Abstract
Dengue virus (DENV) is a zoonotic disease transmitted to humans via mosquito bites. Viral infection is systemic in humans and can lead to organ failure including in the liver. Since there are many molecular changes that precede liver failure, identification of progress at the molecular level is informative. As an RNA virus, DENV perturbs non-coding RNAs including microRNAs. Here we examine microRNA profiles in hepatoma cells using small molecule–mediated annotation of miRNA targets. It was previously shown that treatment with the broadly antiviral oxysterol 25-hydroxycholesterol (25HC) induces the expression of antiviral microRNAs in the liver. Herein, we show that 25HC is potently antiviral against Dengue virus and identify miR-4488 as a microRNA which is overexpressed during infection and downregulated with oxysterol treatment. We also show that miR-4488 is downregulated when the viral levels are lowered using siRNA, suggesting that this microRNA is involved in the host-response to infection. Since miR-4488 levels closely correlate with DENV levels in the liver, it serves as a biomarker for virus infection in the liver and may contribute the overall effects of DENV in this organ.
Introduction
microRNAs are small non-coding RNAs which endogenously regulate gene expression at the post-transcriptional level (Mohr and Mott, 2015; Nelson et al., 2003) with diverse roles in cellular pathways (Inui et al., 2010), metabolism (Rottiers and Näär, 2012; Rotllan et al., 2016), and disease (Calin and Croce, 2006; Croce, 2009). The role of these RNAs in host-pathogen interactions has become increasingly important as several microRNAs promote viral replication or participate in the immune response to infection (Scaria et al., 2006; Zhu et al., 2013). For instance, we have recently shown that microRNA-185 is an important immunomodulatory microRNA upregulated by 25-hydroxycholesterol (25HC) in the liver (Singaravelu et al., 2015). This oxysterol is produced through interferon stimulation of the cholesterol-25-hydroxylase gene as a response to viral infection (Liu et al., 2013), including Hepatitis C virus (HCV) infection. Furthermore, small-molecule inhibition of miR-185 targets was shown to be antiviral against HCV, demonstrating the utility of probing microRNA pathways in host-pathogen interactions (Filip et al., 2021).
The Dengue virus (DENV) is a positive-strand enveloped RNA virus belonging to the same family as HCV, the Flaviviridae. The virus is spread by mosquitos and is most prevalent in tropical regions, causing over 20,000 deaths per year (Roy and Bhattacharjee, 2021). Four serotypes of DENV exist and although primary infection is mild, re-infection with a heterologous serotype increases the risk of Dengue hemorrhagic fever (Diamond and Pierson, 2015). The viral genome of the DENV encodes for a polyprotein which is subsequently cleaved into three structural proteins and seven non-structural proteins including the essential protease (NS3) and an RNA-dependent RNA polymerase (NS5) (Acosta et al., 2014). DENV has been shown to exploit several host factors to facilitate its viral life cycle including inducing glycolysis (Fontaine et al., 2015), altering lipid metabolism (Tongluan et al., 2017; Randall, 2018) and modifying membrane composition (León-Juárez et al., 2016). The virus is also known for causing changes to cellular microRNA expression and both direct and indirect anti-viral microRNAs have been identified (Wong et al., 2020); however, the complete extent of altered microRNA expression during Dengue host-virus interactions remains unclear.
Herein, we use a microRNA screening strategy referred to as small molecule–mediated annotation of miRNA targets (SMART) (Singaravelu et al., 2015) to identify microRNAs which may participate in the immunometabolic response to DENV infection. We identified microRNA-4488 as a microRNA which is upregulated during infection and downregulated by antiviral treatment with 25HC or with an NS5-directed siRNA. This suggests miR-4488 may act as a biomarker for liver infection. When miR-4488 is overexpressed we observe a decrease in DENV by about 25%, suggesting that it may contribute to the cellular immune response. Overall, miR-4488 may be a novel microRNA involved in the immune response to DENV and may serve as a biomarker for infection.
Results and discussion
The immunoregulatory sterol 25HC is antiviral against DENV2
The oxysterol 25-hydroxycholesterol (Figure 1B) is the product of the broadly antiviral interferon-stimulated gene (ISG) cholesterol 25-hydroxylase which has been shown to have antiviral effects against various enveloped viruses by inhibiting viral entry. It is also known to act upon sterol-responsive element binding proteins (SREBPs) and other nuclear receptors (Liu et al., 2013). We sought to validate the antiviral effect of 25HC on the genotype 2 of the Dengue virus. Pre-treatment of Huh7 cells with 25HC 24 h before infection significantly reduced the levels of intracellular virus by approximately 75% after 48 h of infection (Figure 1A). Treating with the oxysterol following infection did not result in a significant reduction of viral RNA (data not shown), suggesting that the effects of 25HC put the cells in a preparatory anti-viral state.
FIGURE 1

25-hydroxycholestrol is antiviral against DENV2. (A) Treatment of Dengue infected Huh7 cells with 25HC reduces the levels of DENV glycoprotein E RNA. Cells were pre-treated for 24 h with 5 μM 25HC in 2% serum media before infection with Dengue at an MOI of 0.1 for 48 h. Levels of DENV2 glycoprotein E RNA were quantified via RT-qPCR and normalized to a non-treated control. n = 3 biologically independent replicates, error bars represent standard deviation. Significance was assessed by two-tailed, unpaired student’s t-test, ***p < 0.001. (B) Structure of 25-hydroxycholesterol.
miR-4488 is upregulated during DENV2 infection and downregulated with 25HC treatment
It is well-established that microRNAs play a crucial role in the immune response to infections (O’connell et al., 2010). To identify microRNAs which may participate in the host-response to DENV infection, we performed a screen on cells infected with the DENV2 virus in addition to cells pre-treated with 25HC. The cells were either infected with the virus, treated with the oxysterol or both, after which they were lysed and the whole RNA was extracted. The RNA was then processed and analyzed using the nanoString nCounter system miRNA assay. The resulting hits were normalized over housekeeping gene levels, resulting in five microRNAs significantly differentially expressed with DENV, eleven with 25HC treatment and two with both infection and 25HC treatment (Table 1).
