- 1Museum of Natural History and Department of Ecology and Evolutionary Biology, University of Colorado, Boulder, CO, United States
- 2Department of Biological Sciences, College of Science, University of Santo Tomas, Manila, NCR, Philippines
- 3Department of Biology and Annis Water Resources Institute, Grand Valley State University, Allendale and Muskegon, MI, United States
- 4Division of Biological Sciences, University of Montana, Missoula, MT, United States
Despite their ecological significance and unique endosymbiotic capabilities, diatoms in the genus Rhopalodia remain poorly represented in genomic databases, particularly with respect to the availability of complete genomes from multiple organellar compartments. This study addresses that gap by presenting, for the first time, the complete chloroplast (133,086 bp), mitochondrial (36,786 bp), and spheroid body (3,024,495 bp) genomes of Rhopalodia sterrenburgii, a nitrogen-fixing diatom with a cyanobacterial endosymbiont. Phylogenetic reconstruction based on five genes from different cellular compartments (18S, 28S, rbcL, psbC, cob) placed R. sterrenburgii as a basal lineage relative to R. gibba (Rhopalodia sensu lato) and the Epithemia sensu stricto species E. argus, E. turgida, and E. sorex, providing new insights into evolutionary relationships within Rhopalodiales. Additionally, pathway analysis revealed the progressive loss of genes involved in vitamin B12 and chlorophyll a biosynthesis in more recently diverged Rhopalodia and Epithemia lineages. Our findings support a pattern of genome reduction in symbiotic diazotrophic diatoms, potentially driven by coevolution with their endosymbionts, and an increasing reliance on integrated metabolic functions between the host and the endosymbiont.
1 Introduction
Members of the Rhopalodiales (Bacillariophyta; Bacillariophyceae), comprised of Rhopalodia O.Müller, Epithemia Brébissonia ex Kützing and Tetralunata Hamsher et al., have the unique feature of possessing intracellular endosymbiotic cyanobacteria, sometimes referred to as spheroid bodies (Geitler, 1932; 1977). Our understanding of the relationship continues to grow, but several aspects of the symbiosis have already been determined. These include: 1) The endosymbiont is derived from a lineage of unicellular marine cyanobacteria and evidence supports a single acquisition of the endosymbiont by the diatom (Nakayama et al., 2011), 2) the cyanobacterium has lost genes for photosynthesis (Nakayama et al., 2014) and other functions but carries out nitrogen fixation (Schvarcz et al., 2022), and 3) the cyanobacterium is vertically inherited through cell division (Nakayama et al., 2011) and sexual reproduction (Kamakura et al., 2021). While a phylogenetics study of the group showed Rhopalodia to be non-monophyletic (Ruck et al., 2016), the authors’ solution to this outcome, combining all Rhopalodia species into the genus Epithemia, has been challenged (Vigneshwaran et al., 2021).
The diatom genus Rhopalodia O.Müller in the United States is not well represented in the diatom flora of the USA, with Patrick and Reimer (1975) listing only six taxa and the website Diatoms of North America listing only three – R. gibba (Ehrenberg) O.Müller, R. musculus (Kützing) O.Müller and R. gibberula (Ehrenberg) O.Müller – within the genus Epithemia (Spaulding et al., 2021). Kociolek et al. (Submitted) provide a listing of all Rhopalodiales reported in the literature from the United States.
Rhopalodia sterrenburgii was described from 49 Palms Oasis in Joshua Tree National Park, in a collection taken in 1985 (Lange-Bertalot and Krammer, 1987). The new species was originally illustrated with three light micrographs and a single SEM, showing a small section of the valve exterior. Subsequently, Krammer (1988a) published two SEM images of the valve exterior of this species. In a separate publication, Krammer (1988b) presented four LM images, though it is debatable whether they represent the same species. He also included one SEM of the valve exterior and another of a small portion of the valve interior. The species has not been treated or reported since. The Rhopalodia genus has been investigated with electron microscopy (Gaul et al., 1993; Henderson and Reimer, 2003) and only one taxon has had complete molecular sequences generated from them - R. gibba (Abresch et al., 2024). There may be some mentions in the literature, but to date, no characterization and fully validated descriptions of the plastome, mitogenome, or spheroid body genome within the genus Rhopalodia have been published in a single paper together with morphological descriptions.
Given the opportunity presented by our re-collection of this taxon from its type locality and having it grow successfully in culture, we offer here new insights into the size diminution series of this species, its valve ultrastructure and the description of its entire plastome, mitogenome and spheroid body genome. In addition, we apply phylogenetics to place this species into the Rhopalodiales tree of life to assess its phylogenetic relationships. We also performed metabolic pathway reconstructions, focusing on the biosynthesis of vitamin B12 and chlorophyll a of the spheroid body genome, to elucidate the molecular mechanisms that enable this species to support and sustain a symbiotic lifestyle.
2 Materials and methods
2.1 Morphology
Freshwater samples were collected from the 49 Palms Oasis at Joshua Tree, California (U.S. National Park Service permit number JOTR-2023-SCI-0012), placed in Whirl-Pak bags and that were then put on ice. Back at the lab, samples were cleaned with nitric acid, and after five settled and rinsing cycles, the cleaned material was air-dried onto glass coverslips. For light microscopy (LM), coverslips with the cleaned material were mounted onto glass microscope slides with Hyrax. LM observations were made on an Olympus BX-51 light microscope with Differential Interference Contrast optics. Digital images were captured with a DP-71 digital camera.
For scanning electron microscopy (SEM), cover glasses with dried material were attached to aluminum stubs with carbon tape, sputter-coated with 4 nm of Pt, and viewed on a Hitachi SU-3500 VP SEM at an accelerating voltage of 15 kV and at a working distance of 5.5 mm at the Colorado Shared Instrumentation in Nanofabrication and Characterization (COSINC) facility at CU-Boulder. Permanent slides and SEM stubs are vouchered in the JPK Diatom Collection at the University of Colorado, Boulder (COLO).
2.2 Isolation and cell culture
Portions of the raw sample were initially diluted in well plates and incubated until the target cells began to increase in number. Individual cells were then isolated under a microscope using a fine-tipped glass pipette and transferred into a sterile culture medium. The cultures were grown in CSi medium (Nakayama et al., 2011) without nitrogen (CSi –N) at room temperature under natural light conditions. To reduce contamination and achieve unialgal cultures, the isolates were subcultured several times and regularly checked under a microscope. Viable, intact cells – identified by their distinct golden-brown color – were then sent out for DNA extraction and sequencing.
2.3 Genome sequencing, assembly and annotation
Frozen cell pellets were shipped on ice to SeqCoast Genomics (Portsmouth, NH, USA) for DNA extraction. Upon receipt, the samples were processed using a bead-beating lysis method to mechanically disrupt cells and release genomic DNA, following the company’s standardized extraction protocol. Extracted DNA was then prepared for short-read sequencing using the Illumina NextSeq2000 platform. To complement the short-read data, long-read sequencing data generated using Oxford Nanopore Technologies were also integrated into the genome assembly. Sequencing was performed on the PromethION 2 Solo platform, equipped with an R10-series FLO-PRO114M flow cell. All resulting sequencing data were provided as FASTQ files for further assembly steps.
Raw sequencing reads underwent quality filtering and trimming using Trimmomatic v0.39 (Bolger et al., 2014). Organelle genomes were assembled de novo using SPAdes v3.13.1 (Nurk et al., 2017). To isolate relevant scaffolds from the assembled data, BLAST searches were performed in the terminal using blastn against a custom database, which was created with makeblastdb command-line tool using Rhopalodiales reference genomes obtained from GenBank. This reference-guided filtering strategy allowed for the selection of contigs based on homology to a closely related species, aiding the identification of chloroplast and mitochondrial genomic sequences for Rhopalodia sterrenburgii.
Chloroplast genomic regions were identified by aligning against annotated sequences from Rhopalodia gibba strain 17Bon1, specifically OR515805.1 (large single-copy region, LSC), OR515806.1 (small single-copy region, SSC), and OR515807.1 (inverted repeat, IR). The mitochondrial genome was similarly extracted using the reference sequence OR515804.1. To verify that assembled contigs formed complete, circular genomes without gaps, a tiling strategy was employed – this involved generating overlapping segments to assess continuity and to allow circularization. This was performed in the terminal using the grep function, and adding or removing sequences based on the raw reads. This process ensured accurate joining of contigs across gaps and validated the structural completeness of the assembly. Once finalized, the assembly quality was verified by aligning it against the raw reads using bwa mem (Li and Durbin, 2009). These alignments were then sorted and indexed using SAMtools tview (Li et al., 2009). Additionally, single nucleotide polymorphisms (SNPs) were identified throughout the whole genomes using a variant calling technique with bcftools mpileup and bcftools call commands (Narasimhan et al., 2016). Post-assembly, error correction was carried out to detect and fix sequencing errors (minimum quality score > 100), thereby improving the accuracy of the final organellar genome reconstructions.
Final plastid and mitochondrial genome annotations were performed using the GeSeq tool (Tillich et al., 2017). Annotations were subsequently reviewed and validated with Sequin v10.3 (NCBI), and genome maps were produced using OGDRAW (Greiner et al., 2019) through the Chlorobox platform (https://chlorobox.mpimp-golm.mpg.de).
The spheroid body (SB) genome was reconstructed by first identifying candidate endosymbiont-derived contigs through BLASTn comparisons against the cyanobacterial endosymbiont genome of Rhopalodia gibba strain 17Bon1 (CP067995.1). Contigs showing strong sequence similarity to the reference were flagged as potential SB sequences. A custom script (Supplementary File S2) was subsequently used to refine the initial selection – eliminating non-SB contigs, determining the relative orientation and order of the retained sequences, and assembling them into a draft genome guided by the structure of the reference genome. In cases where certain genomic regions were absent from the assembly, the script estimated their expected size and position based on the reference genome architecture and inserted placeholder sequences composed of ‘NNNs’ to represent these gaps. This approach facilitated the construction of a continuous, reference-guided draft genome.
The resulting draft SB assembly was annotated using the RASTtk pipeline, a web-based platform designed for the comprehensive annotation of bacterial and archaeal genomes (Brettin et al., 2015) as well as NCBI’s Prokaryotic Genome Annotation Pipeline (PGAP) v6.10. To evaluate assembly and annotation quality, genome summary reports and statistics were generated via the BV-BRC v3.47.11 web interface (https://www.bv-brc.org), which integrates multiple tools for quality assessment and genome analysis (Olson et al., 2023). To analyze the biosynthetic pathways for vitamin B12 and chlorophyll a, we used protein annotation files from our focal SB genome and from previously sequenced SB genomes available in NCBI GenBank. KO (KEGG Orthology) terms were assigned using KofamKOALA (Aramaki et al., 2020), and pathway reconstruction and visualization were carried out in KEGG Mapper. This allowed us to compare the presence and completeness of these pathways across different SB lineages.
