- 1Centro de Biotecnología y Genómica de Plantas, Universidad Politécnica de Madrid (UPM)-Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA), Campus de Montegancedo, Madrid, Spain
- 2Departamento de Biotecnología y Biología Vegetal, ETSI Agronómica, Alimentaria y de Biosistemas, Universidad Politécnica de Madrid, Madrid, Spain
- 3Laboratório de Bioquímica de Microrganismos e Plantas, Faculdade de Ciências Agrárias e Veterinárias de Jaboticabal, Universidade Estadual Paulista Júlio de Mesquita Filho, São Paulo, Brazil
- 4Laboratoire d'Ecologie Microbienne, Faculté des Sciences de la Nature et de la Vie, Université de Bejaia, Bejaia, Algeria
At present, there are numerous examples in which symbiotic nitrogen fixation by rhizobia can totally replace the use of nitrogen fertilizers in legume crops. Over the years, there has been a great effort by research institutions to develop and select rhizobial inoculants adapted for these crops. The symbiotic process is highly dependent on the dynamic exchange of signals and molecular nutrients between partners. Our focus in this review was to discuss the two key determinants in successful symbiotic interactions of rhizobia to nodulate pulses. One of them is the production of exopolysaccharides (EPS) and the other the presence of the type VI secretion system (T6SS). EPS are extracellular polymers weakly associated with the bacterial surface and are abundantly released into acid soils facilitating, among other functions, an adaptation of rhizobia to this environment. On the other hand, different protein secretion systems, involved in symbiosis, have been described in rhizobia. This is not the case with the T6SS. The current availability of various rhizobial genomes offers the possibility of discussing its role in symbiosis. The study of these determinants will be of great utility for the selection of effective inoculants for legumes, a promising way to improve nitrogen acquisition by legumes.
Rhizobia for Pulses
Legumes are of capital importance for human and animal feed and for most ecosystems. This is due, among other factors, to the fact that most species are capable of biologically fixing nitrogen (BNF) by association with soil diazotrophic bacteria called rhizobia. Most legume crops can self-supply the nitrogen they need, this being a fact of great relevance to mitigate the negative effects of nitrogen fertilizer leaching in the environment. All this has led to great interest in the selection of effective inoculants. Brazil is considered a global leader in the use of inoculants and one of the largest producers of pulses worldwide (Santos et al., 2019). A reference is made in the following section to the most commonly selected strains (Bradyrhizobium and Rhizobium), as well as the beneficial role that rhizobial exopolysaccharides (EPS) and T6SS can have in an effective symbiosis and therefore in BNF.
Bradyrhizobium Species in Brazilian Soils
Reports on legume nodulation by Bradyrhizobium strains in Brazil are quite abundant, mainly in terms of legumes and bradyrhizobial isolates diversity. However, there are still a large number of rhizobial isolates waiting to be characterized. In this section, we summarize data on the history and evolution of studies on Bradyrhizobium in soybean (Glycine max).
The SEMIA Rhizobia Collection of Brazil originated in 1950 through the isolation and selection of efficient native strains for legumes of economic importance by professionals from the Agricultural Microbiology Section (SEMIA) of the Agronomic Research Institute (IPAGRO today FEPAGRO). Brazilian soybean crops boosted the beginning of inoculant production in Brazil in the 1960s due to the lack of native bacteria that were able to efficiently nodulate soybean, therefore, supplied mainly with North American strains. The first record of the SEMIA Collection was in 1973. In the 1980s, studies showed enormous genetic and physiological variability between the strains of Bradyrhizobium japonicum, suggesting the subdivision of Bradyrhizobium into two species, B. japonicum and B. elkanii (Kuykendall et al., 1992).
Argentina, Brazil, and the United States are the most important soybean producers. Bradyrhizobium is the most prominent and widely studied in terms of nodulation and nitrogen fixation in soybeans. The Ministry of Agriculture, Livestock and Food (MAPA) of Brazil emitted official reports about the quality of soybean inoculants (number of viable cells, strain identification, and presence of contaminants). In Brazil and South America, today, MAPA authorizes the use of different Bradyrhizobium strains in the formulation of commercial inoculants (Supplementary Table 1), consequently settling these populations in most of the soybean Brazilian soils (Hungria et al., 2001; Delamuta et al., 2013). Since the beginning of inoculant production in the 1980s, seven species that were able to efficiently nodulate soybean have been described, B. japonicum, B. diazoefficiens, B. elkanii, and B. liaoningense (Willems, 2006); and Mesorhizobium tianshanense (Chen et al., 1995); Sinorhizobium fredii and S. xinjiangense (Chen et al., 1988). The number of validated Bradyrhizobium species published has greatly increased in recent years, and ~35% are from South America and another 35% from other regions mainly from China.
The soybean BNF is consolidated though many farmers use synthetic fertilizers. Bradyrhizobium inoculants are alternatives to reduce water pollution and nitrous oxide (N2O) emission (Akiyama et al., 2016). However, Obando et al. (2019) showed the first report that pointed out the incomplete denitrification pathway in five of the most used strains for soybean.
Göttfert et al. (2001) described symbiosis-specific genes in a 410-kilobase DNA region of the B. diazoefficiens USDA110 chromosome and in 2002, the complete genomic sequence was described (Kaneko et al., 2002). The pioneering study reporting the high diversity of Bradyrhizobium genospecies was through a DNA–DNA hybridization conducted by Willems et al. (2001). Bradyrhizobium's genomes are larger than those of other genera of rhizobia, reflecting their vast metabolic, and different life cycles. In addition, high phenotypic and/or genotypic diversity, both intra- and interspecific, between the strains of Bradyrhizobium was observed (Guimarães et al., 2012, 2015; Rufini et al., 2014). Despite this diversity, Bradyrhizobium species have a monophyletic character with few exceptions, particularly the photosynthetic bradyrhizobia (Moulin et al., 2004). Iida et al. (2015) reported the presence of some islands of symbiosis shuffled with abundant insertion sequences in the genomes of extra-slow-growing Bradyrhizobium strains. In the symbiotic islands, there are insertion sequences and groups of symbiotic genes, such as nod, nif, fix, and rhc. nifH and nod Genes are the most used for phylogenetic reconstructions of symbiosis, both transferred vertical and horizontally between different chromosomal backgrounds (Menna and Hungria, 2011). Generally, the transfer of specific symbiotic genes is considered a fundamental mechanism that allows legumes to form symbioses with rhizobia adapted to private soils (Andrews et al., 2017).
Rhizobium Tropici, a Species Resistant to Acidic Growth Conditions
Rhizobium species are widely used as inoculants. R. tropici CIAT899 establishes symbiosis with different legume species, including common beans (Phaseolus vulgaris) (Martínez-Romero et al., 1991). However, the success of this process can be limited by different environmental conditions, such as high temperature, low pH, presence of host legumes, and soil (Martínez-Romero et al., 1991; Hungria et al., 2001; Vinuesa et al., 2003; Puozaa et al., 2017).
When pH is reduced by plant exudates containing protons and organic acids, it provides a limitation for the survival of microorganisms and nodulation, and BNF can be severely affected. Finding acid-tolerant plants and compatible rhizobia are of remarkable agronomic and ecological relevance. R. tropici CIAT899 is more resistant to acidic conditions than most rhizobia. This resistance is related to several factors, such as hydroxylated ornitine lipids, which make the membrane less fluid and less permeable to protons (Vinuesa et al., 2003; Vences-Guzmán et al., 2011). Guerrero-Castro et al. (2018) identified 26 genes in R. tropici CIAT899 involved in the pH stress response, and transcriptomic analysis from cells grown under different pH conditions allowed the identification of 383 genes that are differentially expressed. The genes included response regulators and membrane transporters, enzymes involved in the metabolism of amino acids and carbohydrates, and proton extrusion.
