- 1School of Public Health and Preventive Medicine, Faculty of Medicine, Nursing and Health Sciences, Monash University, Melbourne, VIC, Australia
- 2One Water Laboratory, Department of Civil & Environmental Engineering, Monash University, Clayton, VIC, Australia
- 3Melbourne Water, Melbourne, VIC, Australia
Introduction: Monitoring of drinking water sources is an essential component of broader public health practise. However, routine water monitoring programmes that follow established methodological standards, such as low-volume grab sampling with standard filtration, have limitations in being representative. Particularly for protected source waters where (wildlife introduced) pathogens are in low concentration and are not evenly distributed. Microbial source tracking (MST) offers a promising approach to close this gap, enabling more precise identification of faecal contamination sources and their associated risk. However, as with other culture- and molecular-based approaches, the sensitivity of MST is constrained by sample capture methodology, limited by sample volume, timing, and randomness of grab sampling.
Methods: This study investigated the application of a high-volume sample concentration method (EasyElute ultrafiltration) to enhance microbial recovery from source water. All evaluation was conducted alongside standard grab sampling and filtration methods. Post-concentration analyses combined traditional culture-based quantification of faecal indicator organisms (FIOs) and reference pathogens, with 16S rRNA amplicon MST to provide an integrated approach to surveillance of animal-derived microbial risks in forested water supply catchments.
Results: The results demonstrated that high-volume ultrafiltration enhanced bacterial recovery from source water samples, although turbidity was observed to limit overall efficiency, highlighting potential operational challenges. Comparative analysis demonstrated that amplicon-based MST produced consistent faecal source attribution across both standard and ultrafiltration methods, showing greater sensitivity at increasing volumes.
Discussion: This study advances MST methodology by demonstrating the feasibility and added sensitivity achievable through high-volume, concentrated sample collection approaches. This is particularly relevant where water samples are expected to carry low microbial loads, ultimately offering a practical approach for improving faecal source tracking and risk assessment for water sources to protect public health in water supply catchments.
1 Introduction
Monitoring of drinking water sources is the first step in assessing waterborne public health risk. Catchment-scale water quality monitoring supports sustainable management practises and provides essential information on the potential drivers of risk, including biological, chemical, and/or environmental factors. Amongst biological hazards, human faecal contamination of drinking water is generally considered the greatest risk to human health (Hokajärvi et al., 2024). However, in protected drinking water catchments, faecally derived contamination is predominantly driven by local wildlife populations. This can occur due to direct deposition into water or surface run-off due to climatic events.
Zoonotic risks to drinking water safety are generally regionally specific and dependent on animal populations. However, knowledge of enteric pathogen carriage rates and concentrations is often unknown or is calculated based on similar global studies. Furthermore, regionally specific transport processes (such as surface flow particle association) and external influencing factors (such as UV radiation, temperature, and rainfall) differentially affect the survival and retention of microbes within deposited animal scats. Such factors often reduce, but do not remove, the overall zoonotic-risk profile within nearby surface waters (Derx et al., 2021). Consequently, in order to be informative, the monitoring and quantification methods applied to understand zoonotic risk must have adequate sensitivity and limits of detection, coupled with suitable sampling frequency and spatial coverage. This is to not only alert governing bodies as to changes in risk profile, but also to provide long-term regionally relevant intensive catchment data.
Faecal indicator organisms (FIOs), such as Escherichia coli and enterococci, are applied globally for water quality assessment. These organisms are derived from faeces and can indicate recent faecal contamination events, but due to their widespread distribution as part of mammalian gut microbiota, they do not generally indicate the source of faecal contamination (World Health Organization, 2022). Reference pathogens are a specific subset of microorganisms (viral, bacterial, and protozoa) that are applied to plan and assess the effectiveness of treatment processes (natural, engineered, or chemical) for specific water systems (World Health Organization, 2022). They are often selected due to their potential introduction, within faeces or scats, into the system under investigation (Holcomb and Stewart, 2020). Although FIOs can be applied as a proxy for the presence of pathogens within water, their direct relationship with reference pathogens has been demonstrated to be inconsistent and study specific (Kozak et al., 2025; Korajkic et al., 2018).
In protected water bodies, FIOs and pathogen concentrations are often below method detection limits. However, small changes in overall water quality, as a result of faecal contamination, can influence water quality and should be understood to ensure appropriate treatment barriers are in place and remain effective. These risks can only be characterised where downstream methods are of sufficient sensitivity. Continued collection of at- or below the limit of detection data is not informative for long-term management and is of minimal cost–benefit to the stakeholder. Improved method sensitivity and specificity are therefore required for informed risk mitigation, public health assessment, and economic benefit. Achieving this requires a context-specific understanding of faecal-derived sources of risk, validated methods for the detection and quantification of relevant zoonotic pathogens and faecal indicators, as well as the application of complementary approaches to track underlying changes in source inputs.
Microbial source tracking (MST) offers an approach to the identification of sources of contamination within environments through the application of chemical or biological markers. MST has been widely reviewed (Barrett et al., 2025; Gitter et al., 2023; Scott et al., 2002) as part of public health and water quality monitoring programmes. In general, the application of library-dependent genetic markers has been shown to complement the findings of FIO and reference pathogen quantification, while providing insights into the sources contributing to faeces within catchments (Henry et al., 2016; McCarthy et al., 2017).
Evaluation of large volume ultrafiltration methods for the concentrated capture of FIOs, as well as viruses, protozoa, and Campylobacter spp., has been described previously (Ferrari et al., 2019; Ferguson et al., 2004; Pascual-Benito et al., 2020). However, investigation of the recovery efficiencies, essential knowledge for limiting method bias and over- or under-estimation of risk, of reference bacterial pathogens has had limited investigation. Furthermore, the application of a single uniform capture method to evaluate FIOs, pathogen and genomic-driven MST has not been previously reported, but may provide a platform for advancing microbial water quality surveillance, hazard, and risk assessment.
This study seeks to validate an approach to quantifying microbial hazards, encompassing faecal indicator organisms (FIOs), reference pathogens, and microbial source tracking (MST). A case study using surface water from a protected catchment was applied to demonstrate the applicability of this methodological framework. Limits of detection and recovery efficiencies for standard and large volume ultrafiltration methods are presented for reference bacterial pathogens, with guidance on the application of microbial source tracking tools for evaluation of faecal risk from ultrafiltration outputs. Importantly, this study contributes to the refinement of sampling methodologies for low-contamination settings, demonstrating how integration with enhanced concentration methods can improve microbial surveillance towards supporting operational decision making.
2 Materials and methods
2.1 Preparation of large-scale mixed faeces
To understand faecal recovery using standard filtration and EasyElute ultrafiltration methods, a Master Faeces (MF) sample was constructed, which included contributions from selected representative animal sources (Figure 1). Fresh scat samples were collected as described by Henry et al. (2018) from protected drinking water catchments. Faeces were stored at 4 °C for transport to the One Water Laboratory (OWL; Monash University, Australia) within 4 h of collection. Post receipt, 50–450 g of scat was collected from multiple (>5) representative animals from a single animal source type. These were hand homogenised to mix into a single “Master Source” faeces. From each “Master Source,” 3–30 g was taken and combined to a total weight of 100 g of mixed sources. This mixture was homogenised via stomaching at 250 rpm for 1 min in a single larger container to make the final MF. Secondary homogenisation was then conducted in 100 g batches, to ensure evenly applied mixing, using a stomacher at 250 rpm for 1 min. The final MF was stored at 4 °C across the experimental period of 28 days. The MF was applied to faecal dosing of raw water at 1 day, 14 days, and 28 days post-homogenisation (Figure 1).