2.4 Assembly metrics and genomic output across organelle compartments
Short read sequencing produced approximately 8.5 million paired end reads (2 x 150 bp), while long read sequencing generated 84,129 long reads with an average read length of 5.5 kb (N50 = 8.5 kb). After quality filtering and trimming, all reads were de novo assembled using SPAdes v3.10 using the –1 <short reads R1> –2 <short reads R2> –nano <concatenated long reads> with –meta resulting in a hybrid metagenome assembly of 336.34 Mbp with 691,883 contigs. The hybrid approach improved contiguity and resolved repetitive regions that short-read-only assemblies could not.
Five genes were identified from the assembled contigs after assembly: 1,598 bp 18S gene, 354 bp 28S gene, 1,473 bp rbcL gene, 1,204 bp psbC gene, and 1,005 bp cob gene. All genes were submitted to NCBI GenBank with accession numbers PV764874, PV766016, PV765887, PV765888, and PV765886, respectively. Scaffolds for chloroplast, mitochondrion and spheroid bodies were selected using Rhopalodia gibba strain 17Bon1, with accession numbers OR515805.1 (LSC), OR515806.1 (SSC), and OR515807.1 (IR); mitochondrial sequences were extracted using OR515804.1; and spheroid body sequences were selected using CP067995.1 (Abresch et al., 2024). Plastome, mitogenome and spheroid body genomes were recovered having sizes of 133,086 bp, 36,786 bp and 3,024,495 bp, respectively.
2.5 Phylogenetic analysis
A multi-compartmental 5-gene phylogenetic tree was generated using two genes from the nucleus (18S SSU and 28S LSU), two genes from the chloroplast (rbcL and psbC), and one gene from the mitochondrion (cob). Five genes were extracted from 46 diatom taxa, including 29 outgroup taxa from the genera Halamphora, Thalassiophysa, Entomoneis, Petrodictyon, Surirella, Campylodiscus, Cymatopleura, Protokeelia, and Auricula, and 17 ingroup taxa from the genera Epithemia and Rhopalodia. For some Epithemia taxa, one or two sequences are absent, resulting in 3.91% missing data across the alignment matrix used for phylogenetic reconstruction (Supplementary Table S1). Taxon sampling and gene selection followed the phylogenetic framework and marker set employed by Ruck et al. (2016). Individual alignments for all five genes were performed using MAFFT v7.490 (Katoh and Standley, 2013; Katoh et al., 2002), implemented within Geneious Prime v2025.1.1. Gene alignments were concatenated and subsequently trimmed using TrimAl with the command-line parameters: trimal -in <input alignment file> -out <output filename> -automated1 (Capella-Gutiérrez et al., 2009). The resulting trimmed alignment (Supplementary File S1) was used for model selection and maximum likelihood phylogenetic inference in the IQ-TREE web server (Trifinopoulos et al., 2016), employing the GTR+I+G+F4 substitution model and 1,000 ultrafast bootstrap replicates. The resulting consensus tree was visualized and manually re-rooted using FigTree v1.4.4 (Rambaut, 2018).
3 Results
3.1 Morphological features
Here we detail the morphological features of the glass cell walls as seen with light and scanning electron microscopy. These features are important for the identification of this unique, endemic species.
Light microscopy (Figure 1A). Frustules are segment-like, similar to the segment of a citrus fruit, The frustules include the two valves and girdle bands, each band possessess a row of distinct poroids. Valves have strongly convex dorsal margins, with a notch present at the center; ventral margins are weakly concave, with apices decidedly deflected ventrally and subrostrate. Length 17–31 µm, breadth 6–9 µm. The canal raphe resides in a distinct keel, positioned at the mid-line of the dorsal margin. Costate fibulae are distinct, slightly radiate, 5–6/10 µm. The fibulae define distinct sectors on the valve face, wider along the dorsal margin narrowing towards the ventral margin, 2–5 thickened striae between adjacent fibulae, 12–24/10 µm. Individual areolae are not visible in LM. Distinct circles near the apices.
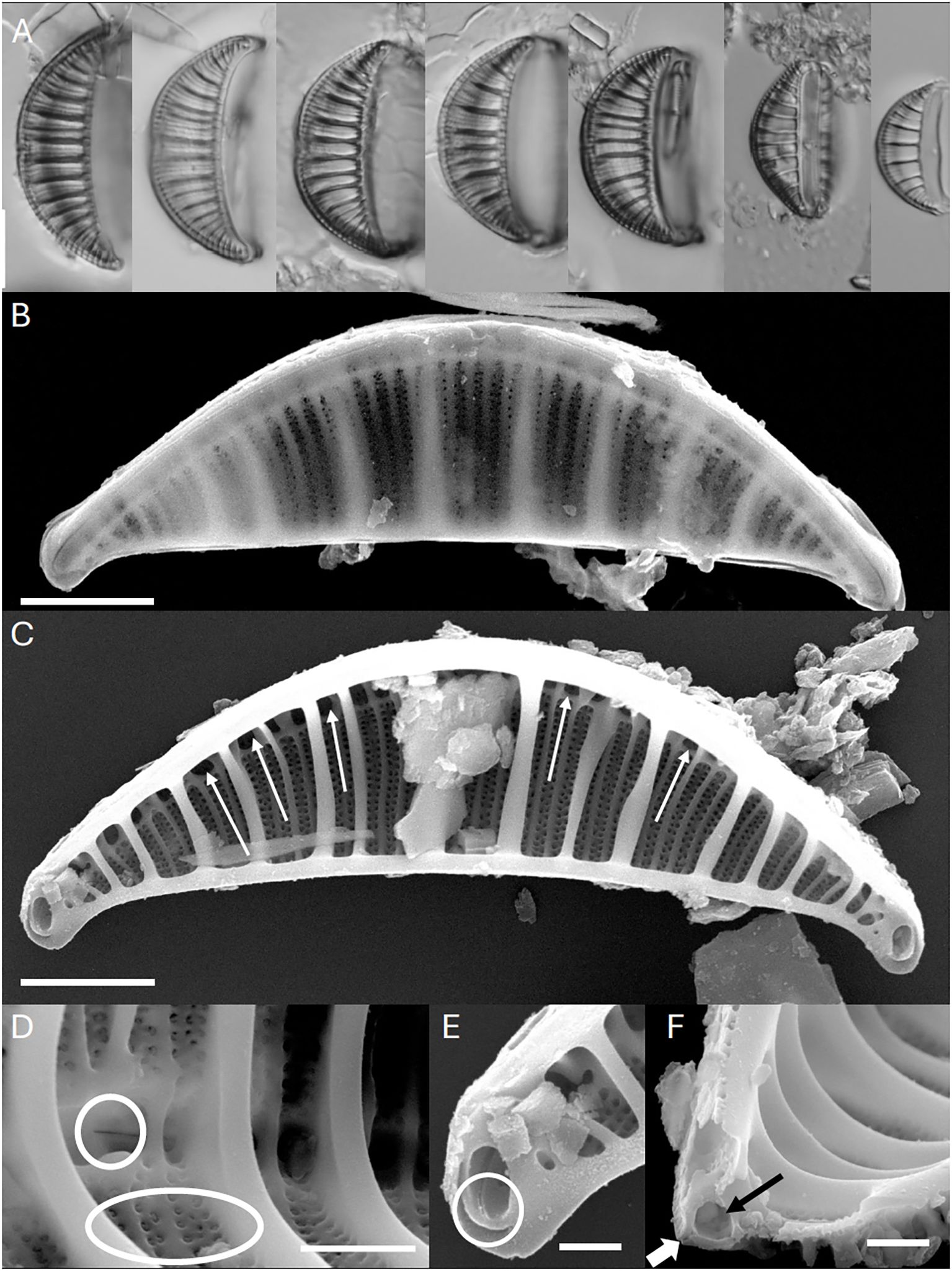
Figure 1. Rhopalodia sterrenburgii. (A) Light microscopy (LM). Valve views showing the size diminution series of this species. Scale bar = 10 µm. (B) Scanning Electron Microscopy (SEM). External view, whole valve. Scale bar = 3 µm. (C) SEM. Internal view, whole valve. Arrows indicate portulae of the canal raphe. Scale bar = 3 µm. (D) SEM. Internal views. Valve center. Robust costate fibulae are evident. Circle shows interrupted raphe at thecentral nodule. Ellipse shows areolae with internal volate occlusions. Scale bar = 1.5 µm. (E) SEM. Internal views. Apex of the valve with internally raised circular structure. Raphe terminus lacks a helictoglossa. Scale bar = 1 µm. (F) SEM. Internal views. Transverse section of the valve with robust costate fibulae evident. Black arrow indicates canal raphe. White arrow indicated raphe slit. Scale bar = 1 µm.
In the SEM (Figure 1B), the valve exterior has the valve face on one side of the raphe being longer and with a more gentle slope towards the thin mantle and a shorter side that is at a more acute angle relative to the raphe. The wide canal raphe is positioned in an elevated keel. The keel is buttressed by thick ridges that extend across the valve face. Proximal and distal raphe ends are deflected towards the ventral margin.
Internally, the valve is dominated by thick costate fibulae, between which are 2–3 secondary fibulae (Figure 1C). The raphe canal has rounded to ovoid portulae, and the central nodule has a wider opening (Figure 1D). At the apices the raphe terminates in a very delicate helictoglossa (Figure 1E). The raphe is interrupted at the center. Areolae are in lines of 2–3 striae between the costate fibulae. At the apices the costate fibulae are modified to form a rounded structure that is elevated internally (Figures 1C, E). Each areola is occluded by volae, that are positioned internally and afford a c-shaped opening (Figure 1D). A transverse section of the raphe shows it to be tubular, with a narrow ridge bordering the raphe slit (Figure 1F).
3.2 Multi-compartmental phylogenetics
R. sterrenburgii is nested within a well-supported clade (bootstrap = 100%) composed of various Epithemia and Rhopalodia species, suggesting a close evolutionary relationship between these two genera (Figure 2). The groups containing E. catenata and E. pelagica represent the earliest diverging lineages (Basal Rhopalodiales), branching off prior to the diversification of the remaining Epithemia and Rhopalodia taxa. Epithemia but not Rhopalodia was recovered as a monophyletic group in this analysis. We refer to the groups shown in the figure as “Basal Rhopalodiales,” Rhopalodia sensu lato”, and “Epithemia sensu stricto” for ease of reference. The terms “Basal Rhopalodiales” and “Rhopalodia sensu lato” are used for convenience only, given the nomenclatural confusion introduced for this group - but they are non-monophyletic groups. The third group, Epithemia sensu stricto, represents the traditional grouping of the genus which is a monophyletic group.
