- 1Division of Molecular and Cellular Function, School of Biological Sciences, Faculty of Biology Medicine and Health, The University of Manchester, Manchester, United Kingdom
- 2Division of Evolution and Genomic Sciences, School of Biological Sciences, Faculty of Biology Medicine and Health, The University of Manchester, Manchester, United Kingdom
Fibroblast Growth Factor 10 (FGF10) is a multifunctional mesenchymal-epithelial signaling growth factor, which is essential for multi-organ development and tissue homeostasis in adults. Furthermore, FGF10 deregulation has been associated with human genetic disorders and certain forms of cancer. Upon binding to FGF receptors with heparan sulfate as co-factor, FGF10 activates several intracellular signaling cascades, resulting in cell proliferation, differentiation, and invasion. FGF10 activity is modulated not only by heparan sulfate proteoglycans in the extracellular matrix, but also by hormones and other soluble factors. Despite more than 20 years of research on FGF10 functions, context-dependent regulation of FGF10 signaling specificity remains poorly understood. Emerging modes of FGF10 signaling regulation will be described, focusing on the role of FGF10 trafficking and sub-cellular localization, heparan sulfate proteoglycans, and miRNAs. Systems biology approaches based on quantitative proteomics will be considered for globally investigating FGF10 signaling specificity. Finally, current gaps in our understanding of FGF10 functions, such as the relative contribution of receptor isoforms to signaling activation, will be discussed in the context of genetic disorders and tumorigenesis.
Introduction
The Fibroblast Growth Factor 10 (Fgf10) gene has been identified in all examined vertebrates (Emoto et al., 1997). It belongs to the FGF7 subfamily of FGFs (Figure 1A) which was generated from a common ancestral gene during the early evolution of vertebrates and shares amino acids sequence similarities and biochemical functions (Ornitz and Itoh, 2015; Figure 1B).
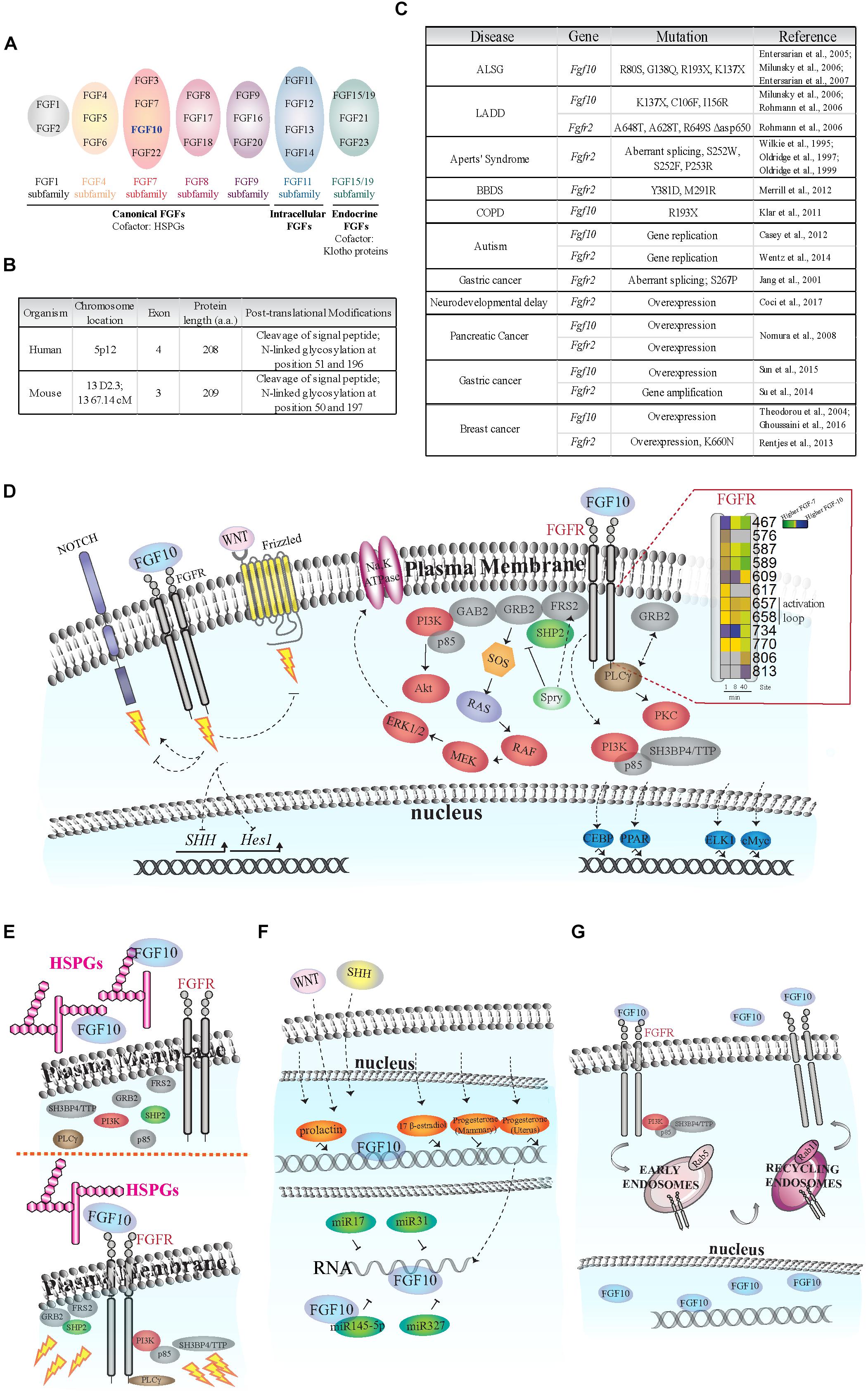
FIGURE 1. Fibroblast of Growth Factor (FGF) signaling activation and regulation. (A) Schematic of FGF subfamilies (see Ornitz and Itoh, 2015). FGF10 belongs to the FGF7 subfamily and is highlighted in blue. (B) Comparison of FGF10 gene/protein in human and mouse. The human FGF10 gene has an extra exon; and the protein length in the two species differs by one amino acid. FGF10 is secreted via the canonical ER-Golgi secretory pathway, as demonstrated by the cleavage of the signal peptide, and have two known glycosylation sites (Source: UniProt). (C) Known disease-causing mutations on Fgf10 and Fgfr2 genes. The rare developmental disorders Aplasia of Lacrimal and major Salivary Glands (ALSG), Lacrimo-Auricuo-Dentro-Digital (LADD) syndrome, Aperts’ Syndrome and Bent Bone Dysplasia Syndrome (BBDS) result from either the loss of key receptor-binding sites on FGF10 or mutations in the receptor kinase (TK) domains or IgII linker regions (Wilkie et al., 1995; Oldridge et al., 1997, 1999; Entesarian et al., 2005, 2007; Milunsky et al., 2006; Rohmann et al., 2006; Merrill et al., 2012). Interestingly, cases of ALSG caused by the R193X mutation also coincide with Chronic Obstructive Pulmonary Disease (COPD) (Klar et al., 2011). More recently, chromosomal translocation and duplication events at the loci of the Fgf10 and Fgfr2 genes have been associated with neurological disorders such as developmental delay and autism (Casey et al., 2012; Wentz et al., 2014). The role of single nucleotide polymorphisms and de novo point mutations causing oncogenic expression of Fgf10 and Fgfr2 are also becoming clearer, particularly in pancreatic, gastric, and breast cancers (Jang et al., 2001; Theodorou et al., 2004; Nomura et al., 2008; Reintjes et al., 2013; Su et al., 2014; Sun et al., 2015; Ghoussaini et al., 2016; Coci et al., 2017). (D) FGF10-dependent activation of FGFR intracellular tyrosine residues (Y; insert), adaptor proteins (gray), protein kinases (red), and transcription factors (blue). Insert on the right: each square represents a phosphorylated tyrosine residue (Y); the numbers correspond to phosphorylated Y residues on FGFR2b identified by proteomics; the color blue indicates higher phosphorylation at a given time point upon FGF10 stimulation of epithelial cells; modified from Francavilla et al. (2013). Dashed arrows represent FGF10-specific activation or inhibition of signaling. The lightning bolt represents the activation of signaling cascades. (E) Left, FGF10 bound to heparan sulfate proteoglycans (HSPGs) in the ECM does not activate FGFR and intracellular signaling. Right, FGF10 bound to HSPGs and FGFR induces the recruitment of protein adaptors to FGFR and signaling activation (represented by lightning bolts). (F) Schematic representation of FGF10 regulation in response to hormones (orange), miRNAs (dark green), WNT and SHH proteins. (G) FGF10 induces FGFR2b internalization into early endosomes and sorting to recycling endosomes and plasma membrane. FGF10 has also been found in the nucleus of certain cell types.
