- 1Graduate School of Public Health, Seoul National University, Seoul, South Korea
- 2N-Bio, Seoul National University, Seoul, South Korea
- 3KoBioLabs, Inc., Seoul, South Korea
- 4Department of Agricultural Biotechnology, Seoul National University, Seoul, South Korea
- 5Center for Human and Environmental Microbiome, Seoul National University, Seoul, South Korea
- 6Institute of Health and Environment, Seoul National University, Seoul, South Korea
Gut microbiota play an important role in immune responses and energy metabolism. In this study, we evaluated whether administration of Lactobacillus fermentum (L. fermentum) KBL375 isolated from healthy Korean feces improves the atopic dermatitis using the house dust mite (Dermatophagoides farinae)-induced atopic dermatitis (AD) mouse model. Administration of L. fermentum KBL375 significantly decreased dermatitis score, ear and dorsal thickness, and serum immunoglobulin E level in AD-induced mice. Significant reductions in mast cells and eosinophils were discovered in skin tissues from L. fermentum KBL375-treated mice. T helper 2 cell-related cytokines interleukin (IL)-4, IL-5, IL-13, and IL-31 significantly decreased, and anti-inflammatory cytokine IL-10 or transforming growth factor-β increased in skin tissues from L. fermentum KBL375-treated mice. In addition to phenotypic changes in skin tissues, L. fermentum KBL375 treatment induced an increase in the CD4+CD25+Foxp3+ cell population in mesenteric lymph nodes. Taxonomic and functional analyses of gut microbiota showed significantly higher cecum bacterial diversities and abundances including genus Bilophila, Dorea, and Dehalobacterium in L. fermentum KBL375-treated mice. Metabolic analysis of the cecum also showed significant changes in the levels of various amino acids including methionine, phenylalanine, serine, and tyrosine, as well as short chain fatty acids such as acetate, butyrate, and propionate in AD-induced mice due to L. fermentum KBL375 treatment. These altered metabolites in AD-induced mice returned to the levels similar to those in control mice when treated with L. fermentum KBL375. Therefore, L. fermentum KBL375 could be useful for AD treatment by modulating the immune system and inducing various metabolites.
Introduction
Clinically, atopic dermatitis (AD) is characterized by chronic and abnormal inflammation of the skin with compromised skin barrier integrity, high inflammatory responses toward stimulants, and a reduction in antimicrobial responses (Rather et al., 2016). The incidence rate of AD has increased globally (Huang et al., 2017), but an effective treatment for AD has not yet been suggested. During the acute phase of AD, an increase in T helper (Th) 2 cells occurs in skin lesions (Meagher et al., 2002; Peng and Novak, 2015). The level of IL-4, the major Th2-related cytokine in inflammatory processes due to AD, is increased, and subsequently, there is an increase in IL-5 levels, which can induce B cells and Immunoglubulin (Ig) E synthesis (Kishimoto and Hirano, 1988). Moreover, the over-expression of thymic stromal lymphopoietin (TSLP), an IL-7-like cytokine, plays an important role in the activation of dendritic cell (DC)-mediated Th2 inflammatory responses, which can occur in keratinocytes (Liu, 2006). The concentrations of Th2-related chemokines, including macrophage-derived chemokine (MDC) and thymus and activation regulated chemokine (TARC), associated with the infiltration of inflammatory cells into skin lesions, also show a strong correlation with the severity of AD (Nakazato et al., 2008; Kim et al., 2013). Th1 and Th17 cells gradually increase during the chronic development of AD (Meagher et al., 2002; Peng and Novak, 2015), and Foxp3+ T regulatory cell (Treg) populations can inhibit AD development via production of anti-inflammatory cytokine IL-10 and transforming growth factor (TGF)-β (Zhang et al., 2014).
Gut microbiota can convey various beneficial effects for inflammatory diseases via modulating the host immune system (Sommer and Bäckhed, 2013; Zeng et al., 2017). To understand the effects of gut microbiota for the host, various animal models such as mouse were used. Mouse has significant similarities with human in their chromosomal genes and gut microbiota (Yue et al., 2014; Hugenholtz and de Vos, 2018). To evaluate the intervention methods for various diseases, abundant laboratory mouse models, such as wild-type and knock-out mice, are available (Perlman, 2016). Therefore, mouse models have been widely used for biological researches even though the clear differences in two species exist. Probiotics, defined as live microorganisms conferring health benefits to the host, can restore function of the gut microbiome and stimulate the production of metabolites such as short-chain fatty acids (SCFAs) (Hemarajata and Versalovic, 2013; Hwang, 2016). SCFAs are mainly generated by anaerobic bacteria in the host's intestine and can play important roles in adjusting host metabolism and regulating of the immune response (Vonk et al., 2011). Amino acids (AAs) are the major components of proteins and are involved in energy regulation in organisms, and are also used for SCFA production (Dai, 2011; Neis et al., 2015). Previous studies have reported that Lactobacillus spp., major probiotics known for their lactic acid production, have strong effects on the host's immune responses such as decreases in Th1, Th2, and Th17-related cytokines or increases in IL-10 or CD4+CD25+ regulatory T cells (Park et al., 2008; Lim et al., 2017; Kwon et al., 2018; Jang et al., 2019; Kim et al., 2019). Moreover, Lactobacillus spp. can alleviate AD via modulation of gut microbiota (Kwon et al., 2018), and affecting host metabolic pathways (Jang et al., 2019; Kim et al., 2019).
Therefore, we evaluated the effects of Lactobacillus fermentum (L. fermentum) KBL375, isolated from the feces of healthy donors (Jang et al., 2019), for AD treatment using an in vivo house dust mite (Dermatophagoides farinae, DFA) extract-induced AD mouse model using NC/Nga mice. After the treatment of L. fermentum KBL375, we assessed the ameliorations of AD symptoms, and immunomodulation effects such as the expression of cytokines or chemokines, and changes in gut microbiota in AD-induced mice for further applications to humans.
Materials and Methods
Preparation of L. fermentum KBL375
L. fermentum KBL375, isolated from healthy Korean feces, was cultured as previously described with minor modifications (Jang et al., 2019; Kim et al., 2019). Briefly, L. fermentum KBL375 was cultivated in Lactobacilli MRS Agar (BD Difco, Sparks, MD, USA) supplemented with 0.05% L-cysteine-hydrochloride at 37°C for 24 h under anaerobic conditions using the Anaeropack (Mitsubishi Gas Chemical Company, Inc., Tokyo, Japan). The concentration of L. fermentum KBL375 was measured using the cultivation method (Unit: Colony forming unit; CFU) and the optical density (OD)-based method at 600 nm. After cultivation, bacterial cells were harvested using centrifugation (1,200 × g) and washed twice with 1 × phosphate-buffered saline (PBS) prior to administration to mice.
AD-Induced in vivo Mouse Model
All experimental procedures for the AD-induced in vivo mouse model were approved by the Institutional Animal Care and Use Committee (IACUC: SNU-160928-1-1) of Seoul National University, Republic of Korea. The AD-induced in vivo mouse model was performed as previously described with minor modifications (Kang et al., 2015). Briefly, 5 week-old male NC/Nga mice (Central Lab Animals Inc., Seoul, Republic of Korea), bred from one production colony, were prepared and grouped with nine mice, per experimental condition. Three mice shared a cage. For induction of AD in skin, we removed the ear and dorsal skin hair using electric clipper and hair removal cream and applied 150 μL of 4% sodium dodecyl sulfate (SDS) in the ear and dorsal skin for 3 h to disrupt skin barrier. Then, we administered 100 mg of Dermatophagoides farinae extract (DFE) cream (Biostir® AD; Biostir, Inc, Hiroshima, Japan) twice a week for 21 days. Next, we administered approximately 1 × 109 CFUs of L. fermentum KBL375 in 200 μL of PBS to mice via oral gavage daily for 28 days with weekly application of DFE cream. After treatment with L. fermentum KBL375, mice were sacrificed and cecum, mesenteric lymph nodes (MLNs), serum, and skin were collected for further analyses.