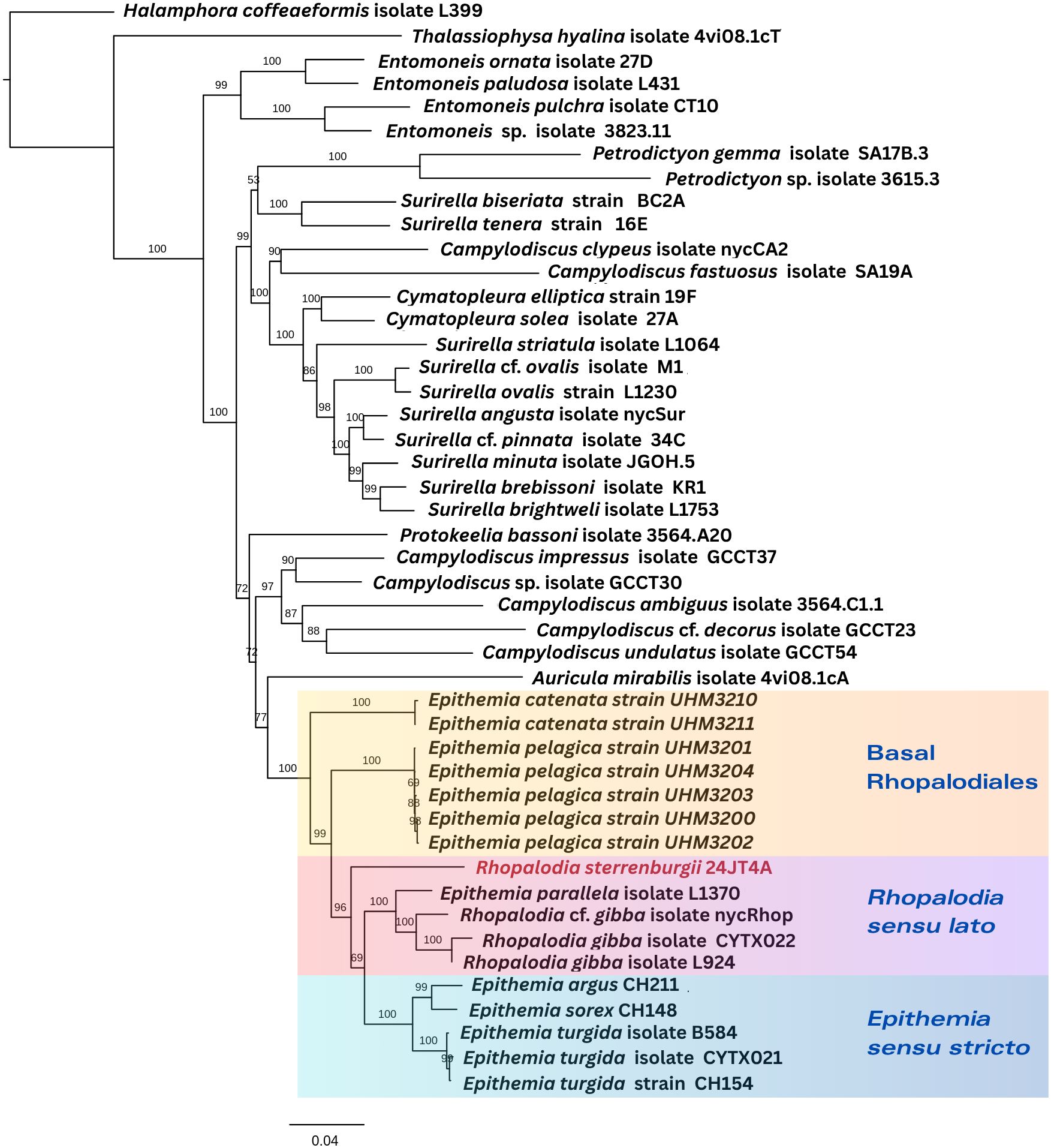
Figure 2. Phylogenetic placement of R. sterrenburgii (highlighted in red) based on concatenated five genes from the nuclear genome (18S and 28S genes), chloroplast genome (rbcL and psbC genes) and mitogenome (cob gene). Numbers at the nodes indicate bootstrap support values based on 1000 replicates. Higher bootstrap values (closer to 100) indicate stronger support for the corresponding clade, while lower values suggest less confidence in the inferred relationship.
3.3 The chloroplast genome
The circularized plastome of Rhopalodia sterrenburgii (133,086 bp) exhibits a typical quadripartite structure (Figure 3A). The complete genome contains 127 protein-coding genes, 29 tRNAs, one tmRNA, three rRNAs, and one pseudogene (Table 1, Supplementary Table S2). Two tRNAs occur in duplicates (trnA-UGC, trnI-GAU), one in each inverted repeat (IR). Two non-functional partial genes are present, psaB in the IRB and rps16 in the IRA, both of which have a full-length functional copy in the opposite IR. Three introns were detected in the plastome, within the petB, rpoA, and groEL genes. Two non-conserved ORFs are present, ORF1, found in duplicates (one in each IR), with 75.94% similarity with ORF134a in Haslea silbo QUS63768, and ORF27, which shares 57.46% similarity with ORF27 in Halamphora calidilacuna NC_044464.
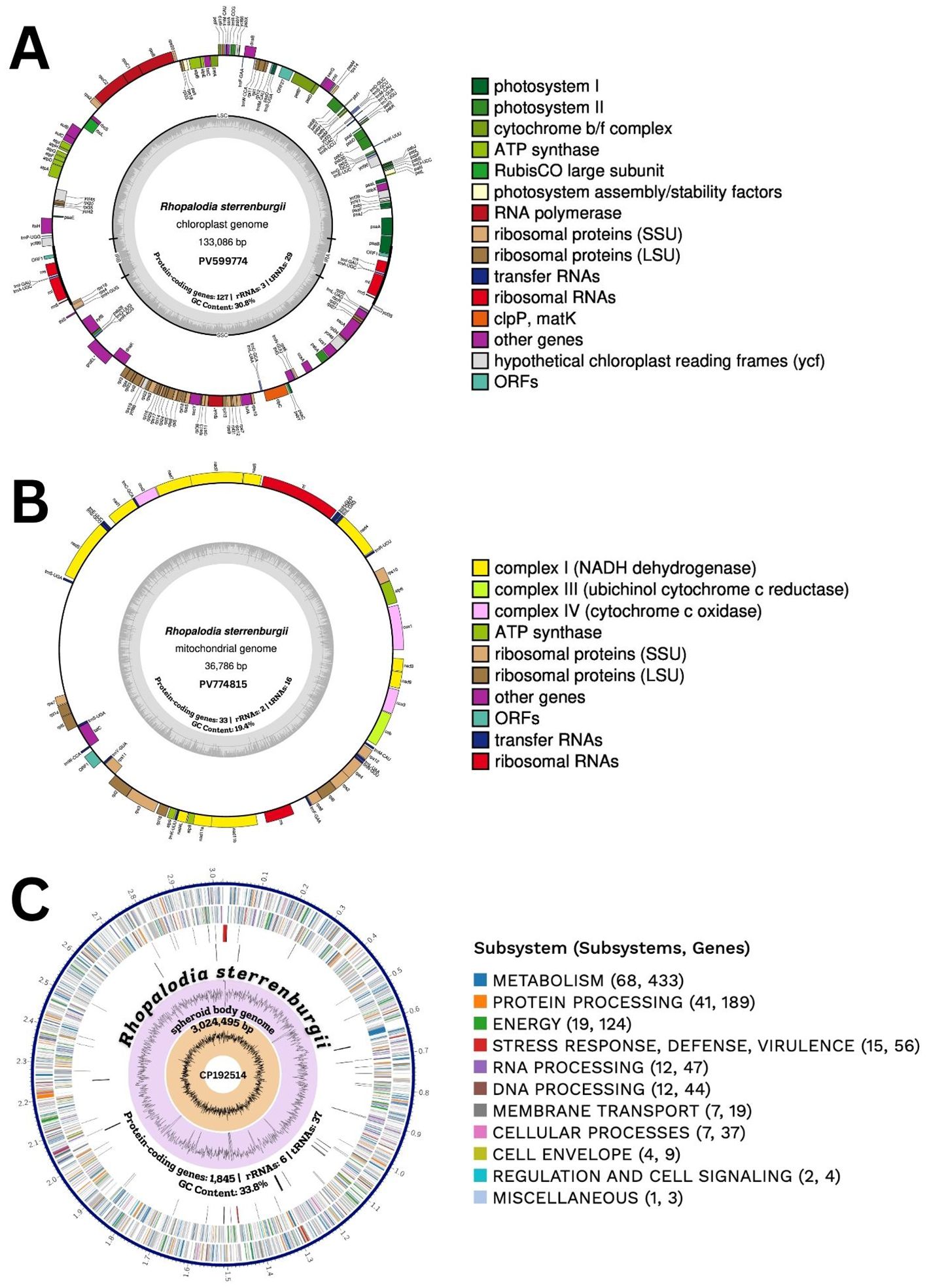
Figure 3. (A) Chloroplast, (B) mitochondrial and (C) spheroid body circular genome maps of Rhopalodia sterrenburgii. In the middle of circular maps are the NCBI GenBank accession numbers, number of protein-coding genes, rRNAs, tRNAs, and GC content. The genome figures are color-coded based on their assigned functional subsystems. (A, B) Genes positioned on the outside of the circle are transcribed in the clockwise (forward) direction, while those on the inside are transcribed counterclockwise (reverse strand). Introns are depicted as interrupted gene boxes, and pseudogenes are indicated with asterisks. (C) The circular map includes, from the outermost to the innermost rings: assembled contigs, coding sequences (CDSs) on the forward strand, CDSs on the reverse strand, RNA genes, CDSs with homology to known antimicrobial resistance genes, CDSs with homology to known virulence factors, GC content, and GC skew.
