- 1 Department of Cell Signaling, A.N. Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, Moscow, Russia
- 2 Biochemistry Group, Department of Biology and Environmental Sciences, University of Oldenburg, Oldenburg, Germany
- 3 Institute for Biological Instrumentation of the Russian Academy of Sciences, Pushchino, Moscow Region, Russia
Phosphorylation of photoactivated rhodopsin by rhodopsin kinase (RK or GRK1), a first step of the phototransduction cascade turnoff, is under the control of Ca2+/recoverin. Here, we demonstrate that calmodulin, a ubiquitous Ca2+-sensor, can inhibit RK, though less effectively than recoverin does. We have utilized the surface plasmon resonance technology to map the calmodulin binding site in the RK molecule. Calmodulin does not interact with the recoverin-binding site within amino acid residues M1-S25 of the enzyme. Instead, the high affinity calmodulin binding site is localized within a stretch of amino acid residues V150-K175 in the N-terminal regulatory region of RK. Moreover, the inhibitory effect of calmodulin and recoverin on RK activity is synergetic, which is in agreement with the existence of separate binding sites for each Ca2+-sensing protein. The synergetic inhibition of RK by both Ca2+-sensors occurs over a broader range of Ca2+-concentration than by recoverin alone, indicating increased Ca2+-sensitivity of RK regulation in the presence of both Ca2+-sensors. Taken together, our data suggest that RK regulation by calmodulin in photoreceptor cells could complement the well-known inhibitory effect of recoverin on RK.
Introduction
Key aspects of the neuronal activity from neurotransmission to gene expression are regulated by changes in the intracellular free calcium concentration. Effects of calcium on various processes in the cell are mediated by Ca2+ sensor proteins. The important property of these proteins is the ability to change their conformation upon Ca2+-binding, thereby transmitting the signal to the effector enzymes. The effects of calcium in different cells of the most tissues are mediated primarily by ubiquitous calcium sensor calmodulin (Hoeflich and Ikura, 2002), belonging to the large group of EF-hand calcium-binding proteins. In addition to calmodulin, the other EF-hand calcium-sensitive proteins of the neuronal calcium sensor (NCS) and caldendrin/calneuron families are expressed in neurons (McCue et al., 2010). These proteins regulate different aspects of neuronal function including neurotransmitter release, channel and receptor regulation, control of gene transcription, neuronal growth, and survival. NCS proteins provide a fine-tuning of regulatory mechanisms triggered by changes in calcium level in neurons while calmodulin operates more as an universal sensor of calcium signals. Some of the protein targets are common for both calmodulin and NCS proteins and in a number of cases they were shown to compete for the same binding sites in target molecules (Schaad et al., 1996; Haynes et al., 2006; Few et al., 2011). The concentration of calmodulin in neurons (including rod outer segments, ROS) is normally 10–25 μM (Liebman, 1972; Kohnken et al., 1981; Kakiuchi et al., 1982) and is comparable to the level of NCS family proteins (maximum intracellular concentration of 30–40 μM was observed in the case of hippocalcin in hippocampal neurons and recoverin in photoreceptors; Klenchin et al., 1995; McCue et al., 2010; Mikhaylova et al., 2011). In this case, one of the possible mechanisms underlying the effective functioning of the NCSs is based on the different affinities of NCSs and calmodulin to calcium. This is for example realized when calmodulin and NCS proteins regulate a specific target protein in different ranges of cytoplasmic calcium (for review, see Burgoyne, 2007; McCue et al., 2010).
In addition to competitive regulation based on the variable Ca2+-sensitivity, the effective functioning of NCSs, and calmodulin can be implemented through their interaction with target proteins using separate binding sites. The examples of simultaneous regulation of the effector enzymes by several Ca2+-binding proteins are quite common. In photoreceptor cells, retinal guanylyl cyclase 1 contains independent binding sites for four Ca2+-binding proteins: guanylyl cyclase activating protein 1 (GCAP1; Lange et al., 1999), GCAP2 (Duda et al., 2005), S100B (Duda et al., 2002), and neurocalcin (Venkataraman et al., 2008). Four Ca2+ binding EF-hand proteins are involved in regulation of sodium/proton exchanger 1 (NHE) via different sites on either juxtamembrane region of NHE1 (calcineurin homologous proteins 1, 2, and tescalcin) or C-terminal regulatory domain (calmodulin; Köster et al., 2011). Another example includes Ca2+-dependent regulation of chimeric calmodulin-dependent protein kinase from plants (Sathyanarayanan et al., 2000). This enzyme contains visinin-like domain, which serves as an intramolecular NCS-like regulator. Such a structure allows the enzyme to complement the external regulation of its activity by calmodulin with intramolecular regulation utilizing the visinin-like domain.
Recoverin is an NCS protein that is primarily expressed in the photoreceptor cells, where it plays a key role in the recovery phase of phototransduction by regulating the activity of rhodopsin kinase (RK; Gorodovikova and Philippov, 1993; Kawamura, 1993; Gorodovikova et al., 1994a,b; Chen et al., 1995; Klenchin et al., 1995; Senin et al., 1995; Makino et al., 2004). Recoverin consists of two domains each containing a pair (functional and non-functional) of EF-hand-type Ca2+-binding sites (Flaherty et al., 1993). The N-terminus of the protein is acylated with myristic acid (Dizhoor et al., 1992). Upon binding of two calcium ions, recoverin undergoes a series of conformational changes allowing the exposure of the amino acid residues of its hydrophobic pocket intended for interaction with RK (Tanaka et al., 1995; Ames et al., 1997, 2006; Komolov et al., 2005). In parallel, the myristoyl group of recoverin becomes exposed imparting the interaction with photoreceptor membranes (so-called Ca2+/myristoyl switch mechanism; Zozulya and Stryer, 1992; Dizhoor et al., 1993). In the photoreceptor cells, recoverin inhibits RK in the dark while this inhibition is relieved upon illumination and the subsequent drop of the intracellular calcium level (Senin et al., 2002b). It allows RK to phosphorylate bleached receptor, rhodopsin, which is the first step to its desensitization (Wilden et al., 1986).
RK (or GRK1) is a member of the G protein-coupled receptor-kinase (GRK) family that specifically phosphorylate agonist-occupied G protein-coupled receptors (GPCRs) and initiate the recruitment of arrestins that induces further receptor desensitization and internalization (Pronin et al., 2002). The common feature of GRKs is the central catalytic domain flanked by two sections of the regulatory RH-domain [from regulator of G protein signaling (RGS) homology] (Singh et al., 2008; Boguth et al., 2010). A C-terminal cysteine residue in RK is modified by a farnesyl group which is involved both in the binding of the enzyme to membranes and in the interaction with its substrate rhodopsin (Inglese et al., 1992; McCarthy and Akhtar, 2000). The recoverin-binding site in RK is localized in the N-terminal portion of the molecule upstream of the RH-domain and consists of the first 25 amino acid residues of the enzyme (Ames et al., 2006; Higgins et al., 2006). Binding of recoverin to this site does not prevent receptor-kinase interaction and thus formation of a transient ternary complex between the kinase, recoverin, and the receptor. Instead, recoverin affects a conformational transition in the RK molecule that is necessary for the phosphorylation of the receptor (Komolov et al., 2009). Previous work has provided some quantitative characteristics of RK regulation by recoverin. The equilibrium dissociation constant (KD) of recoverin–RK complex is about 6 μM (Satpaev et al., 1998). The half-maximal inhibition of rhodopsin phosphorylation occurs at 3–6 μM recoverin and at 1.5–3 μM [Ca2+]free (Klenchin et al., 1995; Senin et al., 1997; Weiergräber et al., 2006; Zernii et al., 2011).
While retina-specific GRK1 (RK) and GRK7 are regulated by Ca2+/recoverin, other GRKs (GRK2-6) are found to be inhibited by the universal mediator of Ca2+ signaling calmodulin (Chuang et al., 1996; Pronin et al., 1997). It was shown that calmodulin binding sites in GRK2 and GRK5 are located at both the N- and C-terminal regions of their RH domains and seem to be independent (Levay et al., 1998). Another report suggests that the activity of RK can be regulated not only by recoverin, but also by some additional NCSs as well as by the other Ca2+-binding proteins presented in the photoreceptor cell (De Castro et al., 1995). Among the latter is calmodulin (Kohnken et al., 1981) which in surface plasmon resonance (SPR) studies binds to RK in a Ca2+-dependent manner. The apparent KD of that complex was 0.1 μM and the half-maximal binding was observed at 3 μM Ca2+ (Levay et al., 1998). Since calmodulin and recoverin are colocalized with RK in photoreceptor cells (Kohnken et al., 1981; Dizhoor et al., 1991; Palczewski et al., 1993; Eckmiller, 2002), the actual process of the Ca2+-dependent regulation of rhodopsin phosphorylation by the enzyme could involve both Ca2+-sensors. In the present study, we demonstrate that a Ca2+-dependent regulation of the RK activity is indeed jointly mediated by recoverin and calmodulin. In particular, we show the synergetic inhibition of the enzyme activity by these Ca2+-sensitive regulators. Additionally, we study the structural basis of this process by mapping the calmodulin binding site in the RK molecule.