TABLE 1
| microRNA | Fold change | P-value |
|---|---|---|
| Mock infected vs. DENV2 infected | ||
| hsa-miR-548ar-5p | 3.4 | 0.003 |
| hsa-miR-4488 | 4.1 | 0.007 |
| hsa-miR-3144-3p | 2.8 | 0.013 |
| hsa-miR-302d-3p | 3.5 | 0.018 |
| hsa-miR-612 | 2.8 | 0.022 |
| Vehicle treated vs. 25HC treated | ||
| hsa-miR-1246 | 3.5 | 0.003 |
| hsa-miR-1180-3p | −2.4 | 0.019 |
| hsa-miR-29b-3p | 2.6 | 0.024 |
| hsa-miR-126-3p | 1.8 | 0.024 |
| hsa-miR-25-5p | 1.8 | 0.029 |
| hsa-miR-448 | 1.4 | 0.034 |
| hsa-miR-520c-3p | 1.4 | 0.034 |
| hsa-miR-585-3p | 1.4 | 0.034 |
| hsa-miR-183-5p | 1.4 | 0.038 |
| hsa-miR-7-5p | 1.4 | 0.039 |
| hsa-miR-574-3p | 1.5 | 0.049 |
| DENV2 infected, vehicle treated vs. DENV2 infected, 25HC treated | ||
| hsa-miR-548ar-5p | −2.0 | 0.015 |
| hsa-miR-4488 | −2.5 | 0.030 |
microRNAs differentially regulated by DENV2 infection, 25HC treatment, or both.
Huh7s were pre-treated with 5 μM 25HC, or EtOH, vehicle for 24 h prior to infection (or mock infection) with DENV2 (MOI, 0.1) for 48 h. RNA, was extracted and then quantified by nanoString nCounter system. miRNA values upregulated during infection but downregulated during infection with 25HC are bolded. The average fold change of n = 3 biologically independent replicates is indicated.
The results obtained from the microRNA screen show two microRNAs which are represented in the DENV2 infected and in the infected, 25HC treated group: miR-548ar-5p and miR-4488. Interestingly, these microRNAs are upregulated with infection but downregulated with oxysterol treatment. We sought to validate these results using RT-qPCR. While expression levels for miR-548ar-5p were quite low, we were able to confirm a trending increase in miR-4488 levels with DENV2 infection and a pronounced, significant decrease in the miR-4488 expression with 25HC treatment (Figures 2A,B).
FIGURE 2
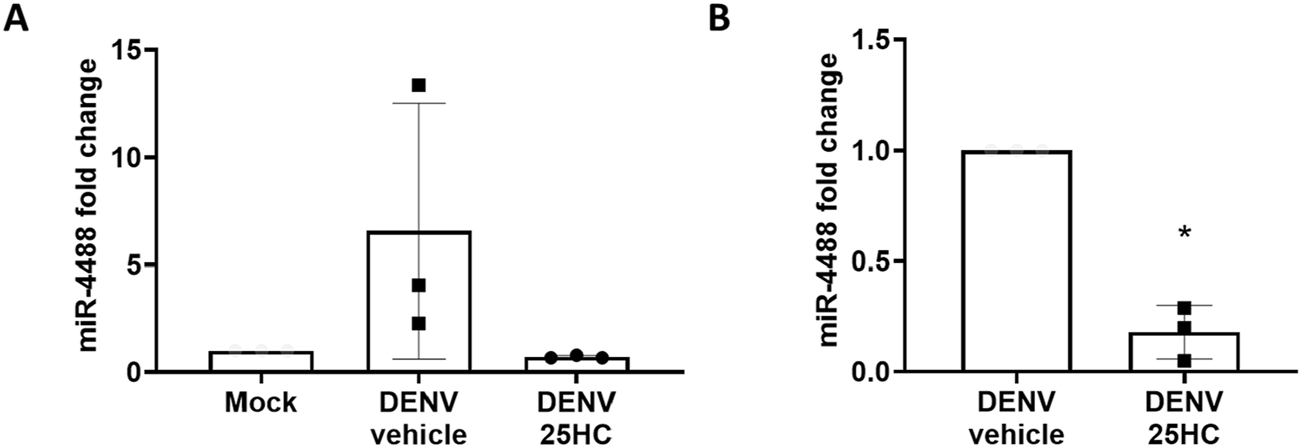
Huh7 cells infected with Dengue virus have higher levels of miR-4488 while cells pre-treated with 25HC pre-infection show reduced levels of miR-4488. Cells were pre-treated for 24 h with 5 μM 25HC or mock (ethanol) in 2% serum media before infection with Dengue at an MOI of 0.1 for 48 h. Levels of miR-4488 were quantified via Taqman RT-qPCR and normalized to non-infected mock (A) or to an infected and vehicle treated control (B). n = 3 biologically independent replicates, error bars represent standard deviation. Significance was assessed by two-tailed, unpaired student’s t-test, *p < 0.05.
