- 1College of Physical, Sungshin University, Seoul, Republic of Korea
- 2College of Science and Technology, Zhengzhou University, Zhengzhou, China
- 3Department of Physical Education, University of Science and Technology Beijing, Beijing, China
- 4School of Agriculture, Zhengzhou University, Zhengzhou, China
- 5College of Physical, Kookmin University, Seoul, Republic of Korea
Exercise-induced fatigue refers to a temporary decline in physiological function resulting from prolonged or high-intensity exercise, which is characterized by decreased muscle strength, diminished exercise performance, and heightened subjective feelings of fatigue. The study of exercise fatigue holds significant importance not only in competitive sports and public health, but also extends to medicine, military applications, and occupational safety. MicroRNA (miRNA) represents a class of non-coding RNA that plays a pivotal role in the regulation of gene expression. The involvement of miRNAs in exercise-induced fatigue has garnered increasing attention within the scientific community. This article provides an overview of fundamental concepts and biological functions associated with miRNAs, defines and classifies exercise fatigue while outlining its physiological changes, emphasizes alterations in miRNA expression during episodes of exercise-induced fatigue, and conducts an in-depth analysis regarding the mechanisms through which miRNAs influence this phenomenon via modulation of energy metabolism, inflammatory responses, and oxidative stress. Furthermore, this article anticipates future research directions as well as potential clinical applications for miRNAs concerning exercise-induced fatigue. This review holds significant importance for elucidating the molecular mechanisms underlying exercise-related fatigue while fostering advancements within sports medicine and rehabilitation science.
1 Introduction
Exercise-induced fatigue typically refers to the phenomenon of reduced exercise capacity resulting from a temporary imbalance in physiological functions during continuous or high-intensity physical activity (Gandevia, 2001). It is characterized by decreased contractility of skeletal muscles, decreased efficiency of neural drive, and increased subjective fatigue perception. Exercise fatigue can be categorized into peripheral fatigue (such as dysfunction within the muscular system) and central fatigue (such as abnormal regulation by the nervous system), based on its location. These two forms of fatigue interact dynamically through the neuromuscular coupling system (Carroll et al., 2017). There are several factors contributing to the phenomenon of exercise-induced fatigue. The most prominent reasons include an imbalance between the resynthesis rate and consumption rate of adenosine triphosphate (ATP), disturbances in the internal environment of muscle cells resulting from high-intensity exercise, dynamic fluctuations in neurotransmitter levels within pathways involving the prefrontal cortex, basal ganglia, and cerebellum, as well as heat stress (Renaud et al., 2023).
MicroRNAs (miRNAs) are a class of endogenous small non-coding RNA molecules, typically ranging from 18 to 24 nucleotides in length. In 1993, researchers first identified the microRNA lin-4 in the nematode Caenorhabditis elegans, which regulates larval development by inhibiting the translation of its target gene lin-14 through incomplete base pairing (Lee et al., 1993). In 2000, a second microRNA, let-7, was discovered; it also plays a crucial role in regulating developmental timing in nematodes and has been shown to be highly conserved across different species (Reinhart et al., 2000). The 2024 nobel prize in physiology or medicine has been awarded jointly to Victor Ambros and Gary Ruvkun for their groundbreaking discovery of microRNAs—small non-coding RNA molecules that regulate gene expression. Their seminal work, beginning with the identification of lin-4 (Lee et al., 1993) and let-7 (Reinhart et al., 2000) in C. elegans, revealed an entirely new layer of genetic regulation conserved across species. This paradigm-shifting finding not only transformed our understanding of developmental timing and cellular communication but also opened new avenues for diagnosing and treating diseases, including cancer and neurological disorders. In 2001, researchers formally introduced the term “microRNA” and predicted the existence of hundreds of such molecules in animals (Lagos-Quintana et al., 2001). By 2002, it was established that microRNAs mediate target mRNA degradation or translational repression via the RNA-induced silencing complex (RISC) (Hutvágner and Zamore, 2002). In 2005, the miRBase database was launched as an authoritative resource for miRNA annotation. In the past decade, microRNAs (miRNAs) have emerged as significant regulators of gene expression and have gradually gained recognition in the field of exercise physiology. During physical activity, miRNAs influence various cellular metabolic pathways, including energy acquisition, inflammation, oxidative stress, and numerous other cellular processes. Additionally, they play crucial roles in cell proliferation, development, and anti-apoptotic mechanisms. These diverse functions underscore the importance of miRNAs as essential molecules for understanding exercise-induced fatigue (Safdar et al., 2016).
Previous studies have established that microRNAs (miRNAs) play a significant role in the onset and progression of exercise-induced fatigue. The alterations in the expression levels of miRNA-1 and miRNA-21 following exercise fatigue are negatively correlated with muscle damage and repair (Meeusen et al., 2021). Specifically, miR-23a may exacerbate exercise-induced fatigue by targeting PGC-1α, thereby inhibiting mitochondrial biosynthesis (Wada et al., 2011). Conversely, miR-146a mitigates inflammation by suppressing TRAF6/NF-κB signaling pathways post-exercise (Baggish et al., 2011). Furthermore, miRNAs are frequently utilized as biomarkers for assessing exercise fatigue (Guescini et al., 2015; Bye et al., 2013). Collectively, these findings suggest that miRNAs represent key molecular players in elucidating the mechanisms underlying exercise fatigue through their regulation of muscle repair, energy metabolism, and inflammatory responses (Baek et al., 2024).
Exercise fatigue is a multi-dimensional and complex phenomenon that involves physiological, metabolic, and nervous systems. As key regulators of gene expression, miRNAs target genes associated with muscle repair, energy metabolism, inflammatory responses, and central nervous system function. They elucidate their regulatory networks at various levels, thereby contributing to the construction of a more comprehensive molecular mechanism regulatory network. This article reviews the research advancements concerning miRNAs in exercise fatigue. Such insights not only enhance our understanding of exercise fatigue but also provide theoretical support and technical groundwork for fields such as sports medicine, rehabilitation science, and the health industry. Looking ahead, with the progression of precision medicine and molecular diagnostic technologies, miRNA-based monitoring and intervention strategies for fatigue are anticipated to emerge as one of the core research directions within sports science.
2 Basic concepts and biological functions of miRNA
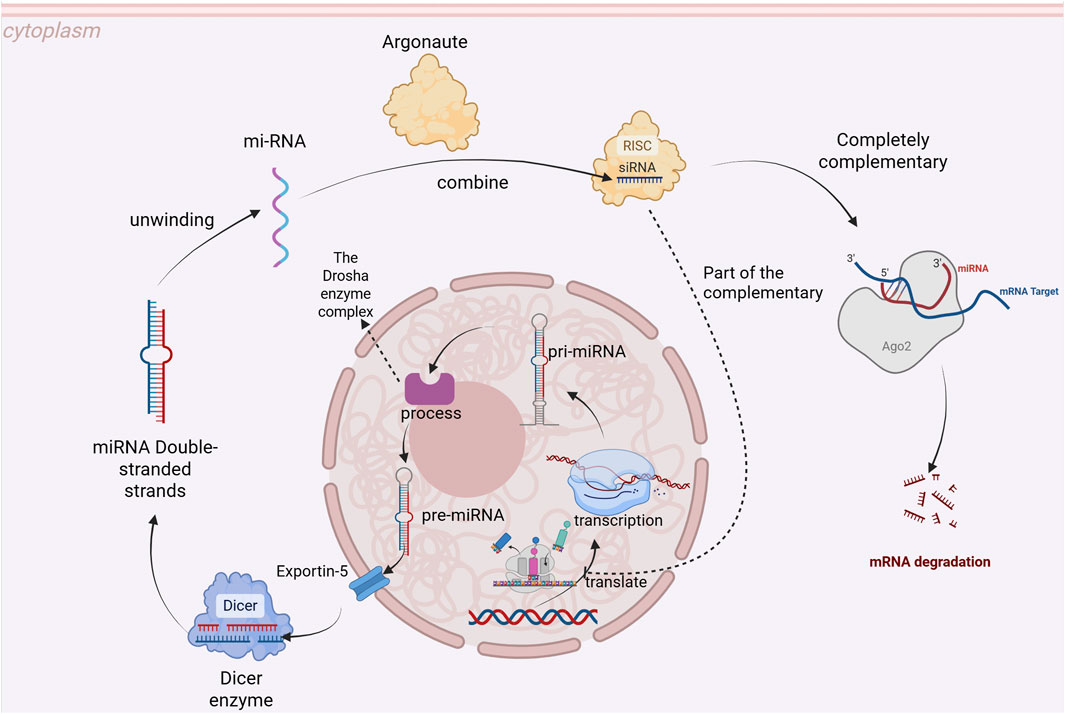
MicroRNAs are widely distributed across both plant and animal kingdoms and play crucial roles in various biological processes by regulating gene expression. miRNAs can be categorized based on their genomic location into intergenic miRNA and intragenic miRNA, as well as based on evolutionary conservation into conserved miRNA and non-conserved miRNA. The genes encoding miRNAs are transcribed by RNA polymerase II to produce primary miRNAs (pri-miRNAs), which are subsequently processed by the Drosha enzyme complex into precursor miRNAs (pre-miRNAs). These pre-miRNAs are then transported to the cytoplasm via Exportin-5, where they undergo cleavage by Dicer enzymes to generate mature miRNA duplexes. The resulting double-stranded mature miRNA unwinds, with one strand—known as the guide strand—binding to Argonaute proteins to form the RNA-induced silencing complex (RISC). miRNAs induce mRNA degradation when there is full complementarity with their target mRNA, while partial complementarity leads to inhibition of mRNA translation (Figure 1). Numerous studies have demonstrated that miRNAs participate in diverse cellular activities such as cell proliferation, differentiation, and programmed cell death. Additionally, they play significant roles in various disease-related pathophysiological processes including cancer, cardiovascular diseases, metabolic disorders (Ambros, 2004; Bartel, 2004; He and Hannon, 2004; Falabrègue et al., 2021; Ma et al., 2021).
