- 1Department of Biology, University of Central Florida, Orlando, FL, United States
- 2Department of Biology, The Pennsylvania State University, University Park, PA, United States
- 3U.S. Geological Survey, Wetland and Aquatic Research Center, Lafayette, LA, United States
- 4U.S. Geological Survey, Wetland and Aquatic Research Center, Gainesville, FL, United States
- 5Department of Herpetology, Audubon Zoo, New Orleans, LA, United States
Understanding the origin and spread of invasive species is critical for predicting when and where new introductions will establish, and impact native species. However, due to the complexity of contributing factors such as multiple introductions, dispersal method, genetic admixture in founding populations, and variable propagule pressure, genetic patterns observed in invasive species may not always conform to a single theoretical expectation. Cuban treefrogs (Osteopilus septentrionalis) are invasive in peninsular Florida and sporadically in the Florida panhandle. Though O. septentrionalis has been occasionally reported in Louisiana since the 1990s, established populations were not present until the discovery of a breeding population in New Orleans in 2017. In this study we investigated the source of this novel population using existing and newly generated cytochrome B (cyt-b) mitochondrial gene sequences from the native and invasive range of O. septentrionalis. We recovered a total of 14 cyt-b haplotypes, nine novel and five previously published. Within the 95 Louisiana invasion samples, we recovered seven haplotypes including five novel haplotypes. The haplotypes most common in Louisiana were shared exclusively with west and east Florida localities in central Florida, indicating a possible source population. The presence of haplotypes private to the Louisiana locality suggests other unsampled localities may also be contributing to the Louisiana settlement. Metrics of genetic diversity across native and invasive localities did not significantly differ. Furthermore, the Louisiana samples had higher genetic diversity than any single location sampled within Florida. Thus, genetic diversity and our haplotype connectivity suggest the Louisiana population is derived from multiple introductions from Florida. Our study highlights how demographic and genetic analyses can be utilized to understand the source and future expansion potential of invasive populations.
1 Introduction
Invasive herpetofauna pose a threat to biodiversity through disruption of native species interactions (Rice et al., 2011; Richter-Boix et al., 2013; Culbertson and Herrmann, 2019; Hill et al., 2019), displacement of native species through predation (Dove et al., 2011; Piquet and López-Darias, 2021), and the spread of infectious diseases (Rivera et al., 2019; Atkinson and Savage, 2023). While over 780 introductions of non-native herpetofauna were documented globally as of 2015, only 250 species have established as independent populations (Kraus, 2015). The disparity between introduced and established invasive species underscores the importance of life history, environmental variation, propagule pressure and genetic diversity on the ability of an invasive population to establish and thrive. Typically, generalist species are more likely to successfully invade and establish compared to specialists that have more specific habitat requirements (Pitt et al., 2005), and environments more like the invasive species native range tend to be conducive to establishment (Bomford et al., 2009). Introductions of non-native herpetofauna largely result from anthropomorphic activity, such as the pet trade (Willson et al., 2011), farming (Laufer et al., 2008), bio-control efforts (Shanmuganathan et al., 2010), and stowaways in cargo shipments (Richmond et al., 2015). Thus, the numerous factors influencing which species become introduced combined with the complexity of novel ecosystems present a challenge for predicting whether and which non-native amphibian and reptile species will become established.
Invasive populations can arise from novel founder events or expansions of pre-existing invasive ranges, and each of these mechanisms should produce distinct patterns of genetic diversity in the invasive population. Founder events are generally expected to decrease genetic diversity and increase inbreeding depression (Frankham, 1998), which has been observed in the case of the coqui (Eleutherodactylus coqui) invasion in Hawaii (Peacock et al., 2009) and the American bullfrog (Rana catesbeiana) invasion in China (Bai et al., 2012). Contrary to this expectation, invasive populations founded by multiple introduction events or with high propagule pressure (i.e., many individuals introduced in one or more release events) (Rius and Darling, 2014) can maintain genetic diversity at adaptive loci (Selechnik et al., 2019; LaFond et al., 2022) or even show higher overall genetic diversity relative to native populations (Allendorf and Lundquist, 2003; Kolbe et al., 2004). A genetically admixed invasive population has increased adaptive potential and often is capable of faster range expansions (Wagner et al., 2017) and adaptation to novel environmental conditions (Krehenwinkel and Tautz, 2013; Rius and Darling, 2014).
Mitochondrial DNA (mtDNA) is commonly used to establish phylogenetic relationships between populations and trace the origins of invasive populations (Avise et al., 1987; Hamner et al., 2007; Colangelo et al., 2015) because it has a higher mutation rate than nuclear loci, allowing for increased resolution of genetic variation (Gray et al., 1989). Additionally, mtDNA is easy to isolate, has a relatively simple genetic structure (i.e., lacking transposable elements, pseudogenes and introns) and exhibits a straightforward mode of inheritance (Avise et al., 1987). The mitochondrial gene cytochrome b (cyt-b) codes for a central redox catalytic subunit involved in the electron transport chain that is present in nearly all eukaryotic organisms. The cyt-b gene is used to evaluate genetic diversity, trace geographic origins, and distinguish cryptic invasive species across vertebrate groups (Kochzius et al., 2003; Hamner et al., 2007; Colangelo et al., 2015; Jackson et al., 2015). Cyt-b data have been used to investigate the origins of several invasive herpetofauna, including the novel invasion of two subspecies of Boa constrictor in Puerto Rico (Reynolds et al., 2013), the origin and identity of cryptic Xenopus species in Chile (Lobos et al., 2014) and Italy (Lillo et al., 2013), the invasion of the greenhouse frog (Eleutherodactylus planirostris) in Florida (Heinicke et al., 2011) and China (Hong et al., 2022), and the invasive American bullfrog (R. catesbeiana) across the western United States (Bai et al., 2012; Kamath et al., 2016; LaFond et al., 2022).
The Cuban treefrog (Osteopilus septentrionalis) is one of the most successful invasive amphibians in the state of Florida (Meshaka, 2001). Invasive populations negatively influence native Florida treefrogs through predation (Wyatt and Forys, 2004), and by influencing pathogen dynamics (Galt et al., 2021; Atkinson and Savage, 2023). The native O. septentrionalis range spans Cuba, the Cayman Islands and the Bahamas, with invasive populations first establishing in the Florida Keys in the late 1800s to early 1900s (Duellman and Schwartz, 1958; Heinicke et al., 2011). Subsequently, O. septentrionalis spread to peninsular Florida by the mid-1900s via multiple anthropogenic introductions (Heinicke et al., 2011) and has now spread throughout much of central and northern Florida and in some areas of the Florida panhandle (Figure 1; Johnson, 2023). Sporadic sightings of O. septentrionalis have been recorded along the southern coastline of Alabama and Mississippi (Morningstar et al., 2024), but no established populations have been reported. Cyt-b from O. septentrionalis has been used to evaluate the ecological drivers of haplotype and species diversity (Rodríguez et al., 2015). Akin to the previous herpetological invasions mentioned above, the origin of invasive O. septentrionalis in Florida was investigated using cyt-b genetic analysis (Heinicke et al., 2011). The resulting mitochondrial phylogeny revealed a deep phylogenetic split among haplotypes of O. septentrionalis, with both clades represented throughout the native range in Cuba and the invasive range in Florida. This finding indicated that current populations are descended from at least two source populations (Heinicke et al., 2011).
