- 1Cell and Molecular Biology Laboratory, Department of Zoology (DST-FIST Sponsored), Soban Singh Jeena University Campus, Almora, India
- 2Department of Biotechnology, Sir J C Bose Technical Campus, Kumaun University, Bhimtal, India
- 3Department of Zoology, Kumaun University, Nainital, India
- 4Department of Agricultural and Biological Engineering, PurdueUniversity, West Lafayette, IN, United States
Soil naturally comprises heavy metals but due to the rapid industrialization and anthropogenic events such as uncontrolled use of agrochemicals their concentration is heightened up to a large extent across the world. Heavy metals are non-biodegradable and persistent in nature thereby disrupting the environment and causing huge health threats to humans. Exploiting microorganisms for the removal of heavy metal is a promising approach to combat these adverse consequences. The microbial remediation is very crucial to prevent the leaching of heavy metal or mobilization into the ecosystem, as well as to make heavy metal extraction simpler. In this scenario, technological breakthroughs in microbes-based heavy metals have pushed bioremediation as a promising alternative to standard approaches. So, to counteract the deleterious effects of these toxic metals, some microorganisms have evolved different mechanisms of detoxification. This review aims to scrutinize the routes that are responsible for the heavy metal(loid)s contamination of agricultural land, provides a vital assessment of microorganism bioremediation capability. We have summarized various processes of heavy metal bioremediation, such as biosorption, bioleaching, biomineralization, biotransformation, and intracellular accumulation, as well as the use of genetically modified microbes and immobilized microbial cells for heavy metal removal.
Introduction
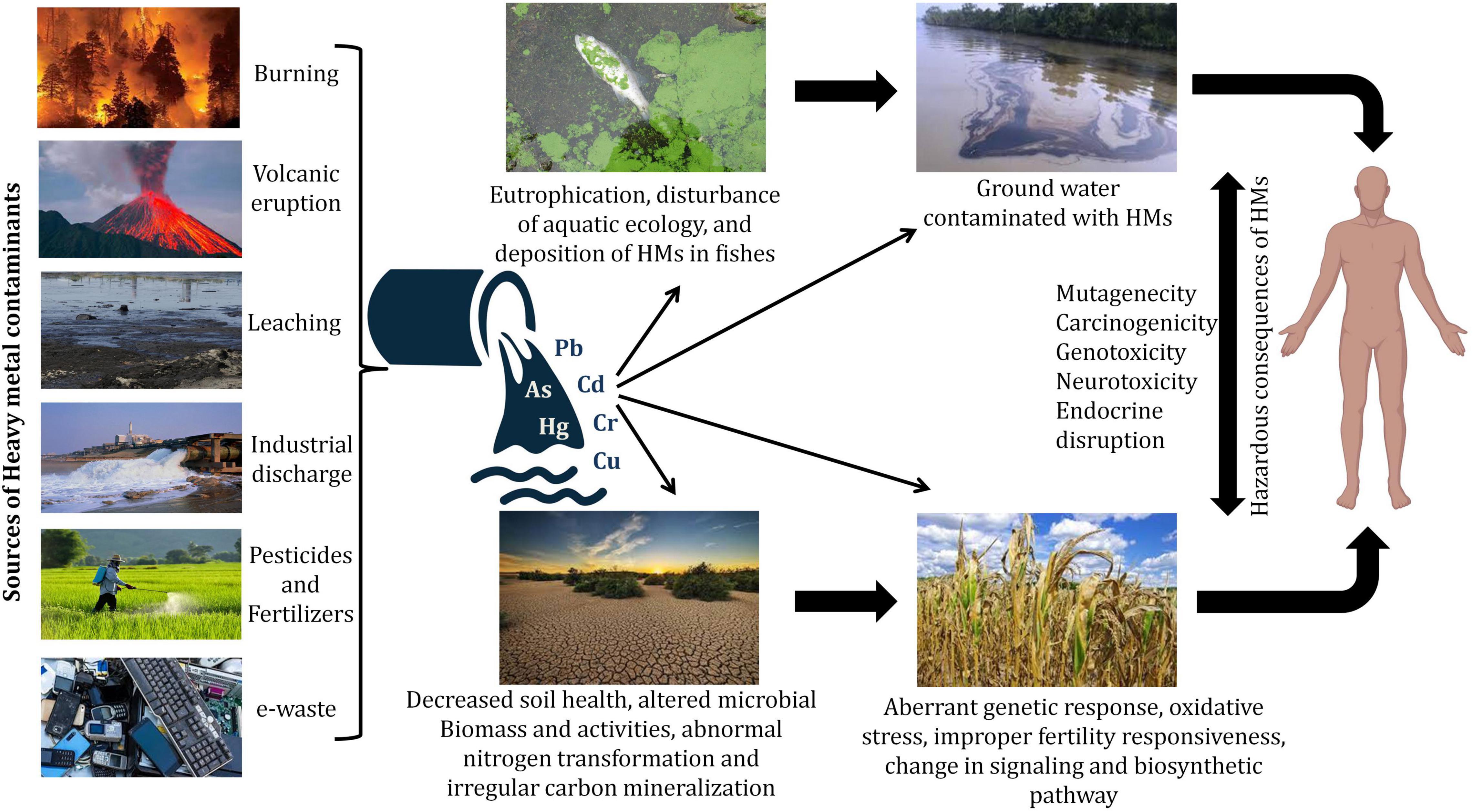
Contamination of heavy metals (HMs) has widely spread all over the world and therefore is a primary matter of concern as it poses threat to animals, plants as well as humans and disturbs the environment. HMs, like other metals and metalloids, are present in the earth’s crust, however, the recalcitrant nature of HMs makes them resistant to degradation. Bioaccumulation of HMs and metalloids via different sources like air, water, causes them to infiltrate plants, animals, and humans, as well as the advancement of the food chain over time (Briffa et al., 2020). Several natural and man-made processes could release these HMs into the environment (Dembitsky and Rezanka, 2003). Due to the growing usage of agrochemicals and inorganic fertilizers, modern agricultural methods have resulted in agricultural pollution, resulting in the ecosystem and environmental destruction (Malik et al., 2017). HMs are also introduced into agricultural systems through the use of sewage sludge and organic waste manure, industrial wastes, and wastewater irrigation (Srivastava et al., 2016; Sharma et al., 2017) (Figure 1). Extraction of HMs from their ores occurs during the processing of minerals and throughout this process, some portions are left out in the open and get relocated to different places due to flood and wind thus causing serious environmental hazards. The essential nourishment of food crops is the soil and therefore agrarian soil is of huge concern owing to its linkage with the production of food, which could affect the health of living organisms. Despite being part of the soil, HMs cause serious harm to the soil as well as plants in their concentrated form. Thus, they are considered to be hazardous (Osmani et al., 2015). HMs are responsible for not only changing the composition of soil but also forming the basis of stress in the plants resulting in the failure of the crop. Biological molecules like lipids, nucleic acids, proteins, and enzymes get damaged due to the production of free radicals by the HMs thus increasing intracellularly the reactive oxygen species (ROS) levels thereby leading to oxidative stress. The failure in all of these biological substances creates several physiological issues, including, cell damage, DNA damage, and enzyme inhibition, all of which can lead to the plant’s death (Wu et al., 2016). HM pollution in modern agriculture has become a severe challenge in most emerging and underdeveloped countries due to a variety of social-economical, scientific, and developmental difficulties. Discovering environmentally safe, long-term solutions to the HM contamination problem is a serious task. Currently, the use of microorganisms or functional biocatalysts in the remediation of soil contaminated with HMs entails the integration of genomes, transcriptomics, proteomics, signaling systems, and synthetic biology knowledge (Hemmat-Jou et al., 2018). These strategies offer a new vista in biotechnology, allowing for the creation of a complex biological system to produce a better microbial system capable of combating HMs contamination (Sayqal and Ahmed, 2021). HMs affect the soil microbiology and modify rhizospheric connections between plants and microorganisms, influencing soil characteristics, plant growth, vegetation type, and agricultural land production, among other things. Different microbial communities with specific metabolic capacities reside in the soil, e.g., organic substances are formed by certain microorganisms while interacting with toxic metals whereas some other assists in the formation of natural nanoparticles thus reducing HMs (Wang and Chen, 2006). Owing to their high surface area to volume ratio, which is associated directly with their increased reactivity, nanotechnology-based materials have also been explored for HMs micro-remediation (Vijayaraj et al., 2019). The approaches and successes of biotechnological applications for environmental protection, decontamination, and the elimination of HMs and metalloids have thus been covered in this study.
The advancement of biotechnological applications and strategies for environmental protection, detoxification, and the removal of HMs and metalloids are the subject of this review. The goal of this review is to compile a list of key findings on HM contamination in modern agriculture, as well as to sketch a probable research roadmap for the future. The review explored the depth information about the mechanism and impacts of the HMs in microbial systems.
Heavy Metal Pollution in Agroecosystem: Consequences and Plant Responses
Effect on Soil Health, Fertility, and Microbial Dynamics
Soil biology plays a vital part in maintaining healthy soil quality, which is crucial for sustainable agriculture. The anthropogenic activities are central in contaminating soil with HMs, e.g., industrial, mining, and agricultural operations as the s present in mining waste, sewage sludge, inorganic fertilizers, and pesticides, tend to disturb soil microbes by percolating into the soil environment (Gupta et al., 2010; Tóth et al., 2016; Sharma et al., 2017). With rising amounts of HM pollution, microbial viability declines. Yuan et al. (2015) showed that microbial survivability was found to be negatively correlated with prolonged Pb exposure. According to de Quadros et al. (2016), coal mining operations; result in a decrease in microbial biomass, abundance, and variability. Nayak et al. (2015) reported that up to 40 and 100% fly ash amendments resulted in better microbial population dynamics with increased concentrations of Zn, Fe, Cu, Mn, Cd, and Cr in agricultural soils. Total microbial activity, as determined by the fluoresceindiacetate (FDA) test, and denitrifiers, on the contrary, exhibited an increasing tendency of up to 40% fly ash addition. The application of fly ash, on the other hand, reduced the activity of both acid and alkaline phosphatase. Various types of HM toxicity and their harmful effects on soil, plants, and humans are presented in Table 1.
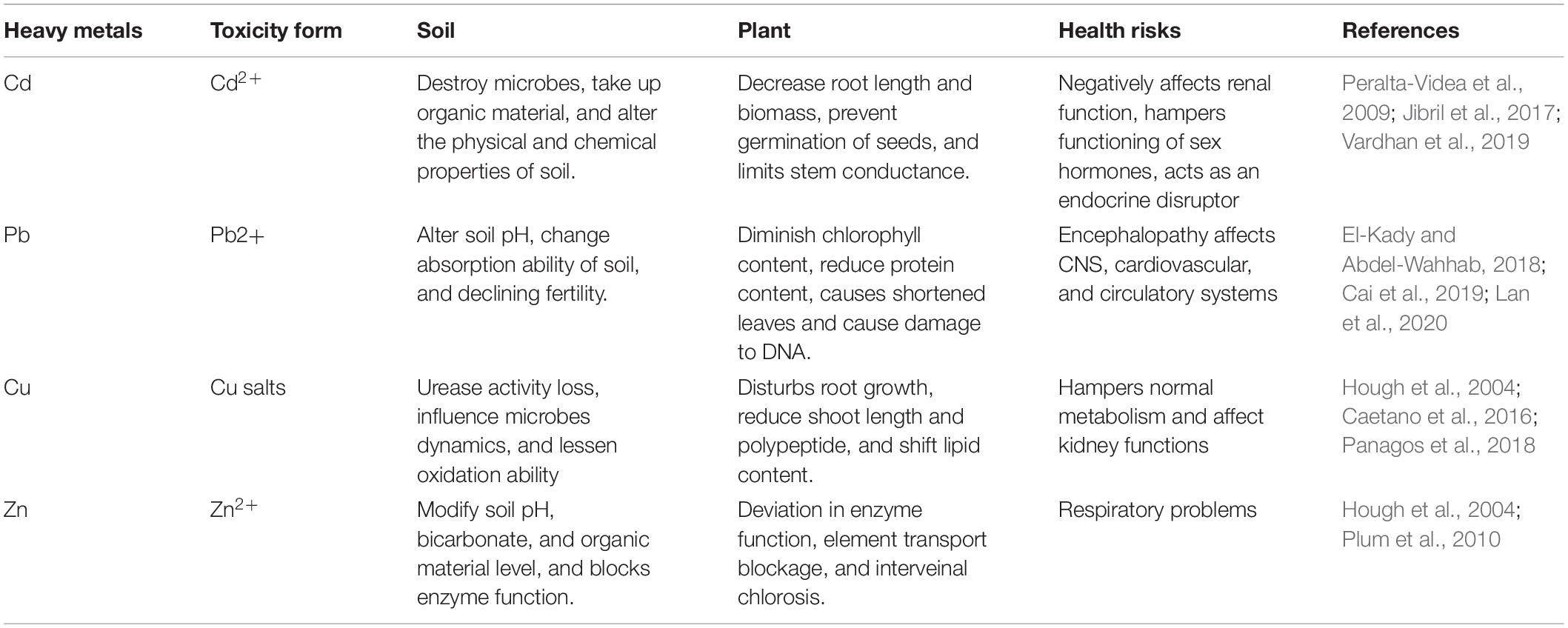
Table 1. Various types of heavy metal toxicity and their harmful effects on soil, plants, and humans.
Effect on Soil Microbial Functions and Processes
Due to HM toxicity, litter breakdown is slowed, resulting in an uneven litter deposit on the soil (Illmer and Schinner, 1991; Giller et al., 1998; Marschner and Kalbitz, 2003). Kozlov and Zvereva (2015) investigated the breakdown rate of mountain birch (Betula pubescens ssp. czerepanovii) leaves in a significantly contaminated industrial setting close to the nickel-copper smelter in Monchegorsk. During 2 years of exposure, there was a substantial reduction of 49% in the relative weight of native leaves compared to the loss observed in the unpolluted forest. Furthermore, anthropogenic HM contamination has been found in a number of studies to have a negative impact on stream litter decomposition (Carlisle and Clements, 2005; Hogsden and Harding, 2012; Ferreira et al., 2016). In both ecological toxicology and ecological tracking investigations, the rate of soil organic carbon mineralization has been routinely utilized as a test for metal toxicity (Giller et al., 1998). Carbon mineralization may be measured using the soil respiration rate. A negative association was found between soil microbial respiration and HM concentration by Nwuche and Ugoji (2008). From an average rate of 2.51–2.56 g of C/g at the start of the trial, the rate of soil microbial respiration was lowered to 0.98, 1.08, and 1.61 g of C/g in the Cu: Zn, Cu, and Zn treated soils, respectively. Because of differences in the experimental designs, fluctuations in soil characteristics, and substrate concentrations, HM exposure can either stimulate or impede N-mineralization. HM pollution disrupts nitrogen transformation pathways, which ultimately affects N-mineralization (Dai et al., 2004; Vásquez-Murrieta et al., 2006; Hamsa et al., 2017). HM pollution has a similar impact on both N mineralization as well as nitrification i.e., both processes tend to decrease with an increasing amount of HM pollutants (deCatanzaro and Hutchinson, 1985). Furthermore, nitrification is more susceptible to HM contamination than N mineralization (Rother et al., 1982; Bewley and Stotzky, 1983).