The miRNAs play a crucial role in the regulation of various cellular processes, including proliferation, apoptosis, differentiation, metabolism, immune response, stress response, migration and invasion, as well as aging (Figure 1). miR-21 enhances cell proliferation and inhibits apoptosis by downregulating the expression of the tumor suppressor gene PTEN (Esquela-Kerscher and Slack, 2006). Conversely, the miR-34 family promotes apoptosis through modulation of the p53 pathway, thereby inhibiting tumor growth (Garzon et al., 2010). Moreover, miR-145 is involved in smooth muscle cell differentiation while miR-133 regulates cardiomyocyte differentiation. Additionally, both miR-1 and miR-133 are essential for skeletal muscle differentiation (Chen et al., 2006; Ivey and Srivastava, 2010). The miR-33 family influences cholesterol metabolism by regulating sterol regulatory element-binding protein (SREBP). Notably, miR-122 is highly expressed in the liver and participates in lipid metabolism and cholesterol synthesis regulation (Rayner et al., 2010; Rottiers and Näär, 2012). Furthermore, miR-155 plays a significant role in B cell and T cell immune responses. miR-146a mitigates inflammatory responses by modulating the NF-kB pathway (Taganov et al., 2006; O’Connell et al., 2007). miR-210 is implicated in hypoxic conditions through its regulation of the HIF-1α pathway (Crosby et al., 2009). Similarly, miR-34a promotes cellular apoptosis following DNA damage via its influence on the p53 pathway (Kulshreshtha et al., 2007). The members of the miR-200 family inhibit tumor cell migration and invasion by obstructing epithelial-mesenchymal transition (EMT) through suppression of ZEB1/2 expression (Gregory et al., 2008). Conversely, miR-9 enhances tumor metastasis by regulating E-cadherin expression levels (Ma et al., 2010). Furthermore, miR-34a facilitates cellular senescence via SIRT1 modulation while the miR17–92 cluster delays senescence through inhibition of p21 (Boehm and Slack, 2005; Bhaumik et al., 2009). In summary, miRNAs play a crucial role in various cellular physiological processes by regulating gene expression. Future research is expected to further elucidate the role of miRNAs in the onset and progression of diseases and to provide new targets for disease diagnosis and treatment.
3 Physiological mechanisms of exercise fatigue
3.1 Definition and classification of exercise fatigue
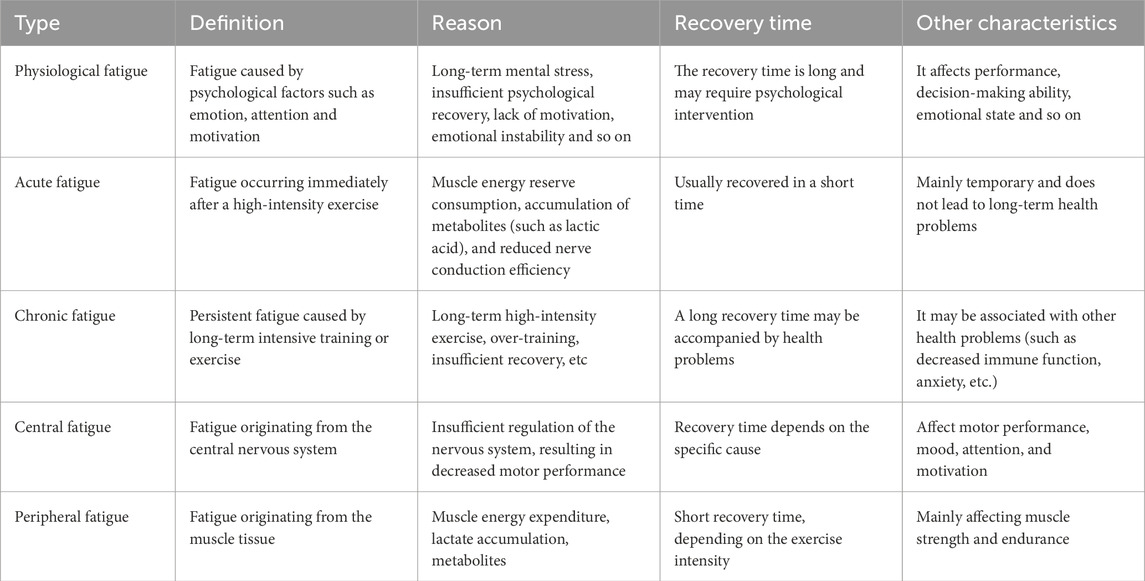
Exercise fatigue is typically defined as a temporary decline in physical function following engagement in sports activities, which results in diminished athletic performance. This form of fatigue can manifest as either physical or psychological. Physical exercise fatigue arises from the depletion of muscle energy reserves, the accumulation of metabolites (such as lactic acid), and a reduction in nerve conduction efficiency. In contrast, psychological exercise fatigue is associated with various psychological factors including emotion, attention, and motivation. Based on different classification criteria for exercise fatigue, it can be categorized into acute exercise fatigue and chronic exercise fatigue (Table 1). Acute exercise fatigue refers to the sensation of tiredness experienced immediately after a single bout of high-intensity exercise; this type of fatigue generally resolves within a short period. Conversely, chronic exercise fatigue denotes persistent exhaustion resulting from prolonged high-intensity training or activity, characterized by an extended recovery time and potentially accompanied by other health issues (Lehninger et al., 2005). Furthermore, according to the site of origin, exercise fatigue can also be classified into central exercise fatigue (Table 1) (originating from the central nervous system) and peripheral exercise fatigue (stemming from muscle tissue). There are notable differences in physiological mechanisms between these two types of exercise-induced fatigues. The following is a comprehensive summary of the classification and characteristics associated with exercise-induced fatigue.
3.2 Physiological changes of exercise fatigue
Exercise fatigue is a phenomenon characterized by a decline in bodily function following prolonged or high-intensity exercise. The physiological changes associated with this condition involve multiple systems, including interactions among the nervous system, muscular system, and endocrine system (Table 2). During physical activity, energy depletion serves as the primary factor contributing to exercise fatigue; the muscle’s energy supply predominantly relies on the oxidation of glycogen and fat. When exercise fatigue manifests, it primarily presents as a reduction in muscle glycogen levels and ATP consumption, which leads to decreased muscle contractility (Bergström et al., 1967; Fitts, 1994). Additionally, the accumulation of metabolites such as lactate and hydrogen ions can lower muscle pH levels and disrupt both muscle contraction and neural signaling (Sahlin et al., 1998; McKenna et al., 2008). Furthermore, diminished neuromuscular function—including central nervous system fatigue and reduced muscle excitability—further compromises motor performance (Gandevia, 2001). Electrolyte imbalances, particularly losses of potassium and sodium ions, also adversely affect both muscular and neurological functions (Maughan et al., 2007). Concurrently, exercise-induced oxidative stress results in an increase in free radicals while weakening antioxidant defenses that are crucial for maintaining cellular structure and function (Ji, 1995; Powers and Jackson, 2008). Finally, hormonal fluctuations—such as elevated cortisol levels coupled with decreased testosterone—may suppress immune responses as well as hinder muscle repair processes (Ji, 1995). The cumulative effects of these physiological alterations ultimately lead to exercise fatigue that negatively impacts athletic performance and recovery capacity. A comprehensive understanding of the physiological mechanisms underlying exercise fatigue is not only advantageous for athlete training and rehabilitation but also holds significant implications for health management within the general population. The following is a concise overview of the physiological mechanisms underlying exercise-induced fatigue.
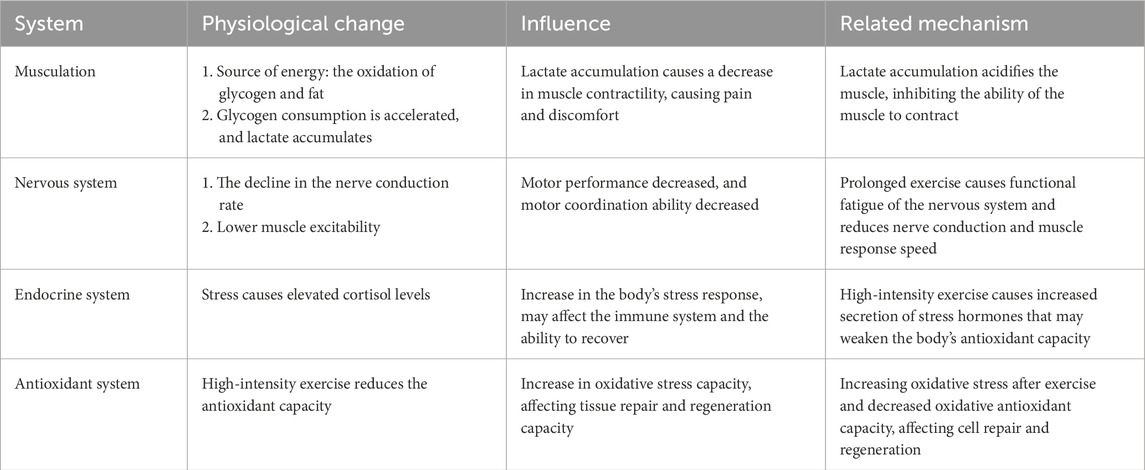
Table 2. A list of physiological changes, impacts, and mechanisms of various systems during exercise fatigue.
