- Longyan First Affiliated Hospital of Fujian Medical University, Longyan, China
Metagenomic next-generation sequencing (mNGS) demonstrates high sensitivity, rapid diagnostic capabilities, and the potential to identify complex pathogens in periprosthetic joint infection (PJI) following arthroplasty, particularly when conventional culture methods are limited. mNGS enables the detection of polymicrobial infections and rare/fastidious pathogens, along with the ability to predict antimicrobial resistance (AMR) genes; however, the concordance between genotypic predictions and phenotypic resistance profiles requires further validation. In clinical practice, mNGS overcomes biofilm-related diagnostic barriers, facilitating early targeted antibiotic therapy and potentially reducing unnecessary revision surgeries, thereby lowering overall healthcare costs and improving patient outcomes. Nevertheless, its widespread adoption is hindered by high costs, lack of standardization, and risks of false-positive/false-negative results. Future research priorities include optimizing sample processing protocols, host DNA depletion, establishing diagnostic thresholds, and validating mNGS through integration with conventional methods. This review synthesizes recent advances in the diagnostic accuracy and clinical utility of mNGS for PJI, aiming to provide evidence-based insights for therapeutic decision-making and enhance the prevention and management of PJI.
1 Introduction
Periprosthetic joint infection (PJI) following arthroplasty remains one of the most formidable complications in joint replacement surgery (1), with its incidence and mortality rates demonstrating a progressive annual increase (2–4). PJI not only imposes severe physiological and psychological burdens on patients but also substantially elevates healthcare costs and socioeconomic expenditures (5–8). The diversity and inconsistent application of diagnostic criteria for PJI represent a major challenge in current clinical practice and research. The JS-BACH classification system categorizes patients into “uncomplicated,” “complex,” and “limited treatment options” based on five dimensions: joint specificity, extent of bone involvement, antibiotic options, soft tissue coverage, and host status. This framework comprehensively covers key factors influencing PJI prognosis (9, 10). Although this classification shows promise in initial studies, its external validity and generalizability require further validation (9). In contrast, the EBJIS criteria classify PJI into “confirmed,” “likely,” and “unlikely,” relying primarily on microbiological evidence, histopathology, synovial fluid analysis, and imaging results (11). These criteria perform particularly well in distinguishing chronic PJI, though their sensitivity depends on comprehensive microbiological testing (12, 13). Additionally, the diagnostic criteria proposed by MSIS are widely used for the clinical identification of PJI, yet their accuracy and applicability remain controversial (14). For example, in periprosthetic shoulder infection, the MSIS criteria may fail to reliably detect infection, with 30% of cases requiring diagnosis based on other clinical parameters (15). The use of different diagnostic standards leads to significant variation in reported PJI incidence rates (21–32%) (16, 17). In recent years, important progress has been made in the prevention, diagnosis, and treatment of PJI. Nevertheless, numerous challenges remain, such as antibiotic resistance due to biofilm formation, diversity of pathogens, lack of uniform diagnostic criteria, and the absence of personalized treatment strategies (18–20).
The reported incidence of PJI following primary arthroplasty is approximately 0. 4%, which rises to 1. 6% after revision surgery (21). Conventional pathogen detection methods suffer from low sensitivity, prolonged processing times, and limited microbial detection capabilities (22). This is particularly evident in culture-negative PJI (CN PJI), where approximately 48% of infections fail to yield identifiable pathogens via culture (23). Moreover, recent antibiotic use heightens the risk of false-negative results, exacerbating diagnostic challenges (24). The advent of metagenomic next-generation sequencing (mNGS) has recently emerged as a transformative strategy for precise PJI diagnosis.
2 Principles and optimization of mNGS technology
mNGS demonstrates significant advantages in infection diagnosis (25). By rapidly identifying pathogens and their genomic characteristics, mNGS provides timely and precise decision-making support for clinical treatment, thereby improving patient outcomes. Particularly in complex infections and public health emergencies, mNGS exhibits diagnostic value unmatched by conventional methods. The core principle of mNGS involves random fragmentation and sequencing of nucleic acids extracted from samples without prior knowledge of pathogens (25). Its workflow primarily includes: (1) sample processing; (2) nucleic acid extraction; (3) library preparation; (4) high-throughput sequencing; and (5) bioinformatic analysis. The greatest strength of this method lies in its ‘hypothesis-free’ detection capability, enabling unbiased identification of all microbial components in a sample, including bacteria, fungi, viruses, and rare pathogens (26). However, clinical application of mNGS still faces challenges, such as high costs, lack of standardized protocols, false-positive risks due to host nucleic acid interference (23), and the need for further optimization of detection workflows across different specimen types (27). Despite these limitations with continuous advancements in bioinformatics tools and sequencing technologies, mNGS is poised to become a vital tool in routine clinical diagnostics.
3 Diagnostic accuracy of mNGS for PJI
In recent years, mNGS has demonstrated transformative potential in the pathogen diagnosis of PJI, particularly serving as an effective tool for analyzing microbial communities in culture-negative and complex infections (28, 29). mNGS is being increasingly applied in detecting pathogens for periprosthetic joint infections (PJI), sepsis, and other infectious diseases (23). By synthesizing existing research, this article explores the unique value of mNGS in PJI to provide evidence-based guidance for clinical practice.
3.1 Comparative analysis of mNGS versus conventional culture methods
Culture-negative periprosthetic joint infection (CN-PJI) represents a major challenge in the diagnosis and treatment of PJI. As the gold standard, conventional microbial culture suffers from limitations including low sensitivity, time-consuming procedures, susceptibility to antibiotic interference, and inability to identify causative pathogens (24, 30), posing significant challenges for clinicians when formulating treatment strategies.