Biosynthesis of EPS encoded in the R. tropici CIAT899 genome has been associated with acid tolerance, and implication of EPS in symbiosis is discussed in the next section.
Importance of Exopolysaccharides Production in Symbiosis
Rhizobial EPS are biopolymers of high–and low–molecular weight, secreted in the environment both in free living and in symbiosis (Skorupska et al., 2006). EPS contribute to tolerance of rhizobia against unfavorable conditions: reactive oxygen species, detergents, salt, acidic pH, drought, antimicrobial agents, etc. (Staehelin et al., 2006; Geddes et al., 2014; Naseem et al., 2018; Sun et al., 2020). Also, they participate in biofilm formation and attachment to abiotic or plant surfaces (Russo et al., 2006; Schäper et al., 2019). In the Rhizobium–legume interactions, EPS affect pre-infection events with the host, such as root hair curvature (Downie, 2010; Janczarek, 2011) and suppress the defense responses of the host plant (Jones et al., 2007). EPS can be perceived by plant LysM kinase receptors impairing or facilitating symbiosis with rhizobia depending on the polysaccharide composition, in L. japonicus, EPR3 receptor recognizes EPS controlling bacterial infection (Kawaharada et al., 2017; Kelly et al., 2017; Wong et al., 2020). EPS can have a negative effect on symbiosis as seen in S. fredii HH103/Glycine max, where flavonoids activate transcription of nodulation genes but repress EPS production (Acosta-Jurado et al., 2016).
EPS biosynthesis is unknown in most of the inoculants authorized in Brazil (Supplementary Table 1), particularly in Bradyrhizobium strains, however, it has been well-studied in Sinorhizobium meliloti and Rhizobium leguminosarum and required exo, exs, exp, and muc genes, which are mainly grouped and conserved in different strains (Janczarek et al., 2015). They are distributed on the chromosome or in symbiotic plasmids (Ivashina and Ksenzenko, 2012; Ormeño-Orrillo et al., 2012). EPS contain mostly D-glucose and D-galactose (Castellane et al., 2014, 2017, 2018, 2019), uronic acids and non-sugar substitutions can also be very important (López-Baena et al., 2016). Enzymes such as glycosyltransferase (PssA) and galactosyltransferase (PssJ) are responsible for assembling EPS sub-units in R. leguminosarum (Marczak et al., 2020).
Rhizobia are capable of producing two EPS forms, type I (succinoglycans) involved in S. meliloti nodule development (Reuber et al., 1991; Becker et al., 2002; Skorupska et al., 2006) and type II (galactoglycans) required for root hair attachment and biofilm formation (Rüberg et al., 1999; Becker et al., 2002; Skorupska et al., 2006; Sorroche et al., 2010). Also, R. leguminosarum EPS protect against zinc stress in the symbiosis with Trifolium (Kopycińska et al., 2018). In the symbiosis of S. fredii HH103/Glycyrrhiza uralensis, EPS were not strictly necessary (Margaret-Oliver et al., 2012) but Mesorhizobium tianshanense non-mucoid mutants were defective in nodulation with the same legume (Wang et al., 2008). R. tropici CIAT899 EPS are not required for bean nodulation but it is involved in competitiveness (Milner et al., 1992). A R. leguminosarum pssZ mutant showed reduced EPS production and nodule formation and affected bacteroids development (Lipa et al., 2018). Different amounts of EPS do not seem to interfere with the host, but with the ability to survive in the rhizosphere (Donati et al., 2013). S. meliloti Rm8530 produced three times more EPS than an expR mutant (Primo et al., 2020), and both had the same efficiency in the symbiosis with Medicago sativa (Pellock et al., 2002). Despite all these studies, the role of EPS in symbiosis remains unclear (Skorupska et al., 2006; Marczak et al., 2017).
In acidic soils, rhizobia from different hosts, such as Cicer, Phaseolus, Lens, and Leucaena can produce a considerable quantity of EPS (Cunningham and Munns, 1984). Secreted EPS are increased under abiotic stress (Hirsch, 1996; Sorroche et al., 2018; Primo et al., 2020). However, at acid pH R. tropici CIAT899 showed a lower production of biopolymers (Avelar Ferreira et al., 2012), whereas EPS production was not affected in R. favelukesii LPU83 (Nilsson et al., 2021).
The EPS in P. vulgaris induce genes involved in carbon metabolism, transcriptional regulation, circadian cycle, and phytohormone production (Via et al., 2015).
Type VI Secretion System (T6SS) in Rhizobia
The T6SS secretion system has been hardly related to symbiosis. It was described in 2006 in Vibrio cholerae and Pseudomonas aeruginosa (Mougous et al., 2006; Pukatzki et al., 2006). Since then it has been characterized in numerous bacteria, mainly Proteobacteria (Boyer et al., 2009). The T6SS is a nano-syringe that enables bacteria to inject proteins called effectors into both eukaryotic and prokaryotic cells (Basler and Mekalanos, 2012).
T6SS biosynthesis usually requires 13 core structural proteins encoded by tss genes and accessories named tag (type six accessory gene) (Shalom et al., 2007; Silverman et al., 2012).
Its structure contains three elements: a trans-membrane complex (TssMLJ), a cytoplasmic baseplate (TssEFGK), and a double tube, the outer one formed by a contractile sheath (TssBC) and the inner one by Hcp (TssD) ending with the puncturing device VgrG (TssI)-PAAR (Wang et al., 2019) (Figure 1). The PAAR domain (proline–alanine–alanine–arginine) is a sharp conical extension on the VgrG spike, is similar to DUF4150 domain, and facilitates secretion of a broad range of toxins (Shneider et al., 2013). The assembly of the inner tube and the contractile sheath requires TssA. After contraction, the T6SS tube is recycled by TssH (ClpV), an ATPase that disassembles the structure (Wang et al., 2019).
