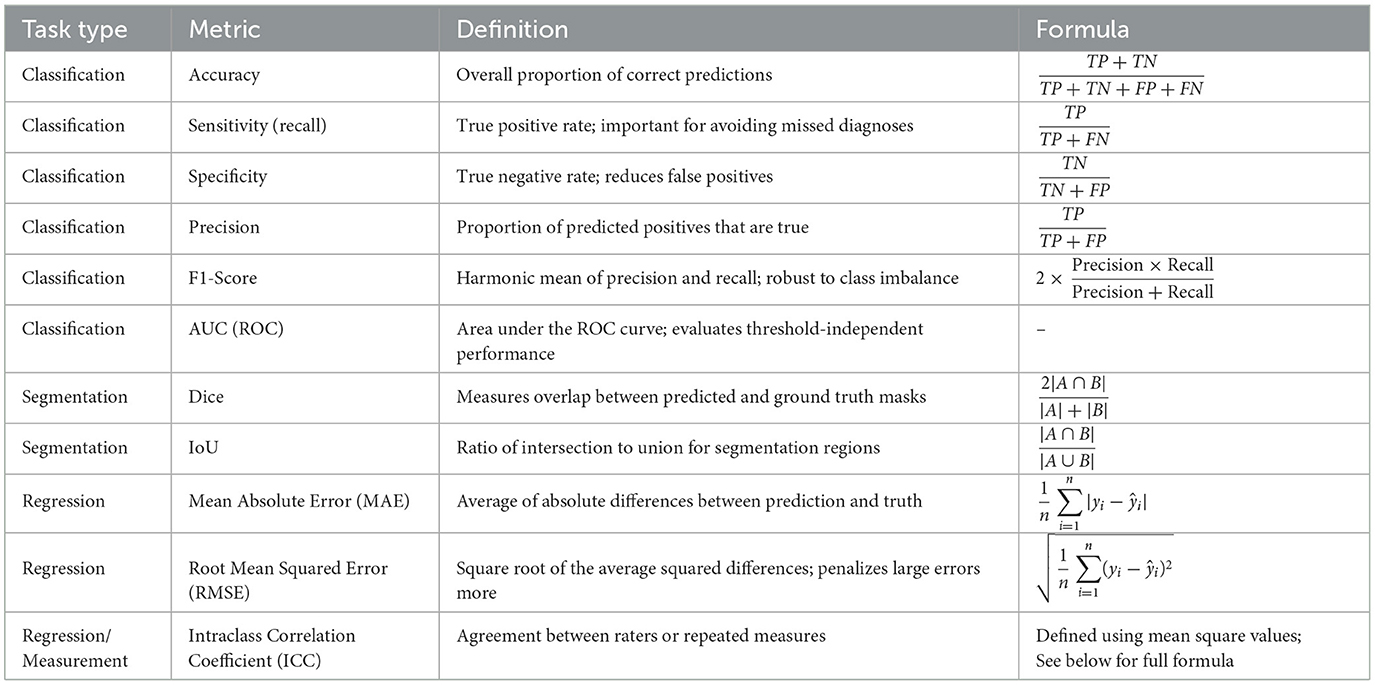
Quantitative evaluation metrics are essential tools used to objectively assess and compare the performance of AI-based diagnostic methods. The following summarizes the commonly used metrics in classification, segmentation, and regression tasks. To enhance clarity and compactness, the metrics are presented in Table 4, accompanied by unified symbol definitions.
To ensure clarity and consistency in interpreting the evaluation metrics presented above, the key symbols and variables used in the formulas are defined as follows:
• TP (True positive): number of positive cases correctly predicted as positive.
• TN (True negative): number of negative cases correctly predicted as negative.
• FP (False positive): number of negative cases incorrectly predicted as positive.
• FN (False negative): number of positive cases incorrectly predicted as negative.
• A, B: in segmentation tasks, A denotes the set of predicted pixels (or regions), and B denotes the ground truth set.
• yi, ŷi: The ground truth and predicted continuous values for the i-th sample, respectively.
• n: total number of samples or observations in the dataset.
• k: number of raters or measurement repetitions in reliability assessments.
• MSR: mean square for rows (typically subjects) in the ICC calculation.
• MSC: mean square for columns (typically raters) in the ICC calculation.
• MSE: mean square error term in the ICC formulation, representing residual variance.
The Intraclass Correlation Coefficient (ICC) is a widely adopted statistical measure for assessing the reliability of quantitative measurements made by different raters or systems. In the context of intelligent diagnosis and clinical research, ICC is commonly used to evaluate either consistency or absolute agreement between observers. Various forms of ICC exist, depending on the statistical model employed, e.g., one-way vs. two-way analysis of variance(ANOVA), the nature of the raters (fixed vs. random effects), and whether the evaluation is based on single or average measurements.
Among these, ICC(2,1) is frequently utilized due to its suitability for assessing absolute agreement under a two-way random-effects model, in which both raters and subjects are assumed to be random samples. This variant is particularly appropriate in studies where generalization to a broader population of raters is desired. The corresponding formula is derived from an ANOVA decomposition that partitions the observed variance into components attributable to subjects, raters, and residual error. It is expressed as:
where MSR, MSC, and MSE denote the mean square values for rows (subjects), columns (raters), and residuals, respectively.
In regression-based medical AI applications, mean absolute error (MAE) and root mean squared error (RMSE) are among the most frequently used evaluation metrics. MAE quantifies the average magnitude of prediction errors, offering direct interpretability in clinical units such as millimeters or severity grades. RMSE, due to its squared term, places greater emphasis on larger errors, making it more sensitive to outliers and thus useful in safety-critical predictions.
These metrics are particularly informative when assessing models that predict continuous clinical scores. For example, in scar severity prediction tasks, MAE values close to 1.0 and RMSE values around 1.4 may indicate that model outputs typically differ from expert-assigned scores by approximately one severity level, reflecting strong alignment with clinical judgment (78).
For classification tasks, the receiver operating characteristic (ROC) curve serves as a standard method to visualize model performance across varying decision thresholds. It plots the true positive rate (sensitivity) against the false positive rate (1− specificity), thus illustrating the trade-off between sensitivity and specificity.
The area under the ROC curve (AUC) condenses this information into a single scalar metric ranging from 0.5 (random performance) to 1.0 (perfect discrimination). AUC offers a threshold-independent assessment of a classifier's ability to distinguish between positive and negative cases. While the numerical value provides a summary, its interpretation is often more intuitive when supported by ROC visualizations.
As illustrated in Figure 8, simulated ROC curves demonstrate how classifiers of varying quality (e.g., Model A, B, and C) differ in performance. Curves that approach the top-left corner correspond to higher AUCs and stronger discriminative power. Such visual representations are especially helpful when comparing models or assessing robustness across thresholds.
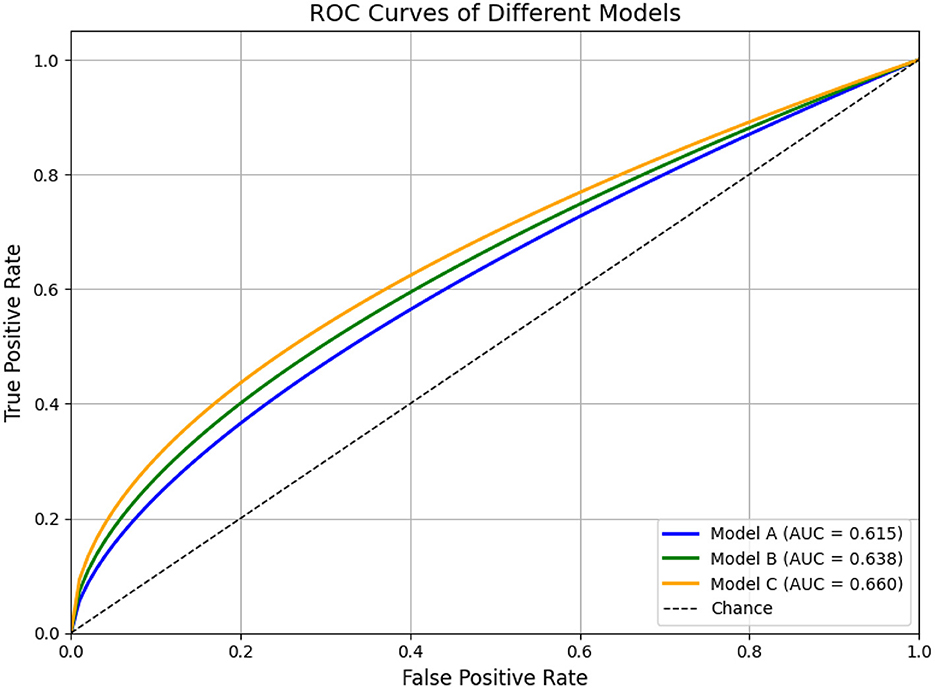
Figure 8. ROC curves for three models, showing the True Positive Rate versus False Positive Rate. Model A (blue) has an AUC of 0.615, Model B (green) 0.638, and Model C (orange) 0.660. A diagonal line represents chance. Simulated ROC curves of three hypothetical classification models (Model A, B, and C). Model A (blue) illustrates moderate classification performance (AUC = 0.80), Model B (green) shows improved overall discrimination (AUC = 0.88), and Model C (orange) demonstrates near-optimal performance (AUC = 0.97). The diagonal dashed line represents random classification (AUC = 0.5). This figure is intended for illustrative purposes to demonstrate how ROC curves and AUC values reflect the ability of models to distinguish between classes across various thresholds.
In segmentation tasks, the Dice similarity coefficient and Intersection over Union (IoU) are commonly employed to evaluate spatial overlap between predicted and ground truth regions. While related, these metrics serve different purposes and are not directly interchangeable.
Dice is particularly advantageous in scenarios with pronounced class imbalance—such as lesion or scar segmentation—where the target region occupies a small fraction of the image. It gives proportionally more weight to correctly identified positive pixels, making it sensitive to small structure detection.
In contrast, IoU is a stricter metric that penalizes both over- and under-segmentation. It is better suited for applications requiring precise boundary delineation, such as organ contouring or multi-class anatomical segmentation.
Therefore, metric selection should align with the clinical goal: Dice is more appropriate for detecting small or subtle targets, whereas IoU is preferred when spatial accuracy and structure completeness are prioritized.
These definitions provide a standardized interpretation of each metric, facilitating consistent comparison and critical evaluation of intelligent diagnostic systems across studies.
6.3 Unsupervised traditional machine learning methods
Unsupervised methods for scar segmentation and measurement typically exploit clustering or rule-based cues to delineate regions of interest without requiring annotated data. Ma et al. (79) presented a saliency-based segmentation framework for skin scars: Gaussian pyramid feature maps are clustered to produce saliency maps, which are then thresholded to isolate scar regions. Khan et al. (80) developed a segmentation pipeline based on fuzzy C-means clustering with an intelligent cluster-selection mechanism; they demonstrated that the Q (YIQ) and I3 (I1I2I3) chrominance components yield optimal cluster separation, achieving 92.63% segmentation accuracy on a set of 50 images. Chantharaphaichi et al. (81) proposed a rule-based image-processing scheme for acne lesion detection: grayscale and HSV(Hue, Saturation, Value) transformations are combined with brightness subtraction and size filtering to generate candidate lesion regions, which are then bounded with minimal operator intervention. Lastly, Jiang et al. (82) used unlabeled smartphone images of keloids to reconstruct three-dimensional models via parallel computing and extracted the maximum diameter, thickness, and volume. These measures showed excellent agreement with manual caliper and ultrasound assessments (ICCs > 0.95), indicating a highly repeatable, fully unsupervised measurement protocol.
6.4 Supervised traditional machine learning methods
Supervised approaches leverage hand-crafted feature extraction followed by classical classifiers trained on labeled examples. Liu et al. (83) combined local binary pattern (LBP) operators with wavelet-based texture analysis on multiphoton fluorescence microscopy images of scars; the resulting features were fed into a support vector machine (SVM) to distinguish scar tissue. Heflin et al. (84) introduced an automatic detection and classification system for scars, marks, and tattoos in unconstrained, forensic-style images by training classifiers on annotated samples from real-world scenarios. Abas et al. (85) fused entropy-based region-of-interest extraction with dual-tree wavelet frame (DWF) and gray-level cooccurrence matrix (GLCM) texture features, then employed decision trees to classify six types of acne lesions, achieving 85.5% accuracy. Alamdari et al. (86) implemented a mobile application that segments lesions via k-means and classifies them with fuzzy logic and SVMs, reporting 100% accuracy in acne detection and up to 80% in scar classification. Kittigul and Uyyanonvara (87) extracted speeded-up robust features (SURF) descriptors along with hue mean, RGB (Red, Green, Blue) standard deviations, and circularity, and used a k-nearest neighbors (k-NN) classifier to achieve 73% sensitivity, 84% precision, and 68% overall accuracy. Al-Tawalbeh et al. (88) built a three-class skin-lesion classifier (benign, melanoma, seborrheic keratosis) using 71 color and texture features across multiple color spaces and Gabor filters; a second-order polynomial SVM yielded 95.8% overall accuracy and 99.7% precision for seborrheic keratosis on non-segmented images. Finally, Maroni et al. (89) combined Haar-cascade body-part detection with random-forest skin segmentation (using multimodal features), CIELab heat-mapping, adaptive thresholding, and Laplacian-of-Gaussian blob detection to count acne lesions and monitor severity under real-world conditions.
As shown in Table 5, traditional machine learning approaches can be broadly classified into unsupervised and supervised methods. Unsupervised techniques (e.g., saliency-based clustering and fuzzy C-means) require no labeled data and offer automated segmentation and measurement, but they can be sensitive to parameter choices and imaging variation. In contrast, supervised methods (e.g., LBP + SVM, SURF + k-NN, Gabor + multiclass SVM) learn from annotated examples to achieve higher classification accuracy, though their performance depends heavily on the quality and diversity of the training dataset.