Fibroblast Growth Factor 10 is a paracrine signaling growth factor of 215 amino acids with a typical signal sequence for secretion and plays an essential role during development and tissue homeostasis in adults (Itoh, 2016). Fgf10 knockout (KO) mice die at birth with defects in multiple organ development, including the limb, lung, kidney, salivary gland and adipose tissue (Ohuchi et al., 2000). Fgf10 gene mutations have been associated with diseases, such as aplasia of lacrimal and salivary glands (ALSG) (Entesarian et al., 2007) and lacrimo-auriculo-dento-digital (LADD) syndrome (Rohmann et al., 2006); chronic obstructive pulmonary disease (Klar et al., 2011) and certain cancer types, including breast (Theodorou et al., 2004; Ghoussaini et al., 2016), pancreatic (Nomura et al., 2008), and gastric (Sun et al., 2015) cancers (Figure 1C).
Fibroblast Growth Factor 10 activates key intracellular signaling pathways in several cell types leading to the modulation of organ branching and cell proliferation, differentiation, and migration during development; wound healing and tissue repair; maintenance of stem cells compartment; and cancer cell invasion and proliferation (Itoh, 2016). Here, we will summarize known FGF10-dependent intracellular signaling pathways and cellular responses, before focusing on how Fgf10 expression and activity are modulated in different cellular contexts. Mechanisms underlying FGF10-dependent control of signaling specificity will be discussed and novel technologies to study the multiple roles of FGF10 will be introduced.
FGF10-Dependent Regulators of Intracellular Signaling
Early Signaling Players
The paracrine actions of FGF10 secreted by mesenchymal cells are mediated by the activation of epithelial FGF receptors with extracellular, transmembrane, and cytoplasmic tyrosine kinase domains (Ornitz and Itoh, 2015), and by heparan sulfate proteoglycans (HSPGs) in the extracellular matrix (ECM) (Patel et al., 2008). There are four Fibroblast Growth Factor receptor (Fgfr) genes (Fgfr1, 2, 3, 4). Fgfr1–3 are alternatively spliced into “b” and “c” isoforms, which differ in their extracellular domain and ligand binding specificity and which are expressed by epithelial or mesenchymal cells, respectively (Ornitz and Itoh, 2015). FGF10 has been shown to bind and selectively activate FGFR1b and 2b (Ornitz et al., 1996; Zhang et al., 2006).
Upon FGF10 binding, FGFRs dimerize and several intracellular tyrosine (Y) residues are trans-autophosphorylated. The sequential order of tyrosine residue phosphorylation has been reported for FGFR1 (Furdui et al., 2006). The catalytic tyrosine Y653 is phosphorylated first, followed by Y583, Y463, Y585, and Y654 phosphorylation, resulting in full receptor activation. Finally, Y677 and Y766 are phosphorylated, allowing the binding of adaptor molecules to the receptor (Furdui et al., 2006). FGF10-dependent dynamic phosphorylation of FGFR2b intracellular tyrosine residues has been studied in epithelial cells using quantitative mass spectrometry (MS)-based phosphoproteomics (Francavilla et al., 2013; Figure 1D, insert). This study showed that FGF10-dependent phosphorylation of Y734 on FGFR2b (or Y730 on FGFR1) specifically induced cell migration (Francavilla et al., 2013). These findings highlight the importance of the sequence surrounding phosphorylated residues on the activated receptor for ligand-dependent activation of downstream signaling pathways.
Fibroblast Growth Factor receptors engage multiple signaling pathways via adaptor proteins (Figure 1D). FGF-regulated substrate 2 (FRS2) binds the juxtamembrane domain of FGFRs independently of receptor activation, and is phosphorylated upon ligand binding, enabling the recruitment of other scaffold proteins, such as tyrosine-protein phosphatase non-receptor type 11 (PTPN11/SHP2) and growth factor receptor-bound protein 2 (GRB2) (Ong et al., 2000). FRS2, SHP2, and GRB2 are necessary to activate the extracellular regulated kinases (ERK1/2) pathway upon FGF10 stimulation in several examined cell types (Hadari et al., 1998; Ong et al., 2000; Upadhyay et al., 2003a) and during the growth of prostate xenografts in mice (Li et al., 2018a). Furthermore, FGF10 can induce the direct recruitment of the negative ERK1/2 signaling regulator Sprouty2 to FRS2 in lung epithelial cells (Tefft et al., 2002). Sprouty2 negatively regulates FGF10-dependent trophoblast invasion (Natanson-Yaron et al., 2007), otic placode size (Mahoney Rogers et al., 2011), and lung branching (Zhao and O’Brien, 2015). GRB2 has been shown to control basal FGFR2 activation (Lin et al., 2012) by competing with the binding of 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1 (PLCgγ) to FGFR2 (Timsah et al., 2014). PLCgγ binds to Y769 of FGFR2 (or Y766 on FGFR1) in the presence of FGF10 and is phosphorylated (Marchese et al., 2001), resulting in activation of protein kinase C (PKC) and calcium release (Figure 1D). Finally, the regulatory subunit of phosphatidylinositol 4,5-bisphosphate 3-kinase (PI3K) p85 which is known to be indirectly recruited to FGFR via the FRS2/GRB2/GAB1 complex (Ong et al., 2001), has been shown to bind to phosphorylated Y734 on FGFR2b upon FGF10 stimulation in epithelial breast cancer cells (Francavilla et al., 2013).
These results suggest context-dependent and fine-tuned modulation of downstream signaling pathways upon FGF10 binding to its receptors. FGF10 acts as a canonical FGFR ligand in recruiting the adaptor proteins FRS2, GRB2, or PLCγ to the receptor, but it can also induce the formation of cell type-specific signaling complexes (e.g., centered on p85). Either structural rearrangements of the receptor or the presence of cell type specific co-activators may explain these two different modes of FGF10 signaling initiation.
Kinases: ERK1/2 and PI3K
Fibroblast Growth Factor receptors signal through ERK1/2 during development (Corson et al., 2003). For instance, FGF10 and ERK1/2 signaling is necessary during duct elongation of submandibular glands (Steinberg et al., 2005), epithelium tooth growth (Cho et al., 2009), and determination of vaginal epithelial cell fate in Müllerian duct epithelium (Terakawa et al., 2016). In human diseases, FGF10 is capable of stimulating the growth of endometrial carcinoma cells by activating the ERK1/2 pathway in a paracrine manner (Taniguchi et al., 2003) and is involved in the growth of ameloblastoma – an epithelial benign tumor of the odontogenic apparatus – partially signaling through ERK1/2 (Nakao et al., 2013). It has also been suggested that FGF10 has a potential therapeutic use in lung edema, as FGF10 up-regulates Na,K-ATPase activity in alveolar epithelial cells via the ERK1/2 pathway (Upadhyay et al., 2003b). Finally, the crosstalk between FGF10 and the ERK1/2 pathway has been extensively studied in cell lines, in which either manipulating the Sprouty2/FRS2 complex which controls ERK1/2 activation (Tefft et al., 2002), or inhibiting upstream activators of ERK1/2, such as MEK, resulted in decreased FGF10-induced cellular responses (Taniguchi et al., 2003; Upadhyay et al., 2003b; Francavilla et al., 2013).