Materials and Methods
Purification of Recoverin and Calmodulin
Heterologous expression and purification of myristoylated wild-type recoverin has been described in detail in a previous study (Senin et al., 2002a). Calmodulin was purified from bovine brain according to Gopalakrishna and Anderson (1982) with some modifications. Briefly, 100 g of brain tissue was extracted with 200 ml of 50 mM Tris–HCl pH 7.5, 1 mM EDTA, 1 mM 2-mercaptoethanol, 0.5 mM phenylmethylsulfonyl fluoride (PMSF). The extract was subjected to isoelectric precipitation at pH 4.3 followed by heat denaturation of the dissolved pellet at 100°C. Further, renaturated calmodulin was loaded onto a phenyl-Sepharose 6B column (GE Healthcare) equilibrated with 50 mM Tris–HCl pH 7.5, 1 mM 2-mercaptoethanol, 0.1 mM CaCl2. The column was washed with the same buffer containing 0.5 M NaCl, and calmodulin fractions were eluted with 1 mM EGTA. The resulted fractions were pooled and loaded onto a 5-ml HiTrap DEAE FF anion exchange chromatography column (GE Healthcare) pre-equilibrated with 50 mM Tris–HCl pH 7.5, 1 mM 2-mercaptoethanol at 1 ml/min. After washing the column with the same buffer (∼10 column volumes), calmodulin was eluted by 30 ml of a linear ionic strength gradient (0–500 mM NaCl) at 0.5 ml/min. Calmodulin containing fractions as revealed by SDS-PAGE were pooled, dialyzed overnight versus 20 mM Tris–HCl pH 8.0, concentrated, and stored at −80°C.
Preparation of Rod Outer Segments and Urea-Washed ROS Membranes
Bovine ROS were prepared from frozen retinae according to a well-established procedure (Papermaster and Dreyer, 1974) with some modifications. All procedures were performed under dim red light. 200 frozen retinae were thawed in 200 ml of buffer A [20 mM Tris–HCl pH 8.0, 0.1 mM EDTA, 1 mM dithiothreitol (DTT)] containing 45% (w/v) sucrose and the resulted mixture was vigorously shaken for 10 min. The suspension was then centrifuged (3,000 g, 4°C) for 20 min and the pellet was discarded. The ROS containing supernatant was diluted twice with buffer A and centrifuged (8,600 g, 4°C) for 30 min. The supernatant was discarded and the pellet containing crude ROS was resuspended in minimal volume of buffer A with addition of 34% (w/v) sucrose, layered over a freshly prepared 34–36% sucrose density step gradient and centrifuged (72,000 g, 4°C) for 2 h. The centrifugation resulted in formation of the compact band containing ROS fraction, which was carefully removed, diluted twice with buffer A and centrifuged for 15 min at 27,200 g, 4°C. The resulting intact ROS pellet was stored at −80°C. Urea-washed ROS membranes were prepared by homogenizing ROS in 5 M urea followed by five-times washing with buffer A as previously described (Senin et al., 2002a).
Preparation of RK Constructs and Native RK
Standard cloning techniques were applied to generate seven RK constructs from a full-length RK cDNA template in a pT7-7 plasmid. DNA fragments corresponding to the RK amino terminal region (N-RK: M1-G183), RK carboxy terminal region (C-RK: I455-S561) as well as to RK amino terminal region stretches M1-S25, M1-L102, L102-G183, D100-V150, and F125-K175 were PCR-amplified and inserted into the pGEX-5x-1 vector to achieve expression as glutathione S-transferase (GST) fusion proteins. Procedures for heterologous expression and affinity chromatography purification of GST–RK constructs were the same as previously described (Komolov et al., 2009). Native RK was purified from ROS as described in detail elsewhere (Senin et al., 2011).
Phosphorylation Assay
RK activity was measured using an in vitro rhodopsin phosphorylation assay essentially as described (Weiergräber et al., 2006). Briefly, the assay was performed in 50 μl reaction mixture containing 10 μM rhodopsin (urea-washed ROS membranes), 20 mM Tris–HCl pH 7.5, 2 mM MgCl2, 1 mM γ-32P-ATP (30–100 dpm/pmol), 1 mM DTT, 1 mM PMSF, and 0.3–0.5 units of rhodopsin kinase with addition of either 200 μM CaCl2 or 1 mM EGTA. Recoverin and/or calmodulin at concentrations indicated in the figure legends was added, as appropriate. 1,2-bis (o-amino-5-bromophenoxy) ethane-N,N,N′,N′-tetraacetic acid tetrapotassium salt (5,5′-dibromo-BAPTA) was used as a Ca2+-buffer in Ca2+-titration experiments according to our previous work (Senin et al., 2002a). The reaction was initiated by addition of ATP and samples were incubated under continuous light for 30 min at 37°C. Incubation was terminated by adding 1 ml of 10% (w/v) trichloroacetic acid. The resulting precipitate was collected by centrifugation and washed three times with 1 ml of 10% trichloroacetic acid; the final pellet was used for Cherenkov counting. The data were analyzed in SigmaPlot 11 (Systat Software) by plotting relative RK activity [assuming 100% activity at no inhibitor(-s) added] versus Ca2+-sensing protein(-s) or free Ca2+ concentration and by fitting the plot to four parameter Hill sigmoidal (Eq. 1):

where y is the relative RK activity and x is the recoverin and/or calmodulin or free calcium concentration. The resulting values b (Hill coefficient, n) and c (half-maximal inhibition, IC50) were expressed as best-fit value ± SE of the fit.
Surface Plasmon Resonance
Surface plasmon resonance measurements were performed on a BIACORE 2000 instrument (GE Healthcare) at 25°C. Details of the operation principle, the immobilization procedures and of the evaluation of sensorgrams had been described before (Koch, 2000; Komolov et al., 2006). The analysis of how various GST–RK constructs interact with calmodulin was performed as described recently for recoverin (Komolov et al., 2009) with some modifications. GST–RK fusion proteins were captured on the surface of a CM5 sensor chip (GE Healthcare) with pre-immobilized goat anti-GST antibodies (GE Healthcare) resulting in a surface density of approximately 1 ng/mm2. Calmodulin was applied in the mobile phase in running buffer (20 mM Tris–HCl pH 7.5, 100 mM NaCl, 2 mM CaCl2, 1 mM MgCl2, 0.005% Tween 20) at 10 μl/min flow rate. Calmodulin concentration in steady-state affinity analysis was varied from 10 to 150 μM. Each calmodulin response was double-referenced by subtracting a blank (buffer) and control (injection over GST-coated surface lacking the RK fragments) response (Myszka, 1999). The sensorgrams were also normalized to the amount of immobilized GST–RK. The apparent KDs for calmodulin–RK peptide complexes were determined by equilibrium analysis as follows: the SPR amplitudes recorded at steady-state of the association phase of each calmodulin injection versus calmodulin concentrations in the mobile phase were plotted using SigmaPlot 11 software and the plots were fitted to the one site saturation ligand-binding model (Eq. 2):

where KD is the apparent equilibrium dissociation constant and Bmax is a maximal SPR signal. The resulting KD values were expressed as best-fit value ± SE of the fit.
Analytical Procedures
Rhodopsin content in urea-washed ROS membranes was determined as described previously (Senin et al., 1995). Concentrations of purified recoverin and calmodulin were determined spectrophotometrically assuming ε280 of 27,500 and 3,030 M−1 cm−1, respectively (Wallace et al., 1983; Johnson et al., 1997). Concentrations of GST–RK constructs were measured using Bradford protein assay (Bio-Rad Laboratories) calibrated with bovine serum albumin. The purity of all the proteins obtained was assessed by SDS-PAGE with Coomassie Brilliant Blue R250 staining.
Results
Ca2+-Dependent Inhibition of Rhodopsin Kinase by Calmodulin
It is well-known that recoverin inhibits RK in vitro in the micromolar range of free Ca2+ concentrations ([Ca2+]free). Taking into account previous data (Pronin et al., 1997; Levay et al., 1998) it can be supposed that the RK activity can be additionally regulated by calmodulin. We performed a detailed investigation of the ability of calmodulin to affect rhodopsin phosphorylation by RK in a Ca2+-dependent manner. For this purpose we applied the in vitro phosphorylation assay in the reconstituted system consisting of 0.3–0.5 units of RK purified from ROS, 10 μM dark-adapted rhodopsin in the content of urea-washed ROS membranes, and various concentrations of calmodulin ranging from 0.01 to 100 μM (Figure 1A). The experiment was conducted at high (200 μM [Ca2+]free) or low (1 mM EGTA) Ca2+ levels.
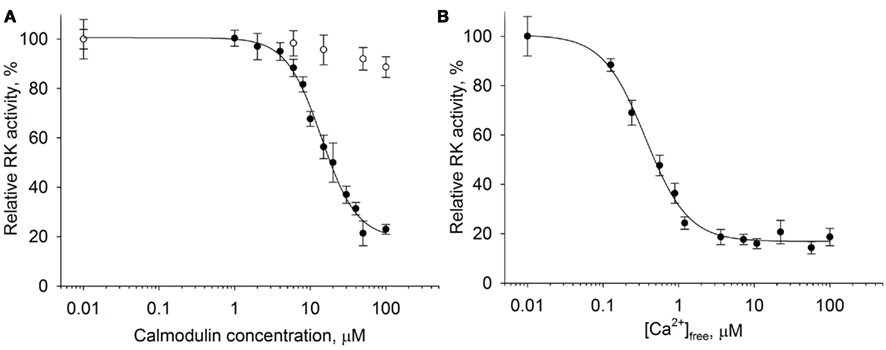
Figure 1. Ca2+-dependent inhibition of rhodopsin kinase by calmodulin. Rhodopsin kinase activity was measured as a function of either calmodulin or free Ca2+ concentrations by in vitro phosphorylation assay. (A) Inhibition of RK activity by increasing calmodulin concentrations at saturating Ca2+ concentration (200 μM, ●) or at 1 mM EGTA (○). Fitting of the data as described in Section “Materials and Methods” yielded IC50 = 14.1 ± 0.9 μM and n = 1.85 ± 0.18. (B) Inhibition of RK activity by 30 μM calmodulin at different concentrations of free Ca2+. Half-maximal inhibition was observed at [Ca2+]free of 0.36 ± 0.03 μM with a Hill coefficient of 1.53 ± 0.13. Data points represent mean ± SE of the mean from three independent experiments.