6.5 Unsupervised CNN-based methods
To date, there have been no purely unsupervised CNN architectures applied to scar recognition or diagnosis in the literature covered; all deep-learning approaches rely on annotated data and end-to-end supervised training to learn feature representations or perform segmentation. However, recent work has explored deep reinforcement learning (DRL) as a form of weakly supervised segmentation that does not require pixel-wise annotation during inference. Usmani et al. (90) cast lesion delineation as a Markov decision process and train an agent via deep deterministic policy gradient (DDPG) to “draw” segmentation masks in a continuous action space, using only global reward signals derived from expert-provided ground-truth masks. Their method achieved accuracy of 96.33% on naevus, 95.39% on melanoma, and 94.27% on seborrheic keratosis in the ISIC 2017 dataset, and comparable performance on HAM10000 and PH2 (96.3%, 95.4%, and 94.3%, respectively). Although this approach still depends on ground-truth masks to compute rewards during training, it eliminates the need for dense, step-by-step pixel annotations and thus represents a promising direction toward unsupervised—or more accurately, weakly supervised—deep segmentation methods for skin lesions.
6.6 Supervised CNN-based methods
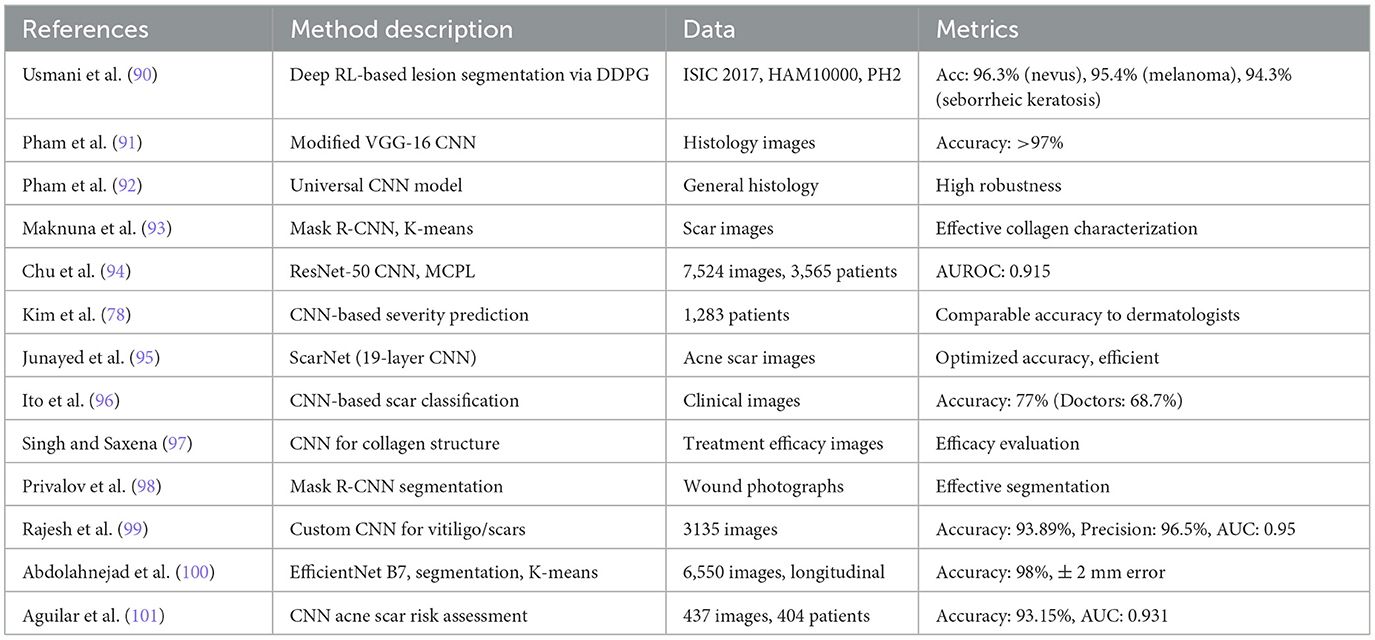
Convolutional neural networks (CNNs) have rapidly become the state of the art in scar recognition and diagnosis by learning hierarchical feature representations directly from data. Table 6 summarizes key CNN architectures, datasets, and performance metrics reported in the literature, while Figure 9 illustrates a prototypical multi-task VGG-based network that combines classification and segmentation branches. In the following section, we examine these supervised CNN approaches by discussing their design innovations, clinical datasets, and quantitative outcomes, and then outline the challenges they face in generalization, interpretability, and computational demands.
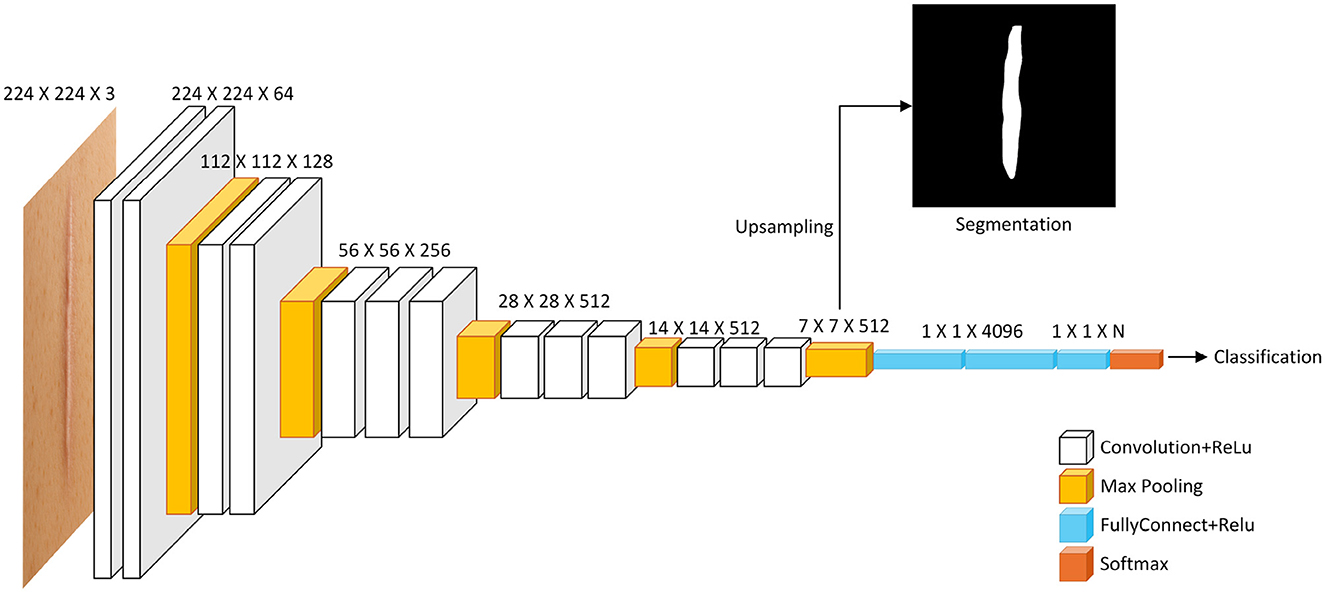
Figure 9. Diagram illustrating a deep learning model structure for image processing. It shows layers including Convolution and ReLU, Max Pooling, Fully Connected with ReLU, and Softmax. The input is a 224 x 224 x 3 image. Outputs include a segmentation image through upsampling and a classification result. Layer dimensions decrease from left to right, with specified sizes at each stage. A multi-task CNN architecture based on VGG. The shared convolutional backbone feeds into two branches: one for image classification via fully connected layers, and another for semantic segmentation. The segmentation path requires upsampling (e.g., transposed convolution) to restore spatial resolution.
Figures 10, 11 summarize the performance metrics of the methods discussed in this chapter, corresponding to classification and segmentation tasks, respectively. These visual comparisons aim to provide readers with a convenient overview of how different approaches perform under commonly used evaluation criteria. The figures include only studies that report standard quantitative metrics; methods employing less conventional evaluations (e.g., mask dimension differences in segmentation) are not represented. Similarly, studies focusing on tasks such as scar characterization or analysis are excluded, as these often lack universally adopted quantitative benchmarks.
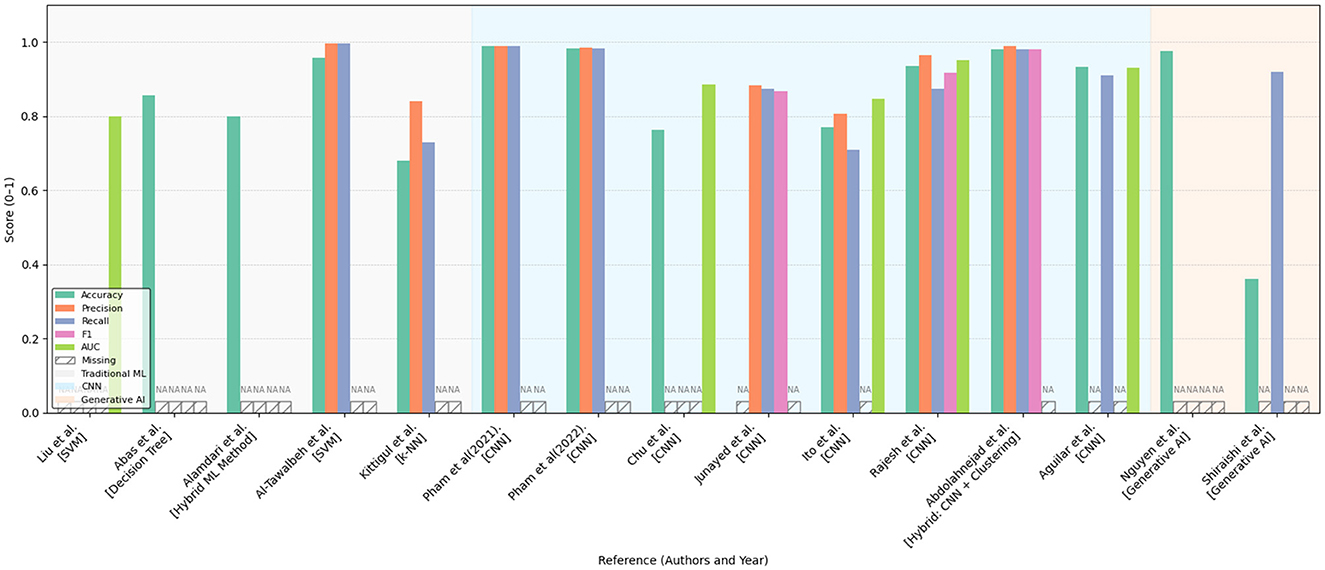
Figure 10. Bar chart comparing various models and methods in terms of accuracy, precision, recall, and F1 score. Different colors represent different metrics. Some bars have data labeled as “NA,” indicating missing information for certain methods. Categories include traditional ML, hybrid ML, GNN, and generative AI. Performance comparison of various machine learning and deep learning approaches across multiple evaluation metrics (Accuracy, Precision, Recall, F1-score, and AUC) as reported in existing literature. The methods are grouped by model type: traditional machine learning (gray-shaded background), convolutional neural networks (light blue background), and generative AI models (light orange background). Missing values are indicated as “NA.” The figure highlights the relative strengths and limitations of each approach within a normalized score range (0–1), providing a comprehensive overview of their effectiveness in the reviewed studies.
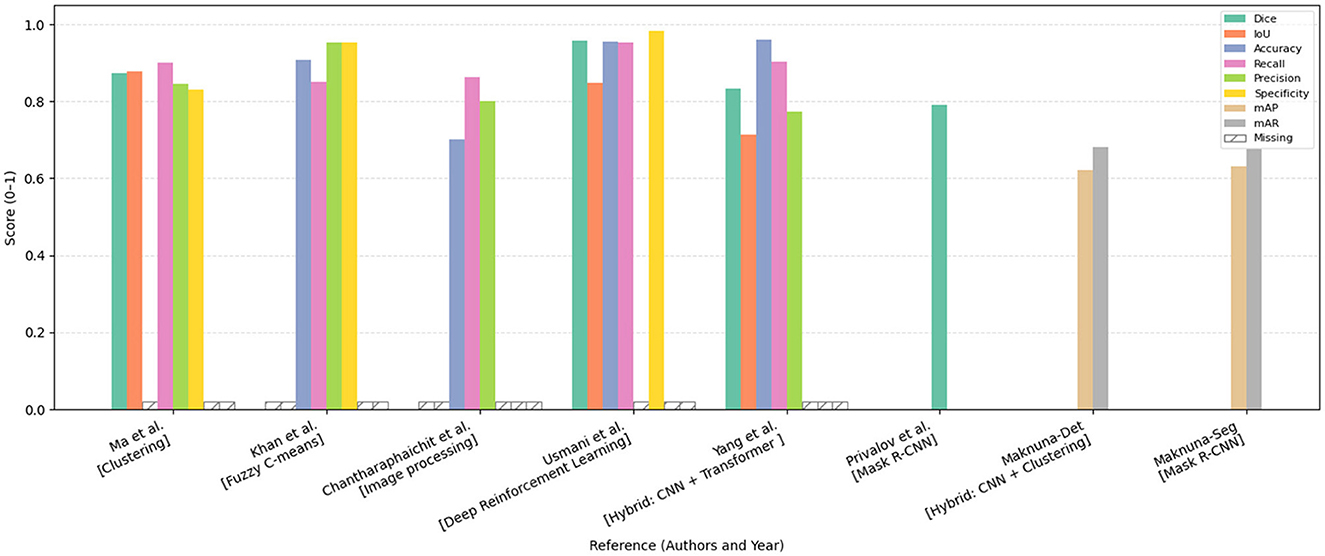
Figure 11. Bar chart comparing the performance of various methods, including clustering, fuzzy C-means, image processing, deep reinforcement learning, CNN, and Mask R-CNN. The chart displays scores across metrics Dice, IoU, accuracy, recall, precision, specificity, mAP, and mAR. Each category has colored bars representing these metrics, with most scores ranging from 0.8 to 1.0. The missing data is marked with a hatch pattern. Comparative performance analysis of segmentation and detection methods reported in this chapter, evaluated across various metrics including Dice coefficient, Intersection over Union (IoU), Accuracy, Recall, Precision, Specificity, mean Average Precision (mAP), and mean Average Recall (mAR). The models are categorized by methodological approach. Missing metric values are denoted as “Missing.” All scores are normalized within the range [0, 1], facilitating a standardized comparison of model effectiveness across diverse approaches.
Due to differences in datasets, sample sizes, and experimental protocols, the reported metrics should be interpreted with caution. Readers are advised to consult the preceding tables and method descriptions for contextual understanding. For studies evaluated on multiple datasets, the plotted results reflect weighted averages based on dataset size to ensure consistent comparison.
