- 1Department of Dermatology, Tianjin First Central Hospital, Tianjin, China
- 2Department of Dermatology and Venereology, The First Affiliated Hospital of Gannan Medical University, Ganzhou, China
- 3Research Department 3, Nanjing Research Institute of Electronic Engineering, Nanjing, China
- 4School of Microelectronics, Tianjin University, Tianjin, China
- 5Tianjin Navigation Instruments Research Institute, Tianjin, China
Electrocardiogram (ECG) is a graphical representation of the electrical activity of the heart and plays a crucial role in diagnosing heart disease and assessing cardiac function. In the context of infectious diseases, ECG classification is particularly critical, as many infections, such as viral myocarditis and sepsis, can cause significant cardiac complications. Early detection of infection-induced cardiac abnormalities through ECG can provide timely intervention and improve patient outcomes. However, current ECG processing methods often overlook the impact of confounding factors caused by statistical associations, which can compromise classification accuracy, especially in infection-related cardiac conditions. To address this, we propose an innovative approach to causal reasoning based on attention mechanisms. By employing backdoor adjustment for each cardiac lead, our method effectively eliminates confounding factors and models the true causal relationship between ECG patterns and underlying cardiac abnormalities caused by infectious diseases. Furthermore, our approach integrates the concept of entropy with causal inference to enhance ECG classification. By quantifying the information content and variability in ECG signals, we can better identify patterns and anomalies associated with infection-induced cardiac conditions. Experimental results demonstrate that our method achieves significant improvements in classification accuracy and robustness across four benchmark ECG datasets, outperforming existing methods. This work provides a novel perspective on the interplay between infection and cardiac function, offering valuable insights into the detection and understanding of infection-related cardiac complications.
1 Introduction
Cardiovascular disease (CVD) remains the leading cause of mortality worldwide, accounting for approximately 20.5 million deaths in 2021, which represents about one-third of all global deaths (Federation, 2023). While high-income countries have made significant strides in cardiovascular health, this progress remains uneven, particularly in low- and middle-income countries, where over 75% of CVD-related deaths occur. These regions often face limited access to specialized cardiology services, further exacerbating the challenge of timely diagnosis and intervention. Moreover, infectious diseases such as COVID-19, HIV, and other viral or bacterial infections have been shown to exacerbate or directly contribute to cardiovascular complications, including myocarditis and sepsis-induced cardiac dysfunction. This dual burden of CVD and infectious diseases underscores the urgent need for accurate, automated tools to interpret electrocardiograms (ECG) and support diagnosis in resource-limited settings. In recent years, machine learning (ML) has played a pivotal role in advancing ECG signal classification (Jekova et al., 2008; Ince et al., 2009), offering new possibilities for understanding the interplay between infectious diseases and cardiovascular health. The ECG, as a critical test in the medical field, reveals the heart’s electrical activity from multiple dimensions. Machine learning algorithms, through their ability to analyze large datasets, identify complex patterns, and classify a wide range of cardiac conditions, are transforming cardiovascular care (Andreao et al., 2006; Martis et al., 2013; Karimifard et al., 2006). For regions heavily impacted by infectious diseases, these algorithms hold promise for detecting infection-induced cardiac abnormalities, such as myocarditis caused by viral infections. Additionally, real-time monitoring systems incorporating unsupervised learning and anomaly detection methods, such as auto-encoders, provide continuous analysis of ECG data, enabling timely warnings for healthcare providers. These technologies not only support early detection of cardiovascular issues but also offer valuable insights into the cardiac complications associated with infectious diseases, facilitating better clinical outcomes and resource allocation.
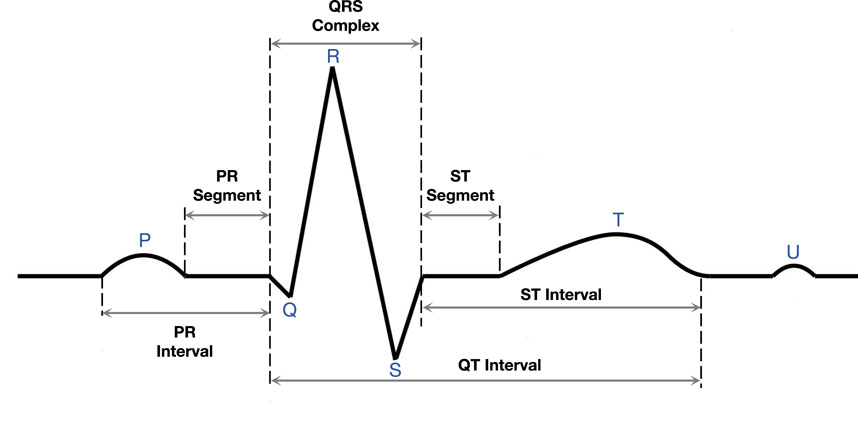
In the traditional diagnosis, a healthcare professional performs a detailed analysis of the ECG, focusing on the shape, duration, and temporal characteristics of electrical waveforms, including P waves, QRS wave groups, and T waves, which are illustrated in Figure 1. Through careful observation and interpretation of these characteristics, medical personnel can deeply understand the heart condition of patients, and then provide an accurate basis for the diagnosis and treatment of cardiovascular problems. ECG signals play a crucial role in diagnosing various cardiac disorders, including those induced by infectious diseases such as viral myocarditis and sepsis-related myocardial injury (Siontis et al., 2021). In these conditions, the infectious agents or systemic inflammatory responses can directly or indirectly affect the heart, leading to characteristic changes in ECG waveforms. For instance, viral myocarditis may manifest as ST-segment elevation, T-wave inversion, or arrhythmias, while sepsis-related myocardial injury often results in prolonged QT intervals or reduced heart rate variability. The detection and classification of these ECG patterns are essential for early diagnosis and timely intervention in infection-related cardiac disorders.
In the field of computer vision, the judgment of the model is often disturbed by a variety of confounding factors, which makes accurate image analysis more complex and challenging (Nie et al., 2023a). Confounding factors can include illumination change, shadow, noise, occlusion, and other environmental or sensor-related artifacts. These factors not only affect the quality of the image but also confuse the model’s correct understanding of the image content. Similarly, in the context of ECG signal analysis, confounding factors manifest in different yet equally impactful ways. For instance, variations in time-scale features, inter-lead differences, basic physiological characteristics of patients, and prior information on disease types can all act as confounders (Nie et al., 2023b). Time-scale features, such as short-term fluctuations and long-term trends, are directly observable and extracted using multi-scale convolutional kernels. Inter-lead differences, particularly in multi-lead ECG signals like 12-lead ECG, reflect the varying characteristics of signals from different leads, which can significantly influence classification results. Basic physiological features, such as heart rate, QRS complex morphology, and T-wave morphology, are inherent components of ECG signals and may simultaneously affect the signals and the classification outcomes. Additionally, prior information on disease types, derived from the labels in the training data, allows us to construct a confounding dictionary by computing the average features for each class, representing the typical confounding characteristics of different classes (Liu et al., 2024; Nie et al., 2024).
