- 1Department of Biology, High Point University, High Point, NC, United States
- 2Department of Biology, Washington University, St. Louis, MO, United States
Agro-ecosystems are dominated by crop plants and the weedy species that thrive under agricultural conditions. Weedy crop relatives are some of the most difficult weeds to manage and can dramatically reduce crop yields when left unchecked. Weedy rice has resulted from multiple de-domestication events from crop rice in different rice growing regions. Interestingly, both South Korea and the United States harbor weedy rice populations that share ancestry with indica cultivars and temperate japonica cultivars. Here we compare weedy rice populations from South Korea and the United States on order to identify if they are the result of the same de-domestication events. We find that weedy rice populations in South Korea are genetically distinct from weedy rice found in the USA and are therefore the result of two unique de-domestication events. Low levels of genetic diversity among Korean weedy rice accessions (haplotype diversity = 0.0188 and 0.0324) indicate recent de-domestication events from crop relatives.
Introduction
Agricultural weeds account for approximately one third of all crop yield loss (Oerke, 2006), contributing to food shortages worldwide. Understanding the population structure and mechanisms of adaption in weedy plants informs best management practices in agro-ecosystems. In particular, weedy crop relatives have played a longstanding role in agro-ecosystem dynamics, driving both the evolution of crops as well as the development of new management strategies (Kwit et al., 2011; Li and Olsen, 2020). A well-documented example is weedy rice, a conspecific weed of cultivated rice (Oryza sativa L.). Weedy rice has a distribution that spans nearly all rice growing regions around the world. Infestations of this species can cause up to an 80% loss in harvest for cultivated rice and is often cited as a major limiting factor for rice production. In the United States alone, estimates of production loss due to weedy rice could feed an additional 12 million people annually (Durand-Morat et al., 2018). Management efforts for weedy rice have ranged from manual removal to large scale herbicide application.
There are five main cultivated rice subtypes that are genetically and phenotypically distinguishable: indica, aus, aromatic, tropical japonica, and temperate japonica. Weedy rice has arisen through de-domestication events from at least 3 of the 5 cultivated subtypes (Qiu et al., 2020). In the United States which has no wild Oryza species, there are three main weedy rice subgroups: (1) straw-hulled weedy rice from the southern US (SH weeds) that are genetically similar to indica type cultivated rice, (2) black-hulled weedy rice from the southern US (BH weeds) that are genetically similar to aus type cultivated rice, and (3) California weedy rice (CA weeds) that is genetically similar to temperate japonica cultivated rice. All three of these subgroups seem to have evolved from de-domestication events in each progenitor cultivar group (Reagon et al., 2010; Kanapeckas et al., 2016; Qiu et al., 2020). In other world regions where wild rice (Oryza rufipogon) is common, weedy rice populations have genetic contributions from both wild rice and cultivated rice populations (Vigueira et al., 2019; Qiu et al., 2020).
Weedy rice from South Korea (Korean weedy rice) is composed of two main subgroups based on population structure analysis: indica-like and temperate japonica-like (Vigueira et al., 2019). Using whole genome sequencing, other researchers have also placed Korean weedy rice into the same two subgroups (He et al., 2017). Like the United States, South Korea is not in the natural range of wild Oryza species. Therefore, all rice crops have been imported into the region for cultivation. Weedy rice populations in these countries have therefore either been introduced with cultivated rice seed or have evolved in place since rice cultivation began (Reagon et al., 2010; Kanapeckas et al., 2016; Qiu et al., 2020).
Here, we aim to more closely examine the genetic similarities of weedy rice from South Korea and the United States. We have used both candidate genes (Rc, controlling pericarp color; Bh4, controlling hull color; and sh4, controlling seed shattering) as well as genome-wide neutral genetic markers (Sequence Tagged Sites) to better understand the evolutionary history and population structure of weedy rice from these two regions. We find that Korean weedy rice is genetically distinct from US weedy rice populations despite their phenotypic similarities, indicating that these weeds were the result of unique de-domestication events.