The role of FGF10 in signaling regulation through PI3K and downstream kinases like protein kinase B (AKT) is less clear (Figure 1D). The PI3K/AKT pathway is important for FGF10-dependent survival of hepatoblasts during early stages of hepatogenesis (Mavila et al., 2012) and for lens development (Chaffee et al., 2016). A role for FGF10/FGFR2/PI3K in neuroprotection after cerebral ischemia has also recently been described (Chen et al., 2017).
Even though the role of other kinases in FGF10 signaling specification remains to be determined, FGF10 is a versatile growth factor that enables epithelial cell growth and migration via ERK1/2 and controls cell survival via PI3K/AKT signaling during development and in several pathological conditions.
Transcription Factors
Fibroblast Growth Factor 10 is known to control cellular outputs through several transcription factors (Figure 1D). FGF10 plays crucial roles in adipogenesis by dynamically modulating the expression of members of the CCAAT/enhancer binding protein (C/EBP) and peroxisome proliferator activated receptor (PPAR) families of transcription factors (Sakaue et al., 2002). ELK-1 and c-MYC, but surprisingly not c-FOS, are regulated by FGF10 in endometrial carcinoma (Taniguchi et al., 2003). Finally, FGF10 controls the switch between vaginal and uterine epithelial cells fate via runt-related transcription factor 1 (RUNX1) in Müllerian duct epithelium (Terakawa et al., 2016).
As FGF10 activates both ERK1/2 and PI3K all the known transcription factors that depend on these kinases (e.g., ATF2, ELK1, FOS and FOXO, NFkB, CREB, respectively) (Yang et al., 2013; Mantamadiotis, 2017) should play a role in FGF10-dependent responses. However, it is not the case for ERK1/2-regulated activation of c-FOS (Taniguchi et al., 2003), suggesting that transcriptional regulation in response to FGF10/ERK1/2 signaling is complex and requires further investigation.
FGF10 Crosstalk With Other Signaling Pathways
Among several other important players, we will focus on four families of proteins with a context-specific role in FGF10 signaling (Figure 1D).
Neurogenic locus notch homolog protein 1 (NOTCH1) is a receptor controlling cell signaling via several ligands and mechanisms (Bray and Gomez-Lamarca, 2018). FGF10 activates NOTCH1 signaling during pancreatic development (Hart et al., 2003; Norgaard et al., 2003) but inhibits NOTCH1-dependent regulation of the gene Hes1 in the adult small intestine (Al Alam et al., 2015).
The functions of WNT ligands, which bind to the frizzled family of seven transmembrane receptors (Zeng et al., 2018), are modulated by FGF10 signaling during stomach (Nyeng et al., 2007) and lung (Volckaert and De Langhe, 2015) development. FGF10 also regulates Bone Morphogenetic Protein 4 (BMP4) during lung branching (Weaver et al., 2000).
The expression of Sonic hedgehog protein (SHH), which plays crucial roles during vertebrate development (Fernandes-Silva et al., 2017), is enhanced by FGF10 during the development of limb (Yokoyama et al., 2001), prostate gland (Huang et al., 2005), and stomach (Nyeng et al., 2007).
Thus, FGF10 modulates a great variety of cellular responses during development and in pathological conditions, through both conventional and ligand-specific signaling players.
Modulation of FGF10 Expression and Activity
Besides known transcription factors (e.g., Tbx4/5, Isl1, Etv1/Ewsv1) (Cebra-Thomas et al., 2003; Yamamoto-Shiraishi et al., 2014; Ching et al., 2018), Fgf10 expression and function are regulated by other factors, including HSPGs and soluble molecules (Figure 1E).
Heparan sulfate proteoglycans are a family of glycoproteins composed of a variety of heparan sulfate moieties (HS) attached to a core protein which play critical roles during organ branching and morphogenesis (Patel et al., 2017). Cleavage of HSPGs during ECM remodeling can release FGF10 from the ECM affecting epithelial cell proliferation and organ development (Figure 1E). FGF10 released from HS in the basement membrane increases salivary and lacrimal gland branching morphogenesis (Patel et al., 2008; Qu et al., 2011), whereas FGF10 binding to FGFR2b regulates the extent of the response to morphogenetics gradients (Makarenkova et al., 2009). Furthermore, the HSPG gradient pattern greatly affects FGF10 functions in the developing lung (Izvolsky et al., 2003) and during stomach morphogenesis (Huang et al., 2018). At a molecular level, FGF10 has a higher affinity for heparan compared to other FGFs (Lu et al., 1999) and it has a preference for certain patterns of sulfation and oligosaccharide length (Li et al., 2016). These unique biophysical properties of FGF10 may explain the great variety of FGF10 roles depending on cellular microenvironment, and may form the basis for the therapeutic control of FGF10 activities in vivo.
Fibroblast Growth Factor 10 gene expression is regulated by several hormones (Figure 1F). In mouse mammary gland 17 beta-estradiol, but not progesterone, increased the expression of Fgf10, whereas prolactin significantly induced Fgf10 gene expression during pregnancy (Cui and Li, 2008). In ovine uterus, progesterone regulates Fgf10 gene expression resulting in improved endometrial functions (Satterfield et al., 2008). These findings might be important not only to better refine FGF10 roles during development, but also to improve hormone-dependent cancer therapies. The latter possibility requires further studies to correlate hormones and FGF10 levels in human tumors, such as prostate or breast cancers.
Other important regulators of Fgf10 gene expression during organ branching are WNT and SHH (Figure 1F). For instance, members of the WNT family are crucial for FGF10-dependent signaling in lung and limb morphogenesis (Kawakami et al., 2001; Goss et al., 2011; Volckaert et al., 2013, 2017). SHH inhibits FGF10 localized expression during lung budding (Pepicelli et al., 1998). It is worth noticing that FGF10, WNT, and SHH proteins regulate each other through the establishment of feedback loops in different cells and in a spatio-temporal regulated manner during the development of branching organs (see “FGF10 Crosstalk With Other Signaling Pathways” section above and Figure 1D), thus confirming the importance of growth factors signaling crosstalk and dynamic regulation in human development and physiology.
Fibroblast Growth Factor 10 expression can also be controlled by micro-RNAs (miRNAs), which are crucial regulators of gene expression (Dragomir and Calin, 2018; Figure 1F). The mir-17 family of miRNAs is important for FGF10/FGFR2b downstream signaling during lung bud morphogenesis (Carraro et al., 2009) and miR-31 negatively regulates expression of Fgf10 during hair follicle growth and hair fiber formation (Mardaryev et al., 2010). More recently, it has been suggested that the miR-327/FGF10/FGFR2 signaling axis may be a therapeutic target for treatment of obesity and metabolic diseases (Fischer et al., 2017) and that the crosstalk between miR-145-5p and FGF10 expression regulates vascular smooth muscle cells proliferation and migration (Shi et al., 2018).
Although a comprehensive picture of Fgf10 expression and activity regulation has clearly emerged, a conundrum in FGF10 signaling still remains: how is FGF10 signaling specificity controlled in different cellular contexts? The importance of subcellular localization in modulating specific aspects of FGF10 responses will be discussed in the next section.
Subcellular Localization
The FGF10 receptor FGFR2b is internalized via clathrin-coated pits into intracellular vesicles (early endosomes) (Figure 1G), and then sorted to recycling endosomes, rather than to canonical late endosomes for degradation (Belleudi et al., 2007; Francavilla et al., 2013). Therefore, once FGFR2b is recycled back to the plasma membrane it may bind its ligands again and activate signaling in a sustained manner (Francavilla et al., 2013). Either the lack of receptor ubiquitination, which is a signal for degradation (Belleudi et al., 2007), or the recruitment of the adaptor proteins p85 and SH3BP4/TTP to phosphorylated FGFR2b (Francavilla et al., 2013) have been suggested as possible mechanisms underlying FGF10-dependent FGFR2b recycling to the plasma membrane. In either case, the sorting route of FGF10-activated receptors affects downstream signaling activation and cellular outputs, by inducing mitogenic responses in keratinocytes (Belleudi et al., 2007) or breast cancer cell migration and mouse embryonic lung branching (Francavilla et al., 2013). It would be interesting to study the endocytic route followed by FGF10 receptors in other cell types and how this affects downstream responses.