Impact on Soil Enzymes
Metal composition, pH of the soil, organic matter, and clay content are important factors regulating the biological availability of metals in the soil. HMs affect soil enzymatic activity such as alkaline phosphatase, arylsulfatase, β-glucosidase, cellulase, dehydrogenase, invertase, protease, and urease (Oliveira and Pampulha, 2006; Burges et al., 2015; Xian et al., 2015). Pan and Yu (2011) reported that HMs (Cd or/and Pb) reduce the activity of soil enzymes such as acid phosphatase, dehydrogenase, and urease, as well as the soil microbial community. Some researchers investigated the combined impact of HMs and soil characteristics on soil functions and concluded that arylsulfatase is the most sensitive soil enzyme that might be utilized as a marker of soil toxicity (Xian et al., 2015).
Heavy Metals Responses in Plant System
Heavy metal contamination is a modern ecological issue that pollutes air, water, and soil. This not only results in significant crop yield losses but also raises health risks. In order to mitigate the damage caused by HM contamination, the antioxidative machinery of plants gets triggered.
Oxidative Stress and Reactive Oxygen Species
“Reactive oxygen species (ROS)” are reactive chemical species produced from molecular oxygen. Various diverse ROS are present momentarily among all aerobes which include: (a) oxygen-derived non-radicals viz. singlet oxygen (1/2O2), organic hydroperoxide (ROOH), hydrogen peroxide (H2O2); and (b) oxygen-derived free radicals viz. superoxide anion (O2–), alkoxyl (RO⋅) radicals, peroxyl (RO2⋅), and hydroxyl (HO⋅) (Circu and Aw, 2010; Shahid et al., 2014; Tamás et al., 2017). Plants tend to produce more ROS, after exposure to HMs as they can disrupt the electron transport chain of the mitochondrial and chloroplast membrane. The increased load of ROS interrupts the redox balance of the cell by causing plasma membrane damage and ion leakage (Dingjan et al., 2016; Anjum et al., 2017), lipid peroxidation, and the disintegration of cellular macromolecules (Carrasco-Gil et al., 2012; Chen et al., 2012; Venkatachalam et al., 2017). The increased amount of Cr in two maize genotypes reduced the content of soluble protein and elevated the level of phenol, proline, and other soluble sugars (Anjum et al., 2017).
Genotoxicity
The pathways regulating metal-induced genotoxicity are intricate and are understudied (Cuypers et al., 2011). However, it is clear that HM-induced genotoxicity/DNA damage happens indirectly via the generation of ROS during oxidative stress (Barbosa et al., 2010; Shahid et al., 2014; Aslam et al., 2017). HM-induced nucleic acid impairments have previously been identified in plants such as Helianthus annuus (Chakravarty and Srivastava, 1992), Vicia faba (Pourrut et al., 2011; Arya et al., 2013; Arya and Mukherjee, 2014), Solanum tuberosum, and Nicotiana tabacum (Gichner et al., 2006), and Allium cepa (Arya et al., 2013; Arya and Mukherjee, 2014; Qin et al., 2015). The oxidation state of HM, its amount, and duration of exposure greatly affects the genotoxic response of any plant (Aslam et al., 2017). The extremely reactive species among ROS is the hydroxyl radical (OH⋅), which can react to and damage all of the DNA molecule’s components (Jones et al., 2011). When ROS react with DNA, it can cause deletion, and modification of nitrogenous bases, breakage of strands, damage to cross-links, and generation of nucleotide dimmers (Gastaldo et al., 2008). When Pb and Cd react with DNA, Yang et al. (1999) detected the formation of 8-hydroxydeoxyguanosine (8-OHdG) adducts, which resulted in the breaking of the strand. Further, Hirata et al. (2011) reported Cr and As-triggered translesion DNA synthesis as a consequence of 8-OHdG production. Several recent studies were made on the genotoxic effects of copper and lead (Qin et al., 2015; Silva et al., 2017; Venkatachalam et al., 2017).
Interference With Signaling Pathways
Deregulation of signaling pathways mediated by HM interactions is the main cause behind HM toxicity by influencing G-proteins, growth factor receptors, and receptor tyrosine kinases (Harris and Shi, 2003). HMs also boost H2O2 production in plants by increasing the synthesis of salicylic acid (SA), jasmonic acid (JA), and ethylene (ET), which interferes with the cell signaling mechanism (Maksymiec, 2007; Schellingen et al., 2014; van de Poel et al., 2015). Plants exposed to As, have higher levels of JA, which stimulates the expression of several signaling and stress-response genes such as MAPK, CDC25, and genes regulating glutathione metabolism (Thapa et al., 2012; Islam et al., 2015).
Physiological and Biochemical Response
The anti-oxidative enzymatic machinery of plants such as ascorbate peroxidase (APX), catalase (CAT), glutathione (GSH), glutathione reductase (GR), guaiacol peroxidase (GPX), peroxidase (POX), and superoxide dismutase (SOD) plays a critical role in neutralizing extra ROS (Zhang and Klessig, 2001; Venkatachalam et al., 2017). Increased MDA generation due to enhanced ROS in the cell was observed when two mangrove plants were exposed to HMs (Zhang and Gu, 2007). Similarly, superoxide dismutase (SOD) level was dramatically increased in leaves and roots at low metal concentrations, but dropped drastically at greater concentrations, suggesting a reduction in SOD scavenging ability. Abelmoschus esculentus plant grown on sewage sludge reflected an initial increase in chlorophyll content but fall sharply in later stages. The apparent decrease in chlorophyll content might be due to the accumulation of HMs in plants at later stages (Singh and Agrawal, 2008). Similar findings were made in Vigna radiata (Singh and Agrawal, 2010) and Oryza sativa (Singh and Agrawal, 2010).
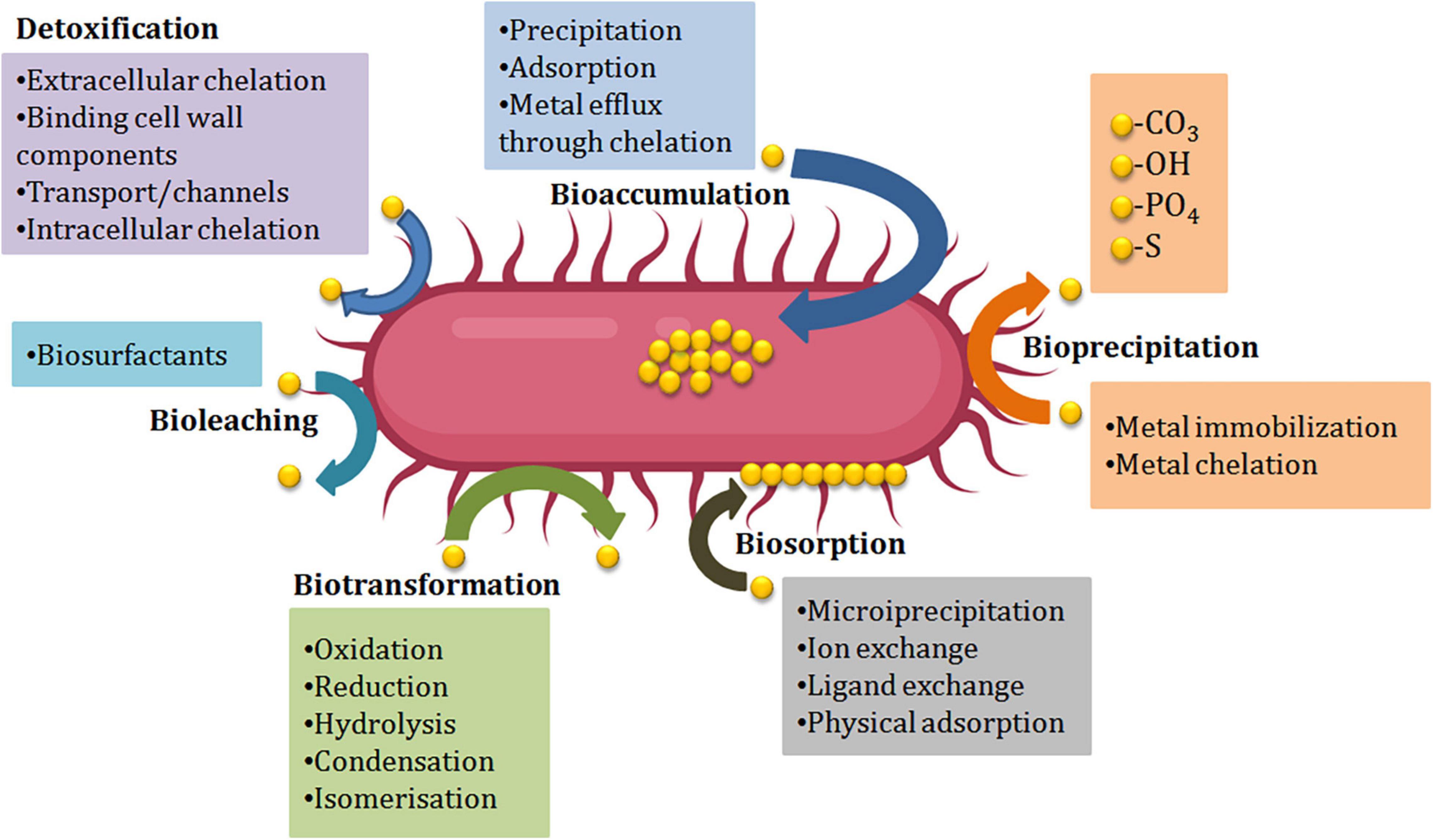
Microbial Resistance to Heavy Metals and Their Mechanisms
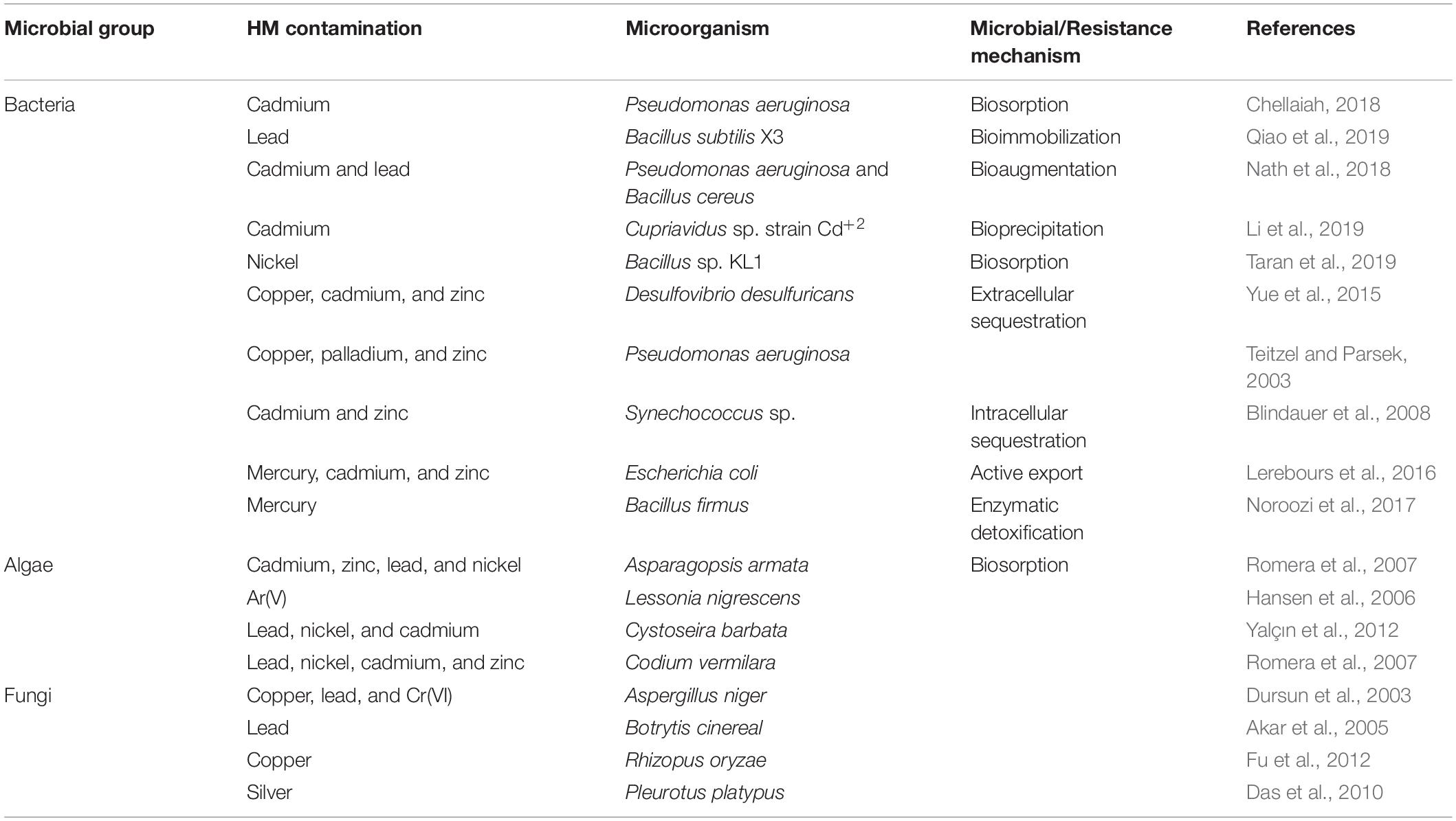
During stress situations developed by HMs, microorganisms either dies of the toxicity caused by the metal or thrive the situation by evolving mechanism of resistance against metals. For the selection of potent bioremediation agents, microorganisms should develop the mechanism of resistance against the toxicity of metals. Different resistance mechanisms developed by microorganisms like Extracellular barriers, extracellular and intracellular sequestration, active transport of metal ions, and enzymatic detoxification are discussed below (Figure 2). Barriers like cell walls, plasma membrane, and other structures present at the surface like EPS, biofilms restrict the passage of HMs into the cells of bacteria. Microbes’ cell surfaces have a variety of characteristics that prevent metal ions from entering by adsorbing them on their surface and functioning as barriers. E.g., a study by Kumar et al. (2014) showed that isolates of fungi and bacteria can cause biosorption of HMs like copper, lead, and chromium. Tolerance against numerous HMs like copper, zinc, iron, nickel, lead, and cadmium was shown by Cellulosimicrobium sp. Chemisorption sites were involved in this resistance mechanism (Bhati et al., 2019). The Biofilms produced by microbes are made up of extracellular polymers that are capable of accumulating metal ions and therefore protect the cells present inside them. The tolerance against lead, zinc, and copper ions has been displayed by the biofilm of Pseudomonas aeruginosa (Teitzel and Parsek, 2003). The efficiency to eliminate metal was enhanced from 91.71 to 95.35% by the biofilm of Rhodotorula mucilaginosa (Grujić et al., 2017). In addition to biofilms and cell walls, EPS has also been shown to be a barrier against metals, e.g., Adsorption of lead ions was reported in P. aeruginosa, Acinetobacter junii L. Pb1, and Azotobacter chroococcum XU1 (Bramhachari et al., 2007; Rasulov et al., 2013; Kushwaha et al., 2017).
