- 1Department of General Medicine, The First People’s Hospital of Zhengzhou, Zhengzhou, Henan, China
- 2First Clinical Medical College, Ningxia Medical University, Yinchuan, Ningxia, China
- 3Department of Ultrasound, The First People’s Hospital of Zhengzhou, Zhengzhou, Henan, China
Background: With global aging and lifestyle changes, carotid atherosclerotic plaques are a major cause of cerebrovascular disease and ischemic stroke. However, ultrasound images suffer from high noise, low contrast, and blurred edges, making it difficult for traditional image processing methods to accurately extract plaque information.
Objective: To establish a deep learning-based DualPlaqueNet model for semantic segmentation and size prediction of plaques in carotid ultrasound images, thereby providing comprehensive and accurate auxiliary information for clinical risk assessment and personalized diagnosis and treatment.
Methods: DualPlaqueNet uses a dual-branch architecture combined with attention mechanisms and joint loss functions to optimize segmentation and regression. Notably, a multi-layer one-dimensional convolutional structure is introduced within the Efficient Channel Attention (ECA) module. The original dataset contained 287 carotid ultrasound images from patients at Zhengzhou First People’s Hospital, which were divided into training, validation, and test sets. Model training, validation, and testing were performed after preprocessing and data augmentation of the training set. Its performance was compared with three other models.
Results: In the plaque semantic segmentation task, DualPlaqueNet outperformed the other three models across all metrics, achieving MIoU of 88.91 ± 1.027 (%), IoU (excluding background) of 88.22 ± 1.065 (%), DSC of 89.95 ± 1.102 (%), and Accuracy of 95.98 ± 0.073 (%). For plaque size prediction, this model demonstrated lower MSE and MAE, along with a higher coefficient of determination R2, proving its ability to accurately extract plaque size information from ultrasound images.
Conclusion: The dual-branch design and attention mechanisms of DualPlaqueNet effectively address the challenges of ultrasound images, achieving precise segmentation and size prediction, demonstrating its potential as an auxiliary tool for future clinical applications.
1 Introduction
In recent years, as global aging accelerates and lifestyles change, cardiovascular diseases have gradually emerged as a major public health threat (Azeez, 2023; Bayoumi and Karasik, 2021; Goldsborough et al., 2022). The formation and progression of carotid atherosclerotic plaques are considered to be key pathological foundations for cerebrovascular diseases and ischemic stroke, and their accurate detection and quantitative analysis are of great significance for clinical prevention, risk assessment, and treatment decision-making (Ihle-Hansen et al., 2023; Miao et al., 2022). Carotid plaques not only reflect the severity of systemic arteriosclerosis but also provide individualized health management recommendations for patients (Hou et al., 2024; van Dam-Nolen et al., 2022). Currently, the diagnosis of carotid plaques requires ultrasound examinations. However, ultrasound physicians must spend long hours in front of display screens, which can lead to occupational ailments such as eye strain and back pain. Moreover, diagnoses made by different sonographers are prone to subjective errors due to varying levels of clinical expertise, and even the same physician may demonstrate different diagnostic efficiency depending on their level of fatigue. Therefore, automatically and accurately segmenting plaque regions from ultrasound images while predicting plaque size and improving efficiency has become a critical issue that urgently needs to be addressed in the field of carotid ultrasound image analysis.
Ultrasound imaging, due to its non-invasive, real-time, cost-effective, and widely applicable nature, has been extensively used for the clinical detection of carotid plaques. However, inherent limitations of ultrasound images—such as high noise levels, low contrast, and blurred edges—make traditional image processing algorithms prone to interference when segmenting plaques, and they struggle to capture the subtle morphological features of plaques (Luo et al., 2021; Singh et al., 2023). Specifically, the main challenges in ultrasound image analysis of carotid plaques are: (1) intense speckle noise and echo attenuation result in very low contrast between the plaque and surrounding tissue; (2) calcified plaques produce strong shadowing effects, causing fragmented boundaries and distorted morphology; (3) considerable variability in vessel anatomy and plaque types across patients makes model generalization difficult; and (4) probe motion and arterial pulsation introduce dynamic artifacts, further degrading segmentation accuracy. In recent years, deep learning techniques, particularly convolutional neural networks (CNNs), have achieved remarkable success in medical image segmentation, greatly advancing the automation of medical image analysis. At present, in addition to the research on cardiac and breast ultrasound, many scholars are also focusing on carotid plaque image segmentation, as shown in Table 1; (Huang et al., 2023). For instance, Zhou et al. proposed a deep - learning - based method for automatically measuring the total plaque area in B- mode ultrasound images. Trained on a small dataset with the UNet++ integrated algorithm, it can efficiently and accurately measure the total plaque area (TPA) and has shown good generalization ability on datasets acquired from different devices (Zhou et al., 2021).
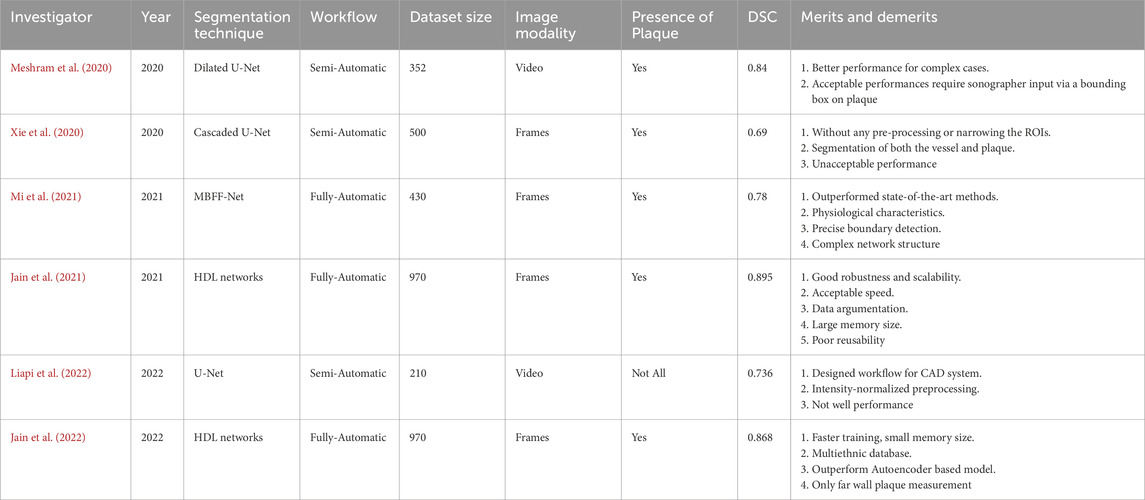
Table 1. Relevant DL-based plaque segmentation techniques and their main features: investigators, references, publication year, segmentation techniques used, workflow type (semi-automatic or fully automatic), dataset size, data type (frames or video), plaque presence (“Yes” or “Not All”), performance metrics, and major advantages and disadvantages of the techniques.
