Abstract
Among over 170 known RNA modifications, N6-methyladenosine (m6A) stands out as the most ubiquitous and extensively studied modification, found in different cellular RNA. The m6A plays a pivotal role in regulating RNA nuclear export, stability, secondary structure, translation, and degradation eventually determining the gene expression. The m6A modification is controlled by three classes of enzymes: “writers”, “erasers”, and “readers” which not only play a role in adding and removing the methyl group but also help in recognizing and interpreting the methylation marks. Although m6A has been widely studied in the context of metabolic diseases and cancers, its influence on infectious diseases remains under-explored. The infection cycle of several pathogens, including viruses, bacteria, and parasites is regulated by the m6A methylation machinery. In this review, we will not only explore the existing knowledge about m6A methylation but also its influence in shaping the pathogen life cycle paving our way to therapeutic aspects to mitigate infectious diseases.
1 Introduction
There are various RNA modifications that involve chemical alterations in RNA molecules, without changing the underlying sequences (Helm and Motorin, 2017), that regulate their expression and stability (Wang et al., 2015a). These modifications are collectively known as “epitranscriptomic modifications” (Helm and Motorin, 2017). With the recent advancement in technologies, more than 170 distinct RNA chemical modifications have been identified so far, in both coding and non-coding RNAs (Cappannini et al., 2024). Among these, some of the most extensively studied RNA modifications include N6-methyladenosine (m6A) found on eukaryotic mRNA (Desrosiers et al., 1974), 5-methylcytosine(m5C) found on rRNA, tRNA, ncRNA and mRNA (Wei et al., 1975a), Pseudouridine(ψ) observed in tRNAs, rRNAs, and snRNA (Schwartz et al., 2014a), N1-methyladenosine (m1A) predominantly found on tRNA and rRNA (Wu et al., 2022), and N4-acetylcytidine (ac4C) detected in tRNA, rRNA, and recently on mRNA (Arango et al., 2018).
Among these, N6-methyladenosine (m6A) is the most common internal modification (Desrosiers et al., 1974), found in the higher eukaryotic RNAs (Horowitz et al., 1984). The pioneering research conducted by Desrosiers, Friderici, and Rottman in 1974 led to the discovery of m6A methylation in polyadenylated RNA from Novikoff hepatoma cells (Desrosiers et al., 1974). The presence and biological significance of m6A methylation was further confirmed in 1975 by Perry and Kelly (Perry et al., 1975). Herein, adenosine is methylated at the N6 position (Desrosiers et al., 1975), which is incorporated both co-transcriptionally (Zhou et al., 2019) and post-transcriptionally (Wang et al., 2015a). m6A is highly conserved ranging from higher eukaryotes, plants, yeasts, and viruses (Gokhale et al., 2016a; Horowitz et al., 1984). The initial discovery methods for m6A methylation include radiolabelled nucleosides, oligo(dT)-cellulose chromatography, poly(A) (−)RNA fractionation, enzymatic degradation, and resolution of nucleosides (Desrosiers et al., 1974; Perry et al., 1975). Recent progression in m6A detection involves the use of m6A-specific antibodies by methods such as methylated RNA immunoprecipitation sequencing (MeRIP-seq or m6A-seq) (Schwartz et al., 2013), m6A individual-nucleotide-resolution cross-linking and immunoprecipitation (miCLIP) (Linder et al., 2015), qPCR-based methods (Zhang J. et al., 2019), mass spectroscopy (Kim et al., 2021) and many more. These transcriptome-wide mappings have shown that the majority of the m6A residues exhibit significant enrichment at the 3′ UTR and in proximity to stop codon (Dimock and Stoltzfus, 1977; Horowitz et al., 1984; Meyer et al., 2012; Schwartz et al., 2013). The predicted frequency of m6A in RNAs is approximately ∼three to five m6A sites in every mRNA of mammals (Desrosiers et al., 1974). The consensus sequence for m6A modification is characterized by the 5′-DRACH-3′(D = A/G/U, R = A/G, H = U/A/C) identified by the methyltransferase complex (Dominissini et al., 2012; Linder et al., 2015). m6A plays a critical role in various biological processes including the regulation of mRNA stability (Wang Y. et al., 2014), control of mRNA translation (Meyer et al., 2015; Xiao et al., 2016), RNA splicing (Dominissini et al., 2012; Ping et al., 2014), mRNA export from the nucleus to cytoplasm facilitating their translation (Wickramasinghe and Laskey, 2015), embryonic development (Ping et al., 2014; Xiao et al., 2016), and in maintaining stem cell pluripotency and differentiation (Batista et al., 2014).
This multifaceted role of m6A modification in regulating a variety of gene expressions has also been linked to the development and progression of multiple diseases. Dysregulation and abnormal cross-talk between m6A writers (Li et al., 2019a), erasers (Hu et al., 2022; Huang et al., 2019; Zhang Z. et al., 2019), and readers (Chen H. et al., 2022; Christiansen et al., 2009) can lead to implications in myriad pathological disorders including cancer (Barbieri et al., 2017; Yang et al., 2019; Zhang et al., 2016), neurological (Chokkalla et al., 2019; Zhang S. et al., 2022) and metabolic disorders (Christiansen et al., 2009; Regué et al., 2019), aging-associated (Liu et al., 2021a), pulmonary (Ma et al., 2021a), cardiovascular diseases (Mathiyalagan et al., 2019; Xing-Bo et al., 2018), and a variety of infectious diseases. In recent years, extensive research has explored various diseases, but the impact of m6A alterations in infectious diseases has not received much attention. This review will focus on recent advancements in understanding “the role of m6A RNA methylation in infectious diseases” and provide compelling insights into m6A as a potential therapeutic target. We will explore the diverse aspects of the pathogenicity of several infectious diseases including viral, bacterial, and parasitic infections along with their host-pathogen interactions. Exploring m6A, in the context of infectious diseases may open new avenues in studying immune responses which holds promising diagnostic strategies, clinical implications, and potential treatment. To understand the intricate cross-talk within m6A machinery, it is essential first to understand its main components.
2 Mechanism of the m6A methylation
In eukaryotes, adenosine methylation at the N6 position is predominantly carried out by three main protein complexes: “writers” (methyltransferases) (Agarwala et al., 2012; Bokar et al., 1994; 1997), “erasers” (demethylases) (Jia et al., 2011; Zheng et al., 2013), and “readers” (m6A effector complex) (Dominissini et al., 2012; Theler et al., 2014; Xiao et al., 2016). “Writers” are responsible for adding the methyl group (Bokar et al., 1994), while “erasers” have an opposite function as they remove the methyl group by oxidative demethylation (Jia et al., 2011). “Readers” bind to the methylated mRNAs and alter their functions (Dominissini et al., 2012). They all work together and play an essential role in regulating the m6A RNA methylome.
2.1 m6A methyltransferase “writer” complex
The methyltransferase enzyme complex mainly includes METTL3, METTL14, and WTAP which is accountable for m6A methylation. Inside the nuclear speckle, METTL3 serves as the core catalyst that transfers the methyl group from S-adenosylmethionine (SAM) to the targeted adenosine of mRNA (Bokar et al., 1997; Narayan and Rottman, 1988). METTL14 amplifies the catalytic efficiency of METTL3 by providing an RNA-binding scaffold(Wang et al., 2016). WTAP, though lacking catalytic function, but functions as an adapter and helps in the localization of methyltransferase complex (Agarwala et al., 2012; Ping et al., 2014). Several other enzymes such as METTL5 (van Tran et al., 2019), METTL16 (Pendleton et al., 2017), KIAA1429 (VIRMA) (Schwartz et al., 2014a), RNA binding motif15 (RBM15)/RBM15B (Patil et al., 2016), HAKAI, Zinc finger CCCH domain-containing 13 (ZC3H13) (Wen et al., 2018), and ZCCHC4 (Ma et al., 2019) have their individual role in the methylation process.
2.1.1 The METTL family
The METTL (Methyltransferase-like) family comprises of a group of enzymes that primarily work in the methylation of nucleic acids. The key members of the METTL family include METTL3 and METTL14 (Śledź and Jinek, 2016; Wang et al., 2016; Wang et al., 2014b). METTL3 was discovered in 1994 (Bokar et al., 1994), and subsequently, METTL14 was identified as an important component of methyltransferase complex (Liu et al., 2014a; Wang et al., 2016). Initially, METTL14 was thought to be a standalone methyl transferase catalysing m6A methylation besides METTL3 (J. Liu et al., 2014b) however, subsequent research have clarified that METTL3 and METTL14 assemble into a 1:1 single heterodimeric complex (1 MDa) inside the nuclear speckle. Both METTL3 and METTL14 work cooperatively to enhance methyltransferases activity where only METTL3 is responsible for catalytic reaction typically occurring co-transcriptionally while the mRNA is being synthesized (Bokar et al., 1997; Ping et al., 2014; Sommer et al., 1978) and its interaction with mRNA is stabilised by the coiled-coil N-terminus of METTL14 (Śledź and Jinek, 2016; Wang et al., 2016; Wang et al., 2014b). In a study, knocking out either METTL14 or METTL3 in HeLa and 293FT cells results in decreased levels of m6A methylation, indicating their importance in this reaction (Dominissini et al., 2012; Liu et al., 2014b). Additional members of the METTL family which participate in m6A RNA methylation processes are METTL4, METTL5, METTL7A and METTL16, etc. METTL16 methylates U6 snRNA, lncRNAs, and MAT2A mRNA, an enzyme that is crucial for SAM synthesis (Pendleton et al., 2017). It has the ability to methylate hairpin structures in MAT2A pre-mRNA, forming m6A in a C-m6A-G context regulating SAM homeostasis. It requires a specific RNA structure and the UACAGAGAA nonamer for its activity (Pendleton et al., 2017; Warda et al., 2017). METTL5, on the other hand, methylates the 18s rRNA at m6A by forming a heterodimeric complex with TRMT112, vital for ribosome biogenesis and function. It is located in the nucleolus and impacts the translation of protein and cell growth (Ignatova et al., 2020; van Tran et al., 2019).
2.1.2 WTAP (Wilms Tumour-1-associated protein)
WTAP is universally expressed as a nuclear protein (Horiuchi et al., 2013) that plays an essential role in regulating the m6A level in mRNA by interacting with METTL3 and METTL14 enzyme complex (Agarwala et al., 2012; Liu et al., 2014b; Ping et al., 2014; Zhong et al., 2008). It was initially identified in Arabidopsis (plant homolog, FIP37) (Zhong et al., 2008) followed by its characterization in yeast (yeast homolog, mum2) (Agarwala et al., 2012). Subsequently, the association between METTL3 and WTAP was identified in various types of mammalian cells (Liu et al., 2014b; Ping et al., 2014; Schwartz et al., 2014b). Although WTAP does not have catalytic activity, it is crucial for proper localization of METTL3 and METTL14 on the nuclear speckle as its knockout decreases m6A content and causes tissue-specific defects, leading to apoptosis (Liu et al., 2014b; Ping et al., 2014). WTAP has also been linked to cancer, where its upregulation in over 30% of cases causes acute myeloid leukaemia (AML) (Bansal et al., 2014; Weng et al., 2018).
2.2 m6A demethylases “eraser” proteins
The regulation of m6A plays a pivotal role in RNA metabolism and function. This dynamic system involves a delicate interplay between methyltransferases, responsible for adding methyl groups (discussed earlier), and demethylases like FTO (Fat Mass and Obesity-associated Protein) (Jia et al., 2011) and ALKBH5 (α-Ketoglutarate-dependent Dioxygenase AlkB Homolog 5), (Zheng et al., 2013), which effectively remove them. These demethylases, alongside methyltransferases, determine the m6A level and its functions (Meyer et al., 2012).
2.2.1 FTO
FTO, a protein related to obesity and an α-ketogluterate dependent Dioxygenase, was first identified as a demethylating enzyme for 3-meT (N3-methylthymidine) in ssDNA (Gerken et al., 2007) and 3-meU (N3-methyluridine) in RNA (Jia et al., 2008). It was later found to demethylate m6A in mRNA (Jia et al., 2011) although, its activity on m6A is exceptionally low compared to its resilient demethylation in m6A.m. (Hess et al., 2013; Jia et al., 2011; Mauer et al., 2017) which is another modified nucleotide found at the 5′UTR in mRNA and snRNA (Mauer et al., 2017; Wei C.-M. et al., 1975). Knockout studies showed that the depletion of FTO significantly increases m6A.m. but not m6A levels (Hess et al., 2013). FTO participates in various biological functions, including the regulation of body mass, tumorigenesis, and pre-adipocyte differentiation (Cui et al., 2017; Fischer et al., 2009).
2.2.2 ALKBH5
ALKBH5, a component of the AlkB family, functions as an enzyme for demethylating m6A modification in mRNA (Zheng et al., 2013). As indicated by its nuclear localization, its function in m6A demethylation is crucial for mRNA biogenesis within the nucleus (Linder et al., 2015). ALKBH5 regulates m6A levels in mRNA, with knockout increasing and overexpression decreasing the m6A content respectively (Garcia-Campos et al., 2019). It also has an essential role in germ cell development, as evidenced by impaired spermatogenesis and fertility in ALKBH5-deficient male mice (Tang et al., 2018; Zheng et al., 2013). Notably, ALKBH5 is significant in viral response by regulating m6A modification on viral transcripts and influencing host immune responses (Zheng et al., 2017). ALKBH5 also selectively demethylates m6A on RNA-RNA hybrids like FOXM1 mRNA and its antisense transcript AS-FOXM1 (Zhang et al., 2017). Although changes in m6A stoichiometry are subtle, the role of ALKBH5 in m6A regulation is critical for specific biological processes.
2.3 m6A “reader” proteins
The discovery of “writers” and “erasers” pioneered advancement in understanding RNA metabolism regulation. Subsequently, identification of m6A binding proteins, known as “reader” proteins including YT521-B homology (YTH) domain proteins like YTHDF1 (Wang et al., 2015b), YTHDF2 (Wang et al., 2014a), YTHDF3 (Li et al., 2017a; Shi et al., 2017), YTHDC1, and YTHDC2 as well as hnRNPs (Alarcón, Goodarzi, et al., 2015a; Liu et al., 2015; 2017) and IGF2BP1-3 (Huang et al., 2018; Zhou and Pan, 2018a) significantly expanded our knowledge (Stoilov et al., 2002; Theler et al., 2014). The ability of “readers” to specifically recognize m6A-modified RNA highlights their importance as key regulators in cellular biology.
2.3.1 DF family proteins
The DF family proteins are involved in cytosolic mRNA regulation, but their specific functions remain debated due to conflicting study results. DF2 is associated with mRNA decay by mediating the instability of m6A containing mRNAs, with its depletion increasing half-lives of mRNAs through transient P-body interactions (Wang X. et al., 2014). Despite having similar binding sites and RNA preferences (Patil et al., 2016), studies have shown that DF1 promotes translation by engaging with DF3, eIF3, and various other initiation factors, without significantly affecting mRNA stability (Li H.-B. et al., 2017; Shi et al., 2017; Wang et al., 2015b). More recent research suggests that all DF proteins may share similar functions, including promoting mRNA degradation and deadenylation (Du et al., 2016). This idea is further supported by studies on Zika and HIV viruses, where DF proteins caused the degradation of virus-encoded m6A transcripts (Gokhale et al., 2016a; Lichinchi et al., 2016a; Tirumuru et al., 2016a) highlighting their functional redundancy and a broad impact on RNA stability and translation regulation.
2.3.2 DC family proteins
DC1, originally recognized as a nuclear splicing regulator, uses its YTH domain to bind m6A-modified RNA and control alternative splicing (Xiao et al., 2016; Zhang et al., 2010). It also mediates an epigenetic role in gene silencing on the X chromosome through the action of noncoding RNA XIST, which relies on m6A sites (Patil et al., 2016). Whereas, the function of DC2 (cytosolic) is less understood but includes enhancing the translation of hypoxia-inducible factor-1 α (HIF1α) mRNA, and degradation (Tanabe et al., 2016). It is not yet known if these roles involve m6A recognition.
2.3.3 Heterogenous nuclear RNPs (hnRNPs)
Another class of m6A readers includes HNRNPC (Liu et al., 2015), HNRNPG (Liu et al., 2017), and HNRNPA2B1 (Alarcón, Goodarzi, et al., 2015b) which recognizes and binds to m6A-modified RNA through a “m6A switch” mechanism. This process involves m6A-induced structural changes in the stem-loop structure which weakens the base pairing within the m6A consensus sequence, exposing RNA binding motifs for these enzymes (Kierzek and Kierzek, 2003; Liu et al., 2015). HNRNPC interacts with the newly accessible uridine track, while HNRNPG binds to purine-rich regions. Knock-out study of HNRNPC suggests that it plays a significant role in maintaining stability and alternative splicing of target RNA while, HNRNPG also contributes to the formation of cellular granules, essential for processing and localization of RNA (Liu et al., 2015; Liu et al., 2017). The role of HNRNPA2B1 is debated as it binds through the m6A “switch mechanism”57 but, also involved in pri-miRNA maturation by interacting with DGCR8 protein complex, highlighting its dual function (Alarcón, Goodarzi, et al., 2015b; Alarcón, Lee, et al., 2015).
2.3.4 IGF2BP1-3
Insulin-like growth factor 2 mRNA binding proteins 1, 2, and 3 (IGF2BP1–3) are a group of m6A reader proteins that bind to the methylated mRNA through their KH (K Homology) domain, rather than typical YTH domain found in other m6A readers (Huang et al., 2018). IGF2BP1-3 has a role in stabilizing the m6A-modified mRNA, preventing degradation, and controlling gene expression. These proteins also play a significant role in cancer by enhancing the expression of oncogenic mRNA like MYC supporting the growth and survival of cancer cells (Huang et al., 2018; Zhou and Pan, 2018b). In upcoming sections, we discussed and highlighted the importance of m6A methylation in infectious diseases.
