- 1Penn Vet Working Dog Center, Clinical Sciences and Advanced Medicine, School of Veterinary Medicine, University of Pennsylvania, Philadelphia, PA, United States
- 2Department of Orthopaedics, Geisel School of Medicine at Dartmouth, Lebanon, NH, United States
- 3Center for One Health Research Edward Via College of Osteopathic Medicine, Blacksburg, VA, United States
- 4Rothman Orthopaedic Institute, Philadelphia, PA, United States
- 5Department of Orthopaedics, Harvard Medical School, Brigham and Women's Hospital, Harvard University, Boston, MA, United States
- 6Department of Clinical Studies New Bolton Center, School of Veterinary Medicine, University of Pennsylvania, Kennett Square, PA, United States
Introduction: The study investigated the utilization of odor detection dogs to identify the odor profile of Staphylococcus aureus (S. aureus) biofilms in pure in vitro samples and in in vivo biosamples from animals and humans with S. aureus periprosthetic joint infection (PJI). Biofilms form when bacterial communities aggregate on orthopedic implants leading to recalcitrant infections that are difficult to treat. Identifying PJI biofilm infections is challenging, and traditional microbiological cultures may yield negative results even in the presence of clinical signs.
Methods: Dogs were trained on pure in vitro S. aureus biofilms and tested on lacrimal fluid samples from an in vivo animal model (rabbits) and human patients with confirmed S. aureus PJI.
Results: The results demonstrated that dogs achieved a high degree of sensitivity and specificity in detecting the odor profile associated with S. aureus biofilms in rabbit samples. Preliminary results suggest that dogs can recognize S. aureus volatile organic compounds (VOCs) in human lacrimal fluid samples.
Discussion: Training odor detection dogs on in vitro S. aureus, may provide an alternative to obtaining clinical samples for training and mitigates biosecurity hazards. The findings hold promise for culture-independent diagnostics, enabling early disease detection, and improved antimicrobial stewardship. In conclusion, this research demonstrates that dogs trained on in vitro S. aureus samples can identify the consistent VOC profile of PJI S. aureus biofilm infections. The study opens avenues for further investigations into a retained VOC profile of S. aureus biofilm infection. These advancements could revolutionize infectious disease diagnosis and treatment, leading to better patient outcomes and addressing the global challenge of antimicrobial resistance.
1 Introduction
Each year in the United States, an estimated 750,000 people undergo hip or knee arthroplasty. According to the National Joint Registry, up to 12% of the hip and 22% of the knee arthroplasties will require a revision surgery due to a periprosthetic joint infection (PJI) (1). PJIs are the leading cause of failure due to their ability to form bacterial biofilms in or around the orthopedic implants (1–3). Biofilms are microbe-derived sessile communities attached to orthopedic implants (1, 4, 5). Once attached, the bacterial biofilms produce a barrier of extracellular polymeric substances (EPS) known as a “slime layer”. The magnitude of virulence is dictated by the specific characteristics of the causative agent(s) (6–8). Staphylococcus aureus (S. aureus) is the most common biofilm in PJI (1, 3, 4, 9). S. aureus virulence is demonstrated in the evasion of the immune system, antibiotic resistance, and ability to disperse via detachment of planktonic bacteria (4, 10).
Identifying the causative organism(s) in musculoskeletal infection, such as fracture related injections (FRI) or PJI, is a challenging task and recent work continues to demonstrate the shortcomings of traditional microbiological cultures. Approximately 20%–50% of musculoskeletal infection patients have negative cultures despite clinical signs and laboratory evidence being consistent with PJI or FRI (11). These findings have produced a new clinical entity designated to be “culture-negative infections”. Moreover, biofilm research has intensified and with the growing burden of biofilm infection, clinical microbiology has undergone a paradigm shift from traditional culture-based to molecular-based methods (12–14). The clinical dilemma of the culture-negative patient is not limited to PJI or FRI, but is a common clinical entity across many areas of medicine (15). Consequently, infectious disease physicians are confronted with empirical treatment choices often requiring multiple courses of broad spectrum antimicrobial regimes when treating culture negative patients. These practices continue to compromise antimicrobial stewardship guidelines amplifying the global threat of antimicrobial resistance (16).
Despite considerable efforts to overcome the diagnostic challenges related to biofilm infections and culture negative results by maximizing microbiological culture yield, the sensitivity of culture still ranges from 39%–70% (11). This comes as no surprise since it is now accepted, following the arrival of polymerase chain reaction (PCR), that <2% of all pathogens that exist are culturable (17). There is convincing data from other medical fields that molecular methods enhance the detection of pathogens missed by traditional cultures and can lead to favorable clinical outcomes with targeted antimicrobial therapy (18–20). In the context of PJI and FRI, the rapidly evolving field of molecular techniques for infectious disease diagnosis will undoubtedly have an important clinical implication. While PCR is a promising tool in the detection of biofilm infections, research demonstrates PCR is not superior to culture (18). As such, it is important to investigate emerging and potentially complementary methods for biofilm detection.
Recently, harnessing the field of molecular techniques, an area of interest has emerged: characterizing specific volatile organic compound (VOC) profiles associated with disease. VOCs are organic chemicals produced as byproducts of metabolism in all living organisms, from bacteria to humans (21, 22). As potential biomarkers, unique VOC signatures have been associated with numerous infectious, metabolic, and neoplastic diseases (21–24). The VOCs associated with disease may originate from altered metabolism of the host, the metabolism of the etiologic agent itself or an interaction between the two (25). These VOCs can be detected in a wide array of biosamples, such as blood, sweat, lacrimal fluid (fluid secreted by the lacrimal glands to lubricate the eye), breath, urine, feces, and tissue. The VOC profile from a single sample can be assessed using chemosensors (e.g., gas chromatography-mass-spectroscopy), electrosensors (e.g., DNA coated nanosensors) and biosensors (e.g., dogs, rats) (23, 26–28).
The focus of this study is utilizing trained dogs to provide proof-of-concept information as to whether S. aureus biofilms have a detectable similar odor profile across both in vitro samples, derived from cultured S. aureus, and in vivo biosamples from animals with experimental infections and humans with spontaneous infections. VOCs may be used as culture-independent biomarker profiles to identify the infection etiology based on a known volatile metabolome profile for a specific infection. To that effect, this method may eliminate the uncertainty of successful microbial growth and the lengthy time required for generating pure diagnostic cultures. The detection of the S. aureus biofilm profile may also be achieved through a canine biosensor. Dogs have been shown to be effective disease biosensors due to their remarkable olfactory sensitivity in identifying, and specificity in discriminating, VOC signatures (26, 28–31). For example, dogs have been trained to detect VOCs associated with a wide range of diseases, including both noninfectious diseases such as cancer, and infectious diseases caused by bacteria, viruses, and prions (30–35). Compared to currently available chemosensors, dogs are capable of rapidly identifying VOC signatures amongst a complex array of volatiles which may lead to the potential to identify diseases at early stages when treatment options may be more effective (28). Dogs may additionally compliment approaches of analytical-organic chemistry, and “e-noses”, as has been demonstrated previously where dogs were trained to detect human ovarian cancer from blood plasma samples, alongside single stranded DNA-coated carbon nanotube sensors (36). However, an obstacle to the wide use of disease detection dogs lies in the reliance on a large number of samples from affected individuals and appropriate controls, which can pose challenges in achieving adequate sample diversity and generalization of subtle odors across a “noisy” background (37). One approach to simplify the training could be to train the dogs on the pure source of the VOCs [e.g., cultured tumor cells (38) or cultured pathogens (30, 31, 39)]. However, it is not currently known whether cultures suitably represent biosamples in practice. In the case of S. aureus, it is important to establish whether dogs can detect an odor associated with S. aureus (indicating that an odor profile exists), and whether dogs trained on cultured samples can generalize this information to non-human animal or human derived biosamples where the complexity of host-microbe interactions are well established. To achieve this, it is beneficial to create a well-controlled experimental test system using non-human animal models as a source of biosamples, so that the strain and volume of S. aureus biofilm can be monitored. Furthermore, the number of colony forming units (CFU (indicative of the bioburden of S. aureus bacteria present in the sample) is of interest as a specific threshold of recognition by the dog may fluctuate based on the number of S. aureus CFU emanating the VOC profile. Zhou et al. reported that only a subset (25%–34%) of in vitro VOCs reliably translate to in vivo detection, and their animal model studies have shown that breath VOCs can be used to identify infection etiology, even down to the strain level for the bacterial pathogen (40). Alongside questions regarding the level of infection required for recognition, it is of interest and clinical relevance to explore if dogs trained on in vitro S. aureus biofilm can continue to detect this VOC profile in experimental S. aureus biofilm infections and in clinical patients of confirmed S. aureus infections with PJI or FRI.
As such, this study sought to answer three research questions to establish whether dogs can identify an odor profile associated with S. aureus biofilms in in vivo biosamples (lacrimal fluid) and to assess the utility of S. aureus cultures as a training aid. The research questions are: (1) can dogs trained on in vitro S. aureus (106 CFU – moderate infection load) identify biosamples (lacrimal fluid) taken from an in vivo animal model (rabbit) infected with moderate S. aureus biofilm, (2) can dogs trained on in vitro S. aureus (104 CFU – mild infection load) identify biosamples (lacrimal fluid) taken from an in vivo animal model (rabbit) infected with mild S. aureus biofilm, and (3) can the dogs involved in research questions 1 and 2 identify S. aureus in biosamples (lacrimal fluid) taken from humans with confirmed S. aureus PJI.