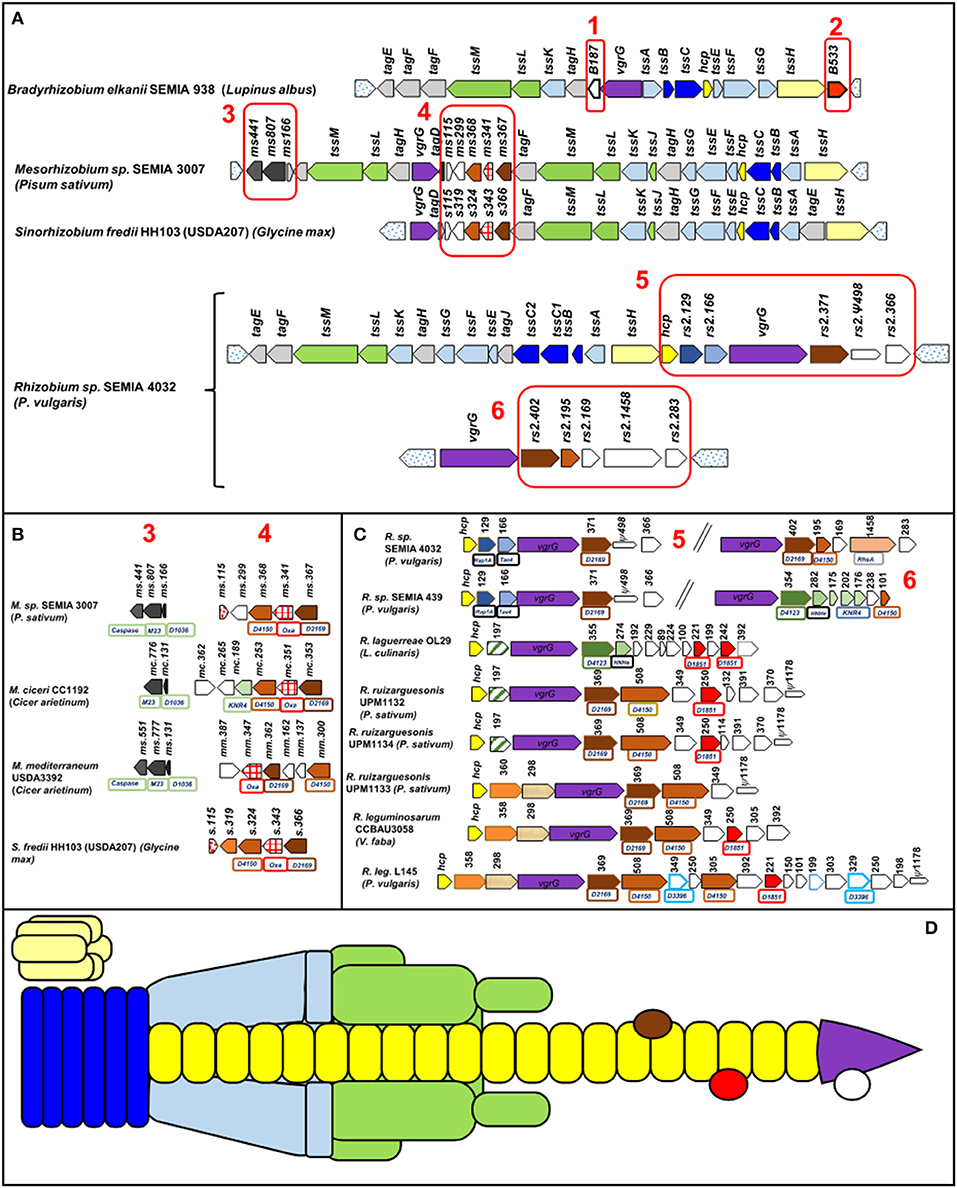
Figure 1. T6SS gene cluster organization of different rhizobia. In parentheses is the host legume. Orthologous genes show the same color and pattern. The numbered Boxes 1–6 contain mainly not tss or tag genes. Box 6 corresponds to strains harboring a second cluster with the vgrG gene. Uncolored genes encode proteins with no known domains. Pseudogenes are indicated by thinner arrows. Dotted arrows have not been considered part of the T6SS. The numbers above the genes indicate the number of amino acids of the corresponding protein and boxes under the genes indicate identified domains (D: DUF, Oxa: oxoacyl-ACP synthases. (A) General organization in four rhizobia genera, (B) detailed gene organization of Boxes 3,4 of Mesorhizobium and Sinorhizobium strains, (C) detailed gene organization of Boxes 5, 6 of Rhizobium strains, and (D) structure of a T6SS. The colors are the same as those of the genes that encode these units in (A–C). The effectors are symbolized by ovoid shapes.
Many of the known effectors targeting bacteria have a toxic function (lipases, DNases, and peptidoglycanases) killing potential competitors (Coulthurst, 2019). Toxic effector genes are adjacent to cognate immunity genes that prevent self-toxicity or toxicity of sibling cells, and they are known as effector–immunity pairs (E/I pairs) (Yang et al., 2018). It is relevant that the presence of genes coding for possible effectors not accompanied by their cognate immunity gene and called orphan genes could encode for effectors whose target was eukaryotic cells (Trunk et al., 2018). Sometimes there is a third gene that codes a chaperone or adaptor for loading the effector onto the cognate VgrG. Adaptors contain conserved domains as DUF2169 or DUF4123 (Lien and Lai, 2017). Genes encoding effectors are often close to hcp and vgrG genes (Ma et al., 2014; Bernal et al., 2017).
The relationship between T6SS and rhizobial symbiosis was studied before the description of the independent role of T6SS. In 2003, a group of genes impairing pea nodulation of Rhizobium leguminosarum RBL5523 were identified (Bladergroen et al., 2003). These genes corresponded to what we already know as tss. Since then, a few researchers have studied the role of T6SS in symbiosis beyond mentioning its presence. In two such studies, it was found that mutations in T6SS genes do not affect symbiotic efficiency but symbiotic competitiveness (De Campos et al., 2017; Lin et al., 2018), and in another study, a positive effect on symbiosis was demonstrated (Salinero-Lanzarote et al., 2019).
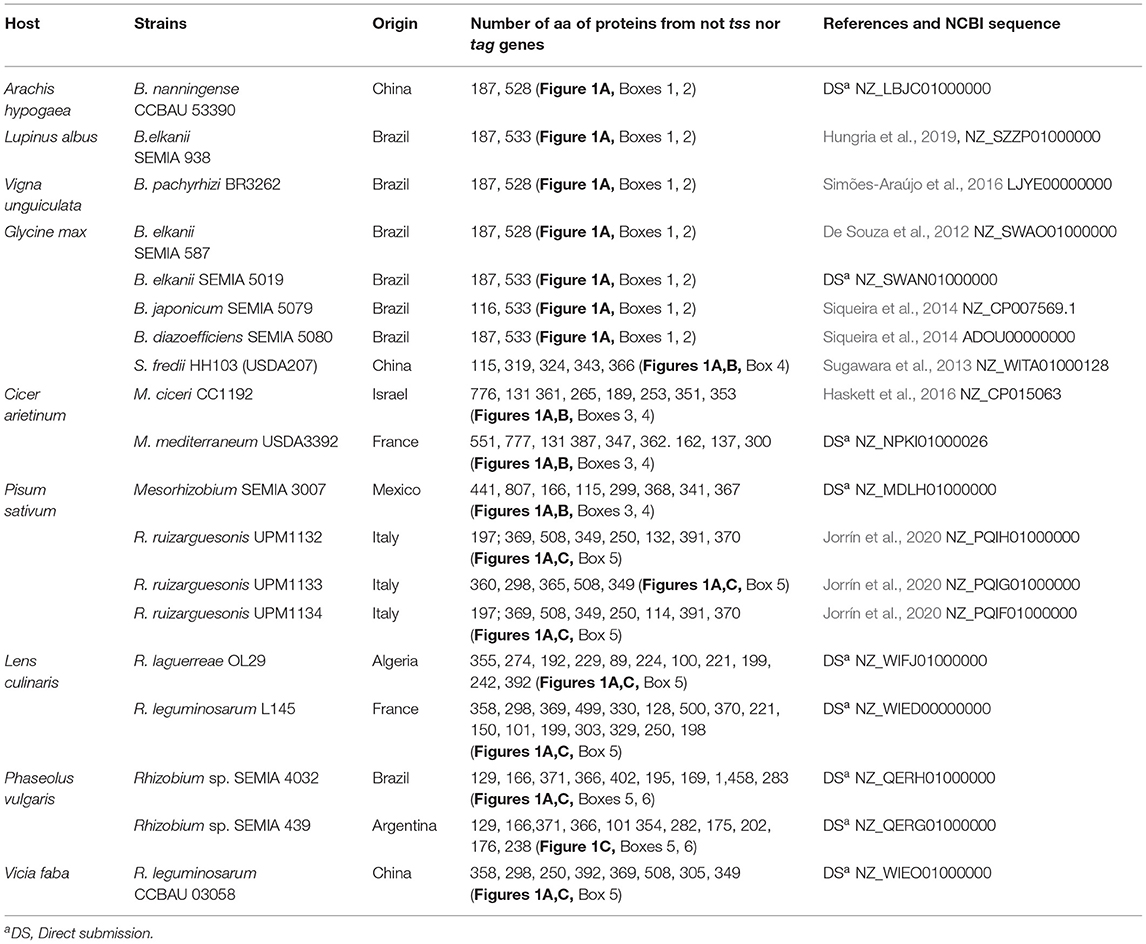
So far the symbiotic importance of any T6SS of recommended inoculants such as those included in Supplementary Table 1 is not known. Figure 1 shows the organization of 19 T6SSs of strains representing the main genera of rhizobia that nodulate pulses. When possible the strains were chosen from Supplementary Table 1.
