Abstract
Natural product libraries are a source of diverse 3D molecular features furnishing an array of biological functions in drug discovery research. In this paper we will oversee the use of affinity selection mass spectrometry (AS-MS) for identifying ligands to a variety of biological targets using natural products as the molecular library. The assay modes used in solution or with the immobilized target and their pros and cons will be presented. Additionally, the required experiments and investigation for accurate chemical annotation of the disclosed ligands in a non-target assay are discussed.
1 Introduction
Natural product library is a source of diverse 3D-structural molecular features furnishing an array of biological functions and are resourceful in furnishing scaffolds for drug discovery research (Newman and Cragg, 2020; Qin et al., 2024). Due to its complexity and broader chemical space, prospecting natural active molecules is classically performed by bio-guided isolation, however, this is an over intensive work and can be hampered by false positive results and loss of activity by multiple fractionation step and repetitive bioassays (Nothias et al., 2018; Quiros-Guerrero et al., 2024). To this end, affinity selection mass spectrometry (AS-MS), which is a consolidate high-throughput screening (HTS) for synthetic libraries, has been efficiently used to disclose ligands from natural products extracts (Muchiri and van Breemen, 2021b; Prudent et al., 2021; Almeida and Cass, 2023).
AS-MS interrogates non-covalent target-ligand complex as a non-functional assay. A variety of target has been used such as soluble or membrane protein, nucleic acid and nucleic acid–protein complexes (Prudent et al., 2023). It is a label-free biophysical method, and it discloses the binders solely by mass spectrometry data providing conditions for chemical annotation of the identified ligands. Moreover, identifies several ligands exhibiting multiple mechanisms of action against the same target, including orthosteric and allosteric ligands (McLaren et al., 2021; de Oliveira et al., 2023).
Despite a few recent reviews on AS-MS related to natural products libraries (Muchiri and van Breemen, 2021a; 2021b; Guo et al., 2023; Qin et al., 2024) the herein reported review will cover broader aspects of this bioprospection technology especially regarding the annotation of the disclosed ligands in a nontarget metabolite assay. Meanwhile, we are going to start this review calling attention to the many used notations and thus, to the difficulty in accessing some of the seminal work due to their used terminology.
1.1 Notations
Different nomenclature has been used for AS-MS, mainly based either if the biomolecular target is in-solution or immobilized on to a carrier. It is named also based on how the ligands are dissociated from the target complex. For the in-solution methods, the notations are most of the time based on the dissociation process, such as: size exclusion chromatography, ALLIS technology (for online SEC-LC-MS), vacuum filtration, gel filtration, and ultrafiltration (O’Connell et al., 2014; Almeida and Cass, 2023; Prudent et al., 2023). For immobilized targets, ligand-fishing is probably the term most used (de Moraes et al., 2014; Zhuo et al., 2016; Zhao et al., 2018; Qin et al., 2024), but names such as, affinity capture MS (AC-MS), magnetic microbeads affinity selection screening (MagMass) (Muchiri and van Breemen, 2021a; Muchiri et al., 2022) and paper (Ablat et al., 2024; Chen et al., 2024; Hu et al., 2024) are widely used. The diversity of acronyms causes difficulties in searching for references; however, the most intricate notation is when the term chromatography is used to denote experiments of AS-MS. The term chromatography is used in the case of some off-line devices (de Moraes et al., 2019), just because it uses solid-supported proteins, i.e., “functional chromatography” (Kang et al., 2014).
It should be said that although zonal and frontal bioaffinity chromatography efficiently disclose ligands through retention times or breakthrough curves, respectively, these approaches differ fundamentally from the static incubation processes characteristic of AS-MS set-ups (de Moraes et al., 2014; Ciesla et al., 2016). The multiplicity of terms accentuates the need for standardization, as inconsistencies complicate literature searches and data interpretation.
1.2 Workflow of the AS-MS assay
As will be discussed herein, while the methods differ by a variety of models, the assays are all based on a static incubation step (1) and involves three other major stages (2–4) (Muchiri and van Breemen, 2021b; Almeida and Cass, 2023; Prudent et al., 2023) (Figure 1).
FIGURE 1
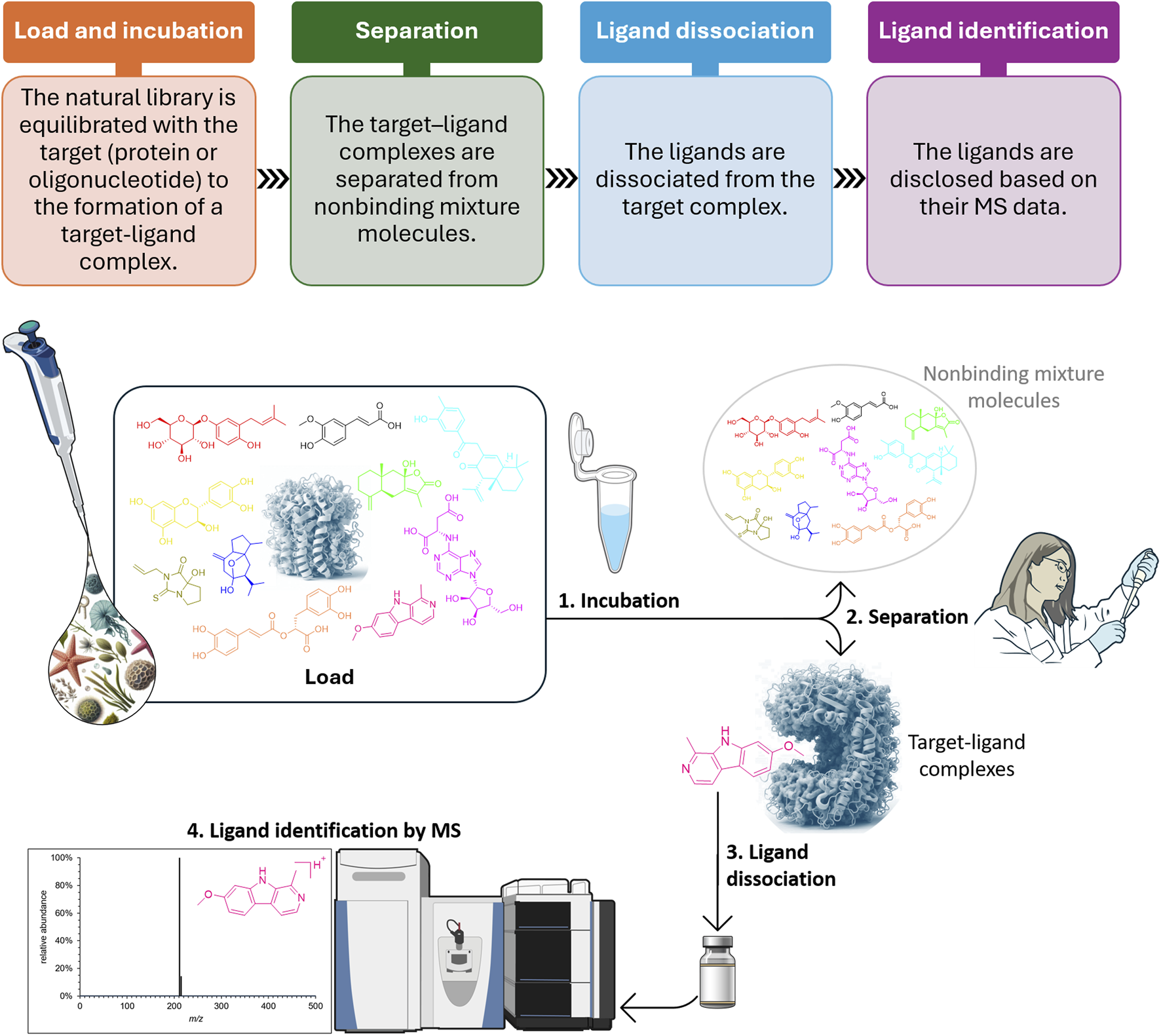
Schematic representation of the AS-MS workflow (NIAID Visual and Medical Arts, 2024).
AS-MS is a biological target-based assay approach, and the assay’s model is the first decision made. For each target and type of molecular library, all stages of the AS-MS platform should be carefully developed.
1. The equilibrium time represented by the incubation stage needs to be investigated and clearly it is influenced by the target type and by the molecular library. It is important to pay attention to molecules with rapid off-rates since they may be lost during stages 1–3. To avoid competition, usually the target is used in molar excess of the small molecules in the library (Almeida and Cass, 2023; Prudent et al., 2023), what is a limiting factor when the library is a natural product extract, since the concentrations of the molecules varies immensely.
2. The separation from nonbinding mixture molecules is carried out in accordance with the selected assay mode, and the washing is also a condition to be tuned.
3. The dissociation step is usually adjusted in accordance with the target. For the in-solution assays, denaturation of the target (protein) is usually preferred (Prudent et al., 2021). When the target is immobilized and thus, can be recycled, the dissociation step can be obtained by displacement with a high affinity ligand, with a change in pH or using organic solvent in a percentage that does not cause denaturation (Vanzolini et al., 2015; do Amaral et al., 2021).
4. For disclosing the ligands different mass spectrometer instrumentation has been used. For that, data processing and curation is necessary and can be done with open source or proprietary software (Prudent et al., 2023). The ligands are identified by affinity or index ratio that are calculated by control experiments and for that the ligands are identified by their mass-to-charge ratio (Wang et al., 2019; Almeida and Cass, 2023). For synthetic libraries, the correlation between the MS data and the molecular structure of the disclosed ligand is direct. Moreover, the singlets can be used to validate the ligands. However, to infer the molecular structure of the disclosed ligands in natural product libraries one needs fragmentation experiments, spectral libraries and molecular networking to deconvolute the obtained mass spectral data. When possible, reference standard is used (Zhao et al., 2018; do Amaral et al., 2021; Lima et al., 2021). These pitfalls will all be discussed in detail in this review.
2 Solution-based AS-MS assays
2.1 Ultrafiltration
Ultrafiltration is a technique that separates molecules based on size. In 1861, Schmidt observed that when solutions containing proteins were passed through a natural bovine pericardium, the resulting filtrate had a reduced concentration of macromolecules compared to the initial solution. Later, in 1906, Bechhold formally introduced the term “ultrafiltration” and developed specialized membranes with controlled porosity by treating filter paper with acetic acid collodion. His work highlighted the critical role of factors like pore size and molecular adsorption in selective filtration (Ferry, 1936).
Modern ultrafiltration membranes are generally designed to retain molecules with hydraulic diameters between 1 and 100 nm, which correlates to molecular weights in the range of 500 to 500,000 Da (Lutz, 2015) – sufficient for most proteins, nucleic acids, and macromolecular complexes. Depending on the setup, separation can be driven by centrifugal force, vacuum, or applied pressure. Through the process, careful control of the filtration rate and pressure is essential to achieve optimal separation without damaging the membrane.
Therefore, ultrafiltration membranes act as selective barriers, allowing smaller molecules and solvents to pass through while retaining larger solutes, such as protein-ligand complexes, making ultrafiltration ideal for separating these complexes from unbound molecules in solution-based AS-MS assays.
In a typical ultrafiltration-based AS-MS experiment, the ligand-protein binding event is initiated by incubating a target protein with a library of molecules. The incubation occurs at low micromolar concentrations, which are optimal for detecting high-affinity ligands while ensuring specific binding interactions. Once equilibrium is reached, ultrafiltration is applied to separate the resulting ligand-protein complexes from unbound molecules. Then, the ligands are dissociated by employing denaturing conditions, such as adding an organic solvent or altering the pH, to disrupt non-covalent bonds between the ligand and protein. Methanol or acetonitrile with a volatile organic acid like formic acid are commonly used for effective dissociation while remaining compatible with subsequent mass spectrometry analysis. If protein reuse is desired, non-denaturing conditions may be preferred to maintain the protein’s structural integrity. The dissociated ligands are then injected into a LC-MS system for further separation and analysis.
Ultrafiltration AS-MS was applied to explore 5-lipoxygenase (5-LOX) ligands in Inonotus obliquus, leading to the identification of three promising molecules: botulin (2.1), lanosterol (2.2), and quercetin (2.3) (Yun et al., 2024) (Figure 2). These potential inhibitors were identified based on their binding affinity with 5-LOX, a result further validated by molecular docking, which detailed their interactions with the enzyme’s active site. Complementary methods, including semi-preparative liquid chromatography and high-speed countercurrent chromatography, were then applied to isolate these molecules with high purity allowing complete chemical characterization. This study demonstrates how ultrafiltration AS-MS can speed up the identification of active molecules from complex natural mixtures, improving efficiency in bioactive molecules research.
