- Department of Biological Sciences, University of Southern California, Los Angeles, CA, United States
Radiolarians are a group of ubiquitous, yet poorly understood, large protists that often harbor photosymbionts. We studied the solitary radiolarian Thalassicolla nucleata by analyzing the transcriptome of its holobiont. We found that T. nucleata contained two dinoflagellate symbionts, one photosymbiont Brandtodinium sp., and one putative Peridiniales parasite. Through comparisons of gene expressions of Brandtodinium sp. and those of a close relative from a free-living culture, we found that the Brandtodinium sp. maintained its photosynthetic activities, but altered its carbon metabolism dramatically in hospite. Gene expression data also suggested carbon and nitrogen exchange between the host and photosymbiont and that lectin-glycan interaction might play an important role in host-symbiont recognition.
Introduction
Radiolarians are single-celled, heterotrophic protists from the phylum Retaria of the supergroup Rhizaria. They are abundant and widespread throughout broad expanses of pelagic oceanic ecosystems, and can account for a significant portion of the planktonic community (Dennett et al., 2002; Biard et al., 2016). Defining characteristics for these species are very large cell sizes ranging from tens of micrometers to 1 mm (colonial species can form gelatinous ribbons > 1 m in length), skeletons (when present) composed of silica or strontium sulfate, and complex pseudopodial networks (Suzuki and Not, 2015). Though radiolarians are predatory (Anderson et al., 1984; Swanberg and Caron, 1991), they often harbor photosynthetic symbionts including a variety of organisms including dinoflagellates (Gast and Caron, 1996), prasinophytes (Gast et al., 2000), and haptophytes (Decelle et al., 2012). Radiolarians containing photosynthetic symbionts have been shown to have exceptionally high rates of photosynthesis, and it has been demonstrated that photosynthate from the photosymbionts is translocated to the host cytoplasm (Anderson et al., 1983; Caron et al., 1995). These studies imply that symbiont-derived photosynthates are an important source of nutrition for their hosts. It is estimated that roughly half of the radiolarian species in oceanic surface waters harbor photosynthetic symbionts, and primary production by the symbionts can account for up to 80% of the carbon budgets of the holobionts (Caron et al., 1995). Besides photosynthetic symbionts, heterotrophic protists (parasites) can also be found in radiolarian species. The most commonly observed parasites belong to marine alveolates Syndiniales, such as Merodinium, Solenodinium, and Syndinium (Anderson, 1983; Coats, 1999; Dolven et al., 2007).
Despite their ubiquitous and conspicuous presence in the open ocean, radiolarians and their symbionts have been poorly studied, in part because of the large size and highly delicate nature of their cell structure. The most common and well-known symbionts are a group of dinoflagellates originally named Scrippsiella spp., phylogenetically related to the photosymbionts found in the hydrozoan, Velella velella (Gast and Caron, 1996, 2001). Probert et al. (2014) found that photosymbionts from 30 polycystine radiolarian samples across three different orders collected worldwide all had nearly identical 18S rRNA sequences with each other, and with previously described symbionts of radiolarian hosts. These photosymbionts formed a new genus separate from free-living Scrippsiella spp., named Brandtodinium (Probert et al., 2014).
The mechanisms of the host-symbiont interaction in radiolarians are largely unknown. Because radiolarians are notoriously difficult to cultivate, knowledge on this topic is very limited compared to other common symbiosis systems involving algae such as those between cnidarians and Symbiodiniaceae. However, there have been at least two attempts to study radiolarian holobionts using genetic methods. Gast et al. (2003) recovered a few genes related to photosymbiosis from the solitary radiolarian, Thalassicolla nucleata, using suppression subtractive hybridization. More recently, Balzano et al. (2015) used high-throughput sequencing to obtain transcriptomes of three radiolarian holobionts bearing photosymbionts. The latter authors recovered thousands of genes, and concluded that the interactions of glycans and c-type lectins might be important in host/photosymbiont recognition in radiolarians.
In this study, we obtained the transcriptome of the T. nucleata holobiont using Next-Generation sequencing technologies. We observed not one but two dinoflagellates in single host cells, a photosymbiont and a putative parasite. Transcripts of the photosymbiont in hospite were bioinformatically separated and compared to those of a free-living culture of the same species. We found that genes related to cell growth and most carbon metabolism pathways were down-regulated in hospite, even though expression of photosynthesis genes were largely unchanged. We found evidence suggesting nutrient exchange, of both carbon and nitrogen, between the host and photosymbiont.
Materials and Methods
Collection of T. nucleata Holobionts
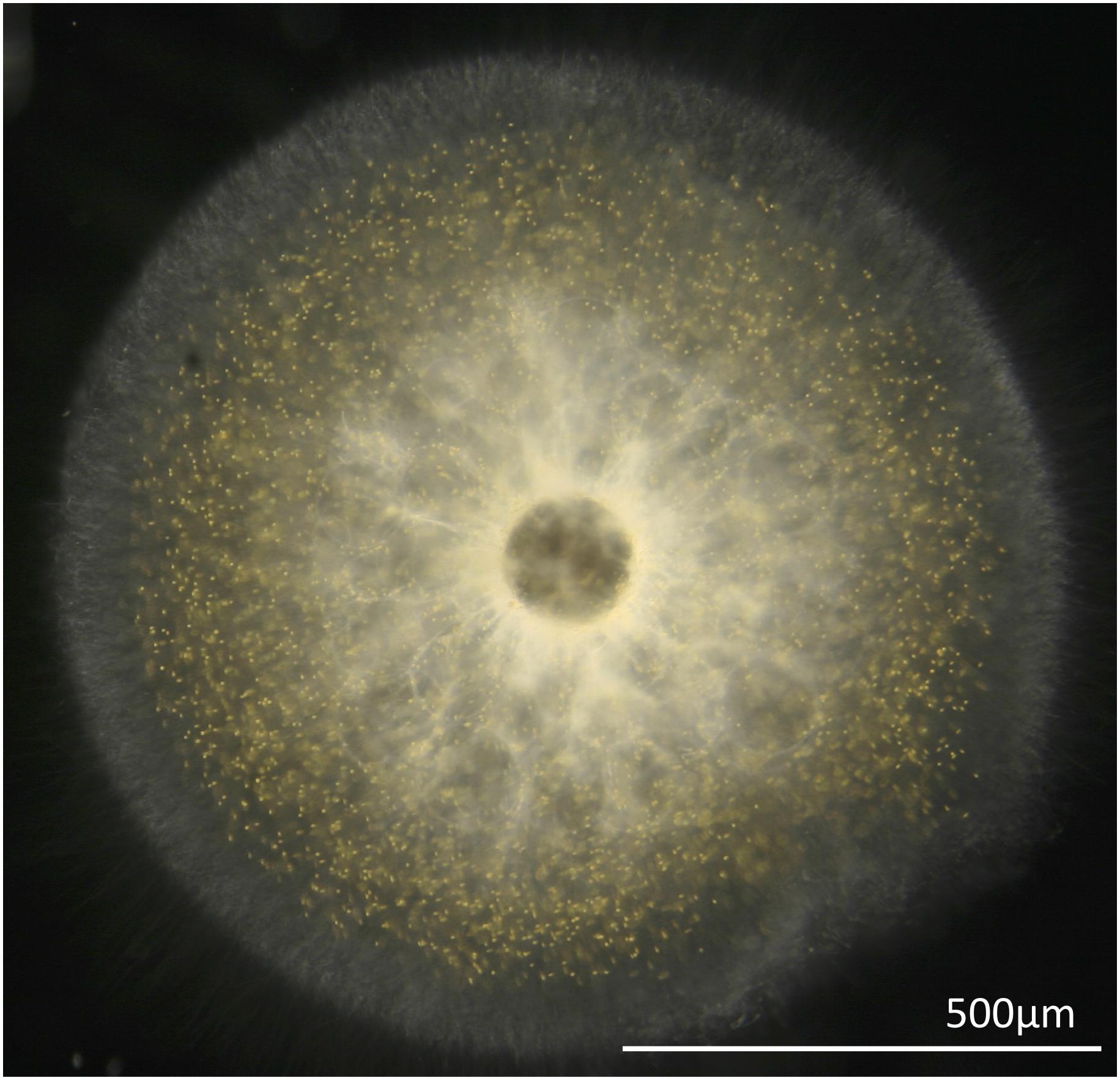
Single T. nucleata cells were collected from Station ALOHA (A Long-Term Oligotrophic Habitat Assessment; 2° 45′N, 158° 00′W) located 100 km north of Oahu, Hawaii aboard the R/V Kilo Moana. Plankton cells were gently collected by conducting drift tows (ship not underway) using a 1 mm mesh plankton net (Sea-Gear Corporation, Melbourne, FL, United States) from the side of the vessel for a duration of 10 min. Plankton cells were sorted manually from the plankton net tow using a dissecting microscope (Leica Wild M10 Stereo Dissecting Microscope). Solitary radiolarians were individually transferred into a multi-welled culture dish filled with 0.22 um-filtered seawater, incubated for 12 h, during which time they were moved twice to newly filtered seawater. This procedure allowed hosts to clear themselves of any prey and other organisms that might cause contamination in downstream analysis. All specimens subsequently used for sequencing recovered fully in the lab and exhibited large numbers of photosymbionts present in their pseudopodial networks (Figure 1).
