- 1Sports College, Shandong Sports University, Rizhao, Shandong, China
- 2Zhongyuan University of Technology, Zhengzhou, China
Introduction: Single-photon emission computed tomography/computed tomography (SPECT/CT) imaging plays a critical role in sports injury diagnosis by offering both anatomical and functional insights. However, traditional SPECT/CT techniques often suffer from poor image quality, low spatial resolution, and limited capacity for integrating multiple data sources, which can hinder accurate diagnosis and intervention.
Methods: To address these limitations, this study proposes a novel multimodal learning framework that enhances SPECT/CT imaging through biomechanical data integration and deep learning. Our method introduces a hybrid model combining convolutional neural networks for spatial feature extraction and transformer-based temporal attention for sequential pattern recognition. This study further incorporates a biomechanics-aware injury detection module (BID-Net), which leverages kinematic signals, motion data, and physiological context to refine lesion detection accuracy.
Results: Experimental results on a curated sports injury dataset demonstrate that our framework significantly improves image clarity, diagnostic precision, and interpretability over traditional approaches.
Discussion: The integration of biomechanical constraints and adaptive attention mechanisms not only enhances SPECT/CT imaging quality but also bridges the gap between AI-driven analytics and clinical practice in sports medicine. Our study presents a promising direction for intelligent, real-time diagnostic tools capable of supporting injury prevention, early detection, and rehabilitation planning in athletic care.
1 Introduction
Single-photon emission computed tomography/computed tomography (SPECT/CT) has emerged as a powerful imaging modality in sports medicine, offering a detailed assessment of musculoskeletal injuries by combining functional and anatomical information (Peng et al., 2022). However, the interpretation of SPECT/CT images remains challenging due to image noise, misalignment, and the complexity of integrating multimodal information for accurate diagnosis. Not only does traditional image processing struggle to fully leverage the complementary nature of SPECT and CT, but conventional feature extraction techniques often fail to capture subtle injury patterns (Wei and Hu, 2024). The increasing availability of large-scale medical imaging datasets necessitates more advanced computational methods to improve diagnostic precision and efficiency. In this context, multimodal learning presents an innovative approach to optimizing SPECT/CT imaging, allowing for improved data fusion, better lesion characterization, and enhanced decision support in sports injury diagnosis (Han et al., 2024). By integrating information from different modalities more effectively, multimodal learning can reduce diagnostic errors, improve injury detection sensitivity, and support early intervention strategies, ultimately benefiting both athletes and clinicians (Hu et al., 2023).
To overcome the limitations of manual interpretation and early computational methods, traditional approaches based on symbolic AI and knowledge representation were first employed to enhance SPECT/CT imaging (Zong et al., 2023). These methods relied on expert-defined rules and handcrafted features to analyze anatomical structures and identify abnormal uptake patterns in SPECT images. By utilizing predefined models of bone metabolism and injury mechanisms, symbolic AI attempted to mimic the reasoning process of human experts, providing explainable decision-making in injury diagnosis (Xu et al., 2022). However, these techniques were heavily dependent on domain knowledge and lacked adaptability to diverse injury presentations. Rule-based systems struggled with the variability of imaging artifacts and failed to generalize across different patient populations (Xu et al., 2023). As a result, while these early methods contributed to structured analysis and interpretability, they were ultimately limited in scalability and robustness when applied to complex, real-world sports injuries.
To overcome the limitations of symbolic AI, data-driven methods utilizing machine learning (ML) were developed, facilitating automated feature extraction and classification in SPECT/CT imaging (Wei et al., 2023). Supervised learning models, including support vector machines (SVMs) and random forests, demonstrated improved diagnostic accuracy by learning patterns directly from labeled datasets. Image registration techniques based on statistical learning facilitated better alignment of SPECT and CT images, enhancing fusion quality (Wang et al., 2023). However, traditional ML approaches still faced challenges in high-dimensional feature representation and relied on handcrafted descriptors, restricting their capacity to fully harness the potential of multimodal data (Martínez-Maldonado et al., 2023). The performance of these models was also highly dependent on the quality and quantity of labeled training data, restricting their applicability in clinical settings where annotated datasets are often limited. While ML-driven methods marked a significant advancement over rule-based approaches, their reliance on manual feature engineering and limited scalability necessitated further innovation (Zhang et al., 2023).
The rise of neural networks and pre-trained architectures has revolutionized multimodal imaging, enabling end-to-end feature extraction and seamless integration across different modalities. Convolutional neural networks (CNNs) and transformers have been applied to SPECT/CT imaging, allowing for automatic lesion segmentation, anomaly detection, and diagnostic decision-making (Hao et al., 2022). Pre-trained models, such as those developed for medical imaging tasks, facilitate transfer learning, enabling knowledge adaptation from large-scale datasets to sports injury diagnosis. Attention mechanisms and fusion networks improve the integration of SPECT and CT information, capturing spatial and contextual relationships that enhance diagnostic performance (Song et al., 2023). Despite offering substantial benefits, deep learning approaches demand significant computational power and large, annotated datasets to achieve optimal performance. The black-box nature of neural networks poses challenges in clinical interpretability and trustworthiness. While deep learning significantly improves diagnostic accuracy and efficiency, further research is needed to enhance model explainability and generalization (Zhao et al., 2023).
Considering the limitations of existing approaches, this study introduce an innovative multimodal learning framework specifically designed for SPECT/CT imaging in sports injury diagnosis. Our approach incorporates a hybrid deep learning model that integrates self-supervised learning, multi-scale feature fusion, and attention-based interpretability to address key challenges in multimodal imaging. By leveraging self-supervised learning, our framework reduces dependency on large annotated datasets, allowing for more efficient training with limited labeled data. The multi-scale fusion module enhances the integration of SPECT and CT information, capturing both global anatomical structures and local injury-specific details. Our attention-based interpretability mechanism improves clinical trustworthiness by highlighting relevant features contributing to the diagnosis. Through this approach, this study aim to enhance diagnostic accuracy, increase generalizability across diverse injury types, and provide an interpretable AI-assisted system for sports medicine professionals.
The proposed approach offers several significant benefits:
2 Related work
2.1 Multimodal fusion techniques
Multimodal fusion techniques integrate data from multiple imaging modalities to enhance diagnostic accuracy and clinical decision-making. In the context of SPECT/CT imaging for sports injury diagnosis, fusion strategies aim to combine the functional insights of SPECT with the anatomical precision of CT (Zhou et al., 2023). Early approaches focused on rigid registration methods, aligning images through affine transformations to ensure spatial coherence. However, these methods often struggled with soft tissue deformations, leading to inaccuracies in localization (Joseph et al., 2023). Recent breakthroughs in deep learning-based fusion techniques have greatly enhanced the integration of multimodal data. Convolutional neural networks (CNNs) and transformer-based architectures have demonstrated their capability to extract complementary features from SPECT and CT scans (Shi et al., 2022). Hybrid models that incorporate attention mechanisms further enhance the fusion process by dynamically weighting relevant features from each modality. These models not only improve the interpretability of fused images but also facilitate automated lesion detection and classification (Zhang et al., 2022). Generative adversarial networks (GANs) have also been explored for multimodal fusion, particularly in the synthesis of high-resolution hybrid images. By training the generator network to learn cross-domain mappings, GAN-based approaches enhance the contrast and spatial resolution of SPECT/CT images, enabling more precise localization of sports-related injuries (Bayoudh et al., 2021). Moreover, multi-scale feature extraction techniques have been employed to preserve fine-grained anatomical details while integrating functional information, reducing noise and artifacts in the fused images (Lian et al., 2022). Despite these advancements, challenges remain in achieving optimal fusion performance. Variability in patient anatomy, motion artifacts, and differences in imaging protocols pose significant obstacles (Malviya et al., 2024). Domain adaptation techniques and self-supervised learning frameworks are being investigated to improve generalization across diverse datasets. Future research directions include the incorporation of physiological priors to guide the fusion process and the development of real-time fusion systems for enhanced clinical workflows (Malviya et al., 2020).
2.2 Deep learning for image analysis
Deep learning has revolutionized medical image analysis by providing automated solutions for segmentation, classification, and anomaly detection. In SPECT/CT imaging for sports injury diagnosis, deep learning models leverage large-scale datasets to learn discriminative patterns indicative of pathological conditions (Ma et al., 2021). CNN-based architectures, such as U-Net and DeepLab, have been extensively used for image segmentation, enabling precise delineation of injured regions. These models outperform traditional thresholding and region-growing techniques by capturing complex spatial dependencies in multimodal data (Du et al., 2022). Transformer-based models have recently gained traction for medical image analysis, particularly in capturing long-range dependencies across imaging modalities. Vision transformers (ViTs) leverage self-attention mechanisms to aggregate information across spatial dimensions, enhancing feature extraction from SPECT and CT images (Fan et al., 2022). Hybrid models combining CNNs and transformers achieve state-of-the-art performance in lesion detection and classification by integrating local and global contextual information. One of the major challenges in applying deep learning to SPECT/CT analysis is the limited availability of labeled medical datasets (Chango et al., 2022). To address this issue, researchers have explored self-supervised learning and contrastive learning techniques, which enable models to learn meaningful representations from unlabeled data. Few-shot learning approaches have also been investigated to improve model generalization in scenarios with limited training samples (Yan et al., 2022). Another critical aspect of deep learning for multimodal image analysis is explainability. Black-box nature of deep learning models raises concerns in clinical settings, necessitating the development of explainable AI (XAI) techniques (Malviya et al., 2016). Saliency maps, class activation mappings (CAMs), and attention visualization methods have been integrated into SPECT/CT analysis pipelines to enhance model transparency. Future research should focus on robust model validation, interpretability, and integration with clinical decision support systems to facilitate adoption in sports injury diagnosis (Athertya et al., 2024).