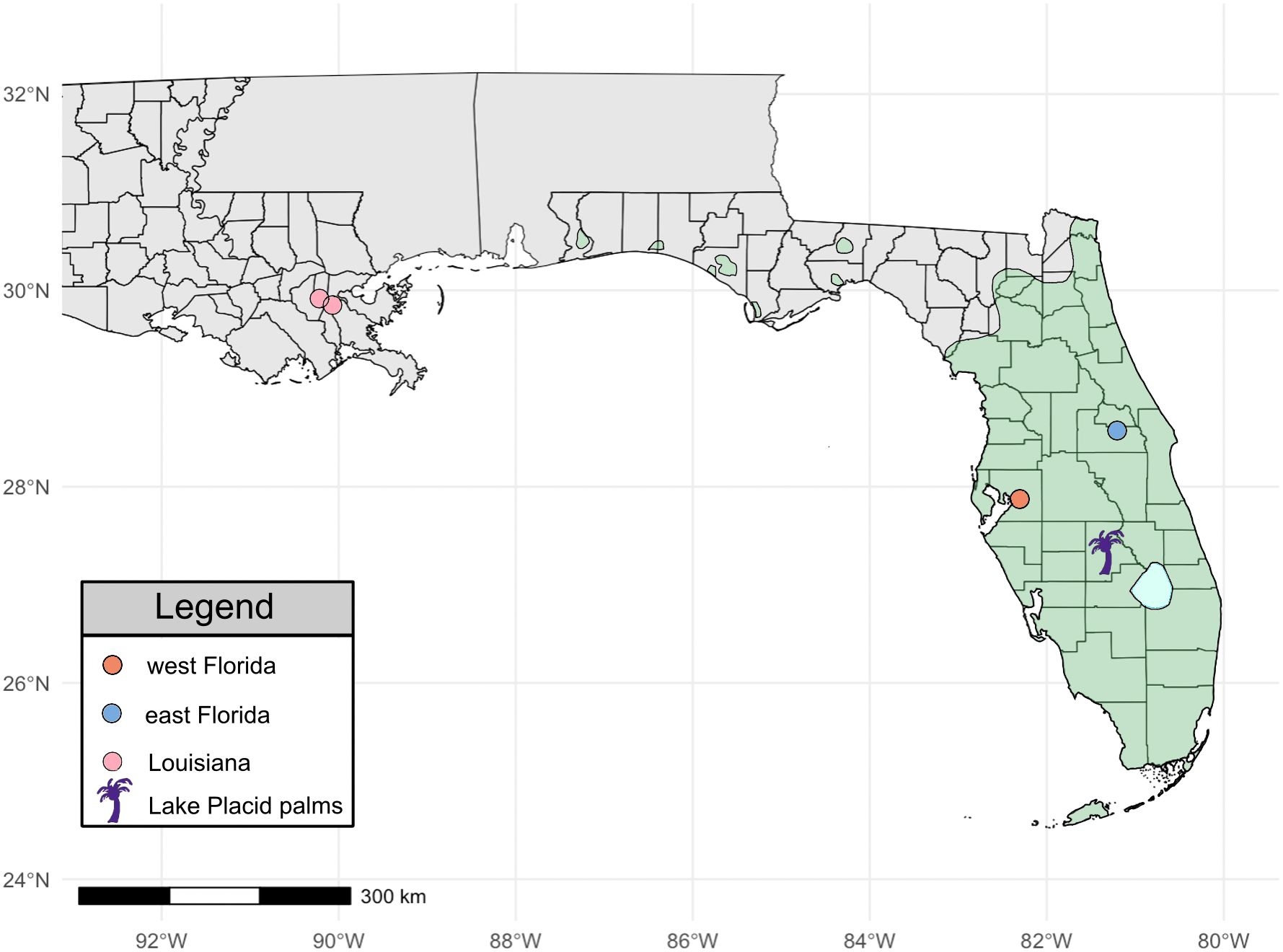
Figure 1. Louisiana and Florida O. septentrionalis sampling locations. Sampling locations are denoted with circles. Louisiana populations are in pink, east Florida samples in blue, and west Florida samples in orange. Lake Placid, the origin of Audubon Zoo palms, is denoted with a purple palm. Green shading signifies the known O. septentrionalis distribution as of January 2023 (Credits: Tracey Bryant, UF/IFAS). Light blue shading represents Lake Okeechobee.
In 2017, the first established breeding population of O. septentrionalis in Louisiana, USA, was observed in New Orleans (Glorioso et al., 2016, 2018c). Although O. septentrionalis had been sporadically documented in Louisiana since the 1990s, no breeding colonies had been observed prior to this event (Chatfield and Vance, 2014; Glorioso et al., 2018b, 2018c). In contrast, the discovery of individuals in different age classes, ranging from tadpoles to metamorphs, were found in Audubon Zoo and the adjacent Riverview Park beginning in 2016, leading to further investigation of this invasion event (Glorioso et al., 2018c). Subsequently, a second breeding population was located in St. Charles Parish (Glorioso et al., 2018a). While the novel New Orleans invasion coincided with the arrival of palms that were brought from Lake Placid, Florida for an Audubon Zoo exhibit (Glorioso et al., 2018c), given the occasional observations of O. septentrionalis prior to this event, and that O. septentrionalis are regularly dispersed via anthropogenic activities (Palis et al., 2021), a definitive source for this invasion remains unclear.
Here, we used cyt-b mitochondrial genetics to evaluate genetic variation in the recently established O. septentrionalis population at the Riverview Park and St. Charles Parish sites in Louisiana and assess likely routes of introduction. We sampled and generated cyt-b haplotypes from 31 east Florida individuals and 40 west Florida individuals within central Florida, and 95 individuals in New Orleans, Louisiana, then combined these new data with previously generated sequences from throughout the native and invasive range (Heinicke et al., 2011; Rodríguez et al., 2015). We conducted phylogenetic and haplotype network analyses and compared metrics of genetic diversity in Louisiana to other native and invasive populations. We used these data to assess whether the Louisiana invasion represents a single introduction event or a more complex history of population establishment.
2 Materials and methods
We collected and processed a total of 166 O. septentrionalis between 2017 and 2022. Of those, 31 were from east Florida, 40 from west Florida, and 95 individuals were from Louisiana populations. East Florida samples were collected from one site in Orange County and west Florida samples were collected from one site in Hillsborough County. Louisianan samples originated from two sampling sites; 85 individuals were from Audubon Riverview Park and the adjacent Audubon Zoo in Orleans Parish, while 10 individuals were from 15 kilometers west in St. Charles Parish (Figure 1).
2.1 Louisiana field sampling
As a means to collect O. septentrionalis, artificial PVC refugia were set at the Riverview Park site in 2019 to capture juvenile metamorphs and adults (Glorioso and Waddle, 2014). Additional samples were collected by hand via standardized visual surveys. Toe clips from metamorphosed anurans were collected in 99% molecular grade ethanol. Scissors were sterilized after each toe clip by alcohol immersion. O. septentrionalis were euthanized via 20% benzocaine. Samples were placed in 99% molecular grade ethanol and stored at -80 °C prior to DNA extraction.
2.2 Florida field sampling
Juvenile metamorphs and adult frog samples were collected by hand via standardized visual surveys. Tadpoles were collected via standardized dip netting and minnow trapping. Tail clips from tadpoles and toe clips from metamorphosed anurans were collected with flame sterilized scissors and placed in 99% molecular grade ethanol. Per Florida regulations on invasive species, O. septentrionalis were euthanized via MS-222 (tricane mesylate). Tissues were preserved in 99% molecular grade ethanol and stored at -20 °C prior to DNA extraction.
2.3 Laboratory methods
We performed DNA extractions on toe or tail tissue samples using a Qiagen DNeasy Blood and Tissue Kit (Qiagen Corporation, Germantown, Maryland, USA) following manufacturer instructions. Extractions were stored at -20 °C prior to further molecular analysis. PCR was performed on all samples to amplify the cyt-b mitochondrial gene using the forward and reverse primers CBL17 (5’-TAGCCTTYTCATCCGTYGCCCATAT-3’) and CBH18 (5’-GTTGATAATGCAACCCTRACCCGATT-3’) (Heinicke et al., 2011). Each 25µL reaction consisted of 16.4µL of molecular grade water, 5µL of NEB OneTaq Buffer, 0.5µL of 10mM dNTPs, 0.5µL each of 10uM forward and reverse primers, 0.13µL of NEB One Taq DNA polymerase and 2µL of template DNA or controls. Thermocycling conditions for PCR consisted of an initial denaturation at 94 °C for 5 minutes, followed by 40 cycles of 94 °C for 30 seconds, 46 °C for 30 seconds, and 72 °C for 60 seconds, with a final elongation step of 72 °C for one minute (Heinicke et al., 2011). PCR products were sent to Eurofins Genomics LLC for Sanger sequencing in the forward and reverse direction. Our newly generated sequences were trimmed of primers and manually inspected and edited to resolve errors and ambiguities by analyzing sanger sequencing graphic outputs. Any sequences that were shorter than 670 bp or had ambiguities, such as stop codons, were excluded from the analyses.
2.4 Phylogenetic analyses
We reconstructed a Bayesian gene tree of O. septentrionalis cyt-b using our sequences (N = 166) and a mix of sequences from native and invasive populations available in GenBank (N = 210; Heinicke et al., 2011; Rodríguez et al., 2015). We used Rana septentrionalis (AY083273), Hyla molleri (MK172098), Osteopilus marianae (HQ831741), Osteopilus vastus (HQ831742), Osteopilus dominicensis (HQ831743), and Osteopilus ocellatus (HQ831744) as outgroups. The untruncated sequences of 903 bp were aligned using Clustal Omega (Sievers et al., 2011) with default parameters in Geneious v. 9.0.5 software (Kearse et al., 2012). To find a model of evolution that best fit the data and to use in the phylogenetic reconstruction, we used jmodeltest2 v. 2.1.1 (Darriba et al., 2012) on the CIPRES Science Gateway server (Miller et al., 2010). The best model according to the Akaike information criterion for small sample sizes (AICc) was the Hasegawa-Kishino-Yano (HKY) model with invariant sites and a gamma distribution. We reconstructed a Bayesian gene tree using MrBayes v. 3.2.7a (Ronquist et al., 2012) on the CIPRES Science Gateway server (Miller et al., 2010). The phylogenetic reconstruction consisted of two independent runs of 1.0 × 107 generations and four chains each. We sampled every 500th generation and the first 100,000 generations were discarded as burnin. We used Tracer v. 1.7 (Rambaut et al., 2018) to confirm Markov chain Monte Carlo convergence and adequate sampling of the posterior distribution. We also reconstructed a maximum likelihood gene tree with IQ-TREE with 1000 ultrafast bootstraps and SH-aLRT branch tests on the IQ-TREE webserver (Trifinopoulos et al., 2016) to compare its topology to that of MrBayes. We visualized the Bayesian gene tree and geographic locations where each haplotype has been found using the R package ggtree v3.3.0.900 in RStudio using R v. 4.1.2 (Yu et al., 2017; RStudio Team, 2021).