RNA Extraction, Library Construction, and Sequencing
After undergoing a thorough wash with filtered seawater, four individual T. nucleata cells were transferred into a single sterile 2.0 ml cyrovial tube. Approximately 1.5 ml of RLT buffer (Qiagen, #79216, add 1% β-Mercaptoethanol) was added to the cyrovial, along with 0.2 μl of sterile beads (BioSpec, #1107915) and flash frozen in liquid nitrogen. Cryovials were stored at −80°C before processing. Further homogenization was facilitated through mechanical bead beating for 5 min and 20 shakes/min (TissueLyser II, Qiagen, #85300). Immediately following cell lysis, the lysate was transferred to a Qiagen RNeasy Micro Column and total RNA was isolated from the sample using Qiagen RNeasy Micro Kit following the manufacturer’s protocol. Total RNA integrity was assessed using a Fragment Analyzer (Advanced Analytical Technology). Total RNA had a RNA Quality Number (RQN) of 6.3 and quantified with Qubit 3.0 RNA High Sensitivity Assay. Total RNA was stored at −80°C until further processing.
cDNA synthesis and amplification were performed following the manufacturer’s protocol for the SMART-Seq V4 Ultra Low Input RNA kit (Clontech) with 18 cycles in the primary PCR. Most of the rRNA was removed during the process via poly-A tail selection. The resulting cDNA library was quality checked with Agilent 2100 Bioanalyzer using a High Sensitivity DNA chip and quantified with Qubit 3.0 DNA High Sensitivity Assay. The cDNA library was multiplexed and prepared for Illumina sequencing using Nextera XT Index Kit (Ilumina, # FC-131-1001) followed by a bead clean up (AMPure, Beckman Coulter #A63881). The sequencing was performed on a single lane of an Illumina HiSeq-2500 sequencer (UPC Genome Core, USC, Los Angeles, CA, United States). A total of 58.3 million paired-end reads were generated. The original data can be accessed through the Sequence Read Archive using the accession number SRP154215.
Assembly and Annotation
All reads were quality trimmed and filtered; Illumina adapter sequences were removed using Trimmomatic v. 0.32 (Bolger et al., 2014). Reads that could be mapped onto the PhiX genome were also removed (artifact from sequencing). All remaining reads were assembled using Trinity v. 2.1.1 (Grabherr et al., 2011) with minimum transcript length set as 250 bp. Reads were mapped back to the assembly and read counts were calculated using RSEM v. 1.2.29 (Li and Dewey, 2011) with bowtie2 (Langmead and Salzberg, 2012). Transcripts with less than 5 aligned reads were discarded. Ribosomal RNA transcripts were identified by BLASTN against the SILVA database release 123. Chloroplast and mitochondrial transcripts were identified by BLASTX and BLASTN against Heterocapsa triquetra, Breviolum minutum (formerly known as Symbiodinium minutum) and Scrippsiella spp. chloroplast and mitochondrial DNA and protein sequences found in Genbank (see Supplementary Material for sequences). These sequences were set aside and not analyzed (see Supplementary Table 1 for summary statistics of different types of transcripts).
The remaining sequences, presumably mRNA transcripts, were searched against a custom database containing all Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP) transcriptomes and additional aquatic protistan genomes and transcriptomes (details described in Hu et al., 2018) using DIAMOND v. 0.8.28 (Buchfink et al., 2015). For each transcript, all hits with a bitscore >90% of the highest bitscore were kept. If all those hits were from dinoflagellate sources, the transcript was considered a dinoflagellate transcript. The same procedure was performed to obtain radiolarian transcripts. The GC content of the resulting transcripts were plotted to show a clear difference between dinoflagellate and radiolarian transcripts.
A Gaussian Mixture Model with two components was fitted to the distribution of GC contents of all mRNA transcripts. All transcripts were then predicted to belong to one of the two components based on the model. Only predictions with >99% confidence were accepted, the rest were deemed uncertain and were not analyzed further.
A chimera identification and removal procedure was carried out for all dinoflagellate transcripts using methods and scripts described in Yang and Smith (2013) and using 30 Peridiniales transcriptomes found in MMETSP as reference. This procedure removed 468 transcripts.
Dinoflagellate transcripts were then aligned to Brandtodinium nutricula RCC 3387 (MMETSP1462) transcriptome assembly (Johnson et al., 2018) with BLASTN using a 90% identity cutoff and 50% coverage cutoff for the shorter sequence. Transcripts aligned to the B. nutricula transcriptome were considered Brandtodinium sequences. These cutoffs were determined using test transcriptomes of related dinoflagellates such as Scrippsiella spp., Heterocapsa spp., and Azadinium spinosum to ensure that the false positive rate of being identified as Brandtodinium was low (around 1%) while maximizing the number of transcripts identified as Brandtodinium (see Supplementary Table 2). Sequences not aligned to B. nutricula most likely belonged to a parasitic dinoflagellate, as described below, but could also contain some Brandtodinium transcripts since recruitment of Brandtodinium sequences using B. nutricula transcriptome was unlikely to be complete.
Coding and protein sequences were predicted from all mRNA transcripts using TransDecoder v. 2.0 (Haas et al., 2013). Functional annotation of the predicted proteins were obtained by HMM searches against Pfam and TIGRFAM databases, and by searches against KEGG database using KAAS server (Moriya et al., 2007).
18S rRNA Sequence Analysis
To validate the identity of the host and symbionts, nearly full length 18S rRNA sequences of the three organisms were PCR amplified from the extracted holobiont cDNA using primers 5′-GTACAAAGGACAGGGACGCA-3′ and 5′-TCCTGCCAGTAGTCATACGC-3′ for T. nucleata, 5′-ACACGGCAAAACTGCGAATG-3′ and 5′-GCGGCAGCTTTCAGGAACT-3′ for Brandtodinium, and 5′-ATACGGCGAAACTGCGAATG-3′ and 5′-TCCGCAGAAAAACTGGGTAA-3′ for the putative parasite. Primers were designed based on transcriptome assemblies. Amplicons were sequenced using Sanger sequencing. Sequences were aligned with other relevant dinoflagellate and radiolarian sequences that were also close to full length using MUSCLE (Edgar, 2004). The alignment was manually curated to remove poorly aligned positions defined as positions where more than half of the sequences were gaps. A phylogenetic tree was generated using the maximum likelihood method with default settings using MEGA7 (Kumar et al., 2016).
Comparison With B. nutricula Transcriptome
Original reads of the B. nutricula RCC3387 (MMETSP1462) transcriptome were downloaded from iMicrobe1. Its transcriptome assembly were downloaded from Johnson et al. (2018). The same pipeline of read alignment, read count generation, rRNA/chloroplast/mitochondria sequence identification, protein prediction and annotation described above was also applied to the MMETSP1462 transcriptome. To compare gene expression of the two different Brandtodinium species/strains, homologous gene clusters were first generated by using the MCL algorithm (Enright et al., 2002). Both transcriptome assemblies were combined, an all-vs.-all BLASTN was conducted with 90% identity cutoff and 50% coverage cutoff of the shorter sequence. Alignment identity multiplied by coverage of the shorter sequence was used as edge weights and an inflation parameter of 1.5 was used (Fischer et al., 2011). Only clusters with transcripts from both organisms were extracted and analyzed. Read counts of transcripts from the same organism and cluster were summed and used as input for differential gene expression analysis using edgeR v. 3.6 (Robinson et al., 2010) with a BCV of 0.4 (Chen et al., 2014). Gene clusters that were significantly (FDR < 0.05) differentially expressed between the two organisms were extracted.