According to our data, calmodulin inhibited RK in the presence of Ca2+; the inhibition was half-maximal at 14 μM protein concentration. In the absence of Ca2+, calmodulin was ineffective. We further examined rhodopsin phosphorylation by RK as a function of free Ca2+ concentration in the presence of saturating concentration (30 μM) of calmodulin (Figure 1B). Inhibition reached half maximum at 0.36 μM [Ca2+]free with a Hill coefficient of 1.53. In summary, these findings indicate the ability of calmodulin to operate in vitro as a Ca2+-dependent inhibitor of RK.
Mapping of the Calmodulin Binding Site in Rhodopsin Kinase
The observed ability of calmodulin to inhibit RK in a Ca2+-dependent manner poses a question whether it binds to the same site in the RK molecule as recoverin does or whether these two Ca2+-sensors interact with separate specific sites in the enzyme. We employed a biosensor-based technique, the SPR spectroscopy, to study the interaction of recoverin and calmodulin with GST-fused N-terminal RK peptide M1-S25 containing the recoverin-binding site (Higgins et al., 2006; here and below, see Figure 2A for a schematic representation of RK fusions constructed and tested). The RK peptide was captured on the anti-GST antibody-coated sensor chip and the binding of Ca2+-loaded forms of calmodulin and recoverin (both at concentration of 40 μM) to the immobilized RK fragment was monitored in real-time resulting in sensorgrams shown at Figure 2B.
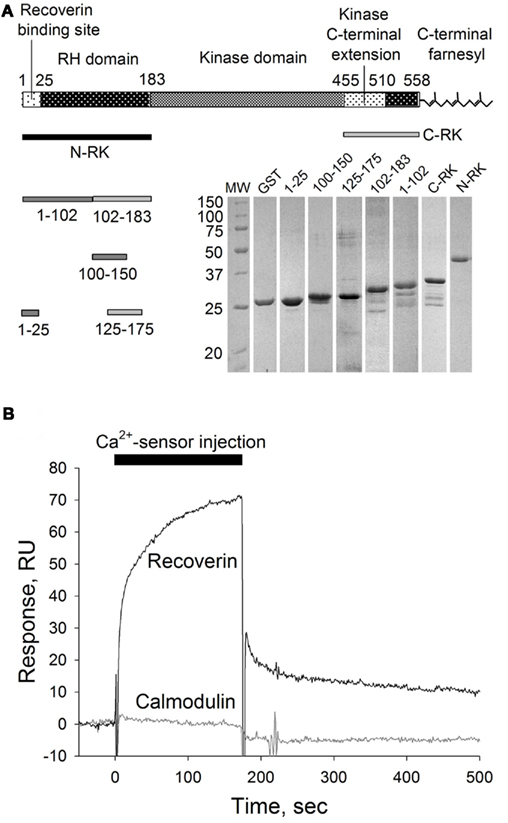
Figure 2. SPR analysis of calmodulin interaction with recoverin-binding site in RK. (A) Schematic representation of RK polypeptide according to Singh et al. (2008) and corresponding GST–RK fusion proteins used in this study. Inset: Coomassie-stained SDS-PAGE of the GST–RK fusion proteins obtained. (B) Overlay of sensorgrams recorded during consecutive injections of Ca2+-loaded recoverin or calmodulin at 40 μM concentration over the sensor chip surface with immobilized RK fragment corresponding to the residues M1-S25 fused with GST. The reference signal recorded from the surface with immobilized GST was subtracted.
As expected, recoverin interacted with the RK fragment containing the first 25 amino acids of the enzyme (Figure 2B). The apparent KD of recoverin for M1-S25 fragment was about 17 μM, as determined in titration series (data not shown). The interaction of calmodulin with the same RK peptide was negligible (Figure 2B). These results suggest the existence of separate binding sites for calmodulin and recoverin. To identify calmodulin binding site(s), we constructed two recombinant GST fusion fragments that encompass amino terminal (N-RK: M1-G183) and carboxy terminal (C-RK: I455-S561) regulatory regions of RK. Only the N-RK fragment readily captured supplied calmodulin (Figure 3A), while the calmodulin binding to C-RK fragment was almost absent. To further localize the calmodulin binding site within N-RK, we tested the ability of calmodulin to interact with N-RK fragments of smaller size: M1-L102 and L102-G183. The interaction occurred in both cases but the response was much more pronounced for M102-G183 (Figure 3B). Moreover, we showed that within the RK sequence L102-G183 the calmodulin binding site is presumably localized within a short stretch of 25 amino acids present in positions 150–175 since the N-RK fragment F125-K175 bound calmodulin, but the fragment D100-V150 did not (Figure 3C). The additional low affinity site for calmodulin is localized within the RK sequence S26-L102 since the interaction of RK fragment M1-S25 with calmodulin was negligible (Figures 2B and 3B). To evaluate the quantitative characteristics of the effects observed, we carried out steady-state affinity analysis of calmodulin interaction to those GST–RK constructs, which displayed a significant binding to the Ca2+-sensor. The analysis was performed by recording series of sensorgrams at different calmodulin concentrations (in the range of 10–150 μM) and measuring SPR signal amplitudes at steady-state (Figure 3D). The apparent equilibrium dissociation constants were determined as concentrations required for half-maximal saturation of the binding signal. The obtained KD values were 63 μM for the full-length N-RK, 54 μM for the M1-L102 fragment, 23 μM for the L102-G183 fragment, and 17 μM for the F125-K175 fragment.
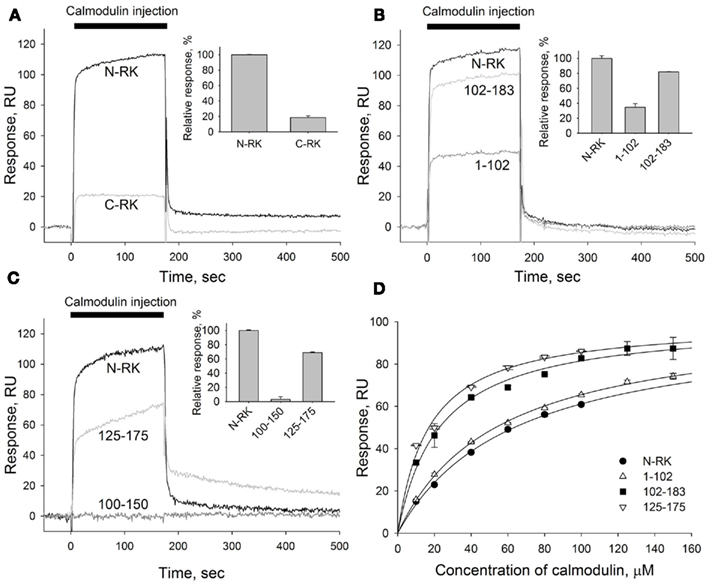
Figure 3. Mapping of the calmodulin binding site in the N-terminal region of RK. (A–C) A representative overlay of SPR sensorgrams showing real-time binding of calmodulin to RK fragments anchored on the sensorchip surface via GST tags using anti-GST antibodies covalently coupled to the dextran matrix. Sixty micromolar calmodulin in running buffer containing 2 mM Ca2+ was injected over the surfaces with immobilized N-terminal or C-terminal regions of RK (termed N-RK and C-RK, respectively) (A), N-RK and its fragments corresponding to the residues M1-L102 and L102-G183 (B), or N-RK and its fragments corresponding to the residues D100-V150 and F125-K175 (C). The sensorgrams were normalized to the amounts of immobilized fragments, and the reference signal from a control surface with immobilized GST was subtracted. Insets show relative responses at the end of the injection observed for each GST–RK fragment indicating the amount of calmodulin bound. (D) Steady-state affinity analysis of calmodulin binding to the full-length N-RK or its fragments M1-L102, L102-G183, or F125-K175. Binding of calmodulin was recorded at concentrations ranging from 10 to 150 μM. The amplitudes of binding signals at equilibrium were determined, normalized, and are shown as a function of calmodulin concentration. Apparent KDs were determined as half-maximal calmodulin concentrations required for saturation of SPR amplitudes as described under Section “Materials and Methods.” The obtained values for N-RK, M1-L102, L102-G183, and F125-K175 are 63 ± 3, 54 ± 2, 23 ± 2, and 17.0 ± 1.5 μM, respectively. Data points represent mean ± SE of the mean from two independent experiments. The calculated KDs are expressed as best-fit value ± SE of the fit.
