- 1School of Marine Sciences, Ningbo University, Ningbo, China
- 2Agricultural Center, Louisiana State University, Baton Rouge, LA, USA
The biological functions of microRNAs (miRNAs) have been studied in a number of eukaryotic species. Recent studies on vertebrate animals have demonstrated critical roles of miRNA in immune and metabolic activities. However, studies on the functions of miRNA in invertebrates are very limited. Here, we demonstrated that miR-31 from Apostichopus japonicus disrupts the balance of lipid metabolism, thus resulting in cell apoptosis by targeting complement C1q tumor necrosis factor-related protein 9 (AjCTRP9), a novel adipokine with pleiotropic functions in immunity and metabolism. Lipidomic analysis suggested that the intercellular lipid metabolites were markedly altered, and three ceramide (Cer) species synchronously increased in the AjCTRP9-silenced coelomocytes. Moreover, exogenous Cer exposure significantly induced apoptosis in the coelomocytes in vivo, in agreement with findings from miR-31 mimic- or AjCTRP9 small-interfering RNA-transfected coelomocytes. Furthermore, we found that the imbalance in sphingolipid metabolism triggered by the overproduction of Cers ultimately resulted in the activation of the apoptosis initiator caspase-8 and executioner caspase-3. Our findings provide the first direct evidence that miR-31 negatively modulates the expression of AjCTRP9 and disturbance of Cer channels, thus leading to caspase-3- and caspase-8-dependent apoptosis, during the interactions between pathogens and host.
Introduction
After the invasion of pathogens, the innate immune response reacts immediately as the first barrier to defend the host. Various cellular and humoral defense pathways are simultaneously activated through different signaling networks. Efforts have been made to identify the roles of immune-related molecules during immune responses. However, studies on the regulatory networks of these molecules and their interactions in host defense processes are lacking. Emerging studies deciphering the various cellular and signaling networks linking the immune defense with metabolism have contributed to the understanding of disease pathogenesis and the development of therapeutic strategies (1). It is therefore no surprise that the dissection of the interface connecting the immune system to metabolic pathways including glycolysis, lipid, and amino acid metabolism has recently gained interest (2, 3). This metabolic regulation is largely controlled by extracellular signals that direct the uptake, storage, and utilization of substrates (glucose, amino acids, and fatty acids) under immune responses. Among them, lipid metabolism has been shown to be closely related to the immune response, because the lipids of the membrane bilayer form the first line of barrier between a cell and its environment (4), and lipid metabolism generates a variety of bioactive signaling molecules, including phosphatidic acid, triacylglycerol (TAG), sphingosine, and ceramide (Cer), which participate in the regulation of many cellular processes such as cell growth, membrane homeostasis, inflammation, and apoptosis (5). Imbalances in cellular lipid metabolism increase the susceptibility of the host to pathogenic invasions and ultimately lead to cell death in the host (6, 7). Therefore, the regulation of lipid metabolism may also change the host immune response to pathogenic challenge. Manipulation of lipid metabolism pathways may contribute to the enhancement of host immune systems against pathogens.
microRNAs (miRNAs) are highly conserved small non-coding RNAs (~22 nucleotides) that regulate gene expression primarily by binding to the 3’-untranslated regions of target mRNAs, thus decreasing target protein synthesis and sometimes resulting in mRNA degradation (8, 9). As one of the most abundant classes of gene regulators, miRNAs are involved in many biological processes, such as cell proliferation (10), tumorigenesis (11), apoptosis (12), and inflammation (13). Recent studies have indicated that miRNAs also play novel roles in regulating metabolic pathways through the control of target genes involved in metabolic processes (14, 15), and especially in controlling lipid metabolic homeostasis and host inflammatory responses in cardiovascular diseases (16, 17).
To date, more than 30 miRNAs have been identified to play crucial roles in the regulation of lipid metabolism through various pathways, such as fatty acid oxidation, and biosynthesis of fatty acids, cholesterol, and TAG (5, 18, 19). The first evidence linking specific miRNAs to critical pathways in lipid metabolism has been found in the fruit fly Drosophila melanogaster, thus indicating that dme-miR-14 regulates lipid metabolism by downregulation of the production of TAG and diacylglycerol (20). Later, another miRNA, miR-33, which has two isoforms (miR-33a and miR-33b) and is highly conserved across species, was identified from a number of cell types including macrophages, hepatocytes, and endothelial cells (21). The overexpression of miR-33a/b decreases both fatty acid β-oxidation and insulin signaling in radioactively labeled hepatic cell lines, whereas the inhibition of endogenous miR-33a/b increases these two metabolic pathways, thereby suggesting that miR-33 plays important roles in regulating lipid homeostasis in concert with the host genes (22). Moreover, a study by Ouimet et al. has revealed that macrophage-specific miR-33 deletion increases oxidative respiration and decreases atherosclerosis progression by promoting the M2 macrophage polarization-associated gene profile, thus suggesting that silencing miR-33 may be an effective therapeutic approach in the treatment of chronic inflammatory diseases (17).
Multiple miRNAs may also share the same target mRNA in the regulating lipid metabolism. Both miR-370 and miR-122 directly target the transcription factor sterol regulatory element binding protein 1c in Hep2G cells, thereby regulating fatty acid and triglyceride biosynthesis (23). Other miRNAs, such as miR-130 and miR-335, are also important players in the regulation of lipid metabolism (24, 25). Although connections between lipid metabolism and the host inflammatory responses through miRNA have been studied (26), the molecular mechanisms of how the miRNAs modulate lipid metabolism in response to pathogenic diseases remain unclear. Systemic studies on the network of lipid metabolism and host defenses in cells are still needed.
The invertebrate sea cucumber, Apostichopus japonicus (Echinodermata, Holothuroidea), has an innate immune system and is one of the most important marine economic species in the Chinese aquaculture. Unfortunately, the natural sources of A. japonicus in China have declined drastically, owing to outbreaks of various viral and bacterial diseases (27). Among these diseases, skin ulceration syndrome (SUS) causes high infectious and lethal rates and has received substantial attention (27, 28). In our previous work, we have found that miR-31 and miR-2008 expression levels are markedly increased in sea cucumbers with SUS by high-throughput analyses, thus indicating that these molecules play crucial roles in this host–pathogen interaction (29). Our other work has also demonstrated that the samples with SUS exhibit significant metabolic differences compared with the metabolic profiles from healthy A. japonicus (30). Undoubtedly, miRNAs might play essential roles in mediating the innate immunity and metabolic pathways in the sea cucumbers with SUS. The connection between miRNAs and the innate immune response is largely unknown to the best of our knowledge, especially in non-model organisms such as the sea cucumber. The present study aimed at exploring the potential of regulation of the complement c1q tumor necrosis factor-related protein 9 gene (AjCTRP9), a novel adipocyte-derived cytokine, by miR-31. CTRP9 is an adipokine that has pleiotropic functions in immunity and metabolism (31, 32) and was selected as a promising target for functional validation. Our results clearly demonstrated that miR-31, a multifunctional regulatory molecule, negatively modulates the expression of AjCTRP9 and induces cell apoptosis by disturbing the lipid metabolism balance, thus resulting in the overproduction of Cers. The findings of the current study provide new insights into the mechanisms of pathogenesis of infectious diseases in A. japonicus from the perspective of miRNA-mediated cross-talk between the immune response and lipid metabolism.
Materials and Methods
Prediction of miR-31 Targets
The potential targets of miR-31 were predicted by using the miRanda v3.01 toolbox by screening of our previous transcriptome data (33, 34). The parameters were set as follows: single–residue pair scores less than the threshold value of 90 and a minimum free energy lower than −17 kcal/mol. Subsequently, the promising candidate metabolism-associated genes with the highest scores and lowest free energy values was selected as potential targets for further analysis.
Animals and Challenge Experiments
Healthy adult sea cucumbers A. japonicus (weighing 150 ± 15 g) were obtained from the Dalian Pacific Aquaculture Company (Dalian, China) and acclimatized in 30 L aerated natural seawater (salinity 28, temperature 16°C) for 3 days. The pathogen Vibrio splendidus was initially isolated from sea cucumbers with SUS from the indoor farms of Jinzhou Hatchery in May 2013, preserved in glycerol, and stored at −80°C. For the immune challenge, the sea cucumbers were randomly divided into five tanks, each containing 10 individuals. Four experimental groups were immersed with live V. splendidus at a final concentration of 107 CFU mL−1. An identical number of individuals in another tank were kept as controls. The coelomic fluids from five individuals were collected after exposure for 0, 6, 24, 48, and 72 h and then centrifuged at 800 g for 5 min to harvest the coelomocytes for the time course expression analysis.
Cloning and Characterization of Full-Length CTRP9 from A. japonicus
The partial sequence of the CTRP9 gene was generated by screening the A. japonicus transcriptome database (34). BLASTx analysis of the fragment revealed that the sequence contained the complete 5’-end compared with that of other reported sequences. No polyA tail was detected in the AjCTRP9 fragment. Therefore, gene-specific primers for AjCTRP9 (Table S1 in Supplementary Material) were designed on the basis of the acquired unigenes, and the full-length cDNA sequence was subsequently cloned using a 3’-Full RACE Kit (TaKaRa), by following the manufacturer’s instructions. The desired PCR products were purified and then cloned into the pMD19-T vector. Three positive clones for each product were sequenced at Sangon (Shanghai, China). Subsequently, the full-length cDNA sequence of AjCTRP9 was analyzed using the BLAST program at National Center for Biotechnology Information1 and the deduced amino acid sequence was analyzed with the expert protein analysis system.2 The signal peptide protein domain features were predicted with the Simple Modular Architecture Research Tool.3 The molecular mass and theoretical isoelectric point (pI) of the protein were calculated on the basis of the deduced amino acid sequence with the ProtParam tool.4 Multiple alignment analysis of each protein was performed using the ClustalW2 multiple alignment program.5
3’-UTR Luciferase Reporter Assays
The complete 3’-UTR of wild-type AjCTRP9 containing the putative target sites of miR-31 was amplified with gene-specific primers (Table S1 in Supplementary Material) to generate fragments and cloned into the pMD19-T simple vector (TaKaRa). The putative miRNA binding sites were mutated with a PCR approach with mutagenic primers (Table S1 in Supplementary Material) and served as the mutant AjCTRP9. The resulting clones were then digested with MluI and HindIII and inserted into MluI/HindIII-digested pMIR-REPORT luciferase plasmid (Promega, USA). These plasmids were sequenced to ensure correct orientation, and each plasmid was purified with an EZNA™ Plasmid Mini Kit (OMEGA, USA). For the transfection experiment, HEK293-T cells were seeded in 96-well plates with 100 μL medium and grown to approximately 70% confluence. Then, two solutions were added to each well as follows: the first solution contained 0.2 μg of pMIR-REPORT with either the wild-type or mutated AjCTRP9 3’-UTR and 0.01 μg of pRL-CMV constructs (the internal control) with 0.25 μL of the Lipofectamine 2000 transfection reagent (Invitrogen, USA). The second solution contained 100 nM miR-31 mimics (Table S1 in Supplementary Material) and 0.25 μL of the transfection reagent. Twenty-five microliters of each solution, diluted in the medium, were mixed before being added to the wells, and the plates were incubated at room temperature for 20 min. Subsequently, the solutions were replaced with 50 μL of medium in each well. The cells co-transfected with 100 nM miR-31 mimics served as the negative control (NC) (NCM; Table S1 in Supplementary Material), and wild-type or mutated constructs served as the positive control. After 48 h of transfection, the cells were prepared for luciferase activity assays using a dual-luciferase reporter assay kit (Promega, USA). The luciferase signal was expressed as a ratio of firefly to Renilla luciferase activities. All of the experiments were performed in six replicates.
