Abstract
Exosomes are extracellular membrane-encapsulated vesicles that are released into the extracellular space or biological fluids by many cell types through exocytosis. As a newly identified form of intercellular signal communication, exosomes mediate various pathological and physiological processes by exchanging various active substances between cells. The incidence and mortality of liver diseases is increasing worldwide. Therefore, we reviewed recent studies evaluating the role of exosomes from various sources in the diagnosis and treatment of liver diseases.
Introduction
Liver diseases have a high incidence and mortality worldwide, accounting for approximately 3.5% of all global deaths annually (Byass, 2014; Asrani et al., 2019; Xiao et al., 2019a). In China, approximately 300 million people suffer from liver diseases. Liver diseases also affect 30 million people each year in the United States. In the European Union, 29 million people are affected by liver diseases (Ma et al., 2019). Liver diseases comprise drug-induced liver injury (DILI), hepatic ischemia reperfusion injury (HIRI), hepatic fibrosis (HF), liver failure (LF) and liver cancer, among others (Llovet et al., 2021). DILI is a common and serious adverse drug reaction that can lead to acute liver failure (ALF) and death (Katarey and Verma, 2016; Shen et al., 2019). Pathological types of DILI include inflammatory necrosis, cholestasis, steatosis and steatohepatitis, vascular injury, and mild lesion types (Gasmi and Kleiner, 2020). HIRI is the main cause of liver dysfunction and LF after transplantation (Ingram et al., 2022) and can lead to hepatocyte necrosis and distant organ damage (Hua et al., 2019), which are associated with significant mortality (Cornide-Petronio et al., 2020). HF is the ultimate common pathway for chronic or persistent liver injury and often progresses to life-threatening liver cirrhosis and liver cancer in advanced stages. Mass deposition of extracellular matrix (ECM) is an important feature of HF, with destruction of the normal structure and function of the liver (Aydin and Akcali, 2018). LF is characterized by heptatocytic injury and decreased synthetic function and may be caused by a range of factors (Arshad et al., 2020). Histological analysis of LF typically demonstrates new or old necrotic lesions (Dong et al., 2020). Mortality from liver cancer ranks third in the world for cancer deaths (Sung et al., 2021). The histological types of primary liver cancer are divided into hepatocellular, bile duct epithelial, and mixed types, of which hepatocellular carcinoma is the most common (accounting for more than 90%) (Villanueva, 2019). Without early intervention, liver diseases may rapidly progress and have a dismal prognosis (Asrani et al., 2019). Therefore, there is a clinical need for novel biomarkers related to liver diseases.
There is increasing scientific interest in the role of exosomes in human disease. Exosomes are a specialized type of extracellular vesicle with a diameter of 30–150 nm (Turturici et al., 2014; Yanez-Mo et al., 2015; Tkach and Thery, 2016). Exosomes observed by transmission electron microscopy are typically disc-shaped or hemispherical with a concave surface. Exosomes are formed by cells through the process of “endocytosis-fusion-discharge” (Corrado et al., 2013). Exosomes have been shown to be present in plasma, urine, saliva, and ascites (Hu et al., 2021; Wen et al., 2021; Nafar et al., 2022). A range of cell types secrete exosomes, including tumor, dendritic, and stem cells (Wu et al., 2021a; Wu et al., 2021b; Lyu et al., 2021; Shao et al., 2021). Various surface molecules of exosomes can directly activate cell receptors and are involved in the exchange of substances between cells (Jones et al., 2018; Yu et al., 2021). Furthermore, exosomes can participate in intercellular signal transduction by carrying proteins, nucleic acids, lipids, and other signaling molecules (Eldh et al., 2010; Yu et al., 2021; Osawa et al., 2022).
The liposome membrane of exosomes can prevent degradation of carried contents, of which nucleic acids and proteins are key mediators of downstream functions (Koga et al., 2011; Wang et al., 2021a; Zhao et al., 2021a). Increasing number of studies have shown that nucleic acids and proteins carried by exosomes are involved in the pathogenesis and progression of liver diseases, including roles in tumor growth, cell migration, fibrosis, and regeneration of hepatocytes (Wu et al., 2018; Jiao et al., 2021a; Jiao et al., 2021b; Shi et al., 2021). Exosomes may have potential as biomarkers in liver diseases. Thus, this article summarizes the role of nucleic acids and proteins carried by exosomes in liver diseases (DILI, LIRI, HF, LF, and liver cancer) (Figure 1).
FIGURE 1
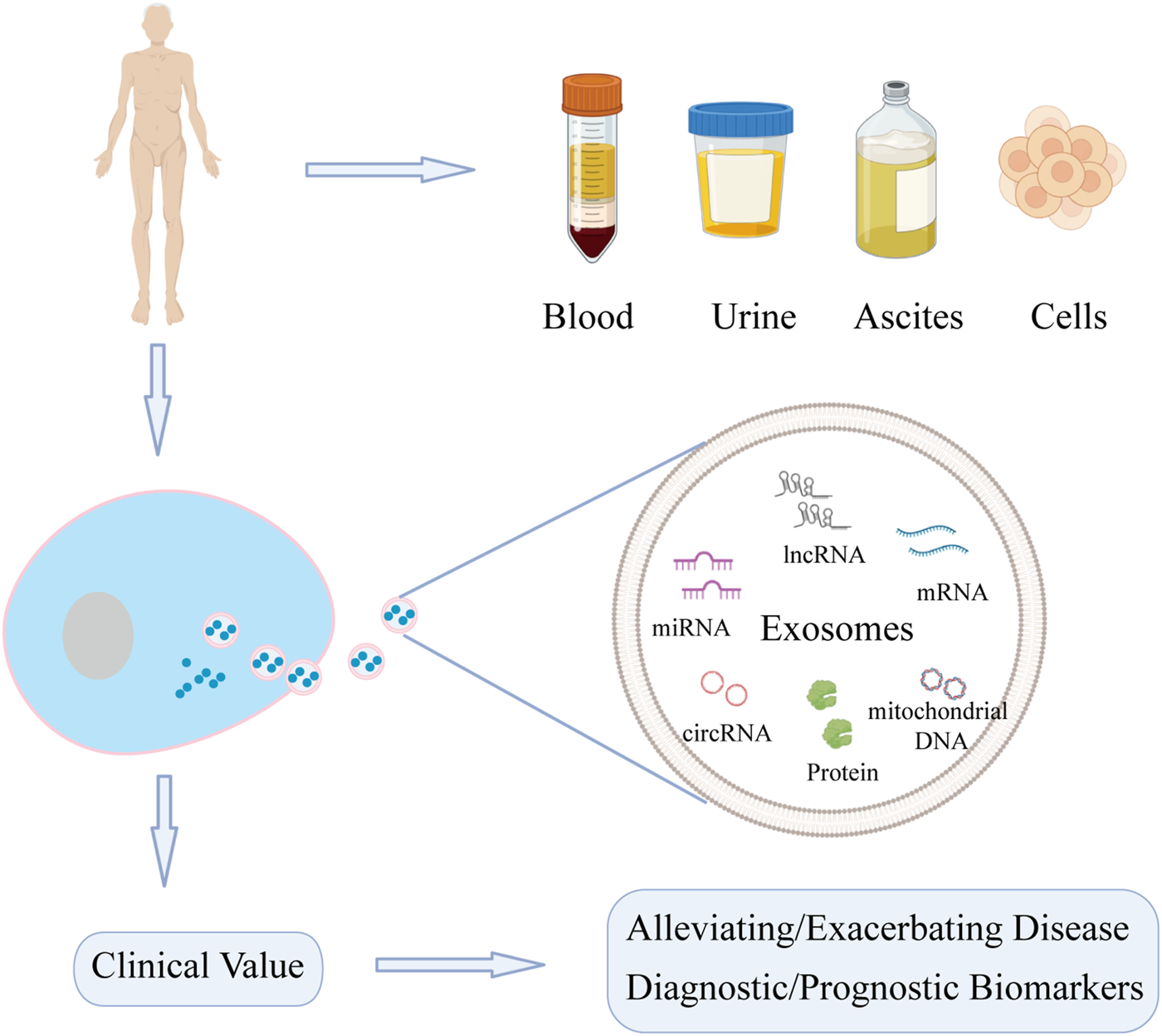
Function of nucleic acids and proteins carried by exosomes from different sources on liver diseases. Exosomes can be extracted from various sources. Nucleic acids and proteins carried by exosomes can not only act as biomarkers for diagnosis and treatment, but also may alleviate or exacerbate liver diseases.
Exosomes and drug-induced liver injury
DILI is defined as liver dysfunction induced by drugs or metabolites and characterized by toxic damage to liver cells or liver injury (Garcia-Cortes et al., 2020). The incidence of DILI has increased in recent years, now accounting for 3%–9% of adverse drug reactions globally (Saithanyamurthi and Faust, 2017). As DILI is predominantly a diagnosis exclusion, there is an urgent need for biomarkers of DILI. DILI has been posited to cause changes in exosomes, with detection of exosomes shown to have utility in the early diagnosis of DILI (Zhao et al., 2021b). In addition, biological molecules carried by exosomes can be delivered to nearby or distant cells, thereby altering their functions and affecting the progression of DILI (Usui and Naisbitt, 2017; Zhao et al., 2021b). Figure 2 and Table 1 show the roles of nucleic acids and proteins carried by exosomes in DILI.
FIGURE 2
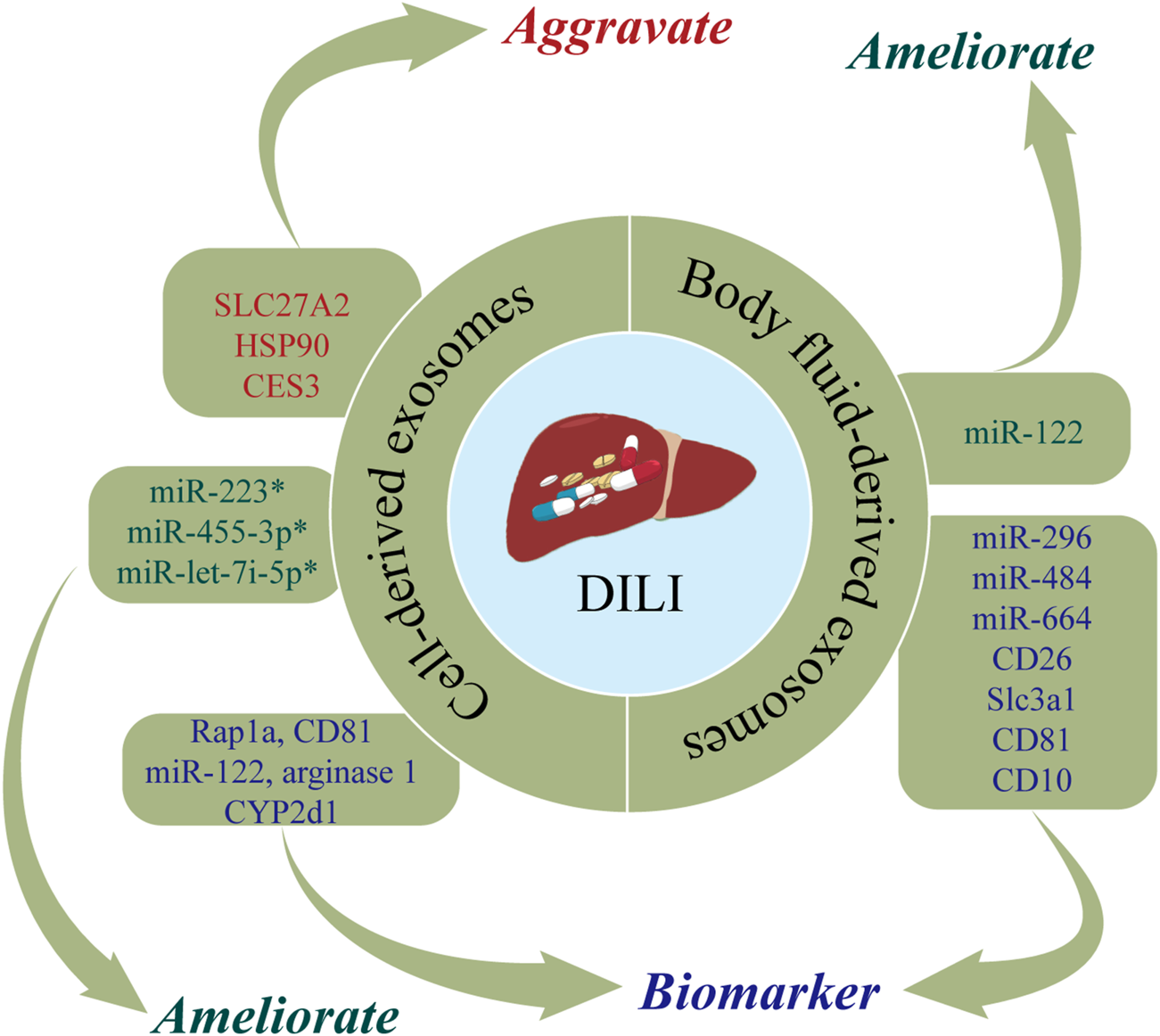
The roles of nucleic acids and proteins carried by exosomes in drug-induced liver injury (DILI). Nucleic acids or proteins that may aggravate DILI are highlighted in red, ameliorating DILI are highlighted in green, and biomarkers are highlighted in purple. * refers to nucleic acids and proteins carried by exosomes from mesenchymal stem cell.