To address these issues, this paper proposes a novel multi-task joint learning model—DualPlaqueNet. The model adopts a dual-branch network architecture that is specifically designed for the tasks of plaque semantic segmentation and size prediction, and it achieves information sharing and collaborative optimization between the two tasks through a cross-fusion mechanism. Specifically, one branch of DualPlaqueNet is dedicated to extracting global semantic features to capture the overall morphology of plaques in complex backgrounds, while the other branch focuses on local detailed features to precisely delineate plaque edges and size information. By designing a joint loss function, the model is able to simultaneously optimize both segmentation and size prediction tasks during training, allowing these tasks to complement each other and collectively enhance the overall performance and robustness of the model.
Based on the research work of the DualPlaqueNet model, this paper aims to establish a multi-task joint optimization framework capable of performing both plaque semantic segmentation and size prediction simultaneously. This framework not only enhances the accuracy of plaque detection but also provides clinicians with richer and more intuitive diagnostic information, ultimately reducing the physicians’ workload.
2 Materials and methods
2.1 Data collection and grouping
In this study, a total of 523 patients underwent carotid ultrasound examination. Based on inclusion and exclusion criteria, 287 patients were ultimately selected, with one high-quality image (manually screened) chosen from each patient’s ultrasound images. These patients were from the outpatient and inpatient departments of Zhengzhou First People’s Hospital, and their carotid ultrasound images constituted the original image dataset for this study.
Inclusion Criteria: Patients who underwent carotid ultrasound examinations and were found to have carotid plaques.
Exclusion Criteria: (1) Patients whose ultrasound reports did not indicate the location of the plaques; (2) Patients whose ultrasound reports did not describe the long or short diameters of the plaques; (3) Patients who did not sign the informed consent form.
This study was conducted in accordance with the Declaration of Helsinki and received approval from the Hospital Ethics Committee (Ethics Review Committee of the First People’s Hospital of Zhengzhou, No. 2024-069). Prior to collecting the carotid ultrasound images, all participants or their guardians signed a consent form, ensuring the ethical compliance of the study.
Two physicians with 10 years of experience in ultrasound confirmed the plaque locations and sizes in the carotid ultrasound reports, and they manually annotated the plaques in the ultrasound images. In cases of disagreement, the two physicians consulted with a senior physician with over 25 years of clinical experience until consensus was reached.
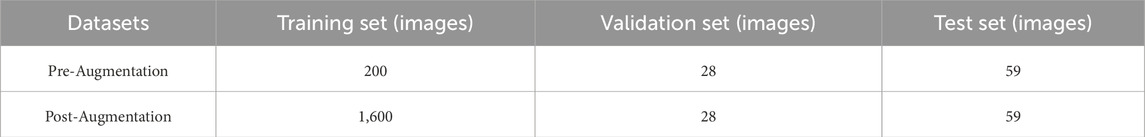
To effectively train, optimize, and evaluate the model, the ultrasound image dataset was randomly divided into training, validation, and test sets at a ratio of 7:1:2, ensuring the scientific and reliable process of model training, validation, and evaluation.
2.2 Data preprocessing
Prior to preprocessing, patients’ personal information was removed from the ultrasound images to protect privacy. Our ultrasound brand involves two types, namely, Mindray-R7, China and Siemens AG-ACUSON Seguoia, Germany. For the convenience of subsequent analysis, the ultrasound images of these two brands were subjected to the same preprocessing steps to standardize them. Considering factors such as segmentation accuracy and training speed, the images were normalized and enhanced for contrast to improve detail representation. Additionally, to enhance the model’s generalizability and robustness, data augmentation was performed on the training set (Table 2). Specific augmentation techniques included elastic deformation, rotation, scaling, and flipping operations. These data augmentation methods not only effectively expanded the training set and prevented model overfitting, but also simulated different clinical scenarios and equipment variations, thereby improving the model’s adaptability in practical applications (Yan et al., 2024; Piao et al., 2022). Figure 1 illustrates an example of the data preprocessing process.