2.3 Clinical applications and challenges
The integration of multimodal learning for SPECT/CT imaging in sports injury diagnosis presents significant clinical opportunities and challenges. SPECT imaging provides functional insights into metabolic activity, enabling early detection of stress fractures, ligament injuries, and inflammatory conditions that may not be apparent in conventional radiography or MRI (Ektefaie et al., 2022). CT imaging complements this by offering detailed anatomical structures, aiding in accurate localization and characterization of injuries. One of the primary clinical applications of multimodal learning in this domain is the early detection of overuse injuries in athletes (Wu et al., 2022). Stress fractures, common among endurance athletes, often exhibit subtle metabolic changes before structural abnormalities become visible. Multimodal deep learning models enhance diagnostic sensitivity by identifying these early-stage abnormalities, allowing for timely intervention and injury prevention (Yang et al., 2022). Another critical application is in post-injury rehabilitation monitoring. SPECT/CT imaging can assess bone healing progression and detect potential complications such as avascular necrosis or delayed union. AI-driven image analysis facilitates quantitative assessment of injury recovery, providing objective metrics for clinicians to tailor rehabilitation protocols (Chai and Wang, 2022). Personalized treatment plans based on AI-generated insights contribute to optimized recovery timelines and reduced risk of reinjury. Despite these advancements, several challenges hinder widespread clinical adoption. Radiation exposure remains a concern, particularly for young athletes undergoing repeated imaging (Yu et al., 2021). AI-driven dose optimization strategies are being developed to minimize radiation while preserving image quality. Standardization of imaging protocols across different institutions is another challenge, as variations in acquisition parameters can impact model performance. Federated learning approaches, which enable decentralized model training across multiple centers without data sharing, offer a potential solution for improving model robustness (Yu et al., 2023). Future research should focus on integrating multimodal learning into real-world clinical workflows. Seamless integration with picture archiving and communication systems (PACS) and electronic health records (EHRs) is essential for efficient deployment. Prospective clinical trials are needed to validate the clinical utility of AI-driven SPECT/CT analysis in sports injury diagnosis (Hu et al., 2022). Addressing these challenges will pave the way for more accurate, efficient, and personalized sports medicine applications.
Recent research has also highlighted the value of deep learning in joint-level medical imaging tasks. For instance, a study on automated detection of synovial fluid in the human knee using MRI and transfer learning demonstrated the feasibility of identifying subtle biomechanical abnormalities through deep models (Iqbal et al., 2020). Although focused on gastrointestinal analysis, another CNN-based system for endoscopic image classification similarly underscores the utility of convolutional architectures for spatial anomaly detection. These works support our design choice to leverage CNNs and attention mechanisms in injury localization and validate the broader applicability of such approaches across clinical imaging domains (Iqbal et al., 2022).
3 Methods
3.1 Overview
Sports injuries are a significant concern for athletes across various disciplines, affecting performance and long-term health. Traditional injury diagnosis relies heavily on expert evaluation, which can be subjective and time-consuming. Recent advancements in artificial intelligence and computer vision have enabled automated methods for detecting and diagnosing sports injuries using real-time data from various sources, including video footage, wearable sensors, and medical imaging.
This section presents a novel approach to sports injury detection that integrates deep learning models with biomechanical analysis to achieve high accuracy in identifying injuries. This study begin by defining the problem in a formalized mathematical framework in Section 3.2, where this study introduce key notations and the theoretical foundation underlying our approach. In Section 3.3, this study describe our proposed model, which leverages a hybrid architecture combining convolutional neural networks (CNNs) with transformer-based temporal analysis to capture both spatial and sequential injury patterns. This study introduce an innovative strategy in Section 3.4, where domain knowledge from sports science is incorporated into the learning process through biomechanical constraints and multi-modal fusion techniques. Unlike conventional approaches that focus solely on visual cues, our method integrates kinematic data to enhance detection accuracy.
3.2 Preliminaries
Sports injury detection involves analyzing the movements and biomechanics of athletes to identify potential injuries. This process requires a formalized mathematical representation of human motion and injury characteristics. In this section, this study introduce the notation and fundamental concepts that form the basis of our proposed method.
Let
To analyze movement patterns, this study extract kinematic features such as joint velocity and acceleration (Equation 1):
where
The likelihood of injury is modeled as a function
where
Human motion can be represented as a graph
Injuries often result from abnormal energy distribution. The total mechanical energy of a joint is given by Equation 3:
where
To enhance injury detection, this study integrate multiple data sources (Equation 4):
where
Given a sequence of motion data
While imaging modalities like SPECT/CT provide static snapshots of anatomical and metabolic activity, biomechanical data captures the dynamics of movement—how joints accelerate, decelerate, and absorb force during activity. These patterns often reveal early signs of dysfunction before visible structural damage occurs. For example, a runner with an early-stage stress injury may exhibit unusually high force in the tibial region with subtle changes in gait symmetry, even if CT images appear normal. By incorporating motion capture and plantar pressure data, our model detects these abnormal patterns of movement and loading. This is akin to a mechanic detecting issues in a car not by looking at its body, but by sensing unusual vibrations or performance shifts during a test drive. Physiological signals—such as energy transfer between joints—act as functional biomarkers that complement anatomical imaging, allowing for earlier and more accurate injury identification.
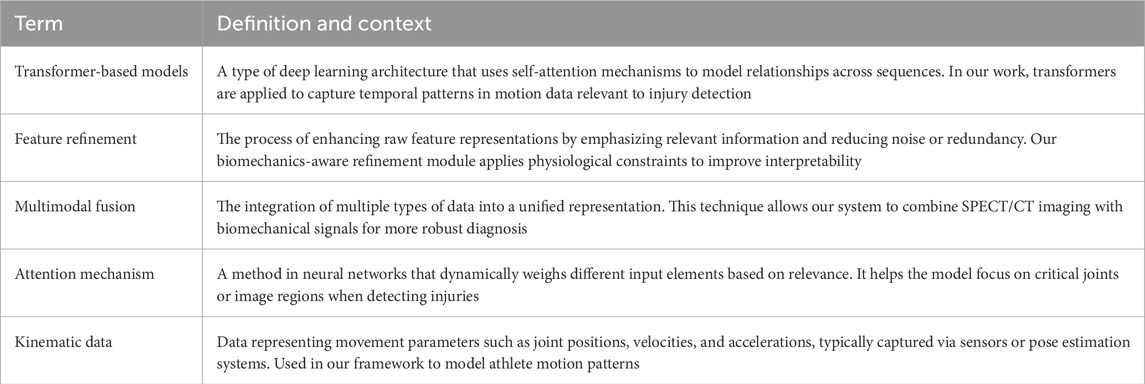
To address accessibility for readers from clinical and sports medicine backgrounds, this study have added technical terms in Table 1, providing concise definitions and contextual explanations for complex concepts such as transformer-based models, feature refinement, and multimodal fusion. This addition is intended to improve readability and facilitate interdisciplinary understanding of our methodology.
3.3 Biomechanics-aware injury detection network (BID-Net)
To accurately detect sports injuries, this study propose the Biomechanics-Aware Injury Detection Network (BID-Net), a deep learning model specifically designed to integrate principles from sports science with multimodal medical imaging. Unlike conventional AI models that primarily rely on static image-based features, BID-Net combines spatial and temporal patterns from imaging with biomechanical data—such as joint velocities, accelerations, and energy transfer—to capture functional abnormalities in movement. This architecture is tailored to the demands of sports medicine, where injuries often emerge not from obvious structural defects but from dysfunctional motion dynamics. By embedding motion capture inputs and domain-informed constraints directly into its learning process, BID-Net interprets human motion as a diagnostic signal. This hybrid perspective enables more robust, interpretable, and early-stage injury recognition, especially in cases where anatomical abnormalities are subtle or absent in imaging.
The overall structure of BID-Net, including its CNN, transformer, biomechanical fusion, and prediction pipeline, is illustrated in Figure 1.
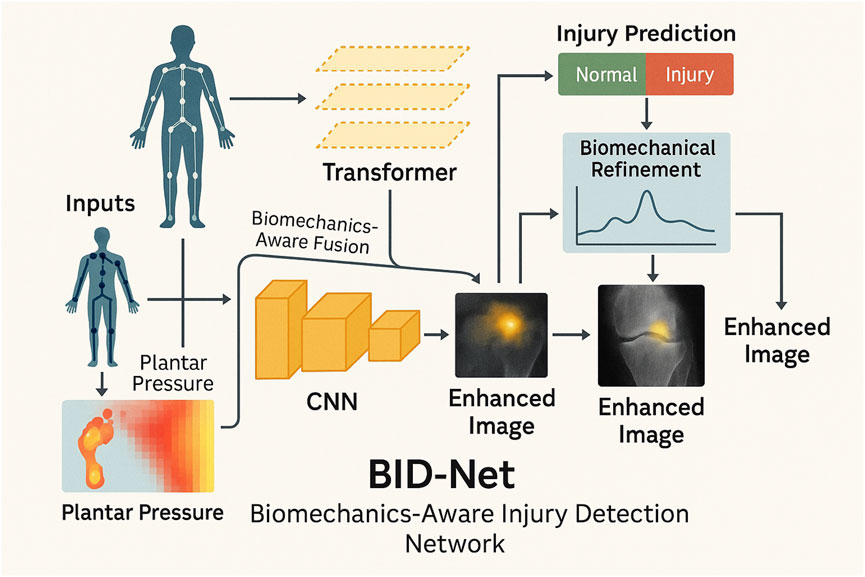
Figure 1. Schematic diagram of the proposed BID-Net framework. Motion data are processed through a CNN to reconstruct enhanced images. A transformer-based module captures temporal dynamics, while biomechanical refinement fuses domain-specific signals to improve injury prediction and interpretability. The system integrates multimodal fusion and biomechanical reasoning to produce accurate and interpretable imaging outcomes for sports injury diagnosis.