2 Materials and methods
2.1 Sample types
2.1.1 In vitro training samples
2.1.1.1
Moderate bacterial load in vitro S. aureus samples were obtained from S. aureus (106 CFU) cultured on 316l stainless steel chips from orthopedic fracture plates and incubated for 16 h at 37°C in Mueller Hinton Broth (MHB) in a 50 ml culture tube. Following the incubation period, each batch of samples was separately cultured to ensure no contamination occurred during the incubation process. The cotton plugs from the headspace of each culture tube (Fisherbrand™) were aseptically cut into smaller sample sizes (1/2, 1/4, 1/8, 1/16, 1/32) of the original cotton plug, transferred into sterile glass jars with lids, and stored at −80°C until use.
2.1.1.2
Mild in vitro S. aureus samples were obtained by placing a cotton plug in the headspace of S. aureus (104 CFU) cultured on 316l stainless steel chips from orthopedic fracture plates and incubated for 16 h at 37°C in Mueller Hinton Broth (MHB) in a 50 ml culture tube and processed using the same methods as for S. aureus (106 CFU).
2.1.1.3
Control in vitro VOC samples were obtained by placing a cotton plug in the headspace of 50 ml sterile culture tubes containing 316l stainless steel chips from orthopedic fracture plates and incubated for 16 h at 37°C in Mueller Hinton Broth (MHB). Following the incubation period, a small portion of each batch of samples were re-cultured to ensure no contamination occurred during the incubation process. Other controls/distractors in this stage: fresh cotton plugs that were not exposed to bacteria or media, unused Schirmer tear strips (Merck & Co., Inc., Rahway, NJ), laboratory nitrile gloves, and isopropyl alcohol (2 ml), were placed in the glass jars used for odor presentation.
2.1.2 In vivo testing samples
2.1.2.1 In vivo S. aureus lacrimal fluid samples from rabbits in cohorts 1 and 2
As part of an IACUC (protocol #805734) approved study, a periprosthetic joint infection model was induced in the right stifle joints of skeletally mature New Zealand White male castrated and female rabbits (n = 24 cohort 1, n = 16 cohort 2; 50:50 ratio male castrated vs. female). The rabbits were housed in stainless steel cages having auditory, olfactory, and visual contact with each other. Their diet consisted of a pelleted diet and a small amount of fresh roughage, with free access to hay and water. Animals were housed in a room with a 12:12 h light cycle and controlled temperature (70.82 ± 0.1 °F) and humidity (35.9% ± 6.2%). All animals were monitored daily by a veterinarian from the principal investigator's laboratory throughout the entire study.
Anesthesia: On the day of surgery, rabbits fasted for 12 h, were induced with an intramuscular injection of ketamine (35 mg/kg) (Ketamine HCl, Covetrus) and xylazine (2.5 mg/kg) (Xylazine Injection, Covetrus). Propofol (PropoFloTM 28, Zoetis) was administered as intravenous bolus (0.5–1 mg/kg) if anesthetic depth was not sufficient to perform the intubation procedure. Glycopyrrolate (0.01 mg/kg) (Glycopyrrolate injection, BluePoint) was administered intravenously if bradycardia (defined as heart rate less than 120 beats per minute) with concurrent hypotension (defined as mean arterial pressure less than 60 mmHg) occurred. Approximately ten minutes after premedication, the right or left auricular vein and artery of the same ear were catheterized with a 24 Ga (artery) and a 22 Ga catheter (vein) (Surflo, Terumo) respectively. Then a v-gel® (v-gel®, Docsinnovent), was placed and confirmed by capnography, a lubricated 3.5 French (∼1.2 mm external diameter), 40 cm polypropylene catheter (Sovereign Urinary Catheter, Henry Schein Animal Health) was inserted through the v-gel® airway channel. The v-gel® was then removed and a 3 mm internal diameter (ID) cuffed ETT (Sheridan/CF®, Teleflex Medical) was threaded over the catheter. After insertion of the ETT, the polypropylene catheter was removed. The cuff was inflated to 20 mmHg (19) measured with an aneroid manometer connected via a three-way stopcock to the cuff balloon. After intubation, the animals were connected to a circle breathing system designed for animals under 7 kg body weight (Anesthesia WorkStation, Hallowell EMC) and maintained on inhalation anesthesia using 1%–5% Isoflurane in oxygen delivered through a circle breathing circuit at 1l/min. Fraction of inspired oxygen (FiO2), respiratory rate (RR) and end tidal concentration of CO2 (ET CO2) were recorded every five minutes. FiO2 and end tidal CO2 were measured with sidestream capnography and gas analysis. Pulse oximetry, ECG, invasive blood pressure (IBP), FiO2, end-tidal carbon dioxide (ETCO2) and respiratory rate (RR) were recorded throughout each anesthetic event using a multiparameter monitor (Datex Ohmeda S/5, GE). Rectal temperatures were recorded throughout the anesthetic session. The rabbits were maintained in dorsal recumbency with the forelimbs and hind limbs extended and head aligned with the spine. Topical 2% lidocaine (0.2 ml) (Lidocaine 2%, VetOne) was applied via atomizer to the larynx two minutes prior to intubation. The v-gel® was lubricated with lidocaine gel (Lidocaine Hydrochloride Oral Topical Solution 2%, Akorn) mixed with regular sterile lubricant (VetOne® OB Lube), and the ETT was lubricated (VetOne® OB Lube).
Surgery: With animals is left lateral recumbency, the right stifle region was clipped, aseptically prepped, and draped. A ∼1.5 cm–2 cm arthrotomy incision was made medial and in parallel to the middle patellar ligament. Synovial fluid was sampled. The fossa intercondylaris was identified, and a 2 mm K-wire was used to open the femoral canal. The PJI model utilized a 3.5 mm cortex stainless-steel screw in combination with an ultra-high molecular weight polyethylene (UHMWPE) washer placed in the intercondylar fossa of the distal femur extending 1/3 into the medullary canal of the femur. The joint capsule and subcutaneous tissues were closed using resorbable sutures. The skin was closed using a subcuticular pattern followed by surgical glue. Ten days later, under the same general anesthesia protocol, the right femorotibial joint was samples for synovial fluid and inoculated with 0.1 ml saline containing 1 × 106 CFU S. aureus for cohort 1 and 1 × 104 CFU S. aureus for cohort 2. No systemic antimicrobials were given at any time. Thirty-eight days after inoculation the rabbits were sacrificed with an overdose of a commercially available euthanasia solution (Pentobarbital 1 ml/5 kg) according to the guidelines set forth by the current AVMA Panel on Euthanasia and ex vivo analyses consisted of bacteriology, imaging of the implant surface topography using confocal microscopy and scanning electron microscopy for confirmation of S. aureus biofilm. Lacrimal fluid was collected preoperative for one week, on the day of surgery and every day postoperative for the duration of the study from both the right and left eye using a sterile Schirmer tear strip (Merck & Co., Inc., Rahway, NJ) using mild physical restraint throughout the duration of the study by one handler while a 2nd handler performed the tear film collection. The tear strip was placed in the medial canthus of each eye for 1 min and then immediately transferred to non-evacuated blood tubes and stored at −80°C until use.
2.1.2.2 Control lacrimal fluid samples from rabbits in cohorts 1 and 2
Lacrimal fluid collected preoperatively for one week from both the right and left eyes using a Schirmer tear strip. The tear strip was placed in the medial canthus of each eye for 1 min and then transferred to non-evacuated blood tubes and stored at −80°C until use. These samples were used as uninfected biologic controls for the in vivo testing for research questions one and two of the study. Background controls for this stage were unused sterile Schirmer tear strips. Other controls/distractors in this stage: fresh cotton plugs that were not exposed to bacteria or media, laboratory nitrile gloves, and isopropyl alcohol (2 ml), were placed in the glass jars used for odor presentation. A summary of samples used in this stage can be seen in Table 1.
2.1.3 Human biosamples (lacrimal fluid)
2.1.3.1 S. aureus lacrimal fluid from human PJI patients
Human biosamples (lacrimal fluid) from patients presenting to the Rothman Institute at Thomas Jefferson University, Philadelphia with a confirmed microbiological culture of S. aureus positive orthopedic implant infections were obtained. Participants were able to approve via standard of care consent form and were informed of the study aims and methods and were reminded of their right to withdraw from the study at any time (IRB #16D.634). Lacrimal fluid samples were obtained via sterile Schirmer tear strips placed in the medial canthus of each eye prior to general anesthesia and revision surgery. Samples were processed using the Guidance Regarding Methods for Deidentification of Protected Health Information in Accordance with the Health Insurance Portability and Accountability Act (HIPAA) Privacy Rule and immediately stored at −80°C until use.
