- Laboratory of Functional Genomics and Molecular Toxicology, Division of Toxicology, CSIR-Central Drug Research Institute, Lucknow, India
Neurodegenerative Parkinson’s disease (PD) is a multi-factorial disorder lacking complete cure. Understanding the complete mechanism of initiation and progression of this disease has been quite challenging; however, progress has been made toward deciphering certain genetic aspects related to the disease condition. Genetics studies have provided clues toward the role of microRNAs (miRNAs) in various disease conditions. One of the crucial miRNA molecules, let-7, is highly conserved miRNA and is known to regulate important functions of development and viability; its altered expression has been reported in C. elegans model of PD. We carried out studies with let-7, employing transgenic C. elegans model expressing ‘human’ alpha-synuclein and developed a let-7 loss-of-function model toward studying the downstream effects related to PD. We observed that let-7 miRNA was upregulated in C. elegans model of PD and figured that loss of let-7 miRNA leads to decreased alpha-synuclein expression, increased autophagy, increased Daf-16 expression, increased oxidative stress and increased lipid content with no effect on dopaminergic/acetylcholinergic neurons. Our findings indicate that let-7 miRNA regulates PD-associated pathways. Our study provides insight toward the role of let-7 in regulating expression of genes associated with these pathways which might have implications on the multi-factorial nature of PD. Potential pharmacological agents modulating the expression of let-7 could be studied toward targeting the multi-factorial aspect of PD.
Introduction
Neurodegenerative Parkinson’s disease (PD) is an age-related disorder, and is characterized by the accumulation of Lewy bodies and Lewy neurites in substantia nigra pars compacta region of the brain. Lewy bodies are composed of alpha-synuclein protein in high proportion (Chaudhuri et al., 2015). PD affects 1–2% of the population and is the second most common among all neurodegenerative diseases (NDs). The symptoms of this disease include bradykinesia, muscle rigidity, cognition defects, tremors, as well as personality and behavioral disorders (Wong and Nass, 2012). PD is an incurable, multi-factorial disease which is associated with aggregation of misfolded proteins, alteration in levels of neurotransmitter dopamine, increase in neuronal cell death and disturbance in the ubiquitin–proteasome system (Gao and Hong, 2011). PD may result from genetic mutations, environmental exposure to toxins and is most commonly associated with old age. PD occurs most commonly in sporadic form rather than familial form. The familial form of PD is caused by the mutation in any of the proteins α-synuclein, perkin, PINK1, UCHL1, DJ1, or LRRK2 genes. The familial form accounts only for 5–10% of patients (Douglas et al., 2007; Shulman et al., 2011) whereas, rest of the cases are sporadic in nature. No single or independent factor is known that could be targeted for combating PD, and for its multifactorial nature, the disease lacks a complete cure. It could be possible that studying miRNA molecules may help identify targets that may be helpful in treating multiple pathways that go awry in PD patients.
MicroRNAs are endogenous evolutionarily conserved 20–25 nucleotide long non-coding RNAs. These molecules function via being trans-acting factors and regulate gene expression machinery at post-transcriptional level. miRNAs inhibit protein synthesis either by degradation of targeted mRNA or by inhibition of its translation. Some recent studies report that miRNAs regulate gene expression at transcriptional level (Selbach et al., 2008; Bicchi et al., 2013). Alterations in the functions/biogenesis of miRNAs have been linked to multiple ailments that include neurodegenerative diseases (NDs), cancer, cardiovascular disease, and diabetes mellitus (Sonntag, 2010; Kumar et al., 2012; Dimmeler and Nicotera, 2013; Kocerha et al., 2014). Like most coding genes, miRNA genes are transcribed through RNA polymerase II transcriptional activity, generating hairpin like structure called primary transcript. Within nucleus these primary transcript pri-miRNAs, are processed by microprocessor complex protein resulting in precursor miRNAs (pre-miRNAs). After that, pre-miRNAs are transported into cytoplasm via expotin-5 from nucleus. Cytoplasmic pre-miRNAs are further processed to generate mature miRNAs which are incorporated into RNA-induced silencing complex followed by inhibition of target mRNA either by degradation of mRNA or by repression of translation (Ling et al., 2013; Chaudhuri et al., 2016; Shamsuzzama et al., 2016). miRBase21 database1 illustrates the presence of 434 predicted mature miRNAs in C. elegans, and 2,588 mature miRNAs in Homo sapiens, although an actual number of miRNAs may be higher.
In mammals, miRNAs play important role in the development of brain, neuronal specification, function, and maintenance (Krichevsky et al., 2003, 2006; Sempere et al., 2004; Schratt et al., 2006). Most of the specific genes required for cellular identity are regulated by miRNAs thus suggesting that miRNAs may have significant role in the development of complex tissue and organs of higher organisms (Lee et al., 2006; Lu et al., 2007).
Let-7 miRNA is 22 nt long non-coding RNA, which was first discovered in C. elegans. It is highly conserved across animal species and the let-7 family consists of 9, 14, and 13 members in C. elegans, mouse and humans, respectively (Shamsuzzama et al., 2016). Let-7 miRNA is found to be downregulated in different types of cancer including lung cancer, breast cancer, colon cancer, gastric cancer, and Burkitt’s lymphoma. Let-7 miRNA acts as tumor suppressing miRNA and may well come up as an interesting target for various cancers (Barh et al., 2010). Let-7 directly regulates oncogenic genes that are involved in signaling pathways in tumor progression. Oncogenes that are regulated by let-7 are ras, hgma2, myc, NIRF and JAK-STAT3 pathway molecules (Wang et al., 2012). There is very little that is known about the role of let-7 miRNA in the progression of PD. However, some studies have shown that its expression levels were altered in C. elegans model of PD (Asikainen et al., 2010), which implies that let-7 miRNA networking pathways may be playing a critical role in PD development.
In order to investigate the role of let-7 miRNA in PD and its associated factors we designed RNAi feeding bacterial clone of let-7 miRNA toward knocking down let-7 miRNA in the nematodes and studied its effect on disease model for various endpoints, including investigation of alpha-synuclein protein expression, lipid content, oxidative stress, quantification of autophagy/apoptosis marker genes, dopaminergic neurodegeneration and associated phenotypes. We found that loss of let-7 miRNA leads to decreased alpha-synuclein expression, increased autophagy, increased Daf-16 expression, increased oxidative stress and increased fat content with no effect on dopaminergic/acetylcholinergic neurons. Our study provides understanding of the role of miRNA let-7 in PD and confirms that absence of let-7 miRNA leads to decrease in accumulation of alpha-synuclein protein in transgenic C. elegans. Our studies further provide evidence that let-7 possibly decreases alpha-synuclein expression via increasing autophagy and increasing daf-16 forkhead box O (FOXO) transcription factor.
Materials and Methods
C. elegans Culture and Maintenance
C. elegans strains were cultured using standard techniques as described previously (Brenner, 1974). Escherichia coli (E. coli) strain-OP50 (uracil auxotroph) was used as standard food. To obtain age synchronized animals, procedure described previously was followed (Stiernagle, 2006). In brief, worms were washed with M9 buffer and then treated with axenizing solution (5 mL of 1M sodium hydroxide solution and 2 mL of sodium hypochlorite) until the eggs were released from the dissolved worm bodies.
C. elegans Strains
Strains employed in this study were N2, wild-type Bristol; NL5901, pkIs2386 [unc-54p::alpha-synuclein::YFP + unc-119(+)]; BZ555, egIs1 [dat-1p::GFP]; LX929, vsIs48[unc-17::GFP]; DA2123, adIs2122 [lgg-1p::GFP+ rol-6(su1006)]. All the strains were procured from the Caenorhabditis Genetics Center (University of Minnesota).
Genomic DNA Isolation
Genomic DNA from mixed population of C. elegans (N2 Bristol) was isolated using PureLink® Genomic DNA Kit (Invitrogen, cat no. K1820-01) as described in the manufacturer’s manual. Briefly, worms were washed with M9 buffer, lysed by adding 180 μl pure link genomic digestion buffer, 20 μl proteinase K and kept at 55°C in water bath for 3 h. Afterward, 200 μl pure link genomic lysis/binding buffer, 200 μl 100% ethanol were added, mixed well and transferred to pure link spin column. This was followed by centrifugation at 10,000 × g for 1 min at RT. Column was washed with provided wash buffers and eluted with pure link genomic elution buffer.
Plasmid Constructs
Plasmids were constructed using standard techniques as described previously (Fraser et al., 2000). In brief let-7 miRNA gene sequence was retrieved from WormBase (sequence number C05G5.6). The 99 bp full length was amplified using standard PCR with a set of primers having sacI and kpnI restriction sites. A 25 μl reaction mixture containing 50 ng of C. elegans genomic DNA, 400 nanomolar forward and reverse primer, 10 mM dNTPs was prepared followed by incubation at 95°C for 10 min (1 cycle), 94°C for 30 s, 55°C for 30 s, 72°C for 30 s (30 cycle), and 72°C for 5 min (1 cycle). Amplified product was subcloned in TA (pCR® 2.1 vector) (Invitrogen cat no. 450046) and then cloned employing Timmons and Fire feeding vector L4440 (Addgene plasmid 1654), and transformed into HT115 (DE3), an RNase III-deficient E. coli strain with IPTG inducible T7 polymerase activity. Colonies containing correct sized insert were confirmed with double digestion by sacI and kpnI restriction digestion.
Forward primer – GAG CTC TAC ACT GTG GAT CCG GTG AGG T (Tm- 59.2)
Reverse primer – GGT ACC TCG AAG AGT TCT GTC TCC GGT A (Tm- 57.8).
let-7 sequence (C05G5.6);
tacactgtggatccggtgaggtagtaggttgtatagtttggaatattaccaccggtgaactatgcaattttctaccttaccggagacagaactcttcga.
Isolation of Non-coding Small RNA Using mirVanaTM miRNA Isolation Kit
The extraction of non-coding was carried out employing standard procedure, briefly, water treated with 0.2% diethyl pyrocarbonate (DEPC-Sigma, Cat. No.-D5758) was used to remove adhering bacteria from age synchronized N2 and let-7 silenced groups. miRNA was isolated using mirVanaTM miRNA isolation kit (Ambion P/N AM1561) as per instruction provided within the manufacturer’s manual. Briefly, 250 μl lysis/binding buffer was added followed by homogenization and addition of 1/10 volume miRNA homogenate additive, the solution was mixed well and kept on ice for 10 min. After that equal amount of acid-phenol:chloroform was added to the lysate, it was mixed well and centrifuged at 10,000 × g for 5 min., the aqueous phase was removed, and 1.25 times volume of 100% ethanol was added followed by filtration via passing through filter cartridge, washing, and elution.
Reverse Transcriptase Reaction
Isolated non-coding small RNAs were converted into cDNA using TaqMan® MicroRNA Reverse Transcription kit (Applied Biosystem cat no. P/N 4366596). The 15 μl reactions were incubated in an Agilent sure Cycler 8800 for 30 min at 16°C, for 30 min at 42°C, 5 min at 85°C and then held at 4°C.
TaqMan miRNA Assay
Quantification of miRNA was carried out using TaqMan® Universal Master MixII (Applied Biosystem cat no. 4440040). The 20 μl maxima contain 1X TaqMan® Universal Master MixII no UNG, 1X TaqMan® assay and 100 ng cDNA template. The program for amplification was 95°C for 10 min (1 cycle), followed by 95°C for 15 s and 60°C for 1 min (40 cycles). Experiment of each sample was carried out in triplicate sets. Fold change of all samples were analyzed using comparative 2-ΔΔCT. U18 was used as endogenous control for normalization of miRNAs expression.
Prediction of Targets and Pathway Analysis of Let-7 miRNA Molecules
We used DIANA TOOLS – mirPath v.3 for target prediction and pathway analysis. DIANA TOOLS works based on specifically designed algorithms and database toward mining available information pertaining to test molecules and associated pathways. Enrichment analysis of target genes of miRNA to Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways is performed by this software thus providing with an overview of pathway which might be regulated by specified miRNAs.