miR-4488 is upregulated during DENV2 infection and downregulated when the virus is inhibited with siRNA
Since miR-4488 did not change significantly with 25HC treatment alone, we considered whether the microRNA was indeed modulated by the oxysterol, but only in the context of infection, or whether its decrease with 25HC treatment was due to the reduction in DENV levels. For that purpose, we used an siRNA designed by Villegas et al. (2018) targeting the NS5 region of the DENV2 genome to inhibit DENV2 replication and measured levels of miR-4488. First, we tested the siRNA in our infectious system and saw a robust downregulation of DENV levels similar to the 25HC treatment (Figure 3A). Interestingly, the levels of miR-4488 followed a similar trend with a strong increase during DENV infection with control siRNA and a significant downregulation in the samples transfected with the NS5 siRNA (Figures 3B,C). This suggests that miR-4488 is not a 25HC downregulated miRNA, but is upregulated during infection with DENV2 and downregulated when the infection is resolved. Thus, it is a biomarker that can track progression of DENV2 expression in the liver.
FIGURE 3
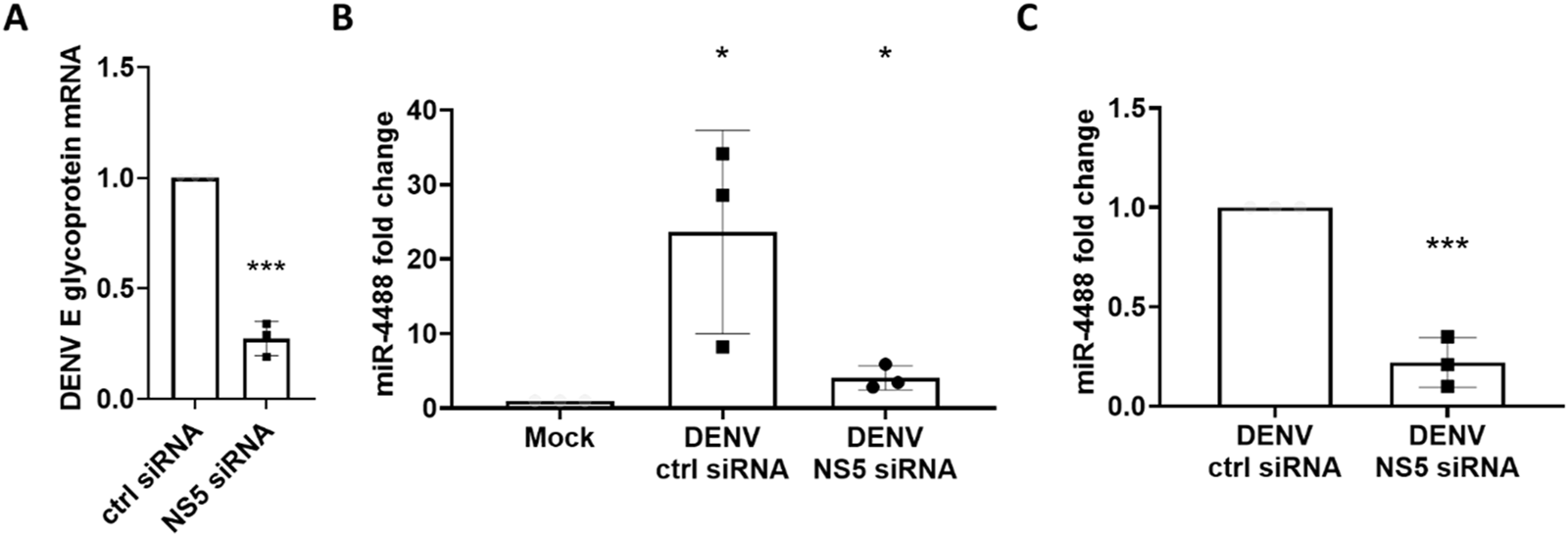
An NS5-targetting siRNA reduces levels of DENV2 virus and miR-4488. (A) An NS5-targeting siRNA is antiviral against DENV2. Cells were pre-transfected with 100 nM siRNA for 6 h before infection with Dengue at an MOI of 0.01 for 72 h (B) Huh7 cells infected with Dengue virus have higher levels of miR-4488 while cells pre-transfected with an NS5-targeting siRNA pre-infection show reduced levels of miR-4488. Levels of miR-4488 were quantified via Taqman RT-qPCR and normalized to non-infected mock (B) or vehicle treated controls (C). n = 3 biologically independent replicates, error bars represent standard deviation. Significance was assessed by two-tailed, unpaired student’s t-test, *p < 0.05, ***p < 0.001.
miR-4488 transfection reduces the levels of DENV RNA
Following evidence that miR-4488 is modulated by the levels of DENV2 virus, we sought to determine whether this microRNA influences DENV2 replication. We transfected Huh7 cells with a miR-4488 mimic or inhibitor 24, 48 or 72 h prior to infection with DENV2. The cells were then infected for 48 h, after which they were lysed and the RNA was extracted for RT-qPCR. We observed a statistically significant decrease of approximately 25% in the levels of DENV glycoprotein E in the samples transfected with the miR-4488 mimic for 24 or 48 h before infection (Figure 4A). However, transfection with the miR-4488 inhibitor did not show a significant increase in DENV2 levels (Figure 4B). This suggests that miR-4488 may be a microRNA involved in innate immunity with a modest antiviral effect.