2.1.4 Control lacrimal fluid from human patients
Human lacrimal fluid from patients presenting to the Rothman Institute at Thomas Jefferson University, Philadelphia for an elective joint surgical procedure such as Total Knee Arthroplasty (TKA) or Total Hip Arthroplasty (THA). Participants were able to approve via standard of care consent form and were informed of the study aims and methods and were reminded of their right to withdraw from the study at any time. Lacrimal fluid samples were obtained via sterile Schirmer tear strips placed in the medial canthus of each eye prior to surgery. Samples were processed using the Guidance Regarding Methods for De-identification of Protected Health Information in Accordance with the HIPAA Privacy Rule and immediately stored at −80°C until use.
2.2 Odor detection dogs
Following Institutional Animal Care and Use Approval (protocols #805676 and 806052) and informed client consent, four Penn Vet Working Dog Center (PVWDC) dogs and four privately owned dogs were enrolled in this study. Dogs varied in breed and sex and ranged in age from 1 to 5 years old (Mean = 3.0). Prior to this study, all PVWDC dogs were trained on a non-biological training odor: Universal Detection Calibrant (UDC) (39) and all privately owned dogs had participated in the American Kennel Club Scent-Work™ odor detection sport or similar organized odor detection training. Information on the individual dogs is described in Table 2.
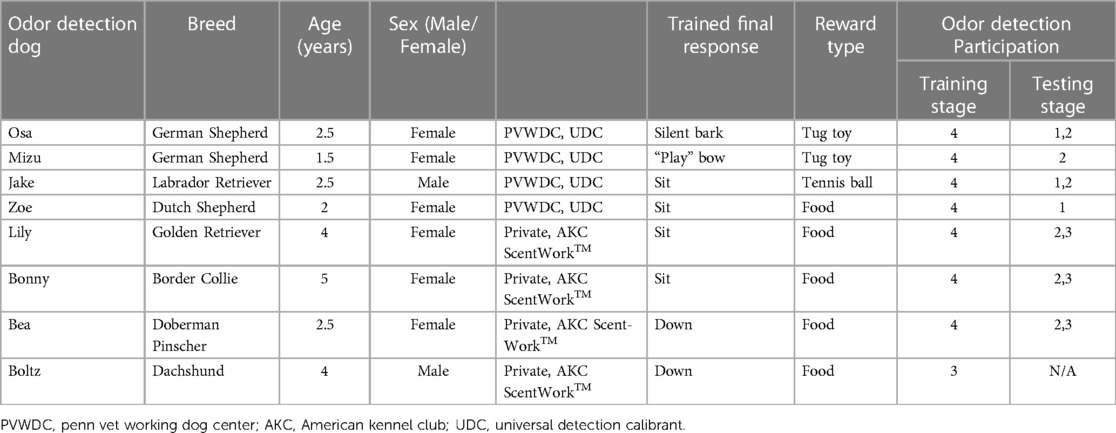
Table 2. Odor detection dog information including trained final response and stage of odor detection training.
2.3 Training of odor detection dogs with in vitro S. aureus samples
2.3.1 Training stages
2.3.1.1 Stage 1 of in vitro S. aureus
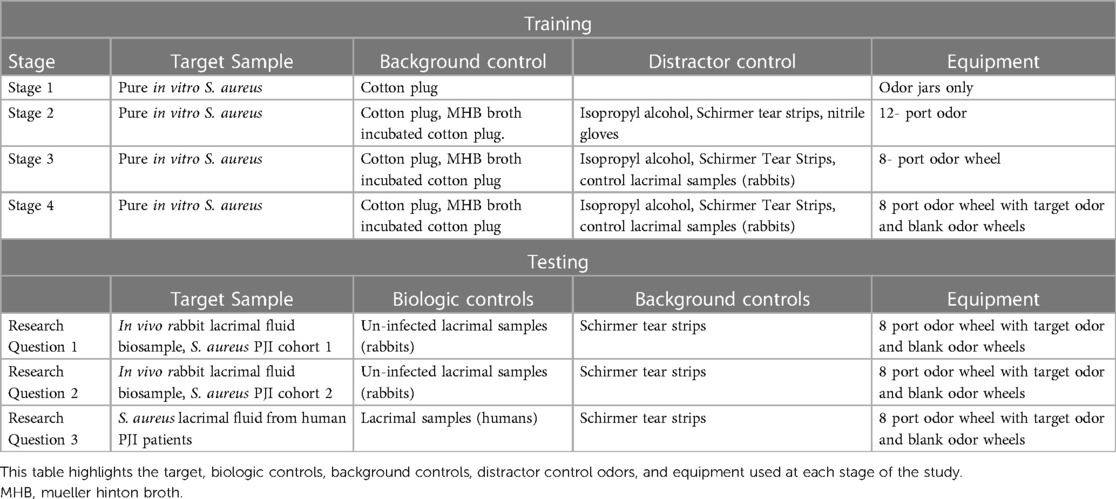
The operant odor discrimination training involved pairing the odor of interest (cotton incubated with cultured S. aureus) with a trained final response, such as sit or down (see Table 1). Positive reinforcement in the form of a reward, such as a dog treat or toy, was used to reinforce the correct final response. Trained final response behavior to known negative samples was not rewarded. Table 2 highlights the training stages used in this study. The initial training began by exposing the dogs to the whole cotton plug incubated with S. aureus contained in a glass odor jar and rewarding the dog for sniffing the sample, establishing a positive association between the odor of interest and the reward. The dogs were trained to perform a trained final response upon identifying the target sample. The final responses were reinforced using a conditioned secondary reinforcer (“clicker”) to mark the correct response. This training was conducted in one morning and one evening session (within the same day), with each dog receiving 10 odor exposures per session. Once the dogs exhibited the trained final response within five seconds of sniffing the target sample 90% of the time in two consecutive training sessions the dogs were moved to training stage 2.
2.3.1.2 Stage 2 of in vitro S. aureus
Stage 2 of training introduced the use of an odor wheel. The protocol for sample placement within the odor wheel consisted of a randomized assignment of target (cultured S. aureus), background controls, and control/distractor odor locations with each port designated by its' number space on the wheel (https://www.randomizer.org/). Each trial was documented on a written record and video recorded on a mounted tripod for review of the dogs' behavior at each port. Nitrile gloves were used and changed between handling of samples and whenever interacting with the odor wheel. The ports of the wheel were wiped with 70% isopropyl alcohol between each trial, the entire wheel was wiped with 70% isopropyl alcohol between each dog, and the floor beneath the wheel was cleaned with 70% isopropyl alcohol each day. A trial was defined as each time the dog was sent to search the wheel. A session was defined as all the consecutive trials completed by the dog in one training or testing session. Training sessions contained ten trials and did not occur more than twice per day. A dog's response at each port was scored as a “1” for a negative response, indicating a dog sniffed a port and moved on, a “2” for a positive response, indicating the dog sniffed a port and gave their trained final response, or a “3” for a hesitation, indicating a change in behavior without the dog demonstrating their trained final response. Stage 2 of training utilized a 12-port odor wheel. The 12-port odor wheel was used in this stage to introduce the background controls and the control/distractor samples for odor discrimination between these samples and the target odor. Between each trial, the 12-port carousel was rotated so that the location of the target sample was moved whereas the 8-port wheel described below allowed the samples to be removed and replaced to create a novel order of samples for each trial. Once the dogs exhibited the trained final response at the target sample with a 90% sensitivity and at the controls with a 95% specificity in three consecutive training sessions the dogs were moved to training stage 3.
2.3.1.3 Stage 3 of in vitro S. aureus
The dogs were transitioned to a newly acquired odor wheel with 8-ports in training stage 3. This wheel was utilized as it had some advantages over the 12-port wheel, namely, the samples could be moved independently (allowing the order of samples presented to be changed each trial), and additionally had stainless-steel barriers between each port, aiding in keeping the odor cones from each sample separated. The dogs were trained to search in a clockwise direction of the 8-port circular odor wheel, sniffing each port in order, correctly sniffing, and passing both the background and distractor control samples, and exhibiting their trained final response behavior when they detected the target odor: cultured S. aureus. Training sessions contained ten trials and did not occur more than twice per day. A dog's response at each port was scored as a “1” for a negative response, indicating a dog sniffed a port and moved on, a “2” for a positive response, indicating the dog sniffed a port and gave their trained final response, or a “3” for a hesitation, indicating a change in behavior without the dog demonstrating their trained final response. For training stage 3, the same randomized sample placement described above was used to determine the target sample port location for each trial. Each port was designated by its' number space on the wheel (1–8) in the random generator. Once the dogs exhibited the trained final response at the target sample with 90% sensitivity and at the controls with 95% specificity in three consecutive training sessions the dogs were moved to training stage 4.
2.3.1.4 Stage 4 of in vitro S. aureus
Training stage 4 introduced the dogs to the concept that the target odor may not be present in the wheel, where they were required to check and pass each port, performing a sit or down response on a low platform outside of the wheel space to indicate an “all clear” (referred to as a “blank wheel”). For training stage 4, the same randomized sample placement described above was used to determine the target sample port location for each trial or if the trial was a blank trial (no target odor). Each port was designated by its' number space on the wheel (1–8), a blank wheel was indicated as a number “9” in the random generator. Training stage 4 sessions contained ten trials once a day. A dog's response at each port was scored as a “1” for a negative response, indicating a dog sniffed a port and moved on, a “2” for a positive response, indicating the dog sniffed a port and gave their trained final response, or a “3” for a hesitation, indicating a change in behavior without the dog demonstrating their trained final response. Once the dogs reached 90% sensitivity and 95% specificity including blank wheels in three consecutive training sessions the dogs were moved to testing.
