Abstract
Microbial natural products are central to drug discovery, yet many biosynthetic gene clusters remain transcriptionally silent under standard laboratory conditions. Conventional screening workflows—based on ex situ cultivation and metabolite extraction—can be labor-intensive and often fail to capture ecologically relevant microbial interactions. To address these limitations, we propose the aNP-TRAP (Activity-guided Natural Product Triaging and Recognition Assay Platform), a conceptual, field-deployable device designed to integrate in situ microbial cultivation with functional detection of bioactivity. The system consists of a honeycomb array of cultivation wells, semipermeable and gradient membranes to permit directional metabolite diffusion, and a detection layer containing biosensors responsive to antibacterial, antifungal, or quorum-sensing–inhibitory compounds. Three detection strategies are envisioned: Escherichia coli JW5503-1 with resazurin for antibacterial activity, Candida albicans for antifungal screening, and Chromobacterium violaceum CV026 for quorum-sensing inhibition. Microbial metabolites diffusing through the membranes interact with the biosensor matrices, potentially generating colorimetric or pigment-based signals. This platform is conceptual and currently lacks empirical validation; all performance expectations derive from simulation-based reasoning. In brief, simulations suggested a 0.2 µm membrane equilibrates nutrients within ∼2–6 h, directional metabolite flux achieves >95% reflux suppression within ∼6–10 h, and biosensor responses become detectable within ∼4–10 h at representative inhibitory ranges. Although unvalidated, this integrated configuration may support early-stage triaging of microbial isolates and help guide the discovery of bioactive compounds from under-explored microbial communities. The platform should be viewed as a hypothesis-generating concept rather than a validated tool.
1 Introduction
Environmental microorganisms are an abundant and underexplored source of chemically diverse natural products that have led to life-saving therapeutics (Berdy, 2012; Newman and Cragg, 2020). Yet a substantial fraction of biosynthetic gene clusters (BGCs) remains silent under conventional cultivation owing to the absence of native cues and interactions (Rutledge and Challis, 2015; Ling et al., 2015; Ziemert et al., 2016; Bauman et al., 2021; Alain and Querellou, 2009; Kaeberlein et al., 2002). Traditional ex situ workflows—isolating organisms, cultivating them under artificial conditions, extracting metabolites, and screening—struggle to access this hidden potential and often rediscover known compounds (Berdy, 2012; Nichols et al., 2010; Epstein, 2013).
Recent methodological reviews reinforce the need for approaches that deliberately couple ecological context to functional detection and to distinguish field-deployable concepts from lab-only biosensor formats (e.g., Hossain, 2024). Here we introduce aNP-TRAP as a conceptual (“simulations only”) in situ device that couples cultivation to embedded functional detection, explicitly distinguishing itself from lab-only biosensor and microfluidic formats by prioritizing field-deployable, low-infrastructure operation. Throughout, we emphasize that the present work is hypothetical pending experimental validation.
2 Device concept, workflow, and simulation-based feasibility
2.1 Device concept
The aNP-TRAP platform is conceived as a modular, field-deployable system enabling
in situmicrobial cultivation with simultaneous functional screening of diffusing metabolites. The device comprises three stacked layers and an acrylic base for readout:
i. Cultivation layer: 56 hexagonal wells (17 mm edge-to-edge, 4 mm depth) containing ∼0.75 mL semisolid medium; the top is sealed with a 0.2 µm semipermeable membrane to allow nutrient influx while preventing microbial escape.
ii. Intermediate layer: a gradient-porosity membrane that favors downward metabolite diffusion and resists upward reflux to promote directional mass transfer.
iiii. Detection layer: 56 biosensor matrices responsive to antibacterial, antifungal, or quorum-sensing (QS) inhibitory signals, aligned with the cultivation wells.
iv. Acrylic base: provides visual readout of colorimetric/pigment changes.
Components are held together with lateral bolts and gaskets to ensure structural integrity and sterility; the architecture accommodates sensor materials without compressive stress. While the standard top membrane is 0.2 µm, an optional anisotropic upper film can further restrict outward metabolite escape to enhance signal accumulation. The conceptual architecture and functional layers are illustrated in Figures 1A–C.