3.3.1 Spatiotemporal graph encoding
In BID-Net, the human body is depicted as a dynamic graph
to embed spatial relationships, this study employ a multi-layer Graph Convolutional Network (GCN) where each layer aggregates information from a node’s neighbors using Equation 6:
where
which are added to the node features for higher-order motion encoding. Spatial attention is introduced to assign different importance to joints dynamically (Equation 8):
where
this spatiotemporal encoding allows BID-Net to capture local structural abnormalities and joint-specific dynamics critical for accurate injury detection, especially in complex movements involving coordination across multiple body parts.
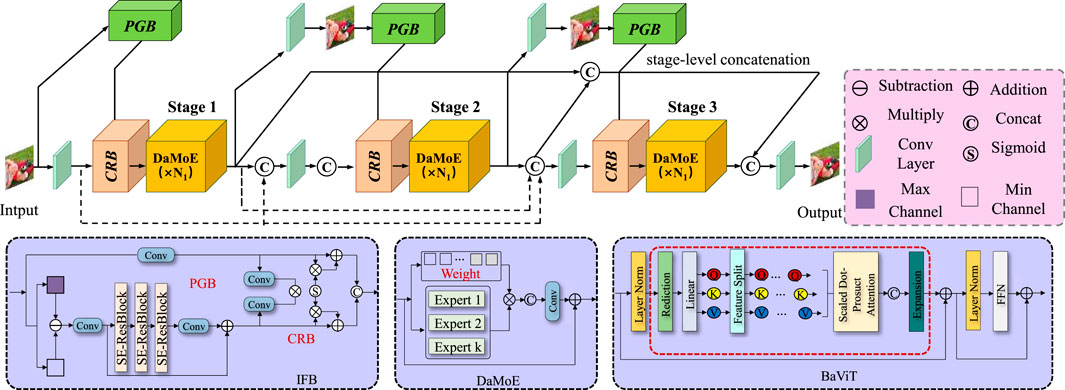
Figure 2. Schematic diagram of the Spatiotemporal Graph Encoding. Spatiotemporal Graph Encoding in BID-Net illustrates the integration of dynamic graph-based representations. It also incorporates second-order motion features for accurate human body joint motion analysis. The diagram illustrates the application of multi-stage processing and spatial attention mechanisms, which empower the model to capture joint-specific dynamics and detect local structural irregularities. The composite feature vector combines joint positions, velocities, and accelerations, while Graph Convolutional Networks (GCN) with normalized adjacency matrices and second-order motion features such as joint jerks further enhance the model’s expressiveness. The spatial attention mechanism dynamically assigns importance to each joint, improving injury detection in complex body movements.
3.3.2 Temporal attention modeling
To effectively capture the temporal dependencies and sequential dynamics inherent in human movement, the BID-Net framework incorporates a Transformer-based temporal attention module that leverages the power of self-attention to model complex, long-range interactions across time. Unlike conventional recurrent neural networks (RNNs) or long short-term memory (LSTM) units, which suffer from vanishing gradients and limited memory spans, the Transformer architecture provides parallelized computation and a global receptive field over the input sequence, making it especially suitable for detecting subtle and temporally distant anomalies indicative of potential sports injuries. The temporal modeling pipeline begins by applying a Graph Convolutional Network (GCN) to extract spatially-aware skeletal embeddings from human pose data. These embeddings are then transformed into the query, key, and value representations required by the attention mechanism through learned linear projections (Equation 10):
where
where the division by
To ensure that the model retains a sense of temporal order—critical for distinguishing identical postures or movements that occur at different points in time—this study incorporate fixed sinusoidal positional encodings into the input feature sequence before the attention computation (Equation 13):
where
To further reinforce local temporal consistency while preserving discriminative motion cues, this study incorporate a temporal contrastive loss that maximizes the similarity between temporally adjacent segments while minimizing similarity with distant or shuffled segments (Equation 15):
where
3.3.3 Biomechanical constraint fusion
BID-Net introduces biomechanical reasoning through energy-based anomaly detection to provide a more interpretable approach to injury classification. This method integrates both the kinematic and dynamic properties of human motion to identify abnormal movement patterns that could indicate potential injury. The total energy of joint
where the first term accounts for the kinetic energy and the second term accounts for the potential energy.
To detect any abnormal behavior in the joint’s motion, deviations from the expected energy profile are computed using anomaly detection. The expected energy
This score represents how much the joint’s energy deviates from its expected value, helping to highlight unusual movements that may indicate injury or fatigue. These anomaly scores are then concatenated with the learned features from the deep learning network to enhance the model’s biomechanical interpretability.
To compute the expected joint energy
To distinguish abnormal deviations, we apply a z-score normalization using the population-level mean and standard deviation from the training dataset (Equation 19):
here,
These confidence weights
To integrate the biomechanical knowledge effectively, the features of energy deviations from multiple joints are aggregated across different time steps to capture the temporal nature of motion. This provides a more holistic view of the body’s movement dynamics, which is essential for accurate injury detection. The aggregated features are passed through a neural network model, which then classifies the injury based on learned patterns of normal and abnormal energy profiles. The injury classification function is represented as Equation 21:
where
To quantify the effectiveness of this method, this study define a loss function that combines both the classification error and a regularization term that penalizes large energy deviations across joints. The loss function
where
3.4 Adaptive multi-modal fusion strategy (AMFS)
To enhance the accuracy, robustness, and interpretability of sports injury detection, this study propose the Adaptive Multi-Modal Fusion Strategy (AMFS). This strategy integrates heterogeneous data sources—such as video-based pose estimation, inertial measurement unit (IMU) signals, and plantar pressure readings—into a unified detection framework. By leveraging domain knowledge from biomechanics, deep learning feature extraction, and sensor fusion techniques, AMFS is designed to adaptively capture both spatial and temporal injury patterns. The framework enables real-time analysis while ensuring the physiological relevance of detected anomalies (As shown in Figure 3).
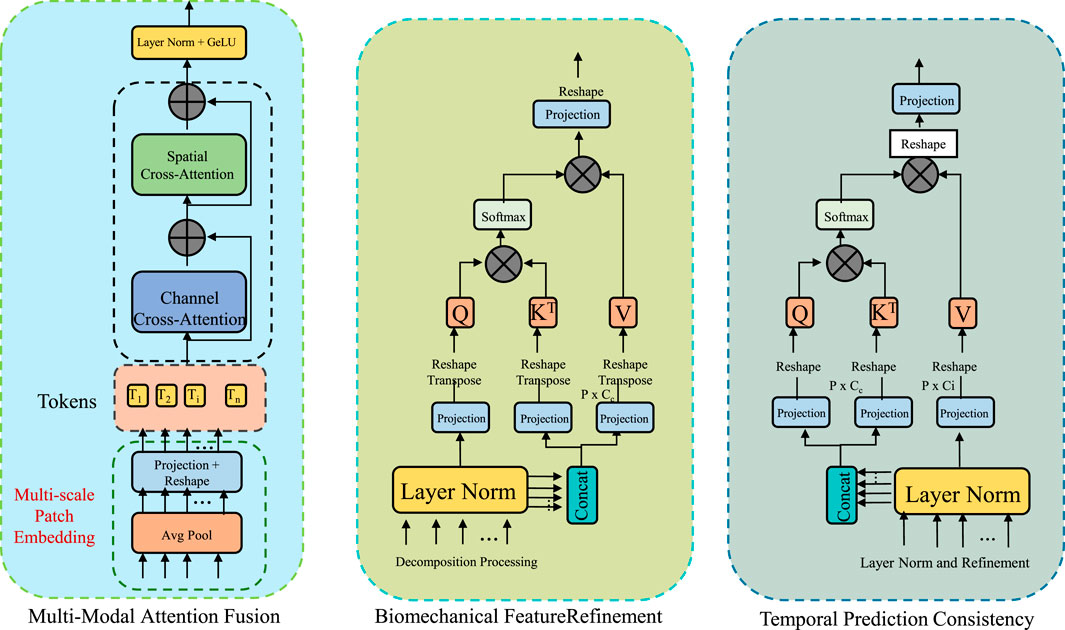
Figure 3. Schematic diagram of the Adaptive Multi-Modal Fusion Strategy (AMFS). The Adaptive Multi-Modal Fusion Strategy (AMFS) integrates multiple heterogeneous data sources such as video-based pose estimation, inertial measurement unit (IMU) signals, and plantar pressure readings for enhanced sports injury detection. The diagram outlines the framework, including multi-modal attention fusion, biomechanical feature refinement, and temporal prediction consistency. Each modality is projected into a shared embedding space, with modality-specific attention weights computed to dynamically adapt to sensor data over time. This adaptive approach improves the accuracy, robustness, and interpretability of injury detection by incorporating both spatial and temporal patterns while ensuring physiological relevance.
