- Division of Integrative Physiology, Department of Physiology, Jichi Medical University, Shimotsuke, Japan
The HCN4 channel is essential for heart rate regulation in vertebrates by generating pacemaker potentials in the sinoatrial node. HCN4 channel abnormality may cause bradycardia and sick sinus syndrome, making it an important target for clinical research and drug discovery. The zebrafish is a popular animal model for cardiovascular research. They are potentially suitable for studying inherited heart diseases, including cardiac arrhythmia. However, it has not been determined how similar the ion channels that underlie cardiac automaticity are in zebrafish and humans. In the case of HCN4, humans have one gene, whereas zebrafish have two ortholog genes (DrHCN4 and DrHCN4L; ‘Dr’ referring to Danio rerio). However, it is not known whether the two HCN4 channels have different physiological functions and roles in heart rate regulation. In this study, we characterized the biophysical properties of the two zebrafish HCN4 channels in Xenopus oocytes and compared them to those of the human HCN4 channel. We found that they showed different gating properties: DrHCN4L currents showed faster activation kinetics and a more positively shifted G-V curve than did DrHCN4 and human HCN4 currents. We made chimeric channels of DrHCN4 and DrHCN4L and found that cytoplasmic domains were determinants for the faster activation and the positively shifted G-V relationship in DrHCN4L. The use of a dominant-negative HCN4 mutant confirmed that DrHCN4 and DrHCN4L can form a heteromultimeric channel in Xenopus oocytes. Next, we confirmed that both are sensitive to common HCN channel inhibitors/blockers including Cs+, ivabradine, and ZD7288. These HCN inhibitors successfully lowered zebrafish heart rate during early embryonic stages. Finally, we knocked down the HCN4 genes using antisense morpholino and found that knocking down either or both of the HCN4 channels caused a temporal decrease in heart rate and tended to cause pericardial edema. These findings suggest that both DrHCN4 and DrHCN4L play a significant role in zebrafish heart rate regulation.
Introduction
The heart, an organ that pumps blood throughout the body, uses action potentials to contract the atria and ventricles regularly. A region called the sinoatrial node acts as a “pacemaker” by spontaneously generating its action potentials, leading to atria and ventricle action potentials. In pacemaker cells of the sinoatrial node that generate spontaneous action potentials, the pacemaker current or the funny current (If) is activated by hyperpolarization (Brown and Difrancesco, 1980; Yanagihara and Irisawa, 1980; Baruscotti et al., 2005). Its molecular entity is a hyperpolarization-activated cyclic nucleotide-gated (HCN) channel. Therefore, HCN channels are also known as pacemaker channels because they control heart rate by generating pacemaker potentials (Ludwig et al., 1999; Moosmang et al., 2001; Brioschi et al., 2009). In addition to the heart, HCN channels are expressed in the central nervous system (Santoro et al., 2000) and the gastrointestinal nervous system (Fujii et al., 2020), where they play a role in the regulation of neuronal excitability and synaptic transmission (Beaumont and Zucker, 2000).
There are four known HCN genes in humans and other mammals: HCN1, HCN2, HCN3, and HCN4. All of the HCN channels are voltage-gated channels activated by membrane hyperpolarization (Ludwig et al., 1998). Each gene encodes a six-transmembrane α subunit, and four subunits combine to form a single HCN channel. The intracellular region contains a cyclic nucleotide-binding domain (CNBD), where cyclic nucleotides bind. Cyclic nucleotide (primarily cAMP in physiological condition) binding can facilitate activation and shift the voltage dependence of the HCN channels. For the HCN2 and HCN4 channels, the cAMP binding is thought to be an important molecular mechanism by which sympathetic activation increases heart rate. Furthermore, the HCN4 channel is mainly expressed in the heart, and mutations in the HCN4 gene may cause sick sinus syndrome and other cardiac disorders (Milanesi et al., 2006; Verkerk and Wilders, 2015). It has also been reported that knockdown of HCN4 channels in mice leads to bradycardia and cardiac block (Herrmann et al., 2007; Baruscotti et al., 2011).
There are some common inhibitors/blockers available for HCN channels including ivabradine (Bucchi et al., 2006, 2002), ZD7288 (BoSmith et al., 1993), and Cs+ (Sartiani et al., 2017). In a previous study using rabbits, it was shown that ivabradine can reduce heart rate by inhibiting pacemaker current If (Bucchi et al., 2002). In a clinical setting, ivabradine has been approved for treatment of chronic heart failure and reduction of heart rate. It has been reported that administration of ivabradine to patients reduces heart rate by about 20% (Swedberg et al., 2010).
The zebrafish (Danio rerio) is a small freshwater fish that is native to India, and it has a body length of 3–5 cm as an adult. It is easy to rear, has rapid embryonic development, and is highly fertile. The body is transparent for about 1 week after fertilization, allowing direct observation of tissues and organs. In addition, zebrafish are vertebrates having organs like humans and are frequently used as model animals in various fields including genetics and developmental biology. Transparency in the early developmental stages has attracted attention in the fields of cardiovascular research, toxicology, and drug discovery because it is possible to directly observe beating hearts (Milan et al., 2006; MacRae and Peterson, 2015; Narumanchi et al., 2021; Maciag et al., 2022). In zebrafish, ionic currents that underlie cardiac automaticity and excitability have also been studied in detail (Nemtsas et al., 2010; Vornanen and Hassinen, 2016; Ravens, 2018; Stoyek and Quinn, 2018). While most ion channel genes are common in humans and zebrafish, different ion channel genes are sometimes used in zebrafish hearts; for example, KCNH6, in addition to KCNH2, underlies a hERG like current in zebrafish (Vornanen and Hassinen, 2016). In addition, due to the genome duplication in teleost evolution, the zebrafish genome sometimes has two orthologs to one gene in mammals (Amores et al., 1998; Glasauer and Neuhauss, 2014). In the case of HCN4, there is only one HCN4 gene in the human genome, HsHCN4 (Hs: Homo sapiens), whereas the zebrafish genome contains two HCN4 channel genes, DrHCN4 (Dr: Danio rerio) and DrHCN4L (Fujii et al., 2020; von der Heyde et al., 2020). It has been shown that DrHCN4 is expressed in the heart and gastrointestinal tract (Fujii et al., 2020; Stoyek et al., 2022, 2015). Cardiac automaticity in zebrafish utilizes membrane mechanism along with calcium clock mechanisms as in mammals (Marchant and Farrell, 2019; Stoyek et al., 2022). In zebrafish, HCN4 is one of the key components in the membrane clock mechanisms as well (Stoyek et al., 2022, 2015). However, there have been no reports on the biophysical, pharmacological, and functional differences of these two zebrafish HCN4 channels.
In this study, we expressed these two zebrafish HCN4 channels and human HCN4 channels in Xenopus oocytes and compared their biophysical and pharmacological properties. We also studied the physiological role of these two zebrafish HCN4 channels in developing zebrafish embryos by administering inhibitors and knocking down the gene with antisense morpholino.
Materials and Methods
Reverse Transcription-PCR
Total RNA was extracted and purified from zebrafish larvae on days 1, 3, and 7 and from the heart and brain of adult fish using NucleoSpin RNA Plus (Takara Bio, Shiga, Japan). Reverse transcription was performed using PrimeScript™ RT Master Mix (Takara Bio). The obtained cDNA was stored at −20°C and later used for PCR as a template. Partial cDNA fragments of DrHCN4, DrHCN4L, and β-actin (control) were amplified using the primers listed below.
DrHCN4-Fw: AGTGGACAACTTCAACGAGGTGCTG.
DrHCN4-Rv: AGGGAGCCCGTTTGAGGGTGTTTTGGTGG.
DrHCN4L-Fw: TGTGGACCATTTTAACGAGGTGTTGGAGG.
DrHCN4L-Rv: GGATGCGAACTGTAGGGAGCCCGTTTGAGGGTG.
β-actin-Fw: TAATACACAGCCATGGATGAGG.
β-actin-Rv: GGGAGCAATGATCTTGATCTTC.
Preparation of cRNA for Expression in Xenopus laevis Oocytes
The zebrafish HCN4 genes DrHCN4 (NCBI Reference sequence number: XM_680986.7) and DrHCN4L (XM_679638.8) were synthesized by GenScript (Piscataway, NJ, United States). They were amplified by PCR using primers containing the Kozak sequence (GCCACC) near the start codon and subcloned into a Xenopus laevis oocyte expression vector (pGEMHE) (Liman et al., 1992) using the In-Fusion HD Cloning Kit (Takara Bio). The human HCN4 gene, HsHCN4 (NP_005468.1), was purchased from Promega (Madison, WI, United States) and amplified with primers containing the N-terminal EcoRI site with the Kozak sequence and the C-terminal HindIII site. The PCR fragments were subcloned between the EcoRI and HindIII sites of the pGEMHE vector. These constructs were used as templates for cRNA synthesis after linearization with NheI. cRNA was synthesized using the mMESSAGE mMACHINE™ T7 Transcription Kit (Thermo Fisher Scientific, Waltham, MA, United States).
Xenopus laevis Oocyte Injection
Female frogs (Xenopus laevis) were anesthetized in water containing 0.1% tricaine (Sigma-Aldrich, St. Louis, MO, United States, E10521), and oocytes were surgically obtained. To remove the follicular cell layer, oocytes were treated with 2 mg/ml collagenase (Sigma-Aldrich, C0130) dissolved in MBSH solution (88 mM NaCl, 1 mM KCl, 2.4 mM NaHCO3, 10 mM HEPES, 0.3 mM Ca(NO3)2, 0.41 mM CaCl2, 0.82 mM MgSO4, pH 7.6) for 6 h at room temperature. Defolliculated oocytes from stages V to VI were selected for cRNA injection. The synthetic cRNA (1–10 ng) was injected into each oocyte using Nanoject II (Drummond Scientific Company, Broomall, PA, United States). cRNA-injected oocytes were incubated in MBSH solution supplemented with 0.1% penicillin-streptomycin (Sigma-Aldrich, P4333) at 18°C for 2–3 days. All animal experiments using Xenopus laevis were approved by the Animal Experiment Committee of Jichi Medical University (Approval No. 18027).