2.4 Testing for S. aureus profile of in vivo and human samples
2.4.1 Testing protocols used for all research questions
The odor wheel searching pattern, cleaning, and randomization protocols used in training stages 3 and 4 were used for all testing stages. Only dogs that passed stage 4 of training proceeded to testing. Testing trials were performed once a day in a laboratory setting utilizing the 8-port odor wheel used during training (Figure 1). One dog was tested at a time and the order in which the dogs were to search during that session was randomized prior to the study day. Target, background and distractor control samples used for testing were removed from the 80°C storage twenty minutes prior to the testing session. Researchers changed gloves between handling of all samples thorough the session to avoid contamination. The odor wheel was located behind a barrier and monitored via a video feed so that the dogs could not see their handler, the recorder, or any observers during the testing trials. Testing trials were double blinded so that the dog and handler did not know where the target sample was placed on the wheel; however, the researcher who was out of the wheel room was aware. Because the handler did not know the location of the target, they were informed once the dog had made a response whether it was correct or incorrect by the researcher (out of sight from both the handler and dog) and the dog was either called away to reset for the next trial within the session (if incorrect) or, if correct, marked by a conditioned secondary reinforcer (a clicker) by the researcher and given a reward by the handler. Each testing session included 10 trials at least one of the trials was a blank wheel and a session was performed once per day. If a biologic control or background control was located in a port location beyond the target sample (example port 1) and the dog correctly alerted to the target sample, then the biologic and background controls were set to be presented in the blank wheel trial for the dog to evaluate the samples and provide a first pass response. First pass for each testing sample was recorded as a “1” for a negative response, indicating a dog sniffed a port and moved on (false negative), a “2” for a positive response, indicating the dog sniffed a port and gave their trained final response (true positive), and used for data analysis.
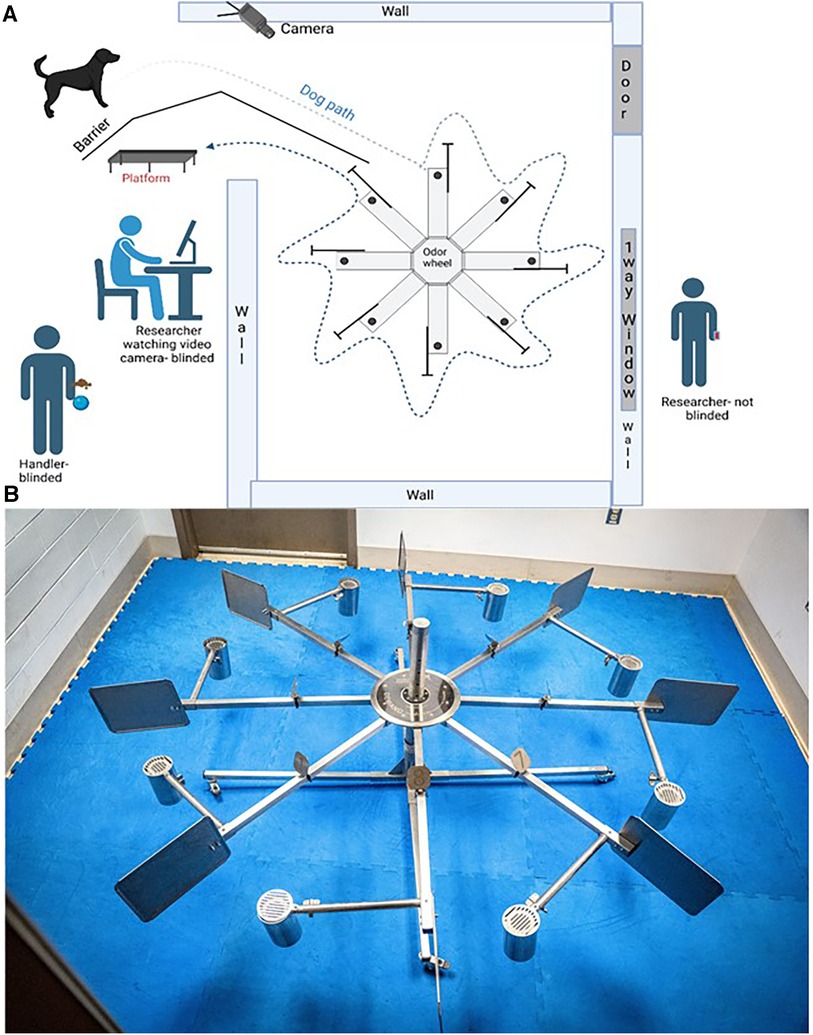
Figure 1. The laboratory setting for this study. (A) is the diagram of the laboratory set up. (B) is a photograph of the 8-port odor wheel.
2.4.2 Research question 1: can dogs trained on in vitro S. aureus (106 CFU – moderate bacterial load) identify biosamples (lacrimal fluid) taken from an in vivo animal model (rabbit) infected with moderate S. aureus biofilm?
To address this research question, the cotton plug samples from the cultured S. aureus were replaced with the experimental PJI rabbit model biosamples (lacrimal fluid from infected rabbits). The biological controls were the lacrimal samples from uninfected rabbits. The background control samples for this phase consisted of a Schirmer tear strips. The distractor control odors were cotton plugs, MHB broth incubated cotton plug, isopropyl alcohol, and nitrile gloves (Table 2). The target samples for research questions two and three are highlighted below.
2.4.3 Research question 2: can dogs trained on in vitro S. aureus (104 CFU – mild bacterial load) identify biosamples (lacrimal fluid) taken from an in vivo animal model (rabbit) infected with mild S. aureus biofilm?
To answer this research question, the target samples were the biosamples (lacrimal fluid) obtained from the second cohort of infected rabbits which were inoculated with 1 × 104 CFU S. aureus. The biological controls were the lacrimal samples from uninfected rabbits. The background control samples for this phase consisted of a Schirmer tear strips. The distractor control odors were cotton plugs, isopropyl alcohol, and nitrile gloves (Table 2).
2.4.4 Research question 3: can the dogs involved in research questions 1 and 2 identify S. aureus in biosamples (lacrimal fluid) taken from human patients with confirmed S. aureus PJI?
To answer this research question, the target samples were the lacrimal fluid samples obtained from human patients presenting for revision surgery secondary to a confirmed S. aureus PJI. The biological controls included for this stage were control lacrimal samples (humans). These biological control lacrimal fluid samples were obtained preoperatively from patients undergoing routine orthopedic surgery. The background control samples of this stage were Schirmer tear strips. The distractor control samples were cotton plugs, isopropyl alcohol, Schirmer Tear Strips, and biological control lacrimal samples (rabbits),.
2.5 Statistical analysis
Sensitivity, specificity, positive predictive value, and negative predictive value were used to evaluate the performance of the odor detection dogs as a diagnostic test or screening tool for S. aureus positive and control samples. Sensitivity was determined as the true positives divided by true positives plus false negatives. Specificity was determined as specificity the number of true negatives divided by the number of true negatives plus the number of false positives. Positive predictive value (PPV) is defined as the true positives divided by true positives plus false positives and negative predictive value (NPV) as the number of true negatives divided by the number of true negatives plus the number of false negatives. A true positive was a final alert on the S. aureus target samples on the first pass of the port. A true negative was the correct omission of a final alert on the biological controls and the background control samples on the first pass of the ports. During the testing phase, only the dogs “first pass” response on the first presentation of the target S. aureus sample was considered for analysis. The probability of a correct trial by chance, where the dog exhibited their trained final response on the new target port and performed no false alert behaviors on the incorrect ports is 12.5%.
3 Results
3.1 Sample collection
3.1.1 Training in vitro S. aureus samples
S. aureus samples were successfully cultured for this study without contamination of other bacteria as evidenced by the quality control cultures performed on each batch by the clinical microbiology laboratory at Penn Vet New Bolton Center following the incubation process.
3.1.2 Testing in vivo S. aureus lacrimal fluid samples from rabbits
Biosamples obtained from the experimental PJI rabbits were obtained for this study are described in Table 3. In cohort 1, two rabbits developed sepsis and were euthanized and removed from the study on day 6 post inoculation. One of the rabbits in cohort 1 and cohort 2 developed ocular conjunctivitis prior the last sample collection and therefore only one sample was collected on day 7 and day 8 of those cohorts. Table 3 highlights the available samples for each day post inoculation. In cohort 1 and 2 a total of 43 and 47 positive S. aureus lacrimal fluid samples were obtained for screening by the dogs. 168 biological controls uninfected lacrimal fluid were collected in cohort 1 and 112 in cohort 2. For testing, the biological control sample was matched to the infected rabbit sample. The other biologic control samples were presented during blank wheel trials.