TABLE 1
| Types | Exosomal contents | Source of exosomes | Reference | Functions |
|---|---|---|---|---|
| miR-455-3p | MSC | Shao et al.47 | Reducing inflammation and liver damage | |
| miR-223 | MSC | Chen et al.48 | Exerting the liver protective effect | |
| miR-let-7i-5p | MSC | Chang et al.49 | Ameliorating DILI | |
| miR-122 | Hepatocyte | Holman et al.43 | Evaluating drug hepatotoxicity | |
| Starckx et al.44 | ||||
| miR-122 | Serum/Plasma | Bakshi et al.51 | Ameliorating DILI | |
| miR-192 | Cho et al.52 | Predicting the drug hepatotoxicity | ||
| miRNAs-122a-5p | Serum | Motawi et al.53 | Biomarkers for liver damage | |
| Nucleic acids | miRNAs-192-5p | |||
| miRNAs-193a-3p | ||||
| miR-296 | Urine | Yang et al.55 | Biomarkers for diagnosing DILI | |
| miR-484 | ||||
| miR-434 | ||||
| miR-664 | ||||
| miR-20b-3p | ||||
| miR-34c | ||||
| miR-330 | ||||
| miR-185 | ||||
| miR-291a-5p | ||||
| miR-433 | ||||
| catecholamine-methyl transferase | Hepatocyte | Palomo et al.45 | Biomarkers for diagnosing DILI | |
| arginase 1 | ||||
| CYP2d1 | ||||
| HSP90 hpsa5 | ||||
| fibronectin | ||||
| fibrinogens | ||||
| integrin 1b | ||||
| integrin-linked kinase | ||||
| CD81 angio-poietin-like 4 | ||||
| Rap1a | ||||
| Proteins | CES3 | Hepatocyte | Eva et al.46 | Evaluating the liver injury or aggravating DILI |
| SLC27A2 | ||||
| HSP90 | ||||
| HSP70 | ||||
| FRIL1 | ||||
| PrPc | Urine | Conde-Vancells et al.54 | Biomarkers for diagnosing DILI | |
| CD26 | ||||
| Slc3a1 | ||||
| CD81 | ||||
| CD10 |
Function of nucleic acids and proteins carried by exosomes on drug-induced liver injury.
Note: DILI, drug-induced liver injury; MSC, mesenchymal stem cell.
Cell-derived exosomes
Nucleic acids or proteins carried by exosomes may have utility as biomarkers for predicting DILI. Intrahepatic cell-derived exosomes are significantly changed during DILI. Holman et al. (2016) reported that miR-122 levels were significantly increased in hepatocyte-derived exosomes following acetaminophen treatment. Changes in miR-122 can be detected earlier than traditional liver injury markers, allowing early detection of drug hepatotoxicity (Starckx et al., 2013). In a rat model of DILI, proteomic analysis of hepatocyte-derived exosomes revealed increased levels of enzymes associated with liver injury (catecholamine-methyltransferase, arginase 1, and CYP2d1) and translation-related proteins (Hsp90 and Hpsa5) and decreased levels of apoptosis-regulating proteins (fibronectin, fibrinogen, integrin 1b, integrin-linked kinase, CD81, angiopoietin-like 4, and the RAS-associated protein, Rap1a) (Palomo et al., 2018). A separate study demonstrated increased expression levels of CES3, SLC27A2, HSP90, HSP70, and FRIL1 in hepatocyte-derived exosomes from rats treated with galactosamine (Rodriguez-Suarez et al., 2014).
Nucleic acids or proteins carried by exosomes may alleviate or aggravate DILI. Mesenchymal stem cells (MSCs) are important sources of exosomes. Shao et al. (2020) developed a mouse model of toxin-induced acute liver injury and demonstrated that miR-455-3p in human umbilical cord MSC-derived exosomes (hUC-MSC-exos) reduced the infiltration of macrophages and inflammatory factors, thereby alleviating liver injury. Bone marrow MSC-derived exosomes (BMSC-exos) with high expression of miR-223 markedly reversed S100 and LPS/ATP-induced liver injury by downregulating cytokines, NLRP3, and caspase-1, which further exerted a hepatoprotective effect (Chen et al., 2018). In addition, Chang et al. (2021a) found that miR-let-7i-5p in human placental chorionic MSC-derived exosomes (pcMSC-exos) inhibited apoptosis in hepatocytes, attenuated hepatic inflammatory responses, decreased liver injury scores, and ultimately improved liver injury. In summary, exosomes extracted from a range of intra- and extrahepatic cell types have important roles in DILI.
Body fluid-derived exosomes
Body fluids are carriers of exosomes secreted by distant cell types, thereby allowing exosomes to exert paracrine effects. DILI can cause changes in body fluid-derived exosomes. Nucleic acids or proteins in body fluid-derived exosomes may have utility as biomarkers for predicting DILI. Several studies have demonstrated that miRNAs in humoral-derived exosomes can act as markers of liver injury and inflammation (Arrese et al., 2015). In serum-derived exosomes from patients with DILI induced by anti-tuberculosis drugs (isoniazid, rifampicin, pyrazinamide), miR-122 and miR-192 levels were substantially increased indicating their potential utility as predictors or therapeutic targets for liver damage caused by anti-tuberculosis drugs (Bakshi et al., 2021). A previous study reported that miR-122 and miR-192 were increased in plasma-derived exosomes from acetaminophen-induced DILI in rats, while the opposite results were obtained after adding N-acetylcysteine (Cho et al., 2017). Motawi et al. (2018) reported that serum exosomal miRNAs-122a-5p, 192-5p, and 193a-3p were associated with liver injury indicating their potential utility as markers of liver damage or in determining the etiology of liver injury. Notably, exosomal miRNA-122a-5p had stronger diagnostic performance.
Urine, as an excretory material, also contains abundant exosomes. Conde-Vancells et al., 2010 analyzed protein levels of urine-derived exosomes from a rat model of DILI and found that PrPc, Cd26, Slc3a1, Cd81, and Cd10 had utility as biomarkers for diagnosing DILI. Yang et al. (2012) identified ten urinary miRNAs (miR-296, miR-484, miR-434, miR-664, miR-20b-3p, miR-34c, miR-330, miR-185, miR-291a-5p, and miR-433) in rats treated with acetaminophen or carbon tetrachloride. These miRNAs may be transported in exosomes and urinary miRNAs may help distinguish liver injury due to hepatotoxic drugs from injury due to non-hepatotoxic causes. In addition, urine can be collected in bulk and this procedure is noninvasive, which is a unique advantage of diagnostic tests based on urine-derived exosomes. Taken together, these findings demonstrate that proteins or nucleic acids carried by humoral-derived exosomes may have utility as predictors of hepatotoxicity.
In summary, nucleic acids and proteins carried by exosomes from different sources are important players in DILI and may have utility in predicting the hepatotoxicity of drugs or as therapeutic targets for DILI.
Exosomes and hepatic ischemia reperfusion injury
HIRI is a phenomenon in which hepatocytes are damaged due to transient ischemia, with liver damage further aggravated when blood flow is restored (Konishi and Lentsch, 2017). HIRI typically occurs after traumatic shock, liver surgery, or liver transplantation (LT) (Zhang et al., 2022a). At present, the most commonly used methods for preventing and treating HIRI are ischemic preconditioning (IPC), reducing the ischemia time, and inhibiting the inflammatory response after reperfusion (Wu et al., 2021c). However, due to the poor tolerance of liver tissue to hypoxia, there is an urgent clinical need for novel therapeutic strategies for HIRI. Exosomes are posited to exert protective and regenerative effects in HIRI (Zheng et al., 2018; Zhang et al., 2022b). Figure 3 and Table 2 show the roles of nucleic acids and proteins carried by exosomes in HIRI.
FIGURE 3
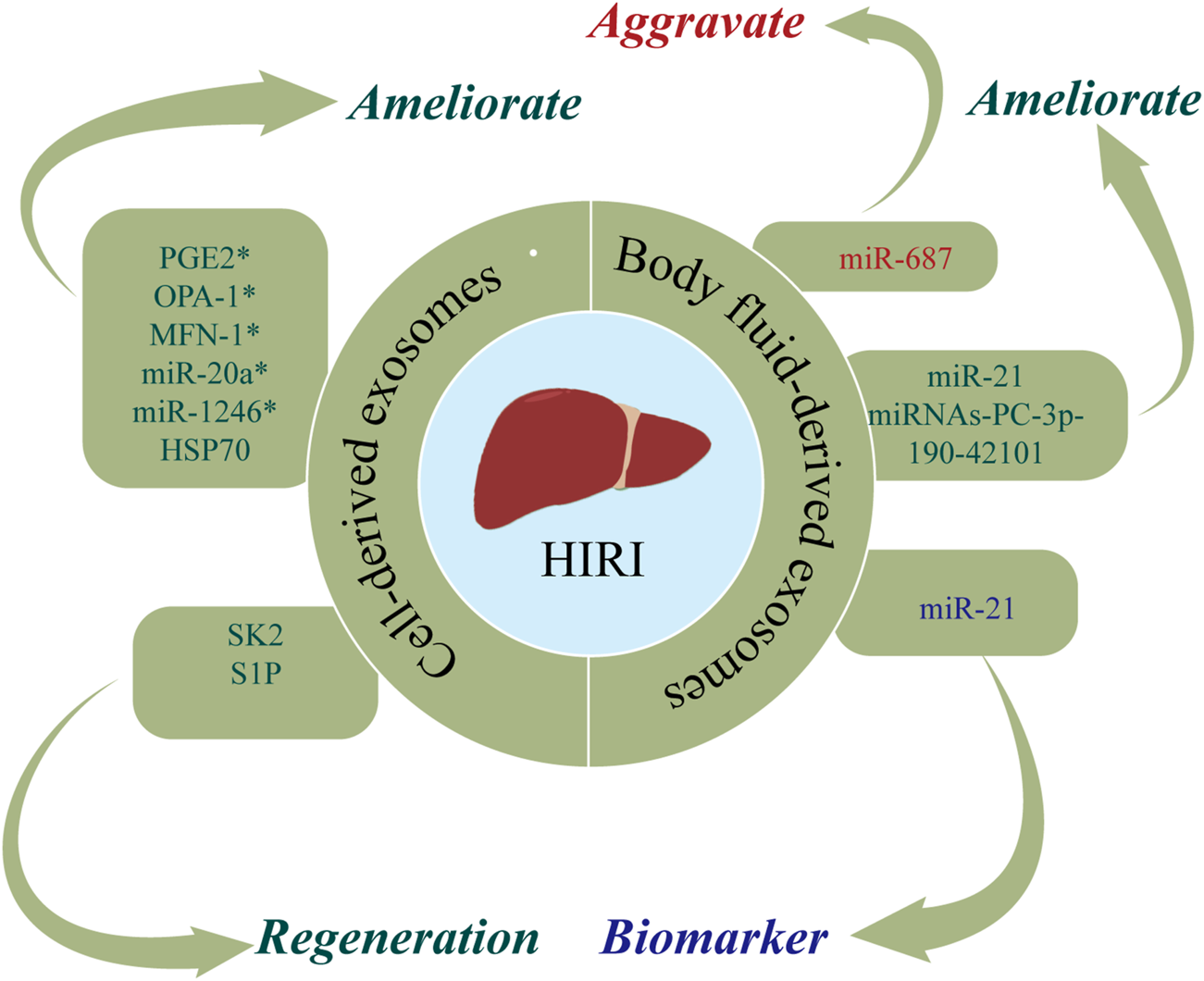
The roles of nucleic acids and proteins carried by exosomes in hepatic ischemia reperfusion injury (HIRI). Nucleic acids or proteins that may aggravate HIRI are highlighted in red, ameliorating HIRI are highlighted in green, and biomarkers are highlighted in purple. * refers to nucleic acids and proteins carried by exosomes from mesenchymal stem cell.
TABLE 2
| Types | Exosomal contents | Source of exosomes | Reference | Functions |
|---|---|---|---|---|
| Nucleic acids | miR-1246 | MSC | Xie et al.63 | Alleviating HIRI |
| Xie et al.64 | ||||
| miR-20a | MSC | Zhang et al.65 | Alleviating HIRI | |
| miRNAs-PC-3p-190-42101 | Plasma | Fei et al.69 | Protect the liver from HIRI | |
| miR-687 | Plasma | Ashour et al.70 | Aggravating HIRI | |
| miR-21 | Serum | Jia et al.71 | Biomarkers for diagnosing | |
| HIRI or alleviating HIRI | ||||
| Proteins | PGE2 | MSC | Zhang et al.60 | Alleviating HIRI |
| OPA-1 | MSC | Zhang et al.66 | Ameliorating HIRI | |
| MFN-1 | ||||
| MFN-2 | ||||
| PGC-1α | ||||
| NRF-1 | ||||
| TFAM | ||||
| LC3-II | MSC | Yang et al.67 | Alleviating HIRI | |
| SK2 | MSC | Du et al.68 | Accelerating liver | |
| SIP | regeneration | |||
| HSP70 | DC | Zheng et al.59 | Alleviating HIRI |
Function of nucleic acids and proteins carried by exosomes on hepatic ischemia reperfusion injury.
Note: HIRI, hepatic ischemia reperfusion injury; MSC, mesenchymal stem cell; DC: dendritic cell.
Cell-derived exosomes
Nucleic acids or proteins carried by exosomes from different cells may alleviate HIRI. MSCs have emerged as a new therapeutic option for HIRI with effects including directed differentiation, induction of angiogenesis, tissue repair, and anti-inflammatory and anti-apoptotic activities (Phinney and Pittenger, 2017; Li et al., 2019). A previous study (Xie et al., 2019a) revealed that hUC-MSCs-exos regulated the GSK3β-mediated Wnt/β-catenin pathway by transporting miR-1246 after hypoxia/reoxygenation in LO2 cells, thereby inhibiting apoptosis and promoting cell proliferation to alleviate HIRI. Xie et al. (2019b) found that hUC-MSCs-exos could also regulate the balance between Tregs and Th17 cells through the miR-1246-mediated IL-6-gp130-STAT3 axis, thereby alleviating HIRI. Moreover, miR-20a in hUC-MSCs-exos can combine with the 3′UTR of Beclin-I and FAS to inhibit their expression, thereby ameliorating apoptosis in HIRI (Zhang et al., 2020a).