Primary Coelomocytes Culture and LPS Exposure
Sea cucumbers (weighing 150 ± 15 g) were dissected with sterilized scissors on ice, as previously described (35, 36). In brief, the coelomic fluids were filtered through a 300-mesh CellCribble to remove large tissue debris, mixed with the anticoagulant solution (0.02 M EGTA, 0.48 M NaCl, 0.019 M KCl, 0.068 M Tri–HCl, pH 7.6) in a 1:1 (V:V) ratio, and then centrifuged at 800 g, 16°C for 10 min. The harvested cells were washed twice with isotonic buffer (0.001 M EGTA, 0.53 M NaCl, 0.01 M Tris–HCl, pH 7.6) and resuspended in Leiboviz’s L-15 cell culture medium (Invitrogen, USA) containing penicillin (100 U mL−1) and streptomycin sulfate (100 mg mL−1); NaCl (0.39 M) was added to adjust the osmotic pressure. The cell suspension was diluted to 106 cells mL−1 and transferred into 24-well culture microplates and incubated at 16°C for 12 h before exposure to lipopolysaccharides (Sigma, USA). For the LPS challenge, the cells were stimulated with 10 μg mL−1 LPS for 0, 3, 6, 12, and 24 h. The untreated cells served as the control. The cells were collected in TRIzol and used for expression analysis.
Expression Analysis of miR-31 and mRNAs under Immune Challenge
The expression levels of mRNAs and miR-31 were analyzed using a Rotor-Gene 6000 real-time PCR detection system. Total RNA was isolated from the coelomocytes using TRIzol (Takara), and cDNA was synthesized using a PrimeScript miRNA RT-PCR Kit (Takara). The primers used are listed in Table S1 in Supplementary Material. RNU6B and Ajβ-actin served as internal controls to normalize the miRNA or the target, respectively, for quantification. Each amplification was carried out in a 20 μL reaction volume containing 8 μL of the 1:50 diluted cDNA, 1 μL of each of the primers, and 10 μL of SYBR Green Mix (Takara). The reaction mixture was incubated for 5 min at 95°C, and this was followed by 40 amplification cycles of 15 s at 95°C, 20 s at 60°C, and 20 s at 72°C. The 2−ΔΔCT method was used to analyze the relative expression levels of both miRNA and mRNA (37), and the values obtained denoted the fold difference relative to the calibrator. The data are presented as the relative expression levels (mean ± SD, n = 5). Differences were considered significant at p < 0.05.
Functional Analysis of miR-31 In Vivo
The miR-31 mimics and inhibitor with the 2’-OMe modification were synthesized by GenePharma (Shanghai, China) and are shown in Table S1 in Supplementary Material. These reagents were dissolved in RNase-free water to obtain a 20 μM working solution. We mixed 10 μL (2 μM) of miR-31 mimics or inhibitor from each working solution with 10 μL of transfection reagent and 80 μL of phosphate buffered solution (PBS) at pH 7.6 to serve as the transfection solution. Ten sea cucumbers (100 ± 10 g) were infected with 100 μL of the above transfection solution. Twenty-four hours after the injection, the control and treated coelomocytes were collected for further RNA and protein extraction.
Silencing of AjCTRP9 In Vivo
Three specific small-interfering RNAs (siRNAs) targeting AjCTRP9 with the extra 2’-OMe modification were designed and synthesized by GenePharma (Shanghai, China). Another siRNA (NC) that did not target any of the genes from the sea cucumber transcriptome data served as a NC. The detailed sequences are shown in Table S1 in Supplementary Material. Each siRNA and the NC siRNA were dissolved in RNase-free water to obtain a 20 μM working solution. For RNA interference, we mixed 10 μL of each siRNA or the NC siRNA with 10 μL of transfection reagent and 80 μL of PBS to serve as the transfection solution. Ten sea cucumbers (100 ± 10 g) were injected with 100 μL of the transfection solution into the tentacles. Twenty-four hours after the injection, the control and treated coelomocytes were collected for further RNA, protein, and lipid extraction. AjCTRP9 expression was analyzed as described below, and the interference efficiency was taken as the average after three siRNA transfections.
AjCTRP9 Recombinant Protein Expression and Polyclonal Antibody Preparation
The partial cDNA sequence of AjCTRP9 was cloned with specific primers (Table S1 in Supplementary Material) then double digested with EcoRI and NotI and ligated into the vector pGEX-4T-2 (GE, USA). Ajβ-actin was used as a control, as described in our previous work (38). The recombinant plasmid (pGEX-4T-2-AjCTRP9) was transformed into Escherichia coli cells of Transetta (DE3) (TransGen, China), and the DNA was sequenced to ensure correct orientation. The positive transformants were subsequently incubated in LB medium containing 50 μg mL−1 of ampicillin at 37°C and 200 rpm. Then, the expression of the GST-tagged fusion protein was induced in pGEX-4T-2-AjCTRP9 with 1 mM isopropyl-b-d-thiogalactopyranoside (IPTG) at 37°C for 4 h, and the protein was collected after cells were disrupted with ultrasonic waves. The GST-Sefinose™ Resin was used for the purification, according to the manufacturer’s protocol (Sangon, China). The concentration of the purified soluble protein was quantified with a Protein Assay Kit (Sangon, China). The soluble target protein, after dialysis, was injected into a 4-week-old mouse to acquire polyclonal antibody according to a previously described protocol (38).
Western Blot Analysis
For the western blot assay, the samples were washed twice in ice-cold PBS and lysed with lysis buffer (Beyotime Biotechnology, China). After lysis, the cells were centrifuged at 10,000 g for 5 min, the supernatant containing the total protein was collected, and the protein concentration was measured using a BCA Protein Assay Kit (Sangon, China). Each sample (25 μg) was resolved with a 12% SDS-PAGE gel and then transferred to polyvinylidene fluoride membranes at 300 mA for 1 h (Millipore, USA) in transfer buffer (25 mM Tris, 192 mM Glycine, 20% [v/v] E-OH). After being blocked with 5% fat-free dry milk in TBST (25 mM Tris–HCl, pH 7.4, 137 mM NaCl, 2.7 mM KCl, 0.1% [v/v] Tween 20) for 1 h at room temperature, the membranes were incubated overnight with AjCTRP9 or Ajβ-actin polyclonal antibodies diluted to 1:500 in 5% fat-free dry milk in TBST. The membranes were washed three times with TBST and subsequently incubated at room temperature for 1 h with goat anti-mouse (Beyotime Biotechnology, China) diluted at 1:3,000 in 5% fat-free dry milk in TBST. After being washed three times with TBST for 10 min each, the membranes were incubated in Western Lightning-ECL substrate (Perkin Elmer) before exposure onto X-OMAT AR X-Ray films (Eastman Kodak, Rochester, NY, USA).
Liquid Chromatography and Mass Spectrometry (LC-MS) Assay after AjCTRP9 Silencing
After AjCTRP9 silencing, 50 mg of coelomocytes (freeze-dried) was extracted in chloroform/methanol/water (1:2:0.8, v/v/v) containing 0.05% butylated hydroxytoluene (BHT), according to a modified version of the Bligh and Dyer method (39). Briefly, the obtained extract was evaporated on a rotary evaporator, and the lipid residue was redissolved in 1 mL of methanol after centrifugation at 12,000 rpm for 5 min at 4°C. The methanol solution of the lipids was filtered with a 0.22 μm ultrafiltration membrane (Millipore, USA) to remove the undissolved residues and subjected to LC-MS analysis using the previously published standard method (40–42).
Chromatographic separations were performed on a Waters ACQUITY UPLC system using an ACQUITY UPLC BEH C8 analytical column (i.d. × l 2.1 × 100 mm, particle size 1.7 μm). Briefly, lithium acetate (0.01%) and formic acid (15 mM) were added to the mobile phase as the electrolyte for the ESI positive mode analysis, and ammonium hydroxide (15 mM) was added for the negative ion mode analysis. To obtain efficient separation of the total lipids, water/acetonitrile (1:2, v/v) was used as mobile phase A, and acetonitrile/isopropyl alcohol/tetrahydrofuran (1:1:1, v/v/v) was used as mobile phase B. The mobile phase B was changed from 0 to 50% in 15 min, then to 80% in 30 min and held for 2 min and finally to the initial 0 in 2 min and equilibrated for 11 min. The temperature of the sample chamber was set at 4°C, and the injection volume was 5 μL for each analysis. Twenty-five percent of the flow out of the column was subjected to mass spectrometry.
Mass spectrometry was performed on a Waters Q-TOF Premier mass spectrometer operating in both positive and negative ESI modes. The nebulization gas was set to 400 L h−1 at a temperature of 300°C, the cone gas was set to 0 L h−1, and the source temperature was set to 120°C. For the ESI+ mode, the capillary voltage was set at 3.0 kV and the sampling cone voltage was set at 80 V. For the ESI− mode, the capillary voltage was set at 2.5 kV, and the sampling cone voltage was set at 60 V. MS2 analysis was performed at a collision energy range of 20–40 V. A time-of-flight (TOF) analyzer was used in a V mode and tuned for maximum resolution (>10,000 resolving power at m/z 1,000). The instrument was previously calibrated with sodium formate, and the lock mass spray for precise mass determination was set with leucine-enkephalin, thus generating an [M + H]+ ion at 556.2771 Da and an [M − H]− ion at 554.2615 Da in the ESI+ mode and ESI− mode, respectively. The lock spray frequency was set at 10 s.
The raw data files obtained from ultra-performance liquid chromatography-quadrupole time-of-flight mass spectrometry runs were analyzed using a MassLynx 4.1 data processing system (Waters, Milford, MA, USA). The resulting multivariate dataset consisting of the peak number (based on the retention time and m/z), sample name, and the normalized peak intensity was exported and analyzed with principal component analysis (PCA), the projection to latent structures was analyzed with discriminant analysis (PLS-DA), and the orthogonal projection to latent structures was analyzed with discriminant analysis (OPLS-DA) using SIMCA-P-12.0 software (Umetrics, Sweden). The loading scatter S-plots and contribution lists were used to describe the candidate markers that were significantly different between the two groups. One-way analysis of variance (ANOVA) was conducted on the A. japonicus between the NC and siAjCTRP9 groups to determine the statistical significance (p < 0.05) of the candidate markers.