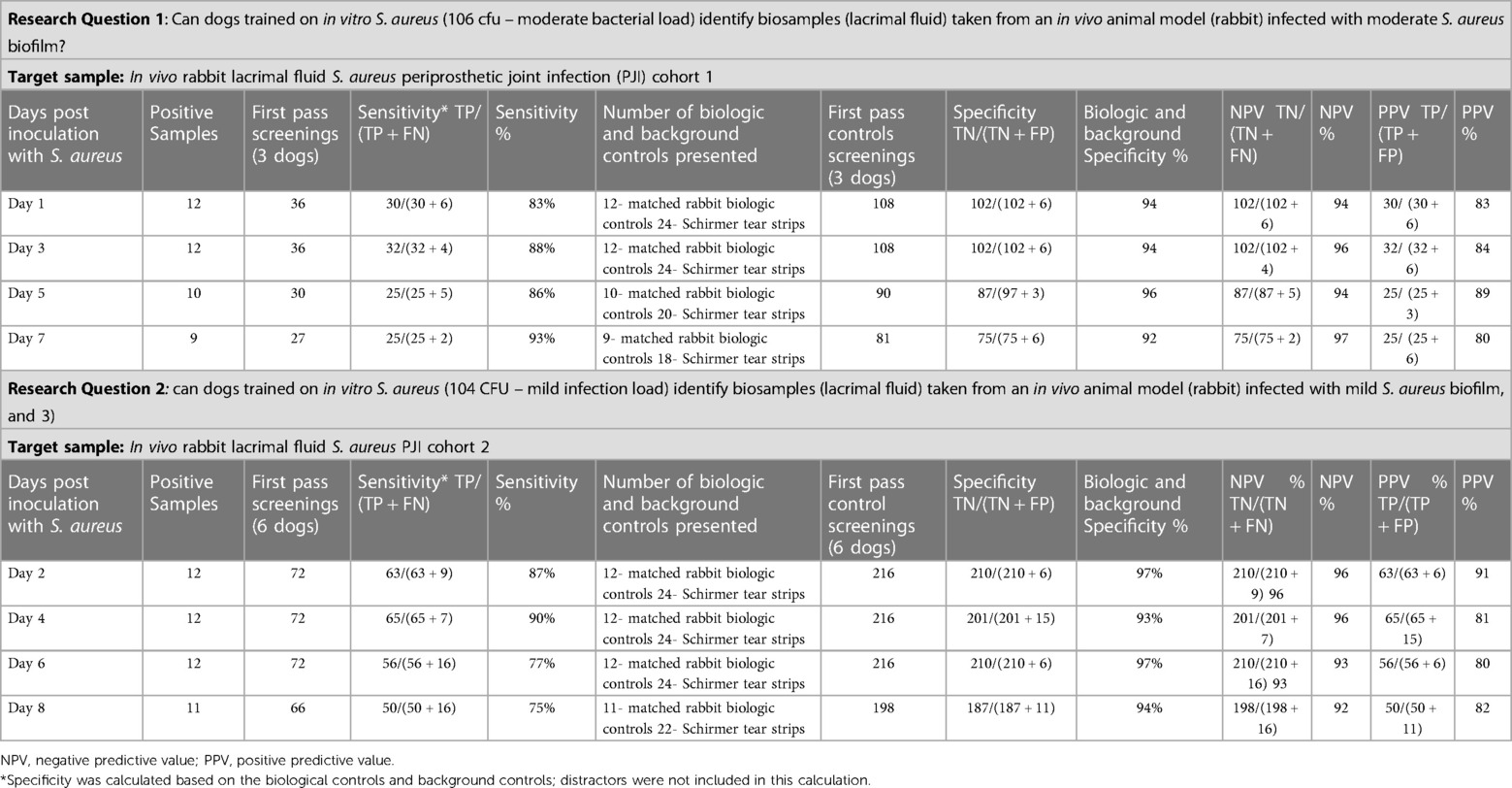
Table 3. The sensitivity, specificity, negative predictive value, and positive predictive value for research questions one and two.
3.1.3 Testing lacrimal fluid samples from human patients
Eight control human lacrimal samples (left or right eye) were obtained from eight healthy patients. Three lacrimal fluid samples (left or right eye) were obtained from three PJI methicillin sensitive Staphylococcus aureus (MSSA) patients as confirmed by culture following surgical revision.
3.2 Detection dogs
3.2.1 Detection dog training
Eight odor detection dogs were recruited for this study. Seven out of the eight dogs completed all four stages of training as outlined in Table 1. Boltz failed to complete training stage four due to lack of indication on the platform when searching the blank wheel. Boltz tried to re-search the wheel after reaching port 8 instead of indicating on the platform and was subsequently removed from further training or progressing to testing.
3.2.2 Detection dog testing
Of the seven dogs, three participated in the first research question testing (Osa, Jake, and Zoe), six dogs participated in research question two testing (Osa, Mizu, Jake, Lily, Bonny, and Bea), and three dogs participated in research question three (Lily, Bonny, and Bea). Zoe was excluded after research question one because she was purchased for a career in law enforcement from the PVWDC program prior to the start of testing research question two. Mizu was trained to replace Zoe for testing research question two. Osa, Mizu, Jake, and Zoe were excluded from testing research question three because they were purchased for their working dog careers prior to the start of testing. For research questions one and two, analysis for differences between each dog and between dogs and specific rabbits were analyzed. There was no significant difference amongst dogs or towards a particular rabbit sample and therefore analysis of days post inoculation was investigated.
3.3 Research question one
The sensitivity and specificity of the three dogs (Osa, Jake, and Zoe) tested for research question one (target odor was the lacrimal fluid from mild S. aureus inoculated rabbits) are greater than 83% sensitivity and 92% specificity. The sensitivity improved as the days post-inoculation increased (days 1–7). PPV was greater than 80% and NPV was greater the 94%. Table 3 summarizes the sensitivity, specificity, negative predictive value, and positive predictive.
3.4 Research question two
The sensitivity and specificity of the six dogs (Osa, Mizu, Jake, Lily, Bonny, and Bea) tested for research question two (where the target odor was the lacrimal fluid from the rabbits inoculated with mild S. aureus) are greater than 75% sensitivity and 93% specificity. PPV was greater than 80% and NPV was greater the 92%. Table 3 summarizes the sensitivity, specificity, negative predictive value, and positive predictive value.
3.5 Research question three
Lily, Bonny, and Bea were tested on each human target (n = 3 target samples) and biological control sample during one session (n = 8 control samples). Unfortunately, due to the COVID-19 pandemic, the testing of human samples was limited to one session on 17 March 2020 in which the three dogs were tested on the three target samples. Both Lily and Bonny gave final responses on patients B and C. Bea did not give a final response on any of the three target odors (Patients A, B or C) (Table 4). Patient A's sample was not correctly identified on first pass by any of the three dogs. The screening of further samples was not continued after 2020 due to withdrawal of the three detection dogs by their owners.
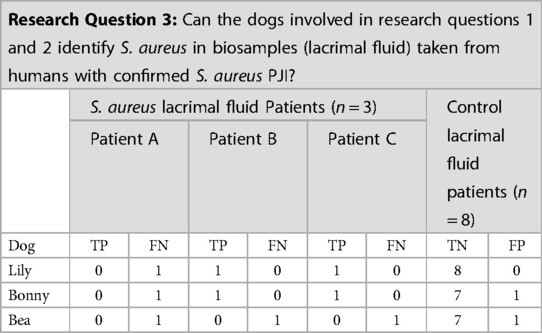
Table 4. Results of the three detection dogs for the S. aureus lacrimal fluid from human periprosthetic joint infection (PJI) patients target odor.
4 Discussion
This study investigated the potential of training odor detection dogs using cultured S. aureus biofilms to detect biofilm-associated infections with a particular interest in the culture-negative patient. The results demonstrate that dogs trained on VOCs from cultured S. aureus biofilms display a high degree of sensitivity and specificity in recognizing S. aureus associated with PJI biofilm in lacrimal fluid from infected rabbits. Furthermore, because dogs trained on in vitro cultured S. aureus were able to generalize to biosamples obtained from a controlled rabbit model, it suggests that training dogs (or machines) on cultured S. aureus biofilms may provide an adequate VOC profile indicative of S. aureus, which may reduce logistical issues surrounding the acquisition of biological samples. Our preliminary results further suggest that this odor profile is consistent and appears to be detectable in human samples, however further testing is required. Our findings are interesting and suggest that there is a common VOC profile that is preserved across S. aureus biofilm from pure in vitro cultures and from lacrimal fluid biosamples obtained from rabbits with PJI secondary to S. aureus infection and human samples of naturally occurring S. aureus infection. This is intriguing since in the context of infections, the S. aureus VOC metabolome reflects complex host-microbial interactions throughout the processes of infection, yet this subset of metabolites detectable by the canine nose seems to be conserved. Moreover, our observations with odor detection dogs in the present study strongly support the potential of metabolomics-based diagnostics that can capture consistent metabolites for infection diagnosis, characterization, and monitoring (41). Diagnostic breath analysis is an area of intense research and its recent rapid advances have provided critical insight in how VOCs can differentiate closely related pathogens, including the capacity to establish antimicrobial sensitivity profiles (41). We elected lacrimal fluid as our source for biosamples because tear fluid contains high concentrations of proteins, is recognized as a potential source of biomarkers for other systemic disorders and collecting tear fluid via Schirmer strips is current standard of care when diagnosing dry eye disease (42). In addition, blood draws are more invasive, it carries VOC metabolites from all tissues and organs systems to the lungs and into exhaled breath potentially expanding the complexity of the VOC profile.