In the following, we review these supervised CNN approaches—highlighting their design innovations, clinical datasets, and quantitative outcomes—as well as the challenges they face in generalization, interpretability, and resource demands.
Pham et al. (91) developed a deep learning-based method using a modified VGG-16 CNN to classify and quantify collagen fiber organization in burn-induced scar tissue from Masson's Trichrome (MT)-stained histology images. The model achieves over 97% classification accuracy and effectively extracts collagen density and directional variance, revealing significant structural differences between scar and normal tissue. While demonstrating robustness across multi-scale images, limitations include sensitivity to tissue heterogeneity and restriction to MT staining. Afterwards, they further proposed a universal CNN model that does not rely on specific histological staining processes to classify and characterize collagen fiber structures in burn-induced scar tissue (92). Maknuna et al. (93) employed machine learning techniques for the automated structural analysis and quantitative feature description of scar tissue. Using Mask R-CNN and K-means algorithms, the study effectively predicted and characterized scar images, such as collagen density and directional variation. Chu et al. (94) proposes a deep learning-based approach for the classification of post-thyroidectomy scar subtypes using a ResNet-50 CNN and a novel multiple clinical photography learning (MCPL) method. A dataset of 7,524 clinical photographs from 3565 patients was used to train and validate the model. The MCPL method, which leverages multiple images of the same scar per patient, improved model robustness and classification accuracy compared to a baseline model, achieving an AUC of up to 0.915 for hypertrophic scars. Kim et al. (78) developed an AI model to predict the severity of post-surgical scars. Using data from 1,283 patients (1,043 in the main dataset and 240 in the external dataset), the model demonstrated comparable accuracy to that of 16 dermatologists. Junayed et al. (95) developed a deep CNN model named ScarNet for the automatic classification of acne scars. ScarNet employs a 19-layer deep learning architecture, with optimizations made to the activation functions, optimization algorithms, loss functions, kernel sizes, and batch sizes to improve classification performance while reducing computational costs. Ito et al. (96) developed a computer vision algorithm based on automated machine learning for diagnosing four types of scars: immature scars, mature scars, hypertrophic scars, and keloids. Compared to doctors' diagnoses, the algorithm achieved an average accuracy of 77%, while doctors' average accuracy was 68.7%. Singh and Saxena (97) developed an image processing algorithm using CNN to evaluate treatment efficacy by analyzing collagen fiber structures in scar images. Privalov et al. (98) validated an automated wound segmentation and measurement method based on Mask R-CNN for processing wound photographs. Rajesh et al. (99) proposes a deep learning-based approach for classifying vitiligo and scar images using a customized CNN with six convolutional layers and three fully connected layers. A dataset of 3,135 images was used, augmented to improve generalization. The model achieved a training accuracy of 93.89%, precision of 96.50%, and an AUC score of 0.95, outperforming existing architectures such as ResNet-50, InceptionV3, and VGG-16. Abdolahnejad et al. (100) introduces a machine learning pipeline for the automated assessment and longitudinal tracking of keloid scars, integrating EfficientNet B7-based CNN for classification, segmentation techniques for lesion boundary detection, and K-Means clustering for colorimetric analysis. The model was trained on a dataset of 6,550 images, achieving a classification accuracy of 98%, with segmentation refined using fiducial markers and contour-based detection. The pipeline was validated through 5–6 months of follow-up imaging, effectively capturing changes in keloid size and pigmentation with a measurement error margin of ± 2 mm. Despite its high accuracy, the method demonstrated limitations in detecting early-stage keloids and challenges in segmenting lesions on darker skin tones due to reduced contrast. Aguilar et al. (101) explores the feasibility of using CNN for automated acne scar risk assessment. A dataset of 437 clinical images from 404 acne patients was annotated by dermatologists and categorized using the four-item Acne-Scar Risk Assessment Tool (4-ASRAT) into low-, moderate-, and high-risk groups. A custom CNN model was trained for both binary (risk/no risk) and three-class classification, achieving 93.15% accuracy and an AUC of 0.931 for the binary classification task. However, performance on the three-class classification was poor (68.26% accuracy) due to the lack of clear separation between mild and severe scarring categories.
6.7 Key limitations and failure modes of CNN-based diagnostic models
Convolutional neural networks (CNNs) have emerged as the cornerstone of numerous state-of-the-art diagnostic systems, offering remarkable performance improvements over traditional machine learning and rule-based methods. Their ability to automatically learn hierarchical representations from raw medical images has led to substantial gains in tasks such as disease classification, lesion detection, and image segmentation. Particularly in domains like radiology, dermatology, and ophthalmology, CNN-based models have approached or even exceeded expert-level diagnostic accuracy in controlled settings. Furthermore, CNNs are highly adaptable to diverse imaging modalities (e.g., CT, MRI, histopathology), and benefit from transfer learning, making them broadly applicable across medical subfields.
However, despite these strengths, CNN-based diagnostic models are not without significant limitations. Their practical deployment in clinical environments is hindered by a series of non-trivial challenges, which compromise model robustness, reliability, and trustworthiness. Below, we summarize key failure modes and systemic limitations of CNNs in biomedical applications.
6.7.1 Overfitting and limited generalization in small clinical datasets
While CNNs excel at learning from large-scale annotated corpora, clinical datasets are often limited in size and suffer from class imbalance, institutional bias, and acquisition variability. This mismatch between model capacity and data availability can lead to overfitting, where CNNs memorize dataset-specific artifacts rather than learning disease-relevant features, As shown in Figure 12. Despite the architectural regularization imposed by convolutional layers and weight sharing, CNNs still require extensive regularization strategies—such as data augmentation, dropout, weight decay, transfer learning, and early stopping—to mitigate this issue and improve generalization to unseen data (102, 103).
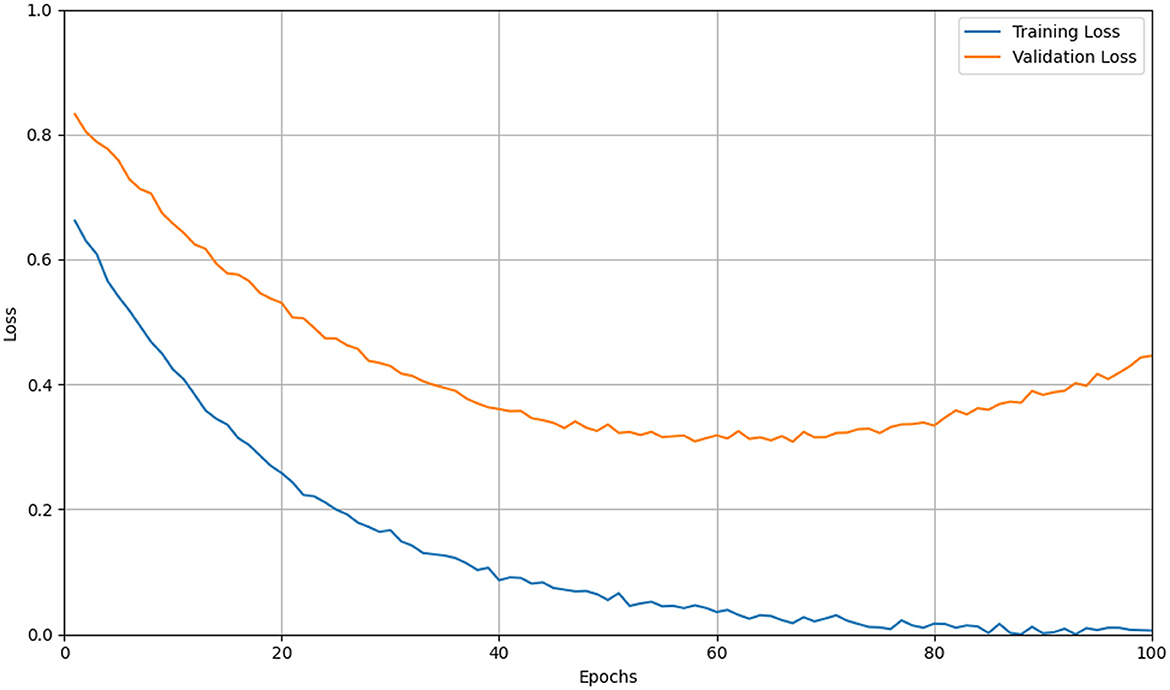
Figure 12. Line graph showing training and validation loss over 100 epochs. Training loss (blue) decreases steadily, while validation loss (orange) decreases initially but rises after 50 epochs, indicating potential overfitting. Simulation of overfitting in CNN training on limited data. The yellow curve traces the training loss steadily decreasing, while the orange curve shows the validation loss initially falling but then rising after mid-training. The divergence between these curves indicates the model's tendency to memorize training-specific noise and lose generalization capability as epochs progress.
6.7.2 Vulnerability to adversarial perturbations and distributional shifts
CNN-based diagnostic systems are highly susceptible to adversarial attacks—minute, often imperceptible modifications to input images that can drastically alter model predictions. Finlayson et al. (104) demonstrated that adversarial examples could significantly impair CNN performance across multiple medical domains under both white-box and black-box threat models. Moreover, CNNs often fail to maintain accuracy when exposed to distributional shifts, such as changes in imaging protocols, hardware, or patient demographics. Recent studies have emphasized the need for robust training paradigms, including adversarial training, domain adaptation, and input validation, to ensure model reliability under real-world deployment scenarios (105, 106).
6.7.3 Sensitivity to batch size and optimization-induced generalization gaps
Optimization dynamics in CNN training are significantly influenced by batch size. Keskar et al. (107) showed that large-batch (LB) training (e.g., >1,000 samples) tends to converge to sharp local minima in the loss landscape, which are associated with poor generalization performance. In contrast, small-batch (SB) training (e.g., 32–128 samples) introduces stochasticity that encourages convergence to flatter minima, yielding more robust models, as shown in Figure 13. Masters and Luschi (108) further demonstrated that extremely small batches (as few as 2–32 samples) often achieve the best generalization even on large datasets like ImageNet and CIFAR-10/100. This highlights the importance of tuning batch size as a hyperparameter and considering its interaction with learning rate schedules in biomedical applications.

Figure 13. Line graph depicting accuracy over epochs for training and testing sets. SB-Training in dashed blue and SB-Testing in solid orange both increase steeply, reaching over 95 and 75 respectively. LB-Training in dashed green and LB-Testing in solid red increase more gradually, reaching about 90 and 70 respectively. Simulation of batch-size sensitivity in CNN training on CIFAR-10. Dashed lines represent training accuracy for small-batch (SB, blue) and large-batch (LB, red) regimes, while solid lines show the corresponding test accuracy. The chart illustrates that larger batch sizes yield reduced gradient noise, converge to sharper minima, and exhibit a substantially wider generalization gap compared to smaller batches.
6.7.4 Lack of interpretability and opaqueness of decision processes
The black-box nature of CNNs raises major concerns in high-stakes domains such as medicine. While techniques like Grad-CAM and other saliency-based methods attempt to visualize decision rationales, they often produce inconsistent or misleading explanations—e.g., focusing on irrelevant regions, highlighting only dominant lesions in multi-lesion cases, or failing basic sanity checks like weight randomization and reproducibility tests (109, 110). This lack of reliable interpretability undermines clinical trust, impairs model debugging, and complicates regulatory approval, emphasizing the need for more principled and faithful explanation methods.
6.7.5 Summary and future directions
In summary, while CNN-based models have revolutionized image-based diagnosis through automated feature learning and superior classification performance, their real-world clinical utility remains constrained by several critical limitations. These include susceptibility to overfitting on small and heterogeneous clinical datasets, vulnerability to adversarial perturbations and domain shifts, sensitivity to training dynamics such as batch size, and persistent issues surrounding interpretability and transparency. Such failure modes not only challenge the robustness of CNNs but also raise ethical and regulatory concerns, particularly in safety-critical applications.
Addressing these challenges calls for a multifaceted research agenda. On the data side, collaborative efforts to curate large-scale, diverse, and well-annotated medical image repositories—ideally spanning multiple institutions and patient demographics—are essential to improve generalization and fairness. In terms of algorithmic development, future work should prioritize robust optimization strategies that are resilient to data shifts and adversarial noise, such as distributionally robust learning, self-supervised pretraining, and uncertainty-aware inference. Furthermore, integrating domain knowledge (e.g., anatomical priors or clinical guidelines) into model design may offer inductive biases that enhance generalizability and interpretability.
Finally, the development of inherently interpretable CNN architectures and rigorous post hoc explanation tools remains a pressing need. These efforts should be coupled with standardized benchmarks and clinical evaluation protocols to quantify explanation reliability and diagnostic value. Bridging the gap between algorithmic performance and clinical trustworthiness will be vital to transition CNN-based diagnostic systems from promising prototypes to dependable tools in routine medical practice.
6.8 Other advanced methods
6.8.1 Methods combining 3D reconstruction and deep learning
integrating 3D reconstruction with deep learning techniques allows for comprehensive analysis and quantification of scars in three-dimensional space, particularly beneficial for precise measurements and assessments on complex skin surfaces. The combination of 3D modeling with deep neural networks opens new avenues for highly accurate, non-contact scar evaluation. Below we discuss recent developments in this promising research direction, and relevant references are also listed in Table 7.