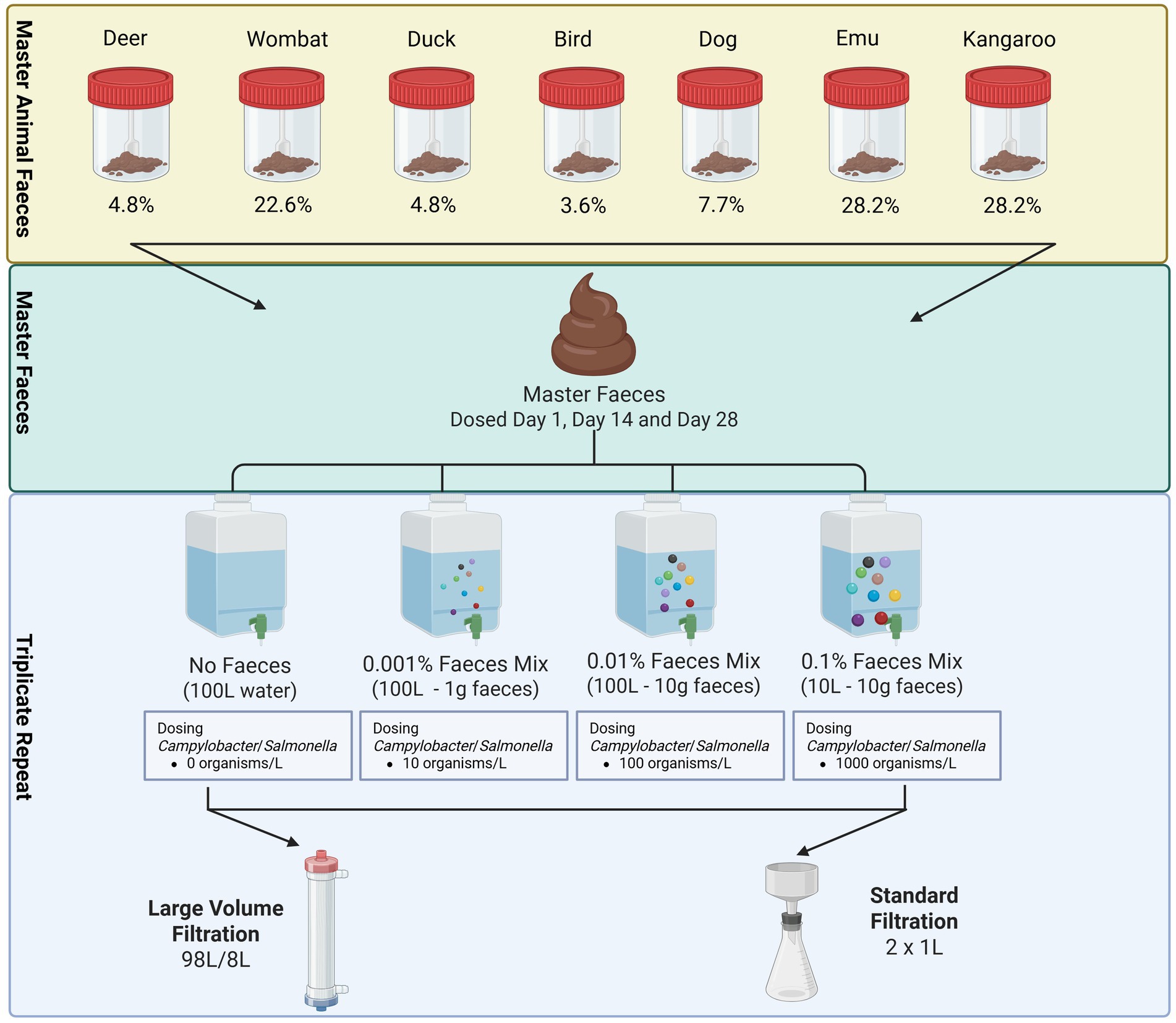
Figure 1. Experimental design for faecal dosing of source water samples for evaluation of EasyElute ultrafiltration system. Weights and percent contribution for each master animal faeces into the final MF are provided under each sample category.
2.2 Preparation and quantification of faecal indicators and reference pathogens
Reference microbes and the methods by which they were quantified are listed in Table 1. Salmonella and Campylobacter were derived from environmental isolates, strains Salmonella_1359 and Campylobacter_20240212, held in the One Water Biobank (Monash University, Clayton, Australia). Prior to and on the day of experiments (at both start and end of experiments), concentrations of both organisms were quantified as CFU/mL using serial dilution following Australian standard methods (Table 1). Campylobacter quantification was confirmed via haemocytometer cell counting and culture-based Most Probable Number (MPN) analysis (Bolton et al., 1982). Briefly, three series of 10x dilutions of suspension from the stock were prepared. The cell suspensions were spread plated (100 μL) over selective Charcoal Agar plates (duplicate per concentration) and incubated at 42 °C for 48 h in microaerophilic conditions. MPN was then calculated. Faecal indicators E. coli and enterococci were quantified to determine background concentrations within the MF (Table 1). Control samples of source water and faecal mixture underwent quantification as negative and positive controls, respectively, for all organisms (Table 1).
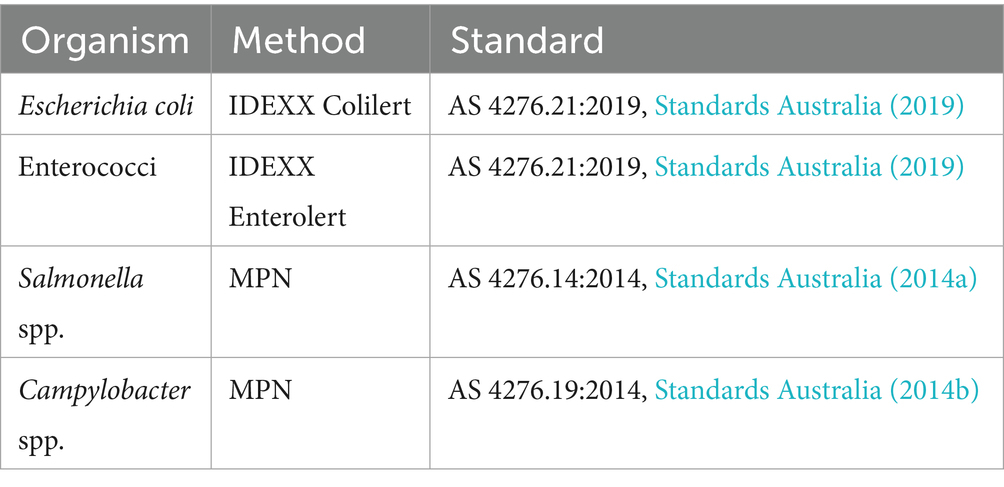
Table 1. Methods applied for the quantification of faecal indicators and reference pathogens in this study.
2.3 Faecal dosing of source water and EasyElute ultrafiltration
At day 0, day 13, and day 27, a volume of 400 L of source water was collected from a single forested catchment reservoir and transported at room temperature to the OWL. The water was held at 20 °C ± 3 °C for a period of no longer than 24 h prior to experimentation, where it was subsequently subdivided into 3× 100 L and 1× 10 L volumes (Figure 1). A 100 L water sample was retained as a source water negative control, with subsequent 100 L samples receiving a 10-fold increasing concentration of MF. This was added, with agitation, to reach a total faeces contribution of 0.001% (1 g) and 0.01% (10 g) (Figure 1). To the 10 L raw water, 10 g of faeces was added with mixing, to reach a contribution of 0.1%. This was informed by initial experiments, which demonstrated limitations to volumes able to be filtered at higher turbidity. Prior to each experiment, a 2 g aliquot of the MF was removed and stored at −80 °C for downstream DNA extraction and 16S rRNA amplicon analysis.
The source-water-faeces mixture was then spiked with reference pathogens (Salmonella and Campylobacter) to a target final concentration ranging from 10 organisms/L (0.001% faecal dose) to 1,000 organisms/L of water (0.1% faecal dose). The dosed mixture was consistently stirred throughout the experiment to ensure homogeneity was maintained. Once dosed, a 2 L sub-sample was removed into a 2 L food-grade vessel, representing a standard “grab” sample. The remaining 98 L (or 8 L) underwent ultrafiltration and subsequent elution using the EasyElute-PBS method (Innovaprep, United States) as per the manufacturer’s directions. Final elution volumes for each of the three experiments are provided in Supplementary Table S1.
Quantification of E. coli, enterococci, Salmonella, and Campylobacter was conducted using the methods outlined in Table 1. Detection limits, as calculated by MPN, for 2 L grab and EasyElute (Innovaprep, United States) approaches are presented in Supplementary Table S1 for each sample type. Water samples were processed for downstream DNA extraction using the method described in Henry et al. (2016). Briefly, for 2 L grab samples, water to a volume of 1 L was sub-sampled and cellular material collected onto 5× 0.22 μM filters (Millipore, Germany). The filters of the same sample type were combined and stored at −80 °C prior to DNA extraction. For EasyElute derived samples, a volume of concentrated eluate equivalent to 1 L was collected on 1× 0.22 μm filters (Millipore, Germany). All filters were stored at −80 °C prior to DNA extraction.
2.4 Recovery efficiency and statistical analysis
Percent recovery of FIOs, Salmonella and Campylobacter was calculated to assess the efficiency of detection across treatments. Recovery was expressed as:
where the expected concentration was determined by:
Observed concentrations (MPN/L) were measured from processed water samples, while the inoculum concentration and associated dilution factor were used to calculate the expected concentration of organisms, based on dosage applied at each of the three experimental time points. This enabled quantitative comparison of recovery efficiency across methods and across faecal dosing levels. Due to the small sample size, a Student’s t-test (Student, 1908) was applied to determine significant differences between expected and observed recoveries.