3 m6A methylation in infectious diseases
Infectious diseases are the major global health challenge causing approximately 13 million deaths every year (Gray and Sharara, 2022; Zumla, 2010). A range of harmful microorganisms including viruses, bacteria, parasites, and fungi are responsible for causing these ailments. Their mode of transmission can be airborne, waterborne, vector-borne, or via contaminated food and direct contact (Drexler and Institute of Medicine (US), 2010). Ranging from the Ebola epidemic to the recent COVID pandemic, infectious diseases have become a critical global concern.
In recent years, the involvement of m6A in infectious diseases has garnered significant attention. Several breakthroughs in infectious disease pathogenesis have highlighted the significance of m6A methylation in the interplay between host and pathogen, modulating the lifecycle and pathogenicity of infectious agents, thus emerging as a key player revealing novel therapeutic targets.
3.1 Viral infections
Apart from the presence of m6A in cellular mRNA, its presence was also discovered in viral transcripts about half a century ago in 1985 (Lavi and Shatkin, 1975a), which opened new chapters in the field of molecular biology. Simian virus 40 (SV40) is one of the first viruses in which m6A has been detected yielding almost 75% of the radioactivity (Lavi and Shatkin, 1975a). Subsequently, many more viral RNAs have been identified that contain m6A methylation, including Rous Sarcoma virus (Kane and Beemon, 1985), influenza virus (Krug et al., 1976), herpes simplex virus (Moss et al., 1977), and many other viral families. Numerous studies have documented the effect of m6A upregulation and deletion on viral replication, life cycle, and expression (summarized in Figure 1).
FIGURE 1

m6A Methylation in Viral Infection: The figure illustrates the role of m6A modification machinery components in modulating viral replication, protein expression, and latency across a variety of viruses. Each virus is associated with different m6A writers (METTL3, METTL14, METTL16), erasers (FTO, ALKBH5), and readers (YTHDF1-3, YTHDC1), which can either promote or inhibit viral replication. Green () arrows indicate the upregulation and red () arrows indicate the downregulation of m6A machinery. Each box represents viruses belonging to one family. *In HIV-1 infection knockout study on METTL3 showed reduced viral replication. Figure Created in BioRender, https://BioRender.com/n85v091
3.1.1 m6A in HIV-1
The Human immunodeficiency virus type 1 (HIV-1), a member of the Retroviridae family, involves an enveloped, positive-sense, single-stranded RNA virus which is a primary causative agent of acquired immunodeficiency syndrome (AIDS) (Humans, 2012). The first study to report the presence of m6A modification in HIV-1 RNA was conducted by Lichinchi et al. (2016). They showed that there is approximately a 30% increase in m6A levels, reported in cellular poly(A) RNAs that were infected with HIV-1. Its topological analysis identified 14 m6A peaks in both coding and non-coding sequences (Lichinchi, Gao, et al., 2016b). These results suggest that viral infections significantly increase the m6A levels in both viral and host cell RNA (Lichinchi, Gao, et al., 2016b). However, the exact demonstration of this regulation was still lacking. To gain insight, a study conducted by Tirumuru and Wu (2019), claimed that the HIV-1 envelope proteins are responsible for the m6A upregulation in CD4+ T-cells even without the virus actively replicating, suggesting that the presence of virus alone triggers the rise of m6A levels. They found a significant ∼3-7-fold increase in m6A levels upon infection with recombinant HIV-1 gp120 (Tirumuru and Wu, 2019).
To comprehend the impact of m6A machinery, studies have utilized gain and loss of function strategies to observe their corresponding effects. The knockdown of m6A writers, i.e., METTL3 or METTL14 resulted in reduced replication of the viral genome, as compared to control cells. Moreover, the concurrent depletion of both enzymes resulted in enhanced hindrance in viral replication (Lichinchi, Gao, et al., 2016b). Whereas, depleting the eraser enzyme ALKBH5 exhibited an increase in the same. Ultimately their findings demonstrated that both writers and erasers significantly affect the HIV-1 replication machinery (Lichinchi, Gao, et al., 2016b; Tirumuru et al., 2016b). Furthermore, a recent study reported that some small molecules activate the methyltransferase activity of METTL3/METTL14/WTAP complexes which also leads to an increase in m6A levels on both HIV-1 and host RNAs (Selberg et al., 2019; 2021). In 2016 two studies on m6A readers presented contradictory findings. Study conducted by Tirumuru et al. found that depletion of YTHDF proteins increased HIV-1 infection, while upregulation decreased it. Thus, YTHDF1-3 proteins function as negative regulators, suppressing the HIV-1 infection after viral entry by interfering with reverse transcription (Tirumuru et al., 2016b). Contrarily, a study by Kennedy et al. demonstrated that the silencing of YTHDF significantly reduced and overexpression boosted HIV-1 RNA expression and viral replication. Thus dedicating YTHDF proteins, especially YTHDF2 as positive regulators (Kennedy et al., 2016). This disparity in results might be due to methodological differences. However, the findings of other studies suggest a conflict with Tirumuru et al.’s statements and show that virions get degraded by viral protease and hence they cannot interfere with reverse transcription (Jurczyszak et al., 2020). Thus YTHDF proteins are considered to have a promoting effect on HIV-1 RNA expression (Jurczyszak et al., 2020; Kennedy et al., 2016; Lichinchi, Gao, et al., 2016b; Tsai et al., 2021).
3.1.2 m6A in Flaviviridae
The members Flaviviridae family involve enveloped, positive-sense, ssRNA viruses that impact both human and animal life (Holbrook, 2017). This family of vector-borne viruses includes a variety of members (Holbrook, 2017), notably, the presence of m6A methylation and its modulation has been identified in viruses including zika virus (ZIKV), dengue virus (DENV), yellow fever virus (YFV), hepatitis C virus (HCV), and japanese encephalitis virus (JEV) (Gokhale et al., 2016b; Yao et al., 2024).
Gokhale et al. (2016a), revealed an inhibitory effect on HCV production and replication (Gokhale et al., 2016b; Kim and Siddiqui, 2021). That means, the knockdown of MTases (METTL3+METTL14) increases the viral production while depletion of FTO decreased their levels, but no significant effect of ALKBH5 was found on the viral titer (Gokhale et al., 2016b). The impact of YTHDF1-3 proteins was found to increase HCV production on depletion16.
A further study by Lichinchi et al. (2016) (Lichinchi, Zhao, et al., 2016) reported the same negative regulatory effect of m6A methylation in ZIKV infection, as seen in HCV. Its topology showed 12 m6A peaks in the ZIKV genome. The knockdown of methylases (METTL3+METTL14) and demethylases (ALKBH5) resulted in increased and decreased viral titer respectively, and showed opposite effects when over-expressed. Likewise, silencing YTHDF1-3 led to increased viral replication and expression, with YTHDF2 showing the greatest effect (Lichinchi, Zhao, et al., 2016). A recent exploration of the JEV exemplified a somewhat opposite effect of m6A in its infection. The study claims that m6A methylation has a positive regulatory role in JEV infection (Yao et al., 2024), which is different from the rest of the family members. They identified five m6A peaks in the JEV RNA and upon infection attained more peaks. The METTL3, YTHDF1-3 silencing showed decreased JEV replication and expression. Whereas no significant effect was seen when writers and readers were overexpressed (Yao et al., 2024).
Furthermore, the relationship between m6A methylation and DENV has also been investigated in mosquito (Aedes aegypti) cells (Dai et al., 2022) with a positive regulatory role in DENV infection by modulating writers and readers. The depletion of METTL3 and METTL14 in DENV-infected cells showed a substantial decrease (about ∼three to four folds) in DENV RNA levels. A similar depletion of YTHDF3 caused decreased DENV production (Dai et al., 2022).
3.1.3 m6A in SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the primary causative agent of the COVID-19 pandemic, has had unprecedented global impact. It is a member of the Coronaviridae family and Betacoronavirus genera (Liu P. et al., 2021), which is characterized by enveloped, positive-sense single-stranded RNA viruses (Wu et al., 2020; Zhou et al., 2020). m6A profiling of SARS-CoV-2 has confirmed the presence of m6A modifications in the viral genome (Becker et al., 2024; Liu et al., 2021). A study conducted by Zhang T. et al. (2021) identified 13 m6A peaks in cells infected with SARS-CoV-2 (Zhang et al., 2021a). Additionally, research by Liu et al. (2021) concluded that SARS RNA gets methylated progressively during infection in host cells (Liu et al., 2021).
The interaction between m6A-related proteins and SARS-CoV-2 infection has been widely investigated. Research led by Liu and co-workers found that viral infection caused increased m6A methylation levels in both viral and host RNA (Liu C. et al., 2022). The knockdown of METTL3, METTL14, and YTHDF2 followed by SARS-CoV-2 infection resulted in increased viral replication and propagation. However, on ALKBH5 depletion, decreased viral replication was observed. Thus, concluding that m6A methylation controlled by the host’s cellular machinery has a negative regulatory role in SARS infection (Liu et al., 2021).
Augmenting the above results, a study by Zhang and co-workers (2021) found that overexpression of METTL3 decreases viral replication while FTO promotes it (Zhang et al., 2021b). Moreover, simultaneous knockdown of YTHDF1-3 significantly reduced positive cells by 72% (Burgess et al., 2021) suggesting that the m6A readers contribute to viral replication and could be a promising target for antiviral drug development (Burgess et al., 2021; Liu et al., 2021).
3.1.4 m6A in Herpesviridae
The herpesviruses possess a linear double-stranded DNA (dsDNA) genome (Feng et al., 2022b), which are characterized by their remarkable ability to establish lifelong infections and their biphasic life cycle, transitioning between both lytic and latent phases (Röder et al., 2022; Tan et al., 2018). Researchers have performed transcriptome-wide mapping of the m6A methylation and identified its presence in Herpes simplex virus (HSV) (Moss et al., 1977), Kaposi’s associated herpesvirus (KSHV) (Tan et al., 2018), Epstein-Barr virus (EBV) (Lang et al., 2019) and more recently in Pseudorabies virus (PRV) (Yu et al., 2023) in both lytic and latent transcripts.
Although the identification of m6A in HSV-1 was done in 1977 (Moss et al., 1977), its role in antiviral innate response of the host did not get much attention. However, in recent years, m6A modification was found to augment HSV-1 replication and infection (Feng et al., 2022b; Xu et al., 2023), while inhibiting m6A by 3-deazaadenosine (DAA), a methylation inhibitor, blocked the viral reproduction (Z. Feng et al., 2022b). Similarly, in HeLa and human rhabdomyosarcoma (RD) cells, enhanced viral infection resulted in increased expression of METTL3, METTL14, and YTHDF1-3 in an early stage of infection whereas decreased the FTO and ALKBH5 expression (Feng et al., 2022b). Knockdown assay of writers and readers suppressed the viral replication and reproduction (Feng et al., 2022b; Xu et al., 2023).
The KSHV has developed a way to hijack the m6A machinery of the host to enhance the lytic replication, thus indicating the crucial role of m6A in regulating gene expression during lytic phase of viral life cycle (Ye et al., 2017). The m6A reader, YTHDC1 along with SRSF3 and SRSF10 (serine/arginine-rich splicing factor) regulates the splicing process on ORF50 pre-mRNA, which is highly involved in the viral gene expression and successful lytic phase (Ye et al., 2017). Silencing METTL3 and YTHDF2 in the KSHV-infected iSLK.219 (recombinant KSHV.219) and iSLK.BAC16 (rKSHV Bacterial artificial chromosome 16) cell lines show reduced levels of viral lytic activator ORF50 and impaired new virions production, respectively. However, these results showed disparity when cell type was changed. In TREX-BCBL-1 cell lines (B-cell), the knockdown of METTL3 and YTHDF2 had an opposite effect with increased ORF50 levels. Thus, signifying that m6A regulation in KSHV infection is cell-type-specific (Hesser et al., 2018). Depletion of the other m6A writer METTL16 in iSLK-RGB-BAC16 cells enhanced the KSHV lytic replication by promoting viral gene expression. That means, in a reduced scenario virus switches from a latent state to a lytic state. These results highlight the vital role of METTL16 in inhibiting the KSHV lytic replication (Zhang et al., 2023).
Recent studies have uncovered significant involvement of m6A in EBV infection and its associated oncogenicity (Lang et al., 2019). The m6A modification has been documented to play a dual role by promoting latency and suppressing lytic gene expression. METTL14 plays a crucial role in maintaining EBV latency, as higher METTL14 levels correlate with latent phase stability. Reducing METTL14 expression disrupts latency and promotes lytic gene expression (Lang et al., 2019). METTL3 enhances the expression of EBV nuclear antigen 2 (EBNA2) (a transcriptional activator) in the Raji cell, thus promoting its stability and translation. Depleting METTL3, decreased the expression of EBNA2. Moreover, the knockdown of FTO resulted in increased gene expression of EBNA2. The impact of m6A readers on EBNA2 expression varied significantly. When YTHDF1 was reduced, EBNA2 levels dropped considerably, whereas reducing YTHDF2&3 led to increased expression levels (Zheng et al., 2021).
Another member of the Herpesvirus family, PRV is primarily known to infect porcine, but recent cases have shown to cause encephalitis in humans (Liu et al., 2021b). Yu et al. (2023) revealed a positive regulatory effect of m6A in PK15 cells (porcine kidney epithelial cells) with increased m6A levels during the infection. The knockdown of METTL3 and METTL14 resulted in decreased PRV replication, with METTL14 having more impact. Conversely, overexpression of METTL14 resulted in increased viral replication. Additionally, knockdown of ALKBH5 showed increased viral replication whereas FTO showed no significant effect. m6A readers also endorsed viral replication (Yu et al., 2023).
3.1.5 m6A in HBV
Hepatitis B virus (HBV), a member of Hepadnaviridae family, contains a double-stranded circular DNA and is a major human pathogen that is responsible for chronic liver diseases and capable of developing hepatocellular carcinoma (HCC) (Kim B. et al., 2020; Yang et al., 2023). HBV has been identified to contain m6A modifications that regulate the viral life cycle (Imam et al., 2018). As previously discussed, m6A modification occurs at specific consensus motifs, called “DRACH” sites. However, transcriptome-wide mapping of m6A on HBV RNAs revealed that these consensus motifs are located within the epsilon (ε) stem-loop at both 5′ and 3′ terminus, and have dual effects (Imam et al., 2018). Methylation at the 5′ terminus is beneficial for the reverse transcription of pgRNA (pregenomic RNA) whereas methylation at 3′ end obstructs the RNA stability and thus is more prone to degradation. Substituting adenosine by cytosine at these motifs resulted in disrupted methylation, which led to decreased efficiency of reverse transcription and production of viral proteins (Imam et al., 2018).
Moreover, both m6A methyltransferases and readers have been found to have a negative regulatory role in HBV protein expression. Depletion of METTL3, METTL14, YTHDF2, or YTHDF3 in HBV-infected cells showed increased viral protein (HBs and HBc) expression. However, the knockdown of erasers, FTO, and ALKBH5 had opposite effects and resulted in decreased expression (Imam et al., 2018). FTO suppression also resulted in the inhibition of HCC cell proliferation (Li et al., 2019b). O-GlcNAcylation (O-linked β-N-acetylglucosamine modification) of YTHDF2 is found significantly elevated in the HBV-infected cells which is responsible for the progression of hepatocellular carcinoma (Yang et al., 2023).
A recent study has uncovered the mechanism by which viruses evade the host’s innate immune system using m6A modification. Retinoic acid-inducible gene I (RIG-I) is a crucial detector of the viral RNA, as it provokes the production of type 1 interferon (IFN 1) and activates the innate immune response in the host cells. The study revealed that the presence of m6A can regulate and impair the ability of RIG-I to recognize the viral RNA by masking the methylated RNA and eventually hindering the RIG-I signalling due to increased binding of YTHDF2. Silencing the METTL3 and METTL14 activated the innate immune response after the robust detection of the viral RNA by RIG-1 (Kim B. et al., 2020). This strategy can be exploited to develop a potent antiviral vaccine, wherein depleting the m6A in viral RNA will promote enhanced immune recognition and trigger a protective response against viral infections.
3.1.6 m6A in Adenovirus
Adenoviruses (AdV) belong to the Adenoviridae family that have non-enveloped double-stranded DNA genome (Henquell et al., 2009). It causes various diseases ranging from the common cold, conjunctivitis, respiratory infections, and gastrointestinal illnesses (Hajikhezri et al., 2023; Lu and Erdman, 2006). Identification of m6A in adenoviral transcripts dates back to the 1970s which revealed its presence in both viral and cellular mRNAs (Sommer et al., 1976). In 2020, Price et al. utilized a combination of meRIP-seq and direct RNA long-read sequencing and identified m6A sites in both early and late adenoviral genes, as well as in host cellular transcripts (Price et al., 2020). They found unaltered levels of METTL3, METTL14, WTAP, YTHDC1, YTHDF1 and YTHDF2. Conversely, they observed a modest increase in FTO and ALKBH5. Depleting METTL3, negatively affected the splicing efficiency of adenoviral transcripts and thus impacted viral expression in late genes, whereas early genes remain relatively less affected (Price et al., 2020). This implies that the loss of METTL3 has a greater effect during the later stages of the viral life cycle and can be considered as a potent therapeutic target to control viral replication.