2.5 Haplotype network
We reconstructed a haplotype network of O. septentrionalis cyt-b utilizing all sequences we generated from our west and east Florida sampling, as well as all additional sequences sampled from Florida O. septentrionalis that were available in GenBank, resulting in the addition of a south Florida locality. Sequences in our alignment were truncated to produce uniform 712 bp long sequences. Cyt-b sequences generated in previous studies that were shorter than 330 bp were excluded from the haplotype network to preserve the quality of our analysis. We used the program PopArt v. 1.7 (Leigh and Bryant, 2015) and the TCS algorithm (Clement et al., 2000) to visualize haplotype connectivity and distribution with a haplotype network analysis.
2.6 Genetic diversity
Untruncated sequence lengths of 903 bp were used to compare genetic diversity among native and invasive locations utilizing our east and west Florida samples, and all O. septentrionalis that were available in GenBank. We separated cyt-b samples into those occurring in Louisiana, central Florida, west Florida, south Florida, west Cuba, central Cuba, east Cuba, and the Caribbean. For each location, we calculated the number of segregating sites (S) and nucleotide diversity (pi) for the preceding locations using pegas v. 1.2 (Paradis, 2010) and ape v. 5.7.1 (Paradis and Schliep, 2019) in RStudio. We generated the distribution of pi with 95% confidence intervals by employing a bootstrap resampling technique with 10,000 iterations using the boot function from boot package v. 1.3.28.1 in R (Davison and Hinkley, 1997; Canty, 2021). For reproducibility, we set the random number generation seed to 500 using the set.seed function in R.
3 Results
We generated robust cyt-b sequences from all 166 frogs we sampled in west Florida, east Florida, and Louisiana, recovering 14 total haplotypes. Of these, nine are novel (GenBank Accession numbers: PP320231.1 – PP320239.1) and five were previously published (Heinicke et al., 2011; Table 1). Novel haplotype se86 was the most common sequence found in our sampling, occurring in 49% of all frogs (Table 1).
In Louisiana we recovered seven haplotypes from 95 individuals, including two previously published haplotypes (Table 1) and five novel haplotypes. Among the five novel haplotypes, se89, se90, and se94 were private alleles recovered exclusively from Louisiana, each from a single individual (Table 1). Novel haplotype se87 was the most common haplotype in Louisiana (N = 37 individuals) and it was also found in one east Florida frog. Novel haplotype se86 was the second most common haplotype in Louisiana (N = 36 individuals), and we also found it at high frequency in west and east Florida (Figure 2). All ten individuals collected from the St. Charles Parish population had this haplotype. Haplotypes se3 (N = 17) and se29 (N = 1) were recovered exclusively in Florida in previous studies. None of the haplotypes present in Louisiana occurred in frogs from Cuba or the Caribbean (electronic supplemental material, Supplementary Table S1).
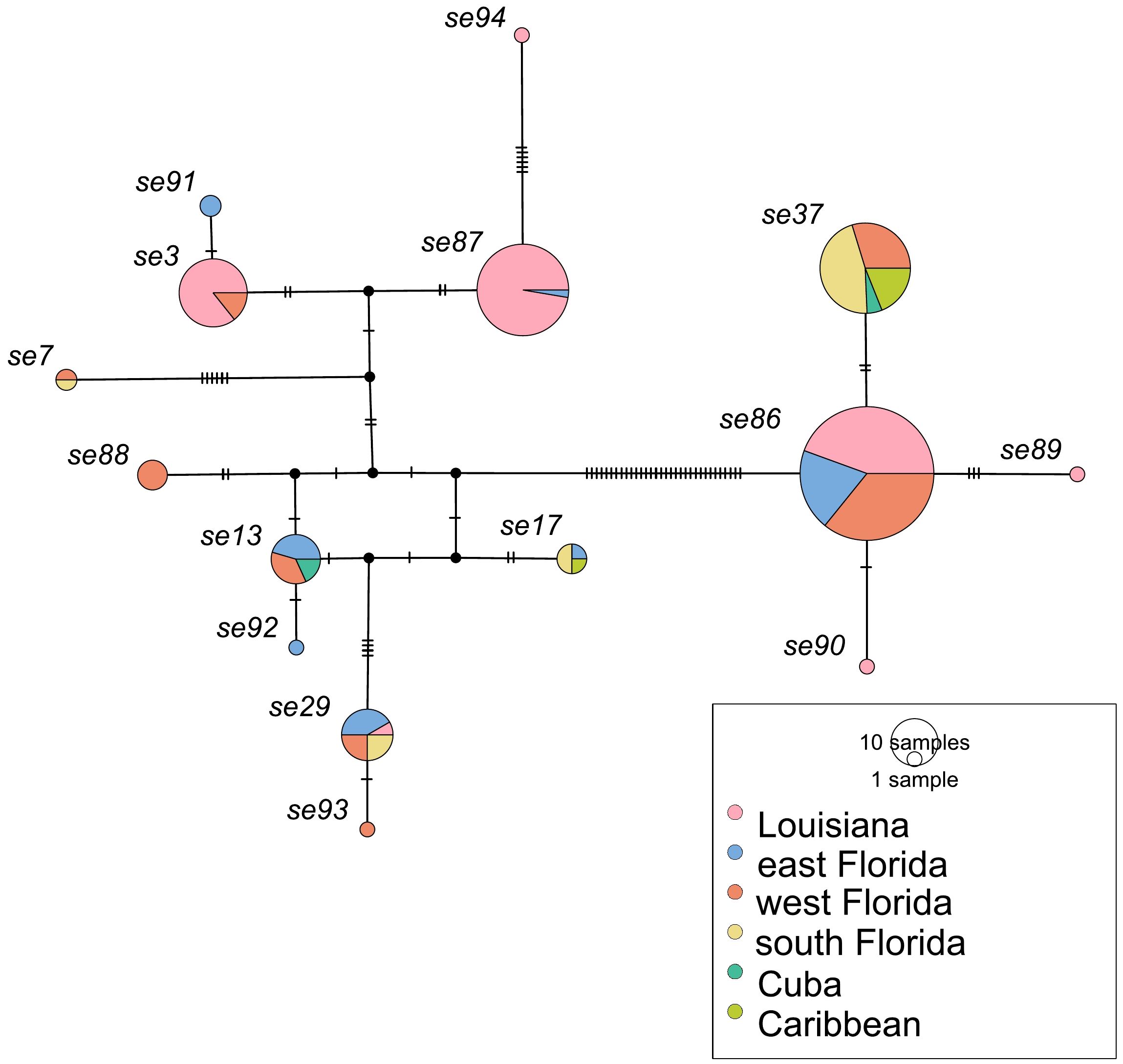
Figure 2. Cyt-b haplotype network of all sequences recovered in O. septentrionalis individuals sampled in Florida and Louisiana, USA. Circle size corresponds to sample size and color denotes sampling location. Tick marks between haplotypes represent the number of nucleotide changes between haplotypes.
The 31 frogs we sampled from east Florida had seven haplotypes, including four novel haplotypes (se92, se92, se86, se87) and three previously published haplotypes (se13, se17, se29). Only two individuals had the novel private haplotype se91, and one individual had the novel private haplotype se92. Most east Florida individuals had the novel haplotype se86 (N = 16), which was also found in west Florida and Louisiana. One east Florida individual had the novel haplotype se87, which was the most common haplotype in Louisiana. The three haplotypes se13, se17, and se29 were previously recovered from west Florida and west Cuba, south Florida and the Caribbean, and west Florida and south Florida, respectively (electronic supplemental material, Supplementary Table S1). Here, we recovered all three of these haplotypes the first time in east Florida.
In west Florida, we recovered seven haplotypes from 40 individuals: three novel haplotypes (se86, se88, se93) and four previously published haplotypes (se3, se7, se13, se29). A single individual had the private novel haplotype se93 and four individuals had the private novel haplotype se88. The novel haplotype se86 was the most common haplotype in west Florida, similar to its high frequency in Louisiana and east Florida. Se3 was previously recovered only in west Florida, but here we found it in one west Florida frog and in 17 Louisiana frogs. We found se7 in west Florida for the first time, but only in one frog. This haplotype was previously found in a single south Florida individual (electronic supplemental material, Supplementary Table S1). We found two other low-frequency haplotypes (se13 and se29) that had previously been detected in Florida and Cuba (electronic supplemental material, Supplementary Table S1).
Our haplotype network illustrates that almost all frogs sampled in Louisiana had haplotypes se3, se86, or se87 (Figure 2). These sequences are only found in Louisiana, east Florida (se86 and se87), and west Florida (se3 and se86). Only one Louisiana frog had a haplotype that is recovered more broadly throughout Florida (se29), and this sequence was many mutational steps away from all other haplotypes found in Louisiana. Private Louisiana haplotypes se89 and se90 were respectively three and one mutational steps away from se86. Likewise, private Louisiana haplotype se94 is six mutational steps from se87, a haplotype only rarely found outside of Louisiana. Collectively these patterns suggest the evolution of new haplotypes within the invasive Florida and/or Louisiana populations.
Phylogenetic analysis of O. septentrionalis mitochondrial sequences revealed minimal geographic structure (Figure 3). We recovered a well-supported basal split between two sequences only found in the Caribbean and all other sequences. The next split was also well-supported and separated one sequence private to Cuba with everything else. All other sequences were separated into two large clades with moderate support (posterior probability = 0.8917). Both of these clades contained numerous haplotypes found in multiple localities through the native and invasive range. Haplotypes sampled from Louisiana frogs were dispersed throughout the phylogeny.