Thus, both high and low affinity calmodulin binding sites are localized within the N-terminal regulatory part of RK, and they are separated from the recoverin-binding site consisting of M1-S25 sequence of the enzyme. Together, our findings indicate that both Ca2+-sensors, recoverin, and calmodulin, are able to bind RK using separate specific sites, suggesting the possibility of their simultaneous binding to and inhibition of RK.
Synergetic Inhibition of Rhodopsin Kinase by Calmodulin and Recoverin
We have found that calmodulin displays an inhibitory effect on RK at free Ca2+ concentrations ranging from 0.01 to 1 μM (Figure 1B). The range overlaps with the physiological changes of Ca2+ concentrations found in the photoreceptor cell (Gray-Keller and Detwiler, 1994). Moreover, the revealed inhibitory action of calmodulin was observed at Ca2+concentrations even below the Ca2+-sensitivity of recoverin, the well-established RK regulator. Since both proteins colocalize in photoreceptor cells with RK, we asked whether these two Ca2+-sensors could have a joint effect on RK activity. Thus we utilized the in vitro phosphorylation assay examining the ability of calmodulin and recoverin to simultaneously regulate the activity of RK. Urea-washed ROS membranes containing 10 μM rhodopsin were incubated with 0.3–0.5 units of RK in the presence of increasing concentrations of either calmodulin, recoverin, or their 1:1 mixture at the saturating free Ca2+ concentration (200 μM).
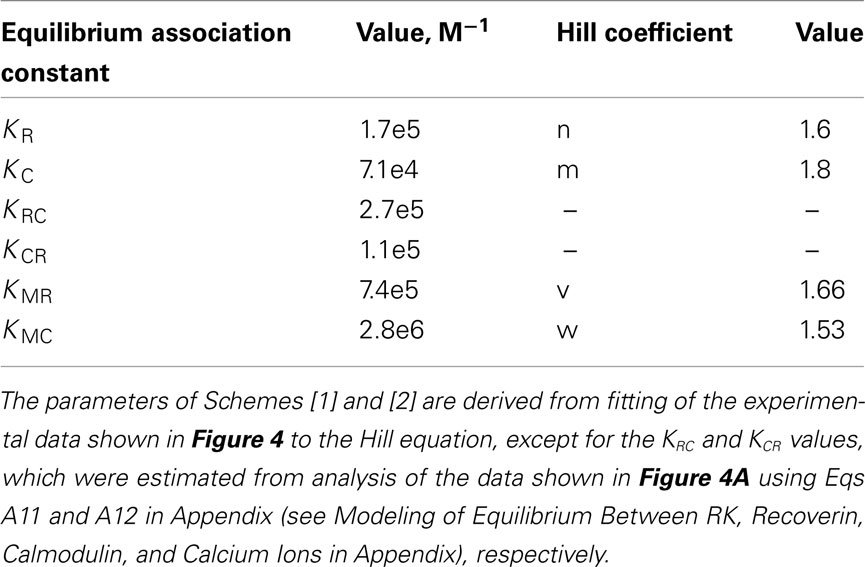
Remarkably, calmodulin and recoverin were able to synergistically inhibit the RK activity (Figure 4A). While the IC50 for calmodulin or recoverin alone were 14.1 and 5.8 μM, respectively, their joint action resulted in half-maximal RK inhibition at 4.6 μM of the total Ca2+-sensing protein concentration in the mixture. Approximation of the experimental data was performed with the Hill equation and revealed that the interaction between RK and recoverin was cooperative with Hill coefficient of 1.6. A good curve fit was also possible when approximating the same data with one site (Langmuir) ligand-binding model (data not shown). Collectively, these results point out that the inhibition of RK by recoverin occurs at equimolar stoichiometric ratio of the proteins. In contrast, the inhibition of RK by calmodulin alone as well as the synergetic inhibition by both Ca2+ sensors had Hill indexes of about 2. The latter fact evidences for the cooperative binding of two molecules of Ca2+-sensor per one RK molecule. The inhibitory effect of calmodulin on RK was found to occur at lower free Ca2+ concentrations than the effect of recoverin. Calmodulin inhibited RK at a half-maximal free Ca2+ concentration of 0.36 μM while the inhibitory effect of recoverin was at 1.36 μM Ca2+ (Figure 4B). The synergetic inhibition of RK by the 30-μM calmodulin/recoverin mixture was observed at a half-maximal free Ca2+ concentration of 1.07 μM. Apparently, the synergetic action of both Ca2+-sensors caused a shift in the affinity of the target–effector interaction and at the same time an extension of the operating Ca2+-range. In Ca2+-titration experiments where the RK inhibition by calmodulin or recoverin was studied, the Hill coefficients were within the range of 1.53–1.66, indicating similar cooperative action of the cation in the case of both Ca2+-sensors. Thus, our data indicate that calmodulin complements the inhibitory action of recoverin on RK and increases the sensitivity of RK inhibition to Ca2+.
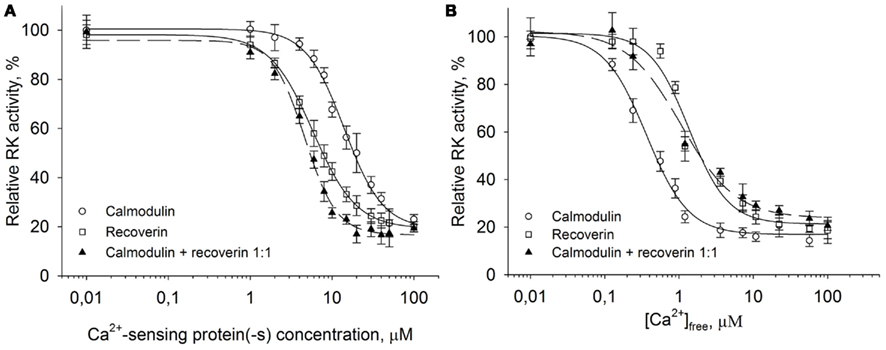
Figure 4. Synergetic inhibition of rhodopsin kinase by recoverin and calmodulin. Rhodopsin kinase activity was measured as a function of Ca2+-sensor protein or free Ca2+ concentration by in vitro phosphorylation assay. (A) Inhibition of RK by increasing concentrations of either calmodulin (○), recoverin (□), or their mixture in the 1:1 ratio (▲) at saturating free Ca2+ concentration (200 μM). Fitting the data to the Hill equation as described in Section “Materials and Methods” resulted in IC50 = 14.1 ± 0.9 μM, n = 1.8 ± 0.2 for calmodulin, IC50 = 5.8 ± 0.7 μM, n = 1.6 ± 0.3 for recoverin, and IC50 = 4.6 ± 0.2 μM, n = 2.2 ± 0.2 for calmodulin/recoverin 1:1 mixture. (B) Inhibition of RK activity by either calmodulin (○), recoverin (□), or their 1:1 mixture (▲) at different free Ca2+ concentrations. Concentration of each Ca2+-sensing protein including their mixture was 30 μM. Fitting the data to the Hill equation as described under Section “Materials and Methods” resulted in values of IC50 = 0.36 ± 0.03 μM, n = 1.53 ± 0.13 for calmodulin, IC50 = 1.36 ± 0.13 μM, n = 1.66 ± 0.23 for recoverin, and IC50 = 1.07 ± 0.15 μM, n = 1.2 ± 0.1 for their 1:1 mixture. The fitted curves are drawn solid for calmodulin or recoverin data sets, and dashed for their mixture. Data points represent mean ± SE of the mean from three independent experiments.
Modeling of Equilibrium between RK, Recoverin, Calmodulin, and Calcium Ions
We further used the parameters obtained to create a model of the synergetic inhibition of RK by calmodulin and recoverin. Since the combined use of both Ca2+-sensors increases efficiency of RK inhibition (Figure 4A), the fully competitive scheme of RK regulation was excluded. The SPR experiments strongly evidence for the presence of separate recoverin- and calmodulin binding sites. The simplified scheme of chemical equilibria describing this case looks as follows (the rationale behind the use of this scheme is explained in the Modeling of Equilibrium Between RK, Recoverin, Calmodulin, and Calcium Ions in Appendix):
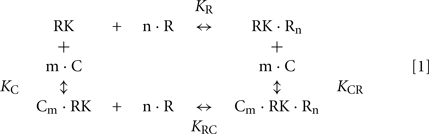
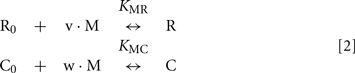
Here, M denotes calcium ion, R and C denote Ca2+-bound states of recoverin and calmodulin, respectively, while R0 and C0 denote their Ca2+-free states. Each of the equilibria is characterized by its own association constant (see Modeling of Equilibrium Between RK, Recoverin, Calmodulin, and Calcium Ions in Appendix; Table 1). Fitting of the experimental data shown in Figure 4 to the Hill equation gives estimates of the most of the constants and binding stoichiometries of Schemes [1] and [2]. Notably, these values implicitly take into account the presence of membranes, which are known to increase the affinity of recoverin to Ca2+ (Zozulya and Stryer, 1992) and to affect the distribution of RK between soluble and membrane-bound forms (Sanada et al., 1996). Therefore, these values should be considered as effective. The analysis of the experimental data shown in Figure 4A (see Modeling of Equilibrium Between RK, Recoverin, Calmodulin, and Calcium Ions in Appendix) enabled us to estimate all parameters of the Scheme [1] (Table 1). According to our analysis, the binding of Ca2+-loaded recoverin molecules to RK facilitates its further association with Ca2+-bound calmodulin, as manifested by 1.55-fold increase in the constant of calmodulin binding to RK. Similarly, association of calmodulin with RK increases its recoverin-binding constant 1.59-fold (Table 1). Thus, the modeling results suggest that recoverin and calmodulin could act in a concerted manner, mutually facilitating inhibition of RK.