Numerous proteins and metabolic products are found in the cell membrane of the microbes that are capable of making complex structures (chelation) with the metal ions. Extracellular sequestration can be defined as the complexation of metal ions as insoluble compounds or metal ions accumulation by the components of the cell in the periplasm. Copper-inducible proteins CopA, CopB (periplasmic proteins), and CopC (outer membrane protein) are produced by copper-resistant Pseudomonas syringae strains that are responsible for the binding of microbial colonies and copper ions. Zinc ions can pass from the cytoplasm and get accumulated into the periplasm of the Synechocystis PCC 6803 strain via the efflux mechanism (Thelwell et al., 1998). Hazardous metals can be reduced by iron and sulfur-reducing bacteria like Desulfuromonas spp. and Geobacter spp. into less or non-hazardous metals. An obligate anaerobe, G. metallireducens, can reduce manganese (Mn) from poisonous Mn (IV) to Mn (II) and uranium (U) from toxic U(VI) to U(II) (IV) (Gavrilescu, 2004). In intracellular sequestration, metal ions are complexed by distinct compounds in the cell cytoplasm. The interaction of metals with the ligands presents in the surface, followed by sluggish transport into the cell, can result in a high concentration of metals within the cells of microorganisms. The ability to accumulate metals intracellularly by bacterial cells has been used in a variety of applications, most notably in waste treatment. With the help of low molecular weight proteins that were rich in cysteine, a cadmium-tolerant Pseudomonas putida strain was able to sequester copper, cadmium, and zinc ions intracellularly (Higham et al., 1986). In Rhizobium leguminosarum cells, glutathione was also found to be involved in the sequestration of cadmium ions intracellularly (Lima et al., 2006). Lipids, chitin, mineral ions, nitrogen-bearing polysaccharide, polyphosphates, and proteins make up the firm cell wall of fungi. The accumulation of metals by numerous fungi into their spores and mycelium helps in decontaminating metal ions by energy uptake, intracellular and extracellular precipitation, and valence exchange. Another strategy to protect against HM stress is to transport HM ions out from the intracellular environment, which can happen through efflux mechanisms that can effectively regulate intracellular HM ion concentrations (Remenar et al., 2018). Efflux systems have been discovered in a variety of microbes, particularly those isolated from metal contaminated surroundings. Metal exporting proteins, such as ABC transporters, P-type efflux ATPase, cation diffusion facilitator, and proton-cation antiporters are widely distributed in the cell membrane to achieve HM ion efflux. For the export of Cu (II), Cd (II), and Zn (II), Gram-positive bacteria utilize P-type efflux ATPase. With the help of ATPase, an exporting protein on the cell membrane regulates arsenite outflow (Yang et al., 2012; Soto et al., 2019). ABC transporters which also called traffic ATPases can assist microorganisms to survive the stress caused due to HMs by mediating membrane translocation of HM ions (Al-Gheethi et al., 2015; Lerebours et al., 2016; Zammit et al., 2016).
The resistance to HMs ions in microbes is also contributed by the enzymes that biologically transform or chemically modify the HM ions from highly hazardous form to less toxic form (Liu et al., 2017). HM ions’ toxicity can be effectively reduced by changing their redox state via reduction or oxidation reactions (Giovanella et al., 2016). Detoxification enzymes can influence this defensive mechanism, which is also controlled by microbe resistance genes. Through mercuric ion reductase, bacteria like Bacillus sp. display resistance to mercury ions (Noroozi et al., 2017). Mercuric reductase transforms the mercuric ion into metallic mercury, which is then discharged into the environment via the cell membrane (Zhang et al., 2012). To reduce toxicity, bacteria such as Micrococcus sp. and Acinetobacter sp. can oxidize hazardous as (III) into less soluble and non-toxic As (V) (Nagvenkar and Ramaiah, 2010).
Microbial Mechanism Involved in Heavy Metal Bioremediation
For the elimination of HMs from the polluted sites, bioremediation methods are employed (Pratush et al., 2018). Usually, these methods involve the absorption/adsorption of toxic metallic ions, and this alleviates the related side effects (Njoku et al., 2020). Different natural resources like wood bark/dust, coconut husk/shells, agro wastes, microorganisms, seaweeds, seeds, discarded coffee beans, and aquatic plants, etc. are being used constantly to reduce the number of HM ions from the place of their origin, in which microbes (algae, fungi, bacteria, yeasts, etc.) play a considerable role (Mudila et al., 2019). Microorganisms change the HMs’ ionic state that influences the solubility, bioavailability, and movement in the soil as well as in the aquatic surroundings (Ayangbenro and Babalola, 2017). Mobilization or immobilization of HMs aids microbial remediation, which is then proceeded by oxidation-reduction, chelation, modification of the metallic complex, and biomethylation (Pratush et al., 2018). The enzymatic catalysis by microbes solubilizes the metals with higher oxidation state to lower oxidation state, for instance, Thiobacillus ferrooxidans and T. thiooxidans are responsible for the enzymatic oxidation of Uranium (Cumberland et al., 2016). The isolation of microorganisms responsible for the degradation of HMs could occur from aerobic as well as anaerobic locations. However, in comparison to anaerobic microorganisms, aerobic microbes are more willing for bioremediation (Azubuike et al., 2016). Microorganisms carry out the transportation of HMs utilizing membrane-linked transport mechanisms and transform them into non-hazardous forms (Igiri et al., 2018). Microorganisms use processes like biosorption, bioaccumulation, biotransformation, and bioleaching to stay alive in a metal-polluted environment (Figure 2 and Table 2). These techniques have been used in clean-up processes (Gadd, 2000; Lin and Lin, 2005).
Bioaccumulation and Biosorption
Both bioaccumulation and biosorption are the processes utilizing which microbes or biomass gets bound to the HMs and pollutants from the surroundings and concentrates them (Joutey et al., 2015). However, the working manner of the processes differs. Biosorption is a process in which microorganisms use their cellular structure to capture HM ions, which they then sorb onto the cell wall’s binding sites (Malik, 2004). This is a passive uptake process and does not depend upon the metabolic cycle. Two common methods for the bioremediation of HMs are adsorption and absorption onto the cell surface of microbes. Adsorption differs from absorption in that it involves the dissolution or permeation of a fluid (the absorbate) by a liquid or solid (the absorbent) (Jovancicevic et al., 1986). Adsorption, on the other hand, is a surface occurrence, whereas absorption affects the full volume of the substance. Various living creatures have been shown to be possible bio sorbents e.g., bacteria like Magnetospirillum gryphiswaldense, Bacillus subtilis, algae-like marine microalgae, and Chaetomorphalinum, fungi like Rhizopus arrhizus and yeast-like Saccharomyces cerevisiae (Romera et al., 2006; Wang and Chen, 2009; Zhou et al., 2012). Bacteria, on the other hand, are regarded as the most exceptional biosorbents among all other creatures due to their high surface-to-volume ratios and numerous chemosorption active sites in their cell wall, such as teichoic acid (Beveridge, 1989). Dead bacterial strains have also been considered as promising biosorbents, with biosorption abilities that exceed those of living cells of the same strain. 13–20% increased capacity for the biosorption of chromium ions was shown by dead Bacillus sphaericus as compared to the cell of its living strain (Velásquez and Dussan, 2009). However, unlike biosorption, bioaccumulation by microbes is metabolically active and relies upon the import-storage system. In this system, HM ions are transported through the lipid bilayer of the cell membrane into the intracellular spaces or cytoplasm with the help of transporter proteins. This is known as active uptake or bioaccumulation. Endocytosis, ion channels, carrier-mediated transport, complex permeation, and lipid permeation are all involved in HM bioaccumulation in the bacterial membrane (Ahemad, 2012; Geva et al., 2016; Shahpiri and Mohammadzadeh, 2018). Bioaccumulation studies of various metals like lead, nickel, silver, mercury, and cadmium have been reported by Ahemad (2012). The study of cadmium by Rani and Goel (2009) discovered periplasmic and intracellular metal accumulation by P. putida 62 BN, and it was performed using transmission electron microscopy. The growing cells of Bacillus cereus M116 were shown to accumulate about 20% nickel (II) intracellularly, as reported by Naskar et al. (2020). Lead and chromium accumulation by Aspergillus niger and Monodictys pelagic was reported by Sher and Rehman (2019). In the process of bioleaching, metal oxides and sulfides from ores deposits and secondary wastes are solubilized by different microorganisms like fungi and bacteria (Mishra et al., 2005; Jafari et al., 2019). Following solubilization, purification is achieved with the help of appropriate methods like ion exchange, selective precipitation, adsorption, and membrane separation (Rohwerder et al., 2003).
Bioleaching
Bioleaching is performed by an extensive range of microbes and among them, acidophiles are the prominent ones. Acidophiles are chemolithotrophs that oxidize Fe (II) to Fe (III) and/or reduce sulfur to sulfuric acid and flourish in low pH environments, particularly 2.0 or below. Sulfuric acid produces ferric ions and protons, which solubilize metal sulfides and oxides from ores (Srichandan et al., 2014), aiding extraction of metal by segregating metals in the solid phase from the more water-soluble phase. Bioleaching, which uses microorganisms as reduction agents, can also be used to extract and recover heavy metals (Wang and Zhao, 2009). The ability of microorganisms to convert the solid chemical within contaminated soil into a soluble substance that can be removed and recovered determines the efficacy of the recovery process. Due to metal resources being non–renewable, recovering metal from industrial waste water may be a viable option for ensuring heavy metal supply (Atkinson et al., 1998). Bioremediation has been offered by a number of researchers as a way to recover raw materials from effluent (Pollmann et al., 2006; Gadd, 2010). Using an Annona squamosa-based absorbent with 0.1 M HCl, Cd(II) recovery up to 98.7% was achieved (Isaac and Sivakumar, 2013). Using Pseudomonas aeruginosa biomass with 0.1 M HCl, a Cd(II) recovery up to 82% was achieved. Using volcanic rock matrix-immobilized P. putida cells with surface-displayed cyanobacterial metallothioneins at pH 2.35 (Ni et al., 2012), 100% Cu(II) recovery was reported. Using activated sludge at pH 1.0 resulted in a 100% recovery of Cu(II) (Hammaini et al., 2007). The introduction of an indigenous strain Enterobacter sp. J1 resulted in a Cu and Pb recovery of over 90% at pH 2 (Lu et al., 2006).
Biotransformation
Biotransformation is a process in which structurally a chemical compound is altered, thereby relatively a more polar molecular is synthesized (Asha and Vidyavathi, 2009; Pervaiz et al., 2013). In other words, this contact of metal and microorganisms causes toxic metals and organic compounds to get altered to a comparatively less hazardous form. The development of this mechanism in the microorganisms causes them to acclimatize the environmental changes. Microbial transformations can be attained through the production of new carbon bonds, isomerization, introducing functional groups, oxidation, reduction, condensation, hydrolysis, methylation, and demethylation. Transformation of metals by application of microbes has been reported. Micrococcus sp. and Acinetobacter sp. oxidize hazardous As (III) into less soluble and non-toxic As(III) and reduce its toxicity (Nagvenkar and Ramaiah, 2010). Thatoi et al. (2014) reported that Cr (VI)-tolerant Bacillus sp. SFC 500-1E through NADH-dependent reductase has been shown to lessen the hazardous Cr (VI) to less toxic Cr (III).
Influence of Environmental Change on the Remediation of Heavy Metal Contaminants
The pH is important for microbial biosorption, and the optimal pH varies depending on the microbe. Firstly, pH influences the enzymatic activity in bacteria, altering the rate of HM microbial metabolism (Morton-Bermea et al., 2002). Secondly, pH alters the microorganism’s surface charge, affecting its ability to adsorb HM ions (Galiulin and Galiulina, 2008). Besides this, pH has an impact on the hydration and movement of a variety of metal ions in the soil (Dermont et al., 2008). Both Rodríguez-Tirado et al. (2012) and Wierzba (2015) found that the rate of HMs removal by microbe upsurges with an increase in pH across a certain range, but after the pH climbs to a specific level, the elimination rate begins to decline. According to a study the ideal pH range for most bacteria, is 5.5–6.5, however there are exceptions (Wang et al., 2001). For instance, Bacillus jeotgali thrives at a pH of 7. This could be because as the pH rises over a certain point, some metal ions form hydroxide precipitates, which are less prone to microbial adsorption (Hu et al., 2010; Rodríguez-Tirado et al., 2012). Furthermore, the optimal pH for aerobic microbes and anaerobic microorganisms may differ.
The rate of absorption of HM is mostly influenced by ambient temperature, which impacts the growth and multiplication of microorganisms (Fang et al., 2011). Various bacteria have different optimal temperatures (Acar and Malkoc, 2004) for example, Acidianus brierleyi and Sulfolobussolfa-tataricus are very thermophilic bacteria, while, Thiobacillus acidophilus, Thiobacillus tepidarius, and Thiobacillus ferrooxidans are medium temperature bacteria.
When it comes to understanding substrate species, there are three things to keep in mind: HM ions, soil additives, and the type of soil. HM adsorption characteristics on different soils might be quite varied. According to a study, beach tidal soil (Freundlich adsorption constant K = 93.79) has a larger adsorption capacity than black soil (K = 16.41), which is higher than yellow mud (K = 1.17), and that the mean desorption rate of soil is Lithic Ochri-Aquic Cambosols in ascending order (0.67%), Fe-accumulic Gleyic Stagnic Anthrosols (3.62%), and Endogleyic Fe-accumulic Stagnic Anthrosols (35.85%) (Chen et al., 1997). Clearly, soil’s adsorption rate and retention of HM ions result in poor mobility of HM ion, making microbial adsorption difficult to achieve (Hu et al., 2010). HM ion species influence HM elimination by changing microbial generation time. The generation period of Thiobacillus ferrooxidans on sulfur as a substrate is about 10–25 h, which is significantly longer than the 6.5–15 h generation time on Fe. Moreover, the existence of metal ions in the soil affects microbial enrichment (Kapoor and Viraraghavan, 1997). Individual bioavailability of Pb2+, Cd2+, and Zn2+ in the soil is often more than that of several metal ions, according to Park et al. (2016). The adsorption of Cd2+ alone is 11.2 mg/g. Its adsorption is reduced to 3.15 mg/g in the presence of Zn2+ and Pb2+, with similar results for Zn2+ and Pb2+, displaying the reduction from 19.5 and 2.25 to 8.08 and 0.915 mg/g, respectively. Soil additions can considerably boost microbial removal of HMs, and the concentration of additives can have varied impacts on HM ion leaching rates. Tyagi et al. (2014) found that adding 20 g/L FeSO4.7H2O to a solution increased the leaching rate of Zn and Cu by 2 and 1.9 times, respectively, but not when the concentration was larger than 20 g/L. The adsorption rate of microorganisms is also affected by the concentration of HM ions. To estimate the quantification of accumulative properties of a bio-sorbent, adequate assessment is required in general (Cervantes et al., 2001). The Langmuir model, whose parameters are interpretable and primarily explains the adsorption of a single-layer surface, is one of the most commonly used equations to describe the features and another Freundlich model, is mostly employed to the adsorption equilibrium of the adsorption surface equation (Febrianto et al., 2009). Despite the fact that the Freundlich model is simpler, it grows unbounded, hence the Langmuir model has been more extensively employed than the Freundlich model until recently. The Langmuir model was utilized by some researchers to investigate the influence of HM concentration (Ehrlich, 1997; Brunetti et al., 2012). They discovered that depending on the microorganisms and HM ions investigated, the concentrations of HM ions with the highest adsorption rates change. Though, the trend, which is consistent across all examples, implies that adsorption increases to a certain point and then remains constant as HM ion concentrations rise.