Methods
Sampling and Sequencing
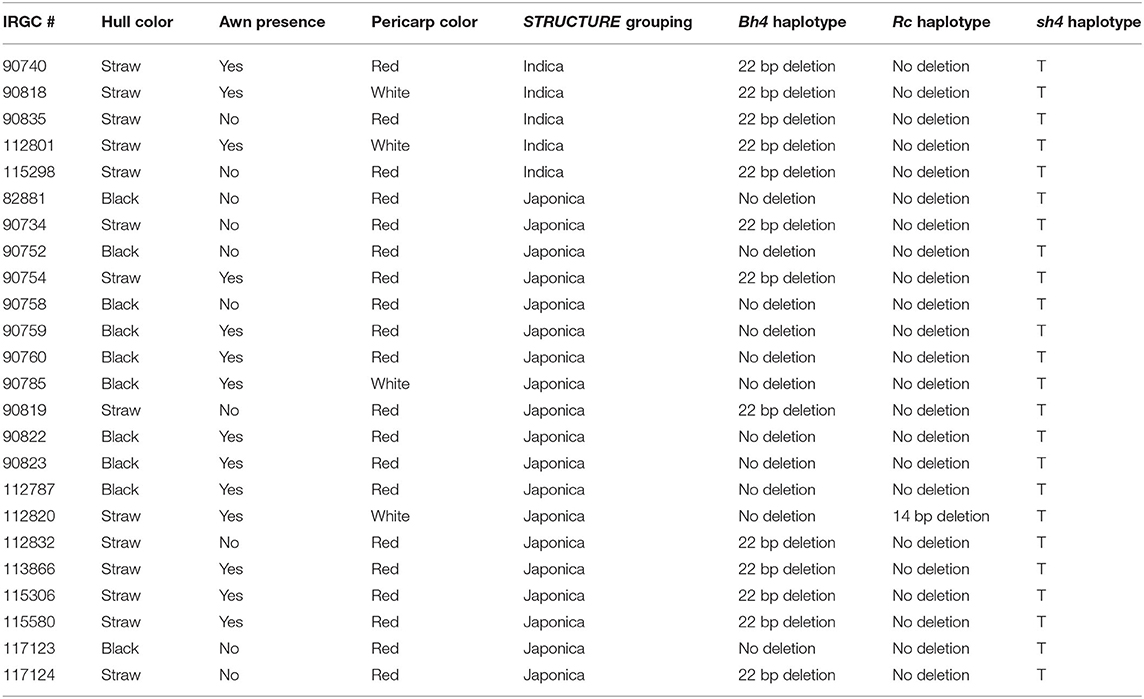
Rice seeds were obtained from the International Rice Germplasm Collection (IRGC) of the International Rice Research Institute (IRRI; Los Baños, Philippines). Twenty-four accessions of weedy rice from South Korea were selected to represent the phenotypic diversity for hull color, pericarp color, and presence of awns (phenotypes are listed in Table 1) from a total sample of 226 accessions (Supplementary Table 1). Eighteen of these samples were previously included in a comparative study with weedy, wild, and cultivated rice from Southeast Asian and the United States (Vigueira et al., 2019). Seeds were germinated and grown to the young seedling stage in the greenhouse. DNA was extracted from young leaf tissue using DNeasy Plant DNA kits (QIAGEN, Hilden, Germany).
Polymerase Chain Reaction (PCR) was carried out using standard conditions to amplify 48 Sequence Tagged Sites (STS loci) as described in Reagon et al. (2010). Due to inconsistent amplification, 7 loci were excluded from the analysis. Regions of the three candidate genes (Rc, Bh4, and sh4) were amplified by PCR using primers and conditions as previously described (Konishi et al., 2006; Sweeney et al., 2006; Zhu et al., 2011). Successful PCR amplification was confirmed using gel electrophoresis and excess primers and dNTPs were removed using Exonuclease I and Antarctic phosphatase treatment. Direct Sanger sequencing in both the forward and reverse direction was carried out by Eurofins Genomics (Louisville, KY, USA).
Sequences were assembled into contiguously aligned sequence “contigs” and aligned using CodonCode Aligner. All sequences were inspected visually for quality and for the presence of heterozygous sites. Low quality sequences were removed from the dataset. Heterozygous base calls were randomly assigned to two pseudo-haplotypes, which were then phased using PHASE version 2.1 (Stephens et al., 2001; Stephens and Scheet, 2005). Due to very low levels of heterozygosity in the data set, haplotypes were inferred with very high probabilities and were consistently assigned across five independent runs. All sequences have been submitted to NCBI GenBank (accession numbers MT976168-MT977030).