It is worth emphasizing that the existence of confounding factors not only makes the judgment of the model fuzzy and uncertain, but these factors themselves may interweave and influence each other to form a more complex visual environment. For example, in ECG signals, patient-specific characteristics such as age, gender, or body composition may interact with recording conditions like electrode quality or environmental noise to create compounded distortions that are difficult to disentangle. However, the discovery and removal of confounding factors in ECG signals is also a challenging task (Sajjan, 2012), as they may take on subtle and imperceptible forms in the signal that are difficult to capture by simple rules or algorithms. If the model deduces based on these confounding features, it is easy to make the wrong judgment because the model does not consider the true causal relationship between the features and the label.
These confounders are considered visible because they can be directly extracted from the ECG signals or data labels without requiring additional assumptions or unobservable variables. Time-scale features and inter-lead differences are extracted using signal processing techniques such as convolutional operations, while basic physiological features are obtained through standard ECG analysis methods. Prior information on disease types is derived directly from the labels in the training data, making it known and observable (Liu et al., 2024). Our approach assumes that these confounders are observable and can be effectively addressed through the construction of a confounding dictionary and causal interventions, such as backdoor adjustment. This assumption is based on the fact that these confounders are directly extracted from the data, eliminating the need for additional unobservable variables, and by constructing a causal graph, we explicitly model the relationships between these confounders, ECG signals, and classification outcomes (Nie et al., 2024; Ribeiro et al., 2020).
Besides, if we ignore time-scale features and only rely on local information to apply the traditional method to ECG signal classification, it will lead to the failure to fully understand the rich information contained in ECG signals (Bender et al., 2023). A single-scale analysis method may lead to an oversimplification of the characteristics of the ECG signal, thus affecting the final classification accuracy. Therefore, in order to interpret ECG signals more comprehensively, we introduce a time-domain features embedding module. By learning and utilizing the features of different time scales, we can better capture the dynamic features of the signal and improve the performance and generalization ability of the classification algorithm. When the ECG signal is used as input, the model is easily disturbed by confounding factors (Miao et al., 2023; Zhan et al., 2023). Confounding factors, such as individual differences, the complexity of the disease, and other physiological disturbances, have a non-negligible impact on the accurate classification of ECG signals. Traditional approaches often fail to fully account for these confounding factors, resulting in models that perform poorly in the face of complexities. Especially in complex clinical settings, the presence of these confounding factors may mask the true causal relationship and reduce the reliability and generalization performance of the classification model.
In order to overcome this challenge, a causal reasoning method was introduced in this study, aiming at deeply mining the correlation in ECG signals, clarifying the causal chain, and effectively eliminating the interference of confounding factors, thus improving the adaptability of the classification model to complex situations (Ghongade and Ghatol, 2007; Nagal and Sharma, 2014). Specifically, we propose a classification network for ECG signals based on causal reasoning. First, in the feature mapping part of the whole network, we use convolution kernels of different sizes. Given the diversity of ECG signals on time scales, the characteristics of different time scales are essential for a comprehensive understanding of the signals. At the same time, the module considers various perspectives of the ECG signal to capture more abundant feature information. This allows the network to consider time dynamics more comprehensively when analyzing ECGs, thus improving the accuracy of the classification algorithm. Second, the causal reasoning module is the de-confounding part of our proposed network. Confounders are often a major obstacle to model performance in ECG signal classification tasks. In the causal reasoning module, we adopt the backdoor adjustment method to eliminate the influence of confounding factors by constructing a causal graph and cutting off the false causal path, so as to realize the identification of a real causal correlation. The causal module ensures that the model is more reliable and stable in the classification process and avoids misjudgments caused by confounding factors.
Given the preceding discussion, the key achievements of the study can be outlined as follows:
• We mitigate the interference of confounding factors in ECG signals by introducing backdoor adjustment, allowing for the preservation of true causal features associated with infection-induced cardiac abnormalities. This approach enhances network classification performance and provides a robust framework for accurately identifying cardiac conditions influenced by infectious diseases.
• We leverage the time-domain diversity of ECG signals to enable the causal reasoning module to effectively address confounding factors arising from temporal variations in ECG data. By eliminating the adverse impact of these factors, our method improves classification accuracy and robustness, particularly in detecting infection-related cardiac anomalies.
• Results of extensive experiments on four multi-label ECG datasets demonstrate that our method surpasses the existing advanced networks across several classification tasks.
2 Related work
2.1 Causal learning for medical images
In the past few years, many research has been devoted to the application of causal learning methods to medical image processing, covering medical image classification, image segmentation, and medical question and answering tasks.
Nie et al. (Nie et al., 2023a; Nie et al., 2023b; Liu et al., 2024) used a variety of causal learning methods in medical image classification. They succeeded in removing visible confounding factors by using backdoor adjustment methods. In addition, they cleverly introduced instrumental variables, extracting information from chest imaging and electronic health record sheets to effectively eliminate invisible confounding factors. Miao et al. (2023) proposed a method of medical image segmentation based on causal graphs, which provides a more interpretable approach for semi-supervised learning of medical image segmentation. In their research, they emphasize the importance of algorithmic independence based on the principle of cause-effect graphs, and design an innovative statistical quantification method to approximate uncomputable algorithmic independence. Zhan et al. (2023) propose an innovative debiased medical visual question-answering (MedVQA) model, which cleverly incorporates counterfactual data during the training phase and directly subtracts the causal effects of linguistic priors, thus successfully migrating linguistic biases in the final MedVQA. This approach not only focuses on the influence of linguistic priors, but also powerfully mitigates bias in training by introducing counterfactual data. However, while existing research has made significant progress in tasks such as medical image classification, segmentation, and intelligent question and answer, the unique nature and data complexity of ECGs make causal reasoning in this field even more complicated.
2.2 Machine learning for ECG signal classification
Jekova et al. (2008) explored morphological features, examining characteristics like the amplitude and width of QRS complex waves. On the other hand, statistical features are obtained using techniques such as wavelet transform or hidden Markov chain (Ince et al., 2009; Andreao et al., 2006). Nagal et al. (Martis et al., 2013) proposed some mathematically based analytical methods to deal with high-dimensional features. In feature classification, algorithms like multilayer perceptron (Karimifard et al., 2006), K-nearest neighbors (Ghongade and Ghatol, 2007), and support vector machines (Nagal and Sharma, 2014) are frequently utilized. Martis et al. (Shimpi et al., 2017) proposed principal component analysis to reduce ECG signal dimensionality, followed by using a visual word bag method, involving feature block extraction, codebook construction, and pooling strategies for generating the final features to the SVM for classification. Lassoued and Ketata, (2018) proposed discrete wavelet transform coefficients for feature extraction, with a fully connected layer serving as the classifier. However, machine learning methods are hindered by their reliance on meticulous data preprocessing, making it challenging to achieve optimal performance.