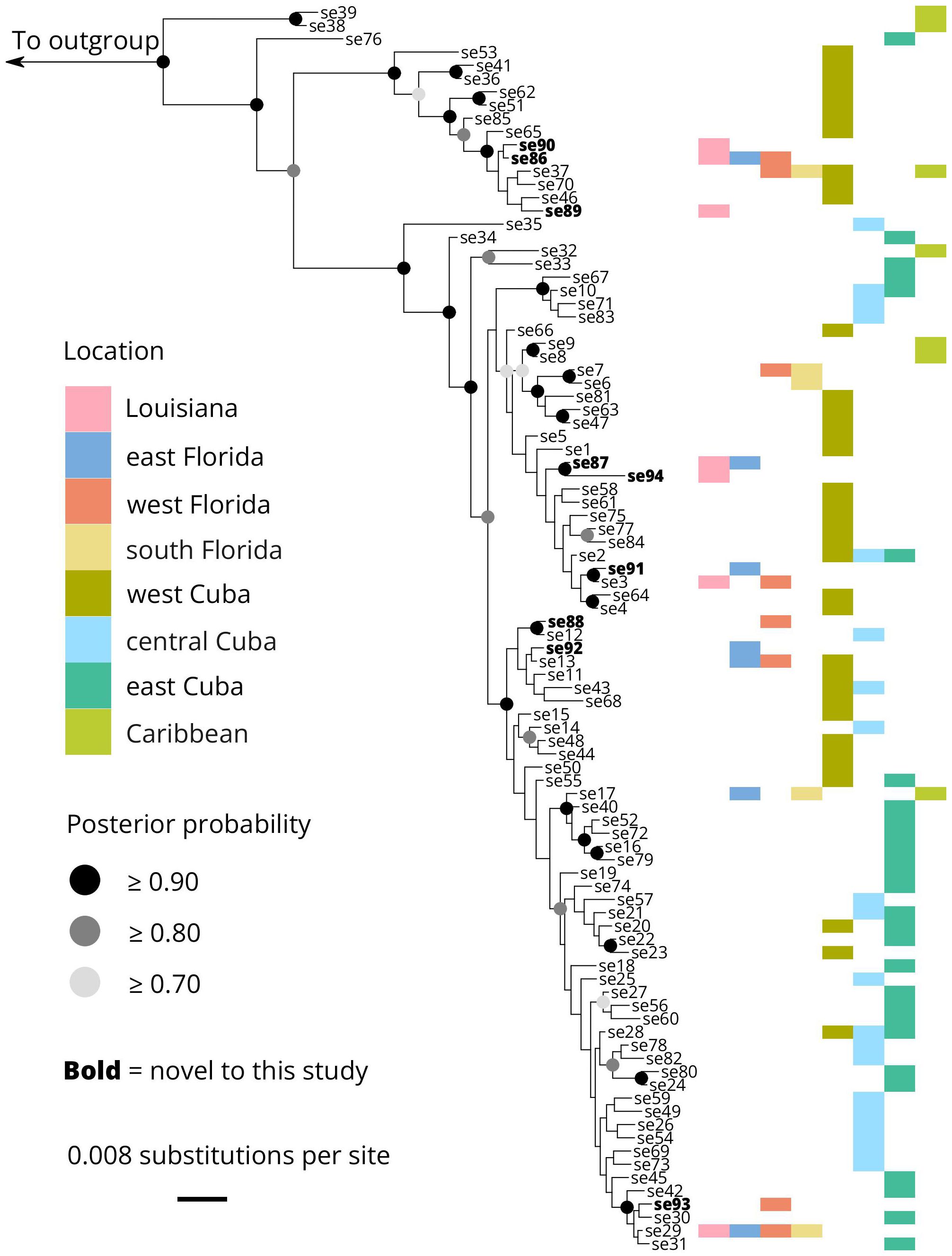
Figure 3. Bayesian gene genealogy of cytochrome-b in O. septentrionalis. Colored horizontal bars denote the different geographic locations where samples were taken. Haplotypes se1 through se39 originated from Heinicke et al. (2011). Haplotypes se40 through se85 originated from Rodríguez et al. (2015). Haplotypes se86 through se94 originate from this study and are denoted in bold.
Among all known cyt-b sequences, Cuba contained the highest number of haplotypes. Frogs sampled in west Cuba had more haplotypes than any other location, followed by east Cuba and central Cuba (Table 2). The Caribbean, Louisiana, and Florida localities each contained similar numbers of haplotypes (Table 2). Native Cuban localities contained similar percentages of private haplotypes, which were high in comparison to the invasive range. Private haplotypes made up 20–30% of the haplotypes recovered in the invasive range localities of east Florida, west Florida, and south Florida whereas 42.86% of Louisiana haplotypes were private (Table 2). Nucleotide diversity in Louisiana was highest among invasive locations in the United States (Figure 4). Nucleotide diversity in Louisiana (0.039; 95% CI = 0.033–0.056) did not overlap with nucleotide diversity of central Cuba (0.017; 95% CI = 0.0092–0.021) or east Cuba (0.018; 95% CI = 0.010–0.023; Table 2; Figure 4). Segregating sites were higher in west Cuba and the Caribbean than within invasive localities in the United States (Table 2).
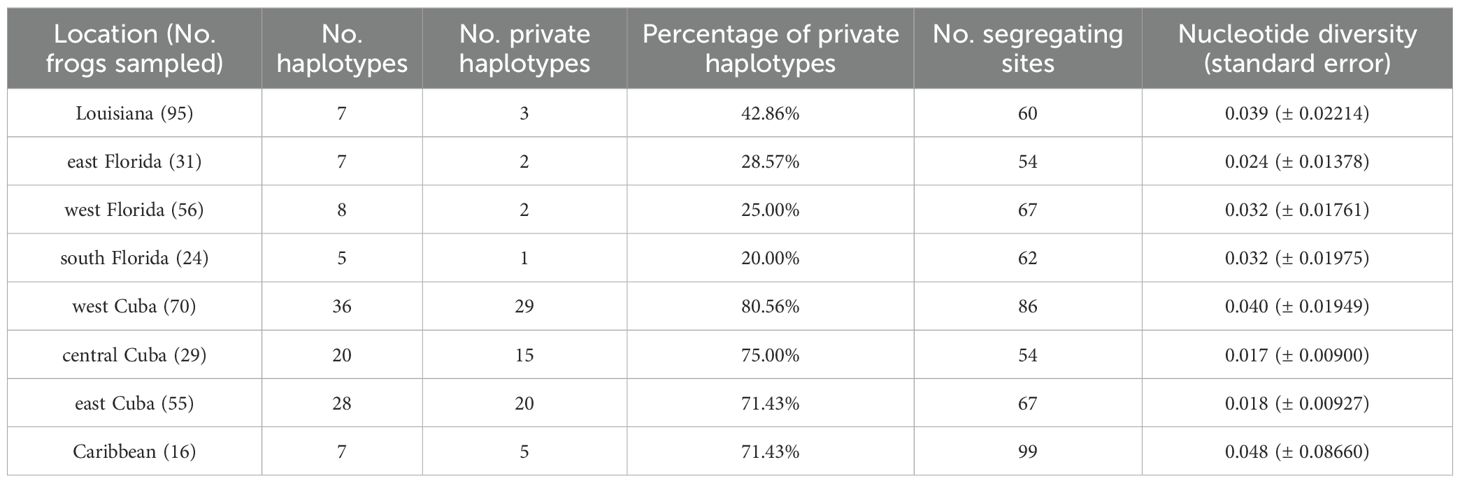
Table 2. Summary of allelic diversity, private alleles, segregating sites, and nucleotide diversity across eight locations.
