- 1Department of Marine Chemistry and Geochemistry, Woods Hole Oceanographic Institution, Woods Hole, MA USA
- 2Department of Biology, Woods Hole Oceanographic Institution, Woods Hole, MA USA
The coccolithophore Emiliania huxleyi forms massive blooms and plays a critical role in global elemental cycles, sequestering significant amounts of atmospheric carbon dioxide on geological time scales via production of calcium carbonate coccoliths and emitting dimethyl sulfoniopropionate (DMSP), which has the potential for increasing atmosph-eric albedo. Because grazing in pelagic systems is a major top-down force structuring microbial communities, the influence of grazers on E. huxleyi populations has been of interest to researchers. Roles of DMSP (and related metabolites) in interactions between E. huxleyi and protist grazers have been investigated, however, little is known about the release of other metabolites that may influence, or be influenced by, such grazing interactions. We used high-resolution mass spectrometry in an untargeted approach to survey the suite of low molecular weight compounds released by four different E. huxleyi strains in response to grazing by the dinoflagellate Oxyrrhis marina. Overall, a strikingly small number of metabolites were detected from E. huxleyi and O. marina cells, but these were distinctly informative to construct metabolic footprints. At most, E. huxleyi strains shared 25% of released metabolites. Furthermore, there appeared to be no unified metabolic response in E. huxleyi strains to grazing; rather, these responses were strain specific. Concentrations of several metabolites also positively correlated with grazer activities, including grazing, ingestion, and growth rates; however, no single metabolite responded uniformly across all strains of E. huxleyi tested. Regardless, grazing clearly transformed the constituents of dissolved organic matter produced by these marine microbes. This study addresses several technical challenges, and presents a platform to further study the influence of chemical cues in aquatic systems and demonstrates the impact of strain diversity and grazing on the complexity of dissolved organic matter in marine systems.
Introduction
Phytoplankton account for 50% of global carbon fixation (Field et al., 1998) and factors influencing their growth and loss processes are critical in modulating global environmental feedback cycles (Falkowski et al., 1998; Strom, 2008). One such phytoplankter is the cosmopolitan marine coccolithophore Emiliania huxleyi, which plays important roles in major biogeochemical cycles. Not only does this species form massive blooms, but E. huxleyi cells also form external calcium carbonate plates, called liths, that make E. huxleyi an important component of the biological pump as blooms terminate and sequester atmospheric carbon in the deep ocean (Bidle and Falkowski, 2004). Additionally, E. huxleyi is a prominent algal producer of dimethyl sulphoniopropionate (DMSP) as well as its degradation products dimethyl sulfide (DMS) and acrylic acid (Wolfe and Steinke, 1996; Alcolombri et al., 2015), with the former acting as nucleation sites for cloud formation and enhancing earth's albedo (Charlson et al., 1987).
The major source of loss for phytoplankton in pelagic systems is consumption by microzooplankton grazers, with grazers consuming, on average, 60–70% of daily primary production (Calbet and Landry, 2004). Therefore, small changes in grazing pressure can have significant consequences for phytoplankton population dynamics (Irigoien et al., 2005). These grazing interactions are highly selective, and consumption of phytoplankton prey is mediated by many factors including prey nutritional quality (Meunier et al., 2012), behavioral responses (Menden-Deuer and Grunbaum, 2006; Harvey and Menden-Deuer, 2011), and structural (Hamm et al., 2003) or chemical defenses (Tillmann, 2004) of prey cells. Together, these phenotypic qualities of prey cells, along with their co-evolutionary history with grazers (Kubanek et al., 2007; Senft-Batoh et al., 2015), can influence their palatability toward both meso- and microzooplankton grazers, thus regulating phytoplankton productivity and population dynamics.
In addition to grazers responding to prey characteristics, prey organisms also respond to grazers in species-specific manners. In some cases, this includes the production of chemical defenses in response to the presence of a grazer (e.g., Selander et al., 2006), or the initiation of morphological changes that may make it more difficult for grazers to locate (Selander et al., 2011) and consume prey (Long et al., 2007). Chemical defenses include induced toxin production (e.g., Bergkvist et al., 2008), and activated defenses, such as oxylipins and polyunsaturated aldehydes (PUAs; reviewed by Pohnert, 2005), which may inhibit grazers during handling and/or after ingestion. Some of these compounds are released into the surrounding environment and can serve to mediate interactions between cells before grazing interactions can take place (Strom et al., 2003a,b). For instance, the production of DMSP, DMS, and acrylic acid has been proposed to serve as a defense mechanism of E. huxleyi against microzooplankton (Wolfe et al., 1997), with one or more of these compounds serving as a toxic compound (Wolfe et al., 1997; Breckels et al., 2011) or as a cue indicating unpalatable prey (Strom et al., 2003a). However, recent studies have shown that DMSP (and related compounds) can also attract potential grazers (Seymour et al., 2010), indicating that there are likely other chemical and structural cues that modulate interactions between E. huxleyi and grazers.
Not only can healthy prey items release inhibitory or defensive cues into the environment, but cell destruction can also release metabolites into the environment that are sensed by other members of the community. These waterborne metabolites can be tracked by grazers looking for prey (Hay, 2009) or by algal conspecifics, in which case these released metabolites may act as alarm cues indicating the presence of a grazer (Lurling, 1998; Van Donk et al., 2011). In some cases, a suite of cues from damaged conspecifics and grazers causes an upregulation of algal defenses (Bergkvist et al., 2008; Senft-Batoh et al., 2015). The presence of such cues can influence other aspects of phytoplankton physiology, not just defensive characteristics. For instance, the presence of the PUA decadienal, which inhibits copepod reproduction (Miralto et al., 1999) also may serve a role in intra-specific signaling, acting as a “stress surveillance system” for diatom populations (Vardi et al., 2006), and other PUAs have been suggested to act as bloom termination signals (Casotti et al., 2005; Vidoudez and Pohnert, 2008).
Metabolomics approaches to characterizing this pool of released algal compounds have been suggested to aid in the identification of not only individual metabolites of ecological interest (e.g., Gillard et al., 2013), but also suites of compounds that together may be indicative of an ecological condition, or physiological state of an organism (Prince and Pohnert, 2010). In the current study, we employed a mass-spectrometry based metabolomics approach to characterize the suite of exuded metabolites released during grazing interactions between four different strains of E. huxleyi and the model dinoflagellate grazer, Oxyrrhis marina. We tested the hypothesis that O. marina grazing will alter the pool of released E. huxleyi metabolites (which may also contain cues or signals). Because strain identity is also a factor influencing metabolite production in phytoplankton, we also analyzed released compounds from four different E. huxleyi strains, which varied in DMSP production and calcification state, to test whether released metabolite pools are specific to strains of E. huxleyi, or if four related strains share a common response to grazers.
Methods
General Culturing Procedures
Four strains of E. huxleyi (National Center for Marine Algae: CCMP379, CCMP374, and Plymouth Algal Culture Collection: DHB607, DHB624) were used in these experiments. All phytoplankton cultures were grown in 0.2-μm sterile-filtered autoclaved natural seawater (FSW), enriched with f/2 nutrients minus silica (Guillard and Hargraves, 1993). All cultures were maintained on a 12:12 h light:dark cycle at 18°C, salinity of ~30, and a light intensity of 85–100 μmol photon m−2 s−1 for the phytoplankton cultures, and 20–30 μmol photon m−2 s−1 for O. marina. The grazer, O. marina (LB1974; UTEX Culture Collection) was cultured in FSW only. Isochrysis galbana (CCMP1323) was used to rear the predator prior to grazing experiments. Although the cultures were maintained using rigorous aseptic technique, the cultures were not received as axenic, nor were they axenic at the time of the experiments performed in this study. Phytoplankton cultures were transferred every 7–10 days to maintain exponential growth. Prior to the grazing experiments, stocks of the predator were maintained on I. galbana (50,000 cells ml−1 final concentration), fed twice weekly. Unless otherwise specified, cell concentrations of both predator and prey cultures were determined by microscope counts using samples fixed with 1% Lugol's solution.
Experimental Set-Up
This experiment was as performed as in the companion study by Harvey et al. (2015). Briefly, the functional grazing response of O. marina to four strains of E. huxleyi was measured by calculating the change in prey abundance over 48 h, at five initial prey concentrations (5000–80,000 cells ml−1; 70–1080 ng C ml−1) for each strain examined. We chose four strains of E. huxleyi that displayed phenotypic variation in calcification states and intracellular DMSP concentration. Initial grazer concentration was ~200 cells mL−1. Seawater-, grazer-, and prey-only controls were also monitored. Prior to the experiment set up, O. marina cultures were starved for 24 h, and the concentration of I. galbana was less than 1000 cells mL−1 at the beginning of the experiments. Organisms were first combined in 2 L cultures at the appropriate concentrations; mixed gently, and then triplicate aliquots were gently poured into 250 mL clear polycarbonate bottles. Sample bottles were placed randomly on a plankton wheel with a rotation of 1 rpm, and maintained on a 12:12 h light:dark cycle at 18°C, and light intensity of 20–30 μmol photon m−2 s−1 for 48 h. The 2 L bottle was sampled for initial measurements and the polycarbonate bottles after 48 h. After 48 h, aliquots were taken for cell abundances to calculate grazing coefficients, ingestion rates, and growth rates of predators (O. marina) and prey species (E. huxleyi; Frost, 1972; Heinbokel, 1978).