Cell-Permeable C6 Cer Treatment
C6 Cer (Biomol, USA), short for C6 Cer, was dissolved in anhydrous DMSO to obtain 5 mg mL−1 C6 Cer stock solution. For Cer treatment, 10 sea cucumbers (100 ± 10 g) were injected with 100 μL of Cer at the concentration of 100 μg mL−1. The sea cucumbers in the control group were given an injection of 100 μL diluted DMSO. Twenty-four hours after the injection, the treated and control coelomocytes were collected for further expression analysis.
Determination of Apoptosis by Flow Cytometry Assay (FACS)
In vivo cell apoptosis rates were determined by flow cytometry. The sea cucumbers were injected with miR-31 mimics, miR-31-inhibitor, AjCTRP9 siRNA, and C6 Cer, as described above. The apoptosis rates of the coelomocytes were measured with an Annexin V-FITC Apoptosis Detection Kit (Beyotime Biotechnology, China). For Annexin V/PI staining, the collected coelomocytes were resuspended in 200 μL Annexin V-FITC binding buffer and 5 μL Annexin V-FITC and 10 μL PI were successively added to 105 cells. Finally, the apoptosis rates were detected with a FACScan (BD, USA) after 10 min incubation in the dark at room temperature.
Measurement of Caspase Activity
Caspase-3/8 activity assays were performed using a Caspase Activity Assay Kit (Beyotime Biotechnology, China). Briefly, the samples were lysed with lysis buffer for 15 min on ice. Then, the cells were centrifuged at 16,000 g for 20 min at 4°C, and the supernatant was immediately used for caspase-3/8 enzyme assays. Caspase activity was measured through the cleavage of a colorless substrate Ac-DEVD-pNA for caspase-3 and Ac-IETD-pNA for caspase-8, thus releasing pNA (p-nitroaniline). Absorbance was measured at OD405 nm using a microplate reader (Thermo Scientific). The protein concentration of the cells was measured using a Bradford Protein Assay Kit (Beyotime Biotechnology, China). Caspases-3/8 activities were calculated using a standard curve. One unit was defined as the amount of enzyme cleaving 1.0 nmol of the colorimetric substrate pNA per hour at 37°C under saturated substrate concentrations. The assays described above were repeated five times.
Statistical Analysis
All experiments were performed in biological replicates. Data are presented as the mean ± SD. The statistical significance was determined by one-way ANOVA followed by Duncan’s multiple range tests to determine the differences in the mean values as compared with the controls. The criterion for significance of the difference between means was p < 0.05.
Results
Target Identification of miR-31
On the basis of the transcriptome data of sea cucumber A. japonicus (33, 34), the target of miR-31 was bioinformatically predicted by the miRanda v3.01 toolbox. Information on the binding sites of miR-31 in the 3’-UTR of AjCTRP9 and the mutated sites is shown in Figure 1A. Imperfect pairing with a GU mismatch was allowed in many instances (43–45). To verify the prediction, we performed dual-luciferase reporter assays in HEK-293T cells transfected with miR-31 or control mimics together with the luciferase reporter plasmids containing either the AjCTRP9 3’-UTR WT or the AjCTRP9 3’-UTR MT (Figure 1B). Overexpression of miR-31, compared with the control mimics, significantly decreased the level of luciferase activity containing the entire 3’-UTR by 33.38% (p < 0.0001), and the results were not significantly different for transfection with the mutant vector, thus indicating that the AjCTRP9 gene may be a target of miR-31.
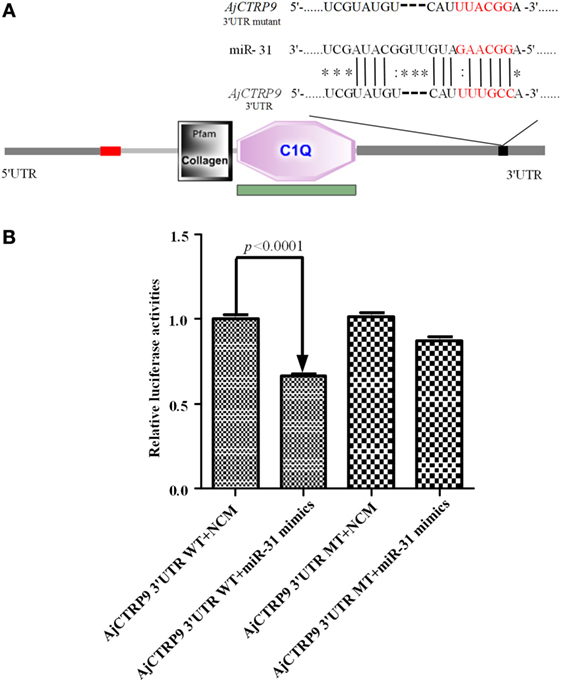
Figure 1. Identification and characterization of the miR-31 binding sites in the 3’-untranslated region (3’-UTR) of AjCTRP9. (A) Schematic representation of putative miR-31 binding sites in the AjCTRP9 3’-UTR and the mutant sites (the protein domain of AjCTRP9 was annotated with SMART at http://smart.embl.de/), the red letters represent “seed” regions. (B) AjCTRP9 luciferase reporter assays conducted in HEK-293T cells carrying the wild-type AjCTRP9 3’-UTR and a mutant AjCTRP9 3’-UTR (final concentration: 100 nM).
Cloning and Sequence Analysis of AjCTRP9
The full-length cDNA of AjCTRP9 was generated by overlapping original fragments from ESTs and using a 3’-RACE approach in the sea cucumber A. japonicus and was deposited in GenBank under accession number KX50326. The cDNA sequence of AjCTRP9 was 1,718 bp in length and contained a 909 bp ORF encoding a predicted product with 302 amino acid residues with a molecular weight of 32.37 kDa and a theoretical pI of 5.37 (Figure S1 in Supplementary Material). Smart analysis showed that the deduced amino acid sequence of AjCTRP9 contains a short N-terminal signal peptides (residues 1–23), a short variable region (residues 25–43), a collagen domain with 34 “Gly-X-Y” repeats (residues 45–155), and an a-terminal globular domain that is homologous to c1q domain immune complement component 1q (C1q) (residues 161–299) (Figure 1A; Figure S1 in Supplementary Material). There were a total of nine proline residues at the third position (Gly-X-Pro) within the collagen domain, which is often hydroxylated to enhance the stability of the collagen triple-helical structure (46). In addition, its collagen domain also had three conserved lysine residues that conformed to the consensus sequence (Gly-X-Lys), which can potentially be hydroxylated and glycosylated. Furthermore, multiple alignments indicated that AjCTRP9 shares 47.3% homology with Saccoglossus kowalevskii CTRP9 (XP_002739445.1), 44.0% homology with Homo sapiens CTRP9 (AAH40438.1), and 40.9% homology with Danio rerio CTRP9 (XP_005162770.1). Because a short N-terminal variable region was present in the CTRP9 from invertebrates and had the lowest similarity to CTRP9 from vertebrates, we speculated that the evolution of invertebrate CTRP9 may have been earlier than that of the other CTRP9 members (Figure S2 in Supplementary Material).
Expression Analysis of AjCTRP9 and miR-31 in Immunity
The temporal expression levels of miR-31 and AjCTRP9 in coelomocytes post V. splendidus challenge and LPS treatment are shown in Figure 2. After the challenge with V. splendidus, the level of miR-31 significantly increased by 1.41-fold (p < 0.05) during the first 6 h, as compared with the control group and reached peak expression at 24 h with 1.85-fold (p < 0.01) increase (Figure 2A). Additionally, after exposure to 10 μg mL−1 LPS, the level of miR-31 sharply increased by 1.65-fold, as compared with the control (p < 0.01) during the first 6 h, and the elevated expression was maintained until 24 h (Figure 2B). However, the AjCTRP9 transcript showed an opposite expression trend from that of miR-31 in response to both V. splendidus challenge and LPS treatment (Figure 2). The transcription of AjCTRP9 was gradually downregulated and exhibited the least expression at 72 h, with a 0.49-fold (p < 0.01) decrease after the V. splendidus challenge (Figure 2A). The minimal expression level of AjCTRP9 after the LPS stimulation was detected at 24 h, with a 0.29-fold (p < 0.01) decrease compared with the control group (Figure 2B).
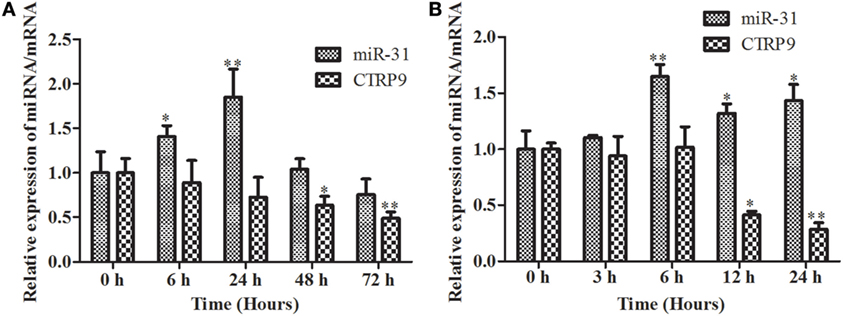
Figure 2. The time course of expression patterns of miR-31 and AjCTRP9. (A) AjCTRP9 mRNA transcripts and miR-31 expression in Vibrio splendidus-challenged Apostichopus japonicus. (B) AjCTRP9 mRNA transcripts and miR-31 expression in LPS-treated coelomocytes. Values are given as the mean ± SD, n = 5. Asterisks indicate significant differences: *p < 0.05, **p < 0.01.
Loss- and Gain-of-Function Assay of miR-31
To further elucidate the functional roles of miR-31 in regulating AjCTRP9, loss-of-function or gain-of-function assays of miR-31 were performed in vivo. As shown in Figure 3, quantitative PCR indicated that the overexpression of miR-31 (Figure 3A) significantly decreased the mRNA expression of AjCTRP9 by 0.46-fold (p < 0.05) (Figure 3B). Accordingly, the inhibition of miR-31 (Figure 3C) resulted in the elevated expression of AjCTRP9 by 1.65-fold (p < 0.05) (Figure 3D). Furthermore, western blot analysis of the aberrant expression of miR-31 revealed that the protein abundance of AjCTRP9 was decreased after miR-31 overexpression and increased after miR-31 inhibition (Figure 3E), in agreement with the AjCTRP9 mRNA expression levels. Our results indicated that miR-31 directly targeted the AjCTRP9 gene.