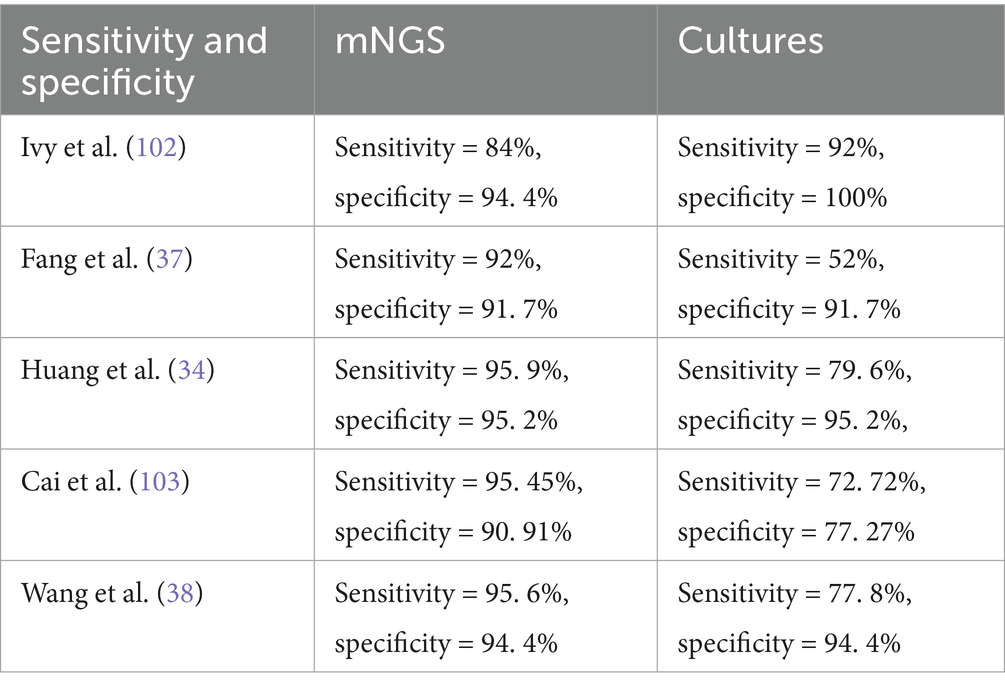
Multiple studies demonstrate that mNGS exhibits significantly higher overall sensitivity than conventional culture (31), particularly in CN PJI, where it detects additional rare pathogens missed by traditional methods (32, 33). However, mNGS may still yield false-negative results, potentially overlooking certain culture-positive pathogens (34). Implementing a “clinician-microbiologist-bioinformatician” tripartite consultation model could mitigate diagnostic inaccuracies. Notably, mNGS detects polymicrobial infections at 1. 5 × the rate of culture, yet its relatively low specificity (60%) necessitates caution against overdiagnosis (35). Among specimen types, sonicate fluid from prosthetic devices shows superior pathogen detection rates via mNGS (27, 35), attributable to its capacity to liberate biofilm-embedded microbes, yielding >10-fold higher sequencing reads and >5-fold greater genome coverage compared to other samples (36, 37). Current technical limitations center on host DNA contamination and bioinformatic complexity, mandating standardized thresholds (e. g., pathogen-specific read counts) to enhance reliability. Furthermore, traditional molecular methods like 16S rDNA PCR concord with mNGS in only 73%-86. 5% of cases, underscoring the necessity of integrating clinical judgment to avoid overreliance on any single technique (38, 39) (Table 1).
4 Comparative evaluation of mNGS versus other molecular diagnostic techniques
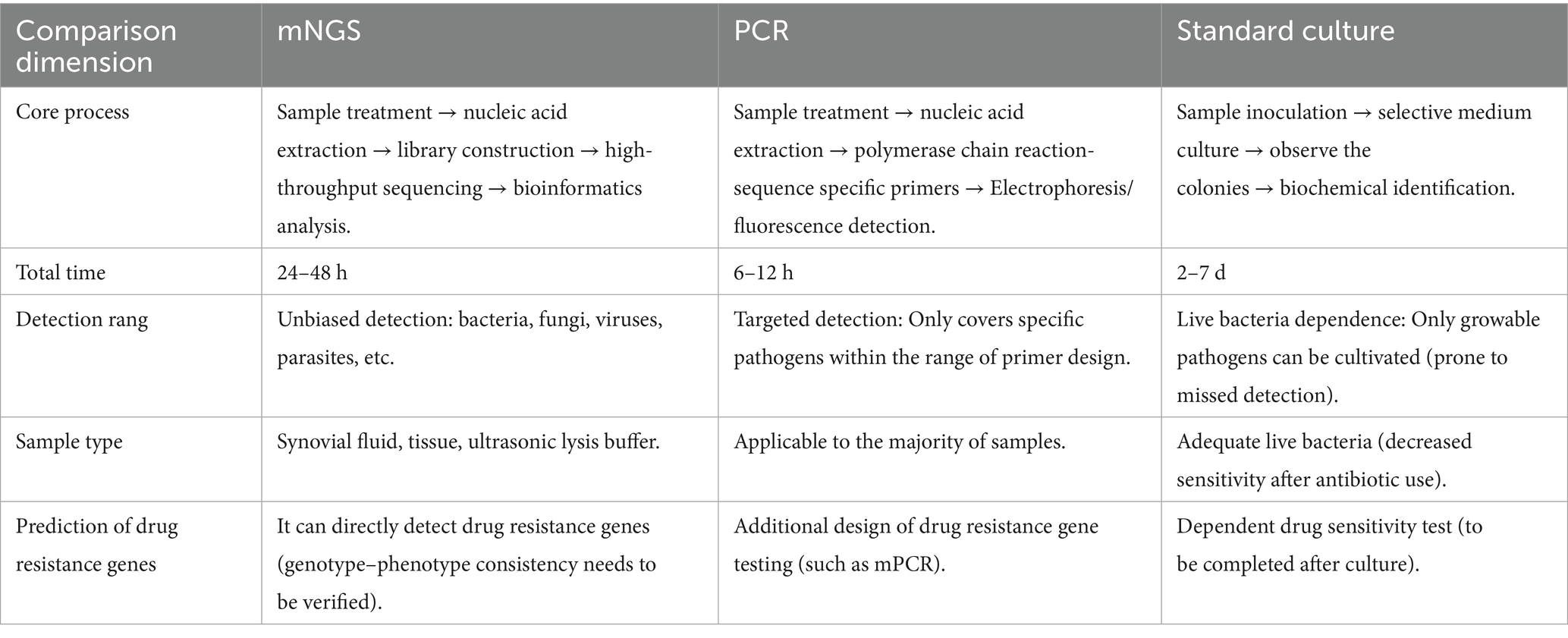
Polymerase chain reaction (PCR) has been introduced into the field of periprosthetic joint infection (PJI) diagnosis due to its rapidity and high sensitivity (40). However, the issue of false-positive results remains a major clinical concern (41, 42). These false positives often arise from various factors such as cross-contamination, non-specific amplification, suboptimal primer design, or improper sample handling. In recent years, multiple studies have focused on reducing the false-positive rate through improvements in detection methodologies and optimization of experimental procedures (43–46). Furthermore, continuous monitoring of melting temperature (47) and integration of culture-based verification (48) are also crucial. These measures are expected to enhance the reliability of PCR-based detection, thereby supporting precision medicine and public health decision-making. Compared with other diagnostic techniques, mNGS also possesses numerous advantages. A primary strength of mNGS is its unbiased sampling capability, enabling broad identification of both known and unexpected pathogens, including novel organisms (49). mNGS can also be coupled with targeted approaches, such as using primers from conserved 16S ribosomal RNA (rRNA) and internal transcribed spacer regions for universal bacterial and fungal detection (50, 51), allowing species-level identification of these organisms. Another advantage of mNGS is its ability to provide strain-level characterization and antimicrobial resistance prediction (52–54). Moreover, the diagnostic accuracy of mNGS significantly surpasses that of conventional PCR. A meta-analysis of 79 studies demonstrated that mNGS exhibits the highest overall diagnostic performance for PJI (39). These findings indicate that while maintaining high specificity, mNGS substantially improves detection rates for PJI, particularly in culture-negative or low-biomass infections. Traditional molecular methods like 16S rDNA PCR show only 73%–86. 5% concordance with mNGS, underscoring the need for clinical correlation to avoid overreliance on any single technique (38, 39). Compared to alternative diagnostic methods, mNGS-guided targeted therapy facilitates faster inflammation control, shorter treatment duration, and potential reduction in hospitalization (55). Furthermore, its diagnostic superiority in complex infections strongly supports broader clinical adoption (Table 2).
5 Pathogen spectrum expansion value of mNGS in PJI
5.1 Superiority in polymicrobial infection detection
mNGS demonstrates superior performance over conventional culture methods in identifying polymicrobial infections. Its high-throughput, culture-independent nature provides a novel approach for rapid and accurate diagnosis of PJI, particularly in mixed infections (56, 57). Notably, in polymicrobial infection cases, mNGS achieves a sensitivity of 72. 23%, compared to merely 27. 27% for culture, underscoring its enhanced capability to detect co-infecting pathogens (35, 56, 58). This high sensitivity renders mNGS a powerful tool for diagnosing culture-negative PJI, especially those caused by fastidious pathogens in mixed infections.