3.3 Changes in miRNA expression during exercise fatigue
Exercise-induced alterations in miRNA expression profiles represent a multivariate dynamic process that encompasses various physiological and biochemical activities. Following exercise, the levels of certain miRNAs within the body undergo significant changes, which are closely associated with energy metabolism, inflammatory responses, oxidative stress, and muscle repair. Investigations into changes in miRNA expression during exercise fatigue underscore their critical role in regulating physiological adaptation and recovery post-exercise (Figure 2). Research has demonstrated that exosomal miRNAs circulating in the bloodstream exhibit substantial modifications following endurance training. For instance, muscle-specific miRNAs such as miR-1, miR-133, and miR-206 are upregulated after exercise and play pivotal roles in modulating muscle differentiation and repair (Hackney, 2006). Additionally, the expressions of both miR-21 and miR-146a increase post-exercise; these changes may be linked to the regulation of inflammatory responses and could aid in mitigating tissue damage resulting from physical activity (Yuasa et al., 2008). Conversely, variations in the expression of miR-23a and miR-486 correlate with oxidative stress and energy metabolism. Exercise intervention has significant tissue specificity for miRNA regulation in adipose tissue. A study in obese women showed that 12 weeks of combined aerobic resistance combination training significantly upregulated the expression of miR-155-5p and miR-329-3p in subcutaneous adipose tissue (GSAT), but not in abdominal subcutaneous fat (ASAT) (Carmen et al., 2022). These specific miRNAs influence mitochondrial function and antioxidant defense mechanisms by regulating target gene expression (Nielsen et al., 2014). Importantly, different types of exercise—such as aerobic versus resistance training—and varying intensities can lead to distinct differences in miRNA expression profiles. This suggests that the regulatory functions of miRNAs concerning exercise fatigue are complex and context-dependent (Russell et al., 2013). Moreover, it is crucial to recognize that the expression of miRNA is influenced by both the level and timing of physical exertion. Of concern is that exercise-induced exosomal miRNA has emerged as a new dimension to uncover fatigue mechanisms. A study in 1,500 m freestyle athletes showed that miR-144-3p, miR-145-3p and miR-509-5p were significantly upregulated in circulating exosomes after fatigue exercise. Their target genes were enriched in vascular endothelial growth factor (VEGF) signaling pathway and glutathione metabolism. This change echoes the mechanism that miR-126 improves angiogenesis by regulating VCAM-1, suggesting that exosomal miRNA may participate in fatigue recovery through oxidative stress regulation and vascular function optimization (Lai et al., 2023). A separate study has demonstrated that it is possible to regulate a specific circulating miRNA (c-miRNA), with fluctuations in its expression correlating with exercise intensity. This observation reflects the body’s adaptive response to various synergistic loads imposed by different forms of physical activity (Safdar et al., 2009). Therefore, exercise-induced alterations in the miRNA expression profile not only enhance our understanding of the effects of exercise on the body but also establish a foundation for future research on exercise interventions.
There are notable differences in miRNA expression associated with various types of exercise, highlighting the complexity of muscle adaptation within the body under different exercise modalities. Research has demonstrated that at the molecular level, resistance training (such as strength training) and aerobic exercise (such as endurance training) are governed by distinct mechanisms involving miRNA regulation. For instance, a study comparing resistance training to aerobic exercise—specifically high-intensity interval training—provided evidence indicating that resistance training significantly modulates levels of miR-23a and miR-206 in muscle tissue compared to high-intensity interval training. This disparity may arise from the differing physiological stress responses elicited by various forms of exercise; resistance training typically results in greater muscle damage and subsequent repair processes, suggesting a potential positive feedback mechanism for specific miRNAs involved in muscle recovery. Moreover, cross-sectional studies examining the effects of exercise have identified gender as a significant factor influencing miRNA responses to different types of physical activity. There exist gender-specific variations in the regulation of miRNA expression post-exercise, which may be linked to underlying sex physiology (Safdar et al., 2009). Consequently, an in-depth investigation into how different types of exercise affect miRNA can not only provide a scientific foundation for developing enhanced intervention and rehabilitation strategies but also offer theoretical insights into preventing the onset and progression of exercise-induced fatigue.
3.4 Mechanism of miRNA in exercise fatigue
3.4.1 Relationship between miRNA and energy metabolism
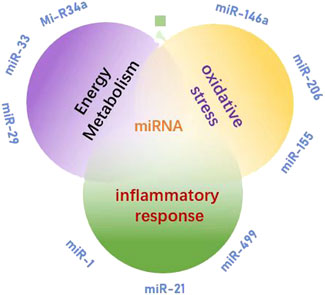
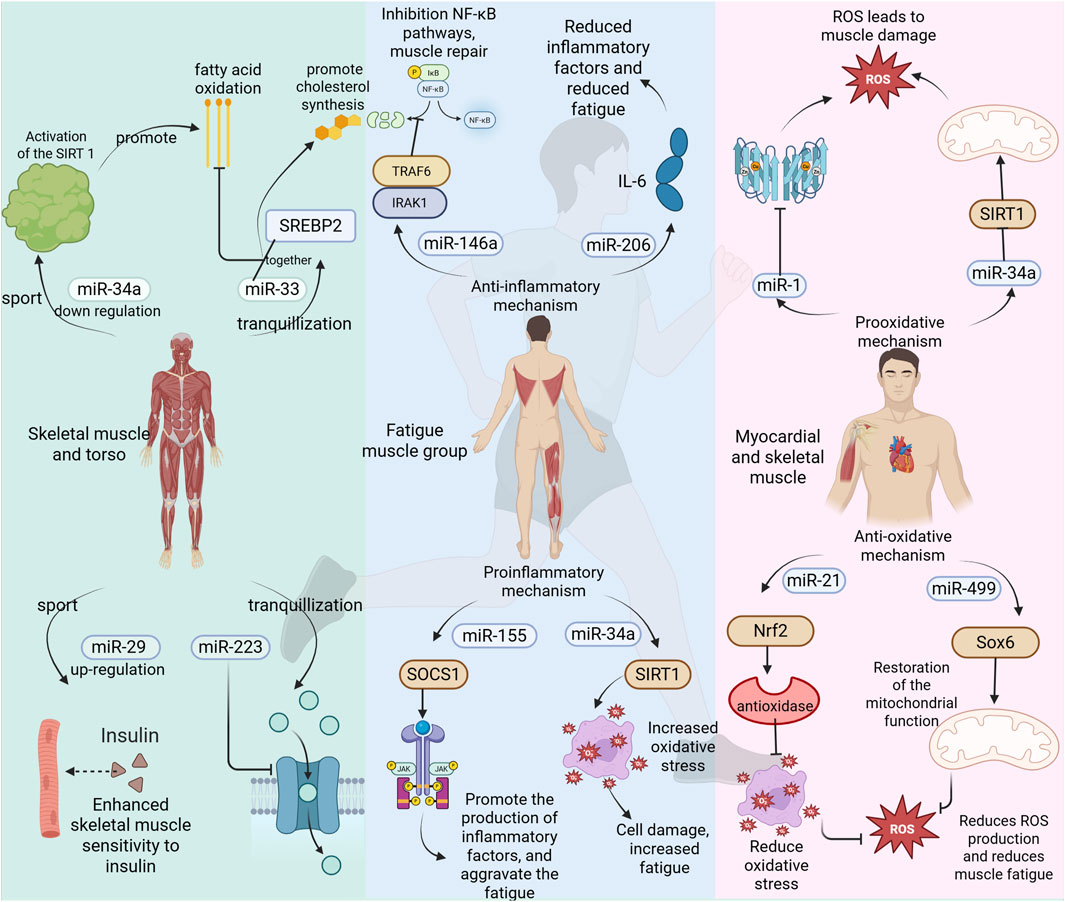
As a type of non-coding RNA, miRNA has garnered significant attention for its role in the regulation of energy metabolism. miR-223 targets glucose transporter 4 (GLUT4) and inhibits its expression, thereby reducing glucose uptake in skeletal muscle (Li et al., 2025). Additionally, miR-223 was implicated in inflammation and the TLR4/NF-κB inflammatory pathway, which was important in exercise induced inflammatory response (M’baya-Moutoula et al., 2018). miR-375 regulates the function of islet β cells and influences insulin secretion; its abnormal expression is closely associated with insulin resistance (Meeusen et al., 2006). Exercise enhances skeletal muscle insulin sensitivity and ameliorates glucose metabolism disorders by upregulating the expression of the miR-29 family (Lu et al., 2010). The co-expression of miR-33 and sterol regulatory element-binding protein 2 (SREBP2) inhibits fatty acid β-oxidation while promoting cholesterol synthesis (Keller and Perez, 2022). Overexpression of miR-122 in the liver can decrease hepatic fat deposition by modulating lipid metabolism-related genes such as fatty acid synthase (FASN) and stearoyl-CoA desaturase 1 (SCD1) (Dalgaard et al., 2022). Furthermore, exercise promotes fatty acid oxidation and mitigates lipid accumulation through downregulation of miR-34a and activation of sirtuin 1 (SIRT1), a deacetylase (Horie et al., 2010). Under hypoxic conditions, upregulation of miR-210 inhibits mitochondrial respiratory chain complex activity while promoting glycolysis—a phenomenon known as the Warburg effect (Guo et al., 2023). Abnormal expression levels of miR-30a-5p may lead to disturbances in myocardial mitochondrial energy metabolism within models of pulmonary hypertensive right heart failure (Chen et al., 2019); however, exercise intervention could potentially restore mitochondrial function by regulating this specific miRNA. Moreover, certain miRNAs enriched in young plasma-derived small extracellular vesicles (sEVs), such as miR-21, activate peroxisome proliferator activated receptor gamma coactivator 1-alpha (PGC-1α), enhance mitochondrial oxidative phosphorylation, and alleviate age-related metabolic decline (Ismaeel et al., 2022). Collectively, these findings underscore that miRNAs play critical roles in regulating glucose metabolism, lipid metabolism, mitochondrial function, and oxidative metabolism (Figure 3). The CRF, which can be divided into “inflammation-driven” (high IL-6/miR-223), “leptin-related” (high leptin/miR-34a) and “depression-related type”, in which miR-223 exacerbates mitochondrial dysfunction by inhibiting PGC-1 α (Schmidt et al., 2024), illustrates the complementary mechanism of mitochondria inhibition of mitochondrial biosynthesis. This suggests mirna intervention strategies targeting different subtypes, such as miR-223 inhibitors for inflammatory type CRF.