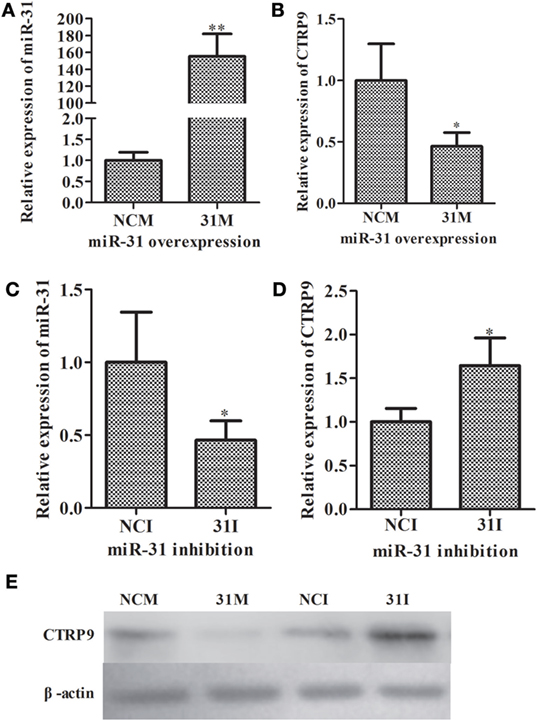
Figure 3. Functional analysis of miR-31 and AjCTRP9 in vivo. (A) Relative miR-31 expression level after transfection with modified microRNA (miRNA) mimics. (B) Relative expression level of AjCTRP9 after transfection with modified miRNA mimics. (C) Relative miR-31 expression level after transfection with modified miRNA inhibitor. (D) Relative expression level of AjCTRP9 after transfection with modified miRNA mimics. (E) Western blot analysis of AjCTRPP protein after transfection with modified miRNA mimics or inhibitor. Values are given as the mean ± SD, n = 5. Asterisks indicate significant differences: *p < 0.05, **p < 0.01.
Lipidomic Analyses of A. japonicus Coelomocytes after siRNA-Mediated AjCTRP9 Silencing
From the above results, we observed that miR-31 directly targets the AjCTRP9 gene. As a CTRP family member, CTRP9 plays important roles in multiple physiological processes, especially in fatty acid oxidation (47). To further investigate the possible biological roles of the AjCTRP9, the lipidomics of coelomocytes were analyzed by LC-MS after AjCTRP9 silencing. Figure 4 shows the expression levels of AjCTRP9 after specific siRNA transfection. Our results showed that the injection of AjCTRP9 siRNA specifically inhibited AjCTRP9 transcript expression (Figure 4A) and protein expression (Figure 4B) in vivo. Therefore, the changes in the lipid metabolites of coelomocytes were identified in the siAjCTRP9 and NC groups.
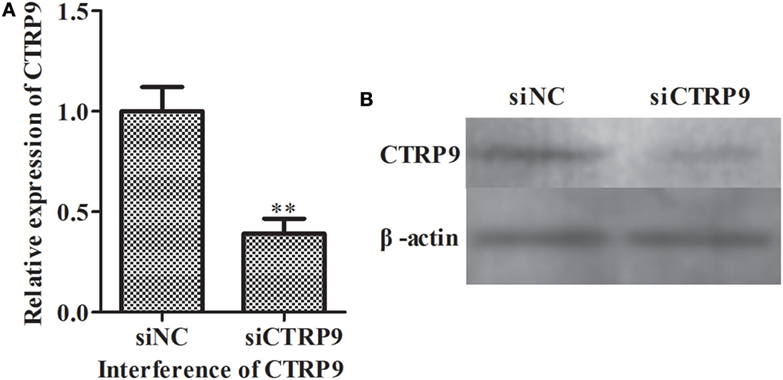
Figure 4. The expression level of AjCTRP9 after transfection with specific small-interfering RNAs (siRNAs). (A) Relative expression level of AjCTRP9 after AjCTRP9 siRNA transfection. (B) Western blot analysis of AjCTRPP protein after AjCTRP9 siRNA transfection. Values are given as the mean ± SD, n = 5. Asterisks indicate significant differences: *p < 0.05, **p < 0.01.
The samples were characterized by LC-MS in both positive and negative ion mode. The mass spectral data were processed by multivariate analysis. The PCA score plots (Figure 5) in both ion modes showed that the two samples of A. japonicus were clearly separated by the first components. A trend of time-dependent cluster formation by the first two components was observed in both ESI+ and ESI− modes, thus explaining 58.1% (Figure 5A) and 62.8% (Figure 5B) of the all variances, respectively. The OPLS-DA score plot showed significant lipidomic differences between the siAjCTRP9 and NC groups (Figure 6). As shown in the positive ion mode (Figure 6A), the parameters R2X, R2Y, and Q2 revealed the high discriminative and predictive ability of the model. Further model validations in PLS-DA, with the number of permutations equal to 200, generated intercepts of R2 = 0.372 and Q2 = −0.178 (Figure 6B). Along with those obtained in negative mode (Figures 6C,D), these results demonstrated that the OPLS-DA models were well fitting and statistically valid. The S-plot visualizes the covariance and correlation among the metabolites, and thus can be used to identify important metabolites (48). The significant metabolites at the top and bottom of the S-plots (Figure 7) were related to the group separation and were selected as potential biomarkers (Table 1) according to parameters of variable importance in the projection (VIP). The relative abundance of significant ions that were higher in the siAjCTRP9 group compared with the NC group in sea cucumber were located in the upper-right quadrant (with >0 of correlation) of the S-plot after the OPLS-DA approach (Figure 7), including seven phosphatidylcholines (PC) (18:1/18:2; 18:2/17:1; 18:1/18:1; 15:0/18:2; 20:1/18:2; 15:0/20:2; 18:2/18:2), three Cers (d15:1/20:0; d15:1/21:0; d17:1/23:1), and TAG (16:0/16:0/16:0), whereas those that were lower in abundance were in the lower-left quadrant (with <0 of correlation), including four PC (17:1/20:5; 17:1/20:4; 15:0/20:5; 22:6/13:0), two phosphatidylethanolamine (PE) (17:1/20:4; 18:1/20:5), two lysophosphatidylcholine (LPE) (20:5; 20:4), three TAG (18:1/18:1/16:0; 18:1/18:1/16:1; 18:1/16:0/20:2), and (LPC) (18:2).
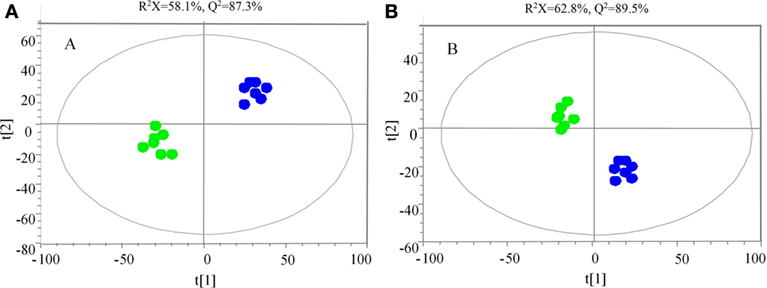
Figure 5. Principal component analysis score scatter plot in ESI+ (A) and ESI− (B) mode for the total lipids of Apostichopus japonicus between the siAjCTRP9 group (green) and the negative control group (blue).
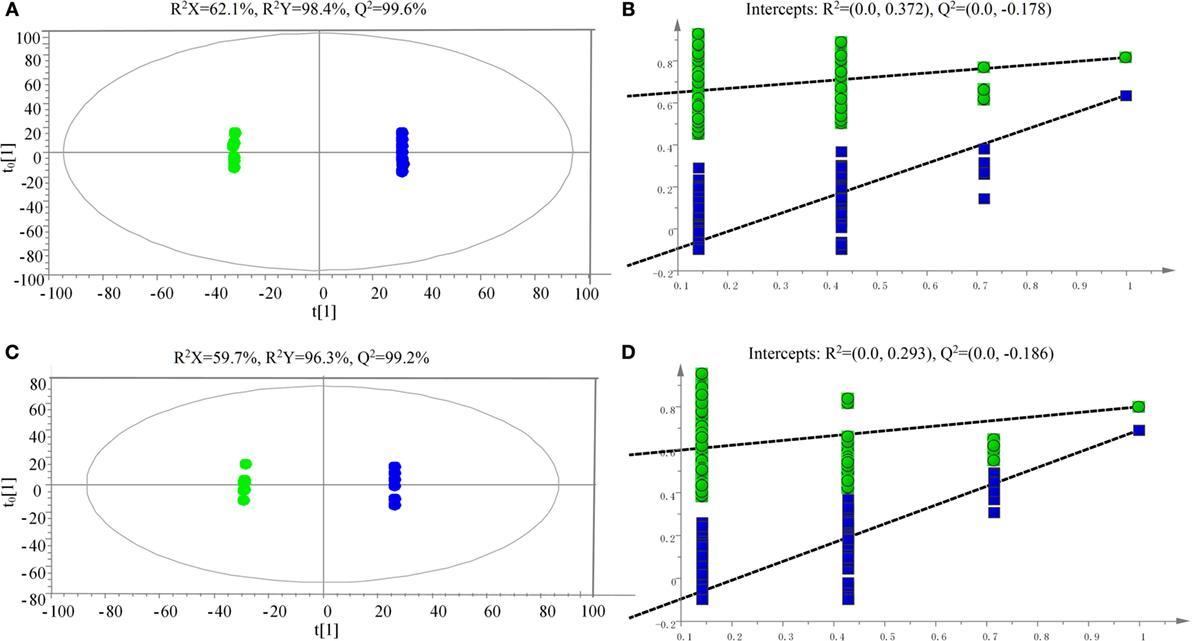
Figure 6. Scatter plot of OPLS-DA scores (A,C) and validation plot (B,D) of the PLS-DA analysis on siAjCTRP9 group in both positive (A,B) and negative (C,D) ion scan modes compared with the negative control group.
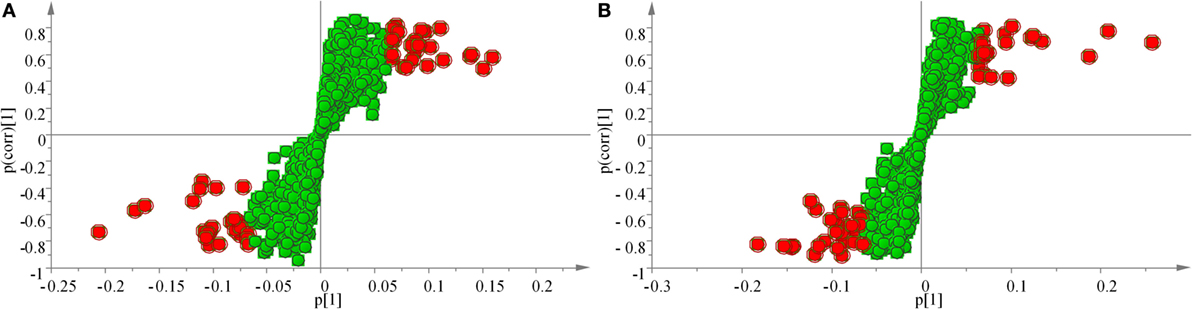
Figure 7. OPLS loading S-plot for the total lipids of the siAjCTRP9 group and the negative control group detected in the ESI+ (A) and ESI− (B) modes. Significant ions are outlined in red and listed in Table 1.