5.2 Identification of rare and fastidious microorganisms
In PJI, beyond common pathogens like Staphylococcus aureus and Staphylococcus epidermidis, mNGS has successfully detected clinically significant Gram-positive bacteria that were rarely reported in PJI due to their fastidious growth requirements (59, 60). The application of mNGS enables identification of these challenging-to-culture anaerobic organisms, including Mycoplasma, Candida parapsilosis, Brucella, and non-tuberculous mycobacteria (24, 61–64), among other rare pathogens. These bacteria demand specialized culture conditions and exhibit slow growth (65). In addition to these major pathogen groups, mNGS has detected even rarer organisms in PJI (26), such as Coxiella burnetii in culture-negative PJI samples where serological testing may initially yield negative results (32, 66). These discoveries significantly expand our understanding of the PJI pathogen spectrum, particularly in detecting rare and fastidious pathogens that conventional cultures often miss (59, 67). These microorganisms span diverse categories, including certain bacteria, fungi, viruses, and parasites (33, 68–71). The implementation of mNGS technology holds critical clinical value for elucidating PJI’s complex etiology, guiding targeted therapy, and improving patient outcomes.
6 Clinical applications of mNGS: enhancing diagnosis and guiding targeted therapy
6.1 Diagnostic advantages of mNGS
mNGS demonstrates significant time efficiency and superior anti-interference capability compared to culture methods, which typically require several days—or even over a week for certain rare pathogens—to yield results (72). In contrast, mNGS reduces the average detection time to 24–48 h (73, 74). Studies indicate (37, 75) that mNGS can complete testing and result interpretation within 48 h of specimen receipt. This temporal advantage is particularly critical for PJI patients requiring urgent treatment, as it mitigates the adverse effects and antimicrobial resistance risks associated with empirical broad-spectrum antibiotic use (31). Notably, mNGS exhibits enhanced diagnostic performance in patients with preoperative antibiotic exposure (24), attributable to its culture-independent nature, which enables detection of residual pathogen DNA post-antibiotic treatment (31, 37). Furthermore, mNGS maintains 92% sensitivity even in low-volume samples (e. g., 1 mL synovial fluid), whereas traditional culture sensitivity drops to 52% under these conditions (37). Approximately 25% of PJI cases show culture-negative results due to antibiotic pretreatment or rare pathogens, leading to significantly prolonged hospitalization compared to culture-positive cases (61). A multicenter retrospective study comparing targeted therapy based on mNGS results (TM group) versus empirical treatment (EM group) demonstrated that mNGS achieved significantly higher diagnostic positivity rates. The TM group exhibited faster postoperative declines in C-reactive protein (CRP) and erythrocyte sedimentation rate (ESR), with significantly shorter normalization times than the EM group (55). Although mNGS incurs costs, its precision diagnostics reduce the burden of erroneous treatments and improve clinical outcomes—such as avoiding unnecessary revision surgeries or prolonged antibiotic courses—justifying its value.
6.2 mNGS-guided treatment strategy optimization
By directly detecting microbial nucleic acid sequences in clinical samples without relying on pathogen cultivation, mNGS effectively identifies pathogens undetectable by conventional methods, thereby directly influencing clinical decision-making. In a case report (76), mNGS analysis of joint aspirate successfully identified Aggregatibacter aphrophilus nucleic acid sequences, with subsequent culture confirming its antibiotic susceptibility profile. For acute PJI, pathogen identification via mNGS achieved a 91. 7% success rate in debridement-antibiotics-implant retention (DAIR) procedures (77). Furthermore, a study of patients with unexplained ESR/CRP elevation revealed that while bacterial cultures were negative, mNGS detected bacterial infections in 63. 63% of cases, guiding postoperative antibiotic management and achieving 100% infection-free survival (78). mNGS enables early detection of resistance genes to optimize antibiotic selection and guide combination therapy (26, 79), a capability absent in traditional methods. For instance, polymicrobial infections may require broad-spectrum regimens covering aerobic, anaerobic, and atypical pathogens rather than single antibiotics (80, 81). Research demonstrates (57) that early mNGS-based diagnosis allowed Mycoplasma PJI cases to be cured with antibiotics alone, avoiding surgery. mNGS-guided therapy improved the 2-year cure rate of sinus tract-associated PJI from 55. 6% to 94. 4%, while reducing antibiotic duration by 11. 5 days (58). Although mNGS unit costs exceed culture, its 22% reduction in treatment modifications may lower overall healthcare expenditures (79, 82). These findings confirm mNGS enhances clinical precision by reducing empirical antibiotic misuse and unnecessary surgeries.
6.3 Limitations of mNGS and corresponding mitigation strategies
While mNGS technology has significantly enhanced pathogen detection capabilities in PJI—particularly demonstrating irreplaceable value for polymicrobial infections, rare pathogens, and culture-negative cases—its clinical interpretation remains controversial. Key challenges include host DNA interference, ambiguous criteria for false-positive/negative determinations, high costs, and clinical decision-making dilemmas when results conflict with culture findings. Multicenter studies and large-scale clinical validation are urgently needed to clarify its optimal applications and refine implementation pathways.
6.4 Technical bottlenecks: sensitivity and specificity limitations
Host DNA depletion, a critical step in enhancing metagenomic sequencing efficiency, requires careful selection based on sample type, characteristics of target microorganisms, and specific application scenarios. Physicochemical methods significantly improve the signal-to-noise ratio (53, 83), while molecular biology-based amplification (84) and bioinformatic filtering (85, 86) offer complementary solutions for low-biomass samples. Furthermore, threshold setting remains highly contentious: some studies propose >10 unique reads as a positive criterion, yet this may lead to under-detection of low-biomass pathogens and over-interpretation of high-abundance commensals (34, 87, 88). Future development should focus on creating more universal host cell lysis reagents, optimizing multiplex molecular enrichment strategies, and establishing standardized performance evaluation frameworks. With continuous technological refinement, the application prospects of mNGS in precision diagnosis and environmental monitoring will broaden considerably.
6.5 Interpretation controversies of mNGS results
Pathogens detected by mNGS require clinical correlation to determine their pathogenicity. Current studies primarily evaluate mNGS using fresh PJI samples, typically including synovial fluid, sonicate fluid from prostheses, or tissue (50). Performance varies significantly across sample types, with issues like false positives/negatives observed (28), though systematic comparisons remain lacking. Additionally, detection of multiple pathogens may obscure true causative organisms, complicating interpretation (89, 90). For instance, while mNGS improves polymicrobial detection in immunocompromised patients (91), distinguishing colonization from true infection becomes more challenging. One study reported 47. 1% of mNGS-positive cases were clinically confirmed as colonization (92). For such discrepancies, multiplex PCR may serve as a rapid screening tool (79, 93). A fundamental limitation of mNGS is its inability to differentiate live/dead bacteria. This could be addressed by integrating microbial viability markers (e. g., Precision Run-On Sequencing, PRO-seq) to assess translational activity (94, 95), potentially complementing mNGS in future diagnostic frameworks. While mNGS serves as a valuable adjunct to conventional methods, its inherent limitations necessitate comprehensive clinical contextualization for accurate result interpretation.