3.1.7 m6A in Ebola Virus
Ebola virus (EBOV) from the Filoviridae family contains a non-segmented, ssRNA genome and is reported to bear m6A. They are responsible for causing haemorrhagic fever in both humans and non-human primates (Martin et al., 2018; Wendt et al., 2023).The robust overexpression of METTL3 mildly inhibits viral processes whereas the knockdown of METTL3 reduced the viral RNA synthesis and protein expression. Other viruses that cause haemorrhagic fever, including Junín virus (JUNV) and Crimean-Congo haemorrhagic fever virus (CCHFV), are also influenced by METTL3, which regulates their transcription and protein expression (Wendt et al., 2023).
3.1.8 m6A in Influenza A Virus
Influenza A virus (IAV), belonging to the family Orthomyxoviridae (Zhu et al., 2023) is unique among RNA viruses for having m6A modifications in their complementary RNA (cRNA). The cRNA is a complimentary copy of viral RNA (vRNA) different from the non-methylated host mRNA. This modification in cRNA of IAV is likely to be mediated by the host nuclear enzymes, thus suggesting a relationship between viral replication and host cellular machinery (Krug et al., 1976). The m6A plays a positive regulatory role, when METTL3 was depleted, it reduced the gene expression and virion production. Conversely, overexpressing the reader YTHDF2 enhanced virion production and also promoted the spread of viral plaques. Interestingly, overexpression of other reader proteins, YTHDF1 and YTHDF3, does not significantly impact viral replication (Courtney et al., 2017). Additionally, YTHDC1 levels were elevated during IAV infection and its silencing repressed the viral replication (Zhu et al., 2023).
3.1.9 m6A in Chikungunya virus
The mosquito-borne Chikungunya virus (CHIKV) is a positive sense, ssRNA virus, belonging to the Togaviridae family, responsible for chikungunya disease characterized by high fever and severe joint pain (Jungfleisch et al., 2022; Kim et al., 2020a). The identification of m6A modification in CHIKV was done by (Kim et al., 2020a), using a novel technique named viral cross-linking and solid phase purification (VIR-CLASP) along with mass spectrometry to explore the relationships between host and viral machinery. These methods indicated the m6A presence and revealed the interaction between YTH-domain proteins and the CHIKV genome. The overexpression of YTHDF1 inhibited the viral replication whereas DF2 promoted it, with DF3 having a marginal influence on the replication (Kim et al., 2020a). However, another study by Baquero-Perez et al. (2024) challenges these findings and shows that the m6A methylation did not affect the replication of CHIKV, using LC-MS/MS, SELECT, m6A-seq, and nanopore sequencing. Moreover, the knockdown of writer or eraser did not impact the CHIKV infection (Baquero-Pérez et al., 2024). The authors also claimed the possibility of false positives achieved by methods like m6A-seq in previous reports (Kim et al., 2020a), or its presence at very low stoichiometry. These contradictory findings raise questions about the presence and functionality of m6A in cytoplasmic viruses like CHIKV. It also emphasizes the need for more robust methodological techniques.
3.1.10 m6A in Enterovirus-71
Enterovirus type 71 (EV71), a positive-sense single-stranded RNA virus belonging to the Picornaviridae family (Hao et al., 2019), is majorly accountable for hand-foot and mouth disease (HFMD) in children (Xiao et al., 2021; Zhu et al., 2021). During EV71 infection, m6A methylation was found to show a positive regulatory effect in the viral replication. Expression levels of METTL3, METTL14, YTHDF1-3, and YTHDC1 levels were enhanced while FTO levels were decreased. Depleting METTL3, YTHDF2, and DF3 showed reduced viral replication while FTO knockdown significantly promoted EV71 replication (Hao et al., 2019). In particular, METTL3 enhanced the translation of the EV71 open reading frame (ORF), promoting viral protein production and its replication. Deploying 3-deazaadenosine (3-DAA) which is an inhibitor of m6A, affected the translation of ORF and eventually the replication. Notably, m6A modification is also implicated in regulating EV71-induced apoptosis and autophagy. During EV71 infection the presence of METTL3 promotes autophagy, while its knockdown in Schwann cells reduces EV71-induced apoptosis which reduced viral titers (Xiao et al., 2021).
3.1.11 m6A in Simian Virus 40
SV40 belongs to the Polyomaviridae family, which is considered a tumorigenic DNA virus. It has been associated with the development of brain and bone cancers, non-Hodgkin’s lymphoma, and mesothelioma (Vilchez et al., 2003). The pioneering study that identified methylation in mRNA was conducted on Simian virus 40 (SV40) (Lavi and Shatkin, 1975b). Research conducted in the 1970s accurately mapped the internal methylation in SV40 mRNAs, revealing an average of three m6A methylation, all located in the coding region (Canaani et al., 1979, p. 40). However, at that time the role of m6A in regulating the viral life cycle was not broadly studied. A study directed by Finkel and Groner (1983), suggested the role of m6A in the transport of mRNA from the nucleus to the cytoplasm and the facilitation of efficient production of late mRNAs (Finkel and Groner, 1983). The m6A methylation in the SV40 infection has a positive regulatory role as the upregulation of the reader enhanced viral replication and its spread. Moreover, overexpressing YTHDF3 resulted in larger plaque formation than the control group. Conversely, when YTHDF2 and METTL3 were depleted it eventually inhibited the viral replication and spread (Tsai et al., 2018).
3.1.12 m6A in Respiratory syncytial virus
Respiratory syncytial virus (RSV) belongs to the Pneumoviridae family, with a non-segmented negative sense (NNS) RNA genome, majorly responsible for respiratory tract infections in children and adults (Xue et al., 2019). Detection of m6A in RSV RNA revealed the presence of 12 peaks, predominantly located in the 3′ end of the RNA (Csepany et al., 1990). m6A peaks were majorly identified on the antigenomic RNA of N, P, G, and F genes, with the highest abundance on the G mRNA (Xue et al., 2019). The m6A methylation in RSV is responsible for regulating viral replication, infection, and suppressing anti-viral host response (Picavet et al., 2024; Xue et al., 2019). The m6A readers (YTHDF1-3) and writers (METTL3 and 14) have a positive regulatory role whereas erasers (FTO and ALKBH5) have a negative regulatory role in gene expression and replication. Thus, the overexpression of writers and readers and the knockdown of erasers can enhance severity of the disease (Xue et al., 2019). However, a recent study suggests that not all readers play a positive regulatory role. For instance, the nuclear reader YTHDC1 plays a negative regulatory role by inhibiting viral entry (Picavet et al., 2024). Research led by Xue et al. (2021) highlights the implication of m6A manipulation for developing attenuated vaccines by promoting robust innate and adaptive responses. Depleting the methyltransferase machinery and thus making an m6A-deficient recombinant RSV RNA induces stronger type I interferon responses, ultimately boosting innate and adaptive immune responses. These mutants provided significant protection against the viral infection in cotton rats. Thus, targeting m6A methylation provides a promising strategy to improve vaccine design against RSV (Xue et al., 2021).
3.1.13 m6A in Vesicular Stomatitis Virus
Vesicular Stomatitis Virus (VSV), has a negative-sense, single-stranded RNA genome that belongs to the family Rhabdoviridae which primarily affects horses and cattle (Lu et al., 2021). The addition of m6A to viral RNA reduced the formation of dsRNA and diminished the ability of host to trigger antiviral signalling pathways including RIG-I and melanoma differentiation-associated gene 5 (MDA5) for helping VSV to evade the detection. The m6A writer METTL3 upregulation in VSV-infected HeLa cells resulted in enhanced viral mRNA production and replication by reducing the antiviral signalling. During VSV infection, METTL3 translocate to the cytoplasm and increases m6A modification thus reducing the host’s ability to recognize the virus which was further validated by knockout studies. These findings highlight a negative regulatory role of METTL3 for the host innate response during VSV infection (Qiu et al., 2021).
3.1.14 m6A in Severe Fever with Thrombocytopenia Syndrome Virus
The negatively stranded RNA genome of Severe Fever with Thrombocytopenia Syndrome Virus (SFTSV) of the family Phenuiviridae, also contains m6A modifications (Wang et al., 2021b). The m6A-seq analysis of SFTSV revealed the presence of approximately 11,520 m6A residues in both infected and healthy patients. These were located in the coding sequence of 3′UTR. However, after infection, there was a significant reduction in the abundance of m6A modifications on about 7,988 genes, suggesting the role of viral infection in the affecting methylation landscape. The FTO has been identified in greater abundance in infected patient samples (Wang et al., 2021b).
3.2 Bacterial infections
The m6A modification was first identified in viral transcripts, leading to a deeper understanding of its role in viral biology. Recently, however, exploration of m6A modifications in bacterial systems has begun to advance (summarized in Figure 2).
FIGURE 2
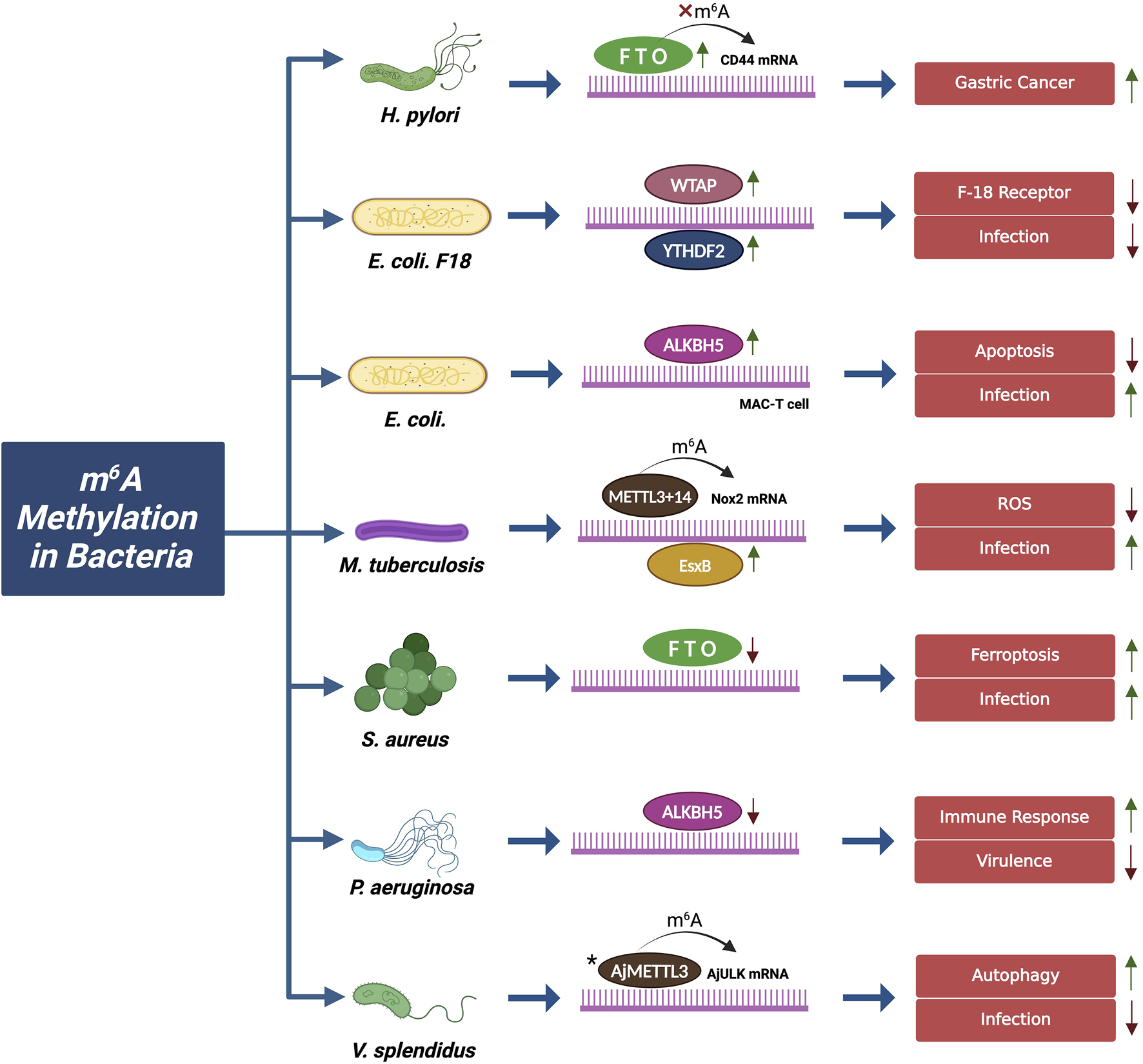
m6A Methylation in Bacterial Infection: The schematic illustrates the role of m6A modification machinery components in modulating various bacterial species and their effect on bacterial infection and pathogenesis. m6A proteins with green (↑) arrows show upregulation and red (↓) arrows show downregulation with their respective enhanced and repressed effects on infection. In V. splendidus, AjULK mRNA shows only its presence, with no upregulation and downregulation study. * Expression status is not studied (J. Liu et al., 2023). Figure Created in BioRender, https://BioRender.com/s09v404
3.2.1 m6A in Helicobacter pylori
Helicobacter pylori is a Gram-negative bacterium with a helical shape that is associated with chronic gastritis, peptic ulcers, intestinal metaplasia, and gastric cancer. The m6A methylation on 3′UTR of Lectin-like Oxidized Low-Density Lipoprotein Receptor 1 (LOX-1), a key receptor in H. pylori adhesion, downregulates its expression which in turn reduces bacterial invasion and adhesion on the gastric epithelial cells (Zeng et al., 2024a). This aligns with a study by Cheng et al. (2023a) that highlights the role of FTO in the malignant transformation of gastric epithelial cells (GES-1) during chronic H. pylori infection. Acute infection with the CagA + strain of H. pylori leads to apoptosis in GES-1 cells, whereas chronic infection results in reduced apoptosis and promotion of tumorigenic traits, such as increased proliferation and invasiveness. This malignant transformation is due to the elevated FTO expression, which further modulates m6A methylation on CD44 mRNA, a recognized marker of cancer stem cells. Silencing FTO reverses these transformations and reduces proliferation, migration, and tumorigenicity. Gastric cancer tissues showed higher levels of FTO and CD44, and elevated levels of FTO are associated with a poor prognosis in patients with gastric cancer. These studies highlight that FTO plays a pivotal role in H. pylori induced carcinogenesis, making FTO a potential therapeutic target for gastric cancer (Cheng et al., 2023a).
3.2.2 m6A in Escherichia coli
Escherichia coli, a Gram-negative pathogen is known to affect both humans and animals globally causing public health issues such as dysentery and mastitis. Research has highlighted how m6A modification influences the host cell response during E. coli infection. In intestinal porcine epithelial cells (IPEC-J2) when infected by E. coli F-18, the WTAP enhances the m6A level on N-acetyl lactosaminide beta-1,6-N-acetylglucosaminyl-transferase (GCNT2) mRNA at the 3′UTR, later destabilized by YTHDF2. This limits the development of E. coli F-18 receptor by controlling the biosynthesis of glycosphingolipids mediated by GCNT2, which in turn enhance E. coli infection (Wu et al., 2022a). Similarly, studies on bovine mammary epithelial cells (MAC-T cells) demonstrated that the exposure to E. coli increased m6A modification on LOC4191 (a lncRNA) to induce m6A-mediated apoptosis that served as a defensive mechanism and limited the E. coli infection by removing compromised cells. However, upregulation of ALKBH5 demethylates the modification on LOC4191 RNA reducing apoptosis and potentially allowing E. coli to evade the host’s defensive apoptotic response and enhances the infection (Xu et al., 2023a).
3.2.3 m6A in Mycobacterium tuberculosis
Mycobacterium tuberculosis (MTB) is an anaerobic, rod-shaped, Gram-positive bacteria responsible for causing tuberculosis (TB) worldwide. Several studies by Zhang et al. examined no association between m6A machinery components and their effect on pulmonary tuberculosis (PTB) susceptibility. However, their expression levels were relatively lower in PTB patients (Li et al., 2022a; Zhang et al., 2022b; Zhang et al., 2022c). But, recently Ma et al. (2024b) further explored the role of METTL14 in MTB infection. Upon MTB infection, p38 kinase get activated in macrophages which phosphorylates METTL14 at Thr72. Phosphorylated METTL14 undergoes liquid-liquid phase separation (LLPS) allowing it to form a complex with METTL3 more effectively. This complex of METTL3 and METTL14 methylates mRNA of the NAPDH oxidase 2 (Nox2) gene responsible for producing reactive oxygen species (ROS) for killing the MTB bacteria. However, an antigen EsxB (Rv3874), secreted by the bacteria, impairs the enzyme by deleting phosphorylation sites on METTL14, therefore, improving the survival of M. tuberculosis and enhancing the severity of the infection (Ma et al., 2024a).
3.2.4 m6A in Staphylococcus aureus
Staphylococcus aureus is another infectious bacterium, known to cause severe infections across species, leading to systemic inflammatory diseases such as sepsis. m6A modification plays a crucial role in S. aureus-induced sepsis by regulating gene expression through m6A-SNPs. A total of 15,720 m6A-cis-eQTLs (expression quantitative trait loci) and 381 m6A-trans-eQTLs were shown to be linked with sepsis, platelet degranulation, and various pathways involved in S. aureus infection. Apart from YTHDF3 (1 eQTL) and YTHDC2 (20 eQTLs), no other m6A regulator was found to be associated with sepsis (Sun et al., 2020). During S. aureus infection in bone marrow mesenchymal stem cells (BMSCs), elevated levels of m6A methylation due to reduced FTO were observed. This reduction in FTO leads to accumulation of Fe2+, lipid peroxide, and malondialdehyde (MDA) which causes oxidative damage and ferroptosis in the cell, augmenting the bacterial infection. But, experimental upregulation of FTO protects the cell from ferroptosis by destabilizing MDM2-TLR4 (mouse double minute two homolog-Toll-like receptor 4) signalling and enhancing the expression of antioxidant proteins like solute carrier family seven members 11 (SLC7A11) and glutathione peroxidase 4 (GPX4), thereby protecting cells from ferroptosis-induced damage (Song et al., 2024). Another similar study showed that S. aureus reduces FTO expression in macrophages which might alter FoxO1/NF-kB signaling to modulate the immune regulation and thus inflammation during infection (Liu et al., 2025). Hence, FTO can be used as potential therapeutic agent in managing S. aureus infections.