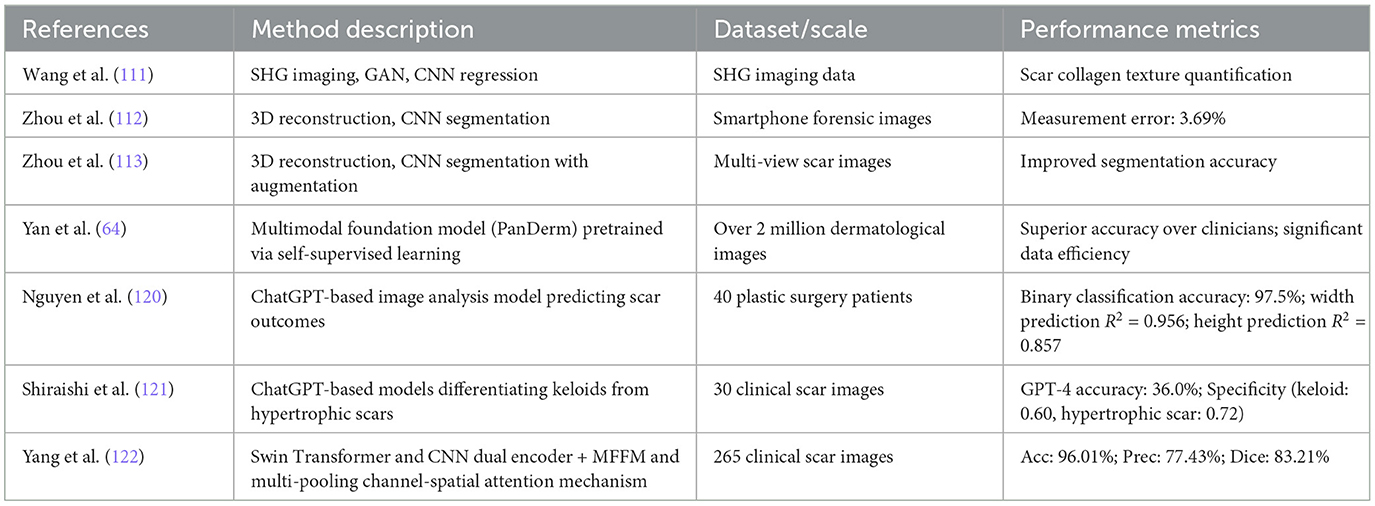
Table 7. Advanced methods combining 3D reconstruction, deep learning, and foundation models for scar diagnosis.
Wang et al. (111) proposed a novel method combining second harmonic generation (SHG) imaging technology and deep learning algorithms. By integrating SHG imaging with GAN and utilizing Tamura texture features, they constructed a regression model to quantitatively analyze collagen textures in human scar tissue and predict scar development. Zhou et al. (112) proposes a deep learning-based method for the automatic measurement of linear scar lengths, particularly for forensic applications. By integrating multiview stereo 3D reconstruction and CNN for image segmentation, the method allows non-contact, automated, and high-accuracy scar measurement using images taken from a smartphone. The model achieved an average measurement error of 3.69%, demonstrating strong agreement with manual measurements. Compared to traditional manual and 2D imaging methods, this approach reduces subjectivity and improves accuracy, especially for scars on curved surfaces. However, limitations include time-consuming 3D reconstruction and reliance on training data quality. Future research should optimize computational efficiency, improve segmentation models, and explore broader clinical applications. Zhou et al. (113) proposed an advanced two-stage deep learning framework for scar segmentation in multi-view images. The first stage includes a novel data augmentation method based on 3D reconstruction and view interpolation to enhance the model's generalization ability.
Methods combining 3D reconstruction with deep learning show substantial promise in enhancing measurement precision and automated analysis of scars, particularly in forensic and clinical contexts requiring high accuracy. Despite their benefits, the significant computational cost, complexity of data acquisition, and reliance on high-quality training data are notable limitations requiring further exploration.
6.8.2 Computational footprint of 3D-reconstruction-driven scar assessment
Although 3D reconstruction noticeably improves geometric fidelity, its clinical roll-out hinges on practical runtime and hardware demands. Table 8 summarizes the details of computational resource requirements for these research works. Wang et al. (111) needed roughly six minutes per case on unspecified hardware, with ScarGAN accounting for most of the 360 s pipeline latency. Zhou et al. (112) executed on a consumer-grade RTX 2060 (8 GB) workstation; even after aggressive image down-sampling, structure-from-motion took a mean ± Standard Deviation (SD) of 111.8 ± 19.9 s, while subsequent measurement added 28.1 ± 8.4 s. To cope with heavier multi-view co-segmentation, Zhou et al. (113) upgraded to an RTX 3090 (24 GB); their MVCSNet contains 31.0 M parameters and incurs 218.7 GFLOPs per forward pass—no end-to-end runtime was disclosed, but the authors note that GPU memory constrained batch-size to 1.
Collectively, these data indicate that current 3-D Reconstruction methods still require 30s–6min per patient and 8–24 GB of GPU memory–tolerable for retrospective analysis yet insufficient for real-time clinical use. Future work should (i) publish full training and inference profiles, (ii) apply pruning, quantisation, and mixed-precision to cut memory below 4 GB, and (iii) replace global multi-View stereo (MVS) with lightweight depth-fusion schemes to bring per-case runtime under 10 s.
6.8.3 Large-scale foundation models in scar diagnosis
Recent advancements in foundation models have demonstrated considerable potential in dermatological diagnostics, including scar recognition and evaluation. A prominent work by Yan et al. (64), published in Nature Medicine, introduced PanDerm, a multimodal vision foundation model pretrained via self-supervised learning on over two million dermatological images collected from 11 clinical institutions, encompassing clinical photography, dermoscopic images, total-body photography, and dermatopathological slides. A general architecture of such large-scale foundation models is illustrated in Figure 14. PanDerm achieved state-of-the-art performance across diverse clinical tasks, notably demonstrating superior data efficiency by surpassing existing methods even when utilizing only 10% of labeled data. Clinical validation confirmed PanDerm's substantial clinical value, notably outperforming clinicians by 10.2% in early-stage melanoma detection and improving diagnostic accuracy across 128 skin conditions by 16.5% among non-specialists. This seminal work highlights the transformative potential of multimodal foundation models in comprehensive dermatological assessments, providing a critical reference point for future intelligent scar diagnostics.

Figure 14. Diagram of a large model represented by a transformer. Inputs include text, image, and multimodal data, with outputs for text generation, classification, and segmentation. Finetuning options like LoRa and Adapter are shown. A generic architecture of large-scale foundation models. The model accepts various types of input (e.g., text, image, or multimodal data), processes them through a unified backbone—typically based on Transformer architecture—and supports a range of downstream tasks such as text generation, classification, and segmentation. Lightweight finetuning methods (e.g., LoRA, adapters) can be employed to adapt the model to specific applications.
PanDerm has demonstrated broad applicability across dermatological tasks through pretraining on over two million multimodal skin disease images. However, fine-tuning for scar evaluation requires addressing the unique morphological, chromatic, and vascular features of scar tissue. First, it is essential to curate a high-quality dataset that includes dermoscopy, optical coherence tomography, and ultrasound images with precise annotations of scar width, height, vascularity, and pigmentation. In the fine-tuning phase, parameter-efficient methods such as layer-wise differentiation of learning rates (freezing early convolutional and Transformer layers while fully training later layers) and Low-Rank Adaptation (LoRA) can significantly reduce trainable parameters without sacrificing performance (114, 115). Moreover, integrating multi-task objectives for scar segmentation, classification, and regression within a shared backbone exploits cross-task synergies, as shown by self-training frameworks leveraging confident pseudo-labels for segmentation (116). To mitigate domain shift between PanDerm's broad pretraining domain and the scar-specific target domain, adversarial domain adaptation techniques such as Domain-Adversarial Neural Networks with gradient reversal layers can promote extraction of scar-invariant features and improve generalization across clinical centers (117, 118). When annotation resources are limited, pseudo-label self-training can expand the training corpus by using confident predictions on unlabeled images. Finally, applying self-supervised pretraining strategies such as masked image modeling or contrastive multimodal learning on scar-centric datasets can further regularize the model, reduce overfitting, and pave the way for federated and few-shot scar assessment systems that support privacy-preserving deployment in diverse clinical settings (119).
Other explorations have investigated generative AI frameworks in scar prognosis and classification. Nguyen et al. (120) assessed the feasibility of a ChatGPT-integrated image analysis model in predicting long-term scar characteristics. Evaluating standardized images from 40 plastic surgery patients, the ChatGPT-based approach achieved remarkable accuracy (97.5%) for binary scar classification. The model performed exceptionally well in predicting static scar attributes such as width (R2 = 0.956) and height (R2 = 0.857), although dynamic features, such as vascularity (R2 = 0.234) and pigmentation (R2 = 0.676), remain challenging. These findings highlight the promising yet currently limited capability of generative AI for objective, long-term scar prediction, especially concerning dynamic scar properties.
Additionally, Shiraishi et al. (121) explored the potential of ChatGPT-based models in distinguishing between keloids and hypertrophic scars through standardized clinical image prompts. Comparing multiple AI chatbots, GPT-4 significantly outperformed others, achieving a higher diagnostic accuracy (36.0% vs. 22.0%) and notably better specificity. Nevertheless, current generative AI models still fall short of clinical standards, underscoring the need for further refinements in accuracy and robustness. This preliminary work provides valuable insights into the potential and limitations of applying large language models for scar diagnosis.
While the above research works exemplifies the potential of large-scale, general-purpose foundation models, task-specific architectures leveraging key components of such models-particularly Transformers-have also shown notable promise. For instance, Yang et al. (122) proposed MFMA-Net, a dual-encoder segmentation network that integrates a CNN and a Swin Transformer to capture both local textures and global context. Through a multi-scale feature fusion module and a multi-pooling channel-spatial attention mechanism, the model achieved state-of-the-art performance on clinical scar segmentation tasks (e.g., 96.01% accuracy, 83.21% Dice coefficient), outperforming classical and Transformer-based baselines alike. Though MFMA-Net is not a foundation model per se, it demonstrates how Transformer-based designs can be effectively adapted for high-precision, task-specific applications in scar assessment.
It is worth noting that, to date, only the aforementioned study has specifically applied Transformer-based or foundation model-inspired architectures to scar diagnosis. However, similar advanced artificial intelligence techniques have been extensively explored in related domains, including the detection, segmentation, and classification of skin lesions and skin cancer (123–149). Beyond these dermatological applications, other biomedical-image analysis tasks have also benefited from novel Transformer architectures. Xiang et al. (150) propose a two-stage Multimodal Masked Autoencoder (Multi-MAE) for vitiligo stage classification. The approach integrates an adaptive masking module that leverages self-attention to dynamically mask and discard non-salient patches, a unified Vision Transformer encoder shared by clinical and Wood's lamp images, and a cross-attention fusion decoder for multimodal reconstruction pre-training. On a modest multimodal dataset, Multi-MAE achieved 95.48% accuracy, outperforming MobileNet, DenseNet, VGG, ResNet-50, BEIT, MaskFeat, SimMIM, and standard MAE by 2.58%–5.16%. Similarly, Song et al. (151) introduce CenterFormer, an end-to-end transformer-based framework for unconstrained dental plaque segmentation. It features a Cluster Center Encoder (CCE) that applies K-means clustering on multi-level feature maps to produce coarse region representations, a pyramid-style Multiple Granularity Perceptions module to fuse local and global contexts, and an MLP decoder for final mask prediction. Evaluated on nearly 3,000 intraoral images, CenterFormer attained an IoU of 60.91% and a pixel accuracy of 76.81%, surpassing SegFormer and other state-of-the-art models by 2.34%–6.08%. Although tailored to vitiligo staging and dental imagery, respectively, both methods employ adaptive, self-supervised pre-training, multimodal fusion, clustering-guided attention, and multi-scale feature integration strategies that are methodologically relevant and technically transferable to scar-related tasks. We expect that such techniques will play an increasingly important role in future developments of intelligent scar analysis.
6.9 Clinical integration
Technical accuracy alone is insufficient to deliver patient-centered value in scar care. Given the documented inter-rater variability of visual/tactile scales, AI systems should be embedded across the care pathway with explicitly defined links to psychosocial wellbeing and satisfaction. Below we outline practical integration points, implementation guardrails, and evaluation strategies, drawing on evidence from dermatology and other clinical domains.
6.9.1 Pre-visit intake and education
Smartphone-guided image capture with automated quality control can reduce uncertainty before clinic visits, while brief electronic patient-reported outcomes (ePROs)-for example, POSAS-patient items, itch/pain numerical rating scales, and short dermatology quality-of-life screens-establish a baseline and flag high-risk psychosocial profiles for clinician review. Educational feedback (expectation setting; capture tips for darker skin tones and various illumination conditions) may lower anxiety and improve perceived preparedness. Teledermatology workflows that combine images with structured questionnaires have shown high patient satisfaction and shorter time-to-advice in multiple evaluations (152, 153).
6.9.2 In-clinic decision support and shared decision-making
AI-aided segmentation, severity scoring, and progression/recurrence risk prediction can be surfaced within the electronic record to support share decision-making. Calibrated risk displays, standardized visual aids (e.g., 3D surface metrics), and curated exemplar galleries help align expectations, reduce decisional conflict, and personalize plans (e.g., prophylaxis for keloid-prone patients). Experience from other specialties (e.g., autonomous diabetic retinopathy screening deployed in primary care) illustrates how validated AI can be safely integrated into routine pathways with defined scopes of use, audit trails, and referral rules (154, 155).
6.9.3 Longitudinal follow-up and remote monitoring
Between visits, remote monitoring can pair periodic photos with short ePROs to track trajectory and symptoms (itch, pain, appearance concerns, sleep impact). Drift-aware alerts notify teams of worsening objective metrics or deteriorating ePROs and can trigger timely intervention or referral to psychological support. Evidence from ePRO programs in other fields shows that structured symptom monitoring can improve quality of life and care experience, and in some settings clinical outcomes (156).
6.9.4 Implementation guardrails and equity
Deployment should adopt human-in-the-loop oversight, clear scope-of-use statements, fail-safes for uncertainty, and governance for updates. Equity safeguards include validated performance across skin tones, scar architectures, and acquisition settings, plus multilingual, accessible capture instructions. Integration with EHRs (e.g., via HL7 FHIR) should store AI outputs, timestamps, and ePROs with audit trails. Privacy and consent must explicitly cover image content and per-image metadata, consistent with international guidance on trustworthy AI in health (157, 158).
In sum, integrating AI into routine scar care can translate technical accuracy into patient-centered value. Embedding decision support at pre-visit, in-clinic, and follow-up stages reduces uncertainty, aligns expectations, and enables timely escalation, while remote monitoring pairs objective image metrics with brief ePROs to track psychosocial needs. Human-in-the-loop oversight, clear scopes of use, and privacy-by-design safeguard safety and trust. Ultimately, success should be judged not only by diagnostic performance but also by improvements in patient-reported outcomes, adherence, and satisfaction, demonstrating tangible clinical benefit.