FIGURE 2
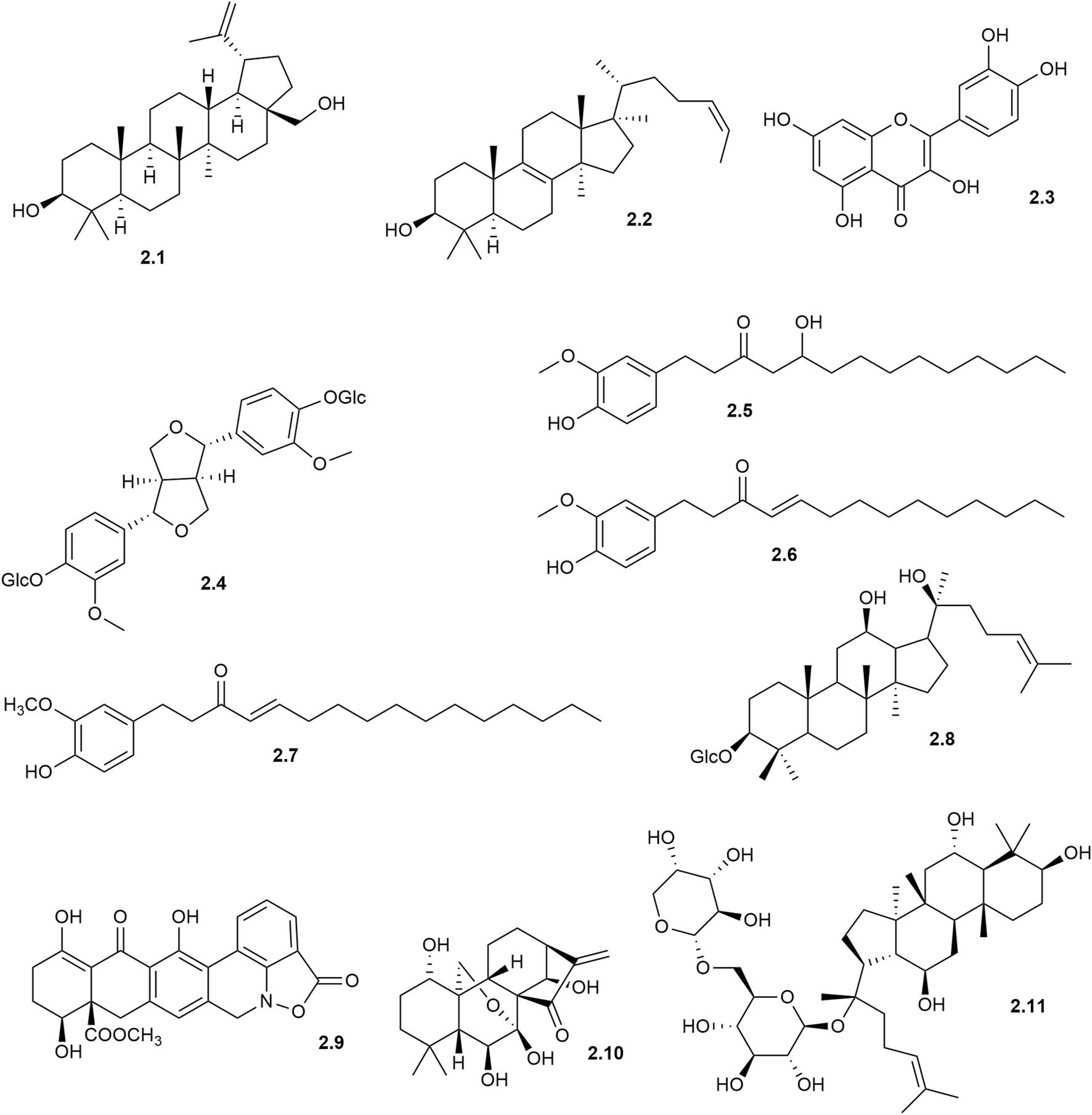
Chemical structures of some ligands disclosed using AS-MS assays with biological targets in solution. These ligands, isolated from natural product extracts, illustrate the applications of ultrafiltration and size-exclusion chromatography (SEC) as the affinity selection setup of the assay.
Hence, in this typical ultrafiltration AS-MS experimental workflow, dissociated ligands are directly disclosed by LC-MS analysis. An alternative approach, however, involves the indirect ligand identification, in which ligand interactions with a target are monitored by analyzing the unbound molecules that pass through the ultrafiltration membrane. In this setup, the presence of an affinity event is inferred by observing a significant reduction in signal intensity for certain molecules in the LC-MS chromatograms compared to a control where the target is denatured. Molecules showing notable decreases in these chromatograms are flagged as potential ligands for the target. While this approach can simplify certain steps by focusing on unbound fractions, it requires careful chromatogram comparison and statistical analysis to reliably identify interactions (Wei et al., 2016).
An example illustrating this indirect ultrafiltration AS-MS assay is the screening of phosphodiesterase (PDE) inhibitors from Eucommia ulmoides bark (Shi et al., 2013). In this approach, ligand interactions with the target enzyme were detected by analyzing the unbound molecules that pass through the ultrafiltration membrane, with ligands disclosed by a notable decrease in their chromatographic peak areas when compared to control assays. For instance, chromatograms in this study highlight selective retention of several lignans (such as (+)-pinoresinol-4,4′-di-O-β-D-glucopyranoside (2.4)) that exhibit a strong affinity for PDE, while the control chromatogram shows evidence of non-specific retention by the ultrafiltration setup itself. This underscores the importance of carefully optimized conditions to minimize background interactions, thereby enhancing the assay’s selectivity and reliability in detecting true target-ligand binding events.
When the incubation and ultrafiltration events are carried out outside the chromatographic system, the assay is referred to as an offline method. In contrast, the online approach, introduced by van Breemen (1997), directly couples an ultrafiltration chamber to a LC-MS system, allowing for continuous processing of ligand-protein complexes. This system, commonly referred to as pulsed ultrafiltration, employs a stirred flow-through chamber constructed from non-binding materials such as polysulfone or PEEK and uses a methylcellulose ultrafiltration membrane to retain high-molecular-weight molecules and ligand-receptor complexes while allowing smaller, unbound molecules to pass through (van Breemen, 1997). This setup enables the chamber contents to be monitored in real time by ESI-MS or LC-MS/MS, facilitating rapid, automated screening of complex mixtures like synthetic or natural libraries. Although this high-throughput, automated approach offers significant advantages in efficiency and selectivity, it requires specialized instrumentation and is therefore currently used mainly within the pioneering research group’s laboratory.
One notable application of online ultrafiltration in ligand screening within natural products is the investigation of ginger roots, which have long been recognized for their anti-inflammatory properties. Ligands targeting cyclooxygenase-2 (COX-2) were screened in a chloroform extract derived from a methanol infusion of ginger using online ultrafiltration-based AS-MS (van Breemen et al., 2011). The study identified ten molecules related to gingerol, including 10-gingerol (2.5), 8-shogaol (2.6), and 10-shogaol (2.7), which specifically bind to the active site of COX-2. These molecules exhibited inhibitory activities against COX-2, with IC50 values of 32 μM, 17.5 μM, and 7.5 μM, respectively, while showing no significant inhibition of COX-1. This selective inhibition is particularly relevant as COX-1 inhibition is often associated with gastrointestinal side effects. The findings suggest that certain gingerol-related molecules could contribute to the anti-inflammatory effects of ginger and serve as marker molecules for standardizing ginger-based dietary supplements.
Independently of the experimental AS-MS approach, a crucial factor for effective and reproducible ultrafiltration separation is choosing the appropriate membrane. The membrane must selectively retain protein-ligand complexes while allowing unbound molecules to pass through, minimizing nonspecific binding and background interference. To ensure accuracy and reduce background signal, control experiments without the protein are often conducted to identify any nonspecifically binding molecules. Additionally, replacing the membrane periodically or after each assay can help reduce potential cross-contamination and improve signal clarity, especially if significant nonspecific binding is observed.
For instance, the ultrafiltration AS-MS approach was explored to investigate Glucagon-like peptide-1 receptor (GLP-1R) agonists from Panax ginseng (Wang et al., 2024). Through this method, five ginsenosides were disclosed as GLP-1R agonists, such as 20(R)-ginsenoside Rh2 (2.8), suggesting a new hypoglycemic mechanism for the anti-diabetic effects of P. ginseng by activating GLP-1R. Notably, the study presented total ion chromatograms (TICs) for both the sample and control analysis (i.e., with and without the target receptor), revealing substantial nonspecific retention of molecules in the control, likely due to interactions with the ultrafiltration device. This highlights the critical importance of carefully selecting experimental conditions in UF AS-MS workflows to minimize nonspecific interactions and improve selectivity for target ligands.
The majority of the ligands identified by ultrafiltration AS-MS are flavonoids (Choi et al., 2011; Wang et al., 2014; Xiao et al., 2015; Li et al., 2016; Chen and Guo, 2017a; 2017b; Liu et al., 2017; Dong et al., 2021; Huang et al., 2024; Ma et al., 2024; Wei et al., 2024), but other classes such as polyphenols (Tian et al., 2022; Ma et al., 2024; Quan et al., 2024), glycosides (Quan et al., 2024), saponins (Chen and Guo, 2017a; Xie et al., 2020; Wei et al., 2024), coumarins (Liang et al., 2019; Tian et al., 2022; Ma et al., 2024), alkaloids (Chen and Guo, 2017a; Li et al., 2019), phenolic acids (Zhang et al., 2017; Huang et al., 2024), and carotenoids (Wang et al., 2018) also have been identified as ligands of various biological targets. These includes proteins like pancreatic lipase (Xiao et al., 2015; Quan et al., 2024; Wei et al., 2024), neuraminidase (Tian et al., 2022; Ma et al., 2024), xanthine oxidase (Liu et al., 2017; Dong et al., 2021; Zhuang et al., 2024), 5-lipoxygenase (Xie et al., 2020), cyclooxygenase-2 (Li et al., 2019; Xie et al., 2020; Huang et al., 2024), thrombin (Zhang et al., 2017), hepatic mitochondria (Liang et al., 2019), α-glucosidase (Wang et al., 2014; Chen and Guo, 2017a), quinone reductase-2 (Choi et al., 2011), hyaluronidase (Huang et al., 2024), topoisomerases I (Chen and Guo, 2017b), lactate dehydrogenase (Li et al., 2016), myeloperoxidase (Wang et al., 2018), among others. By enabling the efficient screening and isolation of bioactive molecules, ultrafiltration AS-MS not only enhances our understanding of the interactions between ligands and their respective targets but also facilitates the discovery of potential therapeutic agents from complex natural libraries.
2.2 Size-exclusion chromatography
Size-exclusion chromatography (SEC) is a well-established technique that exploits differences in the sizes of solute molecules to achieve separation. The primary objectives of SEC include the purification of target molecules from natural sources, as well as the determination of molecular sizes and mass distributions for macromolecules (Štulík et al., 2003). The technique relies on the principle that larger molecules are excluded from the porous stationary phase, allowing them to elute first, while smaller molecules are retained and elute later. This characteristic makes SEC particularly valuable for separating protein-ligand complexes from unbound molecules in analytical workflows.
In SEC-based AS-MS assays, similar to ultrafiltration approaches, the process begins with the incubation of the target with the molecular library. SEC then separates target-ligand complexes from unbound molecules, with the complexes eluting first. To minimize ligand loss from complex dissociation, SEC separations should be performed rapidly and at low temperatures. The protein-ligand complexes are subsequently dissociated using denaturing conditions such as organic solvents, elevated temperatures, or pH adjustments.
SEC was first used in 1997 to isolate protein-ligand complexes from unbound molecules in an AS-MS assay designed to evaluate a synthetic library (Kaur et al., 1997). Following SEC separation, an aliquot of the protein-ligand complex was analyzed by ESI-MS for online desalting and concentration using a reversed-phase cartridge. Chromatographic conditions during elution facilitated the dissociation of the complex, allowing for ligand identification based on its m/z. To confirm the specificity of binding, the assay was repeated with a known high-affinity ligand; specific ligands were displaced by the competitor and thus did not appear in the ESI-MS analysis. Ligands detected in both the absence and presence of the known ligand were interpreted as binding nonspecifically through hydrophobic interactions at sites distinct from the primary binding site.
Beyond its initial applications, the SEC-AS-MS platform has been applied for identifying bioactive molecules in natural products. Methanolic extracts of Allium lusitanicum were screened for ligands of peroxisome proliferator-activated receptors (PPARs) (De Soricellis et al., 2024). Initially, the SEC coupled to LC-MS platform was validated using known PPARα and PPARγ ligands–WY-14643 and rosiglitazone, respectively. Allium lusitanicum extracts are rich in saponins, which are recognized for their metabolic activity and known as PPAR ligands. Saponins are particularly challenging to isolate from natural products due to their amphiphilic structure, which can lead to aggregation and interactions with other compounds in the plant. Despite these challenges, the SEC method effectively isolated saponins as PPAR ligands from the extract.
The SEC column can also be directly connected to a reverse-phase (RP) column, integrating both chromatographic dimensions to create a high-throughput, automated screening assay. In this configuration, a valve directs the excluded protein-ligand complex to the RP column, where the mobile phase, containing organic solvents and acids, dissociates the complex. For instance, an SEC-RP-MS platform was used to disclose parafungins (such as 2.9) as polyadenosine polymerase ligands in acetone extracts from fermentation broths from several fungal strains, including Fusarium larvarum (Adam et al., 2008). UV-based detection monitored the elution of the protein-ligand complex from the SEC column, diverting it to the RP column, where dissociation was achieved using 5% ACN and 0.2% formic acid, followed by gradient elution to separate the parafungins for MS detection.
However, in the SEC-RP-MS setup, directing the protein-ligand complex to the RP column may increase backpressure due to denatured protein accumulation, necessitating periodic column regeneration or replacement. Currently, SEC-based AS-MS assays have predominantly been applied to synthetic libraries, with limited studies focusing on natural products. This highlights a gap and potential area for further research in natural product screening using SEC-AS-MS.
2.3 Collision-induced AS-MS
The collision-induced dissociation affinity selection mass spectrometry (CIAS-MS) technique was recently developed to simplify the AS-MS workflow (Mak et al., 2022). In CIAS-MS, electrospray ionization is used to preserve protein-ligand complexes in their native state. Then, the ionized samples are transferred to the quadrupole for mass selection, effectively trapping protein-ligand complexes and excluding unbound molecules. The complexes then undergo collision-induced dissociation (CID) within the collision cell, releasing the ligands. Subsequently, only the small dissociated ligands reach the mass analyzer, as an ion cyclotron resonance (ICR) cell, for detection. This method eliminates the need for external washing steps, thus reducing the loss of low-affinity ligands or those prone to dissociation during filtration or SEC steps (Gu et al., 2022).
As a proof of concept, CIAS-MS was first employed to investigate known ligands of the SARS-CoV-2 Nsp9 protein (Mak et al., 2022). This method demonstrated its versatility by subsequently screening ligands present in extracts from Rabdosia rubescens, a plant known for its rich content of oridonin (2.10), a known ligand with antiviral activity that has been shown to reduce the viral load of SARS-CoV-2 in Calu-3 cells. The identification of oridonin was further validated through native MS by analyzing the ligand-protein complex.
CIAS-MS has been also applied in recognizing transient receptor potential melastatin 2 (TRPM2) ligands (Gu et al., 2024). This method allowed for the direct visualization of clotrimazole dissociating from its complex with TRPM2. Additionally, the study demonstrated the ability of CIAS-MS to screen a natural library, resulting in the identification of ginsenoside F3 (2.11), a saponin known for its antioxidative properties. By providing insights into the binding dynamics and concentration-dependent effects of ligands, this work highlights the potential of CIAS-MS as a tool for advancing drug discovery efforts.
