- 1Department of Chemistry & Biochemistry, College of Science, George Mason University, Fairfax, VA, United States
- 2Max-Planck-Institute for Heart and Lung Research, Bad Nauheim, Germany
- 3Department of Clinical Research, Amerimmune LLC, McLean, VA, United States
- 4Department of Flowcytometry, Amerimmune LLC, McLean, VA, United States
- 5Center for Molecular Engineering, George Mason University, Manassas, VA, United States
- 6Department of Bioengineering, College of Engineering & Computing, George Mason University, Fairfax, VA, United States
Accurate diagnostic tools for allergic conditions are essential for effective treatment. Traditional methods, such as skin prick tests (SPT) and specific IgE measurements are widely used, but they have limitations in sensitivity and specificity for certain allergens. While the Basophil Activation Test (BAT) offers improved specificity, particularly for allergens such as peanuts and sesame, its practicality and accessibility remain challenges. Mass spectrometry (MS) has recently gained recognition as a promising complementary tool in allergy diagnostics, offering high analytical precision and the capability to detect a wide range of allergen-specific biomarkers. This review explores the integration of MS into allergy diagnostics, emphasizing its potential to enhance BAT applications, particularly for non-responders. We discuss the underlying mechanisms, recent research highlighting its efficacy, and the technical challenges that must be addressed for clinical adoption. Additionally, we examine the standardization requirements and ethical considerations necessary for MS to become a routine diagnostic tool. Finally, we consider the future of allergy diagnostics, highlighting how MS technology could contribute to more precise, personalized, and patient-centered care in allergy management.
1 Introduction
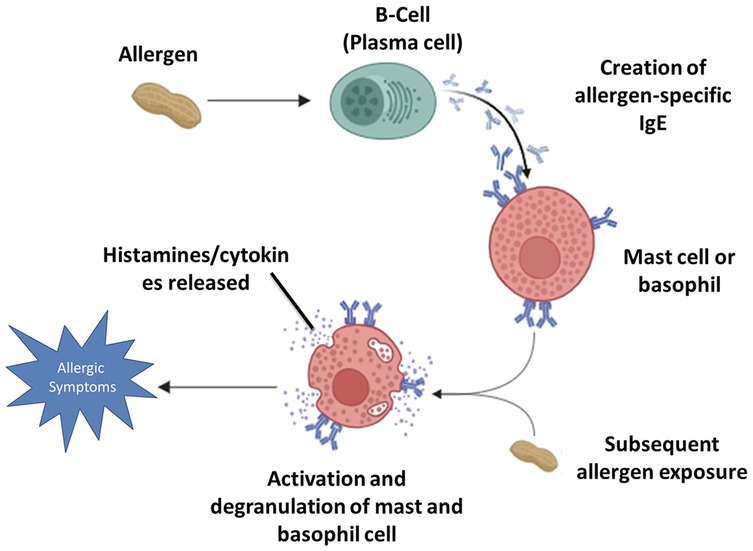
Food allergic reactions begin when the immune system mistakenly identifies a harmless food protein as a threat. This leads to the activation of B-cells (plasma cells), which produce allergen-specific Immunoglobulin E (IgE) antibodies (1–3). These antibodies then bind to high-affinity FcεRI receptors on the surface of mast cells and basophils, a process known as sensitization. Upon subsequent exposure, the allergen cross-links the IgE antibodies on mast cells and basophils, triggering cellular activation and degranulation. During an IgE-mediated allergic reaction, the immune activation leads to the release of both pre-formed mediators stored in granules and newly synthesized molecules that contribute to the inflammatory response. Pre-formed mediators, such as histamine and tryptase, are rapidly released upon mast cell and basophil degranulation. Histamine plays a central role in the immediate hypersensitivity response by inducing vasodilation, increased vascular permeability, smooth muscle contraction, and pruritus. Tryptase, primarily secreted by mast cells, serves as a biomarker of mast cell activation and contributes to tissue remodeling and inflammation. In contrast, newly synthesized mediators, including leukotrienes, cytokines, and prostaglandins, are produced de novo following cellular activation (4–8). Leukotrienes, such as LTC₄, LTD₄, and LTE₄, contribute to prolonged bronchoconstriction, mucus secretion, and increased vascular permeability. Key cytokines, including IL-4, IL-5, IL-13, and TNF-α, regulate immune responses by promoting eosinophil recruitment, enhancing IgE production, and sustaining chronic inflammation. Additionally, prostaglandins, particularly Prostaglandin D₂ (PGD₂), play a role in bronchoconstriction, vasodilation, and immune cell recruitment (9, 10). Collectively, these mediators drive both the early-phase and late-phase allergic responses (11–15). These biochemical changes manifest clinically as allergic symptoms, ranging from mild reactions such as skin rashes, itching, and nasal congestion to severe respiratory distress, gastrointestinal issues, and life-threatening anaphylaxis.
The IgE-mediated responses tend to be immediate, causing a rapid onset of symptoms, while non-IgE-mediated reactions typically manifest later (2, 16, 17). Figure 1 depicts the mechanism of IgE-mediated allergic reactions described above.
The Müller classification categorizes allergic reactions into four grades based on severity, providing a structured framework for clinical assessment. A Grade I reaction includes mild symptoms such as skin flushing, urticaria, and mild angioedema without respiratory or cardiovascular involvement. Grade II reactions are moderate and involve more pronounced symptoms, such as difficulty breathing, wheezing, nausea, and mild hypotension, but without life-threatening manifestations. Grade III reactions are severe and, characterized by life-threatening airway obstruction, bronchospasm, or severe hypotension. Grade IV represents the most critical cases, with circulatory and respiratory failure (16, 18, 19).
The IgE-mediated reactions, in particular, are associated with the most severe cases of food allergies, emphasizing the critical need for accurate diagnosis and effective management to prevent accidental exposure to allergens (20–22). Therefore, IgE-mediated food allergy diagnosis will be our focus in this study.
The gold standard of food allergy diagnosis is the oral food challenge (OFC). The intentional exposure to multiple allergens in OFCs introduces a considerable risk to patients, as it inherently involves provoking allergic reactions (23).
Other diagnostic techniques like the Skin Prick Test (SPT), Specific IgE (sIgE) testing, and the Basophil Activation Test (BAT), are known to be ineffective or inaccurate in some situations. They often require supplementation with patient-reported symptoms or clinical history, which increases the risk of mischaracterizing food sensitivities as true allergies, leading to inappropriate and false-positive diagnoses (24–28).
The SPT test involves the application of small amounts of potential allergens to the skin, typically on the forearm. Although it is generally safe, it can have issues, such as the possibility of false positives, especially with certain foods. While elevated IgE levels alone do not confirm sensitization, higher IgE concentrations are associated with an increased likelihood of clinical allergy and greater severity of allergic reactions. Therefore, it must be used in conjunction with other diagnostic methods to provide a more comprehensive assessment (29–32).
The sIgE testing is a valuable diagnostic tool used to measure allergen-specific immunoglobulin E antibodies in the bloodstream, enabling the identification of sensitization to particular allergens. This method is especially effective for diagnosing IgE-mediated (Type I) hypersensitivity reactions, including those related to food, environmental allergens, and insect venom. Unlike SPT, sIgE assays carry no risk of provoking an allergic response, making them suitable for patients with dermatologic conditions, those on antihistamines, or individuals unable to undergo skin testing. Commonly employed platforms, such as ImmunoCAP and Immulite, provide quantitative assessments of allergen-specific IgE levels, supporting clinicians in the evaluation of allergy severity, monitoring of therapeutic interventions, and formulation of targeted management strategies (33, 34).
The BAT demonstrates high sensitivity and negative predictive value (NPV) across various food allergens, making it a reliable diagnostic tool. For peanut allergy, BAT shows 75% sensitivity and 98% specificity, effectively distinguishing allergic individuals. In cow's milk allergy, it has 89% sensitivity, 83% specificity, and a 96% NPV, strongly predicting tolerance when negative. For egg allergy, BAT's sensitivity ranges from 63%–77%, with 96%–100% specificity, particularly when assessing CD63 expression. These values highlight BAT's role in accurately ruling out allergies and reducing unnecessary dietary restrictions (35–37).
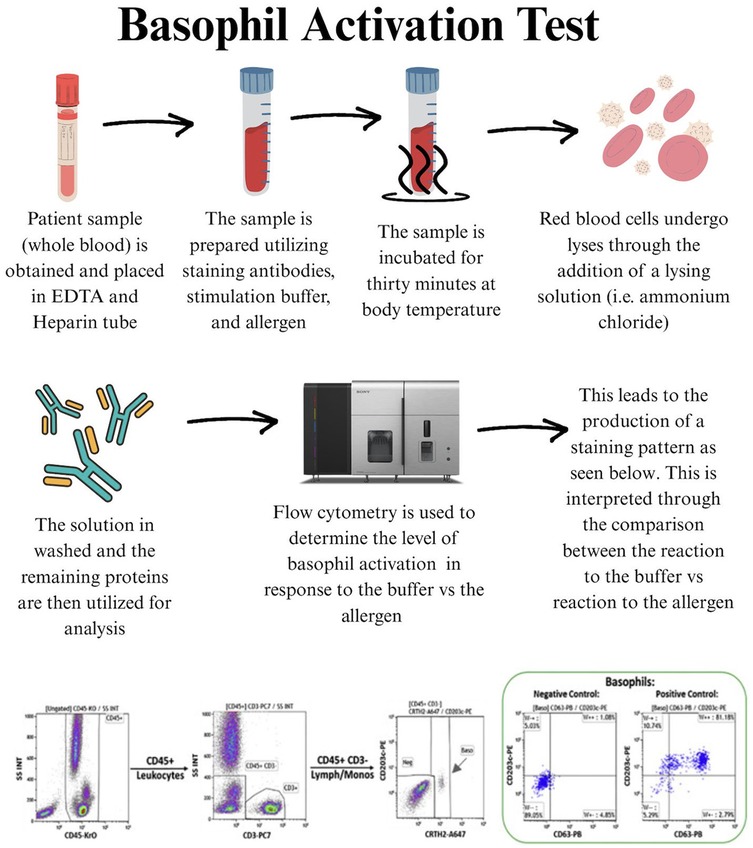
Also, the BAT is a functional diagnostic tool that evaluates the activation of basophils in response to allergens. This test utilizes flow cytometry to measure the translocation of activation markers to the surface of the basophils when exposed to specific allergens. The BAT offers a more specific alternative to sIgE and SPT by directly measuring cellular reactivity to allergens. The BAT has the advantage of reducing false positives and better predicts clinical reactivity, potentially enhancing diagnostic accuracy in food allergy assessments (38–41, 42). The test works by exposing basophils in a blood sample to allergens and detecting surface markers such as CD63 and CD203c via flow cytometry, signaling an allergic response (43, 44). Figure 2 illustrates the typical workflow for the BAT test. Despite the increased specificity of BAT, 15%–20% of people tested have basophils that fail to respond to IgE-mediated stimulation, likely due to a mutation in the promoter of the SYK gene (45).
