- 1Allergy and Clinical Immunology Unit, 2nd Pediatric Clinic, National and Kapodistrian University of Athens, Athens, Greece
- 2Laboratory of Molecular Biology, Department of Biotechnology, School of Food, Biotechnology and Development, Agricultural University of Athens, Athens, Greece
- 3Division of Evolution and Genomic Sciences, University of Manchester, Manchester, United Kingdom
- 4Center for Clinical, Experimental Surgery and Translational Research of the Biomedical Research Foundation of the Academy of Athens, Athens, Greece
- 5Laboratory for Genetics of Microorganisms and Bacteriophages, Eliava Institute of Bacteriophage, Microbiology & Virology, Tbilisi, Georgia
- 6Department of Electrical and Electronic Engineering, University of West Attica, Athens, Greece
- 7Department of Microbiology, P. & A. Kyriakou Children's Hospital, Athens, Greece
The airway epithelium is the primary site where inhaled and resident microbiota interacts between themselves and the host, potentially playing an important role on allergic asthma development and pathophysiology. With the advent of culture independent molecular techniques and high throughput technologies, the complex composition and diversity of bacterial communities of the airways has been well-documented and the notion of the lungs' sterility definitively rejected. Recent studies indicate that the microbial composition of the asthmatic airways across the spectrum of disease severity, differ significantly compared with healthy individuals. In parallel, a growing body of evidence suggests that bacterial viruses (bacteriophages or simply phages), regulating bacterial populations, are present in almost every niche of the human body and can also interact directly with the eukaryotic cells. The triptych of airway epithelial cells, bacterial symbionts and resident phages should be considered as a functional and interdependent unit with direct implications on the respiratory and overall homeostasis. While the role of epithelial cells in asthma pathophysiology is well-established, the tripartite interactions between epithelial cells, bacteria and phages should be scrutinized, both to better understand asthma as a system disorder and to explore potential interventions.
Introduction
Asthma is a chronic inflammatory disease presenting with wheeze, chest tightness and shortness of breath; however these symptoms may vary in appearance, frequency and intensity (1). Underlying mechanisms include genetic and environmental factors acting already from pregnancy and during the first years of life (2, 3), although new cases may appear at any age (4). It is generally accepted that the term “asthma” incorporates a variety of clinical and mechanistic entities (5, 6). Terms such as phenotypes, endotypes, subtypes or clusters are frequently used to subdivide and classify asthma, without however conclusive consensus. Affecting almost 300 million people worldwide, including up to 10% of children (7, 8), asthma has emerged as a significant health problem with huge economic implications on healthcare systems globally (9, 10).
Allergic sensitization of the host and subsequent overreaction to common environmental factors with typical inflammatory and immunological responses are strong determinants for the onset and the development of asthma. The airway epithelium is the primary interphase between inhaled triggers and the host, playing a central role on asthma pathophysiology (development, progression, and exacerbation). The epithelium of the large and small airways of the lungs is constituted by basal and undifferentiated columnar cells as well as ciliated and secretory (large airways) or Clara cells (small airways), forming pseudostratified or columnar and cuboidal structures, respectively. Additionally, distal alveoli are lined by alveolar type I and II epithelial cells (11). The whole spectrum of epithelial lining across the respiratory tract is a physical “tight-sealed” protective barrier against external stressors having also multiple regulating and signaling properties. The epithelial barrier integrity is ensured mainly by the formation of intracellular tight junctions (TJs) and adhesions and secondly by the presence of mucus and periciliary glycocalyx layers produced by adjacent secretory cells. This physical barrier ensures the elimination of noxious agents and their rejection through mucociliary clearance (MCC). This first line of innate immune defense protects the airway tract from incoming pathogens such as bacteria and viruses, as well as other chemical and physical stressors that can act as allergens in the case of atopic asthma.
The asthmatic epithelium has structural and functional abnormalities, probably due to genetic predisposition (12) giving expression of, among other, an “over-reaction” to common environmental allergens. Being under repetitive injury and inflammation, the asthmatic epithelium progressively remodels its structural characteristics (13). Specific allergens or pathogens can induce dysfunction and/or disruption (14–17) of the TJs of the cells and access the airway submucosa through the paracellular way, facilitating sensitization against them. Its demonstrative that in-vitro infection of airway epithelial cells, obtained from children with or without asthma, with human rhinovirus, provokes different expression of TJs proteins. In the case of asthmatic cells it was observed permanent reduction of TJs expression and impairment of epithelial barrier integrity (18). In parallel, changes on the mucus composition and production rates in asthmatics have great impact on the MCC (19, 20). The asthmatic epithelium become more susceptible to infections (21), secreting a cascade of chemokines and cytokines which in turn activate several immune cells (22, 23). In the case of airway epithelial cells, obtained from asthmatic children and infected with human rhinovirus in Air Liquid Interfase cultures (ALIs), it was observed reduced IFN-β and increased inflammatory cytokine production, compared to healthy control (24). Activation of immune cells due to persistent allergic inflammation in asthma progressively alters the bronchial physiology into a pathological state where airway smooth muscle cells hyperplasia, subepithelial fibrosis, extracellular matrix overproduction, continuous angiogenesis, and mucus hypersecretion are detected (25). As a consequence, bronchial wall inflammation and thickening cause airway obstruction resulting in the typical respiratory symptoms of asthma. Strikingly, it has been suggested that epithelial mediation on allergic and immune responses has an impact on epithelial cell memory, establishing a type of epigenetic imprint on them (21).
The human microbiome is a dynamic and diverse entity, composed of an array of microbial genomes from bacteria, phages, fungi, protozoa and viruses. It is affected by immigration, elimination and growth of microbes within and possibly between different niches of each individual (26–28). Despite existing variability, healthy microbial populations share a few common characteristics and abundances that change significantly in the cases of chronic inflammatory diseases, such as asthma (29)where microbial imbalance or “dysbiosis” occurs (30, 31). Indicatively, it is reported that the airways of healthy individuals are dominated mainly by bacteria of the genera Prevotella (Prevotella melaninogenica, Prevotella nanceiensis, Prevotella salivae) and Veilonella (Veillonella alcalescens, Veillonella parvula, Veillonella dispar) (26, 32–34). In contrast, the bacteriome of asthmatic airways is dominated by Haemophilus (Haemophilus influenzae B, non-typable Haemophilus influenzae), Neisseria and Moraxella catarrhalis along with Streptococcus pneumoniae and Staphylococcus aureus (32, 35). Domination of specific bacterial species in asthma raises several questions about a potentially causal role of microbiome imbalance to this disorder (36, 37) and the role of epithelial defense to microbial attachment and invasion (38–41).
Maintenance of the microbial equilibrium/homeostasis is influenced not only by epithelial responses and host immunity, but also and possibly to a greater extent, by bacterial viruses that colonize, along with bacteria, human mucosal surfaces. Phages are capable of controlling microbial populations, expressing high genetic variation and complexity. They are known as the most abundant biological entities on the planet (42) with many references for their presence in the human body and their direct interactions with eukaryotic cells, organs and tissues (43–45). There is a plethora of lytic phages that target bacterial populations, indirectly protecting epithelial surfaces from bacteria colonization and/or overpopulation. On the other hand, there are phages following mainly a lysogenic life cycle integrating their genome on bacterial chromosomes and remaining as prophages in a lethargic state. Induction of prophages may be related with the acquisition of bacterial DNA which includes toxin or antibiotic resistance related genes. Horizontal transfer of these genes (HGT) between bacteria may have an impact on bacterial population fitness across the human body (46–48). The human “phageome” imposes important selective pressure on bacterial populations, influencing and determining along with the eukaryotic host the overall health and well-being (49). It seems that microbe-phage interactions along with epithelium responses participate in a co-evolving game of symbiotic and/or antagonistic relationships thus their simultaneous study could add insight into the pathophysiology of asthma.
Within the context of this review we explore the importance of the triptych: phage-bacterium-respiratory epithelium, toward asthma pathogenicity and development. The current literature around this complex biological system is limited and focused mainly on the gut epithelium and its interactions with local bacteria and phages. Undoubtedly, the information gap around asthma development and the contribution of the triptych remains unbridged. We first describe the current understanding about the role of microbiome in asthma development and exacerbation and then assess the interactions of specific bacterial species with the respiratory epithelium. We further elucidate the direct interactions between phages and human epithelial cells and finally identify the few existent in-vitro and in-vivo studies that have assessed the role of the triptych in respiratory disease models.
Subsections
The Microbiome in Asthma
Healthy adults are breathing ~7000L of air per day (50, 51) and their upper and lower airways are constantly exposed to a variety and large number of microorganisms. The estimated load of inhaled bacteria, viruses and fungi ranges between thousands to millions of particles per day, with the exact number depending upon the environmental exposure (52–54). The nasal cavity, pharynx and paranasal sinuses are part of the upper respiratory tract and their microbial communities along with the oral microbiome determine in a decisive way the lower respiratory tract microbial composition in healthy states (55, 56). The proposed “Adapted Island Model of Lung Biogeography” describes effectively the immigration rates of microbial communities from the upper respiratory tract to the large and small airways and the distal alveoli of the lungs (26, 57). It is assumed that the respiratory tract should be considered as a large and unique ecosystem with different environmental niches affecting decisively the immigration rates and the biodiversity levels of microbial communities in healthy states. Undoubtedly, the whole identity of the airways microbiota is drastically changing in the cases of respiratory diseases such as asthma and the overall physiology and airway function is affected (58–60). Therefore, the delineation of microbial identities in health and asthma could lay new perspectives for future phenotyping and medical interventions.
The microbiome in the gut may also have a role in asthma pathogenesis by modulating the immune system via the induction of immune cells that circulate and reach the lungs, regulating and influencing lung immunity and microbial equilibrium from the first years of life (27, 61–64). The recently emerged lung-gut axis hypothesis was proposed by Schuijt TJ et al. (65), showing that healthy gut microbiota efficiently protect mice lungs from S. pneumoniae infection through the activation of alveolar macrophages and enhanced levels of immunomodulatory cytokines (65). The exact mechanisms by which the gut-lung axis activates innate immune system in health and disease are still unknown. Nevertheless, interactions between respiratory microbiota and the airway mucosa are arguably at least as important in shaping local immunity and dysbiosis in asthma.
In the last decade, culture independent molecular techniques, mostly based on sequencing of the hypervariable regions of the 16S rRNA gene have been used to characterize the airway microbiome in conditions of health and disease (26, 66, 67). It was shown that the bronchial tree is colonized across its whole spectrum by specific microbial phyla thus having its own microbial identity and being protected from infections and chronic respiratory disease development. Hilty et al. analyzed the airway microbiota of the nose, oropharynx and lungs in asthmatic and chronic obstructive pulmonary disease (COPD) patients and compared the results with healthy individuals (35). Using nasal and oropharyngeal swabs as well as bronchoscopic cytology brushings and broncho-alveolar lavage fluid, they performed cladistic analysis to identify the distribution and abundance of airway microbiota across the spectrum of health and disease. They concluded that pathogenic Proteobacteria, particularly with Haemophilus, Neisseria, and Moraxella genera were more frequent in airways of asthmatic and COPD patients compared to healthy individuals and maybe related to increased risk of early asthma development when found in infant's pharynxes. In addition, Staphylococcus and Streptococcus dominance was a common characteristic in children with refractory asthma, while in healthy airways, Prevotella and Veillonella were among the prevalent genera (35). Microbial profiles in sputum samples from severe asthmatics were also identified and compared with healthy individuals in another study. The prevalent bacteria in severe asthmatics were amongst the genera of Haemophilus or Streptococcus and M. catarrhalis species (68). Prevalence of specific bacterial species in the respiratory tract of asthmatics compared to healthy controls seems to reduce the microbial diversity and affect in a decisive yet underexplored way the pathophysiology in asthma.
Observations from wider population studies were generally confirmatory and tried to connect early microbiome fluctuations with asthma development in childhood. A Danish birth cohort of 700 children followed until the age of 6 years showed that airway microbiota dominated by S. pneumoniae and subsequent immune responses were correlated with asthma development risk (69, 70). In another study, nasal samples from 6 to 12 years old asthmatic children had reduced microbiota diversity compared to healthy controls and a notably high abundance of the genus Moraxella. The question raised was whether the loss of abundance in asthmatics is due to prevalence of Moraxella (71). It has been suggested that Moraxella species coming from the upper respiratory system of asthmatic patients are also identified in abundance at the lower airways (72, 73). In another study, infant's nasopharynxes were screened during the first year of life where colonization from common respiratory pathogens occurs. It was proposed that early colonization of nasopharynxes with Moraxella, Staphylococcus, Haemophilus, and/or Streptococcus can provoke upper respiratory tract infections and inflammation that possibly spread to the lower airways, predisposing for future asthma onset in childhood (74, 75). It is apparent that the neonatal airway microbiome in conjunction with genetic predisposition, tissue impairment and/or respiratory infections may act causatively in regard to asthma development from the first years of life and into adulthood. In Table 1 we summarize important clinical studies about the microbiome profile in asthmatic children populations and the role of specific bacterial species found to dominate in their airways.
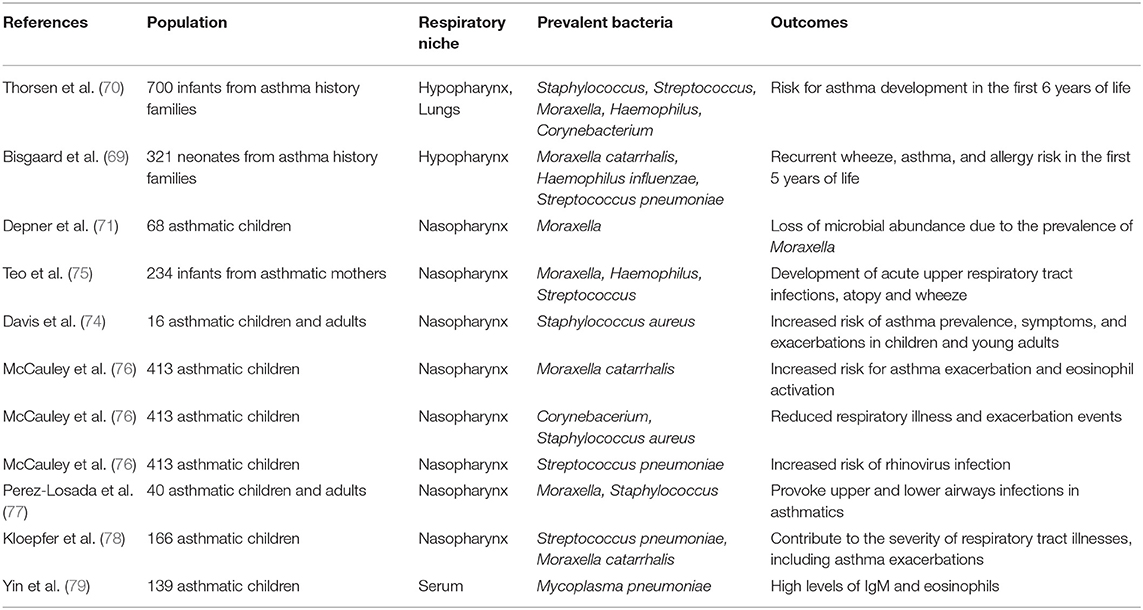
Table 1. Studies of the microbiome profiles of asthmatic infants and children and the assessment of the role of each prevalent bacterium in asthma disease.