Our results for sensitivity and specificity in the detection of a consistent odor profile emanating from S. aureus biofilm infections align with previous studies that specialty trained odor detection dogs can differentiate between a target odor and controls. Although, in our study, the dogs' responses to the biosamples from confirmed S. aureus human patients were less consistent compared to the responses from the infected PJI rabbit samples. An explanation could be that these samples came from a new environment and the human lacrimal sample presented as a new target for the dogs. It is possible that, with additional testing of samples, dogs would have been able to recognize this odor with improving sensitivity and specificity. Many studies of odor detection dogs reveal an impressive sensitivity and specificity in detecting various VOC profiles emanating from several cancers and viruses in biologic samples (22, 30, 35, 39). These studies focused on several different sample types for disease detection in which negative control fluid samples were compared to positive clinical patients of the same substrate type (blood plasma vs. blood plasma from diseasepositive patients) (34) or less invasive samples were compared to more invasive samples [saliva vs. sweat and urine vs. saliva (43)]. Although results from these studies are encouraging, the reliance on samples from patients carrying the disease combined with appropriate controls for training of odor detection dogs may pose limitations in terms of sample acquisition, sample diversity, and generalization. This gap in accessibility dampens the commercial enthusiasm of this diagnostic approach, creating a major barrier in implementing odor detection dogs in a clinical diagnostic setting (26).
To address this gap, we proposed an alternative approach of training odor detection dogs on the VOC profile of the pathogen itself, such as cultured pure in vitro S. aureus biofilms using clinical strains. Training odor detection dogs on cultured samples may eliminate many challenges such as sample acquisition as they can be readily produced in the laboratory. In addition, training odor detection dogs on pathogen specific VOCs rather than on the pathogen itself, mitigates potential biosecurity hazards, and the need for biosafety measures during odor detection training. Recent studies have implemented this approach with different strains of the same bacterial and viral species (30, 31). Koivusalo et al. discovered odor detection dogs can detect and differentiate MSSA and Methicillin Resistant Staphylococcus aureus (MRSA) respectively with 97% sensitivity and 92% specificity (31). Angle et al. successfully trained dogs to differentiate between cultured cells infected with Bovine Viral Diarrheal Virus (BVDV) with a sensitivity of 85% and specificity of 98% (30). The high yield in sensitivity and specificity most likely is a result of sample variation and increased repetition during training amongst appropriate background odors, and further emphasizes the robustness of the canine odor detection test system.
The present study demonstrated encouraging results when utilizing cultured infectious disease pathogens such as S. aureus with a consistent target odor profile for training in preparation for diagnosing clinical samples. However, it is important to recognize that not all diseases lend themselves to in vitro culturing of a specific pathogen for training purpose of odor detection dogs. Murarka et al. explored the utility of odor detection dogs on cultured cell lines of ovarian cancer in hopes it would translate to blood plasma samples from clinical patients (38). Unfortunately, in that study only one dog could differentiate between the cultured ovarian cancer cell line and did not alert on the blood plasma samples when tested on clinical patients with confirmed late-stage ovarian cancer. The conflicting findings and experience with odor detection dogs in the field of cancer and infectious disease emphasize the importance of the underlying pathophysiology of the disease. The hallmark of an infectious disease processes is characterized by a pathogen invading a host triggering a host-microbial interaction. This process affords the training of odor detection dogs on the VOC profiles using pure cultures of a given pathogen. Conversely, the literature is inconclusive for disease processes which originate within the host such as cancer and whether the complexity of the hosts' environment, genetics, and metabolism on a given VOC profile, results in an inconsistent or convolute biomarker signature (34, 35, 44, 45). Further investigation into the interactions between the disease process, host, and source for training is warranted. Indeed, the ongoing controversy in the literature over the lack of well-defined standards and the clinical utility of odor detection dogs in the field of cancer and infectious diseases is perhaps a testament of the intensity of current research over the past 20 years and its considerable progress disentangling the chemistry on olfactory signatures of disease using dogs, advanced VOC analysis and clinical detection.
Our study did not evaluate polymicrobial biofilms, consisting of multiple bacterial species, which may pose challenges in training dogs on cultured biofilms that are composed of one microbial species. Conversely, lacrimal biosamples from non-human animals and human patients as it was the case in our studies are from subjects constantly challenged by microbial attacks. Furthermore, the human patients presenting for revision surgery in our study all underwent different antimicrobial regimes in attempt to treat the underlying clinical problem (PJI or FRI). Thus, while the VOC profiles of biofilm-associated infections may still vary among individuals and species, in our case of known S. aureus biofilm infection, dogs detected a seemingly consistent S. aureus specific odor profile. Undoubtedly, further research is needed to investigate the accuracy and generalizability of odor detection in dogs trained on cultured biofilms for detecting biofilm associated infections from host-derived biological samples. This could involve training dogs on biofilms from different species and strains, as well as comparing the performance of dogs trained on cultured biofilms with those trained on samples from infected individuals and appropriate controls. Additionally, studies comparing the VOC profiles of cultured biofilms with biological samples from infected individuals can provide insights into the similarities and differences in odor profiles and help determine the extent to which cultured biofilms represent the odor profiles of biofilm-associated infections. In fact, the findings by Kuil et al. further amplify the prospect toward differentiating closely-related pathogens of the same genus and species, perhaps even determining antimicrobial resistance and sensitivity based on unique VOC profiles associated with infection (46).
It should also be noted that because this initial study focused on the research question of whether dogs can generalize from S. aureus in vitro cultures to experimental infections in rabbits and naturally occurring PJI infections in human biosamples, we did not include control samples that represented other bacterial infections, or samples taken from people who were unwell with conditions other than S. aureus. Further studies should focus on specifically testing dogs' abilities to discriminate between (1) different types of biofilm infections, and (2) biosamples derived from patients who have confirmed PJI of S. aureus vs. samples taken from patients who are unwell with conditions other than a bacterial infection. This is important as it has been demonstrated in certain disease conditions, that several metabolic changes may occur, impacting an individual's VOC profile, when they become unwell (42). When considering other diseases and infections, previous studies have demonstrated dogs' abilities to specifically detect target samples of interest while ignoring similar conditions or samples indicative of “ill-health” unrelated to the target condition (36, 47, 48). These findings are well aligned with our observations. Our dogs have been trained on in vitro biofilms of S. aureus and continued to detect the target samples in experimental conditions of S. aureus biofilm in rabbits and in naturally occurring PJI of human patients undergoing various antimicrobial therapies, despite significant host-microbial interactions. This is important, since it is well accepted that in vitro and in vivo biofilms are not equivalent and host-microbial interactions significantly change biofilms. Clinical biofilm infections typically evolve with the patient undergoing antimicrobial therapy. He et al. recently demonstrated that quorum cheater emergence can be triggered during S. aureus biofilm infections using certain antimicrobials resulting in a significant increase of the bacterial burden and most importantly, the development of agr mutants (49).
Results of this study inform future developments in the field of culture-independent biofilm diagnostics and treatment. We have shown that not only does S. aureus have a detectable odor profile, but we also demonstrated that in vitro cultured S. aureus can be a viable training aid to be used to imprint dogs on S. aureus. The encouraging findings of odor detection dogs on infection diagnostics (50) combined with high cost and time-consuming training of dogs has inspired chemical analysts and engineers to emulate the dogs' capabilities of analyzing VOCs in biosamples. To this end, various studies have sought to build an electronic nose (28, 51, 52, 55, 56). The goal of an electronic nose is to mimic a bio-detection dog by analyzing VOCs, for example, by using metal-oxide sensors and learning algorithms. When exposed to a breath, blood, or urine sample, it probes a profile or so-called “smell print”. If proven effective, it would be a point-of-care, low-cost, handheld, noninvasive tool that overcomes many of the shortcomings associated with currently available methods. For now, dogs seem to outperform e-noses (53, 54, 57). Research needs to continue to pursue the elite abilities of odor detection dogs to investigate disease detection whilst simultaneously harnessing their skills to refine and enhance the e-nose technology for scalability to bring cultureindependent point-of-care-assays to the clinic floor.