Measurement of Clinical Symptoms
To assess the severity of AD, we anesthetized mice using 2% isoflurane. We then obtained images of the ear or dorsal skin and measured skin thickness (Figure S1A). Dermatitis scores of mice were measured once a week with blinded scoring of four researchers using the following criteria: (1) erythema/hemorrhage, (2) scaling/dryness, (3) edema, and (4) excoriation/erosion (Table S1) (Kang et al., 2015). Total dermatitis scores for mice were calculated with the maximal score of 12. Prior to sacrifice, transepidermal water loss (TEWL), representing the volume of water emitted from the body to the atmosphere through the epidermal layer (skin) by diffusion and evaporation, was measured using a Tewameter TM300 (Courage and Khazaka, Cologene, Germany) at 21–22°C and 50–55% humidity as described previously (Jimbo et al., 2015). The probe was located on the skin for 30 s and the data were measured. Scratching behaviors of mice were also observed for 10 min as described previously with minor modifications (Andoh et al., 2011). Briefly, for acclimation, mice were placed separately in an acrylic case for 1 h. Their behaviors were then recoded individually for 30 min. Total scratching behaviors to the nose, ear, and dorsal skin were counted for 10 min. Repeated and rapid scratching movements over 1 s were counted as one scratching. Blood samples were collected from blood vessels in the eyes of mice. Then, the serum was separated from blood samples by centrifugation at 1,200 × g for 15 min at 4°C. Serum IgE levels were measured using the IgE enzyme-linked immunosorbent assay (ELISA) kit (Komabiotech, Seoul, Korea) according to the manufacturer's instructions.
Histological Analysis
Dorsal skin tissues were fixed in 10% formaldehyde and stained with hematoxylin and eosin to confirm neutrophil infiltration (Kang et al., 2015). Congo red or toluidine blue were also used for staining eosinophils or mast cells in tissue, respectively (Kang et al., 2015). The stained tissues were examined using a Panoramic Viewer as described previously (3DHISTECH, Ltd., Budapest, Hungary) (Jang et al., 2019; Kim et al., 2019).
mRNA Expression of Cytokines or Chemokines in Skin Samples
To measure the mRNA expression of various cytokines or chemokines in the skin, total RNA from homogenates of skin samples was extracted using an Easy-spin Total RNA Extraction Kit (Intron, Seoul, Korea) and reverse-transcribed to complementary DNA (cDNA) using a High-Capacity RNA-to-cDNA Kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer's instructions. Cytokine levels were measured using real-time polymerase chain reaction (real-time PCR) using a Rotor-Gene Q (Qiagen, Hilden, Germany) and a QuantiTect SYBR Green PCR kit (Qiagen) with 0.01 mM primers (Table S2) under the following conditions: initial denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for 5 s and 60°C for 10 s. The relative expression levels of each cytokine or chemokine were calculated using the 2−ΔΔCT method and normalized to the expression levels of hypoxanthine-guanine phosphoribosyltransferase (HPRT), as described previously (Livak and Schmittgen, 2001).
Flow Cytometry Analysis in Treg Populations
Flow cytometry analyses were performed as described previously (Jang et al., 2019; Kim et al., 2019). Briefly, MLN tissues were smashed and filtered using a cell strainer (100 μm pore size (SPL Life Sciences Co., Ltd., Pocheon-si, Gyeonggi-do, Republic of Korea) and isolated T cells were subjected to a FC gamma receptor blocking and surface staining for 30 min at 4°C. To identify the live cells, T cells were stained with Fixable Viability Stain 510 (FVS510; BD Biosciences, San Jose, CA, USA) following the manufacturer's instructions. Moreover, T cell surface were stained with CD3+ fluorescein isothiocyanate (145-2C11; BD Biosciences), CD4+ Percep-Cyanine 5.5 (RM4-5; BD Biosciences) and CD25+ phycoerythrin (PC61; BD Biosciences) following the manufacturer's instructions. T cells were permeabilized with fixation/permeabilization buffer (eBioscience, San Diego, CA, USA) for intracellular staining and were stained with Foxp3+ Alexa Flour 647 (MF23; BD Biosciences) following the manufacturer's instructions. IgG isotypes (BD Biosciences) were used as a control. The CD4+CD25+Foxp3+ T cell population was counted using a BD FACSVerse™ Flow Cytometer (BD Biosciences).
Cecum Microbiota Analysis
Cecum microbiota were analyzed as previously described with minor modifications (Lim et al., 2016; Jang et al., 2019; Kim et al., 2019). Briefly, total DNA was collected using a QIAmp Fast DNA stool mini kit (Qiagen) following the manufacturer's protocol. The V3-4 hypervariable region of the 16S rRNA gene was amplified using the barcoded primers 341F and 805R as described previously with minor modifications. The amplicons were purified subsequently using a QIAquick PCR Purification Kit (Qiagen) and sequenced using a MiSeq platform (Illumina. Inc., San Diego, CA, USA). Data were analyzed by using Quantitative Insights into Microbial Ecology 1.8.0 (QIIME) software (QIIME development team; http://qiime.org/)(Caporaso et al., 2010) and Greengenes version 13_5 database (http://greengenes.secondgemone.com). First, the sequences were clustered using an open-reference operational taxonomic unit (OTU) picking protocol at least 97% nucleotide identity with exclusions of singletons and rare OTUs, which were founded in <10% of samples. Then, the relative abundances of microbial taxa in samples were calculated from the non-rarefied OTU table. Alpha diversity was confirmed using the Chao1 index. Beta diversity was suggested based on the UniFrac distance between samples and visualized based on the weighted principle coordinate analysis (PCoA). Diversities were estimated from a rarefied OTU table, grouped into the mice with different treatments. Different taxa with significance were measured using linear discriminant analysis effect size analysis (LEfSe) (threshold >3.0) with Galaxy ver. 2.1.1 (Hutlab; http://huttenhower.org/galaxy) (Segata et al., 2011) and comparisons of abundances for significantly different microbial taxa among experimental groups were also measured.
Measurement of AAs or SCFAs in Cecum Samples
AAs in cecum samples were analyzed as previously described (Jang et al., 2019). Briefly, approximately 1 mL of cecum extracts (concentration: 20 mg/mL) in liquid chromatography-grade methanol were derivatized by 70 μL of AccQ•Tag™ Ultra Borate Buffer (Waters Corporation., Milford, MA, USA) and 20 μL of AccQ•Tag™ Ultra reagent (Waters Corporation) for 10 min at 55°C. Samples were subsequently analyzed using an Acquity ultra-performance liquid chromatography (UPLC) (Waters Corporation) and a SYNAPT G2-Si mass spectrometer (Waters Corporation) with an ESI probe under the following conditions: 1.5 kV of capillary voltage, 600 and 50 L/h of desolvation or cone gas flow, respectively, and 250°C of desolvation temperature. An AA-S-18 analytical standard mixture (Sigma-Aldrich, St. Louis, MO, USA) was used to identify AAs, and data analyses were performed using MassLynx software 4.1 (Waters Corporation) (Roucher et al., 2013).
To measure SCFAs in samples, ceca were homogenized in distilled water and centrifuged at 13,000 × g for 5 min. Supernatant was collected and internal standards (1% 2-methylpentanoic acid for volatile acids or benzoic acid for non-volatile acids, respectively) were added. Then, extraction solvents (ethyl ether for volatile acids or chloroform for non-volatile acids, respectively) were added to the solution and centrifuged at 13,000 × g for 5 min. The organic layer was collected and analyzed using an Agilent 7890A gas chromatograph (Agilent Technologies, Santa Clara, CA, USA) under the following conditions; 1.5 kV of capillary voltage. Six hundred and fifty liter per hour of desolvation or cone gas flow, respectively, 170°C of oven temperature and 225°C for a flame ionization detector (FID) and an injection port temperature. The standard mixture was used as a reference for retention times and peak areas for extracts (David et al., 2014).
Statistical Analysis
Experimental data were expressed as means ± standard error of the mean (SEM) of experimental groups (nine mice per group). When appropriate, data were analyzed using the Mann-Whitney U test. P-values < 0.05 were considered as statistically significant. GraphPad PRISM 5 (GraphPad software, San Diego, CA) were used for statistical analyses and visualizations.