In respect to asthma exacerbations, nasopharyngeal microbiota dominated by M. catarrhalis and S. aureus in children with asthma were associated with increased risk of asthma exacerbation and eosinophil activation (76, 77). Moreover, in the course of a rhinovirus infection, M. catarrhalis and/or S. pneumoniae were prevalent in the nasal samples of children with or without asthma, contributing to asthma exacerbations (78). Bacterial diversity possibly constitutes a major factor for the maintenance of the physiologic function of the airways and the control of asthma inflammation (59). Studies in patients with neutrophilic asthma (80) or mild asthma (81) advocate that reduced bacterial diversity is associated with neutrophilic inflammation and bronchial hyper responsiveness, respectively. In another study, it was shown that prevalence of H. influenzae, Pseudomonas aeruginosa and S. aureus at the lower airways of severe asthmatics was associated with persistence of asthma and exacerbation events but not with bronchial remodeling (82). P. aeruginosa, isolated from sputum of asthmatic patients was also found to be related with exacerbation (83).
It is reported from various in-vivo studies that Mycoplasma pneumoniae can affect the cilia structure of the respiratory epithelium and promote chronic inflammation, smooth muscle contraction, cytokine and antibody production (84–86), typical reactions during asthma onset and exacerbation (87, 88). M. pneumoniae infection in asthmatic children it was also connected with more IgM production and eosinophils in the blood compared to healthy controls and with, increased susceptibility to asthma symptoms (79). A meta-analysis from Zehua L. et al., confirm the possible implication of M. pneumoniae infections with asthma (79). Several serological studies also suggest that Chlamydia pneumoniae is related with severity phenotypes in asthma (89–91). Although the complexity of the microbiome is high, most of the studies suggest that Proteobacteria more than Firmicutes prevail and are common among the microbiota of patients with lung diseases (92), such as asthma or cystic fibrosis (93). It is therefore plausible that inflammatory signals resulting from such colonization may play an important role in persistent inflammation in asthma.
The Complex Dynamics of Bacteria- Epithelium Interactions in Asthma
Moraxella catarrhalis and Haemophilus influenzae are gram-negative opportunistic pathogens carried as commensals in the respiratory tract. Gram-negative bacteria predominate within the respiratory tract (94) and are mostly colonizing asymptomatically, although they can become causative agents for childhood otitis and other respiratory infections, such as pharyngitis and pneumonia (95). Moreover, they have been implicated in exacerbations of asthma and COPD (96, 97). S. pneumoniae and S. aureus are gram-positive extracellular bacteria colonizing mucosal surfaces of the upper respiratory tract from the early childhood. Infection from S. pneumoniae ranges from asymptomatic pharyngeal carriage and typical mucosal diseases like otitis media and sinusitis up to severe invasive diseases such as pneumonia, endocarditis, meningitis and bacteremia (98). S. aureus infections are also characterized by initial nasal colonization that may lead to subsequent invasive disease of skin or internal organs (99). The microbial identity of the human nasopharynx is unique for each individuals occupation with different serotypes of each species and seasonal modifications are influencing in a decisive way the overall image (100). It has been shown that microbial interactions are affected and change in the context of a respiratory disease (101) where the causative microbe prevail against others (102) and subsequent immune responses of the host alter the environment for all the resident bacteria (103). In asthma, chronic inflammation creates a favorable environment for transmission and colonization of airway bacteria to different respiratory niches (104). It is thus important to understand the dynamics of bacterial colonization and the events of adhesion and invasion to epithelial cells, as the possibility of intervention.
The evaluation of virulence or commensalism nature of M. catarrhalis and H. influenzae can be deemed according to several bacterial components (105), in accordance to their adherence capacity to epithelial surfaces (106) and their resistance to the complement cascade activation. The most significant surface components of M. catarrhalis and H. influenzae belong to the outer membrane proteins (OMP) and lipooligosaccharides (LOS). Serotypes of S. pneumoniae and S. aureus strains are mainly distinguished according to capsular polysaccharides on bacterial isolates that give different antigenicity and immunological reactions (107, 108). S. pneumoniae is classified into >90 different serotypes carrying different chemical identities on their cell wall and expressing distinct specificities across epithelial cells (109, 110). S. pneumoniae serotypes have been used during the last decades for the development of effective vaccination programs in children for the prevention of respiratory diseases caused by this pathogen (94). The abundance of serotypes underpins the complex relationships amongst species and the spectrum of their virulence across epithelial surfaces.
OMPs induce the release of pro-inflammatory cytokines and chemokines, like interleukin 1b (IL-1b), IL-6, IL-8 and prostaglandin E2 that promote adhesion of bacteria to the epithelium. LOS is a major trigger of the inflammatory response and plays a role in both adherence and invasion to the host cells (111, 112). LOS is an outer membrane glycolipid, involved in both adherence (113, 114) and invasion (115) to epithelial cells and in serum resistance (114). In several population studies three differentially distributed serotypes of M. catarrhalis according to LOS type (60–75% A, 20–30% B and 2–6% C) were identified (116–118). However, from these studies there is no clear correlation between serotypes and M. catarrhalis infection events. Nevertheless, a higher frequency of LOS type B and a lower frequency of LOS type A in adults compared to children was observed (117). H. influenzae isolates can be divided into encapsulated (serotypes a-f) and non-encapsulated strains designated also as typable or non-typable isolates accordingly. Colonization of nasopharynx with typable b serotype (<3% of population) causes, in most of the cases, serious invasive disease such as meningitis (119, 120), while colonization with non-typable serotypes (80% of adults and 40% of children) may result in sinusitis, exacerbation of COPD, conjunctivitis and otitis media (121).
Several human cell lines have been utilized to assess adherence and invasion events in-vitro, including A549 (type II pneumocytes), HEp-2 (laryngeal), Chang (conjuctival), Detroit 562 (pharyngeal), NCIH 292 (lung epidermoid), BEAS-2B (bronchial) and 16HBE14o (bronchial) cells (122–124). Recent in-vitro studies suggest that infection with M. catarrhalis provokes more cellular damage and inflammation (IL-33 and IL-8 production) compared to S. aureus (76, 125). In an in-vitro study, A549 were infected with M. catarrhalis and the presence of NOD1 and toll-like receptors (TLR2) were found to play a significant role in bacterial invasion and subsequent IL-8 production (126). In another in-vitro study they examined by electron and confocal microscopy the invasion capacity of M. catarrhalis in BEAS-2B cells, A549 and primary small airway epithelial cells. Formation of lamellipodia and filopodia from the infected epithelium toward M. catarrhalis after adherence was demonstrated, suggesting a macropinocytosis-like mechanism of invasion that comprises a PI3K-dependent contractile mechanism that transforms phagosomes and ruffles into closed intracellular organelles. Bacterial invasion increased steadily up to 6 h after the addition of bacteria and they calculated that ~5% of all infected BEAS-2B were invaded by M. catarrhalis. This suggests that invasion of this extracellular bacterium may be more common than previously thought. Interestingly, an ex-vivo study of primary bronchial epithelial cells obtained from asthmatic children with or without wheeze that did not exhibit any viral or bacterial infection at the time of the sampling, showed that there is an association between the PI3K/Akt pathway and the integrin α5β1 expression with the epithelial cell migration, repair and healing (127). Recurrent wheeze in asthmatic children and exacerbation of asthma are associated with Dysregulation in the PI3K/Akt pathway (128). It is apparent that the delineation of the asthmatic pathophysiology along with its implication with resident microbiome will be promising for controlling, predicting and treating clinical asthma.
Polymeric immunoglobulin receptor (pIgR) on nasopharyngeal epithelium is a key mediator for S. pneumoniae serotypes adherence and translocation from the apical to the basolateral region of the cells and the subsequent release to the bloodstream (129, 130). S. pneumoniae counteracts antimicrobial agents secreted from the lungs by rejecting its capsule with the aid of autolytic enzyme LytA (131) and thus evading killing efficiently. The absence of pneumococcal capsule is not only a protective mechanism against cells but also a strategic plan for bacteria to better reach and interact with epithelial surfaces and promote infection and colonization (132). It is also well-established that genes translated to autolysin, pneumolysin, or hydrogen peroxide are significant virulence factors for S. pneumoniae to survive and cause infection events in the lungs (133–135). S. aureus is typically found in the nasal cavity, where it binds to mucosal epithelial surfaces and specifically to mucin (136, 137). Many adhesins present in bacterium surfaces like surface protein G (SasG) contributing to the high affinity of the species with the nasal cells (138, 139). Keratinized epithelial cells of nasal cavities play also a role in S. aureus adherence and colonization processes (140) through the binding of clumping factor B (ClfB) with cytokeratin type 10 (138). In addition, S. aureus can also adhere to non-mucosal surfaces via polysaccharides that can bind with fibronectin or collagen of extracellular matrix (141). Interestingly it has also been shown that S. aureus adheres better to nasal epithelium of patients with eczema compared to healthy controls (142). There is an unmet need to map the “behavior” of nasopharyngeal commensals on states of health and disease in order to better predict, intervene and cure chronic respiratory diseases.
Clinical studies have monitored the upper respiratory tract of various pediatric populations in order to identify the initial events of bacterial colonization. A number of surveillance studies on pediatric populations suggest that there are antagonistic relationship between the different serotypes of S. pneumoniae and S. aureus (143–149). In contrary, non-typeable H. influenzae (NTHi) and M. catarrhalis populations are strongly correlated with S. pneumoniae during their co-existence in the airways of healthy individuals (145, 146, 150, 151). It has been proposed that NTHi typically antagonizes M. catarrhalis colonization on nasopharyngeal mucosal surfaces as they share a few identical virulent factors (152). A mechanistic study, trying to shed light on the events influencing nasopharyngeal colonization in healthy children, looked into gene polymorphisms on their epithelial cell receptors (153). It was reported that healthy infants with TLR2 polymorphisms on their nasopharyngeal epithelial cells are more likely to be colonized by S. aureus, whereas infants with variants on mannose-binding lectin are simultaneously colonized by S. aureus and S. pneumoniae. Additionally, M. catarrhalis colonization rates are high in infants in which TLR2 and TLR4 polymorphisms were found together. Of note, the biological mechanisms and patterns, characterizing and influencing the co-colonization of respiratory pathogens, are not fully understood and are definitely affected overtime by the states of health and disease (94).
In-vivo experiments in mice report different outcomes related to the interactions of typeable or non-typeable strains of H. influenzae with S. pneumoniae during the co-colonization in the nasopharynx. NTHi and S. pneumoniae typically act with a common mechanism on epithelial surfaces as they both possess neuraminidase activity that enables the use of N-acetyl galactosamine β1-3 galactose on epithelial cells in order to bind and promote infection (154). It has been shown that initial infection with NTHi promotes in a synergistic way the secondary infection with S. pneumoniae in nasopharynxes of mice (155–157). In contrast, initial infection of murine nasopharynxes with a typeable serotype, results in a rapid clearance of subsequent S. pneumoniae addition (158). One possible explanation for this phenomenon is the activation by H. influenzae of a complement-dependent phagocytic mechanism that acts against S. pneumoniae. This study proposed that the innate immune system mediates and determines the competition between commensal bacteria.
Bacterial acquisition is influenced to a great extent by other resident bacteria dynamics (159). In the case of S. aureus it is proposed that accessory gene regulator (agr) and staphylococcal accessory regulator (sar) provoke cross-inhibition of the virulence factors expressed on respiratory pathogens (159). In addition, due to the fact that S. aureus is a stable population expressing high clonality compared to S. pneumoniae (160) we can assume that in the cases of a well-occupied nasopharyngeal niche with S. aureus, it is difficult for another antagonistic respiratory pathogen population to interfere. It is also well-known that S. aureus nasal carriers have more risk for S. aureus infections than infection from other bacteria (161). The above literature demonstrates complex interactions between epithelial cells and bacterial pathogens, when the abundances of the latter are influenced by constant changes and dysbiosis in the case of inflammatory conditions such as asthma.
Targeting Dysbiotic Bacteria With Bacteriophages
Recently, we have proposed the possibility of controlling dysbiotic bacterial populations in asthma through phage interventions (162). New therapeutic modalities for asthma could potentially include targeted and customized re-colonization of the upper airway with lytic phages against key bacteria, including the ability of phages to act as anti-biofilm agents (76). Isolation and characterization of naturally occurring phages against M. catarrhalis, S. aureus, S. pneumoniae and H. influenzae may have prominent importance in this context.
It is well-documented that co-evolution phenomena are taking place between phages and their respective hosts (163). Understanding the molecular and ecological characteristics of M. catarrhalis and its phages is an important target (164, 165). Until now, isolation of lytic phages for M. catarrhalis has not been achieved. Nevertheless, several prophages have been identified and characterized which could be indicative of the microbial ecology and evolution of the species and especially gram-negative bacteria that pre-occupy the respiratory niche as referred previously (166). Davie et al. described phage elements in a M. catarrhalis comparative genomic study of clinical isolates (167), while Ariff et al. also described the presence of at least 32 putative whole prophages harbored in the genomes of 95 M. catarrhalis clinical isolates (168). Although it is yet to be determined if these prophages could be active and able to infect the host if induced, these studies provide evidence of a large variety of prophages and prophage elements, which have interacted and fused their genome with Moraxella hosts over the years. Moreover, host molecular anti-phage strategies, such as CRISPR (169), against lytic phages had been well-documented, a strong indication of ancestral lytic phage-host interaction (167, 170). It is plausible that the large abundances of prophages and prophage elements (171) coupled with the significant presence of CRISPR systems harbored in M. catarrhalis genomes, could act as strong anti-phage strategies, making lytic phage isolation a challenging process.
Haemophilus influenzae is central in the basic phage research for many years (172–174). The binary models of H. influenzae and temperate phages HP1 and HP2 is a valuable tool for researchers to understand the lysis-lysogeny decision of temperate phages and the genes involved in it (175, 176). However, the literature still lacks a well-characterized virulent phage against this genus. Only recently researchers have reported a number of four possible isolates against H. influenzae with the complete characterization of these phages and their lytic efficacy remaining an open question (177).
Staphylococcal species colonize and dominate the human hypopharynx microbiome from early life (178). S. aureushas been suggested to be a serious threat for asthmatic incidents in adult life, through its superantigens (179). Contrary to M. catarrhalis and H. influenzae, staphylococcal lytic phages have been extensively studied as antimicrobial agents for many years against human and non-human bacterial infections. Systematic isolation and characterization of staphylococcal lytic phages has been taking place since 1950s. S. aureus was in the center of attention for the research community being a highly virulent agent with common antibiotic resistance, making it a high priority for discovering novel antimicrobial solutions against it. Interestingly, apart from the phages, lysozymes and endolysins were also tested against antibiotic-resistant strains of S. aureus, especially in strains colonizing skin tissue. Phage treatment has a much better background against S. aureus than other bacteria, offering promising results already (180, 181). Especially, Sb1 and ISP phages, expressing high virulence amongst antibiotic resistant S. aureus strains are used intravenously as therapeutics for many years now by the Eliava Institute of Bacteriophage, Microbiology and Virology (182, 183).
