- 1Department of Spine and Spinal Cord Surgery, Henan Provincial People’s Hospital, Zhengzhou, Henan, China
- 2Department of Medical Imaging, The Third Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan, China
Purpose: Spinal X-ray image segmentation faces several challenges, such as complex anatomical structures, large variations in scale, and blurry or low-contrast boundaries between vertebrae and surrounding tissues. These factors make it difficult for traditional models to achieve accurate and robust segmentation. To address these issues, this study proposes MDWC-Net, a novel deep learning framework designed to improve the accuracy and efficiency of spinal structure identification in clinical settings.
Methods: MDWC-Net adopts an encoder–decoder architecture and introduces three modules—MSCAW, DFCB, and BIEB—to address key challenges in spinal X-ray image segmentation. The network is trained and evaluated on the Spine Dataset, which contains 280 X-ray images provided by Henan Provincial People’s Hospital and is randomly divided into training, validation, and test sets with a 7:1:2 ratio. In addition, to evaluate the model’s generalizability, further validation was conducted on the Chest X-ray dataset for lung field segmentation and the ISIC2016 dataset for melanoma boundary delineation.
Results: MDWC-Net outperformed other mainstream models overall. On the Spine Dataset, it achieved a Dice score of 89.86% ± 0.356, MIoU of 90.53% ± 0.315, GPA of 96.82% ± 0.289, and Sensitivity of 96.77% ± 0.212. A series of ablation experiments further confirmed the effectiveness of the MSCAW, DFCB, and BIEB modules.
Conclusion: MDWC-Net delivers accurate and efficient segmentation of spinal structures, showing strong potential for integration into clinical workflows. Its high performance and generalizability suggest broad applicability to other medical image segmentation tasks.
1 Introduction
The spine, as the structural support for the body and its organs, can develop deformities, cause back pain, or even lead to paralysis when affected by disease (Khalifeh et al., 2024). Accurate spinal segmentation plays a crucial role in the diagnosis and treatment of spinal disorders. It not only enables clinicians to more precisely locate and identify spinal structures, but also provides the foundation for measuring key spinal parameters, spinal registration, and scoliosis classification (Liebmann et al., 2024; Azampour et al., 2024; Sarwahi et al., 2021; Thibodeau-Antonacci et al., 2025; Kim et al., 2022). Such technology is essential for evaluating disease severity, monitoring progression, and planning surgical interventions. Among the various imaging modalities, X-ray technology has become a commonly used clinical tool for spinal disease diagnosis due to its advantages of low radiation exposure, rapid imaging, and cost-effectiveness (Zhang et al., 2020). Consequently, developing automatic segmentation algorithms tailored for spinal X-ray images holds substantial clinical value. In recent years, deep learning—an emerging branch of artificial intelligence—has achieved remarkable progress in image classification, semantic segmentation, and object detection, by learning high-level representations from data (Duan et al., 2025; J. Chen et al., 2025; Zhou et al., 2023; Gui et al., 2024). These advancements offer innovative solutions for accurate segmentation in spinal medical imaging. However, current deep learning-based methods for spinal X-ray image segmentation still face several limitations. First, spinal structures often exhibit complex multi-scale characteristics, and existing methods struggle to capture features at different scales effectively, resulting in suboptimal segmentation performance and loss of fine details. Additionally, many networks suffer from inadequate feature fusion mechanisms, which leads to redundancy and information loss. To address the above challenges, we propose a novel deep learning framework, the Multi-Scale Dynamic-Weighting Context Network (MDWC-Net), for spinal X-ray image segmentation. The network is designed to enhance the extraction of anatomical details across multiple scales, improve the fusion between low- and high-level features, and strengthen the modeling of global contextual information. The main contributions of this work are as follows:
1. A specialized segmentation framework tailored for spinal X-ray images is developed, aiming to provide a reliable and efficient tool to support automatic diagnosis and quantitative analysis in clinical settings.
2. Effective modules are designed to improve multi-scale representation, contextual awareness, and feature interaction within the encoder–decoder architecture.
3. Extensive experiments on spinal and cross-modality datasets demonstrate the superior performance, efficiency, and generalization ability of our proposed method.
2 Materials and methods
2.1 Application of deep learning techniques in medical image segmentation
In recent years, deep learning techniques have made significant progress in various fields. Deep learning automatically extracts features from data through multi-layer neural networks, eliminating the complex process of traditional feature engineering (Talaei Khoei et al., 2023). Medical image segmentation, as one of the key tasks in medical image processing, aims to separate the region of interest from the background in images, helping clinicians with disease diagnosis and treatment. Fully Convolutional Networks (FCNs) are among the earliest deep learning models to achieve significant progress in medical image segmentation tasks (Wang et al., 2022). By replacing the fully connected layers in traditional convolutional neural networks with convolutional layers, FCNs are capable of performing pixel-level classification on input images of any size. FCN-8s is a variant of FCN that fuses feature information from different layers to enhance segmentation accuracy. The DeepLab series is another classic segmentation network model, including DeepLabV1, DeepLabV2, and DeepLabV3 (Yang et al., 2024; Jeong et al., 2024; L. C. Chen et al., 2018). DeepLabV1 extends the receptive field of convolutions through dilated convolutions, effectively improving the segmentation capability for medical images with complex backgrounds or unclear edges. DeepLabV2 builds upon this by incorporating Conditional Random Fields (CRFs) for post-processing, refining the segmentation results. DeepLabV3 further improves the dilated convolution and combines it with an encoder-decoder architecture, enabling it to handle more complex medical image segmentation tasks.
Compared to the classic models mentioned above, U-Net, which is currently the most widely used model in the field of medical image segmentation, was first introduced in 2015 (Falk et al., 2019). It adopts an encoder-decoder architecture and combines low-level features with high-level features through skip connections, preserving the spatial information of the image. Due to its superior segmentation performance, U-Net has also been introduced into industrial fields such as defect detection (Xia et al., 2023; Tulbure et al., 2022) and remote sensing image segmentation (Bai et al., 2023; J. Li et al., 2022). With the deepening of research, more and more scholars have proposed various improvements to address the shortcomings of U-Net, resulting in numerous variant models (Das and Das, 2024; Zhou et al., 2024; Tang et al., 2024; Jisna et al., 2024; Chen et al., 2024; Li et al., 2023) for different segmentation tasks. These network models are widely applied to segmentation tasks in medical images such as those of the heart, liver, blood vessels, and cells (Carneiro et al., 2012; Khan et al., 2022; Gegundez-Arias et al., 2021; Greenwald et al., 2022; Le, 2023). Zhao et al. (2021) introduced a multi-scale up-sampling attention block to enhance feature representation and adopted a nested skip-connection pyramid architecture for feature extraction, applying it to the retinal vessel segmentation task. Li et al. (2023) integrated an attention context encoding module and dual segmentation branches, improving liver segmentation accuracy while keeping the parameter count reasonable. Zhu et al. (2023) used the Swin Transformer framework to extract semantic features and introduced a shift-block labeling strategy during training to achieve more precise brain tumor segmentation. Zhao et al. (2022) focused on two main aspects—sequence encoding and variational information bottlenecks—and proposed an improved model based on different deep learning architectures for peptide toxicity prediction. Although transformer-based models like Swin Transformer have achieved success in brain tumor and peptide segmentation, their application to spinal X-ray segmentation remains limited due to the modality’s lower contrast and structural complexity.