3.2.5 m6A in other bacteria
Infection patterns of other significant pathogens are also influenced by this epitranscriptomic modification. For instance, in Pseudomonas aeruginosa, the m6A modification is found on a distinct 5′-GCCAG-3′ motif compared to 5′-DRACH-3′ motif in the eukaryotes. The m6A-modified mRNAs such as regulator of secondary metabolite gene expression YZ (RsmYZ) and rhamnosyl transferase A and B (rhlAB) were responsible for inducing virulence-like biofilm formation and antibiotic resistance during the infection (Deng et al., 2015). In P. aeruginosa infected RAW264.7 cells, ALKBH5 knockdown led to increased m6A levels on numerous immune-related transcripts, resulting in the upregulation of 340 genes associated with immune responses hence, reduced the bacterial infection (Feng et al., 2022a). Similarly, in Vibrio splendid infection, m6A mRNA methylation plays a crucial role in modulating autophagy in Apostichopus japonicas. The AjMETTL3-mediated modification on Unc-51-like kinase 1(AjULK) mRNA recruits AjYTHDF, which promotes AjULK expression through its interaction with translation elongation factor 1-alpha (AjEEF-1α). This mechanism enhances the autophagic response and reduces the Vibrio splendidus infection (Liu et al., 2023).
3.3 Parasitic infection
Besides viruses and bacteria, m6A has become a significant regulatory mechanism in several parasitic protozoa essential for their development, survival, and infection cycle. Parasites, which rely on one or more hosts to complete their complex life cycle, must adapt to fluctuating environments and escape host immune responses. m6A methylation enhances their stability by promoting effective gene expression in reaction to environmental stress. Several parasitic organisms, such as Plasmodium (Baumgarten et al., 2019), Trypanosoma (Serra et al., 2024a), Toxoplasma (Holmes et al., 2021a), and Leishmania (Zhang et al., 2022a), exhibit this epitranscriptomic modification which contributes to their pathogenicity and survival within the host system (summarized in Figure 3).
FIGURE 3
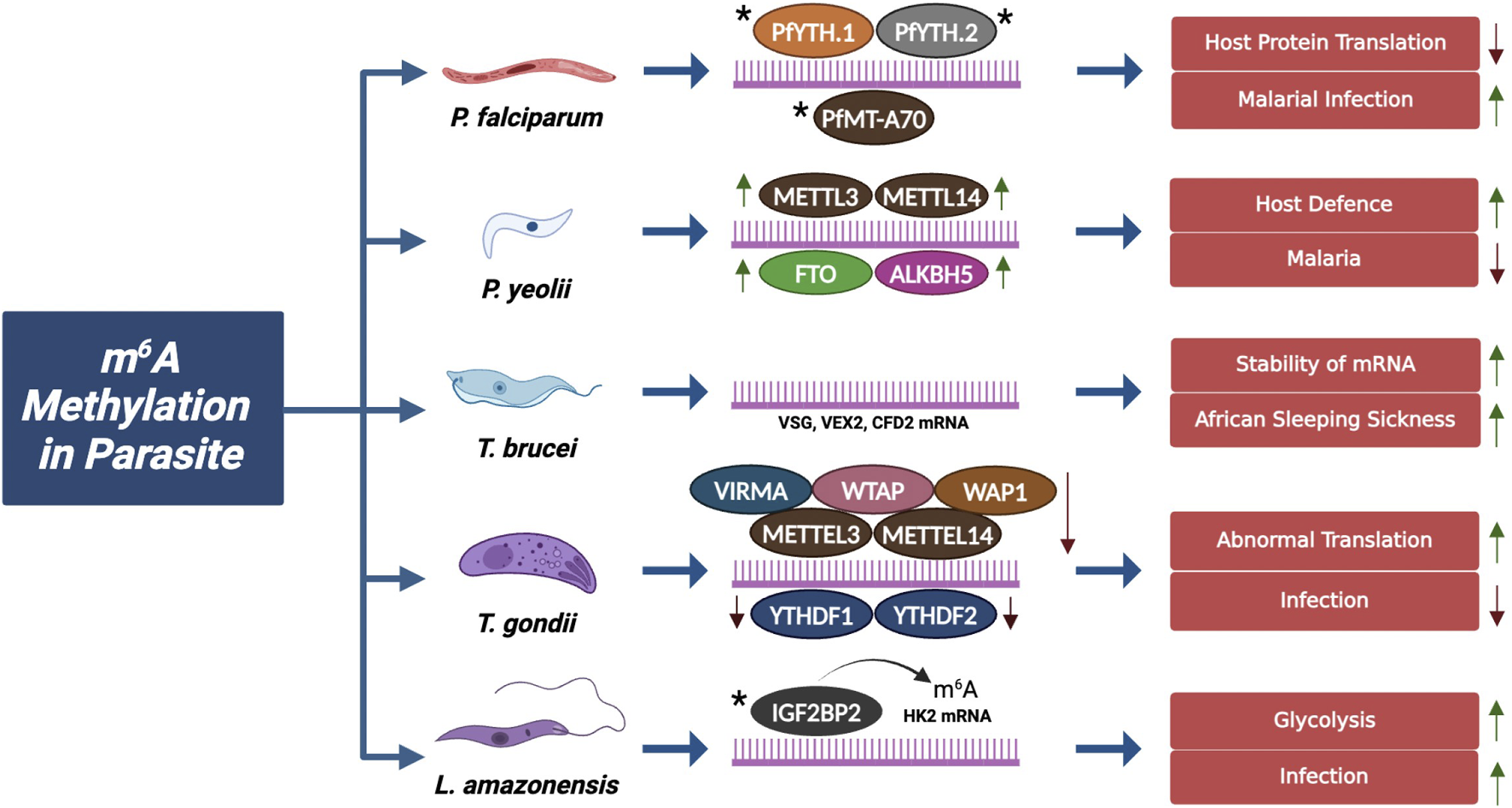
m6A Methylation in Parasitic Infection: The schematic illustrates the role of m6A modification machinery components in modulating various parasite species and their effect in facilitating their infection cycle. Green (↑) arrows indicate the upregulation with enhanced infection and red (↓) arrows indicate downregulation with suppressed infection. In L. amazonensis IGF2BP2 has no knockout studies, it only shows its presence. In Trypanosoma brucei, mRNAs do not comprise any m6A machinery. * Expression status is not studied (Liu et al., 2019; Serra et al., 2024a; Sinha et al., 2021). Figure Created in BioRender, https://BioRender.com/l22s429
3.3.1 m6A in Plasmodium sp.
Recent studies on Plasmodium falciparum have shown the role of m6A mRNA methylation during blood-stage development. PfMT-A70, an orthologue of m6A mRNA methyltransferase, is crucial for adding m6A marks to mRNA, and silencing studies have identified its role in mRNA degradation and lower translational efficiency (Baumgarten et al., 2019). Notably, P. falciparum lacks demethylases but two putative m6A binding YTH proteins (PfYTH.1 and PfYTH.2) were identified (Govindaraju et al., 2020a). Two similar studies by Govindaraju et al. and Sinha et al. characterized PfYTH.2, a key reader that binds specifically to m6A-containing mRNA with the help of F98 residue in the aromatic amino acid cage (Govindaraju et al., 2020a; Sinha et al., 2021). The interaction of PfYTH.2 with eIF3 and poly(A)-binding proteins along with knock out study indicate its role as a host translational repressor in P. falciparum. By binding to the m6A-modified mRNA, PfYTH.2 regulates the recruitment of the translation initiation factor eIF3, thereby controlling the translation of mRNA necessary for the development and survival of the parasite. Knock-out study revealed that disrupting normal cellular localization of PfYTH.2 led to a notable increase in translational efficiency across target transcripts, suggesting that PfYTH.2 normally acts as a host translational repressor (Sinha et al., 2021). A different species of Plasmodium, P. yoelii was also used to study m6A methylation during malarial infection. In infected mice, the m6A/A ratio ranged from 0.52% to 0.55%, compared to 0.23% in non-infected mice. The study found that METTL3, METTL14, ALKBH5, and FTO were upregulated. m6A modification on Ifit2, an interferon-stimulated gene, enhanced its expression in modulating host defence against malaria (Wang et al., 2022a).
3.3.2 m6A in Trypanosoma brucei
Trypanosoma brucei is a single-celled parasite that causes African sleeping sickness in humans and nagana in cattle affecting a large region of tropical Africa. It is found in two stages-bloodstream form (BSF), inside the human host, and procyclic form (PCF) inside the vector, i.e., tsetse fly. Using the method of m6A-CLIP, researchers found 355 m6A peaks across 251 genes in PCF and 95 peaks across 51 genes in BSF (Liu et al., 2019). In the BSF stage of the parasite, 50% of the modification occurs on mRNA encoding for variant surface glycoprotein (VSG), crucial for survival within the human host. The m6A methylation occurs on the poly (A) tail of the VGS transcript to enhance mRNA stability by preventing deadenylation (Viegas et al., 2022). Not only VGS but two important proteins, VSG-exclusion protein 2 (VEX2) and cyclin-like F-box 2 (CFB2), which regulate the gene expression of VGS were also found to be methylated (Serra et al., 2024a). However, the specific roles of m6A “writers”, “erasers”, and “readers” have not been investigated, indicating a significant research gap in this field.
3.3.3 m6A in Toxoplasma gondii
Another parasite responsible for causing food-borne illness and infection in immune-compromised patients is T. gondii. The infection cycle in Toxoplasma gondii is regulated by the m6A “writer” complex consisting of METTL3, METTL14, WTAP, a novel writer-associated protein (WAP1), and an uncharacterized VIRMA but, lacks eraser proteins. The reader proteins, YTH1 and YTH2, bind with m6A methylomes and help in the 3′end formation of mRNA by associating with the polyadenylation mechanism. A knockdown study of “writers” and “readers” showed that m6A is crucial for proper termination of transcription; its absence leads to abnormal gene expression suppressing T. gondii infection (Holmes et al., 2021a).
3.3.4 m6A in Leishmania amazonensis
Leishmania amazonensis is a protozoan parasite responsible for causing leishmaniasis, by invading and residing within the host immune cells such as macrophages. Similar to all other parasites, L. amazonensis utilizes m6A methylation machinery to regulate gene expression for infection and survival within the host. Normally, macrophages are present in either the M1 state (antimicrobial activity by aiding T helper type 1 response) or the M2 state (anti-inflammatory and promote T helper type 2 response), but, in Leishmania-infected macrophages (LIMs) markers of both the states were expressed. This process is controlled by the m6A “reader” protein IGF2BP2 which identifies and binds to m6A-modified transcripts of the glycolytic enzyme hexokinase-2, which increases the stability of these transcripts. This stabilization leads to higher expression of glycolytic enzymes, enhancing glycolysis and providing necessary energy to the parasite, aiding the survival of the pathogen (Zhang et al., 2022a).
4 Therapeutic potential of the m6A methylation in infection biology
Achieving the translation of mRNA in vitro for therapeutic applications took almost 2 decades as we have witnessed the development of mRNA vaccine against SARS-CoV-2 (Karikó, 2022; Sahin et al., 2014). Not only mRNA, drugs based on siRNA such as patisiran, inclisiran, gevosiran, and lumasiran and antisense oligonucleotides like golodisen were approved recently by FDA (Curreri et al., 2023).
Recently, the m6A modification has been exploited for therapeutic purposes to cure or monitor the progression of metabolic diseases such as cardiovascular complications and various types of cancers (Qiu et al., 2023). More than 50 inhibitors have been studied to target m6A methylation machinery, specifically writers METTL3/14, and erasers FTO and ALKBH5. These inhibitors include adenosine analogues, flavenoids such as quercetin and scutellarin, allosteric inhibitors, receptor agonists, and other natural compounds such as Rhein and radicicol, etc. along with amine derivatives like MU06 (Liao et al., 2022; Qiu et al., 2023; Wang et al., 2018; Yan et al., 2018; Yankova et al., 2021). Developing inhibitors against readers (YTH domain-containing proteins) is also of keen interest in regulating the functional consequences of m6A modification. The YTH domain recognizes m6A methylation and the organoselenium compound ebselen disrupts this recognition by interfering with RNA binding properties of YTHDF1-3 but not of YTHDC1 (Micaelli et al., 2022). Another study on the YTHDF1 reader showed that the deletion of YTHDF1 in hepatocellular carcinoma reduced the stemness and drug resistance against lenvatinib and sorafenib anti-cancer drugs. YTHDF1 enhances tumorigenesis by stabilizing m6A methylated NOTCH1 mRNA (Zhang et al., 2024). Interestingly, the anti-cancer drugs either target m6A methylation pathways or are used in combinatorial therapy with m6A methylation targets were shown to be more effective in treating different cancers. For example; in non-small cell lung cancer and cervical cancer, targeting FTO and METTL14 with gefitinib and sorafenib treatments leads to improved tumor suppression respectively (Chen et al., 2022a; Li et al., 2024). The drug resistance against tyrosine kinase inhibitors can be improved by reducing the expression of m6A eraser FTO (Yan et al., 2018). A detailed discussion on the types, purpose, and bedside clinical success is out of the scope for this review but we mentioned the future therapeutic directions particularly related to infectious diseases.
As discussed earlier, developing a vaccine against RSV is still challenging due to host-virus interactions which results in compromised adaptive and innate immune responses after RSV infection. Impairing with m6A methylation either by knockdown of METTL3 or using defective RNA (by removing putative m6A sites) resulted in an activated interferon response pathway which enhanced innate immune response followed by enhanced T cell activation during adaptive response. This study can be implemented in either RNA-based vaccine production or designing m6A defective viral particles to be used in attenuated viral vaccine production (Xue et al., 2021).
A similar study in HSV-1 infection identified the role of m6A methylation machinery in facilitating strong viral replication and propagation at the early stage of the infection. HSV-1 upregulated the expression of METTL3/METTL14 and oppositely reduced the expression of FTO and ALKBH5 for the robust m6A methylation of viral RNAs. Both depletion of m6A methylation machinery components (METTL3 and YTHDF3) and inhibition of m6A methylation by 3-deazaadenosine severely reduced viral replication and further spread, suggesting METTL3 and YTHDF3 mediated therapeutic avenues to be explored (Feng et al., 2022b).
Research on deadly SARS-CoV-2 and less virulent HCoV-OC43 identified that both viruses use m6A methylation for their replication and propagation. SARS-CoV-2 significantly induces the nuclear localization of cytoplasmic METTL3 and YTHDF1-3 to enhance m6A methylation of the viral RNAs, especially for enhanced production of nucleocapsid (N) protein. This study also highlights the importance of exploiting m6A methylation therapeutic purpose to combat the SARS-CoV-2 infection (Burgess et al., 2021).
It would be interesting to note here that bacteria and viruses lack their own m6A methylation machinery genes but parasites such as P. falciparum and T. gondii have their own m6A methylation writers and readers. The P. falciparum writer PfMT-A70 is an orthologue to mammalian METTL3 but readers such as PfYTH1 and PfYTH2 are less similar to mammalian counterparts other than conserved YTH domain which is essential to identify methylated adenosine (Baumgarten et al., 2019; Govindaraju et al., 2020b). Unlike YTHDC1 which prefers guanine at −1 and cytosine at +1 position to bind m6A, PfYTH2 is more permissible for the flanking sequences which is similar to YTHDF1-2 proteins. In case of T. gondii, YTH1 and YTH2 show homology with plant like reader protein known as polyadenylation specificity factors 30 (CPSF30, contains characteristic zinc finger domain) and are required for m6A dependent 3′end processing on mRNA transcripts (Holmes et al., 2021a). Thus, selecting m6A methylation genes for bacterial and viral infections for identifying therapeutic options is easier but future findings are absolute to wisely target parasitic m6A machinery by identifying key similarities and differences in protein sequence and structures.
Overall, RNA therapeutics is a newly emerging field for designing new cost-effective, and personalized medicines. Currently, no therapeutic drugs specifically targeting the m6A network have been approved for clinical use or are in preclinical trials. However, research in this area is promising, and several studies have highlighted the potential of m6A modification as a therapeutic target for various diseases, particularly cancer. In this review, earlier we have mentioned such potent small-molecule inhibitors of m6A machinery (Micaelli et al., 2022; Selberg et al., 2019; Yankova et al., 2021) that are highly feasible for clinical applications in infectious diseases. Technical advancements in next-generation sequencing play a crucial role in uncovering regulatory mechanisms governed by different types of RNAs and open limitless possibilities to add novel RNA-based drugs. The mRNA vaccine against SARS-CoV-2 exploited different RNA modifications such as pseudouridylation, m5A, m1A, etc. to stabilize mRNA and reduce immunogenicity (Granados-Riveron and Aquino-Jarquin, 2021). These modifications are highly understudied in infectious diseases, along with their interplay with m6A methylation as well as with other epigenetic markers such as histone modifications and DNA methylation.