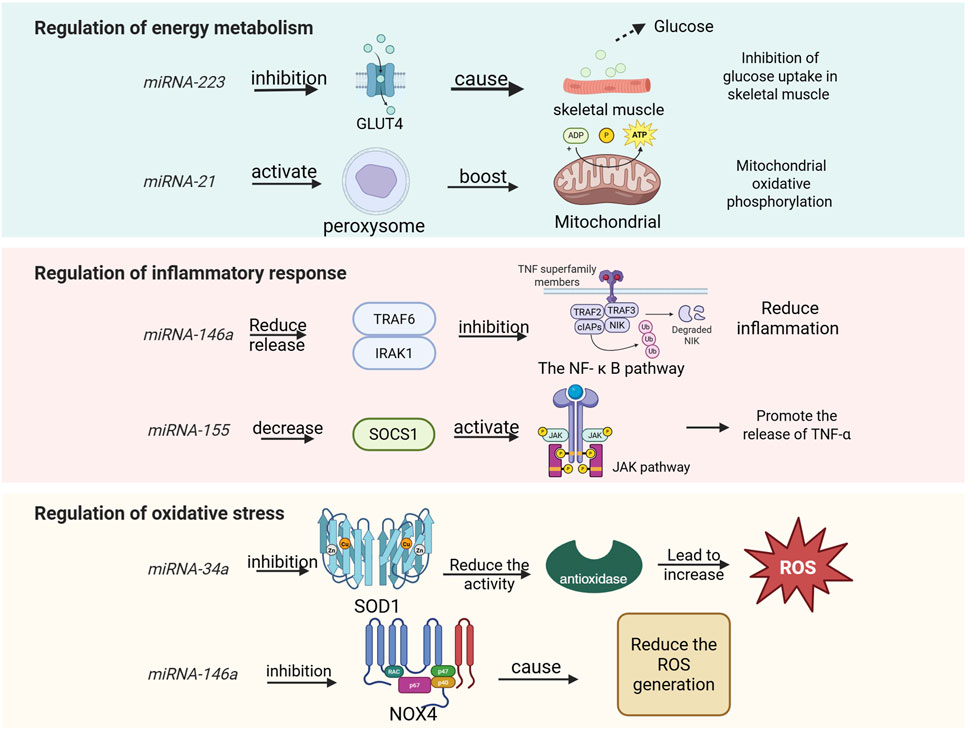
Figure 3. miRNAs involved in human regulation of energy metabolism, inflammatory responses, and oxidative stress.
Given the pivotal role of miRNAs in energy metabolism, and considering that exercise is a process characterized by high energy demand, the body regulates miRNA expression to maintain homeostasis in both energy metabolism and exercise regulation (Lv et al., 2024). Aerobic exercise significantly upregulates miR-1 and miR-133a, which promote glucose uptake and fatty acid oxidation in skeletal muscle (Harmon et al., 2017; Qian et al., 2024). In contrast, resistance exercise enhances the expression of miR-206, inhibiting genes associated with muscle atrophy (such as HDAC4) to sustain metabolic homeostasis within muscle tissue (Foley et al., 2006). Exercise also stimulates the release of exosomes from both muscle and adipose tissues; these exosomes carry specific miRNAs such as miR-30a-5p and miR-486 that can regulate the metabolism of distant target organs (Winbanks et al., 2013; Luu et al., 2020). For instance, exosomal miRNA derived from hypothalamic neural stem cells has been shown to improve systemic energy metabolism by inhibiting the TLR4/NF-κB inflammatory pathway (Huang et al., 2019). Furthermore, physical activity alters miRNA promoter activity through mechanisms involving DNA methylation and histone modification (Wu et al., 2022). The elevated levels of miR-34a induced by a high-fat diet can be reversed through exercise intervention, thereby restoring SIRT1-mediated metabolic balance (Chen et al., 2019). Additionally, miR-208a inhibits the PPARα signaling pathway, reducing the expression of fatty acid transporter CD36 (Tong et al., 2021). This inhibition impedes lipid utilization and forces reliance on anaerobic metabolism, resulting in lactic acid accumulation and subsequent exercise fatigue. Conversely, miR-27b activates the AMPK pathway to promote lipolysis and mitochondrial β-oxidation (Hu et al., 2023); this action enhances fat-derived energy supply efficiency while delaying energy depletion during prolonged physical activity. Studies have demonstrated that adipose-derived exosomal miR-124-3p inhibits hepatic lipid metabolism by targeting PPARγ, which leads to triglyceride accumulation and may indirectly exacerbate energy metabolism disorders during exercise (Zhao et al., 2021). Following high-intensity exercise, the expression patterns of certain miRNAs undergo significant changes. The upregulation of specific miRNAs, such as miR-144-3p and miR-145-3p, is associated with the stimulation of long-term potentiation (LTP) and signaling pathways related to vascular endothelial growth factor (VEGF) (You et al., 2016). Additionally, miR-146a-5p targets Table 1 to inhibit the NF-κB pathway, thereby reducing the release of inflammatory factors (Chen et al., 2022). This action protects mitochondria from oxidative damage and alleviates exercise-induced fatigue. Collectively, these studies suggest that miRNAs play a crucial role in regulating energy metabolism following exercise.
3.4.2 The role of miRNA in inflammatory response
In recent years, an increasing number of studies have demonstrated that miRNAs play a crucial role in the regulation of inflammatory responses. These miRNAs can modulate the activation and inhibition of inflammatory signaling pathways by targeting genes associated with inflammation. For instance, miR-124 facilitates the polarization of macrophages towards the anti-inflammatory M2 phenotype by targeting C/EBP-α (Ponomarev et al., 2011). Similarly, miR-155 promotes the differentiation of Th1 cells while inhibiting the differentiation of Th2 cells through its action on c-Maf (Rodriguez et al., 2007). These findings indicate that miRNAs are integral to inflammatory responses by regulating inflammatory signaling pathways, mediators, and immune cell functions.