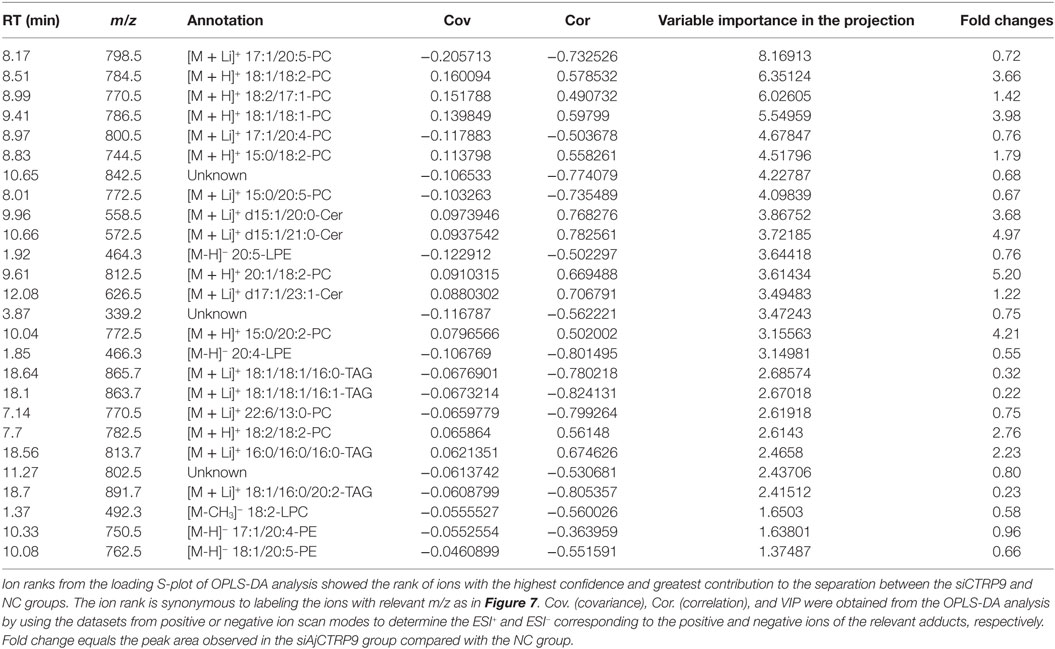
Table 1. Summary of the metabolite ions of Apostichopus japonicus that showed significant changes between the siAjCTRP9 and negative control groups.
AjCTRP9 Knockdown Induces Accumulation of Cer Mass Mainly through Sphingosine Metabolism
According to significantly different metabolites (Table 1), we found that the three identified Cer species (d15:1/20:0; d15:1/21:0; d17:1/23:1) were dramatically increased by 3.68-, 4.97-, and 1.22-fold after AjCTRP9 interference, respectively. Cer is a condensation product of the amino alcohol sphingosine and a fatty acid during an acylation reaction. It is a key intermediate in sphingolipid metabolism and an important signaling molecule in determining cell fate (49). To understand which biological process resulted in the Cer accumulation after AjCTRP9 deletion, we investigated the fluctuations in expression of Cer metabolism-related genes by qPCR (Figure 8A). As shown in Figure 8A, the expression levels of Cer synthase 1 and Cer synthase 6 were significantly increased by 1.44-fold (p < 0.05) and 1.66-fold (p < 0.01) after AjCTRP9 silencing, respectively, whereas the Cer synthase 5 transcript was downregulated by 0.69-fold (p < 0.05). In addition, the mRNA levels of neutral ceramidase 1 and putative neutral sphingomyelinase both sharply decreased by 0.35-fold (p < 0.01) and 0.57-fold (p < 0.05), respectively, after interference. There were no significant changes in serine palmitoyltransferase (SPT) 1 and SPT 2 transcripts after AjCTRP9 knockdown. These results revealed that the increase in the three Cer contents might have represented the Cer synthases after silencing of AjCTRP9, in which different Cer synthases N-acylated sphingosine to form the different Cers.
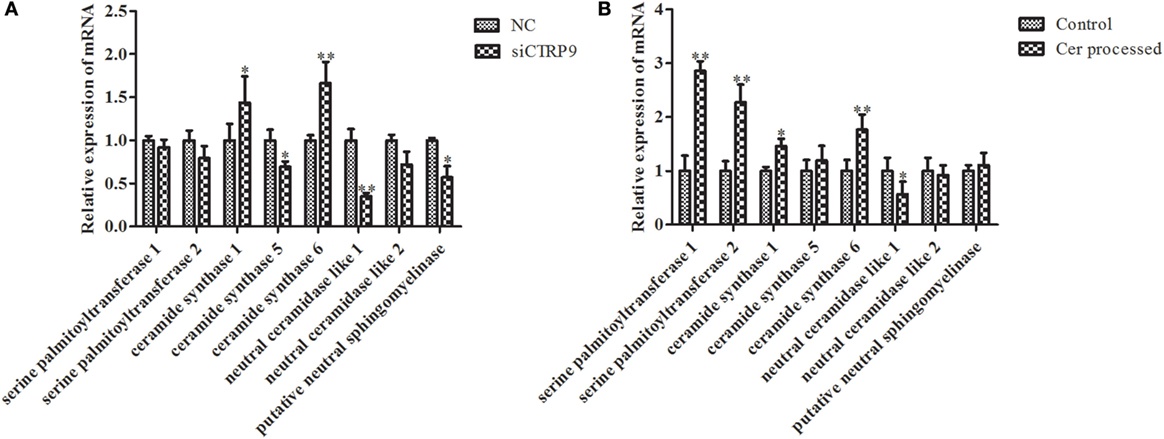
Figure 8. The mRNA expression levels of relative genes after AjCTRP9 silencing (A) and Cer treatment (B). Values are given as the mean ± SD, n = 5. Asterisks indicate significant differences: *p < 0.05, **p < 0.01.
C6 Cer Exposure Increases SPT mRNA Expression and Directly Promotes Cell Apoptosis
To explore the functional effect of Cer on cell survival, we measured cell apoptosis after Cer treatment of A. japonicus in vivo (Figure 9). In response to the stimulation with 100 μg mL−1 Cer, the apoptosis rate was sharply increased by 1.80-fold (p < 0.01) compared with the control (Figures 9A1–A3), thus showing that the disturbance in Cer signaling directly induced cell death in A. japonicus. In addition, we found that the mRNA expression profiles of SPT 1, SPT 2, Cer synthase 1, and Cer synthase 6 were upregulated by 2.86-fold (p < 0.01), 2.28-fold (p < 0.01), 1.46-fold (p < 0.05), and 1.77-fold (p < 0.05), respectively, after the exposure to exogenous Cer (Figure 8B). In contrast, the neutral ceramidase 1 transcript was decreased by 0.56-fold (p < 0.05) compared with the control, thus indicating that the overexpression of short Cer might lead to the enrichment of longer Cers in A. japonicus cells through a de novo synthesis pathway by SPT and that the exposure of exogenous Cer disturbs the sphingolipid metabolism in sea cucumber.
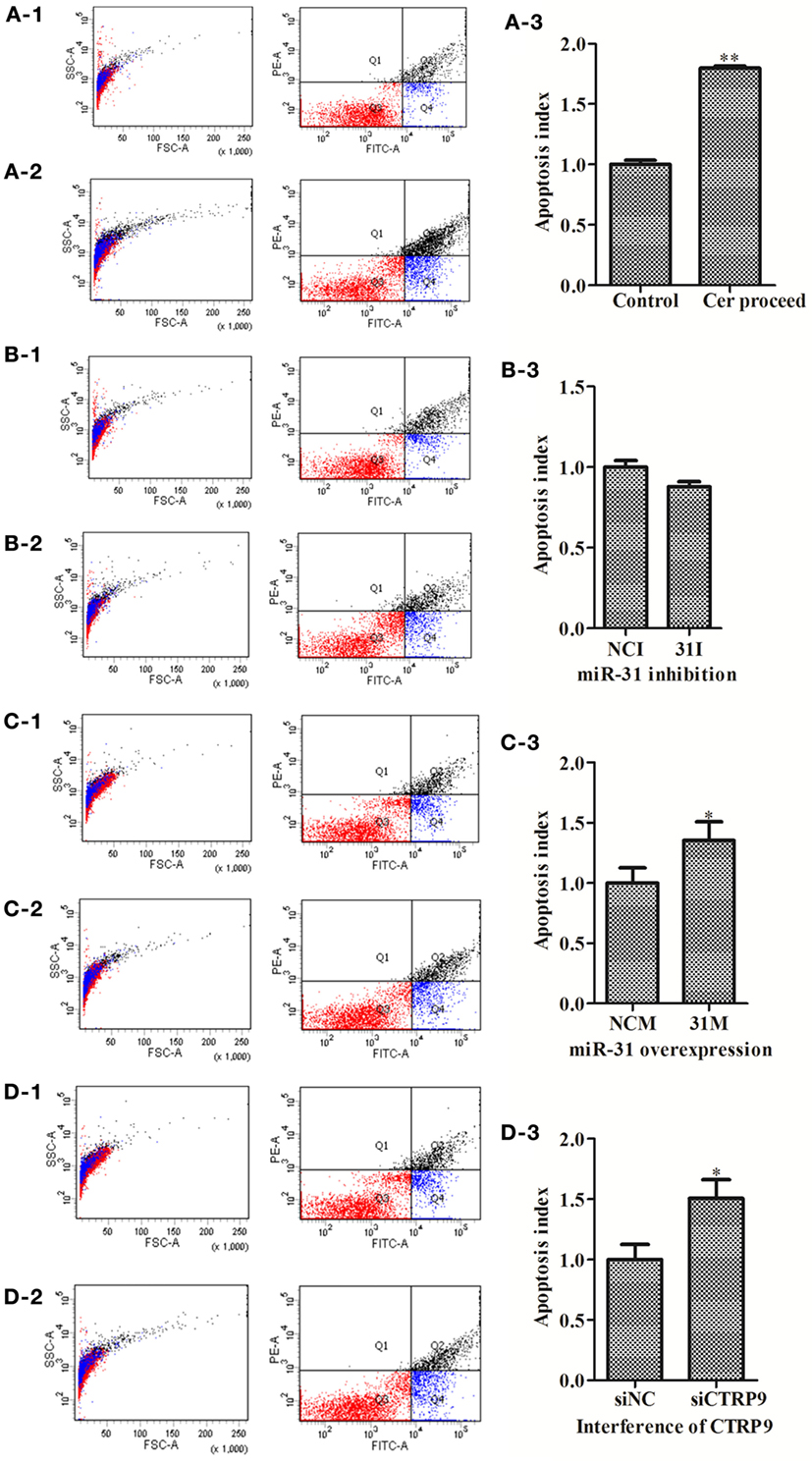
Figure 9. The apoptosis rate of Apostichopus japonicus coelomocytes. (A-1–A-3) Cer treatment; (B-1–B-3) miR-31 inhibition; (C-1–C-3) miR-31 overexpression; (D-1–D-3) AjCTRP9 silencing. Values are given as the mean ± SD, n = 5. Asterisks indicate significant differences: *p < 0.05, **p < 0.01.
miR-31 Regulates Apoptosis in A. japonicus Coelomocytes by Targeting AjCTRP9
To test the hypothesis that miR-31 regulates apoptosis in A. japonicus by targeting AjCTRP9, we further investigated the apoptosis of cells transfected with a series of reagents in vivo (Figure 9). Transfection with miR-31 mimics significantly induced cell apoptosis (1.35-fold, p < 0.05) 24 h post infection (Figures 9C1–C3). Downregulation of AjCTRP9 with specific siRNA also induced apoptosis (1.51-fold, p < 0.05) under the same conditions (Figures 9D1–D3).
miR-31 Targeting AjCTRP9 Modulates Cer Content and Induces Apoptosis through Caspase Cascades
From the results above, we concluded that miR-31 regulates apoptosis by targeting AjCTRP9 in A. japonicus through the overproduction of Cer. To reveal the molecular cascade of apoptosis regulated by signaling of Cer channels, we first examined the expression of apoptotic genes after Cer injection (50) (Figure 10A). We noticed that caspase-3 and caspase-8 transcripts were both markedly upregulated by 1.35-fold (p < 0.05) and 2.42-fold (p < 0.01), respectively, compared with the control, thus indicating that these two molecules play key roles in Cer-induced apoptosis. To accurately evaluate the caspase cascades, the enzymatic activities of caspases were measured from the coelomocytes after transfection with a series of reagents in vivo (Figures 10B,C). As shown in Figure 10B, caspase-8 activity was significantly induced after miR-31 overexpression, AjCTRP9 silencing, and Cer exposure. The caspase-3 activity was also consistent with caspase-8 activity in every condition and had a greater magnitude after AjCTRP9 knockdown, thus suggesting that caspase-8 directly modulates caspase-3 and promotes apoptosis. The regulatory interplay among miR-31, AjCTRP9, Cer, and apoptosis is diagrammed in Figure 11.