2.5 Genomic DNA extraction for water and faecal samples
Total genomic DNA was isolated using the MagMax Wastewater Nucleic Acid kit (Thermofisher Scientific, United States) with garnet bead beating (Capella Science, Australia). Stored 0.22 μm filters were removed from the freezer to thaw briefly. Garnet bead tubes were filled with 800 μL Lysis buffer and 1× 0.22 μm filter paper containing microbial cellular material before being subjected to a bead beating for 45 s at 6.5 m/s. Faecal material (0.25 g in replacement for 1× 0.22 μm filter) was processed equivalently. Automated extraction was performed using a KingFisher Apex system (Thermofisher Scientific, United States), with a 20 min digestion cycle and a 35 min wash cycle. DNA was eluted in 50 μL RNase-Free H2O (Qiagen) prior to storage at -20 °C.
2.6 Short read 16S rRNA amplicon sequencing and data processing
Targeted 16S rRNA amplicon sequencing was undertaken by Monash University Genomics and Bioinformatics Platform (Monash University) as previously described in Henry et al. (2016, 2018). Briefly, 16S rRNA PCR amplicon libraries were prepared by amplifying the V3–4 region of the bacterial rRNA gene in triplicate 50 μL reactions. Reactions contained 1 μm forward (TCGTCGGCAGC GTCAGATGTGTATAAGAGACAGCCTACGGCWGCAG) and reverse (GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGG ACTACHVGGGTATCTAATCC) primers; 5 μL of purified concentration-standardised genomic DNA; 25 μL of 2x HiFi HotStart ReadyMix (Kapa Biosystems, United States). Amplified DNA, derived from the triplicate reactions, was then pooled for each sample and purified using Ampure XP (0.6 V) according to the manufacturer’s protocol. Subsequently, the libraries were pooled in equimolar concentrations and sequenced using an MGITech DNBSEQ-G400RS sequencing instrument with High-Throughput Sequencing Set FCLPE100 chemistry using the manufacturer’s instructions: MGI document SOP-013-B01-082 revision 7. Sequence reads are available on SRA1, Bioproject PRJNA1334114.
Short-read 16S amplicon metagenome sequencing data was processed using QIIME2 v2023.9 (Bolyen et al., 2019). This process began with the import and merging of paired-end sequences. Paired-end sequence reads were first imported, merged, and demultiplexed, and a summary of sequence quality was obtained. Reads were then denoised using the Divisive Amplicon Denoising Algorithm (DADA2) based on their quality information (Callahan et al., 2016). Regions with low-quality scores (below 30) were removed. Prior to the denoising process, FIGARO (Weinstein et al., 2019) and fastp (Chen et al., 2018) were used to determine optimal trimming parameters. As a result, the forward reads were truncated to 294 base pairs and the reverse reads to 256 base pairs, with 7 maximum errors for forward reads, and 6 for reverse reads. The first 20 base pairs of both forward and reverse reads were also trimmed.
Features with counts contributing less than 1% of the sample were filtered out to prepare the data for MST using SourceTracker v2 (Knights et al., 2011). In this framework, all of the animal faecal matter in the sequencing library was assigned as sources; while the MF, direct bacterial spiked water samples, and faecal dosed water samples were assigned as sinks—environments that receive faecal microbial inputs from one or more sources (Table 2). Prior to MST, the quality of sources was assessed using leave-one-out analysis via SourceTracker, to identify if the type of a source sample can be inferred by SourceTracker as the source type when it was initially sampled (Lim et al., 2024). Misclassified sources were removed from the source library. Analysis was then undertaken with the following default parameters: rarefaction depth = 10,000, Gibbs sampling restarts = 10; burn-in iterations = 100; α1 hyperparameter = 0.1; α2 hyperparameter = 0.001; and β hyperparameter = 0.01 (Henry et al., 2016). A total of five SourceTracker replicate runs were conducted. Based on the microbial community contributions, the relative faecal community contributions were calculated by normalising the values to only include contributions from animal faecal sources. The relative standard deviation percentages (%RSD) were calculated for the source contributions, with results containing RSD ≥ 100% being excluded from further analysis.

Table 2. Categories of source and sink samples, including their type and a definition for each of the categories.
To improve the sensitivity for specific detection of faecal contributions, un-spiked water sample controls were included as sources to form a “background” microbial community. The faecally dosed water samples from standard filtration and EasyElute ultrafiltration were then compared against each other to identify differences in predicted faecal contributions. The samples were compared at different spike concentrations through the use of the Kruskal–Wallis test (Cabin and Mitchell, 2000; Kruskal and Wallis, 1952). Faecal contribution analysis was conducted on the MF in order to understand changes in the predicted contributions and composition from different animals (i.e., bird, deer, dog, emu, kangaroo, and wombat) across the experimental period (i.e., P2 and P3). Total faecal contribution and %RSD were calculated as described earlier.
3 Results
3.1 Assessing recovery using high-volume filtration
Culture-based analysis of the MF demonstrated that E. coli and enterococci concentrations remained on average at 2.9 × 105 MPN/g [range of 7.9 × 104–6.1 × 105] and 1.9 × 105 MPN/g [range of 6.0 × 103–4.5 × 105], respectively, over 1 month, at a 4 °C holding temperature. Similarly, taxonomic evaluation specifically targeting E. coli-associated amplicons showed an increase from 0.8 to 11.9% and enterococci from 0.1 to 0.9% across the equivalent period. As temporal variations in the FIO concentrations were not observed, dosing experiments applied the same MF. FIO quantification was conducted for both the standard method and for the ultrafilter eluate at an equivalent volume. Negative control (source water) FIO concentrations were below method detection limits, irrespective of filtration approach (Supplementary Table S1). Recovery of E. coli was comparable between methods, relative to input dose (Figure 2; Supplementary Table S2). This was also observed for enterococci, with the exception of 0.001% faecal input. Overall, the EasyElute method was observed, across experiments, to have significantly higher enterococci recoveries, across experiments and dosing (χ2 = 3.21, df5) than that of the standard filtration method (χ2 = 82.8, df5).
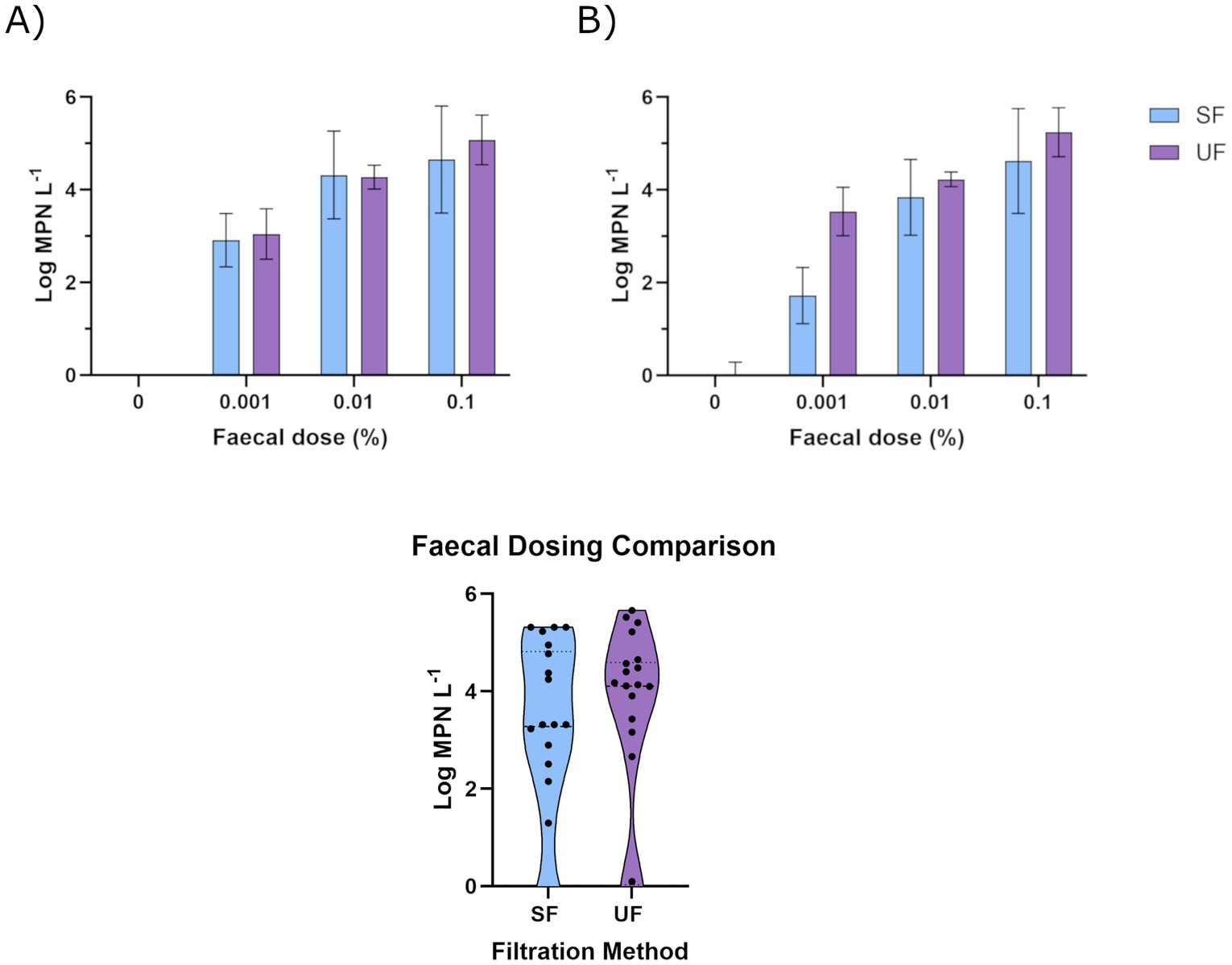
Figure 2. Measured concentration of faecal indicators (Log MPN/L) (A) E. coli and (B) enterococci, using standard filtration (SF, blue) and EasyElute ultrafiltration (UF, purple) at faecal doses of 0–0.1% concentration in 100 L of raw water. Data presented as average and standard deviation of triplicates; where values were non-transformable, they were excluded from analysis (n ≥ 2).