Abundance
To calculate E. huxleyi abundance, 200 μL aliquots were taken from each replicate and pipetted into a 96-well plate and run on a flow cytometer (Guava, Millipore, Billerica MA). Prior to the experiment, the optimal flow cytometer settings for E. huxleyi were determined based on chlorophyll a (692 nm) fluorescence and side scatter. To enumerate O. marina, 5 mL aliquots were preserved using 1% Lugol's solution and enumerated using a Sedgwick Rafter counting chamber and a ZEISS light microscope.
Extraction of Waterborne Compounds
From each replicate at the end of the experiment, 80 mL of culture was decanted into duplicate 50 mL falcon tubes (40 mL of culture in each). Samples were also taken at t = 0 h for the following treatments: prey only, predator only, and three replicates of predator + prey. Cells were removed by gentle centrifugation (3000 rpm, 10 min) and the supernatant was filtered through GF/F filters (GE Healthcare Bio-Sciences, Pittsburgh, PA) and the filtrate was collected and frozen at −20°C until extraction. Dissolved organic constituents in the filtrate were extracted using Waters Oasis HLB columns (200 mg/6 mL) as follows: cartridges were conditioned with 3 mL ethyl acetate followed by 3 mL methanol; then equilibrated with 3 mL of MilliQ water. Samples (80 mL of filtrate per replicate) were loaded onto the column (at a flow rate of ~1 mL min−1), followed by a 4 mL wash of MilliQ water to remove salts. We eluted organic compounds with 2 mL of methanol followed by 2 mL ethyl acetate, repeated once each, resulting ~8 mL final extract volume. Extracts were then dried in vacuo, using a Savant speed vac concentrator at 50°C.
Chromatography and Mass Spectrometry
For metabolic footprinting (Hollywood et al., 2006), extracts were re-dissolved in 95:5 water/methanol and were either normalized to the volume of culture extracted, or the number of E. huxleyi cells. Because we attempted to assess the responses E. huxleyi populations to grazing, extracts of strains CCMP374, DHB607, and DHB624, were brought to equivalent concentrations based on the number of E. huxleyi cells present at the end of the 48 h grazing experiment, (e.g., Poulson-Ellestad et al., 2014; Mausz and Pohnert, 2015), in order to observe per cell amounts of compounds in the environment. For the purposes of this study, we were concerned with only pronounced responses of the E. huxleyi exometabolome to the presence of the grazer O. marina. We therefore combined replicates in silico across E. huxleyi cell concentrations in order to search for chemical features, or metabolites, that varied in concentration due to grazing. Because strain CCMP379 was heavily grazed, samples were brought up to volumetric equivalents, instead of being normalized to cell numbers. Extracts of “grazer only” controls were reconstituted at equivalent O. marina cell concentrations. Seawater only controls were also filtered, extracted, dried, and re-dissolved at the same concentration factor as the most concentrated grazed E. huxleyi samples, in a conservative effort to identify background chemical features present in the filtered seawater medium.
Samples were then profiled as in Whalen et al. (2015) with an untargeted metabolomics approach, using high performance liquid chromatography coupled to time of flight mass spectrometry (Agilent Technologies 6230 Time of Flight MS with a Dual Agilent Jet Stream Electrospray Ionization; HPLC/MS-ToF). Our reversed-phase HPLC solvent scheme was as follows: hold at 5% methanol for 5 min, ramp to 40% methanol over 5 min, and hold at 40% for 2 min. Then ramp to 95% methanol over 5 min, hold at 95% methanol for 3 min, drop to 5% methanol, and hold for 3 min. All solvents were acidified with formic acid, 0.1% (v/v) final concentration. The first 2 min of each run was diverted into waste to avoid contaminating the mass spectrometer with salt, with a 5 min column equilibration time between samples. We used a Kinetex C18 column (2.6 μm; 2.1 × 150 mm; Phenomenex), with a flow rate of 0.2 mL min−1 and injected 3 μL of sample. The column temperature was held at 35°C.
Extracts were run in positive ionization mode, scanning from 20 to 3000 m/z, with mass correction ions of 922.0098 and 121.0509 m/z that were injected at the ESI source. Gas and sheath gas temperatures were 350°C, drying gas flow rate was 8 L min−1 and sheath gas flow rate was 10 L min−1. Nebulizer pressure was 40 psi, with capillary voltage of 6500 V and a nozzle voltage of 1000 V. Fragmenter and skimmer voltages were 135 and 65 V, respectively. To minimize the impacts of batch variability since each grazing experiment was performed separately, one strain at a time; we analyzed strains in independent runs and only statistically compared profiles within each strain of E. huxleyi.
Preprocessing of Mass Spectra for Metabolomics Analysis
Spectral processing was performed in MZmine 2.11 (Katajamaa and Oresic, 2005) and included peak picking (minimum spectral count of 5000), chromatogram building, retention time normalization, and spectral alignment. All MZmine settings were as reported in Whalen et al. (2015) and Macintyre et al. (2014). This resulted in a peak list containing unique m/z-retention time pairs, referred to as chemical features, as well as peak areas from each sample. After generating a peak list, the list was filtered to only include features that were (1) present in two of three replicates in at least one treatment group. (2) present at abundances >10 times that of the seawater blanks, and (3) present at concentrations greater than twice that of the “grazer only” controls. In such cases where polyethylene glycol (PEG) was present in our extracts (particularly in strains DHB607 and DHB624), PEG ions were easily detected and manually removed due to their retention times between 15 and 18 min, and characteristic in-source fragmentation pattern with neutral losses of 44 a.m.u. Molecular formula generation was performed on selected molecular ions using the molecular formulae generator in Mass Hunter Workstation version B0.4.00. Molecular formulae were then searched against the Metlin database for metabolite annotation (Smith et al., 2005).
Statistical Analyses
Before multivariate statistical analyses were performed, data were centered and variance stabilized by autoscaling (van Den Berg et al., 2006) in the PLS Toolbox in Matlab version 8.2.0.7. Multivariate analyses including principal component analysis (PCA) and orthagonalized partial least squares-discriminant analysis (OPLS-DA, cross validated with venetian blinds with six splits) were performed in PLS Toolbox (Poulson-Ellestad et al., 2014 and citations within). Loadings of chemical features from PCA were examined to determine their influence on the differences between exometabolomes (Robertson et al., 2007). Univariate unpaired t-tests comparing relative peak areas with a false discovery rate (FDR) correction (< 0.05; Benjamini and Hochberg, 1995) were performed in Microsoft Excel, while unpaired t-tests on principal component scores were performed in Graph Pad Prism v. 6.05. We also performed 2-way ANOVAs with Sidak's multiple comparisons test in order to determine the influence of E. huxleyi cell concentrations on chemical footprints. Pearson correlation coefficients were calculated, in Graph Pad Prism, in order to determine correlations between metabolite peak areas and predator activities; specifically grazing coefficients, predator ingestion rates, and predator growth rates.
Analysis of Pure Standard
To aid in the annotation of a particular chemical feature (m/z 144.0479) we also analyzed authentic standard of the metabolite 4-methyl-5-thiazoleethanol (Sigma-Aldrich Cat. 190675-25G), diluted to 8 μM in 95:5 water/methanol, with the same chromatographic and spectral methods described above.
Results
Grazing Results
We pooled all replicates across E. huxleyi cell concentrations as a single treatment in order to facilitate our search for metabolites that varied in concentration due to grazing (Figure 1), but see Harvey et al. (2015) for important discussions regarding the response of O. marina as a function of E. huxleyi concentration. We observed different grazing coefficients as well as predator growth rates (μ; Supplementary Figure 1), when O. marina was fed four different E. huxleyi strains. When comparing grazing on noncalcified E. huxleyi, O. marina grazed less on strain CCMP374 than CCMP379, with grazing at rates of 0.84 (± 0.16) d−1 and 1.6 (±0.33) d−1, respectively, (ANOVA with Tukey post-test; p < 0.0001, n = 10–13), when grazing coefficients were averaged across all concentrations of E. huxleyi tested (Figure 1). O. marina also grazed on the two calcified strains of DHB607 and DHB624 with significantly different rates of 0.27 (±0.20) d−1 and 0.80 (±0.13) d−1, respectively, indicating that O. marina does not graze all strains of E. huxleyi equally (p < 0.0001; n = 13; Figure 1). Strains DHB624 and CCMP374 were grazed at nearly identical rates (p = 0.97, Figure 1).