5 Conclusion
Herein we demonstrated that dogs can be trained to detect a consistent odor profile associated with in vitro and in vivo S. aureus biofilms. Research questions one and two were determined to be successful in that dogs trained on in vitro S. aureus were able to generalize to in vivo lacrimal fluid from a non-human animal model (rabbits) at both a mild and moderate infection loads of S. aureus biofilms. Our findings are suggestive that host-microbial interactions do not alter certain VOC profile characteristics of S. aureus biofilm. Results discovered in research question three, further suggests that dogs could also generalize to S. aureus VOCs in human lacrimal fluid in patients presenting for orthopedic revision surgery. Taken together, these findings provide insight into S. aureus biofilm production as it relates to detectable VOC biomarkers that are conserved both in vitro and in vivo despite host-microbial interactions in experimental models of infection (rabbits) and naturally occurring biofilm infection in human patients. These findings may be critical in the design of continued research and development efforts into non-invasive, culture independent diagnostics for early disease detection, refined antimicrobial treatment, and improved antimicrobial stewardship.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by Institutional Review Board, Rothman Orthopaedic Institute. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. The animal studies were approved by Institutional Animal Care & Use Committee, Office of Animal Welfare, University of Pennsylvania. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
MR: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing, Funding acquisition. GC: Conceptualization, Formal Analysis, Funding acquisition, Methodology, Writing – review & editing. CW: Writing – original draft, Writing – review & editing. JG: Conceptualization, Data curation, Investigation, Resources, Writing – review & editing. JK: Conceptualization, Writing – review & editing. JP: Conceptualization, Writing – review & editing. AC: Conceptualization, Writing – review & editing. CO: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing. TS: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Writing – review & editing.
Funding
The authors declare financial support was received for the research, authorship, and/or publication of this article.
Research reported in this publication was supported by AOTrauma North America's Resident Research Grant and the National Center for Advancing Translational Sciences of the National Institutes of Health under award number TL1TR001880. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health and by the Institute for Medical Translation New Bolton Center.
Acknowledgments
The authors would like to acknowledge the support of Dan Bruce, Aubrey Vena in support of the in vivo work and Kier Blevins and Keenan Sobol for the acquisition of the human tear fluid samples. The authors would like to acknowledge the training and business staff, interns and volunteers of the University of Pennsylvania School of Veterinary Medicine Working Dog Center, and the work of graduate students.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Mooney JA, Pridgen EM, Manasherob R, Suh G, Blackwell HE, Barron AE, et al. Periprosthetic bacterial biofilm and quorum sensing. J Orthop Res. (2018) 36(9):2331–9. doi: 10.1002/jor.24019
2. Grimberg AW, Grupp TM, Elliott J, Melsheimer O, Jansson V, Steinbrück A. Ceramic coating in cemented primary total knee arthroplasty is not associated with decreased risk of revision due to early prosthetic joint infection. J Arthroplasty. (2021) 36(3):991–7. doi: 10.1016/j.arth.2020.09.011
3. Wildeman P, Tevell S, Eriksson C, Lagos AC, Söderquist B, Stenmark B. Genomic characterization and outcome of prosthetic joint infections caused by Staphylococcus aureus. Sci Rep. (2020) 10(1):5938. doi: 10.1038/s41598-020-62751-z
4. Unnanuntana A, Bonsignore L, Shirtliff ME, Greenfield EM. The effects of farnesol on Staphylococcus aureus biofilms and osteoblasts. J Bone Joint Surg Am. (2009) 91(11):2683–92. doi: 10.2106/JBJS.H.01699
5. Neut D, Mei HCVD, Bulstra SK, Busscher HJ. The role of small-colony variants in failure to diagnose and treat biofilm infections in orthopedics. Acta Orthop. (2007) 78(3):299–308. doi: 10.1080/17453670710013843
6. Costerton W, Veeh R, Shirtliff M, Pasmore M, Post C, Ehrlich G. The application of biofilm science to the study and control of chronic bacterial infections. J Clin Invest. (2003) 112(10):1466–77. doi: 10.1172/JCI20365
7. Hall-Stoodley L, Stoodley P. Evolving concepts in biofilm infections. Cell Microbiol. (2009) 11(7):1034–43. doi: 10.1111/j.1462-5822.2009.01323.x
8. Costerton JW, Post JC, Ehrlich GD, Hu FZ, Kreft R, Nistico L, et al. New methods for the detection of orthopedic and other biofilm infections. FEMS Immunol Med Mic. (2011) 61(2):133–40. doi: 10.1111/j.1574-695X.2010.00766.x
9. Crawford EC, Singh A, Gibson TWG, Weese JS. Biofilm-associated gene expression in Staphylococcus pseudintermedius on a variety of implant materials: biofilm gene expression on implant surfaces. Vet Surg. (2016) 45(4):499–506. doi: 10.1111/vsu.12471
10. Brady RA, Leid JG, Camper AK, Costerton JW, Shirtliff ME. Identification of Staphylococcus aureus proteins recognized by the antibody-mediated immune response to a biofilm infection. Infect Immun. (2006) 74(6):3415–26. doi: 10.1128/IAI.00392-06
11. Tan TL, Kheir MM, Shohat N, Tan DD, Kheir M, Chen C, et al. Culture-negative periprosthetic joint infection. JB JS Open Access. (2018) 3(3):e0060. doi: 10.2106/JBJS.OA.17.00060
12. Parvizi J, Fassihi SC, Enayatollahi MA. Diagnosis of periprosthetic joint infection following hip and knee arthroplasty. Orthop Clin North Am. (2016) 47(3):505–15. doi: 10.1016/j.ocl.2016.03.001
13. Longo UG, Budhiparama NC, Lustig S, Becker R, Espregueira-Mendes J. Infection in Knee Replacement. Springer International Publishing. (2021). doi: 10.1007/978-3-030-81553-0
14. Kalbian I, Park JW, Goswami K, Lee YK, Parvizi J, Koo KH. Culture-negative periprosthetic joint infection: prevalence, aetiology, evaluation, recommendations, and treatment. Int Orthop (SICOT). (2020) 44(7):1255–61. doi: 10.1007/s00264-020-04627-5
15. Kolpen M, Kragh KN, Enciso JB, Faurholt-Jepsen D, Lindegaard B, Egelund GB, et al. Bacterial biofilms predominate in both acute and chronic human lung infections. Thorax. (2022) 77(10):1015–22. doi: 10.1136/thoraxjnl-2021-217576
16. Zhang R, Eggleston K, Rotimi V, Zeckhauser RJ. Antibiotic resistance as a global threat: evidence from China, Kuwait and the United States. Global Health. (2006) 2:6. doi: 10.1186/1744-8603-2-6
17. Wade W. Unculturable bacteria—the uncharacterized organisms that cause oral infections. J R Soc Med. (2002) 95(2):81–3.11823550
18. Zegaer BH, Ioannidis A, Babis GC, Ioannidou V, Kossyvakis A, Bersimis S, et al. Detection of bacteria bearing resistant biofilm forms, by using the universal and specific PCR is still unhelpful in the diagnosis of periprosthetic joint infections. Front Med. (2014) 1:30. doi: 10.3389/fmed.2014.00030
19. Marín M, Garcia-Lechuz JM, Alonso P, Villanueva M, Alcalá L, Gimeno M, et al. Role of universal 16S rRNA gene PCR and sequencing in diagnosis of prosthetic joint infection. J Clin Microbiol. (2012) 50(3):583–9. doi: 10.1128/JCM.00170-11
20. Wilson MR, Sample HA, Zorn KC, Arevalo S, Yu G, Neuhaus J, et al. Clinical metagenomic sequencing for diagnosis of meningitis and encephalitis. N Engl J Med. (2019) 380(24):2327–40. doi: 10.1056/NEJMoa1803396
21. Lough F, Perry JD, Stanforth SP, Dean JR. Detection of exogenous VOCs as a novel in vitro diagnostic technique for the detection of pathogenic bacteria. TrAC, Trends Anal Chem. (2017) 87:71–81. doi: 10.1016/j.trac.2016.12.004
22. Mandy M, Cornelia F, Malgorzata L, Oliver S, Achim S, Dorothee S. Volatile organic compounds (VOCs) in exhaled breath of patients with breast cancer in a clinical setting. Ginekol Pol. (2012) 83(10):730–6. PMID: 2338355723383557
23. Timm CM, Lloyd EP, Egan A, Mariner R, Karig D. Direct growth of bacteria in headspace vials allows for screening of volatiles by gas chromatography mass spectrometry. Front Microbiol. (2018) 9:491. doi: 10.3389/fmicb.2018.00491