Dosing of Salmonella and Campylobacter was applied at concentrations ranging from zero (source water) to 1,026 MPN/100 L (Log10 3.0) and 760 MPN/100 L (Log10 2.9), respectively. It was noted that the results for both organisms were highly variable between experiments, as demonstrated by the large observed standard deviations (Figure 3). Campylobacter recoveries averaged 115 and 125% for EasyElute and standard filtration samples, respectively, when no MF was dosed, and averaged 158 and 128% respectively, across MF-dosed samples. In contrast, for Salmonella, there was no detection of the organism by either method below 0.01% faecal dosing, indicative of a < 1% recovery efficiency. A difference was observed at 0.1% (1,026 MPN/100 L), with EasyElute recovery efficiency reaching 47%. It should be noted, however, that despite having the highest recovery, only 10 L of the 0.1% raw water-faecal mixture could be processed, unlike the 100 L which was possible for all other dilutions. This was directly associated with sample turbidity (averaging 36.8 Nephelometric Turbidity Units), resulting in blockage of the EasyElute filter and preventing further processing. However, recovery efficiency improved for EasyElute and standard filtration when no MF contribution was present (>76% recovery; 0 + spike). These differences between Campylobacter and Salmonella recoveries, observed between samples that were dosed with MF, compared to those that were not dosed, suggested organism-specific particle attachment and associated removal through the filtration process.
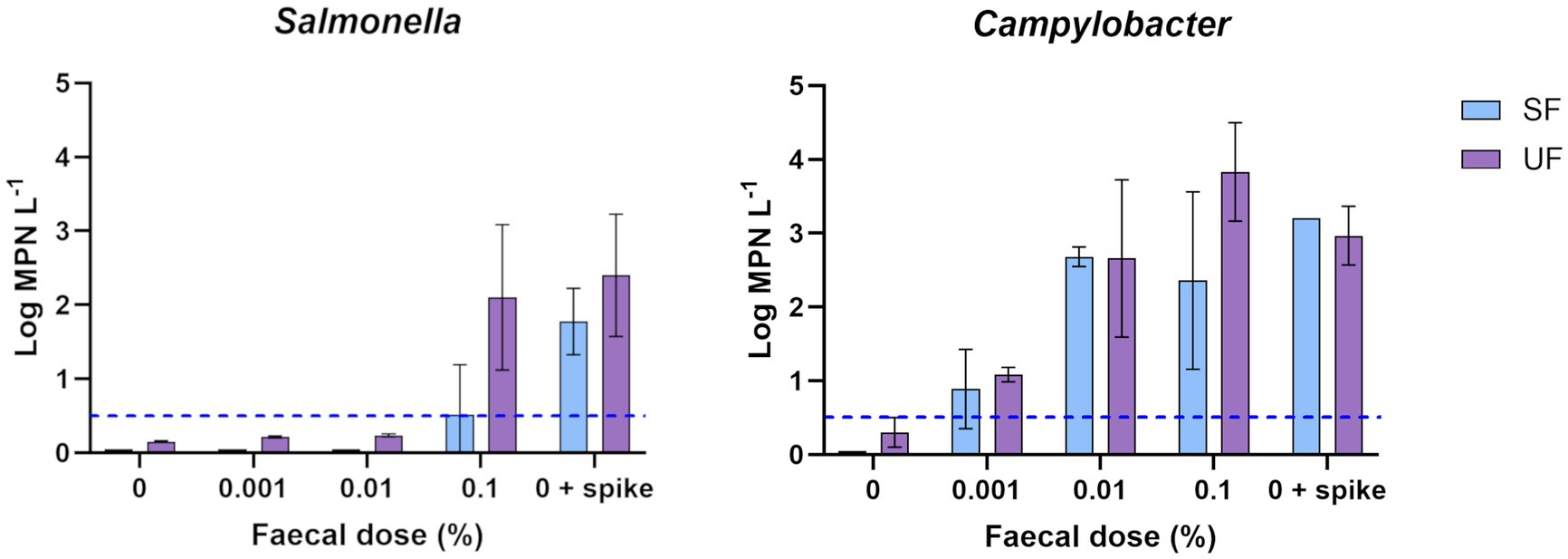
Figure 3. Measured concentration of target pathogens (Log MPN/L) using standard filtration (SF, blue) and EasyElute ultrafiltration (UF, purple) at faecal doses to a final contribution between 0 to 0.1% in raw water. Samples labelled as “0 + spike” no faecal source water control, with Campylobacter and Salmonella to equivalent concentrations to 0.1%. Data presented as average and standard deviation of triplicates, where values were non-transformable, they were excluded from analysis (n ≥ 2). Where source water MPN/L was below detection, value was divided by 2 prior to Log transformation. Where MPN was over-detection, the value was set to the maximum detectable value prior to the Log transformation. The average lower detection limit is indicated by the blue dotted line.
3.2 Assessment of the application of microbial source tracking to high-volume filtrate eluates
Bacterial community analysis and MST were conducted on the amalgamated MF to investigate temporal changes in bacterial composition prior to water analysis. MST results indicated that as faecal age increased, the accuracy of identifying the original source of the scat declined (Figure 4A). Unknown contributions increased from ~20% at 24 h to >80% at 14 days. At 14 days, significant source-specific fingerprints were still detectable, contributing on average 29.1% of the total fingerprint. By 28 days, this declined to 18.5%, with the majority of the fingerprint attributed to wombat (Vombatidae; 16.5%) and emu (Dromaiidae; 1.6%). Increases in unknown contributions were predicted to be associated with corresponding increases in the abundance of Pseudomonas spp., Flavobacterium spp., and Comamonas spp. populations within the MF, alongside decreases in broader bacterial community diversity (Figure 4B).
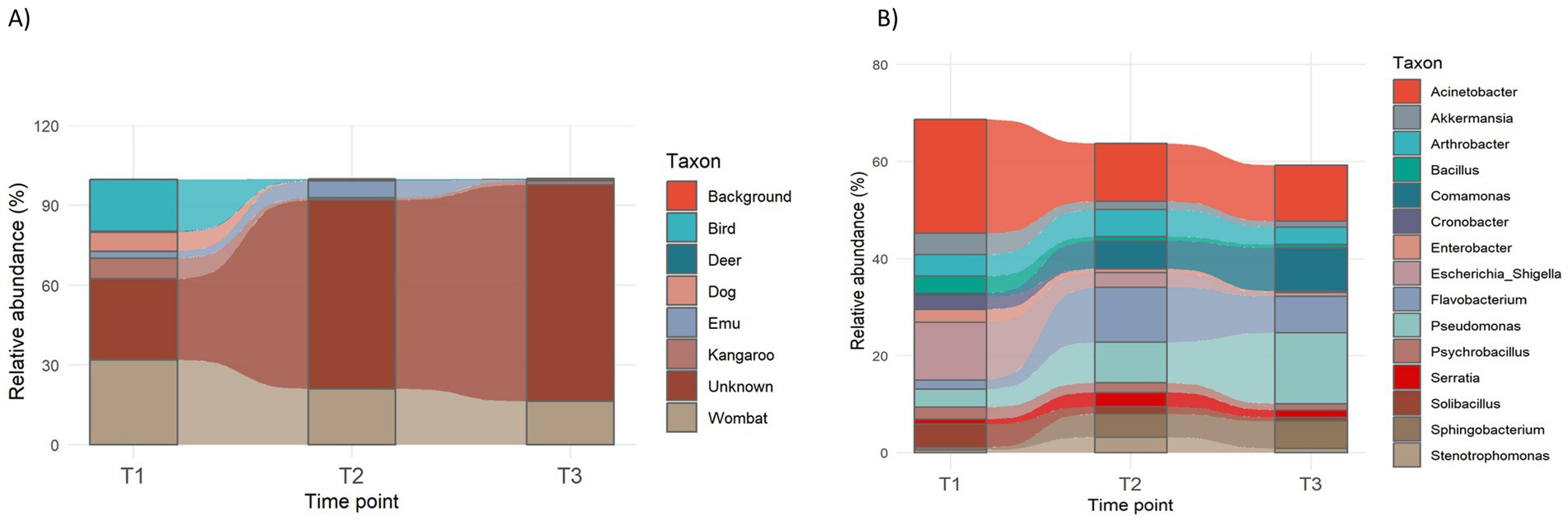
Figure 4. Average source-specific faecal contribution (A) and (B) bacteria contributing >50% of the MF bacterial composition at 24 h (T1), 14 days (T2), and 28 days (T3) post-storage at 4 °C.