Exercise-induced fatigue can be categorized into central fatigue and peripheral fatigue, with the latter being closely associated with muscle damage and inflammatory responses. High-intensity or prolonged exercise can trigger an inflammatory response in muscle tissue, characterized by the release of inflammatory factors (such as IL-6 and TNF-α) and the infiltration of immune cells. This inflammatory response is not only a necessary process for the body to repair damage but may also exacerbate fatigue and delay recovery. For instance, NF-κB serves as a central transcription factor in the inflammatory response. miR-146a negatively regulates the NF-κB signaling pathway and inhibits inflammation by targeting TRAF6 and IRAK1 (Taganov et al., 2006). The upregulation of miR-146a may mitigate inflammation-mediated muscle damage during exercise-induced fatigue. Similarly, miR-21 reduces activation of the MAPK signaling pathway by targeting PDCD4, thereby diminishing the inflammatory response (Sheedy et al., 2010). Additionally, miR-206 inhibits inflammation through its action on IL-6 (McCarthy et al., 2007). In cases of exercise-induced fatigue, increased expression of miR-206 and miR-210 may alleviate symptoms by reducing the release of pro-inflammatory factors. These microRNAs have potential therapeutic roles in addressing exercise-related fatigue by modulating either inflammatory responses or the secretion of inflammatory mediators. However, it is important to note that certain miRNAs can also contribute to exacerbating exercise-induced fatigue via their influence on inflammation. The miR-155 enhances the JAK/STAT signaling pathway and promotes the production of inflammatory factors by targeting SOCS1 (O’Connell et al., 2007). In the context of exercise-induced fatigue, the upregulation of miR-155 expression may exacerbate the inflammatory response, thereby contributing to increased fatigue. Conversely, miR-126 inhibits monocyte migration to sites of inflammation by targeting VCAM-1 (Harris et al., 2008). During exercise fatigue, elevated levels of miR-126 may mitigate inflammation-mediated muscle damage through a reduction in immune cell infiltration. Additionally, miR-34a elevates oxidative stress and inflammation by targeting SIRT1(Yamakuchi et al., 2008). In cases of exercise fatigue, an increase in miR-34a expression could worsen oxidative stress and inflammatory responses, further intensifying feelings of fatigue. It is worth noting that miRNA also shows the potential for cross-system regulation in neurological disease-related fatigue. For example, in multiple sclerosis (MS), miR-126 suppresses immune cell infiltration into the CNS by targeting vascular cell adhesion molecule 1 (VCAM-1), thereby reducing neuroinflammation-mediated fatigue. A study based on wearable sensors showed that fatigue in ms patients was positively correlated with sympathetic activity (e.g., reduced heart rate variability), but negatively with miR-126 expression level. This mechanism is highly similar to the role of mir-126 in alleviating muscle inflammatory injury by inhibiting vcam-1 during exercise fatigue (Moebus et al., 2024) In post-infection ME or CFS patients, HHV-6 viral load was positively correlated with mir-155 expression, which promotes pro-inflammatory factor (e.g., TNF- α) release through activation of the JAK or STAT pathway (Gravelsina et al., 2022). Also confirmed the mechanism by which miR-155 enhances the inflammatory response by targeting socs1, suggesting that miR-155 can be used as an early warning marker for fatigue associated with viral infection. Moreover, miR-21 is found to be upregulated in patients with chronic fatigue syndrome (CFS) and facilitates inflammatory responses by targeting PDCD4 (Bjersing et al., 2015). Both miR-1 and miR-133 are also upregulated following muscle injury due to exercise; they play crucial roles in regulating muscle repair processes by targeting genes associated with inflammation (Chen et al., 2006).
Overall, miRNA significantly influence exercise-related fatigue through their regulation of inflammatory signaling pathways and mediators. Abnormal expression patterns of these miRNA are closely linked to both the onset and progression of exercise-induced fatigue. Investigating the specific regulatory mechanisms employed by miRNA within the context of inflammation during exercise-induced fatigue presents a promising avenue for research within sports medicine and rehabilitation.
3.4.3 Interaction between miRNA and oxidative stress
Oxidative stress is a condition characterized by an imbalance between the production and clearance of intracellular reactive oxygen species (ROS), which is closely associated with the onset and progression of various diseases, including cardiovascular diseases, neurodegenerative disorders, and cancer. The miRNAs play a crucial role in oxidative stress by modulating the expression of genes related to oxidative stress and influencing both the production and elimination of ROS. For instance, miR-34a and miR-1 are implicated in regulating ROS production, while miR-146a and miR-21 are involved in ROS clearance (Figure 4). Additionally, miR-133a and miR-499 regulate mitochondrial function. Furthermore, miRNAs significantly contribute to oxidative stress-related diseases. The miR-210 inhibits mitochondrial function, increases ROS production, and exacerbates myocardial ischemia-reperfusion injury by targeting ISCU (Chan et al., 2009). Similarly, miR-155 impairs the antioxidant defense system, elevates oxidative stress levels, and facilitates the progression of Alzheimer’s disease through its interaction with Nrf2 (Guedes et al., 2014). By targeting ZEB1, miR-200c diminishes the activity of antioxidant enzymes, enhances ROS generation, and promotes apoptosis in cancer cells (Magenta et al., 2011). These studies underscore the pivotal role that miRNAs play in oxidative stress as well as their underlying mechanisms; moreover, abnormal expression patterns of these molecules are closely linked to the development and advancement of various diseases.
During exercise, particularly during high-intensity or prolonged activities, the activity of the mitochondrial electron transport chain is enhanced, resulting in an excessive production of ROS. When ROS levels exceed the body’s antioxidant capacity, oxidative stress is triggered. This can lead to lipid peroxidation, protein oxidation, and DNA damage within cell membranes, ultimately causing muscle fatigue and functional decline. The miRNAs play a crucial role in regulating both the production and clearance of ROS by targeting genes associated with oxidative stress; thus, they influence the onset and progression of exercise-induced fatigue. There are conflicting reports regarding the impact of miRNAs on exercise fatigue. The following miRNAs may exacerbate its occurrence and development: miR-34a targets SIRT1 and inhibits its expression, leading to mitochondrial dysfunction and an overproduction of reactive oxygen species (ROS) (Yamakuchi et al., 2008). In the context of exercise-induced fatigue, the upregulation of miR-34a expression may exacerbate oxidative stress and worsen feelings of fatigue. Similarly, miR-1 reduces the activity of antioxidant enzymes by targeting superoxide dismutase 1 (SOD1) and superoxide dismutase 2 (SOD2), thereby increasing ROS production (Chen et al., 2006). During episodes of exercise-related fatigue, elevated levels of miR-1 may further intensify oxidative stress, resulting in muscle damage and heightened sensations of fatigue. Furthermore, miR-133a impairs mitochondrial biosynthesis and function through its action on peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α), contributing to excessive ROS generation (Safdar et al., 2009). In scenarios involving exercise-induced fatigue, increased expression levels of miR-133a could aggravate both mitochondrial dysfunction and oxidative stress. The following miRNA may mitigate the onset and progression of exercise-induced fatigue. miR-146a inhibits NADPH oxidase activity and reduces reactive oxygen species (ROS) production by targeting NOX4 (Cheng et al., 2013). In the context of exercise fatigue, upregulation of miR-146a expression may alleviate oxidative stress and diminish fatigue symptoms. Notably, miRNA exhibits a similar neural-oxidative stress-inflammation interaction regulatory mechanism in neurodegenerative diseases (e.g., Alzheimer’s disease AD, Parkinson’s disease PD). For example, miR-124-3p relieves neuronal damage in the ad model by targeting the MAPK pathway to inhibit microglial activation, reduce ROS generation and the release of neuroinflammatory factors (e.g., IL-6), while mir-132 enhances mitochondrial antioxidant enzyme activity by regulating BDNF expression and improves dopaminergic neuronal function in the PD model (Azam et al., 2024). Similarly, miR-21 enhances the Nrf2 signaling pathway and promotes the expression of antioxidant enzymes by targeting PDCD4 (Rodriguez et al., 2007). During episodes of exercise fatigue, increased levels of miR-21 may help reduce oxidative stress through bolstering the antioxidant defense system. Furthermore, miR-499 facilitates mitochondrial function recovery while decreasing ROS production by targeting Sox6 (Wang et al., 2011) (Figure 4). In cases of exercise fatigue, elevated expression of miR-499 could enhance mitochondrial functionality and lower oxidative stress. Additionally, miRNAs play a significant role in diseases associated with exercise-related fatigue. For instance, in patients with chronic fatigue syndrome, downregulation of miR-16 leads to inhibited activity of antioxidant enzymes and heightened oxidative stress via its target SOD2 (Bjersing et al., 2015). Conversely, after muscle injury induced by exercise, there is an upregulation of miR-206 which mitigates inflammation and oxidative stress through its action on IL-6 (Sheedy et al., 2010). Oxidative stress represents one critical mechanism underlying exercise-induced fatigue; it arises from excessive ROS production coupled with an imbalance in the antioxidant defense system. Thus, miRNA plays a pivotal role in regulating both ROS generation and clearance as well as maintaining the integrity of the antioxidant defense system; any aberration in their expression is closely linked to the emergence and progression of exercise-related fatigue.
3.5 Future research directions and clinical applications
As a class of small non-coding RNA molecules, miRNAs play a crucial role in the life processes of eukaryotes. miRNAs can inhibit the translation and degradation of target mRNA by binding to it, and they can also influence gene expression through the regulation of DNA methylation and histone modification. Consequently, miRNAs are integral to various cellular functions including proliferation, differentiation, apoptosis, metabolism, and stress responses. Moreover, miRNAs regulate the onset and progression of exercise-induced fatigue by modulating oxidative stress, inflammatory responses, energy metabolism, and neuroendocrine pathways. For instance, miR-34a targets SIRT1 to regulate cellular oxidative stress by inhibiting mitochondrial function and increasing reactive oxygen species (ROS) production (Yamakuchi et al., 2008). Similarly, miR-146a inhibits the NF-κB signaling pathway by targeting TRAF6 and IRAK1, thereby reducing inflammation (Taganov et al., 2006). Additionally, miR-133a regulates energy metabolism through its action on PGC-1α while inhibiting mitochondrial biosynthesis (Safdar et al., 2009). These specific miRNAs are pivotal regulators in the context of exercise fatigue. To further elucidate the functional mechanisms underlying these effects of miRNA on exercise fatigue, high-throughput sequencing combined with bioinformatics analysis will be employed to identify relevant miRNAs along with their target genes associated with this condition. Furthermore, verification of their functional mechanisms is essential. The intricate regulatory network involving miRNA in relation to oxidative stress response as well as inflammatory reactions warrants additional investigation. In terms of epigenetic regulation mechanisms related to exercise fatigue mediated by miRNA continue to be explored actively. Moreover, attention has been directed towards understanding how factors such as exercise intensity, duration, and type impact the expression levels of specific miRNAs. miRNA only one class of small non-coding RNA molecules, there are other type non-coding RNA play a important role in exercise. Such as long non-coding RNAs (lncRNAs), which transcripts >200 nucleotides with no protein-coding potential in mediating the body’s response to exercise. These molecules regulate gene expression epigenetically, transcriptionally, and post-transcriptionally, influencing metabolic, cardiovascular, and musculoskeletal adaptations.