FIGURE 1
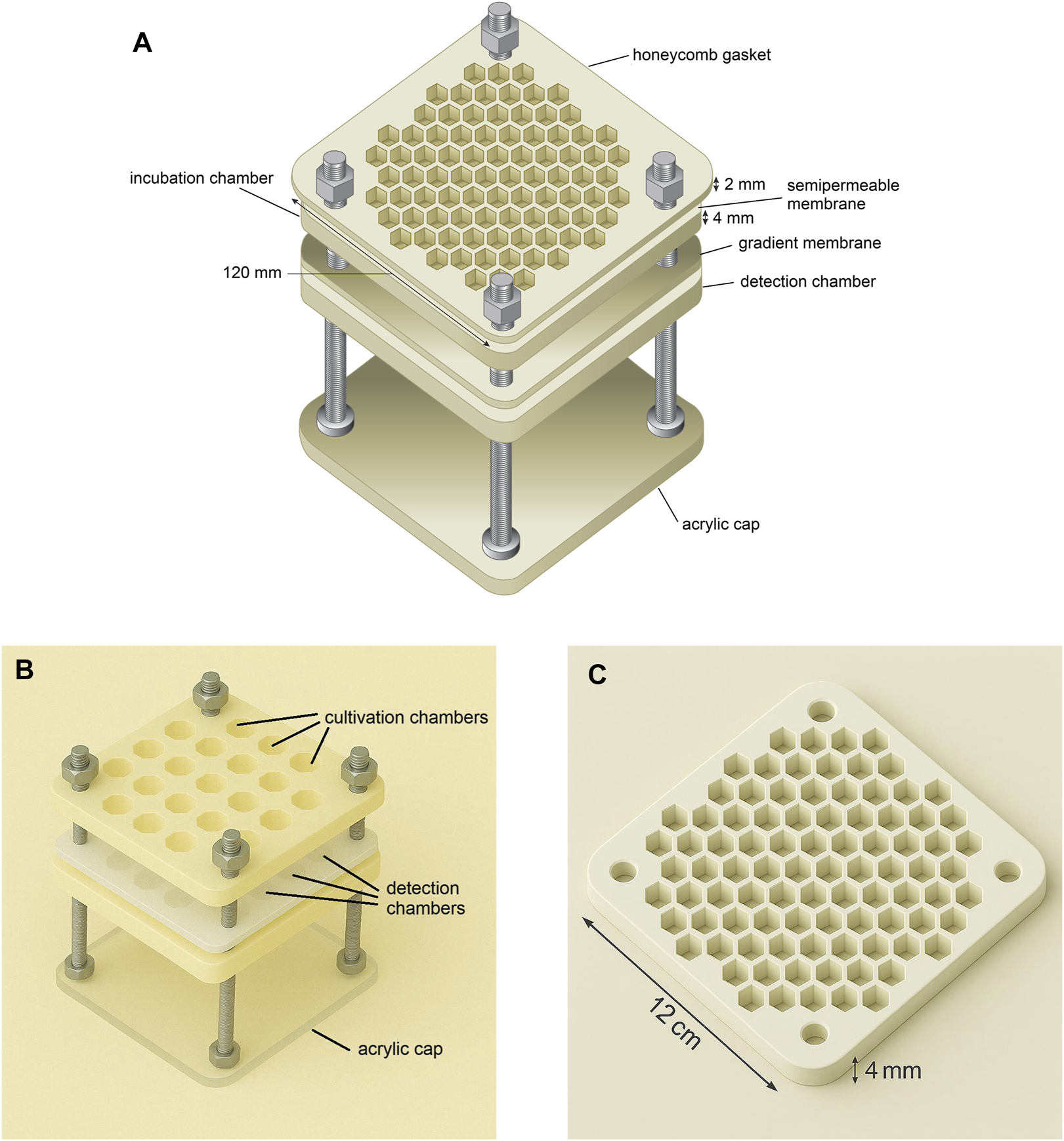
Exploded views of the aNP-TRAP device illustrating its modular configuration and functional components. (A) Conceptual exploded diagram showing the main structural layers of the device, including membranes, cultivation and detection modules, and acrylic base for visual readout. (B) Realistic exploded rendering illustrating the hexagonal cultivation wells, hexagonal biosensor detection layer, and the acrylic base. (C) Detailed view of a single honeycomb layer, highlighting hexagonal chambers that can serve for either microbial incubation or functional detection. Caption placement: per Reviewer #2, the full caption will appear immediately below the figure in the final layout.