7 Conclusion
This review systematically summarized and analyzed recent advancements in intelligent recognition and diagnosis methods for skin scars, encompassing traditional machine learning methods, convolutional neural network (CNN)-based approaches, and hybrid methods integrating 3D reconstruction and deep learning. Traditional clinical scar assessment methods, although valuable in clinical practice, inherently suffer from subjectivity and inconsistencies arising from varied expertise levels among clinicians (159). In contrast, intelligent diagnostic methods leveraging artificial intelligence (AI) provide objective, reproducible, and efficient tools that can significantly enhance clinical assessments and patient care.
Traditional machine learning methods, such as clustering algorithms, texture analysis techniques, and rule-based models, have demonstrated efficacy primarily due to their simplicity, computational efficiency, and interpretability. These methods typically involve handcrafted feature extraction strategies tailored to specific scar characteristics, enabling successful segmentation and classification tasks even with limited computational resources. However, the heavy reliance on manual feature engineering and the susceptibility of these methods to variations in image quality and environmental conditions restrict their generalization capability and applicability across diverse clinical scenarios.
CNN-based methods address many limitations inherent in traditional approaches by automatically extracting hierarchical features from extensive datasets, resulting in higher accuracy, improved robustness, and greater adaptability. CNN architectures such as ResNet, VGG, EfficientNet, and Mask R-CNN have demonstrated impressive results in tasks ranging from scar severity assessment and subtype classification to detailed collagen structure analysis. Despite their superior performance, CNN-based methods face challenges, including substantial computational demands, dependence on large-scale, high-quality annotated datasets, and concerns regarding interpretability and transparency, which remain critical barriers to widespread clinical adoption (160).
Hybrid approaches integrating 3D reconstruction techniques with deep learning represent a promising research frontier in scar assessment, offering precise quantitative analysis and non-contact measurement capabilities. These methods have shown exceptional promise for applications requiring accurate dimensional analysis, particularly in complex surface evaluations such as forensic investigations (112). However, the computational complexity, time-consuming data acquisition, and reliance on high-quality 3D data continue to pose significant practical challenges that need addressing through optimized computational strategies and improved imaging protocols.
Despite significant progress, several critical challenges persist in intelligent scar recognition research. A paramount limitation is the scarcity of standardized, publicly accessible scar image datasets, compounded by patient privacy and ethical considerations. The current dataset availability is insufficient to fully support robust model training and validation, impeding the reproducibility and generalization of research outcomes. Future advancements critically depend on establishing comprehensive, ethically sourced datasets, adopting standardized image acquisition protocols, and developing innovative data augmentation techniques to mitigate dataset limitations.
Additionally, future research should focus on addressing the interpretability and transparency of AI models to build trust among clinicians and ensure the practical applicability of these tools in real-world medical environments. Enhanced interpretability would allow clinicians not only to understand model predictions but also facilitate the integration of AI-based diagnostic tools within routine clinical workflows, ultimately improving patient care quality.
To address these challenges and propel the field forward, we propose several key research directions:
• Development of hybrid models: future research should focus on developing hybrid approaches that combine traditional image processing and deep learning techniques, such as linear-transformation-based dehazing to enhance scar image contrast and suppress illumination artifacts (161), and spectrum-based image enhancement methods that convert RGB images into narrowband or hyperspectral-like representations to boost dermatological lesion classification and detection performance (162–164). By leveraging the interpretability and low computational cost of classical preprocessing alongside the powerful feature extraction of modern networks, these hybrids can improve lesion boundary separability and enable more accurate, real-time scar diagnosis.
• Improvement of dataset accessibility and quality: the establishment of large-scale, publicly available datasets specifically curated for scar analysis is essential. Encouraging collaboration between clinical institutions, research communities, and regulatory bodies can facilitate the creation of diverse, ethically sourced datasets that adhere to strict privacy standards.
• Clinical validation and trials: rigorous clinical validation through well-designed prospective studies and multicenter clinical trials is crucial to evaluate the real-world effectiveness, reliability, and safety of intelligent diagnostic systems (165). Such validation efforts would ensure that AI technologies meet clinical standards and demonstrate tangible patient benefits.
• Advancement in ethical AI application: ethical considerations and patient privacy must be integral to the development and deployment of AI-based diagnostic tools. Implementing transparent, explainable AI practices and stringent data governance frameworks can address ethical concerns and foster broader acceptance among patients and healthcare providers (166).
In conclusion, the integration of intelligent diagnostic technologies into scar recognition and management signifies a transformative shift toward more objective, efficient, and patient-centered healthcare solutions. While substantial progress has been achieved, ongoing efforts in methodological innovation, dataset improvement, clinical validation, and ethical governance are essential for fully realizing the potential of AI in dermatological care. By addressing these critical aspects, future research will undoubtedly pave the way for the broader and more effective adoption of intelligent diagnostic systems, ultimately improving the quality of life for individuals affected by scars.
Author contributions
FH: Writing – original draft, Investigation, Writing – review & editing, Data curation, Methodology, Visualization, Conceptualization. YS: Conceptualization, Writing – original draft, Writing – review & editing, Methodology, Data curation, Visualization, Investigation. JuL: Writing – review & editing, Methodology, Data curation, Investigation. JiL: Methodology, Data curation, Writing – review & editing, Investigation. XX: Investigation, Writing – review & editing, Methodology, Data curation. KS: Data curation, Writing – review & editing, Visualization. YZ: Writing – review & editing, Visualization, Data curation. JZ: Project administration, Supervision, Writing – review & editing, Funding acquisition. XW: Project administration, Funding acquisition, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (Grant Nos. 12301676 and 12371429), the Zhejiang Provincial Natural Science Foundation of China (Grant No. LQ24A010018), the Jinhua Key Science and Technology Program Project (Grant No. 2024-3-031), and the industry-university research cooperation platform of Hangzhou Plastic Surgery Hospital.
Acknowledgments
The authors would like to thank the editors and reviewers for their valuable comments. This work was supported by the National Natural Science Foundation of China (Grant Nos. 12301676 and 12371429), the Zhejiang Provincial Natural Science Foundation of China under Grant No. LQ24A010018, and the Jinhua Key Science and Technology Program Project (Grant No. 2024-3-031). The authors sincerely appreciate the financial support from these funding agencies.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Lin X, Lai Y. Scarring skin: mechanisms and therapies. Int J Mol Sci. (2024) 25:1458. doi: 10.3390/ijms25031458
2. Gauglitz GG, Korting HC, Pavicic T, Ruzicka T, Jeschke MG. Hypertrophic scarring and keloids: pathomechanisms and current and emerging treatment strategies. Mol Med. (2011) 17:113–25. doi: 10.2119/molmed.2009.00153
3. Ud-Din S, Bayat A. Classification of distinct endotypes in human skin scarring: SCAR—A novel perspective on dermal fibrosis. Adv Wound Care. (2022) 11:109–20. doi: 10.1089/wound.2020.1364
4. Berman B, Maderal A, Raphael B. Keloids and hypertrophic scars: pathophysiology, classification, and treatment. Dermatol Surg. (2017) 43:S3-S18. doi: 10.1097/DSS.0000000000000819
5. Cortés H, Rojas-Márquez M, Del Prado-Audelo ML, Reyes-Hernández OD, González-Del Carmen M, Leyva-Gómez G. Alterations in mental health and quality of life in patients with skin disorders: a narrative review. Int J Dermatol. (2022) 61:783–91. doi: 10.1111/ijd.15852
6. Gibson JA, Dobbs TD, Griffiths R, Song J, Akbari A, Bodger O, et al. The association of anxiety disorders and depression with facial scarring: population-based, data linkage, matched cohort analysis of 358 158 patients. BJPsych Open. (2023) 9:e212. doi: 10.1192/bjo.2023.547
7. Van Loey NE, Van Son MJ. Psychopathology and psychological problems in patients with burn scars: epidemiology and management. Am J Clin Dermatol. (2003) 4:245–72. doi: 10.2165/00128071-200304040-00004
8. Shen W, Chen L, Tian F. Research progress of scar repair and its influence on physical and mental health. Int J Burns Trauma. (2021) 11:442.
9. Huang C, Murphy GF, Akaishi S, Ogawa R. Keloids and hypertrophic scars: update and future directions. Plastic Reconstr Surg-Glob Open. (2013) 1:e25. doi: 10.1097/GOX.0b013e31829c4597
10. Du-Harpur X, Watt F, Luscombe N, Lynch M. What is AI? Applications of artificial intelligence to dermatology. Br J Dermatol. (2020) 183:423–30. doi: 10.1111/bjd.18880
11. Castiglioni I, Rundo L, Codari M, Di Leo G, Salvatore C, Interlenghi M, et al. AI applications to medical images: From machine learning to deep learning. Phys Med. (2021) 83:9–24. doi: 10.1016/j.ejmp.2021.02.006
12. Li Y, Shen L. Skin lesion analysis towards melanoma detection using deep learning network. Sensors. (2018) 18:556. doi: 10.3390/s18020556
13. Daghrir J, Tlig L, Bouchouicha M, Sayadi M. Melanoma skin cancer detection using deep learning and classical machine learning techniques: a hybrid approach. In: 2020 5th International Conference on Advanced Technologies for Signal and Image Processing (ATSIP). Sousse: IEEE (2020). p. 1–5. doi: 10.1109/ATSIP49331.2020.9231544
14. Li Z, Jiang S, Xiang F, Li C, Li S, Gao T, et al. White patchy skin lesion classification using feature enhancement and interaction transformer module. Biomed Signal Process Control. (2025) 107:107819. doi: 10.1016/j.bspc.2025.107819
15. Wang G, Ma Q, Li Y, Mao K, Xu L, Zhao Y. A skin lesion segmentation network with edge and body fusion. Appl Soft Comput. (2025) 170:112683. doi: 10.1016/j.asoc.2024.112683
16. Wang S, Dong B, Xiong J, Liu L, Shan M, Koch AW, et al. Phase manipulating Fresnel lenses for wide-field quantitative phase imaging. Opt Lett. (2025) 50:2683–6. doi: 10.1364/OL.555558
17. Wu X, Zou B, Lu C, Wang L, Zhang Y, Wang H. Dynamic security computing framework with zero trust based on privacy domain prevention and control theory. IEEE J Sel Areas Commun. (2025) 43:2266–78. doi: 10.1109/JSAC.2025.3560036
18. Goodman GJ. Postacne scarring: a review of its pathophysiology and treatment. Dermatol Surg. (2000) 26:857–71. doi: 10.1046/j.1524-4725.2000.99232.x
19. Fabbrocini G, Annunziata MC, D' Arco V, De Vita V, Lodi G, Mauriello M, et al. Acne scars: pathogenesis, classification and treatment. Dermatol Res Pract. (2010) 2010:893080. doi: 10.1155/2010/893080
20. Beanes SR, Dang C, Soo C, Ting K. Skin repair and scar formation: the central role of TGF-β. Expert Rev Mol Med. (2003) 5:1–22. doi: 10.1017/S1462399403005817
21. Knapp T, Daniels R, Kaplan E. Pathologic scar formation. Morphologic and biochemical correlates. Am J Pathol. (1977) 86:47.
22. Baryza MJ, Baryza GA. The Vancouver Scar Scale: an administration tool and its interrater reliability. J Burn Care Rehabil. (1995) 16:535–8. doi: 10.1097/00004630-199509000-00013
23. Draaijers LJ, Tempelman FR, Botman YA, Tuinebreijer WE, Middelkoop E, Kreis RW, et al. The patient and observer scar assessment scale: a reliable and feasible tool for scar evaluation. Plast Reconstr Surg. (2004) 113:1960–5. doi: 10.1097/01.PRS.0000122207.28773.56
24. Nedelec B, Correa JA, Rachelska G, Armour A, LaSalle L. Quantitative measurement of hypertrophic scar: interrater reliability and concurrent validity. J Burn Care Res. (2008) 29:501–11. doi: 10.1097/BCR.0b013e3181710881
25. Lee KC, Bamford A, Gardiner F, Agovino A, Ter Horst B, Bishop J, et al. Investigating the intra-and inter-rater reliability of a panel of subjective and objective burn scar measurement tools. Burns. (2019) 45:1311–24. doi: 10.1016/j.burns.2019.02.002
26. Hamet P, Tremblay J. Artificial intelligence in medicine. Metabolism. (2017) 69:S36–40. doi: 10.1016/j.metabol.2017.01.011
27. Jordan MI, Mitchell TM. Machine learning: trends, perspectives, and prospects. Science. (2015) 349:255–60. doi: 10.1126/science.aaa8415
28. Caruana R, Niculescu-Mizil A. An empirical comparison of supervised learning algorithms. In: Proceedings of the 23rd International Conference on Machine Learning. New York, NY: ACM (2006). p. 161–8. doi: 10.1145/1143844.1143865
29. Barlow HB. Unsupervised learning. Neural Comput. (1989) 1:295–311. doi: 10.1162/neco.1989.1.3.295
30. Zhu XJ. Semi-Supervised Learning Literature Survey. Madison, WI: Computer Science, University of Wisconsin-Madison (2005).
31. Su X, Yan X, Tsai CL. Linear regression. Wiley Interdiscip Rev Comput Stat. (2012) 4:275–94. doi: 10.1002/wics.1198
32. Hearst MA, Dumais ST, Osuna E, Platt J, Scholkopf B. Support vector machines. IEEE Intell Syst Appl. (1998) 13:18–28. doi: 10.1109/5254.708428
33. Beyer K, Goldstein J, Ramakrishnan R, Shaft U. When is “nearest neighbor” meaningful? In: Database Theory—ICDT'99: 7th International Conference Jerusalem, Israel, January 10-12, 1999 Proceedings 7. Cham: Springer (1999). p. 217–35. doi: 10.1007/3-540-49257-7
35. Mahbod A, Schaefer G, Ellinger I, Ecker R, Pitiot A, Wang C. Fusing fine-tuned deep features for skin lesion classification. Comput Med Imaging Graph. (2019) 71:19–29. doi: 10.1016/j.compmedimag.2018.10.007
36. Xu R, Wunsch D. Survey of clustering algorithms. IEEE Trans Neural Netw. (2005) 16:645–78. doi: 10.1109/TNN.2005.845141
37. Van Der Maaten L, Postma EO, Van Den Herik HJ, et al. Dimensionality reduction: a comparative review. J Mach Learn Res. (2009) 10:13.