Modern Approaches in Microbe-Intervened Biotechnologies
Rhizoremediation: The Phyto-Microbial Remediation System
Rhizoremediation combines two methods for cleansing polluted substrates: phytoremediation and bioaugmentation. Rhizoremediation is the process of using microorganisms found in the rhizosphere of plants that are involved in the phytoremediation process. Application of plants and plant growth-promoting bacteria (PGPB) is being assessed as an effective and environmentally acceptable way for soil renewal and HMs elimination, among the several integrated techniques (Ali et al., 2013; Sati et al., 2022). Many microorganisms in the rhizosphere, such as mycorrhizal fungi and other rhizospheric organisms, can help plants absorb or adsorb HMs (Bojórquez et al., 2016). Joner and Leyval (1997) found that mycorrhizal plants uptake 90, 127, and 131% more Cd than non-mycorrhizal plants, when the concentration of Cd2+ in the soil is 1, 10, and 100 mg/kg, respectively. Bissonnette et al. (2010) displayed that mycorrhiza inoculation improves the ability to absorb Cu2+, Cd2+, and Zn2+. Mycorrhizal fungi possess mycelia that grow into the soil, thereby increasing the surface area of plant roots (Trellu et al., 2016). For metal extraction with plants, PGPR including Azospirillum, Alcaligenes, Agrobacterium, Arthrobacter, Burkholderia, Bacillus, Pseudomonas, Rhizobium, and Serratia, are commonly utilized (Carlot et al., 2002; Glick, 2003). Metal transformation, immobilization, chelation, or solubilization is aided by the production of exopolysaccharides by PGPB, like oxidases, reductases, siderophores, and organic acids which promotes phytoremediation of HMs. PGPB reduces the pH of the soil by producing organic acids, which aids in the removal of HM ions. Metal resistant siderophore-producing bacteria found near the rhizosphere supply nutrients to the plants namely iron, perhaps reducing the negative consequences of metal contamination (Dimkpa et al., 2008; Sinha and Mukherjee, 2008). Siderophore is also responsible for the formation of stable complexes with radionuclides and metals concerning environment like Cd, Ga, Al, Cu, Zn, In, and Pb (Neubauer et al., 2000; Rajkumar et al., 2010). The synergistic effects of bioaugmentation and phytoremediation leading to rhizoremediation may overcome the difficulties that arise when both processes are employed distinctly. Moreover, the remediation of HM with the help of higher plants has also been reported. Wang et al. (2021) also found that planting Salix in Cd-polluted soil improved the diversity of beneficial microbes, such as the bacteria genera Arthrobacter and Bacillus. Anaeromyxobacter, Novosphingobium, Niabella, Niastella, Flavobacterium, Thermomonas, Lysobacter, Pedomicrobium, Solitalea, Devosia, Flavisolibacter, Mesorhizobium, Nitrospira, Rmlibacter, and Rubrivivax. Phyllobacterium and mycorrhizal genera of fungi include Amanita, Cryptococcus, Conocytes, Actinomucor, Ramicandelaber, Spizellomyces, Xylaria, Rhodotorula, Umbilicaria, Sporobolomyces, Tilletiopsis, Claroideoglomus, and Cirrenaliain plant rhizosphere.
Genetically Engineered Organisms and Modern Molecular Biology
Bioremediation using microorganisms can degrade and dissipate chemicals of complex substances, making it a long-term solution for reducing HMs contamination in soil (Mosa et al., 2016; Bhatt et al., 2020a,b). Recent advances in genetic engineering, as well as the adequacy of genetically engineered microorganisms/biocatalysts for the restoration of the environment, have shown that they are more capable than natural microbes, particularly for the removal of persistent compounds under natural environments (de Lorenzo, 2009; Bhatt et al., 2020c). By the application of various genetic and metabolic engineering approaches, the genetic material of microbes is modified, and engineered microorganisms are produced which are more efficient thus resulting in enhanced bioremediation. Single-gene editing, pathway construction, and change of existing gene sequences i.e., both coding as well as controlling sequences are included in the aspects of engineering, with a focus on the modification of rate-limiting stages of the metabolic processes (Diep et al., 2018). HMs such as Fe, Cd, As, Cu, Hg, and Ni can now be eliminated with the help of engineered bacteria (D’Souza, 2001; Verma and Singh, 2005; Azad et al., 2014). The rate of degradation, on the other hand, is determined by the catalytic efficiency of enzymes present in the cells or those stimulated to a specific substrate (Kang, 2014). Using recombinant DNA technology, foreign genes from another creature of the same or other species are put into the genome of genetically engineered microorganisms (GEMs). The utilization of genetically modified Pseudomonas putida and Escherichia coli strain M109 harboring the merA gene to successfully remove Hg from polluted soils and sediments has been reported (Chen and Wilson, 1997; Barkay et al., 2003; Deckwer et al., 2004). Azad et al. (2014) provided a thorough evaluation of the application of genetically modified bacteria and plants in the bioremediation of HMs and other organic pollutant-contaminated environments. According to a study, the addition of the mer operon from Escherichia coli, which codes for the reduction of Hg2+, into the genetically modified bacterium Deinococcus geothemalis provides the ability to microorganism to lessen the Hg pollution at high temperatures by mer genes (Dixit et al., 2015). Cupriavidus metallidurans strain MSR33, which was genetically engineered with a pTP6 plasmid that provided genes merB and merG which regulate the biodegradation of Hg as well as the production of merB and merA, i.e., organomercurial lyase protein and mercuric reductase, was able to reduce Hg contamination from polluted sites (Rojas et al., 2011; Dixit et al., 2015). The insertion of novel genes into Pseudomonas cultures using the pMR68 plasmid has also resulted in Hg resistance (Sone et al., 2013). Specific genes in n-alkane-degrading microbes, such as alkB, alkB1, alkB2, alkM, aromatic hydrocarbons: xylE, and polycyclic aromatic hydrocarbons: nidA, ndoB, are frequently found on plasmids that allow horizontal gene transfer and are employed as markers to identify microbial biodegradation (Wolejko et al., 2016). Microbial membrane transporters can be genetically modified to improve the bioremediation of HMs in the environment. Transporters and binding mechanisms play crucial roles in this context of HMs remediation (Manoj et al., 2020). Channels, secondary carriers, and primary active transporters are the three principal types of transporters that are usually emphasized. Their location is in the inner lipid membrane such as Fps, Mer T/P, and GlpF in channel transporters; Hxt7, NixA, and Pho84 in secondary carriers; and cdtB/Ip_3327, MntA, TcHMA3, and CopA in primary active transporters. Some of them, such as the porin channels transporters, may also be found in the outer lipid membrane (Jain et al., 2011). As soon as HMs comes inside the cell, numerous phytochelatins, metallothioneins, and polyphosphates work together for the sequestration of the HMs and changing microorganisms’ HM import-storage systems could boost their ability to extract HMs from water and soil (Diep et al., 2018). Thus, in the fight against harmful compounds in the environment, the use of GEMs to speed up the restoration process is crucial. For the successful implementation of GEMs for bioremediation in adverse environmental conditions, the preservation of recombinant bacterial population in the soil is essential, with appropriate environmental conditions prepared and the recombinant bacteria should be capable to endure antagonism from native bacterial species (Dixit et al., 2015). Consequently, further novel molecular approaches for the screening and isolation of microbes for HM bioremediation should be investigated. Multi-omics comprising genomics, metagenomics, metabolomics, proteomics and transcriptomics, and computational biology techniques have been successfully employed in gene mining that supports system biology research of microorganisms at the genetic level concerning bioremediation of HMs (Subashchandrabose et al., 2018; Pande et al., 2020; Sayqal and Ahmed, 2021). Novel genes implicated in the biodegradation processes of several HM contaminants have been discovered because of high-throughput and next-generation sequencing. New technologies involve gene-editing tools like CRISPR-Cas which possesses the ability to enhance the process of bioremediation by engineering microorganisms with genes engaged in the degradation of, particularly recalcitrant substances. When compared to conventional low-throughput ZFNs and TALENs, CRISPR might be utilized to transmit a preferred set of instructions into the genome of microbe in a straightforward manner because it is a programmable, next-generation approach for high-throughput genetic manipulation (Miglani, 2017). A CRISPR segment is likely for the bioremediation by the application of gRNA-guided dCas9 to control the expression of a gene. As a result, fusing transcription factors with dCas9 can either suppress boost or suppress RNA polymerase transcription, which can cause either upregulation or downregulation of the gene expression or a set of genes of interest. Although CRISPER-based approaches can be employed on a variety of mycobacteria and fungi, further applied research in the area of microbe-based removal of HMs from the environment is needed (Shapiro et al., 2018). With the help of multi-omics, biotechnology has developed a large number of strains. The following are some examples Arthrobacter, Chlorella (Gong et al., 2018), Stenotrophomonas maltophilia (Cho et al., 2018), Rhodococcus wratislaviensis, Mycobacterium (Gołêbiewski et al., 2014), Alcaligenes eutrophus, Pseudomonas putida (Cycon and Piotrowska-Seget, 2016), Cyanobacterium synechocystis, Saccharomyces cerevisiae, Populus sp. (Cai et al., 2019), Candida pelliculosa strain S-02, Streptomyces aureus strain HPS-0, Aspergillus niger (Kumar et al., 2018), Sphingomonas sp., and Pseudomonas putida strain KT2440 (Liu et al., 2019).
Nanotechnology in Microbial Bioremediation
With the application of chemical or biological methods, several types of nanoparticles have been effectively produced and studied for bioremediation of HMs, over the last decade (Baragaño et al., 2020). The advantages of nano-biosorbents with an ultrafine arrangement and a great surface area include (1) enhancing chemical activity and capacity of adsorption, (2) boosting surface binding energy, and (3) lowering internal diffusion resistance (Khati et al., 2017). As a result, nano-biosorbents could be used as a replacement for traditional biosorbents (Abdi and Kazemi, 2015; Alviz-Gazitua et al., 2019). Latest advancements in the nanobiosorption model have resulted in a number of sophisticated ways that improve the complete efficiency of a conventional biosorption process while also ensuring its economic viability (Devatha et al., 2018). Different functional groups, such as –NH2, –COOH, and –OH, are intrinsically present in nanoparticles, and tailoring the appropriate functional groups by activating physically/chemically or by modifying surface has proven to improve elimination of HMs. Bacterial strains can also produce nanoparticles that can aid in the bioremediation of HMs (Arshad et al., 2019). Nanomaterials are combined with microbes to improve reduction of HMs, which makes them more effective as compared to their independent application. Factors in determining the interaction between nanomaterials and microbes include (1) the chemical properties of the nanomaterial, its particle size, coating characteristics, and shape, (2) the chemical properties of the nanomaterial, along with the shape, size of the particle, and its coating characteristics, (3) method of metabolism, (4) the nanomaterial’s crystalline phase, (5) the extent of contamination and, lastly (6) the resistance of nanomaterials to the hazardous contaminant and the prevalent ecological conditions. Microbial biostimulation, bioaccumulation, and biotransformation activities are enhanced by nanocomposites, which increase absorption, adsorption, and the number of chemical processes for the reduction of HMs (Tan et al., 2018). Microbes are trapped within nanomaterials to create a nanocomposite; for example, immobilization of gram-negative Halomonas sp. within polyvinyl pyrrolidone-coated iron oxide nanoparticles was confirmed to eliminate Pb (II) and Cd (II) (Cao et al., 2020). On the other hand, the microbe can function as a nanoparticle synthesizer, a method known as green synthesis. Though separating or recovering HMs from nanomaterials, is a time-consuming/laborious technique and hence magnetic nanoparticles have gained considerable attention in recent years, wherein surface amendment, coating of iron/iron oxides, and encapsulation focused for simple separation or retrieval of HMs.
Conclusion
Throughout the world, HM contamination causes severe environmental problems. In this review, several technical strategies i.e., microbe-based as well as hybrid have been discussed, that are currently being employed to mitigate HM contamination in soils and other contaminated surroundings. Because of the contribution in the regulation of biogeochemical cycles that influence climate, soil structure, and fertility, the environmental microbiome is thought to play a critical role. Microbe-mediated bioremediation should be given high attention from a practical standpoint since microbes have a variety of natural roles and mechanisms that considers them a great candidate for the clean-up of the polluted site, management of wastes, and sustainable agriculture. Although microbes are being employed to improve the effectiveness of HM removal from the soil, there is still room for improvement.
Directions to the Future Research
To create approaches that support better tolerance of HMs in microbes, more emphasis should be placed on understanding the physical, chemical, and biological characteristics of microbes in the prevalence of HMs in soil, water, and gaseous surroundings (Njoku et al., 2020). Furthermore, the application of additives in bioremediation, such as surfactants, might expand the region of the interphase between microorganisms and pollutants, pushing microbes beyond their bioremediation limits. Recently, yeast has been genetically modified to have plant-like properties and to act as hyperaccumulators of several HMs in the aqueous environment (Sun et al., 2020). Other bacteria could be developed in the same way to help in clean-up of HMs. More emphasis should be paid to algae in future study studies, as it is considered an effective microbe for the sorption of HMs from the soil. Because of the better genetic abilities and tolerance to HMs, omics-based techniques are advantageous for the production of improved industrial strains that are tolerant to the prevalent environmental surroundings (Hemmat-Jou et al., 2018). Furthermore, as mentioned in this study, the application of nano- and nano (bio)technologies has enormous ability to promote the use of microbial technologies to deal with HMs pollution. When nanotechnology and microbe-based technology are coupled in environmental restoration procedures, the nanoparticles will promote the elimination of greater pollutant loads, reducing the toxicity-based inhibition of the contaminant on the microbe (Ma and Zhang, 2008). As a result, combining various traditional procedures and current technology could be a potential choice if they could improve relevant material qualities and speed up the restoration process. All life forms and the natural ecosystem are in danger from pollution caused by HMs in soil, water, and agrarian land. Sustainable policies have been developed and revised regularly; nevertheless, awareness of the negative effects, as well as knowledge of how to reduce HMs contaminantion in the soil, should be expanded.