Results
Effects of L. fermentum KBL375 on Clinical Symptoms or Immune Cells Using the in vivo AD Model
After 28 days of treatment, AD symptoms including hypertrophy and hyperkeratosis in the DFE + L. fermentum KBL375-treated mice were decreased with the reduction in ear or dorsal skin thickness compared to DFE + PBS-treated mice (P < 0.01 or P < 0.05, respectively) (Figure S1). Moreover, the DFE + L. fermentum KBL375-treated mice showed significant decreases in dermatitis score, with an average of six, compared to the DFE + PBS-treated group (P < 0.05) (Figure 1A). TEWL, scratching behaviors in 10 min, and serum IgE levels recovered after the L. fermentum KBL375 treatment (Figures 1B–D). Figure 2 also shows that the number of eosinophils and mast cells of dorsal skin tissues from DFE + L. fermentum KBL375-treated mice were significantly decreased compared to those in DFE + PBS-treated mice (P < 0.05).
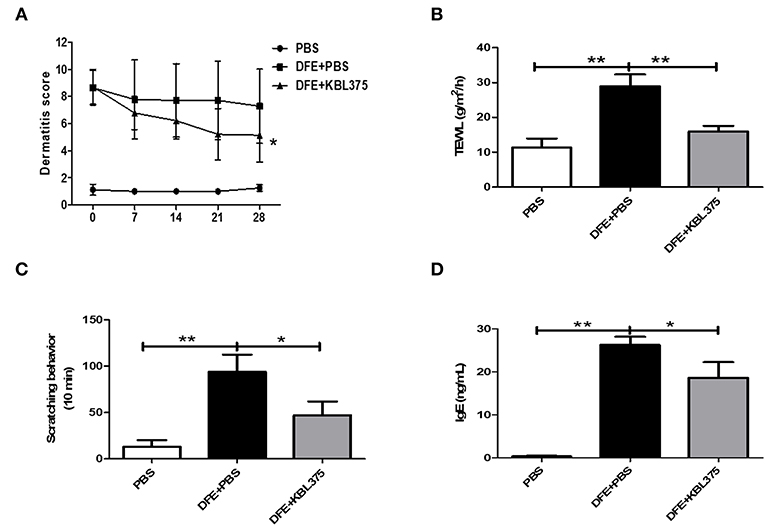
Figure 1. The effects of Lactobacillus fermentum (L. fermentum) KBL375 on clinical symptoms and immunoglobulin E (IgE) levels in an in vivo atopic dermatitis (AD) mouse model. (A) Dermatitis score; (B) Transepidermal water loss (TEWL); (C) Scratching behavior; (D) Serum IgE level. Firstly, we induced AD in male NC/Nga mice using 100 mg of Dermatophagoides farinae extract (DFE) cream twice a week for 21 days. Then, approximately 1 × 109 colony forming units (CFU) of L. fermentum in 200 μL of PBS was administered to mice via oral gavage daily for 28 days, with weekly application of DFE cream. Dermatitis scores for mice were measured once a week. Prior to sacrifice, the TEWL scores and scratching behaviors in 10 min were measured. After 28 days of L. fermentum treatment, the serum IgE levels were measured using an enzyme-linked immunosorbent assay (ELISA). Data were expressed as the means ± SEM of experimental groups (nine mice per group). Asterisks indicate a statistical significance (*P < 0.05; **P < 0.01; Mann-Whitney U test compared to mice treated with DFE + PBS as a positive control).
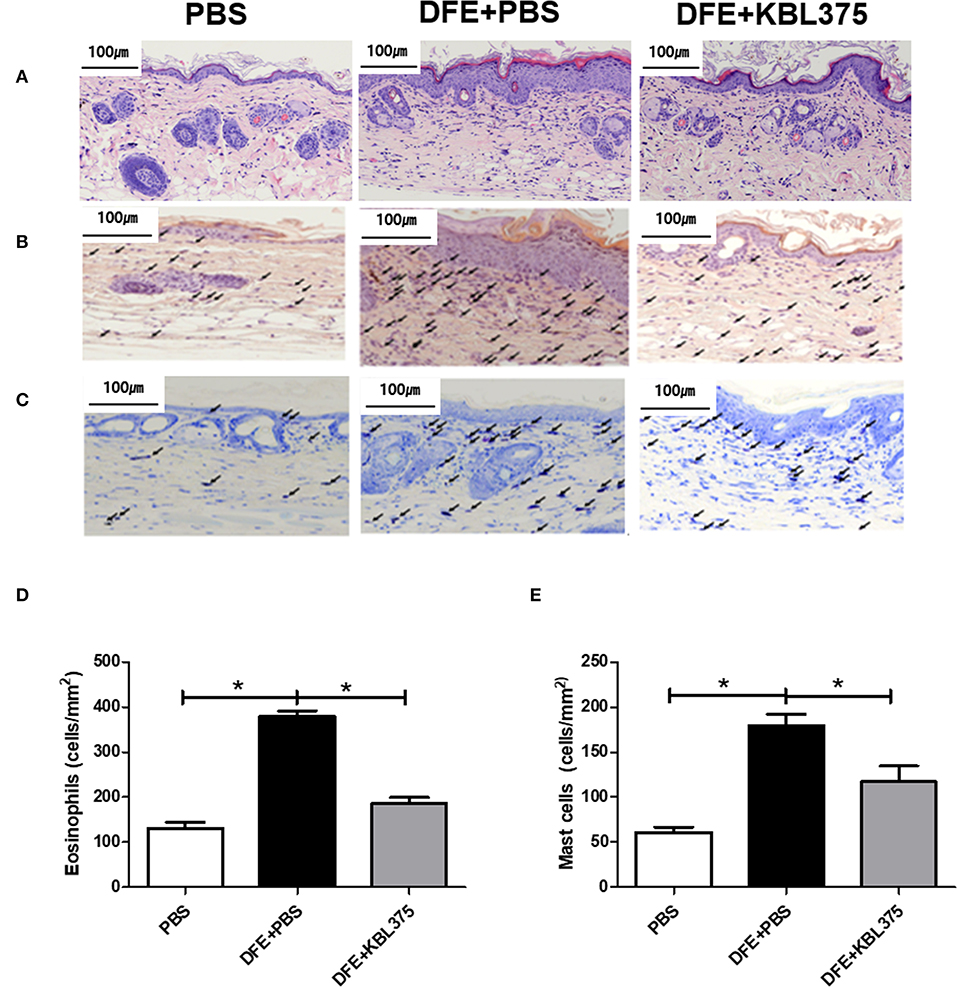
Figure 2. Improvements in dorsal skin inflammation via L. fermentum KBL375 treatment in the in vivo AD model. (A) Hematoxylin and eosin staining; (B) Congo red-staining; (C) Toluidine blue staining; (D) Eosinophils; (E) Mast cells. First, the skin samples were fixed using 10% formaldehyde and stained. Congo red or toluidine blue were used for staining eosinophils or mast cells, respectively. Black arrows indicate eosinophils or mast cells in skin samples. When appropriate, data were expressed as the means ± SEM of experimental groups (nine mice per group). Asterisks indicate a statistical significance (*P < 0.05; Mann-Whitney U test compared to mice treated with DFE + PBS as a positive control).
Effects of L. fermentum KBL375 on the Expression of Cytokines or Chemokines in Skin From the in vivo AD Model
Figure 3 shows the effects of L. fermentum KBL375 treatment on the expression of innate cytokine TSLP or chemokines including MDC or TARC in skin in the in vivo AD model. Skin samples from AD-induced mice treated with L. fermentum KBL375 showed significant reductions in mRNA levels of TSLP, MDC, or TARC compared to controls. Moreover, L. fermentum KBL375 showed strong down-regulatory effects on Th2-related cytokines including IL-4, IL-5, IL-13 and IL-31, and significant up-regulatory effects on anti-inflammatory cytokine IL-10 in AD-induced mice (P < 0.05) (Figure 4). Treatment with L. fermentum KBL375 also resulted in significant down-regulation of Th1-or Th17-related cytokines including TNF, IL-6, interferon (IFN)-γ, and IL-17A in AD-induced mice (P < 0.05) (Figure S2).