Wang et al. (2021) introduced a method that integrates a 33-layer CNN with a attention module. This CNN is adept at extracting spatial and channel features, while the non-local attention mechanism effectively captures long-range dependencies across these dimensions. Chen et al. (2020) proposed an effective ECG classification network comprising five CNN blocks, bidirectional GRU with an attention mechanism. Yang et al. (2023) believed that by extracting the features of ECG signals from multiple views, higher quality features can be obtained compared to a single view. Finally, multiple features are fused through a multi-view fusion module to get the final feature. Zhang et al. (2023) proposed an efficient method to learn multi-scale features by using a two-branch structure. Han et al. (2023) proposed a fusion module for ECG signal classification based on multi-modal and attention mechanisms, aiming at preprocessing methods that are easy to lead to the loss of key information. In addition, in order to avoid extracting only a single feature which makes it difficult for the model to learn the complete ECG information, they use both the time domain information and the visual domain information of the ECG through a multi-modal method.
3 Methods
3.1 Method overview
In this section, we will describe our ECG classification method in detail. Unlike traditional medical image classification tasks, an ECG is not an image, but a signal pattern. The ECG signal consists of multiple waveforms, including the P wave, QRS wave group, and T wave (see Figure 1). The classification basis of the model is to classify diseases according to the changes and anomalies of these waveforms. The whole classification process includes signal preprocessing, feature extraction, feature decoupling, and disease classification. Unlike traditional medical images, an ECG consists of sub-images from multiple perspectives. This feature provides more abundant information for classification tasks, but also inevitably introduces the influence of confounding factors.
Therefore, as shown in Figure 2, we propose a causal inference classification framework based on attention mechanisms. First, we use convolutional layers to extract preliminary features, and time-domain features embedding module consisting of convolution kernel of different sizes are used to further extract time-domain features of ECG signals. The separated temporal features are then fed into their respective causal reasoning modules, utilizing novel attention mechanisms to capture true causal relationships. After causal learning, the characteristics of causal information are selectively retained according to causal relationships. Finally, the classification task is completed by the classification layer, and the result is predicted according to the causal characteristics.
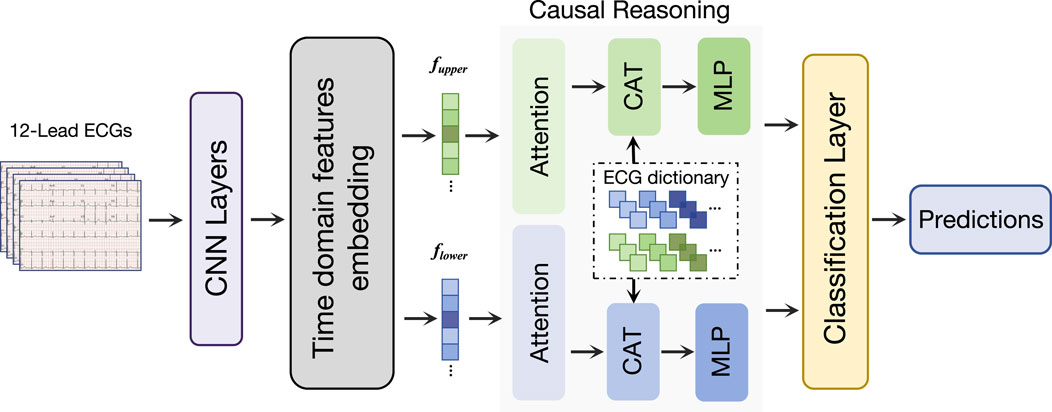
Figure 2. Overview of our proposed network. Our network first processes the ECG signals through the convolutional layer, and then obtains the time-scale features through the time domain features embedding module. We designed a causal reasoning module for ECG classification to perform feature decoupling. Finally, a classification layer is used to make classification predictions.
3.2 Convolution layers
We use the entire ECG as input, so the input consists of signals from multiple angles, represented as
The
where
In an ECG, the convolution kernel is responsible for detecting specific waveform features, such as P waves, QRS wave groups, and T waves. This convolution operation is carried out on the whole signal, and through the mechanism of weight sharing, the convolution kernel learns and recognizes the same local features at different locations.
3.3 Time domain features embedding
We emphasize that in order to extract time information from the ECG, we have adopted an efficient method of using only a few convolution kernel of different sizes without the need to introduce a timing network. Various scale features in ECG reflect the information of different time scales. Oversimplifying the ECG to a single feature block will lead to ignoring the correlation between the features in a specific time frame. Therefore, we introduce the time domain features embedding module, which can be viewed as a time-scale adaptive feature extractor.
The time domain features embedding module obtains more comprehensive time domain features by using convolution kernels of different sizes. Smaller convolution kernel are able to capture short-term signal fluctuations sensitively, while larger convolution kernel are better able to capture long-term signal trends. This feature extraction method helps the network to better understand the complex time domain structure of the ECG signal and improve the sensitivity of the heart condition. It is worth noting that the module can not only avoid the information loss caused by a single feature extraction method, but also enable the network to consider and analyze the signal features at different times more comprehensively. The time-scale embedding process can be expressed as:
where
Specifically, we use two one-dimensional cores of different sizes, namely,
Therefore, the role of this module in ECG classification is mainly reflected in its ability to effectively capture time domain diversity, improve the network’s perception of signal details and global features, and thus provide a more accurate and comprehensive feature representation for the final classification task. In this time domain features embedding module, we use different sizes of cores because the ECG data is one-dimensional, and using convolution cores of different sizes in the one-dimensional domain is better for collecting features on different time scales. For example, larger kernels provide a larger receptive field, enhancing their ability to capture features.
3.4 Causal learning
As shown in Figure 3a, when there are no confounders in ECG signals,
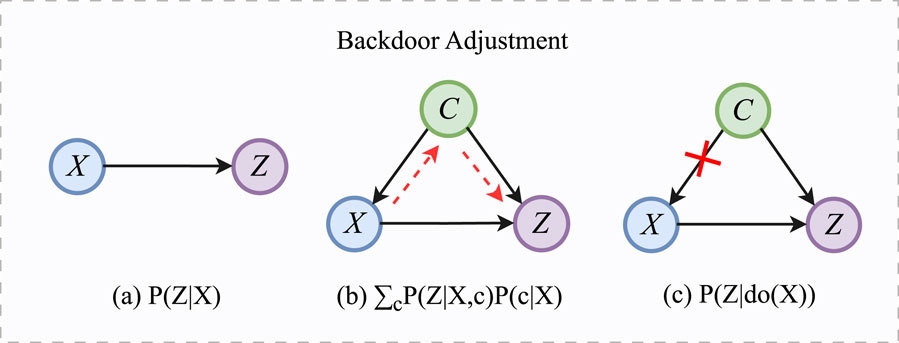
Figure 3. The process of backdoor adjustment. (a)
In Figure 3b,
where the confounder
Specifically, in the ECG classification task, we want to understand the influence of confounding factors
To eliminate the influence of confounding factors, it is crucial to close the backdoor path. For the backdoor path
where
After obtaining
To estimate the probability distribution of a function
where
where
The NWGM normalizes the WGM to ensure that the output is a valid probability distribution, suitable for use in classification tasks.