2.2 Application of deep learning techniques in spinal image segmentation tasks
The segmentation of spinal images aims to assist doctors in better understanding the patient’s condition. H. Li et al. (2021) improved model accuracy by embedding a dual-branch multi-scale attention module. This method achieves the segmentation of vertebrae, laminae, and the dural sac from lumbar MRI images, thereby providing assistance in diagnosing lumbar spinal stenosis. Shi et al. (2022) designed a novel dual-path network based on an attention gate (AGNet). This model consists of a context path and an edge path, aiming to extract semantic and boundary information from the spinal and vertebral regions. A multi-scale supervision mechanism is employed to explore comprehensive features, and an edge-aware fusion mechanism is used to combine the features extracted from both paths, enhancing segmentation performance. Chen et al. (2024) combined U-Net and Mask R-CNN to achieve automatic segmentation and labeling of vertebrae in lateral cervical and lumbar X-ray images, with accuracy improved through rule-based strategies. Deng et al. (2024) proposed a complementary network that integrates the advantages of U-Net and BiseNet for spinal segmentation in MRI images. The network uses strip pooling (SP) blocks to replace the spatial extraction path in the BiseNet framework and employs an attention refinement module to fuse the extracted features, thereby improving segmentation accuracy. While multi-scale and attention-based methods have shown success in MRI and CT segmentation, their direct application to spinal X-rays is limited due to lower soft-tissue contrast, overlapping anatomical structures, and higher noise. Although a few studies have begun exploring such strategies in X-ray contexts, their effectiveness remains constrained. To address the challenges in spinal X-ray image segmentation, we propose MDWC-Net, which integrates task-specific feature weighting and structure-aware fusion strategies. The model enhances both local detail capture and global contextual understanding.
2.3 Overall network architecture
The task of spinal image segmentation often faces numerous challenges, including the complexity of spinal structures, noise interference in images, and the inability to effectively fuse features of different scales. Due to the diverse presentation of the spine in X-ray images and the presence of similar backgrounds, the segmentation process struggles to accurately capture details and boundaries. Furthermore, the lack of effective utilization of features at different scales can lead to a decrease in segmentation accuracy.
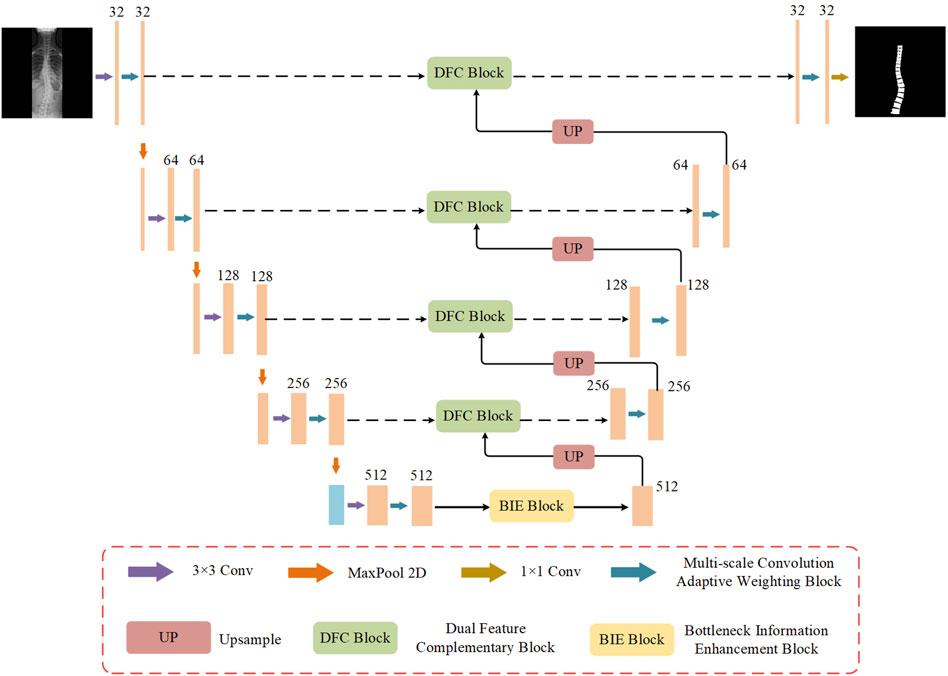
To address these issues, Multi-Scale Dynamic-Weighting Context Network (MDWC-Net) is proposed. As shown in Figure 1, MDWC-Net mainly consists of four parts: the encoder structure, decoder structure, skip connections, and bottleneck structure. MDWC-Net utilizes a multi-scale convolutional adaptive weighting block to perform feature extraction and target reconstruction. By jointly learning different channels of multi-scale feature maps, it dynamically adjusts the importance of different regions. The developed dual feature complementarity block enables effective fusion of high-level semantic information from the encoder structure and low-level spatial information from the decoder structure, enhancing the network’s ability to capture spinal detail information. Furthermore, a bottleneck information enhancement block is designed at the bottleneck layer of the network, allowing the network to more fully capture and utilize global contextual information, thereby strengthening the representation of key information.
2.4 Multi-scale convolution adaptive weighting block
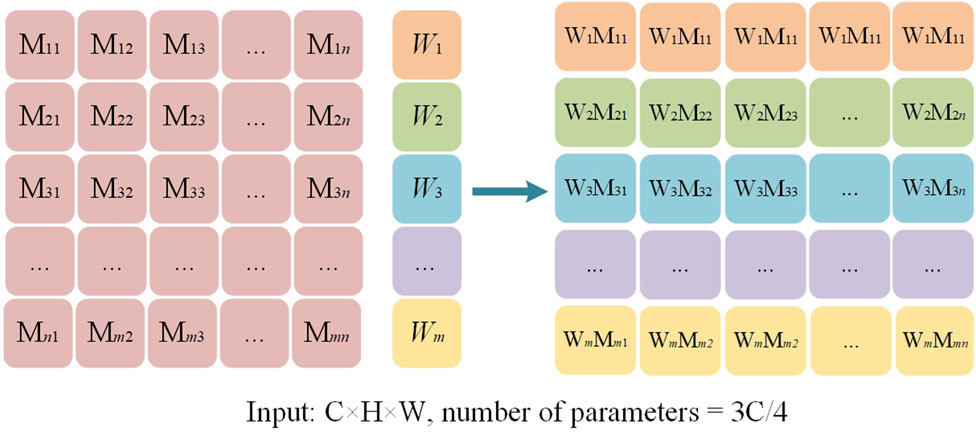
Figure 2 shows the structure of the multi-scale convolutional adaptive weighting block. Different scales of convolutional kernels are selected to extract multi-scale information from the input image, addressing the issue of insufficient multi-scale detail extraction capability in spinal image segmentation tasks. At the same time, an adaptive weighting block is constructed to dynamically adjust channel weights based on the feature information of different input images. Through the construction of multi-scale depthwise separable convolutions and the adaptive weighting block, the model is enabled to efficiently and thoroughly extract multi-scale features from the image while dynamically adjusting the weights of different features based on the input. This enables more accurate segmentation of the regions of interest in the spine.