Results
Holobiont Identities Based on 18S rRNA Sequences
Three 18S rRNA sequences with significant read counts were found in the de novo transcriptome assembly. In addition to the two expected sequences, which were those of T. nucleata and of Brandtodinium sp., the 18S rRNA sequence of a second dinoflagellate species was also found. The sequence of this second dinoflagellate is almost identical to those of the non-photosynthetic, potentially parasitic dinoflagellate previously found in T. nucleata (Gast, 2006). Additionally, the closest (98–99% similar) known relatives of this putative parasite were those found in two nassellarian species, Androcyclas gamphonyca and Ceratospyris hyperborea (Dolven et al., 2007). Phylogenetic analysis revealed that these putative parasites were not related to Syndiniales, which are widespread and well-known protistan parasites (Guillou et al., 2008), but are more closely related to other Peridiniales (Figure 2A). The 18S rRNA sequence of the photosynthetic symbiont was almost identical to those of B. nutricula (formerly known as Scrippsiella nutricula) found in radiolarians (Gast and Caron, 1996; Figure 2A). Sequences of many Brandtodinium species from the Probert et al. (2014) study were shorter and therefore not included in our phylogenetic tree, but they were also more than 99% similar to our photosymbiont sequence (Figure 2A). The 18S rRNA sequences of the two dinoflagellates within T. nucleata were ∼97% similar to each other.
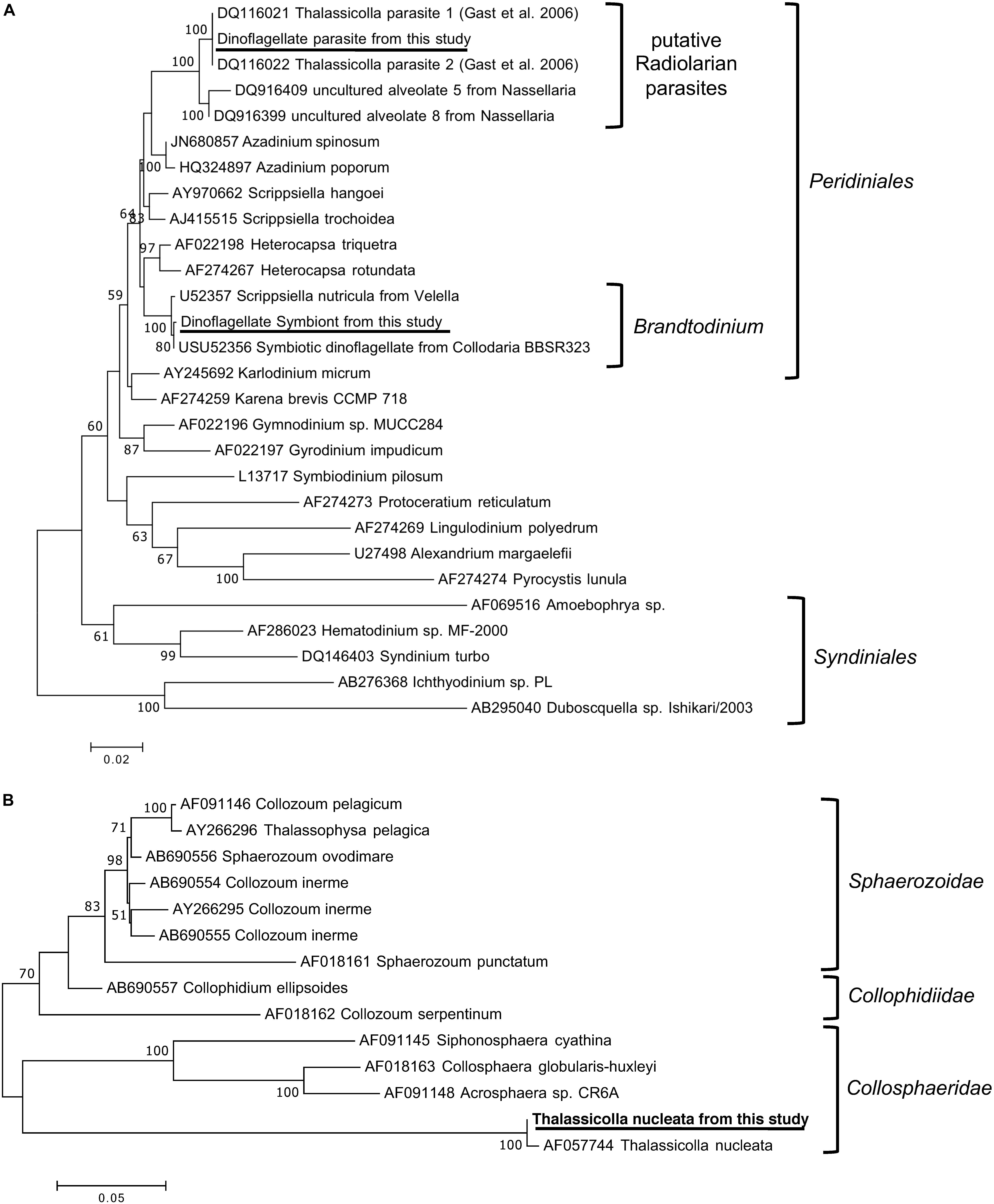
Figure 2. Phylogenetic tree of the 18S rRNA genes from (A) the two dinoflagellate symbionts and (B) T. nucleata, and related organisms. The trees were inferred by the maximum likelihood method based on the Tamura–Nei model. Branch lengths were drawn to scale, with branch lengths measured in the number of substitutions per site. The percentage of trees supporting the tree topology among 100 bootstrap trees were shown next to nodes, support below 50% was not shown. The analyses involved 28 (A) and 14 (B) nucleotide sequences and there were a total of 1561 (A) and 1145 (B) positions in the dataset. Sequences from this study were underlined. The reference sequences and taxonomic groupings illustrated in the figures were adapted from Gast (2006) and Biard et al. (2015).
The 18S rRNA sequence of T. nucleata was >99% similar to that of T. nucleata described in Zettler et al. (1998). We compared these two sequences with reference sequences of the three main groups of polycystine radiolarians according to Biard et al. (2015). T. nucleata did not appeared to be closely related to any of the reference sequences or any of the Thalassicolla sequences generated by Biard et al. (2015). Out of the three main groups, T. nucleata was more similar to Collosphaeridae than the other two (Figure 2B).