Author Contributions
VP conceptualized and wrote the manuscript. SP wrote the manuscript and made the diagrams. DS wrote the manuscript. PB and MS supervised, helped in writing, reviewing, and editing the manuscript. All authors contributed to the article and approved the submitted version.
Funding
We are thankful to the Department of Zoology, Soban Singh Jeena University, Almora (Uttarakhand), India, and DST FIST grant SR/FST/LS-I/2018/131 for providing the facility for this work.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdi, O., and Kazemi, M. (2015). A review study of biosorption of heavy metals and comparison between different biosorbents. J. Mater Environ. Sci. 6, 1386–1399.
Acar, F. N., and Malkoc, E. (2004). The removal of chromium (VI) from aqueous solutions by Fagus orientalis L. Bioresour. Technol. 94, 13–15. doi: 10.1016/j.biortech.2003.10.032
Ahemad, M. (2012). Implications of bacterial resistance against heavy metals in bioremediation: a review. J. Inst. Integr. Omics Appl. Biotechnol. IIOAB 3, 39–46.
Akar, T., Tunali, S., and Kiran, I. (2005). Botrytis cinerea as a new fungal biosorbent for removal of Pb (II) from aqueous solutions. Biochem. Eng. J. 25, 227–235. doi: 10.1016/j.bej.2005.05.006
Al-Gheethi, A. A. S., Lalung, J., Noman, E. A., Bala, J. D., and Norli, I. (2015). Removal of heavy metals and antibiotics from treated sewage effluent by bacteria. Clean Technol. Environ. Policy 17, 2101–2123. doi: 10.1007/s10098-015-0968-z
Ali, H., Khan, E., and Sajad, M. A. (2013). Phytoremediation of heavy metals—concepts and applications. Chemosphere 91, 869–881. doi: 10.1016/J.CHEMOSPHERE.2013.01.075
Alviz-Gazitua, P., Fuentes-Alburquenque, S., Rojas, L. A., Turner, R. J., Guiliani, N., and Seeger, M. (2019). The response of Cupriavidus metallidurans CH34 to cadmium involves inhibition of the initiation of biofilm formation, decrease in intracellular c-di-GMP levels, and a novel metal regulated phosphodiesterase. Front. Microbiol. 10:1499. doi: 10.3389/fmicb.2019.01499
Anjum, S. A., Ashraf, U., Imran, K., Tanveer, M., Shahid, M., Shakoor, A., et al. (2017). Phyto-toxicity of chromium in maize: oxidative damage, osmolyte accumulation, anti-oxidative defense and chromium uptake. Pedosphere 27, 262–273. doi: 10.1016/s1002-0160(17)60315-1
Arshad, F., Selvaraj, M., Zain, J., Banat, F., and Haija, M. A. (2019). Polyethylenimine modified graphene oxide hydrogel composite as an efficient adsorbent for heavy metal ions. Separat. Purif. Technol. 209, 870–880. doi: 10.1016/j.seppur.2018.06.035
Arya, S. K., and Mukherjee, A. (2014). Sensitivity of Allium cepa and Vicia faba towards cadmium toxicity. J. Soil Sci. Plant Nutr. 14, 447–458.
Arya, S. K., Basu, A., and Mukherjee, A. (2013). Lead induced genotoxicity and cytotoxicity in root cells of Allium cepa and Vicia faba. Nucleus 56, 183–189. doi: 10.1007/s13237-013-0099-z
Asha, S., and Vidyavathi, M. (2009). Cunninghamella–a microbial model for drug metabolism studies–a review. Biotechnol. Adv. 27, 16–29. doi: 10.1016/j.biotechadv.2008.07.005
Aslam, R., Bhat, T. M., Choudhary, S., and Ansari, M. Y. K. (2017). An overview on genotoxicity of heavy metal in a spice crop (Capsicum annuum L.) in respect to cyto-morphological behaviour. Caryologia 70, 42–47. doi: 10.1080/00087114.2016.1258884
Atkinson, B. W., Bux, F., and Kasan, H. C. (1998). Considerations for application of biosorption technology to remediate metal-contaminated industrial effluents. Water SA 24, 129–135.
Ayangbenro, A. S., and Babalola, O. O. (2017). A new strategy for heavy metal polluted environments: a review of microbial biosorbents. Int. J. Environ. Res. Public Health 14:94. doi: 10.3390/ijerph14010094
Azad, M. A. K., Amin, L., and Sidik, N. M. (2014). Genetically engineered organisms for bioremediation of pollutants in contaminated sites. Chin. Sci. Bull. 59, 703–714. doi: 10.1007/s11434-013-0058-8
Azubuike, C. C., Chikere, C. B., and Okpokwasili, G. C. (2016). Bioremediation techniques–classification based on site of application: principles, advantages, limitations and prospects. World J. Microbiol. Biotechnol. 32:180. doi: 10.1007/s11274-016-2137-x
Baragaño, D., Forján, R., Welte, L., and Gallego, J. L. R. (2020). Nanoremediation of As and metals polluted soils by means of graphene oxide nanoparticles. Sci. Rep. 10:1896. doi: 10.1038/s41598-020-58852-4
Barbosa, J. S., Cabral, T. M., Ferreira, D. N., Agnez-Lima, L. F., and Batistuzzo de Medeiros, S. R. (2010). Genotoxicity assessment in aquatic environment impacted by the presence of heavy metals. Ecotoxicol. Environ. Saf. 73, 320–325. doi: 10.1016/j.ecoenv.2009.10.008
Barkay, T., Miller, S. M., and Summers, A. O. (2003). Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol. Rev. 27, 355–384. doi: 10.1016/S0168-6445(03)00046-9
Beveridge, T. J. (1989). Role of cellular design in bacterial metal accumulation and mineralization. Annu. Rev. Microbiol. 43, 147–171. doi: 10.1146/annurev.mi.43.100189.001051
Bewley, R. J. F., and Stotzky, G. (1983). Effects of cadmium and zinc on microbial activity in soil; influence of clay minerals. Part I: metals added individually. Sci. Total Environ. 31, 41–55. doi: 10.1016/0048-9697(83)90055-4
Bhati, T., Gupta, R., Yadav, N., Singh, R., Fuloria, A., Waziri, A., et al. (2019). Assessment of bioremediation potential of Cellulosimicrobium sp. for treatment of multiple heavy metals. Microbiol. Biotechnol. Lett. 47, 269–277. doi: 10.4014/mbl.1808.08006
Bhatt, P., Bhatt, K., Huang, Y., Lin, Z., and Chen, S. (2020a). Esterase is a powerful tool for the biodegradation of pyrethroid insecticides. Chemosphere 244:125507. doi: 10.1016/J.CHEMOSPHERE.2019.125507
Bhatt, P., Huang, Y., Zhang, W., Sharma, A., and Chen, S. (2020b). Enhanced cypermethrin degradation kinetics and metabolic pathway in Bacillus thuringiensis strain SG4. Microorganisms 8:223. doi: 10.3390/microorganisms8020223
Bhatt, P., Sethi, K., Gangola, S., Bhandari, G., Verma, A., Adnan, M., et al. (2020c). Modeling and simulation of atrazine biodegradation in bacteria and its effect in other living systems. J. Biomol. Struct. Dyn. 40, 3285–3295. doi: 10.1080/07391102.2020.1846623
Bissonnette, L., St-Arnaud, M., and Labrecque, M. (2010). Phytoextraction of heavy metals by two Salicaceae clones in symbiosis with arbuscular mycorrhizal fungi during the second year of a field trial. Plant Soil 332, 55–67. doi: 10.1007/s11104-009-0273-x
Blindauer, C., Harrison, M., Parkinson, J., Robinson, N., and Sadler, P. (2008). Isostructural replacement of zinc by cadmium in bacterial metallothionein. Metal Ions Biol. Med. 10, 167–173. doi: 10.1016/0300-483x(92)90110-z
Bojórquez, C., Frías Espericueta, M. G., and Voltolina, D. (2016). Removal of cadmium and lead by adapted strains of Pseudomonas aeruginosa and Enterobacter cloacae. Rev. Int. Contaminación Ambiental 32, 407–412. doi: 10.20937/rica.2016.32.04.04
Bramhachari, P. V., Kishor, P. B. K., Ramadevi, R., Rao, B. R., and Dubey, S. K. (2007). Isolation and characterization of mucous exopolysaccharide (EPS) produced by Vibrio furnissii strain VB0S3. Microbiol. Biotechnol. 17, 44–51.
Briffa, J., Sinagra, E., and Blundell, R. (2020). Heavy metal pollution in the environment and their toxicological effects on humans. Heliyon 6:e04691. doi: 10.1016/j.heliyon.2020.e04691
Brunetti, G., Farrag, K., Soler-Rovira, P., Ferrara, M., Nigro, F., and Senesi, N. (2012). The effect of compost and Bacillus licheniformis on the phytoextraction of Cr, Cu, Pb and Zn by three brassicaceae species from contaminated soils in the Apulia region, Southern Italy. Geoderma 170, 322–330. doi: 10.1016/j.geoderma.2011.11.029
Burges, A., Epelde, L., and Garbisu, C. (2015). Impact of repeated single-metal and multi-metal pollution events on soil quality. Chemosphere 120, 8–15. doi: 10.1016/j.chemosphere.2014.05.037
Caetano, A. L., Marques, C. R., Gonçalves, F., da Silva, E. F., and Pereira, R. (2016). Copper toxicity in a natural reference soil: ecotoxicological data for the derivation of preliminary soil screening values. Ecotoxicology 25, 163–177. doi: 10.1007/s10646-015-1577-7
Cai, P., Gao, J., and Zhou, Y. (2019). CRISPR-mediated genome editing in non-conventional yeasts for biotechnological applications. Microb. Cell Fact. 18:63. doi: 10.1186/s12934-019-1112-2
Cao, X., Alabresm, A., Chen, Y. P., Decho, A. W., and Lead, J. (2020). Improved metal remediation using a combined bacterial and nanoscience approach. Sci. Total Environ. 704:135378. doi: 10.1016/j.scitotenv.2019.135378
Carlisle, D. M., and Clements, W. H. (2005). Leaf litter breakdown, microbial respiration and shredder production in metal-polluted streams. Freshwater Biol. 50, 380–390. doi: 10.1111/j.1365-2427.2004.01323.x
Carlot, M., Giacomini, A., and Casella, S. (2002). Aspects of plant-microbe interactions in heavy metal polluted soil. Acta Biotechnol. 22, 13–20. doi: 10.1002/1521-3846(200205)22:1/2<13::aid-abio13>3.0.co;2-9
Carrasco-Gil, S., Estebaranz-Yubero, M., Medel-Cuesta, D., Millán, R., and Hernández, L. E. (2012). Influence of nitrate fertilization on Hg uptake and oxidative stress parameters in alfalfa plants cultivated in a Hg-polluted soil. Environ. Exp. Bot. 75, 16–24. doi: 10.1016/j.envexpbot.2011.08.013
Cervantes, C., Campos-García, J., Devars, S., Gutiérrez-Corona, F., Loza-Tavera, H., Torres-Guzmán, J. C., et al. (2001). Interactions of chromium with microorganisms and plants. FEMS Microbiol. Rev. 25, 335–347. doi: 10.1111/j.1574-6976.2001.tb00581.x
Chakravarty, B., and Srivastava, S. (1992). Toxicity of some heavy metals in vivo and in vitro in Helianthus annuus. Mutat. Res. Lett. 283, 287–294. doi: 10.1016/0165-7992(92)90061-L
Chellaiah, E. R. (2018). Cadmium (heavy metals) bioremediation by Pseudomonas aeruginosa: a minireview. Appl. Water Sci. 8, 1–10. doi: 10.9734/ajsspn/2017/36868
Chen, L., Hu, J. Y., and Wang, S. Q. (2012). The role of antioxidants in photoprotection: a critical review. J. Am. Acad. Dermatol. 67, 1013–1024. doi: 10.1016/j.jaad.2012.02.009
Chen, S., and Wilson, D. B. (1997). Genetic engineering of bacteria and their potential for Hg2+ bioremediation. Biodegradation 8, 97–103. doi: 10.1023/A:1008233704719
Chen, T. B., Wong, J. W. C., Zhou, H. Y., and Wong, M. H. (1997). Assessment of trace metal distribution and contamination in surface soils of Hong Kong. Environ. Pollut. 96, 61–68. doi: 10.1016/s0269-7491(97)00003-1
Cho, S., Shin, J., and Cho, B.-K. (2018). Applications of CRISPR/Cas system to bacterial metabolic engineering. Int. J. Mol. Sci. 19:1089. doi: 10.3390/ijms19041089
Circu, M. L., and Aw, T. Y. (2010). Reactive oxygen species, cellular redox systems, and apoptosis. Free Radic. Biol. Med. 48, 749–762. doi: 10.1016/j.freeradbiomed.2009.12.022
Cumberland, S. A., Douglas, G., Grice, K., and Moreau, J. W. (2016). Uranium mobility in organic matter-rich sediments: a review of geological and geochemical processes. Earth Sci. Rev. 159, 160–185. doi: 10.1016/j.earscirev.2016.05.010
Cuypers, A., Karen, S., Jos, R., Kelly, O., Els, K., Tony, R., et al. (2011). The cellular redox state as a modulator in cadmium and copper responses in Arabidopsis thaliana seedlings. J. Plant Physiol. 168, 309–316. doi: 10.1016/j.jplph.2010.07.010
Cycon, M., and Piotrowska-Seget, Z. (2016). Pyrethroid-degrading microorganisms and their potential for the bioremediation of contaminated soils: a review. Front. Microbiol. 7:1463. doi: 10.3389/fmicb.2016.01463
D’Souza, S. F. (2001). Microbial biosensors. Biosens. Bioelectron. 16, 337–353. doi: 10.1016/S0956-5663(01)00125-7
Dai, J., Becquer, T., Rouiller, J. H., Reversat, G., Bernhard-Reversat, F., and Lavelle, P. (2004). Influence of heavy metals on C and N mineralisation and microbial biomass in Zn-, Pb-, Cu-, and Cd-contaminated soils. Appl. Ecol. 25, 99–109. doi: 10.1016/j.apsoil.2003.09.003
Das, D., Das, N., and Mathew, L. (2010). Kinetics, equilibrium and thermodynamic studies on biosorption of Ag (I) from aqueous solution by macrofungus Pleurotus platypus. J. Hazard. Mater. 184, 765–774. doi: 10.1016/j.jhazmat.2010.08.105
de Lorenzo, V. (2009). Recombinant bacteria for environmental release: what went wrong and what we have learnt from it. Clin. Microbiol. Infect. 15, 63–65. doi: 10.1111/j.1469-0691.2008.02683.x
de Quadros, P. D., Zhalnina, K., Davis-Richardson, A. G., Drew, J. C., Menezes, F. B., Flávio, A., et al. (2016). Coal mining practices reduce the microbial biomass, richness and diversity of soil. Appl. Soil Ecol. 98, 195–203. doi: 10.1016/j.apsoil.2015.10.016
deCatanzaro, J. B., and Hutchinson, T. C. (1985). Effects of nickel addition on nitrogen mineralization, nitrification, and nitrogen leaching in some boreal forest soils. Water Air Soil Pollut. 24, 153–164.