In our proposed method, the conditional probability distribution
where
By combining the backdoor adjustment formula Equation 4 with the NWGM approximation Equations 7 and 8, we derive the interventional probability distribution
where
During the classification process, we combine the obtained causal intervention probability
3.5 Causal reasoning
In this study, we use an attention mechanism for causal reasoning (CR) to select true causal relationships. CR establishes connections among feature channels to reassess their significance and effectively capture spatial information within the feature space (Hou et al., 2021).
Attention mechanisms play a crucial role in enhancing network performance. Squeeze-and-Excitation (SE) attention (Hu et al., 2018), a widely used attention mechanism, is recognized for its efficiency with a minimal parameter count and outstanding performance. While channel attention focuses on constructing relationships between feature channels to reweight their importance, it often overlooks spatial information. However, spatial information is essential for generating spatially selective attention maps.
In our study, we applied an attention fusion module to the task of ECG classification, coupled with causal inference for backdoor adjustment. This step is particularly crucial when dealing with ECG data, as it often suffers from confounding factors such as patients’ physiological states and cardiac histories. We first input the features extracted from the ECG data as queries
After time-scale embedding, the features are divided into upper features and lower features. These features pass through the causal reasoning module, in which true causal features are selected. The extraction process of causal features is as follows:
In Equation 10,
In terms of feature decoupling, we use an Attention Fusion (AF) module for causal reasoning. As shown in Figure 4, given the input, the attention operation in the AF module can be expressed as:
In Equation 11,
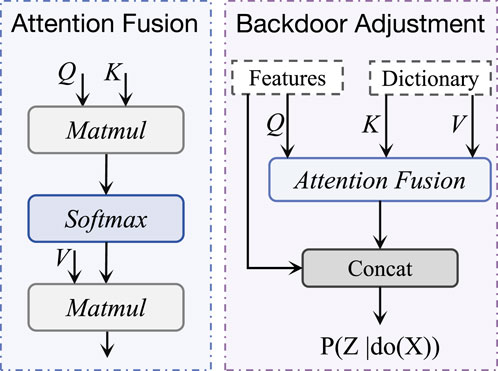
Figure 4. The structure of backdoor adjustment, which is based on the attention fusion module. It takes ECG features and confounding dictionaries as inputs to enable causal reasoning processes.
The confounding dictionary is constructed by computing the average feature vector
where
In the AF module, the keys
By leveraging the confounder dictionary in this way, the AF module explicitly accounts for class-specific confounding factors, enhancing the model’s ability to learn robust and generalizable representations. This transformation of the confounder dictionary into keys and values is a critical component of our causal intervention strategy, enabling the model to disentangle causal relationships from confounding biases.
3.6 Training strategy
In causal reasoning, we want to estimate the true causal characteristics, so we use the cross entropy loss:
In Equation 12,
In Equation 13,
4 Experiment
4.1 Datasets
The PTB-XL dataset (29), which is a recently introduced and comprehensive collection of 12-lead ECG dataset, consists of 21,837 clinical ECG recordings. Each recording spans a duration of 10 s and is sourced from a total of 18,885 individual patients. Table 1 provides an overview of the amount of ECG recordings in various tasks within this dataset. Table 1 summarizes the distribution of ECG recordings across various tasks in the PTB-XL dataset, including rhythm, form, super-diagnosis (super-diag.), sub-diagnosis (sub-diag.), diagnosis (diag.), and all. For each task, the table provides the number of classes, as well as the number of recordings in the training, validation, and test sets, along with the total count. The tasks are designed to capture different levels of ECG interpretation, ranging from broad rhythm classification to detailed diagnostic categorization. This table serves as a comprehensive reference for understanding the dataset’s structure and facilitates the development and evaluation of models across diverse ECG analysis tasks. The CPSC (Liu et al., 2018) comprises 6,877 12-lead ECG recordings, each spanning from 6 to 60 s. The HFHC (Anonymous, 2019) comprises 20,335 medical ECG samples, categorized into 34 classes, and each sample is equipped with 8 leads. The Chapman (Zheng et al., 2020), consisting of 12-lead ECG recordings from 10,646 patients, is utilized in this study.
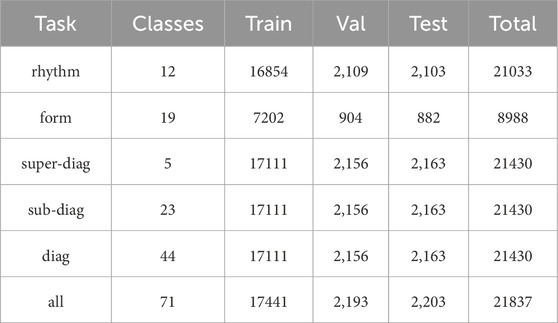
Table 1. Amount of ECG recordings in the training set, validation set, and test set in various tasks on the PTB-XL dataset.
4.2 Implementation details
For each dataset, the samples are randomly divided following an 8:1:1 ratio into training, validation, and test sets. Approximately 80% of the data is used for training the model and capturing latent patterns within the ECG signals. Around 10% of the data is reserved for validation, which is utilized for model selection and hyperparameter tuning. The remaining 10% is set aside as an independent test set to assess the model’s generalization ability. For each dataset, we partition the data into 10 non-overlapping groups. Specifically, groups 1 to 8 are used as the training set to train the network and learn latent patterns from the data. Group 9 is employed as the validation set for model selection and hyperparameter tuning. Group 10 serves as an independent test set to evaluate the performance of the model on unseen data. All experiments are conducted with standardized training settings to ensure consistency and comparability across datasets. The optimizer used is Adam, with an initial learning rate of 0.001. The learning rate is scheduled to decay following a cosine annealing strategy during training. The batch size is set to 64 for all experiments. Cross-entropy loss is adopted as the loss function. The implementation is based on the PyTorch framework. All experiments are performed on a single NVIDIA RTX 3090 GPU with 24 GB memory. This standardized setup facilitates reproducibility and fair comparison across different datasets.
4.3 Experimental results
In the experiment of ECG multi-label classification task, we obtained the results in Table 2 by comparing the performance of our proposed method with other advanced methods on the PTB-XL dataset. In the table, we show the classification performance of various methods on different tasks (rhythm, form, super-diag., sub-diag., diag., all), using sensitivity (SEN) and area under the curve (AUC). For the rhythm task, our method achieves 91.68% on SEN and 97.02% on AUC, respectively, which is significantly superior to other methods, indicating that our model has excellent performance for the rhythm classification task of ECG. Similarly, in the form task, our method has a SEN of 58.81% and an AUC of 88.22%. Compared with other methods, our model also performs well in the task of form detection. On the comprehensive tasks of super-diag., sub-diag., diag., and all, our approach achieves the best results on both SEN and AUC. It achieves 80.17%, 73.28%, 68.54%, 72.96% (SEN) and 92.90%, 92.88%, 93.78%, 93.14% (AUC), respectively, showing comprehensive and stable performance. It is worth noting that in the comparative experiment, our method achieved higher sensitivity and AUC on multiple tasks compared with other models, indicating that our model has better processing ability for different ECG tasks.