FIGURE 4
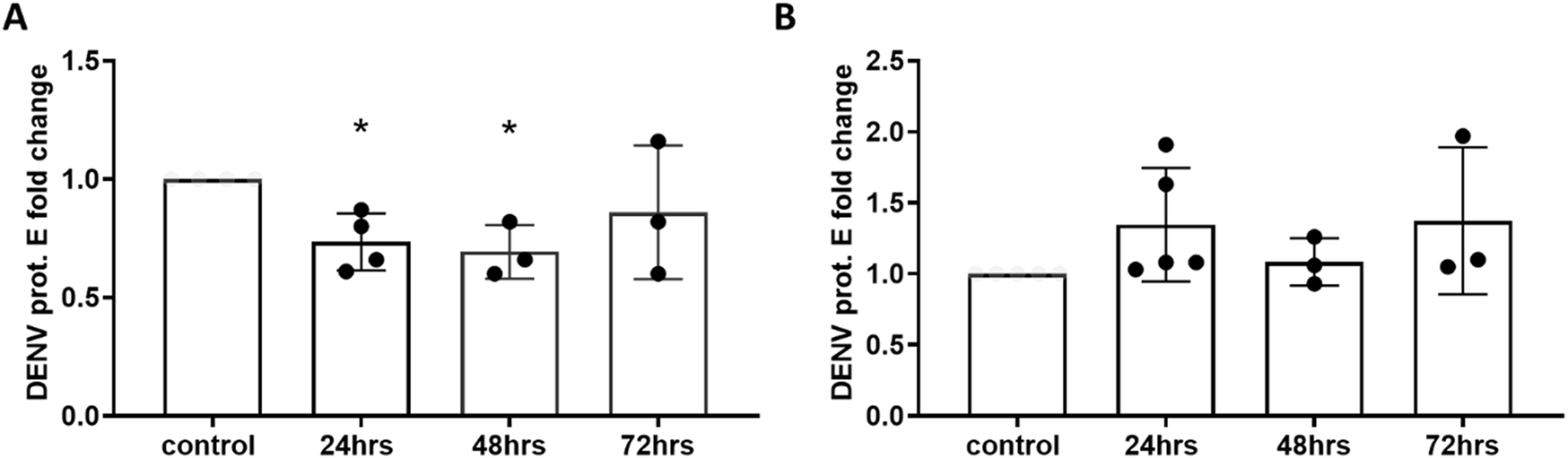
miR-4488 transfection reduces the levels of DENV RNA but inhibition does not significantly increase DENV RNA levels. Cells were transfected with either miR-4488 mimic (A) or inhibitor (B) for 24, 48 or 72 h before infection with DENV2 at an MOI of 0.1 for 48 h. Levels of DENV2 glycoprotein E RNA were quantified via RT-qPCR and normalized to a non-treated control. n = 3 biologically independent replicates. Significance was assessed by two-tailed, unpaired student’s t-test, *p < 0.05.
Transcriptomics and bioinformatic profiling reveals potential targets of miR-4488
To better understand how the microRNA elicits its modest antiviral effect, we transfected Huh7 cells with the miR-4488 mimic for 48 h, lysed the cells, extracted the RNA and performed transcriptomic analysis. Based on the finding that 48 h of microRNA-4488 transfection resulted in the largest DENV viral RNA downregulation, we chose this time point for our transcriptomic analysis. Transcriptional profiling yielded a diverse list of genes with decreased or increased RNA expression after transfection with miR-4488 (Supplementary Table S1).
Interestingly, a differential regulation of the immune system was observed during miR-4488 transfection with a notable 4.2-fold decrease in the interferon alpha 1 (IFNA1) gene, as well as decreases in defensin alpha 6 (DEFA6) and the immunoglobulin domains, IGHV3-72, IGKV1-27/IGKC and IGHV3-38. Interferons are potent regulators of the innate immune system, activated during viral infections to induce expression of antiviral genes (Taniguchi and Takaoka, 2001), defensins are anti-microbial peptides secreted by innate immune cells (Jarczak et al., 2013) and immunoglobulins are key contributors in the humoral immune response (Sun et al., 2020). Conversely, miR-4488 led to a modest increase in ubiquitin D which is regulated by tumor necrosis factor α (TNF-α) in the innate immune response (Kawamoto et al., 2018). Interestingly, interferon alpha 5 (IFN5) was also seen to decrease in transcript abundance by 1.7-fold. Given these findings, and that miR-4488 increases with DENV2, it would suggest that miR-4488 may act to reduce or limit induction of immune response during infection.
To determine if miR-4488 may directly regulate these genes, target prediction was performed with bioinformatic software miRDB (Chen and Wang, 2019). Interestingly, the immune system targets identified in transcriptomic analysis were not predicted targets, suggesting these are likely indirectly regulated or result from larger pathway consequences of direct miR-4488 effectors. To provide a better understanding of these direct effects, PANTHER Gene Ontology (GO) (Mi and Thomas, 2009; Thomas et al., 2022) of pathways (Figure 5A) and biological processes (Figure 5B) was performed. Interestingly, pathways of inflammation mediated by cytokines and chemokines were identified, mirroring the involvement of the immune system observed in the microarray data supporting an impact of miR-4488 on the immune system. The most common pathway regulated by miR-4488 was angiogenesis, consistent with previous findings that exovesicular miR-4488 regulates angiogenesis (Zheng et al., 2020). In addition, several metabolic intensive pathways such as Wnt and VEGF signaling were identified, complementing the top hits for biological processes GO: metabolic processes, biological regulation and cellular processes. Viruses like DENV heavily modulate the metabolome (Ahmed et al., 2022), supporting that miR-4488 is also a biomarker for DENV infection, coinciding with viral proliferation and the metabolic demands. These findings support the previous notion that miR-4488 serves as a biomarker for DENV infection and that miR-4488 may exert its modest antiviral effects through immune modulation.