Deckwer, W.-D., Becker, F. U., Ledakowicz, S., and Wagner-Döbler, I. (2004). Microbial removal of ionic mercury in a three-phase fluidized bed reactor. Environ. Sci. Technol. 38, 1858–1865. doi: 10.1021/es0300517
Dembitsky, V. M., and Rezanka, T. (2003). Natural occurrence of arseno compounds in plants, lichens, fungi, algal species, and microorganisms. Plant Sci. 165, 1177–1192. doi: 10.1016/j.plantsci.2003.08.007
Dermont, G., Bergeron, M., Mercier, G., and Richer-Laflèche, M. (2008). Soil washing for metal removal: a review of physical/chemical technologies and field applications. J. Hazard. Mater. 152, 1–31. doi: 10.1016/j.jhazmat.2007.10.043
Devatha, C. P., Jagadeesh, K., and Patil, M. (2018). Effect of green synthesized iron nanoparticles by Azardirachta indica in different proportions on antibacterial activity. Environ. Nanotechnol. Monit. Manag. 9, 85–94. doi: 10.1016/j.enmm.2017.11.007
Diep, P., Mahadevan, R., and Yakunin, A. F. (2018). Heavy metal removal by bioaccumulation using genetically engineered microorganisms. Front. Bioeng. Biotechnol. 6:157. doi: 10.3389/fbioe.2018.00157
Dimkpa, C. O., Svatoš, A., Dabrowska, P., Schmidt, A., Boland, W., and Kothe, E. (2008). Involvement of siderophores in the reduction of metal-induced inhibition of auxin synthesis in Streptomyces spp. Chemosphere 74, 19–25.
Dingjan, I., Verboogen, D. R., Paardekooper, L. M., Revelo, N. H., Sittig, S. P., Visser, L. J., et al. (2016). Lipid peroxidation causes endosomal antigen release for cross-presentation. Sci. Rep. 6:22064. doi: 10.1038/srep22064
Dixit, R., Malaviya, D., Pandiyan, K., Singh, U. B., Sahu, A., Shukla, R., et al. (2015). Bioremediation of heavy metals from soil and aquatic environment: an overview of principles and criteria of fundamental processes. Sustainability 7, 2189–2212. doi: 10.3390/su7022189
Dursun, A. Y., Uslu, G., Cuci, Y., and Aksu, Z. (2003). Bioaccumulation of copper (II), lead (II) and chromium (VI) by growing Aspergillus niger. Process Biochem. 38, 1647–1651. doi: 10.1016/s0032-9592(02)00075-4
El-Kady, A. A., and Abdel-Wahhab, M. A. (2018). Occurrence of trace metals in foodstuffs and their health impact. Trends Food Sci. Technol. 75, 36–45. doi: 10.1016/j.tifs.2018.03.001
Fang, L., Zhou, C., Cai, P., Chen, W., Rong, X., Dai, K., et al. (2011). Binding characteristics of copper and cadmium by cyanobacterium Spirulina platensis. J. Hazard. Mater. 190, 810–815. doi: 10.1016/j.jhazmat.2011.03.122
Febrianto, J., Kosasih, A. N., Sunarso, J., Ju, Y.-H., Indraswati, N., and Ismadji, S. (2009). Equilibrium and kinetic studies in adsorption of heavy metals using biosorbent: a summary of recent studies. J. Hazard. Mater. 162, 616–645. doi: 10.1016/j.jhazmat.2008.06.042
Ferreira, V., Koricheva, J., Duarte, S., Niyogi, D. K., and Guérold, F. (2016). Effects of anthropogenic heavy metal contamination on litter decomposition in streams – a meta-analysis. Environ. Pollut. 210, 261–270. doi: 10.1016/j.envpol.2015.12.060
Fu, Y.-Q., Li, S., Zhu, H.-Y., Jiang, R., and Yin, L.-F. (2012). Biosorption of copper (II) from aqueous solution by mycelial pellets of Rhizopus oryzae. Afr. J. biotechnol. 11, 1403–1411.
Gadd, G. M. (2000). Bioremedial potential of microbial mechanisms of metal mobilization and immobilization. Curr. Opin. Biotechnol. 11, 271–279. doi: 10.1016/s0958-1669(00)00095-1
Gadd, G. M. (2010). Metals, minerals and microbes: geomicrobiology and bioremediation. Microbiology 156, 609–643. doi: 10.1099/mic.0.037143-0
Galiulin, R. V., and Galiulina, R. A. (2008). Removing heavy metals from soil with plants. Herald Russ. Acad. Sci. 78, 141–143. doi: 10.1134/s1019331608020044
Gastaldo, J., Viau, M., Bouchot, M., Joubert, A., Charvet, A.-M., and Foray, N. (2008). Induction and repair rate of DNA damage: a unified model for describing effects of external and internal irradiation and contamination with heavy metals. J. Theor. Biol. 251, 68–81. doi: 10.1016/j.jtbi.2007.10.034
Gavrilescu, M. (2004). Removal of heavy metals from the environment by biosorption. Eng. Life Sci. 4, 219–232. doi: 10.1002/elsc.200420026
Geva, P., Kahta, R., Nakonechny, F., Aronov, S., and Nisnevitch, M. (2016). Increased copper bioremediation ability of new transgenic and adapted Saccharomyces cerevisiae strains. Environ. Sci. Pollut. Res. 23, 19613–19625. doi: 10.1007/s11356-016-7157-4
Gichner, T., Patková, Z., Száková, J., and Demnerová, K. (2006). Toxicity and DNA damage in tobacco and potato plants growing on soil polluted with heavy metals. Ecotoxicol. Environ. Saf. 65, 420–426. doi: 10.1016/j.ecoenv.2005.08.006
Giller, K. E., Witter, E., and Mcgrath, S. P. (1998). Toxicity of heavy metals to microorganisms and microbial processes in agricultural soils: a review. Soil Biol. Biochem 30, 1389–1414. doi: 10.1016/s0038-0717(97)00270-8
Giovanella, P., Cabral, L., Bento, F. M., Gianello, C., and Camargo, F. A. O. (2016). Mercury (II) removal by resistant bacterial isolates and mercuric (II) reductase activity in a new strain of Pseudomonas sp. B50A. New Biotechnol. 33, 216–223. doi: 10.1016/j.nbt.2015.05.006
Glick, B. R. (2003). Phytoremediation: synergistic use of plants and bacteria to clean up the environment. Biotechnol. Adv. 21, 383–393. doi: 10.1016/s0734-9750(03)00055-7
Gołêbiewski, M., Deja-Sikora, E., Cichosz, M., Tretyn, A., and Wróbel, B. (2014). 16S rDNA pyrosequencing analysis of bacterial community in heavy metals polluted soils. Microb. Ecol. 67, 635–647. doi: 10.1007/s00248-013-0344-7
Gong, T., Xu, X., Dang, Y., Kong, A., Wu, Y., Liang, P., et al. (2018). An engineered Pseudomonas putida can simultaneously degrade organophosphates, pyrethroids and carbamates. Sci. Total Environ. 628–629, 1258–1265. doi: 10.1016/j.scitotenv.2018.02.143
Grujić, S., Vasić, S., Radojević, I., Čomić, L., and Ostojić, A. (2017). Comparison of the Rhodotorula mucilaginosa biofilm and planktonic culture on heavy metal susceptibility and removal potential. Water Air Soil Pollut. 228, 73.
Gupta, A. K., Singh, R. P., Singh, A., and Ibrahim, M. H. (2010). “Effects of heavy metal and metalloid contamination on the soil microbial response: an overview,” in Microbial Ecology of Tropical Soils, eds A. S. F. de Araújo and M. D. V. B. Figueiredo (Hauppauge, NY: Nova Science Publisher), 1–16. doi: 10.1002/ieam.1513
Hammaini, A., González, F., Ballester, A., Blázquez, M. L., and Munoz, J. A. (2007). Biosorption of heavy metals by activated sludge and their desorption characteristics. J. Environ. Manag. 84, 419–426. doi: 10.1016/j.jenvman.2006.06.015
Hamsa, N., Yogesh, G. S., Koushik, U., and Patil, L. (2017). Nitrogen transformation in soil: effect of heavy metals. Int. J. Curr. Microbiol. Appl. Sci 6, 816–832. doi: 10.20546/ijcmas.2017.605.092
Hansen, H. K., Ribeiro, A., and Mateus, E. (2006). Biosorption of arsenic (V) with Lessonia nigrescens. Miner. Eng. 19, 486–490. doi: 10.1016/j.mineng.2005.08.018
Harris, G. K., and Shi, X. (2003). Signaling by carcinogenic metals and metal-induced reactive oxygen species. Mutat. Res. 533, 183–200. doi: 10.1016/j.mrfmmm.2003.08.025
Hemmat-Jou, M. H., Safari-Sinegani, A. A., Mirzaie-Asl, A., and Tahmourespour, A. (2018). Analysis of microbial communities in heavy metals-contaminated soils using the metagenomic approach. Ecotoxicology 27, 1281–1291. doi: 10.1007/s10646-018-1981-x
Higham, D. P., Sadler, P. J., and Scawen, M. D. (1986). Cadmium-binding proteins in Pseudomonas putida: pseudothioneins. Environ. Health Perspect. 65, 5–11. doi: 10.1289/ehp.86655
Hirata, A., Corcoran, G. B., and Hirata, F. (2011). Carcinogenic heavy metals, As3+ and Cr6+, increase affinity of nuclear mono-ubiquitinated annexin A1 for DNA containing 8-oxo-guanosine, and promote translesion DNA synthesis. Toxicol. Appl. Pharmacol. 252, 159–164. doi: 10.1016/j.taap.2011.01.022
Hough, R. L., Breward, N., Young, S. D., Crout, N. M., Tye, A. M., Moir, A. M., et al. (2004). Assessing potential risk of heavy metal exposure from consumption of home produced vegetables by urban populations. Environ. Health Perspect. 112, 215–221. doi: 10.1289/ehp.5589
Hogsden, K. L., and Harding, J. S. (2012). Consequences of acid mine drainage for the structure and function of benthic stream communities: a review. Freshw. Sci. 31, 108–120. doi: 10.1899/11-091.1
Hu, N., Luo, Y., Song, J., Wu, L., and Zhang, H. (2010). Influences of soil organic matter, pH and temperature on Pb sorption by four soils in Yangtze River Delta. Acta Pedol. Sin. 47, 246–252.
Igiri, B. E., Okoduwa, S. I. R., Idoko, G. O., Akabuogu, E. P., Adeyi, A. O., and Ejiogu, I. K. (2018). Toxicity and bioremediation of heavy metals contaminated ecosystem from tannery wastewater: a review. J. Toxicol. 2018:2568038. doi: 10.1155/2018/2568038
Illmer, P., and Schinner, F. (1991). Effects of lime and nutrient salts on the microbiological activities of forest soils. Biol. Fertil. Soils 11, 261–266. doi: 10.1007/bf00335845
Isaac, C. P. J., and Sivakumar, A. (2013). Removal of lead and cadmium ions from water using Annona squamosa shell: kinetic and equilibrium studies. Desalination Water Treat. 51, 7700–7709. doi: 10.1080/19443994.2013.778218
Islam, E., Khan, M. T., and Irem, S. (2015). Biochemical mechanisms of signaling: perspectives in plants under arsenic stress. Ecotoxicol. Environ. Saf. 114, 126–133. doi: 10.1016/j.ecoenv.2015.01.017
Jafari, M., Abdollahi, H., Shafaei, S. Z., Gharabaghi, M., Jafari, H., Akcil, A., et al. (2019). Acidophilic bioleaching: a review on the process and effect of organic–inorganic reagents and materials on its efficiency. Miner. Process. Extract. Metall. Rev. 40, 87–107. doi: 10.1080/08827508.2018.1481063
Jain, P. K., Gupta, V. K., Bajpai, V., Lowry, M., and Jaroli, D. P. (2011). “GMO’s: perspective of bioremediation,” in Recent Advances in Environmental Biotechnology, eds P. K. Jain, V. K. Gupta, and V. Bajpai (Saarbrücken: LAP Lambert Academic Publishing AG and Co. KG), 6–23.
Jibril, S. A., Hassan, S. A., Ishak, C. F., and Megat Wahab, P. E. (2017). Cadmium toxicity affects phytochemicals and nutrient elements composition of lettuce (Lactuca sativa L.). Adv. Agric. 2017:1236830. doi: 10.1155/2017/1236830
Joner, E. J., and Leyval, C. (1997). Uptake of 109Cd by roots and hyphae of a Glomus mosseae/Trifolium subterraneum mycorrhiza from soil amended with high and low concentrations of cadmium. New Phytol. 135, 353–360. doi: 10.1046/j.1469-8137.1997.00633.x
Jones, G. C., Corin, K. C., van Hille, R. P., and Harrison, S. T. (2011). The generation of toxic reactive oxygen species (ROS) from mechanically activated sulphide concentrates and its effect on thermophilic bioleaching. Miner. Eng. 241, 1198–1208. doi: 10.1016/j.mineng.2011.05.016
Joutey, N. T., Sayel, H., Bahafid, W., and El Ghachtouli, N. (2015). Mechanisms of hexavalent chromium resistance and removal by microorganisms. Rev. Environ. Contam. Toxicol. 233, 45–69. doi: 10.1007/978-3-319-10479-9_2
Jovancicevic, V., Bockris, J., Carbajal, J. L., Zelenay, P., and Mizuno, T. (1986). Adsorption and absorption of chloride ions on passive iron systems. J. Electrochem. Soc. 133:2219. doi: 10.1149/1.2108377
Kang, J. W. (2014). Removing environmental organic pollutants with bioremediation and phytoremediation. Biotechnol. Lett. 36, 1129–1139. doi: 10.1007/s10529-014-1466-9
Kapoor, A., and Viraraghavan, T. (1997). Heavy metal biosorption sites in Aspergillus niger. Bioresour. Technol. 61, 221–227. doi: 10.1016/s0960-8524(97)00055-2
Khati, P., Sharma, A., Gangola, S., Kumar, R., Bhatt, P., and Kumar, G. (2017). Impact of agri-usable nanocompounds on soil microbial activity: an indicator of soil health. CLEAN Soil Air Water 45:1600458. doi: 10.1002/clen.201600458
Kozlov, M. V., and Zvereva, E. L. (2015). Decomposition of birch leaves in heavily polluted industrial barrens: relative importance of leaf quality and site of exposure. Environ. Sci. Pollut. Res. 22, 9943–9950. doi: 10.1007/s11356-015-4165-8
Kumar, R., Sharma, A. K., Singh, P., Dhir, B., and Mehta, D. (2014). Potential of some fungal and bacterial species in bioremediation of heavy metals. J. Nucl. Phys. Mater. Sci. Radiat. Appl. 1, 213–223.
