Abstract
Macrocyclic compounds have emerged in the 21st century, among which cyclic peptides are of particular interest. Cyanobactins are ribosomally synthesized and post-translationally modified peptides (RiPPs), many of which exist as cyclic peptides with a prenyl moiety, and prenylation can improve their structural stability and biological activity. This mini-review highlights the recently discovered cyanobactins and cyanobactin prenyltransferases from 2021 to 2024. Cyanobactin prenyltransferases will allow access to unique prenylated natural products for applications in drug discovery.
1 Introduction
Ribosomally synthesized and post-translationally modified peptides (RiPPs) are a rapidly growing class of natural products defined by their post-translational modifications, with approximately 20 classes of RiPPs reported in 2013 and 40 classes in 2021 (Arnison et al., 2013; Montalbán-López et al., 2021). Cyanobactins are diverse RiPPs isolated from symbiotic and free-living cyanobacteria and possess diverse biological activities such as antimalarial, cytotoxicity and antimicrobial properties (Martins and Vasconcelos, 2015; Gu et al., 2019). The typical biosynthetic gene clusters of cyanobactin contain seven genes encoding precursor peptide (E protein), subtilisin-like serine protease (A/G protein), unknown function short protein (B/C protein), YcaO cyclodehydratase (D protein) and prenyltransferase (F protein) (Gu et al., 2019). The key features of cyanobactin include macrocyclization, heterocyclization (thiazole, oxazole, thiazoline and oxazoline), and prenylation (Gu et al., 2019). Only a few linear cyanobactins reported (Leikoski et al., 2013), most of them exist in the form of macrocyclic peptides.
Cyanobactin prenyltransferases are ABBA-type prenyltransferases that displace pyrophosphate group in the prenyl donor by a carbon, nitrogen or oxygen atom from the prenylated amino acid (Zheng et al., 2022; Zhang et al., 2023a). Cyanobactin prenyltransferases are highly selective for isoprenyl donors and amino acids involved in prenylation, but are relatively less selective for amino acids in macrocyclic peptides that are not involved in prenylation (Zheng et al., 2022; Zhang et al., 2023a). This feature has attracted a lot of attention from researchers because it can offer a versatile toolkit for peptide prenylation. To date, 14 cyanobactin prenyltransferases have been biochemically characterized, and one cyanobactin prenyltransferase MonF has been characterized based on genome analysis and the identification of its prenylated product, resulting in a total of 15 cyanobactin prenyltransferases (Figure 1A), LynF from Lyngbya aestuarii (McIntosh et al., 2011; McIntosh et al., 2013), PatF from Prochloron didemni (Bent et al., 2013; Tianero et al., 2016), TruF from Lissoclinum patella (Tianero et al., 2016), KgpF from Microcystis aeruginosa NIES-88 (Parajuli et al., 2016; Inoue et al., 2024), PagF from Oscillatoria agardhii (Hao et al., 2016), AgeMTPT from M. aeruginosa PCC 9432 (Sardar et al., 2017), PirF from M. aeruginosa PCC 7005 (Estrada et al., 2018; Morita et al., 2018), SphF from Sphaerospermopsis sp. LEGE 00249 (Martins et al., 2018), AcyF from Anabaena sp. UHCC-0232 (Dalponte et al., 2018), MusF1/2 from Nostoc spp. PCC 7906 and UHCC 0398 (Mattila et al., 2019), TolF from Tolypothrix sp. PCC 7601 (Purushothaman et al., 2021), AgcF from M. aeruginosa NIES-88 (Phan et al., 2021), AutF from Phormidium autumnale CCAP1446/10 (Clemente et al., 2022), LimF from Limnothrix sp. CACIAM 69days (Zhang et al., 2022), and MonF from Microcoleaceae cyanobacterium LEGE 16532 (Castelo-Branco et al., 2025). The cyanobactin prenyltransferases including (1) biosynthesis of cyanobactins and (2) discovery, biochemical characterization and bioengineering of cyanobactin prenyltransferases have been extensively reviewed elsewhere (Zheng et al., 2022; Zhang et al., 2023a). While this mini-review highlights the recently discovered prenylated cyanobactins and cyanobactin prenyltransferases from 2021 to 2024.
