- Department of Life Science and Multidisciplinary Unit of Research on Translational Initiatives (MURTI), GITAM School of Science, GITAM (Deemed to be University), Visakhapatnam, Andhra Pradesh, India
This study investigated and analysed the presence and antibiotic resistance profiles of Enterobacter strains from municipal water supply and groundwater sources (open wells and handpumps) in a low-income neighbourhood of urban Visakhapatnam, India. Low-income neighbourhoods within the industrialised area of Gajuwaka were found to show high levels of groundwater contamination with multidrug-resistant bacterial strains. Two of these isolates were identified to be multidrug-resistant (MDR) strains, Enterobacter cloacae DSM 30054 (V1) and Enterobacter quasihormaechei WCHEs120003 (V2). The two isolates showed a similar profile with resistance towards penicillins, carbapenems, early and third-generation cephalosporins (such as cephalothin and ceftazidime, respectively), aztreonam, nitrofurantoin, and chloramphenicol. Susceptibility was noticed against cefepime (4th-generation cephalosporin), fluoroquinolones, and piperacillin-tazobactam. This profile suggests a combination of various intrinsic and acquired resistance mechanisms involving chromosomal AmpC β-lactamases and acquired resistance genes, including ESBLs and carbapenemases, potentially facilitated by porin loss. The isolates showcased high MAR index values above 0.5, indicating an environment with high resistance-inducing factors. Since higher levels of contamination were observed in the groundwater than in the municipality’s tap water samples, a type of advanced oxidation process (AOP) called Fenton’s reaction was evaluated for its efficacy in clearing the contamination from the groundwater samples of Visakhapatnam. Reduction in bacterial coverage levels on agar plates with different ratios of Fenton’s reaction regents was noticed during this experiment, going as high as 89.50% reduction (0.2 mM Fe2+ from ferrous ammonium sulphate, 0.5 mM H2O2, and pH 3). The findings of this study observed contamination of drinking water sources in vulnerable communities and highlighted the role of environmental water sources as reservoirs for resistance in Enterobacter spp. This investigation also provided an evaluation of the Fenton-based AOP as a viable technology for the remediation of contaminated groundwater in resource-limited settings of low-income neighbourhoods. There is a need for stringent monitoring of all water sources to maintain universal access to safe drinking water for every individual.
1 Introduction
Maintaining a safe drinking water supply through municipal and groundwater sources is key to securing public health, especially in the case of low-income settlements in urban settings (Chathuranika et al., 2023; Grönwall and Danert, 2020). Contamination of these systems by bacterial species belonging to the Enterobacteriaceae family has emerged as a significant risk in various developed, developing and underdeveloped nations, with a harsh impact felt among vulnerable populations of these countries (Najjembe et al., 2025; Singh and Reddy, 2025c; Dhengesu et al., 2022). Despite the Sustainable Development Goals (SDG) 6 emphasising the importance of universal access to safe drinking water, this goal remains elusive for many communities due to a lack of WASH awareness, limited facilities, overlooked negligence in existing water treatment plants (WTPs), and various environmental factors contributing towards this complicated problem (United Nations General Assembly, 2010; Evaristo et al., 2023; Żywiec et al., 2024; Onyango et al., 2015). Overlooked or poorly managed water systems, whether in the case of municipal supply or even groundwater sources, could lead to the spread of waterborne diseases within overpopulated settlements (Ferreira et al., 2021; Bagordo et al., 2024).
Among various bacterial contaminants, the Enterobacteriaceae family is home to many gram-negative bacterial species. These bacteria can act as causative agents for waterborne diseases, opportunistic or otherwise, and have been observed to be responsible for many gastroenteric infections (Ramirez and Giron, 2025; Akinjogunla et al., 2023). Enterobacter, Klebsiella, Escherichia coli, and Citrobacter species are some of the prominent Enterobacteriaceae pathogens associated with coliform contamination of drinking water sources (Tanner et al., 2019).
Microbial contamination of municipal drinking water sources could arise from poorly maintained or older water treatment systems, negligent or inadequately trained technical staff, recontamination, and natural calamities such as storms and floods, causing widespread contamination (Rafa et al., 2021). Increasing antibiotic resistance in some of these pathogens, also categorised as ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumanii, Pseudomonas aeruginosa, and Enterobacter species), has been flagged as a significant threat to public health (Denissen et al., 2022).
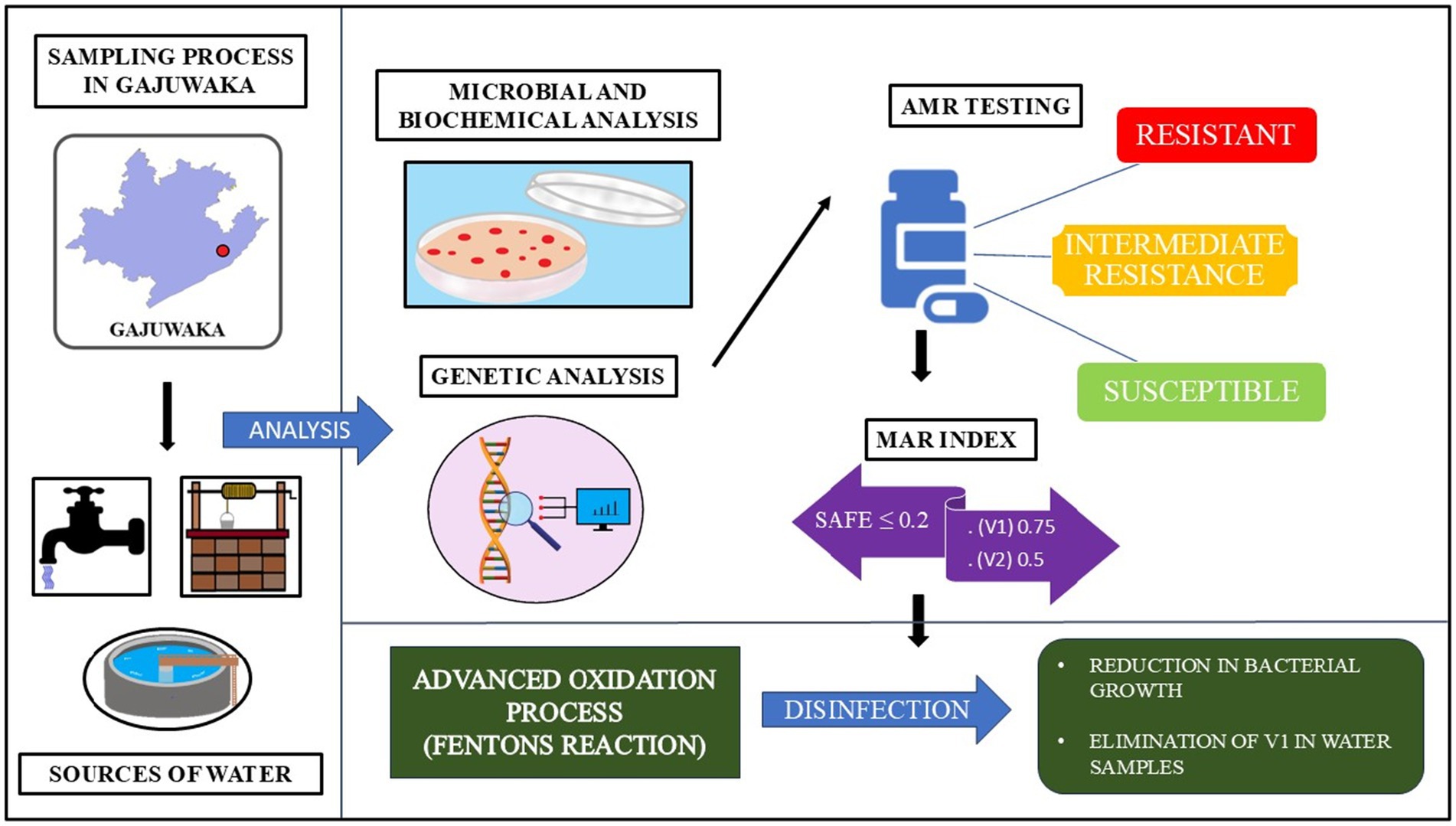
After random sampling, resistant Enterobacter strains were detected during our study in municipal tap water and groundwater sources, hinting at bacterial contamination. This study was conducted in low-income neighbourhoods within urban Visakhapatnam. In particular, the presence of MDR bacteria in Gajuwaka tap and groundwater samples was found to be of higher concern. Microbial and genetic analysis revealed the presence of two crucial isolates, Enterobacter cloacae DSM 30054 (V1) and Enterobacter quasihormaechei WCHEs120003 (V2). V1 and V2 were studied for their antibiotic resistance profiles and found to possess multidrug resistance (MDR) against different classes of antibiotics (Singh and Reddy, 2025a,b).
Despite its unprotected and contaminated state, groundwater sources like open wells were crucial as an alternative to municipal-supplied tap water in low-income neighbourhoods of Visakhapatnam. Hence, a need to improve these localised contamination hotspots was observed during this study. Through various studies, traditional disinfection techniques were reported to have disadvantages such as high operational costs, disinfection byproduct concerns, and variable efficiency in reducing resistant bacteria and resistant genes within water bodies (Chen et al., 2021). Techniques such as chlorination were also reported to be selective for resistant species due to their disinfection byproducts and diminishing residual concentrations in waterlines over time (Zhao et al., 2025; Adefisoye and Olaniran, 2022). Recent studies have evaluated advanced oxidation processes (AOP) such as ozonation, ultraviolet radiation, or Fenton based reaction, as either an alternative or an addition to the existing treatment methods in order to improve the treatment efficacy in terms of pollutants in different waste water or industrial effluent samples (Ayoub, 2022; Chen et al., 2021; Cuerda-Correa et al., 2019; Ponnusami et al., 2023). Only a few studies have evaluated the efficacy of AOPs, especially Fenton-based reactions, on resistant bacterial isolates in natural water matrices, especially groundwater (Mosaka et al., 2023; Vilela et al., 2021; Ahmed et al., 2020; Castaño-Henao et al., 2024).