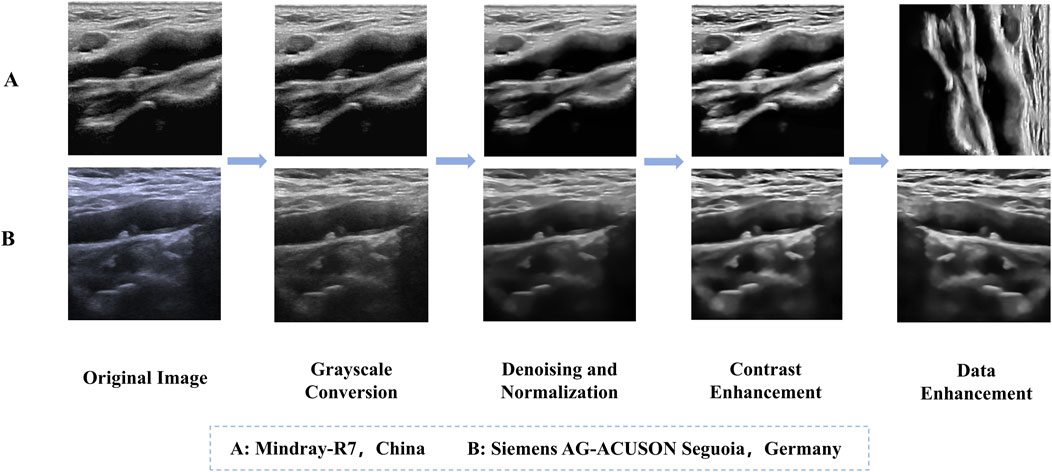
Figure 1. Example of the data preprocessing process. (A) Mindray-R7, China, (B) Siemens AG-ACUSON Seguoia, Germany.
2.3 Model construction
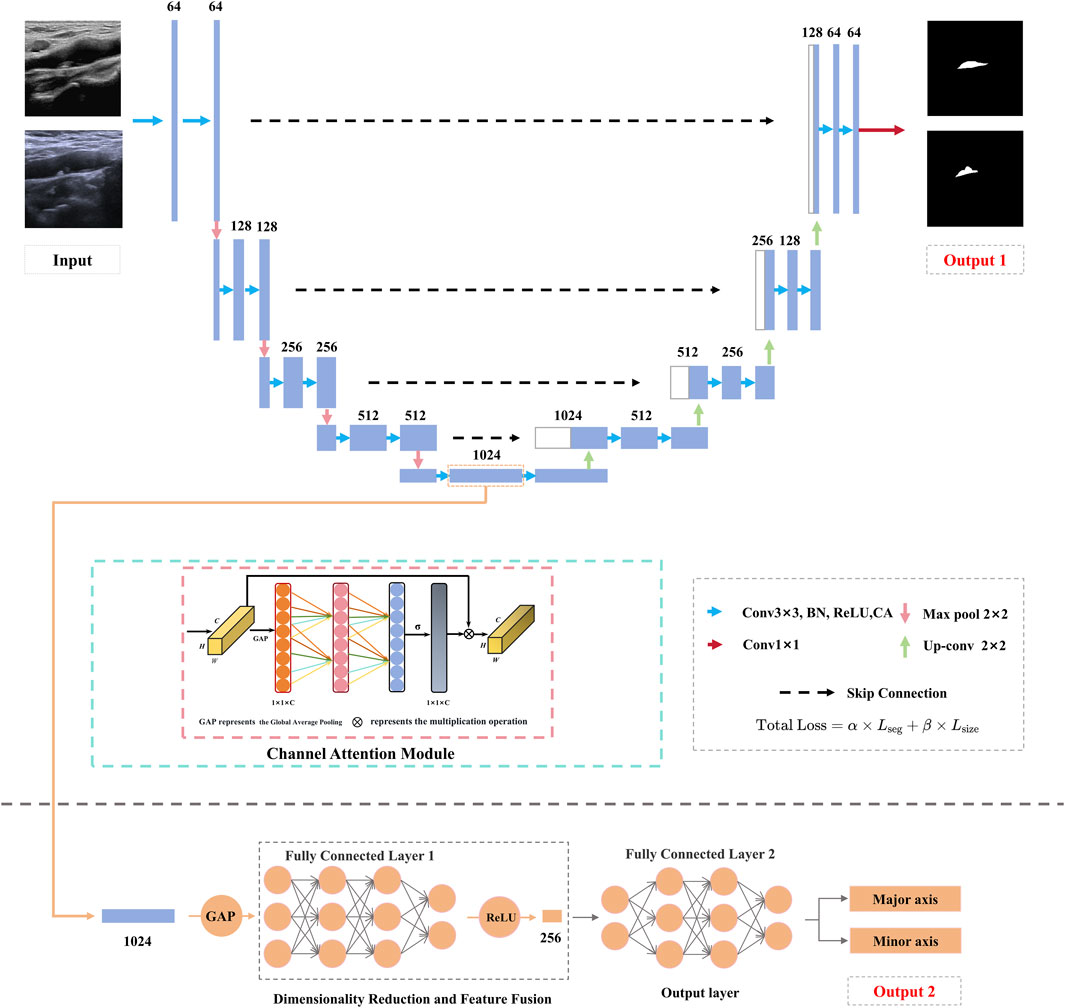
The proposed DualPlaqueNet model (Figure 2) introduces innovative improvements based on the traditional U-Net architecture, aiming to address the semantic segmentation of carotid plaque ultrasound images and the prediction of plaque size. The model first adopts the U-Net encoder-decoder structure (Tseng et al., 2023; Yi et al., 2023), extracting multi-scale features through down-sampling and integrating low-level details with high-level semantic information via up-sampling and skip connections. Additionally, an attention mechanism (Alshomrani et al., 2023; Sheng et al., 2022) is incorporated to achieve precise segmentation of plaque regions. In this study, we adopted and improved the Efficient Channel Attention (ECA) module to enhance the model’s performance in plaque region segmentation. Moreover, we deployed the ECA module at every feature extraction layer in the encoder. The ECA module generates channel weights through local cross-channel interaction, helping the network more precisely capture feature information from different channels, thereby improving segmentation performance. In the original ECA module, a single one-dimensional convolution layer was used to compute channel weights. We introduced a multi-layer one-dimensional convolution structure to extract feature information at different levels layer by layer, further optimizing the channel weight computation process and enhancing the model’s ability to capture complex image features.