The changes in RK-inhibiting activity shown in Figure 4B reflect association of the Ca2+-sensing proteins with calcium ions. These processes can be approximated by the Hill equation based model (Scheme [2]), which is widely used for simplified description of such complex processes. Alternatively, the Ca2+-dependence of interaction between calmodulin and its target could be described by the model of sequential filling of EF-hands (Dagher et al., 2011). However, this approach requires the introduction into the Scheme [2] of additional chemical equilibria, which would drastically complicate modeling of the system. According to our analysis based on Scheme [2] (see Modeling of Equilibrium Between RK, Recoverin, Calmodulin, and Calcium Ions in Appendix), the half-maximal synergetic inhibition of RK by calmodulin and recoverin occurs at free Ca2+ concentration of 0.71 μM. This value is between the respective values obtained for calmodulin (0.36 μM) and recoverin (1.36 μM). The corresponding experimental value equals to 1.07 μM (Figure 4B) which is close to the theoretical estimate.
Discussion
In the present work we have demonstrated that the ubiquitous Ca2+-sensor calmodulin is able to function as an inhibitor of RK. Moreover, we found that the inhibition of RK by calmodulin can complement the inhibitory action of the well-known RK regulator recoverin. Previous attempts to study the ability of calmodulin to regulate GRKs, including GRK1 (RK), were performed using relatively low concentrations of calmodulin (not exceeding 10 μM; Chuang et al., 1996; Pronin et al., 1997) at which its inhibitory effect could not be seen to the full extent. According to our data, the inhibition of RK by calmodulin at saturating Ca2+ concentrations was observed in the range of calmodulin concentration from 2 to 30 μM and was half-maximal at 14 μM of the Ca2+-sensor, whereas recoverin was a more effective inhibitor, as it showed a half-maximal inhibitory effect on the RK activity at 5.8 μM. The calcium range of RK inhibition by calmodulin (0.05–1 μM of [Ca2+]free) overlapped with the physiological range of the cation concentration (0.05–0.55 μM of [Ca2+]free) in ROS (Gray-Keller and Detwiler, 1994). The synergetic action of calmodulin and recoverin on the RK activity was within 0.05–1 μM of [Ca2+]free. These values are slightly above the physiological range of Ca2+ concentration in ROS. However, the recoverin affinity for Ca2+ is significantly increased in the presence of RK and ROS membranes (Zozulya and Stryer, 1992; Ames et al., 1995; Senin et al., 1995, 2004). Similarly, the affinity of calmodulin for Ca2+ increases upon binding to target enzymes (Shifman et al., 2006). Therefore, extrapolation to the in vivo conditions (including high membrane content and presence of target proteins) indicates that the apparent Ca2+-dependence of the synergetic action of calmodulin and recoverin on RK activity could reach the physiological (i.e., submicromolar) range of the calcium concentration. Our experimental results were supplemented with theoretical validation of the ability of calmodulin and recoverin to synergistically bind RK. Different approaches could be applied to modeling of equilibria between RK, recoverin, calmodulin, and calcium ions. Full description of the system would require characterization of each step of the calcium-binding to the calcium-sensing proteins (Dagher et al., 2011), as well as characterization of all protein–protein interactions in the system. Considering that membranes are known to affect the distribution of RK and recoverin between soluble and membrane-bound fractions, as well as to modulate calcium affinity of recoverin (Zozulya and Stryer, 1992; Senin et al., 1995; Sanada et al., 1996), it can be concluded that the total number of chemical equilibria is significantly higher (see Modeling of Equilibrium Between RK, Recoverin, Calmodulin, and Calcium Ions in Appendix). This objective difficulty can be compensated via usage of the Hill model-based representation of the system, which describes the multiple processes of ligand-binding as a single event. Despite being phenomenological, this approach enabled us to estimate the synergy of binding of calmodulin and recoverin to RK. According to our analysis (see Modeling of Equilibrium Between RK, Recoverin, Calmodulin, and Calcium Ions), binding of either calcium sensor to RK results in 1.5-fold increase of the affinity of the complex to another Ca2+-sensing protein. The analysis also showed that the half-maximal synergetic inhibition of RK by calmodulin and recoverin occurs at free Ca2+ concentration of 0.71 μM, which is close to the corresponding experimental value of 1.07 μM.
The structural aspects underlying the synergetic regulation of RK by calmodulin and recoverin can be identified from the experiments on mapping of the calmodulin binding site in RK using a biosensor-based approach (SPR spectroscopy). Our findings indicate that the interaction of calmodulin and recoverin with RK occurs by means of separate binding sites in the N-terminal region of the enzyme, which makes the co-inhibitory action of the two Ca2+-sensors possible. The recoverin-binding site is known to be located in the first 25 amino acid residues of RK (Ames et al., 2006; Higgins et al., 2006). According to our data, RK contains two calmodulin binding sites. The first, low affinity site is located within the sequence S26-L102 of N-terminal regulatory region of RK, which is in agreement with a previous report (Levay et al., 1998). The second high affinity site was identified within the residues V150-K175 of the same RK region. In other studies this fragment was not tested for the ability to bind calmodulin. The C-terminal regulatory region of RK (I455-S561) did not interact with calmodulin both in the current and in the earlier studies. In addition, the report by Levay et al. (1998) suggested another calmodulin binding site within the residues A341-E400. This site is located in the large lobe of the catalytic domain of the enzyme (Singh et al., 2008). However, calmodulin apparently has no effect on the catalytic activity of RK since in our experiments it did not affect the ability of the enzyme to phosphorylate the C-terminal peptide of rhodopsin containing phosphorylation sites (data not shown). Thus, the suggested interaction of calmodulin with A341-E400 site seems to be out of the physiological relevance. Moreover, it was shown that the recombinant fragment corresponding to the full-length catalytic domain of RK (E184-D454) is not able to interact with rhodopsin (Komolov et al., 2009), although the domain function should include this interaction. The latter suggests that this fragment does not fully acquire its native structure. These observations led us not to consider parts of the catalytic domain in the context of Ca2+-dependent regulation of RK by calmodulin.
It should be mentioned that the affinities of calmodulin and recoverin to the RK peptides determined in our SPR experiments were lower than in the previous work (Levay et al., 1998). The possible reason of this discrepancy can be the different strategies applied for the immobilization of the interaction partners on the sensor chip. Levay et al. (1998) immobilized calmodulin and recoverin on the chip via cysteine residues. Calmodulin was biotinylated via the cysteine residue and further captured on the streptavidin-coated surface while recoverin was directly attached to the sensor chip surface via thiol coupling with its cysteine residue. These cysteine residues were shown to play a critical role in the Ca2+-dependent operating of both proteins (Lukas et al., 1984; Permyakov et al., 2007, 2012). Thus, the functional state of calmodulin and recoverin on the sensor chip could be significantly affected upon the immobilization procedure. We used an experimental configuration opposite to their approach with RK fragments being retained on the sensor chip surface and recoverin/calmodulin being supplied in the mobile phase. RK fragments in this case were captured on the surface under mild conditions using antigen–antibody interaction. This strategy might better preserve the protein integrity in BIACORE experiments and therefore affinity constants determined by SPR for calmodulin interaction with its binding site (17 μM) correlate better with the IC50 for calmodulin-mediated RK inhibition (14 μM).
Based on the location of the recoverin and calmodulin binding sites in RK one may suggest the putative mechanisms underlying the intrinsic inhibition activity in the case of each Ca2+-sensor. As it was previously mentioned, recoverin inhibits RK by affecting the conformational transitions of the enzyme required for the formation of the efficient enzyme-substrate complex between RK and rhodopsin C-terminus. Such a mechanism is due to the ability of recoverin to bind to the N-terminal site M1-S25 of RK that is critical for the function of the enzyme (Higgins et al., 2006; Huang et al., 2009; Komolov et al., 2009; Zernii et al., 2011). It is likely that the mechanism of RK inhibition by calmodulin involves a Ca2+-dependent interference with the interaction between RK and photoexcited rhodopsin. This is possible, if the calmodulin binding site in RK overlaps with the receptor docking site. The sequence F150-K175 in RK identified in our work as the high affinity calmodulin binding site contains amino acid residues Y167, Q173, W174, L177 which could be involved in rhodopsin recognition since the corresponding residues in the GRKs 5 and 6 were recently found to be critical for the phosphorylation of the activated receptors (Baameur et al., 2010). Thus, calmodulin may be suggested to inhibit RK by blocking sterically the association of the enzyme with the responsive sites in its receptor substrate rhodopsin. Preliminary observations made by us further support this conclusion, namely that the fragment F150-K175 of RK is indeed capable of direct binding to rhodopsin in a light-dependent manner. Such a mechanism does not contradict the current hypothesis of the formation of the heteromeric complex between RK, rhodopsin, and Ca2+-sensor(s) (Komolov et al., 2009), since it is known that the receptor binding involves not only the N-terminal regulatory region of RK containing the above mentioned fragment but also the C-terminal farnesyl modification of the enzyme (McCarthy and Akhtar, 2000).