As well as in endosomes, FGF10 has been detected in the cytoplasm of cultured prostate stroma cells (Lu et al., 1999) and in the nucleus of urothelial cells (Bagai et al., 2002; Figure 1G). These findings suggest that different subcellular localization of FGF10 may underlie the specificity of FGF10 signaling in different cell types. The importance of FGF10 intracellular localization has been confirmed in studies about the molecular mechanisms underlying the LADD and ALSG human syndromes, which are characterized by mutations in the Fgf10 gene (Rohmann et al., 2006; Entesarian et al., 2007; Figure 1C). Mutated FGF10 failed to translocate into the nucleus. This might attenuate FGF10 intracrine functions, possibly explaining the phenotype observed in LADD or ALSG patients (Mikolajczak et al., 2016).
Understanding how FGF10 regulates signaling specificity in different cell types depending on its subcellular localization and how Fgf10 expression is modulated during organ morphogenesis and in human physiology may have therapeutic implications for cell- and growth factor-based personalized medicine.
System Biology Approaches to Study FGF10 Signaling Specificity
To improve our global understanding of FGF10 signaling specificity, ‘omics approaches might be useful. MS-based quantitative proteomics has become a powerful technology for investigating proteome function, composition, and post-translational modifications (PTMs) (Aebersold and Mann, 2016). In a typical shotgun proteomic workflow, proteins from tissues or cells are digested followed by peptide separation using liquid chromatography (LC) and peptide identification using tandem mass spectrometry (LC-MS/MS) (Aebersold and Mann, 2016). To identify, quantify and localize PTMs an enrichment step is introduced at the peptide level given the low stoichiometry of PTMs, such as phosphorylation (Figure 2A). In combination with functional assays phosphoproteomics has been successfully employed to study changes in intracellular signaling in tissues or perturbed cells (von Stechow et al., 2015) and one “functional proteomics” study has analyzed global FGF10 signaling in epithelial cells (Francavilla et al., 2013).
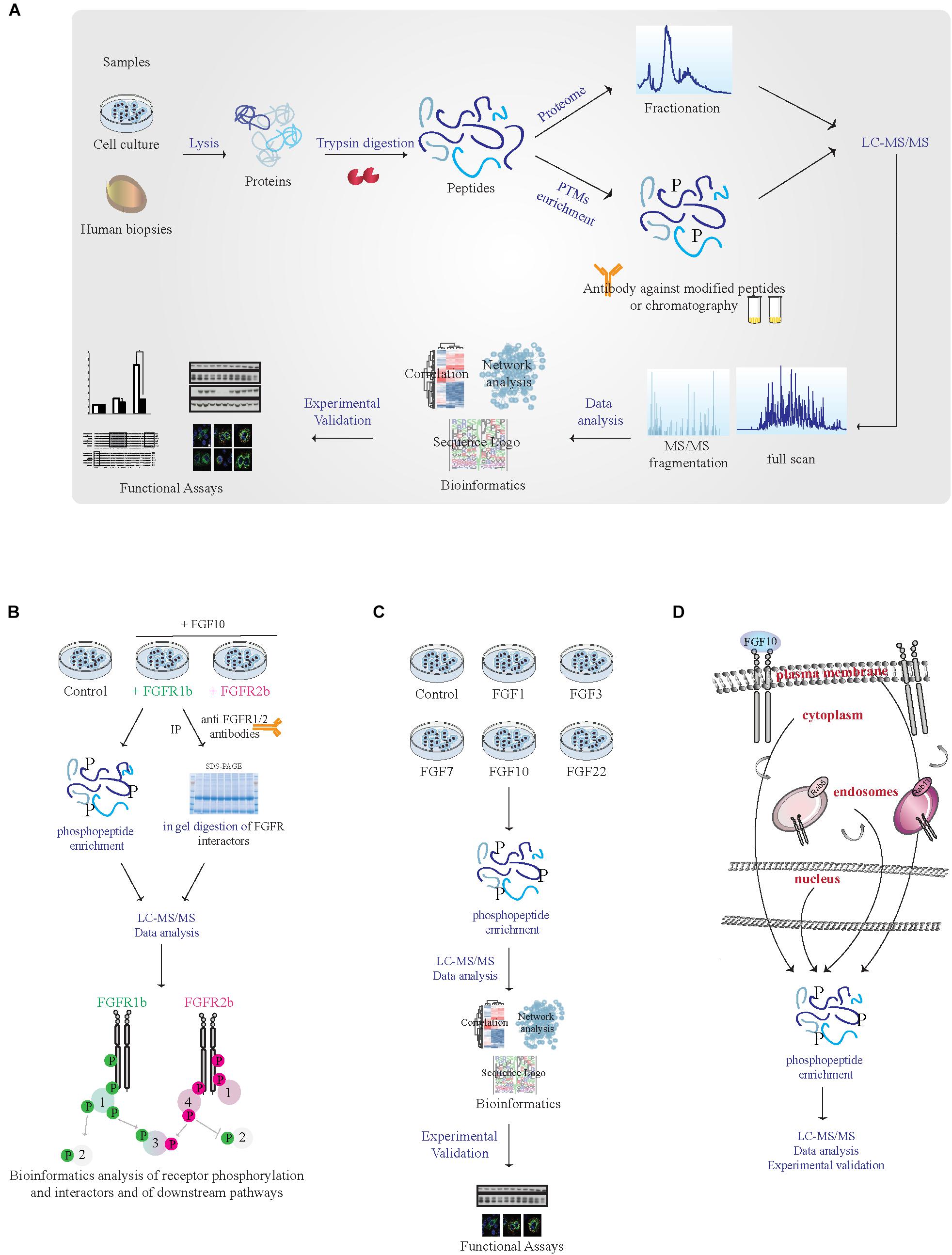
FIGURE 2. MS-based proteomic analysis of FGF10 signaling specificity. (A) Workflow of a typical shotgun proteomic experiment. Samples are lysed and proteins are digested into peptides. Peptides are then either fractionated to reduce sample complexity for the analysis of the whole cellular proteome or enriched for PTMs like phosphorylation using specific antibodies and chromatographic-based methods. Peptides are then separated and analyzed in the mass spectrometer. High resolution full scan and tandem MS/MS spectra are generated. Data are then analyzed by available software and bioinformatics tools before experimental validation of the most interesting hits. (B) Workflow of a proteomic experiment aiming at comparing FGFR1b and FGFR2b signaling in response to FGF10 stimulation. Combining phosphorylated peptide enrichment and immunoprecipitation of FGFRs followed by SDS–PAGE separation and protein in-gel digestion may result in the identification of receptor isoform-specific PTMs, protein interactors, and downstream signaling players. (C) Workflow of a proteomic experiment aiming at comparing signaling activation in response to different FGFs. Phosphoproteomics followed by mass spectrometry and bioinformatics will allow uncovering ligand-specific signaling cascades. (D) Workflow of a proteomic experiment aiming at deciphering subcellular compartment-specific signaling activation upon FGF10 stimulation. Phosphoproteomics is followed by mass spectrometry analysis and bioinformatics.
We suggest a few proteomic approaches to study FGF10 signaling specificity in an unbiased manner:
(1) Given the lack of available isoform-specific antibodies, FGF10 signaling can be compared in cells or organs expressing exclusively FGFR1b or 2b, using CRISPR-Cas9-based techniques or transgenic mice, by quantitative interactomics combined with phosphoproteomics. This will (a) increase our understanding of the relative contribution of FGFR1b and 2b isoforms to FGF10 response, as the phenotype of the FGFR2b (but not FGFR1b) KO mice resembles that of FGF10 KO mice (Ornitz and Itoh, 2015); (b) dissect which signaling players depend of FGFR1-2 activation in response to FGF10, based on recent data showing receptor-specific activation of SRC in prostate cancer (Li et al., 2018b); (c) uncover the potential role of receptor heterodimerization given the reciprocal auto-phosphorylation of FGFR1 and 2 (Bellot et al., 1991; Brewer et al., 2016); and (d) allow studying ligand-dependent receptor interactors and PTMs on the receptor (Figure 2B).