The SPT and specific sIgE tests serve as first-line diagnostic tools for food allergies; however, both have limitations in specificity, which can lead to false-positive results. The BAT, a second-line diagnostic tool, offers improved specificity by directly assessing cellular reactivity, yet it may still be affected by basophil non-responsiveness in certain individuals. The introduction of component-resolved diagnostics (CRD) has significantly enhanced specificity by identifying allergen-specific IgE at the molecular level, thereby reducing misclassification and improving diagnostic accuracy when used in conjunction with BAT tests.
The Basophil Activation Test (BAT) does not always exhibit high sensitivity, with values varying across allergens. For Example, peanut allergy sensitivity is around 75%, cow's milk allergy reaches 89%, while egg allergy can be as low as 63% (46). However, BAT's specificity is particularly high often exceeding 95%, making it highly valuable when skin prick tests (SPT) and specific IgE levels yield inconclusive results in the “gray zone.” For peanut allergy, BAT has 98% specificity, for egg allergy 96%–100%, and for cow's milk allergy around 83%, ensuring fewer false positive results and reducing the need for oral food challenges (OFC), which, while effective, can be burdensome and carry inherent risks (39, 47, 48).
Mass Spectrometry (MS) technology is increasingly employed in medical diagnostics due to its high sensitivity, specificity, and ability to analyze complex biological samples with great precision. For instance, tandem mass spectrometry (MS/MS) plays a vital role in newborn screening by analyzing dried blood spots to detect metabolic disorders such as phenylketonuria (PKU) and other inborn errors of metabolism with high accuracy and sensitivity (49–51). Furthermore, MS played a vital role during the COVID-19 pandemic by aiding in the structural analysis of SARS-CoV-2 proteins, which contributed to the development of diagnostics and therapeutics, showcasing its versatility in addressing emerging global health challenges (52–54).
One promising application of MS is in food allergy diagnostics, where its enhanced sensitivity, specificity, and high-throughput analytical capabilities could significantly improve the detection and characterization of allergenic reactions. It is estimated that more than 10% of the US population suffers from at least one food allergy, with rates continuing to rise globally (7). Given the challenges of current diagnostic tools, there is a dire need for more accurate and reliable methods for allergy diagnosis which represents an opportunity to leverage MS's unparalleled specificity (55). Through direct identification and quantification of allergens and specific antibodies such as IgE based on their unique mass-to-charge ratios, MS has the potential to offer insights into individual sensitivities and immune responses.
Although clinical applications of MS are still developing, it holds promise for advancing the management of food allergies. This review examines the potential application of MS in food allergy diagnostics, focusing on its role in quantifying immunoglobulins such as IgE and its advantages over current assays. Additionally, we explore how MS could enhance diagnostic precision and enable personalized allergy management, addressing existing gaps in sensitivity, specificity, and clinical applicability in food allergy testing.
2 Time dependent response of basophils
It was demonstrated that the BAT, performed by measuring basophil surface markers and intracellular phosphorylation of signaling molecules over a 20-min period at intervals ranging from seconds to several minutes (e.g., 1, 3, 5, 10, and 20 min, or other combinations), correlates with the severity of allergic reactions. Allergens are applied at varying concentrations ranging from 0.1 to 100,000 ng/ml.
First, whole blood is collected from subjects and incubated with the allergen for varying durations and concentrations. Blood cells are subsequently analyzed for the expression of cell surface markers, such as CD203c and CD63, and intracellular phosphorylation markers, including phospho-Lyn, phospho-Fyn, and phospho-IgE receptors. Remarkably, the phosphorylated targets, including the IgE receptor, Fyn, and Lyn, are proximal to Syk kinase and remain unaffected by the absence of Syk. This characteristic enables the assessment of patients with non-reactive basophils based on conventional surface markers (e.g., CD63 or CD203c), who might otherwise not respond to traditional BAT. This method is proposed for use in diagnosing or monitoring allergies and evaluating the effectiveness of therapeutic interventions based on the responsiveness of an individual being treated (37).
A key advantage of this approach is its ability to address the issue of non-releaser or low-responder basophils, observed in 10%–20% of the population (35, 56). Additionally, measurement of the rate of upregulation of these markers over short time intervals introduces a novel aspect, enhancing the sensitivity and specificity of BAT for determining clinical reactivity. Furthermore, an inhibitor of basophil activation (btk inhibitor Ibrutinib) can be used. Btk inhibitors inhibit the activation of the IgE stimulation pathway. This assesses if basophil activation induced by the allergen is specific, as the inhibitor solely impacts the IgE pathway but not the fMLP (N-Formylmethionyl-leucyl-phenylalanine) induced basophil activation. FMLP is a positive control used to assess the response of basophils to a non-IgE pathway stimulation, and overall basophil reactivity. Figure 3 shows the phosphorylation of basophil phospho-proteins upon stimulation by PBS or anti-IgE (3 A-D) and CD63 upregulation over a range of incubation times (3 E). We are currently exploring the value of phospho-markers as well as the time course of the BAT assay in larger cohorts of milk, peanut, and egg allergy.
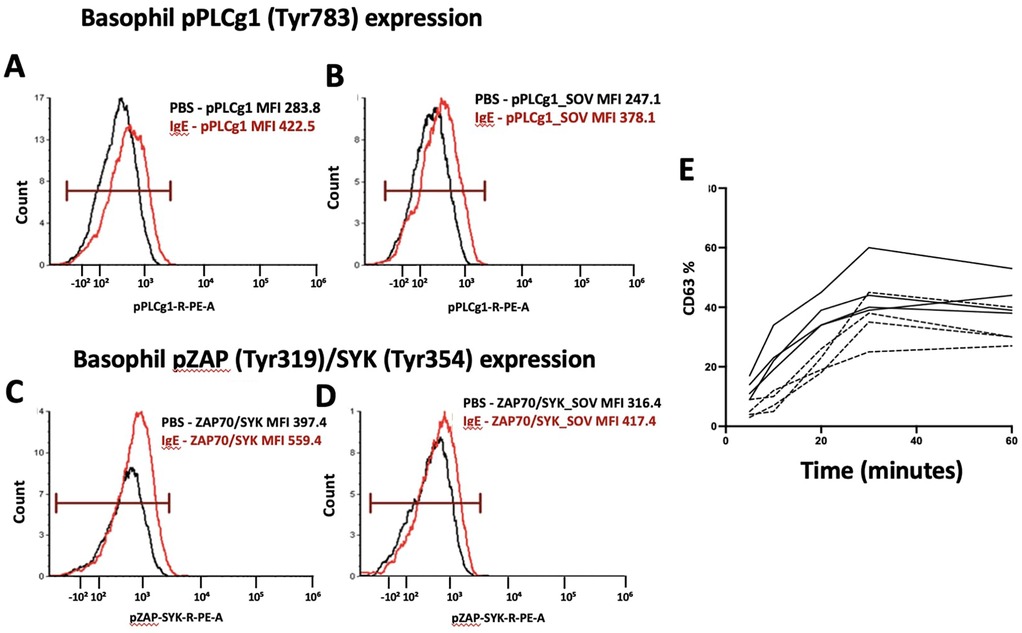
Figure 3. Phosphorylation of signaling proteins in basophils after IgE stimulation. (A,B) Expression of phosphorylated PLCγ1 (pPLCγ1, Tyr783) in basophils treated with PBS (black) vs. IgE (red). (C,D) Expression of phosphorylated ZAP70/SYK (pZAP, Tyr319/Tyr354) in basophils treated with PBS (black) vs. IgE (red). (E) Basophil CD63 upregulation in patients with mild peanut allergy with perioral hives (dashed line) vs. full anaphylactic reaction (solid line) requiring epinephrine.
Basophil activation through the IgE pathway can be inhibited by the btk inhibitor Ibrutinib. This allows the assessment for the non-specific (e.g., non-IgE-dependent) stimulation of basophils, as fMLP stimulation of basophils is not affected by btk inhibitor ibrutinub and other btk inhibitors at concentrations above 0.01 µM. Figure 4. demonstrates the inhibitory effect of Ibrutinib, a BTK inhibitor, on basophil activation as measured by CD63 surface expression. The percentage of CD63-positive basophils decreases with increasing Ibrutinib concentrations (zero to one µM) across various stimuli, including anti-IgE, fMLP, and food allergens (almond, peanuts, and wheat). At higher concentrations (one µM), Ibrutinib significantly reduces allergen-induced basophil activation, indicating its potential to suppress allergic responses.
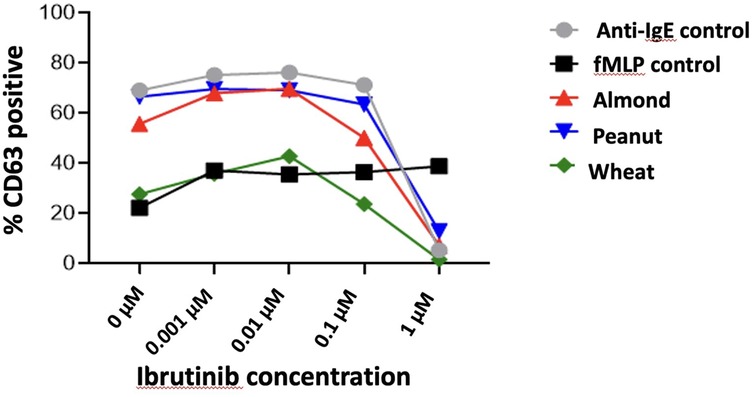
Figure 4. Effect of Ibrutinib on basophil activation and CD63 surface expression across different stimuli.
3 Mass spectrometry in allergy diagnosis
Mass spectrometry (MS) is emerging as a powerful tool in allergy diagnosis, offering high sensitivity, specificity, and multiplexing capabilities to detect allergen-specific biomarkers. Unlike traditional immunoassays, MS enables precise identification and quantification of allergenic proteins, peptides, and immune mediators directly from clinical samples such as serum, plasma, or nasal secretions. It can be used to detect specific IgE-bound allergenic epitopes, characterize allergen isoforms, and measure released mediators like histamine and leukotrienes during allergic reactions. Additionally, MS allows for the detailed profiling of post-translational modifications that may influence allergenicity. Its ability to provide absolute quantification and molecular-level resolution makes MS a valuable complement to standard diagnostic tools such as skin prick testing and ELISA, particularly in complex or ambiguous cases.