Considering the above speculations the question arises what could be the mechanism underlying the synergetic inhibition of RK by both Ca2+-sensors. The SPR experiments strongly evidence that the recoverin- and calmodulin binding sites in RK are separate. Since combined action of recoverin and calmodulin increases efficiency of RK inhibition, the competitive scheme of the enzyme regulation by two Ca2+-sensors is unlikely. The modeling of equilibrium between RK, recoverin, calmodulin, and calcium ions revealed that the binding of Ca2+/recoverin to RK results in 1.5-fold facilitation of binding of the enzyme to Ca2+/calmodulin, and vice versa. Taking into account the higher affinity of recoverin to RK and its higher intracellular concentration as compared to calmodulin it can be suggested that the binding of Ca2+/recoverin to the enzyme precedes that of Ca2+/calmodulin. Presumably, the binding of recoverin to the first 25 amino acid residues of RK allosterically increases the availability of the high affinity calmodulin binding site V150-K175 for the interaction with calmodulin. This is consistent with the fact that the presence of recoverin shifts the binding of calmodulin to RK (and consequent inhibition of the enzyme) to the lower calmodulin concentration. The suggested sequential filling of Ca2+-sensor binding sites in RK, namely recoverin followed by calmodulin, is in agreement with the fact that the Ca2+-dependence of the synergetic inhibition effect is close to the recoverin Ca2+-dependent inhibition profile. Thus, the following consecutive mechanism of RK regulation by recoverin and calmodulin in photoreceptors can be suggested. At high calcium concentration (0.5–0.6 μM of [Ca2+]free) present in dark-adapted photoreceptors, RK is strongly inhibited by the synergetic action of both Ca2+-sensors. Shortly after the light absorption, when the Ca2+-concentration has not yet decreased, recoverin still allosterically inhibits phosphorylation of rhodopsin by RK however not affecting the interaction between the enzyme and the cytoplasmic loops of the receptor. In the same time calmodulin prevents this interaction thus allowing the receptor to effectively activate transducin. This is important for the ability of rod photoreceptors to respond to even low illumination levels. During the lowering of the intracellular Ca2+ concentration associated with phototransduction, recoverin dissociates from the complex with RK, which decreases the affinity of the enzyme to calmodulin. In these conditions calmodulin alone can still inhibit the enzyme though less effectively than it does in combination with recoverin. Further light-induced decrease in free Ca2+ concentration down to 0.025–0.05 μM triggers the dissociation of calmodulin from RK switching the enzyme to the full active form, which causes desensitization of the receptor.
In summary, our study suggested a novel mechanism of regulation of rhodopsin desensitization by rhodopsin kinase involving Ca2+-sensor proteins. Further studies are required for firm establishment of the physiological role of this phenomenon and unraveling the molecular details of the proposed mechanism.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Daniele Dell’Orco for the assistance with the BIACORE measurements. This work was supported by the grants from the Russian Foundation for Basic Research ## 09-04-01778-a (to Ivan I. Senin), 09-04-00666-a (to Evgeni Yu. Zernii), 09-04-00395, and ofi-m-11-04-12108 (to Pavel P. Philippov); the President of Russia # MD-4423.2010.4 (to Ivan I. Senin); the Deutsche Forschungsgemeinschaft ## KO948/7-1, KO948/7-2 (to Karl-Wilhelm Koch); the Program of the Russian Academy of Sciences « Molecular and Cellular Biology» (to Sergei E. Permyakov).
References
Ames, J. B., Ishima, R., Tanaka, T., Gordon, J. I., Stryer, L., and Ikura, M. (1997). Molecular mechanics of calcium-myristoyl switches. Nature 389, 198–202.
Ames, J. B., Levay, K., Wingard, J. N., Lusin, J. D., and Slepak, V. Z. (2006). Structural basis for calcium-induced inhibition of rhodopsin kinase by recoverin. J. Biol. Chem. 281, 37237–37245.
Ames, J. B., Porumb, T., Tanaka, T., Ikura, M., and Stryer, L. (1995). Amino-terminal myristoylation induces cooperative calcium binding to recoverin. J. Biol. Chem. 270, 4526–4533.
Baameur, F., Morgan, D. H., Yao, H., Tran, T. M., Hammitt, R. A., Sabui, S., McMurray, J. S., Lichtarge, O., and Clark, R. B. (2010). Role for the regulator of G-protein signaling homology domain of G protein-coupled receptor kinases 5 and 6 in beta 2-adrenergic receptor and rhodopsin phosphorylation. Mol. Pharmacol. 77, 405–415.
Boguth, C. A., Singh, P., Huang, C. C., and Tesmer, J. J. (2010). Molecular basis for activation of G protein-coupled receptor kinases. EMBO J. 29, 3249–3259.
Burgoyne, R. D. (2007). Neuronal calcium sensor proteins: generating diversity in neuronal Ca2+ signalling. Nat. Rev. Neurosci. 8, 182–193.
Chen, C. K., Inglese, J., Lefkowitz, R. J., and Hurley, J. B. (1995). Ca(2+)-dependent interaction of recoverin with rhodopsin kinase. J. Biol. Chem. 270, 18060–18066.
Chuang, T. T., Paolucci, L., and De Blasi, A. (1996). Inhibition of G protein-coupled receptor kinase subtypes by Ca2+/calmodulin. J. Biol. Chem. 271, 28691–28696.
Dagher, R., Peng, S., Gioria, S., Fève, M., Zeniou, M., Zimmermann, M., Pigault, C., Haiech, J., and Kilhoffer, M. C. (2011). A general strategy to characterize calmodulin-calcium complexes involved in CaM-target recognition: DAPK and EGFR calmodulin binding domains interact with different calmodulin-calcium complexes. Biochim. Biophys. Acta 1813, 1059–1067.
De Castro, E., Nef, S., Fiumelli, H., Lenz, S. E., Kawamura, S., and Nef, P. (1995). Regulation of rhodopsin phosphorylation by a family of neuronal calcium sensors. Biochem. Biophys. Res. Commun. 216, 133–140.
Dizhoor, A. M., Chen, C. K., Olshevskaya, E., Sinelnikova, V. V., Phillipov, P., and Hurley, J. B. (1993). Role of the acylated amino terminus of recoverin in Ca(2+)-dependent membrane interaction. Science 259, 829–832.
Dizhoor, A. M., Ericsson, L. H., Johnson, R. S., Kumar, S., Olshevskaya, E., Zozulya, S., Neubert, T. A., Stryer, L., Hurley, J. B., and Walsh, K. A. (1992). The NH2 terminus of retinal recoverin is acylated by a small family of fatty acids. J. Biol. Chem. 267, 16033–16036.
Dizhoor, A. M., Ray, S., Kumar, S., Niemi, G., Spencer, M., Brolley, D., Walsh, K. A., Philipov, P. P., Hurley, J. B., and Stryer, L. (1991). Recoverin: a calcium sensitive activator of retinal rod guanylate cyclase. Science 251, 915–918.
Duda, T., Fik-Rymarkiewicz, E., Venkataraman, V., Krishnan, R., Koch, K. W., and Sharma, R. K. (2005). The calcium-sensor guanylate cyclase activating protein type 2 specific site in rod outer segment membrane guanylate cyclase type 1. Biochemistry 44, 7336–7345.
Duda, T., Koch, K. W., Venkataraman, V., Lange, C., Beyermann, M., and Sharma, R. K. (2002). Ca(2+) sensor S100beta-modulated sites of membrane guanylate cyclase in the photoreceptor-bipolar synapse. EMBO J. 21, 2547–2556.
Eckmiller, M. S. (2002). Calmodulin immunolocalization in outer segments of Xenopus laevis photoreceptors. Cell Tissue Res. 308, 439–442.
Few, A. P., Nanou, E., Scheuer, T., and Catterall, W. A. (2011). Molecular determinants of CaV2.1 channel regulation by calcium binding protein-1. J. Biol. Chem. 286, 41917–41923.
Flaherty, K. M., Zozulya, S., Stryer, L., and McKay, D. B. (1993). Three-dimensional structure of recoverin, a calcium sensor in vision. Cell 75, 709–716.
Gopalakrishna, R., and Anderson, W. B. (1982). Ca2+-induced hydrophobic site on calmodulin: application for purification of calmodulin by phenyl-Sepharose affinity chromatography. Biochem. Biophys. Res. Commun. 104, 830–836.
Gorodovikova, E. N., Gimelbrant, A. A., Senin, I. I., and Philippov, P. P. (1994a). Recoverin mediates the calcium effect upon rhodopsin phosphorylation and cGMP hydrolysis in bovine retina rod cells. FEBS Lett. 349, 187–190.
Gorodovikova, E. N., Senin, I. I., and Philippov, P. P. (1994b). Calcium-sensitive control of rhodopsin phosphorylation in the reconstituted system consisting of photoreceptor membranes, rhodopsin kinase and recoverin. FEBS Lett. 353, 171–172.
Gorodovikova, E. N., and Philippov, P. P. (1993). The presence of a calcium-sensitive p26-containing complex in bovine retina rod cells. FEBS Lett. 335, 277–279.
Gray-Keller, M. P., and Detwiler, P. B. (1994). The calcium feedback signal in the phototransduction cascade of vertebrate rods. Neuron 13, 849–861.
Haynes, L. P., Fitzgerald, D. J., Wareing, B., O’Callaghan, D. W., Morgan, A., and Burgoyne, R. D. (2006). Analysis of the interacting partners of the neuronal calcium-binding proteins L-CaBP1, hippocalcin, NCS-1 and neurocalcin delta. Proteomics 6, 1822–1832.