(2) System-level analysis of cellular signaling in response to different ligands for FGFR1-2b based on quantitative phosphoproteomics would reveal whether or not cellular responses are encoded by the identity of the ligand (Figure 2C). Two of the FGFR2b ligands, FGF7 and FGF10, are known to induce cell proliferation and lung cyst-like growth or cell migration and lung cell branching, respectively (Bellusci et al., 1997; Francavilla et al., 2013). However, the contribution of FGF1, 3 or 22 – which also bind to FGFR2b (Ornitz and Itoh, 2015) – has never been studied in detail. Quantitative phosphoproteomics would allow dissecting ligand-specific activation of intracellular pathways in different cell-types.
(3) MS-based organelles proteomics has been recently used to map protein subcellular localization (Itzhak et al., 2017). FGF10 signaling from different sub-cellular compartments might be dissected using a similar approach at a cellular level upon organelle enrichment followed by phosphoproteomics (Figure 2D).
Conclusion
Fibroblast Growth Factor 10-dependent responses range from cell proliferation, migration, and invasion, to multi-organ development, cancer or genetic disease progression. Despite the enormous increase in our understanding of FGF10 signaling and regulation, gaps in our knowledge of FGF10 specificity depending on cellular and extracellular environment still exist. Systems biology approaches, including MS-based quantitative proteomics or high-content microscopy (not discussed here due to space limitations) will contribute to a full understanding of FGF10 signaling. Moving toward personalized treatments for human diseases, this knowledge will be fundamental to develop novel therapies. For instance, recombinant FGF10 or antibodies against FGF10 may be developed to modulate FGF10 signaling depending on cellular context.
Author Contributions
All authors listed have contributed to the work and approved it for publication.
Funding
Research in the lab of CF is supported by Wellcome Trust Sir Henry Dale fellowship (107636/Z/15/Z) and by the Biotechnology and Biological Sciences Research Council (BBSRC, BB/R015864/1). JW is supported by the BBSRC Doctoral Training Partnership (BB/M011208/1).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all members of the lab and Dr. Dorey, Dr. Plusa, Dr. Kurinna, Prof. Kadler, and Prof. Sharrocks (The University of Manchester) for helpful discussion and for reading the manuscript. We also thank members of the Bio-MS facility, FBMH, The University of Manchester. We apologize to authors whose work could not be cited due to space limitations.
References
Aebersold, R., and Mann, M. (2016). Mass-spectrometric exploration of proteome structure and function. Nature 537, 347–355. doi: 10.1038/nature19949
Al Alam, D., Danopoulos, S., Schall, K., Sala, F. G., Almohazey, D., Fernandez, G. E., et al. (2015). Fibroblast growth factor 10 alters the balance between goblet and Paneth cells in the adult mouse small intestine. Am. J. Physiol. Gastrointest. Liver Physiol. 308, G678–G690. doi: 10.1152/ajpgi.00158.2014
Bagai, S., Rubio, E., Cheng, J. F., Sweet, R., Thomas, R., Fuchs, E., et al. (2002). Fibroblast growth factor-10 is a mitogen for urothelial cells. J. Biol. Chem. 277, 23828–23837. doi: 10.1074/jbc.M201658200
Belleudi, F., Leone, L., Nobili, V., Raffa, S., Francescangeli, F., Maggio, M., et al. (2007). Keratinocyte growth factor receptor ligands target the receptor to different intracellular pathways. Traffic 8, 1854–1872. doi: 10.1111/j.1600-0854.2007.00651.x
Bellot, F., Crumley, G., Kaplow, J. M., Schlessinger, J., Jaye, M., and Dionne, C. A. (1991). Ligand-induced transphosphorylation between different FGF receptors. EMBO J. 10, 2849–2854. doi: 10.1002/j.1460-2075.1991.tb07834.x
Bellusci, S., Grindley, J., Emoto, H., Itoh, N., and Hogan, B. L. (1997). Fibroblast growth factor 10 (FGF10) and branching morphogenesis in the embryonic mouse lung. Development 124, 4867–4878.
Bray, S. J., and Gomez-Lamarca, M. (2018). Notch after cleavage. Curr. Opin. Cell Biol. 51, 103–109. doi: 10.1016/j.ceb.2017.12.008
Brewer, J. R., Mazot, P., and Soriano, P. (2016). Genetic insights into the mechanisms of Fgf signaling. Genes Dev. 30, 751–771. doi: 10.1101/gad.277137.115
Carraro, G., El-Hashash, A., Guidolin, D., Tiozzo, C., Turcatel, G., Young, B. M., et al. (2009). miR-17 family of microRNAs controls FGF10-mediated embryonic lung epithelial branching morphogenesis through MAPK14 and STAT3 regulation of E-Cadherin distribution. Dev. Biol. 333, 238–250. doi: 10.1016/j.ydbio.2009.06.020
Casey, J. P., Magalhaes, T., Conroy, J. M., Regan, R., Shah, N., Anney, R., et al. (2012). A novel approach of homozygous haplotype sharing identifies candidate genes in autism spectrum disorder. Hum. Genet. 131, 565–579. doi: 10.1007/s00439-011-1094-6
Cebra-Thomas, J. A., Bromer, J., Gardner, R., Lam, G. K., Sheipe, H., and Gilbert, S. F. (2003). T-box gene products are required for mesenchymal induction of epithelial branching in the embryonic mouse lung. Dev. Dyn. 226, 82–90. doi: 10.1002/dvdy.10208
Chaffee, B. R., Hoang, T. V., Leonard, M. R., Bruney, D. G., Wagner, B. D., Dowd, J. R., et al. (2016). FGFR and PTEN signaling interact during lens development to regulate cell survival. Dev. Biol. 410, 150–163. doi: 10.1016/j.ydbio.2015.12.027
Chen, J., Wang, Z., Zheng, Z., Chen, Y., Khor, S., Shi, K., et al. (2017). Neuron and microglia/macrophage-derived FGF10 activate neuronal FGFR2/PI3K/Akt signaling and inhibit microglia/macrophages TLR4/NF-kappaB-dependent neuroinflammation to improve functional recovery after spinal cord injury. Cell Death Dis. 8:e3090. doi: 10.1038/cddis.2017.490
Ching, S. T., Infante, C. R., Du, W., Sharir, A., Park, S., Menke, D. B., et al. (2018). Isl1 mediates mesenchymal expansion in the developing external genitalia via regulation of Bmp4, Fgf10 and Wnt5a. Hum. Mol. Genet. 27, 107–119. doi: 10.1093/hmg/ddx388
Cho, K. W., Cai, J., Kim, H. Y., Hosoya, A., Ohshima, H., Choi, K. Y., et al. (2009). ERK activation is involved in tooth development via FGF10 signaling. J. Exp. Zool. B Mol. Dev. Evol. 312, 901–911. doi: 10.1002/jez.b.21309
Coci, E. G., Auhuber, A., Langenbach, A., Mrasek, K., Riedel, J., Leenen, A., et al. (2017). Novel unbalanced translocations affecting the long arms of chromosomes 10 and 22 cause complex syndromes with very severe neurodevelopmental delay, speech impairment, autistic behavior, and epilepsy. Cytogenet. Genome Res. 151, 171–178. doi: 10.1159/000471501