Analysis of STS Loci
Phased haplotypes were aligned with STS sequences from 27 weedy rice accessions collected in California (Kanapeckas et al., 2016; GenBank accessions KT441140-KT443009) as well as from a diverse sampling of 206 accessions that includes Southern US weedy as well as wild and cultivated rice representing major Oryza varieties and species (Reagon et al., 2010; GenBank accessions GQ999668-GQ999777). Population structure was inferred using STRUCTURE 2.3.4 (Pritchard et al., 2000). Initial runs included all samples in the dataset. To limit the number of possible subpopulations with proposed ancestry to Korean weedy rice and therefore better resolve differences between closely related groups, we reduced the data set to include the following groups: all Korean weedy rice (24 accessions), weedy rice from California (27 accessions), weedy rice from the southern US (58 accessions of BH and SH weedy rice), and the five cultivar groups (75 accessions representing indica, aus, aromatic, temperate japonica, and tropical japonica). The number of populations (K) was tested with five permutations each between values of K = 1 to K = 10. Each permutation had a burn-in period of 100,000 steps and a MCMC chain length of 500,000 steps after the burn-in. STRUCTURE HARVESTER (Earl and vonHoldt, 2012) was used to calculate Delta K (Evanno et al., 2005) and determine the K value that maximized the marginal likelihood. DISTRUCT version 1.1 (Rosenberg, 2003) was used to produce the graphical display of structure results. As a complement to STRUCTURE analysis, principal component analysis (PCA) was run on haplotype data using the ML model in JMP. PCAs were produced for the full dataset and the subset of data used in the STRUCTURE analysis. The PCA produced from data in our STRUCTURE analysis separated California weedy rice from the rest of the groups. Therefore, a PCA was also run with California weedy rice removed from the dataset.
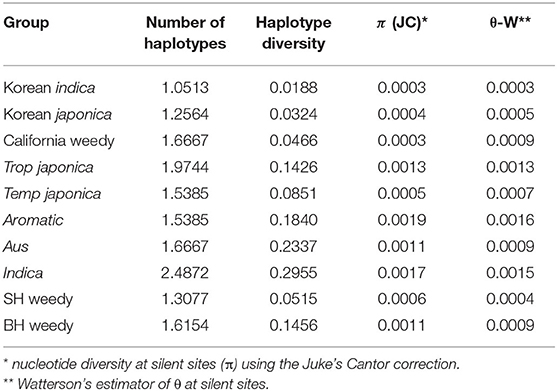
Summary statistics for each STS locus, including nucleotide diversity at silent sites (π) using the Juke's Cantor correction (Jukes and Cantor, 1969), Watterson's estimator of θ at silent sites (Watterson, 1975), number of segregating sites S, and haplotype diversity were calculated in DnaSP version 5.0 (Librado and Rozas, 2009). Averages for these statistics across all STS loci were calculated in Excel.
Candidate Gene Analysis
Candidate gene sequences were aligned with rice sequences from previous studies (Gross et al., 2010; Thurber et al., 2010; Vigueira et al., 2013). Genetic variants were determined by identifying haplotypes and mutations shared between Korean weedy rice and wild, weedy, or cultivated rice varieties.
Results
Population Structure
Analyses of neutral STS markers grouped Korean weedy rice groups two distinct genetic subpopulations. STRUCTURE analysis comparing Korean weeds with the five major cultivars and weedy rice from the United States partitions Korean weeds into an indica-like group and a temperate japonica-like group (Figure 1). STRUCTURE plots are shown for K = 4 based on Delta K results and K = 5 because temperate japonica was distinguishable from tropical japonica at that number of subpopulations. Principal Component Analysis reveals the same genetic groupings as found in STRUCTURE analysis. PCA was performed with and without California weedy rice (Figure 2), as it was the most genetically distinct group. Due to consistent genetic grouping, we analyzed the Korean weedy rice as two separate populations, indica-like and temperate japonica-like weedy rice, for the remainder of the analysis.