Despite the promise of CIAS-MS in ligand screening, few studies have explored its full potential, and none so far have reported the disclosure of a novel ligand directly from a crude natural product extract, with its chemical structure annotation. A key challenge in this approach lies in controlling experimental conditions to ensure reliable detection of the dissociated ligand–a task complicated by the vast structural diversity of natural products. Achieving this level of control becomes even more complex when dealing with unknown ligands, as it requires carefully optimized dissociation, and detection settings to capture low-affinity or structurally unique molecules effectively.
3 Immobilized-based AS-MS assays
Immobilized target methods refer to any technique in which the target is immobilized on a solid support, such as silica, magnetic beads, microfluidic chips, or other materials (Jonker et al., 2011). Early developments focused on ligand-target and protein-protein interactions, especially for protein purification (Cuatrecasas et al., 1968). Immobilization of targets offers several advantages over methodologies that use targets in solution, including easy recovery, its continuous or repeated reuse. When enzymes are used it has been noticed improved enzyme activity, enhanced stability against organic solvents, pH and temperature variations, and increased selectivity (Mateo et al., 2007). Over the years, numerous experimental methods have been developed for immobilizing biological targets, primarily based on physical or chemical binding to an insoluble support. Both covalent and non-covalent immobilization methods can be used (Trindade Ximenes et al., 2021). Compared to solution-based AS-MS, immobilized-based AS-MS requires an additional step: target immobilization. This step involves selecting the appropriate support, choosing the immobilization method, and evaluating the effects of immobilization on kinetic activity when enzymes are used as targets. The most common immobilization modes are adsorption, entrapment, covalent coupling, and cross-linking (Cao, 2006; Hanefeld et al., 2013).
After immobilization process, the biological target-MBs are incubated with complex samples followed by the separation of the supernatant containing molecules that have no affinity for the target. An advantage of using MBs is that the separation step is simplified by employing small external magnets. For other supports, e.g., filter paper, this step involves taken out the paper disk-immobilized target with a tweezer from the solution, followed by washes step (Li et al., 2022). For porous solid supports a filtration step could be used. The ligands are dissociated from the bioreactor in the extraction step and the desorbate analyzed by LC-MS (Zhuo et al., 2016). Control experiments using denatured enzymes (negative control) are used to calculate the affinity or index ratio (Almeida and Cass, 2023; Lima et al., 2021; Wang et al., 2019). A competition assay using a high affinity known orthosteric ligand can determine whether a disclosed ligand is orthosteric or allosteric. Orthosteric ligands will cause a change in binding by competing with the reference ligand, while no change in binding will be observed if the ligand is allosteric (Muchiri and van Breemen, 2021a; Muchiri et al., 2022; Yan et al., 2022).
3.1 Covalent immobilization methods
Protein immobilization methods that rely on the formation of covalent bonds are among the most widely used approaches. This process involves two chemical steps: (1) the activation of the functional groups on the support using specific reagents, such as glutaraldehyde, and (2) the reaction of the activated functional groups with the functional groups on the biomolecule. The functional groups typically involved in this bond include amino, epoxy, carboxyl, diol, and phenolic groups. These functional groups can participate in a variety of reactions, such as diazotization, amide bond formation, arylation, and Schiff’s base formation. The most frequently involved amino acid side chains are lysine (ε-amino group), cysteine (thiol group), and aspartic and glutamic acids (carboxyl groups) (Girelli and Mattei, 2005). The immobilization of biological targets on solid supports is widely used in various fields, including the manufacturing of industrial products in the pharmaceutical, chemical, and food industries (Basso and Serban, 2019). Several solid-support and their applications have been described in the literature (de Moraes et al., 2019; De Simone et al., 2019).
3.2 Magnetic beads as solid support
Magnetic beads (MBs) are a versatile solid support for target immobilization (Bilal et al., 2018; Trindade Ximenes et al., 2021) and were originally developed for protein isolation and purification (Safarik and Safarikova, 2004). The immobilization of human serum albumin (HSA) on MB was first reported in 2007 by (Moaddel et al., 2007) to “fish out” known binders (warfarin (3.1), AZT (3.2), and naproxen (3.3) from nonbinders (nicotine (3.4), fenoterol (3.5), and labetalol (3.6)) using both manual and automated approaches. The assay was referred as ligand fishing. In 2008, Choi and van Breemen reported a magnetic microbead affinity selection screening (MagMASS) followed by LC-MS analysis. They immobilized estrogen receptors (ERs) onto chemically functionalized magnetic particles by covalent and non-covalent immobilization, and screened extracts of Trifolium pratense L. (red clover) or Humulus lupulus L. (hops) for ligands to ER-β. The phytoestrogens genistein (3.7) and daidzein (3.8) were identified in the red clover extract, and the estrogen-like 8-prenylnaringenin (3.9) was identified in the hop extract.
Immobilized protein on MB surfaces has been successfully applied for ligand fishing assays allowing for direct identification of active ligands from natural products (Zhuo et al., 2016; Lima et al., 2021; Muchiri and van Breemen, 2021a; Chi et al., 2022; de Faria et al., 2022; Guo et al., 2024; van Breemen and Muchiri, 2024) and synthetic libraries (Zhao et al., 2018). The versatility of applications of MBs is specially based on significant characteristics such as their high surface-area-to-volume ratios, which provide efficient interactions with analytes and allow for high immobilization yields. MBs also enable simple functionalization of their surfaces with different functional groups and exhibit superparamagnetism, which allows them to be separated from aqueous solutions or complex mixtures using external magnetic field (Bilal et al., 2018). They can be commercially acquired or synthesized with varied properties, such as different sizes (from micrometer-to nanometer-sized), surface modifications, coatings, and core compositions. The most used MBs are iron oxide-based particles, such as magnetite (Fe3O4) and maghemite (γ-Fe2O3). Although there are various methods for synthesizing and coating MBs, the most used MBs for ligand fishing assays are those prepared by co-precipitation and coated with silane derivatives (de Lima et al., 2020). Studies have been conducted to understand the role of these properties for ligand fishing assays. For example, the effect of MB size on the ligand fishing assay for acetylcholinesterase (AChE) from Electrophorus electricus was evaluated comparing four commercially available amine-terminated magnetic particles with diameters ranging from 4.5 nm to 106 μm (MB1 to MB4) to fish out galantamine (a standard AChE inhibitor), from an aqueous solution (de Lima et al., 2020). All MBs covalently immobilized the enzyme via glutaraldehyde bonding. The particles with diameters of about 1 μm (small microparticles) presented a higher protein mass capacity per milligram of particle than did those with diameters of about 4.5 nm (nanoparticles) and those with diameters of about 25–106 μm (large microparticles). The results provided evidence that for activity-based assays, nanoparticles (4.5 nm, MB4) offer an advantage, showing the highest specific activity. This could be explained by the greater average surface area of these nanoparticles, resulting in higher catalytic capacity and dispersion. On the other hand, for AS-MS assays, a limiting factor for achieving a successful assay is the amount of immobilized protein, as previously noted by Cieśla and Moaddel (2016). This was corroborated by the low affinity ratio (1.8 ± 0.1) observed when MB4, containing ∼3 µg of protein, was used. For large microparticles (1 μm, MB1 and MB2) and MB3 (25–106 µm), containing ∼70 µg of protein, the affinity ratios were 23 ± 1.5, 7.0 ± 0.7, and 6.0 ± 1.3, respectively. Additionally, this study observed how the undesirable higher interaction of AChE control MB2 with galantamine negatively affected the assay, stressing the importance of the control assay. In another study, the immobilization efficiency of two commonly available MBs, amine- and carboxyl-terminated MBs, was comparatively evaluated based on the activity and amount of the immobilized monoamine oxidase A (MAOA). The specific binding and nonspecific adsorption on these MBs were also investigated using a model mixture. The results showed that carboxyl-terminated MBs with immobilized MAOA (MBs@COOH@MAOA) were the better approach, as they were successfully used to fish out ligands from the alkaloid extract of Hunteria zeylanica (Liu et al., 2024).
3.3 AS-MS based magnetic beads and in other supports
In the context of COVID-19, immobilized-based AS-MS assays have been used to identify natural ligands to SARS-CoV-2 proteins. Yonamine et al. (2023) reported the expression and purification of SARS-CoV-2 Nsp4, which is involved in the replication/transcription complex (RTC) within double-membrane vesicles (DMVs). Inhibition of Nsp4 disrupts the virus’s infectious cycle. SARS-CoV-2 Nsp4 were covalently immobilized on magnetic particles (Nsp4-MB) to screen an ethanolic extract of Hippeastrum aulicum, identifying three alkaloid ligands: haemanthamine (3.10), albomaculine (3.11), and aulicine (3.12). The identification was based on comparison with LC-MS data from standard alkaloids. Additionally, from a mixture of 10 alkaloids, four were identified as ligands: specifically, 2-α-7-dimethoxyhomolycorine (3.13), haemanthamine (3.10), albomaculine (3.11), and tazettine (3.14) (Figure 3). A control assay was performed, and affinity ratios (>1.89) suggested the presence of strong ligands. From an extract of Glycyrrhiza inflata. Muchiri et al. (2022) identified licochalcone A (3.15) as a ligand for SARS-CoV-2 spike protein Subunit 1 (S1) though dereplication and comparison with standards. In another study of Cannabis sativa (van Breemen et al., 2022) the authors identified and ranked several cannabinoid ligands by their affinity for the spike protein (S1). Two of these, cannabinoid acid (CBDA) (3.16) and cannabigerolic acid (CBGA) (3.17) showed the highest affinities for the spike protein and were confirmed to block infection of the original alive SARSCO-Cov-2 virus variants of concern, including the B.1.1.7 (Alpha, first detected in the United Kingdom) and B.1.351 (Beta, first detected in South Africa).
FIGURE 3
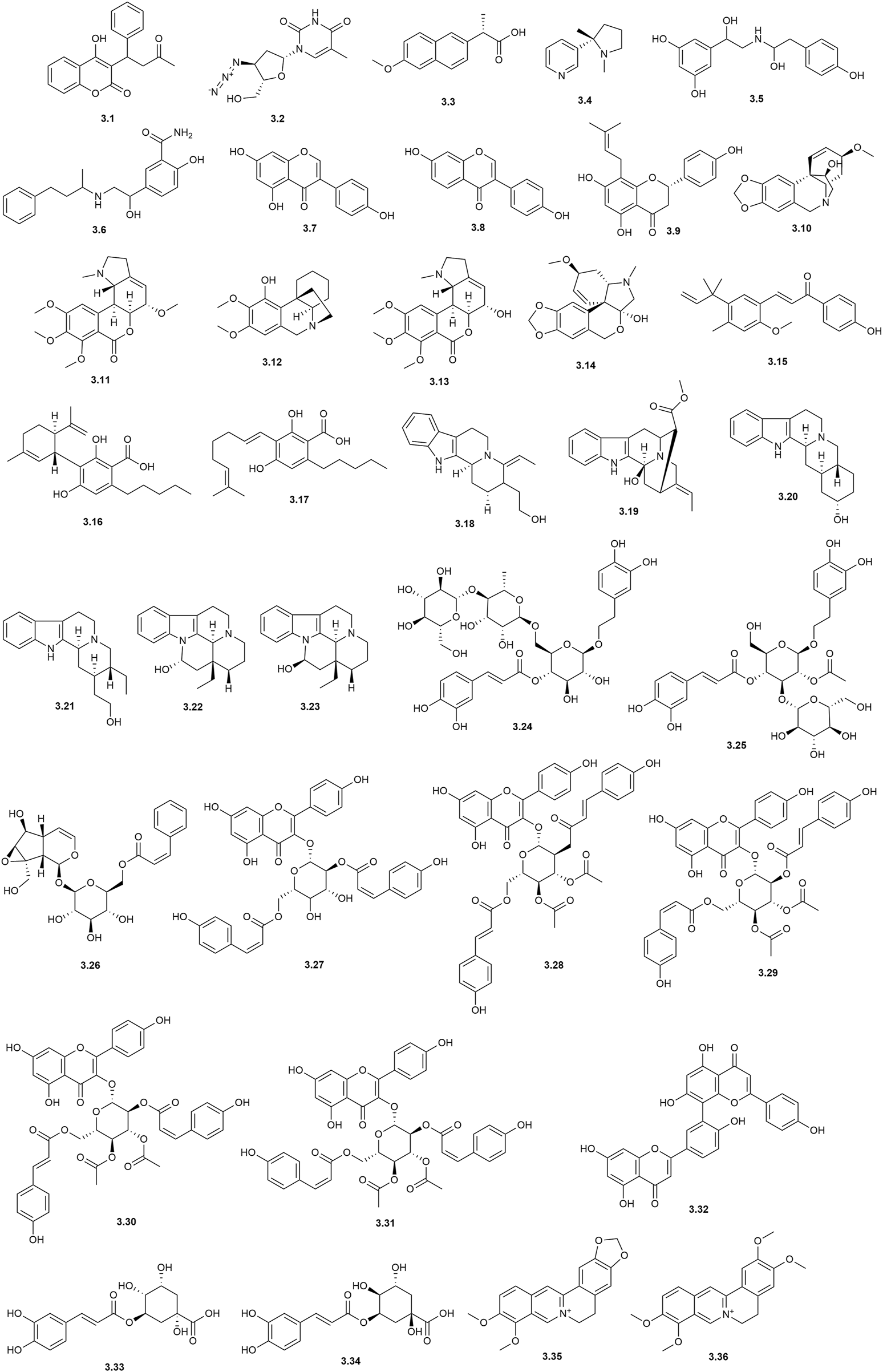
Chemical structures characterized by immobilized-based AS-MS assays.