Two-Electrode Voltage Clamp
Ionic currents were measured by using an OC-725C amplifier (Warner Instruments, Hamden, CT, United States) with a two-electrode voltage clamp. Generation of voltage-clamp protocols and data acquisition were performed using a Digidata 1550 interface (Molecular Devices, San Jose, CA, United States) controlled by Clampex 10.7 software (Molecular Devices). Data were sampled at 10 kHz and low-pass filtered at 1 kHz using Clampex 10.7 software (Molecular Devices). All experiments were performed at room temperature. A glass electrode with a resistance of 0.2–0.5 MΩ was prepared from a borosilicate glass capillary (GC150TF-10, Harvard Apparatus, Holliston, MA, United States) using a micropipette puller (P-1000, Sutter Instrument, Novato, CA, United States). The glass electrode was filled with 3 M KCl. ND66 solution (66 mM NaCl, 32 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2, 5 mM HEPES, pH 7.6) was used as the extracellular solution (Decher et al., 2003; Liu et al., 2020).
Data Analysis
Activation curves (G-V curves) obtained by plotting the tail current at -120 mV were fitted using Clampfit 10.7 software (Molecular Devices) to a single Boltzmann function.
where Gmin is minimum tail current, Gmax is maximum tail current, z is effective charge, V1/2 is half activation potential, F is Faraday constant, R is gas constant, T is absolute temperature.
To analyze the rate of activation, a current of -130 mV was fitted to a single exponential function (equation below) using pClamp 10.7 software to obtain the time constant (τ).
HCN Channel Inhibitors
The HCN inhibitors ZD7288 (Z3777) and ivabradine hydrochloride (SML 0281) were purchased from Sigma-Aldrich. ZD7288 and ivabradine were dissolved in distilled water, and stock solutions of 100 mM or 50 mM were stored at −20°C. Cesium chloride was dissolved in distilled water, and 1 M of the solution was stored at room temperature. The stock solution was diluted with ND66 solution to achieve the final concentration on the day of the experiment.
For zebrafish, the HCN inhibitors at a final concentration of 0.01–1 mM were applied to egg water in which the fish were kept, starting at 24 h post-fertilization (hpf). Heart rates were recorded before (0 h) and at 24 h (24 h) and 48 h (48 h) after the inhibitors had been applied.
Fish Maintenance and Egg Collection
Wild-type zebrafish (RIKEN-WT) were used. Zebrafish were kept in water at 28°C with 14 h of light (8:00–22:00) and 10 h of darkness (22:00–8:00). On the day before spawning, male and female fish were placed in a mating tank separated by a partition. On the morning of the spawning day, the partition was removed. Immediately after spawning, eggs were collected in a 10 cm dish with egg water (0.006% sea salt (Aquarium Systems, Sarrebourg, France) and 0.01% methylene blue). Animal experiments using zebrafish were approved by the Animal Experiment Committee of Jichi Medical University (Approval No. 18037).
Heart Rate Measurement
The heart rate of zebrafish embryos was measured under an Olympus MVX10 microscope at room temperature. After zebrafish embryos were acclimated to the new environment (room temperature, light), heart rate was counted by eye.
Gene Knockdown by Morpholinos
Morpholino antisense oligos (MO) were purchased from Gene Tools (Philomath, OR, United States) (Fujii et al., 2020). Zebrafish 1-cell embryos were injected with approximately 5 nL of MO solution at a concentration of 200 μM (approximately 10 ng/embryo).
The MO sequences used are listed below.
Negative Control (5′-CCTCTTACCTCAGTTACAATTTATA -3′).
DrHCN4 (5′-GTAATTACTGCCACCGTGCACCACA-3′).
DrHCN4L (5′-GGCGACGCTGGCTGAAAAATAGGTC -3′).
Statistical Analysis
Student’s t-tests were used to compare two groups, and the Dunnett or Tukey multiple comparison tests were used for multiple comparisons using EZR software (EZR version 1.54) (Kanda, 2013). p < 0.05 was considered as a significant difference. p < 0.05, <0.01, and <0.001 are denoted in the figures by *, **, and ***, respectively. All exact p values are listed in the tables.
Results
DrHCN4 and DrHCN4L Encode Functional Hyperpolarization-Activated Channels
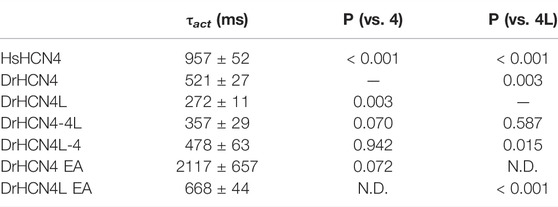
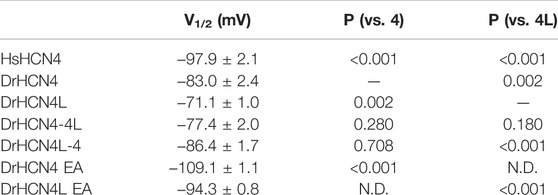
The zebrafish genome contains two HCN4 genes, DrHCN4 on chromosome 18 and DrHCN4L on chromosome 25. Although it is well established that HCN4 channels of other species are hyperpolarization-activated channels, the biophysical properties of the zebrafish HCN4 channels have not been reported. Therefore, we first compared the biophysical properties of zebrafish HCN4 channels (DrHCN4 and DrHCN4L) with those of the human HCN4 channel (HsHCN4). Hyperpolarization-activated inward currents were observed in oocytes expressing DrHCN4 and DrHCN4L, similar to HsHCN4 currents (Figure 1A). The activation time constants (τact) of HsHCN4, DrHCN4, and DrHCN4L at −130 mV were 957 ± 52 ms, 521 ± 28 ms, and 272 ± 11 ms, respectively (Figure 1B and Table 1). Therefore, the zebrafish HCN4 channels had faster activation kinetics than the human HCN4 channel, and DrHCN4L showed the fastest activation kinetics. The G-V (conductance-voltage) relationships are shown in Figure 1C. V1/2 values of the G-V relationships for HsHCN4, DrHCN4, and DrHCN4L were −97.9 ± 2.1 mV, −83.0 ± 2.4 mV, and −71.1 ± 1.0 mV, respectively (Figure 1C and Table 2). Again, the opening of zebrafish HCN4 channels requires less hyperpolarization than that of the human HCN4, and DrHCN4L showed the most positively-shifted G-V relationship among them.
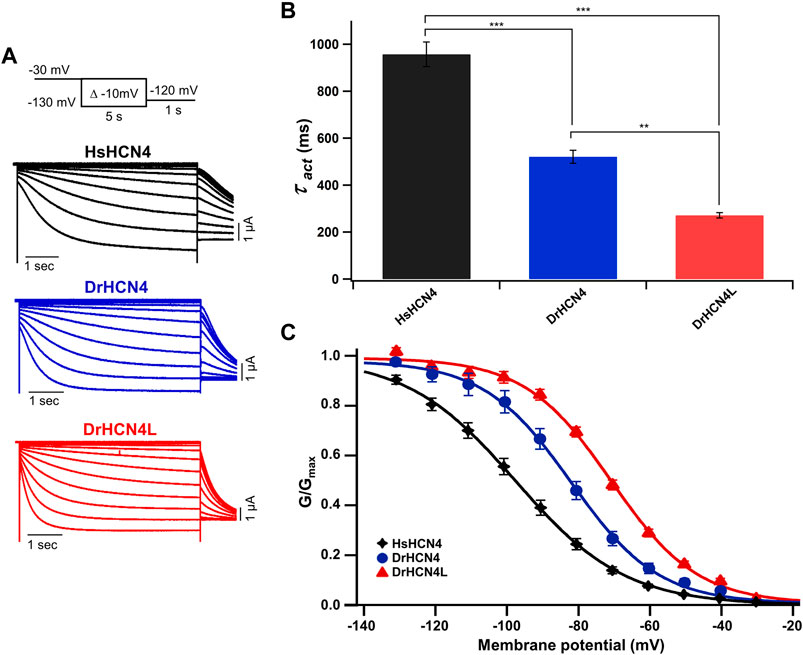
FIGURE 1. Zebrafish HCN4 channels are hyperpolarization-activated ion channels. (A) Representative currents of the human HCN4 channel HsHCN4 (black) and zebrafish HCN4 channels DrHCN4 (blue) and DrHCN4L (red). The holding potential was stepped from −30 mV to −130 mV in 10-mV decrements and held for 5 s at each potential. The inset shows the stimulation protocol. (B) Time constants obtained by fitting the activating currents of HsHCN4 (black), DrHCN4 (red) and DrHCN4L (blue) at −130 mV. (C) Activation curves (G–V curves) of human and zebrafish HCN4 channels (n = 5 for each).
The Cytoplasmic C-Terminal Domain Determines the Voltage Dependence
The voltage sensor movement confers the voltage dependence of HCN channels like other voltage-gated ion channels (Männikkö et al., 2002; Dai et al., 2019). In addition, all HCN channels contain a cyclic nucleotide-binding domain (CNBD) in the intracellular C-terminal region where cyclic nucleotides, primarily cAMP in physiological condition, binds and thereby shifts the G-V relationship of HCN channels towards a depolarized potential. This mechanism is how the activation of Gs-coupled β1-adrenergic receptors potentiates HCN channels. To determine which part of the zebrafish HCN4 channels is responsible for the different voltage dependence, we made two chimera channels (DrHCN4-4L and DrHCN4L-4) by exchanging the cytoplasmic C-terminal regions of DrHCN4 and DrHCN4L. The molecular design and ionic currents of the chimera channels under a two-electrode voltage clamp are shown in Figure 2A and Supplementary Figure S1 (a red arrow). Activation kinetics of DrHCN4-4L, which has an N-terminal region and a transmembrane region of DrHCN4 with the C-terminal region of DrHCN4L, was 357 ± 29 ms and comparable to that of DrHCN4L (p = 0.587), rather than that of DrHCN4 (p = 0.070). On the other hand, the activation kinetics of DrHCN4L-4 was 478 ± 63 ms and comparable to that of DrHCN4 (p = 0.942), rather than that of DrHCN4L (p = 0.015) (Figure 2B and Table 1). The C-terminal exchange of DrHCN4 and DrHCN4L also resulted in a shift of the G-V relationships; V1/2 of DrHCN4L-4 chimera was −86.4 ± 1.7 mV and negatively shifted from V1/2 of DrHCN4L (−71.1 ± 1.0 mV; p < 0.001). V1/2 of DrHCN4-4L (−77.4 ± 2.0 mV) was not significantly shifted from that of DrHCN4 (−83.0 ± 2.4 mV; p = 0.280), though. (Figure 2C and Table 2). These results suggest that the C-terminal region determines the voltage dependence.