It is of significant clinical importance to investigate the role of miRNA in exercise-induced fatigue. miRNA has the potential to serve as a diagnostic biomarker for exercise fatigue, allowing for the assessment of both the severity and recovery from such fatigue through the analysis of miRNA expression profiles in blood, urine, or saliva. Various combinations of miRNAs have been screened to enhance the accuracy and specificity of diagnosing exercise-related fatigue. Moreover, miRNAs can be targeted therapeutically for managing exercise fatigue; specific agonists (e.g., miR-30e agonists) and inhibitors (e.g., miR-34a inhibitors) have been developed to modulate signaling pathways associated with exercise fatigue. Utilizing CRISPR/Cas9 technology enables precise editing of miRNA genes, rendering them effective targets for addressing exercise-induced fatigue. Gene therapy using adeno-associated virus (AAV) vectors has emerged as a leading approach for delivering therapeutic genes, while microRNAs (miRNAs) offer precise gene regulation. Combining these technologies enables targeted, durable treatments for exercise related diseases (Wang, et al., 2019). Personalized exercise regimens may be formulated based on individual miRNA profiles, facilitating tailored training programs that optimize performance while minimizing feelings of exhaustion post-exercise. Additionally, research has explored the involvement of miRNAs in sports nutrition interventions—specifically regarding antioxidants and anti-inflammatory agents. The investigation into the role of miRNAs in exercise-related fatigue offers novel insights into both diagnosis and treatment strategies. Future research directions should focus on elucidating functional mechanisms, regulatory processes governing expression levels, and individual variability among different subjects concerning their respective miRNA profiles. In clinical practice, leveraging miRNAs as diagnostic markers and therapeutic targets presents a promising strategy for achieving.
4 Conclusions and outlook
Studies in the field of sports fatigue have increasingly focused on the circulation features of miRNA, thereby further confirming its significant biological value. Current literature indicates that miRNA is involved in regulating various cellular processes, including energy metabolism, inflammatory responses, and oxidative stress. These mechanisms not only provide an objective explanation for the physiological causes of exercise-induced fatigue but also open new avenues for understanding this concept.
The findings from recent studies demonstrate a correlation between miRNA levels and exercise fatigue. This suggests that different types of miRNAs may serve distinct regulatory functions across various forms of exercise-related fatigue; therefore, when investigating biomarkers associated with exercise fatigue, it is essential to consider these diverse aspects and relevant findings in a coordinated manner. Future research should focus on establishing causal relationships between miRNA and exertion, which is crucial for comprehending the specific roles this molecule plays in regulating adaptation and recovery processes related to exercise. In a similar vein, the challenges associated with the potential of miRNA as a biomarker should not be underestimated. The analysis of specific miRNAs in human serum has the capacity to predict both the degree of exercise-induced fatigue and recovery status. This provides a scientific foundation for athlete training and rehabilitation, facilitating the development of individualized training programs that enhance athletic performance and mitigate sports injuries. However, it is crucial to acknowledge that while the properties of miRNA appear advantageous for investigating exercise fatigue, further empirical research is necessary to validate its prospects in clinical practice. Continued emphasis on scientific inquiry into the physiological functions of miRNA related to exercise is essential, as this will help expand its application potential and future developments concerning exercise fatigue.
Autophagy, a critical cellular recycling process, has emerged as a key mechanism in exercise-induced fatigue regulation. During prolonged exercise, autophagy clears damaged organelles (e.g., dysfunctional mitochondria) and protein aggregates, maintaining cellular homeostasis. miRNAs intricately regulate autophagy pathways: for instance, miR-30e suppresses autophagy initiation by targeting BECN1 (beclin-1), while miR-223 enhances autophagic flux via FOXO3 modulation. Impairments in exercise-triggered autophagy exacerbate oxidative stress and energy depletion, accelerating fatigue onset. Notably, endurance training upregulates miR-30e, promoting mitochondrial biogenesis through PGC-1α activation while fine-tuning autophagic activity to balance clearance and conservation of cellular resources. Future studies should explore miRNA-mediated autophagy as a therapeutic target to delay fatigue and enhance recovery (Safdar et al., 2016). The gut-muscle axis represents a novel frontier in exercise fatigue research. Gut microbiota dysbiosis during intense exercise impairs intestinal barrier integrity, increasing systemic inflammation and oxidative stress. miRNAs modulate this axis: miR-146a downregulates TLR4/NF-κB signaling in gut epithelial cells, reducing inflammation-induced fatigue, while miR-21 targets PDCD4 to maintain mucosal homeostasis. Conversely, microbial metabolites (e.g., short-chain fatty acids) influence host miRNA expression—butyrate upregulates *miR-200c*, enhancing mitochondrial function in skeletal muscle. Exercise-induced shifts in Firmicutes/Bacteroidetes ratio correlate with circulating miR-486 levels, suggesting microbiota-driven miRNA regulation of energy metabolism. Harnessing this crosstalk via probiotics or miRNA-targeted interventions may mitigate fatigue (Li et al., 2025).
In conclusion, a deeper understanding of the functions and regulatory mechanisms governing miRNAs will provide more robust theoretical support and effective references for advancing sports medicine.
Author contributions
SN: Writing – review and editing, Writing – original draft. XY: Writing – review and editing, Software, Writing – original draft. QC: Writing – review and editing. KH: Writing – review and editing, Writing – original draft. ZD: Writing – review and editing, Writing – original draft. JC: Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by grants from the National Natural Science Foundation in China (32200934).
Acknowledgments
Thanks for the graduate student Juan Liu’s help in picture revised.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Azam H. M. H., Rößling R. I., Geithe C., Khan M. M., Dinter F., Hanack K., et al. (2024). MicroRNA biomarkers as next-generation diagnostic tools for neurodegenerative diseases: a comprehensive re-view. Front. Mol. Neurosci. 17, 1386735. doi:10.3389/fnmol.2024.1386735
Baek S. H., Baek S., Kim G., Chi A. S., Kim E., Hwang D. H., et al. (2024). Possible effect of exercise with anti-fatigue nutrition on ROS-induced depression and suicide risk: a review. Phys. Act. Nutr. 28, 52–58. doi:10.20463/pan.2024.0016
Baggish A. L., Hale A., Weiner R. B., Lewis G. D., Systrom D., Wang F., et al. (2011). Dynamic regulation of circulating microRNA during acute exhaustive exercise and sustained aerobic exercise training. J. Physiol. 589, 3983–3994. doi:10.1113/jphysiol.2011.213363
Bartel D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi:10.1016/s0092-8674(04)00045-5
Bergström J., Hermansen L., Hultman E., Saltin B. (1967). Diet, muscle glycogen and physical performance. Acta Physiol. Scand. 71, 140–150. doi:10.1111/j.1748-1716.1967.tb03720.x
Bhaumik D., Scott G. K., Schokrpur S., Patil C. K., Orjalo A. V., Rodier F., et al. (2009). MicroRNAs miR-146a/b negatively modulate the senescence-associated inflammatory mediators IL-6 and IL-8. Aging (Albany NY) 1, 402–411. doi:10.18632/aging.100042
Bjersing J. L., Bokarewa M. I., Mannerkorpi K. (2015). Profile of circulating microRNAs in fibromyalgia and their relation to symptom severity: an exploratory study. Rheumatol. Int. 35, 635–642. doi:10.1007/s00296-014-3139-3
Boehm M., Slack F. (2005). A developmental timing microRNA and its target regulate life span in C. elegans. Science 310, 1954–1957. doi:10.1126/science.1115596