Genomic Organization of Rhizobia T6SS Clusters
Organization and arrangement of T6SS genes with structural, regulatory, or accessory functions called tss or tag genes are totally conserved in all the species of each genus included in Table 1 and Figure 1. Genes not identified as tss or tag, match mostly to hypothetical proteins and among them are potential effectors as discussed below. These genes have been marked in Figure 1 by boxes numbered 1–6 and they are not as conserved as the previous ones, especially when different genera are compared. The T6SSs of all the Bradyrhizobium strains in Table 1 have identical gene organization (Figure 1A). The three Mesorhizobium T6SSs presented slight differences among them as shown in Boxes 3, 4 of Figure 1. Sinorhizobium T6SS is similar to that of Mesorhizobium. The T6SSs of the Rhizobium strains show the greatest diversity in genes of Boxes 5, 6. Genomes of R. SEMIA 439 and R. SEMIA 4032 have an extra copy of gene vgrG (Figure 1C, Box 6).
Putative Effectors in Rhizobia T6SS
Numerous effectors are found in the vicinity of the structural genes vgrG and hcp, and it is common to find three genes in tandem so that the first corresponds to an adaptor with DUF2169 or DUF 4123 domains, the second to a toxic effector, and the third to an immunity protein. Other possibilities are E/I pair or orphan effectors (Ma et al., 2014; Bondage et al., 2016; Trunk et al., 2018; Yang et al., 2018). Many effectors have an N-terminal DUF4150 or PAAR domain followed by a C-terminal region of variable function and some immunity proteins have a similar structure to KNR4 proteins or contain DUF1851 domains (Zhang et al., 2011; Ma et al., 2014). Possible effectors are discussed below based on their location in T6SS or on domains noted in Pfam protein families database (El-Gebali et al., 2019) (Supplementary Table 2).
Two conserved genes in Bradyrhizobium T6SS could encode orphan effectors (Figure 1, Boxes 1, 2). Proteins from these genes were designated B.187 and B.528. Their number of amino acids depends on the strain (Table 1). No functional domain has been identified in B.187. The other, B.528, has a methyltransferase domain. The role of these proteins is unknown. Other orphan effector (197 aa) could be encoded by the gene between hcp and vrgG in R. laguerrae and R. ruizarguesonis UPM1132 and UPM 1134 (Figure 1C).
Box 3 in Figures 1A,B contains genes for three types of Mesorhizobium proteins, Ms166 has the DUF1036 domain mainly present in membrane proteins of alphaproteobacteria. Ms808 is homologous to M23 metallopeptidases that lyse bacterial cell wall peptidoglycans (Lewis et al., 2019). And the third, Ms441, not present in M. ciceri, presents a caspase domain. Caspases (cysteine–aspartic proteases) participate in programmed cell death in animal tissues; however, the function of most bacterial caspase homologs are unknown (Asplund-Samuelsson, 2015). Box 4 of Figures 1A,B includes genes encoding proteins with DUF2169, DUF4150, or oxoacyl-ACP synthases domains, but their roles are unknown.
Pairs E/I are present in R. sp. SEMIA 4032 and SEMIA 439 as Rap1/Tae4 pair (Figure 1C). Rap1 is homologous to an immunity protein that neutralizes the amidase Tae4 able to cleave muropeptides of peptidoglycans (English et al., 2012; Srikannathasan et al., 2013; Zhang et al., 2013). Other E/I pair could correspond to the genes between hcp and vgrG in the Rhizobium strains UPM1133, CCBAU3058, and L145, although they do not have similarity with any E/I pair described.
Proteins with N-terminal PAAR-like/DUF4150 domain followed by a C-terminal region that contains a putative effector domain (Bondage et al., 2016) are present in Boxes 4–6 of Figure 1. No function for these proteins has been identified with the exception of M. ciceri Mc253 that contains a DNase_NucA_NucB domain (Figure 1B, Box 4). Rse.1458 from R. sp. SEMIA4032, Figure 1 Box 6, has two toxic RhsA domains often present in effectors secreted by T6SS (Pei et al., 2020).
Conclusions
It is desirable that collections of rhizobia, such as the Brazilian SEMIA, have and provide well-characterized rhizobia capable of maximizing nitrogen fixation in legumes.
Rhizobial EPS role in plant signaling and in bacterial protection against different stresses should be elucidated because they play a crucial role in symbiosis. EPS can promote competitiveness, the development of nodules, and therefore influence the effectiveness of inoculants. The connection between different symbiotic signals and regulation of EPS and other relevant surface polysaccharide expression in different rhizobia should also be considered.
The presence of T6SS in inoculants with high nitrogen-fixing capacity can lead to (i) better competitiveness against native soil endosymbionts able to nodulate the same host legume; (ii) biocontrol against pathogens/microorganisms in the rhizosphere as demonstrated by Pseudomonas preventing phytopathogenic bacteria (Bernal et al., 2017, 2018) and by the antifungal activity of Serratia marcescens (Trunk et al., 2018); and (iii) targeting effectors to host legumes as revealed by some nodulation external proteins (Nops). Nops are type III secretion system–dependent effectors with a positive, negative, or neutral effect on symbiosis (Deakin and Broughton, 2009; Miwa and Okazaki, 2017; Kusakabe et al., 2020). It has been identified that T6SS anti-eukaryotic effectors enable infectious bacteria to survive against the immune response of their hosts (Monjarás- Feria and Valvano, 2020).
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
LR was supported by the Ministerio de Ciencia, Innovación y Universidades, Spain (RTI2018-094985-B-100). TC and EL were supported by the National Council for Scientic and Technological Development (CNPq) (Proceeding No. 401886/2016-1) and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) (Proceeding No. 19/13493-1). BD was supported by a scholarship from CNPq-GDE-204842/2018-2. LT was supported by scholarships from the National Exceptional Program (PNE), Ministry of Higher Education and Scientific Research of Algeria, and from the Erasmus+ Programme project 2019-1-ES01-KA107-063778.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This paper is dedicated to the memory of Prof. Tomás Ruiz-Argüeso (1943–2020) whose teachings and friendship will always be with us.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2021.661468/full#supplementary-material
References
Acosta-Jurado, S., Alias-Villegas, C., Navarro-Gómez, P., Zehner, S., Murdoch, P. D., RodrÃíguez-Carvajal, M. A., et al. (2016). The Sinorhizobium fredii HH103 MucR1 global regulator is connected with the nod regulon and is required for efficient symbiosis with Lotus burttii and Glycine max cv. Williams. Mol. Plant Microbe Interact. 29, 700–712. doi: 10.1094/mpmi-06-16-0116-r
Akiyama, H., Hoshino, Y. T., Itakura, M., and Shimomura, Y. (2016). Mitigation of soil N2O emission by inoculation with a mixed culture of indigenous Bradyrhizobium diazoefficiens. Nat. Publ. Gr. 6:32869. doi: 10.1038/srep32869
Andrews, M., and Andrews, M. E. (2017). Specificity in legume-rhizobia symbioses. Int. J. Mol. Sci. 18:705. doi: 10.3390/ijms18040705
Asplund-Samuelsson, J. (2015). The art of destruction: revealing the proteolytic capacity of bacterial caspase homologs. Mol. Microbiol. 98, 1–6. doi: 10.1111/mmi.13111
Avelar Ferreira, P. A., Bomfeti, C. A., Lima Soares, B., and Moreira, F. M. (2012). Efficient nitrogen-fixing Rhizobium strains isolated from amazonian soils are highly tolerant to acidity and aluminium. World J. Microbiol. Biotechnol. 28, 1947–1959. doi: 10.1007/s11274-011-0997-7
Basler, M., and Mekalanos, J. (2012). Type 6 secretion dynamics within and between bacterial cells. Science 337, 815–815. doi: 10.1126/science.1222901
Becker, A., Ruberg, S., Baum Garth, B., Bertram-Drogatz, P. A., Quester, I., and Pühler, A. (2002). Regulation of succinoglycan and galactoglucan biosynthesis in Sinorhizobium meliloti. J. Mol. Microbiol. Biotechnol. 4, 187–190.