5 Discussion and conclusion
The methylation is catalyzed by a methyltransferase-like (METTL) family of proteins having S-adenosyl methionine (SAM) binding domain on DNA, RNA, and proteins, as their molecular substrates. A plethora of functions of different METTL proteins have been identified in cancer and stem cell biology. METTL3-METTL14 (act as heterodimer), METTL16, METTL4, METTL7A, and METTL5 all catalyze m6A methylation on mRNA, rRNA, tRNA, miRNA, snRNA, snoRNA, lncRNA, and mitochondrial DNA (Tooley et al., 2023). As we have observed in above sections, HIV, HCV-1, PRV, IAV, EV71, SV40, RSV, VSV, and adenovirus increase the m6A methylation either by activation of m6A writers METTL3 and METTL14 or by increase in readers such as YTHDF1-3 or by modulation of erasers like ALKBH5 for their viral replication (Kennedy et al., 2016; Lichinchi, Gao, et al., 2016b; Selberg et al., 2021; Tirumuru et al., 2016b). Additionally, there are evidences that YTH reader proteins recruit RNA-binding proteins to influence infection in certain viruses. In HBV infection, YTHDF2 recruits 3′-5′ exonuclease Interferon-stimulated gene 20 (ISG20) and binds to the epsilon-stem loop structure on the viral RNA for its degradation (Imam et al., 2018; Liu et al., 2017). Whereas, in KSHV infection, YTHDC2 protects the cellular mRNA by interacting with the m6A modified SOX resistance element in the Interleukin-6 (IL-6) transcript to prevent the degradation from the viral endoribonuclease ORF37 (SOX) (Macveigh-Fierro et al., 2022).
Oppositely, in HCV, ZIKA, SARS-CoV-2, HBV, and SFTSV infection, m6A methylation has the reverse role as it reduces the viral replication. The silencing of METTL3 and METTL14 increased the viral production in contrast FTO deletion decreased the viral load (Gokhale et al., 2016b; Imam et al., 2018; Lichinchi et al., 2016; Liu et al., 2021; Wang et al., 2021b; Yao et al., 2024).
The role of METTL4, METTL7A, and METTL5 is not identified in any of the above-mentioned infections. Other family members such as METTL1 add methyl group on guanine, METTL6, METTL8, METTL17 on cytosine, and METTL19 on uracil on different types of RNA. Interestingly METTL3, METTL10, METTL11, METTL12, and METTL22 are also responsible for methyl group catalysis on various protein substrates (Tooley et al., 2023). The methylation specificity (m6A, m5C, m6A.m., m7G) and substrate identification (type of RNA) are still under investigation. Most pathogens usually inactivate, break, deceive, silence, or escape the innate and adaptive immune system pathways for their survival, proliferation, and spread. The role of m6A methylation and other METTLs in infection biology has not been explored which is absolutely needed to fill the gap of immune regulatory mechanisms. As shown in other diseases, m6A methylation is involved in anti-tumor, anti-inflammatory, and anti-viral immunity. The m6A writer METTL3, readers YTHDF1-3, and erasers FTO and ALKBH5 have been reported to regulate key immune pathways like macrophage polarization, dendritic cell activation, and antigen presentation (Li et al., 2017b; Ma et al., 2021b).
In bacterial infections a few cell line studies on m6A methylation during E. coli, M. tuberculosis, S. aureus, P. aeruginosa, and H. pylori infections have identified specific regulatory roles either benefiting bacterial growth or inhibiting bacterial virulence. For example,; during H. pylori infection with gastric epithelial cells, m6A methylation of LOX-1 and CD44 transcripts reduced the bacterial pathogenesis whereas the methylation enhances the tumorigenic properties of epithelial cells post H. pylori infection (Cheng et al., 2023b; Zeng et al., 2024b). Similarly, in M. tuberculosis pathogenesis, METTL14 phosphorylation is suppressed by bacteria to decrease ROS production thus supporting bacterial survival (Ma et al., 2024b). During E. coli F-18 infection in IPEC-J2 cells, m6A methylation enhances the host response by increasing m6A levels of GCNT2 which in turn limits infection (Wu et al., 2022b). Another study with MAC-T cells showed that E. coli infection increases m6A modification on a long noncoding RNA, LOC4191 which enhances the cell apoptosis and thus reduces the severity of infection (Xu et al., 2023b). Additionally, YTHDF1 shows interaction with DEAD domain in the host factor DDX60 (RNA helicase) to bind with the m6A-modified mRNA and increase translation of TRAF6 molecule involved in nuclear factor-κB (NF-κB) signalling enhancing innate immune response (Zong et al., 2021). In S. aureus infection, m6A methylations of bacterial rRNA and host circRNA play a crucial role in regulating infection events such as inflammation and cell apoptosis (Xu et al., 2022).
m6A methylation is extensively discussed for its application as potential biomarkers in various diseases such as diabetes, cardiovascular disease and cancer (Li et al., 2022b; Li et al., 2023; Liu et al., 2022a; Liu et al., 2022b; Tang et al., 2023). Though we have limited research in infectious diseases but very recently in human patient’s datasets, four genes including YTHDF1, HNRNPC, LRPPRC, and ELAVL1 have been identified and suggested as biomarkers for M. tuberculosis infection. These results were experimentally validated using RT-PCR and the expression of all four genes was significantly lower in MTB infected patients compared to the healthy control (Ding et al., 2024). Similarly, another computational analysis in healthy and SARS-CoV-2-infected patients has identified 18 m6A machinery regulators that were differentially expressed and reported that eight genes could be used for predicting COVID-19 infection. Among these, VIRMA, HNRNPA2B1, and RBM15B were shown to be protective factors whereas elevation of METTL3, RBM15, IGFBP3, FMR1 and ELAVL1 were associated with increased pathogenicity of the virus (Qing et al., 2022). The two m6A readers HNRNPA2B1 and RBM15 were shown to be important for predicting the prognosis of hepatitis B virus mediated hepatocellular carcinoma (Fang and Chen, 2020). Though, the experimental validation and further efforts for clinical applications for these studies are still awaited.
The m6A regulation is highly dynamic in parasitic infections. Apicomplexa parasites such as P. falciparum and T. brucei have their own m6A machinery. The Plasmodium m6A reader protein, PfYTH.2 interacts with eIF3 and poly(A)-binding proteins and reduces the host protein translation to promote parasite survival (Sinha et al., 2021). P. yoelii infection also increases m6A methylation but this pathogen enhances the expression of host m6A writers and easers. Now, this intrigues an interesting question that the increase in the writer’s expression results in a 52% increase in the m6A/A ratio which should be reduced after an increase in eraser expression. It is possible that P. yoelli might be directing easers in other pathways to support parasitic infection (Wang et al., 2022b).
During T. gondii infection m6A methylation gets upregulated by its own writer complex having WAP1 as a novel component compared to other eukaryotes. Identifying similar novel components will help to understand the site and stimulus-specific regulation of m6A methylation. Hypermethylation of RNA targets helps T. gondii in establishing the infection as the knockdown of m6A machinery suppresses the pathogenesis (Holmes et al., 2021b; Serra et al., 2024b). Leishmania uses the reader protein IGF2BP2 to bind with hypermethylated transcripts such as hexokinase-2 two which in turn supports the parasite survival in the host macrophage (Wang et al., 2021a).
So far, no RNA based therapeutic drug targets have been identified in parasites. The absence of DNA methylation in certain parasites may suggest a dependence on RNA modifications like m6A for gene regulation, but this area is unexplored to provide sufficient evidence to draw definitive conclusions.
Since 1928, infectious diseases have been efficiently treated with the help of antibiotics (Fleming, 1929). However, the widespread excessive use of antibiotics has led to antibiotic resistance in bacterial species, leading to severe health threats worldwide (Park et al., 2010). One of the major causes for resistance development is due to m6A methylation on the bacterial 23S rRNA catalysed by Erm (erythromycin resistance methylase)-type rRNA methyltransferases on nucleotide A2058 (Weisblum, 1995; Graham and Weisblum, 1979; Stojković et al., 2016). This modification is present in E. coli, S. aureus, Streptomyces spp. Bacillus spp. etc., providing resistance towards macrolides, lincosamides, and streptogramin B (MLS) antibiotics (Weisblum, 1995; Graham and Weisblum, 1979; Sharkey et al., 2022; Svetlov et al., 2021). The basic mechanism involves the addition of two methyl groups on the N6 amine of A2058 by Erm methyltransferase leading to steric hindrance between MLS antibiotics and ribosome, rendering the antibiotics ineffective to inhibit translation of bacterial protein, immune evasion by preventing recognition by Toll-like receptors (TLRs) and contributing to bacterial fitness (Sharkey et al., 2022; Shields et al., 2024; Svetlov et al., 2021). Hence, understanding the molecular and structural basis of such resistance mechanisms is important for developing novel RNA based antimicrobial strategies.
Our keen observation on the role of m6A methylation in infection biology leads us to conclude that every pathogen modulates m6A methylation in their unique way thus suggesting the stimuli-specific regulatory mechanism. Some pathogens modulate the writer complex; the others modify either readers or easers. The complex interactome and reason behind this selection is unknown. This certainly warns extensive research to control deadly infections and generate a healthy therapeutic armoury to overcome the exponentially increasing drug resistance.
Statements
Author contributions
NY: Conceptualization, Validation, Writing–original draft, Writing–review and editing. ReK: Conceptualization, Visualization, Writing–original draft, Writing–review and editing. SG: Software, Validation, Visualization, Writing–original draft, Writing–review and editing. RaK: Conceptualization, Formal Analysis, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Agarwala S. D. Blitzblau H. G. Hochwagen A. Fink G. R. (2012). RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLOS Genet.8 (6), e1002732. 10.1371/journal.pgen.1002732
2
Alarcón C. Goodarzi H. Lee H. Liu X. Tavazoie S. Tavazoie S. F. (2015a). HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cell162 (6), 1299–1308. 10.1016/j.cell.2015.08.011
3
Alarcón C. Goodarzi H. Lee H. Liu X. Tavazoie S. Tavazoie S. F. (2015b). HNRNPA2B1 is a mediator of m(6)a-dependent nuclear RNA processing events. Cell162 (6), 1299–1308. 10.1016/j.cell.2015.08.011
4
Alarcón C. R. Lee H. Goodarzi H. Halberg N. Tavazoie S. F. (2015). N6-methyladenosine marks primary microRNAs for processing. Nature519 (7544), 482–485. 10.1038/nature14281
5
Arango D. Sturgill D. Alhusaini N. Dillman A. A. Sweet T. J. Hanson G. et al (2018). Acetylation of cytidine in mRNA promotes translation efficiency. Cell175 (7), 1872–1886.e24. 10.1016/j.cell.2018.10.030
6
Bansal H. Yihua Q. Iyer S. P. Ganapathy S. Proia D. A. Penalva L. O. et al (2014). WTAP is a novel oncogenic protein in acute myeloid leukemia. Leukemia28 (5), 1171–1174. 10.1038/leu.2014.16
7
Baquero-Pérez B. Yonchev I. D. Delgado-Tejedor A. Medina R. Puig-Torrents M. Sudbery I. et al (2024). N6-methyladenosine modification is not a general trait of viral RNA genomes. Nat. Commun.15 (1), 1964. 10.1038/s41467-024-46278-9
8
Barbieri I. Tzelepis K. Pandolfini L. Shi J. Millán-Zambrano G. Robson S. C. et al (2017). Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature552 (7683), 126–131. 10.1038/nature24678
9
Batista P. J. Molinie B. Wang J. Qu K. Zhang J. Li L. et al (2014). M(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell15 (6), 707–719. 10.1016/j.stem.2014.09.019
10
Baumgarten S. Bryant J. M. Sinha A. Reyser T. Preiser P. R. Dedon P. C. et al (2019). Transcriptome-wide dynamics of extensive m6A mRNA methylation during Plasmodium falciparum blood-stage development. Nat. Microbiol.4 (12), 2246–2259. 10.1038/s41564-019-0521-7
11
Becker M. A. Meiser N. Schmidt‐Dengler M. Richter C. Wacker A. Schwalbe H. et al (2024). m6A methylation of transcription leader sequence of SARS-CoV-2 impacts discontinuous transcription of subgenomic mRNAs. Chem. – A Eur. J.30 (42), e202401897. 10.1002/chem.202401897
12
Bokar J. A. Rath-Shambaugh M. E. Ludwiczak R. Narayan P. Rottman F. (1994). Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J. Biol. Chem.269 (26), 17697–17704. 10.1016/s0021-9258(17)32497-3
13
Bokar J. A. Shambaugh M. E. Polayes D. Matera A. G. Rottman F. M. (1997). Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA3 (11), 1233–1247.
14
Burgess H. M. Depledge D. P. Thompson L. Srinivas K. P. Grande R. C. Vink E. I. et al (2021). Targeting the m6A RNA modification pathway blocks SARS-CoV-2 and HCoV-OC43 replication. Genes & Dev.35 (13–14), 1005–1019. 10.1101/gad.348320.121
15
Canaani D. Kahana C. Lavi S. Groner Y. (1979). Identification and mapping of N 6 -methyladenosine containing sequences in Simian Virus 40 RNA. Nucleic Acids Res.6 (8), 2879–2899. 10.1093/nar/6.8.2879
16
Cappannini A. Ray A. Purta E. Mukherjee S. Boccaletto P. Moafinejad S. N. et al (2024). MODOMICS: a database of RNA modifications and related information. 2023 update. Nucleic Acids Res.52 (D1), D239–D244. 10.1093/nar/gkad1083
17
Chen H. Jia B. Zhang Q. Zhang Y. (2022a). Meclofenamic acid restores gefinitib sensitivity by downregulating breast cancer resistance protein and multidrug resistance protein 7 via FTO/m6A-Demethylation/c-Myc in non-small cell lung cancer. Front. Oncol.12, 870636. 10.3389/fonc.2022.870636
18
Chen J. Wei X. Wang X. Liu T. Zhao Y. Chen L. et al (2022b). TBK1-METTL3 axis facilitates antiviral immunity. Cell Rep.38 (7), 110373. 10.1016/j.celrep.2022.110373
19
Cheng S. Li H. Chi J. Zhao W. Lin J. Liu X. et al (2023a). FTO-mediated m6A modification promotes malignant transformation of gastric mucosal epithelial cells in chronic Cag A+ Helicobacter pylori infection. J. Cancer Res. Clin. Oncol.149 (10), 7327–7340. 10.1007/s00432-023-04684-4
20
Cheng S. Li H. Chi J. Zhao W. Lin J. Liu X. et al (2023b). FTO-mediated m6A modification promotes malignant transformation of gastric mucosal epithelial cells in chronic Cag A+ Helicobacter pylori infection. J. Cancer Res. Clin. Oncol.149 (10), 7327–7340. 10.1007/s00432-023-04684-4
21
Chokkalla A. K. Mehta S. L. Kim T. Chelluboina B. Kim J. Vemuganti R. (2019). Transient focal ischemia significantly alters the m6A epitranscriptomic tagging of RNAs in the brain. Stroke50 (10), 2912–2921. 10.1161/STROKEAHA.119.026433
22
Christiansen J. Kolte A. M. Hansen T. v. O. Nielsen F. C. (2009). IGF2 mRNA-binding protein 2: biological function and putative role in type 2 diabetes. J. Mol. Endocrinol.43 (5), 187–195. 10.1677/JME-09-0016
23
Courtney D. G. Kennedy E. M. Dumm R. E. Bogerd H. P. Tsai K. Heaton N. S. et al (2017). Epitranscriptomic enhancement of influenza A virus gene expression and replication. Cell Host & Microbe22 (3), 377–386.e5. 10.1016/j.chom.2017.08.004
24
Csepany T. Lin A. Baldick C. J. Beemon K. (1990). Sequence specificity of mRNA N6-adenosine methyltransferase. J. Biol. Chem.265 (33), 20117–20122. 10.1016/S0021-9258(17)30477-5
25
Cui Q. Shi H. Ye P. Li L. Qu Q. Sun G. et al (2017). m6A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep.18 (11), 2622–2634. 10.1016/j.celrep.2017.02.059
26
Curreri A. Sankholkar D. Mitragotri S. Zhao Z. (2023). RNA therapeutics in the clinic. Bioeng. & Transl. Med.8 (1), e10374. 10.1002/btm2.10374
27
Dai Z. Etebari K. Asgari S. (2022). N6-methyladenosine modification of the Aedes aegypti transcriptome and its alteration upon dengue virus infection in Aag2 cell line. Commun. Biol.5 (1), 607. 10.1038/s42003-022-03566-8
28
Deng X. Chen K. Luo G.-Z. Weng X. Ji Q. Zhou T. et al (2015). Widespread occurrence of N6-methyladenosine in bacterial mRNA. Nucleic Acids Res.43 (13), 6557–6567. 10.1093/nar/gkv596
29
Desrosiers R. Friderici K. Rottman F. (1974). Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci.71 (10), 3971–3975. 10.1073/pnas.71.10.3971
30
Desrosiers R. C. Friderici K. H. Rottman F. M. (1975). Characterization of Novikoff hepatoma mRNA methylation and heterogeneity in the methylated 5’ terminus. Biochemistry14 (20), 4367–4374. 10.1021/bi00691a004
31
Dimock K. Stoltzfus C. M. (1977). Sequence specificity of internal methylation in B77 avian sarcoma virus RNA subunits. Biochemistry16 (3), 471–478. 10.1021/bi00622a021
32
Ding S. Gao J. Huang C. Zhou Y. Yang Y. Cai Z. (2024). Identification of diagnostic biomarkers and molecular subtype analysis associated with m6A in Tuberculosis immunopathology using machine learning. Sci. Rep.14 (1), 29982. 10.1038/s41598-024-81790-4
33
Dominissini D. Moshitch-Moshkovitz S. Schwartz S. Salmon-Divon M. Ungar L. Osenberg S. et al (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature485 (7397), 201–206. 10.1038/nature11112
34
Du H. Zhao Y. He J. Zhang Y. Xi H. Liu M. et al (2016). YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun.7, 12626. 10.1038/ncomms12626
35
Fang Q. Chen H. (2020). The significance of m6A RNA methylation regulators in predicting the prognosis and clinical course of HBV-related hepatocellular carcinoma. Mol. Med.26 (1), 60. 10.1186/s10020-020-00185-z
36
Feng J. Zhang T. Sorel O. Meng W. Zhang X. Lai Z. et al (2022a). Global profiling reveals common and distinct N6-methyladenosine (m6A) regulation of innate immune responses during bacterial and viral infections. Cell Death & Dis.13 (3), 234. 10.1038/s41419-022-04681-4
37
Feng Z. Zhou F. Tan M. Wang T. Chen Y. Xu W. et al (2022b). Targeting m6A modification inhibits herpes virus 1 infection. Genes & Dis.9 (4), 1114–1128. 10.1016/j.gendis.2021.02.004
38
Finkel D. Groner Y. (1983). Methylations of adenosine residues (m6A) in pre-mRNA are important for formation of late simian virus 40 mRNAs. Virology131 (2), 409–425. 10.1016/0042-6822(83)90508-1
39
Fischer J. Koch L. Emmerling C. Vierkotten J. Peters T. Brüning J. C. et al (2009). Inactivation of the Fto gene protects from obesity. Nature458 (7240), 894–898. 10.1038/nature07848
40
Fleming A. (1929). On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. Influenzæ. Br. J. Exp. Pathology10 (3), 226–236.