Higgins, M. K., Oprian, D. D., and Schertler, G. F. (2006). Recoverin binds exclusively to an amphipathic peptide at the N terminus of rhodopsin kinase, inhibiting rhodopsin phosphorylation without affecting catalytic activity of the kinase. J. Biol. Chem. 281, 19426–19432.
Hoeflich, K. P., and Ikura, M. (2002). Calmodulin in action: diversity in target recognition and activation mechanisms. Cell 108, 739–742.
Huang, C. C., Yoshino-Koh, K., and Tesmer, J. J. (2009). A surface of the kinase domain critical for the allosteric activation of G protein-coupled receptor kinases. J. Biol. Chem. 284, 17206–17215.
Inglese, J., Glickman, J. F., Lorenz, W., Caron, M. G., and Lefkowitz, R. J. (1992). Isoprenylation of a protein kinase. Requirement of farnesylation/alpha-carboxyl methylation for full enzymatic activity of rhodopsin kinase. J. Biol. Chem. 267, 1422–1425.
Johnson, W. C. Jr., Palczewski, K., Gorczyca, W. A., Riazance-Lawrence, J. H., Witkowska, D., and Polans, A. S. (1997). Calcium binding to recoverin: implications for secondary structure and membrane association. Biochim. Biophys. Acta 1342, 164–174.
Kakiuchi, S., Yasuda, S., Yamazaki, R., Teshima, Y., Kanda, K., Kakiuchi, R., and Sobue, K. (1982). Quantitative determinations of calmodulin in the supernatant and particulate fractions of mammalian tissues. J. Biochem. 92, 1041–1048.
Kawamura, S. (1993). Rhodopsin phosphorylation as a mechanism of cyclic GMP phosphodiesterase regulation by S-modulin. Nature 362, 855–857.
Klenchin, V. A., Calvert, P. D., and Bownds, M. D. (1995). Inhibition of rhodopsin kinase by recoverin. Further evidence for a negative feedback system in phototransduction. J. Biol. Chem. 270, 16147–16152.
Koch, K. W. (2000). Identification and characterization of calmodulin binding sites in cGMP-gated channel using surface plasmon resonance spectroscopy. Meth. Enzymol. 315, 785–797.
Kohnken, R. E., Chafouleas, J. G., Eadie, D. M., Means, A. R., and McConnell, D. G. (1981). Calmodulin in bovine rod outer segments. J. Biol. Chem. 256, 12517–12522.
Komolov, K. E., Senin, I. I., Kovaleva, N. A., Christoph, M. P., Churumova, V. A., Grigoriev, I. I., Akhtar, M., Philippov, P. P., and Koch, K. W. (2009). Mechanism of rhodopsin kinase regulation by recoverin. J. Neurochem. 110, 72–79.
Komolov, K. E., Senin, I. I., Philippov, P. P., and Koch, K. W. (2006). Surface plasmon resonance study of G protein/receptor coupling in a lipid bilayer-free system. Anal. Chem. 78, 1228–1234.
Komolov, K. E., Zinchenko, D. V., Churumova, V. A., Vaganova, S. A., Weiergräber, O. H., Senin, I. I., Philippov, P. P., and Koch, K. W. (2005). One of the Ca2+ binding sites of recoverin exclusively controls interaction with rhodopsin kinase. Biol. Chem. 386, 285–289.
Köster, S., Pavkov-Keller, T., Kühlbrandt, W., and Yildiz, O. (2011). Structure of human Na+/H+ exchanger NHE1 regulatory region in complex with calmodulin and Ca2+. J. Biol. Chem. 286, 40954–40961.
Lange, C., Duda, T., Beyermann, M., Sharma, R. K., and Koch, K. W. (1999). Regions in vertebrate photoreceptor guanylyl cyclase ROS-GC1 involved in Ca(2+)-dependent regulation by guanylyl cyclase-activating protein GCAP-1. FEBS Lett. 460, 27–31.
Levay, K., Satpaev, D. K., Pronin, A. N., Benovic, J. L., and Slepak, V. Z. (1998). Localization of the sites for Ca2+-binding proteins on G protein-coupled receptor kinases. Biochemistry 37, 13650–13659.
Liebman, P. A. (1972). “Microspectrophotometry of photoreceptors,” in Handbook of Sensory Physiology, ed. H. J. A. Dartnall (New York: Springer-Verlag), 7, 481–528.
Lukas, T. J., Iverson, D. B., Schleicher, M., and Watterson, D. M. (1984). Structural characterization of a higher plant calmodulin: Spinacia oleracea. Plant Physiol. 75, 788–795.
Makino, C. L., Dodd, R. L., Chen, J., Burns, M. E., Roca, A., Simon, M. I., and Baylor, D. A. (2004). Recoverin regulates light-dependent phosphodiesterase activity in retinal rods. J. Gen. Physiol. 123, 729–741.
McCarthy, N. E., and Akhtar, M. (2000). Function of the farnesyl moiety in visual signalling. Biochem. J. 347, 163–171.
McCue, H. V., Haynes, L. P., and Burgoyne, R. D. (2010). The diversity of calcium sensor proteins in the regulation of neuronal function. Cold Spring Harb. Perspect. Biol. 2, a004085.
Mikhaylova, M., Hradsky, J., and Kreutz, M. R. (2011). Between promiscuity and specificity: novel roles of EF-hand calcium sensors in neuronal Ca2+ signalling. J. Neurochem. 118, 695–713.
Palczewski, K., Buczylko, J., Lebioda, L., Crabb, J. W., and Polans, A. S. (1993). Identification of the N-terminal region in rhodopsin kinase involved in its interaction with rhodopsin. J. Biol. Chem. 268, 6004–6013.
Papermaster, D. S., and Dreyer, W. J. (1974). Rhodopsin content in the outer segment membranes of bovine and frog retinal rods. Biochemistry 13, 2438–2444.
Permyakov, S. E., Nazipova, A. A., Denesyuk, A. I., Bakunts, A. G., Zinchenko, D. V., Lipkin, V. M., Uversky, V. N., and Permyakov, E. A. (2007). Recoverin as a redox-sensitive protein. J. Proteome Res. 6, 1855–1863.
Permyakov, S. E., Zernii, E. Y., Knyazeva, E. L., Denesyuk, A. I., Nazipova, A. A., Kolpakova, T. V., Zinchenko, D. V., Philippov, P. P., Permyakov, E. A., and Senin, I. I. (2012). Oxidation mimicking substitution of conservative cysteine in recoverin suppresses its membrane association. Amino Acids 42. doi: 10.1007/s00726-011-0843-0
Pronin, A. N., Loudon, R. P., and Benovic, J. L. (2002). Characterization of G protein-coupled receptor kinases. Meth. Enzymol. 343, 547–559.
Pronin, A. N., Satpaev, D. K., Slepak, V. Z., and Benovic, J. L. (1997). Regulation of G protein-coupled receptor kinases by calmodulin and localization of the calmodulin binding domain. J. Biol. Chem. 272, 18273–18280.
Sanada, K., Shimizu, F., Kameyama, K., Haga, K., Haga, T., and Fukada, Y. (1996). Calcium-bound recoverin targets rhodopsin kinase to membranes to inhibit rhodopsin phosphorylation. FEBS Lett. 384, 227–230.
Sathyanarayanan, P. V., Cremo, C. R., and Poovaiah, B. W. (2000). Plant chimeric Ca2+/calmodulin-dependent protein kinase. Role of the neural visinin-like domain in regulating autophosphorylation and calmodulin affinity. J. Biol. Chem. 275, 30417–30422.
Satpaev, D. K., Chen, C. K., Scotti, A., Simon, M. I., Hurley, J. B., and Slepak, V. Z. (1998). Autophosphorylation and ADP regulate the Ca2+-dependent interaction of recoverin with rhodopsin kinase. Biochemistry 37, 10256–10262.
Schaad, N. C., De Castro, E., Nef, S., Hegi, S., Hinrichsen, R., Martone, M. E., Ellisman, M. H., Sikkink, R., Rusnak, F., Sygush, J., and Nef, P. (1996). Direct modulation of calmodulin targets by the neuronal calcium sensor NCS-1. Proc. Natl. Acad. Sci. U.S. A. 93, 9253–9258.
Senin, I. I., Dean, K. R., Zargarov, A. A., Akhtar, M., and Philippov, P. P. (1997). Recoverin inhibits the phosphorylation of dark-adapted rhodopsin more than it does that of bleached rhodopsin: a possible mechanism through which rhodopsin kinase is prevented from participation in a side reaction. Biochem. J. 321(Pt 2), 551–555.
Senin, I. I., Fischer, T., Komolov, K. E., Zinchenko, D. V., Philippov, P. P., and Koch, K. W. (2002a). Ca2+-myristoyl switch in the neuronal calcium sensor recoverin requires different functions of Ca2+-binding sites. J. Biol. Chem. 277, 50365–50372.
Senin, I. I., Koch, K. W., Akhtar, M., and Philippov, P. P. (2002b). Ca2+-dependent control of rhodopsin phosphorylation: recoverin and rhodopsin kinase. Adv. Exp. Med. Biol. 514, 69–99.