Real-time PCR (qPCR) Assay
Age synchronized control and let-7 knockdown worms were washed twice with 0.2% DEPC (Sigma, Cat. No.-D5758) treated water to remove adhering bacteria, following which total RNA was isolated using RNAzol® RT method (Sigma, Cat. No. R4533), and quantified through NanoDrop (Thermo, Quawell, UV-Vis Spectrophotometer, Q5000). About 5 μg of total RNA was used for the synthesis of cDNA using RevertAid First Strand cDNA synthesis kit (Thermo Scientific, Cat. No. #K1622). Quantification of mRNA level was carried out using SYBR® Select Master Mix (Applied Biosystems cat. No.4472908) chemistry as described previously (Jadiya et al., 2012). In brief cDNA equivalent to 125 ng was amplified in 20 μl maxima using Agilent MX3005P-detection system (Agilent Technologies). The program for amplification was, 50°C for 2 min 95°C for 10 min (1 cycle), followed by 95°C for 30 s, 55°C for 30 s and 60°C for 30 s (40 cycle) and melting curve detection (95°C for 5 s, 65°C for 1 min). Experiment of each sample was carried out in duplicate sets. Fold change of all samples was analyzed using comparative 2-ΔΔCT. Integrated DNA Technologies (IDT) software was used for designing of primers of desired genes. act-1 mRNA was used as endogenous control for normalization. Primers sequences of genes used are as follows:-
act-1 forward: TTA CTC TTT CAC CAC CAC CGC TGA
act-1 reverse: TCG TTT CCG ACG GTG ATG ACT TGT
lgg-1 forward: AAC AAC TTT GAG AAG CGT CGT GCC
lgg-1 reverse: TCT TCT GGA CGA AGT TGG ATG CGT
atg-5 forward: TGA TGA AAG ACG AGT CGG CAC AGT
atg-5 reverse: GTT TGG CAG TGA TTA GGG CCT GTT
atg-7 forward: CGC TTG GAT GTA ACA TTG CCC GTT
atg-7 reverse: AAT GCG TTG GAT AGC AGC TTG TGC
atg-13 forward: ACT CCA GAA GAC AAA GAG CCA ACG
atg-13 reverse: TTG GCG CAC CAC CGA AAT CTG ATA
vps-34 forward: TGG ATC CCT TTG CAT CAC GAC GTA
vps-34 reverse: CGA AAC AAT CCC AAC ACC ACC GTT
bec-1 forward: AGG AGC TGG AGC AAC AGT TGA AGA
bec-1 reverse: ATA TTG ACG TTC GGC TTC CAG CGA
ced-4 forward: AGT GCT CTT GCT TTC GCA GTT GTG
ced-4 reverse: TGA GAA GAG CTC CAC GTT TGC TGA
cep-1 forward: AGT CGT CTT CAT GGA TGC GTT CCT
cep-1 reverse: TTG CGT CGG AAC CCA AGT GTA TCT
lin-45 forward: AGT CTG CCG AGA TGT GCT TCT TCA
lin-45 reverse: TTG TCA CTT GTT CCT GCT CCT CCA
nsy-1 forward: TCT TCA TTC CAC GTT GTG CCA TGC
nsy-1 reverse: ACC CTC CAG AAT TTG CTT CCC GTA
jnk-1 forward: TGG CTG GTT CCA TCA TCA TCT GGT
jnk-1 reverse: CGT TTG AGA ACA ACC ATC TGC GCT
jkk-1 forward: GAA GCT GCT GCG TCG CAT TTA TCA
jkk-1 reverse: ACA CAG CTT TAC ATT GCC GCT GTC
daf-16 forward: GCG AAT CGG TTC CAG CAA TTC CAA
daf-16 reverse: ACA CGA TCC ACG GAC ACT GTT CAA
daf-12 forward: GCT CCT GGT ATG AAT GGG TATC
daf-12 reverse: ACT CTC TTC GCT GGA GTC TAA
Silencing of Let-7 miRNA
The knockdown of let-7 miRNA was achieved by employing standard feeding protocol as described previously (Fraser et al., 2000). dsRNA expressing bacterial clone targeted for C. elegans miRNA, was cultured for 6–8 h in LB containing 50 μg/ml ampicillin, then seeded onto NGM plates with 5 mM IPTG and 25 mg/L carbenicillin followed by an overnight incubation at 22°C to induce expression. Synchronous populations of embryos were transferred onto these plates and control (EV) for further studies.
Assay for Alpha-synuclein Protein Accumulation
Expression of alpha-synuclein protein was examined in control and let-7 miRNA knockdown worms of the NL5901 strains as described previously (Jadiya et al., 2012). In brief after 48 h of treatment, worms were washed with M9 buffer up to three or four times until all adhering bacteria were removed. Worms were immobilized with 100 mM sodium azide (Sigma, cat no. 71289) and placed onto slides with agar pads (2% agarose) followed by sealing with cover slip. Imaging of immobilized worms was carried out using fluorescence microscope (Carl Zeiss) to monitor alpha-synuclein protein expression. In each group minimum 10 images were taken for analysis and each experiment was repeated thrice. ImageJ software (ImageJ, National Institutes of Health, Bethesda, MD, United States) was used to quantify the expression of protein by measuring fluorescence intensity.
Assay for Autophagy in Worms
Transgenic strain DA2123 (LGG-1::GFP) was used for assessment of autophagy. LGG-1 is associated with autophagosomal membrane and has been widely used for autophagy detection (Jenzer et al., 2014; Manil-Segalen et al., 2014). LGG-1 is the ortholog of the mammalian LC3 and Saccharomyces cerevisiae Atg8 protein. In this study, control and let-7 miRNA silenced worms were washed with M9 buffer to remove any adhering bacteria. Worms were immobilized with 100 mM sodium azide (Sigma, cat no. 71289) onto 2% agar padded slides and sealed with a cover slip. Imaging of live (immobilized) worms was done using fluorescence microscope (Carl Zeiss) at 63× oil magnification to monitor punctate GFP. Experiments were repeated three times and five worms were taken for analysis for each individual group.
Assay for Motility of Worms
Motility defect due to neurotransmitter imbalance is one of symptoms of PD. Motility of worms was quantified through thrashing assay in which number of thrashes was counted as per previously described method (Buckingham and Sattelle, 2009). One thrash was defined as complete bending of the body one way to the outermost angle and back to the initial posture. In this study, worms from different assay conditions were washed with M9 buffer to eliminate adhering bacteria; a single worm was placed on a drop of M9 buffer. The timer was set at 30 s and sigmoidal body bends were counted under stereo-zoom microscope (Leica). Ten worms were counted for every group; experiments were repeated thrice.
Estimation of Reactive Oxygen Species (ROS)
In order to explore the effect of let-7 miRNA on the level of oxidative stress, we carried out estimation of ROS levels using 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) assay following the protocol as described by Kaur et al. (2012). Worms of control and let-7 miRNA silenced groups were washed thrice with M9 buffer and twice with phosphate buffer saline (PBS). An approximate number of 100 worms/100 μl assay solution, were transferred to assay wells of OptiPlate-96 F (Perkin Elmer) with each group being assayed in triplicates. A volume of 100 μl H2DCFDA (Cat. No. D399, Invitrogen) from an ethanol stock of 100 μM, was added to each well. Fluorescence from each well was quantified at three time points – (i) before addition of the dye, (ii) immediately after addition of the dye, and (iii) post 1 h incubation of addition of the dye. The Fluorescence intensity measurements were carried out using Multimode plate reader (Perkin Elmer, VICTORTM X3), at excitation wavelength 485 nm and an emission wavelength 520 nm. The change in fluorescence was calculated by subtracting initial reading from the final reading; the numbers were presented as fluorescence intensity per worm and plotted as mean ± SE. Statistical significance was calculated by student’s t-test using GraphPad Prism software package.
Assay for Fat Content in Nematodes
The effect of let-7 miRNA silencing on fat content in nematodes was studied by staining worms with Nile red (MP Biomedicals cat no. 151744) a fat staining dye. Nile red was mixed with control (EV)/let-7 miRNA RNAi clone and seeded onto NGM-IPTG plates followed the protocol as described previously (Ashrafi et al., 2003). Synchronous aged embryos derived from sodium hypochlorite treatment were transferred onto Nile red pre-mixed plates and kept for 48 h at 22°C. After 48 h, synchronized worms were washed off with M9 buffer two to three times to eliminate adhering bacteria. Worms were mounted with 100 mM sodium azide (Sigma, cat no. 71289) using agar padded cover slip on a glass slide. The extent of fat content was analyzed using fluorescence microscope (Carl Zeiss). Fluorescence intensity was quantified using ImageJ software (ImageJ, National Institutes of Health, Bethesda, MD, United States). Five worms were analyzed for each group and experiments were repeated thrice.
Studies on Acetylcholinergic and Dopaminergic Neurons
Here, we employed transgenic C. elegans strain LX929 (unc-17::GFP; expressing GFP under the influence of the unc-17 promoter specifically in cholinergic neurons) and BZ555 (Pdat-1::GFP; expressing GFP under the influence of the dat-1 promoter specifically in the dopaminergic neurons) for assaying the effect of let-7 miRNA silencing on acetylcholinergic and dopaminergic neurons (Pu and Le, 2008; Barbagallo et al., 2010). Worms of different groups were washed with M9 buffer to remove adhering bacteria. The 100 mM sodium azide (Sigma, cat no. 71289) was used for anesthetizing the worms and mounted onto glass slide with agar padded cover slip. Fluorescence intensity measurement for GFP was carried out using fluorescence microscope (Carl Zeiss). Experiments were repeated thrice and imaging was carried out for a minimum 10 worms in each individual group.
Effect on Dopamine Associated Function
Dopamine is a neurotransmitter of phenethylamine and catecholamine families. It plays important role in the regulation of motor behavior of C. elegans and also plays important role in functions like olfaction. Response of C. elegans to volatile attractants and repellents depends upon the levels and functioning of dopamine. Any alteration in dopamine leads to defects in motor function in response to foods and repellents (Sharples et al., 2014). We studied effect of dopamine in worms employing well established assay, i.e., nonanol repulsion assay (Jadiya et al., 2016). A single worm was placed in NGM plate and a drop of 1-nonanol was put near the head of the worm. The time taken by the worm in responding to the repellent by its ‘repulsion response’ was assayed for 10 worms of each group. The mean time of repulsion (in seconds) was calculated and the statistical significance of test groups was calculated with reference to control.
Statistical Analysis
Statistical analysis was carried out using GraphPad software package; statistical significance of data was calculated employing student’s t-test. All data are presented as mean ± standard error of the mean.
Results
Let-7 miRNA Was Over-expressed in C. elegans Model of PD
Impaired miRNA expression is known to be associated with the development and progression of neurodegenerative PD (Wong and Nass, 2012). We employed C. elegans model of PD, (NL5901) for quantification of the expression level of let-7 miRNA. NL5901 is the transgenic strain expressing wild type “human” alpha-synuclein protein in body wall muscle under the control of unc-54 promoter. This strain is designed in such a way that the alpha-synuclein expression is driven to muscle cells via unc-54 promoter. Considering the fact that neuronal cells are largely refractory to RNAi mediated silencing, studying such effects in muscles makes it effective and the YFP expression can be detected via microscopy. Transgenic strains expressing either wild type or mutant alpha-synuclein protein under pan neuronal promoter have shown similar effect on locomotion (Lakso et al., 2003). In order to quantify the expression level of let-7 miRNA, we carried out TaqMan based real-time PCR studies for let-7 miRNA in wild type (N2) and alpha-synuclein expressing strain (NL5901) of C. elegans. We observed that let-7 miRNA was overexpressed in PD model by 75% (p < 0.001) as compared to that of control group (Figure 1A). This, intriguingly, is in contrast to the findings gathered studying ‘mutant’ alpha-synuclein, as against wild type species studied by us. Asikainen et al. (2010) reported that let-7 miRNA was downregulated in transgenic strain expressing mutant alpha-synuclein (A53T). This opposite effect could be attributed to the fact that the previous studies reported data on “mutant’ alpha-synuclein species whereas our studies were conducted on the ‘wild type’ alpha-synuclein.