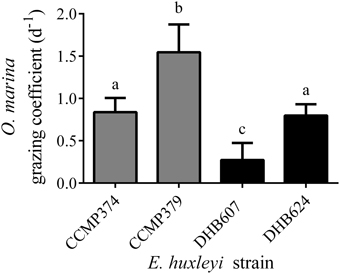
Figure 1. Grazing of Oxyrrhis marina LB1974 on four Emiliania huxleyi strains over 48 h. Grazing is expressed as mean grazing coefficient (d−1), averaged over multiple concentrations of E. huxleyi. Error bars represent ±1 S.D. E. huxleyi strains include non-calcified strains CCMP374 and CCMP379 (denoted by gray bars) as well as calcified strains DHB607 and DHB624 (black bars). Letters (a–c) indicate statistical equivalency from ANOVA with Tukey post-test (n = 10–13).
Grazing Alters Chemical Profiles of E. huxleyi Strains
From strain CCMP374, which was heavily grazed, we limited our analysis to 24 chemical features (i.e., distinct m/z –retention time pairs), that remained after applying our selection criteria. Therefore, although we initially detected 553 chemical features we focused our efforts on less than 5% of these features.
In order to explore the overall response of the CCMP374 exometabolome to grazing, we analyzed the peak areas of the 24 metabolites from strain CCMP374 using PCA, and found a striking and significant separation between exometabolomes from grazed cultures and prey only control cultures along the first principal component (unpaired t-test, p < 0.0001; Figure 2). Additional analyses revealed that there was a significant influence of grazing, but not of E. huxleyi cell concentration on the PC1 scores (2-way ANOVA with Sidak's multiple comparison test; effect of grazing p < 0.0001, cell concentration p = 0.6145; Supplementary Table 1, Supplementary Figure 2). There was a significant impact of the interaction between grazing and E. huxleyi cell concentration (grazing x cell concentration p = 0.0128; Supplementary Table 1). The PCA model indicated that there were nine metabolites with increased concentrations as a result of grazing (features with loadings >0.2 on principal component 1; Supplementary Table 2). Additionally, there was a group of five metabolites that were found to be more prevalent in prey only samples, (features with loadings < -0.15, Supplementary Table 2), indicating that these chemical features may reflect normal activity of CCMP374 populations. Of all metabolites with concentrations significantly altered by the presence of grazers in the PCA model, three were not found in the grazer only treatment, indicating that these are most likely produced by E. huxleyi cells in response to O. marina.
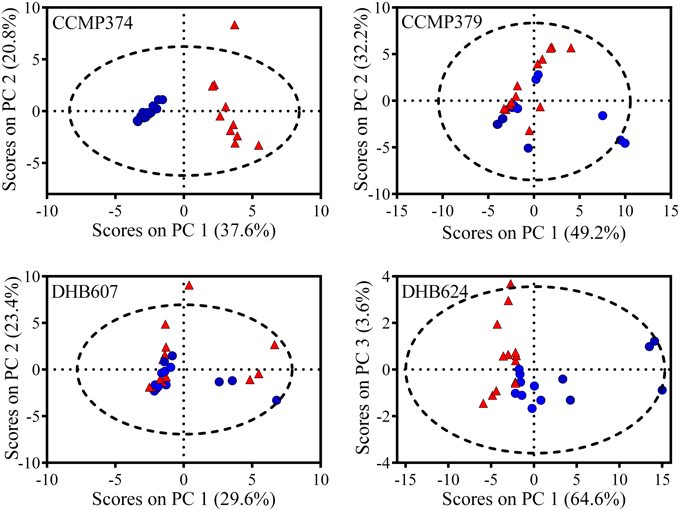
Figure 2. PCA Scores plot comparing exometabolomes of Emiliania huxleyi grazed by Oxyrrhis marina (red triangles) and E. huxleyi only (blue circles) control cultures (n = 10–13, depending on E. huxleyi strain). Four different strains (CCMP374, CCMP379, DHB607, and DHB624) of E. huxleyi were grazed for 48 h before extraction of exometabolomes. Dashed line represents 95% confidence interval.
We detected several metabolites that could be candidates for biomarkers of protist grazing on CCMP374. Twelve metabolites had significantly varying concentrations as a result of grazing: ten metabolites had positive fold changes from 2.55 to 6.36 and two metabolites with −1.82 and −2.54-fold reductions in concentration (t-test with FDR < 0.05 correction; Figure 3; Supplementary Table 2). Only four of these metabolites were not detected in the exometabolomes of grazer only cultures, three of which (m/z 251.1500, 329.1104, and 400.1467; potential molecular formulae C12H18N4O2, C13H20N4O2S, and C15H21N5O8, respectively) were not detected in E. huxleyi only cultures, indicating that these metabolites are E. huxleyi stress response compounds or O. marina-specific digestion metabolites (Supplementary Table 2). Additionally, one metabolite (m/z 399.2340, retention time 19.1 min with potential molecular formulae C17H30N6O5 or C16H34N2O9) that was also absent in extracts of O. marina cultures, experienced a −2.54-fold reduction in relative concentration in grazed cultures, suggesting this chemical feature is an indicator of non-stressed E. huxleyi strain CCMP374.
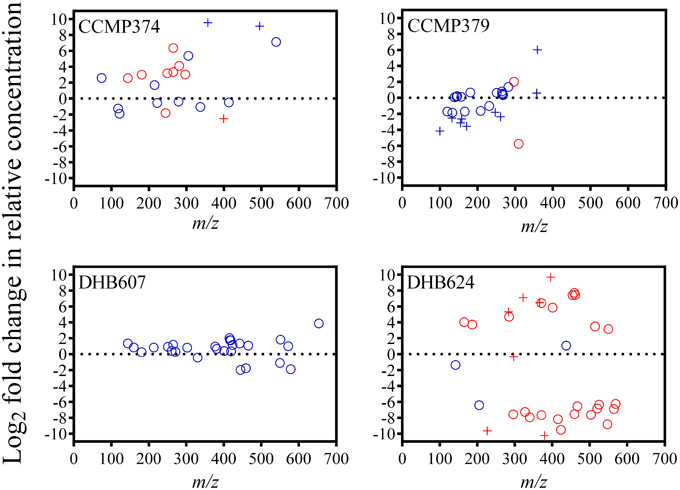
Figure 3. Fold changes of chemical features (circles) detected in the exometabolomes of four Emiliania huxleyi strains (CCMP374, CCMP379, DHB607, and DHB624) exposed to the predator Oxyrrhis marina, arranged according to m/z. Fold change was calculated as the Log2 of the ratio between feature peak area in grazed vs. control (i.e., not grazed) extracts. Features with peak areas that were significantly different (unpaired t-test with FDR < 0.05 correction) in extracts of grazed and control cultures are colored red. Pluses (+) indicate features that were not present in extracts of grazer only cultures.
From the highly grazed strain CCMP379, we detected 29 features that were adequate (as discussed above) for further analyses. For this strain, we normalized extract concentrations to volume extracted vs. final E. huxleyi cell concentration, as done with the other strains, due to minimal E. huxleyi cells remaining at the end of the experiment. PCA on these 29 chemical features revealed a significant separation (p = 0.0052; Figure 2) between grazed and control exometabolomes on the second principal component of a three-component model (88.3% of variance captured). When assessing the relative impact of cell concentrations vs. grazing on exometabolomes, we found significant impacts of grazing (p = 0.0003), E. huxleyi concentration (p = 0.0022) and the interaction of the two factors (p = 0.030; Supplementary Table 3). A Sidak's multiple comparisons test revealed that there was a significant difference between control and grazed samples at only the highest concentration of E. huxleyi tested (p = 0.0003, Supplementary Figure 2). There were 11 metabolites with loadings >0.15 on the second principal component, (Supplementary Table 4), suggesting their concentrations increased as a result of grazing; whereas the relative concentrations of 12 metabolites appeared to be reduced by grazing (i.e., enhanced concentrations in controls relative to grazed samples), with loadings < -0.15 on principal component 2 (Supplementary Table 4). However, the concentrations of only two features were significantly altered by grazing (Figure 3). Concentrations of one of these features, m/z 297.1015 (C12H16N4O3S, retention time 3.9 min) significantly increased 1.89-fold whereas the concentrations of the other feature, m/z 309.2049 (C18H28O4, retention time 20.0 min), significantly decreased by −5.76-fold as a result of grazing (Figure 3). Both features were also present in grazer only extracts, indicating that they are possibly O. marina metabolites.
Our analysis revealed that there were still several candidate metabolites specific to E. huxleyi CCMP379, despite detecting only a few compounds that varied in concentration due to grazing. Eleven features were not found in extracts of the T0 and/or T48 h grazer only cultures, indicating that they are either E. huxleyi specific, or are only produced by O. marina during grazing (Supplementary Table 4). Of these 11, eight had negative loadings in the PCA model and negative fold changes, suggesting that together this suite of compounds may be characteristic of healthy E. huxleyi CCMP379. Additionally, three of these 11 features were found to have increased concentrations in response to grazing — all had positive loadings on PC2, but fold change was not calculated for one (m/z 227.1064, C15H14O2) because it was not present in CCMP379 only cultures (Supplementary Table 4).