Bernal, P., Allsopp, L. P., Filloux, A., and Llamas, M. A. (2017). The Pseudomonas putida T6SS is a plant warden against phytopathogens. ISME J. 11, 972–987. doi: 10.1038/ismej.2016.169
Bernal, P., Llamas, M. A., and Filloux, A. (2018). Type VI secretion systems in plant-associated bacteria. Environ. Microbiol. 20, 1–15. doi: 10.1111/1462-2920.13956
Bladergroen, M. R., Badelt, K., and Spaink, H. P. (2003). Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol. Plant Microbe Interact. 16, 53–64. doi: 10.1094/MPMI.2003.16.1.53
Bondage, D. D., Lin, J. S., Ma, L. S., Kuo, C. H., and Lai, E. M. (2016). VgrG C terminus confers the type VI effector transport specificity and is required for binding with PAAR and adaptor-effector complex. Proc. Natl. Acad. Sci. U.S.A. 113, E3931–E3940. doi: 10.1073/pnas.1600428113
Boyer, F., Fichant, G., Berthod, J., Vandenbrouck, Y., and Attree, I. (2009). Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics. 10:104. doi: 10.1186/1471-2164-10-104
Castellane, T. C. L., Campanharo, J. C., Colnago, L. A., Coutinho, I. D., Lopes, É. M., Lemos, M. V. F., et al. (2017). Characterization of new exopolysaccharide production by Rhizobium tropici during growth on hydrocarbon substrate. Int. J. Biol. Macromol. 96, 361–369. doi: 10.1016/j.ijbiomac.2016.11.123
Castellane, T. C. L., de Sousa, B. F. S., and Lemos, E. G. D. M. (2019). “Exopolysaccharide Produced from Rhizobium spp. An interesting product for industry and environment”. Curr. Appl. Polym. Sci. 3:157. doi: 10.2174/2452271603666191016143811
Castellane, T. C. L., Lemos, M. V. F., and Lemos, E. G. D. M. (2014). Evaluation of the biotechnological potential of Rhizobium tropici strains for exopolysaccharide production. Carbohydr. Polym. 111, 191–197. doi: 10.1016/j.carbpol.2014.04.066
Castellane, T. C. L., Lemos, M. V. F., and Lemosa, E. G. M. (2018). Exploring and utilization of some bacterial exopolysaccharide. Biopolym. Res. 2:1000106.
Chen, W., Wang, E., Wang, S., Li, Y., Chen, X., and Li, Y. (1995). Characteristics of Rhizobium tianshanense sp. nov., a moderately and slowly growing root nodule bacterium isolated from an arid saline environment in Xinjiang, People's Republic of China. Int. J. Syst. Evol. Microbiol. 45, 153–159. doi: 10.1099/00207713-45-1-153
Chen, W. X., Yan, G. H., and Li, J. L. (1988). Numerical taxonomic study of fast-growing soybean rhizobia and a proposal that Rhizobium fredii be assigned to Sinorhizobium gen. nov. Int. J. Syst. Evol. Microbiol. 38, 392–397. doi: 10.1099/00207713-38-4-392
Coulthurst, S. (2019). The Type VI secretion system: a versatile bacterial weapon. Microbiology 165, 503–515. doi: 10.1099/mic.0.000789
Cunningham, S. D., and Munns, D. N. (1984). The correlation between extracellular polysaccharide production and acid tolerance in Rhizobium. Soil Sci. Soc. Am. J. 48:1273. doi: 10.2136/sssaj1984.0361599500480006001
De Campos, S. B., Lardi, M., Gandolfi, A., Eberl, L., and Pessi, G. (2017). Mutations in two Paraburkholderia phymatum type VI secretion systems cause reduced fitness in interbacterial competition. Front. Microbiol. 8, 2473. doi: 10.3389/fmicb.2017.02473
De Souza, J. A., Tieppo, E., Magnani Gde, S., Alves, L. M., Cardoso, R. L., Cruz, L. M., et al. (2012). Draft genome sequence of the nitrogen-fixing symbiotic bacterium Bradyrhizobium elkanii 587. J. Bacteriol. 194, 3547–3548. doi: 10.1128/JB.00563-12
Deakin, W. J., and Broughton, W. J. (2009). Symbiotic use of pathogenic strategies: rhizobial protein secretion systems. Nat. Rev. Microbiol. 7, 312–320. doi: 10.1038/nrmicro2091
Delamuta, J. R. M., Ribeiro, R. A., Ormeno-Orrillo, E., Melo, I. S., Martínez-Romero, E., and Hungria, M. (2013). Polyphasic evidence supporting the reclassification of Bradyrhizobium japonicum group Ia strains as Bradyrhizobium diazoefficiens sp. nov. Int. J. Syst. Evol. Microbiol. 63, 3342–3351. doi: 10.1099/ijs.0.049130-0
Donati, A. J., Lee, H. I., Leveau, J. H., and Chang, W. S. (2013). Effects of indole-3-acetic acid on the transcriptional activities and stress tolerance of Bradyrhizobium japonicum. PLoS ONE 8:e76559. doi: 10.1371/journal.pone.0076559
Downie, J. A. (2010). The roles of extracellular proteins, polysaccharides and signals in the interactions of rhizobia with legume roots. FEMS Microbiol. Rev. 34, 150–170. doi: 10.1111/j.1574-6976.2009.00205.x
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432. doi: 10.1093/nar/gky995
English, G., Trunk, K., Rao, V. A., Srikannathasan, V., Hunter, W. N., and Coulthurst, S. J. (2012). New secreted toxins and immunity proteins encoded within the Type VI secretion system gene cluster of Serratia marcescens. Mol. Microbiol. 86, 921–936. doi: 10.1111/mmi.12028
Geddes, B. A., González, J. E., and Oresnik, I. J. (2014). Exopolysaccharide production in response to medium acidification is correlated with an increase in competition for nodule occupancy. Mol. Plant Microbe Interact. 27, 1307–1317. doi: 10.1094/MPMI-06-14-0168-R
Göttfert, M., Röthlisberger, S., Kündig, C., Beck, C., Marty, R., and Hennecke, H. (2001). Potential symbiosis-specific genes uncovered by sequencing a 410-kilobase DNA region of the Bradyrhizobium japonicum chromosome. J. Bacteriol. 183 1405–1412. doi: 10.1128/JB.183.4.1405-1412.2001
Guerrero-Castro, J., Lozano, L., and Sohlenkamp, C. (2018). Dissecting the acid stress response of Rhizobium tropici CIAT 899. Front. Microbiol. 9:846. doi: 10.3389/fmicb.2018.00846
Guimarães, A. A., Florentino, L. A., Almeida, K. A., Lebbe, L., Silva, K. B., Willems, A., et al. (2015). High diversity of Bradyrhizobium strains isolated from several legume species and land uses in Brazilian tropical ecosystems. Syst. Appl. Microbiol. 38, 433–441. doi: 10.1016/j.syapm.2015.06.006
Guimarães, A. A., Jaramillo, P. M. D., Nóbrega, R. S. A., Florentino, L. A., Silva, K. B., and de Souza Moreira, F. M. (2012). Genetic and symbiotic diversity of nitrogen-fixing bacteria isolated from agricultural soils in the western Amazon by using cowpea as the trap plant. Appl. Environ. Microbiol. 78, 6726–6733. doi: 10.1128/AEM.01303-12
Haskett, T., Wang, P., Ramsay, J., O'Hara, G., Reeve, W., Howieson, J., et al. (2016). Complete genome sequence of Mesorhizobium ciceri strain CC1192, an efficient nitrogen-fixing microsymbiont of Cicer arietinum. Genome Announc. 4:16. doi: 10.1128/genomeA.00516-16
Hirsch, P. R. (1996). Population dynamics of indigenous and genetically modified rhizobia in the field. New Phytol. 133, 159–171. doi: 10.1111/j.1469-8137.1996.tb04351.x
Hungria, M., Campo, R. J., and Mendes, I. D. C. (2001). Fixação Biológica Do Nitrogênio Na Cultura Da Soja. Londrina: Embrapa Soja-Circular Técnica (INFOTECA-E).