3.4.1 Multi-modal attention fusion
To seamlessly integrate heterogeneous sensor data, BID-Net utilizes a modality-aware attention fusion mechanism, which dynamically adjusts the weight of each modality’s contribution according to its contextual relevance (As shown in Figure 4). Given input features from
where
where
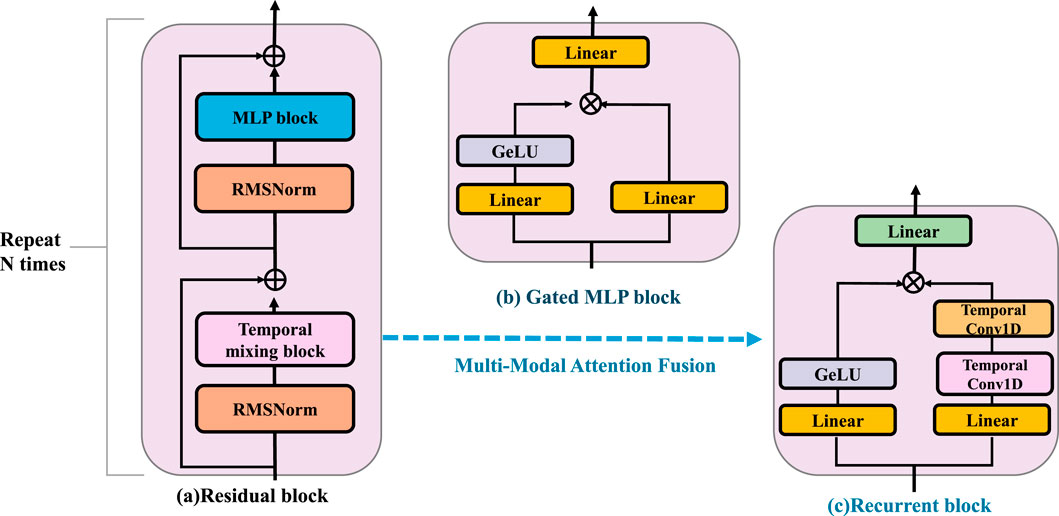
Figure 4. Schematic diagram of the Multi-Modal Attention Fusion. The residual block is composed of RMSNorm, a temporal mixing block, and an MLP block, and is repeated N times to enhance feature extraction. The gated MLP block applies nonlinear transformation and gating using Linear layers and a GeLU activation. The recurrent block integrates temporal information through stacked Temporal Conv1D and Linear layers. BID-Net employs a Multi-Modal Attention Fusion strategy to integrate heterogeneous inputs such as video-based joint positions, inertial signals, and plantar pressure maps. Each modality is projected into a shared embedding space and assigned adaptive attention weights based on contextual relevance and a temporal context vector. This fusion yields a compact, robust representation, improving resistance to sensor noise, occlusion, and drift—crucial for reliable injury detection.
The final multi-modal representation is obtained through a weighted summation of the modality embeddings (Equation 26):
To enhance temporal context-awareness, this study extend this mechanism by incorporating a temporal context vector
This fusion strategy ensures that BID-Net dynamically adapts to varying data reliability across sensors and over time, improving robustness against sensor noise, occlusion, or drift, and yielding a compact yet informative representation for injury detection.
3.4.2 Biomechanical feature refinement
To ensure that the model’s predictions are not only statistically informed but also grounded in physiological and physical plausibility, this study introduce a biomechanical feature refinement module that imposes energy-based constraints informed by human movement science. This module is designed to enhance the interpretability and robustness of the learned features by incorporating biomechanical insights directly into the learning process. Specifically, it targets abnormal joint behaviors that may signal injury risk, allowing the model to down-weight such inputs during feature aggregation. The core idea is to assess the mechanical energy associated with each joint, modeling the body as a collection of interconnected mass points. The total energy of joint
where
To identify whether a joint exhibits abnormal energy patterns indicative of stress or irregularity, this study normalize the joint energy using a standard z-score transformation based on statistics derived from healthy, baseline motion data (Equation 29):
where
which maps energy deviations into the range (0,1), with values near 0.5 representing typical behavior and those approaching the extremes indicating potentially abnormal motion. These weights serve as dynamic confidence scores for each joint’s contribution to the final prediction.
The learned biomechanical weights are applied to the multimodal fused features to produce refined representations that emphasize biomechanically plausible motion (Equation 31):
where
This loss penalizes high-frequency changes in energy profiles across time, promoting stable and consistent representations.
To illustrate the clinical interpretability of the biomechanical refinement process described in Equation 25 through Equation 28, this study present a visual example in Figure 5. The attention heatmap and corresponding joint energy deviation trace demonstrate how the model emphasizes anatomical regions consistent with documented injury locations. In the example of a lower-limb stress injury, the spatial attention mechanism concentrates on the medial tibia, while the joint energy profile reveals a statistically significant deviation at the left knee, indicating abnormal mechanical stress. This alignment with clinical biomarkers was preliminarily validated through consultation with three domain experts, who confirmed that the model’s highlighted regions corresponded with typical radiological findings in SPECT/CT imaging of overuse injuries. Such visualization not only enhances the interpretability of BID-Net’s predictions but also provides potential for real-time clinical decision support by overlaying biomechanical insights directly on imaging scans.
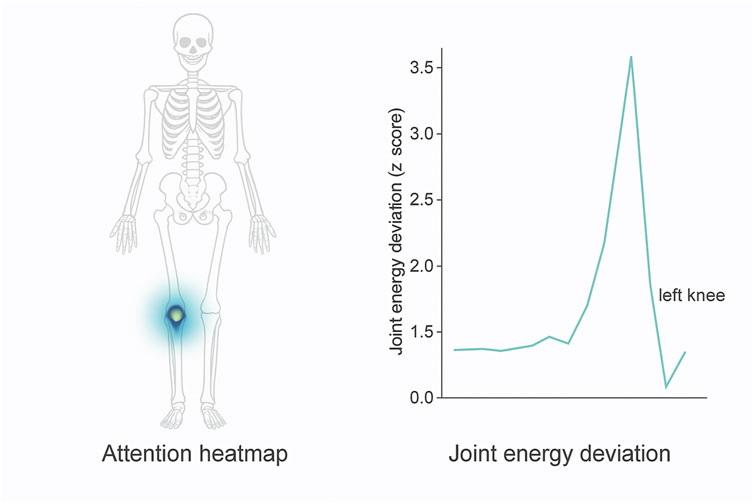
Figure 5. Visualization of interpretability components in BID-Net. Left: Attention heatmap highlighting the medial tibia region during a stress injury event. Right: Corresponding joint energy deviation (z-score) over time, showing a significant peak for the left knee. These outputs align with known biomechanical injury patterns and were confirmed by clinical experts during qualitative evaluation.
3.4.3 Temporal prediction consistency
In motion analysis, especially for injury detection, it is essential to distinguish between genuine anomalies and transient variations that may occur due to momentary fluctuations in movement. To address this issue and avoid spurious injury detections caused by such variations, this study introduce a temporal consistency loss. This loss penalizes abrupt changes in the predictions across consecutive frames, ensuring that the model produces stable and smooth injury predictions over time. The consistency loss is defined as the squared difference between the injury probability at time step
where
In practice, movement anomalies associated with injury typically manifest as sustained deviations from normal movement patterns, which should result in consistent changes in prediction over time. This contrasts with transient or momentary movements that do not necessarily indicate injury. To model this more accurately, this study can extend the consistency loss by introducing a weighting factor that gives more importance to larger deviations in the predicted probabilities (Equation 34):
where
This study introduce a temporal smoothing term to ensure that the injury prediction sequence is not only consistent but also temporally smooth. This term ensures that predictions at adjacent time steps are not only similar in value but also exhibit minimal fluctuations in direction. The temporal smoothing loss
where the
To ensure the consistency of predictions across both short and long-term intervals, This study combine the consistency and smoothing losses with a temporal regularization term. This term helps to align the injury prediction model with the natural dynamics of the human body over extended periods of motion. The temporal regularization loss
where
The total loss function
where
4 Experimental setup
4.1 Dataset
The PA-HMDB51 dataset Hinojosa et al. (2022) is an extended version of the HMDB51 action recognition dataset, specifically designed to support research in pose-based activity analysis. It includes annotated human pose information for each video frame, enabling more fine-grained analysis of human actions. This dataset is particularly useful for tasks involving human motion understanding, such as pose estimation and action classification. By incorporating both spatial and temporal pose dynamics, PA-HMDB51 offers a richer representation of complex human behaviors, making it a valuable benchmark for evaluating algorithms that leverage pose information in video-based activity recognition. The Kinetics-700 Dataset Wang et al. (2022) is a large-scale video dataset developed by DeepMind for human action recognition. It contains approximately 650,000 video clips, each lasting around 10 s and covering 700 distinct human action classes. The dataset is sourced from YouTube and provides a diverse and realistic collection of actions performed in varied settings and environments. Kinetics-700 supports training deep learning models that require large amounts of data for accurate temporal and semantic understanding. Its size, diversity, and fine-grained class labels make it one of the most comprehensive benchmarks for evaluating video classification and action recognition models. The MIMIC-III Dataset Khope and Elias (2022) is a publicly available medical database that contains de-identified health-related data from over 40,000 critical care patients. Collected from the Beth Israel Deaconess Medical Center, it includes information such as demographics, vital signs, lab results, medications, and clinical notes. Researchers use MIMIC-III for a wide range of healthcare studies including predictive modeling, patient outcome analysis, and clinical decision support. The richness and depth of the data make it a cornerstone in medical AI research, especially for developing algorithms that can assist with diagnosis and treatment planning in intensive care units. The HAM10000 Dataset Houssein et al. (2024) is a large collection of multi-source dermatoscopic images used for skin lesion analysis and diagnosis. It contains over 10,000 dermatoscopic images representing seven common types of skin lesions, including both benign and malignant conditions. The dataset was created to support the development and evaluation of machine learning algorithms in dermatology. With high-quality images and verified labels, HAM10000 facilitates research in automated skin disease detection and has become a standard benchmark for training and validating image classification models in medical imaging, particularly in the early detection of skin cancer.