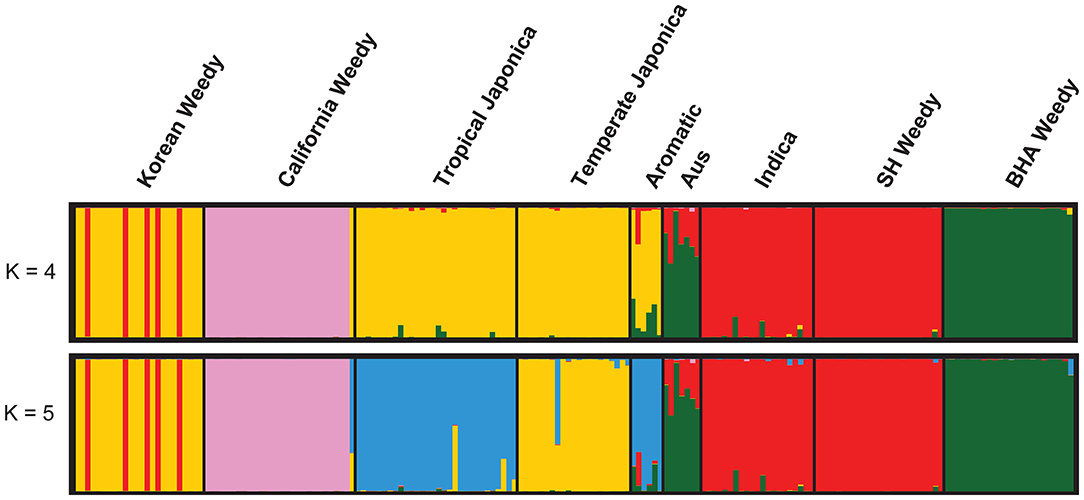
Figure 1. STRUCTURE plot of five cultivated rice subgroups and four weedy rice groups. K = 4 was chosen using Delta K, while K = 5 was included as it separated temperate and tropical japonica. Korean weedy rice groups with indica (red) and temperate japonica (yellow) cultivars.
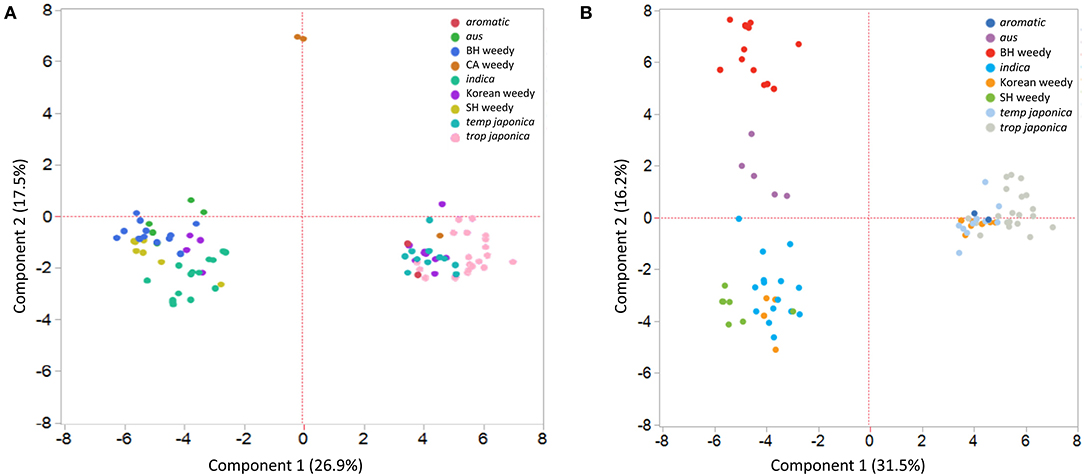
Figure 2. Principal Component Analysis (PCA) of cultivated and weedy rice including California weedy rice (A) and without California weedy rice. (B) Korean weedy rice groups with temperate japonica cultivars and indica cultivars.
Diversity of Korean Weedy Rice
Summary statistics indicated very low genetic diversity within Korean weedy rice sub-groups, consistent with a population bottleneck during de-domestication from cultivated rice (Table 2). Average haplotype diversity and nucleotide diversity is lower in weedy rice groups compared to cultivated rice. Korean weedy rice has even lower values of average haplotype diversity (0.0188 and 0.0324) than SH weedy rice (0.0515), BH weedy rice (0.1456) and California weedy rice (0.0466). In addition, Korean weedy rice had an average pairwise nucleotide diversity (π) of about half the average value found in SH and BH weedy rice from the Southern US (Table 2). This level of genetic diversity is similar to that found in California weedy rice. Low levels of genetic diversity in Korean weedy rice could be a result of a very recent genetic bottleneck associated with a recent de-domestication event.
Candidate Gene Alleles
Korean weedy rice phenotypes and candidate gene allele information can be found in Table 1. Of the 24 Korean weedy rice accessions included in our analysis, 14 had a straw colored hull and 10 had a black colored hull. All straw colored hull accessions with the exception of one (IRGC #112820) contained the 22 bp causal deletion in Bh4 that is found in most cultivated rice. As expected, black hulled Korean weeds carried the ancestral wild-type allele (lacking the 22 bp deletion) which is found in black hull weedy and wild rice.