FIGURE 5
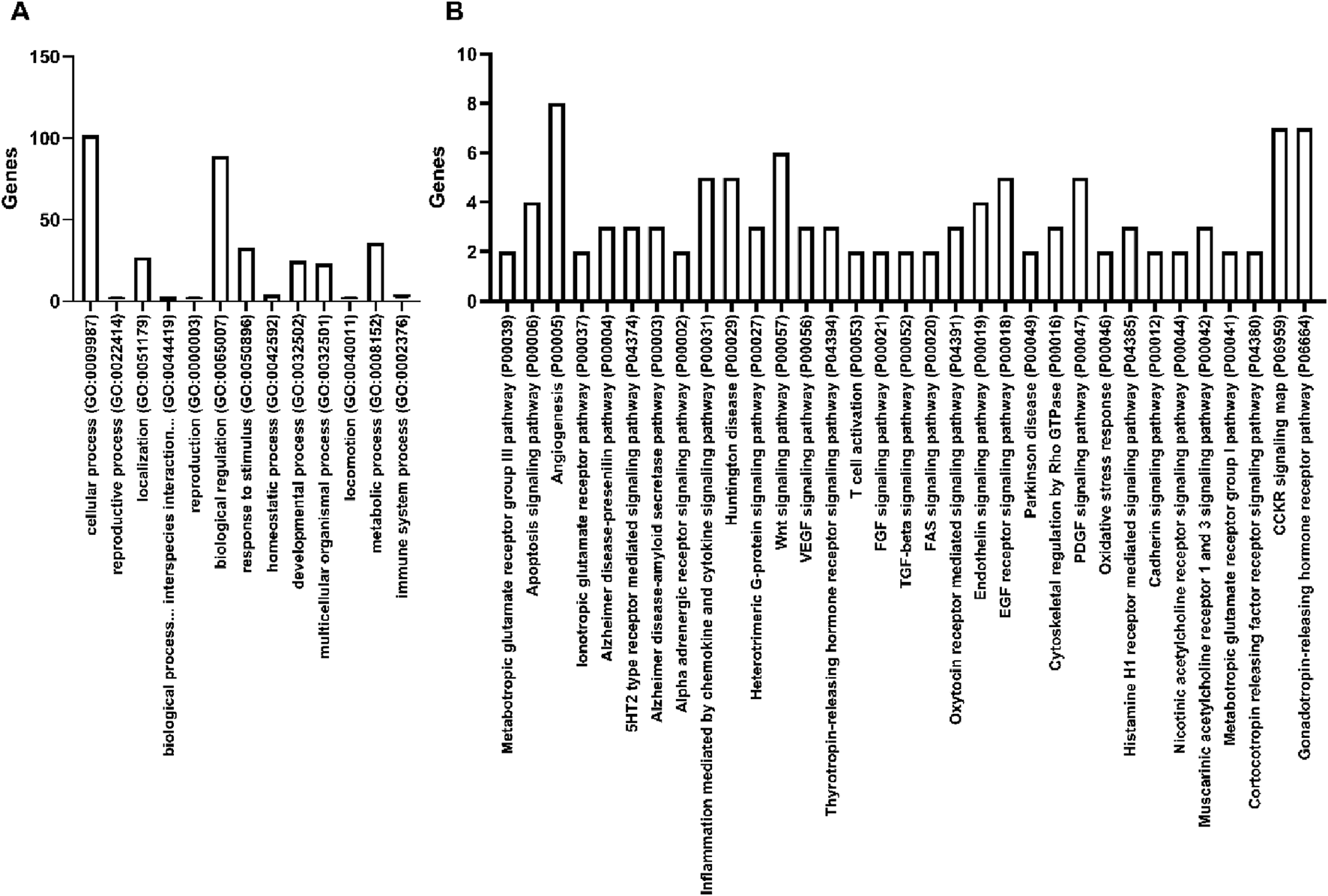
Gene Ontology (GO) analysis of miR-4488 predicted targets reveals several regulated pathways. miRDB was used to predict miR-4488 targets, yielding 208 genes. Panther GO was then performed on these predicted targets for biological processes (A) and pathways (B). Unspecified hits were removed. For pathway analysis, only pathways with regulated by >1 gene were included for clarity.
We have shown that miR-4488 is significantly overexpressed during DENV2 infection and that a resolution of the infection downregulates the microRNA's expression. This was the case with both 25HC treatment and with an DENV NS5-targeting siRNA. Transfecting the microRNA prior to DENV2 infection reduced viral RNA by approximately 25% and the preliminary transcriptomics analysis identified various immune related genes as potential genes of interest. These findings were then supported by bioinformatic analysis of predicted targets from high throughput crosslinking data and gene ontology. The data is also consistent with work that has recently shown miR-4488 to regulate macrophages in the immune response (Lu et al., 2024). This group specifically looked at the liver, which is impacted by severe DENV infection (Diamond and Pierson, 2015), and parallels other findings that miR-4488 is a biomarker for non-alcoholic fatty liver disease (Shen et al., 2023). Considering this information with our results suggests that miR-4488 may have focused utility as a biomarker for DENV progression and damage to the liver. Specific miRNAs for DENV-related liver damage have been explored in far less detail than differentially regulated genes, suggesting this could fill a beneficial clinical role. Future work characterizing liver tissue during in vivo DENV infection in the liver could offer deeper insight on the use of miR-4488 as a biomarker for viral progression and clearance.