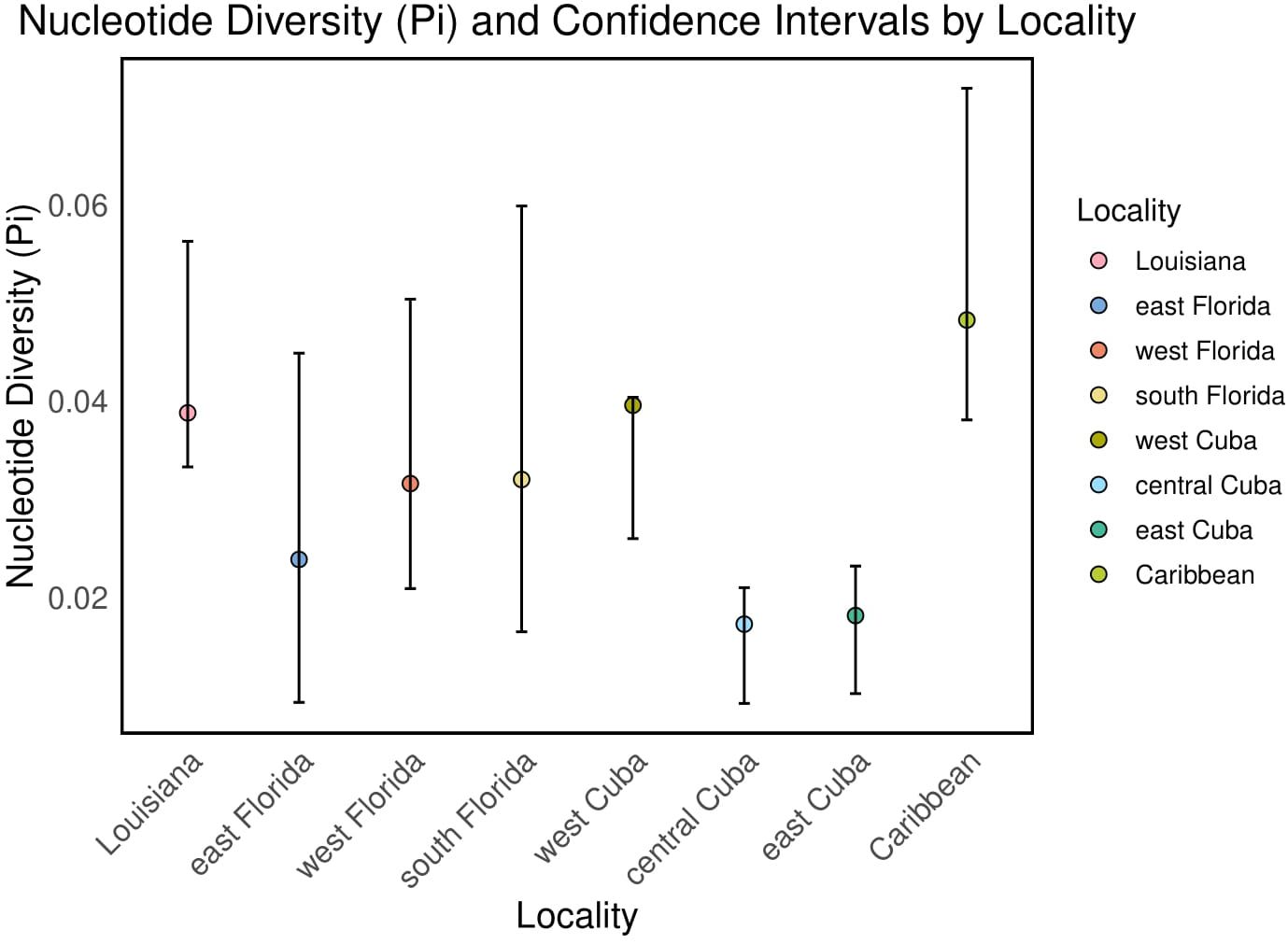
Figure 4. Nucleotide diversity (pi) of O. septentrionalis by locality with 95% confidence intervals.
4 Discussion
Our study is the first to investigate the origins of the establishment of O. septentrionalis in Louisiana, the first documented establishment of O. septentrionalis in the contiguous U.S. outside of Florida (Glorioso et al., 2018c). We aimed to determine whether this invasion was the result of one or more founding events from remote locations. Our results indicate both Louisiana O. septentrionalis populations are the result of multiple introduction events, likely from central Florida localities. Four of seven haplotypes found in Louisiana individuals were present only in Florida and Louisiana, while the remaining three were rare and only found in Louisiana. This pattern excludes Caribbean locations as direct sources of the novel population and strongly points toward Florida origins. One haplotype lending the most support to our introduction hypothesis is se86, which is novel to our study (Table 1). It was the most abundant haplotype we recovered overall and was at high frequency in our Florida and Louisiana sample sites, consistent with the hypothesis that central Florida is a source of the Louisiana O. septentrionalis.
Though we did not sample any localities in the Florida panhandle, the likelihood that frogs spread naturally from Florida to Louisiana in a range expansion is minimal. The high nucleotide diversity of haplotypes in Louisiana is inconsistent with the expectation for a range expansion. Range expansions of invasive species typically result in decreased genetic diversity as the leading edge expands (Wilson et al., 2005; Davis et al., 2011). Instead, we see higher nucleotide diversity in Louisiana compared to all locations except for west Cuba and the Caribbean (Figure 4), which is where the species originated (Heinicke et al., 2011; Johnson, 2023). Notably, the nucleotide diversity in Louisiana is nearly equivalent to values found in west Cuba. Furthermore, there is no evidence for a leading edge, as established populations of O. septentrionalis between the Florida panhandle and the Louisiana population have not yet been documented (Johnson, 2023). Future work that samples O. septentrionalis from the Florida panhandle and the Gulf Coast of Alabama and Mississippi, and analyzes their genetic composition could enable assessment of potential new population establishments and identification of their source populations.
High genetic diversity metrics in the Louisiana population suggests a high propagule pressure invasion and/or multiple introduction events (Kolbe et al., 2008; Simberloff, 2009; Wang et al., 2019). Given that human transport is the most plausible route of invasion, multiple founding events is the most likely scenario explaining the patterns we recovered in Louisiana. For example, repeated transport of one or a few individuals traveling on ornamental plants purchased from locations within Florida and planted in New Orleans is a plausible source of the novel invasion. Anecdotal accounts of O. septentrionalis being found within many purchased plants from Florida (Johnson, 2023), the scale and economic importance of the nursery industry in Florida (Khachatryan et al., 2022), and low efficacy of state-level biosecurity measures in preventing the spread of established invasive species (Paini et al., 2010) lend support to this hypothesis. Therefore, it is plausible that the known planting of ornamental palms from Lake Placid, Florida in the Audubon Zoo contributed to this invasion (Glorioso et al., 2018c). Sampling in or around the nursery in Lake Placid—which lies farther south than our sampling sites—to verify the presence of haplotypes shared with Louisiana could strengthen support for this hypothesis. Additionally, increased sampling at or near ornamental horticultural nurseries and retailers in top economic contributing regions, such as central and south Florida (Hodges et al., 2016), could give greater insight into the impacts of this industry on the spread of O. septentrionalis.
Human traffic near the invasion site has the potential to facilitate introductions from distant source populations. New Orleans is a large city that is visited by nearly 20 million tourists annually, with many visitors arriving via vehicular transportation or large cruise ships that depart from docks approximately 5 miles from the invasion site (Adams, 2024). Florida is a frequent point of origin, ranking as the 3rd most common origin of interstate travel entering Louisiana in 2023 (Adams, 2024). As vehicular transport is the primary mode of invasion of O. septentrionalis (Johnson, 2023), it is plausible that multiple instances of individual frogs hitch-hiking within vehicles departing from central Florida have contributed to the New Orleans invasion. Additionally, Interstate 10 (I-10), a major thoroughfare linking Florida to Louisiana, provides direct access to New Orleans and is heavily utilized by Floridians during hurricane evacuations (Ghorbanzadeh et al., 2021). While hurricane evacuees likely are minor contributors in comparison to tourists, increasing hurricane severity and resulting evacuation efforts may increase the frequency of these individual translocation events. Additionally, cruise ships departing and arriving at the Port of New Orleans frequent many Caribbean islands and various Floridian ports. Caribbean sampling of O. septentrionalis is minimal, leaving unclear resolution on what haplotypes are present in this region. Increased Caribbean sampling may reveal haplotypes from this study found exclusively in Louisiana may also occur in the Caribbean and may be derived from there. Overall increased sampling efforts throughout Florida, as well as the Caribbean could provide increased clarity on the contribution of translocation events to the founding of the Louisianan population.
Heinicke et al. (2011) previously reported two divergent clades of O. septentrionalis to be phylogeographically grouped based on relation to the Guanahacabibes peninsula in remote west Cuba. They additionally reported Bahamian haplotypes form a basal monophyletic group. With the additional samples of Rodríguez et al. (2015) as well as our novel sequences, our phylogenetic analysis groups together west Cuban haplotypes within and outside of the Guanahacabibes peninsula together, indicating this peninsular boundary does not impede gene flow to other localities in Cuba. Our study does not expand on the Heinicke et al., 2011 finding of Bahamian samples forming a monophyly, as we did not include additional Bahamian sampling outside of Heinicke et al. (2011) providing another reason for additional sampling in the Caribbean and specifically the Bahamas.
We expect that O. septentrionalis will continue to negatively affect native species through resource use and influences on local pathogen dynamics, but changes in the genetic composition of O. septentrionalis populations are a potential cause for concern. Anecdotal sightings of O. septentrionalis outside their established invasive territory are plentiful and range from Georgia to as far north as Vermont along the east coast of the United States (Morningstar et al., 2024). These sightings are likely a result of anthropogenic transport either via horticulture or other items transported from their invasive range to wider regions of the U.S. and are not yet representative of established breeding colonies. Cold temperatures play a large role in limiting the northern distribution of O. septentrionalis, which have been observed dead at temperatures below 0 °C (Haggerty and Crisman, 2015), but shifting global temperatures threaten this climatic barrier to expansion. With fewer freeze events occurring in regions north of the current range of O. septentrionalis, these populations may persist and establish farther north than previously expected (Osland et al., 2021).
Admixed populations at range limits may also benefit from an increased evolutionary potential resulting from heterosis, thereby increasing the extent of dispersal and probability of permanent establishment (Forsman, 2013; Wagner et al., 2017). Novel genotypes resulting from heterosis (Verhoeven et al., 2011) may allow for adaptation to colder temperatures. Evidence shows O. septentrionalis from the northern latitudinal limits of their Florida range had critical thermal minima trending lower than their southern counterparts and that in general O. septentrionalis has a higher tolerance for cold weather than previously believed (Simpson, 2013). Though cold tolerance tends to result from phenotypic plasticity (McCann et al., 2014; Mittan and Zamudio, 2019), it is possible that exposure to freeze events and the existence of novel genotypes within northern populations result in adapted cold tolerance. Though we observed patterns suggesting local adaptation of the cyt-b mitochondrial gene in Florida and Louisiana, more work could help to assess adaptation in functionally relevant genes. Future experimental studies on mitochondrial gene transcription associated with thermal regulation across invasive populations could provide deeper insight into thermal adaptation (Hong et al., 2024).