In regard to S. pneumoniae, lytic phages have also been isolated for many years now (184). S. pneumoniae demonstrates severe antibiotic resistance and apart from asthma-related incidents, it is a major causative agent for several respiratory diseases worldwide. Thus, novel approaches such as phage therapy could potentially have important role against it. Just as in S. aureus, potential therapeutic candidates are phage-originated murein hydrolases, which are able to degrade the outer thick peptidoglycan layer, the first line of defense for gram-positive bacteria, and induce lysis, without the use of phages (185–187).
Bacteriophages and the Epithelium
Bacteriophages on Airway Mucosal Surfaces
Phages are part of the human virome; collectively, also referred as “phageome,” they regulate the populations of bacteria through prey-predator interactions (44). Recent studies show that phages are present in every niche of the human body including oral (188, 189), pulmonary (190, 191) and intestinal (192, 193) cavities and also the urinary tract (194), the skin (195, 196) and the blood (43, 197). The phageome constitutes the majority of viruses in a healthy lung and play a potentially crucial role to lung health and immunity, framing the bacterial population (45, 198). Bacteriophages have also been shown to reprogram the eukaryotic microbiome (199) and play an important role, as part of the immune system of mammals (200). The microbial load in healthy airways appears to be reduced as we proceed from the upper to the lower respiratory tract and except from the anatomical differences, it is also affected by the presence of different phage communities in different niches (26).
The surface of the nasal and bronchial epithelium is covered, as previously mentioned by mucus, while the surface of the alveolar epithelium is covered by pulmonary surfactant secreted from alveolar type II cells. Mucus contains mucin glycoproteins and nutrients that provide a favorable environment for commensal bacteria and phage symbionts. The most abundant types of mucin glycoproteins found in human airways are MUC5AC and MUC5B. In-vivo experiments in mice showed that MUC5B play a critical role in airway defense and bacterial clearance through MCC (30). In-vitro experiments with ALI differentiated bronchial epithelial cells from asthmatics, COPD and healthy control individuals, infected with rhinovirus-A1, showed no alteration of MUC5AC protein levels in any group (23). Nevertheless, the alteration of mucus patterns and production rates in many cases of clinical asthma provoke lung inflammation and multiple bacterial species colonization. In parallel, it is established that phage communities are more abundant in mucosal surfaces compared to adjacent non-mucosal areas (201, 202). The overall dysfunction of mucus secretions in asthma may influence not only the MCC mechanism but also the phage communities that regulate and control bacterial populations.
Mucus is an important intermediate in phage-bacteria interactions. In one related study they used T4 phages expressing or not an Ig-like protein (Hoc) in their capsid that binds to mucin glycoproteins. They performed in-vitro testing on mucus producing and mucus knockdown A549 lung epithelial cells. The cells were pretreated with hoc+ and hoc– T4 phages and then infected with Escherichia coli strains. They observed that mucus-producing cells pretreated with hoc+ phages, were protected from bacterial infection and cell death, in contrast to the mucus knockdown cells. In a microfluidic device simulating the constant air flow and mucin secretion dynamics, they showed the persistence of T4 phages and their subdiffusive motion on mucosal surfaces contributes to efficient phage-host encounters (201, 203). Although, recent literature about the interaction of phages with mucus is still limited, it is possible that future studies may unravel important mediation of these viruses in microbial equilibrium at the healthy airways.
During the last decade it was shown that several phages from the Myoviridae, Siphoviridae, and Podoviridae families, approximately one quarter of them, are carrying capsids with Ig-like proteins on their surfaces thus having the ability to complementary bind with mucin glycoproteins (204–206). In addition, it has been reported that there are large and diverse phage populations in mucosal surfaces compared with resident bacteria populations and with adjacent non-mucosal areas (201). We can assume that the direct association of phages with mucosal surfaces is providing a non-host-mediated mechanism of immunity (201) or an enhanced mechanism of host innate immunity that may induce a long-lasting adaptive immune response. In case of an infection, epithelial cells respond by secreting antimicrobial agents and mucin glycoproteins, thus giving the possibility for more phages to be attached and protect the surfaces from further bacterial colonization. The presence of phages in mucosal surfaces may also stimulate weak immune responses from the host thus inducing the continuous production of low concentrations of cytokines that may act protectively in the case of an infection (207, 208). Of note, it has been demonstrated in-vitro that Ig-like domains facilitate phage adsorption to the bacterial host and give a proliferation advantage against bacterial symbionts in mucosal areas (205, 206). It is becoming increasingly apparent that phages are key players for the overall homeostasis of human mucosal surfaces; further research may unravel new mechanisms and models for this complex symbiosis.
Bacteriophages and Epithelial Cell Interactions
The direct implication of phages with epithelial cells is a recently proposed concept, enhanced by several in-vitro studies. Lehti et al. propose that E. coli phage PK1A2, belonging to Podoviridae family, could directly interact with eukaryotic neuroblastoma epithelial cells. They used fluorescent and electron microscopy to observe the specific binding of phages with mammalian cells and follow the internalization process through the endolysosomal mechanism. Internalized phages remained intact for 24 h into the cytoplasm without affecting the viability of cells and then appeared in the lysosomes where their proteins and nucleic acids degraded from the cell (209). Recent reports, explain also the pivotal interactions that phages can develop with mammalian cells, including entering mammalian cells similarly to mammalian viruses, killing bacteria not only in the periplasmic space, but also both in the phagosome and the cytosol areas and finally express their genetic information in the nucleus (210). In-vitro experiments with bronchial primary epithelial cells, obtained from asthmatic children attempt to understand, as previously mentioned, the mechanisms of epithelial cell repairing and migration after wounding treatment (127). Future in-vitro and in-vivo studies could be focused on asthmatic epithelial repair during bacterial infection and/or phage presence. These hypotheses emphasize the critical role of phages in human environments and raise many questions about their undefined role, as protective, anti-inflammatory, biocontrol or nutrient agents for the cells.
Neuroblastoma cells express polysialic acid on their surfaces that shows high homology with polysaccharide of the bacterial host E. coli K1 of the phage. Additional molecules in airway epithelial surfaces have molecular homology with bacterial components, such as hyaluronan capsule of group A (211) and sialylated capsular polysaccharide of group B (212) S. pneumoniae and sialylated lipooligosaccharide of Neisseria meningitides (213). It appears that eukaryotic and prokaryotic cells exchange genetic material through HGT mainly after internalization of bacteria and/or phages in human cells. In addition, phages appear to have intrinsic tropism not only to bacterial cells but also to eukaryotic cells (214, 215). It is proposed that the molecular mimicry between bacteria and host cells may have an impact on phage's interaction with human cells.
Considering the enormous diversity of phage symbionts, a relevant number of epithelial epitopes may be capable to induce efficient intake of phages into the epithelial human cells. Specifically, it has been assumed that the human body can absorb ~1010 phage particles per day which can be transported through epithelial surfaces and reach the blood circulation (209, 216). Internalization and deposition of phages to the lymphatic and circulatory systems may also contribute to the remission of strong intracellular immunity signals that potentially make the cells resistant against microbial invaders (217). Typically, antigen recognition is a well-characterized ability of specific immune cells of the adaptive system; nevertheless Charles A. Janeway proposed that germline-encoded receptors on cells can sense bacterial conserved patterns and activate intracellular pathways for bacterial destruction after the cell invasion (218). We can assume that endocytosed phages in epithelial cells could be implicated in similar intracellular pathways that give the advantage for rapid response in the case of bacterial invasion.
Intra-Body Phages
Phages pass through epithelial mucosal surfaces via transcytosis (216), adhere to specific cell receptors and be endocytosed from the apical epithelial surface (215, 219). If they successfully escape lysosome degradation, phages are finally exocytosed from the basal side of the human cells and getting into the bloodstream from where they can reach distal areas (216, 220). This phenomenon was described for several phages such as T4, T5, T7, P22, SP01, and SPP1 (216). It was also reported that phages provided intraperitoneally or intranasally can be found even in the brain (221), suggesting the high motility and utterly different dynamic properties characterizing these viruses.
In an in-vitro study they used a series of different human epithelial cells from the colon (CaCo2 and T84), lung (A549), liver (Huh7), kidney (MDCK), and brain (hBMec) to monitor different phage families (Podoviridae, Myoviridae, and Siphoviridae) and to assess the ability of transcytosis across the confluent epithelial cell monolayers. Approximately 10% of epithelial cells expressed specific phage receptors and contained membrane-bound vesicles that allowed the internalization and transcytosis of phages (216). In another in-vitro study they used bovine mammary epithelial cells infected with S. aureus JYG2 in order to test the ability of virulent vB_SauM_JS25 phage to enter and lyse intracellular bacteria. After 12 h incubation with phages, they observed via confocal microscopy the presence of phages near the nuclei of infected cells and they established the progressive elimination of intracellular S. aureus in a time and dose dependent manner (222). In addition, It is reported that several phages such as Φ29, Bam35, Cp-1, Nf, PRD1 possess terminal proteins with inherent nuclear transmission signals (47) that enable the phage absorption to the eukaryotic nuclei and the effective delivery of proteins and genes into the eukaryotic genome (46). Transcytosis of phages enables the effective HGT between phages and eukaryotic cells and raise the question of whether endocytosed phages can replicate inside the cells and/or protect them from inflammation and cell death after bacterial invasion (210). Results from further experiments showed that phages can reduce inflammation in MAC-T bovine mammary epithelial cells suppressing the LPS-induced phosphorylation of NF-kB p65 protein from S. aureus (223).
The ability of phages to enter the cells and pass through internal organs is a new and yet not fully described phenomenon. The plethora of phages and the variety of human cells that can be used render the aforementioned experiments just the beginning for a new field of research. The characterization of the phenomenon demands the expansion of the related in-vitro and in-vivo studies in order to establish a good predictive model of the phage addition in the human body. Phages could play a significant role to host immunity and microbial balance and can be used as a novel therapeutic and anti-inflammatory tool in many chronic airway diseases. The future introduction of phages in clinical trials will be a new and promising step for the whole medical community and pharmaceutical industry, dealing with chronic and refractory respiratory diseases.
Tripartite Interactions of Bacteria, Phages and the Airway Epithelium
Phages Develop Synergistic and/or Antagonistic Relationships With Bacteria on Airway Epithelial Surfaces Regulating Chronic Airway Inflammation
Although airway inflammation differs between asthma and cystic fibrosis (CF) important lessons can be learnt from the latter. Phage populations within the lungs of healthy individuals differ significantly compared with those suffering from CF (190). Different areas of explanted lungs of late-stage CF patients were examined with metagenomic analysis in order to identify and quantify different viral populations. They interestingly observed the absence of phages related to CF common pathogen P. aeruginosa, S. aureus, and H. influenzae at the left and right upper lobes (224), suggesting that phage communities play an important role in the homeostasis and overall health of the airways. Phage communities express similarities between CF patients and have a specific metabolic profile adapted to CF pathophysiology that differs significantly from the healthy state (225).
Recently, it has been reported that phage populations within the lungs of healthy individuals differ significantly compared with those suffering from asthma (162) and/or CF (190). The absence of phages related to common pathogens in asthma (162) and CF (224) suggests an important role of phage symbionts in airways homeostasis and healthy state. In an in-vitro study primary bronchial epithelial cells were isolated from children with CF and healthy controls and different concentrations of purified P. aeruginosa phage preparations were tested on submerged cell cultures in order to investigate the cytotoxic and/or inflammatory effect of phages. Results showed that the viability of cells was maintained, while no production of inflammatory cytokines IL-6 or IL-8 was observed. Of note, the apoptosis of cells was reduced in the presence of phages compared to untreated control cells (225). In contrast, in-vivo induction of filamentous phage Pf4 from P. aeruginosa biofilms enhances bacteria persistence in CF patient's lungs, promoting adhesion of the bacteria to mucus and inhibiting cell invasion (226). Therefore, in-vivo production of Pf4 phage leads to reduced lung injury, giving the fitness benefit to bacterial population to escape from phagocytosis and immune response. It is apparent that induction of prophage in this case changes the inflammation human response and lead to chronic persistence of bacterial population in CF lungs (227). From the aforementioned experiments we can expect that phages related to common pathogens in asthma may induce diverse behaviors in-vitro and in-vivo, favoring or eliminating bacterial populations.
Co-culture of S. pneumoniae with human cells can provoke the induction of streptococcal pyrogenic exotoxin C (SpeC), a well-characterized phage gene product. Interestingly this induction occurred without the addition of any phage induction reagents, like mytomycin C or application of UV radiation and just occurred during the co-culture. Broudy T. et al. examined the supernatants of the culture with electron microscopy to validate the existence of phage particles and they identified whole phage viruses during the co-culture of group A streptococci with Detroit 562 human pharyngeal cells (228). These results suggest the existence of a soluble yet uncharacterized phage inducing factor secreted from epithelial cells. This induction may occur independently from bacterial contact with human cells leading to the lysis of some streptococci and the subsequent release of cytoplasmic virulence factors. Subsequent in-vitro and in-vivo experiments in mice showed that the presence of mammalian cells may play a significant role in phage induction, implying that phages have obtained through natural selection a system where induction occurred at eukaryotic niches where bacteria are becoming more virulent (229).
The presence of prophage genetic elements with phage-encoded lysins in ~70% of clinically isolated S. pneumoniae strains (230) is a well-described phenomenon but the role of prophages during bacterial colonization is mainly undefined. In an in-vivo study, it was shown that the presence of Spn1 prophage element at S. pneumoniae provoked changes on bacterial cell wall and had a negative effect on nasopharyngeal colonization in mice (231). In another study, SV1 prophage elements were correlated with S. pneumoniae biofilm formation and promotion of chronic lung infection (232). Adherence of another nasopharyngeal human commensal N. meningitides to the epithelial surfaces was enhanced by the existence of filamentous prophage MDAΦ in an in-vitro study. This finding suggests that MDAΦ colonization of its host bacterium may increase occurrence of meningitis. Interestingly, this effect was not observed on endothelial cells, but only on epithelial cells tested (233).
In another in-vitro study, phiCDHS1 lytic phages appeared more active against Clostridium difficile in the presence of HT-29 epithelial cells than without them. They performed subsequent experiments to audit whether there are specific HT-29 secretions that enhance phage activity, culturing phages with fresh or spend HT-29 culture medium. They concluded that phage lysis activity is not affected by the substances on cell culture media but it is promoted by the adherence of phages to the epithelial monolayer facilitating subsequent adherence with C. difficile (234).