Adipose-derived mesenchymal stem cells (ADSCs) have latent effects on HIRI. Zhang et al. (2022b) confirmed that PGE2 in ADSC-derived exosomes (ADSCs-exos) mediated the phosphorylation of ERK1/2 and GSK-3β, upregulated Bcl-2, downregulated Bax, and reduced the levels of reactive oxygen species, thereby ameliorating inflammation, inhibiting apoptosis, and effectively protecting the liver from HIRI. A separate study (Zhang et al., 2021a) found that ADSCs-exos upregulated mitochondrial-associated proteins (OPA-1, MFN-1, MFN-2, PGC-1α, NRF-1, and TFAM), thereby maintaining mitochondrial homeostasis and ameliorating liver dysfunction in a rat model of HIRI. Yang et al. (2020) injected MSC-derived hepatocyte-like cell exosomes (MSC-Heps-exos) into the tail vein of HIRI mice and demonstrated increased levels of LC3-II, a marker of autophagy activity, increased hepatocyte autophagy, and decreased levels of circulating liver enzymes, thereby alleviating HIRI. Du et al. (2017) demonstrated that human-induced pluripotent stem cell-derived MSC-derived exosomes (hiPSC-MSCs-exos) could fuse with hepatocytes, thereby facilitating the synthesis of sphingosine kinase 2 (SK2) and sphingosine-1-phosphate (S1P), improving the tolerance of hepatocytes to hypoxia, increasing cell proliferation, and accelerating liver regeneration after HIRI. Moreover, they found that hepatocytes could activate the S1P pathway in the same way to promote cell proliferation. A further study (Zheng et al., 2018) confirmed that bone marrow-derived dendritic cell-derived exosomes (DEXs) transferred HSP70 into T cells and activated the PI3K/mTOR pathway to regulate the balance between Tregs and Th17 cells, ultimately having a protective effect on HIRI.
Body fluid-derived exosomes
Nucleic acids or proteins in body fluid-derived exosomes may alleviate or aggravate HIRI. Fei et al. (2021) performed next-generation sequencing (NGS) of miRNAs in plasma-derived exosomes and verified that miRNAs-PC-3p-190-42101 in exosomes could reduce the levels of inflammatory factors and protect the liver from HIRI. In a rat model of HIRI, Ashour et al. (2021) reported that plasma exosomes had increased levels of miR-687, in addition to increased levels of liver tissue inflammatory markers and caspase-3. Inhibiting the expression of exosomal miR-687 may protect against hepatic injury. Therefore, exosomal miR-687 may play an important role in inducing HIRI. Furthermore, Jia et al. (2017) confirmed that serum exosomal miR-21 can inhibit the activity of NF-κb, downregulate programmed cell death protein 4, and upregulate bcl-2, thereby inhibiting apoptosis and reducing inflammation. Accordingly, serum exosomal miR-21 may represent a potential therapeutic target for HIRI.
Taken together, these results indicate that nucleic acids and proteins carried by exosomes play important roles in HIRI and may have utility in predicting HIRI. However, different molecules carried by exosomes may exert deleterious or protective effects on HIRI.
Exosomes and hepatic fibrosis
HF is a pathological process of excessive deposition and abnormal distribution of ECM after liver injury due to a range of etiologies (including alcohol, viruses, and autoimmune reactions) (Roehlen et al., 2020). The central link in HF is the activation of hepatic stellate cells (HSCs) (Sun and Kisseleva, 2015). HF may progress to cirrhosis that confers increased risks of liver cancer, LF, and death (Seki and Brenner, 2015). At present, there are no effective anti-fibrotic drug therapies for HF, with clinical treatment predominantly focusing on managing the underlying etiology and symptoms. Therefore, there is a need for studies of the effect of exosomes in HF. Figure 4 and Table 3 show the roles of nucleic acids and proteins carried by exosomes in HF.
FIGURE 4
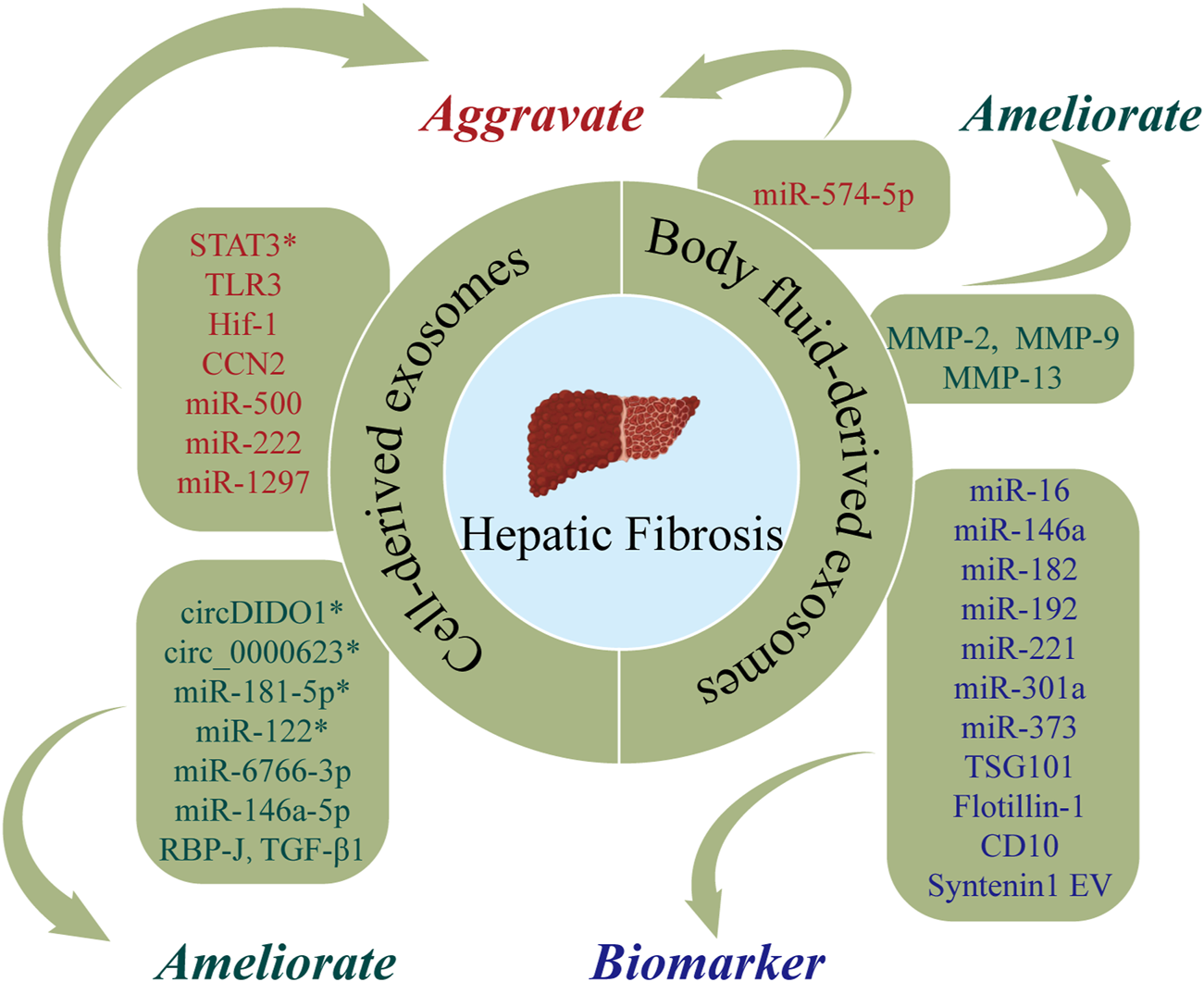
The roles of nucleic acids and proteins carried by exosomes in hepatic fibrosis (HF). Nucleic acids or proteins that may aggravate HF are highlighted in red, ameliorating HF are highlighted in green, and biomarkers are highlighted in purple. * refers to nucleic acids and proteins carried by exosomes from mesenchymal stem cell.
TABLE 3
| Types | Exosomal contents | Source of exosomes | Reference | Functions |
|---|---|---|---|---|
| Nucleic acids | circDIDO1 | MSC | Ma et al.75 | Alleviating HF |
| mmu_circ_0000623 | MSC | Zhu et al.76 | Alleviating HF | |
| miR-181-5p | MSC | Qu et al.77 | Alleviating HF | |
| miR-122 | MSC | Lou et al.78 | Alleviating HF | |
| miR-6766-3p | hESC | Wang et al.79 | Alleviating HF | |
| miR-500 | Macrophage | Chen et al.83 | Aggravating HF | |
| miR-146a-5p | HLSC | Chiabotto et al.84 | Alleviating HF | |
| miR-222 | Hepatocyte | Zhang et al.86 | Aggravating HF | |
| miR-1297 | Hepatocyte | Luo et al.87 | Aggravating HF | |
| lncRNA-H19 | Cholangiocyte | Li et al.88 | Aggravating HF | |
| Liu et al.89 | ||||
| miR-574-5p | Serum | Zhou et al.92 | Aggravating HF | |
| miR-122 | Serum | Chang et al.93 | Prognostic biomarker for HF | |
| circDIDO1 | Serum | Ma et al.75 | Alleviating HF | |
| lncRNA-H19 | Serum | Xiao et al.94 | Aggravating HF | |
| miR-16 | Plasma | Frundt et al.97 | Being diagnostic and prognostic markers for HF | |
| miR-146a | ||||
| miR-192 | ||||
| miR-221 | ||||
| miR-182 | Ascites | Muhammad et al.98 | Prognostic biomarker for HF | |
| miR-301a | ||||
| miR-373 | ||||
| Proteins | RBP-J | HEK293T cell | He et al.80 | Alleviating HF |
| TGF-β1 | NK cell | Wang et al.81 | Alleviating HF | |
| STAT3 | MSC | Tang et al.82 | Aggravating HF | |
| TLR3 | Hepatocyte | Seo et al.85 | Aggravating HF | |
| Hif-1 | HSC | Wan et al.90 | Aggravating HF | |
| CCN2 | HSC | Charrier et al.91 | Aggravating HF | |
| MMP-2 | Plasma | Huang et al.95 | Alleviating HF | |
| MMP-9 | ||||
| MMP-13 | ||||
| TSG101 | Urine | Gonzalez et al.96 | Diagnostic biomarkers for HF | |
| Flotillin-1 | ||||
| CD10 | ||||
| Syntenin1 EV |
Function of nucleic acids and proteins carried by exosomes on hepatic fibrosis.
Note: HF, hepatic fibrosis; MSC, mesenchymal stem cell; hESC, human embryonic stem cell; HLSC, human liver stem cell; HEK293T, human embryonic kidney cell; NK, cell, natural killer cell; HSC, hepatic stellate cell.
Cell-derived exosomes
Nucleic acids or proteins carried by exosomes from different cells may alleviate or aggravate HF. HSCs play important roles in the pathogenesis of HF. Ma et al. (2022) revealed that MSC-derived exosomes delivered circDIDO1 to HSCs, which lead to inhibition of HSC activation through the miR-141-3p/PTEN/AKT pathway, thereby alleviating HF. ADSC-exos modified with mmu_circ_0000623 ameliorated HF by promoting autophagy (Zhu et al., 2020). Similarly, miR-181-5p-modified ADSC-exos activated autophagy and downregulated Stat3 and Bcl-2 in HST-T6 cells, thereby preventing HF (Qu et al., 2017). Lou et al. (2017) reported that MiR-122-modified ADSC-exos inhibited HSC activation, reduced collagen deposition, and ameliorated HF. A further study (Wang et al., 2021b) revealed that miR-6766-3p in 3D cultured human embryonic stem cell-derived exosomes (3D-hESC-exos) repressed the SMAD pathway by downregulating TGFβRII, thereby inhibiting HSC activation and slowing the progression of HF. He et al. (2022) demonstrated that HEK293T-derived exosomes effectively inhibited the Notch pathway in macrophages by delivering the transcription factor, RBP-J, thereby attenuating HF.
Wang et al. (2020) isolated exosomes from NK-92MI cells (NK-exos) and demonstrated that NK-exos inhibited the proliferation and activation of HSCs by downregulating TGF-β1, which further alleviated HF in mice. One study (Tang et al., 2021) designed fibroblast-like MSC-derived exosomes to carry siRNA or antisense oligonucleotides (ASOs) targeting STAT3, demonstrating that iExosiRNA-STAT3 or iExo-mASO-STAT3 downregulated STAT3, reduced ECM deposition, and ameliorated HF. Chen et al. (2021) demonstrated that macrophage-derived exosomal miR-500 promoted the proliferation and activation of HSCs by targeting MFN2, thereby aggravating HF. Chiabotto et al. (2021) concluded that human liver stem cell-derived extracellular vesicles (HLSC-EVs) attenuated the activation of HSCs by delivering miR-146a-5p. Seo et al. (2016) reported that hepatocyte-derived exosomes mediated the activation of TLR3, which upregulated IL-17A and aggravated HF. A recent study found that exosomal miR-222 from HBV-infected hepatocytes accelerated HF by inhibiting the transferrin receptor (TFRC) and ferroptosis (Zhang et al., 2022c). Lipotoxic hepatocyte-derived exosomal miR-1297 can promote the activation of HSCs via activation of the PTEN/PI3K/AKT signaling pathway (Luo et al., 2021). In addition, Li et al. (Li et al., 2018) and Liu et al. (2019) demonstrated that cholangiocyte-derived exosomes promoted the activation of HSCs by delivering lncRNA-H19 and accelerating the progression of cholestatic HF. Wan et al. (2019) revealed that hypoxia-inducible factor 1 (HIF-1) in activated HSC-derived exosomes mediated the transmission of glycolysis-related proteins (GLUT1 and PKM2) and enhanced glycolysis, thereby exacerbating HF. Moreover, Charrier et al. (2014) confirmed that HSC-derived exosomes can also accelerate the activation of HSCs by transmitting connective tissue growth factor (CCN2).