From the DHB624, the concentrations of 50 chemical features were analyzed. The relative concentrations of 47 metabolites were significantly altered in response to grazing (t-test with FDR < 0.05; Figure 3; Supplementary Table 5). Additionally 21 of these 50 features were not present in extracts of grazer only cultures at the final sampling point (t = 48 h), while five of these were also not present in grazer only cultures at t = 0 h. Of these five, four had significantly greater concentrations in grazed samples, with fold changes ranging from 5.32 to 9.69. These features included m/z 283.1163 (C11H10N10, retention time 14.2 min), 322.1852 (C11H19N11O, retention time 14.8 min), 366.2118 (C13H23N11O2, retention time 15.5 min); and 396.2582 (C15H29N11O2, retention time 18.6 min). The concentrations of one feature, m/z 297.1316 (C16H16N4O2, retention time 18.8 min) were significantly reduced by -0.33-fold in grazed samples (Supplementary Table 5).
When we analyzed these 50 metabolites with PCA, we determined that there were significant separations of chemical profiles from grazed and control cultures on the first and third principal components (p = 0.0017 and p = 0.0402, respectively; Figure 2). This was a three-component model accounting for 92.3% of total variance in the dataset. When considering the influence of E. huxleyi cell concentration on these scores from PC1, we found significant effects of E. huxleyi concentration (p < 0.0001), grazing (p < 0.0001), and the interaction of both factors (p < 0.0001, Supplementary Table 6). Results of the multiple comparisons test showed that there were significant differences between grazed and prey only controls at the three lowest concentrations of E. huxleyi tested (p < 0.0001 for all; Supplementary Figure 2). There were 14 features that had small, negative loadings on principal component one (loadings ≤ −0.05), and had relative concentrations that were significantly greater in grazed samples (Supplementary Table 5). In contrast, PCA revealed 32 metabolites with reduced concentrations in grazed samples compared to control samples. Based on the separation obtained on the third principal component, there was one feature with significantly greater concentrations in the exometabolomes of grazed cultures (m/z 549.7926; retention time 17.4 min; fold change 3.17) and that was also present in extracts of strain DHB607, however this metabolite did not appear discriminatory on the first principal component (Supplementary Table 5).
In order to narrow down which features may have predictive or discriminatory power for distinguishing between grazed DHB624 and control exometabolomes, we analyzed these data with an orthagonalized PLS-DA model, with grazed and control treatments as our two classes. This model was partially successful in classifying grazed and prey only exometabolomes, with a 23% cross validation error rate. The variance captured on the first latent variable was 34.3%. From this model, 15 features had selectivity ratios ≥1.0, indicating that they may have predictive power in distinguishing between grazed and control extracts (Rajalahti et al., 2009). The relative concentrations of 14 features were greater in extracts of grazed cultures (Supplementary Table 5). Only one feature decreased in concentrations in response to grazing (fold change −6.43). We found that the same four features that were never detected in extracts of grazer only cultures, but had concentrations significantly increased due to grazing, had selectivity ratios >1.0 (Supplementary Table 5), and are most likely E. huxleyi response compounds or exuded waste metabolites of O. marina.
The presence of a grazer did not alter the exometabolomes of strain DHB607, which is highly calcified and poorly grazed by O. marina (Figure 1). None of the 29 metabolites we analyzed had concentrations that were significantly altered in response to grazing or the presence of the grazer (t-test with FDR < 0.05; Figure 3, Supplementary Table 7). In addition, only two features were not present in the predator only extracts (Supplementary Table 7). A five PC model (87.0% of cumulative variance explained), demonstrated that there was a small, but significant separation between grazed and control exometabolomes along the second principal component, which accounts for 23.4% of total variance (p = 0.045; Figure 2). The 2-way ANOVA revealed significant effects of both grazing (p = 0.0002) and E. huxleyi concentration (p < 0.0001), with a significant impact of interaction between the two factors (p = 0.023; Supplementary Table 8). Sidak's multiple comparisons test revealed that the effect of grazing is driven by the significant differences of the chemical profiles of grazed and controls cultures at the second lowest E. huxleyi concentration tested (p = 0.0006; Supplementary Figure 2). Based on this model, we found 11 features with increased relative concentrations due to grazing, (loadings >0.18; fold changes in relative concentration ranged from 0.29 to 2.03). We also found eight features with negative fold changes and negative loadings (< −0.07) on the second principal component indicating that this might be a suite of compounds that are characteristic of non-stressed E. huxleyi DHB607. However, it is important to note that the individual relative concentrations of these features were not significantly altered due to grazing, despite being relevant in the PC model. In addition, OPLS-DA was unable to sufficiently classify grazed and prey only exometabolomes, with 38% cross validation error, and no features from this model had selectivity ratios >1.0.
Exometabolomic Profiles E. huxleyi Strains are Diverse
One striking result was that there was no metabolite, which remained after our stringent feature filters, common to all E. huxleyi strains. However, a small number of chemical features were shared by more than one strain (Figure 4). The strains that shared the largest number of features were the non-calcified strains CCMP379 and CCMP374, which shared six features, while both strains shared four of these features with strain DHB607 (Figure 4). Concentrations of five of these shared features significantly increased in response to grazing in strains CCMP374 (while only one increased in concentration in CCMP379). However, all six (including the four metabolites also detected in extracts of DHB607) were additionally found in O. marina only cultures, as well as E. huxleyi only cultures (Supplementary Tables 2, 4, 7), suggesting they are common to marine eukaryotic cells.

Figure 4. Venn diagram displaying shared metabolites (chemical features) detected in exometabolomes of four Emiliania huxleyi strains. Exometabolomes were generated independently for each E. huxleyi strain.
When we considered the effects of grazer-specific rates on the concentrations of shared metabolites, we found several that correlated with grazer activities. For instance, concentrations of one feature, m/z 144.0479, (retention time 5 min) were positively correlated to O. marina grazing coefficients on CCMP374 (Pearson correlation coefficient r = 0.6355, p = 0.048), CCMP379 (r = 0.6860, p = 0.0096), and DHB607 (r = 0.9387, p < 0.0001). Concentrations of this metabolite also significantly increased in response to grazing in strains CCMP374 and CCMP379 when pooled across all treatments. After determining the molecular formula of this feature as C6H9NOS (molecular ion [M+H]+), we then annotated this metabolite as 4-methyl-5-thiazoleethanol, based on both retention time and molecular weight matches (mass accuracy 0.0–1.38 delta ppm for all strains) to the authentic standard (Supplementary Figure 3). Although concentrations of this metabolite correlated with grazing coefficients for three of our four E. huxleyi strains, it displayed variable expression patterns when considering other O. marina activities. For example, in strain CCMP374 concentration of this metabolite did not correlate with O. marina ingestion (r = 0.4893, p = 0.1512) nor growth rates (r = 0.6160, p = 0.058), yet was positively correlated with these O. marina rates in CCMP379 (r = 0.8728–0.9342, p < 0.0001 for both). However, concentrations negatively correlated with O. marina ingestion rates on strain DHB607 (r = −0.7046, p = 0.0072), indicating that concentrations of this metabolite are not altered uniformly across all strains in response to grazing pressure.
Strain specific responses to variable grazing pressures were also observed when considering the other shared metabolites between these three strains. Concentrations of three metabolites, m/z 181.1089 (C8H12N4O, retention time 2 min), m/z 265.1295 (C12H16N4O3, retention time 4 min), and m/z 297.1015 (C12H16N4O3S, retention time 4 min), positively correlated with O. marina grazing coefficients in E. huxleyi strains CCMP374 (r = 0.7409−0.9502, p < 0.02 for all) and CCMP379 (r = 0.5817–0.6759, p < 0.02 for all), and were either positively or not correlated to O. marina ingestion and growth rates. Concentrations of one feature, m/z 265.1294 (retention time 5 min) was positively correlated with grazing, ingestion, and growth rates (r = 0.6320–0.9074, p < 0.02 for all rates) in CCMP379 while being negatively correlated with these rates in extracts of CCMP374 (r = −0.8263 − −0.9398, p < 0.003 for all). The calcified strains DHB607 and DHB624 shared one common feature m/z 549.7961; retention time 17 min; Figure 4), concentrations of which were significantly greater in extracts of grazed DHB624 cultures, but were unaffected in strain DHB607. Due to low ion abundances and isobaric interferences in this region of the spectra, we were unable to determine a plausible chemical formula for this feature. Concentrations of this feature also positively correlated with O. marina grazing rates on both DHB607 and DHB624 (r = 0.6769, p = 0.011; r = 0.7019, p = 0.0075, respectively) suggesting that this metabolite may have been produced by grazing O. marina.
We also detected chemical features that appeared to be E. huxleyi specific, although they had concentrations that were not impacted by grazing. One feature (m/z 357.2734, C15H32N8O2, retention time 18 min), common to both CCMP379 and CCMP374, was not found in the exometabolomes of O. marina cultures indicating that this feature is specific to E. huxleyi. However, concentrations of this metabolite did not significantly differ (p = 0.047, p = 0.046 t-test with FDR correction) between grazed and control cultures, despite a sometimes large, positive fold change in response to grazing (fold change 0.59 and 9.55). Additionally, peak areas of this metabolite did not correlate with grazing coefficients, ingestion rates, or predator growth rates (p > 0.20 for all correlations) for either strain. There was one metabolite shared between CCMP379 and DHB624, however retention time of this feature experienced a 2 min shift between strains. Concentrations of this metabolite also did not differ between grazed and control exometabolomes; however this feature was not found to be produced by O. marina monocultures, qualifying it as a candidate for an E. huxleyi specific metabolite.