41
Garcia-Campos M. A. Edelheit S. Toth U. Safra M. Shachar R. Viukov S. et al (2019). Deciphering the “m6A code” via antibody-independent quantitative profiling. Cell178 (3), 731–747.e16. 10.1016/j.cell.2019.06.013
42
Gerken T. Girard C. A. Tung Y.-C. L. Webby C. J. Saudek V. Hewitson K. S. et al (2007). The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Sci. (New York, N.Y.)318 (5855), 1469–1472. 10.1126/science.1151710
43
Gokhale N. S. McIntyre A. B. R. McFadden M. J. Roder A. E. Kennedy E. M. Gandara J. A. et al (2016a). N6-Methyladenosine in Flaviviridae viral RNA genomes regulates infection. Cell Host & Microbe20 (5), 654–665. 10.1016/j.chom.2016.09.015
44
Gokhale N. S. McIntyre A. B. R. McFadden M. J. Roder A. E. Kennedy E. M. Gandara J. A. et al (2016b). N6-Methyladenosine in Flaviviridae viral RNA genomes regulates infection. Cell Host & Microbe20 (5), 654–665. 10.1016/j.chom.2016.09.015
45
Govindaraju G. Kadumuri R. V. Sethumadhavan D. V. Jabeena C. A. Chavali S. Rajavelu A. (2020a). N6-Adenosine methylation on mRNA is recognized by YTH2 domain protein of human malaria parasite Plasmodium falciparum. Epigenetics & Chromatin13 (1), 33. 10.1186/s13072-020-00355-7
46
Govindaraju G. Kadumuri R. V. Sethumadhavan D. V. Jabeena C. A. Chavali S. Rajavelu A. (2020b). N6-Adenosine methylation on mRNA is recognized by YTH2 domain protein of human malaria parasite Plasmodium falciparum. Epigenetics & Chromatin13 (1), 33. 10.1186/s13072-020-00355-7
47
Graham M. Y. Weisblum B. (1979). 23S ribosomal ribonucleic acid of macrolide-producing streptomycetes contains methylated adenine. J. Bacteriol.137 (3), 1464–1467. 10.1128/jb.137.3.1464-1467.1979
48
Granados-Riveron J. T. Aquino-Jarquin G. (2021). Engineering of the current nucleoside-modified mRNA-LNP vaccines against SARS-CoV-2. Biomed. & Pharmacother.142, 111953. 10.1016/j.biopha.2021.111953
49
Gray A. Sharara F. (2022). Global and regional sepsis and infectious syndrome mortality in 2019: a systematic analysis. Lancet Glob. Health10, S2. 10.1016/S2214-109X(22)00131-0
50
Hajikhezri Z. Kaira Y. Schubert E. Darweesh M. Svensson C. Akusjärvi G. et al (2023). Fragile X-related protein FXR1 controls human adenovirus capsid mRNA metabolism. J. Virology97 (2), e0153922–22. 10.1128/jvi.01539-22
51
Hao H. Hao S. Chen H. Chen Z. Zhang Y. Wang J. et al (2019). N 6-methyladenosine modification and METTL3 modulate enterovirus 71 replication. Nucleic Acids Res.47 (1), 362–374. 10.1093/nar/gky1007
52
Helm M. Motorin Y. (2017). Detecting RNA modifications in the epitranscriptome: predict and validate. Nat. Rev. Genet.18 (5), 275–291. 10.1038/nrg.2016.169
53
Henquell C. Bœuf B. Mirand A. Bacher C. Traore O. Déchelotte P. et al (2009). Fatal adenovirus infection in a neonate and transmission to health-care workers. J. Clin. Virology45 (4), 345–348. 10.1016/j.jcv.2009.04.019
54
Hess M. E. Hess S. Meyer K. D. Verhagen L. A. Koch L. Bronneke H. S. et al (2013). The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat. Neurosci.16 (8), 1042–1048. 10.1038/nn.3449
55
Hesser C. R. Karijolich J. Dominissini D. He C. Glaunsinger B. A. (2018). N6-methyladenosine modification and the YTHDF2 reader protein play cell type specific roles in lytic viral gene expression during Kaposi’s sarcoma-associated herpesvirus infection. PLoS Pathog.14 (4), e1006995. 10.1371/journal.ppat.1006995
56
Holbrook M. R. (2017). Historical perspectives on flavivirus research. Viruses9 (5), 97. 10.3390/v9050097
57
Holmes M. J. Padgett L. R. Bastos M. S. Sullivan W. J. (2021a). m6A RNA methylation facilitates pre-mRNA 3’-end formation and is essential for viability of Toxoplasma gondii. PLoS Pathog.17 (7), e1009335. 10.1371/journal.ppat.1009335
58
Holmes M. J. Padgett L. R. Bastos M. S. Sullivan W. J. (2021b). m6A RNA methylation facilitates pre-mRNA 3’-end formation and is essential for viability of Toxoplasma gondii. PLoS Pathog.17 (7), e1009335. 10.1371/journal.ppat.1009335
59
Horiuchi K. Kawamura T. Iwanari H. Ohashi R. Naito M. Kodama T. et al (2013). Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J. Biol. Chem.288 (46), 33292–33302. 10.1074/jbc.M113.500397
60
Horowitz S. Horowitz A. Nilsen T. W. Munns T. W. Rottman F. M. (1984). Mapping of N6-methyladenosine residues in bovine prolactin mRNA. Proc. Natl. Acad. Sci.81 (18), 5667–5671. 10.1073/pnas.81.18.5667
61
Hu Y. Gong C. Li Z. Liu J. Chen Y. Huang Y. et al (2022). Demethylase ALKBH5 suppresses invasion of gastric cancer via PKMYT1 m6A modification. Mol. Cancer21 (1), 34. 10.1186/s12943-022-01522-y
62
Huang H. Weng H. Sun W. Qin X. Shi H. Wu H. et al (2018). Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol.20 (3), 285–295. 10.1038/s41556-018-0045-z
63
Huang Y. Su R. Sheng Y. Dong L. Dong Z. Xu H. et al (2019). Small-molecule targeting of oncogenic FTO demethylase in acute myeloid leukemia. Cancer Cell35 (4), 677–691.e10. 10.1016/j.ccell.2019.03.006
64
Humans I. W. (2012). “HUMAN IMMUNODEFICIENCY VIRUS-1,” in Biological agents. International agency for research on cancer. Available online at: https://www.ncbi.nlm.nih.gov/books/NBK304351/.
65
Ignatova V. V. Stolz P. Kaiser S. Gustafsson T. H. Lastres P. R. Sanz-Moreno A. et al (2020). The rRNA m6A methyltransferase METTL5 is involved in pluripotency and developmental programs. Genes & Dev.34 (9–10), 715–729. 10.1101/gad.333369.119
66
Imam H. Khan M. Gokhale N. S. McIntyre A. B. R. Kim G.-W. Jang J. Y. et al (2018). N6-methyladenosine modification of hepatitis B virus RNA differentially regulates the viral life cycle. Proc. Natl. Acad. Sci.115 (35), 8829–8834. 10.1073/pnas.1808319115
67
Drexler M. , and Institute of Medicine (US). (2010). What You Need to Know About Infectious Disease. United states: National Academies Press. Available online at: http://www.ncbi.nlm.nih.gov/books/NBK209706/
68
Jia G. Fu Y. Zhao X. Dai Q. Zheng G. Yang Y. et al (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol.7 (12), 885–887. 10.1038/nchembio.687
69
Jia G. Yang C.-G. Yang S. Jian X. Yi C. Zhou Z. et al (2008). Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett.582 (23–24), 3313–3319. 10.1016/j.febslet.2008.08.019
70
Jungfleisch J. Böttcher R. Talló-Parra M. Pérez-Vilaró G. Merits A. Novoa E. M. et al (2022). CHIKV infection reprograms codon optimality to favor viral RNA translation by altering the tRNA epitranscriptome. Nat. Commun.13 (1), 4725. 10.1038/s41467-022-31835-x
71
Jurczyszak D. Zhang W. Terry S. N. Kehrer T. Bermúdez González M. C. McGregor E. et al (2020). HIV protease cleaves the antiviral m6A reader protein YTHDF3 in the viral particle. PLOS Pathog.16 (2), e1008305. 10.1371/journal.ppat.1008305
72
Kane S. E. Beemon K. (1985). Precise localization of m6A in Rous sarcoma virus RNA reveals clustering of methylation sites: implications for RNA processing. Mol. Cell. Biol.5 (9), 2298–2306. 10.1128/mcb.5.9.2298
73
Karikó K. (2022). Developing mRNA for therapy. Keio J. Med.71 (1), 31. 10.2302/kjm.71-001-ABST
74
Kennedy E. M. Bogerd H. P. Kornepati A. V. R. Kang D. Ghoshal D. Marshall J. B. et al (2016). Posttranscriptional m(6)A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host & Microbe19 (5), 675–685. 10.1016/j.chom.2016.04.002
75
Kierzek E. Kierzek R. (2003). The thermodynamic stability of RNA duplexes and hairpins containing N6-alkyladenosines and 2-methylthio-N6-alkyladenosines. Nucleic Acids Res.31 (15), 4472–4480. 10.1093/nar/gkg633
76
Kim B. Arcos S. Rothamel K. Jian J. Rose K. L. McDonald W. H. et al (2020a). Discovery of widespread host protein interactions with the pre-replicated genome of CHIKV using VIR-CLASP. Mol. Cell78 (4), 624–640.e7. 10.1016/j.molcel.2020.04.013
77
Kim G.-W. Imam H. Khan M. Siddiqui A. (2020b). N6-Methyladenosine modification of hepatitis B and C viral RNAs attenuates host innate immunity via RIG-I signaling. J. Biol. Chem.295 (37), 13123–13133. 10.1074/jbc.RA120.014260
78
Kim G.-W. Siddiqui A. (2021). N6-methyladenosine modification of HCV RNA genome regulates cap-independent IRES-mediated translation via YTHDC2 recognition. Proc. Natl. Acad. Sci. U. S. A.118 (10), e2022024118. 10.1073/pnas.2022024118
79
Kim K. L. van Galen P. Hovestadt V. Rahme G. J. Andreishcheva E. N. Shinde A. et al (2021). Systematic detection of m6A-modified transcripts at single-molecule and single-cell resolution. Cell Rep. Methods1 (5), 100061. 10.1016/j.crmeth.2021.100061
80
Krug R. M. Morgan M. A. Shatkin A. J. (1976). Influenza viral mRNA contains internal N6-methyladenosine and 5’-terminal 7-methylguanosine in cap structures. J. Virology20 (1), 45–53. 10.1128/JVI.20.1.45-53.1976
81
Lang F. Singh R. K. Pei Y. Zhang S. Sun K. Robertson E. S. (2019). EBV epitranscriptome reprogramming by METTL14 is critical for viral-associated tumorigenesis. PLOS Pathog.15 (6), e1007796. 10.1371/journal.ppat.1007796
82
Lavi S. Shatkin A. J. (1975a). Methylated simian virus 40-specific RNA from nuclei and cytoplasm of infected BSC-1 cells. Proc. Natl. Acad. Sci.72 (6), 2012–2016. 10.1073/pnas.72.6.2012
83
Lavi S. Shatkin A. J. (1975b). Methylated simian virus 40-specific RNA from nuclei and cytoplasm of infected BSC-1 cells. Proc. Natl. Acad. Sci.72 (6), 2012–2016. 10.1073/pnas.72.6.2012
84
Li A. Chen Y.-S. Ping X.-L. Yang X. Xiao W. Yang Y. et al (2017a). Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res.27 (3), 444–447. 10.1038/cr.2017.10
85
Li F. Yi Y. Miao Y. Long W. Long T. Chen S. et al (2019a). N6-Methyladenosine modulates nonsense-mediated mRNA decay in human glioblastoma. Cancer Res.79 (22), 5785–5798. 10.1158/0008-5472.CAN-18-2868
86
Li H.-B. Tong J. Zhu S. Batista P. J. Duffy E. E. Zhao J. et al (2017b). m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature548 (7667), 338–342. 10.1038/nature23450
87
Li H.-M. Tang F. Wang L.-J. Huang Q. Pan H.-F. Zhang T.-P. (2022a). Association of N6-methyladenosine readers’ genes variation and expression level with pulmonary tuberculosis. Front. Public Health10, 925303. 10.3389/fpubh.2022.925303
88
Li J. Zhu L. Shi Y. Liu J. Lin L. Chen X. (2019b). m6A demethylase FTO promotes hepatocellular carcinoma tumorigenesis via mediating PKM2 demethylation. Am. J. Transl. Res.11 (9), 6084–6092.