Among them, the multi-scale convolution operation is the core method for achieving multi-scale feature extraction. To capture information at different spatial scales, we designed multiple convolution kernels of different sizes (1 × 1, 3 × 3, 5 × 5, 7 × 7) and performed the computations using depthwise separable convolutions. Unlike the fully connected convolutions in traditional convolutional neural networks, depthwise separable convolutions break down the convolution operation into two steps: first, performing a channel-wise convolution on each individual channel, and then performing a pointwise convolution across channels. This strategy not only effectively reduces computational complexity but also enables the simultaneous capture of small-scale features at the fine detail level and large-scale features at the global level. It provides a rich feature foundation for subsequent feature fusion and dynamic weighting.
To further optimize the utilization of features, this study designs an adaptive weighting block based on the extraction of multi-scale features. The block contains learnable weight parameters, which are optimized during the training process according to the specific feature requirements of the image. By dynamically adjusting the weights of features at different scales, MDWC-Net can more flexibly adapt to different input features, thus achieving higher accuracy in the spine region of interest segmentation task. The weighting process is shown in Figure 3, where the feature extraction with convolution kernels of sizes 1 × 1, 3 × 3, 5 × 5, and 7 × 7 corresponds to the dynamic weighted feature vectors M, N, R, and Q, respectively. Using a parallel approach, M, N, R, and Q are merged to form the multi-scale convolution adaptive weighting block.
For the adaptive weighting layer shown in Figure 3, the weight
Where L represents the loss function, and
The weight update can be expressed as (Equation 2):
Where l represents the number of parameter updates,
The entire module computation process is as follows (Equations 3–6):
Where
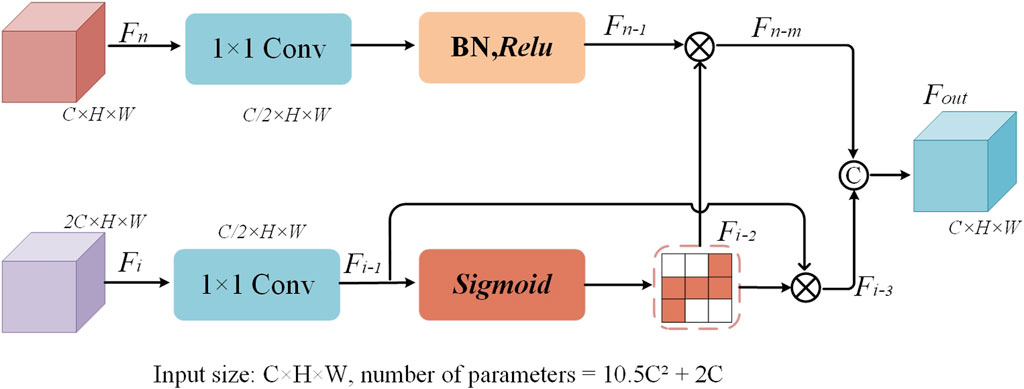
2.5 Dual feature complementary block
The traditional U-Net uses skip connections that directly concatenate the feature maps from the encoder and decoder parts to recover lost spatial details during the decoding process. However, this approach often leads to insufficient information fusion, especially when it comes to recovering fine details. To address this issue, an innovative dual feature complementary block has been designed, which optimizes the traditional skip connection method through a series of processing steps. This enables the network to more effectively complement and fuse the feature maps from the encoder and decoder. The structure of the dual feature complementary block is shown in Figure 4.
The core idea of the dual feature complementary block is to enhance the interaction and information transfer between feature maps by progressively optimizing the feature map fusion process. Specifically, the dual feature complementary block independently processes each feature map from the skip connections, including operations such as convolution for dimensionality reduction, batch normalization, and nonlinear activation. Then, pixel-wise multiplication is applied to enhance the mutual influence between feature maps. Finally, the results of both feature maps are fused and concatenated to form a more refined feature map representation. When processing the encoder feature map, the first step is to reduce the number of channels of feature map
Where M, N, and P represent the 1 × 1 convolution operation, the Relu activation function, and the exponential function, respectively.
When processing the decoder feature map
Where
After independently processing the feature maps, the dual feature complementary block performs a pixel-wise multiplication operation between the processed encoder feature map
Where
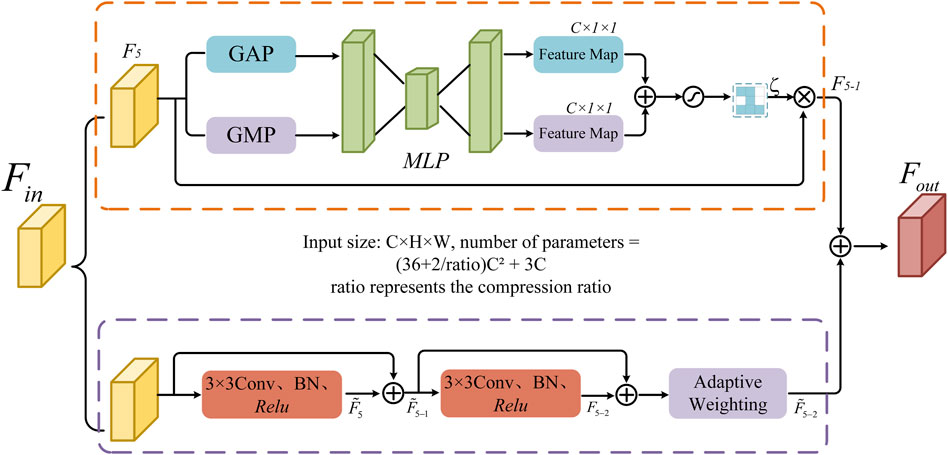
2.6 Bottleneck information enhancement block
Convolutional neural networks often encounter issues such as feature information loss and insufficient global information capture when processing high-dimensional feature maps. To address this problem, this study designs a bottleneck information enhancement block, as shown in Figure 5. This block is constructed using parallel dual branches.
The upper branch first applies global average pooling and global max pooling operations to capture global average information and extract the maximum value from each local region. These operations provide a smooth representation of the entire image’s features and help capture prominent local features. The spatial dimensions of feature map
Where
The lower branch utilizes two consecutive 3 × 3 convolutional layers to extract feature information. The output fused after the first convolution is fed into the second convolutional layer. Through residual connections, more information is retained during feature propagation, which helps avoid information loss in deep networks and facilitates the stable transmission of information flow. Subsequently, an adaptive weighting mechanism is introduced to enhance the key information in the feature map, resulting in feature map
where
Finally, the output results from the upper and lower branches are fused to obtain the final output feature map
Where,
3 Experiment and results
3.1 Dataset
The spine X-ray dataset used in this study was provided by the Department of Spine Surgery at Henan Provincial People’s Hospital, consisting of 280 PNG-format images. Prior to the study, all images underwent de-identification processing to protect patient privacy and were named the “Spine Dataset.” The spine regions in the images were precisely annotated by two spine surgeons using the Labelme annotation tool. To ensure annotation consistency, two spine surgeons jointly defined annotation standards and performed quality cross-checks on randomly selected samples, confirming a high level of agreement. The annotated dataset was randomly divided into training, validation, and test sets in a 7:1:2 ratio. In addition, to validate the generalization ability of MDWC-Net across different segmentation tasks, experiments were conducted using the ISIC 2016 (Gutman et al., 2016) dataset for skin lesion segmentation and the Chest X-ray dataset (Jaeger et al., 2014; Candemir et al., 2014) for lung field segmentation. Both datasets were randomly split into training, validation, and test sets using a 7:1:2 ratio. All images were uniformly resized to 256 × 256 pixels according to a proportional scaling principle.