Corson, L. B., Yamanaka, Y., Lai, K. M., and Rossant, J. (2003). Spatial and temporal patterns of ERK signaling during mouse embryogenesis. Development 130, 4527–4537. doi: 10.1242/dev.00669
Cui, Y., and Li, Q. (2008). Effect of mammogenic hormones on the expression of FGF7, FGF10 and their receptor in mouse mammary gland. Sci. China C Life Sci. 51, 711–717. doi: 10.1007/s11427-008-0092-y
Dragomir, M., and Calin, G. A. (2018). Circular RNAs in cancer - lessons learned from microRNAs. Front. Oncol. 8:179. doi: 10.3389/fonc.2018.00179
Emoto, H., Tagashira, S., Mattei, M. G., Yamasaki, M., Hashimoto, G., Katsumata, T., et al. (1997). Structure and expression of human fibroblast growth factor-10. J. Biol. Chem. 272, 23191–23194. doi: 10.1074/jbc.272.37.23191
Entesarian, M., Dahlqvist, J., Shashi, V., Stanley, C. S., Falahat, B., Reardon, W., et al. (2007). FGF10 missense mutations in aplasia of lacrimal and salivary glands (ALSG). Eur. J. Hum. Genet. 15, 379–382. doi: 10.1038/sj.ejhg.5201762
Entesarian, M., Matsson, H., Klar, J., Bergendal, B., Olson, L., Arakaki, R., et al. (2005). Mutations in the gene encoding fibroblast growth factor 10 are associated with aplasia of lacrimal and salivary glands. Nat. Genet. 37, 125–127. doi: 10.1038/ng1507
Fernandes-Silva, H., Correia-Pinto, J., and Moura, R. S. (2017). Canonical sonic hedgehog signaling in early lung development. J. Dev. Biol. 5:3. doi: 10.3390/jdb5010003
Fischer, C., Seki, T., Lim, S., Nakamura, M., Andersson, P., Yang, Y., et al. (2017). A miR-327-FGF10-FGFR2-mediated autocrine signaling mechanism controls white fat browning. Nat. Commun. 8:2079. doi: 10.1038/s41467-017-02158-z
Francavilla, C., Rigbolt, K. T., Emdal, K. B., Carraro, G., Vernet, E., Bekker-Jensen, D. B., et al. (2013). Functional proteomics defines the molecular switch underlying FGF receptor trafficking and cellular outputs. Mol. Cell 51, 707–722. doi: 10.1016/j.molcel.2013.08.002
Furdui, C. M., Lew, E. D., Schlessinger, J., and Anderson, K. S. (2006). Autophosphorylation of FGFR1 kinase is mediated by a sequential and precisely ordered reaction. Mol. Cell 21, 711–717. doi: 10.1016/j.molcel.2006.01.022
Ghoussaini, M., French, J. D., Michailidou, K., Nord, S., Beesley, J., Canisus, S., et al. (2016). Evidence that the 5p12 variant rs10941679 confers susceptibility to estrogen-receptor-positive breast cancer through FGF10 and MRPS30 regulation. Am. J. Hum. Genet. 99, 903–911. doi: 10.1016/j.ajhg.2016.07.017
Goss, A. M., Tian, Y., Cheng, L., Yang, J., Zhou, D., Cohen, E. D., et al. (2011). Wnt2 signaling is necessary and sufficient to activate the airway smooth muscle program in the lung by regulating myocardin/Mrtf-B and Fgf10 expression. Dev. Biol. 356, 541–552. doi: 10.1016/j.ydbio.2011.06.011
Hadari, Y. R., Kouhara, H., Lax, I., and Schlessinger, J. (1998). Binding of Shp2 tyrosine phosphatase to FRS2 is essential for fibroblast growth factor-induced PC12 cell differentiation. Mol. Cell Biol. 18, 3966–3973. doi: 10.1128/MCB.18.7.3966
Hart, A., Papadopoulou, S., and Edlund, H. (2003). Fgf10 maintains notch activation, stimulates proliferation, and blocks differentiation of pancreatic epithelial cells. Dev. Dyn. 228, 185–193. doi: 10.1002/dvdy.10368
Huang, L., Pu, Y., Alam, S., Birch, L., and Prins, G. S. (2005). The role of Fgf10 signaling in branching morphogenesis and gene expression of the rat prostate gland: lobe-specific suppression by neonatal estrogens. Dev. Biol. 278, 396–414. doi: 10.1016/j.ydbio.2004.11.020
Huang, M., He, H., Belenkaya, T., and Lin, X. (2018). Multiple roles of epithelial heparan sulfate in stomach morphogenesis. J. Cell Sci. 131:jcs210781. doi: 10.1242/jcs.210781
Itoh, N. (2016). FGF10: a multifunctional mesenchymal-epithelial signaling growth factor in development, health, and disease. Cytokine Growth Factor Rev. 28, 63–69. doi: 10.1016/j.cytogfr.2015.10.001
Itzhak, D. N., Davies, C., Tyanova, S., Mishra, A., Williamson, J., Antrobus, R., et al. (2017). A mass spectrometry-based approach for mapping protein subcellular localization reveals the spatial proteome of mouse primary neurons. Cell Rep. 20, 2706–2718. doi: 10.1016/j.celrep.2017.08.063
Izvolsky, K. I., Zhong, L., Wei, L., Yu, Q., Nugent, M. A., and Cardoso, W. V. (2003). Heparan sulfates expressed in the distal lung are required for Fgf10 binding to the epithelium and for airway branching. Am. J. Physiol. Lung. Cell Mol. Physiol. 285, L838–L846. doi: 10.1152/ajplung.00081.2003
Jang, J. H., Shin, K. H., and Park, J. G. (2001). Mutations in fibroblast growth factor receptor 2 and fibroblast growth factor receptor 3 genes associated with human gastric and colorectal cancers. Cancer Res. 61, 3541–3543.
Kawakami, Y., Capdevila, J., Buscher, D., Itoh, T., Rodriguez Esteban, C., and Izpisua Belmonte, J. C. (2001). WNT signals control FGF-dependent limb initiation and AER induction in the chick embryo. Cell 104, 891–900. doi: 10.1016/S0092-8674(01)00285-9
Klar, J., Blomstrand, P., Brunmark, C., Badhai, J., Hakansson, H. F., Brange, C. S., et al. (2011). Fibroblast growth factor 10 haploinsufficiency causes chronic obstructive pulmonary disease. J. Med. Genet. 48, 705–709. doi: 10.1136/jmedgenet-2011-100166
Li, Q., Alsaidan, O. A., Ma, Y., Kim, S., Liu, J., Albers, T., et al. (2018a). Pharmacologically targeting the myristoylation of the scaffold protein FRS2alpha inhibits FGF/FGFR-mediated oncogenic signaling and tumor progression. J. Biol. Chem. 293, 6434–6448. doi: 10.1074/jbc.RA117.000940
Li, Q., Ingram, L., Kim, S., Beharry, Z., Cooper, J. A., and Cai, H. (2018b). Paracrine fibroblast growth factor initiates oncogenic synergy with epithelial fgfr/src transformation in prostate tumor progression. Neoplasia 20, 233–243. doi: 10.1016/j.neo.2018.01.006
Li, Y., Sun, C., Yates, E. A., Jiang, C., Wilkinson, M. C., and Fernig, D. G. (2016). Heparin binding preference and structures in the fibroblast growth factor family parallel their evolutionary diversification. Open Biol. 6: 150275. doi: 10.1098/rsob.150275
Lin, C. C., Melo, F. A., Ghosh, R., Suen, K. M., Stagg, L. J., Kirkpatrick, J., et al. (2012). Inhibition of basal FGF receptor signaling by dimeric Grb2. Cell 149, 1514–1524. doi: 10.1016/j.cell.2012.04.033
Lu, W., Luo, Y., Kan, M., and McKeehan, W. L. (1999). Fibroblast growth factor-10. A second candidate stromal to epithelial cell andromedin in prostate. J. Biol. Chem. 274, 12827–12834. doi: 10.1074/jbc.274.18.12827