Transcript Binning
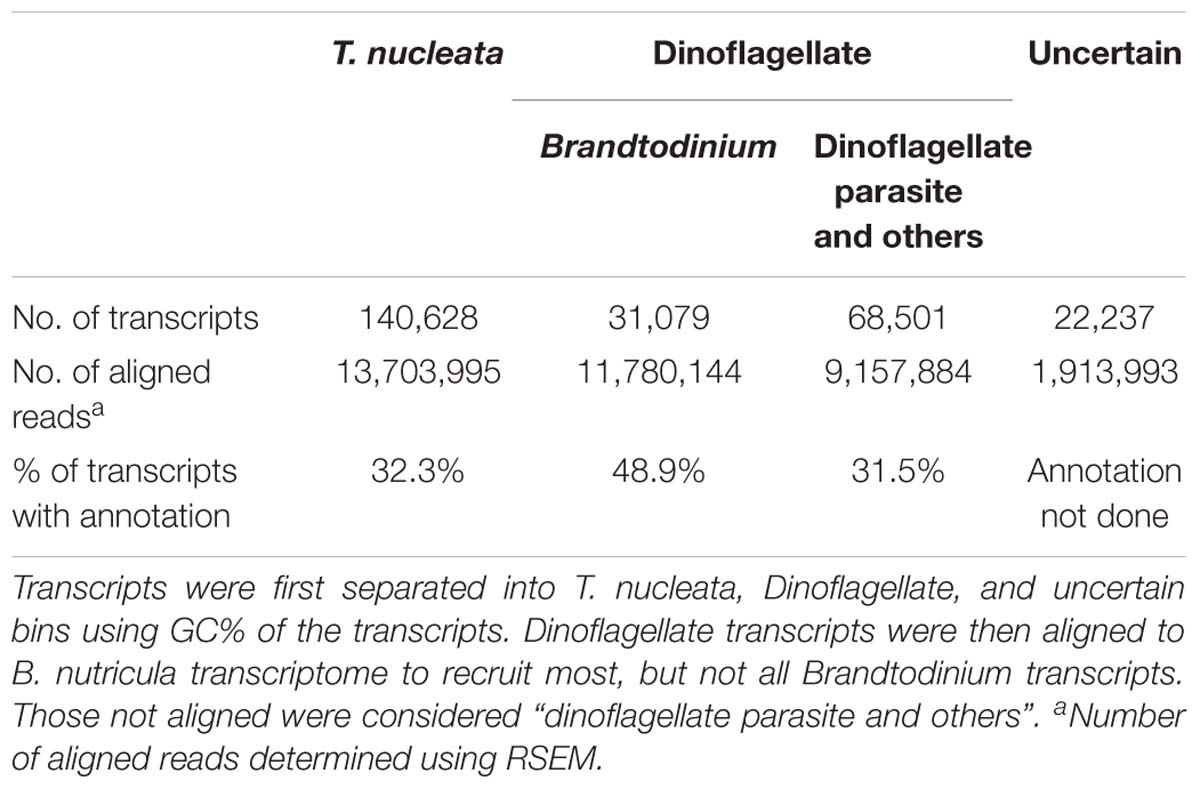
De novo assembly of the holobiont transcriptome generated 435,985 transcripts, totaling 310,563,069 bp, with a N50 of 996 bp. After filtering out transcripts with low read support, and separating transcripts of rRNA, chloroplast and mitochondrial origin, 262,445 transcripts remained. The GC content of filtered transcripts clearly exhibited a bimodial distribution (Figure 3A). The likely taxonomy of the origin of these transcripts was obtained by database searches. Transcripts that were most similar to dinoflagellates in the database had much higher GC% (66.0%) than those most similar to radiolarians (41.8%), and their combined GC% distributions were similar to that of the whole transcriptome (Figure 3B). Therefore, transcripts were statistically separated into three bins: the high GC% dinoflagellate bin, which consisted of 99,580 sequences, and the low GC% T. nucleata bin, which consisted of 140,628 sequences, and a third bin whose source could not be confidently assigned (Table 1).
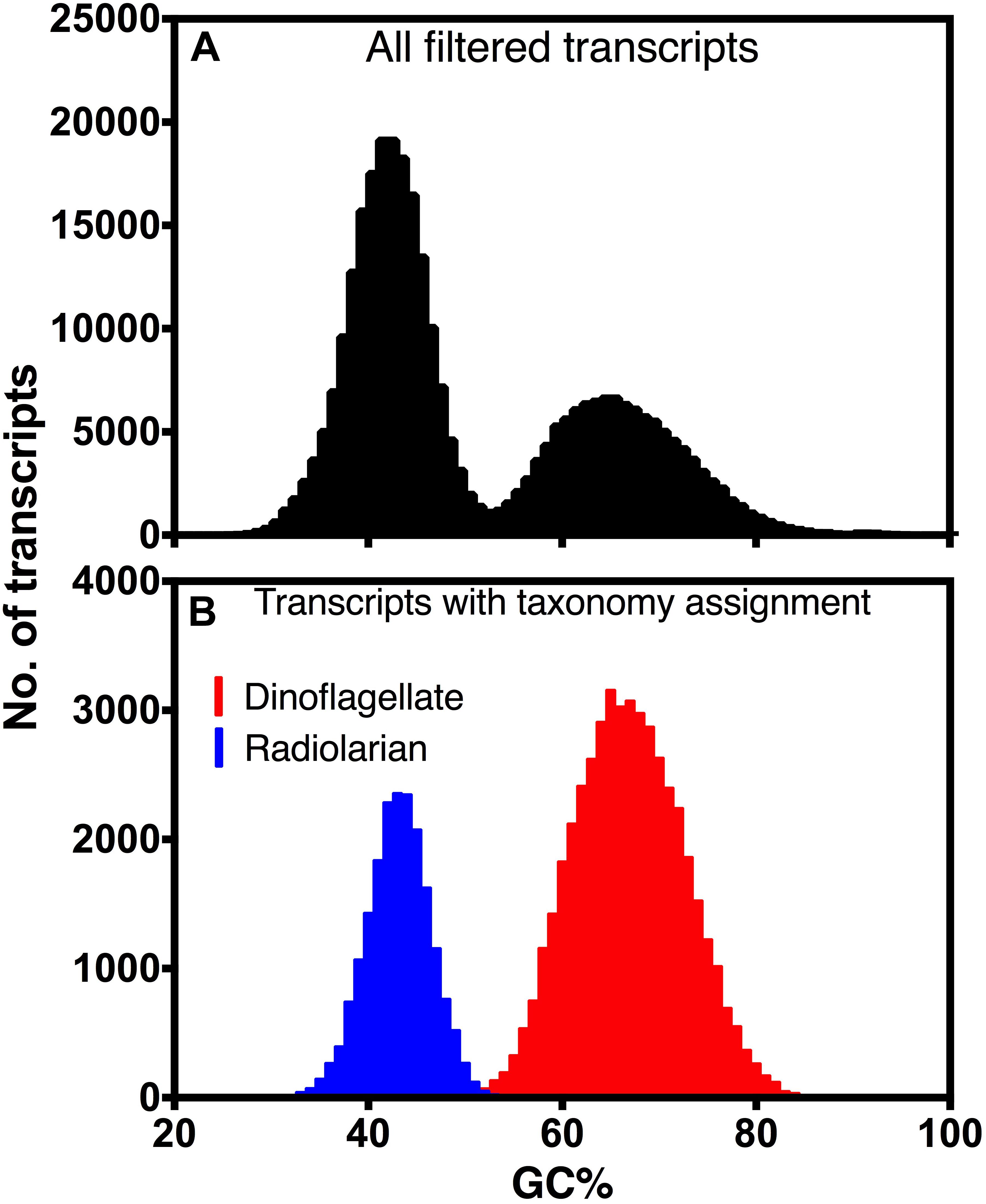
Figure 3. Distribution of GC content of (A) all assembled transcripts that met filtering criteria and (B) transcripts with taxonomy assignments of either dinoflagellate or radiolarian.
Brandtodinium transcripts were further separated from the dinoflagellate bin by aligning sequences to the transcriptome assembly of B. nutricula RCC 3387 in its free-living state. Sequences (31,079) that were highly similar (>90%) to those of B. nutricula RCC 3387 were considered Brandtodinium transcripts. The recruitment of sequences to the Brandtodinium reference is thought to be an underestimate as the reference transcriptome originates from a separate strain cultivated in a different living condition. Therefore, the sequences that were not recruited were considered a mixture of transcripts from Brandtodinium and the putative parasite. A number of other bioinformatic approaches including expression level-based and tetranucleotide frequency-based methods were attempted to separate the transcripts of the two dinoflagellates, but were unsuccessful because of the nature of transcriptome assembly (shorter sequences) and the similarity between the two organisms. A summary of the statistics of the different transcript bins is listed in Table 1.
Differential Gene Expression of Brandtodinium sp. in hospite
Relative gene expression levels of Brandtodinium sp. were compared with those of B. nutricula in the free-living state. Significant differences were observed in several pathways and functions. In all, 19,987 homologous gene clusters were compared between the two transcriptomes, and 8,598 of them had significantly different expression levels between the two (FDR < 0.05). Among them, 5,091 gene clusters had lower expression levels in hospite, while 3,507 had higher expression levels in hospite.