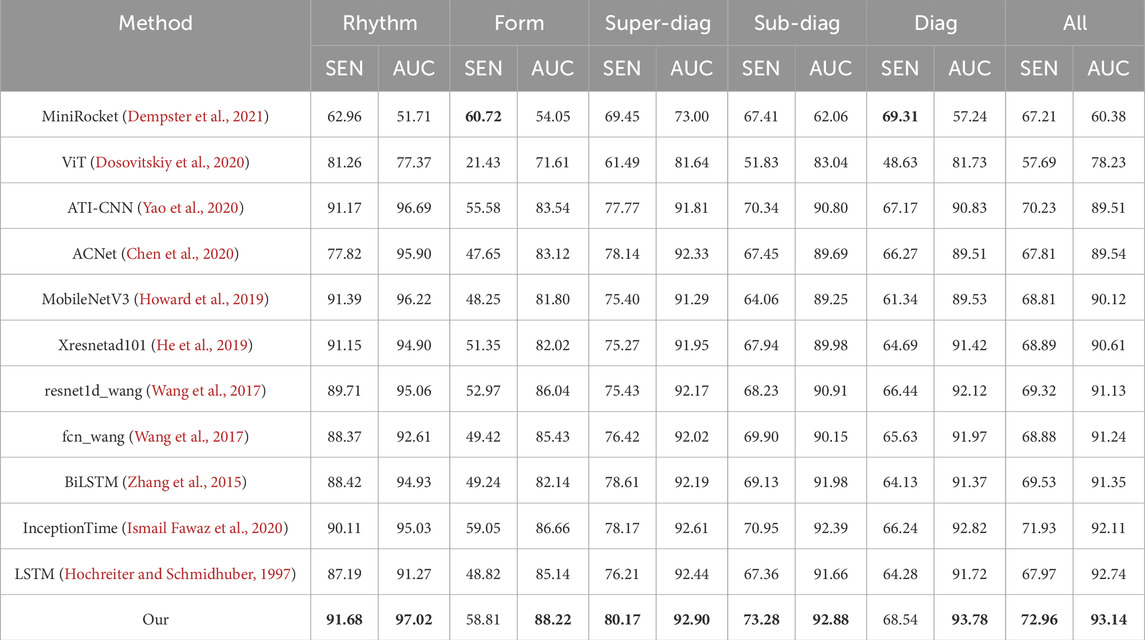
Table 2. Comparison of various methods with our proposed method across multiple tasks of the PTB-XL. Bold values indicate the best results.
Table 3 shows the performance of our proposed method compared with other advanced methods on CPSC2018 and HFHC datasets. On the CPSC2018 dataset, our method achieved 95.88% and 84.49% on AUC and SEN, respectively, showing significant advantages compared with other methods. In particular, compared with the suboptimal method, our model improves AUC by at least 0.65% and SEN by at least 3.25%. This shows that our model has a stronger classification ability on the CPSC2018 dataset and can identify the relevant features of ECG more accurately. On the HFHC dataset, our method also achieved excellent performance, with AUC and SEN reaching 94.96% and 95.73%, respectively. Compared to other methods, our model improves at least 2.18% on AUC and 3.26% on SEN. This further validates the superiority of our model on HFHC datasets with higher sensitivity and better classification performance.
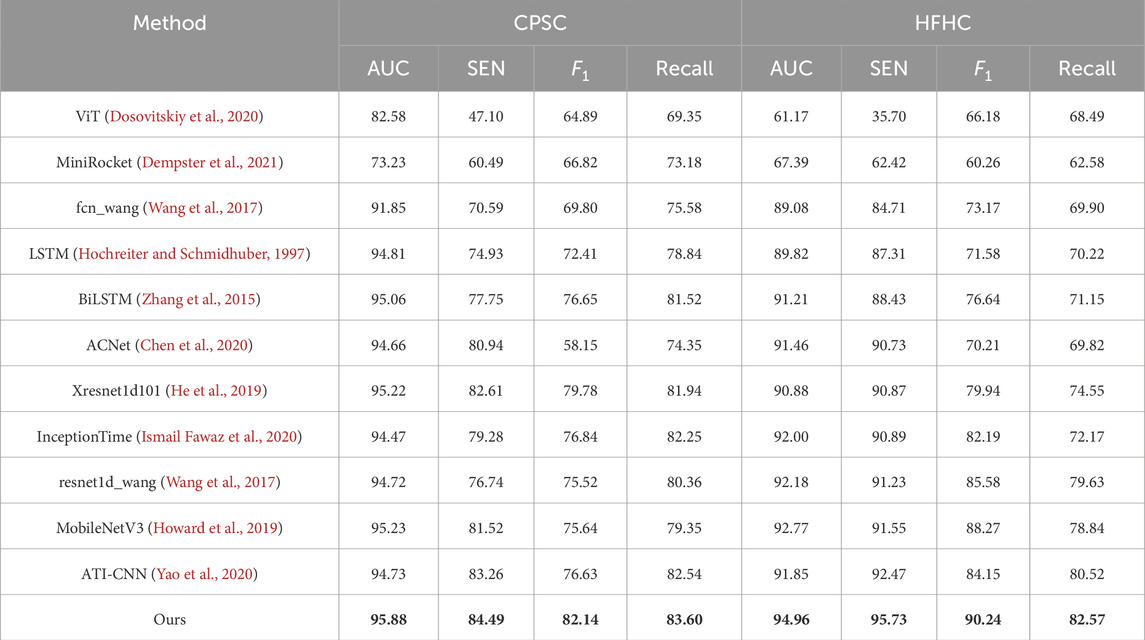
Table 3. Comparison of various methods with our proposed method across multiple tasks of the CPSC2018 dataset and the HFHC dataset. Bold values indicate the best results.
Table 4 presents a comparison of our proposed method with other networks on various tasks within the Chapman dataset. Our model showcases remarkable performance, achieving a notable

Table 4. Comparison of
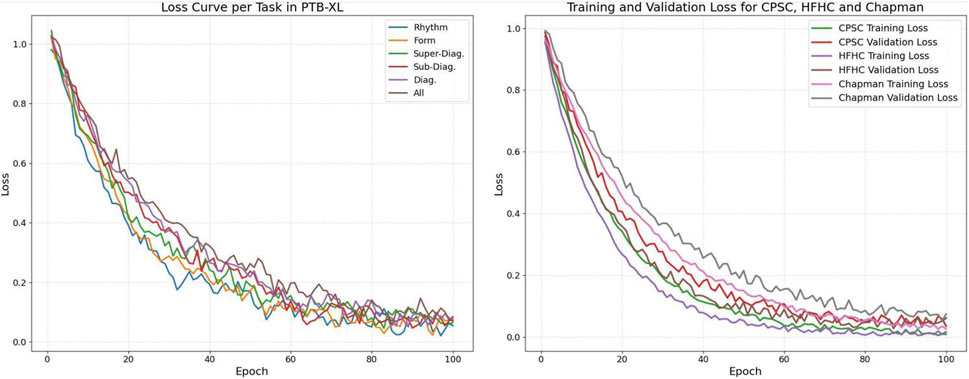
Figure 5 illustrates the loss curves across different datasets and tasks, providing a comprehensive view of the model’s learning dynamics. As shown in the left subfigure, the training losses for the six PTB-XL subtasks—including Rhythm, Form, Super-Diagnostic, Sub-Diagnostic, Diagnostic, and All—exhibit a consistent downward trend with moderate fluctuations, indicating stable convergence. The right subfigure presents both training and validation losses for CPSC, HFHC, and Chapman datasets. The clear and steadily decreasing validation curves, along with the narrowing gaps between training and validation loss, suggest that the model effectively avoids overfitting and generalizes well across datasets.