FIGURE 1
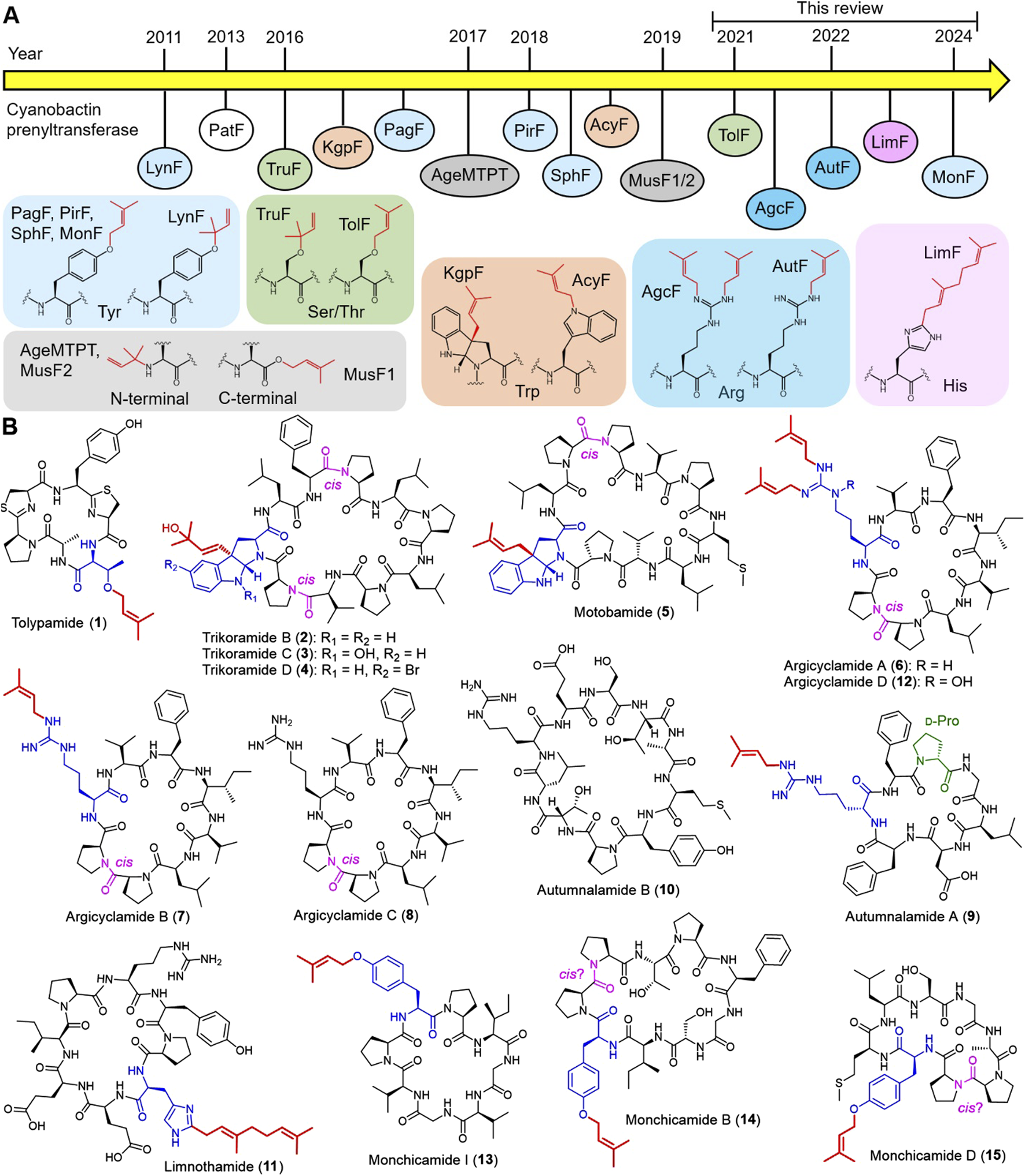
(A) Discovery timeline of cyanobactin prenyltransferases and their chemical transformations. (B) Structures of cyanobactins from 2021 to 2024. Prenyl groups are colored in red. Prenylated residues are colored in blue. cis amide bonds between Pro/Pro and Pro/X residues are colored in purple. d-amino acids are colored in green.
2 Discovery of cyanobactins and cyanobactin prenyltransferases from 2021 to 2024
A review published a decade ago reported a total of 57 cyanobactins (Martins and Vasconcelos, 2015), this number has not been updated since then, but a rough estimate is between 80 and 100 cyanobactins today. In 2021, a genome mining approach was used to prioritize cyanobacterial strains containing cyanobactin prenyltransferase from uncharacterized cluster in sequence-function space. This led to the isolation of tolypamide (1) and biochemical characterization of cyanobactin prenyltransferase TolF from Tolypothrix sp. PCC 7601 (Figure 1B) (Purushothaman et al., 2021). Tolypamide (1) showed no activity against six cancer cell lines (DU145, A549, HeLa CCL2, HepG2, and MDA-MB 231) or three bacterial strains (Escherichia coli ATCC 25922, Staphylococcus aureus ATCC 29737 and Pseudomonas aeruginosa ATCC 9027). The substrate scope of TolF showed a certain degree of tolerant towards non-native peptide substrates of different lengths, sequence compositions and ring sizes. In the same year, two natural products studies reported cyanobactin-like structures without identifying their biosynthetic gene clusters. Three Trp-prenylated cyanobactins, trikoramides B-D (2–4) were isolated from Symploca hydnoides collected at Bintan Island, Indonesia (Phyo et al., 2021). Trikoramides B-D (2–4) possessed unique structures with additional hydroxylation or/and bromination on prenylated Trp residue (Figure 1B), but the enzymes involved in this chemical transformation are remains unknown. Trikoramides B (2) and D (4) showed cytotoxicity against acute lymphoblastic leukemia cell line (MOLT-4) with IC50 5.2 and 4.7 µM, respectively. One Trp-prenylated cyanobactin, motobamide (5) was isolated from Leptolyngbya sp. collected at Okinawa, Japan (Takahashi et al., 2021). In cyclic peptides, the peptide bond geometry between two adjacent Pro residues was orientated in a cis conformation (Figure 1B), as observed in motobamide (5) where the cis amide bond was determined based on the (1) 13C nuclear magnetic resonance (NMR) chemical shift differences between the Cβ and Cγ positions, and (2) NOESY correlations. Interestingly, two non-adjacent Pro residues in trikoramides (2–4) were assigned as cis in a similar manner. Motobamide (5) showed inhibitory activity against Trypanosoma brucei rhodesiense at IC50 2.3 μM.