Advanced oxidation processes (AOPs) have emerged as a transformative solution for addressing complex water quality challenges in dense, resource-limited urban settings (Satyam and Patra, 2025). AOPs are a set of chemical and photochemical treatment processes designed to generate highly reactive hydroxyl radicals, capable of non-selectively degrading a broad spectrum of organic contaminants and even mineralising them into harmless end products like carbon dioxide and water (Gaur et al., 2022; Palma, 2021). Unlike conventional chemical oxidants or disinfection agents, hydroxyl radicals produced by AOP possess exceedingly high oxidation potential, enabling them to break down even the most persistent organic pollutants, including pharmaceutical residues, pesticides, industrial solvents, and endocrine disruptors that pose substantial risks for human health and ecological stability in marginalised urban areas. A variety of AOP configurations exist, with practical options for urban deployment including UV/Hydrogen Peroxide, Fenton and Photo-Fenton (hydrogen peroxide + iron and UV light), Ozone-based systems, and combinations like UV/Ozone. More recent advances involve catalytic wet peroxide oxidation, microwave/hydrogen peroxide, and even supervisorial water oxidation, demonstrating remarkable contamination removal in challenging conditions (Quintanilla and Munoz, 2019).
During this investigation, an experiment was carried out to study the effect of Fenton-based AOP for groundwater treatment in low-income settings. Millimolar concentrations of Fe2+ and Hydrogen Peroxide (H2O2) were used under acidic conditions (pH 3) for the treatment (Walling et al., 2021; Ayoub, 2022). The results showed significant microbial inactivation, making this process potentially suitable for improving contaminated and unprotected groundwater sources. No other peer-reviewed studies were found on the isolation of MDR Enterobacter strains from municipality and unprotected groundwater sources in Gajuwaka, nor on the reduction of bacterial content from natural groundwater samples from the study area. Moreover, to the best of available evidence, a gap was also observed in the investigation of Fe2+ and H2O2 based classic Fenton reaction on culture-based reduction of bacteria in natural groundwater sources, especially from unprotected wells and handpumps. This study urgently calls for monitoring resistance in environmental sources, such as surface and groundwater, to achieve universal access to safe drinking water.
2 Experimental section
2.1 Sampling site
Gajuwaka area (17.7004°N, 83.2168°E) is a major industrial zone with proximity to various large-scale industries like the Visakhapatnam steel plant, Visakha dairy plant, heavy machinery manufacturing units, pharmaceutical manufacturing units, and numerous other large and small engineering and metal works (Hariharan and Begum, 2023; Srinivasa Rao et al., 2015). Many pockets of lower economic neighbourhoods or slums exist within sub-localities such as Mindi and Akkireddypalem, which mostly house daily wage workers of nearby industries. Despite rapid industrialisation and urbanisation, the lower economic neighbourhoods often face pollution and water scarcity-related issues (Venkata Ramana et al., 2018). Residents of low-income areas of Gajuwaka exclusively depend on municipal taps for daily drinking water requirements, in the absence of which they resort to unprotected groundwater sources like public handpumps and wells (Hariharan and Begum, 2023). The Meghadrigedda reservoir is utilised as the source for municipality taps after purification by the municipality water treatment plant (Narendra and Rao, 2006). The residential study area boasted a dense population with houses present wall to wall and narrow roads of 6 to 10 feet. These areas also faced the challenge of having open drains in close proximity to water taps, wells, and handpumps, as illustrated in Figure 1. Monsoon and cyclonic rains often lead to inundation of roads and seepage from open drains, polluting the water sources. Since the Visakhapatnam region experiences the highest amount of rainfall during this season, with added effects of cyclones from the Bay of Bengal, it becomes essential to closely monitor the water sources in order to control the spread of water contamination-associated diseases.
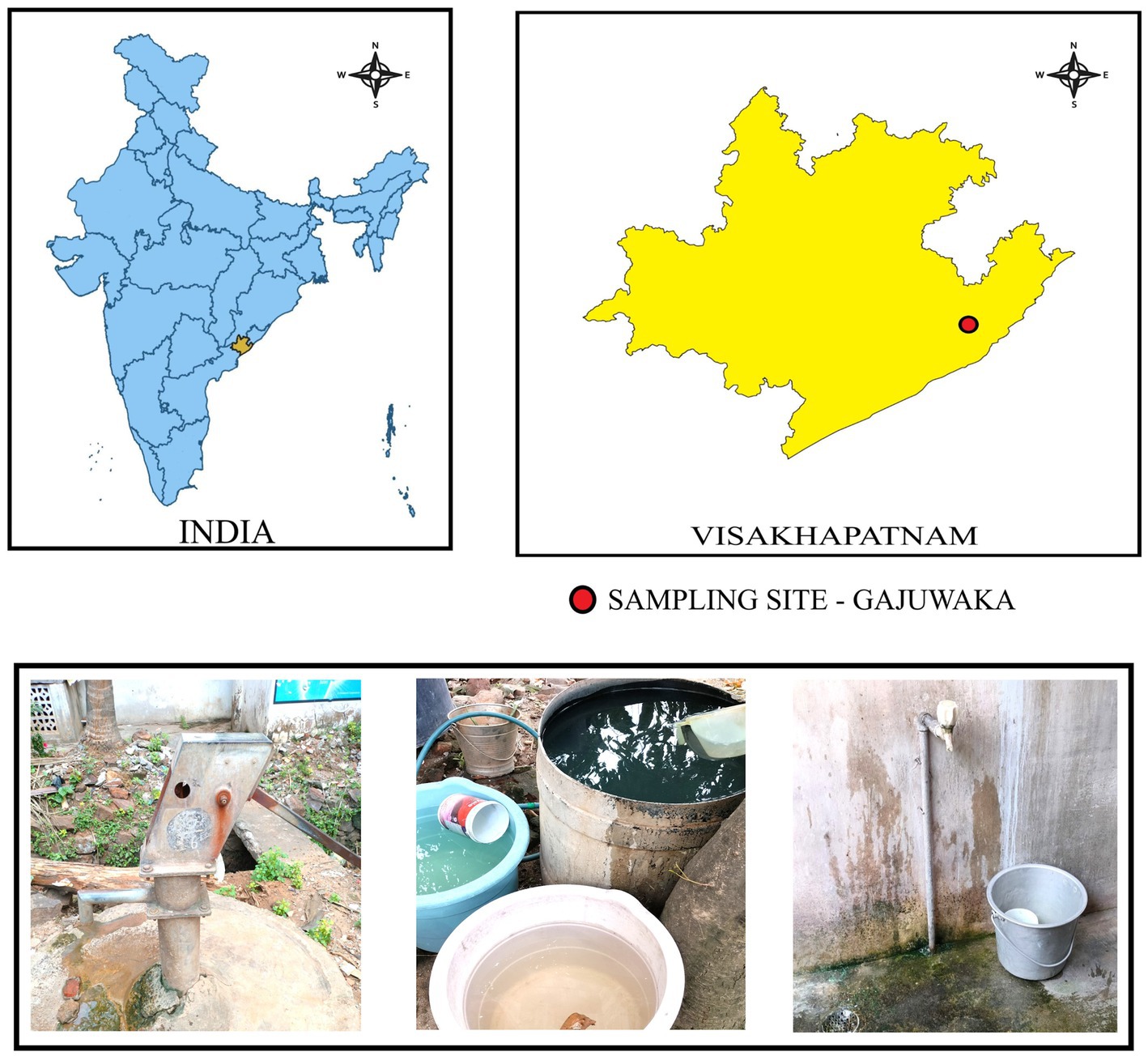
Figure 1. Sampling site map describing the location of Gajuwaka, India, and the sample collection points involving municipality taps and groundwater sources such as handpumps within low-income neighbourhoods of Gajuwaka.
2.2 Sample collection
This study was carried out in the monsoon season of 2023 (October–November period) in selected low-income settlements of Gajuwaka, Visakhapatnam, India. The chosen localities were primarily inhabited by low-income residents, relying on municipal taps and groundwater sources such as handpumps for their drinking water requirements. A total of 30 sites were selected for same-day triplicates within the region, including residential municipality taps, groundwater sources, and the local water treatment plant (WTP). The samples of WTP origin were accessed from the raw water source, post-chlorination tank, and the post-treatment overhead tank. Triplicate sampling was performed on randomly chosen groundwater and municipal taps in sterile polypropylene bottles, which were then transferred to the laboratory in a cooler containing ice packs.