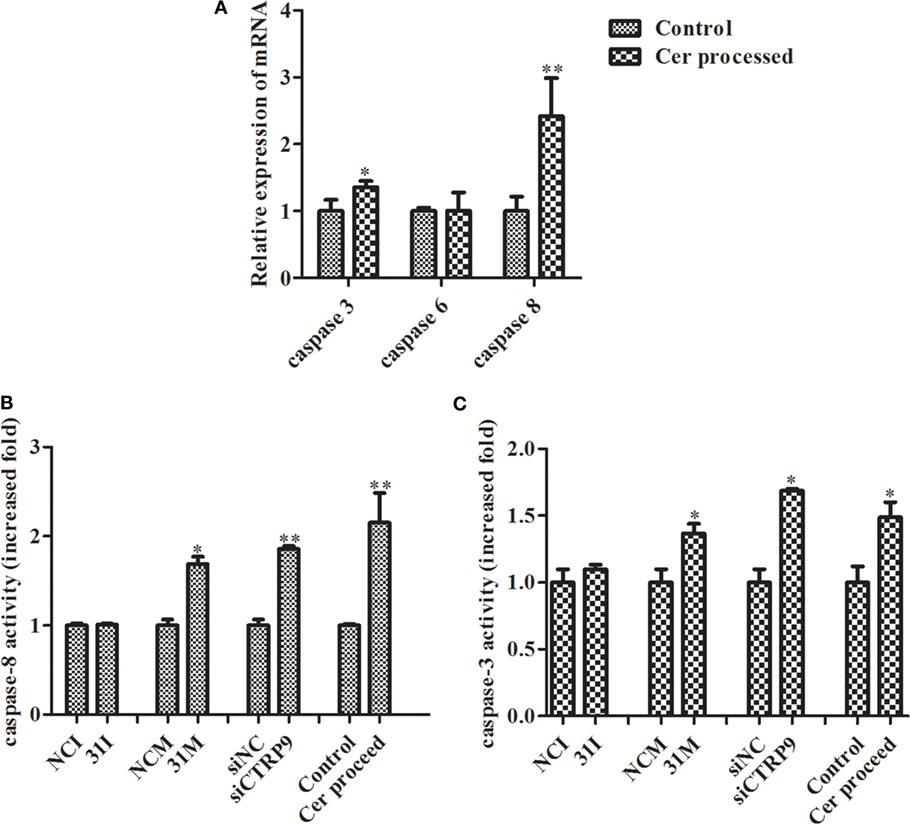
Figure 10. Effects of Cer treatment on caspase activity in the coelomocytes of Apostichopus japonicus. (A) The mRNA expression level of caspase-3/-6/-8; (B) caspase-8 activity; (C) caspase-3 activity. Values are given as the mean ± SD, n = 5. Asterisks indicate significant differences: *p < 0.05, **p < 0.01.
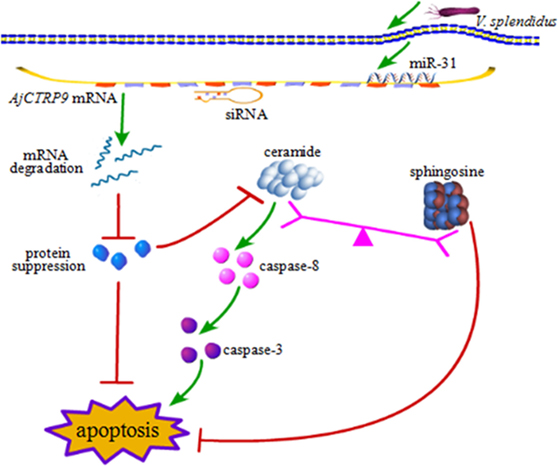
Figure 11. Schematic representation of the involvement of miR-31 in disturbing the lipid metabolism balance by targeting AjCTRP9.
Discussion
microRNAs are powerful regulators of gene expression under normal physiological processes or in pathological states (51). In our previous work, we have revealed that miR-31 modulates respiratory burst by targeting NF-κB (p105) during pathological development in sea cucumbers (52). Currently, accumulating evidence suggests that miRNAs are key players in the regulation of inflammation and metabolism (16, 53). A given miRNA may have multiple mRNA targets that are involved in nearly every biological process (54). miR-31 belongs to a highly evolutionarily conserved miRNA “seed family” that participates in the immune response, and whose abnormal expression or function in the immune system has been linked to multiple diseases (55–57). However, the limitations of these studies restrict the extrapolation to effects on immune defense and cell homeostasis after host–pathogen interactions. In the current study, we discovered a novel role of miR-31 in promoting cell apoptosis in A. japonicus by targeting AjCTRP9, which encodes an important adipokine involved in multiple physiological processes. The most salient finding of this paper is the identification of the role of miR-31 in regulating lipid-mediated signal transduction events during the interactions between bacterial pathogens and host cells.
Metabolism and immunity are often liked by proteins of dual function. CTRP9, a member of the CTRP family, has attracted much interest because of its anti-inflammatory and metabolic effects, especially in glucose metabolism and fatty acid oxidation (46, 58). In the present study, we found that the mRNA level of AjCTRP9 was significantly decreased and showed opposite expression trends from those of miR-31 after V. splendidus challenge and LPS exposure in primary coelomocytes (Figure 2). Moreover, our results showed that the overexpression of miR-31 or silencing of AjCTRP9 in vivo resulted in an increase in the apoptotic rates (Figure 9), thus suggesting that miR-31 negatively targets AjCTRP9 and induces cell apoptosis during the immune response. However, the apoptosis rate was not significantly changed after transfection with the miR-31 inhibitor (Figures 9B1–B3). In humans, CTRP9 levels are negatively associated with the amount of visceral fat and are positively associated with a favorable glucose or metabolic phenotype (31, 59). When overexpressed, CTRP9 significantly ameliorates palmitate- or tunicamycin-induced hepatic steatosis and apoptosis (60). Additionally, CTRP9 protects against acute cardiac damage in response to pathological stimuli by suppressing inflammatory reactions through adiponectin receptors (61). However, targeted deletion of CTRP9 increases food intake, decreases insulin sensitivity, and promotes hepatic steatosis in mice (58). These studies have clearly demonstrated that CTRP9 plays pivotal roles in modulating metabolism during inflammation. More importantly, the complementary gain-of-function and loss-of-function results support a role for CTRP9 in regulating lipid metabolism. In addition, our previous data obtained from high-throughput sequencing have revealed that the AjCTRP9 transcript is sharply downregulated after miR-31 overexpression, and significant metabolic differences are also present in sea cucumbers with SUS (29, 30). On the basis of these results, we speculate that the abnormal expression of miR-31 targeting the AjCTRP9-induced apoptosis might perturb the lipid balance in the cells and may have contributed to SUS outbreaks in A. japonicus.
To explore the miR-31-mediated apoptosis resulting from perturbed cell lipid metabolism, we further investigated the lipidomics of coelomocytes analyzed by LC-MS after AjCTRP9 silencing. The results indicated remarkable differences in lipid profiles of the two groups (Figures 5–7), and some lipid molecules including PC, LPC, TAG, PE, LPE, and Cer were significantly increased or decreased after AjCTRP9 knockdown (Table 1). These lipids have previously been shown to have multiple critical roles in cellular functions, such as cell growth, signal transduction, membrane homeostasis, and apoptosis (5, 62, 63). From these results, we concluded that the miR-31-targeted modulation of AjCTRP9 expression may be an extremely important mechanism in lipid metabolism. Notably, three types of identified Cers (d15:1/20:0; d15:1/21:0; d17:1/23:1) were synchronously increased after AjCTRP9 interference, thus indicating that the Cer species might be promising biomarkers for lipid metabolism imbalance. Cers are important intermediators of sphingolipid metabolism and function not only as structural components of cell membranes but also as key mediators of apoptosis triggered by various stimuli in most cells (64, 65). Accumulating evidence indicates that excessive levels of Cer directly promote cell apoptosis (66, 67). In this study, we also observed that the apoptosis rate was significantly increased after Cer exposure (Figure 9). Hence, we confirmed that the downregulation of AjCTRP9 by miR-31 mainly results in the production of Cers and further induces apoptosis in coelomocytes. Cers are mainly produced from the de novo synthesis pathway regulated by the enzyme SPT (68, 69). Alternatively, they can also be generated by the sphingosine salvage pathway through the action of Cer synthases or the reverse activity of neutral ceramidase (70). Ogretmen et al. have determined that exogenous short-chain C6 Cer induces the production of endogenous long-chain Cer in the A549 human adenocarcinoma cell line through the recycling of the sphingosine backbone (71). Grether-Beck et al. have reported that stimulation of keratinocytes with exogenous C6 Cer induces de novo Cer synthesis in human keratinocytes through the induction and activation of the SPT mRNA (72). In our case, there were no significant changes in the SPT transcript after silencing AjCTRP9, but its mRNA expression was markedly increased after Cer injection (Figure 8), thus indicating that treatment with short Cers might result in the synthesis of longer Cers. In addition, sphingomyelin or sphingosine hydrolysis in cell membranes also contributes to the production of Cers by different metabolic enzymes (69). We found that the mRNA levels of Cer synthase 1 and Cer synthase 6 were both notably upregulated, whereas neutral ceramidase 1 and putative neutral sphingomyelinase transcripts were both significantly downregulated after AjCTRP9 knockdown. Clearly, the overproduction of Cers after AjCTRP9 interference occurs mainly as a result of sphingosine hydrolysis. Moreover, Cer synthase 1, Cer synthase 6, and neutral ceramidase 1 transcript levels after Cer challenge displayed almost identical expression profiles with the levels after silencing AjCTRP9. A study by Osawa et al. has indicated that the dynamic balance between the intracellular levels of Cers and sphingosine may determine cell survival (73). Hence, the aberrant expressions of metabolic enzymes regulate Cer channel formation and induce cell apoptosis. Our present findings indicated that miR-31 negatively modulates the expression of AjCTRP9 and the disturbance in Cer channels, thereby initiating apoptosis. Apoptosis is a very complex multistep process dependent on several apoptosis-related molecules, but its signaling is triggered by either the extrinsic pathway or the intrinsic pathway (74). The primary regulators of apoptosis are a family of cysteine-aspartic specific proteases known as caspases (75). Our results showed that the levels of caspase-3 and caspase-8 mRNAs were significantly induced after Cer treatment (Figure 10A). Furthermore, the enzymatic activities of the initiator caspase-8 and the executioner caspase-3 were both clearly upregulated after miR-31 overexpression and AjCTRP9 silencing, as well as exogenous Cer exposure (Figures 10B,C), thus suggesting that the sequential activation of caspases plays critical roles in the final steps of apoptosis.