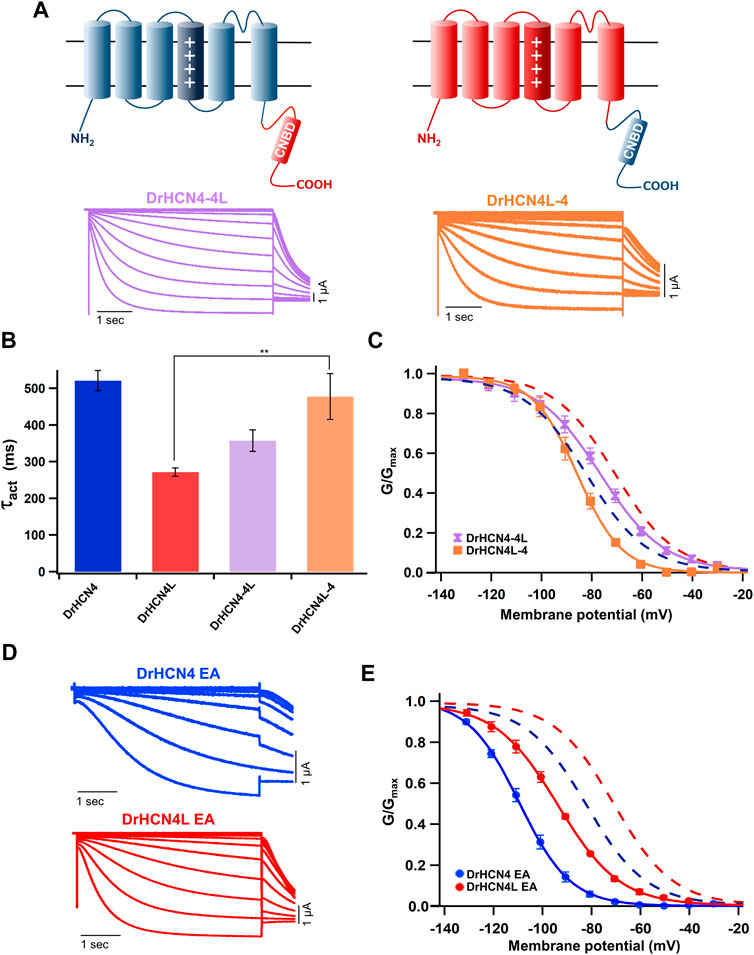
FIGURE 2. The cytoplasmic C-terminal region of DrHCN4 and DrHCN4L determines the voltage dependence. (A) Molecular designs and representative currents of chimeric channels: DrHCN4-4L (left, purple) and DrHCN4L (right, orange). (B) Activation time constants (τact) at −130 mV of DrHCN4 (blue), DrHCN4L (red), DrHCN4-4L (purple) and DrHCN4L-4 (orange). The currents were fitted with a single exponential. (C) Activation curves of the zebrafish chimeric HCN4 channels (n = 5). The Boltzmann function was used to fit the curves. Blue and red dashed curves are the activation curves of DrHCN4 and DrHCN4L from Figure 1C. (D) Representative current traces of the EA mutants, DrHCN4 EA and DrHCN4L EA. (E) Activation curves of the EA mutant channels (n = 5). The Boltzmann function was used to fit the curves. Blue and red dashed curves are the activation curves of DrHCN4 and DrHCN4L from Figure 1C.
Because the C-terminal region contains the CNBD domain, where cAMP (or other cyclic nucleotides) binds and modulates the gating properties, we hypothesized that the difference in the voltage dependence could be due to a difference in cAMP sensitivity. As it is known that Xenopus oocytes contain a high concentration of cAMP, a possible cAMP sensitivity difference could have a high impact on the gating of zebrafish HCN4 channels. To examine if the cAMP sensitivities are different between DrHCN4 and DrHCN4L, we mutated two critical amino acid residues for cAMP binding (R630 and T631 in DrHCN4; R628 and T629 in DrHCN4L) to glutamate and alanine (DrHCN4 EA and DrHCN4L EA). This mutant is known to lose a sensitivity to cAMP (Magee et al., 2015; Fenske et al., 2020). By assuming DrHCN4 EA and DrHCN4L EA lost cAMP-dependent regulation, we expressed them in Xenopus oocytes and examined their voltage-dependence and the gating behavior. The EA mutants showed slower activation kinetics and negatively shifted G-V relationships (Figures 2D,E). The G-V shifts were by more than 20 mV, from −83.0 ± 2.4 mV to −109.1 ± 1.1 mV in DrHCN4 EA (p < 0.001), and from −71.1 ± 1.0 mV to −94.3 ± 0.8 mV in DrHCN4L EA (p < 0.001) (Figure 2E and Table 2). These results suggest that both DrHCN4 and DrHCN4L are markedly sensitive to cAMP.
In conclusion, the gating properties of DrHCN4 and DrHCN4L are primarily dependent on the C-terminal region; however, it is not due to a difference in cAMP sensitivity.
DrHCN4 and DrHCN4L can Form a Heterotetramer
There are four known isoforms of HCN channels (HCN1-4). A heterotetramer can be formed among different isoforms, and the formation of a heterotetramer diversifies their physiological functions (Chen et al., 2001; Sartiani et al., 2017; Rivolta et al., 2020). For example, the HCN1-HCN2 heteromer is implicated in the pathology of temporal lobe epilepsies (Noam et al., 2011). The HCN1-HCN4 heteromer has been suggested to form native pacemaker channels in the rabbit sinoatrial node (Altomare et al., 2003). Therefore, it is reasonable to speculate that DrHCN4 and DrHCN4L can form a heterotetramer. To verify whether DrHCN4 and DrHCN4L constitute a heterotetramer, a mixture of DrHCN4 and DrHCN4L cRNAs in a ratio of 1:1 (Figure 3A, green) or 3:1 (Figure 3A, light green) was injected into Xenopus oocytes. In the G-V relationships, the mixtures of DrHCN4 and DrHCN4L showed intermediate properties between DrHCN4 alone and DrHCN4L alone and could be fitted with a single Boltzmann function (Figure 3B and Table 3).
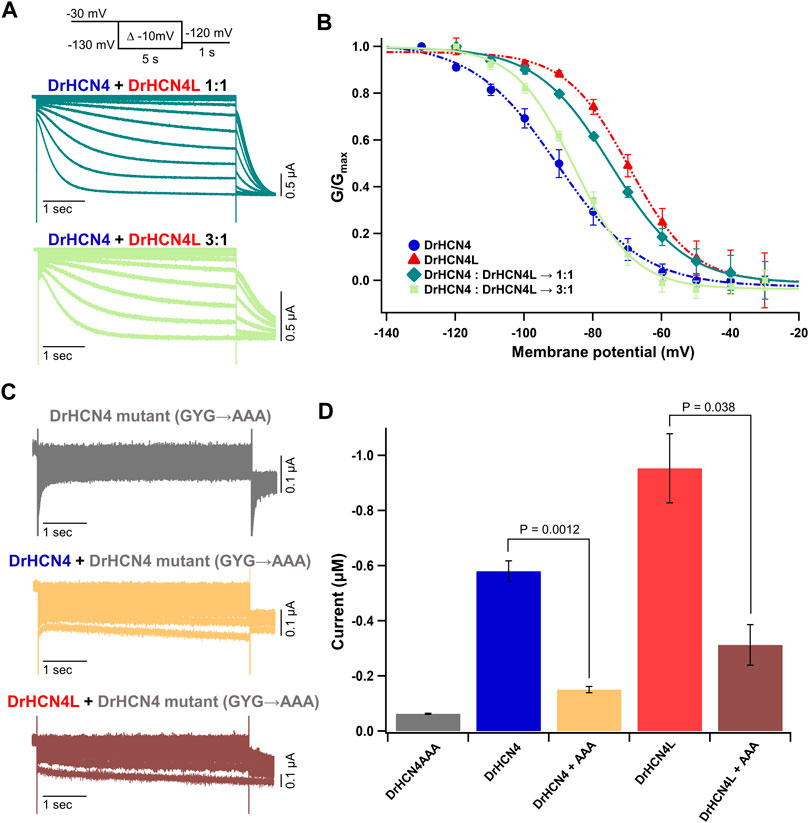
FIGURE 3. DrHCN4 and DrHCN4L can form a heterotetrametric channel. (A) Representative current traces of a mixture of DrHCN4 and DrHCN4L mRNA. The mixed mRNA concentrations were 1:1 (green) and 3:1 (light green). Membrane potential was varied from −30 mV to −130 mV in 10-mV decrements and held for 5 s at each potential. The inset shows the stimulation protocol. (B) Activation curves of the mixed channels of DrHCN4 and DrHCN4L. The mixed mRNA concentrations were 1:1 (green) and 3:1 (light green). Blue and red curves are the activation curves of DrHCN4 and DrHCN4L, respectively. The Boltzmann function fits the activation curves (n = 5). All data are from the same batch of oocytes. Therefore, the data sets of DrHCN4 and DrHCN4L are different from the datasets used in Figures 1, 2. (C) Representative current traces of a dominant-negative mutant with or without wild-type DrHCN4 or DrHCN4L. From top to bottom: DrHCN4 mutant (DrHCN4AAA) only, DrHCN4 (wild-type) + DrHCN4AAA, DrHCN4L (wild-type) + DrHCN4AAA. Wild-type and mutant cRNAs were mixed in a 1:1 ratio with 5 ng each. (D) Averaged current amplitudes at −100 mV were plotted as bar graphs [n = 5 for each except DrHCN4AAA alone (n = 4)].
To further demonstrate the presence of the heterotetramer, we made a DrHCN4 mutant “DrHCN4AAA” by replacing glycine-tyrosine-glycine (GYG) amino acid residues located at the selectivity filter with triple alanine (AAA) residues (Supplementary Figure S1, orange box). This mutant is known to function as the dominant-negative mutant in HCN and potassium channels (Xue et al., 2002; Pai et al., 2017; Pitcairn et al., 2017). If more than one DrHCN4AAA mutant subunit(s) is/are included in a DrHCN4L tetramer, it/they will inhibit the ionic current. If that is not the case, wild-type DrHCN4L and DrHCN4AAA mutants independently constitute a channel, and the ionic currents would be observed as if DrHCN4L is expressed alone. First, we confirmed that the DrHCN4AAA mutant alone did not generate ionic currents (Figure 3C, gray). Next, when the DrHCN4AAA mutant was co-expressed with DrHCN4, the current was almost completely suppressed, confirming that the DrHCN4AAA mutant was a dominant-negative mutant (Figure 3C, yellow). Finally, we co-expressed the DrHCN4AAA mutant with DrHCN4L and found that the DrHCN4L current was significantly inhibited (Figures 3C,D). These results further support that DrHCN4 and DrHCN4L can form a heterotetramer.