Interestingly, the broader context of miR-4488-virus interactions is poorly understood. This microRNA has previously been identified as down-regulated in adeno-associated virus (AAV) infected cells, and transfection with the microRNA in Huh7 cells increased viral transduction (Arumugam et al., 2020). This supports the association between miR-4488 immunity involvement and host-pathogen interactions, but suggests there may be differential function depending on the pathogen. Future work exploring the effect of miR-4488 in the context of different viral infections, in addition to different pathogens, could enable deeper understanding into its role.
Taken together, this work establishes miR-4488 as a biomarker for DENV2 infection in the liver and has potential interesting mechanistic links to the management of DENV infection in this organ. The information has potential to aid in diagnosing disease severity and may offer insight into how to better therapeutically target this increasingly relevant virus.
Materials and methods
Cell culture
Human hepatoma cells (Huh7) were grown at 37°C in DMEM (Gibco, Thermo Fisher) supplemented with 10% fetal bovine serum (FBS) (Gibco, Thermo Fisher) and 10 mM non-essential amino acids (Thermo Fisher).
Virus infections and 25HC cell treatments
For the initial 25HC assays and miRNA panels, cells were seeded in six well plates in full media. The next day, select wells were treated with 5 μM 25HC or EtOH vehicle in DMEM with 2% FBS. The cells were incubated for 24 h, after which the media was replaced with DMEM containing infectious DENV particles at an MOI of 0.1, or just DMEM, and incubated for 2 h. Afterwards, the media was replaced with full DMEM, 10% FBS and incubated for 48 h. The cells were then lysed in RL buffer for RNA extraction. The total RNA isolation kit (Norgen) was used to perform RNA isolation as per manufacturer’s protocol.
MicroRNA profiling
Huh7 cells were cultured and treated as described above. The total RNA isolation kit (Norgen) was used to perform RNA isolation as per manufacturer’s protocol with on-column DNA digestion. The RNA samples were prepared for miRNA assay using the nanoString nCounter system as per manufacturer’s protocol. Briefly, the RNA was annealed to the nCounter miRNA tags, ligated and purified. The samples were then subjected to hybridisation with capture and reporter probes and post-hybridisation processing as well as data collection was performed using the nCounter Analysis system (nanostring). The resulting raw data was analysed using the nSolver (nanostring) software. The microRNA values were normalised over all housekeeping genes and fold changes were calculated relative to non-infected and/or vehicle treated samples.
NS5 siRNA transfections and infection
The NS5-targetting siRNA (5′-AGAGAAAGTGGACACGAGA-3′) was described by Villegas et al. (2018). The siRNA oligonucleotide was custom ordered (Sigma), reconstituted and annealed in annealing buffer (100 mM potassium acetate, 30 mM Hepes, and 2 mM magnesium acetate, ph7.5) by heating the sample to 95°C for 2 min and allowing it to gradually cool down at a rate of 1°C/min. Huh7 cells were seeded in six-well plates and incubated for 24 h. The next day, 100 nM of NS5 siRNA or non-targeting control was transfected using lipofectamine RNAimax (ThermoFisher Scientific) in Optimem (Gibco, Life Technologies) as per manufacturer’s protocol and the cells were incubated for 6 h. Afterwards, the siRNA mixture was removed and replaced with DMEM containing infectious DENV particles at an MOI of 0.01, or just DMEM, and incubated for 2 h. Following infection, the media was replaced with full DMEM, 10% FBS and incubated for 72 h after which the cells were lysed in RL buffer for RNA extraction using the total RNA isolation kit (Norgen) as per manufacturer’s protocol.
miR-4488 transfections and infection
Huh7 cells were seeded in six-well plates and incubated for 24 h. The next day, hsa-miRNA-4488 miRNA mimic, inhibitor or control miRNA (miRVana, ThermoFisher Scientific) were combined with Lipofectamine RNAiMAX transfection reagent (ThermoFisher Scientific) in Optimem (Gibco, Life Technologies) medium as per manufacturer’s protocol. The mixtures were added to the growth media of cells for a final concentration of 100 nM of miRNA. The cells were incubated for 24, 48 or 72 h after which the media was removed and replaced with DMEM containing infectious DENV particles at an MOI of 0.1, or just DMEM, and incubated for 2 h. Following infection, the media was replaced with full DMEM, 10% FBS and incubated for 48 h after which the cells were lysed in RL buffer for RNA extraction using the total RNA isolation kit (Norgen) as per manufacturer’s protocol.
Quantitative PCR
Reverse transcription was performed using the iScript cDNA Synthesis Kit (Bio-Rad) using 500 ng of RNA as per manufacturer’s protocol. Quantitative PCR (qPCR) for the DENV2 glycoprotein E was performed using SYBR Green Supermix (Bio-Rad) as per manufacturer’s protocol on the CFX Real-Time PCR Detection System (Bio-Rad). The primer sequences are as follows: fwrd 5′-TTGAGTAAACTGTGCAGCCTGTAGCTC-3′, rev 5′-GGGTCTCCTCTAACCTCTAGTCCT-3′. 18S rRNA was used for normalisation and expression fold changes relative to mock treatments were calculated using the 2−ΔΔCT method. For quantitative PCR of microRNA-4488, reverse transcription and qPCR was performed using the TaqMan MicroRNA assay (ThermoFisher) as per manufacturer’s protocol and the RNU6B target was used for normalisation.