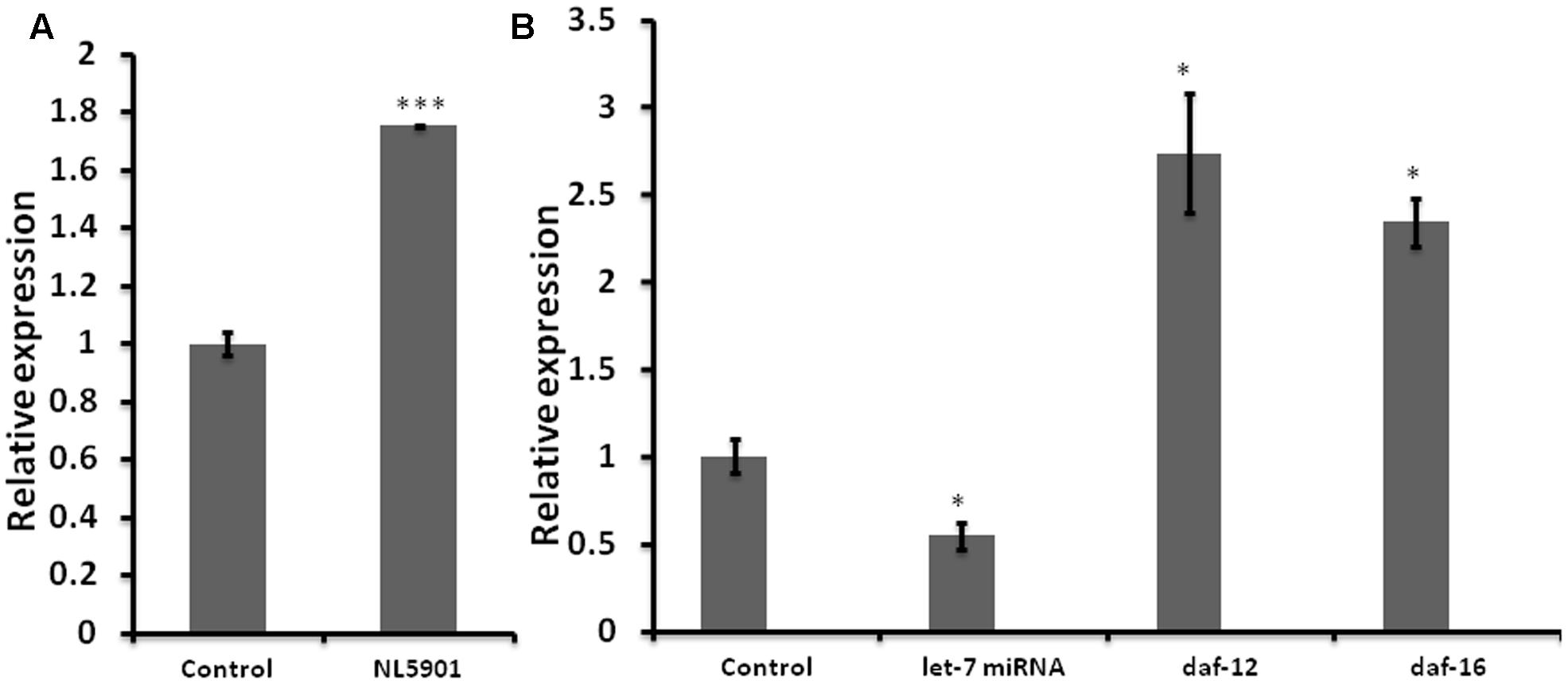
FIGURE 1. Graph depicting relative expression of let-7 miRNA and its targets, studied through real-time PCR (qPCR). (A) Expression level of let-7 miRNA in PD model vs. wild type. (B) Expression level of let-7 miRNA and their targets in let-7 knockdown worms. ∗p < 0.05, ∗∗∗p < 0.001.
Mature Let-7 miRNAs Were Downregulated in Let-7 miRNA Silenced Worms
Reverse genetics is a widely used method in functional genomics research. Knockdown of genes via RNA interference has been well established and widely accepted tool for studying the function of target genes in C. elegans. Keeping this in mind we created RNAi feeding bacterial clone for let-7 miRNA in order to decipher its function. For the validation of RNAi mediated inhibition we carried out TaqMan miRNA assay toward quantification of let-7 miRNA levels under untreated and let-7 miRNA silenced conditions. We observed that let-7 miRNA was reduced by 46% (p < 0.05) in let-7 miRNA silenced worms as shown in Figure 1B.
RNAi of Let-7 miRNA Resulted in Upregulation of Downstream Target Genes
miRNAs are endogenous short nucleotide targets of mRNA which suppress target gene function either by its degradation or repression of translation. Single mature miRNA could target hundreds of mRNA molecules at the same time. We carried out quantitative real-time PCR studies toward quantification of the mRNA levels of daf-12 and daf-16 in worms of control and let-7 miRNA silenced groups. DAF-12 is a nuclear hormone receptor; a member of steroid hormone receptor superfamily. daf-12 mRNA is negatively regulated by let-7 miRNA. DAF-12 affects dauer formation and aging via TGF and insulin signaling pathway (Grosshans et al., 2005). DAF-16 homolog of mammalian FOXO transcription factor plays important role in neuroprotection (Tehrani et al., 2014). We observed that there was a significant 174% (p < 0.05), and 134% (p < 0.05) upregulation of daf-12 and daf-16, respectively, as compared to control (Figure 1B) that validated the role of let-7 miRNA in the regulation of daf-12 (previously reported; Grosshans et al., 2005) and predicted daf-16 genes (miRBase21)2.
Pathway Analysis of Let-7 miRNA
It is well known that single miRNA molecule might regulate multiple genes simultaneously (Qiu et al., 2011) thereby regulating multiple pathways. In order to investigate which biological pathways are affected by let-7 miRNA, we applied miRPath v.3, a miRNA pathway analysis tool. According to findings gathered from this tool, let-7 miRNA is involved in pathways of apoptosis, autophagy, cell cycle regulation, glycolysis/gluconeogenesis, MAPK signaling pathway and P13K-Akt signaling pathway (Figure 2).
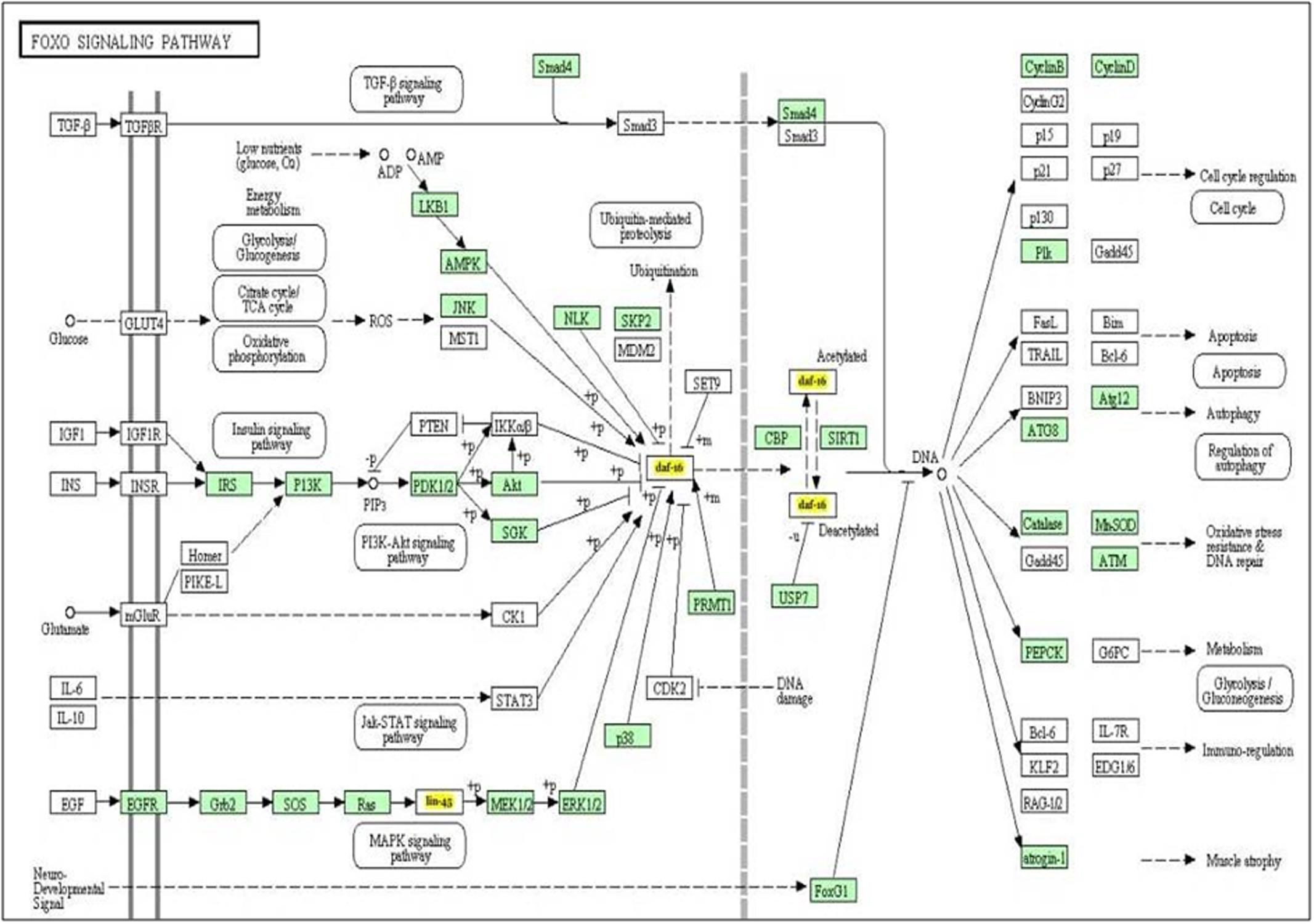
FIGURE 2. The KEGG pathway “FOXO signaling pathway” is regulated by let-7 miRNA. Target genes of let-7 miRNA are denoted by yellow color.
Knockdown of Let-7 miRNA Led to Reduced Expression of Alpha-synuclein Protein
Silencing let-7 in NL5901 transgenic strain led to decreased accumulation of alpha-synuclein. Accumulation of protein was deliberated as YFP expression pattern in the transgenic strain NL5901. Worms in the control group (NL5901 fed on EV) expressed optimal level of alpha-synuclein protein (Figure 3A), while let-7 miRNA silenced worms showed reduction in the level of alpha-synuclein protein (Figure 3B). The knockdown of let-7 miRNA decreased the fluorescence intensity of alpha-synuclein::YFP by 2.82-fold (p < 0.001) when compared to control worms; with mean fluorescence intensity for the control group 31.57 ± 0.5497 (N = 10) arbitrary units and that for let-7 miRNA knockdown worms was 11.18 ± 0.2047 (N = 10) arbitrary units (Figure 3C).
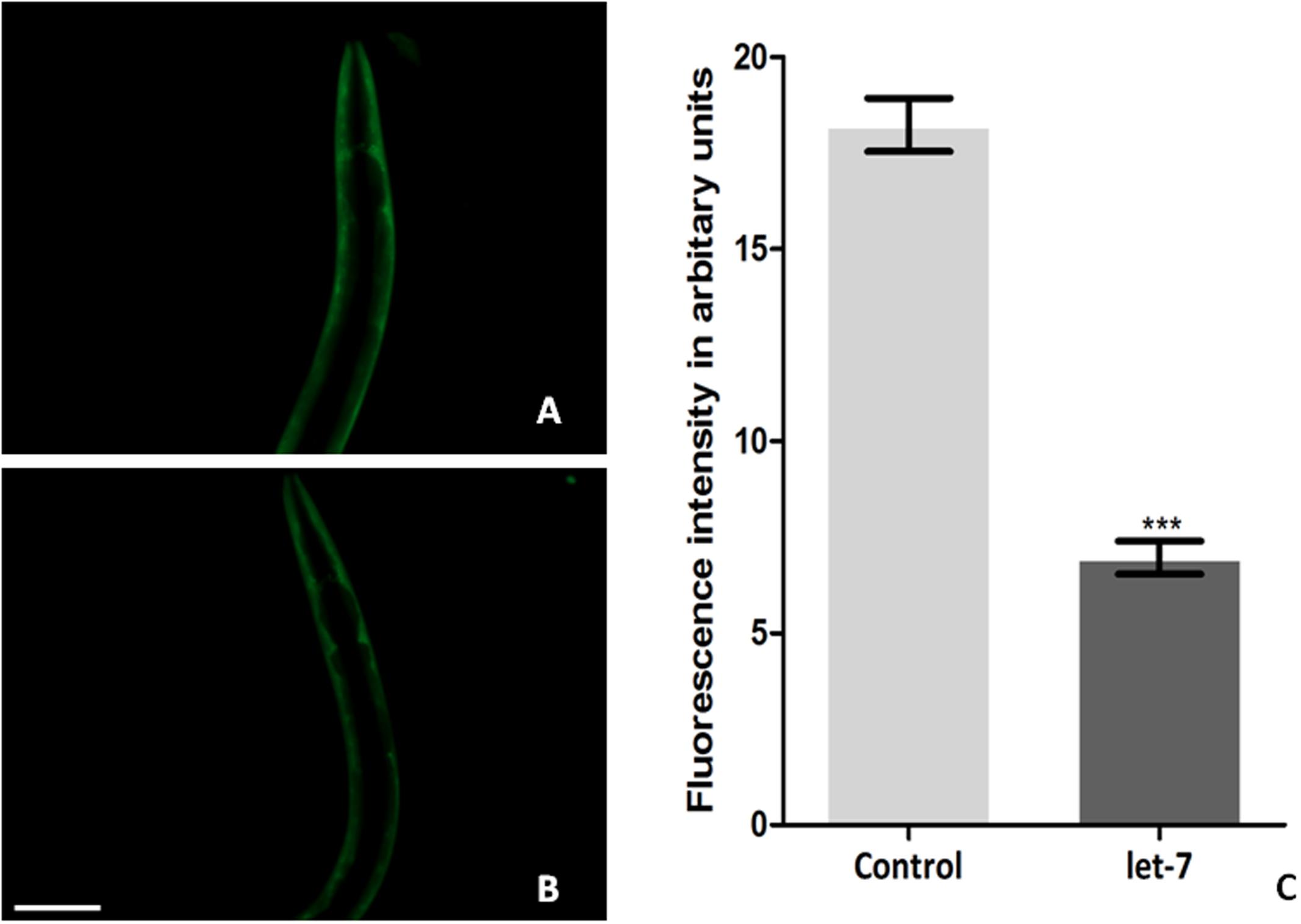
FIGURE 3. Alpha-synuclein expression in NL5901 strain of C. elegans (studied through fluorescence microscopy) fed on control (A) and let-7 miRNA knockdown condition (B). (C) Graphical representation of fluorescence intensity as quantified using ImageJ software (∗∗∗p < 0.001). Scale bar: 50 μm.