Sensitivity of DrHCN4 and DrHCN4L to HCN Inhibitors
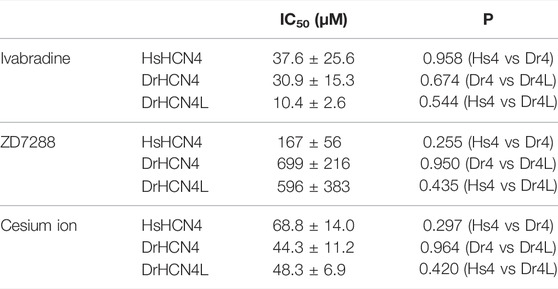
We next analyzed the inhibition of DrHCN4 and DrHCN4L by known HCN inhibitors [ivabradine, ZD7288, and cesium ions (Cs+)]. Human and zebrafish HCN4 channels were expressed in Xenopus oocytes, and the inhibitors were applied to the recording bath. The concentrations of the inhibitors were in the range of 1–100 μM (ivabradine) or 1–1000 μM (ZD7288 and Cs+). In the case of ivabradine, the currents of HsHCN4, DrHCN4, and DrHCN4L were similarly inhibited (Figures 4A–C). The IC50 values were 38 ± 26 μM for HsHCN4, 31 ± 15 μM for DrHCN4, and 10 ± 3 μM for DrHCN4L (Figure 4D; Table 4). In the case of ZD7288, the IC50 values were 167 ± 56 μM for HsHCN4, 699 ± 216 μM for DrHCN4, and 596 ± 382 μM for DrHCN4L (Figures 4E–H; Table 4). In the case of Cs+, the IC50 values were 69 ± 14 μM for HsHCN4, 44 ± 11 μM for DrHCN4, and 48 ± 7 μM for DrHCN4L (Figures 4I–L; Table 4).
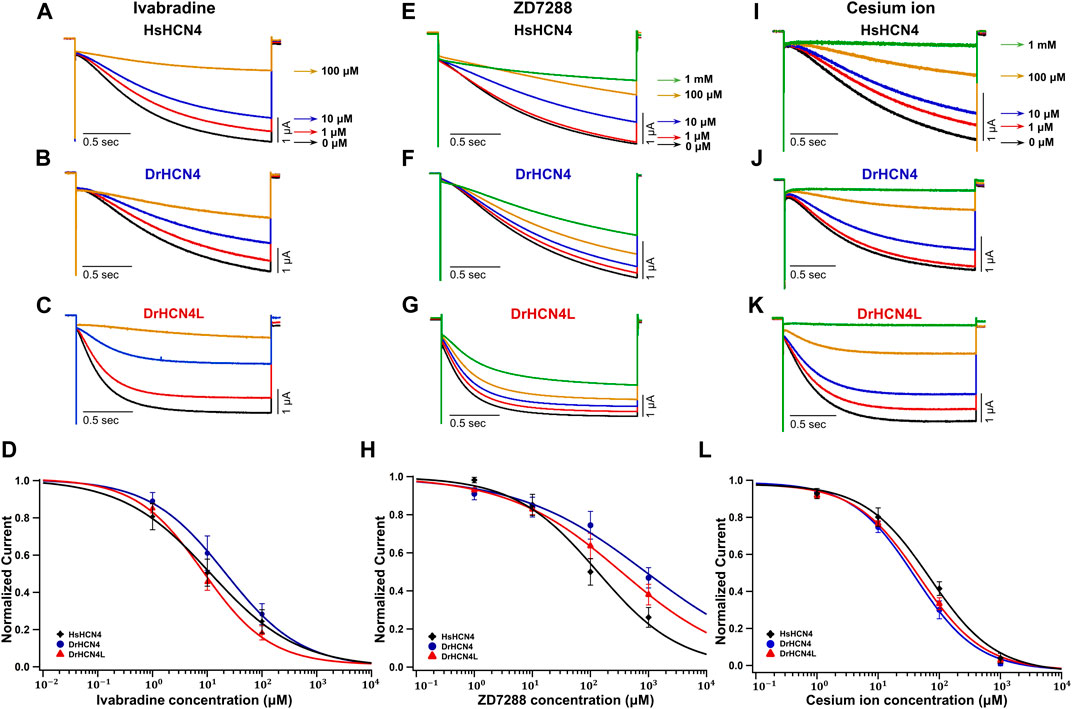
FIGURE 4. Sensitivities of DrHCN4 and DrHCN4L to HCN inhibitors. (A–C) Representative current traces sequentially applied with 0–100 µM of ivabradine. The holding potential was −30 mV and was hyperpolarized to −120 mV for 2 s. (D) Ivabradine inhibition curves based on currents of (A–C). DrHCN4 (blue; n = 5), DrHCN4L (red; n = 4) and HsHCN4 (black; n = 4). (E–G) Representative current traces sequentially applied with 0 μM–1 mM of ZD7288. The holding potential was −30 mV and was hyperpolarized to −120 mV for 2 s. (H) ZD7288 inhibition curves based on currents of (E–G). DrHCN4 (blue; n = 5), DrHCN4L (red; n = 5) and HsHCN4 (black; n = 5). (I–K) Representative currents sequentially applied with 0 μM–1 mM of cesium ion. The holding potential was −30 mV and was hyperpolarized to −120 mV for 2 s. (L) Cesium ion inhibition curves based on currents of (I–K). DrHCN4 (blue; n = 5), DrHCN4L (red; n = 5) and HsHCN4 (black; n = 5).
In conclusion, the HCN inhibitors also inhibit zebrafish HCN4 channels as well.
HCN Inhibitors Reduce the Heart Rate of Developing Zebrafish Embryos
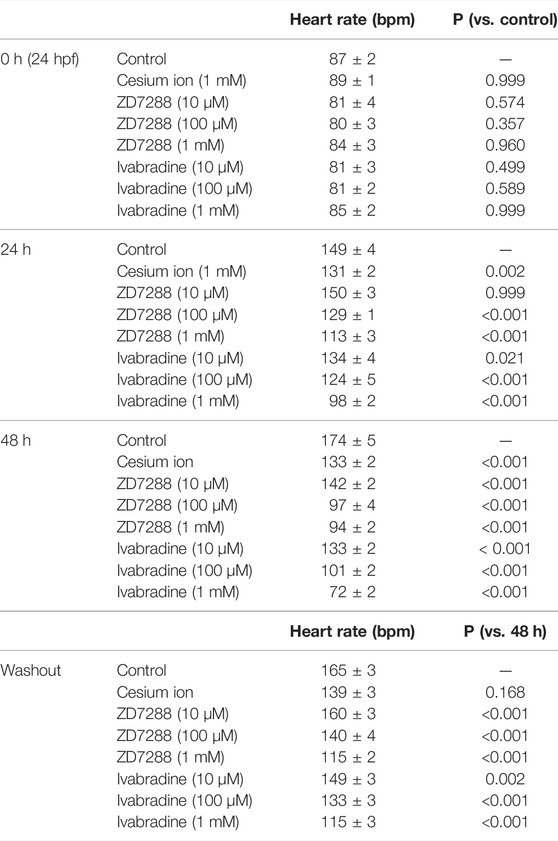
Ivabradine is used to treat chronic heart failure and other disorders as it slows heart rate by inhibiting HCN channel currents in the sinoatrial node (Swedberg et al., 2010). Next, we applied the HCN inhibitors, including ivabradine, to developing zebrafish embryos to see if they were equally efficient in lowering heart rate as they are humans. The HCN inhibitors (Cs+, ZD7288, and ivabradine) at a final concentration of 1 mM were applied to the egg water in which fish were kept, starting at 24 h post-fertilization (hpf). Heart rates were recorded before (0 h) and at 24 h (24 h) and 48 h (48 h) after the application of the inhibitors. Zebrafish embryos were then brought back to the inhibitor-free water, and their heart rates were recorded after 6 h (54 h: “washout”) (Figure 5A). In the case of developing zebrafish embryos without an inhibitor (control), heart rate increased from 87 ± 2 bpm [0 h (24 hpf)] to 149 ± 4 bpm [24 h (48 hpf)] and 174 ± 5 bpm [48 h (72 hpf)] during the span (Figure 5B, gray bars). All HCN inhibitors significantly reduced the heart rate of zebrafish at 24 and 48 h after drug application (Figure 5B). After the subsequent washout of HCN inhibitors, the heart rates in both the ZD7288 and ivabradine groups (but not the Cs+ group) recovered (Figure 5B and Table 5). We then examined the concentration dependence of the effects of ivabradine and ZD7288 (Figures 5C,D). ZD7288 significantly lowered heart rate, except for a concentration of 10 μM at 24 h (Figure 5C). Ivabradine significantly suppressed heart rate at all concentrations and time points, and the effect was more potent at higher concentrations and longer time (Figure 5D). Furthermore, the heart rates in all ZD7288- and ivabradine-treated zebrafish were significantly recovered by washing out the inhibitors (Figures 5C,D and Table 5).