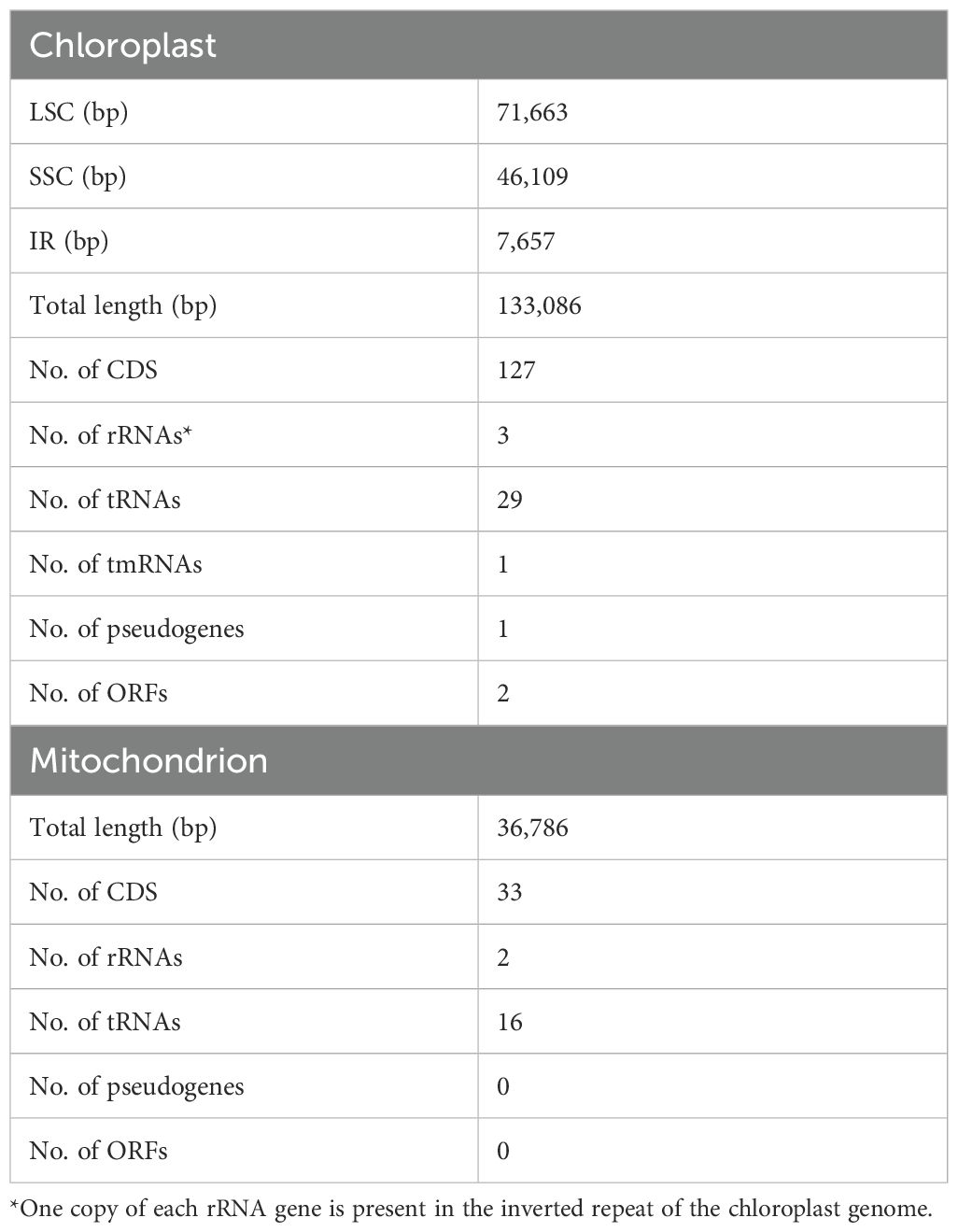
Table 1. Summary of basic features of Rhopalodia sterrenburgii chloroplast and mitochondrial genome.
The large single-copy (LSC) region spans 71,663 bp and encodes 74 protein-coding genes, one pseudogene (ycf42), 18 tRNAs, and one tmRNA. The small single-copy (SSC) spans 46,109 bp, encoding 52 protein-coding genes and seven tRNAs. Each inverted repeat (IRA, IRB) ranges from 7,657 bp and encodes two tRNAs, and three rRNAs. The overall GC content is 30.8%. The plastid genome annotations are available on NCBI (Accession PV599774).
3.4 The mitochondrial genome
The mitogenome of R. sterrenburgii is 36,786 bp in length (Figure 3B). It contains complete copies of 33 protein coding DNA sequences, 16 transfer RNAs, and the large and small ribosomal RNA genes (Table 1; Supplementary Table S3). The overall GC content is 19.4%. It contains a highly repetitive region, presumed to contain the regulatory D-loop, spanning roughly 4,000 bp. The mitogenome is available on NCBI (Accession PV774815).
3.5 The spheroid body genome
The number of spheroid bodies (SB) in Rhopalodiales usually ranges from one to four per cell (Kneip et al., 2008; Schvarcz et al., 2024). In R. sterrenburgii, two spheroid bodies per diatom cell were observed. The cyanobacterial endosymbiont of R. sterrenburgii has a genome size of approximately 3,024,495 bp and a GC content of 33.8% (Figure 3C). Although the assembly contained 22 unresolved regions, the draft genome is 99.3% complete based on gene content, and its overall size falls within the typical range (2.4 Mbp to 3 Mbp) observed in other spheroid body genomes. Genome annotation identified 1,845 protein-coding sequences, 6 rRNA genes (3 × 5S, 1 × 16S, 2 × 23S), 37 tRNAs, 4 non-coding RNAs, and 74 pseudogenes. All the genes needed for nitrogen fixation were found, but genes for light-dependent photosynthesis were not present. The complete circular SB genome is under the accession number CP192514 (NCBI Genbank). Figures 4 and 5 present the genes involved in the biosynthesis of vitamin B12 and chlorophyll a, both of which are members of the tetrapyrrole biosynthetic network along with heme. The analysis includes SB genomes of eight symbiotic diatom taxa within the order Rhopalodiales – seven previously described spheroid body (SB) genomes available in NCBI, along with R. sterrenburgii, described here for the first time. Pathway reconstructions of SB genomes revealed that taxa representing the earliest divergences (basal Rhopalodiales and Rhopalodia sensu lato) – E. catenata, E. clementina, E. pelagica, R. gibberula, R. sterrenburgii, R. gibba – retained a complete set of genes involved in vitamin B12 coenzyme biosynthesis (Figure 4) (Nakayama and Inagaki, 2017). In contrast, more recently diverged taxa (Epithemia sensu stricto) – E. adnata and E. turgida – showed extensive gene loss, resulting in incomplete pathways. Even more extensive gene losses were observed in the chlorophyll a biosynthesis pathway (Figure 5), where all SB taxa lost the capacity to produce chlorophyll a. All pathways were truncated mid-way, with the loss of half or more of the associated genes.
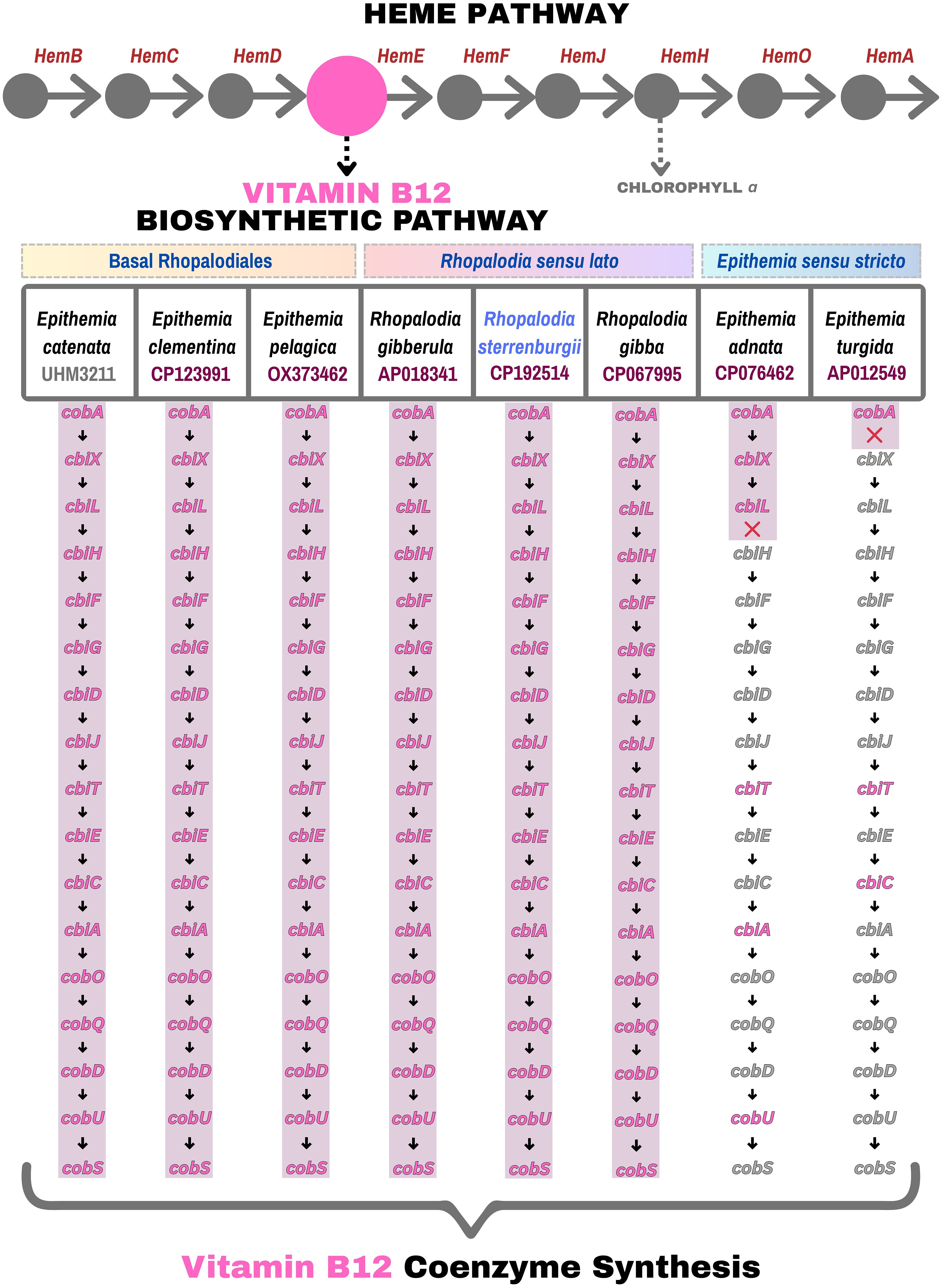
Figure 4. Pathway maps comparing eight Rhopalodiales taxa arranged from the earliest to most recent diverged taxa (left to right), highlighting the presence (pink) and absence (grey) of genes involved in vitamin B12 coenzyme biosynthesis.