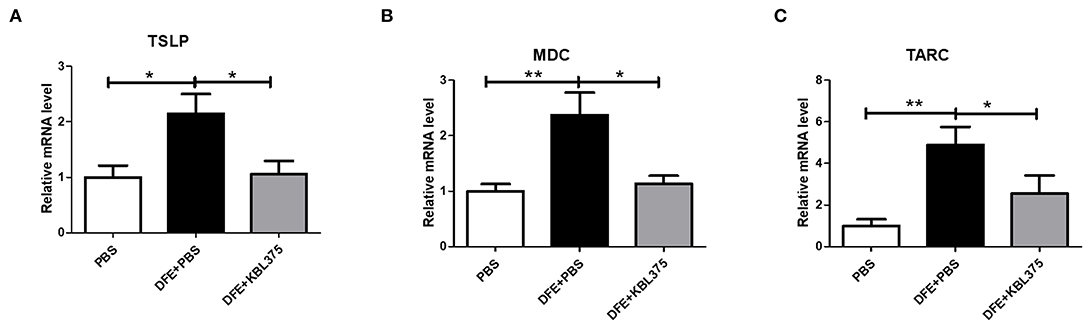
Figure 3. The effects of L. fermentum KBL375 on the expression of innate cytokines or chemokines in skin. (A) Thymic stromal lymphopoietin (TSLP); (B) Macrophage-derived chemokine (MDC); (C) Thymus and activation regulated chemokine (TARC). Briefly, total RNA from homogenates of skin samples was extracted and reverse-transcribed to complementary DNA (cDNA). Then, cytokine levels were measured using real-time polymerase chain reaction (real-time PCR) and normalized using hypoxanthine-guanine phosphoribosyltransferase (HPRT). Data were expressed as the means ± SEM of experimental groups (nine mice per group). Asterisks indicate a statistical significance (*P < 0.05; **P < 0.01; Mann-Whitney U test compared to mice treated with DFE + PBS as a positive control).
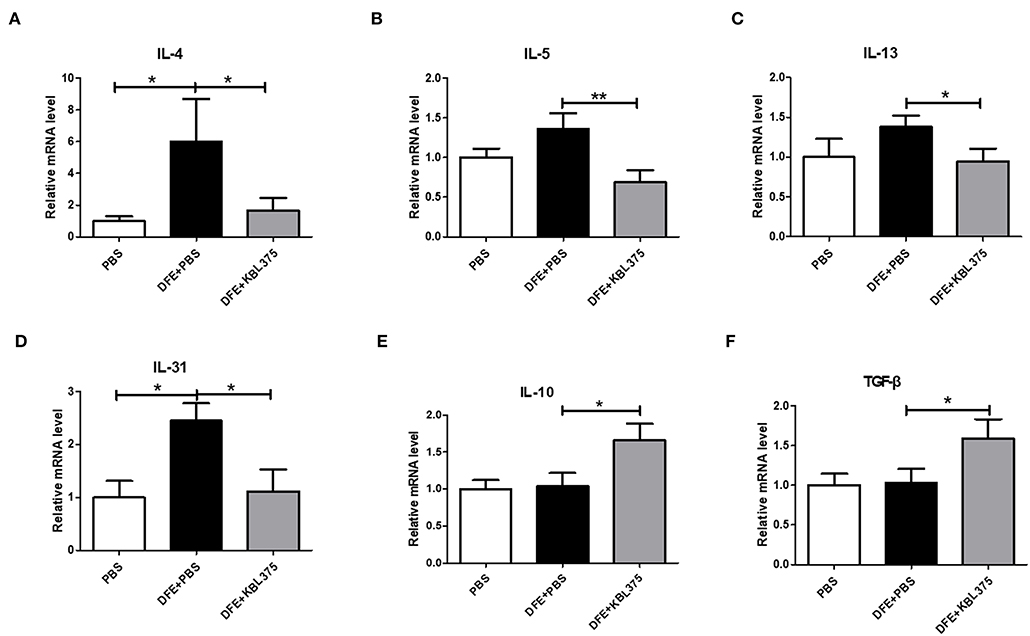
Figure 4. The effects of L. fermentum KBL375 on cytokine expression related to T helper (Th) 2 cells, IL-10, or transforming growth factor (TGF)-β. (A) Interleukin (IL)-4; (B) IL-5; (C) IL-13; (D) IL-31; (E) IL-10; (F) TGF-β. Cytokine levels were measured by real-time PCR and normalized using HPRT. Data were expressed as the means ± SEM of experimental groups (nine mice per group). Asterisks indicate a statistical significance (*P < 0.05; **P < 0.01; Mann-Whitney U test compared to mice treated with DFE + PBS as a positive control).
Changes in Treg Populations in MLN Samples and Foxp3+ Expression of Skin Tissues After the L. fermentum KBL375 Treatment
Figure 5A shows the CD4+CD25+Foxp3+ Treg populations in MLN samples from AD-induced mice. Compared to DFE + PBS-treated mice, CD4+CD25+Foxp3+ Treg populations in MLN were significantly increased in L. fermentum KBL375 treated mice (P < 0.05). Moreover, Foxp3+ expression in skin tissues also significantly increased in DFE + L. fermentum KBL375-treated mice compared to DFE + PBS-treated mice (P < 0.05) (Figure 5B).
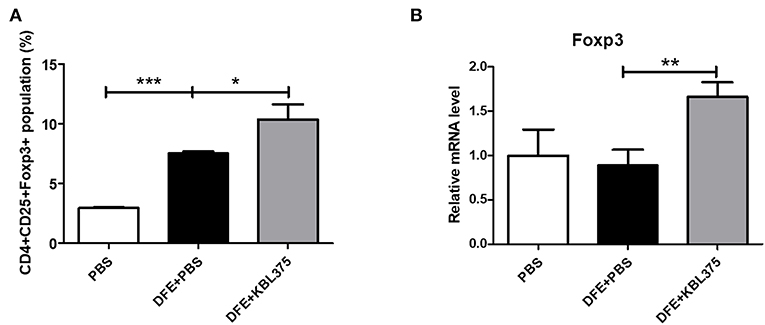
Figure 5. Flow cytometry analyses of T cells in mesenteric lymph nodes (MLNs) and Foxp3+ expression of skin tissues. (A) CD4+CD25+Foxp3+ T cell population; (B) Foxp3+ expression in skin samples in RNA level. T cells were collected in MLN tissues from sacrificed mice and initially stained using Fixable Viability Stain 510. Then, T cell surfaces were stained with CD3+ fluorescein isothiocyanate, CD4+ Percep-Cyanine5.5, and CD25+ phycoerythrin. Permeabilized T cells were also stained with Foxp3+ Alexa Fluor 647 for intracellular staining. The CD4+CD25+Foxp3+ T cell population was analyzed using a flow cytometer and the Foxp3+ mRNA levels were measured using real-time PCR and normalized using HPRT. Data were expressed as the means ± SEM of experimental groups (9 mice per each group). Asterisks indicate a statistical significance (*P < 0.05; **P < 0.01; ***P < 0.001; Mann-Whitney U test compared to mice treated with DFE + PBS as a positive control).
Changes in Cecum Microbiota in the in vivo AD Model After the L. fermentum KBL375 Treatment
Figure 6 shows changes in bacterial communities in the cecum from AD-induced mice. PCoA plots showed that the cecum microbiota from DFE + L. fermentum KBL375-treated mice were similar to those from PBS-treated mice, which were clearly distinguishable from those of with DFE + PBS-treated mice (Figure 6A). S24-7 family was the dominant bacterial group in PBS-treated or DFE + L. fermentum KBL375-treated mice (24.4 and 29.8%, respectively) (Figure 6B). However, in DFE + PBS-treated mice, the abundance of S24-7 family was clearly decreased (10.7%), and a high proportion of genus Bacteroides was observed (8.9%) (Figure 6B). The cecum samples from PBS-treated and DFE + L. fermentum KBL375-treated mice showed significantly higher relative abundance of genus Bilophila, Dehalobacterium, and Dorea than DFE + PBS-treated samples (Figures 6C–E). On the other hand, cecum microbiota from DFE + PBS-treated mice showed a higher proportion of genus Bacteroides, Mucisprillum, and Sutterella compared to the other experimental groups (Figures 6C–E).