Regarding the choice of ECA over other more advanced attention mechanisms, this is mainly due to its efficiency and low computational overhead. ECA uses one-dimensional convolution to compute channel weights, making it have lower computational complexity compared to other attention mechanisms (such as multi-head self-attention or Manhattan attention). When processing medical images, especially segmentation tasks for small targets like plaque regions, ECA can maintain efficient inference speed while effectively improving performance through relatively low computational overhead. Although mechanisms like multi-head self-attention and Manhattan attention can provide stronger feature capture capabilities, they typically have high computational overhead, especially when processing high-resolution medical images, which may lead to slower training and inference speeds. Therefore, selecting the ECA module can improve model performance while ensuring efficient computational efficiency.
Although measuring dimensions on plaque segmentation results is a feasible approach, this method may overlook the complexity of the dimension prediction task. Dimension prediction is not merely simple post-processing based on segmentation results; it involves comprehensive understanding of multiple factors such as plaque morphology, boundaries, and position. If the model relies solely on segmentation results for dimension measurement, it may ignore the detailed features of plaques, thus affecting the accuracy of dimension prediction. Through joint training, we enable the model to learn the low-level features and semantic information required for dimension prediction while performing plaque segmentation. This design allows the model to simultaneously optimize both tasks, capture the interconnections between them, and enhance the model’s comprehensive understanding of plaques. Therefore, DualPlaqueNet introduces a novel branch dedicated to plaque size prediction. This branch extracts plaque morphological information from the deeper features of the encoder and, through a series of convolutional and fully-connected layers, regresses the plaque’s long and short diameters. To enable multi-task collaborative learning, a joint loss function is employed, with an automatic parameter tuning method used to determine the values of parameters α and β, thus balancing the semantic segmentation loss and regression loss to promote mutual optimization between the two tasks. In this study, we used Cross-Entropy Loss and Mean Squared Error Loss (MSE Loss) as the loss functions for the two main tasks. We used α and β to control the relative importance of the segmentation task and the size prediction task. See Equations 1, 2 for details.
Where
2.4 Evaluation metrics
In this study, the prediction performance of DualPlaqueNet was compared with that of U-Net, ResUnet, and TransUNet. For the segmentation of carotid ultrasound images, the plaque region is considered the positive sample, while the non-plaque region is treated as the negative sample. These are categorized as true positives (TP), false positives (FP), true negatives (TN), and false negatives (FN). In this study, Accuracy, Mean Intersection over Union (MIoU), and Dice Similarity Coefficient (DSC) are used as evaluation metrics. Accuracy (ACC) reflects the ratio of correctly predicted pixels to the total number of pixels, with higher values indicating more precise segmentation. The Dice coefficient quantifies the similarity between the model’s predictions and the ground truth annotations. We have introduced the “background excluded mIoU” calculation method, which excludes background pixels (with a value of 0) in the mIoU calculation and only considers the IoU of the plaque area. This method avoids the influence of background areas on the evaluation results and more accurately reflects the segmentation performance of the model in patch areas. MIoU provides a more comprehensive evaluation of the model’s performance by averaging the IoU values for each class. The calculation Equations 3–5 for each evaluation metric are as follows:
Among these, TP, FP, TN, FN, and k represent true positive, false positive, true negative, false negative, and the number of classes, respectively.
For the prediction of plaque size in carotid ultrasound images, this study employs the following three statistical metrics to evaluate the predictive performance on the test set. Mean Squared Error (MSE) is the mean of the squared differences between the predicted and actual values, while Mean Absolute Error (MAE) is the average of the absolute differences between the predicted and actual values. The smaller the MSE and MAE, the more accurate the predictions; R2 measures the model’s ability to explain the variability of the data, and the closer R2 is to 1, the stronger the model’s predictive performance. The calculation Equations 6–8 for these three statistical metrics are as follows:
Here, n represents the number of samples,
3 Experiments and results
3.1 Experimental environment
The models in this study were implemented using Python 3.12.6 and PyTorch 2.4.1, and trained on an NVIDIA RTX 4060 GPU. The Adam optimizer (Aamir et al., 2023; Abirami et al., 2025) was used with an initial learning rate of 0.001. All models were trained with a batch size of 16 for 100 epochs.
3.2 Image segmentation
After image preprocessing, the DualPlaqueNet model was trained on the data-augmented training set, while three other models were simultaneously trained for comparison (Figure 3). The validation set was used to tune hyperparameters and prevent overfitting during the training process. The test set was used to evaluate model performance on the region segmentation task and generate automatic segmentation result images of target regions.
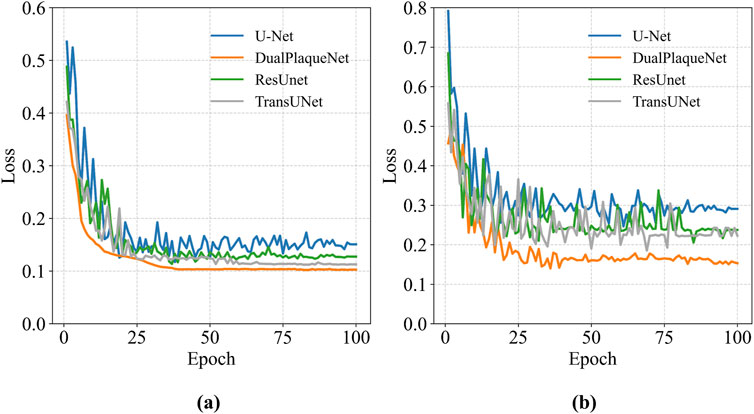
Figure 3. Loss curves of each model on the training set and validation set. (a) The loss curve of the training set, (b) The loss curve of the validation set.
We performed 10 repeated training sessions for DualPlaqueNet, U-Net, ResUnet, and TransUNet. After each training session, the optimal network parameters were saved, and then the average values and corresponding standard deviations of evaluation metrics for the 4 networks on the same test set were calculated. These results are shown in Table 3. The reason for conducting 10 repeated training sessions was primarily to evaluate the model’s stability and generalization ability, reducing the impact of random factors (such as parameter initialization and data order) during the training process on the final results. Due to these random factors, each training session may lead to different training results. Through multiple repeated training sessions and saving the optimal network parameters that performed best on the validation set in each training session, we were able to calculate the average performance and standard deviation of the model across multiple training sessions, thereby more reliably evaluating the model’s overall performance.