2.2 Sample preparation and chamber inoculation
Device assembly is performed under sterile conditions, drawing on precedents such as the iChip and diffusion chambers (Nichols et al., 2010; Berdy et al., 2017; Jung et al., 2020). Environmental samples (e.g., soil, rhizosphere, sediments) are suspended in native moisture or sterile buffer (PBS/Ringer). Preprocessing with mild bead agitation and mesh filtration reduces debris while preserving viability. Wells receive low-nutrient semisolid medium (1.5%–2% agar/gellan, cooled to 40°C–45 °C). Inoculation can target limiting dilutions for clonal isolation or be performed in bulk for consortia studies.
2.3 Device assembly and environmental deployment
After inoculation, the bottom of the cultivation layer is sealed with the gradient-porosity membrane and the top with a 0.2 µm membrane. Compression sealing with gaskets and fasteners yields a robust, portable unit. The device can be embedded 5–10 cm in the target matrix or suspended in water using tethers, enabling passive nutrient/signal exchange under near-native conditions. Alternative pore sizes can accommodate fungi/yeasts (Berdy et al., 2017). A lateral schematic of mass transfer and sensing modalities is provided in Figure 2.
FIGURE 2
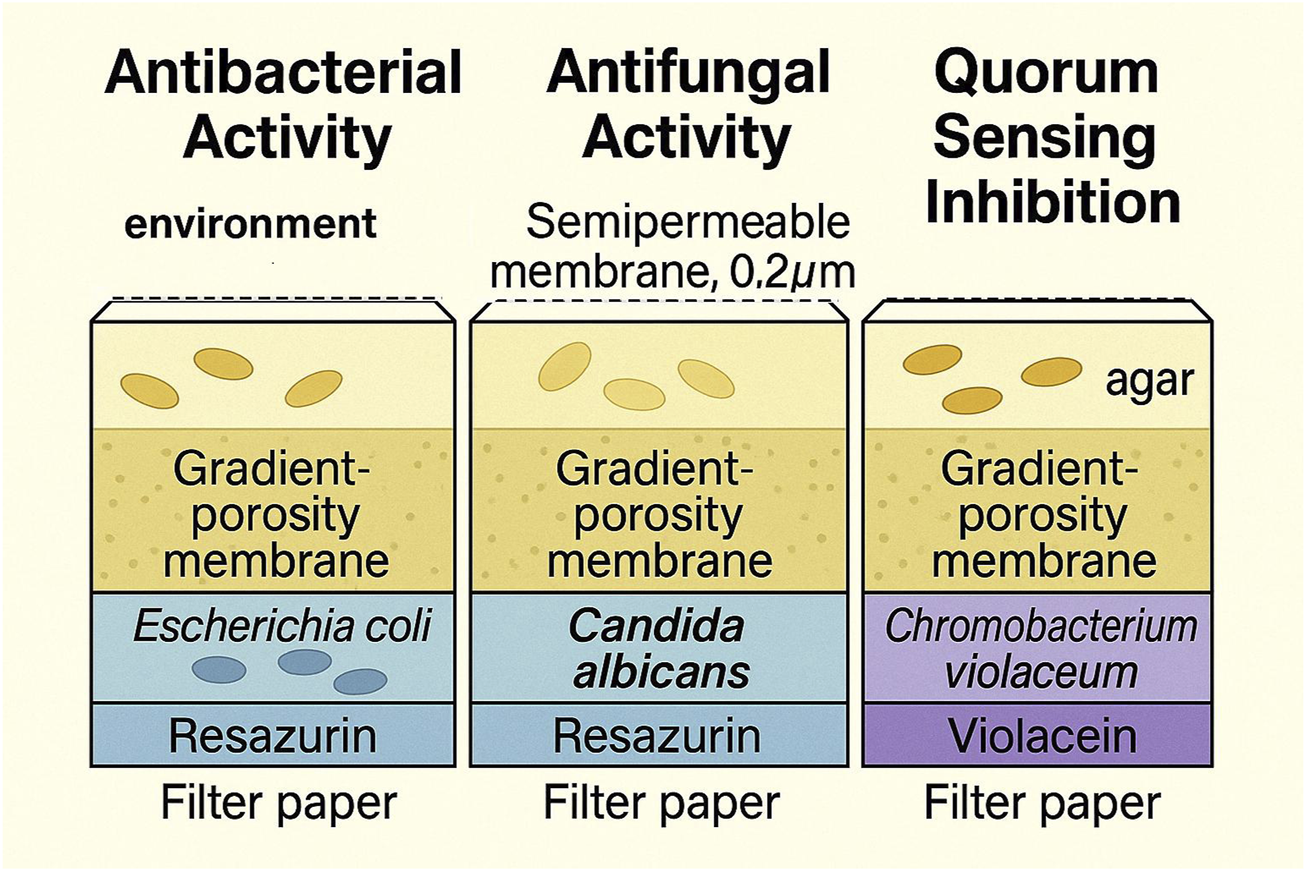
Lateral schematic view of the aNP-TRAP device showing the layered configuration and expected metabolite diffusion starting from the semipermeable membrane, followed by the cultivation chamber, gradient-porosity membrane, and reaching the hexagonal biosensor detection layer. The figure also illustrates the three initially proposed sensing systems: antibacterial (Escherichia coli JW5503-1 with resazurin), antifungal (Candida albicans with resazurin), and quorum-sensing inhibition (Chromobacterium violaceum CV026). Caption placement: per Reviewer #2, the full caption will appear immediately below the figure in the final layout.