2.3 Microbial analysis
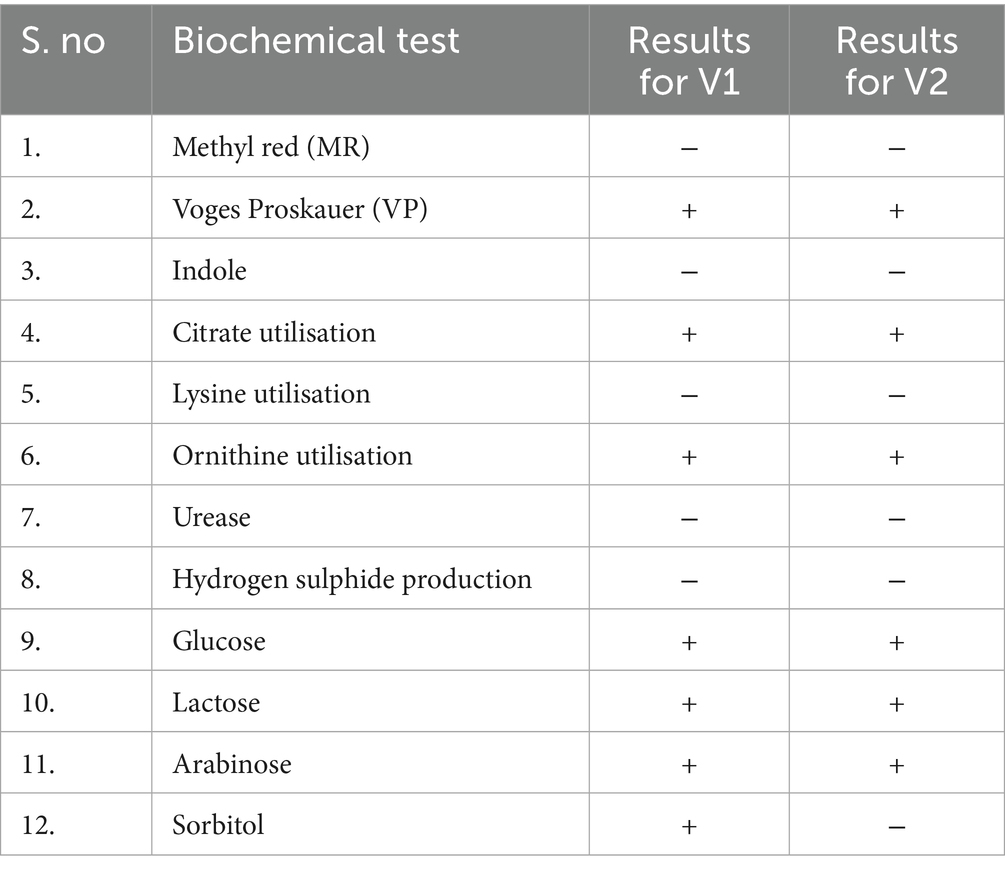
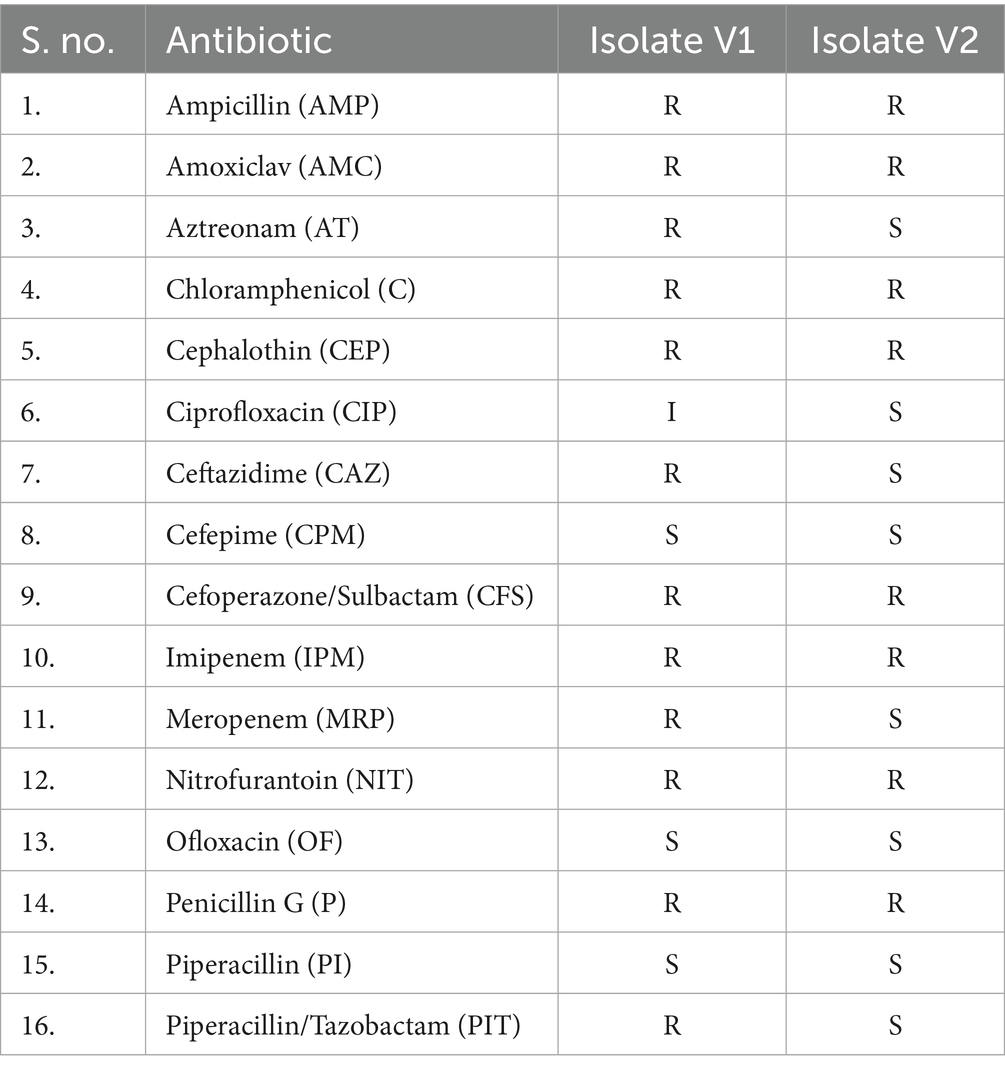
Microbial analysis of water samples was conducted using various selective and differential culture media and kits obtained from HiMedia Laboratories, India. The agar plates, such as deoxycholate citrate agar (DCA) and eosin methylene blue (EMB) agaṛ were used to isolate gram-negative coliforms such as Enterobacter species, were prepared under aseptic conditions and inoculated using the spread plate method, wherein 0.1 mL of the microbial suspension was evenly distributed across the agar surface using a sterile glass spreader (Lal and Cheeptham, 2007; Wang and Wu, 2024; Singh and Reddy, 2025c). The plates were incubated at 37 °C for 24 to 48 h in an incubator (KEMI Lab Equipments, India), awaiting the growth of bacterial cultures. Chosen isolates were pure-cultured and analysed using the Kirby-Bauer method for antimicrobial susceptibility and biochemical tests to identify the isolates (Hudzicki, 2009). The multiple antibiotic resistance (MAR) index was determined for the isolates based on their resistance profiles (Mir et al., 2022). Tests such as Gram staining, hydrogen sulphide production, indole test, Voges-Proskauer test, methyl red test, and citrate utilisation test were performed to understand the metabolism of the isolates as listed in Table 1 (Uppal, 2019; Moyes et al., 2009). During the analysis, ATCC bacterial cultures of Escherichia coli strain (ATCC 25922) were used as a positive control, and the sterile medium was used as a negative control. The isolates V1 and V2 were also viewed under a scanning electron microscope to visualise their physical structure (FESEM TESCAN MIRA S6123).
2.4 Genetic analysis
Genetic identification for the isolates was carried out using the 16S rDNA sequencing method and NCBI BLAST. First, DNA was isolated from the cultures, and its purity was determined using 1.0% agarose gel electrophoresis. A single DNA band with high molecular weight was obtained, signifying its purity. It was followed by 16S rDNA amplification with 27F and 1,492 R primers, producing a 1,500 bp amplicon for forward and reverse sequencing. A BDT v3.1 cycle kit was used on an ABI 3730xl genetic analyser to complete the sequencing. Consensus sequences of the isolates were generated using the aligner software and used in the NCBI BLAST nucleotide tool to check for similarity within the GenBank database (NCBI BLAST, 2025; Clark et al., 2016). During this process, match or mismatch scores of 1 and −2 were utilised while the scoring parameters were set at linear gap costs. The Clustal W program was used to align the first ten sequences obtained from the BLAST search, and phylogenetic trees with 1,000 replicates were created using the MEGA 7 software (Kumar et al., 2016). In this regard, the Kimura 2-parameter model was utilised to surmise the evolutionary data of the isolates through the maximum likelihood method (Kimura, 1980). The trees were made to be bootstrap consensus, and only visualised branches showing higher than 50% reproduction of bootstrap replicates (Felsenstein, 1985). During the preparation of phylogenetic trees, the maximum composite likelihood approach was used with Neighbour-Join and BioNJ algorithms to procure pairwise distance matrices. The percentage of replicated trees with the clustering of associated taxa was represented within the figures of phylogenetic trees, which were prepared while keeping the superior log likelihood values in mind. During the preparation of the final dataset, gaps and missing values were removed. A total of 11 nucleotide sequences were used for the analysis, and 1,434 positions were present in the final datasets of the isolates. Sequences for the isolates V1 and V2 were submitted to the NCBI GenBank database with accession numbers PV739642 and PV739646, respectively.
2.5 Fentons reaction on water samples
The Fenton reaction was applied as an advanced oxidation process to evaluate its microbial inactivation potential in natural groundwater collected from open wells and handpumps. Triplicate samples were treated with ferrous ammonium sulphate and hydrogen peroxide at different millimolar concentrations (0.1–0.5 mM) and maintained at an optimum acidic pH 3 using 0.3 mL of HCl, as supported by literature establishing this condition for effective hydroxyl radical generation (Ayoub, 2022):
The reactions, as described in Equations 1–3, were conducted at ambient temperature and treated versus untreated samples were analysed at intervals of 10–50 min using nutrient agar to quantify microbial load reduction. While this study primarily focused on microbial inactivation efficiency, physicochemical parameters such as pH and dissolved oxygen were monitored prior to and after treatment to ensure experimental control. Future work aims to extend these measurements to include total organic carbon (TOC), chemical oxygen demand (COD), and UV absorbance (SUVA254) indices, as recommended by recent AOP benchmarking studies (Krupińska, 2024; Tubić et al., 2013), to better capture the mechanistic link between organic matter degradation and microbial reduction effectiveness.
3 Results
3.1 Microbial analysis
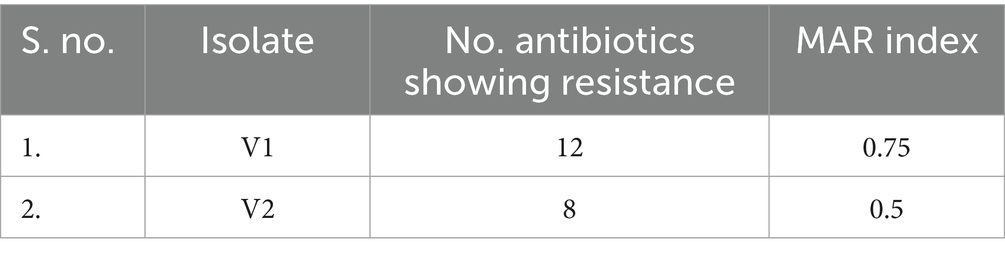
The two isolates, V1 and V2, were grown on EMB and DCA agar, following which pure cultures were obtained for microbial and biochemical analysis. Biochemical tests on the isolates showed similar metabolic activities except for the sorbitol test, where V1 showed a positive result, unlike V2. These results are listed in Table 1. Both isolates were observed to be Gram-negative, rod-shaped bacilli. It was further corroborated by images obtained from SEM analysis. The isolates from V1 and V2 samples were measured to lengths between 1.3 μm and 2.5 μm, falling within the general size range of Enterobacter species and represented in Figure 2. A resistance profile was obtained by the Kirby-Bauer method for V1 and V2. Both isolates were found to show MDR against multiple classes of antibiotics, including penicillins, cephalosporins, and carbapenems. These results have been detailed in Tables 2–4. V1 displayed an alarmingly high MAR index value of 0.75 as it was resistant to 12 antibiotics. V2 also showed a high MAR index of 0.5 due to resistance against eight antibiotics. V1 showed resistance against ampicillin, amoxiclav, penicillin G (penicillins), cephalothin and ceftazidime (cephalosporins), imipenem and meropenem (carbapenems), nitrofurantoin (nitrofuran), aztreonam (monobactam), chloramphenicol (amphenicol), cefoperazone/sulbactam (cephalosporin + beta-lactamase inhibitor), and piperacillin/tazobactam (penicillin + beta-lactamase inhibitor). Intermediate resistance was also shown against ciprofloxacin (fluoroquinolone). V2 showed resistance against ampicillin, amoxiclav, penicillin G, chloramphenicol, cephalothin, cefoperazone/sulbactam, imipenem, and nitrofurantoin.