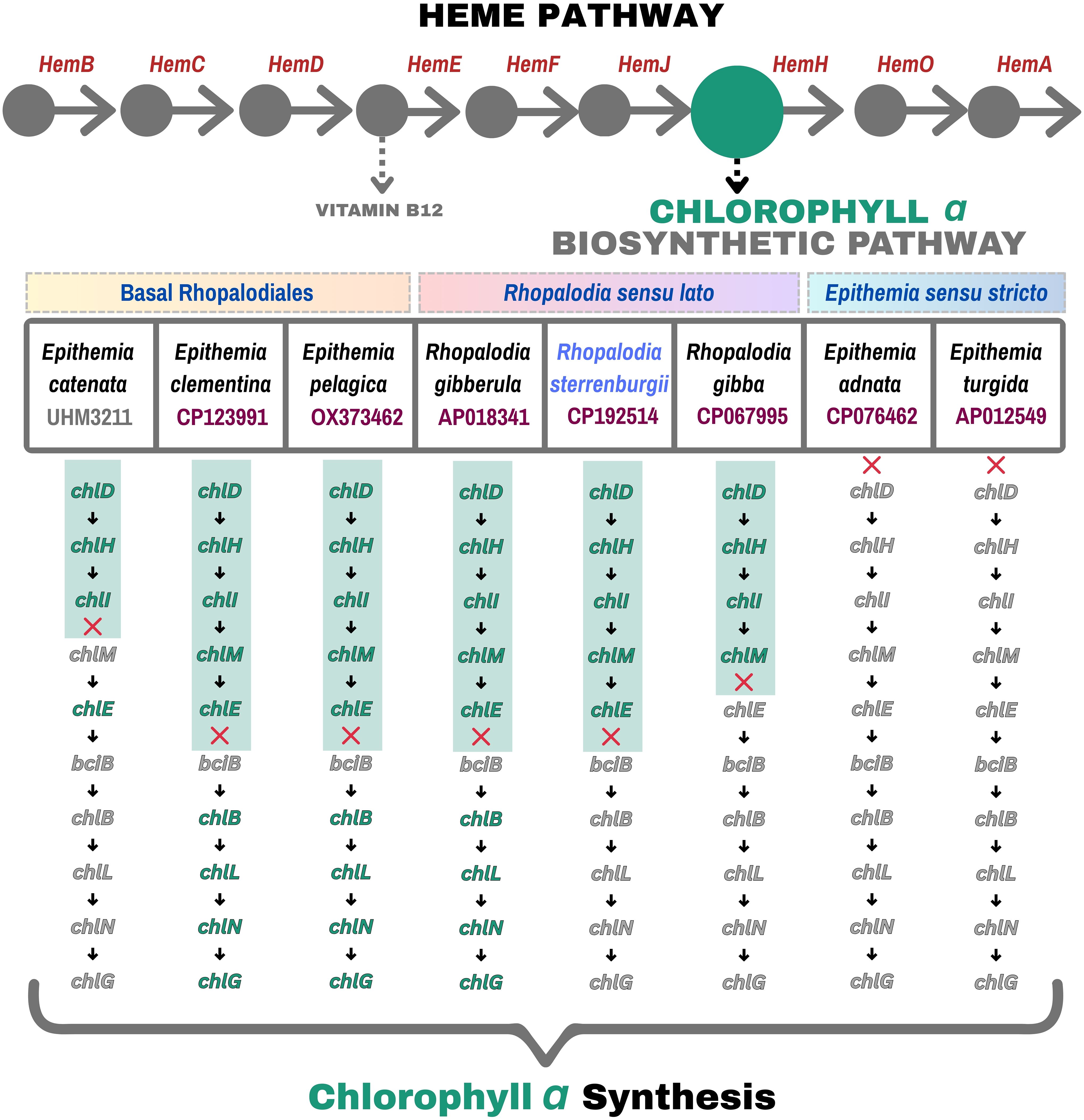
Figure 5. Pathway maps comparing eight Rhopalodiales taxa arranged from the earliest to most recent diverged taxa (left to right), highlighting the presence (green) and absence (grey) of genes involved in chlorophyll a biosynthesis.
4 Discussion
4.1 Morphological observations
Rhopalodia sterrenburgii is morphologically similar to R. gibba (Ehrenberg) O.Müller, the generitype of the genus Rhopalodia, by virtue of the organization of the frustule, position and structure of the raphe, and, importantly, by possessing costate fibulae, a feature some have used to diagnose the Rhopalodiales (Sims, 1983; Ruck et al., 2016). In the latter feature, it is distinguished from species that have been assigned to the group by virtue of possessing cyanobacterial endosymbionts, but otherwise lacking many of the valve features found in the Rhopalodiales (e.g. Schvarcz et al., 2022). The elevated tubular canal raphe observed in R. sterrenburgii is similar to other species assigned to the genus, for example, R. gibberula (Ehrenberg) O.Müller, R. constricta (Kützing) O. Müller and R. wetzelii Hustedt (Krammer, 1988a, b; Kociolek et al., 2024; Kociolek et al., Submitted, though this feature is reduced in R. gibba and its allies (Moore et al., 2025). A raphe in an elevated keel is not found in the genus Epithemia (Sims, 1983; Kobayasi and Kobayashi, 1988; Kociolek et al., 2025a). Rhopalodia sterrenburgii differs from other Rhopalodia species by apparently lacking any areolar occlusions. Such occlusions are found in other species assigned to both Rhopalodia (Lange-Bertalot and Krammer, 1987; Krammer, 1988a, b; Krammer and Lange-Bertalot, 1988; Kociolek et al., 2025b) an Epithemia (Sims, 1983; Kobayasi and Kobayashi, 1988; Kociolek et al., 2025a). Morphological evidence supports the molecular phylogeny where R. sterrenburgii is positioned within the Rhopalodiales, but distinct from R. gibba and Epithemia sensu stricto.
4.2 Phylogenetic inference using multi-compartmental genes
The phylogenetic analysis based on concatenated multi-compartmental genes shed new light on the evolutionary relationships within Rhopalodiales, particularly among Epithemia and Rhopalodia species. The tree topology revealed that E. catenata and E. pelagica formed the earliest diverging lineage within the order, branching off prior to the diversification of all other Epithemia and Rhopalodia taxa. This basal placement, strongly supported by high bootstrap values (=100%), suggested that these two species represent distinct lineages within the group.
The genus names applied to taxa with molecular data in the Rhopalodiales introduces confusion into the interpretation of relationships in the Order. Less than 10 years ago the separation of Rhopalodia and Epithemia was clear, based on extensive morphological evidence compiled over a century with light (Müller, 1895; F. Fricke in Schmidt, 1904, 1905; Hustedt, 1930; Patrick and Reimer, 1975; Krammer and Lange-Bertalot, 1988) and scanning electron microscopy (Krammer, 1988a, b; Sims, 1983; Kobayasi et al., 1993). A third genus, Tetralunata (Hamsher et al., 2014) was proposed for the group and a morphological phylogeny showed it to be more closely allied to Epithemia than Rhopalodia. Ruck et al. (2016) showed with molecular data that unnamed taxa they assigned to Rhopalodia did not form a monophyletic group with the generitype, R. gibba, and its allies, (a conclusion reached 30 years prior by Krammer, 1988b based on morphology). They proposed: 1) a putative synapomorphy and diagnostic feature for the entire order was the presence of costate fibulae and 2) due to the non-monophyly of taxa previously assigned to Rhopalodia, all taxa of that genus (>250 species, varieties and forms) of that genus should be transferred to Epithemia. Ruck et al. (2016) argued this approach was more conservative than creating two new genera to accommodate the taxa in Rhopalodia not in the R. gibba clade. Vigneshwaran et al. (2021) rebutted the conclusions of Ruck et al., noting the radical nature of their proposal. Following Ruck et al.’s (2016) proposal, Schvarcz et al. (2022) described “Epithemia” catenata and “Epithemia” pelagica even though these taxa lack the diagnostic feature of the Order (as suggested by Ruck et al., 2016), let alone the features of Epithemia as typified by E. turgida. Our analysis here on the phylogenetic position of these two taxa shows them far outside Epithemia sensu stricto. Kociolek et al. (2025a) showed tremendous morphological variability amongst taxa within Epithemia sensu stricto, identifying 5 groups within the genus. Lange-Bertalot and Krammer (1987), Krammer (1988a, b) and more recently Kociolek et al. (2024) documented significant morphological variability amongst taxa assigned to Rhopalodia, leading to the description of a new genus for marine, fossil taxa (Kociolek et al., 2025b).
The work presented herein on R. sterrenburgii adds to the list of features for taxa previously assigned to the genus Rhopalodia by documenting the presence of internally-placed volate occlusions in the areolae. And the significant differences in the presence and absence of genes in the biosynthetic pathways of vitamin B12 and chlorophyll a between Epithemia sensu stricto species and others within the Rhopalodiales adds additional support to the approach to separate out taxa based on morphology and molecular data.
The topological incongruences observed here may also be due to the limited phylogenetic resolution of past morphology-based groupings and emphasizes the value of multi-compartmental gene data for resolving complex evolutionary relationships, particularly in diatoms. However, for better resolution within Rhopalodiales, broader and more representative taxon sampling, especially from underrepresented clades, will be essential.
4.3 Insights into organelle genomes of Rhopalodia sterrenburgii
The 133,086-bp complete plastid of Rhopalodia sterrenburgii falls within the typical size range for diatom chloroplasts, resembling the chloroplast of Epithemia pelagica OX459761 (130,746 bp), a species sister to Rhopalodia sterrenburgii + Rhopalodia gibberula + Epithemia sensu stricto species. Because R. sterrenburgii represents the first fully characterized chloroplast genome in the Rhopalodiales, comparisons are presented here with closely related lineages outside the order. Smaller genomes were found in Entomoneis umbratica OR464352 (128,818-bp; Jeong and Lee, 2024), Entomoneis sp. MG755800 (122,056-bp; Yu et al., 2018), and Halamphora coffeaeformis MK045452 (121,927-bp; Hamsher et al., 2019). A similar genome size is reported for H. vantushpaensis PP962255 (133,866-bp; Yılmaz et al., 2024), while larger sizes were described for H. americana MK045450 (142,551-bp; Hamsher et al., 2019) and H. calidilacuna MK045451 (150,738-bp; Hamsher et al., 2019). The observed variations can be attributed to extended inverted repeats, the presence of intron-encoded endonucleases and hypothetical ORFs, features that typically increase organellar genome size substantially (Hamsher et al., 2019; Pogoda et al., 2019).
An opposite trend is observed in the GC content, with a slightly lower percentage in Rhopalodia sterrenburgii (30.8%) compared to Epithemia pelagica (31.5%), Entomoneis species (31.1–32.4%) and Halamphora species (31.7–32.3%). In diatom chloroplasts, low GC content may result from codon usage bias favoring A and T base pairs, which enhances the stability and efficiency of protein synthesis (Yao et al., 2024). Additional sequencing of complete chloroplast genomes within the Rhopalodiales could help clarify whether the variability in genome size and GC content is affected by having an endosymbiont.