2.4 Functional detection modules
The detection layer contains biosensors immobilized in paper/hydrogel/agarose matrices aligned to the wells. Three modalities are envisioned:
Antibacterial: Escherichia coli JW5503-1 + resazurin in hydrogel (PVA or low-melting agarose). Inhibitory activity suppresses metabolic reduction (blue → pink), generating a retained-blue signal (Allen et al., 2022; Sarker et al., 2007).
Antifungal: Candida albicans embedded in redox-sensitive hydrogel with resazurin. Depending on screening goals, alternative fungal sensors (e.g., Saccharomyces cerevisiae, Aspergillus nidulans) may be used.
Quorum-sensing inhibition: Chromobacterium violaceum CV026 in hydrogel with 10–20 µM C6-HSL as inducer; inhibitors suppress violacein pigmentation (McClean et al., 1997; Duddy and Bassler, 2021; Miller and Bassler, 2001). If CV026 stability is suboptimal, wild-type C. violaceum is a viable alternative. The layered configuration and mass flow are depicted in Figure 2.
Viability/specificity in mixed settings: No experiments were performed; feasibility and specificity remain hypothetical and are extrapolated from prior biosensor uses under controlled conditions.
2.5 Incubation and signal monitoring
In situ incubation is expected for 3–10 days under ambient conditions (shorter windows help dye/signal stability (Demir et al., 2024)). Monitoring can be visual or via portable imaging; simple image analysis (e.g., ImageJ) can extract RGB/hue measures. Internal controls include uninoculated wells and calibration wells with known compound concentrations. If weak signals are observed, devices may be redeployed to allow signal maturation and microbial proliferation. Thresholds and quantification steps follow standard colorimetric workflows and are specified in Table 1 (Imaging and quantification).