Hungria, M., Delamuta, J. R. M., Ribeiro, R. A., and Nogueira, M. A. (2019). Draft genome sequence of Bradyrhizobium elkanii Strain SEMIA 938, used in commercial inoculants for Lupinus spp. in Brazil. Microbiol. Resour. Announc. 8, e00546–e00519. doi: 10.1128/MRA.00546-19
Iida, T., Itakura, M., Anda, M., Sugawara, M., Isawa, T., Okubo, T., et al. (2015). Symbiosis island shuffling with abundant insertion sequences in the genomes of extra-slow-growing strains of soybean bradyrhizobia. Appl. Environ. Microbiol. 81, 4143–4154. doi: 10.1128/AEM.00741-15
Ivashina, T. V., and Ksenzenko, V. N. (2012). “Exopolysaccharide biosynthesis in Rhizobium leguminosarum: from genes to functions,” in The Complex World of Polysaccharides, Desiree Nedra Karunaratne (IntechOpen), 99–127. doi: 10.5772/51202
Janczarek, M. (2011). Environmental signals and regulatory pathways that influence exopolysaccharide production in rhizobia. Int. J. Mol. Sci. 12, 7898–7933. doi: 10.3390/ijms12117898
Janczarek, M., Rachwał, K., and Kopcińska, J. (2015). Genetic characterization of the Pss region and the role of PssS in exopolysaccharide production and symbiosis of Rhizobium leguminosarum bv. trifolii with clover. Plant Soil 396, 257–275. doi: 10.1007/s11104-015-2567-5
Jones, K. M., Kobayashi, H., Davies, B. W., Taga, M. E., and Walker, G. C. (2007). How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat. Rev. Microbiol. 5, 619–633. doi: 10.1038/nrmicro1705
Jorrín, B., Palacios, J. M., Peix, A., and Imperial, J. (2020). Rhizobium ruizarguesonis sp. nov., isolated from nodules of Pisum sativum L. Syst. Appl. Microbiol. 43:126090. doi: 10.1016/j.syapm.2020.126090
Kaneko, T., Nakamura, Y., Sato, S., Minamisawa, K., Uchiumi, T., Sasamoto, S., et al. (2002). Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9, 189–197. doi: 10.1093/dnares/9.6.189
Kawaharada, Y., Nielsen, M., Kelly, S., et al. (2017). Differential regulation of the Epr3 receptor coordinates membrane-restricted rhizobial colonization of root nodule primordia. Nat. Commun. 8:14534. doi: 10.1038/ncomms14534
Kelly, S., Radutoiu, S., and Stougaard, J. (2017). Legume LysM receptors mediate symbiotic and pathogenic signalling. Curr. Opin. Plant Biol. 39, 152–158. doi: 10.1016/j.pbi.2017.06.013
Kopycińska, M., Lipa, P., Cieśla, J., Kozie,ł, M., and Janczarek, M. (2018). Extracellular polysaccharide protects Rhizobium leguminosarum cells against zinc stress in vitro and during symbiosis with clover. Environ. Microbiol. Rep. 10, 355–368. doi: 10.1111/1758-2229.12646
Kusakabe, S., Higasitani, N., Kaneko, T., Yasuda, M., Miwa, H., Okazaki, S., et al. (2020). Lotus accessions possess multiple checkpoints triggered by different type III secretion system effectors of the wide-host-range symbiont Bradyrhizobium elkanii USDA61. Microbes Environ. 35:ME19141. doi: 10.1264/jsme2.ME19141
Kuykendall, L. D., Saxena, B., Devine, T. E., and Udell, S. E. (1992). Genetic diversity in Bradyrhizobium japonicum Jordan 1982 and a proposal for Bradyrhizobium elkanii sp. nov. Can. J. Microbiol. 38, 501–505. doi: 10.1139/m92-082
Lewis, J. M., Deveson Lucas, D., Harper, M., and Boyce, J. D. (2019). Systematic identification and analysis of Acinetobacter baumannii type VI secretion system effector and immunity components. Front. Microbiol. 10:2440. doi: 10.3389/fmicb.2019.02440
Lien, Y. W., and Lai, E. M. (2017). Type VI secretion effectors: methodologies and biology. Front. Cell. Infect. Microbiol. 7:254. doi: 10.3389/fcimb.2017.00254
Lin, H. H., Huang, H. M., Yu, M., Lai, E. M., Chien, H. L., and Liu, C. T. (2018). Functional exploration of the bacterial type VI secretion system in mutualism: Azorhizobium caulinodans ORS571–Sesbania rostrata as a research model. Mol. Plant Microbe Interact. 31, 856–867. doi: 10.1094/MPMI-01-18-0026-R
Lipa, P., Vinardell, J. M., Kopcińska, J., Zdybicka-Barabas, A., and Janczarek, M. (2018). Mutation in the pssZ gene negatively impacts exopolysaccharide synthesis, surface properties, and symbiosis of Rhizobium leguminosarum bv. trifolii with clover. Gene 97:369. doi: 10.3390/genes9070369
López-Baena, F. J., Ruiz-Sainz, J. E., Rodríguez-Carvajal, M. A., and Vinardell, J. M. (2016). Bacterial molecular signals in the Sinorhizobium fredii-soybean symbiosis. Int. J. Mol. Sci. 17:755. doi: 10.3390/ijms17050755
Ma, L. S., Hachani, A., Lin, J. S., Filloux, A., and Lai, E. M. (2014). Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta. Cell Host Microbe 16, 94–104. doi: 10.1016/j.chom.2014.06.002
Marczak, M., Mazur, A., Koper, P., Zebracki, K., and Skorupska, A. (2017). Synthesis of rhizobial exopolysaccharides and their importance for symbiosis with legume plants. Genes 8:360. doi: 10.3390/genes8120360
Marczak, M., Wójcik, M., Zebracki, K., Turska-Szewczuk, A., Talarek, K., Nowak, D., et al. (2020). PssJ is a terminal galactosyltransferase involved in the assembly of the exopolysaccharide subunit in Rhizobium leguminosarum bv. trifolii. Int. J. Mol. Sci. 21:7764. doi: 10.3390/ijms21207764
Margaret-Oliver, I., Lei, W., Parada, M., Rodríguez-Carvajal, M. A., Crespo-Rivas, J. C., Hidalgo, Á., et al. (2012). Sinorhizobium fredii HH103 does not strictly require KPS and/or EPS to nodulate Glycyrrhiza uralensis, an indeterminate nodule-forming legume. Arch. Microbiol. 194, 87–102. doi: 10.1007/s00203-011-0729-2
Martínez-Romero, E., Segovia, L., Mercante, F. M., Franco, A. A., Graham, P., and Pardo, M. A. (1991). Rhizobium tropici, a novel species nodulating Phaseolus vulgaris L. beans and Leucaena sp. trees. Int. J. Syst. Evol. Microbiol. 41, 417–426. doi: 10.1099/00207713-41-3-417
Menna, P., and Hungria, M. (2011). Phylogeny of nodulation and nitrogen-fixation genes in Bradyrhizobium: supporting evidence for the theory of monophyletic origin, and spread and maintenance by both horizontal and vertical transfer. Int. J. Syst. Evol. Microbiol. 61, 3052–3067. doi: 10.1099/ijs.0.028 803-0
Milner, J. L., Araujo, R. S., and Handelsman, J. (1992). Molecular and symbiotic characterization of exopolysaccharide-deficient mutants of Rhizobium tropici strain CIAT899. Mol. Microbiol. 6, 3137–3147. doi: 10.1111/j.1365-2958. 1992.tb01770.x