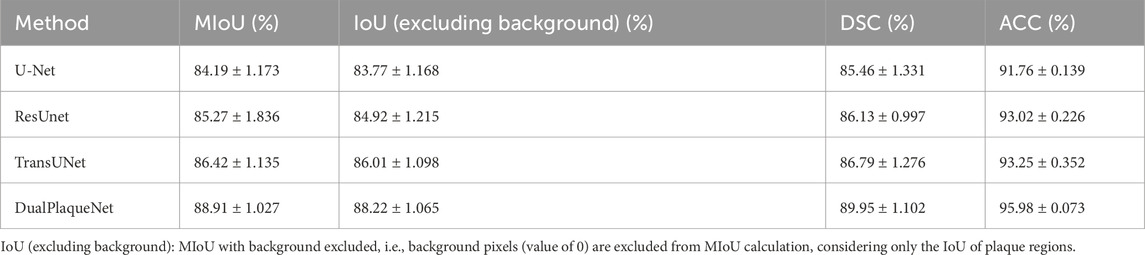
Table 3. Comparison of evaluation metrics between DualPlaqueNet and other network models on the same test set.
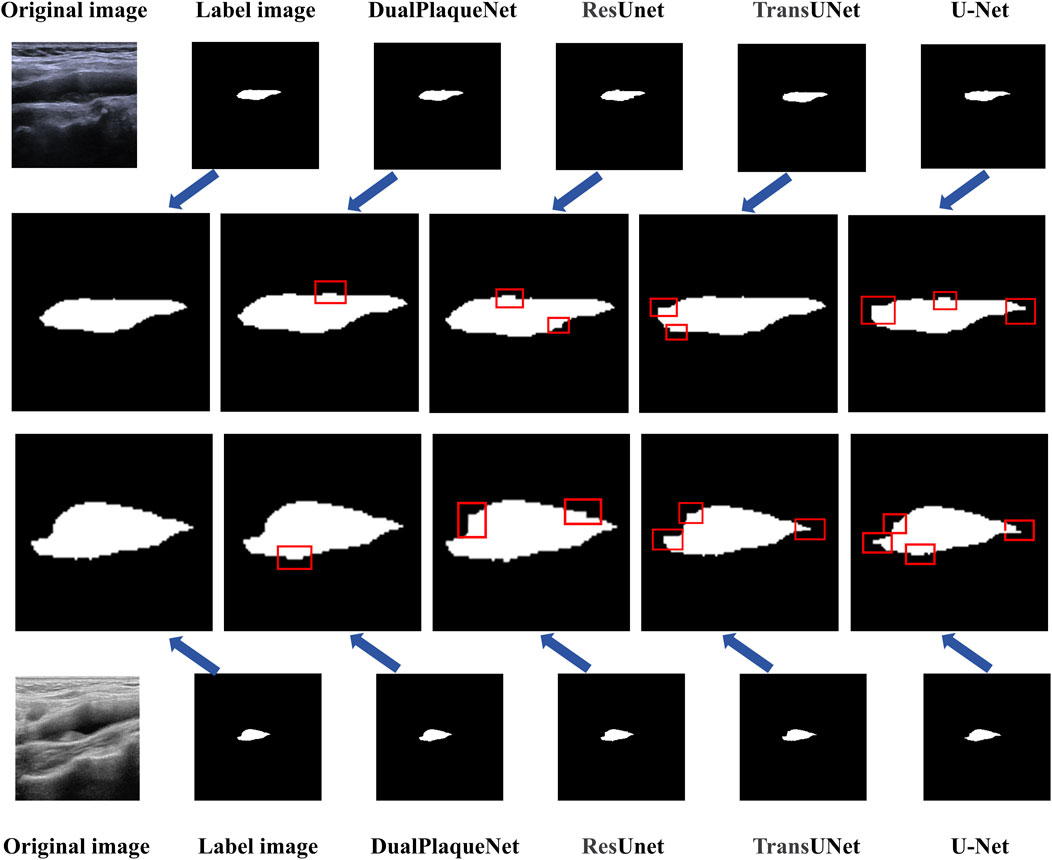
Regarding the overfitting issue, we used the validation set during the training process to select optimal parameters and ensured that the final performance evaluation was conducted on the test set to validate the model’s generalization ability. Saving optimal network parameters does not mean the model has overfitted, because these optimal parameters were selected based on performance on the validation set, rather than solely relying on performance on the training set. This approach better ensures the model’s generalization ability and stability. Additionally, we also used techniques such as early stopping during the experimental process to prevent model overfitting, further ensuring that overfitting would not occur during the training process. Compared with the other three network models, DualPlaqueNet’s segmentation results were highly similar to doctors’ manual labels (Figure 4). This figure demonstrates that DualPlaqueNet is more sensitive to boundary information and closer to the true label images.