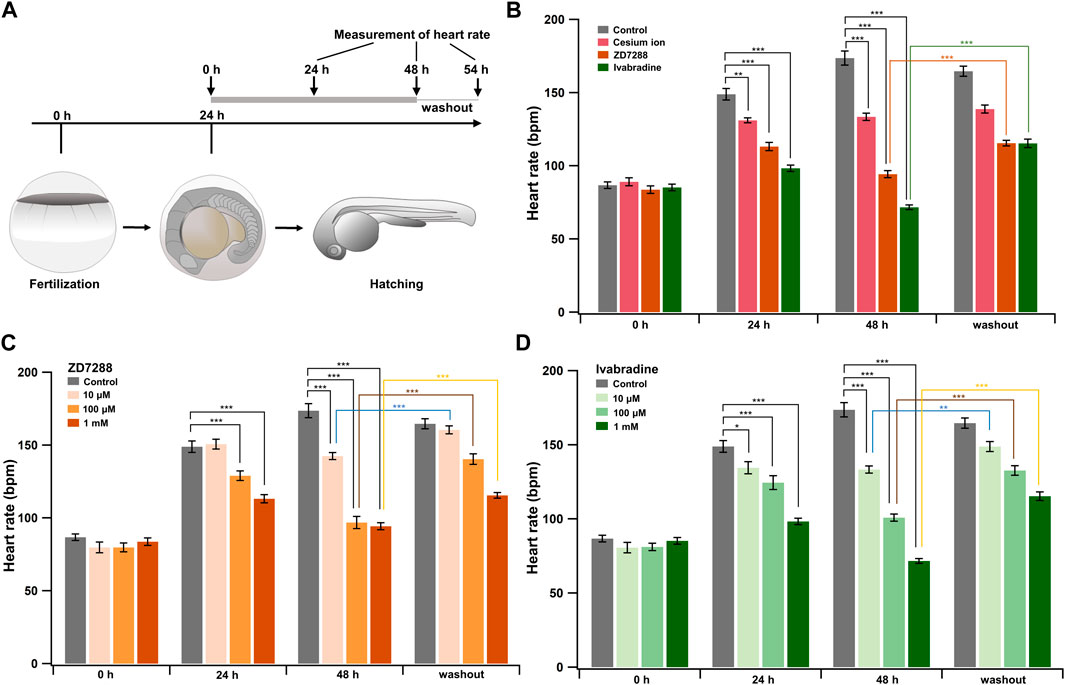
FIGURE 5. HCN inhibitors reduce heart rate in developing zebrafish embryos. (A) Experimental method. Inhibitors were administered in the egg water at 24 hpf, and this time point was designated as “0 h”. After administering the inhibitors, heart rate was measured at 24 h (24 h) and 48 h (48 h). Zebrafish were then transferred back to the inhibitor-free egg water, and their heart rates were measured 6 h later (washout). (B) Changes in heart rate with time in zebrafish before (control; grey; n = 10) and after treatment with 1 mM cesium ion (pink; n = 10), ZD7288 (orange; n = 10) and ivabradine (green; n = 10) inhibitors. Heart rates before (0 h) and at 24 h (24 h) and 48 h (48 h), after application of the inhibitors and at 6 h after washing out the inhibitors (washout) are shown as bar graphs. Dunnett’s test was used for statistical analysis, and Student’s t-test was used to compare before and after “washout.” (C) Changes in heart rate with time in zebrafish treated with ZD7288 before (gray; n = 10) and after treatment with 10 μM (light orange; n = 10), 100 μM (orange; n = 10), and 1 mM (dark orange; n = 10). Dunnett’s test was used for statistical analysis, and Student’s t-test was used to compare before and after washout. (D) Changes in heart rate with time in zebrafish treated with ivabradine before (gray; n = 10) and after treatment with 10 μM (thin green; n = 10), 100 μM (light green; n = 10) and 1 mM (dark green; n = 10). Dunnett’s test was used for statistical analysis, and Student’s t-test was used to compare before and after washout.
These results suggest that ivabradine and ZD7288 reduce heart rate in developing zebrafish embryos. The inhibitory effect of cesium ions on zebrafish heart rate was less than that expected from the results for Xenopus oocytes.
HCN4 Channel Knockdown Transiently Reduces Heart Rate and Induces Pericardial Edema in Zebrafish
Since DrHCN4 and DrHCN4L have similar sensitivities to the HCN inhibitors, individual gene knockdowns would be required for evaluating each channel’s role in heart rate regulation. Before knocking down each gene, we first confirmed that both HCN4 channels (DrHCN4 and DrHCN4L) were expressed in day 1, day 3, and day 7 embryos and in the heart and brain of adult fish (Figure 6A). We next investigated whether the knockdown of each gene affects heart rate in zebrafish. The expression of each gene was knocked down by using antisense morpholino (MO), which is commonly used in zebrafish (Blum et al., 2015; Jou et al., 2017; Fujii et al., 2020). MO solutions were injected into 1-cell embryos of zebrafish. First, we confirmed that water or control MO injection did not affect heart rate (Supplementary Figure S2). Embryos were injected with control MO, DrHCN4 MO, and DrHCN4L MO at concentrations of 200 μM or with a mixture of DrHCN4 MO and DrHCN4L MO (200 μM for each). At 24 h after injection of MO, there was a slight decrease of heart rate in embryos injected with DrHCN4 MO (44 ± 2 bpm, p = 0.049) and DrHCN4L MO (45 ± 2 bpm, p = 0.092) compared to the heart rate in embryos injected with control MO (54 ± 1 bpm). The heart rate of zebrafish injected with a mixture of DrHCN4 MO and DrHCN4L MO was significantly reduced to 23 ± 6 bpm (p < 0.001; Figure 6B and Table 6). After 48 h, heart rate was considerably lower in all three groups compared to that in the control group (135 ± 4 bpm): DrHCN4 MO (110 ± 3 bpm, p < 0.001), DrHCN4L MO (111 ± 1 bpm, p = 0.001), and mixture of DrHCN4 MO and DrHCN4L MO (116 ± 7 bpm, p = 0.014). However, at 72 h after MO injection, heart rate in all three groups had returned to the same level as that in the control group (145 ± 5 bpm): DrHCN4 MO (136 ± 3 bpm, p = 0.509), DrHCN4L MO (141 ± 5 bpm, p = 0.921), and mixture of DrHCN4 MO and DrHCN4L MO (147 ± 7 bpm, p = 0.991) (Figure 6B and Table 6).
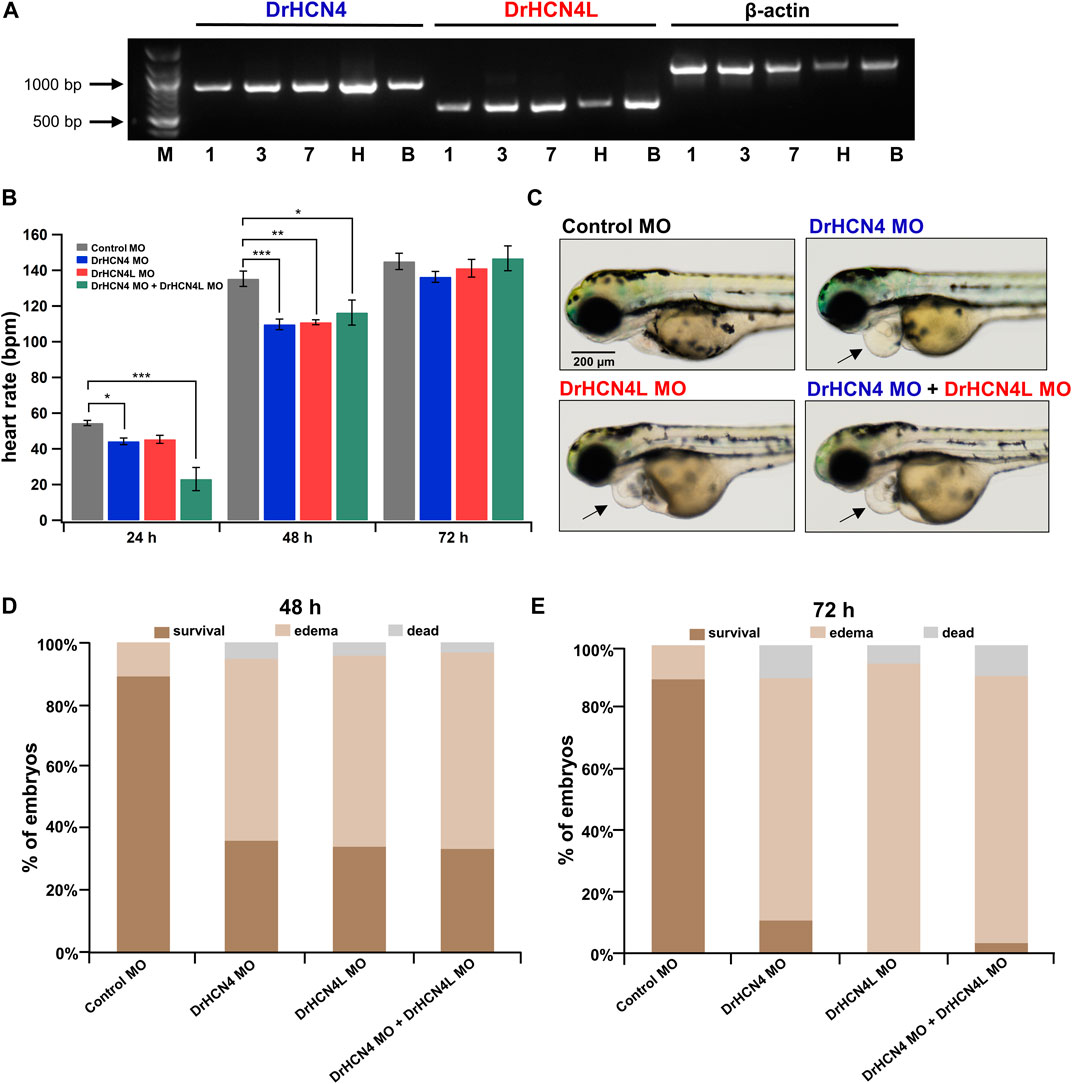
FIGURE 6. Effects of HCN4 channel knockdown on zebrafish. (A) Expression of DrHCN4 and DrHCN4L in one (1), three (3), and seven (7) day embryos/larvae as well as in the heart (H) and brain (B) of adult fish was confirmed by RT-PCR. Arrows indicate the DNA ladder markers of 1,000 bp and 500 bp. (B) Heart rates at 24, 48, and 72 h are shown in bar graphs for 200 μM MO-injected zebrafish embryos: control MO (n = 20), DrHCN4 MO (n = 10), DrHCN4L MO (n = 10), and mixed DrHCN4 and DrHCN4L MO (n = 10). Statistical analysis was performed by Dunnett’s test. (C) Representative examples of zebrafish injected with control MO, DrHCN4 MO, DrHCN4L MO, and mixed DrHCN4 and DrHCN4L MO. Zebrafish with DrHCN4 MO, DrHCN4L MO, and mixed DrHCN4 and DrHCN4L MO showed pericardial edema (black arrows). Bar, 200 μm. (D) Ratios of normal embryos, pericardial edema, and death at 48 h after injection of 200 µM MO: control MO (n = 136), DrHCN4 MO (n = 75), DrHCN4L MO (n = 34), and mixed DrHCN4 and DrHCN4L MO (n = 30). (E) Pericardial edema and death ratios at 72 h after injection of 200 µM MO. Control MO (n = 136), DrHCN4 MO (n = 75), DrHCN4L MO (n = 34), and mixed DrHCN4 and DrHCN4L MO (n = 30).