4.4 Ablation studies
In this section, we conduct ablation experiments to evaluate the effects of the time domain features embedding module versus the causal reasoning module. At the same time, we investigate the effects of the convolution kernel size of the time domain features embedding module on ECG classification performance.
4.4.1 Impact of kernel size
Table 5 shows the performance comparison of different convolution kernel combinations in time-domain features embedding, covering both the PTB-XL dataset and the Chapman dataset. We focused on several key metrics to assess the impact of different convolution kernel combinations on feature extraction in the time domain.
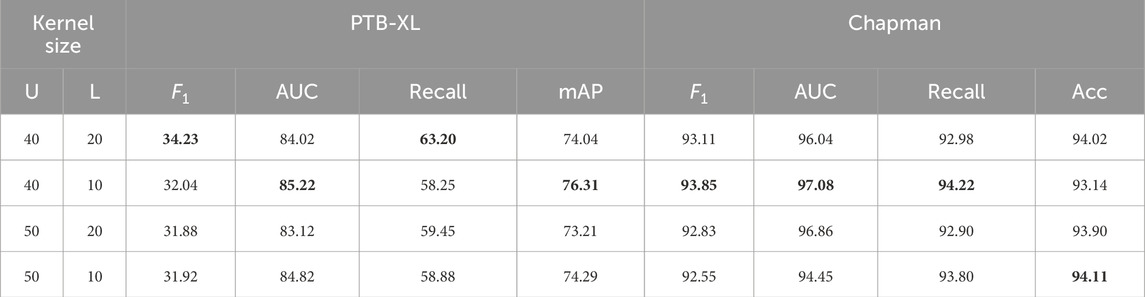
Table 5. Performance of various kernel sizes in Time Domain Features Embedding on the PTB-XL dataset and the Chapman dataset. Bold values indicate the best results.
On the PTB-XL dataset, the convolution kernel combinations 40/20 perform best in terms of
In general, the choice of convolution kernel combination is very important for the extraction of time domain features. This reminds us that different combinations of convolution kernel may be required on different tasks and data sets to optimize the performance of feature extraction in the time domain.
4.4.2 Module ablation experiment
Table 6 shows the performance comparison of the time domain embedding module (TDFE), causal inference module (CR), and their combination (TDFE + CR) under different tasks. In the rhythm task, the TDFE module achieved 91.74% and 86.39% on AUC and SEN, and the CR module achieved 91.22% and 88.42% on AUC and SEN, respectively. However, the TDFE + CR combination performed the best, with AUC and SEN reaching 97.02% and 91.68%, respectively, a significant improvement over any single module. This shows that both TDFE and CR modules play an active role in rhythm tasks, and their combination effect is better.
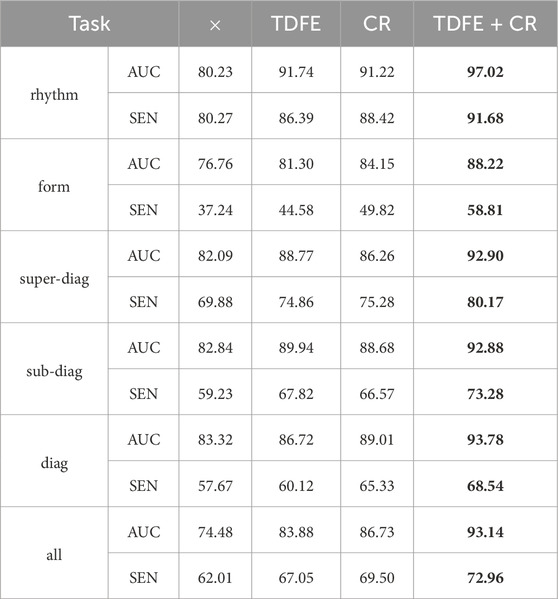
Table 6. Performance of Time Domain Features Embedding (TDFE) and Causal Reasoning module (CR), as well as their combination (TDFE + CR). Bold values indicate the best results.
In the form task, the TDFE module achieved 81.30% and 44.58% on AUC and SEN, and the CR module achieved 84.15% and 49.82% on AUC and SEN, respectively. The TDFE + CR combination achieved 88.22% and 58.81% on AUC and SEN, respectively, significantly outperforming a single module. This shows that both TDFE and CR modules are beneficial in form tasks, and their combination is more significant. Similar trends are observed in other tasks (super-diag., sub-diag., diag., all). Overall, the TDFE and CR modules had a positive impact across the tasks, and their combination was even more significant, providing more powerful performance for the ECG classification task. This further validates the effectiveness of comprehensive treatment of confounders by obtaining image features and temporal domain features and discovering true causal associations.
4.4.3 Parameters analysis
As shown in Table 7, our model performs well on both the number of parameters and the inference time. Specifically, we had the second smallest number of participants at 0.25 million. The inference time was only 2.27 milliseconds, the second fastest. Compared to other methods, the number of parameters in our model is much lower than most other methods, only 0.25 million. This means that our model has a more concise structure, and fewer parameters help to reduce the complexity of the model, reduce the risk of overfitting, and save computing resources. Second, our model’s inference time is also excellent, at just 2.27 milliseconds. Although there are some methods with shorter inference times, our model is still at a faster level. This means that our model has high real-time and efficiency in practical applications, and can process data quickly. This indicates that our model is less complex and the cost of implementing causal reasoning is not high.
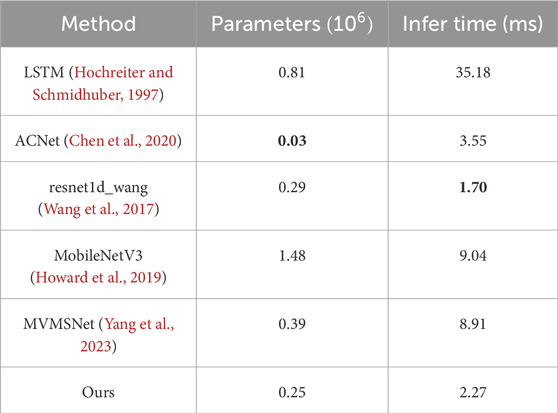
Table 7. The number of parameters and inference time for each method. Bold values indicate the best results.