6.6 Economic and temporal constraints in mNGS implementation
Although mNGS has shortened the turnaround time for some diagnostics (84), its high cost limits routine clinical application, particularly in resource-limited healthcare settings (29). The substantial expenses associated with mNGS primarily arise from sample preparation (96), the sequencing process (79), and the complexity of bioinformatic analysis (25). These technical steps require specialized equipment and trained personnel, contributing to both direct financial burden and time investment (54, 97). Studies indicate (79, 82) that the cost of a single mNGS test is approximately $260, compared to less than $50 for culture. However, evidence regarding the balance between direct detection expenses and indirect savings—such as reduced reoperation rates—remains insufficient, underscoring the need for further research to quantify the overall cost-effectiveness of mNGS.
6.7 Absence of unified standards for mNGS implementation
Current mNGS lacks standardized experimental protocols and bioinformatics analysis criteria. Significant variations exist across centers in sample collection volumes for synovial fluid, tissue, or prosthesis sonicate fluid (27, 31, 36), potentially due to differences in DNA extraction methods or sequencing platforms (97). Since mNGS can detect microbial contaminants from samples, processing reagents, or laboratory environments, result interpretation becomes complicated. Even biopsies from theoretically sterile sites may be inadvertently contaminated during routine clinical sampling. Therefore, strict adherence to quality control procedures for reagents and workflows is essential to maintain an aseptic, nucleic acid-free testing environment and prevent cross-contamination that could lead to false-positive results (28, 98, 99). Furthermore, bioinformatics analysis remains unstandardized. For instance, resistance gene detection matched phenotypic resistance in only 37. 5% of culture-positive PJIs (79, 100), with insufficient accuracy limiting its therapeutic guidance value (32, 101). While international consensus meetings recommend incorporating mNGS as an adjunct criterion in MSIS diagnostic standards, most supporting evidence remains level II-III (28, 39). Future efforts should establish expert consortiums to develop standardized mNGS operational guidelines for PJI.
7 Discussion
mNGS is reshaping the diagnostic paradigm for PJI, with its core value lying in overcoming the limitations of traditional culture methods to achieve rapid and comprehensive pathogen identification. However, its clinical application remains constrained by issues such as technical performance (inability to determine whether detected sequences originate from live or dead pathogens), cost-effectiveness, and lack of standardization (absence of standardized procedures and criteria to prevent nucleic acid contamination from specimen collection through processing and environmental exposure). At this stage, the technology cannot yet fully replace conventional microbial culture. Targeted sequencing (tNGS) and nanopore sequencing (mONS) represent important directions for the development of mNGS. tNGS offers lower costs, shorter turnaround times, and the ability to simultaneously detect resistance genes. Nanopore sequencing enables real-time pathogen identification and resistance analysis, although the issue of host DNA interference remains to be resolved. Furthermore, the combination of multiplex PCR (mPCR) with mNGS can shorten pathogen detection time, making it suitable for rapid screening. Currently, mNGS should serve as a complementary approach to traditional culture, with a focus on cases of culture-negative PJI (CN-PJI), patients with preoperative antibiotic exposure, and suspected polymicrobial infections. The establishment of standardized protocols is a core prerequisite for its broader clinical adoption. Future research should focus on three major aspects: first, developing multicenter standardized testing protocols based on sonicate fluid from explanted prostheses to address operational heterogeneity; second, creating efficient host DNA depletion techniques to improve pathogen nucleic acid capture efficiency; and third, advancing clinical validation of mNGS-guided personalized antibiotic therapy and exploring its translational potential in resistance prediction. Optimizing sample processing, establishing tiered reporting standards, and integrating multimodal diagnostic results are essential to balance detection breadth with clinical utility. Dedicated to establishing a more unified definition and diagnostic criteria for PJI, through the accumulation of high-quality evidence from multicenter studies and large-sample validation can mNGS be integrated into the standard PJI diagnosis and treatment framework. This integration will ultimately achieve the goals of improving patient outcomes, curbing antibiotic misuse, and establishing a closed-loop mNGS-driven system for rapid diagnosis and targeted therapy in PJI. mNGS holds promise to advance from an auxiliary diagnostic tool to a new gold standard for pathogen diagnosis in PJI.
Author contributions
HH: Investigation, Methodology, Project administration, Writing – original draft, Writing – review & editing. YT: Data curation, Formal analysis, Investigation, Writing – review & editing. XH: Methodology, Project administration, Supervision, Writing – review & editing. F-kL: Project administration, Supervision, Writing – review & editing. RC: Conceptualization, Investigation, Methodology, Project administration, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Plate, JF, Zuskov, A, and Seyler, TM. Use of adjunct antiseptic agents in periprosthetic joint infections. J Am Acad Orthop Surg. (2021) 29:e1151–8. doi: 10.5435/JAAOS-D-21-00154
2. Gallo, J, and Nieslanikova, E. Prevention of prosthetic joint infection: from traditional approaches towards quality improvement and data mining. J Clin Med. (2020) 9:2190. doi: 10.3390/jcm9072190