Mahoney Rogers, A. A., Zhang, J., and Shim, K. (2011). Sprouty1 and Sprouty2 limit both the size of the otic placode and hindbrain Wnt8a by antagonizing FGF signaling. Dev. Biol. 353, 94–104. doi: 10.1016/j.ydbio.2011.02.022
Makarenkova, H. P., Hoffman, M. P., Beenken, A., Eliseenkova, A. V., Meech, R., Tsau, C., et al. (2009). Differential interactions of FGFs with heparan sulfate control gradient formation and branching morphogenesis. Sci. Signal. 2:ra55. doi: 10.1126/scisignal.2000304
Mantamadiotis, T. (2017). Towards targeting PI3K-dependent regulation of gene expression in brain cancer. Cancers 9:E60. doi: 10.3390/cancers9060060
Marchese, C., Felici, A., Visco, V., Lucania, G., Igarashi, M., Picardo, M., et al. (2001). Fibroblast growth factor 10 induces proliferation and differentiation of human primary cultured keratinocytes. J. Invest. Dermatol. 116, 623–628. doi: 10.1046/j.0022-202x.2001.01280.x
Mardaryev, A. N., Ahmed, M. I., Vlahov, N. V., Fessing, M. Y., Gill, J. H., Sharov, A. A., et al. (2010). Micro-RNA-31 controls hair cycle-associated changes in gene expression programs of the skin and hair follicle. FASEB J. 24, 3869–3881. doi: 10.1096/fj.10-160663
Mavila, N., James, D., Utley, S., Cu, N., Coblens, O., Mak, K., et al. (2012). Fibroblast growth factor receptor-mediated activation of AKT-beta-catenin-CBP pathway regulates survival and proliferation of murine hepatoblasts and hepatic tumor initiating stem cells. PLoS One 7:e50401. doi: 10.1371/journal.pone.0050401
Merrill, A. E., Sarukhanov, A., Krejci, P., Idoni, B., Camacho, N., Estrada, K. D., et al. (2012). Bent bone dysplasia-FGFR2 type, a distinct skeletal disorder, has deficient canonical FGF signaling. Am. J. Hum. Genet. 90, 550–557. doi: 10.1016/j.ajhg.2012.02.005
Mikolajczak, M., Goodman, T., and Hajihosseini, M. K. (2016). Interrogation of a lacrimo-auriculo-dento-digital syndrome protein reveals novel modes of fibroblast growth factor 10 (FGF10) function. Biochem. J. 473, 4593–4607. doi: 10.1042/BCJ20160441
Milunsky, J. M., Zhao, G., Maher, T. A., Colby, R., and Everman, D. B. (2006). LADD syndrome is caused by FGF10 mutations. Clin. Genet. 69, 349–354. doi: 10.1111/j.1399-0004.2006.00597.x
Nakao, Y., Mitsuyasu, T., Kawano, S., Nakamura, N., Kanda, S., and Nakamura, S. (2013). Fibroblast growth factors 7 and 10 are involved in ameloblastoma proliferation via the mitogen-activated protein kinase pathway. Int. J. Oncol. 43, 1377–1384. doi: 10.3892/ijo.2013.2081
Natanson-Yaron, S., Anteby, E. Y., Greenfield, C., Goldman-Wohl, D., Hamani, Y., Hochner-Celnikier, D., et al. (2007). FGF 10 and Sprouty 2 modulate trophoblast invasion and branching morphogenesis. Mol. Hum. Reprod. 13, 511–519. doi: 10.1093/molehr/gam034
Nomura, S., Yoshitomi, H., Takano, S., Shida, T., Kobayashi, S., Ohtsuka, M., et al. (2008). FGF10/FGFR2 signal induces cell migration and invasion in pancreatic cancer. Br. J. Cancer 99, 305–313. doi: 10.1038/sj.bjc.6604473
Norgaard, G. A., Jensen, J. N., and Jensen, J. (2003). FGF10 signaling maintains the pancreatic progenitor cell state revealing a novel role of Notch in organ development. Dev. Biol. 264, 323–338. doi: 10.1016/j.ydbio.2003.08.013
Nyeng, P., Norgaard, G. A., Kobberup, S., and Jensen, J. (2007). FGF10 signaling controls stomach morphogenesis. Dev. Biol. 303, 295–310. doi: 10.1016/j.ydbio.2006.11.017
Ohuchi, H., Hori, Y., Yamasaki, M., Harada, H., Sekine, K., Kato, S., et al. (2000). FGF10 acts as a major ligand for FGF receptor 2 IIIb in mouse multi-organ development. Biochem. Biophys. Res. Commun. 277, 643–649. doi: 10.1006/bbrc.2000.3721
Oldridge, M., Lunt, P. W., Zackai, E. H., McDonaldMcGinn, D. M., Muenke, M., Moloney, D. M., et al. (1997). Genotype-phenotype correlation for nucleotide substitutions in the IgII-IgIII linker of FGFR2. Hum. Mol. Genet. 6, 137–143. doi: 10.1093/hmg/6.1.137
Oldridge, M., Zackai, E. H., McDonald-McGinn, D. M., Iseki, S., Morriss-Kay, G. M., Twigg, R. F., et al. (1999). De novo Alu-element insertions in FGFR2 identify a distinct pathological basis for Apert syndrome. Am. J. Hum. Genet. 64, 446–461. doi: 10.1086/302245
Ong, S. H., Guy, G. R., Hadari, Y. R., Laks, S., Gotoh, N., Schlessinger, J., et al. (2000). FRS2 proteins recruit intracellular signaling pathways by binding to diverse targets on fibroblast growth factor and nerve growth factor receptors. Mol. Cell. Biol. 20, 979–989. doi: 10.1128/MCB.20.3.979-989.2000
Ong, S. H., Hadari, Y. R., Gotoh, N., Guy, G. R., Schlessinger, J., and Lax, I. (2001). Stimulation of phosphatidylinositol 3-kinase by fibroblast growth factor receptors is mediated by coordinated recruitment of multiple docking proteins. Proc. Natl. Acad. Sci. U.S.A. 98, 6074–6079. doi: 10.1073/pnas.111114298
Ornitz, D. M., and Itoh, N. (2015). The fibroblast growth factor signaling pathway. Wiley Interdiscip. Rev. Dev. Biol. 4, 215–266. doi: 10.1002/wdev.176
Ornitz, D. M., Xu, J., Colvin, J. S., McEwen, D. G., MacArthur, C. A., Coulier, F., et al. (1996). Receptor specificity of the fibroblast growth factor family. J. Biol. Chem. 271, 15292–15297. doi: 10.1074/jbc.271.25.15292
Patel, V. N., Likar, K. M., Zisman-Rozen, S., Cowherd, S. N., Lassiter, K. S., Sher, I., et al. (2008). Specific heparan sulfate structures modulate FGF10-mediated submandibular gland epithelial morphogenesis and differentiation. J. Biol. Chem. 283, 9308–9317. doi: 10.1074/jbc.M709995200
Patel, V. N., Pineda, D. L., and Hoffman, M. P. (2017). The function of heparan sulfate during branching morphogenesis. Matrix Biol. 5, 311–323. doi: 10.1016/j.matbio.2016.09.004
Pepicelli, C. V., Lewis, P. M., and McMahon, A. P. (1998). Sonic hedgehog regulates branching morphogenesis in the mammalian lung. Curr. Biol. 8, 1083–1086. doi: 10.1016/S0960-9822(98)70446-4
Qu, X., Carbe, C., Tao, C., Powers, A., Lawrence, R., van Kuppevelt, T. H., et al. (2011). Lacrimal gland development and Fgf10-Fgfr2b signaling are controlled by 2-O- and 6-O-sulfated heparan sulfate. J. Biol. Chem. 286, 14435–14444. doi: 10.1074/jbc.M111.225003
Reintjes, N., Li, Y., Becker, A., Rohmann, E., Schmutzler, R., and Wollnik, B. (2013). Activating somatic FGFR2 mutations in breast cancer. PLoS One 8:e60264. doi: 10.1371/journal.pone.0060264
Rohmann, E., Brunner, H. G., Kayserili, H., Uyguner, O., Nurnberg, G., Lew, E. D., et al. (2006). Mutations in different components of FGF signaling in LADD syndrome. Nat. Genet. 38, 414–417. doi: 10.1038/ng1757
Sakaue, H., Konishi, M., Ogawa, W., Asaki, T., Mori, T., Yamasaki, M., et al. (2002). Requirement of fibroblast growth factor 10 in development of white adipose tissue. Genes Dev. 16, 908–912. doi: 10.1101/gad.983202