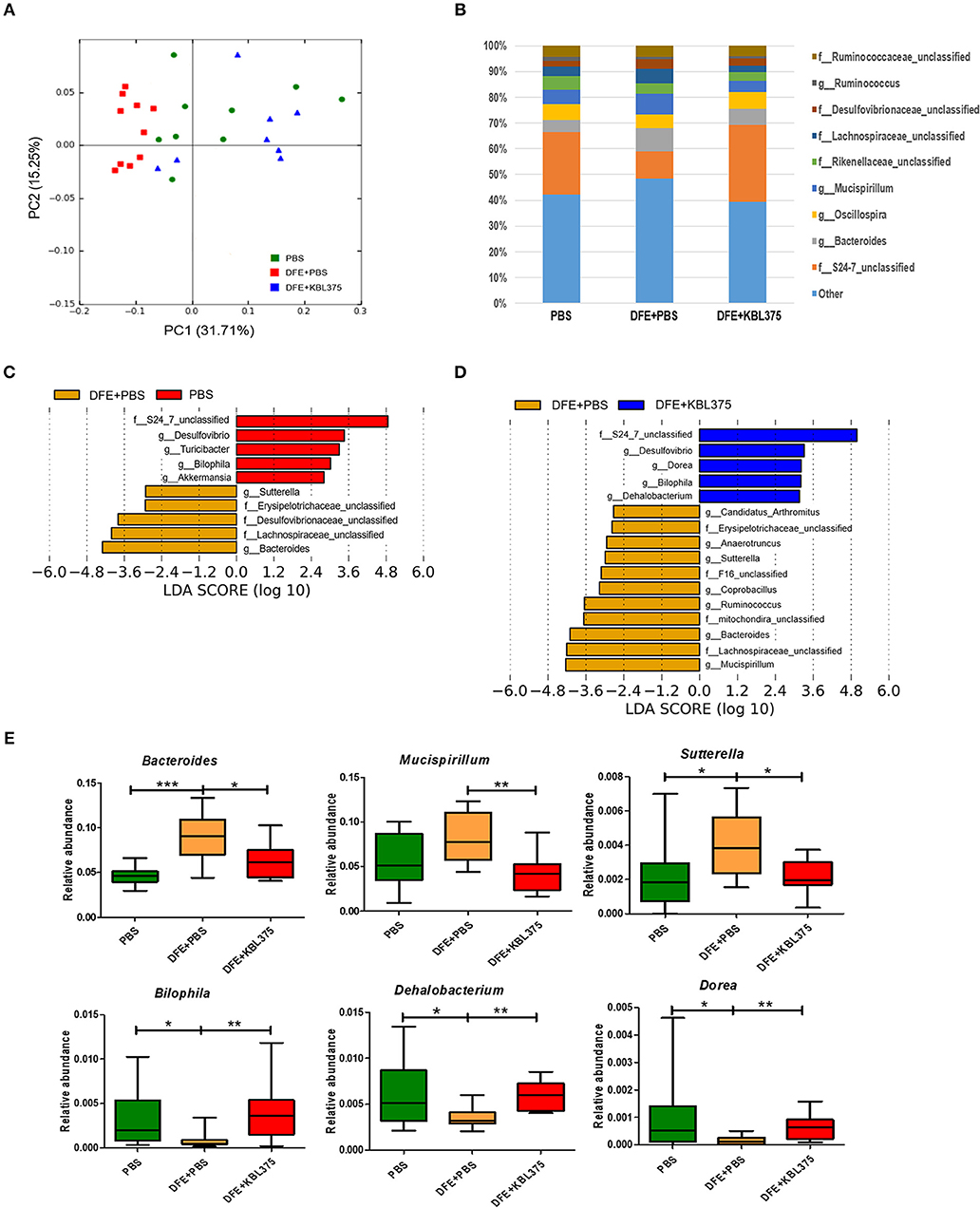
Figure 6. The effects of L. fermentum KBL375 on cecum microbiota of mice. (A) Principal coordinate analysis (PCoA) plot of the cecal microbiota with weighted UniFrac distance; (B) Taxonomic shifts in cecum microbiota for L. fermentum KBL375 treatment at the genus level; (C) Different taxa with significance measured using linear discriminant analysis effect size analysis for mice treated with PBS compared to mice treated with DFE + PBS (threshold >2.5); (D) Different taxa with significance measured using linear discriminant analysis effect size analysis for mice treated with DFE + L. fermentum KBL375 compared to mice treated with DFE + PBS (threshold >2.5); (E) Comparison of abundances of significantly different microbial taxa. Asterisks indicate a statistical significance (*P < 0.05; **P < 0.01; ***P < 0.001; Mann-Whitney U test compared to mice treated with DFE + PBS as a positive control).
Changes in Metabolic Pathways Predicted Using Microbial Compositions or Metabolites in the Cecum of the in vivo AD Model After the L. fermentum KBL 375 Treatment
Figure 7 shows the predicted changes in metabolic pathways after the L. fermentum KBL375 treatment in the ceca of AD-induced mice. The concentrations of various AAs, including methionine, phenylalanine, serine, and tyrosine, were significantly lower in ceca from AD-induced mice after L. fermentum KBL375 treatment in ceca from DSS + PBS-treated mice (Figure 7A). Significantly higher concentrations of three SCFAs, including acetate, butyrate and propionate were also measured in the ceca with the L. fermentum KBL375 treatment, similar to the results in PBS-treated mice (Figure 7B).
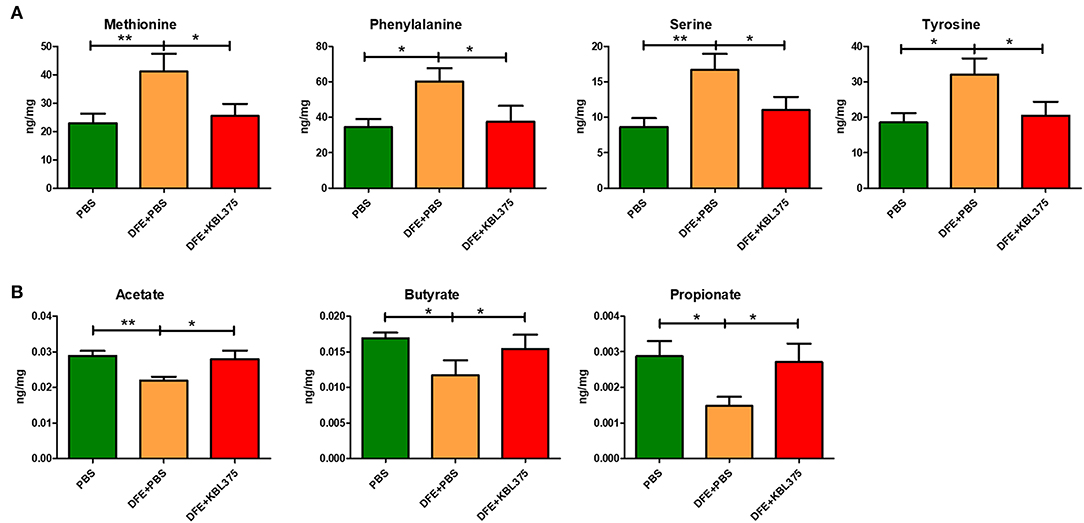
Figure 7. The effects of L. fermentum KBL375 on functional alterations related to various metabolites. (A) Amino acids; (B) Short-chain fatty acids. Amino acids or short-chain fatty acids in cecum samples were measured appropriately using an Acquity ultra performance liquid chromatograph with a SYNAPT G2-Si mass spectrometer or an Agilent 7890A gas chromatograph, respectively. Asterisks indicate a statistical significance (*P < 0.05; **P < 0.01; Mann-Whitney U test compared to mice treated with DFE + PBS as a positive control).