TABLE 1
| Benchmarking step | Metric/readout | Minimal success criteria | Notes/tools |
|---|---|---|---|
| Benchtop mock-up and sterile hold | Mechanical integrity, leak tests, contamination check | No leaks; sterile hold ≥72 h | Gaskets/bolts; agar plates and sterility swabs |
| Mass-transfer directionality | Tracer flux ratio (downward vs. upward) | ≥90–95% reflux suppression | Use dye or fluorescent tracer; quantify intensity ratio |
| Sensor dose–response | Signal vs. concentration curve | Monotonic response; LOD within intended range | Resazurin or violacein readouts; plate standards |
| Signal stability | Dye/AHL stability over time | ≤20% signal loss over 24–72 h | Vary pH, temperature, and light exposure |
| Cross-sensor specificity | Off-target responses | ≤10–15% unintended cross-response | Compare antibacterial/antifungal/QS assays |
| Pilot environmental deployment | Field signal vs. controls; culture recovery | Reproducible positives with traceable recovery | Barcode/grid map; LC–MS/MS dereplication |
| Imaging and quantification | RGB/hue extraction; thresholding pipeline | Consistent segmentation; SNR >3 | Portable imaging; ImageJ or equivalent |
Proposed early benchmarking steps and minimal success criteria (mock-up integrity and sterile hold; sensor dose-response/LOD; ≥90–95% reflux suppression for tracers; ≤20% signal loss over 24–72 h; ≤10–15% unintended cross-responses; reproducible field signals vs. controls with traceable culture recovery). Includes imaging/quantification thresholds and use of antimicrobial/AHL standards.
2.6 Recovery and characterization of positive cultures
Wells exhibiting reproducible biosensor responses (e.g., retained blue, pigment loss) are prioritized for isolation and scale-up. Downstream characterization includes MALDI-TOF MS, LC-MS/MS, and 16S/ITS sequencing for dereplication and producer prioritization (Ling et al., 2015). Each well’s position is traceable via barcoding or a grid map, and active fractions are dereplicated to prioritize novel candidates. For early validation, we recommend using antimicrobial standards (e.g., ampicillin, nystatin) and AHL analogs to calibrate sensor response functions.
2.7 In silico modeling of diffusion and biosensor response
Clarification on “simulations”: In this manuscript, “simulations” refers to conceptual, order-of-magnitude estimates and internal consistency checks (e.g., dimensional analysis, spreadsheet-level parametric sweeps) derived from literature parameters—not to the execution of numerical solvers (no FEM/CFD, COMSOL®, or MATLAB® runs were performed). Reported values are illustrative and for design guidance only.
To evaluate theoretical feasibility, estimates were structured following the logic typical of COMSOL®/MATLAB® model setups (again, not executed):
Nutrient diffusion: A 0.2 µm polyethersulfone (PES) membrane (∼100 µm thick) with D ≈ 5–7 × 10-6 cm2 s-1 supports equilibration within ∼2–6 h for common nutrients (Stewart, 2003).
Directional transfer: Anisotropic transport (forward D = 5 × 10−7; reverse D = 5 × 10−9 cm2 s-1) accumulates metabolites (e.g., violacein, rifamycin) in the detection zone within ∼6–10 h, with >95% reflux suppression (Zhao et al., 2022; Hou et al., 2019).
Containment: Hydrophobic antibiotics (logP >1; MW > 400 Da) show >98% retention at 24 h; smaller/hydrophilic molecules exhibit polarity/size-dependent back-diffusion.
Sensor kinetics (illustrative): antibacterial—>50% viability-proxy drop in 4–6 h at ≥10 μg mL-1; antifungal—>80% metabolic signal decline within ∼8 h at ∼25 μg mL-1; QS inhibition—∼70% violacein repression within ∼10 h for IC50 ≈ 5–20 μg mL-1.
Table 2 Key modeling assumptions and parameter ranges (membrane pore size/thickness; diffusion coefficients; anisotropy targets; retention criteria by logP/MW; inducer dosing for CV026; time-to-signal windows; field incubation ranges), compiled for transparency and to guide early prototyping.