In the same year, LC-MS approach was used to search for prenylated cyanobactins by targeting a mass difference of 68 Da (isoprene), as this represented the mass difference between non-prenylated and prenylated cyanobactins. This led to the discovery of bis-prenylated, mono-prenylated and non-prenylated cyanobactins, argicyclamides A-C (6–8) and biochemical characterization of cyanobactin prenyltransferase AgcF from Microcystis aeruginosa NIES-88 (Figure 1B) (Phan et al., 2021). Previously, a cyanobactin prenyltransferase KgpF involved in the biosynthesis of Trp-prenylated cyanobactin, kawaguchipeptin A was characterized from the same strain (Parajuli et al., 2016). However, the putative precursor peptide AgcE was not found in the genome of M. aeruginosa NIES-88 (ASM157807v1), although the strain could produce both argicyclamides and kawaguchipeptins. Combining long-read and short-read re-sequencing of M. aeruginosa NIES-88 (ASM1970427v1) revealed argicyclamide biosynthetic gene clusters. Argicyclamides A-C (6–8) showed no activity against two cancer cell lines (P388 and MCF-7) but interestingly, their antibacterial activity was significantly improved based on the number of prenyl groups at Arg residue, argicyclamide A (6) has a MIC of 3.12–6.25 µM against S. aureus ATCC 12600, methicillin-resistant S. aureus ATCC 43300 and Bacillus subtilis ATCC 6051. For the substrate scope study of AgcF, several non-native peptide substrates were designed by exchanging the prenylated residue Arg to Trp, Tyr, Ser, Thr and Lys, but no prenylation activity was detected, indicating that AgcF is selective for Arg prenylation. A year later, a study involved solving the structure of the enzyme-substrate complex of LimF proposed that His167 in AgcF correlated to His172 in LimF is the catalytic residue for Arg-Nω prenylation (Zhang et al., 2022). The discovery of cyanobactin prenyltransferase AgcF expands the biocatalytic toolbox of this protein family, enabling them to catalyze prenylation on the amino acid with charged side chain. Prior to the discovery of AgcF, prenylation of this protein family are restricted to only amino acids with hydrophobic (Tyr and Trp) and uncharged side chains (Ser and Thr). The patent application on AgcF has been published (PCT/JP 2022/004501).
In 2022, cyanobactin prenyltransferase AutF was biochemical characterized from P. autumnale CCAP1446/10 (Clemente et al., 2022), a producer of autumnalamides A and B (9 and 10) (Figure 1B) (Sánchez et al., 2017). In the same year, a genome mining approach was used to explore sequence-function space of cyanobactin prenyltransferases and found an uncharacterized cyanobactin prenyltransferase LimF from Limnothrix sp. CACIAM 69d (Zhang et al., 2022). The NMR characterization of the in vitro generated product, limnothamide (11) confirmed a His-prenylated cyanobactin (Figure 1B). Limnothrix sp. CACIAM 69d was not used for the isolation of limnothamide (11). Among the cyanobactin prenyltransferases discovered between 2021 and 2024, LimF has the greatest potential to be developed into a biocatalyst, where LimF (1) can prenylate the His residue of any non-native peptide substrates, regardless of their length, overall sequence composition, and ring size; (2) has secondary function to prenylated Tyr residue that has not been seen in other cyanobactin prenyltransferases; (3) crystal structure in complex with substrate have been solved, PDB 7VMW and 7VMY; (4) key catalytic residues have been identified; and (5) can prenylated FDA-approved His-containing peptide/non-peptide drugs such as leuprorelin, pramlintide and cimetidine. The patent application on LimF has been published (PCT/JP 2022/038924).
In 2023, a re-visit of M. aeruginosa NIES-88 and found argicyclamide D (12) from the culture condition at 37°C without ammonium ferric citrate (iron source) supplemented in BG-11 media (Figure 1B) (Ballo et al., 2023), while argicyclamides A-C (6–8) were previously isolated from the culture condition at 25°C with ammonium ferric citrate supplemented in BG-11 media.