Discussion
This study clearly demonstrated that the L. fermentum KBL375 treatment showed various protective effects against AD, such as improvements of clinical symptoms, immunomodulation of the host, and changes in metabolic pathways due to gut microbiota restoration. The DFE-induced AD mice showed severe physiological dysfunction, including skin barrier disruption, thickness, and dryness as previously suggested (Figure 1 and Figure S1) (Saeki et al., 2009). However, after 28 days of L. fermentum KBL375 administration, clinical features including dermatitis score, TEWL, scratching behavior, and serum IgE levels, which are considered typical abnormalities induced by AD, improved significantly (Figure 1) (Brandt, 2011; Lee and Yu, 2011; Zheng et al., 2011; Galli and Tsai, 2013). Previous studies reported that IgE-mediated activation of mast cells subsequently led to infiltration of eosinophils into skin lesions, which can contribute to the development of clinical features of AD (Matsuda et al., 1997; Galli and Tsai, 2013). Our results indicated that treatment of L. fermentum KBL375 inhibited the infiltration of eosinophils and mast cells (Figure 2).
Moreover, the DFE + L. fermentum KBL375-treated mice showed reductions in innate cytokine TSLP or chemokine MDC or TARC levels in skin (Figure 3). TSLP is known to be an activation factor for DCs, and activated DCs can boost the production of Th2-attracting chemokines including TARC and MDC, which can mediate Th2 recruitment into skin lesions (Pineiro and Stanton, 2007; Olszak et al., 2012; Tatsuno et al., 2015). Our results showed that L. fermentum KBL375 treatment can decrease the expression of not only Th2-related cytokines, including IL-4, IL-5, IL-13 and IL-31 but also other cytokines related to pro-inflammation, such as Th1- or Th17 (Figure 4 and Figure S2). Pro-inflammatory cytokine TNF and IL-6 contribute to inflammatory responses due to AD (Chang et al., 2013; Kim et al., 2013) and the responses of Th-related cytokines are important in the pathogenesis of AD (Grewe et al., 1998; Takaoka et al., 2006; Peng and Novak, 2015). On the other hand, L. fermentum KBL375 treatment showed the up-regulation of anti-inflammatory cytokine IL-10 and TGF-β in AD-induced mice indicating that the suppression of immune responses were occurred (Figure 4) (Agrawal et al., 2011; Kosaka et al., 2011; Purchiaroni et al., 2013). Therefore, L. fermentum KBL375-induced modulations in various cytokines or chemokines could be one of the major protective effects against AD.
Additionally, L. fermentum KBL375 treatment significantly increased CD4+CD25+Foxp3+ Treg populations in MLNs and skin tissues (Figure 5). As an effective suppressor of inflammation, CD4+Foxp3+ Treg populations are migrate and increase anti-inflammatory cytokines (Kim and Ji, 2012). Previous studies have also reported that probiotics can induce the generation of tolerogenic DCs, which are highly associated with IL-10 production and Treg differentiation (Kwon et al., 2010). Therefore, oral administration of L. fermentum KBL375 can induce strong protective effects for AD through immunomodulation.
Previous studies have shown that the interaction between a host and their microbiota is important to maintain the healthy status of the host (Sommer and Bäckhed, 2013). In this study, we investigated the changes in bacterial abundance and diversities in AD-induced mice after L. fermentum KBL375 treatment and confirmed that DFE + L. fermentum KBL375-treated mice had similar microbiota to PBS-treated mice, indicated that the restoration or protection of cecum microbiota were occurred by the L. fermentum KBL375 treatments in AD-induced mice (Figures 6A,B). Especially, S24-7 family, showed a clear reduction in a DFE + PBS-treated mice, but was dominant in ceca from both DFE + L. fermentum KBL375-treated and PBS-treated mice (Figure 6B). High relative abundances of genus Bilophila, which showed a negative correlation with lipopolysaccharide [LPS]- or Candida albicans-induced TNF-α production (Biancheri and Watson, 2017), and Dehalobacterium, which is associated with a protective effect against atherosclerosis (Chan et al., 2016), were observed in DFE + L. fermentum KBL375-treated mice (Figures 6C–E). Moreover, the genus Bacteroides, Mucispirillum, and Sutterella were significantly more abundant in DFE + PBS-treated mice than in DFE + L. fermentum KBL375-treated mice (Figures 6C–E). The abundance of genus Bacteroides is highly correlated with the occurrence of colitis (Lucke et al., 2006). Previous studies have also reported that genus Mucispirillum showed the an association with chemical-induced or Citrobacter rodentium-induced colitis in mice (Loy et al., 2017) and Sutterella species were associated with low secretory IgA levels due to their ability to degrade of both IgA and IgA-associated peptides (Levy et al., 2017). Taken together, these results indicate that L. fermentum KBL375 treatment could have positive effects in cecum microbiota altered by AD.
Our results elucidated that L. fermentum KBL375 treatment has shown the increases of metabolites such as AAs and SCFAs (Figure 7). Microbial metabolites such as AAs from intestinal microbiota play important roles in host physiology and immunity (Dai, 2011; Rooks and Garrett, 2016). Moreover, AAs, which are produced by instestinal microbiota, can serve as precursors for SCFA synthesis (Neis et al., 2015). SCFAs are used as energy sources, substrates or signal molecules, and are strongly associated with the metabolism of lipids, glucose, and cholesterols (Kim et al., 2017; McNabney and Henagan, 2017). Especially, butyrate, an important SCFA that modulates immune responses in macrophages or promotes the formation of Tregs, can be produced by family Lachnospiraceae or Ruminococcaceae including genus Dorea or Oscillospira, respectively (Cushing et al., 2015; Blacher et al., 2017; Kang et al., 2017). Therefore, L. fermentum KBL375 treatment can induce the increases of AAs, including methionine, phenylalanine, serine, and tyrosine, and SCFAs including acetate, butyrate, and propionate, in the intestines of AD-induced mice. However, further longitudinal studies with a large number of animal or human subjects and various study designs should be performed to elucidate the effects of L. fermentum KBL375 on microbiota or microbiota-related metabolites.
In conclusion, L. fermentum KBL375 treatment showed strong anti-AD effects in the in vivo mouse model via modulation of the immune system and induction of various metabolites. Therefore, L. fermentum KBL375 could be a useful to ameliorate AD through with various applications such as functional foods or drugs.
Data Availability Statement
The datasets for this study can be found in the: https://www.ncbi.nlm.nih.gov/bioproject/558163.
Ethics Statement
All experimental procedures for AD-induced in vivo mouse models were approved by the Institutional Animal Care and Use Committee (IACUC: SNU-160928-1-1) of Seoul National University, Republic of Korea.
Author Contributions
W-KK, YJ, and GK mainly designed the study, and other authors contributed to data acquisition and manuscript preparation. GK supervised this study.
Funding
This work was supported by a grant of National Research Foundation of Korea (NRF) (NRF-2018R1A2A1A05078258) funded by the Korean government (MSIP), Republic of Korea, and the Technology Innovation Program (No. 10076742) funded by the Ministry of Trade, Industry & Energy (MOTIE), Republic of Korea.