3.1 Application of MS in immunoglobulin quantification: advancing IgG and IgE detection for allergy diagnostics
The direct quantification of IgE antibodies using MS in allergy diagnostics remains a largely unexplored area, with few studies specifically addressing this application. To date, most MS-based research on immunoglobulin quantification has focused on immunoglobulin G (IgG), which is present in higher concentrations and has more established analytical methods in human samples. The limited exploration of IgE quantification via MS can be attributed to its significantly lower serum concentration and the widespread use of standardized immunoassays, such as Enzyme-Linked Immunosorbent Assay (ELISA) for IgE detection. Both IgG and IgE are immunoglobulins with different serum concentrations and functions. The IgG is more abundant and is responsible for long-term immunity, whereas IgE is involved in allergic responses and is present at much lower concentrations in serum. Despite these differences, the methodologies used for IgG quantification via MS can be employed to quantify IgE (57–59).
Studies on IgG quantification via MS have demonstrated that specific proteotypic peptides unique to the IgG molecule can be used as proxies for the whole antibody, allowing precise measurement in biological samples. One example is the MASCALE (Mass Spectrometry Enabled Conversion to Absolute Levels of ELISA Antibodies) method, which was developed to convert ELISA-derived values into absolute concentrations of IgG by calibrating the mass spectrometry signal of IgG-specific peptides (60). In this method, peptides such as VVSVLTVLHQDWLNGK, which are unique to the IgG subclass (IgG1, IgG3, and IgG4), were used to establish calibration curves that correlate the MS signal (peak area ratio) with the actual concentration of IgG antibodies in serum samples. This method has shown high accuracy and precision (60). It was suggested that MASCALE must be used alongside ELISA to improve the interpretation of immune responses, offering more precise and standardized quantification of antibodies in clinical trials (60).
Furthermore, microflow LC-MS/MS-PRM was recruited to quantify glycopeptides in serum, targeting specific N-glycoforms of IgG, and demonstrating the method's capability to distinguish between eight different N-glycoforms of IgG together with two O-glycoforms of hemopexin (HPX) in relation to disease biomarkers. This study introduced a targeted microflow liquid chromatography-tandem mass spectrometry in parallel reaction monitoring mode (LC–MS/MS-PRM) method for quantifying multiple glycopeptides in unfractionated serum samples, enabling precise and simultaneous quantification of these glycoforms. The application of this assay to patients with hepatitis C virus (HCV)-induced liver fibrosis demonstrated that specific IgG and HPX glycoforms could effectively detect fibrotic disease at varying stages (61). These findings highlight the potential of LC–MS/MS-PRM assays for rapid and reproducible biomarker assessments, targeting both N- and O-glycoforms of peptides, thereby advancing the clinical application of LC–MS/MS-based diagnostics (61). Additionally, a complementary study introduced a chemoenzymatic strategy for synthesizing isotopically labeled glycopeptides of IgG1, which were incorporated into an optimized LC-MS/MS-PRM workflow. The use of stable isotope-labeled N-acetylglucosamine facilitated highly accurate quantification of IgG glycoforms, with minimal variability and enhanced sensitivity through both Electron Transfer Dissociation (ETD) and Higher-energy Collisional Dissociation (HCD) workflows (62). This approach was exemplified in a rapid (13-minute) quantification of IgG1 Fc glycoforms in COVID-19 patients. Together, these methodologies underscore the versatility and efficiency of MS in high-throughput immunoglobulin quantification, offering valuable insights and a potential framework for adapting similar approaches to IgE analysis (61, 62).
In a different approach, immunoaffinity capillary electrophoresis (IACE) coupled with Matrix-Assisted laser Desorption Ionization MALDI-MS was employed to diagnose cow's milk allergy by quantifying the Specific IgE in the serum of allergic patients (63). In MALDI-MS, the sample is co-crystallized with a matrix—a small organic compound that absorbs the laser energy, allowing for the gentle ionization of large molecules without fragmentation. Upon laser irradiation, the matrix absorbs energy and transfers it to the analyte, facilitating its ionization and subsequent detection by a Time Of Flight (TOF) mass analyzer (64). It is widely used in proteomics, biomarker discovery, and clinical diagnostics due to its high throughput, minimal sample preparation, and ability to analyze complex biological mixtures. Moreover, it is particularly advantageous for detecting intact proteins and post-translational modifications, making it a valuable tool for IgG quantification and characterization in serological studies (63).
Magnetic beads (MBs) coated with anti-IgE antibodies were used to isolate and quantify total IgE, providing a general allergy diagnosis. Subsequently, specific allergens, such as bovine serum albumin, Lactoferrin, and α-casein were identified as key allergens responsible for eliciting the allergic reaction (65). By chemically cross-linking the immunocomplex formed during the IgE quantification phase, these allergenic proteins were then detected using MALDI-MS, which directly identified the molecular masses and structures of the allergens. This method not only enabled precise identification of the allergens responsible for the allergic reaction but also allowed for the use of actual food extracts, which could lead to more tailored allergy diagnostics and epitope mapping. The detection was sensitive and required 2 µl of blood serum, making MALDI-MS a highly efficient tool in allergy diagnostics.
Similarly, Ultra-Performance Liquid Chromatography coupled with mass spectrometry (UPLC-MS) was recruited for the quantification of total IgE in human serum (66). The UPLC-MS uses columns packed with smaller particle sizes (typically ≤2 µm) and operates at higher pressures, leading to faster analysis, improved peak resolution, and greater sensitivity. When coupled with magnetic beads (MS), UPLC enables precise identification and quantification of small molecules, peptides, proteins, and metabolites (66).
The MBs coupled with anti-IgE antibodies were used to extract and quantify IgE via a signal peptide after digestion. This method demonstrated high sensitivity, with limits of detection and quantification at 400 ng/ml and 800 ng/ml, respectively (66). By evaluating the binding capacity of the extracted IgE with different allergens, the study successfully identified allergens that induced allergic responses in patients. This MS-based method quantified total IgE and pinpointed the specific allergens involved, offering an advanced tool for allergy diagnosis with high precision. The method's effectiveness was shown by its application in analyzing serum samples from allergic and healthy individuals, further underscoring its clinical relevance in improving allergy detection and improving therapeutic decision-making. Figure 5 displays a general workflow for the clinical diagnosis of allergens, exemplifying advanced methodologies for sensitive and selective allergen detection utilizing MBs and UPLC-MS/MS technology.
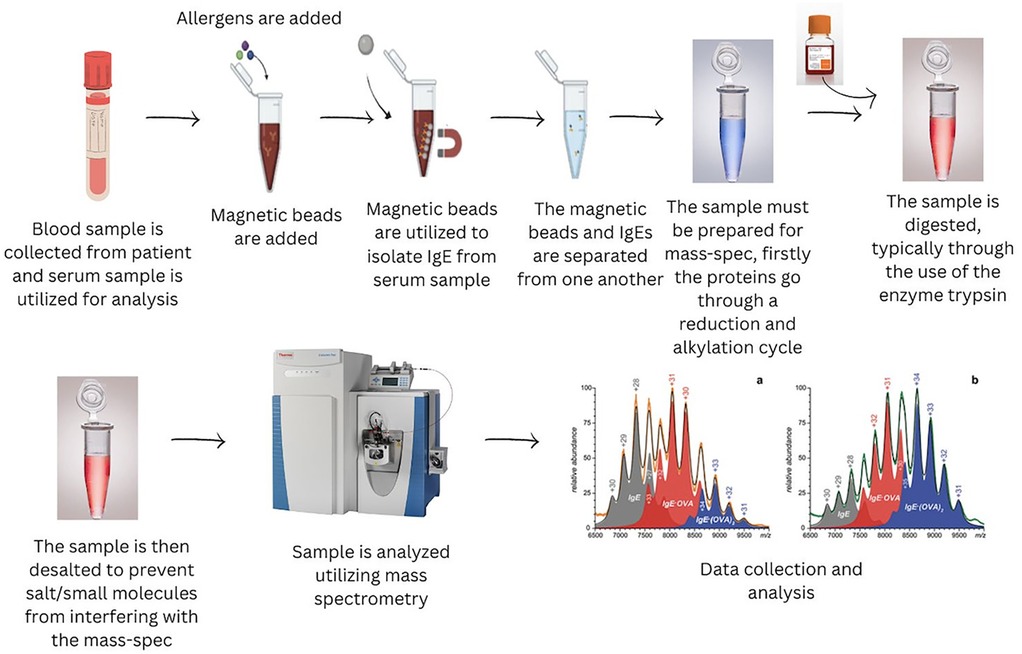
Figure 5. A workflow incorporating extraction of allergens with magnetic beads for UPLC –MS/MS analysis.
3.2 Using MS to identify other early phosphoproteins in determining basophil activation
A critical component of the signaling pathway leading to basophil activation is spleen tyrosine kinase (Syk). Syk is essential for transmitting signals from the high-affinity IgE receptor (FcεRI) upon allergen binding, culminating in basophil degranulation and the release of inflammatory mediators (67, 68). Phosphorylation of Syk propagates downstream signaling events that result in calcium mobilization, degranulation, and the release of inflammatory mediators, ultimately driving the basophil's role in allergic responses. However, studies have discovered that a subset of individuals, referred to as “non-releasers,” possess basophils that fail to release histamine upon IgE receptor cross-linking due to the absence or significant reduction of Syk protein expression. This deficiency impairs the FcεRI signaling pathway, leading to false-negative results in BAT and challenging the reliability of standard basophil-based allergy diagnostics for these individuals (69, 70). To overcome these challenges, alternative methods have been explored to assess basophil activation in Syk-deficient individuals. One approach involves using non-IgE-dependent stimulants, such as N-formyl-methionyl-leucyl-phenylalanine (fMLP) or interleukin-3 (IL-3), which can activate basophils through alternative pathways (71). Additionally, measuring intracellular signaling markers like p38 MAPK and ERK1/2, or assessing the release of mediators such as histamine or leukotriene C4 (LTC4) by MS, may provide alternative indicators of basophil responsiveness (72, 73). Furthermore, ex vivo whole blood stimulation assays using cytokines like IL-3 or IL-33 have been proposed to characterize basophil activation when standard BAT markers fail. These alternative methods aim to provide a more comprehensive assessment of allergic responses, especially in individuals with atypical basophil activation profiles (74).