Knockdown of Let-7 miRNA Influenced the Expression of Autophagy Marker Genes
Clearance of damaged organelles and protein aggregates is associated with autophagy. Mutation in autophagy regulating genes is known to be related with NDs (Nixon, 2013). To explore the function of let-7 miRNA in autophagy mediated neuroprotection, we studied known autophagy marker genes (Yue et al., 2009) and quantified their mRNA levels using quantitative real-time PCR (qPCR) in normal and let-7 miRNA silenced condition. We found that expression level of lgg-1 and atg-13 mRNAs was significantly upregulated by 19% (p < 0.05) and 47% (p < 0.05) while mRNA levels of atg-5 and atg-7 were significantly downregulated by 35% (p < 0.05) and 33% (p < 0.05), respectively, when compared to that of worms from control group (Figure 4A). lgg-1 is an ortholog of Saccharomyces cerevisiae Atg8p and mammalian MAP-LC3, which is required for degradation of cellular components, atg-13 is an autophagy related gene required for autophagosome formation. The process of autophagy is known either to be dependent or independent of atg-5/atg-7 pathway (Nishida et al., 2009). We observed that knocking down of let-7 miRNA led to increase in lgg-1 and atg-13 whereas it led to decrease in atg-5 and atg-7 expression. These results suggest that absence of let-7 miRNA exerts its effects via atg-5/atg-7 independent alternative pathway for clearance of misfolded aggregated proteins.
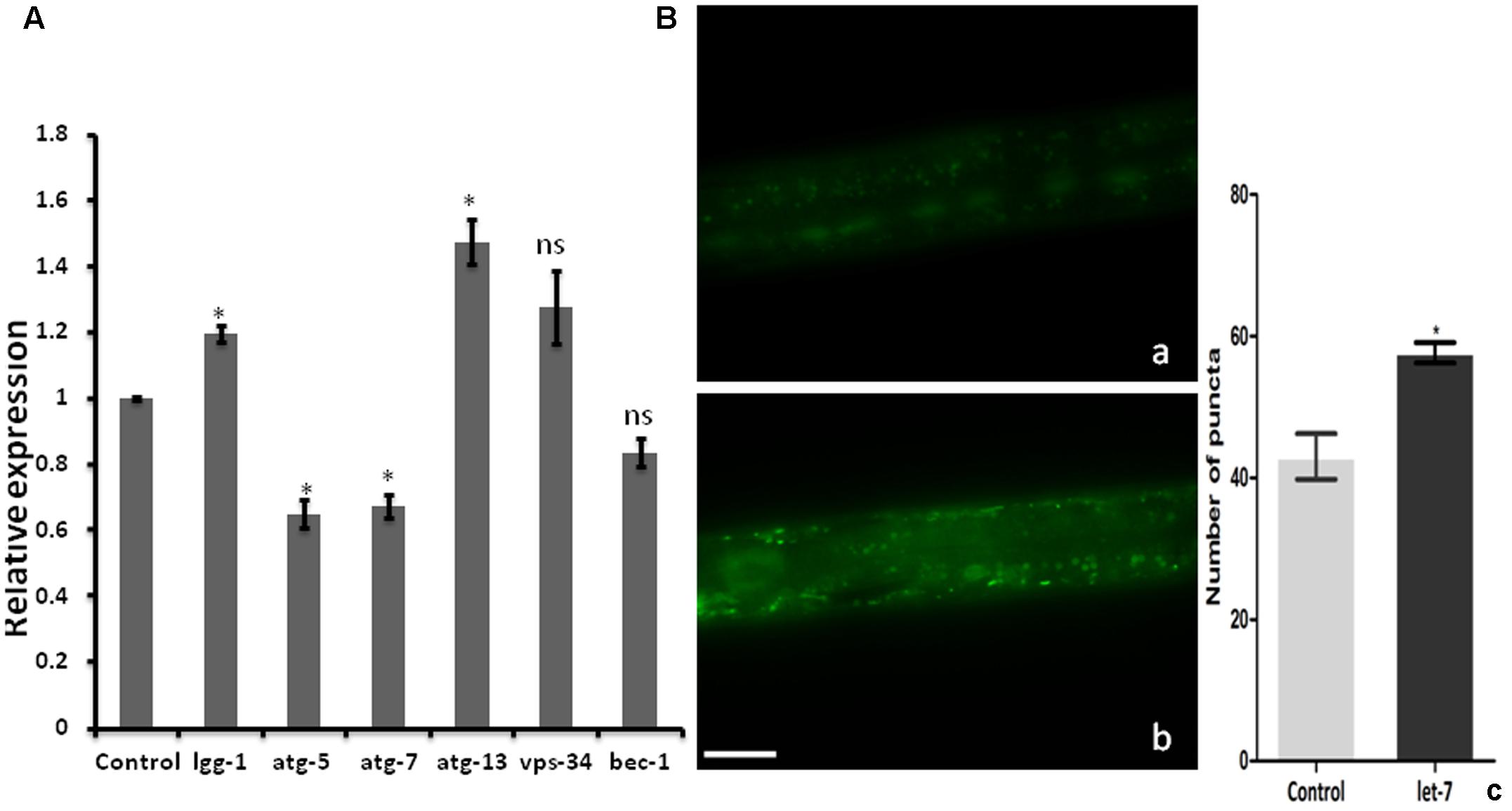
FIGURE 4. Assay for autophagy marker genes in C. elegans; (A) relative expression of autophagy marker genes studied through real-time PCR after let-7 miRNA silencing (B): Expression pattern of LGG-1::GFP in DA2123 using fluorescence microscope; control (a), let-7 knockdown (b), number of puncta as quantified using ImageJ software (c). ∗p < 0.05, ns, not significant. Scale bar: 50 μm.
GFP::LGG-1 Was Increased in Let-7 miRNA Silenced Condition
As we observed a significant induction in expression of autophagy related genes, we went on to study the formation of autophagosomal vesicles employing a GFP::LGG-1 strain in which increased GFP puncta reflect an increase in autophagosome formation (Alberti et al., 2010). We observed an increase in punctae by 34.11% (∗p < 0.05) in let-7 miRNA silenced worms as compared to that control worms. The mean punctae for the let-7 miRNA silenced worms was 57.67 ± 1.453 (N = 5) whereas it was 43.00 ± 3.215 (N = 5) for the control group (Figure 4B).
Expression of Apoptosis Marker Genes Was Altered in Let-7 miRNA Silenced Worms
Neurodegenerative diseases including Alzheimer’s disease, PD Amyotrophic lateral sclerosis, and Huntington disease are characterized by neuronal cell death (Mattson, 2000). Wishing to delineate the possible role of let-7 in multifactorial aspect of PD, we further examined the effect of let-7 miRNA knockdown on the expression level of genes associated with cell death. We carried out quantitative real-time PCR of some of previously reported genes of cell death (Cecconi et al., 1998; Kuan et al., 1999; Hayakawa et al., 2011; Rutkowski et al., 2011; Jiang and Wu, 2014) under control and let-7 miRNA silenced condition. We observed that expression levels of ced-4 and jnk-1 mRNA were significantly downregulated by 26% (p < 0.05) and 30% (p < 0.05), respectively, whereas mRNA level of lin-45 was upregulated by 42% (p < 0.05) as compared to that control group (Figure 5). ced-4 mediates programmed cell death by activating ced-3 (Aballay and Ausubel, 2001). jnk-1 encodes a serine/threonine kinase required for apoptotic signaling in both extrinsic and intrinsic pathways (Dhanasekaran and Reddy, 2008). lin-45 encodes an ortholog of vertebrate protein RAF which is required for vulval differentiation (Han et al., 1993). In our study, we found that let-7 miRNA silenced worms showed downregulation of ced-4 and jnk-1 while upregulation of lin-45 mRNA. Thus knocking down of let-7 miRNA protects cell death by reducing the expression level of ced-4 and jnk-1 as well as via maintaining vulval viability by increasing the expression level of lin-45.
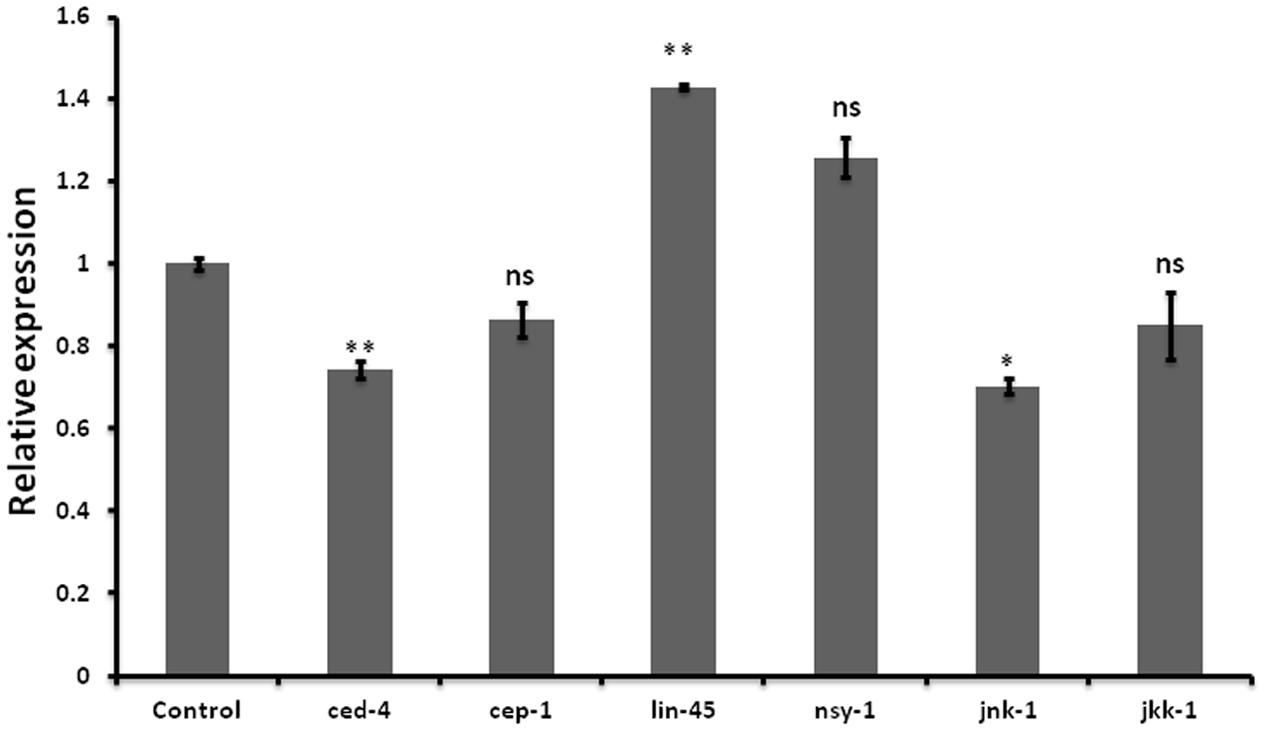
FIGURE 5. Graphical representation of relative mRNA expression of apoptosis pathway genes after let-7 miRNA silencing using qPCR analysis in C. elegans. ∗∗p < 0.01, ∗p < 0.05, ns, not significant.