38. Jain AK, Murty MN, Flynn PJ. Data clustering: a review. ACM Comput Surv. (1999) 31:264–323. doi: 10.1145/331499.331504
39. Murtagh F, Contreras P. Algorithms for hierarchical clustering: an overview. Wiley Interdiscipl Rev Data Min Knowl Discov. (2012) 2:86–97. doi: 10.1002/widm.53
40. Abdi H, Williams LJ. Principal component analysis. Wiley Interdiscip Rev Comput Stat. (2010) 2:433–59. doi: 10.1002/wics.101
41. Lopez C, Tucker S, Salameh T, Tucker C. An unsupervised machine learning method for discovering patient clusters based on genetic signatures. J Biomed Inform. (2018) 85:30–9. doi: 10.1016/j.jbi.2018.07.004
42. Jiao R, Zhang Y, Ding L, Xue B, Zhang J, Cai R, et al. Learning with limited annotations: a survey on deep semi-supervised learning for medical image segmentation. Comput Biol Med. (2024) 169:107840. doi: 10.1016/j.compbiomed.2023.107840
43. Sarker IH. Deep learning: a comprehensive overview on techniques, taxonomy, applications and research directions. SN Comput Sci. (2021) 2:420. doi: 10.1007/s42979-021-00815-1
44. Voulodimos A, Doulamis N, Doulamis A, Protopapadakis E. Deep learning for computer vision: a brief review. Comput Intell Neurosci. (2018) 2018:7068349. doi: 10.1155/2018/7068349
45. Otter DW, Medina JR, Kalita JK. A survey of the usages of deep learning for natural language processing. IEEE Trans Neural Netw Learn Syst. (2020) 32:604–24. doi: 10.1109/TNNLS.2020.2979670
46. Ching T, Himmelstein DS, Beaulieu-Jones BK, Kalinin AA, Do BT, Way GP, et al. Opportunities and obstacles for deep learning in biology and medicine. J R Soc Interface. (2018) 15:20170387. doi: 10.1098/rsif.2017.0387
47. Shen D, Wu G, Suk HI. Deep learning in medical image analysis. Annu Rev Biomed Eng. (2017) 19:221–48. doi: 10.1146/annurev-bioeng-071516-044442
48. Li LF, Wang X, Hu WJ, Xiong NN, Du YX, Li BS. Deep learning in skin disease image recognition: a review. IEEE Access. (2020) 8:208264–80. doi: 10.1109/ACCESS.2020.3037258
49. Liu X, Song L, Liu S, Zhang Y. A review of deep-learning-based medical image segmentation methods. Sustainability. (2021) 13:1224. doi: 10.3390/su13031224
50. Fliorent R, Fardman B, Podwojniak A, Javaid K, Tan IJ, Ghani H, et al. Artificial intelligence in dermatology: advancements and challenges in skin of color. Int J Dermatol. (2024) 63:455–61. doi: 10.1111/ijd.17076
51. López-Pérez M, Hauberg S, Feragen A. Are generative models fair? A study of racial bias in dermatological image generation. In: Scandinavian Conference on Image Analysis. Cham: Springer. (2025). p. 389–402. doi: 10.1007/978-3-031-95918-9_27
52. Almuzaini AA, Dendukuri SK, Singh VK. Toward fairness across skin tones in dermatological image processing. In: 2023 IEEE 6th International Conference on Multimedia Information Processing and Retrieval (MIPR). Singapore: IEEE (2023). p. 1–7. doi: 10.1109/MIPR59079.2023.00030
53. Sullivan T, Smith J, Kermode J, Mclver E, Courtemanche D. Rating the burn scar. J Burn Care Rehabil. (1990) 11:256–60. doi: 10.1097/00004630-199005000-00014
54. Fogelberg K, Chamarthi S, Maron RC, Niebling J, Brinker TJ. Domain shifts in dermoscopic skin cancer datasets: evaluation of essential limitations for clinical translation. N Biotechnol. (2023) 76:106–17. doi: 10.1016/j.nbt.2023.04.006
55. Branciforti F, Meiburger KM, Zavattaro E, Veronese F, Tarantino V, Mazzoletti V, et al. Impact of artificial intelligence-based color constancy on dermoscopical assessment of skin lesions: a comparative study. Skin Res Technol. (2023) 29:e13508. doi: 10.1111/srt.13508
56. Yoo MG, Kim IH. Keloids and hypertrophic scars: characteristic vascular structures visualized by using dermoscopy. Ann Dermatol. (2014) 26:603. doi: 10.5021/ad.2014.26.5.603
57. Hayyawi HH, Al-Hamamy HR. Dermoscopic features of hypertrophic scars and keloids. J Pak Assoc Dermatol. (2023) 33:991–6.
58. Peake M, Pan K, Rotatori RM, Powell H, Fowler L, James L, et al. Incorporation of 3D stereophotogrammetry as a reliable method for assessing scar volume in standard clinical practice. Burns. (2019) 45:1614–20. doi: 10.1016/j.burns.2019.05.005
59. Machado BHB, De Melo E, Silva ID, Pautrat WM, Frame J, Najlah M. Scientific validation of three-dimensional stereophotogrammetry compared to the IGAIS clinical scale for assessing wrinkles and scars after laser treatment. Sci Rep. (2021) 11:12385. doi: 10.1038/s41598-021-91922-9
60. Chiou AS, Omiye JA, Gui H, Swetter SM, Ko JM, Gastman B, et al. Multimodal image dataset for AI-based skin cancer (MIDAS) benchmarking. NEJM AI. (2025) 2:AIdbp2400732. doi: 10.1056/AIdbp2400732
61. Gong P, McLaughlin RA, Liew YM, Munro PR, Wood FM, Sampson DD. Assessment of human burn scars with optical coherence tomography by imaging the attenuation coefficient of tissue after vascular masking. J Biomed Opt. (2014) 19:021111–021111. doi: 10.1117/1.JBO.19.2.021111
62. Huang SY, Xiang X, Guo RQ, Cheng S, Wang LY, Qiu L. Quantitative assessment of treatment efficacy in keloids using high-frequency ultrasound and shear wave elastography: a preliminary study. Sci Rep. (2020) 10:1375. doi: 10.1038/s41598-020-58209-x
63. Alipour N, Burke T, Courtney J. Skin type diversity in skin lesion datasets: a review. Curr Dermatol Rep. (2024) 13:198–210. doi: 10.1007/s13671-024-00440-0
64. Yan S, Yu Z, Primiero C, Vico-Alonso C, Wang Z, Yang L, et al. A multimodal vision foundation model for clinical dermatology. Nat Med. (2025). p. 1–12. doi: 10.1038/s41591-025-03747-y
65. Guan H, Yap PT, Bozoki A, Liu M. Federated learning for medical image analysis: A survey. Pattern Recognit. (2024) 151:110424. doi: 10.1016/j.patcog.2024.110424
66. Adnan M, Kalra S, Cresswell JC, Taylor GW, Tizhoosh HR. Federated learning and differential privacy for medical image analysis. Sci Rep. (2022) 12:1953. doi: 10.1038/s41598-022-05539-7
67. Rehman MHU, Hugo Lopez Pinaya W, Nachev P, Teo JT, Ourselin S, Cardoso MJ. Federated learning for medical imaging radiology. Br J Radiol. (2023) 96:20220890. doi: 10.1259/bjr.20220890
68. Haripriya R, Khare N, Pandey M. Privacy-preserving federated learning for collaborative medical data mining in multi-institutional settings. Sci Rep. (2025) 15:12482. doi: 10.1038/s41598-025-97565-4
69. Frid-Adar M, Diamant I, Klang E, Amitai M, Goldberger J, Greenspan H. GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing. (2018) 321:321–31. doi: 10.1016/j.neucom.2018.09.013
70. Sundaram S, Hulkund N. Gan-based data augmentation for chest X-ray classification. arXiv [Preprint]. (2021) arXiv:2107.02970. doi: 10.48550/arXiv.2107.02970
71. Shin HC, Tenenholtz NA, Rogers JK, Schwarz CG, Senjem ML, Gunter JL, et al. Medical image synthesis for data augmentation and anonymization using generative adversarial networks. In: International Workshop on Simulation and Synthesis in Medical Imaging. Cham: Springer. (2018). p. 1–11. doi: 10.1007/978-3-030-00536-8_1
72. Figueira A, Vaz B. Survey on synthetic data generation, evaluation methods and GANs. Mathematics. (2022) 10:2733. doi: 10.3390/math10152733
73. Nampalle KB, Singh P, Narayan UV, Raman B. Vision through the veil: differential privacy in federated learning for medical image classification. arXiv [Preprint]. (2023) arXiv:2306.17794. doi: 10.48550/arXiv.2306.17794
74. Munjal K, Bhatia R. A systematic review of homomorphic encryption and its contributions in healthcare industry. Complex Intell Syst. (2023) 9:3759–86. doi: 10.1007/s40747-022-00756-z
75. Dutil F, See A, Di Jorio L, Chandelier F. Application of homomorphic encryption in medical imaging. arXiv [preprint]. (2021) arXiv:2110.07768. doi: 10.48550/arXiv.2110.07768
76. Roumpies F, Kakarountas A. A review of homomorphic encryption and its contribution to the sector of health services. In: Proceedings of the 27th Pan-Hellenic Conference on Progress in Computing and Informatics. New York, NY: ACM (2023). p. 237–242. doi: 10.1145/3635059.3635096
77. Zhu Y, Yin X, Wee-Chung Liew A, Tian H. Privacy-preserving in medical image analysis: a review of methods and applications. In: International Conference on Parallel and Distributed Computing: Applications and Technologies. Cham: Springer. (2024). p. 166–78. doi: 10.1007/978-981-96-4207-6_15
78. Kim J, Oh I, Lee YN, Lee JH, Lee YI, Kim J, et al. Predicting the severity of postoperative scars using artificial intelligence based on images and clinical data. Sci Rep. (2023) 13:13448. doi: 10.1038/s41598-023-40395-z
79. Ma C, He T, Gao J. Skin scar segmentation based on saliency detection. Vis Comput. (2023) 39:4887–99. doi: 10.1007/s00371-022-02635-7
80. Khan J, Malik AS, Kamel N, Dass SC, Affandi AM. Segmentation of acne lesion using fuzzy C-means technique with intelligent selection of the desired cluster. In: 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). Milan: IEEE. (2015). p. 3077–80. doi: 10.1109/EMBC.2015.7319042
81. Chantharaphaichi T, Uyyanonvara B, Sinthanayothin C, Nishihara A. Automatic acne detection for medical treatment. In: 2015 6th International Conference of Information and Communication Technology for Embedded Systems (IC-ICTES). Hua Hin: IEEE. (2015). p. 1–6. doi: 10.1109/ICTEmSys.2015.7110813
82. Jiang W, Guo L, Wu H, Ying J, Yang Z, Wei B, et al. Use of a smartphone for imaging, modelling, and evaluation of keloids. Burns. (2020) 46:1896–902. doi: 10.1016/j.burns.2020.05.026
83. Liu Y, Gong H, Li Y, Zhu X, Chen G. Texture features for classification of skin scar multi-photon fluorescence microscopic images. In: 2014 7th International Congress on Image and Signal Processing. Dalian: IEEE. (2014). p. 726–30. doi: 10.1109/CISP.2014.7003873
84. Heflin B, Scheirer W, Boult TE. Detecting and classifying scars, marks, and tattoos found in the wild. In: 2012 IEEE Fifth International Conference on Biometrics: Theory, Applications and Systems (BTAS). Arlington, VA: IEEE. (2012). p. 31–8. doi: 10.1109/BTAS.2012.6374555
85. Abas FS, Kaffenberger B, Bikowski J, Gurcan MN. Acne image analysis: lesion localization and classification. In: Medical Imaging 2016: Computer-Aided Diagnosis, Vol. 9785. Bellingham, WA: SPIE. (2016). p. 64–72. doi: 10.1117/12.2216444
86. Alamdari N, Tavakolian K, Alhashim M, Fazel-Rezai R. Detection and classification of acne lesions in acne patients: a mobile application. In: 2016 IEEE International Conference on Electro Information Technology (EIT). Grand Forks, ND: IEEE. (2016). p. 0739–43. doi: 10.1109/EIT.2016.7535331
87. Kittigul N, Uyyanonvara B. Acne detection using speeded up robust features and quantification using K-Nearest neighbors algorithm. In: Proceedings of the 6th International Conference on Bioinformatics and Biomedical Science. New York, NY: ACM (2017). p. 168–71. doi: 10.1145/3121138.3121168
88. Al-Tawalbeh J, Alshargawi B, Al-Daraghmeh M, Alquran H, Mustafa WA, Al-Dolaimy F, et al. Automated classification of skin lesions using different classifiers. In: 2023 6th International Conference on Engineering Technology and its Applications (IICETA). Al-Najaf: IEEE (2023). p. 103–7. doi: 10.1109/IICETA57613.2023.10351388
89. Maroni G, Ermidoro M, Previdi F, Bigini G. Automated detection, extraction and counting of acne lesions for automatic evaluation and tracking of acne severity. In: 2017 IEEE Symposium Series on Computational Intelligence (SSCI). Honolulu, HI: IEEE. (2017). p. 1–6. doi: 10.1109/SSCI.2017.8280925
90. Usmani UA, Watada J, Jaafar J, Aziz IA, Roy A. A reinforcement learning algorithm for automated detection of skin lesions. Appl Sci. (2021) 11:9367. doi: 10.3390/app11209367
91. Pham TTA, Kim H, Lee Y, Kang HW, Park S. Deep learning for analysis of collagen fiber organization in scar tissue. IEEE Access. (2021) 9:101755–64. doi: 10.1109/ACCESS.2021.3097370
92. Pham TTA, Kim H, Lee Y, Kang HW, Park S. Universal convolutional neural network for histology-independent analysis of collagen fiber organization in scar tissue. IEEE Access. (2022) 10:34379–92. doi: 10.1109/ACCESS.2022.3162272
93. Maknuna L, Kim H, Lee Y, Choi Y, Kim H, Yi M, et al. Automated structural analysis and quantitative characterization of scar tissue using machine learning. Diagnostics. (2022) 12:534. doi: 10.3390/diagnostics12020534
94. Chu Y, Jung SW, Lee S, Lee SG, Heo YW, Lee SH, et al. Deep learning algorithms for assessment of post-thyroidectomy scar subtype. Dermatol Ther. (2025) 2025:4636142. doi: 10.1155/dth/4636142
95. Junayed MS, Islam MB, Jeny AA, Sadeghzadeh A, Biswas T, Shah AS. ScarNet: development and validation of a novel deep CNN model for acne scar classification with a new dataset. IEEE Access. (2021) 10:1245–58. doi: 10.1109/ACCESS.2021.3138021
96. Ito H, Nakamura Y, Takanari K, Oishi M, Matsuo K, Kanbe M, et al. Development of a novel scar screening system with machine learning. Plast Reconstr Surg. (2022) 150:465e–72e. doi: 10.1097/PRS.0000000000009312
97. Singh P, Saxena V. Assessing the scar images to check medical treatment effectiveness. In: 2018 5th International Conference on Signal Processing and Integrated Networks (SPIN). Noida: IEEE. (2018). p. 624–9. doi: 10.1109/SPIN.2018.8474247
98. Privalov M, Beisemann N, Barbari JE, Mandelka E, Müller M, Syrek H, et al. Software-based method for automated segmentation and measurement of wounds on photographs using mask R-CNN: a validation study. J Digit Imaging. (2021) 34:788–97. doi: 10.1007/s10278-021-00490-x