Analysis of the Rhopalodia sterrenburgii chloroplast genome revealed a typical set of genes present in diatom plastids, including those involved in photosynthesis, ATP synthesis, and protein synthesis (Nakayama et al., 2011; Yu et al., 2018). This suggests that it retains a high degree of metabolic autonomy, regardless of its symbiotic relationship with spheroid bodies (Abresch et al., 2024). For instance, the presence of fully functional photosystems I and II (PSI and PSII) in the chloroplast indicates that the diatom host is still responsible for light-dependent reactions essential for energy and carbohydrate production (Prechtl et al., 2004; Bothe et al., 2010; Moulin et al., 2024). Because this capability remains unchanged and spheroid bodies appear to be shifting toward a specialized N-fixation scheme adapted to an intracellular lifestyle, a gradually increasing level of SB-diatom integration is presumed (Adler et al., 2014; Kneip et al., 2008; Nakayama and Inagaki, 2014; Nakayama et al., 2014; Caputo et al., 2019). Moreover, our data suggest that there is no gene transfer from the spheroid bodies to the diatom plastid, as the only non-conserved coding regions in R. sterrenburgii are two ORFs that share similarities with hypothetical proteins found in non-endosymbiont-bearing diatoms (ORF134 in Haslea silbo QUS63768, and ORF27 in Halamphora calidilacuna NC_044464). In this complementary system, the diatom chloroplast is likely supplying and maintaining their spheroid bodies with energetic products generated through photosynthesis, and, in exchange, directly or indirectly receiving a usable form of nitrogen, as previously reported for other taxa in the Rhopalodiales (Abresch et al., 2024; Moulin et al., 2024).
The mitogenome of R. sterrenburgii encodes ribosomal protein S7 (rps7), which is missing from other unrelated diatom mitogenomes such as Campylodiscus clypeus (PV266141) and Plagiotropis lepidoptera (PV266140). The presence of mitoribosomal protein subunits (MRPs) encoded on the mitochondrial genome is highly variable across eukaryotes, with some lineages, like mammals, encoding all subunits of the mitoribosome in the nuclear genome (Boengler et al., 2011), which are subsequently imported into the mitochondrion. Other lineages, such as apicomplexans and some diatoms encode some of their mitoribosomal proteins on the mitochondrial genome (Gupta et al., 2014).
The mitoribosomal RNAs, which are catalytic RNA molecules that form the mitoribosomal complex, are encoded in tandem in many diatom mitogenomes (see Rhopalodia spp. PV553663, PV553664, PV553665; Plagiotropis lepidoptera PV266140; Campylodiscus clypeus PV266141; Halamphora spp. MF997424, MF997420; Thalassiosira spp. NC_007405, NC_060383.1). In some published lineages, the large and small ribosomal RNAs are not encoded in tandem, such as Thalassiosira profunda NC_060383.1 and Thalassiosira nordensioeldii NC_057471, as well as in R. sterrenburgii. It is plausible that the tandem-encoded large and small subunit RNAs are regulated under different mechanisms than those that are not tandem-encoded, suggesting that the regulatory mechanisms for ribosomal RNA expression are evolving at within-genus timescales.
4.4 Retention of nitrogen fixation and loss of photosynthesis in the spheroid body
The cyanobacterial endosymbionts found in diatoms play a key role in nitrogen fixation – a function that has been well established (Nakayama and Inagaki, 2017). This function remains conserved in R. sterrenburgii which possesses the core nif genes – nifB, nifD, nifE, nifH, nifK, nifN – essential for carrying out nitrogen fixation.
On the contrary, the complete loss of photosystem I and II (psa and psb) genes in the R. sterrenburgii endosymbiont – also reported in other cyanobacterial endosymbionts, also referred to as spheroid bodies, of diatoms (Nakayama et al., 2014; Nakayama and Inagaki, 2017) – indicates that these spheroid bodies are no longer capable of performing active photosynthesis. However, this loss does not appear detrimental to the host–endosymbiont relationship. Since the diatom host retains functional chloroplasts for photosynthesis, the cyanobacterium endosymbiont’s role may have shifted toward a more specialized function, which is to fix nitrogen for the host. This functional shift likely complements the mutualistic relationship, as the spheroid body provides a critical nutrient while relying on the host for carbon and energy sources derived from the host’s chloroplasts.
4.5 Loss of vitamin B12 and chlorophyll a biosynthetic pathway in the spheroid body
Evidence of host–endosymbiont integration is seen at the genomic level where significant changes are observed in endosymbiotic cyanobacteria. Free-living cyanobacteria typically have larger genomes and greater gene content compared to their endosymbiotic counterparts. They have genome sizes ranging from approximately 4.6 Mbp in Rippkaea orientalis (NC_011726.1) to around 9.0 Mbp in Nostoc punctiforme (NZ_JAOEFB000000000.1). Well below the expected range (~1.4 Mbp) is found in the marine haptophyte alga Braarudosphaera bigelowii, which harbors a highly reduced cyanobacterial organelle known as the nitroplast, or Candidatus Atelocyanobacterium thalassa UCYN-A (Tripp et al., 2010). This supports the idea that genome reduction in terms of size and gene content most likely occurs when cyanobacteria transition to a symbiotic lifestyle. This phenomenon has already been well-documented in diatom spheroid bodies, particularly within various lineages of the order Rhopalodiales (Kneip et al., 2008; Nakayama et al., 2014; Nakayama and Inagaki, 2017; Schvarcz et al., 2024). Earlier research has demonstrated that after cyanobacteria become integrated as endosymbionts within diatom hosts, they undergo significant gene loss. This reduction often affects genes involved in light-dependent photosynthesis – as mentioned in the previous section – as well as those required for the biosynthesis of vitamin B12 (cobalamin) and chlorophyll a, indicating changes in metabolic roles within the partnership (Nakayama et al., 2014; Abresch et al., 2024).
These genomic changes in the spheroid body are also evident in R. sterrenburgii. As shown in Figure 4, the vitamin B12 pathway, derived from a branch point in the heme biosynthesis pathway (Grossman, 2016), is largely conserved among several endosymbionts of basal Rhopalodiales and Rhopalodia sensu lato diatoms. These diatom hosts retain a complete set of cobalamin biosynthesis genes in their SBs, including early steps (cobA, cbiX, cbiL) (Frank et al., 2005), intermediate modifications (cbiT, cbiE, cbiC) (Moore et al., 2013), and coenzyme assembly components (cobD, cobU, cobS) (Lawrence and Roth, 1995). In contrast, E. adnata and E. turgida exhibit varying degrees of gene loss. Notably, E. turgida lacks the pathway-initiating cobA gene and all downstream genes, indicating a complete disruption of vitamin B12 biosynthesis.
It seems to suggest that genes in the biosynthetic pathway for vitamin B12 are no longer essential within the intracellular host environment for some Epithemia SBs, that is why B12-associated genes are frequently lost or transferred to the nucleus. However, Nakayama and Inagaki (2017) noted the presence of an intact metH gene in the R. gibberula SB, which encodes the cobalamin-dependent methionine synthase (Croft et al., 2006). The metH gene is important because it makes an enzyme called methionine synthase, which helps turn a molecule called homocysteine into methionine, an essential building block for proteins. This process cannot happen without vitamin B12, which acts as a helper for the enzyme. Without the metH gene, the enzyme wouldn’t be made, and without vitamin B12, the enzyme wouldn’t work – so both are needed (Froese et al., 2019). This reinforces the functional relevance of the synthesized B12 within the endosymbiont. R. sterrenburgii also retains a functional 3,579 bp metH gene, confirming the presence of this enzyme in the Rhopalodia ‘gibberula’ group. On the contrary, the SB of E. turgida, which lacks the complete B12 biosynthetic pathway, also retains an intact metH gene. This suggests that even spheroid bodies who lost the B12 pathway may still rely on vitamin B12, raising the question of how SBs acquire it. A possible explanation is that the endosymbiont acquires vitamin B12 from external sources via uptake mechanisms. However, genes associated with the BtuCD-F ABC transporter system, which mediates vitamin B12 import (Balabanova et al., 2021), were not detected in the R. sterrenburgii SB. B12 uptake genes were also not identified in either the R. gibberula SB or Epithemia turgida SB genomes, so Nakayama and Inagaki (2017) proposed two possibilities: (1) a yet-unknown transporter intrinsic to the spheroid body, or (2) a host-derived transporter mechanism. The latter scenario implies potential endosymbiotic gene transfer (EGT), in which genes originally from the cyanobacterial symbiont were transferred to the host diatom genome and now functionally compensate by targeting proteins to the endosymbiont membrane (Jiroutová et al., 2010). Alternatively, the host may have evolved or modified a transporter of non-cyanobacterial origin to facilitate B12 delivery to the endosymbiont (Nakayama and Inagaki, 2017).
A different pattern emerges in the chlorophyll a biosynthesis pathway among the analyzed cyanobacterial endosymbionts, showing substantially greater pathway degradation compared to the vitamin B12 pathway. While early biosynthetic genes such as chlD, chlH, chlI, and chlM are often retained, genes involved in later stages, including chlE, bciB, chlB, chlL, chlN, and chlG, are frequently missing or inconsistently present across SB taxa. This widespread gene loss strongly suggests that light-dependent photosynthesis is no longer a primary function of these endosymbionts. For instance, E. adnata and E. turgida exhibit extensive losses, losing key genes necessary for magnesium chelation (chlD) (Sousa et al., 2013) and chlorophyll ring modifications (chlE, chlG) (Proctor et al., 2022). These observations align with previous reports of reduced or lost photosynthetic capacity in cyanobacterial endosymbionts of diatoms, where the host compensates by performing photosynthesis while the endosymbiont shifts its metabolic focus toward nitrogen fixation and other supportive roles (Moulin et al., 2024; Nakayama and Inagaki, 2017).
Supporting this metabolic specialization, Moulin et al. (2024) demonstrated that the E. clementina SB, exhibits continuous nitrogenase activity throughout day and night, different from its free-living relative Crocosphaera subtropica and Cyanothece, which temporally separates nitrogen fixation from photosynthesis (Reddy et al., 1993; Prechtl et al., 2004; Moulin et al., 2024). The metabolism of E. clementina SB is tightly integrated with the host, relying on host-derived carbohydrates rather than its own glycogen stores to fuel nitrogenase activity. This close connection allows the endosymbiont to use carbohydrates from the host efficiently (Prechtl et al., 2004), keeping only the key pathways needed to produce energy (ATP) and reducing power for nitrogen fixation while controlling oxygen levels (Moulin et al., 2024). Some lineages such as R. sterrenburgii still maintain partially complete chlorophyll a biosynthesis gene set, which may indicate a transitional evolutionary state. Overall, these patterns show that functions of diatom endosymbionts have moved away from photosynthesis and focus more on nitrogen fixation – closely coordinating their metabolic activities with the host to support this transition.