3.2 Experiment setup and evaluation metrics
3.2.1 Experiment setup
All models in this study were implemented using the PyTorch deep learning framework and Python 3.7, with computations performed on an NVIDIA RTX 2080Ti GPU. The batch size was set to 8, and the cross-entropy loss function was adopted. Random horizontal flipping was applied as a data augmentation technique to the training dataset. The SGD optimizer was used for model training, with an initial learning rate of 0.001. The total number of training epochs was set to 100, and the learning rate was reduced by a factor of 10 every 20 epochs.
3.2.2 Evaluation metrics
To better assess the network performance, this study employs four commonly used image segmentation evaluation metrics: Global Pixel Accuracy (GPA), Dice Coefficient (Dice), Mean Intersection over Union (MIoU), and Sensitivity. Global Pixel Accuracy measures the proportion of correctly classified pixels overall. The Dice Similarity Coefficient provides a comprehensive evaluation of the overlap between the segmentation results and the ground truth labels. Mean Intersection over Union considers the degree of overlap between the predicted and ground truth labels for each class, while Sensitivity reflects the model’s ability to recognize positive class regions. In addition, this study also reports the number of parameters (Params), floating-point operations (FLOPs), and Training_time for each model to evaluate their computational efficiency. The specific expressions for these metrics are as follows (Equations 20–23):
Where TP refers to true positives, FP represents false positives, TN denotes true negatives, FN refers to false negatives, and m stands for the total number of different classes.
3.3 Experimental results
In this study, MDWC-Net was compared with other deep learning-based segmentation algorithms, including FCN-8S, DeeplabV3+ (Baban and Chaari, 2023), PSPNet (H. S. Zhao et al., 2017), U-Net, ResU-Net (Tang et al., 2024), Attention U-Net (Falk et al., 2019), TransU-Net (Chen et al., 2024), and PLU-Net (Song et al., 2024), through extensive experiments on the Spine Dataset. The experimental results were thoroughly analyzed, and a series of ablation experiments were conducted on the proposed blocks to validate their effectiveness.
3.3.1 Experimental results on the spine dataset
The performance of MDWC-Net on the Spine Dataset is shown in Table 1. From the experimental results in Table 1, it can be observed that MDWC-Net demonstrates superior performance across multiple evaluation metrics. Specifically, the Dice and MIoU scores of MDWC-Net reach 89.86% and 90.53%, respectively. The Dice score is 7.58, 4.34, and 3.58 percentage points higher than those of FCN-8S, DeeplabV3+, and U-Net, indicating that MDWC-Net exhibits a stronger capability in distinguishing between classes. Furthermore, compared to the second-best model in Table 1, MDWC-Net also demonstrates its superiority, achieving a 1.65% and 2.71% increase in Dice and MIoU, respectively. In terms of two key metrics, GPA and Sensitivity, MDWC-Net also demonstrates remarkable performance, achieving excellent scores of 96.82% and 96.77%, respectively. Moreover, MDWC-Net achieves these results with fewer Params, lower FLOPs, and shorter Training_time. This fully demonstrates the efficiency and accuracy of MDWC-Net in segmentation tasks. The experimental data indicate that MDWC-Net not only identifies target regions more accurately in the spine segmentation task but also delineates the edges of the targets more precisely, effectively reducing instances of missed and incorrect segmentations.
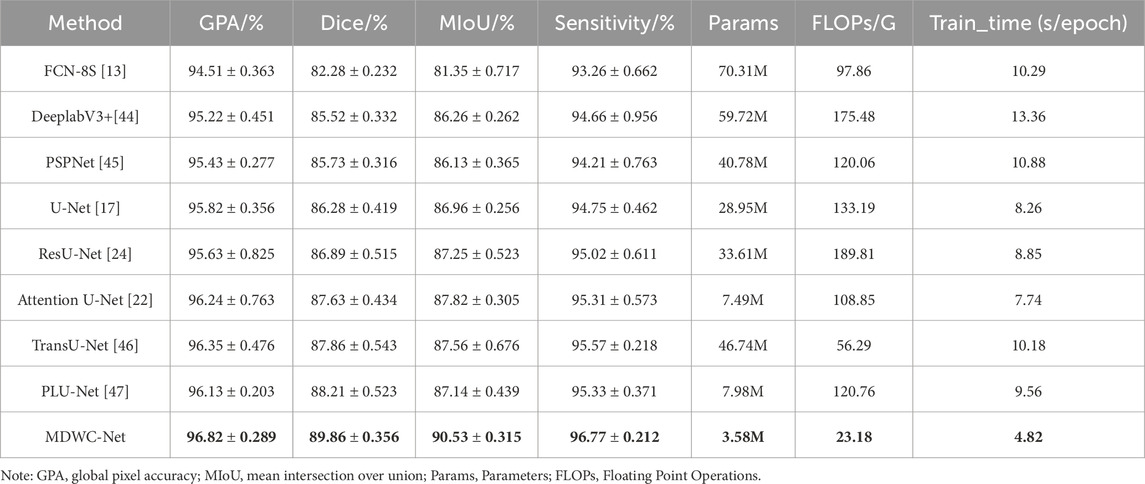
Table 1. Quantitative performance of different models on spine dataset (highest score indicated in bold font).
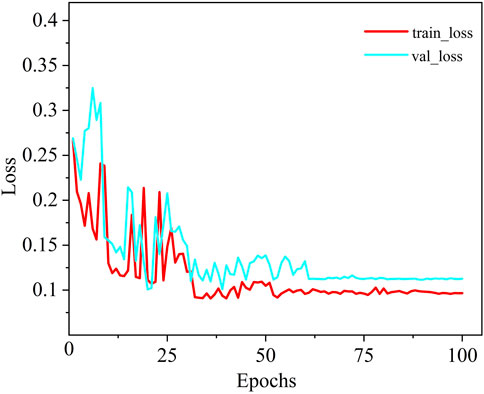
As shown in Figure 6, a comparison of the training loss convergence and Dice scores on the test set for each model is presented. After 100 training epochs, the MDWC-Net model demonstrates a more stable and efficient convergence speed. Moreover, as the number of iterations increases, the efficient convergence of MDWC-Net further proves that effective parameter optimization can be achieved even without pre-training. This achievement is attributed to its network design, which integrates multi-scale and information-complementary feature representations. Figure 7 shows the detailed training and validation curves of MDWC-Net. The training loss decreases steadily from approximately 0.2667 to 0.096, while the validation loss follows a similar trend, stabilizing around 0.112 after 61 epochs. The close alignment between training and validation curves indicates that MDWC-Net achieves good generalization without significant overfitting issues.
Additionally, to visually assess the accuracy of spine region segmentation, Figure 8 presents a comparative visualization of the segmentation results from different algorithms on the Spine Dataset. As shown in Figure 8, more accurate segmentation results are achieved by MDWC-Net. Specifically, other algorithms generally exhibit significant loss of the spine region in the segmentation output, particularly at the edges and finer structures of the spine. This issue is primarily attributed to the limitations of these algorithms in capturing spine location information beyond the receptive field and in handling finer details within the images.