24. Belizário JE, Faintuch J, Malpartida MG. Breath biopsy and discovery of exclusive volatile organic compounds for diagnosis of infectious diseases. Front Cell Infect Microbiol. (2021) 10:564194. doi: 10.3389/fcimb.2020.564194
25. Filipiak W, Beer R, Sponring A, Filipiak A, Ager C, Schiefecker A, et al. Breath analysis for in vivo detection of pathogens related to ventilator-associated pneumonia in intensive care patients: a prospective pilot study. J Breath Res. (2015) 9(1):016004. doi: 10.1088/1752-7155/9/1/016004
26. Edwards TL, Browne CM, Schoon A, Cox C, Poling A. Animal olfactory detection of human diseases: guidelines and systematic review. J Vet Behav. (2017) 20:59–73. doi: 10.1016/j.jveb.2017.05.002
27. Bean HD, Rees CA, Hill JE. Comparative analysis of the volatile metabolomes of Pseudomonas aeruginosa clinical isolates. J Breath Res. (2016) 10(4):047102. doi: 10.1088/1752-7155/10/4/047102
28. Brooks SW, Moore DR, Marzouk EB, Glenn FR, Hallock RM. Canine olfaction and electronic nose detection of volatile organic compounds in the detection of cancer: a review. Cancer Investig. (2015) 33(9):411–9. doi: 10.3109/07357907.2015.1047510
29. Mallikarjun A, Collins A, Verta A, Hanadari-Levy A, Chaskes MB, Rosen MR, et al. Canine scent detection of sinonasal-inverted papilloma in blood plasma and nasal secretions. J Vet Behav. (2023) 62:29–38. doi: 10.1016/j.jveb.2023.02.008
30. Craig Angle T, Passler T, Waggoner PL, Fischer TD, Rogers B, Galik PK, et al. Real-time detection of a virus using detection dogs. Front Vet Sci. (2016) 2(JAN). doi: 10.3389/fvets.2015.00079
31. Koivusalo M, Vermeiren C, Yuen J, Reeve C, Gadbois S, Katz K. Canine scent detection as a tool to distinguish meticillin-resistant Staphylococcus aureus. J Hosp Infect. (2017) 96(1):93–5. doi: 10.1016/j.jhin.2017.03.005
32. Mallikarjun A, Swartz B, Kane SA, Gibison M, Wilson I, Collins A, et al. Canine detection of chronic wasting disease (CWD) in laboratory and field settings. Prion. (2023) 17(1):16–28. doi: 10.1080/19336896.2023.2169519
33. Seo I-S, Lee H-G, Koo B, Koh CS, Park H-Y, Im C, et al. Cross detection for odor of metabolic waste between breast and colorectal cancer using canine olfaction. PLoS One. (2018) 13(2):1–9. doi: 10.1371/journal.pone.0192629
34. Hackner K, Pleil J. Canine olfaction as an alternative to analytical instruments for disease diagnosis: understanding “dog personality” to achieve reproducible results. J Breath Res. (2017) 11(1):012001. doi: 10.1088/1752-7163/aa5524
35. Horvath G, Andersson H, Nemes S. Cancer odor in the blood of ovarian cancer patients: a retrospective study of detection by dogs during treatment, 3 and 6 months afterward. BMC Cancer. (2013) 13(1):396. doi: 10.1186/1471-2407-13-396
36. Cornu JN, Cancel-Tassin G, Ondet V, Girardet C, Cussenot O. Olfactory detection of prostate cancer by dogs sniffing urine: a step forward in early diagnosis. Eur Urol. (2011) 59(2):197–201. doi: 10.1016/j.eururo.2010.10.006
37. Kybert N, Prokop-Prigge K, Otto CM, Ramirez L, Joffe E, Tanyi J, et al. Exploring ovarian cancer screening using a combined sensor approach: a pilot study. AIP Adv. (2020) 10:035213. doi: 10.1063/1.5144532
38. Otto CM, Sell TK, Veenema TG, Hosangadi D, Vahey RA, Connell ND, et al. The promise of disease detection dogs in pandemic response: lessons learned from COVID-19. Disaster Med Public Health Prep. (2023) 17:e20. doi: 10.1017/dmp.2021.183
39. Murarka M, Vesley-Gross ZI, Essler JL, Smith PG, Hooda J, Drapkin R, et al. Testing ovarian cancer cell lines to train dogs to detect ovarian cancer from blood plasma: a pilot study. J Vet Behav. (2019) 32:42–8. doi: 10.1016/j.jveb.2019.04.010
40. Bryce E, Zurberg T, Zurberg M, Shajari S, Roscoe D. Identifying environmental reservoirs of clostridium difficile with a scent detection dog: preliminary evaluation. J Hosp Infect. (2017) 97(2):140–5. doi: 10.1016/j.jhin.2017.05.023
41. Azad RK, Shulaev V. Metabolomics technology and bioinformatics for precision medicine. Brief Bioinform. (2018) 20(6):1957–71. doi: 10.1093/bib/bbx170
42. Kuil SD, Hidad S, Schneeberger C, Singh P, Rhodes P, de Jong MD, et al. Susceptibility testing by volatile organic compound detection direct from positive blood cultures: a proof-of-principle laboratory study. Antibiotics. (2022) 11(6):705. doi: 10.3390/antibiotics11060705
43. Calonge M, Pinto-Fraga J, González-García MJ, Enríquez-de-Salamanca A, López-de la Rosa A, Fernández I, et al. Severity, therapeutic, and activity tear biomarkers in dry eye disease: an analysis from a phase III clinical trial. Ocul Surf. (2018) 16(3):368–76. doi: 10.1016/j.jtos.2018.05.001
44. Essler JL, Kane SA, Nolan P, Akaho EH, Berna AZ, DeAngelo A, et al. Discrimination of SARS-CoV-2 infected patient samples by detection dogs: a proof of concept study. PLoS One. (2021) 16(4):e0250158. doi: 10.1371/journal.pone.0250158
45. Bauër P, Leemans M, Audureau E, Gilbert C, Armal C, Fromantin I. Remote medical scent detection of cancer and infectious diseases with dogs and rats: a systematic review. Integr Cancer Ther. (2022) 21:15347354221140516. doi: 10.1177/15347354221140516
46. Gordon RT, Schatz CB, Myers LJ, Kosty M, Gonczy C, Kroener J, et al. The use of canines in the detection of human cancers. J Altern Complement Med. (2008) 14(1):61–7. doi: 10.1089/acm.2006.6408
47. Schmidt K, Podmore I. Current challenges in volatile organic compounds analysis as potential biomarkers of cancer. J Biomark. (2015) 2015:981458. doi: 10.1155/2015/981458
48. Kane SA, Lee YE, Essler JL, Mallikarjun A, Preti G, Plymouth VL, et al. Canine discrimination of ovarian cancer through volatile organic compounds. Talanta. (2022) 250:123729. doi: 10.1016/j.talanta.2022.123729
49. Cambau E, Poljak M. Sniffing animals as a diagnostic tool in infectious diseases. Clin Microbiol Infect. (2020) 26(4):431–5. doi: 10.1016/j.cmi.2019.10.036
50. He L, Lv H, Wang Y, Jiang F, Liu Q, Zhang F, et al. Antibiotic treatment can exacerbate biofilm-associated infection by promoting quorum cheater development. npj Biofilms Microbiomes. (2023) 9(1):26. doi: 10.1038/s41522-023-00394-4
51. Bomers MK, van Agtmael MA, Luik H, van Veen MC, Vandenbroucke-Grauls CMJE, Smulders YM. Using a dog’s superior olfactory sensitivity to identify Clostridium difficile in stools and patients: proof of principle study. Br Med J. (2012) 345(dec13 8):e7396. doi: 10.1136/bmj.e7396
52. Waltman CG, Marcelissen TAT, Van Roermund JGH. Exhaled-breath testing for prostate cancer based on volatile organic compound profiling using an electronic nose device (aeonose™): a preliminary report. Eur Urol Focus. (2020) 6(6):1220–5. doi: 10.1016/j.euf.2018.11.006
53. Jendrny P, Twele F, Meller S, Osterhaus ADME, Schalke E, Volk HA. Canine olfactory detection and its relevance to medical detection. BMC Infect Dis. (2021) 21:838. doi: 10.1186/s12879-021-06523-8
54. Catala A, Grandgeorge M, Schaff JL, Cousillas H, Hausberger M, Cattet J. Dogs demonstrate the existence of an epileptic seizure odour in humans. Sci Rep. (2019) 9:4103. doi: 10.1038/s41598-019-40721-4
55. Zhu J, Bean HD, Wargo MJ, Leclair LW, Hill JE. Detecting bacterial lung infections: in vivo evaluation of in vitro volatile fingerprints. J Breath Res. (2013) 7(1):016003. doi: 10.1088/1752-7155/7/1/016003
Keywords: canine detection, Staphylococcus aureus, biofilm, volatile organic compound (VOC), olfaction, periprosthetic joint infection (PJI), culture-negative infection
Citation: Ramos MT, Chang G, Wilson C, Gilbertie J, Krieg J, Parvizi J, Chen AF, Otto CM and Schaer TP (2024) Dogs can detect an odor profile associated with Staphylococcus aureus biofilms in cultures and biological samples. Front. Allergy 5:1275397. doi: 10.3389/falgy.2024.1275397
Received: 9 August 2023; Accepted: 23 January 2024;
Published: 13 February 2024.
Edited by:
Gabriele Magna, University of Rome Tor Vergata, ItalyReviewed by:
Leticia Gomes De Pontes, University of São Paulo, BrazilAdee Schoon, Animal Detection Consultancy, Netherlands
© 2024 Ramos, Chang, Wilson, Gilbertie, Krieg, Parvizi, Chen, Otto and Schaer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cynthia M. Otto Y21vdHRvQHZldC51cGVubi5lZHU= Thomas P. Schaer dHBzY2hhZXJAdmV0LnVwZW5uLmVkdQ==
 Meghan T. Ramos
Meghan T. Ramos Gerard Chang
Gerard Chang Clara Wilson
Clara Wilson Jessica Gilbertie
Jessica Gilbertie James Krieg4
James Krieg4 Javad Parvizi
Javad Parvizi Antonia F. Chen
Antonia F. Chen Cynthia M. Otto
Cynthia M. Otto Thomas P. Schaer
Thomas P. Schaer