Disparities in size between northern and southern populations of O. septentrionalis have also been recorded (McGarrity and Johnson, 2009). Studies do not yet indicate if this shift to smaller sizes in colder latitudes is genetically based or if these changes result from environmental effects such as shifts in food availability or metabolic constraints resulting from cold temperatures. It is possible that smaller body size and increasing urban development may aid in their spread, as O. septentrionalis can exploit urban refugia to escape or minimize exposure to thermal temperature limits (Haggerty and Crisman, 2015; Meshaka, 2001).
Invasive species present a unique opportunity to study evolution in action by assessing novel selection pressures in situ. These systems can serve as natural experiments, with introduced populations functioning as treatment groups and native populations acting as controls. Invasive O. septentrionalis have the potential to serve as a powerful model for studying rapid adaptation to novel environmental conditions, especially in the recently established Louisiana population. While mitochondrial genes markers have historically served as a valuable tool in determining the origin of invasive species, next-generation sequencing (NGS) techniques offer better resolution to fine scale population structure and more robust findings on this recent population divergence (North et al., 2021). Whole mitochondrial genome analysis can expand on findings derived from single genes for more comprehensive insights as demonstrated with Rhinella marina, the invasive cane toad (Cheung et al., 2024). Like many amphibian species, O. septrentrionalis has not yet had its nuclear or mitochondrial genome sequenced, due in part to the difficulty of assemblage (Kosch et al., 2024; O’Connell et al., 2024) inhibiting the depth of evolutionary questioning utilizing this species.
Our study concludes that the newly established Louisiana population of O. septentrionalis is derived from multiple introductions, primarily from central Florida. These introductions likely occurred via anthropogenic means (Johnson, 2023), though we cannot conclude with certainty whether the transport of flora from Florida to Louisiana contributed to this establishment. To fully resolve the origin of Louisiana’s population, future work could prioritize increased sampling across the Florida panhandle, Caribbean, and key Floridian localities. By identifying the source populations and the degree of admixture within the Louisiana population, we gain insight into where management efforts could be prioritized and understanding of the genetic variation available for selection to act upon. In the case of O. septentrionalis, this knowledge can be used not only for reconstructing invasion history, but could also anticipate the species’ capacity to adapt to colder climates, urban environments, and other novel pressures.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was approved by Institutional Animal Care and Use Committee. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
EB: Investigation, Writing – review & editing, Visualization, Formal analysis, Project administration, Writing – original draft, Data curation. KP: Data curation, Investigation, Writing – review & editing. KM: Investigation, Writing – review & editing, Formal analysis, Visualization, Data curation, Project administration. MA: Data curation, Project administration, Investigation, Funding acquisition, Writing – review & editing, Writing – original draft, Conceptualization. BG: Methodology, Investigation, Resources, Data curation, Project administration, Funding acquisition, Conceptualization, Writing – review & editing. JW: Conceptualization, Methodology, Resources, Data curation, Investigation, Writing – review & editing, Project administration, Funding acquisition. RM: Investigation, Data curation, Writing – review & editing, Resources. AS: Writing – review & editing, Funding acquisition, Investigation, Supervision, Resources, Project administration, Methodology.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. USGS, NSF and FWC contributed funds supporting research. Article processing charges were provided in part by the UCF College of Graduate Studies Open Access Publishing Fund.
Acknowledgments
Thanks to the Conservation, Restoration and Communication REU, and students from the Savage lab including Maddie Thurber, Siena Krullis, Ivan Santana, Patricia Hernandez, and Savannah Freeman who assisted in catching or processing samples. Thanks to Maryam Ghoojaei for her guidance on implementing bootstrapping analyses in RStudio. Animals in Florida were captured under Florida Fish and Wildlife Conservation Commission Scientific Collecting Permits and handled in accordance with the approved IACUC protocols. We thank Lindy J. Muse, Nicole D. Jennings, Brittany R. Maldonado, Raymond P. Kidder, Melanie Litton, CJ Hillard, Katie Everett, and many volunteers and a few private landowners for Louisiana tissue collection. We captured animals under annual Louisiana Department of Wildlife and Fisheries Scientific Collecting Permits and handled all animals in accordance with approved IACUC protocols (USGS WARC FY2008–1). Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government. This is contribution number 959 of the U.S. Geological Survey Amphibian Research and Monitoring Initiative (ARMI).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/famrs.2025.1646714/full#supplementary-material
References
Adams A. (2024). “Louisiana visitor profile calendar year 2023,” in MMGY travel intelligence. Mclean, VA: MMGY Travel Intelligence. Available online at: https://www.explorelouisiana.com/sites/default/files/2024-07/2023%20Louisiana%20Visitor%20Profile%20Report.pdf (Accessed March 30, 2025).
Allendorf F. W. and Lundquist L. L. (2003). Introduction: population biology, evolution, and control of invasive species. Conserv. Biol. 17, 24–30. doi: 10.1046/j.1523-1739.2003.02365.x
Atkinson M. S. and Savage A. E. (2023). Invasive amphibians alter host-pathogen interactions with primarily negative outcomes for native species. Biol. Conserv. 286, 110310. doi: 10.1016/j.biocon.2023.110310
Avise J. C., Arnold J., Ball R. M., Bermingham E., Lamb T., Neigel J. E., et al. (1987). Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annu. Rev. Ecol. Syst. 18, 489–522. doi: 10.1146/annurev.es.18.110187.002421
Bai C., Ke Z., Consuegra S., Liu X., and Li Y. (2012). The role of founder effects on the genetic structure of the invasive bullfrog (Lithobates catesbeianaus) in China. Biol. Invasions. 14, 1785–1796. doi: 10.1007/s10530-012-0189-x
Bomford M., Kraus F., Barry S. C., and Lawrence E. (2009). Predicting establishment success for alien reptiles and amphibians: a role for climate matching. Biol. Invasions. 11, 713–724. doi: 10.1007/s10530-008-9285-3
Chatfield M. W. H. and Vance M. (2014). Geographic distribution. Osteopilus septentrionalis (Cuban Treefrog). Herpetol. Rev. 45 (2), 278. Available online at: https://ssarherps.org/herpetological-review-pdfs/.
Cheung K., Amos T. G., Shine R., DeVore J. L., Ducatez S., Edwards R. J., et al. (2024). Whole-mitogenome analysis unveils previously undescribed genetic diversity in cane toads across their invasion trajectory. Ecol. Evol. 14, e11115. doi: 10.1002/ece3.11115
Clement M., Posada D., and Crandall K. A. (2000). TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9, 1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x
Colangelo P., Abiadh A., Aloise G., Amori G., Capizzi D., Vasa E., et al. (2015). Mitochondrial phylogeography of the black rat supports a single invasion of the western Mediterranean basin. Biol. Invasions. 17, 1859–1868. doi: 10.1007/s10530-015-0842-2
Culbertson K. A. and Herrmann N. C. (2019). Asymmetric interference competition and niche partitioning between native and invasive Anolis lizards. Oecologia. 190, 811–820. doi: 10.1007/s00442-019-04466-1
Darriba D., Taboada G. L., Doallo R., and Posada D. (2012). jModelTest 2: more models, new heuristics and high-performance computing. Nat. Methods 9, 772. doi: 10.1038/nmeth.2109
Davis M. C., Novak S. J., and Hampikian G. (2011). Mitochondrial DNA analysis of an immigrant Basque population: loss of diversity due to founder effects. Am. J. Phys. Anthropol. 144, 516–525. doi: 10.1002/ajpa.21432
Davison A. C. and Hinkley D. V. (1997). Bootstrap methods and their Application (Cambridge, UK: Cambridge University Press).
Dove C. J., Snow R. W., Rochford M. R., and Mazzotti F. J. (2011). Birds consumed by the invasive Burmese python (Python molurus bivittatus) in Everglades National Park, Florida, USA. Wilson. J. Ornithol. 123, 126–131. doi: 10.1676/10-092.1
Duellman W. and Schwartz A. (1958). Amphibians and reptiles of southern Florida. Bull. Fla. Mus. Nat. Hist. 3, 181–324. doi: 10.58782/flmnh.gtne5332
Forsman A. (2013). Effects of genotypic and phenotypic variation on establishment are important for conservation, invasion, and infection biology. Proc. Natl. Acad. Sci. 111, 302–307. doi: 10.1073/pnas.1317745111
Frankham R. (1998). Inbreeding and extinction: island populations. Conserv. Biol. 12, 665–675. doi: 10.1046/j.1523-1739.1998.96456.x
Galt N., Atkinson M., Glorioso B., Waddle H., Litton M., and Savage A. (2021). Widespread ranavirus and perkinsea infections in Cuban treefrogs (Osteopilus septentrionalis) invading New Orleans, USA. Herpetol. Conserv. Biol. 16, 17–29. doi: 10.5061/DRYAD.7WM37PVS8
Ghorbanzadeh M., Burns S., Rugminiamma L. V. N., Erman Ozguven E., and Huang W. (2021). Spatiotemporal analysis of highway traffic patterns in Hurricane Irma evacuation. Transp. Res. Rec. 2675, 321–334. doi: 10.1177/03611981211001870
Glorioso B. M., Macedo K., Maldonado B. R., Hillard C. J., Morenc I. N., and Grimes E. S. (2018a). Geographic distribution: Osteopilus septentrionalis (Cuban treefrog). Herpetol. Rev. 49 (4), 709. Available online at: https://ssarherps.org/herpetological-review-pdfs/.