Discussion
Exometabolomes of E. huxleyi strains were found to be more different than they were alike, as determined from comparing the retention times and m/z of the chemical features detected in extracts. Overall, only a small number of common metabolites were shared among E. huxleyi strains, and in fact no features were shared among all four strains. Despite this, non-calcified strains (CCMP374 and CCMP379) produced more similar exometabolomes than calcified cells, with six shared chemical features (24–25% of metabolites found for strains CCMP374 and CCMP379, respectively). While previous studies have reported large chemical diversity in metabolomes and exometabolomes of phytoplankton (Barofsky et al., 2009; Paul et al., 2009; Huseby et al., 2012; Becker et al., 2014), by reason, more closely related organisms produce more similar exometabolomes. For instance, Becker et al. (2014) found that roughly 12–15% of metabolites detected in diatom exudates from two different Thalassiosira species were shared, while only 2% of all metabolites detected were shared between Thalassiosira and another diatom species. Further, < 15% of metabolites were shared among exometabolomes of different cyanobacterial strains, and cyanobacterial exometabolomes were more similar (in terms of shared metabolites) to each other than to diatom exometabolomes (Becker et al., 2014). Huseby et al. (2012) showed that metabolomes of 19 different Chaetoceros socialis strains are distinguishable based on isolate source (in this case North Atlantic vs. the Mediterranean Sea); however, these authors compared internal metabolite pools as opposed to exometabolomes.
The majority of the features detected in our experimental extracts were found in the seawater controls, representing 50–97% of detected features, depending on E. huxleyi strain, echoing the importance of carefully subtracting seawater background. Similarly, when applying stringent requirements, Macintyre et al. (2014) found that up to 70% of detected metabolites could be traced to media controls. Clearly, aspects such as the choice of growth medium, mass spectrometry platform (impacting resolution and mass accuracy as well as ionization efficiencies), spectral processing, and contamination removal criteria greatly influences the number of observed signals in each sample, highlighting the absolute requirement for appropriate medium blanks in each metabolomics study. We hypothesize that the small number of compounds described in these exometabolomes is also related to selective pressures that may exist to reduce a cell's chemical signature in the water column. For instance, the release of chemical compounds into the environment, particularly metabolites that signal to conspecifics, carries the ecological cost of attracting predators (Wolfe, 2000; Strauss et al., 2002; Seymour et al., 2010). These ecological costs have been studied in terrestrial predator-prey interactions (e.g., Wertheim et al., 2005), and have been suggested to exist in marine planktonic environments (Pohnert et al., 2007).
To our knowledge, studies comparing E. huxleyi exometabolomes have not been performed, however, E. huxleyi strains are known to vary in DMSP-lyase activity, resulting in differential production of DMS and acrylate, which appears to influence grazer preference on different E. huxleyi strains (Wolfe and Steinke, 1996; Wolfe et al., 1997; Strom et al., 2003a). Lipidomic studies have also shown differences in lipid composition among E. huxleyi strains (Rose et al., 2014) which appear to correlate with sensitivity of E. huxleyi strains to viral infection (Fulton et al., 2014). In addition, strains of E. huxleyi demonstrate large variability in gene expression based on strain identity and life stage (haploidy vs. diploidy, for example) (Rokitta et al., 2011), response to abiotic stressors, such as nitrogen limitation (Rokitta et al., 2014) and acidification (Rokitta et al., 2012). Our current study, along with Harvey et al. (2015), were designed to maximize phenotypic variability in E. huxleyi by choosing strains with varying levels of both calcification and DMSP production. Given this experimental design, as well as the inherent phenotypic variability within this species, it is not surprising that we also detected inter-strain variability in E. huxleyi metabolic footprints. Future work could further describe the variability of these exometabolomes within a strain over time, as well as the consistency of these responses to grazing. Potentially this variability could relate to the ecological success of E. huxleyi in the global ocean.
In addition to describing unique exometabolomes among E. huxleyi strains, we found that exposure to grazing influences the composition of metabolites released into the environment for at least two strains of E. huxleyi, DHB624, and CCMP374 (Figures 2, 3), which experienced similar levels of moderate grazing (Figure 1). Exometabolomes of strains CCMP379 and DHB607 were less variable (Figures 2, 3), and these strains experienced high and low levels of grazing, respectively, (Figure 1). There may be several strain-specific explanations for this lack of variability observed between grazed and not grazed samples. For CCMP379, the high levels of grazing observed on this resulted in an unusual technical challenge—there simply were not enough E. huxleyi cells remaining to normalize samples to cell number, resulting in our volumetric normalization approach (detailed in methods). Separate analyses revealed that a normalization approach had little impact on the overall differences in exometabolomes among strains (i.e., each strain still responds uniquely to grazing, Supplementary Figure 4), however, a volumetric normalization approach may have shielded us from detecting differences in exometabolomes of CCMP379. For strain DHB607, many E. huxleyi cells were present at the time of sampling. It is possible that the lack of grazing resulted in minimal alterations in DHB607 exometabolomes, due to reduced stress of DHB607 and minimal damage to E. huxleyi cells. Additionally the relative contribution of O. marina metabolites would be minimal in extracts from grazed DHB607, as O. marina populations either did not grow or declined during the course of the experiment (Supplementary Figure 1). Thus, it appears that most chemical features detected in DHB607 extracts are indeed E. huxleyi metabolites.
Our grazing results suggest E. huxleyi strain DHB607 is defended from grazing, as it was grazed the least of all four strains tested, and O. marina populations experience poor growth in response to diets of pure DHB607 (Supplementary Figure 1; Harvey et al., 2015). Because we did not observe significant differences in the exometabolomes of grazed DHB607 vs. control DHB607 cultures, it is likely that any E. huxleyi metabolites that influenced this grazing interaction were either constitutively expressed or maintained intracellularly. However, our preliminary tests exposing O. marina to both exudates and cellular extracts of DHB607 did not inhibit O. marina growth, nor grazing on palatable strains of E. huxleyi (data not shown). An alternative hypothesis is that DHB607 does not produce the particular cue(s) that O. marina may use to detect prey, essentially allowing DHB607 cells to “fly under the radar,” and avoid predation. Yet, because grazing on DHB607 occurred mainly at the lowest E. huxleyi concentrations tested (when encounter rates between predator and prey were minimal), and because diets of DHB607 occasionally caused O. marina to perish, it is more likely that DHB607 is chemically defended from grazing. Although the role that calcification plays in E. huxleyi—protist grazing is still unclear, grazing on calcified strains has been shown to inhibit grazer growth rates (Harvey et al., 2015). In light of the fact that grazing on calcified strains still occurs, synergistic or additive effects of calcification and toxicity cannot be discounted, and this phenomenon warrants further study. Indeed, several other studies have shown that inter-strain variation in phytoplankton chemical defenses exist (Teegarden et al., 2008), and that strains of E. huxleyi vary in calcification, DMSP production, and palatability (e.g., Hansen et al., 1996; Wolfe and Steinke, 1996; Strom et al., 2003a).
Strikingly, there appears to be little similarity in how multiple metabolic profiles of the E. huxleyi strains changed in response to O. marina grazing. The limited number of shared chemical features suggests that there are few candidate biomarkers for protist grazing on E. huxleyi. Nevertheless, among the exometabolomes of the non-calcified strains CCMP374 and CCMP379, there appear to be five metabolites that are indicative of O. marina grazing on non-calcified strains. The relative concentrations of some of these potential biomarkers also correlated with O. marina activity (i.e., grazing coefficients, ingestion rates, and growth rates), suggesting that they could be derived from grazing interactions. For instance, 4-methyl-5-thiazoleethanol, a thiamine precursor produced by E. huxleyi (Mcrose et al., 2014; Rose et al., 2014; Supplementary Figure 3), was observed in exometabolomes of strains CCMP374, CCMP379, and DHB607, and concentrations of this metabolite positively correlated with grazing rate. Although E. huxleyi can produce 4-methyl-5-thiazoleethanol, this may be a strain dependent trait (Read et al., 2013) and E. huxleyi is unable to grow when provided with the B1 vitamin precursor alone (Mcrose et al., 2014; Rose et al., 2014). Therefore, the role of this molecule is unknown. Regardless, this metabolite appears to be a direct indicator of grazing, and may simply act as a marker for E. huxleyi cell death, or is a transformation product produced by O. marina. It is interesting to consider that grazing on E. huxleyi releases vitamin precursors to the upper ocean, potentially recycling key organic nutrients through the water column. Regardless, this observation does highlight once again that E. huxleyi strains respond uniquely to grazing, and that ultimately, there may be genetic underpinnings to the distribution of particular components of the marine DOM pool.