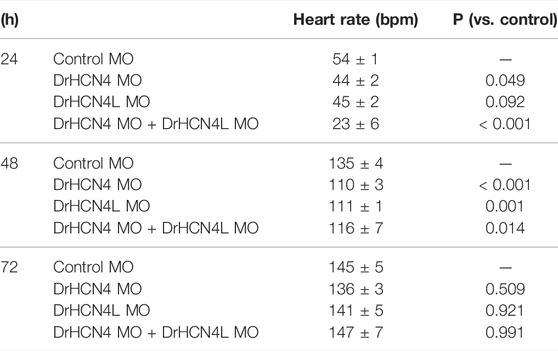
TABLE 6. Inhibitory effects of antisense MO on heart rates (bpm) and p-values in developing zebrafish embryos.
We noticed that zebrafish embryos injected with HCN4 MO tended to exhibit pericardial edema (Figure 6C). Pericardial edema is often observed in embryonic zebrafish with heart failure (Narumanchi et al., 2021). We determined the incidence of edema in zebrafish injected with 200 μM MO. We only examined zebrafish embryos that survived for the first 24 h after MO injection, and the rates of edema occurrence were determined at 48 and 72 h after injection. At 48 h after injection, 59, 62, and 63% of the embryos injected with DrHCN4 MO, DrHCN4L MO, and a mixture of DrHCN4 MO and DrHCN4L MO, respectively, showed pericardial edema (Figure 6D). After 72 h, the proportions of embryos with edema increased to 79, 94, and 87% (Figure 6E). These results suggested that both DrHCN4 and DrHCN4L are required for proper cardiac function at early stages.
Discussion
Biophysical Properties of DrHCN4 and DrHCN4L Channels
In this study, we first analyzed the biophysical properties of DrHCN4 and DrHCN4L and compared them to the biophysical properties of HsHCN4. DrHCN4L showed the fastest activation kinetics, DrHCN4 showed the second-fastest activation kinetics, and HsHCN4 showed the slowest activation kinetics (Figure 1B). The G-V curves showed similar tendencies; DrHCN4L showed the most positively shifted G-V curve (Figure 1C). The chimera experiments suggest that the cytoplasmic region might determine their voltage dependence (Figure 2). It is known that the intracellular cyclic nucleotides cAMP activates HCN channels (Zagotta et al., 2003; Alvarez-Baron et al., 2018; Saponaro et al., 2021). According to the EA mutant experiments, however, the difference in the voltage dependence is not due to different cAMP sensitivities (Figures 2D,E). Although we did not further explore the causes of different gating properties here, we noticed some unique amino acid residues only in DrHCN4L around the CNBD domain (Supplementary Figure S1). Those unique amino acid residues might change the gating properties and accelerate the activation kinetics and positively shift the voltage dependence in DrHCN4L.
Mammalian HCN1, HCN2, and HCN4 are known to form a heterotetramer (Chen et al., 2001; Rivolta et al., 2020). Therefore, it is highly likely that DrHCN4 and DrHCN4L form a heterotetramer. By generating a dominant-negative mutant (DrHCN4AAA) (Figure 3C), we confirmed that the zebrafish HCN4 channels (DrHCN4 and DrHCN4L) are also capable of forming a heterotetramer, at least in Xenopus oocytes. Using a similar HCN4 dominant-negative mutant, it has been shown that HCN4 channels play an important role in the early development of Xenopus laevis (Pai et al., 2017; Pitcairn et al., 2017). This dominant-negative mutant of DrHCN4 can be used as a tool to elucidate the early developmental and physiological roles of DrHCN4 and DrHCN4L in zebrafish, as in the case of Xenopus laevis.
Pharmacological Properties of Zebrafish HCN4 Channels
We compared the sensitivities of HCN4 channels (HsHCN4, DrHCN4, and DrHCN4L) to common HCN channel inhibitors (ZD7288, ivabradine, and Cs+) (Figure 4). Cs+, ZD7288, and ivabradine similarly inhibited zebrafish HCN4 channels and human HCN4 channels expressed in Xenopus oocytes (Figure 4 and Table 4).
Using developing zebrafish embryos, we investigated whether the administration of HCN inhibitors (ZD7288, ivabradine, and Cs+) in the egg water affected heart rate (Figure 5). The results showed that HCN inhibitors, especially ivabradine, effectively reduced the heart rate of zebrafish. However, the effects of these blockers were not immediately apparent, and a significant difference was seen only after several hours. In some previous studies using other compounds, there was also a waiting period of 4–24 h before cardiotoxicity was observed, partly because compounds must be taken up through the epidermis without mouth opening at the early stages (Bauer et al., 2021). In recent pharmacological experiments using adult zebrafish, intraperitoneal injection of zatebradine reduces maximum heart rate by 38–65% depending on the test temperatures (Marchant and Farrell, 2019). Isolated adult zebrafish heart is also effectively blocked by bath-applications of ivabradine and Cs+ to by 86 and 73%, respectively (Stoyek et al., 2022). Therefore, uptake (or excretion) might be the reason for ineffectiveness of Cs+ here.
We have to mention that the HCN inhibitors may affect other cardiac ion channels such as voltage-gated sodium channels and hERG, especially at high concentration (Wu et al., 2012; Haechl et al., 2019; Hackl et al., 2022). Inhibiting these “off-target” ion channels by the HCN inhibitors could affect heart rate and might explain why ivabradine and ZD7288 had larger effects than cesium or than the morpholino experiments.
Physiological Functions of DrHCN4 and DrHCN4L in the Heart
To discriminate possible physiological functions and roles of DrHCN4 and DrHCN4L in the heart, we tried to suppress the expression of each gene. In the present study, we used antisense morpholino (MO) to knock down DrHCN4 and DrHCN4L genes because MO knockdown is commonly used in zebrafish (Blum et al., 2015; Fujii et al., 2020; Minhas et al., 2021).
Injection of MO into zebrafish 1-cell embryos resulted in a transient reduction in heart rate (Figure 6). In this knockdown experiment performed using MO, heart rate was temporarily reduced but returned to almost the same level as that in the wild type at 72 hpf. One possible explanation of this transient nature of heart rate reduction is due to compensation mechanism by other HCN channel genes or other pacemaker-related ion channel genes such as T-type Ca2+ channels. Another possibility is degradation of MO. Unfortunately, we could not exclude the latter possibility due to lack of detection tools like antibodies. Therefore, we discuss the following section by assuming MO is effective throughout the experimental time window (∼72 hpf).
Jou et al. previously examined the MO against zebrafish HCN4 and observed a similar heart rate reduction at 36–40 hpf (Jou et al., 2017). However, they did not examine the MO-injected embryos at later stages; therefore, they do not describe the heart rate recovery we observed at 72 h. In addition, the MO used in their study was only for DrHCN4, while ours is the first study in which heart rate was assessed by using an MO targeting both DrHCN4 and DrHCN4L. At 48 hpf, the heart rate was similarly reduced regardless of the knockdown gene (Figure 6B). However, other HCN channels, such as HCN1 and HCN2, may be expressed in zebrafish hearts (Ludwig et al., 1999). In addition, the “Ca2+ clocks” mechanism also contributes to the automaticity of the zebrafish heart (Stoyek et al., 2022). Therefore, heart rate may not be determined only by HCN4 and HCN4L expression. von der Heyde et al. used CRISPR/Cas9 targeting DrHCN4 and DrHCN4L to examine the effects of mutations on heart rate and heart rate variability in zebrafish (von der Heyde et al., 2020). They “unexpectedly” observed a higher heart rate at 2 days post-fertilization (dpf) and 5 dpf in zebrafish with nonsense mutations in both DrHCN4 alleles and speculated that the higher heart rate was facilitated by compensatory expression of HCN channels (von der Heyde et al., 2020). Although we do not know why they did not observe heart rate reduction with DrHCN4 knockout by CRISPR/Cas9, the heart rate recovery we observed may be due to a similar compensation mechanism by HCN expressions, or other pacemaker mechanisms including T-type Ca2+ channels or calcium clock-related proteins.
We also found that knockdown of the HCN4 genes by MO tended to induce pericardial edema (Figures 6D,E). Therefore, DrHCN4 and DrHCN4L may play an important role in controlling heart rate but also in normal function of the heart in the early stages. On the other hand, no pericardial edema was observed in zebrafish treated with ivabradine or ZD7288. Unlike the MO knockdown experiment, the HCN4 channel proteins are present in the plasma membrane during inhibition. Therefore, the presence of the HCN4 channel protein, not the ionic currents, might be required in normal function of the heart at the early stages.
One of the limitations of this study is that our findings may only be applied to embryonic zebrafish. Because zebrafish are transparent during the early stages, it is a great advantage to use them for exploring the physiological functions of organs like the heart in vivo. However, the physiological functions we studied here could differ in adult fish. At least, it has been reported that HCN4 proteins are expressed in putative pacemaker cells in adult zebrafish (Stoyek et al., 2015). We ourselves confirmed that both DrHCN4 and DrHCN4L were expressed in the adult heart and brain (Figure 6A). Still, future works would be required to verify that DrHCN4 and/or DrHCN4L are also physiologically important in adult fish.
Another possible limitation of this study is temperature. As we performed all experiments at room temperature, including two-electrode voltage-clamp and heart rate measurement. However, it is known that temperature has a substantial effect on heart rate and action potentials in zebrafish (Lin et al., 2014; Rayani et al., 2018). It is also known that HCN channels are somewhat temperature-dependent: the value of Q10 (folds of increase for every 10°C) of activation and deactivation kinetics is 3–5, and the Q10 of current amplitude is approximately 2 (Magee, 1998; Elinder et al., 2006). Our experimental conditions (approximately 25°C) are about 3°C lower than the breeding condition (28°C), which could slow activation kinetics up to 1.6 -fold and reduce amplitude to about 80%. Therefore, temperature differences must be considered if the present study’s data is to be translated into in vivo study.
In conclusion, the results of our study suggest that DrHCN4 and DrHCN4L play functional roles in heart rate regulation and possibly normal cardiac function during early stages. Therefore, when using zebrafish to create transgenic lines or disease models of HCN4 channels, it should be taken into consideration that two HCN4 channels are functional in zebrafish.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
The animal study was reviewed and approved by the Animal Experiment Committee of Jichi Medical University.