3. Natsuhara, KM, Shelton, TJ, Meehan, JP, and Lum, ZC. Mortality during total hip periprosthetic joint infection. J Arthroplast. (2019) 34:S337–s42. doi: 10.1016/j.arth.2018.12.024
4. Fischbacher, A, and Borens, O. Prosthetic-joint infections: mortality over the last 10 years. J Bone Joint Infect. (2019) 4:198–202. doi: 10.7150/jbji.35428
5. Kennedy, IW, Haddad, FS, Abdel, MP, Garbuz, DS, Gant, V, Meek, RMD, et al. Periprosthetic joint infection: development of a core outcome set. Bone Joint J. (2025) 107-b:455–60. doi: 10.1302/0301-620X.107B4.BJJ-2024-1727
6. Puhto, T, Puhto, AP, Vielma, M, and Syrjälä, H. Infection triples the cost of a primary joint arthroplasty. Infect Dis. (2019) 51:348–55. doi: 10.1080/23744235.2019.1572219
7. Musil, D, Šnorek, M, Gallo, J, Jahoda, D, and Stehlík, J. Economic analysis of the costs of hospital stay of patients with infection as a complication of Total replacements - part 2: Total hip arthroplasty. Acta Chir Orthop Traumatol Cechoslov. (2019) 86:241–8. doi: 10.55095/achot2019/041
8. Yin, JM, Liu, ZT, Zhao, SC, and Guo, YJ. Diagnosis, management, and prevention of prosthetic joint infections. Front Biosci. (2013) 18:1349–57. doi: 10.2741/4184
9. Hotchen, AJ, Wismayer, MG, Robertson-Waters, E, McDonnell, SM, Kendrick, B, Taylor, A, et al. The joint-specific BACH classification: a predictor of outcome in prosthetic joint infection. EClinicalMedicine. (2021) 42:101192. doi: 10.1016/j.eclinm.2021.101192
10. Unsworth, A, Young, B, Scarborough, M, and McNally, M. A comparison of causative pathogens in bone and prosthetic joint infections: implications for antimicrobial therapy. Antibiotics (Basel, Switzerland). (2024) 13:1125. doi: 10.3390/antibiotics13121125
11. Papalini, C, Pucci, G, Cenci, G, Mencacci, A, Francisci, D, Caraffa, A, et al. Prosthetic joint infection diagnosis applying the three-level European bone and joint infection society (EBJIS) approach. Eur J Clin Microbiol Infect Dis. (2022) 41:771–8. doi: 10.1007/s10096-022-04410-x
12. Ribeiro, TC, Honda, EK, Daniachi, D, Cury, RPL, da Silva, CB, Klautau, GB, et al. The impact of sonication cultures when the diagnosis of prosthetic joint infection is inconclusive. PLoS One. (2021) 16:e0252322. doi: 10.1371/journal.pone.0252322
13. Velasquez, AXB, Klautau, GB, Kurihara, MNL, Santos, INM, Campos, LB, Silva, MM, et al. Improving the microbiological diagnosis of fracture-related infection and prosthetic joint infection through culturing sonication fluid in Bactec blood culture bottles. Brazilian J Microbiol. (2024) 55:3591–601. doi: 10.1007/s42770-024-01545-1
14. Frangiamore, SJ, Siqueira, MB, Saleh, A, Daly, T, Higuera, CA, and Barsoum, WK. Synovial cytokines and the MSIS criteria are not useful for determining infection resolution after periprosthetic joint infection explantation. Clin Orthop Relat Res. (2016) 474:1630–9. doi: 10.1007/s11999-016-4710-x
15. Kabore, C, Reda, M, Mahieu, X, and Thirion, T. The MusculoSkeletal infection society diagnostic criteria are insufficient to diagnose shoulder Periprosthetic infection: a retrospective study case and literature review. Acta Orthop Belg. (2025) 91:45–50. doi: 10.52628/91.1.11390
16. Streck, LE, Sterneder, CM, Haralambiev, L, Brenneis, M, Chiu, YF, and Boettner, F. Significant differences in the rate of periprosthetic joint infections in revision hip and knee arthroplasty depending on the applied definition. Arch Orthop Trauma Surg. (2025) 145:441. doi: 10.1007/s00402-025-05994-7
17. Boelch, SP, Rüeckl, K, Streck, LE, Szewczykowski, V, Weißenberger, M, Jakuscheit, A, et al. Diagnosis of chronic infection at total hip arthroplasty revision is a question of definition. Biomed Res Int. (2021) 2021:8442435. doi: 10.1155/2021/8442435
18. Goswami, K, Parvizi, J, and Maxwell Courtney, P. Current recommendations for the diagnosis of acute and chronic PJI for hip and knee-cell counts, alpha-defensin, leukocyte esterase, next-generation sequencing. Curr Rev Musculoskelet Med. (2018) 11:428–38. doi: 10.1007/s12178-018-9513-0
19. Chisari, E, and Parvizi, J. Accuracy of blood-tests and synovial fluid-tests in the diagnosis of periprosthetic joint infections. Expert Rev Anti-Infect Ther. (2020) 18:1135–42. doi: 10.1080/14787210.2020.1792771
20. Parikh, MS, and Antony, S. A comprehensive review of the diagnosis and management of prosthetic joint infections in the absence of positive cultures. J Infect Public Health. (2016) 9:545–56. doi: 10.1016/j.jiph.2015.12.001
21. Lenguerrand, E, Whitehouse, MR, Beswick, AD, Jones, SA, Porter, ML, and Blom, AW. Revision for prosthetic joint infection following hip arthroplasty: evidence from the National Joint Registry. Bone Joint Res. (2017) 6:391–8. doi: 10.1302/2046-3758.66.BJR-2017-0003.R1
22. Foddai, ACG, and Grant, IR. Methods for detection of viable foodborne pathogens: current state-of-art and future prospects. Appl Microbiol Biotechnol. (2020) 104:4281–8. doi: 10.1007/s00253-020-10542-x
23. Hong, HL, Flurin, L, Thoendel, MJ, Wolf, MJ, Abdel, MP, Greenwood-Quaintance, KE, et al. Targeted versus shotgun metagenomic sequencing-based detection of microorganisms in sonicate fluid for Periprosthetic joint infection diagnosis. Clin Infect Dis. (2023) 76:e1456–62. doi: 10.1093/cid/ciac646
24. Yu, Y, Wang, S, Dong, G, and Niu, Y. Diagnostic performance of metagenomic next-generation sequencing in the diagnosis of prosthetic joint infection using tissue specimens. Infection and drug resistance. (2023) 16:1193–201. doi: 10.2147/IDR.S397260
25. Zhao, Y, Zhang, W, and Zhang, X. Application of metagenomic next-generation sequencing in the diagnosis of infectious diseases. Front Cell Infect Microbiol. (2024) 14:1458316. doi: 10.3389/fcimb.2024.1458316
26. Huang, C, Ding, H, Lin, Y, Zhang, Z, Fang, X, Chen, Y, et al. Diagnosis of Coxiella burnetii prosthetic joint infection using mNGS and ptNGS: a case report and literature review. Orthop Surg. (2023) 15:371–6. doi: 10.1111/os.13600
27. Tan, J, Wu, L, Zhan, L, Sheng, M, Tang, Z, Xu, J, et al. Optimal selection of specimens for metagenomic next-generation sequencing in diagnosing periprosthetic joint infections. Front Cell Infect Microbiol. (2024) 14:1356804. doi: 10.3389/fcimb.2024.1356804