99. Rajesh E, Madanagopal C, John R, Thanikaiselvan V, Tamizharasi T, Amirtharajan R. A deep learning approach for classification of vitiligo and scar images. In: 2024 10th International Conference on Communication and Signal Processing (ICCSP). Melmaruvathur: IEEE. (2024). p. 1051–6. doi: 10.1109/ICCSP60870.2024.10543467
100. Abdolahnejad M, Zandi A, Wong J, Chan HO, Lin V, Jeong H, et al. A prototype machine learning pipeline for assessing and tracking keloid scars. medRxiv. (2024). p. 2024–09. doi: 10.1101/2024.09.30.24314501
101. Aguilar J, Benítez D, Peréz N, Estrella-Porter J, Camacho M, Viteri M, et al. Towards the development of an acne-scar risk assessment tool using deep learning. In: 2022 IEEE International Autumn Meeting on Power, Electronics and Computing (ROPEC), Vol. 6. Ixtapa: IEEE (2022). p. 1–6. doi: 10.1109/ROPEC55836.2022.10018763
102. Sarvamangala D, Kulkarni RV. Convolutional neural networks in medical image understanding: a survey. Evol Intell. (2022) 15:1–22. doi: 10.1007/s12065-020-00540-3
103. Yamashita R, Nishio M, Do RKG, Togashi K. Convolutional neural networks: an overview and application in radiology. Insights Imaging. (2018) 9:611–29. doi: 10.1007/s13244-018-0639-9
104. Finlayson SG, Chung HW, Kohane IS, Beam AL. Adversarial attacks against medical deep learning systems. arXiv [Preprint]. (2018) arXiv:1804.05296. doi: 10.48550/arXiv.1804.05296
105. Dong J, Chen J, Xie X, Lai J, Chen H. Survey on adversarial attack and defense for medical image analysis: methods and challenges. ACM Comput Surv. (2024) 57:1–38. doi: 10.1145/3702638
106. Bortsova G. González-Gonzalo C, Wetstein SC, Dubost F, Katramados I, Hogeweg L, et al. Adversarial attack vulnerability of medical image analysis systems: unexplored factors. Med Image Anal. (2021) 73:102141. doi: 10.1016/j.media.2021.102141
107. Keskar NS, Mudigere D, Nocedal J, Smelyanskiy M, Tang PTP. On large-batch training for deep learning: generalization gap and sharp minima. arXiv [Preprint]. (2016) arXiv:1609.04836. doi: 10.4850/arXiv.1609.04836
108. Masters D, Luschi C. Revisiting small batch training for deep neural networks. arXiv [Preprint]. (2018). arXiv:1804.07612. doi: 10.48550/arXiv.1804.07612
109. Houssein EH, Gamal AM, Younis EM, Mohamed E. Explainable artificial intelligence for medical imaging systems using deep learning: a comprehensive review. Cluster Comput. (2025) 28:469. doi: 10.1007/s10586-025-05281-5
110. Arun N, Gaw N, Singh P, Chang K, Aggarwal M, Chen B, et al. Assessing the trustworthiness of saliency maps for localizing abnormalities in medical imaging. Radiol Artif Intell. (2021) 3:e200267. doi: 10.1148/ryai.2021200267
111. Wang Q, Liu W, Chen X, Wang X, Chen G, Zhu X. Quantification of scar collagen texture and prediction of scar development via second harmonic generation images and a generative adversarial network. Biomed Opt Express. (2021) 12:5305–19. doi: 10.1364/BOE.431096
112. Zhou J, Zhou Z, Chen X, Shi F, Xia W. A deep learning-based automatic tool for measuring the lengths of linear scars: forensic applications. Forensic Sci Res. (2023) 8:41–9. doi: 10.1093/fsr/owad010
113. Zhou J, Dai Y, Liu D, Zhu W, Xiang D, Chen X, et al. Improving deep learning based segmentation of scars using multi-view images. Biomed Signal Process Control. (2024) 94:106254. doi: 10.1016/j.bspc.2024.106254
114. Hu EJ, Shen Y, Wallis P, Allen-Zhu Z, Li Y, Wang S, et al. Lora: low-rank adaptation of large language models. ICLR. (2022) 1:3.
115. Lian C, Zhou HY, Yu Y, Wang L. Less could be better: parameter-efficient fine-tuning advances medical vision foundation models. arXiv [Preprint]. (2024) arXiv:240.112215. doi: 10.48550/arXiv.2401.12215
116. Dzieniszewska A, Garbat P, Piramidowicz R. Improving skin lesion segmentation with self-training. Cancers. (2024) 16:1120. doi: 10.3390/cancers16061120
117. Ganin Y, Ustinova E, Ajakan H, Germain P, Larochelle H, Laviolette F, et al. Domain-adversarial training of neural networks. J Mach Learn Res. (2016) 17:1–35.
118. Gilani SQ, Umair M, Naqvi M, Marques O, Kim HC. Adversarial training based domain adaptation of skin cancer images. Life. (2024) 14:1009. doi: 10.3390/life14081009
119. Wang H, Ahn E, Bi L, Kim J. Self-supervised multi-modality learning for multi-label skin lesion classification. Comput Methods Programs Biomed. (2025) 265:108729. doi: 10.1016/j.cmpb.2025.108729
120. Nguyen AT, Li RA, Galiano RD. Assessing the predictive accuracy of ChatGPT-based image analysis in forecasting long-term scar characteristics from 3-month assessments-a pilot study. J Plastic Reconstr Aesthet Surg. (2025) 104:200–8. doi: 10.1016/j.bjps.2025.03.021
121. Shiraishi M, Miyamoto S, Takeishi H, Kurita D, Furuse K, Ohba J, et al. The potential of chat-based artificial intelligence models in differentiating between keloid and hypertrophic scars: a pilot study. Aesthetic Plast Surg. (2024) 48:5367–72. doi: 10.1007/s00266-024-04380-9
122. Yang W, Wang X, Chen G, Wen J, Kong D, Zhang J, et al. A dual encoder network with multiscale feature fusion and multiple pooling channel spatial attention for skin scar image segmentation. Sci Rep. (2025) 15:22810. doi: 10.1038/s41598-025-05239-y
123. Xin C, Liu Z, Zhao K, Miao L, Ma Y, Zhu X, et al. An improved transformer network for skin cancer classification. Comput Biol Med. (2022) 149:105939. doi: 10.1016/j.compbiomed.2022.105939
124. He X, Tan EL Bi H, Zhang X, Zhao S, Lei B. Fully transformer network for skin lesion analysis. Med Image Anal. (2022) 77:102357. doi: 10.1016/j.media.2022.102357
125. Moon CI, Kim EB, Baek YS, Lee O. Transformer based on the prediction of psoriasis severity treatment response. Biomed Signal Process Control. (2024) 89:105743. doi: 10.1016/j.bspc.2023.105743
126. Huang K, Sun K, Li J, Wu Z, Wu X, Duan Y, et al. Intelligent strategy for severity scoring of skin diseases based on clinical decision-making thinking with lesion-aware transformer. Artif Intell Rev. (2025) 58:95. doi: 10.1007/s10462-024-11083-9
127. Mohan J, Sivasubramanian A, Sowmya V, Ravi V. Enhancing skin disease classification leveraging transformer-based deep learning architectures and explainable AI. Comput Biol Med. (2025) 190:110007. doi: 10.1016/j.compbiomed.2025.110007
128. Salam AA, Asaf MZ, Akram MU, Ali A, Mashallah MI, Rao B, et al. Skin whole slide image segmentation using lightweight-pruned transformer. Biomed Signal Process Control. (2025) 106:107624. doi: 10.1016/j.bspc.2025.107624
129. Krishna GS, Supriya K, Mallikharjuna Rao K, Sorgile M. Lesionaid: vision transformers-based skin lesion generation and classification. arXiv [Preprint]. (2023) arXiv:2302.01104. doi: 10.48550/arXiv.2302.01104
130. Cai G, Zhu Y, Wu Y, Jiang X, Ye J, Yang D. A multimodal transformer to fuse images and metadata for skin disease classification. Vis Comput. (2023) 39:2781–93. doi: 10.1007/s00371-022-02492-4
131. Radiah F, Rahman K, Asadullah L, Sohan MSR, Ahmed J. Explainable AI (XAI) Driven Skin Cancer Detection Using Transformer and CNN Based Architecture. Dhaka: Brac University (2023).
132. Nikitin V, Danilov V. Transformer vs. mamba as skin cancer classifier: preliminary results. KPI Sci News. (2024) 137:26–30. doi: 10.20535/kpisn.2024.1-4.301028
133. Pacal I, Alaftekin M, Zengul FD. Enhancing skin cancer diagnosis using swin transformer with hybrid shifted window-based multi-head self-attention and SwiGLU-based MLP. J Imaging Inform Med. (2024) 37:1–19. doi: 10.1007/s10278-024-01140-8
134. Vishwakarma R, Sood D. Artificial intelligence for image classification of skin diseases with convolution transformer. In: 2023 4th International Conference on Intelligent Technologies (CONIT). Bangalore: IEEE. (2024). p. 1–6. doi: 10.1109/CONIT61985.2024.10626497
135. Bozorgpour A, Sadegheih Y, Kazerouni A, Azad R, Merhof D. Dermosegdiff: a boundary-aware segmentation diffusion model for skin lesion delineation. In: International Workshop on Predictive Intelligence in Medicine. Cham: Springer (2023). p. 146–58. doi: 10.1007/978-3-031-46005-0_13
136. Wang J, Yang J, Zhou Q, Wang L. Medical boundary diffusion model for skin lesion segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Cham: Springer (2023). p. 427–36. doi: 10.1007/978-3-031-43901-8_41
137. Hu M, Yan S, Xia P, Tang F, Li W, Duan P, et al. Diffusion model driven test-time image adaptation for robust skin lesion classification. arXiv [Preprint]. (2024) arXiv:2405.11289. doi: 10.48550/arXiv.2405.11289
138. Farooq MA, Yao W, Schukat M, Little MA, Corcoran P. Derm-t2im: harnessing synthetic skin lesion data via stable diffusion models for enhanced skin disease classification using VIT and CNN. In: 2024 46th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). Orlando, FL: IEEE. (2024). p. 1–5. doi: 10.1109/EMBC53108.2024.10781852
139. Huang Z, Li J, Mao N, Li J. BADM: boundary-assisted diffusion model for skin lesion segmentation. Eng Appl Artif Intell. (2024) 137:109213. doi: 10.1016/j.engappai.2024.109213
140. Ivanovici M, Stoica D. Color diffusion model for active contours-an application to skin lesion segmentation. In: 2012 Annual International Conference of the IEEE Engineering in Medicine and Biology Society. San Diego, CA: IEEE. (2012). p. 5347–50. doi: 10.1109/EMBC.2012.6347202
141. Jin Q, Cui H, Sun C, Meng Z, Su R. Cascade knowledge diffusion network for skin lesion diagnosis and segmentation. Appl Soft Comput. (2021) 99:106881. doi: 10.1016/j.asoc.2020.106881
142. Du S, Wang X, Lu Y, Zhou Y, Zhang S, Yuille A, et al. Boosting dermatoscopic lesion segmentation via diffusion models with visual and textual prompts. In: 2024 IEEE International Symposium on Biomedical Imaging (ISBI). Athens: IEEE (2024). p. 1–5. doi: 10.1109/ISBI56570.2024.10635486
143. Guo Y, Cai Q. BGDiffSeg: a fast diffusion model for skin lesion segmentation via boundary enhancement and global recognition guidance. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Cham: Springer (2024). p. 150–9. doi: 10.1007/978-3-031-72114-4_15
144. Jing X, Yang S, Zhou H, Wang G, Mao K. SkinDiff: a novel data synthesis method based on latent diffusion model for skin lesion segmentation. In: International Conference on Intelligent Computing. Cham: Springer (2024). p. 179–91. doi: 10.1007/978-981-97-5603-2_15
145. Patcharapimpisut P, Khanarsa P. Generating synthetic images using stable diffusion model for skin lesion classification. In: 2024 16th International Conference on Knowledge and Smart Technology (KST). Krabi: IEEE. (2024). p. 184–9. doi: 10.1109/KST61284.2024.10499667
146. Shuai Z, Chen Y, Mao S, Zho Y, Zhang X. Diffseg: a segmentation model for skin lesions based on diffusion difference. arXiv [Preprint]. (2024) arXiv:2404.16474. doi: 10.48550/arXiv.2404.16474
147. Munia N, Imran AAZ. DermDiff: generative diffusion model for mitigating racial biases in dermatology diagnosis. arXiv [Preprint]. (2025) arXiv:2503.17536. doi: 10.48550/arXiv.2503.17536
148. Mittal A, Kalkhof J, Mukhopadhyay A, Bhavsar A. Medsegdiffnca: diffusion models with neural cellular automata for skin lesion segmentation. arXiv [Preprint]. (2025). doi: 10.48550/arXiv.2501.02447
149. Barcelos CAZ, Pires V. An automatic based nonlinear diffusion equations scheme for skin lesion segmentation. Appl Math Comput. (2009) 215:251–61. doi: 10.1016/j.amc.2009.04.081
150. Xiang F, Li Z, Jiang S, Li C, Li S, Gao T, et al. Multimodal masked autoencoder based on adaptive masking for vitiligo stage classification. J Imaging Inform Med. (2025). p. 1–14. doi: 10.1007/s10278-025-01521-7
151. Song W, Wang X, Guo Y, Li S, Xia B, Hao A. Centerformer: a novel cluster center enhanced transformer for unconstrained dental plaque segmentation. IEEE Trans Multimed. New York, NY: ACM (2024). doi: 10.1109/TMM.2024.3428349
152. Finnane A, Dallest K, Janda M, Soyer HP. Teledermatology for the diagnosis and management of skin cancer: a systematic review. JAMA Dermatol. (2017) 153:319–27. doi: 10.1001/jamadermatol.2016.4361
153. Abràmoff MD, Lavin PT, Birch M, Shah N, Folk JC. Pivotal trial of an autonomous AI-based diagnostic system for detection of diabetic retinopathy in primary care offices. NPJ Digit Med. (2018) 1:39. doi: 10.1038/s41746-018-0040-6
154. Grzybowski A, Singhanetr P, Nanegrungsunk O, Ruamviboonsuk P. Artificial intelligence for diabetic retinopathy screening using color retinal photographs: from development to deployment. Ophthalmol Ther. (2023) 12:1419–37. doi: 10.1007/s40123-023-00691-3
155. Vasey B, Nagendran M, Campbell B, Clifton DA, Collins GS, Denaxas S, et al. Reporting guideline for the early stage clinical evaluation of decision support systems driven by artificial intelligence: DECIDE-AI. BMJ. (2022) 377:e070904. doi: 10.1136/bmj-2022-070904
156. Basch E, Stover AM, Schrag D, Chung A, Jansen J, Henson S, et al. Clinical utility and user perceptions of a digital system for electronic patient-reported symptom monitoring during routine cancer care: findings from the PRO-TECT trial. JCO Clin Cancer Inform. (2020) 4:947–57. doi: 10.1200/CCI.20.00081
157. Rivera SC, Liu X, Chan AW, Denniston AK, Calvert MJ, Ashrafian H, et al. Guidelines for clinical trial protocols for interventions involving artificial intelligence: the SPIRIT-AI extension. Lancet Digit Health. (2020) 2:e549–60. doi: 10.1136/bmj.m3210