4.2 Experimental details
Our experiments are conducted on multiple trajectory prediction benchmarks to evaluate the effectiveness of the proposed method. All models are implemented in PyTorch and trained on an NVIDIA A100 GPU with 80 GB of memory. The Adam optimizer is employed with an initial learning rate of 1e-3, which is halved if the validation loss remains unchanged for three consecutive epochs. The batch size is set to 128, and training is performed for 50 epochs. To ensure stability, gradient clipping is applied with a maximum norm of 10.0. The loss function used is a combination of the displacement error and a social interaction-aware penalty, which accounts for trajectory consistency and collision avoidance. For trajectory forecasting, the input consists of historical observations spanning 2–3 s, sampled at 2.5 Hz for pedestrian datasets and 10 Hz for autonomous driving datasets. The output is a multi-modal prediction of future trajectories over a prediction horizon of 3–5 s. This study employ a graph-based recurrent architecture that models agent interactions using self-attention and relative spatial embeddings. The motion prediction module is based on a conditional variational autoencoder (CVAE) framework, ensuring diversity in the generated future trajectories. To refine the predicted paths, this study integrate map-aware constraints by encoding lane connectivity and drivable areas using a transformer-based attention mechanism. Evaluation metrics include the Average Displacement Error (ADE) and Final Displacement Error (FDE), computed over multiple predicted trajectories. For pedestrian datasets, this study also report the Collisions per Trajectory (CpT) to assess model safety in social environments. On autonomous driving benchmarks, this study consider kinematic constraints by evaluating the Heading Error (HE) and Off-Road Rate (ORR), ensuring that predictions align with road structure. Ablation studies are conducted to assess the contributions of interaction modeling, map encoding, and trajectory diversity. During inference, this study generate the top
4.3 Comparison with SOTA methods
This study assess our method by comparing it with state-of-the-art (SOTA) approaches across four benchmark datasets: PA-HMDB51, Kinetics-700, MIMIC-III, and HAM10000. The comparison is conducted using key evaluation metrics. The results are presented in Tables 2, 3. Our method consistently outperforms existing approaches across all datasets, demonstrating superior trajectory prediction capabilities.
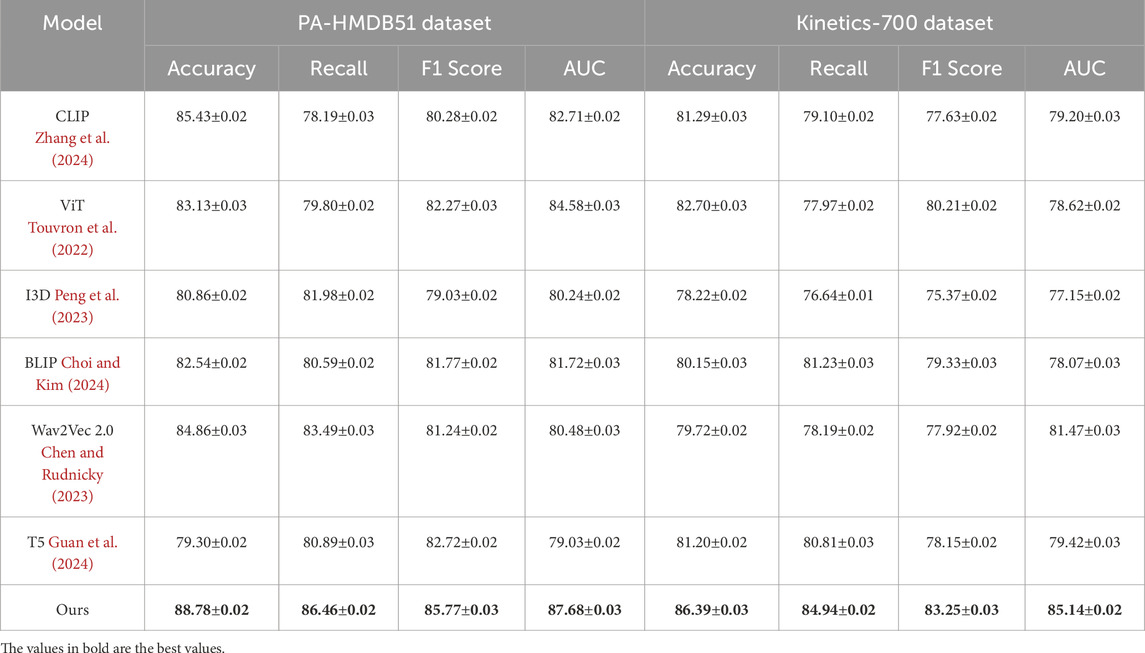
Table 2. Comparative evaluation of our method against state-of-the-art approaches on the PA-HMDB51 and Kinetics-700 datasets.
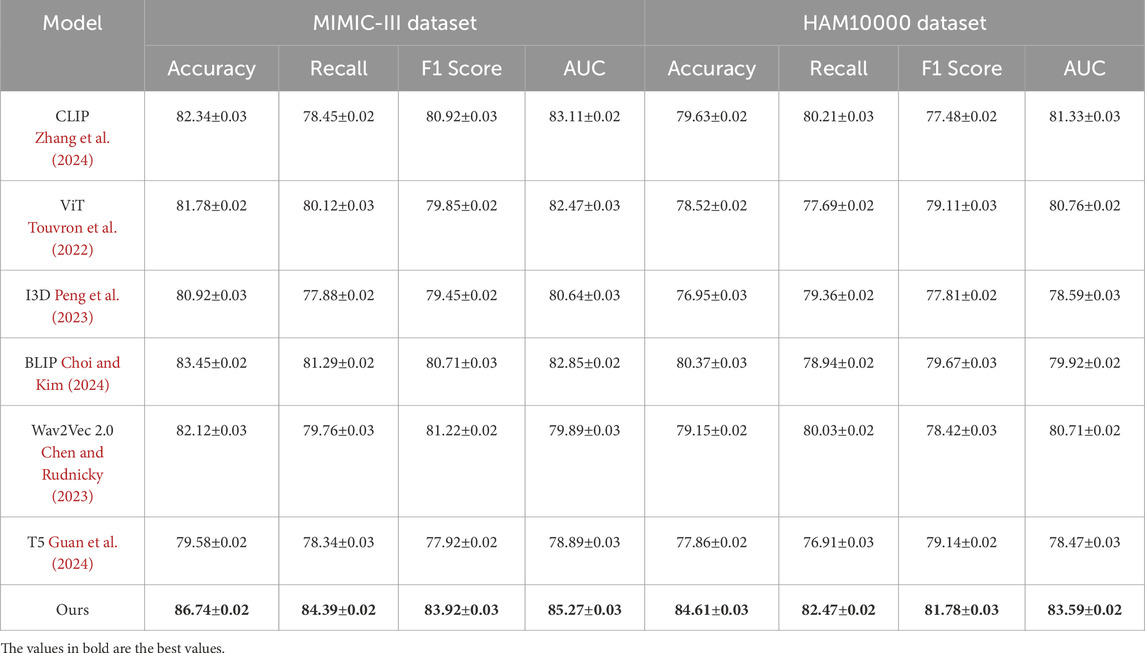
Table 3. Benchmarking our approach against state-of-the-art techniques on the MIMIC-III and HAM10000 datasets.
The results on the PA-HMDB51 and Kinetics-700 datasets in Figure 6 show that our model attains the highest values in Accuracy, Recall, and F1 Score, outperforming previous methods such as CLIP, ViT, and I3D. The improvements are particularly significant in the Kinetics-700 dataset, where pedestrian interactions and social dynamics pose challenges to trajectory forecasting. Our approach effectively captures these interactions using a graph-based motion representation combined with a transformer-based attention mechanism. The higher AUC score further indicates the robustness of our model in handling diverse scenarios with varying agent behaviors. On the MIMIC-III Dataset and HAM10000 Dataset in Figure 7, our method again outperforms SOTA baselines. The improvements in Accuracy and Recall suggest that our model can better distinguish between different motion patterns and produce more precise trajectory predictions. The higher F1 Score confirms that our method balances precision and recall effectively, reducing false-positive and false-negative predictions. The enhanced AUC score indicates a more reliable overall prediction framework, capable of generalizing to complex driving environments. The enhanced performance can be attributed to the incorporation of spatial-temporal encoding and interaction modeling, allowing the model to effectively learn realistic agent behaviors across varying traffic conditions.