Body fluid-derived exosomes
Nucleic acids or proteins in body fluid-derived exosomes may alleviate or aggravate HF and serve as biomarkers for predicting HF. A previous study (Zhou et al., 2022) extracted serum exosomes from healthy adults and patients with liver cirrhosis and co-cultured exosomes with a human hepatic stellate cell line, LX-2. This study found higher levels of serum exosomal miR-574-5p in patients with liver cirrhosis. Furthermore, serum exosomal miR-574-5p levels have been shown to be positively correlated with collagen deposition and α-SMA in liver tissue of HF mice. Chang et al. (2021b) extracted serum exosomes from 71 patients with HF for NGS, finding that exosomal miR-122 was negatively correlated with the degree of HF and serum exosomal miR-122 could act as a noninvasive predictor for HF. Conversely, downregulation of miR-122 accelerated the progression of HF. Ma et al. (2022) revealed that circDIDO1 in serum exosomes was associated with HF. Xiao et al. (2019b) reported that serum exosomal lncRNA-H19 promoted HF via the S1PR2/SphK2 and let-7/HMGA2 pathways, indicating that serum exosomal lncRNA-H19 may represent a novel therapeutic target for cholestatic HF. A separate study (Huang et al., 2021) found that upregulation of matrix metalloproteinases (MMP-2, MMP-9, MMP-13) in human umbilical cord blood plasma-derived exosomes (hUCB-exos) inhibited the accumulation of ECM and the progression of HF. Gonzalez et al. (2021) performed proteomic analysis of urinary extracellular vesicles (uEVs) from normal adults and patients with liver cirrhosis and identified 1,304 proteins. The levels of 90 proteins (such as TSG101, flotillin-1, CD10, and syntenin 1) were significantly altered, and these proteins were proposed as potential diagnostic biomarkers for liver cirrhosis. Fründt et al. (2021) posited that plasma exosomal miR-16, miR-146a, miR-192, and miR-221 are promising diagnostic and prognostic markers for liver cirrhosis. A further study (Muhammad Yusuf et al., 2020) showed that levels of ascites-derived exosomal miR-182, miR-301a, and miR-373 were elevated in patients with liver cirrhosis, indicating that these miRNAs may have utility as biomarkers in patients with liver cirrhosis.
Taken together, these studies demonstrate that nucleic acids and proteins carried by exosomes from different sources may aggravate or alleviate HF in certain conditions and have utility as biomarkers for predicting HF.
Exosomes and liver failure
LF is a clinical syndrome characterized by severe liver damage, coagulation disorders, jaundice, hepatic encephalopathy, and ascites (Liver, 2019). It has a high incidence, high mortality, and low cure rate (Putignano et al., 2018; Jalan et al., 2021). There are currently no specific pharmaceutical treatments for LF, with liver transplantation representing the only curative treatment option (Liver, 2019). However, LT is constrained by organ shortages, high costs, and the use of immunosuppressive drugs (Trebicka et al., 2020). Accordingly, there is an urgent need for novel treatments for LF. Figure 5 and Table 4 show the roles of nucleic acids and proteins carried by exosomes in LF.
FIGURE 5
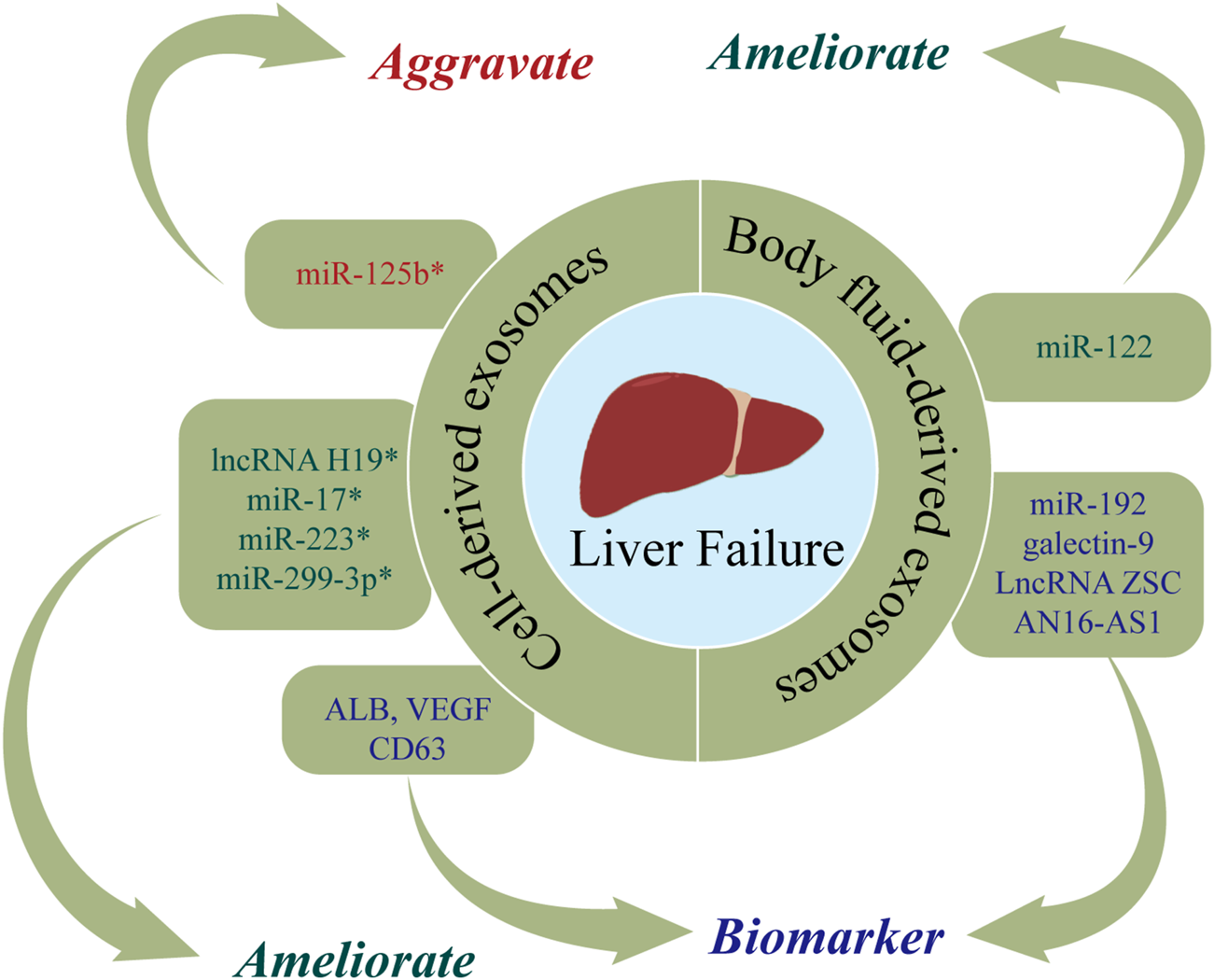
The roles of nucleic acids and proteins carried by exosomes in liver failure (LF). Nucleic acids or proteins that may aggravate LF are highlighted in red, ameliorating LF are highlighted in green, and biomarkers are highlighted in purple. * refers to nucleic acids and proteins carried by exosomes from mesenchymal stem cell.
TABLE 4
| Types | Exosomal contents | Source of exosomes | Reference | Functions |
|---|---|---|---|---|
| Nucleic acids | miR-17 | MSC | Liu et al.103 | Alleviating LF |
| lncRNA H19 | MSC | Jin et al.104 | Alleviating LF | |
| miR-223 | MSC | Chen et al.48 | Alleviating LF | |
| miR-122 | MSC | Lou et al.78 | Alleviating LF | |
| miRNA-125b | MSC | Hyun et al.107 | Aggravating LF | |
| miR-299-3p | MSC | Zhang et al.108 | Alleviating LF | |
| miR-20a-5p | Hepatocyte | Zhang et al.111 | Alleviating LF | |
| NOX1 mRNA | Serum | Chen et al.113 | Biomarkers for predicting LF | |
| LncRNA ZSCAN16-AS1 | ||||
| miR-122 | Plasma | Baker et al.114 | Prognostic markers for predicting LF | |
| miR-192 | ||||
| Proteins | GPX1 | MSC | Yan et al.105 | Alleviating LF |
| ERK1/2 | MSC | Wu et al.106 | Alleviating LF | |
| PI3K | ||||
| CRP | MSC | Jun et al.109 | Alleviating LF | |
| ICAM-1 angiopoietin-2 | MenSC | Chen et al.110 | Alleviating LF | |
| Axl | ||||
| Angiogenin | ||||
| IGFBP-6 | ||||
| Osteoprotegerin | ||||
| IL-6 | ||||
| IL-8 | ||||
| ALB | Hepatocyte | Jiao et al.38 | Prognostic biomarkers for LF | |
| CD63 | ||||
| VEGF | ||||
| Galectin-9 | Plasma | Zhang et al.112 | Being a prognostic marker for LF |
Function of nucleic acids and proteins carried by exosomes on liver failure.
Note: LF, liver failure; MSC, mesenchymal stem cell; MenSC, human menstrual blood stem cell.
Cell-derived exosomes
Nucleic acids or proteins carried by exosomes from mesenchymal stem cells may alleviate or aggravate LF. Liu et al. (2018) found that ADSC-exos had therapeutic efficacy in ALF, while the effect was abolished after miR-17 knockout. MiR-17 ameliorated GalN/TNF-α-induced ALF by blocking the activation of NLRP3 in macrophages through targeting of TXNIP. Jin et al. (2018) injected exosomes derived from human adipose stem cells (hASCs) into ALF rats and observed that lncRNA H19 was upregulated, which in turn promoted the proliferation of hepatocytes and improved the survival rate of rats. The survival rate was reduced to 40% when lncRNA-H19 was silenced. A separate study (Yan et al., 2017) found that glutathione peroxidase 1 (GPX1) in hUC-MSCs-exos could counteract the toxic effects of CCl4 or H2O2, thereby reducing oxidative stress and exerting protective effects on LF. When GPX1 was knocked out, the protective effect was correspondingly attenuated. Wu et al. (2021d) found that hUC-MSCs-exos suppressed apoptosis and improved ALF by upregulating ERK1/2 and PI3K/AKT pathways. The opposite results were obtained with the addition of a PI3K or ERK1/2 inhibitor. Chen et al. (2018) infected BMSCs with pre-miR-223 and extracted BMSCs-exomiR-223 (+), demonstrating that BMSCs-exomiR-223 (+) decreased serum levels of ALT and AST, downregulated NLRP3 and caspase-1, and reversed liver injury. Furthermore, miR-122-modified ADSCs-exos attenuated collagen deposition by inhibiting the activation of HSCs (Lou et al., 2017). Hyun et al. (Hyun et al., 2015) revealed that inhibiting miRNA-125b in exosomes extracted from chorionic plate-derived MSCs (CP-MSCs) resulted in upregulation of Hh in HSCs, which in turn exacerbated LF. A further study (Zhang et al., 2020b) found that hUC-MSCs-exos inhibit activation of the NLRP3 pathway by delivering miR-299-3p, thereby reducing inflammation and promoting tissue repair. Jun et al. (2020) demonstrated that exosomal CRP from placental MSCs upregulated factors related to the Wnt pathway and angiogenesis in a rat model of LF, thereby promoting angiogenesis and liver regeneration. Chen et al. (2017) reported that exosomes extracted from human menstrual blood stem cells (MenSC-exos) inhibited apoptosis by promoting the expression of cytokines (ICAM-1, angiopoietin-2, Axl, angiogenin, IGFBP-6, osteoprotegerin, IL-6, and IL-8), and downregulating caspase-3.
Nucleic acids or proteins carried by exosomes from hepatocytes may alleviate LF and serve as important markers for predicting LF. Jiao et al. (2021b) extracted exosomes from hepatocytes of patients with acute-on-chronic LF (ACLF), demonstrating increased levels of ALB, CD63, and VEGF, which may represent more accurate prognostic indicators than alpha-fetoprotein (AFP). A separate study (Zhang et al., 2021b) indicated that miR-20a-5p was downregulated in hepatocyte-derived exosomes from ACLF mice, which in turn led to the upregulation of CXCL8 and increased inflammation. However, CXCL8 levels were decreased and liver injury was markedly alleviated after upregulation of exosomal miR-20a-5p.
Body fluid-derived exosomes
Nucleic acids or proteins in body fluid-derived exosomes may have utility as markers for predicting LF. Zhang et al. (2019) revealed that plasma levels of exosomal galectin-9 in LF patients with acute cellular rejection were associated with poor prognosis, indicating that galectin-9 may be a predictor of rejection after liver transplantation. Chen et al. (2020) performed RNA sequencing of serum exosomes from normal adults and patients with ACLF caused by HBV (HBV-ACLF). They found that NOX1 mRNA and LncRNA ZSCAN16-AS1 were upregulated, indicating their potential utility as predictors for HBV-ACLF. Furthermore, Baker et al. (2015) reported that plasma exosomal miR-122 and miR-192 levels were increased at the onset of ALF, indicating their potential efficacy in predicting LF.
In conclusion, nucleic acids and proteins carried by exosomes from different cells may alleviate or aggravate LF and serve as important markers for predicting LF.
Exosomes and liver cancer
Hepatocellular carcinoma (HCC) accounts for 75–85% of all liver cancers (Bray et al., 2018). It typically has an insidious onset, rapid development, and high mortality (Kulik and El-Serag, 2019). More than 800,000 people die from HCC each year, with a 5-years survival rate of approximately 6% (Raees et al., 2021). Therefore, there is an urgent clinical need for biomarkers that may contribute to the diagnosis and treatment of HCC. Increasing evidence indicates that the contents of exosomes are linked with tumor invasiveness and the tumor microenvironment and may influence the occurrence and development of HCC through related signaling pathways (An et al., 2018; Hwang and Yang, 2021). Figure 6 and Table 5 show the roles of nucleic acids and proteins carried by exosomes in liver cancer.