In 2024, a genome mining approach was used to prioritize cyanobacterial strains containing cyanobactin prenyltransferase from uncharacterized cluster in sequence-function space. This led to the detection of 11 cyanobactins, monchicamides A-K, and identification of cyanobactin prenyltransferase MonF from Microcoleaceae cyanobacterium LEGE 16532 (Castelo-Branco et al., 2025). Only monchicamide I (13) was isolated and characterized by NMR, whereas the structures of the other cyanobactins were proposed based on LC-MS/MS data, which revealed that monchicamides B (14), D (15), F, G and K were prenylated cyanobactins. Interestingly, a trans amide bond between two adjacent Pro residues was proposed in the cyclic peptides of monchicamides B and D (14 and 15) (Figure 1B). However, if monchicamides B and D (14 and 15) were measured by NMR, the peptide bond between two adjacent Pro residues is most likely a cis geometry, and many similar cis amide bonds have been reported between Pro/Pro residues in cyclic peptides, such as 5-7, noducyclamides and phakellistatins (Kwon et al., 2018; Phan et al., 2021; Takahashi et al., 2021; Mehjabin et al., 2024). Monchicamide I (13) showed no activity against three cancer cell lines (HepG2, HCT 116 and SH-SY5Y), four bacterial strains (S. aureus ATCC 29213, B. subtilis ATCC 6633, E. coli ATCC 25922 and Salmonela typhimurium ATCC 25241), one yeast (Candida albicans ATCC 10231) and three amoeba strains (Acanthamoeba castellanii, Acanthamoeba polyphaga and Dictyostelium discoideum).
3 Protein engineering of cyanobactin prenyltransferases from 2021 to 2024
Rational engineering of enzyme complexes in nonribosomal peptide synthetases (NRPS) and polyketide synthases (PKS) is a next-generation technology for natural products or small molecule drug discovery (Bozhüyük et al., 2024; Mabesoone et al., 2024). Many engineering studies in RiPPs have focused on the precursor peptides (Goto and Suga, 2018; Do and Link, 2023; Zhong et al., 2022). A relatively few engineering studies focused on the post-translational modification enzymes to alter or expand enzyme substrate scope (Phan and Morinaka, 2024b; Barrett et al., 2025). Cyanobactin prenyltransferases are known for their strict selectivity for the prenyl donors, with LimF and PirF only accept the GPP (C10, geranyl pyrophosphate) but not the DMAPP (C5, dimethylallyl pyrophosphate), while AutF, KgpF, PagF and TolF only accept the DMAPP but not the GPP, but AgcF can accept both the DMAPP and GPP (Figure 2D).
FIGURE 2
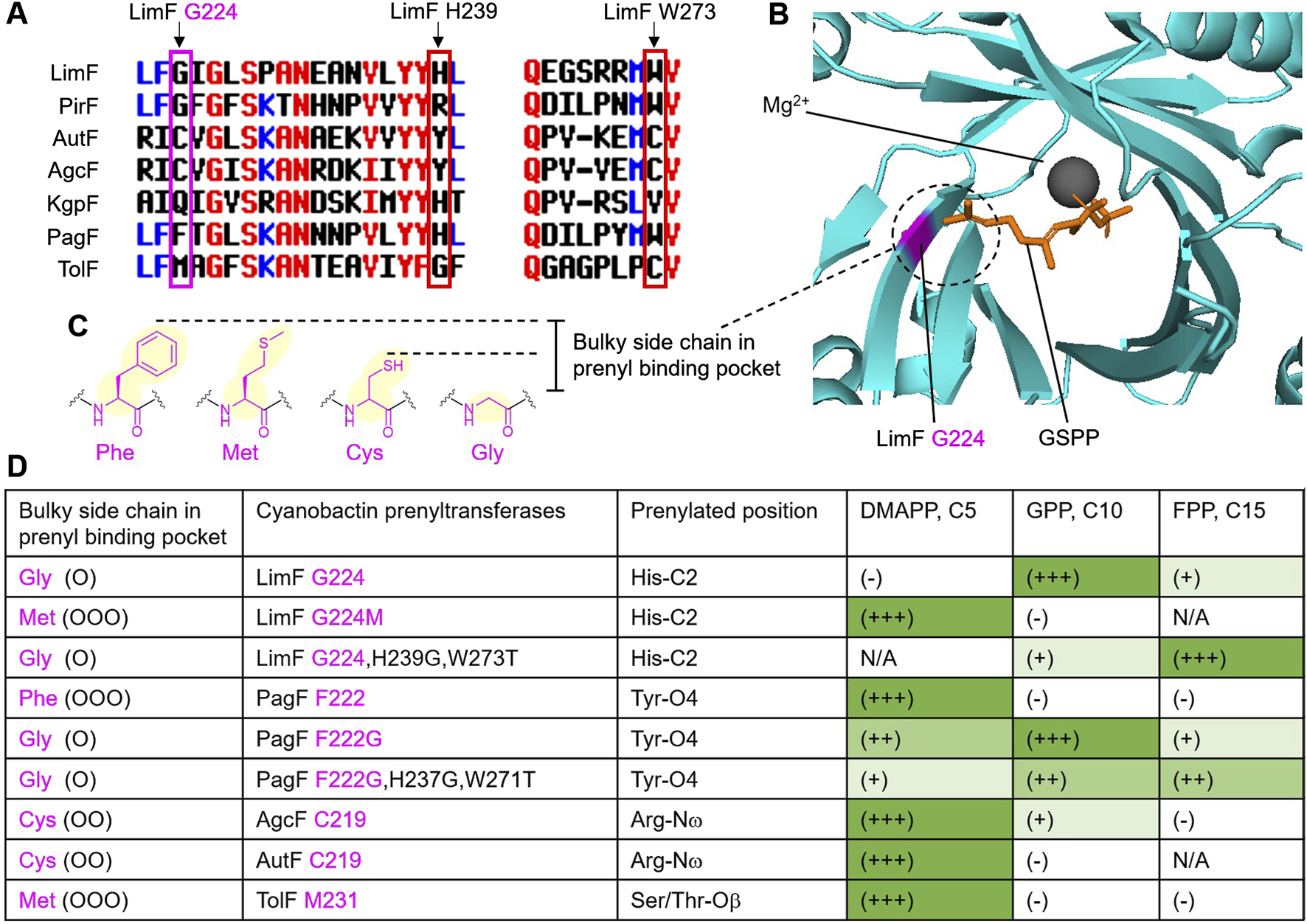
(A) Sequence alignment of representative cyanobactin prenyltransferases, highlighting the active sites correlated to G224, H239 and W273 in LimF. (B) Complex structure of LimF-GSPP (PDB:7VMW), showing the space occupied by G224 in the prenyl binding pocket. (C) Bulky side chain residues that affect pocket sizes are colored in purple. (D) Prenylation activities. High, moderate, and low prenylation activities are indicated as (+++), (++) and (+), respectively. No reactions and not tested are indicated as (−) and N/A, respectively. Large, moderate, and small bulky side chains are indicated as (OOO), (OO) and (O), respectively.