5 Conclusion
This is the first study to present and characterize the morphology, plastid, mitochondrial, and spheroid body genomes of the genus Rhopalodia together in a single paper. Our phylogenetic reconstruction based on nuclear, chloroplast, and mitochondrial genes revealed a basal placement of Rhopalodia sterrenburgii relative to R. gibba and its allies as well as the Epithemia species E. argus, E. turgida, and E. sorex, providing new insights into evolutionary relationships within this group of diazotrophic diatoms. The analysis also supports the need for re-evaluating the genus-level classification in the Rhopalodiales, which must be guided by integrative approaches that combine robust molecular, morphological, and ecological data – as ecological factors such as salinity, nutrient availability, pH, temperature, and light conditions strongly influence diatom distribution, physiology, and genome evolution. The inferred phylogenetic relationships revealed here provide a strong foundation for future taxonomic revisions and offered new insights into diatom diversification and evolution. Furthermore, we demonstrate that key biosynthetic pathways, specifically those involved in vitamin B12 and chlorophyll a synthesis, are being progressively lost in the more recently diverged lineages of Rhopalodia sensu lato and Epithemia sensu stricto. These findings suggest ongoing genome reduction and specialization among symbiotic diazotrophic diatoms.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
AC: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Project administration, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. MA: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Project administration, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. KK: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Project administration, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. MG: Data curation, Formal Analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. JL: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing. SH: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing. SM: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing. JK: Conceptualization, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was partially supported by the National Science Foundation (NSF) Grant No. 2222944.
Acknowledgments
We gratefully acknowledge financial support from the National Science Foundation (NSF). We also extend our thanks to the University of Colorado Boulder – Museum of Natural History and the Department of Ecology and Evolutionary Biology (EBIO) for their invaluable support and resources that made this research possible.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frpro.2025.1663791/full#supplementary-material
References
Abresch H., Bell T., and Miller S. R. (2024). Diurnal transcriptional variation is reduced in a nitrogen-fixing diatom endosymbiont. ISME J. 18, wrae064. doi: 10.1093/ismejo/wrae064
Adler S., Trapp E. M., Dede C., Maier U. G., and Zauner S. (2014). Rhopalodia gibba: the first steps in the birth of a novel organelle? Endosymbiosis 330, 167–179. doi: 10.1007/978-3-7091-1303-5_9
Aramaki T., Blanc-Mathieu R., Endo H., Ohkubo K., Kanehisa M., Goto S., et al. (2020). KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 36, 2251–2252. doi: 10.1093/bioinformatics/btz859
Geneious Prime 2025.1.1. Available online at: https://www.geneious.com (Accessed September 17, 2024).
Balabanova L., Averianova L., Marchenok M., Son O., and Tekutyeva L. (2021). Microbial and genetic resources for cobalamin (vitamin B12) biosynthesis: From ecosystems to industrial biotechnology. Int. J. Mol. Sci. 22, 4522. doi: 10.3390/ijms22094522
Boengler K., Heusch G., and Schulz R. (2011). Nuclear-encoded mitochondrial proteins and their role in cardioprotection. Biochim. Biophys. Acta (BBA)-Molecular Cell Res. 1813, 1286–1294. doi: 10.1016/j.bbamcr.2011.01.009
Bolger A. M., Lohse M., and Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bothe H., Tripp H. J., and Zehr J. P. (2010). Unicellular cyanobacteria with a new mode of life: the lack of photosynthetic oxygen evolution allows nitrogen fixation to proceed. Arch. Microbiol. 192, 783–790. doi: 10.1007/s00203-010-0621-5
Brettin T., Davis J. J., Disz T., Edwards R. A., Gerdes S., Olsen G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5, 8365. doi: 10.1038/srep08365
Capella-Gutiérrez S., Silla-Martínez J. M., and Gabaldón T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Caputo A., Nylander J. A., and Foster R. A. (2019). The genetic diversity and evolution of diatom-diazotroph associations highlights traits favoring symbiont integration. FEMS Microbiol. Lett. 366, fny297. doi: 10.1093/femsle/fny297
Croft M. T., Warren M. J., and Smith A. G. (2006). Algae need their vitamins. Eukaryotic Cell 5, 1175–1183. doi: 10.1128/ec.00097-06
Frank S., Brindley A. A., Deery E., Heathcote P., Lawrence A. D., Leech H. K., et al. (2005). Anaerobic synthesis of vitamin B12: characterization of the early steps in the pathway. Biochem. Soc. Trans. 33, 811–814. doi: 10.1042/bst0330811
Froese D. S., Fowler B., and Baumgartner M. R. (2019). Vitamin B12, folate, and the methionine remethylation cycle—biochemistry, pathways, and regulation. J. inherited Metab. Dis. 42, 673–685. doi: 10.1002/jimd.12009
Gaul U., Geissler U., Henderson M., Mahoney R., and Reimer C. W. (1993). Bibliography on the fine-structure of diatom frustules (Bacillariophyceae). Proc. Acad. Natural Sci. Philadelphia 144, 69–238. Available online at: https://www.jstor.org/stable/4065006.
Geitler L. (1977). Zur Entwicklungsgeschichte der Epithemiaceen Epithemia, Rhopalodia und Denticula (Diatomophyceae) und ihre vermutlich symbiotischen Sphäroidkörper. Plant Systematics Evol. 128, 259–275. doi: 10.1007/BF00984562
Greiner S., Lehwark P., and Bock R. (2019). OrganellarGenomeDRAW (OGDRAW) version 1.3. 1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47, W59–W64. doi: 10.1093/nar/gkz238
Grossman A. (2016). Nutrient acquisition: the generation of bioactive vitamin B12 by microalgae. Curr. Biol. 26, R319–R321. doi: 10.1016/j.cub.2016.02.047
Gupta A., Shah P., Haider A., Gupta K., Siddiqi M. I., Ralph S. A., et al. (2014). Reduced ribosomes of the apicoplast and mitochondrion of Plasmodium spp. and predicted interactions with antibiotics. Open Biol. 4, 140045. doi: 10.1098/rsob.140045
Hamsher S. E., Graeff C. L., Stepanek J. G., and Kociolek J. P. (2014). Frustular morphology and polyphyly in freshwater Denticula (Bacillariophyceae) species, and the description of Tetralunata gen. nov.(Epithemiaceae, Rhopalodiales). Plant Ecol. Evol. 147, 346–365. doi: 10.5091/plecevo.2014.990
Hamsher S. E., Keepers K. G., Pogoda C. S., Stepanek J. G., Kane N. C., and Kociolek J. P. (2019). Extensive chloroplast genome rearrangement amongst three closely related Halamphora spp. (Bacillariophyceae), and evidence for rapid evolution as compared to land plants. PloS One 14, e0217824. doi: 10.1371/journal.pone.0217824
Henderson M. V. and Reimer C. W. (2003). “Bibliography on the Fine Structure of Diatom Frustules (Bacillariophyceae): (deletions, addenda und corrigenda for bibliography I),” in Diatom Monographs, vol. 2. (Ruggell, Lichtenstein: Gantner).
Hustedt F. (1930). “Bacillariophyta (Diatomeae),” in Die Süsswasser-Flora Mitteleuropas, vol. 10 . Ed. Pascher A. (Zweite Auflage. Heft), 466 pp. Gustav Fischer, Jena.
Jeong Y. and Lee J. (2024). Comparative analysis of organelle genomes provides conflicting evidence between morphological similarity and phylogenetic relationship in diatoms. Front. Mar. Sci. 10. doi: 10.3389/fmars.2023.1283893
Jiroutová K., Kořený L., Bowler C., and Oborník M. (2010). A gene in the process of endosymbiotic transfer. PloS One 5, e13234. doi: 10.1371/journal.pone.0013234
Kamakura S., Mann D. G., Nakamura N., and Sato S. (2021). Inheritance of spheroid body and plastid in the raphid diatom Epithemia (Bacillariophyta) during sexual reproduction. Phycologia 60, 265–273. doi: 10.1080/00318884.2021.1909399
Katoh K., Misawa K., Kuma K. I., and Miyata T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Katoh K. and Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kneip C., Voβ C., Lockhart P. J., and Maier U. G. (2008). The cyanobacterial endosymbiont of the unicellular algae Rhopalodia gibba shows reductive genome evolution. BMC evolutionary Biol. 8, 1–16. doi: 10.1186/1471-2148-8-30
Kobayasi H. and Kobayashi H. (1988). A study of Epithemia amphicephala. In Östr.) comb. et stat. nov. and E. reticulate Kütz., with special reference to the areolar occlusion. Proc. 19 Int. Diat. Symp. Bristol, 459–468.
Kobayasi H., Nagumo T., and Tanaka (1993). “Rhopalodia iriomotensis sp. nov., a brackish diatom with shoehorn-shaped projections on the canal raphe (Bacillariophyceae),” in Progress in diatom studies, Contributions to taxonomy, ecology and nomenclature, vol. 106 . Ed. Sims P. A. (Koenigstein, Germany: Nova Hedwigia, Beiheft), 133–141. Special volume in honour of Robert Ross on the occasion of his 80th Birthday.
Kociolek J. P., Buczkó K., Greenwood M., Hamsher S., Miller S., and Li J. (2025b). Observations and typification of diatoms of the order Rhopalodiales (Bacillariophyta) in the Josef Pantocsek collection, Hungarian Natural History Museum. I. Epithemia species from Élesd. Diatom Res. 40, 1–8. doi: 10.1080/0269249x.2025.2480138
Kociolek J. P., Greenwood M., Hamsher S. E., Miller S., and Li J. (2024). Valve ultrastructure of Rhopalodia constricta (W. Smith) Krammer (Rhopalodiales, Bacillariophyceae) and a consideration of its systematic placement. Diatom Res. 39, 51–60. doi: 10.1080/0269249x.2024.2378769
Kociolek J. P., Greenwood M., Vouilloud A. A., Guerrero J., Sala S. E., Serino F., et al. Valve ultrastructure and systematic position of Rhopalodia wetzelii Hustedt (Rhopalodiales, Bacillariophyceae). Proc. Acad. Nat. Sci. Phil.
Kociolek J. P., Sala S. E., Guerrero J., Uyua N., Hamsher S. E., Miller S., et al. (2025a). Valve ultrastructure, systematics, and diversity of the Rhopalodiales. I. Introduction and consideration of morphological groups within the genus Epithemia Brébisson ex Kützing. Nova Hedwigia 120, 109–185. doi: 10.1127/nova_hedwigia/2025/1016
Krammer K. (1988a). “The gibberula-group in the genus Rhopalodia O. Müller (Bacillariophyceae),” in I: Observations on the valve morphology, vol. 46. (Heidleberg, Germany: Nova Hedwigia), 277–303.
Krammer K. (1988b). “The gibberula-group in the genus Rhopalodia O. Muller (Bacillariophyceae),” in II. Revision of the group and new taxa, vol. 47. (Heidleberg, Germany: Nova Hedwigia), 159–205.