FIGURE 6
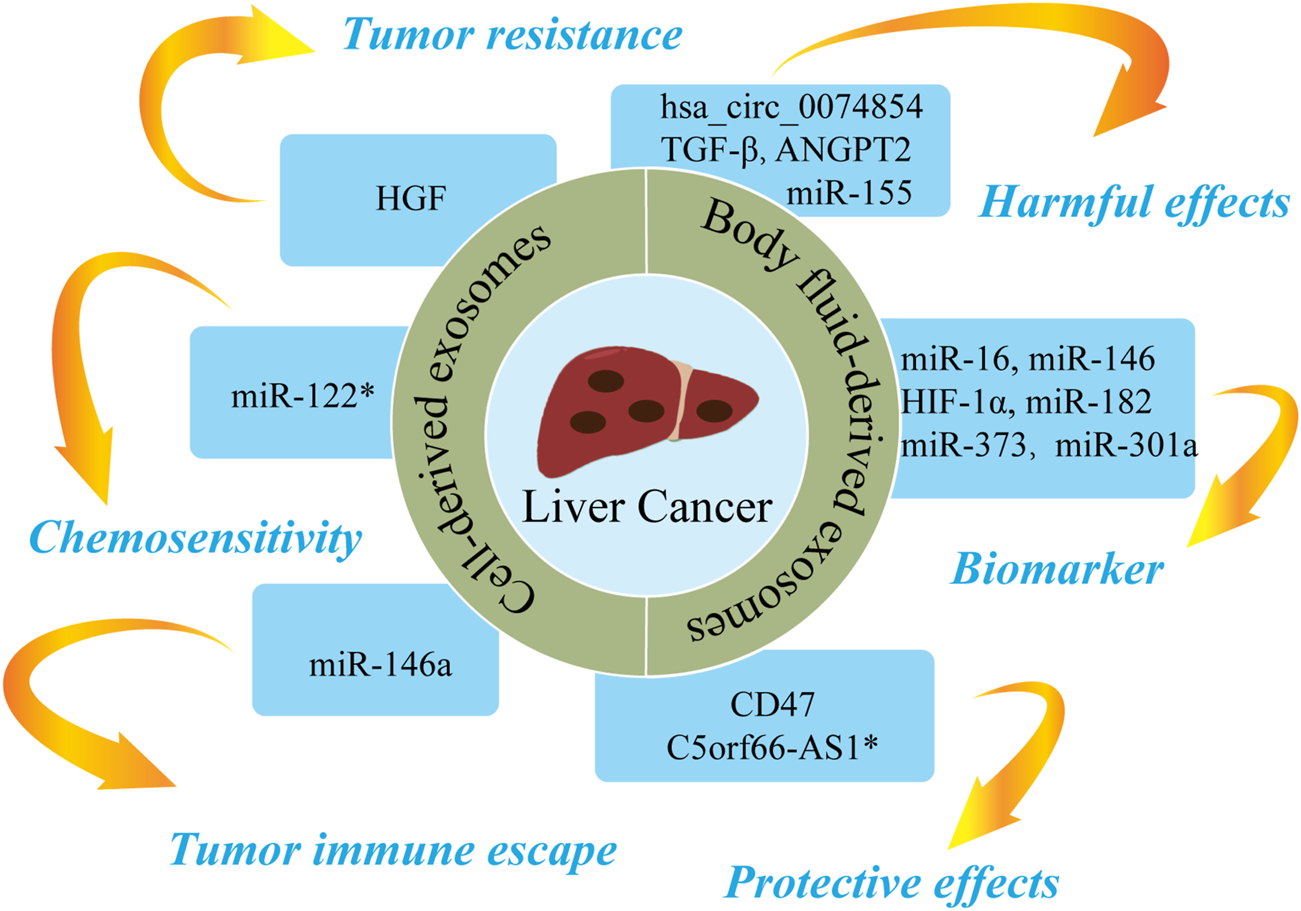
The roles of nucleic acids and proteins carried by exosomes in liver cancer. Nucleic acids or proteins carried by exosomes can exert harmful/protective effects on liver cancer and may also be predictive biomarkers for liver cancer. Some nucleic acids or proteins may be involved in tumor resistance, tumor immune escape, or chemosensitivity. * refers to nucleic acids and proteins carried by exosomes from mesenchymal stem cell.
TABLE 5
| Types | Exosomal contents | Source of exosomes | Reference | Functions |
|---|---|---|---|---|
| Nucleic acids | miRNA-210 | Cancer cell/serum | Lin et al.121 | Promoting cancer |
| miR-155 | Cancer cell/plasma | Matsuura et al.122 | Promoting cancer | |
| miR-146a | Cancer cell | Han et al.130 | Driving tumor immune escape | |
| hsa_circ_0074854 | Cancer cell | Wang et al.131 | Promoting cancer | |
| miR-660-5p | Macrophage | Tian et al.132 | Promoting cancer | |
| miR-27a-3p | Macrophage | Li et al.133 | Promoting cancer | |
| C5orf66-AS1 | MSC | Gu et al.134 | Suppressing cancer | |
| miR-122 | MSC | Lou et al.135 | Enhancing sensitivity to chemotherapy drugs | |
| miR-12 | Serum | Mjelle et al.138 | Prognostic predictors for liver cancer | |
| let-7 miRNA | ||||
| miR-141 | ||||
| miR-146 | ||||
| miR-718 | Serum | Sugimachi et al.139 | Promoting cancer | |
| miR-16 | Plasma | Frundt et al.97 | Being diagnostic and prognostic biomarkers for liver cancer | |
| miR-146a | ||||
| miR-192 | ||||
| miR-221 | ||||
| miR-182 | Ascites | Muhammad et al.98 | Being biomarkers for the diagnosis and prognosis of liver cancer | |
| miR-301a | ||||
| miR-373 | ||||
| Proteins | CLEC3B | Cancer cell | Xie et al.123 | Promoting cancer |
| HGF | Cancer cell | Qu et al.124 | Promoting tumor resistance | |
| TGF-β | Cancer cell | Qu et al.125 | Promoting cancer | |
| ANGPT2 | Cancer cell | Xie et al.126 | Promoting cancer | |
| AKT | Cancer cell | Wang et al.127 | Promoting cancer | |
| STAT5α | ||||
| ERK1/2 | ||||
| GSK3β | ||||
| Shh | Cancer cell/plasma | Li et al.128 | Promoting cancer | |
| S100A4 | Cancer cell/plasma | Sun et al.129 | Promoting cancer | |
| CD11b/CD18 | Macrophage | Wu et al.25 | Promoting cancer | |
| CD47 | HEK293T cell | Du et al.136 | Suppressing cancer | |
| AFP | DC | Lu et al.137 | Suppressing cancer | |
| HIF-1α | Serum | Xu et al.140 | Promoting cancer | |
| CPE | Serum | Hareendran et al.141 | Promoting cancer | |
| TGF-β1 | Ascites | Wei et al.142 | Promoting cancer |
Function of nucleic acids and proteins carried by exosomes on liver cancer.
Note: MSC, mesenchymal stem cell; HEK293T, human embryonic kidney cell; DC, dendritic cell.
Cell-derived exosomes
Nucleic acids or proteins carried by exosomes from different cells may aggravate or alleviate HCC. Tumor cells can regulate the function of endothelial cells by releasing exosomal contents, thereby increasing vascular permeability and promoting angiogenesis and tumor metastasis (Aslan et al., 2019). Lin et al. (Lin et al., 2018) confirmed that miRNA-210 in HCC cell-derived exosomes (HCC-exos) can be transmitted to endothelial cells to downregulate SMAD4 and STAT6, thereby promoting angiogenesis. A study (Matsuura et al., 2019) revealed that miR-155 in HCC-exos induces angiogenesis and promotes tumor recurrence under hypoxic conditions. A further study (Xie et al., 2020a) demonstrated that CLEC3B in HCC-exos could inhibit the AMPK pathway and upregulate VEGF, thereby promoting angiogenesis and tumor progression. Qu et al. (2016) concluded that HCC-exos increases tumor resistance by upregulating HGF, activating the c-Met/Akt pathway, and repressing apoptosis. HCC-exos can also promote epithelial-mesenchymal transition (EMT) through the TGF-β/Smad pathway, thereby accelerating tumor metastasis (Qu et al., 2019). Similarly, HCC-exos has been shown to transport ANGPT2 to human umbilical vein endothelial cells via endocytosis, thereby promoting EMT via the Tie2-independent pathway (Xie et al., 2020b). HCC-exos can also activate phosphokinases (AKT, STAT5α, ERK1/2, and GSK3β), which in turn activate the NF-κB pathway to accelerate angiogenesis and cell migration (Wang et al., 2018). Li et al. (2021a) reported that HCC-exos secrete Shh and thereby activate the Hedgehog pathway, which increase cancer stem cells (CSCs) and promote cell proliferation. Sun et al. (2021) confirmed that S100A4 in HCC-exos promoted metastasis by activating STAT3 phosphorylation and upregulating OPN. A further study (Han et al., 2019) confirmed that miR-146a in HCC-exos could promote polarization of M2 macrophages and inhibit T-cell functions, thereby driving tumor immune escape. Furthermore, HCC-exos can transfer hsa_circ_0074854 to macrophages, with downregulation of hsa_circ_0074854 shown to repress the polarization of M2 macrophages and cell migration (Wang et al., 2021c).
Moreover, a previous study (Wu et al., 2021b) found that M2 macrophage-derived exosomes (M2-exos) delivered CD11b/CD18 to activate MMP-9 pathway and promote tumor metastasis. MiR-660-5p-modified M2-exos downregulated KHF3 to promote EMT and the development of HCC (Tian et al., 2021). Li et al. (2021b) concluded that miR-27a-3p in M2-exos downregulated TXNIP, thereby enhancing the stemness, drug resistance, and invasiveness of HCC cells. A separate study (Gu et al., 2021) reported that MSC-derived exosomes upregulated C5orf66-AS1 to activate the miR-127-3p/DUSP1/ERK axis and inhibit the malignant behavior of CSCs in HCC. Furthermore, miR-122-modified ADSCs-exos enhanced the sensitivity of HCC cells to chemotherapeutic drugs by regulating miR-122 (Lou et al., 2015). Du et al. (2021) confirmed that CD47-modified HEK293T cell-derived exosomes can induce ferroptosis, which may represent a novel therapeutic target for HCC. A further study (Lu et al., 2017) revealed that AFP-enriched DEX elicited antitumor immune responses and remodeled the tumor microenvironment, a mechanism that may have utility in immunotherapy for HCC.
Body fluid-derived exosomes
Nucleic acids or proteins in body fluid-derived exosomes can alleviate or aggravate HCC and act as biomarkers for predicting HCC. Mjelle et al. (2019) performed RNA sequencing on serum from HCC patients. High levels of miR-12, let-7 miRNA, miR-141, and miR-146 in serum exosomes were found to be associated with poor survival, indicating that these miRNAs may have utility as prognostic predictors for HCC. In addition, miR-210 in serum exosomes was associated with microvessel density in HCC tissues (Lin et al., 2018). Serum exosomal miR-718 has been shown to be associated to tumor aggressiveness (Sugimachi et al., 2015), and plasma exosomal miR-155 has been shown to be associated with HCC recurrence (Matsuura et al., 2019). High levels of plasma exosomal Shh are associated with later tumor stage, higher histological grade, and higher recurrence in HCC (Li et al., 2021a). A further study (Sun et al., 2021) proposed exosomal S100A4 as a novel target for HCC metastasis as high levels of S100A4 in plasma exosomes were found to be associated with poor prognosis. Xu et al. (2019) found that HIF-1α in serum exosomes from HCC patients activated the PI3K/AKT pathway, thereby promoting angiogenesis and cell proliferation. Hareendran et al. (2022) reported that serum exosomal CPE was increased in HCC patients and HCC-exos loaded with CPE-shRNA inhibited cell proliferation by downregulating cyclin D1 and c-MYC. A further study (Frundt et al., 2021) proposed plasma exosomal miR-16, miR-146a, miR-192, and miR-221 as potential diagnostic and prognostic indicators in HCC patients. Furthermore, Muhammad Yusuf et al. (2020) demonstrated that ascites-derived exosomal miR-182, miR-301a, and miR-373 were upregulated in HCC, which may have utility in the diagnosis and prognosis of HCC patients. Wei et al. (2017) confirmed that ascites-derived exosomal TGF-β1 from HCC patients can promote the transformation of mesothelial cells into carcinoma-associated fibroblasts, thereby promoting peritoneal metastasis.
Taken together, these findings indicate that nucleic acids and proteins carried by exosomes from different sources can not only predict the recurrence and metastasis of HCC but may also play deleterious or protective roles in HCC.
Conclusion and perspectives
Nucleic acids and proteins carried by exosomes may represent promising biomarkers for the diagnosis and treatment of liver diseases by facilitating early diagnosis and prognostication. Interestingly, some nucleic acids or proteins were identified in more than one study. Table 6 summarizes studies identifying nucleic acids and proteins carried by exosomes.
TABLE 6
| Types | Exosomal contents | Liver diseases and references | Functions |
|---|---|---|---|
| Nucleic acids | miR-122 | DILI43-44,51–52 | Evaluating drug hepatotoxicity or ameliorating DILI |
| HF78,93 | Acting as a non-invasive predictor for HF or ameliorating HF | ||
| LF78,114 | Predicting the prognosis of LF or ameliorating LF | ||
| Liver cancer135 | Enhancing the sensitivity of cancer cells to chemotherapeutic drugs | ||
| miR-223 | DILI48 | Exerting the liver protective effect | |
| LF48 | Reversing liver injury | ||
| miR-192 | DILI51-52 | Predicting the drug hepatotoxicity | |
| HF97 | Being diagnostic and prognostic biomarker | ||
| LF114 | Predicting and assessing prognosis | ||
| Liver cancer97 | Being diagnostic and prognostic biomarker | ||
| lncRNA-H19 | HF88-89,94 | Accelerating the progression of cholestatic HF, being the diagnostic biomarker and potential therapeutic target for cholestatic HF. | |
| LF104 | Improving the survival rate and being a potential therapeutic target for LF. | ||
| miR-221 | HF97 | Being a diagnostic and prognostic biomarker | |
| Liver cancer97 | Being a diagnostic and prognostic biomarker | ||
| miR-146a | HF84,97 | Being a diagnostic and prognostic biomarker, or alleviating HF | |
| Liver cancer97 | Being a diagnostic and prognostic biomarker | ||
| miR-20a | HIRI65 | Exerting the liver protective effect | |
| LF111 | Aggravated liver inflammation | ||
| Proteins | HSP90 | DILI45-46 | Evaluating the liver injury |
| CD81 | DILI45,54 | Biomarkers for the diagnosis of liver injury | |
| HSP70 | DILI46 | Evaluating the liver injury | |
| HIRI59 | Exerting the liver protective effect | ||
| CD10 | DILI54 | Biomarkers for the diagnosis of liver injury | |
| HF96 | Biomarkers for the diagnosis of HF | ||
| ERK1/2 | LF106 | Improving APAP-induced LF | |
| Liver cancer127 | Promoting tumor metastasis | ||
| TGF-β | HF81 | Aggravating HF | |
| Liver cancer125,142 | Promoting tumor metastasis |
Overview of the roles of nucleic acids and proteins carried by exosomes in different liver diseases.