Four Korean weedy rice accessions had a white pericarp color and twenty had red pericarps. Of the four, one contained a previously determined white pericarp allele (14 bp deletion) found in nearly all cultivated rice. Interestingly, this accession is the same accession (IRGC #112820) that does not have the 22 bp deletion at the Bh4 locus despite having a straw hull. The other three had no obvious deletions or loss of function mutations.
All 24 Korean weedy rice accessions contained the “T” reduced-shattering allele found in cultivated rice at the sh4 locus. This allele is also present in weedy rice from other world regions (Thurber et al., 2010), which further supports the gain of shattering phenotype in weedy rice was acquired during de-domestication.
Discussion
Korean weedy rice groups most closely with two distinct cultivated rice subtypes: indica and temperate japonica. These weeds likely originated from two distinct de-domestication events from cultivated varieties. These de-domestication events are likely recent, given low genetic divergence from cultivated groups and low genetic diversity within weedy rice groups. We find patterns consistent with cultivated rice de-domestication at both STS loci as well as candidate genes for weedy traits (Bh4, Rc, and sh4). These patterns have also been found in weedy rice from the United States (Gross et al., 2010; Thurber et al., 2010; Vigueira et al., 2013).
Weedy rice subtypes from the southern US include straw-hulled (indica-like) and black-hulled (aus-like). In California, there is a distinct weedy rice population that groups most closely with temperate japonica cultivars based on coalescent modeling in Kanapeckas et al. (2016). Our PCA results from the California weedy rice group may better resolve this grouping. One weedy accession from California groups closely with japonica cultivars in our PCA, while the other accessions are genetically distinct from all other rice subtypes. This is an interesting discovery that warrants additional sampling from the California weedy rice population. Weedy rice from Korea does not seem to share recent ancestry with any of the US weedy rice populations, further supporting the previous findings that this group is the result of two distinct de-domestication events from indica and japonica cultivars.
Although this study does not provide definitive evidence for the location of Korean weedy rice de-domestication, recent studies of world-wide samples of weedy and cultivated rice points to other de-domestication events from rice cultivars found in the Korean peninsula (Qiu et al., 2020). Interestingly, kinship analysis results for Korean weeds identified closest cultivar relatives from Korea, Japan, China, India, and Egypt (Qiu et al., 2020). Taken together, it seems that Korean weedy rice may have multiple origins possibly including de-domestication events from cultivars in situ.
Data Availability Statement
The datasets generated for this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI GenBank (accession numbers MT976168-MT977030).
Author Contributions
CV, PV, and KO designed the study. CV, PV, CW, and ZC collected the data. CV and PV analyzed the data. CV wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by High Point University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2020.602612/full#supplementary-material
References
Durand-Morat, A., Nalley, L. L., and Thoma, G. (2018). The implications of red rice on food security. Glob. Food Sec. 18, 62–75. doi: 10.1016/j.gfs.2018.08.004
Earl, D. A., and vonHoldt, B. M. (2012). Structure harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361. doi: 10.1007/s12686-011-9548-7
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Gross, B. L., Reagon, M., Hsu, S.-C., Caicedo, A. L., Jia, Y., and Olsen, K. M. (2010). Seeing red: the origin of grain pigmentation in US weedy rice. Mol. Ecol. 19, 3380–3393. doi: 10.1111/j.1365-294X.2010.04707.x
He, Q., Kim, K. W., and Park, Y. J. (2017). Population genomics identifies the origin and signatures of selection of Korean weedy rice. Plant Biotechnol. J. 15, 357–366. doi: 10.1111/pbi.12630