Miwa, H., and Okazaki, S. (2017). How effectors promote beneficial interactions. Curr. Opin. Plant Biol. 38, 148–154. doi: 10.1016/j.pbi.2017.05.011
Monjarás- Feria, J., and Valvano, M. A. (2020). An overview of anti-eukaryotic T6SS effectors. Front. Cell. Infect. Microbiol. 10:617. doi: 10.3389/fcimb.2020.584751
Mougous, J. D., Cuff, M. E., Raunser, S., Shen, A., Zhou, M., Gifford, C. A., et al. (2006). A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312, 1526–1530. doi: 10.1126/science.1128393
Moulin, L., Béna, G., Boivin-Masson, C., and Stepkowski, T. (2004). Phylogenetic analyses of symbiotic nodulation genes support vertical and lateral gene co-transfer within the Bradyrhizobium genus. Mol. Phylogenet. Evol. 30, 720–732. doi: 10.1016/S1055-7903(03)00255-0
Naseem, H., Ahsan, M., Shahid, M. A., and Khan, N. (2018). Exopolysaccharides producing rhizobacteria and their role in plant growth and drought tolerance. J. Basic Microbiol. 58, 1009–1022. doi: 10.1002/jobm.201800309
Nilsson, J. F., Castellani, L. G., Draghi, W. O., Mogro, E. G., Wibberg, D., Winkler, A., et al. (2021). Global transcriptome analysis of Rhizobium favelukesii LPU83 in response to acid stress. FEMS Microbiol. Ecol. 97:235. doi: 10.1093/femsec/fiaa235
Obando, M., Correa-Galeote, D., Castellano-Hinojosa, A., Gualpa, J., Hidalgo, A., Alché, J. D. D., et al. (2019). Analysis of the denitrification pathway and greenhouse gases emissions in Bradyrhizobium sp. strains used as biofertilizers in South America. J. Appl. Microbiol. 127, 739–749. doi: 10.1111/jam.14233
Ormeño-Orrillo, E., Menna, P., Almeida, L. G., Ollero, F. J., Nicolás, M. F., Pains Rodrigues, E., et al. (2012). Genomic basis of broad host range and environmental adaptability of Rhizobium tropici CIAT 899 and Rhizobium sp. PRF 81 which are used in inoculants for common bean (Phaseolus vulgaris L.). BMC Genom. 13:735. doi: 10.1186/1471-2164-13-735
Pei, T. T., Li, H., Liang, X., Wang, Z. H., Liu, G., Wu, L. L., et al. (2020). Intramolecular chaperone-mediated secretion of an Rhs effector toxin by a type VI secretion system. Nat. Commun. 11, 1–13. doi: 10.1038/s41467-020-15774-z
Pellock, B. J., Teplitski, M., Boinay, R. P., Bauer, W. D., and Walker, G. C. (2002). A LuxR homolog controls production of symbiotically active extracellular polysaccharide II by Sinorhizobium meliloti. J. Bacteriol. 184, 5067–5076. doi: 10.1128/jb.184.18.5067-5076.2002
Primo, E. D., Cossovich, S., Nievas, F., Bogino, P., Humm, E. A., Hirsch, A. M., et al. (2020). Exopolysaccharide production in Ensifer meliloti laboratory and native strains and their effects on alfalfa inoculation. Arch. Microbiol. 202, 391–398. doi: 10.1007/s00203-019-01756-3
Pukatzki, S., Ma, A. T., Sturtevant, D., Krastins, B., Sarracino, D., Nelson, W. C., et al. (2006). Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U.S.A. 103, 1528–1533. doi: 10.1073/pnas.0510322103
Puozaa, D. K., Jaiswal, S. K., and Dakora, F. D. (2017). African origin of Bradyrhizobium populations nodulating Bambara groundnut (Vigna subterranea L. Vedic) in Ghanaian and South African soils. PLoS ONE. 12:e0184943. doi: 10.1371/journal.pone.0184943
Reuber, T. L., Reed, J., Glazebrook, J., Glucksmann, M. A., Ahmann, D., Marra, A., et al. (1991). Rhizobium meliloti exopolysaccharides: genetic analyses and symbiotic importance. Biochem. Soc. Trans. 19, 636–644. doi: 10.1042/bst0190636
Rüberg, S., Pühler, A., and Becker, A. (1999). Biosynthesis of the exopolysaccharide galactoglucan in Sinorhizobium meliloti is subject to a complex control by the phosphate-dependent regulator PhoB and the proteins ExpG and MucR. Microbiology. 145, 603–611. doi: 10.1099/13500872-145-3-603
Rufini, M., Da Silva, M. A. P., Ferreira, P. A. A., de Souza Cassetari, A., Soares, B. L., De Andrade, M. J. B., et al. (2014). Symbiotic efficiency and identification of rhizobia that nodulate cowpea in a Rhodic Eutrudox. Biol. Fert. Soils. 50, 115–122. doi: 10.1007/s00374-013-0832-4
Russo, D. M., Williams, A., Edwards, A., Posadas, D. M., Finnie, C., Dankert, M., et al. (2006). Proteins exported via the PrsD-PrsE type I secretion system and the acidic exopolysaccharide are involved in biofilm formation by Rhizobium leguminosarum. J. Bacteriol. 188, 4474–4486. doi: 10.1128/JB.00246-06
Salinero-Lanzarote, A., Pacheco-Moreno, A., Domingo-Serrano, L., Durán, D., Ormeño-Orrillo, E., Martínez-Romero, E., et al. (2019). The Type VI secretion system of Rhizobium etli Mim1 has a positive effect in symbiosis. FEMS Microbiol. Ecol. 95:54. doi: 10.1093/femsec/fiz054
Santos, M. S., Nogueira, M. A., and Hungria, M. (2019). Microbial inoculants: reviewing the past, discussing the present and previewing an outstanding future for the use of beneficial bacteria in agriculture. AMB Express. 9, 1–22. doi: 10.1186/s13568-019-0932-0
Schäper, S., Wendt, H., Bamberger, J., Sieber, V., Schmid, J., and Becker, A. (2019). A bifunctional UDP-sugar 4-epimerase supports biosynthesis of multiple cell surface polysaccharides in Sinorhizobium meliloti. J. Bacteriol. 201, e00801–e00818. doi: 10.1128/JB.00801-18
Shalom, G., Shaw, J. G., and Thomas, M. S. (2007). In vivo expression technology identifies a type VI secretion system locus in Burkholderia pseudomallei that is induced upon invasion of macrophages. Microbiology 153, 2689–2699. doi: 10.1099/mic.0.2007/006585-0
Shneider, M. M., Buth, S. A., Ho, B. T., Basler, M., Mekalanos, J. J., and Leiman, P. G. (2013). PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 15, 350–353. doi: 10.1038/nature12453
Silverman, J. M., Brunet, Y. R., Cascales, E., and Mougous, J. D. (2012). Structure and regulation of the type VI secretion system. Annu. Rev. Microbiol. 66, 453–472. doi: 10.1146/annurev-micro-121809-151619
Simões-Araújo, J. L., Leite, J., Marie Rouws, L. F., Passos, S. R., Xavier, G. R., Rumjanek, N. G., et al. (2016). Draft genome sequence of Bradyrhizobium sp. strain BR 3262, an effective microsymbiont recommended for cowpea inoculation in Brazil. Braz. J. Microbiol. 47, 783–784. doi: 10.1016/j.bjm.2016.03.001