5 Discussions
Causal reasoning plays a pivotal role in ECG classification, particularly in scenarios where cardiac abnormalities may be influenced by infectious diseases. By leveraging causal reasoning, we gain deeper insights into the causal relationships between features in ECG signals and their connections to classification outcomes, thereby enhancing both the accuracy and interpretability of classification models.
In this study, causal reasoning helps uncover the underlying causal chains in ECG signals, revealing how different physiological events and pathological states, such as those induced by infections, interact with cardiac function. This understanding is crucial for medical professionals, as it provides a clearer perspective on the patient’s condition, aiding in the timely and precise diagnosis and treatment of heart conditions potentially linked to infectious diseases. By identifying these cause-and-effect relationships, causal reasoning allows us to design more effective feature extraction and selection methods. These methods improve the model’s sensitivity to clinically relevant information while minimizing its susceptibility to noise and confounding effects. Moreover, causal reasoning contributes to building more reliable and generalizable models by reducing overfitting and ensuring a clearer direction of causality during model training. This ultimately has a direct impact on the early detection and treatment of heart diseases, particularly those influenced by infectious agents, improving the quality of medical services.
Our study specifically addresses the removal of confounding factors in ECG classification tasks by employing backdoor adjustment techniques. This approach enhances classification performance by re-weighting attention mechanisms and refining the representation of confounding factors. Experimental results demonstrate that the proposed causal attention module effectively mitigates the impact of confounders, significantly improving classification accuracy. By dynamically adjusting attention weights and optimizing the confounding dictionary representation, our method directs the model’s focus toward features most relevant to ECG classification, particularly in infection-related cases. This confounding removal strategy not only boosts model performance but also improves its interpretability and reliability, providing a valuable tool for clinical applications. Such advancements are essential in aiding healthcare professionals in diagnosing and managing cardiac conditions, especially those influenced by infectious diseases, with greater precision and confidence.
6 Conclusion
In this study, we propose an innovative ECG signal classification network that integrates temporal feature embedding and attention-based causal inference, addressing the challenges of multi-label ECG classification in medical scenarios, including infectious diseases. The network employs a convolutional architecture and dual temporal feature embedding modules to efficiently learn temporal patterns in ECG signals, enabling accurate classification in complex clinical settings. To enhance causal reasoning, we introduce confounding dictionaries and attention mechanisms, allowing the network to capture high-quality ECG information while mitigating the effects of confounding factors. Specifically, the proposed causal reasoning framework models the causal relationships in ECG signals, identifies true causal connections, and eliminates confounding effects, which are critical for distinguishing cardiac abnormalities potentially caused by infections, such as myocarditis or sepsis-induced cardiac dysfunction. Experimental results demonstrate that our network achieves competitive performance, highlighting its potential in diagnosing infectious disease-related cardiac complications.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
MW: Conceptualization, Methodology, Supervision, Writing – original draft. CY: Data curation, Software, Validation, Writing – original draft. WZ: Formal Analysis, Visualization, Writing – review and editing. ZX: Formal Analysis, Writing – original draft. QiL: Methodology, Project administration, Writing – review and editing. QaL: Writing – review and editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Andreao R. V., Dorizzi B., Boudy J. (2006). ECG signal analysis through hidden markov models. IEEE Trans. Biomed. Eng. 53 (8), 1541–1549. doi:10.1109/TBME.2006.877103
Anonymous (2019). Hefei high-tech cup dataset, 2019. Available online at: https://tianchi.aliyun.com/competition/entrance/231754/information.
Bender T., Beinecke J. M., Krefting D., Muller C., Dathe H., Seidler T., et al. (2023). Analysis of a deep learning model for 12-lead ECG classification reveals learned features similar to diagnostic criteria. IEEE J. Biomed. health Inf. 28 (4), 1848–1859. doi:10.1109/JBHI.2023.3271858
Chen T.-M., Huang C.-H., Shih E. S., Hu Y.-F., Hwang M.-J. (2020). Detection and classification of cardiac arrhythmias by a challenge-best deep learning neural network model. Iscience 23 (3), 100886. doi:10.1016/j.isci.2020.100886
Dempster A., Schmidt D. F., Webb G. I. (2021). “Minirocket: a very fast (almost) deterministic transform for time series classification,” in Proceedings of the 27th ACM SIGKDD conference on knowledge discovery and data mining, 248–257.
Dosovitskiy A., Beyer L., Kolesnikov A., Weissenborn D., Zhai X., Unterthiner T., et al. (2020). An image is worth 16x16 words: transformers for image recognition at scale.
Gao S.-H., Cheng M.-M., Zhao K., Zhang X.-Y., Yang M.-H., Torr P. (2019). Res2net: a new multi-scale backbone architecture. IEEE Trans. pattern analysis Mach. Intell. 43 (2), 652–662. doi:10.1109/TPAMI.2019.2938758
Ghongade R., Ghatol A. (2007). “Performance analysis of feature extraction schemes for artificial neural network based ECG classification,” in International conference on computational intelligence and multimedia applications (ICCIMA) (IEEE), 2, 486–490. doi:10.1109/iccima.2007.221
Han H., Lian C., Zeng Z., Xu B., Zang J., Xue C. (2023). Multimodal multi-instance learning for long-term ECG classification. Knowledge-Based Syst. 270, 110555. doi:10.1016/j.knosys.2023.110555
He T., Zhang Z., Zhang H., Zhang Z., Xie J., Li M. (2019). “Bag of tricks for image classification with convolutional neural networks,” in Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, 558–567.
Hochreiter S., Schmidhuber J. (1997). Long short-term memory. Neural Comput. 9 (8), 1735–1780. doi:10.1162/neco.1997.9.8.1735
Hou Q., Zhou D., Feng J. (2021). “Coordinate attention for efficient mobile network design,” in Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, 13 713–13 722.
Howard A., Sandler M., Chu G., Chen L.-C., Chen B., Tan M., et al. (2019). “Searching for mobilenetv3,” in Proceedings of the IEEE/CVF international conference on computer vision, 1314–1324.
Hu J., Shen L., Sun G. (2018). “Squeeze-and-excitation networks,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 7132–7141.
Ince T., Kiranyaz S., Gabbouj M. (2009). A generic and robust system for automated patient-specific classification of ECG signals. IEEE Trans. Biomed. Eng. 56 (5), 1415–1426. doi:10.1109/TBME.2009.2013934
Ismail Fawaz H., Lucas B., Forestier G., Pelletier C., Schmidt D. F., Weber J., et al. (2020). Inceptiontime: finding alexnet for time series classification. Data Min. Knowl. Discov. 34 (6), 1936–1962. doi:10.1007/s10618-020-00710-y
Jekova I., Bortolan G., Christov I. (2008). Assessment and comparison of different methods for heartbeat classification. Med. Eng. and Phys. 30 (2), 248–257. doi:10.1016/j.medengphy.2007.02.003
Karimifard S., Ahmadian A., Khoshnevisan M., Nambakhsh M. S. (2006). “Morphological heart arrhythmia detection using hermitian basis functions and KNN classifier,” in 2006 international conference of the IEEE engineering in medicine and biology society (IEEE), 1367–1370.