3.3 Quantifying signaling thresholds and post-translational modifications in basophil activation research using MS
One of the most promising applications of MS in basophil activation research is its capacity to quantify signaling thresholds and post-translational modifications (PTMs) of key regulatory proteins. A critical target in this context is CD45, a transmembrane protein tyrosine phosphatase that modulates Src family kinase (SFK) activity and plays an essential role in determining basophil activation potential. Dysregulated CD45 function can disrupt phosphorylation cascades downstream of FcεRI engagement, potentially leading to hypo-responsiveness in certain individuals. By utilizing MS-based phosphoproteomics, it is possible to detect differential phosphorylation patterns of CD45, providing mechanistic insights into its role in either facilitating or inhibiting early basophil activation events (75, 76).
Immunoreceptor Tyrosine-Based Activation Motifs (ITAMs) are conserved sequences found in the cytoplasmic tails of various immune receptors. They play a crucial role in signal transduction by recruiting and activating tyrosine kinases upon receptor engagement. MS offers a highly precise approach for characterizing ITAMs within FcεRI-associated signaling complexes, which play a crucial role in initiating allergic immune responses. Also, ITAM phosphorylation acts as a key regulatory checkpoint, governing the recruitment and activation of Syk kinase, a central mediator of intracellular signaling cascades downstream of FcεRI engagement. Disruptions in the phosphorylation dynamics of these motifs can significantly alter downstream signaling, contributing to immune dysregulation and hypo-responsiveness in allergic conditions (35, 77, 78). By leveraging targeted MS techniques such as selected reaction monitoring (SRM) and parallel reaction monitoring (PRM), researchers can achieve highly sensitive and quantitative measurements of ITAM phosphorylation states. This enables the detection of subtle alterations in FcεRI-mediated signal transduction, particularly in cases where basophils exhibit reduced responsiveness. Such quantitative insights are invaluable for identifying regulatory disruptions that may underlie immune tolerance, aberrant signaling in allergic diseases, or therapeutic resistance in conditions modulated by FcεRI activation.
3.4 Expanding the scope: early phosphoproteins in basophil activation
Beyond ITAM phosphorylation, MS can be employed to investigate a broader network of early phosphoproteins involved in basophil activation. LAT (Linker for Activation of T cells) and SLP-76 (SH2 domain-containing leukocyte protein of 76 kDa) serve as essential scaffolding proteins facilitating signal transduction from FcεRI to downstream effector pathways. Phosphorylation of these adaptor proteins plays a crucial role in orchestrating intracellular signaling events that determine basophil reactivity. Quantitative phosphoproteomics can be recruited to map phosphorylation sites on LAT and SLP-76, identifying alterations in their activation states that may contribute to immune dysregulation in basophilic non-responders (79–84).
Another key signaling molecule is PLCγ (Phospholipase C gamma), which is activated downstream of FcεRI signaling and is responsible for hydrolyzing phosphatidylinositol 4, 5-bisphosphate (PIP2) into inositol trisphosphate (IP3) and diacylglycerol (DAG) (85, 86). These second messengers are essential for calcium mobilization and protein kinase C (PKC) activation, both crucial for basophil degranulation. Using MS-based site-specific phosphorylation analysis, we can evaluate the activation state of PLCγ and identify potential dysfunctions that may lead to impaired histamine release and allergen-specific immune responses (80, 85, 86).
Moreover, Grb2-associated binding protein 2 (Gab2) functions as a docking protein that facilitates interactions with PI3 K (Phosphoinositide 3-kinase), influencing downstream Akt/mTOR signaling. Dysregulation of Gab2 phosphorylation may impact basophil survival and cytokine release, providing another avenue for MS-driven biomarker discovery. PRM-MS and phosphosite mapping could help elucidate changes in Gab2 phosphorylation that correlate with distinct basophil activation phenotypes (87–89).
3.5 Alternative activation pathways: IgE-independent basophil activation
Given that not all basophil activation is driven by FcεRI engagement, MS also plays a pivotal role in characterizing IgE-independent signaling pathways. Complement receptor C5aR (CD88) mediates basophil activation in response to C5a-C5aR interactions, serving as an alternative activation axis (90, 91). MS-based approaches allow for precise quantification of C5aR expression levels, the characterization of its PTMs (such as phosphorylation and glycosylation), and the assessment of downstream effector pathways. This is particularly relevant in individuals exhibiting non-responder phenotypes in the BAT test, where FcεRI-mediated activation is impaired, yet complement-driven basophil activation remains functional. By integrating quantitative proteomics, phosphoproteomics, and glycoproteomics, MS can provide a systems-level understanding of the molecular architecture underpinning basophil reactivity. These analyses are critical for refining diagnostic strategies and uncovering novel therapeutic targets in allergic diseases.
3.6 Kinase activity profiling in basophil signaling
MS-based approaches can be used for quantifying kinase activity within basophil signaling networks, enabling precise analysis of activation states and regulatory mechanisms. One particularly relevant kinase in this context is Lyn (Lck/Yes-related novel tyrosine kinase), a key member of the Src family kinases (SFKs). Notably, Lyn remains expressed at near-normal levels in non-releaser basophils, a subset of basophils that exhibit impaired degranulation despite activation stimuli (92).
Lyn plays a dual role in basophil activation, functioning both as a priming kinase that facilitates intracellular signaling and as a negative regulator that modulates FcεRI (high-affinity IgE receptor) and C5aR (complement component 5a receptor)-mediated pathways. Its ability to fine-tune these pathways is critical for maintaining immune balance (93, 94).
By utilizing phosphosite-specific enrichment (a targeted method for isolating phosphorylated peptides) in combination with tandem MS, LC-MS/MS), researchers can map Lyn activation kinetics with high resolution. This approach provides critical insights into dysregulated phosphorylation cascades, particularly in hypo-responsive basophils, where altered kinase activity may underlie impaired immune responses and allergic dysfunction.
Beyond Lyn, MS-based kinase assays can also assess activity levels of Syk, Btk (Bruton's tyrosine kinase), and PI3 K, which are key regulators of intracellular basophil signaling. Dysregulation of these kinases may contribute to impaired degranulation, cytokine release, and cellular priming, offering potential biomarkers for distinguishing between intrinsic activation defects and pathway-specific hypo-responsiveness (95).
3.7 MS in cytokine receptor signaling and alternative immune pathways
In addition to its role in receptor-mediated activation, MS can be used to quantify cytokine receptor signaling pathways involving interleukin-3 (IL-3), interleukin-4 (IL-4), and interleukin-5 (IL-5) (96, 97). These cytokines are crucial for basophil priming, survival, and effector function, and their receptors engage the Janus kinase (JAK)-STAT signaling cascade (98, 99). MS-based proteomic profiling facilitates: the quantification of receptor abundance to determine basophil responsiveness to cytokine stimulation, ligand-binding dynamics assessment to evaluate cytokine-receptor interactions, and downstream phosphorylation event analysis to identify molecular signatures of cytokine responsiveness. Such insights are particularly valuable for identifying alternative activation pathways in patients with atypical allergic phenotypes, where traditional IgE-centric diagnostics fail to capture the full spectrum of immune dysregulation. By expanding our understanding of basophil signaling networks through MS-based methodologies, clinicians and researchers can develop more comprehensive diagnostic tools and targeted therapeutic strategies to manage allergic and inflammatory conditions effectively.
4 Challenges and future directions of mass spectrometry in allergy diagnostics
Despite its immense potential in allergy diagnostics, the integration of MS into routine clinical workflows faces substantial technical, operational, and regulatory challenges that must be addressed to facilitate widespread clinical adoption.
One of the primary hurdles in applying MS to allergy diagnostics is the accurate quantification of proteins. Unlike traditional immunoassay-based techniques, such as ELISA, which measures total protein concentrations directly via antibody-antigen interactions, MS-based approaches typically rely on peptide-level quantification following enzymatic digestion (100). This introduces inherent variability due to factors such as differential peptide ionization efficiency, variations in post-translational modifications (PTMs), and proteolytic cleavage efficiency. These inconsistencies can lead to inaccurate quantification, which is particularly problematic in allergy diagnostics, where precise protein concentration measurements are critical for clinical decision-making (101).
To improve the reliability of MS-based protein quantification, absolute quantification techniques, such as isotope dilution MS (IDMS) and targeted multiple reaction monitoring MS (MRM-MS), have been developed. These methods incorporate stable isotope-labeled internal standards that correct for sample preparation and ionization variability (102). However, further refinement is needed to enhance sensitivity, reproducibility, and robustness, particularly for low-abundance allergenic proteins and IgE antibodies, which are often present in minute concentrations. Additionally, optimizing sample digestion protocols and developing improved enrichment strategies, such as immunoprecipitation-MS or affinity-based purification, could help mitigate issues related to peptide fragmentation variability and enhance detection sensitivity (103).
Mass spectrometers are inherently complex, requiring sophisticated hardware, precise calibration, and highly trained personnel for operation and data interpretation. Unlike immunoassays or flow cytometry-based techniques like the BAT, which are relatively straightforward to implement in clinical laboratories, MS-based workflows involve extensive sample preparation, advanced liquid chromatography separations, and rigorous bioinformatics processing. These factors significantly limit the accessibility of MS for routine allergy diagnostics, particularly in non-specialized clinical settings.
Automation and user-friendly software solutions are critical to overcoming these barriers. Advances in robotic sample preparation, microfluidics-based workflows, and artificial intelligence (AI)-driven data analysis platforms could simplify MS-based diagnostics and reduce reliance on highly trained personnel. AI-powered algorithms capable of automated spectral deconvolution, feature extraction, and real-time data interpretation could improve diagnostic accuracy and reproducibility. Furthermore, the development of ambient ionization techniques, such as paper spray ionization (PSI-MS) and matrix-assisted laser desorption/ionization (MALDI-MS), offers the potential for more streamlined workflows by reducing the need for extensive chromatographic separations and simplifying sample preparation.
For MS to be successfully incorporated into allergy diagnostics, standardization, and regulatory approval are critical (104, 105). Unlike conventional immunoassays, which rely on well-characterized antibodies and established clinical thresholds, MS-based approaches lack universally accepted reference materials, standardized protocols, and validated biomarkers for allergy-related proteins (106). This lack of standardization complicates inter-laboratory reproducibility and poses significant challenges for regulatory approval (107).
To address these challenges, global efforts should focus on developing certified reference materials for allergenic proteins and IgE antibodies, establishing harmonized sample preparation and analytical protocols, and implementing robust quality control (QC) measures across laboratories. Regulatory agencies, such as the FDA and EMA, will require comprehensive validation studies demonstrating the clinical reliability, sensitivity, and specificity of MS-based assays before approving their use in diagnostic settings. Collaborative initiatives between academic institutions, regulatory bodies, and industry partners will be essential in driving these standardization efforts forward.