Note: DILI, drug-induced liver injury; HIRI, hepatic ischemia reperfusion injury; HF: hepatic fibrosis; LF, liver failure.
Despite initial studies of the role of exosomes in liver diseases, the development of exosome-related therapies for liver disease remains at the stage of in vitro and animal experiments, with a substantial amount of further development required to translate this work into clinical practice. Several outstanding challenges and issues have yet to be resolved. First, methods for mass production, isolation, purification, and preservation of exosomes are still in development. Second, the most appropriate and efficacious source of exosomes remains unclear. Third, there is a lack of knowledge regarding the biogenesis, release, targets, and molecular mechanisms of exosomes in the liver. Fourth, the safety and efficacy of exosomes in the treatment of liver disease have yet to be demonstrated. Therefore, further studies are required to overcome the obstacles to the translation of exosome research into clinical practice. With further research and improved technology, exosomes may represent a future therapeutic option for patients with liver diseases.
Statements
Author contributions
Conceptualization, XL; resources, DX and BQ; writing—original draft preparation, DX and BQ; writing—review and editing, XL and DX and BQ. All authors have read and agreed to the published version of the manuscript. DX and BQ contribute equally to this article.
Funding
This work was supported by the National Natural Science Foundation of China (82060119), and Provincial Key Talent Project of Gansu Province ([2020] No. 9), and Special funds for basic scientific research business expenses of Lanzhou University (lzujbky-2021-kb37).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
An M. Zhu J. Wu J. Cuneo K. C. Lubman D. M. (2018). Circulating microvesicles from pancreatic cancer accelerate the migration and proliferation of PANC-1 cells. J. Proteome Res.17 (4), 1690–1699. 10.1021/acs.jproteome.8b00014
2
Arrese M. Eguchi A. Feldstein A. E. (2015). Circulating microRNAs: Emerging biomarkers of liver disease. Semin. Liver Dis.35 (1), 43–54. 10.1055/s-0034-1397348
3
Arshad M. A. Murphy N. Bangash M. N. (2020). Acute liver failure. Clin. Med.20 (5), 505–508. 10.7861/clinmed.2020-0612
4
Ashour H. Rashed L. Elkordy M. A. Abdelwahed O. M. (2021). Remote liver injury following acute renal ischaemia-reperfusion: Involvement of circulating exosomal miR-687 and regulation by thymoquinone. Exp. Physiol.106 (11), 2262–2275. 10.1113/EP089765
5
Aslan C. Maralbashi S. Salari F. Kahroba H. Sigaroodi F. Kazemi T. et al (2019). Tumor-derived exosomes: Implication in angiogenesis and antiangiogenesis cancer therapy. J. Cell. Physiol.234 (10), 16885–16903. 10.1002/jcp.28374
6
Asrani S. K. Devarbhavi H. Eaton J. Kamath P. S. (2019). Burden of liver diseases in the world. J. Hepatol.70 (1), 151–171. 10.1016/j.jhep.2018.09.014
7
Aydin M. M. Akcali K. C. (2018). Liver fibrosis. Turk. J. Gastroenterol.29 (1), 14–21. 10.5152/tjg.2018.17330
8
Baker L. A. Lee K. C. Palacios Jimenez C. Alibhai H. Chang Y. M. Leckie P. J. et al (2015). Circulating microRNAs reveal time course of organ injury in a porcine model of acetaminophen-induced acute liver failure. PloS one10 (5), e0128076. 10.1371/journal.pone.0128076
9
Bakshi S. Kaur M. Saini N. Mir A. A. Duseja A. Sinha S. K. et al (2021). Altered expressions of circulating microRNAs 122 and 192 during antitubercular drug induced liver injury indicating their role as potential biomarkers. Hum. Exp. Toxicol.40 (9), 1474–1484. 10.1177/0960327121997975
10
Bray F. Ferlay J. Soerjomataram I. Siegel R. L. Torre L. A. Jemal A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca. Cancer J. Clin.68 (6), 394–424. 10.3322/caac.21492
11
Byass P. (2014). The global burden of liver disease: A challenge for methods and for public health. BMC Med.12, 159. 10.1186/s12916-014-0159-5
12
Chang C. Y. Chen K. Y. Shih H. J. Chiang M. Huang I. T. Huang Y. H. et al (2021). Let-7i-5p mediates the therapeutic effects of exosomes from human placenta choriodecidual membrane-derived mesenchymal stem cells on mitigating endotoxin-induced mortality and liver injury in high-fat diet-induced obese mice. Pharm. (Basel)15 (1), 36. 10.3390/ph15010036
13
Chang Y. Han J. A. Kang S. M. Jeong S. W. Ryu T. Park H. S. et al (2021). Clinical impact of serum exosomal microRNA in liver fibrosis. PloS one16 (9), e0255672. 10.1371/journal.pone.0255672
14
Charrier A. Chen R. Chen L. Kemper S. Hattori T. Takigawa M. et al (2014). Exosomes mediate intercellular transfer of pro-fibrogenic connective tissue growth factor (CCN2) between hepatic stellate cells, the principal fibrotic cells in the liver. Surgery156 (3), 548–555. 10.1016/j.surg.2014.04.014
15
Chen J. Xu Q. Zhang Y. Zhang H. (2020). RNA profiling analysis of the serum exosomes derived from patients with chronic hepatitis and acute-on-chronic liver failure caused by HBV. Sci. Rep.10 (1), 1528. 10.1038/s41598-020-58233-x
16
Chen L. Huang Y. Duan Z. Huang P. Yao H. Zhou Y. et al (2021). Exosomal miR-500 derived from lipopolysaccharide-treated macrophage accelerates liver fibrosis by suppressing MFN2. Front. Cell Dev. Biol.9, 716209. 10.3389/fcell.2021.716209
17
Chen L. Lu F. B. Chen D. Z. Wu J. L. Hu E. D. Xu L. M. et al (2018). BMSCs-derived miR-223-containing exosomes contribute to liver protection in experimental autoimmune hepatitis. Mol. Immunol.93, 38–46. 10.1016/j.molimm.2017.11.008
18
Chen L. Xiang B. Wang X. Xiang C. (2017). Exosomes derived from human menstrual blood-derived stem cells alleviate fulminant hepatic failure. Stem Cell Res. Ther.8 (1), 9. 10.1186/s13287-016-0453-6
19
Chiabotto G. Ceccotti E. Tapparo M. Camussi G. Bruno S. (2021). Human liver stem cell-derived extracellular vesicles target hepatic stellate cells and attenuate their pro-fibrotic phenotype. Front. Cell Dev. Biol.9, 777462. 10.3389/fcell.2021.777462
20
Cho Y. E. Kim S. H. Lee B. H. Baek M. C. (2017). Circulating plasma and exosomal microRNAs as indicators of drug-induced organ injury in rodent models. Biomol. Ther.25 (4), 367–373. 10.4062/biomolther.2016.174
21
Conde-Vancells J. Rodriguez-Suarez E. Gonzalez E. Berisa A. Gil D. Embade N. et al (2010). Candidate biomarkers in exosome-like vesicles purified from rat and mouse urine samples. Proteomics. Clin. Appl.4 (4), 416–425. 10.1002/prca.200900103
22
Cornide-Petronio M. E. Alvarez-Mercado A. I. Jimenez-Castro M. B. Peralta C. (2020). Current knowledge about the effect of nutritional status, supplemented nutrition diet, and gut microbiota on hepatic ischemia-reperfusion and regeneration in liver surgery. Nutrients12 (2), E284. 10.3390/nu12020284
23
Corrado C. Raimondo S. Chiesi A. Ciccia F. De Leo G. Alessandro R. (2013). Exosomes as intercellular signaling organelles involved in health and disease: Basic science and clinical applications. Int. J. Mol. Sci.14 (3), 5338–5366. 10.3390/ijms14035338
24
Dong V. Nanchal R. Karvellas C. J. (2020). Pathophysiology of acute liver failure. Nutr. Clin. Pract.35 (1), 24–29. 10.1002/ncp.10459
25
Du J. Wan Z. Wang C. Lu F. Wei M. Wang D. et al (2021). Designer exosomes for targeted and efficient ferroptosis induction in cancer via chemo-photodynamic therapy. Theranostics11 (17), 8185–8196. 10.7150/thno.59121
26
Du Y. Li D. Han C. Wu H. Xu L. Zhang M. et al (2017). Exosomes from human-induced pluripotent stem cell-derived mesenchymal stromal cells (hiPSC-MSCs) protect liver against hepatic ischemia/reperfusion injury via activating sphingosine kinase and sphingosine-1-phosphate signaling pathway. Cell. Physiol. biochem.43 (2), 611–625. 10.1159/000480533
27
Eldh M. Ekstrom K. Valadi H. Sjostrand M. Olsson B. Jernas M. et al (2010). Exosomes communicate protective messages during oxidative stress; possible role of exosomal shuttle RNA. PloS one5 (12), e15353. 10.1371/journal.pone.0015353
28
Fei Y. Shao J. Huang G. Wang L. Zou S. Sun H. et al (2021). Effect of edaravone on MicroRNA expression in exosomes after hepatic ischemia-reperfusion injury. Curr. Mol. Pharmacol.15, 870–882. 10.2174/1874467214666211130162152
29
Frundt T. Krause L. Hussey E. Steinbach B. Kohler D. von Felden J. et al (2021). Diagnostic and prognostic value of miR-16, miR-146a, miR-192 and miR-221 in exosomes of hepatocellular carcinoma and liver cirrhosis patients. Cancers (Basel)13 (10), 2484. 10.3390/cancers13102484
30
Garcia-Cortes M. Robles-Diaz M. Stephens C. Ortega-Alonso A. Lucena M. I. Andrade R. J. (2020). Drug induced liver injury: An update. Arch. Toxicol.94 (10), 3381–3407. 10.1007/s00204-020-02885-1
31
Gasmi B. Kleiner D. E. (2020). Liver histology: Diagnostic and prognostic features. Clin. Liver Dis.24 (1), 61–74. 10.1016/j.cld.2019.09.004
32
Gonzalez E. Azkargorta M. Garcia-Vallicrosa C. Prieto-Elordui J. Elortza F. Blanco-Sampascual S. et al (2021). Could protein content of urinary extracellular vesicles be useful to detect cirrhosis in alcoholic liver disease?Int. J. Biol. Sci.17 (8), 1864–1877. 10.7150/ijbs.59725
33
Gu H. Yan C. Wan H. Wu L. Liu J. Zhu Z. et al (2021). Mesenchymal stem cell-derived exosomes block malignant behaviors of hepatocellular carcinoma stem cells through a lncRNA C5orf66-AS1/microRNA-127-3p/DUSP1/ERK axis. Hum. Cell34 (6), 1812–1829. 10.1007/s13577-021-00599-9
34
Han Q. Zhao H. Jiang Y. Yin C. Zhang J. (2019). HCC-derived exosomes: Critical player and target for cancer immune escape. Cells8 (6), E558. 10.3390/cells8060558
35
Hareendran S. Albraidy B. Yang X. Liu A. Breggia A. Chen C. C. et al (2022). Exosomal carboxypeptidase E (CPE) and CPE-shRNA-loaded exosomes regulate metastatic phenotype of tumor cells. Int. J. Mol. Sci.23 (6), 3113. 10.3390/ijms23063113
36
He F. Li W. N. Li X. X. Yue K. Y. Duan J. L. Ruan B. et al (2022). Exosome-mediated delivery of RBP-J decoy oligodeoxynucleotides ameliorates hepatic fibrosis in mice. Theranostics12 (4), 1816–1828. 10.7150/thno.69885
37
Holman N. S. Mosedale M. Wolf K. K. LeCluyse E. L. Watkins P. B. (2016). Subtoxic alterations in hepatocyte-derived exosomes: An early step in drug-induced liver injury?Toxicol. Sci.151 (2), 365–375. 10.1093/toxsci/kfw047
38
Hu H. Ling B. Shi Y. Wu H. Zhu B. Meng Y. et al (2021). Plasma exosome-derived SENP1 may Be a potential prognostic predictor for melanoma. Front. Oncol.11, 685009. 10.3389/fonc.2021.685009
39
Hua S. Ma M. Fei X. Zhang Y. Gong F. Fang M. (2019). Glycyrrhizin attenuates hepatic ischemia-reperfusion injury by suppressing HMGB1-dependent GSDMD-mediated kupffer cells pyroptosis. Int. Immunopharmacol.68, 145–155. 10.1016/j.intimp.2019.01.002
40
Huang Y. J. Cao J. Lee C. Y. Wu Y. M. (2021). Umbilical cord blood plasma-derived exosomes as a novel therapy to reverse liver fibrosis. Stem Cell Res. Ther.12 (1), 568. 10.1186/s13287-021-02641-x
41
Hwang S. Yang Y. M. (2021). Exosomal microRNAs as diagnostic and therapeutic biomarkers in non-malignant liver diseases. Arch. Pharm. Res.44 (6), 574–587. 10.1007/s12272-021-01338-2
42
Hyun J. Wang S. Kim J. Kim G. J. Jung Y. (2015). MicroRNA125b-mediated Hedgehog signaling influences liver regeneration by chorionic plate-derived mesenchymal stem cells. Sci. Rep.5, 14135. 10.1038/srep14135
43
Ingram H. Dogan M. Eason J. D. Kuscu C. Kuscu C. (2022). MicroRNAs: Novel targets in hepatic ischemia-reperfusion injury. Biomedicines10 (4), 791. 10.3390/biomedicines10040791
44