28. Fida, M, and Tande, AJ. State-of-the-art metagenomic sequencing and its role in the diagnosis of Periprosthetic joint infections. Infect Dis Clin N Am. (2024) 38:813–25. doi: 10.1016/j.idc.2024.07.011
29. Gamie, Z, Karthikappallil, D, Gamie, E, Stamiris, S, Kenanidis, E, and Tsiridis, E. Molecular sequencing technologies in the diagnosis and management of prosthetic joint infections. Expert Rev Mol Diagn. (2022) 22:603–24. doi: 10.1080/14737159.2021.1894929
30. Hao, L, Bian, W, Qing, Z, Ma, T, Li, H, Xu, P, et al. Will previous antimicrobial therapy reduce the positivity rate of metagenomic next-generation sequencing in periprosthetic joint infections? A clinical study. Front Cell Infect Microbiol. (2023) 13:1295962. doi: 10.3389/fcimb.2023.1295962
31. Zhang, C, Fang, X, Huang, Z, Li, W, Zhang, CF, Yang, B, et al. Value of mNGS in sonication fluid for the diagnosis of periprosthetic joint infection. Arthroplasty (London, England). (2019) 1:9. doi: 10.1186/s42836-019-0006-4
32. Shi, T, Chen, H, Liu, Y, Wu, Y, and Lin, F. Clinical applications of metagenomic next-generation sequencing in the identification of pathogens in periprosthetic joint infections: a retrospective study. J Orthop Surg Res. (2024) 19:301. doi: 10.1186/s13018-024-04745-5
33. Rodino, KG, and Simner, PJ. Status check: next-generation sequencing for infectious-disease diagnostics. J Clin Invest. (2024) 134:8003. doi: 10.1172/JCI178003
34. Huang, Z, Li, W, Lee, GC, Fang, X, Xing, L, Yang, B, et al. Metagenomic next-generation sequencing of synovial fluid demonstrates high accuracy in prosthetic joint infection diagnostics: mNGS for diagnosing PJI. Bone Joint Res. (2020) 9:440–9. doi: 10.1302/2046-3758.97.BJR-2019-0325.R2
35. Mei, J, Hu, H, Zhu, S, Ding, H, Huang, Z, Li, W, et al. Diagnostic role of mNGS in Polymicrobial periprosthetic joint infection. J Clin Med. (2023) 12:1838. doi: 10.3390/jcm12051838
36. He, R, Wang, Q, Wang, J, Tang, J, Shen, H, and Zhang, X. Better choice of the type of specimen used for untargeted metagenomic sequencing in the diagnosis of periprosthetic joint infections. Bone Joint J. (2021) 103-b:923–30. doi: 10.1302/0301-620X.103B5.BJJ-2020-0745.R1
37. Fang, X, Cai, Y, Shi, T, Huang, Z, Zhang, C, Li, W, et al. Detecting the presence of bacteria in low-volume preoperative aspirated synovial fluid by metagenomic next-generation sequencing. Int J Infect Dis. (2020) 99:108–16. doi: 10.1016/j.ijid.2020.07.039
38. Wang, CX, Huang, Z, Fang, X, Li, W, Yang, B, and Zhang, W. Comparison of broad-range polymerase chain reaction and metagenomic next-generation sequencing for the diagnosis of prosthetic joint infection. Int J Infect Dis. (2020) 95:8–12. doi: 10.1016/j.ijid.2020.03.055
39. Olearo, F, Zein, SE, Portillo, ME, Zapf, A, Rohde, H, Berbari, EF, et al. Diagnostic accuracy of 16S rDNA PCR, multiplex PCR and metagenomic next-generation sequencing in periprosthetic joint infections: a systematic review and meta-analysis. Clin Microbiol Infect. (2025) 31:1115–25. doi: 10.1016/j.cmi.2025.02.022
40. Lausmann, C, Kolle, KN, Citak, M, Abdelaziz, H, Schulmeyer, J, Delgado, GD, et al. How reliable is the next generation of multiplex-PCR for diagnosing prosthetic joint infection compared to the MSIS criteria? Still missing the ideal test. Hip Int. (2020) 30:72–7. doi: 10.1177/1120700020938576
41. Maekawa, AS, Santos, LS, Velho, P, and Drummond, MR. Use of a synthetic oligonucleotide to detect false positives caused by cross-contamination in nested PCR. J Microbiol Methods. (2024) 226:107040. doi: 10.1016/j.mimet.2024.107040
42. Leblanc, M, Berry, K, Graciano, S, Becker, B, and Reuter, JD. False-positive results after environmental pinworm PCR testing due to Rhabditid nematodes in corncob bedding. J Am Assoc Lab Anim Sci. (2014) 53:717–24.
43. Pal, N, Block, CC, and Gardner, CAC. A real-time PCR differentiating Pantoea stewartii subsp. stewartii from P. stewartii subsp. indologenes in corn seed. Plant Dis. (2019) 103:1474–86. doi: 10.1094/PDIS-06-18-0936-RE
44. Wang, H, Yang, H, Yang, J, Liu, X, Xie, B, Xu, M, et al. Establishment of a 23S rRNA assay for Brucella and its application in evaluating bacterial growth status. Vet Res Commun. (2025) 49:110. doi: 10.1007/s11259-025-10676-1
45. Bouam, A, Vincent, JJ, Drancourt, M, Raoult, D, and Levy, PY. Preventing contamination of PCR-based multiplex assays including the use of a dedicated biosafety cabinet. Lett Appl Microbiol. (2021) 72:98–103. doi: 10.1111/lam.13375
46. Peeters, B, Herijgers, P, Beuselinck, K, Peetermans, WE, Herregods, MC, Desmet, S, et al. Comparison of PCR-electrospray ionization mass spectrometry with 16S rRNA PCR and amplicon sequencing for detection of Bacteria in excised heart valves. J Clin Microbiol. (2016) 54:2825–31. doi: 10.1128/JCM.01240-16
47. Park, K, Park, B, Won, EJ, Sung, H, and Kim, MN. False-positive results for seasonal coronavirus infections on using the FilmArray pneumonia panel. Microbiol Spect. (2024) 12:e0134324. doi: 10.1128/spectrum.01343-24
48. Navarro-Pérez, D, García-Oreja, S, Tardáguila-García, A, León-Herce, D, Álvaro-Afonso, FJ, and Lázaro-Martínez, JL. Microbiological culture combined with PCR for the diagnosis of onychomycosis: descriptive analysis of 121 patients. Mycoses. (2023) 66:1045–9. doi: 10.1111/myc.13648
49. Chiu, CY. Viral pathogen discovery. Curr Opin Microbiol. (2013) 16:468–78. doi: 10.1016/j.mib.2013.05.001
50. Wang, CX, Huang, Z, Fang, W, Zhang, Z, Fang, X, Li, W, et al. Preliminary assessment of nanopore-based metagenomic sequencing for the diagnosis of prosthetic joint infection. Int J Infect Dis. (2020) 97:54–9. doi: 10.1016/j.ijid.2020.05.044
51. Gu, W, Crawford, ED, O'Donovan, BD, Wilson, MR, Chow, ED, Retallack, H, et al. Depletion of abundant sequences by hybridization (DASH): using Cas9 to remove unwanted high-abundance species in sequencing libraries and molecular counting applications. Genome Biol. (2016) 17:41. doi: 10.1186/s13059-016-0904-5