Kumar, S., Kaushik, G., Dar, M. A., Nimesh, S., Lopez-Chuken, U. J., and Villarreal-Chiu, J. F. (2018). Microbial degradation of organophosphate pesticides: a review. Pedosphere 28, 190–208. doi: 10.1016/S1002-0160(18)60017-7
Kushwaha, A., Rani, R., Kumar, S., Thomas, T., David, A. A., and Ahmed, M. (2017). A new insight to adsorption and accumulation of high lead concentration by exopolymer and whole cells of lead-resistant bacterium Acinetobacter junii L. Pb1 isolated from coal mine dump. Environ. Sci. Pollut. Res. 24, 10652–10661. doi: 10.1007/s11356-017-8752-8
Lan, M. M., Liu, C., Liu, S. J., Qiu, R. L., and Tang, Y. T. (2020). Phytostabilization of cd and Pb in highly polluted farmland soils using ramie and amendments. Int. J. Environ. Res. Public Health 17:1661. doi: 10.3390/ijerph17051661
Lerebours, A., To, V. V., and Bourdineaud, J. (2016). Danio rerio ABC transporter genes abcb3 and abcb7 play a protecting role against metal contamination. J. Appl. Toxicol. 36, 1551–1557. doi: 10.1002/jat.3313
Li, F., Zheng, Y., Tian, J., Ge, F., Liu, X., Tang, Y., et al. (2019). Cupriavidus sp. strain Cd02-mediated pH increase favoring bioprecipitation of Cd2+ in medium and reduction of cadmium bioavailability in paddy soil. Ecotoxicol. Environ. Saf. 184:109655.
Lima, A. I. G., Corticeiro, S. C., Figueira, E. M., and de, A. P. (2006). Glutathione-mediated cadmium sequestration in Rhizobium leguminosarum. Enzyme Microb. Technol. 39, 763–769. doi: 10.1016/j.enzmictec.2005.12.009
Lin, C.-C., and Lin, H.-L. (2005). Remediation of soil contaminated with the heavy metal (Cd2+). J. Hazard. Mater. 122, 7–15.
Liu, S.-H., Zeng, G.-M., Niu, Q.-Y., Liu, Y., Zhou, L., Jiang, L.-H., et al. (2017). Bioremediation mechanisms of combined pollution of PAHs and heavy metals by bacteria and fungi: a mini review. Bioresour. Technol. 224, 25–33. doi: 10.1016/j.biortech.2016.11.095
Liu, X.-X., Hu, X., Cao, Y., Pang, W.-J., Huang, J.-Y., Guo, P., et al. (2019). Biodegradation of phenanthrene and heavy metal removal by acid-tolerant Burkholderia fungorum FM-2. Front. Microbiol. 10:408. doi: 10.3389/fmicb.2019.00408
Lu, W.-B., Shi, J.-J., Wang, C.-H., and Chang, J.-S. (2006). Biosorption of lead, copper and cadmium by an indigenous isolate Enterobacter sp. J1 possessing high heavy-metal resistance. J. Hazard. Mater. 134, 80–86. doi: 10.1016/j.jhazmat.2005.10.036
Ma, L., and Zhang, W. (2008). Enhanced biological treatment of industrial wastewater with bimetallic zero-valent iron. Environ. Sci. Technol. 42, 5384–5389. doi: 10.1021/es801743s
Maksymiec, W. (2007). Signaling responses in plants to heavy metal stress. Acta Physiol. Plant. 29:177. doi: 10.1007/s11738-007-0036-3
Malik, A. (2004). Metal bioremediation through growing cells. Environ. Int. 30, 261–278. doi: 10.1016/j.envint.2003.08.001
Malik, Z., Ahmad, M., Abassi, G. H., Dawood, M., Hussain, A., and Jamil, M. (2017). “Agrochemicals and soil microbes: interaction for soil health,” in Xenobiotics in the Soil Environment, eds M. Hashmi, V. Kumar, and A. Varma (Cham: Springer), 139–152. doi: 10.1007/978-3-319-47744-2_11
Manoj, S. R., Karthik, C., Kadirvelu, K., Arulselvi, P. I., Shanmugasundaram, T., Bruno, B., et al. (2020). Understanding the molecular mechanisms for the enhanced phytoremediation of heavy metals through plant growth promoting rhizobacteria: a review. J. Environ. Manag. 254, 109779. doi: 10.1016/j.jenvman.2019.109779
Marschner, B., and Kalbitz, K. (2003). Controls of bioavailability and biodegradability of dissolved organic matter in soils. Geoderma 113, 211–235. doi: 10.1016/s0016-7061(02)00362-2
Miglani, G. S. (2017). Genome editing in crop improvement: present scenario and future prospects. J. Crop Improv. 31, 453–559. doi: 10.1080/15427528.2017.1333192
Mishra, D., Kim, D.-J., Ahn, J.-G., and Rhee, Y.-H. (2005). Bioleaching: a microbial process of metal recovery; a review. Metals Mater. Int. 11, 249–256. doi: 10.1007/bf03027450
Morton-Bermea, O., Álvarez, E. H., Gaso, I., and Segovia, N. (2002). Heavy metal concentrations in surface soils from Mexico City. Bull. Environ. Contam. Toxicol. 68, 383–388. doi: 10.1007/s001280265
Mosa, K. A., Saadoun, I., Kumar, K., Helmy, M., and Dhankher, O. P. (2016). Potential biotechnological strategies for the cleanup of heavy metals and metalloids. Front. Plant Sci. 7:303. doi: 10.3389/fpls.2016.00303
Mudila, H., Prasher, P., Kumar, M., Kapoor, H., Kumar, A., Zaidi, M. G. H., et al. (2019). An insight into cadmium poisoning and its removal from aqueous sources by graphene adsorbents. Int. J. Environ. Health Res. 29, 1–21. doi: 10.1080/09603123.2018.1506568
Nagvenkar, G. S., and Ramaiah, N. (2010). Arsenite tolerance and biotransformation potential in estuarine bacteria. Ecotoxicology 19, 604–613. doi: 10.1007/s10646-009-0429-8
Naskar, A., Majumder, R., and Goswami, M. (2020). Bioaccumulation of Ni (II) on growing cells of Bacillus sp.: response surface modeling and mechanistic insight. Environ. Technol. Innov. 20:101057. doi: 10.1016/j.eti.2020.101057
Nath, S., Deb, B., and Sharma, I. (2018). Isolation of toxic metal-tolerant bacteria from soil and examination of their bioaugmentation potentiality by pot studies in cadmium- and lead-contaminated soil. Int. Microbiol. 21, 35–45. doi: 10.1007/s10123-018-0003-4
Nayak, A. K., Raja, R., Rao, K. S., Shukla, A. K., Mohanty, S., Shahid, M., et al. (2015). Effect of fly ash application on soil microbial response and heavy metal accumulation in soil and rice plant. Ecotoxicol. Environ. Saf. 114, 257–262. doi: 10.1016/j.ecoenv.2014.03.033
Neubauer, U., Furrer, G., Kayser, A., and Schulin, R. (2000). Siderophores, NTA, and citrate: potential soil amendments to enhance heavy metal mobility in phytoremediation. Int. J. Phytoremediation 2, 353–368. doi: 10.1080/15226510008500044
Ni, H., Xiong, Z., Ye, T., Zhang, Z., Ma, X., and Li, L. (2012). Biosorption of copper (II) from aqueous solutions using volcanic rock matrix-immobilized Pseudomonas putida cells with surface-displayed cyanobacterial metallothioneins. Chem. Eng. J. 204, 264–271. doi: 10.1016/j.cej.2012.05.029
Njoku, K. L., Asunmo, M. O., Ude, E. O., Adesuyi, A. A., and Oyelami, A. O. (2020). The molecular study of microbial and functional diversity of resistant microbes in heavy metal contaminated soil. Environ. Technol. Innov. 17:100606. doi: 10.1016/j.eti.2020.100606
Noroozi, M., Amoozegar, M. A., Pourbabaee, A. A., Naghavi, N. S., and Nourmohammadi, Z. (2017). Isolation and characterization of mercuric reductase by newly isolated halophilic bacterium, Bacillus firmus MN8. Glob. J. Environ. Sci. Manag. 3:427.
Nwuche, C. O., and Ugoji, E. O. (2008). Effects of heavy metal pollution on the soil microbial activity. Int. J. Environ. Sci. Technol. 5, 409–414.
Oliveira, A., and Pampulha, M. E. (2006). Effects of long-term heavy metal contamination on soil microbial characteristics. J. Biosci. Bioeng. 102, 157–161. doi: 10.1263/jbb.102.157
Osmani, M., Bani, A., and Hoxha, B. (2015). Heavy metals and ni phytoextractionin in the metallurgical area soils in Elbasan. Albanian J. Agric. Sci. 14:414.
Pan, J., and Yu, L. (2011). Effects of Cd or/and Pb on soil enzyme activities and microbial community structure. Ecol. Eng. 37, 1889–1894. doi: 10.1016/j.ecoleng.2011.07.002
Panagos, P., Ballabio, C., Lugato, E., Jones, A., Borrelli, P., Scarpa, S., et al. (2018). Potential sources of anthropogenic copper inputs to European agricultural soils. Sustainability 10:2380. doi: 10.3390/su10072380
Pande, V., Pandey, S., Sati, D., Pande, V., and Samant, M. (2020). Bioremediation: an emerging effective approach towards environment restoration. Environ. Sustain. 3, 91–103. doi: 10.1007/s42398-020-00099-w
Park, J. H., Lee, S.-J., Lee, M.-E., and Chung, J. W. (2016). Comparison of heavy metal immobilization in contaminated soils amended with peat moss and peat moss-derived biochar. Environ. Sci. Process. Impacts 18, 514–520. doi: 10.1039/c6em00098c
Peralta-Videa, J. R., Lopez, M. L., Narayan, M., Saupe, G., and Gardea-Torresdey, J. (2009). The biochemistry of environmental heavy metal uptake by plants: implications for the food chain. Int. J. Biochem. Cell Biol. 41, 1665–1677. doi: 10.1016/j.biocel.2009.03.005
Pervaiz, I., Ahmad, S., Madni, M. A., Ahmad, H., and Khaliq, F. H. (2013). Microbial biotransformation: a tool for drug designing. Appl. Biochem. Microbiol. 49, 437–450. doi: 10.7868/s0555109913050097
Plum, L. M., Rink, L., and Haase, H. (2010). The essential toxin: impact of zinc on human health. Int. J. Environ. Res. Public Health 7, 1342–1365. doi: 10.3390/ijerph7041342
Pollmann, K., Raff, J., Merroun, M., Fahmy, K., and Selenska-Pobell, S. (2006). Metal binding by bacteria from uranium mining waste piles and its technological applications. Biotechnol. Adv. 24, 58–68. doi: 10.1016/j.biotechadv.2005.06.002
Pourrut, B., Jean, S., Silvestre, J., and Pinelli, E. (2011). Lead-induced DNA damage in Vicia faba root cells: potential involvement of oxidative stress. Mutat. Res. 726, 123–128. doi: 10.1016/j.mrgentox.2011.09.001
Pratush, A., Kumar, A., and Hu, Z. (2018). Adverse effect of heavy metals (As, Pb, Hg, and Cr) on health and their bioremediation strategies: a review. Int. Microbiol. 21, 97–106. doi: 10.1007/s10123-018-0012-3
Qiao, W., Zhang, Y., Xia, H., Luo, Y., Liu, S., Wang, S., et al. (2019). Bioimmobilization of lead by Bacillus subtilis X3 biomass isolated from lead mine soil under promotion of multiple adsorption mechanisms. R. Soc. Open Sci. 6:181701. doi: 10.1098/rsos.181701
Qin, R., Wang, C., Chen, D., Björn, L. O., and Li, S. (2015). Copper-induced root growth inhibition of Allium cepa var. agrogarum L. involves disturbances in cell division and DNA damage. Environ. Toxicol. Chem. 34, 1045–1055. doi: 10.1002/etc.2884
Rajkumar, M., Ae, N., Prasad, M. N. V., and Freitas, H. (2010). Potential of siderophore-producing bacteria for improving heavy metal phytoextraction. Trends Biotechnol. 28, 142–149. doi: 10.1016/j.tibtech.2009.12.002
Rani, A., and Goel, R. (2009). “Strategies for crop improvement in contaminated soils using metal-tolerant bioinoculants,” in Microbial Strategies for Crop Improvement, eds M. Khan, A. Zaidi, and J. Musarrat (Berlin: Springer), 85–104. doi: 10.1007/978-3-642-01979-1_5
Rasulov, B. A., Yili, A., and Aisa, H. A. (2013). Biosorption of metal ions by exopolysaccharide produced by Azotobacter chroococcum XU1. J. Environ. Protect. 4:36802.