TABLE 2
| Parameter | References value(s)/range | Notes/assumptions | Representative sources |
|---|---|---|---|
| Top semipermeable membrane pore size | 0.2 µm | Prevents cell egress; permits ingress of nutrients and small metabolites | Nichols et al., 2010; Berdy et al., 2017 |
| Membrane thickness | ∼100 µm (PES) | Used to estimate characteristic diffusion times | Stewart (2003) |
| Diffusion coefficient (small nutrients in membrane) | D ≈ 5–7 × 10−6 cm2⋅s−1 | Basis for equilibration-time estimates across the top membrane | Stewart (2003) |
| Nutrient equilibration time | ∼2–6 h | Order-of-magnitude estimate for small molecules | Derived from parameters above |
| Intermediate anisotropic (gradient-porosity) layer | Forward D ≈ 5 × 10−7; reverse D ≈ 5 × 10−9 cm2⋅s−1 | Target >95% reflux suppression within ∼6–10 h | Hou et al., 2019; Zhao et al., 2022 |
| Downward metabolite accumulation window | ∼6–10 h | Representative for typical antimicrobial/metabolite sizes | Conceptual estimate |
| Containment of hydrophobic antibiotics | >98% retention at 24 h for logP >1; MW > 400 Da | Smaller/hydrophilic molecules may back-diffuse depending on polarity and size | Conceptual estimate |
| Antibacterial sensor configuration | E. coli JW5503-1 + resazurin | >50% viability-proxy drop in 4–6 h at ≥10 μg⋅mL−1 | Allen et al., 2022; Sarker et al., 2007 |
| Antifungal sensor configuration | Candida albicans + resazurin | >80% metabolic signal decline ∼8 h at ∼25 μg⋅mL−1 | Conceptual estimate; Demir et al., 2024 |
| QS-inhibition sensor configuration | Chromobacterium violaceum CV026 | 10–20 µM C6-HSL inducer; ∼70% violacein repression ∼10 h (IC50 ≈ 5–20 μg⋅mL−1) | McClean et al., 1997; Duddy and Bassler, 2021 |
| Field incubation window | 3–10 days | Balances growth/signal development with dye/AHL stability constraints | Nichols et al., 2010; Berdy et al., 2017 |
Key modeling assumptions and parameter ranges (membrane pore size/thickness; diffusion coefficients; anisotropy targets; retention criteria by logP/MW; inducer dosing for CV026; time-to-signal windows; field incubation ranges).
3 Discussion
aNP-TRAP is advanced here strictly as a conceptual innovation: a single, modular device that merges in situ cultivation with embedded functional detection, aiming to overcome limitations of ex situ workflows (Nichols et al., 2010; Berdy, 2012). Unlike the iChip—which excels at environmental cultivation without built-in detection (Nichols et al., 2010; Ling et al., 2015)—aNP-TRAP integrates a biosensor layer designed to report locally diffusing small molecules (<∼1,000 Da) while restricting microbial translocation (Billings et al., 2015). In contrast to lab-only microfluidic/droplet systems (Aoi et al., 2009; Burmeister and Grünberger, 2020; Barakat et al., 2025), the focus here is field-deployability with minimal infrastructure.
To avoid any overstatement, we reiterate that aNP-TRAP is untested and hypothetical; empirical prototyping and validation are required before performance claims can be made. Potential confounders (e.g., environmental pigments/phenolics, AHL hydrolysis, dye photoreduction) warrant rigorous controls and cross-sensor comparison. An initial benchmarking plan is summarized in Table 1, including standards-based calibration (ampicillin, nystatin, C6-HSL) to quantify dose-response, LOD, and specificity. As an alternative readout, GFP-based reporters may provide fluorescence-based viability signals (Chalfie et al., 1994; Andersen et al., 1998). Preserving ecological signals may prime otherwise silent pathways during in situ interactions (Traxler et al., 2013).
4 Conclusion
aNP-TRAP is presented as a hypothesis-generating, conceptual, untested device that integrates in situ cultivation with functional detection to help triage microbial producers under ecologically relevant conditions. Conceptual “simulations” outline plausible timescales for nutrient equilibration, downward metabolite flux, and biosensor response, but these remain illustrative. The system should be viewed as unvalidated and requiring experimental prototyping and benchmarking as a next step.