52. Hasan, MR, Rawat, A, Tang, P, Jithesh, PV, Thomas, E, Tan, R, et al. Depletion of human DNA in spiked clinical specimens for improvement of sensitivity of pathogen detection by next-generation sequencing. J Clin Microbiol. (2016) 54:919–27. doi: 10.1128/JCM.03050-15
53. Ji, XC, Zhou, LF, Li, CY, Shi, YJ, Wu, ML, Zhang, Y, et al. Reduction of human DNA contamination in clinical cerebrospinal fluid specimens improves the sensitivity of metagenomic next-generation sequencing. J Mol Neurosci. (2020) 70:659–66. doi: 10.1007/s12031-019-01472-z
54. He, Y, Fang, K, Shi, X, Yang, D, Zhao, L, Yu, W, et al. Enhanced DNA and RNA pathogen detection via metagenomic sequencing in patients with pneumonia. J Transl Med. (2022) 20:195. doi: 10.1186/s12967-022-03397-5
55. Zhang, G, Zhang, H, Hu, X, Xu, D, Tang, B, Tang, M, et al. Clinical application value of metagenomic next-generation sequencing in the diagnosis of spinal infections and its impact on clinical outcomes. Front Cell Infect Microbiol. (2023) 13:1076525. doi: 10.3389/fcimb.2023.1076525
56. Zhou, Z, Song, Y, Yan, Y, and Zheng, Y. Metagenomic next-generation sequencing improves the diagnosis efficiency of mixed Periprosthetic joint infections. Infect Drug Resist. (2025) 18:2165–74. doi: 10.2147/IDR.S516650
57. Lin, Z, Chen, Y, Yu, Z, Zhang, Z, Lin, Y, Zhang, W, et al. Early and accurate pathogen identification based on mNGS: key to timely therapy for Mycoplasma prosthetic joint infection. Orthop Surg. (2025) 17:1995–2003. doi: 10.1111/os.70069
58. Yuan, X, Wang, Q, Ding, H, Li, H, Fang, X, Zhang, W, et al. The role of applying metagenomic next-generation sequencing (mNGS) in Periprosthetic joint infection with sinus tract: a retrospective study. Int J Gen Med. (2025) 18:3787–95. doi: 10.2147/IJGM.S531444
59. Huang, Z, Zhang, C, Li, W, Fang, X, Wang, Q, Xing, L, et al. Metagenomic next-generation sequencing contribution in identifying prosthetic joint infection due to Parvimonas micra: a case report. J Bone Joint Inf. (2019) 4:50–5. doi: 10.7150/jbji.30615
60. Wang, X, Guo, X, Liu, H, Wang, B, Wu, J, Chen, S, et al. Augmented pathogen detection in brain abscess using metagenomic next-generation sequencing: a retrospective cohort study. Microbiol Spect. (2024) 12:e0032524. doi: 10.1128/spectrum.00325-24
61. Lin, L, Li, J, Zhang, C, Li, J, Wu, B, Huang, Z, et al. Comprehensive analysis of culture-negative periprosthetic joint infection with metagenomic next-generation sequencing. Front Cell Infect Microbiol. (2025) 15:1564488. doi: 10.3389/fcimb.2025.1564488
62. Auñon, A, Salar-Vidal, L, Mahillo-Fernandez, I, Almeida, F, Pereira, P, Lora-Tamayo, J, et al. Prosthetic joint infections caused by Mycobacterium tuberculosis complex-an ESGIAI-ESGMYC multicenter, retrospective study and literature review. Microorganisms. (2024) 12:849. doi: 10.3390/microorganisms12050849
63. Chang, Y, Jiang, K, Zhang, L, Yang, F, and Huang, J. Application of next-generation sequencing technology in the detection of pathogenic bacteria of the periprosthetic joint infection after arthroplasty. Int Wound J. (2023) 20:2121–8. doi: 10.1111/iwj.14087
64. Arkun, R, and Mete, BD. Musculoskeletal brucellosis. Semin Musculoskelet Radiol. (2011) 15:470–9. doi: 10.1055/s-0031-1293493
65. Liu, H, Li, Q, Ouyang, X, Li, Q, Min, Y, and Dai, L. Diagnosis of early neurobrucellosis using metagenomic next-generation sequencing of the cerebrospinal fluid in nonepidemic zone: case report and lecture review. Medicine. (2025) 104:e41481. doi: 10.1097/MD.0000000000041481
66. Tissot-Dupont, H, and Raoult, D. Q fever. Infect Dis Clin N Am. (2008) 22:505–14, ix. doi: 10.1016/j.idc.2008.03.002
67. Chen, X, Zhu, J, Liu, Z, Ye, J, Yang, L, and Zhang, Z. Mixed infection of three nontuberculous mycobacteria species identified by metagenomic next-generation sequencing in a patient with peritoneal dialysis-associated peritonitis: a rare case report and literature review. BMC Nephrol. (2023) 24:95. doi: 10.1186/s12882-023-03156-8
68. Shi, P, Liu, J, Liang, A, Zhu, W, Fu, J, Wu, X, et al. Application of metagenomic next-generation sequencing in optimizing the diagnosis of ascitic infection in patients with liver cirrhosis. BMC Infect Dis. (2024) 24:503. doi: 10.1186/s12879-024-09396-9
69. Lin, Q, Yao, Y, Li, X, Zhang, S, Guo, H, Ma, X, et al. The application of nanopore targeted sequencing for pathogen diagnosis in bronchoalveolar lavage fluid of patients with pneumonia: a prospective multicenter study. Infect Dis. (2024) 56:128–37. doi: 10.1080/23744235.2023.2276785
70. Chien, JY, Yu, CJ, and Hsueh, PR. Utility of metagenomic next-generation sequencing for etiological diagnosis of patients with Sepsis in intensive care units. Microbiol Spect. (2022) 10:e0074622. doi: 10.1128/spectrum.00746-22
71. Carrega, G, Cavagnaro, L, Basso, M, Riccio, G, Ronca, A, Salomone, C, et al. Azole-resistant Candida albicans prosthetic joint infection treated with prolonged administration of anidulafungin and two-stage exchange with implant of a mega-prosthesis. J Chemother. (2017) 29:386–8. doi: 10.1080/1120009X.2016.1199409
72. Mu, S, Hu, L, Zhang, Y, Liu, Y, Cui, X, Zou, X, et al. Prospective evaluation of a rapid clinical metagenomics test for bacterial pneumonia. Front Cell Infect Microbiol. (2021) 11:684965. doi: 10.3389/fcimb.2021.684965
73. Li, N, Cai, Q, Miao, Q, Song, Z, Fang, Y, and Hu, B. High-throughput metagenomics for identification of pathogens in the clinical settings. Small Methods. (2021) 5:2000792. doi: 10.1002/smtd.202000792
74. Wu, H, Cao, H, Gao, X, Shi, C, Wang, L, and Gao, B. The role of metagenomic next-generation sequencing in diagnosing and managing post-kidney transplantation infections. Front Cell Infect Microbiol. (2024) 14:1473068. doi: 10.3389/fcimb.2024.1473068
75. Han, D, Li, Z, Li, R, Tan, P, Zhang, R, and Li, J. mNGS in clinical microbiology laboratories: on the road to maturity. Crit Rev Microbiol. (2019) 45:668–85. doi: 10.1080/1040841X.2019.1681933
76. Cai, YQ, Fang, XY, Huang, ZD, Zhang, CF, Li, WB, Yang, B, et al. Rare Aggregatibacter aphrophilus infection after total hip arthroplasty: a case report and literature review. Chin J Infect Control. (2021) 20:269–72.