Jalan R. Gustot T. Fernandez J. Bernal W. (2021). Equity' and 'justice' for patients with acute-on chronic liver failure: A call to action. J. Hepatol.75 (5), 1228–1235. 10.1016/j.jhep.2021.06.017
45
Jia P. Wu X. Dai Y. Teng J. Fang Y. Hu J. et al (2017). MicroRNA-21 is required for local and remote ischemic preconditioning in multiple organ protection against sepsis. Crit. Care Med.45 (7), e703–e710. 10.1097/CCM.0000000000002363
46
Jiao Y. Lu W. Xu P. Shi H. Chen D. Chen Y. et al (2021). Hepatocyte-derived exosome may be as a biomarker of liver regeneration and prognostic valuation in patients with acute-on-chronic liver failure. Hepatol. Int.15 (4), 957–969. 10.1007/s12072-021-10217-3
47
Jiao Y. Xu P. Shi H. Chen D. Shi H. (2021). Advances on liver cell-derived exosomes in liver diseases. J. Cell. Mol. Med.25 (1), 15–26. 10.1111/jcmm.16123
48
Jin Y. Wang J. Li H. Gao S. Shi R. Yang D. et al (2018). Extracellular vesicles secreted by human adipose-derived stem cells (hASCs) improve survival rate of rats with acute liver failure by releasing lncRNA H19. EBioMedicine34, 231–242. 10.1016/j.ebiom.2018.07.015
49
Jones L. B. Bell C. R. Bibb K. E. Gu L. Coats M. T. Matthews Q. L. (2018). Pathogens and their effect on exosome biogenesis and composition. Biomedicines6 (3), E79. 10.3390/biomedicines6030079
50
Jun J. H. Kim J. Y. Choi J. H. Lim J. Y. Kim K. Kim G. J. (2020). Exosomes from placenta-derived mesenchymal stem cells are involved in liver regeneration in hepatic failure induced by bile duct ligation. Stem Cells Int.2020, 5485738. 10.1155/2020/5485738
51
Katarey D. Verma S. (2016). Drug-induced liver injury. Clin. Med.16 (6), s104–s109. 10.7861/clinmedicine.16-6-s104
52
Koga Y. Yasunaga M. Moriya Y. Akasu T. Fujita S. Yamamoto S. et al (2011). Exosome can prevent RNase from degrading microRNA in feces. J. Gastrointest. Oncol.2 (4), 215–222. 10.3978/j.issn.2078-6891.2011.015
53
Konishi T. Lentsch A. B. (2017). Hepatic ischemia/reperfusion: Mechanisms of tissue injury, repair, and regeneration. Gene Expr.17 (4), 277–287. 10.3727/105221617X15042750874156
54
Kulik L. El-Serag H. B. (2019). Epidemiology and management of hepatocellular carcinoma. Gastroenterology156 (2), 477–491. 10.1053/j.gastro.2018.08.065
55
Li L. Wang R. Jia Y. Rong R. Xu M. Zhu T. (2019). Exosomes derived from mesenchymal stem cells ameliorate renal ischemic-reperfusion injury through inhibiting inflammation and cell apoptosis. Front. Med.6, 269. 10.3389/fmed.2019.00269
56
Li L. Zhao J. Zhang Q. Tao Y. Shen C. Li R. et al (2021). Cancer cell-derived exosomes promote HCC tumorigenesis through Hedgehog pathway. Front. Oncol.11, 756205. 10.3389/fonc.2021.756205
57
Li W. Xin X. Li X. Geng J. Sun Y. (2021). Exosomes secreted by M2 macrophages promote cancer stemness of hepatocellular carcinoma via the miR-27a-3p/TXNIP pathways. Int. Immunopharmacol.101 (1), 107585. 10.1016/j.intimp.2021.107585
58
Li X. Liu R. Huang Z. Gurley E. C. Wang X. Wang J. et al (2018). Cholangiocyte-derived exosomal long noncoding RNA H19 promotes cholestatic liver injury in mouse and humans. Hepatology68 (2), 599–615. 10.1002/hep.29838
59
Lin X. J. Fang J. H. Yang X. J. Zhang C. Yuan Y. Zheng L. et al (2018). Hepatocellular carcinoma cell-secreted exosomal MicroRNA-210 promotes angiogenesis in vitro and in vivo. Mol. Ther. Nucleic Acids11, 243–252. 10.1016/j.omtn.2018.02.014
60
Liu R. Li X. Zhu W. Wang Y. Zhao D. Wang X. et al (2019). Cholangiocyte-derived exosomal long noncoding RNA H19 promotes hepatic stellate cell activation and cholestatic liver fibrosis. Hepatology70 (4), 1317–1335. 10.1002/hep.30662
61
Liu Y. Lou G. Li A. Zhang T. Qi J. Ye D. et al (2018). AMSC-derived exosomes alleviate lipopolysaccharide/d-galactosamine-induced acute liver failure by miR-17-mediated reduction of TXNIP/NLRP3 inflammasome activation in macrophages. EBioMedicine36, 140–150. 10.1016/j.ebiom.2018.08.054
62
Liver F. (2019). Artificial liver group CSoIDCMA, severe liver D, artificial liver group CSoHCMA. [Guideline for diagnosis and treatment of liver failure]. Zhonghua Gan Zang Bing Za Zhi27 (1), 18–26.
63
Llovet J. M. Kelley R. K. Villanueva A. Singal A. G. Pikarsky E. Roayaie S. et al (2021). Hepatocellular carcinoma. Nat. Rev. Dis. Prim.7 (1), 6. 10.1038/s41572-020-00240-3
64
Lou G. Song X. Yang F. Wu S. Wang J. Chen Z. et al (2015). Exosomes derived from miR-122-modified adipose tissue-derived MSCs increase chemosensitivity of hepatocellular carcinoma. J. Hematol. Oncol.8, 122. 10.1186/s13045-015-0220-7
65
Lou G. Yang Y. Liu F. Ye B. Chen Z. Zheng M. et al (2017). MiR-122 modification enhances the therapeutic efficacy of adipose tissue-derived mesenchymal stem cells against liver fibrosis. J. Cell. Mol. Med.21 (11), 2963–2973. 10.1111/jcmm.13208
66
Lu Z. Zuo B. Jing R. Gao X. Rao Q. Liu Z. et al (2017). Dendritic cell-derived exosomes elicit tumor regression in autochthonous hepatocellular carcinoma mouse models. J. Hepatol.67 (4), 739–748. 10.1016/j.jhep.2017.05.019
67
Luo X. Luo S. Z. Xu Z. X. Zhou C. Li Z. H. Zhou X. Y. et al (2021). Lipotoxic hepatocyte-derived exosomal miR-1297 promotes hepatic stellate cell activation through the PTEN signaling pathway in metabolic-associated fatty liver disease. World J. Gastroenterol.27 (14), 1419–1434. 10.3748/wjg.v27.i14.1419
68
Lyu T. Zhang B. Li M. Jiao X. Song Y. (2021). Research progress on exosomes derived from mesenchymal stem cells in hematological malignancies. Hematol. Oncol.39 (2), 162–169. 10.1002/hon.2793
69
Ma L. Wei J. Zeng Y. Liu J. Xiao E. Kang Y. et al (2022). Mesenchymal stem cell-originated exosomal circDIDO1 suppresses hepatic stellate cell activation by miR-141-3p/PTEN/AKT pathway in human liver fibrosis. Drug Deliv.29 (1), 440–453. 10.1080/10717544.2022.2030428
70
Ma Z. Zhang B. Fan Y. Wang M. Kebebe D. Li J. et al (2019). Traditional Chinese medicine combined with hepatic targeted drug delivery systems: A new strategy for the treatment of liver diseases. Biomed. Pharmacother.117, 109128. 10.1016/j.biopha.2019.109128
71
Matsuura Y. Wada H. Eguchi H. Gotoh K. Kobayashi S. Kinoshita M. et al (2019). Exosomal miR-155 derived from hepatocellular carcinoma cells under hypoxia promotes angiogenesis in endothelial cells. Dig. Dis. Sci.64 (3), 792–802. 10.1007/s10620-018-5380-1
72
Mjelle R. Dima S. O. Bacalbasa N. Chawla K. Sorop A. Cucu D. et al (2019). Comprehensive transcriptomic analyses of tissue, serum, and serum exosomes from hepatocellular carcinoma patients. BMC Cancer19 (1), 1007. 10.1186/s12885-019-6249-1
73
Motawi T. K. Mohamed M. R. Shahin N. N. Ali M. A. M. Azzam M. A. (2018). Time-course expression profile and diagnostic potential of a miRNA panel in exosomes and total serum in acute liver injury. Int. J. Biochem. Cell Biol.100, 11–21. 10.1016/j.biocel.2018.05.002
74
Muhammad Yusuf A. N. Raja Ali R. A. Muhammad Nawawi K. N. Mokhtar N. M. (2020). Potential biomarkers in NASH-induced liver cirrhosis with hepatocellular carcinoma: A preliminary work on roles of exosomal miR-182, miR-301a, and miR-373. Malays. J. Pathol.42 (3), 377–384.
75
Nafar S. Nouri N. Alipour M. Fallahi J. Zare F. Tabei S. M. B. (2022). Exosome as a target for cancer treatment. J. Investig. Med.70, 1212–1218. 10.1136/jim-2021-002194
76
Osawa M. Matsuda Y. Sakata J. Wakai T. (2022). Exosomal p38 mitogen-activated protein kinase promotes tumour repopulation in TP53-mutated bile duct cancer cells. Anticancer Res.42 (2), 745–757. 10.21873/anticanres.15533
77
Palomo L. Mleczko J. E. Azkargorta M. Conde-Vancells J. Gonzalez E. Elortza F. et al (2018). Abundance of cytochromes in hepatic extracellular vesicles is altered by drugs related with drug-induced liver injury. Hepatol. Commun.2 (9), 1064–1079. 10.1002/hep4.1210
78
Phinney D. G. Pittenger M. F. (2017). Concise review: MSC-derived exosomes for cell-free therapy. Stem Cells35 (4), 851–858. 10.1002/stem.2575
79
Putignano A. Figorilli F. Alabsawy E. Agarwal B. Jalan R. (2018). Long-term outcome in patients with acute liver failure. Liver Int.38 (12), 2228–2238. 10.1111/liv.13914
80
Qu Y. Zhang Q. Cai X. Li F. Ma Z. Xu M. et al (2017). Exosomes derived from miR-181-5p-modified adipose-derived mesenchymal stem cells prevent liver fibrosis via autophagy activation. J. Cell. Mol. Med.21 (10), 2491–2502. 10.1111/jcmm.13170
81
Qu Z. Feng J. Pan H. Jiang Y. Duan Y. Fa Z. (2019). Exosomes derived from HCC cells with different invasion characteristics mediated EMT through TGF-β/Smad signaling pathway.Onco. Targets. Ther.12, 6897–6905. 10.2147/OTT.S209413
82
Qu Z. Wu J. Wu J. Luo D. Jiang C. Ding Y. (2016). Exosomes derived from HCC cells induce sorafenib resistance in hepatocellular carcinoma both in vivo and in vitro. J. Exp. Clin. Cancer Res.35 (1), 159. 10.1186/s13046-016-0430-z
83
Raees A. Kamran M. Ozkan H. Jafri W. (2021). Updates on the diagnosis and management of hepatocellular carcinoma. Euroasian J. Hepatogastroenterol.11 (1), 32–40. 10.5005/jp-journals-10018-1335
84
Rodriguez-Suarez E. Gonzalez E. Hughes C. Conde-Vancells J. Rudella A. Royo F. et al (2014). Quantitative proteomic analysis of hepatocyte-secreted extracellular vesicles reveals candidate markers for liver toxicity. J. Proteomics103, 227–240. 10.1016/j.jprot.2014.04.008
85
Roehlen N. Crouchet E. Baumert T. F. (2020). Liver fibrosis: Mechanistic concepts and therapeutic perspectives. Cells9 (4), E875. 10.3390/cells9040875
86
Saithanyamurthi H. Faust A. J. (2017). Drug-induced liver disease: Clinical course. Clin. Liver Dis.21 (1), 21–34. 10.1016/j.cld.2016.08.007
87
Seki E. Brenner D. A. (2015). Recent advancement of molecular mechanisms of liver fibrosis. J. Hepatobiliary. Pancreat. Sci.22 (7), 512–518. 10.1002/jhbp.245
88
Seo W. Eun H. S. Kim S. Y. Yi H. S. Lee Y. S. Park S. H. et al (2016). Exosome-mediated activation of toll-like receptor 3 in stellate cells stimulates interleukin-17 production by γδ T cells in liver fibrosis.Hepatology64 (2), 616–631. 10.1002/hep.28644
89
Shao B. Dang Q. Chen Z. Chen C. Zhou Q. Qiao B. et al (2021). Effects of tumor-derived exosome programmed death ligand 1 on tumor immunity and clinical applications. Front. Cell Dev. Biol.9, 760211. 10.3389/fcell.2021.760211
90
Shao M. Xu Q. Wu Z. Chen Y. Shu Y. Cao X. et al (2020). Exosomes derived from human umbilical cord mesenchymal stem cells ameliorate IL-6-induced acute liver injury through miR-455-3p. Stem Cell Res. Ther.11 (1), 37. 10.1186/s13287-020-1550-0
91
Shen T. Liu Y. Shang J. Xie Q. Li J. Yan M. et al (2019). Incidence and etiology of drug-induced liver injury in mainland China. Gastroenterology156 (8), 2230–2241. 10.1053/j.gastro.2019.02.002
92
Shi Y. Du L. Lv D. Li Y. Zhang Z. Huang X. et al (2021). Emerging role and therapeutic application of exosome in hepatitis virus infection and associated diseases. J. Gastroenterol.56 (4), 336–349. 10.1007/s00535-021-01765-4
93
Starckx S. Batheja A. Verheyen G. R. Jonghe S. D. Steemans K. Dijck B. V. et al (2013). Evaluation of miR-122 and other biomarkers in distinct acute liver injury in rats. Toxicol. Pathol.41 (5), 795–804. 10.1177/0192623312464436
94
Sugimachi K. Matsumura T. Hirata H. Uchi R. Ueda M. Ueo H. et al (2015). Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br. J. Cancer112 (3), 532–538. 10.1038/bjc.2014.621
95
Sun H. Wang C. Hu B. Gao X. Zou T. Luo Q. et al (2021). Exosomal S100A4 derived from highly metastatic hepatocellular carcinoma cells promotes metastasis by activating STAT3. Signal Transduct. Target. Ther.6 (1), 187. 10.1038/s41392-021-00579-3