Let-7 miRNA Silenced Worms Exhibited Enhanced Motor Function in Transgenic Strain NL5901
Knockdown of let-7 miRNA exhibited no marked effect on motility in wild type strain N2. However, motility was significantly increased after knockdown of let-7 miRNA in NL5901 strain as compared to that of N2 and NL5901. We observed a mean thrashing count of 45.10 ± 0.8622 (N = 10) and 36.90 ± 0.9481 (N = 10) in the worms of control group N2 and NL5901, respectively. Whereas let-7 miRNA NL5901 silenced worms exhibited a thrashing count 51.80 ± 1.583 (N = 10), thus exhibiting 12.93% (p < 0.01) and 28.76% (p < 0.001) increased motility as compared to N2 and NL5901 strains (Figure 6).
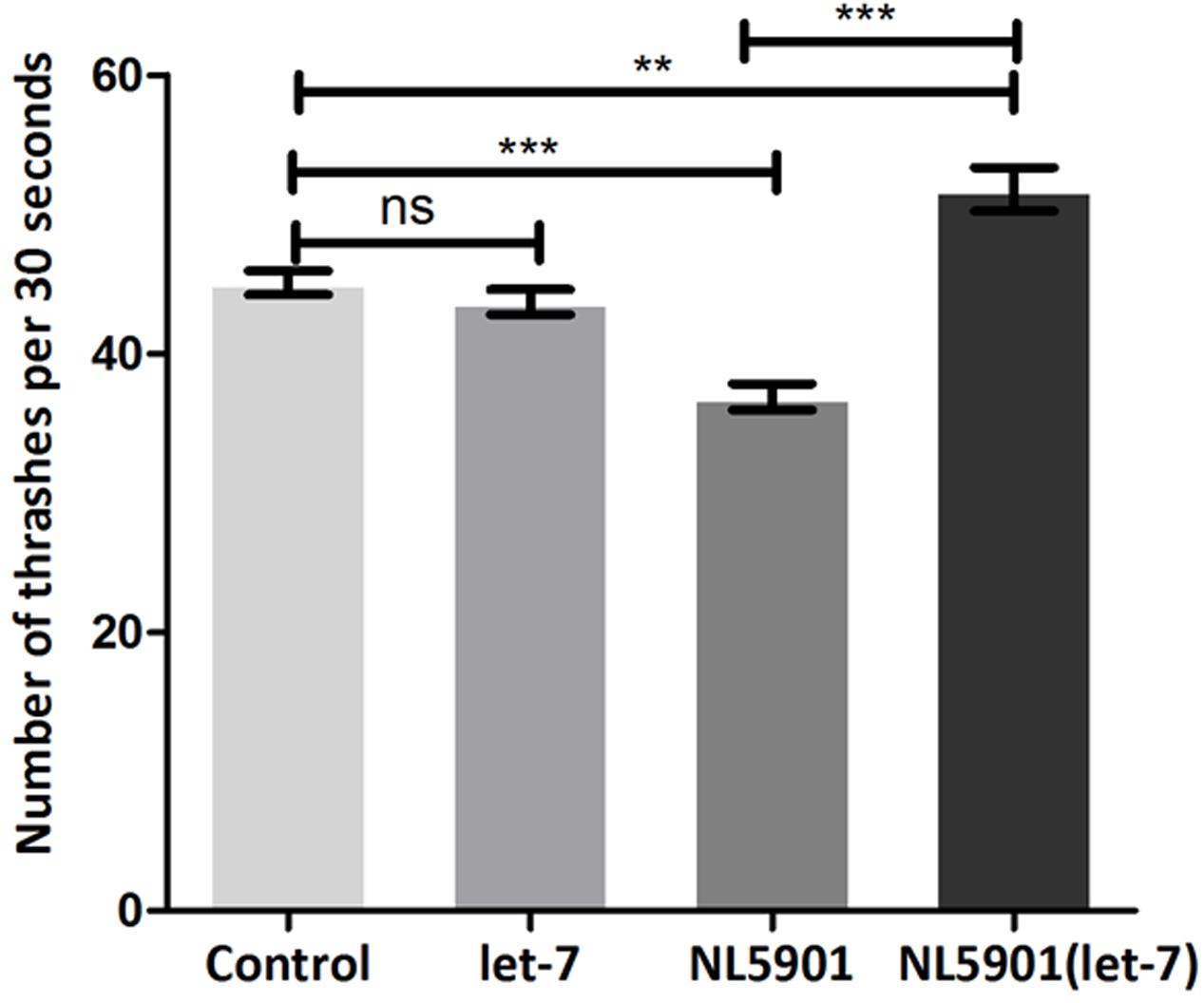
FIGURE 6. Graphical representation of thrashing pattern in wild type N2 strain and human alpha-synuclein expressing NL5901 strain. ∗∗∗p < 0.001, ∗∗p < 0.01, ns, non-significant.
Knockdown of Let-7 miRNA Increases Oxidative Stress
Neurodegenerative PD is characterized by mitochondrial dysfunction. Altered mitochondrial function is implicated in increased reactive oxygen species (ROS) and cellular energy impairment (Guo et al., 2013). We studied the effect of let-7 miRNA knockdown on the alteration of ROS level. Employing H2DCFDA assay we checked ROS level in control and let-7 miRNA knockdown groups. We observed fluorescence intensity of 3.636 ± 0.3434 relative fluorescence intensity units (RFU) per worm in control group whereas let-7 miRNA knockdown worms exhibited fluorescence intensity of 7.300 ± 0.5500 RFU per worm, thereby displaying 50.19% (p < 0.05) increased ROS level with respect to that of control group (Figure 7).
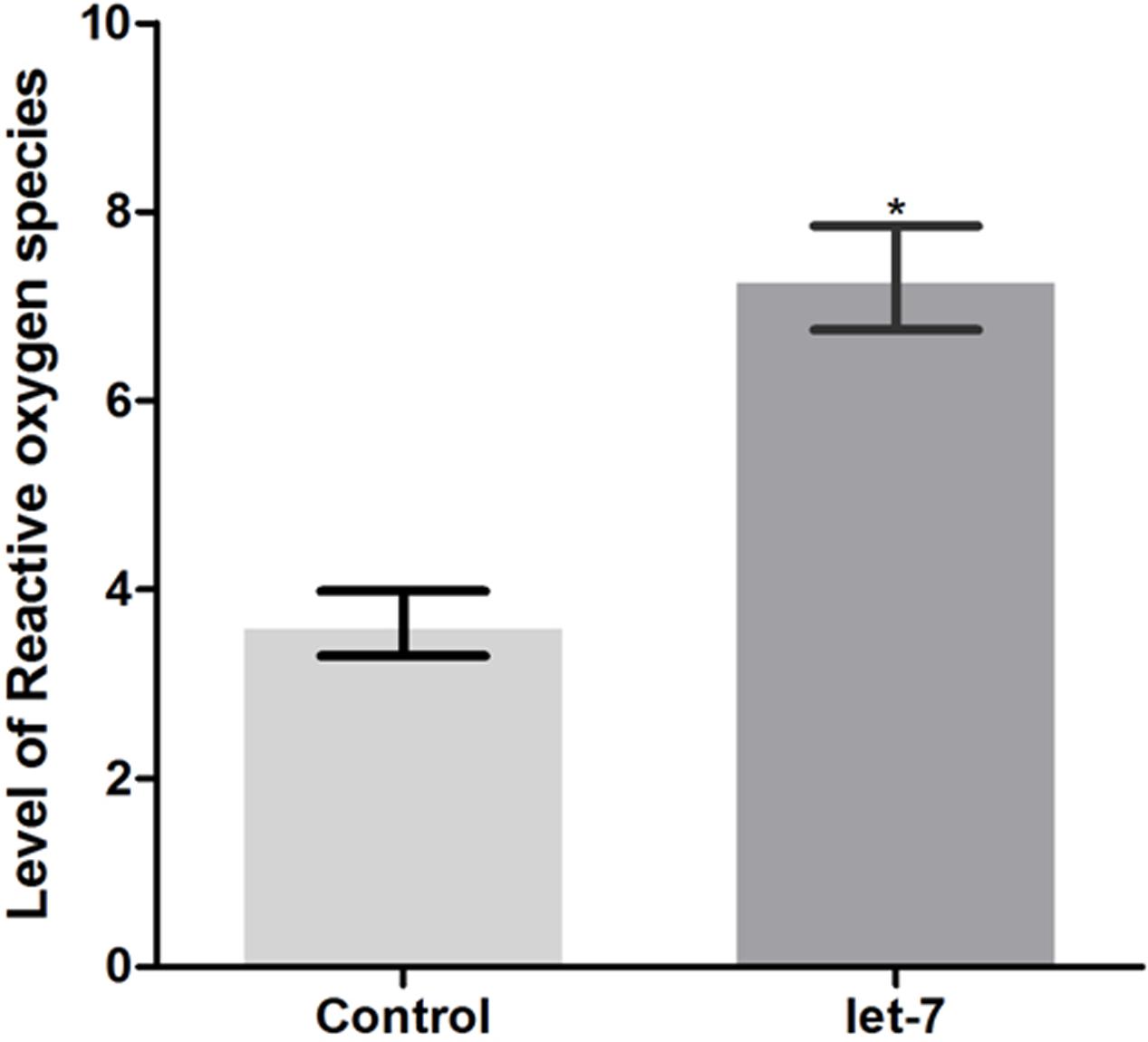
FIGURE 7. ROS production levels as estimated by H2DCFDA assay in wild type strain N2 (control and let-7 knockdown condition). ∗p < 0.05.
Let-7 miRNA Silenced Worms Displayed Enhanced Fat Content
Low level of fat deposition is known to associate with PD (Lorefalt et al., 2009; Bernhardt et al., 2016). In order to delineate the effect of let-7 miRNA in fat deposition, we employed Nile red for assaying the fat deposition in worms of control and let-7 miRNA silenced group. Fluorescent intensity of stained fat was analyzed through fluorescence microscopy and quantified using ImageJ software. We observed optimal level of fat content in worms of control group (Figure 8A) where the mean fluorescence intensity was 6.174 ± 0.2050 (N = 5) arbitrary units. Staining intensity in the worms of let-7 miRNA silenced group was increased (Figure 8B) and mean fluorescence intensity was 17.04 ± 0.3631 (N = 5), thereby exhibiting 2.75-fold increased (p < 0.001) fat content as compared to that of control group (Figure 8C).
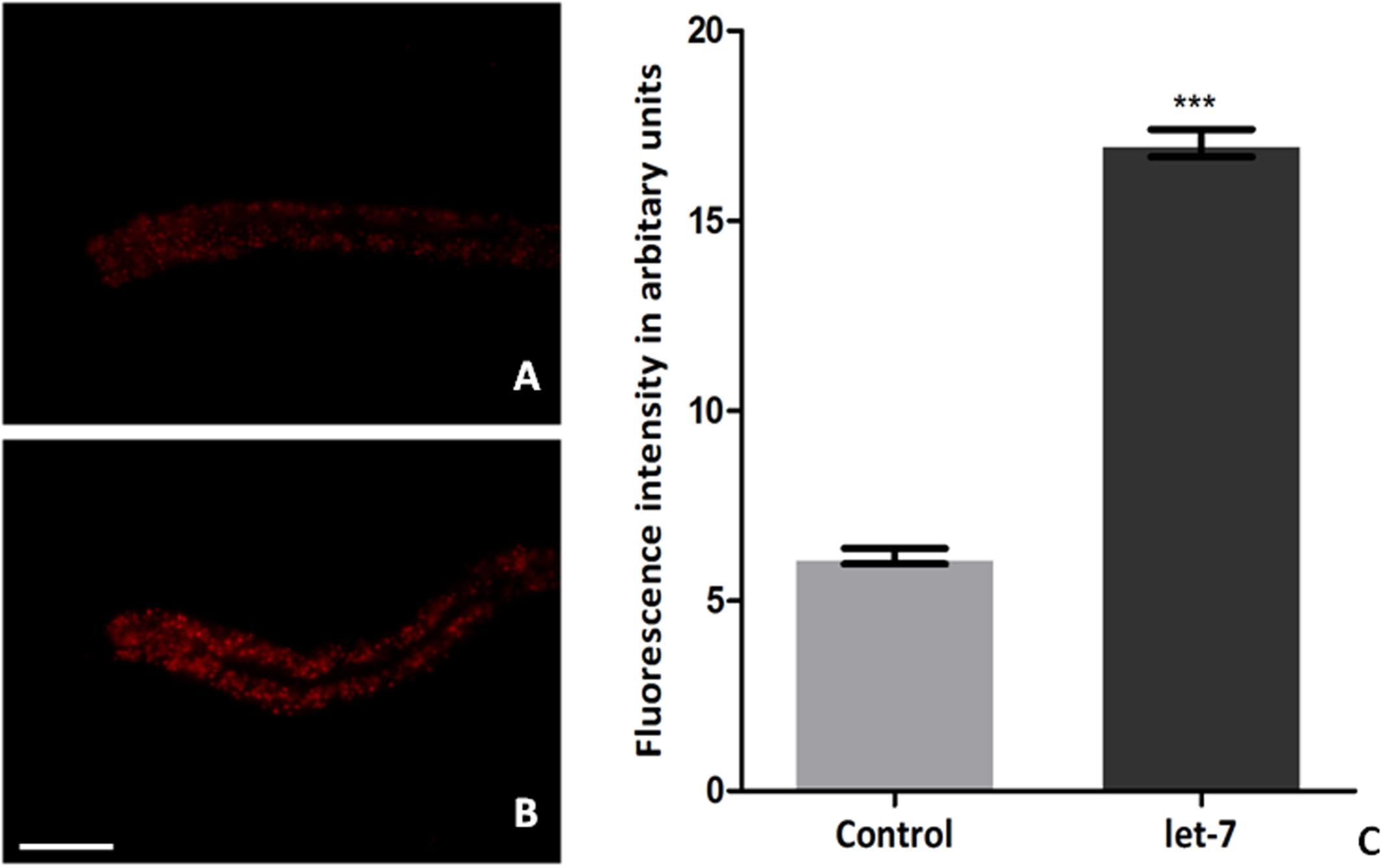
FIGURE 8. Nile red staining for fat content in C. elegans from control (A), let-7 miRNA silenced group (B), and graphical representation for fluorescence intensity of the worms as quantified using ImageJ software (C). ∗∗∗p < 0.001. Scale bar: 50 μm.