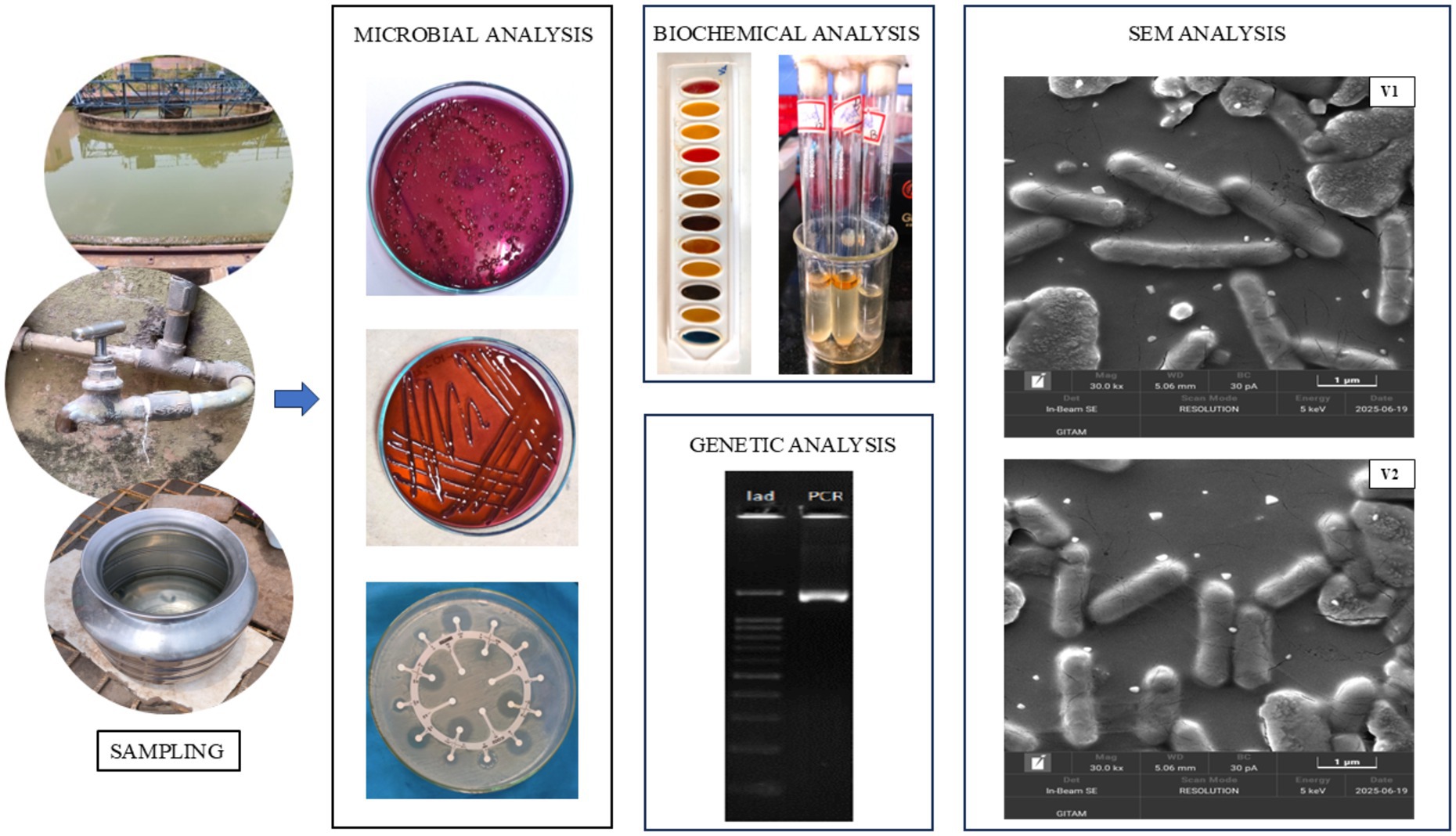
Figure 2. The study on drinking water involved sampling followed by various analyses involving microbial work (bacterial isolation on EMB agar, pure culturing, and Kirby-Bauer method), biochemical tests, genetic analysis, and SEM analysis for the isolates V1 and V2.
3.2 Genetic analysis
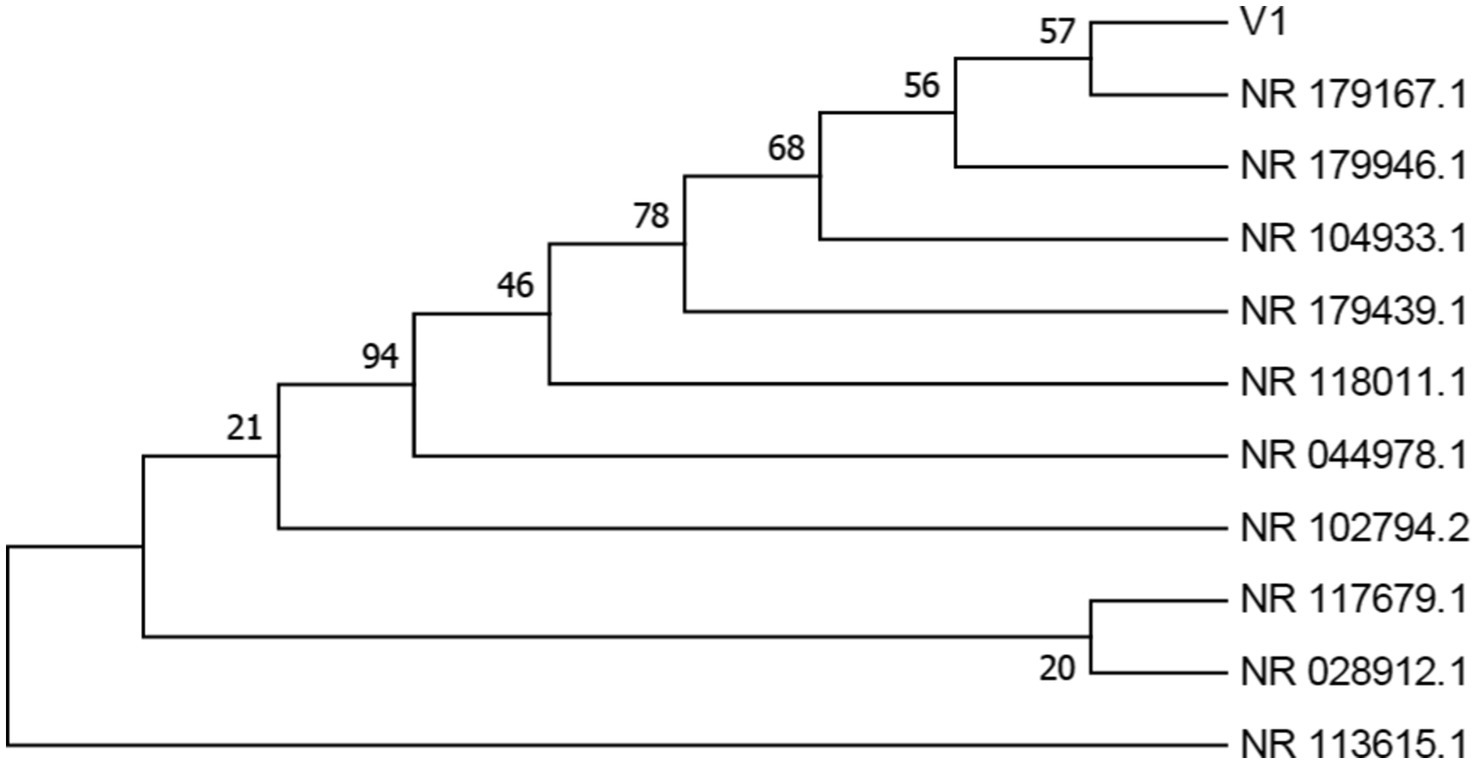
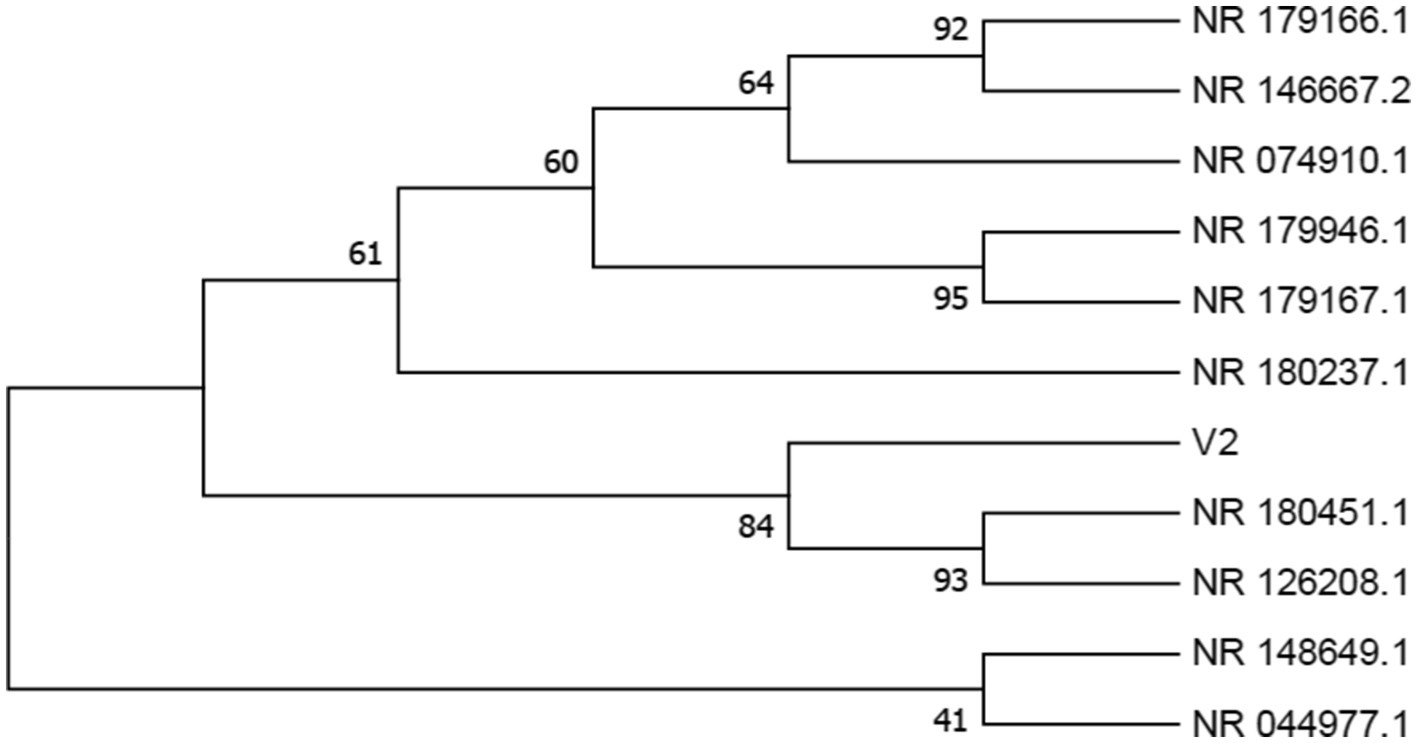
The isolated samples V1 and V2 were identified by rRNA sequencing, followed by NCBI BLAST and phylogenetic analysis, which are depicted in Figures 3, 4, S1, and S2. V1 showed maximum similarity with Enterobacter cloacae strain DSM 30054, and V2 with Enterobacter quasihormaechei strain WCHEs120003. These bacteria belonged to the Enterobacteriaceae family and occupied positions in the Enterobacter cloacae complex (ECC).