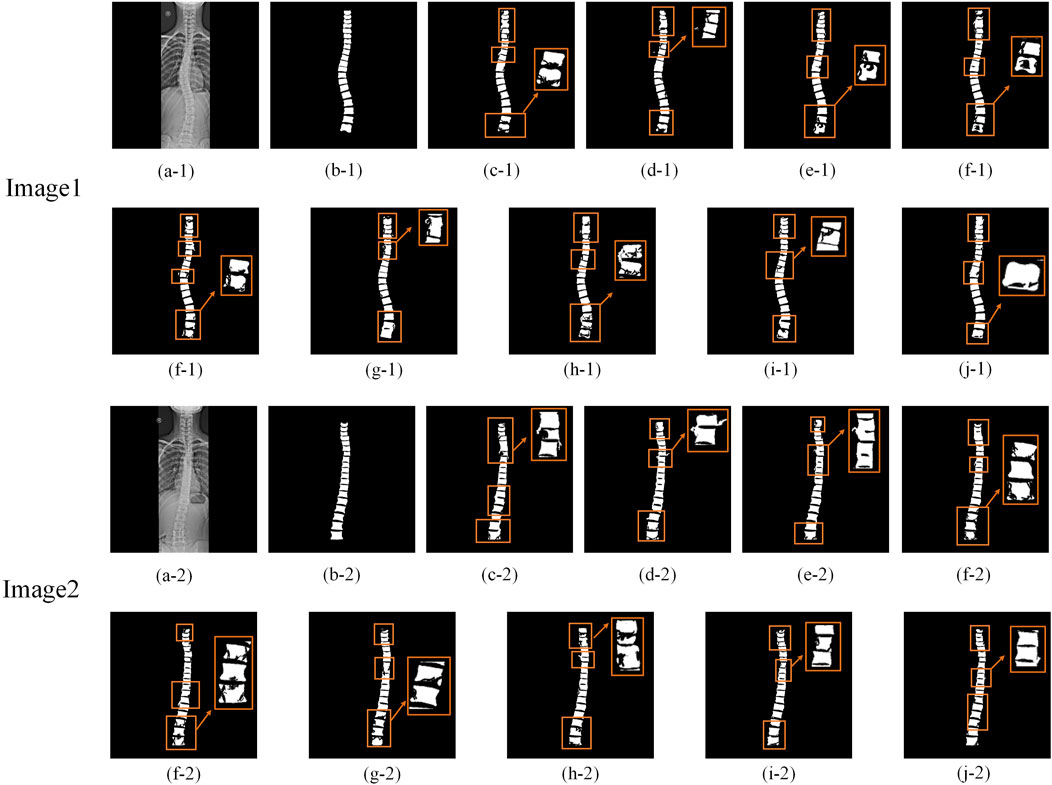
Figure 8. Comparison of segmentation results. The rectangular boxes highlight significant segmentation differences among models. (a) Original image; (b) Ground Truth (GT); (c–j) Segmentation results of FCN-8s, DeepLabV3+, PSPNet, U-Net, ResU-Net, Attention U-Net, TransU-Net, PLU-Net, and MDWC-Net, respectively, on the Spine dataset.
In contrast, the MDWC-Net algorithm effectively alleviates this problem by integrating complementary high-level semantic features and low-level texture features, resulting in more refined and accurate spine region segmentation. In Figure 8, compared to MDWC-Net, other algorithms demonstrate more pronounced over-segmentation and under-segmentation issues during the segmentation process. Over-segmentation occurs when unnecessary details are incorrectly labeled as part of the spine region, while under-segmentation leads to the omission of key spinal structures. The network’s ability to focus on relevant features and utilize global information is enhanced by MDWC-Net through the innovative introduction of a multi-scale adaptive weighting block, a dual-feature complementary block, and a bottleneck information enhancement block. As a result, MDWC-Net outperforms other algorithms in terms of both segmentation accuracy and completeness, demonstrating exceptional performance in the spine X-ray image segmentation task.
3.3.2 Significance testing of segmentation performance
To verify whether the performance improvement of the proposed MDWC-Net over other models (such as DeeplabV3+, PSPNet, U-Net, Attention U-Net, TransU-Net, and PLU-Net) is statistically significant, we conducted paired t-tests on the Dice and MIoU metrics across these models on the Spine dataset. The significance level was set to α = 0.05. As shown in Table 2, all p-values are much smaller than 0.01, indicating that the improvements of MDWC-Net in both Dice and MIoU are statistically significant. These results further demonstrate the effectiveness and robustness of the proposed method.
3.3.3 Ablation experiment
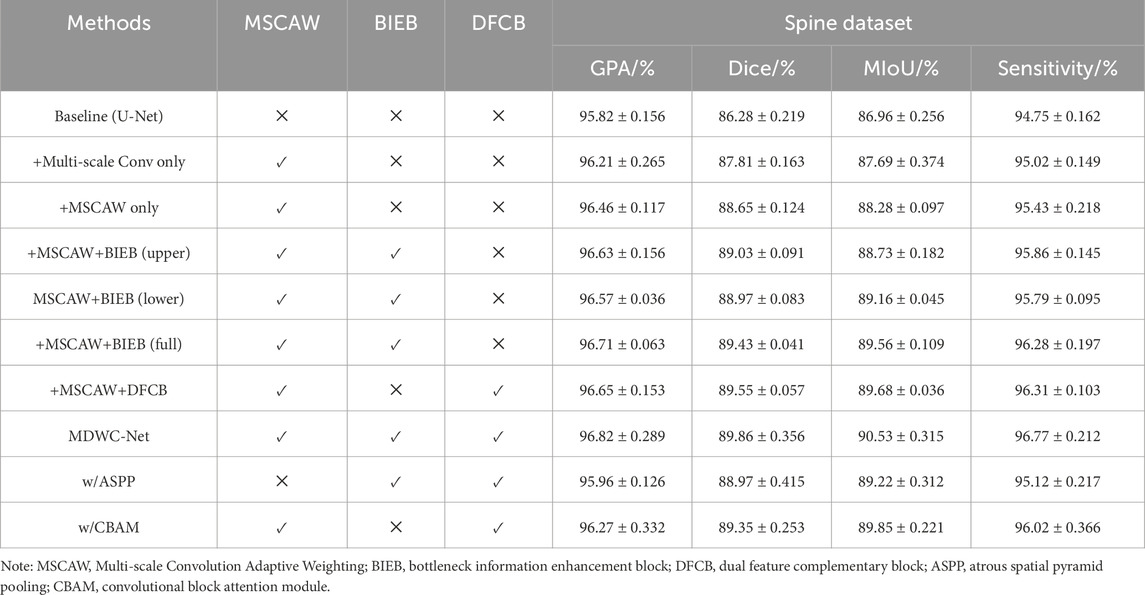
To further explore the contribution of each block in this study, we conducted a series of ablation experiments using the Spine Dataset. First, the U-Net model was used as the baseline. Then, the multi-scale convolution block with the adaptive weighting mechanism removed was integrated into the baseline, referred to as “+Multi-scale Conv only.” The complete Multi-scale Convolution Adaptive Weighting (MSCAW) block was then integrated into the baseline, named “+MSCAW only.” Next, to verify the effectiveness of different branches in the Bottleneck Information Enhancement Block (BIEB), we tested configurations with “+MSCAW+BIEB (upper)” using only the upper branch of BIEB, and “+MSCAW+BIEB (lower)” using only the lower branch. The “+MSCAW+BIEB (full)” configuration integrates both the MSCAW block and the complete BIEB. Finally, “+MSCAW+DFCB” combines the MSCAW block with the Dual Feature Complementary Block. In addition, to further validate the effectiveness and advantages of the proposed modules, we designed replacement experiments: “w/ASPP” replaces the MSCAW module with the classical ASPP module, and “w/CBAM” replaces the BIEB module with the CBAM module. The relevant experimental results are shown in Table 3.