An interesting example of nasopharyngeal microbiome regulation by phages has been reported regarding S. pneumoniae and S. aureus interactions. It was reported that S. aureus may be restricted from the nasopharynx in the presence of S. pneumoniae (235). H2O2 produced by S. pneumoniae promotes the production of hyperoxides via Fenton chemical reaction. Hyperoxides are able to induce SOS response in S. aureus that is typically a reaction that promotes DNA restoration, causing the activation of resident prophages from S. aureus lysogenic strains. Subsequent lysis of S. aureus promotes the S. pneumoniae prevalence at the nasopharynx. In contrary, S. pneumoniae lysogenic strains do not undergo SOS-response thus having a prevalence asset against S. aureus symbionts. The induction of prophages in neighboring organisms is a strategy for releasing active phages into the human body. This effect can cause subsequent spread of virulence genes and mobile genetic elements into different cells thus reshaping the existent equilibrium (236).
From the aforementioned literature it is clear that phages possess high immune modulatory capacities (207, 237) in CF (238, 239), COPD (240, 241), and other respiratory disease conditions. To the extent of our knowledge there are limited relevant published studies about the role that phages own on asthmatic epithelium. It cannot be excluded that microbial imbalance in asthma may be related and/or caused by underlying phage imbalance (162). In the near future, with the continuous research on that field, we expect the establishment of new mechanisms of phages-bacteria-eukaryote cells interactions in order to better orchestrate the triptych symbioses model (Figure 1).
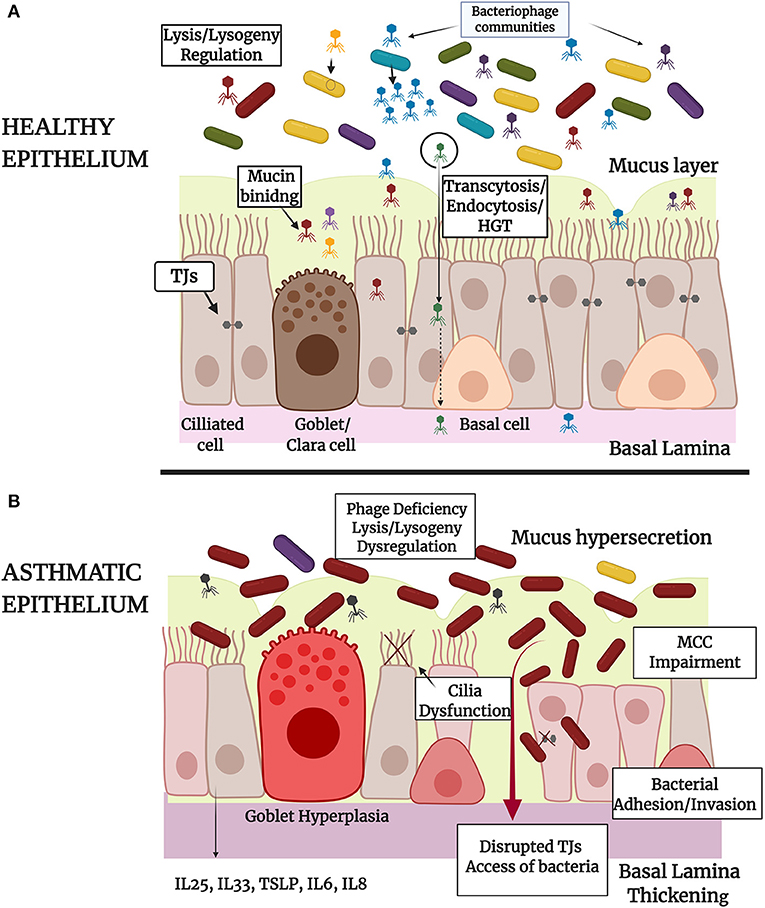
Figure 1. Schematic representation of the tripartite symbiosis model between bacteriophages, bacteria and epithelium within healthy (A) and asthmatic (B) airways. (A) Healthy epithelium of large and small airways of the lungs is constituted by basal, ciliated and secretory Goblet (large airways) or Clara cells (small airways) that form pseudostratified or columnar and cuboidal structures, respectively. Epithelial barrier integrity is ensured by the formation of intracellular Tight Junctions (TJs) and by the secretion of mucus by Goblet or Clara cells. Healthy epithelium physically controls bacterial populations and eliminates bacterial invasion through mucociliary clearance (MCC). In healthy airways there is a diverse population of bacteria and bacteriophage communities that interact directly through lysis-lysogeny mechanisms and horizontal gene transfer as represented in the figure. Bacteriophages interact also directly with epithelium through adhesion to mucus or through specifically biding to cellular receptors (endocytosis) and non-specific binding to membrane vesicles (transcytosis). Bacteriophage-epithelium uptake protects the cells from bacterial invasion and proliferation and enables the delivery of bacteriophage nucleic acids (HGT) and proteins into the nucleus. (B) Asthmatic epithelium is characterized by Goblet hyperplasia, mucus hypersecretion, MCC impairment and disrupted TJs that lead to bacterial adherence and invasion into the human cells or access of bacteria into the lamina paracellularly. In the case of asthma, bacteriophage communities are deregulated, as it is presented in the figure, and this may be associated with an altered microbiota profile with limited bacterial diversity and dominance of specific bacterial species. The role of prophage elements in bacterial genomes has also an influence on the ability of pathogens to provoke infection and epithelial damage. Irritated asthmatic epithelium secretes a range of chemokines and cytokines, contributing to immune system activation and chronic inflammation in asthma. The image was created using BioRender.
Discussion
Current literature related to the triptych bacteria–phages–airway epithelium highlights a number of aspects of this dynamic system. The new era of precision medicine demands the construction of a well-established system for personalized monitoring (28). Emerging molecular and screening techniques could give the possibility to stratify the microbiome of an individual from infancy until adulthood. In this way we may be able to map dynamic microbiome profiles thus following and understanding the alterations occurring during an infection or in the onset of a chronic disease. Novel lytic phages could play a pivotal role in restoring bacteria pathogens abundances, thus making their isolation and characterization a priority challenge. Still, literature lacks well-characterized lytic phages for important bacterial species that colonize the human hypopharynx, such as M. catarrhalis and H. influenzae.
Researchers have already pinpointed the importance of studying the biology of the wider system when phage therapy is taking place, rather than only the binary interaction of bacterial host and virus (242–244). The incidences of phage-bacteria interaction on epithelial surfaces are influenced to a great extent by the subdiffusive motion of phages on mucosal surfaces (203). In addition, another important parameter is the motility rate of bacterial hosts on specific environments where phages can be added. Specifically high bacterial motility rates are related with more frequent phage encountering independently of the phage motion in the mucus (244). We must also take into account the ability of some bacteria to form biofilms on human airway niches and thus further evaluate the anti-biofilm capacity of important lytic phages (245, 246).
There is still limited number of studies on phage-bacteria population interactions in-vivo. The existent studies are using mainly mice models in order to predict and evaluate the effect of phage addition on the gut microbiome (247–249). The potential changes related with phage intervention on the airways are mainly unexplored. The addition of lytic phages on the mouse gut can affect not only the sensitive bacteria but also other resident bacteria populations in a cascade (249). It is thus important to attempt to predict the new dynamics that will be created following potential phage therapy at the respiratory tract.
In this review, we attempt to shed light to a new field of basic research, focused on the tripartite interactions between respiratory epithelial cells, bacteria and phages and to connect its results with asthma pathophysiology. Concepts emerging through this review need to be tested in the clinic. In Table 2 we list a number of such concepts, suggesting targets for future clinical studies.
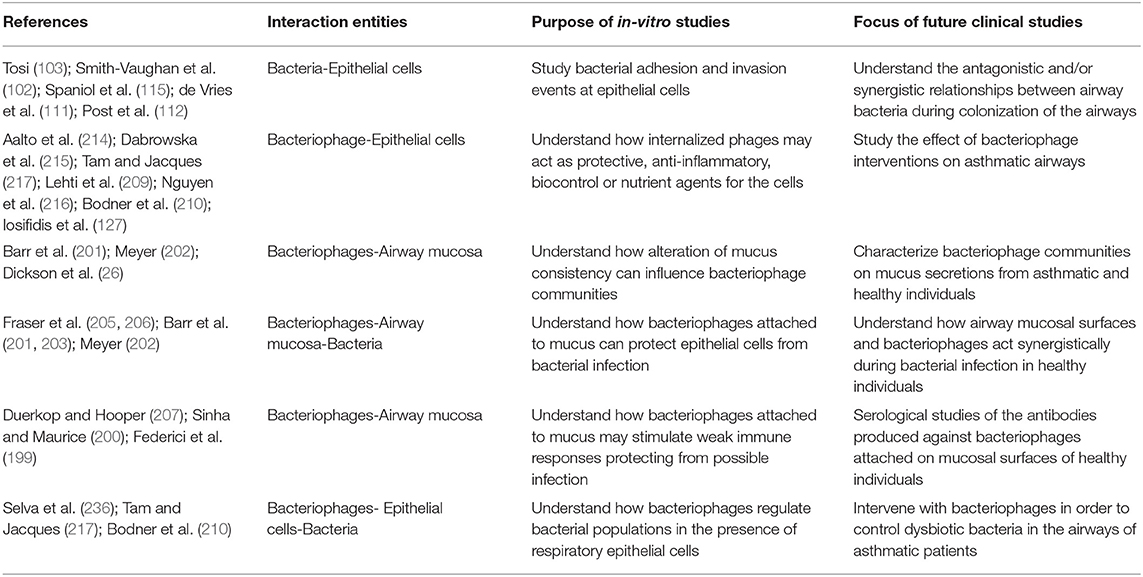
Table 2. Novel ideas that are coming up from current in-vitro studies focused on the interaction between bacteriophages, bacteria and airway mucosa and proposed future in-vivo studies that can be significant for clinical asthma.
Author Contributions
PT-T designed and wrote the manuscript. DS contributed textually and designed along with PT-T the Figure. NP and SM conceptualized the main idea. NP coordinated the whole review. All authors contributed critically on reviewing and making substantial comments on the final article.
Funding
Funding for this work provided by European Union (Horizon 2020) in the context of the CURE Programme Constructing a Eubiosis Reinstatement Therapy for Asthma.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work is conducted in the context of the CURE programme Constructing a Eubiosis Reinstatement Therapy for Asthma, financed by the European Union (Horizon 2020) under agreement No. 767015. The figure was created with BioRender.com.
References
1. Pelaia G, Vatrella A, and Maselli R. The potential of biologics for the treatment of asthma. Nat Rev Drug Discov. (2012) 11:958–72. doi: 10.1038/nrd3792
2. Martinez FD. Genes, environments, development and asthma: a reappraisal. Eur Respir J. (2007) 29:179–84. doi: 10.1183/09031936.00087906
3. Desai M, and Oppenheimer J. Elucidating asthma phenotypes and endotypes: progress towards personalized medicine. Ann Allergy Asthma Immunol. (2016) 116:394–401. doi: 10.1016/j.anai.2015.12.024
4. Ferreira MAR, Mathur R, Vonk JM, Szwajda A, Brumpton B, Granell R, et al. Genetic architectures of childhood- and adult-onset asthma are partly distinct. Am J Hum Genet. (2019) 104:665–84. doi: 10.1016/j.ajhg.2019.02.022
5. Agache I, and Akdis CA. Precision medicine and phenotypes, endotypes, genotypes, regiotypes, and theratypes of allergic diseases. J Clin Invest. (2019) 129:1493–503. doi: 10.1172/JCI124611
6. Kuruvilla ME, Lee FE, and Lee GB. Understanding asthma phenotypes, endotypes, and mechanisms of disease. Clin Rev Allergy Immunol. (2019) 56:219–33. doi: 10.1007/s12016-018-8712-1
7. Croisant S. Epidemiology of asthma: prevalence and burden of disease. Adv Exp Med Biol. (2014) 795:17–29. doi: 10.1007/978-1-4614-8603-9_2
8. Dharmage SC, Perret JL, and Custovic A. Epidemiology of asthma in children and adults. Front Pediatr. (2019) 7:246. doi: 10.3389/fped.2019.00246
9. Nunes C, Pereira AM, and Morais-Almeida M. Asthma costs and social impact. Asthma Res Pract. (2017) 3:1. doi: 10.1186/s40733-016-0029-3
10. Yaghoubi M, Adibi A, Safari A, FitzGerald JM, and Sadatsafavi M. The projected economic and health burden of uncontrolled asthma in the United States. Am J Respir Crit Care Med. (2019) 200:1102–12. doi: 10.1164/rccm.201901-0016OC
11. Crystal RG, Randell SH, Engelhardt JF, Voynow J, and Sunday ME. Airway epithelial cells: current concepts and challenges. Proc Am Thorac Soc. (2008) 5:772–7. doi: 10.1513/pats.200805-041HR
12. Bijanzadeh M, Mahesh PA, and Ramachandra NB. An understanding of the genetic basis of asthma. Indian J Med Res. (2011) 134:149–61. Available online at: https://www.ijmr.org.in/text.asp?2011/134/2/149/84727
13. Wu J, Dong F, Wang R, Wang J, Zhao J, Yang M, et al. Central role of cellular senescence in TSLP-induced airway remodeling in asthma. PLoS ONE. (2013) 8:e77795. doi: 10.1371/journal.pone.0077795
14. Wan H, Winton HL, Soeller C, Tovey ER, Gruenert DC, Thompson PJ, et al. Der p 1 facilitates transepithelial allergen delivery by disruption of tight junctions. J Clin Invest. (1999) 104:123–33. doi: 10.1172/JCI5844
15. Sajjan U, Wang Q, Zhao Y, Gruenert DC, and Hershenson MB. Rhinovirus disrupts the barrier function of polarized airway epithelial cells. Am J Respir Crit Care Med. (2008) 178:1271–81. doi: 10.1164/rccm.200801-136OC
16. Post S, Nawijn MC, Jonker MR, Kliphuis N, Berge M, Oosterhout AJM, et al. House dust mite-induced calcium signaling instigates epithelial barrier dysfunction and CCL20 production. Allergy. (2013) 68:1117–25. doi: 10.1111/all.12202
17. Rezaee F, DeSando SA, Ivanov AI, Chapman TJ, Knowlden SA, Beck LA, et al. Sustained protein kinase D activation mediates respiratory syncytial virus-induced airway barrier disruption. J Virol. (2013) 87:11088–95. doi: 10.1128/JVI.01573-13
18. Looi K, Buckley AG, Rigby PJ, Garratt LW, Iosifidis T, Zosky GR, et al. Effects of human rhinovirus on epithelial barrier integrity and function in children with asthma. Clin Exp Allergy. (2018) 48:513–24. doi: 10.1111/cea.13097
19. Welsh KG, Rousseau K, Fisher G, Bonser LR, Bradding P, Brightling CE, et al. MUC5AC and a glycosylated variant of MUC5B alter mucin composition in children with acute asthma. Chest. (2017) 152:771–9. doi: 10.1016/j.chest.2017.07.001
20. Gon Y, and Hashimoto S. Role of airway epithelial barrier dysfunction in pathogenesis of asthma. Allergol Int. (2018) 67:12–7. doi: 10.1016/j.alit.2017.08.011
21. Frey A, Lunding LP, Ehlers JC, Weckmann M, Zissler UM, and Wegmann M. More than just a barrier: the immune functions of the airway epithelium in asthma pathogenesis. Front Immunol. (2020) 11:761. doi: 10.3389/fimmu.2020.00761
22. Fehrenbach H, Wagner C, and Wegmann M. Airway remodeling in asthma: what really matters. Cell Tissue Res. (2017) 367:551–69. doi: 10.1007/s00441-016-2566-8
23. Veerati PC, Troy NM, Reid AT, Fung Li N, Nichol KS, and Kaur P. Airway epithelial cell immunity is delayed during rhinovirus infection in asthma and COPD. Front Immunol. (2020) 11:974. doi: 10.3389/fimmu.2020.00974