Almost all genes encoding eukaryotic ribosomal proteins were expressed at a lower level in hospite compared to the free-living state (Figure 4A). Other pathways related to RNA and protein production such as spliceosome, and aminoacyl-tRNA biosynthesis were also mostly down-regulated in hospite (Figures 4B,C). In contrast to eukaryotic ribosomal genes, genes encoding chloroplast and mitochondrial ribosomes had similar expression levels between the two lifestyles (Figure 4D). Among photosynthesis genes, more gene clusters were up-regulated (13) than were down-regulated (6) in hospite (Figure 4E). The Calvin cycle, which is responsible for photosynthetic carbon fixation, shares a lot of genes with other carbon metabolism pathways such as glycolysis and pentose phosphate pathway. Most Calvin cycle genes were expressed at lower levels in hospite, however, the most important and most highly expressed gene in the pathway, rbcL encoding RuBisCO, was unchanged between the two living states (labeled in Figure 4F). Most of the genes involved in carbon metabolism were expressed at a lower level in hospite. Specifically, most glycolysis/gluconeogenesis genes, including the fructose-1,6-bisphosphatase (fbp) which is the specific gene for gluconeogenesis, were down-regulated (Figure 4G). The same was true for most TCA cycle genes including citrate synthase (CS, Figure 4H). The majority of fatty acid biosynthesis genes exhibited the same pattern (Figure 4I).
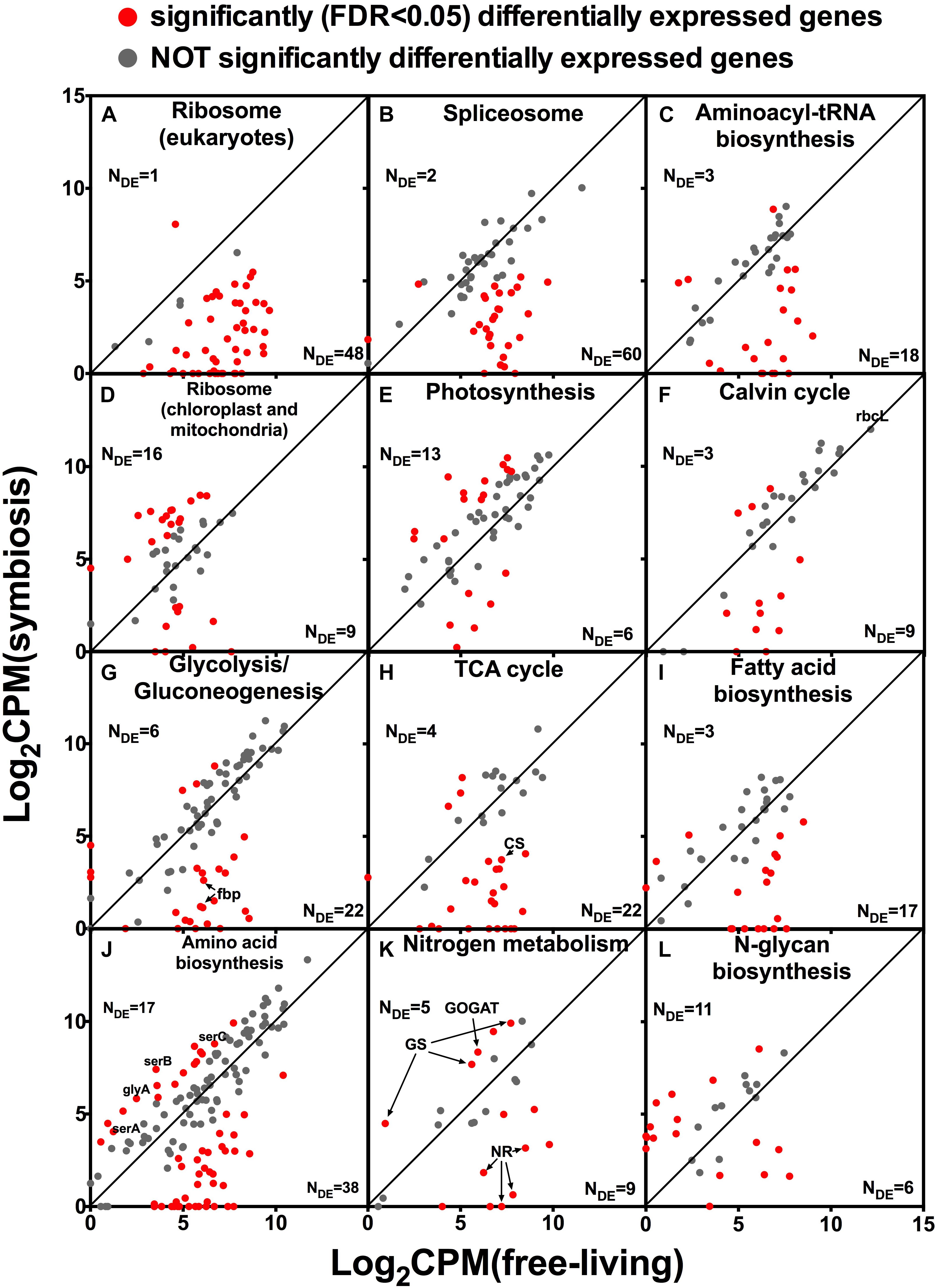
Figure 4. Relative expression levels of genes of selected pathways in Brandtodinium sp. in symbiosis with T. nucleata compared to those of B. nutricula in the free-living state. Log 2 values of counts per million (CPM) were plotted. Red dots represent gene clusters that were significantly differentially expressed between the two conditions. Gray dots indicate those that were not. The number of gene clusters that were either up-regulated or down-regulated (NDE) are shown. rbcL, Ribulose bisphosphate carboxylase large subunit; fbp, fructose-1,6-bisphosphatase; CS, citrate synthase; serA, 2-oxoglutarate reductase; serB, phosphoserine phosphatase; serC, phosphoserine aminotransferase; glyA, glycine hydroxymethyltransferase; NR, nitrate reductase; GS, glutamine synthetase; GOGAT, glutamate synthase.
Genes involved in amino acid biosynthesis were mostly down-regulated in hospite (Figure 4J), but a small number of them displayed increased expression levels, including the three genes responsible for serine biosynthesis, serABC, and glyA; the latter is responsible for glycine biosynthesis (Figures 4J, 5). Genes involved in nitrogen metabolism had varied expression patterns which appeared to be related to the different substrate with which they interact. Nitrate transporter, nitrate reductase, and nitrite reductase (NR) were all down-regulated in hospite. In contrast, genes responsible for ammonium uptake, namely glutatmine synthetase and glutamate synthase (GS and GOGAT), were up-regulated (Figure 4K). Among genes involved in glycan biosynthesis, for example, N-glycan biosynthesis (Figure 4L), more genes were up-regulated than were down-regulated. A similar trend was observed for O-glycan and glycan precursor biosynthesis (data not shown).