The future of MS in allergy diagnostics lies in miniaturization, high-throughput adaptation, and integration with emerging technologies. One promising direction involves the development of portable and benchtop MS systems that can be deployed in clinical laboratories with minimal infrastructure requirements. Innovations in microfluidics and lab-on-a-chip technologies could further reduce sample volume requirements and improve assay throughput.
Ion mobility spectrometry (IMS), when coupled with MS, offers a refined analytical approach for distinguishing isobaric and isomeric ions-molecules with identical masses but differing in structure or conformation (108). This capability is particularly valuable in allergy diagnostics, where complex biological matrices often contain structurally related compounds that can confound accurate analyte identification (109). The IMS separates ions based on their size, shape, and charge in the gas phase prior to mass analysis, thereby enhancing molecular resolution and analytical specificity. This allows for more accurate detection and quantification of allergy-relevant biomarkers, such as specific immunoglobulins, lipid mediators, and signaling peptides implicated in hypersensitivity reactions. The incorporation of ion mobility into diagnostic workflows has the potential to significantly improve the accuracy, reliability, and clinical utility of MS-based assays in the evaluation of allergic diseases (109, 110).
5 Predicting severity of allergic reactions
BAT is a functional assay that measures the upregulation and activation markers, such as CD63 and CD203c, on circulating basophils following in vitro allergen stimulation. This test provides a direct assessment of immune cell activation, and its results correlate with the severity of allergic reactions observed during clinical food challenges. One of BAT's primary advantages is its ability to evaluate immediate hypersensitivity reactions, offering valuable insights into anaphylaxis risk. Additionally, it has shown a strong association between basophil activation levels and allergic reaction severity, making it a promising tool for identifying high-risk patients. BAT is also non-invasive and rapid, requiring only a small blood sample, with results available within hours. However, the test has some drawbacks, including the need for fresh blood samples, as basophils degrade quickly, necessitating same-day analysis. Furthermore, patient-specific variability influenced by factors such as medication use (e.g., antihistamines) and immune modulation can impact results. Additionally, standardization remains a challenge, as differences in laboratory protocols and reagents can affect reproducibility across institutions.
In contrast, MS-based allergy diagnosis offers a molecular-level analysis of allergic responses by quantifying allergenic proteins, analyzing immune complex formation (IgE and IgG-allergen binding), and profiling post-translational modifications (PTMs) that influence allergenicity. Furthermore, MS-based phosphoproteomic analysis can identify JAK-STAT and FcεRI signaling pathway activation, providing mechanistic insights into allergic severity. The advantages of MS include its ability to perform comprehensive molecular profiling, offering a detailed view of allergenic proteins, immune complexes, and intracellular signaling events linked to allergic responses. It is also highly objective and reproducible, as it eliminates variability due to cellular responsiveness, producing standardized quantitative data. Additionally, MS can identify phosphorylation patterns in effector cells, such as mast cells and basophils, which may serve as biomarkers for severe allergic reactions. However, MS has significant disadvantages, including high costs and the need for specialized instrumentation and expertise, limiting its accessibility in clinical settings. Furthermore, MS is not yet standardized for routine allergy diagnostics, as ongoing research is needed to validate its clinical utility. Another limitation is that MS does not directly measure real-time basophil degranulation or immune cell activation, as BAT does, but rather infers reaction severity through biomolecular signatures.
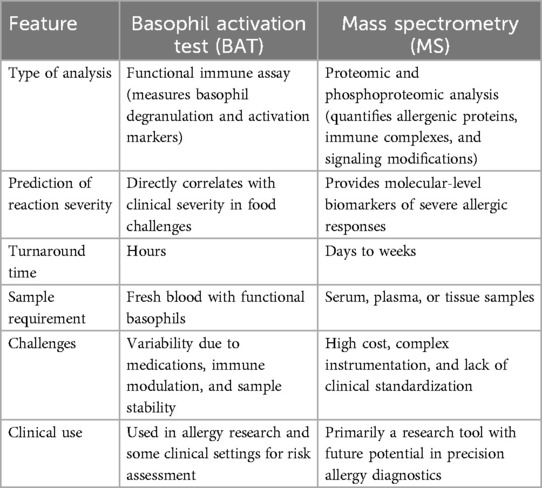
When comparing BAT and MS for predicting allergy severity, BAT is a functional immune assay that directly measures basophil degranulation and activation markers, while MS is a proteomic and phosphoproteomic tool that quantifies allergenic proteins, immune complexes, and signaling modifications. BAT has a fast turnaround time (hours), whereas MS-based approaches take days to weeks. BAT requires fresh blood with functional basophils, whereas MS can be performed on serum, plasma, or tissue samples. The challenges of BAT include variability due to medications, immune modulation, and sample stability, while MS is constrained by high costs, complex instrumentation, and lack of standardization. BAT is already used in some clinical settings for risk assessment, whereas MS remains primarily a research tool with the potential for precision allergy diagnostics in the future. Table 1 demonstrates a comparison of BAT vs. MS for Predicting Allergy Severity.
In a nutshell, BAT is currently the more clinically applicable method, as it provides a real-time functional assessment of basophil activation and correlates well with clinical reaction severity. It is also faster, more affordable, and more accessible than MS, making it a valuable tool for allergy diagnosis and risk stratification. However, MS offers deeper molecular insights by profiling allergenic proteins, immune complexes, and phosphorylation signatures. While MS has the potential to identify novel biomarkers and enhance precision medicine in allergy diagnostics, its high cost and complexity currently limit routine clinical use. Moving forward, a combined approach that integrates BAT's functional immune assessment with MS's molecular precision may provide the most comprehensive predictive tool for assessing allergy severity. Standardizing MS-based biomarkers and making them more clinically accessible could revolutionize personalized allergy management in the future.
6 Sensitivity and selectivity across major allergy diagnostic tests
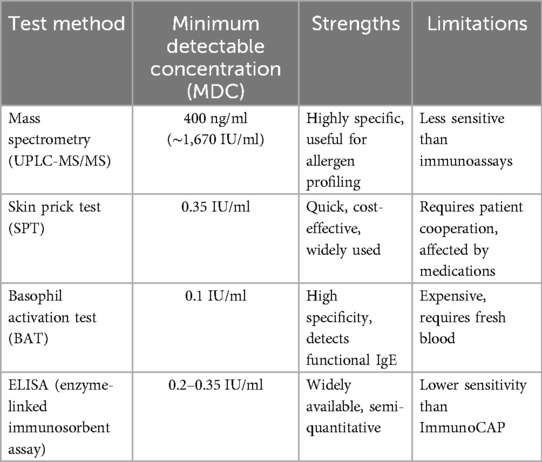
To date, MS is the least sensitive method for IgE detection, with a limit of detection (LOD) of 400 ng/ml (∼1,670 IU/ml), which is significantly higher than that of immunoassay-based techniques. In contrast, SPT and ELISA can detect IgE at levels as low as 0.35 IU/ml, making them far more sensitive for routine allergy diagnostics (66, 111). The most sensitive method, BAT, can detect IgE concentrations as low as 0.1 IU/ml, providing greater accuracy in identifying allergen-specific sensitization (39, 46). While MS offers high specificity and precise molecular characterization, its poor sensitivity limits its use for detecting low IgE concentrations in clinical allergy testing. Consequently, SPT and BAT remain the preferred methods for diagnosing allergies, as they are better suited for detecting and quantifying IgE at clinically relevant levels (46). Table 2 represents a Comparison of Minimum Detectable Concentration of IgE Tests.
7 Conclusion
Allergy diagnostics rely on a variety of methods, each with distinct advantages and limitations. The SPT and ELISA are widely used due to their cost-effectiveness and sensitivity, detecting IgE levels as low as 0.35 IU/ml. The BAT test offers higher specificity and provides functional and quantitative allergen-specific IgE data, making it a gold-standard technique for precise allergy profiling. However, these immunoassay-based approaches, while highly sensitive, may suffer from cross-reactivity and variability in results across different platforms.
Mass spectrometry has proven its value in allergy diagnostics through various applications, particularly in identifying and quantifying allergenic proteins in food and immunoglobulins in biological samples. Several studies highlighted the potential of MS to significantly enhance allergy diagnostics. However, further research is necessary to increase the practicality of MS and optimize its techniques for broader clinical applications. Although early studies have demonstrated its ability to quantify allergen-specific IgE and IgG, translating these methodologies into routine diagnostic tools for diverse allergic conditions remains a challenge. Future efforts are likely to focus on improving the sensitivity and throughput of MS technologies for detecting low-abundance targets including IgEs and addressing issues related to sample preparation and matrix effects. Despite these hurdles, integrating MS into clinical practice holds great promise. By integrating MS into existing diagnostic workflows alongside SPT, BAT, and ImmunoCAP, clinicians could achieve a more comprehensive, accurate, and personalized assessment of allergic responses, ultimately improving patient management and risk assessment.
Author contributions
NW: Writing – original draft, Writing – review & editing, Formal analysis, Investigation, Resources. MS: Writing – original draft, Writing – review & editing. MP: Formal analysis, Investigation, Resources, Writing – original draft, Writing – review & editing, Methodology, Supervision, Project administration. MP: Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Resources, Supervision, Writing – original draft, Writing – review & editing. NE: Writing – original draft, Writing – review & editing. LR: Writing – original draft, Writing – review & editing. AA: Writing – original draft, Writing – review & editing. HS: Writing – original draft, Writing – review & editing. DL: Writing – original draft, Writing – review & editing. SU: Writing – original draft, Writing – review & editing. OA: Writing – original draft, Writing – review & editing, Formal analysis, Investigation, Methodology, Project administration, Resources, Supervision, Data curation, Funding acquisition, Validation. MG: Conceptualization, Formal analysis, Investigation, Methodology, Project administration, Resources, Supervision, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Science Foundation (Award: 2413335), the Center for Applied Proteomics and Molecular Medicine, the Center for Molecular Engineering, and in part by the National Institute of Health NIH Grants U01CA230692.