3.3 Effect of Fenton’s reaction on bacterial plate coverage percentage
The results of this experiment showed significant microbial inactivation accompanied by a reduction in total dissolved solids (TDS) values, making this process potentially suitable for improving contaminated and unprotected groundwater sources like open wells. Initial contamination levels, as indicated by the untreated ‘blank’ samples, were high, with bacterial plate coverage ranging from 73.44 to 98.32%. Upon treatment, a rapid and significant reduction in microbial viability was observed, typically within the first 20 min. For instance, in trial #10 (0.2 mM FAS, 0.5 mM H2O2), bacterial coverage plummeted from 82.53% to just 8.66%. The disinfection efficacy was particularly pronounced at higher oxidant doses; combinations using 0.5 mM FAS with H2O2 concentrations of 0.3 mM and higher consistently yielded strong results. The results were listed out in detail in Table 5, Figure 5. Gajuwaka area samples showed better reduction in coverage of the plates when compared with other sampling sites during this study. It was also observed that the bacterial isolate diversity from the water samples declined after the treatments based on Fentons reaction and V1 isolates were completely eliminated.
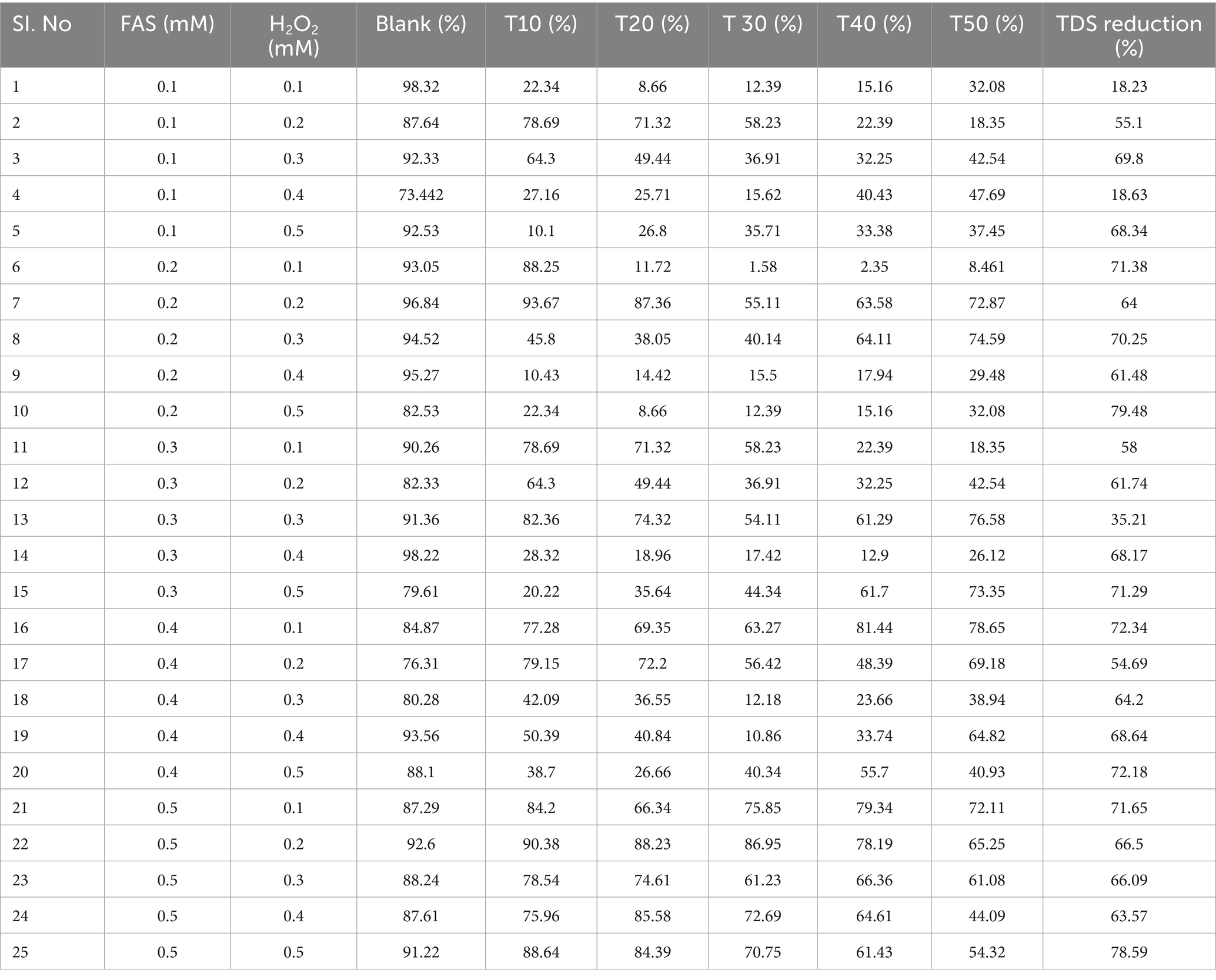
Table 5. Effect of Fenton-based AOP (FAS–H2O2, pH 3) on plate growth coverage (%) and TDS of groundwater samples from Visakhapatnam.
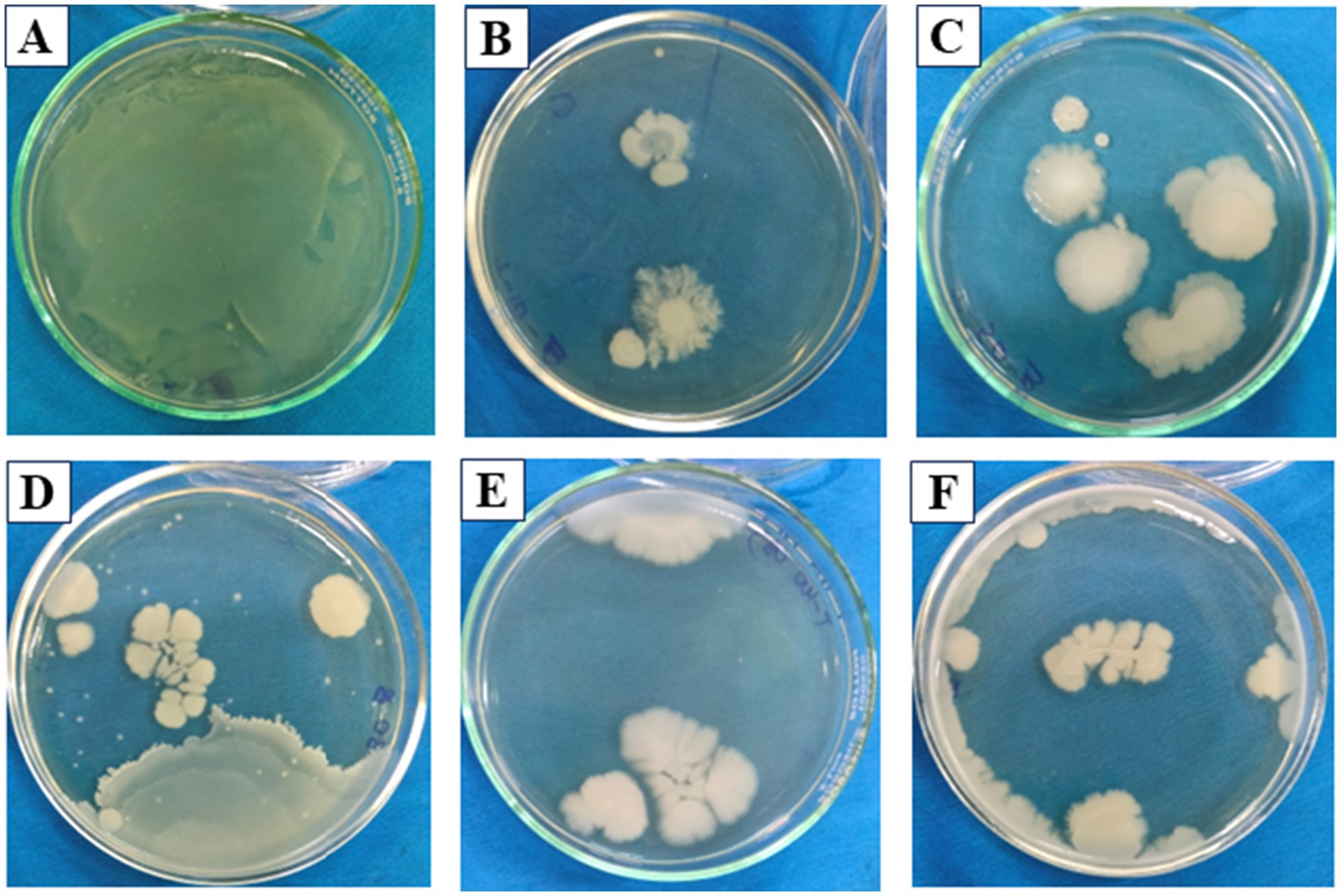
Figure 5. Bacterial growth coverage after Fenton reaction on Gajuwaka sample—FAS (0.1 mM)—H2O2 (0.5 mM), representing (A) Blank, (B) T10, (C) T20, (D) T30, (E) T40, (F) T50.