Krammer K. and Lange-Bertalot H. (1988). “Bacillariophyceae,” in 2. Teil: Bacillariaceae, Epithemiaceae, Surirellaceae. Eds. Ettl H., Gerloff J., and Mollenhauer D. (Publisher location is: Jena, Germany: Band 2/2, Gustav Risher, Jena). Susswasserflora von Mitteleuropa.
Lange-Bertalot H. and Krammer K. (1987). Bacillariaceae, Epithemiaceae, Surirellaceae. Neue und wenig bekannte taxa, neue kombinationen und synonyme sowie bemerkungen und ergânzungen zu den Naviculaceae. Bibliotheca diatomologica 15, 1–289.
Lawrence J. G. and Roth J. R. (1995). The cobalamin (coenzyme B12) biosynthetic genes of Escherichia coli. J. bacteriology 177, 6371–6380. doi: 10.1128/jb.177.22.6371-6380.1995
Li H. and Durbin R. (2009). Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Moore M., Greenwood M., Kociolek P., Hamsher S., Miller S., and Li J. (2025). A new fossil species of Rhopalodia O. Müller (Bacillariophyceae, Rhopalodiales) from Mexico. Diatom Res. 40, 1–8. doi: 10.1080/0269249x.2025.2478955
Moore S. J., Lawrence A. D., Biedendieck R., Deery E., Frank S., Howard M. J., et al. (2013). Elucidation of the anaerobic pathway for the corrin component of cobalamin (vitamin B12). Proc. Natl. Acad. Sci. 110, 14906–14911. doi: 10.1073/pnas.1308098110
Moulin S. L., Frail S., Braukmann T., Doenier J., Steele-Ogus M., Marks J. C., et al. (2024). The endosymbiont of Epithemia clementina is specialized for nitrogen fixation within a photosynthetic eukaryote. ISME Commun. 4, ycae055. doi: 10.1093/ismeco/ycae055
Müller O. (1895). “Rhopalodia, ein Genus der Bacillariaceen,” in Botanische Jahrbücher für Systematik, Pflanzengeschichte und Pflanzengeographie Leipzig, vol. 22, 54–71.
Nakayama T., Ikegami Y., Nakayama T., Ishida K. I., Inagaki Y., and Inouye I. (2011). Spheroid bodies in rhopalodiacean diatoms were derived from a single endosymbiotic cyanobacterium. J. Plant Res. 124, 93–97. doi: 10.1007/s10265-010-0355-0
Nakayama T. and Inagaki Y. (2014). Unique genome evolution in an intracellular N2-fixing symbiont of a rhopalodiacean diatom. Acta Societatis Botanicorum Poloniae 83, 2–8. doi: 10.5586/asbp.2014.046
Nakayama T. and Inagaki Y. (2017). Genomic divergence within non-photosynthetic cyanobacterial endosymbionts in rhopalodiacean diatoms. Sci. Rep. 7, p.13075. doi: 10.1038/s41598-017-13578-8
Nakayama T., Kamikawa R., Tanifuji G., Kashiyama Y., Ohkouchi N., Archibald J. M., et al. (2014). Complete genome of a nonphotosynthetic cyanobacterium in a diatom reveals recent adaptations to an intracellular lifestyle. Proc. Natl. Acad. Sci. 111, 11407–11412. doi: 10.1073/pnas.1405222111
Narasimhan V., Danecek P., Scally A., Xue Y., Tyler-Smith C., and Durbin R. (2016). BCFtools/RoH: a hidden Markov model approach for detecting autozygosity from next-generation sequencing data. Bioinformatics 32, 1749–1751. doi: 10.1093/bioinformatics/btw044
Nurk S., Meleshko D., Korobeynikov A., and Pevzner P. A. (2017). metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27, 824–834. doi: 10.1101/gr.213959.116
Olson R. D., Assaf R., Brettin T., Conrad N., Cucinell C., Davis J. J., et al. (2023). Introducing the bacterial and viral bioinformatics resource center (BV-BRC): a resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. 51, D678–D689. doi: 10.1093/nar/gkac1003
Patrick R. M. and Reimer C. W. (1975). “Entomoneidaceae, cymbellaceae, gomphonemaceae, epithemiaceae,” in Monographs of the Academy of Natural Sciences of Philadelphia, vol. II/1. (The Diatoms of the United States, Philadelphia), 213p.
Pogoda C. S., Keepers K. G., Hamsher S. E., Stepanek J. G., Kane N. C., and Kociolek J. P. (2019). Comparative analysis of the mitochondrial genomes of six newly sequenced diatoms reveals group II introns in the barcoding region of cox1. Mitochondrial DNA Part A 30, 43–51. doi: 10.1080/24701394.2018.1450397
Prechtl J., Kneip C., Lockhart P., Wenderoth K., and Maier U. G. (2004). Intracellular spheroid bodies of Rhopalodia gibba have nitrogen-fixing apparatus of cyanobacterial origin. Mol. Biol. Evol. 21, 1477–1481. doi: 10.1093/molbev/msh086
Proctor M. S., Sutherland G. A., Canniffe D. P., and Hitchcock A. (2022). The terminal enzymes of (bacterio) chlorophyll biosynthesis. R. Soc. Open Sci. 9, p.211903. doi: 10.1098/rsos.211903
Rambaut A. (2018). FigTree–Tree Figure Drawing Tool Version v. 1.4. 4 (Edinburgh: Institute of Evolutionary Biology, University of Edinburgh).
Reddy K. J., Haskell J. B., Sherman D. M., and Sherman L. A. (1993). Unicellular, aerobic nitrogen-fixing cyanobacteria of the genus Cyanothece. J. bacteriology 175, 1284–1292. doi: 10.1128/jb.175.5.1284-1292.1993
Ruck E. C., Nakov T., Alverson A. J., and Theriot E. C. (2016). Phylogeny, ecology, morphological evolution, and reclassification of the diatom orders Surirellales and Rhopalodiales. Mol. Phylogenet. Evol. 103, 155–171. doi: 10.1016/j.ympev.2016.07.023
Schvarcz C. R., Stancheva R., Turk-Kubo K. A., Wilson S. T., Zehr J. P., Edwards K. F., et al. (2024). The genome sequences of the marine diatom Epithemia pelagica strain UHM3201 (Schvarcz, Stancheva & Steward 2022) and its nitrogen-fixing, endosymbiotic cyanobacterium. Wellcome Open Res. 9, 232. doi: 10.12688/wellcomeopenres.21534.1
Schvarcz C. R., Wilson S. T., Caffin M., Stancheva R., Li Q., Turk-Kubo K. A., et al. (2022). Overlooked and widespread pennate diatom-diazotroph symbioses in the sea. Nat. Commun. 13, 799. doi: 10.1038/s41467-022-28065-6
Sims P. A. (1983). A taxonomic study of the genus. Epithemia with special reference to the type species E. turgida (Ehrenb.) Kütz. Bacillaria 6, 211–235.
Sousa F. L., Shavit-Grievink L., Allen J. F., and Martin W. F. (2013). Chlorophyll biosynthesis gene evolution indicates photosystem gene duplication, not photosystem merger, at the origin of oxygenic photosynthesis. Genome Biol. Evol. 5, 200–216. doi: 10.1093/gbe/evs127
Spaulding S. A., Potapova M. G., Bishop I. W., Lee S. S., Gasperak T. S., Jovanoska E., et al. (2021). Diatoms. org: supporting taxonomists, connecting communities. Diatom Res. 36, 291–304. doi: 10.1080/0269249x.2021.2006790
Tillich M., Lehwark P., Pellizzer T., Ulbricht-Jones E. S., Fischer A., Bock R., et al. (2017). GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45, W6–W11. doi: 10.1093/nar/gkx391
Trifinopoulos J., Nguyen L. T., von Haeseler A., and Minh B. Q. (2016). W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235. doi: 10.1093/nar/gkw256
Tripp H. J., Bench S. R., Turk K. A., Foster R. A., Desany B. A., Niazi F., et al. (2010). Metabolic streamlining in an open-ocean nitrogen-fixing cyanobacterium. Nature 464, pp.90–pp.94. doi: 10.1038/nature08786
Vigneshwaran A., Liu Y., Kociolek J. P., and Karthick B. (2021). A new species of Epithemia Kützing (Bacillariophyceae, Rhopalodiales) from the Mula river, Western Ghats, India, with comments on the phylogenetic position of Rhopalodia and Epithemia. Phytotaxa 489, 171–181. doi: 10.11646/phytotaxa.489.2.5
Yao H., Xu Y., Lan Y., Xiang D., Jiao P., Xu H., et al. (2024). Codon usage bias analysis in the chloroplast genomes of diatoms within the family Thalassiosiraceae and Skeletonemataceae. doi: 10.21203/rs.3.rs-5343164/v1
Yılmaz E., Gastineau R., Solak C. N., Górecka E., Trobajo R., Turmel M., et al. (2024). Morphological and molecular characterization of Halamphora vantushpaensis (Bacillariophyceae, Amphipleuraceae), a new diatom species widely dispersed on the shores of the soda Lake Van (Türkiye). PhytoKeys 249, 95. doi: 10.3897/phytokeys.249.133205
Keywords: diatoms, symbiosis, plastomes, mitogenomes, spheroid body, morphology
Citation: Chang ACG, Amaral MWW, Keepers KG, Greenwood M, Li J, Hamsher SE, Miller SR and Kociolek JP (2025) Integrative taxonomy, whole organelle genomes and endosymbiosis in Rhopalodia sterrenburgii Krammer. Front. Protistol. 3:1663791. doi: 10.3389/frpro.2025.1663791
Received: 10 July 2025; Accepted: 07 October 2025;
Published: 20 October 2025.
Edited by:
Daria Tashyreva, University of Warsaw, PolandReviewed by:
Ab. Matteen Rafiqi, Bezmialem Vakıf University, TürkiyeTimothy G Stephens, The State University of New Jersey, United States
Copyright © 2025 Chang, Amaral, Keepers, Greenwood, Li, Hamsher, Miller and Kociolek. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aimee Caye G. Chang, YWltZWUuY2hhbmdAY29sb3JhZG8uZWR1; J. Patrick Kociolek, cGF0cmljay5rb2Npb2xla0Bjb2xvcmFkby5lZHU=
 Aimee Caye G. Chang
Aimee Caye G. Chang Mailor W. W. Amaral
Mailor W. W. Amaral Kyle G. Keepers
Kyle G. Keepers Megan Greenwood1
Megan Greenwood1 Jingchun Li
Jingchun Li Sarah E. Hamsher
Sarah E. Hamsher Scott R. Miller
Scott R. Miller J. Patrick Kociolek
J. Patrick Kociolek