Conflict of Interest
GK was the founder of KoBioLabs, Inc., and SP was employed by KoBioLabs, Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer TU declared a past co-authorship with one of the authors DH to the handling editor.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2019.00092/full#supplementary-material
References
Agrawal, R., Wisniewski, J. A., and Woodfolk, J. A. (2011). The role of regulatory T cells in atopic dermatitis. Karger 41, 112–124. doi: 10.1159/000323305
Andoh, T., Haza, S., Saito, A., and Kuraishi, Y. (2011). Involvement of leukotriene B4 in spontaneous itch-related behaviour in NC mice with atopic dermatitis-like skin lesions. Exp. Dermatol. 20, 894–898. doi: 10.1111/j.1600-0625.2011.01346.x
Biancheri, P., and Watson, A. J. M. (2017). The relative contributions of the gut microbiome, host genetics, and environment to cytokine responses to microbial stimulation. Gastroenterology 152, 2068–2070. doi: 10.1053/j.gastro.2017.04.037
Blacher, E., Levy, M., Tatirovsky, E., and Elinav, E. (2017). Microbiome-modulated metabolites at the interface of host immunity. J. Immunol. 198, 572–580. doi: 10.4049/jimmunol.1601247
Brandt, E. B. (2011). Th2 cytokines and atopic dermatitis. J. Clin. Cell. Immunol. 2:110. doi: 10.4172/2155-9899.1000110
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chan, Y. K., Brar, M. S., Kirjavainen, P. V., Chen, Y., Peng, J., Li, D., et al. (2016). High fat diet induced atherosclerosis is accompanied with low colonic bacterial diversity and altered abundances that correlates with plaque size, plasma A-FABP and cholesterol: a pilot study of high fat diet and its intervention with Lactobacillus rhamnosus GG (LGG) or telmisartan in ApoE−/− mice. BMC Microbiol. 16:264. doi: 10.1186/s12866-016-0883-4
Chang, H.-H., Miaw, S.-C., Tseng, W., Sun, Y.-W., Liu, C.-C., Tsao, H.-W., et al. (2013). PTPN22 modulates macrophage polarization and susceptibility to dextran sulfate sodium-induced colitis. J. Immunol. 191, 2134–2143. doi: 10.4049/jimmunol.1203363
Cushing, K., Alvarado, D. M., and Ciorba, M. A. (2015). Butyrate and mucosal inflammation: new scientific evidence supports clinical observation. Clin. Transl. Gastroenterol. 6:e108. doi: 10.1038/ctg.2015.34
Dai, Z.-L. (2011). Amino acid metabolism in intestinal bacteria: links between gut ecology and host health. Front. Biosci. 16, 1768–1786. doi: 10.2741/3820
David, L. A., Maurice, C. F., Carmody, R. N., Gootenberg, D. B., Button, J. E., Wolfe, B. E., et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563. doi: 10.1038/nature12820
Galli, S. J., and Tsai, M. (2013). IgE and mast cells in allergic disease. Nat. Med. 18, 693–704. doi: 10.1038/nm.2755
Grewe, M., Bruijnzeel-Koomen, C. A., Schöpf, E., Thepen, T., Langeveld-Wildschut, A. G., Ruzicka, T., et al. (1998). A role for Th1 and Th2 cells in the immunopathogenesis of atopic dermatitis. Immunol. Today 128, 2569–2571. doi: 10.1016/S0167-5699(98)01285-7
Hemarajata, P., and Versalovic, J. (2013). Effects of probiotics on gut microbiota: mechanisms of intestinal immunomodulation and neuromodulation. Ther. Adv. Gastroenterol. 6, 39–51. doi: 10.1177/1756283X12459294
Huang, R., Ning, H., Shen, M., Li, J., Zhang, J., and Chen, X. (2017). Probiotics for the treatment of atopic dermatitis in children: a systematic review and meta-analysis of randomized controlled trials. Front. Cell. Infect. Microbiol. 7:392. doi: 10.3389/fcimb.2017.00392
Hugenholtz, F., and de Vos, W. M. (2018). Mouse models for human intestinal microbiota research: a critical evaluation. Cell. Mol. Life. Sci. 75, 149–160. doi: 10.1007/s00018-017-2693-8
Hwang, S. W. (2016). “Helicobacter pylori,” in Probiotics, Vol. 47, ed N. Kim (Singapore: Springer Science Business Media), 479–485. doi: 10.1007/978-981-287-706-2_47
Jang, Y. J., Kim, W. K., Han, D. H., Lee, K., and Ko, G. (2019). Lactobacillus fermentum species ameliorate dextran sulfate sodium-induced colitis by regulating the immune response and altering gut microbiota. Gut Microbes 3, 1–16. doi: 10.1080/19490976.2019.1589281
Jimbo, N., Kawada, C., and Nomura, Y. (2015). Herb extracts and collagen hydrolysate improve skin damage resulting from ultraviolet-induced aging in hairless mice. Biosci. Biotechnol. Biochem. 79, 1624–1628. doi: 10.1080/09168451.2015.1046362
Kang, C., Wang, B., Kaliannan, K., Wang, X., Lang, H., Hui, S., et al. (2017). Gut microbiota mediates the protective effects of dietary capsaicin against chronic low-grade inflammation and associated obesity induced by high-fat diet. MBio 8, e00900–e00917. doi: 10.1128/mBio.00470-17
Kang, H., Lee, C. H., Kim, J. R., Kwon, J. Y., Seo, S. G., Han, J. G., et al. (2015). Chlorella vulgaris attenuates dermatophagoides farinae-induced atopic dermatitis-like symptoms in NC/Nga mice. Int. J. Mol. Sci. 16, 21021–21034. doi: 10.3390/ijms160921021
Kim, D., Zeng, M. Y., and Núñez, G. (2017). The interplay between host immune cells and gut microbiota in chronic inflammatory diseases. Exp. Mol. Med. 49:e339. doi: 10.1038/emm.2017.24
Kim, J. Y., Park, B. K., Park, H. J., Park, Y. H., Kim, B. O., and Pyo, S. (2013). Atopic dermatitis-mitigating effects of new Lactobacillus strain, Lactobacillus sakei probio 65 isolated from Kimchi. J. Appl. Microbiol. 115, 517–526. doi: 10.1111/jam.12229
Kim, N. Y., and Ji, G. E. (2012). Effects of probiotics on the prevention of atopic dermatitis. Korean J. Pediatr. 55, 193–201. doi: 10.3345/kjp.2012.55.6.193
Kim, W.-K., Jang, Y. J., Seo, B., Han, D. H., Park, S., and Ko, G. (2019). Administration of Lactobacillus paracasei strains improves immunomodulation and changes the composition of gut microbiota leading to improvement of colitis in mice. J. Funct. Foods 52, 565–575. doi: 10.1016/j.jff.2018.11.035
Kishimoto, T., and Hirano, T. (1988). Molecular regulation of B lymphocyte response. Annu. Rev. Immunol. 6, 485–512. doi: 10.1146/annurev.iy.06.040188.002413
Kosaka, S., Tamauchi, H., Terashima, M., Maruyama, H., Habu, S., and Kitasato, H. (2011). IL-10 controls Th2-type cytokine production and eosinophil infiltration in a mouse model of allergic airway inflammation. Immunobiology 16, 811–820. doi: 10.1016/j.imbio.2010.12.003
Kwon, H.-K., Lee, C.-G., So, J.-S., Chae, C.-S., Hwang, J.-S., Sahoo, A., et al. (2010). Generation of regulatory dendritic cells and CD4+Foxp3+ T cells by probiotics administration suppresses immune disorders. Proc. Natl. Acad. Sci. U.S.A. 107, 2159–2164. doi: 10.1073/pnas.0904055107
Kwon, M. S., Lim, S. K., Jang, J. Y., Lee, J., Park, H. K., Kim, N., et al. (2018). Lactobacillus sakei WIKIM30 ameliorates atopic dermatitis-like skin lesions by inducing regulatory T cells and altering gut microbiota structure in mice. Front. Immunol. 9:1905. doi: 10.3389/fimmu.2018.01905
Lee, C. H., and Yu, H. S. (2011). Biomarkers for itch and disease severity in atopic dermatitis. Curr. Probl. Dermatol. 41, 136–148. doi: 10.1159/000323307