Method comparison was undertaken to determine whether genomic MST performed equivalently between methods (Figure 5). Raw water was included as a source, alongside animal community fingerprints, to account for background water bacterial contributions in MST analysis. As expected, predicted raw water contributions declined markedly across faecal doses—from ~64.5% and ~69.3% for standard and EasyElute at 0.001% dose, to 1.14 and 1.37%, respectively, at 0.1%. Comparison of predicted source contributions at equivalent sample volumes showed no significant method-specific differences (Figure 5; p > 0.99). However, at the 0.001% faecal dose, significant contributions from kangaroo (Macropodidae) and avian were detected at higher proportions than in the MF (Figures 5A,B). This may indicate that dilution can improve sensitivity for detecting specific sources in mixed faecal deposits. Overall, predicted faecal contributions were comparable regardless of the age of faeces used for dosing. These findings suggest that method-specific differences in elution and recovery do not significantly affect conclusions drawn from amplicon-based MST analysis.
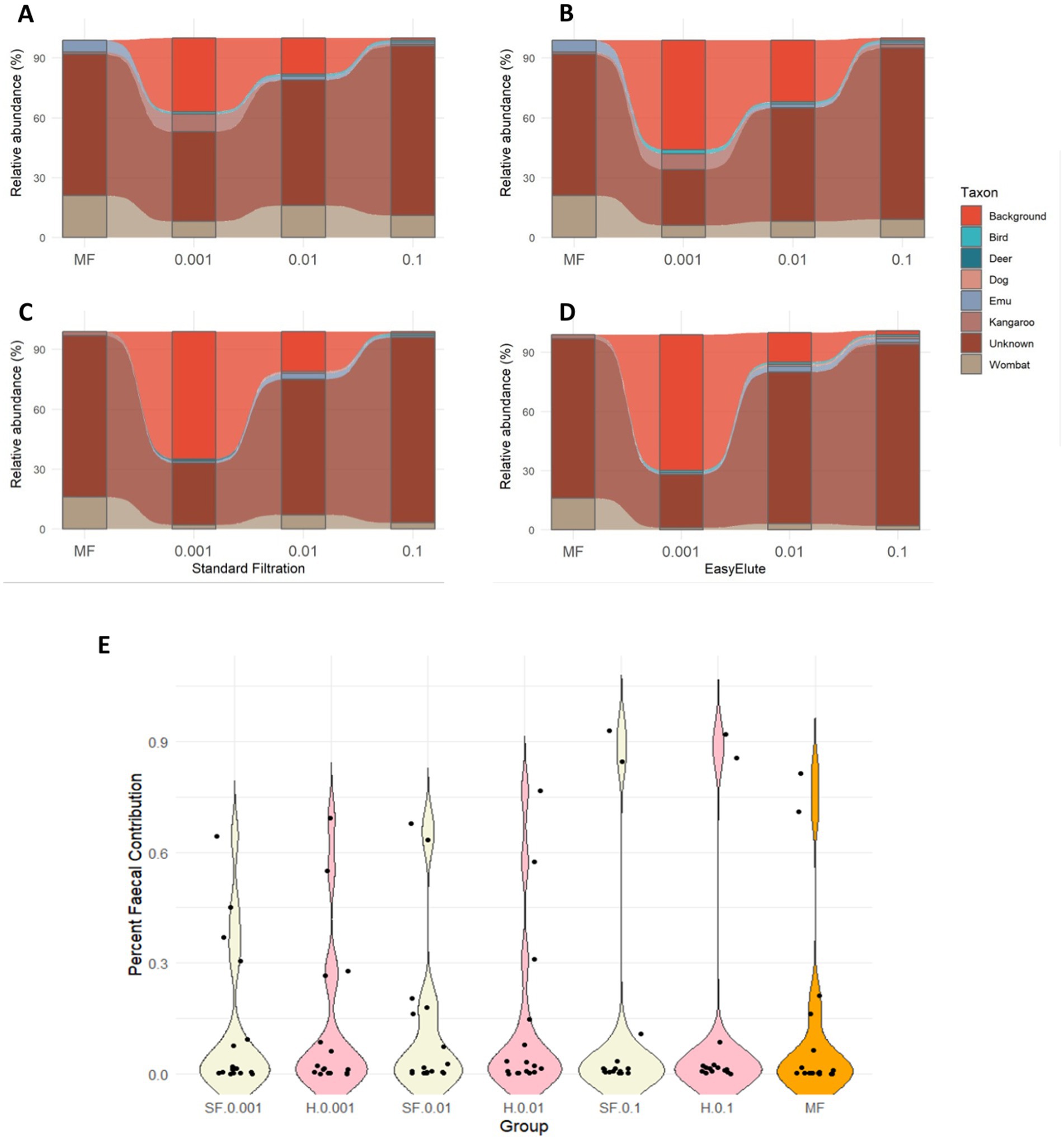
Figure 5. Predicted faecal contributions (A–D) within EasyElute ultrafilters (B,D), standard filtration (A,C), and master faeces (MF) across three dosing concentrations (0.001, 0.01 and 0.1%) at 14 days (A,B) and 28 days (C,D) post faecal storage at 4 °C. Distribution of predicted faecal contributions (E) across all replicates and sample configurations (standard filtration = SF, EasyElute = H) irrespective of the age of MF applied to dosing.
4 Discussion
Monitoring the microbial quality of water sources within drinking water supply catchments is a cornerstone of public health protection (Kroehler et al., 2014). However, current surveillance methods often lack the sensitivity to detect low-prevalence, high-risk pathogens, particularly those shed by local wildlife populations, thus limiting their ability to inform timely and effective interventions (Pluym et al., 2024). In many protected water supply catchments, significant resources are invested in routine water monitoring programmes that follow established methodological standards, such as low-volume grab sampling with standard filtration. While these approaches provide consistency, they inherently constrain detection sensitivity, often representing less than a millionth of 1% of the total supply, and with limited temporal coverage. This mismatch between sampling volume and the spatial–temporal distribution of contamination, particularly from wildlife faecal deposition both on land and water, can lead to underestimation of risk and inefficient use of monitoring resources. Improving both the sensitivity and contextual accuracy of pathogen detection, including differentiation between human and animal sources, is therefore a critical step for developing a more robust understanding of health risk towards supporting strategic risk-management and operational decision-making.
This study applied three faecal dosing thresholds, each exceeding levels typically found in water sampled from protected drinking water catchments. Though, in general, these represented supra-environmental loads, their application was necessary to allow direct comparison and uncertainty assessment between approaches, mirroring other method validation studies (Hubbard et al., 2024; Kataoka et al., 2025; Smith and Hill, 2009). Furthermore, FIO concentrations in surface waters of low-impacted pastoral catchments are often on the order of 102–104 CFU 100 mL−1 (Lim et al., 2022) and higher during E. coli bloom events, which have been reported to occur in open catchments (Bertone et al., 2019). Thus, while the dosing employed exceeds typical environmental levels within forested catchments by one or more orders of magnitude, experimentation under these conditions provided a framework relevant to a wide range of drinking water catchments. The use of a composite master faecal inoculum, incorporating multiple wildlife sources, further reflected the microbial complexity of real-world contamination events, particularly those following rainfall.