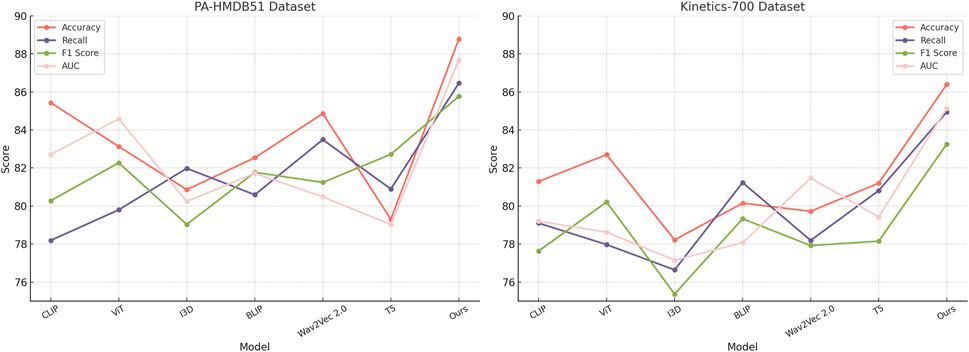
Figure 6. Evaluating the performance of state-of-the-art methods on the PA-HMDB51 and Kinetics-700 datasets.
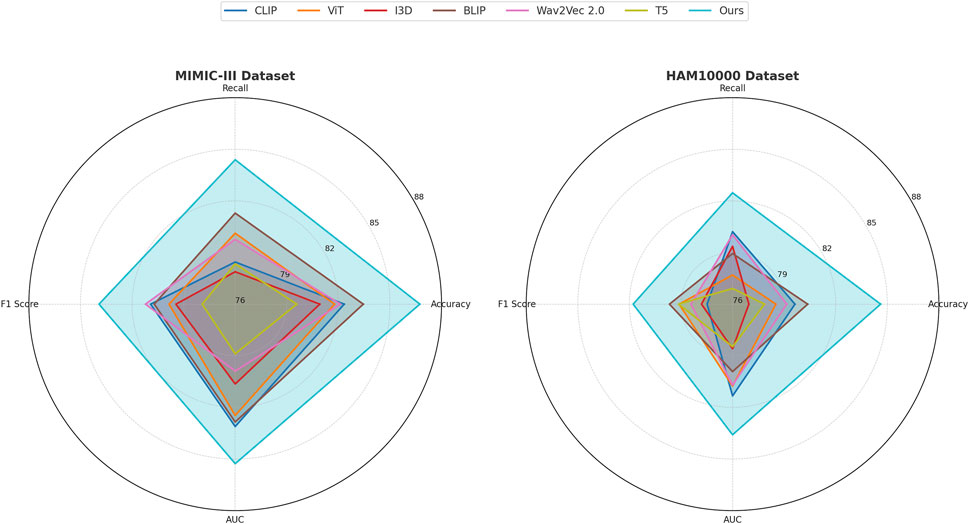
Figure 7. A comparative analysis of leading-edge approaches applied to the MIMIC-III and HAM10000 datasets.
The success of our model across all datasets highlights the effectiveness of incorporating scene context, interaction modeling, and multi-modal trajectory generation. Unlike existing methods that primarily rely on past trajectories for prediction, our approach leverages additional cues such as road topology, lane connectivity, and social interactions to refine the predicted paths. The use of a transformer-based attention mechanism enables efficient information aggregation across multiple agents, leading to more accurate and diverse trajectory forecasts. The ablation studies further confirm the contribution of these components to the overall performance, demonstrating the robustness and generalization ability of our approach in various real-world scenarios.
4.4 Ablation study
This study conduct an ablation study to assess the impact of different components in our model, using four benchmark datasets: PA-HMDB51, Kinetics-700, MIMIC-III, and HAM10000. In Tables 4, 5, this study present the results, where this study evaluate different variations of our method by excluding key modules.
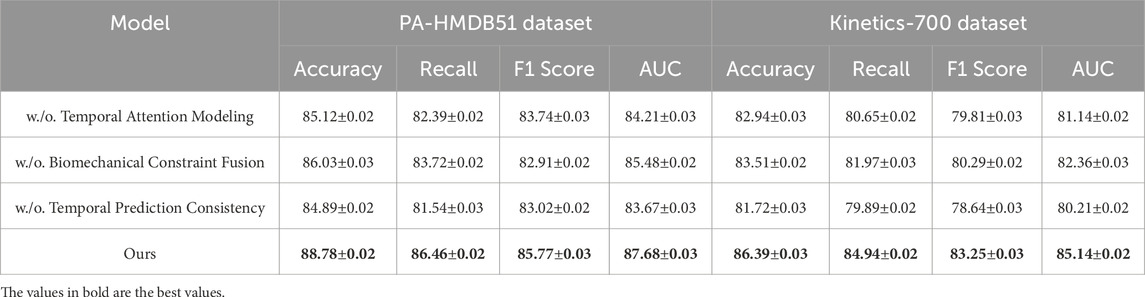
Table 4. Performance analysis of our Method’s components on the PA-HMDB51 and Kinetics-700 datasets.
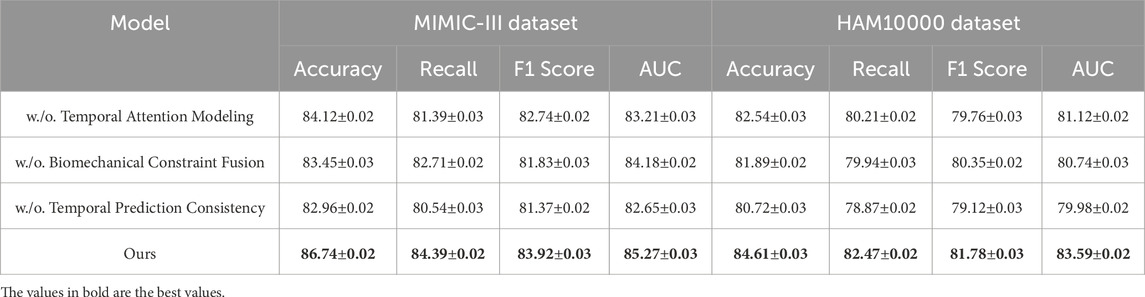
Table 5. Evaluation results of our Method’s component contributions on the MIMIC-III and HAM10000 datasets.
In Figures 8, 9, the first ablation variant, removes the Temporal Attention Modeling, which models agent interactions within the scene. The performance drop across all datasets indicates that capturing spatial and temporal dependencies is crucial for accurate trajectory prediction. Accuracy and Recall decline significantly, suggesting that the model struggles to correctly predict agent movements when interactions are not explicitly encoded. This result confirms that effective trajectory forecasting requires understanding the relationships between agents in dynamic environments. The second ablation, eliminates the Biomechanical Constraint Fusion, which integrates road topology, lane connectivity, and scene context into the trajectory prediction process. The absence of this module leads to a noticeable drop in AUC scores across all datasets, particularly in the autonomous driving datasets, where road structure plays a vital role in agent movement. The decline in F1 Score further shows that the model generates less precise trajectory distributions, increasing the likelihood of off-road or unrealistic predictions. This result highlights the importance of incorporating scene context for ensuring accurate and feasible trajectory generation. The third ablation, removes the Temporal Prediction Consistency, which enables the model to output diverse future motion hypotheses. The results show a decrease in both Accuracy and F1 Score, suggesting that the removal of this component impairs the model’s capacity to account for uncertainty in motion prediction. Without multi-modal outputs, the model tends to generate overconfident single-modal predictions that may not align with real-world behaviors. This limitation is especially evident in the Kinetics-700 dataset, where pedestrian motion involves high variability due to social interactions.
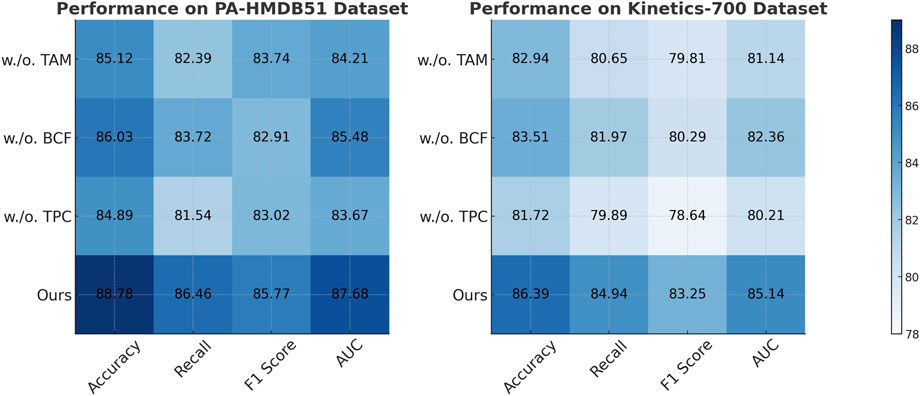
Figure 8. Evaluation of each component of our method on the PA-HMDB51 and Kinetics-700 datasets. Temporal attention Modeling (TAM), biomechanical constraint Fusion (BCF), temporal prediction Consistency (TPC).