AS-MS assays have been successfully used to identify natural ligands for monoamine oxidases isoforms MAO-A (Zhang et al., 2019) and MAO-B (Jiang et al., 2019; Wu et al., 2019). For example, Liu et al. (2024) reported, for the first time, the inhibition of MAO-A by monoterpene indoles alkaloids, which were isolated from H. zeylanica. They covalently immobilized MAO-A on amine- and carboxyl-terminated MBs. The carboxyl-terminated MBs-MAOA were used for screening the alkaloid extract of H. zeylanica. Twelve monoterpene indoles alkaloids were identified as MAO-A ligands, and nine of them were annotated by standards data, as a target strategy. To structure characterize these ligands the authors compared their retention times with those of the reference standards, analyzing their MS and MS/MS data, and referring to relevant literature for proposed patterns. To investigate their potential inhibitory activity, IC50 values for 10 structurally characterized ligands were calculated. Six of these ligands exhibited IC50 values a little higher than positive standard toloxatone (IC50 = 3.26 μM): geissoschizol (3.18) (15.55 ± 5.73), vobasinol (3.19), (17.65 ± 3.53), yohimbol (3.20) (24.58 ± 5.31), dihydrocorynanthenol (3.21) (13.82 ± 1.93), eburnamine (3.22) (14.85 ± 4.99) and (+)-isoeburnamine (3.23) (9.67 ± 1.47 μM). The mechanism of action was determined and docking studies corroborated the observed activity. These results provided insights for future investigations into tryptophan-derived alkaloids as potential drug candidates for MAO-A inhibition.
MAO-B was immobilized on cellulose filter paper to identify ligands in a fractionated n-BuOH extract of Tibetan strawberry (Fragaria nubicola), a plant known for various health benefits, such as neuroprotection and antioxidant properties (Hu et al., 2024). In this study, three new MAO-B inhibitors were isolated and identified, including two known compounds, turrillioside A (3.24) and 2′-O-acetylplantamajoside (3.25), as well as one new compound, nubicolosides A (3.26). This study represents an “nontarget strategy,” since only limited chemical information was available, and the active compounds were isolated for activity evaluation. The inhibitors showed IC50 values of 16.95 ± 0.93, 24.69 ± 0.20, and 46.77 ± 0.78 μM, respectively. Of particular interest, the metabolite 3.24 exhibited neuroprotective effects against 6-OHDA-induced injury in PC12 cells. Notably, the method demonstrates rapidity and effectiveness in screening of MAO-B inhibitors from complex matrices. Similarly, AChE was immobilized on cellulose filter paper for fishing out ligands from Terminalia chebula extracts (Li et al., 2022).
Diabetes is a chronic metabolic disease characterized by insufficient insulin secretion and insulin resistance. A review on the identification of anti-diabetic components from natural products such as Ginkgo biloba, Morus alba, lotus leaves, Pueraria lobata, Prunella vulgaris, and Magnolia cortex using ligand fishing approaches has been published (Guo et al., 2023). These approaches involve various immobilization methods, including entrapment, physical adsorption, covalent binding, affinity immobilization, multienzyme systems, and carrier-free immobilization, utilizing different carriers such as hollow fibers, magnetic materials, microreactors, and metal-organic frameworks.
A paper-based ligand fishing method was also developed to screen for α-glucosidase (GAA) inhibitors from Chinese herbs (Chen et al., 2024). Firstly, by using the same paper immobilized enzyme approach fourteen plants were screened, and the four most active extracts were selected for the ligand fishing approach. The best results were found with the leaves of Quercus variabilis Blume, which resulted in eighteen peaks, most of which were acylated flavonol glycosides. Five compounds named: kaempferol-3-O-(2″,6″-di-O-(Z)-p-coumaroyl)-β-glucopyranoside (3.27) kaempferol-3-O-(3″,4″-di-O-acetyl-2″,6″-di-O-(E)-p-coumaroyl)-β-glucopyranoside (3.28), kaempferol-3-O-(3″,4″-di-O-acetyl-2″-O-(E)-p-coumaroyl-6″-O-(Z)-p-coumaroyl)-β-glucopyranoside (3.29), kaempferol-3-O-(3″,4″-di-O-acetyl-2″-O-(Z)-p-coumaroyl-6″-O-(E)-p-coumaroyl)-β-glucopyranoside (3.30) and kaempferol-3-O-(3″,4″-di-O-acetyl-2″,6″-di-O-(Z)-p-coumaroyl)-β-glucopyranoside (3.31) were successfully isolated for complete characterization and further verification, with their structures elucidated by NMR. The IC50 values of these ligands ranged in 20–70 µM.
In a rapid screening for AChE inhibitors from Selaginella doederleinii Hieron using functionalized magnetic Fe3O4 nanoparticles, four compounds were confirmed to be potent AChE inhibitors. Among these, amentoflavone (3.32) exhibited a stronger AChE inhibitory effect than tacrine (positive control) with an IC50 of 0.73 ± 0.009 μM. Here, the authors (Zhang et al., 2022) used the targeted strategy, identifying the compounds by comparing MS data with standards.
AS-MS-based on immobilized enzyme is a fast, selective and widely used method with many unique features and huge application potentials and has been successfully used as a tool to identify ligands in natural extracts for several biological targets, including cancer related targets (Duarte-Filho et al., 2023), enzymes AChE (Vanzolini et al., 2013; 2015; 2018a; 2018b; Lima et al., 2021), α-glucosidase (Guo et al., 2023; Zhu et al., 2024), cyclooxygenase-2 (van Breemen et al., 2011), 15-lipoxygenase (Rush et al., 2016), and angiotensin (Luo et al., 2022).
AS-MS-based methods have been explored to investigate neurodegenerative diseases associated with misfolded proteins, such as prion diseases. These pathologies result from the conversion of the cellular prion protein (PrPC) into its pathogenic form (PrPSc), leading to progressive and fatal neurodegeneration. In the absence of effective therapies, a recent study has, for the first time, applied AS-MS assays to identify protein aggregation modulators in natural products (Amorim et al., 2025). Using this approach, a hydroethanolic extract from Moringa oleifera leaves was analyzed, resulting in the identification of chlorogenic (3.33) and neochlorogenic acids (3.34) as potent inhibitors of PrP aggregation. These compounds demonstrated significant antiprion activity, with IC50 values of 64.41 ± 12.12 μM and 35.34 ± 7.09 μM, respectively, and were able to inhibit PrPC-to-PrPSc conversion as well as disaggregate preformed PrPSc fibrils in vitro. This study highlights the potential of AS-MS assays to expedite the development of targeted therapies for prion diseases and other amyloid-related disorders.
3.4 Immobilization by streptavidin-biotin interaction
Another widely used approach in immobilized biomolecule assays leverages the streptavidin-biotin interaction. This method allows the biomolecule to remain structurally free to interact with screening molecular library due to its immobilization via the biotin tag. Compared to direct binding methods, this approach better preserves the biomolecule’s structure. The streptavidin-biotin system exhibits an affinity (KD = 10–15) comparable to covalent bond strength (Ozawa et al., 2017), ensuring stable immobilization. In this context, Xu et al. (2012) employed a AS-MS assay to screen triplex DNA binders. The triplex consisted of the sequences d (CCTTCCTCTTCTCT) (T1), AGAGAAGAGGAAGG (T2), and the biotinylated strand biotin-(dT)5-d (TCTCTTCTCCTTCC) (T3). The biotinylated oligonucleotide T3 was immobilized on streptavidin-agarose beads, followed by the addition of T1 and T2 to form triplex DNA. As a control, beads with only the T3 oligonucleotide were used. The DNA-beads were incubated with an ethanolic-aqueous extract of Phellodendron chinense Schneid cortex (phellodendron), washed to remove unbound compounds, and analyzed by LC-MS. This approach identified two alkaloids, berberine (3.35) and palmatine (3.36), as binders based on retention time and MS data compared with authentic standards. Similarly, Yang et al. (2017) immobilized biotinylated triplex DNA (CCTTCCTCTTCTCT (T1), AGAGAAGAGGAAGG (T2), and biotin-(dT)5-d (TCTCTTCTCCTTCC) (T3)) on streptavidin-coated 96-well plates, using the single oligonucleotide (T3) as a control. Parameters for triplex DNA formation, such as incubation time, temperature, and buffer conditions, were optimized using four known binders: coralyne, ethidium bromide, vitexin, and formononetin. The ligand fishing assay was then applied to a Rhizoma Coptidis extract, which was analyzed by LC-MS. Seven metabolites were identified as ligands and ranked by affinity ratio as follows: columbamine > palmatine ≈ epiberberine > an unknown compound at m/z 322.10 > coptisine ≈ jatrorrhizine > berberine.
Despite the vast applications, the identification of new compounds and nontarget approaches remains a challenge. Most of the work reported in the literature uses metabolite targeted approaches, employing standards to define the structural identity of the disclosed ligands from natural products.
4 Dereplication tools and their role in AS-MS for natural product research
The nontarget exploration of natural products extracts by affinity techniques presents a more complex challenge than the screening of synthetic libraries. Natural products, derived from biological sources such as plants, fungi, and marine organisms, tend to exhibit unique, complex scaffolds that often extended beyond the chemical space covered by synthetic libraries (Silva and Emery, 2018; Conrado et al., 2024). This structural diversity is a key reason why natural products continue to play a significant role in bioactive compounds discovery (Newman and Cragg, 2020; Conrado et al., 2024).
In the scope of AS-MS applied to natural products, dereplication has become an essential step for efficiently navigating complex extracts and prioritizing novel bioactive compounds for further isolation. Dereplication is an identification of known molecules and helps to avoid unnecessary re-isolation of already characterized metabolites, allowing researchers to focus on novel compounds (Gaudêncio and Pereira, 2015). One of the most powerful tools for dereplication in natural products research is liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS), which allows for the analysis of complex mixtures by separating the metabolites based on their physical chemical properties, providing detailed molecular weight information and fragmentation patterns which can be compared with a diversity of MS/MS libraries (Gaudêncio et al., 2023) that possesses thousands of chemical structures, such as Global Natural Products Social Molecular Networking (GNPS) (Wang et al., 2016), LipidMaps (Conroy et al., 2024), Metlin (Guijas et al., 2018), MassBank (Horai et al., 2010), and Human Metabolome Database (HMDB) (Wishart et al., 2022), MetaboLights (Yurekten et al., 2024), NIST Mass Spectral Library, etc.
In studies where dereplication is employed before AS-MS, researchers generally aim to characterize the metabolite profile of an extract to reduce the complexity of subsequent affinity-based screening. In contrast, studies that do not use dereplication before AS-MS rely solely on affinity screening to identify ligands. This approach can be advantageous when the primary goal is to discover novel interactions between natural products and macromolecular targets. For example, Jiao et al. (2019) investigated an ethanolic-aqueous extract of Andrographis paniculata for its activity against COX-2 without prior dereplication, using ultrafiltration to identify five affinity-bound compounds, in which andrographidine E demonstrated an IC50 of 19 µM.
Dereplication methods can use manual approach, semi-automated or automated tools such as GNPS (Wang et al., 2016). While all these methods aim to efficiently identify known compounds within complex extracts, they differ in terms of efficiency, accuracy, and the required expertise. Preliminary dereplication of extracts, while valuable, does not ensure that the annotated molecules will match those specifically disclosed by affinity during AS-MS experiments. This mismatch can lead to significant time and effort expended on dereplicating molecules that ultimately lack the desired affinity, highlighting an apparent inefficiency within the workflow. Nonetheless, profiling the extract remains highly informative. This practice not only enriches our understanding of the extract’s chemical landscape but also contributes broadly to the natural products field, where knowledge of the complex and often unique metabolite compositions can guide future explorations and facilitate the discovery of novel bioactive molecules and analogues. While dereplication facilitates the annotation of metabolites within complex extracts, AS-MS distinguishes itself by focusing on molecular annotation of the ligands, emphasizing the importance of distinguishing between metabolite annotation and ligand identification.
Lima et al. (2021) investigated alkaloid extracts from Annona salzmannii bark to identify ligands for AChE. Through dereplication and molecular networking by GNPS, 34 isoquinoline alkaloids were annotated, including 26 compounds newly reported for A. salzmannii and the novel alkaloid N,O-dimethylcoclaurine N-oxide. However, only eight of these were disclosed as AChE ligands. Three other disclosed ligands, due to their lower intensity spectra, were not annotated and only their class was inferred. This was achieved using AChE immobilized onto magnetic beads, with ligand affinity determined by an affinity ratio. Comparisons between active and inactive enzyme bioreactors revealed compounds with affinity ratios exceeding that of the known inhibitor galantamine. The study underscores the meticulous alignment needed between metabolite annotation and affinity-based ligand assay, emphasizing the detailed experimental workflow required.
4.1 Manual dereplication
Manual dereplication can be a challenging and time-consuming process that demands extensive expertise. Researchers need to compare the spectral and chromatographic properties of the metabolites against known data from literature or in-house libraries, requiring a detailed understanding of mass fragmentation patterns, particularly when working with structurally complex natural molecules. For instance, Hsu et al. (2021) investigated xanthine oxidase (XOD) ligands in alfalfa (Medicago sativa) for potential hyperuricemia prevention. Using UHPLC-ESI-Q-TOF-MS/MS to profile the extract, they employed a subtraction method on the XO-incubated extract, enabling manual dereplication of twelve compounds. Molecular docking showed high XO affinity for nine compounds, including salicylic acid, tricin 7-O-glucuronopyranoside, chrysoeriol-7-glucoside, ferulic acid, apigenin 7-O-β-glucuronopyranoside, apigenin, tricin, chrysoeriol, and liquiritigenin. In vitro bioassays confirmed potent XO inhibitory activities for apigenin, chrysoeriol, and liquiritigenin, with IC50 values of 0.25, 0.5, and 1 μM, respectively, surpassing the standard XO inhibitor allopurinol (IC50 = 1.41 µM).
The limitations of manual dereplication become apparent in high-throughput settings, where a lack of automation may result in slower processing times and, occasionally, ambiguous annotation. Despite its challenges, manual dereplication is still employed, particularly in cases where computational tools may not have sufficient reference data for the specific class of compounds under investigation. However, the growing availability of spectral databases and automated tools has led to a decline in the use of purely manual methods.
4.2 GNPS: a collaborative platform for natural product research
The GNPS (Wang et al., 2016) is a powerful platform for natural product dereplication using LC-MS/MS data. GNPS allows researchers to upload MS/MS spectra (in mzXML, mzML, and mgf formats) to identify known metabolites by comparing them with a vast database of public spectra. Compounds are grouped by structural similarities, highlighting chemical relationships through fragmentation patterns in a feature-based molecular network (FBMN). In addition to the classic FBMN workflow, the platform’s flexibility includes integration of FBMN with MZmine, MS-DIAL, XCMS, and OpenMS, Menoscabos, and Progenesis QI. Each tool impacts peak alignment, annotation accuracy, and feature detection sensitivity, offering distinct advantages for specific experimental needs, with GNPS providing access to over 100,000 MS/MS spectra for comparison.