Let-7 miRNA Knockdown Had No Effect on Acetylcholinergic and Dopaminergic Neurons
The effect of let-7 miRNA knockdown on acetylcholinergic and dopaminergic neurons was studied via expression of GFP tagged with unc-17 and dat-1 transporter of acetylcholinergic and dopaminergic neurons, respectively. Transgenic strains LX929 and BZ555 were used for this study and expression of the transgenes was studied via fluorescence microscopy. We observed no significant effect on expression of GFP either in let-7 miRNA knockdown LX929 or BZ555 strain as compared to their respective controls (Figures 9A–D), suggesting that knockdown of let-7 miRNA has no effect on these neuronal subpopulations.
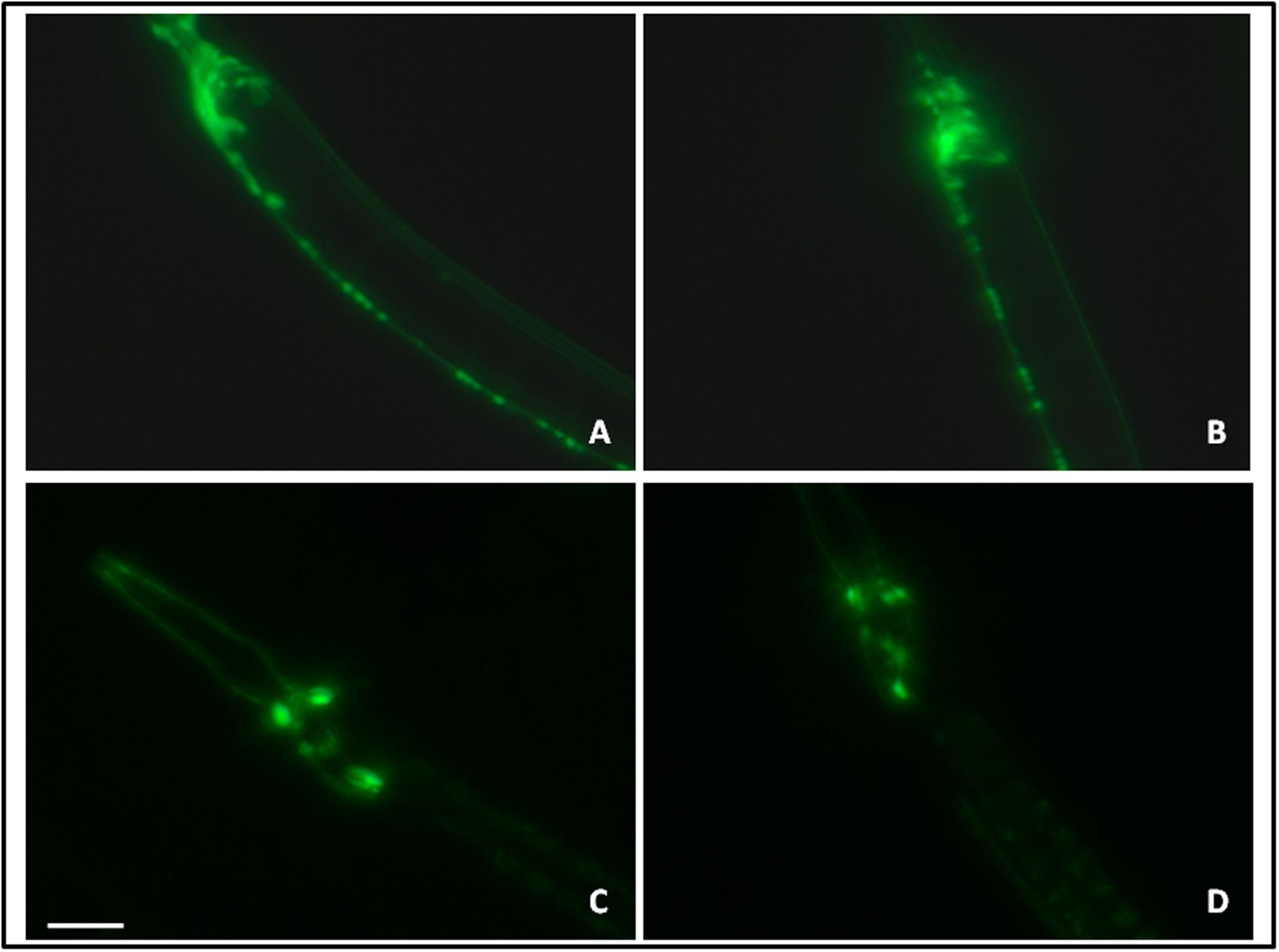
FIGURE 9. GFP expression pattern in the unc-17::GFP strain (A: control, B: lct-7 miRNA knockdown) and dat-l::GFP strain (C: control and D: let-7 miRNA knockdown) using fluorescence microscopy. Scale bar: 50 μm.
Dopamine Associated Function Is Not Affected under Let-7 miRNA Silencing
Various behavioral functions of worms are regulated by neurotransmitter dopamine (Sawin et al., 2000). Alterations in dopamine related functions are associated with PD (Li et al., 2015). To assess the effect of let-7 knockdown on dopamine function, we employed the odor-based repellent assay using 1-nonanol for various conditions. We observed that wild type N2 strain exhibited a mean response time of 1.600 ± 0.2449 s (N = 10) whereas the mean response time of let-7 miRNA knockdown worms was 2.200 ± 0.3742 s (N = 10) (Figure 10A). In contrast transgenic strain NL5901 displayed mean response time of 2.600 ± 0.5099 s (N = 10) and silencing of let-7 miRNA in this strain resulted in a mean response time of 2.000 ± 0.4472 s (N = 10) (Figure 10B). Worms under let-7 silenced condition did not exhibit any significant change in their functions associated with neurotransmitter dopamine as studied via nonanol repulsion assay.
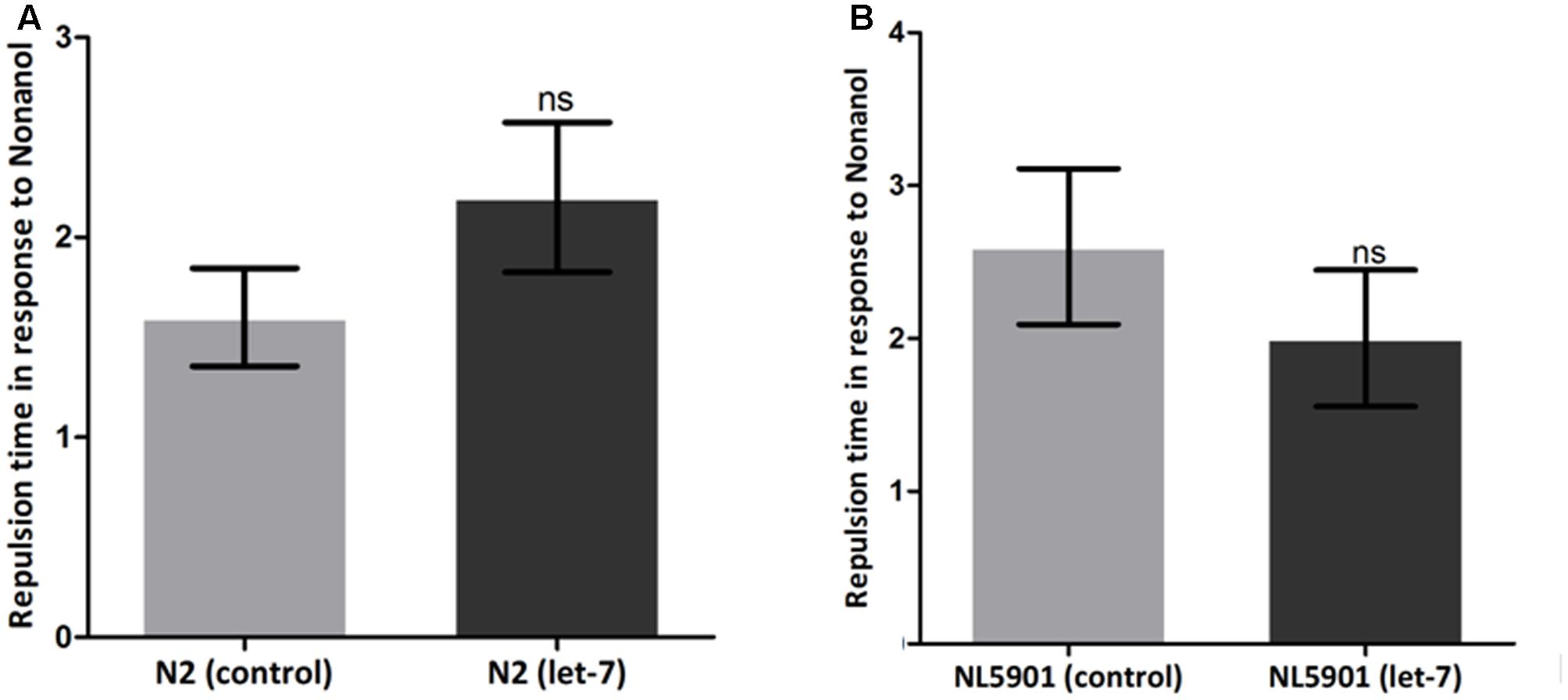
FIGURE 10. Estimation of dopamine content employing nonanol assay. C. elegans wild type strain N2 {control and let-7 knockdown} (A). Transgenic strain NL5901 expressing human alpha-synuclein {control and lct-7 miRNA knockdown} (B). ns, non-significant.