24. Kicic A, Stevens PT, Sutanto EN, Kicic-Starcevich E, Ling KM, Looi K, et al. Impaired airway epithelial cell responses from children with asthma to rhinoviral infection. Clin Exp Allergy. (2016) 46:1441–55. doi: 10.1111/cea.12767
25. Holgate ST, Wenzel S, Postma DS, Weiss ST, Renz H, Sly PD, et al. Asthma. Nat Rev Dis Primers. (2015) 1:15025. doi: 10.1038/nrdp.2015.25
26. Dickson RP, Erb-Downward JR, Freeman CM, McCloskey L, Beck JM, Huffnagle JB, et al. Spatial variation in the healthy human lung microbiome and the adapted island model of lung biogeography. Ann Am Thorac Soc. (2015) 12:821–30. doi: 10.1513/AnnalsATS.201501-029OC
27. Fujimura KE, and Lynch SV. Microbiota in allergy and asthma and the emerging relationship with the gut microbiome. Cell Host Microbe. (2015) 17:592–602. doi: 10.1016/j.chom.2015.04.007
28. Behrouzi A, Nafari AH, and Siadat SD. The significance of microbiome in personalized medicine. Clin Transl Med. (2019) 8:16. doi: 10.1186/s40169-019-0232-y
29. Hufnagl K, Pali-Schöll I, Roth-Walter F, and Jensen-Jarolim E. Dysbiosis of the gut and lung microbiome has a role in asthma. Semin Immunopathol. (2020) 42:75–93. doi: 10.1007/s00281-019-00775-y
30. Roy MG, Livraghi-Butrico A, Fletcher AA, McElwee MM, Evans SE, Boerner RM, et al. Muc5b is required for airway defence. Nature. (2014) 505:412–6. doi: 10.1038/nature12807
31. Hooks KB, and O'Malley MA. Dysbiosis and its discontents. MBio. (2017) 8:e01492–17. doi: 10.1128/mBio.01492-17
32. Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. (2011) 184:957–63. doi: 10.1164/rccm.201104-0655OC
33. Morris A, Beck JM, Schloss PD, Campbell TB, Crothers K, et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med. (2013) 187:1067–75. doi: 10.1164/rccm.201210-1913OC
34. Dickson RP, Erb-Downward JR, Martinez FJ, and Huffnagle GB. The microbiome and the respiratory tract. Annu Rev Physiol. (2016) 78:481–504. doi: 10.1146/annurev-physiol-021115-105238
35. Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, et al. Disordered microbial communities in asthmatic airways. PLoS ONE. (2010) 5:e8578. doi: 10.1371/journal.pone.0008578
36. Di Cicco M, Pistello M, Jacinto T, Ragazzo V, Piras M, Freer G, et al. Does lung microbiome play a causal or casual role in asthma? Pediatr Pulmonol. (2018) 53:1340–5. doi: 10.1002/ppul.24086
37. Kozik AJ, and Huang YJ. The microbiome in asthma: role in pathogenesis, phenotype, and response to treatment. Ann Allergy Asthma Immunol. (2019) 122:270–5. doi: 10.1016/j.anai.2018.12.005
38. Holgate ST. Epithelium dysfunction in asthma. J Allergy Clin Immunol. (2007) 120:1233–44. doi: 10.1016/j.jaci.2007.10.025
39. Schulz BL, Sloane AJ, Robinson LJ, Prasad SS, Lindner RA, Robinson M, et al. Glycosylation of sputum mucins is altered in cystic fibrosis patients. Glycobiology. (2007) 17:698–712. doi: 10.1093/glycob/cwm036
40. Gill DJ, Tham KM, Chia J, Wang SC, Steentoft C, Clausen H, et al. Initiation of GalNAc-type O-glycosylation in the endoplasmic reticulum promotes cancer cell invasiveness. Proc Natl Acad Sci USA. (2013) 110:E3152–E3161. doi: 10.1073/pnas.1305269110
41. Sharma L, Feng J, Britto CJ, and Dela Cruz CS. Mechanisms of epithelial immunity evasion by respiratory bacterial pathogens. Front Immunol. (2020) 11:91. doi: 10.3389/fimmu.2020.00091
43. Brown-Jaque M, Muniesa M, and Navarro F. Bacteriophages in clinical samples can interfere with microbiological diagnostic tools. Sci Rep. (2016) 6:33000. doi: 10.1038/srep33000
44. Manrique P, Bolduc B, Walk ST, Van der Oost J, de Vos WM, and Young MJ. Healthy human gut phageome. Proc Natl Acad Sci USA. (2016) 113:10400–5. doi: 10.1073/pnas.1601060113
45. Barr JJ. A bacteriophages journey through the human body. Immunol Rev. (2017) 279:106–22. doi: 10.1111/imr.12565
46. Redrejo-Rodriguez M, Bolduc B, Walk ST, Van der Oost J, de Vos WM, and Young MJ. Nuclear localization signals in phage terminal proteins provide a novel gene delivery tool in mammalian cells. Commun Integr Biol. (2013) 6:e22829. doi: 10.4161/cib.22829
47. Redrejo-Rodriguez M, and Salas M. Multiple roles of genome-attached bacteriophage terminal proteins. Virology. (2014) 468–470, 322–329. doi: 10.1016/j.virol.2014.08.003
48. Hall JPJ, Brockhurst MA, and Harrison E. Sampling the mobile gene pool: innovation via horizontal gene transfer in bacteria. Philos Trans R Soc Lond, B, Biol Sci. (2017) 372:2–7. doi: 10.1098/rstb.2016.0424
49. Clemente JC, Ursell LK, Parfrey LW, and Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. (2012) 148:1258–70. doi: 10.1016/j.cell.2012.01.035
50. Bowers RM, Ursell LK, Parfrey LW, and Knight R. Sources of bacteria in outdoor air across cities in the midwestern United States. Appl Environ Microbiol. (2011) 77:6350–6. doi: 10.1128/AEM.05498-11
51. O'Dwyer DN, Dickson RP, and Moore BB. The lung microbiome, immunity, and the pathogenesis of chronic lung disease. J Immunol. (2016) 196:4839–47. doi: 10.4049/jimmunol.1600279
52. Prussin AJ, Garcia EB, and Marr LC. Total virus and bacteria concentrations in indoor and outdoor air. Environ Sci Technol Lett. (2015) 2:84–8. doi: 10.1021/acs.estlett.5b00050
53. Tignat-Perrier R, Dommergue A, Thollot A, Keuschnig C, Magand O, Vogel TM, et al. Global airborne microbial communities controlled by surrounding landscapes and wind conditions. Sci Rep. (2019) 9:14441. doi: 10.1038/s41598-019-51073-4
54. Horve PF, Lloyd S, Mhuireach GA, Dietz L, Fretz M, MacCrone G, et al. Building upon current knowledge and techniques of indoor microbiology to construct the next era of theory into microorganisms, health, and the built environment. J Expo Sci Environ Epidemiol. (2020) 30:219–35. doi: 10.1038/s41370-019-0157-y
55. Charlson ES, Diamond JM, Bittinger K, Fitzgerald AS, Yadav A, Haas AR, et al. Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. Am J Respir Crit Care Med. (2012) 186:536–45. doi: 10.1164/rccm.201204-0693OC
56. Wilson MT, and Hamilos DL. The nasal and sinus microbiome in health and disease. Curr Allergy Asthma Rep. (2014) 14:485. doi: 10.1007/s11882-014-0485-x
57. Dickson RP, Erb-Downward JR, and Huffnagle GB. Towards an ecology of the lung: new conceptual models of pulmonary microbiology and pneumonia pathogenesis. Lancet Respir Med. (2014) 2:238–46. doi: 10.1016/S2213-2600(14)70028-1
58. Hauptmann M, and Schaible UE. Linking microbiota and respiratory disease. FEBS Lett. (2016) 590:3721–38. doi: 10.1002/1873-3468.12421
59. Singanayagam A, Ritchie AI, and Johnston SL. Role of microbiome in the pathophysiology and disease course of asthma. Curr Opin Pulm Med. (2017) 23:41–7. doi: 10.1097/MCP.0000000000000333
60. Chung KF. Potential role of the lung microbiome in shaping asthma phenotypes. Ann Am Thorac Soc. (2017) 14:S326–S331. doi: 10.1513/AnnalsATS.201702-138AW
61. Chung KF. Airway microbial dysbiosis in asthmatic patients: a target for prevention and treatment? J Allergy Clin Immunol. (2017) 139:1071–81. doi: 10.1016/j.jaci.2017.02.004
62. Stokholm J, Blaser MJ, Thorsen J, Rasmussen MA, Waage J, Vinding RK, et al. Maturation of the gut microbiome and risk of asthma in childhood. Nat Commun. (2018) 9:141. doi: 10.1038/s41467-017-02573-2
63. Dang AT, and Marsland BJ. Microbes, metabolites, and the gut-lung axis. Mucosal Immunol. (2019) 12:843–50. doi: 10.1038/s41385-019-0160-6
64. Nomura A, Matsubara A, Goto S, Takahata J, Sawada K, Ihara K, et al. Relationship between gut microbiota composition and sensitization to inhaled allergens. Allergol Int. (2020) 69:437–442. doi: 10.1016/j.alit.2019.12.010
65. Schuijt TJ, Lankelma JM, Scicluna BP, de Sousa e Melo F, Roelofs JTH, de Boer JD, et al. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut. (2016) 65:575–583. doi: 10.1136/gutjnl-2015-309728
66. Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA, et al. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS ONE. (2011) 6:e16384. doi: 10.1371/journal.pone.0016384
67. Sze MA, Dimitriu PA, Hayashi S, Elliott WM, McDonough JE, Gosselink JV, et al. The lung tissue microbiome in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. (2012) 185:1073–80. doi: 10.1164/rccm.201111-2075OC
68. Green BJ, Wiriyachaiporn S, Grainge C, Rogers GB, Kehagia V, Lau L, et al. Potentially pathogenic airway bacteria and neutrophilic inflammation in treatment resistant severe asthma. PLoS ONE. (2014) 9:e100645. doi: 10.1371/journal.pone.0100645
69. Bisgaard H, Hermansen MN, Buchvald F, Loland L, Halkjaer LB, Bønnelykke K, et al. Childhood asthma after bacterial colonization of the airway in neonates. N Engl J Med. (2007) 357:1487–95. doi: 10.1056/NEJMoa052632
70. Thorsen J, Rasmussen MA, Waage J, Mortensen M, Brejnrod A, Bønnelykke K, et al. Infant airway microbiota and topical immune perturbations in the origins of childhood asthma. Nat Commun. (2019) 10:5001. doi: 10.1038/s41467-019-12989-7
71. Depner M, Ege MJ, Cox MJ, Dwyer S, Walker AW, Birzele LT, et al. Bacterial microbiota of the upper respiratory tract and childhood asthma. J Allergy Clin Immunol. (2017) 139:826–34 e13. doi: 10.1016/j.jaci.2016.05.050
72. Marsh RL, Kaestli M, Chang AB, Binks MJ, Pope CE, Hoffman LR, et al. The microbiota in bronchoalveolar lavage from young children with chronic lung disease includes taxa present in both the oropharynx and nasopharynx. Microbiome. (2016) 4:37. doi: 10.1186/s40168-016-0182-1
73. Durack J, Huang YJ, Nariya S, Christian LS, Ansel KM, Beigelman A, et al. Bacterial biogeography of adult airways in atopic asthma. Microbiome. (2018) 6:104. doi: 10.1186/s40168-018-0487-3
74. Davis MF, Peng RD, McCormack MC, and Matsui EC. Staphylococcus aureus colonization is associated with wheeze and asthma among US children and young adults. J Allergy Clin Immunol. (2015) 135:811–3 e5. doi: 10.1016/j.jaci.2014.10.052
75. Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. (2015) 17:704–15. doi: 10.1016/j.chom.2015.03.008
76. McCauley K, Durack J, Valladares R, Fadrosh DW, Lin DL, Calatroni A, et al. Distinct nasal airway bacterial microbiotas differentially relate to exacerbation in pediatric patients with asthma. J Allergy Clin Immunol. (2019) 144:1187–97. doi: 10.1016/j.jaci.2019.05.035
77. Perez-Losada M, Alamri L, Crandall KA, and Freishtat RJ. Nasopharyngeal microbiome diversity changes over time in children with asthma. PLoS ONE. (2017) 12:e0170543. doi: 10.1371/journal.pone.0170543
78. Kloepfer KM, Lee WM, Pappas TE, Kang TJ, Vrtis RF, Evans MD, et al. Detection of pathogenic bacteria during rhinovirus infection is associated with increased respiratory symptoms and asthma exacerbations. J Allergy Clin Immunol. (2014) 133:1301–7. doi: 10.1016/j.jaci.2014.02.030
79. Yin SS, Ma FL, and Gao X. Association of Mycoplasma pneumoniae infection with increased risk of asthma in children. Exp Ther Med. (2017) 13:1813–9. doi: 10.3892/etm.2017.4219
80. Simpson JL, Daly J, Baines KJ, Yang IA, Upham JW, Reynolds PN, et al. Airway dysbiosis: Haemophilus influenzae and Tropheryma in poorly controlled asthma. Eur Respir J. (2016) 47:792–800. doi: 10.1183/13993003.00405-2015
81. Huang YJ, Nelson CE, Brodie EL, Desantis TZ, Baek MS, Liu J, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol. (2011) 127:372–81 e1–3. doi: 10.1016/j.jaci.2010.10.048
82. Zhang Q, Illing R, Hui CK, Downey K, Carr D, Stearn M, et al. Bacteria in sputum of stable severe asthma and increased airway wall thickness. Respir Res. (2012) 13:35. doi: 10.1186/1465-9921-13-35
83. Mao B, Yang JW, Lu HW, and Xu JF. Asthma and bronchiectasis exacerbation. Eur Respir J. (2016) 47:1680–6. doi: 10.1183/13993003.01862-2015
84. Marek W, Ozyurt M, Potthast J, and Mensing T. Endotoxin-induced airway hyperresponsiveness in rabbits: contribution of neuropeptides. J Physiol Pharmacol. (2008) 59(Suppl. 6), 421–432. Available online at: http://www.jpp.krakow.pl/journal/archive/12_08_s6/articles/38_article.html
85. Dobbs NA, Zhou X, Pulse M, Hodge LM, Schoeb TR, and Simecka JW. Antigen-pulsed bone marrow-derived and pulmonary dendritic cells promote Th2 cell responses and immunopathology in lungs during the pathogenesis of murine Mycoplasma pneumonia. J Immunol. (2014) 193:1353–63. doi: 10.4049/jimmunol.1301772
86. Eddens T, Campfield BT, Serody K, Manni ML, Horne W, Elsegeiny W, et al. A novel CD4(+) T cell-dependent murine model of pneumocystis-driven asthma-like pathology. Am J Respir Crit Care Med. (2016) 194:807–20. doi: 10.1164/rccm.201511-2205OC