96
Sun M. Kisseleva T. (2015). Reversibility of liver fibrosis. Clin. Res. Hepatol. Gastroenterol.39 (1), S60–S63. 10.1016/j.clinre.2015.06.015
97
Sung H. Ferlay J. Siegel R. L. Laversanne M. Soerjomataram I. Jemal A. et al (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca. Cancer J. Clin.71 (3), 209–249. 10.3322/caac.21660
98
Tang M. Chen Y. Li B. Sugimoto H. Yang S. Yang C. et al (2021). Therapeutic targeting of STAT3 with small interference RNAs and antisense oligonucleotides embedded exosomes in liver fibrosis. FASEB J.35 (5), e21557. 10.1096/fj.202002777RR
99
Tian B. Zhou L. Wang J. Yang P. (2021). miR-660-5p-loaded M2 macrophages-derived exosomes augment hepatocellular carcinoma development through regulating KLF3. Int. Immunopharmacol.101 (1), 108157. 10.1016/j.intimp.2021.108157
100
Tkach M. Thery C. (2016). Communication by extracellular vesicles: Where we are and where we need to go. Cell164 (6), 1226–1232. 10.1016/j.cell.2016.01.043
101
Trebicka J. Sundaram V. Moreau R. Jalan R. Arroyo V. (2020). Liver transplantation for acute-on-chronic liver failure: Science or fiction?Liver Transpl.26 (7), 906–915. 10.1002/lt.25788
102
Turturici G. Tinnirello R. Sconzo G. Geraci F. (2014). Extracellular membrane vesicles as a mechanism of cell-to-cell communication: Advantages and disadvantages. Am. J. Physiol. Cell Physiol.306 (7), C621–C633. 10.1152/ajpcell.00228.2013
103
Usui T. Naisbitt D. J. (2017). Human leukocyte antigen and idiosyncratic adverse drug reactions. Drug Metab. Pharmacokinet.32 (1), 21–30. 10.1016/j.dmpk.2016.11.003
104
Villanueva A. (2019). Hepatocellular carcinoma. N. Engl. J. Med.380 (15), 1450–1462. 10.1056/NEJMra1713263
105
Wan L. Xia T. Du Y. Liu J. Xie Y. Zhang Y. et al (2019). Exosomes from activated hepatic stellate cells contain GLUT1 and PKM2: A role for exosomes in metabolic switch of liver nonparenchymal cells. FASEB J.33 (7), 8530–8542. 10.1096/fj.201802675R
106
Wang L. Wang Y. Quan J. (2020). Exosomes derived from natural killer cells inhibit hepatic stellate cell activation and liver fibrosis. Hum. Cell33 (3), 582–589. 10.1007/s13577-020-00371-5
107
Wang N. Li X. Zhong Z. Qiu Y. Liu S. Wu H. et al (2021). 3D hESC exosomes enriched with miR-6766-3p ameliorates liver fibrosis by attenuating activated stellate cells through targeting the TGFβRII-SMADS pathway.J. Nanobiotechnology19 (1), 437. 10.1186/s12951-021-01138-2
108
Wang S. Dong Y. Gong A. Kong H. Gao J. Hao X. et al (2021). Exosomal circRNAs as novel cancer biomarkers: Challenges and opportunities. Int. J. Biol. Sci.17 (2), 562–573. 10.7150/ijbs.48782
109
Wang S. Xu M. Li X. Su X. Xiao X. Keating A. et al (2018). Exosomes released by hepatocarcinoma cells endow adipocytes with tumor-promoting properties. J. Hematol. Oncol.11 (1), 82. 10.1186/s13045-018-0625-1
110
Wang Y. Gao R. Li J. Tang S. Li S. Tong Q. et al (2021). Downregulation of hsa_circ_0074854 suppresses the migration and invasion in hepatocellular carcinoma via interacting with HuR and via suppressing exosomes-mediated macrophage M2 polarization. Int. J. Nanomedicine16, 2803–2818. 10.2147/IJN.S284560
111
Wei M. Yang T. Chen X. Wu Y. Deng X. He W. et al (2017). Malignant ascites-derived exosomes promote proliferation and induce carcinoma-associated fibroblasts transition in peritoneal mesothelial cells. Oncotarget8 (26), 42262–42271. 10.18632/oncotarget.15040
112
Wen J. Yang T. Mallouk N. Zhang Y. Li H. Lambert C. et al (2021). Urinary exosomal CA9 mRNA as a novel liquid biopsy for molecular diagnosis of bladder cancer. Int. J. Nanomedicine16, 4805–4811. 10.2147/IJN.S312322
113
Wu H. Y. Zhang X. C. Jia B. B. Cao Y. Yan K. Li J. Y. et al (2021). Exosomes derived from human umbilical cord mesenchymal stem cells alleviate acetaminophen-induced acute liver failure through activating ERK and IGF-1R/PI3K/AKT signaling pathway. J. Pharmacol. Sci.147 (1), 143–155. 10.1016/j.jphs.2021.06.008
114
Wu J. Gao W. Tang Q. Yu Y. You W. Wu Z. et al (2021). M2 macrophage-derived exosomes facilitate HCC metastasis by transferring αM β2 integrin to tumor cells.Hepatology73 (4), 1365–1380. 10.1002/hep.31432
115
Wu J. Yu C. Zeng X. Sun C. (2021). The hepatoprotective effect from ischemia-reperfusion injury of remote ischemic preconditioning in the liver related surgery: A meta-analysis. ANZ J. Surg.92, 1332–1337. 10.1111/ans.17236
116
Wu J. Zhao R. Dai J. Lai G. Khan A. U. Yu X. et al (2021). Analysis of differential expression of long noncoding RNAs in exosomes derived from mature and immature dendritic cells. Mol. Med. Rep.23 (2), 132. 10.3892/mmr.2020.11771
117
Wu J. Y. Ji A. L. Wang Z. X. Qiang G. H. Qu Z. Wu J. H. et al (2018). Exosome-Mimetic Nanovesicles from Hepatocytes promote hepatocyte proliferation in vitro and liver regeneration in vivo. Sci. Rep.8 (1), 2471. 10.1038/s41598-018-20505-y
118
Xiao J. Wang F. Wong N. K. He J. Zhang R. Sun R. et al (2019). Global liver disease burdens and research trends: Analysis from a Chinese perspective. J. Hepatol.71 (1), 212–221. 10.1016/j.jhep.2019.03.004
119
Xiao Y. Liu R. Li X. Gurley E. C. Hylemon P. B. Lu Y. et al (2019). Long noncoding RNA H19 contributes to cholangiocyte proliferation and cholestatic liver fibrosis in biliary atresia. Hepatology70 (5), 1658–1673. 10.1002/hep.30698
120
Xie J. Y. Wei J. X. Lv L. H. Han Q. F. Yang W. B. Li G. L. et al (2020). Angiopoietin-2 induces angiogenesis via exosomes in human hepatocellular carcinoma. Cell Commun. Signal.18 (1), 46. 10.1186/s12964-020-00535-8
121
Xie K. Liu L. Chen J. Liu F. (2019). Exosomal miR-1246 derived from human umbilical cord blood mesenchymal stem cells attenuates hepatic ischemia reperfusion injury by modulating T helper 17/regulatory T balance. IUBMB Life71 (12), 2020–2030. 10.1002/iub.2147
122
Xie K. Liu L. Chen J. Liu F. (2019). Exosomes derived from human umbilical cord blood mesenchymal stem cells improve hepatic ischemia reperfusion injury via delivering miR-1246. Cell Cycle18 (24), 3491–3501. 10.1080/15384101.2019.1689480
123
Xie X. W. Jiang S. S. Li X. (2020). CLEC3B as a potential prognostic biomarker in hepatocellular carcinoma. Front. Mol. Biosci.7, 614034. 10.3389/fmolb.2020.614034
124
Xu Q. You R. Yin G. Gu J. (2019). Hepatocellular carcinoma serum derived exosomal HIF-1α induces phosphoinositide-3 kinase/protein kinase B signaling to induce the angiogenesis and proliferation of HCC.Transl. Cancer Res.8 (4), 1550–1559. 10.21037/tcr.2019.08.07
125
Yan Y. Jiang W. Tan Y. Zou S. Zhang H. Mao F. et al (2017). hucMSC exosome-derived GPX1 is required for the recovery of hepatic oxidant injury. Mol. Ther.25 (2), 465–479. 10.1016/j.ymthe.2016.11.019
126
Yanez-Mo M. Siljander P. R. Andreu Z. Zavec A. B. Borras F. E. Buzas E. I. et al (2015). Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles4, 27066. 10.3402/jev.v4.27066
127
Yang B. Duan W. Wei L. Zhao Y. Han Z. Wang J. et al (2020). Bone marrow mesenchymal stem cell-derived hepatocyte-like cell exosomes reduce hepatic ischemia/reperfusion injury by enhancing autophagy. Stem Cells Dev.29 (6), 372–379. 10.1089/scd.2019.0194
128
Yang X. Greenhaw J. Shi Q. Su Z. Qian F. Davis K. et al (2012). Identification of urinary microRNA profiles in rats that may diagnose hepatotoxicity. Toxicol. Sci.125 (2), 335–344. 10.1093/toxsci/kfr321
129
Yu W. Hurley J. Roberts D. Chakrabortty S. K. Enderle D. Noerholm M. et al (2021). Exosome-based liquid biopsies in cancer: Opportunities and challenges. Ann. Oncol.32 (4), 466–477. 10.1016/j.annonc.2021.01.074
130
Zhang A. B. Peng Y. F. Jia J. J. Nie Y. Zhang S. Y. Xie H. Y. et al (2019). Exosome-derived galectin-9 may be a novel predictor of rejection and prognosis after liver transplantation. J. Zhejiang Univ. Sci. B20 (7), 605–612. 10.1631/jzus.B1900051
131
Zhang J. Gao J. Lin D. Xiong J. Wang J. Chen J. et al (2021). Potential networks regulated by MSCs in acute-on-chronic liver failure: Exosomal miRNAs and intracellular target genes. Front. Genet.12, 650536. 10.3389/fgene.2021.650536
132
Zhang L. Song Y. Chen L. Li D. Feng H. Lu Z. et al (2020). MiR-20a-containing exosomes from umbilical cord mesenchymal stem cells alleviates liver ischemia/reperfusion injury. J. Cell. Physiol.235 (4), 3698–3710. 10.1002/jcp.29264
133
Zhang Q. Piao C. Ma H. Xu J. Wang Y. Liu T. et al (2021). Exosomes from adipose-derived mesenchymal stem cells alleviate liver ischaemia reperfusion injury subsequent to hepatectomy in rats by regulating mitochondrial dynamics and biogenesis. J. Cell. Mol. Med.25 (21), 10152–10163. 10.1111/jcmm.16952
134
Zhang Q. Qu Y. Zhang Q. Li F. Li B. Li Z. et al (2022). Exosomes derived from Hepatitis B virus-infected hepatocytes promote liver fibrosis via miR-222/TFRC axis. Cell Biol. Toxicol.1, 1. 10.1007/s10565-021-09684-z
135
Zhang S. Jiang L. Hu H. Wang H. Wang X. Jiang J. et al (2020). Pretreatment of exosomes derived from hUCMSCs with TNF-alpha ameliorates acute liver failure by inhibiting the activation of NLRP3 in macrophage. Life Sci.246, 117401. 10.1016/j.lfs.2020.117401
136
Zhang S. Rao S. Yang M. Ma C. Hong F. Yang S. (2022). Role of mitochondrial pathways in cell apoptosis during He-patic ischemia/reperfusion injury. Int. J. Mol. Sci.23 (4), 2357. 10.3390/ijms23042357
137
Zhang Y. Li Y. Wang Q. Zheng D. Feng X. Zhao W. et al (2022). Attenuation of hepatic ischemia‑reperfusion injury by adipose stem cell‑derived exosome treatment via ERK1/2 and GSK‑3β signaling pathways.Int. J. Mol. Med.49 (2), 13. 10.3892/ijmm.2021.5068
138
Zhao L. Shi J. Chang L. Wang Y. Liu S. Li Y. et al (2021). Serum-derived exosomal proteins as potential candidate biomarkers for hepatocellular carcinoma. ACS Omega6 (1), 827–835. 10.1021/acsomega.0c05408
139
Zhao L. Wang Y. Zhang Y. (2021). The potential diagnostic and therapeutic applications of exosomes in drug-induced liver injury. Toxicol. Lett.337, 68–77. 10.1016/j.toxlet.2020.11.021
140
Zheng L. Li Z. Ling W. Zhu D. Feng Z. Kong L. (2018). Exosomes derived from dendritic cells attenuate liver injury by modulating the balance of treg and Th17 cells after ischemia reperfusion. Cell. Physiol. biochem.46 (2), 740–756. 10.1159/000488733
141
Zhou X. Liang Z. Qin S. Ruan X. Jiang H. (2022). Serum-derived miR-574-5p-containing exosomes contribute to liver fibrosis by activating hepatic stellate cells. Mol. Biol. Rep.49 (3), 1945–1954. 10.1007/s11033-021-07008-2
142
Zhu M. Liu X. Li W. Wang L. (2020). Exosomes derived from mmu_circ_0000623-modified ADSCs prevent liver fibrosis via activating autophagy. Hum. Exp. Toxicol.39 (12), 1619–1627. 10.1177/0960327120931152
Summary
Keywords
liver disease, exosomes, nucleic acid, protein, biomarker
Citation
Xie D, Qian B and Li X (2022) Nucleic acids and proteins carried by exosomes from various sources: Potential role in liver diseases. Front. Physiol. 13:957036. doi: 10.3389/fphys.2022.957036
Received
02 June 2022
Accepted
17 August 2022
Published
23 September 2022
Volume
13 - 2022
Edited by
Chandana B Herath, University of New South Wales, Australia
Reviewed by
Haidi Yin, Shenzhen Bay Laboratory, China
Faezeh Vakhshiteh, Iran University of Medical Sciences, Iran
Updates
Copyright
© 2022 Xie, Qian and Li.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xun Li, lxdr21@126.com
This article was submitted to Gastrointestinal Sciences, a section of the journal Frontiers in Physiology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.