Conflict of interest
Author(s) AA, DL, MP, SU and OA were employed by Amerimmune LLC.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor, RW, declared a past co-authorship with the author OA.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Alvarez-Arango S, Kumar M, Chow TG, Sabato V. Non-IgE-mediated immediate drug-induced hypersensitivity reactions. Allergy Clin Immunol Pract. (2024) 12(5):1109–19. doi: 10.1016/j.jaip.2024.02.019
2. Vitte J, Vibhushan S, Bratti M, Montero-Hernandez JE, Blank U. Allergy, anaphylaxis, and nonallergic hypersensitivity: IgE, mast cells, and beyond. Med Princ Pract. (2022) 31(6):501–15. doi: 10.1159/000527481
3. Tedner SG, Asarnoj A, Thulin H, Westman M, Konradsen JR, Nilsson C. Food allergy and hypersensitivity reactions in children and adults-a review. J Intern Med. (2022) 291(3):283–302. doi: 10.1111/joim.13422
4. Fong M, Crane JS. Histology, mast cells. In: Herbert DBR, editor. StatPearls. Treasure Island (FL): StatPearls Publishing (2025).
5. Dobrzanski MJ. Expanding roles for CD4T cells and their subpopulations in tumor immunity and therapy. Front Oncol. (2013) 3:63. doi: 10.3389/fonc.2013.00063
6. Goldberg YP, Price N, Namdari R, Cohen C, Lamers H, Winters C, et al. Treatment of Na(v)1.7-mediated pain in inherited erythromelalgia using a novel sodium channel blocker. Pain. (2012) 153(1):80–5. doi: 10.1016/j.pain.2011.09.008
7. Krystel-Whittemore M, Dileepan KN, Wood JG. Mast cell: a multi-functional master cell. Front Immunol. (2015) 6:620. doi: 10.3389/fimmu.2015.00620
8. Leich E, Zamo A, Horn H, Haralambieva E, Puppe B, Gascoyne RD, et al. MicroRNA profiles of t(14;18)-negative follicular lymphoma support a late germinal center B-cell phenotype. Blood. (2011) 118(20):5550–8. doi: 10.1182/blood-2011-06-361972
9. Hall S, Agrawal DK. Key mediators in the immunopathogenesis of allergic asthma. Int Immunopharmacol. (2014) 23(1):316–29. doi: 10.1016/j.intimp.2014.05.034
10. Banafea GH, Bakhashab S, Alshaibi HF, Natesan Pushparaj P, Rasool M. The role of human mast cells in allergy and asthma. Bioengineered. (2022) 13(3):7049–64. doi: 10.1080/21655979.2022.2044278
11. Dilley M, Geng B. Immediate and delayed hypersensitivity reactions to antibiotics: aminoglycosides, clindamycin, linezolid, and metronidazole. Clin Rev Allergy Immunol. (2022) 62(3):463–75. doi: 10.1007/s12016-021-08878-x
12. Minaldi E, Phillips EJ, Norton A. Immediate and delayed hypersensitivity reactions to beta-lactam antibiotics. Clin Rev Allergy Immunol. (2022) 62(3):449–62. doi: 10.1007/s12016-021-08903-z
13. Justiz Vaillant AA, Vashisht R, Zito PM. Immediate hypersensitivity reactions (archived). In: StatPearls. Treasure Island (FL): StatPearls Publishing (2024).
14. Komi DEA, Mortaz E, Amani S, Tiotiu A, Folkerts G, Adcock IM. The role of mast cells in IgE-independent lung diseases. Clin Rev Allergy Immunol. (2020) 58(3):377–87. doi: 10.1007/s12016-020-08779-5
15. Ogawa Y, Grant JA. Mediators of anaphylaxis. Immunol Allergy Clin North Am. (2007) 27(2):249–60. doi: 10.1016/j.iac.2007.03.013
16. Abbas M, Moussa M, Akel H. Type I hypersensitivity reaction. In: StatPearls. Treasure Island (FL): StatPearls Publishing (2024).
17. Joshi SR, Khan DA. Non-IgE-mediated drug hypersensitivity reactions. Curr Allergy Asthma Rep. (2021) 21(7):41. doi: 10.1007/s11882-021-01018-7
18. Zuberbier T, Abdul Latiff AH, Abuzakouk M, Aquilina S, Asero R, Baker D, et al. The international EAACI/GA(2)LEN/EuroGuiDerm/APAAACI guideline for the definition, classification, diagnosis, and management of urticaria. Allergy. (2022) 77(3):734–66. doi: 10.1111/all.15090
19. Poziomkowska-Gesicka I, Kurek M. Clinical manifestations and causes of anaphylaxis. Analysis of 382 cases from the anaphylaxis registry in west pomerania province in Poland. Int J Environ Res Public Health. (2020) 17(8). doi: 10.3390/ijerph17082787
20. Scheinpflug J, Pfeiffenberger M, Damerau A, Schwarz F, Textor M, Lang A, et al. Journey into bone models: a review. Genes (Basel). (2018) 9(5):247. doi: 10.3390/genes9050247
21. Penman A, Hill AE, Hewat S, Scarinci N. Speech-language pathology students’ perceptions of simulation-based learning experiences in stuttering. Int J Lang Commun Disord. (2022) 57(2):466–7. doi: 10.1111/1460-6984.12688
22. Swift K, Vujcich E, Matthews T, Reade MC. Caravan explosions: a case series of burns patients at the royal Brisbane and women’s hospital. ANZ J Surg. (2021) 91(1–2):73–6. doi: 10.1111/ans.16436
23. Maeda M, Kuwabara Y, Tanaka Y, Nishikido T, Hiraguchi Y, Yamamoto-Hanada K, et al. Is oral food challenge test useful for avoiding complete elimination of cow’s milk in Japanese patients with or suspected of having IgE-dependent cow’s milk allergy? Allergol Int. (2022) 71(2):214–20. doi: 10.1016/j.alit.2021.09.001
24. NIAID-Sponsored Expert Panel, Boyce JA, Assa’ad A, Burks AW, Jones SM, Sampson HA, et al. Guidelines for the diagnosis and management of food allergy in the United States: report of the NIAID-sponsored expert panel. J Allergy Clin Immunol. (2010) 126(6 Suppl):S1–58. doi: 10.1016/j.jaci.2010.10.007
25. Sicherer SH, Warren CM, Dant C, Gupta RS, Nadeau KC. Food allergy from infancy through adulthood. J Allergy Clin Immunol Pract. (2020) 8(6):1854–64. doi: 10.1016/j.jaip.2020.02.010
26. Cosme-Blanco W, Arroyo-Flores E, Ale H. Food allergies. Pediatr Rev. (2020) 41(8):403–15. doi: 10.1542/pir.2019-0037
27. Foong RX, Dantzer JA, Wood RA, Santos AF. Improving diagnostic accuracy in food allergy. J Allergy Clin Immunol Pract. (2021) 9(1):71–80. doi: 10.1016/j.jaip.2020.09.037
28. Santos AF, Riggioni C, Agache I, Akdis CA, Akdis M, Alvarez-Perea A, et al. EAACI guidelines on the diagnosis of IgE-mediated food allergy. Allergy. (2023) 78(12):3057–76. doi: 10.1111/all.15902
29. Foong RX, Du Toit G, van Ree R, Bahnson HT, Radulovic S, Craven J, et al. Biomarkers of peanut allergy in children over time. Allergy. (2024) 79(10):2775–86. doi: 10.1111/all.16193
30. Conway AE, Golden DBK, Brough HA, Santos AF, Shaker MS. Serologic measurements for peanut allergy: predicting clinical severity is complex. Ann Allergy Asthma Immunol. (2024) 132(6):686–93. doi: 10.1016/j.anai.2024.01.018
31. Mahavar N, Asghari M, Mofatteh M, Jaberi S, Erfanian N, Chahkandi M, et al. Evaluation of the local and systemic pattern of sensitization to allergens in patients with adenotonsillar hypertrophy. Eur Arch Oto-Rhino-Laryngol. (2024) 281(8):4231–9. doi: 10.1007/s00405-024-08550-y
32. Thorpe M, Moverare R, Fischer C, Lidholm J, Rudengren M, Borres MP. History and utility of specific IgE cutoff levels: what is the relevance for allergy diagnosis? J Allergy Clin Immunol Pract. (2023) 11(10):3021–9. doi: 10.1016/j.jaip.2023.05.022
33. Graham F, Begin P, Paradis L, Lacombe-Barrios J, Paradis J, Des Roches A. Comparison of ImmunoCAP and immulite serum specific IgE assays for the assessment of egg allergy. Allergy Asthma Clin Immunol. (2016) 12:29. doi: 10.1186/s13223-016-0134-0
34. Al Hawi Y, Nagao M, Furuya K, Sato Y, Ito S, Hori H, et al. Agreement between predictive, allergen-specific IgE values assessed by ImmunoCAP and IMMULITE 2000 3gAllergy assay systems for milk and wheat allergies. Allergy Asthma Immunol Res. (2021) 13(1):141–53. doi: 10.4168/aair.2021.13.1.141
35. Santos AF, Alpan O, Hoffmann HJ. Basophil activation test: mechanisms and considerations for use in clinical trials and clinical practice. Allergy. (2021) 76(8):2420–32. doi: 10.1111/all.14747
36. Alpan O, Layhadi JA, Ulrik Sonder S, Li H, Shamji MH. Basophil activation test: a diagnostic, predictive and monitoring assay for allergen immunotherapy. Allergy. (2021) 76(5):1321–4. doi: 10.1111/all.14585
37. Sonder SU, Plassmeyer M, Schroeder N, Peyton S, Paige M, Girgis M, et al. Basophil activation test; user’s manual. J Immunol Methods. (2025) 537:113815. doi: 10.1016/j.jim.2025.113815
38. Piletta-Zanin A, Ricci C, Santos AF, Eigenmann PA. BAT And MAT for diagnosis of peanut allergy: a systematic review and meta-analysis. Pediatr Allergy Immunol. (2024) 35(6):e14140. doi: 10.1111/pai.14140
39. Bergmann MM, Santos AF. Basophil activation test in the food allergy clinic: its current use and future applications. Expert Rev Clin Immunol. (2024) 20(11):1297–304. doi: 10.1080/1744666X.2024.2336568
40. Radulovic S, Foong RX, Bartha I, Marques-Mejias A, Krawiec M, Kwok M, et al. Basophil activation test as predictor of severity and threshold of allergic reactions to egg. Allergy. (2024) 79(2):419–31. doi: 10.1111/all.15875
41. Gawinowska M, Specjalski K, Zielinski M, Trzonkowski P, Niedoszytko M, Chelminska M. Basophil activation test is inferior to provocation test in diagnosing aspirin hypersensitivity. Int Arch Allergy Appl Immunol. (2024) 185(10):928–38. doi: 10.1159/000538111