Glorioso B. M., Steece A., Lemann Z. K., Lazare R., and Beck J. W. (2016). Geographic distribution: Osteopilus septentrionalis (Cuban Treefrog). Herpetol. Rev. 47 (2), 249.
Glorioso B. M., Vanbergen P., Roy J., Walter M., Leonpacher L., and Freistak M. (2018b). Geographic distribution: Osteopilus septentrionalis (Cuban Treefrog). Herpetol. Rev. 49 (1), 70–71. Available online at: https://ssarherps.org/herpetological-review-pdfs/.
Glorioso B. M. and Waddle J. H. (2014). A review of pipe and bamboo artificial refugia as sampling tools in anuran studies. Herpetol. Conserv. Biol. 9, 609–625. Available online at: https://www.herpconbio.org/Volume_9/Issue_3/Glorioso_Waddle_2014.pdf.
Glorioso B. M., Waddle J. H., Muse L. J., Jennings N. D., Litton M., Hamilton J., et al. (2018c). Establishment of the exotic invasive Cuban treefrog (Osteopilus septentrionalis) in Louisiana. Biol. Invasions. 20, 2707–2713. doi: 10.1007/s10530-018-1732-1
Gray M. W., Cedergren R., Abel Y., and Sankoff D. (1989). On the evolutionary origin of the plant mitochondrion and its genome. Proc. Natl. Acad. Sci. 86, 2267–2271. doi: 10.1073/pnas.86.7.2267
Haggerty C. J. E. and Crisman T. L. (2015). Pulse disturbance impacts from a rare freeze event in Tampa, Florida on the exotic invasive Cuban treefrog, Osteopilus septentrionalis, and native treefrogs. Biol. Invasions. 17, 2103–2111. doi: 10.1007/s10530-015-0863-x
Hamner R., Freshwater D. W., and Whitfield P. (2007). Mitochondrial cytochrome b analysis reveals two invasive lionfish species with strong founder effects in the western Atlantic. J. Fish Biol. 71, 214–222. doi: 10.1111/j.1095-8649.2007.01575.x
Heinicke M. P., Diaz L. M., and Hedges S. B. (2011). Origin of invasive Florida frogs traced to Cuba. Biol. Lett. 7, 407–410. doi: 10.1098/rsbl.2010.1131
Hill S. A., Beard K. H., Siers S. R., and Shiels A. B. (2019). Invasive coqui frogs are associated with differences in mongoose and rat abundances and diets in Hawaii. Biol. Invasions. 21, 2177–2190. doi: 10.1007/s10530-019-01965-3
Hodges A. W., Khachatryan H., and Rahmani M. (2016). “Economic contributions of the environmental horticulture industry in Florida in 2015,” in Electronic data information source, university of Florida institute of food and agricultural sciences. Gainesville, FL: University of Florida-Institute of Food and Agricultural Sciences Food and Resource Economics Department. Available online at: https://web.archive.org/web/20180426111529id_/http://fred.ifas.ufl.edu/pdf/EconContEnvirHortIndFL2015-11-15-16.pdf.
Hong Y., He Y., Lin Z., Du Y., Chen S., Han L., et al. (2022). Complex origins indicate a potential bridgehead introduction of an emerging amphibian invader (Eleutherodactylus planirostris) in China. NeoBiota. 77, 23–37. doi: 10.3897/neobiota.77.83205
Hong Y. H., Yuan Y.-N., Li K., Storey K. B., Zhang J.-Y., Zhang S.-S., et al. (2024). Differential mitochondrial genome expression of four Hylid frog species under low-temperature stress and its relationship with Amphibian temperature adaptation. Int. J. Mol. Sci. 25, 5967. doi: 10.3390/ijms25115967
Jackson H., Strubbe D., Tollington S., Prys-Jones R., Matthysen E., and Groombridge J. J. (2015). Ancestral origins and invasion pathways in a globally invasive bird correlate with climate and influences from bird trade. Mol. Ecol. 24, 4269–4285. doi: 10.1111/mec.13307
Johnson S. A. (2023). The Cuban treefrog (Osteopilus septentrionalis) in Florida: WEC218/UW259, 2/2023. EDIS. 2023, 1. doi: 10.32473/edis-UW259-2023
Kamath P. L., Sepulveda A. J., and Layhee M. (2016). Genetic reconstruction of a bullfrog invasion to elucidate vectors of introduction and secondary spread. Ecol. Evol. 6, 5221–5233. doi: 10.1002/ece3.2278
Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., et al (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Khachatryan H., Knuth M., Hodges A., and Hall C. (2022). Florida nursery and landscape industry economic contributions report: FE1114/FE1114, 02/2022. EDIS. 2022, 1. doi: 10.32473/edis-fe1114-2022
Kochzius M., Söller R., Khalaf M. A., and Blohm D. (2003). Molecular phylogeny of the lionfish genera Dendrochirus and Pterois (Scorpaenidae, Pteroinae) based on mitochondrial DNA sequences. Mol. Phylogenet. Evol. 28, 396–403. doi: 10.1016/S1055-7903(02)00444-X
Kolbe J. J., Glor R. E., Rodríguez Schettino L., Lara A. C., Larson A., and Losos J. B. (2004). Genetic variation increases during biological invasion by a Cuban lizard. Nat. 431, 177–181. doi: 10.1038/nature02807
Kolbe J. J., Larson A., Losos J. B., and De Queiroz K. (2008). Admixture determines genetic diversity and population differentiation in the biological invasion of a lizard species. Biol. Lett. 4, 434–437. doi: 10.1098/rsbl.2008.0205
Kosch T. A., Torres-Sánchez M., Liedtke H. C., Summers K., Yun M. H., Crawford A. J., et al. (2024). The Amphibian Genomics Consortium: advancing genomic and genetic resources for amphibian research and conservation. BMC Genomics 25, 1025. doi: 10.1186/s12864-024-10899-7
Kraus F. (2015). Impacts from invasive reptiles and amphibians. Annu. Rev. Ecol. Evol. Syst. 46, 75–97. doi: 10.1146/annurev-ecolsys-112414-054450
Krehenwinkel H. and Tautz D. (2013). Northern range expansion of European populations of the wasp spider Argiope bruennichi is associated with global warming–correlated genetic admixture and population-specific temperature adaptations. Mol. Ecol. 22, 2232–2248. doi: 10.1111/mec.12223
LaFond J., Martin K. R., Dahn H., Richmond J. Q., Murphy R. W., Rollinson N., et al. (2022). Invasive bullfrogs maintain MHC polymorphism including alleles associated with chytrid fungal infection. Integr. Comp. Biol. 62, 262–274. doi: 10.1093/icb/icac044
Laufer G., Canavero A., Núñez D., and Maneyro R. (2008). Bullfrog (Lithobates catesbeianus) invasion in Uruguay. Biol. Invasions. 10, 1183–1189. doi: 10.1007/s10530-007-9178-x
Leigh J. W. and Bryant D. (2015). POPART: full-feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116. doi: 10.1111/2041-210X.12410
Lillo F., Dufresnes C., Faraone F., Lo Valvo M., and Stöck M. (2013). Identification and potential origin of invasive clawed frogs Xenopus (Anura: Pipidae) in Sicily based on mitochondrial and nuclear DNA. Ital. J. Zool. 80, 566–573. doi: 10.1080/11250003.2013.847502
Lobos G., Mendez M. A., Cattan P., and Jaksic F. (2014). Low genetic diversity of the successful invasive African clawed frog Xenopus laevis (Pipidae) in Chile. Stud. Neotrop. Fauna Environ. 49, 50–60. doi: 10.1080/01650521.2014.912865
McCann S., Greenlees M. J., Newell D., and Shine R. (2014). Rapid acclimation to cold allows the cane toad to invade montane areas within its Australian range. Funct. Ecol. 28, 1166–1174. doi: 10.1111/1365-2435.12255
McGarrity M. E. and Johnson S. A. (2009). Geographic trend in sexual size dimorphism and body size of Osteopilus septentrionalis (Cuban treefrog): implications for invasion of the southeastern United States. Biol. Invasions. 11, 1411–1420. doi: 10.1007/s10530-008-9349-4
Meshaka W. E. (2001). The Cuban treefrog in Florida: life history of a successful colonizing species (Gainesville, FL: University Press of Florida).
Miller M. A., Pfeiffer W., and Schwartz T. (2010). Creating the CIPRES Science Gateway for inference of large phylogenetic trees., in 2010 gateway computing environments workshop (GCE). IEEE., 1–8. doi: 10.1109/GCE.2010.5676129
Mittan C. S. and Zamudio K. R. (2019). Rapid adaptation to cold in the invasive cane toad Rhinella marina. Conserv. Physiol. 7, coy075. doi: 10.1093/conphys/coy075
Morningstar C. R., Daniel W. M., and Somma L. A. (2024). Cuban Treefrog (Osteopilus septentrionalis) - Species Profile (Gainesville, FL: U.S. Geological Survey). Available online at: https://nas.er.usgs.gov/queries/factsheet.aspx?SpeciesID=57 ((Accessed April 23, 2025)).