3.3 Plaque size prediction
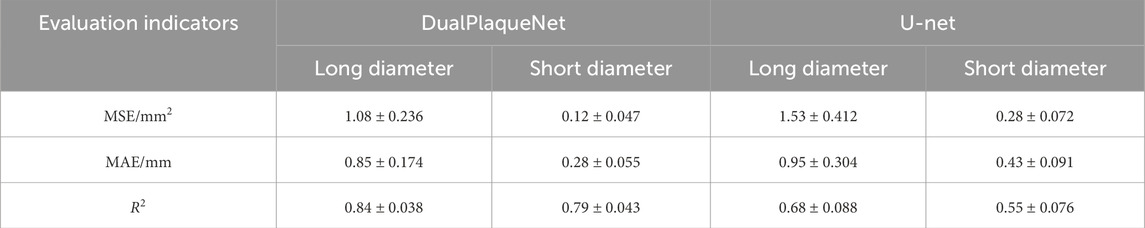
For plaque size prediction, the training procedure is identical to that of image segmentation, using the augmented training set. During the manual annotation process by ultrasound physicians, the manually measured long and short diameters of the plaques were recorded in an Excel sheet and embedded into the metadata of the corresponding image files (written into DICOM private tags). In this study, DualPlaqueNet directly predicts the long and short diameters of the plaques, whereas U-Net first segments the images and then measures the segmented regions to obtain the long and short diameters. We conducted 10 repeated training sessions for both DualPlaqueNet and U-Net, saving the optimal network parameters after each training session. The average values and corresponding standard deviations of MSE, MAE, and R2 on the same test set were calculated; these results are presented in Table 4. DualPlaqueNet achieved lower average MSE and MAE values and a higher average R2 value compared to U-Net, indicating that DualPlaqueNet has a superior capability for predicting plaque size.
4 Discussion
In this study, a DualPlaqueNet model based on a multi-task joint learning framework was developed and validated, aiming to simultaneously achieve semantic segmentation and size prediction of carotid plaques. Experimental results show that, compared with the U-Net, ResUnet, and TransUNet models, DualPlaqueNet achieved significant advantages in MIoU, IoU, DSC, and ACC metrics. In predicting the plaque’s long and short diameters, its mean squared error and mean absolute error were both significantly reduced relative to U-Net, and the R2 value also indicated a higher degree of fit. In this study, we adopted and improved the ECA (Efficient Channel Attention) module to enhance the performance of the model in plaque region segmentation. The design principle of the ECA module is to generate channel weights through local cross channel interactions, reducing computational overhead and achieving higher efficiency. The original ECA module used a layer of one-dimensional convolution to calculate channel weights. In this study, we introduced a multi-layer one-dimensional convolution structure inside the ECA module, which further optimized the calculation process of channel weights by extracting different levels of feature information layer by layer, enhancing the ability to capture complex image features. And, we will add it to the feature extraction section of the encoder. This design approach effectively overcomes the inherent limitations of ultrasound images, such as low contrast, high noise levels, and blurred edges, and significantly improves the model’s sensitivity to subtle changes in plaque characteristics, thereby maintaining high robustness and accuracy even in complex imaging backgrounds.
Currently, both domestic and international scholars have conducted extensive exploration and research in the field of carotid plaque detection and other medical image segmentation tasks (Chen et al., 2021; Flannery et al., 2021; Wang et al., 2021). Traditional image processing-based algorithms often focus on methods such as edge detection, which are limited by their sensitivity to noise and difficulty in characterizing complex lesion areas (Tsantis et al., 2014; Alshayeji et al., 2017; Zheng et al., 2015). In recent years, the introduction of deep learning technologies, such as CNNs, has provided a new breakthrough for addressing segmentation challenges in ultrasound and other medical images. For example, Yanhan Li et al. (Li et al., 2022) proposed a novel deep convolutional neural network model, FRDD-Net, for the automatic segmentation of carotid plaque ultrasound images. By incorporating a feature remapping module and a dense decoding mechanism, this model enhances feature extraction and utilization efficiency, overcoming the limitations of existing methods when dealing with low-quality images and irregular plaques. Experimental results indicate that FRDD-Net performs excellently on multiple datasets, demonstrating its potential and robustness in medical image segmentation tasks. Avesta A et al. (Avesta et al., 2023) proposed a brain image segmentation method based on a 3D Capsule Network (CapsNet), and compared it with traditional U-Net and nnUNet models. The experimental results show that CapsNet demonstrates significant advantages when processing the test set. Its segmentation accuracy is significantly higher than that of U-Net, and there is also a significant improvement in computational efficiency. CapsNet not only effectively segments brain structures but also requires lower memory and trains faster. Dong P et al. (Dong et al., 2024) introduced a UNet++ model enhanced with a dual-path attention mechanism (DPAM-UNet++) for the automatic segmentation of thyroid nodule ultrasound images. By integrating a dual-path attention module into the skip connections of UNet++, the model is able to effectively capture global contextual information, thereby improving the segmentation performance for small nodules and multiple nodules. Experimental results indicate that DPAM-UNet++ outperforms traditional segmentation models across multiple performance metrics, particularly in enhancing boundary precision and handling multiple nodules. Compared to the aforementioned works, this study leverages the advantages of traditional deep learning frameworks while organically integrating plaque semantic segmentation and size prediction through a multi-task joint optimization strategy. This approach enables comprehensive information sharing and complementarity, helping to overcome the limitations of single-task methods in information extraction, thereby providing a more comprehensive and efficient technical means for the quantitative analysis of carotid plaques.
The DualPlaqueNet model presented in this study embodies both foresight and practical value in its design. By introducing a dual-branch structure and a cross-fusion strategy, the model achieves collaborative learning of plaque morphology and size information. This multi-task joint learning approach overcomes the limitations of previous single-objective optimizations, effectively enhancing the model’s performance in the complex environments of ultrasound imaging. Additionally, the embedded attention mechanism allows the model to automatically focus on key feature regions, further improving the extraction of both global semantic information and local detail features, thereby optimizing plaque region segmentation and size prediction. Nevertheless, there are certain limitations to this approach. First, the model’s training and validation were conducted on a single-center dataset with a relatively limited amount of data, which might lead to insufficient generalization performance in multi-center or multi-device application scenarios. Second, the inherent noise and variability in ultrasound images can result in local misjudgments, especially in regions with fuzzy edges or low contrast. Moreover, although multi-task learning facilitates feature sharing to a certain extent, the challenge of balancing the different tasks still requires further investigation. How to adaptively adjust task weights under varying data distributions remains a direction for future research. In summary, DualPlaqueNet shows significant advantages in improving automated plaque detection and quantitative analysis, offering considerable support to ultrasound physicians and enhancing the diagnostic efficiency for carotid plaques. However, for its practical application and broader clinical promotion, continuous optimization is necessary. This includes increasing sample sizes, incorporating multi-center data, and further refining the model architecture to ensure stable and efficient performance in a wider range of clinical scenarios.