42. Eberlein B. Basophil activation as marker of clinically relevant allergy and therapy outcome. Front Immunol. (2020) 11:1815. doi: 10.3389/fimmu.2020.01815
43. Kim T, Yu J, Li H, Scarupa M, Wasserman RL, Economides A, et al. Validation of inducible basophil biomarkers: time, temperature and transportation. Cytometry B Clin Cytom. (2021) 100(6):632–44. doi: 10.1002/cyto.b.21991
44. Brasal-Prieto M, Fernandez-Prades L, Dakhaoui H, Sobrino F, Lopez-Enriquez S, Palomares F. Update on in vitro diagnostic tools and treatments for food allergies. Nutrients. (2023) 15(17):3744. doi: 10.3390/nu15173744
45. MacGlashan D Jr, Gao L. Association of a SYK promoter polymorphism with SYK expression and IgE-mediated histamine release. Clin Exp Allergy. (2023) 53(5):573–6. doi: 10.1111/cea.14289
46. Briceno Noriega D, Teodorowicz M, Savelkoul H, Ruinemans-Koerts J. The basophil activation test for clinical management of food allergies: recent advances and future directions. J Asthma Allergy. (2021) 14:1335–48. doi: 10.2147/JAA.S237759
47. Boobalan M, Amaladasan M, Ramalingam S, Tamilvendan D, Venkatesa Prabhu G, Bououdina M. First principles and DFT supported investigations on vibrational spectra and electronic structure of 2-((phenylamino)methyl)isoindoline-1,3-dione–an antioxidant active mannich base. Spectrochim Acta A Mol Biomol Spectrosc. (2015) 137:962–78. doi: 10.1016/j.saa.2014.08.101
48. Yu H, van Erp N, Bins S, Mathijssen RH, Schellens JH, Beijnen JH, et al. Development of a pharmacokinetic model to describe the complex pharmacokinetics of pazopanib in cancer patients. Clin Pharmacokinet. (2017) 56(3):293–303. doi: 10.1007/s40262-016-0443-y
49. Antunes MV, Charao MF, Linden R. Dried blood spots analysis with mass spectrometry: potentials and pitfalls in therapeutic drug monitoring. Clin Biochem. (2016) 49(13–14):1035–46. doi: 10.1016/j.clinbiochem.2016.05.004
50. Perko D, Groselj U, Cuk V, Iztok Remec Z, Zerjav Tansek M, Drole Torkar A, et al. Comparison of tandem mass spectrometry and the fluorometric method-parallel phenylalanine measurement on a large fresh sample series and implications for newborn screening for phenylketonuria. Int J Mol Sci. (2023) 24(3):2487. doi: 10.3390/ijms24032487
51. Wagner M, Tonoli D, Varesio E, Hopfgartner G. The use of mass spectrometry to analyze dried blood spots. Mass Spectrom Rev. (2016) 35(3):361–438. doi: 10.1002/mas.21441
52. Banerjee A, Pai MGJ, Singh A, Nissa MU, Srivastava S. Mass spectrometry and proteome analysis to identify SARS-CoV-2 protein from COVID-19 patient swab samples. STAR Protoc. (2022) 3(1):101177. doi: 10.1016/j.xpro.2022.101177
53. Wandernoth P, Kriegsmann K, Kriegsmann J, Kriegsmann M. Detection of SARS-CoV-2 by mass spectrometry. Methods Mol Biol. (2022) 2452:183–96. doi: 10.1007/978-1-0716-2111-0_12
54. Viana LG, Lebkuchen A, Schuch RA, Okai GG, Salgueiro JS, Cardozo KHM, et al. Mass spectrometry multiplexed detection of SARS-CoV-2. Methods Mol Biol. (2022) 2511:161–74. doi: 10.1007/978-1-0716-2395-4_12
55. Malucelli M, Farias RJ, Mello RG, Prando C. Biomarkers associated with persistence and severity of IgE-mediated food allergies: a systematic review. J Pediatr (Rio J). (2023) 99(4):315–21. doi: 10.1016/j.jped.2023.02.004
56. Koren A, Korosec P. Multiplex basophil activation tests for allergy diagnosis: present and future applications. Front Allergy. (2024) 5:1515843. doi: 10.3389/falgy.2024.1515843
57. Carmel A, Musa-Lempel N, Tsur D, Ziv-Ukelson M. The worst case complexity of maximum parsimony. J Comput Biol. (2014) 21(11):799–808. doi: 10.1089/cmb.2014.0128
58. Hawasli AH, Chacko R, Szrama NP, Bundy DT, Pahwa M, Yarbrough CK, et al. Electrophysiological sequelae of hemispherotomy in ipsilateral human cortex. Front Hum Neurosci. (2017) 11:149. doi: 10.3389/fnhum.2017.00149
59. Svoboda SM, Park H, Naff N, Dorai Z, Williams MA, Youssef Y. Preventing distal catheter obstruction in laparoscopic ventriculoperitoneal shunt placement in adults: the “falciform technique”. J Laparoendosc Adv Surg Tech A. (2015) 25(8):642–5. doi: 10.1089/lap.2015.0196
60. Tang C, Verwilligen A, Sadoff J, Brandenburg B, Sneekes-Vriese E, van den Kerkhof T, et al. Absolute quantitation of binding antibodies from clinical samples. NPJ Vaccines. (2024) 9(1):8. doi: 10.1038/s41541-023-00793-w
61. Panigrahi A, Zhang L, Benicky J, Sanda M, Ahn J, Goldman R. A multiplexed microflow LC-MS/MS-PRM assay for serologic quantification of IgG N- and HPX O- glycoforms in liver fibrosis. Sci Rep. (2023) 13(1):606. doi: 10.1038/s41598-023-27382-0
62. Sanda M, Yang Q, Zong G, Chen H, Zheng Z, Dhani H, et al. LC-MS/MS-PRM quantification of IgG glycoforms using stable isotope labeled IgG1 fc glycopeptide standard. J Proteome Res. (2023) 22(4):1138–47. doi: 10.1021/acs.jproteome.2c00475
63. Gasilova N, Girault HH. Component-resolved diagnostic of cow’s milk allergy by immunoaffinity capillary electrophoresis-matrix assisted laser desorption/ionization mass spectrometry. Anal Chem. (2014) 86(13):6337–45. doi: 10.1021/ac500525n
64. Hawkinson TR, Sun RC. Matrix-assisted laser desorption/ionization mass spectrometry imaging of glycogen in situ. Methods Mol Biol. (2022) 2437:215–28. doi: 10.1007/978-1-0716-2030-4_15
65. Birse N, Burns DT, Walker MJ, Quaglia M, Elliott CT. Food allergen analysis: a review of current gaps and the potential to fill them by matrix-assisted laser desorption/ionization. Compr Rev Food Sci Food Saf. (2023) 22(5):3984–4003. doi: 10.1111/1541-4337.13216
66. Li Y, Yang Y, Liu Y, Liu J, Yang Y, Zhang J, et al. Combination of magnetic beads extraction and ultraperformance liquid chromatography tandem mass spectrometry detection for the clinical diagnosis of allergies. Anal Chim Acta. (2022) 1221:340157. doi: 10.1016/j.aca.2022.340157
67. Kepley CL, Youssef L, Andrews RP, Wilson BS, Oliver JM. Syk deficiency in nonreleaser basophils. J Allergy Clin Immunol. (1999) 104(2 Pt 1):279–84. doi: 10.1016/S0091-6749(99)70367-2
68. Kepley CL, Youssef L, Andrews RP, Wilson BS, Oliver JM. Multiple defects in fc epsilon RI signaling in syk-deficient nonreleaser basophils and IL-3-induced recovery of syk expression and secretion. J Immunol. (2000) 165(10):5913–20. doi: 10.4049/jimmunol.165.10.5913
69. Min SY, Park CH, Yu HW, Park YJ. Anti-inflammatory and anti-allergic effects of saponarin and its impact on signaling pathways of RAW 264.7, RBL-2H3, and HaCaT cells. Int J Mol Sci. (2021) 22(16):8431. doi: 10.3390/ijms22168431
70. Kashiwakura JI, Yoshihara M, Saitoh K, Kagohashi K, Sasaki Y, Kobayashi F, et al. Propolis suppresses cytokine production in activated basophils and basophil-mediated skin and intestinal allergic inflammation in mice. Allergol Int. (2021) 70(3):360–7. doi: 10.1016/j.alit.2020.11.005
71. Darrasse-Jeze G, Deroubaix S, Mouquet H, Victora GD, Eisenreich T, Yao KH, et al. Feedback control of regulatory T cell homeostasis by dendritic cells in vivo. J Exp Med. (2009) 206(9):1853–62. doi: 10.1084/jem.20090746
72. Yuan NY, Maung R, Xu Z, Han X, Kaul M. Arachidonic acid cascade and eicosanoid production are elevated while LTC4 synthase modulates the lipidomics profile in the brain of the HIVgp120-transgenic mouse model of NeuroHIV. Cells. (2022) 11(13):2123. doi: 10.3390/cells11132123
73. Tang X, Fuchs D, Tan S, Trauelsen M, Schwartz TW, Wheelock CE, et al. Activation of metabolite receptor GPR91 promotes platelet aggregation and transcellular biosynthesis of leukotriene C(4). J Thromb Haemost. (2020) 18(4):976–84. doi: 10.1111/jth.14734
74. Schneider E, Petit-Bertron AF, Bricard R, Levasseur M, Ramadan A, Girard J-P, et al. IL-33 activates unprimed murine basophils directly in vitro and induces their in vivo expansion indirectly by promoting hematopoietic growth factor production. J Immunol. (2009) 183(6):3591–7. doi: 10.4049/jimmunol.0900328
75. Pugliese GM, Latini S, Massacci G, Perfetto L, Sacco F. Combining mass spectrometry-based phosphoproteomics with a network-based approach to reveal FLT3-dependent mechanisms of chemoresistance. Proteomes. (2021) 9(2):19. doi: 10.3390/proteomes9020019
76. Mukai K, Gaudenzio N, Gupta S, Vivanco N, Bendall SC, Maecker HT, et al. Assessing basophil activation by using flow cytometry and mass cytometry in blood stored 24 h before analysis. J Allergy Clin Immunol. (2017) 139(3):889–99. doi: 10.1016/j.jaci.2016.04.060
77. Furlan G, Minowa T, Hanagata N, Kataoka-Hamai C, Kaizuka Y. Phosphatase CD45 both positively and negatively regulates T cell receptor phosphorylation in reconstituted membrane protein clusters. J Biol Chem. (2014) 289(41):28514–25. doi: 10.1074/jbc.M114.574319