Dai et al. (2024) constructed a GNPS-based molecular network from the bark of Cinnamomum cassia, analyzed by LC-MS/MS, categorizing this extract as having significant α-glucosidase inhibitory and antioxidant activities. To disclose the ligands, affinity-based ultrafiltration was employed, resulting in the identification of nine procyanidins with notable antioxidant and enzyme inhibition properties. Among these, procyanidins A1, A2, B1, and C1 were isolated, with their activity validated through functional assays, underscoring their potential as dual inhibitors of α-glucosidase and pancreatic lipase.
Recent advances in machine learning and artificial intelligence have driven the development of tools that enhance prediction, classification, and evaluation in dereplication. Within the GNPS platform, these tools support more efficient metabolite annotation for natural libraries, which is transformative for nontarget metabolomics and AS-MS applications. For instance, DEREPLICATOR+ (Mohimani et al., 2018) specializes in identifying peptides and other complex secondary metabolites from MS/MS spectra through in silico fragmentation, enhancing the annotation of molecular structures. Similarly, MolDiscovery (Cao et al., 2021) is optimized for smaller molecules, improving on DEREPLICATOR+ by leveraging machine learning algorithms to increase the accuracy of identifying novel metabolites in natural products.
Network Annotation Propagation (NAP) (da Silva et al., 2018) further aids in assigning annotations across molecular networks by propagating known information to structurally related but uncharacterized molecules by in silico fragmentation, offering an effective approach to understanding compound analogies within complex mixtures. MS2LDA (Wandy et al., 2018), employing probabilistic modeling, identifies “fragmentation motifs” from conserved fragment and neutral loss features in spectral data, offering an effective classification method for compound families in natural product extracts. The MolNetEnhancer (Ernst et al., 2019) tool enhances molecular networks by integrating MS2LDA and in silico fragmentation (i.g. DEREPLICATOR+) combine with the automated chemical taxonomy classification by ClassyFire (Djoumbou Feunang et al., 2016).
The Mass Spectrometry Search Tool (MASST) (Wang et al., 2020) allows direct MS/MS spectral searches against public datasets to determine the contexts in which a molecule has been previously identified. Recently, MASST has evolved with specialized versions like foodMASST, microbeMASST, plantMASST, and personalcareMASST. Such functionality is particularly valuable for natural product research, as it aids in validating whether a compound annotated in a dereplication workflow genuinely aligns with natural origins or biosynthetic pathways specific to its source.
An already well-established tool is SIRIUS (Dührkop et al., 2019), a suite designed for structural elucidation of small molecules from tandem mass spectrometry data. In combination with CSI:FingerID (Dührkop et al., 2015), SIRIUS generates accurate molecular formula predictions and provides structure elucidation through predicted molecular fingerprints. In the GNPS platform, SIRIUS works synergistically with the ZODIAC (Ludwig et al., 2020) algorithm, which enables the de novo annotation of molecular formulas in complex samples by refining the molecular formula prediction process. CSI:FingerID facilitates high-confidence structural assignments, and CANOPUS (Dührkop et al., 2021) enables the classification of compounds into broader chemical classes, expediting dereplication in natural product libraries by helping researchers identify and categorize unknown compounds more precisely.
Collectively, these GNPS-integrated tools streamline natural product research by facilitating the analysis of complex datasets and accelerating the discovery of bioactive compounds. In AS-MS, this suite enables more efficient identification of specific ligands, such as enzyme inhibitors or other protein-binding metabolites, enhancing dereplication, structural elucidation, and the discovery of bioactivity in natural product extracts.
With the recent launch of GNPS2, the field of AS-MS and natural product dereplication is poised for further advancements. GNPS2 incorporates machine-learning algorithms, improving spectral matching precision and allowing users to upload and search larger datasets. This platform not only builds on the foundational GNPS framework but also enhances data sharing and collaboration among researchers. Additionally, GNPS2 now includes an integrated statistical tool specifically designed to support FBMN for nontarget metabolomics including facilitates data cleaning, normalization, and uni- and multivariate statistical analyses of FBMN outputs, with further visualization support through Cytoscape integration (Pakkir Shah et al., 2024). The option for blank removal is particularly valuable for AS-MS when working with immobilized biological targets. This feature can help address nonspecific interactions, as some compounds may bind to the support itself rather than to the target, potentially leading to misleading results. As more natural products are characterized and entered the GNPS2 database, the tool’s predictive capabilities will become more comprehensive, thus improving dereplication accuracy (Nothias et al., 2020) for AS-MS applications. The integration of advanced algorithms in GNPS2 also points to an era where dereplication can approach near-real-time processing, offering significant advantages in high-throughput AS-MS studies.
4.3 Limitations of dereplication in AS-MS
While automated tools vastly improve dereplication efficiency, their limitations remain, particularly in cases of stereoisomers and when library data for rare or novel compounds is lacking. Manual validation is often necessary to confirm structural accuracy. Despite modern advancements in instrumentation, software, and databases that facilitate high-throughput metabolite annotation, the conclusive identification of ligands often requires the isolation of individual compounds or the use of authentic standards. This challenge becomes particularly pronounced when dealing with stereoisomers. do Amaral et al. (2021) immobilized phosphoenolpyruvate carboxykinase from Trypanosoma cruzi onto magnetic particles to carried out nontarget AS-MS on ethanolic extracts of Brazilian cerrado plants, identifying 11 ligands. Among these, catechin or its epimer, epicatechin, was detected in the Byrsonima coccolobifolia extract. Since both compounds are present in the extract and share identical mass fragmentation patterns, they could not be distinguished by mass spectrometry alone. To allow correct characterization of the ligand, authentic standards were analyzed under the same LC-MS conditions used for the AS-MS desorbate analysis. Retention time comparison confirmed catechin as the ligand. This study underlines the importance of affinity-based interaction specificity between the targets and ligands, as both catechin and epicatechin were present in the extract, but only catechin was selectively fished out.
Native MS were employed to screen for chymotrypsin inhibitors in a marine cyanobacterium Rivularia sp. (Reher et al., 2022), identifying 30 potential cyclodepsipeptide-protease complexes in methanolic extracts. Using tools as SIRIUS, ZODIAC and MS/MS spectrum as indicative of a “cyclic depsipeptide” based on the classification with CANOPUS, they isolated and carried out a complete characterization (1D/2D NMR experiments and manual MS/MS interpretation) of rivulariapeptolides 1185 (4.1), 1155 (4.2), 1121 (4.3), and 989 (4.4), molassamide (4.5) and the novel molassamide B (4.6) (Figure 4). Chymotrypsin inhibition assays showed high efficacy (IC50 < 100 nM) for rivulariapeptolides and molassamide B. These findings prompted further evaluation of the six peptides against two additional serine proteases, elastase and proteinase K, ranking them as some of the most potent Ahp-cyclodepsipeptides reported to date. Remarkably, the authors shared in GNPS the spectral data to collaborate with future studies.
FIGURE 4
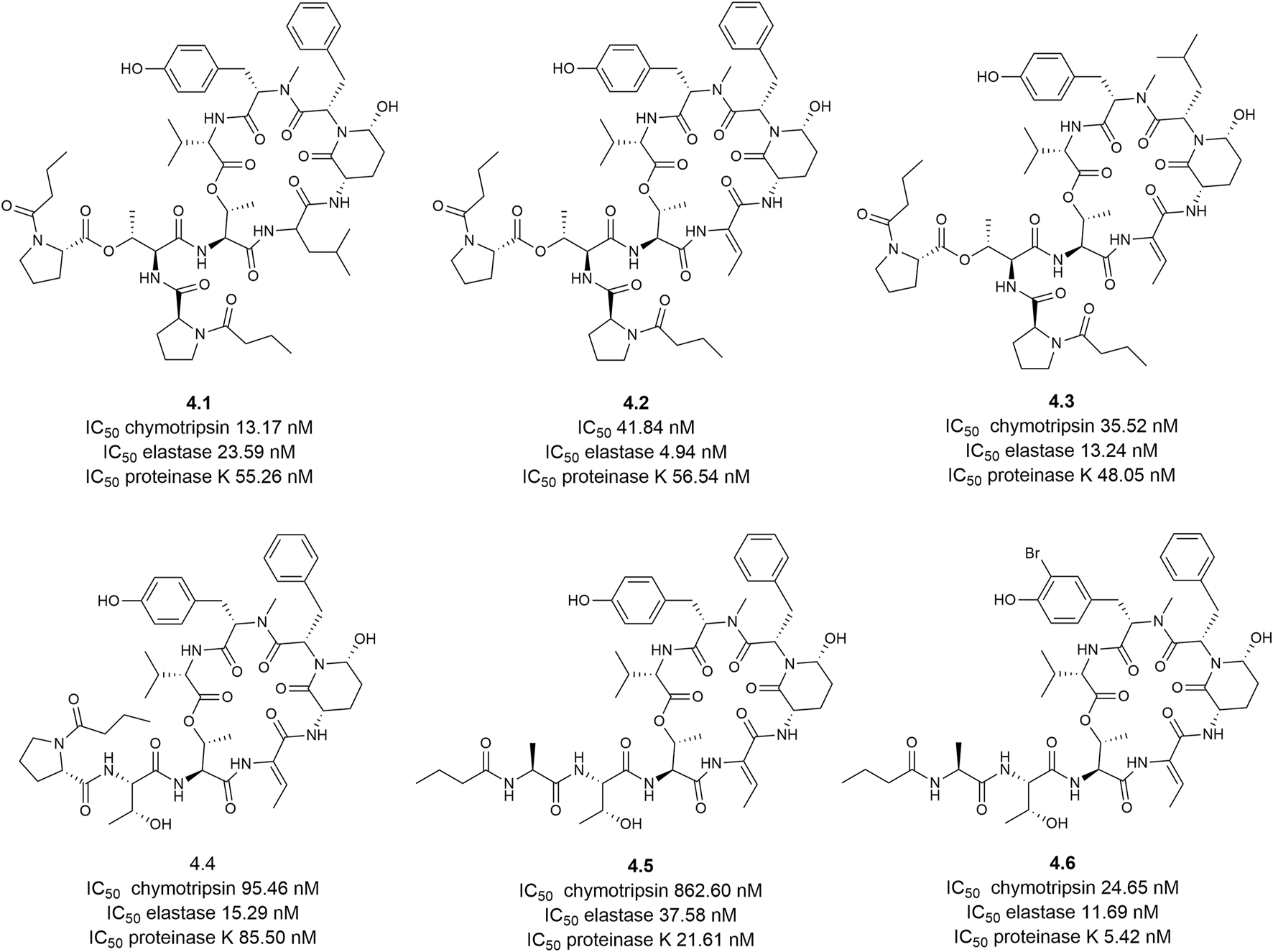
Structures and potency of the isolated rivulariapeptolides and molassamides from an environmental cyanobacteria community for three serine proteases.
Moreover, critically evaluating the annotation of suggested metabolites by automated tools is essential. Researchers must assess whether the proposed structures are truly natural products and if they could feasibly be biosynthesized by the species under study, ensuring that the annotation findings are relevant and biologically accurate. XOD has been investigated for inhibitors in Pterocladiella capillacea, an edible seaweed, using affinity ultrafiltration combined with feature-based molecular networking analysis facilitated by MS-DIAL and MS-FINDER (Wang et al., 2023). While 20 compounds were initially identified as XOD ligands, manual verification revealed that many lacked taxonomic alignments with the seaweed’s lineage, either at the Order or Phylum level. Finally, eight compounds were confirmed as seaweed-derived, with seven unreported for anti-gout activity. Molecular docking studies further suggested these compounds exhibit strong affinity for XOD, highlighting their therapeutic potential.
The use of automated dereplication tools and molecular networking followed by manual validation combines the speed of computational methods with the accuracy of traditional structural elucidation, optimizing AS-MS workflows for comprehensive natural product discovery. Additionally, orthogonal assays are necessary for accurate ligand annotation, especially in the context of structurally similar metabolites and stereoisomers prevalent in natural product extracts. Important to stress that the stereochemistry of annotated ligands cannot be confidently assigned solely through AS-MS workflows and requires further characterization using chiral analytical methods or comparison with well-characterized standards. Accurate stereochemical assignments are essential to ensure biological relevance, as stereoisomers often exhibit distinct binding affinities, activities, and pharmacological properties.
4.4 Target ligand applications of AS-MS
Despite the potential application of AS-MS for nontarget natural products extracts exploration its application frequently mirrors the focused approach of synthetic libraries, in targeted workflows, where predefined known ligands or metabolite classes drive experimental design. This target strategy can simplify assay setups, reducing the complexity of control and facilitating hit elucidation, and enables the validation of specific hypotheses when a well-characterized extract or a natural product library are available. For example, Munigunti et al. (2011) used ultrafiltration and LC-MS to screen 133 structurally diverse natural products from a commercial library against Plasmodium falciparum thioredoxin reductase, identifying nine ligands with binding affinities below 1 µM. Similarly, Vu et al. (2018) investigated 62 Plasmodium proteins potential for malaria drugs using native MS (ESI-FT-ICR-MS) to screen a natural product fragment library containing 643 fragments (most of them compliant with the rule of three), in which 96 fragments were identified as ligands for 32 proteins, with 79 of these demonstrating growth inhibition against P. falciparum.
In addition to using libraries, other works leverage the known bioactivities of extracts to guide ligand identification. van Breemen et al. (2011) applied a chloroform partition of a methanol extract from ginger roots (Zingiber officinale) to detect COX-2 ligands using ultrafiltration and LC-MS justly because ginger presents anti-inflammatory properties, including COX inhibition. The study identified 19 metabolites, being primarily gingerol-related compounds. Among these, 10 were confirmed as COX-2 ligands, with IC50 values determined for 10-gingerol, 8-shogaol, and 10-shogaol, all exhibiting selective COX-2 inhibition. Similarly, Li et al. (2009) investigated a flavonoid extract from hawthorn (Crataegus oxyacantha L.), a plant known for its medicinal properties and rich flavonoid content. An affinity assay combining ultrafiltration and LC-DAD-MS disclosed four flavonoids as ligands for α-glucosidase, including quercetin derivatives and vitexin glycosides. Since three flavonoids are C-glycosylated, other ones were tested (vitexin, isovitexin, orientin, isooriention and their aglycones apigenin and luteolin). Further testing confirmed their inhibitory activity, validating the bioactivity of the extract.