Author Contributions
JL performed experiments and analyzed data. GK assisted with the two-electrode voltage clamp recordings. BZ assisted with the experiments using zebrafish. JL and KN wrote the initial manuscript. All authors edited the manuscript. KN supervised all of the research.
Funding
This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI (Grant No. 17K08552 and 21K06786 to KN).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank F. Ono (Osaka Medical and Pharmaceutical University) and RIKEN BioResource Research Center for providing wild-type zebrafish. We also thank F. Ono and K. Fujii (Kasaoka Daiichi Hospital) for sharing the antisense morpholino oligonucleotides. We thank Seiko Ohkuma at JMU for her excellent care of the zebrafish used in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2022.901571/full#supplementary-material
Supplementary Figure S1 | Sequence alignment of human and zebrafish HCN4 channels. The amino acid sequences were aligned using Clustal Omega (Sievers et al., 2011), and are shown using ESPript3 (Robert and Gouet, 2014). The S1 to S6 segments and CNBD domain from human HCN4 structure (PDB: 6GYO) are labeled above the alignments. The selective filter “GYG motif” is highlighted with an orange square. The residues involved in the cAMP binding and mutated in this study are highlighted with red dots. The red arrow indicates where the chimera constructs (4-4L and 4L-4 in Figure 2) are switched.
Supplementary Figure S2 | Control MO or water injection do not affect heart rate. Averaged heart rates of uninjected, water-injected, and control MO-injected zebrafish embryos (52 hpf).
References
Altomare C., Terragni B., Brioschi C., Milanesi R., Pagliuca C., Viscomi C., et al. (2003). Heteromeric HCN1-HCN4 Channels: a Comparison with Native Pacemaker Channels from the Rabbit Sinoatrial Node. J. Physiology 549, 347–359. doi:10.1113/jphysiol.2002.027698
Alvarez-Baron C. P., Klenchin V. A., Chanda B. (2018). Minimal Molecular Determinants of Isoform-specific Differences in Efficacy in the HCN Channel Family. J. Gen. Physiol. 150, 1203–1213. doi:10.1085/jgp.201812031
Amores A., Force A., Yan Y.-L., Joly L., Amemiya C., Fritz A., et al. (1998). Zebrafish Hox Clusters and Vertebrate Genome Evolution. Science 282, 1711–1714. doi:10.1126/science.282.5394.1711
Baruscotti M., Bucchi A., Difrancesco D. (2005). Physiology and Pharmacology of the Cardiac Pacemaker (“funny”) Current. Pharmacol. Ther. 107, 59–79. doi:10.1016/j.pharmthera.2005.01.005
Baruscotti M., Bucchi A., Viscomi C., Mandelli G., Consalez G., Gnecchi-Rusconi T., et al. (2011). Deep Bradycardia and Heart Block Caused by Inducible Cardiac-specific Knockout of the Pacemaker Channel Gene Hcn4. Proc. Natl. Acad. Sci. U.S.A. 108, 1705–1710. doi:10.1073/pnas.1010122108
Bauer B., Mally A., Liedtke D. (2021). Zebrafish Embryos and Larvae as Alternative Animal Models for Toxicity Testing. Ijms 22, 13417. doi:10.3390/ijms222413417
Beaumont V., Zucker R. S. (2000). Enhancement of Synaptic Transmission by Cyclic AMP Modulation of Presynaptic Ih Channels. Nat. Neurosci. 3, 133–141. doi:10.1038/72072
Blum M., De Robertis E. M., Wallingford J. B., Niehrs C. (2015). Morpholinos: Antisense and Sensibility. Dev. Cell 35, 145–149. doi:10.1016/j.devcel.2015.09.017
BoSmith R. E., Briggs I., Sturgess N. C. (1993). Inhibitory Actions of ZENECA ZD7288 on Whole-Cell Hyperpolarization Activated Inward Current (If) in guinea-pig Dissociated Sinoatrial Node Cells. Br. J. Pharmacol. 110, 343–349. doi:10.1111/j.1476-5381.1993.tb13815.x
Brioschi C., Micheloni S., Tellez J. O., Pisoni G., Longhi R., Moroni P., et al. (2009). Distribution of the Pacemaker HCN4 Channel mRNA and Protein in the Rabbit Sinoatrial Node. J. Mol. Cell. Cardiol. 47, 221–227. doi:10.1016/j.yjmcc.2009.04.009
Brown H., Difrancesco D. (1980). Voltage-clamp Investigations of Membrane Currents Underlying Pace-Maker Activity in Rabbit Sino-Atrial Node. J. Physiol. 308, 331–351. doi:10.1113/jphysiol.1980.sp013474
Bucchi A., Baruscotti M., DiFrancesco D. (2002). Current-dependent Block of Rabbit Sino-Atrial Node if Channels by Ivabradine. J. Gen. Physiol. 120, 1–13. doi:10.1085/jgp.20028593
Bucchi A., Baruscotti M., Nardini M., Barbuti A., Micheloni S., Bolognesi M., et al. (2013). Identification of the Molecular Site of Ivabradine Binding to HCN4 Channels. PLoS One 8, e53132. doi:10.1371/journal.pone.0053132
Bucchi A., Tognati A., Milanesi R., Baruscotti M., DiFrancesco D. (2006). Properties of Ivabradine-Induced Block of HCN1 and HCN4 Pacemaker Channels. J. Physiol. 572, 335–346. doi:10.1113/jphysiol.2005.100776
Chen S., Wang J., Siegelbaum S. A. (2001). Properties of Hyperpolarization-Activated Pacemaker Current Defined by Coassembly of HCN1 and HCN2 Subunits and Basal Modulation by Cyclic Nucleotide. J. Gen. Physiol. 117, 491–504. doi:10.1085/jgp.117.5.491
Cheng L., Kinard K., Rajamani R., Sanguinetti M. C. (2007). Molecular Mapping of the Binding Site for a Blocker of Hyperpolarization-Activated, Cyclic Nucleotide-Modulated Pacemaker Channels. J. Pharmacol. Exp. Ther. 322, 931–939. doi:10.1124/jpet.107.121467
Dai G., Aman T. K., DiMaio F., Zagotta W. N. (2019). The HCN Channel Voltage Sensor Undergoes a Large Downward Motion during Hyperpolarization. Nat. Struct. Mol. Biol. 26, 686–694. doi:10.1038/s41594-019-0259-1
Decher N., Bundis F., Vajna R., Steinmeyer K. (2003). KCNE2 Modulates Current Amplitudes and Activation Kinetics of HCN4: Influence of KCNE Family Members on HCN4 Currents. Pflugers Arch. - Eur. J. Physiol. 446, 633–640. doi:10.1007/s00424-003-1127-7
Elinder F., Männikkö R., Pandey S., Larsson H. P. (2006). Mode Shifts in the Voltage Gating of the Mouse and Human HCN2 and HCN4 Channels. J. Physiol. 575, 417–431. doi:10.1113/jphysiol.2006.110437
Fenske S., Hennis K., Rötzer R. D., Brox V. F., Becirovic E., Scharr A., et al. (2020). cAMP-Dependent Regulation of HCN4 Controls the Tonic Entrainment Process in Sinoatrial Node Pacemaker Cells. Nat. Commun. 11, 5555. doi:10.1038/s41467-020-19304-9
Fujii K., Nakajo K., Egashira Y., Yamamoto Y., Kitada K., Taniguchi K., et al. (2020). Gastrointestinal Neurons Expressing HCN4 Regulate Retrograde Peristalsis. Cell Rep. 30, 2879–2888. e3. doi:10.1016/j.celrep.2020.02.024
Glasauer S. M. K., Neuhauss S. C. F. (2014). Whole-genome Duplication in Teleost Fishes and its Evolutionary Consequences. Mol. Genet. Genomics 289, 1045–1060. doi:10.1007/s00438-014-0889-2
Hackl B., Lukacs P., Ebner J., Pesti K., Haechl N., Földi M. C., et al. (2022). The Bradycardic Agent Ivabradine Acts as an Atypical Inhibitor of Voltage-Gated Sodium Channels. Front. Pharmacol. 13, 809802. doi:10.3389/fphar.2022.809802
Haechl N., Ebner J., Hilber K., Todt H., Koenig X. (2019). Pharmacological Profile of the Bradycardic Agent Ivabradine on Human Cardiac Ion Channels. Cell Physiol. Biochem. 53, 36–48. doi:10.33594/000000119
Herrmann S., Stieber J., Stöckl G., Hofmann F., Ludwig A. (2007). HCN4 Provides a 'depolarization Reserve' and Is Not Required for Heart Rate Acceleration in Mice. EMBO J. 26, 4423–4432. doi:10.1038/sj.emboj.7601868
Jou C. J., Arrington C. B., Barnett S., Shen J., Cho S., Sheng X., et al. (2017). A Functional Assay for Sick Sinus Syndrome Genetic Variants. Cell Physiol. Biochem. 42, 2021–2029. doi:10.1159/000479897
Kanda Y. (2013). Investigation of the Freely Available Easy-To-Use Software 'EZR' for Medical Statistics. Bone Marrow Transpl. 48, 452–458. doi:10.1038/bmt.2012.244
Liman E. R., Tytgat J., Hess P. (1992). Subunit Stoichiometry of a Mammalian K+ Channel Determined by Construction of Multimeric cDNAs. Neuron 9, 861–871. doi:10.1016/0896-6273(92)90239-a
Lin E., Ribeiro A., Ding W., Hove-Madsen L., Sarunic M. V., Beg M. F., et al. (2014). Optical Mapping of the Electrical Activity of Isolated Adult Zebrafish Hearts: Acute Effects of Temperature. Am. J. Physiology-Regulatory, Integr. Comp. Physiology 306, R823–R836. doi:10.1152/ajpregu.00002.2014
Liu Y., Xu X., Gao J., Naffaa M. M., Liang H., Shi J., et al. (2020). A PIP2 Substitute Mediates Voltage Sensor-Pore Coupling in KCNQ Activation. Commun. Biol. 3, 385. doi:10.1038/s42003-020-1104-0
Ludwig A., Zong X., Jeglitsch M., Hofmann F., Biel M. (1998). A Family of Hyperpolarization-Activated Mammalian Cation Channels. Nature 393, 587–591. doi:10.1038/31255