In summary, the present study provides novel evidence that miR-31 plays a key role in Cer channel formation by targeting AjCTRP9, thereby linking immune defense with lipid metabolism (Figure 11). miR-31, a highly conserved and multifunctional regulator that negatively targets AjCTRP9 and disturbs the lipid metabolism balance, further results in Cer channel formation, thus ultimately leading to cell apoptosis during host–pathogen interactions. These new findings may provide new therapeutic perspectives with which to suppress miR-31 expression in sea cucumbers with SUS.
Data Sharing
Requests for access to the data, statistical code, questionnaires, and technical processes may be made by contacting the corresponding author at lichenghua@nbu.edu.cn.
Ethics Statement
The sea cucumbers (A. japonicus) used here were commercially cultured, and all the experiments were conducted in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The study protocol was approved by the Experimental Animal Ethics Committee of Ningbo University, China.
Author Contributions
CL and YS conceived and designed the experiments; YS, PZ, and XZ performed the experiments; YS, CL, and WZ analyzed the data; CL and WZ contributed reagents/materials/analysis tools; YS, CL, and WX wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This work was financially supported by the National Natural Science Foundation of China (31522059, 41576139), the Zhejiang Provincial Natural Science Foundation of China (LR14C190001), Collaborative Innovation Center for Zhejiang Marine High-Efficiency and Healthy Aquaculture, and the K. C. Wong Magna Fund in Ningbo University.
Supplementary Material
The Supplementary Material for this article can be found online at http://journal.frontiersin.org/article/10.3389/fimmu.2017.00263/full#supplementary-material.
Footnotes
References
1. Osborn O, Olefsky JM. The cellular and signaling networks linking the immune system and metabolism in disease. Nat Med (2012) 18:363–74. doi: 10.1038/nm.2627
2. Ganeshan K, Chawla A. Metabolic regulation of immune responses. Annu Rev Immunol (2014) 32:609–34. doi:10.1146/annurev-immunol-032713-120236
3. Brestoff JR, Artis D. Immune regulation of metabolic homeostasis in health and disease. Cell (2015) 161(1):146–60. doi:10.1016/j.cell.2015.02.022
4. Im S, Yousef L, Blaschitz C, Liu JZ, Edwards RA, Young SG, et al. Linking lipid metabolism to the innate immune response in macrophages through sterol regulatory element binding protein-1a. Cell Metab (2011) 13(5):540–9. doi:10.1016/j.cmet.2011.04.001
5. Huang C, Freter C. Lipid metabolism, apoptosis and cancer therapy. Int J Mol Sci (2015) 16(1):924–49. doi:10.3390/ijms16010924
6. Pizarro-Cerdá J, Cossart P. Subversion of phosphoinositide metabolism by intracellular bacterial pathogens. Nat Cell Biol (2004) 6:1026–33. doi:10.1038/ncb1104-1026
7. Feng D, Tang Y, Kwon H, Zong H, Hawkins M, Kitsis RN, et al. High-fat diet-induced adipocyte cell death occurs through a cyclophilin D intrinsic signaling pathway independent of adipose tissue inflammation. Diabetes (2011) 60(8):2134–43. doi:10.2337/db10-1411
8. Bartel DP. microRNAs: genomics, biogenesis, mechanism, and function. Cell (2004) 116(2):281–97. doi:10.1016/S0092-8674(04)00045-5
9. Pasquinelli AE, Ruvkun G. Control of developmental timing by mircoRNAs and their targets. Annu Rev Cell Dev Biol (2002) 18(1):495–513. doi:10.1146/annurev.cellbio.18.012502.105832
10. Yuan Y, Zeng Z, Liu X, Gong DJ, Tao J, Cheng HZ, et al. microRNA-203 inhibits cell proliferation by repressing [Delta]Np63 expression in human esophageal squamous cell carcinoma. BMC Cancer (2011) 11:57. doi:10.1186/1471-2407-11-57
11. Smith AL, Iwanaga R, Drasin DJ, Micalizzi DS, Vartuli RL, Tan AC, et al. The miR-106b-25 cluster targets Smad7, activates TGF-beta signaling, and induces EMT and tumor initiating cell characteristics downstream of Six1 in human breast cancer. Oncogene (2012) 31(50):5162–71. doi:10.1038/onc.2012.11
12. Nie J, Liu L, Zheng W, Chen L, Wu X, Xu Y, et al. microRNA-365, down-regulated in colon cancer, inhibits cell cycle progression and promotes apoptosis of colon cancer cells by probably targeting Cyclin D1 and Bcl-2. Carcinogenesis (2012) 33(1):220–5. doi:10.1093/carcin/bgr245
13. Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res (2008) 79(4):581–8. doi:10.1093/cvr/cvn156
14. Wilfred BR, Wang W, Nelson PT. Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol Genet Metab (2007) 91:209–17. doi:10.1016/j.ymgme.2007.03.011
15. Flowers E, Froelicher ES, Aouizerat BE. microRNA regulation of lipid metabolism. Metabolism (2012) 62(1):12–20. doi:10.1016/j.metabol.2012.04.009
16. Chen T, Huang Z, Wang L, Wang Y, Wu F, Meng S, et al. micro-RNA-125a-5p partly regulates the inflammatory response, lipid uptake, and ORP9 expression in oxLDL-stimulated monocyte/macrophages. Cardiovasc Res (2009) 83(1):131–9. doi:10.1093/cvr/cvp121
17. Ouimet M, Ediriweera HN, Gundra UM, Sheedy FJ, Ramkhelawon B, Hutchison SB, et al. microRNA-33-dependent regulation of macrophage metabolism directs immune cell polarization in atherosclerosis. J Clin Invest (2015) 125(12):4334–48. doi:10.1172/JCI81676
18. Gerin I, Bommer GT, McCoin CS, Sousa KM, Krishnan V, MacDougald OA. Roles for miRNA-378/378* in adipocyte gene expression and lipogenesis. Am J Physiol Endocrinol Metab (2010) 299(2):E198–206. doi:10.1152/ajpendo.00179.2010
19. Aranda JF, Madrigal-Matute J, Rotllan N, Fernández-Hernando C. microRNA modulation of lipid metabolism and oxidative stress in cardiometabolic diseases. Free Radic Biol Med (2013) 64:31–9. doi:10.1016/j.freeradbiomed.2013.07.014
20. Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA miR-14 suppresses cell death and is required for normal fat metabolism. Curr Biol (2003) 13(9):790–5. doi:10.1016/S0960-9822(03)00250-1
21. Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, et al. miR-33 contributes to the regulation of cholesterol homeostasis. Science (2010) 328(5985):1570–3. doi:10.1126/science.1189862
22. Dávalos A, Goedeke L, Smibert P, Ramírez CM, Warrier NP, Andreo U, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci U S A (2011) 108(22):9232–7. doi:10.1073/pnas.1102281108
23. Iliopoulos D, Drosatos K, Hiyama Y, Goldberg IJ, Zannis VI. microRNA-370 controls the expression of microRNA-122 and Cpt1alpha and affects lipid metabolism. J Lipid Res (2010) 51(6):1513–23. doi:10.1194/jlr.M004812
24. Nakanishi N, Nakagawa Y, Tokushige N, Aoki N, Matsuzaka T, Ishii K, et al. The up-regulation of microRNA-335 is associated with lipid metabolism in liver and white adipose tissue of genetically obese mice. Biochem Biophys Res Commun (2009) 385(4):492–6. doi:10.1016/j.bbrc.2009.05.058
25. Lee EK, Lee MJ, Abdelmohsen K, Kim W, Kim MM, Srikantan S, et al. miR-130 suppresses adipogenesis by inhibiting peroxisome proliferator-activated receptor γ expression. Mol Cell Biol (2011) 31(4):626–38. doi:10.1128/MCB.00894-10
26. Rotllan N, Price N, Pati P, Goedeke L, Fernández-Hernando C. microRNAs in lipoprotein metabolism and cardiometabolic disorders. Atherosclerosis (2016) 246:352–60. doi:10.1016/j.atherosclerosis.2016.01.025
27. Deng H, He C, Zhou Z, Liu C, Tan K, Wang N, et al. Isolation and pathogenicity of pathogens from skin ulceration disease and viscera ejection syndrome of the sea cucumber Apostichopus japonicus. Aquaculture (2009) 287:18–27. doi:10.1016/j.aquaculture.2008.10.015
28. Liu H, Zheng F, Sun X, Hong X, Dong S, Wang B, et al. Identification of the pathogens associated with skin ulceration and peristome tumescence in cultured sea cucumbers Apostichopus japonicus (Selenka). J Invertebr Pathol (2010) 105(3):236–42. doi:10.1016/j.jip.2010.05.016
29. Li C, Feng W, Qiu L, Xia C, Su X, Jin C, et al. Characterization of skin ulceration syndrome associated microRNAs in sea cucumber Apostichopus japonicus by deep sequencing. Fish Shellfish Immunol (2012) 33(2):436–41. doi:10.1016/j.fsi.2012.04.013
30. Shao Y, Li C, Ou C, Zhang P, Lu Y, Su X, et al. Divergent metabolic responses of Apostichopus japonicus suffered from skin ulceration syndrome and pathogen challenge. J Agric Food Chem (2013) 61(45):10766–71. doi:10.1021/jf4038776
31. Hwang YC, Oh SW, Park SW, Park CY. Association of serum C1q/TNF-related protein-9 (CTRP9) concentration with visceral adiposity and metabolic syndrome in humans. Int J Obes (2014) 38(9):1207–12. doi:10.1038/ijo.2013.242
32. Forouh N, Saedisomeolia A, Djalali M, Eshraghian MR, Morshedzadeh N, Zabetian-Targhi F, et al. Serum C1q and tumor necrosis factor (TNF)-related protein 9 in women with polycystic ovary syndrome. Diabetes Metab Syndr (2016) 10:S131–4. doi:10.1016/j.dsx.2016.03.012
33. Zhang P, Li C, Zhu L, Su X, Li Y, Jin C, et al. De novo assembly of the sea cucumber Apostichopus japonicus hemocytes transcriptome to identify miRNA targets associated with skin ulceration syndrome. PLoS One (2013) 8(9):e73506. doi:10.1371/journal.pone.0073506
34. Sun X, Lv Z, Li C, Lu M, Zhang P, Zhang W. Identification and characterization of miR-31 potential targets by RNA-seq. Fish Shellfish Immunol (2016) 51:26–32. doi:10.1016/j.fsi.2016.02.014