From the experimental results in Table 3, it can be observed that after integrating the multi-scale convolution adaptive weighting block, the model’s performance was significantly improved, with Dice and MIoU rising to 88.65% and 88.28%, respectively. This improvement is attributed to the key role of this block in feature weighting and decision-making. The block fuses features from different scales and dynamically adjusts the scale weights of each channel based on the regional characteristics of the input image, enabling the model to more accurately capture multi-scale detail information. In addition, the combination of different branches of the Bottleneck Information Enhancement Block with the multi-scale convolution adaptive weighting block led to improvements in the network’s performance across all evaluation metrics. This enhancement is attributed to the bottleneck block’s ability to extract and utilize global contextual feature information.
Furthermore, the integration of the Dual Feature Complementary Block also contributed to the improvement in network performance. Specifically, the embedding of the DFCB resulted in the model’s GPA, Dice, MIoU, and Sensitivity increasing to 96.65%, 89.55%, 89.68%, and 96.31%, respectively. The results show that this block effectively utilizes high-level features to guide low-level features in selecting key information, reducing the loss of important information and interference from irrelevant data. And the proposed modules also outperformed classical counterparts in segmentation performance, further supporting their design rationality and task-specific effectiveness. Finally, through the integration of all blocks, MDWC-Net achieved optimal segmentation performance, and the experimental results strongly validate the effectiveness and practicality of the designed blocks.
4 Discussion
To further evaluate the generalization capability of the proposed MDWC-Net beyond spinal X-ray segmentation, we conducted additional experiments on two publicly available datasets: Chest X-ray dataset for lung field segmentation and ISIC2016 dataset for skin lesion segmentation. These datasets represent two distinct directions of generalization: Chest X-rays are anatomically and radiologically similar to spinal X-rays, while ISIC2016 features highly heterogeneous textures and modalities.
4.1 Experimental results on the chest X-ray dataset
The Chest X-ray dataset provides pixel-level annotations of lung fields. Chest radiographs share similar grayscale distribution and imaging characteristics with spinal X-rays. As shown in Table 4, it can be observed that MDWC-Net achieved a Dice coefficient of 85.32% and MIoU of 86.09%, surpassing baseline methods including U-Net and DeepLabV3+. These results demonstrate that the proposed architecture generalizes effectively not only within the spinal domain but also to other thoracic structures captured by similar imaging modalities. Such findings highlight the potential of MDWC-Net for broader applications in skeletal and soft-tissue segmentation tasks within the domain of radiography.

Table 4. Quantitative performance of each model on the chest X-ray dataset (highest score indicated in bold font, GPA represents Global Pixel Accuracy).
4.2 Experimental results on the ISIC2016 dataset
The ISIC2016 dataset contains various types of skin lesions, including melanoma and basal cell carcinoma, with high-quality pixel-level annotations. Unlike spinal X-rays, skin lesion images exhibit irregular shapes, blurry boundaries, and strong variability in texture and contrast, posing distinct challenges to segmentation models. Applying MDWC-Net to this domain allows us to evaluate its robustness across structurally unrelated medical tasks. As shown in Table 5, MDWC-Net achieved excellent results on the ISIC2016 dataset, with a GPA of 95.96%, Dice coefficient of 87.25%, MIoU of 86.75%, and Sensitivity of 94.61%, outperforming several state-of-the-art models—including a 4.83% and 5.28% improvement in GPA and Sensitivity over FCN-8s, and a 3.28% and 3.33% gain in Dice and MIoU over U-Net. Moreover, compared with the Transformer-based TransUNet, MDWC-Net achieved consistent improvements of 1.58%, 2.12%, and 2.69% in GPA, Dice, and MIoU, respectively.
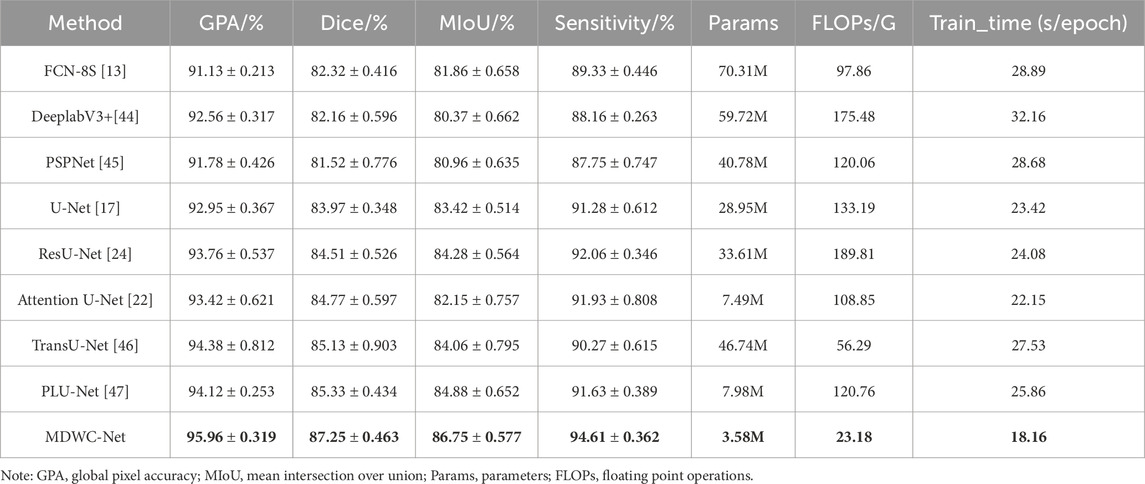
Table 5. Quantitative performance of each model on the ISIC2016 dataset (highest score indicated in bold font, GPA represents Global Pixel Accuracy).
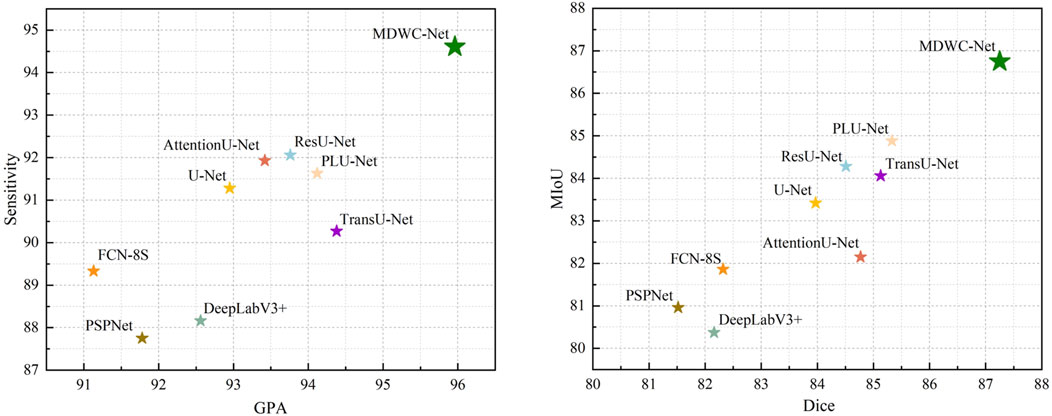
These findings demonstrate that the multi-scale feature modeling and dynamic information fusion mechanisms in MDWC-Net are effective not only for structured anatomical regions like the spine but also for unstructured lesion segmentation tasks. To better visualize model performance, Figure 9 shows that MDWC-Net consistently appears in the top-right region of the GPA–Sensitivity and Dice–MIoU scatter plots, indicating a strong trade-off between accuracy and robustness, and confirming its generalizability across diverse medical image modalities.