87. Nisar N, Guleria R, Kumar S, Chand CT, and Ranjan BN. Mycoplasma pneumoniae and its role in asthma. Postgrad Med J. (2007) 83:100–4. doi: 10.1136/pgmj.2006.049023
88. Hong SJ. The Role of Mycoplasma pneumoniae Infection in Asthma. Allergy Asthma Immunol Res. (2012) 4:59–61. doi: 10.4168/aair.2012.4.2.59
89. Von HLC. Role of persistent infection in the control and severity of asthma: focus on Chlamydia pneumoniae. Eur Respir J. (2002) 19:546–56. doi: 10.1183/09031936.02.00254402
90. Von HLC, Vasankari T, Liippo K, Wahlström E, and Puolakkainen M. Chlamydia pneumoniae and severity of asthma. Scand J Infect Dis. (2002) 34:22–7. doi: 10.1080/00365540110077155
91. Hahn DL, Schure A, Patel K, Childs T, Drizik E, and Webley W. Chlamydia pneumoniae-specific IgE is prevalent in asthma and is associated with disease severity. PLoS ONE. (2012) 7:e35945. doi: 10.1371/journal.pone.0035945
92. Marsland BJ, and Gollwitzer ES. Host-microorganism interactions in lung diseases. Nat Rev Immunol. (2014) 14:827–35. doi: 10.1038/nri3769
93. Francoise A, and Hery-Arnaud G. The microbiome in cystic fibrosis pulmonary disease. Genes. (2020) 11:536. doi: 10.3390/genes11050536
94. Dunne EM, Smith-Vaughan HC, Robins-Browne RM, Mulholland EK, and Satzke C. Nasopharyngeal microbial interactions in the era of pneumococcal conjugate vaccination. Vaccine. (2013) 31:2333–42. doi: 10.1016/j.vaccine.2013.03.024
95. Skevaki CL, Tsialta P, Trochoutsou AI, Logotheti I, Makrinioti H, Taka S, et al. Associations between viral and bacterial potential pathogens in the nasopharynx of children with and without respiratory symptoms. Pediatr Infect Dis J. (2015) 34:1296–301. doi: 10.1097/INF.0000000000000872
96. Murphy TF, and Parameswaran GI. Moraxella catarrhalis, a human respiratory tract pathogen. Clin Infect Dis. (2009) 49:124–31. doi: 10.1086/599375
97. Sriram KB, Cox AJ, Clancy RL, Slack MPE, and Cripps AW. Nontypeable Haemophilus influenzae and chronic obstructive pulmonary disease: a review for clinicians. Crit Rev Microbiol. (2018) 44:125–42. doi: 10.1080/1040841X.2017.1329274
98. Varghese R, Jayaraman R, and Veeraraghavan B. Current challenges in the accurate identification of Streptococcus pneumoniae and its serogroups/serotypes in the vaccine era. J Microbiol Methods. (2017) 141:48–54. doi: 10.1016/j.mimet.2017.07.015
99. Wertheim HF, Melles DC, Vos MC, van Leeuwen W, van Belkum A, Verbrugh HA, et al. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis. (2005) 5:751–62. doi: 10.1016/S1473-3099(05)70295-4
100. Bogaert D, Keijser B, Huse S, Rossen J, Veenhoven R, van Gils E, et al. Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS ONE. (2011) 6:e17035. doi: 10.1371/journal.pone.0017035
101. Xu Q, Almudervar A, Casey JR, and Pichicheroet ME. Nasopharyngeal bacterial interactions in children. Emerging Infect Dis. (2012) 18:1738–45. doi: 10.3201/eid1811.111904
102. Smith-Vaughan H, Byun R, Nadkarni M, Jacques NA, Hunter N, Halpin S, et al. Measuring nasal bacterial load and its association with otitis media. BMC Ear Nose Throat Disord. (2006) 6:10. doi: 10.1186/1472-6815-6-10
103. Tosi MF. Innate immune responses to infection. J Allergy Clin Immunol. (2005) 116:241–9. doi: 10.1016/j.jaci.2005.05.036
104. Weiser JN, Ferreira DM, and Paton JC. Streptococcus pneumoniae: transmission, colonization and invasion. Nat Rev Microbiol. (2018) 16:355–67. doi: 10.1038/s41579-018-0001-8
105. Ren D, and Pichichero ME. Vaccine targets against Moraxella catarrhalis. Expert Opin Ther Targets. (2016) 20:19–33. doi: 10.1517/14728222.2015.1081686
106. Bootsma HJ, van der Heide HG, van de Pas S, Schouls LM, and Mooi FR. Analysis of Moraxella catarrhalis by DNA typing: evidence for a distinct subpopulation associated with virulence traits. J Infect Dis. (2000) 181:1376–87. doi: 10.1086/315374
107. Brito DA, Ramirez M, and de Lencastre H. Serotyping Streptococcus pneumoniae by multiplex PCR. J Clin Microbiol. (2003) 41:2378–84. doi: 10.1128/JCM.41.6.2378-2384.2003
108. Sutter DE, Summers AM, Keys CE, Taylor KL, Frasch CE, Braun LE, et al. Capsular serotype of Staphylococcus aureus in the era of community-acquired MRSA. FEMS Immunol Med Microbiol. (2011) 63:16–24. doi: 10.1111/j.1574-695X.2011.00822.x
109. Hirst RA, Sikand KS, Rutman A, Mitchell TJ, Andrew PW, and O'Callaghanet C. Relative roles of pneumolysin and hydrogen peroxide from Streptococcus pneumoniae in inhibition of ependymal ciliary beat frequency. Infect Immun. (2000) 68:1557–62. doi: 10.1128/IAI.68.3.1557-1562.2000
110. Brugger SD, Troxler LJ, Rüfenacht S, Frey PM, Morand B, Geyer R, et al. Polysaccharide capsule composition of pneumococcal serotype 19A subtypes is unaltered among subtypes and independent of the nutritional environment. Infect Immun. (2016) 84:3152–60. doi: 10.1128/IAI.00474-16
111. de Vries SP, Bootsma HJ, Hays JP, and Hermans PW. Molecular aspects of Moraxella catarrhalis pathogenesis. Microbiol Mol Biol Rev. (2009) 73:389–406. doi: 10.1128/MMBR.00041-09
112. Post DM, Ketterer MR, Coffin JE, Reinders LM, Munson RSJr, Bair T, et al. Comparative analyses of the lipooligosaccharides from nontypeable Haemophilus influenzae and haemophilus haemolyticus show differences in sialic acid and phosphorylcholine modifications. Infect Immun. (2016) 84:765–74. doi: 10.1128/IAI.01185-15
113. Swords WE, Buscher BA, Ver Steeg Ii K, Preston A, Nichols WA, Weiser JN, et al. Non-typeable Haemophilus influenzae adhere to and invade human bronchial epithelial cells via an interaction of lipooligosaccharide with the PAF receptor. Mol Microbiol. (2000) 37:13–27. doi: 10.1046/j.1365-2958.2000.01952.x
114. Peng D, Hong W, Choudhury BP, Carlson RW, and Gu XX. Moraxella catarrhalis bacterium without endotoxin, a potential vaccine candidate. Infect Immun. (2005) 73:7569–77. doi: 10.1128/IAI.73.11.7569-7577.2005
115. Spaniol V, Heiniger N, Troller R, and Aebi C. Outer membrane protein UspA1 and lipooligosaccharide are involved in invasion of human epithelial cells by Moraxella catarrhalis. Microbes Infect. (2008) 10:3–11. doi: 10.1016/j.micinf.2007.09.014
116. Edwards KJ, Schwingel JM, Datta AK, and Campagnari AA. Multiplex PCR assay that identifies the major lipooligosaccharide serotype expressed by Moraxella catarrhalis clinical isolates. J Clin Microbiol. (2005) 43:6139–43. doi: 10.1128/JCM.43.12.6139-6143.2005
117. Verhaegh SJC, Streefland A, Dewnarain JK, Farrell DJ, van Belkum A, and Hays JP. Age-related genotypic and phenotypic differences in Moraxella catarrhalis isolates from children and adults presenting with respiratory disease in 2001-2002. Microbiology. (2008) 154:1178–84. doi: 10.1099/mic.0.2007/015057-0
118. Mitov IG, Gergova RT, and Ouzounova-Raykova VV. Distribution of genes encoding virulence factors ompB2, ompCD, ompE, beta-lactamase and serotype in pathogenic and colonizing strains of Moraxella catarrhalis. Arch Med Res. (2010) 41:530–5. doi: 10.1016/j.arcmed.2010.10.003
119. Jalalvand F, and Riesbeck K. Update on non-typeable Haemophilus influenzae-mediated disease and vaccine development. Expert Rev Vaccines. (2018) 17:503–12. doi: 10.1080/14760584.2018.1484286
120. Delfino Sosa M, Zabala C, Pardo L, Fernández L, Nieves C, Más M, et al. Haemophilus influenzae type b invasive infections in children hospitalized between 2000 and 2017 in a Pediatric Reference Hospital (PRH). Heliyon. (2020) 6:e03483. doi: 10.1016/j.heliyon.2020.e03483
121. Wen S, Feng D, Chen D, Yang L, and Xu Z. Molecular epidemiology and evolution of Haemophilus influenzae. Infect Genet Evol. (2020) 80:104205. doi: 10.1016/j.meegid.2020.104205
122. Pizarro-Cerda J, and Cossart P. Bacterial adhesion and entry into host cells. Cell. (2006) 124:715–27. doi: 10.1016/j.cell.2006.02.012
123. Heiniger RW, Winther-Larsen HC, Pickles RJ, Koomey M, and Wolfgang MC. Infection of human mucosal tissue by Pseudomonas aeruginosa requires sequential and mutually dependent virulence factors and a novel pilus-associated adhesin. Cell Microbiol. (2010) 12:1158–73. doi: 10.1111/j.1462-5822.2010.01461.x
124. Letourneau J, Levesque C, Berthiaume F, Jacques M, and Mourez M. In vitro assay of bacterial adhesion onto mammalian epithelial cells. J Vis Exp. (2011) 51:2783. doi: 10.3791/2783
125. Tan TT, Nordström T, Forsgren A, and Riesbeck K. The respiratory pathogen Moraxella catarrhalis adheres to epithelial cells by interacting with fibronectin through ubiquitous surface proteins A1 and A2. J Infect Dis. (2005) 192:1029–38. doi: 10.1086/432759
126. Slevogt H, Seybold J, Tiwari KN, Hocke AC, Jonatat C, Dietel S, et al. Moraxella catarrhalis is internalized in respiratory epithelial cells by a trigger-like mechanism and initiates a TLR2- and partly NOD1-dependent inflammatory immune response. Cell Microbiol. (2007) 9:694–707. doi: 10.1111/j.1462-5822.2006.00821.x
127. Iosifidis T, Sutanto EN, Buckley AG, Coleman L, Gill EE, Lee AH, et al. Aberrant cell migration contributes to defective airway epithelial repair in childhood wheeze. JCI Insight. (2020) 5:e133125. doi: 10.1172/jci.insight.133125
128. Khoo SK, Read J, Franks K, Zhang G, Bizzintino J, Coleman L, et al. Upper airway cell transcriptomics identify a major new immunological phenotype with strong clinical correlates in young children with acute wheezing. J Immunol. (2019) 202:1845–58. doi: 10.4049/jimmunol.1800178
129. Luton F, Vergés M, Vaerman JP, Sudol M, and Mostov KE. The SRC family protein tyrosine kinase p62yes controls polymeric IgA transcytosis in vivo. Mol Cell. (1999) 4:627–32. doi: 10.1016/S1097-2765(00)80213-0
130. Zhang JR, Mostov KE, Lamm ME, Nanno N, Shimida S, Ohwaki M, et al. The polymeric immunoglobulin receptor translocates pneumococci across human nasopharyngeal epithelial cells. Cell. (2000) 102:827–37. doi: 10.1016/S0092-8674(00)00071-4
131. Kietzman CC, Gao G, Mann B, Myers L, and Tuomanen EI. Dynamic capsule restructuring by the main pneumococcal autolysin LytA in response to the epithelium. Nat Commun. (2016) 7:10859. doi: 10.1038/ncomms10859
132. Loughran AJ, Orihuela CJ, and Tuomanen EI. Streptococcus pneumoniae: invasion and inflammation. Microbiol Spectr. (2019) 7:1–2. doi: 10.1128/microbiolspec.GPP3-0004-2018
133. Canvin JR, Marvin AP, Sivakumaran M, Paton JC, Boulnois GJ, Andrew PW, et al. The role of pneumolysin and autolysin in the pathology of pneumonia and septicemia in mice infected with a type 2 pneumococcus. J Infect Dis. (1995) 172:119–23. doi: 10.1093/infdis/172.1.119
134. Spellerberg B, Cundell DR, Sandros J, Pearce BJ, Idanpaan-Heikkila I, Rosenow C, et al. Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol Microbiol. (1996) 19:803–13. doi: 10.1046/j.1365-2958.1996.425954.x
135. Berry AM, and Paton JC. Additive attenuation of virulence of Streptococcus pneumoniae by mutation of the genes encoding pneumolysin and other putative pneumococcal virulence proteins. Infect Immun. (2000) 68:133–40. doi: 10.1128/IAI.68.1.133-140.2000
136. Shuter J, Hatcher VB, and Lowy FD. Staphylococcus aureus binding to human nasal mucin. Infect Immun. (1996) 64:310–8. doi: 10.1128/IAI.64.1.310-318.1996
137. Weidenmaier C, Kokai-Kun JF, Kulauzovic E, Kohler T, Thumm G, Stoll H, et al. Differential roles of sortase-anchored surface proteins and wall teichoic acid in Staphylococcus aureus nasal colonization. Int J Med Microbiol. (2008) 298:505–13. doi: 10.1016/j.ijmm.2007.11.006
138. O'Brien LM, O'Brien LM, Walsh EJ, Massey RC, Peacock SJ, and Foster TJ. Staphylococcus aureus clumping factor B (ClfB) promotes adherence to human type I cytokeratin 10: implications for nasal colonization. Cell Microbiol. (2002) 4:759–70. doi: 10.1046/j.1462-5822.2002.00231.x
139. Roche FM, Meehan M, and Foster TJ. The Staphylococcus aureus surface protein SasG and its homologues promote bacterial adherence to human desquamated nasal epithelial cells. Microbiology. (2003) 149:2759–67. doi: 10.1099/mic.0.26412-0
140. Bibel DJ, Aly R, Shinefield HR, Maibach HI, and Strauss WG. Importance of the keratinized epithelial cell in bacterial adherence. J Invest Dermatol. (1982) 79:250–3. doi: 10.1111/1523-1747.ep12500072
141. Patti JM, Allen BL, McGavin MJ, and Höök M. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu Rev Microbiol. (1994) 48:585–617. doi: 10.1146/annurev.mi.48.100194.003101