158. Guidance W. Ethics and Governance of Artificial Intelligence for Health. Geneva: World Health Organization (2021).
159. Verhaegen PD, van der Wal MB, Middelkoop E, van Zuijlen PP. Objective scar assessment tools: a clinimetric appraisal. Plast Reconstr Surg. (2011) 127:1561–70. doi: 10.1097/PRS.0b013e31820a641a
160. Mienye ID, Swart TG, Obaido G, Jordan M, Ilono P. Deep convolutional neural networks in medical image analysis: a review. Information. (2025) 16:195. doi: 10.3390/info16030195
161. Wang W, Yuan X, Wu X, Liu Y. Fast image dehazing method based on linear transformation. IEEE Trans Multimed. (2017) 19:1142–55. doi: 10.1109/TMM.2017.2652069
162. Lin TL, Mukundan A, Karmakar R, Avala P, Chang WY, Wang HC. Hyperspectral imaging for enhanced skin cancer classification using machine learning. Bioengineering. (2025) 12:755. doi: 10.3390/bioengineering12070755
163. Huang NC, Mukundan A, Karmakar R, Syna S, Chang WY, Wang HC. Novel snapshot-based hyperspectral conversion for dermatological lesion detection via YOLO object detection models. Bioengineering. (2025) 12:714. doi: 10.3390/bioengineering12070714
164. Lin TL, Lu CT, Karmakar R, Nampalley K, Mukundan A, Hsiao YP, et al. Assessing the efficacy of the spectrum-aided vision enhancer (SAVE) to detect acral lentiginous melanoma, melanoma in situ, nodular melanoma, and superficial spreading melanoma. Diagnostics. (2024) 14:1672. doi: 10.3390/diagnostics14151672
165. Han R, Acosta JN, Shakeri Z, Ioannidis JP, Topol EJ, Rajpurkar P. Randomised controlled trials evaluating artificial intelligence in clinical practice: a scoping review. Lancet Digit Health. (2024) 6:e367–73. doi: 10.1016/S2589-7500(24)00047-5
166. Mennella C, Maniscalco U, De Pietro G, Esposito M. Ethical and regulatory challenges of AI technologies in healthcare: a narrative review. Heliyon. (2024) 10:e26297. doi: 10.1016/j.heliyon.2024.e26297
167. LeCun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proc IEEE. (1998) 86:2278–324. doi: 10.1109/5.726791
168. Chen M, Zhou P, Wu D, Hu L, Hassan MM, Alamri A. AI-Skin: skin disease recognition based on self-learning and wide data collection through a closed-loop framework. Inf Fusion. (2020) 54:1–9. doi: 10.1016/j.inffus.2019.06.005
169. Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. In: Advances in Neural Information Processing Systems. Red Hook, NY: Curran Associates, Inc. (2012). p. 25.
170. Hosny KM, Kassem MA, Foaud MM. Classification of skin lesions using transfer learning and augmentation with Alex-net. PLoS ONE. (2019) 14:e0217293. doi: 10.1371/journal.pone.0217293
171. Kingma DP. Auto-encoding variational bayes. arXiv [Preprint]. (2013). arXiv:1312.6114. doi: 10.48550/arXiv.1312.6114
172. Ravi KM, Kiran M, Umadevi V. Acne classification using deep learning models. In: 2024 1st International Conference on Communications and Computer Science (InCCCS). Bangalore: IEEE (2024). p. 1–6. doi: 10.1109/InCCCS60947.2024.10593603
173. Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, et al. Generative adversarial nets. In: Advances in Neural Information Processing Systems. Red Hook, NY: Curran Associates, Inc. (2014). p. 27.
174. Sharma V, Mehta S. Deep neural networks for dermatology: CNN-GAN in multi-class skin disease detection. In: 2024 5th International Conference on Electronics and Sustainable Communication Systems (ICESC). Coimbatore: IEEE (2024). p. 985–90. doi: 10.1109/ICESC60852.2024.10689851
175. Heenaye-Mamode Khan M, Gooda Sahib-Kaudeer N, Dayalen M, Mahomedaly F, Sinha GR, Nagwanshi KK, et al. Multi-class skin problem classification using deep generative adversarial network (DGAN). Comput Intell Neurosci. (2022) 2022:1797471. doi: 10.1155/2022/1797471
176. Simonyan K. Very deep convolutional networks for large-scale image recognition. arXiv [Preprint]. (2014) arXiv:1409.1556. doi: 10.48550/arXiv.1409.1556
177. AlSuwaidan L. Deep learning based classification of dermatological disorders. Biomed Eng Comput Biol. (2023) 14:11795972221138470. doi: 10.1177/11795972221138470
178. Thomsen K, Christensen AL, Iversen L, Lomholt HB, Winther O. Deep learning for diagnostic binary classification of multiple-lesion skin diseases. Front Med. (2020) 7:574329. doi: 10.3389/fmed.2020.574329
179. Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. In: Medical Image Computing and Computer-Assisted Intervention-MICCAI 2015: 18th International Conference, Munich, Germany, October 5-9, 2015, Proceedings, Part III 18. Cham: Springer (2015). p. 234–41. doi: 10.1007/978-3-319-24574-4_28
180. Tang P, Liang Q, Yan X, Xiang S, Sun W, Zhang D, et al. Efficient skin lesion segmentation using separable-Unet with stochastic weight averaging. Comput Methods Programs Biomed. (2019) 178:289–301. doi: 10.1016/j.cmpb.2019.07.005
181. Wu J, Chen EZ, Rong R, Li X, Xu D, Jiang H. Skin lesion segmentation with C-UNet. In: 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). Berlin: IEEE (2019). p. 2785–8. doi: 10.1109/EMBC.2019.8857773
182. Kibriya H, Siddiqa A, Khan WZ. Melanoma lesion localization using UNet and explainable AI. Neural Comput Appl. (2025) 37:10175–96. doi: 10.1007/s00521-025-11080-1
183. Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, et al. Going deeper with convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. Boston, MA: IEEE (2015). p. 1–9. doi: 10.1109/CVPR.2015.7298594
184. Hirano G, Nemoto M, Kimura Y, Kiyohara Y, Koga H, Yamazaki N, et al. Automatic diagnosis of melanoma using hyperspectral data and GoogLeNet. Skin Res Technol. (2020) 26:891–7. doi: 10.1111/srt.12891
185. Santhiya S, Parvathi M, Jayadharshini P, Harish S, Navin E, Indrajith N. Automated skin disease detection using GoogLeNet and MobileNet to enhance the diagnostic accuracy with convolutional neural networks. In: 2024 2nd International Conference on Advances in Computation, Communication and Information Technology (ICAICCIT), Vol. 1. Faridabad: IEEE (2024). p. 316–9. doi: 10.1109/ICAICCIT64383.2024.10912179
186. Mnih V, Kavukcuoglu K, Silver D, Rusu AA, Veness J, Bellemare MG, et al. Human-level control through deep reinforcement learning. Nature. (2015) 518:529–33. doi: 10.1038/nature14236
187. Jin JK, Dajani K, Kim M, Kim SD, Khan B, Jin DH. Reinforcement learning architecture for facial skin treatment recommender. In: 2024 IEEE/ACIS 22nd International Conference on Software Engineering Research, Management and Applications (SERA). Honolulu, HI: IEEE (2024). p. 47–54. doi: 10.1109/SERA61261.2024.10685645
188. Doğan N, Mayanja A, Taşdemir Ş. Class-weighted reinforcement learning for skin cancer image classification. Expert Syst Appl. (2025) 293:128426. doi: 10.1016/j.eswa.2025.128426
189. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. Las Vegas, NV: IEEE (2016). p. 770–8. doi: 10.1109/CVPR.2016.90
190. Sharma M, Jain B, Kargeti C, Gupta V, Gupta D. Detection and diagnosis of skin diseases using residual neural networks (RESNET). Int J Image Graph. (2021) 21:2140002. doi: 10.1142/S0219467821400027
191. Gouda N, Amudha J. Skin cancer classification using ResNet. In: 2020 IEEE 5th International Conference on Computing Communication and Automation (ICCCA). Greater Noida: IEEE (2020). p. 536–541. doi: 10.1109/ICCCA49541.2020.9250855
192. Mehra A, Bhati A, Kumar A, Malhotra R. Skin cancer classification through transfer learning using ResNet-50. In: Emerging Technologies in Data Mining and Information Security: Proceedings of IEMIS 2020, Volume 2. Cham: Springer (2021). p. 55–62. doi: 10.1007/978-981-33-4367-2_6
193. Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. Honolulu, HI: IEEE (2017). p. 4700–8. doi: 10.1109/CVPR.2017.243
194. De A, Mishra N, Chang HT. An approach to the dermatological classification of histopathological skin images using a hybridized CNN-DenseNet model. PeerJ Comput Sci. (2024) 10:e1884. doi: 10.7717/peerj-cs.1884
195. Adegun AA, Viriri S. FCN-based DenseNet framework for automated detection and classification of skin lesions in dermoscopy images. IEEE Access. (2020) 8:150377–96. doi: 10.1109/ACCESS.2020.3016651
196. Vaswani A. Attention is all you need. In: Advances in Neural Information Processing Systems. Red Hook, NY: Curran Associates, Inc. (2017).
197. Tschandl P, Rosendahl C, Kittler H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci Data. (2018) 5:1–9. doi: 10.1038/sdata.2018.161
198. Combalia M, Codella NC, Rotemberg V, Helba B, Vilaplana V, Reiter O, et al. Bcn20000: dermoscopic lesions in the wild. arXiv [Preprint]. (2019) arXiv:1908.02288. doi: 10.48550/arXiv:1908.02288
199. Sun X, Yang J, Sun M, Wang K. A benchmark for automatic visual classification of clinical skin disease images. In: Computer Vision-ECCV 2016: 14th European Conference, Amsterdam, The Netherlands, October 11-14, 2016, Proceedings, Part VI 14. Cham: Springer. (2016). p. 206–22. doi: 10.1007/978-3-319-46466-4_13
200. Pacheco AG, Lima GR, Salomao AS, Krohling B, Biral IP, de Angelo GG, et al. PAD-UFES-20: a skin lesion dataset composed of patient data and clinical images collected from smartphones. Data Brief . (2020) 32:106221. doi: 10.1016/j.dib.2020.106221
201. Groh M, Harris C, Soenksen L, Lau F, Han R, Kim A, et al. Evaluating deep neural networks trained on clinical images in dermatology with the fitzpatrick 17k dataset. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. Nashville, TN: EEE (2021). p. 1820–8. doi: 10.1109/CVPRW53098.2021.00201
Keywords: artificial intelligence in dermatology, computer vision for skin analysis, dataset, medical image process, large-scale foundation model
Citation: Hu F, Shao Y, Liu J, Liu J, Xiao X, Shi K, Zheng Y, Zhang J and Wang X (2025) Advances in intelligent recognition and diagnosis of skin scar images: concepts, methods, challenges, and future trends. Front. Med. 12:1667087. doi: 10.3389/fmed.2025.1667087
Received: 16 July 2025; Accepted: 18 August 2025;
Published: 04 September 2025.
Edited by:
Arvind Mukundan, National Chung Cheng University, TaiwanReviewed by:
Nitin Goyal, Central University of Haryana, IndiaHsiang-Chen Wang, National Chung Cheng University, Taiwan
Copyright © 2025 Hu, Shao, Liu, Liu, Xiao, Shi, Zheng, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianfeng Zhang, amZ6aGFuZ0B6am51LmVkdS5jbg==; Xuelian Wang, bHF6cndsekAxNjMuY29t
†These authors have contributed equally to this work and share first authorship
 Fuhua Hu1†
Fuhua Hu1† Jianfeng Zhang
Jianfeng Zhang