Lassoued H., Ketata R. (2018). “ECG multi-class classification using neural network as machine learning model,” in 2018 international conference on advanced systems and electric technologies (IC_ASET) (IEEE), 473–478.
Liu F., Liu C., Zhao L., Zhang X., Wu X., Xu X., et al. (2018). An open access database for evaluating the algorithms of electrocardiogram rhythm and morphology abnormality detection. J. Med. Imaging Health Inf. 8 (7), 1368–1373. doi:10.1166/jmihi.2018.2442
Liu H., Li Q., Nie W., Xu Z., Liu A. (2024) “Causal intervention for brain tumor segmentation,” in Medical image computing and computer assisted intervention, 160–170.
Lynn H. M., Pan S. B., Kim P. (2019). A deep bidirectional gru network model for biometric electrocardiogram classification based on recurrent neural networks. IEEE Access 7, 145 395–145 405. doi:10.1109/access.2019.2939947
Martis R. J., Acharya U. R., Min L. C. (2013). Ecg beat classification using PCA, LDA, ICA and discrete wavelet transform. Biomed. Signal Process. Control 8 (5), 437–448. doi:10.1016/j.bspc.2013.01.005
Miao J., Chen C., Liu F., Wei H., Heng P.-A. (2023) “CauSSL: causality-inspired semi-supervised learning for medical image segmentation,” in Proceedings of the IEEE/CVF international conference on computer vision (ICCV), 21426–21437.
Murugesan B., Ravichandran V., Ram K., P. P. S., Joseph J., Shankaranarayana S. M., et al. (2018). “Ecgnet: deep network for arrhythmia classification,” in 2018 IEEE international symposium on medical measurements and applications (MeMeA), 1–6.
Nagal D., Sharma S. (2014). “Simultaneous 12-lead QRS detection by k-means clustering algorithm,” in International conference on recent advances and innovations in engineering (ICRAIE-2014) (IEEE), 1–4.
Nie W., Yu Y., Zhang C., Song D., Zhao L., Bai Y. (2024). Temporal-spatial correlation attention network for clinical data analysis in intensive care unit. IEEE Trans. Biomed. Eng. 71 (2), 583–595. doi:10.1109/tbme.2023.3309956
Nie W., Zhang C., Song D., Bai Y., Xie K., Liu A. (2023a) “Chest X-ray image classification: a causal perspective,” in Medical image computing and computer assisted intervention – miccai, 25–35.
Nie W., Zhang C., Song D., Bai Y., Xie K., Liu A. (2023b). Instrumental variable learning for chest X-ray classification. arXiv:2305.12070, 4506–4512. [eess.IV]. doi:10.1109/smc53992.2023.10394601
Ribeiro A. H., Ribeiro M. H., Paixão G. M. M., Oliveira D. M., Gomes P. R., Canazart J. A., et al. (2020). Automatic diagnosis of the 12-lead ECG using a deep neural network. Nat. Commun. 11 (1), 1760. doi:10.1038/s41467-020-15432-4
Sajjan M. (2012). Learn ECG in a day: a systematic approach. New Delhi, India: Jaypee Brothers Medical Ltd
Shimpi P., Shah S., Shroff M., Godbole A. (2017). “A machine learning approach for the classification of cardiac arrhythmia,” in 2017 international conference on computing methodologies and communication (ICCMC) (IEEE), 603–607.
Siontis K. C., Noseworthy P. A., Attia Z. I., Friedman P. A. (2021). Artificial intelligence-enhanced electrocardiography in cardiovascular disease management. Nat. Rev. Cardiol. 18 (7), 465–478. doi:10.1038/s41569-020-00503-2
Wagner P., Strodthoff N., Bousseljot R.-D., Kreiseler D., Lunze F. I., Samek W., et al. (2020). Ptb-xl, a large publicly available electrocardiography dataset. Sci. data 7 (1), 154. doi:10.1038/s41597-020-0495-6
Wang J., Qiao X., Liu C., Wang X., Liu Y., Yao L., et al. (2021). Automated ecg classification using a non-local convolutional block attention module. Comput. Methods Programs Biomed. 203, 106006. doi:10.1016/j.cmpb.2021.106006
Wang Z., Yan W., Oates T. (2017). “Time series classification from scratch with deep neural networks: a strong baseline,” in 2017 International joint conference on neural networks (IJCNN) (IEEE), 1578–1585.
Yang S., Lian C., Zeng Z., Xu B., Zang J., Zhang Z. (2023). A multi-view multi-scale neural network for multi-label ecg classification. IEEE Trans. Emerg. Top. Comput. Intell. 7, 648–660. doi:10.1109/tetci.2023.3235374
Yao Q., Wang R., Fan X., Liu J., Li Y. (2020). Multi-class arrhythmia detection from 12-lead varied-length ECG using attention-based time-incremental convolutional neural network. Inf. Fusion 53, 174–182. doi:10.1016/j.inffus.2019.06.024
Zhan C., Peng P., Zhang H., Sun H., Shang C., Chen T., et al. (2023) “Debiasing medical visual question answering via counterfactual training,” in Medical image computing and computer assisted intervention – miccai, 382–393.
Zhang M.-L., Zhou Z.-H. (2013). A review on multi-label learning algorithms. IEEE Trans. Knowl. data Eng. 26 (8), 1819–1837. doi:10.1109/tkde.2013.39
Zhang S., Lian C., Xu B., Zang J., Zeng Z. (2023). A token selection-based multi-scale dual-branch CNN-transformer network for 12-lead ECG signal classification. Knowledge-Based Syst. 280, 111006. doi:10.1016/j.knosys.2023.111006
Zhang S., Zheng D., Hu X., Yang M. (2015). “Bidirectional long short-term memory networks for relation classification,” in Proceedings of the 29th Pacific Asia conference on language, information and computation, 73–78.
Keywords: ECG classification, attention, time domain features, causal reasoning, backdoor adjustment, infectious disease diagnosis, cardiac signal variability
Citation: Wang M, You C, Zhang W, Xu Z, Liang Q and Li Q (2025) Causal ECGNet: leveraging causal inference for robust ECG classification in cardiac disorders. Front. Physiol. 16:1543417. doi: 10.3389/fphys.2025.1543417
Received: 13 December 2024; Accepted: 07 May 2025;
Published: 19 May 2025.
Edited by:
Rajesh Kumar Tripathy, Birla Institute of Technology and Science, IndiaReviewed by:
Ming Huang, Chinese Academy of Sciences (CAS), ChinaYehualashet Megersa Ayano, Ethiopian Artificial Intelligence Institute, Ethiopia
Copyright © 2025 Wang, You, Zhang, Xu, Liang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cong You, eW91Y29uZzE5ODdAdG11LmVkdS5jbg==
 Mei Wang1
Mei Wang1 Cong You
Cong You Zibo Xu
Zibo Xu Qiang Li
Qiang Li