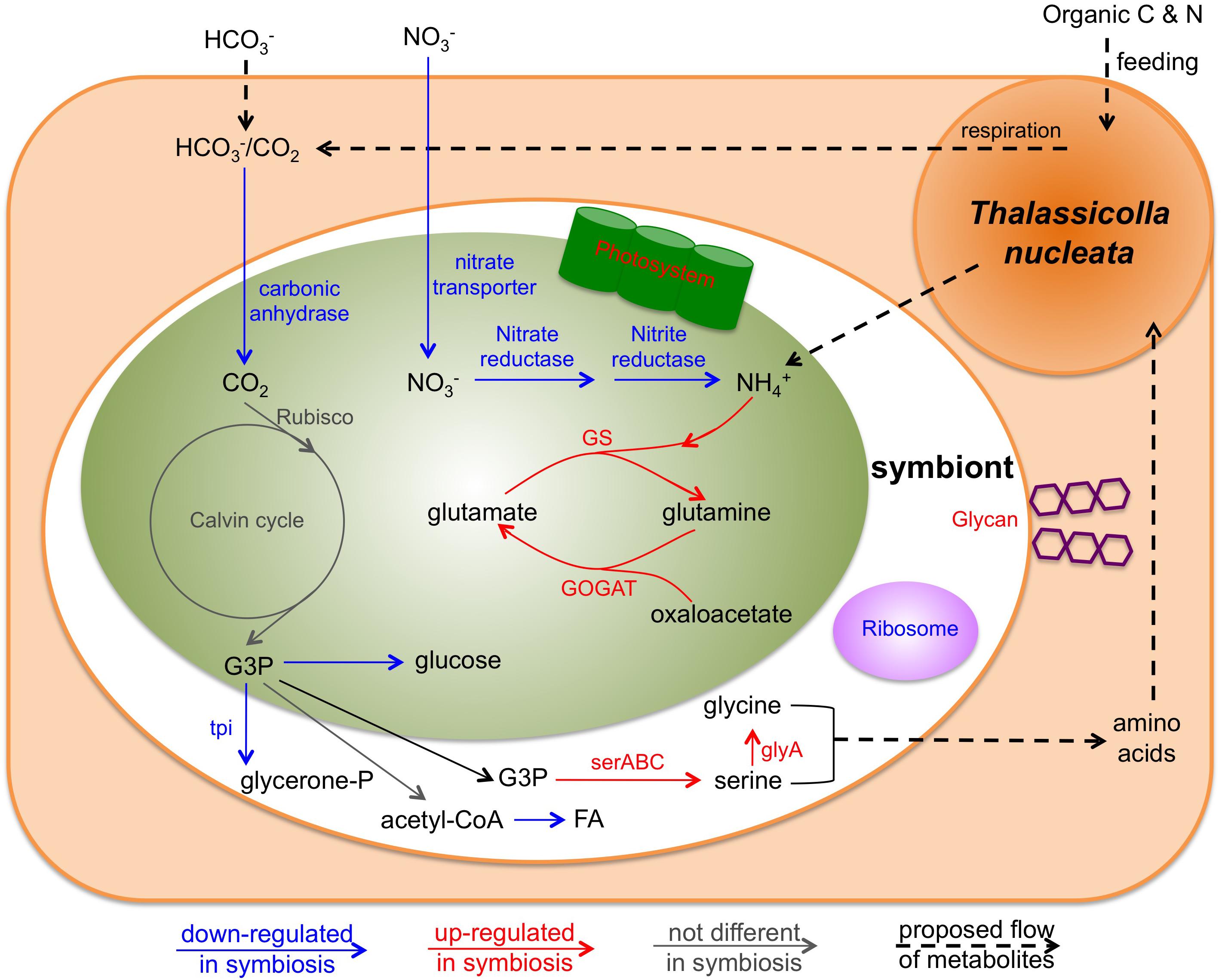
Figure 5. Conceptual model of the interaction of the holobiont involving T. nucleata and Brandtodinium sp. inferred from transcriptome data in this study. Blue arrows and text represent genes that were down-regulated in symbiosis compared to the free-living state; Red ones represent genes that were up-regulated; Gray ones represent those that were unchanged. tpi, Triose phosphate isomerase; serABC, 2-oxoglutarate reductase, phosphoserine phosphatase, and phosphoserine aminotransferase; glyA, glycine hydroxymethyltransferase; GS, glutmine synthetase; GOGAT, glutamate synthase.
T. nucleata Transcripts
Thalassicolla nucleata transcripts accounted for more than half of the filtered assembled transcriptome (Table 1), yet more than half of the transcripts did not have any functional annotation. Genes encoding structural proteins such as tubulin were among the most highly expressed genes. 56 transcripts encoding c-type lectins were found in the T. nucleata transcriptome and one of them was among the 100 most highly expressed transcripts. Several nitrogen metabolism genes including two glutamate dehydrogenases, two ammonium transporters, a glutamine synthetase, and an amino acid transporter were among the 500 most highly expressed transcripts (see Supplementary Table 3). Nitrate reduction genes were not found in the T. nucleata transcriptome.
Discussion
A Holobiont With Two Dinoflagellate Symbionts
The presence of photosymbionts and parasites in T. nucleata has been documented multiple times. Indeed, our identification of the photosymbiont, a Brandtodinium sp., was consistent with previous studies (Gast and Caron, 1996, 2001; Probert et al., 2014), as the rRNA gene sequence of the photosymbiont from the host in this study was nearly identical to sequences obtained in those studies. However, most observations of parasites in T. nucleata and other radiolarians have been most closely related to Syndiniales species, with only one exception. Gast (2006) reported a colorless dinoflagellate being released from disintegrated central capsules of T. nucleata. The putative parasite was not related to Syndiniales, but rather was closer to Peridiniales. That observation was confirmed in our study. The 18S rRNA sequence of this putative parasite was most closely related to two other dinoflagellates found in two different polycystine radiolarians (Dolven et al., 2007). This suggests the existence of a genus within Peridiniales that, similar to some Syndiniales species, specializes as parasites of radiolarians (Figure 2). It is not clear how widespread these parasites are, however, the study of Gast and Caron (1996) was conducted on specimens collected from the North Atlantic while specimens in the present study were collected in the North Pacific, implying a very broad geographic distribution. In an 18S rRNA survey of 30 different polycystine radiolarians, no parasite 18S sequences were reported (Probert et al., 2014), even though the primers used in the study should have amplified 18S rRNA of the parasites. One explanation for this is that the parasites were not detected simply because their particular hosts were not sampled, suggesting the relationships between the parasites and hosts are more specific than those between the photosymbionts and hosts. Nevertheless, our results showed that a Peridiniales parasite is a third player in the radiolarian-dinoflagellate association, at least in some species, and a potentially important interaction that should be considered in future studies.
Identifying the Photosymbiont Transcriptome
Studying the transcriptome of the radiolarian holobiont is a challenge because these associations are difficult to access with their exclusively open ocean distributions. Additionally, specimens are often a complex mixture of organisms (e.g., captured prey in their pseudopodial networks) (Balzano et al., 2015). As a result, we presently lack reference genomes/transcriptomes, especially of the radiolarians. In our study, there was a very large difference of GC% (>24%) between the radiolarian and dinoflagellate sequences. That difference allowed us to separate radiolarian from dinoflagellate sequences with confidence. Depending on the GC% of the host, this approach may be suitable for studying the transcriptomes of other radiolarian holobionts; while the GC% of Brandtodinium spp. in various polycystine hosts is expected to be similar to the 66% observed here, the GC% of radiolarians varies widely from ∼45 to ∼55% in different species (Balzano et al., 2015). Recently, a k-mer based similarity method showed promise in separating transcripts of host and photosymbiont in Collozoum sp. holobiont (Meng et al., 2018). Such approaches may be good alternatives when there is not enough separation in GC%.
Our tranascriptomic analysis was complicated by the presence of a putative parasite that was closely related to Brandtodinium sp. Fortunately, the B. nutricula transcriptome from MMETSP allowed us to recruit Brandtodinium sp. transcripts and compare them to those derived from the transcriptome obtained from a free-living culture. However, this comparison between the two Brandtodinium species had its own challenges. This comparison was in no way an ideal or even typical comparative transcriptome study. The strains compared, the media used, the sample collection and library preparation protocols, and the sequencing platforms in the two studies were all different. These differences could all contribute to differences in gene expression in ways that have nothing to do with symbiosis. For example, the two different strains could have different gene pools and different baseline gene expression; B. nutricula was grown in L1 media, which had far higher nitrate and phosphate concentration than surface ocean waters. In an effort to take these differences into account in our analysis, we used a very large biological coefficient of variation (BCV) when comparing gene expression. A larger BCV is suitable for comparing different individuals of the same species for statistical analyses, allowing us to account for the larger systemic noises and biases caused by those differences. Additionally, we largely drew our inferences based on patterns observed in pathways and groups of genes, not individual genes. When discussing the reasons behind an observed gene expression pattern, we always try to consider the above-mentioned difference first (see below). Nevertheless, we recognize that conclusions of gene expression patterns in this study will need to be verified in better controlled experiments in the future.
Metabolic Differences of the Photosymbiont in hospite
The most remarkable differences in gene expression of Brandtodinium sp. between the free-living and symbiotic states was the dramatic decrease of genes involved in RNA and protein synthesis in hospite. An overwhelming majority of ribosome genes were down-regulated in hospite (48 out of 52 genes, Figure 4A). The expression patterns of spliceosome genes and aminoacyl-tRNA biosynthesis genes were similar to that of ribosome genes in that they were also largely down-regulated (Figures 4B,C). Taken together, these results imply that the growth of Brandtodinium sp. is suppressed inside the host, a finding that is consistent with reports from other host-photosymbiont systems. In cnidaria-Symbiodiniaceae symbiosis, Symbiodiniaceae grow much slower in hospite than in culture, and it is hypothesized that a reduced growth rate might be imposed and regulated by the host (reviewed by Davy et al., 2012). However, it is important to recognize that the laboratory conditions under which the free-living B. nutricula were grown might have been much more favorable for growth and reproduction compared to conditions in the host’s cytoplasm. The growth rates of the symbionts in radiolarian hosts have not been measured, and it remains an interesting question whether intracellular existence in the host is beneficial or detrimental with respect to symbiont growth rates.