The persistence of E. coli and enterococci within the MF, held at 4 °C across 28 days, aligned with previous studies, which demonstrated low decay rates for these organisms at low temperature (Börjesson and Rehnstam-Holm, 2024; Tran et al., 2020; Lowe et al., 2010). Although beyond the scope of the current study, it is noted that faecal indicator persistence, under cooler climatic conditions, complicates their interpretation and application as markers of recent contamination (Nowicki et al., 2021); a fact that should be considered as part of the development of monitoring frameworks. Initial trials also investigated direct pathogen dosing into the faecal matrix to better replicate their natural introduction into catchment waters. However, Campylobacter and Salmonella strains recently isolated from wildlife exhibited rapid die-off (<1 h) when added to the master faeces composite, an effect not observed when the same pathogens were dosed directly into water samples. Comparable reductions in pathogen viability within manure have been reported and often attributed to microbial competition or predation in nutrient-rich environments (Hutchison et al., 2005; García et al., 2010). While this limitation is unlikely to have affected the present findings, it underscores an important methodological consideration for experiments modelling pathogen transport from faecal sources to receiving waters.
Culture-based results for E. coli and enterococci concentrated through the EasyElute system were consistent with previous ultrafiltration studies (Smith and Hill, 2009; Mull and Hill, 2012). For Campylobacter, previous studies have applied large-volume ultrafiltration without explicitly quantifying recovery variability (Ferrari et al., 2019; Ferrari et al., 2019). The current study demonstrated this to be close to 100%, with less variability than the standard method and minimal observable influence from particle association as a result of increased faecal dosing. In contrast, Salmonella results presented a more complex picture. Despite direct dosing, recovery decreased with increasing water turbidity, suggesting faecal particle association and sediment settling effects were occurring rapidly within the sample mixture. The EasyElute did achieve overall higher Salmonella recoveries than what was observed by the standard approach, and in line with previous bulk water dosing studies (Kraft et al., 2023); where >96% recovery was achieved under conditions equivalent to “0 + spike” samples. While there is a lack of studies that have investigated Salmonella dosing within animal faeces prior to entry into waterbodies, particle association and biofilm formation have both been described within the genus (Bardsley et al., 2021; Pardo et al., 2020). Given the time course of the current study, biofilm formation was unlikely to have resulted in the observed recovery as occurred across both standard and EasyElute methods, suggesting particle association and loss through attachment to the filter membrane as a possible cause.
Dead-end ultrafilters, such as the EasyElute, capture microbes and particulates on the filter fibres while allowing liquid to pass through membrane pores <0.1 μm in diameter (Mull and Hill, 2012). Turbidity has been identified in previous studies as a significant factor affecting microbial recovery efficiency (Kataoka et al., 2025; Mull and Hill, 2012; Ferrari et al., 2019; Smith and Hill, 2009; Leskinen et al., 2012; Kataoka et al., 2025). However, this effect is not uniform across organisms. For example, positive correlations with turbidity are often observed for E. coli and enterococci, whereas inverse relationships have been reported for organisms such as Clostridium and MS2 bacteriophage (Mull and Hill, 2012; Smith and Hill, 2009). The observed inter-experiment and turbidity-associated variability highlights the need for further investigation into potential operational constraints, particularly when deploying systems targeting specific organisms. These findings underscore the importance of evaluating system performance under the range of water quality extremes expected in the field and knowledge of target microbe capture under varying turbidity conditions prior to operational deployment.
The findings suggest that application of high-volume ultrafiltration, alongside or as an alternative to standard sampling, can improve overall sensitivity through reduction of method detection limits when applied to organism-specific assays. Overlaying water quality factors, in particular turbidity, will provide an upper limit to volumes that can be processed through the system, and associated microbial recoveries (Pascual-Benito et al., 2020). However, the advantages offered by these systems include potential for deployment over extended sampling periods, allowing improved resolution, at a lower cost, of temporal dynamics that cannot be achieved by standard low-volume sampling alone (Leskinen et al., 2012). For public health response, the demonstrated potential for application during and over high-risk events, as a means of detecting and monitoring outbreaks, represents a significant advantage to current methods (Kataoka et al., 2025).
Genomic approaches using 16S rRNA amplicons have previously been shown to relate to FIO patterns and provide valuable insights into faecal source attribution (Henry et al., 2016; McCarthy et al., 2017). However, their application has been limited to grab samples, with no prior studies linking high-volume concentrates to both source and pathogen data. An unexpected finding of the current study was that, although the sensitivity of source-specific attribution declined with increasing MF age, dilution effects, introduced as a result of source water dosing, were observed to partly mitigate this reduction. In particular, kangaroo, emu, and avian signatures were observed to re-emerge at lower concentrations; however, these were minimised by 28 days. This suggests that ageing effects, which can negatively affect MST resolution (Wong et al., 2016; Devane et al., 2023), can be partially overcome by increased MST sensitivity under low faecal load.
Though this phenomenon has not been previously reported, to the authors’ knowledge, the observed finding is hypothesised to be the result of several interacting mechanisms. Particularly in response to dilution and reduction of faecal biomass, which can improve amplification of low-abundance taxa. Dilution reduces the concentration of PCR inhibitors such as bile salts, humic acids, and polysaccharides, improving amplification of low-abundance DNA from aged faeces (Hunter et al., 2019). It also reduces total biomass per extraction, preventing saturation of extraction binding matrices in initial steps, ensuring more even recovery of minority templates. In addition, abundant taxa in concentrated samples can dominate PCR and sequencing, masking signatures from older or low-abundance organisms (Bender et al., 2018). Overall, these results suggest that controlled dilution may have helped mitigate ageing-related detection biases in MST assays; a finding that warrants further validation across diverse catchments and contamination scenarios.
The findings of the 16S rRNA amplicon study also have broader implications. As the environmental persistence of pathogens is increasingly recognised (Balta et al., 2024), the ability to profile sources from both recent and aged contamination offers a valuable tool for ongoing risk assessment. Fresh faeces represent an immediate hazard, but older deposits can continue to contribute pathogens to waterways, particularly as climate change alters microbial survival and transport dynamics (Liu and Salles, 2024). Effective management, therefore, requires not only systems for detecting current risks but also approaches that capture longer-term, low-level hazards. The integration of high-volume capture with sensitive molecular tools, such as MST, therefore, provides a pathway for this enhanced understanding. Thus, allowing prioritisation of mitigation measures, alongside quantitative data for regulatory FIOs and reference pathogens, in response to wildlife contamination signals. Future investigations should include defining regionally specific and sensitive molecular MST markers to support quantification of risk and future integration as part of quantitative microbial risk assessment evaluation of risk-based thresholds (Barrett et al., 2025; Gitter et al., 2023).
The methodological framework demonstrated here has broad applicability across diverse catchment types and geographic regions, particularly for water supplies where contamination events are infrequent but may pose elevated public health risk. In this context, early detection is essential to understand contamination events, identify sources, and support preventive actions and planning. Integration of the EasyElute method into routine monitoring should consider several operational factors, including assessment of water quality extremes, particularly turbidity and particle loads, to anticipate potential recovery limitations, incorporation of appropriate internal recovery controls matched to target organisms to quantify method-associated losses, and optimisation of sample processing through strategies such as reduced system flow rates to mitigate filter clogging and improve reproducibility. Adaptive monitoring designs could further enhance performance by deploying high-volume ultrafiltration during low-turbidity periods or alongside standard methods, especially for detecting low-prevalence pathogens or performing microbial source tracking. Collectively, these measures will ensure that both sensitivity and operational feasibility are maintained during catchment surveillance.
Further field-based validation, inclusive of organism-specific recoveries, is required to establish if there is a direct correlation between FIOs, pathogens, and MST results. However, future studies should align with broader operational priorities for drinking water quality programmes globally, including (1) Incorporation of pathogen surveillance as direct indicators of health risk, alongside traditional faecal indicator monitoring, to more accurately assess health risks from enteric pathogens. (2) Adopting high-volume ultrafiltration to improve the likelihood of detecting low-prevalence hazards, thereby increasing the robustness of downstream risk assessments (such as quantitative microbial risk assessment). (3) Increasing certainty surrounding pathogen concentrations in source waters to ensure drinking water treatment is commensurate with risk, consistent with the risk-based approach to drinking water management presented in guidelines, such as the Australian Drinking Water Guidelines (NHMRC, NRMMC, 2011), and (4) Expanding the use of molecular and genomic tools, including MST, to identify, quantify, and track sources of faecal contamination, guiding the selection of relevant monitoring targets and thereby improving understanding of animal-related risks. By uniting high-volume capture, sensitive detection, and source attribution in a single workflow, this study provides a transferable and scalable platform for advancing microbial water quality surveillance and risk assessment. Its adoption can support more proactive, evidence-based management of drinking water sources and contribute to safeguarding public health.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because animal scats were obtained post defecation, from within the forested catchments. No animal interaction occurred, or were disturbed as a result of the study.