4 Discussion
The genus Enterobacter has been a critical component of the ESKAPE pathogens group, notorious for its ability to acquire multidrug resistance, especially regarding life-threatening nosocomial infections (Ravi and Singh, 2024). Species, such as Enterobacter cloacae, are also well known for promoting biofilm growth within pipelines, leading to higher chances of developing resistance and increasing contamination levels within the system (Misra et al., 2022; Bitton, 2014). Since this study was conducted in a lower economic neighbourhood, the vulnerability of the residents towards contaminated water represents a higher risk. The residents depend entirely on the daily municipal water supply, available only for a limited time during the day, with local handpumps and open wells as an alternative. Since safer drinking water sources, such as purifiers or mineral water, are not an affordable option, there is a need for stringent quality monitoring of the municipal water supply to uphold the human right to safe drinking water (United Nations General Assembly, 2015). Added to this is the lack of monitoring of groundwater sources like handpumps and open wells. These often show high contamination levels due to a lack of maintenance, sewer seepage, or natural factors like rainfall, cyclones, and flooding (Abanyie et al., 2023; Rivett et al., 2022; Akinola et al., 2022). The isolates from these two sources were studied for their identification and characterisation.
The isolated strains V1 and V2 were identified as Enterobacter cloacae strain DSM 30054 and Enterobacter quasihormaechei strain WCHEs120003, respectively, through 16S rRNA gene sequencing and BLAST analysis. These two isolates are also a part of the Enterobacter cloacae complex (ECC), which is found widely in both environmental and clinical settings. Many studies have observed and documented multifaceted resistance profiles, mechanisms, and adaptability of these species within the ECC (Ganbold et al., 2023; Mustafa et al., 2020). Their abilities in evading multiple classes of antibiotics could be attributed to the production and acquisition of carbapenemases, AmpC β-lactamases, extended-spectrum β-lactamases (ESBLs), and mechanisms involving efflux pumps and porin loss (Davin-Regli et al., 2019). Their capacity to withstand multiple antibiotic classes and disseminate resistance to other bacterial species makes them an essential focus for global monitoring and antibiotic stewardship.
The V1 strain recovered from freshwater samples was found to be resistant to a broad spectrum of antibiotics, including aztreonam, nitrofurantoin, chloramphenicol, and a few in the carbapenem, cephalosporin, and penicillin groups. The isolate was susceptible to cefepime, piperacillin-tazobactam, and fluoroquinolones. This type of resistance profile can be attributed to a combination of acquired and intrinsic mechanisms, such as resistance towards penicillins and early-generation cephalosporins due to chromosomal AmpC β-lactamases (Galgano et al., 2025). Meanwhile, porin loss, acquiring ESBLs, carbapenemases (such as NDM-1), and overproduction of AmpC could have contributed towards protection against advanced cephalosporins and carbapenems (Annavajhala et al., 2019). Numerous studies from India have reported carbapenem resistance in Enterobacteriaceae, especially in water environments that could potentially affect drinking water sources (Stefaniak et al., 2024; Sivalingam et al., 2019; Lamba et al., 2018). The isolated V1 strain showed a high MAR index value of 0.7, which indicates a significantly higher risk of exposure to antibiotics and resistant species within water bodies. It was consistent with studies associating environmental water reserves as reservoirs for MDR species and resistome in the Indian and global context. Enterobacter quasihormaechei strain WCHEs120003 (V2) is a newly added member of the EEC, displaying potentially similar MDR activities to the other complex members. V2 showed resistance towards penicillin G, ampicillin, amoxiclav, chloramphenicol, cephalothin, cefoperazone/sulbactam, imipenem, and nitrofurantoin. This profile again highlights the issue of carbapenemase genes being harboured within freshwater settings. Furthermore, the additional β-lactamase and non-β-lactamase activities against the β-lactam inhibitors and other antibiotics, such as nitrofurantoin, respectively, are a stark reminder of their adaptability against multiple classes of antibiotics and their ability to transfer the resistance to other bacteria within the environment (Caliskan-Aydogan and Alocilja, 2023; Ebomah and Okoh, 2021). The detection of such MDR species in drinking water sources with high MAR index values of 0.75 and 0.5 raises concerns about the circulation of opportunistic pathogens within India’s treated and untreated water systems (Afunwa et al., 2020; Poonia et al., 2014).
The Advanced Oxidation Process using Fenton’s reaction was tested on groundwater samples in triplicate. These sources involved open wells and handpumps, which were untreated despite contamination events in the sampling area involving phenomena such as heavy rains and sewage runoffs. Adding in the factor that a large majority of the residents did not use purification methods such as boiling or filtering pre-consumption, it was essential to investigate an effective method for decentralised disinfection. During the AOP experiment, a rapid inactivation of cells could be observed due to the generation of highly reactive hydroxyl radicals (•OH), a characteristic of Fenton chemistry. During this process, the •OH plays an important role in degrading organic pollutants and resistant bacterial species by direct oxidation. It could lead to damage of the cell surface in bacteria by swelling or rupturing of the cell, causing its internal components to leak outside, resulting in its death (Chen et al., 2021). However, a partial microbial rebound was noted at later time points (30–50 min). For instance, in trial no. 15, plate coverage increased from 20.22% at 10 min to 73.35% by 50 min. This phenomenon likely indicated complex reaction dynamics, such as the scavenging of radicals by groundwater components like natural organic matter and dissolved minerals or the repair and proliferation of bacteria that sustained only sub-lethal injury. This finding emphasises the critical need to optimise both reagent dosage and contact time to effectively prevent microbial regrowth. In addition to its disinfecting action, the Fenton process saw a noticeable reduction in the groundwater’s TDS, ranging from approximately 18–79.5%. The large reductions in TDS were coupled with conditions that achieved disinfection and promoted precipitation, likely involving precipitation of iron oxyhydroxides, followed by co-precipitation and adsorption of dissolved metals like iron and copper in the water. Oxidation and de-complexation of metal–organic species could have occurred at the acidic pH resulting in an increase in the effective availability of catalytic metals, coupled with a decrease in the radical scavenging by organic ligands and intensifying the direct and indirect oxidative injuries to bacterial membranes, proteins, and DNA, thereby increasing the chances of full inactivation rather than sublethal damage (Chen et al., 2021; Cuerda-Correa et al., 2019). After the subsequent neutralisation, Fe (OH)3 flocs would have formed that could capture the metals (Gómez et al., 2023; Faggiano et al., 2023; Kalajdžić et al., 2013). This, coupled with the stripping of CO2, could have shifted carbonate speciation, allowing for a decrease in the dissolved ions, thereby affecting the TDS (Geroni et al., 2012; Omar et al., 2024). During the Fenton reaction, pH, temperature, and concentrations of Fe2+ and •OH would influence the oxidation of organic matter and bacterial cells present within the water. Due to the nature of this reaction, it was essential to maintain the pH in strictly acidic conditions of 3–3.5 to maintain the production of •OH species in the water. An increase in pH value to basic conditions could render the process ineffective since the hydrogen peroxide would be decomposed into water and oxygen by Fe2+ without producing •OH in the water (Chen et al., 2021). This investigation provided a strong, data-driven validation of the Fenton-based AOP as a viable and effective technology for the dual-purpose remediation of contaminated groundwater in resource-limited settings.