In contrast to the eukaryotic ribosomal genes, prokaryotic ribosomal genes (i.e., chloroplast and mitochondrial ribosomal genes) did not exhibit large changes in expression between the free-living and in hospite conditions (Figure 4D). This result suggested that the photosymbiont maintained similar activities in its chloroplast and mitochondrion relative to its free-living state. Consistent with this interpretation, the expression levels of photosynthesis genes were similar between the two conditions, if not slightly higher in hospite (Figure 4E). Expression of carbon fixation genes was more difficult to infer because the Calvin cycle largely overlaps with other carbon metabolism pathways. Yet the expression of the gene encoding RuBisCO, arguably the most important gene expressed in these pathways, remained unchanged between the two conditions (Figure 4F). These data suggested that photosynthetic activities were likely similar between the two conditions.
The significant discordance between the expression patterns of photosynthesis genes and cell growth-related genes suggested a different fate for fixed carbon in hospite. We examined various metabolic pathways that could act as a carbon sink for the photosymbiont. Most of the differentially expressed genes in glycolysis/gluconeogenesis pathway were down-regulated in hospite (Figure 4G), including the key gene in making sugar, fructose-1,6-bisphosphatase, and the key gene in making glycerol, triose phosphate isomerase (Figure 5). These findings imply that neither sugar nor glycerol were the likely destination of carbon fixed by the photosymbiont. Similar patterns (largely down-regulation in hospite) were observed in TCA cycle genes (Figure 4H) and fatty acid biosynthesis genes (Figure 4I), suggesting that respiration and fatty acid/lipid synthesis were also unlikely routes of increased organic carbon flux. While the expression levels of amino acid biosynthesis were lower in hospite overall (Figure 4J), genes involved in the synthesis of specific amino acids including serine and glycine were higher (Figure 5). Collectively, these data seemed to imply that certain amino acids might be produced more than others in the photosymbiont. Though, we caution this deduction was based on the observation of only a few genes, and would require more evidence in future studies.
Nutrient Exchange Between Host and Photosymbiont
The nature of organic carbon and nutrient exchange between hosts and their photosymbionts is one of the most central questions in understanding these associations. It has been concluded in other symbiotic interactions involving dinoflagellates that these photosymbionts released a variety of compounds including glucose, glycerol, small organic acids, amino acids, and lipids to the hosts (Wang and Douglas, 1999; Whitehead and Douglas, 2003; Davy et al., 2012). Similar processes (or some subset of them) are presumably occurring in radiolarian holobionts because these associations have exceptionally high rates of photosynthetic activity, with symbionts contributing up to ∼80% of the overall carbon budgets (Caron et al., 1995). However, as noted above, patterns of gene expression in the present study did not support an important role for sugar, glycerol or fatty acid biosynthesis as major pathways for production and translocation of organic carbon to the host in hospite. There was only limited support for increased production of specific amino acids. We therefore hypothesize that amino acids might be at least one of the predominant forms of translocated compounds to the host in T. nucleata (Figure 5).
If amino acids constitute the major form of translocated carbon from photosymbiont to host in the radiolarian, then a mechanism for transporting nitrogen back into the photosymbiont should also exist. In agreement with this expectation, we observed dramatic changes in the expression of nitrogen metabolism genes of the photosymbiont in hospite. Genes related to nitrate reduction were all down-regulated, but genes involved in ammonium uptake were all up-regulated (Figures 4K, 5). This was at least partially due to the fact that B. nutricula was grown in L1 media with nitrate as the only nitrogen source. Nevertheless, our data indicate that ammonium was an important part of the nitrogen source for Brandtodinium sp. during symbiosis. Ammonium is the most likely form of nitrogen produced from prey digestion by the host. High expression levels of several nitrogen metabolism genes in the host seemed to corroborate this speculation. Oligotrophic surface waters are often nitrogen-limited, and nitrate reduction is expensive. Therefore, ammonium provided by the host is likely a vital source of nitrogen for the intracellular photosymbionts, and a mechanism by which the host modulates the metabolism of its photosymbionts.
Another likely important factor controlling intracellular gene expression and metabolism of the photosymbiont is the high CO2/HCO3− concentration that may be present in the cytoplasm, promoting more efficient photosynthesis by the symbiont. We observed decreases in the expression of carbonic anhydrase genes in the symbiont in hospite (Figure 5). This finding implies a lessened requirement for a carbon-concentrating mechanism (CCM) during symbiosis, presumably due to the close proximity of CO2 produced by host respiration. In some other symbiotic systems such as the sea anemone-Symbiodiniaceae symbiosis, there is evidence that the host also possesses CCMs (Weis and Reynolds, 1999; Davy et al., 2012). We found more than 30 carbonic anhydrase transcripts in the host transcriptome, although none of them were highly expressed. It is therefore unclear if T. nucleata employs CCMs to influence photosymbiont photosynthesis.
Host-Symbiont Recognition
How hosts and symbionts recognize each other, or from a different perspective, how symbionts evade host digestion, is another fundamental question for heterotroph-alga symbiotic associations. Previous studies have concluded that interactions between c-type lectins and glycans play an important role in host-symbiont recognition in cnidarian-Symbiodiniaceae symbiosis (Wood-Charlson et al., 2006; Davy et al., 2012). Many c-type lectin genes have been identified in metazoan (Wood-Charlson and Weis, 2009) and radiolarian hosts (Balzano et al., 2015). We identified 56 c-type lectin transcripts in T. nucleata, with at least one of them being expressed at very high level. We also observed genes involved in glycan biosynthesis in the symbiont expressed at higher levels in hospite (Figure 4L). These findings are consistent with the possibility that recognition between the radiolarian host, T. nucleata, and its photosymbiont, Brandtodinium sp., might also involve lectin/glycan interactions.
Author Contributions
DC designed the study. LM and SH collected the samples and carried out the molecular biology preparations. ZL conducted the bioinformatic studies and wrote the manuscript. All authors read, revised, and approved the final manuscript.