In 2023, a structure-based engineering of the prenyl binding pocket size expansion was achieved in LimF (Zhang et al., 2023b). Three important active sites G224, H239 and W273 in LimF were identified (Figure 2A), particularly G224 located at the apex of the prenyl binding pocket (Figure 2B), which was thought to be the key residue in differentiating the pocket size for preference of C5 or C10 prenyl donors based on the bulky side chain of this amino acid at position 224 (Figure 2C). Mutation of G224 to Met successfully altered the prenyl donor preference from GPP to DMAPP (Figure 2D). Remarkably, the double mutant of LimF H237G,W271T achieved farnesylation accepting FPP (C15, farnesyl pyrophosphate) for the first time in cyanobactin prenyltransferases (Figure 2D). In a previous study, it was reported that the mutant PagF F222G exhibited geranylation activity (Estrada et al., 2018), and only a low farnesylation activity was detected (Zhang et al., 2023b). Interestingly, transfer of these two engineered sites in LimF H237G,W271T to PagF F222G could improve the farnesylation activity (Figure 2D) (Zhang et al., 2023b). In 2024, the mutant LimF I52A based on substrate binding pocket engineering enabled the enzyme to accept substrates with bulky chain side residue Phe preceding to the prenylated residue His, where the wild type substrate has less bulky residue Ala preceding to His (Zhang et al., 2024). Currently, the biological activities of the prenylated products catalyzed by these engineered proteins have not been evaluated. However, these engineering efforts have broadened the chemical space of cyanobactin prenyltransferases and further expanded the biocatalytic toolbox for prenylation.
4 Conclusion
Cyanobactins are a class of natural products that belong to the RiPPs. To date, RiPP is an exciting area of research for the discovery of new chemistry catalyzed by the post-translational modification enzymes (Zhong, 2023; Hubrich et al., 2022; Hubrich et al., 2024; Nguyen et al., 2024; Phan and Morinaka, 2024a; Khan et al., 2025; Kandy et al., 2025; Shi et al., 2025). Although cyanobactin system especially the subtilisin-like serine protease (A/G protein), YcaO cyclodehydratase (D protein) and prenyltransferase (F protein) has been extensively studied, there are still several aspects that can be further explored, for examples (1) unlike the A/G and D proteins, whose mechanisms have been studied (Koehnke et al., 2012; Koehnke et al., 2015; Zheng and Nair, 2023), mechanistic studies of cyanobactin prenyltrasnferases have not been performed; (2) a non-functional cyanobactin prenyltransferase PatF was found in the patellamide gene cluster (Bent et al., 2013), but its role remains unknown; (3) whether more diverse chemistries exist in the sequence-function space of cyanobactin prenyltransferases remains unclear; and (4) another open question is the logic governing the geometry of proline residues in cyclic peptides. While many cyanobactins feature a cis peptide bond between adjacent proline residues, the trikoramides interestingly show a cis configuration between non-adjacent prolines, suggesting the need for further investigation. Cyanobacteria are a rich source of natural products and biosynthetic enzymes (D'Agostino, 2023; Weiss et al., 2025), and cyanobactin prenyltransferases will allow access to unique prenylated natural products for applications in drug discovery.
Statements
Author contributions
AK: Writing – original draft, Writing – review and editing. C-SP: Writing – review and editing, Writing – original draft, Conceptualization.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was funded by EU project No. 101087181 (Natural Products Research at Latvian Institute of Organic Synthesis as a Driver for Excellence in Innovation).