Statements
Author’s note
This manuscript is conceptual and unvalidated experimentally. It was developed with language/figure assistance by AI tools under the author’s direction. All scientific ideas, device logic, and design choices are the author’s responsibility.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
MA: Methodology, Investigation, Conceptualization, Writing – review and editing, Writing – original draft, Formal Analysis.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that Generative AI was used in the creation of this manuscript. The author(s) verify and take full responsibility for the use of generative AI in the preparation of this manuscript. This manuscript was developed with language and figure support from OpenAI's ChatGPT, based on the author's technical input. All scientific content, including the concept, design, and methodology of the device, is original and solely the responsibility of the author. It has not yet been prototyped or tested due to limited technical and financial resources. Collaborative inquiries for development and validation are welcome.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Alain K. Querellou J. (2009). Cultivating the uncultured: limits, advances and future challenges. Extremophiles13, 583–594. 10.1007/s00792-009-0261-3
2
Allen J. L. Kennedy S. J. Shaw L. N. (2022). Colorimetric assays for the rapid and high-throughput screening of antimicrobial peptide activity against diverse bacterial pathogens. Methods Enzymol.663, 131–156. 10.1016/bs.mie.2021.10.008
3
Andersen J. B. Sternberg C. Poulsen L. K. Bjørn S. P. Givskov M. Molin S. (1998). New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol.64, 2240–2246. 10.1128/aem.64.6.2240-2246.1998
4
Aoi Y. Kinoshita T. Hata T. Oyaizu H. Obokata H. Tsuneda S. (2009). Hollow-fiber membrane chamber as a device for in situ environmental cultivation. Appl. Environ. Microbiol.75, 3826–3833. 10.1128/aem.02542-08
5
Barakat H. Bolton M. Li X. (2025). Droplet microfluidics in the antibiotic discovery pipeline: from microbial ecology to resistance mechanisms. ChemRxiv. 10.26434/chemrxiv-2025-6lfdz
6
Bauman K. D. Butler K. S. Moore B. S. Chekan J. R. (2021). Genome mining methods to discover bioactive natural products. Nat. Prod. Rep.38 (11), 2100–2129. 10.1039/d1np00032b
7
Berdy J. (2012). Thoughts and facts about antibiotics: where we are now and where we are heading. J. Antibiotics65, 385–395. 10.1038/ja.2012.27
8
Berdy B. Spoering A. L. Ling L. L. Epstein S. S. (2017). In situ cultivation of previously uncultivable microorganisms using the ichip. Nature protocols12 (10), 2232–2242.
9
Billings N. Birjiniuk A. Samad T. S. Doyle P. S. Ribbeck K. (2015). Material properties of biofilms-a review of methods for understanding permeability and mechanics. Rep. Prog. Phys.78 (3), 036601. 10.1088/0034-4885/78/3/036601
10
Burmeister A. Grünberger A. (2020). Microfluidic cultivation and analysis tools for interaction studies of microbial communities. Curr. Opin. Biotechnol.61, 175–183. 10.1016/j.copbio.2019.09.001
11
Chalfie M. Tu Y. Euskirchen G. Ward W. W. Prasher D. C. (1994). Green fluorescent protein as a marker for gene expression. Science263, 802–805. 10.1126/science.8303295
12
Demir E. Kırboga K. K. Işık M. (2024). “An overview of stability and lifetime of electrochemical biosensors,” Novel nanostructured materials for electrochemical bio-sensing applications, 129–158. 10.1016/B978-0-443-15334-1.00022-5
13
Duddy O. P. Bassler B. L. (2021). Quorum sensing across bacterial and viral domains. PLoS pathogens, 17 (1), e1009074. 10.1371/journal.ppat.1009074
14
Epstein S. S. (2013). The phenomenon of microbial uncultivability. Curr. Opin. Microbiol.16, 636–642. 10.1016/j.mib.2013.08.003
15