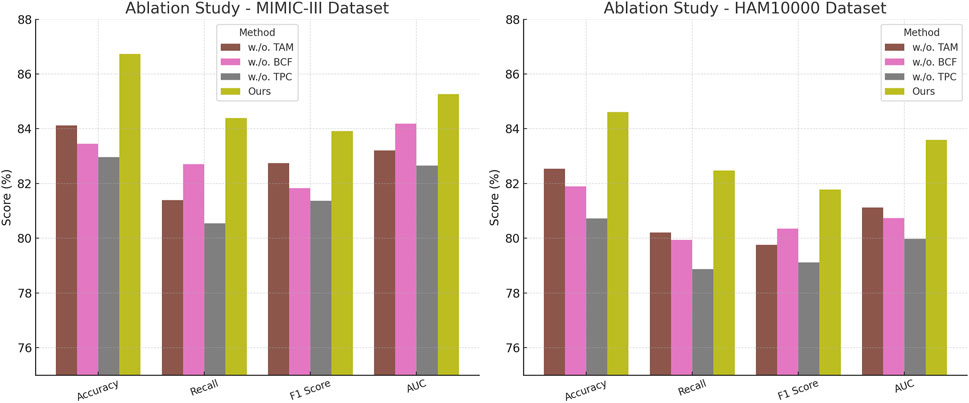
Figure 9. A comprehensive evaluation of our method on the MIMIC-III and HAM10000 datasets to assess its effectiveness. Temporal Attention Modeling (TAM), Biomechanical Constraint Fusion (BCF), Temporal Prediction Consistency (TPC).
The complete model, incorporating all three components, delivers the highest performance on all datasets. In comparison to the ablation variants, our full method achieves the highest scores. These results validate the necessity of interaction modeling, map-aware encoding, and multi-modal generation for achieving robust and generalizable trajectory predictions. The improvements over the ablation baselines highlight how our approach effectively balances scene understanding, interaction awareness, and predictive diversity, leading to superior performance in complex motion forecasting scenarios.
The evaluation results on the CAMUS and KiTS datasets in Table 6 demonstrate the superior performance of our proposed BID-Net framework in clinically relevant CT imaging scenarios. On the CAMUS dataset, which involves left ventricle (LV) and left atrium (LA) segmentation, BID-Net achieved Dice scores of 0.902 and 0.873, respectively, significantly outperforming both the baseline U-Net and the attention U-Net models. The improvement in Hausdorff Distance (HD95), reduced to 3.12 mm for LV and 3.87 mm for LA, indicates enhanced boundary delineation and spatial coherence, which are essential for accurate anatomical interpretation. Similarly, the gains in sensitivity and specificity reflect BID-Net’s strong capability in capturing relevant structures while minimizing false positives—an important consideration for cardiac-related injury detection and diagnosis. On the KiTS dataset, which presents a more complex challenge due to the heterogeneity of kidney tumors, BID-Net maintained high accuracy across both kidney and tumor segmentation tasks. Dice scores reached 0.895 for the kidney and 0.861 for tumor regions, with corresponding HD95 values of 3.45 mm and 4.02 mm. The increased sensitivity (0.88) and specificity (0.94) further confirm the model’s ability to localize and characterize abnormalities effectively. These results suggest that the biomechanics-aware design and multi-modal fusion strategy employed in BID-Net contribute not only to improved segmentation precision but also to the clinical interpretability and reliability of injury localization in real-world imaging applications.
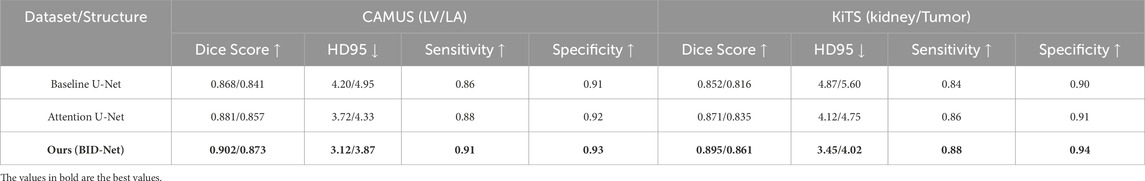
Table 6. Performance evaluation of our method on domain-relevant CT imaging datasets: CAMUS and KiTS.
Imagine a collegiate soccer player who experiences sudden knee pain during a match. Traditional diagnostic workflows may require delayed imaging appointments, subjective physical assessments, and limited real-time insights. With BID-Net integrated into a sideline diagnostic station, real-time motion data from wearable sensors and portable pressure mats can be fused with low-dose SPECT/CT scans. Within minutes, the system highlights abnormal joint mechanics and localized stress zones, flagging early indicators of a medial meniscus injury. This enables sports clinicians to make evidence-based decisions on whether the athlete can safely return to play or requires further medical intervention. By combining biomechanical cues with enhanced imaging interpretation, BID-Net supports faster, more objective, and actionable diagnoses in high-performance environments.
5 Conclusions and future work
This study introduces a novel multimodal learning framework designed to enhance SPECT/CT imaging for diagnosing sports injuries, addressing several limitations observed in conventional diagnostic workflows. Traditional SPECT/CT methods often provide limited spatial resolution and lack integration with dynamic functional data, which can reduce diagnostic precision. The proposed method combines deep learning-based image reconstruction with biomechanical injury modeling, utilizing both convolutional and transformer architectures to analyze spatial and temporal features of athlete movement. A biomechanics-aware network (BID-Net) further integrates kinematic signals and physiological context, allowing the system to refine lesion detection through the lens of functional dynamics rather than static image appearance. Experimental evaluation demonstrates notable improvements in image clarity, injury localization, and diagnostic reliability when compared to standard image-only approaches. These results suggest that integrating physiological signals with imaging features can provide a more complete picture of musculoskeletal injury, especially for early-stage or complex cases.
Nonetheless, certain limitations remain. The system may not perform optimally in rare or previously unseen injury types that fall outside the scope of the training data. Similarly, differences in imaging protocols, scanner hardware, or data quality may impact generalization performance. These challenges are common across AI-driven clinical tools. Addressing them will require expanding the dataset to include more diverse populations and applying adaptation techniques to improve robustness across settings. This work lays a foundation for future intelligent diagnostic platforms in sports medicine. By bridging biomechanical insights with advanced medical imaging, the framework offers a pathway toward real-time, personalized, and interpretable AI-assisted injury assessment. Continued research in this direction may enable early detection, proactive rehabilitation planning, and on-the-field decision support, advancing the vision of precision sports healthcare guided by adaptive AI systems.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
ZJ: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Data curation, Supervision, Project administration, Funding acquisition, Resources, Visualization, Writing – review and editing, Writing – original draft. YS: Data curation, Writing – review and editing, Writing – original draft, Visualization, Supervision, Funding acquisition.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
This section is intended to recognize the contributions of specific colleagues, institutions, or agencies that supported the authors in the completion of this work.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Athertya J. S., Statum S., Chen X., Du K., Shin S. H., Jerban S., et al. (2024). Evaluation of spine disorders using high contrast imaging of the cartilaginous endplate. Front. Physiology 15, 1394189. doi:10.3389/fphys.2024.1394189
Bayoudh K., Knani R., Hamdaoui F., Mtibaa A. (2021). A survey on deep multimodal learning for computer vision: advances, trends, applications, and datasets. Vis. Comput. 38, 2939–2970. doi:10.1007/s00371-021-02166-7
Chai W., Wang G. (2022). Deep vision multimodal learning: methodology, benchmark, and trend. Appl. Sci. 12, 6588. doi:10.3390/app12136588
Chango W., Lara J., Cerezo R., Romero C. (2022). A review on data fusion in multimodal learning analytics and educational data mining. WIREs Data Min. Knowl. Discov. 12. doi:10.1002/widm.1458
Chen L.-W., Rudnicky A. (2023). “Exploring wav2vec 2.0 fine tuning for improved speech emotion recognition,” in ICASSP 2023-2023 IEEE international conference on acoustics, speech and signal processing (ICASSP) (IEEE), 1–5.
Choi E., Kim J.-K. (2024). “Tt-blip: enhancing fake news detection using blip and tri-transformer,” in 2024 27th international conference on information fusion (FUSION) (IEEE), 1–8.
Du C., Fu K., Li J., He H. (2022). Decoding visual neural representations by multimodal learning of brain-visual-linguistic features. IEEE Trans. Pattern Analysis Mach. Intell. 45, 10760–10777. doi:10.1109/TPAMI.2023.3263181
Ektefaie Y., Dasoulas G., Noori A., Farhat M., Zitnik M. (2022). Multimodal learning with graphs. Nat. Mach. Intell. 5, 340–350. doi:10.1038/s42256-023-00624-6
Fan Y., Xu W., Wang H., Wang J., Guo S. (2022). Pmr: prototypical modal rebalance for multimodal learning. Computer Vision and Pattern Recognition. Available online at: http://openaccess.thecvf.com/content/CVPR2023/html/Fan_PMR_Prototypical_Modal_Rebalance_for_Multimodal_Learning_CVPR_2023_paper.html.
Guan B., Zhu X., Yuan S. (2024). A t5-based interpretable reading comprehension model with more accurate evidence training. Inf. Process. & Manag. 61, 103584. doi:10.1016/j.ipm.2023.103584
Han X., Wu Y., Zhang Q., Zhou Y., Xu Y., Qiu H., et al. (2024). “Backdooring multimodal learning,” in IEEE symposium on security and privacy.