Acknowledgments
We would like to acknowledge (1) Eric W. Schmidt and Brandon I. Morinaka; Toshiyuki Wakimoto and Tatsufumi Okino; David P. Fewer and Wael E. Houssen; Toru Sengoku, Yuki Goto and Hiroaki Suga for the discovery and biochemical characterization of cyanobactin prenyltransferases TolF; AgcF; AutF; LimF, and (2) Lik Tong Tan; Kiyotake Suenaga; Pedro N. Leão for the discovery and isolation of cyanobactins trikoramide; motobamide; monchicamide.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Arnison P. G. Bibb M. J. Bierbaum G. Bowers A. A. Bugni T. S. Bulaj G. et al (2013). Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep.30 (1), 108–160. 10.1039/c2np20085f
2
Ballo N. Mehjabin J. J. Phan C.-S. Okino T. (2023). Heat shock and iron limitation modulate the metabolic profile of the cyanobacterium Microcystis aeruginosa NIES-88. Phycol. Res.71 (4), 200–208. 10.1111/pre.12529
3
Barrett S. E. Yin S. Jordan P. Brunson J. K. Gordon-Nunez J. Costa Machado da Cruz G. et al (2025). Substrate interactions guide cyclase engineering and lasso peptide diversification. Nat. Chem. Biol.21 (3), 412–419. 10.1038/s41589-024-01727-w
4
Bent A. F. Koehnke J. Houssen W. E. Smith M. C. Jaspars M. Naismith J. H. (2013). Structure of PatF from Prochloron didemni. Acta Crystallogr. Sect. F. Struct. Biol. Cryst. Commun.69 (Pt 6), 618–623. 10.1107/S1744309113012931
5
Bozhüyük K. A. J. Präve L. Kegler C. Schenk L. Kaiser S. Schelhas C. et al (2024). Evolution-inspired engineering of nonribosomal peptide synthetases. Science383 (6689), eadg4320. 10.1126/science.adg4320
6
Castelo-Branco R. Pereira J. P. Freitas S. Preto M. Vieira A. R. Morais J. et al (2025). Genome-informed discovery of monchicamides A-K: cyanobactins from the Microcoleaceae cyanobacterium LEGE 16532. J. Nat. Prod.88 (1), 86–93. 10.1021/acs.jnatprod.4c01063
7
Clemente C. Johnson N. Ouyang X. Popin R. V. Dall'Angelo S. Wahlsten M. et al (2022). Biochemical characterization of a cyanobactin arginine-N-prenylase from the autumnalamide biosynthetic pathway. Chem. Commun.58 (86), 12054–12057. 10.1039/d2cc01799g
8
Dalponte L. Parajuli A. Younger E. Mattila A. Jokela J. Wahlsten M. et al (2018). N-prenylation of tryptophan by an aromatic prenyltransferase from the cyanobactin biosynthetic pathway. Biochemistry57 (50), 6860–6867. 10.1021/acs.biochem.8b00879
9
D'Agostino P. M. (2023). Highlights of biosynthetic enzymes and natural products from symbiotic cyanobacteria. Nat. Prod. Rep.40 (11), 1701–1717. 10.1039/d3np00011g
10
Do T. Link A. J. (2023). Protein engineering in ribosomally synthesized and post-translationally modified peptides (RiPPs). Biochemistry62 (2), 201–209. 10.1021/acs.biochem.1c00714
11
Estrada P. Morita M. Hao Y. Schmidt E. W. Nair S. K. (2018). A single amino acid switch alters the isoprene donor specificity in ribosomally synthesized and post-translationally modified peptide prenyltransferases. J. Am. Chem. Soc.140 (26), 8124–8127. 10.1021/jacs.8b05187
12
Goto Y. Suga H. (2018). Engineering of RiPP pathways for the production of artificial peptides bearing various non-proteinogenic structures. Curr. Opin. Chem. Biol.46, 82–90. 10.1016/j.cbpa.2018.06.014
13
Gu W. Dong S. H. Sarkar S. Nair S. K. Schmidt E. W. (2019). The biochemistry and structural biology of cyanobactin pathways: enabling combinatorial biosynthesis. Methods Enzymol.604, 113–163. 10.1016/bs.mie.2018.03.002
14
Hao Y. Pierce E. Roe D. Morita M. McIntosh J. A. Agarwal V. et al (2016). Molecular basis for the broad substrate selectivity of a peptide prenyltransferase. Proc. Natl. Acad. Sci. U. S. A.113 (49), 14037–14042. 10.1073/pnas.1609869113
15
Hubrich F. Bösch N. M. Chepkirui C. Morinaka B. I. Rust M. Gugger M. et al (2022). Ribosomally derived lipopeptides containing distinct fatty acyl moieties. Proc. Natl. Acad. Sci. U. S. A.119 (3), e2113120119. 10.1073/pnas.2113120119
16
Hubrich F. Kandy S. K. Chepkirui C. Padhi C. Mordhorst S. Moosmann P. et al (2024). Ribosomal peptides with polycyclic isoprenoid moieties. Chem10 (10), 3224–3242. 10.1016/j.chempr.2024.07.026
17