89
Li L. Zeng J. He S. Yang Y. Wang C. (2024). METTL14 decreases FTH1 mRNA stability via m6A methylation to promote sorafenib-induced ferroptosis of cervical cancer. Cancer Biol. & Ther.25 (1), 2349429. 10.1080/15384047.2024.2349429
90
Li Y. Zhang W. Dai Y. Chen K. (2022b). Identification and verification of IGFBP3 and YTHDC1 as biomarkers associated with immune infiltration and mitophagy in hypertrophic cardiomyopathy. Front. Genet.13, 986995. 10.3389/fgene.2022.986995
91
Li Y.-L. Li L. Liu Y.-H. Hu L.-K. Yan Y.-X. (2023). Identification of metabolism-related proteins as biomarkers of insulin resistance and potential mechanisms of m6A modification. Nutrients15 (8), 1839. 10.3390/nu15081839
92
Liao L. He Y. Li S.-J. Zhang G.-G. Yu W. Yang J. et al (2022). Anti-HIV drug elvitegravir suppresses cancer metastasis via increased proteasomal degradation of m6A methyltransferase METTL3. Cancer Res.82 (13), 2444–2457. 10.1158/0008-5472.CAN-21-4124
93
Lichinchi G. Gao S. Saletore Y. Gonzalez G. M. Bansal V. Wang Y. et al (2016a). Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat. Microbiol.1, 16011. 10.1038/nmicrobiol.2016.11
94
Lichinchi G. Gao S. Saletore Y. Gonzalez G. M. Bansal V. Wang Y. et al (2016b). Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat. Microbiol.1, 16011. 10.1038/nmicrobiol.2016.11
95
Lichinchi G. Zhao B. S. Wu Y. Lu Z. Qin Y. He C. et al (2016). Dynamics of human and viral RNA methylation during zika virus infection. Cell Host & Microbe20 (5), 666–673. 10.1016/j.chom.2016.10.002
96
Linder B. Grozhik A. V. Olarerin-George A. O. Meydan C. Mason C. E. Jaffrey S. R. (2015). Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods12 (8), 767–772. 10.1038/nmeth.3453
97
Liu C. Gu L. Deng W. Meng Q. Li N. Dai G. et al (2022a). N6-Methyladenosine RNA methylation in cardiovascular diseases. Front. Cardiovasc. Med.9, 887838. 10.3389/fcvm.2022.887838
98
Liu D. Yuan M. Wang Z. Sun L. Fang Y. Ma X. et al (2022b). Comprehensive analysis of N6-methyladenosine (m6A) RNA methylation regulators and tumour microenvironment cell infiltration involving prognosis and immunotherapy in gastroesophageal adenocarcinomas. Can. J. Gastroenterology Hepatology2022, 1–17. 10.1155/2022/3506518
99
Liu J. Shao Y. Li D. Li C. (2023). N6-methyladenosine helps Apostichopus japonicus resist Vibrio splendidus infection by targeting coelomocyte autophagy via the AjULK-AjYTHDF/AjEEF-1α axis. Commun. Biol.6 (1), 547. 10.1038/s42003-023-04929-5
100
Liu J. Xu Y.-P. Li K. Ye Q. Zhou H.-Y. Sun H. et al (2021). The m6A methylome of SARS-CoV-2 in host cells. Cell Res.31 (4), 404–414. 10.1038/s41422-020-00465-7
101
Liu J. Yue Y. Han D. Wang X. Fu Y. Zhang L. et al (2014a). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol.10 (2), 93–95. 10.1038/nchembio.1432
102
Liu J. Yue Y. Han D. Wang X. Fu Y. Zhang L. et al (2014b). A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol.10 (2), 93–95. 10.1038/nchembio.1432
103
Liu L. Zeng S. Jiang H. Zhang Y. Guo X. Wang Y. (2019). Differential m6A methylomes between two major life stages allows potential regulations in Trypanosoma brucei. Biochem. Biophysical Res. Commun.508 (4), 1286–1290. 10.1016/j.bbrc.2018.12.043
104
Liu N. Dai Q. Zheng G. He C. Parisien M. Pan T. (2015). N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature518 (7540), 560–564. 10.1038/nature14234
105
Liu N. Zhou K. I. Parisien M. Dai Q. Diatchenko L. Pan T. (2017). N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res.45 (10), 6051–6063. 10.1093/nar/gkx141
106
Liu P. Li F. Lin J. Fukumoto T. Nacarelli T. Hao X. et al (2021a). m6A-independent genome-wide METTL3 and METTL14 redistribution drives the senescence-associated secretory phenotype. Nat. Cell Biol.23 (4), 355–365. 10.1038/s41556-021-00656-3
107
Liu Q. Wang X. Xie C. Ding S. Yang H. Guo S. et al (2021b). A novel human acute encephalitis caused by Pseudorabies virus variant strain. Clin. Infect. Dis.73 (11), e3690–e3700. 10.1093/cid/ciaa987
108
Liu S. Li K. Long C. Lao M. Ma B. Liu C. et al (2025). The role of FTO in m6A RNA methylation and immune regulation in Staphylococcus aureus infection-related osteomyelitis. Front. Microbiol.16, 1526475. 10.3389/fmicb.2025.1526475
109
Lu M. Xue M. Wang H.-T. Kairis E. L. Ahmad S. Wei J. et al (2021). Nonsegmented negative-sense RNA viruses utilize N6-methyladenosine (m6A) as a common strategy to evade host innate immunity. J. Virology95 (9), e01939. 10.1128/jvi.01939-20
110
Lu X. Erdman D. D. (2006). Molecular typing of human adenoviruses by PCR and sequencing of a partial region of the hexon gene. Archives Virology151 (8), 1587–1602. 10.1007/s00705-005-0722-7
111
Ma H. Wang X. Cai J. Dai Q. Natchiar S. K. Lv R. et al (2019). N6-Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat. Chem. Biol.15 (1), 88–94. 10.1038/s41589-018-0184-3
112
Ma L. Zhang X. Yu K. Xu X. Chen T. Shi Y. et al (2021a). Targeting SLC3A2 subunit of system XC- is essential for m6A reader YTHDC2 to be an endogenous ferroptosis inducer in lung adenocarcinoma. Free Radic. Biol. Med.168, 25–43. 10.1016/j.freeradbiomed.2021.03.023
113
Ma M. Duan Y. Peng C. Wu Y. Zhang X. Chang B. et al (2024a). Mycobacterium tuberculosis inhibits METTL14-mediated m6A methylation of Nox2 mRNA and suppresses anti-TB immunity. Cell Discov.10 (1), 36. 10.1038/s41421-024-00653-4
114
Ma M. Duan Y. Peng C. Wu Y. Zhang X. Chang B. et al (2024b). Mycobacterium tuberculosis inhibits METTL14-mediated m6A methylation of Nox2 mRNA and suppresses anti-TB immunity. Cell Discov.10 (1), 36. 10.1038/s41421-024-00653-4
115
Ma Z. Gao X. Shuai Y. Xing X. Ji J. (2021b). The m6A epitranscriptome opens a new charter in immune system logic. Epigenetics16 (8), 819–837. 10.1080/15592294.2020.1827722
116
Macveigh-Fierro D. Cicerchia A. I. Cadorette A. Sharma V. Muller M. Watt A. et al (2022). The m6A reader YTHDC2 is essential for escape from KSHV SOX-induced RNA decay. Proceedings of the National Academy of Sciences of the United States of America, 119 (8), e2116662119. 10.1073/pnas.2116662119
117
Martin S. Chiramel A. I. Schmidt M. L. Chen Y.-C. Whitt N. Watt A. et al (2018). A genome-wide siRNA screen identifies a druggable host pathway essential for the Ebola virus life cycle. Genome Med.10 (1), 58. 10.1186/s13073-018-0570-1
118
Mathiyalagan P. Adamiak M. Mayourian J. Sassi Y. Liang Y. Agarwal N. et al (2019). FTO-dependent N6-methyladenosine regulates cardiac function during remodeling and repair. Circulation139 (4), 518–532. 10.1161/CIRCULATIONAHA.118.033794
119
Mauer J. Luo X. Blanjoie A. Jiao X. Grozhik A. V. Patil D. P. et al (2017). Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature541 (7637), 371–375. 10.1038/nature21022
120
Meyer K. D. Patil D. P. Zhou J. Zinoviev A. Skabkin M. A. Elemento O. et al (2015). 5′ UTR m6A promotes cap-independent translation. Cell163 (4), 999–1010. 10.1016/j.cell.2015.10.012
121
Meyer K. D. Saletore Y. Zumbo P. Elemento O. Mason C. E. Jaffrey S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell149 (7), 1635–1646. 10.1016/j.cell.2012.05.003
122
Micaelli M. Dalle Vedove A. Cerofolini L. Vigna J. Sighel D. Zaccara S. et al (2022). Small-molecule ebselen binds to YTHDF proteins interfering with the recognition of N 6-methyladenosine-modified RNAs. ACS Pharmacol. & Transl. Sci.5 (10), 872–891. 10.1021/acsptsci.2c00008
123
Mo X. B. Lei S. F. Zhang Y. H. Zhang H. (2018). Detection of m6A-associated SNPs as potential functional variants for coronary artery disease. Epigenomics10 (10), 1279–1287. 10.2217/epi-2018-0007
124
Moss B. Gershowitz A. Stringer J. R. Holland L. E. Wagner E. K. (1977). 5’-Terminal and internal methylated nucleosides in herpes simplex virus type 1 mRNA. J. Virology23 (2), 234–239. 10.1128/JVI.23.2.234-239.1977
125
Narayan P. Rottman F. M. (1988). An in vitro system for accurate methylation of internal adenosine residues in messenger RNA. Sci. (New York, N.Y.)242 (4882), 1159–1162. 10.1126/science.3187541
126
Park A. K. Kim H. Jin H. J. (2010). Phylogenetic analysis of rRNA methyltransferases, Erm and KsgA, as related to antibiotic resistance. FEMS Microbiol. Lett.309 (2), 151–162. 10.1111/j.1574-6968.2010.02031.x
127
Patil D. P. Chen C.-K. Pickering B. F. Chow A. Jackson C. Guttman M. et al (2016). m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature537 (7620), 369–373. 10.1038/nature19342
128
Pendleton K. E. Chen B. Liu K. Hunter O. V. Xie Y. Tu B. P. et al (2017). The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell169 (5), 824–835.e14. 10.1016/j.cell.2017.05.003
129
Perry R. P. Kelley D. E. Friderici K. Rottman F. (1975). The methylated constituents of L cell messenger RNA: evidence for an unusual cluster at the 5’ terminus. Cell4 (4), 387–394. 10.1016/0092-8674(75)90159-2
130
Picavet L. W. van Vroonhoven E. C. N. Scholman R. C. Smits Y. T. H. Banerjee R. Besteman S. B. et al (2024). m6A reader YTHDC1 impairs respiratory syncytial virus infection by downregulating membrane CX3CR1 expression. Viruses16 (5), 778. 10.3390/v16050778
131
Ping X.-L. Sun B.-F. Wang L. Xiao W. Yang X. Wang W.-J. et al (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res.24 (2), 177–189. 10.1038/cr.2014.3
132
Price A. M. Hayer K. E. McIntyre A. B. R. Gokhale N. S. Abebe J. S. Della Fera A. N. et al (2020). Direct RNA sequencing reveals m6A modifications on adenovirus RNA are necessary for efficient splicing. Nat. Commun.11 (1), 6016. 10.1038/s41467-020-19787-6
133
Qing X. Chen Q. Wang K. (2022). m6A regulator-mediated methylation modification patterns and characteristics in COVID-19 patients. Front. Public Health10, 914193. 10.3389/fpubh.2022.914193
134
Qiu L. Jing Q. Li Y. Han J. (2023). RNA modification: mechanisms and therapeutic targets. Mol. Biomed.4 (1), 25. 10.1186/s43556-023-00139-x
135
Qiu W. Zhang Q. Zhang R. Lu Y. Wang X. Tian H. et al (2021). N6-methyladenosine RNA modification suppresses antiviral innate sensing pathways via reshaping double-stranded RNA. Nat. Commun.12 (1), 1582. 10.1038/s41467-021-21904-y
136
Regué L. Minichiello L. Avruch J. Dai N. (2019). Liver-specific deletion of IGF2 mRNA binding protein-2/IMP2 reduces hepatic fatty acid oxidation and increases hepatic triglyceride accumulation. J. Biol. Chem.294 (31), 11944–11951. 10.1074/jbc.RA119.008778
137
Röder K. Barker A. M. Whitehouse A. Pasquali S. (2022). Investigating the structural changes due to adenosine methylation of the Kaposi’s sarcoma-associated herpes virus ORF50 transcript. PLoS Comput. Biol.18 (5), e1010150. 10.1371/journal.pcbi.1010150
138
Sahin U. Karikó K. Türeci Ö. (2014). mRNA-based therapeutics—developing a new class of drugs. Nat. Rev. Drug Discov.13 (10), 759–780. 10.1038/nrd4278
139
Schwartz S. Agarwala S. D. Mumbach M. R. Jovanovic M. Mertins P. Shishkin A. et al (2013). High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell155 (6), 1409–1421. 10.1016/j.cell.2013.10.047
140
Schwartz S. Bernstein D. A. Mumbach M. R. Jovanovic M. Herbst R. H. León-Ricardo B. et al (2014a). Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell159 (1), 148–162. 10.1016/j.cell.2014.08.028
141
Schwartz S. Mumbach M. R. Jovanovic M. Wang T. Maciag K. Bushkin G. et al (2014b). Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep.8 (1), 284–296. 10.1016/j.celrep.2014.05.048
142
Selberg S. Blokhina D. Aatonen M. Koivisto P. Siltanen A. Mervaala E. et al (2019). Discovery of small molecules that activate RNA methylation through cooperative binding to the METTL3-14-WTAP complex active site. Cell Rep.26 (13), 3762–3771.e5. 10.1016/j.celrep.2019.02.100
143
Selberg S. Žusinaite E. Herodes K. Seli N. Kankuri E. Merits A. et al (2021). HIV replication is increased by RNA methylation METTL3/METTL14/WTAP complex activators. ACS Omega6 (24), 15957–15963. 10.1021/acsomega.1c01626
144
Serra L. Silva Pereira S. Viegas I. J. Machado H. López-Escobar L. Figueiredo L. M. (2024a). m6A landscape is more pervasive when Trypanosoma brucei exits the cell cycle. Biomed. J., 100728. 10.1016/j.bj.2024.100728
145
Serra L. Silva Pereira S. Viegas I. J. Machado H. López-Escobar L. Figueiredo L. M. (2024b). m6A landscape is more pervasive when Trypanosoma brucei exits the cell cycle. Biomed. J., 100728. 10.1016/j.bj.2024.100728
146
Sharkey R. E. Herbert J. B. McGaha D. A. Nguyen V. Schoeffler A. J. Dunkle J. A. (2022). Three critical regions of the erythromycin resistance methyltransferase, ErmE, are required for function supporting a model for the interaction of Erm family enzymes with substrate rRNA. RNA (New York, N.Y.)28 (2), 210–226. 10.1261/rna.078946.121
147
Shi H. Wang X. Lu Z. Zhao B. S. Ma H. Hsu P. J. et al (2017). YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res.27 (3), 315–328. 10.1038/cr.2017.15
148
Shields K. E. Ranava D. Tan Y. Zhang D. Yap M.-N. F. (2024). Epitranscriptional m6A modification of rRNA negatively impacts translation and host colonization in Staphylococcus aureus. PLoS Pathog.20 (1), e1011968. 10.1371/journal.ppat.1011968
149
Sinha A. Baumgarten S. Distiller A. McHugh E. Chen P. Singh M. et al (2021). Functional characterization of the m6A-dependent translational modulator PfYTH.2 in the human malaria parasite. mBio12 (2), 006611-21–e721. 10.1128/mBio.00661-21
150
Śledź P. Jinek M. (2016). Structural insights into the molecular mechanism of the m6A writer complex. eLife5, e18434. 10.7554/eLife.18434
151
Sommer S. Lavi U. Darnell J. E. (1978). The absolute frequency of labeled N-6-methyladenosine in HeLa cell messenger RNA decreases with label time. J. Mol. Biol.124 (3), 487–499. 10.1016/0022-2836(78)90183-3
152
Sommer S. Salditt-Georgieff M. Bachenheimer S. Darnell J. E. Furuichi Y. Morgan M. et al (1976). The methylation of adenovirus-specific nuclear and cytoplasmic RNA. Nucleic Acids Res.3 (3), 749–766. 10.1093/nar/3.3.749
153
Song M. Lv K. Xu Z. Li J. Sun J. Shi J. et al (2024). N6 methyladenosine eraser FTO suppresses Staphylococcus aureus ‐induced ferroptosis of bone marrow mesenchymal stem cells to ameliorate osteomyelitis through regulating the MDM2/TLR4/SLC7A11 signaling pathway. Cell Biol. Int.48 (4), 450–460. 10.1002/cbin.12115
154
Stoilov P. Rafalska I. Stamm S. (2002). YTH: a new domain in nuclear proteins. Trends Biochem. Sci.27 (10), 495–497. 10.1016/s0968-0004(02)02189-8
155
Stojković V. Noda-Garcia L. Tawfik D. S. Fujimori D. G. (2016). Antibiotic resistance evolved via inactivation of a ribosomal RNA methylating enzyme. Nucleic Acids Res.44 (18), 8897–8907. 10.1093/nar/gkw699
156
Sun X. Dai Y. Tan G. Liu Y. Li N. (2020). Integration analysis of m6A-SNPs and eQTLs associated with sepsis reveals platelet degranulation and Staphylococcus aureus infection are mediated by m6A mRNA methylation. Front. Genet.11, 7. 10.3389/fgene.2020.00007
157
Svetlov M. S. Syroegin E. A. Aleksandrova E. V. Atkinson G. C. Gregory S. T. Mankin A. S. et al (2021). Structure of Erm-modified 70S ribosome reveals the mechanism of macrolide resistance. Nat. Chem. Biol.17 (4), 412–420. 10.1038/s41589-020-00715-0
158
Tan B. Liu H. Zhang S. da Silva S. R. Zhang L. Meng J. et al (2017). Viral and cellular N6-methyladenosine and N6,2′-O-dimethyladenosine epitranscriptomes in the KSHV life cycle. Nat. Microbiol.3 (1), 108–120. 10.1038/s41564-017-0056-8
159
Tanabe A. Tanikawa K. Tsunetomi M. Takai K. Ikeda H. Konno J. et al (2016). RNA helicase YTHDC2 promotes cancer metastasis via the enhancement of the efficiency by which HIF-1α mRNA is translated. Cancer Lett.376 (1), 34–42. 10.1016/j.canlet.2016.02.022
160
Tang C. Klukovich R. Peng H. Wang Z. Yu T. Zhang Y. et al (2018). ALKBH5-dependent m6A demethylation controls splicing and stability of long 3’-UTR mRNAs in male germ cells. Proc. Natl. Acad. Sci. U. S. A.115 (2), E325-E333–E333. 10.1073/pnas.1717794115
161
Tang J. Zhang J. Lu Y. He J. Wang H. Liu B. et al (2023). Novel insights into the multifaceted roles of m6A-modified LncRNAs in cancers: biological functions and therapeutic applications. Biomark. Res.11 (1), 42. 10.1186/s40364-023-00484-7
162