5 Conclusion
This study proposes MDWC-Net, an efficient deep network designed for spinal X-ray image segmentation. By incorporating multi-scale convolution adaptive weighting, dual feature complementary block, and bottleneck information enhancement block, the model demonstrates outstanding segmentation performance with strong generalization capability and deployment potential. Although this work primarily focuses on improving segmentation accuracy, its high-quality structural boundary extraction also provides a reliable basis for downstream clinical tasks such as spinal parameter measurement and preoperative path planning. In addition, the lightweight design and low computational cost make it suitable for integration into radiology-assisted reading systems or surgical planning platforms. While MDWC-Net demonstrates robust performance across diverse imaging conditions, further optimization could enhance its effectiveness in extremely challenging scenarios such as very low-contrast or heavily degraded X-ray images. Future work will incorporate clinical user feedback for prospective validation, optimize model deployment through techniques such as model pruning and knowledge distillation, and focus on enhancing robustness under challenging imaging conditions to meet diverse clinical requirements.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by Ethics Committee of the Henan Provincial People’s Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
ZG: Data curation, Formal Analysis, Validation, Visualization, Writing – original draft, Writing – review and editing. XW: Formal Analysis, Methodology, Visualization, Writing – original draft. BC: Funding acquisition, Resources, Supervision, Validation, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This project was financially supported by the Henan Provincial Medical Science and Technology Research Plan Provincial-Ministry Joint Project (Grant no. SBGJ202403002).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Azampour M. F., Tirindelli M., Lameski J., Gafencu M., Tagliabue E., Fatemizadeh E., et al. (2024). Anatomy-aware computed tomography-to-ultrasound spine registration. Med. Phys. 51 (3), 2044–2056. doi:10.1002/mp.16731
Baban A. E. T. R., Chaari L. (2023). Mid-DeepLabv3+: a novel approach for image semantic segmentation applied to African food dietary assessments. Sensors (Basel) 24 (1), 209. doi:10.3390/s24010209
Bai S., Liang J., Long T., Liang C., Zhou J., Ge W., et al. (2023). An efficient approach to detect and track winter flush growth of litchi tree based on UAV remote sensing and semantic segmentation. Front. Plant Sci. 14, 1307492. doi:10.3389/fpls.2023.1307492
Candemir S., Jaeger S., Palaniappan K., Musco J. P., Singh R. K., Zhiyun X., et al. (2014). Lung segmentation in chest radiographs using anatomical atlases with nonrigid registration. IEEE Trans. Med. Imaging 33 (2), 577–590. doi:10.1109/tmi.2013.2290491
Carneiro G., Nascimento J. C., Freitas A. (2012). The segmentation of the left ventricle of the heart from ultrasound data using deep learning architectures and derivative-based search methods. IEEE Trans. Image Process 21 (3), 968–982. doi:10.1109/tip.2011.2169273
Chen L. C., Papandreou G., Kokkinos I., Murphy K., Yuille A. L. (2018). DeepLab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell. 40 (4), 834–848. doi:10.1109/tpami.2017.2699184
Chen S., Luo C., Liu S., Li H., Liu Y., Zhou H., et al. (2024). LD-UNet: a long-distance perceptual model for segmentation of blurred boundaries in medical images. Comput. Biol. Med. 171, 108120. doi:10.1016/j.compbiomed.2024.108120
Chen Y., Mo Y., Readie A., Ligozio G., Mandal I., Jabbar F., et al. (2024). VertXNet: an ensemble method for vertebral body segmentation and identification from cervical and lumbar spinal X-rays. Sci. Rep. 14 (1), 3341. doi:10.1038/s41598-023-49923-3
Chen J., Mei J., Li X., Lu Y., Yu Q., Wei Q., et al. (2024). TransUNet: rethinking the U-Net architecture design for medical image segmentation through the lens of transformers. Med. Image Anal. 97, 103280. doi:10.1016/j.media.2024.103280
Chen J., Liu Y., Wei S., Bian Z., Subramanian S., Carass A., et al. (2025). A survey on deep learning in medical image registration: new technologies, uncertainty, evaluation metrics, and beyond. Med. Image Anal. 100, 103385. doi:10.1016/j.media.2024.103385
Das N., Das S. (2024). Attention-UNet architectures with pretrained backbones for multi-class cardiac MR image segmentation. Curr. Probl. Cardiol. 49 (1 Pt C), 102129. doi:10.1016/j.cpcardiol.2023.102129
Deng Y. J., Gu F., Zeng D. X., Lu J. Y., Liu H. T., Hou Y. L., et al. (2024). An effective U-Net and BiSeNet complementary network for spine segmentation. Biomed. Signal Process. Control 89, 105682. doi:10.1016/j.bspc.2023.105682
Duan X., Ma X., Zhu M., Wang L., You D., Deng L., et al. (2025). Deep learning-assisted screening and diagnosis of scoliosis: segmentation of bare-back images via an attention-enhanced convolutional neural network. J. Orthop. Surg. Res. 20 (1), 161. doi:10.1186/s13018-025-05564-y
Falk T., Mai D., Bensch R., Çiçek Ö., Abdulkadir A., Marrakchi Y., et al. (2019). U-Net: deep learning for cell counting, detection, and morphometry. Nat. Methods 16 (1), 67–70. doi:10.1038/s41592-018-0261-2
Gegundez-Arias M. E., Marin-Santos D., Perez-Borrero I., Vasallo-Vazquez M. J. (2021). A new deep learning method for blood vessel segmentation in retinal images based on convolutional kernels and modified U-Net model. Comput. Methods Programs Biomed. 205, 106081. doi:10.1016/j.cmpb.2021.106081
Greenwald N. F., Miller G., Moen E., Kong A., Kagel A., Dougherty T., et al. (2022). Whole-cell segmentation of tissue images with human-level performance using large-scale data annotation and deep learning. Nat. Biotechnol. 40 (4), 555–565. doi:10.1038/s41587-021-01094-0
Gui S. X., Song S., Qin R. J., Tang Y. (2024). Remote sensing object detection in the deep learning Era-A review. Remote Sens. 16 (2), 327. doi:10.3390/rs16020327
Gutman D., Codella N. C. F., Celebi E., Helba B., Marchetti M., Mishra N., et al. (2016). Skin lesion analysis toward melanoma detection: a challenge at the international symposium on biomedical imaging (ISBI) 2016, hosted by the international skin imaging collaboration (ISIC).