Funding
This work was supported by a grant from the Simons Foundation (P49802 to DC).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Alyssa G. Gellene for her help during sample collection.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2019.00284/full#supplementary-material
Footnotes
References
Anderson, O. R., Swanberg, N. R., and Bennett, P. (1983). Assimilation of symbiont-derived photosynthesis in some solitary and colonial radiolaria. Mar. Biol. 77, 265–269. doi: 10.1007/bf00395815
Anderson, O. R., Swanberg, N. R., and Bennett, P. (1984). An estimate of predation rate and relative preference for algal versus crustacean prey by a spongiose skeletal radiolarian. Mar. Biol. 78, 205–207. doi: 10.1007/bf00394702
Balzano, S., Corre, E., Decelle, J., Sierra, R., Wincker, P., Da Silva, C., et al. (2015). Transcriptome analysis to investigate symbiotic relationships between marine protists. Front. Microbiol. 6:98. doi: 10.3389/fmicb.2015.00098
Biard, T., Pillet, L., Decelle, J., Poirier, C., Suzuki, N., and Not, F. (2015). Towards an intergrative morpho-molecular classification of the Collodaria (Polycystinea, Radiolaria). Protist 166, 374–388. doi: 10.1016/j.protis.2015.05.002
Biard, T., Stemmann, L., Picheral, M., Mayot, N., Vandromme, P., Hauss, H., et al. (2016). In situ imaging reveals the biomass of giant protists in the global ocean. Nature 532, 504–507. doi: 10.1038/nature17652
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Caron, D. A., Michaels, A. F., Swanberg, N. R., and Howse, F. A. (1995). Primary productivity by symbiont-bearing planktonic sarcodines (Acantharia, Radiolaria, Foraminifera) in surface waters near Bermuda. J. Plankton Res. 17, 103–129. doi: 10.1093/plankt/17.1.103
Chen, Y., Lun, A. T., and Smyth, G. K. (2014). “Differential expression analysis of complex RNA-seq experiments using edgeR,” in Statistical Analysis of Next Generation Sequencing Data, eds D. Nettleton and S. Datta (Cham: Springer International Publishing), 51–74. doi: 10.1007/978-3-319-07212-8_3
Coats, D. W. (1999). Parasitic life styles of marine dinoflagellates. J. Eukaryot. Microbiol. 46, 402–409. doi: 10.1038/ismej.2017.167
Davy, S. K., Allemand, D., and Weis, V. M. (2012). Cell biology of cnidarian-dinoflagellate symbiosis. Microbiol. Mol. Biol. Rev. 76, 229–261. doi: 10.1128/MMBR.05014-11
Decelle, J., Probert, I., Bittner, L., Desdevises, Y., Colin, S., de Vargas, C., et al. (2012). An original mode of symbiosis in open ocean plankton. Proc. Natl. Acad. Sci. U.S.A. 109, 18000–18005. doi: 10.1073/pnas.1212303109
Dennett, M. R., Caron, D. A., Michaels, A. F., Gallager, S. M., and Davis, C. S. (2002). Video plankton recorder reveals high abundances of colonial radiolaria in surface waters of the central North Pacific. J. Plankton Res. 24, 797–805. doi: 10.1093/plankt/24.8.797
Dolven, J. K., Lindqvist, C., Albert, V. A., Bjørklund, K. R., Yuasa, T., Takahashi, O., et al. (2007). Molecular diversity of alveolates associated with neritic North Atlantic radiolarians. Protist 158, 65–76. doi: 10.1016/j.protis.2006.07.004
Edgar, R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 5:113. doi: 10.1186/1471-2105-5-113
Enright, A. J., Van Dongen, S., and Ouzounis, C. A. (2002). An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30, 1575–1584. doi: 10.1093/nar/30.7.1575
Fischer, S., Brunk, B. P., Chen, F., Gao, X., Harb, O. S., Iodice, J. B., et al. (2011). Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr. Protoc. Bioinform. Chapter 6, Unit 6.12.1–19.
Gast, R. J. (2006). Molecular phylogeny of a potentially parasitic dinoflagellate isolated from the solitary radiolarian, Thalassicolla nucleata. J Eukaryot. Microbiol. 53, 43–45. doi: 10.1111/j.1550-7408.2005.00071.x
Gast, R. J., Beaudoin, D. J., and Caron, D. A. (2003). Isolation of symbiotically expressed genes from the dinoflagellate symbiont of the solitary radiolarian Thalassicolla nucleata. Biol. Bull. 204, 210–214. doi: 10.2307/1543561
Gast, R. J., and Caron, D. A. (1996). Molecular phylogeny of symbiotic dinoflagellates from planktonic foraminifera and radiolaria. Mol. Biol. Evol. 13, 1192–1197. doi: 10.1093/oxfordjournals.molbev.a025684
Gast, R. J., and Caron, D. A. (2001). Photosymbiotic associations in planktonic foraminifera and radiolaria. Hydrobiologia 461, 1–7.
Gast, R. J., McDonnell, T. A., and Caron, D. A. (2000). srDNA-based taxonomic affinities of algal symbionts from a planktonic foraminifer and a solitary radiolarian. J Phycol. 36, 172–177. doi: 10.1046/j.1529-8817.2000.99133.x
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Guillou, L., Viprey, M., Chambouvet, A., Welsh, R. M., Kirkham, A. R., Massana, R., et al. (2008). Widespread occurrence and genetic diversity of marine parasitoids belonging to Syndiniales (Alveolata). Environ. Microbiol. 10, 3349–3365. doi: 10.1111/j.1462-2920.2008.01731.x
Haas, B. J., Papanicolaou, A., Yassour, M., Grabherr, M., Blood, P. D., Bowden, J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. doi: 10.1038/nprot.2013.084
Hu, S. K., Liu, Z., Alexander, H., Campbell, V., Connell, P. E., Dyhrman, S. T., et al. (2018). Shifting metabolic priorities among key protistan taxa within and below the euphotic zone. Environ. Microbiol. 20, 2865–2879. doi: 10.1111/1462-2920.14259
Johnson, L. K., Alexander, H., and Brown, C. T. (2018). Re-assembly, quality evaluation, and annotation of 678 microbial eukaryotic reference transcriptomes. bioRxiv giy158. doi: 10.1101/323576
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 12:323. doi: 10.1186/1471-2105-12-323
Meng, A., Marchet, C., Corre, E., Peterlongo, P., Alberti, A., Da Silva, C., et al. (2018). A de novo approach to disentangle partner identity and function in holobiont systems. Microbiome 6:105. doi: 10.1186/s40168-018-0481-9
Moriya, Y., Itoh, M., Okuda, S., Yoshizawa, A. C., and Kanehisa, M. (2007). KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182–W185.
Probert, I., Siano, R., Poirier, C., Decelle, J., Biard, T., Tuji, A., et al. (2014). Brandtodinium gen. nov. and B. nutricula comb. nov. (Dinophyceae), a dinoflagellate commonly found in symbiosis with polycystine radiolarians. J. Phycol. 50, 388–399. doi: 10.1111/jpy.12174
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edger: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Suzuki, N., and Not, F. (2015). “Biology and ecology of Radiolaria,” in Marine Protists, eds S. Ohtsuka, T. Suzaki, T. Horiguchi, N. Suzuki, and F. Not (Tokyo: Springer), 179–222. doi: 10.1007/978-4-431-55130-0_8
Swanberg, N. R., and Caron, D. A. (1991). Patterns of sarcodine feeding in epipelagic oceanic plankton. J. Plankton Res. 13, 287–312. doi: 10.1093/plankt/13.2.287
Wang, J. T., and Douglas, A. E. (1999). Essential amino acid synthesis and nitrogen recycling in an alga-invertebrate symbiosis. Mar. Biol. 135, 219–222. doi: 10.1007/s002270050619
Weis, V. M., and Reynolds, W. S. (1999). Carbonic anhydrase expression and synthesis in the sea anemone Anthopleura elegantissima are enhanced by the presence of dinoflagellate symbiosis. Physiol. Biochem. Zool. 72, 307–316. doi: 10.1086/316674
Whitehead, L. F., and Douglas, A. E. (2003). Metabolite comparisons and the identity of nutrients translocated from symbiotic algae to an animal host. J. Exp. Biol. 206, 3148–3157.
Wood-Charlson, E. M., Hollingsworth, L. L., Krupp, D. A., and Weis, V. M. (2006). Lectin/glycan interactions play a role in recognition in a coral/dinoflagellate symbiosis. Cell. Microbiol. 8, 1985–1993. doi: 10.1111/j.1462-5822.2006.00765.x
Wood-Charlson, E. M., and Weis, V. M. (2009). The diversity of C-type lectins in the genome of a basal metazoan, Nematostella vectensis. Dev. Comp. Immunol. 33, 881–889. doi: 10.1016/j.dci.2009.01.008
Yang, Y., and Smith, S. A. (2013). Optimizing de novo assembly of short-read RNA-seq data for phylogenomics. BMC Genomics 14:328. doi: 10.1186/1471-2164-14-328
Keywords: radiolarian, Brandtodinium, transcriptome, photosymbiosis, holobiont
Citation: Liu Z, Mesrop LY, Hu SK and Caron DA (2019) Transcriptome of Thalassicolla nucleata Holobiont Reveals Details of a Radiolarian Symbiotic Relationship. Front. Mar. Sci. 6:284. doi: 10.3389/fmars.2019.00284
Received: 30 August 2018; Accepted: 16 May 2019;
Published: 04 June 2019.
Edited by:
Virginia M. Weis, Oregon State University, United StatesReviewed by:
Manuel Aranda, King Abdullah University of Science and Technology, Saudi ArabiaJohn Everett Parkinson, SECORE International, United States
Copyright © 2019 Liu, Mesrop, Hu and Caron. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenfeng Liu, emhlbmZlbmcubGl1MUBnbWFpbC5jb20=; David A. Caron, ZGNhcm9uQHVzYy5lZHU=
 Zhenfeng Liu
Zhenfeng Liu Lisa Y. Mesrop
Lisa Y. Mesrop Sarah K. Hu
Sarah K. Hu David A. Caron
David A. Caron