35. Xing J, Leung MF, Chia FS. Quantitative analysis of phagocytosis by amebocytes of a sea cucumber. Holothuria Leucospilota Invertebr Biol (1998) 117(1):13–22. doi:10.2307/3226853
36. Gu M, Ma H, Mai K, Zhang W, Ai Q, Wang X, et al. Immune response of sea cucumber Apostichopus japonicus coelomocytes to several immunostimulants in vitro. Aquaculture (2010) 306(1–4):49–56. doi:10.1016/j.aquaculture.2010.05.024
37. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods (2011) 25(4):402–8. doi:10.1006/meth.2001.1262
38. Zhang P, Li C, Zhang R, Zhang W, Jin C, Wang L, et al. The roles of two miRNAs in regulating the immune response of sea cucumber. Genetics (2015) 201(4):1397–410. doi:10.1534/genetics.115.178871
39. Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol (1959) 37(8):911–7. doi:10.1139/o59-099
40. Li S, Xu J, Jiang Y, Zhou C, Yu X, Zhong Y, et al. Lipidomic analysis can distinguish between two morphologically similar strains of Nannochloropsis oceanica. J Phycol (2015) 51(2):264–76. doi:10.1111/jpy.12271
41. Chen D, Yan X, Xu J, Su X, Li L. Lipidomic profiling and discovery of lipid biomarkers in Stephanodiscus sp. under cold stress. Metabolomics (2013) 9:949–59. doi:10.1007/s11306-013-0515-z
42. Li S, Xu J, Chen J, Chen J, Zhou C, Yan X. The major lipid changes of some important diet microalgae during the entire growth phase. Aquaculture (2014) 428-429:104–10. doi:10.1016/j.aquaculture.2014.02.032
43. Vella MC, Reinert K, Slack FJ. Architecture of a validated microRNA: target interaction. Chem Biol (2004) 11(12):1619–23. doi:10.1016/j.chembiol.2004.09.010
44. Huang Y, Qi Y, Ruan Q, Ma Y, He R, Ji Y, et al. A rapid method to screen putative mRNA targets of any known microRNA. Virol J (2011) 8:8. doi:10.1186/1743-422X-8-8
45. Brodersen P, Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nat Rev Mol Cell Biol (2009) 10(2):141–8. doi:10.1038/nrm2619
46. Wong GW, Krawczyk SA, Kitidis-Mitrokostas C, Ge G, Spooner E, Hug C, et al. Identification and characterization of CTRP9, a novel secreted glycoprotein, from adipose tissue that reduces serum glucose in mice and forms heterotrimers with adiponectin. FASEB J (2009) 23(1):241–58. doi:10.1096/fj.08-114991
47. Peterson JM, Wei Z, Seldin M, Byerly MS, Aja S. CTRP9 transgenic mice are protected from diet-induced obesity and metabolic dysfunction. Am J Physiol Regul Integr Comp Physiol (2013) 305(5):R522–33. doi:10.1152/ajpregu.00110.2013
48. Wiklund S, Johansson E, Sjösttoöm L, Mellerowicz EJ, Edlund U, Shokcor JP, et al. Visualization of GC/TOF-MS-based metobolomics data for identification of biochemically interesting compounds using OPLS class models. Anal Chem (2008) 80(1):115–22. doi:10.1021/ac0713510
49. Ogretmen B. Sphingolipids in cancer: regulation of pathogenesis and therapy. FEBS Lett (2006) 580(23):5467–79. doi:10.1016/j.febslet.2006.08.052
50. Shao Y, Li C, Zhang W, Duan X, Li Y, Jin C, et al. Molecular cloning and characterization of four caspases members in Apostichopus japonicu. Fish Shellfish Immunol (2016) 55:203–11. doi:10.1016/j.fsi.2016.05.039
51. Raisch J, Darfeuille-Michaud A, Nguyen HTT. Role of microRNAs in the immune system, inflammation and cancer. World J Gastroenterol (2013) 19(20):2985–96. doi:10.3748/wjg.v19.i20.2985
52. Lu M, Zhang P, Li C, Zhang W, Jin C, Han Q. miR-31 modulates coelomocytes ROS production via targeting p105 in Vibrio splendidus challenged sea cucumber Apostichopus japonicus in vitro and in vivo. Fish Shellfish Immunol (2015) 45(2):293–9. doi:10.1016/j.fsi.2015.04.024
53. Rottiers V, Näär AM. microRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol (2012) 13(4):239–50. doi:10.1038/nrm3313
54. Bartel DP. microRNAs: target recognition and regulatory functions. Cell (2009) 136(2):215–33. doi:10.1016/j.cell.2009.01.002
55. Laurila EM, Kallioniemi A. The diverse role of miR-31 in regulating cancer associated phenotypes. Genes Chromosome Cancer (2013) 52:1103–13. doi:10.1002/gcc.22107
56. Schmittgen TD. miR-31: a master regulator of metastasis? Future Oncol (2010) 6(1):17–20. doi:10.2217/fon.09.150
57. Stepicheva NA, Song JL. Function and regulation of microRNA-31 in development and disease. Mol Reprod Dev (2016) 83(8):654–74. doi:10.1002/mrd.22678
58. Wei Z, Lei X, Petersen PS, Aja S, Wong GW. Targeted deletion of C1q/TNF-related protein 9 increases food intake, decreases insulin sensitivity, and promotes hepatic steatosis in mice. Am J Physiol Endocrinol Metab (2014) 306(7):E779–90. doi:10.1152/ajpendo.00593.2013
59. Jung CH, Lee MJ, Kang YM, Lee YL, Seol SM, Yoon HK, et al. C1q/TNF-related protein-9 inhibits cytokine-induced vascular inflammation and leukocyte adhesiveness via AMP-activated protein kinase activation in endothelial cells. Mol Cell Endocrinol (2016) 419:235–43. doi:10.1016/j.mce.2015.10.023
60. Jung TW, Hong HC, Hwang H, Yoo H, Baik S, Choi KM. C1q/TNF-related Protein 9 (CTRP9) attenuates hepatic steatosis via the autophagy-mediated inhibition of endoplasmic reticulum stress. Mol Cell Endocrinol (2015) 417:131–40. doi:10.1016/j.mce.2015.09.027
61. Kambara T, Shibata R, Ohashi K, Matsuo K, Hiramatsu-Ito M, Enomoto T, et al. C1q/tumor necrosis factor-related protein 9 protects against acute myocardial injury through an adiponectin receptor I-AMPK-dependent mechanism. Mol Cell Biol (2015) 35(12):2173–85. doi:10.1128/MCB.01518-14
62. Zechner R, Zimmermann R, Eichmann TO, Kohlwein SD, Haemmerle G, Lass A, et al. Fat signals-lipases and lipolysis in lipid metabolism and signaling. Cell Metab (2012) 15(3):279–91. doi:10.1016/j.cmet.2011.12.018
63. Santos CR, Schulze A. Lipid metabolism in cancer. FEBS J (2012) 279(15):2610–23. doi:10.1111/j.1742-4658.2012.08644.x
64. Mullen TD, Obeid LM. Ceramide and apoptosis: exploring the enigmatic connections between sphingolipid metabolism and programmed cell death. Anticancer Agents Med Chem (2012) 12(4):340–63. doi:10.2174/187152012800228661
65. Obeid LM, Linardic CM, Karolak LA, Hannun YA. Programmed cell death induced by ceramide. Science (1993) 259(5102):1769–71. doi:10.1126/science.8456305
66. Lupi R, Dotta F, Marselli L, Del GS, Masini M, Santangelo C, et al. Prolonged exposure to free fatty acids has cytostatic and pro-apoptotic effects on human pancreatic islets: evidence that beta-cell death is caspase mediated, partially dependent on ceramide pathway, and Bcl-2 regulated. Diabetes (2002) 51(5):1437–42. doi:10.2337/diabetes.51.5.1437
67. Kroesen BJ, Pettus B, Luberto C, Busman M, Sietsma H, de Leij L, et al. Induction of apoptosis through B-cell receptor cross-linking occurs via de novo generated C16-ceramide and involves mitochondria. J Biol Chem (2001) 276(17):13606–14. doi:10.1074/jbc.M009517200
68. Wang S, Hojabrpour P, Zhang P, Kolesnick RN, Steinbrecher UP, Gómez-Muñoz A, et al. Regulation of ceramide generation during macrophage apoptosis by ASMase and de novo synthesis. Biochim Biophys Acta (2015) 1851(11):1482–9. doi:10.1016/j.bbalip.2015.08.002
69. Levy M, Futerman AH. Mammalian ceramide synthases. IUBMB Life (2010) 62(5):347–56. doi:10.1002/iub.319
70. Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol (2008) 9(2):139–50. doi:10.1038/nrm2329
71. Ogretmen B, Pettus BJ, Rossi MJ, Wood R, Usta J, Szulc Z, et al. Biochemical mechanisms of the generation of endogenous long chain ceramide in response to exogenous short chain ceramide in the A549 human lung adenocarcinoma cell line. J Biol Chem (2002) 277(15):12960–9. doi:10.1074/jbc.M110699200
72. Grether-Beck S, Timmer A, Felsner I, Brenden H, Brammertz D, Krutmann J. Ultraviolet A-induced signaling involves a ceramide-mediated autocrine loop leading to ceramide de novo synthesis. J Invest Dermatol (2000) 125(3):545–53. doi:10.1111/j.0022-202X.2005.23782.x
73. Osawa Y, Uchinami H, Bielawski J, Schwabe RF, Hannun YA, Brenner DA. Roles for C16-ceramide and sphingosine 1-phosphate in regulating hepatocyte apoptosis in response to tumor necrosis factor-α. J Biol Chem (2005) 280(30):27879–87. doi:10.1074/jbc.M503002200
74. Golstein P, Kroemer G. Cell death by necrosis: towards a molecular definition. Trends Biochem Sci (2007) 32(1):37–43. doi:10.1016/j.tibs.2006.11.001
Keywords: Apostichopus japonicus, miR-31, complement C1q tumor necrosis factor-related protein 9, lipidomics, ceramide, apoptosis
Citation: Shao Y, Li C, Xu W, Zhang P, Zhang W and Zhao X (2017) miR-31 Links Lipid Metabolism and Cell Apoptosis in Bacteria-Challenged Apostichopus japonicus via Targeting CTRP9. Front. Immunol. 8:263. doi: 10.3389/fimmu.2017.00263
Received: 12 January 2017; Accepted: 23 February 2017;
Published: 13 March 2017
Edited by:
Juarez Antonio Simões Quaresma, Federal University of Pará, BrazilReviewed by:
Zeeshan Ahmed, The Jackson Laboratory, USAJose Andres Fernandez, University of the Basque Country, Spain
Copyright: © 2017 Shao, Li, Xu, Zhang, Zhang and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chenghua Li, lichenghua@nbu.edu.cn
 Yina Shao1
Yina Shao1 Chenghua Li
Chenghua Li Wei Xu
Wei Xu Weiwei Zhang
Weiwei Zhang