142. Kluytmans J, van Belkum A, and Verbrugh H. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev. (1997) 10:505–20. doi: 10.1128/CMR.10.3.505
143. Bogaert D, van Belkum A, Sluijter M, Luijendijk A, de Groot R, Rümke HC, et al. Colonisation by Streptococcus pneumoniae and Staphylococcus aureus in healthy children. Lancet. (2004) 363:1871–2. doi: 10.1016/S0140-6736(04)16357-5
144. Regev-Yochay G, Dagan R, Raz M, Carmeli Y, Shainberg B, Derazne E, et al. Association between carriage of Streptococcus pneumoniae and Staphylococcus aureus in Children. JAMA. (2004) 292:716–20. doi: 10.1001/jama.292.6.716
145. Kuo CY, Hwang KP, Hsieh YC, Cheng CH, Huang FL, Shen YH, et al. Nasopharyngeal carriage of Streptococcus pneumoniae in Taiwan before and after the introduction of a conjugate vaccine. Vaccine. (2011) 29:5171–7. doi: 10.1016/j.vaccine.2011.05.034
146. Kwambana BA, Barer MR, Bottomley C, Adegbola RA, and Antonio M. Early acquisition and high nasopharyngeal co-colonisation by Streptococcus pneumoniae and three respiratory pathogens amongst Gambian new-borns and infants. BMC Infect Dis. (2011) 11:175. doi: 10.1186/1471-2334-11-175
147. Quintero B, Araque M, van der Gaast-de Jongh C, Escalona F, Correa M, Morillo-Puente S, et al. Epidemiology of Streptococcus pneumoniae and Staphylococcus aureus colonization in healthy Venezuelan children. Eur J Clin Microbiol Infect Dis. (2011) 30:7–19. doi: 10.1007/s10096-010-1044-6
148. Bae S, Yu JY, Lee K, Lee S, Park B, and Kang Y. Nasal colonization by four potential respiratory bacteria in healthy children attending kindergarten or elementary school in Seoul, Korea. J Med Microbiol. (2012) 61:678–85. doi: 10.1099/jmm.0.040584-0
149. Dukers-Muijrers NH, Stobberingh E, Beisser P, Boesten RC, Jacobs P, and Hoebe CJ. Nasal carriage of Streptococcus pneumoniae serotypes and Staphylococcus aureus in Streptococcus pneumoniae-vaccinated and non-vaccinated young children. Epidemiol Infect. (2013) 141:631–8. doi: 10.1017/S095026881200115X
150. Lewnard JA, Huppert A, Givon-Lavi N, Pettigrew MM, Regev-Yochay G, Dagan R, et al. Density, serotype diversity, and fitness of Streptococcus pneumoniae in upper respiratory tract cocolonization with nontypeable Haemophilus influenzae. J Infect Dis. (2016) 214:1411–20. doi: 10.1093/infdis/jiw381
151. Dunne EM, Murad C, Sudigdoadi S, Fadlyana E, Tarigan R, Indriyani SAK, et al. Carriage of Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis, and Staphylococcus aureus in Indonesian children: a cross-sectional study. PLoS ONE. (2018) 13:e0195098. doi: 10.1371/journal.pone.0195098
152. McMichael JC, and Green BA. Vaccines for Moraxella catarrhalis and non-typeable Haemophilus influenzae. Curr Opin Investig Drugs. (2003) 4:953–8. Available online at: https://europepmc.org/article/med/14508879
153. Vuononvirta J, Toivonen L, Gröndahl-Yli-Hannuksela K, Barkoff AM, Lindholm L, Mertsola J, et al. Nasopharyngeal bacterial colonization and gene polymorphisms of mannose-binding lectin and toll-like receptors 2 and 4 in infants. PLoS ONE. (2011) 6:e26198. doi: 10.1371/journal.pone.0026198
154. Peltola VT, and McCullers JA. Respiratory viruses predisposing to bacterial infections: role of neuraminidase. Pediatr Infect Dis J. (2004) 23:S87–97. doi: 10.1097/01.inf.0000108197.81270.35
155. McCullers JA, and Bartmess KC. Role of neuraminidase in lethal synergism between influenza virus and Streptococcus pneumoniae. J Infect Dis. (2003) 187:1000–9. doi: 10.1086/368163
156. Peltola VT, Murti KG, and McCullers JA. Influenza virus neuraminidase contributes to secondary bacterial pneumonia. J Infect Dis. (2005) 192:249–57. doi: 10.1086/430954
157. Nakamura S, Davis KM, and Weiser JN. Synergistic stimulation of type I interferons during influenza virus coinfection promotes Streptococcus pneumoniae colonization in mice. J Clin Invest. (2011) 121:3657–65. doi: 10.1172/JCI57762
158. Lysenko ES, Ratner AJ, Nelson AL, and Weiser JN. The role of innate immune responses in the outcome of interspecies competition for colonization of mucosal surfaces. PLoS Pathog. (2005) 1:e1. doi: 10.1371/journal.ppat.0010001
159. Lina G, Boutite F, Tristan A, Bes M, Etienne J, and Vandenesch F. Bacterial competition for human nasal cavity colonization: role of Staphylococcal agr alleles. Appl Environ Microbiol. (2003) 69:18–23. doi: 10.1128/AEM.69.1.18-23.2003
160. Feil EJ, Smith JM, Enright MC, and Spratt BG. Estimating recombinational parameters in Streptococcus pneumoniae from multilocus sequence typing data. Genetics. (2000) 154:1439–50. Available online at: https://europepmc.org/article/med/10747043
161. Peacock SJ, Justice A, Griffiths D, de Silva GD, Kantzanou MN, Crook D, et al. Determinants of acquisition and carriage of Staphylococcus aureus in infancy. J Clin Microbiol. (2003) 41:5718–25. doi: 10.1128/JCM.41.12.5718-5725.2003
162. Megremis S, Constantinides B, Xepapadaki P, Bachert C, Neurath-Finotto S, Jartti P, et al. Bacteriophage deficiency characterizes respiratory virome dysbiosis in childhood asthma. BioRxiv [preprint]. (2020) 1:1–46. doi: 10.1101/2020.08.04.236067
163. Koskella B, and Brockhurst MA. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol Rev. (2014) 38:916–31. doi: 10.1111/1574-6976.12072
164. Hays J. Mobile genetic elements in Moraxella catarrhalis. Mob Genet Elements. (2011) 1:155–8. doi: 10.4161/mge.1.2.17632
165. Verhaegh SJC, Snippe ML, Levy F, Verbrugh HA, Jaddoe VWV, Hofman A, et al. Colonization of healthy children by Moraxella catarrhalis is characterized by genotype heterogeneity, virulence gene diversity and co-colonization with Haemophilus influenzae. Microbiology. (2011) 157:169–78. doi: 10.1099/mic.0.042929-0
166. Hay ID, and Lithgow T. Filamentous phages: masters of a microbial sharing economy. EMBO Rep. (2019) 20:e47427.167. doi: 10.15252/embr.201847427
167. Davie JJ, Earl J, de Vries SP, Ahmed A, Hu FZ, Bootsma HJ, et al. Comparative analysis and supragenome modeling of twelve Moraxella catarrhalis clinical isolates. BMC Genomics. (2011) 12:70. doi: 10.1186/1471-2164-12-70
168. Ariff A, Wise MJ, Kahler CM, Tay CY, Peters F, Perkins TT, et al. Novel Moraxella catarrhalis prophages display hyperconserved non-structural genes despite their genomic diversity. BMC Genomics. (2015) 16:860. doi: 10.1186/s12864-015-2104-1
169. Sorek R, Kunin V, and Hugenholtz P. CRISPR–a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. (2008) 6:181–6. doi: 10.1038/nrmicro1793
170. Zomer A, de Vries SP, Riesbeck K, Meinke AL, Hermans PW, and Bootsma HJ. Genome sequence of Moraxella catarrhalis RH4, an isolate of seroresistant lineage. J Bacteriol. (2012) 194:6969. doi: 10.1128/JB.01833-12
171. Dedrick RM, Jacobs-Sera D, Bustamante CA, Garlena RA, Mavrich TN, Pope WH, et al. Prophage-mediated defence against viral attack and viral counter-defence. Nat Microbiol. (2017) 2:16251. doi: 10.1038/nmicrobiol.2016.251
172. Samuels J, and Clarke JK. New bacteriophage of Haemophilus influenzae. J Virol. (1969) 4:797–8. doi: 10.1128/JVI.4.5.797-798.1969
173. Boling ME, Setlow JK, and Allison DP. Bacteriophage of Haemophilus influenzae. I Differences between infection by whole phage, extracted phage DNA and prophage DNA extracted from lysogenic cells. J Mol Biol. (1972) 63:335–48. doi: 10.1016/0022-2836(72)90431-7
174. Boling ME, Allison DP, and Setlow JK. Bacteriophage of Haemophilus influenzae. 3. Morphology, DNA homology, and immunity properties of HPlcl, S2, and the defective bacteriophage from strain Rd. Jirol. (1973) 11:585–91. doi: 10.1128/JVI.11.4.585-591.1973
175. Hwang ES, and Scocca JJ. Interaction of integration host factor from Escherichia coli with the integration region of the Haemophilus influenzae bacteriophage HP1. J Bacteriol. (1990) 172:4852–60. doi: 10.1128/JB.172.9.4852-4860.1990
176. Williams BJ, Golomb M, Phillips T, Brownlee J, Olson MV, and Smith AL. Bacteriophage HP2 of Haemophilus influenzae. J Bacteriol. (2002) 184:6893–905. doi: 10.1128/JB.184.24.6893-6905.2002
177. Gatea Kaabi SA, and Musafer HK. New Phage cocktail against infantile Sepsis bacteria. Microb Pathog. (2020) 148:104447. doi: 10.1016/j.micpath.2020.104447
178. Mortensen MS, Brejnrod AD, Roggenbuck M, Abu Al-Soud W, Balle C, Krogfelt KA, et al. The developing hypopharyngeal microbiota in early life. Microbiome. (2016) 4:70. doi: 10.1186/s40168-016-0215-9
179. Pastacaldi C, Lewis P, and Howarth P. Staphylococci and staphylococcal superantigens in asthma and rhinitis: a systematic review and meta-analysis. Allergy. (2011) 66:549–55. doi: 10.1111/j.1398-9995.2010.02502.x
180. Monk AB, Rees CD, Barrow P, Hagens S, and Harper DR. Bacteriophage applications: where are we now? Lett Appl Microbiol. (2010) 51:363–9. doi: 10.1111/j.1472-765X.2010.02916.x
181. Seth AK, Geringer MR, Nguyen KT, Agnew SP, Dumanian Z, Galiano RD, et al. Bacteriophage therapy for Staphylococcus aureus biofilm-infected wounds: a new approach to chronic wound care. Plast Reconstr Surg. (2013) 131:225–34. doi: 10.1097/PRS.0b013e31827e47cd
182. Vandersteegen K, Mattheus W, Ceyssens PJ, Bilocq F, De Vos D, Pirnay JP, et al. Microbiological and molecular assessment of bacteriophage ISP for the control of Staphylococcus aureus. PLoS ONE. (2011) 6:e24418. doi: 10.1371/journal.pone.0024418
183. Vandersteegen K, Kropinski AM, Nash JH, Noben JP, Hermans K, and Lavigne R. Romulus and Remus, two phage isolates representing a distinct clade within the Twortlikevirus genus, display suitable properties for phage therapy applications. J Virol. (2013) 87:3237–47. doi: 10.1128/JVI.02763-12
184. Lopez R, Garcia E, and Ronda C. Bacteriophages of Streptococcus pneumoniae. Rev Infect Dis. (1981) 3:212–23. doi: 10.1093/clinids/3.2.212
185. Gilmer DB, Schmitz JE, Euler CW, and Fischetti VA. Novel bacteriophage lysin with broad lytic activity protects against mixed infection by Streptococcus pyogenes and methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. (2013) 57:2743–50. doi: 10.1128/AAC.02526-12
186. Vazquez R, Garcia E, and Garcia P. Phage lysins for fighting bacterial respiratory infections: a new generation of antimicrobials. Front Immunol. (2018) 9:2252. doi: 10.3389/fimmu.2018.02252
187. Fischetti VA. Surface proteins on gram-positive bacteria. Microbiol Spectr. (2019) 7:1–9. doi: 10.1128/microbiolspec.GPP3-0012-2018
188. Pride DT, Salzman J, Haynes M, Rohwer F, Davis-Long C, White RA, et al. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J. (2012) 6:915–26. doi: 10.1038/ismej.2011.169
189. Santiago-Rodriguez TM, Naidu M, Abeles SR, Boehm TK, Ly M, and Pride DT. Transcriptome analysis of bacteriophage communities in periodontal health and disease. BMC Genomics. (2015) 16:549. doi: 10.1186/s12864-015-1781-0
190. Willner D, Furlan M, Haynes M, Schmieder R, Angly FE, Silva J, et al. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS ONE. (2009) 4:e7370. doi: 10.1371/journal.pone.0007370
191. Lim YW, Schmieder R, Haynes M, Furlan M, Matthews TD, and Whiteson K. Mechanistic model of Rothia mucilaginosa adaptation toward persistence in the CF lung, based on a genome reconstructed from metagenomic data. PLoS ONE. (2013) 8:e64285. doi: 10.1371/journal.pone.0064285
192. Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. (2010) 466:334–8. doi: 10.1038/nature09199
193. Edwards RA, Vega AA, Norman HM, Ohaeri M, Levi K, Dinsdale EA, et al. Global phylogeography and ancient evolution of the widespread human gut virus crAssphage. Nat Microbiol. (2019) 4:1727–36. doi: 10.1038/s41564-019-0494-6
194. Santiago-Rodriguez TM, Ly M, Bonilla N, and Pride DT. The human urine virome in association with urinary tract infections. Front Microbial. (2015) 6:14. doi: 10.3389/fmicb.2015.00014
195. Foulongne V, Sauvage V, Hebert C, Dereure O, Cheval J, and Gouilh MA. Human skin microbiota: high diversity of DNA viruses identified on the human skin by high throughput sequencing. PLoS ONE. (2012) 7:e38499. doi: 10.1371/journal.pone.0038499
196. Oh J, Byrd AL, Deming C, Conlan S, Kong HH, Segre JA, et al. Biogeography and individuality shape function in the human skin metagenome. Nature. (2014) 514:59–64. doi: 10.1038/nature13786
197. Moustafa A, Xie C, Kirkness E, Biggs W, Wong E, Turpaz Y, et al. The blood DNA virome in 8,000 humans. PLoS Pathog. (2017) 13:e1006292. doi: 10.1371/journal.ppat.1006292