77. Zhang, CF, He, L, Fang, XY, Huang, ZD, Bai, GC, Li, WB, et al. Debridement, antibiotics, and implant retention for acute Periprosthetic joint infection. Orthop Surg. (2020) 12:463–70. doi: 10.1111/os.12641
78. Zhang, Q, Sun, X, Pan, JB, and Kong, G. Feasibility of mNGS in joint replacement patients exhibiting elevated ESR and CRP levels without an underlying diagnosis. Eur J Med Res. (2024) 29:515. doi: 10.1186/s40001-024-02118-6
79. Huang, C, Huang, Y, Wang, Z, Lin, Y, Li, Y, Chen, Y, et al. Multiplex PCR-based next generation sequencing as a novel, targeted and accurate molecular approach for periprosthetic joint infection diagnosis. Front Microbiol. (2023) 14:1181348. doi: 10.3389/fmicb.2023.1181348
80. Lau, A, Nguyen, N, Hui, A, Ong, J, and Salehpour, M. Fournier's gangrene with Streptococcus anginosus in the setting of hidrandenitis suppurativa perineal abscess: a case report. Int J Surg Case Rep. (2024) 116:109319. doi: 10.1016/j.ijscr.2024.109319
81. Jones, RN. Clinical use of beta-lactamase inhibitors in combination with extended-spectrum penicillins. Am J Health Syst Pharm. (1995) 52:S29–33. doi: 10.1093/ajhp/52.6_Suppl_2.S29
82. Tan, J, Liu, Y, Ehnert, S, Nüssler, AK, Yu, Y, Xu, J, et al. The effectiveness of metagenomic next-generation sequencing in the diagnosis of prosthetic joint infection: a systematic review and Meta-analysis. Front Cell Infect Microbiol. (2022) 12:875822. doi: 10.3389/fcimb.2022.875822
83. Kok, NA, Peker, N, Schuele, L, de Beer, JL, Rossen, JWA, Sinha, B, et al. Host DNA depletion can increase the sensitivity of Mycobacterium spp. detection through shotgun metagenomics in sputum. Front Microbiol. (2022) 13:949328. doi: 10.3389/fmicb.2022.949328
84. Thurlow, CM, Joseph, SJ, Ganova-Raeva, L, Katz, SS, Pereira, L, Chen, C, et al. Selective whole-genome amplification as a tool to enrich specimens with low Treponema pallidum genomic DNA copies for whole-genome sequencing. mSphere. (2022) 7:e0000922. doi: 10.1128/msphere.00009-22
85. Guccione, C, Patel, L, Tomofuji, Y, McDonald, D, Gonzalez, A, Sepich-Poore, GD, et al. Incomplete human reference genomes can drive false sex biases and expose patient-identifying information in metagenomic data. Nat Commun. (2025) 16:825. doi: 10.1038/s41467-025-56077-5
86. Babalola, OO, Adebayo, AA, and Enagbonma, BJ. Shotgun metagenomics dataset of the core rhizo-microbiome of monoculture and soybean-precedent carrot. BMC Genomic Data. (2025) 26:26. doi: 10.1186/s12863-025-01320-7
87. Cai, Y, Ding, H, Chen, X, Chen, Y, Huang, C, Zhang, C, et al. Optimization and standardization of mNGS-based procedures for the diagnosis of Mycoplasma periprosthetic joint infection: a novel diagnostic strategy for rare bacterial periprosthetic joint infection. Front Cell Infect Microbiol. (2023) 13:1089919. doi: 10.3389/fcimb.2023.1089919
88. Ebinger, A, Fischer, S, and Höper, D. A theoretical and generalized approach for the assessment of the sample-specific limit of detection for clinical metagenomics. Comput Struct Biotechnol J. (2021) 19:732–42. doi: 10.1016/j.csbj.2020.12.040
89. Long, Y, Xia, X, Feng, H, and Zhao, P. Improving pulmonary infection diagnosis with metagenomic next-generation sequencing of bronchoalveolar lavage fluid. J Med Microbiol. (2024) 73:1808. doi: 10.1099/jmm.0.001808
90. Dong, Y, Chen, Q, Tian, B, Li, J, Li, J, and Hu, Z. Advancing microbe detection for lower respiratory tract infection diagnosis and management with metagenomic next-generation sequencing. Infect Drug Resist. (2023) 16:677–94. doi: 10.2147/IDR.S387134
91. Wang, C, Yin, X, Ma, W, Zhao, L, Wu, X, Ma, N, et al. Clinical application of bronchoalveolar lavage fluid metagenomics next-generation sequencing in cancer patients with severe pneumonia. Respir Res. (2024) 25:68. doi: 10.1186/s12931-023-02654-5
92. Lai, Y, Chen, B, Chen, S, and Shen, Y. Experience of implementing metagenomic next-generation sequencing in patients with suspected pulmonary infection in clinical practice. Sci Rep. (2025) 15:9579. doi: 10.1038/s41598-025-94840-2
93. Ghirardelli, S, Scaggiante, F, Troi, C, Valpiana, P, Cristofolini, G, Aloisi, G, et al. Multiplex PCR in septic arthritis and periprosthetic joint infections microorganism identification: results from the application of a new molecular testing diagnostic algorithm. J Experimental Orthopaedics. (2024) 11:e12097. doi: 10.1002/jeo2.12097
94. Vill, AC, Rice, EJ, De Vlaminck, I, Danko, CG, and Brito, IL. Precision run-on sequencing (PRO-seq) for microbiome transcriptomics. Nat Microbiol. (2024) 9:241–50. doi: 10.1038/s41564-023-01558-w
95. Liu, L, Zhao, Y, Hassett, R, Toneyan, S, Koo, PK, and Siepel, A. Probabilistic and machine-learning methods for predicting local rates of transcription elongation from nascent RNA sequencing data. Nucleic Acids Res. (2025) 53:gkaf092. doi: 10.1093/nar/gkaf092
96. Liu, L, Hakhverdyan, M, Wallgren, P, Vanneste, K, Fu, Q, Lucas, P, et al. An interlaboratory proficiency test using metagenomic sequencing as a diagnostic tool for the detection of RNA viruses in swine fecal material. Microbiol Spect. (2024) 12:e0420823. doi: 10.1128/spectrum.04208-23
97. Zhao, M, Zhang, Y, Chen, L, Yan, X, Xu, T, Fu, M, et al. Nanopore sequencing of infectious fluid is a promising supplement for gold-standard culture in real-world clinical scenario. Front Cell Infect Microbiol. (2024) 14:1330788. doi: 10.3389/fcimb.2024.1330788
98. Wilson, MR, O'Donovan, BD, Gelfand, JM, Sample, HA, Chow, FC, Betjemann, JP, et al. Chronic meningitis investigated via metagenomic next-generation sequencing. JAMA Neurol. (2018) 75:947–55. doi: 10.1001/jamaneurol.2018.0463
99. Schlaberg, R, Chiu, CY, Miller, S, Procop, GW, and Weinstock, G. Validation of metagenomic next-generation sequencing tests for universal pathogen detection. Arch Pathol Lab Med. (2017) 141:776–86. doi: 10.5858/arpa.2016-0539-RA
100. Indelli, PF, Ghirardelli, S, Violante, B, and Amanatullah, DF. Next generation sequencing for pathogen detection in periprosthetic joint infections. EFORT open reviews. (2021) 6:236–44. doi: 10.1302/2058-5241.6.200099
101. Hu, B, Tao, Y, Shao, Z, Zheng, Y, Zhang, R, Yang, X, et al. A comparison of blood pathogen detection among droplet digital PCR, metagenomic next-generation sequencing, and blood culture in critically ill patients with suspected bloodstream infections. Front Microbiol. (2021) 12:641202. doi: 10.3389/fmicb.2021.641202
102. Ivy, MI, Thoendel, MJ, Jeraldo, PR, Greenwood-Quaintance, KE, Hanssen, AD, Abdel, MP, et al. Direct detection and identification of prosthetic joint infection pathogens in synovial fluid by metagenomic shotgun sequencing. J Clin Microbiol. (2018) 56:e00402-18. doi: 10.1128/JCM.00402-18
Keywords: mNGS, PJI, infection, culture-negative, diagnostic accuracy
Citation: Huang H, Tong Y, Hu X, Liao F-k and Chen R (2025) The application value and challenges of metagenomic next-generation sequencing in the diagnosis of periprosthetic joint infection after arthroplasty. Front. Med. 12:1686503. doi: 10.3389/fmed.2025.1686503
Edited by:
Shilong Su, Peking University Third Hospital, ChinaReviewed by:
Changyu Huang, Quanzhou Orthopedic-Traumatological Hospital, ChinaPaul Cool, Keele University Faculty of Medicine and Health Sciences, United Kingdom
Copyright © 2025 Huang, Tong, Hu, Liao and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rijiang Chen, bHlzeGQwNkAxNjMuY29t
 Huahui Huang
Huahui Huang Yan Tong
Yan Tong Xiunian Hu
Xiunian Hu Fa-ke Liao
Fa-ke Liao Rijiang Chen
Rijiang Chen