Author contributions
FL: Data curation, Formal analysis, Project administration, Supervision, Validation, Writing – original draft, Writing – review & editing. TL: Data curation, Formal analysis, Investigation, Validation, Writing – original draft, Writing – review & editing. MellS: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Writing – review & editing. MeliS: Conceptualization, Funding acquisition, Resources, Writing – review & editing. LB: Formal analysis, Investigation, Methodology, Validation, Writing – review & editing. GL: Data curation, Investigation, Writing – review & editing. DD: Data curation, Formal analysis, Methodology, Writing – review & editing. SS: Conceptualization, Funding acquisition, Methodology, Writing – review & editing. NO: Conceptualization, Methodology, Resources, Writing – review & editing. AW: Conceptualization, Funding acquisition, Methodology, Resources, Writing – review & editing. SH: Conceptualization, Funding acquisition, Methodology, Writing – review & editing. C-WT: Formal analysis, Investigation, Methodology, Supervision, Validation, Writing – review & editing. RH: Conceptualization, Funding acquisition, Methodology, Resources, Supervision, Writing – original draft, Writing – review & editing, Formal analysis, Investigation.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The Australian Medical Research Future Fund MRFCRI000195 which supported the salaries of the authors to conduct this research. Melbourne Water for funding the analytical components of this study and providing access to forested catchments and research support for this study.
Acknowledgments
The authors wish to acknowledge the Australian Medical Research Future Fund MRFCRI000195 and Melbourne Water for funding this study. The authors acknowledge the application of Microsoft Copilot, powered by the GPT-4 architecture, to support primary editing, where necessary, of the prepared document.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that Gen AI was used in the creation of this manuscript. We acknowledge the application of Microsoft Copilot, powered by the GPT-4 architecture, to support primary editing, where necessary, of the prepared document.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frwa.2025.1694489/full#supplementary-material
Footnotes
References
Balta, I., Lemon, J., Murnane, C., Pet, I., Vintila, T., McCleery, D., et al. (2024). The one health aspect of climate events with impact on foodborne pathogens transmission. One Health 19:100926. doi: 10.1016/j.onehlt.2024.100926
Bardsley, C. A., Weller, D. L., Ingram, D. T., Chen, Y., Oryang, D., Rideout, S. L., et al. (2021). Strain, soil-type, irrigation regimen and poultry litter influence Salmonella survival and die-off in agricultural soils. Front. Microbiol. 12:590303. doi: 10.3389/fmicb.2021.590303
Barrett, L. R., Beasy, P., Palacios Delgado, Y. M., Boyce, J. D., Leder, K., McCarthy, D. T., et al. (2025). Beyond borders: a systematic review and meta-analysis of human-specific faecal markers across geographical settings. Crit. Rev. Environ. Sci. Technol. 55, 447–464. doi: 10.1080/10643389.2025.2455031
Bender, J. M., Li, F., Adisetiyo, H., Lee, D., Zabih, S., Hung, L., et al. (2018). Quantification of variation and the impact of biomass in targeted 16S rRNA gene sequencing studies. Microbiome 6:155. doi: 10.1186/s40168-018-0543-z
Bertone, E., Kozak, S., and Roiko, A. (2019). Understanding and modelling the occurrence of E. coli blooms in drinking water reservoirs. Water Resour. Res. 55, 10518–10526. doi: 10.1029/2019WR025736
Bolton, F. J., Hinchliffe, P. M., Coates, D., and Robertson, L. (1982). A most probable number method for estimating small numbers of campylobacters in water. J. Hyg. (Lond) 89, 185–190. doi: 10.1017/S0022172400070716
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Börjesson, S. M. E., and Rehnstam-Holm, A. S. (2024). Different persistence among strains of E. Faecalis and E. faecium in sterile treated wastewater microcosms: effects of temperature and ciprofloxacin. J. Water Health 22, 1328–1336. doi: 10.2166/wh.2024.131
Cabin, R. J., and Mitchell, R. J. (2000). To Bonferroni or not to Bonferroni: when and how are the questions. Bull. Ecol. Soc. Am. 81, 246–248.
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884. doi: 10.1093/bioinformatics/bty560
Derx, J., Demeter, K., Linke, R., Cervero-Aragó, S., Lindner, G., Stalder, G., et al. (2021). Genetic microbial source tracking support QMRA modelling for a riverine wetland drinking water resource. Front. Microbiol. 12:668778. doi: 10.3389/fmicb.2021.668778
Devane, M. L., Taylor, W., Dupont, P. Y., Armstrong, B., Weaver, L., and Gilpin, B. J. (2023). Exploring the bacterial community in aged fecal sources from dairy cows: impacts on fecal source tracking. Microorganisms 11:1161. doi: 10.3390/microorganisms11051161
Ferguson, C., Kaucner, C., Krogh, M., Deere, D., and Warnecke, M. (2004). Comparison of methods for the concentration of Cryptosporidium oocysts and Giardia cysts from raw waters. Can. J. Microbiol. 50, 675–682. doi: 10.1139/w04-059
Ferrari, S., Frosth, S., Svensson, L., Fernström, L. L., Skarin, H., and Hansson, I. (2019). Detection of Campylobacter spp. in water by dead-end ultrafiltration and application at farm level. J. Appl. Microbiol. 127, 1270–1279. doi: 10.1111/jam.14379
García, R., Bælum, J., Fredslund, L., Santorum, P., and Jacobsen, C. S. (2010). Influence of temperature and predation on survival of Salmonella enterica serovar typhimurium and expression of inva in soil and manure-amended soil. Appl. Environ. Microbiol. 76, 5025–5031. doi: 10.1128/AEM.00628-10
Gitter, A., Gidley, M., Mena, K. D., Ferguson, A., Sinigalliano, C., Bonacolta, A., et al. (2023). Integrating microbial source tracking with quantitative microbial risk assessment to evaluate site specific risk-based thresholds at two South Florida beaches. Front. Microbiol. 14:1210192. doi: 10.3389/fmicb.2023.1210192
Henry, R., Galbraith, P., Coutts, S., Prosser, T., Boyce, J., McCarthy, D. T., et al. (2018). What’s the risk? Identifying potential human pathogens within grey-headed flying foxes faeces. PLoS One, 13, e0191301.
Henry, R., Schang, C., Coutts, S., Kolotelo, P., Prosser, T., Crosbie, N., et al. (2016). Into the deep: evaluation of SourceTracker for assessment of faecal contamination of coastal waters. Water Res. 93, 242–253. doi: 10.1016/j.watres.2016.02.029
Hokajärvi, A. M., Tiwari, A., Räsänen, P., Wessels, L., Rankinen, K., Juntunen, J., et al. (2024). Campylobacter species, Salmonella serotypes and ribosomal RNA-based fecal source tracking in the Kokemäki River watershed. Sci. Total Environ. 954:176559. doi: 10.1016/j.scitotenv.2024.176559
Holcomb, D. A., and Stewart, J. R. (2020). Microbial indicators of fecal pollution: recent progress and challenges in assessing water quality. Curr. Environ. Health Rep. 7:311. doi: 10.1007/s40572-020-00278-1
Hubbard, L. E., Stelzer, E. A., Poulson, R. L., Kolpin, D. W., Szablewski, C. M., and Givens, C. E. (2024). Development of a large-volume concentration method to recover infectious avian influenza virus from the aquatic environment. Viruses 16:1898. doi: 10.3390/v16121898
Hunter, M. E., Ferrante, J. A., Meigs-Friend, G., and Ulmer, A. (2019). Improving eDNA yield and inhibitor reduction through increased water volumes and multi-filter isolation techniques. Sci. Rep. 9:5259. doi: 10.1038/s41598-019-40977-w
Hutchison, M. L., Walters, L. D., Avery, S. M., and Moore, A. (2005). Decline of zoonotic agents in livestock waste and bedding heaps. J. Appl. Microbiol. 99, 354–362. doi: 10.1111/j.1365-2672.2005.02591.x
Kataoka, A., Wolny, J. L., Guzman, J. R., Battin, A., Wang, S. S., Zaayenga, R., et al. (2025). Validation and use of the dead-end ultrafiltration method for the capture and recovery of Shiga toxin-producing Escherichia coli from surface water. J. Environ. Qual. 54, 1152–1162. doi: 10.1002/jeq2.70035
Knights, D., Kuczynski, J., Charlson, E. S., Zaneveld, J., Mozer, M. C., Collman, R. G., et al. (2011). Bayesian community-wide culture-independent microbial source tracking. Nat. Methods 8, 761–763. doi: 10.1038/nmeth.1650
Korajkic, A., McMinn, B. R., and Harwood, V. J. (2018). Relationships between microbial indicators and pathogens in recreational water settings. Int. J. Environ. Res. Public Health 15:2842. doi: 10.3390/ijerph15122842
Kozak, S., Roiko, A., Gutjahr-Holland, K., Ahmed, W., Veal, C., Fisher, P., et al. (2025). The use of faecal indicator organisms to manage microbial health risks in recreational waterways not impacted by point sources of sewage: a systematic review of the epidemiological evidence. J. Water Health 23, 563–586. doi: 10.2166/wh.2025.304
Kraft, A. L., Wells, J. E., Frye, J. G., Ibekwe, A. M., Durso, L. M., Hiott, L., et al. (2023). A comparison of methods to detect low levels of Salmonella enterica in surface waters to support antimicrobial resistance surveillance efforts performed in multiple laboratories. Sci. Total Environ. 905:167189. doi: 10.1016/j.scitotenv.2023.167189
Kroehler, C. J., Kroehler, C. J., Younos, T., and Grady, C. A.. Potable water quality standards and regulations: a historical and world overview. Handbook of Environmental Chemistry. (2014); 30:1–36. Available online at: https://link.springer.com/chapter/10.1007/978-3-319-06563-2_1 (accessed Feb 26, 2025).