Transcriptomics profiling
Huh7 cells were seeded in six-well plates and incubated for 24 h. The next day, hsa-miRNA-4488 miRNA mimic or control miRNA (miRVana, ThermoFisher Scientific) were combined with Lipofectamine RNAiMAX transfection reagent (ThermoFisher Scientific) in Optimem (Gibco, Life Technologies) medium as per manufacturer’s protocol. The mixtures were added to the growth media of cells for a final concentration of 100 nM of miRNA. The cells were incubated for 48 h after which they were lysed and the RNA was extracted using the total RNA isolation kit (Norgen) as per manufacturer’s protocol. RNA microarray profiling was performed by The Centre for Applied Genomics (TCAG), The Hospital for Sick Children, Toronto, Ontario, Canada. Analysis was performed using the transcriptome analysis console (Thermofisher).
miRNA target prediction and Panther GO
Bioinformatic miRNA prediction tool, miRDB (Chen and Wang, 2019) was used to generate a list of predicted miR-4488 targets. miRDB employs a machine learning model trained on experimental crosslinking immunoprecipitation (CLIP) and RNA-sequencing data to predict mRNA targets (Chen and Wang, 2019). A total of 208 genes met the software minimum threshold (scored >50) and all of these genes were included in subsequent analysis using Panther Gene Ontology (Mi and Thomas, 2009; Thomas et al., 2022). The two types of analysis performed using Panther Gene Ontology were Panther Biological Processes-Slim GO and Panther Pathway analysis. For Pathway analysis, due to the number of pathways identified, only those with >1 gene identified were included.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
RF: Data curation, Formal Analysis, Investigation, Methodology, Resources, Validation, Visualization, Writing – original draft. PE: Formal Analysis, Investigation, Software, Validation, Visualization, Writing – original draft, Writing – review and editing. SU: Investigation, Methodology, Writing – review and editing. RA: Investigation, Writing – original draft. NaA: Investigation, Writing – original draft. NoA: Investigation, Writing – original draft. JP: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by Natural Sciences and Engineering Council of Canada (210719).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frnar.2025.1566829/full#supplementary-material
References
1
Acosta E. G. Kumar A. Bartenschlager R. (2014). Revisiting dengue virus – host cell interaction: new insights into molecular and cellular virology. Adv. Virus Res.88, 1–109. 10.1016/B978-0-12-800098-4.00001-5
2
Ahmed N. Ahmed N. Pezacki J. P. (2022). miR-383 regulates hepatic lipid homeostasis and response to dengue virus infection. ACS Infect. Dis.8, 928–941. 10.1021/acsinfecdis.1c00470
3
Arumugam S. Mary B. Kumar M. Jayandharan G. R. (2020). Analysis of hepatic and retinal cell microRNAome during AAV infection reveals their diverse impact on viral transduction and cellular physiology. Gene724, 144157. 10.1016/j.gene.2019.144157
4
Calin G. a. Croce C. M. (2006). MicroRNA signatures in human cancers. Nat. Rev. Cancer6, 857–866. 10.1038/nrc1997
5
Chen Y. Wang X. (2019). miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res.48, D127–D131. 10.1093/nar/gkz757
6
Croce C. M. (2009). Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet.10, 704–714. 10.1038/nrg2634
7
Diamond M. S. Pierson T. C. (2015). Molecular insight into dengue virus pathogenesis and its implications for disease control. Cell162, 488–492. 10.1016/j.cell.2015.07.005
8
Filip R. Desrochers G. F. Lefebvre D. M. Reed A. Singaravelu R. Cravatt B. F. et al (2021). Profiling of MicroRNA targets using activity-based protein profiling: linking enzyme activity to MicroRNA-185 function. Cell Chem. Biol.28, 202–212.e6. 10.1016/j.chembiol.2020.12.009
9
Fontaine K. A. Sanchez E. L. Camarda R. Lagunoff M. (2015). Dengue virus induces and requires glycolysis for optimal replication. J. Virol.89, 2358–2366. 10.1128/jvi.02309-14
10
Inui M. Martello G. Piccolo S. (2010). MicroRNA control of signal transduction. Nat. Rev. Mol. Cell Biol.11, 252–263. 10.1038/nrm2868
11
Jarczak J. Kosciuczuk E. M. Lisowski P. Strazlkowska N. Jozwik A. Harbanczuk J. et al (2013). Bagnicka. Defensins: Natural component of human innate immunity. Hum. Immunol.74, 1069–1079. 10.1016/j.humimm.2013.05.008
12
Kawamoto A. Nagata S. Anzai S. Takahashi J. Kawai M. Hama M. et al (2018). Ubiquitin D is upregulated by synergy of notch signalling and TNF-α in the inflamed intestinal epithelia of IBD patients. J. Crohn’s Colitis13, 495–509. 10.1093/ecco-jcc/jjy180
13
León-Juárez M. Martínez-Castillo M. Shrivastava G. García-Cordero J. Villegas-Sepulveda N. Mondragón-Castelán M. et al (2016). Recombinant Dengue virus protein NS2B alters membrane permeability in different membrane models. Virol. J.13, 1–11. 10.1186/s12985-015-0456-4