Bye A., Røsjø H., Aspenes S. T., Condorelli G., Omland T., Wisløff U. (2013). Circulating microRNAs and aerobic fitness--the HUNT-Study. PLoS One 8, e57496. doi:10.1371/journal.pone.0057496
Carmen P., Stephanie D., Amy E. M., Babalwa J., Tarryn W., Nasr E., et al. (2022). Changes in subcutaneous adipose tissue microRNA expression in response to exercise training in obese African women. Sci. Rep. 12, 18408. doi:10.1038/s41598-022-23290-x
Carroll T. J., Taylor J. L., Gandevia S. C. (2017). Recovery of central and peripheral neuromuscular fatigue after exercise. J. Appl. Physiol. 122, 1068–1076. doi:10.1152/japplphysiol.00775.2016
Chan S. Y., Zhang Y.-Y., Hemann C., Mahoney C. E., Zweier J. L., Loscalzo J. (2009). MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab. 10, 273–284. doi:10.1016/j.cmet.2009.08.015
Chen J.-F., Mandel E. M., Thomson J. M., Wu Q., Callis T. E., Hammond S. M., et al. (2006). The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38, 228–233. doi:10.1038/ng1725
Chen P., Chen F., Lei J., Li Q., Zhou B. (2019). Activation of the miR-34a-mediated SIRT1/mTOR signaling pathway by urolithin A attenuates D-galactose-induced brain aging in mice. Neurotherapeutics 16, 1269–1282. doi:10.1007/s13311-019-00753-0
Chen X., Li W., Chen T., Ren X., Zhu J., Hu F., et al. (2022). miR-146a-5p promotes epithelium regeneration against LPS-induced inflammatory injury via targeting TAB1/TAK1/NF-κB signaling pathway. Int. J. Biol. Macromol. 221, 1031–1040. doi:10.1016/j.ijbiomac.2022.09.056
Cheng H. S., Sivachandran N., Lau A., Boudreau E., Zhao J. L., Baltimore D., et al. (2013). MicroRNA-146 represses endothelial activation by inhibiting pro-inflammatory pathways. EMBO Mol. Med. 5, 1017–1034. doi:10.1002/emmm.201202318
Crosby M. E., Kulshreshtha R., Ivan M., Glazer P. M. (2009). MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 69, 1221–1229. doi:10.1158/0008-5472.CAN-08-2516
Dalgaard L. T., Sørensen A. E., Hardikar A. A., Joglekar M. V. (2022). The microRNA-29 family: role in metabolism and metabolic disease. Am. J. Physiol. Cell Physiol. 323, C367–C377. doi:10.1152/ajpcell.00051.2022
Esquela-Kerscher A., Slack F. J. (2006). Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer 6, 259–269. doi:10.1038/nrc1840
Falabrègue M., Boschat A.-C., Jouffroy R., Derquennes M., Djemai H., Sanquer S., et al. (2021). Lack of skeletal muscle serotonin impairs physical performance. Int. J. Tryptophan Res. 14, 11786469211003109. doi:10.1177/11786469211003109
Fitts R. H. (1994). Cellular mechanisms of muscle fatigue. Physiol. Rev. 74, 49–94. doi:10.1152/physrev.1994.74.1.49
Foley T. E., Greenwood B. N., Day H. E. W., Koch L. G., Britton S. L., Fleshner M. (2006). Elevated central monoamine receptor mRNA in rats bred for high endurance capacity: implications for central fatigue. Behav. Brain Res. 174, 132–142. doi:10.1016/j.bbr.2006.07.018
Gandevia S. C. (2001). Spinal and supraspinal factors in human muscle fatigue. Physiol. Rev. 81, 1725–1789. doi:10.1152/physrev.2001.81.4.1725
Garzon R., Marcucci G., Croce C. M. (2010). Targeting microRNAs in cancer: rationale, strategies and challenges. Nat. Rev. Drug Discov. 9, 775–789. doi:10.1038/nrd3179
Gravelsina S., Vilmane A., Svirskis S., Rasa-Dzelzkaleja S., Nora-Krukle Z., Vecvagare K., et al. (2022). Biomarkers in the diagnostic algorithm of myalgic encephalomyelitis/chronic fatigue syndrome. Front. Immunol. 13, 928945. doi:10.3389/fimmu.2022.928945
Gregory P. A., Bert A. G., Paterson E. L., Barry S. C., Tsykin A., Farshid G., et al. (2008). The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10, 593–601. doi:10.1038/ncb1722
Guedes J. R., Custódia C. M., Silva R. J., de Almeida L. P., Pedroso de Lima M. C., Cardoso A. L. (2014). Early miR-155 upregulation contributes to neuroinflammation in Alzheimer’s disease triple transgenic mouse model. Hum. Mol. Genet. 23, 6286–6301. doi:10.1093/hmg/ddu348
Guescini M., Canonico B., Lucertini F., Maggio S., Annibalini G., Barbieri E., et al. (2015). Muscle releases alpha-sarcoglycan positive extracellular vesicles carrying miRNAs in the bloodstream. PLoS One 10, e0125094. doi:10.1371/journal.pone.0125094
Guo Z., Li N., Jiang Y., Zhang L., Tong L., Wang Y., et al. (2023). HOXB9 a miR-122-5p regulated gene, suppressed the anticancer effects of brusatol by upregulating SCD1 expression in melanoma. Biomed. Pharmacother. 162, 114650. doi:10.1016/j.biopha.2023.114650
Hackney A. C. (2006). Stress and the neuroendocrine system: the role of exercise as a stressor and modifier of stress. Expert Rev. Endocrinol. Metab. 1, 783–792. doi:10.1586/17446651.1.6.783
Harmon J. L., Gibbs W. S., Whitaker R. M., Schnellmann R. G., Adkins D. L. (2017). Striatal mitochondrial disruption following severe traumatic brain injury. J. Neurotrauma 34, 487–494. doi:10.1089/neu.2015.4395
Harris T. A., Yamakuchi M., Ferlito M., Mendell J. T., Lowenstein C. J. (2008). MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc. Natl. Acad. Sci. U. S. A. 105, 1516–1521. doi:10.1073/pnas.0707493105
He L., Hannon G. J. (2004). MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522–531. doi:10.1038/nrg1379
Horie T., Ono K., Horiguchi M., Nishi H., Nakamura T., Nagao K., et al. (2010). MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc. Natl. Acad. Sci. U. S. A. 107, 17321–17326. doi:10.1073/pnas.1008499107
Hu S., Rong Y., Deng Y., Li L., Hu J., Yuan X., et al. (2023). miR-27b-3p inhibits estrogen secretion of goose granulosa cells by targeting CYP1B1 through the AMPK signaling pathway. Poult. Sci. 102, 102546. doi:10.1016/j.psj.2023.102546
Huang J., Wang S., Feng X., Liu X., Zhao J., Zheng Q., et al. (2019). miRNA transcriptome comparison between muscle and adipose tissues indicates potential miRNAs associated with intramuscular fat in Chinese swamp buffalo. Genome 62, 729–738. doi:10.1139/gen-2018-0178
Hutvágner G., Zamore P. D. (2002). A microRNA in a multiple-turnover RNAi enzyme complex. Science 297, 2056–2060. doi:10.1126/science.1073827
Ismaeel A., Fletcher E., Miserlis D., Wechsler M., Papoutsi E., Haynatzki G., et al. (2022). Skeletal muscle MiR-210 expression is associated with mitochondrial function in peripheral artery disease patients. Transl. Res. 246, 66–77. doi:10.1016/j.trsl.2022.03.003
Ivey K. N., Srivastava D. (2010). MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell 7, 36–41. doi:10.1016/j.stem.2010.06.012
Ji L. L. (1995). Oxidative stress during exercise: implication of antioxidant nutrients. Free Radic. Biol. Med. 18, 1079–1086. doi:10.1016/0891-5849(94)00212-3
Keller D. M., Perez I. G. (2022). Dual regulation of miR-375 and CREM genes in pancreatic beta cells. Islets 14, 139–148. doi:10.1080/19382014.2022.2060688
Kulshreshtha R., Ferracin M., Wojcik S. E., Garzon R., Alder H., Agosto-Perez F. J., et al. (2007). A microRNA signature of hypoxia. Mol. Cell Biol. 27, 1859–1867. doi:10.1128/MCB.01395-06
Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. (2001). Identification of novel genes coding for small expressed RNAs. Science 294, 853–858. doi:10.1126/science.1064921
Lai Z., Lin W., Yan X., Chen X., Xu G. (2023). Fatiguing freestyle swimming modifies miRNA profiles of circulating extracellular vesicles in athletes. Eur. J. Appl. Physiol. 123, 2041–2051. doi:10.1007/s00421-023-05167-7
Lee R., Feinbaum R., Ambros V. (1993). The C-elegans heterochronic gene lin-4 encodes small rnas with antisense complementarity to lin-14. Cell 75, 843–854. doi:10.1016/0092-8674(93)90529-Y
Li Y., Song X., Dai L., Wang Y., Luo Q., Lei L., et al. (2025). Mechanism of action of exercise regulating intestinal microflora to improve spontaneous hypertension in rats. Biomol. Biomed. 25, 648–662. doi:10.17305/bb.2024.11174
Lu H., Buchan R. J., Cook S. A. (2010). MicroRNA-223 regulates Glut4 expression and cardiomyocyte glucose metabolism. Cardiovasc. Res. 86, 410–420. doi:10.1093/cvr/cvq010
Luu B. E., Lefai E., Giroud S., Swenson J. E., Chazarin B., Gauquelin-Koch G., et al. (2020). MicroRNAs facilitate skeletal muscle maintenance and metabolic suppression in hibernating brown bears. J. Cell Physiol. 235, 3984–3993. doi:10.1002/jcp.29294
Lv W., Liu H., Zheng Q., Niu H. (2024). LINC02535 + miR-30a-5p combination enhances proliferation and inhibits apoptosis in metastatic breast Cancer cells. Toxicol. Vitro. 98, 105845. doi:10.1016/j.tiv.2024.105845