Theler D. Dominguez C. Blatter M. Boudet J. Allain F. H.-T. (2014). Solution structure of the YTH domain in complex with N6-methyladenosine RNA: a reader of methylated RNA. Nucleic Acids Res.42 (22), 13911–13919. 10.1093/nar/gku1116
163
Tirumuru N. Wu L. (2019). HIV-1 envelope proteins up-regulate N6-methyladenosine levels of cellular RNA independently of viral replication. J. Biol. Chem.294 (9), 3249–3260. 10.1074/jbc.RA118.005608
164
Tirumuru N. Zhao B. S. Lu W. Lu Z. He C. Wu L. (2016a). N(6)-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. eLife5, e15528. 10.7554/eLife.15528
165
Tirumuru N. Zhao B. S. Lu W. Lu Z. He C. Wu L. (2016b). N(6)-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. eLife5, e15528. 10.7554/eLife.15528
166
Tooley J. G. Catlin J. P. Tooley C. E. S. (2023). METTLing in stem cell and cancer biology. Stem Cell Rev. Rep.19 (1), 76–91. 10.1007/s12015-022-10444-7
167
Tsai K. Bogerd H. P. Kennedy E. M. Emery A. Swanstrom R. Cullen B. R. (2021). Epitranscriptomic addition of m6A regulates HIV-1 RNA stability and alternative splicing. Genes & Dev.35 (13–14), 992–1004. 10.1101/gad.348508.121
168
Tsai K. Courtney D. G. Cullen B. R. (2018). Addition of m6A to SV40 late mRNAs enhances viral structural gene expression and replication. PLOS Pathog.14 (2), e1006919. 10.1371/journal.ppat.1006919
169
van Tran N. Ernst F. G. M. Hawley B. R. Zorbas C. Ulryck N. Hackert P. et al (2019). The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res.47 (15), 7719–7733. 10.1093/nar/gkz619
170
Viegas I. J. de Macedo J. P. Serra L. De Niz M. Temporão A. Silva Pereira S. et al (2022). N6-methyladenosine in poly(A) tails stabilize VSG transcripts. Nature604 (7905), 362–370. 10.1038/s41586-022-04544-0
171
Vilchez R. A. Kozinetz C. A. Arrington A. S. Madden C. R. Butel J. S. (2003). Simian virus 40 in human cancers. Am. J. Med.114 (8), 675–684. 10.1016/S0002-9343(03)00087-1
172
Wang J. Chen L. Qiang P. (2021a). The role of IGF2BP2, an m6A reader gene, in human metabolic diseases and cancers. Cancer Cell Int.21 (1), 99. 10.1186/s12935-021-01799-x
173
Wang L. Wu J. Liu R. Chen W. Pang Z. Zhou F. et al (2022a). Epitranscriptome profiling of spleen mRNA m6A methylation reveals pathways of host responses to malaria parasite infection. Front. Immunol.13, 998756. 10.3389/fimmu.2022.998756
174
Wang L. Wu J. Liu R. Chen W. Pang Z. Zhou F. et al (2022b). Epitranscriptome profiling of spleen mRNA m6A methylation reveals pathways of host responses to malaria parasite infection. Front. Immunol.13, 998756. 10.3389/fimmu.2022.998756
175
Wang P. Doxtader K. A. Nam Y. (2016). Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell63 (2), 306–317. 10.1016/j.molcel.2016.05.041
176
Wang R. Han Z. Liu B. Zhou B. Wang N. Jiang Q. et al (2018). Identification of natural compound radicicol as a potent FTO inhibitor. Mol. Pharm.15 (9), 4092–4098. 10.1021/acs.molpharmaceut.8b00522
177
Wang X. Lu Z. Gomez A. Hon G. C. Yue Y. Han D. et al (2014a). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature505 (7481), 117–120. 10.1038/nature12730
178
Wang X. Zhao B. S. Roundtree I. A. Lu Z. Han D. Ma H. et al (2015a). N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell161 (6), 1388–1399. 10.1016/j.cell.2015.05.014
179
Wang X. Zhao B. S. Roundtree I. A. Lu Z. Han D. Ma H. et al (2015b). N6-methyladenosine modulates messenger RNA translation efficiency. Cell161 (6), 1388–1399. 10.1016/j.cell.2015.05.014
180
Wang Y. Han S. Ran R. Li A. Liu H. Liu M. et al (2021b). A longitudinal sampling study of transcriptomic and epigenetic profiles in patients with thrombocytopenia syndrome. Nat. Commun.12, 5629. 10.1038/s41467-021-25804-z
181
Wang Y. Li Y. Toth J. I. Petroski M. D. Zhang Z. Zhao J. C. (2014b). N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol.16 (2), 191–198. 10.1038/ncb2902
182
Warda A. S. Kretschmer J. Hackert P. Lenz C. Urlaub H. Höbartner C. et al (2017). Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep.18 (11), 2004–2014. 10.15252/embr.201744940
183
Wei C. Gershowitz A. Moss B. (1975a). N6, O2’-dimethyladenosine a novel methylated ribonucleoside next to the 5’ terminal of animal cell and virus mRNAs. Nature257 (5523), 251–253. 10.1038/257251a0
184
Wei C.-M. Gershowitz A. Moss B. (1975b). Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell4 (4), 379–386. 10.1016/0092-8674(75)90158-0
185
Weisblum B. (1995). Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother.39 (3), 577–585. 10.1128/AAC.39.3.577
186
Wen J. Lv R. Ma H. Shen H. He C. Wang J. et al (2018). Zc3h13 regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal. Mol. Cell69 (6), 1028–1038.e6. 10.1016/j.molcel.2018.02.015
187
Wendt L. Pickin M. J. Bodmer B. S. Reiche S. Fénéant L. Hölper J. E. et al (2023). N6-methyladenosine is required for efficient RNA synthesis of Ebola virus and other haemorrhagic fever viruses. Emerg. Microbes & Infect.12 (2), 2223732. 10.1080/22221751.2023.2223732
188
Weng H. Huang H. Wu H. Qin X. Zhao B. S. Dong L. et al (2018). METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m6A modification. Cell Stem Cell22 (2), 191–205.e9. 10.1016/j.stem.2017.11.016
189
Wickramasinghe V. O. Laskey R. A. (2015). Control of mammalian gene expression by selective mRNA export. Nat. Rev. Mol. Cell Biol.16 (7), 431–442. 10.1038/nrm4010
190
Wu F. Zhao S. Yu B. Chen Y.-M. Wang W. Song Z.-G. et al (2020). A new coronavirus associated with human respiratory disease in China. Nature579 (7798), 265–269. 10.1038/s41586-020-2008-3
191
Wu Y. Chen Z. Xie G. Zhang H. Wang Z. Zhou J. et al (2022). RNA m 1 A methylation regulates glycolysis of cancer cells through modulating ATP5D. Proc. Natl. Acad. Sci. U. S. A.119 (28), e2119038119. 10.1073/pnas.2119038119
192
Wu Z. Wang Y. Li T. Yang L. Jin J. Wu S. et al (2022a). Comprehensive analysis revealed the potential roles of N6-methyladenosine (m6A) mediating E. coli F18 susceptibility in IPEC-J2 cells. Int. J. Mol. Sci.23 (21), 13602. 10.3390/ijms232113602
193
Wu Z. Wang Y. Li T. Yang L. Jin J. Wu S. et al (2022b). Comprehensive analysis revealed the potential roles of N6-methyladenosine (m6A) mediating E. coli F18 susceptibility in IPEC-J2 cells. Int. J. Mol. Sci.23 (21), 13602. 10.3390/ijms232113602
194
Xiao W. Adhikari S. Dahal U. Chen Y.-S. Hao Y.-J. Sun B.-F. et al (2016). Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell61 (4), 507–519. 10.1016/j.molcel.2016.01.012
195
Xiao Y. Yang Y. Hu D. (2021). Knockdown of METTL3 inhibits enterovirus 71-induced apoptosis of mouse Schwann cell through regulation of autophagy. Pathogens Dis.79 (6), ftab036. 10.1093/femspd/ftab036
196
Xu H. Lin C. Li T. Zhu Y. Yang J. Chen S. et al (2022). N6-Methyladenosine-Modified circRNA in the bovine mammary epithelial cells injured by Staphylococcus aureus and Escherichia coli. Front. Immunol.13, 873330. 10.3389/fimmu.2022.873330
197
Xu H. Lin C. Wang C. Zhao T. Yang J. Zhang J. et al (2023a). ALKBH5 stabilized N6-methyladenosine—modified LOC4191 to suppress E. Coli-induced apoptosis. Cells12 (22), 2604. 10.3390/cells12222604
198
Xu H. Lin C. Wang C. Zhao T. Yang J. Zhang J. et al (2023b). ALKBH5 stabilized N6-methyladenosine—modified LOC4191 to suppress E. Coli-induced apoptosis. Cells12 (22), 2604. 10.3390/cells12222604
199
Xu J. Qi Y. Ju Q. (2023). Promotion of the resistance of human oral epithelial cells to herpes simplex virus type I infection via N6-methyladenosine modification. BMC Oral Health23 (1), 121. 10.1186/s12903-023-02744-2
200
Xue M. Zhang Y. Wang H. Kairis E. L. Lu M. Ahmad S. et al (2021). Viral RNA N6-methyladenosine modification modulates both innate and adaptive immune responses of human respiratory syncytial virus. PLOS Pathog.17 (12), e1010142. 10.1371/journal.ppat.1010142
201
Xue M. Zhao B. S. Zhang Z. Lu M. Harder O. Chen P. et al (2019). Viral N6-methyladenosine upregulates replication and pathogenesis of human respiratory syncytial virus. Nat. Commun.10 (1), 4595. 10.1038/s41467-019-12504-y
202
Yan F. Al-Kali A. Zhang Z. Liu J. Pang J. Zhao N. et al (2018). A dynamic N6-methyladenosine methylome regulates intrinsic and acquired resistance to tyrosine kinase inhibitors. Cell Res.28 (11), 1062–1076. 10.1038/s41422-018-0097-4
203
Yang F. Jin H. Que B. Chao Y. Zhang H. Ying X. et al (2019). Dynamic m6A mRNA methylation reveals the role of METTL3-m6A-CDCP1 signaling axis in chemical carcinogenesis. Oncogene38 (24), 4755–4772. 10.1038/s41388-019-0755-0
204
Yang Y. Yan Y. Yin J. Tang N. Wang K. Huang L. et al (2023). O-GlcNAcylation of YTHDF2 promotes HBV-related hepatocellular carcinoma progression in an N6-methyladenosine-dependent manner. Signal Transduct. Target. Ther.8 (1), 63. 10.1038/s41392-023-01316-8
205
Yankova E. Blackaby W. Albertella M. Rak J. De Braekeleer E. Tsagkogeorga G. et al (2021). Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature593 (7860), 597–601. 10.1038/s41586-021-03536-w
206
Yao M. Cheng Z. Li X. Li Y. Ye W. Zhang H. et al (2024). N6-methyladenosine modification positively regulate Japanese encephalitis virus replication. Virology J.21 (1), 23. 10.1186/s12985-023-02275-w
207
Ye F. Chen E. R. Nilsen T. W. (2017). Kaposi’s sarcoma-associated herpesvirus utilizes and manipulates RNA N6-adenosine methylation to promote lytic replication. J. Virology91 (16), e00466. 10.1128/jvi.00466-17
208
Yu P.-L. Wu R. Cao S.-J. Wen Y.-P. Huang X.-B. Zhao S. et al (2023). Pseudorabies virus exploits N6-methyladenosine modification to promote viral replication. Front. Microbiol.14, 1087484. 10.3389/fmicb.2023.1087484
209
Zeng J. Xie C. Huang Z. Cho C. H. Chan H. Li Q. et al (2024a). LOX-1 acts as an N6-methyladenosine-regulated receptor for Helicobacter pylori by binding to the bacterial catalase. Nat. Commun.15 (1), 669. 10.1038/s41467-024-44860-9
210
Zeng J. Xie C. Huang Z. Cho C. H. Chan H. Li Q. et al (2024b). LOX-1 acts as an N6-methyladenosine-regulated receptor for Helicobacter pylori by binding to the bacterial catalase. Nat. Commun.15 (1), 669. 10.1038/s41467-024-44860-9
211
Zhang C. Samanta D. Lu H. Bullen J. W. Zhang H. Chen I. et al (2016). Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m⁶A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. U. S. A.113 (14), E2047–E2056. 10.1073/pnas.1602883113
212
Zhang F. Ran Y. Tahir M. Li Z. Wang J. Chen X. (2022). Regulation of N6-methyladenosine (m6A) RNA methylation in microglia-mediated inflammation and ischemic stroke. Front. Cell. Neurosci.16, 955222. 10.3389/fncel.2022.955222
213
Zhang J. Guo S. Piao H.-Y. Wang Y. Wu Y. Meng X.-Y. et al (2019a). ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J. Physiology Biochem.75 (3), 379–389. 10.1007/s13105-019-00690-8
214
Zhang S. Lecoeur H. Varet H. Legendre R. Mahtal N. Proux C. et al (2022a). The m6A reader IGF2BP2 directs immune-metabolic reprogramming in Leishmania amazonensis-infected macrophages. 10.1101/2022.09.08.507100
215
Zhang S. Zhao B. S. Zhou A. Lin K. Zheng S. Lu Z. et al (2017). m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell31 (4), 591–606.e6. 10.1016/j.ccell.2017.02.013
216
Zhang T. Yang Y. Xiang Z. Gao C.-C. Wang W. Wang C. et al (2021a). N6-methyladenosine regulates RNA abundance of SARS-CoV-2. Cell Discov.7 (1), 7–4. 10.1038/s41421-020-00241-2
217
Zhang T.-P. Li R. Wang L.-J. Huang Q. Li H.-M. (2022b). Roles of the m6A methyltransferases METTL3, METTL14, and WTAP in pulmonary tuberculosis. Front. Immunol.13, 992628. 10.3389/fimmu.2022.992628
218
Zhang T.-P. Li R. Wang L.-J. Li H.-M. (2022c). Impact of m6A demethylase (ALKBH5, FTO) genetic polymorphism and expression levels on the development of pulmonary tuberculosis. Front. Cell. Infect. Microbiol.12, 1074380. 10.3389/fcimb.2022.1074380
219
Zhang X. Hao H. Ma L. Zhang Y. Hu X. Chen Z. et al (2021b). Methyltransferase-like 3 modulates severe acute respiratory syndrome coronavirus-2 RNA N6-methyladenosine modification and replication. mBio12 (4), e0106721. 10.1128/mbio.01067-21
220
Zhang X. Meng W. Feng J. Gao X. Qin C. Feng P. et al (2023). METTL16 controls Kaposi’s sarcoma-associated herpesvirus replication by regulating S-adenosylmethionine cycle. Cell Death & Dis.14 (9), 591. 10.1038/s41419-023-06121-3
221
Zhang X. Su T. Wu Y. Cai Y. Wang L. Liang C. et al (2024). N6-Methyladenosine reader YTHDF1 promotes stemness and therapeutic resistance in hepatocellular carcinoma by enhancing NOTCH1 expression. Cancer Res.84 (6), 827–840. 10.1158/0008-5472.CAN-23-1916
222
Zhang Z. Chen L.-Q. Zhao Y.-L. Yang C.-G. Roundtree I. A. Zhang Z. et al (2019b). Single-base mapping of m6A by an antibody-independent method. Sci. Adv.5 (7), eaax0250. 10.1126/sciadv.aax0250
223
Zhang Z. Theler D. Kaminska K. H. Hiller M. de la Grange P. Pudimat R. et al (2010). The YTH domain is a novel RNA binding domain. J. Biol. Chem.285 (19), 14701–14710. 10.1074/jbc.M110.104711
224
Zheng G. Dahl J. A. Niu Y. Fedorcsak P. Huang C.-M. Li C. J. et al (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell49 (1), 18–29. 10.1016/j.molcel.2012.10.015
225
Zheng Q. Hou J. Zhou Y. Li Z. Cao X. (2017). The RNA helicase DDX46 inhibits innate immunity by entrapping m6A-demethylated antiviral transcripts in the nucleus. Nat. Immunol.18 (10), 1094–1103. 10.1038/ni.3830
226
Zheng X. Wang J. Zhang X. Fu Y. Peng Q. Lu J. et al (2021). RNA m6 A methylation regulates virus-host interaction and EBNA2 expression during Epstein-Barr virus infection. Immun. Inflamm. Dis.9 (2), 351–362. 10.1002/iid3.396
227
Zhong S. Li H. Bodi Z. Button J. Vespa L. Herzog M. et al (2008). MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell20 (5), 1278–1288. 10.1105/tpc.108.058883
228
Zhou K. I. Pan T. (2018a). An additional class of m6A readers. Nat. Cell Biol.20 (3), 230–232. 10.1038/s41556-018-0046-y
229
Zhou K. I. Pan T. (2018b). An additional class of m6A readers. Nat. Cell Biol.20 (3), 230–232. 10.1038/s41556-018-0046-y
230
Zhou K. I. Shi H. Lyu R. Wylder A. C. Matuszek Ż. Pan J. N. et al (2019). Regulation of Co-transcriptional pre-mRNA splicing by m6A through the low-complexity protein hnRNPG. Mol. Cell76 (1), 70–81.e9. 10.1016/j.molcel.2019.07.005
231
Zhou P. Yang X.-L. Wang X.-G. Hu B. Zhang L. Zhang W. et al (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature579 (7798), 270–273. 10.1038/s41586-020-2012-7
232
Zhu D. Song Y. Hu D. Li S. Liu G. Li P. et al (2021). Characterization of enterovirus associated m6A RNA methylation in children with neurological symptoms: a prospective cohort study. Front. Neurosci.15, 791544. 10.3389/fnins.2021.791544
233
Zhu Y. Wang R. Zou J. Tian S. Yu L. Zhou Y. et al (2023). N6-methyladenosine reader protein YTHDC1 regulates influenza A virus NS segment splicing and replication. PLoS Pathog.19 (4), e1011305. 10.1371/journal.ppat.1011305
234
Zong X. Xiao X. Shen B. Jiang Q. Wang H. Lu Z. et al (2021). The N6-methyladenosine RNA-binding protein YTHDF1 modulates the translation of TRAF6 to mediate the intestinal immune response. Nucleic Acids Res., 49 (10), 5537–5552. 10.1093/nar/gkab343
235
Zumla A. (2010). Mandell, Douglas, and Bennett’s principles and practice of infectious diseases. Lancet. Infect. Dis.10 (5), 303–304. 10.1016/S1473-3099(10)70089-X
Summary
Keywords
N6-methyladenosine (m6A), writers, erasers, readers, infectious diseases
Citation
Yadav N, Khan R, Goyal S and Kumari R (2025) The role of m6A RNA methylation in infectious diseases. Front. RNA Res. 3:1531242. doi: 10.3389/frnar.2025.1531242
Received
20 November 2024
Accepted
18 March 2025
Published
10 April 2025
Volume
3 - 2025
Edited by
Sunny Sharma, The State University of New Jersey, United States
Reviewed by
Ashutosh Prakash Dubey, University at Buffalo, United States
Gayathri Govindaraju, The State University of New Jersey, United States
Updates
Copyright
© 2025 Yadav, Khan, Goyal and Kumari.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rajesh Kumari, drrajeshkumari@allduniv.ac.in
†These authors have contributed equally to this work
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.