5 Conclusion
This study proposes the DualPlaqueNet model, which integrates a dual-branch structure and attention mechanism. Through comparisons with models such as U-Net, ResUnet, and TransUNet, it was found that DualPlaqueNet demonstrates excellent performance in both semantic segmentation and size prediction tasks for carotid artery plaques, showing promise as a tool to assist in early screening and risk assessment of cerebrovascular diseases.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by the Ethics Review Committee of the First People’s Hospital of Zhengzhou. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin. Written informed consent was obtained from the individual(s), and minor(s)’ legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
Author contributions
LD: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review and editing. XnD: Conceptualization, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review and editing. YS: Investigation, Supervision, Writing – original draft. YW: Resources, Validation, Writing – review and editing. DS: Resources, Validation, Writing – review and editing. XaD: Data curation, Investigation, Resources, Supervision, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This project is financially supported by the 2024 Zhengzhou Municipal Medical and Health Sector Science and Technology Innovation Guidance Project (No. 2024YLZDJH107); Henan Province Science and Technology Research Project (No. 252102311084); Ningxia Key Clinical Pathogenic Microorganisms Laboratory Open Research Project (No. MKLG-2024-06).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aamir F., Aslam I., Arshad M., Omer H. (2023). Accelerated diffusion-weighted MR image reconstruction using deep neural networks. J. Digit. Imaging 36 (1), 276–288. doi:10.1007/s10278-022-00709-5
Abirami S., Ramesh K., Lalitha VaniSree K. (2025). Classification and pixel change detection of brain tumor using adam kookaburra optimization-based shepard convolutional neural network. NMR Biomed. 38 (2), e5307. doi:10.1002/nbm.5307
Alshayeji M., Al-Roomi S. A., Abed S. (2017). Optic disc detection in retinal fundus images using gravitational law-based edge detection. Med. Biol. Eng. Comput. 55 (6), 935–948. doi:10.1007/s11517-016-1563-0
Alshomrani S., Arif M., Al Ghamdi M. A. (2023). SAA-UNet: spatial attention and attention gate UNet for COVID-19 pneumonia segmentation from computed tomography. Diagn. (Basel) 13 (9), 1658. doi:10.3390/diagnostics13091658
Avesta A., Hui Y., Aboian M., Duncan J., Krumholz H. M., Aneja S. (2023). 3D capsule networks for brain image segmentation. AJNR Am. J. Neuroradiol. 44 (5), 562–568. doi:10.3174/ajnr.A7845
Azeez T. A. (2023). Osteoporosis and cardiovascular disease: a review. Mol. Biol. Rep. 50 (2), 1753–1763. doi:10.1007/s11033-022-08088-4
Bayoumi E., Karasik P. (2021). Cardiovascular disease in older women. Clin. Geriatr. Med. 37 (4), 651–665. doi:10.1016/j.cger.2021.05.010
Chen K. B., Xuan Y., Lin A. J., Guo S. H. (2021). Lung computed tomography image segmentation based on U-Net network fused with dilated convolution. Comput. Methods Programs Biomed. 207, 106170. doi:10.1016/j.cmpb.2021.106170
Dong P., Zhang R., Li J., Liu C., Liu W., Hu J., et al. (2024). An ultrasound image segmentation method for thyroid nodules based on dual-path attention mechanism-enhanced UNet+. BMC Med. Imaging 24 (1), 341. doi:10.1186/s12880-024-01521-z
Flannery S. W., Kiapour A. M., Edgar D. J., Murray M. M., Fleming B. C. (2021). Automated magnetic resonance image segmentation of the anterior cruciate ligament. J. Orthop. Res. 39 (4), 831–840. doi:10.1002/jor.24926
Goldsborough E., Osuji N., Blaha M. J. (2022). Assessment of cardiovascular disease risk: a 2022 update. Endocrinol. Metab. Clin. North Am. 51 (3), 483–509. doi:10.1016/j.ecl.2022.02.005
Hou C., Li M. X., He W. (2024). Carotid Plaque-RADS: a novel stroke risk classification system. JACC Cardiovasc Imaging 17 (2), 226. doi:10.1016/j.jcmg.2023.11.007
Huang Q. H., Tian H. Z., Jia L. Z., Li Z. M., Zhou Z. S. (2023). A review of deep learning segmentation methods for carotid artery ultrasound images. Neurocomputing 545, 126298. doi:10.1016/j.neucom.2023.126298
Ihle-Hansen H., Vigen T., Berge T., Walle-Hansen M. M., Hagberg G., Ihle-Hansen H., et al. (2023). Carotid plaque score for stroke and cardiovascular risk prediction in a middle-aged cohort from the general population. J. Am. Heart Assoc. 12 (17), e030739. doi:10.1161/jaha.123.030739
Jain P. K., Sharma N., Giannopoulos A. A., Saba L., Nicolaides A., Suri J. S. (2021). Hybrid deep learning segmentation models for atherosclerotic plaque in internal carotid artery B-mode ultrasound. Comput. Biol. Med. 136, 104721. doi:10.1016/j.compbiomed.2021.104721
Jain P. K., Sharma N., Kalra M. K., Johri A., Saba L., Suri J. S. (2022). Far wall plaque segmentation and area measurement in common and internal carotid artery ultrasound using U-series architectures: an unseen artificial intelligence paradigm for stroke risk assessment. Comput. Biol. Med. 149, 106017. doi:10.1016/j.compbiomed.2022.106017
Li Y., Zou L., Xiong L., Yu F., Jiang H., Fan C., et al. (2022). FRDD-Net: automated carotid plaque ultrasound images segmentation using feature remapping and dense decoding. Sensors (Basel) 22 (3), 887. doi:10.3390/s22030887
Liapi G. D., Kyriacou E., Loizou C. P., Panayides A. S., Pattichis C. S., Nicolaides A. N. (2022). “Deep learning-based segmentation of the atherosclerotic carotid plaque in ultrasonic images,” in Paper presented at the artificial intelligence applications and innovations. AIAI 2022 IFIP WG 12.5 international workshops. Cham.