Inoue S. Thanh Nguyen D. Hamada K. Okuma R. Okada C. Okada M. et al (2024). De novo discovery of pseudo-natural prenylated macrocyclic peptide ligands. Angew. Chem. Int. Ed.63 (36), e202409973. 10.1002/anie.202409973
18
Kandy S. K. Pasquale M. A. Chekan J. R. (2025). Aromatic side-chain crosslinking in RiPP biosynthesis. Nat. Chem. Biol.21, 168–181. 10.1038/s41589-024-01795-y
19
Khan A. H. Haedar J. R. Zīle A. Phan C.-S. (2025). Radical SAM cyclophane synthase catalyzes uniform cyclophane formation at multiple Trp-motifs. Tetrahedron Chem.14, 100128. 10.1016/j.tchem.2025.100128
20
Koehnke J. Bent A. Houssen W. E. Zollman D. Morawitz F. Shirran S. et al (2012). The mechanism of patellamide macrocyclization revealed by the characterization of the PatG macrocyclase domain. Nat. Struct. Mol. Biol.19 (8), 767–772. 10.1038/nsmb.2340
21
Koehnke J. Mann G. Bent A. F. Ludewig H. Shirran S. Botting C. et al (2015). Structural analysis of leader peptide binding enables leader-free cyanobactin processing. Nat. Chem. Biol.11 (8), 558–563. 10.1038/nchembio.1841
22
Kwon O. S. Kim C. K. Byun W. S. Oh J. Lee Y. J. Lee H. S. et al (2018). Cyclopeptides from the sponge Stylissa flabelliformis. J. Nat. Prod.81 (6), 1426–1434. 10.1021/acs.jnatprod.8b00121
23
Leikoski N. Liu L. Jokela J. Wahlsten M. Gugger M. Calteau A. et al (2013). Genome mining expands the chemical diversity of the cyanobactin family to include highly modified linear peptides. Chem. Biol.20 (8), 1033–1043. 10.1016/j.chembiol.2013.06.015
24
Mabesoone M. F. J. Leopold-Messer S. Minas H. A. Chepkirui C. Chawengrum P. Reiter S. et al (2024). Evolution-guided engineering of trans-acyltransferase polyketide synthases. Science383 (6689), 1312–1317. 10.1126/science.adj7621
25
Martins J. Leikoski N. Wahlsten M. Azevedo J. Antunes J. Jokela J. et al (2018). Sphaerocyclamide, a prenylated cyanobactin from the cyanobacterium Sphaerospermopsis sp. LEGE 00249. Sci. Rep.8 (1), 14537. 10.1038/s41598-018-32618-5
26
Martins J. Vasconcelos V. (2015). Cyanobactins from cyanobacteria: current genetic and chemical state of knowledge. Mar. Drugs13 (11), 6910–6946. 10.3390/md13116910
27
Mattila A. Andsten R. M. Jumppanen M. Assante M. Jokela J. Wahlsten M. et al (2019). Biosynthesis of the bis-prenylated alkaloids muscoride A and B. ACS Chem. Biol.14 (12), 2683–2690. 10.1021/acschembio.9b00620
28
McIntosh J. A. Donia M. S. Nair S. K. Schmidt E. W. (2011). Enzymatic basis of ribosomal peptide prenylation in cyanobacteria. J. Am. Chem. Soc.133 (34), 13698–13705. 10.1021/ja205458h
29
McIntosh J. A. Lin Z. Tianero M. D. Schmidt E. W. (2013). Aestuaramides, a natural library of cyanobactin cyclic peptides resulting from isoprene-derived Claisen rearrangements. ACS Chem. Biol.8 (5), 877–883. 10.1021/cb300614c
30
Mehjabin J. J. Phan C. S. Okino T. (2024). Noducyclamides A1-A4, B1, and B2 from the cyanobacterium nodularia sp. NIES-3585. J. Nat. Prod.87 (4), 984–993. 10.1021/acs.jnatprod.3c01272
31
Montalbán-López M. Scott T. A. Ramesh S. Rahman I. R. van Heel A. J. Viel J. H. et al (2021). New developments in RiPP discovery, enzymology and engineering. Nat. Prod. Rep.38 (1), 130–239. 10.1039/d0np00027b
32
Morita M. Hao Y. Jokela J. K. Sardar D. Lin Z. Sivonen K. et al (2018). Post-translational tyrosine geranylation in cyanobactin biosynthesis. J. Am. Chem. Soc.140 (19), 6044–6048. 10.1021/jacs.8b03137
33
Nguyen D. T. Mitchell D. A. van der Donk W. A. (2024). Genome mining for new enzyme chemistry. ACS Catl14 (7), 4536–4553. 10.1021/acscatal.3c06322
34
Parajuli A. Kwak D. H. Dalponte L. Leikoski N. Galica T. Umeobika U. et al (2016). A unique tryptophan C-prenyltransferase from the kawaguchipeptin biosynthetic pathway. Angew. Chem. Int. Ed.55 (11), 3596–3599. 10.1002/anie.201509920
35
Phan C. S. Matsuda K. Balloo N. Fujita K. Wakimoto T. Okino T. (2021). Argicyclamides A-C unveil enzymatic basis for guanidine bis-prenylation. J. Am. Chem. Soc.143 (27), 10083–10087. 10.1021/jacs.1c05732
36
Phan C. S. Morinaka B. I. (2024a). Bacterial cyclophane-containing RiPPs from radical SAM enzymes. Nat. Prod. Rep.41 (5), 708–720. 10.1039/d3np00030c
37