Hossain T. J. (2024). Methods for screening and evaluation of antimicrobial activity: a review of protocols, advantages, and limitations. Eur. J. Microbiol. Immunol. (Bp)14 (2), 97–115. 10.1556/1886.2024.00035
16
Hou L. Wang N. Man X. Cui Z. Wu J. Liu J. et al (2019). Interpenetrating janus membrane for high rectification ratio liquid unidirectional penetration. ACS Nano13 (4), 4124–4132. 10.1021/acsnano.8b08753
17
Jung D. Liu L. He S. (2020). Application of in situ cultivation in marine microbial resource mining. Mar. Life Sci. Technol.3 (2), 148–161. 10.1007/s42995-020-00063-x
18
Kaeberlein T. Lewis K. Epstein S. S. (2002). Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science296, 1127–1129. 10.1126/science.1070633
19
LaSarre B. Federle M. J. (2013). Exploiting quorum sensing to confuse bacterial pathogens. Microbiol. Mol. Biol. Rev.77, 73–111. 10.1128/mmbr.00046-12
20
Ling L. L. Schneider T. Peoples A. J. Spoering A. L. Engels I. Conlon B. P. et al (2015). A new antibiotic kills pathogens without detectable resistance. Nature517, 455–459. 10.1038/nature14098
21
McClean K. H. Winson M. K. Fish L. Taylor A. Chhabra S. R. Camara M. et al (1997). Quorum sensing and chromobacterium violaceum: a model for bioassay of AHL autoinducers. Microbiology143, 3703–3711. 10.1099/00221287-143-12-3703
22
Miller M. B. Bassler B. L. (2001). Quorum sensing in bacteria. Annu Rev Microbiol. 55, 165–99. 10.1146/annurev.micro.55.1.165
23
Newman D. J. Cragg G. M. (2020). Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod.83, 770–803. 10.1021/acs.jnatprod.9b01285
24
Nichols D. Cahoon N. Trakhtenberg E. M. Pham L. Mehta A. Belanger A. et al (2010). Use of iChip for high-throughput in situ cultivation of “uncultivable” microbial species. Appl. Environ. Microbiol.76, 2445–2450. 10.1128/aem.01754-09
25
Rutledge P. J. Challis G. L. (2015). Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol.13, 509–523. 10.1038/nrmicro3496
26
Sarker S. D. Nahar L. Kumarasamy Y. (2007). Microtitre plate-based antibacterial assay incorporating resazurin as an indicator of cell growth, and its application in the in vitro antibacterial screening of phytochemicals. Methods42, 321–324. 10.1016/j.ymeth.2007.01.006
27
Stewart P. S. (2003). Diffusion in biofilms. J. Bacteriol.85 (5), 1485–91. 10.1128/JB.185.5.1485-1491.2003
28
Traxler M. F. Watrous J. D. Alexandrov T. Dorrestein P. C. Kolter R. (2013). Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. mBio4, e00459-13–13. 10.1128/mbio.00459-13
29
Zhao Z. Ning Y. Ben S. Zhang X. Li Q. Yu C. et al (2022). Liquid-Assisted single-layer janus membrane for efficient unidirectional liquid penetration. Adv. Sci.9 (2), e2103765. 10.1002/advs.202103765
30
Ziemert N. Alanjary M. Weber T. (2016). The evolution of genome mining in microbes – a review. Nat. Product. Rep.33, 988–1005. 10.1039/c6np00025h
Summary
Keywords
in situ cultivation, functional biosensing, natural product discovery, modular microenvironment, quorum sensing inhibition, field-deployable screening
Citation
Abegg MA (2025) aNP-TRAP: a conceptual platform for in situ microbial cultivation and functional detection of antimicrobial activity. Front. Nat. Prod. 4:1617079. doi: 10.3389/fntpr.2025.1617079
Received
23 April 2025
Accepted
13 August 2025
Published
04 September 2025
Volume
4 - 2025
Edited by
Louis Pergaud Sandjo, Federal University of Santa Catarina, Brazil
Reviewed by
Ahmed F. Roumia, Menoufia University, Egypt
Suman Tiwari, The University of Texas at Dallas, United States
Updates
Copyright
© 2025 Abegg.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maxwel Adriano Abegg, maxabegg@gmail.com
ORCID: Maxwel Adriano Abegg, orcid.org/0000-0002-0328-1122
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.