78. Zhang E, Liu C. A new antibacterial co-cr-mo-cu alloy: preparation, biocorrosion, mechanical and antibacterial property. Mater Sci Eng C Mater Biol Appl. (2016) 69:134–43. doi: 10.1016/j.msec.2016.05.028
79. Cao L, Ding Y, Hung N, Yu K, Ritz A, Raphael BJ, et al. Quantitative phosphoproteomics reveals SLP-76 dependent regulation of PAG and Src family kinases in T cells. PLoS One. (2012) 7(10):e46725. doi: 10.1371/journal.pone.0046725
80. Cao L, Yu K, Banh C, Nguyen V, Ritz A, Raphael BJ, et al. Quantitative time-resolved phosphoproteomic analysis of mast cell signaling. J Immunol. (2007) 179(9):5864–76. doi: 10.4049/jimmunol.179.9.5864
81. Lappas CM, Rieger JM, Linden J. A2A adenosine receptor induction inhibits IFN-gamma production in murine CD4+ T cells. J Immunol. (2005) 174(2):1073–80. doi: 10.4049/jimmunol.174.2.1073
82. Lawrence EA, Kague E, Aggleton JA, Harniman RL, Roddy KA, Hammond CL. The mechanical impact of col11a2 loss on joints; col11a2 mutant zebrafish show changes to joint development and function, which leads to early-onset osteoarthritis. Philos Trans R Soc Lond B Biol Sci. (2018) 373(1759):20170335. doi: 10.1098/rstb.2017.0335
83. Jacquemyn H, Waud M, Brys R, Lallemand F, Courty PE, Robionek A, et al. Mycorrhizal associations and trophic modes in coexisting orchids: an ecological Continuum between auto- and mixotrophy. Front Plant Sci. (2017) 8:1497. doi: 10.3389/fpls.2017.01497
84. Chen X, Peng M, Fu B, Chen Y, Hua J, Zeng C, et al. Risk factors and survival analysis for central nervous system complications after allogeneic hematopoietic stem cell transplantation. Zhong Nan Da Xue Xue Bao Yi Xue Ban. (2020) 45(10):1176–84. doi: 10.11817/j.issn.1672-7347.2020.200050
85. Ku X, Wang J, Li H, Meng C, Yu F, Yu W, et al. Proteomic portrait of human lymphoma reveals protein molecular fingerprint of disease specific subtypes and progression. Phenomics. (2023) 3(2):148–66. doi: 10.1007/s43657-022-00075-w
86. Wist M, Meier L, Gutman O, Haas J, Endres S, Zhou Y, et al. Noncatalytic bruton’s tyrosine kinase activates PLCgamma(2) variants mediating ibrutinib resistance in human chronic lymphocytic leukemia cells. J Biol Chem. (2020) 295(17):5717–36. doi: 10.1074/jbc.RA119.011946
87. Li X, Mak VCY, Zhou Y, Wang C, Wong ESY, Sharma R, et al. Deregulated Gab2 phosphorylation mediates aberrant AKT and STAT3 signaling upon PIK3R1 loss in ovarian cancer. Nat Commun. (2019) 10(1):716. doi: 10.1038/s41467-019-08574-7
88. Zhang X, Lavoie G, Fort L, Zhang X, Lavoie G, Fort L, Huttlin EL, Tcherkezian J, Galan JA, et al. Gab2 phosphorylation by RSK inhibits Shp2 recruitment and cell motility. Mol Cell Biol. (2013) 33(8):1657–70. doi: 10.1128/MCB.01353-12
89. Yang X, Turke AB, Qi J, Song Y, Rexer BN, Miller TW, et al. Using tandem mass spectrometry in targeted mode to identify activators of class IA PI3K in cancer. Cancer Res. (2011) 71(18):5965–75. doi: 10.1158/0008-5472.CAN-11-0445
90. Reis ES, Chen H, Sfyroera G, Monk PN, Köhl J, Ricklin D, et al. C5a receptor-dependent cell activation by physiological concentrations of desarginated C5a: insights from a novel label-free cellular assay. J Immunol. (2012) 189(10):4797–805. doi: 10.4049/jimmunol.1200834
91. Matsubara D, Yanase Y, Ishii K, Takahagi S, Tanaka A, Ozawa K, et al. Basophils activation of patients with chronic spontaneous urticaria in response to C5a despite failure to respond to IgE-mediated stimuli. Front Immunol. (2022) 13:994823. doi: 10.3389/fimmu.2022.994823
92. Franciosa G, Locard-Paulet M, Jensen LJ, Olsen JV. Recent advances in kinase signaling network profiling by mass spectrometry. Curr Opin Chem Biol. (2023) 73:102260. doi: 10.1016/j.cbpa.2022.102260
93. Scapini P, Pereira S, Zhang H, Lowell CA. Multiple roles of lyn kinase in myeloid cell signaling and function. Immunol Rev. (2009) 228(1):23–40. doi: 10.1111/j.1600-065X.2008.00758.x
94. Shelby SA, Veatch SL, Holowka DA, Baird BA. Functional nanoscale coupling of lyn kinase with IgE-FcepsilonRI is restricted by the actin cytoskeleton in early antigen-stimulated signaling. Mol Biol Cell. (2016) 27(22):3645–58. doi: 10.1091/mbc.E16-06-0425
95. Gilfillan AM, Rivera J. The tyrosine kinase network regulating mast cell activation. Immunol Rev. (2009) 228(1):149–69. doi: 10.1111/j.1600-065X.2008.00742.x
96. Zemba M, Ionescu MA, Pirvulescu RA, Dumitrescu OM, Daniel-Constantin B, Radu M, et al. Biomarkers of ocular allergy and dry eye disease. Rom J Ophthalmol. (2023) 67(3):250–9. doi: 10.22336/rjo.2023.42
97. Chu H, Kim SM, Zhang K, Wu Z, Lee H, Kim JH, et al. Head and neck dermatitis is exacerbated by malassezia furfur colonization, skin barrier disruption, and immune dysregulation. Front Immunol. (2023) 14:1114321. doi: 10.3389/fimmu.2023.1114321
98. Bao J, Zhang P, Wu B, Wang J, Li S, Li A, et al. Gp130 promotes inflammation via the STAT3/JAK2 pathway in allergic conjunctivitis. Invest Ophthalmol Visual Sci. (2023) 64(4):5. doi: 10.1167/iovs.64.4.5
99. Jones N, Vincent EE, Felix LC, Cronin JG, Scott LM, Hole PS, et al. Interleukin-5 drives glycolysis and reactive oxygen species-dependent citric acid cycling by eosinophils. Allergy. (2020) 75(6):1361–70. doi: 10.1111/all.14158
100. Campos D, Girgis M, Sanda M. Site-specific glycosylation of SARS-CoV-2: big challenges in mass spectrometry analysis. Proteomics. (2022) 22(15–16):e2100322. doi: 10.1002/pmic.202100322
101. Bellia C, Sardina DS, Scazzone C, Lio D, Scola L, Uasuf CG. Diagnostic accuracy of lipid transfer proteins (LTPs) specific IgE assay in food allergy: a systematic review. Int J Mol Sci. (2024) 25(23):12925. doi: 10.3390/ijms252312925
102. Villanueva J, Carrascal M, Abian J. Isotope dilution mass spectrometry for absolute quantification in proteomics: concepts and strategies. J Proteomics. (2014) 96:184–99. doi: 10.1016/j.jprot.2013.11.004
103. Jensen P, Patel B, Smith S, Sabnis R, Kaboord B. Improved immunoprecipitation to mass spectrometry method for the enrichment of low-abundant protein targets. Methods Mol Biol. (2021) 2261:229–46. doi: 10.1007/978-1-0716-1186-9_14
104. Spiric J, Schulenborg T, Holzhauser T, Schuler F, Bonertz A, Lauer I, et al. Quality control of allergen products with mass spectrometry part I: positioning within the EU regulatory framework. Allergy. (2024) 79(8):2088–96. doi: 10.1111/all.16080
105. Chapman MD, Briza P. Molecular approaches to allergen standardization. Curr Allergy Asthma Rep. (2012) 12(5):478–84. doi: 10.1007/s11882-012-0282-3
106. Zimmer J, Bridgewater J, Ferreira F, van Ree R, Rabin RL, Vieths S. The history, present and future of allergen standardization in the United States and Europe. Front Immunol. (2021) 12:725831. doi: 10.3389/fimmu.2021.725831
107. Lowenstein H. Characterization and standardization of allergen extracts. Chem Immunol Allergy. (2014) 100:323–32. doi: 10.1159/000359989
108. Dodds JN, Baker ES. Ion mobility spectrometry: fundamental concepts, instrumentation, applications, and the road ahead. J Am Soc Mass Spectrom. (2019) 30(11):2185–95. doi: 10.1007/s13361-019-02288-2
109. Koomen DC, May JC, McLean JA. Insights and prospects for ion mobility-mass spectrometry in clinical chemistry. Expert Rev Proteomics. (2022) 19(1):17–31. doi: 10.1080/14789450.2022.2026218
110. Christofi E, Barran P. Ion mobility mass spectrometry (IM-MS) for structural biology: insights gained by measuring mass, charge, and collision cross section. Chem Rev. (2023) 123(6):2902–49. doi: 10.1021/acs.chemrev.2c00600
Keywords: BAT, mass spectrometry, food allergy, diagnostics, specificity, sensitivity
Citation: Wheeler N, Sanda M, Rasool L, Elemary N, Alpan A, Safi H, Loizou D, Plassmeyer M, Paige M, Ulrik Sonder S, Alpan O and Girgis M (2025) Enhancing allergy diagnosis: mass spectrometry as a complementary technique to the basophil activation test. Front. Allergy 6:1568670. doi: 10.3389/falgy.2025.1568670
Received: 30 January 2025; Accepted: 9 April 2025;
Published: 30 May 2025.
Edited by:
Richard L. Wasserman, Medical City Children's Hospital, United StatesReviewed by:
Carina Silva Pinheiro, Federal University of Bahia (UFBA), BrazilMarcel Bergmann, University of Italian Switzerland, Switzerland
Copyright: © 2025 Wheeler, Sanda, Rasool, Elemary, Alpan, Safi, Loizou, Plassmeyer, Paige, Ulrik Sonder, Alpan and Girgis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael Girgis, bXlhc3NhZ2lAZ211LmVkdQ==
 Nicole Wheeler
Nicole Wheeler Miloslav Sanda
Miloslav Sanda Lanya Rasool
Lanya Rasool Noha Elemary1
Noha Elemary1 Hamed Safi
Hamed Safi Matthew Plassmeyer
Matthew Plassmeyer Soren Ulrik Sonder
Soren Ulrik Sonder Oral Alpan
Oral Alpan Michael Girgis
Michael Girgis