Previous studies have shown that physiological state (e.g., growth phase) and environmental conditions influence the profiles of both exudates and internal metabolites of eukaryotic phytoplankton (e.g., Bölling and Fiehn, 2005; Kluender et al., 2009; Paul et al., 2009; Huseby et al., 2012; Vidoudez and Pohnert, 2012; Mausz and Pohnert, 2015). This highlights the need to consider multiple physiological states of both predators and prey when investigating the response of exometabolomes which are likely to vary drastically in both the composition and relative concentrations of metabolites with perturbation. Biotic factors, including co-culturing with competitor phytoplankton also influence the internal metabolomes of phytoplankton (Poulson-Ellestad et al., 2014) and the profiles of released metabolites (Paul et al., 2009). Therefore, it is not surprising that stress from exposure to grazers as well as the grazing events themselves alter the composition of released metabolites by plankton, as has been demonstrated qualitatively elsewhere (Kujawinski et al., 2004).
Our study presents clear evidence that ecological interactions, such as protist grazing, alter the characteristics of released organic matter from plankton. This is evident despite the fact that extraction techniques for dissolved organic matter from seawater are highly variable (Dittmar et al., 2008; Keil and Neibauer, 2009), and we are unable to report extraction efficiencies for our own experiments. Additionally, we only focused our efforts on low molecular weight (i.e., < 3000 Da), waterborne compounds, limiting our ability to discuss the impacts of grazing on the abundances of membrane-bound and high molecular weight compounds. Presumably grazing would also influence the abundance and distribution of such molecules. Yet even from our untargeted footprinting approach, it is clear that these exuded metabolites vary in size and polarity and likely contain heteroatoms, suggesting that they are produced from a variety of metabolic pathways. With our subtractive approach, we were also able to classify which metabolites O. marina and E. huxleyi cells (or their epibionts) likely produced by comparing profiles of grazed cultures to E. huxleyi and O. marina monocultures. Although we did detect some chemical features that were not produced in control monocultures, it is possible that many of these metabolites could be O. marina biotransformation metabolites, or metabolites that are produced by growing, non-stressed O. marina cells.
We also cannot discount the potential for epibionts, or other bacteria co-cultured with our test organisms, to have been the true source of the metabolites detected in the current study. As stated previously, the strains used in this study were not obtained or maintained axenically, but we did make significant efforts to follow up on possible epibiont mediated influences. For example, later tests have shown that O. marina grazing does not appear to be impacted by bacterial presence in E. huxleyi cultures, as similar grazing results have been observed for axenic cultures (E. Harvey, unpublished). In general, the role of bacteria contributing to these exometabolomes is unknown and requires further study. Other ecological interactions such as competition, viral infection, and bacterial attack will also presumably influence the exometabolomes of E. huxleyi, thereby further altering the pool of dissolved organic matter released by these organisms (Kujawinski, 2011).
In addition to the influence of grazing on these exometabolomes, it is also possible that compounds within these exometabolomes influence ecological interactions. For instance, these exometabolomes may contain compounds that serve as cues for O. marina grazers to locate appropriate food items. In contrast, these exometabolomes could also contain metabolites that may act as kairomones or alarm cues used by E. huxleyi to enhance cellular defenses (e.g., Selander et al., 2006; Senft-Batoh et al., 2015). Assessing these possibilities will provide insights into the selective pressures on marine microbes to produce, and detect, certain metabolites. To that end, at least one study suggests that a haploid E. huxleyi strain is able to induce an unidentified chemical or structural defense against grazing (Kolb and Strom, 2013). Another study found that haploid E. huxleyi cells are able to escape viral infection, likely because they have a different lipid chemistry, preventing viral particles from attaching to the haploid form of E. huxleyi cells (Frada et al., 2008). Together, this may suggest that haploid cells have a very different cell surface chemistry that mediates ecological interactions taking place at the cell surface. From our current study, strain CCMP379 is likely haploid, yet was grazed at a similar rate to other non-calcified E. huxleyi strains (Harvey et al., 2015). We also noted that the exometabolome of CCMP379 was most similar, (based on number of shared metabolites) to CCMP374, a non-calcified diploid strain. In order to understand fully the influence of haploid E. huxleyi metabolites on ecological interactions, future studies should investigate the similarity of haploid E. huxleyi metabolomes by comparing the exudate, and cellular chemistries of multiple haploid strains. Because of the observed inter-strain variability in E. huxleyi physiology and chemistry, this system provides a good model to probe the effects of exudate chemistry on ecological and evolutionary relationships among marine microbes.
Here we demonstrate that protist grazing influences the exometabolomes of E. huxleyi. Although the design of our study limits our ability to determine which metabolites were produced by grazing O. marina or E. huxleyi cells specifically, it is clear that grazing elicited a strain-specific impact on exometabolomes and transformed the constituents of dissolved organic matter. Our results also indicate that the waterborne “scent” of natural E. huxleyi populations is conditional upon both the genetic diversity of blooms as well as the prevalence of active grazers. This study sets a foundation for investigating the potential roles of specific predation cues, chemical defenses, and alarm cues that may mediate predation on E. huxleyi. Such investigations will enhance our predictive capabilities and mechanistic understanding of not only planktonic grazing interactions but also the influence of these interactions on large-scale processes, including nutrient cycling and carbon sequestration.
Author Contributions
KP, EH, MJ, and TM contributed to the experimental design and conception of the project; KP and EH acquired and analyzed data; KP, EH, MJ, and TM worked toward data interpretation and wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank D Beaudoin, K Rathjen, H Ranson, A Aicher, and Dr. K Whalen for assistance in the laboratory. Drs. A Vardi, BAS Van Mooy, and K Bidle provided helpful thoughts and comments. Dr. K Bidle graciously provided strains of E. huxleyi. Funding for this work was provided by the Gordon and Betty Moore Foundation, Grant #3301 awarded to A Vardi, BAS. Van Mooy, K Bidle, MJ, and TM. Additional funding for this work was provided by an award from the Flatley Discovery Lab to TM.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmars.2016.00001
References
Alcolombri, U., Ben-Dor, S., Feldmesser, E., Levin, Y., Tawfik, D. S., and Vardi, A. (2015). Identification of the algal dimethyl sulfide-releasing enzyme: a missing link in the marine sulfur cycle. Science 348, 1466–1469. doi: 10.1126/science.aab1586
Barofsky, A., Vidoudez, C., and Pohnert, G. (2009). Metabolic profiling reveals growth stage variability in diatom exudates. Limnol. Oceanogr. Methods 7, 382–390. doi: 10.4319/lom.2009.7.382
Becker, J. W., Berube, P. M., Follett, C. L., Waterbury, J. B., Chisholm, S. W., Delong, E. F., et al. (2014). Closely related phytoplankton species produce similar suites of dissolved organic matter. Front. Microbiol. 5:111. doi: 10.3389/fmicb.2014.00111
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate - a practical and powerful approach to multiple testing. J. R. Stat. Soc. B Methodol. 57, 289–300.