Satterfield, M. C., Hayashi, K., Song, G., Black, S. G., Bazer, F. W., and Spencer, T. E. (2008). Progesterone regulates FGF10, MET, IGFBP1, and IGFBP3 in the endometrium of the ovine uterus. Biol. Reprod. 79, 1226–1236. doi: 10.1095/biolreprod.108.071787
Shi, L., Tian, C., Sun, L., Cao, F., and Meng, Z. (2018). The lncRNA TUG1/miR-145-5p/FGF10 regulates proliferation and migration in VSMCs of hypertension. Biochem. Biophys. Res. Commun. 501, 688–695. doi: 10.1016/j.bbrc.2018.05.049
Steinberg, Z., Myers, C., Heim, V. M., Lathrop, C. A., Rebustini, I. T., Stewart, J. S., et al. (2005). FGFR2b signaling regulates ex vivo submandibular gland epithelial cell proliferation and branching morphogenesis. Development 132, 1223–1234. doi: 10.1242/dev.01690
Su, X., Zhan, P., Gavine, P. R., Morgan, S., Womack, C., Ni, X., et al. (2014). FGFR2 amplification has prognostic significance in gastric cancer: results from a large international multicentre study. Br. J. Cancer. 110, 967–975. doi: 10.1038/bjc.2013.802
Sun, Q., Lin, P., Zhang, J., Li, X., Yang, L., Huang, J., et al. (2015). Expression of fibroblast growth factor 10 is correlated with poor prognosis in gastric adenocarcinoma. Tohoku J. Exp. Med. 236, 311–318. doi: 10.1620/tjem.236.311
Taniguchi, F., Harada, T., Sakamoto, Y., Yamauchi, N., Yoshida, S., Iwabe, T., et al. (2003). Activation of mitogen-activated protein kinase pathway by keratinocyte growth factor or fibroblast growth factor-10 promotes cell proliferation in human endometrial carcinoma cells. J. Clin. Endocrinol. Metab. 88, 773–780. doi: 10.1210/jc.2002-021062
Tefft, D., Lee, M., Smith, S., Crowe, D. L., Bellusci, S., and Warburton, D. (2002). mSprouty2 inhibits FGF10-activated MAP kinase by differentially binding to upstream target proteins. Am. J. Physiol. Lung. Cell Mol. Physiol. 283, L700–L706. doi: 10.1152/ajplung.00372.2001
Terakawa, J., Rocchi, A., Serna, V. A., Bottinger, E. P., Graff, J. M., and Kurita, T. (2016). FGFR2IIIb-MAPK activity is required for epithelial cell fate decision in the lower mullerian duct. Mol. Endocrinol. 30, 783–795. doi: 10.1210/me.2016-1027
Theodorou, V., Boer, M., Weigelt, B., Jonkers, J., van der Valk, M., and Hilkens, J. (2004). Fgf10 is an oncogene activated by MMTV insertional mutagenesis in mouse mammary tumors and overexpressed in a subset of human breast carcinomas. Oncogene 23, 6047–6055. doi: 10.1038/sj.onc.1207816
Timsah, Z., Ahmed, Z., Lin, C. C., Melo, F. A., Stagg, L. J., Leonard, P. G., et al. (2014). Competition between Grb2 and Plcgamma1 for FGFR2 regulates basal phospholipase activity and invasion. Nat. Struct. Mol. Biol. 21, 180–188. doi: 10.1038/nsmb.2752
Upadhyay, D., Correa-Meyer, E., Sznajder, J. I., and Kamp, D. W. (2003a). FGF-10 prevents mechanical stretch-induced alveolar epithelial cell DNA damage via MAPK activation. Am. J. Physiol. Lung Cell Mol. Physiol. 284, L350–L359.
Upadhyay, D., Lecuona, E., Comellas, A., Kamp, D. W., and Sznajder, J. I. (2003b). Fibroblast growth factor-10 upregulates Na,K-ATPase via the MAPK pathway. FEBS Lett. 545, 173–176.
Volckaert, T., Campbell, A., Dill, E., Li, C., Minoo, P., and De Langhe, S. (2013). Localized Fgf10 expression is not required for lung branching morphogenesis but prevents differentiation of epithelial progenitors. Development 140, 3731–3742. doi: 10.1242/dev.096560
Volckaert, T., and De Langhe, S. P. (2015). Wnt and FGF mediated epithelial-mesenchymal crosstalk during lung development. Dev. Dyn. 244, 342–366. doi: 10.1002/dvdy.24234
Volckaert, T., Yuan, T., Chao, C. M., Bell, H., Sitaula, A., Szimmtenings, L., et al. (2017). Fgf10-hippo epithelial-mesenchymal crosstalk maintains and recruits lung basal stem cells. Dev. Cell 43, 48–59.e5. doi: 10.1016/j.devcel.2017.09.003
von Stechow, L., Francavilla, C., and Olsen, J. V. (2015). Recent findings and technological advances in phosphoproteomics for cells and tissues. Expert Rev. Proteomics 12, 469–487. doi: 10.1586/14789450.2015.1078730
Weaver, M., Dunn, N. R., and Hogan, B. L. (2000). Bmp4 and Fgf10 play opposing roles during lung bud morphogenesis. Development 127, 2695–2704.
Wentz, E., Vujic, M., Karrstedt, E. L., Erlandsson, A., and Gillberg, C. (2014). A case report of two male siblings with autism and duplication of Xq13-q21, a region including three genes predisposing for autism. Eur. Child Adolesc. Psychiatry 23, 329–336. doi: 10.1007/s00787-013-0455-1
Wilkie, A. O. M., Slaney, S. F., Oldridge, M., Poole, M. D., Ashworth, G. J., Hockley, A. D., et al. (1995). Apert syndrome results from localized mutations of Fgfr2 and is allelic with crouzon syndrome. Nat. Genet. 9, 165–172. doi: 10.1038/ng0295-165
Yamamoto-Shiraishi, Y., Higuchi, H., Yamamoto, S., Hirano, M., and Kuroiwa, A. (2014). Etv1 and Ewsr1 cooperatively regulate limb mesenchymal Fgf10 expression in response to apical ectodermal ridge-derived fibroblast growth factor signal. Dev. Biol. 394, 181–190. doi: 10.1016/j.ydbio.2014.07.022
Yang, S. H., Sharrocks, A. D., and Whitmarsh, A. J. (2013). MAP kinase signalling cascades and transcriptional regulation. Gene 513, 1–13. doi: 10.1016/j.gene.2012.10.033
Yokoyama, H., Ide, H., and Tamura, K. (2001). FGF-10 stimulates limb regeneration ability in Xenopus laevis. Dev. Biol. 233, 72–79. doi: 10.1006/dbio.2001.0180
Zeng, C. M., Chen, Z., and Fu, L. (2018). Frizzled receptors as potential therapeutic targets in human cancers. Int. J. Mol. Sci. 19:E1543. doi: 10.3390/ijms19051543
Zhang, X., Ibrahimi, O. A., Olsen, S. K., Umemori, H., Mohammadi, M., and Ornitz, D. M. (2006). Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J. Biol. Chem. 281, 15694–15700. doi: 10.1074/jbc.M601252200
Keywords: fibroblast growth factor 10, FGF receptor, signaling, development, cancer, genetic disorders, mass spectrometry, quantitative proteomics
Citation: Watson J and Francavilla C (2018) Regulation of FGF10 Signaling in Development and Disease. Front. Genet. 9:500. doi: 10.3389/fgene.2018.00500
Received: 23 August 2018; Accepted: 05 October 2018;
Published: 23 October 2018.
Edited by:
Saverio Bellusci, Justus-Liebig-Universität Gießen, GermanyReviewed by:
Jin-San Zhang, Mayo Clinic, United StatesNan Tang Tang, National Institute of Biological Sciences (NIBS), China
Elie El Agha, Justus-Liebig-Universität Gießen, Germany
Copyright © 2018 Watson and Francavilla. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chiara Francavilla, Y2hpYXJhLmZyYW5jYXZpbGxhQG1hbmNoZXN0ZXIuYWMudWs=
 Joanne Watson
Joanne Watson Chiara Francavilla
Chiara Francavilla