Discussion
The expression of miRNA varies for different tissues, and any deviation from its normal state may lead to various conditions including diseases like NDs (Tan et al., 2015; Basak et al., 2016). There are many findings that report on the importance of miRNAs in disease progression (Sempere et al., 2004). The miRNA regulatory event is very complex in nature with many intersecting pathways. miRNAs regulate transcriptional factors and in turn are being regulated by genes that code for RNA binding proteins. The regulatory network and feedback loop is used by miRNA for maintaining optimal level of gene expression pattern during body and organ development such as during the development of nervous system and cardiovascular system (Nam et al., 2011). Loss or over-expression of specific miRNA in elderly vertebrate animals results in neurodegeneration (Bicchi et al., 2013). Let-7 is an evolutionarily conserved miRNA that has been reported to repress multiple oncogenes by affecting key regulators of the cell cycle, cell differentiation, and apoptotic pathways. Let-7 miRNA is differentially expressed in alpha-synuclein transgenic animals and human Parkin ortholog pdr-1 mutant animals (Asikainen et al., 2010). Let-7 miRNA are also downregulated by pathogenic LRRK2 (Gehrke et al., 2010). Let-7 miRNA, by bioinformatics analysis, is known to regulate genes of cell death, autophagy, mTOR and insulin pathway. C. elegans homolog of amyloid precursor protein apl-1 is also reported to be controlled by let-7 miRNA (Yokota et al., 2003; Revuelta et al., 2008). We observed that let-7 miRNA was over expressed in C. elegans model of PD expressing wild type ‘human’ alpha-synuclein protein, while its expression was reduced in C. elegans model expressing mutant alpha-synuclein. Toward our studies of exploring the importance of let-7 miRNA in the context of PD, we constructed an RNAi feeding bacterial clone of let-7 miRNA and studied it employing transgenic C. elegans strain expressing human alpha-synuclein. Our results suggest that loss of function of let-7 miRNA results in significant upregulation of daf-12 and daf-16 gene expression validating the fact that let-7 miRNA controls the expression level of daf-12 and daf-16 mRNA. daf-12 mRNA acts as a downstream target of let-7 miRNA as reported previously (Hammell et al., 2009). daf-12 and daf-16 encode member of the steroid hormone receptor superfamily and FOXO transcriptional factor, respectively, both reported to regulate the pathogenesis associated with PD (Haque and Nazir, 2014; Mahanti et al., 2014). In our studies, we observed that alpha-synuclein accumulation, and end points associated with PD were decreased in the absence of let-7 miRNA indicating the importance of let-7 miRNA directly with the progression of NDs. Over-expression of alpha-synuclein leads to degeneration of dopaminergic neurons (Cao et al., 2010). Therefore, silencing of let-7 miRNA might be protecting the dopaminergic neurons via decreasing the accumulation of alpha-synuclein. Further, since decrease in the accumulation of alpha-synuclein protein is associated with enhanced autophagy, we quantified mRNA levels of different autophagy marker genes. Our findings indicate that mRNA levels of lgg-1 and atg-13 were increased in let-7 silenced worms. GFP::LGG-1 was also increased in let-7 knockdown worms that further validate the previous findings. lgg-1 is an ortholog of Saccharomyces cerevisiae Atg8p and mammalian MAP-LC3, which is required for degradation of cellular components, atg-13 is an autophagy related gene required for autophagosome formation. Our studies suggest that the targets of let-7 miRNA might be involved in autophagy pathway which was increased in the absence of let-7 miRNA.
A defect in locomotion due to imbalance of neurotransmitters is the hallmark of PD. In order to understand the effect of let-7 miRNA knockdown on normal locomotory behavior we employed thrashing assay to quantify motility in the worms. Our studies showed enhanced motility which suggests let-7 miRNA may have role in excitatory neurotransmission. Further, we studied the effect of let-7 miRNA silencing on dopamine function via nonanol repulsion assay. Our results indicate no direct role of let-7 miRNA on functions associated with dopamine content. It suggests that loss of let-7 miRNA function does not have any effect on dopamine synthesis or overall availability. This can be explained by the fact that expression of some miRNAs is very specific and its alteration in expression leads to death of particular cell. Keeping this in mind we evaluated the effect of let-7 miRNA knockdown on dopaminergic and acetylcholinergic neurons. Our findings indicate that absence of let-7 miRNA has no effect on these neurons. This provides a clue toward protective role of let-7 miRNA in cell death. So, we next examined the effect of let-7 miRNA silencing on programmed cell death associated genes. Our studies indicate that knockdown of let-7 miRNA protects cells from death by reducing the ced-4 and jnk-1 mRNA expression. ced-4 is an enzyme required for initiation of programmed cell death by activating ced-3 which encodes a caspase essential for execution of apoptosis (Huang et al., 2013). jnk-1 is the variant of Jun N-terminal kinases (JNKs), which belongs to MAP-kinase superfamily. JNK-1 mediates apoptotic signaling in both extrinsic and intrinsic pathways (Dhanasekaran and Reddy, 2008). Our finding indicates that loss of let-7 miRNA might play protective role in C. elegans.
Parkinson’s disease is known to be associated with oxidative stress. Hence, we studied ROS in the worms at the basal level and after silencing of let-7 miRNA. In our studies, we observed elevated level of ROS. Many research groups have reported that elevated ROS leads to induction of autophagy (Chen et al., 2009; Filomeni et al., 2015). Silencing of let-7 miRNA leads to elevated ROS level and mild increase in ROS level acts as inducer of autophagy pathway. This study provides evidence that any increase in the level of ROS leads to the increase in autophagy that might help in the clearance of misfolded protein aggregates. Mild increases in ROS level also promote the longevity by activating the hif-1 which encodes hypoxia inducible factor (HIF-1), a highly conserved transcription factor that activates survival promoting genes during hypoxia (Lee et al., 2010). Thus absence of let-7 miRNA might help in the reduction of alpha-synuclein protein aggregates in C. elegans model and enhancing life span. Altered lipid content is known to be associated with PD (Lorefalt et al., 2009; Bernhardt et al., 2016). Reduced level of fat content has been seen in the worms expressing human alpha-synuclein protein. Previous study showed that C. elegans model of PD has low level of lipid content (Jadiya et al., 2011). Our study shows fat content was increased by decreasing the expression of alpha-synuclein in let-7 miRNA silenced worms. This suggests that absence of let-7 miRNA might help in maintaining lipid content in worms.
Conclusion
Our study leads to an understanding of the role of C. elegans let-7 miRNA in progression of PD and confirms that absence of let-7 miRNA leads to decrease in accumulation of alpha-synuclein protein in transgenic worms. Our studies further provide a clue toward the role of let-7 miRNA in possibly decreasing alpha-synuclein expression via increasing autophagy and increasing daf-16 FOXO transcription factor. Decrease in the expression of alpha-synuclein protein via Daf-16 has been reported (Pino et al., 2014). Our studies prove that loss of let-7 miRNA did not affect the dopaminergic and acetylcholinergic neurons. Our results also open avenues for further research toward deciphering the importance of let-7 miRNA in the context of various other diseases and may prove to be beneficial target for the treatment of PD in future. Let-7 could particularly be studied for its role in modulating the multi-factorial aspect of neurodegenerative PD.
Author Contributions
Shamsuzzama and AN conceived the studies. Shamsuzzama and LK performed the experiments. Shamsuzzama and AN wrote the paper. AN provided reagents and guidance.
Funding
This work was funded by CSIR-NWP EpiHeD (BSC0118).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Shamsuzzama and LK are supported by Senior Research Fellowship from UGC and CSIR, respectively. Nematode strains used in the study were provided by Caenorhabditis Genetics Center (CGC), University of Minnesota, Minneapolis, MN, United States, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). We thank Dr. Snober S. Mir, Associate Professor, Integral University Lucknow, for her critical inputs during the writing of this paper. CSIR-CDRI communication number is 9568.
Footnotes
References
Aballay, A., and Ausubel, F. M. (2001). Programmed cell death mediated by ced-3 and ced-4 protects Caenorhabditis elegans from Salmonella typhimurium-mediated killing. Proc. Natl. Acad. Sci. U.S.A. 98, 2735–2739. doi: 10.1073/pnas.041613098
Alberti, A., Michelet, X., Djeddi, A., and Legouis, R. (2010). The autophagosomal protein LGG-2 acts synergistically with LGG-1 in dauer formation and longevity in C. elegans. Autophagy 6, 622–633. doi: 10.4161/auto.6.5.12252
Ashrafi, K., Chang, F. Y., Watts, J. L., Fraser, A. G., Kamath, R. S., Ahringer, J., et al. (2003). Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421, 268–272. doi: 10.1038/nature01279
Asikainen, S., Rudgalvyte, M., Heikkinen, L., Louhiranta, K., Lakso, M., Wong, G., et al. (2010). Global microRNA expression profiling of Caenorhabditis elegans Parkinson’s disease models. J. Mol. Neurosci. 41, 210–218. doi: 10.1007/s12031-009-9325-1
Barbagallo, B., Prescott, H. A., Boyle, P., Climer, J., and Francis, M. M. (2010). A dominant mutation in a neuronal acetylcholine receptor subunit leads to motor neuron degeneration in Caenorhabditis elegans. J. Neurosci. 30, 13932–13942. doi: 10.1523/JNEUROSCI.1515-10.2010
Barh, D., Malhotra, R., Ravi, B., and Sindhurani, P. (2010). MicroRNA let-7: an emerging next-generation cancer therapeutic. Curr. Oncol. 17, 70–80. doi: 10.3747/co.v17i1.356
Basak, I., Patil, K. S., Alves, G., Larsen, J. P., and Moller, S. G. (2016). microRNAs as neuroregulators, biomarkers and therapeutic agents in neurodegenerative diseases. Cell. Mol. Life Sci. 73, 811–827. doi: 10.1007/s00018-015-2093-x
Bernhardt, D., Muller, H. P., Ludolph, A. C., Dupuis, L., and Kassubek, J. (2016). Body fat distribution in Parkinson’s disease: an MRI-based body fat quantification study. Parkinsonism Relat. Disord. 33, 84–89. doi: 10.1016/j.parkreldis.2016.09.016
Bicchi, I., Morena, F., Montesano, S., Polidoro, M., and Martino, S. (2013). MicroRNAs and molecular mechanisms of neurodegeneration. Genes 4, 244–263. doi: 10.3390/genes4020244
Buckingham, S. D., and Sattelle, D. B. (2009). Fast, automated measurement of nematode swimming (thrashing) without morphometry. BMC Neurosci. 10:84. doi: 10.1186/1471-2202-10-84
Cao, P., Yuan, Y., Pehek, E. A., Moise, A. R., Huang, Y., Palczewski, K., et al. (2010). Alpha-synuclein disrupted dopamine homeostasis leads to dopaminergic neuron degeneration in Caenorhabditis elegans. PLOS ONE 5:e9312. doi: 10.1371/journal.pone.0009312
Cecconi, F., Alvarez-Bolado, G., Meyer, B. I., Roth, K. A., and Gruss, P. (1998). Apaf1 (CED-4 homolog) regulates programmed cell death in mammalian development. Cell 94, 727–737. doi: 10.1016/S0092-8674(00)81732-8
Chaudhuri, A. D., Choi, D. C., Kabaria, S., Tran, A., and Junn, E. (2016). MicroRNA-7 regulates the function of mitochondrial permeability transition pore by targeting VDAC1 expression. J. Biol. Chem. 291, 6483–6493. doi: 10.1074/jbc.M115.691352
Chaudhuri, A. D., Kabaria, S., Choi, D. C., Mouradian, M. M., and Junn, E. (2015). MicroRNA-7 promotes glycolysis to protect against 1-Methyl-4-phenylpyridinium-induced cell death. J. Biol. Chem. 290, 12425–12434. doi: 10.1074/jbc.M114.625962
Chen, Y., Azad, M. B., and Gibson, S. B. (2009). Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ. 16, 1040–1052. doi: 10.1038/cdd.2009.49
Dhanasekaran, D. N., and Reddy, E. P. (2008). JNK signaling in apoptosis. Oncogene 27, 6245–6251. doi: 10.1038/onc.2008.301
Dimmeler, S., and Nicotera, P. (2013). MicroRNAs in age-related diseases. EMBO Mol. Med. 5, 180–190. doi: 10.1002/emmm.201201986
Douglas, M. R., Lewthwaite, A. J., and Nicholl, D. J. (2007). Genetics of Parkinson’s disease and parkinsonism. Expert Rev. Neurother. 7, 657–666. doi: 10.1586/14737175.7.6.657
Filomeni, G., De Zio, D., and Cecconi, F. (2015). Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ. 22, 377–388. doi: 10.1038/cdd.2014.150
Fraser, A. G., Kamath, R. S., Zipperlen, P., Martinez-Campos, M., Sohrmann, M., and Ahringer, J. (2000). Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408, 325–330. doi: 10.1038/35042517
Gao, H. M., and Hong, J. S. (2011). Gene-environment interactions: key to unraveling the mystery of Parkinson’s disease. Prog. Neurobiol. 94, 1–19. doi: 10.1016/j.pneurobio.2011.03.005
Gehrke, S., Imai, Y., Sokol, N., and Lu, B. (2010). Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature 466, 637–641. doi: 10.1038/nature09191
Grosshans, H., Johnson, T., Reinert, K. L., Gerstein, M., and Slack, F. J. (2005). The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans. Dev. Cell 8, 321–330. doi: 10.1016/j.devcel.2004.12.019
Guo, C., Sun, L., Chen, X., and Zhang, D. (2013). Oxidative stress, mitochondrial damage and neurodegenerative diseases. Neural Regen. Res. 8, 2003–2014.