14
Liu S. Aliyari R. Chikere K. Li G. Marsden M. Smith J. et al (2013). Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity38, 92–105. 10.1016/j.immuni.2012.11.005
15
Lu F. Ye M. Shen Y. Xu Y. Hu C. Chen J. et al (2024). Hypoxic tumor-derived exosomal miR-4488 induces macrophage M2 polarization to promote liver metastasis of pancreatic neuroendocrine neoplasm through RTN3/FABP5 mediated fatty acid oxidation. Int. J. Biol. Sci.20, 3201–3218. 10.7150/ijbs.96831
16
Mi H. Thomas P. (2009). PANTHER pathway: an ontology-based pathway database coupled with data analysis tools. Methods Mol. Biol.563, 123–140. 10.1007/978-1-60761-175-2_7
17
Mohr A. M. Mott J. L. (2015). Overview of microRNA biology. Semin. Liver Dis.35, 003–011. 10.1055/s-0034-1397344
18
Nelson P. Kiriakidou M. Sharma A. Maniataki E. Mourelatos Z. (2003). The microRNA world: small is mighty. Trends Biochem. Sci.28, 534–540. 10.1016/j.tibs.2003.08.005
19
O’connell R. M. Rao D. S. Chaudhuri A. A. Baltimore D. (2010). Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol.10, 111–122. 10.1038/nri2708
20
Randall G. (2018). Lipid droplet metabolism during dengue virus infection. Trends Microbiol.26, 640–642. 10.1016/j.tim.2018.05.010
21
Rotllan N. Price N. Pati P. Goedeke L. Fernández-Hernando C. (2016). microRNAs in lipoprotein metabolism and cardiometabolic disorders. Atherosclerosis246, 352–360. 10.1016/j.atherosclerosis.2016.01.025
22
Rottiers V. Näär A. M. (2012). MicroRNAs in metabolism and metabolic disorders. Nat. Rev. Mol. Cell Biol.13, 239–250. 10.1038/nrm3313
23
Roy S. K. Bhattacharjee S. (2021). Dengue virus: epidemiology, biology, and disease aetiology. Can. J. Microbiol.67, 687–702. 10.1139/cjm-2020-0572
24
Scaria V. Hariharan M. Maiti S. Pillai B. Brahmachari S. K. (2006). Host-virus interaction: a new role for microRNAs. Retrovirology3, 68. 10.1186/1742-4690-3-68
25
Shen N. Tang L. Qian Y. Pan J. Pan J. Miao H. et al (2023). Serum miR-4488 as a potential biomarker of lean nonalcoholic fatty liver disease. Ann. Transl. Med.11, 173. 10.21037/atm-22-6620
26
Singaravelu R. O’Hara S. Jones D. M. Chen R. Taylor N. G. Srinivasan P. et al (2015). MicroRNAs regulate the immunometabolic response to viral infection in the liver. Nat. Chem. Biol.11, 988–993. 10.1038/nchembio.1940
27
Sun Y. Huang T. Hammarstrom L. Zhao Y. (2020). The immunoglobulins: new insights, implications and applications. Annu. Rev. Anim. Biosci.8, 145–169. 10.1146/annurev-animal-021419-083720
28
Taniguchi T. Takaoka A. (2001). A weak signal for strong responses: interferon-alpha/beta revisited. Nat. Rev. Mol. Cell Biol.2, 378–386. 10.1038/35073080
29
Thomas P. D. Ebert D. Muruganujan A. Mushayahama T. Albou L. Mi H. (2022). PANTHER: making genome-scale phylogenetics accessible to all. Protein Sci.31, 8–22. 10.1002/pro.4218
30
Tongluan N. Ramphan S. Wintachai P. Jaresitthikunchai J. Khongwichit S. Wikan N. et al (2017). Involvement of fatty acid synthase in dengue virus infection. Virol. J.14, 28–18. 10.1186/s12985-017-0685-9
31
Villegas P. M. Ortega E. Villa-Tanaca L. Barrón B. L. Torres-Flores J. (2018). Inhibition of dengue virus infection by small interfering RNAs that target highly conserved sequences in the NS4B or NS5 coding regions. Arch. Virol.163, 1331–1335. 10.1007/s00705-018-3757-2
32
Wong R. R. Abd-Aziz N. Affendi S. Poh C. L. (2020). Role of microRNAs in antiviral responses to dengue infection. J. Biomed. Sci.27, 4–11. 10.1186/s12929-019-0614-x
33
Zheng X. Lu S. He Z. Huang H. Yao Z. Miao Y. et al (2020). MCU-dependent negative sorting of miR-4488 to extracellular vesicles enhances angiogenesis and promotes breast cancer metastatic colonization. Oncogene39, 6975–6989. 10.1038/s41388-020-01514-6
34
Zhu S. Pan W. Qian Y. (2013). MicroRNA in immunity and autoimmunity. J. Mol. Med.91, 1039–1050. 10.1007/s00109-013-1043-z
Summary
Keywords
microRNA, dengue virus (DENV), nanoString, 25-hydroxycholesterol, miR-4488
Citation
Filip R, Evers P, Uguccioni SM, Alonzi RC, Ahmed N, Ahmed N and Pezacki JP (2025) microRNA-4488 is differentially regulated during dengue virus infection and clearance of the virus. Front. RNA Res. 3:1566829. doi: 10.3389/frnar.2025.1566829
Received
25 January 2025
Accepted
02 April 2025
Published
16 April 2025
Volume
3 - 2025
Edited by
Simranjeet Kaur, Steno Diabetes Center Copenhagen (SDCC), Denmark
Reviewed by
Yuemei Dong, Johns Hopkins University, United States
Yuquan Tong, The Scripps Research Institute, United States
Updates
Copyright
© 2025 Filip, Evers, Uguccioni, Alonzi, Ahmed, Ahmed and Pezacki.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John P. Pezacki, john.pezacki@uottawa.ca
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.