198. Jankauskaite L, Misevičiene V, Vaideliene L, and Kevalas R. Lower airway virology in health and disease-from invaders to symbionts. Medicina. (2018) 54, 1–10. doi: 10.3390/medicina54050072
199. Federici S, Nobs SP, and Elinav E. Phages and their potential to modulate the microbiome and immunity. Cell Mol Immunol. (2020) 1:1–12. doi: 10.1038/s41423-020-00532-4
200. Sinha A, and Maurice CF. Bacteriophages: uncharacterized and dynamic regulators of the immune system. Mediators Inflamm. (2019) 2019:3730519. doi: 10.1155/2019/3730519
201. Barr JJ, Auro R, Furlan M, Whiteson KL, Erb ML, Pogliano J, et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc Natl Acad Sci USA. (2013) 110:10771–6. doi: 10.1073/pnas.1305923110
202. Meyer JR. Sticky bacteriophage protect animal cells. Proc Natl Acad Sci USA. (2013) 110:10475–6. doi: 10.1073/pnas.1307782110
203. Barr JJ, Auro R, Sam-Soon N, Kassegne S, Peters G, Bonilla N, et al. Subdiffusive motion of bacteriophage in mucosal surfaces increases the frequency of bacterial encounters. Proc Natl Acad Sci USA. (2015) 112:13675–80. doi: 10.1073/pnas.1508355112
204. Halaby DM, and Mornon JP. The immunoglobulin superfamily: an insight on its tissular, species, and functional diversity. J Mol Evol. (1998) 46:389–400. doi: 10.1007/PL00006318
205. Fraser JS, Yu Z, Maxwell KL, and Davidson AR. Ig-like domains on bacteriophages: a tale of promiscuity and deceit. J Mol Biol. (2006) 359:496–507. doi: 10.1016/j.jmb.2006.03.043
206. Fraser JS, Maxwell KL, and Davidson AR. Immunoglobulin-like domains on bacteriophage: weapons of modest damage? Curr Opin Microbiol. (2007) 10:382–7. doi: 10.1016/j.mib.2007.05.018
207. Duerkop BA, and Hooper LV. Resident viruses and their interactions with the immune system. Nat Immunol. (2013) 14:654–9. doi: 10.1038/ni.2614
208. Park K, Cha KE, and Myung H. Observation of inflammatory responses in mice orally fed with bacteriophage T7. J Appl Microbiol. (2014) 117:627–33. doi: 10.1111/jam.12565
209. Lehti TA, Pajunen MI, Skog MS, and Finne J. Internalization of a polysialic acid-binding Escherichia coli bacteriophage into eukaryotic neuroblastoma cells. Nat Commun. (2017) 8:1915. doi: 10.1038/s41467-017-02057-3
210. Bodner K, Melkonian AL, and Covert MW. The enemy of my enemy: new insights regarding bacteriophage-mammalian cell interactions. Trends Microbiol. (2020) 1:349–356. doi: 10.1016/j.tim.2020.10.014
211. Secundino I, Lizcano A, Roupé KM, Wang X, Cole JN, Olson J, et al. Host and pathogen hyaluronan signal through human siglec-9 to suppress neutrophil activation. J Mol Med. (2016) 94:219–33. doi: 10.1007/s00109-015-1341-8
212. Carlin AF, Uchiyama S, Chang YC, Lewis AL, Nizet V, and Varki A. Molecular mimicry of host sialylated glycans allows a bacterial pathogen to engage neutrophil Siglec-9 and dampen the innate immune response. Blood. (2009) 113:3333–6. doi: 10.1182/blood-2008-11-187302
213. Tsai CM. Molecular mimicry of host structures by lipooligosaccharides of Neisseria meningitidis: characterization of sialylated and nonsialylated lacto-N-neotetraose (Galbeta1-4GlcNAcbeta1-3Galbeta1-4Glc) structures in lipooligosaccharides using monoclonal antibodies and specific lectins. Adv Exp Med Biol. (2001) 491:525–42. doi: 10.1007/978-1-4615-1267-7_35
214. Aalto J, Pelkonen S, Kalimo H, and Finne J. Mutant bacteriophage with non-catalytic endosialidase binds to both bacterial and eukaryotic polysialic acid and can be used as probe for its detection. Glycoconj J. (2001) 18:751–8. doi: 10.1023/A:1021147316647
215. Dabrowska K, Switała-Jelen K, Opolski A, Weber-Dabrowska B, and Gorski A. Bacteriophage penetration in vertebrates. J Appl Microbiol. (2005) 98:7–13. doi: 10.1111/j.1365-2672.2004.02422.x
216. Nguyen S, Baker K, Padman BS, Patwa R, Dunstan RA, Weston TA, et al. Bacteriophage transcytosis provides a mechanism to cross epithelial cell layers. mBio. (2017) 8:e01874–17. doi: 10.1128/mBio.01874-17
217. Tam JC, and Jacques DA. Intracellular immunity: finding the enemy within–how cells recognize and respond to intracellular pathogens. J Leukoc Biol. (2014) 96:233–44. doi: 10.1189/jlb.4RI0214-090R
218. Janeway CA Jr. Pillars article: approaching the asymptote? Evolution and revolution in immunology. Cold spring harb symp quant biol. 1989. 54:1-13. J Immunol. (2013) 191:4475–87. doi: 10.1101/SQB.1989.054.01.003
219. Xu J, and Xiang Y. Membrane penetration by bacterial viruses. J Virol. (2017) 91:e00162–17. doi: 10.1128/JVI.00162-17
220. Van Belleghem JD, Dabrowska K, Vaneechoutte M, Barr JJ, and Bollyky PL. Interactions between bacteriophage, bacteria, and the mammalian immune system. Viruses. (2018) 11:1–15. doi: 10.3390/v11010010
221. Frenkel D, and Solomon B. Filamentous phage as vector-mediated antibody delivery to the brain. Proc Natl Acad Sci USA. (2002) 99:5675–9. doi: 10.1073/pnas.072027199
222. Zhang L, Sun L, Wei R, Gao Q, He T, Xu C, et al. Intracellular Staphylococcus aureus control by virulent bacteriophages within MAC-T bovine mammary epithelial cells. Antimicrob Agents Chemother. (2017) 61:e01990–16. doi: 10.1128/AAC.01990-16
223. Zhang L, Hou X, Sun L, He T, Wei R, Pang M, et al. Staphylococcus aureus bacteriophage suppresses LPS-induced inflammation in MAC-T bovine mammary epithelial cells. Front Microbiol. (2018) 9:1614. doi: 10.3389/fmicb.2018.02511
224. Willner D, Haynes MR, Furlan M, Hanson N, Kirby B, Lim YW, et al. Case studies of the spatial heterogeneity of DNA viruses in the cystic fibrosis lung. Am J Respir Cell Mol Biol. (2012) 46:127–31. doi: 10.1165/rcmb.2011-0253OC
225. Trend S, Chang BJ, O'Dea M, Stick SM, Kicic A, and WAERP; AusREC; AREST C.F. Use of a primary epithelial cell screening tool to investigate phage therapy in cystic fibrosis. Front Pharmacol. (2018) 9:1330. doi: 10.3389/fphar.2018.01330
226. Secor PR, Sweere JM, Michaels LA, Malkovskiy AV, Lazzareschi D, Katznelson E, et al. Filamentous bacteriophage promote biofilm assembly and function. Cell Host Microbe. (2015) 18:549–59. doi: 10.1016/j.chom.2015.10.013
227. Secor PR, Michaels LA, Smigiel KS, Rohani MG, Jennings LK, Hisert KB, et al. Filamentous bacteriophage produced by Pseudomonas aeruginosa alters the inflammatory response and promotes noninvasive infection in vivo. Infect Immun. (2017) 85:e00648–16. doi: 10.1128/IAI.00648-16
228. Broudy TB, Pancholi V, and Fischetti VA. Induction of lysogenic bacteriophage and phage-associated toxin from group a streptococci during coculture with human pharyngeal cells. Infect Immun. (2001) 69:1440–3. doi: 10.1128/IAI.69.3.1440-1443.2001
229. Fischetti VA. In vivo acquisition of prophage in Streptococcus pyogenes. Trends Microbiol. (2007) 15:297–300. doi: 10.1016/j.tim.2007.05.003
230. Ramirez M, Severina E, and Tomasz A. A high incidence of prophage carriage among natural isolates of Streptococcus pneumoniae. J Bacteriol. (1999) 181:3618–25. doi: 10.1128/JB.181.12.3618-3625.1999
231. DeBardeleben HK, Lysenko ES, Dalia AB, and Weiser JN. Tolerance of a phage element by Streptococcus pneumoniae leads to a fitness defect during colonization. J Bacteriol. (2014) 196:2670–80. doi: 10.1128/JB.01556-14
232. Carrolo M, Frias MJ, Pinto FR, Melo-Cristino J, and Ramirez M. Prophage spontaneous activation promotes DNA release enhancing biofilm formation in Streptococcus pneumoniae. PLoS ONE. (2010) 5:e15678. doi: 10.1371/journal.pone.0015678
233. Bille E, Meyer J, Jamet A, Euphrasie D, Barnier JP, Brissac T, et al. A virulence-associated filamentous bacteriophage of Neisseria meningitidis increases host-cell colonisation. PLoS Pathog. (2017) 13:e1006495. doi: 10.1371/journal.ppat.1006495
234. Shan J, Ramachandran A, Thanki AM, Vukusic FBI, Barylski J, and Clokie MRJ. Bacteriophages are more virulent to bacteria with human cells than they are in bacterial culture; insights from HT-29 cells. Sci Rep. (2018) 8:5091. doi: 10.1038/s41598-018-23418-y
235. Regev-Yochay G, Trzcinski K, Thompson CM, Malley R, and Lipsitch M. Interference between Streptococcus pneumoniae and Staphylococcus aureus: in vitro hydrogen peroxide-mediated killing by Streptococcus pneumoniae. J Bacteriol. (2006) 188:4996–5001. doi: 10.1128/JB.00317-06
236. Selva L, Viana D, Regev-Yochay G, Trzcinski K, Corpa JM, Lasa I, et al. Killing niche competitors by remote-control bacteriophage induction. Proc Natl Acad Sci USA. (2009) 106:1234–8. doi: 10.1073/pnas.0809600106
237. Mitchell AB, and Glanville AR. Introduction to techniques and methodologies for characterizing the human respiratory virome. Methods Mol Biol. (2018) 1838:111–23. doi: 10.1007/978-1-4939-8682-8_9
238. Willner D, and Furlan M. Deciphering the role of phage in the cystic fibrosis airway. Virulence. (2010) 1:309–13. doi: 10.4161/viru.1.4.12071
239. Fancello L, Desnues C, Raoult D, and Rolain JM. Bacteriophages and diffusion of genes encoding antimicrobial resistance in cystic fibrosis sputum microbiota. J Antimicrob Chemother. (2011) 66:2448–54. doi: 10.1093/jac/dkr315
240. D'Anna SE, Balbi B, Cappello F, Carone M, and Di Stefano A. Bacterial-viral load and the immune response in stable and exacerbated COPD: significance and therapeutic prospects. Int J Chron Obstruct Pulmon Dis. (2016) 11:445–53. doi: 10.2147/COPD.S93398
241. Davies EV, James CE, Kukavica-Ibrulj I, Levesque RC, Brockhurst MA, and Winstanley C. Temperate phages enhance pathogen fitness in chronic lung infection. ISME J. (2016) 10:2553–5. doi: 10.1038/ismej.2016.51
242. Leung CYJ, and Weitz JS. Modeling the synergistic elimination of bacteria by phage and the innate immune system. J Theor Biol. (2017) 429:241–52. doi: 10.1016/j.jtbi.2017.06.037
243. Roach DR, Leung CY, Henry M, Morello E, Singh D, Di Santo JP, et al. Synergy between the host immune system and bacteriophage is essential for successful phage therapy against an acute respiratory pathogen. Cell Host Microbe. (2017) 22:38–47 e4. doi: 10.1016/j.chom.2017.06.018
244. Joiner KL, DeJonckheere M, Whittemore R, and Grey M. Impact of bacteria motility in the encounter rates with bacteriophage in mucus. Sci Rep. (2019) 9:16427. doi: 10.1038/s41598-019-52794-2
245. Abedon ST. Ecology of anti-biofilm agents I: antibiotics versus bacteriophages. Pharmaceuticals. (2015) 8:525–58. doi: 10.3390/ph8030525
246. Abedon ST. Phage therapy of pulmonary infections. Bacteriophage. (2015) 5:e1020260. doi: 10.1080/21597081.2015.1020260
247. Weiss M, Denou E, Bruttin A, Serra-Moreno R, Dillmann ML, and Brüssow H. In vivo replication of T4 and T7 bacteriophages in germ-free mice colonized with Escherichia coli. Virology. (2009) 393:16–23. doi: 10.1016/j.virol.2009.07.020
248. Reyes A, Wu M, McNulty NP, Rohwer FL, and Gordon JI. Gnotobiotic mouse model of phage-bacterial host dynamics in the human gut. Proc Natl Acad Sci USA. (2013) 110:20236–41. doi: 10.1073/pnas.1319470110
Keywords: bacteria, asthma, bacteriophages, airway mucosa, tripartite symbiosis
Citation: Tzani-Tzanopoulou P, Skliros D, Megremis S, Xepapadaki P, Andreakos E, Chanishvili N, Flemetakis E, Kaltsas G, Taka S, Lebessi E, Doudoulakakis A and Papadopoulos NG (2021) Interactions of Bacteriophages and Bacteria at the Airway Mucosa: New Insights Into the Pathophysiology of Asthma. Front. Allergy 1:617240. doi: 10.3389/falgy.2020.617240
Received: 14 October 2020; Accepted: 29 December 2020;
Published: 26 January 2021.
Edited by:
Luminita Aurelia Stanciu, Imperial College London, United KingdomReviewed by:
Lena Uller, Lund University, SwedenAnthony Kicic, University of Western Australia, Australia
Copyright © 2021 Tzani-Tzanopoulou, Skliros, Megremis, Xepapadaki, Andreakos, Chanishvili, Flemetakis, Kaltsas, Taka, Lebessi, Doudoulakakis and Papadopoulos. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Panagiota Tzani-Tzanopoulou, dHphbml0ekBtZWQudW9hLmdy
 Panagiota Tzani-Tzanopoulou
Panagiota Tzani-Tzanopoulou Dimitrios Skliros
Dimitrios Skliros Spyridon Megremis
Spyridon Megremis Paraskevi Xepapadaki
Paraskevi Xepapadaki Evangelos Andreakos
Evangelos Andreakos Nina Chanishvili
Nina Chanishvili Emmanouil Flemetakis
Emmanouil Flemetakis Grigoris Kaltsas
Grigoris Kaltsas Styliani Taka1
Styliani Taka1 Anastassios Doudoulakakis
Anastassios Doudoulakakis Nikolaos G. Papadopoulos
Nikolaos G. Papadopoulos