Bergkvist, J., Selander, E., and Pavia, H. (2008). Induction of toxin production in dinoflagellates: the grazer makes a difference. Oecologia 156, 147–154. doi: 10.1007/s00442-008-0981-6
Bidle, K. D., and Falkowski, P. G. (2004). Cell death in planktonic, photosynthetic microorganisms. Nat. Rev. Microbiol. 2, 643–655. doi: 10.1038/nrmicro956
Bölling, C., and Fiehn, O. (2005). Metabolite profiling of Chlamydomonas reinhardtii under nutrient deprivation. Plant Physiol. 139, 1995–2005. doi: 10.1104/pp.105.071589
Breckels, M. N., Roberts, E. C., Archer, S. D., Malin, G., and Steinke, M. (2011). The role of dissolved infochemicals in mediating predator-prey interactions in the heterotrophic dinoflagellate Oxyrrhis marina. J. Plankton Res. 33, 629–639. doi: 10.1093/plankt/fbq114
Calbet, A., and Landry, M. R. (2004). Phytoplankton growth, microzooplankton grazing, and carbon cycling in marine systems. Limnol. Oceanogr. 49, 51–57. doi: 10.4319/lo.2004.49.1.0051
Casotti, R., Mazza, S., Brunet, C., Vantrepotte, V., Ianora, A., and Miralto, A. (2005). Growth inhibition and toxicity of the diatom aldehyde 2-trans, 4-trans-decadienal on Thalassiosira weissflogii (Bacillariophyceae). J. Phycol. 41, 7–20. doi: 10.1111/j.1529-8817.2005.04052.x
Charlson, R. J., Lovelock, J. E., Meinrat, O. A., and Warren, S. G. (1987). Oceanic phytoplankton, atmopshperic sulphur, cloud albedo and climate. Nature 326, 655–661. doi: 10.1038/326655a0
Dittmar, T., Koch, B., Hertkorn, N., and Kattner, G. (2008). A simple and efficient method for the solid-phase extraction of dissolved organic matter (SPE-DOM) from seawater. Limnol. Oceanogr. Methods 6, 230–235. doi: 10.4319/lom.2008.6.230
Falkowski, P. G., Barber, R. T., and Smetacek, V. (1998). Biogeochemical controls and feedbacks on ocean primary production. Science 281, 200–206. doi: 10.1126/science.281.5374.200
Field, C. B., Behrenfeld, M. J., Randerson, J. T., and Falkowski, P. (1998). Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281, 237–240. doi: 10.1126/science.281.5374.237
Frada, M., Probert, I., Allen, M. J., Wilson, W. H., and de Vargas, C. (2008). The “Cheshire Cate” escape strategy of the coccolithophore Emiliania huxleyi in response to viral infection. Proc. Natl. Acad. Sci. U.S.A. 105, 15944–15949. doi: 10.1073/pnas.0807707105
Frost, B. W. (1972). Effects of size and concentration of food particles on the feeding behavior of the marine planktonic copepod Calanus pacificus. Limnol. Oceanogr. 17, 805–815. doi: 10.4319/lo.1972.17.6.0805
Fulton, J. M., Fredricks, H. F., Bidle, K. D., Vardi, A., Kendrick, B. J., DiTullio, G. R., et al. (2014). Novel molecular determinants of viral susceptibility and resistance in the lipidome of Emiliania huxleyi. Environ. Microbiol. 16, 1137–1149. doi: 10.1111/1462-2920.12358
Gillard, J., Frenkel, J., Devos, V., Sabbe, K., Paul, C., Rempt, M., et al. (2013). Metabolomics enables the structure elucidation of a diatom sex pheromone. Angew. Chem. Int. Ed. Engl. 52, 854–857. doi: 10.1002/anie.201208175
Guillard, R. R. L., and Hargraves, P. E. (1993). Stichochrysis-immobilis is a diatom, not a chyrsophyte. Phycologia 32, 234–236. doi: 10.2216/i0031-8884-32-3-234.1
Hamm, C. E., Merkel, R., Springer, O., Jurkojc, P., Maier, C., Prechtel, K., et al. (2003). Architecture and material properties of diatom shells provide effective mechanical protection. Nature 421, 841–843. doi: 10.1038/nature01416
Hansen, F. C., Witte, H. J., and Passarge, J. (1996). Grazing in the heterotrophic dinoflagellate Oxyrrhis marina: size selectivity and preference for calcified Emiliania huxleyi cells. Aquat. Microb. Ecol. 10, 307–313. doi: 10.3354/ame010307
Harvey, E. L., Bidle, K. D., and Johnson, M. D. (2015). Consequences of strain variability and calcification in Emiliania huxleyi on microzooplankton grazing. J. Plankton Res. 37, 1137–1148. doi: 10.1093/plankt/fbv081
Harvey, E. L., and Menden-Deuer, S. (2011). Avoidance, movement, and mortality: the interactions between a protistan grazer and Heterosigma akashiwo, a harmful algal bloom species. Limnol. Oceanogr. 56, 371–378. doi: 10.4319/lo.2011.56.1.0371
Hay, M. E. (2009). Marine chemical ecology: chemical signals and cues structure marine populations, communities, and ecosystems. Ann. Rev. Mar. Sci. 1, 193–212. doi: 10.1146/annurev.marine.010908.163708
Heinbokel, J. F. (1978). Studies on functional role of Tintinnids in Southern-California Bight.1. Grazing and growth-rates in laboratory cultures. Mar. Biol. 47, 177–189. doi: 10.1007/BF00395638
Hollywood, K., Brison, D. R., and Goodacre, R. (2006). Metabolomics: current technologies and future trends. Proteomics 6, 4716–4723. doi: 10.1002/pmic.200600106
Huseby, S., Degerlund, M., Zingone, A., and Hansen, E. (2012). Metabolic fingerprinting reveals differences between northern and southern strains of the cryptic diatom Chaetoceros socialis. Eur. J. Phycol. 47, 480–489. doi: 10.1080/09670262.2012.741714
Irigoien, X., Flynn, K. J., and Harris, R. P. (2005). Phytoplankton blooms: a 'loophole' in microzooplankton grazing impact? J. Plankton Res. 27, 313–321. doi: 10.1093/plankt/fbi011
Katajamaa, M., and Oresic, M. (2005). Processing methods for differential analysis of LC/MS profile data. BMC Bioinformatics 6:179. doi: 10.1186/1471-2105-6-179
Keil, R. G., and Neibauer, J. A. (2009). Analysis of cooking spices in natural waters. Limnol. Oceanogr. Methods 7, 848–855. doi: 10.4319/lom.2009.7.848
Kluender, C., Sans-Piche, F., Riedl, J., Altenburger, R., Hartig, C., Laue, G., et al. (2009). A metabolomics approach to assessing phytotoxic effects on the green alga Scenedesmus vacuolatus. Metabolomics 5, 59–71. doi: 10.1007/s11306-008-0139-x
Kolb, A., and Strom, S. (2013). An inducible antipredatory defense in haploid cells of the marine microalga Emiliania huxleyi (Prymnesiophyceae). Limnol. Oceanogr. 58, 932–944. doi: 10.4319/lo.2013.58.3.0932
Kubanek, J., Snell, T. W., and Pirkle, C. (2007). Chemical defense of the red tide dinoflagellate Karenia brevis against rotifer grazing. Limnol. Oceanogr. 52, 1026–1035. doi: 10.4319/lo.2007.52.3.1026
Kujawinski, E. B. (2011). The impact of microbial metabolism on marine dissolved organic matter. Ann. Rev. Mar. Sci. 3, 567–599. doi: 10.1146/annurev-marine-120308-081003
Kujawinski, E. B., Del Vecchio, R., Blough, N. V., Klein, G. C., and Marshall, A. G. (2004). Probing molecular-level transformations of dissolved organic matter: insights on photochemical degradation and protozoan modification of DOM from electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Mar. Chem. 92, 23–37. doi: 10.1016/j.marchem.2004.06.038
Long, J. D., Smalley, G. W., Barsby, T., Anderson, J. T., and Hay, M. E. (2007). Chemical cues induce consumer-specific defenses in a bloom-forming marine phytoplankton. Proc. Natl. Acad. Sci. U.S.A. 104, 10512–10517. doi: 10.1073/pnas.0611600104
Lurling, M. (1998). Effect of grazing-associated infochemicals on growth and morphological, development in Scenedesmus acutus (chlorophyceae). J. Phycol. 34, 578–586. doi: 10.1046/j.1529-8817.1998.340578.x
Macintyre, L., Zhang, T., Viegelmann, C., Martinez, I. J., Cheng, C., Dowdells, C., et al. (2014). Metabolomic tools for secondary metabolite discovery from marine microbial symbionts. Mar. Drugs 12, 3416–3448. doi: 10.3390/md12063416
Mausz, M. A., and Pohnert, G. (2015). Phenotypic diversity of diploid and haploid Emiliania huxleyi cells and of cells in different growth phases revealed by comparative metabolomics. J. Plant Physiol. 172, 137–148. doi: 10.1016/j.jplph.2014.05.014
Mcrose, D., Guo, J., Monier, A., Sudek, S., Wilken, S., Yan, S., et al. (2014). Alternatives to vitamin B-1 uptake revealed with discovery of riboswitches in multiple marine eukaryotic lineages. ISME J. 8, 2517–2529. doi: 10.1038/ismej.2014.146
Menden-Deuer, S., and Grunbaum, D. (2006). Individual foraging behaviors and population distributions of a planktonic predator aggregating to phytoplankton thin layers. Limnol. Oceanogr. 51, 109–116. doi: 10.4319/lo.2006.51.1.0109
Meunier, C. L., Hantzsche, F. M., Cunha-Dupont, A. O., Haafke, J., Opperman, B., Malzahn, A. M., et al. (2012). Intraspecific selectivity, compensatory feeding and flexible homeostatsis in the phagatrophic flagellate Oxyrrhis marina: three ways to handle food quality fluctuations. Hydrobiologia 680, 53–62. doi: 10.1007/s10750-011-0900-4