5 Conclusion
Universal access to drinking water has been guaranteed to every person regardless of their identity and economic status. The duty to undertake this massive operation falls on the government. Despite this, there is still a significant need for vigilant monitoring and education of the general public regarding the WASH protocols, especially regarding the growing threat of antibiotic resistance in environmental settings such as drinking water sources. The isolated Enterobacter strains belonged to both ESKAPE and ECC groups, showcasing MDR activities typically associated with them. These isolates showcased resistance against various antibiotics from different classes and generations. The findings of Fenton’s reaction-based AOP demonstrated that the process’s efficacy was critically dependent on the optimisation of key parameters, specifically the reagent concentrations and contact time, to balance the generation of hydroxyl radicals against matrix scavenging effects. Furthermore, the observed phenomenon of microbial regrowth highlighted an important process limitation, underscoring the need to incorporate post-treatment strategies, such as filtration or secondary disinfection, to ensure sustained microbiological safety. The significance of this study lies in its demonstration that strict monitoring of all drinking water-related systems, ranging from water treatment plants, pipelines, and secondary storage, to alternative sources such as handpumps and open wells, is essential to safeguard against the growing threat of antibiotic resistance. Monitoring and effective disinfection of all sources of drinking water is a critical part of ensuring universal access to safe drinking water. This access should be available to all, including those in vulnerable situations. Vulnerability could especially arise due to income-related restrictions, limiting the ability to purchase purified packaged water or domestic purification devices when no other safe drinking water options are available. Moreover, disinfection techniques such as AOPs need to be thoroughly evaluated to improve the efficiency of existing protocols at centralised and community levels to maintain the water quality, especially after contamination events. Hence, this study aimed to raise awareness of critical water quality concerns within vulnerable low-income neighbourhoods, where access to safe municipal and groundwater is paramount for maintaining public health, while also trying to evaluate a decentralised disinfection process for localised contamination hotspots.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
VS: Writing – original draft. MKR: Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
The authors thank the Department of Life Sciences, GITAM School of Sciences, and MURTI Facility, GITAM (Deemed to be University), for providing laboratory facilities. The authors also thank Eurofins Genomics India Pvt. Ltd. for providing support in the genetic analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frwa.2025.1709063/full#supplementary-material
References
Abanyie, K., Apea, O. B., Abagale, S. A., Amuah, E. E. Y., and Sunkari, E. D. (2023). Sources and factors influencing groundwater quality and associated health implications: a review. Emerg. Contam. 9:100207. doi: 10.1016/j.emcon.2023.100207
Adefisoye, M. A., and Olaniran, A. O. (2022). Does chlorination promote antimicrobial resistance in waterborne pathogens? Mechanistic insight into co-resistance and its implication for public health. Antibiotics 11:564. doi: 10.3390/antibiotics11050564
Afunwa, R. A., Ezeanyinka, J., Afunwa, E. C., Udeh, A. S., Oli, A. N., and Unachukwu, M. (2020). Multiple antibiotic resistant index of gram-negative bacteria from bird droppings in two commercial poultries in Enugu, Nigeria. Open J. Med. Microbiol. 10, 171–181. doi: 10.4236/ojmm.2020.104015
Ahmed, Y., Lu, J., Yuan, Z., Bond, P. L., and Guo, J. (2020). Efficient inactivation of antibiotic resistant bacteria and antibiotic resistance genes by photo-Fenton process under visible LED light and neutral pH. Water Res. 179:115878. doi: 10.1016/j.watres.2020.115878
Akinjogunla, O. J., Odeyemi, A. T., Udofia, E. A., Adefiranye, O. O., Yah, C. S., Ehinmore, I., et al. (2023). Enterobacteriaceae isolates from clinical and household tap water samples: antibiotic resistance, screening for extended-spectrum, metallo- and ampC-beta-lactamases, and detection of blaTEM, blaSHVand blaCTX-M in Uyo, Nigeria. Germs 13:50. doi: 10.18683/germs.2023.1366
Akinola, O. T., Onyeaghasiri, F. U., Oluranti, O. O., and Elutade, O. O. (2022). Assessment of well water as a reservoir for extended-spectrum β-lactamases (ESBL) and carbapenem resistant Enterobacteriaceae from Iwo, Osun state, Nigeria. Iran. J. Microbiol. 14:351. doi: 10.18502/ijm.v14i3.9772
Annavajhala, K., Gomez-Simmonds, A., and Uhlemann, A.-C. (2019). Multidrug-resistant Enterobacter cloacae complex emerging as a global, diversifying threat. Front. Microbiol. 10:44. doi: 10.3389/fmicb.2019.00044
Ayoub, M. (2022). Fenton process for the treatment of wastewater effluent from the edible oil industry. Water Sci. Technol. 86, 1388–1401. doi: 10.2166/wst.2022.283
Bagordo, F., Brigida, S., Grassi, T., Caputo, M. C., Apollonio, F., De Carlo, L., et al. (2024). Factors influencing microbial contamination of groundwater: a systematic review of field-scale studies. Microorganisms 12:913. doi: 10.3390/microorganisms12050913
Bitton, G. (2014). “Drinking water distribution systems: biofilm microbiology” in Microbiology of drinking water (Wiley), 91–115. Available online at: https://www.wiley.com/en-us/Microbiology+of+Drinking+Water%3A+Production+and+Distribution-p-9781118743928#aboutauthors-section
Caliskan-Aydogan, O., and Alocilja, E. C. (2023). A review of carbapenem resistance in Enterobacterales and its detection techniques. Microorganisms 11:1491. doi: 10.3390/microorganisms11061491
Castaño-Henao, L., Mendez, D. F. G., Egan, S., and Sanabria, J. (2024). Changes in groundwater and surface water bacterial communities under disinfection processes: chlorination, ozonization, photo-Fenton and ultraviolet radiation. Curr. Res. Microb. Sci. 7:100244. doi: 10.1016/j.crmicr.2024.100244
Chathuranika, I. M., Sachinthanie, E., Zam, P., Gunathilake, M. B., Denkar, D., Muttil, N., et al. (2023). Assessing the water quality and status of water resources in urban and rural areas of Bhutan. J. Hazard. Mater. Adv. 12:100377. doi: 10.1016/j.hazadv.2023.100377
Chen, Y. D., Duan, X., Zhou, X., Wang, R., Wang, S., Ren, N. Q., et al. (2021). Advanced oxidation processes for water disinfection: features, mechanisms and prospects. Chem. Eng. J. 409:128207. doi: 10.1016/j.cej.2020.128207
Clark, K., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J., and Sayers, E. W. (2016). GenBank. Nucleic Acids Res. 44, D67–D72. doi: 10.1093/nar/gkv1276
Cuerda-Correa, E. M., Alexandre-Franco, M. F., and Fernández-González, C. (2019). Advanced oxidation processes for the removal of antibiotics from water: an overview. Water 12:102. doi: 10.3390/w12010102
Davin-Regli, P., Lavigne, J. M., and Pagès, J. M. (2019). Enterobacter spp.: update on taxonomy, clinical aspects, and emerging antimicrobial resistance. Clin. Microbiol. Rev. 32, e00050–e00018. doi: 10.1128/CMR.00002-19
Denissen, J., Reyneke, B., Waso-Reyneke, M., Havenga, B., Barnard, T., Khan, S., et al. (2022). Prevalence of ESKAPE pathogens in the environment: antibiotic resistance status, community-acquired infection and risk to human health. Int. J. Hyg. Environ. Health 244:114006. doi: 10.1016/j.ijheh.2022.114006
Dhengesu, D., Lemma, H., Asefa, L., and Tilahun, D. (2022). Antimicrobial resistance profile of Enterobacteriaceae and drinking water quality among households in Bule Hora town, South Ethiopia. Risk Manag. Healthc. Policy 15, 1569–1580. doi: 10.2147/rmhp.s370149
Ebomah, K. E., and Okoh, A. I. (2021). Enterobacter cloacae harbouring blaNDM-1, blaKPC, and blaOXA-48-like carbapenem-resistant genes isolated from different environmental sources in South Africa. Int. J. Environ. Stud. 78, 151–164. doi: 10.1080/00207233.2020.1778274
Evaristo, J., Jameel, Y., Tortajada, C., Wang, R. Y., Horne, J., Neukrug, H., et al. (2023). Water woes: the institutional challenges in achieving SDG 6. Sustain. Earth Rev. 6:13. doi: 10.1186/s42055-023-00067-2
Faggiano, A., De Carluccio, M., Cerrato, F., Junior, C. A. G., Proto, A., Fiorentino, A., et al. (2023). Improving organic matter and nutrients removal and minimizing sludge production in landfill leachate pre-treatment by Fenton process through a comprehensive response surface methodology approach. J. Environ. Manag. 340:117950. doi: 10.1016/j.jenvman.2023.117950
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.2307/2408678
Ferreira, D. C., Graziele, I., Marques, R. C., and Gonçalves, J. (2021). Investment in drinking water and sanitation infrastructure and its impact on waterborne diseases dissemination: the Brazilian case. Sci. Total Environ. 779:146279. doi: 10.1016/j.scitotenv.2021.146279
Galgano, M., Pellegrini, M., Catalano, F., Capozzi, E., Del Sambro, L., Sposato, L., et al. (2025). Acquired bacterial resistance to antibiotics and resistance genes: from past to future. Antibiotics 14:222. doi: 10.3390/antibiotics14030222
Ganbold, M., Seo, J., Wi, Y. M., Kwon, K. T., and Ko, K. S. (2023). Species identification, antibiotic resistance, and virulence in Enterobacter cloacae complex clinical isolates from South Korea. Front. Microbiol. 14:1122691. doi: 10.3389/fmicb.2023.1122691
Gaur, N., Dutta, D., Singh, A., Dubey, R., and Kamboj, D. V. (2022). Recent advances in the elimination of persistent organic pollutants by photocatalysis. Front. Environ. Sci. 10:872514. doi: 10.3389/fenvs.2022.872514
Geroni, J. N., Cravotta, C. A. III, and Sapsford, D. J. (2012). Evolution of the chemistry of Fe-bearing waters during CO2 degassing. Appl. Geochem. 27, 2335–2347. doi: 10.1016/j.apgeochem.2012.07.017
Gómez, C. A., Gómez-García, M. Á., and Dobrosz-Gómez, I. (2023). Analysis of the capacity of the Fenton process for the treatment of polluted wastewater from the leather dyeing industry. Sci. World J. 2023:4724606. doi: 10.1155/2023/4724606
Grönwall, J., and Danert, K. (2020). Regarding groundwater and drinking water access through a human rights lens: self-supply as a norm. Water 12:419. doi: 10.3390/w12020419