Ma L., Young J., Prabhala H., Pan E., Mestdagh P., Muth D., et al. (2010). miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol. 12, 247–256. doi:10.1038/ncb2024
Ma X., Chen H., Cao L., Zhao S., Zhao C., Yin S., et al. (2021). Mechanisms of physical fatigue and its applications in nutritional interventions. J. Agric. Food Chem. 69, 6755–6768. doi:10.1021/acs.jafc.1c01251
Magenta A., Cencioni C., Fasanaro P., Zaccagnini G., Greco S., Sarra-Ferraris G., et al. (2011). miR-200c is upregulated by oxidative stress and induces endothelial cell apoptosis and senescence via ZEB1 inhibition. Cell Death Differ. 18, 1628–1639. doi:10.1038/cdd.2011.42
Maughan R. J., Shirreffs S. M., Watson P. (2007). Exercise, heat, hydration and the brain. J. Am. Coll. Nutr. 26, 604S–612S. doi:10.1080/07315724.2007.10719666
M’baya-Moutoula E., Louvet L., Molinié R., Guerrera I. C., Cerutti C., Fourdinier O., et al. (2018). A multi-omics analysis of the regulatory changes induced by miR-223 in a monocyte/macrophage cell line. Biochim. Biophys. Acta. Mol. Basis Dis. 1864, 2664–2678. doi:10.1016/j.bbadis.2018.05.010
McCarthy J. J., Esser K. A., Andrade F. H. (2007). MicroRNA-206 is overexpressed in the diaphragm but not the hindlimb muscle of mdx mouse. Am. J. Physiol. Cell Physiol. 293, C451–C457. doi:10.1152/ajpcell.00077.2007
McKenna M. J., Bangsbo J., Renaud J.-M. (2008). Muscle K+, Na+, and Cl disturbances and Na+-K+ pump inactivation: implications for fatigue. J. Appl. Physiol. 104, 288–295. doi:10.1152/japplphysiol.01037.2007
Meeusen R., Van Cutsem J., Roelands B. (2021). Endurance exercise-induced and mental fatigue and the brain. Exp. Physiol. 106, 2294–2298. doi:10.1113/EP088186
Meeusen R., Watson P., Hasegawa H., Roelands B., Piacentini M. F. (2006). Central fatigue: the serotonin hypothesis and beyond. Sports Med. 36, 881–909. doi:10.2165/00007256-200636100-00006
Moebus M., Gashi S., Hilty M., Oldrati P., Holz C. (2024). Meaningful digital biomarkers derived from wearable sensors to predict daily fatigue in multiple sclerosis patients and healthy controls. iScience 27, 108965. doi:10.1016/j.isci.2024.108965
Nielsen S., Åkerström T., Rinnov A., Yfanti C., Scheele C., Pedersen B. K., et al. (2014). The miRNA plasma signature in response to acute aerobic exercise and endurance training. PLoS One 9, e87308. doi:10.1371/journal.pone.0087308
O’Connell R. M., Taganov K. D., Boldin M. P., Cheng G., Baltimore D. (2007). MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. U. S. A. 104, 1604–1609. doi:10.1073/pnas.0610731104
Ponomarev E. D., Veremeyko T., Barteneva N., Krichevsky A. M., Weiner H. L. (2011). MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-α-PU.1 pathway. Nat. Med. 17, 64–70. doi:10.1038/nm.2266
Powers S. K., Jackson M. J. (2008). Exercise-induced oxidative stress: cellular mechanisms and impact on muscle force production. Physiol. Rev. 88, 1243–1276. doi:10.1152/physrev.00031.2007
Qian C., Zhou F., Lu D., Huang J., Sun M. (2024). Exercise intensity and mortality in overweight and obese patients with chronic kidney disease: longitudinal analysis (1999-2016). BMC Public Health 24, 3020. doi:10.1186/s12889-024-20498-6
Rayner K. J., Suárez Y., Dávalos A., Parathath S., Fitzgerald M. L., Tamehiro N., et al. (2010). MiR-33 contributes to the regulation of cholesterol homeostasis. Science 328, 1570–1573. doi:10.1126/science.1189862
Reinhart B. J., Slack F. J., Basson M., Pasquinelli A. E., Bettinger J. C., Rougvie A. E., et al. (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906. doi:10.1038/35002607
Renaud J.-M., ortenblad N., McKenna M. J., Overgaard K. (2023). Exercise and fatigue: integrating the role of K+, Na+ and Cl- in the regulation of sarcolemmal excitability of skeletal muscle. Eur. J. Appl. Physiol. 123, 2345–2378. doi:10.1007/s00421-023-05270-9
Rodriguez A., Vigorito E., Clare S., Warren M. V., Couttet P., Soond D. R., et al. (2007). Requirement of bic/microRNA-155 for normal immune function. Science 316, 608–611. doi:10.1126/science.1139253
Rottiers V., Näär A. M. (2012). MicroRNAs in metabolism and metabolic disorders. Nat. Rev. Mol. Cell Biol. 13, 239–250. doi:10.1038/nrm3313
Russell A. P., Lamon S., Boon H., Wada S., Güller I., Brown E. L., et al. (2013). Regulation of miRNAs in human skeletal muscle following acute endurance exercise and short-term endurance training. J. Physiol. 591, 4637–4653. doi:10.1113/jphysiol.2013.255695
Safdar A., Abadi A., Akhtar M., Hettinga B. P., Tarnopolsky M. A. (2009). miRNA in the regulation of skeletal muscle adaptation to acute endurance exercise in C57Bl/6J male mice. PLoS One 4, e5610. doi:10.1371/journal.pone.0005610
Safdar A., Saleem A., Tarnopolsky M. A. (2016). The potential of endurance exercise-derived exosomes to treat metabolic diseases. Nat. Rev. Endocrinol. 12, 504–517. doi:10.1038/nrendo.2016.76
Sahlin K., Tonkonogi M., Söderlund K. (1998). Energy supply and muscle fatigue in humans. Acta Physiol. Scand. 162, 261–266. doi:10.1046/j.1365-201X.1998.0298f.x
Schmidt M. E., Maurer T., Behrens S., Seibold P., Obi N., Chang-Claude J., et al. (2024). Cancer-related fatigue: towards a more targeted approach based on classification by biomarkers and psychological factors. Int. J. Cancer 154, 1011–1018. doi:10.1002/ijc.34791
Sheedy F. J., Palsson-McDermott E., Hennessy E. J., Martin C., O’Leary J. J., Ruan Q., et al. (2010). Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat. Immunol. 11, 141–147. doi:10.1038/ni.1828
Taganov K. D., Boldin M. P., Chang K.-J., Baltimore D. (2006). NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. U. S. A. 103, 12481–12486. doi:10.1073/pnas.0605298103
Tong L., Hao H., Zhang Z., Lv Y., Liang X., Liu Q., et al. (2021). Milk-derived extracellular vesicles alleviate ulcerative colitis by regulating the gut immunity and reshaping the gut microbiota. Theranostics 11, 8570–8586. doi:10.7150/thno.62046
Wada S., Kato Y., Okutsu M., Miyaki S., Suzuki K., Yan Z., et al. (2011). Translational suppression of atrophic regulators by MicroRNA-23a integrates resistance to skeletal muscle atrophy. J. Biol. Chem. 286, 38456–38465. doi:10.1074/jbc.M111.271270
Wang J-X., Jiao J-Q., Li Q., Long B., Wang K., Liu J-P., et al. (2011). miR-499 regulates mitochondrial dynamics by targeting calcineurin and dynamin-related protein-1. Nat. Med. 17, 71–78. doi:10.1038/nm.2282
Wang S., Huang J., Feng X., Liu X., Zhao J., Zheng Q., et al. (2019). miRNA transcriptome comparison between muscle and adipose tissues indicates potential miRNAs associated with intramuscular fat in Chinese swamp buffalo. Genome 62, 729–738. doi:10.1139/gen-2018-0178
Winbanks C. E., Beyer C., Hagg A., Qian H., Sepulveda P. V., Gregorevic P. (2013). miR-206 represses hypertrophy of myogenic cells but not muscle fibers via inhibition of HDAC4. PLoS One 8, e73589. doi:10.1371/journal.pone.0073589
Wu H., Xia L., Xu H. (2022). Role of FUS-CHOP in myxoid liposarcoma via miR-486/CDK4 Axis. Biochem. Genet. 60, 1095–1106. doi:10.1007/s10528-021-10151-x
Yamakuchi M., Ferlito M., Lowenstein C. J. (2008). miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. U. S. A. 105, 13421–13426. doi:10.1073/pnas.0801613105
You S., Ji F., He F., Tong B. (2016). Effect of transcutaneous electrical acupoint stimulation on red blood cell and hemoglobin in college athletes with exercise fatigue. Rehabil. Med. 26, 43. doi:10.3724/SP.J.1329.2016.04043
Yuasa K., Hagiwara Y., Ando M., Nakamura A., Takeda S., Hijikata T. (2008). MicroRNA-206 is highly expressed in newly formed muscle fibers: implications regarding potential for muscle regeneration and maturation in muscular dystrophy. Cell Struct. Funct. 33, 163–169. doi:10.1247/csf.08022
Keywords: exercise fatigue, microRNA, energy metabolism, inflammatory response, oxidative stress
Citation: Niu S, Yin X, Cao Q, Huang K, Deng Z and Cao J (2025) miRNAs involved in the regulation of exercise fatigue. Front. Physiol. 16:1614942. doi: 10.3389/fphys.2025.1614942
Received: 20 April 2025; Accepted: 10 June 2025;
Published: 23 June 2025.
Edited by:
Pierre-Marie Leprêtre, Université de Rouen, FranceReviewed by:
Laurent Metzinger, University of Picardie Jules Verne, FranceYingfang Wang, GDPU, China
Copyright © 2025 Niu, Yin, Cao, Huang, Deng and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongyuan Deng, ZHp5QHp6dS5lZHUuY24=; Jie Cao, Y2FvamllNzdAbmF2ZXIuY29t
†These authors have contributed equally to this work
 Siyuan Niu1†
Siyuan Niu1† Xiupeng Yin
Xiupeng Yin