Miralto, A., Barone, G., Romano, G., Poulet, S. A., Ianora, A., Russo, G. L., et al. (1999). The insidious effect of diatoms on copepod reproduction. Nature 402, 173–176. doi: 10.1038/46023
Paul, C., Barofsky, A., Vidoudez, C., and Pohnert, G. (2009). Diatom exudates influence metabolism and cell growth of co-cultured diatom species. Mar. Ecol. Prog. 389, 61–70. doi: 10.3354/meps08162
Pohnert, G. (2005). Diatom/copepod interactions in plankton: the indirect chemical defense of unicellular algae. Chembiochem 6, 946–959. doi: 10.1002/cbic.200400348
Pohnert, G., Steinke, M., and Tollrian, R. (2007). Chemical cues, defence metabolites and the shaping of pelagic interspecific interactions. Trends Ecol. Evol. 22, 198–204. doi: 10.1016/j.tree.2007.01.005
Poulson-Ellestad, K. L., Jones, C. M., Roy, J., Viant, M. R., Fernández, F. M., Kubanek, J., et al. (2014). Metabolomics and proteomics reveal impacts of chemically mediated competition on marine plankton. Proc. Natl. Acad. Sci. U.S.A. 111, 9009–9014. doi: 10.1073/pnas.1402130111
Prince, E. K., and Pohnert, G. (2010). Searching for signals in the noise: metabolomics in chemical ecology. Anal. Bioanal. Chem. 396, 193–197. doi: 10.1007/s00216-009-3162-5
Rajalahti, T., Arneberg, R., Berven, F. S., Myhr, K., Ulvik, R. J., and Kvalheim, O. M. (2009). Biomarker discovery in mass spectral profiles by means of selectivity ratio plot. Chemometr. Intell. Lab. Syst. 95, 35–48. doi: 10.1016/j.chemolab.2008.08.004
Read, B. A., Kegel, J., Klute, M. J., Kuo, A., Lefebvre, S. C., Maumus, F., et al. (2013). Pan genome of the phytoplankton Emiliania underpins its global distribution. Nature 499, 209–213. doi: 10.1038/nature12221
Robertson, D. G., Reily, M. D., and Baker, J. D. (2007). Metabonomics in pharmaceutical discovery and development. J. Proteome Res. 6, 526–539. doi: 10.1021/pr060535c
Rokitta, S. D., De Nooijer, L. J., Trimborn, S., De Vargas, C., Rost, B., and John, U. (2011). Transcriptome analyses reveal differential gene expression patterns between the life-cycle stages of Emiliania huxleyi (Haptophyta) and reflect specialization to different ecological niches. J. Phycol. 47, 829–838. doi: 10.1111/j.1529-8817.2011.01014.x
Rokitta, S. D., John, U., and Rost, B. (2012). Ocean acidification affects redox-balance and ion-homeostasis in the life-cycle stages of Emiliania huxleyi. PLoS ONE 7:e52212. doi: 10.1371/journal.pone.0052212
Rokitta, S. D., Von Dassow, P., Rost, B., and John, U. (2014). Emiliania huxleyi endures N-limitation with an efficient metabolic budgeting and effective ATP synthesis. BMC Genomics 15:1051. doi: 10.1186/1471-2164-15-1051
Rose, S. L., Fulton, J. M., Brown, C. M., Natale, F., Van Mooy, B. A. S., and Bidle, K. D. (2014). Isolation and characterization of lipid rafts in Emiliania huxleyi: a role for membrane microdomains in host-virus interactions. Environ. Microbiol. 16, 1150–1166. doi: 10.1111/1462-2920.12357
Selander, E., Jakobsen, H. H., Lombard, F., and Kiørboe, T. (2011). Grazer cues induce stealth behavior in marine dinoflagellates. Proc. Natl. Acad. Sci. U.S.A. 108, 4030–4034. doi: 10.1073/pnas.1011870108
Selander, E., Thor, P., Toth, G., and Pavia, H. (2006). Copepods induce paralytic shellfish toxin production in marine dinoflagellates. Proc. R. Soc. B Biol. Sci. 273, 1673–1680. doi: 10.1098/rspb.2006.3502
Senft-Batoh, C. D., Dam, H. G., Shumway, S. E., Wikfors, G. H., and Schlichting, C. D. (2015). Influence of predator-prey evolutionary history, chemical alarm-cues, and feeding selection on induction of toxin production in a marine dinoflagellate. Limnol. Oceanogr. 60, 318–328. doi: 10.1002/lno.10027
Seymour, J. R., Simó, R., Ahmed, T., and Stocker, R. (2010). Chemoattraction to dimethylsulfoniopropionate throughout the marine microbial food web. Science 329, 342–345. doi: 10.1126/science.1188418
Smith, C. A., O'Maille, G., Want, E. J., Qin, C., Trauger, S. A., Brandon, T. R., et al. (2005). METLIN: a metabolite mass spectral database. Ther. Drug Monit. 27, 747–751. doi: 10.1097/01.ftd.0000179845.53213.39
Strauss, S. Y., Rudgers, J. A., Lau, J. A., and Irwin, R. E. (2002). Direct and ecological costs of resistance to herbivory. Trends Ecol. Evol. 17, 278–285. doi: 10.1016/S0169-5347(02)02483-7
Strom, S. L. (2008). Microbial ecology of ocean biogeochemistry: a community perspective. Science 320, 1043–1045. doi: 10.1126/science.1153527
Strom, S., Wolfe, G., Holmes, J., Stecher, H., Shimeneck, C., Lambert, S., et al. (2003a). Chemical defense in the microplankton I: feeding and growth rates of heterotrophic protists on the DMS-producing phytoplankter Emiliania huxleyi. Limnol. Oceanogr. 48, 217–229. doi: 10.4319/lo.2003.48.1.0217
Strom, S., Wolfe, G., Slajer, A., Lambert, S., and Clough, J. (2003b). Chemical defense in the microplankton II: inhibition of protist feeding by beta-dimethylsulfoniopropionate (DMSP). Limnol. Oceanogr. 48, 230–237. doi: 10.4319/lo.2003.48.1.0230
Teegarden, G. J., Campbell, R. G., Anson, D. T., Ouellett, A., Westman, B. A., and Durbin, E. G. (2008). Copepod feeding response to varying Alexandrium spp. cellular toxicity and cell concentration among natural plankton samples. Harmful Algae 7, 33–44. doi: 10.1016/j.hal.2007.05.010
Tillmann, U. (2004). Interactions between planktonic microalgae and protozoan grazers. J. Eukaryot. Microbiol. 51, 156–168. doi: 10.1111/j.1550-7408.2004.tb00540.x
van Den Berg, R. A., Hoefsloot, H. C. J., Westerhuis, J. A., Smilde, A. K., and van Der Werf, M. J. (2006). Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics 7:142. doi: 10.1186/1471-2164-7-142
Van Donk, E., Ianora, A., and Vos, M. (2011). Induced defences in marine and freshwater phytoplankton: a review. Hydrobiologia 668, 3–19. doi: 10.1007/s10750-010-0395-4
Vardi, A., Formiggini, F., Casotti, R., De Martino, A., Ribalet, F., Miralto, A., et al. (2006). A stress surveillance system based on calcium and nitric oxide in marine diatoms. PLoS Biol. 4:e60. doi: 10.1371/journal.pbio.0040060
Vidoudez, C., and Pohnert, G. (2008). Growth phase-specific release of polyunsaturated aldehydes by the diatom Skeletonema marinoi. J. Plankton Res. 30, 1305–1313. doi: 10.1093/plankt/fbn085
Vidoudez, C., and Pohnert, G. (2012). Comparative metabolomics of the diatom Skeletonema marinoi in different growth phases. Metabolomics 8, 654–669. doi: 10.1007/s11306-011-0356-6
Wertheim, B., van Baalen, E. J. A., Dicke, M., and Vet, L. E. M. (2005). Pheromone-mediated aggregation in nonsocial arthropods: an evolutionary ecological perspective. Annu. Rev. Entomol. 50, 321–346. doi: 10.1146/annurev.ento.49.061802.123329
Whalen, K. E., Poulson-Ellestad, K. L., Deering, R. W., Rowley, D. C., and Mincer, T. J. (2015). Enhancement of antibiotic activity against multidrug-resistant bacteria by the efflux pump inhibitor 3,4-dibromopyrrole-2,5-dione Isolated from a Pseudoalteromonas sp. J. Nat. Prod. 78, 402–412. doi: 10.1021/np500775e
Wolfe, G. V. (2000). The chemical defense ecology of marine unicellular plankton: constraints, mechanisms, and impacts. Biol. Bull. 198, 225–244. doi: 10.2307/1542526
Wolfe, G. V., and Steinke, M. (1996). Grazing-activated production of dimethyl sulfide (DMS) by two clones of Emiliania huxleyi. Limnol. Oceanogr. 41, 1151–1160. doi: 10.4319/lo.1996.41.6.1151
Keywords: dissolved organic matter, environmental metabolomics, grazing, metabolic footprinting, phytoplankton
Citation: Poulson-Ellestad KL, Harvey EL, Johnson MD and Mincer TJ (2016) Evidence for Strain-Specific Exometabolomic Responses of the Coccolithophore Emiliania huxleyi to Grazing by the Dinoflagellate Oxyrrhis marina. Front. Mar. Sci. 3:1. doi: 10.3389/fmars.2016.00001
Received: 19 September 2015; Accepted: 11 January 2016;
Published: 28 January 2016.
Edited by:
Marcelino T. Suzuki, Sorbonne Universities (UPMC) and Centre National de la Recherche Scientifique, FranceReviewed by:
Gerhard J. Josef Herndl, University of Vienna, AustriaIan Salter, Alfred Wegener Institute, Germany
Copyright © 2016 Poulson-Ellestad, Harvey, Johnson and Mincer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tracy J. Mincer, dG1pbmNlckB3aG9pLmVkdQ==
†Present Address: Kelsey L. Poulson-Ellestad, Department of Biology, Chemistry and Physical Sciences, Roosevelt University, Chicago, IL, USA;
Elizabeth L. Harvey, Department of Marine Sciences, Skidaway Institute of Oceanography, University of Georgia, Savannah, GA, USA
 Kelsey L. Poulson-Ellestad
Kelsey L. Poulson-Ellestad Elizabeth L. Harvey2†
Elizabeth L. Harvey2† Tracy J. Mincer
Tracy J. Mincer