Phan C. S. Morinaka B. I. (2024b). Sequence-function space of radical SAM cyclophane synthases reveal conserved active site residues that influence substrate specificity. RSC Chem. Biol.5 (12), 1195–1200. 10.1039/d4cb00227j
38
Phyo M. Y. Goh T. M. B. Goh J. X. Tan L. T. (2021). Trikoramides B-D, bioactive cyanobactins from the marine cyanobacterium Symploca hydnoides. Mar. Drugs19 (10), 548. 10.3390/md19100548
39
Purushothaman M. Sarkar S. Morita M. Gugger M. Schmidt E. W. Morinaka B. I. (2021). Genome-mining-based discovery of the cyclic peptide tolypamide and TolF, a ser/thr forward O-prenyltransferase. Angew. Chem. Int. Ed.60 (15), 8460–8465. 10.1002/anie.202015975
40
Sánchez J. A. Alfonso A. Thomas O. P. Botana L. M. (2017). Autumnalamide targeted proteins of the immunophilin family. Immunobiology222 (2), 241–250. 10.1016/j.imbio.2016.09.016
41
Sardar D. Hao Y. Lin Z. Morita M. Nair S. K. Schmidt E. W. (2017). Enzymatic N- and C-protection in cyanobactin RiPP natural products. J. Am. Chem. Soc.139 (8), 2884–2887. 10.1021/jacs.6b12872
42
Shi Y. Xia Y. Gao W. Wang J. Shi B. Wang H. (2025). Enzymatic crosslinking of histidine side chains in peptide natural products. Nat. Prod. Rep.42, 763–773. advance article. 10.1039/D5NP00001G
43
Takahashi H. Iwasaki A. Kurisawa N. Suzuki R. Jeelani G. Matsubara T. et al (2021). Motobamide, an antitrypanosomal cyclic peptide from a Leptolyngbya sp. marine cyanobacterium. J. Nat. Prod.84 (5), 1649–1655. 10.1021/acs.jnatprod.1c00234
44
Tianero M. D. Pierce E. Raghuraman S. Sardar D. McIntosh J. A. Heemstra J. R. et al (2016). Metabolic model for diversity-generating biosynthesis. Proc. Natl. Acad. Sci. U. S. A.113 (7), 1772–1777. 10.1073/pnas.1525438113
45
Weiss M. B. Borges R. M. Sullivan P. Domingues J. P. B. da Silva F. H. S. Trindade V. G. S. et al (2025). Chemical diversity of cyanobacterial natural products. Nat. Prod. Rep.42 (1), 6–49. 10.1039/d4np00040d
46
Zhang Y. Goto Y. Suga H. (2023a). Discovery, biochemical characterization, and bioengineering of cyanobactin prenyltransferases. Trends biochem. Sci.48 (4), 360–374. 10.1016/j.tibs.2022.11.002
47
Zhang Y. Goto Y. Suga H. (2024). Modulating the acceptor preference of His-C-geranyltransferase LimF. Isr. J. Chem.64, e202300182. 10.1002/ijch.202300182
48
Zhang Y. Hamada K. Nguyen D. T. Inoue S. Satake M. Kobayashi S. et al (2022). LimF is a versatile prenyltransferase for histidine-C-geranylation on diverse non-natural substrates. Nat. Catal.5, 682–693. 10.1038/s41929-022-00822-2
49
Zhang Y. Hamada K. Satake M. Sengoku T. Goto Y. Suga H. (2023b). Switching prenyl donor specificities of cyanobactin prenyltransferases. J. Am. Chem. Soc.145 (44), 23893–23898. 10.1021/jacs.3c07373
50
Zheng Y. Cong Y. Schmidt E. W. Nair S. K. (2022). Catalysts for the enzymatic lipidation of peptides. Acc. Chem. Res.55 (9), 1313–1323. 10.1021/acs.accounts.2c00108
51
Zheng Y. Nair S. K. (2023). YcaO-mediated ATP-dependent peptidase activity in ribosomal peptide biosynthesis. Nat. Chem. Biol.19 (1), 111–119. 10.1038/s41589-022-01141-0
52
Zhong G. (2023). Cytochromes P450 associated with the biosyntheses of ribosomally synthesized and post-translationally modified peptides. ACS Bio and Med Chem Au3 (5), 371–388. 10.1021/acsbiomedchemau.3c00026
53
Zhong G. Wang Z. J. Yan F. Zhang Y. Huo L. (2022). Recent advances in discovery, bioengineering, and bioactivity-evaluation of ribosomally synthesized and post-translationally modified peptides. ACS Bio and Med Chem Au3 (1), 1–31. 10.1021/acsbiomedchemau.2c00062
Summary
Keywords
cyanobacterial natural products, cyanobactins, RiPPs, post-translational modification, prenyltransferases
Citation
Khan AH and Phan C-S (2025) Recent advancement of cyanobactins and cyanobactin prenyltransferases from 2021 to 2024. Front. Nat. Prod. 4:1616031. doi: 10.3389/fntpr.2025.1616031
Received
22 April 2025
Accepted
26 May 2025
Published
12 June 2025
Volume
4 - 2025
Edited by
Robert Alan Burrow, Universidade Federal De Santa Maria, Brazil
Reviewed by
Yuxin Fu, Nankai University, China
Updates
Copyright
© 2025 Khan and Phan.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chin-Soon Phan, chinsoon@osi.lv
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.