Kruskal, W. H., and Wallis, W. A. (1952). Use of ranks in one-criterion variance analysis. J. Am. Stat. Assoc. 47, 583–621.
Leskinen, S. D., Kearns, E. A., Jones, W. L., Miller, R. S., Bevitas, C. R., Kingsley, M. T., et al. (2012). Automated dead-end ultrafiltration of large volume water samples to enable detection of low-level targets and reduce sample variability. J. Appl. Microbiol. 113, 351–360. doi: 10.1111/j.1365-2672.2012.05345.x
Lim, T. J. Y., Delgado, Y. M. P., Lintern, A., McCarthy, D. T., and Henry, R. (2024). Assessing accuracy and specificity of faecal source library for microbial source-tracking, using sourcetracker as case study. Bioinform. Adv. 5:vbaf103. doi: 10.1093/bioadv/vbaf103
Lim, T. J. Y., Sargent, R., Henry, R., Fletcher, T. D., Coleman, R. A., McCarthy, D. T., et al. (2022). Riparian buffers: disrupting the transport of E. coli from rural catchments to streams. Water Res. 222:118897. doi: 10.1016/j.watres.2022.118897
Liu, X., and Salles, J. F. (2024). Bridging ecological assembly process and community stability upon bacterial invasions. ISME J. 18:66. doi: 10.1093/ismejo/wrae066
Lowe, R. M. S., Munns, K., Selinger, L. B., Kremenik, L., Baines, D., McAllister, T. A., et al. (2010). Factors influencing the persistence of Escherichia coli O157:H7 lineages in feces from cattle fed grain versus grass hay diets. Can. J. Microbiol. 56, 667–675. doi: 10.1139/W10-051
McCarthy, D. T., Jovanovic, D., Lintern, A., Teakle, I., Barnes, M., Deletic, A., et al. (2017). Source tracking using microbial community fingerprints: method comparison with hydrodynamic modelling. Water Res. 109, 253–265. doi: 10.1016/j.watres.2016.11.043
Mull, B., and Hill, V. R. (2012). Recovery of diverse microbes in high turbidity surface water samples using dead-end ultrafiltration. J. Microbiol. Methods 91, 429–433. doi: 10.1016/j.mimet.2012.10.001
NHMRC, NRMMC (2011). Australian Drinking Water Guidelines Paper 6 National Water Quality Management Strategy. National Health and Medical Research Council, National Resource Management Ministerial Council, Commonwealth of Australia, Canberra. Available at: https://www.nhmrc.gov.au/about-us/publications/australian-drinking-water-guidelines.
Nowicki, S., DeLaurent, Z. R., De Villiers, E. P., Githinji, G., and Charles, K. J. (2021). The utility of Escherichia coli as a contamination indicator for rural drinking water: evidence from whole genome sequencing. PLoS One 16:e0245910. doi: 10.1371/journal.pone.0245910
Pardo, L., Domínguez-Maqueda, M., Cecilia, J. A., Rodríguez, M. P., Osajima, J., Moriñigo, M. Á., et al. (2020). Adsorption of Salmonella in clay minerals and clay-based materials. Minerals 10:130. doi: 10.3390/min10020130
Pascual-Benito, M., Emiliano, P., Casas-Mangas, R., Dacal-Rodríguez, C., Gracenea, M., Araujo, R., et al. (2020). Assessment of dead-end ultrafiltration for the detection and quantification of microbial indicators and pathogens in the drinking water treatment processes. Int. J. Hyg. Environ. Health 230:113628. doi: 10.1016/j.ijheh.2020.113628
Pluym, T., Waegenaar, F., De Gusseme, B., and Boon, N. (2024). Microbial drinking water monitoring now and in the future. Microb. Biotechnol. 17:e14532. doi: 10.1111/1751-7915.14532
Standards Australia. (2014a). Water microbiology, Method 14: Detection of Salmonella spp. Available online at: https://www.standards.org.au/standards-catalogue/standard-details?designation=as-4276-14-2014 (Accessed August 12, 2025).
Standards Australia. (2014b). Water microbiology, Method 19: Examination for thermophilic Campylobacter spp. Available online at: https://www.standards.org.au/standards-catalogue/standard-details?designation=as-4276-14-2014 (Accessed August 12, 2025).
Standards Australia. (2019). Water microbiology, Method 21: Examination for coliforms and Escherichia coli. Available online at: https://store.standards.org.au/product/as-4276-21-2019?ref=savingsasaservice.com.au (Accessed August 12, 2025)
Scott, T. M., Rose, J. B., Jenkins, T. M., Farrah, S. R., and Lukasik, J. (2002). Microbial source tracking: current methodology and future directions. Appl. Environ. Microbiol. 68:5796. doi: 10.1128/AEM.68.12.5796-5803.2002
Smith, C. M., and Hill, V. R. (2009). Dead-end hollow-fiber ultrafiltration for recovery of diverse microbes from water. Appl. Environ. Microbiol. 75:5284. doi: 10.1128/AEM.00456-09
Tran, D. T. Q., Bradbury, M. I., van Ogtrop, F. F., Bozkurt, H., Jones, B. J., and McConchie, R. (2020). Environmental drivers for persistence of Escherichia coli and Salmonella in manure-amended soils: a meta-analysis. J. Food Prot. 83, 1268–1277. doi: 10.4315/0362-028X.JFP-19-460
Weinstein, M. M., Prem, A., Jin, M., Tang, S., and Bhasin, J. M.. FIGARO: an efficient and objective tool for optimizing microbiome rRNA gene trimming parameters. bioRxiv (2019) 610394. Available online at: https://www.biorxiv.org/content/10.1101/610394v1 (Accessed December 13, 2024).
Wong, K., Shaw, T. I., Oladeinde, A., Glenn, T. C., Oakley, B., and Molina, M. (2016). Rapid microbiome changes in freshly deposited cow feces under field conditions. Front. Microbiol. 7:500. doi: 10.3389/fmicb.2016.00500
World Health Organization. (2022). Guidelines for drinking-water quality: fourth edition incorporating the first and second addenda. Available online at: https://www.who.int/publications/i/item/9789240045064 (Accessed August 14, 2025).
Keywords: faecal indicator organisms (FIOs), reference pathogens, microbial source tracking (MST), ultrafiltration, 16S rRNA amplicon, water quality
Citation: Lynch F, Lim TJY, Steele M, Stevens M, Bata L, Lynch GP, Dewi DAPR, Sharp S, Orr N, Ward A, Haydon S, Tseng C-W and Henry R (2025) Enhanced detection of animal-derived microbial hazards in forested catchments using high-volume ultrafiltration and amplicon-based microbial source tracking. Front. Water. 7:1694489. doi: 10.3389/frwa.2025.1694489
Edited by:
Marthie Magdaleen Ehlers, University of Pretoria, South AfricaReviewed by:
Lisa Paruch, Norwegian Institute of Bioeconomy Research (NIBIO), NorwayMohit Kumar, Harvard Medical School, United States
Copyright © 2025 Lynch, Lim, Steele, Stevens, Bata, Lynch, Dewi, Sharp, Orr, Ward, Haydon, Tseng and Henry. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rebekah Henry, cmViZWthaC5oZW5yeUBtb25hc2guZWR1
†These authors share first authorship
 Fiona Lynch
Fiona Lynch Timothy J. Y. Lim
Timothy J. Y. Lim Mellisa Steele3
Mellisa Steele3 Dewa A. P. Rasmika Dewi
Dewa A. P. Rasmika Dewi Rebekah Henry
Rebekah Henry