North H. L., McGaughran A., and Jiggins C. D. (2021). Insights into invasive species from whole-genome resequencing. Mol. Ecol. 30, 6289–6308. doi: 10.1111/mec.15999
O’Connell L. A., Rodríguez A., Kosch T. A., Kwon T., Bolsoni L., Lourenço H., et al. (2024). ““Genomics: using genomics approaches in amphibian conservation”,” in Amphibian conservation action plan. Eds. Wren S., Borzée A., R. Marcec-Greaves, and Angulo A. (Gland, Switzerland: IUCN SSC Amphibian Specialist Group), 310–334. Available online at: https://www.researchgate.net/profile/Simon-Clulow/publication/382486709_Amphibian_conservation_action_plan_a_status_review_and_roadmap_for_global_amphibian_conservation/links/66d90fefb1606e24c2e1a991/Amphibian-conservation-action-plan-a-status-review-and-roadmap-for-global-amphibian-conservation.pdf#page=314.
Osland M. J., Stevens P. W., Lamont M. M., Brusca R. C., Hart K. M., Waddle J. H., et al. (2021). Tropicalization of temperate ecosystems in North America: The northward range expansion of tropical organisms in response to warming winter temperatures. Glob. Change Biol. 27, 3009–3034. doi: 10.1111/gcb.15563
Paini D. R., Worner S. P., Cook D. C., De Barro P. J., and Thomas M. B. (2010). Threat of invasive pests from within national borders. Nat. Commun. 1, 115. doi: 10.1038/ncomms1118
Palis J. G., Johnson S. A., Marks J. R., and Boehler M. B. (2021). Cuban treefrog (Osteopilus septentrionalis) in illinois. Bull. Chic. Herpetol. Soc 56 (9), 153–155.
Paradis E. (2010). pegas: an R package for population genetics with an integrated–modular approach. Bioinformatics. 26, 419–420. doi: 10.1093/bioinformatics/btp696
Paradis E. and Schliep K. (2019). ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 35, 526–528. doi: 10.1093/bioinformatics/bty633
Peacock M. M., Beard K. H., O’NEILL E. M., Kirchoff V. S., and Peters M. B. (2009). Strong founder effects and low genetic diversity in introduced populations of Coqui frogs. Mol. Ecol. 18, 3603–3615. doi: 10.1111/j.1365-294X.2009.04308.x
Piquet J. C. and López-Darias M. (2021). Invasive snake causes massive reduction of all endemic herpetofauna on Gran Canaria. Proc. R. Soc B: Biol. Sci. 288, 20211939. doi: 10.1098/rspb.2021.1939
Pitt W., Vice D., and Pitzler M. (2005). Challenges of invasive reptiles and amphibians. Wildlife Damage Manage. Conferences - Proc. 84, 112– 119. Available online at: https://digitalcommons.unl.edu/icwdm_wdmconfproc/84 (Accessed July 17, 2024).
Rambaut A., Drummond A. J., Xie D., Baele G., and Suchard M. A. (2018). Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67, 901–904. doi: 10.1093/sysbio/syy032
Reynolds R. G., Puente-Rolón A. R., Reed R. N., and Revell L. J. (2013). Genetic analysis of a novel invasion of Puerto Rico by an exotic constricting snake. Biol. Invasions. 15, 953–959. doi: 10.1007/s10530-012-0354-2
Rice K. G., Waddle J. H., Miller M. W., Crockett M. E., Mazzotti F. J., and Percival H. F. (2011). Recovery of native treefrogs after removal of nonindigenous Cuban Treefrogs, Osteopilus septentrionalis. Herpetologica. 67, 105–117. doi: 10.1655/HERPETOLOGICA-D-10-00020.1
Richmond J. Q., Wood D. A., Stanford J. W., and Fisher R. N. (2015). Testing for multiple invasion routes and source populations for the invasive brown treesnake (Boiga irregularis) on Guam: implications for pest management. Biol. Invasions. 17, 337–349. doi: 10.1007/s10530-014-0733-y
Richter-Boix A., Garriga N., Montori A., Franch M., San Sebastián O., Villero D., et al. (2013). Effects of the non-native amphibian species Discoglossus pictus on the recipient amphibian community: niche overlap, competition and community organization. Biol. Invasions. 15, 799–815. doi: 10.1007/s10530-012-0328-4
Rius M. and Darling J. A. (2014). How important is intraspecific genetic admixture to the success of colonising populations? Trends Ecol. Evol. 29, 233–242. doi: 10.1016/j.tree.2014.02.003
Rivera B., Cook K., Andrews K., Atkinson M. S., and Savage A. E. (2019). Pathogen dynamics in an invasive frog compared to native species. EcoHealth. 16, 222–234. doi: 10.1007/s10393-019-01432-4
Rodríguez A., Börner M., Pabijan M., Gehara M., Haddad C. F. B., and Vences M. (2015). Genetic divergence in tropical anurans: deeper phylogeographic structure in forest specialists and in topographically complex regions. Evol. Ecol. 29, 765–785. doi: 10.1007/s10682-015-9774-7
Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Höhna S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
RStudio Team (2021). RStudio: integrated development environment for R (Boston, MA: RStudio, PBC). Available online at: http://www.rstudio.com/.
Selechnik D., Richardson M. F., Shine R., DeVore J. L., Ducatez S., and Rollins L. A. (2019). Increased adaptive variation despite reduced overall genetic diversity in a rapidly adapting invader. Front. Genet. 10. doi: 10.3389/fgene.2019.01221
Shanmuganathan T., Pallister J., Doody S., McCallum H., Robinson T., Sheppard A., et al. (2010). Biological control of the cane toad in Australia: a review. Anim. Conserv. 13, 16–23. doi: 10.1111/j.1469-1795.2009.00319.x
Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. doi: 10.1038/msb.2011.75
Simberloff D. (2009). The role of propagule pressure in Biol. Invasions. Annu. Rev. Ecol. Evol. Syst. 40, 81–102. doi: 10.1146/annurev.ecolsys.110308.120304
Simpson S. E. (2013). Assessing critical thermal minima to determine the thermal limits of the invasive Cuban treefrog (Osteopilus septentrionalis). [master’s thesis]. University of Florida, Gainesville (FL.
Trifinopoulos J., Nguyen L.-T., von Haeseler A., and Minh B. Q. (2016). W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235. doi: 10.1093/nar/gkw256
Verhoeven K. J., Macel M., Wolfe L. M., and Biere A. (2011). Population admixture, Biol. Invasions and the balance between local adaptation and inbreeding depression. Proc. R. Soc B: Biol. Sci. 278, 2–8. doi: 10.1098/rspb.2010.1272
Wagner N. K., Ochocki B. M., Crawford K. M., Compagnoni A., and Miller T. E. (2017). Genetic mixture of multiple source populations accelerates invasive range expansion. J. Anim. Ecol. 86, 21–34. doi: 10.1111/1365-2656.12567
Wang S., Liu C., Wu J., Xu C., Zhang J., Bai C., et al. (2019). Propagule pressure and hunting pressure jointly determine genetic evolution in insular populations of a global frog invader. Sci. Rep. 9, 448. doi: 10.1038/s41598-018-37007-6
Willson J. D., Dorcas M. E., and Snow R. W. (2011). Identifying plausible scenarios for the establishment of invasive Burmese pythons (Python molurus) in Southern Florida. Biol. Invasions. 13, 1493–1504. doi: 10.1007/s10530-010-9908-3
Wilson G. A., Nishi J. S., Elkin B. T., and Strobeck C. (2005). Effects of a recent founding event and intrinsic population dynamics on genetic diversity in an ungulate population. Conserv. Genet. 6, 905–916. doi: 10.1007/s10592-005-9077-6
Wyatt J. L. and Forys E. A. (2004). Conservation implications of predation by Cuban treefrogs (Osteopilus septentrionalis) on native hylids in Florida. Southeast. Nat. 3, 695–700. doi: 10.1656/1528-7092(2004)003[0695:CIOPBC]2.0.CO;2
Keywords: invasive species, genetics, biological invasion, cytochrome b, Osteopilus septentrionalis
Citation: Brosnan EB, Paniagua Torres KA, Martin KR, Atkinson MS, Glorioso BM, Waddle JH, Mendyk RW and Savage AE (2025) Tracing invasion routes of Cuban treefrogs into Louisiana using mitochondrial DNA. Front. Amphib. Reptile Sci. 3:1646714. doi: 10.3389/famrs.2025.1646714
Received: 13 June 2025; Accepted: 24 September 2025;
Published: 17 October 2025.
Edited by:
Amanda Kissel, United States Geological Survey, United StatesReviewed by:
Steve A. Johnson, University of Florida, United StatesAndrew Parks, US Fish and Wildlife Service, United States
Copyright © 2025 Brosnan, Paniagua Torres, Martin, Atkinson, Glorioso, Waddle, Mendyk and Savage. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Erin B. Brosnan, ZXJpbi5icm9zbmFuQHVjZi5lZHU=
 Erin B. Brosnan
Erin B. Brosnan Karen A. Paniagua Torres
Karen A. Paniagua Torres Katherine R. Martin
Katherine R. Martin Matthew S. Atkinson1
Matthew S. Atkinson1 Brad M. Glorioso
Brad M. Glorioso Robert W. Mendyk
Robert W. Mendyk Anna E. Savage
Anna E. Savage