Siqueira, A. F., Ormeno-Orrillo, E., Souza, R. C., Rodrigues, E. P., Almeida, L. G. P., Barcellos, F. G., et al. (2014). Comparative genomics of Bradyrhizobium japonicum CPAC 15 and Bradyrhizobium diazoefficiens CPAC 7: elite model strains for understanding symbiotic performance with soybean. BMC Genom. 15:420. doi: 10.1186/1471-2164-15-420
Skorupska, A., Janczarek, M., Marczak, M., Mazur, A., and Król, J. (2006). Rhizobial exopolysaccharides: genetic control and symbiotic functions. Microb. Cell Fact. 5, 1–19. doi: 10.1186/1475-2859-5-7
Sorroche, F., Bogino, P., Russo, D. M., Zorreguieta, A., Nievas, F., Morales, G. M., et al. (2018). Cell autoaggregation, biofilm formation, and plant attachment in a Sinorhizobium meliloti lpsB Mutant. Mol. Plant Microbe. Interact. 31, 1075–1082. doi: 10.1094/MPMI-01-18-0004-R
Sorroche, F. G., Rinaudi, L. V., Zorreguieta, Á., and Giordano, W. (2010). EPS II-dependent auto aggregation of Sinorhizobium meliloti planktonic cells. Curr. Microbiol. 61, 465–470. doi: 10.1007/s00284-010-9639-9
Srikannathasan, V., English, G., Bui, N. K., Trunk, K., O'Rourke, P. E., Rao, V. A., et al. (2013). Structural basis for type VI secreted peptidoglycan DL-endopeptidase function, specificity and neutralization in Serratia marcescens. Acta Crystallogr. Sect. D Biol. Crystallogr. 69, 2468–2482. doi: 10.1107/S0907444913022725
Staehelin, C., Forsberg, L. S., D'Haeze, W., Gao, M. Y., Carlson, R. W., Xie, Z. P., et al. (2006). Exo-oligosaccharides of Rhizobium sp. strain NGR234 are required for symbiosis with various legumes. J. Bacteriol. 188, 6168–6178. doi: 10.1128/JB.00365-06
Sugawara, M., Epstein, B., Badgley, B. D., Unno, T., Xu, L., Reese, J., et al. (2013). Comparative genomics of the core and accessory genomes of 48 Sinorhizobium strains comprising five genospecies. Genome Biol. 14:R17. doi: 10.1186/gb-2013-14-2-r17
Sun, Y., Liu, Y., Liu, X., Dang, X., Dong, X., Xie, Z., et al. (2020). Azorhizobium caulinodans c-di-GMP phosphodiesterase Chp1 involved in motility, EPS production, and nodulation of the host plant. Appl. Microbiol. Biotechnol. 104, 2715–2729. doi: 10.1007/s00253-020-10404-6
Trunk, K., Peltier, J., Liu, Y. C., Dill, B. D., Walker, L., Gow, N. A., et al. (2018). The type VI secretion system deploys antifungal effectors against microbial competitors. Nat. Microbiol. 3, 920–931. doi: 10.1038/s41564-018-0191-x
Vences-Guzmán, M. A., Guan, Z., Ormeno-Orrillo, E., González-Silva, N., López-Lara, I. M., Martínez-Romero, E., et al. (2011). Hydroxylated ornithine lipids increase stress tolerance in Rhizobium tropici CIAT899. Mol. Microbiol. 79, 1496–1514. doi: 10.1111/j.1365-2958.2011.07535.x
Via, V. D., Narduzzi, C., Aguilar, O. M., Zanetti, M. E., and Blanco, F. A. (2015). Changes in the common bean transcriptome in response to secreted and surface signal molecules of Rhizobium etli. Plant Physiol. 169, 1356–1370. doi: 10.1104/pp.15.00508
Vinuesa, P., Neumann-Silkow, F., Pacios-Bras, C., Spaink, H. P., Martínez-Romero, E., and Werner, D. (2003). Genetic analysis of a pH-regulated operon from Rhizobium tropici CIAT899 involved in acid tolerance and nodulation competitiveness. Mol. Plant-Microbe Interact. 16, 159–168. doi: 10.1094/MPMI.2003.16.2.159
Wang, J., Brodmann, M., and Basler, M. (2019). Assembly and subcellular localization of bacterial type VI secretion systems. Annu. Rev. Microbiol. 73, 621–638. doi: 10.1146/annurev-micro-020518-115420
Wang, P., Zhong, Z., Zhou, J., Cai, T., and Zhu, J. (2008). Exopolysaccharide biosynthesis is important for Mesorhizobium tianshanense: plant host interaction. Arch. Microbiol. 189, 525–530. doi: 10.1007/s00203-007-0345-3
Willems, A. (2006). The taxonomy of rhizobia: an overview. Plant Soil. 287, 3–14. doi: 10.1007/s11104-006-9058-7
Willems, A., Doignon-Bourcier, F., Goris, J., Coopman, R., de Lajudie, P., De Vos, P., et al. (2001). DNA-DNA hybridization study of Bradyrhizobium strains. Int. J. Syst. Evol. Microbiol. 51, 1315–1322. doi: 10.1099/00207713-51-4-1315
Wong, J. E. M. M., Gysel, K., Birkefeldt, T. G., Vinther, M., Muszyński, A., Azadi, P., et al. (2020). Structural signatures in EPR3 define a unique class of plant carbohydrate receptors. Nat. Commun. 11:3797. doi: 10.1038/s41467-020-17568-9
Yang, X., Long, M., and Shen, X. (2018). Effector–immunity pairs provide the T6SS nanomachine its offensive and defensive capabilities. Molecules 23:1009. doi: 10.3390/molecules23051009
Zhang, D., Iyer, L. M., and Aravind, L. (2011). A novel immunity system for bacterial nucleic acid degrading toxins and its recruitment in various eukaryotic and DNA viral systems. Nucleic Acids Res. 39, 4532–4552. doi: 10.1093/nar/gkr036
Zhang, H., Gao, Z. Q., Wang, W. J., Liu, G. F., Xu, J. H., Su, X. D., et al. (2013). Structure of the type VI effector-immunity complex (Tae4-Tai4) provides novel insights into the inhibition mechanism of the effector by its immunity protein. J. Biol. Chem. 288, 5928–5939. doi: 10.1074/jbc.M112.434357
Keywords: Rhizobium, root nodulating bacteria, effectors, type VI secretion systems, inoculant, exopolysaccharide, symbiosis
Citation: De Sousa BFS, Castellane TCL, Tighilt L, Lemos EGM and Rey L (2021) Rhizobial Exopolysaccharides and Type VI Secretion Systems: A Promising Way to Improve Nitrogen Acquisition by Legumes. Front. Agron. 3:661468. doi: 10.3389/fagro.2021.661468
Received: 30 January 2021; Accepted: 04 June 2021;
Published: 05 July 2021.
Edited by:
Juan Imperial, Consejo Superior de Investigaciones Científicas (CSIC), SpainReviewed by:
Xiancan Zhu, Anhui Normal University, ChinaJose Enrique Ruiz-Sainz, Sevilla University, Spain
Francisco Pérez Montaño, Sevilla University, Spain
Copyright © 2021 De Sousa, Castellane, Tighilt, Lemos and Rey. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luis Rey, bHVpcy5yZXlAdXBtLmVz; Eliana Gertrudes de Macedo Lemos, ZWxpYW5hZy5sZW1vc0BnbWFpbC5jb20=
†These authors share first authorship
 Bruna Fernanda Silva De Sousa
Bruna Fernanda Silva De Sousa Tereza Cristina Luque Castellane
Tereza Cristina Luque Castellane Lilia Tighilt1,4
Lilia Tighilt1,4 Eliana Gertrudes de Macedo Lemos
Eliana Gertrudes de Macedo Lemos Luis Rey
Luis Rey