Remenar, M., Kamlarova, A., Harichova, J., Zámocký, M., and Ferianc, P. (2018). The heavy-metal resistance determinant of newly isolated bacterium from a nickel-contaminated soil in Southwest Slovakia. Pol. J. Microbiol. 67:191. doi: 10.21307/pjm-2018-022
Rodríguez-Tirado, V., Green-Ruiz, C., and Gómez-Gil, B. (2012). Cu and Pb biosorption on Bacillus thioparans strain U3 in aqueous solution: Kinetic and equilibrium studies. Chem. Eng. J. 181, 352–359. doi: 10.1016/j.cej.2011.11.091
Rohwerder, T., Gehrke, T., Kinzler, K., and Sand, W. (2003). Bioleaching review part A. Appl. Microbiol. Biotechnol. 63, 239–248. doi: 10.1007/s00253-003-1448-7
Rojas, L. A., Yáñez, C., González, M., Lobos, S., Smalla, K., and Seeger, M. (2011). Characterization of the metabolically modified heavy metal-resistant Cupriavidus metallidurans strain MSR33 generated for mercury bioremediation. PLoS One 6:e17555. doi: 10.1371/journal.pone.0017555
Romera, E., Gonzalez, F., Ballester, A., Blázquez, M. L., and Munoz, J. A. (2006). Biosorption with algae: a statistical review. Crit. Rev. Biotechnol. 26, 223–235. doi: 10.1080/07388550600972153
Romera, E., González, F., Ballester, A., Blázquez, M. L., and Munoz, J. A. (2007). Comparative study of biosorption of heavy metals using different types of algae. Bioresour. Technol. 98, 3344–3353. doi: 10.1016/j.biortech.2006.09.026
Rother, J. A., Millbank, J. W., and Thornton, I. (1982). Seasonal fluctuations in nitrogen fixation (acetylene reduction) by free-living bacteria in soils contaminated with cadmium, lead and zinc. J. Soil Sci. 33, 101–113. doi: 10.1111/j.1365-2389.1982.tb01751.x
Sati, D., Pande, V., Pandey, S. C., and Samant, M. (2022). Recent advances in PGPR and molecular mechanisms involved in drought stress resistance. J. Soil Sci. Plant Nutr. doi: 10.1007/s42729-021-00724-5
Sayqal, A., and Ahmed, O. B. (2021). Advances in heavy metal bioremediation: an overview. Appl. Bionics Biomech. 2021:1609149. doi: 10.1155/2021/1609149
Schellingen, K., van der Straeten, D., Vandenbussche, F., Prinsen, E., Remans, T., Vangronsveld, J., et al. (2014). Cadmium-induced ethylene production and responses in Arabidopsis thaliana rely on ACS2 and ACS6 gene expression. BMC Plant Biol. 14:214. doi: 10.1186/s12870-014-0214-6
Shahid, M., Pourrut, B., Dumat, C., Nadeem, M., Aslam, M., and Pinelli, E. (2014). Heavy-metal-induced reactive oxygen species: phytotoxicity and physicochemical changes in plants. Rev. Environ. Contam. Toxicol. 232, 1–44. doi: 10.1007/978-3-319-06746-9_1
Shahpiri, A., and Mohammadzadeh, A. (2018). Mercury removal by engineered Escherichia coli cells expressing different rice metallothionein isoforms. Ann. Microbiol. 68, 145–152. doi: 10.1007/s13213-018-1326-2
Shapiro, R. S., Chavez, A., and Collins, J. J. (2018). CRISPR-based genomic tools for the manipulation of genetically intractable microorganisms. Nat. Rev. Microbiol. 16, 333–339. doi: 10.1038/s41579-018-0002-7
Sharma, B., Sarkar, A., Singh, P., and Singh, R. P. (2017). Agricultural utilization of biosolids: a review on potential effects on soil and plant grown. Waste Manag. 64, 117–132. doi: 10.1016/j.wasman.2017.03.002
Sher, S., and Rehman, A. (2019). Use of heavy metals resistant bacteria—a strategy for arsenic bioremediation. Appl. Microbiol. Biotechnol. 103, 6007–6021. doi: 10.1007/s00253-019-09933-6
Silva, S., Silva, P., Oliveira, H., Gaivão, I., Matos, M., Pinto-Carnide, O., et al. (2017). Pb low doses induced genotoxicity in Lactuca sativa plants. Plant Physiol. Biochem. 112, 109–116. doi: 10.1016/j.plaphy.2016.12.026
Singh, R. P., and Agrawal, M. (2008). Potential benefits and risks of land application of sewage sludge. Waste Manag. 28, 347–358. doi: 10.1016/j.wasman.2006.12.010
Singh, R. P., and Agrawal, M. (2010). Variations in heavy metal accumulation, growth and yield of rice plants grown at different sewage sludge amendment rates. Ecotoxicol. Environ. Saf. 73, 632–641. doi: 10.1016/j.ecoenv.2010.01.020
Sinha, S., and Mukherjee, S. K. (2008). Cadmium–induced siderophore production by a high Cd-resistant bacterial strain relieved Cd toxicity in plants through root colonization. Curr. Microbiol. 56, 55–60. doi: 10.1007/s00284-007-9038-z
Sone, Y., Mochizuki, Y., Koizawa, K., Nakamura, R., Pan-Hou, H., Itoh, T., et al. (2013). Mercurial-resistance determinants in Pseudomonas strain K-62 plasmid pMR68. AMB Express 3:41. doi: 10.1186/2191-0855-3-41
Soto, D. F., Recalde, A., Orell, A., Albers, S.-V., Paradela, A., Navarro, C. A., et al. (2019). Global effect of the lack of inorganic polyphosphate in the extremophilic archaeon Sulfolobus solfataricus: a proteomic approach. J. Proteomics 191, 143–152. doi: 10.1016/j.jprot.2018.02.024
Srichandan, H., Pathak, A., Singh, S., Blight, K., Kim, D.-J., and Lee, S. W. (2014). Sequential leaching of metals from spent refinery catalyst in bioleaching–bioleaching and bioleaching–chemical leaching reactor: comparative study. Hydrometallurgy 150, 130–143. doi: 10.1016/j.hydromet.2014.09.019
Srivastava, V., de Araujo, A. S. F., Vaish, B., Bartelt-Hunt, S., Singh, P., and Singh, R. P. (2016). Biological response of using municipal solid waste compost in agriculture as fertilizer supplement. Rev. Environ. Sci. Biotechnol. 15, 677–696. doi: 10.1007/s11157-016-9407-9
Subashchandrabose, S. R., Venkateswarlu, K., Krishnan, K., Naidu, R., Lockington, R., and Megharaj, M. (2018). Rhodococcus wratislaviensis strain 9: an efficient p-nitrophenol degrader with a great potential for bioremediation. J. Hazard. Mater. 347, 176–183. doi: 10.1016/j.jhazmat.2017.12.063
Sun, G. L., Reynolds, E. E., and Belcher, A. M. (2020). Using yeast to sustainably remediate and extract heavy metals from waste waters. Nat. Sustain. 3, 303–311. doi: 10.1038/s41893-020-0478-9
Tamás, L., Mistrík, I., and Zelinová, V. (2017). Heavy metal-induced reactive oxygen species and cell death in barley root tip. Environ. Exp. Bot. 140, 34–40. doi: 10.1016/j.envexpbot.2017.05.016
Tan, W., Peralta-Videa, J. R., and Gardea-Torresdey, J. L. (2018). Interaction of titanium dioxide nanoparticles with soil components and plants: current knowledge and future research needs–a critical review. Environ. Sci. Nano 5, 257–278. doi: 10.1039/c7en00985b
Taran, M., Fateh, R., Rezaei, S., and Gholi, M. K. (2019). Isolation of arsenic accumulating bacteria from garbage leachates for possible application in bioremediation. Iran. J. Microbiol. 11:60.
Teitzel, G. M., and Parsek, M. R. (2003). Heavy metal resistance of biofilm and planktonic Pseudomonas aeruginosa. Appl. Environ. Microbiol. 69, 2313–2320. doi: 10.1128/AEM.69.4.2313-2320.2003
Thapa, G., Sadhukhan, A., Panda, S. K., and Sahoo, L. (2012). Molecular mechanistic model of plant heavy metal tolerance. Biometals 25, 489–505. doi: 10.1007/s10534-012-9541-y
Thatoi, H., Das, S., Mishra, J., Rath, B. P., and Das, N. (2014). Bacterial chromate reductase, a potential enzyme for bioremediation of hexavalent chromium: a review. J. Environ. Manag. 146, 383–399. doi: 10.1016/j.jenvman.2014.07.014
Thelwell, C., Robinson, N. J., and Turner-Cavet, J. S. (1998). An SmtB-like repressor from Synechocystis PCC 6803 regulates a zinc exporter. Proc. Natl. Acad. Sci. 95, 10728–10733. doi: 10.1073/pnas.95.18.10728
Tóth, G., Hermann, T., da Silva, M. R., and Montanarella, L. (2016). Heavy metals in agricultural soils of the European Union with implications for food safety. Environ. Int. 88, 299–309. doi: 10.1016/j.envint.2015.12.017
Trellu, C., Mousset, E., Pechaud, Y., Huguenot, D., van Hullebusch, E. D., Esposito, G., et al. (2016). Removal of hydrophobic organic pollutants from soil washing/flushing solutions: a critical review. J. Hazard. Mater. 306, 149–174. doi: 10.1016/j.jhazmat.2015.12.008
Tyagi, S., Kumar, V., Singh, J., Teotia, P., Bisht, S., and Sharma, S. (2014). Bioremediation of pulp and paper mill effluent by dominant aboriginal microbes and their consortium. Int. J. Environ. Res. 8, 561–568.
van de Poel, B., Smet, D., and van der Straeten, D. (2015). Ethylene and hormonal cross talk in vegetative growth and development. Plant Physiol. 169, 61–72. doi: 10.1104/pp.15.00724
Vardhan, K. H., Kumar, P. S., and Panda, R. C. (2019). A review on heavy metal pollution, toxicity and remedial measures: current trends and future perspectives. J. Mol. Liq. 290:111197. doi: 10.1016/j.molliq.2019.111197
Vásquez-Murrieta, M. S., Migueles-Garduño, I., Franco-Hernández, O., Govaerts, B., and Dendooven, L. (2006). C and N mineralization and microbial biomass in heavy-metal contaminated soil. Eur. J. Soil Biol. 42, 89–98. doi: 10.1016/j.ejsobi.2005.10.002
Velásquez, L., and Dussan, J. (2009). Biosorption and bioaccumulation of heavy metals on dead and living biomass of Bacillus sphaericus. J. Hazard. Mater. 167, 713–716. doi: 10.1016/j.jhazmat.2009.01.044
Venkatachalam, P., Jayalakshmi, N., Geetha, N., Sahi, S. V., Sharma, N. C., Rene, E. R., et al. (2017). Accumulation efficiency, genotoxicity and antioxidant defense mechanisms in medicinal plant Acalypha indica L. under lead stress. Chemosphere 171, 544–553. doi: 10.1016/j.chemosphere.2016.12.092
Verma, N., and Singh, M. (2005). Biosensors for heavy metals. Biometals 18, 121–129. doi: 10.1007/s10534-004-5787-3
Vijayaraj, A. S., Mohandass, C., and Joshi, D. (2019). Microremediation of tannery wastewater by siderophore producing marine bacteria. Environ. Technol. 41, 3619–3632. doi: 10.1080/09593330.2019.1615995
Wang, G., Zhang, Q., Du, W., Ai, F., Yin, Y., Ji, R., et al. (2021). Microbial communities in the rhizosphere of different willow genotypes affect phytoremediation potential in Cd contaminated soil. Sci. Total Environ. 769:145224. doi: 10.1016/j.scitotenv.2021.145224
Wang, J., and Chen, C. (2006). Biosorption of heavy metals by Saccharomyces cerevisiae: a review. Biotechnol. Adv. 24, 427–451. doi: 10.1016/J.BIOTECHADV.2006.03.001
Wang, J., and Chen, C. (2009). Biosorbents for heavy metals removal and their future. Biotechnol. Adv. 27, 195–226. doi: 10.1016/j.biotechadv.2008.11.002
Wang, S., and Zhao, X. (2009). On the potential of biological treatment for arsenic contaminated soils and groundwater. J. Environ. Manag. 90, 2367–2376. doi: 10.1016/j.jenvman.2009.02.001
Wang, Y., Guo, J., and Liu, R. (2001). Biosorption of heavy metals by bacteria isolated from activated sludge. Huan Jing Ke Xue 22, 72–75.
Wierzba, S. (2015). Biosorption of lead (II), zinc (II) and nickel (II) from industrial wastewater by Stenotrophomonas maltophilia and Bacillus subtilis. Pol. J. Chem. Technol. 17, 79–87. doi: 10.1515/pjct-2015-0012
Wolejko, E., Wydro, U., and Loboda, T. (2016). The ways to increase efficiency of soil bioremediation. Ecol. Chem. Eng. 23:155. doi: 10.1515/eces-2016-0011
Wu, X., Cobbina, S. J., Mao, G., Xu, H., Zhang, Z., and Yang, L. (2016). A review of toxicity and mechanisms of individual and mixtures of heavy metals in the environment. Environ. Sci. Pollut. Res. 23, 8244–8259. doi: 10.1007/s11356-016-6333-x
Xian, Y., Wang, M., and Chen, W. (2015). Quantitative assessment on soil enzyme activities of heavy metal contaminated soils with various soil properties. Chemosphere 139, 604–608. doi: 10.1016/j.chemosphere.2014.12.060
Yalçın, S., Sezer, S., and Apak, R. (2012). Characterization and lead (II), cadmium (II), nickel (II) biosorption of dried marine brown macro algae Cystoseira barbata. Environ. Sci. Pollut. Res. 19, 3118–3125. doi: 10.1007/s11356-012-0807-2
Yang, H.-C., Fu, H.-L., Lin, Y.-F., and Rosen, B. P. (2012). Pathways of arsenic uptake and efflux. Curr. Top. Membr. 69, 325–358. doi: 10.1016/b978-0-12-394390-3.00012-4
Yang, J., Wang, L., Chang, C., and Liu, T. (1999). Singlet oxygen is the major species participating in the induction of DNA strand breakage and 8-hydroxydeoxyguanosine adduct by lead acetate. Environ. Mol. Mutagen. 33, 194–201. doi: 10.1002/(sici)1098-2280(1999)33:3<194::aid-em3>3.0.co;2-o
Yuan, L., Zhi, W., Liu, Y., Karyala, S., Vikesland, P. J., Chen, X., et al. (2015). Lead toxicity to the performance, viability, and community composition of activated sludge microorganisms. Environ. Sci. Technol. 49, 824–830. doi: 10.1021/es504207c
Yue, Z.-B., Li, Q., Li, C., Chen, T., and Wang, J. (2015). Component analysis and heavy metal adsorption ability of extracellular polymeric substances (EPS) from sulfate reducing bacteria. Bioresour. Technol. 194, 399–402. doi: 10.1016/j.biortech.2015.07.042
Zammit, C. M., Weiland, F., Brugger, J., Wade, B., Winderbaum, L. J., Nies, D. H., et al. (2016). Proteomic responses to gold (III)-toxicity in the bacterium Cupriavidus metallidurans CH34. Metallomics 8, 1204–1216. doi: 10.1039/c6mt00142d
Zhang, S., and Klessig, D. F. (2001). MAPK cascades in plant defense signaling. Trends Plant Sci. 6, 520–527. doi: 10.1016/s1360-1385(01)02103-3
Zhang, W., Chen, L., and Liu, D. (2012). Characterization of a marine-isolated mercury-resistant Pseudomonas putida strain SP1 and its potential application in marine mercury reduction. Appl. Microbiol. Biotechnol. 93, 1305–1314. doi: 10.1007/s00253-011-3454-5
Zhang, W.-M., and Gu, S.-F. (2007). Catalytic effect of activated carbon on bioleaching of low-grade primary copper sulflde ores. Trans. Nonferrous Metals Soc. China 17, 1123–1127. doi: 10.1016/s1003-6326(07)60236-2
Keywords: heavy metals, bioremediation, biosorption, biotransformation, bioleaching
Citation: Pande V, Pandey SC, Sati D, Bhatt P and Samant M (2022) Microbial Interventions in Bioremediation of Heavy Metal Contaminants in Agroecosystem. Front. Microbiol. 13:824084. doi: 10.3389/fmicb.2022.824084
Received: 28 November 2021; Accepted: 31 March 2022;
Published: 06 May 2022.
Edited by:
Pradipta Saha, University of Burdwan, IndiaReviewed by:
Vineet Kumar, National Environmental Engineering Research Institute (CSIR), IndiaRenitta Jobby, Amity University, Mumbai, India
Vipin Kumar Singh, Banaras Hindu University, India
Copyright © 2022 Pande, Pandey, Sati, Bhatt and Samant. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pankaj Bhatt, pankajbhatt.bhatt472@gmail.com; Mukesh Samant, mukeshsamant@gmail.com
 Veni Pande
Veni Pande Satish Chandra Pandey
Satish Chandra Pandey Diksha Sati
Diksha Sati Pankaj Bhatt
Pankaj Bhatt Mukesh Samant
Mukesh Samant