Studies on Traditional Chinese Medicine (TCM) often adopt a similar approach, aiming known bioactive compounds within herbal mixtures. AS-MS techniques have been employed to investigate interactions between TCM components and specific targets, such as enzymes or receptors. For instance, affinity-based methods have revealed interactions of chlorogenic acids, coumarins, and flavonoids with proteins like plasma albumin and G protein-coupled receptors, aligning traditional knowledge with scientific validation (Jiao et al., 2018; Zhang et al., 2024). Such works illustrate the value of targeted AS-MS in bridging ethnopharmacological insights with molecular mechanisms.
5 Concluding remarks
AS-MS has gained prominence in target-based drug discovery programs and is widely used by the industry to search for small molecules in synthetic libraries. Despite its versatility and broad applications, AS-MS remains underexplored for ligand prospecting in natural product libraries. The complexity of the chemical space, with high concentrations of one class of molecules present alongside others at lower concentrations, can hinder both the formation of ligand complexes and the annotation of ligands present in low concentrations. Additionally, nonselective interactions with the target, such as those caused by Pan-assay interference compounds (PAINS), pose a significant challenge when assaying natural libraries. To fully exploit the potential of AS-MS, future efforts should focus on developing more robust methods for annotating ligands from natural libraries, including enhanced data analysis workflows. As in dereplication workflows, stereoisomers and positional isomers must not be overlooked.
Most AS-MS applications using natural product libraries are still focused on enzyme targets, followed by other protein types, with relatively few studies involving diverse biological targets. Expanding AS-MS to underexplored target types and leveraging its compatibility with natural product libraries could unlock new opportunities in drug discovery. Furthermore, broadening the diversity of biological targets and improving the integration of AS-MS with complementary techniques, such as structural biology and structure-based virtual screening, could further enhance its utility. By addressing these challenges, AS-MS has the potential to bridge the gap between the chemical diversity of natural products and biological function, thus significantly advancing drug discovery.
Statements
Author contributions
BA: Conceptualization, Data curation, Formal Analysis, Writing – original draft, Writing – review and editing. MdM: Conceptualization, Data curation, Formal Analysis, Writing – original draft, Writing – review and editing. Carmen Lucia CC: Conceptualization, Data curation, Formal Analysis, Writing – original draft, Writing – review and editing. QC: Conceptualization, Data curation, Formal Analysis, Writing – original draft, Writing – review and editing, Project administration.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The authors acknowledge São Paulo State Research Foundation (FAPESP–Grant 2022/00432-7). The Coordenação de Aperfeiçoamento de Pessoal de Nível Superior–Brasil (CAPES) – Finance Code 001; Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (Grants: 307108/2021-0; 302464/2022-0) and FAPERJ (Grants E-26/200.172/2023 and E-26/210.017/2024).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Correction note
A correction has been made to this article. Details can be found at: 10.3389/fntpr.2025.1645358.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Ablat A. Li M. J. Zhai X. R. Wang Y. Bai X. L. Shu P. et al (2024). Fast screening of tyrosinase inhibitors in Coreopsis tinctoria Nutt. by ligand fishing based on paper-immobilized tyrosinase. Molecules29, 4018. 10.3390/molecules29174018
2
Adam G. C. Parish C. A. Wisniewski D. Meng J. Liu M. Calati K. et al (2008). Application of affinity selection/mass spectrometry to determine the structural isomer of parnafungins responsible for binding polyadenosine polymerase. J. Am. Chem. Soc.130, 16704–16710. 10.1021/ja805531w
3
Almeida F. Cass Q. (2023). Affinity selection mass spectrometry (AS-MS) as a tool for prospecting target ligands. Braz. J. Anal. Chem.10, 13–16. 10.30744/brjac.2179-3425.letter-almeida-cass
4
Amorim M. S. d Amaral-do-Nascimento M. Severino V. G. P. Silva J. L. da Vieira T. C. R. G. de Moraes M. C. (2025). Identification of chlorogenic acids from Moringa oleifera leaves as modulators of prion aggregation using affinity selection-mass spectrometry. ACS Omega10, 2919–2930. 10.1021/acsomega.4c09150
5
Basso A. Serban S. (2019). Industrial applications of immobilized enzymes—a review. Mol. Catal.479, 110607. 10.1016/j.mcat.2019.110607
6
Bilal M. Zhao Y. Rasheed T. Iqbal H. M. N. (2018). Magnetic nanoparticles as versatile carriers for enzymes immobilization: a review. Int. J. Biol. Macromol.120, 2530–2544. 10.1016/j.ijbiomac.2018.09.025
7
Cao L. (2006). Introduction: immobilized enzymes: past, present and prospects. 1, 1–52. 10.1002/3527607668.ch1
8
Cao L. Guler M. Tagirdzhanov A. Lee Y.-Y. Gurevich A. Mohimani H. (2021). MolDiscovery: learning mass spectrometry fragmentation of small molecules. Nat. Commun.12, 3718. 10.1038/s41467-021-23986-0
9
Chen G. Guo M. (2017a). Rapid screening for α-glucosidase inhibitors from Gymnema sylvestre by affinity ultrafiltration-HPLC-MS. Front. Pharmacol.8, 228–8. 10.3389/fphar.2017.00228
10
Chen G. Guo M. (2017b). Screening for natural inhibitors of topoisomerases I from Rhamnus davurica by affinity ultrafiltration and high-performance liquid chromatography–mass spectrometry. Front. Plant Sci.8, 1521–1611. 10.3389/fpls.2017.01521
11
Chen X. Wu Y. Wu S. Gu Y. Luo J. Kong L. (2024). Paper-based ligand fishing method for rapid screening and real-time capturing of α-glucosidase inhibitors from the Chinese herbs. J. Pharm. Biomed. Anal.242, 116037. 10.1016/j.jpba.2024.116037
12
Chi M. Wang H. Yan Z. Cao L. Gao X. Qin K. (2022). Magnetic ligand fishing using immobilized cyclooxygenase-2 for identification and screening of anticoronary heart disease ligands from Choerospondias axillaris. Front. Nutr.8, 794193. 10.3389/fnut.2021.794193
13
Choi Y. Jermihov K. Nam S. J. Sturdy M. Maloney K. Qiu X. et al (2011). Screening natural products for inhibitors of quinone reductase-2 using ultrafiltration LC-MS. Anal. Chem.83, 1048–1052. 10.1021/ac1028424
14
Choi Y. van Breemen R. (2008). Development of a screening assay for ligands to the estrogen receptor based on magnetic microparticles and LC-MS. Comb. Chem. High. Throughput Screen11, 1–6. 10.2174/138620708783398340
15
Cieśla Ł. Moaddel R. (2016). Comparison of analytical techniques for the identification of bioactive compounds from natural products. Nat. Prod. Rep.33, 1131–1145. 10.1039/c6np00016a
16
Ciesla L. Okine M. Rosenberg A. Dossou K. S. S. Toll L. Wainer I. W. et al (2016). Development and characterization of the α3β4α5 nicotinic receptor cellular membrane affinity chromatography column and its application for on line screening of plant extracts. J. Chromatogr. A1431, 138–144. 10.1016/j.chroma.2015.12.065
17
Conrado G. G. da Rosa R. Reis R. D. Pessa L. R. (2024). Building natural product–based libraries for drug discovery: challenges and opportunities from a Brazilian pharmaceutical industry perspective. Rev. Bras. Farmacogn.34, 706–721. 10.1007/s43450-024-00540-9
18
Conroy M. J. Andrews R. M. Andrews S. Cockayne L. Dennis E. A. Fahy E. et al (2024). LIPID MAPS: update to databases and tools for the lipidomics community. Nucleic Acids Res.52, D1677–D1682. 10.1093/nar/gkad896
19
Cuatrecasas P. Wilchek M. Anfinsen C. B. (1968). Selective enzyme purification by affinity chromatography. Proc. Natl. Acad. Sci. U. S. A.61, 636–643. 10.1073/pnas.61.2.636
20
Dai J. Liu Z. Ma L. Yang C. Bai L. Han D. et al (2024). Identification of procyanidins as α-glucosidase inhibitors, pancreatic lipase inhibitors, and antioxidants from the bark of Cinnamomum cassia by multi-bioactivity-labeled molecular networking. Food Res. Int.192, 114833. 10.1016/j.foodres.2024.114833
21
da Silva R. R. Wang M. Nothias L.-F. van der Hooft J. J. J. Caraballo-Rodríguez A. M. Fox E. et al (2018). Propagating annotations of molecular networks using in silico fragmentation. PLoS Comput. Biol.14, e1006089. 10.1371/journal.pcbi.1006089
22
de Faria R. A. Oliveira P. C. O. de Carvalho M. D. P. Peixoto B. S. Severino V. G. P. Tinoco L. W. et al (2022). High-resolution inhibition profiling and ligand fishing for screening of nucleoside hydrolase ligands in Moringa oleifera Lamarck. J. Pharm. Biomed. Anal.211, 114614. 10.1016/j.jpba.2022.114614
23
de Lima J. M. Furlani I. L. da Silva L. R. G. Valverde A. L. Cass Q. B. (2020). Micro- and nano-sized amine-terminated magnetic beads in a ligand fishing assay. Anal. Methods12, 4116–4122. 10.1039/D0AY01269F
24
de Moraes M. C. Cardoso C. L. Cass Q. B. (2019). Solid-supported proteins in the liquid chromatography domain to probe ligand-target interactions. Front. Chem.7, 752. 10.3389/fchem.2019.00752
25
de Moraes M. C. Vanzolini K. L. Cardoso C. L. Cass Q. B. (2014). New trends in LC protein ligand screening. J. Pharm. Biomed. Anal.87, 155–166. 10.1016/j.jpba.2013.07.021
26
de Oliveira P. C. O. Tinoco L. W. Cardoso C. L. Cass Q. B. de Moraes M. C. (2023). Advances in screening assays for identifying cholinesterase ligands. TrAC Trends Anal. Chem.168, 117362. 10.1016/j.trac.2023.117362
27
De Simone A. Naldi M. Bartolini M. Davani L. Andrisano V. (2019). Immobilized enzyme reactors: an overview of applications in drug discovery from 2008 to 2018. Chromatographia82, 425–441. 10.1007/s10337-018-3663-5
28
De Soricellis G. Rinaldi F. Tengattini S. Temporini C. Negri S. Capelli D. et al (2024). Development of an analytical platform for the affinity screening of natural extracts by SEC-MS towards PPARα and PPARγ receptors. Anal. Chim. Acta1309, 342666. 10.1016/j.aca.2024.342666
29
Djoumbou Feunang Y. Eisner R. Knox C. Chepelev L. Hastings J. Owen G. et al (2016). ClassyFire: automated chemical classification with a comprehensive, computable taxonomy. J. Cheminform8, 61. 10.1186/s13321-016-0174-y
30
do Amaral B. S. da Silva L. R. G. Valverde A. L. de Sousa L. R. F. Severino R. P. de Souza D. H. F. et al (2021). Phosphoenolpyruvate carboxykinase from T. cruzi magnetic beads affinity-based screening assays on crude plant extracts from Brazilian Cerrado. J. Pharm. Biomed. Anal.193, 113710. 10.1016/j.jpba.2020.113710
31
Dong X. Wang B. Cao J. Zheng H. Ye L. H. (2021). Ligand fishing based on bioaffinity ultrafiltration for screening xanthine oxidase inhibitors from citrus plants. J. Sep. Sci.44, 1353–1360. 10.1002/jssc.202000708
32
Duarte-Filho L. A. M. de S. de Oliveira P. C. O. Leal C. E. Y. de Moraes M. C. Picot L. (2023). Ligand fishing as a tool to screen natural products with anticancer potential. J. Sep. Sci., 1–25. 10.1002/jssc.202200964