Hariharan, A. V. L. N. S. H., and Begum, M. (2023). Water quality parametric evaluation in the proximity of Gajuwaka, Vizag district, Andhra Pradesh. Glob. J. Res. Anal. 12, 59–61. doi: 10.36106/gjra/9504248
Hudzicki, J. (2009). Kirby–Bauer disk diffusion susceptibility test protocol. Am. Soc. Microbiol. 15, 1–23. Available online at: https://asm.org/getattachment/2594ce26-bd44-47f6-8287-0657aa9185ad/kirby-bauer-disk-diffusion-susceptibility-test-protocol-pdf.pdf (accessed July 13, 2025)
Kalajdžić, B., Habuda-Stanić, M., Romić, Ž., and Kuleš, M. (2013). Removal of natural organic matter from groundwater using Fenton’s process. Glob. NEST J. 15, 13–20. doi: 10.30955/gnj.000707
Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Krupińska, I. (2024). Application of Fenton’s reaction for removal of organic matter from groundwater. Molecules 29:5150. doi: 10.3390/molecules29215150
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lal, A., and Cheeptham, N. (2007). Eosin-methylene blue agar plates protocol. Am. Soc. Microbiol.. Available online at: https://asm.org/asm/media/protocol-images/eosin-methylene-blue-agar-plates-protocol.pdf (accessed July 13, 2025)
Lamba, M., Gupta, S., Shukla, R., Graham, D. W., Sreekrishnan, T. R., and Ahammad, S. Z. (2018). Carbapenem resistance exposures via wastewaters across New Delhi. Environ. Int. 119, 302–308. doi: 10.1016/j.envint.2018.07.004
Mir, R., Salari, S., Najimi, M., and Rashki, A. (2022). Determination of frequency, multiple antibiotic resistance index and resistotype of Salmonella spp. in chicken meat collected from southeast of Iran. Vet. Med. Sci. 8, 229–236. doi: 10.1002/vms3.647
Misra, T., Tare, M., and Jha, P. N. (2022). Insights into the dynamics and composition of biofilm formed by environmental isolate of Enterobacter cloacae. Front. Microbiol. 13:877060. doi: 10.3389/fmicb.2022.877060
Mosaka, T. B., Unuofin, J. O., Daramola, M. O., Tizaoui, C., and Iwarere, S. A. (2023). Inactivation of antibiotic-resistant bacteria and antibiotic-resistance genes in wastewater streams: current challenges and future perspectives. Front. Microbiol. 13:1100102. doi: 10.3389/fmicb.2022.1100102
Moyes, R. B., Reynolds, J., and Breakwell, D. P. (2009). Differential staining of bacteria: gram stain. Curr. Protoc. Microbiol. 15, A.3C.1–A.3C.8. doi: 10.1002/9780471729259.mca03cs15
Mustafa, A., Ibrahim, M., Rasheed, M. A., Kanwal, S., Hussain, A., Sami, A., et al. (2020). Genome-wide analysis of four Enterobacter cloacae complex type strains: insights into virulence and niche adaptation. Sci. Rep. 10:8150. doi: 10.1038/s41598-020-65001-4
Najjembe, C. A., Aladejana, O. M., Uwanibe, J. N., Happi, C. T., and Folarin, O. A. (2025). Microbiological and molecular characterization of bacterial communities in domestic water sources in Nabuti Village, Mukono District, Central Uganda. Microbiol. Res. 16:99. doi: 10.3390/microbiolres16050099
Narendra, K., and Rao, K. N. (2006). Morphometry of the Meghadrigedda watershed, Visakhapatnam district, Andhra Pradesh using GIS and Resourcesat data. J. Indian Soc. Remote Sens. 34, 101–110. doi: 10.1007/BF02991815
NCBI BLAST (2025). Search nucleotide databases using a nucleotide query. Available online at: https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch (accessed July 13, 2025)
Omar, M., Zyadah, M. A., Ali, M. Y., and El-Sonbati, M. A. (2024). Pre-treatment of composite industrial wastewater by Fenton and electro-Fenton oxidation processes. Sci. Rep. 14:27906. doi: 10.1038/s41598-024-78846-w
Onyango, L. A., Quinn, C., Tng, K. H. T., Wood, J. G., and Leslie, G. (2015). A study of failure events in drinking water systems as a basis for comparison and evaluation of the efficacy of potable reuse schemes. Environ. Health Insights 9, EHI–S31749. doi: 10.4137/EHI.S31749
Palma, D. (2021). Role of colored organic matter in the photochemical fate and removal of contaminants of emerging concern. (doctoral dissertation). Université Clermont Auvergne. Available online at: https://theses.hal.science/tel-03595044/
Ponnusami, A. B., Sinha, S., Ashokan, H., Paul, M. V., Hariharan, S. P., Arun, J., et al. (2023). Advanced oxidation process (AOP) combined biological process for wastewater treatment: a review on advancements, feasibility and practicability of combined techniques. Environ. Res. 237:116944. doi: 10.1016/j.envres.2023.116944
Poonia, S., Singh, T. S., and Tsering, D. C. (2014). Antibiotic susceptibility profile of bacteria isolated from natural sources of water from rural areas of East Sikkim. Indian J. Community Med. 39, 156–160. doi: 10.4103/0970-0218.137152
Quintanilla, A., and Munoz, M. (2019). Editorial catalysts: special issue on trends in catalytic wet peroxide oxidation processes. Catalysts 9:918. doi: 10.3390/catal9110918
Rafa, N., Jubayer, A., and Uddin, S. M. (2021). Impact of cyclone Amphan on the water, sanitation, hygiene, and health (WASH2) facilities of coastal Bangladesh. J. Water Sanit. Hyg. Dev. 11, 304–313. doi: 10.2166/washdev.2021.170
Ramirez, D., and Giron, M. (2025). Enterobacter infections. In: StatPearls. StatPearls Publishing. Available online at: https://www.ncbi.nlm.nih.gov/books/NBK559296/ (accessed July 13, 2025)
Ravi, K., and Singh, B. (2024). ESKAPE: navigating the global battlefield for antimicrobial resistance and defence in hospitals. Bacteria 3, 76–98. doi: 10.3390/bacteria3020006
Rivett, O., Tremblay-Levesque, L. C., Carter, R., Thetard, R. C., Tengatenga, M., Phoya, A., et al. (2022). Acute health risks to community hand-pumped groundwater supplies following cyclone Idai flooding. Sci. Total Environ. 806:150598. doi: 10.1016/j.scitotenv.2021.150598
Satyam, S., and Patra, S. (2025). The evolving landscape of advanced oxidation processes in wastewater treatment: challenges and recent innovations. PRO 13:987. doi: 10.3390/pr13040987
Singh, V., and Reddy, M. K. (2025a). Enterobacter cloacae strain DSM 30054 16S ribosomal RNA gene, partial sequence. NCBI Nucleotide.(PV739642). Available online at: https://www.ncbi.nlm.nih.gov/nuccore/PV739642 (accessed July 6, 2025)
Singh, V., and Reddy, M. K.. (2025b). Enterobacter quasihormaechei strain WCHEs120003 16S ribosomal RNA gene, partial sequence. NCBI Nucleotide.(PV739646). Available online at: https://www.ncbi.nlm.nih.gov/nuccore/PV739646 (accessed July 6, 2025)
Singh, V., and Reddy, M. K. (2025c). Detection of multidrug resistance in Enterobacteriaceae in drinking water. Environ. Sci. Adv. 4, 1594–1598. doi: 10.1039/d5va00117j
Sivalingam, P., Poté, J., and Prabakar, K. (2019). Environmental prevalence of carbapenem resistant Enterobacteriaceae (CRE) in a tropical ecosystem in India: human health perspectives and future directives. Pathogens 8:174. doi: 10.3390/pathogens8040174
Srinivasa Rao, S., Rajamani, N. S., and Reddi, E. U. B. (2015). Dispersal conditions and assimilative capacity of air environment at Gajuwaka industrial hub in Visakhapatnam. Int. J. Sci. Res. Sci. Eng. Technol. 1. Available online at: https://www.academia.edu/download/38680636/366.pdf
Stefaniak, K., Kiedrzyński, M., Korzeniewska, E., Kiedrzyńska, E., and Harnisz, M. (2024). Preliminary insights on carbapenem resistance in Enterobacteriaceae in high-income and low−/middle-income countries. Sci. Total Environ. 957:177593. doi: 10.1016/j.scitotenv.2024.177593
Tanner, W. D., VanDerslice, J. A., Goel, R. K., Leecaster, M. K., Fisher, M. A., Olstadt, J., et al. (2019). Multi-state study of Enterobacteriaceae harboring extended-spectrum beta-lactamase and carbapenemase genes in US drinking water. Sci. Rep. 9:3938. doi: 10.1038/s41598-019-40420-0
Tubić, A., Agbaba, J., Dalmacija, B., Molnar, J., Maletić, S., Watson, M., et al. (2013). Insight into changes during coagulation in NOM reactivity for trihalomethanes and haloacetic acids formation. J. Environ. Manag. 118, 153–160. doi: 10.1016/j.jenvman.2012.11.046
United Nations General Assembly (2010). The human right to water and sanitation. Resolution 64/292. Available online at: https://undocs.org/A/Res/64/292 (accessed March 13, 2023)
United Nations General Assembly (2015). Transforming our world: the 2030 agenda for sustainable development. A/RES/70/1. Available online at: https://www.un.org/en/development/desa/population/migration/generalassembly/docs/globalcompact/A_RES_70_1_E.pdf (accessed July 13, 2025)
Uppal, B. (2019). Biochemical tests—IMViC. Elsevier Health Sciences. Available online at: https://books.google.co.in/books/about/Biochemical_Tests_IMViC.html?id=H9Y6EAAAQBAJ (accessed July 13, 2025)
Venkata Ramana, C., Reddy, M. S., Reddy, T. B., and Ajay, B. (2018). Groundwater quality in and around Gajuwaka area, Visakhapatnam, Andhra Pradesh, India. Res. Rev. Int. J. Multidiscip. 3, 913–918. doi: 10.5281/zenodo.2531844
Vilela, P. B., Neto, R. P. M., Starling, M. C. V., Martins, A. D. S., Pires, G. F., Souza, F. A., et al. (2021). Metagenomic analysis of MWWTP effluent treated via solar photo-Fenton at neutral pH: effects upon microbial community, priority pathogens, and antibiotic resistance genes. Sci. Total Environ. 801:149599. doi: 10.1016/j.scitotenv.2021.149599
Walling, S. A., Um, W., Corkhill, C. L., and Hyatt, N. C. (2021). Fenton and Fenton-like wet oxidation for degradation and destruction of organic radioactive wastes. NPJ Mater. Degrad. 5:50. doi: 10.1038/s41529-021-00192-3
Wang, L., and Wu, Z. (2024). A simple high-throughput technology for microorganism detection and quantitative analysis. Foods 13:2954. doi: 10.3390/foods13182954
Zhao, W., Hou, Y., Wei, L., Wei, W., Zhang, K., Duan, H., et al. (2025). Chlorination-induced spread of antibiotic resistance genes in drinking water systems. Water Res. 274:123092. doi: 10.1016/j.watres.2025.123092
Keywords: multidrug resistance, Enterobacteriaceae, drinking water, disinfection, advanced oxidation process, Fenton’s reaction
Citation: Singh V and Reddy MK (2025) Fenton’s process for removal of resistant bacteria from unprotected aquifer samples. Front. Water. 7:1709063. doi: 10.3389/frwa.2025.1709063
Edited by:
Olivier Aubry, Université d'Orléans, FranceReviewed by:
Pradeep Madhamanchi, CCE-Andhra Pradesh, IndiaP. V. Gayathri, Sree Narayana Gurukulam College of Engineering, India
Barnabas Oluoch, University of KwaZulu-Natal, South Africa
Copyright © 2025 Singh and Reddy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: M. Kiranmai Reddy, a21hamppQGdpdGFtLmVkdQ==
 Visakha Singh
Visakha Singh M. Kiranmai Reddy
M. Kiranmai Reddy