Jaeger S., Karargyris A., Candemir S., Folio L., Siegelman J., Callaghan F., et al. (2014). Automatic tuberculosis screening using chest radiographs. IEEE Trans. Med. Imaging 33 (2), 233–245. doi:10.1109/tmi.2013.2284099
Jeong S., Kim J., Kim S., Min D. (2024). Revisiting domain-adaptive semantic segmentation via knowledge distillation. IEEE Trans. Image Process 33, 6761–6773. doi:10.1109/tip.2024.3501076
Jisna V. A., Ajay A. P., Jayaraj P. B. (2024). Using attention-UNet models to predict protein contact maps. J. Comput. Biol. 31 (7), 691–702. doi:10.1089/cmb.2023.0102
Khalifeh K., Brown N. J., Pennington Z., Pham M. H. (2024). Spinal robotics in adult spinal deformity surgery: a systematic review. Neurospine 21 (1), 20–29. doi:10.14245/ns.2347138.569
Khan R. A., Luo Y., Wu F. X. (2022). RMS-UNet: residual multi-scale UNet for liver and lesion segmentation. Artif. Intell. Med. 124, 102231. doi:10.1016/j.artmed.2021.102231
Kim H. J., Yang J. H., Chang D. G., Lenke L. G., Suh S. W., Nam Y., et al. (2022). Adult spinal deformity: a comprehensive review of current advances and future directions. Asian Spine J. 16 (5), 776–788. doi:10.31616/asj.2022.0376
Le N. Q. K. (2023). Predicting emerging drug interactions using GNNs. Nat. Comput. Sci. 3 (12), 1007–1008. doi:10.1038/s43588-023-00555-7
Li H., Luo H., Huan W., Shi Z., Yan C., Wang L., et al. (2021). Automatic lumbar spinal MRI image segmentation with a multi-scale attention network. Neural Comput. Appl. 33 (18), 11589–11602. doi:10.1007/s00521-021-05856-4
Li J., Wang H., Zhang A., Liu Y. (2022). Semantic segmentation of hyperspectral remote sensing images based on PSE-UNet model. Sensors (Basel) 22 (24), 9678. doi:10.3390/s22249678
Li X., Qin X., Huang C., Lu Y., Cheng J., Wang L., et al. (2023). SUnet: a multi-organ segmentation network based on multiple attention. Comput. Biol. Med. 167, 107596. doi:10.1016/j.compbiomed.2023.107596
Li Y., Zou B., Dai P., Liao M., Bai H. X., Jiao Z. (2023). AC-E network: attentive context-enhanced network for liver segmentation. IEEE J. Biomed. Health Inf. 27 (8), 4052–4061. doi:10.1109/jbhi.2023.3278079
Liebmann F., von Atzigen M., Stütz D., Wolf J., Zingg L., Suter D., et al. (2024). Automatic registration with continuous pose updates for marker-less surgical navigation in spine surgery. Med. Image Anal. 91, 103027. doi:10.1016/j.media.2023.103027
Sarwahi V., Galina J., Atlas A., Gecelter R., Hasan S., Amaral T. D., et al. (2021). Scoliosis surgery normalizes cardiac function in adolescent idiopathic scoliosis patients. Spine (Phila Pa 1976) 46 (21), e1161–e1167. doi:10.1097/brs.0000000000004060
Shi W., Xu T., Yang H., Xi Y., Du Y., Li J., et al. (2022). Attention gate based dual-pathway network for vertebra segmentation of X-ray spine images. IEEE J. Biomed. Health Inf. 26 (8), 3976–3987. doi:10.1109/jbhi.2022.3158968
Song W., Yu H., Wu J. (2024). PLU-Net: extraction of multiscale feature fusion. Med. Phys. 51 (4), 2733–2740. doi:10.1002/mp.16840
Talaei Khoei T., Ould Slimane H., Kaabouch N. (2023). Deep learning: systematic review, models, challenges, and research directions. Neural Comput. Appl. 35 (31), 23103–23124. doi:10.1007/s00521-023-08957-4
Tang Y., Cao Z., Guo N., Jiang M. (2024). A Siamese Swin-Unet for image change detection. Sci. Rep. 14 (1), 4577. doi:10.1038/s41598-024-54096-8
Thibodeau-Antonacci A., Popovic M., Ates O., Hua C. H., Schneider J., Skamene S., et al. (2025). Trade-off of different deep learning-based auto-segmentation approaches for treatment planning of pediatric craniospinal irradiation autocontouring of OARs for pediatric CSI. Med. Phys. 52 (6), 3541–3556. doi:10.1002/mp.17782
Tulbure A. A., Tulbure A. A., Dulf E. H. (2022). A review on modern defect detection models using DCNNs - deep convolutional neural networks. J. Adv. Res. 35, 33–48. doi:10.1016/j.jare.2021.03.015
Wang S., Liu C., Zhang Y. H. (2022). Fully convolution network architecture for steel-beam crack detection in fast-stitching images. Mech. Syst. Signal Process. 165, 108377. doi:10.1016/j.ymssp.2021.108377
Xia Y., Han S. W., Kwon H. J. (2023). Image generation and recognition for railway surface defect detection. Sensors (Basel) 23 (10), 4793. doi:10.3390/s23104793
Yang T., Wei J., Xiao Y., Wang S., Tan J., Niu Y., et al. (2024). LT-DeepLab: an improved DeepLabV3+ cross-scale segmentation algorithm for Zanthoxylum bungeanum Maxim leaf-trunk diseases in real-world environments. Front. Plant Sci. 15, 1423238. doi:10.3389/fpls.2024.1423238
Zhang B., Yu K., Ning Z., Wang K., Dong Y., Liu X., et al. (2020). Deep learning of lumbar spine X-ray for osteopenia and osteoporosis screening: a multicenter retrospective cohort study. Bone 140, 115561. doi:10.1016/j.bone.2020.115561
Zhao H. S., Shi J. P., Qi X. J., Wang X. G., Jia J. Y. (2017). “Pyramid scene parsing network,” in Paper presented at the 30th IEEE/CVF conference on computer vision and pattern recognition (CVPR), Honolulu, HI.
Zhao R. H., Li Q., Wu J. R., You J. N. (2021). A nested U-shape network with multi-scale upsample attention for robust retinal vascular segmentation. Pattern Recognit. 120, 107998. doi:10.1016/j.patcog.2021.107998
Zhao Z., Gui J., Yao A., Le N. Q. K., Chua M. C. H. (2022). Improved prediction model of protein and peptide toxicity by integrating channel attention into a convolutional neural network and gated recurrent units. ACS Omega 7 (44), 40569–40577. doi:10.1021/acsomega.2c05881
Zhou H., Sun C., Huang H., Fan M., Yang X., Zhou L. (2023). Feature-guided attention network for medical image segmentation. Med. Phys. 50 (8), 4871–4886. doi:10.1002/mp.16253
Zhou H., Leung H., Balaji B. (2024). AR-UNet: a deformable image registration network with cyclic training. IEEE/ACM Trans. Comput. Biol. Bioinform 21 (4), 692–700. doi:10.1109/tcbb.2023.3284215
Keywords: convolutional neural networks, spinal image segmentation, multi-scale convolutional adaptive weighting, dual feature complementary block, bottleneck information enhancement block
Citation: Gu Z, Wang X and Chen B (2025) MDWC-Net: a multi-scale dynamic-weighting context network for precise spinal X-ray segmentation. Front. Physiol. 16:1651296. doi: 10.3389/fphys.2025.1651296
Received: 21 June 2025; Accepted: 17 August 2025;
Published: 29 August 2025.
Edited by:
Rajesh Kumar Tripathy, Birla Institute of Technology and Science, IndiaReviewed by:
Nguyen Quoc Khanh Le, Taipei Medical University, TaiwanHongwei Ning, Anhui Science and Technology University, China
Swapnil Singh, MicroStrategy Incorporated, United States
Copyright © 2025 Gu, Wang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Baojun Chen, enp1NjE1QDEyNi5jb20=
 Zhongzheng Gu
Zhongzheng Gu Xuan Wang2
Xuan Wang2