Ludwig A., Zong X., Stieber J., Hullin R., Hofmann F., Biel M. (1999). Two Pacemaker Channels from Human Heart with Profoundly Different Activation Kinetics. EMBO J. 18, 2323–2329. doi:10.1093/emboj/18.9.2323
Maciag M., Wnorowski A., Bednarz K., Plazinska A. (2022). Evaluation of β-adrenergic Ligands for Development of Pharmacological Heart Failure and Transparency Models in Zebrafish. Toxicol. Appl. Pharmacol. 434, 115812. doi:10.1016/j.taap.2021.115812
MacRae C. A., Peterson R. T. (2015). Zebrafish as Tools for Drug Discovery. Nat. Rev. Drug Discov. 14, 721–731. doi:10.1038/nrd4627
Magee J. C. (1998). Dendritic Hyperpolarization-Activated Currents Modify the Integrative Properties of Hippocampal CA1 Pyramidal Neurons. J. Neurosci. 18, 7613–7624. doi:10.1523/JNEUROSCI.18-19-07613.1998
Magee K. E. A., Madden Z., Young E. C. (2015). HCN Channel C-Terminal Region Speeds Activation Rates Independently of Autoinhibition. J. Membr. Biol. 248, 1043–1060. doi:10.1007/s00232-015-9816-7
Männikkö R., Elinder F., Larsson H. P. (2002). Voltage-sensing Mechanism Is Conserved Among Ion Channels Gated by Opposite Voltages. Nature 419, 837–841. doi:10.1038/nature01038
Marchant J. L., Farrell A. P. (2019). Membrane and Calcium Clock Mechanisms Contribute Variably as a Function of Temperature to Setting Cardiac Pacemaker Rate in Zebrafish Danio rerio. J. Fish. Biol. 95, 1265–1274. doi:10.1111/jfb.14126
Milan D. J., Jones I. L., Ellinor P. T., MacRae C. A. (2006). In Vivo recording of Adult Zebrafish Electrocardiogram and Assessment of Drug-Induced QT Prolongation. Am. J. Physiology-Heart Circulatory Physiology 291, H269–H273. doi:10.1152/ajpheart.00960.2005
Milanesi R., Baruscotti M., Gnecchi-Ruscone T., DiFrancesco D. (2006). Familial Sinus Bradycardia Associated with a Mutation in the Cardiac Pacemaker Channel. N. Engl. J. Med. 354, 151–157. doi:10.1056/NEJMoa052475
Minhas R., Loeffler-Wirth H., Siddiqui Y. H., Obrębski T., Vashisht S., Nahia K. A., et al. (2021). Transcriptome Profile of the Sinoatrial Ring Reveals Conserved and Novel Genetic Programs of the Zebrafish Pacemaker. BMC Genomics 22, 715. doi:10.1186/s12864-021-08016-z
Moosmang S., Stieber J., Zong X., Biel M., Hofmann F., Ludwig A. (2001). Cellular Expression and Functional Characterization of Four Hyperpolarization-Activated Pacemaker Channels in Cardiac and Neuronal Tissues. Eur. J. Biochem. 268, 1646–1652. doi:10.1046/j.1432-1327.2001.02036.x10.1046/j.1432-1033.2001.02036.x
Narumanchi S., Wang H., Perttunen S., Tikkanen I., Lakkisto P., Paavola J. (2021). Zebrafish Heart Failure Models. Front. cell Dev. Biol. 9, 662583. doi:10.3389/fcell.2021.662583
Nemtsas P., Wettwer E., Christ T., Weidinger G., Ravens U. (2010). Adult Zebrafish Heart as a Model for Human Heart? an Electrophysiological Study. J. Mol. Cell. Cardiol. 48, 161–171. doi:10.1016/j.yjmcc.2009.08.034
Noam Y., Bernard C., Baram T. Z. (2011). Towards an Integrated View of HCN Channel Role in Epilepsy. Curr. Opin. Neurobiol. 21, 873–879. doi:10.1016/j.conb.2011.06.013
Pai V. P., Willocq V., Pitcairn E. J., Lemire J. M., Paré J.-F., Shi N.-Q., et al. (2017). HCN4 Ion Channel Function Is Required for Early Events that Regulate Anatomical Left-Right Patterning in a Nodal- and Lefty Asymmetric Gene Expression-independent Manner. Biol. Open 6, 1445–1457. doi:10.1242/bio.025957
Pitcairn E., Harris H., Epiney J., Pai V. P., Lemire J. M., Ye B., et al. (2017). Coordinating Heart Morphogenesis: A Novel Role for Hyperpolarization-Activated Cyclic Nucleotide-Gated (HCN) Channels during Cardiogenesis inXenopus Laevis. Commun. Integr. Biol. 10, e1309488. doi:10.1080/19420889.2017.1309488
Ravens U. (2018). Ionic Basis of Cardiac Electrophysiology in Zebrafish Compared to Human Hearts. Prog. Biophysics Mol. Biol. 138, 38–44. doi:10.1016/j.pbiomolbio.2018.06.008
Rayani K., Lin E., Craig C., Lamothe M., Shafaattalab S., Gunawan M., et al. (2018). Zebrafish as a Model of Mammalian Cardiac Function: Optically Mapping the Interplay of Temperature and Rate on Voltage and Calcium Dynamics. Prog. Biophysics Mol. Biol. 138, 69–90. doi:10.1016/j.pbiomolbio.2018.07.006
Rivolta I., Binda A., Masi A., DiFrancesco J. C. (2020). Cardiac and Neuronal HCN Channelopathies. Pflugers Arch. - Eur. J. Physiol. 472, 931–951. doi:10.1007/s00424-020-02384-3
Robert X., Gouet P. (2014). Deciphering Key Features in Protein Structures with the New ENDscript Server. Nucleic Acids Res. 42, W320–W324. doi:10.1093/nar/gku316
Santoro B., Chen S., Lüthi A., Pavlidis P., Shumyatsky G. P., Tibbs G. R., et al. (2000). Molecular and Functional Heterogeneity of Hyperpolarization-Activated Pacemaker Channels in the Mouse CNS. J. Neurosci. 20, 5264–5275. doi:10.1523/JNEUROSCI.20-14-05264.2000
Saponaro A., Bauer D., Giese M. H., Swuec P., Porro A., Gasparri F., et al. (2021). Gating Movements and Ion Permeation in HCN4 Pacemaker Channels. Mol. Cell 81, 2929–2943. e6. doi:10.1016/j.molcel.2021.05.033
Sartiani L., Mannaioni G., Masi A., Novella Romanelli M., Cerbai E. (2017). The Hyperpolarization-Activated Cyclic Nucleotide-Gated Channels: from Biophysics to Pharmacology of a Unique Family of Ion Channels. Pharmacol. Rev. 69, 354–395. doi:10.1124/pr.117.014035
Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., et al. (2011). Fast, Scalable Generation of High‐quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 7, 539. doi:10.1038/msb.2011.75
Stoyek M. R., Croll R. P., Smith F. M. (2015). Intrinsic and Extrinsic Innervation of the Heart in Zebrafish (Danio rerio). J. Comp. Neurol. 523, 1683–1700. doi:10.1002/cne.23764
Stoyek M. R., MacDonald E. A., Mantifel M., Baillie J. S., Selig B. M., Croll R. P., et al. (2022). Drivers of Sinoatrial Node Automaticity in Zebrafish: Comparison with Mechanisms of Mammalian Pacemaker Function. Front. Physiol. 13, 818122. doi:10.3389/fphys.2022.818122
Stoyek M. R., Quinn T. A. (2018). One Fish, Two Fish, Red Fish, Blue Fish*: Zebrafish as a Model for Cardiac Research. Prog. Biophysics Mol. Biol. 138, 1–2. doi:10.1016/j.pbiomolbio.2018.11.003
Swedberg K., Komajda M., Böhm M., Borer J. S., Ford I., Dubost-Brama A., et al. (2010). Ivabradine and Outcomes in Chronic Heart Failure (SHIFT): a Randomised Placebo-Controlled Study. Lancet 376, 875–885. doi:10.1016/S0140-6736(10)61198-1
Verkerk A., Wilders R. (2015). Pacemaker Activity of the Human Sinoatrial Node: an Update on the Effects of Mutations in HCN4 on the Hyperpolarization-Activated Current. Ijms 16, 3071–3094. doi:10.3390/ijms16023071
von der Heyde B., Emmanouilidou A., Mazzaferro E., Vicenzi S., Höijer I., Klingström T., et al. (2020). Translating GWAS-Identified Loci for Cardiac Rhythm and Rate Using an In Vivo Image- and CRISPR/Cas9-based Approach. Sci. Rep. 10, 11831. doi:10.1038/s41598-020-68567-1
Vornanen M., Hassinen M. (2016). Zebrafish Heart as a Model for Human Cardiac Electrophysiology. Channels 10, 101–110. doi:10.1080/19336950.2015.1121335
Wu X., Liao L., Liu X., Luo F., Yang T., Li C. (2012). Is ZD7288 a Selective Blocker of Hyperpolarization-Activated Cyclic Nucleotide-Gated Channel Currents? Channels 6, 438–442. doi:10.4161/chan.22209
Xue T., Marbán E., Li R. A. (2002). Dominant-Negative Suppression of HCN1- and HCN2-Encoded Pacemaker Currents by an Engineered HCN1 Construct. Circulation Res. 90, 1267–1273. doi:10.1161/01.res.0000024390.97889.c6
Yanagihara K., Irisawa H. (1980). Inward Current Activated during Hyperpolarization in the Rabbit Sinoatrial Node Cell. Pflugers Arch. 385, 11–19. doi:10.1007/BF00583909
Keywords: HCN4 channels, zebrafish, heart rate, morpholino, two-electrode voltage-clamp, ivabradine
Citation: Liu J, Kasuya G, Zempo B and Nakajo K (2022) Two HCN4 Channels Play Functional Roles in the Zebrafish Heart. Front. Physiol. 13:901571. doi: 10.3389/fphys.2022.901571
Received: 22 March 2022; Accepted: 31 May 2022;
Published: 30 June 2022.
Edited by:
John Bankston, University of Colorado Anschutz Medical Campus, United StatesReviewed by:
Colin Heath Peters, University of Colorado Anschutz Medical Campus, United StatesT. Alexander Quinn, Dalhousie University, Canada
Copyright © 2022 Liu, Kasuya, Zempo and Nakajo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Koichi Nakajo, a25ha2Fqb0BqaWNoaS5hYy5qcA==
 Jiaying Liu
Jiaying Liu Go Kasuya
Go Kasuya Buntaro Zempo
Buntaro Zempo Koichi Nakajo
Koichi Nakajo