33
Dührkop K. Fleischauer M. Ludwig M. Aksenov A. A. Melnik A. V. Meusel M. et al (2019). SIRIUS 4: a rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods16, 299–302. 10.1038/s41592-019-0344-8
34
Dührkop K. Nothias L.-F. Fleischauer M. Reher R. Ludwig M. Hoffmann M. A. et al (2021). Systematic classification of unknown metabolites using high-resolution fragmentation mass spectra. Nat. Biotechnol.39, 462–471. 10.1038/s41587-020-0740-8
35
Dührkop K. Shen H. Meusel M. Rousu J. Böcker S. (2015). Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proc. Natl. Acad. Sci. U. S. A.112, 12580–12585. 10.1073/pnas.1509788112
36
Ernst M. Kang K. B. Caraballo-Rodríguez A. M. Nothias L.-F. Wandy J. Chen C. et al (2019). MolNetEnhancer: enhanced molecular networks by integrating metabolome mining and annotation tools. Metabolites9, 144. 10.3390/metabo9070144
37
Ferry J. D. (1936). Ultrafilter membranes and ultrafiltration. Chem. Rev.18, 373–455. 10.1021/cr60061a001
38
Gaudêncio S. P. Bayram E. Lukić Bilela L. Cueto M. Díaz-Marrero A. R. Haznedaroglu B. Z. et al (2023). Advanced methods for natural products discovery: bioactivity screening, dereplication, metabolomics profiling, genomic sequencing, databases and informatic tools, and structure elucidation. Mar. Drugs21, 308. 10.3390/md21050308
39
Gaudêncio S. P. Pereira F. (2015). Dereplication: racing to speed up the natural products discovery process. Nat. Prod. Rep.32, 779–810. 10.1039/C4NP00134F
40
Girelli A. M. Mattei E. (2005). Application of immobilized enzyme reactor in on-line high performance liquid chromatography: a review. J. Chromatogr. B Anal. Technol. Biomed. Life Sci.819, 3–16. 10.1016/j.jchromb.2005.01.031
41
Gu Y. Liu M. Ma L. Quinn R. J. (2024). Identification of ligands for ion channels: TRPM2. ChemBioChem25, e202300790. 10.1002/cbic.202300790
42
Gu Y. Liu M. Quinn R. J. (2022). Metabolite-protein interactions: native mass spectrometry and collision induced affinity selection mass spectrometry in natural product screening. Front. Anal. Sci.2, 1–11. 10.3389/frans.2022.1014017
43
Guijas C. Montenegro-Burke J. R. Domingo-Almenara X. Palermo A. Warth B. Hermann G. et al (2018). METLIN: a technology platform for identifying knowns and unknowns. Anal. Chem.90, 3156–3164. 10.1021/acs.analchem.7b04424
44
Guo H. Guo L. Yu J. Zhao F. Yang W. Li J. et al (2024). Magnetic nanoparticles immobilized thrombin ligand fishing to screen thrombin inhibitors in natural products. J. Pharm. Biomed. Anal.243, 116110. 10.1016/j.jpba.2024.116110
45
Guo S. Wang S. Meng J. Gu D. Yang Y. (2023). Immobilized enzyme for screening and identification of anti-diabetic components from natural products by ligand fishing. Crit. Rev. Biotechnol.43, 242–257. 10.1080/07388551.2021.2025034
46
Hanefeld U. Cao L. Magner E. (2013). Enzyme immobilisation: fundamentals and application. Chem. Soc. Rev.42, 6211. 10.1039/c3cs90042h
47
Horai H. Arita M. Kanaya S. Nihei Y. Ikeda T. Suwa K. et al (2010). MassBank: a public repository for sharing mass spectral data for life sciences. J. Mass Spectrom.45, 703–714. 10.1002/jms.1777
48
Hsu S.-J. Verpoorte R. Lin S.-M. Lee C.-K. (2021). Fast dereplication of xanthine oxidase-inhibiting compounds in alfalfa using comparative metabolomics. Food Res. Int.141, 110170. 10.1016/j.foodres.2021.110170
49
Hu Y. K. Ma C. Li M. J. Bai X. L. Liu Y. M. Liao X. (2024). Screening of monoamine oxidase B inhibitors in Tibetan strawberry by ligand fishing based on enzyme functionalized cellulose filter paper. Microchem. J.203, 110838. 10.1016/j.microc.2024.110838
50
Huang C. Xiong X. Zhang D. Ruan Q. Jiang J. Wang F. et al (2024). Targeted screening of multiple anti-inflammatory components from Chrysanthemi indici Flos by ligand fishing with affinity UF-LC/MS. Front. Pharmacol.15, 1272087–1272112. 10.3389/fphar.2024.1272087
51
Jiang X. Yuan Y. Chen L. Liu Y. Xiao M. Hu Y. et al (2019). Monoamine oxidase B immobilized on magnetic nanoparticles for screening of the enzyme’s inhibitors from herbal extracts. Microchem. J.146, 1181–1189. 10.1016/j.microc.2019.02.049
52
Jiao J. Yang Y. Wu Z. Li B. Zheng Q. Wei S. et al (2019). Screening cyclooxygenase-2 inhibitors from Andrographis paniculata to treat inflammation based on bio-affinity ultrafiltration coupled with UPLC-Q-TOF-MS. Fitoterapia137, 104259. 10.1016/j.fitote.2019.104259
53
Jiao Q. Wang R. Jiang Y. Liu B. (2018). Study on the interaction between active components from traditional Chinese medicine and plasma proteins. Chem. Cent. J.12, 48. 10.1186/s13065-018-0417-2
54
Jonker N. Kool J. Irth H. Niessen W. M. A. (2011). Recent developments in protein-ligand affinity mass spectrometry. Anal. Bioanal. Chem.399, 2669–2681. 10.1007/s00216-010-4350-z
55
Kang M. J. Wu T. Wijeratne E. M. K. Lau E. C. Mason D. J. Mesa C. et al (2014). Functional chromatography reveals three natural products that target the same protein with distinct mechanisms of action. ChemBioChem15, 2125–2131. 10.1002/cbic.201402258
56
Kaur S. McGuire L. Tang D. Dollinger G. Huebner V. (1997). Affinity selection and mass spectrometry-based strategies to identify lead compounds in combinatorial libraries. J. Protein Chem.16, 505–511. 10.1023/A:1026369729393
57
Li H. Song F. Xing J. Tsao R. Liu Z. Liu S. (2009). Screening and structural characterization of α-glucosidase inhibitors from hawthorn leaf flavonoids extract by ultrafiltration LC-DAD-MSn and SORI-CID FTICR MS. J. Am. Soc. Mass Spectrom.20, 1496–1503. 10.1016/j.jasms.2009.04.003
58
Li S. Li S. Tang Y. Liu C. Chen L. Zhang Y. (2016). Ultrafiltration-LC–MS combined with semi-preparative HPLC for the simultaneous screening and isolation of lactate dehydrogenase inhibitors from Belamcanda chinensis. J. Sep. Sci.39, 4533–4543. 10.1002/jssc.201600703
59
Li S. Zhang Y. Shi D. Hou W. Xia J. Liu C. (2019). Screening and isolation of cyclooxygenase-2 inhibitors from the stem bark of Phellodendron amurense Ruprecht by ultrafiltration with liquid chromatography and tandem mass spectrometry, and complex chromatography. J. Sep. Sci.42, 1905–1914. 10.1002/jssc.201801262
60
Li Y. J. He F. Q. Zhao H. H. Li Y. Chen J. (2022). Screening and identification of acetylcholinesterase inhibitors from Terminalia chebula fruits by immobilized enzyme on cellulose filter paper coupled with ultra-performance liquid chromatography-quadrupole time-of-flight mass spectrometry and molecular docking. J. Chromatogr. A1663, 462784. 10.1016/j.chroma.2021.462784
61
Liang L. Li F. J. Liu X. Mu J. K. Wang X. Dong J. C. et al (2019). Identification of mitochondrial ligands with hepatoprotective activity from Notopterygii Rhizoma et Radix using affinity ultrafiltration/liquid chromatography/mass spectrometry. Biomed. Res. Int.2019, 1–9. 10.1155/2019/5729263
62
Lima J. M. Leme G. M. Costa E. V. Cass Q. B. (2021). LC-HRMS and acetylcholinesterase affinity assay as a workflow for profiling alkaloids in Annona salzmannii extract. J. Chromatogr. B1164, 122493. 10.1016/j.jchromb.2020.122493
63
Liu H. Wang J. Yang S. Li Z. Song M. Zhang X. et al (2024). A magnetic beads-based ligand fishing method for rapid discovery of monoterpene indoles as monoamine oxidase A inhibitors from Hunteria zeylanica. J. Chromatogr. A1722, 464896. 10.1016/j.chroma.2024.464896
64
Liu L. Xiao A. Ma L. Li D. (2017). Analysis of xanthine oxidase inhibitors from Puerariase flos using centrifugal ultrafiltration coupled with HPLC-MS. J. Braz. Chem. Soc.28, 360–366. 10.5935/0103-5053.20160185
65
Ludwig M. Nothias L.-F. Dührkop K. Koester I. Fleischauer M. Hoffmann M. A. et al (2020). Database-independent molecular formula annotation using Gibbs sampling through ZODIAC. Nat. Mach. Intell.2, 629–641. 10.1038/s42256-020-00234-6
66
Luo H. Qian Y. Qi J. Liu X. (2022). A novel strategy for screening angiotensin-converting enzyme inhibitors from natural products based on enzyme-immobilized ligand fishing combined with active-site blocking and directional enrichment. J. Chromatogr. B Anal. Technol. Biomed. Life Sci.1195, 123203. 10.1016/j.jchromb.2022.123203
67
Lutz H. (2015). “Fundamentals,” in Ultrafiltration for bioprocessing. Editor LutzH. (Cambridge, UK: Woodhead Publishing), 1–6. 10.1016/B978-1-907568-46-6.00001-X
68
Ma Z. Qin Y. Wang X. Zhang G. Zhang X. Jiang H. et al (2024). Identification of chemical compounds of Schizonepeta tenuifolia Briq. and screening of neuraminidase inhibitors based on AUF-MS and SPR technology. J. Pharm. Biomed. Anal.237, 115787. 10.1016/j.jpba.2023.115787
69
Mak T. Rossjohn J. Littler D. R. Liu M. Quinn R. J. (2022). Collision-induced affinity selection mass spectrometry for identification of ligands. ACS Bio Med Chem Au2, 450–455. 10.1021/acsbiomedchemau.2c00021
70
Mateo C. Palomo J. M. Fernandez-Lorente G. Guisan J. M. Fernandez-Lafuente R. (2007). Improvement of enzyme activity, stability and selectivity via immobilization techniques. Enzyme Microb. Technol.40, 1451–1463. 10.1016/j.enzmictec.2007.01.018
71
McLaren D. G. Shah V. Wisniewski T. Ghislain L. Liu C. Zhang H. et al (2021). High-throughput mass spectrometry for hit identification: current landscape and future perspectives. SLAS Discov.26, 168–191. 10.1177/2472555220980696
72
Moaddel R. Marszałł M. P. Bighi F. Yang Q. Duan X. Wainer I. W. (2007). Automated ligand fishing using human serum albumin-coated magnetic beads. Anal. Chem.79, 5414–5417. 10.1021/ac070268+
73
Mohimani H. Gurevich A. Shlemov A. Mikheenko A. Korobeynikov A. Cao L. et al (2018). Dereplication of microbial metabolites through database search of mass spectra. Nat. Commun.9, 4035. 10.1038/s41467-018-06082-8
74
Muchiri R. N. Kibitel J. Redick M. A. van Breemen R. B. (2022). Advances in magnetic microbead affinity selection screening: discovery of natural ligands to the SARS-CoV-2 spike protein. J. Am. Soc. Mass Spectrom.33, 181–188. 10.1021/jasms.1c00318
75
Muchiri R. N. van Breemen R. B. (2021a). Affinity selection–mass spectrometry for the discovery of pharmacologically active compounds from combinatorial libraries and natural products. J. Mass Spectrom.56, e4647. 10.1002/jms.4647
76
Muchiri R. N. van Breemen R. B. (2021b). Drug discovery from natural products using affinity selection-mass spectrometry. Drug Discov. Today Technol.40, 59–63. 10.1016/j.ddtec.2021.10.005
77
Munigunti R. Mulabagal V. Calderón A. I. (2011). Screening of natural compounds for ligands to PfTrxR by ultrafiltration and LC–MS based binding assay. J. Pharm. Biomed. Anal.55, 265–271. 10.1016/j.jpba.2011.01.033
78
Newman D. J. Cragg G. M. (2020). Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod.83, 770–803. 10.1021/acs.jnatprod.9b01285
79
NIAID Visual and Medical Arts (2024). BIOART - category: equipment (vaccine vial, pipette, eppendorf tube, LCMS and female scientist pipetting). Available online at: https://bioart.niaid.nih.gov/discover?bioartCategory=500043 (Accessed January 13, 2025).