Jukes, T. H., and Cantor, C. R. (1969). “Evolution of protein molecules,” in Mammalian Protein Metabolism, ed H. N. Munro (New York, NY: Academic Press). 21–132. doi: 10.1016/B978-1-4832-3211-9.50009-7
Kanapeckas, Kimberly, L., Vigueira, C. C., Ortiz, A., Gettler, K. A., Burgos, N. R., Fischer, A. J., et al. (2016). Escape to ferality: the endoferal origin of weedy rice from crop rice through de-domestication. PLoS ONE 11:e0162676. doi: 10.1371/journal.pone.0162676
Konishi, S., Izawa, T., Lin, S. Y., Ebana, K., Fukuta, Y., Sasaki, T., et al. (2006). An SNP caused loss of seed shattering during rice domestication. Science 312, 1392–1396. doi: 10.1126/science.1126410
Kwit, C., Moon, H. S., Warwick, S. I., and Stewart, C. N. (2011). Transgene introgression in crop relatives: Molecular evidence and mitigation strategies. Trends Biotechnol. 29, 284–293. doi: 10.1016/j.tibtech.2011.02.003
Li, L.-F., and Olsen, K. M. (2020). “Population genomics of weedy crop relatives: insights from weedy rice,” in Population Genomics, eds O. P. Rajora (Cham: Springer). doi: 10.1007/13836_2020_77
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Oerke, E. C. (2006). Crop losses to pests. J. Agric. Sci. 144, 31–43. doi: 10.1017/S0021859605005708
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics, 155, 945–959. doi: 10.1111/j.1471-8286.2007.01758.x
Qiu, J., Jia, L., Wu, D., Weng, X., Chen, L., Sun, J., et al. (2020). Diverse genetic mechanisms underlie worldwide convergent rice feralization. Genome Biol. 21:70. doi: 10.1186/s13059-020-01980-x
Reagon, M., Thurber, C. S., Gross, B. L., Olsen, K. M., Jia, Y., and Caicedo, A. L. (2010). Genomic patterns of nucleotide diversity in divergent populations of U.S. weedy rice. BMC Evol. Biol. 10:180. doi: 10.1186/1471-2148-10-180
Rosenberg, N. A. (2003). Distruct: a program for the graphical display of population structure. Mol. Ecol. Notes 4, 137–138. doi: 10.1046/j.1471-8286.2003.00566.x
Stephens, M., and Scheet, P. (2005). Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am. J. Hum. Genet. 76, 449–462. doi: 10.1086/428594
Stephens, M., Smith, N. J., and Donnelly, P. (2001). A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68, 978–989. doi: 10.1086/319501
Sweeney, M. T., Thomson, M. J., Pfeil, B. E., and Mccouch, S. (2006). Caught red-handed : rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell 18, 283–294. doi: 10.1105/tpc.105.038430
Thurber, C. S., Reagon, M., Gross, B. L., Olsen, K. M., Jia, Y., and Caicedo, A. L. (2010). Molecular evolution of shattering loci in U.S. weedy rice. Mol. Ecol. 19, 3271–3284. doi: 10.1111/j.1365-294X.2010.04708.x
Vigueira, C. C., Li, W., and Olsen, K. M. (2013). The role of Bh4 in parallel evolution of hull colour in domesticated and weedy rice. J. Evol. Biol. 26, 1738–1749. doi: 10.1111/jeb.12171
Vigueira, C. C., Qi, X., Song, B.-K., Li, L.-F., Caicedo, A. L., Jia, Y., et al. (2019). Call of the wild rice: Oryza rufipogon shapes weedy rice evolution in Southeast Asia. Evol. Appl. 12:eva12581. doi: 10.1111/eva.12581
Watterson, G. A. (1975). On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7, 256–276. doi: 10.1016/0040-5809(75)90020-9
Keywords: oryza, weedy crop relatives, de-domestication, agricultural weeds, populaton genomics
Citation: Vigueira PA, Olsen KM, Wagner CR, Chittick ZB and Vigueira CC (2020) Weedy Rice From South Korea Arose From Two Distinct De-domestication Events. Front. Agron. 2:602612. doi: 10.3389/fagro.2020.602612
Received: 03 September 2020; Accepted: 13 October 2020;
Published: 13 November 2020.
Edited by:
Lauren M. Lazaro, Louisiana State University Agricultural Center, United StatesReviewed by:
Md Asaduzzaman, New South Wales Department of Primary Industries, AustraliaPrasanta Kumar Subudhi, Louisiana State University, United States
Copyright © 2020 Vigueira, Olsen, Wagner, Chittick and Vigueira. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cynthia C. Vigueira, Y3ZpZ3VlaXJAaGlnaHBvaW50LmVkdQ==
 Patrick A. Vigueira
Patrick A. Vigueira Kenneth M. Olsen
Kenneth M. Olsen Christopher R. Wagner
Christopher R. Wagner Zoey B. Chittick
Zoey B. Chittick Cynthia C. Vigueira
Cynthia C. Vigueira