Levy, M., Kolodziejczyk, A. A., Thaiss, C. A., and Elinav, E. (2017). Dysbiosis and the immune system. Nat. Rev. Immunol. 17, 219–232. doi: 10.1038/nri.2017.7
Lim, M.Y., You, H.J., Yoon, H.S., Kwon, B., Lee, J.Y., Lee, S., et al. (2016). The effect of heritability and host genetics on the gut microbiota and metabolic syndrome. Gut 66, 1031–1038. doi: 10.1136/gutjnl-2015-311326
Lim, S. K., Kwon, M. S., Lee, J., Oh, Y. J., Jang, J. Y., Lee, J. H., et al. (2017). Weissella cibaria WIKIM28 ameliorates atopic dermatitis-like skin lesions by inducing tolerogenic dendritic cells and regulatory T cells in BALB/c mice. Sci. Rep. 9:40040. doi: 10.1038/srep40040
Liu, Y.-J. (2006). Thymic stromal lymphopoietin: master switch for allergic inflammation. J. Exp. Med. 203, 269–273. doi: 10.1084/jem.20051745
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Loy, A., Pfann, C., Steinberger, M., Hanson, B., Herp, S., Brugiroux, S., et al. (2017). Lifestyle and horizontal gene transfer-mediated evolution of Mucispirillum schaedleri, a core member of the murine gut microbiota. mSystems 2, e00717–e00176. doi: 10.1128/mSystems.00171-16
Lucke, K., Miehlke, S., Jacobs, E., and Schuppler, M. (2006). Prevalence of Bacteroides and Prevotella spp. in ulcerative colitis. J. Med. Microbiol. 55, 617–624. doi: 10.1099/jmm.0.46198-0
Matsuda, H., Watanabe, N., Geba, G. P., Sperl, J., Tsudzuki, M., Hiroi, J., et al. (1997). Development of atopic dermatitis-like skin lesion with IgE hyperproduction in NC/Nga mice. Int. Immunol. 9, 461–466. doi: 10.1093/intimm/9.3.461
McNabney, S. M., and Henagan, T. M. (2017). Short chain fatty acids in the colon and peripheral tissues: a focus on butyrate, colon cancer, obesity and insulin resistance. Nutrients 9:E1348. doi: 10.3390/nu9121348
Meagher, L. J., Wines, N. Y., and Cooper, A. J. (2002). Atopic dermatitis: review of immunopathogenesis and advances in immunosuppressive therapy. Australas. J. Dermatol. 43, 247–254. doi: 10.1046/j.1440-0960.2002.00610.x
Nakazato, J., Kishida, M., Kuroiwa, R., Fujiwara, J., Shimoda, M., and Shinomiya, N. (2008). Serum levels of Th2 chemokines, CCL17, CCL22, and CCL27, were the important markers of severity in infantile atopic dermatitis. Pediatr. Allergy Immunol. 19, 605–613. doi: 10.1111/j.1399-3038.2007.00692.x
Neis, E. P., Dejong, C. H., and Rensen, S. S. (2015). The role of microbial amino acid metabolism in host metabolism. Nutrients 7, 2930–2946. doi: 10.3390/nu7042930
Olszak, T., An, D., Zeissig, S., Vera, M. P., Richter, J., Franke, A., et al. (2012). Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336, 489–493. doi: 10.1126/science.1219328
Park, C. W., Youn, M., Jung, Y.-M., Kim, H., Jeong, Y., Lee, H.-K., et al. (2008). New functional probiotic Lactobacillus sakei probio 65 alleviates atopic symptoms in the mouse. J. Med. Food 11, 405–412. doi: 10.1089/jmf.2007.0144
Peng, W., and Novak, N. (2015). Pathogenesis of atopic dermatitis. Clin. Exp. Allergy 45, 566–574. doi: 10.1111/cea.12495
Perlman, R.L. (2016). Mouse models of human disease: an evolutionary perspective. Evol. Med. Public Health 2016, 170–176. doi: 10.1093/emph/eow014
Pineiro, M., and Stanton, C. (2007). Probiotic bacteria: legislative framework– requirements to evidence basis. J. Nutr. 137, 850S−853S. doi: 10.1093/jn/137.3.850S
Purchiaroni, F., Tortora, A., Gabrielli, M., Bertucci, F., Gigante, G., Ianiro, G., et al. (2013). The role of intestinal microbiota and the immune system. Eur. Rev. Med. Pharmacol. 17, 323–333.
Rather, I. A., Bajpai, V. K., Kumar, S., Lim, J., Paek, W. K., and Park, Y. H. (2016). Probiotics and atopic dermatitis: an overview. Front. Microbiol. 36, 899–906. doi: 10.3389/fmicb.2016.00507
Rooks, M. G., and Garrett, W. S. (2016). Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 16, 341–352. doi: 10.1038/nri.2016.42
Roucher, V. F., Desnots, E., Naël, C., Agnoux, A. M., Alexandre-Gouabau, M. C., Darmaun, D., et al. (2013). Use of UPLC-ESI-MS/MS to quantitate free amino acid concentrations in micro-samples of mammalian milk. Springerplus 2:622. doi: 10.1186/2193-1801-2-622
Saeki, H., Furue, M., Furukawa, F., Hide, M., Ohtsuki, M., Katayama, I., et al. (2009). Guidelines for management of atopic dermatitis. J. Dermatol. 36, 563–577. doi: 10.1111/j.1346-8138.2009.00706.x
Segata, N., Izard, J., Walron, L., Gevers, D., Miropolsky, L., Garrett, W.S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Sommer, F., and Bäckhed, F. (2013). The gut microbiota-masters of host development and physiology. Nat. Rev. Microbiol. 11, 227–238. doi: 10.1038/nrmicro2974
Takaoka, A., Arai, I., Sugimoto, M., Honma, Y., Futaki, N., Nakamura, A., et al. (2006). Involvement of IL-31 on scratching behavior in NC/Nga mice with atopic-like dermatitis. Exp. Dermatol. 15, 161–167. doi: 10.1111/j.1600-0625.2006.00405.x
Tatsuno, K., Fujiyama, T., Yamaguchi, H., Waki, M., and Tokura, Y. (2015). TSLP directly interacts with skin-homing Th2 cells highly expressing its receptor to enhance IL-4 production in atopic dermatitis. J. Invest. Dermatol. 135, 3017–3024. doi: 10.1038/jid.2015.318
Vonk, R. J., Priebe, M., Meijer, K., Venema, K., and Roelofsen, H. (2011). The interaction of short-chain fatty acids (SCFA) with adipose tissue; relevance for systemic inflammation. Gastroenterology 140:860. doi: 10.1016/S0016-5085(11)63571-3
Yue, F., Cheng, Y., Breschi, A., Viestra, J., Wu, W., Ryba, T., et al. (2014). A comparative encyclopedia of DNA elements in the mouse genome. Nature 515, 355–364. doi: 10.1038/nature13992
Zeng, M. Y., Inohara, N., and Nuñez, G. (2017). Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 10, 18–26. doi: 10.1038/mi.2016.75
Zhang, H., Kong, H., Zeng, X., Guo, L., Sun, X., and He, S. (2014). Subsets of regulatory T cells and their roles in allergy. J. Transl. Med. 12:125. doi: 10.1186/1479-5876-12-125
Keywords: atopic dermatitis, Lactobacillus fermentum, immunomodulation, microbiota, metabolome, microbiome
Citation: Kim W-K, Jang YJ, Han DH, Seo B, Park S, Lee CH and Ko G (2019) Administration of Lactobacillus fermentum KBL375 Causes Taxonomic and Functional Changes in Gut Microbiota Leading to Improvement of Atopic Dermatitis. Front. Mol. Biosci. 6:92. doi: 10.3389/fmolb.2019.00092
Received: 27 May 2019; Accepted: 11 September 2019;
Published: 27 September 2019.
Edited by:
Christopher Staley, University of Minnesota Twin Cities, United StatesReviewed by:
Joshua Michael Albert Stough, The University of Tennessee, Knoxville, United StatesTatsuya Unno, Jeju National University, South Korea
Copyright © 2019 Kim, Jang, Han, Seo, Park, Lee and Ko. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: GwangPyo Ko, Z2tvQHNudS5hYy5rcg==
†These authors have contributed equally to this work as co-first authors
 Woon-Ki Kim
Woon-Ki Kim You Jin Jang
You Jin Jang Dae Hee Han
Dae Hee Han Boram Seo
Boram Seo SungJun Park
SungJun Park Chang Hyung Lee
Chang Hyung Lee GwangPyo Ko
GwangPyo Ko