80
Nothias L. F. Nothias-Esposito M. Da Silva R. Wang M. Protsyuk I. Zhang Z. et al (2018). Bioactivity-based molecular networking for the discovery of drug leads in natural product bioassay-guided fractionation. J. Nat. Prod.81, 758–767. 10.1021/acs.jnatprod.7b00737
81
Nothias L.-F. Petras D. Schmid R. Dührkop K. Rainer J. Sarvepalli A. et al (2020). Feature-based molecular networking in the GNPS analysis environment. Nat. Methods17, 905–908. 10.1038/s41592-020-0933-6
82
O’Connell T. N. Ramsay J. Rieth S. F. Shapiro M. J. Stroh J. G. (2014). Solution-based indirect affinity selection mass spectrometry—a general tool for high-throughput screening of pharmaceutical compound libraries. Anal. Chem.86, 7413–7420. 10.1021/ac500938y
83
Ozawa M. Ozawa T. Nishio M. Ueda K. (2017). The role of CH/π interactions in the high affinity binding of streptavidin and biotin. J. Mol. Graph Model75, 117–124. 10.1016/j.jmgm.2017.05.002
84
Pakkir Shah A. K. Walter A. Ottosson F. Russo F. Navarro-Diaz M. Boldt J. et al (2024). Statistical analysis of feature-based molecular networking results from non-targeted metabolomics data. Nat. Protoc.20, 92–162. 10.1038/s41596-024-01046-3
85
Prudent R. Annis D. A. Dandliker P. J. Ortholand J. Y. Roche D. (2021). Exploring new targets and chemical space with affinity selection-mass spectrometry. Nat. Rev. Chem.5, 62–71. 10.1038/s41570-020-00229-2
86
Prudent R. Lemoine H. Walsh J. Roche D. (2023). Affinity selection mass spectrometry speeding drug discovery. Drug Discov. Today28, 103760. 10.1016/j.drudis.2023.103760
87
Qin Y. Yan Y. Zhou J. Gao X. Zhao L. Guan X. (2024). Advancements in screening systems for active ingredients in traditional Chinese medicines using ligand fishing technique. TrAC Trends Anal. Chem.180, 117983. 10.1016/j.trac.2024.117983
88
Quan S. Wen M. Xu P. Chu C. Zhang H. Yang K. et al (2024). Efficient screening of pancreatic lipase inhibitors from Rheum palmatum by affinity ultrafiltration–high-performance liquid chromatography combined with high-resolution inhibition profiling. Phytochem. Anal.35, 540–551. 10.1002/pca.3311
89
Quiros-Guerrero L. M. Marcourt L. Chaiwangrach N. Koval A. Ferreira Queiroz E. David B. et al (2024). Integration of Wnt-inhibitory activity and structural novelty scoring results to uncover novel bioactive natural products: new bicyclo[3.3.1]non-3-ene-2,9-diones from the leaves of Hymenocardia punctata. Front. Chem.12, 1371982. 10.3389/fchem.2024.1371982
90
Reher R. Aron A. T. Fajtová P. Stincone P. Wagner B. Pérez-Lorente A. I. et al (2022). Native metabolomics identifies the rivulariapeptolide family of protease inhibitors. Nat. Commun.13, 4619. 10.1038/s41467-022-32016-6
91
Rush M. D. Walker E. M. Burton T. Van Breemen R. B. (2016). Magnetic microbead affinity selection screening (MagMASS) of botanical extracts for inhibitors of 15-lipoxygenase. J. Nat. Prod.79, 2898–2902. 10.1021/acs.jnatprod.6b00693
92
Safarik I. Safarikova M. (2004). Magnetic techniques for the isolation and purification of proteins and peptides. Biomagn. Res. Technol.2, 7. 10.1186/1477-044X-2-7
93
Shi S. Y. Peng M. J. Zhang Y. P. Peng S. (2013). Combination of preparative HPLC and HSCCC methods to separate phosphodiesterase inhibitors from Eucommia ulmoides bark guided by ultrafiltration-based ligand screening. Anal. Bioanal. Chem.405, 4213–4223. 10.1007/s00216-013-6806-4
94
Silva D. G. Emery F. da S. (2018). Strategies towards expansion of chemical space of natural product-based compounds to enable drug discovery. Braz. J. Pharm. Sci.54, 1–16. 10.1590/s2175-97902018000001004
95
Štulík K. Pacáková V. Tichá M. (2003). Some potentialities and drawbacks of contemporary size-exclusion chromatography. J. Biochem. Biophys. Methods56, 1–13. 10.1016/S0165-022X(03)00053-8
96
Tian Z. Sun L. Chi B. Du Z. Zhang X. Liu Y. et al (2022). Affinity ultrafiltration and UPLC-HR-Orbitrap-MS based screening of neuraminidase inhibitors from Angelica pubescens. J. Chromatogr. B Anal. Technol. Biomed. Life Sci.1208, 123398. 10.1016/j.jchromb.2022.123398
97
Trindade Ximenes I. A. de Oliveira P. C. O. Wegermann C. A. de Moraes M. C. (2021). Magnetic particles for enzyme immobilization: a versatile support for ligand screening. J. Pharm. Biomed. Anal.204, 114286. 10.1016/j.jpba.2021.114286
98
van Breemen R. B. Huang C. R. Nikolic D. Woodbury C. P. Zhao Y. Z. Venton D. L. (1997). Pulsed ultrafiltration mass spectrometry: a new method for screening combinatorial libraries. Anal. Chem.69, 2159–2164. 10.1021/ac970132j
99
van Breemen R. B. Muchiri R. N. (2024). Affinity selection-mass spectrometry in the discovery of anti-SARS-CoV-2 compounds. Mass Spectrom. Rev.43, 39–46. 10.1002/mas.21800
100
van Breemen R. B. Muchiri R. N. Bates T. A. Weinstein J. B. Leier H. C. Farley S. et al (2022). Cannabinoids block cellular entry of SARS-CoV-2 and the emerging variants. J. Nat. Prod.85, 176–184. 10.1021/acs.jnatprod.1c00946
101
van Breemen R. B. Tao Y. Li W. (2011). Cyclooxygenase-2 inhibitors in ginger (Zingiber officinale). Fitoterapia82, 38–43. 10.1016/j.fitote.2010.09.004
102
Vanzolini K. L. Ainsworth S. Bruyneel B. Herzig V. Seraus M. G. L. Somsen G. W. et al (2018a). Rapid ligand fishing for identification of acetylcholinesterase-binding peptides in snake venom reveals new properties of dendrotoxins. Toxicon152, 1–8. 10.1016/j.toxicon.2018.06.080
103
Vanzolini K. L. da R. Leme G. M. de V. R. Vilela A. F. L. Cardoso C. L. et al (2018b). Acetylcholinesterase affinity-based screening assay on Lippia gracilis Schauer extracts. J. Pharm. Biomed. Anal.153, 232–237. 10.1016/j.jpba.2018.02.035
104
Vanzolini K. L. Jiang Z. Zhang X. Vieira L. C. C. Corrêa A. G. Cardoso C. L. et al (2013). Acetylcholinesterase immobilized capillary reactors coupled to protein coated magnetic beads: a new tool for plant extract ligand screening. Talanta116, 647–652. 10.1016/j.talanta.2013.07.046
105
Vanzolini K. L. Vieira L. C. C. Corrêa A. G. Moaddel R. Cass Q. B. (2015). Acetylcholinesterase immobilized on modified magnetic beads as a tool for screening a compound library. Microchim. Acta182, 2209–2213. 10.1007/s00604-015-1562-0
106
Vu H. Pedro L. Mak T. McCormick B. Rowley J. Liu M. et al (2018). Fragment-based screening of a natural product library against 62 potential malaria drug targets employing native mass spectrometry. ACS Infect. Dis.4, 431–444. 10.1021/acsinfecdis.7b00197
107
Wandy J. Zhu Y. van der Hooft J. J. J. Daly R. Barrett M. P. Rogers S. (2018). Ms2lda.org: web-based topic modelling for substructure discovery in mass spectrometry. Bioinformatics34, 317–318. 10.1093/bioinformatics/btx582
108
Wang H. P. Lin Z. Z. Yin Q. Du J. (2024). Screening of GLP-1r agonists from natural products using affinity ultrafiltration screening coupled with UPLC-ESI-Orbitrap-MS technology: a case study of Panax ginseng. J. Asian Nat. Prod. Res.0, 176–188. 10.1080/10286020.2024.2378821
109
Wang J. Liu S. Li S. Song F. Zhang Y. Liu Z. et al (2014). Ultrafiltration LC-PDA-ESI/MS combined with reverse phase-medium pressure liquid chromatography for screening and isolation potential α-glucosidase inhibitors from Scutellaria baicalensis Georgi. Anal. Methods6, 5918–5924. 10.1039/c4ay01077a
110
Wang M. Carver J. J. Phelan V. V. Sanchez L. M. Garg N. Peng Y. et al (2016). Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat. Biotechnol.34, 828–837. 10.1038/nbt.3597
111
Wang M. Jarmusch A. K. Vargas F. Aksenov A. A. Gauglitz J. M. Weldon K. et al (2020). Mass spectrometry searches using MASST. Nat. Biotechnol.38, 23–26. 10.1038/s41587-019-0375-9
112
Wang Y. Hong F. Li D. Qi J. Liu X. (2018). A novel strategy for evaluation of natural products acting on the myeloperoxidase/hypochlorous acid system by combining high-performance liquid chromatography-photodiode array detection-chemiluminescence and ultrafiltration-mass spectrometry techniques. J. Sep. Sci.41, 4222–4232. 10.1002/jssc.201800658
113
Wang Y. Zhou L. Chen M. Liu Y. Yang Y. Lu T. et al (2023). Mining xanthine oxidase inhibitors from an edible seaweed Pterocladiella capillacea by using in vitro bioassays, affinity ultrafiltration LC-MS/MS, metabolomics tools, and in silico prediction. Mar. Drugs21, 502. 10.3390/md21100502
114
Wang Z. Kim U. Liu J. Cheng C. Wu W. Guo S. et al (2019). Comprehensive TCM molecular networking based on MS/MS in silico spectra with integration of virtual screening and affinity MS screening for discovering functional ligands from natural herbs. Anal. Bioanal. Chem.411, 5785–5797. 10.1007/s00216-019-01962-4
115
Wei H. Zhang X. Tian X. Wu G. (2016). Pharmaceutical applications of affinity-ultrafiltration mass spectrometry: recent advances and future prospects. J. Pharm. Biomed. Anal.131, 444–453. 10.1016/j.jpba.2016.09.021
116
Wei Y. Chen T. Song H. Wang S. Shen C. Wang X. et al (2024). Rapidly screening of pancreatic lipase inhibitors from Clematis tangutica using affinity ultrafiltration-HPLC-QTOFMS technique combined with targeted separation, in vitro validation, and molecular docking. Phytochem. Anal., 1–12. 10.1002/pca.3422
117
Wishart D. S. Guo A. Oler E. Wang F. Anjum A. Peters H. et al (2022). HMDB 5.0: the human metabolome database for 2022. Nucleic Acids Res.50, D622–D631. 10.1093/nar/gkab1062
118
Wu G. F. Jiang X. L. Gong Y. Z. Hu Y. D. Bai X. L. Liao X. (2019). Ligand fishing of anti-neurodegenerative components from Lonicera japonica using magnetic nanoparticles immobilised with monoamine oxidase B. J. Sep. Sci.42, 1289–1298. 10.1002/jssc.201801255
119
Xiao S. Yu R. Ai N. Fan X. (2015). Rapid screening natural-origin lipase inhibitors from hypolipidemic decoctions by ultrafiltration combined with liquid chromatography-mass spectrometry. J. Pharm. Biomed. Anal.104, 67–74. 10.1016/j.jpba.2014.11.022
120
Xie L. Lee D. Y. Shang Y. Cao X. Wang S. Liao J. et al (2020). Characterization of spirostanol glycosides and furostanol glycosides from Anemarrhenae Rhizoma as dual targeted inhibitors of 5-lipoxygenase and Cyclooxygenase-2 by employing a combination of affinity ultrafiltration and HPLC/MS. Phytomedicine77, 153284. 10.1016/j.phymed.2020.153284
121
Xu N. Yang H. Cui M. Wan C. Liu S. (2012). High-performance liquid chromatography–electrospray ionization-mass spectrometry ligand fishing assay: a method for screening triplex DNA binders from natural plant extracts. Anal. Chem.84, 2562–2568. 10.1021/ac202796v
122
Yan T. Cao J. Ye L. (2022). Recent advances on discovery of enzyme inhibitors from natural products using bioactivity screening. J. Sep. Sci.45, 2766–2787. 10.1002/jssc.202200084
123
Yang H. Yao W. Wang Y. Shi L. Su R. Wan D. et al (2017). High-throughput screening of triplex DNA binders from complicated samples by 96-well pate format in conjunction with peak area-fading UHPLC-Orbitrap MS. Analyst142, 670–675. 10.1039/C6AN01974A
124
Yonamine D. K. Narciso dos Reis V. E. Feu A. E. de Souza Borges W. Cardoso C. L. Dinamarco T. M. (2023). Ligand fishing approach to explore Amaryllidaceae alkaloids as potential antiviral candidates targeting SARS-CoV-2 Nsp4. J. Pharm. Biomed. Anal.240, 115935. 10.1016/j.jpba.2023.115935
125
Yun H. Liu Z. Hou W. Liu Q. Nong Y. Li S. et al (2024). Rapid screening and isolation of 5-lipoxygenase inhibitors in Inonotus obliquus and mechanism of action in the treatment of asthma. J. Sep. Sci.47, e2300647. 10.1002/jssc.202300647
126
Yurekten O. Payne T. Tejera N. Amaladoss F. X. Martin C. Williams M. et al (2024). MetaboLights: open data repository for metabolomics. Nucleic Acids Res.52, D640–D646. 10.1093/nar/gkad1045
127
Zhang F. Li S. Liu C. Fang K. Jiang Y. Zhang J. et al (2022). Rapid screening for acetylcholinesterase inhibitors in Selaginella doederleinii Hieron by using functionalized magnetic Fe3O4 nanoparticles. Talanta243, 123284. 10.1016/j.talanta.2022.123284
128
Zhang Q. Yang Y. X. Li S. Y. Wang Y. L. Yang F. Q. Chen H. et al (2017). An ultrafiltration and high performance liquid chromatography coupled with diode array detector and mass spectrometry approach for screening and characterizing thrombin inhibitors from Rhizoma Chuanxiong. J. Chromatogr. B Anal. Technol. Biomed. Life Sci.1061–1062, 421–429. 10.1016/j.jchromb.2017.07.050
129
Zhang T. An W. You S. Chen S. Zhang S. (2024). G protein-coupled receptors and traditional Chinese medicine: new thinks for the development of traditional Chinese medicine. Chin. Med.19, 92. 10.1186/s13020-024-00964-4
130
Zhang Y. Wang Q. Liu R. Zhou H. Crommen J. Moaddel R. et al (2019). Rapid screening and identification of monoamine oxidase-A inhibitors from Corydalis Rhizome using enzyme-immobilized magnetic beads based method. J. Chromatogr. A1592, 1–8. 10.1016/j.chroma.2019.01.062
131
Zhao Y. M. Wang L. H. Luo S. F. Wang Q. Q. Moaddel R. Zhang T. T. et al (2018). Magnetic beads-based neuraminidase enzyme microreactor as a drug discovery tool for screening inhibitors from compound libraries and fishing ligands from natural products. J. Chromatogr. A1568, 123–130. 10.1016/j.chroma.2018.07.031
132
Zhu P. Zhou L. Lin Y. Wang Y. Han Y. Cai S. (2024). A magnetic beads-based ligand fishing method coupled with UHPLC-QTOF MS for screening and identification of α-glucosidase inhibitors from Houttuynia cordata Thunb. Talanta270, 125583. 10.1016/j.talanta.2023.125583
133
Zhuang S. Yun H. Zhou X. Li Y. Li S. Liu C. et al (2024). Screening, isolation, and activity evaluation of potential xanthine oxidase inhibitors in Poria Cum Radix Pini and mechanism of action in the treatment of gout disease. J. Sep. Sci.47, e2300505. 10.1002/jssc.202300505
134
Zhuo R. Liu H. Liu N. Wang Y. (2016). Ligand fishing: a remarkable strategy for discovering bioactive compounds from complex mixture of natural products. Molecules21, 1516.
Summary
Keywords
ligand discovery, immobilized targets, solution-based assays, chemical annotation, non-functional assays, molecular library
Citation
do Amaral BS, de Moraes MC, Cardoso CL and Cass QB (2025) Affinity selection mass spectrometry (AS-MS) for prospecting ligands in natural product libraries. Front. Nat. Prod. 4:1562501. doi: 10.3389/fntpr.2025.1562501
Received
17 January 2025
Accepted
05 May 2025
Published
21 May 2025
Corrected
09 July 2025
Volume
4 - 2025
Edited by
Robert Alan Burrow, Universidade Federal De Santa Maria, Brazil
Reviewed by
Afif Felix Monteiro, University of São Paulo, Brazil
Adriana Lopes, University of Ribeirão Preto, Brazil
Updates
Copyright
© 2025 do Amaral, de Moraes, Cardoso and Cass.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Quezia B. Cass, qcass@ufscar.br
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.