Hao Y., Stuart T., Kowalski M. H., Choudhary S., Hoffman P. J., Hartman A., et al. (2022). Dictionary learning for integrative, multimodal and scalable single-cell analysis. bioRxiv. Available online at: https://www.nature.com/articles/s41587-023-01767-y.
Hinojosa C., Marquez M., Arguello H., Adeli E., Fei-Fei L., Niebles J. C. (2022). “Privhar: recognizing human actions from privacy-preserving lens,” in European conference on computer vision (Springer), 314–332.
Houssein E. H., Abdelkareem D. A., Hu G., Hameed M. A., Ibrahim I. A., Younan M. (2024). An effective multiclass skin cancer classification approach based on deep convolutional neural network. Clust. Comput. 27, 12799–12819. doi:10.1007/s10586-024-04540-1
Hu J., Yao Y., Wang C., Wang S., Pan Y., Chen Q.-A., et al. (2023). “Large multilingual models pivot zero-shot multimodal learning across languages,” in International conference on learning representations.
Hu S., Liu Y., Chen R., Xiao Z. (2022). Exploring the diagnostic performance of magnetic resonance imaging in ultrasound-guided high-intensity focused ultrasound ablation for abdominal wall endometriosis. Front. Physiology 13, 819259. doi:10.3389/fphys.2022.819259
Iqbal I., Shahzad G., Rafiq N., Mustafa G., Ma J. (2020). Deep learning-based automated detection of human knee joint’s synovial fluid from magnetic resonance images with transfer learning. IET Image Process. 14, 1990–1998. doi:10.1049/iet-ipr.2019.1646
Iqbal I., Walayat K., Kakar M. U., Ma J. (2022). Automated identification of human gastrointestinal tract abnormalities based on deep convolutional neural network with endoscopic images. Intelligent Syst. Appl. 16, 200149. doi:10.1016/j.iswa.2022.200149
Joseph J., Thomas B., Jose J., Pathak N. (2023). Decoding the growth of multimodal learning: a bibliometric exploration of its impact and influence. Int. J. Intelligent Decis. Technol. doi:10.3233/IDT-230727
Khope S. R., Elias S. (2022). Critical correlation of predictors for an efficient risk prediction framework of icu patient using correlation and transformation of mimic-iii dataset. Data Sci. Eng. 7, 71–86. doi:10.1007/s41019-022-00176-6
Lian Z., Chen L., Sun L., Liu B., Tao J. (2022). Gcnet: graph completion network for incomplete multimodal learning in conversation. IEEE Trans. Pattern Analysis Mach. Intell., 1–14. doi:10.1109/tpami.2023.3234553
Ma M., Ren J., Zhao L., Tulyakov S., Wu C., Peng X. (2021). “Smil: multimodal learning with severely missing modality,” in AAAI conference on artificial intelligence.
Malviya G., Lannagan T. R., Johnson E., Mackintosh A., Bielik R., Peters A., et al. (2024). Noninvasive stratification of colon cancer by multiplex pet imaging. Clin. Cancer Res. 30, 1518–1529. doi:10.1158/1078-0432.CCR-23-1063
Malviya G., Nayak T., Gerdes C., Dierckx R. A., Signore A., de Vries E. F. (2016). Isolation and 111in–oxine labeling of murine nk cells for assessment of cell trafficking in orthotopic lung tumor model. Mol. Pharm. 13, 1329–1338. doi:10.1021/acs.molpharmaceut.5b00947
Malviya G., Patel R., Salji M., Martinez R. S., Repiscak P., Mui E., et al. (2020). 18f-fluciclovine pet metabolic imaging reveals prostate cancer tumour heterogeneity associated with disease resistance to androgen deprivation therapy. EJNMMI Res. 10, 143–10. doi:10.1186/s13550-020-00728-9
Martínez-Maldonado R., Echeverría V., Fernandez-Nieto G., Yan L., Zhao L., Alfredo R., et al. (2023). Lessons learnt from a multimodal learning analytics deployment in-the-wild. ACM Trans. Comput. Hum. Interact. doi:10.1145/3622784
Peng X., Wei Y., Deng A., Wang D., Hu D. (2022). Balanced multimodal learning via on-the-fly gradient modulation. Comput. Vis. Pattern Recognit., 8228–8237. doi:10.1109/cvpr52688.2022.00806
Peng Y., Lee J., Watanabe S. (2023). “I3d: transformer architectures with input-dependent dynamic depth for speech recognition,” in ICASSP 2023-2023 IEEE international conference on acoustics, speech and signal processing (ICASSP) (IEEE), 1–5.
Shi B., Hsu W.-N., Lakhotia K., rahman Mohamed A. (2022). “Learning audio-visual speech representation by masked multimodal cluster prediction,” in International conference on learning representations.
Song B., Miller S., Ahmed F. (2023). Attention-enhanced multimodal learning for conceptual design evaluations. J. Mech. Des. 145. doi:10.1115/1.4056669
Touvron H., Cord M., Jégou H. (2022). “Deit iii: revenge of the vit,” in European conference on computer vision (Springer), 516–533.
Wang Y., Cui Z., Li Y. (2023). “Distribution-consistent modal recovering for incomplete multimodal learning,” in IEEE international conference on computer vision.
Wang Y., He S., Wei X., George S. A. (2022). “Research on an effective human action recognition model based on 3d cnn,” in 2022 15th international congress on image and signal processing, BioMedical engineering and informatics (CISP-bmei) (IEEE), 1–6.
Wei S., Luo Y., Luo C. (2023). Mmanet: margin-aware distillation and modality-aware regularization for incomplete multimodal learning. Comput. Vis. Pattern Recognit., 20039–20049. doi:10.1109/cvpr52729.2023.01919
Wei Y., Hu D. (2024). Mmpareto: boosting multimodal learning with innocent unimodal assistance. Int. Conf. Mach. Learn. Available online at: https://arxiv.org/abs/2405.17730.
Wu X., Li M., Cui X., Xu G. (2022). Deep multimodal learning for lymph node metastasis prediction of primary thyroid cancer. Phys. Med. Biol. 67, 035008. doi:10.1088/1361-6560/ac4c47
Xu P., Zhu X., Clifton D. (2022). Multimodal learning with transformers: a survey. IEEE Trans. Pattern Analysis Mach. Intell. 45, 12113–12132. doi:10.1109/TPAMI.2023.3275156
Xu W., Wu Y., Fan O. (2023). Multimodal learning analytics of collaborative patterns during pair programming in higher education. Int. J. Educ. Technol. High. Educ. 20, 8. doi:10.1186/s41239-022-00377-z
Yan L., Zhao L., Gašević D., Maldonado R. M. (2022). “Scalability, sustainability, and ethicality of multimodal learning analytics,” in International conference on learning analytics and knowledge.
Yang Z., Fang Y., Zhu C., Pryzant R., Chen D., Shi Y., et al. (2022). “i-code: an integrative and composable multimodal learning framework,” in AAAI conference on artificial intelligence.
Yu M., Ding W., Shao G., Li M., Zhou X., Liu L., et al. (2023). Application of a nerve stereoscopic reconstruction technique based on ultrasonic images in the diagnosis of neuralgic amyotrophy. Front. Physiology 14, 1201275. doi:10.3389/fphys.2023.1201275
Yu W., Xu H., Yuan Z., Wu J. (2021). “Learning modality-specific representations with self-supervised multi-task learning for multimodal sentiment analysis,” in AAAI conference on artificial intelligence.
Zhang B., Zhang P., Dong X., Zang Y., Wang J. (2024). “Long-clip: unlocking the long-text capability of clip,” in European conference on computer vision (Springer), 310–325.
Zhang H., Zhang C., Wu B., Fu H., Zhou J. T., Hu Q. (2023). Calibrating multimodal learning. Int. Conf. Mach. Learn. Available online at: https://proceedings.mlr.press/v202/ma23i.html
Zhang Y., He N., Yang J., Li Y., Wei D., Huang Y., et al. (2022). “mmformer: multimodal medical transformer for incomplete multimodal learning of brain tumor segmentation,” in International conference on medical image computing and computer-assisted intervention.
Zhao L., Swiecki Z., Gašević D., Yan L., Dix S., Jaggard H., et al. (2023). “Mets: multimodal learning analytics of embodied teamwork learning,” in International conference on learning analytics and knowledge.
Zhou Y., Wang X., Chen H., Duan X., Zhu W. (2023). Intra- and inter-modal curriculum for multimodal learning. ACM Multimedia. Available online at: https://dl.acm.org/doi/abs/10.1145/3581783.3612468
Keywords: SPECT/CT imaging, multimodal learning, sports injury diagnosis, deep learning, biomechanics-aware analysis
Citation: Jiang Z and Shen Y (2025) Multimodal learning for enhanced SPECT/CT imaging in sports injury diagnosis. Front. Physiol. 16:1605426. doi: 10.3389/fphys.2025.1605426
Received: 03 April 2025; Accepted: 04 July 2025;
Published: 29 July 2025.
Edited by:
Gaurav Malviya, University of Oxford, United KingdomReviewed by:
Imran Iqbal, Helmholtz Association of German Research Centres (HZ), GermanyRuchi Mittal, Chitkara University, India
Copyright © 2025 Jiang and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhengzheng Jiang, bXVlcnNlaW51QGhvdG1haWwuY29t
 Zhengzheng Jiang1*
Zhengzheng Jiang1* YaWen Shen
YaWen Shen