Luo Y., Huang W., Zeng K., Zhang C., Yu C., Wu W. (2021). Intelligent noise reduction algorithm to evaluate the correlation between human fat deposits and uterine fibroids under ultrasound imaging. J. Healthc. Eng. 2021, 5390219. doi:10.1155/2021/5390219
Meshram N. H., Mitchell C. C., Wilbrand S., Dempsey R. J., Varghese T. (2020). Deep learning for carotid plaque segmentation using a dilated U-Net architecture. Ultrason. Imaging 42 (4-5), 221–230. doi:10.1177/0161734620951216
Mi S., Bao Q., Wei Z., Xu F., Yang W. (2021). “MBFF-Net: multi-branch feature fusion network for carotid plaque segmentation in ultrasound,” in Paper presented at the medical image computing and computer assisted intervention – Miccai 2021. Cham.
Miao M., Zhou G., Bao A., Sun Y., Du H., Song L., et al. (2022). Triglyceride-glucose index and common carotid artery intima-media thickness in patients with ischemic stroke. Cardiovasc Diabetol. 21 (1), 43. doi:10.1186/s12933-022-01472-1
Piao C., Lv M., Wang S., Zhou R., Wang Y., Wei J., et al. (2022). Multi-objective data enhancement for deep learning-based ultrasound analysis. BMC Bioinforma. 23 (1), 438. doi:10.1186/s12859-022-04985-4
Sheng M., Xu W., Yang J., Chen Z. (2022). Cross-attention and deep supervision UNet for lesion segmentation of chronic stroke. Front. Neurosci. 16, 836412. doi:10.3389/fnins.2022.836412
Singh H., Ahmed A. S., Melandsø F., Habib A. (2023). Ultrasonic image denoising using machine learning in point contact excitation and detection method. Ultrasonics 127, 106834. doi:10.1016/j.ultras.2022.106834
Tsantis S., Spiliopoulos S., Skouroliakou A., Karnabatidis D., Hazle J. D., Kagadis G. C. (2014). Multiresolution edge detection using enhanced fuzzy c-means clustering for ultrasound image speckle reduction. Med. Phys. 41 (7), 072903. doi:10.1118/1.4883815
Tseng W., Liu H., Yang Y., Liu C., Furutani K., Beltran C., et al. (2023). Performance assessment of variant UNet-based deep-learning dose engines for MR-Linac-based prostate IMRT plans. Phys. Med. Biol. 68 (17), 175004. doi:10.1088/1361-6560/aceb2c
van Dam-Nolen D. H. K., Truijman M. T. B., van der Kolk A. G., Liem M. I., Schreuder F., Boersma E., et al. (2022). Carotid plaque characteristics predict recurrent ischemic stroke and TIA: the PARISK (plaque at RISK) study. JACC Cardiovasc Imaging 15 (10), 1715–1726. doi:10.1016/j.jcmg.2022.04.003
Wang H., Minnema J., Batenburg K. J., Forouzanfar T., Hu F. J., Wu G. (2021). Multiclass CBCT image segmentation for orthodontics with deep learning. J. Dent. Res. 100 (9), 943–949. doi:10.1177/00220345211005338
Xie M., Li Y., Xue Y., Huntress L., Beckerman W., Rahimi S. A., et al. (2020). “Two-stage and dual-decoder convolutional U-Net ensembles for reliable vessel and plaque segmentation in carotid ultrasound images,” in Paper presented at the 2020 19th IEEE international conference on machine learning and applications (ICMLA).
Yan S., Liu R., Zhang Y., Yao X., Yang Y., Wang Q., et al. (2024). Investigation and application of data balancing and combined discriminant model in rock burst severity prediction. Sci. Rep. 14 (1), 29657. doi:10.1038/s41598-024-81307-z
Yi X., Wang J., Wu P., Wang G., Mo L., Lou X., et al. (2023). AC-UNet: an improved UNet-based method for stem and leaf segmentation in Betula luminifera. Front. Plant Sci. 14, 1268098. doi:10.3389/fpls.2023.1268098
Zheng Y., Zhou Y., Zhou H., Gong X. (2015). Ultrasound image edge detection based on a novel multiplicative gradient and canny operator. Ultrason. Imaging 37 (3), 238–250. doi:10.1177/0161734614554461
Keywords: carotid plaque, semantic segmentation, carotid ultrasound, image analysis, deep learning
Citation: Deng L, Duan X, Sun Y, Wang Y, Song D and Duan X (2025) DualPlaqueNet with dual-branch structure and attention mechanism for carotid plaque semantic segmentation and size prediction. Front. Physiol. 16:1629637. doi: 10.3389/fphys.2025.1629637
Received: 20 May 2025; Accepted: 03 July 2025;
Published: 15 July 2025.
Edited by:
Dalin Tang, Worcester Polytechnic Institute, United StatesReviewed by:
Xiaoya Guo, Nanjing University of Posts and Telecommunications, ChinaRongpu Cui, Tsinghua University, China
Dongwei Wang, Zhengzhou Central Hospital, China
Copyright © 2025 Deng, Duan, Sun, Wang, Song and Duan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaokai Duan, YTI1NTgwNDAyMjFAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Lili Deng
Lili Deng Xingyu Duan
Xingyu Duan Yongxiang Sun1
Yongxiang Sun1 Xiaokai Duan
Xiaokai Duan