Hammell, C. M., Karp, X., and Ambros, V. (2009). A feedback circuit involving let-7-family miRNAs and DAF-12 integrates environmental signals and developmental timing in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 106, 18668–18673. doi: 10.1073/pnas.0908131106
Han, M., Golden, A., Han, Y., and Sternberg, P. W. (1993). C. elegans lin-45 raf gene participates in let-60 ras-stimulated vulval differentiation. Nature 363, 133–140. doi: 10.1038/363133a0
Haque, R., and Nazir, A. (2014). SMAD transcription factor, Sma-9, attunes TGF-beta signaling cascade towards modulating amyloid beta aggregation and associated outcome in transgenic C. elegans. Mol. Neurobiol. 53, 109–119. doi: 10.1007/s12035-014-8988-y
Hayakawa, T., Kato, K., Hayakawa, R., Hisamoto, N., Matsumoto, K., Takeda, K., et al. (2011). Regulation of anoxic death in Caenorhabditis elegans by mammalian apoptosis signal-regulating kinase (ASK) family proteins. Genetics 187, 785–792. doi: 10.1534/genetics.110.124883
Huang, W., Jiang, T., Choi, W., Qi, S., Pang, Y., Hu, Q., et al. (2013). Mechanistic insights into CED-4-mediated activation of CED-3. Genes Dev. 27, 2039–2048. doi: 10.1101/gad.224428.113
Jadiya, P., Fatima, S., Baghel, T., Mir, S. S., and Nazir, A. (2016). A systematic RNAi screen of neuroprotective genes identifies novel modulators of alpha-synuclein-associated effects in transgenic Caenorhabditis elegans. Mol. Neurobiol. 53, 6288–6300. doi: 10.1007/s12035-015-9517-3
Jadiya, P., Khan, A., Sammi, S. R., Kaur, S., Mir, S. S., and Nazir, A. (2011). Anti-Parkinsonian effects of Bacopa monnieri: insights from transgenic and pharmacological Caenorhabditis elegans models of Parkinson’s disease. Biochem. Biophys. Res. Commun. 413, 605–610. doi: 10.1016/j.bbrc.2011.09.010
Jadiya, P., Mir, S. S., and Nazir, A. (2012). Effect of various classes of pesticides on expression of stress genes in transgenic C. elegans model of Parkinson’s disease. CNS Neurol. Disord. Drug Targets 11, 1001–1005. doi: 10.2174/1871527311211080009
Jenzer, C., Manil-Segalen, M., Lefebvre, C., Largeau, C., Glatigny, A., and Legouis, R. (2014). Human GABARAP can restore autophagosome biogenesis in a C. elegans lgg-1 mutant. Autophagy 10, 1868–1872. doi: 10.4161/auto.29745
Jiang, H. S., and Wu, Y. C. (2014). LIN-3/EGF promotes the programmed cell death of specific cells in Caenorhabditis elegans by transcriptional activation of the pro-apoptotic gene egl-1. PLOS Genet. 10:e1004513. doi: 10.1371/journal.pgen.1004513
Kaur, S., Sammi, S. R., Jadiya, P., and Nazir, A. (2012). RNAi of cat-2, a putative tyrosine hydroxylase, increases alpha synuclein aggregation and associated effects in transgenic C. elegans. CNS Neurol. Disord. Drug Targets 11, 387–394. doi: 10.2174/187152712800792811
Kocerha, J., Xu, Y., Prucha, M. S., Zhao, D., and Chan, A. W. (2014). microRNA-128a dysregulation in transgenic Huntington’s disease monkeys. Mol. Brain 7:46. doi: 10.1186/1756-6606-7-46
Krichevsky, A. M., King, K. S., Donahue, C. P., Khrapko, K., and Kosik, K. S. (2003). A microRNA array reveals extensive regulation of microRNAs during brain development. RNA 9, 1274–1281. doi: 10.1261/rna.5980303
Krichevsky, A. M., Sonntag, K. C., Isacson, O., and Kosik, K. S. (2006). Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells 24, 857–864. doi: 10.1634/stemcells.2005-0441
Kuan, C. Y., Yang, D. D., Samanta Roy, D. R., Davis, R. J., Rakic, P., and Flavell, R. A. (1999). The Jnk1 and Jnk2 protein kinases are required for regional specific apoptosis during early brain development. Neuron 22, 667–676. doi: 10.1016/S0896-6273(00)80727-8
Kumar, M., Nath, S., Prasad, H. K., Sharma, G. D., and Li, Y. (2012). MicroRNAs: a new ray of hope for diabetes mellitus. Protein Cell 3, 726–738. doi: 10.1007/s13238-012-2055-0
Lakso, M., Vartiainen, S., Moilanen, A. M., Sirvio, J., Thomas, J. H., Nass, R., et al. (2003). Dopaminergic neuronal loss and motor deficits in Caenorhabditis elegans overexpressing human alpha-synuclein. J. Neurochem. 86, 165–172. doi: 10.1046/j.1471-4159.2003.01809.x
Lee, C. T., Risom, T., and Strauss, W. M. (2006). MicroRNAs in mammalian development. Birth Defects Res. C Embryo Today 78, 129–139. doi: 10.1002/bdrc.20072
Lee, S. J., Hwang, A. B., and Kenyon, C. (2010). Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Curr. Biol. 20, 2131–2136. doi: 10.1016/j.cub.2010.10.057
Li, J., Li, D., Yang, Y., Xu, T., Li, P., and He, D. (2015). Acrylamide induces locomotor defects and degeneration of dopamine neurons in Caenorhabditis elegans. J. Appl. Toxicol. 36, 60–67. doi: 10.1002/jat.3144
Ling, H., Fabbri, M., and Calin, G. A. (2013). MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat. Rev. Drug Discov. 12, 847–865. doi: 10.1038/nrd4140
Lorefalt, B., Toss, G., and Granerus, A. K. (2009). Weight loss, body fat mass, and leptin in Parkinson’s disease. Mov. Disord. 24, 885–890. doi: 10.1002/mds.22466
Lu, Y., Thomson, J. M., Wong, H. Y., Hammond, S. M., and Hogan, B. L. (2007). Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev. Biol. 310, 442–453. doi: 10.1016/j.ydbio.2007.08.007
Mahanti, P., Bose, N., Bethke, A., Judkins, J. C., Wollam, J., Dumas, K. J., et al. (2014). Comparative metabolomics reveals endogenous ligands of DAF-12, a nuclear hormone receptor, regulating C. elegans development and lifespan. Cell Metab. 19, 73–83. doi: 10.1016/j.cmet.2013.11.024
Manil-Segalen, M., Lefebvre, C., Jenzer, C., Trichet, M., Boulogne, C., Satiat-Jeunemaitre, B., et al. (2014). The C. elegans LC3 acts downstream of GABARAP to degrade autophagosomes by interacting with the HOPS subunit VPS39. Dev. Cell 28, 43–55. doi: 10.1016/j.devcel.2013.11.022
Mattson, M. P. (2000). Apoptosis in neurodegenerative disorders. Nat. Rev. Mol. Cell Biol. 1, 120–129. doi: 10.1038/35040009
Nam, Y., Chen, C., Gregory, R. I., Chou, J. J., and Sliz, P. (2011). Molecular basis for interaction of let-7 microRNAs with Lin28. Cell 147, 1080–1091. doi: 10.1016/j.cell.2011.10.020
Nishida, Y., Arakawa, S., Fujitani, K., Yamaguchi, H., Mizuta, T., Kanaseki, T., et al. (2009). Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 461, 654–658. doi: 10.1038/nature08455
Nixon, R. A. (2013). The role of autophagy in neurodegenerative disease. Nat. Med. 19, 983–997. doi: 10.1038/nm.3232
Pino, E., Amamoto, R., Zheng, L., Cacquevel, M., Sarria, J. C., Knott, G. W., et al. (2014). FOXO3 determines the accumulation of alpha-synuclein and controls the fate of dopaminergic neurons in the substantia nigra. Hum. Mol. Genet. 23, 1435–1452. doi: 10.1093/hmg/ddt530
Pu, P., and Le, W. (2008). Dopamine neuron degeneration induced by MPP+ is independent of CED-4 pathway in Caenorhabditis elegans. Cell Res. 18, 978–981. doi: 10.1038/cr.2008.279
Qiu, C., Wang, J., and Cui, Q. (2011). miR2Gene: pattern discovery of single gene, multiple genes, and pathways by enrichment analysis of their microRNA regulators. BMC Syst. Biol. 5(Suppl. 2):S9. doi: 10.1186/1752-0509-5-S2-S9
Revuelta, G. J., Rosso, A., and Lippa, C. F. (2008). Association between progranulin and beta-amyloid in dementia with Lewy bodies. Am. J. Alzheimers Dis. Other Demen. 23, 488–493. doi: 10.1177/1533317508321910
Rutkowski, R., Dickinson, R., Stewart, G., Craig, A., Schimpl, M., Keyse, S. M., et al. (2011). Regulation of Caenorhabditis elegans p53/CEP-1-dependent germ cell apoptosis by Ras/MAPK signaling. PLOS Genet. 7:e1002238. doi: 10.1371/journal.pgen.1002238
Sawin, E. R., Ranganathan, R., and Horvitz, H. R. (2000). C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron 26, 619–631. doi: 10.1016/S0896-6273(00)81199-X
Schratt, G. M., Tuebing, F., Nigh, E. A., Kane, C. G., Sabatini, M. E., Kiebler, M., et al. (2006). A brain-specific microRNA regulates dendritic spine development. Nature 439, 283–289. doi: 10.1038/nature04367
Selbach, M., Schwanhausser, B., Thierfelder, N., Fang, Z., Khanin, R., and Rajewsky, N. (2008). Widespread changes in protein synthesis induced by microRNAs. Nature 455, 58–63. doi: 10.1038/nature07228
Sempere, L. F., Freemantle, S., Pitha-Rowe, I., Moss, E., Dmitrovsky, E., and Ambros, V. (2004). Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 5:R13. doi: 10.1186/gb-2004-5-3-r13
Shamsuzzama, Kumar, L., Haque, R., and Nazir, A. (2016). Role of microRNA Let-7 in modulating multifactorial aspect of neurodegenerative diseases: an overview. Mol. Neurobiol. 53, 2787–2793. doi: 10.1007/s12035-015-9145-y
Sharples, S. A., Koblinger, K., Humphreys, J. M., and Whelan, P. J. (2014). Dopamine: a parallel pathway for the modulation of spinal locomotor networks. Front. Neural Circuits 8:55. doi: 10.3389/fncir.2014.00055
Shulman, J. M., De Jager, P. L., and Feany, M. B. (2011). Parkinson’s disease: genetics and pathogenesis. Annu. Rev. Pathol. 6, 193–222. doi: 10.1146/annurev-pathol-011110-130242
Sonntag, K. C. (2010). MicroRNAs and deregulated gene expression networks in neurodegeneration. Brain Res. 1338, 48–57. doi: 10.1016/j.brainres.2010.03.106
Tan, L., Yu, J. T., and Tan, L. (2015). Causes and consequences of microRNA dysregulation in neurodegenerative diseases. Mol. Neurobiol. 51, 1249–1262. doi: 10.1007/s12035-014-8803-9
Tehrani, N., Del Rosario, J., Dominguez, M., Kalb, R., and Mano, I. (2014). The insulin/IGF signaling regulators cytohesin/GRP-1 and PIP5K/PPK-1 modulate susceptibility to excitotoxicity in C. elegans. PLOS ONE 9:e113060. doi: 10.1371/journal.pone.0113060
Wang, X., Cao, L., Wang, Y., Wang, X., Liu, N., and You, Y. (2012). Regulation of let-7 and its target oncogenes (Review). Oncol. Lett. 3, 955–960.
Wong, G., and Nass, R. (2012). miRNAs and their putative roles in the development and progression of Parkinson’s disease. Front. Genet. 3:315. doi: 10.3389/fgene.2012.00315
Yokota, T., Sugawara, K., Ito, K., Takahashi, R., Ariga, H., and Mizusawa, H. (2003). Down regulation of DJ-1 enhances cell death by oxidative stress, ER stress, and proteasome inhibition. Biochem. Biophys. Res. Commun. 312, 1342–1348. doi: 10.1016/j.bbrc.2003.11.056
Keywords: let-7, microRNA, C. elegans, Parkinson’s disease, RNAi
Citation: Shamsuzzama, Kumar L and Nazir A (2017) Modulation of Alpha-synuclein Expression and Associated Effects by MicroRNA Let-7 in Transgenic C. elegans. Front. Mol. Neurosci. 10:328. doi: 10.3389/fnmol.2017.00328
Received: 28 February 2017; Accepted: 28 September 2017;
Published: 13 October 2017.
Edited by:
Gaiti Hasan, National Centre for Biological Sciences, IndiaReviewed by:
Beena Pillai, Institute of Genomics and Integrative Biology (CSIR), IndiaSandhya Padmanabhan Koushika, Tata Institute of Fundamental Research, India
Copyright © 2017 Shamsuzzama, Kumar and Nazir. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aamir Nazir, YW5hemlyQGNkcmkucmVzLmlu
 Shamsuzzama
Shamsuzzama Lalit Kumar
Lalit Kumar Aamir Nazir
Aamir Nazir