Senin, I. I., Höppner-Heitmann, D., Polkovnikova, O. O., Churumova, V. A., Tikhomirova, N. K., Philippov, P. P., and Koch, K. W. (2004). Recoverin and rhodopsin kinase activity in detergent-resistant membrane rafts from rod outer segments. J. Biol. Chem. 279, 48647–48653.
Senin, I. I., Tikhomirova, N. K., Churumova, V. A., Grigoriev, I. I., Kolpakova, T. A., Zinchenko, D. V., Philippov, P. P., and Zernii, E. Y. (2011). Amino acid sequences of two immune-dominant epitopes of recoverin are involved in Ca2+/recoverin-dependent inhibition of phosphorylation of rhodopsin. Biochemistry Mosc. 76, 332–338.
Senin, I. I., Zargarov, A. A., Alekseev, A. M., Gorodovikova, E. N., Lipkin, V. M., and Philippov, P. P. (1995). N-myristoylation of recoverin enhances its efficiency as an inhibitor of rhodopsin kinase. FEBS Lett. 376, 87–90.
Shifman, J. M., Choi, M. H., Mihalas, S., Mayo, S. L., and Kennedy, M. B. (2006). Ca2+/calmodulin-dependent protein kinase II (CaMKII) is activated by calmodulin with two bound calciums. Proc. Natl. Acad. Sci. U.S.A. 103, 13968–13973.
Singh, P., Wang, B., Maeda, T., Palczewski, K., and Tesmer, J. J. (2008). Structures of rhodopsin kinase in different ligand states reveal key elements involved in G protein-coupled receptor kinase activation. J. Biol. Chem. 283, 14053–14062.
Tanaka, T., Ames, J. B., Harvey, T. S., Stryer, L., and Ikura, M. (1995). Sequestration of the membrane-targeting myristoyl group of recoverin in the calcium-free state. Nature 376, 444–447.
Venkataraman, V., Duda, T., Ravichandran, S., and Sharma, R. K. (2008). Neurocalcin delta modulation of ROS-GC1, a new model of Ca(2+) signaling. Biochemistry 47, 6590–6601.
Wallace, R. W., Tallant, E. A., and Cheung, W. Y. (1983). Assay of calmodulin by Ca2+-dependent phosphodiesterase. Meth. Enzymol. 102, 39–47.
Weiergräber, O. H., Senin, I. I., Zernii, E. Y., Churumova, V. A., Kovaleva, N. A., Nazipova, A. A., Permyakov, S. E., Permyakov, E. A., Philippov, P. P., Granzin, J., and Koch, K. W. (2006). Tuning of a neuronal calcium sensor. J. Biol. Chem. 281, 37594–37602.
Wilden, U., Hall, S. W., and Kühn, H. (1986). Phosphodiesterase activation by photoexcited rhodopsin is quenched when rhodopsin is phosphorylated and binds the intrinsic 48-kDa protein of rod outer segments. Proc. Natl. Acad. Sci. U.S.A. 83, 1174–1178.
Zernii, E. Y., Komolov, K. E., Permyakov, S. E., Kolpakova, T., Dell’Orco, D., Poetzsch, A., Knyazeva, E. L., Grigoriev, I. I., Permyakov, E. A., Senin, I. I., Philippov, P. P., and Koch, K. W. (2011). Involvement of the recoverin C-terminal segment in recognition of the target enzyme rhodopsin kinase. Biochem. J. 435, 441–450.
Zozulya, S., and Stryer, L. (1992). Calcium-myristoyl protein switch. Proc. Natl. Acad. Sci. U.S.A. 89, 11569–11573.
Appendix
Modeling of Equilibrium between RK, Recoverin, Calmodulin, and Calcium Ions
In the simplest case the process of binding of two ligand (L) molecules by a protein (P) is described with the “four-states” scheme:
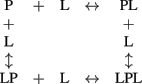
This scheme may be approximated by the scheme of sequential filling of the ligand-binding sites of the protein upon certain ratio of the equilibrium binding constants, describing the scheme. In the case of equilibrium between RK, recoverin, calmodulin, and calcium ions the following equilibria should be taken into account: (1) two four-states schemes, describing the calmodulin–calcium and RK–calmodulin interactions (overall, eight equilibria); (2) one sequential binding scheme, describing the recoverin–calcium interaction (two equilibria); (3) one equilibrium between RK and recoverin; (4) additional equilibria related to the presence of mixed RK forms with simultaneously bound calmodulin and recoverin molecule(s). Overall, the number of chemical equilibria describing this system exceeds 11. Furthermore, the experiments were performed in the presence of membranes, which affect the distribution of recoverin and RK between soluble and membrane-bound forms (Zozulya and Stryer, 1992; Sanada et al., 1996). Hence, the number of equilibria describing this system is actually much higher. The complexity of this system prevents from usage of microscopic constants for its description. The alternative approach is the use of effective equilibrium constants, describing separate stages of the process. Although this approach is phenomenological, it greatly facilitates description of the system and enables to establish the synergy between calmodulin- and recoverin-binding sites of RK.
The following simplified scheme of chemical equilibria describing the interactions between RK, recoverin, calmodulin, and calcium ions can be suggested:
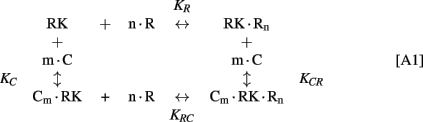
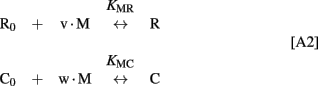
Here, M denotes calcium ion, R and C denote Ca2+-bound states of recoverin and calmodulin, respectively, while R0 and C0 denote their Ca2+-free states. Each of the equilibria is characterized by its own effective association constant:
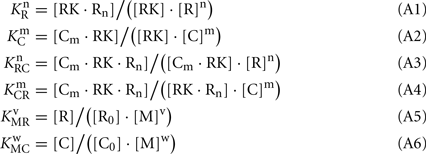
Combining the Eqs A1–A4, the following relationship between the parameters of the Scheme [A1] can be derived:

The changes in RK-inhibiting activity (see Figure 4A) reflect association of the Ca2+-sensing proteins with RK under saturating Ca2+ level, so at the middle of the transition the following condition takes place:
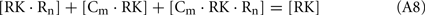
Using Eqs A1–A3, the latter equation can be rewritten:
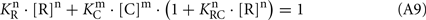
Assuming that the synergetic inhibition of RK by recoverin/calmodulin mixture occurs at [C] = [R] = 4.6 μM (see Figure 4A; other numerical values are taken from Table 1),
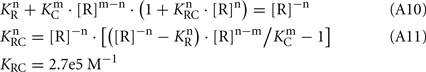
Hence, from Eq. 7,
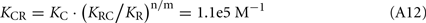
The Ca2+-dependence of RK-inhibiting activity of recoverin and calmodulin (see Figure 4B) reflects association of the Ca2+-sensing proteins with calcium ions, so at the middle of the transition the following equation takes place:
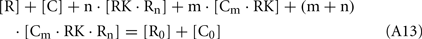
Since total concentration of RK was about 20 nM, the concentrations of all RK states may be neglected in comparison with concentrations of the RK-free forms of recoverin and calmodulin:

Considering the material balance in the system,

where R0 and C0 are total concentrations of recoverin and calmodulin, respectively.
The system of Eqs A14–A16 and A5–A6 can be solved with respect to [M].
Substituting Eqs A5–A6 into Eqs A14–A16,
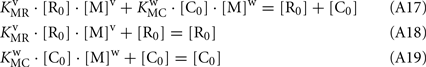
From Eqs A17–A19:
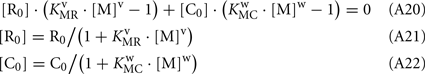
Substituting Eqs A21–A22 into Eq. 20, and considering that R0 = C0:
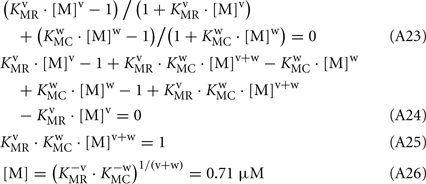
Keywords: rhodopsin kinase, calmodulin, recoverin, phosphorylation, surface plasmon resonance
Citation: Grigoriev II, Senin II, Tikhomirova NK, Komolov KE, Permyakov SE, Zernii EY, Koch K-W and Philippov PP (2012) Synergetic effect of recoverin and calmodulin on regulation of rhodopsin kinase. Front. Mol. Neurosci. 5:28. doi: 10.3389/fnmol.2012.00028
Received: 05 January 2012; Accepted: 17 February 2012;
Published online: 08 March 2012.
Edited by:
Michael R. Kreutz, Leibniz-Institute for Neurobiology, GermanyReviewed by:
Karl Heinz Braunewell, Southern Research Institute, USAJacques Haiech, University of Strasbourg, France
Copyright: © 2012 Grigoriev, Senin, Tikhomirova, Komolov, Permyakov, Zernii, Koch and Philippov. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Ivan I. Senin and Evgeni Yu. Zernii, Department of Cell Signaling, A.N. Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, Leninskie Gory 1, Building 40, Moscow 119992, Russia. e-mail:c2VuaW5AYmVsb3plcnNreS5tc3UucnU=;emVybmlAYmVsb3plcnNreS5tc3UucnU=
†Present address: Konstantin E. Komolov, Department of Biochemistry and Molecular Biology, Thomas Jefferson University, Philadelphia, PA 19107, USA.


 Natalya K. Tikhomirova1
Natalya K. Tikhomirova1